ABSTRACT
Family with sequence similarity of 83D (FAM83D) is overexpressed in various cancers. However, no pan-cancer analysis is presently available. In the present study, we used a bioinformatics analysis to explore the diagnostic and prognostic value of FAM83D expression levels in human cancers. The GEPIA 2, TIMER 2.0, ENCORI, and DriverDBV3 databases were used to evaluate FAM83D expression levels. The potential prognostic value of FAM83D expression was analyzed using the GEPIA 2, UALCAN, and TISIB databases. The driver gene and promoter methylation levels regarding FAM83D were evaluated using the TIMER 2.0 and UALCAN databases. To further analyze interactive networks for FAM83D, FAM83D-binding proteins and related genes were determined using STRING and Gene MANIA analytic tools. Highly expressed FAM83D could be associated with mutated TP53 and promoter DNA methylation. Relative network analysis suggested that FAM83D was mainly involved in the progesterone-mediated oocyte maturation pathway, cell cycle regulation, and several other signaling pathways. Therefore, the differential expression of FAM83D could serve as a diagnostic and prognostic biomarker for various cancers. Our study revealed useful information about the differential expression of FAM83D, prognostic values, and potential functional networks in a variety of cancers, providing valuable substantive and methodological information to explore the underlying mechanisms.
Abbreviations: BP: Biological processes; CC: Cellular components; DAVID: Database for Annotation, Visualization, and Integrated Discovery; DFS: Disease-free survival; ENCORI: Encyclopedia of RNA Interactomes; FAM83: Family with sequence similarity 83; FAM83D: Family with sequence similarity of 83D; GEO: Gene Expression Omnibus; GEPIAx2: Gene Expression Profiling Interactive Analysis 2; GO: Gene Ontology; GTEx: Genotype-Tissue Expression; KEGG: Kyoto Encyclopedia of Genes and Genomes; KIRC: Kidney renal clear cell carcinoma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; MF: Molecular functions miRNA: microRNA; OS: Overall survival; PAAD: Pancreatic adenocarcinoma; PPI: Protein – protein interaction; RNA-seq: RNA-sequencing; TCGA: The Cancer Genome Atlas; TIMER 2.0: Tumor Immune Estimation Resource 2.0; UALCAN: University of Alabama at Birmingham Cancer; UCEC: Uterine corpus endometrial carcinoma.
KEYWORDS: FAM83D, bioinformatics analysis, pan-cancer, differential expression, prognostic value, biomarker
Introduction
Cancer, a major and often devastating public health problem, is the leading cause of morbidity and mortality worldwide [1,2]. The GLOBOCAN 2020 report estimated that there were approximately 19.3 million cancer cases and 10.0 million cancer deaths worldwide in 2020 [3]. Despite rapid improvements in cancer diagnosis and anti-cancer therapies in recent decades, they are still not satisfactory, as evidenced by the continued morbidity, mortality, and severe social and economic burdens imposed by cancers [4]. Therefore, it is important to explore novel diagnostic and predictive cancer biomarkers to develop more effective screening, diagnosis, and therapeutic efforts.
Previous studies have reported on the oncogene family FAM83 (family with sequence similarity 83) [5]. The FAM83 family consists of eight genes: FAM83A – FAM83 H, all of which share a highly conserved domain of unknown function (DUF1669) near the N-terminal [6]. The FAM83 family not only takes part in several important biological processes but is also closely related to tumorigenesis and disease progression [5].
FAM83D, a microtubule-associated protein, is a member of the FAM83 family. In recent years, evidence has been accumulated regarding the role of FAM83D in various cancers [7–9]. Emerging evidence has indicated that FAM83D overexpression is associated with ovarian cancer and hepatocellular carcinoma [10,11]. However, the biological function, prognostic value, and molecular mechanisms regarding FAM83D have not been fully elucidated for most cancers. Thus, the role of FAM83D in most cancers is still unclear to some extent, and additional studies are needed on the relationships among FAM83D expression, survival, and prognosis.
In this study, we comprehensively investigated FAM83D gene expression and prognostic value in pan-cancer using numerous databases, with a focus on The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets [12,13]. Beyond that, to explore the potential mechanisms of FAM83D in pan-cancer, we used an online tool to analyze the functional network of FAM83D. The findings of the present study provide useful information on the correlations among FAM83D overexpression, survival status, and prognostic value and suggest a potential candidate biomarker for the diagnosis and prognosis of several cancers.
Methods
Gene Expression Profiling Interactive Analysis 2 (GEPIA 2) database analysis
GEPIA 2 (http://gepia2.cancer-pku.cn/#index) is a popular interactive website used for analyzing gene expression levels based on 9,736 tumor and 8,587 normal tissue samples from TCGA and Genotype-Tissue Expression (GTEx) databases [14]. We selected the “Box Plot” module and entered “GENE: FAM83D” to analyze FAM83D expression in all cancer datasets. Furthermore, we used the “Stage Plot” module to acquire FAM83D expression data at different tumor stages. We obtained overall survival (OS) and disease-free survival (DFS) data for all cancers using the “Survival Map” module and the “Survival Analysis” module to acquire OS and DFS data for candidate cancers. We compared hazard ratios using the log-rank test. P value <0.05 was considered to indicate significance.
Tumor Immune Estimation Resource 2.0 (TIMER 2.0) database analysis
TIMER 2.0 (http://timer.cistrome.org/) is a comprehensive resource used for analyzing gene expression and correlations across cancers [15]. We used the “Gene_DE” module of TIMER 2.0 to determine FAM83D expression levels in diverse cancers. The significance of the differential expression between tumor and adjacent normal tissues was computed using the Wilcoxon test. Furthermore, we used the “Gene-Corr” module to study the correlation between FAM83D and related candidate gene expression in various cancer types, with Spearman’s Rho representing the degree of correlation. Subsequently, we used the “Gene_Mutation” module to analyze differential gene expression between TP53 mutation and FAM83D expression, and constructed heatmaps showing findings for log2 fold changes with regard to differential expression in each cancer type. P value <0.05 was considered to indicate significance.
Encyclopedia of RNA Interactomes (ENCORI) database analysis
The ENCORI (https://starbase.sysu.edu.cn/index.php) is an open-source platform for exploring differential gene expression data for 32 types of tumors, derived from 10,882 RNA-sequencing (RNA-seq) and 10,546 mi RNA-seq datapoints [16]. We adopted the “Pan-Cancer” platform to analyze differential gene expression for FAM83D in pan-cancer. P value <0.05 was considered to indicate significance.
DriverDBV3 database analysis
DriverDBv3 (http://driverdb.tms.cmu.edu.tw/) is a cancer omics database that incorporates data on somatic mutations, RNA expression, microRNA (miRNA) expression, and methylation [17]. We entered the term “GENE: FAM83D” and used the “Expression” module to obtain differential gene expression data for FAM83D across multiple cancer types. We only kept the “Primary Solid Tumor” and “Solid Tissue Normal” data. P value <0.05 was considered to indicate significance.
TISIDB database analysis
The TISIDB online data portal (http://cis.hku.hk/TISIDB/) not only provides data on tumor-immune system interactions but also provides data relevant to clinical analysis (OS and tumor stage) and gene expression across molecular subtypes [18]. We entered the term “Gene Symbol: FAM83D” and separately selected the “Clinical-Cancer Stage” and “Subtype-Molecular Subtype” modules to analyze FAM83D expression data for different tumor stages and various molecular subtypes of candidate cancers. In addition, we obtained OS data via the “Clinical-Overall Survival Analysis” module. The results for cancer stage, molecular subtype, and OS were evaluated using the Spearman correlation, Kruskal – Wallis, and log rank tests, respectively. P value <0.05 was considered to indicate significance.
University of Alabama at Birmingham Cancer (UALCAN) database analysis
The UALCAN data analysis portal (http://ualcan.path.uab.edu/) is a comprehensive, user-friendly, and interactive resource for exploring cancer data [19,20]. We used UALCAN to explore OS, promoter DNA methylation, and TP53 mutation status in candidate cancers according to FAM83D expression. In addition, the relationship between FAM83D expression and the clinicopathologic features of patients was compared in liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and uterine corpus endometrial carcinoma (UCEC). P value <0.05 was considered to indicate significance.
STRING analysis
STRING (https://cn.string-db.org/) is a database of known and predicted protein – protein interaction (PPI) networks that currently covers 24,584,628 proteins from 5,090 organisms [21]. The STRING database was used to analyze the PPI network of FAM83D. The results of this analysis were displayed as follows: the network type (full STRING network), meaning of the network edges (evidence), active interaction sources (all), minimum required interaction score (low confidence, 0.150), and maximum number of interactors for display (no more than 10 interactors).
Gene MANIA analysis
Gene MANIA (http://genemania.org/) is an open-access online tool for the analysis of PPI networks [22]. The Gene MANIA tool was used to analyze the PPI network of FAM83D using bioinformatics methods, including physical gene interactions, prediction, gene co-expression, co-localization, genetic interactions, pathways, and shared protein domains. The term “FAM83D” was entered, and the top 20 gene networks were visualized.
Metascape analysis
Metascape (https://metascape.org/gp/index.html#/main/step1), a website-based tool, is a tool used to analyze gene/protein lists and inform better data-driven decisions [23]. Genes interacting with FAM83D, determined using STRING and Gene MANIA, were input into Metascape for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The GO term analysis comprised biological processes (BP), molecular functions (MF), and cellular components (CC). “H. sapiens” (Homo sapiens) was input as the species. We entered the term “Custom Analysis” and selected the “Enrichment” module. The Metascape analysis results were displayed with minimum overlap (3), a P-value cutoff of 0.05, a minimum enrichment of 1.5, and a reset analysis (selecting GO Molecular Functions, GO Biological Processes, GO Cellular Components, and KEGG Pathway Analysis).
Database for Annotation, Visualization, and Integrated Discovery (DAVID) analysis
DAVID (https://david.ncifcrf.gov/home.jsp) is a comprehensive bioinformatics resource for functional enrichment analysis and functional annotation of gene lists [24,25]. We selected the “Shortcut to DAVID Tools” and the “Functional Annotation” module. The genes interacting with FAM83D, determined using STRING and Gene MANIA, were then added to the “Gene List” for GO and KEGG pathway analyses. The identifier was “OFFICIAL-GENE-SYMBOL”, the input species was “H.sapiens”, and the type was “Gene List”. GO terms, including BP, CC, and MF, and KEGG pathways were analyzed using the R statistical software (v.4.2.0, The R Project for Statistical Computing, Vienna, Austria) via the “ggplot2” package. P value <0.05 was considered to indicate significance.
Results
FAM83D differential expression levels in pan-cancer
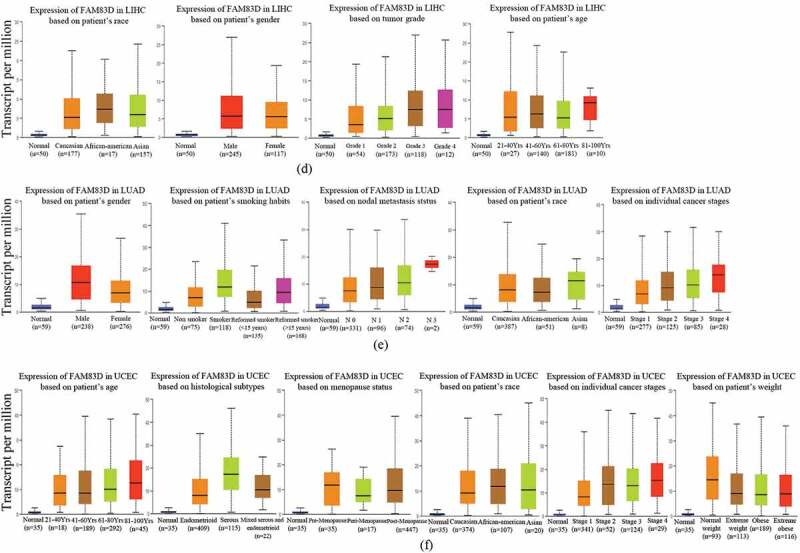
To find differences in FAM83D expression between tumor and adjacent normal tissue samples, we firstly studied the differential FAM83D expression levels in four different databases: GEPIA 2, TIMER 2.0, ENCORI, and DriverDBV3. FAM83D overexpression was discovered in over 10 cancer types (Figure 1(a)). Then, we analyzed the prognostic value of FAM83D in three different databases: GEPIA 2, UALCAN, and TISIB. We selected five candidate tumors, including kidney renal clear cell carcinoma (KIRC), LIHC, LUAD, pancreatic adenocarcinoma (PAAD), and UCEC, that had higher FAM83D expression levels compared to normal tissue samples in at least two different databases (Figure 1(b) and Figure 1(c)) and also had prognostic value in at least two different databases among these tumors. Meanwhile, we further considered FAM83D expression as a diagnostic criterion regarding different tumor stages and molecular subtypes. We analyzed FAM83D expression levels in different clinical stages of KIRC, LIHC, LUAD, and PAAD (Figure 2(a)) and in UCEC no significant differences were found in the GEPIA 2 database. In addition, the correlation between the clinical stage of UCEC and FAM83D expression levels was determined using the TISIDB database (Figure 2(b)). The differential expression of FAM83D may be related to the molecular subtypes of LIHC (including iCluster 1, iCluster 2, and iCluster 3) and UCEC (including CN_HIGH, CN_LOW, MSI, and POLE) (Figure 2(c)). The prognosis of iCluster 1 was the worst among the molecular subtypes of LIHC, and this molecular subtype showed the highest expression level of FAM83D. Similarly, the prognosis of CN_HIGH was the worst, and this subtype showed the highest expression level of FAM83D in UCEC. In addition, correlation was observed between the level of FAM83D overexpression and clinicopathologic features using the UALCAN database. The results showed potential a correlation between differential FAM83D expression and race, gender, tumor grade, and age in LIHC (Figure 2(d)). A correlation was also observed between differential FAM83D expression and gender, smoking habits, nodal metastasis status, race, and individual cancer stages in LUAD (Figure 2(e)). Moreover, a correlation existed between differential FAM83D expression and age, histological subtypes, menopause status, race individual cancer stages, and weight in UCEC (Figure 2(f)). These findings strongly support the possibility that FAM83D is a carcinogen in many aspects and lay the foundation for further research on the carcinogenic effect of FAM83D.
Figure 1.
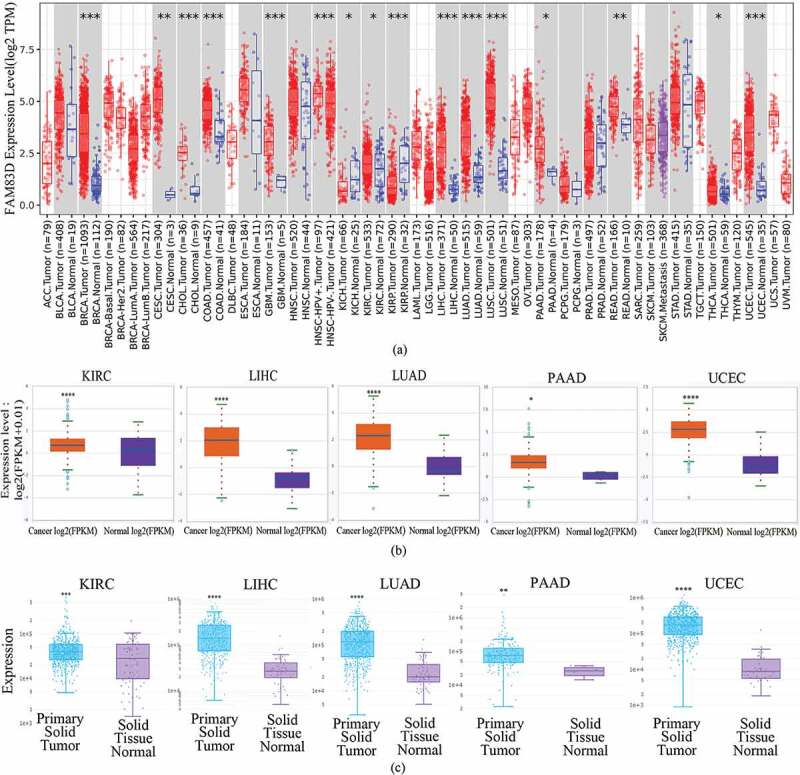
Differential expression of family with sequence similarity of 83D (FAM83D) in many types of human tumors. (a) Differential expression of FAM83D in all tumors or specific tumor subtypes was analyzed via the TIMER 2.0 database. (b) FAM83D expression was different between normal tissues and primary tissues among candidate tumors in the ENCORI database. (c) FAM83D expression was different between normal tissues and primary tissues among candidate tumors in the DriverDbv3 database as well. *P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
Figure 2.
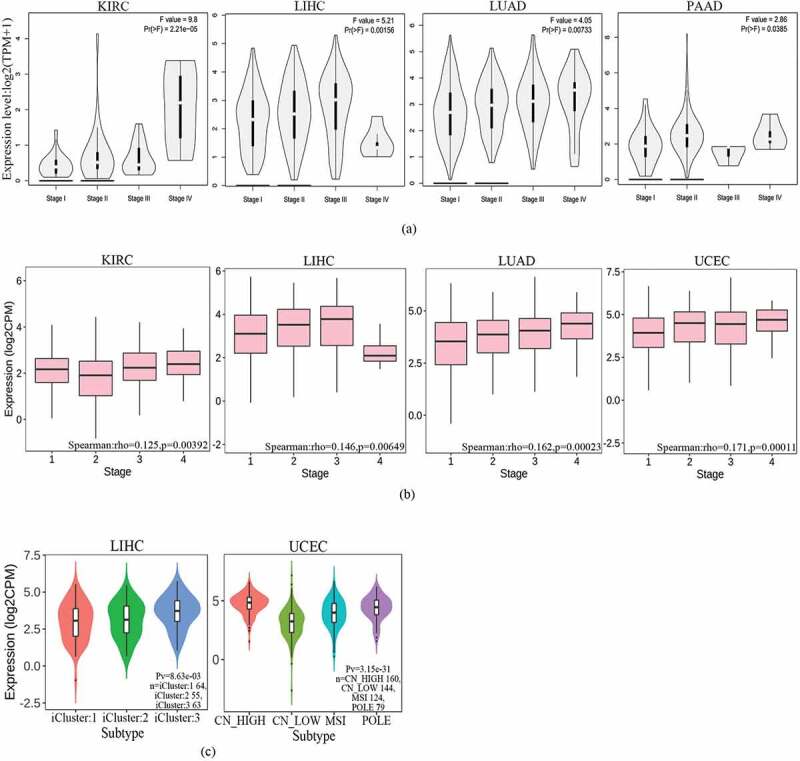
Family with sequence similarity of 83D (FAM83D) expression levels according to the clinical features of candidate tumors. (a) FAM83D expression levels across clinical stages of kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and pancreatic adenocarcinoma (PAAD) were analyzed using the GEPIA 2 database. (b) FAM83D expression levels across tumor stages of KIRC, LIHC, LUAD, and uterine corpus endometrial carcinoma (UCEC) were analyzed using the TISIDB database. (c) FAM83D expression levels across molecular subtypes of LIHC and UCEC were analyzed using the TISIDB database. (d) FAM83D overexpression levels were associated with clinical features in LIHC using the UALCAN database. (e) FAM83D overexpression levels were associated with clinical features in LUAD using the UALCAN database. (f) FAM83D overexpression levels were associated with clinical features in UCEC using the UALCAN database.
Figure 2.
(continued).
Correlation between prognostic value and FAM83D expression
Based on the relationship between FAM83D expression and clinical features, we speculated that the differential expression of FAM83D may be related to tumor prognostic value. The GEPIA 2 database, which provided a survival map for all cancers regarding OS and DFS, was used to identify the correlation between FAM83D overexpression and prognostic value in patients with cancer (Figure 3(a)). Our analysis showed that FAM83D overexpression was associated with OS and DFS in most of the evaluated tumors. Interestingly, the GEPIA 2 database revealed that highly-expressed FAM83D was correlated with negative prognosis for several cancers, including LIHC, LUAD, and PAAD (Figure 3(b)). Furthermore, high-expressed FAM83D showed a significant association with reduced OS of patients with UCEC in the UALCAN database (Figure 3(c)), but not in the GEPIA 2 database. Moreover, FAM83D expression was correlated with poor prognosis in patients with all candidate tumors in the TISIB database (Figure 3(d)). Therefore, the above data demonstrated that high FAM83D expression, which was correlated with poor prognosis in the present analysis, may serve as a potential prognostic biomarker for specific tumor types.
Figure 3.
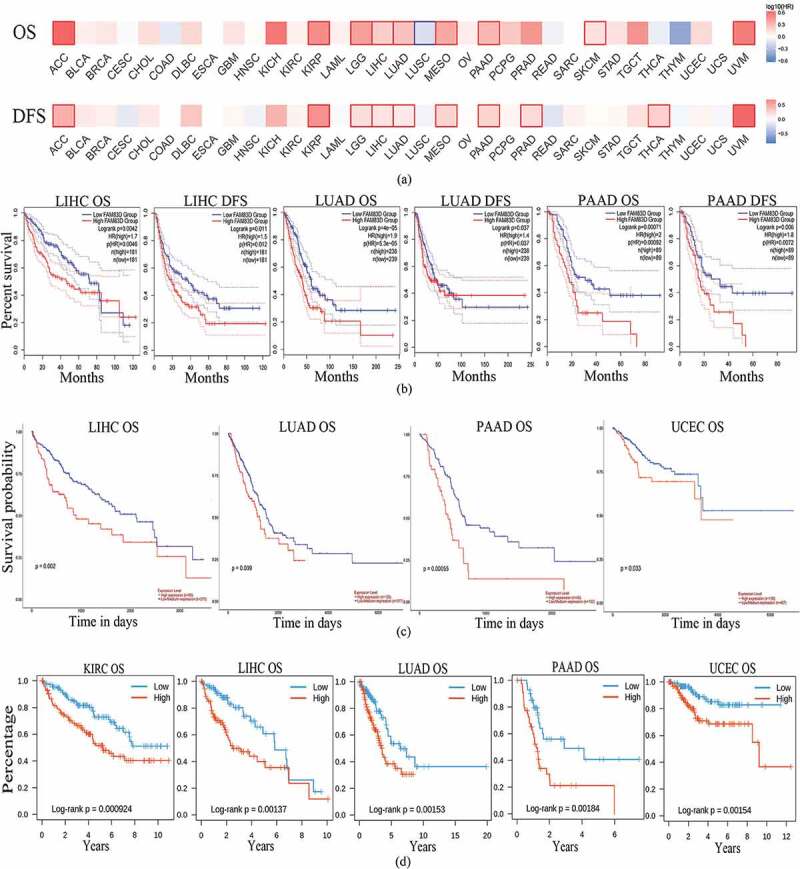
Correlation between family with sequence similarity of 83D (FAM83D) expression levels and survival curves in a range of cancers. (a) Survival map of all cancers with regard to overall survival (OS) and disease-free survival (DFS) were analyzed through the GEPIA 2 database. (b) Correlation between FAM83D expression levels and survival value in the GEPIA 2 database. (c) the OS of liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), pancreatic adenocarcinoma (PAAD), and uterine corpus endometrial carcinoma (UCEC) in the UALCAN database. (d) the OS of kidney renal clear cell carcinoma (KIRC), LIHC, LUAD, PAAD, and UCEC in the TISIDB database.
Correlation between gene mutation and FAM83D expression
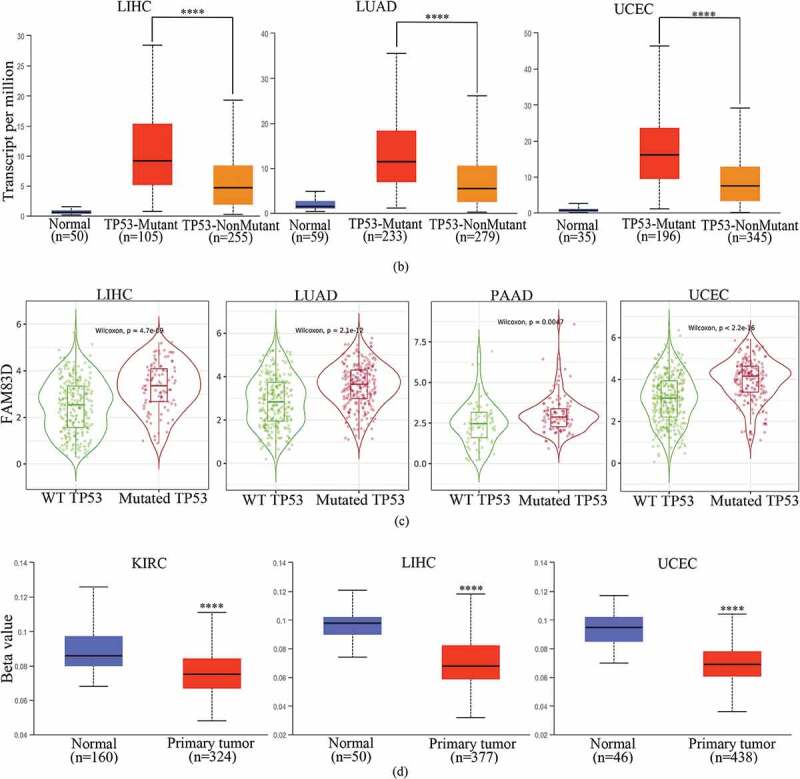
To better understand the reasons for FAM83D overexpression in oncogenesis and progression, we explored the correlation between FAM83D expression and important driver genes across mutation status and different cancers using the TIMER 2.0 database, including the APC, BRAF, KRAS, POLE, and TP53 genes (Figure 4(a)). We observed that mutated TP53 was detected in most tumors. Thus, the TP53 mutation driver gene could be a causative factor of FAM83D overexpression in oncogenesis and progression. In the UALCAN database, the TP53 mutation was closely associated with various candidate tumors, including LIHC, LUAD, and UCEC (Figure 4(b)). The TP53 mutation was also significantly correlated with FAM83D expression in LIHC, LUAD, PAAD, and UCEC in the TIMER 2.0 database, further validating the abovementioned results (Figure 4(c)). To better understand the cause of highly expressed FAM83D in KIRC, we evaluated whether KIRC was associated with DNA methylation within the UALCAN database (Figure 4(d)). In addition, an analysis within the UALCAN database showed FAM83D was closely associated not only with mutated TP53 but also with DNA methylation in LIHC and UCEC, suggesting another possible causative factor (Figure 4(d)).
Figure 4.
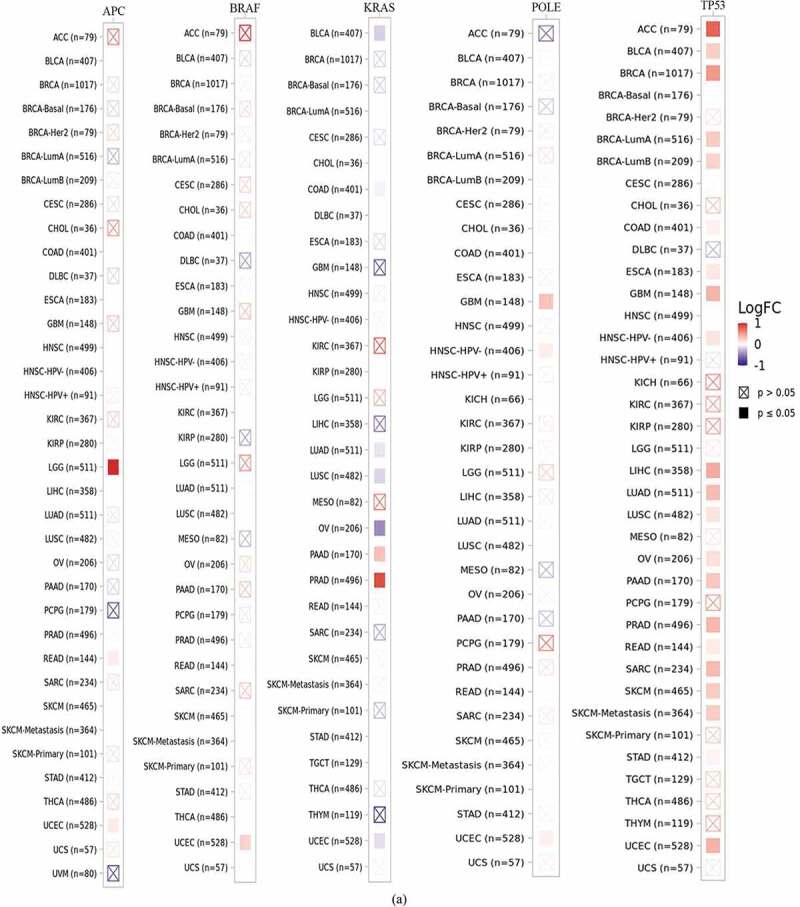
Correlation between family with sequence similarity of 83D (FAM83D) expression levels and driver gene and promoter DNA methylation in a range of cancers. (a) Correlation between FAM83D expression and mutated driver genes in pan-cancer within the TIMER 2.0 database. (b) Correlation between FAM83D expression and mutated TP53 in liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and uterine corpus endometrial carcinoma (UCEC) by the UALCAN database. (c) the correlation between FAM83D expression and mutated TP53 in LIHC, LUAD, pancreatic adenocarcinoma (PAAD), and UCEC in the TIMER 2.0 database. (d) Using the UALCAN database, highly-expressed FAM83D was analyzed with regard to promoter DNA methylation in kidney renal clear cell carcinoma (KIRC), LIHC, and UCEC. **** P < 0.0001.
Figure 4.
(continued).
Correlation between molecular pathways and FAM83D expression
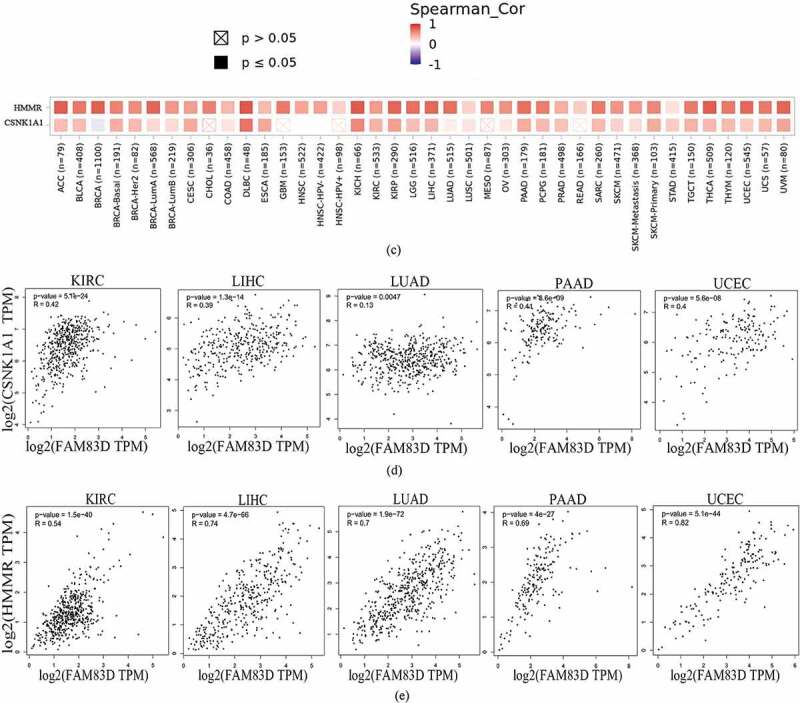
To analyze the molecular mechanisms underlying the role of FAM83D in tumorigenesis, we determined FAM83D-binding proteins and genes associated with FAM83D. Using the STRING online tool, we obtained 10 genes that showed correlations with FAM83D (see Figure 5(a)). We also explored data regarding the PPI network of FAM83D using the Gene MANIA tool. The list of top 20 genes correlated with FAM83D was discovered using Gene MANIA (Figure 5(b)). Analysis of the above datasets showed two common members: CSNK1A1 and HMMR. We determined the correlation between the network of core genes linked to FAM83D (CSNK1A1 and HMMR) and FAM83D expression across cancers using TIMER 2.0 (Figure 5(c)). Subsequently, the GEPIA 2 analysis showed that FAM83D expression was positively associated with CSNK1A1 and HMMR in candidate tumors (Figure 5(d) and Figure 5(e)). Finally, to predict functional enrichment, information from combining the two datasets representing the PPI network for FAM83D was used for GO enrichment and KEGG pathway analyses in the Metascape online tool (Figure 6(a) and (Figure 6(b)) and the DAVID database (Figure 6(c) and Figure 6(d)). The results showed that the progesterone-mediated oocyte maturation pathway and cell cycle regulation may be the primary pathways involved in the functional mechanisms of FAM83D (Figure 6(b) and Figure 6(d)).
Figure 5.
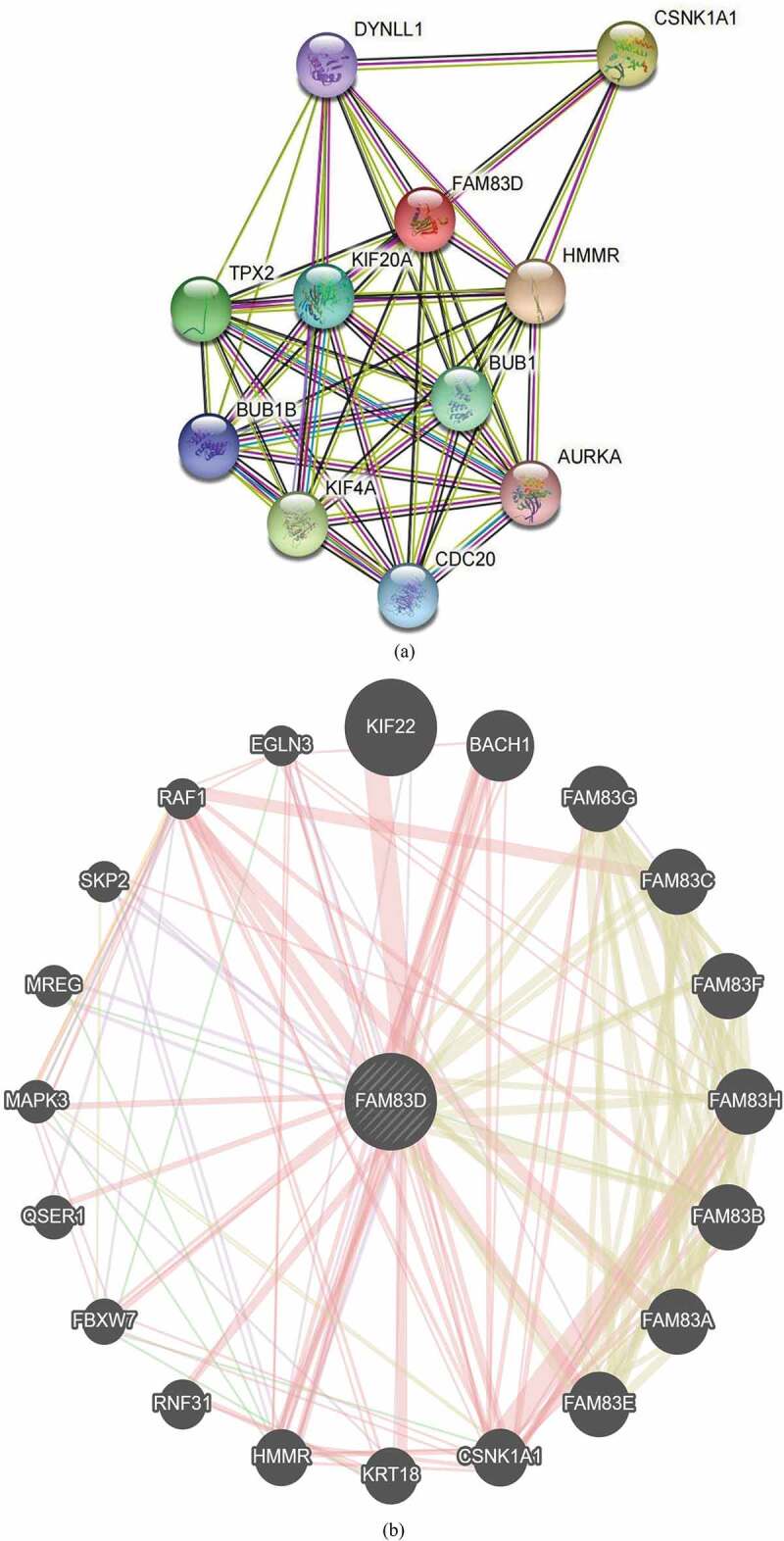
Family with sequence similarity of 83D (FAM83D)-related gene enrichment analysis. (a) and (b) FAM83D-binding proteins and genes related to FAM83D were individually analyzed via the STRING database (a) and the Gene MANIA online tool (b). (c) Correlation between the primary FAM83D-related genes (CSNK1A1 and HMMR) and FAM83D expression in pan-cancer, obtained via the TIMER 2.0. (d) and (e) FAM83D expression was associated with CSNK1A1 (d) and HMMR (e) in candidate tumors within the GEPIA 2 database.
Figure 5.
(continued).
Figure 6.
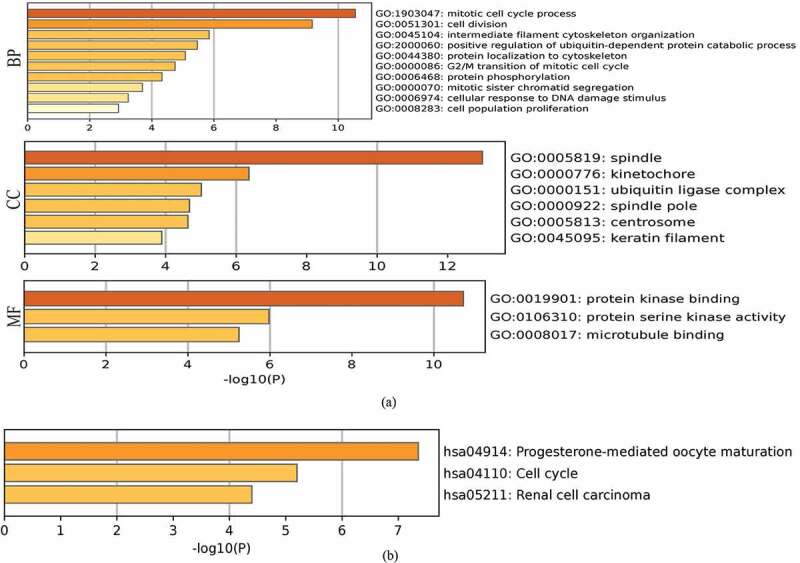
Heatmaps for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses across family with sequence similarity of 83D (FAM83D) interactive genes. (a) and (b) GO enrichment (a) and KEGG pathway analysis (b) for FAM83D were performed using the Metascape online tool. (c) and (d) GO enrichment (c) and KEGG pathway analyses (d) for FAM83D were performed via the DAVID database.
Figure 6.
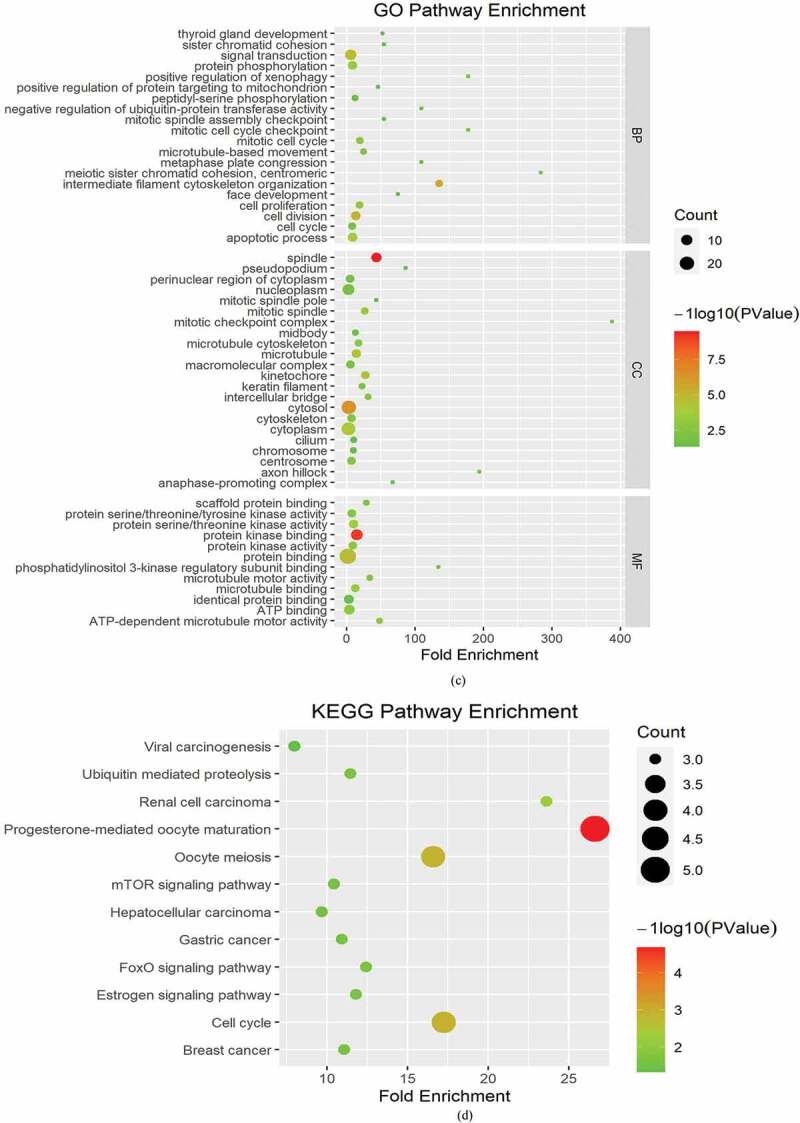
(continued).
Discussion
Despite rapid improvements in early cancer diagnosis and anti-cancer therapies in recent decades, cancer is still a threat to human survival worldwide [4]. Therefore, it is important to explore novel biomarkers that may be beneficial for early diagnosis, prognostics, and risk assignment for cancer therapy. Previous studies have shown that the FAM83 family is closely related to tumorigenesis, and FAM83D is an important member of the FAM83 family [5]. In recent years, increasing studies have demonstrated that FAM83D is correlated with several tumors [7,8,26]. Based on bioinformatics data, we performed a pan-cancer analysis of the role of FAM83D in tumors from the view of overall tumorigenesis.
In this study, we simultaneously investigated the overexpression of FAM83D in at least two different databases and evaluated the potential of FAM83D as a diagnostic biomarker. With LIHC, LUAD, and UCEC, we also analyzed the relationship between FAM83D overexpression and clinicopathologic features. Our results showed that FAM83D expression levels were closely related to tumor occurrence. FAM83D also has the potential to be a prognostic biomarker. In addition, the correlation between FAM83D expression and prognostic values was analyzed in candidate tumors, including KIRC, LIHC, LUAD, PAAD, and UCEC. FAM83D overexpression was associated with poor survival in candidate tumors, further emphasizing its potential as a prognostic biomarker. Regarding the molecular mechanism and functional enrichment analysis underlying the role of FAM83D in tumors, our results showed that high expression of FAM83D was affected by multiple driver genes, especially TP53, which showed the strongest relationship with FAM83D expression in all candidate tumors. It is critical to further study the mechanisms underlying the association between FAM83D expression and TP53 mutations in future research. In addition, we detected a correlation between FAM83D expression and DNA methylation of the FAM83D promoter, providing evidence of a pathway leading toward highly expressed FAM83D. As the final step of our analysis, we combined information on the FAM83D-binding protein and FAM83D-related genes for enrichment analysis. We found that the functional mechanisms of FAM83D could be relevant to the progesterone-mediated oocyte maturation pathway and cell cycle regulation.
Our study used the most popular bioinformatic databases to perform a comprehensive analysis between genes and tumors. This methodology has several advantages, such as large numbers of samples, lower costs, and a clearer genomic and functional analysis. However, this methodology has many drawbacks, including the limitations inherent in online databases. For example, different databases may produce different results due to distinct collected samples. Therefore, as our study presented bioinformatics analyses based on different databases, further experimental verification is required, including reverse transcription-polymerase chain reaction and western blotting.
Conclusion
FAM83D is a member of the FAM83 family, the differential expression of which is strongly linked to tumorigenesis and tumor progression [7,8,26]. In this study, we systematically analyzed the correlation between highly expressed FAM83D and prognostic value in candidate cancers. Our results showed that FAM83D overexpression could serve as a biomarker in the diagnosis and prognosis of patients with KIRC, LIHC, LUAD, PAAD, and UCEC. Furthermore, through a comprehensive analysis using online databases, we found several enrichment pathways significantly correlated with FAM83D, including the progesterone-mediated oocyte maturation pathway, cell cycle regulation, and several other signaling pathways. These results indicate the importance of FAM83D in carcinogenesis and its potential role as a diagnostic and prognostic biomarker in tumors. Subsequent research efforts should focus on further proving the molecular mechanisms underlying FAM83D gene expression in cancers through applied experimental research. In this paper, we have indicated a relationship between FAM83D and tumorigenesis using a large amount of data, which will provide a key reference for promoting medical development.
Acknowledgments
We acknowledge TCGA and GEO databases for providing platforms and contributors for uploading their datasets.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethical approval
Our study is based on the open-source data, so the IRB (Institutional Review Board) review was exempted.
Data availability statement
All databases are freely available as public resources.
References
- [1].Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. [DOI] [PubMed] [Google Scholar]
- [2].Xia C, Dong X, Li H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135(5):584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- [4].Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. [DOI] [PubMed] [Google Scholar]
- [5].Snijders AM, Lee SY, Hang B, et al. FAM83 family oncogenes are broadly involved in human cancers: an integrative multi-omics approach. Mol Oncol. 2017;11(2):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fulcher LJ, Bozatzi P, Tachie-Menson T, et al. The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci Signal. 2018;11(531). DOI: 10.1126/scisignal.aao2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang D, Han S, Peng R, et al. FAM83D activates the MEK/ERK signaling pathway and promotes cell proliferation in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;458(2):313–320. [DOI] [PubMed] [Google Scholar]
- [8].Yin C, Lin X, Wang Y, et al. FAM83D promotes epithelial-mesenchymal transition, invasion and cisplatin resistance through regulating the AKT/mTOR pathway in non-small-cell lung cancer. Cell Oncol. 2020;43(3):395–407. [DOI] [PubMed] [Google Scholar]
- [9].Li X, Sun C, Chen J, et al. Suppression of FAM83D inhibits glioma proliferation, invasion and migration by regulating the AKT/mTOR signaling pathway. Transl Oncol. 2022;22:101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang Q, Yu S, Lok SIS, et al. FAM83D promotes ovarian cancer progression and its potential application in diagnosis of invasive ovarian cancer. J Cell Mol Med. 2019;23(7):4569–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liao W, Liu W, Liu X, et al. Upregulation of FAM83D affects the proliferation and invasion of hepatocellular carcinoma. Oncotarget. 2015;6(27):24132–24147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Blum A, Wang P, Zenklusen JC.. SnapShot: tCGA-Analyzed Tumors. Cell. 2018;173(2):530. [DOI] [PubMed] [Google Scholar]
- [13].Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu SH, Shen PC, Chen CY, et al. DriverDbv3: a multi-omics database for cancer driver gene research. Nucleic Acids Res. 2020;48(D1):D863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. [DOI] [PubMed] [Google Scholar]
- [19].Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- [25].Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhu H, Diao S, Lim V, et al. FAM83D inhibits autophagy and promotes proliferation and invasion of ovarian cancer cells via PI3K/AKT/mTOR pathway. Acta Biochim Biophys Sin (Shanghai). 2019;51(5):509–516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All databases are freely available as public resources.


