Abstract
Polygenic risk scores aggregate an individual’s burden of risk alleles to estimate the overall genetic risk for a specific trait or disease. Polygenic risk scores derived from genome-wide association studies of European populations perform poorly for other ancestral groups. Given the potential for future clinical utility, underperformance of polygenic risk scores in South Asian populations has the potential to reinforce health inequalities. To determine whether European-derived polygenic risk scores underperform at multiple sclerosis prediction in a South Asian-ancestry population compared with a European-ancestry cohort, we used data from two longitudinal genetic cohort studies: Genes & Health (2015–present), a study of ∼50 000 British–Bangladeshi and British–Pakistani individuals, and UK Biobank (2006–present), which is comprised of ∼500 000 predominantly White British individuals. We compared individuals with and without multiple sclerosis in both studies (Genes & Health: NCases = 42, NControl = 40 490; UK Biobank: NCases = 2091, NControl = 374 866). Polygenic risk scores were calculated using clumping and thresholding with risk allele effect sizes obtained from the largest multiple sclerosis genome-wide association study to date. Scores were calculated with and without the major histocompatibility complex region, the most influential locus in determining multiple sclerosis risk. Polygenic risk score prediction was evaluated using Nagelkerke’s pseudo-R2 metric adjusted for case ascertainment, age, sex and the first four genetic principal components. We found that, as expected, European-derived polygenic risk scores perform poorly in the Genes & Health cohort, explaining 1.1% (including the major histocompatibility complex) and 1.5% (excluding the major histocompatibility complex) of disease risk. In contrast, multiple sclerosis polygenic risk scores explained 4.8% (including the major histocompatibility complex) and 2.8% (excluding the major histocompatibility complex) of disease risk in European-ancestry UK Biobank participants. These findings suggest that polygenic risk score prediction of multiple sclerosis based on European genome-wide association study results is less accurate in a South Asian population. Genetic studies of ancestrally diverse populations are required to ensure that polygenic risk scores can be useful across ancestries.
Keywords: multiple sclerosis, genetics, ethnicity
Breedon et al. report that polygenic risk scores for multiple sclerosis derived from the European-ancestry genetic studies underperform in a South Asian-ancestry population. Their results stress the need for ancestrally diverse genetic studies of multiple sclerosis risk to ensure the generalizability of polygenic risk scores.
Graphical Abstract
Graphical abstract.
Introduction
An individual’s risk of developing multiple sclerosis is influenced by common variation across the genome.1,2 Multiple sclerosis is a typical complex disease in which the genetic contribution to risk is governed by a large number of susceptibility alleles with individually weak effects. Variation within the major histocompatibility complex (MHC) has the greatest impact on individual risk [odds ratio (OR) associated with DRB1*1501 3.1 and 6.2 for heterozygous and homozygous carriage, respectively].2,3 Genome-wide association studies (GWAS) of multiple sclerosis susceptibility have demonstrated at least 200 risk alleles outside the MHC locus, each with a small incremental effect (OR per allele ≤1.3).2 There is no convincing evidence for monogenic forms of multiple sclerosis in the general population.4
Predicting who is likely to develop multiple sclerosis in the future has potential utility for research studies. Accurate disease prediction could facilitate the design of trials for candidate preventive strategies, such as an Epstein–Barr virus (EBV) vaccine or a vitamin D supplementation trial. As multiple sclerosis is a relatively rare disease, such trials will only have the power to demonstrate a risk reduction if the trial population is sufficiently enriched with people at high risk of multiple sclerosis, effectively increasing the proportion likely to develop the disease.5 Furthermore, identifying those at highest risk of disease may allow treatment during the ‘prodromal’ period, prior to overt clinical manifestations.6
Polygenic risk scores (PRS) summarize an individual’s cumulative burden of genetic risk alleles to approximate their overall disease risk. Most PRS are calculated by weighting the individual’s burden of risk alleles by the estimated effect of each allele on risk—these estimates are usually obtained from GWAS. In two large cohort studies—UK Biobank (UKB) and University of California San Francisco (UCSF) Expression, Proteomics, Imaging, Clinical (EPIC)—PRS have been empirically demonstrated to distinguish multiple sclerosis cases from controls at a population level.7-9
PRS perform poorly in non-European ancestral groups, a phenomenon largely due to differences in linkage disequilibrium (LD) and allele frequencies between populations.10-12 It is now clear that multiple sclerosis affects individuals of all ethnic backgrounds and that, broadly speaking, the genetic architecture of multiple sclerosis susceptibility overlaps considerably between ancestral groups.13-22 We therefore sought to evaluate the performance of multiple sclerosis PRS in ∼50 000 individuals of South Asian ancestry from the Genes & Health (G&H) cohort to determine the applicability of PRS in this population.
Methods
Cohort description and phenotype definition
We used data from the July 2021 data freeze of G&H, a longitudinal genetic cohort study of ∼50 000 British–Bangladeshi and British–Pakistani individuals.23 Genotypes and clinical data for 44 396 participants were included in this study. After exclusion of principal component analysis (PCA) outliers (n = 206), samples with >10% missing genotypes (n = 3452) and samples without corresponding phenotype/covariate data (n = 206), 40 532 individuals were retained for analysis. Of this final cohort, 42 individuals had a coded diagnosis of multiple sclerosis and 40 490 did not (Fig. 1). Cases were defined using linked electronic health records from primary care, hospital episode statistics (HES) data and local hospital recording of admissions and outpatient encounters. Healthcare data were harmonized across International Classification of Diseases Revision 10 (ICD10) codes, Systematized Nomenclature of Medicine (SNOMED) description IDs and SNOMED concept IDs into a consistent three-digit ICD10 coding system. Individuals with at least one multiple sclerosis diagnostic code (ICD10 code G35) in their records were considered cases, and those without were considered non-multiple sclerosis controls. Details of phenotype definitions can be found in Supplementary File 1. An online version of this file is continuously updated and can be viewed here.
Figure 1.
Flow diagram of individual quality control in G&H. From an initial 44 396 individuals with genotype data, 40 532 individuals were retained for analysis comprising 42 multiple sclerosis cases and 40 490 controls
Genotype data and target data quality control
Genomic DNA was obtained from saliva samples using Oragene-600 kits. Individuals were genotyped using the Illumina Global Screening Array chip (version 3) with extra multi-disease content.
Genotypes were imputed using the multi-ancestral Trans-Omics for Precision Medicine Program (TOPMed)-R2 panel. Variant quality control was performed to remove low-quality variants using the following filters: call rate >90%, imputation quality INFO >0.7, minor allele frequency (MAF) >0.01, no deviation from Hardy–Weinberg equilibrium (HWE) at P < 1−10 and genotype missingness <10%. Sex chromosomes and mitochondrial variants were excluded. Individual quality control was performed to remove PCA outliers and individuals with high missingness. Full details of genotype data quality control can be found in the Supplementary Methods.
Polygenic risk score calculation
PRS were derived using PRSice-2, which uses a clumping-and-thresholding approach.9 We used external weights from the discovery-stage International Multiple Sclerosis Genetics Consortium (IMSGC) 2019 GWAS meta-analysis (cases: 14 802, controls: 26 703).2 We used LD-based clumping to determine independent signals using European samples from the 1000 Genomes Project (n = 503).1 We harmonized single nucleotide polymorphisms (SNPs) between the multiple sclerosis GWAS and the G&H cohort imputed genotype data. LiftOver was used to convert the coordinates to hg38. We excluded SNPs with incompatible alleles and restricted to non-palindromic, biallelic SNPs.
We generated 224 different (but overlapping) PRS by varying the clumping R2 and P-value threshold for variant selection. Many of the SNPs incorporated in these PRS overlap, and so while these PRS are distinct, they are correlated with each other. Specifically, we used clumping R2 thresholds of 0.001, 0.01, 0.05, 0.1, 0.2, 0.4 and 0.6 and P-value thresholds of 1 × 10−8, 5 × 10−8, 1 × 10−7, 5 × 10−7, 1 × 10−6, 5 × 10−6, 1 × 10−5, 5 × 10−5, 1 × 10−4, 5 × 10−4, 1 × 10−3, 5 × 10−3, 0.01, 0.05, 0.1 and 0.5. For each combination of threshold P-value and clumping R2 value, we derived PRS both including and excluding the MHC region (chromosome 6: 25 000 000–35 000 000 in hg38). In order to isolate the contribution of the MHC, we generated a further 112 scores including just the MHC region. Overall, we generated 336 PRS (112 with MHC, 112 without and 112 just MHC). A null model was generated using the covariates alone [age, sex and genetic principal components (PCs) 1–4]. PRS were calculated using the sum of the weighted allelic burden for each individual, i.e. for the jth SNP and the ith individual, where G is the genotype dosage and β is the effect size of the SNP:
| (1) |
Missing genotypes were centred so as to contribute a mean of 0 to the overall score. Discriminative performance was evaluated using Nagelkerke’s pseudo-R2 metric adjusted for case ascertainment (assuming a population prevalence of 0.002) and corresponding P-values, with adjustment for age, sex and the first four genetic principal components. The PRS with the lowest model fit P-value was considered the ‘optimal score’, and the statistical significance of this score was evaluated using the P-value for the model fit.24
Logistic regression was used to determine the OR of multiple sclerosis in each quartile of PRS (compared to the lowest quartile as reference). In all regression models, age, sex and genetic PCs 1–4 were included as covariates. To determine the area under the curve (AUC) discrimination statistics of each PRS and to determine the calibration, we used the fitted probabilities of the logistic models including the PRS as a covariate. We compared the performance of each PRS with null models comprising only age, sex and PCs 1–4.
UK Biobank replication
To compare the performance of the multiple sclerosis PRS across ancestries, we repeated the analysis using UKB, a longitudinal cohort study of predominantly White British adults aged >40.25 We used largely similar methods for SNP and individual quality control. We restricted the analysis to individuals of genetically European ancestry (UKB field ID 22006) determined using principal components. We excluded one of each pair of highly related individuals (kinship coefficient > 0.0884). We generated and tested a variety of PRS using the same methods as in G&H.
To formally compare PRS performance between the two cohorts controlling for sample size bias, we randomly sub-sampled the UKB cohort to have the same number of cases and controls in our G&H analyses (42 cases, 40 490 controls). For each iteration, we determined the optimal PRS and the estimated Nagelkerke’s pseudo-R2. We calculated an empirical P-value for the hypothesis that the liability explained by the PRS was lower in the South Asian-ancestry (SAS) cohort:
| (2) |
where Niter is the total number of iterations (1000) and NUKB < GH is the number of iterations in which the observed Nagelkerke’s pseudo-R2 in the UKB sub-sample was lower than the observed value in G&H. As a comparator, we also evaluated the performance of multiple sclerosis PRS in the whole cohort (without splitting into training and test sets), comprising 2091 multiple sclerosis cases and 374 866 controls. For analyses in UKB, we used the same clumping R2 values that were optimal in G&H.
Power calculations
We performed post hoc power calculations to determine our statistical power to detect a difference in multiple sclerosis PRS between cases and controls in G&H. To do so, we simulated a normally distributed PRS in 42 cases and 40 490 controls. We varied the difference in the mean of the case and control distributions from 0 to 3 standard deviations. We performed 1000 bootstrap iterations for each scenario and evaluated the power as the proportion of iterations yielding a Wald test P-value of <0.05. These simulations showed that, given this number of cases and controls, we would have 91% power to detect a difference of 0.5 standard deviations in the PRS. For context, in UKB participants of European ancestry, the difference in mean PRS between cases and controls is ∼0.7 standard deviations for the MHC PRS and 0.5 standard deviations for the non-MHC PRS.
Statistical analysis and computing
Analysis of G&H data was conducted within the dedicated Google Cloud Trusted Research Environment. Target data QC was performed using PLINK version >2.26 PRS were calculated using PRSice version 2.3.5.24 Statistical analysis was performed using R version 4.2.0. Analysis of UKB data was conducted using the Apocrita High Performance Cluster based at Queen Mary University of London.27
Ethical approval
This research was conducted under an approved application to use the G&H resource (‘Brain consortium’ application). G&H was approved by the London South East NRES Committee of the Health Research Authority (14/LO/1240). Replication in UKB was performed under approved application 43101. This research was undertaken under UKB’s existing ethical approval (REC approval 11/NW/0382; North West Multi-centre Research Ethics Committee).
Results
Following quality control, we analysed data from 40 532 individuals of South Asian ancestry in the G&H cohort, comprising 42 multiple sclerosis cases and 40 490 controls (Table 1). Demographics of included participants are shown in Table 1.
Table 1.
Genes & Health cohort characteristics
| Control (N = 40 490) | Case (N = 42) | |
|---|---|---|
| Sex, n (%) | ||
| Female | 22 493 (55.6) | 30 (71.4) |
| Male | 17 997 (44.4) | 12 (28.6) |
| Age at recruitment, mean (SD) | 41.2 (14.2) | 41.0 (0.8) |
| Genetic ancestry, n (%) | ||
| British–Bangladeshi | 22 900 (56.6) | 11 (26.2) |
| British–Pakistani | 17 590 (43.4) | 31 (73.8) |
PRS derived from European-ancestry (EUR) multiple sclerosis GWAS were associated with multiple sclerosis in the G&H cohort of British South Asian individuals (Nmultiple sclerosis = 42, Ncontrol = 40 490) (Fig. 2A and B). The optimal PRS containing the MHC region (PRSMHC) explained ∼1.1% of the liability to multiple sclerosis in this cohort (adjusted Nagelkerke’s pseudo-R2 0.011, P = 0.033, NSNP = 1356, clumping R2 0.05, threshold P-value 0.001). The optimal PRS excluding the MHC region (PRSNon-MHC) performed similarly, explaining ∼1.5% of the liability to multiple sclerosis (adjusted Nagelkerke’s pseudo-R2 0.015, P = 0.015, NSNP = 1965, clumping R2 0.4, threshold P-value 0.001). The difference in performance of the PRSNon-MHC and PRSMHC was not statistically significant (likelihood ratio P-value = 1). PRS using variants only lying within the MHC did not correlate with multiple sclerosis disease status (P = 0.19).
Figure 2.
Multiple sclerosis PRS performance in the G&H cohort of South Asian-ancestry individuals. (A and B) Density plots showing the distribution of PRS for PRS with (A) and without (B) the MHC locus in multiple sclerosis cases and controls. (C) Odds ratio quartile plots for individual PRS scores. ORs were calculated relative to the lower quartile. (D) Receiver operating characteristic (ROC) curves for the MHC PRS model, non-MHC model and the null model, with corresponding AUC scores. (E and F) Calibration plots showing the absolute multiple sclerosis disease probabilities (prevalence) for each PRS quartile versus mean fitted probabilities within each quartile from the PRS models. Plots shown for MHC PRS model, non-MHC PRS model, null model and the observed multiple sclerosis risk in each quartile. Odds ratios and AUC values are derived from multivariable logistic regression models
The predicted risk of multiple sclerosis based on PRS was reasonably well-calibrated to absolute risk (Fig. 2C). Individuals in the top 25% of PRSMHC were nominally more likely to have multiple sclerosis than those in the lowest 25% (OR 2.72, 95% CI 0.99–7.50), although our statistical confidence in this result is tempered by the small number of cases leading to wide confidence intervals which just cross the null. We observed a similar effect for the PRSNon-MHC (OR 1.96, 95% CI 0.74–5.24), again with wide confidence intervals crossing the null. Both the PRSMHC and PRSNon-MHC demonstrated reasonable discrimination between cases and controls at a population level (AUCMHC 0.70, AUCnon-MHC 0.71), but it is important to note that age, sex and genetic principal components alone account for much of this discriminative power (AUCnull 0.664) (Fig. 2D). In models without any covariates, the PRSMHC and PRSNon-MHC have weaker discriminative ability (AUCMHC 0.63, AUCnon-MHC 0.60) but still perform better than chance.
For the PRSMHC, in the lowest quartile, 5/10 133 participants had multiple sclerosis (0.05%), as opposed to 15/10 133 in the highest quartile (0.15%). For the PRSNon-MHC, 6/10 133 (0.06%) and 12/10 133 (0.12%) had multiple sclerosis in the lowest and highest quartiles, respectively (Fig. 2E and F).
In order to directly compare the performance of the PRS in this cohort with a European-ancestry cohort, we then applied the same methods to UKB (Fig. 3). Using the entire cohort of unrelated EUR-ancestry individuals in UKB (Nmultiple sclerosis = 2091, NControl = 374 866), both the PRSMHC and PRSNon-MHC performed better than in G&H, explaining 4.4% and 2.3% of liability, respectively (PRSMHC: adjusted Nagelkerke’s pseudo-R2 = 0.044, P = 2 × 10−211, NSNP = 235454, clumping R2 0.4, threshold P-value 0.5; PRSNon-MHC: adjusted Nagelkerke’s pseudo-R2 = 0.023, P = 1.9 × 10−104, NSNP = 42759, clumping R2 0.6, threshold P-value 0.05).
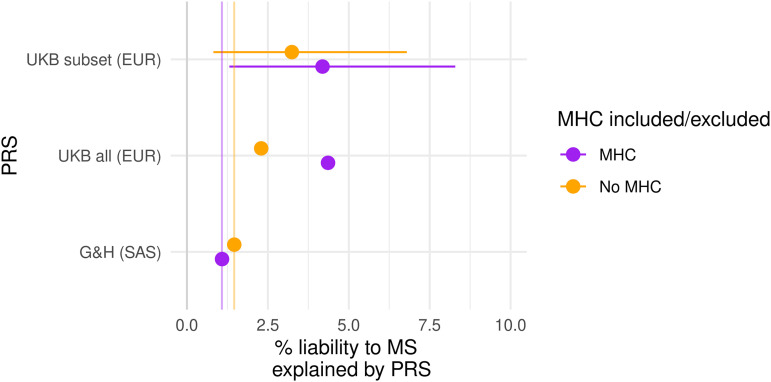
Figure 3.
Estimates of PRS performance in EUR UKB participants and SAS G&H participants. Each point represents the estimated liability explained by the optimal PRS, with 95% confidence intervals for the sub-samples of UKB. The vertical lines indicate the performance of each score in G&H. PRS containing the MHC are coloured in purple, and those without coloured in orange. Estimates reflect Nagelkerke’s pseudo-R2 statistic adjusted for disease prevalence, which is derived from multivariable logistic regression models. ‘UKB all’ refers to scores calculated in all EUR-ancestry UKB participants. To control for effects of sample size, we resampled subsets of UKB to have equivalent case and control numbers to G&H (42 cases, 40 490 controls). ‘UKB subset’ refers to estimates derived from 1000 replicates of this random sampling procedure, with empirical 95% confidence intervals
To mitigate the effects of sample size, we randomly sampled 1000 sets of 42 multiple sclerosis cases and 40 490 controls of European ancestry. We used the optimal clumping R2 thresholds derived from G&H (0.05 and 0.4 for the PRSMHC and PRSNon-MHC, respectively). For each iteration, we compared the liability explained in UKB with the observed values in G&H (1.1% for PRSMHC and 1.5% for PRSNon-MHC), thus obtaining empirical estimates for the sampling distribution and model fit in UKB.
Using this permutation-based approach, both the PRSMHC and PRSNon-MHC remained strongly associated with multiple sclerosis disease status. In the UKB population, the performance of the PRSMHC was substantially greater than that of the PRSNon-MHC, reflecting the large portion of heritability accounted for by this locus. The PRSMHC explained more liability to multiple sclerosis in European-ancestry UKB participants than in G&H (UKB adjusted R2 4.3%, 95% CI 1.5–8.5%; G&H adjusted R2 1.1%, P = 0.01). The difference in the performance of the PRSNon-MHC was less pronounced but also suggestive of weaker performance in the South Asian cohort (UKB adjusted R2 3.2%, 95% CI 0.9–6.9%; G&H adjusted R2 1.5%, P = 0.10), although the confidence intervals span the G&H estimate, and so we cannot reject the possibility that the performance of the PRSNon-MHC is similar in both cohorts.
Discussion
PRS derived from European GWAS perform poorly in non-European populations across a range of traits and disorders.10,12 We report evidence to suggest that this drop-off in PRS performance also applies to multiple sclerosis prediction in a large cohort of South Asian ancestry. We demonstrate that although the European-derived PRS performs relatively poorly in this setting, it does still have some predictive power, consistent with significant overlap in the genetic architecture of multiple sclerosis risk between populations.13-22
The lower predictive power of multiple sclerosis PRS we report in an ancestrally South Asian cohort is likely driven by differences in the minor allele frequency of variants and LD structures between European and South Asian populations, rather than due to differences in causal variants.28 If variants included in the PRS are not causal themselves but tag causal variants in Europeans, it does not follow that they will tag the causal variant in other populations, diminishing the accuracy of the score. Previous genetic analyses of multiple sclerosis risk in non-European populations—including small studies of South Asian populations—argue that, broadly speaking, the genetic architecture of multiple sclerosis risk between populations is highly correlated.14,16,17,29 Our finding that a European multiple sclerosis PRS has some accuracy in a South Asian cohort, but less so than in Europeans, is entirely consistent with this view.
It is notable that the inclusion of the MHC locus did not improve the PRS in the South Asian cohort. This result could be due to limited statistical power, different causal human leukocyte antigen (HLA) alleles and/or poor tagging of causal HLA alleles by the European GWAS variants. It is important to note that available data suggest that the major HLA risk alleles in Europeans have similar effects in South Asians, and so in our view, it is primarily differences in LD (in addition to the limited case numbers) that drive this unexpected result in the cohort, as well as the statistical imprecision of the effect estimates due to the small number of cases in G&H. Larger studies are required to clarify whether this is merely a power issue.
These results should be interpreted with some degree of caution given the relatively small number of multiple sclerosis cases in the G&H cohort (and the resulting wide confidence intervals), the potential inaccuracies of using electronic health records to ascertain cases (including the possibility of missed cases) and the lack of an external validation cohort. Due to the number of multiple sclerosis cases in G&H, we fitted and evaluated the PRS on the same dataset, which increases the risk of overfitting and therefore may produce an inflated estimate of how well the PRS models disease risk in the population. Furthermore, while we aim to compare PRS performance in UKB and G&H, it is important to note that these cohorts were genotyped on different chips and imputed with different panels (TOPMed versus Haplotype Reference Consortium).25,30 Therefore, although we use the same external reference panel to perform LD clumping, the SNPs included in the PRS for any given set of clumping-and-thresholding parameters are not identical between cohorts. The mean age in the G&H cohort is also less than that in UKB, raising the possibility of individuals in the G&H control group going on to develop multiple sclerosis in the future. We aimed to mitigate the effect of sample size by sampling the UKB dataset to an equivalent size.
Given the potential uses of a multiple sclerosis PRS in both clinical care and trial design, the limited cross-ancestry transferability of European-derived PRS is concerning and may reinforce pre-existing health inequalities between different ethnic and ancestral groups. Although advances in statistical methods for applying PRS across populations are likely to enhance transferability,11,31 there is an unmet need for ancestrally diverse GWAS of multiple sclerosis risk to ensure that genetics can play a useful role in risk stratification.
Supplementary Material
Acknowledgements
B.M.J. and R.D. conceived the study. The primary analysis was performed by J.B. and B.M.J. The initial manuscript was drafted by J.B. All authors provided input into critical revisions of the manuscript prior to submission. B.M.J. and J.B. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. We thank S.H. for his help with constructing the lay video abstract. We thank Social Action for Health, Centre of The Cell, members of our Community Advisory Group and staff who have recruited and collected data from volunteers. We thank the NIHR National Biosample Centre (UK Biocentre), the Social Genetic and Developmental Psychiatry Centre (King’s College London), Wellcome Sanger Institute and Broad Institute for sample processing, genotyping, sequencing and variant annotation. We thank Barts Health NHS Trust, NHS Clinical Commissioning Groups (City and Hackney, Waltham Forest, Tower Hamlets, Newham, Redbridge, Havering, Barking and Dagenham), East London NHS Foundation Trust, Bradford Teaching Hospitals NHS Foundation Trust, Public Health England (especially David Wyllie), Discovery Data Service/Endeavour Health Charitable Trust (especially David Stables) and NHS Digital—for their GDPR-compliant data sharing backed by individual written informed consent. Most of all we thank all of the volunteers participating in G&H.
Abbreviations
- AUC =
area under the curve
- EBV =
Epstein–Barr virus
- EPIC =
Expression, Proteomics, Imaging, Clinical
- G&H =
Genes & Health
- GWAS =
genome-wide association study(ies)
- HES =
hospital episode statistics
- HLA =
human leukocyte antigen
- HWE =
Hardy–Weinberg equilibrium
- ICD =
International Classification of Diseases
- IMSGC =
International Multiple Sclerosis Genetics Consortium
- LD =
linkage disequilibrium
- MAF =
minor allele frequency
- MHC =
major histocompatibility complex
- OR =
odds ratio
- PCA =
principal component analysis
- PRS =
polygenic risk score(s)
- SNOMED =
Systematized Nomenclature of Medicine
- SNP =
single nucleotide polymorphism
- TOPMed =
Trans-Omics for Precision Medicine Program
- UCSF =
University of California San Francisco
- UKB =
UK Biobank
Appendix: Current Genes & Health Research Team (in alphabetical order by surname)
Shaheen Akhtar, Mohammad Anwar, Elena Arciero, Omar Asgar, Samina Ashraf, Gerome Breen, Raymond Chung, Charles J Curtis, Shabana Chaudhary, Maharun Chowdhury, Grainne Colligan, Panos Deloukas, Ceri Durham, Faiza Durrani, Fabiola Eto, Sarah Finer, Ana Angel Garcia, Chris Griffiths, Joanne Harvey, Teng Heng, Qin Qin Huang, Matt Hurles, Karen A Hunt, Shapna Hussain, Kamrul Islam, Benjamin M Jacobs, Ahsan Khan, Amara Khan, Cath Lavery, Sang Hyuck Lee, Robin Lerner, Daniel MacArthur, Daniel Malawsky, Hilary Martin, Dan Mason, Mohammed Bodrul Mazid, John McDermott, Sanam McSweeney, Shefa Miah, Sabrina Munir, Bill Newman, Elizabeth Owor, Asma Qureshi, Samiha Rahman, Nishat Safa, John Solly, Farah Tahmasebi, Richard C Trembath, Karen Tricker, Nasir Uddin, David A van Heel, Caroline Winckley, John Wright.
Contributor Information
Joshua R Breedon, Preventive Neurology Unit, Wolfson Institute of Population Health, Queen Mary University of London, London EC1M 6BQ, UK.
Charles R Marshall, Preventive Neurology Unit, Wolfson Institute of Population Health, Queen Mary University of London, London EC1M 6BQ, UK; Department of Neurology, Royal London Hospital, London E1 1FR, UK.
Gavin Giovannoni, Preventive Neurology Unit, Wolfson Institute of Population Health, Queen Mary University of London, London EC1M 6BQ, UK; Department of Neurology, Royal London Hospital, London E1 1FR, UK; Blizard Institute, Queen Mary University of London, London E1 2AT, UK.
David A van Heel, Blizard Institute, Queen Mary University of London, London E1 2AT, UK.
Ruth Dobson, Preventive Neurology Unit, Wolfson Institute of Population Health, Queen Mary University of London, London EC1M 6BQ, UK; Department of Neurology, Royal London Hospital, London E1 1FR, UK.
Benjamin M Jacobs, Preventive Neurology Unit, Wolfson Institute of Population Health, Queen Mary University of London, London EC1M 6BQ, UK; Department of Neurology, Royal London Hospital, London E1 1FR, UK.
Genes & Health Research Team:
Shaheen Akhtar, Mohammad Anwar, Elena Arciero, Omar Asgar, Samina Ashraf, Gerome Breen, Raymond Chung, Charles J Curtis, Shabana Chaudhary, Maharun Chowdhury, Grainne Colligan, Panos Deloukas, Ceri Durham, Faiza Durrani, Fabiola Eto, Sarah Finer, Ana Angel Garcia, Chris Griffiths, Joanne Harvey, Teng Heng, Qin Qin Huang, Matt Hurles, Karen A Hunt, Shapna Hussain, Kamrul Islam, Benjamin M Jacobs, Ahsan Khan, Amara Khan, Cath Lavery, Sang Hyuck Lee, Robin Lerner, Daniel MacArthur, Daniel Malawsky, Hilary Martin, Dan Mason, Mohammed Bodrul Mazid, John McDermott, Sanam McSweeney, Shefa Miah, Sabrina Munir, Bill Newman, Elizabeth Owor, Asma Qureshi, Samiha Rahman, Nishat Safa, John Solly, Farah Tahmasebi, Richard C Trembath, Karen Tricker, Nasir Uddin, David A van Heel, Caroline Winckley, and John Wright
Funding
This work was performed at the Preventive Neurology Unit, which is funded by the Barts Charity. B.M.J. is supported by a Medical Research Council Clinical Research Training Fellowship (Grant Reference MR/V028766/1) which is co-funded by the UK Multiple Sclerosis Society. Genes & Health is/has recently been core-funded by Wellcome (WT102627, WT210561), the Medical Research Council (UK) (M009017, MR/X009777/1), Higher Education Funding Council for England Catalyst, Barts Charity (845/1796), Health Data Research UK (for London substantive site) and research delivery support from the National Health Service National Institute for Health Research Clinical Research Network (North Thames). Genes & Health is/has recently been funded by Alnylam Pharmaceuticals, Genomics PLC and a Life Sciences Industry Consortium of Astra Zeneca PLC, Bristol-Myers Squibb Company, GlaxoSmithKline Research and Development Limited, Maze Therapeutics Inc., Merck Sharp & Dohme LLC, Novo Nordisk A/S, Pfizer Inc. and Takeda Development Centre Americas Inc.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain Communications online.
Data availability
G&H data are available on application via https://www.genesandhealth.org/research/scientists-using-genes-health-scientific-research. UK Biobank data can be accessed on application via https://www.ukbiobank.ac.uk/. IMSGC. GWAS summary statistics are available on application to the Data Access Committee of IMSGC at https://imsgc.net/. All code used in this study is available on GitHub at https://github.com/benjacobs123456/PRS_UKB_cross_ancestry.
References
- 1.International Multiple Sclerosis Genetics Consortium (IMSGC), et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Multiple Sclerosis Genetics Consortium . Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moutsianas L, Jostins L, Beecham AH, et al. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat Genet. 2015;47:1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawcer S, Ban M, Maranian M, et al. A high-density screen for linkage in multiple sclerosis. Am J Hum Genet. 2005;77:454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hone L, Giovannoni G, Dobson R, Jacobs BM. Predicting multiple sclerosis: Challenges and opportunities. Front Neurol. 2022;12:761973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wijnands JMA, Kingwell E, Zhu F, et al. Health-care use before a first demyelinating event suggestive of a multiple sclerosis prodrome: A matched cohort study. Lancet Neurol. 2017;16:445–451. [DOI] [PubMed] [Google Scholar]
- 7.Shams H, Shao X, Santaniello A, et al. Polygenic risk score association with multiple sclerosis susceptibility and phenotype in Europeans. Brain. 2023;146:645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs BM, Noyce AJ, Bestwick J, Belete D, Giovannoni G, Dobson R. Gene-environment interactions in multiple sclerosis: A UK Biobank Study. Neurol Neuroimmunol Neuroinflamm. 2021;8:e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs BM, Watson C, Marshall C, Noyce A, Dobson R. No evidence for association between polygenic risk of multiple sclerosis and MRI phenotypes in ∼30,000 healthy adult UK Biobank participants. Multiple Sclerosis Journal. 2022;28:1656–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Privé F, Aschard H, Carmi S, et al. Portability of 245 polygenic scores when derived from the UK biobank and applied to 9 ancestry groups from the same cohort. Am J Hum Genet. 2022;109:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissbrod O, Kanai M, Shi H, et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat Genet. 2022;54:450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan L, Shen H, Gelaye B, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs BM, Peter M, Giovannoni G, Noyce AJ, Morris HR, Dobson R. Towards a global view of multiple sclerosis genetics. Nat Rev Neurol. 2022;18:613–623. [DOI] [PubMed] [Google Scholar]
- 14.Pandit L, Malli C, Singhal B, et al. HLA Associations in South Asian multiple sclerosis. Mult Scler. 2016;22:19–24. [DOI] [PubMed] [Google Scholar]
- 15.Pandit L, Ban M, Beecham AH, et al. European Multiple sclerosis risk variants in the south Asian population. Mult Scler. 2016;22:1536–1540. [DOI] [PubMed] [Google Scholar]
- 16.Isobe N, Madireddy L, Khankhanian P, et al. An ImmunoChip study of multiple sclerosis risk in African Americans. Brain. 2015;138:1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beecham AH, Amezcua L, Chinea A, et al. The genetic diversity of multiple sclerosis risk among Hispanic and African American populations living in the United States. Mult Scler. 2020;26:1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi C, Shao X, Rhead B, et al. Admixture mapping reveals evidence of differential multiple sclerosis risk by genetic ancestry. PLoS Genet. 2019;15:e1007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langer-Gould AM, Gonzales EG, Smith JB, Li BH, Nelson LM. Racial and ethnic disparities in multiple sclerosis prevalence. Neurology. 2022;98:e1818–e1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs B, Hospital RL, Dobson R. Ethnicity and multiple sclerosis - moving beyond preconceptions. Adv Clin Neurosci Rehabil. 2021;20. [Google Scholar]
- 21.Amezcua L, Smith JB, Gonzales EG, Haraszti S, Langer-Gould A. Race, ethnicity, and cognition in persons newly diagnosed with multiple sclerosis. Neurology. 2020;94:e1548–e1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romanelli RJ, Huang Q, Lacy J, Hashemi L, Wong A, Smith A. Multiple sclerosis in a multi-ethnic population from Northern California: A retrospective analysis, 2010-2016. BMC Neurol. 2020;20:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finer S, Martin HC, Khan A, et al. Cohort profile: East London Genes & Health (ELGH), a community-based population genomics and health study in British Bangladeshi and British Pakistani people. Int J Epidemiol. 2020;49:20–21i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, Butcher & Zalewski . Apocrita-high performance computing cluster for Queen Mary University of London. Zenodo; 2017. [Google Scholar]
- 28.Wang Y, Guo J, Ni G, Yang J, Visscher PM, Yengo L. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat Commun. 2020;11:3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandit L, Ban M, Sawcer S, et al. Evaluation of the established non-MHC multiple sclerosis loci in an Indian population. Mult Scler. 2011;17:139–143. [DOI] [PubMed] [Google Scholar]
- 30.Luo Y, Kanai M, Choi W, et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat Genet. 2021;53:1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruan Y, Lin Y-F, Feng Y-CA, et al. Improving polygenic prediction in ancestrally diverse populations. Nat Genet. 2022;54:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
G&H data are available on application via https://www.genesandhealth.org/research/scientists-using-genes-health-scientific-research. UK Biobank data can be accessed on application via https://www.ukbiobank.ac.uk/. IMSGC. GWAS summary statistics are available on application to the Data Access Committee of IMSGC at https://imsgc.net/. All code used in this study is available on GitHub at https://github.com/benjacobs123456/PRS_UKB_cross_ancestry.