Summary
Background
Pseudomonas aeruginosa is an opportunistic pathogen consisting of three phylogroups (hereafter named A, B, and C). Here, we assessed phylogroup-specific evolutionary dynamics across available and also new P. aeruginosa genomes.
Methods
In this genomic analysis, we first generated new genome assemblies for 18 strains of the major P. aeruginosa clone type (mPact) panel, comprising a phylogenetically diverse collection of clinical and environmental isolates for this species. Thereafter, we combined these new genomes with 1991 publicly available P. aeruginosa genomes for a phylogenomic and comparative analysis. We specifically explored to what extent antimicrobial resistance (AMR) genes, defence systems, and virulence genes vary in their distribution across regions of genome plasticity (RGPs) and “masked” (RGP-free) genomes, and to what extent this variation differs among the phylogroups.
Findings
We found that members of phylogroup B possess larger genomes, contribute a comparatively larger number of pangenome families, and show lower abundance of CRISPR-Cas systems. Furthermore, AMR and defence systems are pervasive in RGPs and integrative and conjugative/mobilizable elements (ICEs/IMEs) from phylogroups A and B, and the abundance of these cargo genes is often significantly correlated. Moreover, inter- and intra-phylogroup interactions occur at the accessory genome level, suggesting frequent recombination events. Finally, we provide here the mPact panel of diverse P. aeruginosa strains that may serve as a valuable reference for functional analyses.
Interpretation
Altogether, our results highlight distinct pangenome characteristics of the P. aeruginosa phylogroups, which are possibly influenced by variation in the abundance of CRISPR-Cas systems and are shaped by the differential distribution of other defence systems and AMR genes.
Funding
German Science Foundation, Max-Planck Society, Leibniz ScienceCampus Evolutionary Medicine of the Lung, BMBF program Medical Infection Genomics, Kiel Life Science Postdoc Award.
Keywords: Pangenome, Pseudomonas aeruginosa, Regions of genome plasticity, Antibiotic resistance, CRISPR-Cas systems, Defence systems
Research in context.
Evidence before this study
To date, pangenome studies exploring the epidemiology and evolutionary dynamics of bacterial pathogens have been limited due to the use of gene frequencies across whole species dataset without accounting for biased sampling or the population structure of the genomes in the dataset. We searched PubMed without language restrictions for articles published before September 1, 2021, that investigated the phylogroup-specific evolutionary dynamics across bacterial species. In this literature search we used the search terms “pangenome” and “phylogroup” or “uneven”, which returned 14 results. Of these, only one study used a population structure-aware approach to explore pangenome dynamics in a bacterial species consisting of multiple phylogroups with an uneven number of available genomes per phylogroup.
Added value of this study
To our knowledge, this study is the first to assess phylogroup-specific evolutionary dynamics in a collection of genomes belonging to the nosocomial pathogen Pseudomonas aeruginosa. Using a refined analysis approach, we found specific signatures for each of the three phylogroups. We demonstrate that members of phylogroup B contribute a comparatively larger number of pangenome families, have larger genomes, and a lower prevalence of CRISPR-Cas systems. Additionally, we observed that antibiotic resistance and defence systems are pervasive in regions of genome plasticity and integrative and conjugative/mobilizable elements from phylogroups A and B, and that antibiotic resistance and defence systems are often significantly correlated in these mobile genetic elements.
Implications of all the available evidence
These results indicate that biases inherent in traditional pangenome analysis approaches can obscure the true distribution of important cargo genes in a bacterial species with a complex population structure. Furthermore, our findings highlight the specific characteristics of distinct phylogroups of the opportunistic human pathogen P. aeruginosa and shed new light on the role that integrative and conjugative/mobilizable elements may play in protecting the host from foreign DNA.
Introduction
Pseudomonas aeruginosa is a ubiquitous metabolically versatile γ-proteobacterium. This Gram-negative bacterium is an opportunistic human pathogen commonly linked to life-threatening acute and chronic infections.1 It belongs to the ESKAPE pathogens collection,2 highlighting its major contribution to nosocomial infections across the globe and its ability to “escape” antimicrobial therapy because of the widespread evolution of antimicrobial resistance (AMR).3 This species is also often found to be multi- as well as extensively drug resistant (MDR and XDR, respectively),4 making it difficult and in some cases even impossible to treat. For this reason, P. aeruginosa has been placed by the World Health Organization (WHO) in the top priority group of most critical human pathogens, for which new treatment options are urgently needed.5 These efforts rely on an in-depth understanding of the species’ biology and its evolutionary potential, which may be improved through a functional analysis of whole genome sequencing data.
The combined pool of genes belonging to the same bacterial species is commonly referred to as the pangenome. Often, only a small fraction of these genes is shared by all members of the species (the core genome). On the contrary, a substantial fraction of the total gene pool is heterogeneously distributed across the members (the accessory genome). Following Koonin and Wolf,6 the pangenome can be divided into 3 categories: i) the persistent or softcore genome, for gene families present in the majority of the genomes; ii) the shell genome, for those present at intermediate frequencies and which are gained and lost rather slowly; iii) the cloud genome, for gene families present at low frequencies in all genomes and which are rapidly gained and lost.7 Clusters of genes that are part of the accessory genome (i.e, the shell and cloud genome) are often located in so-called regions of genome plasticity (RGPs), genomic loci that appear to be prone to insertion of foreign DNA. By harbouring divergent accessory DNA in different strains, these loci can represent highly variable genomic regions. The shell and cloud genomes are also characterized by mobile genetic elements (MGEs) that can be transferred laterally between bacterial cells, including plasmids, integrative and conjugative/mobilizable elements (ICEs/IMEs), and prophages.8,9 These MGEs can mediate the exchange of cargo genes that may provide a selective advantage to the recipient cell, such as resistance to antibiotics, increased pathogenicity, and defence systems against foreign DNA.10, 11, 12
Most pangenome studies described to date have characterized gene frequencies across the whole species dataset without accounting for biased sampling or the population structure of the genomes in the dataset. This is particularly relevant for species consisting of multiple phylogroups with unevenly distributed members. As recently reported for Escherichia coli,13 genes classified as part of the accessory genome using traditional pangenome approaches are actually core to specific phylogroups. Since P. aeruginosa is composed of three different-sized phylogroups (hereafter referred to as phylogroups A, B, and C as per the nomenclature proposed by Ozer et al.14; see also Results), characterized by high intraspecific functional variability,15,16 it is likely that evolution in these phylogroups is driven by specific sets of genes found in the majority of members within the groups, but not across groups.
The aim of the current study is to enhance our understanding of the pangenome of the human pathogen P. aeruginosa by specifically assessing phylogroup-specific characteristics and genome dynamics, including data from more than 2000 genomes. We explore to what extent particular groups of cargo genes, such as those encoding AMR, virulence, and defence systems, vary in their distribution across RGPs and “masked” (RGP-free) genomes, and to what extent this variation differs among the phylogroups. Our data set includes new complete genome sequences of a representative set of P. aeruginosa strains, the ‘major P. aeruginosa clone type’ (mPact) strain panel. This set of strains was previously isolated from both clinical and environmental samples and made available by the Tümmler lab (Hanover, Germany).17 This mPact panel encompasses the most common clone types in the contemporary population18, 19, 20 and provides a manageable, focused resource for in-depth functional analysis.
Methods
Sequencing and hybrid assembly of the mPact strain panel
Genomic DNA from 18 strains of the mPact panel17 were extracted using the Macherey–Nagel NucleoSpin Tissue kit, according to the standard bacteria support protocol from the manufacturer. We used Nanodrop 1000 for DNA quantification and quality control (260/280 and 260/230 ratios), followed by measurements in Qubit for a more precise quantification. The Agilent TapeStation and the FragmentAnalyzer Genomic DNA 50 KB kit served to control fragment size. Sequencing libraries were prepared with the Illumina Nextera DNA flex and Pacific Bioscience (PacBio) SMRTbell express template prep kit 2.0. Libraries were sequenced on the Illumina MiSeq at 2 × 300bp or the PacBio Sequel II, respectively. Illumina reads were quality checked using FastQC v0.11.9 and trimmed with Trim Galore v0.6.6, using the paired-end mode with default parameters and a quality Phred score cutoff of 10. Both datasets were then combined using the Unicycler v0.4.8 assembly pipeline.21 We used the default normal mode in Unicycler to build the assembly graphs of most strains, except of the mPact strains H02, H14, H15, H18, and H19, where we used the bold mode. The assemblies were visually inspected using the assembly graph tool Bandage v0.8.1.22
Bacterial collection
We downloaded a total of 5468 P. aeruginosa genomes from RefSeq's NCBI database using PanACoTA v1.2.0.23 After quality control to remove low-quality assemblies, 2704 were retained and 2764 genomes with more than 100 contigs were discarded (Table S1). Next, 713 genomes were discarded by the distance filtering step, using minimum (1e-4) and maximum (0.05) mash distance cut-offs to remove duplicates and misclassified assemblies at the species level,24 respectively. This resulted in 1991 publicly available genomes. The 18 genomes sequenced in this study from the mPact panel17 passed both filtering steps, resulting in a pruned collection of 2009 genomes in total. Multi-locus sequence typing (MLST) profiles were determined with mlst v2.19.0 (https://github.com/tseemann/mlst).
Pangenome and phylogenomics
The average nucleotide identity (ANI) between the 2009 genomes was calculated with fastANI v1.33.24 We used the genome sequences to generate a pangenome with the panrgp subcommand of PPanGGOLiN v1.1.136.25,26 We built a softcore-genome alignment (threshold 95%), followed by inference of a maximum likelihood tree with the General Time Reversible model of nucleotide substitution in IQ-TREE v2.1.2.27 To detect recombination events in our collection and account for them in phylogenetic reconstruction, we used ClonalFrameML v1.12.28 Phylogenetic trees were plotted in iTOL v6 (https://itol.embl.de/)29 and related to cluster genomes according to the phylogroup. Due to differences in sample size, we subsequently focused the analysis on each phylogroup separately. Pangenome analysis was performed for each phylogroup, using the panrgp subcommand of PPanGGOLiN. Core and accessory genes were classified across genomes from different phylogroups with a publicly available R script (https://github.com/ghoresh11/twilight).13 We used the gene presence/absence output from the whole collection's pangenome and the grouping of our genomes according to the phylogroup.
Identification of RGPs and ICEs/IMEs
To mask all the genomes, we used the RGP coordinates determined by panrgp for each individual genome as input to bedtools maskfasta v2.30.0.30 We extracted the RGP nucleotide sequences with the help of bedtools getfasta. All genomes were annotated with prokka v1.4.6.31 To search for ICEs/IMEs on complete genomes, we used the genbank files generated by prokka as input in the standalone-version of ICEfinder.32
Annotation of functional categories
We retrieved the annotated proteins for the RGPs and masked genomes across the three phylogroups. We clustered each of the six groups of proteins with MMseqs2 v13.4511133 and an identity cut-off of 80%. These clustered proteins were scanned for functional categories in eggNOG-mapper v2,34 using the built-in database for clusters of orthologous groups.35 We calculated the relative abundance of these categories by dividing the absolute counts for each category by the total number of clustered proteins found in each of the six groups. CRISPR-Cas systems were identified with the help of CRISPRCasTyper v1.2.3.36 AMRFinder v3.10.1837 served to locate AMR genes and resistance-associated point mutations. Virulence genes were characterized with the pre-downloaded database from VFDB38 (updated on the 12-05-2021 and including 3867 virulence factors) in abricate v1.0.1 (https://github.com/tseemann/abricate). Finally, we searched for defence systems using the protein sequences generated by prokka as input in defense-finder v0.0.11,39 a tool developed to identify known defence systems in prokaryotic genomes, for which at least one experimental evidence of the defence function is available.
Network-based analysis of RGPs and ICEs/IMEs
As a first step, we calculated the Jaccard Index between the RGPs with the help of BinDash v0.2.140 with k-mer size equal to 21 bp. In detail, we used the sketch subcommand to reduce multiple sequences into one sketch, followed by the dist subcommand, to estimate distance (and relevant statistics) between RGPs in query sketch and RGPs in target-sketch. The Jaccard Index between ICEs/IMEs was similarly obtained with BinDash. We used the mean () function in R to calculate the arithmetic mean of the Jaccard Index. Only Jaccard Index values equal to or above the mean were considered, and the mutation distances served as edge attributes to plot the networks with Cytoscape v3.9.1 under the prefuse force directed layout (https://cytoscape.org/). Based on the Analyzer function in Cytoscape, we computed a comprehensive set of topological parameters, such as the clustering coefficient, the network density, the centralization, and the heterogeneity. Clusters in our networks were identified with the AutAnnotate and clusterMaker apps available in Cytoscape, using the connected components as the clustering algorithm.
Statistical analysis
The correlation matrix was ordered using the hclust function in R. Statistical comparison of the variation between groups was always based on non-parametric tests, thereby taking into account that the compared groups varied in data distributions (e.g., at least one group with a skewed distribution) and/or showed unequal variances. Moreover, as non-parametric tests are usually considered to be conservative, the thus identified significant test results should indicate trustworthy differences between groups. In particular, the three phylogroups (e.g., genome size, GC content) were generally compared using the Kruskal-Wallis test. The unpaired two-sample Wilcoxon test was used for multiple comparisons between two independent groups of samples (RGPs vs. masked genomes, CRISPR-Cas positive vs negative genomes). For both tests, p-values were adjusted using the Holm–Bonferroni method. Values above 0.05 were considered as non-significant (ns). We used the following convention for symbols indicating statistical significance: ∗ for p ≤ 0.05, ∗∗ for p ≤ 0.01, ∗∗∗ for p ≤ 0.001, and ∗∗∗∗ for p ≤ 0.0001.
Role of funding source
The funders (Max-Planck Society, German Science Foundation, German ministry for education and research, Kiel university) had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data and take final responsibility for the decision to submit for publication. None of the contributing authors was denied access to the data of this study.
Results
The P. aeruginosa phylogeny is composed of three phylogroups
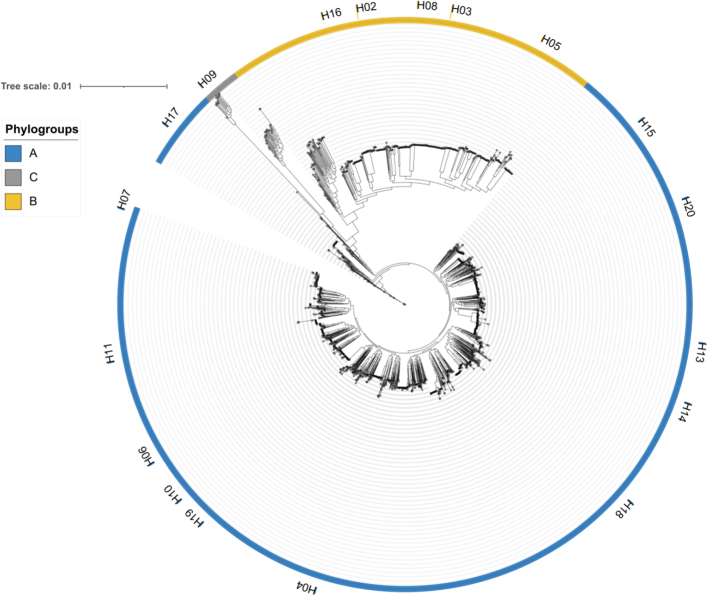
Our phylogenomic characterization was based on 2009 assembled P. aeruginosa genomes belonging to 519 MLST profiles and including 1991 publicly available genomes (following quality control and distance filtering, Table S2) and an additional 18 genomes for the mPact strain panel (Table S3).17 Analysis of the ANI values (Fig. S1) and the softcore-genome alignment of these genomes identified three phylogroups, as previously reported14,41 (Fig. 1). The two main reference isolates are part of the larger phylogroups: PAO142 belongs to phylogroup A (n = 1531), while the PA14 strain falls into phylogroup B (n = 435). Phylogroup C contains a substantially smaller number of members (n = 43) (Table S2). Members of the phylogroup C have recently been subdivided into either 214 or 3 clusters, including the distantly related PA7 cluster.41 In this work, however, the PA7 cluster was excluded, and we focused our analysis only on the remainder of phylogroup C, because the PA7 cluster genomes were too distantly related to the other genomes. Indeed, the PA7 strain was first described as a taxonomic outlier of this species,43 and genomes belonging to this cluster have recently been proposed to belong to a new Pseudomonas species.44 To test the effect of recombination on the softcore-genome alignment, we used ClonalFrameML to reconstruct the phylogenomic tree with corrected branch lengths. The segregation of P. aeruginosa into three phylogroups was maintained, resulting in a tree with decreased branch lengths and with identical number of members assigned to each phylogroup (Fig. S2). Genomes from the mPact panel sequenced in this study were widely distributed across the P. aeruginosa phylogeny, with 12 strains in phylogroup A, 5 in phylogroup B, and 1 in phylogroup C (Fig. 1 and Table S3). Our results show that P. aeruginosa consists of three asymmetric phylogroups and that the segregation of the 2009 genomes into phylogenetically distinct groups is not an artefact of recombination.
Fig. 1.
Maximum-likelihood tree of the softcore-genome alignment of all P. aeruginosa isolates used in this study (n = 2009). The scale bar represents the genetic distance. Arcs in blue represent phylogroup A, yellow B, and grey C. The phylogenetic placement of the major P. aeruginosa clone type (mPact) strain panel, sequenced in this study, is highlighted in the tree, with the strain name (the “H” before each number stands for Hanover, referring to the location of the Tuemmler lab and the study that first described this collection17) next to strips coloured according to the phylogroup.
Phylogroup B contributes comparatively more gene families to the pangenome than the other two phylogroups
We next built a pangenome for the entire species, and separate pangenomes for each of the three phylogroups. This latter approach is important to account for phylogenetic subdivisions of the species, which is additionally critical because the three phylogroups in our collection have substantially different sample sizes. We observed that the number of persistent gene families in the larger phylogroups A and B was similar to that found in the species as a whole, while the phylogroup C contained a substantially smaller number of persistent gene families (Table S4).
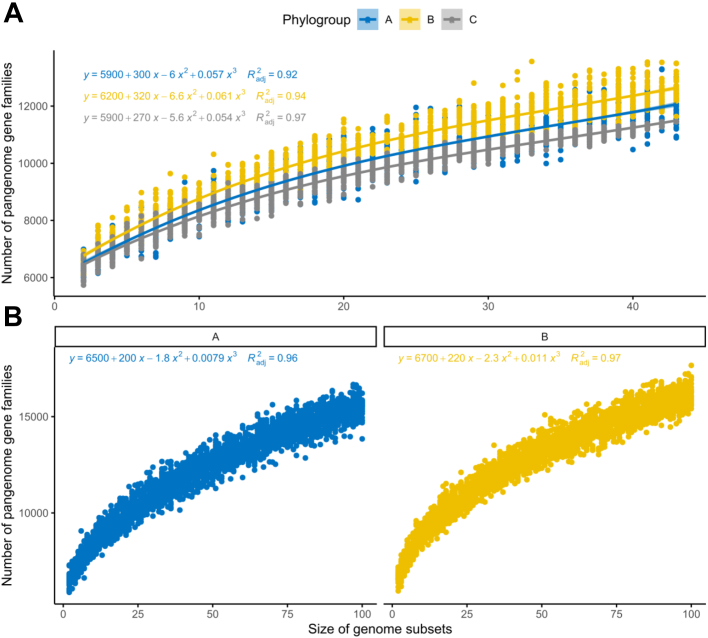
The pangenome of bacterial species is usually classified into two types: open and closed pangenomes.45 Since P. aeruginosa is an example of a bacterial species with an open pangenome14 (i.e., the sequencing of new genomes will increase the size of the pangenome), we explored the contribution of each phylogroup to the pangenome. To ensure comparability among the three phylogroups in our first analysis, we randomly sampled 43 genomes from each phylogroup (thus, including the total sample size of the smallest phylogroup C), and observed that there is more diversity in the accessory genes of phylogroup B in terms of the functions contributed by the acquired genes (Fig. 2A and Table S5). In our second analysis, we focused only on the two larger phylogroups A and B, for which we randomly sampled 100 genomes each, and found the trend unchanged (Fig. 2B and Table S6).
Fig. 2.
Rarefaction curves of the pangenome gene families for each phylogroup. All curves were inferred using polynomial regression lines. Curves in blue represent phylogroup A, yellow B, and grey C. A) The curves were generated by randomly re-sampling 43 genomes from each phylogroup several times and then plotting the average number of pangenome families found on each genome. B) Rarefaction curves were plotted with 100 random genomes from phylogroups A and B.
We then explored if specific gene families were pervasive across single or multiple phylogroups. We found 14 phylogroup-specific softcore gene families in phylogroup C, and one gene family each was exclusively found in the softcore genomes of phylogroups A and B, respectively (Fig. S3 and Table S7). Most gene families uniquely found on the softcore genome of phylogroup C were part of the Xcp type-II secretion system (T2SS), which is one of two complete and functionally distinct T2SS present in this species (Table S8). The Xcp system is encoded in a cluster containing 11 genes (xcpP–Z), plus an additional xcpA/pilD gene found elsewhere in the genome.46 These genes were also found in the majority of the genomes from phylogroups A and B (Table S9), but the encoded proteins were too distantly related to those from phylogroup C. A similar pattern was observed for the two gene families indicated exclusively for either phylogroup A or B, for which we also found distantly related orthologues in phylogroup C. Taken together, these results highlight that phylogroup B differs from the other two in that it contributes a comparatively larger number of gene families to the pangenome, possibly suggesting that phylogroup B members have larger genomes.
Phylogroup B genomes are significantly larger and most carry no CRISPR-Cas systems
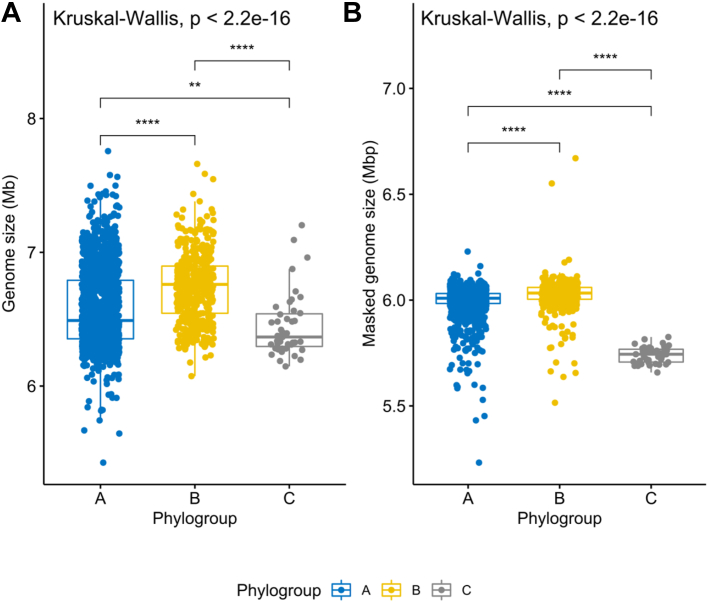
A comparison of genome lengths revealed significantly larger genome sizes for phylogroup B than the other two phylogroups (Fig. 3A, p-value <2.2e-16). We then extracted the RGPs from each phylogroup, and found a total of 57901 RGPs across the three phylogroups. The RGPs from phylogroup B were significantly larger than those from phylogroup A (Fig. S4), thus at least contributing to the overall size difference. Nevertheless, after removing the RGPs, the resulting “masked” genomes from phylogroup B were still significantly larger than those from the other two phylogroups (Fig. 3B, p-value <2.2e-16). Additionally, we found that genomes from phylogroup B were still significantly larger than those from the other two phylogroups, even when the sample sizes of the phylogroups were adjusted to the sample size of the smallest group, phylogroup C (with 43 genomes; Fig. S5, p-value 3.2e-07). These results point to a potentially higher number of genes conserved across genomes from phylogroup B. Still, the difference in genome size between phylogroups A and B is mainly explained by differences in accessory genome size (Fig. S4). Masked genomes from phylogroup C are significantly smaller than genomes from the other two phylogroups, which is consistent with the smaller number of persistent gene families identified in this phylogroup (Table S4). We further explored variation in GC content and observed that the GC content from phylogroup B genomes was significantly lower than that from other phylogroups (Fig. S6, p-value < 2.2e-16).
Fig. 3.
Boxplots representing the variation in genome size (A) and masked genome size (B) across the three phylogroups. Values above 0.05 were considered as non-significant (ns). Stars indicate significance level: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. Boxplots in blue represent phylogroup A, yellow B, and grey C.
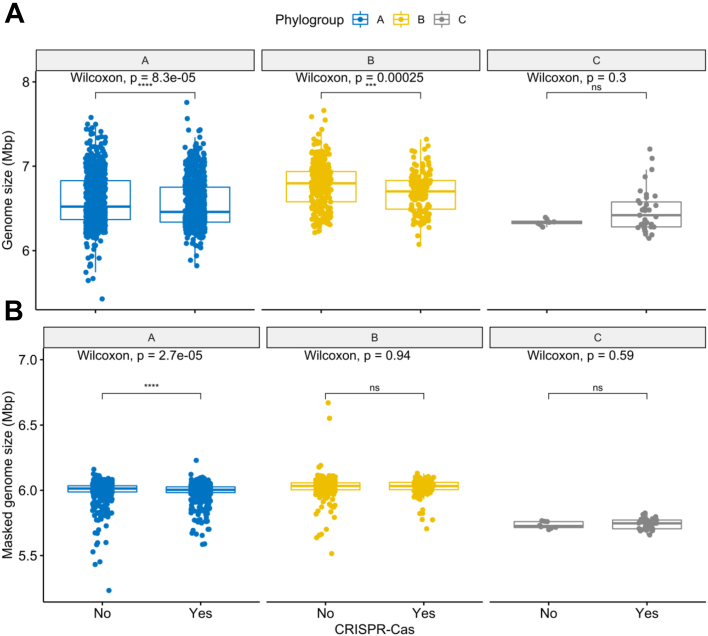
We next assessed whether presence of the defence CRISPR-Cas system was associated with genome size variation. Since CRISPR-Cas systems are important to defend bacteria against foreign DNA,12,47 we expected that genomes carrying these systems would be smaller, whereas those lacking these systems would accumulate mobile elements and hence be larger. We subdivided genomes from each of the three phylogroups into two groups depending on whether they contain or lack CRISPR-Cas systems, respectively (CRISPR-Caspos, CRISPR-Casneg). We indeed found that genomes with CRISPR-Cas systems are significantly smaller than those without (Fig. 4A, p-values 8.3e-05 and 0.00025 for the phylogroup A and B comparisons, respectively), supporting the hypothesis that CRISPR-Cas systems may restrict horizontal gene transfer in P. aeruginosa.48, 49, 50 While the number of CRISPR-Caspos and CRISPR-Casneg genomes in phylogroups A and C is evenly distributed, phylogroup B genomes without CRISPR-Cas (n = 279) were almost twice as abundant as those that carried these systems (n = 156, Table S2). Interestingly, the masked genome size of CRISPR-Caspos and CRISPR-Casneg phylogroup B isolates was no longer significantly different from one another (Fig. 4B). In line with this finding, we observed that the cumulative size of all RGPs was higher in genomes without CRISPR-Cas systems across phylogroups A and B (Fig. S7). The absence of these defence systems in most genomes from phylogroup B may help to explain the observed larger size.
Fig. 4.
Boxplots showing the variation in genome size (A) and masked genome size (B) across pairs of conspecific genomes from the same phylogroup with and without CRISPR-Cas systems. Values above 0.05 were considered as non-significant (ns). Stars indicate significance level: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. Boxplots in blue represent phylogroup A, yellow B, and grey C.
We observed a greater diversity of CRISPR-Cas systems in genomes from phylogroup A, including I–C, I–E, I–F, IV-A1, and IV-A2 (Fig. S8 and Table S10). These CRISPR-Cas subtypes were all found in genomes from phylogroup B, with the exception of the IV-A2. Curiously, only subtypes I-E and I–F were present in phylogroup C. Type IV CRISPR-Cas systems were found almost exclusively on plasmids, and recent work has shown that they contribute to plasmid–plasmid warfare.12,51 The type I–C CRISPR–Cas subtype is typically encoded on ICEs and is also involved in competition dynamics between mobile elements.49,52 Overall, our findings show that phylogroup B genomes are significantly larger and have a wider pool of accessory genes than those from the other two phylogroups, possibly driven by the lower prevalence of CRISPR-Cas systems in phylogroup B.
AMR and defence systems are overrepresented in RGPs from phylogroups A and B
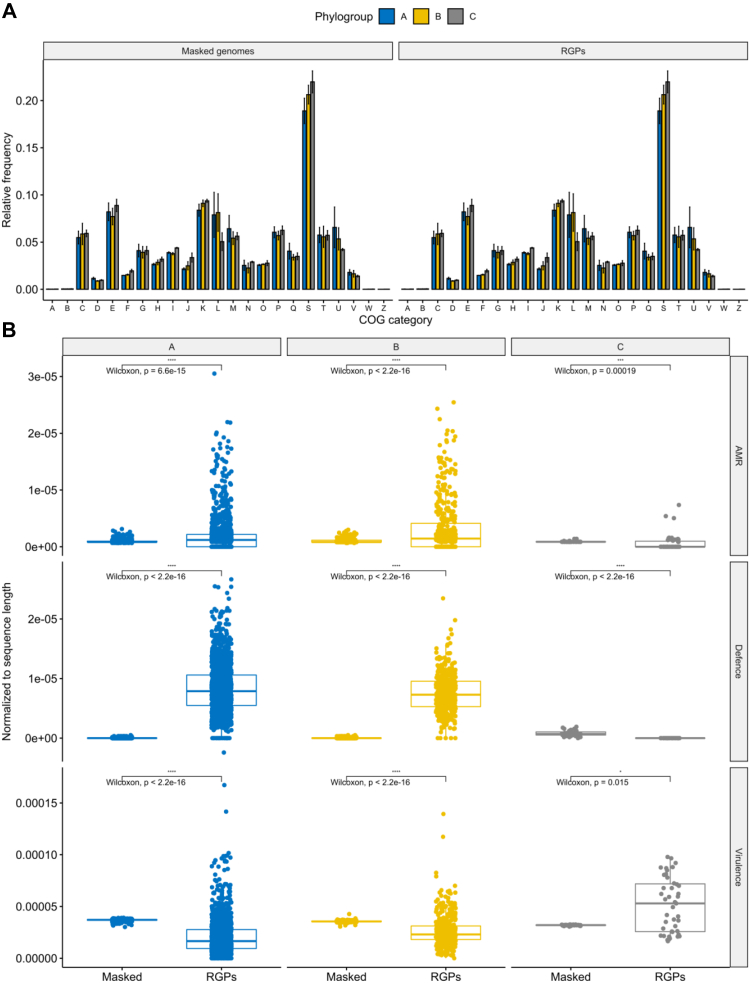
We next assessed variation in the relative abundance of proteins encoded in RGPs from different phylogroups. We observed that most functional categories are conserved across phylogroups. However, proteins coding for replication, recombination and repair functions were more prevalent in phylogroups A and B RGPs than those from phylogroup C (Fig. 5A). Since these proteins are often involved in mobilization, this finding may suggest that genomes in these phylogroups have more functional mobile elements, with the ability to be horizontally transferred, whereas the RGPs in phylogroup C may be derived from remnants of mobile elements that can no longer be mobilized.
Fig. 5.
Distribution of functional categories across RGPs and masked genomes from the different phylogroups. Bar and boxplots in blue represent phylogroup A, yellow B, and grey C. (A) Relative frequencies of cluster of orthologous groups categories. The relative frequencies were calculated by dividing the absolute counts for each category by the total number of clustered proteins found in each of the six groups. Error bars indicate the degree of variation across each COG category from each phylogroup across RGPs and masked genomes. The functional categories are indicated by capital letters, including: A, RNA processing and modification; B, chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control and mitosis; E, amino acid metabolism and transport; F, nucleotide metabolism and transport; G, carbohydrate metabolism and transport; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelop biogenesis; N, cell motility; O, post-translational modification, protein turnover, chaperone functions; P, inorganic ion transport and metabolism; Q, secondary structure; R, general functional prediction only; S, function unknown; T, signal transduction; U, intracellular trafficking and secretion; V, defence mechanisms; W, extracellular structures; Z, cytoskeleton. (B) Boxplots of the variation in the number of AMR genes, defence systems, and virulence genes found in RGPs and masked genomes across the three phylogroups. Absolute counts of genes and systems were normalized to RGP and masked genome sequence lengths in each strain. Values above 0.05 were considered as non-significant (ns). Stars indicate significance level: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
We next assessed to what extent RGPs and masked genomes vary in prevalence of genes for three types of functions, which are often encoded on MGEs, including virulence, defence systems, and AMR. Since the cumulative size of all RGPs is substantially smaller than that of masked genomes (Table S2), the number of virulence genes, defence systems, and AMR genes was normalized to the sequence length of the RGPs and masked genomes for each strain. We observed that the gene prevalence for these functions is conserved across masked genomes from different phylogroups, whereas they are unevenly distributed in RGPs (Fig. 5B).
Two important virulence factors were only present in some genomes from phylogroup C, and absent from the other two phylogroups (Table S9). These genes (exlA and exlB) encode hemolysins, and when genomes from phylogroup C carry these genes, the typical type-III secretion system (T3SS) machinery found in most bacteria (encoding the toxins ExoS, ExoY, ExoT, and ExoU) is absent from these genomes, supporting previous reports that these are mutually exclusive.53 In agreement with previous findings,14 we further found that two important genes encoding T3SS effector proteins (exoS and exoU) were unevenly distributed across the phylogroups: the exoS gene was pervasive among genomes from phylogroup A (99.5%, 1524/1531) and the majority of phylogroup C strains (28/43), while the exoU gene was overrepresented in genomes from phylogroup B (408/435) and nearly absent in genomes from the other two phylogroups (Table S9). Surprisingly, we also found 23 genomes with the atypical exoS+/exoU+ genotype, all belonging to phylogroup A (Table S9). A high frequency of this genotype has recently been reported in patients from the Brazilian Amazon and Peruvian hospitals.54,55 As expected,56 some virulence genes were exclusively found on RGPs (i.e., absent from masked genomes): flagellar-associated proteins fleI/flag, flgL, fliC and fliD, as well as wzy, which codes for an O-antigen chain length regulator. All these virulence genes were found in RGPs from both phylogroups A and B.
In agreement with the important role of MGEs as vectors for AMR genes in P. aeruginosa,9,57 we found that AMR genes were overrepresented in RGPs from phylogroups A and B (Fig. 5B and S9). We then calculated the relative proportion of different AMR classes across RGPs from the three phylogroups, revealing that most AMR classes were overrepresented across RGPs from phylogroup B (Fig. S10). This result is consistent with our finding that RGPs play a significant role in the larger genome sizes from this phylogroup (Fig. S4). Point mutations linked to resistance to beta-lactams and quinolones were observed for all phylogroups (Table S11).
A wide array of defence systems has recently been characterized in P. aeruginosa to show a patchy distribution in closely related and distantly related strains, suggesting high rates of horizontal gene transfer.39 According to this hypothesis, we would expect to observe an abundance of defence systems in RGPs, when compared with masked genomes. Similar to our results for AMR genes, we found that defence systems are indeed overrepresented in RGPs from phylogroups A and B (Fig. 5B). Defence systems such as the globally distributed restriction-modification and CRISPR-Cas systems were common in RGPs from both phylogroups. Some rarer systems such as cyclic-oligonucleotide-based anti-phage signalling systems (CBASS),58 Zorya, Gabija, Druantia,59 abortive infection,60 and bacteriophage exclusion (BREX)61 were also observed in RGPs from phylogroups A and B (Fig. S11 and Table S12). In contrast, dGTPases were absent from both phylogroups. Finally, we also observed that AMR and defence systems were overrepresented in certain MLST profiles, including the high-risk clones ST111 and ST233 (Fig. S12).1 Our results revealed that AMR and defence systems are pervasive in RGPs from both phylogroups A and B, and that the majority of AMR classes are overrepresented in RGPs from phylogroup B.
AMR and defence systems are prevalent in ICEs/IMEs from phylogroups A and B
Given that the distribution and clustering of defence systems in P. aeruginosa does not depend on the phylogenetic distance between all strains,39 and considering the high prevalence of ICEs/IMEs in this species,62 we explored the potential role of these elements as defence islands. To accurately detect these MGEs, we focused our analysis on complete genomes. We noted that 12.6% of our collection consisted of complete genomes (254/2009), including 172 genomes from phylogroup A, 78 from phylogroup B, and 4 genomes from phylogroup C (Table S2). 215 out of the 254 complete genomes harboured a total of 477 ICEs and 76 IMEs (Table S13). These ICEs/IMEs were present in 136 genomes from phylogroup A, 77 from phylogroup B, and 2 from phylogroup C. Thus, ICEs/IMEs were pervasive in strains from phylogroup B (77/78) and in the majority of strains from phylogroup A (136/172).
Almost half of the ICEs/IMEs carried at least one AMR gene (228/553), with the ciprofloxacin-modifying crpP gene and the sulphonamide-resistance sul1 gene being most frequent (Table S14). Indeed, the crpP gene has recently been shown to be widely distributed across ICEs from P. aeruginosa.63 About one third of the ICEs/IMEs (193/553) carried at least one defence system, resulting in a total of 250 defence systems across the ICEs/IMEs and including 27 different types (Fig. S13 and Table S14). The most frequent defence subtypes were CBASS-III and restriction-modification type-II.58,60 Virulence genes were present in a smaller proportion of the ICEs/IMEs (99/553) and showed higher variation in abundance across ICEs/IMEs than AMR genes and defence systems do (Fig. S14). The exoU gene encoding for the effector protein and the spcU gene encoding for its chaperone were the most frequent virulence genes, all in ICEs/IMEs from phylogroup B (Table S14).
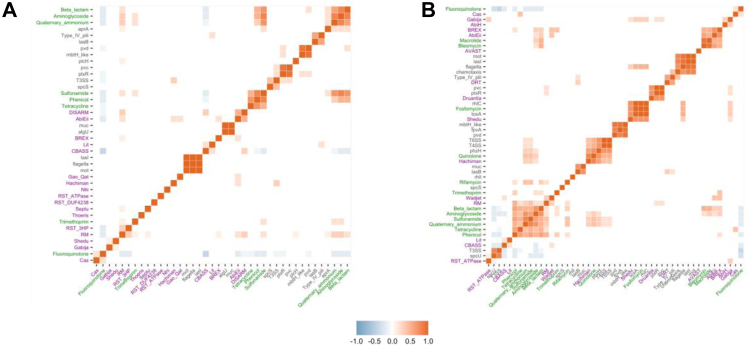
We next explored to what extent the prevalence of these three functional groups is correlated across ICEs/IMEs from the two larger phylogroups A and B. We observed that genes encoding resistance to fluoroquinolones were negatively correlated with genes involved in resistance to other antibiotic classes, and also with specific defence systems as restriction-modification and CBASS (Fig. 6A). ICEs/IMEs from phylogroup B carrying fluoroquinolone-encoding resistance genes were also negatively associated with genes from the type-III secretion systems (Fig. 6B). In contrast, genes encoding resistance to distinct antibiotic classes (e.g., beta-lactams, aminoglycosides, and sulphonamides) were often positively correlated in the ICEs/IMEs from both phylogroups, consistent with the previous observations that these genes tend to be co-localized in genetic structures named integrons.64 Virulence genes involved in flagellar motility were also often correlated, either additionally with (phylogroup B) or without (phylogroup A) genes involved in chemotaxis.65 Defence systems BREX and AbiEii60,61 were positively correlated in ICEs/IMEs from phylogroup B. AMR and defence systems showed a high density in ICEs/IMEs from phylogroups A and B, and their frequencies were positively correlated in both phylogroups.
Fig. 6.
Correlation plots between AMR classes, virulence genes, and defence systems across ICEs/IMEs from phylogroup A (A) and phylogroup B (B). The distribution of cargo genes across ICEs/IMEs was converted into a presence/absence matrix. Correlation matrices were ordered using the hierarchical clustering function. Positive correlations are shown in different shades of red, while negative correlations are shown in different shades of blue. AMR genes and point mutations encoding resistance to particular AMR classes are part of the AMRFinder database,37 defence systems of defense-finder,39 and virulence genes of the VFDB.38 Virulence gene labels are coloured in black, AMR in green, and defence systems in purple.
ICEs/IMEs and RGPs from different phylogroups share high genetic similarity
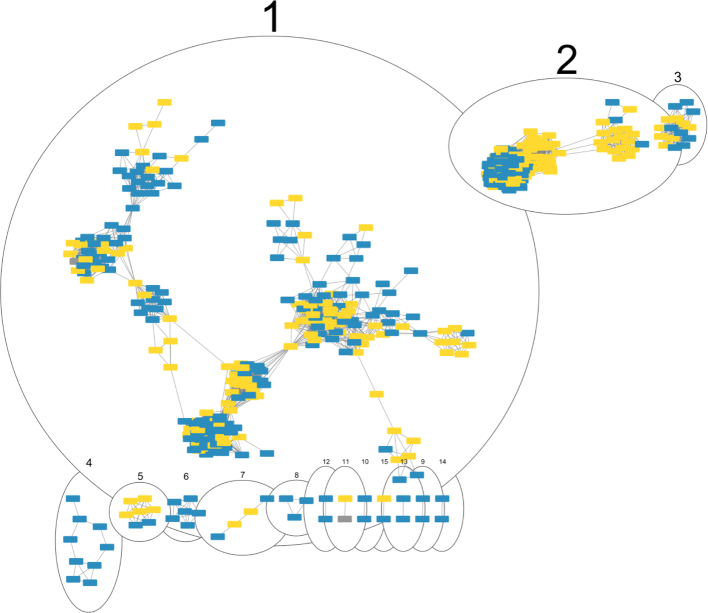
We next used an alignment-free sequence similarity comparison of the ICEs/IMEs to infer an undirected network. The density plot showed a right-skewed distribution of pairwise distance similarities where the vast majority of ICE/IME pairs shared little similarity, with a Jaccard Index value below 0.5 (Fig. S15), in accordance with the high diversity frequently observed across MGEs.66 To reduce the density and increase the sparsity of the network, we used the mean Jaccard Index between all pairs of RGPs as a threshold (0.12184). The network assigned 95.8% (530/553) of the ICEs/IMEs into 15 clusters (Fig. 7). Almost half of the ICEs/IMEs were grouped in cluster 1 (259/530, Table S15), which includes representatives of the three phylogroups.
Fig. 7.
Network of clustered ICEs/IMEs from the three phylogroups, using the mean Jaccard Index between all pairs of ICEs/IMEs as a threshold. Each ICE/IME is represented by a node, connected by edges according to the pairwise distances between all ICE/IME pairs. Numbered ellipses represent ICEs/IMEs that belong to the same cluster. The network has a clustering coefficient of 0.794, a density of 0.099, a centralization of 0.217, and a heterogeneity of 0.785. ICEs/IMEs from phylogroup A are coloured in blue, from phylogroup B in yellow, and from phylogroup C in grey.
We then focused our analysis on the RGPs we extracted from all phylogroups (57901 RGPs in total). We filtered out RGPs smaller than 10 kb, and calculated the Jaccard Index between all pairs of the resulting 32744 RGPs. To reduce the density and increase the sparsity of the network, we used as a threshold the mean value (0.0919429) of the estimated pairwise distances between the 32744 RGPs identified in this study. The network assigned 99.7% (32651/32744) of the RGPs larger than 10 kb into 51 clusters (Fig. S16). While the majority of the RGP clusters were homogeneous for a given phylogroup, we also observed similar DNA regions across different phylogroups. These findings suggest that RGPs and ICEs/IMEs from different P. aeruginosa phylogroups share high genetic identity.
Discussion
In this work, we explored the pangenome of the opportunistic human pathogen P. aeruginosa in consideration of its three main phylogroups. This approach allowed us to characterize the defining properties of each phylogroup. In particular, we identified genes that are prevalent in the small phylogroup C and absent from members of the two larger phylogroups. These genes would have been classified as part of the accessory genome in conventional analyses of the pangenome of the species as a whole. In contrast, our refined approach suggests that these genes have an evolutionary advantage in a specific genetic context that is particular to this phylogroup.67 Moreover, phylogroup C is also clearly distinct from the other two phylogroups A and B in having a significantly smaller genome size and a low relative abundance of AMR and defence systems across RGPs. In addition, our results indicate an inverse association between the size of the phylogroup B accessory genome and the presence of CRISPR-Cas systems. This association could (but need not) be causal, such that a low prevalence of CRISPR-Cas defence systems may possibly favour an increase in the size of the accessory genome. Remarkably, genomes devoid of CRISPR-Cas systems in phylogroups A and B were generally significantly larger than those with these systems, a trend that was no longer observed when only considering the non-RGP (“masked”) genomes. This observation is consistent with the hypothesis that CRISPR-Cas systems can restrict horizontal gene transfer in P. aeruginosa,48, 49, 50,68 at least for genomes belonging to the larger phylogroups.
The three phylogroups vary substantially in the distribution of AMR genes, defence systems, and virulence genes. This variation is particularly evident in the separate analyses of RGPs and masked genomes. While the length of RGPs is substantially smaller than that of masked genomes, the absolute counts of most defence systems were higher in RGPs than in masked genomes across the three phylogroups (Fig. S11). Curiously, representatives of the recently described set of defence systems that are part of Doron's seminal study,59 such as Zorya, Wadjet, and Hachiman systems, were exclusively found in RGPs across the three phylogroups. In Doron's study, the authors demonstrated that the Wadjet system provides protection against plasmid transformation in Bacillus subtilis, while the Zorya and Hachiman systems mediate defence against bacteriophages. These findings highlight the important role of defence systems encoded in RGPs in protecting genomes from infection by foreign DNA and their contribution to MGE–MGE conflict. Moreover, AMR and defence systems are rare in RGPs from phylogroup C, which may suggest that these strains are more often subject to infection by foreign DNA. Assuming that there is no sampling bias across the three phylogroups, then the smaller number of phylogroup C members in public databases could thus be a consequence of the weaker arsenal of AMR and defence systems. Alternatively, phylogroup C strains may indeed be underrepresented, for example if they mainly occur in non-clinical habitats, which are usually less well sampled. Collecting P. aeruginosa samples from distinct geographic regions and environments may further help us reconstruct variation in metabolic competences and their connection to origin.69
In general, our results highlight the role of ICEs/IMEs as vectors not only of AMR genes,57 but also of defence systems. Indeed, most of these systems show nonrandom clustering in defence islands and are often co-localized with mobilome genes.59,70, 71, 72 Co-occurrence of genes alone, however, does not demonstrate an ecological interaction between them.73 Recently, it has been proposed that the accessory genome of the genus Pseudomonas is influenced by natural selection, showing a higher level of genetic structure than would be expected if neutral processes governed the pangenome formation.74 This suggests that coincident genes in ICEs/IMEs are more likely to act together for the benefit of the host or to ensure their own maintenance.9,11 ICEs/IMEs, in particular, provide abundant material for the experimental study of bacterial defence systems. For example, SXT ICEs in Vibrio cholerae, which are also involved in AMR, consistently encode defence systems localized to a single hotspot of genetic shuffling.75 Additionally, ICEs in Acidithiobacillia carry type-IV CRISPR-Cas systems with remarkable evolutionary plasticity, which are often involved in MGE–MGE warfare.76 Moreover, a recent study proposed that size constraints may account for the low abundance of large defence systems on prophages.39 In turn, the comparatively larger size of ICEs/IMEs (when compared to prophages)77 may then explain that they commonly harbor large systems such as BREX and defence island system associated with restriction–modification (DISARM)78 across our dataset (Fig. S13). Even though the CBASS systems are not as prevalent as restriction-modification and CRISPR-Cas systems across the bacterial phylogeny,39 three types of this system were found across ICEs/IMEs from the larger phylogroups.
For our analyses, we used complete and draft genome assemblies retrieved from public databases. However, incomplete genome assemblies likely impact RGP definition, due to highly fragmented genomes, that might have inadvertently split RGPs into multiple contigs. With this in mind, we subsampled the complete genomes from our collection and used them to accurately delineate ICEs/IMEs. By comparing sequence similarity between all pairs of ICEs/IMEs found in this study, as well as between all pairs of RGPs, we were able to explore interactions between these elements, suggesting that members of the same and of different phylogroups frequently undergo DNA shuffling events. Importantly, this network-based approach using pairwise genetic distances of alignment-free k-mer sequences between MGE pairs has bypassed the exclusion of non-coding elements, providing a more comprehensive picture of MGE populations and dynamics.49,79 Nevertheless, with the current advances in sequencing technology, especially including long-read sequencing, we anticipate a much larger number of fully assembled P. aeruginosa genomes in the future, which will then improve the reliable assessment of the RGP composition and the role of particular MGEs or gene functions in shaping this species’ genome characteristics.
To conclude, our work has used a refined approach to explore phylogroup-specific and pangenome dynamics in P. aeruginosa. Members of phylogroup B contribute a comparatively larger number of pangenome families, have larger genomes, and have a lower prevalence of CRISPR-Cas systems. AMR and defence systems are widespread in RGPs and ICEs/IMEs from phylogroups A and B, and these two functional groups are often significantly correlated, including both positive and negative correlations. We also observed multiple interaction events between the accessory genome contents, both between and within phylogroups, suggesting that recombination events are frequent. Our conclusions are contingent on the current range of sequenced genomes for P. aeruginosa. We cannot exclude the possibility that some groups, such as phylogroup C and possibly its subgroups, are not fully represented in the currently available data. Future sequencing efforts are likely to rectify such a possible problem, thus allowing to test the findings from our study. Finally, our work provides a representative set of phylogenetically diverse P. aeruginosa strains, the mPact strain panel, which should prove useful as a reference set for future functional analyses. Such functional analyses may help to experimentally assess the underlying reasons for some of the correlations identified in our study, for example the role of specific defence systems in RGP size expansion or in mediating conflict between different MGE types.
Contributors
JB conceptualized and designed the work, acquired and analysed the data, interpreted the data, and wrote the original draft of the manuscript. LT conceptualized and designed the work, acquired the data, and interpreted the data. JF, FB, CU, JK, SN, and BT acquired and analysed the data. HS conceptualized and designed the work, interpreted the data, and contributed to writing of the original draft of the manuscript. HS, JB, SN, and BT acquired funding for this work. All authors read, revised, and approved the final manuscript.
Data sharing statement
Scripts for reproducing the analyses performed in this work are available at https://gitlab.gwdg.de/botelho/pa_pangenome. The representative set of P. aeruginosa genomes and the input file used for the network analysis in Fig. 7 are available at the Figshare project https://figshare.com/projects/P_aeruginosa_pangenome/155021. Analyses were made with a combination of shell and R 4.0.3 scripting. Sequencing performed in this project were deposited in NCBI under the Bioproject accession number PRJNA810040.
Declaration of interests
We declare no competing interests.
Acknowledgments
We acknowledge financial support from the German Science Foundation (Deutsche Forschungsgemeinschaft, DFG) (grant SCHU 1415/12-2 to HS, funding under Germany’s Excellence Strategy EXC 2167-390884018 as well as the Research Training Group 2501 TransEvo to HS and SN, and funding within the SFB 900 TP A2 to BT), the Max Planck Society (Max-Planck fellowship to HS), the Leibniz ScienceCampus Evolutionary Medicine of the Lung (EvoLUNG, to HS and SN), and the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) program Medical Infection Genomics (AZ 0315827A to BT). This work was supported by the Kiel Life Science of Kiel University Postdoc Award to JB and by the DFG Research Infrastructure NGS_CC (project 407495230) as part of the Next Generation Sequencing Competence Network (project 423957469). NGS was carried out at the Competence Centre for Genomic Analysis (Kiel). This research was supported in part through high-performance computing resources available at the Kiel University Computing Centre.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104532.
Contributor Information
João Botelho, Email: botelho@evolbio.mpg.de.
Hinrich Schulenburg, Email: hschulenburg@zoologie.uni-kiel.de.
Appendix A. Supplementary data
References
- 1.Botelho J., Grosso F., Peixe L. Antibiotic resistance in Pseudomonas aeruginosa – mechanisms, epidemiology and evolution. Drug Resist Updates. 2019;44:100640. doi: 10.1016/j.drup.2019.07.002. https://www.sciencedirect.com/science/article/pii/S1368764619300238 [cited 2019 Sep 25]. Available from: [DOI] [PubMed] [Google Scholar]
- 2.De Oliveira D.M.P., Forde B.M., Kidd T.J., et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev. 2020;33(3) doi: 10.1128/CMR.00181-19. https://cmr.asm.org/content/33/3/e00181-19 00181–19 [cited 2020 Aug 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray C.J., Ikuta K.S., Sharara F., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. http://www.thelancet.com/article/S0140673621027240/fulltext [cited 2022 Mar 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horcajada J.P., Montero M., Oliver A., et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4) doi: 10.1128/CMR.00031-19. https://journals.asm.org/doi/abs/10.1128/CMR.00031-19 00031–19 [cited 2022 Mar 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tacconelli E., Carrara E., Savoldi A., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. http://www.ncbi.nlm.nih.gov/pubmed/29276051 [cited 2018 Dec 21]. Available from: [DOI] [PubMed] [Google Scholar]
- 6.Koonin E.V., Wolf Y.I. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36(21):6688–6719. doi: 10.1093/nar/gkn668. https://academic.oup.com/nar/article/36/21/6688/2410005 [cited 2022 Feb 9]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins R.E., Higgs P.G. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol Biol Evol. 2012;29(11):3413–3425. doi: 10.1093/molbev/mss163. https://academic.oup.com/mbe/article/29/11/3413/1155627 [cited 2022 Feb 9]. Available from: [DOI] [PubMed] [Google Scholar]
- 8.Arnold B.J., Huang I.-T., Hanage W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Microbiol. 2021;20(4):206–218. doi: 10.1038/s41579-021-00650-4. https://www.nature.com/articles/s41579-021-00650-4 [cited 2021 Nov 15]. Available from: [DOI] [PubMed] [Google Scholar]
- 9.Botelho J., Schulenburg H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2020;29(1):8–18. doi: 10.1016/j.tim.2020.05.011. https://www.cell.com/trends/microbiology/fulltext/S0966-842X(20)30137-2 [cited 2020 Jun 12]. Available from: [DOI] [PubMed] [Google Scholar]
- 10.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4) doi: 10.1128/CMR.00088-17. http://www.ncbi.nlm.nih.gov/pubmed/30068738 [cited 2018 Aug 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocha E.P.C., Bikard D. Microbial defenses against mobile genetic elements and viruses: who defends whom from what? PLoS Biol. 2022;20(1):e3001514. doi: 10.1371/journal.pbio.3001514. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001514 [cited 2022 Jan 14]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinilla-Redondo R., Russel J., Mayo-Muñoz D., et al. CRISPR-Cas systems are widespread accessory elements across bacterial and archaeal plasmids. Nucleic Acids Res. 2021;50(8) doi: 10.1093/nar/gkab859. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab859/6381142 4315–4328. [cited 2021 Oct 5]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horesh G., Taylor-Brown A., McGimpsey S., et al. Different evolutionary trends form the twilight zone of the bacterial pan-genome. Microb Genom. 2021;7(9):670. doi: 10.1099/mgen.0.000670. https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000670 [cited 2021 Sep 25]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozer E.A., Nnah E., Didelot X., Whitaker R.J., Hauser A.R. The population structure of Pseudomonas aeruginosa is characterized by genetic isolation of exoU+ and exoS+ lineages. Genome Biol Evol. 2019;11(1):1780–1796. doi: 10.1093/gbe/evz119. http://www.ncbi.nlm.nih.gov/pubmed/31173069 [cited 2019 Jun 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trouillon J., Imbert L., Villard A.-M., Vernet T., Attrée I., Elsen S. Determination of the two-component systems regulatory network reveals core and accessory regulations across Pseudomonas aeruginosa lineages. Nucleic Acids Res. 2021;49(20):11476–11490. doi: 10.1093/nar/gkab928. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab928/6413601 [cited 2021 Nov 1]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trouillon J., Han K., Attrée I., Lory S., Kook H. The core and accessory Hfq interactomes across Pseudomonas aeruginosa lineages. Nat Commun. 2022;13(1):1258. doi: 10.1038/s41467-022-28849-w. https://www.nature.com/articles/s41467-022-28849-w [cited 2022 Mar 10]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilker R., Munder A., Klockgether J., et al. Interclonal gradient of virulence in the P seudomonas aeruginosa pangenome from disease and environment. Environ Microbiol. 2015;17(1):29–46. doi: 10.1111/1462-2920.12606. http://www.ncbi.nlm.nih.gov/pubmed/25156090 [cited 2018 Aug 20]. Available from: [DOI] [PubMed] [Google Scholar]
- 18.Wiehlmann L., Cramer N., Tümmler B. Habitat-associated skew of clone abundance in the Pseudomonas aeruginosa population. Environ Microbiol Rep. 2015;7(6):955–960. doi: 10.1111/1758-2229.12340. https://pubmed.ncbi.nlm.nih.gov/26419222/ [cited 2022 Mar 12]. Available from: [DOI] [PubMed] [Google Scholar]
- 19.Wiehlmann L., Wagner G., Cramer N., et al. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104(19):8101–8106. doi: 10.1073/pnas.0609213104. https://www.pnas.org/doi/full/10.1073/pnas.0609213104 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer S., Dethlefsen S., Klockgether J., Tümmler B. Phenotypic and genomic comparison of the two most common ExoU-positive Pseudomonas aeruginosa clones, PA14 and ST235. mSystems. 2020;5(6) doi: 10.1128/mSystems.01007-20. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7743143/ 01007–20 [cited 2022 Mar 16]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6) doi: 10.1371/journal.pcbi.1005595. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005595 [cited 2018 Oct 17]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wick R.R., Schultz M.B., Zobel J., Holt K.E. Bandage: interactive visualization of de novo genome assemblies: fig. 1. Bioinformatics. 2015;31(20):3350–3352. doi: 10.1093/bioinformatics/btv383. https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btv383 [cited 2018 Oct 17]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrin A., Rocha E.P.C. PanACoTA: a modular tool for massive microbial comparative genomics. NAR Genom Bioinform. 2021;3(1) doi: 10.1093/nargab/lqaa106. https://academic.oup.com/nargab/article/3/1/lqaa106/6090162 lqaa106. [cited 2021 Jan 27]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain C., Rodriguez -R.L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. http://www.nature.com/articles/s41467-018-07641-9 [cited 2019 Jun 8]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautreau G., Bazin A., Gachet M., et al. PPanGGOLiN: depicting microbial diversity via a partitioned pangenome graph. PLoS Comput Biol. 2020;16(3) doi: 10.1371/journal.pcbi.1007732. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007732 [cited 2020 Jul 31]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazin A., Gautreau G., Médigue C., Vallenet D., Calteau A. panRGP: a pangenome-based method to predict genomic islands and explore their diversity. Bioinformatics. 2020;36(2):i651–i658. doi: 10.1093/bioinformatics/btaa792. https://pubmed.ncbi.nlm.nih.gov/33381850/ [cited 2021 Feb 10]. Available from: [DOI] [PubMed] [Google Scholar]
- 27.Minh B.Q., Schmidt H.A., Chernomor O., et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. https://pubmed.ncbi.nlm.nih.gov/32011700/ [cited 2022 Mar 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Didelot X., Wilson D.J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11(2):e1004041. doi: 10.1371/journal.pcbi.1004041. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004041 [cited 2021 Oct 1]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. https://academic.oup.com/nar/article/49/W1/W293/6246398 [cited 2021 Dec 30]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. http://code.google.com/p/bedtools [cited 2021 Jun 8]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btu153 [cited 2018 Aug 11]. Available from: [DOI] [PubMed] [Google Scholar]
- 32.Liu M., Li X., Xie Y., et al. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2018;47(D1):D660–D665. doi: 10.1093/nar/gky1123. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gky1123/5165266 [cited 2018 Dec 21]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinegger M., Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35(11):1026–1028. doi: 10.1038/nbt.3988. https://www.nature.com/articles/nbt.3988 [cited 2022 Mar 3]. Available from: [DOI] [PubMed] [Google Scholar]
- 34.Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 2021;38(12):5825–5829. doi: 10.1093/molbev/msab293. https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msab293/6379734 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. doi: 10.1093/nar/28.1.33. https://academic.oup.com/nar/article/28/1/33/2384317 [cited 2022 Mar 15]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russel J., Pinilla-Redondo R., Mayo-Muñoz D., Shah S.A., Sørensen S.J. CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-cas loci. CRISPR J. 2020;3(6):462–469. doi: 10.1089/crispr.2020.0059. www.liebertpub.com [cited 2021 Jun 8]. Available from: [DOI] [PubMed] [Google Scholar]
- 37.Feldgarden M., Brover V., Haft D.H., et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63(11) doi: 10.1128/AAC.00483-19. http://www.ncbi.nlm.nih.gov/pubmed/31427293 00483–19 [cited 2019 Nov 15]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B., Zheng D., Jin Q., Chen L., Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D692. doi: 10.1093/nar/gky1080. https://pubmed.ncbi.nlm.nih.gov/30395255/ [cited 2022 Mar 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tesson F., Hervé A., Mordret E., et al. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat Commun. 2022;13(1):2561. doi: 10.1038/s41467-022-30269-9. https://www.nature.com/articles/s41467-022-30269-9 [cited 2022 May 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X. BinDash, software for fast genome distance estimation on a typical personal laptop. Bioinformatics. 2019;35(4):671–673. doi: 10.1093/bioinformatics/bty651. https://academic.oup.com/bioinformatics/article/35/4/671/5058094 [cited 2020 Oct 29]. Available from: [DOI] [PubMed] [Google Scholar]
- 41.Freschi L., Vincent A.T., Jeukens J., et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer and pathogenicity. Genome Biol Evol. 2018;11(1):109–120. doi: 10.1093/gbe/evy259. http://www.ncbi.nlm.nih.gov/pubmed/30496396 [cited 2018 Dec 21]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stover C.K., Pham X.Q., Erwin A.L., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. http://www.nature.com/articles/35023079 [cited 2018 Aug 17]. Available from: [DOI] [PubMed] [Google Scholar]
- 43.Roy P.H., Tetu S.G., Larouche A., et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One. 2010;5(1):e8842. doi: 10.1371/journal.pone.0008842. https://pubmed.ncbi.nlm.nih.gov/20107499/ [cited 2022 Feb 18]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morimoto Y., Tohya M., Aibibula Z., Baba T., Daida H., Kirikae T. Re-identification of strains deposited as Pseudomonas aeruginosa, Pseudomonas fluorescens and Pseudomonas putida in GenBank based on whole genome sequences. Int J Syst Evol Microbiol. 2020;70(11):5958–5963. doi: 10.1099/ijsem.0.004468. https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijsem.0.004468 [cited 2020 Sep 17]. Available from: [DOI] [PubMed] [Google Scholar]
- 45.Brockhurst M.A., Harrison E., Hall J.P.J., Richards T., McNally A., MacLean C. The ecology and evolution of pangenomes. Curr Biol. 2019;29(20):R1094–R1103. doi: 10.1016/j.cub.2019.08.012. https://www.sciencedirect.com/science/article/abs/pii/S0960982219310280 [cited 2019 Dec 11]. Available from: [DOI] [PubMed] [Google Scholar]
- 46.Filloux A. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol. 2011;2:155. doi: 10.3389/fmicb.2011.00155. https://www.frontiersin.org/articles/10.3389/fmicb.2011.00155/full Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koonin E.V., Makarova K.S., Wolf Y.I., Krupovic M. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet. 2019;21(2):119–131. doi: 10.1038/s41576-019-0172-9. http://www.nature.com/articles/s41576-019-0172-9 [cited 2019 Oct 19]. Available from: [DOI] [PubMed] [Google Scholar]
- 48.Wheatley R.M., MacLean R.C. CRISPR-Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2020;15(5):1420–1433. doi: 10.1038/s41396-020-00860-3. http://www.nature.com/articles/s41396-020-00860-3 [cited 2020 Dec 23]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Botelho J., Cazares A., Schulenburg H. The ESKAPE mobilome contributes to the spread of antimicrobial resistance and CRISPR-mediated conflict between mobile genetic elements. Nucleic Acids Res. 2023;51(1):236–252. doi: 10.1093/nar/gkac1220. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkac1220/6970226 [cited 2023 Jan 9]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pursey E., Dimitriu T., Paganelli F.L., Westra E.R., van Houte S. CRISPR-Cas is associated with fewer antibiotic resistance genes in bacterial pathogens. Philos Trans R Soc B. 2022;377(1842):20200464. doi: 10.1098/rstb.2020.0464. https://royalsocietypublishing.org/doi/abs/10.1098/rstb.2020.0464 [cited 2021 Nov 29]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinilla-Redondo R., Mayo-Muñoz D., Russel J., et al. Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 2020;48(4):2000–2012. doi: 10.1093/nar/gkz1197. https://academic.oup.com/nar/article/48/4/2000/5687823 [cited 2021 Jul 2]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.León L.M., Park A.E., Borges A.L., Zhang J.Y., Bondy-Denomy J. Mobile element warfare via CRISPR and anti-CRISPR in Pseudomonas aeruginosa. Nucleic Acids Res. 2021;49(4):2114–2125. doi: 10.1093/nar/gkab006. https://academic.oup.com/nar/article/49/4/2114/6129318 [cited 2021 Jul 6]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reboud E., Basso P., Maillard A.P., Huber P., Attrée I. Exolysin shapes the virulence of Pseudomonas aeruginosa clonal outliers. Toxins. 2017;9(11):364. doi: 10.3390/toxins9110364. https://pubmed.ncbi.nlm.nih.gov/29120408/ [cited 2021 Nov 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horna G., Amaro C., Palacios A., Guerra H., Ruiz J. High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant “high-risk clones” of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci Rep. 2019;9(1):10874. doi: 10.1038/s41598-019-47303-4. https://pubmed.ncbi.nlm.nih.gov/31350412/ [cited 2023 Jan 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodrigues Y.C., Furlaneto I.P., Pinto Maciel A.H., et al. High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon. PLoS One. 2020;15(9):e0238741. doi: 10.1371/journal.pone.0238741. https://pubmed.ncbi.nlm.nih.gov/32911510/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arora S.K., Bangera M., Lory S., Ramphal R. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc Natl Acad Sci U S A. 2001;98(16):9342–9347. doi: 10.1073/pnas.161249198. https://www.pnas.org/doi/full/10.1073/pnas.161249198 [cited 2022 Apr 20]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Botelho J., Mourão J., Roberts A.P., Peixe L. Comprehensive genome data analysis establishes a triple whammy of carbapenemases, ICEs and multiple clinically relevant bacteria. Microb Genom. 2020;6(10):mgen000424. doi: 10.1099/mgen.0.000424. https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000424 [cited 2020 Aug 27]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen D., Melamed S., Millman A., et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature. 2019;574(7780):691–695. doi: 10.1038/s41586-019-1605-5. http://www.nature.com/articles/s41586-019-1605-5 [cited 2019 Oct 11]. Available from: [DOI] [PubMed] [Google Scholar]
- 59.Doron S., Melamed S., Ofir G., et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359(6379):eaar4120. doi: 10.1126/science.aar4120. http://www.ncbi.nlm.nih.gov/pubmed/29371424 [cited 2019 Apr 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315. https://www.nature.com/articles/nrmicro2315 [cited 2022 Mar 12]. Available from: [DOI] [PubMed] [Google Scholar]
- 61.Goldfarb T., Sberro H., Weinstock E., et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34(2):169–183. doi: 10.15252/embj.201489455. https://pubmed.ncbi.nlm.nih.gov/25452498/ [cited 2022 Mar 12]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guglielmini J., Quintais L., Garcillán-Barcia M.P., de la Cruz F., Rocha E.P.C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7(8) doi: 10.1371/journal.pgen.1002222. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1002222 [cited 2018 Aug 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Botelho J., Grosso F., Peixe L. ICEs are the main reservoirs of the ciprofloxacin-modifying crpP gene in Pseudomonas aeruginosa. Genes. 2020;11(8):889. doi: 10.3390/genes11080889. https://pubmed.ncbi.nlm.nih.gov/32759827/ [cited 2020 Aug 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghaly T.M., Geoghegan J.L., Tetu S.G., Gillings M.R. The peril and promise of integrons: beyond antibiotic resistance. Trends Microbiol. 2020;28(6):455–464. doi: 10.1016/j.tim.2019.12.002. https://www.cell.com/trends/microbiology/fulltext/S0966-842X(19)30317-8 Available from: [DOI] [PubMed] [Google Scholar]
- 65.Matilla M.A., Martín-Mora D., Gavira J.A., Krell T. Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol Mol Biol Rev. 2021;85(1) doi: 10.1128/MMBR.00151-20. https://pubmed.ncbi.nlm.nih.gov/33441490/ 00151–20 [cited 2022 Mar 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cury J., Oliveira P.H., de la Cruz F., Rocha E.P.C. Host range and genetic plasticity explain the coexistence of integrative and extrachromosomal mobile genetic elements. Mol Biol Evol. 2018;35(9):2230–2239. doi: 10.1093/molbev/msy123. https://academic.oup.com/mbe/article/35/9/2230/5037826 [cited 2019 May 4]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lassalle F., Muller D., Nesme X. Ecological speciation in bacteria: reverse ecology approaches reveal the adaptive part of bacterial cladogenesis. Res Microbiol. 2015;166(10):729–741. doi: 10.1016/j.resmic.2015.06.008. https://pubmed.ncbi.nlm.nih.gov/26192210/ Available from: [DOI] [PubMed] [Google Scholar]
- 68.van Belkum A., Soriaga L.B., LaFave M.C., et al. Phylogenetic distribution of CRISPR-cas systems in antibiotic-resistant Pseudomonas aeruginosa. mBio. 2015;6(6) doi: 10.1128/mBio.01796-15. https://pubmed.ncbi.nlm.nih.gov/26604259/ [cited 2018 Aug 21]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saati-Santamaría Z., Baroncelli R., Rivas R., García-Fraile P. Comparative genomics of the genus Pseudomonas reveals host- and environment-specific evolution. Microbiol Spectr. 2022;10(6) doi: 10.1128/spectrum.02370-22. https://pubmed.ncbi.nlm.nih.gov/36354324/ [cited 2022 Nov 14]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Makarova K.S., Wolf Y.I., Snir S., Koonin E.V. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol. 2011;193(21):6039–6056. doi: 10.1128/JB.05535-11. https://journals.asm.org/doi/abs/10.1128/JB.05535-11 [cited 2022 Mar 2]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hussain F.A., Dubert J., Elsherbini J., et al. Rapid evolutionary turnover of mobile genetic elements drives bacterial resistance to phages. Science. 2021;374(6566):488–492. doi: 10.1126/science.abb1083. https://www.science.org/doi/abs/10.1126/science.abb1083 [cited 2021 Oct 28]. Available from: [DOI] [PubMed] [Google Scholar]
- 72.van Vliet A.H.M., Charity O.J., Reuter M. A Campylobacter integrative and conjugative element with a CRISPR-Cas9 system targeting competing plasmids: a history of plasmid warfare? Microb Genom. 2021;7(11):000729729. doi: 10.1099/mgen.0.000729. https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000729 [cited 2021 Nov 15]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blanchet F.G., Cazelles K., Gravel D. Co-occurrence is not evidence of ecological interactions. Ecol Lett. 2020;23(7):1050–1063. doi: 10.1111/ele.13525. https://onlinelibrary.wiley.com/doi/abs/10.1111/ele.13525 [cited 2020 Oct 30]. Available from: [DOI] [PubMed] [Google Scholar]
- 74.Whelan F.J., Hall R.J., McInerney J.O. Evidence for selection in the abundant accessory gene content of a prokaryote pangenome. Mol Biol Evol. 2021;38(9):3697–3708. doi: 10.1093/molbev/msab139. https://academic.oup.com/mbe/article/38/9/3697/6272232 [cited 2021 Jun 3]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.LeGault K.N., Hays S.G., Angermeyer A., et al. Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts. Science. 2021;373(6554):eabg2166. doi: 10.1126/science.abg2166. https://science.sciencemag.org/content/373/6554/eabg2166 [cited 2021 Jul 30]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moya-Beltrán A., Makarova K.S., Acuña L.G., et al. Evolution of type IV CRISPR-cas systems: insights from CRISPR loci in integrative conjugative elements of Acidithiobacillia. CRISPR J. 2021;4(5):656–672. doi: 10.1089/crispr.2021.0051. https://www.liebertpub.com/doi/10.1089/crispr.2021.0051 [cited 2021 Sep 29]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cury J., Touchon M., Rocha E.P.C. Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Res. 2017;45(15):8943–8956. doi: 10.1093/nar/gkx607. http://www.ncbi.nlm.nih.gov/pubmed/28911112 [cited 2018 Aug 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ofir G., Melamed S., Sberro H., et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol. 2017;3(1):90–98. doi: 10.1038/s41564-017-0051-0. https://www.nature.com/articles/s41564-017-0051-0 [cited 2022 Mar 16]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Acman M., van Dorp L., Santini J.M., Balloux F. Large-scale network analysis captures biological features of bacterial plasmids. Nat Commun. 2020;11(1):2452. doi: 10.1038/s41467-020-16282-w. http://www.nature.com/articles/s41467-020-16282-w [cited 2020 May 19]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.