Abstract
During influenza virus entry, the hemagglutinin (HA) protein binds receptors and causes membrane fusion after endosomal acid activation. To improve vaccine efficiency and pandemic risk assessment for currently-dominant H3N2 influenza viruses, we investigated HA stability of 6 vaccine reference viruses and 42 circulating viruses. Recent vaccine reference viruses had destabilized HA proteins due to egg-adaptive mutation HA1-L194P. Virus growth in cell culture was independent of HA stability. In ferrets, the vaccine reference viruses and circulating viruses required a relatively stable HA (activation and inactivation pH < 5.5) for airborne transmissibility. The recent vaccine reference viruses with destabilized HA proteins had reduced infectivity, had no airborne transmissibility unless reversion to HA1-P194L occurred, and had skewed antigenicity away from the studied viruses and circulating H3N2 viruses. Other vaccine reference viruses with stabilized HAs retained infectivity, transmissibility, and antigenicity. Therefore, HA stabilization should be prioritized over destabilization in vaccine reference virus selection to reduce mismatches between vaccine and circulating viruses.
Stabilized hemagglutinin proteins should be prioritized over destabilized ones in influenza vaccine reference virus selection.
INTRODUCTION
Influenza A viruses (IAVs) cause pandemics and annual epidemics (1). Diverse IAVs circulate in wild aquatic birds and occasionally infect domestic poultry and swine (2, 3). Since the 1968 Hong Kong pandemic, H3N2 viruses cause seasonal outbreaks in humans. Influenza circulation was suppressed during the onset of the coronavirus disease 2019 (COVID-19) pandemic (4). However, the detection of influenza viruses has been increasing since November 2021 and appears to have increased to pre–COVID-19 pandemic levels in several countries and regions (5, 6). The H3N2 subtype is the current dominant strain in humans (5, 6), and it consists of seven clades (clades 1 to 7) and multiple subclades (7). Clade 2a and clade 3a are the predominant clades circulating in humans (7). In North American swine, H3N2 IAVs emerged in the late 1990s and quickly evolved into clusters I, II, III, and IV (8). Swine H3N2 IAVs have also been detected in humans (9–11). Thus, currently-circulating and -emerging H3N2 pose a threat to public health.
It is challenging to identify animal-origin IAVs that pose the greatest risk of causing a human pandemic. Efficient airborne transmissibility is an essential characteristic of pandemic viruses (12, 13). The IAV hemagglutinin (HA) surface glycoprotein plays key roles in the virus replication cycle and transmission. The HAs of human IAVs bind with higher affinity to host cell α2,6-linked receptors, while those of avian IAVs prefer α2,3-linked (1). After virion endocytosis, the HA protein undergoes acid-dependent, irreversible conformational changes that promote the fusion between viral and cellular membranes (2, 3, 14). The threshold of acidic pH required to activate the HA protein has been defined as HA activation pH (2, 15, 16). Multiple assays can be used to measure the HA activation pH value. These include various cell-to-cell fusion assays (e.g., syncytia formation, dye transfer, or reporter gene expression), trypsin susceptibility, and binding to conformation-specific monoclonal antibodies using flow cytometry [reviewed in (2)]. Numerous human H1N1 pandemic isolates and H3N2 isolates have been demonstrated to have HA activation pH values ≤ 5.6 (17–19). Swine gamma H1N1 viruses had HA activation pH values ranging from 5.5 to 5.9 (17), and swine H3N2 isolates (of unknown phylogenetic information) had pH values from 5.3 to 5.8 (18, 20). Acid-triggered HA confirmational changes outside of a target cell leads to virion inactivation. The pH at which virions lose infectivity outside of cells is defined as virus inactivation pH (2, 21). Virus inactivation pH is measured by aliquoting virus samples, exposing the aliquots to buffers of a range of pH, neutralizing the samples, and measuring the remaining virus infectivity [reviewed in (2)]. Seven human H3N2 isolates before 2013 had acid inactivation pH ≤ 5.4, while 2 swine H3N2 isolates had pH values of 5.7 to 5.9 (22, 23). Overall, infectious virions need HA to be activated to undergo replication and transcription intracellularly while resisting activation to protect from losing infectivity extracellularly (24–27). The pH values of HA activation and virus inactivation may not be always equal, and limited studies have been performed to compare these two values. Five human H3N2 and four swine H1N1 gamma viruses had similar pH values between HA activation and virus inactivation (21), whereas several other swine H3N2 and H1N1 isolates had varied values between these two (18). It is necessary to investigate the relationship between HA activation and virus inactivation pH on a larger scale.
Ferrets are the primary animal model for investigating IAV transmission (28, 29). H1N1 and H5N1 viruses contain structural group 1 HA proteins (30, 31). For these subtypes, a stable HA protein (activation pH < 5.6) is necessary for airborne transmissibility in ferrets (17, 20, 21, 32–35). H3N2 viruses have structural group 2 HA proteins (30, 36). Limited information is known about the impact of HA stability on group 2 virus transmissibility in ferrets. Airborne transmission in ferrets was reduced for a cell-adapted H3N2 virus with modified HA and NA (23). It is unclear whether this isolate can be a representative for circulating seasonal H3N2 viruses. Four swine H3N2v isolates with activation pH ≤ 5.6 had varied airborne transmission (19). It is unknown whether human H3N2 viruses require a pH threshold of HA stability for airborne transmissibility. Such knowledge is necessary to inform pandemic risk assessment algorithms.
Vaccination is the most effective way to protect against infectious diseases. Vaccine effectiveness against recent H3N2 IAVs has typically been low (<33%) (37), in part, due to mismatches between vaccine and circulating strains. These are caused by antigenic changes in circulating H3N2 and, to a lesser extent, the egg-culturing systems used for vaccine production (37, 38). H3N2 evolution leads to the emergence of antigenically novel viruses approximately every 2 to 5 years (39). Understanding the determinants of H3N2 transmissibility could potentially inform models aiming at predicting forward evolution. Vaccine growth in eggs often introduces egg-adapted HA variations. Human- and egg-adapted H3N2 IAVs preferentially bind α2,6-linked and α2,3-linked receptors, respectively (40). Therefore, egg-adaptation variations to switch receptor-binding specificity have been reported, including HA1-H156Q (41), HA1-T160K (42), HA1-G186V (43), and HA1-L194P (44, 45). However, it remains unclear whether these adaptive mutations may emerge in the circulating viruses and alter virus characteristics (i.e., HA stability, replication, and antigenicity).
Here, we conducted a comprehensive investigation on H3N2 influenza vaccine reference viruses and circulating viruses to address the contributions of HA stability to vaccine reference virus selection and pandemic risk assessment. We identified mismatched HA stability between the recent vaccine reference viruses and human circulating viruses. We demonstrated that virus growth in the cell cultures used in commercial vaccine production was independent of HA stability, whereas virus initial growth in ferrets was associated with HA stability. Furthermore, we revealed that the recent vaccine reference viruses having destabilized HA proteins due to the egg-adaptive mutation HA1-L194P had skewed antigenicity away from the airborne transmitted viruses in ferrets and circulating viruses. Other vaccine reference viruses that acquired egg-culturing adaptation HA1-G186V retained a stable HA protein and had no changes in virus replication, antigenicity, and transmission. Thus, we concluded (i) future selection of H3N2 vaccine reference viruses should prioritize stabilized HA proteins over destabilized ones, and (ii) H3N2 viruses with stabilized HAs have greater pandemic potential.
RESULTS
Recent H3N2 vaccine reference viruses have destabilized HA proteins
Phylogenetic analyses on the HA genes of 22 human and 26 swine H3N2 IAVs (2012–2019) showed that the 22 human IAVs were distributed in clades 3C.2a, 3C.3a, and 3C (fig. S1). Eight human-like swine IAVs were also in clade 3C, and the remaining 18 swine isolates belonged to swine cluster IV. The isolates were widely distributed within these clades and taken as genetically diverse and representative. The H3N2 human isolates had seasonal non-HA segments, while those from swine were from the triple-reassortant internal gene constellation and/or pdm09 lineages (Table 1). All isolates had human-adaptive receptor-binding HA1 residues D190, I226, P227, and S228, except cluster IV swine, which contained HA1 human-adaptive (D190 and S228) and avian-like (V226 and S227) receptor-binding residues (Table 1). The human isolates had more potential glycosylation sites and human-adaptive polymerase substitutions when compared to the swine isolates (Table 1).
Table 1. Genetic analyses of human and swine IAVs.
| Genetic characteristics | Human | Human-like swine | Cluster IV swine |
|---|---|---|---|
| (n = 22) | (n = 8) | (n = 18) | |
| Gene constellation | |||
| PB2 | Seasonal | TRIG/P* | TRIG |
| PB1 | Seasonal | TRIG/P | TRIG |
| PA | Seasonal | TRIG/P | TRIG/P |
| HA | Seasonal | Human like | Cluster IV |
| NP | Seasonal | TRIG/P | TRIG/P |
| NA | Seasonal | 2002 N2 | 2002 N2 |
| M | Seasonal | P | P |
| NS | Seasonal | TRIG/P | TRIG |
| HA receptor binding | |||
| Human-like | |||
| HA1-D190 | D† | D | D |
| HA1-I226 | I | I | – |
| HA1-P227 | P | P | – |
| HA1-S228 | S | S | S |
| Avian-like | |||
| HA1-V226 | – | – | V |
| HA1-S227 | – | – | S |
| No. of glycosylation sites (mean ± SD) | 10.2(±0.8) | 6.9(±0.6) | 8.0 |
| PB2 human-adaptive substitutions | |||
| PB2-N9 | N | – | – |
| PB2-T/L147 | T | T | T |
| PB2-S199 | S | – | – |
| PB2-G249 | G | G | – |
| PB2-A271 | A | A | A |
| PB2-D309 | D | D | D |
| PB2-R526 | R | – | – |
| PB2-I/V588 | I (5/22)‡ | – | – |
| PB2-S590/R591 | – | SR§ | SR |
| PB2-K627 | K | – | – |
| PB2-A661 | A (1/22) | – | – |
| PB2-T683 | T | T | T |
| PB2-S684 | S | – | – |
| PB2-R702 | R | – | – |
| PA human-adaptive substitutions | |||
| PA-S/Y321 | Y | – | – |
| PA-M336 | – | – | M (1/18) |
| PA-R356 | R | – | R (1/18) |
| PA-S409 | S | S (1/8) | – |
| PA-I669 | I (2/22) | – | – |
*TRIG, Triple-reassortant internal gene constellation lineage; P, pdm09.
†Predominant amino acids for the indicated clades.
‡X/Y refers to number of isolates having the indicated residue divided by the total number of isolates in that clade.
§SR, S590/R591.
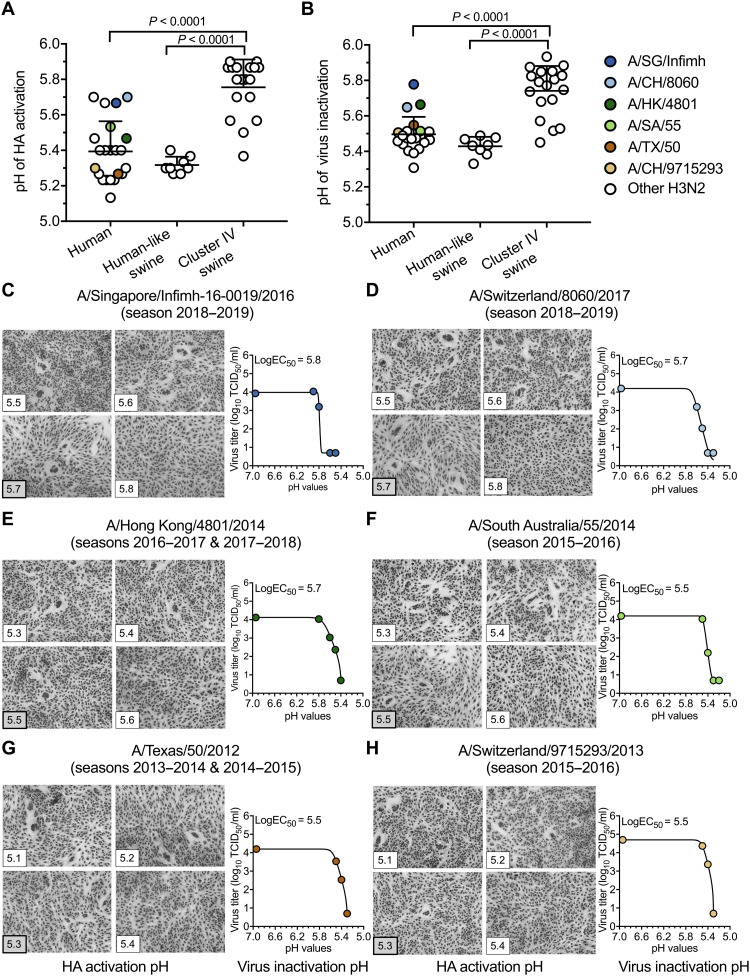
The human isolates had HA activation and virus inactivation pH values ranging from 5.1 to 5.7 and from 5.3 to 5.8, respectively (Fig. 1, A and B). Six wild-type human isolates were World Health Organization (WHO)–recommended H3N2 vaccine reference viruses from 2013 to 2019 (table S1). Three egg-based vaccine reference viruses [i.e., A/Singapore/Infimh-16-0019/2016 (A/SG/Infimh), A/Switzerland/8060/2017 (A/CH/8060), and A/Hong Kong/4801/2014 (A/HK/4801)] had egg-adaptive mutations HA1-T160K and HA1-L194P (table S1). These three viruses had the least stable HA proteins (pH 5.5 to 5.8) of the human viruses (Fig. 1, C to E). Three other egg-based vaccine reference viruses [i.e., A/South Australia/55/2014 (A/SA/55), A/Texas/50/2012 (A/TX/50), and A/Switzerland/9715293/2013 (A/CH/9715293)] had alternate egg-adaptive mutation HA1-G186V (table S1), and these viruses had more stable HA proteins (pH 5.3 to 5.5) (Fig. 1, F to H). Of note, the other egg-based wild-type vaccine reference viruses or reassortant vaccine seed viruses of A/SG/Infimh, A/CH/8060, and A/HK/4801 also had HA1-T160K and HA1-L194P egg-adaptive mutations, and those of A/TX/50 and A/CH/9715293 had HA1-G186V (table S1) (45, 46). Cell-based A/TX/50 and A/HK/5738 did not have egg-adaptive mutations and had relatively stable HA proteins (pH 5.2 to 5.5) (Table 2). Overall, egg-adaptive mutation HA1-G186V had minimal effect on HA stability, whereas HA1-L194P and/or HA1-T160K may be associated with HA destabilization.
Fig. 1. HA activation pH and virus inactivation pH values for human and swine H3N2 IAVs.
(A) HA activation pH values. Vero cells were infected with IAVs (human, n = 22; human-like swine, n = 8; and cluster IV swine, n = 18) at a multiplicity of infection (MOI) of 3 plaque-forming units (PFU) per cell. Virus HA activation pH values were measured by syncytia assay. Each symbol represents the mean highest pH of syncytia formation for an individual isolate. (B) Virus inactivation pH values. Virus aliquots were treated with phosphate-buffered saline (PBS) buffers having the indicated pH. Residual infectivity was measured by TCID50 and analyzed using nonlinear regression. (C to H) Representative syncytia and virus inactivation assays for the six vaccine reference viruses. Six vaccine reference viruses—A/SG/Infimh (season 2018–2019) (C), A/CH/8060 (season 2018–2019) (D), A/HK/4801 (seasons 2016–2017 and 2017–2018) (E), A/SA/55 (season 2015–2016) (F), A/TX/50 (seasons 2013–2014 and 2014–2015) (G), and A/CH/9715293 (season 2015–2016) (H)—were indicated by colored symbols shown in the legend. LogEC50 was reported as virus HA inactivation pH. All experiments were independently performed at least twice (means ± SD). P values were determined according to Mann-Whitney U test.
Table 2. Summary of characterization of selected human H3N2 IAVs before and after infection and transmission in ferrets.
| Virus | A/SG/Infimh | A/CH/8060 | A/HK/4801* | A/SA/55 | A/TX/50/2012* | A/CH/971523 | A/HK/5738 | A/MEM/2 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Season of vaccine strain | 2018–2019 | 2018–2019 | 2016–2018 | 2015–2016 | 2013–2015 | 2015–2016 | A/HK/4801-like virus† | N/A‡ | |||
| HA stability | HA activation pH | 5.7 | 5.7 | 5.5 | 5.5 | 5.3 | 5.3 | 5.2 | 5.3 | ||
| Virus inactivation pH | 5.8 | 5.7 | 5.7 | 5.5 | 5.5 | 5.5 | 5.4 | 5.3 | |||
| Passage History | Egg, MDCK, or MDCK-SIAT§ | E5/E2/E1/C1 | E7/E1/C1 | E5/E2/E2/E1/C1 | E5/E2/E2/E1 | E5/E2/E1/C1 | C1/C2/C2 | E6 | E4/E3/C1 | C1/C2/SIAT2/C2 | C3 |
| Egg-adaptive mutation║ | HA1 mutation | T160K L194P D225G |
T160K L194P T203I |
N96S T160K L194P |
S145N G186V D225G |
None | G186V S219F |
G186V | None | None | |
| Peak titers in vitro (log10 TCID50)¶ | MDCK cells | 7.59(±0.5) | 7.55(±0.4) | 8.11(±0.2) | n.d.# | 8.30(±0.1) | 7.07(±0.8) | n.d. | 8.28(±0.2) | 7.82(±0.2) | 7.82(±0.4) |
| Vero cells | 7.32(±0.2) | 7.55(±0.6) | 7.57(±0.5) | n.d. | 7.82(±0.5) | 6.57(±0.5) | n.d. | 7.07(±0.2) | 7.19 | 6.70(±0.4) | |
| Donor ferrets | Viruses isolated | 3/3 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 3/3 |
| Seroconversion | 3/3 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 3/3 | |
| Virus titers on initial day (log10 TCID50) | 2.20(±0.9) | n.d. | 3.88(±0.5) | 3.13(±0.6) | n.d. | 5.67(±0.6) | 4.75(±0.4) | n.d. | 4.76(±0.4) | 6.02(±0.7) | |
| Day of peak titer | 3 | n.d. | 5 | 4 | n.d. | 1 | 3(±1.15) | n.d. | 9 | 1 | |
| Peak titer (log10 TCID50) | 5.78(±0.5) | n.d. | 4.76(±0.6) | 4.86(±0.9) | n.d. | 5.67(±0.6) | 5.00(±0.3) | n.d. | 6.72(±0.3) | 6.02(±0.2) | |
| Temperature ≥ 40°C** | 0/3 | n.d. | 0/3 | 0/3 | n.d. | 0/3 | 0/4 | n.d. | 2/3 | 2/3 | |
| Major HA mutation(s) arising | P1941L P2271S |
n.d. | P1941L T2031I D2251N/G |
P1941 T2031I D2251N/G |
n.d. | None | None | n.d. | None | K1601T | |
| Contact ferrets | Viruses isolated | 3/3 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 3/3 |
| Seroconversion | 3/3 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 4/4 | n.d. | 3/3 | 3/3 | |
| Initial day of virus shedding | 6.7(±3.06) | n.d. | 4 | 4.5(±1) | n.d. | 4 | 2.5(±1) | n.d. | 4 | 4 | |
| Peak titers (log10 TCID50) |
5.51(±1.1) |
n.d. |
5.09(±0.7) |
4.13(±1.2) |
n.d. | 4.53(±0.3) | 4.87 (±0.6) | n.d. |
7.92(±0.2) |
6.14(±0.4) | |
| Major HA mutation(s) arising | P1941L N1221D P2271S |
n.d. | P1941L T2031I D2251N/G |
P1941L T2031I D2251N/G |
n.d. | None | None | n.d. | None | None | |
| Airborne ferrets | Viruses isolated | 1/3 | n.d. | 0/3 | 0/4 | n.d. | 2/3 | 2/4 | n.d. | 3/3 | 3/3 |
| Seroconversion | 1/3 | n.d. | 0/3 | 0/4 | n.d. | 2/3 | 2/4 | n.d. | 3/3 | 3/3 | |
| Peak titers (log10 TCID50) | 5.20 | n.d. | None | None | n.d. | 4.25(±1.1) | 5.13(±0.5) | n.d. | 6.64(±0.6) | 6.37(±0.2) | |
| HA activation pH | 5.2 | n.d. | n.d. | n.d. | n.d. | 5.2–5.4 | 5.2–5.4 | n.d. | 5.2–5.4 | 5.2–5.4 | |
| Virus inactivation pH | 5.4–5.5 | n.d. | n.d. | n.d. | n.d. | 5.4–5.5 | 5.4–5.5 | n.d. | 5.3–5.5 | 5.2–5.3 | |
| Major HA mutation arising | P1941L | n.d. | n.d. | n.d. | n.d. | None | None | n.d. | None | None | |
*Two trials of experiments were conducted with A/HK/4801/2014 and A/TX/50/2012. For these viruses, data from each experiment are listed separately.
†A/HK/5738 is similar to A/HK/4801, which is a WHO-recommended vaccine reference virus during 2016–2018 influenza seasons.
‡N/A, not applicable.
§Egg passage (E), MDCK cell passage (C), MDCK-SIAT1 cell passage (SIAT). Numbers listed represent the number of passages.
║HA adaptive mutations are listed that emerged during egg culturing before all experiments, and detailed information is shown in table S1.
¶Only isolates that had been passaged in MDCK cells at least once were investigated.
#n.d., not determined.
**Ferret body weight and body temperature are shown in figs. S8 and S9.
Human-like swine viruses had relatively stable HA proteins with HA activation and virus inactivation pH values ranging from 5.2 to 5.4 and from 5.3 to 5.5, respectively (Fig. 1 and table S2). In contrast, cluster IV swine viruses had HA activation and virus inactivation pH ranging from 5.3 to 5.9 and from 5.4 to 5.9, respectively, which were significantly higher than the human and human-like swine viruses (Fig. 1 and table S2).
Human isolates had lower growth in vitro
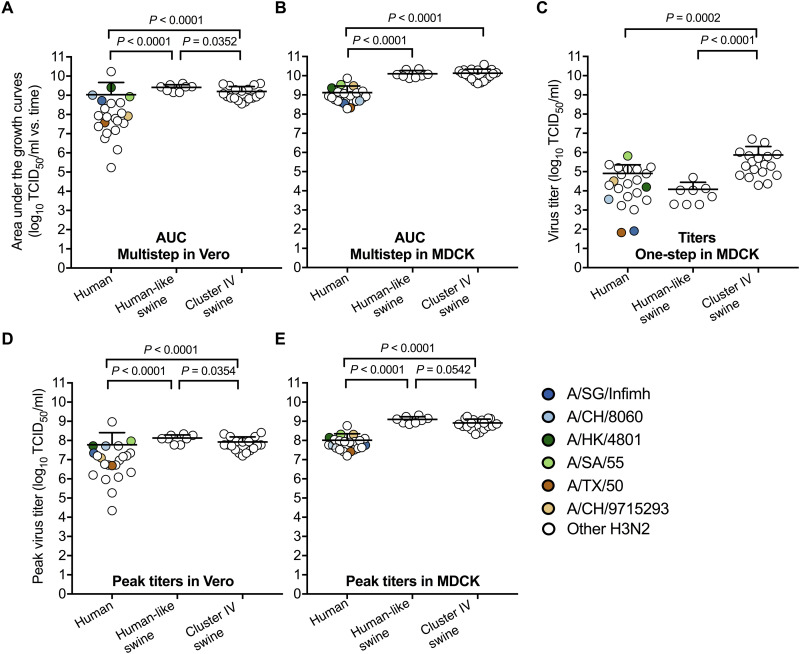
We measured virus growth in African green monkey kidney (Vero) and Madin-Darby canine kidney (MDCK) cells, which are used for influenza vaccine production (47). The human viruses had lower total amount of infectious viruses [assessed by area under the growth curves (AUC) or virus titers] than the swine viruses in both cell types (Fig. 2, A to C, and fig. S2). Average peak titers of the human viruses reached approximately 108 median tissue culture infectious dose (TCID50)/ml with a range of 104 to 109 TCID50/ml in Vero and 107 to 109 TCID50/ml in MDCK cells (Fig. 2D). The six vaccine reference viruses grew to peak titers greater than 6.6 × 106 TCID50/ml in both Vero and MDCK cells (Fig. 2D and Table 2).
Fig. 2. Virus replication in Vero and MDCK cells.
(A to B) Area under the growth curves (AUC) of virus multistep replication in Vero and MDCK cells (MOI of 0.01 PFU per cell). Cell culture supernatants in Vero cells were collected at 16, 24, 48, and 60 hours postinfection (hpi), and those in MDCK cells were collected at 16, 24, and 36 hpi. The samples were titrated by TCID50. AUC of the total amount of infectious viruses for an individual virus was calculated. (C) Virus titers of one-step growth in MDCK cells (MOI of 2 PFU per cell). Cell culture supernatants were collected at 6 hpi. (D to E) Peak virus titers in Vero and MDCK cells. Six vaccine reference viruses—A/SG/Infimh, A/CH/8060, A/HK/4801, A/SA/55, A/TX/50, and A/CH/9715293—were indicated by colored symbols shown in the legend. All experiments were independently performed at least twice (means ± SD). P values were determined according to Mann-Whitney U test.
Relationships between HA stability, virus stability, and replication in vitro
We investigated the relationships between HA stability, virus stability, and in vitro replication. Virus inactivation pH correlated poorly with HA activation pH for the 22 human H3N2 viruses compared to the 18 cluster IV swine viruses (Fig. 3A). This suggested that HA activation may be an important, albeit nonexclusive, correlation of H3N2 virus resistance to inactivation by mild acid. In MDCK and Vero cells, virus yields at earlier time points increased as the HA activation pH or inactivation pH values increased from 5.2 to 5.9 (Fig. 3, B to E). However, virus titers at all time points and either HA activation pH or virus inactivation pH were poorly correlated [coefficient of determination (R2) < 0.24 in Vero and R2 < 0.35 in MDCK] (Fig. 3, B to E). MDCK cells produced higher virus yields than Vero cells at earlier time points (Fig. 3F).
Fig. 3. Relationships between HA activation pH, virus inactivation pH, and virus growth in Vero and MDCK cells.
(A) Virus inactivation pH as a function of HA activation pH. (B) Virus growth in Vero cells as a function of HA activation pH. (C) Virus growth in Vero cells as a function of virus inactivation pH. (D) Virus growth in MDCK cells as a function of HA activation pH. The MOI was 0.01 PFU per cell for the left three panels and 2 PFU per cell for the right panel. (E) Virus growth in MDCK cells as a function of virus inactivation pH. (F) Virus growth (three left panels) and peak titers (right panel) in MDCK cells versus Vero cells. Six vaccine reference viruses—A/SG/Infimh, A/CH/8060, A/HK/4801, A/SA/55, A/TX/50, and A/CH/9715293—were indicated by colored symbols shown in the legend. Titers at indicated time points are the average titer of an individual isolate (log10TCID50/ml). A linear regression model using Python seaborn regplot package was applied for the scatter plots. The regression lines and R2 values are shown.
Human H3N2 infection and transmission in ferrets
We infected ferrets with five human H3N2 isolates. Two viruses (i.e., A/SG/Infimh and A/HK/4801) were egg-based WHO-recommended vaccine reference viruses containing HA1-L194P that had virus inactivation pH values of 5.8 and 5.7, respectively. Both cell-based and egg-based (containing HA1-G186V) A/TX/50 had virus inactivation pH values of 5.5. Cell-based A/HK/5738 (inactivation pH of 5.4) and seasonal human A/MEM/2 (inactivation pH of 5.3) had the most stable HAs.
On day 0, we intranasally inoculated naïve (donor) ferrets with 106 plaque-forming units (PFU) of viruses. On day 1, we introduced naïve (contact) ferrets into the same cage and naïve (airborne) ferrets into adjacent noncontact compartments separated by perforated dividers. We monitored ferret body weight and temperatures daily, performed nasal washes every other day until day 14, and collected blood on day 21.
In brief, all viruses transmitted by contact route at 100%. In the egg-based A/SG/Infimh and A/HK/4801 groups that had HA1-L194P, the reversion mutation HA1-P194L arose in donor, contact, and the single airborne-transmitted ferret (Table 2). In contrast, egg-based A/TX/50 containing HA1-G186V airborne transmitted in two of the three ferrets without reversion. Cell-based A/HK/5738 and A/MEM/2 had 100% airborne transmission (three of three) (Table 2). Detailed results were as follows.
Egg-based A/SG/Infimh
A/SG/Infimh contained egg-adaptive mutations HA1-T160K, HA1-L194P, and HA1-D225G (Fig. 4A and Table 2). After 1 day in donors, A/SG/Infimh produced a nasal titer of 2.2 log10TCID50/ml in one animal and was not detected in the other two (Fig. 4B). A/SG/Infimh grew to peak titers (5.78 ± 0.5 log10TCID50/ml) by day 3 in all three donors (Fig. 4B and Table 2). Transmission was first detected in contact animals on days 4, 6, and 10 (Fig. 4B and Table 2). Airborne transmission was first detected on day 12 in ferret A2 (Fig. 4B and Table 2).
Fig. 4. Replication and transmission of egg-based vaccine reference virus A/SG/Infimh containing egg-adaptive mutation HA1-L194P in ferrets.
Three donor ferrets were intranasally infected on day 0 with egg-based A/SG/Infimh. After 1 day, three naïve contact ferrets were co-caged with the donor ferrets, and three naïve airborne ferrets were introduced into adjacent cages. Ferret nasal washes were collected every other day until day 14. (A) HA amino acid variations between the A/SG/Infimh isolates cultured in eggs and in MDCK or MDCK-SIAT1 (MDCK/SIAT) cells. N/A, not applicable. (B) Nasal virus titers of egg-based A/SG/Infimh in donor, contact, and airborne ferrets. (C) Proportions of HA1-P194 and HA1-L194 as determined by next-generation sequencing. All reads with a frequency higher than 0% are reported. (D) HA activation pH and virus inactivation pH values (means ± SD) of airborne-transmitted virus samples collected from ferret A2. (E) HA activation and virus inactivation pH (means ± SD) of A/SG/Infimh carrying HA1-P194 or HA1-L194. A/SG/Infimh carrying HA1-L194 was plaque-purified from airborne transmitted viruses and tested along with the inoculum containing HA1-P194. (F) Cross-reactive HA inhibition (HAI) responses. Infected/exposed ferret sera were grouped according to the variants detected, HA1-P194 and HA1-L194, and a mixture of HA1-P194 and HA1-L194 (HA1-P/L194). HAI assay was performed using viruses A/SG/Infimh carrying HA1-P194 and HA1-L914. The experiments were independently performed at least twice (means ± SD). P values were determined according to Welch’s t test.
Next-generation sequencing on ferret nasal washes showed the A/SG/Infimh inoculum had 100% egg-adaptive mutation HA1-P194 (Fig. 4C). Reversion (HA1-P194L) was detected in two of the three donor ferrets by day 5 (Fig. 4C and fig. S3). The HA1-P194L reversion variant transmitted to one of the three contacts and one of the three airborne ferrets (Fig. 4C and fig. S3). Airborne-transmitted viruses had lower HA activation and virus inactivation pH (5.2 and 5.4, respectively) than inoculum (Fig. 4D). We plaque-purified and verified A/SG/Infimh contained the HA1-P194L reversion but no other mutations. HA1-P194L had HA activation and virus inactivation pH values that were 0.4 pH units less than that of HA1-P194 (Fig. 4E). Both viruses bound preferentially to red blood cells (RBCs) expressing α2,6-linked sialic acid, and HA1-P194 retained binding to turkey RBCs expressing α2,3-linked sialic acid (table S3). In summary, A/SG/Infimh did not transmit via the airborne route until it acquired the HA-stabilizing reversion mutation HA1-P194L.
We collected sera from A/SG/Infimh-infected ferrets and segregated serum samples into three groups based on the next-generation sequencing results of nasal washes: (i) sera from ferrets infected with HA1-P194, (ii) sera from ferrets infected with reversion variant HA1-L194 after transmission, and (iii) sera from ferrets with a mixed infection of HA1-P194 and HA1-L194 (Fig. 4F and fig. S3). As expected, infection with virus containing egg-adaptive variation HA1-P194 elicited significantly lower HA inhibition (HAI) responses toward HA1-L194 virus, infection with HA1-L194 elicited greater HA1 responses toward HA1-L194 virus, and sera from dual-infected ferrets was equally reactive to both viruses (Fig. 4F). Overall, in the context of A/SG/Infimh, the egg-adaptive HA1-L194P variation destabilized the HA protein, prevented airborne transmission in ferrets until the stabilizing reversion mutation HA1-P194L occurred, and reduced reactivity to antibodies generated after infection with viruses with HA1-L194, a residue observed in the airborne transmitted virus and circulating human H3N2 viruses.
Egg-based A/HK/4801 and cell-based A/HK/5738
A/HK/4801 contained egg-adaptive mutations HA1-P194, HA1-K160, and HA1-G255 (Fig. 5A and Table 2). The virus retained these mutations after passage in MDCK cells (fig. S4). We used viruses with or without a final cell passage in MDCK cells in ferret transmission experiments (trial 1 and trial 2, respectively). Both trials showed 100% contact and 0% airborne transmission (Fig. 5B and Table 2). Next-generation sequencing showed that six of the seven donors (three of three in trial 1 and three of four in trial 2) and seven of the seven contacts had the reversion mutation HA1-P194L, albeit never exceeding 60% (Fig. 5C and fig. S4).
Fig. 5. Replication and transmission of egg-based vaccine reference virus A/HK/4801 containing HA1-L194P, and cell-based vaccine-like virus A/HK/5738 containing HA1-L194 in ferrets.
The experiment was performed as in Fig. 4 with either three or four ferrets per group. (A) HA variations between A/HK/4801 isolates cultured in eggs and in MDCK cells. (B) Nasal virus titers of egg-based A/HK/4801 in donor, contact, and airborne ferrets. (C) Proportions of HA1-P194 and HA1-L194 in the inoculum, donor, contact, and airborne ferrets for A/HK/4801. Trial 1 and trial 2 ferret experiments were performed using A/HK/4801 with passage histories of E5/E2/E2/E1/C1 and E5/E2/E2/E1, respectively. Trial 1 used three donor, contact, and airborne ferrets each. Trial 2 used four donor, contact, and airborne ferrets each. (D) Nasal virus titers of A/HK/4801-like virus, cell-based A/HK/5738 in donor, contact, and airborne ferrets. (E) Proportions of HA1-L194 and HA1-P194 for A/HK/5738. (F) HA activation pH and virus inactivation pH values (means ± SD) of airborne-transmitted viruses for A/HK/5738. (G) Cross-reactive HAI responses. Sera from ferrets infected or exposed to A/HK/5738 (carrying HA1-L194) and A/HK/4801 carrying HA1-P194 (L194 < 16%) were tested using viruses A/HK/5738 and A/HK/4801. The experiments were independently performed twice (means ± SD). P values were determined according to Welch’s t test.
A/HK/5738 differed from A/HK/4801 by 12 amino acids, 4 of which were in HA1 (table S4). A/HK/5738 contained stabilizing residue HA1-L194 and had HA activation pH of 5.2 (fig. S5, A and B). Despite A/HK/5738 and A/HK/4801 having similar peak titers in MDCK and Vero cells (Table 2), A/HK/5738 replicated to higher titers in donors on day 1 (Fig. 5D and Table 2). A/HK/5738 donor and contact peak titers were also higher than A/HK/4801 (Table 2). A/HK/5738 had 100% airborne transmissibility (three of three), while A/HK/4801 had 0% (Fig. 5, B and D, and Table 2). A/HK/5738 retained 100% HA1-L194 in all ferrets (Fig. 5E and fig. S5C). Airborne-transmitted A/HK/5738 isolated from nasal washes had HA activation pH of 5.3 (Fig. 5F). Infection with A/HK/5738 (HA1-L194–containing) resulted in significantly higher HAI titers to their matching antigen than against A/HK/4801 (HA1-P194–containing) (Fig. 5G). Overall, A/HK/5738 containing HA1-L194 had a relatively stable HA, was 100% airborne transmissible in ferrets, and yielded ferret sera more reactive against virus containing HA1-L194 than virus containing HA1-P194.
Cell-based A/TX/50 versus egg-based A/TX/50
Egg-based vaccine reference viruses A/SA/55, ACH/9715293, and A/TX/50 contained egg-adaptive mutation HA1-G186V rather than HA1-L194P (Table 2). They also had relatively stable HAs (pH 5.3 to 5.5) (Table 2). Cell- and egg-based A/TX/50 had similar HA stability and specificity for binding α2,6-linked sialic acid receptors (Fig. 6A and table S3). Cell- and egg-based A/TX/50 also had similar replication and transmission in ferrets (Fig. 6B). Ferret nasal wash samples showed no significant variants in HA (Fig. 6C and fig. S6), and both viruses retained stable HAs after airborne transmission (Fig. 6D). No antigenic differences were detected between A/TX/50 cultured in eggs and cells (Fig. 6E). Overall, egg-adaptive variation HA1-G186V, which impedes the emergence of HA1-L194P in egg culture (45) and in ferrets as shown in this study, did not alter HA stability, receptor binding specificity, airborne transmissibility in ferrets, or antigenicity in ferrets.
Fig. 6. Replication and transmission of vaccine reference virus A/TX/50 with and without the egg-adaptive mutation HA1-G186V in ferrets.
Trial 1 and trial 2 ferret experiments were performed using A/TX/50 cultured in MDCK/SIAT cells and in eggs, respectively. Trial 1 had three donor, contact, and airborne ferrets per group, while trial 2 had four donor, contact, and airborne ferrets per group. Both trials were performed as described in Fig. 4. (A) Genotype and phenotype variations between A/TX/50 isolates cultured in eggs and in MDCK/SIAT cells. (B) Nasal virus titers of A/TX/50 in donor, contact, and airborne ferrets. (C) Proportions of amino acids at HA1-186, HA1-219, and HA1-194. (D) HA activation pH and virus inactivation pH values (means ± SD) of airborne-transmitted viruses. (E) Cross-reactive HAI responses. Sera from infected ferrets (trial 1, n = 8; and trial 2 n = 6 with the absence of the donor sera) were tested by using A/TX/50 isolates cultured in cells (HA1-G186/S219) and in eggs (HA1-V186/F219). The experiments were independently performed twice (means ± SD).
A/MEM/2
Cell-based A/MEM/2 was a non–vaccine reference virus with HA activation and virus inactivation pH values of 5.3 (Fig. 7A). Inoculated A/MEM/2 virus contained 7.9% HA1-T160 and 92.1% HA1-K160 (fig. S7). HA1-K160 modifies the HA1-N158 glycosylation site and is present in recent human H3N2 vaccine reference viruses including A/SG/Infimh, A/CH/8060, and A/HK/4801. A/MEM/2 grew to relatively high titers in ferrets and 100% transmitted by contact and airborne (Fig. 7B and Table 2). Only HA1-K160, the major variant, transmitted (Fig. 7C and fig. S7), and all airborne-transmitted viruses retained stable HAs (Fig. 7D). Thus, the HA1-K160 glycosylation-deletion variation supported efficient replication and transmission in ferrets and was unlikely responsible for loss-of-function airborne transmission of A/SG/Infimh and A/HK/4801.
Fig. 7. Replication and transmission of human seasonal H3N2 virus A/MEM/2 with a mixed population of HA1-160 K/T in ferrets.
The experiment was performed as in Fig. 4. (A) HA activation pH and virus inactivation pH values. (B) Nasal virus titers in donor, contact, and airborne ferrets. (C) Proportions of HA1-K160 and HA1-T160. (D) HA activation pH and virus inactivation pH values (means ± SD) of airborne-transmitted viruses.
Relationships between HA stability, virus stability, and replication in ferrets
In ferrets, egg-based vaccine reference viruses containing HA1-L194P (i.e., A/SG/Infimh and A/HK/4801) had lower initial titers in donors than A/TX/50, A/HK/5738, and A/MEM/2 (Fig. 8A). HA stabilization, represented by decreasing HA activation or virus inactivation pH, correlated with increased initial nasal titers (R2 = 0.77) but not virus shed from the ferrets (quantified using AUC) (R2 ≤ 0.07) (Fig. 8, A and B).
Fig. 8. Relationships between HA activation pH, virus inactivation pH, and virus growth in ferrets.
(A) Virus initial titers in ferrets as a function of HA acid stability. Initial nasal titers in donor ferrets versus HA activation pH (first panel) and virus inactivation pH (second panel). (B) AUC of virus growth in ferrets as a function of HA acid stability. AUC of virus growth in donors versus HA activation pH (first panel) and virus inactivation pH (second panel). The scatter plots were summarized in table S5. The regression lines and R2 values are shown.
DISCUSSION
H3N2 IAVs cause severe epidemics (7). Seasonal H3N2 influenza vaccines have low effectiveness (<33%), in part, due to antigenic mismatches between vaccine and circulating viruses, which are exacerbated by egg-adaptive mutations arising during manufacturing (37, 38, 44, 45, 48, 49). Here, we showed that recent vaccine reference viruses had unstable HA proteins compared to those from circulating human H3N2 viruses, and we mapped this to destabilizing egg-adaptive mutation HA1-L194P. These recent vaccine reference viruses with unstable HA proteins had reduced virus replication, impaired airborne transmission unless the stabilizing HA1-P194L reversion mutation occurred, and skewed antigenicity in ferrets. In contrast, vaccine reference viruses with egg-adaptive mutations that did not destabilize HA (i.e., HA1-V186) retained infectivity, transmissibility, and antigenicity in ferrets. Moreover, HA1-V186 impeded the emergence of destabilizing HA1-L194P in egg culture (45) and in ferrets in this study, suggesting that it should be prioritized over HA1-L194P during H3N2 vaccine strain selection. In addition, HA stabilization was shown to be required for H3N2 airborne transmission in ferrets and should be considered explicitly in formal assessments of pandemic risk.
Most seasonal influenza vaccines are manufactured in embryonated chicken eggs (47), and many contemporary human H3N2 IAVs rapidly acquire adaptive mutations when passaged in eggs (40). Egg-adaptive H3N2 HA mutations often alter receptor binding and, consequently, alter antigenicity due to the presence of antigenic sites at the periphery of the receptor-binding pocket (43, 50–52). HA1 residues 194 and 186 are located at the rim of the receptor-binding pocket (45) and in H3 epitope B (39). HA1-L194P and -G186V increase H3N2 growth in eggs by increasing binding to α2,3-linked receptors or disrupting avidity toward long-chain α2,6-linked receptors (43, 50–53). In 2018 to 2019 H3N2 vaccine reference virus A/SG/Infimh, the present study showed that, in addition to increasing binding to RBCs expressing α2,3-linked sialic acid receptors, the HA1-L194P variation also destabilizes the HAs. HA destabilization of pH1N1 has been shown to reduce live attenuated influenza vaccine (LAIV) stability and decrease infectivity in ferrets (54). FluMist is manufactured in embryonated chicken eggs (55). The present and previous results suggest that LAIV production in cell culture instead of eggs might increase vaccine stability and in vivo infectivity and immunogenicity, albeit at greater manufacturing cost, by avoiding egg-adaptive mutations like HA1-L194P that alter HA receptor binding and HA stability. It is unknown whether HA destabilization due to the presence of egg-adaptive mutations would decrease antibody responses of intramuscularly injected inactivated vaccines, but this could be tested.
MDCK and Vero cells are also used to manufacture influenza vaccines (47). In the present study, H3N2 virus yields in Vero and MDCK cells did not vary as a function of HA stability, although virus replication at initial stages in ferrets did. MDCK cells appear highly attractive for influenza vaccine virus production, as H3N2 growth in MDCK cells is (i) independent of HA stability, (ii) higher than in Vero cells for a subset of isolates, and (iii) more genetically stable when compared with virus growth in eggs.
Human-adapted IAVs have HA stability values that range from pH 5.0 to 5.5 (16), leading to the hypothesis that a stabilized HA protein is required for human pandemic potential (2). HA stabilization promotes airborne transmission by preventing virion inactivation outside of the host and modulating antiviral responses intracellularly (24, 26, 27). Here, 3 of the 22 human seasonal H3N2 viruses had virus inactivation pH values above 5.6, which was unexpected. However, all three viruses with high inactivation pH (i.e., A/SG/Infimh, A/CH/8060, and A/HK/4801) were egg-adapted vaccine reference viruses containing the destabilizing HA1-L194P mutation, and these viruses were incapable of airborne transmission in ferrets without reversion to HA1-P194L and HA stabilization. For pH1N1, a stabilized HA protein (activation pH of 5.5 or less) was associated with adaptation to humans and was shown to be required for airborne transmission from ferret to ferret and swine to ferret (20, 35). An HA activation pH of 5.5 or less was also shown to be necessary for airborne transmission of H5N1 between ferrets (32–34). With respect to swine viruses, gamma clade H1N1 viruses with relatively unstable HA proteins (activation pH > 5.6) were outcompeted by minority populations (<17%) containing stabilized HA within 3 days of growth in ferrets, and only viruses with a relatively stable HA protein (pH 5.6 or lower) airborne transmitted (17, 21). Now, HA stability is not considered explicitly in formal risk assessment tools of pandemic potential by emerging IAVs (i.e., IRAT and TIPRA). As described above, evidence supporting inclusion of HA stability in pandemic risk assessment tools includes the following: (i) human-adapted IAVs contain relatively stable HA proteins; and (ii) a stabilized HA protein has been shown to be required for ferret airborne transmission of human pH1N1, swine H1N1, H5N1, and, now, human H3N2 viruses.
Swine may serve as an intermediate host within which avian-like IAVs can acquire mutations that switch receptor-binding specificity to the human-preferred form (α2,6-linked) and stabilize the HA protein (20). Several clades of swine IAVs recently diverged from human seasonal viruses have relatively stable HA proteins. These include swine H1N1 pandemic lineage (17) and swine H3N2 human-like (reported here) that have ranges of HA activation pH from 5.1 to 5.5 and from 5.2 to 5.4, respectively. Other clades of swine IAVs have, on average, less stable HA proteins including classical swine H1N1 (5.5 to 5.8), Eurasian avian-like H1N1 (5.8 to 6.0), North American triple-reassortant H1N1 (5.4 to 5.8), gamma H1N1 (5.5 to 5.9), and cluster IV H3N2 (5.3 to 5.9), as reported in previous (17, 20, 35) or the present work. Clades with stabilized HA proteins may present a higher risk for the emergence of pandemic influenza if most humans do not have preexisting immunity. Of note, pH1N1 isolates collected at the start of the 2009 pandemic had HA activation pH values of approximately 5.5 to 5.6 (35), while isolates from 2010 to present have lower HA activation pH values (17, 35, 54). This does not rule out the possibility that an influenza virus with a relatively high HA activation pH may acquire one or more HA-stabilizing mutations, enabling efficient human-to-human spread. Such an evolutionary pathway has been observed experimentally in the selection of airborne-transmissible variants in ferrets (17, 21, 32, 33, 35).
With the resumption of epidemic influenza since November 2021 (5, 6), there is an urgent need for improvement of influenza vaccine efficiency. This work showed that the presence of reversion mutation HA1-P194L stabilizes the HA protein in ferrets and skews antigenicity of vaccine reference viruses away from circulating viruses. Future studies are needed to examine the potential of other HA-stabilizing mutations to enhance influenza vaccine stability and antigenicity. In addition, a relatively stable HA protein was necessary for airborne transmission of human H3N2 viruses in ferrets, just as has been observed for human H1N1 (20, 35), swine H1N1 (17, 21), and avian H5N1 viruses (32, 33). This further supports the hypothesis that a stable HA protein is a trait that enhances the pandemic potential of emerging influenza viruses. Along with population immunity, pandemic preparedness for emerging IAVs will need to consider multiple viral properties including HA-NA balance, polymerase activity, and other unknown viral traits. It will also be important to understand whether a deficiency in one human-adaptive trait may be overcome by an enhancement of another.
MATERIALS AND METHODS
Cells
MDCK cells and Vero cells were maintained in minimum essential medium (MEM; Thermo Fisher Scientific) and Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies), respectively. All culturing media were supplemented with 10% HyClone standard fetal bovine serum (FBS; Life Technologies) and 1% penicillin/streptomycin (Thermo Fisher Scientific). All cells were grown at 37°C with 5% CO2 (56, 57).
Virus preparation
Forty-eight human and swine H3N2 IAVs (2012–2019) were retrieved from St. Jude Children’s Research Hospital, Center of Excellence for Influenza Research and Response, or provided by the WHO’s Global Influenza Surveillance and Response System. Of note, the six vaccine reference viruses were nonreassortant H3N2 wild-type viruses provided by Centers for Disease Control and Prevention (Atlanta, GA, USA), WHO Influenza Centre, MRC National Institute for Medical Research (London, UK), and Francis Crick Institute (London, UK). They contain all eight gene segments of H3N2 and not the six internal PR8 segments of influenza vaccine viruses. Detailed virus passage history information is listed in table S2. These viruses were then propagated in MDCK cells (unless otherwise stated) with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)–treated trypsin (1 μg/ml), as described previously (56, 57).
Phylogenic analysis of virus HA segments
Phylogenic analyses of virus HA segments were generated using a previously reported workflow (17). Briefly, whole genomes of viruses used in this study (2012–2019) were obtained by next-generation sequencing. Full-length HA nucleotide sequences of representative human and swine IAVs (2012–2019) were retrieved from Global Initiative on Sharing All Influenza Data (www.gisaid.org). In-house HA sequences were aligned with representative human and swine representative sequences using MAFFT v7.490. Phylogenic analyses of virus HA segments were conducted using FastTree 2.1 with a generalized time-reversible model and a single rate approximation for each site (the CAT approximation). The generated tree was annotated by an R package, ggtree, for better visualization.
Virus growth in MDCK and Vero cells
Virus multistep growth was assessed by infecting MDCK or Vero cells in six-well plates at a multiplicity of infection (MOI) of 0.01 PFU per cell. The inocula were removed after 1 hour of incubation at 37°C. The infected cells were washed twice by phosphate-buffered saline (PBS) (pH ~7.4) and were cultured using 3 ml of MEM culture medium supplemented with TPCK-treated trypsin (1 μg/ml). Infected cells were then maintained at 37°C. At indicated hours postinfection (hpi), 300 μl of cell culture supernatant was collected and titrated by TCID50 on MDCK cells (17, 21). For one-step virus growth, viruses were inoculated into MDCK cells in 24-well plates at an MOI of 2 PFU per cell. The infected culture supernatant was harvested at 6 hpi and titrated by TCID50 (21, 27).
HA activation pH assay
HA activation pH was measured by syncytia assay. Briefly, viruses were inoculated into Vero cells in 24-well plates at an MOI of 3 PFU per cell. After 1 hour of incubation, the inocula were removed and replaced with 1 ml of MEM culture medium. At 16 hpi, the infected cells were treated with 0.3 ml of DMEM supplemented with TPCK-treated trypsin (5 μg/ml) for 15 min. After that, the infected cells were washed by PBS twice and maintained with pH-adjusted PBS buffers ranging from 4.8 to 6.2 for 15 min. The pH-adjusted PBS solutions were aspirated, and Vero cells were incubated in DMEM supplemented with 5% FBS for 3 hours at 37°C. The cells were then fixed and stained using a Hema 3 Fixative and Solutions (Thermo Fisher Scientific). Syncytia were recorded using a light microscope (17, 21). HA activation pH values were reported as the highest treated pH at which the cells contained more than two syncytia with at least five nuclei (17, 21).
Virus inactivation pH assay
Viruses were mixed with pH-adjusted PBS buffers in a ratio of 1:100 in 96-well plates and incubated at 37°C for 1 hour. After that, the remaining infectivity was measured by TCID50 on MDCK cells (35, 58). Virus titers were analyzed by using GraphPad Prism software (version 7). Nonlinear regression (curve fit) with an equation of log (agonist) versus response-variable slope (four parameters) was applied, and best-fit value logEC50 was reported as virus HA inactivation pH (21).
Ferret transmission experiment
Sixty-nine Fitch ferrets (male, 5 to 6 months old) were purchased from Triple F Farms (Sayre, PA) and verified to be serologically negative for now circulating influenza viruses and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by performing HAI assay and enzyme-linked immunosorbent assay, respectively. Ferret transmission studies were performed as described previously (17, 21). On day 0, randomly allocated naïve ferrets (three or four in each cubicle as indicated in the study) were intranasally inoculated with 106 PFU of viruses in 500 μl and were caged separately. These ferrets were donor ferrets. On day 1, naïve ferrets (three or four each) were cohoused with the donor ferrets and designated as contact ferrets. At the same time, three or four naïve ferrets were introduced in adjacent cages and taken as airborne ferrets. Aerosol exposure was only permitted among ferrets within each cubicle. Until day 14, ferret body weight and temperature were monitored daily, and ferret nasal washes were collected every other day. On day 21, all ferrets were euthanized to collect the whole blood. Ferret sera were extracted and subjected to HAI assay. During the experiments, cubicle temperature and humidity were controlled to 22°C and 40 to 60%, respectively. Animal studies were performed in compliance with the St. Jude Children’s Research Hospital Animal Care and Use Committee guidelines.
Virus genome sequencing analysis
Virus whole-genome sequences were determined by the next-generation sequencing (17, 21). Briefly, viral RNAs were extracted from cell-free culturing supernatants or ferret nasal washes using the QIAamp Viral RNA Kits (QIAGEN). Subsequently, viral complementary DNAs were obtained by reverse transcription polymerase chain reaction (PCR) using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific). Next, PCR amplicons were generated using the Phusion High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs). The resulting DNA libraries were purified by the QIAquick Gel Extraction Kit (QIAGEN) and were submitted to St. Jude Harwell Center for whole-genome sequencing. After retrieving raw sequencing data, CLC Genomics Workbench (version 22.0) was used to perform data analyses. Variant parameters were set as >10 reads and >5% minimum frequency unless otherwise stated. The frequencies of a single-nucleotide variant (SNV) were calculated by the percent of read counts of a nucleotide variant to the whole nucleotide read counts at a specific position. The frequencies of SNVs were visualized by heat maps and parts of whole, which were generated by using GraphPad Prism software (version 7).
HAI assay
Ferret sera were obtained from ferret whole blood and treated with receptor-destroying enzymes (Denka Seiken Co. Ltd.) according to the provided protocol. Subsequently, the samples were diluted and added into 96-well plates (25 μl per well), which contained standard antigens 25 μl per well (HA titer 8). After mixing and maintaining in room temperature for 45 min, 0.5% turkey RBCs were added into the 96-well plates (50 μl per well). The plates were incubated at room temperature for 30 min. The highest dilutions that completely inhibited hemagglutination were reported as HAI titers (17, 20).
Hemagglutination assay with regular or modified RBCs
Modified turkey RBCs expressing either α2,6-linked sialic acids (α2,6-turkey RBCs) or α2,3-linked SA (α2,3-turkey RBCs) were prepared with modifications as described previously (59). Briefly, α2,6-turkey RBCs were generated by incubating 500 μl of 20% turkey RBCs in PBS with 400 U of α2,3 Neuraminidase S, 10× GlycoBuffer, and 100× purified bovine serum albumin (New England Biolabs) at 37°C for 3 hours. To generate α2,3-turkey RBCs, various sialic acids on cell surfaces were removed by incubating 62.5 μl of 20% turkey RBCs with 50 mM Vibrio cholerae neuraminidase (sialidase) (Roche) at 37°C for 1.5 hours. Subsequently, resialylation was performed by incubating the resulting turkey RBCs in 75 μl with 0.5 mU of α2,3-sialyltransferase from Pasteurella multocida (Sigma-Aldrich) and 1.5 mM cytidine 5′-monophosphate (CMP)–sialic acid (Sigma-Aldrich) at 37°C for 2.5 hours. After that, HA assays were performed by using 50 μl of twofold diluted tested viruses mixed with 50 μl of regular turkey RBCs (0.5%), regular guinea pig RBCs (0.6%), modified α2,6-turkey RBCs (0.5%), and modified α2,3-turkey RBCs. After 45 min, HA titers were reported as the highest dilutions that presented RBC agglutination. A/TN/1-560/2009 (H1N1) and recombinant A/PR/08/1934 with the HA of A/mallard/Alberta/383/2009 (H5N1) virus were used as control viruses with specific binding toward α2,6-linked sialic acids and α2,3-linked sialic acids, respectively (17, 35).
Analysis of HA activation pH, virus inactivation pH, and virus replication
Area under the growth curves (AUC) was calculated by using GraphPad Prism software (version 7). HA activation pH, virus inactivation pH, virus titers (log10), and AUC (log10) were fitted in a linear regression model using Python seaborn regplot package (0.12.0). The 99% confidence interval (unless otherwise stated) for the regression estimate was shown using translucent bands. The R2 values were calculated.
Statistical analysis
All data analyses were performed using GraphPad Prism software (version 7). Mann-Whitney U test, Welch’s t test, or two-way analysis of variance (ANOVA) followed by Tukey’s test was used to determine statistical significances. P < 0.05 was considered significant.
Acknowledgments
We thank J. Carol Crumpton and St. Jude Animal Resources Center for help with the ferret experiments. We thank D. Walker for help with virus collection. We also thank the Hartwell Center DNA Sequencing and Genotyping, Hartwell Center Functional Genomics, and the Hartwell Center Genome Sequencing Facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding: This work was supported by the National Institute of Allergy and Infectious Diseases under Collaborative Influenza Vaccine Innovation Centers (CIVICs) contract no. 75N93019C00052 (C.J.R.), Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400006C (C.J.R.), and Centers of Excellence for Influenza Research and Response (CEIRR) contract no. 75N93021C00016 (R.J.W.); the St. Jude Children’s Research Hospital (R.J.W. and C.J.R.); and the American Lebanese Syrian Associated Charities (ALSAC) (R.J.W. and C.J.R.).
Author contributions: Conceptualization: M.H., C.J.R., and R.J.W. Methodology: M.H., C.J.R., R.J.W., L.K., S.L., and L.L. Investigation: M.H., C.K., B.B., C.R.O., J.C.J., and L.L. Visualization: M.H. and S.L. Supervision: C.J.R. and R.J.W. Writing—original draft: M.H. and C.J.R. Writing—review and editing: M.H., C.J.R., R.J.W., and J.C.J.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All sequences of viruses in the paper have been submitted to Global Initiative on Sharing All Influenza Data (GISAID) (www.gisaid.org) and accession numbers are shown in tables S1 and S2. The viruses can be provided by St. Jude Children’s Research Hospital pending scientific review and a completed material transfer agreement. Requests for the viruses should be submitted to R.J.W.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S5
REFERENCES AND NOTES
- 1.E. J. Schrauwen, R. A. Fouchier, Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes Infect. 3, e9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. J. Russell, M. Hu, F. A. Okda, Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 26, 841–853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J. S. Long, B. Mistry, S. M. Haslam, W. S. Barclay, Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 17, 67–81 (2019). [DOI] [PubMed] [Google Scholar]
- 4.V. Dhanasekaran, S. Sullivan, K. M. Edwards, R. Xie, A. Khvorov, S. A. Valkenburg, B. J. Cowling, I. G. Barr, Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat. Commun. 13, 1721 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention, Weekly U.S. Influenza Surveillance Report, updated 21 October 2022; www.cdc.gov/flu/weekly/index.htm.
- 6.World Health Organization, Influenza Update, 3 October 2022; www.who.int/publications/m/item/influenza-update-n-429.
- 7.J. D. Allen, T. M. Ross, H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation. Hum. Vaccin. Immunother. 14, 1840–1847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. S. Rajao, R. R. Walia, B. Campbell, P. C. Gauger, A. Janas-Martindale, M. L. Killian, A. L. Vincent, Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J. Virol. 91, e01763–01716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. B. Pearce, A. Jayaraman, C. Pappas, J. A. Belser, H. Zeng, K. M. Gustin, T. R. Maines, X. Sun, R. Raman, N. J. Cox, R. Sasisekharan, J. M. Katz, T. M. Tumpey, Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc. Natl. Acad. Sci. U.S.A. 109, 3944–3949 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.B. S. Kaplan, J. B. Kimble, J. Chang, T. K. Anderson, P. C. Gauger, A. Janas-Martindale, M. L. Killian, A. S. Bowman, A. L. Vincent, Aerosol transmission from infected swine to ferrets of an H3N2 virus collected from an agricultural fair and associated with human variant infections. J. Virol. 94, e01009–01020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. S. Bowman, R. R. Walia, J. M. Nolting, A. L. Vincent, M. L. Killian, M. M. Zentkovich, J. N. Lorbach, S. E. Lauterbach, T. K. Anderson, C. T. Davis, N. Zanders, J. Jones, Y. Jang, B. Lynch, M. R. Rodriguez, L. Blanton, S. E. Lindstrom, D. E. Wentworth, J. Schiltz, J. J. Averill, T. Forshey, Influenza A(H3N2) virus in swine at agricultural fairs and transmission to humans, Michigan and Ohio, USA, 2016. Emerg. Infect. Dis. 23, 1551–1555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention, Influenza Risk Assessment Tool (IRAT), reviewed 27 November 2020; www.cdc.gov/flu/pandemic-resources/national-strategy/risk-assessment.htm.
- 13.World Health Organization, Tool for Influenza Pandemic Risk Assessment (TIPRA), May 2016; https://apps.who.int/iris/handle/10665/250130.
- 14.M. A. Benhaim, V. Mangala Prasad, N. K. Garcia, M. Guttman, K. K. Lee, Structural monitoring of a transient intermediate in the hemagglutinin fusion machinery on influenza virions. Sci. Adv. 6, eaaz8822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J. J. Skehel, D. C. Wiley, Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 (2000). [DOI] [PubMed] [Google Scholar]
- 16.S. E. Galloway, M. L. Reed, C. J. Russell, D. A. Steinhauer, Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: Implications for host range and adaptation. PLOS Pathog. 9, e1003151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu M., Yang G., DeBeauchamp J., Crumpton J. C., Kim H., Li L., Wan X. F., Kercher L, Bowman A. S., Webster R. G., Webby R. J., Russell C. J., HA stabilization promotes replication and transmission of swine H1N1 gamma influenza viruses in ferrets. eLife. 9, e56236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.G. Yang, C. R. Ojha, C. J. Russell, Relationship between hemagglutinin stability and influenza virus persistence after exposure to low pH or supraphysiological heating. PLOS Pathog. 17, e1009910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.X. Sun, J. A. Pulit-Penaloza, J. A. Belser, C. Pappas, M. B. Pearce, N. Brock, H. Zeng, H. M. Creager, N. Zanders, Y. Jang, T. M. Tumpey, C. T. Davis, T. R. Maines, Pathogenesis and transmission of genetically diverse swine-origin H3N2 variant influenza A viruses from multiple lineages isolated in the United States, 2011–2016. J. Virol. 92, e00665-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Russier, G. Yang, A. Marinova-Petkova, P. Vogel, B. S. Kaplan, R. J. Webby, C. J. Russell, H1N1 influenza viruses varying widely in hemagglutinin stability transmit efficiently from swine to swine and to ferrets. PLOS Pathog. 13, e1006276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu M., Jones J. C., Banoth B., Ojha C. R., Crumpton J. C., Kercher L., Webster R. G., Webby R. J., Russell C. J., Swine H1N1 influenza virus variants with enhanced polymerase activity and HA stability promote airborne transmission in ferrets. J. Virol 96, e0010022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Scholtissek, Stability of infectious influenza A viruses at low pH and at elevated temperature. Vaccine 3, 215–218 (1985). [DOI] [PubMed] [Google Scholar]
- 23.V. Le Sage, K. A. Kormuth, E. Nturibi, J. M. Lee, S. A. Frizzell, M. M. Myerburg, J. D. Bloom, S. S. Lakdawala, Cell-culture adaptation of H3N2 influenza virus impacts acid stability and reduces airborne transmission in ferret model. Viruses 13, 719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. Singanayagam, J. Zhou, R. A. Elderfield, R. Frise, J. Ashcroft, M. Galiano, S. Miah, L. Nicolaou, W. S. Barclay, Characterising viable virus from air exhaled by H1N1 influenza-infected ferrets reveals the importance of haemagglutinin stability for airborne infectivity. PLOS Pathog. 16, e1008362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. Singanayagam, M. Zambon, W. S. Barclay, Influenza virus with increased pH of hemagglutinin activation has improved replication in cell culture but at the cost of infectivity in human airway epithelium. J. Virol. 93, e00058-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T. Gerlach, L. Hensen, T. Matrosovich, J. Bergmann, M. Winkler, C. Peteranderl, H.-D. Klenk, F. Weber, S. Herold, S. Pohlmann, M. Matrosovich, pH optimum of hemagglutinin-mediated membrane fusion determines sensitivity of influenza a viruses to the interferon-induced antiviral state and IFITMs. J. Virol. 91, e00246-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. Russier, G. Yang, B. Briard, V. Meliopoulos, S. Cherry, T.-D. Kanneganti, S. Schultz-Cherry, P. Vogel, C. J. Russell, Hemagglutinin stability regulates H1N1 influenza virus replication and pathogenicity in mice by modulating type I interferon responses in dendritic cells. J. Virol. 94, e01423-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. A. Belser, E. H. Y. Lau, W. Barclay, I. G. Barr, H. Chen, R. A. M. Fouchier, M. Hatta, S. Herfst, Y. Kawaoka, S. S. Lakdawala, L. Y. Y. Lee, G. Neumann, M. Peiris, D. R. Perez, C. Russell, K. Subbarao, T. C. Sutton, R. J. Webby, H. Yang, H.-L. Yen; Working group on the standardization of the ferret model for influenza risk assessment , Robustness of the ferret model for influenza risk assessment studies: A cross-laboratory exercise. mBio 13, e0117422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. V. Forst, L. Martin-Sancho, S. Tripathi, G. Wang, L. G. Dos Anjos Borges, M. Wang, A. Geber, L. Lashua, T. Ding, X. Zhou, C. E. Carter, G. Metreveli, A. Rodriguez-Frandsen, M. D. Urbanowski, K. M. White, D. A. Stein, H. Moulton, S. K. Chanda, L. Pache, M. L. Shaw, T. M. Ross, E. Ghedin, A. Garcia-Sastre, B. Zhang, Common and species-specific molecular signatures, networks, and regulators of influenza virus infection in mice, ferrets, and humans. Sci. Adv. 8, eabm5859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. G. Joyce, A. K. Wheatley, P. V. Thomas, G. Y. Chuang, C. Soto, R. T. Bailer, A. Druz, I. S. Georgiev, R. A. Gillespie, M. Kanekiyo, W.-P. Kong, K. Leung, S. N. Narpala, M. S. Prabhakaran, E. S. Yang, B. Zhang, Y. Zhang, M. Asokan, J. C. Boyington, T. Bylund, S. Darko, C. R. Lees, A. Ransier, C.-H. Shen, L. Wang, J. R. Whittle, X. Wu, H. M. Yassine, C. Santos, Y. Matsuoka, Y. Tsybovsky, U. Baxa; NISC Comparative Sequencing Program, J. C. Mullikin, K. Subbarao, D. C. Douek, B. S. Graham, R. A. Koup, J. E. Ledgerwood, M. Roederer, L. Shapiro, P. D. Kwong, J. R. Mascola, A. B. McDermott, Vaccine-induced antibodies that neutralize group 1 and group 2 influenza a viruses. Cell 166, 609–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. Li, J. Chen, Q. Zheng, W. Xue, L. Zhang, R. Rong, S. Zhang, Q. Wang, M. Hong, Y. Zhang, L. Cui, M. He, Z. Lu, Z. Zhang, X. Chi, J. Li, Y. Huang, H. Wang, J. Tang, D. Ying, L. Zhou, Y. Wang, H. Yu, J. Zhang, Y. Gu, Y. Chen, S. Li, N. Xia, Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses. Nat. Commun. 13, 5182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Herfst, E. J. Schrauwen, M. Linster, S. Chutinimitkul, E. de Wit, V. J. Munster, E. M. Sorrell, T. M. Bestebroer, D. F. Burke, D. J. Smith, G. F. Rimmelzwaan, A. D. Osterhaus, R. A. Fouchier, Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Imai, T. Watanabe, M. Hatta, S. C. Das, M. Ozawa, K. Shinya, G. Zhong, A. Hanson, H. Katsura, S. Watanabe, C. Li, E. Kawakami, S. Yamada, M. Kiso, Y. Suzuki, E. A. Maher, G. Neumann, Y. Kawaoka, Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Linster, S. van Boheemen, M. de Graaf, E. J. A. Schrauwen, P. Lexmond, B. Manz, T. M. Bestebroer, J. Baumann, D. van Riel, G. F. Rimmelzwaan, A. Osterhaus, M. Matrosovich, R. A. M. Fouchier, S. Herfst, Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157, 329–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. Russier, G. Yang, J. E. Rehg, S. S. Wong, H. H. Mostafa, T. P. Fabrizio, S. Barman, S. Krauss, R. G. Webster, R. J. Webby, C. J. Russell, Molecular requirements for a pandemic influenza virus: An acid-stable hemagglutinin protein. Proc. Natl. Acad. Sci. U.S.A. 113, 1636–1641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.X. Sun, C. Liu, X. Lu, Z. Ling, C. Yi, Z. Zhang, Z. Li, M. Jin, W. Wang, S. Tang, F. Wang, F. Wang, S. Wangmo, S. Chen, L. Li, L. Ma, Y. Zhang, Z. Yang, X. Dong, Z. Qian, J. Ding, D. Wang, Y. Cong, B. Sun, Unique binding pattern for a lineage of human antibodies with broad reactivity against influenza A virus. Nat. Commun. 13, 2378 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Q. McLean, E. A. Belongia, Influenza vaccine effectiveness: New insights and challenges. Cold Spring Harb. Perspect. Med. 11, a038315 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R. K. Zimmerman, M. P. Nowalk, J. Chung, M. L. Jackson, L. A. Jackson, J. G. Petrie, A. S. Monto, H. Q. McLean, E. A. Belongia, M. Gaglani, K. Murthy, A. M. Fry, B. Flannery; US Flu VE Investigators; U.S. Flu VE Investigators , 2014–2015 influenza vaccine effectiveness in the United States by vaccine type. Clin. Infect. Dis. 63, 1564–1573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D. J. Smith, A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, R. A. Fouchier, Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004). [DOI] [PubMed] [Google Scholar]
- 40.F. Broszeit, R. J. van Beek, L. Unione, T. M. Bestebroer, D. Chapla, J.-Y. Yang, K. W. Moremen, S. Herfst, R. A. M. Fouchier, R. P. de Vries, G.-J. Boons, Glycan remodeled erythrocytes facilitate antigenic characterization of recent A/H3N2 influenza viruses. Nat. Commun. 12, 5449 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.H. Jin, H. Zhou, H. Liu, W. Chan, L. Adhikary, K. Mahmood, M. S. Lee, G. Kemble, Two residues in the hemagglutinin of A/Fujian/411/02-like influenza viruses are responsible for antigenic drift from A/Panama/2007/99. Virology 336, 113–119 (2005). [DOI] [PubMed] [Google Scholar]
- 42.S. J. Zost, K. Parkhouse, M. E. Gumina, K. Kim, S. Diaz Perez, P. C. Wilson, J. J. Treanor, A. J. Sant, S. Cobey, S. E. Hensley, Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U.S.A. 114, 12578–12583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.L. Parker, S. A. Wharton, S. R. Martin, K. Cross, Y. Lin, Y. Liu, T. Feizi, R. S. Daniels, J. W. McCauley, Effects of egg-adaptation on receptor-binding and antigenic properties of recent influenza A (H3N2) vaccine viruses. J. Gen. Virol. 97, 1333–1344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.N. C. Wu, S. J. Zost, A. J. Thompson, D. Oyen, C. M. Nycholat, R. McBride, J. C. Paulson, S. E. Hensley, I. A. Wilson, A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLOS Pathog. 13, e1006682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.N. C. Wu, H. Lv, A. J. Thompson, D. C. Wu, W. W. S. Ng, R. U. Kadam, C. W. Lin, C. M. Nycholat, R. McBride, W. Liang, J. C. Paulson, C. K. P. Mok, I. A. Wilson, Preventing an antigenically disruptive mutation in egg-based H3N2 seasonal influenza vaccines by mutational incompatibility. Cell Host Microbe 25, e835– 844.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.W. Liang, T. J. C. Tan, Y. Wang, H. Lv, Y. Sun, R. Bruzzone, C. K. P. Mok, N. C. Wu, Egg-adaptive mutations of human influenza H3N2 virus are contingent on natural evolution. PLOS Pathog. 18, e1010875 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. Perez Rubio, J. M. Eiros, Cell culture-derived flu vaccine: Present and future. Hum. Vaccin. Immunother. 14, 1874–1882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Z. Chen, H. Zhou, H. Jin, The impact of key amino acid substitutions in the hemagglutinin of influenza A (H3N2) viruses on vaccine production and antibody response. Vaccine 28, 4079–4085 (2010). [DOI] [PubMed] [Google Scholar]
- 49.S. Nakowitsch, M. Wolschek, A. Morokutti, T. Ruthsatz, B. M. Krenn, B. Ferko, N. Ferstl, A. Triendl, T. Muster, A. Egorov, J. Romanova, Mutations affecting the stability of the haemagglutinin molecule impair the immunogenicity of live attenuated H3N2 intranasal influenza vaccine candidates lacking NS1. Vaccine 29, 3517–3524 (2011). [DOI] [PubMed] [Google Scholar]
- 50.J. Stevens, L.-M. Chen, P. J. Carney, R. Garten, A. Foust, J. Le, B. A. Pokorny, R. Manojkumar, J. Silverman, R. Devis, K. Rhea, X. Xu, D. J. Bucher, J. C. Paulson, N. J. Cox, A. Klimov, R. O. Donis, Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J. Virol. 84, 8287–8299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.L. Mochalova, A. Gambaryan, J. Romanova, A. Tuzikov, A. Chinarev, D. Katinger, H. Katinger, A. Egorov, N. Bovin, Receptor-binding properties of modern human influenza viruses primarily isolated in Vero and MDCK cells and chicken embryonated eggs. Virology 313, 473–480 (2003). [DOI] [PubMed] [Google Scholar]
- 52.H. Powell, H. Liu, A. Pekosz, Changes in sialic acid binding associated with egg adaptation decrease live attenuated influenza virus replication in human nasal epithelial cell cultures. Vaccine 39, 3225–3235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.B. Lu, H. Zhou, D. Ye, G. Kemble, H. Jin, Improvement of influenza A/Fujian/411/02 (H3N2) virus growth in embryonated chicken eggs by balancing the hemagglutinin and neuraminidase activities, using reverse genetics. J. Virol. 79, 6763–6771 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.C. R. Cotter, H. Jin, Z. Chen, A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLOS Pathog. 10, e1003831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.B.-C. Childress, J. D. Montney, E. A. Albro, Making evidence-based selections of influenza vaccines. Hum. Vaccin. Immunother. 10, 2729–2732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. Hu, H. Chu, K. Zhang, K. Singh, C. Li, S. Yuan, B. K. Chow, W. Song, J. Zhou, B.-J. Zheng, Amino acid substitutions V63I or A37S/I61T/V63I/V100A in the PA N-terminal domain increase the virulence of H7N7 influenza A virus. Sci. Rep. 6, 37800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Hu, S. Yuan, K. Zhang, K. Singh, Q. Ma, J. Zhou, H. Chu, B.-J. Zheng, PB2 substitutions V598T/I increase the virulence of H7N9 influenza A virus in mammals. Virology 501, 92–101 (2017). [DOI] [PubMed] [Google Scholar]
- 58.H. Zaraket, O. A. Bridges, S. Duan, T. Baranovich, S.-W. Yoon, M. L. Reed, R. Salomon, R. J. Webby, R. G. Webster, C. J. Russell, Increased acid stability of the hemagglutinin protein enhances H5N1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J. Virol. 87, 9911–9922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. Chutinimitkul, D. van Riel, V. J. Munster, J. M. van den Brand, G. F. Rimmelzwaan, T. Kuiken, A. D. Osterhaus, R. A. Fouchier, E. de Wit, In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J. Virol. 84, 6825–6833 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S5