Abstract
With the rapid spread of the new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen agent of COVID-19 pandemic created a serious threat to global public health, requiring the most urgent research for potential therapeutic agents. The availability of genomic data of SARS-CoV-2 and efforts to determine the protein structure of the virus facilitated the identification of potent inhibitors by using structure-based approach and bioinformatics tools.
Many pharmaceuticals have been proposed for the treatment of COVID-19, although their effectiveness has not been assessed yet. However, it is important to find out new-targeted drugs to overcome the resistance concern. Several viral proteins such as proteases, polymerases or structural proteins have been considered as potential therapeutic targets. But the virus target must be essential for host invasion match some drugability criterion.
In this Work, we selected the highly validated pharmacological target main protease Mpro and we performed high throughput virtual screening of African Natural Products Databases such as NANPDB, EANPDB, AfroDb, and SANCDB to identify the most potent inhibitors with the best pharmacological properties.
In total, 8753 natural compounds were virtually screened by AutoDock vina against the main protease of SARS-CoV-2. Two hundred and five (205) compounds showed high-affinity scores (less than − 10.0 Kcal/mol), while fifty-eight (58) filtered through Lipinski’s rules showed better affinity than known Mpro inhibitors (i.e., ABBV-744, Onalespib, Daunorubicin, Alpha-ketoamide, Perampanel, Carprefen, Celecoxib, Alprazolam, Trovafloxacin, Sarafloxacin, Ethyl biscoumacetate…). Those promising compounds could be considered for further investigations toward the developpement of SARS-CoV-2 drug development.
Keywords: SARS-CoV-2, Main Protease, African Natural Products, Virtual Screening
1. Introduction
On January 30th, 2020, the World Health Organization (WHO) has declared the outbreak of the spread of coronavirus (Covid-19) as an international public health emergency [1]. In March 2020 the WHO has classified the COVID-19 as a pandemic [2] showing the rapid spread of the threat due to the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2).
This infection has already infected more than 667,000,000 people and caused more than 6,7 million deaths worldwide (https://coronavirus.jhu.edu/map.html) in January 2023. Although not spared, the Africa continent had more than 258.000 deaths and more than 12,700,000 cases at the beginning of August 2021 (https://www.worldometers.info/ ), and those number are constantly increasing. This rapid spread of the pandemic causes a serious threat to public health, requiring rapid research into therapeutic agents. The development of a new drug requires far too much time and money by conventional methods [3] for situations of extreme urgency such as this; with Omics technologies and innovative Bioinformatics tools, the time and costs are considerably reduced for a better understanding of the mechanisms of action of the involved biomolecules. The availability of viral genomic data and protein structure determination methods has facilitated the identification of potential therapeutic targets and inhibitors using bioinformatics tools.
Thus, many pharmaceuticals have been proposed for the treatment of COVID-19, although their efficiency has not been evaluated yet [4],[5],[6]. Several viral proteins such as proteases, polymerases or structural proteins have been considered as potential therapeutic targets [7].
Natural products (NPs) are broadly defined as chemical substances produced by living organisms. More precised definitions of NPs exist, but they still do not make consensus. Some include all small molecules resulting from metabolic reactions, others classify as “NPs” only the secondary or non-essential metabolism products [8],[9]. In this document, the term Natural Products will designate extracted active compounds from Plants. They have benefited mankind in food, pesticides, cosmetic products, and especially in drugs [10],[8]. The crude extracts from Plants used in traditional medicine contain many pharmacological active compounds [11],[12]. They have shown their healing power in reducing diseases since ancient civilization [13]. These crude medicines can lead to the discovery of other active molecules and eventually to the development of chemical pure drugs that have real beneficial effects. Currently, many prescribed medicines are derived from investigations on natural products. The oldest examples of drugs based on natural products are analgesics, for which willow bark was used to relieve pain, due to salicin, a natural product hydrolyzed into salicylic acid; acetylsalicylic acid, better known as aspirin, is a synthetic derivate used as an analgesic [13]. Some of the notable approved drugs, either from pure or derived NPs, include lefamulin, the aminoglycoside antibiotic plazomicin; tafenoquine succinate, an antimalarial agent; and aplidine, an anticancer agent [14]. These findings are positive proofs that natural products can be used to find efficient drugs.
After the first SARS-CoV epidemic in the early 2000s, the main protease (Mpro, nsp5) also named chymotrypsin-like protease (3CLpro) [15], has been the object of particular attention. Many studies have demonstrated that this protease is a precious therapeutic target due to its essential role in viral replication [16],[17]. Other important coronaviral therapeutic targets include spike protein (S), RNA-dependent RNA polymerase (RdRp, nsp12), NTPase/helicase (nsp13), and papain-like protease (PLpro, part of nsp3) [18], [19]. Mpro plays a central role in mediating viral replication and transcription functions through extensive proteolytic processing of two replication polyproteins, pp1a (486 kDa) and pp1ab (790 kDa) [20]. It exists only in viruses and is not present in humans. Interestingly, it is the most conserved enzyme among SARS-CoV-2 related viruses [21]. The sequence of the Mpro enzyme shows high identity (> 96%) with the SARS-CoV, except for one key residue (Ala285Thr), which may contribute to the high infectivity of the SARS-CoV-2 virus [22],[23]. The functional centrality of Mpro in the viral life cycle makes it an interesting target for drug development against SARS and other CoV infections. Therefore, its inhibition may block the production of infectious virus particles and thus alleviate disease symptoms [16],[24]. By capitalizing on this knowledge, Mpro is one of the most attractive viral targets for antiviral drug discovery against SARS.
That is why, in this study, we carried out investigations of potential inhibitors of the main protease [25] of SARS-CoV-2 by high-throughput virtual screening of African natural products Databases.
2. Materials And Methods
2.1. Screening data:
In this study, we used a dataset of compounds from databases of African natural products: AfroDb [26], EANPDB [9], NANPDB [27] and SANCDB [10],[28]. Because these databases did not have the same formats, they were prepared differently. The molecules of the AfroDb and EANPDB databases were prepared using PyRx – Python Prescription 0.8 while those of SANCDB and NANPDB were prepared by Open Babel [29] using homemade scripts. Then all compounds in SDF (Standard Delay Format) format were converted to the pdbqt format. The compounds (Alpha-ketoamide 13b, Daunorubicin, Onalespib, and ABBV-744), have been prepared using AutoDockTools to be used as controls.
2.2. Protein preparation:
The main protease (Code PBD: 6Y2F), in complex with alpha-ketoamide 13b (αk-13b) was used as the receptor file. The coordinate file was loaded into PyMol to delete the ligand [6],[30]. Then the protein structure without the ligand was prepared with AutoDockTool-1.5.7 (ADT) [31]. ADT removed non-polar Hydrogens, added the Gasteiger charges, and assigned Solvation parameters and Atom Types. The Computed Atlas of Surface Topography of protein (CASTp) server is an online server that locates and measures pockets and voids on 3D protein structures [32],[33]. It was used to determine the ligand-binding pocket size. Based on the pocket size and taking into account of the alpha-ketoamide 13b position in the crystal structure, the grid box coordinates were set: center_x = −0.000, center_y = −0.704, center_z = −0.000 and size_x = 40, size_y = 40, size_z = 40.
2.3. Virtual screening:
Virtual screening is a widely used technique for identifying the top compounds against specific proteins from a library of thousands of compounds [34]. The sum of 8741 molecules was virtually screened with the main protease Mpro using AutoDock Vina 1.1.2 [35] in Command Line. After the preparation of the protein and ligands in pdbqt format, all the files were put in the same folder, with a configuration file containing the receptor name, the grid box coordinates, the list of ligands. The Vina script was launched with default parameters.
2.4. Lipinski rules of five verifications:
Lipinski’s rule helps to distinguish drug-like from non-drug-like molecules [36]. It states that a drug-like molecule must have at least two of the following rules: Molecular weight less than 500 Dalton, high lipophilicity (expressed as LogP less than 5), less than 5 hydrogen bond donors; less than 10 hydrogen bond acceptors; molar refractivity must be between 40 and 130 [37],[36]. These parameters have been verified using the SWISS ADMET server (http://www.swissadme.ch/) [38] .
2.5. Protein-ligand interaction determination:
The interactions between small molecules with the highest affinity scores with Mpro was determined using PyMol-2.0 [39] and the Ligplot + v.2.2.4 software [40].
3. Results
3.1. Natural Compounds from African Databases:
AfroDb is a collection of natural products from African medicinal plants with known bioactivities [26]. It represents the largest diversified collection of 3D structures of natural products covering the entire African continent. These structures can be easily downloaded and used in virtual screening studies (http://african-compounds.org/about/afrodb/). The compounds with a large number of tested biological activities are included in the ZINC database (http://zinc.docking.org/catalogs/afronp/). The South African Natural Compounds Database (SANCDB) is a fully referenced database of natural compounds from sources in South Africa ( https://sancdb.rubi.ru.ac.za/ ) [10]. The Northern African Natural Products Database (NANPDB) is the largest collection of natural compounds produced by indigenous organisms of North Africa (http://african-compounds.org/nanpdb/) [41]. The Eastern Africa Natural Products Database (http://african-compounds.org) containing the structural and bioactivity information of 1870 unique isolated molecules from about 300 source species from the Eastern African region [9]. In total, 8741 natural compounds were obtained from these different databases (Table 1).
Table 1.
Natural Compounds from African databases used for virtual screening
3.2. SARS-CoV2 active site:
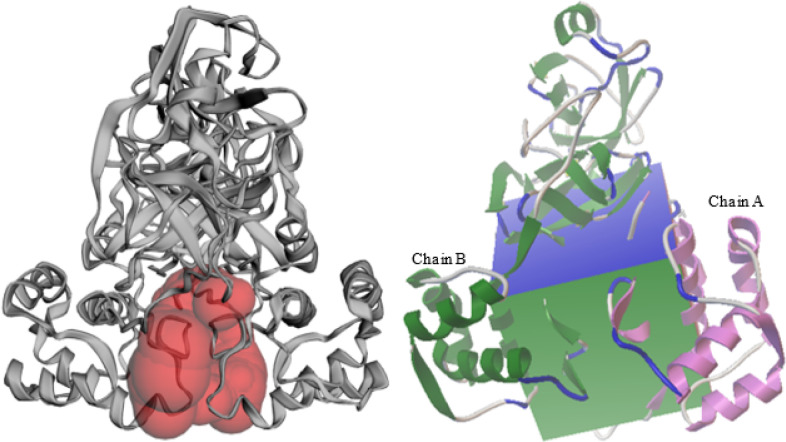
The structure of SARS-CoV-2 (code PDB: 6Y2F) was used for the virtual screening. The binding pocket determined by the CASTp server was presented in Fig. 1.
Figure 1.
Binding pocket (Red surface) of SARS-CoV-2 Mpro determined by CASTp server (left), highlighted in the Grid box, AutoDockTools (right) highlights also the two chains of the protein (chain A in purple and chain B in green).
The structure was constituted of two protomers (A and B) and a catalytic dyad (His41-Cys145) per protomer, very similar to that of the SARS protease [42]. The enzyme was composed of three domains: the domain I (residues 1–101), domain II (residues 102–184) are mainly made of antiparallel β-sheets, and an α-helical domain III (residues 201–301) [43],[44]. The catalytic domain III contains the Ser284-Thr285-Ile286 segment, an additional domain far from the catalytic dyad. One major difference with SARS-CoV-I is the substitution of the Thr285 residue by Ala [23].
The Fig. 1 left side showed the binding pocket of Mpro represented here by the red surface. The molecular surface area of the pocket is 1738.8 Å3. This pocket was located between the two protomers. It was used to define a grid box covering the amino acids of the binding site by AutoDockTools (Fig. 1, Right). The grid box volume was made large enough to allow a number of Natural Compounds to dock with the protein.
Indeed, a molecule able to strongly interact between the two protomers could inactivate the enzyme activity, even preventing the protein dimerization [45].
3.3. Identification of promising inhibitors from the virtual screening:
In total, 8741 molecules were screened against the SARS-CoV-2 Mpro among which two hundred and five (205) molecules have presented affinity scores that varied between − 12.1 Kcal/mol and − 10.0 Kcal/mol.
Among them, fifty-eight (58) were passed through the Lipinski’s rules [37] and twelve (12) did not present any violation of the rule. Those molecules got affinity scores ranging from − 11.2 Kcal/mol and − 10.0 Kcal/mol. Those molecules were considered as promising inhibitors of SARS-CoV-2 (Table 2). The molecules ABBV-744, Daunorubicin and Onalespib described as potent inhibitors of Mpro [6] were also screened by using the same docking parameters as controls (Table 2).
Table 2.
List of the most promising potent inhibitors of SARS-CoV-2 Mpro from African Natural Compounds with improved drug-like properties and standard inhibitors
| No | Compounds | Molecular weight (Dalton) | Log(P) | Hydrogen donor | Hydrogen acceptor | Affinity (Kcal/mol) |
|---|---|---|---|---|---|---|
| 1 | Sphaeropsidin A (NA) | 346.42 | 2.27 | 2 | 5 | −11.2 |
| 2 | Gypsogenic acid (NA) | 486.68 | 2.98 | 3 | 5 | −10.6 |
| 3 | Yardenone (SANC00595) | 488.7 | 4.51 | 0 | 5 | −10.5 |
| 4 | A-homo-3a-oxa-5beta-olean-12-en-3- one-28-oic acid (EA) | 470.691 | −3.503 | 1 | 5 | −10.4 |
| 5 | Epigallocatechin (NA) | 306.27 | 0.42 | 6 | 7 | −10.4 |
| 6 | Neopellitorine B (NA) | 235.37 | 3.60 | 0 | 1 | −10.4 |
| 7 | Caretroside A (NA) | 402.48 | 2.20 | 3 | 6 | −10.3 |
| 8 | Pallidol (NA) | 454.47 | 2.51 | 5 | 6 | −10.3 |
| 9 | Maslinic acid (NA) | 472.70 | 3.38 | 3 | 4 | −10.2 |
| 10 | Cabralealactone (NA) | 414.62 | 3.86 | 0 | 3 | −10.2 |
| 11 | Olibanumol H (NA) | 460.73 | 4.30 | 3 | 3 | −10.1 |
| 12 | Clionamine D (SANC00290) | 401.54 | 2.43 | 2 | 5 | −10.0 |
| Known Mpro Inhibitors used as controls | ||||||
| 1 | Alpha-ketoamide 13b | 593.7 | 3.96 | 4 | 7 | −9.2 |
| 2 | Daunorubicin | 527.5 | 2.66 | 5 | 11 | −9.1 |
| 3 | Onalespib | 409.5 | 3.84 | 2 | 5 | −8.4 |
| 4 | ABBV-744 | 491.6 | 3.65 | 3 | 5 | −8.0 |
NA = NANPDB; EA = EANPDB and SANC00290, SANC00595 = compounds id in SANCDB.
The Table 2 showed for each molecule, the parameters of Lipinski’s rules which are molecular weight, logP, number of hydrogen bonds donor and number of hydrogen bonds acceptor. The best twelve molecules identified as SARS-CoV-2 Mpro inhibitors did not shown any violation of Lipinski’s rules indicating improved pharmaceutical properties. Moreover, those compounds have shown higher affinity score than standard Mpro inhibitors used as controls [6].
3.4. SARS-CoV-2 interactions with identified potent inhibitors
For better understanding of the interaction’s details of Mpro with the molecules in Table 2, Ligplot and PyMol were used to show interactions (Table 3). The inhibitors showed specific interactions with key residues of Mpro.
Table 3.
Interactions between with M pro and the thirteen potent inhibitors.
| Molecules | Interactions with Protein via | |
|---|---|---|
| Chain A | Chain B | |
| Sphaeropsidin A | Arg-4, Lys-5 | Glu-288 |
| Gypsogenic acid | Arg-4 | Glu-283 |
| Yardenone | Arg-4, Gly-283 | Arg-4 |
| A-homo-3a-oxa-5beta-olean-12-en-3- one-28-oic acid | None | Arg-4, Glu-288 |
| Epigallocatechin | Lys-137, 2Glu-288, Asp-289, Glu-290 | Arg-4 |
| Neopellitorine B | Lys-137, Thr-169, Leu-287, Asp-289 | Thr-280, 2Glu-283 |
| Caretroside A | Glu-288 | Trp-207, Gly-283, Lys-5 |
| Pallidol | Phe-3, Asp-289 | Lys-5, Glu-288 |
| Maslinic acid | Phe-3 | Leu-282 |
| Cabralealactone | Arg-4 | Leu-282 |
| Olibanumol H | Arg-4, Gly-138 | Lys-5, 2Phe-3 |
| Clionamine D | Phe-3, Leu-282 | Phe-3, Leu-282 |
The Table 3 presented the details of the interactions between Mpro and the twelve identified compounds. The analysis of the interactions showed that the inhibitors interact mostly with residues Arg4, Leu282, Gly283, Glu288.
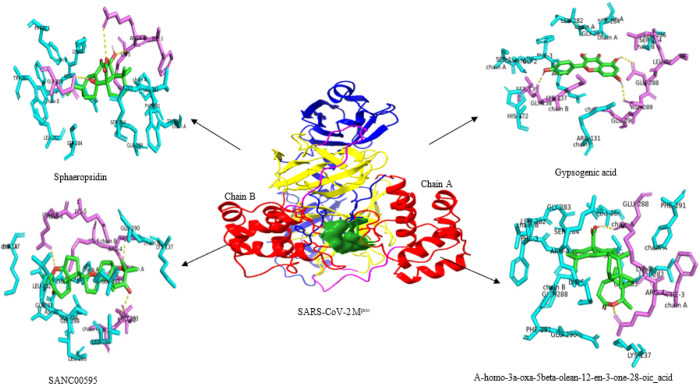
The Fig. 2 showed the interactions between Mpro and the top four inhibitors based on their affinity scores, docked poses and interactions with the protein key amino acids. The details of the other compounds are presented on supplementary data (Fig. 3).
The LigPlot + program was used to map the 2D interactions between Mpro and the top four identified compounds (supplementary data; Fig. 4).
The analysis of the interaction’s details was summarized in Table 4.
The Table 4 revealed that the main residues of Mpro interacting with the ligands were Arg 4, Lys 5 Glu 283, Gly 283 and Glu 288 involving the catalytic domain surrounding the active site dyad (His41-Cys145).
4. Discussion
The discovery of new drugs capable of inhibiting the infection caused by SARS-CoV-2 is a global priority in order to put a definitive end to this health emergency. Many studies have demonstrated that the Mpro protein was an attractive target against SARS-CoV-2 because of its important role in virus replication, its conservation among other related viruses, and its cleavage specificity different from that of Human proteases [20],[21]. Many crystallographic structures were available for this interesting target which made it very suitable for structure-based drug design [47],[17]. Africa being a rich continent in plant diversity, the first recourse for care is constituted of natural products that are easy to access and less expensive [8]. African natural products have demonstrated antiviral, antifungal and antibacterial properties [46]. These molecules are suitable candidates for high throughput virtual screening against validated drug targets.
In this study, we screened out 8741 compounds from African natural product databases: AfroDb (880), EANPDB (1815), NANPDB (4912) and SANCDB (1134) with the SARS-CoV-2 Mpro (code PDB: 6Y2F). The results showed that twelve (12) compounds (Table 2): Sphaeropsidin A, Gypsogenic acid, Yardenone, A-homo-3a-oxa-5beta-olean-12-en-3-one-28-oic acid, Epigallocatechin, NeopellitorinB, Caretroside A, Pallidol, Maslinic acid, Cabralealactone, Tribulus saponin aglycone 3, Olibanumol H and Clionamine D presented high-affinity scores scores ranging from − 11.2 to −10.0 Kcal/mol with SARS-CoV-2 Mpro. Those molecules presented better affinity scores than standard Mpro inhibitors such as 6-Deaminosinefungin (−8.1 Kcal/mol) and UNII-O9H5KY11SV (−8.4 Kcal/mol) proposed by Mohammad and al. [4] and Daunorubicin (−9.33 Kcal/mol), Onalespib (−8.21 Kcal/mol), ABBV-744 (−7.79 Kcal/mol) identified by Fakhar and al. [6] for the same target in similar screening conditions (Table 2). Moreover, those molecules showed improved pharmacological properties proved by Lipinski’s parameters values. The 12 identified natural products have demonstrated their potential usage as interesting drug precursors of SARS-CoV-2.
The analysis of their interactions with Mpro revealed that they bind mainly to protein residues Arg 4, Lys 5 Glu 283, Gly 283 and Glu 288 involving the catalytic domain surrounding the active site dyad (His41-Cys145). Hence the selected compounds could actually prevent the activity of the enzyme. It would be interesting to pursue experimental assays with those promising inhibitors and the SARS-Cov-2 Protein.
Many of these molecules have already shown efficiency in other studies : Sphaeropsidine A, against drug-resistant cancer cells [49]; Gypsogenic acid against Bacillus subtilis and Bacillus thrungiensis also as potential antitumor agents [50]; Yardenone against hypoxia-inducing factor 1 (HIF-1) activation in breast and prostate tumor cells [51]; Epigallocatechin extracted from Acacia karroo, was used medicinally to treat diarrhea, colds, dysentery, conjunctivitis and hemorrhages. Acacia karroo and other local plant species such as Artemisia afra, Ziziphus mucronata and Eucomis autumnalis, have been widely used for the treatment of symptoms related to listeriosis [52]; A-homo-3a-oxa-5beta-olean-12-en-3- one-28-oic Acid, extracted from Albizia gummifera, was used in the indigenous medical system for various nutrients [53].
This study’s findings imply that the inhibitors identified were promising inhibitors of SARS-Cov-2 main protease Mpro, interesting for the development of efficient drugs against SARS-Cov-2. Further investigations are needed to deepen these findings in the process of drug development.
5. Conclusion
In this study, we identified twelve (12) compounds (Sphaeropsidin A, Gypsogenic acid, Yardenone, A-homo-3a-oxa-5beta-olean-12-en-3- one-28-oic acid, Epigallocatechin, Neopellitorine B, Caretroside A, Pallidol, Maslinic acid, Cabralealactone, Tribulus saponin aglycone 3, Olibanumol H, Clionamine D) from African Natural Product which showed high-affinity and validated interesting pharmacological properties against the main protease Mpro, the most attractive target of SARS-CoV-2. Further investigations have to be pursued with the identified compounds to foster the way into the development of new drugs against the COVID-19 virus.
Figure 2.
Details of Mpro interactions with top four potential inhibitors: - top left, Sphaeropsidin A; - top right, Gypsogenic acid; - bottom left, Yardenone (SANC00595); and - bottom right, A-homo-3a-oxa-5beta-olean-12-en-3- one-28-oic acid. The ligands are illustrated in green stick surrounded by polar residues (magenta) establishing hydrogen bonds(yellow) with the protein and non-polar residues (cyan). Mpro is illustrated according to its different domains: domain I in Medium Blue; domain II in Yellow; domain III in red, the catalytic dyad in surface representation (Forest green). A long loop connects domain II to the domain III C-terminal (Magenta).
Table 4.
Summary of interactions between Mpro and the top four compounds:
| Number of H Bonds | Mpro Residues involved in | ||
|---|---|---|---|
| H Bonds | VdW interactions | ||
| Sphaeropsidin A | 3 | Arg-4, Lys-5 (Chain A) Glu-288 (Chain B) |
Phe-291, Phe-3, Ser-284, Glu-288, Leu-282, (Chain A) Lys-5, Trp-207, Phe-291 (Chain B) |
| Gypsogenic acid | 2 | Arg-4 (chain A) Glu-283 (chain B) |
Gly-283, Phe-3, Gly-2, Leu-282 (Chain A) Leu-286, Lys-137 (Chain B) |
| Yardenone | 3 | Arg-4, Gly-283 (Chain A) Arg-4 (chain B) |
Lys-5, Leu-282, Glu-290, Glu-288, Gly-283 (Chain A) Glu-290, Gly-283, Leu-282, Lys-5, Lys-137, Glu-288 (Chain B) |
| A-homo-3a-oxa-5beta-olean-12-en-3-one-28-oic acid | 2 | Arg-4, Glu-288 (chain B) | Leu-282, Gly-283, Arg-4, Phe-3, Phe-291, Lys-5 (Chain A) Phe-291, Phe-3, Lys-5, Glu-288 (Chain B) |
Acknowledgments:
We thank the African Center of Excellence in Bioinformatics of Bamako (ACE-B)/USTTB for conducting and monitoring the project and the University of Clinical Research Center (UCRC)/USTTB, Mali for the technical support and the Department of Biostatistics and Data Science/Tulane University and the African Computational Genomics (TACG) Research group for manuscript review.
Funding:
NIH Cooperative Agreements U2R TW010673 supported this study for West African Center of Excellence for Global Health Bioinformatics Research Training. The study team members received financial support from H3ABioNet (U24HG006941), Fogarty International Center D43TW008652, and the West African International Centers for Excellence in Malaria Awards U19 AI 089696 and U19 AI 129387.
Footnotes
Competing Interests:
The authors have no competing interests to declare that are relevant to the content of this article.
Contributor Information
Oudou DIABATE, University of Sciences, Technics and Technologies of Bamako (USTTB).
Cheickna CISSE, University of Sciences, Technics and Technologies of Bamako (USTTB).
Mamadou SANGARE, The African Computational Genomics (TACG) Research group.
Opeyemi Soremekun, The African Computational Genomics (TACG) Research group.
Segun Fatumo, University of Sciences, Technics and Technologies of Bamako (USTTB).
Jeffrey G. SHAFFER, Tulane University School of Public Health and Tropical Medicine
Seydou DOUMBIA, University of Sciences, Technics and Technologies of Bamako (USTTB).
Mamadou WELE, University of Sciences, Technics and Technologies of Bamako (USTTB).
Data Availability
The databases used during the current study were available in http://african-compounds.org/about/afrodb/ for AfroDB; https://sancdb.rubi.ru.ac.za/ for SANCDB; http://african-compounds.org/nanpdb/ for NANPDB and EANPDB.
References
- 1.Anwar S., Ara Shamsad I., Amirul Morshed A., et al. Farzana F., « Clinical Profile of Child COVID-19 Patients of Bangladesh », AJP, vol. 7, no 1, p. 5, doi: 10.11648/j.ajp.20210701.12. [DOI] [Google Scholar]
- 2.Cucinotta D. et al. Vanelli M., « WHO Declares COVID-19 a Pandemic », Acta Bio Medica Atenei Parmensis, vol. 91, no 1, p. 157–160, doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grinter S. et al. Zou X., « Challenges, Applications, and Recent Advances of Protein-Ligand Docking in Structure-Based Drug Design », Molecules, vol. 19, no 7, p. 10150–10176, doi: 10.3390/molecules190710150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammad T. et al. , « Identification of high-affinity inhibitors of SARS-CoV-2 main protease: Towards the development of effective COVID-19 therapy », Virus Research, vol. 288, p. 198102, doi: 10.1016/j.virusres.2020.198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwivedi D. R., « The ability of natural flavonoids to inhibit », https://www.news-medical.net/news/20211031
- 6.Fakhar Z., Khan S., AlOmar S. Y., Alkhuriji A., et al. Ahmad A., « ABBV-744 as a potential inhibitor of SARS-CoV-2 main protease enzyme against COVID-19 », Sci Rep, vol. 11, no 1, p. 234, doi: 10.1038/s41598-020-79918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaurav A. et al. Al-Nema M., « Chapter 10 - Polymerases of Coronaviruses: Structure, Function, and Inhibitors », in Viral Polymerases, , p. 271–300. doi: 10.1016/B978-0-12-815422-9.00010-3. [DOI] [Google Scholar]
- 8.Sorokina M. et al. Steinbeck C., « Review on natural products databases: where to find data in 2020 », J Cheminform, vol. 12, no 1, p. 20, doi: 10.1186/s13321-020-00424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simoben C. V. et al. , « Pharmacoinformatic Investigation of Medicinal Plants from East Africa », Mol. Inf., vol. 39, no 11, p. 2000163, doi: 10.1002/minf.202000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatherley R. et al. , « SANCDB: a South African natural compound database », J Cheminform, vol. 7, no 1, p. 29, déc. 2015, doi: 10.1186/s13321-015-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. K, « Fundulopanchax gardneri Test: A Convenient Method of Bioassay for Active Constituents of Natural Products », Nat Prod Chem Res 2014, vol. 2, no 4, 2014, doi: 10.4172/2329-6836.1000133. [DOI] [Google Scholar]
- 12.Mamadou A. K. et al. , « Evaluation of the biological activities of leaf and bark extracts of Ficus platiphylla Delile, a medicinal plant used in Mali », J. Med. Plants Res., vol. 14, no 3, p. 118–128, doi: 10.5897/JMPR2019.6874. [DOI] [Google Scholar]
- 13.Chintoju N., Konduru P., Kathula R. L., et al. Remella R., « Research and Reviews: Journal of Hospital and Clinical Pharmacy », vol. 1, no 1, 2015. [Google Scholar]
- 14.Newman D. J. et al. Cragg G. M., « Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019 », J. Nat. Prod., vol. 83, no 3, p. 770–803, doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Liu Q., et al. Guo D., « Emerging coronaviruses: Genome structure, replication, and pathogenesis », J Med Virol, vol. 92, no 4, Art. no 4, doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., et al. Hilgenfeld R., « Coronavirus Main Proteinase (3CL pro ) Structure: Basis for Design of Anti-SARS Drugs », Science, vol. 300, no 5626, p. 1763–1767, doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 17.Yang H. et al. , « The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor », Proc. Natl. Acad. Sci. U.S.A., vol. 100, no 23, p. 13190–13195, doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziebuhr J., « Molecular biology of severe acute respiratory syndrome coronavirus », Current Opinion in Microbiology, vol. 7, no 4, p. 412–419, doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiel V. et al. , « Mechanisms and enzymes involved in SARS coronavirus genome expression », Journal of General Virology, vol. 84, no 9, p. 2305–2315, doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 20.« Du Q.S., Wang S.Q., Wei D.Q., Zhu Y., Guo H., Sirois S. and Chou K.C. (2004) Polyprotein Cleavage Mechanism of SARS CoV Mpro and Chemical Modification of Octapeptide. Peptides, 25, 1857–1864. - References - Scientific Research Publishing ». https://www.scirp.org/(S(351jmbntvnsjt1aadkozje))/reference/referencespapers.aspx?referenceid=2698795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimeno A. et al. , « Prediction of Novel Inhibitors of the Main Protease (M-pro) of SARS-CoV-2 through Consensus Docking and Drug Reposition », International Journal of Molecular Sciences, vol. 21, no 11, Art. no 11, doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gimeno A. et al. , « Prediction of Novel Inhibitors of the Main Protease (M-pro) of SARS-CoV-2 through Consensus Docking and Drug Reposition », IJMS, vol. 21, no 11, p. 3793, doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J., « 2019-nCoV 3C-Like Protease carries an activity-enhancing T285 /A variation which may contribute to its high infectivity », Open Science Framework, preprint, doi: 10.31219/osf.io/skznv. [DOI] [Google Scholar]
- 24.Kuo C.-J. et al. Liang P.-H., « Characterization and Inhibition of the Main Protease of Severe Acute Respiratory Syndrome Coronavirus », ChemBioEng Reviews, vol. 2, no 2, p. 118–132, doi: 10.1002/cben.201400031. [DOI] [Google Scholar]
- 25.Zhang L. et al. , « Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors », Science, vol. 368, no 6489, p. 409–412, doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ntie-Kang F. et al. , « AfroDb: a select highly potent and diverse natural product library from African medicinal plants », PloS one, vol. 8, no 10, p. e78085, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ntie-Kang F. et al. , « NANPDB: A Resource for Natural Products from Northern African Sources », J. Nat. Prod., vol. 80, no 7, p. 2067–2076, doi: 10.1021/acs.jnatprod.7b00283. [DOI] [PubMed] [Google Scholar]
- 28.Diallo B. N., Glenister M., Musyoka T. M., Lobb K., et al. Tastan Bishop Ö., « SANCDB: an update on South African natural compounds and their readily available analogs », J Cheminform, vol. 13, no 1, p. 37, doi: 10.1186/s13321-021-00514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., et al. Hutchison G. R., « Open Babel: An open chemical toolbox », J Cheminform, vol. 3, no 1, p. 33, déc. 2011, doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.« Support | pymol.org ». https://pymol.org/2/support.html?#citing.
- 31.Eberhardt J., Santos-Martins D., Tillack A. F., et al. Forli S., « AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings », J. Chem. Inf. Model., vol. 61, no 8, p. 3891–3898, doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., et al. Liang J., « CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues », Nucleic Acids Research, vol. 34, no Web Server, p. W116–W118, doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binkowski T. A., « CASTp: Computed Atlas of Surface Topography of proteins », Nucleic Acids Research, vol. 31, no 13, p. 3352–3355, doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cosconati S., Forli S., Perryman A. L., Harris R., Goodsell D. S., et al. Olson A. J., « Virtual screening with AutoDock: theory and practice », Expert Opinion on Drug Discovery, vol. 5, no 6, p. 597–607, doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trott O. et al. Olson A. J., « AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading », J. Comput. Chem., p. NA–NA, doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipinski C. A., Lombardo F., Dominy B. W., et al. Feeney P. J., « Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settingsq », Advanced Drug Delivery Reviews, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Lipinski C. A., « Lead- and drug-like compounds: the rule-of-five revolution », Drug Discovery Today: Technologies, vol. 1, no 4, p. 337–341, déc. 2004, doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Daina A., Michielin O., et V. Zoete, « SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules », Sci Rep, vol. 7, no 1, p. 42717, doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrödinger, LLC, « The PyMOL Molecular Graphics System, Version 1.8 ».
- 40.Laskowski R. A. et al. Swindells M. B., « LigPlot+: multiple ligand-protein interaction diagrams for drug discovery », J Chem Inf Model, vol. 51, no 10, p. 2778–2786, doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 41.Ntie-Kang F. et al. , « NANPDB: A Resource for Natural Products from Northern African Sources », J. Nat. Prod., vol. 80, no 7, p. 2067–2076, juill. 2017, doi: 10.1021/acs.jnatprod.7b00283. [DOI] [PubMed] [Google Scholar]
- 42.Citarella A., Scala A., Piperno A., et al. Micale N., « SARS-CoV-2 Mpro: A Potential Target for Peptidomimetics and Small-Molecule Inhibitors », Biomolecules, vol. 11, no 4, p. 607, avr. doi: 10.3390/biom11040607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentile D., Patamia V., Scala A., Sciortino M. T., Piperno A., et al. Rescifina A., « Putative Inhibitors of SARS-CoV-2 Main Protease from A Library of Marine Natural Products: A Virtual Screening and Molecular Modeling Study », Marine Drugs, vol. 18, no 4, p. 225, doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Günther S. et al. , « X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease », Science, vol. 372, no 6542, p. 642–646, doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goyal B. et al. Goyal D., « Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy », ACS Combinatorial Science, doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 46.Balasubramaniam B. et al. , « Antibacterial and Antiviral Functional Materials: Chemistry and Biological Activity toward Tackling COVID-19-like Pandemics », ACS Pharmacol. Transl. Sci., vol. 4, no 1, p. 8–54, doi: 10.1021/acsptsci.0c00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L. et al. , « Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors », Science, vol. 368, no 6489, p. 409–412, doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.« OSF Preprints | 2019-nCoV 3C-Like Protease carries an activity-enhancing T285 /A variation which may contribute to its high infectivity ». https://osf.io/skznv.
- 49.Mathieu V. et al. , « Sphaeropsidin A shows promising activity against drug-resistant cancer cells by targeting regulatory volume increase », Cell. Mol. Life Sci., vol. 72, no 19, p. 3731–3746, doi: 10.1007/s00018-015-1902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emirdağ-Öztürk S., Karayıldırım T., Çapcı-Karagöz A., Alankuş-Çalışkan Ö., Özmen A., et E. Poyrazoğlu-Çoban, « Synthesis, antimicrobial and cytotoxic activities, and structure-activity relationships of gypsogenin derivatives against human cancer cells », Eur J Med Chem, vol. 82, p. 565–573, doi: 10.1016/j.ejmech.2014.05.084. [DOI] [PubMed] [Google Scholar]
- 51.Dai J., Fishback J. A., Zhou Y.-D., et al. Nagle D. G., « Sodwanone and Yardenone Triterpenes from a South African Species of the Marine Sponge Axinella Inhibit Hypoxia-Inducible Factor-1 (HIF-1) Activation in Both Breast and Prostate Tumor Cells », J. Nat. Prod., vol. 69, no 12, p. 1715–1720, doi: 10.1021/np060278q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyila M. A., Leonard C. M., Hussein A. A., et al. Lall N., « Activity of South African medicinal plants against Listeria monocytogenes biofilms, and isolation of active compounds from Acacia karroo », South African Journal of Botany, vol. 78, p. 220–227, doi: 10.1016/j.sajb.2011.09.001. [DOI] [Google Scholar]
- 53.Debella A. et al. , « Triterpenoid saponins and sapogenin lactones from Albizia gummifera », Phytochemistry, vol. 53, no 8, p. 885–892, doi: 10.1016/S0031-9422(99)00464-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The databases used during the current study were available in http://african-compounds.org/about/afrodb/ for AfroDB; https://sancdb.rubi.ru.ac.za/ for SANCDB; http://african-compounds.org/nanpdb/ for NANPDB and EANPDB.