Abstract
Spinal cord ischemia-reperfusion (IR) injury (SCIRI) is a significant secondary injury that causes damage to spinal cord neurons, leading to the impairment of spinal cord sensory and motor functions. Excessive reactive oxygen species (ROS) production is considered one critical mechanism of neuron damage in SCIRI. Nonetheless, the molecular mechanisms underlying the resistance of neurons to ROS remain elusive. Our study revealed that the deletion of Git1 in mice led to poor recovery of spinal cord motor function after SCIRI. Furthermore, we discovered that Git1 has a beneficial effect on neuron resistance to ROS production. Mechanistically, Git1 interacted with PGK1, regulated PGK1 phosphorylation at S203, and affected the intermediate products of glycolysis in neurons. The influence of Git1 on glycolysis regulates the dimerization of Keap1, which leads to changes in Nrf2 ubiquitination and plays a role in resisting ROS. Collectively, we show that Git1 regulates the Keap1/Nrf2 axis to resist ROS in a PGK1-dependent manner and thus is a potential therapeutic target for SCIRI.
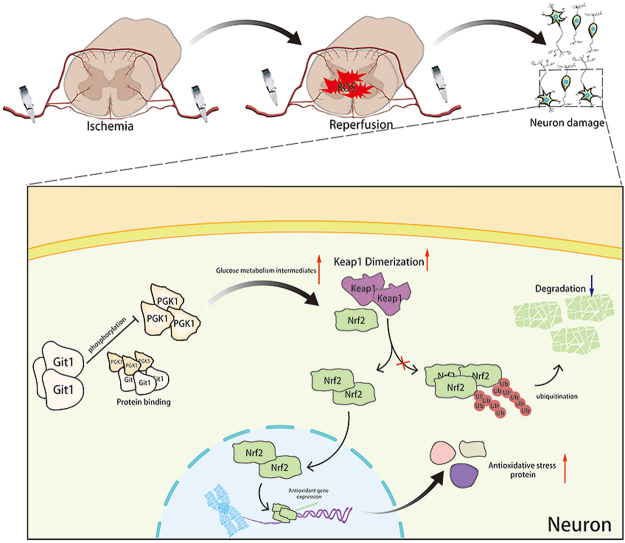
Graphical abstract
Author contributions
Guoyong Yin, Jian Chen and Tao Xu designed and supervised this study. Tao Xu and Peng Gao conducted the majority of the experiments and completed the manuscript. Yifan Huang and Shujie Zhao analyzed the data. Mengyuan Wu, Jiang Yi, Zheng Zhou, Xuan Zhao, Tao Jiang, Hao Liu, Tao Qin, Zhenqi Yang, Xiaowei Wang and Tianyi Bao participated in the experiments and manuscript writing.
All authors approved the final version of the manuscript.
1. Introduction
Spinal cord ischemic-reperfusion (IR) injury (SCIRI) is a serious condition that can cause significant impairment of both sensory and motor functions in the spinal cord [1]. Clinically, ischemia-reperfusion (IR) is a common complication after thoracoabdominal aortic surgery or spinal cord decompression surgery, and can also be directly caused by spinal cord trauma and spinal canal tumor compression [[2], [3], [4]]. Spinal cord functional recovery is restricted due to limited regenerative potential of the mammalian central nervous system (CNS) and limited clinical treatments [[5], [6], [7]]. Therefore, it is particularly important to identify feasible and effective treatment methods by exploring the molecular mechanism of SCIRI.
Neurons underlie spinal cord sensory and motor function [8,9]. Neurons are injured and undergo apoptosis following spinal cord ischemia-reperfusion [[10], [11], [12]]. The most important mechanism is reactive oxygen species (ROS) generation after reperfusion. The ROS generated by reperfusion can induce neuronal apoptosis, resulting in severe impairment of spinal cord function [[13], [14], [15], [16], [17]]. The weak antioxidant reserve of neurons renders them highly vulnerable to ROS damage [18,19]. Effectively resisting the ROS produced by the body can alleviate ROS-induced neuronal apoptosis, which can promote the recovery of spinal cord function [20,21]. Therefore, exploring the mechanism of neuronal resistance to ROS will be the key in the study of SCIRI.
Metabolism is the foundation and core of all biological processes and functions, providing living organisms with energy and essential substances [22]. Glycolysis is a major metabolic pathway that provides cells with energy, and a variety of enzymes are involved in this biochemical process [23]. Glucose metabolism can also affect the cellular microenvironment through its own metabolic intermediates and regulate cellular signaling pathways [24]. It has been shown that after ischemia-reperfusion, the metabolic level of glycolysis in endothelial cells, cardiomyocytes, fibroblasts, and other cells changes, which affects the metabolism and state of cells. The glycolytic state can also modulate the distribution and secretion of immune cells and inflammatory cells throughout the body [[25], [26], [27]]. This cellular-level metabolite alteration determines the activation/repression of signaling pathways, epigenetic and post-transcriptional regulation of inflammatory genes, and post-translational modification of proteins [[28], [29], [30], [31]]. Phosphoglycerate kinase 1 (PGK1) is a crucial enzyme in glycolysis by catalyzing the conversion of 1,3-bisphosphoglycerate (1,3-BPG) to 3-phosphoglycerate (3-PG). One of the two ATP-producing enzymes in glycolysis is PGK1, which participates in regulating energy homeostasis the regulation of energy homeostasis [32,33]. In addition, PGK1 was found to regulate the KEAP1–NRF2 pathway, which couples glucose metabolism with states of oxidative stress [[34], [35], [36]]. The regulation of cellular redox status by glycolytic intermediates has been emphasized. Nevertheless, the involvement of glycolysis in SCIRI remains unexplored.
G‐protein‐coupled receptor (GPCR) kinase 2‐interacting protein‐1 (Git1) is a GTPase-activating protein [37,38]. Different domains of Git1 bind various proteins and regulate their localization and function [39,40]. Git1 exerts a physiological regulatory ability by affecting the state of intracellular signaling pathways [[41], [42], [43]]. Git1 is a postsynaptic structural protein that promotes spine maturation; Git1 also regulates neuronal metabolism, affecting the excitability of postsynaptic neurons [[44], [45], [46]]. Neuronal aging is closely related to the expression of Git1 [47,48]. Although the role of Git1 in neuroscience has been extensively studied, the mechanism of Git1 in the pathogenesis of SCIRI remains to be investigated.
In this investigation, a close association exists between glycolysis and SCIR. The Git1-PGK1 interaction acts as an adaptive mechanism to cope with oxidative stress-targeting neurons in SCIRI. This finding provides an intimate link between redox signaling and Git1-PGK1 interaction in neuronal cells.
2. Materials and methods
2.1. Animals
As previously described [49], Git1 Knock-out (KO) mice on a C57BL/6 background were generated by researchers at the Aab Cardiovascular Research Institute and Department of Medicine at the University of Rochester in Rochester, NY, USA. The mice were kindly gifted by Bradford C. Berk. For this study, both Git1 knock-out (KO) mice and their wild-type littermates (WT) were utilized. The pups underwent genotyping through both PCR and western blotting. The samples were also subjected to sequencing analysis (Figs. S1B and C). The Institutional Animal Care and Use Committee (IACUC) at the First Affiliated Hospital of Nanjing Medical University approved all animal-related experiments.
2.2. Spinal cord ischemia-reperfusion (IR) injury (SCIRI) mouse model
The method for establishing the SCIRI mouse model was previously described [19]. In brief, 8-week-old male C57BL/6 mice were used for the model [50,51]. All mouse-related animal experiments conducted in this study were approved by the Institutional Animal Care and Use Committee of Nanjing Medical University. All procedures carried out in this study were in accordance with the Health Animal Laboratory Animal Care and Use Guidelines. Briefly, a mixture of isoflurane and oxygen was used to anesthetize the mice for the experiment. We utilized a midline abdominal incision to access the left kidney via the peritoneum, and separated the abdominal aorta near the left renal artery. In the experimental group, we applied a clip to the abdominal aorta below the outlet of the left renal artery, while this was not done in the sham-operated group. We removed the clip after 60 min of occlusion, and restored blood perfusion to the affected area. This resulted in ischemia-reperfusion injury to the spinal cord at the L1-L3 level. Following the surgery, each mouse was housed in a separate cage and the bladder was massaged at intervals of 8 h until the bladder reflex was regained.
2.3. Motor function analysis
Before functional testing, all mice were allowed to acclimate to the testing arena or apparatus for a period of 1 h. The recovery of neuromotor function was evaluated using the Basso Mouse Scale (BMS) score at 6 h, 12 h, 1 day, 3 days, 7 days, and 14 days following the SCIRI procedure. The BMS score was utilized to monitor alterations in the motor function of mice, and recovery of motor function was assessed by corresponding scores, including primary and secondary scoring systems. The key parameters assessed included trunk stability, tail position, the range of motion of the posterior ankle joint and the tactile sensation of the sole and instep. In normal mice, a BMS score of 9 corresponds to intact motor function, while a score of 0 signifies total paralysis. The experimenter used a double-blind method for scoring [52].
Rotarod tests were performed as previously described [53]. To assess motor ability and coordination, the rotarod test was used, which involved placing the mouse on a rotating rod that could be accelerated from 0 to 40 rpm, and measuring the time taken for the mouse to fall off the rotarod. Each mouse was given a single practice trial, followed by two test trials after a single exercise session, with a 20-min interval between the tests. The average of the two test times was recorded as the evaluation time.
The methodology for the footprint analysis was carried out in accordance with previously described studies [19]. Blue and red dyes were used to mark the forepaws and hind paws of the mice, respectively. The footprints of the mice walking on the paper were observed and the collected footprint patterns were analyzed to evaluate the coordination and motor recovery.
The swimming test was employed to evaluate the restoration of motor function in mice following SCIRI [54]. Briefly, the movements of the front and rear limbs, trunk coordination and tail position were observed during swimming after SCIRI. The main scoring points of the Louisville Swim Scale (LSS) are hindlimb swing and movement, forelimb dependence, body angle, and trunk stability, with a total of 18 levels corresponding to 0–17 points.
2.4. Preparation of frozen tissue sections
The mice were first perfused with 0.9% saline solution, followed by perfusion with 4% paraformaldehyde. Spinal cords (L1–L3) were dissected, removed, and fixed in 4% paraformaldehyde solution overnight. The spinal cord tissue was dehydrated using 20% and 30% sucrose solutions before being embedded in optimal cutting temperature compound (OCT, Sakura, CA, USA) and sectioned into slices that were 10 μm thick.
2.5. Nissl staining
Nissl bodies are unique cellular structures of neurons that are basophilic and can be stained with basic dyes such as tar violet. Frozen sections were soaked in distilled water, chloroform, 100% alcohol, 95% alcohol, and 75% alcohol for 1 min. Spinal cord sections were stained with cresyl violet (FD Neuro Technologies, MD, USA) (37 °C, 10 min), washed twice with distilled water, and decolorized twice with 95% ethanol. Finally, 85%, 80%, and 75% ethanol were used in sequence to dehydrate the sections. The dehydrated sections were air-dried, and the tissue was sealed with xylene and neutral gum, and air-dried for observation using an optical microscope.
2.6. Antibodies
The following antibodies were utilized for immunoblotting: anti-H2A.X (1:2000, Abcam), anti-phospho-H2A.X (1:2000, Abcam), anti-eIF-2α (1:2000, Cell Signaling Technology), anti-phospho-eIF-2α (1:2000, Cell Signaling Technology), anti-β-actin (1:5000, Cell Signaling Technology), anti-Git1 (1:2000, Cell Signaling Technology), anti-Git1 (1:2000, Novus), anti-PGK1 (1:500, Santa Cruz Biotechnology), anti-PGK1 pS203 (1:2000, Signalway Antibody), anti-HA (1:2000, Cell Signaling Technology), anti-FLAG (1:2000, Cell Signaling Technology), anti-Keap1 (1:2000, Abcam), anti-Nrf2 (1:2000, Cell Signaling Technology), anti-Ubiquitin (1:2000, Cell Signaling Technology), anti-HO1 (1:2000, Santa Cruz Biotechnology), anti-NQO1 (1:2000, Cell Signaling Technology), anti-Thioredoxin1 (1:2000, Abcam), anti-H3 (1:2000, Cell Signaling Technology), anti-Cleaved Caspase-3 (1:2000, Cell Signaling Technology), anti-Bcl2 (1:2000, Proteintech), anti-Bax (1:2000, Proteintech), and anti-Myc (1:2000, Cell Signaling Technology).
The following antibodies were used for immunofluorescence staining: anti-NeuN (1:200, Abcam), anti-Neurofilament 200 (NF-200) (1:200, Abcam), anti-phospho-eIF-2α (1:200, Cell Signaling Technology), anti-phospho-H2A.X (1:200, Abcam), anti-Glial fibrillary-associated protein (GFAP) (1:200, Cell Signaling Technology), anti-Git1 (1:200, Novus), anti-MAP2 (1:200, Abcam), and anti-TAU (1:200, Novus).
2.7. Immunofluorescence staining
The cells or spinal cord sections were treated with 4% paraformaldehyde for 30 min to fix them, followed by permeabilization with 0.3% Triton X-100 for 30 min. Next, they were blocked with 10% goat serum for 2 h, and finally incubated overnight at 4 °C with primary antibodies. Samples were washed with phosphate-buffered saline (PBS) after primary antibody removal, and incubated with fluorescent secondary antibodies (Alexa Fluor® 488 and 594) (Jackson ImmunoResearch, West Grove, PA, USA) for 2 h at room temperature conditions. Finally, nuclei were stained with DAPI (Thermo Fisher Scientific, Waltham, MA, USA) after washing and images were acquired using the same exposure time and conditions.
2.8. Dihydroethidium staining
Dihydroethidium (DHE) (Beyotime, China) was intravenously injected into mice at a concentration of 0.05 mg DHE/g of mouse body weight. DHE can be oxidized by ROS in tissues and exhibit red fluorescence. After 30 min, mice were perfused with 4% paraformaldehyde to obtain spinal cords. DHE-labeled spinal cord sections were stored in the dark and subjected to immunofluorescence staining.
2.9. Extraction and cultivation of primary mouse neurons
Primary mouse neurons were extracted from embryonic mice aged E16-E18. Spinal cords were dissected out and dissociated neurons were digested using 0.25% trypsin-EDTA solution (Thermo Fisher Scientific, MA, USA). The response was halted by using horse serum (Sigma-Aldrich). After centrifugation at 4 °C for 5 min (1000 rpm), A cell suspension was obtained by resuspending the cells in DMEM/F-12 medium supplemented with 10% horse serum, penicillin (100 IU/mL), streptomycin (100 mg/mL; Thermo Fisher Scientific), and glutamine (0.5 mm; Thermo Fisher Scientific). Cells were counted and seeded on poly-l-lysine-coated boards (Corning, NY, USA). After cells adhered, it was replaced with neurobasal medium (Thermo Fisher Scientific) supplemented with 2% B27, 1% penicillin–streptomycin, and 1% glutamine (Gibco Laboratory, Grand Island, NY). Half of the medium was replaced every 2 days.
2.10. Protocol for inducing oxygen and glucose deprivation and subsequent reperfusion
To simulate in vivo ischemia-reperfusion (IR), an oxygen glucose deprivation and reperfusion (OGD/R) model was created in vitro, as described previously [19]. The OGD/R model consisted of a control group (control), which did not undergo oxygen and glucose deprivation, and an experimental group (OGD/R), which underwent oxygen and glucose deprivation followed by reglucose and reoxygenation treatment. Briefly, primary neurons or HEK293T cells were transferred to a sugar- and serum-free medium and incubated in a hypoxic incubator at 37 °C. The incubator was filled with a gas mixture of 5% CO2 and 95% N2, with no added oxygen. After 1 h of hypoxia, cells were retrieved and the medium was substituted with regular medium. Neurons or cells were then cultured in a regular incubator for further analysis.
2.11. Reactive oxygen species analysis
According to previous studies [55,56], the levels of reactive oxygen species (ROS) in cells were assessed using a ROS Assay Kit (ab113851, Abcam) using the ROS-sensitive fluorescent probe H2DCFDA, and analyzed by a fluorescence microscope (STELLARIS STED, Lecia) and flow cytometry (FACSCalibur, BD Biosciences).
2.12. Determination of lipid peroxidation
Following the manufacturer's instructions, Click-iT Lipid Peroxidation Imaging Kit (C10446, C10447, Thermo Fisher Scientific) to assess lipid peroxidation and lipid peroxidation-induced protein denaturation in cells was used. The Lipid Peroxidation MDA Assay Kit (Beyotime, China) detects lipid oxide levels in cells.
2.13. Mitochondrial superoxide detection
The MitoSOX™ Red Mitochondrial Superoxide Indicator (M36008, Thermo Fisher Scientific) was utilized to detect superoxide in live cells within mitochondria, following the manufacturer's protocol. After treating the cells according to the experimental requirements, Hoechst (BL803A, Lot:22075473, Biosharp Life sciences) was used to stain the nuclei and observation of the cells was conducted using a confocal microscope.
2.14. Western blotting
Protein extraction was carried out from tissues or cells to obtain the total protein. The NuCLEAR™ Extraction Kit (Millipore) was utilized for the extraction of nuclear protein. Protein concentration was measured with a BCA Protein Assay Kit (Thermo Fisher Scientific). Following separation by SDS-polyacrylamide gel electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Millipore). The protein-printed membranes were then blocked with 5% BSA for 2 h at room temperature conditions and incubated with primary antibodies overnight at 4 °C. After washing, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG (1:5000, Thermo Fisher Scientific, USA) for 2 h. To visualize the immunolabeled proteins, enhanced chemiluminescence reagents (Thermo Fisher Scientific, USA) were subsequently employed. Protein expression analysis was performed using the ImageJ software. The dimerization of KEAP1 occurs as a consequence of metabolic changes in the cell. This dimerized KEAP1 is called high-molecular mass KEAP1 (HMM- KEAP1). As for HMM-KEAP1 analysis, HMM-KEAP1 was detected by non-denaturing SDS–PAGE [34,[57], [58], [59]].
2.15. Adeno-associated virus (AAV) infection
AAV HBAD-Adeasy-Git1 shRNA (AAV-Git1) was used to knock down Git1 in neurons in vitro, and AAV HBAD-Adeasy- NC shRNA (AAV-Vec) was used as a control vector (HanBio Biotechnology, Shanghai, China). AAV HBAD-Adeasy-overexpressing Git1 shRNA (Git1-OE) was used to overexpress Git1 in neurons in vitro, and AAV HBAD-Adeasy-Vector shRNA (Git1-Vec) was used as a control vector (HanBio Biotechnology, Shanghai, China).
Git1-Overexpress (AAV) 2/9-Syn-MCS-Git1- 3xFlag (AAV-Syn-Flag-Git1, Git1-OE) was used to specifically overexpress Git1 in neurons in the spinal cord. AAV 2/9-Syn-MCS - 3xFlag (AAV-Syn-Flag, Git1-Vec) was used as a control vector (HanBio Biotechnology, Shanghai, China).
2.16. Transfection of plasmids
The Flag-, Myc-, and HA tagged pcDNA3.1 vectors (Thermo Fisher Scientific) were used for subcloning the full-length sequences of human Git1, Nrf2, PGK1, and ubiquitin into the EcoRI and NotI sites. Lipofectamine 2000 (Invitrogen) was used to transfect HEK 293T cells with the expression vectors for the specified proteins.
2.17. Immunoprecipitation
Cell lysis was performed using cell immunoprecipitation (IP) lysis buffer that included protease inhibitors (Millipore). The collected cell lysate was sequentially subjected to the addition of corresponding antibodies and protein A/G magnetic beads (Thermo Fisher Scientific), followed by slow shaking and overnight incubation at 4 °C. Following IP, the magnetic beads were removed from the cell lysate and bound proteins were eluted by boiling the magnetic beads and subjected to Western blot analysis. To detect polyubiquitinated proteins, proteins were harvested after 4 h of MG132 (10 μM) treatment followed by immunoprecipitation.
2.18. Pull down polyubiquitinated proteins with tandem ubiquitin binding entities (TUBEs)
The neuronal cells were treated with 5 μM proteasome inhibitor MG-132 (HY-13259, Medchem Express) prior to cell harvesting, as per the manufacturer's instructions [60]. Next, cells were lysed using TUBEs lysis buffer containing protease inhibitors (Roche, Switzerland). Then, 100 μL of Magnetic-TUBE2 (UM402 M, LifeSensors, USA) was added to the cell lysate with a total protein of 2 mg, and the volume was brought up to 500 μL. After 4 h of incubation on a rotary shaker at 4 °C, the polyubiquitinated proteins adsorbed on the magnetic beads were collected and the magnetic beads were resuspended in SDS buffer. After boiling for 5 min, the samples were subjected to western blotting to visualize polyubiquitinated proteins. The statistical analysis of quantified polyubiquitinated proteins was based on previous references [[61], [62], [63], [64]].
2.19. IP coupled with liquid chromatography tandem mass spectrometry
Cellular total protein was extracted and subjected to IP using the designated antibodies and magnetic beads, following the aforementioned protocol. Subsequently, the eluted immunoprecipitates from the magnetic beads were analyzed using liquid chromatography (LC) tandem mass spectrometry (LC-MS/MS).
2.20. Proximity ligation assay
The binding of two proteins in neurons was detected by a Duolink detection kit and Detection Reagents Red (Sigma‐Aldrich, DUO94004). In brief, cells were permeabilized, blocked, and then incubated with primary antibodies overnight at 4 °C. The corresponding proteins were labeled with antibodies of different species, and then labeled with proximity ligation assay (PLA) PLUS and PLA MINUS probes of the corresponding species. Cells were incubated with a fluorescent secondary antibody and observed by confocal microscopy.
2.21. Quantitative metabolomic profiling
Metabolic profiling was performed using a Dionex Ultimate 3000 RS UHPLC coupled with a Q-Exactive quadrupole-Orbitrap mass spectrometer equipped with a heated electrospray ionization (ESI) source (Thermo Fisher Scientific, Waltham, MA, USA) in both ESI positive and ESI negative ion modes. The cells were washed with PBS at 4 °C after removing the medium. Then, cells were scraped with a methanol-water (V:V = 4:1) mixture, and the cell suspension was sonicated (Fisher Scientific FB-505) in a centrifuge tube. Internal standard (L-2-chlorophenylalanine) was added and centrifuged at 16,000 g for 10 min. The N2 gas was used to evaporate the supernatant and obtain a dry sample. The dried metabolites were subsequently resuspended in an appropriate solvent, and the resulting solution was loaded onto the instrument for further analysis.
2.22. Statistical analyses
Data analysis was conducted using GraphPad Prism 7 software (GraphPad Software, CA, USA). The data were presented as mean ± standard deviation (SD), and each group of data included at least three independent experiment replicates. Multiple groups of experimental data were compared using one-way ANOVA analysis, and the Tukey's post hoc test of variance was used. Comparison between two groups was performed using the t-test. P < 0.05 was considered statistically significant.
3. Results
3.1. Git1 deletion impairs functional recovery and aggravates neuronal loss after I/R
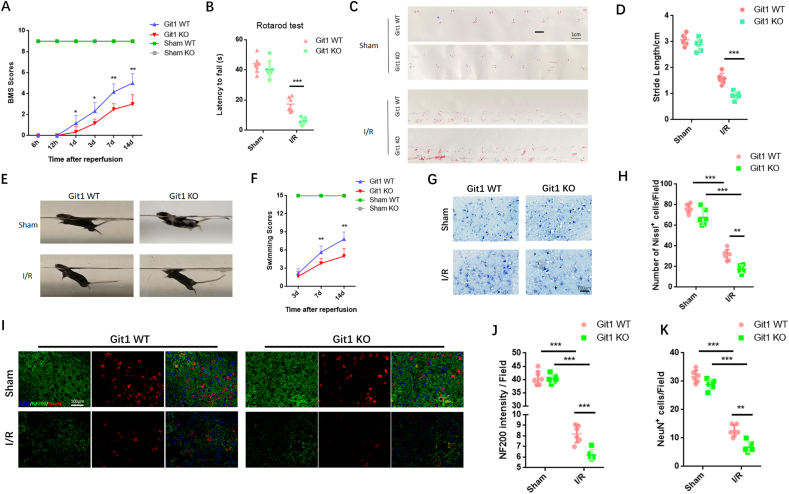
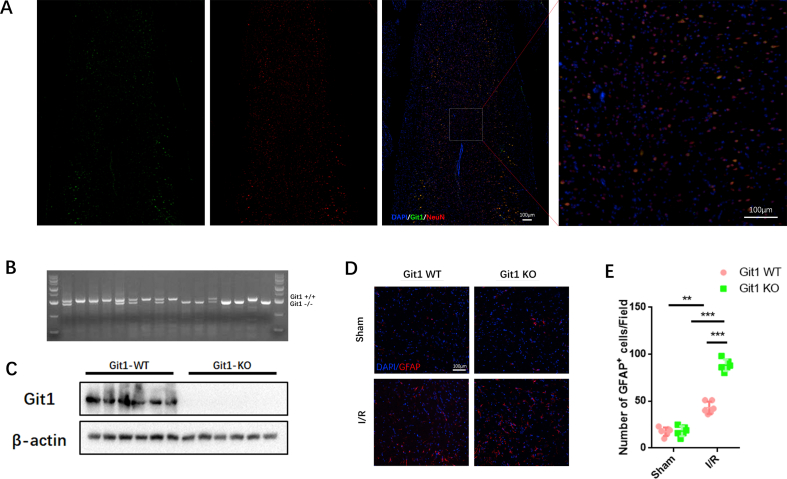
Given the important role of Git1 in the nervous system, we investigated the role of Git1 in SCIRI [44,46,48,65]. Immunofluorescence staining of mouse spinal cords showed that Git1 co-localized with NeuN, and Git1 was mainly expressed in neurons (Fig. S1A). To evaluate the effects of physiological deficiencies of Git1 in vivo, we employed Git1 knockout mice. Git1 knockout was successfully achieved in the spinal cord (Figs. S1B and C). Next, spinal cord ischemia-reperfusion (I/R) surgery in Git1 WT and KO mice was performed, and motor functional behavior analysis after SCIRI was assessed by Basso mouse scale (BMS) score. The results showed no significant difference between Git1 WT and KO in the sham group. The BMS score of the Git1 KO group was significantly lower than that of the Git1 WT group at 1, 3, 7 and 14 days after I/R (Fig. 1A). Additional motor function behavioral tests (rotarod test, footprint analysis, and swimming test) further assess motor function recovery after I/R (Fig. 1B, C, E). The results of these tests showed that Git1 WT mice had significantly better motor function recovery compared to Git1 KO mice (Fig. 1B–F).
Fig. 1.
Knockout of Git1 impairs functional recovery and aggravates neuron loss in SCIRI. A BMS scores at 6 h, 12 h, 1 day, 3 days, 7 days, and 14 days after ischemia-reperfusion (IR) in Git1 WT and KO mice (n = 6). The two sham operation groups had the same score (Sham WT (green line) and Sham KO (gray line)). B–D Rotarod test and footprint analysis 14 days after I/R demonstrated worse functional recovery of Git1 KO mice (scale bar = 1 cm, n = 6). E, F Swim test to assess trunk stability and hindlimb motor function at 3, 7, and 14 days of reperfusion, and statistical analysis of the Louisville Swim Scale scores between the Git1 WT and KO groups (n = 6). The two sham operation groups had the same score (Sham WT (green line) and Sham KO (gray line)). G, H Normal Nissl bodies of neurons were observed in Nissl-stained sections of the sham group in both Git1 WT and KO mice, while Git1 KO mice indicated significantly fewer Nissl bodies at 12 h after reperfusion compared to WT mice (scale bar = 100 μm, n = 6). I Representative immunofluorescence images of NeuN (Red) and NF200 (Green) in spinal cord of Git1 WT and KO mice at 12 h post-reperfusion. Nuclei were labeled with DAPI (blue) (scale bar = 100 μm). J, K Quantification of NeuN+ neurons and NF200 relative intensity in the spinal cord (n = 6). *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To explore the cause of impaired motor function in SCIRI, staining analysis of spinal cord tissue sections was performed. After 12 h of reperfusion, Nissl staining showed that Git1 KO mice exhibited significant morphological changes in Nissl-positive neurons, which were reduced compared to WT mice (Fig. 1G and H). There was no significant difference in the immunofluorescence staining of NeuN and NF200 neurons in the spinal cord sections between WT and KO mice in the sham group (Fig. 1I). After 12 h of reperfusion, compared with the WT group, the KO group had a notable decrease in the number of NeuN-positive cells, as well as nerve fiber (NF200-labeled) loss and morphological disorder (Fig. 1I–K). In addition, 12 h after reperfusion, the KO group had more significant glial scar hyperplasia (Figs. S1D and E). Excessive glial scarring indicates poor neuronal cell survival and regeneration. Our results indicate that Git1 has a positive regulatory effect on SCIRI.
3.2. Git1 affects the accumulation of ROS in neuronal cells
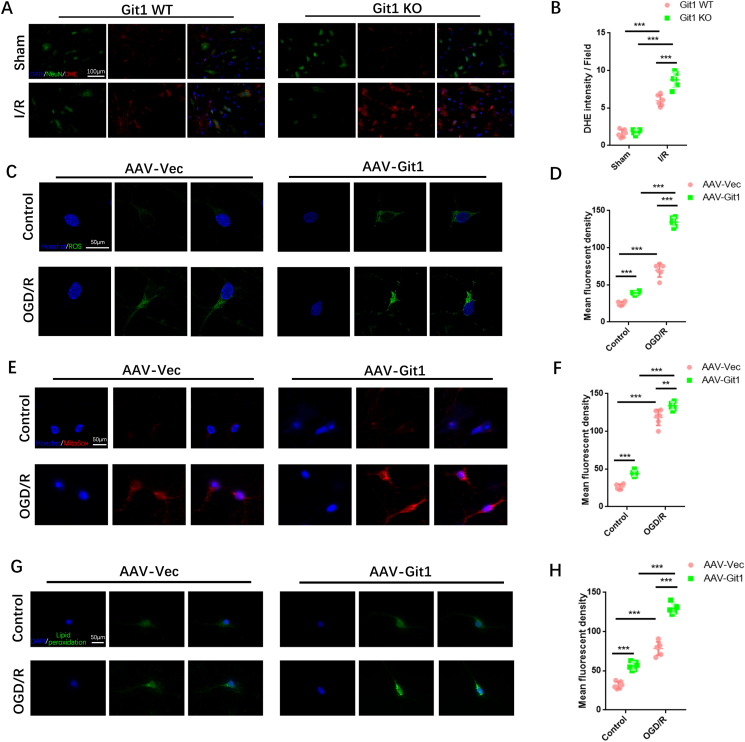
ROS are considered an important mechanism in secondary spinal cord injury. Abnormal accumulation of ROS is an important stage in the progression of SCIRI injury [13,14,16,19]. Therefore, we next studied the relationship between Git1 and ROS. The level of ROS in spinal cord tissue was explored using the DHE fluorescent probe. At 4 h after reperfusion, there was significant DHE fluorescence in spinal cord tissue, and it was mainly localized in NeuN+ cells (Fig. 2A). Git1 KO mice had stronger DHE fluorescence levels than WT mice (Fig. 2B).
Fig. 2.
Regulation of ROS levels in neurons by GIT1 in vitro and in vivo. A ROS levels in the spinal cord were detected by DHE staining (scale bar = 100 μm). B Quantification of DHE fluorescence intensity in the spinal cord of Git1 WT and KO mice in the sham group and I/R group (4 h of reperfusion, n = 6). C Cell immunofluorescence detection of ROS levels in neurons (scale bar = 50 μm). D Quantification of mean fluorescence intensity of ROS in neurons (n = 6). E,F Immunofluorescence detection and quantification of mitochondrial superoxide (MitoSox, red) in neurons (scale bar = 50 μm, n = 6). G,H Immunofluorescence detection and quantification of lipid peroxidation (green) in neurons (scale bar = 50 μm, n = 6) (OGD/R, reperfusion 4 h). *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
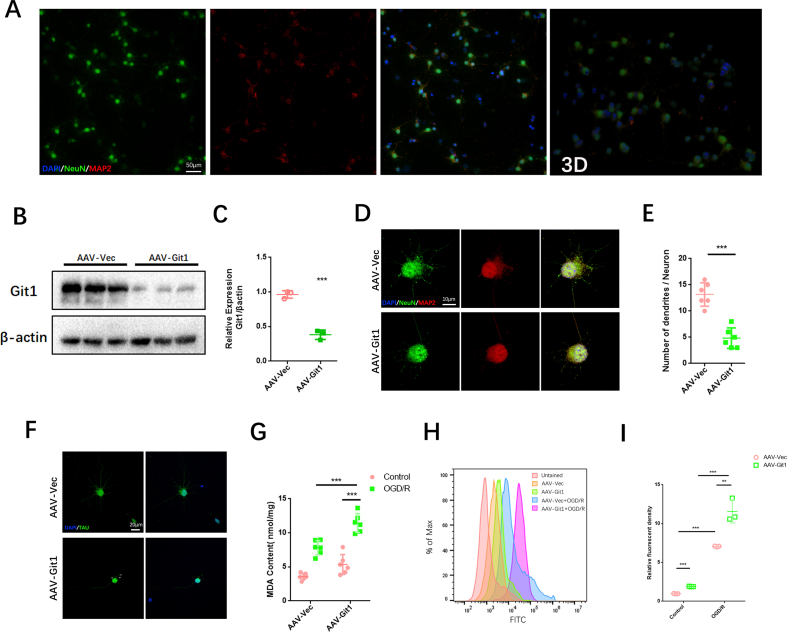
The effect of Git1 on ROS in neuronal cells after ischemia-reperfusion was further verified in vitro. Git1 expression was knocked down in neurons by adenovirus in vitro (Figs. S2A–F). Flow cytometry analysis and immunofluorescence staining showed that in the control group (without OGD/R), ROS in neurons was significantly upregulated after Git1 knockdown. After OGD/R (reperfusion for 4 h) treatment, ROS upregulation following Git1 knockdown in neurons was more pronounced (Fig. 2C, D, Figs. S2H and I). Mitochondrial superoxide levels in neurons were further analyzed. We found that mitochondrial superoxide levels were significantly increased after Git1 knockdown using mitochondrial superoxide fluorescent reagent (MitoSox). This phenomenon was significantly upregulated after OGD/R (reperfusion for 4 h), and the Git1 knockdown group (AAV-Git1) was significantly higher than the Git1 control group (AAV-Vec) (Fig. 2E and F). In addition, lipid peroxides were labeled by cell fluorescence staining; the fluorescence intensity of lipid peroxides in neurons was enhanced after Git1 knockdown. After OGD/R (reperfusion for 4 h), lipid peroxides were significantly elevated after Git1 knockdown and higher than those in the Git1 control group (Fig. 2G and H). Lipid peroxidation malondialdehyde (MDA) assay also indicated that after OGD/R, lipid peroxides in the AAV-Git1 group were higher than in the AAV-Vec group (Fig. S2G). These results suggest that Git1 may play a critical role in I/R-induced ROS levels in neuronal cells.
3.3. Git1 deletion aggravates endoplasmic reticulum stress and DNA damage in neurons after I/R and OGD/R
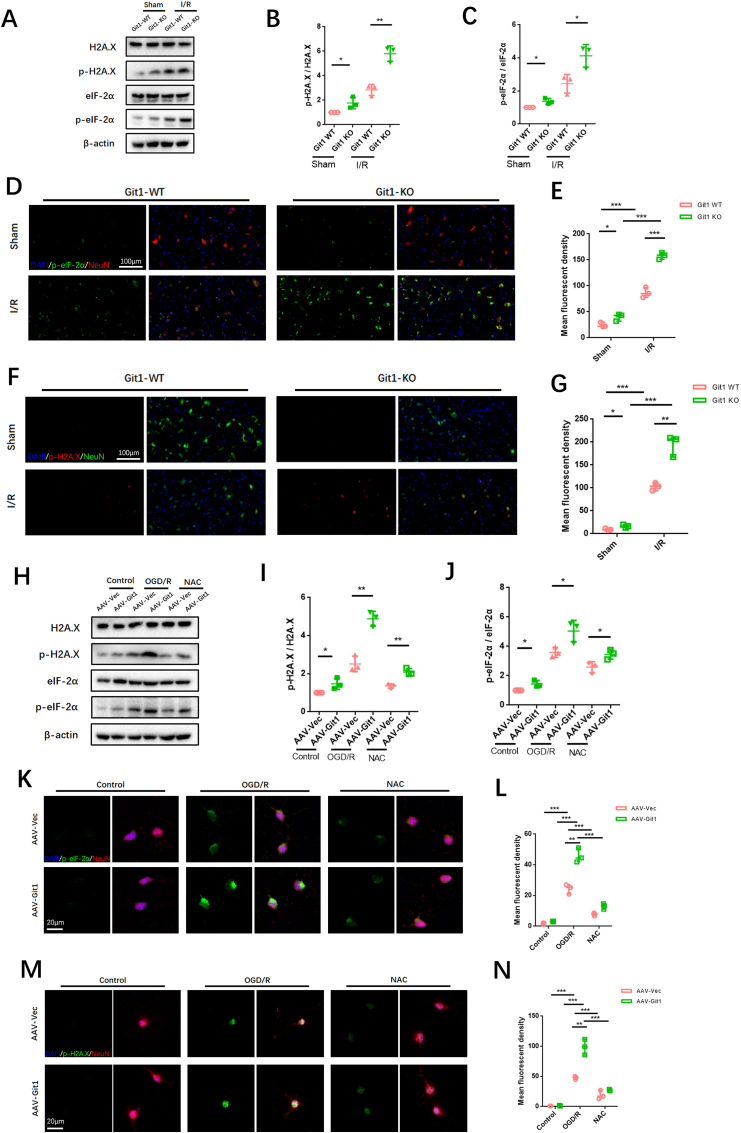
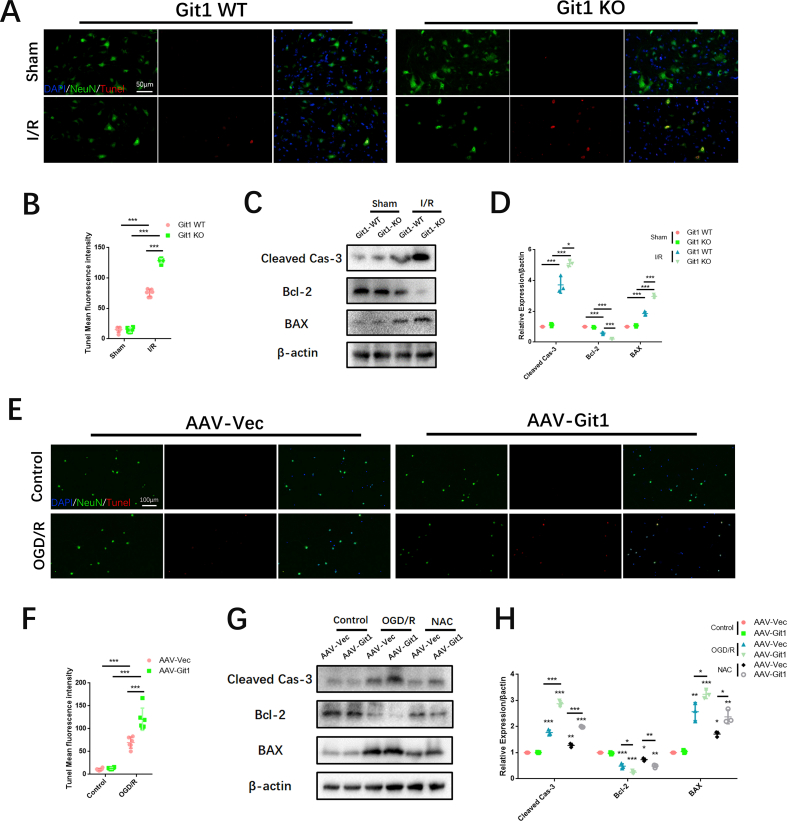
Previous research has demonstrated that the endoplasmic reticulum (ER) is vulnerable to reactive oxygen species (ROS) stimulation, which leads to the accumulation of excessive misfolded and unfolded proteins in the ER, resulting in endoplasmic reticulum stress (ERS) [66,67]. In addition, ROS can directly cause DNA damage [68]. Given the close relationship between Git1 and ROS, we explored ERS and DNA damage in the spinal cord after SCIRI. Western blot analysis revealed increased levels of phosphorylated eIF-2α and H2A.X in the spinal cord of Git1 KO mice as compared to the WT mice (Fig. 3A). This difference was more pronounced in the injured spinal cord after I/R (reperfusion 4 h) (Fig. 3B and C). Immunofluorescence staining further verified ERS and DNA lesion in the spinal cord, and the results suggested that phosphorylation of eIF-2α and H2A.X in the spinal cord of both Git1 WT and KO mice in the sham group were at lower levels and mainly localized in neurons (Fig. 3D–G). Furthermore, both phosphorylated eIF-2α and H2A.X were significantly increased in the spinal cord of Git1 KO mice compared to WT mice. After I/R (reperfusion 4 h), phosphorylation eIF-2α and H2A.X in the spinal cord of Git1 WT and KO mice was increased to a certain extent; phosphorylation of eIF-2α and H2A.X in spinal cord of Git1 KO mice was significantly higher than that of WT mice (Fig. 3D–G). ERS and DNA damage caused by ROS often lead to apoptosis. We observed apoptosis indicators by immunofluorescence staining and immunoblotting. The apoptosis level of spinal cord tissue increased significantly after I/R (reperfusion 4 h), and Git1 knockout significantly aggravated the level of apoptosis (Figs. S3A–D).
Fig. 3.
Git1 regulates endoplasmic reticulum stress and DNA damage in neurons after I/R and OGD/R. A–C Western blotting analysis of the expression of ERS (phosphorylated eIF-2α) and DNA damage (phosphorylated H2A.X)-related proteins in spinal cord after I/R (reperfusion 4 h) (n = 3). D, E Immunofluorescence analysis shows ERS levels (phosphorylated eIF-2α) in neurons in the spinal cord (scale bar = 100 μm, n = 3) (I/R, reperfusion 4 h). F, G Immunofluorescence analysis shows DNA damage levels (phosphorylated H2A.X) in neurons in the spinal cord (scale bar = 100 μm, n = 3). H–J Western blot analysis of the levels of ERS (phosphorylated eIF-2α) and DNA damage (phosphorylated H2A.X)-related proteins in neurons after OGD/R or NAC treatment (with OGD/R) (n = 3). K–N Immunofluorescence of phosphorylated eIF-2α and H2A.X expression levels in neurons after OGD/R or NAC treatment (during reperfusion) (scale bar = 20 μm, n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
In vitro, we further investigated the correlation between ERS and DNA damage and ROS levels associated with Git1 in neurons. Immunoblotting showed significant upregulation of phosphorylated eIF-2α and H2A.X after Git1 knockdown (Fig. 3H–J). In addition, after OGD/R (reperfusion 4 h), the levels of phosphorylated eIF-2α and H2A.X were increased in both the AAV-Vec and AAV-Git1 groups. The AAV-Git1 group phosphorylation level was significantly higher than the AAV-Vec group phosphorylation level (Fig. 3H–J). Besides that, such changes in ERS and DNA damage levels can be significantly inhibited by N-acetyl cysteine (NAC), a ROS scavenger (Fig. 3H–J). Immunofluorescence staining also indicated that after OGD/R (reperfusion 4 h), expression levels of phosphorylated eIF-2α and H2A.X were significantly increased after Git1 knockdown, compared with the control group (Fig. 3K–N). This level of phosphorylation can be inhibited by NAC (Fig. 3K–N). The above results indicate that the correlation between ROS and Git1 can significantly affect ERS and DNA damage in neurons. ERS and DNA damage in neurons can lead to apoptosis [69,70]. Immunofluorescence and western blotting indicated that the apoptosis level was significantly up-regulated in Git1 knockdown neurons after OGD/R (reperfusion 4 h), and this apoptosis level could be inhibited by NAC (Figs. S3E–H). Overall, these results suggest that ROS accumulation following Git1 deletion in SCIRI can damage neurons, leading to ERS and DNA damage in neurons. ERS and DNA damage influence neuronal apoptosis in SCIRI.
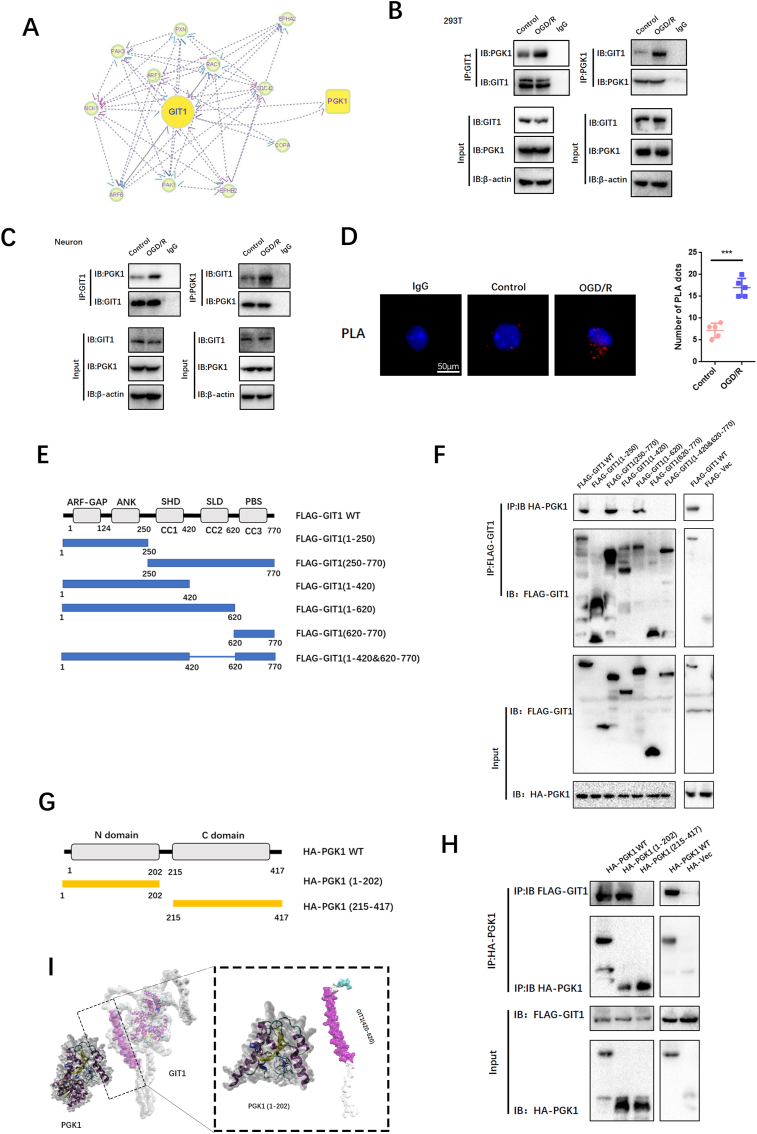
3.4. Git1 interacts with PGK1 in neurons
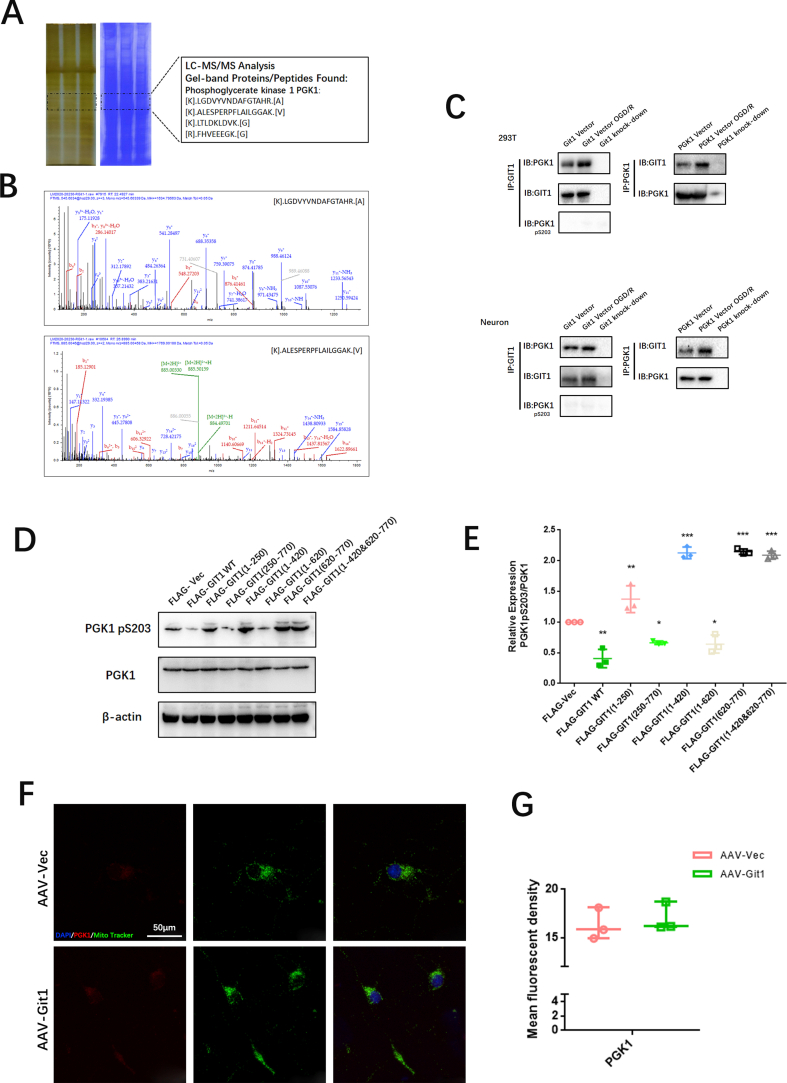
Research conducted in the past has revealed that Git1 is a multifaceted shuttle protein, with its primary role being the interaction with various proteins to exert its biological functions. This has been demonstrated in various studies [37,38]. To study the underlying mechanisms by which Git1 modulates ROS, we used immunoprecipitation coupled with LC-MS/MS to study the Git1 binding proteins. LC–MS/MS identified PGK1 as a novel protein bound by Git1 (Fig. 4A, Figs. S4A and B). To further validate the LC-MS/MS findings, Co-Immunoprecipitation (Co-IP) analysis was carried out, which demonstrated that Git1 interacted with PGK1 in 293T cells under physiological conditions as well as in response to OGD/R (reperfusion 4 h). Furthermore, it was observed that the OGD/R treatment led to an increased interaction between Git1 and PGK1 (Fig. 4B). Reverse Co-Immunoprecipitation (Co-IP) analysis also validated that Git1 significantly precipitated PGK1 (Fig. 4B). We further confirmed by Co-IP that endogenous Git1 also interacts with PGK1 in neurons, which is consistent with that in 293T cells (Fig. 4C). Further Co-IP experiments demonstrated that endogenous Git1 did not bind to PGK1 (pS203) (Fig. S4C). Proximity Ligation Assay (PLA) was employed to further establish the binding between Git1 and PGK1 in neurons under both physiological conditions and in response to OGD/R. The results obtained through the assay provided additional evidence of the binding between Git1 and PGK1 (Fig. 4D). These data demonstrate that Git1 interacts with PGK1. Git1 is a multi-domain protein containing three coiled-coil (CC)-rich domains [40,41]. PGK1 can be primarily divided into two domains: the N-terminal domain, which is rich in coiled-coils, and the C-terminal domain [[71], [72], [73]]. In 293T cells, we utilized full-length protein as well as a series of truncated Flag-labeled Git1 and HA-labeled PGK1 fragments to investigate the binding region (Fig. 4E, G). Our results revealed that the interaction between Git1 and PGK1 occurs through the synaptic localization domain (SLD) structure, which includes the coiled-coil 2 (CC2) domain (Fig. 4F). Additional Co-Immunoprecipitation (Co-IP) analysis revealed that Git1 binds to the N-terminal domain of PGK1, suggesting that the interaction may be dependent on the coiled-coil structure present in the N-terminal domain (Fig. 4H and I).
Fig. 4.
Git1 interacts with PGK1. A Interaction between Git1 and other proteins. B, C Co-immunoprecipitation detection of endogenous Git1 binding to PGK1 in untreated or OGD/R-treated neurons and 293T cells (OGD/R reoxygenation for 4 h, n = 3). D Proximity ligation assay (PLA) detects the binding of Git1 to PGK1 in neurons under physiologic conditions or in response to OGD/R treatment (reperfusion 4 h) (scale bar = 50 μm, n = 6). E Functional domains of Git1. F Flag-Git1-WT or Flag-Git1 deletion mutants were co-transfected with HA-PGK1 in HEK 293T cells, and were then subjected to OGD/R for 4 h. Immunoprecipitation (IP) was conducted using Flag antibody, followed by probing with HA to detect the interaction between PGK1 and Git1 or Git1 deletion mutants (n = 3). G Functional domains of PGK1. H HEK 293T cells were co-transfected with Flag-Git1 and either full-length HA-labeled PGK1 or its deletion mutant. The cell lysate was subsequently analyzed by IP (n = 3). I Three-dimensional structures of Git1 and PGK1 proteins.
3.4.1. Git1 regulates the Keap1-Nrf2 signaling pathway by affecting the phosphorylation of PGK1
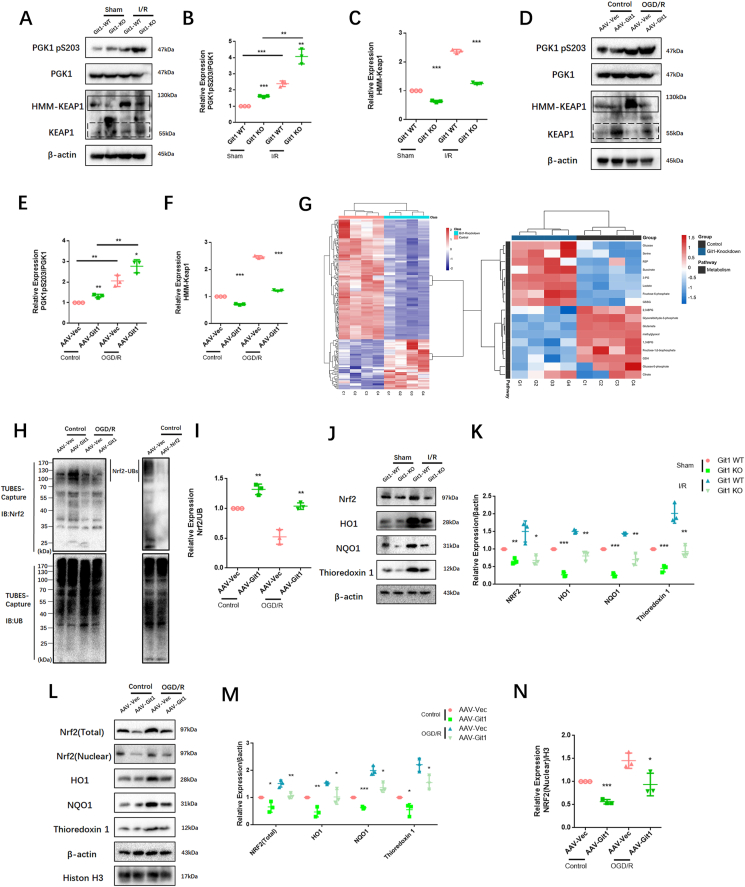
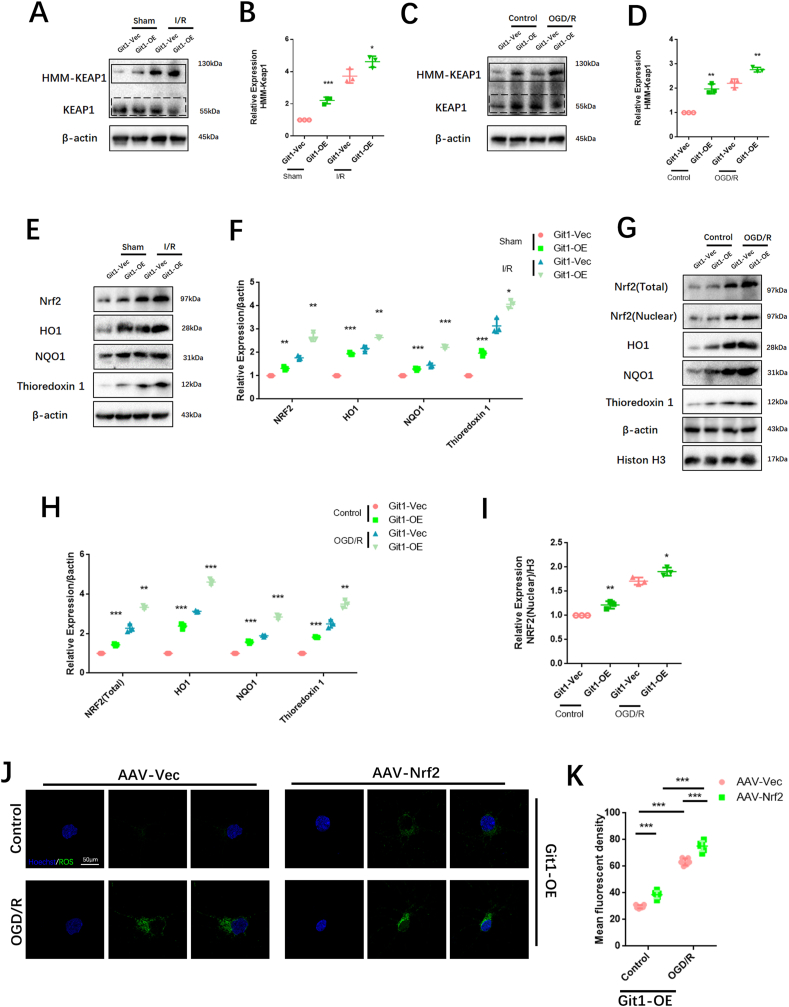
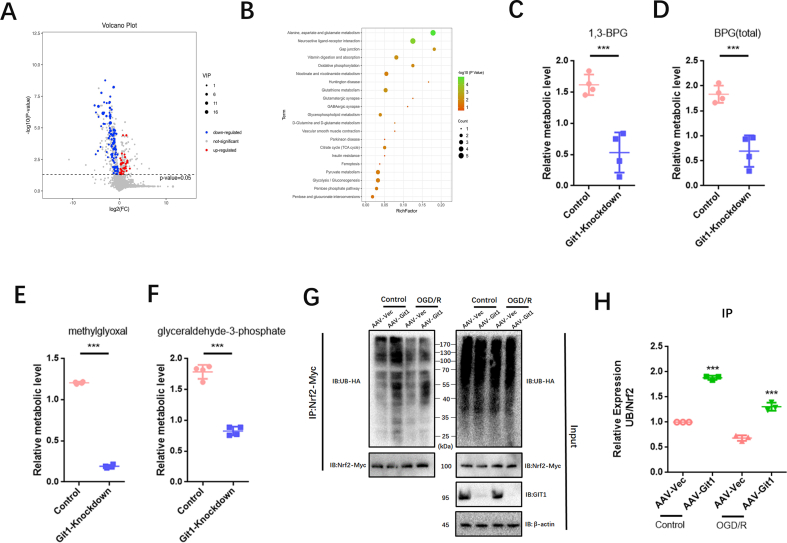
PGK1 is a critical enzyme involved in glycolysis, and its activity can affect Keap1-Nrf2 signaling, resulting in changes in cellular oxidative stress status [34]. Western blotting showed that the phosphorylation level of PGK1 S203 in spinal cord of Git1 KO mice was higher than that of Git1 WT mice in the sham-operated and ischemia-reperfusion groups (Fig. 5A and B). Git1 deletion variants that bind to PGK1 also significantly inhibited PGK1 phosphorylation (Figs. S4D and E). Git1 deletion variants unable to bind to PGK1 significantly promoted PGK1 phosphorylation (Figs. S4D and E). The results further indicated that the binding of Git1 to PGK1 could significantly inhibit the phosphorylation of PGK1. After ischemia-reperfusion, the level of PGK1 S203 phosphorylation in the spinal cord was significantly higher than that of the sham-operated group (Fig. 5A and B). Meanwhile, in both the sham-operated group and the ischemia-reperfusion group, the level of HMM-Keap1 (high-molecular mass Keap1, dimerization of Keap1) in the spinal cord of Git1 KO mice was significantly lower than that of WT mice, which correlated with the phosphorylation level of PGK1 (Fig. 5A–C). In response to control treatment and OGD/R, Git1 knockdown neurons (AAV-Git1) also had higher PGK1 phosphorylation levels and lower HMM-Keap1 expression compared to Git1 control neurons (AAV-Vec) (Fig. 5D–F). Immunofluorescence staining further verified Git1 knockdown did not affect the distribution and expression of PGK1 (Figs. S4F and G). PGK1 can affect the level of intracellular metabolites with electrophilic groups that can affect the dimerization of Keap1 [34,74,75]. Therefore, relevant metabolites before and after Git1 knockdown in neurons were analyzed by quantitative metabolomic profiling. Git1 knockdown results in dramatic differences in metabolites in neurons (Fig. S5A). As expected, based on the differences in metabolites, associated metabolic pathway analysis indicated the involvement of several neuronal metabolism, glucose metabolism and oxidative stress pathways, including oxidative phosphorylation glutathione metabolism and glycolysis (Fig. S5B). Metabolomic analysis specifically targeting neurons revealed a significant reduction in levels of metabolites upstream of PGK1, including methylglyoxal, 1,3-and total-bisphosphoglycerate (BPG) and glyceraldehyde-3-phosphate (GAP), and accumulation of downstream metabolites, such as lactate and 3-phosphoglycerate, in Git1-knockdown neurons, compared to the control group (Fig. 5G, Figs. S5C–F). Taken together, these findings suggest that the binding between Git1 and PGK1 can modulate glycolytic intermediates, whose changes are linked to the expression of HMM-Keap1, and could potentially act as a signal to the Nrf2 signaling pathway.
Fig. 5.
Git1 inhibits phosphorylation of PGK1 and activates Nrf2 signaling. A Representative western blots of PGK1, phosphorylation of PGK1 S203, Keap1, and HMM-Keap1 expression in the spinal cords of Git1 wild-type (WT) and knockout (KO) mice exposed to either sham operation or ischemia/reperfusion (I/R) injury for 4 h. Keap1: black dotted line box, HMM-Keap1: black solid line box. B, C Quantification of phosphorylation of PGK1 S203 and HMM-Keap1 in the spinal cord (n = 3). D Representative western blots of phosphorylation of PGK1 S203, PGK1, Keap1, and HMM-Keap1 expression in control and Git1 knockdown neurons under control and OGD/R treatment (reperfusion 4 h). Keap1: black dotted line box, HMM-Keap1: black solid line box. E, F Quantification of phosphorylation of PGK1 S203 and HMM-Keap1 in neurons (n = 3). G Heat map visualization of metabolite levels in control and Git1-knockdown neurons. R5P, ribose-5-phosphate; 3-PG; 3-phosphoglycerate; GSSG, glutathione disulfide; BPG, bisphosphoglycerate; GSH, glutathione-SH. H, I Co-immunoprecipitation analysis and quantitation of the polyubiquitination of Nrf2 in neurons in AAV-Vec (vector adenovirus) and AAV-Git1 (Git1 knockdown adenovirus) groups under control and OGD/R (reperfusion 4 h) (n = 3). AAV-Nrf2 (Nrf2 knockdown adenovirus). J, K Representative western blots of Nrf2, HO1, NQO1, and thioredoxin 1 expression in the spinal cords of Git1 wild-type (WT) and knockout (KO) mice exposed to either sham operation or ischemia/reperfusion (I/R) injury for 4 h (n = 3). L–N Representative western blots of Nrf2 (total), Nrf2 (nuclear), HO1, NQO1, and thioredoxin 1 expression in control and Git1 knockdown neurons under control and OGD/R treatment (reperfusion 4 h) (n = 3).
The Nrf2 signaling axis is an important oxidative stress protection mechanism, and ubiquitination of Nrf2 is regulated by Keap1. When Keap1 dimerization increases, Nrf2 ubiquitination decreases, Nrf2 expression increases, and Nrf2 enters the nucleus to play an antioxidant role [[76], [77], [78]]. Co-immunoprecipitation showed that Nrf2 ubiquitination in the Git1 knockdown group in 293T cells under control and OGD/R treatment increased compared with the Git1 control group (Figs. S5G and H). Further, using magnetic beads (magnetic-tandem ubiquitin binding entities, TUBES), it was also found that Git1 could affect polyubiquitination in neurons (Fig. 5H and I). In response to sham surgery and I/R, knockout of Git1 downregulated the expression levels of Nrf2, as well as Nrf2 target genes such as HO1, NQO1, and thioredoxin 1 in the spinal cord (Fig. 5J and K). Immunoblot analysis of Git1 knockdown neurons under control and OGD/R treatment in vitro further indicated that Git1 can affect the expression levels of Nrf2 and Nrf2-mediated antioxidant genes and the nuclear import of Nrf2 (Fig. 5L–N). These above experimental results clarified that Git1 regulates the phosphorylation level of PGK1, affects the level of glucose metabolism intermediates, leading to changes in Keap1 dimerization and ultimately changing the expression of antioxidant Nrf2 axis-related proteins. This elucidates the mechanism by which Git1 resists ROS in neurons.
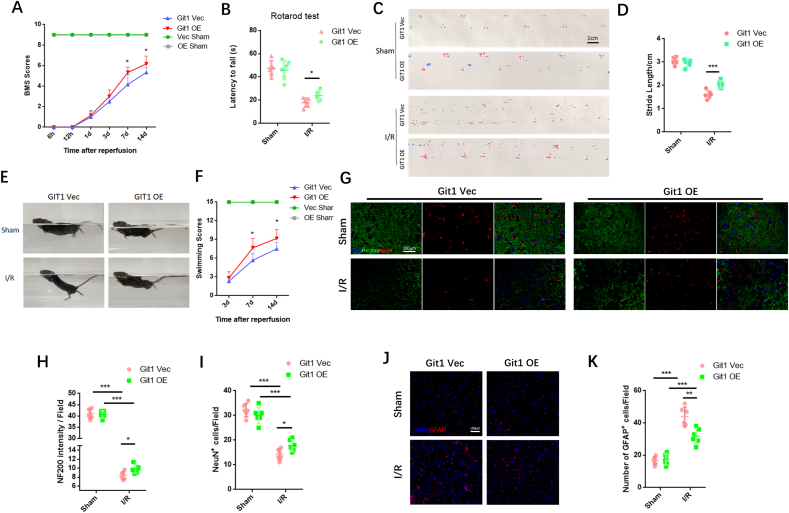
3.5. Neuronal overexpression of Git1 enhances functional recovery and reduces neuronal loss following SCIRI
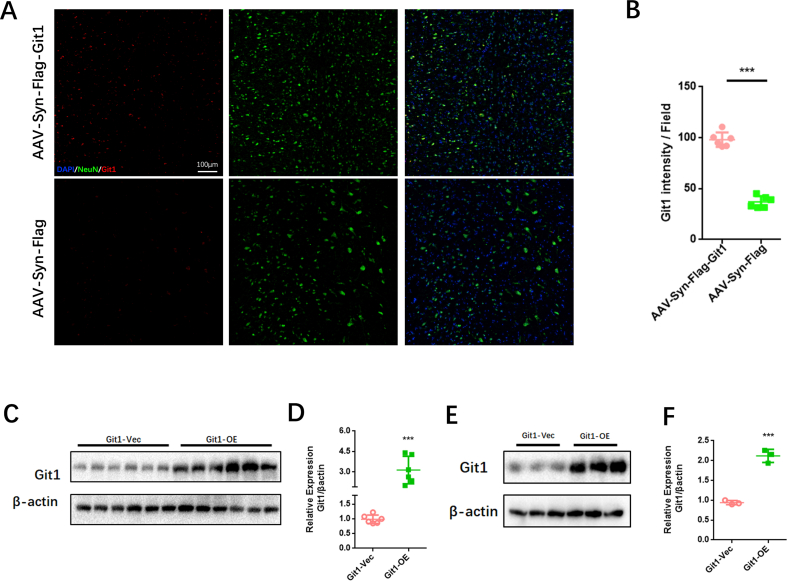
Given the essential role of Git1 in SCIRI, we injected neuron-specific adeno-associated virus into the spinal cord to overexpress Git1. Immunofluorescence staining showed that Git1 was significantly more expressed in spinal cord neurons in the overexpression virus-injected group (AAV-Syn-Flag-Git1) than in the control virus-injected group (AAV-Syn-Flag) (Figs. S6A and B). In addition, western blotting indicated that the expression of Git1 in neurons in the overexpression group (Git1-OE) was significantly higher than that in the control group (Git1-Vec) (Figs. S6C and D). The BMS score and swimming test revealed that injection of AAV-Git1 led to improved functional recovery compared to the control vector (Git1-Vec) on days 7 and 14 post-I/R (Fig. 6A, E, F). Further motor function tests, including the rotarod test and footprint analysis, provided additional evidence that overexpression of Git1 in neurons led to enhanced motor recovery (Fig. 6B–D). Immunofluorescence staining showed no significant difference in neuron number and nerve fiber morphology between Git1 OE and Git1 Vec mice in the sham-operated group. After I/R (12 h of reperfusion), mice overexpressing Git1 had more viable neurons and better morphological nerve fibers than Git1-Vec mice injected with AAV-Vector (Fig. 6G–I). Furthermore, Git1-OE mice had less gliosis scarring than Git1-Vec mice after I/R (12 h of reperfusion) (Fig. 6J and K). Collectively, these findings indicate that targeted upregulation of Git1 in neurons facilitates functional recovery and reduces neuronal loss in the setting of SCIRI.
Fig. 6.
Enhanced expression of Git1 in neurons promotes functional recovery and mitigates loss of neurons after SCIRI. A BMS scores at various time points in Git1-OE (Adeno-associated virus overexpressing Git1) and Git1-Vec (Adeno-associated virus control vector) mice after I/R (n = 6). The two sham operation groups had the same score (Vec Sham (green line) and OE Sham (gray line)). B Rotarod test at day 14 after I/R demonstrated better functional recovery in Git1-OE mice (n = 6). C, D Footprint assay to assess functional recovery at day 14 after I/R in Git1-OE and Git1-Vec mice (scale bar = 1 cm, n = 6). E, F Swimming test to assess trunk stability and hindlimb motor function at days 3, 7, and 14 after I/R, and statistical analysis of the Louisville Swim Scale data in Git1 Vec and Git1 OE mice (n = 6). G Representative immunofluorescence images of NeuN (red) and NF200 (green) in the spinal cord tissue sections of Git1 Vec and Git1 OE mice at 12 h post-reperfusion. Nuclei were highlighted using DAPI fluorescence (in blue) (scale bar = 100 μm, n = 6). H, I Quantification of NeuN+ neurons and NF200 relative intensity in the spinal cord (n = 6). J Representative immunofluorescence images of GFAP (red) and DAPI (blue) in spinal cord tissues of Git1 Vec and Git1 OE mice at 12 h post-reperfusion (scale bar = 100 μm, n = 6). D Quantification of GFAP+ cells (astrocytes) in the spinal cord (n = 6). *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Overexpression of Git1 in neurons reduces ROS levels after SCIRI
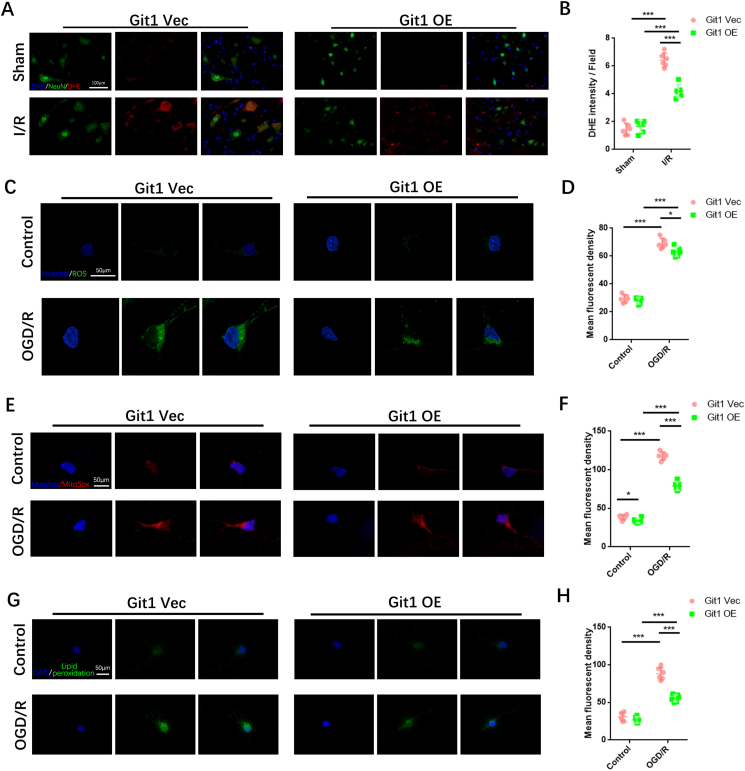
Given the possible relationship between Git1 and ROS, we observed ROS levels after I/R in mice overexpressing Git1. Immunofluorescence staining analysis revealed a notable increase in levels of reactive oxygen species (ROS) in neurons located in the spinal cord of both Git1-OE mice and control (Git1-Vec) mice 4 h after experiencing ischemia/reperfusion (I/R) injury. Interestingly, the levels of ROS in Git1-OE mice were found to be significantly lower compared to those observed in Git1-Vec mice (Fig. 7A and B). Overexpression of Git1 in neurons in vitro was used to investigate ROS levels after OGD/R (Figs. S6E and F). Immunofluorescence staining showed that ROS levels in neurons overexpressing Git1 (Git1-OE) were significantly lower than those in control neurons (Git1-Vec) 4 h after OGD/R (Fig. 7C and D). Further research found that mitochondrial superoxide in neurons was also affected by Git1. Mitochondrial superoxide was significantly decreased in Git1-overexpressing neurons after OGD/R (4 h after reperfusion) (Fig. 7E and F). Likewise, lipid peroxides were labeled by immunofluorescence staining, and the results revealed that levels of lipid peroxides in neurons of Git1-OE mice were significantly lower when compared to those observed in Git1-Vec mice after OGD/R (4 h after reperfusion) (Fig. 7G and H). The above results suggest that Git1 overexpression can reduce ischemia-reperfusion-induced ROS in neurons.
Fig. 7.
Overexpression of Git1 reduces reperfusion-induced ROS levels in neurons. A Detection of neuronal ROS levels in spinal cord by immunofluorescence staining (scale bar = 100 μm). B Quantification of DHE fluorescence intensity in the spinal cord of Git1 Vec and Git1 OE mice after sham-operated and I/R (4 h of reperfusion, n = 6). C, D Representative immunofluorescence images of ROS levels in Git1 Vec and Git1 OE neurons and quantification of mean fluorescence intensity of ROS (scale bar = 50 μm, n = 6). E, F Immunofluorescence and quantification of mitochondrial superoxide (MitoSox labeled, red) in Git1 Vec and Git1 OE neurons (scale bar = 50 μm, n = 6). G, H Immunofluorescence detection and quantification of lipid peroxidation (green) in Git1 Vec and Git1 OE neurons (scale bar = 50 μm, n = 6). (OGD/R, reperfusion 4 h). *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Overexpression of Git1 in neurons reduces reperfusion-induced ROS levels via enhancing Nrf2 signaling
To further confirm that Git1 affects ROS levels by modulating the Keap1-Nrf2 signaling axis, we investigated the expression of the Keap1-Nrf2 signaling pathway after I/R in mice overexpressing Git1. Western blotting analysis revealed a significant increase in the level of high molecular weight Keap1 (HMM-Keap1) in the spinal cord of mice overexpressing Git1 (Git1-OE) compared to that observed in control mice (Git1-Vec) (Fig. 8A and B). Meanwhile, in response to control treatment and OGD/R, Git1-OE neurons also had higher HMM-Keap1 expression compared to Git1-Vec neurons in vitro (Fig. 8C and D). In addition, in response to sham surgery and I/R, overexpression of Git1 upregulated the expression levels of Nrf2, and Nrf2 target genes HO1, NQO1, and thioredoxin 1 in the spinal cord (Fig. 8E and F). Immunoblot analysis of neurons overexpressing Git1, both under normal conditions and following OGD/R treatment in vitro, revealed that Git1 can modulate the expression of Nrf2 and Nrf2-related antioxidant genes, as well as the nuclear import of Nrf2 (Fig. 8G–I). To further verify the regulatory mechanism of Git1 on Nrf2, Nrf2 expression was knocked down in Git1-overexpressing neurons. Immunofluorescence staining indicated that ROS levels in Git1-OE neurons after Nrf2 knockdown (AAV-Nrf2) were significantly higher than those in Git1-OE neurons without Nrf2 knockdown (AAV-Vec) 4 h after OGD/R (Fig. 8J and K). The above results clarified that overexpression of Git1 in neurons reduces reperfusion-induced ROS levels via enhancing Nrf2 signaling. This further elucidated the mechanism of Git1 resists ROS in neurons by promoting Keap1 dimerization and activating Nrf2 antioxidant pathway (see Fig. 9).
Fig. 8.
Overexpression of Git1 in neurons enhances Keap1-Nrf2 signaling pathway. A Western blot analysis of Keap1 and HMM-Keap1 expression in the spinal cords of Git1 Vec and OE mice following either a sham operation or ischemia/reperfusion (I/R) injury for 4 h. Keap1: black dotted line box, HMM-Keap1: black solid line box. B Quantification of HMM-Keap1 in the spinal cord (n = 3). C Representative western blots of Keap1 and HMM-Keap1 expression in Git1 Vec and OE neurons under control and OGD/R treatment (reperfusion 4 h). Keap1: black dotted line box, HMM-Keap1: black solid line box. D Quantification of phosphorylation of HMM-Keap1 in neurons (n = 3). E, F Representative western blots of Nrf2, HO1, NQO1, and thioredoxin 1 expression levels in the spinal cords of Git1 Vec and OE mice subjected to sham operation or ischemia/reperfusion (I/R) injury for 4 h (n = 3). G-I Representative western blots of Nrf2 (total), Nrf2 (nuclear), HO1, NQO1, and thioredoxin 1 expression in Git1 Vec and OE neurons under control and OGD/R treatment (reperfusion 4 h) (n = 3). J, K Representative immunofluorescence images of ROS levels in AAV-Vec (Nrf2 vector adenovirus) and AAV-Nrf2 (Nrf2 knockdown adenovirus) Git1 OE-neurons and quantification of mean fluorescence intensity of ROS (scale bar = 50 μm, n = 6). *P < 0.05; **P < 0.01; ***P < 0.001.
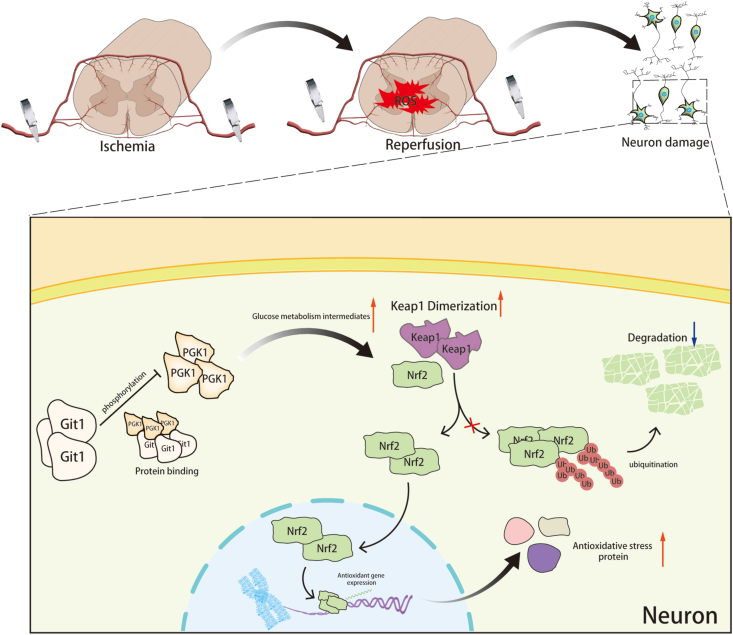
Fig. 9.
Git1-PGK1 interaction results in self-protection against spinal cord ischemia-reperfusion injury by modulating KEAP1–Nrf2 signaling. Binding of Git1 to PGK1 restricts PGK1 phosphorylation leading to increased glucose metabolism intermediates, ultimately promoting dimerization of Keap1 and reducing ubiquitination of Nrf2, leading to better antioxidant capacity of neurons and better functional recovery after SCIRI.
4. Discussion
We first found that Git1 has a protective role in SCIRI. Deletion of Git1 inhibits functional recovery in mice after SCIRI. We also found that Git1 interacts with PGK1 to regulate glucose metabolites in neurons. Furthermore, the role of Git1 in glucose metabolism affects oxidative stress in neurons, which has not been reported in previous studies. The interaction of Git1 with PGK1 reduces phosphorylation of S203 in PGK1, resulting in the accumulation of upstream glucose metabolites. Such metabolites can upregulate the dimerization of Keap1, thereby reducing ubiquitination and degradation of Nrf2, and increasing expression and nuclear entry of Nrf2, enhancing the antioxidant capacity of neurons. Git1 couples glucose metabolism to oxidative stress in neurons. We innovatively demonstrate the molecular mechanism of Git1 resistance to ROS in SCIRI. Our findings suggest that Git1 represents a compelling therapeutic target for ameliorating SCIRI, highlighting its potential as a promising avenue for treatment.
Git1 (G‐protein‐coupled receptor (GPCR) kinase 2‐interacting protein‐1) is a shuttle protein with multiple biological functions. Different domains of Git1 bind different proteins and regulate their activity [37,38]. Within the central nervous system, Git1 exerts a beneficial function. Previous studies have suggested that Git1 is a postsynaptic structural protein that promotes spine maturation. Git1 also plays a regulatory role in neuronal metabolism and can regulate the excitability of postsynaptic neurons [44,46]. The expression of Git1 is intricately tied to neuronal aging, and a multitude of neurodegenerative disorders have been linked to Git1 as well [48,79,80].
Previous studies have shown that Git1 is mainly expressed in neurons in the spinal cord [81], consistent with our immunofluorescence results. We found that loss of Git1 aggravated neuronal apoptosis and impaired functional recovery after SCIRI. SCIRI is a common and important type of secondary spinal cord injury, and its damage mechanism is mainly due to an imbalance of the homeostasis of oxidative stress in the spinal cord microenvironment, resulting in severe impairment of sensory and motor functions [82]. Therefore, studying the molecular mechanisms of oxidative stress in SCIRI is important for clinically controllable and treatable spinal cord injury (SCI) [4,83]. Considering the protective role of Git1 in SCIRI, immunofluorescence staining of spinal cord sections showed that Git1-knockout spinal cord mice had a more severe oxidative stress state after SCIRI. In vitro, we also found that knockdown of Git1 resulted in excessive production of ROS, mitochondrial superoxide, and lipid peroxides in neurons after OGD/R. Git1 is tightly linked to oxidative stress in neurons. Our immunofluorescence staining and Western blot results indicated that loss of Git1 can lead to severe ROS-related endoplasmic reticulum stress and DNA damage after SCIRI or OGD/R. These results further elucidate the molecular mechanism by which Git1 protects neurons in SCIRI.
To further analyze the mechanism by which Git1 regulates oxidative stress in neurons, we identified a new protein, PGK1, which interacts with Git1 in neurons and HEK293T cells by immunoprecipitation and IP coupled with LC-MS/MS. PGK1 is an important enzyme in glucose metabolism and plays a role in regulating energy homeostasis [31,32,84]. Intracellular glucose metabolism represents a key metabolic pathway responsible for furnishing cellular energy [24]. In addition, the state of glucose metabolism in cells can affect oxidative stress and immune function [23,26]. Intracellular glucose metabolites are closely related to activation/inhibition of signaling pathways and epigenetic and post-transcriptional regulation of genes [28,29,85]. Therefore, the binding of PGK1 and Git1 may be an important regulatory mechanism of cellular metabolism. Immunoblotting indicated that Git1 affects phosphorylation of S203 of PGK1. Quantitative metabolomic profiling showed that after Git1 knockdown, metabolite levels upstream of PGK1 decreased, mainly including 1,3- and total-bisphosphoglycerate (BPG), glyceraldehyde-3-phosphate (GAP), and methylglyoxal. These metabolites can affect the dimerization of Keap1, resulting in altered levels of Nrf2 ubiquitination. Immunoblotting showed that reduced levels of metabolites following Git1 knockdown resulted in reduced Keap1 dimerization and increased Nrf2 ubiquitination. This indicates that Git1 regulates the Nrf2 pathway by affecting glucose metabolites and exerts an antioxidant effect. Clinically, there is a certain correlation between elevated blood glucose and prognosis in patients with spinal cord injury [[86], [87], [88]]. In addition, an increasing number of studies have shown that alterations in glycolysis levels influence the prognosis of many neurological diseases [25,89,90]. In this study, it was found that Git1 coupled glucose metabolism and oxidative stress in neurons. This provides evidence for the role of Git1 in neuronal antioxidant activity, and also reveals the possible mechanism of Git1 in neurodegenerative diseases.
We further injected AAV into the spinal canal to specifically overexpress Git1 in spinal cord neurons. From analysis of motor function experiments, we clearly showed that overexpression of Git1 notably leads to a marked improvement in motor function recovery in mice following SCIRI. In addition, both in vivo and in vitro experiments showed that overexpression of Git1 can inhibit the production of ROS and other peroxides after I/R or OGD/R injury, and reduce the oxidative stress damage of neurons. Collectively, these findings implicate Git1 as a crucial mediator of oxidative stress. Immunoblot analysis of neurons overexpressing Git1 showed that overexpression of Git1 in neurons enhanced Nrf2 signaling. These results indicate that the neuroprotective function and antioxidant capacity after Git1 overexpression are closely related to the further activation of Nrf2 pathway. Git1 plays an important role in the regulation of Nrf2 signaling pathway and is an important target for the regulation of cellular antioxidation. The close molecular link between Git1 and Nrf2 may be an important target for clinical treatment of SCIRI. Neurons with Git1 overexpression were able to inhibit the antioxidant function of Git1 after Nrf2 knockdown. This further reveals the antioxidant mechanism of Git1 in neurons. The regulation of intracellular oxidative stress levels is multi-mechanism. Although this study elucidates the mechanism by which Git1 couples glucose metabolism to regulate oxidative stress, the mechanism of oxidative stress in neurons may be multifactorial, and further exploration is warranted to investigate other possible mechanisms.
In summary, Git1 plays an important role in neurons after SCIRI. Oxidative stress is an important injury mechanism in SCIRI and an important target for therapy. The interaction of Git1 and PGK1 is a key molecular event that triggers changes in glucose metabolism in neurons, regulates Keap1 dimerization and Nrf2 ubiquitination, and affects intracellular oxidative stress. Comprehending the molecular underpinnings and signaling pathways involved in Git1-associated perturbations in metabolism may pave the way for novel diagnostic and therapeutic modalities aimed at modulating the survival and demise of neuronal cells post-SCIRI. Our results underscore the critical role of Git1-mediated PGK1 phosphorylation and oxidative stress modulation in neuronal cells, highlighting the potential of Git1 targeting as a viable therapeutic avenue for SCIRI. Further investigations are warranted to engineer Git1-specific recombinant proteins for managing SCIRI, presenting a promising avenue for future therapeutic development.
Funding
The authors acknowledge financial support from the National Natural Science Foundation of China, China (Grant Nos. 81902211, 82030069, 81772351, and 8151001184), the Jiangsu Natural Science Foundation, China (Grant Nos. BK20191061), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant nos. JX10213715) and the Jiangsu Committee of Science and Technology–Social Development Plan, China (Grant Nos. BE2017755).
Ethics approval and consent to participate
All animal procedures were performed under the guidelines of the institutional review board and the ethics committee of Nanjing Medical University.
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102682.
Contributor Information
Jian Chen, Email: cbccj@sina.com.
Shujie Zhao, Email: zhaoshujie@njmu.edu.cn.
Guoyong Yin, Email: guoyong_yin@sina.com.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
Fig. S5.
Fig. S6.
Data availability
Data will be made available on request.
References
- 1.Smith P.D., Puskas F., Meng X., Lee J.H., Cleveland J.C., Jr., Weyant M.J., et al. The evolution of chemokine release supports a bimodal mechanism of spinal cord ischemia and reperfusion injury. Circulation. 2012;126:S110–S117. doi: 10.1161/CIRCULATIONAHA.111.080275. [DOI] [PubMed] [Google Scholar]
- 2.Bell M.T., Puskas F., Agoston V.A., Cleveland J.C., Jr., Freeman K.A., Gamboni F., et al. Toll-like receptor 4-dependent microglial activation mediates spinal cord ischemia-reperfusion injury. Circulation. 2013;128:S152–S156. doi: 10.1161/CIRCULATIONAHA.112.000024. [DOI] [PubMed] [Google Scholar]
- 3.Gu C., Li L., Huang Y., Qian D., Liu W., Zhang C., et al. Salidroside ameliorates mitochondria-dependent neuronal apoptosis after spinal cord ischemia-reperfusion injury partially through inhibiting oxidative stress and promoting mitophagy. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/3549704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wynn M.M., Acher C.W. A modern theory of spinal cord ischemia/injury in thoracoabdominal aortic surgery and its implications for prevention of paralysis. J. Cardiothorac. Vasc. Anesth. 2014;28:1088–1099. doi: 10.1053/j.jvca.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Huang H., Young W., Skaper S., Chen L., Moviglia G., Saberi H., et al. Clinical neurorestorative therapeutic guidelines for spinal cord injury (IANR/CANR version 2019) J Orthop Translat. 2020;20:14–24. doi: 10.1016/j.jot.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron E.G., Goldberg J.L. NEUROREGENERATION. Promoting CNS repair. Science. 2016;353:30–31. doi: 10.1126/science.aag3327. [DOI] [PubMed] [Google Scholar]
- 7.Cameron E.G., Kapiloff M.S. Intracellular compartmentation of cAMP promotes neuroprotection and regeneration of CNS neurons. Neural Regen Res. 2017;12:201–202. doi: 10.4103/1673-5374.200797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen J.B. Human spinal motor control. Annu. Rev. Neurosci. 2016;39:81–101. doi: 10.1146/annurev-neuro-070815-013913. [DOI] [PubMed] [Google Scholar]
- 9.Abraira V.E., Ginty D.D. The sensory neurons of touch. Neuron. 2013;79:618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Zhou W., Fan J., Ren Y., Yin G. G-protein-coupled receptor kinase interactor-1 serine 419 accelerates premature synapse formation in cortical neurons by interacting with Ca(2+)/calmodulin-dependent protein kinase IIβ. Brain Res. Bull. 2013;95:70–77. doi: 10.1016/j.brainresbull.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Fan J., Long H., Li Y., Liu Y., Zhou W., Li Q., et al. Edaravone protects against glutamate-induced PERK/EIF2α/ATF4 integrated stress response and activation of caspase-12. Brain Res. 2013;1519:1–8. doi: 10.1016/j.brainres.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 12.Fan J., Xu G., Nagel D.J., Hua Z., Zhang N., Yin G. A model of ischemia and reperfusion increases JNK activity, inhibits the association of BAD and 14-3-3, and induces apoptosis of rabbit spinal neurocytes. Neurosci. Lett. 2010;473:196–201. doi: 10.1016/j.neulet.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 13.Yang C., Ren Y., Liu F., Cai W., Zhang N., Nagel D.J., et al. Ischemic preconditioning suppresses apoptosis of rabbit spinal neurocytes by inhibiting ASK1-14-3-3 dissociation. Neurosci. Lett. 2008;441:267–271. doi: 10.1016/j.neulet.2008.06.037. [DOI] [PubMed] [Google Scholar]
- 14.Latimer H.R., Veal E.A. Peroxiredoxins in regulation of MAPK signalling pathways; sensors and barriers to signal transduction. Mol. Cell. 2016;39:40–45. doi: 10.14348/molcells.2016.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarkovic K. 4-hydroxynonenal and neurodegenerative diseases. Mol. Aspect. Med. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 16.Al Mamun A., Wu Y., Monalisa I., Jia C., Zhou K., Munir F., et al. Role of pyroptosis in spinal cord injury and its therapeutic implications. J. Adv. Res. 2021;28:97–109. doi: 10.1016/j.jare.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J., Ritzel R.M., Wu J. Functions and mechanisms of the voltage-gated proton channel Hv1 in brain and spinal cord injury. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.662971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Zhang B., Yao W., Zhang C., Wan L., Zhang Y. APC-Cdh1 regulates neuronal apoptosis through modulating glycolysis and pentose-phosphate pathway after oxygen-glucose deprivation and reperfusion. Cell. Mol. Neurobiol. 2019;39:123–135. doi: 10.1007/s10571-018-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong Y., Fan J., Ji C., Wang Z., Ge X., Wang J., et al. USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1. Cell Death Differ. 2021;29:1164–1175. doi: 10.1038/s41418-021-00907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B., Jiang W., Liu H., Wang J., Zheng K., Cui P., et al. Upregulation of neuronal PGC-1α ameliorates cognitive impairment induced by chronic cerebral hypoperfusion. Theranostics. 2020;10:2832–2848. doi: 10.7150/thno.37119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z., Min L., Chen H., Deng W., Tan M., Liu H., et al. Iron overload in the motor cortex induces neuronal ferroptosis following spinal cord injury. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto-Heredero G., Gómez de Las Heras M.M., Gabandé-Rodríguez E., Oller J., Mittelbrunn M. Glycolysis - a key player in the inflammatory response. FEBS J. 2020;287:3350–3369. doi: 10.1111/febs.15327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan V.P., Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11:963–964. doi: 10.1080/15548627.2015.1042195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang B.L. Glucose, glycolysis, and neurodegenerative diseases. J. Cell. Physiol. 2020;235:7653–7662. doi: 10.1002/jcp.29682. [DOI] [PubMed] [Google Scholar]
- 25.Rabbani N., Thornalley P.J. Hexokinase-2 glycolytic overload in diabetes and ischemia-reperfusion injury. Trends Endocrinol. Metabol. 2019;30:419–431. doi: 10.1016/j.tem.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 26.O' Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buck M.D., Sowell R.T., Kaech S.M., Pearce E.L. Metabolic instruction of immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D., Shao N.Y., Moonen J.R., Zhao Z., Shi M., Otsuki S., et al. ALDH1A3 coordinates metabolism with gene regulation in pulmonary arterial hypertension. Circulation. 2021;143:2074–2090. doi: 10.1161/CIRCULATIONAHA.120.048845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diskin C., Ryan T., O' Neill L. Modification of proteins by metabolites in immunity. Immunity. 2021;54:19–31. doi: 10.1016/j.immuni.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Shakespear M.R., Iyer A., Cheng C.Y., Das Gupta K., Singhal A., Fairlie D.P., et al. Lysine deacetylases and regulated glycolysis in macrophages. Trends Immunol. 2018;39:473–488. doi: 10.1016/j.it.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., Yu G., Chu H., Wang X., Xiong L., Cai G., et al. Macrophage-associated PGK1 phosphorylation promotes aerobic glycolysis and tumorigenesis. Mol. Cell. 2018;71:201–215.e7. doi: 10.1016/j.molcel.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Qian X., Li X., Shi Z., Xia Y., Cai Q., Xu D., et al. PTEN suppresses glycolysis by dephosphorylating and inhibiting autophosphorylated PGK1. Mol. Cell. 2019;76:516–527.e7. doi: 10.1016/j.molcel.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Bollong M.J., Lee G., Coukos J.S., Yun H., Zambaldo C., Chang J.W., et al. A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature. 2018;562:600–604. doi: 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang J., Zhang X.Y., Zhen Y.F., Chen C., Tan H., Hu J., et al. PGK1 depletion activates Nrf2 signaling to protect human osteoblasts from dexamethasone. Cell Death Dis. 2019;10:888. doi: 10.1038/s41419-019-2112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Q., Maksimovic I., Upad A., David Y. Non-enzymatic covalent modifications: a new link between metabolism and epigenetics. Protein Cell. 2020;11:401–416. doi: 10.1007/s13238-020-00722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Premont R.T., Claing A., Vitale N., Freeman J.L., Pitcher J.A., Patton W.A., et al. beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claing A., Perry S.J., Achiriloaie M., Walker J.K., Albanesi J.P., Lefkowitz R.J., et al. Multiple endocytic pathways of G protein-coupled receptors delineated by GIT1 sensitivity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1119–1124. doi: 10.1073/pnas.97.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W., Li X., Premont R.T. Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J. Cell Sci. 2016;129:1963–1974. doi: 10.1242/jcs.179465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoefen R.J., Berk B.C. The multifunctional GIT family of proteins. J. Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 41.Yin G., Zheng Q., Yan C., Berk B.C. GIT1 is a scaffold for ERK1/2 activation in focal adhesions. J. Biol. Chem. 2005;280:27705–27712. doi: 10.1074/jbc.M502271200. [DOI] [PubMed] [Google Scholar]
- 42.Yin G., Haendeler J., Yan C., Berk B.C. GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Mol. Cell Biol. 2004;24:875–885. doi: 10.1128/MCB.24.2.875-885.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang J., Xu X., Wang X., Majumder S., Wang J., Korshunov V.A., et al. G-protein-coupled receptor kinase interacting protein-1 mediates intima formation by regulating vascular smooth muscle proliferation, apoptosis, and migration. Arterioscler. Thromb. Vasc. Biol. 2013;33:999–1005. doi: 10.1161/ATVBAHA.112.300966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Webb D.J., Asmussen H., Niu S., Horwitz A.F. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiuza M., González-González I., Pérez-Otaño I. GluN3A expression restricts spine maturation via inhibition of GIT1/Rac1 signaling. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20807–20812. doi: 10.1073/pnas.1312211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H., Webb D.J., Asmussen H., Horwitz A.F. Synapse formation is regulated by the signaling adaptor GIT1. J. Cell Biol. 2003;161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M.J., Biag J., Fass D.M., Lewis M.C., Zhang Q., Fleishman M., et al. Functional analysis of rare variants found in schizophrenia implicates a critical role for GIT1-PAK3 signaling in neuroplasticity. Mol. Psychiatr. 2017;22:417–429. doi: 10.1038/mp.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Gastel J., Boddaert J., Jushaj A., Premont R.T., Luttrell L.M., Janssens J., et al. GIT2-A keystone in ageing and age-related disease. Ageing Res. Rev. 2018;43:46–63. doi: 10.1016/j.arr.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Yin G., Sheu T.J., Menon P., Pang J., Ho H.C., Shi S., et al. Impaired angiogenesis during fracture healing in GPCR kinase 2 interacting protein-1 (GIT1) knock out mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galvan M.D., Luchetti S., Burgos A.M., et al. Deficiency in complement C1q improves histological and functional locomotor outcome after spinal cord injury. J. Neurosci. 2008;28:13876–13888. doi: 10.1523/JNEUROSCI.2823-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirshblum S., Snider B., Eren F., et al. Characterizing natural recovery after traumatic spinal cord injury. J. Neurotrauma. 2021;38:1267–1284. doi: 10.1089/neu.2020.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basso D.M., Fisher L.C., Anderson A.J., Jakeman L.B., McTigue D.M., Popovich P.G. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J. Neurotrauma. 2006;23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- 53.Hamm R.J., Pike B.R., O' Dell D.M., Lyeth B.G., Jenkins L.W. The rotarod test: an evaluation of its effectiveness in assessing motor deficits following traumatic brain injury. J. Neurotrauma. 1994;11:187–196. doi: 10.1089/neu.1994.11.187. [DOI] [PubMed] [Google Scholar]
- 54.Smith R.R., Burke D.A., Baldini A.D., Shum-Siu A., Baltzley R., Bunger M., et al. The Louisville Swim Scale: a novel assessment of hindlimb function following spinal cord injury in adult rats. J. Neurotrauma. 2006;23:1654–1670. doi: 10.1089/neu.2006.23.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Q., Zhang G., Zheng Y., Yang Y., Chen C., Xia J., et al. SLC27A5 deficiency activates NRF2/TXNRD1 pathway by increased lipid peroxidation in HCC. Cell Death Differ. 2020;27:1086–1104. doi: 10.1038/s41418-019-0399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee D.H., Park J.S., Lee Y.S., Han J., Lee D.K., Kwon S.W., et al. SQSTM1/p62 activates NFE2L2/NRF2 via ULK1-mediated autophagic KEAP1 degradation and protects mouse liver from lipotoxicity. Autophagy. 2020;16:1949–1973. doi: 10.1080/15548627.2020.1712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu D., Xia Y., Li S., et al. Iso-seco-tanapartholide activates Nrf2 signaling pathway through Keap1 modification and oligomerization to exert anti-inflammatory effects. Free Radic. Biol. Med. 2022;178:398–412. doi: 10.1016/j.freeradbiomed.2021.12.259. [DOI] [PubMed] [Google Scholar]
- 58.Zhang M., Wang L., Zhao X., et al. TRAF-interacting protein (TRIP) negatively regulates IFN-β production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J. Exp. Med. 2012;209:1703–1711. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi Q., Jin X., Zhang P., et al. SPOP mutations promote p62/SQSTM1-dependent autophagy and Nrf2 activation in prostate cancer. Cell Death Differ. 2022;29:1228–1239. doi: 10.1038/s41418-021-00913-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopitz-Otsoa F., Rodriguez-Suarez E., Aillet F., Casado-Vela J., Lang V., Matthiesen R., et al. Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs) J. Proteonomics. 2012;75:2998–3014. doi: 10.1016/j.jprot.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Lang V., Pallara C., Zabala A., et al. Tetramerization-defects of p53 result in aberrant ubiquitylation and transcriptional activity. Mol Oncol. 2014;8:1026–1042. doi: 10.1016/j.molonc.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jena K.K., Kolapalli S.P., Mehto S., et al. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng C., Zhan J., Chen D., et al. The deubiquitinase USP11 regulates cell proliferation and ferroptotic cell death via stabilization of NRF2 USP11 deubiquitinates and stabilizes NRF2. Oncogene. 2021;40:1706–1720. doi: 10.1038/s41388-021-01660-5. [DOI] [PubMed] [Google Scholar]
- 65.Segura I., Essmann C.L., Weinges S., Acker-Palmer A. Grb4 and GIT1 transduce ephrinB reverse signals modulating spine morphogenesis and synapse formation. Nat. Neurosci. 2007;10:301–310. doi: 10.1038/nn1858. [DOI] [PubMed] [Google Scholar]
- 66.Tu B.P., Weissman J.S. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tse G., Yan B.P., Chan Y.W., Tian X.Y., Huang Y. Reactive oxygen species, endoplasmic reticulum stress and mitochondrial dysfunction: the link with cardiac arrhythmogenesis. Front. Physiol. 2016;7:313. doi: 10.3389/fphys.2016.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguiar P.H., Furtado C., Repolês B.M., Ribeiro G.A., Mendes I.C., Peloso E.F., et al. Oxidative stress and DNA lesions: the role of 8-oxoguanine lesions in Trypanosoma cruzi cell viability. PLoS Neglected Trop. Dis. 2013;7:e2279. doi: 10.1371/journal.pntd.0002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H., Wu F., Kong X., Yang J., Chen H., Deng L., et al. Nerve growth factor improves functional recovery by inhibiting endoplasmic reticulum stress-induced neuronal apoptosis in rats with spinal cord injury. J. Transl. Med. 2014;12:130. doi: 10.1186/1479-5876-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sproul A.A., Xu Z., Wilhelm M., Gire S., Greene L.A. Cbl negatively regulates JNK activation and cell death. Cell Res. 2009;19:950–961. doi: 10.1038/cr.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X., Zhao C., Li X., Wang T., Li Y., Cao C., et al. Terazosin activates Pgk1 and Hsp90 to promote stress resistance. Nat. Chem. Biol. 2015;11:19–25. doi: 10.1038/nchembio.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gondeau C., Chaloin L., Lallemand P., Roy B., Périgaud C., Barman T., et al. Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase. Nucleic Acids Res. 2008;36:3620–3629. doi: 10.1093/nar/gkn212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szilágyi A.N., Ghosh M., Garman E., Vas M. A 1.8 A resolution structure of pig muscle 3-phosphoglycerate kinase with bound MgADP and 3-phosphoglycerate in open conformation: new insight into the role of the nucleotide in domain closure. J. Mol. Biol. 2001;306:499–511. doi: 10.1006/jmbi.2000.4294. [DOI] [PubMed] [Google Scholar]
- 74.Hipkiss A.R. Energy metabolism and ageing regulation: metabolically driven deamidation of triosephosphate isomerase may contribute to proteostatic dysfunction. Ageing Res. Rev. 2011;10:498–502. doi: 10.1016/j.arr.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 75.de Bari L., Scirè A., Minnelli C., Cianfruglia L., Kalapos M.P., Armeni T. Interplay among oxidative stress, methylglyoxal pathway and S-glutathionylation. Antioxidants. 2020;10 doi: 10.3390/antiox10010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 77.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020:40. doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bellezza I., Giambanco I., Minelli A., Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:721–733. doi: 10.1016/j.bbamcr.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 79.Blanco-Luquin I., Acha B., Urdánoz-Casado A., Sánchez-Ruiz De Gordoa J., Vicuña-Urriza J., Roldán M., et al. Early epigenetic changes of Alzheimer's disease in the human hippocampus. Epigenetics. 2020;15:1083–1092. doi: 10.1080/15592294.2020.1748917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 81.Russ D.E., Cross R., Li L., Koch S.C., Matson K., Yadav A., et al. A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat. Commun. 2021;12:5722. doi: 10.1038/s41467-021-25125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Griffin J.M., Bradke F. Therapeutic repair for spinal cord injury: combinatory approaches to address a multifaceted problem. EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.201911505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venkatesh K., Ghosh S.K., Mullick M., Manivasagam G., Sen D. Spinal cord injury: pathophysiology, treatment strategies, associated challenges, and future implications. Cell Tissue Res. 2019;377:125–151. doi: 10.1007/s00441-019-03039-1. [DOI] [PubMed] [Google Scholar]
- 84.Boyd P.J., Tu W.Y., Shorrock H.K., Groen E., Carter R.N., Powis R.A., et al. Bioenergetic status modulates motor neuron vulnerability and pathogenesis in a zebrafish model of spinal muscular atrophy. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phan A.T., Goldrath A.W., Glass C.K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46:714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phang I., Zoumprouli A., Papadopoulos M.C., Saadoun S. Microdialysis to optimize cord perfusion and drug delivery in spinal cord injury. Ann. Neurol. 2016;80:522–531. doi: 10.1002/ana.24750. [DOI] [PubMed] [Google Scholar]
- 87.Sala F., Menna G., Bricolo A., Young W. Role of glycemia in acute spinal cord injury. Data from a rat experimental model and clinical experience. Ann. N. Y. Acad. Sci. 1999;890:133–154. doi: 10.1111/j.1749-6632.1999.tb07989.x. [DOI] [PubMed] [Google Scholar]
- 88.Kobayakawa K., Kumamaru H., Saiwai H., Kubota K., Ohkawa Y., Kishimoto J., et al. Acute hyperglycemia impairs functional improvement after spinal cord injury in mice and humans. Sci. Transl. Med. 2014;6:256ra137. doi: 10.1126/scitranslmed.3009430. [DOI] [PubMed] [Google Scholar]
- 89.Sun M., Li M., Huang Q., Han F., Gu J.H., Xie J., et al. Ischemia/reperfusion-induced upregulation of TIGAR in brain is mediated by SP1 and modulated by ROS and hormones involved in glucose metabolism. Neurochem. Int. 2015;80:99–109. doi: 10.1016/j.neuint.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 90.Cai R., Zhang Y., Simmering J.E., Schultz J.L., Li Y., Fernandez-Carasa I., et al. Enhancing glycolysis attenuates Parkinson's disease progression in models and clinical databases. J. Clin. Invest. 2019;129:4539–4549. doi: 10.1172/JCI129987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.