Abstract
The discovery of asymmetric histone inheritance during asymmetrically dividing Drosophila melanogaster male germline stem cells indicates a mechanism for introducing cellular diversity. This process is proposed to occur in three steps: First, establishment of histone asymmetry between sister chromatids; second, recognition of sister chromatids carrying asymmetric epigenetic information; and third, execution of the asymmetric epigenome in the resulting daughter cells. Histone chaperones and replisome components influence replication-coupled histone assembly onto sister chromatids, which is important for maintaining epigenetic memory and genomic integrity. Recognition of the asymmetric epigenome involves a sister centromere asymmetry-centered ‘mitotic drive’ mechanism, where differences in centromere protein levels and asynchronized microtubule activity bias inheritance of epigenetically distinct sister chromatids. Finally, differences in epigenomes differentially influence cell cycle progression and likely gene expression in the daughter cells. This review discusses current knowledge for each step and how this process contributes to the cell fate determination in multicellular organisms.
Keywords: epigenetic inheritance, DNA replication-coupled histone assembly, epigenetic memory, nonrandom chromatid segregation, mitotic drive, nucleosome density
Introduction
During multicellular organism development, DNA replication in a mother cell duplicates the genome. Next, mitosis produces two genetically identical daughter cells. How this process contributes to cellular diversity in a multicellular organism remains a fundamental question in developmental biology. One symmetry-breaking event that contributes to cellular diversity is asymmetric cell division (ACD). During development and adult tissue homeostasis, ACD produces two genetically identical daughter cells with distinct cell fates. It is thought that cell fate determination is mediated in part through epigenetic mechanisms that influence properties of the genome, but do not change the underlying DNA sequences. A critical epigenetic factor is the nucleosome, which is composed of a histone octamer structure containing canonical histones H3, H4, H2A, and H2B. Histone post-translational modifications (PTMs) and histone variants both influence chromatin compaction and recruitment of factors, which regulate many cellular processes like gene transcription, DNA replication, and DNA repair (63). It is possible that during ACD, the identities of the two daughter cells are regulated by inheriting distinct epigenetic states.
Evidence of asymmetric histone inheritance was first demonstrated during ACD of the Drosophila melanogaster male germline stem cells (GSCs) (138). Here histones synthesized in the previous cell cycle (parental/old) are retained in the self-renewing GSC, whereas histones synthesized in the current cell cycle (new) are inherited by the differentiating daughter gonialblast (138, 148). To understand the generality of this phenomenon, asymmetric histone inheritance has since been investigated in diverse systems. As a result, varying degrees of asymmetric histone inheritance during asymmetric stem cell division have been reported (Figure 1). The D. melanogaster intestinal stem cells and male GSCs demonstrate global inheritance (138, 148, 163), while the D. melanogaster female GSCs display local asymmetries at genomic regions hosting genes required for either stemness or differentiation (57). Distinct local asymmetries were similarly detected in mouse embryonic stem cells (ESCs) induced to undergo ACD (80), likely related to epigenome and transcriptome changes (137). Interestingly, in vivo asymmetric histone inheritance was investigated using a chemical method to label old histones in Mus musculus adult skeletal muscle stem cells. While the authors claimed no asymmetry for old histone inheritance, their published images show an asymmetric pattern particularly in daughter cells with drastic gene expression changes manifested by the reporter for either stemness or differentiation genes (34). However, further investigation into the conservation of this phenomenon in different adult stem cell lineages and during development are warranted.
Figure 1. Modes of asymmetric histone inheritance:
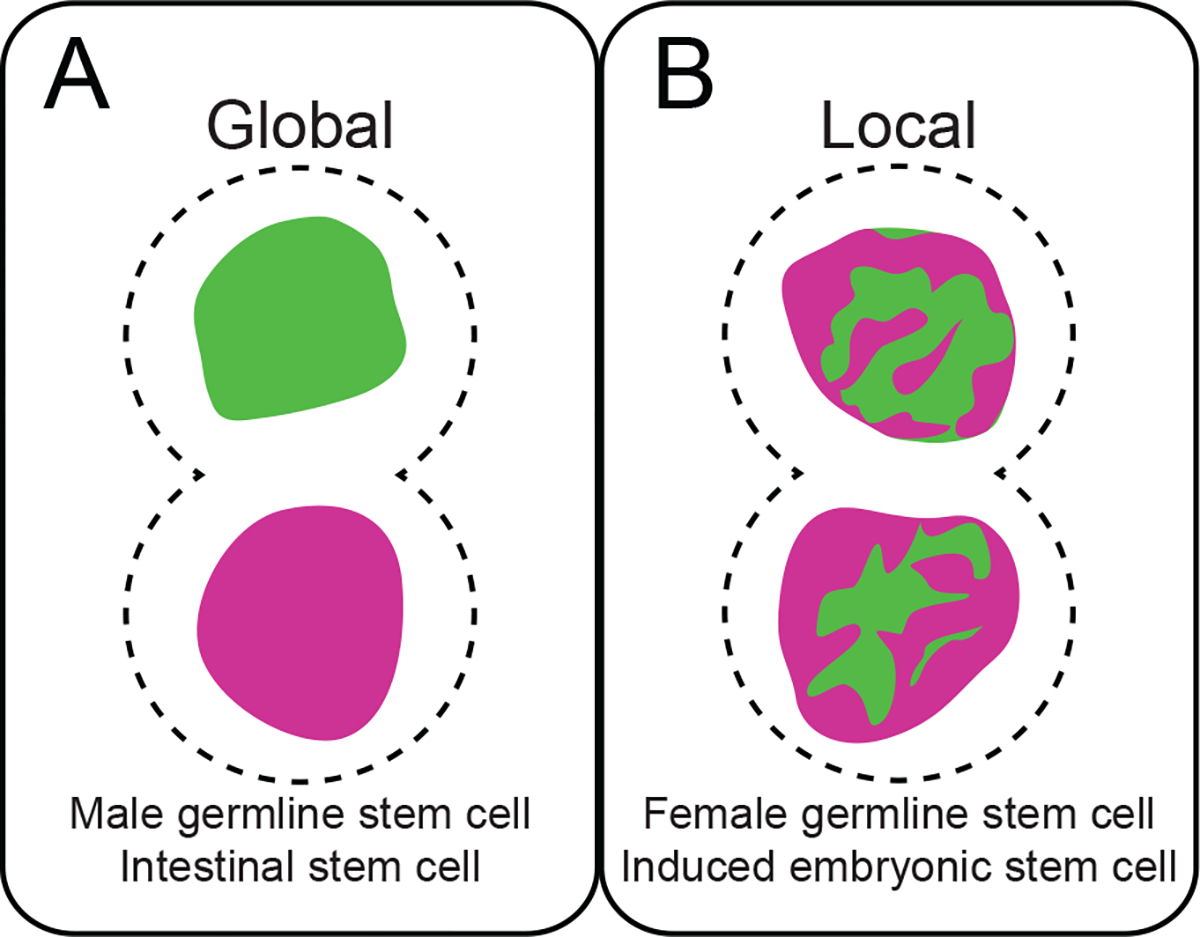
(A) Global asymmetric histone inheritance presents as segregation of predominantly old or new histone-enriched sisters to daughter cells. (B) In contrast, local histone inheritance patterns are large-scale domains distinctly enriched for old or new histones.
Evidence from the D. melanogaster male GSC system suggests that histone asymmetries between sister chromatids are established during DNA replication; old histones are enriched in one set of sister chromatids whereas new histones are incorporated into the other set of sister chromatids (148). Together with the observed asymmetric histone segregation patterns during mitosis, it was proposed that asymmetric histone inheritance involves minimally three steps with molecular and cellular mechanisms published that support this model (115, 116, 148). In Step 1, histone asymmetry is established via DNA replication-coupled histone incorporation. Step 2 involves asymmetry recognition of epigenetically distinct sister chromatids by the mitotic machinery (115). In Step 3, inherited histone asymmetries initiate distinct cellular programs in the resulting daughter cells. It is plausible that differing degrees of asymmetry in histone inheritance (Figure 1) reflect differences in the first step. Thus, DNA replication is critical to either maintain or change epigenetic information between sister chromatids at either global or local scales. This decision could underlie the dynamic versus mild cell fate changes observed during multicellular organism development, tissue homeostasis, and disease progression.
I. Establishing histone asymmetry
1. Replication-coupled chromatin assembly of old and new histones
1.1. Replication-dependent incorporation of canonical histones
Canonical histone biosynthesis and incorporation are linked to DNA replication to accommodate the reformation of chromatin following DNA duplication. Because of this, canonical histones are referred to as replication-dependent histones. On the other hand, the expression and incorporation of histone variants like H3.3 and centromere-specific H3 variant, Centromere protein A (CENP-A), are not linked to DNA replication. These variants are referred to as replication-independent histones (85). Some features of chromatin incorporation are common for both types of histones. However, replication-dependent histone incorporation is the focus of this discussion.
Chromatin poses a large obstacle to DNA replication machinery. It must be deconstructed in front of the DNA replication fork, granting DNA Polymerases access to the underlying DNA for duplication. Behind the replication fork, chromatin is restored through concerted efforts of recycling old histones and incorporating new histones. Histone chaperones and specific components of the replication machinery contribute to the process of chromatin disassembly and reassembly onto newly replicated DNA. Recent studies have identified the molecules that determine symmetric or asymmetric histone incorporation patterns between sister chromatids. While the molecular details of these highly orchestrated events are described elsewhere (135, 159), the process is summarized here.
1.2. Chromatin disassembly
Removal of H2A-H2B dimers by the Facilitates Chromatin Transcription (FACT) complex initiates parental chromatin disassembly (Figure 2) (112, 119). Subsequently, Minichromosome Maintenance 2 (MCM2) and Anti-silencing Factor 1 (Asf1) remove and dissociate parental (H3-H4)2 to form a 1:1:1:1 MCM2-H3-H4-Asf1 complex (117). During this process, MCM2 interacts with H3-H4 dimers through a conserved histone binding fold at its amino-terminus (117, 143). Meanwhile, Asf1 binds to the H3-H4 tetramer interface, inhibiting (H3-H4)2 reformation and preventing erroneous H2A-H2B binding (50, 117, 143). The MCM helicase next unwinds the DNA double helix, exposing ssDNA in preparation for synthesis.
Figure 2. Replication-coupled chromatin assembly and restoration:
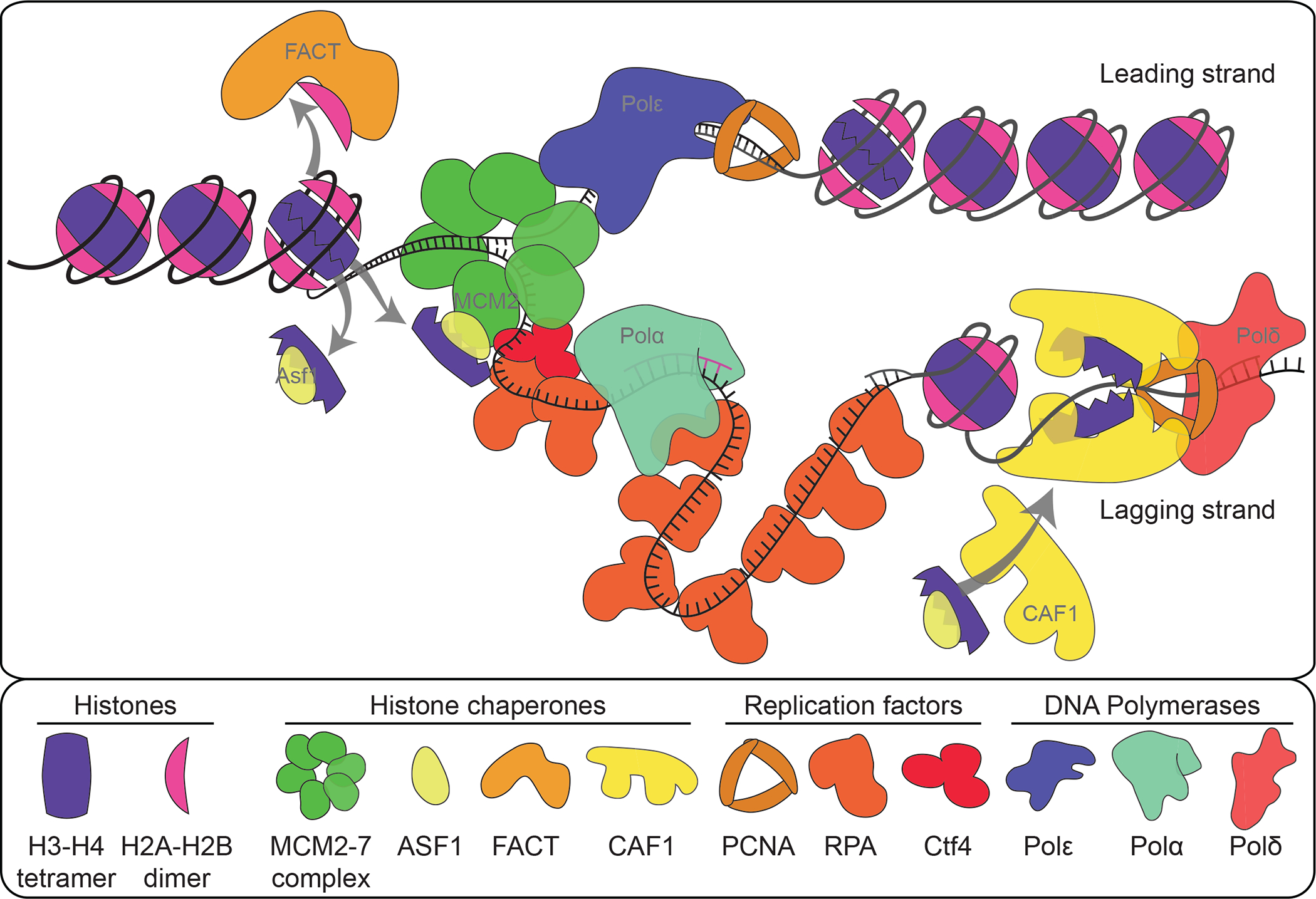
Histone chaperones and replisome components orchestrate disassembly of parental chromatin ahead of the replication fork. Concurrently, parental histones are shuttled behind the fork while new histones are recruited to stimulate chromatin restoration on newly synthesized DNA.
1.3. Parental histone recycling
Following chromatin disassembly and DNA unwinding, Replication Protein A (RPA) binds to the exposed ssDNA. RPA is located at the juncture between ssDNA and dsDNA, where it interacts with H3-H4 (75). Thus, RPA may help guide (H3-H4)2 reformation on newly synthesized dsDNA. Parental histone transfer behind the replication fork is also facilitated by replisome components (7). The two smallest subunits of the leading strand DNA Polymerase ε (Polε) form a stable dimer that directly binds to H3-H4 and promotes tetramer incorporation into DNA (7, 152). Recycling old histones onto the lagging strand is likely mediated through an interaction involving MCM2, Ctf4, and DNA Polymerase α (Polα). The amino-terminus of the catalytic Pol1 subunit of Polα contains a conserved histone binding motif whose mutation abolishes the interaction between Polα and histones (35). Although disrupting the interaction between Ctf4 and Polα has no effect on DNA synthesis, it does impair epigenome restoration potentially by inhibiting histone recycling to the lagging strand (35, 73).
1.4. New histone incorporation during replication
More current knowledge is about the mechanisms underlying new histone incorporation. Following their protein synthesis in the cytoplasm, new H3-H4 dimers are translocated into the nucleus where all non-chromosomal bound histones interact with Asf1 (43). Once in the nucleus, the Saccharomyces cerevisiae histone acetyltransferase Rtt109 (named CBP/p300 in fly and human) acetylates H3 at the 56th lysine (H3K56ac) when H3 binds to Asf1 (28, 46). Asf1 then delivers H3-H4 dimers to the heterotrimeric Chromatin Assembly Factor 1 (Caf1) complex. In yeast, in vitro studies showed that H3-H4 binding is mediated by a composite interface formed between the two largest Caf1 subunits, Cac1 and Cac2 (87). The conserved Asf1-H3-H4-Caf1 interaction is transient and mediated through the second largest subunit of the Caf1 complex (76, 93, 141).
At the replication fork, Caf1 associates with Proliferating Cell Nuclear Antigen (PCNA). The PCNA homotrimer is a component of the replication fork that supports DNA polymerase processivity (161). The largest Caf1 subunit interacts with PCNA and DNA through the PCNA Interacting Peptide (PIP) and Winged-Helix Domain (WHD) structures, respectively (139, 158). The interaction between Caf1 and PCNA brings Caf1-bound H3-H4 dimers into proximity of the newly synthesized DNA. There, two Caf1-H3-H4 complexes associate to initiate (H3-H4)2 tetramer incorporation in a DNA-dependent manner (88).
1.5. Chaperone interactions between specific histone populations
1.5.1. Histone variants
Histone variants provide flexibility in chromatin composition and distinguish functionally distinct epigenomic domains. Variants differ from their canonical counterparts due to slight variations in their primary amino acid sequences that alter nucleosomal structure and function. For example, the H3 histone variant CENP-A epigenetically marks the centromere regions. Certain histone chaperones recognize these unique differences, making them histone variant specific. Holliday Junction recognition protein (HJURP) is one example that specifically facilitates CENP-A deposition (86). Additionally, incorporation of H3.3 is mediated by Histone regulator A (HIRA) at transcriptionally active genes or by Death domain associated protein (DAXX) and ATRX at pericentromeric and telomeric regions (42). Other histone chaperones, like Asf1 and MCM2, recognize identical surfaces shared between canonical histones and histone variants. This may be essential for faithful epigenome restoration following DNA replication. For example, interaction between CENP-A, MCM2, and HJURP is required to retain centromere composition in the proper genomic context following DNA replication (155).
1.5.2. Post-translational modifications (PTMs)
Histone PTMs regulate histone structural properties to influence interactions with DNA, other nucleosomes, and other chromatin-associated factors as well as histone nuclear import (12, 160). PTMs such as acetylation, methylation, phosphorylation, and ubiquitylation, confer distinct molecular properties and can be different between old versus new histones. For example, acetylation of H4 at lysine 5 and 12 (H4K5,12ac) is enriched on new histones in many species. This modification regulates histone association with importin and mutants at these residues are defective in nuclear import (32, 157). Recently, this modification was used as a proxy for new histones in epigenomic studies to understand new histone deposition patterns at the replication fork (110). On the other hand, old histones are enriched with methylation marks such as H3K27me3 (3). Formation of PTMs like H3K9me3 and H3K27me3 begins during S-phase but become fully matured beyond one cell cycle (156). Thus, these methylation marks have been used as proxies for old histones.
As certain PTMs can distinguish populations of old versus new histones, these modifications could influence interactions between histones and their chaperones, a topic that has been reviewed previously (12). One notable PTM-mediated interaction is between H3 acetylation at the 56th lysine (H3K56ac) and chaperone Caf1. In S. cerevisiae, this modification increases binding affinity between newly synthesized H3 and Caf1 (71). Unlike other histone methylations, H3K9me1 is found in both cytoplasmic and nuclear histone fractions, suggesting it is associated with new H3 (78). Interestingly, an HP1Į-Caf1-SetDB1 complex facilitates mono-methylation of H3K9 (79). This interaction suggests a replication-coupled mechanism to provide H3K9me1 substrate for subsequent trimethylation in pericentric regions, which could ensure faithful propagation of pericentric heterochromatin following DNA replication. Whether and how other old versus new histone-specific modifications influence binding dynamics with their chaperones need further studies.
2. Phenotypes associated with compromised histone chaperone function
2.1. Impaired genome integrity in Chromatin Assembly Factor 1 mutants:
Studies on histone chaperone function are motivated by the hypothesis that unfaithful replication-coupled histone assembly could impair epigenetic memory and dysregulate cell fate. Since its discovery, the role of Caf1-mediated replication-coupled histone assembly has been investigated in many systems (132). One of the most fascinating phenotypes of compromised Caf1-mediated chromatin assembly is demonstrated in Caenorhabditis elegans (98). In this organism, mutation of H3-encoding his-9 gene at the His 113 residue (H113D) disrupts (H3-H4)2 formation and subsequent Caf1-mediated histone assembly. In the presence of the H113D mutant histone expression or when Caf1 is reduced by RNAi, specification of the MI neuronal fate is lost, likely due to impaired asymmetric epigenetic regulation. In other systems where Caf1-mediated chromatin assembly has been studied, dysregulation of heterochromatin is the most common phenotype. In Arabidopsis thaliana, the genes that encode homologs for the two larger replication-specific components of Caf1 are FASCIATA1 and FASCIATA2 (FAS1 and FAS2) (60, 105). In A. thaliana, loss of either gene has pleiotropic effects in the shoot and root apical meristems. These structures host highly organized adult stem cell populations capable of division and growth that give rise to the plant body and root tissue, respectively. In fasciata mutants, the ordered cellular organization of each meristem is disrupted with a loss of stable stem cell population (60). Within both the shoot and root apical meristems, abnormal expression of cell type-specific genes WUSCHEL (shoot) and SCARECROW (root) was observed, suggesting random gain/loss of gene expression that deteriorated over time (60). These results led to the hypothesis that the FAS complex is required for stable propagation of epigenetic information through DNA replication-coupled chromatin assembly. Later studies show that without FAS, silenced genes located in heterochromatin are derepressed (104). Further, fas1 and fas2 mutants exhibit an increase in DNA damage response and chromosomal aberrations like decondensed mitotic chromatin, chromosome bridges, fusions, and fragments. Altogether, these results demonstrate that the Caf1 complex plays an essential role in genome integrity in plants (142).
Compromised heterochromatin silencing is also observed in D. melanogaster mutants of the largest Caf1 subunit, Caf1-p180, an essential gene for development (133). Loss of one copy of Caf1-p180 enhances Polycomb mutant phenotypes and disrupts localization of the pericentric heterochromatin modification H3K9me3 (51, 133). In addition, these flies have increased sensitivity to double-strand DNA breaks (DSBs) and compromised DNA repair. A tissue-specific role for Caf1 was investigated in D. melanogaster adult ovaries. RNAi-mediated reduction of Caf1-p180 in female germ cells activates the DNA damage checkpoint, attributed in part to an overabundance of ssDNA at the rDNA locus. Interestingly, germ cells with reduced Caf1-p180 form cell fusions between the female GSCs and the differentiating germ cells, with the latter cells displaying mixed features of both stem and committed cell identities (23). The authors hypothesize that these fused cells arise from premature differentiation that causes impaired cell abscission during cytokinesis. However, under-replicated regions, like that at the rDNA locus, could form ultra-fine DNA bridges. These structures are DNA connections between sister chromatids that are undetectable by DNA dyes, which may underlie the abscission defect in the female germline (17).
Unlike other eukaryotes, the Caf1 complex is not essential in S. cerevisiae. However, loss of mating-type locus and telomere silencing is observed in Caf1 mutant yeast (33, 58). This phenotype is also detected in the pol30 mutant that affects PCNA function (161). The PCNA-Caf1 interaction facilitates histone assembly onto newly synthesized DNA in yeast. Thus, loss of this interaction could impede faithful chromatin reassembly during DNA synthesis, leading to such an epigenetic memory loss phenotype.
2.2. Replisome mutations and epigenetic memory
Structure-based studies have been integral to characterizing direct interactions between replisome components and histones. These studies motivated genome editing strategies to create precise mutations that impair histone binding with minimal effect on overall replication progression. This approach has improved our understanding of in vivo replication-coupled histone assembly by providing a molecular perspective on histone inheritance during DNA replication. One such example is the identification and subsequent site-directed mutagenesis of two Tyr residues in MCM2 that are necessary for histone binding (50, 117). Biochemical experiments identified an interaction between the two smallest subunits of Polε and H3-H4 that is conserved in humans and S. cerevisiae (POLE3-POLE4 in human and Dpb3-Dpb4 in yeast) (7). Finally, the Pol1 subunit of Polα interacts with H3-H4 via a highly conserved domain, where histone binding can be abolished by mutating just two residues within this domain (35, 73).
In S. cerevisiae, the CRASH (Cre-reported altered states of heterochromatin) assay is an elegant method for detecting transient expression of silenced genes. It relies on integration of the Cre recombinase sequence into the transcriptionally silent HML locus. Loss of silencing at the HML transiently expresses Cre, leading to site-specific recombination and a heritable RFP to GFP reporter switch. This assay has been used to test whether faithful inheritance of parental (H3-H4)2 is required to maintain the silenced state. Mutation of three Tyr residues to Ala in MCM2 (mcm2–3A) increases loss of the silenced state (122). The mcm2–3A and dpb3 double mutant has an even stronger effect. In another study, the CRASH assay measured silencing loss in mcm2–3A and pol1–2A2 mutant strains, where loss of silencing was detected in the single mutants but no enhancement was observed in the double mutant, suggesting that MCM2 and Pol1 function in the same pathway (73).
A role for replisome components in establishment of silenced states has also been studied. Here yeast cells are sorted into two populations: HMR silenced or expressed. Monitoring these populations until they reached an equilibrium of both states determines rates of silencing loss or gain. Again, the mcm2–3A and dpb3 double mutants have higher rates of silencing loss than the single mutants. Interestingly, dpb3 but not mcm2–3A also functions to establish the silenced state (122). In another study, memory of nucleosome position was tested in both the mcm2–3A and dpb3 mutant strains (123). For both mutants, a loss in nucleosome positional memory was observed. Altogether these studies indicate that faithful histone inheritance is mediated in part through replisome components and that this regulation is required to maintain genome integrity.
2.3. Replisome-mediated histone chaperoning at the replication fork
Genomic approaches provide a molecular view on how replisome components mediate parental histone transfer during replication-coupled chromatin assembly (see sidebar 1). Two analogous methods investigate the strand bias of histone deposition during DNA replication: sister chromatids after replication by DNA sequencing (SCAR-seq) and enrichment and sequencing of protein-associated nascent DNA (eSPAN) (110, 152, 153). These were the first studies to demonstrate replisome-mediated strand-specific incorporation of old versus new histones onto distinct replicating DNA strands. In mouse ESCs, SCAR-seq revealed MCM2 mediates deposition of parental H3 onto the lagging strand to produce balanced inheritance of old H3 between leading and lagging strands. In the mcm2–2A mutant, a pronounced bias in parental histone deposition to the leading strand was detectable (110). In contrast, using eSPAN in S. cerevisiae it has been found that Polİ facilitates parental histone transfer to the leading strand. Studies using eSPAN in both S. cerevisiae and mouse ESCs showed this leading strand bias is disrupted by knocking out the dpb3-dpb4 subunits of Polε (73, 153).
Sidebar 1:
Protein dynamics during DNA replication
DNA replication studies use incorporation of a thymidine analog like ethynyl-2’-deoxyuridine (EdU) to label nascent DNA. Combined with chromatin immunoprecipitation (ChIP), these methods define behavior of chromatin-bound proteins during DNA replication. Approaches like Isolation of proteins on nascent DNA, DNA-mediated Chromatin Pull-down, and Nascent Chromatin Capture identify proteins associated with replicating DNA (4, 62, 130). Through nascent DNA enrichment followed by protein detection, these studies defined spatial and temporal protein dynamics at forks, nascent, and mature chromatin. Inversely, protein enrichment with subsequent nascent DNA purification underlies SCAR-seq, eSPAN, and ChIP-N-ChaP (110, 152, 162). These methods identified strand inheritance biases in parental versus new histones and RNAPII. Using super-resolution microscopy, the chromatin fiber method corroborated genomic findings by visualizing proteins at a single replication fork in cells derived from a multicellular organism(148). Altogether, genomic methods provide insight into the replication proteome and histone dynamics. However, these approaches have yet to be applied in tissue due to low cell number, cell heterogeneity, and limited knowledge of replication origins. Tagmentation and low-input strand-specific DNA sequencing approaches make these studies amenable for small cell populations(74). Thus, characterizing protein dynamics in tissue during DNA replication, particularly throughout development and disease progression, is on the horizon.
Experiments in S. cerevisiae describe an axis of parental histone transfer to the lagging strand involving MCM2-Ctf4-PolĮ (38). The Ctf4 adapter protein is thought to connect the CMG helicase on the leading strand to PolĮ on the lagging strand. Loss of Ctf4 phenocopies the mcm2–3A mutant, showing impaired transfer of old H3 to the lagging strand. Further, mutations in Ctf4 that disrupt binding to Pol1, or mutations in Pol1 that abolish Ctf4 binding, also compromise parental H3-H4 transfer like that of the mcm2–3A mutant. However, these studies did not address whether PolĮ itself also facilitates parental histone transfer. This was recently tested using a modified eSPAN protocol that includes simultaneous chromatin digestion and adapter ligation (tagmentation) mediated by protein A-tagged transposase Tn5 (pA-Tn5) (14, 59). This study in mouse ESCs revealed that PolĮ facilitates (H3-H4)2 deposition onto lagging strands through a direct interaction. This interaction can be abolished via mutation of two conserved amino acids to Ala (POLA1–2A) (73). Here, the impact of replication complex-mediated histone transfer on mammalian chromatin integrity was also tested. In the POLA1–2A mutant, endogenous retroviral elements (ERVs), which are located in H3K9me3-enriched chromatin and typically silenced in mammalian cells (73), show increased expression corresponding to a decrease of H3K9me3. Notably, total H3K9me3 levels or expression of enzymes that regulate this PTM are unaffected, demonstrating for the first time in a mammalian cell type that replication components with inefficient histone binding negatively impact genomic integrity.
3. Mechanism and outcome of choosing symmetric versus asymmetric histone deposition
3.1. What factors influence this choice?
Context-specific interactions between histones and other proteins is a simple model explaining how histone incorporation mode is determined. In support of this, the mcm2–2A histone binding mutant alters histone deposition from a symmetric distribution between leading and lagging strands to biased leading strand incorporation (110). A similar outcome is found in the pol1–2A2 and POLA1–2A histone binding mutants (73). These results suggest MCM2 and PolĮ actively ensure balanced histone incorporation onto both strands. However, it is unlikely that in asymmetric systems, biased histone incorporation arises through mutational manipulation of these critical replisome factors. While the structural integrity of the replisome is critical, it is likely that a manipulatable and benign mechanism is involved: one that allows context-specific biased sister chromatid histone inheritance but does not dramatically affect the overall replication program.
While histone chaperones mediate histone shuttling at the replication fork, upstream mechanisms likely regulate histone incorporation pattern. Ongoing studies now illuminate that replication speed contributes to histone distribution between leading and lagging strands, raising a possible mechanism that couples nucleosome assembly and replication progression. Indeed, Caf1-mediated nucleosome assembly onto the lagging strand is required for Okazaki fragment maturation, as nucleosomes prevent continued processivity of DNA polymerase delta (Polį. In the absence of Caf1 or in Caf1 DNA-binding mutants, Polį is not halted by newly incorporated nucleosomes, causing increased strand displacement and longer Okazaki fragments (88, 131). Two recent studies provide support that replication fork speed is regulated by new histone supply. RNAi-mediated histone reduction results in benign fork slowing, which can progress to DNA damage if left uncorrected. DNA replication progression is also halted when the transfer of new histones from Asf1 to Caf1 to DNA is blocked by HIRA-B peptide expression (92). Additionally, single-molecule imaging of replicating DNA molecules from Xenopus laevis oocyte extracts shows that the efficiency of parental histone recycling depends on free histone availability such that high recycling efficiency occurs with low to no new histone supply (44).
It has long been considered that the orientation of gene transcription with respect to DNA replication can either impede or facilitate fork progression. A recent study demonstrates additional support that the direction of fork progression with respect to transcription orientation determines replication fork speed and chromatin-associated protein inheritance patterns. In this study, RNA Polymerase II (RNAPII) inheritance onto nascent daughter strands was investigated using Chromatin IP-Nascent Chromatin avidin Pulldown (ChIP-NChaP) (162). This study identified that old histones and RNAP II are more likely to be inherited by the strand that was first replicated. The strand replicated second is more likely to inherit new histones. Hence, genes transcribed in the same direction as DNA replication are synthesized first, enriched for old histones, and inherit RNAP II (162). This study proposes a two-step model where strands are synthesized asynchronously, depending on the direction of transcription. Old histones out-compete new histones for reassembly onto the strand synthesized first. RNAP II is also recycled to the first-replicated strand. Next, new histones repopulate the sister that is replicated second, at which point RNAP II also shifts to this strand.
If DNA synthesis rate affects histone deposition, then uncoupling leading versus lagging strand synthesis could alter the histone incorporation pattern between sister chromatids. A potential condition to uncouple leading versus lagging strand synthesis is replication stress. In support of this, strand-specific nascent DNA sequencing shows measurable differences in leading versus lagging strand synthesis rates following replication stress (125). Using the double-click method, a recent study found support for lagging strand biased new histone incorporation. The double-click method involves co-translational incorporation of methionine surrogate azidohomoalanine (AHA) into newly synthesized histones. This modified amino acid can be conjugated with biotin in a pull-down assay of new histones. Subsequently, nascent DNA is purified for strand-specific sequencing. This method provides a view of new histone incorporation pattern in the context of replication orientation (164). Using this strategy, lagging-strand bias of new histone incorporation was found to invert upon treatment with the replication inhibitor hydroxyurea (HU), which uncouples helicase movement with Polİ activity. The authors note that the observed differences in parental histone strand preference between mouse ESCs (leading strand, no HU) and S. cerevisiae (lagging strand, with HU), may be explained by differences in HU treatment (110, 153, 164). Therefore, further exploration of old versus new histone incorporation patterns under normal conditions will address how replication speed of leading versus lagging strand contributes to epigenome establishment during DNA replication.
3.2. What is the outcome from different choices at the replication fork?
How chromatin modifying complexes and other chromatin-associated factors behave during genome duplication have been tested in D. melanogaster embryos. Here it was found that components of the Trithorax (Trx) and Polycomb (Pc) group complexes remain associated with replicative DNA. Trx group protein Trx, as well as Pc group proteins Pc and E(z), all associate with PCNA at replicating loci in early embryos. Interestingly, parental histones containing H3K4me3 or H3K27me3 are replaced with unmethylated H3 at the fork. These results argue that histone modifying enzymes but not modified histones are important for epigenetic inheritance (109). However, early D. melanogaster embryonic cells are pluripotent and have a specialized cell cycle composed only of S- and M-phases, raising the possibility that this phenomenon is likely applicable to this unique developmental stage.
Gene regulatory factors may leverage differences in chromatin environments on sister chromatids to influence cell fate. For example, it is hypothesized that the asymmetric cell identities in C. elegans results from replication-mediated differential nucleosome densities on gene loci required to specify neuronal cell fate (98). Though not formally tested in this system, selective inheritance of sister chromatids was implicated. Recently, it was discovered in the D. melanogaster male GSCs that sister chromatids with distinct nucleosome densities are inherited asymmetrically (116). Further, in the early stages of mouse ESC differentiation, accumulation of post-replication H3K27me3 is delayed. It was proposed that because H3K27me3 condenses chromatin, delayed H3K27me3 accumulation allows lineage-specific chromatin factors access to DNA (108). Thus, differences in nucleosome density between sister chromatids established during DNA replication could underlie asymmetric cell fate decisions through differential accessibility of regulators for transcription and other chromatin-mediated biological processes.
II. Recognition of histone asymmetry
1. Centromere: cis-asymmetry driving biased chromosomal segregation
Centromeres are epigenetically defined chromosomal regions that provide an assembly platform for kinetochore proteins and attachment region for spindle microtubules (Figure 3A) (89). Centromeres ensure equal segregation of genetic material to daughter cells during mitosis (Figure 3B). They are critical for genome stability and normal development (89) as compromised centromere function could lead to chromosome segregation defects, genome instability, and nondisjunction, a major cause of various diseases (99). While the centromere has been long known, its molecular identity has only been revealed recently. The centromere is epigenetically defined by a H3 variant called Centromere protein A [CENP-A in human; centromere identifier (CID) in D. melanogaster, Histone H3 like centromere protein (HCP3) in C. elegans and centromeric histone H3 (CenH3) in A. thaliana] (11, 89, 95). For simplicity, we use CENP-A in this review, which is necessary and sufficient for defining the centromere and assembling the kinetochore. The huge kinetochore complexes are assembled on the centromere with two major parts, the inner kinetochore, and the outer kinetochore (Figure 3A). The inner kinetochore interacts with the centromere, whereas the outer kinetochore binds to microtubules ensuring faithful sister chromatid attachment and separation (Figure 3A–B). Centromeres also contribute to selective homologous chromosomal inheritance during meiosis, which will be discussed further.
Figure 3. Centromere structure:
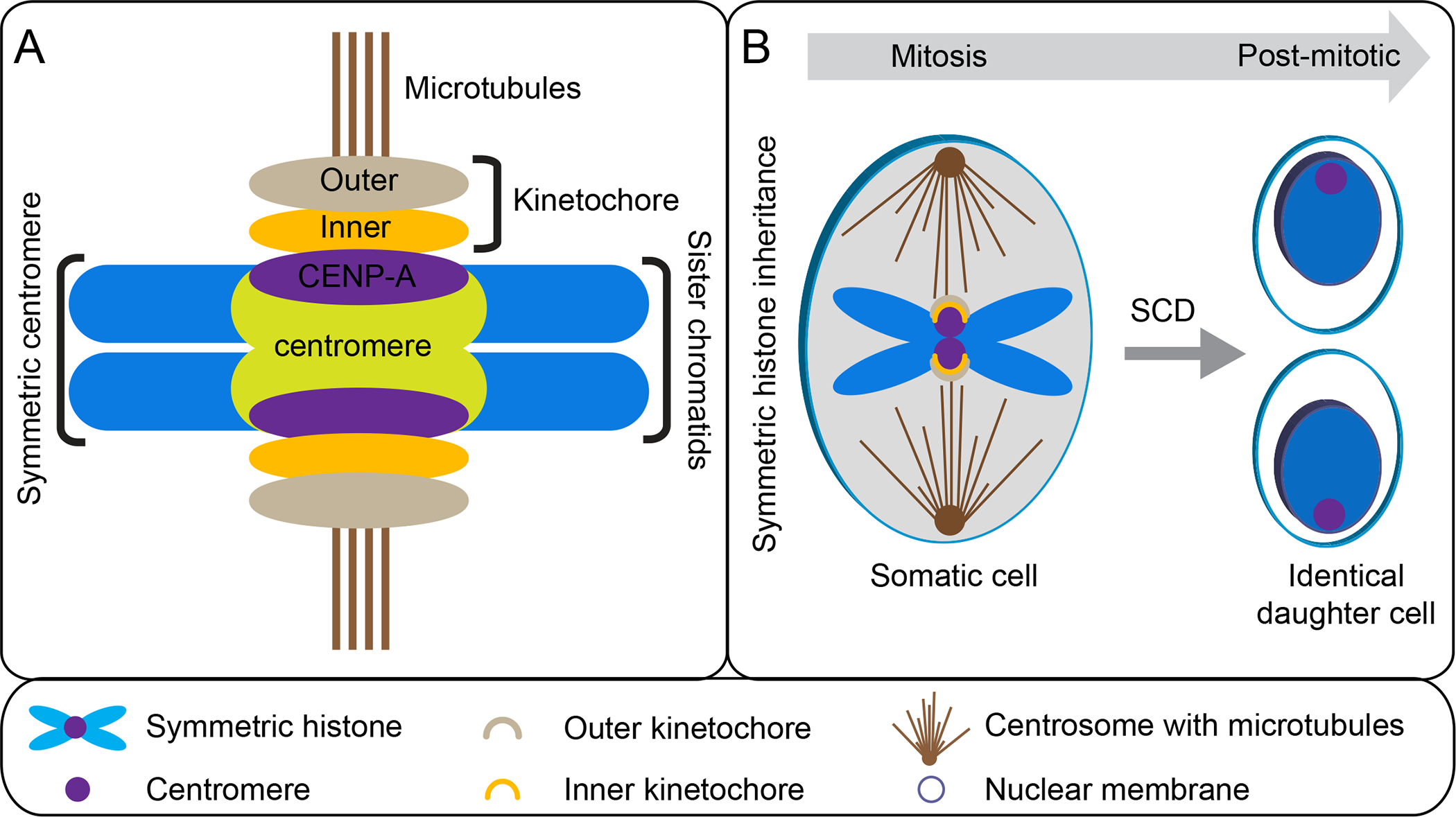
(A) Schematic of centromere, kinetochore, and microtubule in somatic cells. (B) The canonical function of the centromere during symmetric cell division (SCD).
Recent studies have revealed that centromeres are involved in diverse functions, such as (i) meiotic drive (ii) stem cell mitotic drive (iii) sensing and signaling, and (iv) chromosome scaling and shaping.
1.1. Centromeres in meiosis
1.1.1. Meiotic drive: Driving karyotype evolution
In 1957, Sandler & Novitski introduced the term “meiotic drive” during gametogenesis (121), whereby selfish genetic elements bias their transmission to gametes at a rate greater than predicted by Mendelian ratios (13). It is thought that meiotic drive is engaged in an evolutionary arms race where the genome is under pressure to evolve mechanisms that suppress the drive and prevent the deleterious effects of selfish elements(145, 154).
Gametogenesis is a critical process for selfish genetic elements to achieve this “drive” (13). In female oogenesis, completion of the second meiotic division produces a single gamete (i.e., egg) and three non-gamete polar bodies, which often undergo disintegration. Female meiotic drive occurs when a selfish genetic element increases its likelihood of transmission to the egg (10, 30). This process depends on three conditions: (1) Asymmetry in cell fates, such as egg versus polar body, offering the opportunity to have one ‘winner’(i.e., egg) and three ‘losers’ (i.e., polar bodies); (2) Heterozygosity, which refers to an individual with two different alleles; (3) Asymmetry in cellular structure such as the meiotic spindle.
1.1.2. Centromere drive
When the selfish genetic element discussed above involves centromere DNA, it is called “centromere drive” (67). During karyotype evolution, Robertsonian fusions form metacentric chromosomes when two telocentric chromosomes fuse at their centromeres. In D. americana, fused metacentric chromosomes preferentially segregate to the egg (136). In contrast, M. musculus domesticus heterozygotes with Robertsonian fusions bias transmission of the two unfused telocentric chromosomes to the egg (30). Further study has shown that more kinetochore proteins are recruited to the centromere of the telocentric chromosome compared with the metacentric chromosome, leading to their preferential segregation to the egg (21). This suggests that biased segregation is due to the ability to recruit kinetochore proteins to the centromere (Figure 4A).
Figure 4. Meiotic drive in mice:
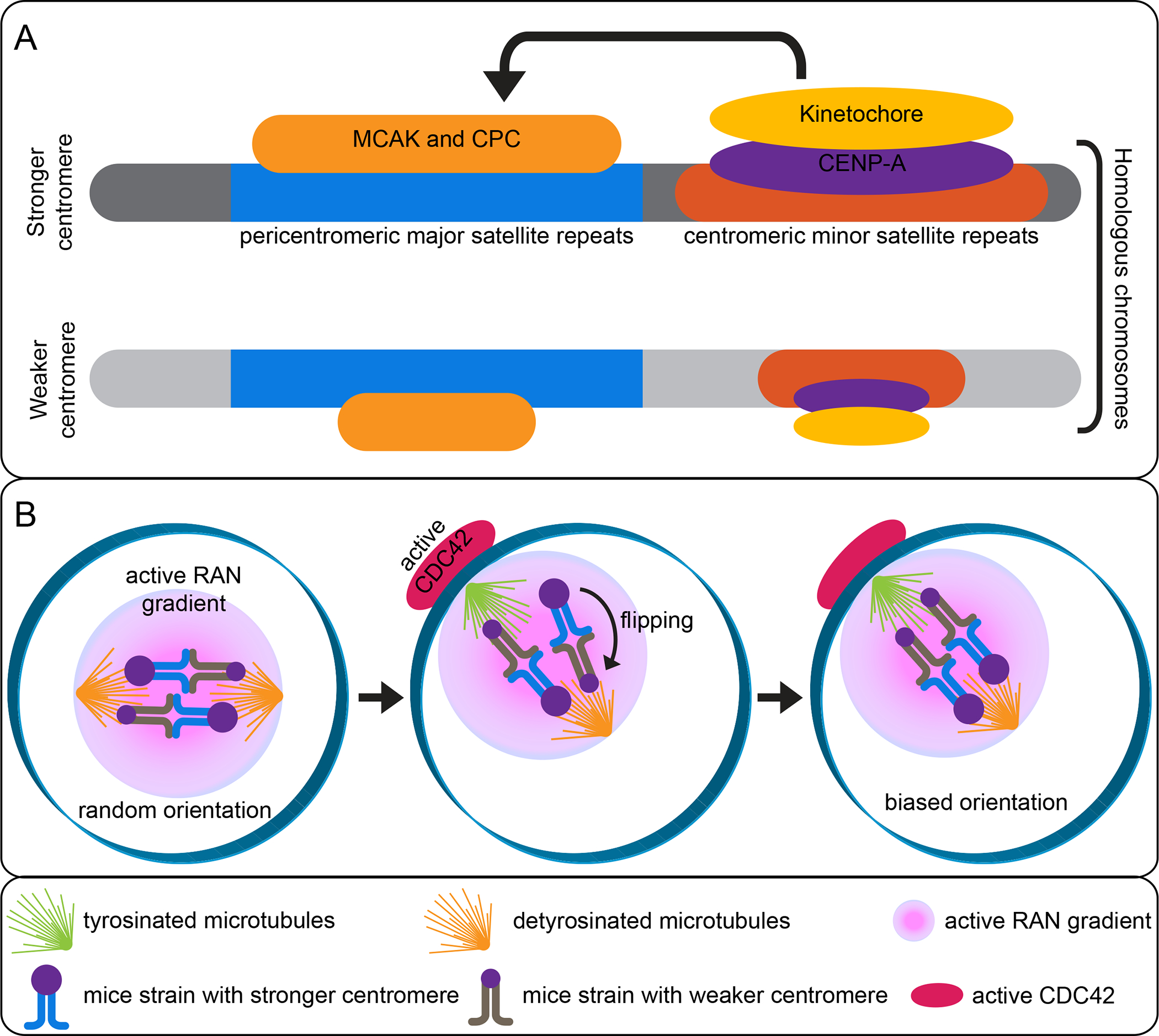
Mechanisms of centromere drive. (A) Stronger centromeres have more minor satellite repeats and build larger kinetochores that recruit more destabilizers than weaker centromeres. (B) The cortical positioning of the spindle induces microtubule tyrosination, leading to directional flipping until the stronger centromeres are preferentially oriented towards the egg side.
To understand how centromeres regulate kinetochore protein recruitment, intraspecific crosses were made between M. musculus domesticus strains carrying strong versus weak centromeres (52). In the hybrid progeny, the strong centromere recruits more kinetochore proteins and is preferentially segregated to the egg (Figure 4B). Importantly, the strong centromere contains 6–10 fold more minor satellite repeats than the weak centromere (Figure 4A), suggesting that the expansion of centromere repeats are partly responsible for recruiting more kinetochore proteins (52). It was found that the strong kinetochore interacts with the meiotic spindle to bias homologous chromosomal segregation (1). After the spindle is positioned close to the cortex, tyrosinaWHGĮ-tubulin becomes enriched toward the cortical side (Figure 4B). At this moment, a chromatin-based RAN activity gradient is established, producing cortical polarization and enrichment of active CDC42 (45, 72). Constitutively active or dominant negative mutations of either RAN or CDC42 abolishes spindle asymmetry without cortical polarization. Further, spindle asymmetry is essential for the strong centromere to preferentially orient towards the egg pole. These results suggest that the cortical CDC42 signals serve as a cue for the selfish centromere to distinguish the cortical pole and bias its inheritance to the egg during the first meiotic division. The strong centromere located toward the cortical side also has unstable microtubule attachments that facilitate flipping the bivalents towards the egg side (Figure 4B) (1). It was subsequently shown that more microtubule destabilizing factors, such as the chromosome passenger complex (CPC) and mitotic centromere-associated kinesin (MCAK), are recruited to the strong centromeres compared with the weak centromeres (Figure 4A) (2). Due to the unstable nature of tyrosinated microtubules compared with their de-tyrosinated counterparts, attachment of the strong centromere by the tyrosinated microtubules increases its likelihood to be flipped toward the cortical side (107, 129). Together, these studies provide a comprehensive molecular and cellular view on the mechanisms of centromere drive involving differences in centromere DNA sequence and spindle microtubules.
1.2. Stem cell mitotic drive: driving distinct cell fates
Introduced by Ranjan et al., stem cell “mitotic drive” refers to a bias in centromere protein transmission that occurs during asymmetric stem cell division resulting in asymmetric epigenetic inheritance (115). Mitotic drive could enable non-random sister chromatid segregation to bias inheritance of specific epigenetic information to the resulting daughter cells. It is proposed that this drive contributes to distinct cell fate determination.
The ‘silent sister hypothesis’ (SSH) proposes that the two sister chromatids have different epigenetic marks, particularly at the centromeres, to segregate non-randomly during ACD (68). According to this hypothesis, asymmetric epigenetic inheritance may lead to differential gene expression in the resulting two daughter cells, such as the expression of stemness genes and the silencing of differentiation genes in the stem cells. Some recent studies support this idea: For example, asymmetric histone inheritance could be global, such as in D. melanogaster male GSCs and intestinal stem cells (ISCs) (Figure 1A, 5A) (138, 163), or local, such as in D. melanogaster female GSCs and induced mouse ESCs (Figure 1B, 5B) (57, 80). In addition to the canonical histones, CENP-A also segregates asymmetrically during the ACD of D. melanogaster male and female GSCs as discussed above, as well as D. melanogaster ISCs that produces an ISC and differentiating enteroblast (EBs) (20, 39).
Figure 5. Asymmetric histone inheritance (global vs local):
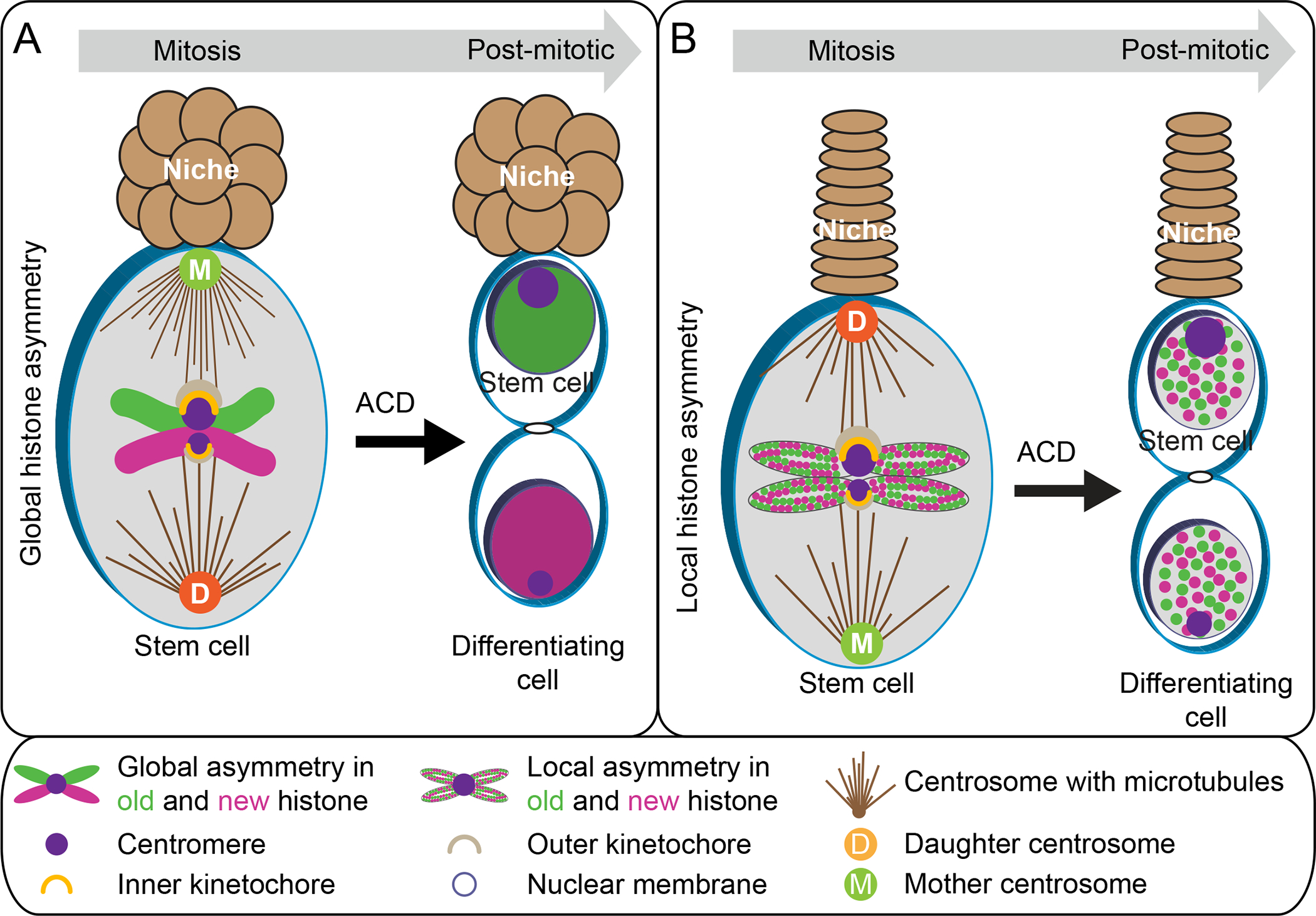
(A) Representation of the global asymmetric inheritance of histone H3 and H4 in D. melanogaster male GSC and ISC. (B) Representation of the local asymmetric inheritance of histone H3 and H4 in D. melanogaster female GSC and induced mouse ESCs.
1.2.1. Centromere: CENP-A asymmetry is critical for mitotic drive
Recent studies on non-random histone segregation during ACD of D. melanogaster male GSCs revealed that sister centromeres have different amounts of CENP-A in prometaphase. This makes one sister stronger than the other (Figure 5A, 6A) (115). Conversely, symmetrically dividing progenitor germ cells do not display such an asymmetry, suggesting that this is a stem cell and/or ACD specific phenomenon. During ACD, the strong centromeres are inherited by the self-renewing stem daughter cell, while the weak centromeres are inherited by the differentiating daughter cell gonialblast. Further, the strong centromere assembles more outer kinetochore component Ndc80 with an even higher degree of asymmetry compared to the asymmetry detected for centromere, indicating a relay mechanism from centromere asymmetry to kinetochore asymmetry (Figure 6A) (115). These observations gave the first direct evidence that sister centromeres with epigenetic differences could bias sister chromatid segregation.
Figure 6. Mitotic drive in D. melanogaster GSCs:
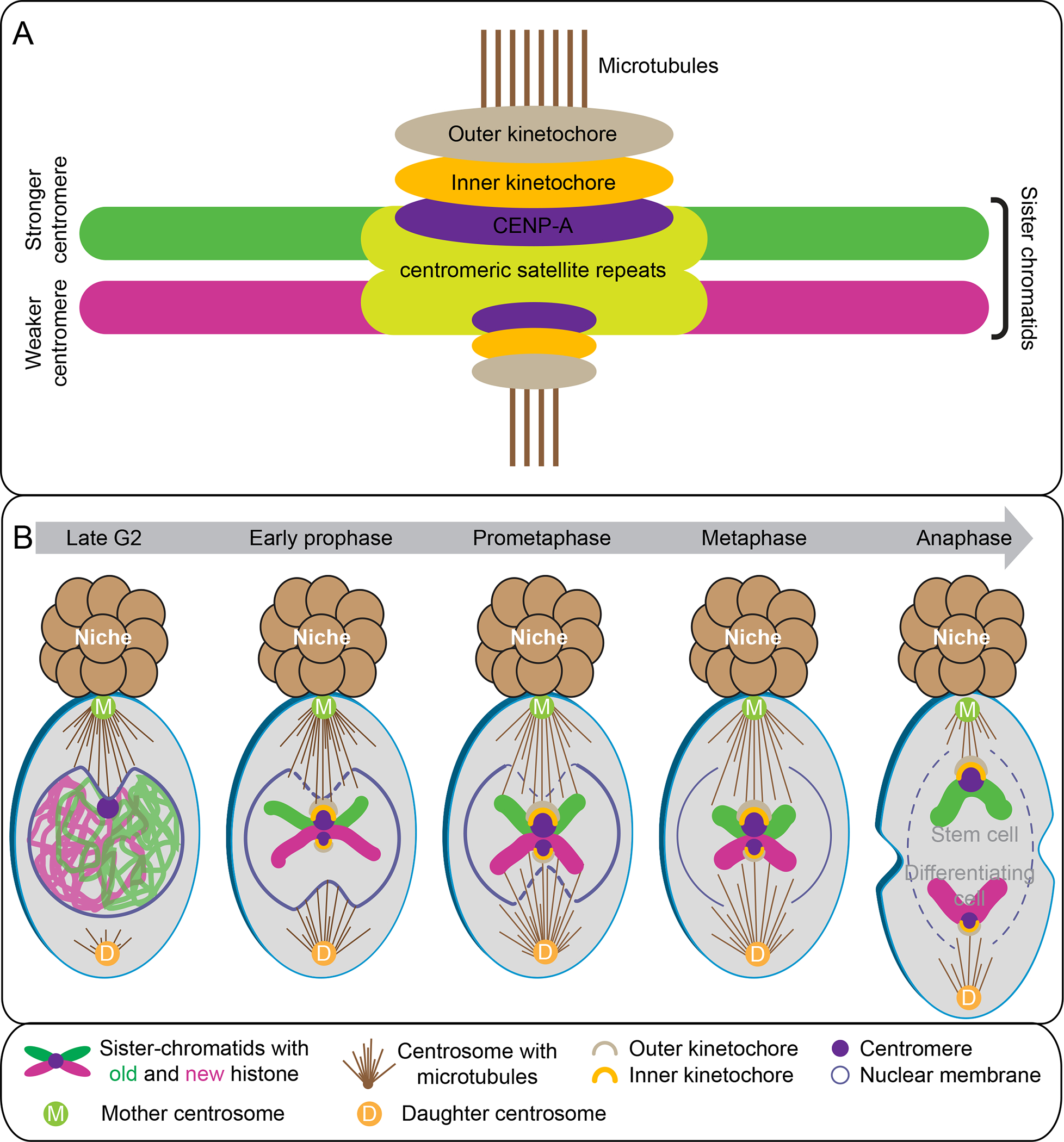
Mechanisms of the mitotic drive. (A) Stronger centromeres build larger kinetochores that bind more microtubules compared with weaker centromeres. (B) Sister chromatids with asymmetric histone epigenome in male GSCs. A temporal asymmetry of microtubules activity, NEBD, and CENP-A epigenetic asymmetry tightly coordinate to ensure non-random segregation.
Consistent with the male GSC results, asymmetric CENP-A between sister chromatids was also detected in D. melanogaster female GSCs with more CENP-A-containing sister chromatids inherited by the self-renewed GSCs (29). Similar to male GSCs, an asymmetric assembly of the inner kinetochore protein CENP-C was detected in female GSCs (15, 29). Further, knockdown of the CENP-A histone chaperone CAL1 disrupts CENP-A asymmetry in male GSCs, resulting in symmetric centromere formation and GSC loss (115). In female GSCs, overexpression of CENP-A and CAL1 together or CENP-A alone promotes GSC self-renewal while CAL1 overexpression promotes differentiation (29). Consistently, CENP-A asymmetry in female GSCs was lost following CENP-A and CAL1 co-overexpression. In addition, depletion of CENP-C in female GSCs enhances the centromere asymmetry and shift GSCs towards self-renewal tendency (15). These observations revealed CAL1 and CENP-C play important roles in asymmetric centromere assembly as well as in cell fate determination (15, 29, 115).
1.2.2. New CENP-A: incorporation timing matters for mitotic drive
New CENP-A must be incorporated during each cell cycle to maintain centromere identity following each cell division (89). After genome duplication, old CENP-A is diluted by half (53). However, unlike the canonical histones that are incorporated in a DNA replication-dependent manner, newly synthesized CENP-A is incorporated in a replication-independent manner, with different timing in different systems (126). In Hela cells, new CENP-A incorporation occurs in late telophase to early G1-phase (53). In D. melanogaster embryonic cells, new CENP-A is incorporated at anaphase (124). In D. melanogaster cultured cells, new CENP-A is incorporated at metaphase or early G1-phase (31, 94). Recently, new CENP-A incorporation in male D. melanogaster GSCs was shown to occur from mid-G2 phase to early mitosis and between DNA replication and prophase in female GSCs (29, 115).
CENP-A levels can be significantly reduced in human cells without disrupting mitotic functions, such as kinetochore assembly, microtubule attachment, or faithful sister chromatid segregation (9). One study revealed that once kinetochore assembly is complete, CENP-A at the centromere region is dispensable for mitosis (49). For these reasons, it was hypothesized that new CENP-A incorporation functions to maintain centromere identity, which should happen after mitosis but before centromere duplication in S-phase. Therefore, it is unclear why D. melanogaster GSCs incorporate new CENP-A prior to mitosis. One possible explanation is that GSCs use this assembly time point to establish or enhance centromere asymmetry, which would ensure asymmetric kinetochore establishment and mitotic drive. Consistent with this speculation, knocking down CAL1 in the adult male D. melanogaster GSCs leads to GSC loss, since in the absence of new CENP-A incorporation symmetric sister centromeres form (115). Since mitotic functions are not affected when compromising CAL1, it is proposed that the new CENP-A incorporation timing might only be critical for the mitotic drive in asymmetrically dividing cells.
Recent X-ray crystallography studies show that CAL1 binds to both CENP-A and CENP-C simultaneously. The N-terminus of CAL1 binds to CENP-A/H4 through multiple physical contacts and the C-terminus of CAL1 binds to CENP-C (91, 118). CAL1 is recruited to chromatin through an interaction with CENP-C that is bound to old CENP-A-containing nucleosomes. Subsequently, CAL1 recruits a new CENP-A-H4 dimer, promoting tetramer formation onto DNA. In this process, CAL1 also recruits CENP-C to bind the newly formed nucleosome. This positive feedback loop may help establish centromere asymmetry between sister chromatids.
1.3. Sensing and signaling: Centromere regulates chromosome condensation
A recent study in yeast demonstrates that chromosomes without centromeres (CEN−) and non-chromosomal DNA circles (cen− DNA circles) are unable to condense in mitosis (64). To investigate whether centromeres instruct chromosomal condensation in mitosis, the centromere on chromosome IV was flanked with loxP recombination sites to allow its excision and form CEN4*. Prior to excision, Chr IV condenses during mitosis; however, after CEN4* excision the chromosome failed to condense. This suggests that centromeres play a critical role in regulating chromosome condensation in mitosis. Furthermore, this study shows that centromeres promote chromosome condensation by recruiting kinases, such as Bub1 and Aurora B, which trigger chromosomal condensation through phosphorylation of key histone residues such as Ser121 of H2A and Ser10 of H3, respectively. To test the potential direct role of Aurora B recruitment in chromosome condensation, TetR-Aurora B fusion protein was targeted to a single TetO array on the CEN− Chr IV. Interestingly, ectopic recruitment of Aurora B fully rescued both contraction and compaction defects of CEN− Chr IV. Moreover, Aurora B-TetR promotes condensation of both CEN+ and CEN− Chr IV, irrespective of the cell-cycle stage. These data suggest that the recruitment of Aurora B onto a chromosome is sufficient for chromosomal condensation in the absence of centromere. In addition, Bub1 kinase, which is also required for chromosomal condensation, phosphorylates histone H2A on S121 (H2A-pS121). This recruits Shugoshin (Sgo1) to the pericentromeric chromatin. In PP2A rts1 mutant cells, PP2A lacks the B’ regulatory subunit and loses interaction with Sgo1. Since Sgo1 directly binds to and recruits phosphatase PP2A (84), the rts1 mutant cells produce constitutively condensed Chr IV due to derepression of the Bub1-independent function of Sgo1 in condensation. It has also been shown that Sgo1 and the deacetylase Hst2 facilitate spreading of the condensation signal to the chromosome arms for coordinated condensation. These results suggest that yeast cells license chromosome condensation in a centromere dependent manner. Only non-chromosomal DNA containing centromeres can recruit Aurora B activity and condense in mitosis. This work pinpoints an intriguing possibility that the centromere may control organization of an entire chromosome (64).
1.4. Chromosome scaling and shaping: Centromere scale chromosome in accordance with cell size
During metazoan development, early embryonic cells decrease in size due to multiple rounds of cleavages without growth of the embryo. Despite this, the genome size remains the same. The maximum length of the mitotic chromosomes has been shown to be no more than half of the spindle length (47), suggesting that the mitotic chromosomes scale in response to decreased cell size and shorter mitotic spindle. Studies using an RNA interference screen in a C. elegans strain carrying an exceptionally long chromosome identified CENP-A as a candidate modulator of chromosome size (66). Compromising CENP-A levels resulted in shorter chromosomes than controls. In addition, downregulation of KNL-2, the centromere licensing factor (81), reduced chromatin-associated CENP-A without affecting the entire CENP-A protein levels and led to abnormally short chromosomes. Because CENP-A is incorporated at discrete domains distributed periodically along the entire length of each chromosome (134), a linear array is formed that contributes to the rigidity of mitotic chromosomes in C. elegans. A severe loss of chromatin-associated CENP-A results in collapsed and round chromosomes instead of rod-shaped (82, 103), whereas a partial loss produced abnormally short but still rod-shaped chromosomes. These results showed a linear relationship between CENP-A levels and chromosome length, the less CENP-A the shorter chromosomes. Further, CENP-A incorporation is inversely correlated to germline transcription in C. elegans embryos (40). If germline transcription is disrupted by depleting the Argonaut protein CSR-1 (22), increased CENP-A is incorporated into individual chromosomes in early embryos and results in increased chromosome length. In addition, reducing CSR-1 resulted in increased CENP-A amount at the centromeric regions. Overall, these data suggest that CENP-A-containing nucleosomes can be modulated at the centromeric regions, as opposed to ectopic CENP-A deposition at the neocentromeric loci. Taken together, CENP-A could act as a ruler that regulates the length of chromosomes.
1.5. Centromere sequence: evolving for diverse functions of centromere
Previous studies have shown that the centromere is composed of tandemly repeated DNA sequences called satellite DNA, whose sequences differ widely among species (48). Human centromeres are composed of alpha satellite DNA Į6DW sequences with ~171-bp monomers, which are repeated to span over millions of base pairs (146). Different monomer subtypes form higher-order repeats (HORs); for instance, subtypes of monomers a,b,c can be repeated as abc-abc-abc (90). Each array can contain thousands of HORs, but kinetochore proteins only bind to a subset of HORs on each chromosome (90). HORs evolve rapidly, generating a high degree of polymorphism across individuals (96). Centromeric sequences and active kinetochore protein-binding sites are also found at pericentromeric regions, including smaller αSat monomer arrays that lack HORs (127).
Due to technical limitations and the inability to assemble long, repetitive sequences using short DNA sequencing reads, the human genome assembly has excluded about 10% of the sequences, most of which are at or around the highly repetitive centromere regions (102). The recent development of long-read sequencing and assembly methods have made it possible to produce the first complete human genome (102). Hence the composition of the human centromeric sequence on individual chromosomes has been deciphered (77, 97). More recently, detailed maps of previously unassembled centromeric and pericentromeric regions have also been studied using oligo-FISH, CRISPR-based experiments, and NTRprism, a newly developed versatile algorithm to discover and visualize the periodicity of satellite repeat (5). This study identified large- and small-scale variations in the organization and composition of active centromeres, which provide evidence for a layered expansion model of centromere evolution.
A recent study in D. melanogaster also revealed the sequence composition and organization of centromeres by combining long-read sequencing, chromatin immunoprecipitation for CENP-A, and high-resolution chromatin fiber imaging (18). In contrast to the previous model that the satellite repeats are the major functional components, this study revealed that centromeres form on islands of DNA sequences enriched in retroelements, which are flanked by large arrays of satellite repeats. While each centromere shows distinct size and arrangement of DNA elements, it has been shown that the G2/Jockey-3 retroelement is highly enriched in CENP-A chromatin and is the only shared element among all centromeres.
2. Pericentromere: a modulator of centromere function
In D. melanogaster male GSCs, phosphorylation of histone H3 at Thr 3 (H3T3P), a pericentromeric mitotic mark, plays a critical role in asymmetric histone inheritance (149). In particular, the old histone-enriched sister chromatid inherited by the GSC has more H3T3P than the new histone-enriched sister chromatid inherited by the differentiating daughter cell. Disrupting phosphorylation of H3T3 by introducing a dominant negative non-phosphorylatable Ala residue at this position (H3T3A) randomized old versus new H3 inheritance. Moreover, expression of the mutant H3T3A leads to the loss of GSCs and early germline tumor phenotypes, suggesting that proper histone inheritance is essential for both stem cell maintenance and proper differentiation of the daughter cell. These phenotypes could be enhanced by mutating the Haspin kinase responsible for phosphorylating Thr3 of H3. Consistently, another study showed that downregulation of Haspin (27) results in a 65% increase of CENP-A levels at centromeres and loss of CENP-A asymmetry in female GSCs (29). Together, these reports indicate that pericentromeric H3T3P could regulate centromere assembly, providing an additional mechanism that regulate asymmetric sister chromatid inheritance in GSCs.
3. Centrosome: establishment of trans asymmetry
The centrosome is the microtubule organization center (MTOC) in a cell. Centrosomes contain two centrioles and are surrounded by several pericentriolar matrix (PCM) proteins, which are necessary to establish the MTOC. Centrioles replicate once during interphase, where a daughter centriole forms in perpendicular to the mother centriole. Later, the two centrioles separate to produce two mature MTOCs that form a bipolar spindle (26). Consequently, such a replicative cycle results in an intrinsic asymmetry in centriole and centrosome age (54). During ACD, the centrosome shows an asymmetric inheritance pattern in different systems, with male D. melanogaster GSCs and M. musculus neural glial progenitor cells inheriting the mother centrosome (Figure 5A) (144, 151). In contrast, D. melanogaster female GSCs (120) and neuroblasts inherit the daughter centrosome (Figure 5B) (25, 54). While the D. melanogaster male and female GSCs inversely inherit the mother versus daughter centrosomes, both male and female GSCs inherit the centrosome with a higher MTOC activity. However, how increased MTOC activity is established at the stem cell side in both male and female GSCs, independent of centrosome age, remains unknown (Figure 6B). Studies in D. melanogaster neuroblasts show that the mitotic kinase Polo/Plk1 regulates differential MTOC activity through phosphorylating PCM proteins (24, 36, 101). The daughter centriole maintains Polo and the MTOC activity. In contrast, the mother centriole downregulates Polo and loses MTOC activity (55, 70, 113, 128). In addition, Polo-like kinase 4 (Plk4) contributes to biased MTOC activity, which regulates centriole duplication and triggers centriole activity (37). Interestingly, when the mother centriole initiates maturation and forms a second active MTOC, biased MTOC activity disappears in the mitotic neuroblasts.
It is unclear whether and how asymmetric inheritance of centrosomes contributes to asymmetric histone inheritance. Therefore, ongoing efforts are towards understanding the mechanism by which the mitotic machinery, in particular microtubules, recognizes sister centromere asymmetry to ensure non-random sister chromatid segregation. It was revealed that microtubule activity is temporally asymmetric in D. melanogaster male GSCs (115). In late G2 phase GSC, microtubules near the niche side are highly active, leading to a polarized nuclear envelope breakdown (NEBD) on this side at G2-M phase (Figure 6B). Later, the centrosome located toward the differentiating daughter cell side becomes active in prophase and induces NEBD at prometaphase. This polarized NEBD could promote preferential anchoring of the earlier active microtubules at the stem cell side to the stronger centromere (115). A superresolution live snapshot (SRLS) method was developed allowing high spatial and temporal imaging on live cells to visualize highly dynamic cellular processes, such as microtubule-kinetochore attachment (114). SRLS of D. melanogaster male GSCs revealed that the stronger sister centromere is attached by more microtubules emanating from the mother centrosome at the stem cell side in prometaphase (Figure 6A–B), likely due to the higher affinity of the stronger centromere with microtubules. Disruption of microtubule asymmetry or sister centromere asymmetry results in randomized sister chromatid segregation and GSC maintenance defects. These results indicate that the temporal asymmetry in microtubule activity and polarized NEBD facilitate biased microtubule-kinetochore attachment. Unlike meiotic drive where spindle re-orientation occurs (Figure 4B and table 1), in mitotic drive, the stronger centromere is stably anchored with more microtubules from one side of the spindle (1, 115). How this preferential attachment between stronger centromere and microtubules is maintained and how the spindle assembly checkpoint acts in this context are to be studied. Similar results have been reported in female GSCs (29). Together, these studies demonstrate that non-random sister chromatid segregation in D. melanogaster GSCs is mediated by asymmetries in the centromere, kinetochore, and microtubules (15, 29, 115). With the current knowledge, stem cell mitotic drive involves (i) centromere epigenetic asymmetry, and (ii) mitotic machinery asymmetry, such as microtubules, and kinetochore (Figure 6B and table 1) (16, 147). Future studies need to investigate the underlying molecular mechanism involved in the mitotic drive, such as the role of spindle assembly checkpoint proteins and microtubule destabilizers, such as CPC and MCAK, and more importantly, how histone asymmetries coordinate with the centromere asymmetry to bias sister chromatid segregation.
Table 1.
Comparison of mitotic and meiotic drive.
| Driving Factors | Meiotic Drive | Mitotic Drive |
|---|---|---|
| Chromosomal feature | Occurs between homologous chromosomes | Occurs between sister chromatids |
| Centromere feature | Asymmetric centromere sequence length | Symmetric centromere sequence length |
| Reason for centromere asymmetry | Due to more number of minor satellite repeats at centromere | Due to asymmetric level of CENP-A on sister-centromere |
| Type of selfish elements | Genetic: Satellite repeats and heterochromatin repeats | Epigenetic: CENP-A |
| Structural asymmetries | Post-translational modification asymmetry in microtubules | Temporal and quantitative asymmetry in microtubules |
| Cell division | Meiosis | Mitosis |
| Centromere orientation mechanisms | Uses destabilizers to flip and reorient centromeres, e.g MCAK and CPC | Remain elusive |
| Non-centromeric region involved | Heterochromatin containing repeats act as a neocentromere, e.g knob domain in Maize | Remain elusive |
| Frequency of occurrence | Occasionally when mouse lines with distinct centromere size breed | Almost always when stem cell undergoes ACD |
| Biological significance | Karyotype evolution | Cell fate determination and tissue homeostasis |
Recent studies highlight the functional consequences of biased centrosome inheritance. Biased centrosome inheritance was initially reported in S. cerevisiae where the old spindle pole body (SPB; the yeast equivalent of centrosome) was inherited by the mother cell instead of the daughter cell (bud) (106). Using a genetically engineered strain that inverts MTOC fate, compromised distribution of protein aggregates and aged mitochondria accumulate, leading to reduced replicative lifespan of the buds (83). In D. melanogaster male GSCs, disruption of the preferential attachment between microtubules and sister centromere leads to randomized segregation of sister chromatids (as described above). More recently, a strong monoastral spindle apparatus without a centrosome was observed in the early 8-cell stage M. musculus embryo. This monoastral spindle regulates an ACD producing the first lineage specification in M. musculus embryo between trophectoderm and inner cell mass of the blastocyst. In addition, asymmetric inheritances of microtubules and microtubule regulators were detected in this ACD. Disruption of the monoastral spindle formation leads to an imbalance of the inner and outer cell mass as well as defective lineage specification (111).
III. Execution of the asymmetric epigenome
1. Chromatin statuses: Nucleosome density differences and differential chromosomal compaction lead to distinct readout in the resulting daughter cells
Histones H3 and H4 carry a majority of the known PTMs that regulate many cellular functions, including 3D genome organization (8, 41, 56, 160). Therefore, it is conceivable that asymmetric H3 and H4 inheritance could regulate distinct cellular behaviors and properties. However, the downstream biological consequences of asymmetric histone inheritance were not well understood until recently. It has been demonstrated that old histone-enriched sister chromatids in D. melanogaster male GSCs have a higher nucleosome density compared to new histone-enriched sister chromatids. In addition, old histone-enriched sister chromatids condense further and prior to the new histone-enriched sister chromatids (116). This differential condensation is correlated with differential histone H3S10 phosphorylation, a modification known to be required for chromosomal condensation (65, 100). In D. melanogaster male GSCs during mitosis, the Ser 10 residue of old H3 is more phosphorylated than the Ser 10 on new H3 (116). Interestingly, these different chromatin features make the new histone-enriched sister chromatids more accessible to critical cell cycle regulators, such as the DNA replication initiation component Cdc6. Differential Cdc6 association promotes asynchronous cell cycle progression in the two resulting daughter cells: the differentiating daughter cell enters S-phase prior to the self-renewed GSC (116). However, it is unclear whether such differences in nucleosome density and condensation regulate other cellular processes that are critical for cell fate, such as gene expression (see sidebar 2). A very recent study in D. melanogaster male GSCs investigated the functional impact of locus specific homologous chromosome pairing at the stemness gene stat92E (6), which is required for GSC establishment and maintenance (61, 69, 140). Using OligoPaint fluorescent in situ hybridization (FISH), the interaction between homologous stat92E region was found to be tightly paired in GSCs but immediately loosened in gonialblasts and remained unpaired in differentiating germ cells (6). This change in pairing state at the stat92E locus is required for the downregulation of its expression during germ cell differentiation. Further, mis-regulation of asymmetric histone inheritance by expressing the H3T3A mutant histone, or by knocking down cal1 and haspin as described above (115, 149), leads to aberrant homologous chromosomal interaction at the stat92E gene locus and its abnormal expression. These results suggest that homologous chromosomal pairing is an intrinsically programmed process during ACD. Importantly, changed local pairing status may be common to alter gene activity during stem cell differentiation, providing a new paradigm for how inter-chromosomal interactions regulate gene activity.
Sidebar 2:
Visualizing chromatin accessibility with new technologies
The recent development of a transposase-based technique to visualize chromatin accessibility allows us to understand chromatin dynamics during development. Recently, an assay of transposase-accessible chromatin with visualization (ATAC- see) has been developed to directly image the accessible epigenome in situ, followed by deep sequencing to reveal the molecular identity of the accessible regions(19). ATAC-see has revealed cell-type-specific spatial organization of the accessible genome and cell-cycle dependent chromatin accessibility. More recently, a three-dimensional assay for transposase-accessible chromatin-photoactivated localization microscopy (3D ATAC-PALM) has been developed to image the accessible genome at a nanometer scale in situ(150). This method combines ATAC-see, PALM superresolution imaging, and lattice light-sheet microscopy. The study using 3D ATAC-PALM combined with genomic data showed that spatially segregated accessible chromatin domains (ACDs) enclose active chromatin and transcribed genes. In the future, combining imaging-based techniques with epigenomics would provide a general and scalable approach to decipher the spatiotemporal architecture of chromatin for gene regulation.
In summary, we proposed “cell cycle reprogramming” process that establishes, recognizes, and executes epigenetically distinct sister chromatids carrying different histones, histone variants, or other chromatin-associated factors, for producing distinct daughter cells. This process is initiated as early as in DNA replication and mitosis acts to distinguish such an asymmetry, with both steps occurring within the same cell. Only upon ACD, epigenetically distinct sister chromatids are inherited by the two daughter cells for executing different cellular and molecular functions: one of which is to reprogram the cell cycle along with differential gene expression. This hypothesis is opposed to the thought that cell fate determination is controlled by extrinsic cues that occur only after cells exit mitosis. However, exploring this intrinsic mechanism along with extrinsic cues will be an interesting topic for future studies.
Summary Points:
Varying degrees of asymmetric histone inheritance have been observed in diverse systems. Some demonstrate globally distinct inheritance patterns while other display regionalized inheritance patterns.
Chaperones facilitate stable propagation of epigenetic states through DNA-replication coupled chromatin restoration. The most common phenotypes of impaired histone-chaperone mediate chromatin assembly are dysregulation of heterochromatin domains, expression of silenced genes, and impaired genome integrity.
Direct interactions between histones and replisome components are required for parental histone recycling and may be involved in directing biased histone inheritance patterns onto sister chromatids.
Factors such as availability of histones at the replication fork and replication speed have been demonstrated to modulate histone incorporation into new DNA.
Centromere drive during meiosis involves differences in centromere DNA sequence and spindle asymmetry. Increased microtubule destabilizing factors are enriched on the stronger centromeres and cortical polarization of CDC42 serve as a signal to selfish elements to facilitate chromosome flipping, increasing likelihood of orienting the selfish element toward the future egg.
Stem cell mitotic drive: Asymmetric epigenetic inheritance may involve differences in sister centromere CENP-A proteins and temporally regulated microtubule activity, resulting in non-random sister chromatid inheritance. Adult stem cells inherit sister chromatids enriched with more CENP-A and kinetochore proteins.
Timing of CENP-A incorporation is cell type-specific and may be critical for mitotic drive in asymmetrically dividing cells resulting in asymmetric epigenetic inheritance.
Differences in nucleosome density produces differentially condensed sister chromatids during mitosis that may influence recruitment of gene regulatory factors to specific genomic loci during and after ACD.
Future Issues (8):
Is asymmetric histone inheritance a general mechanism to introduce cell diversity in adult tissue homeostasis and development?
What molecular factors contribute to biased histone segregations during chromatin assembly and what mechanisms do they employ to manipulate asymmetric versus symmetric outcomes onto sister chromatids?
How do differences in histone tails and modifications influence chaperone interactions and is this utilized during chromatin assembly to incorporate distinct populations of histones in appropriate genomic locations?
What is the relationship between replication fork speed, leading versus lagging strand synthesis, and histone segregation patterns on sister chromatids? How does histone supply contribute to this process?
How do asymmetries in histone patterns coordinate with centromere assembly to direct non-random sister chromatid inheritance?
How do asymmetries in centrosome age contribute to asymmetric histone inheritance and how is asymmetric microtubule activity established irrespective of mother or daughter centrosome?
How do asymmetric sister centromeres form on genetically identical sequence between sister chromatids in GSCs?
How do differences in nucleosome density and chromatin condensation during ACD regulate distinct cellular processes critical to cell fate determination?
Acknowledgements
We thank Chen lab members for insightful suggestions. Supported by ACS grant# 133950-PF-19-131-01-DMC (J.U.), NIGMS/NIH R35GM127075, NICHD/NIH R01HD102474, and the Howard Hughes Medical Institute (X.C.).
Terms and Definitions List:
- Asymmetric cell division
cellular division producing two genetically identical daughter cells with fates influenced by the information they inherit and microenvironment they reside
- Replication-coupled chromatin assembly
The process whereby parental and new histones incorporate into newly synthesized DNA during DNA replication to form nucleosomes
- Replication-dependent histones
Canonical histones H3, H4, H2A, and H2B, whose biosynthesis and incorporation into chromatin are linked to S-phase
- Replication-independent histones
Histone variants, like CENP-A and H3.3, whose synthesis and incorporation into chromatin is not linked to DNA replication
- Parental (old) histones
Histones synthesized during S-phase of the previous cell cycle, having experienced G2 and M-phase.
- New histones
Histones synthesized during S-phase of the current cell cycle
- Histone recycling
The process of deconstructing chromatin to facilitate parental histone transfer into newly synthesized DNA
- Epigenetic memory
Heritable regulation of gene expression that does not alter DNA sequence but is maintained through successive rounds of cell division
- Ultra-fine DNA bridges
Unresolved DNA entanglements connecting sister chromatids during mitotic segregation. They cannot be detected by typical DNA dyes
- Centromere
Epigenetically defined regions that serve as platforms for microtubule attachment to equally segregate genetic material
- Kinetochore
A huge protein complex that assembles on the centromere, which serves to attach the spindle during mitosis
- Meiotic drive
When a chromosomal element biases transmission to offspring during meiosis at higher frequencies than expected
- Polar body
Haploid cells formed as byproducts during oocyte meiosis that typically are not fertilized
- Robertsonian fusion
Chromosome rearrangement whereby the long arms of two telocentric chromosomes fuse producing one metacentric chromosome, losing the two short arms
- Stem cell mitotic drive
Non-random sister chromatid segregation during stem cell division involving asymmetric temporal microtubule activation and centromere attachment ensuring biased epigenetic inheritance
- Chromatin statuses
The 3D organization of chromatin can be local: gene or domain-specific; or global: chromatid or cell type-specific
- Chromosome condensation
Varies from highly decondensed and accessible in S-phase to a highly compact and inaccessible state in metaphase
- Nucleosome density
The distance between nucleosomes generally reflects nucleosome density, which could regulate chromatin state and gene expression
Footnotes
Competing financial interests: The authors declare no competing financial interests.
Literature cited:
- 1.Akera T, Chmátal L, Trimm E, Yang K, Aonbangkhen C, et al. 2017. Spindle asymmetry drives non-Mendelian chromosome segregation. Science (80-. ). 358(6363):668–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akera T, Trimm E, Lampson MA. 2019. Molecular Strategies of Meiotic Cheating by Selfish Centromeres. Cell. 178(5):1132–1144.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alabert C, Barth TK, Reverón-Gómez N, Sidoli S, Schmidt A, et al. 2015. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29(6):585–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, et al. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014 163. 16(3):281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altemose N, Logsdon GA, Bzikadze AV., Sidhwani P, Langley SA, et al. 2021. Complete genomic and epigenetic maps of human centromeres. bioRxiv. 2021.07.12.452052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antel M, Masoud M, Raj R, Pan Z, Li S, et al. 2021. Interchromosomal interaction of homologous Stat92E alleles regulates transcriptional switch during stem-cell differentiation. bioRxiv. 2021.11.08.467622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellelli R, Belan O, Pye VE, Clement C, Maslen SL, et al. 2018. POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol. Cell. 72(1):112–126.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhanu NV, Sidoli S, Garcia BA. 2016. Histone modification profiling reveals differential signatures associated with human embryonic stem cell self-renewal and differentiation. Proteomics. 16(3):448–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black BE, Brock MA, Bédard S, Woods VL, Cleveland DW. 2007. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. 104(12):5008–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandvain Y, Coop G. 2015. Sperm should evolve to make female meiosis fair. Evolution (N. Y). 69(4):1004–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. 1999. A histone-H3-like protein in C. elegans. Nature. 401(6753):547–48 [DOI] [PubMed] [Google Scholar]
- 12.Burgess RJ, Zhang Z. 2013. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burt A, Trivers RCN-Q 8. SB 2006. 2006. Genes in conflict: the biology of selfish genetic elements. Cambridge, Mass: Belknap Press of Harvard University Press. 602 pp. [Google Scholar]
- 14.Carter B, Ku WL, Kang JY, Hu G, Perrie J, et al. 2019. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun. 10(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carty BL, Dattoli AA, Dunleavy EM. 2020. CENP-C regulates centromere assembly, asymmetry and epigenetic age in Drosophila germline stem cells. Cell Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carty BL, Dunleavy EM. 2020. Centromere assembly and non-random sister chromatid segregation in stem cells. Essays Biochem. 64(2):223–32 [DOI] [PubMed] [Google Scholar]
- 17.Chan YW, West SC. 2018. A new class of ultrafine anaphase bridges generated by homologous recombination. Cell Cycle. 17(17):2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, et al. 2019. Islands of retroelements are major components of Drosophila centromeres. PLOS Biol. 17(5):e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, et al. 2016. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods. 13(12):1013–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Zion E. 2020. Asymmetric Histone Inheritance Regulates Stem Cell Fate in Drosophila Midgut. Social Science Research Network, Rochester, NY [Google Scholar]
- 21.Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, et al. 2014. Centromere Strength Provides the Cell Biological Basis for Meiotic Drive and Karyotype Evolution in Mice. Curr. Biol. 24(19):2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, et al. 2009. The Argonaute CSR-1 and Its 22G-RNA Cofactors Are Required for Holocentric Chromosome Segregation. Cell. 139(1):123–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clémot M, Molla-Herman A, Mathieu J, Huynh JR, Dostatni N. 2018. The replicative histone chaperone CAF1 is essential for the maintenance of identity and genome integrity in adult stem cells. Dev. 145(17): [DOI] [PubMed] [Google Scholar]
- 24.Conduit PT, Feng Z, Richens JH, Baumbach J, Wainman A, et al. 2014. The Centrosome-Specific Phosphorylation of Cnn by Polo/Plk1 Drives Cnn Scaffold Assembly and Centrosome Maturation. Dev. Cell. 28(6):659–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conduit PT, Raff JW. 2010. Cnn Dynamics Drive Centrosome Size Asymmetry to Ensure Daughter Centriole Retention in Drosophila Neuroblasts. Curr. Biol. 20(24):2187–92 [DOI] [PubMed] [Google Scholar]
- 26.Conduit PT, Wainman A, Raff JW. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16(10):611–24 [DOI] [PubMed] [Google Scholar]
- 27.Dai J, Sultan S, Taylor SS, Higgins JMG. 2005. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19(4):472–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das C, Lucia MS, Hansen KC, Tyler JK. 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 459(7243):113–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dattoli AA, Carty BL, Kochendoerfer AM, Morgan C, Walshe AE, Dunleavy EM. 2020. Asymmetric assembly of centromeres epigenetically regulates stem cell fate. J. Cell Biol. 219(4): Centromere epigenetic differences bias sister chromatid segregation during ACD in female GSCs.
- 30.de Villena FP-M, Sapienza C. 2001. Female Meiosis Drives Karyotypic Evolution in Mammals. Genetics. 159(3):1179–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH. 2012. The Cell Cycle Timing of Centromeric Chromatin Assembly in Drosophila Meiosis Is Distinct from Mitosis Yet Requires CAL1 and CENP-C. PLOS Biol. 10(12):e1001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. 2011. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol. Biol. Cell. 22(2):245–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enomoto S, Berman J. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12(2):219–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evano B, Khalilian S, Le Carrou G, Almouzni G, Tajbakhsh S. 2020. Dynamics of Asymmetric and Symmetric Divisions of Muscle Stem Cells In Vivo and on Artificial Niches. Cell Rep. 30(10):3195–3206.e7 [DOI] [PubMed] [Google Scholar]
- 35.Evrin C, Maman JD, Diamante A, Pellegrini L, Labib K. 2018. Histone H2A-H2B binding by Pol α in the eukaryotic replisome contributes to the maintenance of repressive chromatin. EMBO J. 37(19): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Z, Caballe A, Wainman A, Johnson S, Haensele AFM, et al. 2017. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell. 169(6):1078–1089.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gambarotto D, Pennetier C, Ryniawec JM, Buster DW, Gogendeau D, et al. 2019. Plk4 Regulates Centriole Asymmetry and Spindle Orientation in Neural Stem Cells. Dev. Cell. 50(1):11–24.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan H, Serra-Cardona A, Hua X, Zhou H, Labib K, et al. 2018. The Mcm2-Ctf4-Pol α Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol. Cell. 72(1):140–151.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García del Arco A, Edgar BA, Erhardt S. 2018. In Vivo Analysis of Centromeric Proteins Reveals a Stem Cell-Specific Asymmetry and an Essential Role in Differentiated, Non-proliferating Cells. Cell Rep. 22(8):1982–93 [DOI] [PubMed] [Google Scholar]
- 40.Gassmann R, Rechtsteiner A, Yuen KW, Muroyama A, Egelhofer T, et al. 2012. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 484(7395):534–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldberg AD, Allis CD, Bernstein E. 2007. Epigenetics: A Landscape Takes Shape. Cell. 128(4):635–38 [DOI] [PubMed] [Google Scholar]
- 42.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 140(5):678–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 17(2):301–11 [DOI] [PubMed] [Google Scholar]
- 44.Gruszka DT, Xie S, Kimura H, Yardimci H. 2020. Single-molecule imaging reveals control of parental histone recycling by free histones during DNA replication. Sci. Adv. 6(38):330–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halet G, Carroll J. 2007. Rac Activity Is Polarized and Regulates Meiotic Spindle Stability and Anchoring in Mammalian Oocytes. Dev. Cell. 12(2):309–17 [DOI] [PubMed] [Google Scholar]
- 46.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 315(5812):653–55 [DOI] [PubMed] [Google Scholar]
- 47.Hara Y, Iwabuchi M, Ohsumi K, Kimura A. 2013. Intranuclear DNA density affects chromosome condensation in metazoans. Mol. Biol. Cell. 24(15):2442–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henikoff S, Ahmad K, Malik HS. 2001. The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA. Science (80-. ). [DOI] [PubMed] [Google Scholar]
- 49.Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, et al. 2016. CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep. 17(9):2394–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Strømme CB, Saredi G, Hödl M, Strandsby A, et al. 2015. A unique binding mode enables MCM2 to chaperone histones H3–H4 at replication forks. Nat. Struct. Mol. Biol. 22(8):618–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H, Yu Z, Zhang S, Liang X, Chen J, et al. 2010. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J. Cell Sci. 123(Pt 16):2853–61 [DOI] [PubMed] [Google Scholar]
- 52.Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, et al. 2017. Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Curr. Biol. 27(15):2365–2373.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jansen LET, Black BE, Foltz DR, Cleveland DW. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176(6):795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Januschke J, Llamazares S, Reina J, Gonzalez C. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2(1):243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Januschke J, Reina J, Llamazares S, Bertran T, Rossi F, et al. 2013. Centrobin controls mother–daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15(3):241–48 [DOI] [PubMed] [Google Scholar]
- 56.Jenuwein T, Allis CD. 2001. Translating the Histone Code. Science (80-. ). 293(5532):1074–80 [DOI] [PubMed] [Google Scholar]
- 57.Kahney EW, Zion EH, Sohn L, Viets-Layng K, Johnston R, Chen X. 2021. Characterization of histone inheritance patterns in the Drosophila female germline. EMBO Rep. 22(7):e51530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaufman PD, Kobayashi R, Stillman B. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11(3):345–57 [DOI] [PubMed] [Google Scholar]
- 59.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, et al. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. 2001. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 104(1):131–42 [DOI] [PubMed] [Google Scholar]
- 61.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. 2001. Stem Cell Self-Renewal Specified by JAK-STAT Activation in Response to a Support Cell Cue. Science (80-. ). 294(5551):2542–45 [DOI] [PubMed] [Google Scholar]
- 62.Kliszczak AE, Rainey MD, Harhen B, Boisvert FM, Santocanale C. 2011. DNA mediated chromatin pull-down for the study of chromatin replication. Sci. Reports 2011 11. 1(1):1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kouzarides T 2007. Chromatin Modifications and Their Function. Cell. 128(4):693–705 [DOI] [PubMed] [Google Scholar]
- 64. Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y. 2018. Centromeres License the Mitotic Condensation of Yeast Chromosome Arms. Cell. 175(3):780–795.e15 The centromere regulates chromosome condensation state during mitosis.
- 65.Ladouceur A-M, Ranjan R, Maddox PS. 2011. Cell Size: Chromosomes Get Slapped by a Midzone Ruler. Curr. Biol. 21(10):R388–90 [DOI] [PubMed] [Google Scholar]
- 66. Ladouceur A-M, Ranjan R, Smith L, Fadero T, Heppert J, et al. 2017. CENP-A and topoisomerase-II antagonistically affect chromosome length. J. Cell Biol. 216(9):2645–55 CENP-A nucleosome level can be manipulated within the centromere region.
- 67.Lampson MA, Black BE. 2017. Cellular and Molecular Mechanisms of Centromere Drive. Cold Spring Harb. Symp. Quant. Biol. 82:249–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lansdorp PM. 2007. Immortal Strands? Give Me a Break. Cell. 129(7):1244–47 [DOI] [PubMed] [Google Scholar]
- 69.Leatherman JL, Di Nardo S. 2008. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 3(1):44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lerit DA, Rusan NM. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202(7):1013–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, et al. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 134(2):244–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li R, Albertini DF. 2013. The road to maturation: somatic cell interaction and selforganization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 14(3):141–52 [DOI] [PubMed] [Google Scholar]
- 73.Li Z, Hua X, Serra-Cardon A, Xu X, Gan S, et al. 2020. DNA polymerase α interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci. Adv. 6(35): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z, Hua X, Serra-Cardon A, Xu X, Gan S, et al. 2020. DNA polymerase α interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci. Adv. 6(35):eabb5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu S, Xu Z, Leng H, Zheng P, Yang J, et al. 2017. RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science (80-. ). 355(6323):415–20 [DOI] [PubMed] [Google Scholar]
- 76.Liu WH, Roemer SC, Port AM, Churchill ME. 2017. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 45(16):9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Logsdon GA, Vollger MR, Hsieh P, Mao Y, Liskovykh MA, et al. 2021. The structure, function and evolution of a complete human chromosome 8. Nature. 593(7857):101–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. 2006. PTMs on H3 Variants before Chromatin Assembly Potentiate Their Final Epigenetic State. Mol. Cell. 24(2):309–16 [DOI] [PubMed] [Google Scholar]
- 79.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, et al. 2009. The HPlα–CAF1–SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10(7):769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma B, Trieu TJ, Cheng J, Zhou S, Tang Q, et al. 2020. Differential Histone Distribution Patterns in Induced Asymmetrically Dividing Mouse Embryonic Stem Cells. Cell Rep. 32(6): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. 2007. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176(6):757–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maddox PS, Portier N, Desai A, Oegema K. 2006. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc. Natl. Acad. Sci. 103(41):15097–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manzano-López J, Matellán L, Álvarez-Llamas A, Blanco-Mira JC, Monje-Casas F. 2019. Asymmetric inheritance of spindle microtubule-organizing centres preserves replicative lifespan. Nat. Cell Biol. 21(8):952–65 [DOI] [PubMed] [Google Scholar]
- 84.Marston AL. 2014. Shugoshins: Tension-Sensitive Pericentromeric Adaptors Safeguarding Chromosome Segregation. Mol. Cell. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008 911. 9(11):843–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mattiroli F, D’Arcy S, Luger K. 2015. The right place at the right time: chaperoning core histone variants. EMBO Rep. 16(11):1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mattiroli F, Gu Y, Balsbaugh JL, Ahn NG, Luger K. 2017. The Cac2 subunit is essential for productive histone binding and nucleosome assembly in CAF-1. Sci. Rep. 7(1):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mattiroli F, Gu Y, Yadav T, Balsbaugh JL, Harris MR, et al. 2017. DNA-mediated association of two histone-bound complexes of yeast chromatin assembly factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. Elife. 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKinley KL, Cheeseman IM. 2016. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17(1):16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McNulty SM, Sullivan BA. 2018. Alpha satellite DNA biology: finding function in the recesses of the genome. Chromosom. Res. An Int. J. Mol. Supramol. Evol. Asp. Chromosom. Biol. 26(3):115–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Medina-Pritchard B, Lazou V, Zou J, Byron O, Abad MA, et al. 2020. Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1. EMBO J. 39(7):e103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, et al. 2014. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 204(1):29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mello JA, Silljé HHW, Roche DMJ, Kirschner DB, Nigg EA, Almouzni G. 2002. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3(4):329–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. 2011. Assembly of Drosophila Centromeric Chromatin Proteins during Mitosis. PLOS Genet. 7(5):e1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. 2011. Drosophila CENH3 Is Sufficient for Centromere Formation. Science (80-. ). [DOI] [PubMed] [Google Scholar]
- 96.Miga KH. 2019. Centromeric Satellite DNAs: Hidden Sequence Variation in the Human Population. Genes (Basel). 10(5):E352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, et al. 2020. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 585(7823):79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakano S, Stillman B, Horvitz HR. 2011. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell. 147(7):1525–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Naylor RM, van Deursen JM. 2016. Aneuploidy in Cancer and Aging. Annu. Rev. Genet. 50:45–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neurohr G, Naegeli A, Titos I, Theler D, Greber B, et al. 2011. A Midzone-Based Ruler Adjusts Chromosome Compaction to Anaphase Spindle Length. Science (80-. ). 332(6028):465–68 [DOI] [PubMed] [Google Scholar]
- 101.Novak ZA, Wainman A, Gartenmann L, Raff JW. 2016. Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes. Dev. Cell. 37(6):545–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV., et al. 2021. The complete sequence of a human genome. bioRxiv. 2021.05.26.445798 [Google Scholar]
- 103.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. 2001. Functional Analysis of Kinetochore Assembly in Caenorhabditis elegans. J. Cell Biol. 153(6):1209–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ono T, Kaya H, Takeda S, Abe M, Ogawa Y, et al. 2006. Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells. 11(2):153–62 [DOI] [PubMed] [Google Scholar]
- 105.Ottoline Leyser HM, Furner IJ. 1992. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 116(2):397–403 [Google Scholar]
- 106.Pereira G, Tanaka TU, Nasmyth K, Schiebel E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p–Bub2p checkpoint protein complex. EMBO J. 20(22):6359–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peris L, Wagenbach M, Lafanechère L, Brocard J, Moore AT, et al. 2009. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185(7):1159–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Petruk S, Cai J, Sussman R, Sun G, Kovermann SK, et al. 2017. Delayed Accumulation of H3K27me3 on Nascent DNA Is Essential for Recruitment of Transcription Factors at Early Stages of Stem Cell Differentiation. Mol. Cell. 66(2):247–257.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, et al. 2012. TrxG and PcG Proteins but Not Methylated Histones Remain Associated with DNA through Replication. Cell. 150(5):922–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, et al. 2018. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science. 361(6409):1389–92 [DOI] [PubMed] [Google Scholar]
- 111.Pomp O, Lim HYG, Skory RM, Moverley AA, Tetlak P, et al. 2022. A monoastral mitotic spindle determines lineage fate and position in the mouse embryo. Nat. Cell Biol, pp. 1–13 [DOI] [PubMed] [Google Scholar]
- 112.Ramachandran S, Henikoff S. 2015. Replicating nucleosomes. Sci. Adv. 1(7):e1500587–e1500587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramdas Nair A, Singh P, Salvador Garcia D, Rodriguez-Crespo D, Egger B, Cabernard C. 2016. The Microcephaly-Associated Protein Wdr62/CG7337 Is Required to Maintain Centrosome Asymmetry in Drosophila Neuroblasts. Cell Rep. 14(5):1100–1113 [DOI] [PubMed] [Google Scholar]
- 114.Ranjan R, Chen X. 2021. Super-resolution live cell imaging of subcellular structures. J. Vis. Exp. 2021(167):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ranjan R, Snedeker J, Chen X. 2019. Asymmetric Centromeres Differentially Coordinate with Mitotic Machinery to Ensure Biased Sister Chromatid Segregation in Germline Stem Cells. Cell Stem Cell. 25(5):666–81 Centromere epigenetic differences bias sister chromatid segregation during ACD in male GSCs.
- 116. Ranjan R, Snedeker J, Wooten M, Chu C, Bracero S, et al. 2021. Differential condensation of sister chromatids coordinates with Cdc6 to ensure distinct cell cycle progression in Drosophila male germline stem cell lineage. bioRxiv. 2021.03.08.434490 Global nucleosome density and chromosome condensation dictate cell cycle progression in daughter cells.
- 117.Richet N, Liu D, Legrand P, Velours C, Corpet A, et al. 2015. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 43(3):1905–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Roure V, Medina-Pritchard B, Lazou V, Rago L, Anselm E, et al. 2019. Reconstituting Drosophila Centromere Identity in Human Cells. Cell Rep. 29(2):464–479.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rowlands H, Dhavarasa P, Cheng A, Yankulov K. 2017. Forks on the Run: Can the Stalling of DNA Replication Promote Epigenetic Changes? Front. Genet. 8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salzmann V, Chen C, Chiang CYA, Tiyaboonchai A, Mayer M, Yamashita YM. 2014. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol. Biol. Cell. 25(2):267–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sandler L, Novitski E. 1957. Meiotic Drive as an Evolutionary Force. Am. Nat. 91(857):105–10 [Google Scholar]
- 122.Saxton DS, Rine J. 2019. Epigenetic memory independent of symmetric histone inheritance. Elife. 8:1–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schlissel G, Rine J. 2019. The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 116(41):20605–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schuh M, Lehner CF, Heidmann S. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into Centromeres during Early Embryonic Anaphase. Curr. Biol. 17(3):237–43 [DOI] [PubMed] [Google Scholar]
- 125.Serra-Cardona A, Yu C, Zhang X, Hua X, Yao Y, et al. 2021. A mechanism for Rad53 to couple leading- And lagging-strand DNA synthesis under replication stress in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 118(38): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shelby RD, Monier K, Sullivan KF. 2000. Chromatin Assembly at Kinetochores Is Uncoupled from DNA Replication. J. Cell Biol. 151(5):1113–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA. 2009. The Evolutionary Origin of Man Can Be Traced in the Layers of Defunct Ancestral Alpha Satellites Flanking the Active Centromeres of Human Chromosomes. PLOS Genet. 5(9):e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Singh P, Ramdas Nair A, Cabernard C. 2014. The Centriolar Protein Bld10/Cep135 Is Required to Establish Centrosome Asymmetry in Drosophila Neuroblasts. Curr. Biol. 24(13):1548–55 [DOI] [PubMed] [Google Scholar]
- 129.Sirajuddin M, Rice LM, Vale RD. 2014. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16(4):335–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25(12):1320–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smith DJ, Whitehouse I. 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 483(7390):434–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Smith S, Stillman B. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 58(1):15–25 [DOI] [PubMed] [Google Scholar]
- 133.Song Y, He F, Xie G, Guo X, Xu Y, et al. 2007. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev. Biol. 311(1):213–22 [DOI] [PubMed] [Google Scholar]
- 134.Steiner FA, Henikoff S. 2014. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. Elife. 3:e02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stewart-Morgan KR, Petryk N, Groth A. 2020. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 2020 224. 22(4):361–71 [DOI] [PubMed] [Google Scholar]
- 136.Stewart NB, Ahmed-Braimah YH, Cerne DG, McAllister BF. 2019. Female meiotic drive preferentially segregates derived metacentric chromosomes in Drosophila [Google Scholar]
- 137.Sun Z, Tang Y, Zhang Y, Fang Y, Jia J, et al. 2021. Joint single-cell multiomic analysis in Wnt3a induced asymmetric stem cell division. Nat. Commun. 2021 121. 12(1):1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tran V, Lim C, Xie J, Chen X. 2012. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science (80-. ). 338(6107):679–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsirkas I, Dovrat D, Lei Y, Kalyva A, Lotysh D, et al. 2021. Cac1 WHD and PIP domains have distinct roles in replisome progression and genomic stability. Curr. Genet. 67(1):129–39 [DOI] [PubMed] [Google Scholar]
- 140.Tulina N, Matunis E. 2001. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science (80-. ). 294(5551):2546–49 [DOI] [PubMed] [Google Scholar]
- 141.Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, et al. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21(19):6574–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Varas J, Santos JL, Pradillo M. 2017. The absence of the arabidopsis chaperone complex CAF-1 produces mitotic chromosome abnormalities and changes in the expression profiles of genes involved in DNA repair. Front. Plant Sci. 8:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang H, Wang M, Yang N, Xu RM. 2015. Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell. 6(9):693–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang X, Tsai J-W, Imai JH, Lian W-N, Vallee RB, Shi S-H. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 461(7266):947–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Werren JH. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. 108(Supplement 2):10863–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Willard HF. 2011. The genomics of long tandem arrays of satellite DNA in the human genome. Genome [DOI] [PubMed] [Google Scholar]
- 147. Wooten M, Ranjan R, Chen X. 2020. Asymmetric Histone Inheritance in Asymmetrically Dividing Stem Cells. Trends Genet. 36(1):30–43 D. melanogaster testis chromatin fibers display strand-specific histone incorporation at replication forks.
- 148.Wooten M, Snedeker J, Nizami ZF, Yang X, Ranjan R, et al. 2019. Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat. Struct. Mol. Biol. 26(8):732–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xie J, Wooten M, Tran V, Chen B-C, Pozmanter C, et al. 2015. Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell. 163(4):920–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xie L, Dong P, Chen X, Hsieh T-HS, Banala S, et al. 2020. 3D ATAC-PALM: superresolution imaging of the accessible genome. Nat. Methods. 17(4):430–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science (80-. ). 315(5811):518–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu C, Gan H, Han J, Zhou Z-X, Jia S, et al. 2014. Strand-Specific Analysis Shows Protein Binding at Replication Forks and PCNA Unloading from Lagging Strands when Forks Stall. Mol. Cell. 56(4):551–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, et al. 2018. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science. 361(6409):1386–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zanders SE, Unckless RL. 2019. Fertility Costs of Meiotic Drivers. Curr. Biol. 29(11):R512–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zasadzińska E, Huang J, Bailey AO, Guo LY, Lee NS, et al. 2018. Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP. Dev. Cell. 47(3):348–362.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zee BM, Britton L-MP, Wolle D, Haberman DM, Garcia BA. 2012. Origins and Formation of Histone Methylation across the Human Cell Cycle. Mol. Cell. Biol. 32(13):2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang H, Han J, Kang B, Burgess R, Zhang Z. 2012. Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. J. Biol. Chem. 287(9):6573–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang K, Gao Y, Li J, Burgess R, Han J, et al. 2016. A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks. Nucleic Acids Res. 44(11):5083–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zhang W, Feng J, Li Q. 2020. The replisome guides nucleosome assembly during DNA replication Detailed review on replisome factors that contribute to replication-coupled histone assembly.
- 160.Zhang Y, Sun Z, Jia J, Du T, Zhang N, et al. 2021. Overview of Histone Modification. Adv. Exp. Med. Biol. 1283:1–16 [DOI] [PubMed] [Google Scholar]
- 161.Zhang Z, Shibahara KI, Stillman B. 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nat. 2000 4086809. 408(6809):221–25 [DOI] [PubMed] [Google Scholar]
- 162. Ziane R, Camasses A, Radman-Livaja M. 2022. The asymmetric distribution of RNA polymerase II and nucleosomes on replicated daughter genomes is caused by differences in replication timing between the lagging and the leading strand. Genome Res. Parental histones and RNAPII are deposited onto the strand that was replicated first.
- 163.Zion E, Chen X. 2020. Asymmetric histone inheritance regulates stem cell fate in Drosophila midgut. bioRxiv. 2020.08.15.252403 [Google Scholar]
- 164. Zwinderman MRH, Lobo TJ, Van Der Wouden PE, Spierings DCJ, Van Vugt MATM, et al. 2021. Deposition Bias of Chromatin Proteins Inverts under DNA Replication Stress Conditions. ACS Chem. Biol. 16(11):2193–2201 Leading or lagging strand bias of parental histones switches when treated with HU.
Related resources:
- Fernandez-Casanas M., and Chan K. 2018. The unresolved problem of DNA bridging. Genes. 9(12):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D. 2017. Proteomic analyses of the eukaryotic replication machinery. Methods in Enzymology. 591:33–53 [DOI] [PMC free article] [PubMed] [Google Scholar]


