ABSTRACT
Strain specificity (within-species variation) of microorganisms occurs widely in nature. It might affect microbiome construction and function in a complex microbial environment. Tetragenococcus halophilus, a halophilic bacterium that generally is used in high salt food fermentation, consists of two histamine-producing and non-histamine-producing subgroups. It is unclear whether and how the strain specificity of histamine-producing capacity influences the microbial community function during food fermentation. Here, based on systematic bioinformatic analysis, histamine production dynamic analysis, clone library construction analysis, and cultivation-based identification, we identified that T. halophilus is the focal histamine-producing microorganism during soy sauce fermentation. Furthermore, we discovered that a larger number and ratio of histamine-producing subgroups of T. halophilus significantly contributed more histamine production. We were able to artificially decrease the ratio of histamine-producing to non-histamine-producing subgroups of T. halophilus in complex soy sauce microbiota and realized the reduction of histamine by 34%. This study emphasizes the significance of strain specificity in regulating microbiome function. This study investigated how strain specificity influenced microbial community function and developed an efficient technique for histamine control.
IMPORTANCE Inhibiting the production of microbiological hazards under the assumption of stable and high-quality fermentation is a critical and time-consuming task for the food fermentation industry. For spontaneously fermented food, it can be realized theoretically by finding and controlling the focal hazard-producing microorganism in complex microbiota. This work used histamine control in soy sauce as a model and developed a system-level approach to identify and regulate the focal hazard-producing microorganism. We discovered that the strain specificity of focal hazard-producing microorganisms had an important impact on hazard accumulation. Microorganisms frequently exhibit strain specificity. Strain specificity is receiving increasing interest since it determines not only microbial robustness but also microbial community assembly and microbiome function. This study creatively explored how the strain specificity of microorganisms influenced microbiome function. In addition, we believe that this work provides an excellent model for microbiological hazard control which can promote future work in other systems.
KEYWORDS: strain specificity, Tetragenococcus halophilus, histamine, biogenic amines, soy sauce, spontaneous fermentation
INTRODUCTION
Dependent on cultural-based approaches, microorganisms were found with characteristics of strain diversity (within-species variety) at the single-clone level (1). With the development of a high-resolution metagenomic approach, the strain diversity of bacteria has been discovered and analyzed at the natural microecosystem level (1). The strain specificity of microorganisms generally occurs due to the vertical accumulation of mutations or the horizontal acquisition of genes (1, 2). In complex microbial ecosystems, strain specificity can improve microbial responsiveness to external disturbance (3) or influence microbial community function (4, 5) at the subspecies level. However, it is unclear how the strain diversity of microorganisms influences the microbial community function.
Biogenic amines (BAs), including histamine, tyramine, and putrescine, are a kind of toxic low-molecular nitrogen-containing compound that is formed by microbial decarboxylation of corresponding amino acids (6). They are usually detected in high-protein fermented foods, such as cheese (7), fermented fish (8), and soy sauce (9), bringing a huge risk to food safety. Histamine is regarded as the most toxic BA, and excessive intake will cause serious pathological reactions (10). Thus, concentration limits for histamine in certain foods have been established to protect consumers from possible safety risks. In 2003, the European Commission stipulated that the amount of histamine in foodstuffs should not exceed 100 mg/kg of body weight (11). Furthermore, in 2005, the United States Food and Drug Administration set the maximum concentration limit of histamine at 50 mg/kg in aquatic products (12).
To reduce the biogenic amine (especially histamine) accumulation in fermented foods, multiple approaches have been proposed and developed. On one hand, as BAs are synthesized by the microbial decarboxylation reaction, approaches that can inhibit microbial proliferation or reduce decarboxylase activity, such as lowering the temperature, improving the salinity, and inoculating microbial starter (13, 14), might play roles in reducing BA accumulation. On the other hand, BAs can be degraded by adding amine oxidases or relevant microorganisms (15, 16), which can convert BAs into aldehyde, ammonia, and hydrogen peroxide (17). However, BA degradation will result in nitrogen loss through ammonia volatilization. A new BA control strategy that can realize the process of controlling of BAs by rationally inhibiting the growth of focal BA-producing microorganisms was needed urgently.
Soy sauce, with an intense umami taste, pleasant aroma, and bioactive compounds, is one of the most popular condiments in Asian countries and is increasingly consumed in the Western world. However, at least one BA was detected in commercial soy sauce samples (9). Furthermore, more than one-quarter of soy sauce samples contain more than 0.2 mg/mL of histamine, which may cause histamine intolerance in susceptible people (18). Soybean (or soybean meal) is the primary raw material used in soy sauce production, which means that the fermentation process contains a high concentration of BA biosynthetic substrates and amino acids. In addition, Chinese soy sauce ferments spontaneously and has a complex microbial community structure (19, 20). These phenomena illustrate the necessity and difficulty of identifying the focal BA-producing microorganisms from complex soy sauce microbiota and subsequently controlling BA accumulation.
In this study, we identified T. halophilus as the focal histamine-producing strain during spontaneous soy sauce fermentation. T. halophilus consists of histamine-producing and non-histamine-producing subgroups. We found that the ratio of histamine-producing to non-histamine-producing subgroups of T. halophilus determines the histamine accumulation. By artificially decreasing the ratio of histamine-producing to non-histamine-producing subgroups of T. halophilus, the histamine concentration decreased by 34%. This study provided an effective strategy for histamine control. We highlighted the importance of strain specificity on microbiome function. It also investigated how bacterial strain diversity influences complex microbial community function.
RESULTS
Accumulation patterns of biogenic amines during soy sauce fermentation.
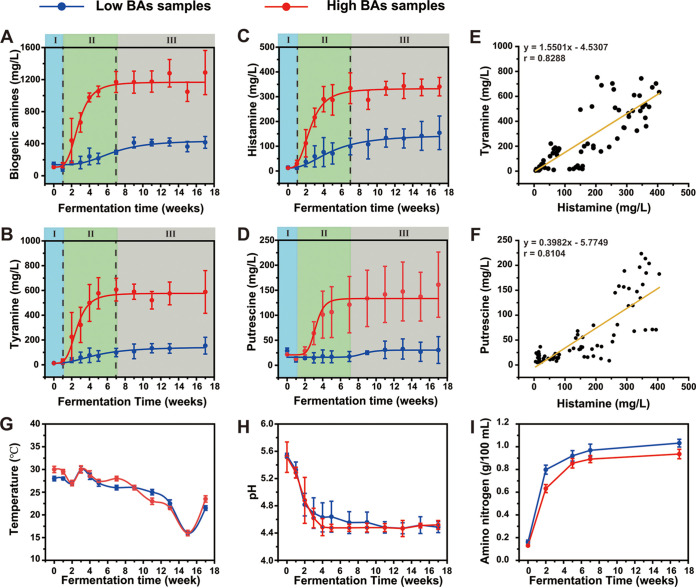
To determine if and when biogenic amines (BAs) accumulate during soy sauce fermentation, we took weekly samples of fermented soy sauce moromi and measured their BA contents. Six kinds of BAs, including phenethylamine, putrescine, cadaverine, histamine, tyramine, and spermidine, were detected (Fig. 1; see Fig. S1 and S2 in the supplemental material). Tyramine and histamine accounted for more than 70% of the total BAs in soy sauce samples, whereas four other BAs accounted for less than 30% (Fig. S1). The majority of BAs were accumulated between the first and seventh fermentative weeks (Fig. 1; see Fig. S3 in the supplemental material). Based on this finding, we divided the soy sauce fermentation into three stages, as follows: stage I, BA-delay period (0 to 1 week); stage II, BA-accumulation period (1 to 7 week); and stage III, BA-stable period (7 to 17 week) (Fig. 1A to D). We discovered that the BA concentrations of various samples were considerably different from the start of stage II, even though their fermentation parameters, such as fermentation temperature and pH, were similar (Fig. 1G to I). Based on the BA content at the fermentation endpoint, three soy sauce samples were identified as high-BA samples with BA concentrations above 1,000 mg/L (with an average concentration of BAs of 1,287.66 mg/L), and the other three soy sauce samples were identified as low-BA samples with BA concentrations below 500 mg/L (with an average concentration of BAs of 416.66 mg/L). These phenomena enabled us to investigate the focal BA-producing microorganisms, for example, by comparing the microbial differences of high- and low-BA samples during the BA-accumulation phases. Different BAs have different synthesis processes, and some synthesis pathways of BA are quite complicated (7). Therefore, we have studied only one major BA at this time due to the high workload. Histamine is considered one of the most toxic BAs and is particularly relevant for food safety (21, 22). Moreover, histamine is one of the prominent BAs during soy sauce fermentation (Fig. 1C; Fig. S1). It is worth noting that the histamine production is closely correlated with that of tyramine and putrescine, which are two other major BAs (Fig. 1E and F; Fig. S2C and D). Therefore, we focused primarily on the focal histamine-producing microorganisms and histamine control in the following studies, aiming to achieve simultaneous control of multiple BAs.
FIG 1.
Biogenic amines and fermentation parameters during the soy sauce fermentation. (A to D) Dynamic of biogenic amine accumulation in high-BA (red line) and low-BA (blue line) samples. The total biogenic amines (A), tyramine (B), histamine (C), and putrescine (D). Correlation analysis of tyramine (E) and putrescine (F) with histamine. (G to I) Dynamic of fermentation parameter in high-BA (red line) and low-BA (blue line) samples. Fermentation temperature (G), fermentation pH (H), and amino acid concentration (I). Data are indicated as the average of three samples ± standard deviation.
Potential histamine-producing bacterial studies by using bioinformatics analysis.
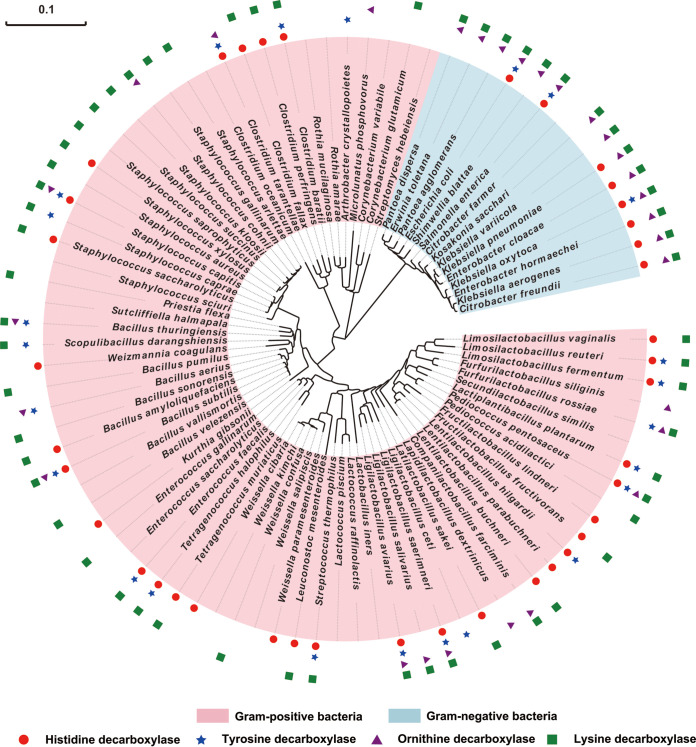
Traditional Chinese soy sauce is made by spontaneous fermentation, which implies that a complex microbial community participates in the fermentation of soy sauce. Therefore, we analyzed microorganisms involved in the soy sauce fermentation process and their ability to produce BAs. We collected and analyzed the soy sauce microorganisms reported in the previous literature (20, 23–27). A total of 88 bacteria were reported during soy sauce fermentation, among which 73 bacteria belong to the Gram-positive group and 15 bacteria belong to the Gram-negative group. As shown in Fig. 2, the phylogenetic tree of soy sauce bacteria was constructed using the 16S rRNA gene sequence.
FIG 2.
BA production capability of soy sauce microorganisms. The phylogenetic tree was constructed based on the 16S rRNA genes sequence of the type species of soy sauce microorganisms. Red dot, microorganisms containing histidine decarboxylase (potential histamine production capability); blue star, microorganisms containing tyrosine decarboxylase (potential tyramine production capability); purple triangle, microorganisms containing ornithine decarboxylase (potential putrescine production capability); green square, microorganisms containing lysine decarboxylase (potential cadaverine production capability).
To investigate whether the soy sauce bacteria have the histamine production capability, we searched the histidine decarboxylase gene (and three other amino acid decarboxylase genes for biosynthesis of tyramine, putrescine, and cadaverine) from genomes by using NCBI BLAST. As shown in Fig. 2, 86.36% (76/88) of the bacteria have relevant amino acid decarboxylase genes and thus have the potential to produce one or more BAs. Most strains may produce multiple BAs simultaneously. For example, nine bacteria, including Bacillus subtilis, Escherichia coli, Klebsiella oxytoca, Ligilactobacillus aviarius, Furfurilactobacillus rossiae, Ligilactobacillus saerimneri, Pediococcus pentosaceus, Salmonella enterica, and Staphylococcus capitis, contain four amino acid decarboxylases. Also, 13 bacteria contain 3 amino acid decarboxylases simultaneously (Fig. 2). These results might explain why histamine production is closely related to the accumulation of the other BAs. In those soy sauce bacteria, 44.32% (39/88) of soy sauce bacteria have histidine decarboxylase genes and thus might produce histamine. This result indicates that many soy sauce bacteria have the potential to produce histamine, and it is difficult to study focal histamine-producing microorganisms from complex soy sauce microbiota.
Except for the Tetragenococcus genus, all Gram-positive bacteria revealed a relatively weak histamine-producing capability.
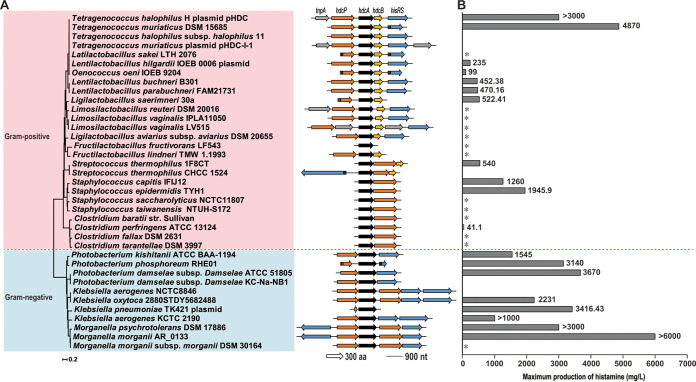
To reveal the focal histamine-producing microorganisms during soy sauce fermentation, we analyzed the structure of histidine biosynthesis gene clusters and the reported maximum histamine concentration produced by these strains. As shown in Fig. 3A, the structure of the histidine decarboxylase gene cluster varied between strains, and a complete histamine biosynthesis gene cluster contains at least one histidine decarboxylase gene hdcA and one histidine-histamine exchanger gene hdcP. In Gram-negative bacteria, a complete histamine biosynthesis gene cluster also contains a histidyl-tRNA synthetase gene, hisRS. Concerningly, some Gram-negative bacteria, such as members of the Morganella and Klebsiella genera, have several hdcP and/or hisRS genes for unknown reasons. In most Gram-positive bacteria, the histamine biosynthesis gene cluster contains genes hdcP, hdcA, hdcB, and hisRS. The gene hdcB contributes to the maturation of histidine decarboxylase hdcA (28).
FIG 3.
Histamine-biosynthesis gene cluster analysis (A) and the maximum histamine production capacity (B) in histamine-producing bacteria. The phylogenetic tree was constructed based on the hdcA gene sequence of histamine-producing microorganisms. Arrows indicate open reading frames (ORFs) and transcription directions. Lines indicate intergenic regions (IGRs). *, Histamine production capacity has not been reported. Note: Information on the maximum histamine production capacity in histamine-producing bacteria is shown in Table S1.
Diverse gene clusters often indicate a wide range of histamine-production capabilities. We collected information on the highest reported histamine concentrations produced by the bacteria listed above, which were cultured in culture media or fermentation systems (see Table S1 in the supplemental material). As shown in Fig. 3B, based on reported data on the histamine-producing capacity of bacteria, we found that Gram-negative bacteria usually exhibited greater histamine production than Gram-positive bacteria, ranging from 1,000 mg/L to 6,000 mg/L, although the histamine-producing capacity of many Gram-positive bacteria has not been investigated (labeled with asterisk in Fig. 3B). Gram-positive bacteria, particularly lactic acid bacteria, typically produce low concentrations of histamine (<1,000 mg/L), except Tetragenococcus halophilus and Tetragenococcus muriaticus, which belong to the Tetragenococcus genus and can produce a maximum of 3,000 and 4,870 mg/L of histamine, respectively (Fig. 3B).
Soy sauce fermentation is usually carried out in a hypertonic environment with a salt concentration of more than 20%. As reported previously, we found that the majority of microorganisms in soy sauce fermentation are Gram-positive bacteria (Fig. 2). However, some Gram-negative bacteria were also reported in soy sauce fermentation, such as Klebsiella pneumoniae and Enterobacter hormaechei (20), which may be able to produce histamine (Fig. 3B). Combined with the histamine-production capability, it is confusing and worth studying whether Gram-negative or Gram-positive bacteria play key roles in histamine production during spontaneous soy sauce fermentation.
T. halophilus is the focal histamine-producing microorganism present during soy sauce fermentation.
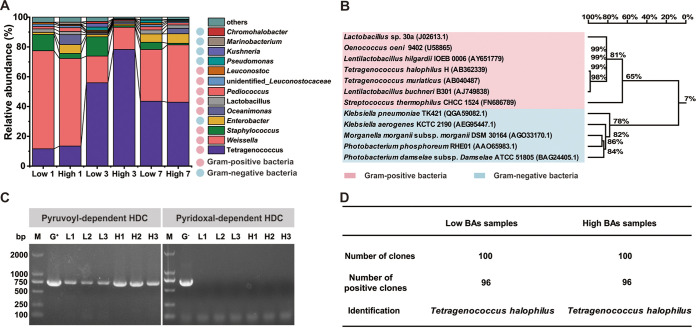
As shown in Fig. 1, both high-histamine and low-histamine soy sauce samples were present during the same period (from the first to seventh weeks), with the highest histamine-accumulation rate occurring during the third fermentation week (Fig. S3C). To determine why and what microorganisms contributed to the high rate of histamine accumulation, we first analyzed the bacterial community diversity of the high-histamine and low-histamine fermentation samples at three time points (1, 3, and 7 fermentation weeks) via high-throughput sequencing. We obtained 1,827,706 high-quality reads and thus generated 125 bacterial genera from 18 soy sauce samples by analyzing the V3-V4 hypervariable region of bacterial 16S rRNA gene sequences. It was found that Gram-positive bacteria occupied the main microbial community during the soy sauce fermentation, with the total relative abundance of more than 90%, which was consistent with our previous hypothesis (Fig. 4A). Tetragenococcus and Weissella were the predominant genera in soy sauce samples. In addition, the relative abundance dynamic of Tetragenococcus was consistent with the production dynamic of histamine, which increased significantly during the growth period of histamine. However, we could not find obvious differences in microbial community between high-histamine and low-histamine soy sauce fermented samples.
FIG 4.
Analysis of the focal histamine-producing microorganism during soy sauce fermentation. (A) Bacterial communities of soy sauce fermentation samples were collected at the 1st, 3rd, and 7th fermentation weeks. Low 1/3/7, bacterial community of low-histamine soy sauce samples which were collected at 1st, 3rd, and 7th fermentation weeks, respectively. High 1/3/7, bacterial community of high-histamine soy sauce samples which were collected at 1st, 3rd, and 7th fermentation weeks, respectively. (B) Analysis of histidine decarboxylase (HDC) protein sequence similarity between Gram-positive and Gram-negative bacteria. (C) Gene amplification of two kinds of histidine decarboxylase genes in low- and high-histamine soy sauce samples. M, marker; G+, gene amplification by using a Gram-positive bacterium genome as the template; G−, gene amplification by using the hdcA gene of Gram-negative bacteria added to fermented soy sauce moromi as the template; L1 to L3, three parallel soy sauce samples of low-histamine production; H1 to H3, three parallel soy sauce samples of high-histamine production. (D) Clone library analysis of hdcA gene in different soy sauce samples.
Histidine decarboxylase (HDC) (EC. 4.1.1.22) is a crucial enzyme in the synthesis of histamine by bacteria. Based on protein sequences, we compared the similarity of histidine decarboxylase (HDC) in Gram-positive and Gram-negative bacteria. As shown in Fig. 4B, the HDC in Gram-positive bacteria showed a high homology of 65% to 99%, whereas they exhibited a low similarity of 7% with HDC in Gram-negative bacteria. This result is consistent with the previous report that HDC in bacteria is classified into the following two types: pyridoxal-phosphate-dependent HDC found in Gram-negative bacteria and pyruvoyl-dependent HDC found in Gram-positive bacteria (29). This phenomenon allows us to investigate whether Gram-positive or Gram-negative bacteria participate in soy sauce histamine production.
As shown above, histamine accumulated fastest in the third fermentation week (Fig. 1C; Fig. S3C). Also, to determine the focal histamine-producing microorganisms during soy sauce fermentation, we extracted total genomic DNA from third-week soy sauce moromi samples and amplified the conserved region of two histidine decarboxylase genes (hdcA) by using those genomic DNA as the PCR template (about 100 ng/μL). As shown in Fig. 4C, we effectively amplified the pyruvoyl-dependent HDC gene (in Gram-positive bacteria) but not the pyridoxal phosphate-dependent HDC gene (in Gram-negative bacteria). Furthermore, hdcA amplification bands from high-histamine soy sauce samples were brighter than those from low-histamine soy sauce samples. These phenomena suggested that Gram-positive bacteria, rather than Gram-negative bacteria, play a key role in histamine production.
To investigate which Gram-positive bacterium plays a key role in histamine accumulation, the clone library analysis was conducted in this study. We first amplified the histidine decarboxylase gene hdcA by using soy sauce genomic DNA as the template. Then, the amplified hdcA gene was ligated to pEASY-Blunt simple plasmid and was transformed into E. coli DH 5α competent cells. A total of 200 clones were picked and sequenced. Statistical analysis showed that, except for 8 clones that were not sequenced successfully, 192 clones were successfully sequenced and identified. Sequence alignment showed that the sequenced hdcA gene in clones has more than 99% similarity to that of the genera Tetragenococcus, Lactobacillus, and Oenococcus. Fortunately, there are differences in specific bases among hdcA genes of various genera, allowing us to identify the real microbial species by sequence alignment of hdcA genes. The hdcA gene differed between Tetragenococcus and other genera (including Lactobacillus and Oenococcus) at base 591, which was G in the Tetragenococcus genus and T in other genera (see Fig. S4A in the supplemental material). Furthermore, the hdcA gene differed between T. halophilus and Tetragenococcus muriaticus at base 708, which was A in T. halophilus and C in T. muriaticus (Fig. S4B). As shown in Fig. 4D, except for 8 clones that were not sequenced successfully, all of the other 192 clones were sequenced successfully and were identified as T. halophilus. Combining the above bioinformatics analysis that T. halophilus, as a Gram-positive bacterium, can maximumly produce histamine at the same level as Gram-negative bacteria, we considered that T. halophilus might be the focal histamine-producing bacterium present during soy sauce fermentation.
Ratio of histamine-producing/non-histamine-producing subgroups of T. halophilus determines the histamine accumulation.
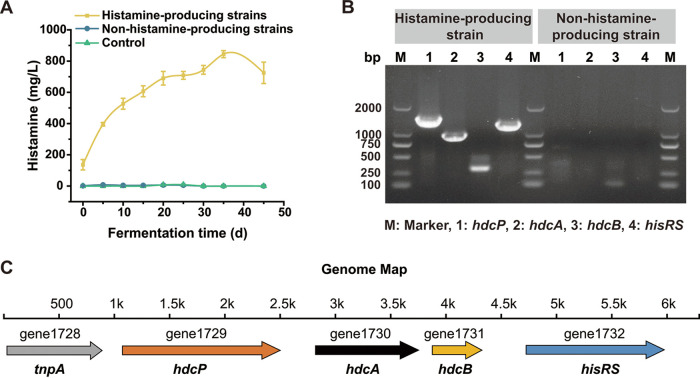
To further investigate the focal histamine-producing bacteria, 205 isolates from high-histamine samples and 205 isolates from low-histamine samples were acquired. By using 16S rRNA gene sequencing, all isolates were identified as T. halophilus. Furthermore, we detected the histamine-producing capability of these strains. As shown in Table S2 in the supplemental material, 37 T. halophilus strains with hdcA-gene positivity and histamine synthesis capacity can biosynthesize histamine, of which 24 were isolated from high-histamine samples and 13 were isolated from low-histamine samples. When we added the histamine-producing T. halophilus as the starter, histamine accumulated continuously (maximum 844 mg/L histamine) during simulated soy sauce fermentation. On the contrary, when the non-histamine-producing T. halophilus was added as the starter in simulated soy sauce fermentation, no histamine was produced (Fig. 5A).
FIG 5.
Strain specificity of the histamine-producing capacity of T. halophilus. (A) Histamine production of T. halophilus in simulated soy sauce fermentation; control, no microorganism inoculated. (B) The amplification of the genes, parts of histamine-biosynthesis gene cluster, by using histamine-producing or non-histamine-producing T. halophilus strain genomes as the templates. PCR products have been verified by gene sequencing, but the data were not shown. (C) Genome map of histamine-biosynthesis gene cluster in histamine-producing T. halophilus strain.
As shown above, the histamine-producing gene operon in lactic acid bacteria includes the following four genes: hdcP, hdcA, hdcB, and hisRS (Fig. 3A). By using PCR amplification, hdcP, hdcA, hdcB, and hisRS genes were detected in 37 histamine-producing T. halophilus strains but not in 10 randomly selected non-histamine-producing strains (Fig. 5B, Table 1). Furthermore, we randomly selected two T. halophilus strains, of which one could produce histamine and the other could not, and we sequenced their whole genomes by using the Illumina sequencing platform. We discovered a complete HDC gene cluster in the histamine-producing T. halophilus genome (Fig. 5C) but not in the non-histamine-producing strain. Moreover, a transposase gene (tnpA) was located at the side of the histamine-biosynthesis gene cluster, indicating that horizontal gene transfer may occur in this gene cluster under specific conditions. Thus, we concluded that T. halophilus was the focal histamine-producing strain, and the histamine production capacity of T. halophilus was strain specific. However, it is uncertain if a “super”-histamine-producing T. halophilus or a higher ratio of histamine-producing to non-histamine-producing T. halophilus is responsible for the increased histamine production.
TABLE 1.
Primers used in this study
| Name | Sequence (5′–3′) | Use | Reference |
|---|---|---|---|
| L-F2 | TGATGGTATTGTTTCGTATGACC | Amplification of hdcA in Gram-positive bacteria | In this study |
| L-R4 | TAACCCATCTTGTCVAGCCATTCTG | ||
| Hdc-f | TCHATYARYAACTGYGGTGACTGGRG | Amplification of hdcA in Gram-negative bacteria | 59 |
| Hdc-r | CCCACAKCATBARWGGDGTRTGRCC | ||
| Hdc-1 | TTGACCGTATCTCAGTGAGTCCAT | Quantification of hdcA in Gram-positive bacteria | 60 |
| Hdc-2 | ACGGTCATACGAAACAATACCATC | ||
| TetRT-F1 | AGCTCAAAGGCGCTTTACAG | Quantification of T. halophilus | In this study |
| TetRT-R1 | CCTCTTTCTCCTGTTCTTTGCTG | ||
| 27F | AGAGTTTGATCCTGGCTCAG | Species identification | 61 |
| 1492R | CGGTTACCTTGTTACGACTT | ||
| HdcA-F | ATGGAGGTAACAATTATGTCAGAG | Amplification of histamine-biosynthesis gene cluster in T. halophilus | In this study |
| HdcA-R | TTAATAATTGATGTTTCCACCCTTAG | ||
| HdcB-F | ACTGCACATGGAATCTCCGC | In this study | |
| HdcB-R | TACTTACCAGCCTTGGGCAG | ||
| HdcP-F | ATGGACGACGCTGAACGCGCTAAAAAG | In this study | |
| HdcP-R | TTAAGCCATTAATGCATAAATCCCATAAAC | ||
| HisRS-F | ATGACTGATGTAAATTACCAAAGACT | In this study | |
| HisRS-R | TTAGCTGAGGAATTTAGTTGGATTATC |
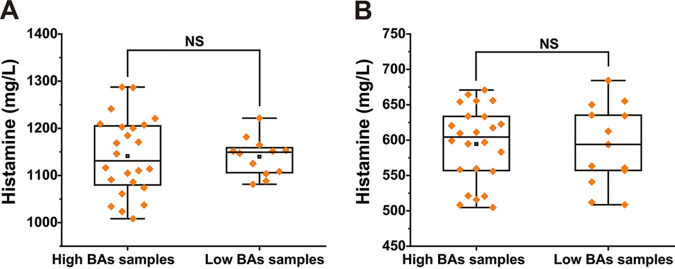
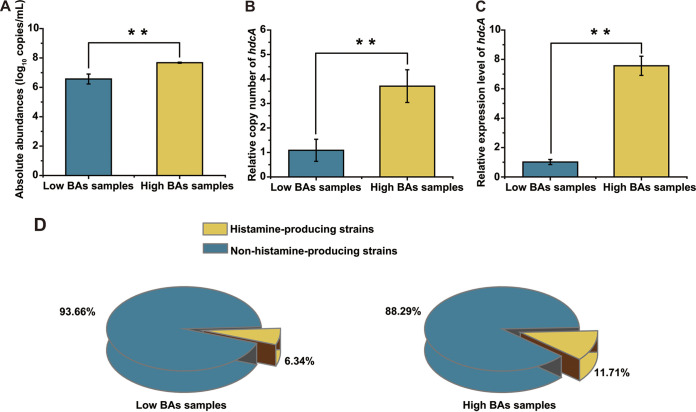
We compared the histamine production capability of 37 T. halophilus strains isolated from high-histamine samples and 13 T. halophilus strains isolated from low-histamine samples, which were detected in MRS medium (add histidine) and simulated soy sauce fermentation system. As shown in Fig. 6, there is no significant difference in histamine production capability between strains screened from high- or low-histamine samples (Fig. 6; Table S2). Second, we detected the absolute number of T. halophilus bacteria in high- and low-histamine soy sauce samples (third fermentation week) by quantitative real-time PCR analysis. As shown in Fig. 7A, T. halophilus was detected in low-histamine samples at a concentration of 6.5 ± 0.34 log10 copies/mL, while that in high-histamine samples was at a concentration of 7.68 ± 0.03 log10 copies/mL. Meanwhile, we determined the relative copy number of the histidine decarboxylase gene (hdcA) in low-histamine samples and high-histamine samples. As shown in Fig. 7B, the relative copy number of the hdcA gene in high-histamine samples was significantly higher than that in low-histamine samples (about 3.5-fold). Third, we also determined the relative transcriptional level of hdcA (RNA level) and found that hdcA maintained much higher transcription levels in high-histamine samples than those in low-histamine samples (about 7-fold) (Fig. 7C).
FIG 6.
Histamine production capacity of T. halophilus screened from high- and low-BA soy sauce samples. (A) MRS medium (adding histidine). (B) Simulated soy sauce fermentation system. NS, not significant.
FIG 7.
Quantitative analysis of histamine-producing microorganisms and HDC-encoding gene in low- or high-BA soy sauce samples. (A) The absolute abundance of T. halophilus in high- and low-BA soy sauce samples. (B) Relative quantitative analysis of hdcA between the high- and low-BA samples. The relative copy number was normalized to the copy number of the 16S rRNA gene of T. halophilus. (C) The relative expression level of hdcA gene between the high- and low-BA samples. The relative expression levels of hdcA were normalized to the expression level of the 16S rRNA gene of T. halophilus. (**, P < 0.01; determined by t-test). (D) The ratio of histamine-producing to non-histamine-producing strains in high- and low-BA samples.
In addition, the ratio of histamine-producing to non-histamine-producing T. halophilus in soy sauce samples (third fermentation week) was analyzed (Fig. 7D). A total of 205 T. halophilus strains were isolated from low-histamine samples, of which 13 (6.34%) strains were capable of producing histamine. Meanwhile, 205 T. halophilus strains were isolated from high-histamine samples, of which 24 (11.71%) strains were capable of producing histamine. These phenomena suggested that no “super”-histamine-producing T. halophilus strains were present in high-histamine soy sauce samples. Also, a larger amount of histamine-producing T. halophilus and more proportion of histamine-producing to non-histamine-producing T. halophilus are responsible for the increased histamine accumulation in high-histamine soy sauce samples.
Artificially decreasing the ratio of histamine-producing/non-histamine-producing T. halophilus and realized histamine control.
Based on the above research results, we attempted to verify the above speculation and study a control strategy for the inhibition of histamine production. In the laboratory, we conducted a simulated fermentation of soy sauce by inoculating the in situ microflora (1%) of a high-BA sample as the starter. Then, we divided the simulated soy sauce fermentation into two groups. One group was named experimental group, which was inoculated with 1.0 × 107 CFU/mL of non-histamine-producing T. halophilus. The other group was named control group, which was inoculated with the same volume of phosphate-buffered saline (PBS) buffer. After the above treatment, we sampled every 5 days and then detected their histamine concentrations.
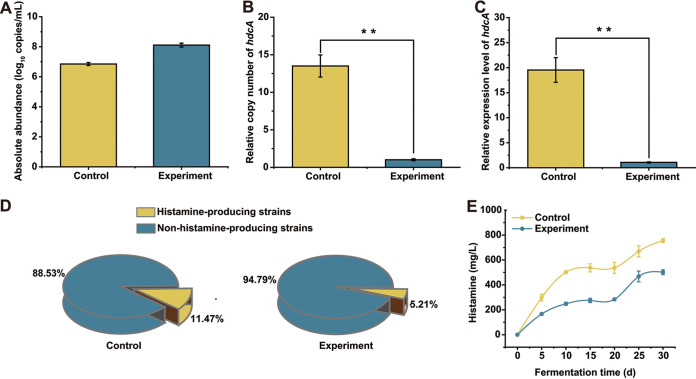
After 30 days of fermentation, the biomass of T. halophilus in the experimental group was significantly higher than that in the control group (Fig. 8A). We detected the hdcA gene by quantitative PCR (qPCR) and found that the copy number of the hdcA gene in the experimental group (6.5 log10 copies/mL) was significantly lower than that in the control group (8.0 log10 copies/mL) (Fig. 8B). In addition, the transcription amount of hdcA detected in the experimental group was significantly lower (1:20) than that in the control group as well (Fig. 8C). Finally, the ratio of histamine-producing to non-histamine-producing T. halophilus in the simulated soy sauce fermentation system was analyzed (Fig. 8D). Two hundred isolates were selected randomly from the simulated soy sauce fermentation system, and 96 strains of T. halophilus were identified from the experimental group, and 96 strains of T. halophilus were identified from the control group. The histamine production capability was detected by hdcA gene amplification. We found that the ratio of histamine-producing to non-histamine-producing T. halophilus in the experimental group was 5.21% (5/96), while that in the control group was 11.47% (11/96). This phenomenon suggested that the ratio of within-species variants can be changed artificially.
FIG 8.
Histamine control by artificially modifying the ratio of histamine-producing to non-histamine-producing T. halophilus strains during soy sauce fermentation. Experimental group was inoculated with 1.0 × 107 CFU/mL of non-histamine-producing T. halophilus at the initial stage of fermentation. Control group was inoculated with the same volume of PBS buffer. (A) The absolute abundances of T. halophilus at 30 days of fermentation in the control and experimental group. (B) Relative quantitative analysis of hdcA in the control and experimental group at 30 days of fermentation. The relative copy number was normalized to the copy number of the 16S rRNA gene of T. halophilus. (C) The relative expression level of hdcA gene in the control and experimental group at 30 days of fermentation. The relative expression levels of hdcA were normalized to the expression level of the 16S rRNA gene of T. halophilus. (**, P < 0.01; determined by t-test). (D) The ratio of histamine-producing to non-histamine-producing strains in the control group and experimental group at 30 days of fermentation. (E) Dynamic of histamine accumulation in the control and experimental group during the 30 days of fermentation. Data were obtained from three parallel trials. Figure 8D was obtained statistically after mixing three parallel samples of equal mass.
We collected the simulated soy sauce fermentation samples at an interval time and detected their BA (especially histamine) concentrations. As shown in Fig. 8E, compared with the control group, the experimental group produced a lower concentration of histamine due to a lower ratio of histamine-producing to non-histamine-producing strains (from 755.67 mg/L to 502.02 mg/L). Meanwhile, we found that the experimental group also accumulated lower concentrations of other biogenic amines, including tyramine, putrescine, phenethylamine, and cadaverine (see Fig. S5 in the supplemental material). These results suggested that the addition of non-histamine-producing T. halophilus as the starter will reduce the proportion of histamine-producing/non-histamine-producing T. halophilus strains and thus help reduce histamine production. In addition, the addition of non-histamine-producing T. halophilus as the starter is always helpful for the reduction of other BAs.
DISCUSSION
In this study, we found that T. halophilus was the focal histamine-producing microorganism during soy sauce fermentation. More importantly, a difference in the histamine production capability of T. halophilus has been reported, which was named strain specificity or within-species variants. Strain specificity might be an important characteristic of BA-producing microorganisms (30). Lucas et al. (31) found that the histidine decarboxylase gene of Lactobacillus hilgardii IOEB 0006 was located on an 80-kb plasmid, which was proven to be unstable (the strain could retain or lose the plasmid depending on culture conditions). Satomi et al. (32) analyzed the diversity of plasmids encoding the histidine decarboxylase gene in 19 Tetragenococcus sp. strains, which were isolated from Japanese fish sauce, suggesting that each histamine producer harbored a different histamine-related plasmid.
Strain specificity is a universal phenomenon in nature. The importance of strain specificity in the context of pathogenicity has been particularly well studied, and many species have been found to have both pathogenic and commensal subtypes (for example, Propionibacterium acnes [33] and Staphylococcus aureus [34]). Furthermore, strain specificity has been discovered in a variety of other areas, including drug response (3), biofilm formation (35), nutrient cycling (36), carbohydrate utilization (37), and toxin production (38). Mutations are the most direct reason for intraspecific diversity. The transfer of genetic variation from one population to another through, for example, horizontal gene transfer, homologous recombination, and plasmid acquisition, can cause rapid and large-scale additions or rearrangements of genomic regions. Given the existence of the transposase gene (tnpA) in a histamine-biosynthesis gene cluster (32) and the foundation of plasmid-based gene cluster (39), T. halophilus might obtain the histamine production capability through horizontal gene transfer or plasmid acquisition.
Rather than existing as a single colony, most microorganisms live in complex ecosystems. It was found that the strain specificity of microorganisms has an important ecological significance in affecting microbial community assembly (4), ecological function (5, 35), and niche construction (40). In 2016, Johnson et al. (41) found that strain-level differences of Propionibacterium acnes in porphyrin production are associated with skin disease. In the same year, Lee et al. (42) found that the strain-specific features of extracellular polysaccharides have an impact on Lactobacillus pantarum-host interactions. In addition, researchers found that microorganisms can respond to external environmental disturbances at the strain level (5, 43). In 2020, Brittany et al. (4) characterized the strain-level diversity across cheese rinds and determined its implications for community assembly and function. However, researchers do not know which strain-diversity parameters impact microbiome function.
This study found that T. halophilus is the focal histamine-producing microorganism during soy sauce fermentation and fully described the strain-specific characteristics of T. halophilus in histamine production. The results showed that the ratio of histamine-producing to non-histamine-producing T. halophilus plays a key role in histamine accumulation. Furthermore, by artificially changing the ratio, the concentration of histamine produced by soy sauce microbiota decreased. This report is the first one about how strain specificity influences microbiome function. Previously, some researchers achieved reduction of BAs in fermented foods by adding starters, including T. halophilus (14). This study explained, from the perspective of microbial strain-specific impacts on community functions, why the addition of T. halophilus, which could not degrade BAs, resulted in the decrease of BAs.
To test our hypothesis that the ratio of histamine-producing to non-histamine-producing T. halophilus is a critical factor in histamine accumulation, we introduced microbiota from industrial high-BA-production soy sauce to control samples of a simulated soy sauce control experiment. As predicted, a high ratio of histamine-producing to non-histamine-producing T. halophilus and high concentrations of histamine were detected in the control samples (Fig. 8D and E). In the experimental samples, non-histamine-producing T. halophilus was introduced at the beginning of a simulated soy sauce fermentation. As expected, we achieved histamine reduction by inoculating non-histamine-producing T. halophilus to reduce the ratio of histamine-producing to non-histamine-producing T. halophilus in the experimental group (Fig. 8E). As a positive control, samples inoculated with histamine-producing T. halophilus resulted in high-histamine production in simulated fermentation experiments (Fig. 5A). These phenomena verified our suggestion that the ratio of histamine-producing to non-histamine-producing T. halophilus is a critical factor in histamine accumulation. In fact, T. halophilus is beneficial in high-salt food fermentation for the production of aroma-active and umami-taste compounds (44). Besides histamine production, T. halophilus has genomic diversity and strain specificity in carbohydrate metabolism, amino acid degradation, and lactate production (45). It is important to investigate whether and how strain specificity in physiological metabolism can impact microbiome function when T. halophilus is used as a starter in food fermentation.
Biogenic amines (BAs) are a kind of endogenous harmful compound which exist widely in various fermented foods (46). When BAs in fermented foods are ingested by humans, portions of BAs usually can be metabolized by amine oxidase in the human gut (47). However, when the BAs are ingested excessively, or the human body appears to have inadequate detoxification action (for genetic reasons or inhibitory effects from some medicines or alcohol), excess BAs can enter the human systemic circulation and then trigger the release of adrenaline and noradrenaline, as well as provoking gastric acid secretion, increasing cardiac output, and elevating blood pressure (48). Furthermore, soluble nitrogen concentration (or free amino acid concentration) is an essential quality indicator for high-protein fermented foods, and the formation of BAs might result in nitrogen loss and amino acid concentration decline (Fig. 1I). Thus, it is vital to prevent the production of BAs in fermented food.
Taking soy sauce as an example, it is difficult to completely inhibit BA production. On the one hand, soy sauce itself is rich in amino acids, which provide abundant precursors for the biosynthesis of biogenic amines. On the other hand, soy sauce usually ferments spontaneously, which means that a complex microbiota, probably coming from Koji, the brewing environment, and raw materials, participates in the soy sauce production. The microbiota during soy sauce fermentation was studied using next-generation sequencing, and an average of 1,360 operational taxonomic units (OTUs) were discovered, with 22 microbial genera. In our study, we found 277 bacterial OTUs, with 29 bacterial OTUs at a relative abundance greater than 0.1% in our soy sauce samples.
A variety of microorganisms are positive for BA production because BA biosynthesis has important physiological implications. In prokaryotic cells, the decarboxylation reaction (from amino acid) is one of the defense mechanisms used by bacteria to compete with the acidic environment (49, 50). Some reports suggest that the biosynthesis of BAs can help some respiratory-chain-negative microorganisms, such as lactic acid bacteria (LAB), to obtain more energy (51). Furthermore, BA production in bacteria may mediate other physiological functions, such as osmotic and oxidative stress responses (52, 53). This information is consistent with our findings that 86.36% of soy sauce bacteria have relevant amino acid decarboxylase genes and can thus produce at least one kind of BA. The complex microbial community structure and strain-specific characteristics of BA production make histamine control increasingly difficult.
Biogenic amines (BAs) are produced by microbial decarboxylation reactions. Identification of the focal BA-producing microorganisms is of great significance for understanding the mechanism of BA production and for controlling BAs. However, it is difficult to study the microbial function in complex multispecies fermentation systems. Traditional methods for identifying the BA-producing microorganisms include molecular methods and chemical methods, depending on microorganism cultivation (54). The development of omics technology has promoted the study of functional microorganisms. Hu et al. (55) found the bacteria that are responsible for producing biogenic amines in sufu by using metagenome sequencing. Kim et al. (56) identified BA-producing microbes during ganjang fermentation through metagenomic and metatranscriptomic analyses. However, as shown above, given the diversity of amino acid decarboxylase genes, the structural complexity of genetic clusters, and the uncertainty of relevant enzyme activity, there are some difficulties in identifying the focal BA-producing microorganisms by culture-independent approaches. In this study, focal microorganisms for histamine production during soy sauce fermentation were identified by combining bioinformatics analysis, high-throughput sequencing, gene amplification, clone library construction, and microbial cultivation methods.
During soy sauce fermentation, the pH dropped visibly from 6.5 to 4.5 (Fig. 1H). Also, the production of BAs helps microbes to resist an acidic environment (50). This information might explain why decarboxylase genes are present in most soy sauce microbes and why many microbes can produce multiple BAs simultaneously (Fig. 2). Furthermore, almost all BAs, including histamine, tyramine, putrescine, phenethylamine, and cadaverine, are produced via the decarboxylation of corresponding amino acids. The transcriptional expression of all these amino acid decarboxylases is stimulated by acidic pH (50, 57). This information might explain why the production of various BAs has strong correlations (Fig. 1E and F; Fig. S2C and D). Meanwhile, we inhibited the growth of BA-producing microorganisms in this study, reducing the production of other BAs.
In conclusion, this study systematically analyzed the focal histamine-producing microorganisms of high-salt liquid-state soy sauce. Bioinformatics analysis showed that more than 80% of soy sauce bacteria have amino acid decarboxylase genes and thus have the potential to produce one or more BAs. After the genetic analysis, clone library construction, and strain cultivation, we found that T. halophilus is the focal histamine-producing microorganism during soy sauce fermentation. The histamine production capacity of T. halophilus is strain specific. The ratio of histamine-producing to non-histamine-producing subgroups of T. halophilus determines the histamine accumulation during the spontaneous fermentation of soy sauce. Finally, we investigated BA control strategies during soy sauce fermentation based on the strain-specific characteristics of BA-producing microorganisms. This study provided a systematic method and a new strategy for the process control of BAs and highlighted the importance of strain specificity in microbiome function.
MATERIALS AND METHODS
Sample collection.
Soy sauce samples were collected from a typical Chinese traditional brewing soy sauce factory in China. The initial concentration of salt water added in soy sauce fermentation was 18 to 19°Bé (21.62 g to 23.11 g sodium chloride per 100 mL brine). A total of six samples (100 mL per sample) were collected at different fermentation times (0, 1, 2, 3, 4, 5, 7, 9, 11, 13, 15, and 17 fermentation weeks) in August 2019. Cells were collected by centrifugation at 8,000 rpm for 30 min at 4°C and then stored at −80°C for DNA and RNA extraction. The supernatant was stored at −20°C for BA and physicochemical parameter determination.
Biogenic amine determination.
Seven BAs (histamine, tyramine, putrescine, cadaverine, phenethylamine, spermine, and spermidine) were detected by reverse-phase ultra-performance liquid chromatography (UPLC) (Waters H-Class; Waters Co., Milford, MA) that was equipped with an Acquity UPLC BEH C18 analytical column (2.1 by 100 mm, 1.7 μm; Waters) and a UV detector, with derivatization with dansyl chloride (Sigma, USA) according to a previous report (9). The gradient elution system includes acetonitrile (A) and ultrapure water (B) containing 0.1% formic acid. The gradient elution procedure was optimized and operated at a flow rate of 0.3 mL/min as follows: 30% A at 2 min, 50% A at 2.5 min, 60% A at 8.5 min, 75% A at 13 min, 100% A at 15 min, and 30% A at 17 min. The column temperature was set to 30°C, and the injection volume was 5 μL.
The BA-producing capability analysis of soy sauce bacteria.
We collected and listed the reported soy sauce bacteria and analyzed their potential BA (histamine, tyramine, putrescine, and cadaverine)-production capability by using bioinformatic analysis. We investigated whether the soy sauce bacteria have the amino acid decarboxylase, including lysine decarboxylase (LDC), histidine decarboxylase (HDC), tyrosine decarboxylase (TDC), and ornithine decarboxylase (ODC) by searching these proteins in the protein and nucleotide databases of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). We assumed that species with the corresponding amino acid decarboxylase have the potential to produce BAs. In addition, the neighbor-joining phylogenetic tree of soy sauce bacteria was constructed based on the 16S rRNA gene sequences by MEGA 7.0.
Histamine-biosynthesis gene cluster analysis.
We collected the gene sequences of histidine decarboxylase (hdcA) and histidine/histamine antiporter (hdcP) from NCBI. Then, using the hdcA gene and hdcP gene as target genes, we searched via BLAST the histamine-biosynthesis gene clusters from different soy sauce bacteria based on the nucleotide database.
Total DNA and RNA extraction.
The total DNA was extracted from 100 mL of soy sauce sample using the phenol-chloroform method as described by Song et al. (58). The total RNA of the soy sauce sample was extracted using a soil RNA kit (Omega Bio-Tek, Norcross, GA) following the manufacturer’s instructions and reversely transcribed into cDNA (Vazyme, Nanjing, China). Genomic DNA of the strain was extracted using a genomic DNA purification kit (Promega, Madison, USA) following the manufacturer’s instructions. Total DNA and RNA were quantified by using a NanoDrop ND-1000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The quality of DNA and RNA was determined by agarose gel electrophoresis. DNA was stored at −20°C, and RNA was stored at −80°C before analysis.
16S rRNA amplicon sequencing and sequence processing.
The V3-V4 region of the bacterial 16S rRNA gene sequence was amplified by PCR (Bio-Rad, USA) using TransStart Fastpfu DNA polymerase (TransGen AP221-02; TransGen Biotech Co., Beijing, China). The amplification primers used were universal primers 338F/806R, and the reaction system and amplification method were carried out according to the manufacturer’s instructions. Then the amplified products were purified using an Agencourt AMPure XP nucleic acid purification kit, and the concentrations were determined by the Thermo Scientific NanoDrop 8000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE). High-throughput sequencing was performed on the MiSeq instrument (Illumina, San Diego, CA) for 2 × 300-bp paired-end (PE) sequencing.
All MiSeq-generated paired-end (PE) sequence data were processed by using the QIIME platform. Briefly, the raw Fastq sequence was quality controlled by Trimmomatic (v0.36) and Pear (v0.9.6) software, and the sequences with average quality scores of <20, with lengths of <120 bp, and containing N base were trimmed. Forward and reverse sequences were merged with FLASH using a minimum overlap of 10 bp and 0.1 allowed mismatch ratio. Chimera sequences were removed using the UCHIME algorithm for the known database and using the Denovo method for the unknown database. After that step, the high-quality sequence was clustered into operational taxonomic units (OTUs) with a 97% identity threshold by the QIIME uparse pipeline.
Amplification of the histidine decarboxylase (HDC) gene from genomes of soy sauce samples.
The conserved region of histidine decarboxylase genes (hdcA) was amplified by the following PCR procedure: 95°C for 2 min; followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 20 s; and a final extension of 72°C for 5 min. Genomic DNA of soy sauce samples with a similar concentration (about 100 ng/μL) was used as the PCR template. The Gram-positive bacterium T. halophilus with histamine-producing capacity screened from soy sauce moromi was used as a positive control for pyruvoyl-dependent HDC gene amplification. Gram-negative bacteria with histamine-producing capacity were not obtained in this study, so we added an engineered bacterium (E. coli) containing a pyridoxal phosphate-dependent HDC gene plasmid as the positive control for this HDC gene amplification. The pyruvoyl-dependent HDC gene sequence was amplified by primers L-F2 and L-R4 with a length of 744 bp (Table 1). The pyridoxal-phosphate-dependent HDC gene sequence was amplified by primers Hdc-f and Hdc-r with a length of 709 bp (Table 1) (59). The TransStart Fastpfu DNA polymerase (TransGen AP221-02, TransGen Biotech Co., Beijing, China) was used for sequence amplification, and the amplified products were detected with a 1% agarose gel.
Clone library construction and analysis.
The hdcA gene sequence was amplified by primers L-F2 and L-R4. The PCR was performed using the total genomic DNA extracted from high- or low-BA soy sauce samples as the templates. The amplified PCR products were purified by using the FastPure gel DNA extraction minikit (Vazyme, Nanjing, China). Then the purified PCR products were ligated with pEASYblunt plasmid (TransGen CB101, Beijing, China), and then transformed into E. coli DH5α competent cells. A total of 200 recombinant clones were selected randomly from each sample and sent to Sangon Biotech (Shanghai, China) for sequencing. Nucleotide sequences were aligned through BLASTx to determine their functional annotation and taxonomic classification.
Quantitative analysis of absolute abundance and hdcA gene of T. halophilus by using qPCR.
The qPCRs were carried out using the StepOnePlus instrument (Applied Biosystems, Foster City, CA). The reactions were performed by using 10 μL AceQ universal SYBR green qPCR master mix (Vazyme, Nanjing, China), 0.4 μL of each primer, 1 μL template, and 7.2 μL double-distilled water (ddH2O). To determine the absolute abundance of T. halophilus, the species-specific primers TetRT-F1 and TetRT-R1 were designed based on the 16S rRNA gene (Table 1). The specificity of the primers TetRT-F1 and TetRT-R1 was validated by cross-PCR and melting curves of qPCR (see Fig. S6 in the supplemental material). The quantitative standard curve was shown in Fig. S7 in the supplemental material. The copy number and expression level of hdcA gene of T. halophilus were analyzed by qPCR, with Hdc-1/Hdc-2 as primers (Table 1) (60). The relative copy number and relative expression level for hdcA gene were calculated with the threshold cycle (2−ΔΔCT) method, using the low-histamine samples (or experiment group) as the reference sample. The relative copy number and relative expression level for the hdcA gene were normalized to the copy number and expression level of the 16S rRNA gene of T. halophilus, respectively. Each reaction consisted of three technical replicates.
Isolation and identification of histamine-producing microorganisms.
The isolation of lactic acid bacteria (LAB) used de Man Rogosa Sharpe (MRS) medium (Difco, Detroit, MI) containing 10% NaCl and 1% histidine (MRSH medium). One milliliter of sauce mush samples, which fermented for 3 weeks, were diluted to 10−4 to 10−5 with a 0.9% NaCl solution. Then we took 0.1 mL of diluted samples and spread them onto the MRSH medium plate. The plates were incubated at 30°C for 5 to 7 days under anaerobic conditions. Randomly selected colonies were inoculated in liquid MRSH medium, cultured anaerobically at 30°C for 5 days, and then stored in glycerol. The histamine-producing capability of the strain was detected using three methods. (i) The first method was PCR amplification of the HDC gene. (ii) The second method was the detection of the histamine-producing capability of strains in MRS medium. The strains were inoculated into MRSH medium and cultured anaerobically at 30°C for 5 days, and then the histamine concentration was detected by UPLC. (iii) The third method was the detection of the histamine-producing capability of strains in simulated soy sauce fermentation. The strains were inoculated into simulated soy sauce fermentation system and cultured anaerobically at 30°C for 30 days, and then the histamine concentration was detected by UPLC. Species identification was performed by sequencing the partial 16S rRNA gene of the strain. The 16S rRNA genes were amplified by PCR using the universal bacterial primers 27F and 1492R (Table 1) and sent to Sangon Biotech (Shanghai, China) for sequencing.
Simulated fermentation of soy sauce.
It was performed according to Zhang et al. (44) described and improved.
(i) Preparation of starter culture. The strains were inoculated in MRS broth supplemented with 10% (wt/vol) NaCl (pH 7.0) and static cultured at 30°C for 5 to 7 days until the bacterial concentration reached 109 CFU/mL. The bacteria were collected by centrifugation at 5,000 rpm at 4°C for 5 min. The collected cells were washed twice with sterile phosphate-buffed saline (PBS, pH7.0), resuspended in PBS, and counted with a hemocytometer for soy sauce fermentation. The soy sauce microflora was collected from high-BA soy sauce samples fermented in the factory for 3 weeks. The fermented grains were treated with sterile PBS and then filtered with four layers of sterile gauze. The filtrate was then centrifuged at 5,000 rpm at 4°C for 5 min. The collected soy sauce microflora was suspended in PBS.
(ii) Preparation of soy sauce Koji. Whole soybeans were selected and soaked in warm water for 6 to 7 h before steaming and then cooled to room temperature. The steamed soybean was mixed with flour in a ratio of 1:0.4 (wt:wt) and inoculated with a prescribed ratio (approximately 0.04% of the dry weight of soybeans) of seed koji (Aspergillus oryzae 3.042), and fermented for 44 to 48 h. During the process of making koji, the temperature was controlled between 28 to 40°C and the relative humidity was controlled between 75 to 95%. At 16-h and 24-h fermentation time, Koji material was turned over to control temperature and oxygen. The koji was yellowish-green, was full of spores, and had no peculiar smell, indicating the success of koji production.
(iii) Soy sauce fermentation. The mature koji was mixed with 19°Bé saltwater in a ratio of 1:2.2, then put into a 500-mL fermentation flask, inoculated with soy sauce fermentation microbiota and starter, and fermented at 30°C. The simulated fermentation in this study used the same koji and soy sauce microbiota. Each group consisted of three parallel trials.
(iv) Analysis of total acid and amino nitrogen. Total acid and amino acid nitrogen were detected according to GB 5009.235—2016 Determination of amino acid nitrogen in foodstuffs (China National Standards for Food Safety).
Data availability.
The bacterial raw sequence data were submitted to the NCBI Sequence Read Archive (SRA) under accession number PRJNA826566.
ACKNOWLEDGMENTS
This work was financially supported by National Key R&D Program of China (2018YFC1604100).
J.M., L.Z., Y.N., and Y.X. conceived and designed the experiments. J.M. performed the experiments. J.M. and L.Z. analyzed the data. J.M. and L.Z. wrote the manuscript. All the authors have reviewed the manuscript.
There are no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Lijie Zhang, Email: zhanglj@jiangnan.edu.cn.
Yan Xu, Email: yxu@jiangnan.edu.cn.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Van Rossum T, Ferretti P, Maistrenko OM, Bork P. 2020. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol 18:491–506. 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito IL. 2021. Examining horizontal gene transfer in microbial communities. Nat Rev Microbiol 19:442–453. 10.1038/s41579-021-00534-7. [DOI] [PubMed] [Google Scholar]
- 3.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555:623–628. 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niccum BA, Kastman EK, Kfoury N, Robbat A, Jr, Wolfe BE. 2020. Strain-level diversity impacts cheese rind microbiome assembly and function. mSystems 5:e00149-20. 10.1128/mSystems.00149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu GJ, Zhang CH, Wu H, Wang RR, Shen J, Wang LH, Zhao YF, Pang XY, Zhang XJ, Zhao LP, Zhang MH, Zhou JZ. 2017. Genomic microdiversity of bifidobacterium pseudocatenulatum underlying differential strain-level responses to dietary carbohydrate intervention. mBio 8:e02348-16. 10.1128/mBio.02348-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Liu Y, Huang X, Xiao Z, Yang Y, Yu Q, Chen S, He L, Liu A, Liu S, Zou L, Yang Y. 2021. A review on mechanistic overview on the formation of toxic substances during the traditional fermented food processing. Food Rev Int:1–18. 10.1080/87559129.2021.1933021. [DOI] [Google Scholar]
- 7.Benkerroum N. 2016. Biogenic amines in dairy products: origin, incidence, and control means. Compr Rev Food Sci Food Saf 15:801–826. 10.1111/1541-4337.12212. [DOI] [PubMed] [Google Scholar]
- 8.Sivamaruthi BS, Kesika P, Chaiyasut C. 2021. A narrative review on biogenic amines in fermented fish and meat products. J Food Sci Technol 58:1623–1639. 10.1007/s13197-020-04686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu YM, Chen XH, Jiang M, Lv X, Rahman N, Dong MS, Yan GJ. 2009. Biogenic amines in Chinese soy sauce. Food Control 20:593–597. 10.1016/j.foodcont.2008.08.020. [DOI] [Google Scholar]
- 10.Feng C, Teuber S, Gershwin ME. 2016. Histamine (scombroid) fish poisoning: a comprehensive review. Clin Rev Allergy Immunol 50:64–69. 10.1007/s12016-015-8467-x. [DOI] [PubMed] [Google Scholar]
- 11.European Commission. 2003. Commission recommendation of 10 January 2003 concerning a coordinated programme for the official control of foodstuffs for 2003 (2003/10). Official J the European Commission 7:76–81. [Google Scholar]
- 12.FDA. 2012. CPG Sec. 540.525. Decomposition and histamine raw, frozen tuna and mahi-mahi; canned tuna; and related species. FDA, Silver Spring, MD. [Google Scholar]
- 13.Li J, Jiang K, Huang H, Cheng H, Ye X, Zhi Z. 2020. Process improvement to prevent the formation of biogenic amines during soy sauce brewing. Food Chem 331:127347. 10.1016/j.foodchem.2020.127347. [DOI] [PubMed] [Google Scholar]
- 14.Qi Q, Huang J, Zhou R, Jin Y, Wu C. 2022. Abating biogenic amines and improving the flavor profile of Cantonese soy sauce via co-culturing Tetragenococcus halophilus and Zygosaccharomyces rouxii. Food Microbiol 106:104056. 10.1016/j.fm.2022.104056. [DOI] [PubMed] [Google Scholar]
- 15.Zhao JD, Niu CT, Du SY, Liu CF, Zheng FY, Wang JJ, Li Q. 2020. Reduction of biogenic amines formation during soybean paste fermentation by using Staphylococcus carnosus M43 and Pediococcus acidilactici M28 as starter culture. LWT 133:109917. 10.1016/j.lwt.2020.109917. [DOI] [Google Scholar]
- 16.Guo J, Luo W, Fan J, Suyama T, Zhang WX. 2020. Co-inoculation of Staphylococcus piscifermentans and salt-tolerant yeasts inhibited biogenic amines formation during soy sauce fermentation. Food Res Int 137:109436. 10.1016/j.foodres.2020.109436. [DOI] [PubMed] [Google Scholar]
- 17.Guarcello R, De Angelis M, Settanni L, Formiglio S, Gaglio R, Minervini F, Moschetti G, Gobbetti M. 2016. Selection of amine-oxidizing dairy lactic acid bacteria and identification of the enzyme and gene involved in the decrease of biogenic amines. Appl Environ Microbiol 82:6870–6880. 10.1128/AEM.01051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menne A, Bodmer S, Amon U. 2001. Der Sektprovokationstest in der Diagnostik einer enteralen Histaminose. Akt Dermatol 27:58–62. 10.1055/s-2001-11493. [DOI] [Google Scholar]
- 19.Sulaiman J, Gan H, Yin W, Chan K. 2014. Microbial succession and the functional potential during the fermentation of Chinese soy sauce brine. Front Microbiol 5:556. 10.3389/fmicb.2014.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Y, Qian Y, Ji F, Chen J, Han B. 2013. Microbial composition during Chinese soy sauce koji-making based on culture dependent and independent methods. Food Microbiol 34:189–195. 10.1016/j.fm.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Schirone M, Visciano P, Tofalo R, Suzzi G. 2016. Histamine food poisoning. Handb Exp Pharmacol 241:217–235. 10.1007/164_2016_54. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Perez S, Comas-Baste O, Veciana-Nogues MT, Latorre-Moratalla ML, Vidal-Carou MC. 2021. Low-histamine diets: is the exclusion of foods justified by their histamine content? Nutrients 13:1395. 10.3390/nu13051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Deng Y, Jin Y, Liu Y, Xia B, Sun Q. 2017. Dynamics of microbial community during the extremely long-term fermentation process of a traditional soy sauce. J Sci Food Agric 97:3220–3227. 10.1002/jsfa.8169. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka Y, Watanabe J, Mogi Y. 2012. Monitoring of the microbial communities involved in the soy sauce manufacturing process by PCR-denaturing gradient gel electrophoresis. Food Microbiol 31:100–106. 10.1016/j.fm.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ST, Zhou B, Tong X, Zhou QY. 2019. Research progress on analysis methods of microbial diversity in soy sauce brewing process. China Condiment 44:193–200. [Google Scholar]
- 26.Geng YH, Xie XH, Li GJ. 2015. Dynamic changes in bacterial community structure during soy sauce fermentation. Mod Food Sci Technol 31:83–87. [Google Scholar]
- 27.Deng Y, Yang Y, Liang LJ, Chi YL, Sun Q. 2021. Study on microbial diversity and flavor of soy sauce brewed by traditional technology. China Condiment 46:104–108. [Google Scholar]
- 28.Trip H, Mulder NL, Rattray FP, Lolkema JS. 2011. HdcB, a novel enzyme catalysing maturation of pyruvoyl-dependent histidine decarboxylase. Mol Microbiol 79:861–871. 10.1111/j.1365-2958.2010.07492.x. [DOI] [PubMed] [Google Scholar]
- 29.Landete JM, De las Rivas B, Marcobal A, Munoz R. 2008. Updated molecular knowledge about histamine biosynthesis by bacteria. Crit Rev Food Sci Nutr 48:697–714. 10.1080/10408390701639041. [DOI] [PubMed] [Google Scholar]
- 30.Jeong DW, Heo S, Lee JH. 2017. Safety assessment of Tetragenococcus halophilus isolates from doenjang, a Korean high-salt-fermented soybean paste. Food Microbiol 62:92–98. 10.1016/j.fm.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Lucas PM, Wolken WAM, Claisse O, Lolkema JS, Lonvaud-Funel A. 2005. Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006. Appl Environ Microbiol 71:1417–1424. 10.1128/AEM.71.3.1417-1424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satomi M, Furushita M, Oikawa H, Yano Y. 2011. Diversity of plasmids encoding histidine decarboxylase gene in Tetragenococcus spp. isolated from Japanese fish sauce. Int J Food Microbiol 148:60–65. 10.1016/j.ijfoodmicro.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 33.Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, Li H. 2013. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio 4:e00003-13. 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BO. 2016. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci USA 113:E3801–E3809. 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landis EA, Fogarty E, Edwards JC, Popa O, Eren AM, Wolfe BE. 2022. Microbial diversity and interaction specificity in kombucha tea fermentations. mSystems 7:e00157-22. 10.1128/msystems.00157-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. 2018. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J 12:185–198. 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuhren J, Rosch C, Ten Napel M, Schols HA, Kleerebezem M. 2020. Synbiotic matchmaking in Lactobacillus plantarum: substrate screening and gene-trait matching to characterize strain-specific carbohydrate utilization. Appl Environ Microbiol 86:e01081-20. 10.1128/AEM.01081-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. 2013. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4:2601. 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satomi M, Furushita M, Oikawa H, Yoshikawa-Takahashi M, Yano Y. 2008. Analysis of a 30 kbp plasmid encoding histidine decarboxylase gene in Tetragenococcus halophilus isolated from fish sauce. Int J Food Microbiol 126:202–209. 10.1016/j.ijfoodmicro.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, LaSota ED, Cecere AG, LaPenna KB, Larios-Valencia J, Wollenberg MS, Miyashiro T. 2016. Intraspecific competition impacts vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl Environ Microbiol 82:3082–3091. 10.1128/AEM.04143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson T, Kang D, Barnard E, Li H. 2016. Strain-level differences in porphyrin production and regulation in Propionibacterium acnes elucidate disease associations. mSphere 1:e00023-15. 10.1128/mSphere.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee IC, Caggianiello G, van S, II, Taverne N, Meijerink M, Bron PA, Spano G, Kleerebezem M. 2016. Strain-specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host Interactions. Appl Environ Microbiol 82:3959–3970. 10.1128/AEM.00306-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filippis FD, Storia AL, Villani F, Ercolini D. 2019. Strain-level diversity analysis of Pseudomonas fragi after In situ pangenome reconstruction shows distinctive spoilage-associated metabolic traits clearly selected by different storage conditions. Appl Environ Microbiol 85:e02212-18. 10.1128/AEM.02212-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Zhang L, Xu Y. 2020. Effects of Tetragenococcus halophilus and Candida versatilis on the production of aroma-active and umami-taste compounds during soy sauce fermentation. J Sci Food Agric 100:2782–2790. 10.1002/jsfa.10310. [DOI] [PubMed] [Google Scholar]
- 45.Chun BH, Han DM, Kim KH, Jeong SE, Park D, Jeon CO. 2019. Genomic and metabolic features of Tetragenococcus halophilus as revealed by pan-genome and transcriptome analyses. Food Microbiol 83:36–47. 10.1016/j.fm.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Spano G, Russo P, Lonvaud-Funel A, Lucas P, Alexandre H, Grandvalet C, Coton E, Coton M, Barnavon L, Bach B, Rattray F, Bunte A, Magni C, Ladero V, Alvarez M, Fernandez M, Lopez P, de Palencia PF, Corbi A, Trip H, Lolkema JS. 2010. Biogenic amines in fermented foods. Eur J Clin Nutr 64:S95–S100. 10.1038/ejcn.2010.218. [DOI] [PubMed] [Google Scholar]
- 47.Kusche J, Lorenz W, Schmidt J. 1975. Oxidative deamination of biogenic amines by intestinal amine oxidases: histamine is specifically inactivated by diamine oxidase. Hoppe-Seylers Z Physiol Chem 356:1485–1486. 10.1515/bchm2.1975.356.2.1485. [DOI] [PubMed] [Google Scholar]
- 48.Ladero V, Calles-Enríquez M, Fernández M, Alvarez MA. 2010. Toxicological effects of dietary biogenic amines. Curr Nutr Food Sci 6:145–156. 10.2174/157340110791233256. [DOI] [Google Scholar]
- 49.Eun RJ, Haeng RJ, Youl RP, Ho CS. 2002. Identification of the cadBA operon from Vibrio vulnificus and its influence on survival to acid stress. FEMS Microbiol Lett 208:245–251. 10.1111/j.1574-6968.2002.tb11089.x. [DOI] [PubMed] [Google Scholar]
- 50.Diaz M, Del Rio B, Ladero V, Redruello B, Fernández M, Martin MC, Alvarez MA. 2020. Histamine production in Lactobacillus vaginalis improves cell survival at low pH by counteracting the acidification of the cytosol. Int J Food Microbiol 321:108548. 10.1016/j.ijfoodmicro.2020.108548. [DOI] [PubMed] [Google Scholar]
- 51.Molenaar D, Bosscher JS, ten Brink B, Driessen AJ, Konings WN. 1993. Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J Bacteriol 175:2864–2870. 10.1128/jb.175.10.2864-2870.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schiller D, Kruse D, Kneifel H, Krämer R, Burkovski A. 2000. Polyamine transport and role of potE in response to osmotic stress in Escherichia coli. J Bacteriol 182:6247–6249. 10.1128/JB.182.21.6247-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tkachenko A, Nesterova L, Pshenichnov M. 2001. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Arch Microbiol 176:155–157. 10.1007/s002030100301. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Zhu XJ, Xu RT, Gao Q, Wang DP, Zhang Y. 2018. Isolation and identification of histamine-producing Enterobacteriaceae from Qu fermentation starter for Chinese rice wine brewing. Int J Food Microbiol 281:1–9. 10.1016/j.ijfoodmicro.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 55.Hu M, Dong J, Tan GL, Li XY, Zheng ZY, Li M. 2021. Metagenomic insights into the bacteria responsible for producing biogenic amines in sufu. Food Microbiol 98:103762. 10.1016/j.fm.2021.103762. [DOI] [PubMed] [Google Scholar]
- 56.Kim KH, Chun BH, Kim J, Jeon CO. 2021. Identification of biogenic amine-producing microbes during fermentation of ganjang, a Korean traditional soy sauce, through metagenomic and metatranscriptomic analyses. Food Control 121:107681. 10.1016/j.foodcont.2020.107681. [DOI] [Google Scholar]
- 57.Linares DM, Fernandez M, Martin MC, Alvarez MA. 2009. Tyramine biosynthesis in Enterococcus durans is transcriptionally regulated by the extracellular pH and tyrosine concentration. Microb Biotechnol 2:625–633. 10.1111/j.1751-7915.2009.00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Z, Du H, Zhang Y, Xu Y. 2017. Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing. Front Microbiol 8:1294. 10.3389/fmicb.2017.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi H, Kimura B, Yoshikawa M, Fujii T. 2003. Cloning and sequencing of the histidine decarboxylase genes of gram-negative, histamine-producing bacteria and their application in detection and identification of these organisms in fish. Appl Environ Microbiol 69:2568–2579. 10.1128/AEM.69.5.2568-2579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernández M, del Río B, Linares DM, Martín MC, Alvarez MA. 2006. Real-time polymerase chain reaction for quantitative detection of histamine-producing bacteria: use in cheese production. J Dairy Sci 89:3763–3769. 10.3168/jds.S0022-0302(06)72417-1. [DOI] [PubMed] [Google Scholar]
- 61.Rochelle PA, Fry JC, Parkes RJ, Weightman AJ. 1992. DNA extraction for 16S rRNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol Lett 100:59–65. 10.1111/j.1574-6968.1992.tb14019.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.01884-22-s0001.pdf, PDF file, 0.9 MB (944.6KB, pdf)
Data Availability Statement
The bacterial raw sequence data were submitted to the NCBI Sequence Read Archive (SRA) under accession number PRJNA826566.