Abstract
A cluster of three genes CELSR2, PSRC1, and SORT1 has been associated with cardiovascular diseases. Thus, the aim of this study was (i) to perform a systematic review and updated meta-analysis of the association of three polymorphisms (rs646776, rs599839, and rs464218) of this cluster with cardiovascular diseases, and (ii) to explore by PheWAS signals of the three SNPs in cardiovascular diseases and to evaluate the effect of rs599839 with tissue expression by in silico tools. Three electronic databases were searched to identify eligible studies. The meta-analysis showed that the rs599839 (allelic OR 1.19, 95% CI 1.13–1.26, dominant OR 1.22, 95% CI 1.06–1.39, recessive OR 1.23, 95% CI 1.15–1.32), rs646776 (allelic OR 1.46, 95% CI 1.17–1.82) polymorphisms showed an increased risk for cardiovascular diseases. PheWas analysis showed associations with coronary artery disease and total cholesterol. Our results suggest a possible involvement of the CELSR2-PSRC1-SORT1 cluster variants in the risk association of cardiovascular diseases, particularly coronary artery disease.
Keywords: CELSR2, PSRC1, SORT1, cardiovascular diseases, meta-analysis
1. Introduction
Nowadays, cardiovascular diseases (CVDs) are defined as the “pathological processes involving the cardiovascular system such as the heart, the blood vessels, or the pericardium” by the Medical Subject Headings criteria (MeSH). The American Heart Association, until 2020, reported that cardiovascular diseases represented the leading cause of mortality worldwide, with approximately 18.6 million deaths and 523 million cases since records began [1]. In this sense, despite technological advances in the diagnosis and treatment of CVDs, they are the principal contributor to disabilities and decreased quality of life in the population [2].
Among the cardiovascular diseases of atherosclerotic origin is mainly coronary artery disease [3]. Therefore, it is important to study several risk factors for CVDs, including dyslipidemias, metabolic syndrome and type 2 diabetes mellitus, and genetics [4,5,6].
Since 2007, various genome-wide association studies (GWAS) have identified risk loci and candidate genes involved in the development of CVDs [7]. In this way, the 1p13.3 locus is of interest because it harbors a group of three candidate genes CELSR2-PSRC1-SORT1 that could participate in mechanisms that involve the alteration of serum lipids and plasmatic cholesterol levels and increase susceptibility for the development of CVD [8,9].
The CELSR2 (Cadherin EGF LAG Seven-Pass G-Type Receptor 2) is a member of the flamingo subfamily, part of the cadherin superfamily. This gene encodes a cadherin responsible for contact-mediated cell adhesion and ligand-receptor cell interactions [10]. The PSRC1 (proline/serine-rich coiled-coil 1) gene is partly responsible for microtubule destabilization [11,12]. The SORT1 (sortilin) is a gene that encodes the sortilin 1 protein, implicated in several lipid-associated functions such as VLDL secretion, atherosclerotic lesions development, LDL-cholesterol metabolism, and PCSK9 secretion [13]. It also participates in other pathophysiological mechanisms, including inflammation, dyslipidemia, vascular calcification, insulin resistance, and the formation of foam cells [14]. Some of these SORT1-associated functions lead to an increased risk of cardiovascular diseases. However, the results regarding these three genes and their participation in CVDs are so far contradictory and not well understood [8,15,16]. Likewise, previously reported associations show the possible role of these genetic variants. However, it is necessary to know the behavior of these polymorphisms in other populations and with updated studies.
Therefore, this study aimed to perform a systematic review and updated meta-analysis of the association of CELSR2-PSRC1-SORT1 (rs646776, rs599839, and rs464218) with cardiovascular diseases. Finally, we explored prior genome-wide association signals of the rs599839 polymorphism with other phenotypes related to cardiovascular diseases and evaluated the effect of rs599839 on gene expression in the liver using in silico tools.
2. Materials and Methods
2.1. Databases and Literature Sources
The present work followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [17] (Supplementary Material S1). This study was also registered and authorized by PROSPERO (International prospective register of systematic reviews), endorsed by the University of York with protocol ID: CRD42021284700.
A systematic literature search was performed up to December 2021 to identify relevant articles published on three widely used electronic literature databases (PubMed, Web of Science, and Scopus). First, the following keywords were combined to obtain the eligible articles: (i) “rs646776”, “rs599839”, or “rs464218” and (ii) “coronary artery disease”, “cardiovascular disease”, “coronary heart disease”, “cerebrovascular disease”, “myocardial infarction”, “peripheral arterial”, “atherosclerosis”, or “ischemic stroke”. To perform a more detailed search, we also included some cardiovascular risk factors: (i) “rs646776”, “rs599839”, or “rs464218” and (ii) “hypertension”, “lipids”, “metabolic syndrome”, “glucose”, “insulin”, “type 2 diabetes mellitus”, “abdominal obesity”, or “dyslipidemia” in combination with the gene polymorphisms previously mentioned (Supplementary Material S2).
2.2. Eligibility Criteria
Only studies meeting the following inclusive selection criteria were eligible: (1) studies that evaluated human subjects; (2) case-control study designs; (3) studies investigating the association between the genetic variants of CELSR2-PSRC1-SORT1 (rs646776, rs599839, and rs464218) and cardiovascular disease susceptibility; (4) studies in which genotypes in cases and controls were available or (5) had sufficient data to calculate an odds ratio (OR) with a 95% confidence interval (95% CI); and (6) peer-reviewed studies written in the English language.
The exclusion criteria were meta-analyses, reviews, case reports, abstracts, editorial articles, or duplicate publications with overlapping data. For meta-analyses only, we excluded studies in which the control group had a significant Hardy–Weinberg equilibrium (HWE) p-value < 0.05.
Fourteen studies were eligible for systematic review and twelve for meta-analysis following the abovementioned search.
2.3. Assessment of Study Quality
The quality of the articles included in our study was evaluated using the NOS (Newcastle-Ottawa quality assessment) scale. The evaluation was based on three aspects: the selection (total score of 4), the comparability between groups (total score of 2), and exposure factors (total score of 3), with a total score of 9 points. Studies with a score ≥6 were considered high quality for inclusion in this analysis.
2.4. Data Extraction
Two authors (RGCA and TBGC) searched, applied the eligibility criteria and quality assessment, and extracted data from eligible studies. Two other investigators (JMRP and NPH) resolved discrepancies by consensus.
The information obtained was:
last name of the leading author;
year of publication;
self-reported ancestry and country of study population;
number of cases and controls;
diagnostic criteria of cases and controls;
allele and genotype frequencies.
2.5. Methods for Quantitative Synthesis and Statistical Analysis
We calculated the odds ratio and 95% CI to estimate the association between genetic variants of CELSR2-PSRC1-SORT1 (rs646776, rs599839, and rs464218) and the risk of cardiovascular diseases. Statistical significance was established at a Z-test p-value < 0.05. We used the main models of inheritance reported in statistical analyses of genetic polymorphisms in epidemiological studies [18]:
1. Allelic
Allele 1 (Reference)
Allele 2;
2. Codominant
11 (Reference)
12
22;
3. Dominant
11 (Reference)
12 + 22;
4. Recessive
11 + 12 (References)
22.
We evaluated statistical heterogeneity between studies by a chi-square-based Cochran Q test. A Higgins I-squared statistic p-value < 0.1 or I2 > 50% indicated heterogeneity. We used the random effects model unless otherwise stated, in which case we used the fixed effects model. For a better understanding of CVDs, we performed sub-analyses based on:
overall, coronary artery disease, myocardial infarction, acute coronary artery syndrome, ischemic stroke, and peripheral arterial disease;
studies including only coronary artery disease;
studies including only healthy controls;
Asian populations.
We replicated the analyses, excluding one study at a time as a sensitivity analysis for the stability of the pooled results. We visualized publication bias by funnel plots, and the quantitative method of Begg/Egger linear regression (p-value < 0.05 suggests bias) identified any bias. Chi-squared statistics evaluated Hardy–Weinberg equilibrium (HWE) in each study. All data were analyzed using Comprehensive Meta-Analysis Software version 2. In addition, we duplicated the searches in the different databases: (a) manually by the assigned researchers (RGCA, TBGC, JMRP, and NPH), and (b) by using the COVIDENCE software (https://www.covidence.org/) (accessed on 29 December 2021) to verify searches and articles found. The p-value adjusted by the Bonferroni method was considered, and significance was set at p-value < 0.004 (p-value correction = 0.05/12), and the results were expressed in scientific notation.
2.6. Prior Genome-Wide Association Signals with Phenotypes Related to Cardiovascular Diseases
We, additionally, explored previous genome-wide association signals of rs599839 with other cardiovascular disease-related phenotypes considering that this genetic variant has shown the highest number of associations in previous sub-analyses. Therefore, we performed a query of prior associations of rs599839 with related phenotypes in the web tool PheWAS of the GWAS Atlas portal in order to identify any association of this variant with other CAD risk factors [19]. GWAS Atlas is a curated web repository of prior genome-wide associated variants to different phenotypes. We filtered the association based on p-values and phenotypes previously associated with cardiovascular diseases.
2.7. Effect of Genetic Variants in the Expression of CELSR2-PSRC1-SORT1 Cluster Transcripts in the Liver
Liver expression of the CELSR2-PSRC1-SORT1 genes is essential for lipid metabolism. Therefore, we explored the GTeX portal for associations of the rs599839 with the transcripts of the CELSR2-PSRC1-SORT1 cluster.
3. Results
3.1. Systematic Review
Study Characteristics
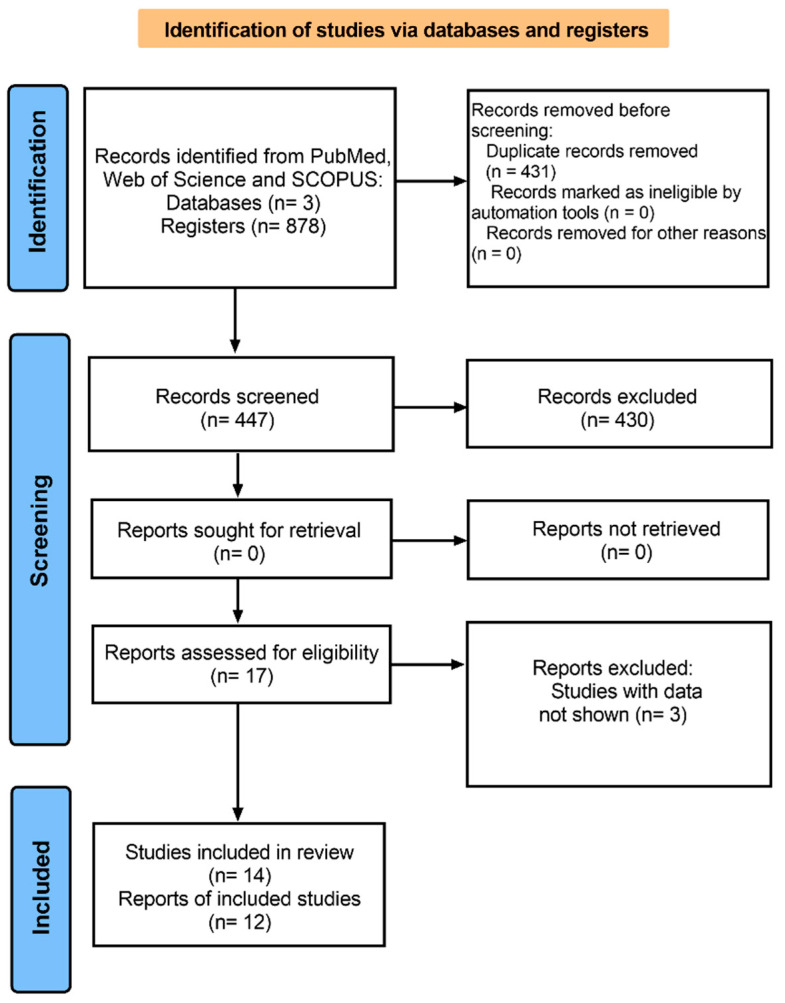
A total of 878 articles (306 from PubMed, 305 from Web of Science, and 267 from Scopus) were identified. The flow chart for the study selection is shown in Figure 1. The detailed characteristics of the studies included in the systematic review are shown in Table 1. Of the 14 included studies, based on the country, we observed the following distribution: 4 in China [15,20,21,22], 4 in Japan [23,24,25,26], 1 in New Zealand [27], 1 in Germany [28], 1 in Arabia [29], 1 in Stockholm [30], 1 in the USA [31], and 1 in Pakistan [32]. We found a differential distribution of sample sizes and ages for each SNP. The analysis of the rs599839 polymorphism included 21,553 cases and 29,985 controls. The observed mean age was 61.37 for cases and 59.36 years for controls. The analysis of the rs646776 polymorphism included 2356 cases and 2505 controls, with a mean age of 55.24 years for cases and 53.42 for controls. Finally, for the rs464218 polymorphism, 1315 cases and 691 controls were included.
Figure 1.
Flow chart showing study inclusion and exclusion details.
Table 1.
Systematic review of cases and controls about the association between rs599839, rs646776, and rs464218 polymorphisms and cardiovascular diseases.
| First Author | Country | Cases | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diagnostic | NOS | N | Male | Female | Age * | Diagnostic | N | Male | Female | Age * | ||
| rs599839 polymorphism | ||||||||||||
| Zhou, Y.J., 2015 [15] | China | CAD | 7 | 561 | 417 | 144 | 62.1 ± 10.5 | HC | 590 | 431 | 159 | 61.3 ± 9.7 |
| China | IS | 7 | 527 | 383 | 144 | 62.7 ± 12.3 | - | - | - | - | - | |
| Zhou, L., 2011 [20] | China | CHD | 8 | 1269 | 971 | 298 | 60.5 ± 10.4 | HC | 2745 | 1844 | 901 | 60.6 ± 8.5 |
| Qin, J., 2018 [21] | China | DM+PAD | 7 | 440 | 223 | 217 | 66.7 ± 7.5 | NC | 442 | 246 | 196 | 60.4 ± 7.9 |
| Ueyama, C., 2015 [23] | Japan | MetS | 8 | 1822 | 1228 | 594 | 64.4 ± 10.3 | HC | 1096 | 635 | 461 | 63.4 ± 11.7 |
| Matsuoka, R., 2015 [24] | Japan | MI | 7 | 1824 | 1468 | 356 | 64.6 ± 10.3 | HC | 2329 | 1029 | 1300 | 62.3 ± 11.8 |
| Fujimaki, T., 2015 [25] | Japan | AH | 8 | 3348 | 2139 | 1209 | 66.2 ± 10.2 | HC | 2112 | 1162 | 950 | 61.9 ± 12.1 |
| Ellis, K.L., 2011 [27] | New Zealand | CD | 8 | 1794 | 1272 | 525 | 66.5 ± 12.3 | HC | 1649 | 1093 | 622 | 62.8 ± 10.8 |
| Kleber, M.E., 2010 [28] | Germany | CAD | 8 | 2508 | 1881 | 627 | 64 ± 10 | HC | 681 | 354 | 327 | 58 ± 12 |
| Germany | CAD, MI | 8 | 1575 | 1197 | 378 | 64 ± 10 | - | - | - | - | - | |
| Rizk, N.M., 2015 [29] | Arabia | ACS | 8 | 136 | 110 | 26 | 56.3 ± 10.5 | NC | 91 | 68 | 23 | 56.86 ± 9.3 |
| Han, W., 2020 [22] | China | CHD | 8 | 227 | NA | NA | 51.2 ± 6.9 | HC | 101 | NA | NA | 49.93 ± 8.0 |
| Gigante, B., 2012 [30] | Stockholm | MI | 8 | 1213 | 852 | 361 | 60 (53–65) | HC | 1561 | 1054 | 507 | 61 (54–66) |
| Bressler, J., 2010 [31] | USA | CHD | 7 | 397 | 197 | 200 | 54.8 | HC | 3206 | 1131 | 2075 | 53.1 |
| USA | CHD | 7 | 1362 | 931 | 431 | 55.5 | HC | 8710 | 3643 | 5067 | 53.9 | |
| Abe S, 2015 [26] | Japan | High-LDL-C | 8 | 1174 | 672 | 500 | 63.7 ± 10.7 | NC | 3296 | 2146 | 1150 | 64.5 ± 11.0 |
| Overall | 21,553 | 14,941 | 6386 | 61.37 | - | 29,985 | 15,794 | 14,156 | 59.36 | |||
| rs646776 polymorphism | ||||||||||||
| Ansari, W.M., 2019 [32] | Pakistan | PCAD | 8 | 340 | 329 | 11 | 42 ± 3.8 | HC | 310 | 298 | 12 | 39 ± 7.8 |
| Qin, J., 2018 [21] | China | DM+PAD | 7 | 440 | 223 | 217 | 66.7 ± 7.5 | NC | 442 | 246 | 196 | 60.4 ± 7.9 |
| Rizk, N.M., 2015 [29] | Arabia | ACS | 8 | 136 | 110 | 26 | 56.3 ± 10.5 | NC | 91 | 68 | 23 | 56.8 ± 9.3 |
| Han, W., 2020 [22] | China | CHD | 8 | 227 | NA | NA | 51.2 ± 6.9 | HC | 101 | NA | NA | 49.9 ± 8.0 |
| Gigante, B., 2012 [30] | Stockholm | MI | 8 | 1213 | 852 | 361 | 60 (53–65) | HC | 1561 | 1054 | 507 | 61 (54–66) |
| Overall | 2356 | 1514 | 615 | 55.24 | - | 2505 | 1666 | 738 | 53.42 | |||
| rs464218 polymorphism | ||||||||||||
| Zhou, Y.J., 2015 [15] | China | CAD | 7 | 561 | 417 | 144 | 62.1 ± 10.5 | HC | 590 | 431 | 159 | 61.3 ± 9.7 |
| China | IS | 7 | 527 | 383 | 144 | 62.7 ± 12.3 | - | - | - | - | - | |
| Han, W., 2020 [22] | China | CHD | 8 | 227 | NA | NA | 51.2 ± 6.9 | HC | 101 | NA | NA | 49.9 ± 8.0 |
| Overall | 1315 | 800 | 288 | 58.6 | - | 691 | 431 | 159 | 55.6 | |||
* Mean ± standard deviation or median (interquartile range). CAD: coronary artery disease; IS: ischemic stroke; CHD: coronary heart disease; DM: type 2 diabetes mellitus; PAD: peripheral arterial disease; MetS: metabolic syndrome; MI: myocardial infarction; AH: arterial hypertension; CD: coronary disease; ACS: acute coronary artery syndrome; High-LDL-C: hyper-LDL-cholesterolemia. HC: healthy control; NC: Non-cases (<50% stenosis, some comorbidities).
All the included studies were of quality on the NOS scale (score > 6).
3.2. Meta-Analysis
The quantitative analysis excluded two studies, by Ellis, K.L., 2011 and by Rizk, N. M., 2015, because the control group had a p-value < 0.05 in HWE. Subsequently, twelve studies were taken into consideration for further analysis.
3.2.1. rs599839 Polymorphism and Susceptibility to Cardiovascular Diseases
From the twelve studies included in this meta-analysis, eleven reported genotype data for the rs599839. We performed various sub-analyses for the rs599839 polymorphism as a risk of CVDs. The first analysis was for the overall cardiovascular diagnoses after excluding heterogeneity; we found statistically significant associations in the following genetic models: allelic (OR: 1.19, 95% CI: 1.13–1.26, p-value 1 × 10−4, Q test p-value: 0.175, I2: 30.38), dominant (OR: 1.22, 95% CI: 1.06–1.39, p-value 3 × 10−3, Q test p-value: 0.141, I2: 34.63), and recessive (OR: 1.23, 95% CI: 1.15–1.32), p-value 1 × 10−4, Q test p-value: 0.423, I2: 1.30) (Table 2).
Table 2.
Association between rs599839 polymorphism and cardiovascular diseases.
| Model | Random Effect | Z p-Value | Q Test p-Value | I2 | Egger Test p-Value |
|---|---|---|---|---|---|
| Overall | |||||
| G | Reference | ||||
| A | 1.27 (1.11–1.44) | 1 × 10−4 | 1 × 10−4 | 80.82 | 0.100 |
| A * | 1.19 (1.13–1.26) | 1 × 10−4 | 0.175 | 30.38 | 0.108 |
| GG c | Reference | ||||
| GA c | 1.11 (0.92–1.35) | 0.28 | 0.155 | 31.70 | 0.100 |
| AA c | 1.33 (1.00–1.75) | 0.045 | 0.009 | 58.90 | 0.100 |
| GG d | Reference | ||||
| GA+AA d | 1.25 (0.98–1.59) | 0.064 | 0.019 | 53.15 | 0.300 |
| GA+AA d* | 1.22 (1.06–1.39) | 3 × 10−3 | 0.141 | 34.63 | 0.446 |
| GG+GA r | Reference | ||||
| AA r | 1.29 (1.12–1.49) | 1 × 10−4 | 1 × 10−4 | 76.65 | 0.103 |
| AA r* | 1.23 (1.15–1.32) | 1 × 10−4 | 0.423 | 1.30 | 0.175 |
| CAD | |||||
| G | Reference | ||||
| A | 1.30 (1.11–1.51) | 1 × 10−4 | 1 × 10−4 | 81.22 | 0.100 |
| A * | 1.17 (1.10–1.24) | 1 × 10−4 | 0.265 | 21.54 | 0.310 |
| GG c | Reference | ||||
| GA c | 1.18 (1.00–1.39) | 0.039 | 0.340 | 11.7 | 0.100 |
| AA c | 1.51 (1.14–2.00) | 4 × 10−3 | 0.041 | 54.2 | 0.100 |
| AA c* | 1.39 (1.18–1.65) | 1 × 10−4 | 0.325 | 13.66 | 0.614 |
| GG d | Reference | ||||
| GA+AA d | 1.39 (1.08–1.79) | 0.010 | 0.037 | 52.97 | 0.104 |
| GA+AA d* | 1.24 (1.08–1.42) | 2 × 10−3 | 0.153 | 36.11 | 0.301 |
| GG+GA r | Reference | ||||
| AA r | 1.32 (1.12–1.56) | 1 × 10−3 | 1 × 10−4 | 75.99 | 0.100 |
| AA r* | 1.20 (1.11–1.29) | 1 × 10−4 | 0.645 | 0.000 | 0.386 |
| Asian population | |||||
| G | Reference | ||||
| A | 1.32 (1.12–1.57) | 1 × 10−3 | 1 × 10−4 | 80.70 | 0.290 |
| A * | 1.27 (1.18–1.36) | 1 × 10−4 | 0.494 | 0.000 | 0.236 |
| GG c | Reference | ||||
| GA c | 0.84 (0.60–1.16) | 0.288 | 0.600 | 0.000 | 0.340 |
| AA c | 1.29 (1.29–1.30) | 0.260 | 0.060 | 43.48 | 0.320 |
| AA c* | 1.07 (0.77–1.49) | 0.676 | 0.424 | 1.201 | 0.653 |
| GG d | Reference | ||||
| GA+AA d | 1.12 (0.81–1.55) | 0.466 | 0.112 | 37.12 | 0.323 |
| GG+GA r | Reference | ||||
| AA r | 1.34 (1.14–1.59) | 1 × 10−4 | 1 × 10−4 | 77.79 | 0.324 |
| AA r* | 1.30 (1.21–1.40) | 1 × 10−4 | 0.629 | 0.000 | 0.333 |
c: codominant; d: dominant; r: recessive. * Model without heterogeneity.
In a sub-analysis, we took into consideration the studies that specifically evaluated coronary artery disease and its relation to the rs599839 polymorphism. The findings without heterogeneity revealed a statistical association with CAD in all the genetic models: allelic (OR: 1.17, 1.10–1.24, p-value 1 × 10−4, Q test p-value: 0.265, I2: 21.54), codominant (OR: 1.39, 1.18–1.65, p-value 1 × 10−4, Q test p-value: 0.325, I2: 13.66), dominant (OR: 1.24, 1.08–1.42, p-value 2 × 10−3, Q test p-value: 0.153, I2: 36.11), and recessive (OR: 1.20, 1.11–1.29, p-value 1 × 10−4, Q test p-value: 0.645, I2: 0.000) (Table 2).
Lastly, in an additional sub-analysis, we explored the association of rs599839 polymorphism and CVDs in Asian populations. In this sub-analysis without the presence of heterogeneity, we observed a statistically significant relation in the following genetic models: allelic (OR: 1.27, 1.18–1.36, p-value 1 × 10−4, Q test p-value: 0.494, I2: 0.000), and recessive (OR: 1.30, 1.21–1.40, p-value 1 × 10−4, Q test p-value: 0.629, I2: 0.000) (Table 2).
3.2.2. rs646776 Polymorphism and Susceptibility to Cardiovascular Diseases
For the studies that genotyped the rs646776 polymorphism, we first evaluated the overall cardiovascular diagnoses. However, the findings revealed no statistical association in the allelic genetic model (OR: 1.17, 95% CI: 0.86–1.59, p-value 0.290, Q test p-value: 0.040, I2: 68.81). Then, in another sub-analysis, we evaluated studies performed in the Asian population. In this sense, the outcomes evidenced a statistical association in the following model: allelic (OR: 1.46, 95% CI: 1.17–1.82, p-value 1 × 10−3, Q test p-value: 0.643, I2: 0.00) (Table 3).
Table 3.
Association between rs646776 polymorphism and cardiovascular diseases.
| Model | Random Effect | Z p-Value | Q Test p-Value | I2 | Egger Test p-Value |
|---|---|---|---|---|---|
| Overall | |||||
| C | Reference | ||||
| T | 1.17 (0.86–1.59) | 0.290 | 0.040 | 68.81 | 0.420 |
| Asian Population | |||||
| C | Reference | ||||
| T | 1.46 (1.17–1.82) | 1 × 10−3 | 0.643 | 0.00 | 0.462 |
3.2.3. rs464218 Polymorphism and Susceptibility to Cardiovascular Diseases
In the analysis of the studies that included the rs464218 polymorphism, we only analyzed the overall diagnoses due to the limited available reports. In this analysis, we found a statistically significant association in the recessive genetic model (OR: 2.03, 95% CI: 1.19–3.47, p-value 9 × 10−3, Q test p-value: 8 × 10−3, I2: 79.47) (Table 4). However, after correcting the p-value, it does not remain significant.
Table 4.
Association between rs464218 polymorphism and cardiovascular diseases.
| Model | Random Effect | Z p-Value | Q Test p-Value | I2 | Egger Test p-Value |
|---|---|---|---|---|---|
| Overall | |||||
| G | Reference | ||||
| A | 1.10 (0.85–1.41) | 0.448 | 0.020 | 74.58 | 0.101 |
| GG c | Reference | ||||
| GA c | 0.847 (0.37–1.92) | 0.692 | 1 × 10−4 | 94.4 | 0.100 |
| AA c | 1.94 (0.76–4.92) | 0.163 | 1 × 10−4 | 92.2 | 0.100 |
| GG d | Reference | ||||
| GA+AA d | 1.10 (0.48–2.48) | 0.817 | 1 × 10−4 | 95.27 | 0.893 |
| GG+GA r | Reference | ||||
| AA r | 2.03 (1.19–3.47) | 9 × 10−3 | 8 × 10−3 | 79.47 | 0.951 |
c: codominant; d: dominant; r: recessive.
3.3. Publication Bias and Sensitive Analysis
Funnel plots visualized publication bias; nonetheless, no evidence of publication bias was found in the pooled analyses. On the other hand, a sensitivity analysis was conducted to assess the influence of one study on the pooled ORs value (allele and dominant genetic models) and whether the results can be reverted by eliminating the individual study. The result did not change after the leaving-one-out analysis, indicating the stability of the results of this meta-analysis.
3.4. Bioinformatic Analysis
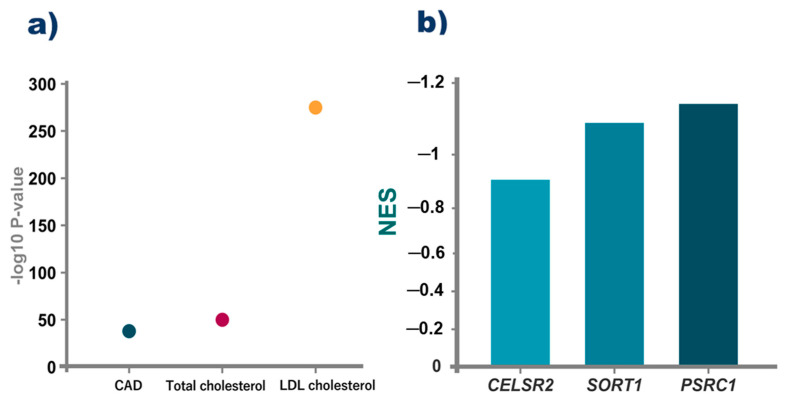
We performed the bioinformatic analysis only on rs599839, because it showed the highest number of significant associations in the cluster. The results from the PheWAS indicate that this genetic variant had been associated with intermediate phenotypes for CAD, total cholesterol, and LDL levels (Figure 2). Additionally, data from GTeX shows that this variant could change the expression of the three cluster genes in the liver.
Figure 2.
Previous associations of rs599839 with related phenotypes. (a) p-values for the prior association of rs599839 with coronary artery disease (CAD), total cholesterol, and LDL cholesterol obtained from the PheWAS atlas. (b) NES value for the rs599839 on the transcripts liver expression of the genes included on the cluster of CELSR2-PSRC1-SORT1. NES is the normalized effect size, which is the slope of the linear regression of the alternative compared to the reference allele.
4. Discussion
Cardiovascular diseases have a complex multifactorial etiology involving various risk factors that contribute to their development, including a genetic component. In this regard, several studies have suggested that genes that together form the CELSR2-PSRC1-SORT1 gene cluster and are located on chromosome 1 could be involved in the development of cardiovascular diseases through mechanisms related to serum lipid levels and plasma cholesterol levels [8,15].
Therefore, we analyzed three polymorphisms (rs646776, rs599839, and rs464218) in the CELSR2-PSRC1-SORT1 cluster in 25224 cases and 33181 controls, investigating the association between these genetic variants and susceptibility to develop cardiovascular diseases through a systematic review and updated meta-analysis.
First, after discarding heterogeneity in the included studies, we observed that the rs599839 could increase the risk of developing any cardiovascular disease (coronary heart disease, myocardial infarction, or acute coronary artery syndrome) by 1.19 to 1.23 times in the allelic, dominant, and recessive models.
Also, when we analyzed the rs599839 polymorphism and coronary artery disease only, we found a strong risk association that ranged from 1.17 to 1.39 times more presenting coronary artery disease in all genetic models evaluated: allelic, codominant, dominant, and recessive. These results suggest that the rs599839 polymorphism may be associated with increased susceptibility to developing coronary artery disease. Previous studies have explored the relationship between the rs599839 polymorphism and coronary artery disease [15,28]. Although we found a strong risk association, Rodríguez-Arellano ME, 2020, reported a protective association with CAD in the Mexican population [16]. These contradictory results could be due to variables that affect the genotype–phenotype association, such as age, ancestry, gene–environment interactions, sex, and sample size. However, we must consider that our CAD sub-analysis included various Asian, American, and European populations, not just one population [15,27,28,31].
Finally, we carried out a specific analysis of Asian populations. When we ruled out heterogeneity, we found that carriers of the allelic and recessive models had 1.27 and 1.30 more times, respectively, the risk of developing cardiovascular diseases. In this regard, Matsuoka, R., 2015 found an association between the rs599839 polymorphism (PSRC1, FDR = 0.0118) and myocardial infarction in the Asian population [24]. Our results suggest that the rs599839 polymorphism of the PSRC1 gene could participate in the development of cardiovascular diseases and mainly in the genetic predisposition to coronary artery disease.
We also analyzed the rs646776 polymorphism of the CELSR2 gene. In the first analysis, we evaluated the overall diagnoses, and we did not find statistical significance in the allelic model. Then, we performed a second analysis evaluating the Asian population in the included studies, and we observed in the allelic model a higher risk of developing cardiovascular diseases. In this regard, Kathiresan et al., 2009 [33] reported a significant association between rs646776 and early-onset myocardial infarction in 560 controls and 1231 patients from the following countries: Italy, Finland, Boston and Seattle in the United States, Spain, and Sweden. Our results suggest that the rs646776 polymorphism could be associated with an increased risk of cardiovascular diseases in the Asian population.
Moreover, we analyzed the rs464218 polymorphism of the SORT1 gene. We observed an association that suggests the risk of developing CVDs (overall diagnoses) under the recessive model. There are few studies described in the literature on this polymorphism, and the results are controversial. Therefore, additional studies are required to elucidate the participation of this genetic variant in cardiovascular diseases [15,22]. Although we observed statistical significance, it is important to mention that because of the limited number of studies included in this sub-analysis, we could not rule out heterogeneity. In addition, after correcting p-value, the significance does not remain. Therefore, it is possible that the results are biased. Furthermore, this sub-analysis only included the Asian population due to the lack of studies to include other populations.
On the other hand, our work has important strengths: this is the first meta-analysis involving three genetic variants (rs646776, rs599839, and rs464218) contained in the CELSR2-PSRC1-SORT1 cluster, and we found an association with different cardiovascular diseases. Second, this work shows a strong risk association between the rs599839 polymorphism and coronary artery disease. Third, in this meta-analysis, we included studies in the main electronic databases and multiple searches to gather as much information as possible.
We recognize that we have limitations in our study: First, due to the few studies reported in different populations, we were only able to perform a sub-analysis in the Asian population. Second, due to the small sample size for some subgroup analyses, it will be necessary to conduct additional studies with larger samples that include different populations in order to improve the reliability and stability of the meta-analysis. Third, because only studies published in the English language were included, there could be a language bias. Fourth, despite the few studies included in the present meta-analysis, the p-value was adjusted by Bonferroni correction. However, this correction could be improved in future studies with more diverse and larger sample sizes, considering other approaches applied to candidate gene studies. Fifth, we did not perform a haplotype analysis due to the few studies included in this meta-analysis. Sixth, we suggest other computational tools and approaches that could be more stringent in calculating statistical significance for future studies.
5. Conclusions
In conclusion, our results suggest an association of the CELSR2-PSRC1-SORT1 cluster variants with an increased risk of cardiovascular diseases, particularly coronary artery disease. However, our findings warrant replication in larger sample sizes due to several limitations.
Acknowledgments
The authors are grateful to Universidad Juárez Autónoma de Tabasco and the Department of Molecular Biology from Instituto Nacional de Cardiología Ignacio Chavez for all support received for the present research. Moreover, the authors are grateful to Laura E. Martínez-Gómez from the Instituto Nacional de Rehabilitación Luis Guillermo Ibarra Ibarra for her support in the critical review of this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jcdd10030091/s1, Supplementary Material S1: PRISMA 2020 Checklist; Supplementary Material S2: Search formulas in the databases included.
Author Contributions
Conceptualization, R.G.C.-A., T.B.G.-C., C.A.T.-Z., N.P.-H. and J.M.R.-P.; methodology, R.G.C.-A., T.B.G.-C., I.E.J.-R., M.L.L.-N. and V.M.B.-C.; software, T.B.G.-C. and J.J.M.-M.; validation, R.G.C.-A., T.B.G.-C. and V.M.B.-C.; formal analysis, T.B.G.-C. and J.J.M.-M.; investigation, R.G.C.-A., T.B.G.-C., C.A.T.-Z., N.P.-H., J.J.M.-M., V.M.B.-C. and J.M.R.-P.; data curation, T.B.G.-C.; writing—original draft preparation, R.G.C.-A., T.B.G.-C., C.A.T.-Z., N.P.-H. and J.M.R.-P.; writing—review and editing, R.G.C.-A., T.B.G.-C., C.A.T.-Z., N.P.-H. and J.M.R.-P.; visualization, P.I.A.-V., M.L.L.-N. and I.E.J.-R.; supervision, P.I.A.-V., T.B.G.-C., C.A.T.-Z., N.P.-H. and J.M.R.-P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Virani S.S., Alonso A., Aparicio H.J., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Cheng S., Delling F.N., et al. Heart Disease and Stroke Statistics-2021 Update: A Report from the American Heart Association. Circulation. 2021;143:e254–e743. doi: 10.1161/cir.0000000000000950. [DOI] [PubMed] [Google Scholar]
- 2.Roth G.A., Mensah G.A., Johnson C.O., Addolorato G., Ammirati E., Baddour L.M., Barengo N.C., Beaton A.Z., Benjamin E.J., Benziger C.P., et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020;76:2982–3021. doi: 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flora G.D., Nayak M.K. A Brief Review of Cardiovascular Diseases, Associated Risk Factors and Current Treatment Regimes. Curr. Pharm. Des. 2019;25:4063–4084. doi: 10.2174/1381612825666190925163827. [DOI] [PubMed] [Google Scholar]
- 4.Tzoulaki I., Elliott P., Kontis V., Ezzati M. Worldwide Exposures to Cardiovascular Risk Factors and Associated Health Effects: Current Knowledge and Data Gaps. Circulation. 2016;133:2314–2333. doi: 10.1161/CIRCULATIONAHA.115.008718. [DOI] [PubMed] [Google Scholar]
- 5.Shah V.N., Bailey R., Wu M., Foster N.C., Pop-Busui R., Katz M., Crandall J., Bacha F., Nadeau K., Libman I., et al. Risk Factors for Cardiovascular Disease (CVD) in Adults with Type 1 Diabetes: Findings from Prospective Real-life T1D Exchange Registry. J. Clin. Endocrinol. Metab. 2020;105:e2032–e2038. doi: 10.1210/clinem/dgaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusuf S., Joseph P., Rangarajan S., Islam S., Mente A., Hystad P., Brauer M., Kutty V.R., Gupta R., Wielgosz A., et al. Modifiable risk factors, cardiovascular disease, and mortality in 155 722 individuals from 21 high-income, middle-income, and low-income countries (PURE): A prospective cohort study. Lancet. 2020;395:795–808. doi: 10.1016/S0140-6736(19)32008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Møller P.L., Rohde P.D., Winther S., Breining P., Nissen L., Nykjaer A., Bøttcher M., Nyegaard M., Kjolby M. Sortilin as a Biomarker for Cardiovascular Disease Revisited. Front. Cardiovasc. Med. 2021;8:652584. doi: 10.3389/fcvm.2021.652584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arvind P., Nair J., Jambunathan S., Kakkar V.V., Shanker J. CELSR2-PSRC1-SORT1 gene expression and association with coronary artery disease and plasma lipid levels in an Asian Indian cohort. J. Cardiol. 2014;64:339–346. doi: 10.1016/j.jjcc.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Samani N.J., Braund P.S., Erdmann J., Götz A., Tomaszewski M., Linsel-Nitschke P., Hajat C., Mangino M., Hengstenberg C., Stark K., et al. The novel genetic variant predisposing to coronary artery disease in the region of the PSRC1 and CELSR2 genes on chromosome 1 associates with serum cholesterol. J. Mol. Med. 2008;86:1233–1241. doi: 10.1007/s00109-008-0387-2. [DOI] [PubMed] [Google Scholar]
- 10.Xu M., Zhu S., Xu R., Lin N. Identification of CELSR2 as a novel prognostic biomarker for hepatocellular carcinoma. BMC Cancer. 2020;20:313. doi: 10.1186/s12885-020-06813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Eitan L.N., Elsaqa B.Z., Almasri A.Y., Aman H.A., Khasawneh R.H., Alghamdi M.A. Influence of PSRC1, CELSR2, and SORT1 Gene Polymorphisms on the Variability of Warfarin Dosage and Susceptibility to Cardiovascular Disease. Pharm. Pers. Med. 2020;13:619–632. doi: 10.2147/PGPM.S274246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei M., Li P., Guo K. The impact of PSRC1 overexpression on gene and transcript expression profiling in the livers of ApoE(-/-) mice fed a high-fat diet. Mol. Cell. Biochem. 2020;465:125–139. doi: 10.1007/s11010-019-03673-x. [DOI] [PubMed] [Google Scholar]
- 13.Kjolby M., Nielsen M.S., Petersen C.M. Sortilin, encoded by the cardiovascular risk gene SORT1, and its suggested functions in cardiovascular disease. Curr. Atheroscler. Rep. 2015;17:496. doi: 10.1007/s11883-015-0496-7. [DOI] [PubMed] [Google Scholar]
- 14.Goettsch C., Kjolby M., Aikawa E. Sortilin and Its Multiple Roles in Cardiovascular and Metabolic Diseases. Arterioscler. Thromb. Vasc. Biol. 2018;38:19–25. doi: 10.1161/ATVBAHA.117.310292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y.J., Hong S.C., Yang Q., Yin R.X., Cao X.L., Chen W.X. Association of variants in CELSR2-PSRC1-SORT1 with risk of serum lipid traits, coronary artery disease and ischemic stroke. Int. J. Clin. Exp. Pathol. 2015;8:9543–9551. [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez-Arellano M.E., Solares-Tlapechco J., Costa-Urrutia P., Cárdenas-Hernández H., Vallejo-Gómez M., Granados J., Salas-Padilla S. Association of the PSRC1 rs599839 Variant with Coronary Artery Disease in a Mexican Population. Medicina. 2020;56:427. doi: 10.3390/medicina56090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iniesta R., Guinó E., Moreno V. Statistical analysis of genetic polymorphisms in epidemiological studies. Gac. Sanit. 2005;19:333–341. doi: 10.1157/13078029. [DOI] [PubMed] [Google Scholar]
- 19.Tian D., Wang P., Tang B., Teng X., Li C., Liu X., Zou D., Song S., Zhang Z. GWAS Atlas: A curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020;48:D927–D932. doi: 10.1093/nar/gkz828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L., Ding H., Zhang X., He M., Huang S., Xu Y., Shi Y., Cui G., Cheng L., Wang Q.K., et al. Genetic variants at newly identified lipid loci are associated with coronary heart disease in a Chinese Han population. PLoS ONE. 2011;6:e27481. doi: 10.1371/journal.pone.0027481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J., Tian J., Liu G., Zhang Y., Tian L., Zhen Y., Zhang H., Xu J., Sun X., Fang H. Association between 1p13 polymorphisms and peripheral arterial disease in a Chinese population with diabetes. J. Diabetes Investig. 2018;9:1189–1195. doi: 10.1111/jdi.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han W., Wei Z., Zhang H., Geng C., Dang R., Yang M., Zhang J., Wang C., Jiang P. The Association between Sortilin and Inflammation in Patients with Coronary Heart Disease. J. Inflamm. Res. 2020;13:71–79. doi: 10.2147/JIR.S240421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueyama C., Horibe H., Yamase Y., Fujimaki T., Oguri M., Kato K., Arai M., Watanabe S., Murohara T., Yamada Y. Association of FURIN and ZPR1 polymorphisms with metabolic syndrome. Biomed. Rep. 2015;3:641–647. doi: 10.3892/br.2015.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka R., Abe S., Tokoro F., Arai M., Noda T., Watanabe S., Horibe H., Fujimaki T., Oguri M., Kato K., et al. Association of six genetic variants with myocardial infarction. Int. J. Mol. Med. 2015;35:1451–1459. doi: 10.3892/ijmm.2015.2115. [DOI] [PubMed] [Google Scholar]
- 25.Fujimaki T., Oguri M., Horibe H., Kato K., Matsuoka R., Abe S., Tokoro F., Arai M., Noda T., Watanabe S., et al. Association of a transcription factor 21 gene polymorphism with hypertension. Biomed. Rep. 2015;3:118–122. doi: 10.3892/br.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abe S., Tokoro F., Matsuoka R., Arai M., Noda T., Watanabe S., Horibe H., Fujimaki T., Oguri M., Kato K., et al. Association of genetic variants with dyslipidemia. Mol. Med. Rep. 2015;12:5429–5436. doi: 10.3892/mmr.2015.4081. [DOI] [PubMed] [Google Scholar]
- 27.Ellis K.L., Frampton C.M., Pilbrow A.P., Troughton R.W., Doughty R.N., Whalley G.A., Ellis C.J., Skelton L., Thomson J., Yandle T.G., et al. Genomic risk variants at 1p13.3, 1q41, and 3q22.3 are associated with subsequent cardiovascular outcomes in healthy controls and in established coronary artery disease. Circ. Cardiovasc. Genet. 2011;4:636–646. doi: 10.1161/CIRCGENETICS.111.960336. [DOI] [PubMed] [Google Scholar]
- 28.Kleber M.E., Renner W., Grammer T.B., Linsel-Nitschke P., Boehm B.O., Winkelmann B.R., Bugert P., Hoffmann M.M., März W. Association of the single nucleotide polymorphism rs599839 in the vicinity of the sortilin 1 gene with LDL and triglyceride metabolism, coronary heart disease and myocardial infarction. The Ludwigshafen Risk and Cardiovascular Health Study. Atherosclerosis. 2010;209:492–497. doi: 10.1016/j.atherosclerosis.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 29.Rizk N.M., El-Menyar A., Egue H., Souleman Wais I., Mohamed Baluli H., Alali K., Farag F., Younes N., Al Suwaidi J. The Association between Serum LDL Cholesterol and Genetic Variation in Chromosomal Locus 1p13.3 among Coronary Artery Disease Patients. BioMed Res. Int. 2015;2015:678924. doi: 10.1155/2015/678924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gigante B., Leander K., Vikström M., Ye S., de Faire U. Chromosome 1p13 genetic variants antagonize the risk of myocardial infarction associated with high ApoB serum levels. BMC Cardiovasc. Disord. 2012;12:90. doi: 10.1186/1471-2261-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bressler J., Folsom A.R., Couper D.J., Volcik K.A., Boerwinkle E. Genetic variants identified in a European genome-wide association study that were found to predict incident coronary heart disease in the atherosclerosis risk in communities study. Am. J. Epidemiol. 2010;171:14–23. doi: 10.1093/aje/kwp377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ansari W.M., Humphries S.E., Naveed A.K., Khan O.J., Khan D.A., Khattak E.H. Effect of Coronary Artery Disease risk SNPs on serum cytokine levels and cytokine imbalance in Premature Coronary Artery Disease. Cytokine. 2019;122:154060. doi: 10.1016/j.cyto.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., Schunkert H., et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.