Abstract
Glioblastoma multiforme is the most common, highly aggressive malignant brain tumor which is marked by highest inter and intra-tumoral heterogeneity. Despite, immunotherapy, and combination therapies developed; the clinical trials often result into large number of failures. Often cancer cells are known to communicate with surrounding cells in tumor microenvironment (TME). Extracellular vesicles (EVs) consisting of diverse cargo mediates this intercellular communication and is believed to modulate the immune function against GBM. Tumor-associated microglia (TAM), though being the resident innate immune cell of CNS is known to attain pro-tumorigenic M2 phenotype, and this immunomodulation is aided by extracellular vesicle mediated transfer of oncogenic, immunomodulatory molecules. Besides, oncogenic proteins, long non-coding RNAs (lncRNAs) are believed to carry oncogenic potential and therefore understanding the mechanism leading to microglial dysregulation mediated by GBM-derived extracellular vesicle (GDEV) lncRNAs becomes crucial. This review focuses on current understanding of role of GDEV and lncRNA in microglial dysfunction and its potential as a therapeutic target.
Keywords: Intra-tumoral heterogeneity, tumor microenvironment (TME), tumor-associated microglia (TAM), extracellular vesicles, long non-coding RNA (lncRNA)
1. Introduction:
Glioblastoma multiforme of all the primary brain tumor malignancies is the most common tumor found in an age group of 40–70 years exhibiting the highest incidence rate (47%) [1]. The current limitations in the therapeutic approaches with the median survival of less than 15 months [2] surely demands for in depth understanding of mechanisms leading to GBM survival. This complexity of GBM is attributed to its highest inter and intra-tumoral heterogeneity [3] with tumor microenvironment (TME) adding a layer of diversity. The GBM microenvironment consists of an array of non-neoplastic cells, stromal cells, resident and infiltrating immune cells, glial cells and glioma stem cells (GSCs) which are now considered to be the key players for GBM progression [3, 4]. Resident microglia and infiltrating macrophages constitute a population called tumor associated microglia/macrophages (TAMs) in TME constituting 30–40% of total tumor mass [5]. Tumor cells often exhibit immune evasion strategy to escape immune surveillance mechanism by TAMs and instead reprogram them to facilitate highly immunosuppressive pro-tumorigenic environment.
The complex interaction between the tumor cells and TAMs have been widely studied and this intercellular communication is known to be mediated either by the release of several chemokines C-C motif chemokine Ligand 2 (CCL2), C-X3-C motif ligand-1 (CX3CL1), cytokines interleukin-6 (IL-6), transforming growth factor β (TGF-β), growth factors epidermal growth factor (EGF), vascular endothelial growth factor (VEGF) and matrix metalloproteinase (MMP-2, MMP-1) [6–8] or through direct cell contact via adhesion molecules and gap junctions in turn facilitating tumor cell survival and progression [9]. The release of membrane bound vesicles termed as extracellular vesicles (EVs) are also now emerging as novel mechanism driving the intercellular communication.
The discovery of EVs dates to 1940s by scientist Chargaff and West, where they originally reported the presence of EVs in blood, it was initially believed to be the platelet dust particles [10] separated by ultracentrifugation rich in lipid content [10, 11]. Later in early 1980s two independent groups reported the formation of vesicles and exocytosis during reticulocyte differentiation [12, 13] which were termed as exosomes (derived from endolysosomal pathway) and later in 1990s Raposo et al reported vesicle release by B-lymphocytes stimulating T-cells by presenting antigenic peptide-MHC complex [14]. EVs are heterogeneous membrane bound broad range of vesicles (differing in size and biogenesis) that are mainly classified as- exosomes (50–150nm) [15] released through multivesicular bodies (MVBs), microvesicles (100–1000nm) [16] budding off from plasma membrane, apoptotic bodies (100–5000nm) [17, 18] released from apoptotic cells in the extracellular space and large oncosomes (>1μm) [19] from cancer cells. The complex cargo carrying ability and relative stability of EVs makes it a unique delivery vesicle known to control many normal cellular functions and physiological processes. Bebelman and colleagues have highlighted the diverse role of EV in cancer by reprogramming stromal and immune cells to support tumor angiogenesis, immune suppression, tumor invasion, pre-metastatic niche formation and treatment resistance [20]. Furthermore, EVs contain immunomodulatory molecules (peptides, proteins, mRNA, miRNA, lncRNAs) that when transferred to (TAMs) changes its phenotype leading to immune privilege microenvironment in the brain. There has been an increase in the studies to elucidate the involvement and immunomodulatory role of diverse EV cargoes with a special focus on long non-coding RNAs as recent findings report the regulatory role of lncRNAs in gene expression at transcriptional and translational level, DNA synthesis and gene rearrangement [21]. This necessarily points at its immunoregulatory role in cancer driving immunosuppression within TME and aiding tumor progression [22]. Hence, understanding the GBM-derived extracellular vesicle (GDEV) biology in modulating immune response will pave way for development of novel therapeutics against GBM. Therefore, in this article we have reviewed the role of GDEVs in modulating TAMs response and polarization to tumor supportive phenotype and possible implications of EVs in therapy against GBM.
2. Extracellular vesicles size and morphology:
Traditionally EVs based on the size and biogenesis pathway, were primarily classified into 3 types- exosomes (endosomal origin), microvesicles (outward blebbing of plasma membrane) and apoptotic bodies (released specifically from dying cells). Furthermore, oncosomes derived specifically from tumor cells form a separate class of EVs ranging in size from 1 to 10μm [23, 24]. However, due to overlapping sizes and lack of specific surface markers, the nomenclature for EVs still remains a debate in the field of EV research. As per the recent guidelines by the international society of extracellular vesicles (ISEV), MISEV 2018 recommends on using Extracellular Vesicles as the “generic term for particles naturally released from the cell that are delimited by lipid bilayer and cannot replicate” [25] and can be termed more carefully based on clear, measurable characteristics such as size- small EVs (sEVs <100nm or <200nm) and medium/large EVs (m/lEVs >200nm), density (low, middle, high), biochemical composition (CD63+/CD81+/Annexin A5 stained), experimental conditions (hypoxic, normoxic), cells of origin (large oncosomes, apoptotic bodies) [26].
EVs when analyzed as whole mounted vesicle deposited on EM grid and embedded in uranyl acetate and methyl cellulose, displayed cup shaped appearance [14, 27, 28]. However, EM has its own caveat with respect to sample preparation that makes it more complex to relate the observed structure with the native morphology of cells and EVs [29]. Nevertheless, this never ending quest to study EV morphology has led to the development of cryogenic EM technique in order to prevent sample dehydration and observed results using cryo-EM display EVs as round shaped [27, 29]. Recent study by Emelyanov and colleagues [30] described different subpopulations of EVs through cryoelectron microscopy suggesting that different and specific functions may exist. Due to such smaller size of EVs, the most preferred techniques to characterize its morphology are transmission electron microscopy (TEM), scanning electron microscopy (SEM) and cryoelectron microscopy (Cryo-EM). However, these methods have its own caveats too. Several other articles have detailed the characterization of extracellular vesicles [15, 29, 31–34].
3. Tumor cell derived EVs and its nucleic acid composition:
EV mediated intercellular communication and its crucial role in various cellular processes under physiological and pathophysiological state is owned by its diverse cargo content that are selectively sorted into EVs, depending on the mode of biogenesis, source cell and surrounding microenvironment. Unlike non-tumor cell derived EVs, tumor cells are known to release more EVs and carry distinct bioactive molecules (oncoproteins, lipids, nucleic acids). However, common EV composition consists of proteins (tetraspannins- CD9, CD63, CD81), endosomal sorting complex (ESCRT) proteins (Rab GTPases- Rab27a, Rab27b, Rab11), Alg-2 interacting protein X (ALIX), tumor susceptibility gene-101(TSG101), antigen presenting molecules (MHC I, MHC II), RNA binding proteins (RBPs), ribonucleoproteins, signalling receptors (epidermal growth factor receptor EGFR), integrins, lipids (sphingomyelin, cholesterol, ganglioside GM3, phosphatidylserine, ceramide) [19, 32], coding (mRNAs) [35] and an array of noncoding RNAs- (miRNAs) [36], small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), piwi-interacting RNAs (piRNAs), vault RNA, Y-RNA, circular RNA, long noncoding RNA (lncRNAs) [18, 37]. A detailed and updated list on EV content (in EXOCARTA database) enlists the presence of total 9769 proteins, 3408 mRNAs, 2838 miRNAs, 1116 lipids [38, 39].
Recent studies have revealed the potential role of nucleic acid transfer (fusion gene mRNA such as echinoderm microtubule-associated protein like 4-anaplastic lymphoma kinase -EML4-ALK, oncogenic lncRNAs) [40] to recipient cells (specifically ncRNAs) in remodelling the tumor microenvironment to more protumorigenic. As RNA content within EVs encode biological state specific information of source cell and microenvironment, remains protected within EVs and are useful as potential biomarkers, its analysis is of paramount importance specifically in context of cancer diagnosis, prevalence, and pathophysiology. Tumor derived EVs are found to be enriched in mRNAs (for eg: the human telomerase reverse transcriptase hTERT mRNA) [41, 42] that promote tumor growth by inducing telomerase activity in the recipient fibroblasts. Furthermore, a study by Al Nedawi et al [43] reported the presence of mutant form of EGFR mRNA in GDEVs and its transfer to GBM cells lacking EGFRvIII promotes anchorage independent growth of tumor cells [41, 44].
Besides this, EV-non-coding RNAs (ex-ncRNAs) also mediate this tumor cell-non-tumor cell crosstalk by its function as a gene transcription regulator. Functional delivery of many oncogenic miRNAs (miR-9, miR-21, miR-195, miR-203, miR486–5P, miR-451a, miR-4257) trafficked into EVs aids in immunosuppression, angiogenesis, cancer cell stemness, proliferation, migration and invasion [18] in glioblastoma, breast cancer, rhabdomyosarcoma and lung cancer [45–48].
Other noncoding RNAs such as piRNAs, snoRNAs [49, 50] are also aberrantly expressed in human tumors that get sorted as EV cargo and mediate genome stability, DNA methylation and regulate the gene expression [51]. Apart from this, long noncoding RNAs (>200 nucleotides) has now extensively been studied for its role in cancer progression, angiogenesis, metastasis [52], drug resistance [53]. A study by Gezer et al, reported the presence of several lncRNAs- Metastasis associated lung adenocarcinoma transcript-1 (MALAT1), Hox antisense intergenic RNA (HOTAIR), lncRNAp21, growth arrest specific-5 (Gas5), Taurine upregulated gene-1 (TUG1), cyclin D1 (CCND1) in EVs derived from human cervical and breast carcinomas [54]. In vitro and in vivo studies have further confirmed the presence of many other lncRNAs in EVs and their oncogenic potential such as plasmacytoma variant translocation-1 (PVT1), zinc finger antisense-1 (ZFAS1), zeb-1 antisense 1 (ZEB1-AS1), lncRNA TUC339, colon-cancer associated transcript-2 (CCAT2), lncRNA POU3F3, urothelial carcinoma associated 1 (UCA1), lncRNA ATB in various other cancers [42, 55] as detailed in Table 1. However, the heterogeneity in EV subtypes, tumor types and distinct tumor microenvironment makes the RNA network complex within EVs which demands for deeper investigation that could reveal reliable EV markers and cargo contents that will ease the cancer diagnosis and treatment in future.
Table 1:
Extracellular vesicle derived known long noncoding RNAs in Tumor-associated Macrophage/microglia (TAMs) polarization in different types of cancer.
| Long noncoding RNA (lncRNA) | Cancer type | Expression in cancer cells | Target/signalling pathways in TAMs polarization | References |
|---|---|---|---|---|
| ANCR | Gastric cancer | Upregulated | Inhibits FOXO1 and aids in macrophage M2 polarization | [56] |
| BCRT1 | Breast cancer | Upregulated | Competitively binds to miR-1303 thereby preventing the degradation of PTBP3 and aids M2 macrophage polarization | [57] |
| CCAT1 | Prostate cancer | Downregulated | Increases miR-148a expression favouring M2 macrophage polarization | [58] |
| linc-RNA COX2 | Hepatocellular carcinoma | Downregulated | Promotes M2 macrophage polarization | [59] |
| CASC2C | GBM | Downregulated | Regulates Coagulation Factor X expression that promotes M2 polarization | [60] |
| lncSNHG15 | GBM | Upregulated | Promotes M2 polarization of microglia in TMZ-R GBM cells | [61] |
| lnc-TALC | GBM | Upregulated | Promoting microglia to M2 state corelated with the secretion of complement components C5/C5a | [62] |
| GAS5 | Hepatocellular carcinoma | Downregulated | is a negative regulator for M2 TAMs, its overexpression inhibits M2 polarization via PTEN activation | [63] |
| GNAS-AS1 | Breast cancer and NSCLC | Upregulated | Sponging of miR-433–3P thereby aiding M2 polarization of TAMs | [64, 65] |
| LINC00662 | Hepatocellular carcinoma | Upregulated | acts as ceRNA that binds to miR-15a, miR-16, and miR-107, inhibiting their action on the target genes and inducing the activation of Wnt/β-catenin signalling | [66] |
| Linc00514 | Breast cancer cells | Upregulated | Recruits JAK2 that phosphorylates STAT3, which increases JAGGED 1 expression in nucleus, activating NOTCH-1 signalling | [67] |
| MALAT1 | Hepatocellular carcinoma | Upregulated | Molecular sponge to miR-140 in HCC inducing angiogenesis ultimately leading to M2 macrophage polarisation | [68] |
| NEAT1 | Multiple myeloma, Endometrial carcinoma | Upregulated | molecular sponging of miR-214 and regulates the expression of B7-H3 (immune checkpoint regulator) thus aiding M2 macrophage polarization | [69, 70] |
| NIFK-AS1 | Endometrial cancer | Downregulated | Via miR-146a/NOTCH 1 axis activation | [71] |
| LincRNA-P21 | Breast cancer, NSCLC | Upregulated | Directly interacts with p53 and inhibits MDM2 degradation which results into TAMs polarization to M2 eventually | [72, 73] |
| lnc-RPPH1 | Colorectal cancer | Upregulated | Via TUBB3/SNAIL pathway activation promoting M2 macrophage activation | [74] |
| lncRNARP11–361F155.2 | Osteosarcoma | Upregulated | Sponging miR-30c-5p that increases the expression of CPEB4 ultimately leading to M2 polarization of TAMs | [75] |
| TUC339 | Hepatocellular carcinoma | Upregulated | Downregulates FcR mediated phagocytosis pathway, reduces pro-inflammatory cytokine production leading to M2 phenotype | [76] |
| XIST | Lung cancer | Upregulated | TCF4 mediated XIST overexpression promoting TAMs M2 | [77] |
ANCR: antidifferentiation noncoding RNA; BCRT1: breast cancer-related transcript 1; PTBP3: polypyrimidine tract binding protein 3; CCAT1: colon cancer related transcript 1; CASC2C: cancer susceptibility candidate 2c; GAS5: growth arrest specific 5; NSCLC: non-small cell lung cancer; ceRNA: competing endogenous RNA; HCC: hepatocellular carcinoma; MALAT1: metastasis associated lung adenocarcinoma transcript 1; NEAT1: nuclear enriched abundant transcript 1; TUBB3: beta-tubulin 3; CBEP4: cytoplasmic polyadenylation binding protein 4; XIST: X-inactive specific transcript; TCF4: transcription factor 4.
4. Glioblastoma-derived extracellular vesicles (GDEVs):
The highly aggressive and heterogeneous nature (at genetic and cellular level) of glioblastoma multiforme imposes a serious challenge in defining its characteristic features to develop an effective treatment and hence it still holds poor prognosis among GBM patients. This constant rate of failure in various therapeutic treatments and tumor recurrence is attributed to the highly immunosuppressive tumor microenvironment (TME) aided by an active interaction between GBM and surrounding non tumor cells. In addition to various known soluble factors and chemokines, EVs are unique intercellular delivery vehicle mediating GBM proliferation, migration, invasion, angiogenesis, immune evasion, metabolic alterations and therapy resistance [78] (Fig.1). Also, a clinical comparative study shows the higher number of EVs in the plasma of GBM patients to that of healthy individuals as reported by Osti et al which necessarily points towards EVs as a crucial mediator in GBM pathogenesis [79].
Fig 1. Glioblastoma-derived EVs in tumor cell proliferation, angiogenesis, and TAMs polarization.
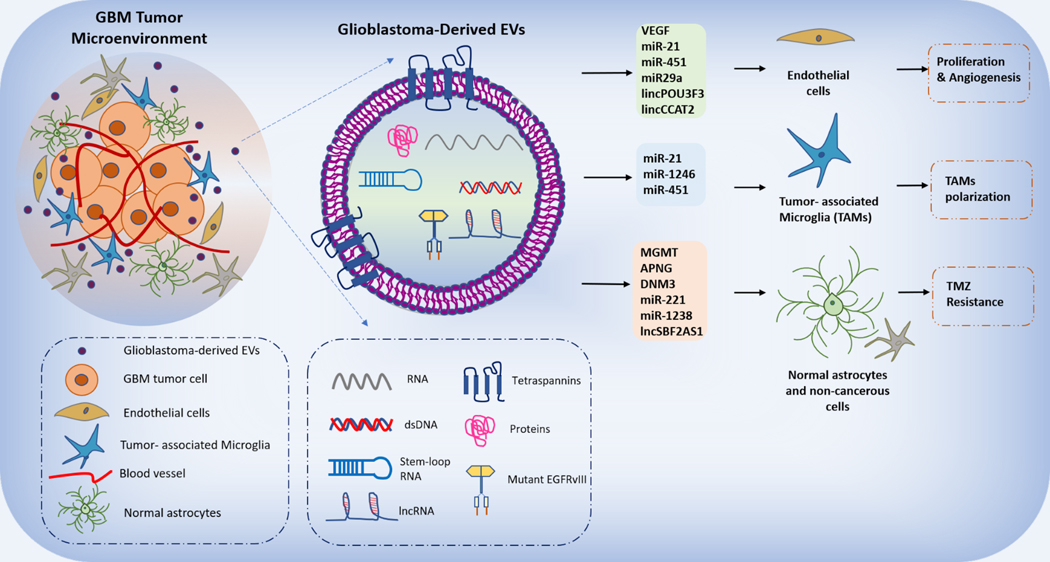
Glioblastoma-derived extracellular vesicles (GDEVs) consist of diverse array of oncogenic proteins, mRNA transcripts, ncRNAs (miRNAs and lncRNAs) when functionally transferred to recipient endothelial cells, tumor-associated microglia (TAMs) and other non-cancerous glial cells (astrocytes) drives key GBM hallmarks.
GDEVs carry oncogenic mutant EGFR mRNA which is delivered to the surrounding non-cancerous cells resulting in the activation of downstream signalling PI3K/AKT/MAPK pathways leading to GBM proliferation and survival [44, 78]. Other mRNA transcripts of isocitrate dehydrogenase (IDH1) [80], O-(6)-methylguanine DNA methyltransferase (MGMT) [81], alkylpurine-DNA N-glycosylase (APNG) [82], p65, DNM3, metalloproteinases such as MMP2, MMP9, plasminogen activators such as tissue type plasminogen activator (tPA), urokinase types plasminogen activator (uPA) [83], VEGF, Tenascin C, and Erb-B2 receptor tyrosine kinase-2 (ErbB2) are also reported to be elevated in GDEVs found in the CSF of GBM patients involved in mediating TMZ resistance, angiogenesis and GBM proliferation [78]. Among noncoding RNAs, wide array of miRNAs are reported to be either functioning as oncogene or tumor suppressor [84] miR-21 [85], miR-451, miR-29a, miR-222, miR-30a, miR-92b, miR-221, miR-23, miR-9, miR-10b, miR-1238) in GBM and are significantly found to be elevated in GDEVs enhancing cell proliferation, angiogenesis, inhibition of apoptotic pathway, TMZ resistance [18, 78]. miR-21, the most abundantly expressed oncomir in GBM, inhibits apoptosis by affecting the expression levels of tumor suppressive proteins (PTEN, RECK and PDCD4) [78]. Also, the functional transfer of miR-21 from GDEVs to surrounding endothelial cells and microglia enhances the tubular formation (in ECs), angiogenesis and downregulates the Btg2 gene expression (in microglia) thereby promoting cell cycle progression [86]. The detailed list of other oncomir dysregulated in GBM and present in GDEVs activating PI3K/AKT/mTOR, NOTCH, MAPK and Wnt/β-catenin pathway is listed in other articles [55, 87, 88]. In addition to miRNAs, short non-coding RNA sequences of GDEVs (from differentiated GBM cells) reveal the presence of total 712 non-coding RNAs including short RNAs such as piRNA, y-RNA, snRNAs, rRNA and snoRNA [89].
Next generation sequencing of GDEV-RNA significantly shows the presence and involvement of many long noncoding RNAs in GBM pathogenesis which are aberrantly expressed. Long noncoding RNA form the largest class of noncoding RNA with approximately 10,000 lncRNA genes and significantly lacking open reading frames (ORFs) [90]. In GBM, various lncRNAs for example: linc-POU3F3, linc-CCAT2, antisense transcript of hypoxia-inducible factor-1α (AHIF), set-binding factor 2 antisense RNA1 (SBF2-AS1), activated by TGF-β (ATB), HOTAIR, maternally imprinted gene (H19), Gas5, lncGRS1, PVT1, small nucleolar RNA host gene 15 (SNHG15), colorectal neoplasia differentially expressed (CRNDE), MALAT1, TP73-AS1, lncRNA TMZ-associated lncRNA in GBM recurrence (lnc-TALC), lnc-UCA1 are found to be overexpressed aiding cancer cell migration, invasion, angiogenesis, TMZ and radiation resistance and immunosuppression [87]. Although many lncRNAs are listed to be overexpressed in GBM tumor, only few lncRNAs (POU3F3, CCAT2, SBF2AS1, AHIF, ATB, TALC) are reported to be trafficked into GDEVs and exerts its profound protumorigenic effect on surrounding non-cancerous cells. GDEVs mediated transfer of linc-POU3F3 [91], linc-CCAT2 [92] to endothelial cells increase the expression of pro-angiogenic factor VEGF and inhibit the expression of pro-apoptotic proteins (Bcl-2, Bax and caspase 3). LncRNA SBF2-AS1 enhances TMZ resistance to GBM tumor cells by inhibiting miR-151A-3P and XRCC4 [93]. An in vitro study by Bian et al in U251 cells revealed the over expression of Exosomal lncRNA ATB that activated recipient normal astrocytes to a pro-tumorigenic state via suppression of miR-204–3p. These reactive astrocytes in turn promotes GBM tumor cell invasion [94]. A wide number of studies in GBM with respect to lncRNA expression reveals its crucial role mainly in context of angiogenesis and therapy resistance to the cancer cells while very less is known about lncRNA involvement in altering the microglia state within TME resulting in highly immunosuppression. LncRNA SNHG15 is significantly shown to be overexpressed in TMZ resistant clinical GBM samples and associated with GBM malignant properties- a critical onco-RNA. LncRNA SNHG15 in GBM increased the oncogenic and stemness markers SOX2, β-catenin, EGFR and CDK6 which caused the increased ability of TMZ-R GBM cells to generate M2 tumor-associated microglia through suppression of tumor-suppressor miR-627 [95]. Another upregulated lncRNA TALC incorporated into exosomes is transmitted to TAMs and promotes M2 polarization of microglia which is followed by the secretion of complement components C5/C5a that reduces tumor sensitivity to TMZ chemotherapy [62]. In contrast to this, macrophage polarization to M2 is inhibited by the over expression of lncRNA CASC2C that negatively regulates the coagulation factor X and miR-338–3p important for macrophage recruitment and polarization thereby acting as the tumor suppressor lncRNA.
Additionally, the detailed list of other lncRNAs dysregulated in GBM are well explained in recent articles [87, 90, 96–98].
Moreover, hypoxia adds a layer of diversity to the GBM tumor and affects the crosstalk between cancerous and non-cancerous cells in GBM TME. It is noteworthy to mention the influence of hypoxia on EV release [99] and cargo leading to the alterations in tumor metabolism, immune response, TMZ sensitivity, and blood vessel dynamics [100, 101]. Hypoxic EVs from GBM cells are enriched in pro-angiogenic proteins VEGF, interleukin-8 (IL8), insulin like growth factor binding protein (IGFBP1, IGFBP3); MMP-9, pentraxin-3 (PTX3), PDGF-AB/AA, CD26, plasminogen activator inhibitor-1 (PAI1), caveolin-1 (CAV1), tissue factor (TF) [102] and mRNA adrenomedullin (ADM), lysyl oxidase (LOX), IGF binding protein (IGFPB), B-cell lymphoma (BCL), BCL/2 adenovirus E18–19kDa interacting protein-3 (BNIP3), N- myelocytomatosis viral related oncogene (myc) downstream regulated 1 (NDRG1), pro-collagen-lysine-2-oxoglutarate 5-dioxygenase 2 (PLOD2), (PAI1) [103].
MiR-301a from hypoxic GDEVs, inhibit transcription elongation factor A-like 7 (TCEAL7) and thereby leads to the activation of Wnt/β-catenin pathway making normoxic GBM cells resistant to radiation. Few lncRNAs such as AHIF, H19 are actively found to be released from hypoxic GDEVs conferring radioresistance, invasion and stemness to cancer cells [104, 105]. LncRNA ROR1-AS1/miR-4686 axis contributes to tumor progression [106]. However, the complexity of GBM tumor and GDEV heterogeneity limits the complete analysis of GDEV cargo in context of lncRNA specifically, and therefore it is of paramount importance to determine lncRNA expression and its role mediated via GDEVs to unveil the underlying mechanism of GBM pathogenesis.
5. Glioblastoma-derived EVs and tumor microenvironment:
Although immunotherapy has been developed for GBM, its limited success under clinical trials increases the need to better understand the failures. These failures are because of the continuous local and systemic immunosuppression that establishes a tumor supportive environment surrounding tumor. There occurs a vast heterogeneity of immune cells infiltrating the GBM microenvironments revealed through histopathological, glow and mass cytometry analysis, single cell-RNA sequencing analysis (sc-RNA seq) in various human and rodent diseased models [107–110]. Myeloid cells constitute about one third of the total tumor mass and exert pro-tumorigenic functions in the GBM microenvironment [111]. The cells of the myeloid origin in GBM includes bone-marrow derived macrophages (peripheral macrophages), resident microglia, myeloid-derived suppressor cells, dendritic cells, and neutrophils [112]. Till the discovery of resident innate immune microglia by Pio del Rio Hortega, brain was thought to be an immune privilege site. However, studies have shown that CNS is immune competent as well as actively interactive with the peripheral immune system. These findings suggest a putative role of resident and infiltrating immune cells in CNS. Innate immune microglia exhibit two different activated states- M1 proinflammatory and M2 anti-inflammatory. In cancer, these TAMs are known to be in their tumor supportive phenotype which is anti-inflammatory (M2). The recruitment and subsequent polarization of microglia to M2 state is mediated by the release of various soluble factors and polarizing cytokines from cancer cells, which in turn causes altered microglia to release tumor supportive factors (EGF, TGF-β) aiding GBM progression. This constant urge to decode the underlying mechanisms in the tumor biology has led to the discovery of EVs as a novel mediator in driving cancer immunosuppression.
The fact that single GBM cells can secrete as many as 10,000 EVs over the period of 48 hrs suggests its significant role in tumor-stromal cell crosstalk and key mechanisms driving GBM [44]. GDEVs contribute to immune suppression by the expression and release of various immunosuppressive molecules (Cytotoxic T lymphocyte antigen-4, Fas Ligand, CD39, TNF-related apoptosis inducing ligand, ncRNAs) [113]. The interaction between one such immune checkpoint molecules- programmed death-1 (PD-1) and its ligand programmed death ligand-1 (PD-L1) is known to be negatively regulating the immune response, inhibiting the anti-tumor T-cell activation and aiding immune evasion in cancer cells [114]. Recent study reveals the clinical and prognostic significance of PD-L1 in glioblastoma, suggesting the higher expression of PD-L1 associated with poor prognosis and survival of GBM patients [115]. In context of EV research, the active understanding on expression of PD-L1 on EV reveals its involvement in inhibiting antitumor response contributing to immunosuppression within TME [116, 117]. Glioblastoma stem cell-derived EVs (GSC-derived EVs) expressing PD-L1 are reported to suppress T-cell activation, one of the possible immune evasion strategy adapted by GBM cells [118]. Additionally, monocytes readily take up GSC-derived EVs and once internalized changes its morphology, phenotype that leads to monocyte polarization to immunosuppressive M2 state [119, 120]. GDEV mediated functional transfer of miR-21 and miR-451 to microglia and macrophages in vitro as well as to macrophages in vivo resulted in the downregulation of c-Myc expression and its target gene BTG2 and subsequent polarization to M2 phenotype of microglia [86, 121, 122]. The downregulation of BTG2 transcription factor resulted in an increased microglial proliferation, tumor growth and formation of hypoxic microenvironment. Hypoxic GDEVs consisting of miR-1246 contribute to M2 macrophage polarization by inhibiting NF-kβ and activating STAT3 pathway [123, 124].
GDEVs also functionally alter other myeloid cells such as dendritic cells (DCs) by the interaction between galectin-9 expressed on GDEVs and Tim-3. This interaction reduces the antigen presenting ability of DCs in vitro, ultimately causing the significant inactivation of CD8+ T-cells [125]. Recent data reveals that GDEVs do not directly inhibit T-cell activation but instead it induces the formation of inhibitory non-classical monocytes expressing PD-1 thus skewing T-cell activation [126]. Inhibition of T-cell activation by EVs is also known to be myeloid cell dependent [127]. Further in context of myeloid-derived suppressor cells (MDSCs), its role in cancer progression and inhibition of T-cell activation is well studied promoting tumor angiogenesis, invasion, and metastasis [128]. A recent study investigates the underlying mechanism of immunosuppression through GDEVs and concludes that hypoxic GDEVs activate MDSCs by the transfer of miR-10a and miR-21 activating downstream NF-kβ and PTEN/PI3K pathway [129] and expansion of MDSCs via miR-29a/Hbp1 and miR-92a/Prkar1a [130] ultimately leading to the inhibition of T-cell activation. These findings suggest the pathophysiological role of GDEVs in modulating immune microenvironment in GBM.
6. GBM-microglia crosstalk through EVs: a two-way communication:
As discussed, GDEVs are known to carry immunomodulatory molecules (ncRNAs) that when functionally transferred to TAMs, reprogram them to tumor supportive state as summarized in Table 2. The efficient release and uptake of GDEVs by microglia is confirmed by intravital microscopy imaging in vivo, suggesting the interaction of cancer cells locally and systemically through EVs. In vitro study reports the alterations in the cytokine secretion from tumor associated microglia (TAMs) on exposure to GBM-derived EVs for 5 days resulting in the upregulation of many pro-tumorigenic cytokines (CXCL10, CXCL1, CCL2, CCL5, IL-6 and TIMP1) and downregulation of anti-tumorigenic cytokines (IL-16, IL-23, IL-27) [131]. The aberrantly expressed miR-214–5p in U87MG cells, selectively sorted into exosomes when shuttled to microglia modulated the inflammatory response by inhibiting CXCR5 [132]. A recent in vivo study reported an increase in the levels of miR-21 in microglia after the intracranial tumor injection in syngeneic mouse model that resulted in the downregulation of Btg2 gene, an anti-proliferative factor 2, thereby increasing the microglial proliferation [86]. Hypoxic GDEVs containing miR-1246 were demonstrated to facilitate the formation of an immunosuppressive GBM TME through inducing M2 macrophage polarisation in targeting telomeric repeat-binding factor 2-interacting protein 1 (TERF2IP), thereby activating the signal transducer and activator of transcription 3 (STAT3) pathway [121].
Table 2:
GDEV cargo (miRNAs and lncRNA) in tumor-associated microglia/macrophages (TAMs) reprogramming and immunosuppression
| GDEV cargo | Recipient cell | Effect on Recipient cell | Reference |
|---|---|---|---|
| miR-451 | Microglia | Downregulating c-Myc mRNA and enhancing M2 phenotype | [139] |
| miR-21 | Microglia | Downregulating of Btg2 gene and enhancing microglial proliferation and consecutive polarisation to M2 | [86] |
| miR-29a, miR92a | MDSCs | Promotes Differentiation and proliferation of MDSCs via Hbp1/ Prkar1a/PKA/P-STAT3 axis leading to immunosuppression | [129, 130] |
| miR-1246 | Macrophages | Targets TERF2IP and activates STAT3 pathway and inhibiting NF-kβ | [124] |
| MALAT-1 | Microglia | Inhibits miR-129–5P and activating HMGB1 leading to pro-tumorigenic cytokine secretion (IL-6, IL-8 and TNF-α | [136] |
MDSCs: myeloid derived suppressor cells; c-MYC: cellular MYC; BTG2: B-cell translocation gene 2; Prkar1a: c-AMP dependent protein kinase type-I alpha regulatory subunit; TERF2IP: telomeric repeat-binding factor 2; NFkβ: nuclear factor kappa-light-chain- enhancer of activated B-cells; HMGB1: high mobility group box protein 1.
It is equally important to understand the role of hypoxic GDEV lncRNAs in microglial dysregulation as several ongoing studies have demonstrated its potential use as a prognostic and molecular typing marker for precision treatment against GBM [133–135]. In context to this, EV lncRNA MALAT1 from glioma stem cells modulates the inflammatory response in recipient microglia cells by inhibiting the miR-129–5p and upregulating high mobility group box protein-1 (HMGB1) expression leading to IL-6, IL-8 and TNF-α secretion thereby promoting glioma progression [136]. Although it is evident that glioma EVs skew the microglial response to tumor supportive phenotype, the complete aspect of EV-derived lncRNA in microglial dysregulation is still unknown.
Growing evidence suggests that microglia and other supporting glial cells also shed EVs in response to their activation in the GBM TME thereby carrying out bilateral intercellular communication. Grimaldi and colleagues demonstrated that the EVs released by BV2 microglial cells in response to lipopolysaccharide (LPS) and interferon gamma (IFN-γ) stimulation exerts the anti-tumor property by modulating their inflammatory state in glioma, thus re-establishing the brain homeostasis [137]. Furthermore, the potential therapeutic role of microglia derived EVs is reported as it suppresses the invasive behaviour of tumor cells in 3D spheroid glioma culture [138]. Taken together, these results suggest the protective role of microglia derived EVs in cancer by reprogramming the TAMs phenotype. However, the anti-tumor property of immune cell EVs requires further clarification.
7. Therapeutic implications of EVs for GBM: current updates
With the increase in EV research and several clinical and preclinical evidence reported, EVs are a potential target and therapeutic tool for treatment against GBM, besides being a source of biomarker. Large number of studies report the increase in EV count in plasma samples of the cancer patients than compared to healthy individuals due to characteristic factors such as metabolic reprogramming in cancer cells, autophagy, invasive behavior, pH of the TME and hypoxia [140] and corelates with the patient’s survival in melanoma [141]. Therefore, owing to these factors and its several advantages: such as high selective targetability, size distribution, stability, extravasation capacity, less immunogenic than the parental cells, inherent tissue repair property, its ability to penetrate blood-brain barrier (BBB) and diverse cargo carrying capacity at a time [18, 142, 143] enables EVs as a promising target in the therapeutics. Furthermore, unlike cells, EVs do not replicate after injection and present less risk of developing tumor.
Emerging therapeutic strategies for EVs include 1. EVs as therapeutic target 2. EVs as therapeutic agent 3. EVs in drug delivery [142, 144].
1. EVs as therapeutic target:
Strategies aiming at targeting EVs include inhibiting its various functions such as EV biogenesis, release, uptake, altering its composition, removal of EVs [145].
Inhibition of EV biogenesis and release:
As EV biogenesis is greatly enhanced in cancer [28] depending upon the physiological conditions, targeting its biogenesis machineries has gained wider attention and involves inhibiting ESCRT, Rab proteins (Rab 2a/b) [146, 147], blocking tetraspannins [148], sphingomyelinase [149], membrane bound heat-shock protein (HSP70, HSP72, HSP90) [150]. Rab proteins are crucial for cellular endosomal trafficking [151] and also mediate cancer cell progression and survival [152]. A study by Bobrie et al highlighted the pro-tumoral function of Rab27a expression by mouse metastatic breast carcinoma and its inhibition led to the reduced number of EVs and inhibition of lung metastasis of 4T1 breast cancer cells in vivo [153]. In addition, the use of pharmacological inhibitors [154] either affect the EV trafficking- such as manumycin-A inhibiting Ras and its farnesyltransferases and other ribonucleoproteins, thus significantly reducing EV production by 50–60% in prostate cancer cell lines [155] or affect lipid metabolism such as GW4869- a sphingomyelinase inhibitor. Panigrahi et al reported the reduction in cell viability of prostate cancer cells treated with GW4869 in hypoxia [156]. Furthermore in vitro and in vivo studies highlight the use of other EV inhibiting drug such as dimethyl amiloride- an anti-hypotensin drug that inhibits calcium channels and impairs the intracellular calcium release [157] resulting into reduced EV release into culture medium and blood serum respectively [158] which culminates the STAT3 phosphorylation in MDSCs and its immunosuppressive activity. Peptidylarginine deiminase (PAD) is reported to be crucial for EV biogenesis in range of cancers including GBM [159–164] and known to be upregulated in response to hypoxia in malignant gliomas. An in vitro study shows the effect of pan-PAD inhibitor- Cl-Amidine [163] and PAD- isozyme specific inhibitor [165] on EV release in two GBM cell lines LN18 and LN229 thus concluding the key role of PAD-mediated pathway for EV biogenesis in GBM with a potential therapeutic target. EV biogenesis mechanism is largely understood; however, it still remains to be fully elucidated as the involvement of various protein in EV biogenesis and release vary from cell type and the cellular state.
Inhibition of EV uptake:
Inhibition of EV uptake by recipient cells is another strategy to target EVs as number of uptake mechanisms have been proposed such as receptor-mediated endocytosis, phagocytosis, and micropinocytosis [166]. Many surface proteins- tetraspannins, integrins, immunoglobulins, proteoglycans, lectins, SNAREs, Rab GTPases, Sec1/Munc-18 related proteins (SM-proteins) interact with the receptors on the recipient cell to facilitate the docking of EVs [167]. As EV uptake by cancer cells has been reported to be one of the major contributing factors for chemoresistant in tumor cells, the use of EV-uptake inhibitors is shown to reduce the EV uptake by chemosensitive cells thus retaining their sensitivity to the chemotherapy. As per several in vitro studies, these EV uptake inhibitors either block the clathrin dependent endocytosis e.g., chlorpromazine, dynasore, ikarugamycin studied in lung cancer cell lines (H1299, HCC366, H137) or inhibit clathrin-independent endocytosis e.g., methyl-β-cyclodextrin (MβCD), genistein, heparin studied in ovarian cancer cells, urothelial carcinoma [168]. Two in vitro studies on investigating the significant role of heparin-sulfate proteoglycans (HSPGs) and caveolin in EV uptake demonstrate the reduction in EV-mediated cell migration and EV internalization in heparin [169] and MβCD [170] treated GBM (U87MG) cells due to cholesterol depletion in a dose dependent manner. Treatment of microglia with alkalinizing pharmacological inhibitors as Bafilomycin-A, monensin and chloroquine significantly inhibited the vesicle internalisation in microglia by reducing vacuolar acidification required for macropinocytosis [171]. Blocking of phosphatidyl serine (PS) with annexin V [172] or inhibiting integrin [173] by disintegrin inhibitor present on the surface of EVs also notably inhibited the EV uptake into microglia and other recipient cells. Furthermore, in context to this, the capturing and removal of tumor cell derived EVs by specific antibodies to block ligand-receptor interaction is a novel strategy to reduce the oncogenic function of EVs [174]. In vitro and in vivo studies in human breast cancer xenograft mouse model suggested the elimination of anti-CD9 and anti-CD63 tagged EVs by macrophages ultimately leading to the inhibition of pro-metastatic effects of tumor cell derived EVs [175]. Despite the ongoing quest to use EVs as therapeutic targets, there occur challenges and limitations to these strategies. These strategies currently lack any pre-clinical and clinical investigations as most of the EV inhibitor studies are in context of its biogenesis, release and uptake are tested under in vitro conditions. Most of the EV inhibitors lack specificity and efficacy due to multiple targets and pathways involved in the EV biology. Hence, targeting EVs need a better and elaborative understanding of the underlying pathways.
2. EVs as therapeutic agents:
Extracellular vesicles because of its wide involvement in normal physiological as well as pathophysiological processes, it is also used as a potential therapeutic agent in the regenerative medicine and tissue repair [144]. EVs have an innate therapeutic potential because of the presence of bioactive cargoes. EVs derived from multipotent stem cells drive angiogenesis, cell proliferation, and inhibit apoptosis by reprogramming the recipient cells and hence are useful in the regenerative medicine. Mesenchymal stem cells derived exosomes are known to drive GBM tumorigenesis and impart stemness to the glioma cells [176]. A study reported the increased sensitivity to TMZ on functional delivery of anti miR-9 via MSC-derived exosomes [177]. Besides stem cells, EVs derived from differentiated cell types such as immune cells are also one of the potential therapeutic agents in used for immunomodulation. Natural killer (NK) cells derived exosomes target GBM cells and inhibit its growth by specifically localizing to the tumor site as NK cell-derived exosomes contain FasL, perforin, granzyme-B and TNF-α [178]. Microglial-derived EVs suppressed the tumor invasion in a time dependent manner in a 3D-spheroid glioma model [138]. A study by Liu and colleagues, shows that the co-delivery of tumor derived exosomes with α-galactosyl ceramide in a DC vaccine induces a strong activation and proliferation of tumor-specific T-lymphocytes [179].
3. EVs in drug delivery system:
The characteristic property of EVs such as small size, biocompatibility, high penetration and selectivity, cargo delivery and presence of adhesive molecules on its surface enables EVs to be an efficient drug delivery vehicle.
Furthermore, the most intriguing property of EVs is the functional transfer of miRNAs to recipient cells and therefore many miRNA inhibitors, sponge constructs (miR-21 sponge constructs) packaged into exosomes have emerged as a new therapeutic approach [180]. Therefore, EV mediated drug or gene delivery is considered to be the optimal therapeutic strategy against cancer. EVs derived from mesenchymal cells, tumor cells and immune cells are reported to be used as nanocarriers for the transport of chemotherapeutic drugs such as doxorubicin (DOX), paclitaxel (PTX) in the treatment of various cancers. In vitro study by yang et al suggested that DOX-loaded exosomes derived from MCF-7 cancer cells showed increased cytotoxic effect against tumor cells with a significant reduction in the tumor growth. In addition to this, another anti-mitotic chemotherapeutic drug PTX has also been used in the treatment of ovarian cancer, lung cancer and GBM. Brain endothelium cell derived (bEND.3) exosomes loaded with paclitaxel and DOX crossed the BBB and significantly reduced the GBM tumor growth which was evident by the decreased fluorescent intensity in xenotransplanted zebrafish cancer model [181–184]. Various studies [185–187] on human bone marrow derived exosomes (hBMSCs) containing miRNA and lncRNA have reported decrease in glioma cell proliferation, invasion and enhanced chemosensitivity to temozolomide (TMZ). As EVs carry immunomodulatory molecules, in vivo studies confirmed that exosomes from DCs loaded with chaperone rich cell-lysates elicits a potential T-cell immune response against intracranial glioma in mice [188]. Besides this, human neural stem cells derived (hNSCs) EVs and microvesicles have also been shown to impart neuroprotective effect in the irradiated brain following radiation therapy for the treatment of brain cancers [189, 190]. The major limitation to this approach is the reduced scalability and lower loading efficiency of anti-cancer drugs into the EVs. Recent studies have demonstrated the large scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation [191]. In addition to this, the loading efficiency of exosomes loaded with doxorubicin and paclitaxel is increased via microfluidics and inhibits glioma cell proliferation [192]. Furthermore, focused ultrasound hyperthermia is a non-invasive technique to improve drug delivery and immunological recognition of tumor cells which is also now known to augment the release of glioma derived EVs capable of causing innate immune activation [193].
Besides scalability and loading efficiency, other caveats to using EVs as therapeutic targets include high heterogeneity, reduced endogenous yield, reproducibility, biodistribution, absence of methods to isolate homogeneous population, cost-ineffective technologies, off-site accumulation, decreased efficiency and lack of complete understanding on EV biology and its role in disease pathology altogether demands for the development of engineered EVs to overcome the above-mentioned limitations Modified EVs generated are known by different terms such as bioengineered EVs, artificial EVs, EV mimetics, exosome-like-nanovesicles, exosome-based-mimetics, however till date there is no clear criterion for their classification and hence it is suggested to name them as artificial EVs for the systematic classification [194]. Artificial EVs are engineered directly or indirectly and are categorized into as a. fully synthetic and b. semi-synthetic EVs. Fully synthetic EVs are the synthetic analogs that mimic the properties of natural EVs popularly known as EV mimetics while semi-synthetic EVs are the ones generated from the natural substrate-cells that are modified pre or post isolation.
Synthetic EVs or EV mimetics: The possible advantage of designing the synthetic EV mimetic over modifying the natural EVs is increasing the scalability of EVs to use in clinical studies, sterility, and uniformity in size [195]. Till date, three sub-types of EV mimetics are classified- i. Artificial EV mimetics ii. Physical-origin EV mimetics and iii. Hybrid EV mimetics [195]. EV-mimetics are manufactured by two broadly classified techniques- top-down and bottom-up method. Top-down nanotechnology method uses the larger substrate that is reduced to smaller units for the formation of nano-vesicles while bottom-up nanotechnology method uses the smaller individual fragments that self-assemble into higher order structures with tunable compositions [194]. Top-down nanotechnology method includes strategies that rely on the self-assembly of lipids and lipid membranes into spherical structures and encapsulation of cargo into the nanovesicle cavity. Methods such as extrusion, microfluidic, cell-slicing by using SixNy blades, nitrogen cavitation based, sonication-based, cell-bleb based are included in the generation of artificial EVs through top-down technique [196, 197]. The other emerging alternative to EV mimetics is the hybrid-EVs that enables the fusing of EVs with synthetic liposomes to increase the stability, shelf-life in circulation and decrease the immunogenicity for the improved therapeutic use. The bottom-up technique uses the methods that enables the generation of fully artificial synthetic EVs by assembling individual cargo molecules such as lipids, proteins into complex structure to resemble the EV membrane [194]. The most commonly used and reviewed EV mimic are the liposomes (100nm) [198] and since not all methodologies yield uniform EV mimics, only small unilamellar vesicles (SUV) are ideal precursors. The common approaches for SUV preparation include ether injection, ethanol injection, reverse phase evaporation method, thin-film hydration method, and microfluidics [194]. The details for these techniques are reviewed thoroughly in [194]. Despite these advancements in EV based therapeutics, EV mimics are still challenging to use in settings due to certain disadvantages over natural or semi-synthetic EVs such as membrane deformation, random packaging, lack of definite biomarker.
Semi-synthetic EVs: Semi-synthetic EVs make use of strategies that modify extracellular vesicles in order to augment their innate properties (functional transfer of cargo) for the improved therapeutic use [199]. The methods employed for semi-synthetic EV preparation include direct methods involving modification of EVs post-isolation or indirect by genetically engineering donor cells. EVs as nanocarriers are manipulated for the cargo loading either exogenously or endogenously. Endogenous cargo loading involves reprogramming the parental cells to overexpress the protein or nucleic acid of interest which is then packaged into EVs and delivered to the targeted recipient cell. In addition to this, exogeneous stimulation of cells by additional growth factors, cytokines and drugs into the culture medium, environmental stimulation, fusogenic liposomes, 3D culture platforms and bioreactors are alternative methods to re-engineer parental cells to enhance EV yield, scalability, reproducibility, and therapeutic potency [200]. Furthermore, methods to modify EVs post isolation are broadly categorized into active and passive methods. Active methods are physical methods involving electroporation, extrusion, saponin assisted loading, sonication, hypotonic dialysis, and freeze-thawing and chemically induced methods include hydrophobic insertion, click chemistry and non-covalent modification, while passive methods include co-incubation of isolated EV with the target drug [194]. All these methods enable the modification of EVs on its surface and despite these advancements in the EV engineering it still remains a challenge in the field of therapeutics and regenerative medicine. The major challenge in employing these methods require controlled reaction conditions such as pH, temperature in order to avoid the EV membrane disruption [200]. GDEVs hold a great promise for future therapeutics, however it still requires more clinical and preclinical studies to understand its potential in GBM and an elaborative understanding on GBM associated EV cargo contents that modulates the microglial response preferably to M2 phenotype.
8. Future Directions:
The role of GDEVs in TAMs polarization is poorly understood and highly immunosuppressive state within TME remains to be a big challenge in the field of therapeutics. The characteristic heterogeneous population of EV subtypes, its stability and yield depend largely on the cell types from which it is secreted and surrounding physiological conditions that shape up the EV cargo and hence impose a big challenge for EV-based therapeutics. Moreover, the isolation of EVs also affect its size, specificity, cargo content, yield and hence it demands for optimizing the EV isolation technique to yield homogeneous EV population. The biodistribution kinetics of EVs under in vivo is also poorly understood and hence more of in vivo model studies are required in future to aid in the efficacy of EV therapeutics and subsequently help in understanding the tumor-glial cell crosstalk. Moreover, how significantly do the peripheral EVs circulating in the body influence the CNS function (both in normal and in diseased state) remains to be determined. Furthermore, it also becomes important to determine whether immune modulation in TAMs is imparted by single lncRNA or in association with diverse EV cargo selectively packaged under hypoxia. A detailed characterisation of GDEV cargoes will help in better understanding of underlying mechanisms. In conclusion, future work in the field of EV-based therapy for GBM requires better understanding of EV biology and its role in pathogenesis in context of immune dysregulation and TAMs polarization and its use in clinical applications.
9. Concluding remarks:
This review summarizes the current understanding on GBM-microglia crosstalk mediated through EVs and its potential as a therapeutic target for development of more robust and effective immunotherapy. Most of the immunotherapies for GBM focuses on the importance of T-cells, however, much lesser is known for TAMs. TAMs are believed to be the central drivers aiding GBM proliferation and survival. Despite the presence of innate immune cells in the TME, the inability of microglial cells to elicit an immune response calls for further investigations driving it to the altered tumor supportive state. As EVs released by tumor cells are one of the key mediators in the intercellular communication it is of paramount importance to study the EV biology in GBM-microglia crosstalk. Other than known protein, lipids and ncRNA as EV cargoes, lncRNAs are also recently being emerged as a crucial mediator in cancer. However, microglial dysregulation mediated through GDEV-lncRNA is yet to be understood and its in-depth understanding will provide us important cues about non-cancerous, glial factors contributing to GBM proliferation and survival.
Acknowledgements:
The first author, Ms. Sangati Pancholi is highly thankful for the Junior Research Scholarship (JRS) award from Lady Tata Memorial Trust, Mumbai for supporting her doctoral thesis work. The authors would like to sincerely thank the laboratory members, Ms. Pampa Pain and Ms. Swagatama Mukherjee for participating in the discussions during the review manuscript preparation.
Funding:
The authors do not have anything to disclose regarding funding source for preparing this manuscript.
Footnotes
Ethics Approval statement:
This review manuscript has been prepared based on exhaustive literature survey and we have also discussed new conceptual advancement in the field. Hence, ethical approval is not applicable.
Declaration of competing interest:
The authors have declared that no competing interests exist.
Consent to participate and Consent for publication:
Not applicable
Data availability statement:
Not applicable as the current review manuscript is solely based on literature survey and discusses the future road map for the research problem discussed.
References:
- 1.Ostrom QT, et al. , CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro-oncology, 2020. 22(Supplement_1): p. iv1-iv96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koshy M, et al. , Improved survival time trends for glioblastoma using the SEER 17 population-based registries. Journal of neuro-oncology, 2012. 107(1): p. 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z.and Hambardzumyan D, Immune microenvironment in glioblastoma subtypes. Frontiers in immunology, 2018. 9: p. 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matarredona ER and Pastor AM, Extracellular Vesicle-Mediated Communication between the Glioblastoma and Its Microenvironment. Cells, 2020. 9(1): p. 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutmann DH and Kettenmann H, Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron, 2019. 104(3): p. 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prionisti I, et al. , Harnessing microglia and macrophages for the treatment of glioblastoma. Frontiers in pharmacology, 2019. 10: p. 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hambardzumyan D, Gutmann DH, and Kettenmann H, The role of microglia and macrophages in glioma maintenance and progression. Nature neuroscience, 2016. 19(1): p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho da Fonseca AC and Badie B, Microglia and macrophages in malignant gliomas: recent discoveries and implications for promising therapies. Clinical and Developmental Immunology, 2013. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vader P, Breakefield XO, and Wood MJ, Extracellular vesicles: emerging targets for cancer therapy. Trends in molecular medicine, 2014. 20(7): p. 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf P, The nature and significance of platelet products in human plasma. British journal of haematology, 1967. 13(3): p. 269–288. [DOI] [PubMed] [Google Scholar]
- 11.Chargaff E.and West R, The biological significance of the thromboplastic protein of blood. Journal of Biological Chemistry, 1946. 166(1): p. 189–197. [PubMed] [Google Scholar]
- 12.Harding C, Heuser J, and Stahl P, Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. Journal of cell biology, 1983. 97(2): p. 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnstone RM, et al. , Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). Journal of Biological Chemistry, 1987. 262(19): p. 9412–9420. [PubMed] [Google Scholar]
- 14.Raposo G, et al. , B lymphocytes secrete antigen-presenting vesicles. Journal of Experimental Medicine, 1996. 183(3): p. 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Niel G, d’Angelo G, and Raposo G, Shedding light on the cell biology of extracellular vesicles. Nature reviews Molecular cell biology, 2018. 19(4): p. 213–228. [DOI] [PubMed] [Google Scholar]
- 16.Heijnen HF, et al. , Activated Platelets Release Two Types of Membrane Vesicles: Microvesicles by Surface Shedding and Exosomes Derived From Exocytosis of Multivesicular Bodies and▪-Granules. Blood, The Journal of the American Society of Hematology, 1999. 94(11): p. 3791–3799. [PubMed] [Google Scholar]
- 17.Mathieu M, et al. , Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nature cell biology, 2019. 21(1): p. 9–17. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien K, et al. , RNA delivery by extracellular vesicles in mammalian cells and its applications. Nature reviews Molecular cell biology, 2020. 21(10): p. 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaborowski MP, et al. , Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience, 2015. 65(8): p. 783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bebelman MP, et al. , Biogenesis and function of extracellular vesicles in cancer. Pharmacology & therapeutics, 2018. 188: p. 1–11. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, et al. , Noncoding RNAs: the shot callers in tumor immune escape. Signal transduction and targeted therapy, 2020. 5(1): p. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marar C, Starich B, and Wirtz D, Extracellular vesicles in immunomodulation and tumor progression. Nature Immunology, 2021. 22(5): p. 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meehan B, Rak J, and Di Vizio D, Oncosomes–large and small: what are they, where they came from? Journal of extracellular vesicles, 2016. 5(1): p. 33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Vizio D, et al. , Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer research, 2009. 69(13): p. 5601–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witwer KW and Théry C, Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. 2019, Taylor & Francis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Théry C, et al. , Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles, 2018. 7(1): p. 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colombo M, Raposo G, and Théry C, Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual review of cell and developmental biology, 2014. 30: p. 255–289. [DOI] [PubMed] [Google Scholar]
- 28.Raposo G.and Stoorvogel W, Extracellular vesicles: exosomes, microvesicles, and friends. Journal of Cell Biology, 2013. 200(4): p. 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartjes TA, et al. , Extracellular vesicle quantification and characterization: common methods and emerging approaches. Bioengineering, 2019. 6(1): p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emelyanov A, et al. , Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid. PLoS One, 2020. 15(1): p. e0227949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Théry C, et al. , Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology, 2006. 30(1): p. 3.22. 1–3.22. 29. [DOI] [PubMed] [Google Scholar]
- 32.Veziroglu EM and Mias GI, Characterizing extracellular vesicles and their diverse RNA contents. Frontiers in genetics, 2020. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aatonen MT, et al. , Isolation and characterization of platelet-derived extracellular vesicles. Journal of extracellular vesicles, 2014. 3(1): p. 24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholl JN, et al. , Characterization and antiproliferative activity of glioma-derived extracellular vesicles. Nanomedicine, 2020. 15(10): p. 1001–1018. [DOI] [PubMed] [Google Scholar]
- 35.Batagov AO and Kurochkin IV, Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biology direct, 2013. 8(1): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, et al. , & Kohli M(2013). Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC genomics. 14(1): p. 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolte-’t Hoen EN, et al. , Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic acids research, 2012. 40(18): p. 9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai J, et al. , Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduction and Targeted Therapy, 2020. 5(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson RJ, Kalra H, and Mathivanan S, ExoCarta as a resource for exosomal research. Journal of extracellular vesicles, 2012. 1(1): p. 18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu R, et al. , Extracellular vesicles in cancer—implications for future improvements in cancer care. Nature reviews Clinical oncology, 2018. 15(10): p. 617–638. [DOI] [PubMed] [Google Scholar]
- 41.Kim S, Jeon OH, and Jeon Y-J, Extracellular RNA: Emerging roles in cancer cell communication and biomarkers. Cancer Letters, 2020. 495: p. 33–40. [DOI] [PubMed] [Google Scholar]
- 42.Xavier CP, et al. , The role of extracellular vesicles in the hallmarks of cancer and drug resistance. Cells, 2020. 9(5): p. 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Nedawi K, et al. , Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature cell biology, 2008. 10(5): p. 619–624. [DOI] [PubMed] [Google Scholar]
- 44.Skog J, et al. , Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature cell biology, 2008. 10(12): p. 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghamloush F, et al. , The PAX3-FOXO1 oncogene alters exosome miRNA content and leads to paracrine effects mediated by exosomal miR-486. Scientific reports, 2019. 9(1): p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucero R, et al. , Glioma-derived miRNA-containing extracellular vesicles induce angiogenesis by reprogramming brain endothelial cells. Cell reports, 2020. 30(7): p. 2065–2074. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q, Peng F, and Chen J, The role of exosomal microRNAs in the tumor microenvironment of breast cancer. International journal of molecular sciences, 2019. 20(16): p. 3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen M, et al. , Chemotherapy-induced extracellular vesicle miRNAs promote breast cancer stemness by targeting ONECUT2. Cancer research, 2019. 79(14): p. 3608–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esteller M, Non-coding RNAs in human disease. Nature reviews genetics, 2011. 12(12): p. 861–874. [DOI] [PubMed] [Google Scholar]
- 50.Cheng J, et al. , piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clinica chimica acta, 2011. 412(17–18): p. 1621–1625. [DOI] [PubMed] [Google Scholar]
- 51.Redzic JS, et al. Extracellular RNA mediates and marks cancer progression. in Seminars in cancer biology. 2014. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang M-C, et al. , Emerging roles of lncRNA in cancer and therapeutic opportunities. American journal of cancer research, 2019. 9(7): p. 1354. [PMC free article] [PubMed] [Google Scholar]
- 53.Liu K, et al. , Long non-coding RNAs regulate drug resistance in cancer. Molecular cancer, 2020. 19(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gezer U, et al. , Long non‐coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell biology international, 2014. 38(9): p. 1076–1079. [DOI] [PubMed] [Google Scholar]
- 55.Cheng J, et al. , Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Molecular Cancer, 2020. 19(1): p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie C, Guo Y, and Lou S, LncRNA ANCR promotes invasion and migration of gastric cancer by regulating FoxO1 expression to inhibit macrophage M1 polarization. Digestive diseases and sciences, 2020. 65(10): p. 2863–2872. [DOI] [PubMed] [Google Scholar]
- 57.Liang Y, et al. , LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Molecular cancer, 2020. 19(1): p. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Liu J, et al. , Long non‐coding RNA CCAT1/miR‐148a/PKCζ prevents cell migration of prostate cancer by altering macrophage polarization. The Prostate, 2019. 79(1): p. 105–112. [DOI] [PubMed] [Google Scholar]
- 59.Ye Y, et al. , Long non‐coding RNA cox‐2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. Journal of Cellular Biochemistry, 2018. 119(3): p. 2951–2963. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. , Coagulation factor X regulated by CASC2c recruited macrophages and induced M2 polarization in glioblastoma multiforme. Frontiers in immunology, 2018. 9: p. 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, et al. , Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. Journal of Experimental & Clinical Cancer Research, 2019. 38(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z, et al. , Glioblastoma cell-derived lncRNA-containing exosomes induce microglia to produce complement C5, promoting chemotherapy resistance. Cancer Immunology Research, 2021. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, et al. , Long non‐coding RNA GAS5 overexpression inhibits M2‐like polarization of tumour‐associated macrophages in SMCC‐7721 cells by promoting PTEN expression. International Journal of Experimental Pathology, 2020. 101(6): p. 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S-Q, et al. , LncRNA GNAS-AS1 facilitates ER+ breast cancer cells progression by promoting M2 macrophage polarization via regulating miR-433–3p/GATA3 axis. Bioscience Reports, 2020. 40(7): p. BSR20200626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, et al. , GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Functional & integrative genomics, 2020. 20(1): p. 17–28. [DOI] [PubMed] [Google Scholar]
- 66.Li N, et al. , Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur Rev Med Pharmacol Sci, 2019. 23(9): p. 3688–3698. [DOI] [PubMed] [Google Scholar]
- 67.Tao S, et al. , Linc00514 promotes breast cancer metastasis and M2 polarization of tumor-associated macrophages via Jagged1-mediated notch signaling pathway. Journal of Experimental & Clinical Cancer Research, 2020. 39(1): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou Z-H, et al. , Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. American Journal of Physiology-Cell Physiology, 2020. 318(3): p. C649–C663. [DOI] [PubMed] [Google Scholar]
- 69.Gao Y, et al. , LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Molecular immunology, 2020. 117: p. 20–28. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, et al. , Regulation of NEAT1/miR-214–3p on the growth, migration and invasion of endometrial carcinoma cells. Archives of gynecology and obstetrics, 2017. 295(6): p. 1469–1475. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Y. x., et al. , Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. The international journal of biochemistry & cell biology, 2018. 104: p. 25–33. [DOI] [PubMed] [Google Scholar]
- 72.Castellano JJ, et al. , LincRNA-p21 impacts prognosis in resected non–small cell lung Cancer patients through angiogenesis regulation. Journal of Thoracic Oncology, 2016. 11(12): p. 2173–2182. [DOI] [PubMed] [Google Scholar]
- 73.Zhou L, et al. , LincRNA-p21 knockdown reversed tumor-associated macrophages function by promoting MDM2 to antagonize* p53 activation and alleviate breast cancer development. Cancer Immunology, Immunotherapy, 2020: p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang Z. x., et al. , LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell death & disease, 2019. 10(11): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang D, et al. , LncRNA RP11–361F15. 2 promotes osteosarcoma tumorigenesis by inhibiting M2-Like polarization of tumor-associated macrophages of CPEB4. Cancer letters, 2020. 473: p. 33–49. [DOI] [PubMed] [Google Scholar]
- 76.Kogure T, et al. , Extracellular vesicle–mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes & cancer, 2013. 4(7–8): p. 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Y.and Xu J, TCF-4 regulated lncRNA-XIST promotes M2 polarization of macrophages and is associated with lung cancer. OncoTargets and therapy, 2019. 12: p. 8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yekula A, et al. , Extracellular Vesicles in Glioblastoma Tumor Microenvironment. Frontiers in Immunology, 2019. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Osti D, et al. , Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clinical Cancer Research, 2019. 25(1): p. 266–276. [DOI] [PubMed] [Google Scholar]
- 80.Chen WW, et al. , BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Molecular Therapy-Nucleic Acids, 2013. 2: p. e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu T, et al. , Delivery of MGMT mRNA to glioma cells by reactive astrocyte-derived exosomes confers a temozolomide resistance phenotype. Cancer letters, 2018. 433: p. 210–220. [DOI] [PubMed] [Google Scholar]
- 82.Shao H, et al. , Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nature communications, 2015. 6(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giusti I, et al. , From glioblastoma to endothelial cells through extracellular vesicles: messages for angiogenesis. Tumor Biology, 2016. 37(9): p. 12743–12753. [DOI] [PubMed] [Google Scholar]
- 84.Floyd D.and Purow B, Micro-masters of glioblastoma biology and therapy: increasingly recognized roles for microRNAs. Neuro-oncology, 2014. 16(5): p. 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masoudi MS, Mehrabian E, and Mirzaei H, MiR‐21: A key player in glioblastoma pathogenesis. Journal of cellular biochemistry, 2018. 119(2): p. 1285–1290. [DOI] [PubMed] [Google Scholar]
- 86.Abels ER, et al. , Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell reports, 2019. 28(12): p. 3105–3119. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shahzad U, et al. , Noncoding RNAs in Glioblastoma: Emerging Biological Concepts and Potential Therapeutic Implications. Cancers, 2021. 13(7): p. 1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quezada C, et al. , Role of extracellular vesicles in glioma progression. Molecular aspects of medicine, 2018. 60: p. 38–51. [DOI] [PubMed] [Google Scholar]
- 89.de Mooij T, et al. , Short non-coding RNA sequencing of glioblastoma extracellular vesicles. Journal of neuro-oncology, 2020. 146(2): p. 253–263. [DOI] [PubMed] [Google Scholar]
- 90.Chaudhary R, Potential of long non-coding RNAs as a therapeutic target and molecular markers in glioblastoma pathogenesis. Heliyon, 2021. 7(3): p. e06502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lang H, et al. , Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci, 2017. 21(5): p. 959–972. [PubMed] [Google Scholar]
- 92.Lang H-L, et al. , Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncology reports, 2017. 38(2): p. 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Z, et al. , Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. Journal of Experimental & Clinical Cancer Research, 2019. 38(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bian EB, et al. , Exosomal lncRNA‑ATB activates astrocytes that promote glioma cell invasion. International journal of oncology, 2019. 54(2): p. 713–721. [DOI] [PubMed] [Google Scholar]
- 95.Li Z, et al. , Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. Journal of Experimental & Clinical Cancer Research, 2019. 38(1): p. 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DeSouza PA, et al. , Long, noncoding RNA dysregulation in glioblastoma. Cancers, 2021. 13(7): p. 1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rezaei O, et al. , Emerging Role of Long Non-Coding RNAs in the Pathobiology of Glioblastoma. Frontiers in oncology, 2021. 10: p. 3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yadav B, et al. , Long non-coding RNAs Associated with Glioblastoma: From Transcriptional Noise to Novel Regulators with a Promising Role in Therapeutics. Molecular Therapy-Nucleic Acids, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.King HW, Michael MZ, and Gleadle JM, Hypoxic enhancement of exosome release by breast cancer cells. BMC cancer, 2012. 12(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Colwell N, et al. , Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells. Neuro-oncology, 2017. 19(7): p. 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shao C, et al. , Role of hypoxia-induced exosomes in tumor biology. Molecular Cancer, 2018. 17(1): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Svensson KJ, et al. , Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2–mediated heparin-binding EGF signaling in endothelial cells. Proceedings of the National Academy of Sciences, 2011. 108(32): p. 13147–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kucharzewska P, et al. , Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proceedings of the National Academy of Sciences, 2013. 110(18): p. 7312–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dai X, et al. , AHIF promotes glioblastoma progression and radioresistance via exosomes. International journal of oncology, 2019. 54(1): p. 261–270. [DOI] [PubMed] [Google Scholar]
- 105.Schweiger MW and Tannous BA, Small but Fierce: Tracking the Role of Extracellular Vesicles in Glioblastoma Progression and Therapeutic Resistance. Advanced biosystems, 2020. 4(12): p. 2000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chai Y, et al. , Exosomal lncRNA ROR1-AS1 Derived from Tumor Cells Promotes Glioma Progression via Regulating miR-4686. International Journal of Nanomedicine, 2020. 15: p. 8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gentles AJ, et al. , The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature medicine, 2015. 21(8): p. 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quail DF, et al. , The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science, 2016. 352(6288). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klemm F, et al. , Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell, 2020. 181(7): p. 1643–1660. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Friebel E, et al. , Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell, 2020. 181(7): p. 1626–1642. e20. [DOI] [PubMed] [Google Scholar]
- 111.Locarno CV, et al. , Role of myeloid cells in the immunosuppressive microenvironment in gliomas. Immunobiology, 2020. 225(1): p. 151853. [DOI] [PubMed] [Google Scholar]
- 112.De Leo A, Ugolini A, and Veglia F, Myeloid Cells in Glioblastoma Microenvironment. Cells, 2021. 10(1): p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Azambuja JH, et al. , Molecular profiles and immunomodulatory activities of glioblastoma-derived exosomes. Neuro-oncology advances, 2020. 2(1): p. vdaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han Y, Liu D, and Li L, PD-1/PD-L1 pathway: current researches in cancer. American journal of cancer research, 2020. 10(3): p. 727. [PMC free article] [PubMed] [Google Scholar]
- 115.Hao C, et al. , PD-L1 expression in glioblastoma, the clinical and prognostic significance: a systematic literature review and meta-analysis. Frontiers in Oncology, 2020. 10: p. 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen G, et al. , Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature, 2018. 560(7718): p. 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lawler SE, et al. , Immune Escape Mediated by Exosomal PD‐L1 in Cancer. Advanced biosystems, 2020. 4(12): p. 2000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ricklefs FL, et al. , Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Science advances, 2018. 4(3): p. eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gabrusiewicz K, et al. , Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. Oncoimmunology, 2018. 7(4): p. e1412909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Vrij J, et al. , Glioblastoma‐derived extracellular vesicles modify the phenotype of monocytic cells. International journal of cancer, 2015. 137(7): p. 1630–1642. [DOI] [PubMed] [Google Scholar]
- 121.Simon T, Jackson E, and Giamas G, Breaking through the glioblastoma micro-environment via extracellular vesicles. Oncogene, 2020: p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maas SL, et al. , Glioblastoma hijacks microglial gene expression to support tumor growth. Journal of neuroinflammation, 2020. 17: p. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tankov S.and Walker PR, Glioma-Derived Extracellular Vesicles–Far More Than Local Mediators. Frontiers in Immunology, 2021. 12: p. 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qian M, et al. , Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene, 2020. 39(2): p. 428–442. [DOI] [PubMed] [Google Scholar]
- 125.Wang M, et al. , Exosomal LGALS9 in the cerebrospinal fluid of glioblastoma patients suppressed dendritic cell antigen presentation and cytotoxic T-cell immunity. Cell death & disease, 2020. 11(10): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Himes BT, et al. , The role of extracellular vesicles and PD-L1 in glioblastoma-mediated immunosuppressive monocyte induction. Neuro-oncology, 2020. 22(7): p. 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Domenis R, et al. , Systemic T cells immunosuppression of glioma stem cell-derived exosomes is mediated by monocytic myeloid-derived suppressor cells. PLoS One, 2017. 12(1): p. e0169932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Condamine T, et al. , Regulation of tumor metastasis by myeloid-derived suppressor cells. Annual review of medicine, 2015. 66: p. 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guo X, et al. , Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene, 2018. 37(31): p. 4239–4259. [DOI] [PubMed] [Google Scholar]
- 130.Guo X, et al. , Glioma exosomes mediate the expansion and function of myeloid‐derived suppressor cells through microRNA‐29a/Hbp1 and microRNA‐92a/Prkar1a pathways. International journal of cancer, 2019. 144(12): p. 3111–3126. [DOI] [PubMed] [Google Scholar]
- 131.Spellicy SE and Stice SL, Tissue and stem cell sourced extracellular vesicle communications with microglia. Stem Cell Reviews and Reports, 2021. 17(2): p. 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang J. k., et al. , Exosomal miR-214–5p Released from Glioblastoma Cells Modulates Inflammatory Response of Microglia after Lipopoly-saccharide Stimulation through Targeting CXCR5. CNS & Neurological Disorders-Drug Targets (Formerly Current Drug Targets-CNS & Neurological Disorders), 2019. 18(1): p. 78–87. [DOI] [PubMed] [Google Scholar]
- 133.Chen W, et al. , MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget, 2017. 8(14): p. 22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou X, et al. , HOTAIR is a therapeutic target in glioblastoma. Oncotarget, 2015. 6(10): p. 8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Peng Z, Liu C, and Wu M, New insights into long noncoding RNAs and their roles in glioma. Molecular cancer, 2018. 17(1): p. 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang J, et al. , Extracellular vesicle lncRNA metastasis-associated lung adenocarcinoma transcript 1 released from glioma stem cells modulates the inflammatory response of microglia after lipopolysaccharide stimulation through regulating miR-129–5p/high mobility group box-1 protein axis. Frontiers in immunology, 2020. 10: p. 3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grimaldi A, et al. , Microglia-derived microvesicles affect microglia phenotype in glioma. Frontiers in cellular neuroscience, 2019. 13: p. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Murgoci AN, et al. , Brain‐cortex microglia‐derived exosomes: Nanoparticles for glioma therapy. ChemPhysChem, 2018. 19(10): p. 1205–1214. [DOI] [PubMed] [Google Scholar]
- 139.van der Vos KE, et al. , Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro-oncology, 2015. 18(1): p. 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bebelman MP, et al. , The forces driving cancer extracellular vesicle secretion. Neoplasia, 2021. 23(1): p. 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cappello F, et al. , Exosome levels in human body fluids: a tumor marker by themselves? European Journal of Pharmaceutical Sciences, 2017. 96: p. 93–98. [DOI] [PubMed] [Google Scholar]
- 142.Shi J, et al. , Role of Exosomes in the Progression, Diagnosis, and Treatment of Gliomas. Medical science monitor: international medical journal of experimental and clinical research, 2020. 26: p. e924023–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Murphy DE, et al. , Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Experimental & molecular medicine, 2019. 51(3): p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Andaloussi SE, et al. , Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews Drug discovery, 2013. 12(5): p. 347–357. [DOI] [PubMed] [Google Scholar]
- 145.Sun H, et al. , Extracellular vesicles in the development of cancer therapeutics. International Journal of Molecular Sciences, 2020. 21(17): p. 6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hendrix A.and De Wever O, Rab27 GTPases distribute extracellular nanomaps for invasive growth and metastasis: implications for prognosis and treatment. International journal of molecular sciences, 2013. 14(5): p. 9883–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li Z, et al. , Functional implications of Rab27 GTPases in cancer. Cell Communication and Signaling, 2018. 16(1): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Andreu Z.and Yáñez-Mó M, Tetraspanins in extracellular vesicle formation and function. Frontiers in immunology, 2014. 5: p. 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Menck K, et al. , Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. Journal of extracellular vesicles, 2017. 6(1): p. 1378056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Albakova Z, et al. , Extracellular heat shock proteins and cancer: New perspectives. Translational Oncology, 2021. 14(2): p. 100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ostrowski M, et al. , Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature cell biology, 2010. 12(1): p. 19–30. [DOI] [PubMed] [Google Scholar]
- 152.Jin H, et al. , Rab GTPases: Central Coordinators of Membrane Trafficking in Cancer. Frontiers in Cell and Developmental Biology, 2021. 9: p. 1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bobrie A, et al. , Rab27a supports exosome-dependent and-independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer research, 2012. 72(19): p. 4920–4930. [DOI] [PubMed] [Google Scholar]
- 154.Catalano M.and O’Driscoll L, Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. Journal of extracellular vesicles, 2020. 9(1): p. 1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Datta A, et al. , Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells. Cancer letters, 2017. 408: p. 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Panigrahi GK, et al. , Hypoxia-induced exosome secretion promotes survival of African-American and Caucasian prostate cancer cells. Scientific reports, 2018. 8(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Savina A, et al. , Exosome release is regulated by a calcium-dependent mechanism in K562 cells. Journal of Biological Chemistry, 2003. 278(22): p. 20083–20090. [DOI] [PubMed] [Google Scholar]
- 158.Chalmin F, et al. , Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. The Journal of clinical investigation, 2010. 120(2): p. 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kholia S, et al. , A novel role for peptidylarginine deiminases in microvesicle release reveals therapeutic potential of PAD inhibition in sensitizing prostate cancer cells to chemotherapy. Journal of extracellular vesicles, 2015. 4(1): p. 26192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lange S, et al. , Peptidylarginine deiminases—roles in cancer and neurodegeneration and possible avenues for therapeutic intervention via modulation of exosome and microvesicle (EMV) release? International journal of molecular sciences, 2017. 18(6): p. 1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lange S, Peptidylarginine deiminases and extracellular vesicles: Prospective drug targets and biomarkers in central nervous system diseases and repair. Neural Regeneration Research, 2021. 16(5): p. 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kosgodage US, et al. , Chloramidine/bisindolylmaleimide-I-mediated inhibition of exosome and microvesicle release and enhanced efficacy of cancer chemotherapy. International journal of molecular sciences, 2017. 18(5): p. 1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kosgodage US, et al. , Peptidylarginine deiminases post-translationally deiminate prohibitin and modulate extracellular vesicle release and MicroRNAs in glioblastoma multiforme. International journal of molecular sciences, 2019. 20(1): p. 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Uysal-Onganer P, et al. , Peptidylarginine Deiminase Inhibitor Application, Using Cl-Amidine, PAD2, PAD3 and PAD4 Isozyme-Specific Inhibitors in Pancreatic Cancer Cells, Reveals Roles for PAD2 and PAD3 in Cancer Invasion and Modulation of Extracellular Vesicle Signatures. International Journal of Molecular Sciences, 2021. 22(3): p. 1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Uysal-Onganer P, et al. , Peptidylarginine deiminase isozyme-specific PAD2, PAD3 and PAD4 inhibitors differentially modulate extracellular vesicle signatures and cell invasion in two glioblastoma multiforme cell lines. International journal of molecular sciences, 2020. 21(4): p. 1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mulcahy LA, Pink RC, and Carter DRF, Routes and mechanisms of extracellular vesicle uptake. Journal of extracellular vesicles, 2014. 3(1): p. 24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.French KC, Antonyak MA, and Cerione RA. Extracellular vesicle docking at the cellular port: Extracellular vesicle binding and uptake. in Seminars in cell & developmental biology. 2017. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hayatudin R, et al. , Overcoming Chemoresistance via Extracellular Vesicle Inhibition. Frontiers in Molecular Biosciences, 2021. 8: p. 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Christianson HC, et al. , Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proceedings of the National Academy of Sciences, 2013. 110(43): p. 17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Svensson KJ, et al. , Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. Journal of Biological Chemistry, 2013. 288(24): p. 17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fitzner D, et al. , Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of cell science, 2011. 124(3): p. 447–458. [DOI] [PubMed] [Google Scholar]
- 172.Yuyama K, et al. , Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. Journal of Biological Chemistry, 2012. 287(14): p. 10977–10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Altei WF, et al. , Inhibition of αvβ3 integrin impairs adhesion and uptake of tumor-derived small extracellular vesicles. Cell Communication and Signaling, 2020. 18(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kogure A, Yoshioka Y, and Ochiya T, Extracellular vesicles in cancer metastasis: Potential as therapeutic targets and materials. International Journal of Molecular Sciences, 2020. 21(12): p. 4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nishida-Aoki N, et al. , Disruption of circulating extracellular vesicles as a novel therapeutic strategy against cancer metastasis. Molecular Therapy, 2017. 25(1): p. 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Federici C, et al. , Natural-killer-derived extracellular vesicles: Immune sensors and interactors. Frontiers in immunology, 2020. 11: p. 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Munoz JL, et al. , Delivery of functional anti-miR-9 by mesenchymal stem cell–derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Molecular Therapy-Nucleic Acids, 2013. 2: p. e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhu L, et al. , Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics, 2017. 7(10): p. 2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu H, et al. , Co-delivery of tumor-derived exosomes with alpha-galactosylceramide on dendritic cell-based immunotherapy for glioblastoma. Cancer letters, 2017. 411: p. 182–190. [DOI] [PubMed] [Google Scholar]
- 180.Monfared H, et al. , Potential therapeutic effects of exosomes packed with a miR-21-sponge construct in a rat model of glioblastoma. Frontiers in oncology, 2019. 9: p. 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang T, et al. , Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharmaceutical research, 2015. 32(6): p. 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Salarpour S, et al. , Paclitaxel incorporated exosomes derived from glioblastoma cells: comparative study of two loading techniques. DARU Journal of Pharmaceutical Sciences, 2019. 27(2): p. 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Del Fattore A, et al. , Differential effects of extracellular vesicles secreted by mesenchymal stem cells from different sources on glioblastoma cells. Expert opinion on biological therapy, 2015. 15(4): p. 495–504. [DOI] [PubMed] [Google Scholar]
- 184.Salarpour S, et al. , Exosome-loaded Paclitaxel: Preparation and toxicity evaluation on two glioblastoma cell lines. Nanomedicine Research Journal, 2019. 4(4): p. 239–246. [Google Scholar]
- 185.Yu L, et al. , Exosomes derived from microRNA-199a-overexpressing mesenchymal stem cells inhibit glioma progression by down-regulating AGAP2. Aging (Albany NY), 2019. 11(15): p. 5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ding X, et al. , Human Bone Marrow-Derived Mesenchymal Stem Cell-Secreted Exosomes Overexpressing Microrna-124–3p Inhibit DLBCL Progression By Downregulating NFATc1. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Deng S-Z, et al. , Human marrow stromal cells secrete microRNA-375-containing exosomes to regulate glioma progression. Cancer gene therapy, 2020. 27(3): p. 203–215. [DOI] [PubMed] [Google Scholar]
- 188.Bu N, et al. , Exosomes from dendritic cells loaded with chaperone-rich cell lysates elicit a potent T cell immune response against intracranial glioma in mice. Journal of Molecular Neuroscience, 2015. 56(3): p. 631–643. [DOI] [PubMed] [Google Scholar]
- 189.Smith SM, et al. , Functional equivalence of stem cell and stem cell‐derived extracellular vesicle transplantation to repair the irradiated brain. Stem cells translational medicine, 2020. 9(1): p. 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Baulch JE, et al. , Cranial grafting of stem cell-derived microvesicles improves cognition and reduces neuropathology in the irradiated brain. Proceedings of the National Academy of Sciences, 2016. 113(17): p. 4836–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang Z, et al. , Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nature biomedical engineering, 2020. 4(1): p. 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Thakur A, et al. , Inhibition of Glioma Cells’ Proliferation by Doxorubicin-Loaded Exosomes via Microfluidics. International journal of nanomedicine, 2020. 15: p. 8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sheybani ND, et al. , Focused ultrasound hyperthermia augments release of glioma-derived extracellular vesicles with differential immunomodulatory capacity. Theranostics, 2020. 10(16): p. 7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.García-Manrique P, et al. , Therapeutic biomaterials based on extracellular vesicles: classification of bio-engineering and mimetic preparation routes. Journal of extracellular vesicles, 2018. 7(1): p. 1422676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sil S, et al. , Strategies for the use of Extracellular Vesicles for the Delivery of Therapeutics. Journal of Neuroimmune Pharmacology, 2020. 15(3): p. 422–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jia X, et al. , Recent Progress of Extracellular Vesicle Engineering. ACS Biomaterials Science & Engineering, 2021. 7(9): p. 4430–4438. [DOI] [PubMed] [Google Scholar]
- 197.Li Y-J, et al. , Artificial exosomes for translational nanomedicine. Journal of Nanobiotechnology, 2021. 19(1): p. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.De Jong OG, et al. , Extracellular vesicles: potential roles in regenerative medicine. Frontiers in immunology, 2014. 5: p. 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ramasubramanian L, Kumar P, and Wang A, Engineering extracellular vesicles as nanotherapeutics for regenerative medicine. Biomolecules, 2020. 10(1): p. 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Man K, et al. , Engineered extracellular vesicles: tailored-made nanomaterials for medical applications. Nanomaterials, 2020. 10(9): p. 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable as the current review manuscript is solely based on literature survey and discusses the future road map for the research problem discussed.


