Abstract
The blueprints of developing organs are preset at the early stages of embryogenesis. Transcriptional and epigenetic mechanisms are proposed to preset developmental trajectories. However, we reveal that the competence for the future cardiac fate of human embryonic stem cells (hESCs) is preset in pluripotency by a specialized mRNA translation circuit controlled by RBPMS. RBPMS is recruited to active ribosomes in hESCs to control the translation of essential factors needed for cardiac commitment program, including Wingless/Integrated (WNT) signaling. Consequently, RBPMS loss specifically and severely impedes cardiac mesoderm specification, leading to patterning and morphogenetic defects in human cardiac organoids. Mechanistically, RBPMS specializes mRNA translation, selectively via 3′UTR binding and globally by promoting translation initiation. Accordingly, RBPMS loss causes translation initiation defects highlighted by aberrant retention of the EIF3 complex and depletion of EIF5A from mRNAs, thereby abrogating ribosome recruitment. We demonstrate how future fate trajectories are programmed during embryogenesis by specialized mRNA translation.
A specialized mRNA translation circuit instated in pluripotency presets the competence for cardiogenesis in humans.
INTRODUCTION
Embryonic development relies on precise and coordinated cell fate decisions, a complex process that sculpts an entire organism from a single totipotent cell (1–4). The success of developmental cell fate decisions requires timely, specific, accurate, and efficient rewiring of the regulatory proteome to support rapid cellular identity changes (5–7). In this regard, most research efforts over the past decades have concentrated on morphogen signaling, epigenetic, and transcriptional mechanisms (8–12). However, selective translational control is arguably the primary determinant of regulatory protein abundance in mammals and thus proposed as a central regulator of embryonic cell fate decisions (6, 13–17). Yet, how the developmental transcriptome is selectively translated to authorize cell fate decisions is a fundamental question that remains largely unclear.
The relevance of translational control in embryonic cell fate decisions is highlighted by the following evidence and concepts. While poorly understood, mRNA abundance does not reflect protein abundance at the systems level across evolution, especially during cell fate decisions (6, 18, 19). Regulation at the level of translation would allow an accurate and restricted subcellular abundance of fate-regulatory proteins, thus enabling spatiotemporal precision in gene function without a need for de novo mRNA synthesis (20). In addition, because translation is the most energy-demanding cellular process, decoupling mRNA abundance from protein abundance via selective translational control would ensure rapid and efficient responses during early embryogenesis when energy availability is rate-limiting (21). In support of this notion, studies using mouse embryonic stem cells (mESCs) proposed an immediate and substantial increase in protein synthesis upon induction of differentiation, indicative of a systems-wide yet poorly understood reprogramming of the “translatome” (22, 23). In vivo studies in mice have suggested that, at the exit from pluripotency, the mesoderm lineage is particularly susceptible to translational control, while the regulatory mechanisms remain unclear (24, 25). Recently, an elegant proteomics-based study analyzing ribosome composition using tagged ribosomal subunits in mESCs suggested a role for proteins associated with ribosomal complexes [loosely termed “ribosome-associated proteins” (RAPs)] in the selective control of translation, while the role of RAPs in humans remains to be investigated (26).
Together, despite the proposed prominent role of translational control in the developmental cell fate decisions, a systematic systems-wide understanding of the regulators, the molecular mechanism(s), and the principles by which the developmental transcriptome is differentially translated in time and space to allow cell fate specification remain largely elusive, especially in humans (27). We address these outstanding questions using human embryonic stem cell (hESC)–based cell fate decision models as a paradigm.
RESULTS
ARC-MS identifies proteins recruited to translationally active ribosomes during mesoderm commitment
We hypothesized that the competence for embryonic cell fate decisions is translationally controlled by cell fate–specific RAPs, which control the selective and privileged translation of developmental regulators. However, the identification of functionally relevant RAPs recruited to ribosomes is challenging due to a number of reasons. Current methods used for identifying RAPs (i) either require the generation of engineered ribosomal proteins (27), (ii) suffer from contamination of unrelated protein complexes (26), or (iii) cannot distinguish between translationally active and inert ribosomes (22, 23). During early embryogenesis, a substantial fraction of ribosomes are inert and ribosomal proteins are generated in excess (22). Therefore, prioritizing RAPs recruited to active ribosomes could be beneficial for identifying those that are functional in a cell fate-, context-, or stimulus-specific manner.
To cumulatively address such bottlenecks in the faithful identification of RAPs, we established active ribosome capture–mass spectrometry (ARC-MS), a versatile and easy-to-implement method tailored for the identification of RAPs recruited to translationally active ribosomes (Fig. 1A, detailed protocol in Materials and Methods). Briefly, ARC-MS involves labeling of de novo synthesized proteins via a brief pulse (5 min) of cell-permeable, “clickable” methionine analog [i.e., a derivative of noncanonical l-azidohomoalanine (AHA)], followed by the stable anchoring of labeled nascent peptides to ribosomes using an anisomycin derivative (28). Active ribosomal complexes are isolated by “clicking” nascent peptides harboring AHA directly to dibenzocyclooctyne beads. Next, RAPs, along with ribosomal proteins and translation factors, are biochemically separated from de novo synthesized nascent proteins (which remain covalently linked to the beads) and quantitatively detected independently via liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Fig. 1A, workflow). RAPs are identified by filtering out known ribosomal proteins, translation factors, and de novo synthesized proteins (detected independently after on-bead digestion of peptides). Because of the short AHA pulse labeling, ARC-MS captures active ribosomal complexes at the early stages of translation, arguably the rate-limiting and most regulated stage of protein synthesis, further increasing the probability of identifying functional RAPs. The ability to experimentally separate de novo synthesized proteins from ribosomal complexes allows added stringency in the identification of RAPs.
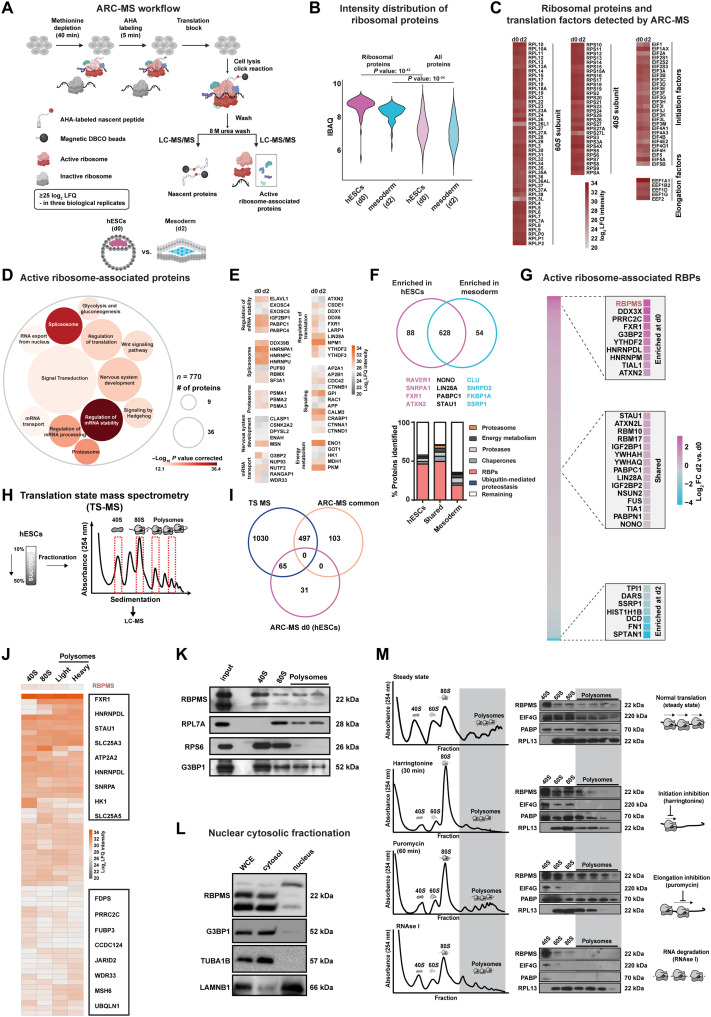
Fig. 1. ARC-MS identifies proteins recruited to translationally active ribosomes during mesoderm commitment.
(A) Schematic of ARC-MS workflow. ARC-MS was performed in hESCs (d0) and hESC-derived mesoderm progenitors (d2). (B) Violin plots depicting intensity-based absolute quantification (IBAQ) values of ribosomal proteins and all identified proteins from ARC-MS data from hESCs versus mesoderm progenitors. (C) Heatmap showing the enrichment of ribosomal proteins and translation factors (EIFs and EEFs) detected by ARC-MS in hESCs and mesoderm progenitors. (D) GO-based functional enrichment analysis for proteins (excluding ribosomal proteins and translation factors) reliably identified by ARC-MS. (E) Heatmap depicting log2 LFQ of significantly enriched representative proteins recruited on active ribosomes from major GO term categories identified by ARC-MS. (F) Venn diagram summarizing the distribution of proteins on active ribosomes in hESCs and mesoderm progenitors. Identified proteins, categorized based on molecular function, are depicted as a percentage of the total in the bar graphs below. (G) Heatmap displaying the enrichment of RBPs identified by ARC-MS between d0 and d2. (H) Schematic outline of the underlying strategy used for TS-MS. (I) Overlap of proteins enriched at d0 and d2 ARC-MS with proteins detected via TS-MS in hESCs. (J) Distribution of proteins recruited on active ribosomes selectively in hESCs identified by ARC-MS that are overlapping with TS-MS on indicated ribosomal fraction. (K) Confirmation of RBPMS enrichment in ribosomal fractions by polysome profiling followed by immunoblotting. RPL7A, RPS6, and G3BP1 = controls. (L) RBPMS is predominantly a cytosolic protein in hESCs, evaluated by Western blot analysis upon nuclear/cytosolic fractionation, G3BP1, and TUBA cytosolic control, LAMINB1 nuclear control. (M) Residence of RBPMS on ribosomal complexes confirmed by polysome profiling upon treatment with the indicated translation inhibitors. Error bars represent ±SEM; P values are calculated using Student’s t test; biological replicates n = 3.
As proof of principle for the systematic identification of RAPs regulating cell fate decisions, we focused on the transition from pluripotency to mesoderm. We reasoned that the discord between active ribosomes and ribosomal abundance in pluripotency and the documented susceptibility of mesoderm lineage to translational control renders this transition as an ideal platform for identifying cell fate–regulating RAPs using ARC-MS. Therefore, we performed ARC-MS in hESCs (d0) and hESC-derived mesoderm (d2) (fig. S1A) and only significantly detected proteins found in all three replicates [with log2 label-free quantification (LFQ) ≥ 25 in all replicates, false discovery rate (FDR) ≤ 0.01] were considered for further analysis (fig. S1B and table S1, sheet 1 contains data for all proteins detected by ARC-MS at d0 and d2). To ensure successful and specific isolation of ribosomal complexes, we first calculated the intensity distribution of ribosomal proteins. They were the most abundant among the detected proteins (Fig. 1B). Ribosomal proteins, translational initiation, and elongation factors constituted the majority of top enriched proteins, demonstrating successful isolation of active ribosomes (Fig. 1, B and C). The even distribution of ribosomal proteins on active ribosomes in both pluripotency and mesoderm exemplified the faithful isolation of ribosomal complexes (Fig. 1C). Notably, a few ribosomal protein isoforms (e.g., RPL3L, RPL26L1, and RPL27L) and translation factors (e.g., EIF3 complex) showed differential enrichment, indicating functional heterogeneity in line with recent studies (29–31).
Next, we filtered out ribosomal proteins and known translation factors to identify RAPs recruited on active ribosomes. In total, we identified 770 such RAPs in hESCs and hESC-derived mesoderm progenitors (table S1, sheet 1). Indicating direct cross-talk and synergy between the different stages of the mRNA life cycle and ribosomes, RAPs included proteins known to regulate pre-mRNA splicing, mRNA processing, stability, transport, or export apart from known translation regulators [Fig. 1D and table S1, sheet 2 for details on all Gene Ontology (GO) terms]. We identified proteins previously suggested to bind ribosomal complexes to selectively regulate translation, such as FXR1, LIN28A, LARP1, ATXN2, DDX1, and PKM1 (Fig. 1E) (26, 27). Notably, we identified various known regulators of embryonic development, energy metabolism, protein homeostasis, and components of morphogen signaling central to embryonic cell fate decisions on active ribosomes, including mediators of Wingless/Integrated (WNT) signaling (Fig. 1E). Previous reports using mESCs reported membrane proteins, centrosomes, clathrin complexes, and the Vault complex to be present as potential contaminants upon isolating ribosomal complexes to study associated proteins (26). However, components of these complexes were scarce in ARC-MS data and were duly filtered out, although we cannot completely rule out transient interactions or shuttling of factors between complexes, which might be functionally relevant (fig. S1G). Thus, ARC-MS allows for the robust identification of RAPs recruited on actively translating ribosomes. Our hESC-to-mesoderm RAPs align with emerging hypotheses derived from bacteria, yeast, and mice that, rather than constitutive protein synthesis factories, ribosomes can act as control hubs for cellular decision-making (26, 32, 33).
Cell fate–specific recruitment of RAPs on active ribosomal complexes was revealed by ARC-MS
We identified 88 proteins to be preferentially recruited to active ribosomes in hESCs [fold change (FC) ≥ 2, FDR ≤ 0.05] as opposed to 54 in the mesoderm, and 628 were present in both states (Fig. 1F and table S1, sheet 1). RBPs were the largest class of proteins (~50%) among them (Fig. 1F, bottom). Considering the direct role of RBPs in the regulation of translation, we focused on RBPs recruited onto active ribosomes for further investigation (27). Among those shared between hESCs and mesoderm progenitors included known translational regulators, including LIN28A, IGF2BP1, STAU1, PABPC1, FUS, and TIA1 (34–36). Mesoderm-specific ones were among the least known for their direct role in controlling selective translation, and it included RBPs such as LSM2, DARS, and HTATSF1 (Fig. 1G). Because we were interested in identifying regulators of selective translation controlling the transition from pluripotency to the mesoderm, we focused on RAPs enriched on ribosomes in hESCs. RBPMS (RNA binding protein with multiple splicing) was among the top enriched RBPs recruited to active ribosomes in hESCs (d0) along with known regulators of selective translation, like FXR1, G3BP2, YTHDF2, TIAL1, and ATXN2. This consolidates that our identification and prioritization criteria yielded bona fide translational regulators (Fig. 1G and table S1, sheet 4) (27, 36). RBPMS has never been reported as a regulator of translation but has been proposed to be a potential regulator of embryogenesis, making it an ideal candidate for further investigation (37, 38).
To independently verify the recruitment of identified proteins to ribosomal complexes, we sought a global analysis of proteins that differentially associate with ribosomal complexes. To achieve this, we isolated ribosomal fractions from hESCs corresponding to 40S, 80S (monosome), and polysomes (light polysome fraction and heavy polysome fraction) by polysome profiling and subjected them to LC-MS/MS [translation state mass spectrometry (TS-MS)] (Fig. 1H, detailed protocol in Materials and Methods). The occurrence of the different ribosomal proteins in their expected fractions across all the samples indicated that the isolated fractions represented the presumed ribosomal complexes (fig. S1C). After the removal of ribosomal proteins and known translation factors, we identified 1408 proteins on ribosomal complexes, of which 600 were RBPs in line with our findings from ARC-MS (table S1, sheet 7). Similarly, proteins identified by TS-MS belonged to similar functional categories as ARC-MS (fig. S1, D to F). Next, by intersecting the data from ARC-MS and TS-MS, we identified 65 proteins that are selectively recruited to ribosomal complexes in hESCs (Fig. 1I). RBPMS was also among the identified RBPs by TS-MS, thus confirming its recruitment on ribosomal complexes at the state of pluripotency (Fig. 1J).
RBPMS is preferentially recruited to active ribosomes in hESCs
Our candidate RAP in hESCs, RBPMS, is evolutionarily conserved across vertebrates and carries a single RNA recognition motif (37, 39). While it has been suggested as a potential regulator of embryonic development based on studies in xenopus and zebrafish, as well as binucleation of cardiomyocytes in mice, the function of RBPMS in human embryonic cell fate decisions, ribosome association, and translational control remains unknown (38, 40, 41). In line with a role in the regulation of translation, RBPMS sediments with ribosomal fractions and is enriched on the 40S ribosomal subunit (Fig. 1K), similar to G3BP1, a regulator of selective translation known to associate with the 40S ribosomal subunit (42). It is also predominantly cytosolic in hESCs. Three isoforms of RBPMS are expressed in hESCs, of which two of the cytosolic isoforms account for the majority of the protein (Fig. 1L).
To validate the association of RBPMS with ribosomal complexes, we used the following orthogonal approaches. We reasoned that if RBPMS associates with the translation machinery in hESCs, then upon treatment with specific translation inhibitors, it should show a characteristic shift in sedimentation commensurate to the inhibited step of translation. First, we used the specific translation initiation inhibitor harringtonine (2 μg/ml for 30 min). RBPMS was depleted from polysomes and concomitantly enriched in initiation fractions. The characteristic changes in the enrichment of bona fide components of the translation machinery, EIF4G, PABP, and RPL13 serve as controls (Fig. 1M, top two panels). We next treated hESCs with an inhibitor of elongation, puromycin (1 μg/ml for 1 hour), to induce translational arrest. This led to the redistribution of RBPMS across fractions (Fig. 1M, third panel). Last, we used ribonuclease (RNase) I (5 U, 30 min) treatment, which led to the disruption of ribosomal complexes and the accumulation of RBPMS in the 40S fraction (Fig. 1M, last panels). This suggests the role of RBPMS in mediating translation initiation. To our knowledge, there are no other complexes that would show a similar sedimentation profile as ribosomal complexes and simultaneously show such characteristic change upon treatment with specific translation inhibitors. Considering its enrichment on the 40S complex in steady state and upon various treatments with translation inhibitors, notably upon RNase I, we can rule out contamination by nascent RBPMS polypeptides. Thus, RBPMS is an active RAP in hESCs.
RBPMS loss causes global translation inhibition without affecting self-renewal in hESCs
To investigate the functional role of RBPMS, we generated a complete CRISPR-Cas9–mediated knockout in hESCs (hereafter RBPMS-KO) by targeting the exon-intron boundary of exon 1 with two guide RNAs (gRNAs) (Fig. 2A and fig. S2A). Exon 1 was specifically targeted because it is shared between all annotated isoforms of RBPMS; thus, its disruption will ensure the complete loss of RBPMS. Homozygous deletion of this exon-intron boundary disrupted the natural open reading frame of RBPMS, resulting in a complete loss of expression, which we confirmed both at the RNA and protein levels (Fig. 2B and fig. S2B). Considering the recruitment of RBPMS on active ribosomes in hESCs, we next examined global translation in RBPMS-KO compared to wild type (WT) using polysome profiling. We observed a severe reduction in the abundance and distribution of ribosomal complexes in RBPMS-KO hESCs where heavy polysomes (the most translationally active fraction) were nearly absent (Fig. 2C). This was followed by a 50% reduction in global de novo protein synthesis, reflected in the substantial reduction of newly synthesized proteins detected by short-term puromycin labeling evaluated using either an anti-puromycin antibody or fluorescent azide–conjugated O-propargyl-puromycin (OPP) to avoid any detection biases (Fig. 2D and fig. S3, A and B). We used an independent RBPMS-KO clone (RBPMS-KO cl.2) to confirm these data (fig. S2, E to G) further. This severe global inhibition of translation upon loss of RBPMS did not alter the levels of pluripotency markers or self-renewal factors (fig. S2, C and D) over a period of >20 passages. Critically, the global nascent transcriptional output of RBPMS-KO hESCs was only marginally affected (fig. S3C). Similarly, mitochondrial integrity (fig. S3, D and E), overall mitochondrial metabolism (fig. S3G), and cell cycle (fig. S3F) remain largely unaffected, while glycolysis was marginally affected (fig. S3H) in RBPMS-KO. Together, our data reveal that loss of RBPMS exclusively inhibits mRNA translation without affecting other fundamental molecular processes in hESCs.
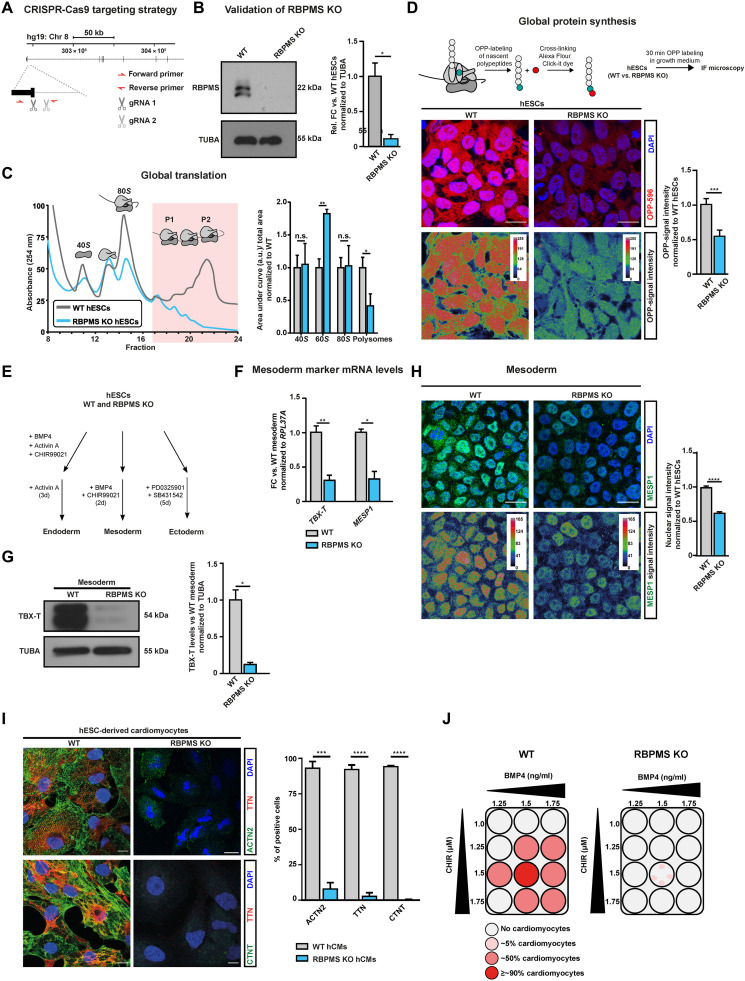
Fig. 2. RBPMS loss causes global translation inhibition in hESCs without affecting self-renewal and selectively impedes cardiac mesoderm specification.
(A) Schematic representation of RBPMS locus in humans and the CRISPR-Cas9–based targeting strategy used to generate homozygous RBPMS-KO (B) confirmed via immunoblot. (C) Loss of RBPMS impedes translation in hESCs, indicated by representative polysome profiles of RBPMS-KO hESCs with respect to isogenic WT along with quantification of the area under the indicated ribosomal fractions on the right. (D) De novo protein synthesis is inhibited upon RBPMS loss, evaluated by measuring puromycin incorporation on nascent polypeptides in RBPMS-KO compared to WT by measuring uptake of OPP (quantifications on the right). (E) Schematic of lineage differentiation approaches used to determine the competence of RBPMS-KO hESCs to undergo germline commitment. (F) Mesoderm commitment is severely impaired upon loss of RBPMS as indicated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for TBX-T (mesoderm) and MESP1 (cardiac mesoderm), as well as (G) Western blot for TBX-T (quantification on the right). (H) Representative images of MESP1 staining upon mesoderm induction of RBPMS-KO hESCs compared to WT (quantification on the right). (I) Immunofluorescence images for cardiac-specific ACTN2 and TTN. The bar graph shows normalized expression levels of indicated cardiomyocyte markers. (J) Schematic summarizing the cardiac differentiation efficiency along the cardiac corridor for WT and RBPMS-KO, indicating the inability of hESCs to terminally differentiate to cardiomyocytes upon RBPMS loss. Error bars represent ±SEM; P values calculated using Student’s t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n = 3).
RBPMS loss in pluripotency selectively impedes cardiac mesoderm specification
Because RBPMS loss abrogated translation homeostasis in hESCs, we reasoned that its loss would hamper cell fate decisions enabling lineage commitment, a process heavily dependent on the de novo synthesis of fate commitment factors. To investigate the role of RBPMS in this process, we used a defined and directed differentiation method toward the three primary germ layers: ectoderm, mesoderm, and endoderm, recapitulating the early embryonic cell fate decisions (Fig. 2E) (43).
Loss of RBPMS severely and specifically inhibited mesoderm commitment without affecting endoderm and ectoderm differentiation (Fig. 2F and fig. S4, A to D). It abolished the ability of hESCs to effectively activate TBX-T (BRACHYURY), a master regulator of mesoderm commitment, and MESP1, a key early cardiac mesoderm marker (Fig. 2, F to H, and fig. S2G) (44, 45). In addition, upon mesoderm induction, RBPMS-KO cells still expressed pluripotency factors aberrantly (fig. S4F), indicative of the inability of RBPMS-KO cells to efficiently exit pluripotency and undergo mesoderm lineage commitment upon mesoderm instructive morphogen signaling. To confirm that mesoderm commitment defects due to RBPMS loss were not a result of disrupted timing, we analyzed the expression dynamics of key mesoderm markers at close intervals. Markers such as T and MIXL1, as well as WNT signaling mediators, failed to activate in RBPMS-KO cells over the course of 24 hours of mesoderm induction (fig. S4E) (46–48).
Next, we tested whether the impaired differentiation of RBPMS-KO hESCs toward the mesodermal lineage detrimentally affects terminal fate choices. In this regard, we chose defined differentiation to cardiomyocytes because it is a robust, high-efficiency method allowing near-synchronous differentiation to a functional terminal fate (43, 47). Loss of RBPMS severely affected the formation of cardiac progenitor cells while aberrantly retaining signatures of pluripotency (fig. S5J). RBPMS-KO cells failed to properly activate key genes defining cardiac identity, including aberrant expression of cardiac-specific transcription factors and sarcomeric genes, and to produce cardiomyocytes in contrast to WT cells that constantly yielded homogeneous populations of cardiomyocytes (Fig. 2I and fig. S2, H, I, and G). In addition, RBPMS-KO cells failed to produce cardiomyocytes across the “cardiac corridor” (Fig. 2J), which is a bone morphogenetic protein (BMP)/WNT concentration grid for testing the ability of pluripotent stem cells to give rise to cardiomyocytes.
Last, to test whether the loss of RBPMS in pluripotency affects only the cardiac mesoderm or other closely related lineages, we systematically tested the ability of RBPMS-KO hESCs to differentiate into paraxial mesoderm, endothelium, definitive endoderm, and mesenchymal stem cells. We observed only a marginal effect in paraxial mesoderm differentiation, while the RBPMS-KO showed a higher propensity to commit to definitive endoderm, indicated by the significantly higher expression of SOX17 and FOXA2 (fig. S5, E to G). Furthermore, RBPMS loss did not affect the differentiation to endothelial or mesenchymal lineages (fig. S5, H and I). Thus, we conclude that RBPMS is essential for accurate cell fate decisions, allowing cardiac mesoderm commitment and cardiac differentiation of hESCs.
RBPMS is essential for cardiac mesoderm patterning and morphogenesis in human cardioids
Development of cardiomyocytes in the heart requires complex, rapid patterning and morphogenesis events in the cardiac mesoderm and the developing heart [events that cannot be recapitulated using directed two-dimensional (2D) differentiation methods] (46). To understand whether loss of RBPMS in hESCs impedes cardiac mesoderm patterning, morphogenesis, and 3D organization (molecularly less understood processes in humans compared to cardiac cell fate decisions), we resorted to a recently introduced human cardiac organoid (cardioid) model (Fig. 3A and fig. S5, A to C). Cardioids closely recapitulate otherwise hard-to-study cellular complexity, patterning, and stratification of a developing human heart, including the molecular contributions from intercellular signaling and morphogenetic events, like chamber formation.
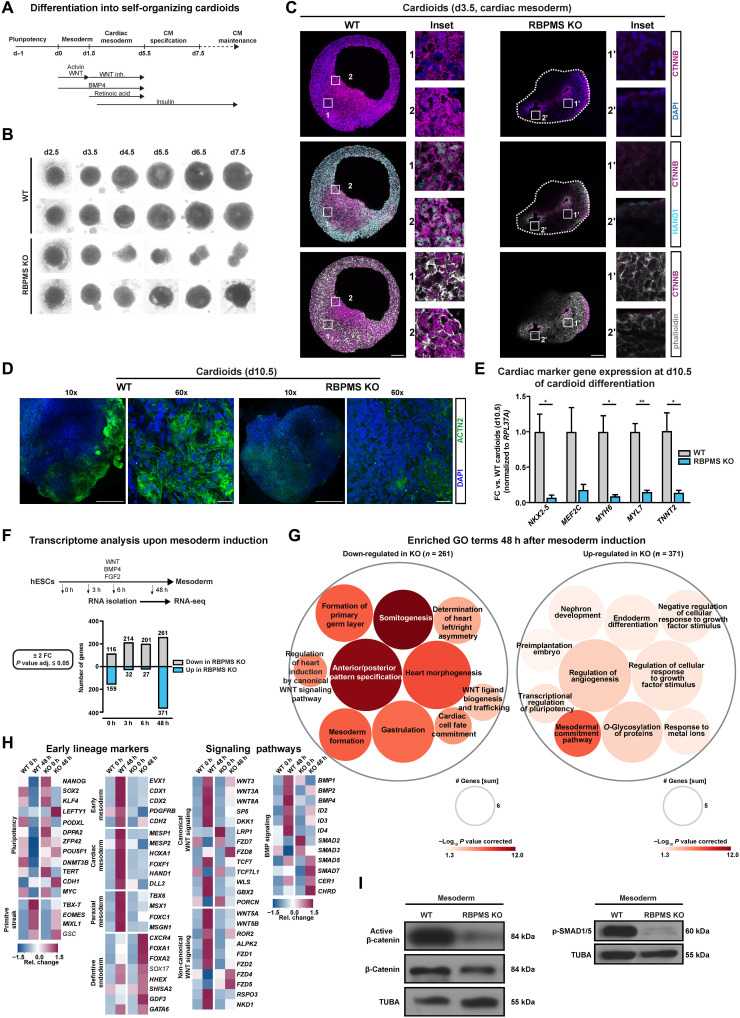
Fig. 3. RBPMS is essential for cardiac mesoderm patterning and morphogenesis in human cardioids.
(A) Schematic of cardioid generation method. (B) Loss of RBPMS impairs cardioid formation at early stages of cardiogenesis as indicated by bright-field images taken at indicated days during cardiac induction. (C) Whole-organoid confocal imaging for HAND1, CTNNB, and phalloidin at cardiac mesoderm stage cardioids (d3.5) derived from RBPMS-KO and WT hESCs. Stitched images of the whole organoid acquired by 63× objective are shown. (D) Whole-organoid imaging for cardiomyocyte-specific ACTN2 in WT and RBPMS-KO cardioids (d10.5). (E) RT-qPCR analysis for mRNA levels of cardiac-specific transcription factors and sarcomeric proteins in the indicated samples, performed on cardioids at d10.5. (F) Systematic identification of differentially expressed genes at indicated early and late time points during mesoderm induction upon RBPMS loss (FC ± 2, P ≤ 0.05, n = 3) evaluated by RNA-seq, represented as a bar graph. (G) GO-based analysis of genes down-regulated and up-regulated at 48 hours after mesoderm induction indicates defects in mesoderm and cardiac mesoderm cell fate specification and WNT signaling in RBPMS-KO cells w.r.t WT. (H) Heatmaps showing the expression of cell fate markers (first two panels) and the components of WNT and BMP signaling in pluripotency and upon mesoderm induction of WT and RBPMS-KO. (I) Absence of RBPMS impairs WNT signaling activity, indicated by active β-catenin levels, and BMP signaling, indicated by pSMAD1/5, upon cardiac mesoderm induction of WT and RBPMS-KO. Error bars represent ±SEM; P values calculated using Student’s t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n = 3).
To investigate the effects on cardiac mesoderm patterning and cardiac morphogenesis by RBPMS loss in hESCs, we generated cardioids from RBPMS-KO along a closely spaced time window between mesoderm commitment and cardiomyocyte specification (Fig. 3B) and compared them to those derived from isogenic WT hESCs. RBPMS-KO cardioids were consistently smaller, lacking lumen and chambers, which often collapsed prematurely and disintegrated before reaching cardiac specification stages in line with their mesoderm commitment defects influencing patterning and morphogenesis (Fig. 3, B and C).
To further evaluate the role of RBPMS in cardiac mesoderm patterning, we analyzed cardiac mesoderm–stage cardioids (d3.5). We focused on the distribution of cardiac mesodermal cells, lumen formation, and the WNT-BMP signaling axis that is central to cardiac mesoderm patterning and morphogenesis (46, 47, 49). First, WTs displayed a large lumen; RBPMS-KO often had multiple small lumens, which later did not fuse (Fig. 3C). Second, indicating severe impairment in WNT signaling dynamics, the β-catenin levels were low in RBPMS-KO, indicating severe impairment in WNT signaling dynamics (Fig. 3C, top). In WT d3.5 cardioids, the regions close to the periphery displayed a mosaic distribution of cells in terms of nuclear/cell membrane localization of β-catenin (Fig. 3C, top inset 1), while in the dense inner regions, cells displayed uniform nuclear localization of β-catenin (Fig. 3C, top inset 2). β-Catenin was only present in small patches in RBPMS-KO and was near-exclusively localized to the cell membrane (Fig. 3C, top insets 1′ and 2′), indicating aberrant distribution and cardiac mesoderm patterning defects. In support of this, cells positive for HAND1 expression (a key WNT-BMP target marking cardiac mesoderm) were severely depleted in RBPMS-KO cardioids (Fig. 3C, middle, insets 1, 2, and 1′, 2′). The limited number of HAND1-positive cells in the RBPMS-KO scenario was distributed along the inner lumen compared to WTs. In WT cardioids, the inner part of the cardioids showed a mosaic distribution of HAND1-positive and HAND1-negative cells (Fig. 3C, middle, inset 1), while toward the periphery, the cells were evenly HAND1-positive (Fig. 3C, middle, inset 2). However, such a distinction was absent in RBPMS-KO, which only showed patches of HAND1-positive cells near the collapsed lumens (Fig. 3C, middle, insets 1′ and 2′), following the aberrant distribution and subcellular localization of β-catenin. This points to severe cardiac mesoderm patterning defects in RBPMS-KO. In agreement with the inability of RBPMS-KO to generate cardiomyocytes efficiently, the few RBPMS-KO cardioids that survived specification showed severe depletion of cardiomyocytes compared to corresponding WT (Fig. 3D), which was accompanied by a near lack of expression of cardiac-specific transcription factors and sarcomere genes (Fig. 3E). Together, our data reveal that RBPMS is central to cardiac mesoderm patterning and cardiomyocyte specification of hESCs.
RBPMS is essential to activate the regulatory program instructing cardiac mesoderm
To carefully evaluate why RBPMS-KO fails to commit to cardiac mesoderm, we performed a whole-transcriptome analysis of RBPMS-KO compared to WT [n = 3 biological replicates, poly(A)-selected mRNAs, paired-end 150–base pair (bp) stranded libraries, ~50 × 106 clean reads per sample] in a closely spaced window of mesoderm commitment differentiation (Fig. 3F and table S2, sheet 1). The time points were chosen to evaluate the transcriptional differences at the stage of pluripotency (0 hours), upon receiving the mesoderm commitment cue (3 hours), early mesoderm (6 hours), and upon mesoderm commitment (48 hours).
First, at the transcriptome level, RBPMS-KO and WTs were comparatively similar except for 275 differentially expressed genes [log2FC ≥ (+/−) 1, P value adjusted ≤ 0.05], arguing that the striking difference in the global translation upon RBPMS loss is not due to changes in mRNA levels. In addition, the differentially expressed genes did not have a direct connection to translational control or mesoderm/cardiac mesoderm commitment (Fig. 3F, graph below, and table S2, sheet 1). We observed the most significant change in developmentally relevant gene expression signature at the 48-hour time point, aligning the cardiac mesoderm differentiation defect. Notably, GO term analysis on differentially expressed genes revealed that RBPMS-KO cells, upon mesoderm induction, were unable to activate the WNT signaling network, as well as those regulating gastrulation, mesoderm, cardiac cell fate commitment, and heart morphogenesis (Fig. 3G and table S2, sheet 2). In contrast, RBPMS-KO cells retain aberrant pluripotency gene expression while also activating definitive endodermal and ectodermal genes upon mesoderm induction (Fig. 3G). Pluripotency factors, including OCT4, NANOG, ESSRB, and ZFP42, failed to be silenced in RBPMS-KO even after 48 hours, while primitive streak (e.g., TBX-T and MIXL1), early mesoderm, paraxial mesoderm, and cardiac mesoderm markers (e.g., EVX1, PDGFRB, MESP1, HAND1, TBX6, MSX, and FOXC1) were significantly down-regulated compared to corresponding WT (Fig. 3H). Notably, RBPMS-KO aberrantly activates early endodermal (e.g., FOXA2 and GDF3) and ectodermal genes (e.g., OTX2) upon mesoderm induction, suggesting that RBPMS loss disturbs germ layer decisions in hESCs (fig. S5D).
To derive molecular insights into why RBPMS-KO cells fail to efficiently generate cardiac mesoderm, we evaluated the key morphogen signal transduction machinery programming cardiac mesoderm, namely, WNT, BMP, transforming growth factor (TGF), and NOTCH (Fig. 3H and fig. S5D). Notably, a substantial number of genes involved in both canonical and noncanonical WNT signaling (e.g., WNT3, WNT8A, WNT3A, FZD7/8, CTNNB1, TCF7, DK.K.1, FZD1/2, WNT5A, WNT5B, and ROR2) failed to be activated in RBPMS-KO upon mesoderm induction. Similarly, the key mediators of BMP, NOTCH, and TGF signaling also failed to be activated, while the inhibitors of BMP signaling (e.g., CHRD) were aberrantly expressed in RBPMS-KO at the 48-hour time point (Fig. 3H and fig. S5D). This goes hand in hand with the reduced active β-catenin levels as well as reduced phosphorylation of SMAD1/5 in RBPMS-KO upon cardiac mesoderm induction (Fig. 3I). Of note, cardiac mesoderm is particularly sensitive to the duration and timing of WNT-BMP signaling axis compared to other mesodermal lineages (8, 47, 50, 51). Thus, we reason that the severe impairment of cardiac mesoderm specification, patterning, and subsequent cardiogenesis defects observed in RBPMS-KO is due to this severe imbalance in WNT, BMP, and TGF signal transduction components upon mesoderm induction. Collectively, RBPMS is essential for hESCs to activate the gene regulatory network and signal transduction machinery specifying cardiac mesoderm.
mRNAs encoding cardiac mesoderm–instructive gene regulatory network are targeted by RBPMS via 3′UTR binding
The repertoire of mRNAs targeted and regulated by RBPMS in hESCs is currently unknown. To comprehensively and stringently identify the network of mRNAs regulated by RBPMS in hESCs, we used enhanced ultraviolet (UV) cross-linking and immunoprecipitation of ribonucleoprotein complex followed by massively parallel sequencing (eCLIP-seq) (Fig. 4A and fig. S6, A and B) (52). Following the removal of polymerase chain reaction (PCR) duplicates and normalization relative to size-matched input (SMI) controls from four independent replicates, we compiled transcriptome-wide, nucleotide-resolution, and high-confidence binding maps displaying >80% overlap of target mRNAs between replicates (Fig. 4B; fig. S6, C to E; and table S3, sheet 1). Only statistically significantly enriched targets (fold enrichment over SMI ≥ 2, P ≤ 0.05) present in all four replicates were considered for further analysis.
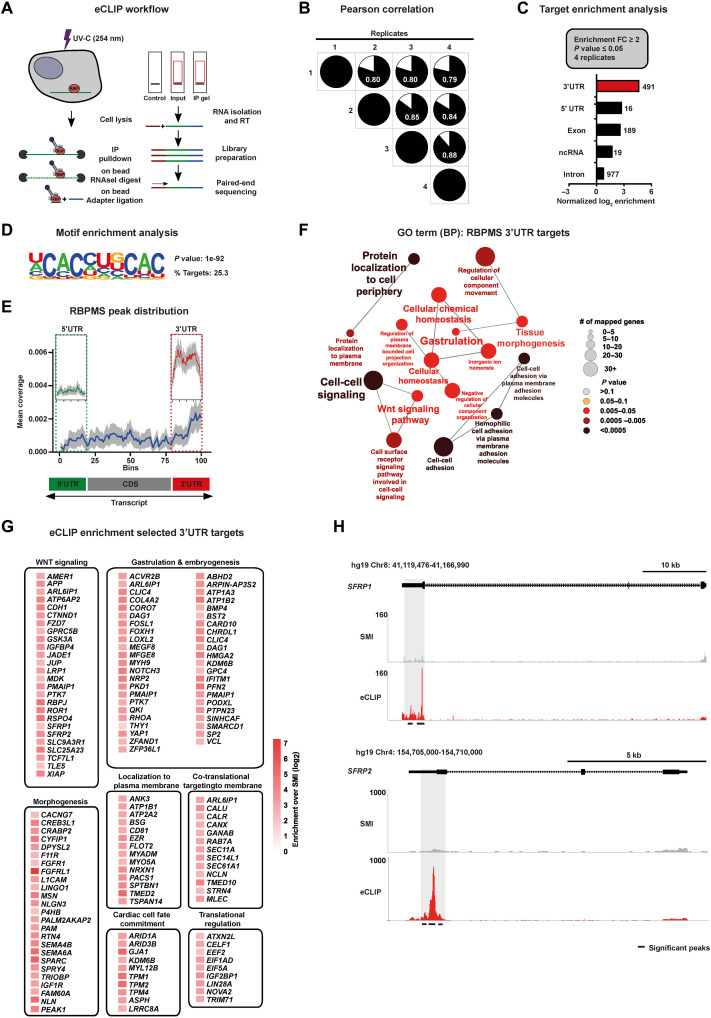
Fig. 4. mRNAs encoding cardiac mesoderm regulators are targeted by RBPMS via 3′UTR binding.
(A) Schematic of the eCLIP-seq approach used to faithfully generate a transcriptome-wide direct binding map for RBPMS at single-nucleotide resolution. (B) Biological quadruplicates of RBPMS eCLIP-seq show at least 80% overlap. Pie charts show the correlation of statistically significant uniquely mapped reads for each replicate over SMInput. (C) RBPMS reliably binds predominantly the 3′UTR of transcripts, demonstrated here by the distribution of the significantly enriched eCLIP peaks against the paired SMInput (FC ≥ 2; P ≤ 0.05 in all four replicates). (D) Top sequence motif significantly bound by RBPMS. (E) Metagene plot visualizing the RBPMS peak distribution over SMInput illustrating prominent 3′UTR binding. (F) 3′UTR targets of RBPMS regulate molecular processes central to mesoderm/cardiac commitment, including WNT signal transduction, depicted by significantly enriched GO terms. (G) A curated set of RBPMS 3′UTR targets grouped based on their proven role in the indicated cellular, developmental, and functional process, depicted as a heatmap of fold enrichment over SMInput. (H) Representative read density tracks show read density for RBPMS across the gene body of SFRP1 and SFRP2, a representative target.
RBPMS was predominantly found to bind 3′ untranslated regions (3′UTRs) of mRNAs (Fig. 4C and table S3, sheet 2) on a bipartite CAC motif (Fig. 4D and fig. S6F). RBPMS showed a higher degree of enrichment in the 3′UTR of target mRNAs compared to other regions, supported by normalized peak enrichment (Fig. 4C). This was further evident when calculating RBPMS peak distribution in metagene plots, which showed substantial 3′UTR bias (Fig. 4E), and in the representative loci of SFRP1 and SFRP2 (Fig. 4H).
Indicative of the direct role of RBPMS in regulating cardiac mesoderm fate of hESCs, the functional annotation of 3′UTR-bound mRNA targets revealed significant enrichment for key mediators of gastrulation, WNT signaling, as well as cell surface and secreted proteins involved in developmental signaling (Fig. 4F and table S3, sheet 3). Furthermore, the curation of 3′UTR targets based on high signal-over-input enrichment followed by grouping based on their molecular and developmental functions revealed that RBPMS targets mRNAs involved in early embryonic cell fate decisions and cardiac mesoderm development. This included regulators of WNT signaling (e.g., FZD7, GSK3A, SFRP1, and SFRP2), gastrulation and embryogenesis (e.g., FOXH1, MYH9, YAP1, BMP4, SMARCD1, and SP2), morphogenesis (e.g., FGFR1, MSN, IGF1R, and NLN), and cardiac cell fate commitment/cardiac identity (e.g., ARID1A, KDM6B, MYL12B, and TPM1) (Fig. 4G). In addition, mRNAs encoding translational regulators were also bound by RBPMS at their 3′UTR (e.g., CELF1, EEF2, EIF5A, IGFBP1, and LIN28A), thereby offering an explanation of how the loss of RBPMS affects global translation in hESCs (Fig. 4G). The 3′UTR targets of RBPMS encode proteins mostly localized to all subcellular locations, with a bias for membrane and secreted proteins (fig. S6G and table S3, sheet 4). Together, we found that RBPMS directly targets a network of mRNAs encoding central regulators of early embryonic cell fate decisions, especially those critical for mesoderm instructive morphogen signaling, core components of the translation machinery, and regulators of mRNA translation.
RBPMS controls mRNA translation of factors that are essential to initiate cardiac mesoderm commitment
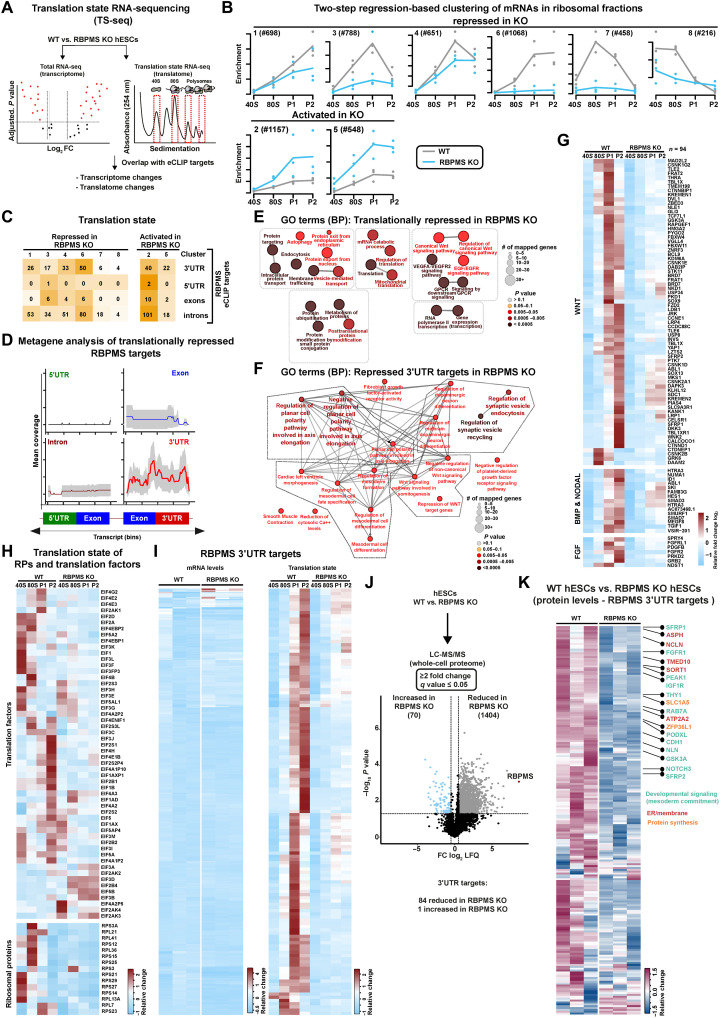
To determine how the loss of RBPMS causes translation inhibition in hESCs and prevents cardiac mesoderm commitment, we first applied translation state RNA sequencing (TS-seq) to investigate transcriptome-wide changes in the occupancy of ribosomal complexes upon RBPMS loss in hESCs. To this end, transcripts associated with ribosomal complexes (the 40S, 80S, light, and heavy polysomes) were isolated after ribosome fractionation, enriched for poly-adenylated transcripts, and subjected to transcriptome sequencing in parallel with total RNA from RBPMS-KO and isogenic WT hESCs (Fig. 5A). To correct for technical variability and allow data normalization, two different sets of spike-ins were added to each fraction of our three biological replicates, after lysis and after polysome fractionation, respectively. We obtained 20 million to 30 million clean reads per ribosomal fraction per replicate to ensure reliable quantification of ribosome occupancy differences in low- to medium-expressed transcripts.
Fig. 5. RBPMS controls mRNA translation of factors essential to initiate cardiac mesoderm commitment.
(A) Schematic of the TS-seq strategy used to evaluate the translational status of RBPMS-KO compared to WT (n = 3). (B) Global impact of the loss of RBPMS on ribosome occupancy in hESCs, revealed by two-step regression analysis of the mRNAs enriching on indicated ribosomal fractions derived from TS-seq. (C) Translation state of mRNAs bound by RBPMS in RBPMS-KO compared to WT, grouped based on RBPMS binding coordinates, in the indicated translationally affected clusters identified by TS-seq. (D) Metagene plot revealing RBPMS 3′UTR binding bias for translationally repressed RBPMS targets. (E) Functional analysis of all translationally repressed mRNAs and (F) translationally repressed 3′UTR targets in RBPMS-KO versus WT hESCs illustrated as a significantly enriched, curated list of GO terms. (G) Loss of RBPMS severely inhibits translation of the components of vital mesoderm specifying signal transduction networks (WNT, BMP, NODAL, and FGF signaling), as well as (H) translation factors and ribosomal proteins. (I) mRNAs bound by RBPMS at the 3′UTR are depleted from ribosomes in RBPMS-KO without affecting the transcript levels. The heatmap on the left depicts mRNA levels of RBPMS 3′UTR targets, while the heatmap on the right depicts their ribosome occupancy. (J) Changes in total proteome between WT hESCs and RBPMS-KO hESCs depicted as a volcano plot derived from whole-cell proteomics analysis. (K) Heatmap depicting protein levels (as log2 LFQ values) of RBPMS 3′UTR targets in WT and RBPMS-KO hESCs.
The loss of RBPMS resulted in severe translational inhibition (Fig. 2), which makes the evaluation of changes in ribosome occupancy on specific mRNAs cumbersome. Therefore, a two-step regression-based clustering approach was used to identify meaningful differences in ribosome occupancy after normalization with dual spike-in controls. This approach allows for the identification of clusters of transcripts that were significantly different in their distribution of ribosomes, taking into account their occupancy across ribosomal complexes in the RBPMS-KO hESCs in comparison to isogenic WTs.
We identified 8 mRNA clusters harboring >5500 mRNAs that exhibit a significant difference in their translation status following RBPMS-KO. Notably, upon RBPMS loss, ribosomal complexes were severely depleted in six clusters that harbored most of the translationally affected transcripts, while two clusters showed enrichment (Fig. 5B and table S4, sheet 2). Translationally repressed genes were crucial ones for cardiac cell fate commitment and protein and mRNA metabolism (Fig. 5E), while translationally activated genes were involved in neurogenesis and endoderm and ectoderm development (fig. S7B and table S4, sheets 5 and 9). Integrative analysis of transcriptomics and TS-seq data revealed that transcripts only showing transcriptional changes were not directly implicated in morphogen signaling or cardiac mesoderm development, the processes detrimentally affected by the loss of RBPMS (fig. S7A and table S4), in agreement with our transcriptome analysis of RBPMS-KO versus WT hESCs (Fig. 3F and table S2, sheet 7).
Next, we investigated the translation status of RBPMS targets we identified in hESCs in relation to where it binds on the mRNAs (binding coordinates). In line with the prominent 3′UTR binding on developmentally relevant genes, a substantial number of 3′UTR-bound RBPMS targets were depleted from ribosomes in RBPMS-KO (Fig. 5, C and I; fig. S7D; and table S4, sheets 3 and 4). Metagene analysis of translationally affected RBPMS targets revealed that the transcripts bound by RBPMS at the 3′UTR are depleted from ribosomes. In contrast, those bound at the 5′UTR, exons, and introns were not significantly affected. To further ensure that the depletion of ribosomes from its 3′UTR targets in the RBPMS-KO is not due to any indirect effects on mRNA levels, we systematically compared the transcript levels of 3′UTR targets and their ribosome occupancy upon loss of RBPMS. The substantial majority of the 3′UTR targets were translationally repressed in RBPMS-KO (Fig. 5I, heatmap on the right), while the respective transcript levels of these targets remain largely unaffected (Fig. 5I, heatmap on the left). These data further reveal that RBPMS binding at the 3′UTR determines the translational status of its target mRNAs (Fig. 5D).
Translationally inhibited 3′UTR targets of RBPMS encode regulators central to mesoderm specification, cell fate commitment, and morphogen signaling, including WNT (Fig. 5F and table S4, sheet 6). Because morphogen signaling, particularly by WNT, BMP/NODAL, and fibroblast growth factor (FGF), defines mesoderm commitment from pluripotency, we then investigated the ribosome occupancy on mRNAs relevant to these processes (8, 46, 53, 54). Notably, most WNT signal transduction components and those of BMP/NODAL and FGF signaling were severely depleted from active translational compartments upon RBPMS-KO (Fig. 5G and table S4). Of note, a subset of 3′UTR targets of RBPMS was translationally activated. They regulate processes not related to translation and cardiac mesoderm commitment (fig. S7C).
Unexpectedly, we observed a depletion of mRNAs encoding core translation machinery, including translation initiation factors and ribosomal proteins in RBPMS-KO (Fig. 5H and table S4). However, only a few of these were directly bound by RBPMS. This could further explain the global inhibition of translation upon loss of RBPMS (Figs. 2, D and E, and 5B). This supports a model by which RBPMS selectively regulates the translation of its client mRNAs through 3′UTR binding and globally influences translation in an mRNA binding–independent fashion. Its role in general translation could be through its direct interaction with translation machinery as indicated by ARC-MS and its association with the 40S complex (Fig. 1, G and M). RBPMS does not affect transcript stability (assessed for a selection of pluripotency factors and direct RBPMS 3′UTR targets involved in WNT signaling following actinomycin D treatment to inhibit transcription; fig. S7E).
RBPMS was suggested to regulate splicing in smooth muscle cells, extrapolated from targets identified by overexpression of RBPMS in human embryonic kidney (HEK) 293T cells, which do not naturally express RBPMS (55). A cursory analysis revealed minimal overlap between the targets reported in HEK293T cells and those we identified in hESCs, implying cell type specificity. Nevertheless, despite the 3′UTR binding bias in RBPMS eCLIP data, we also detected low-affinity binding at introns (Fig. 4C and table S3)
To address whether RBPMS contributes to splicing in both hESCs (0 hours) and during mesoderm commitment (3, 6, and 48 hours after mesoderm induction), we computed splicing changes in RBPMS-KO. Our sequencing depth (~50 million clean reads/sample/replicate from stranded poly-A selected library, n = 3 biological replicates) allowed reliable investigation in changes in mRNA splicing (56, 57). Briefly, we determined ψ scores for WT and RBPMS-KO at the state of pluripotency and along the mesoderm commitment time course, denoting significant changes with an FDR ≤ 0.01. A minimum of 10 junction reads were counted to compute inclusion/exclusion events to robustly call splicing changes. Using a relaxed cutoff of Δψ ≤ −0.5 (inclusion) or Δψ ≥ 0.5 (exclusion) to detect a maximum number of splicing changes, we could only find minimal differences regardless of the time point investigated. These few changes cannot explain the profound effects on mRNA translation or cardiac mesoderm commitment in RBPMS-KO cells (fig. S8, A and B, and table S5). In d0 data, none of the 3′UTR and 5′UTR targets of RBPMS were affected at the level of splicing, while only two of its intronic and one of its exonic targets were among the genes whose splicing was affected. Similar trends were observed upon mesoderm induction time course, thus confirming our initial hypothesis that RBPMS controls mRNA translation without directly influencing mRNA splicing in hESCs and during mesoderm commitment (fig. S8, A and B, and table S5).
Last, we confirmed that the depletion of ribosomal complexes globally and from RBPMS 3′UTR targets results in a significant reduction in the abundance of corresponding proteins by performing an in-depth whole proteome analysis using LC-MS comparing WT and RBPMS-KO hESCs (Fig. 5, J and K). Notably, 1404 proteins were reduced in RBPMS-KO, while only 70 were increased, confirming that RBPMS is essential for protein homeostasis in hESCs. Specifically, the protein levels of the 3′UTR targets of RBPMS were significantly reduced in RBPMS-KO, including WNT signal transduction components and mesoderm regulators (Fig. 5K and fig. S7F), thus confirming the pivotal role of RBPMS in controlling their protein abundance in hESCs.
Together, our data reveal that RBPMS is a central regulator of mRNA translation in hESCs, primarily controlling the components of mesoderm-instructive morphogen signaling, regulators of cell fate decisions, and mRNA translation (Fig. 5, E to G). Thus, RBPMS primes the selective translation of factors essential to initiate cardiac fate programming in hESCs, revealing that the competence for committing to the cardiac fate is predetermined by the RBPMS-mediated translation circuit already instated in pluripotency.
RBPMS specializes mRNA translation in pluripotency, selectively via 3′UTR binding and globally by controlling translation initiation and ribosome recruitment
Our data so far suggest that the role of RBPMS in translation could be two-pronged: (i) as an activator of selective translation of mesoderm instructive cell fate regulators via 3′UTR binding, and (ii) as a general regulator of translation in hESCs via recruitment to ribosomal complexes.
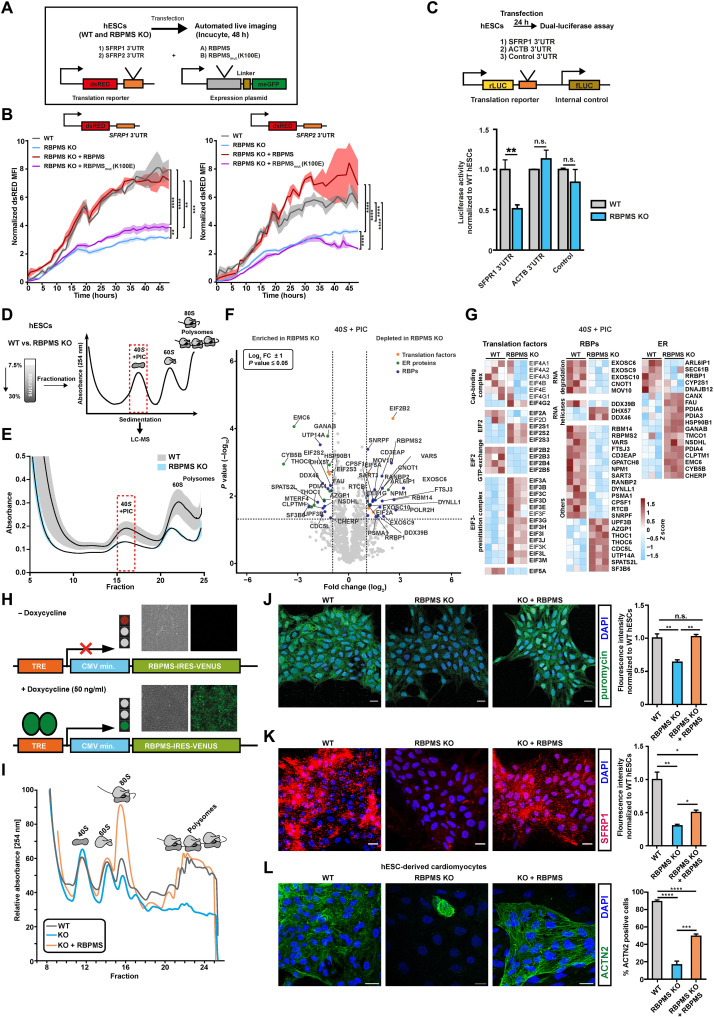
Therefore, we asked whether RBPMS binding at the 3′UTR can selectively control translation in hESCs. To this end, we first generated a destabilized dsRED-based reporter system carrying the 3′UTRs of two RBPMS targets, SFRP1 and SFRP2, inferred by RBPMS eCLIP-seq in hESCs (Fig. 6A). The translation state and protein levels of both SFRP1 and SFRP2 are RBPMS dependent in hESCs (fig. S7, D and G). As evidenced by time-lapse microscopy, dsRED signal intensity was significantly reduced in RBPMS-KO compared to WT hESCs (Fig. 6B). This reduction could be rescued by ectopically expressing WT RBPMS, but not by RBPMS mutant carrying a point mutation (K100E) abolishing the RNA binding ability (Fig. 6B). To confirm the ability of RBPMS to selectively control translation in a 3′UTR-dependent manner, we generated a set of dual luciferase-based bicistronic reporter constructs with or without RBPMS binding sites in the 3'UTR (with SFRP1 or ACTB 3′UTR, respectively) (Fig. 6C, illustration). RBPMS loss led to a significant reduction in luciferase activity for SFRP1-3′UTR fusions, while ACTB-3′UTR and luciferase-only controls remained unaffected (Fig. 6C). Collectively, these reporter-based assays reveal that RBPMS selectively activates translation of mRNAs carrying its binding sites at the 3′UTR.
Fig. 6. RBPMS specializes mRNA translation in pluripotency, selectively via 3′UTR binding and globally by controlling translation initiation and ribosome recruitment.
(A) Schematic of the reporter system and the experimental workflow used to investigate the 3′UTR binding motif–dependent regulation of translation by RBPMS in hESCs. (B) RBPMS activates translation of reporter mRNA carrying RBPMS binding motifs in the 3′UTR, evaluated by time-lapse microscopy. (C) The presence of the RBPMS binding motif is required for 3′ binding–dependent translation activation by RBPMS evaluated using indicated luciferase-based bicistronic reporters. (D) Schematic outlining the translation complex profiling–based isolation of 40S and preinitiation complex (PIC), followed by proteomics analysis in WT and RBPMS-KO hESCs. (E) Translation complex profiling traces of WT and RBPMS-KO hESCs (shades represent SEM). 40S + PIC fractions were subjected to LC-MS/MS. (F) Proteins significantly changing in the 40S + PIC fraction between WT and RBPMS-KO hESCs represented as volcano plot. Dashed lines indicate significance thresholds (−log10 P ≥ 1.3 and log2 FC ± 2) (selected translation factors, ER proteins, and RBPs are highlighted by orange, green, and blue dots, respectively). (G) Heatmap depicting differentially enriched translation factors, RBPs, and translation-associated ER proteins in the 40S + PIC fraction between WT and RBPMS-KO hESCs (significantly changing proteins are highlighted in bold). (H) Illustration of the PiggyBac-based strategy used to reexpress RBPMS in RBPMS-KO. Representative microscopy images in the inlets before and after induction. Timely reconstitution of RBPMS in RBPMS-KO hESCs rescues (I) translation defects, (J) protein synthesis defects, (K) translation defect of representative 3′UTR target of RBPMS, SFRP1, and (L) cardiac differentiation defect. Quantification of the microscopy images on the right side as bar graphs. Error bars represent ±SEM; P values calculated using Student’s t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; n = 3).
To obtain a further mechanistic understanding of RBPMS-mediated global translational control, we first performed immunoprecipitation of RBPMS from hESCs, followed by proteomics analysis after prolonged RNase I treatment (to avoid indirect RNA-mediated associations) (fig. S9, A to C, and table S6, sheet 1). RBPMS co-purified with translational regulators, ribosomal proteins, and proteins involved in endoplasmic reticulum (ER)–mediated translation, including canonical regulators of translation initiation (fig. S9, C and D). Notably, EIF3 complex components, EIF5A and EIF4G, along with multiple ribosomal subunits, were specifically and significantly enriched with RBPMS, suggesting that RBPMS can influence the residence of key regulatory components on the translation apparatus of hESCs. This is in line with the enrichment of RBPMS on 40S complexes upon treatment with translation inhibitors and RNase (Fig. 1M). Next, we evaluated the distribution translation initiation factors interacting with RBPMS on the ribosomal complexes upon RBPMS loss by polysome profiling followed by Western blotting (fig. S9E). First, we specifically examined the enrichment of EIF4G (that mediates the cross-talk of the 43S preinitiation complex with EIF4F complexes) and of the poly(A)-binding protein PABP. Both displayed comparable levels, indicating that RBPMS loss does not influence the predisposition of mRNAs to be translated (fig. S9E). However, a key component of the 43S preinitiation complex and a regulatory hub for global translation, EIF2A, showed aberrant retention across ribosomal fractions following RBPMS loss (fig. S9E) (58). In addition, two key EIF3 complex components, EIF3E (involved in selective translation) and EIF3H (reported to be involved in selective translation during embryonic development), were aberrantly retained in polysomal fractions and markedly depleted from the 40S complex in RBPMS-KO cells, respectively (fig. S9E) (59, 60). Notably, EIF5A, essential for translation initiation, elongation, error resolution at ribosomal pause sites, and termination, was sequestered in the 40S ribosomal fraction in the absence of RBPMS (fig. S9E) (61). The total levels of these factors remain unchanged and do not reflect the change in their distribution pattern in ribosomal fractions in RBPMS-KO hESCs (fig. S9F). Motivated by these data, we next asked whether the loss of RBPMS disrupts the abundance and assembly of translation initiation complexes on translationally engaged mRNAs in hESC. To this end, we used translation complex profiling (Fig. 6, D and E), a variant of polysome profiling that is specifically designed to analyze translation initiation complexes (62). To have an unbiased evaluation of the effect of the loss of RBPMS on the translation initiation complexes, we subjected the fraction containing 40S and the preinitiation complex to LC-MS–based proteomics analysis (Fig. 6D and table S6, sheet 2). The loss of RBPMS disrupted the composition of translation initiation complexes, including the cap-binding complex, EIF2 complex, and EIF3 complex (Fig. 6G). Notably, components involved in guanosine 5′-triphosphate exchange in the EIF2 complex were significantly depleted, while the majority of the EIF3 complex where aberrantly retained on the initiation fractions upon RBPMS loss. As observed by the polysome fractionation-based analysis (fig. S9E), EIF5A was significantly depleted (Fig. 6G). Notably, crucial mediators of ER-associated translation, such as SEC61B and RRBP1, were significantly reduced due to the loss of RBPMS. In addition, several RBPs, including RANBP2, known to enhance the translation of secretory proteins, and the components of RNA degradation machinery were depleted upon RBPMS loss, while RNA helicases such as DHX57 and DDX46 were enriched in the absence of RBPMS (Fig. 6G). Thus, our data show that loss of RBPMS causes translation initiation defects highlighted by aberrant retention of the EIF3 complex and depletion of EIF5A from mRNAs, revealing its role in regulating global translation in hESCs. Collectively, our data confirm RBPMS as a functional RAP essential for global translation in hESCs and a selective translation activator of target mRNAs via 3′UTR binding.
Next, to confirm that RBPMS determines the translation status in hESCs, its competence to mesoderm and cardiac commitment, and to rule out discrepancies stemming from genome engineering “off-target” effects in RBPMS-KO hESCs, we knocked in an inducible copy of RBPMS using PiggyBac transposon-based genomic insertion (hereafter RBPMS-KO + RBPMS) (Fig. 6H). Timely reexpression of the cytosolic isoform of RBPMS that is most abundant in hESCs (fig. S9G) in RBPMS-KO cells fully restored both ribosome occupancy defects (Fig. 6I) and protein synthesis (Fig. 6J), including SFRP1 protein levels, an RBPMS 3′UTR target, and WNT signaling mediator (Fig. 6K). Notably, RBPMS reexpression also restored mesoderm commitment capacity, now allowing RBPMS-KO cells to generate terminally differentiated cardiomyocytes (Fig. 6L). To assess whether the absence of RBPMS beyond initial mesoderm induction influences terminal differentiation into cardiomyocytes, we performed a controlled reconstitution of RBPMS, restricted either to cardiac mesoderm commitment or during the entire duration of cardiac commitment. Reconstitution of RBPMS only during the cardiac mesoderm stage was sufficient to rescue the cardiomyocyte differentiation defects in RBPMS-KO (fig. S9H). Furthermore, reexpression of RBPMS in RBPMS-KO could rescue the cardiac commitment defects in our cardioid model (fig. S9I). However, the expression levels and distribution of cardiomyocyte markers could only be reinstated partially, compared to the WTs. In summary, we show that the competence of hESCs to commit to cardiac lineage program is preset by RBPMS-mediated selective mRNA translation circuit.
DISCUSSION
Appropriate abundance and balance of cell fate–determining morphogen signaling components at the state of pluripotency is vital for the ability of hESCs to undergo accurate lineage decisions. This is especially important for mesoderm and cardiac mesoderm differentiation and patterning, where early exposure to WNT-BMP-NODAL signaling dosage encountered by cells committing to mesoderm that transgresses the primitive streak is postulated to predetermine their ability to commit to future cardiac lineages both in vitro and in vivo (8, 47, 50, 51, 63). How this unique predisposition to a future terminal fate is molecularly regulated is currently unknown. Our work reveals that the competence of hESCs for future cardiac commitment is already predetermined at the state of pluripotency in a specialized mRNA translational circuit controlled by RBPMS. Highlighting its key role in early embryonic cell fate decisions, RBPMS selectively primes the translation of regulatory components essential to initiate the cardiac commitment program, especially WNT signaling. The RBPMS-mediated selective mRNA translation circuit licenses the abundance of “morphogen signaling infrastructure” necessary to authorize cardiac mesoderm in hESCs. Thus, RBPMS presets the future cardiac commitment competence of hESCs by programming selective mRNA translation. On the basis of our findings, we propose that RBPMS is a translation specialization factor.
We propose translation specialization as a regulatory mechanism that primes ribosomes to control translation temporally and/or spatially for a set of mRNAs necessary for future events in response to particular stimuli or fate transitions. This allows efficient division of labor among the ~10 M ribosomes present in each cell, which are tasked with synthesizing ~2 M proteins/min, so the flow of information is streamlined and, as we show, specialized.
Mechanistically, on the one hand, RBPMS associates with components of translation initiation complexes, and its loss abrogates translation initiation and ribosome recruitment, primarily by disrupting EIF2 and EIF3 complexes. RBPMS loss severely depletes the translation apparatus of the key “surveillance” factor EIF5A. These points to a role for RBPMS in shaping translation initiation in hESCs as its absence leads to global inhibition of translation. On the other hand, RBPMS selectively regulates translation of mesoderm instructive signal transduction components in hESCs by binding target 3′UTRs via its specific mRNA binding motif. Our reporter assays showed that the insertion of RBPMS recognition elements in the 3′UTR suffices for boosting translation. Thus, reinstating RBPMS levels rescues translation defects and restores the cardiac mesoderm specification capacity of hESCs. Notably, not all RBPMS targets are translationally repressed. This is not uncommon for selective regulators of mRNA translation. For instance, the loss of EIF3D, a translation factor enabling selective translation upon stress, leads to both activation and inhibition of mRNAs in a context-dependent manner, in addition to its role as a general translation initiation factor (64, 65).
Same RBPs can affect distinct (or multiple) aspects of RNA processing, including mRNA splicing, localization, and translation in a cell type, developmental stage/context-dependent fashion (36, 57). For example, QKI is a regulator of mRNA splicing in cardiomyocytes, while it is reported to regulate translation/mRNA localization in astrocytes and germline lineages (66, 67). Thus, it is also possible that RBPMS controls distinct mRNA processing and regulatory mechanisms in a cell type/context-dependent fashion, as reported in smooth muscle cells and recent mouse cardiomyocytes (41, 55). The mRNA targets of RBPMS and its proposed role in pre-mRNA splicing in terminally differentiated mice cardiomyocytes remain to be experimentally determined. In contrast, in hESCs, we reveal its role as a RAP selectively recruited in active ribosomes and controlling mRNA translation without directly influencing transcription, mRNA splicing, or mRNA abundance.
In summary, we propose a model by which the state of pluripotency is translationally poised for differentiation into future lineages via specialized translation of the regulators of embryonic cell fate. Our work reveals that cell fate–specific translation specialization factors selectively program the translation of mRNAs encoding key developmental regulators that are essential for initiating future cell fate choices, akin to how pioneering transcription factors program specific transcriptional networks allowing cell fate decisions. Therefore, we reveal a pivotal role for translational specialization in sculpting cellular identity during early developmental lineage decisions and propose that ribosomes act as a unifying hub for cellular decision-making rather than a constitutive protein synthesis factory.
MATERIALS AND METHODS
Human pluripotent stem cells and culture conditions
HuES6 (genotype:female) cells [shared by B. Greber’s laboratory, Max Planck Institute for Molecular Medicine (Muenster, Germany)], derived from inner cell mass of blastocysts, were used for the in vitro differentiation experiments. Cardiomyocyte reporter human-induced pluripotent stem cell (hiPS)C line, WTC MYL7-GFP iPSC, was a gift from the Mendjan laboratory, IMBA Vienna. Authenticated cell lines were provided by the indicated providers. The work on human embryonic stem cells (hESCs) was done with approval from the Robert Koch Institute (permission number: AZ: 3.04.02/0145). Human pluripotent stem cells (hPSCs) were maintained on Matrigel-coated six-well dishes in FTDA medium. FTDA medium contains Dulbecco’s modified Eagle’s medium (DMEM)/F-12 supplemented with 1× penicillin/streptomycin/glutamine, 1× insulin/transferrin/selenic acid, 0.1% human serum albumin, lipid mix (1:100), FGF2 (50 ng/ml), TGFβ1 (0.2 ng/ml), 50 nM dorsomorphin, and activin A (4 ng/ml). For passaging, cells were washed with phosphate-buffered saline (PBS) and dissociated with Accutase, supplemented with 10 μM ROCK inhibitor (Y-27632) for 10 min at 37°C. Accutase was blocked with DMEM/F-12, and the desired number of cells was centrifuged for 2 min at 300g at room temperature (RT). The cell pellet was resolved in 2 ml of FTDA medium, supplemented with 10 μM Y-27632, and pipetted onto fresh Matrigel-coated plates, followed by 24-hour incubation at 37°C and 5% CO2. After 24 hours, the medium was changed to 2.5 ml of fresh FTDA medium. Medium change was repeated daily with increasing FTDA volumes, until cells reached confluency.
Methods details
RBPMS-KO using CRISPR-Cas9 in hESCs
For the generation of CRISPR-Cas9–mediated RBPMS-KO, gRNA pairs targeting exon-intron boundary of exon 1 of RBPMS (rationally designed accounting for off-target effects and tested for targeting efficiency) were cloned into px330A and transfected into HuES6 hESCs. Independent RBPMS-KO clones were selected, picked, and expanded as we previously reported (43).
Generation of stable inducible RBPMS expression lines
For the generation of doxycycline-inducible RBPMS expression in RBPMS-KO hESCs, PiggyBac-based transposon-mediated genomic insertion was performed as we previously reported (43).
Mesoderm differentiation protocol
For mesoderm differentiation, we adapted the protocol (43). Briefly, we seeded cells in FTA medium (FTDA without dorsomorphin) supplemented with 10 μM Y-27632 (see maintenance of hPSCs), which was lacking dorsomorphin. Upon reaching 70% confluency, cells were treated with mesoderm induction medium [FTA + 5 μM CHIR99021 and BMP4 (5 ng/ml)] for 24 hours at 37°C. Cells were collected for downstream experiments 24 hours after mesoderm induction.
Cardiomyocyte differentiation
Cardiomyocyte differentiation was performed as previously described (43, 47).
Cardioid differentiation
For the generation of cardiac organoids (cardioids), media and conditions were adapted from (49). Briefly, hPSCs were grown in FTDA to approximately 70% confluency. For cardioid formation, 7500 cells per well were seeded into ultralow-attachment 96-well plates (Corning) and centrifuged for 5 min at 200g. After 24 hours, cells were induced with FLyABCH medium [Cardiac Differentiation Medium (CDM) containing FGF2 (30 ng/ml; Proteintech), activin A (50 ng/ml; Miltenyi Biotech), BMP4 (10 ng/ml; R&D Systems), 3 μM CHIR (Tocris), 5 μM LY294002 (Tocris), and insulin (1 μg/ml; Roche)] for 36 to 40 hours. After this, cells were treated with BFIIWPRa medium [CDM, containing FGF2 (8 ng/ml), BMP4 (10 ng/ml), 1 μM IWR-1 (Tocris), and 0.5 μM retinoic acid (Sigma-Aldrich)] for 96 hours with medium change every 24 hours. Following this, medium was changed to BFI [CDM, containing BMP4 (10 ng/ml), FGF2 (8 ng/ml), and insulin (10 μg/ml)] for 48 hours with medium change after 24 hours. After 48 hours in BFI, cells were kept in CDM + I (10 μg/ml of insulin) until being harvested or used for imaging. For RNA isolation, three organoids were pooled per replicate and transferred into 250 μl of TRIzol solution. For imaging, organoids were fixed for 15 min in 4% paraformaldehyde (PFA), washed three times in PBS, and kept in at 4°C until further processing.
OPP and puromycin labeling
For OPP labeling, the reagents were prepared according to the manufacturer’s instructions (Thermo Fisher Scientific). Cells were seeded onto coverslips in FTDA. The following day, cells were treated for 30 min at 37°C with 2 μM OPP added to the growth medium. After 30 min, cells were washed with PBS and fixed by 3.7% PFA for 15 min at RT. Following fixation, cells were permeabilized with 0.5% Triton X-100 for 15 min at RT. Afterward, cells were washed once with PBS and treated with OPP reaction cocktail for 30 min. After 30 min, cells were washed once with rinse buffer, and nuclei were stained using NuclearMask Blue Stain for 30 min at RT in the dark. After washing, the cells were ready for imaging analysis.
For puromycin labeling, cells were washed once with PBS, and the medium was changed to growth medium containing puromycin (0.5 μg/ml), while the addition of cycloheximide (0.1 μg/ml) was used as a negative control. After 15- to 30-min incubation at 37°C, cells were washed once with PBS and then harvested and flash-frozen for Western blot or fixed with 4% PFA for immunostainings, respectively.
Active ribosome capture mass spectrometry
For ARC-MS, the AHARIBO protein module (Immagina) was adapted to simultaneously isolate both nascent peptides and ribosomes. Briefly, hESCs were grown in FTDA to 70% confluency, washed once with PBS, and treated for 40 min with methionine-free growth medium (Thermo Fisher Scientific) containing 10% fetal bovine serum and 0.8 mM l-leucine to deplete methionine reserves. After 40 min, 10 μl of AHA reagent was added to the medium and cells were incubated at 37°C for 5 min, following addition of 2.6 μl of sBlock for 5 min at 37°C. Afterward, cells were placed on ice and washed once with 1 ml of cold PBS. PBS was removed with a pipette, and cells were lysed using 40 μl of cold lysis buffer using a cell scraper. Cell lysate was transferred to a 1.5-ml microcentrifuge tube, and cell debris was pelleted by centrifugation at 20,000g for 5 min at 4°C. The supernatant was transferred to a new tube and kept it on ice for 10 min. Absorbance was measured by NanoDrop at 260 nm with lysis buffer as blank subtraction. For the capture of active ribosome complexes, 0.4 absorbance units (AU) was transferred to a new tube and the volume was adjusted to 100 μl with freshly prepared W buffer. To this, 100 μl of dBeads was added and the mixture was incubated for 60 min at 4°C on a rotating wheel. After 60 min, the supernatant was removed and the beads were washed twice with UWS buffer. The supernatant of these washes (containing active ribosomes and associated proteins) was combined and stored at 4°C until MS preparation via solution digest. For validation of AHA capture, two more washes in UWS were performed and beads were resuspended in 200 μl of distilled water. For proteomics analysis, an on-bead digestion was performed.
Enhanced eCLIP-seq
Enhanced cross-linking with immunoprecipitation was performed as previously described (52, 68). Briefly, hPSCs (20 million cells) were UV-crosslinked (400 mJ/cm2 constant energy), lysed in iCLIP lysis buffer, and sonicated (BioRuptor). Lysates were treated with RNase I (Ambion, AM2294) to fragment RNA, after which RBPMS protein-RNA complexes were immunoprecipitated using the indicated antibody. In addition to the RBP-IPs, a parallel SMI library was generated for each sample; these samples were not immunoprecipitated with anti-RBPMS antibodies but were otherwise treated identically. Stringent washes were performed as described in iCLIP, during which RNA was dephosphorylated with FastAP enzyme (Fermentas) and T4 PNK (NEB, M0201S). Subsequently, a 3′ RNA adaptor was ligated onto the RNA with T4 RNA ligase (NEB, M0242S). Protein-RNA complexes were run on an SDS–polyacrylamide gel electrophoresis gel and transferred to nitrocellulose membranes, and RNA was isolated off the membrane identically to standard iCLIP. After precipitation, RNA was reverse-transcribed with AffinityScript reverse transcriptase (Agilent, 600107), free primer was removed with ExoSap-IT (Thermo Fisher Scientific, 78201.1.ML), and a 3′ DNA adaptor was ligated onto the complementary DNA product with T4 RNA ligase (NEB). Libraries were then amplified with 2× Q5 PCR mix (NEB). Purified libraries were then sequenced via HiSeq 3000 (Illumina) with 75-bp paired-end reads at the Cologne Center for Genomics (CCG) and analyzed using the eCLIP-seq data analysis pipeline.
Microscopy imaging
Cells were seeded on Matrigel- or gelatin-coated coverslips or chamber slides (Ibidi) in respective growth medium. For fixation, cells were washed one time with PBS and fixed with 4% PFA for 10 min at RT. After fixation, the PFA was removed and the cells were washed three times with PBS. Cells in each chamber were treated for 10 min with 1% Triton X-100 (in PBS) for permeabilization. Cells were then incubated with the blocking solution containing 2% bovine serum albumin (BSA) and 2% glycine in phosphate-buffered saline with Tween 20 (PBS-T) or tris-buffered saline with Tween 20 (TBS-T) for phosphor antibodies. After blocking, the blocking solution was removed and cells were washed one time with PBS-T/TBS-T. Cells were incubated with either single or double primary antibodies (different host species) in 0.5% BSA in PBS-T overnight at 4°C. Primary and secondary antibodies were incubated for 1 hour at RT or overnight at 4°C. After incubation in the secondary antibody, cells were washed with PBS, while 4′,6-diamidino-2-phenylindole (DAPI) was added to the second wash. Antibodies used were listed in Key Resources Table. After washing, samples were rinsed with water and mounted with ProLong Gold mounting solution. Images were acquired with Leica SP7 or SP8 confocal microscopes with 3× line averaging bidirectional scanning using 63× oil objectives.
For live-cell imaging, 50,000 cells per well were seeded in 24-well dishes coated with Matrigel (Corning). The fluorescence readout was measured in IncuCyte S3. Sixteen areas in each well were imaged every 1 hour over a period of 48 hours after transfection.
RNA-seq and analysis
RNA was sequenced in the CCG and prepared according to the Illumina RNA Sequencing library preparation kit protocol. Libraries were sequenced on HiSeq 3000 or NovaSeq Sequencers (Illumina), with stranded paired-end reads of 75-bp read length (eCLIP-seq and TS-seq) or 150 bp (for poly-A sequencing for the short time course of mesoderm differentiation) with a depth of at least ~30 million reads per library for eCLIP-seq, ~25 million clean reads per library after ribosomal RNA depletion for TS-seq, and ~50 million clean reads per library for poly-A sequencing.
Differential gene expression analyses were performed on the RNA-sequencing (RNA-seq) data during indicated differentiation, including total RNA-seq and poly-A RNA-seq. Reads were aligned to the hg19 genome using Star, with the Gencode annotation as the reference transcriptome. Sequences aligned to tRNA and rRNA genes were removed. Differential expression analyses and gene quantification were performed with Deseq2. Differential expression analyses and gene quantification were performed with Deseq2 (FDR < 0.01 and FC > 2). For analysis, genes filtered by Deseq automatic independent filtering for low normalized read counts were discarded from analysis.
For TS-seq, reads were aligned and mapped in R using the kallisto package using hg19 as reference genome. For clustering, the R package maSigPro was used (69). Number of clusters was manually set to 8 to account for the most meaningful differences observed in the dataset.
Significant differential alternative splicing was identified with rMATS (version 4.1.2) (70). Analysis of inclusion level across time points was performed by recalculating percent inclusion (PSI) using junction-spanning reads only, requiring the read extend at least 10 nucleotides into the exon regions on both sides of the junction. Unless otherwise noted, at least 20 junction-spanning reads were required for calculating PSI values and only events meeting these criteria in at least two replicates were included in further analysis.
Quantification and statistical analysis
All data presented here are from at least three independent experiments. Graphs were plotted using GraphPad Prism and R Stats packages. Student’s t test was used to test for significance. Mean values ± SEM are shown. Differences between more than two groups were tested by one-way analysis of variance (ANOVA) analysis. Symbols representing P value cutoffs in the figures, i.e., ∗, ∗∗, and ∗∗∗, refer to P values of ≤0.05, ≤0.01, and ≤0.001, respectively.
Acknowledgments
We acknowledge the central facilities at CECAD and CMMC for technical assistance and Annibaldi laboratory and Lucardi laboratory for advice in various stages of this project. We thank G. Yeo (UCSD) and E. Van Nostrand (Baylor College) for sharing the eCLIP technology, Van Nostrand laboratory for the mRNA splicing analysis, and L. S. Valášek (The Czech Academy of Sciences) for sharing the translation complex profiling (TCP)-profiling method. We would like to thank L. Lauterjung and C. Walter for assisting in various stages of the project and J. Altmueller (CCG-Cologne) for advice and assistance.
Funding: The Kurian laboratory was supported by the NRW Stem Cell Network Independent Group Leader Grant (grant nos. 3681000801 and 2681101801), Else Kröner-Fresenius-Stiftung (grant no. 3640062621), Deutsche Forschungsgemeinschaft (DFG) (grant nos. KU 3511/4-1 and KU 3511/10-1), Center for Molecular Medicine Cologne (CMMC; ZMMK grant no. 3622801511), and European Research Council (ERC) Consolidator Grant (TRANSCEND, project number: GA 101043645).
Author contributions: Conceptualization: L.K. Methodology: D.B., G.A., K.K., J.-W.L, J.H., T.W., M.C., S.M., and L.K. Investigation: D.B. and L.K. Visualization: D.B. Supervision: L.K. Writing—original draft: L.K. Writing—review and editing: L.K., D.B., H.B., and A.P.
Competing interests: The Institute for Molecular Biotechnology (IMBA) filed a patent application (EP20164637.9) on cardioids with S.M. named as coinventor. The rest of the authors declare no competing interests.
Data and materials availability: Data from all proteomics experiments, including ARC-MS, TS-MS, and TCP-MS, are available via ProteomeXchange with the identifier PXD032045. The RNA-seq dataset, related to Fig. 3, is available in Gene Expression Omnibus (GEO): GSE205300, eCLIP-seq data in GSE205794, and TS-seq data in GSE205793. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S9
Legends for tables S1 to S6
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.J. Newport, M. Kirschner, A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 (1982). [DOI] [PubMed] [Google Scholar]
- 2.J. Newport, M. Kirschner, A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 (1982). [DOI] [PubMed] [Google Scholar]
- 3.H. Spemann, H. Mangold, Induction of embryonic primordia by implantation of organizers from a different species. 1923. Int. J. Dev. Biol. 45, 13–38 (2001). [PubMed] [Google Scholar]
- 4.L. Xiang, Y. Yin, Y. Zheng, Y. Ma, Y. Li, Z. Zhao, J. Guo, Z. Ai, Y. Niu, K. Duan, J. He, S. Ren, D. Wu, Y. Bai, Z. Shang, X. Dai, W. Ji, T. Li, A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 577, 537–542 (2020). [DOI] [PubMed] [Google Scholar]
- 5.D. van Hoof, J. Krijgsveld, C. Mummery, Proteomic analysis of stem cell differentiation and early development. Cold Spring Harb. Perspect. Biol. 4, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. Lu, F. Markowetz, R. D. Unwin, J. T. Leek, E. M. Airoldi, B. D. MacArthur, A. Lachmann, R. Rozov, A. Ma’ayan, L. A. Boyer, O. G. Troyanskaya, A. D. Whetton, I. R. Lemischka, Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462, 358–362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Gao, X. Liu, B. Tang, C. Li, Z. Kou, L. Li, W. Liu, Y. Wu, X. Kou, J. Li, Y. Zhao, J. Yin, H. Wang, S. Chen, L. Liao, S. Gao, Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969 (2017). [DOI] [PubMed] [Google Scholar]
- 8.K. M. Loh, A. Chen, P. W. Koh, T. Z. Deng, R. Sinha, J. M. Tsai, A. A. Barkal, K. Y. Shen, R. Jain, R. M. Morganti, N. Shyh-Chang, N. B. Fernhoff, B. M. George, G. Wernig, R. E. A. Salomon, Z. Chen, H. Vogel, J. A. Epstein, A. Kundaje, W. S. Talbot, P. A. Beachy, L. T. Ang, I. L. Weissman, Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell 166, 451–467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.G. Peng, S. Suo, G. Cui, F. Yu, R. Wang, J. Chen, S. Chen, Z. Liu, G. Chen, Y. Qian, P. P. L. Tam, J. D. J. Han, N. Jing, Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 572, 528–532 (2019). [DOI] [PubMed] [Google Scholar]
- 10.R. Stadhouders, G. J. Filion, T. Graf, Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354 (2019). [DOI] [PubMed] [Google Scholar]
- 11.K. W. Rogers, A. F. Schier, Morphogen gradients: From generation to interpretation. Annu. Rev. Cell Dev. Biol. 27, 377–407 (2011). [DOI] [PubMed] [Google Scholar]
- 12.C. A. Gifford, M. J. Ziller, H. Gu, C. Trapnell, J. Donaghey, A. Tsankov, A. K. Shalek, D. R. Kelley, A. A. Shishkin, R. Issner, X. Zhang, M. Coyne, J. L. Fostel, L. Holmes, J. Meldrim, M. Guttman, C. Epstein, H. Park, O. Kohlbacher, J. Rinn, A. Gnirke, E. S. Lander, B. E. Bernstein, A. Meissner, Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D. Harnett, M. C. Ambrozkiewicz, U. Zinnall, A. Rusanova, E. Borisova, A. N. Drescher, M. Couce-Iglesias, G. Villamil, R. Dannenberg, K. Imami, A. Münster-Wandowski, B. Fauler, T. Mielke, M. Selbach, M. Landthaler, C. M. T. Spahn, V. Tarabykin, U. Ohler, M. L. Kraushar, A critical period of translational control during brain development at codon resolution. Nat. Struct. Mol. Biol. 29, 1277–1290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. Schwanhausser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, W. Chen, M. Selbach, Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 15.F. K. Teixeira, R. Lehmann, Translational control during developmental transitions. Cold Spring Harb. Perspect. Biol. 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. R. Kristensen, J. Gsponer, L. J. Foster, Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 9, 689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. Kong, P. Lasko, Translational control in cellular and developmental processes. Nat. Rev. Genet. 13, 383–394 (2012). [DOI] [PubMed] [Google Scholar]
- 18.C. Buccitelli, M. Selbach, mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 21, 630–644 (2020). [DOI] [PubMed] [Google Scholar]
- 19.C. Vogel, E. M. Marcotte, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Baser, M. Skabkin, S. Kleber, Y. Dang, G. S. Gülcüler Balta, G. Kalamakis, M. Göpferich, D. C. Ibañez, R. Schefzik, A. S. Lopez, E. L. Bobadilla, C. Schultz, B. Fischer, A. Martin-Villalba, Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104 (2019). [DOI] [PubMed] [Google Scholar]
- 21.J. Hausser, A. Mayo, L. Keren, U. Alon, Central dogma rates and the trade-off between precision and economy in gene expression. Nat. Commun. 10, 68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. Sampath, D. K. Pritchard, L. Pabon, H. Reinecke, S. M. Schwartz, D. R. Morris, C. E. Murry, A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460 (2008). [DOI] [PubMed] [Google Scholar]
- 23.N. T. Ingolia, L. F. Lareau, J. S. Weissman, Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. Fujii, Z. Shi, O. Zhulyn, N. Denans, M. Barna, Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat. Commun. 8, 14443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.N. Guzzi, M. Cieśla, P. C. T. Ngoc, S. Lang, S. Arora, M. Dimitriou, K. Pimková, M. N. E. Sommarin, R. Munita, M. Lubas, Y. Lim, K. Okuyama, S. Soneji, G. Karlsson, J. Hansson, G. Jönsson, A. H. Lund, M. Sigvardsson, E. Hellström-Lindberg, A. C. Hsieh, C. Bellodi, Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 (2018). [DOI] [PubMed] [Google Scholar]
- 26.D. Simsek, G. C. Tiu, R. A. Flynn, G. W. Byeon, K. Leppek, A. F. Xu, H. Y. Chang, M. Barna, The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Z. Shi, M. Barna, Translating the genome in time and space: Specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 31, 31–54 (2015). [DOI] [PubMed] [Google Scholar]
- 28.L. Minati, C. Firrito, A. del Piano, A. Peretti, S. Sidoli, D. Peroni, R. Belli, F. Gandolfi, A. Romanel, P. Bernabo, J. Zasso, A. Quattrone, G. Guella, F. Lauria, G. Viero, M. Clamer, One-shot analysis of translated mammalian lncRNAs with AHARIBO. eLife 10, e59303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Z. Shi, K. Fujii, K. M. Kovary, N. R. Genuth, H. L. Röst, M. N. Teruel, M. Barna, Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.H. Li, Y. Huo, X. He, L. Yao, H. Zhang, Y. Cui, H. Xiao, W. Xie, D. Zhang, Y. Wang, S. Zhang, H. Tu, Y. Cheng, Y. Guo, X. Cao, Y. Zhu, T. Jiang, X. Guo, Y. Qin, J. Sha, A male germ-cell-specific ribosome controls male fertility. Nature 612, 725–731 (2022). [DOI] [PubMed] [Google Scholar]
- 31.N. R. Genuth, Z. Shi, K. Kunimoto, V. Hung, A. F. Xu, C. H. Kerr, G. C. Tiu, J. A. Oses-Prieto, R. E. A. Salomon-Shulman, J. D. Axelrod, A. L. Burlingame, K. M. Loh, M. Barna, A stem cell roadmap of ribosome heterogeneity reveals a function for RPL10A in mesoderm production. Nat. Commun. 13, 5491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. V. Vo, J. das, M. J. Meyer, N. A. Cordero, N. Akturk, X. Wei, B. J. Fair, A. G. Degatano, R. Fragoza, L. G. Liu, A. Matsuyama, M. Trickey, S. Horibata, A. Grimson, H. Yamano, M. Yoshida, F. P. Roth, J. A. Pleiss, Y. Xia, H. Yu, A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell 164, 310–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. Butland, J. M. Peregrín-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, A. Emili, Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 (2005). [DOI] [PubMed] [Google Scholar]
- 34.F. E. Tan, S. Sathe, E. C. Wheeler, J. K. Nussbacher, S. Peter, G. W. Yeo, A transcriptome-wide translational program defined by LIN28B expression level. Mol. Cell 73, 304–313.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. Cho, H. Chang, S. C. Kwon, B. Kim, Y. Kim, J. Choe, M. Ha, Y. K. Kim, V. N. Kim, LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765–777 (2012). [DOI] [PubMed] [Google Scholar]
- 36.M. W. Hentze, A. Castello, T. Schwarzl, T. Preiss, A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018). [DOI] [PubMed] [Google Scholar]
- 37.T. A. Farazi, C. S. Leonhardt, N. Mukherjee, A. Mihailovic, S. Li, K. E. A. Max, C. Meyer, M. Yamaji, P. Cekan, N. C. Jacobs, S. Gerstberger, C. Bognanni, E. Larsson, U. Ohler, T. Tuschl, Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets. RNA 20, 1090–1102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.T. Aguero, Y. Zhou, M. Kloc, P. Chang, E. Houliston, M. King, Hermes (Rbpms) is a critical component of RNP complexes that sequester germline RNAs during oogenesis. J. Dev. Biol. 4, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Teplova, T. A. Farazi, T. Tuschl, D. J. Patel, Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS. Q. Rev. Biophys. 49, e1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.W. V. Gerber, S. A. Vokes, N. R. Zearfoss, P. A. Krieg, A role for the RNA-binding protein, hermes, in the regulation of heart development. Dev. Biol. 247, 116–126 (2002). [DOI] [PubMed] [Google Scholar]
- 41.P. Gan, Z. Wang, M. G. Morales, Y. Zhang, R. Bassel-Duby, N. Liu, E. N. Olson, RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev. Cell 57, 959–973.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.C. Meyer, A. Garzia, P. Morozov, H. Molina, T. Tuschl, The G3BP1-family-USP10 deubiquitinase complex rescues ubiquitinated 40S subunits of ribosomes stalled in translation from lysosomal degradation. Mol. Cell 77, 1193–1205.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 43.S. Frank, G. Ahuja, D. Bartsch, N. Russ, W. Yao, J. C.-C. Kuo, J.-P. Derks, V. S. Akhade, Y. Kargapolova, T. Georgomanolis, J.-E. Messling, M. Gramm, L. Brant, R. Rehimi, N. E. Vargas, A. Kuroczik, T.-P. Yang, R. G. A. Sahito, J. Franzen, J. Hescheler, A. Sachinidis, M. Peifer, A. Rada-Iglesias, M. Kanduri, I. G. Costa, C. Kanduri, A. Papantonis, L. Kurian, yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24, 318–327.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 44.A. Bondue, G. Lapouge, C. Paulissen, C. Semeraro, M. Iacovino, M. Kyba, C. Blanpain, Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 3, 69–84 (2008). [DOI] [PubMed] [Google Scholar]
- 45.J. Tosic, G. J. Kim, M. Pavlovic, C. M. Schröder, S. L. Mersiowsky, M. Barg, A. Hofherr, S. Probst, M. Köttgen, L. Hein, S. J. Arnold, Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 21, 1518–1531 (2019). [DOI] [PubMed] [Google Scholar]
- 46.T. Brade, L. S. Pane, A. Moretti, K. R. Chien, K. L. Laugwitz, Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 3, a013847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Rao, M. J. Pfeiffer, S. Frank, K. Adachi, I. Piccini, R. Quaranta, M. Araúzo-Bravo, J. Schwarz, D. Schade, S. Leidel, H. R. Schöler, G. Seebohm, B. Greber, Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell 18, 554–556 (2016). [DOI] [PubMed] [Google Scholar]
- 48.S. Mazzotta, C. Neves, R. J. Bonner, A. S. Bernardo, K. Docherty, S. Hoppler, Distinctive roles of canonical and noncanonical Wnt signaling in human embryonic cardiomyocyte development. Stem Cell Rep. 7, 764–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.P. Hofbauer, S. M. Jahnel, N. Papai, M. Giesshammer, A. Deyett, C. Schmidt, M. Penc, K. Tavernini, N. Grdseloff, C. Meledeth, L. C. Ginistrelli, C. Ctortecka, Š. Šalic, M. Novatchkova, S. Mendjan, Cardioids reveal self-organizing principles of human cardiogenesis. Cell 184, 3299–3317.e22 (2021). [DOI] [PubMed] [Google Scholar]
- 50.H. Kempf, R. Olmer, A. Haase, A. Franke, E. Bolesani, K. Schwanke, D. Robles-Diaz, M. Coffee, G. Göhring, G. Dräger, O. Pötz, T. Joos, E. Martinez-Hackert, A. Haverich, F. F. R. Buettner, U. Martin, R. Zweigerdt, Bulk cell density and Wnt/TGFβ signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat. Commun. 7, 13602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C. L. Jiang, Y. Goyal, N. Jain, Q. Wang, R. E. Truitt, A. J. Coté, B. Emert, I. A. Mellis, K. Kiani, W. Yang, R. Jain, A. Raj, Cell type determination for cardiac differentiation occurs soon after seeding of human-induced pluripotent stem cells. Genome Biol. 23, 90 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.E. L. Van Nostrand, G. A. Pratt, A. A. Shishkin, C. Gelboin-Burkhart, M. Y. Fang, B. Sundararaman, S. M. Blue, T. B. Nguyen, C. Surka, K. Elkins, R. Stanton, F. Rigo, M. Guttman, G. W. Yeo, Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. J. Marvin, G. Di Rocco, A. Gardiner, S. M. Bush, A. B. Lassar, Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 15, 316–327 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Z. Steinhart, S. Angers, Wnt signaling in development and tissue homeostasis. Development 145, (2018). [DOI] [PubMed] [Google Scholar]
- 55.E. E. Nakagaki-Silva, C. Gooding, M. Llorian, A. G. Jacob, F. Richards, A. Buckroyd, S. Sinha, C. W. J. Smith, Identification of RBPMS as a mammalian smooth muscle master splicing regulator via proximity of its gene with super-enhancers. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.B. A. Yee, G. A. Pratt, B. R. Graveley, E. L. Van Nostrand, G. W. Yeo, RBP-maps enables robust generation of splicing regulatory maps. RNA 25, 193–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E. L. Van Nostrand, G. A. Pratt, B. A. Yee, E. C. Wheeler, S. M. Blue, J. Mueller, S. S. Park, K. E. Garcia, C. Gelboin-Burkhart, T. B. Nguyen, I. Rabano, R. Stanton, B. Sundararaman, R. Wang, X.-D. Fu, B. R. Graveley, G. W. Yeo, Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol. 21, 90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R. C. Wek, Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Y. Lin, F. Li, L. Huang, C. Polte, H. Duan, J. Fang, L. Sun, X. Xing, G. Tian, Y. Cheng, Z. Ignatova, X. Yang, D. A. Wolf, eIF3 associates with 80S ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. Mol. Cell 79, 575–587.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 60.A. Choudhuri, U. Maitra, T. Evans, Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 9818–9823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. P. Schuller, C. C. Wu, T. E. Dever, A. R. Buskirk, R. Green, eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. Wagner, A. Herrmannová, V. Hronová, S. Gunišová, N. D. Sen, R. D. Hannan, A. G. Hinnebusch, N. E. Shirokikh, T. Preiss, L. S. Valášek, Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol. Cell 79, 546–560.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. Ghimire, V. Mantziou, N. Moris, A. Martinez Arias, Human gastrulation: The embryo and its models. Dev. Biol. 474, 100–108 (2021). [DOI] [PubMed] [Google Scholar]
- 64.A. S. Lee, P. J. Kranzusch, J. H. Cate, eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.A. S. Lee, P. J. Kranzusch, J. A. Doudna, J. H. Cate, eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.X. Chen, Y. Liu, C. Xu, L. Ba, Z. Liu, X. Li, J. Huang, E. Simpson, H. Gao, D. Cao, W. Sheng, H. Qi, H. Ji, M. Sanderson, C. L. Cai, X. Li, L. Yang, J. Na, K. Yamamura, Y. Liu, G. Huang, W. Shou, N. Sun, QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function. Nat. Commun. 12, 89 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.K. Sakers, Y. Liu, L. Llaci, S. M. Lee, M. J. Vasek, M. A. Rieger, S. Brophy, E. Tycksen, R. Lewis, S. E. Maloney, J. D. Dougherty, Loss of quaking RNA binding protein disrupts the expression of genes associated with astrocyte maturation in mouse brain. Nat. Commun. 12, 1537 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.E. L. Van Nostrand, P. Freese, G. A. Pratt, X. Wang, X. Wei, R. Xiao, S. M. Blue, J.-Y. Chen, N. A. L. Cody, D. Dominguez, S. Olson, B. Sundararaman, L. Zhan, C. Bazile, L. P. B. Bouvrette, J. Bergalet, M. O. Duff, K. E. Garcia, C. Gelboin-Burkhart, M. Hochman, N. J. Lambert, H. Li, M. P. Mc Gurk, T. B. Nguyen, T. Palden, I. Rabano, S. Sathe, R. Stanton, A. Su, R. Wang, B. A. Yee, B. Zhou, A. L. Louie, S. Aigner, X.-D. Fu, E. Lécuyer, C. B. Burge, B. R. Graveley, G. W. Yeo, A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.M. J. Nueda, S. Tarazona, A. Conesa, Next maSigPro: Updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30, 2598–2602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. Shen, J. W. Park, Z. X. Lu, L. Lin, M. D. Henry, Y. N. Wu, Q. Zhou, Y. Xing, rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S9
Legends for tables S1 to S6
Tables S1 to S6