ABSTRACT
The tumor-suppressive activity of p53 is largely attributed to its ability to induce cell death, including apoptosis, through transcription-dependent and transcription-independent mechanisms. On the one hand, nuclear p53 transcriptionally activates the expression of a myriad of pro-apoptotic BCL-2 family genes, such as NOXA, PUMA, BID, BAD, BIK, BAX, etc., whereas it inactivates the expression of anti-apoptotic BCL-2, BCL-XL, and MCL1, leading to mitochondrial apoptosis. On the other hand, cytoplasmic p53 also promotes mitochondrial apoptosis by directly associating with multiple BCL-2 family proteins in the mitochondria. Apoptosis-related protein in TGF-β signaling pathway (ARTS), a mitochondria-localized pro-apoptotic protein encoded by an alternative spliced variant of the SEPT4 gene, triggers apoptosis by facilitating proteasomal degradation of BCL-2 and XIAP upon pro-apoptotic stimuli. We recently identified SEPT4/ARTS as a new p53 target gene in response to genotoxic stress. ARTS in turn binds to p53, drives its mitochondrial localization, and enhances the interaction between p53 and BCL-XL, thereby promoting mitochondrial apoptosis. This review will illustrate the mechanisms of p53-induced mitochondrial apoptosis, offer some recently discovered new insights into the functions of ARTS in regulating mitochondrial cell death, and discuss the clinical significance of ARTS in cancer and non-cancer diseases.
Keywords: p53, ARTS, SEPT4, BCL-2 family, apoptosis, cancer therapy
Introduction
The fine balance between cell survival and death is vital for organism growth and development. However, disruption of this balance leads to various diseases, including cancer. Programmed cell death was first observed in the development of toads in the 1840s (Cotter, 2009). One of the major forms of programmed cell death is apoptosis, which can be initiated through either the extrinsic or the intrinsic pathway. The protease activity of caspases is crucial to the morphological and biochemical changes of apoptotic cells (Marino et al., 2014; Ichim and Tait, 2016). In the extrinsic apoptotic pathway, the death receptors upon binding by their cognate ligands can activate the initiator caspases, caspase-8 and caspase-10, which in turn mediate the cleavage of the effectors, caspase-3, caspase-6, and caspase-7, consequently leading to apoptosis (Ichim and Tait, 2016). The intrinsic apoptotic pathway, also known as mitochondrial apoptosis, involves a complex interplay between the pro-apoptotic and anti-apoptotic BCL-2 family proteins (Singh et al., 2019). Upon various stress signals, the pro-apoptotic BCL-2 homology domain 3 (BH3)-only proteins, including BCL-2-associated agonist of cell death (BAD), BH3-interacting domain death agonist (BID), BCL-2-interacting killer (BIK), BCL-2-interacting mediator of cell death (BIM), BCL-2-modifying factor (BMF), activator of apoptosis harakiri (HRK), phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1, also known as NOXA), and p53-upregulated modulator of apoptosis (PUMA), can be triggered to interact with and activate the pore-forming proteins, BCL-2-associated X protein (BAX) and BCL-2 antagonist/killer 1 (BAK, also known as BAK1), at the outer mitochondrial membrane, leading to mitochondrial outer membrane permeabilization (MOMP) and the release of cytochrome c, second mitochondria-derived activator of caspases (SMAC), and serine protease HTRA2/OMI. Cytochrome c associates with apoptotic peptidase activating factor 1 (APAF1) in the cytoplasm to form the apoptosome and facilitate caspase-9 activation, while SMAC and HTRA2/OMI suppress the anti-apoptotic X-linked inhibitor of apoptosis protein (XIAP). These signals are committed to mediating the cleavage of the effector caspases and triggering apoptosis (Singh et al., 2019). To circumvent apoptosis and sustain their own survival and propagation, cancer cells employ the multi-BH domain-containing anti-apoptotic BCL-2 family proteins, such as BCL-2, BCL-XL (the longer isoform of BCL2L1), and MCL1, to repress the pro-apoptotic BCL-2 family proteins by directly associating with the latter (Green, 2022). This complicated apoptotic process is finely tuned by various regulators in cancer cells.
One of the important regulators is the tumor suppressor p53. p53 is regarded as the ‘guardian of the genome’ because of its important role in preventing tumorigenesis and inhibiting cancer progression (Levine, 2020). In response to various stresses, p53 is activated through different post-translational modifications (PTMs) (Liu et al., 2019; Wen and Wang, 2022) to act as either a transcription factor in the nucleus (Riley et al., 2008) or an apoptosis inducer in the cytoplasm (Green and Kroemer, 2009). On the one hand, p53 suppresses cancer development by transcriptionally regulating the expression of myriad genes (Riley et al., 2008). These downstream target genes are responsible for different biological processes, such as cell cycle arrest, DNA repair, and apoptosis (Levine, 2019, 2020). On the other hand, the cytoplasmic p53 protein induces apoptosis by directly interacting with multiple BCL-2 family proteins (Green and Kroemer, 2009). However, p53-dependent apoptosis is causative for developmental abnormalities when p53 is inappropriately activated during embryonic and postnatal development (Bowen and Attardi, 2019). For instance, germline mutations or haploinsufficiency of ribosome biogenesis-associated genes cause ribosomopathies that compose a group of developmental disorders characterized by reduced viability and population of erythroid precursors, neural crest cells, or other tissue-specific cell types (Zhou et al., 2015; Calo et al., 2018). Consistently, multiple mouse strains with increased p53 activity due to conventional or conditional inactivation of Mdm2 and/or Mdmx, which encode two master inhibitors of p53, display diverse developmental defects (Bowen and Attardi, 2019). The pro-apoptotic function of p53 was first described by the Oren group in 1991 (Yonish-Rouach et al., 1991), which was validated later in mouse embryonic fibroblasts (MEFs) (Lowe et al., 1993) and cancer cells (Aubrey et al., 2018) and will be further discussed below (Figure 1). Recently, we have identified a new target gene of p53, SEPT4/ARTS, which encodes a pro-apoptotic protein that directly binds to p53 and enhances the interaction between p53 and BCL-XL at the mitochondria, resulting in augmented apoptosis (Hao et al., 2021a). In this review, we illustrate the mechanisms underlying p53 regulation of apoptosis, provide an updated overview of the function of apoptosis-related protein in TGF-β signaling pathway (ARTS) as a critical component of the p53-mediated apoptotic pathway, and discuss the clinical relevance of ARTS in cancer and non-cancer diseases.
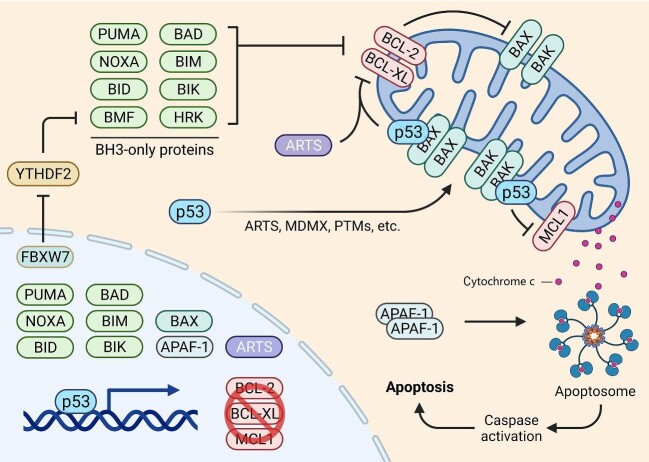
Figure 1.
p53-dependent mitochondrial apoptotic signaling network. Nuclear p53 transcriptionally activates the expression of PUMA, NOXA, BID, BAD, BIM, BIK, BAX, and APAF1, but represses the expression of BCL-2, BCL-XL, and MCL1. In addition, p53 may induce BMF expression through the FBXW7–YTHDF2 cascade. With the aid of ARTS, MDMX, and PTMs, cytoplasmic p53 can translocate to the mitochondria to interact with BAX, BAK, BCL-2, BCL-XL, and MCL1, consequently inducing apoptosis.
p53-dependent transcriptional regulation of the BCL-2 family genes
p53’s function is mainly executed through its transcriptional activity, because it can induce or repress gene expression by binding to the responsive elements (REs) on gene promoters. A typical RE sequence is degenerate and composed of two ‘half-sites’, 5′-RRRCWWGYYY-3′, which are separated by a spacer of 0–21 base pairs (Riley et al., 2008). NOXA was the first BH3-only gene identified as a direct transcriptional target of p53 (Oda et al., 2000). The expression of Noxa was found to be upregulated by X-ray irradiation in MEFs and thymocytes but not in p53-deficient (p53−/−) murine cells. After induction, Noxa translocates to the mitochondria and interacts with the anti-apoptotic BCL-2 members, leading to caspase-9 activation and apoptosis. Depletion of Noxa renders significant resistance of MEFs to DNA damage-induced and p53-mediated apoptosis (Shibue et al., 2003). PUMA, also known as BBC3, was later found to be another BH3-only gene critical for p53-induced apoptosis by three independent groups (Han et al., 2001; Nakano and Vousden, 2001; Yu et al., 2001). PUMA binds to and inhibits BCL-2 and BCL-XL at the mitochondria, consequently leading to cytochrome c release and APAF1-dependent apoptosis (Han et al., 2001; Nakano and Vousden, 2001; Yu et al., 2001). Genetic studies using Noxa or Puma knockout mice revealed that Puma acts as a more powerful mediator of p53-dependent apoptosis, as only loss of Puma can protect lymphocytes and hematopoietic cells from apoptosis induced by DNA damage stress (Jeffers et al., 2003; Villunger et al., 2003). Another study using combined knockout mouse models demonstrated that, although both Noxa and Puma are required for p53-induced apoptosis in murine fibroblasts and thymocytes, Puma has a predominant pro-apoptotic function in many other cell types (Michalak et al., 2008). Consistently, p53-dependent PUMA induction leads to chemotherapy-induced intestinal injury, while inhibition of PUMA protects intestinal stem cells against apoptosis (Leibowitz et al., 2018). The critical role of PUMA in p53-mediated apoptosis may be also reflected by the fact that the pseudo-caspase FLIP(L) suppresses apoptosis by selectively inhibiting p53 induction of PUMA, but not other pro-apoptotic target genes, such as NOXA and BAX (Lees et al., 2020). An exception is that loss of Noxa can inhibit UV radiation-induced apoptosis in murine fibroblasts and keratinocytes more dramatically than loss of Puma (Naik et al., 2007), suggesting that NOXA and PUMA may coordinate with each other to regulate apoptosis in the context of different cell types and cellular stresses (El-Saafin et al., 2022). These two BH3-only proteins are both critically important to p53-induced apoptosis, because their combined depletion can prevent murine cells from apoptosis as effectively as knockout of p53 (Michalak et al., 2008).
Besides these two pro-apoptotic genes, p53 can transcriptionally induce more BH3-only genes, including BID (Sax et al., 2002), BIK (Mathai et al., 2002), and BAD (Jiang et al., 2006). BIM was initially considered an indirect p53 target gene, as its expression could only be induced after 6 h treatment of etoposide, which was much later than the induction of PUMA and NOXA (Happo et al., 2010). However, a later study suggested that p53 may directly regulate BIM expression via a potential p53-RE on its promoter region through a genome-wide analysis (Kenzelmann Broz et al., 2013). BMF could be upregulated by acetylated p53, as IFN-γ-mediated HDAC1 deacetylation of p53 leads to reduced expression of BMF (Contreras et al., 2013). We recently found that the E3 ubiquitin ligase FBXW7 promotes proteolytic degradation of the m6A-binding protein YTHDF2, leading to stabilization of m6A-modified BMF mRNA (Xu et al., 2021). Given that p53 transcriptionally activates FBXW7 expression (Mao et al., 2004; Perez-Losada et al., 2005), our study implies that BMF may be an indirect target gene of p53. In addition, p53 represses the transcription of the anti-apoptotic members, BCL-2, BCL-XL, and MCL1, through indirect mechanisms (Miyashita et al., 1994; Sugars et al., 2001; Pietrzak and Puzianowska-Kuznicka, 2008; Aubrey et al., 2022). Collectively, p53 can promote mitochondrial apoptosis by either activating the expression of the pro-apoptotic BCL-2 family genes or inhibiting the expression of the anti-apoptotic genes.
Inhibition of the anti-apoptotic BCL-2 proteins leads to the activation of the pore-forming proteins, BAX and BAK, which is a prerequisite for MOMP and the release of cytochrome c (Oltvai et al., 1993; Chittenden et al., 1995; Farrow et al., 1995; Kiefer et al., 1995). BCL-2 and BCL-XL inactivate the pore-forming proteins by forming heterodimers with the latter (Yin et al., 1994; Farrow et al., 1995). When cells are under apoptotic stimulation, however, the BH3-only proteins bind to BAX and BAK to facilitate their homo-oligomerization, leading to the pore formation at the mitochondrial outer membrane and consequent cytochrome c release (Czabotar et al., 2013). Interestingly, a putative p53-RE was found on the BAX gene promoter, which was responsible for BAX transcription upon genotoxic stress in a p53-dependent fashion (Miyashita et al., 1994; Selvakumaran et al., 1994; Zhan et al., 1994; Miyashita and Reed, 1995). Recently, we unveiled an additional mechanism accounting for p53-induced BAX transcription (Liao et al., 2016). The transcription elongation factor TFIIS.h, which is encoded by the TCEA3 gene that is transcriptionally induced by p53, specifically associates with the genomic DNA and the transcripts of BAX, thereby enhancing its transcription (Liao et al., 2016). Thus, p53 can induce BAX mRNA expression by activating its transcription initiation and enhancing its transcription elongation via distinct mechanisms. Finally, p53 transcriptionally activates the APAF1-encoding gene, whose protein product can serve as a scaffold for apoptosome assembly and caspase activation (Soengas et al., 1999; Fortin et al., 2001; Kannan et al., 2001; Moroni et al., 2001; Robles et al., 2001). Taken together, these studies demonstrate that p53 can regulate the expression of a wide range of genes involved in the multistep mitochondrial apoptosis, including activation of pro-apoptotic and inhibition of anti-apoptotic BCL-2 proteins, MOMP, cytochrome c release, apoptosome assembly, and caspase activation (Figure 1).
Regulation of the BCL-2 family proteins by cytosolic p53
The transcription-independent pro-apoptotic activity of p53 was first described in the middle of the 1990s, as evidenced by the fact that several p53 mutants with deficiencies in transcription activity could still trigger apoptosis (Caelles et al., 1994; Haupt et al., 1995). Stress-activated p53 was found to translocate to the mitochondria, thereby eliciting cytochrome c release and caspase activation in both primary and cancer cells (Marchenko et al., 2000; Mihara et al., 2003). Mechanistically, mitochondrial p53, like many other BH3-only proteins, can form complexes with and, as such, inhibit the apoptotic antagonists, BCL-2 and BCL-XL (Mihara et al., 2003; Wei et al., 2021). This action may be facilitated by the MDM2 homolog, MDMX, as it promotes the translocation of cytoplasmic p53 to the mitochondria and enhances p53 interaction with BCL-2 (Mancini et al., 2009). Interestingly, the polymorphic variants of p53 were shown to have distinct accessibility to the mitochondria—the arginine-72 variant of p53 exhibits greater potential to trigger cytochrome c release than the proline-72 variant (Dumont et al., 2003; Almeida et al., 2021). Conversely, overexpression of the anti-apoptotic BCL-2 members can inhibit p53-induced apoptosis as well (Strasser et al., 1994; Chipuk et al., 2005). BCL-XL was found to bind to the DNA-binding domain (DBD) of p53 (Follis et al., 2014) and block cytoplasmic p53 from triggering apoptosis upon inhibition of EGFR-driven glucose metabolism, which leads to an effective combination therapy by targeting EGFR and pharmacologically stabilizing p53 (Mai et al., 2017). In addition, p53 directly binds to and activates the pore-forming protein, BAX, and depletion of BAX completely abrogates cytoplasmic p53-induced apoptosis in cancer cells and MEFs (Chipuk et al., 2003, 2004). p53 was also found to promote the translocation of BAX from the cytoplasm to the mitochondria through their direct interaction (Dubrez et al., 2001; Schuler et al., 2003). A later study showed that cis–trans isomerization of proline 47 within the p53 protein catalyzed by the prolyl isomerase PIN1 is required for BAX activation (Follis et al., 2015). Another pore-forming protein, BAK, is also critical for the pro-apoptotic function of cytoplasmic p53. p53 interacts with BAK at the mitochondria to facilitate homo-oligomerization of the latter through perturbation of the MCL1/BAK interaction, resulting in MOMP and cytochrome c release (Leu et al., 2004), while TRAF6-mediated p53 ubiquitination prevents its mitochondrial translocation and the interaction between p53 and MCL1/BAK (Zhang et al., 2016). Additionally, p53 can mediate the proteasomal degradation of MCL1, though the underlying mechanism is not well understood. Activation of p53 by the MDM2 inhibitor, RG7388, modulates phosphorylation of MCL1 and promotes its proteasomal degradation, thus overcoming apoptosis resistance of acute myeloid leukemia (Pan et al., 2017). Recently, p53 was also found to promote MCL1 degradation, resulting in the release of BAK and induction of apoptosis, in MYC-driven B-cell lymphomas (Domostegui et al., 2021). An interesting observation is that cancer-associated mutations in the DBD of p53 impair the pro-apoptotic activity of cytoplasmic p53 (Zhang et al., 2020), as they disrupt the interactions of BCL-XL and MCL1 with p53’s DBD (Mihara et al., 2003; Leu et al., 2004; Pietsch et al., 2008). Together, cytoplasmic p53 participates in mitochondrial apoptosis by physically interacting with and regulating BCL-2 family proteins (Figure 1).
The role of ARTS in promoting mitochondrial apoptosis
ARTS protein, encoded by an alternative spliced variant of the SEPT4 gene, is located at the outer membrane of the mitochondria (Figure 2; Larisch et al., 2000; Mandel-Gutfreund et al., 2011). ARTS was originally identified as a TGF-β-responsive protein by retroviral insertional mutagenesis screening, because overexpression of ARTS enhanced, whereas depletion of ARTS inhibited, TGF-β-induced caspase activation and apoptosis (Larisch et al., 2000). Later studies showed that ARTS can also be activated by a variety of pro-apoptotic stimuli, including staurosporine, arabinoside, 5-azacytidine, etoposide, and UV irradiation (Elhasid et al., 2004; Gottfried et al., 2004; Edison et al., 2012b). One of the mechanisms underlying the pro-apoptotic activity of ARTS is through inhibition of XIAP (Gottfried et al., 2004). Upon pro-apoptotic stimuli, a portion of ARTS molecules are released from the mitochondria to the cytoplasm, where they bind to the BIR1 and BIR3 domains of XIAP (Gottfried et al., 2004; Bornstein et al., 2011). Also, ARTS binds to and recruits the E3 ubiquitin ligase SIAH1 to promote ubiquitination and degradation of XIAP (Garrison et al., 2011). The translocation of ARTS to the cytoplasm precedes the release of cytochrome c and SMAC, which is therefore considered a critical step for apoptosis initiation (Edison et al., 2012b). Recently, ARTS was found to directly associate with the BH3 domain of BCL-2 and promote XIAP-induced BCL-2 proteasomal degradation (Edison et al., 2017). The physiological function of ARTS has also been extensively studied in multiple genetic mouse models, demonstrating that this apoptosis inducer plays essential roles in spermatogenesis, development of hematopoietic and intestinal stem cells, and skin regeneration (Kissel et al., 2005; Garcia-Fernandez et al., 2010; Fuchs et al., 2013; Koren et al., 2018). ARTS expression is controlled by surveillance mechanisms that maintain a low level of ARTS to support cell survival, while the ubiquitin-mediated degradation of ARTS is repressed when cells are under pro-apoptotic stimuli or DNA damage stress (Lotan et al., 2005). The apoptosis inhibitor, XIAP, which is repressed and degraded by ARTS (Gottfried et al., 2004; Bornstein et al., 2011; Garrison et al., 2011), was identified as an E3 ubiquitin ligase targeting ARTS for proteasomal degradation, thus forming a negative feedback circuit (Bornstein et al., 2012). In addition, the E3 ubiquitin ligase, Parkin, which is encoded by the Parkinson's disease-associated gene, PRKN, was found to promote ARTS ubiquitination and degradation in neuronal cells and rat brains (Kemeny et al., 2012). This finding may partially explain why mutations of PRKN result in neuronal cell death and neurodegeneration.
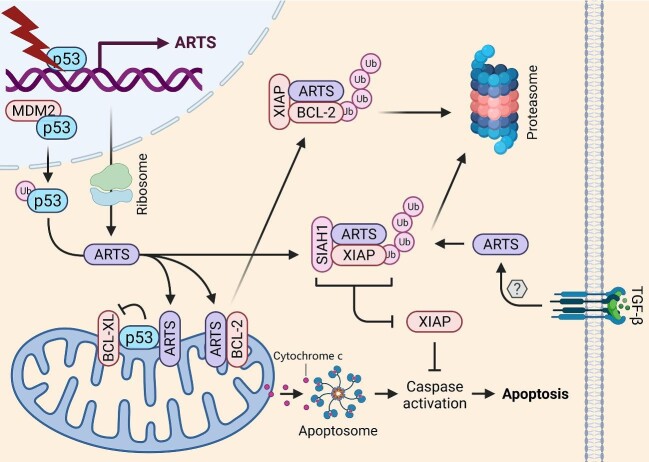
Figure 2.
The role of ARTS in promoting mitochondrial apoptosis. ARTS is encoded by a p53-target gene whose expression is responsive to diverse apoptotic stimuli, such as DNA damage stress and TGF-β activation. There are three mechanisms for ARTS-induced mitochondrial apoptosis: (i) ARTS facilitates the mitochondrial localization of p53 and enhances the p53–BCL-XL interaction at the mitochondria; (ii) ARTS suppresses XIAP and promotes its proteasomal degradation by recruiting SIAH1 as a ternary complex; and (iii) ARTS bridges BCL-2 and XIAP by directly binding to them, allowing for XIAP-mediated proteasomal degradation of BCL-2.
Recently, we identified SEPT4/ARTS as a p53-responsive gene that plays a critical role in p53-dependent mitochondrial apoptosis (Figure 2; Hao et al., 2021a). To elucidate the tumor-suppressive functions of p53, we performed a microarray analysis to screen the significant differentially expressed genes in colon cancer HCT116 cells treated with or without the p53-inducing agent, Inauhzin (Liao et al., 2012; Zhang et al., 2012). This screening led to the identification of several important transcriptional target genes of p53, such as NGFR (Zhou et al., 2016), PHLDB3 (Chao et al., 2016), TCEA3 (Liao et al., 2016), and SEPT4/ARTS (Hao et al., 2021a). To support this, treatment of cancer cells with various p53-inducing agents, including cisplatin, doxorubicin, 5-fluorouracil, and Nutlin-3, or overexpression of p53 in cancer cells, resulted in the increase of both mRNA and protein levels of ARTS. Additionally, γ-irradiation dramatically boosted Sept4/Arts expression in thymuses and spleens of p53+/+, but not p53−/−, mice. Importantly, a p53-RE at −2279 bp upstream of the transcription initiation site was validated through the luciferase reporter assay and the chromatin IP assay, demonstrating SEPT4/ARTS as a bona fide p53 target gene. Intriguingly, we accidentally found that mutant p53 might bind to a peptide (KLQDQALKE) encoded by the SEPT4 gene in ovarian cancer in another study (Chen et al., 2019). This prompted us to test whether wild-type p53 can also bind to ARTS, because both mutant and wild-type p53 share many common binding partners (Hao et al., 2020). Indeed, we further confirmed that p53 interacts with ARTS in cancer cells and, more importantly, at the mitochondria. Unlike NGFR (Zhou et al., 2016) and PHLDB3 (Chao et al., 2016), ARTS did not regulate p53 protein stability, as enforced expression of ARTS failed to affect the expression of exogenous and endogenous p53 in H1299 and HCT116 cells, respectively. Instead, ARTS facilitated p53 translocation to the mitochondria and increased the interaction of p53 with BCL-XL, consequently leading to inhibition of BCL-XL and augmented apoptosis. Remarkably, our study also revealed ARTS as a biomarker for tumor chemosensitivity, dependent on p53, because overexpression of ARTS enhanced, whereas depletion of ARTS impaired, chemotherapy-induced apoptosis in wild-type p53-harboring cancer cells (Hao et al., 2021a). Together, our findings unveil ARTS as a new p53 target and partner in the p53-mediated mitochondrial apoptotic pathway.
Relevance of ARTS in cancer and non-cancer diseases
Since ARTS is an apoptosis inducer by antagonizing the anti-apoptotic proteins, such as XIAP, BCL-2, and BCL-XL, as described above (Figure 2), it might act as a tumor suppressor. Supporting this speculation are the following lines of evidence. First, ARTS was found to be underexpressed in lymphoblasts of >70% of childhood acute lymphoblastic leukemia (ALL) patients (Elhasid et al., 2004). This cancer-associated deficiency was specific to the ARTS isoform, as the expression of H5, another variant encoded by SEPT4, was not affected, suggesting that an RNA splicing mechanism might be dysregulated in these patients. After treatment with chemotherapy, the levels of ARTS in lymphocytes were increased by 2–30 folds, which was correlated with disease remission. Also, leukemic cells lacking ARTS expression are resistant to chemotherapy-induced apoptosis. These findings support the idea that ARTS acts as both a tumor suppressor and a prognostic biomarker, at least, for ALL. The tumor-suppressive role of ARTS was further validated by a study using a Sept4-deficient mouse model (Garcia-Fernandez et al., 2010). Deletion of Sept4 led to an increased number of hematopoietic stem cells or tumor-initiating cells, upregulation of XIAP, resistance to apoptosis, and, consequently, accelerated development of leukemias and lymphomas. The findings suggest that ARTS mimetics or activators might serve as potential therapeutic agents for cancers with high levels of XIAP, BCL-2, or BCL-XL (Shahar and Larisch, 2020). The C-terminus of ARTS contains a unique 27-residue peptide, which is distinct from other known proteins (Edison et al., 2012a). Subdivision of this peptide revealed that a 9-residue peptide is sufficient for binding to the BIR3 domain of XIAP and inducing caspase activation and apoptosis. Recently, a candidate compound that mimics ARTS to specifically bind to XIAP was identified through a structure-based computational screen (Mamriev et al., 2020). The compound can induce apoptosis by promoting the degradation of both XIAP and BCL-2. In addition, loss of ARTS expression in leukemia is partially due to epigenetic silencing by DNA methylation (Elhasid et al., 2004). The methylation inhibitor, 5-azacytidine, was shown to elevate the expression of ARTS in both leukemic cell lines and ALL patients. Furthermore, genotoxic agents or p53-inducing agents can induce the expression of ARTS, consistent with our findings as described above (Hao et al., 2021a). It is thus speculated that the combination of methylation inhibitors, which remove DNA methylation from the SEPT4/ARTS promoter, with genotoxic agents, which induce p53 activation (Hao et al., 2021b), could be a more effective approach for the treatment of cancers, such as leukemia, which sustain wild-type p53 and lower levels of ARTS, by inducing ARTS expression and ARTS-dependent apoptosis. Hence, further exploring this translational potential in the near future would be tremendously conducive to the development of a new strategy for anti-cancer therapies.
In addition, ARTS induction may contribute to developmental defects caused by aberrant p53 activation, as depletion of Sept4/Arts promotes the development and renewal of several types of stem cells, including germinal, hematopoietic, and cutaneous stem cells, by inhibiting apoptosis (Kissel et al., 2005; Garcia-Fernandez et al., 2010; Fuchs et al., 2013). Thus, the development of ARTS antagonists could be helpful for treating subsets of ribosomopathies that are caused by tissue-specific activation of p53 (Zhou et al., 2015; Calo et al., 2018). Another possible clinical application of ARTS inhibition might prevent intestinal injury triggered by cancer chemotherapies. Most chemotherapeutic agents can cause intestinal dysfunction or enterotoxicity that is associated with intestinal crypt apoptosis (Leibowitz et al., 2018). Since knockout of Sept4/Arts promotes the renewal of Lgr5+ intestinal stem cells and thus the regeneration of crypts (Koren et al., 2018), targeting intestinal ARTS could be a promising strategy for alleviating normal intestinal damage without affecting the pro-apoptotic activity of p53 in cancer cells.
Acknowledgements
We thank the innovative research team of the high-level local university in Shanghai.
Contributor Information
Qian Hao, Fudan University Shanghai Cancer Center and Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Jiaxiang Chen, Department of Physiology, Medical College of Nanchang University, Nanchang 330006, China; Jiangxi Provincial Key Laboratory of Reproductive Physiology and Pathology, Nanchang University, Nanchang 330006, China.
Hua Lu, Department of Biochemistry & Molecular Biology and Tulane Cancer Center, Tulane University School of Medicine, New Orleans, LA 70112, USA.
Xiang Zhou, Fudan University Shanghai Cancer Center and Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China; Key Laboratory of Breast Cancer in Shanghai, Fudan University Shanghai Cancer Center, Fudan University, Shanghai 200032, China; Shanghai Key Laboratory of Medical Epigenetics, International Co-laboratory of Medical Epigenetics and Metabolism (Ministry of Science and Technology), Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China.
Funding
This study was supported by the National Natural Science Foundation of China (82072879 and 82273098 to X.Z., 82173022 to Q.H., and 82060278 to J.C.) and the Reynolds and Ryan Families Chair Fund in Transitional Cancer at Tulane to H.L.
Conflict of interest: none declared.
Author contributions: Q.H., J.C., and X.Z. drafted the manuscript; H.L. and X.Z. revised the manuscript.
References
- Almeida A., Sanchez-Moran I., Rodriguez C. (2021). Mitochondrial–nuclear p53 trafficking controls neuronal susceptibility in stroke. IUBMB Life 73, 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey B.J., Brennan M.S., Diepstraten S.T.et al. (2022). Loss of TRP53 reduces but does not overcome dependency of lymphoma cells on MCL-1. Cell Death Differ. 29, 1074–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey B.J., Kelly G.L., Janic A.et al. (2018). How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 25, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein B., Edison N., Gottfried Y.et al. (2012). X-linked inhibitor of apoptosis protein promotes the degradation of its antagonist, the pro-apoptotic ARTS protein. Int. J. Biochem. Cell Biol. 44, 489–495. [DOI] [PubMed] [Google Scholar]
- Bornstein B., Gottfried Y., Edison N.et al. (2011). ARTS binds to a distinct domain in XIAP-BIR3 and promotes apoptosis by a mechanism that is different from other IAP-antagonists. Apoptosis 16, 869–881. [DOI] [PubMed] [Google Scholar]
- Bowen M.E., Attardi L.D. (2019). The role of p53 in developmental syndromes. J. Mol. Cell Biol. 11, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C., Helmberg A., Karin M. (1994). p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370, 220–223. [DOI] [PubMed] [Google Scholar]
- Calo E., Gu B., Bowen M.E.et al. (2018). Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 554, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T., Zhou X., Cao B.et al. (2016). Pleckstrin homology domain-containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53. Nat. Commun. 7, 13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Hao Q., Wang J.et al. (2019). Ubiquitin ligase TRIM71 suppresses ovarian tumorigenesis by degrading mutant p53. Cell Death Dis. 10, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk J.E., Bouchier-Hayes L., Kuwana T.et al. (2005). PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309, 1732–1735. [DOI] [PubMed] [Google Scholar]
- Chipuk J.E., Kuwana T., Bouchier-Hayes L.et al. (2004). Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303, 1010–1014. [DOI] [PubMed] [Google Scholar]
- Chipuk J.E., Maurer U., Green D.R.et al. (2003). Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell 4, 371–381. [DOI] [PubMed] [Google Scholar]
- Chittenden T., Harrington E.A., O'Connor R.et al. (1995). Induction of apoptosis by the Bcl-2 homologue Bak. Nature 374, 733–736. [DOI] [PubMed] [Google Scholar]
- Contreras A.U., Mebratu Y., Delgado M.et al. (2013). Deacetylation of p53 induces autophagy by suppressing Bmf expression. J. Cell Biol. 201, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter T.G. (2009). Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer 9, 501–507. [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Westphal D., Dewson G.et al. (2013). Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531. [DOI] [PubMed] [Google Scholar]
- Domostegui A., Peddigari S., Mercer C.A.et al. (2021). Impaired ribosome biogenesis checkpoint activation induces p53-dependent MCL-1 degradation and MYC-driven lymphoma death. Blood 137, 3351–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrez L., Coll J.L., Hurbin A.et al. (2001). Caffeine sensitizes human H358 cell line to p53-mediated apoptosis by inducing mitochondrial translocation and conformational change of BAX protein. J. Biol. Chem. 276, 38980–38987. [DOI] [PubMed] [Google Scholar]
- Dumont P., Leu J.I., Della Pietra A.C. IIIet al. (2003). The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33, 357–365. [DOI] [PubMed] [Google Scholar]
- Edison N., Curtz Y., Paland N.et al. (2017). Degradation of Bcl-2 by XIAP and ARTS promotes apoptosis. Cell Rep. 21, 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edison N., Reingewertz T.H., Gottfried Y.et al. (2012a). Peptides mimicking the unique ARTS–XIAP binding site promote apoptotic cell death in cultured cancer cells. Clin. Cancer Res. 18, 2569–2578. [DOI] [PubMed] [Google Scholar]
- Edison N., Zuri D., Maniv I.et al. (2012b). The IAP-antagonist ARTS initiates caspase activation upstream of cytochrome C and SMAC/Diablo. Cell Death Differ. 19, 356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Saafin F., Bergamasco M.I., Chen Y.et al. (2022). Loss of TAF8 causes TFIID dysfunction and p53-mediated apoptotic neuronal cell death. Cell Death Differ. 29, 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhasid R., Sahar D., Merling A.et al. (2004). Mitochondrial pro-apoptotic ARTS protein is lost in the majority of acute lymphoblastic leukemia patients. Oncogene 23, 5468–5475. [DOI] [PubMed] [Google Scholar]
- Farrow S.N., White J.H., Martinou I.et al. (1995). Cloning of a bcl-2 homologue by interaction with adenovirus E1B 19 K. Nature 374, 731–733. [DOI] [PubMed] [Google Scholar]
- Follis A.V., Llambi F., Merritt P.et al. (2015). Pin1-induced proline isomerization in cytosolic p53 mediates BAX activation and apoptosis. Mol. Cell 59, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis A.V., Llambi F., Ou L.et al. (2014). The DNA-binding domain mediates both nuclear and cytosolic functions of p53. Nat. Struct. Mol. Biol. 21, 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin A., Cregan S.P., MacLaurin J.G.et al. (2001). APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J. Cell Biol. 155, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y., Brown S., Gorenc T.et al. (2013). Sept4/ARTS regulates stem cell apoptosis and skin regeneration. Science 341, 286–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez M., Kissel H., Brown S.et al. (2010). Sept4/ARTS is required for stem cell apoptosis and tumor suppression. Genes Dev. 24, 2282–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison J.B., Correa R.G., Gerlic M.et al. (2011). ARTS and Siah collaborate in a pathway for XIAP degradation. Mol. Cell 41, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried Y., Rotem A., Lotan R.et al. (2004). The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J. 23, 1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R. (2022). The mitochondrial pathway of apoptosis part II: the BCL-2 protein family. Cold Spring Harb. Perspect. Biol. 14, a041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R., Kroemer G. (2009). Cytoplasmic functions of the tumour suppressor p53. Nature 458, 1127–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Flemington C., Houghton A.B.et al. (2001). Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl Acad. Sci. USA 98, 11318–11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Chen J., Liao J.et al. (2021a). p53 induces ARTS to promote mitochondrial apoptosis. Cell Death Dis. 12, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Chen Y., Zhou X. (2020). The Janus face of p53-targeting ubiquitin ligases. Cells 9, 1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Lu H., Zhou X. (2021b). A potential synthetic lethal strategy with PARP inhibitors: perspective on ‘inactivation of the tumor suppressor p53 by long noncoding RNA RMRP’. J. Mol. Cell Biol. 13, 690–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happo L., Cragg M.S., Phipson B.et al. (2010). Maximal killing of lymphoma cells by DNA damage-inducing therapy requires not only the p53 targets Puma and Noxa, but also Bim. Blood 116, 5256–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Rowan S., Shaulian E.et al. (1995). Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 9, 2170–2183. [DOI] [PubMed] [Google Scholar]
- Ichim G., Tait S.W. (2016). A fate worse than death: apoptosis as an oncogenic process. Nat. Rev. Cancer 16, 539–548. [DOI] [PubMed] [Google Scholar]
- Jeffers J.R., Parganas E., Lee Y.et al. (2003). Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328. [DOI] [PubMed] [Google Scholar]
- Jiang P., Du W., Heese K.et al. (2006). The Bad guy cooperates with good cop p53: bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol. Cell. Biol. 26, 9071–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K., Kaminski N., Rechavi G.et al. (2001). DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene 20, 3449–3455. [DOI] [PubMed] [Google Scholar]
- Kemeny S., Dery D., Loboda Y.et al. (2012). Parkin promotes degradation of the mitochondrial pro-apoptotic ARTS protein. PLoS One 7, e38837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D., Spano Mello S., Bieging K.T.et al. (2013). Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 27, 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer M.C., Brauer M.J., Powers V.C.et al. (1995). Modulation of apoptosis by the widely distributed Bcl-2 homologue Bak. Nature 374, 736–739. [DOI] [PubMed] [Google Scholar]
- Kissel H., Georgescu M.M., Larisch S.et al. (2005). The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev. Cell 8, 353–364. [DOI] [PubMed] [Google Scholar]
- Koren E., Yosefzon Y., Ankawa R.et al. (2018). ARTS mediates apoptosis and regeneration of the intestinal stem cell niche. Nat. Commun. 9, 4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larisch S., Yi Y., Lotan R.et al. (2000). A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat. Cell Biol. 2, 915–921. [DOI] [PubMed] [Google Scholar]
- Lees A., McIntyre A.J., Crawford N.T.et al. (2020). The pseudo-caspase FLIP(L) regulates cell fate following p53 activation. Proc. Natl Acad. Sci. USA 117, 17808–17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz B.J., Yang L., Wei L.et al. (2018). Targeting p53-dependent stem cell loss for intestinal chemoprotection. Sci. Transl. Med. 10, eaam7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J.I., Dumont P., Hafey M.et al. (2004). Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat. Cell Biol. 6, 443–450. [DOI] [PubMed] [Google Scholar]
- Levine A.J. (2019). The many faces of p53: something for everyone. J. Mol. Cell Biol. 11, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J. (2020). p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480. [DOI] [PubMed] [Google Scholar]
- Liao J.M., Cao B., Deng J.et al. (2016). TFIIS.h, a new target of p53, regulates transcription efficiency of pro-apoptotic bax gene. Sci. Rep. 6, 23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J.M., Zeng S.X., Zhou X.et al. (2012). Global effect of Inauhzin on human p53-responsive transcriptome. PLoS One 7, e52172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tavana O., Gu W. (2019). p53 modifications: exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 11, 564–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R., Rotem A., Gonen H.et al. (2005). Regulation of the proapoptotic ARTS protein by ubiquitin-mediated degradation. J. Biol. Chem. 280, 25802–25810. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Ruley H.E., Jacks T.et al. (1993). p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74, 957–967. [DOI] [PubMed] [Google Scholar]
- Mai W.X., Gosa L., Daniels V.W.et al. (2017). Cytoplasmic p53 couples oncogene-driven glucose metabolism to apoptosis and is a therapeutic target in glioblastoma. Nat. Med. 23, 1342–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamriev D., Abbas R., Klingler F.M.et al. (2020). A small-molecule ARTS mimetic promotes apoptosis through degradation of both XIAP and Bcl-2. Cell Death Dis. 11, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F., Di Conza G., Pellegrino M.et al. (2009). MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J. 28, 1926–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y., Kosti I., Larisch S. (2011). ARTS, the unusual septin: structural and functional aspects. Biol. Chem. 392, 783–790. [DOI] [PubMed] [Google Scholar]
- Mao J.H., Perez-Losada J., Wu D.et al. (2004). Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432, 775–779. [DOI] [PubMed] [Google Scholar]
- Marchenko N.D., Zaika A., Moll U.M. (2000). Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275, 16202–16212. [DOI] [PubMed] [Google Scholar]
- Marino G., Niso-Santano M., Baehrecke E.H.et al. (2014). Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 15, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathai J.P., Germain M., Marcellus R.C.et al. (2002). Induction and endoplasmic reticulum location of BIK/NBK in response to apoptotic signaling by E1A and p53. Oncogene 21, 2534–2544. [DOI] [PubMed] [Google Scholar]
- Michalak E.M., Villunger A., Adams J.M.et al. (2008). In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 15, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M., Erster S., Zaika A.et al. (2003). p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11, 577–590. [DOI] [PubMed] [Google Scholar]
- Miyashita T., Krajewski S., Krajewska M.et al. (1994). Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9, 1799–1805. [PubMed] [Google Scholar]
- Miyashita T., Reed J.C. (1995). Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80, 293–299. [DOI] [PubMed] [Google Scholar]
- Moroni M.C., Hickman E.S., Lazzerini Denchi E.et al. (2001). Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3, 552–558. [DOI] [PubMed] [Google Scholar]
- Naik E., Michalak E.M., Villunger A.et al. (2007). Ultraviolet radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J. Cell Biol. 176, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Vousden K.H. (2001). PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694. [DOI] [PubMed] [Google Scholar]
- Oda E., Ohki R., Murasawa H.et al. (2000). Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- Oltvai Z.N., Milliman C.L., Korsmeyer S.J. (1993). Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74, 609–619. [DOI] [PubMed] [Google Scholar]
- Pan R., Ruvolo V., Mu H.et al. (2017). Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell 32, 748–760.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Losada J., Mao J.H., Balmain A. (2005). Control of genomic instability and epithelial tumor development by the p53–Fbxw7/Cdc4 pathway. Cancer Res. 65, 6488–6492. [DOI] [PubMed] [Google Scholar]
- Pietrzak M., Puzianowska-Kuznicka M. (2008). p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol. Chem. 389, 383–393. [DOI] [PubMed] [Google Scholar]
- Pietsch E.C., Perchiniak E., Canutescu A.A.et al. (2008). Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J. Biol. Chem. 283, 21294–21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T., Sontag E., Chen P.et al. (2008). Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412. [DOI] [PubMed] [Google Scholar]
- Robles A.I., Bemmels N.A., Foraker A.B.et al. (2001). APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res. 61, 6660–6664. [PubMed] [Google Scholar]
- Sax J.K., Fei P., Murphy M.E.et al. (2002). BID regulation by p53 contributes to chemosensitivity. Nat. Cell Biol. 4, 842–849. [DOI] [PubMed] [Google Scholar]
- Schuler M., Maurer U., Goldstein J.C.et al. (2003). p53 triggers apoptosis in oncogene-expressing fibroblasts by the induction of Noxa and mitochondrial Bax translocation. Cell Death Differ. 10, 451–460. [DOI] [PubMed] [Google Scholar]
- Selvakumaran M., Lin H.K., Miyashita T.et al. (1994). Immediate early up-regulation of bax expression by p53 but not TGFβ1: a paradigm for distinct apoptotic pathways. Oncogene 9, 1791–1798. [PubMed] [Google Scholar]
- Shahar N., Larisch S. (2020). Inhibiting the inhibitors: targeting anti-apoptotic proteins in cancer and therapy resistance. Drug Resist. Updat. 52, 100712. [DOI] [PubMed] [Google Scholar]
- Shibue T., Takeda K., Oda E.et al. (2003). Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 17, 2233–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Letai A., Sarosiek K. (2019). Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas M.S., Alarcon R.M., Yoshida H.et al. (1999). Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science 284, 156–159. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Jacks T.et al. (1994). DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell 79, 329–339. [DOI] [PubMed] [Google Scholar]
- Sugars K.L., Budhram-Mahadeo V., Packham G.et al. (2001). A minimal Bcl-x promoter is activated by Brn-3a and repressed by p53. Nucleic Acids Res. 29, 4530–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A., Michalak E.M., Coultas L.et al. (2003). p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038. [DOI] [PubMed] [Google Scholar]
- Wei H., Qu L., Dai S.et al. (2021). Structural insight into the molecular mechanism of p53-mediated mitochondrial apoptosis. Nat. Commun. 12, 2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Wang D. (2022). Deciphering the PTM codes of the tumor suppressor p53. J. Mol. Cell Biol. 13, 774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Li J., Ni M.et al. (2021). FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2. Mol. Cancer 20, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.M., Oltvai Z.N., Korsmeyer S.J. (1994). BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 369, 321–323. [DOI] [PubMed] [Google Scholar]
- Yonish-Rouach E., Resnitzky D., Lotem J.et al. (1991). Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352, 345–347. [DOI] [PubMed] [Google Scholar]
- Yu J., Zhang L., Hwang P.M.et al. (2001). PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682. [DOI] [PubMed] [Google Scholar]
- Zhan Q., Fan S., Bae I.et al. (1994). Induction of bax by genotoxic stress in human cells correlates with normal p53 status and apoptosis. Oncogene 9, 3743–3751. [PubMed] [Google Scholar]
- Zhang C., Liu J., Xu D.et al. (2020). Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 12, 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zeng S.X., Zhang Y.et al. (2012). A small molecule Inauhzin inhibits SIRT1 activity and suppresses tumour growth through activation of p53. EMBO Mol. Med. 4, 298–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li C.F., Zhang L.et al. (2016). TRAF6 restricts p53 mitochondrial translocation, apoptosis, and tumor suppression. Mol. Cell 64, 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Hao Q., Liao P.et al. (2016). Nerve growth factor receptor negates the tumor suppressor p53 as a feedback regulator. eLife 5, e15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao W.J., Liao J.M.et al. (2015). Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]