Abstract
Tuberculosis (TB), an infectious airborne disease caused by Mycobacterium tuberculosis (Mtb), is a serious public health threat reported as the leading cause of morbidity and mortality worldwide. South Africa is a high-TB-burden country with TB being the highest infectious disease killer. This study investigated the distribution of Mtb mutations and spoligotypes in rural Eastern Cape Province. The Mtb isolates included were 1157 from DR-TB patients and analysed by LPA followed by spoligotyping of 441 isolates. The distribution of mutations and spoligotypes was done by spatial analysis. The rpoB gene had the highest number of mutations. The distribution of rpoB and katG mutations was more prevalent in four healthcare facilities, inhA mutations were more prevalent in three healthcare facilities, and heteroresistant isolates were more prevalent in five healthcare facilities. The Mtb was genetically diverse with Beijing more prevalent and largely distributed. Spatial analysis and mapping of gene mutations and spoligotypes revealed a better picture of distribution.
Keywords: tuberculosis, spatial analysis, mutations, spoligotypes, heteroresistance
1. Introduction
Tuberculosis (TB), a chronic inflammatory infectious disease caused by Mycobacterium tuberculosis (Mtb), is a serious public health threat and is reported as the leading cause of morbidity and mortality worldwide. It is easily transmitted from one person to another by airborne droplet nuclei [1,2]. In 2021, the most current global TB data reported the incidence of TB to be 10.6 million new cases with 1.6 million attributable fatalities globally [3]. The distribution of TB differs geographically both within counties and on different continents of the world. Africa accounts for 29% and 34% of all TB cases and deaths, respectively worldwide with the highest recorded incidence rate of 275 cases per 100,000 people [4,5].
South Africa is a high TB burden country with TB being the highest infectious disease killer. According to the global burden of disease study, TB is the fifth leading cause of years of life lost (YLL) and disability-adjusted life years (DALY) in the country [6]. With an estimated population of 60.6 million by the end of June 2022 [7], South Africa (SA) shares borders with 6 other nations, including Botswana, Lesotho, Namibia, Mozambique, Swaziland, and Zimbabwe, where TB is also endemic. In 2019, the TB incidence in South Africa was estimated to be 615 cases per 100,000 population, ranging from 427 to 835 cases per 100,000 and with estimated 360,000 people who developed TB [8]. SA had the second-highest absolute number of notified rifampicin (RIF)-resistant and multidrug-resistant (MDR) cases globally with 18,734 cases reported in 2015 [9]. Eastern Cape is one of the three provinces in South Africa that have the highest TB incidence rates [1].
A better understanding of local geographic heterogeneity in routinely identified TB cases and the correlation of that heterogeneity with the location of undiagnosed prevalent cases may, therefore, be useful in directing active case-finding interventions to high-risk areas [10]. Furthermore, there is a need for accurate and early detection of drug-resistant TB (DR-TB) for minimizing the development of drug resistance, effective patient care, and preventing the spread of DR strains [11].
Our knowledge of the epidemiology of TB on a local and global level has been substantially improved by the development and use of genotyping methods for Mtb; likewise, with the help of geospatial analytical technologies, the understanding of public health issues can be enhanced [12,13]. Spoligotyping combined with geospatial analytical methods, such as geographic information systems (GIS) and directly observed treatment short-course (DOTS) strategy, can help assess the transmission of Mtb strains and are promising essential tools for helping to understand the distribution of drug-resistant strains as well as the drivers of drug resistance [14]. Previous spatial studies have used GIS, whole genome sequencing (WGS), and spatial statistics to identify transmission hotspot areas with elevated risk for prioritisation of control and intervention measures [15,16,17]. These spatial studies have also shown that MDR-TB is clustered in specific geographical areas associated with location, socio-economic status, and population density [4]. Even though there is an increase in the number of studies that use geospatial analytical methods to understand TB and other public health problems [18,19,20], however, the transmission dynamics of prevalent Mtb strains in rural Eastern Cape are not well understood.
Understanding such spatial variations in TB prevalence is crucial for improved targeting of interventions and resources for the prevention and management of TB in a particular area. The geographical distribution of MDR-TB in high TB burden settings is very important for the effective control of TB epidemics. This can inform a basis for understanding DR gene migrations within populations since the frequency of mutations varies geographically. In this study, we sought to investigate the spatial patterns of Mtb in order to determine the transmission patterns and mixed-strain infections. We present the first spatial analysis of DR-TB and mutations associated with RIF and isoniazid (INH) causing heteroresistance and spoligotype distributions of Mtb in healthcare facilities (HCFs) of Mthatha and surrounding areas.
2. Methods
2.1. Study Design and Setting
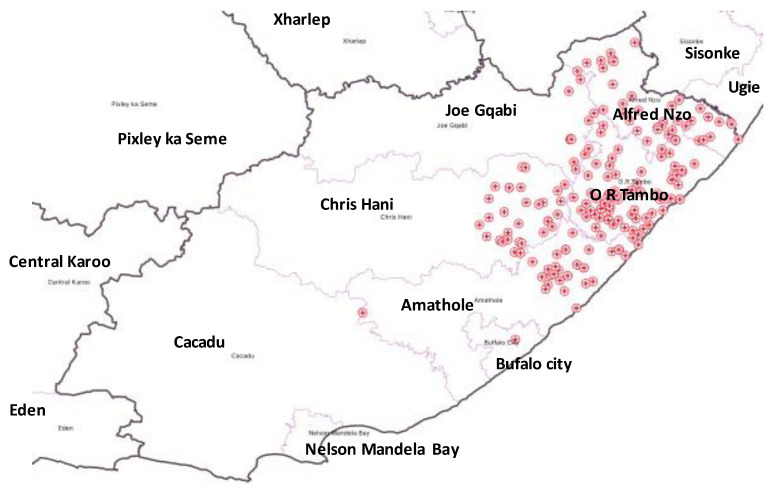
This was an ecological study design conducted in 4 districts, namely Oliver Reginald Tambo (O. R. Tambo), Alfred Nzo, Amatole, and Chris Hani, and 1 metropolitan municipality (Buffalo City), with 101 healthcare facilities in rural Eastern Cape Province (ECP), South Africa; the distribution of the healthcare facilities is as shown (Figure 1).
Figure 1.
Distribution of healthcare facilities in the study area.
Eastern Cape province is the second largest province in the country and serves a population of 7,130,480. O. R. Tambo district is 1 of the 7 districts of the ECP located on the coastline. The seat of O. R. Tambo is in Mthatha. The population is about 1,676,463. Five local municipalities, namely King Sabata Dalindyebo, Nyandeni, Mhlontlo, Port St. Johns, and Ingquza, form O. R. Tambo district municipality.
2.2. Study Population and Sampling Strategy
The study population included all DR-TB cases registered and living in the catchment area of the five municipalities’ TB treatment centres and Nelson Mandela Academic Hospital National Health Laboratory Services (NHLS) catchment areas, registered between the years 2018 and 2020.
2.3. Data Collection and Isolates Identification
Sputum samples were collected from consecutive clinically diagnosed pulmonary TB patients reporting to 101 selected health facilities within the district municipalities named above. The samples were submitted to NHLS TB Laboratory for diagnostic testing. A convenience sample of 1157 Mtb isolates was selected to include: (1) INH monoresistance (IMR-TB) defined as resistance to a single first-line drug, isoniazid, (2) RIF monoresistance (RMR-TB) defined as resistance to a single first-line drug, rifampicin, (3) MDR-TB defined as resistant to at least isoniazid and rifampin, the two most potent TB drugs, and (4) heteroresistance defined by the coexistence of susceptible and resistant organisms to anti-tuberculosis drugs in the same patient (defined by the hybridisation to both the wild type (WT) and mutant (MUT) probes on the MTBDRplus assay). The information of patients on the laboratory requisition form was recorded for spatial analysis in order to determine the distribution of mutations in rpoB, katG, and inhA genes and their spoligotypes among isolates. Mutations were determined by the presence of the binding of amplicons to probes targeting the most commonly occurring mutation probes or inferred by the lack of hybridization (i.e., lack of binding) of the amplicons to the corresponding WT probes. Isolates were collected over a 3-year period and investigated using first-line probe assay (LPA).
2.4. Isolates Analysis
The GenoType MTBDRplus VER 2.0 is a DNA-strip based in vitro assay for identifying the Mtb complex (MTBC) and its resistance to RIF and INH from smear-positive pulmonary sputum samples and positive culture samples. DNA was extracted using Genolyse® kit (Hain Life Science GmbH, Nehren, Germany). The extracted DNA was processed by the LPA using DRplus [21] to detect MTBC and RIF and/or INH resistance according to the manufacturer’s instructions. Out of 1157 isolates, 441 were conveniently selected to include IMR-TB, RMR-TB, and MDR-TB and heteroresistant isolates for spoligotyping to determine genotypes of Mtb isolates circulating in Mthatha and surrounding areas. Spoligotyping was performed using microbeads from TB-SPOL Kit (Beamedex®, Orsay, France), and the fluorescence intensity was measured using Luminex 200® (Austin, TX, USA). Generated patterns were assigned to families using the SITVIT2 international database of the Pasteur Institute of Guadeloupe and compared [22].
The LPA and spoligotype results of Mtb isolates were analysed for distribution of the mutations and spoligotypes using the QGIS 3.14 software. LPA score and banding patterns were used to determine the type of DR-TB and heteroresistance. Clinics within hospitals with the same coordinates were merged in the analysis. We assessed the distribution of mutations in rpoB, katG, and inhA, as well as the distribution of heteroresistant strains and spoligotypes.
3. Results
Distribution of Mutations, Lineages of Isolates
A total of 1157 DR-TB and heteroresistant clinical isolates were isolated from the different healthcare facilities.
RMR was represented by the LPA score of rpoB MUT/katG WT/inhA WT, while IMR was represented by rpoB WT/katG WT/inhA MUT or rpoB WT/katG MUT/inhA WT and rpoB WT/katG MUT/inhA MUT LPA score. The MDR-TB strains were represented by LPA score of rpoB MUT/katG WT/inhA MUT or rpoB MUT/katG MUT/inhA WT and rpoB MUT/katG MUT/inhA MUT. Heteroresistant strains were represented by LPA score where there were both mutation and wildtype bands on any of the three genes (rpoB, katG, and inhA).
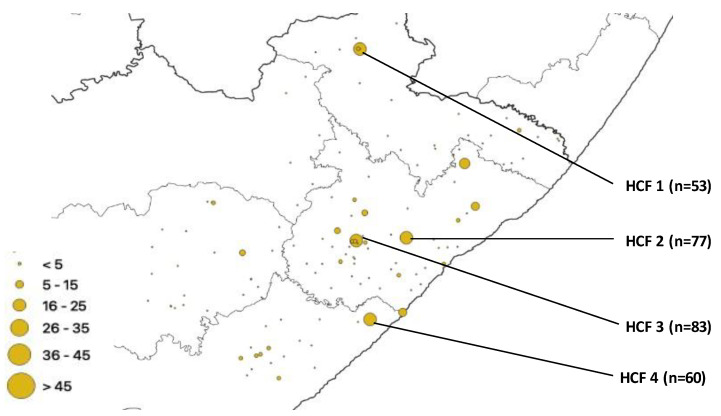
The number of isolates from 6 HCFs that had a higher proportion of gene mutations and spoligotypes was HCF1 (78/1157), HCF 2 (94/1157), HCF 3 (114/1157), HCF 4 (83/1157), HCF 5 (73/1157), and HCF 6 (55/1157). All three (rpoB, katG and inhA) genes under investigation had one or more mutations from each isolate. The total number of mutations that occurred in the rpoB gene was 761, representing the highest number of mutations, followed by the katG gene with 683 mutations, while the inhA gene accounted for 286 mutations. The distribution of these rpoB mutations is shown in Figure 2. A total of 4 healthcare facilities had more gene mutations, namely HCF 3 (n = 83), HCF 2 (n = 77), HCF 4 (n = 60), and HCF 1 (n = 53).
Figure 2.
rpoB mutation distribution.
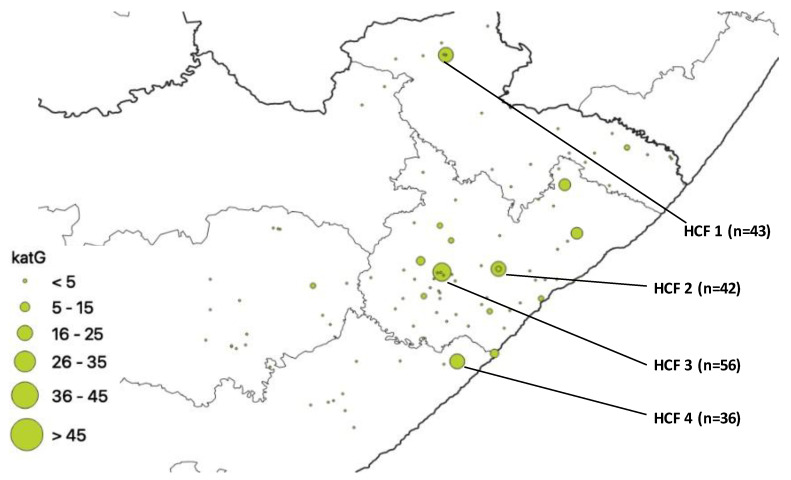
katG gene mutations are shown below (Figure 3). A total of 4 healthcare facilities had more mutations namely HCF 4, n =36, HCF 2, n = 42, HCF 1, n = 43, and HCF 3, n = 56.
Figure 3.
katG mutation distribution.
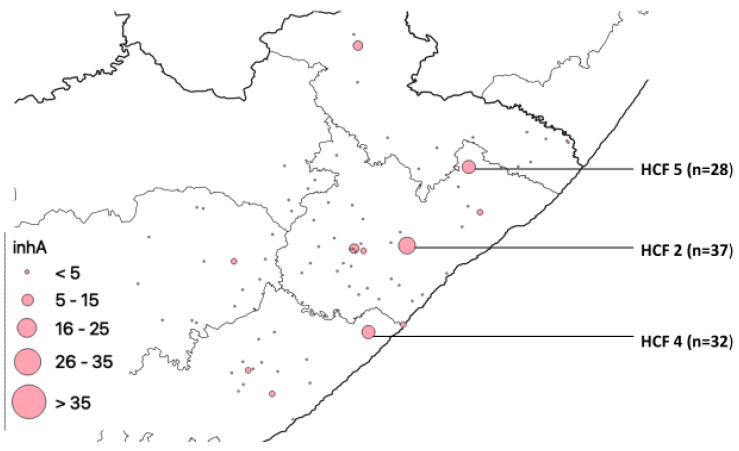
There were 3 healthcare facilities that had the highest number of inhA gene mutations, HCF 5, HCF 4, and HCF 2, with 28, 32, and 37 mutations, respectively, indicated in Figure 4.
Figure 4.
inhA mutation distribution.
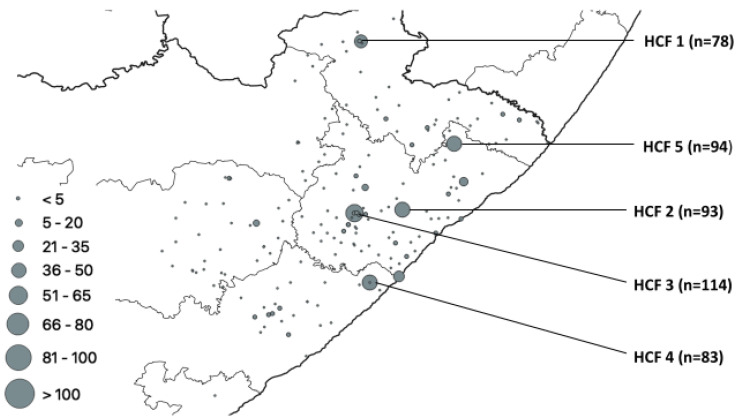
The total number of heteroresistant isolates was 207, which was 17.9% of the total number of isolates. These isolates had one or more mutations in different genes. The following clinics had the most mutations in heteroresistant isolates, HCF 1, HCF 4, HCF 2, HCF 5, and HCF 3, with 78, 83, 93, 94, and 114 mutations, respectively (Figure 5).
Figure 5.
Geospatial distribution of heteroresistance in rural ECP.
The prevalent gene mutations in HCF within three municipalities were captured in the table below (Table 1).
Table 1.
Distribution of prevalent gene mutations in HCF in rural ECP.
| HCF | Municipality | rpoB | katG | inhA | Heteroresistant Genes |
|---|---|---|---|---|---|
| 1 | Alfred Nzo | 53 | 43 | 0 | 78 |
| 2 | O. R. Tambo | 77 | 42 | 37 | 93 |
| 3 | O. R. Tambo | 83 | 56 | 0 | 114 |
| 4 | Amathole | 60 | 36 | 32 | 83 |
| 5 | O. R. Tambo | 0 | 0 | 28 | 94 |
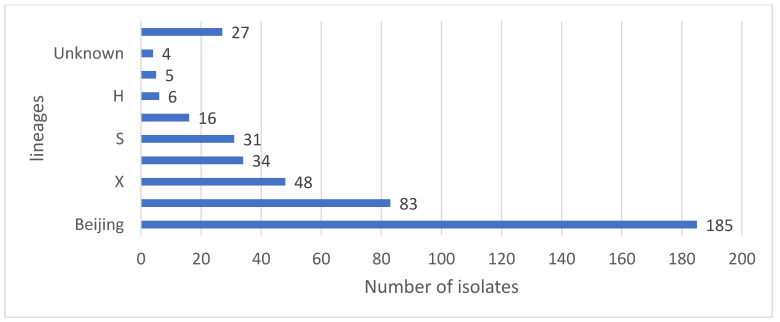
Based on the 441 spoligotyping results, 8 families were identified. Of 441 isolates queried for the lineage assignment, 59 (81.9%) were classified into the previously known lineages, and 13 (18.1%) were not assigned to any known lineages (Table 2). The Beijing family was the predominant group, representing 42.0% (n = 185) of all isolates, followed by the LAM family, 18.8% (n = 83), X family, 10.9% (n = 48), T family, 7.7% (n = 34), S family, 7.0% (n = 31), EAI family, 3.6% (n = 16), H, 1.4% (n = 6), and CAS family, 1.1% (n = 5) (Figure 6). Only 4 (0.9%) isolates showed unknown patterns that were not assigned to any known major lineages in the SITVIT2 database, and 27 (6.1%) had no results.
Table 2.
Spoligotyping Patterns.
| Lineage | No of Isolates | Sublineage | No of Isolates | SIT | Spoligo Pattern | No of Isolates |
|---|---|---|---|---|---|---|
| Beijing | 185 | 1 |

|
185 | ||
| LAM | 83 | LAM3 | 62 | 33 |

|
36 |
| 719 |

|
17 | ||||
| ORPHAN |

|
2 | ||||
| 4 |

|
1 | ||||
| 130 |

|
1 | ||||
| 376 |

|
1 | ||||
| 2014 |

|
1 | ||||
| 2284 |

|
1 | ||||
| 2302 |

|
1 | ||||
| not in SITVIT |

|
1 | ||||
| LAM4 | 15 | 60 |
|
15 | ||
| LAM5 | 2 | 93 |

|
1 | ||
| 136 |

|
1 | ||||
| LAM9 | 2 | 42 |

|
2 | ||
| LAM11-ZWE | 2 | 811 |

|
1 | ||
| 1873 |

|
1 | ||||
| X | 48 | X1 | 18 | 2022 |

|
10 |
| 2226 |

|
4 | ||||
| 119 |

|
3 | ||||
| 336 |

|
1 | ||||
| X2 | 3 | 18 |

|
1 | ||
| 137 |

|
1 | ||||
| 2016 |

|
1 | ||||
| X3 | 27 | 92 |

|
21 | ||
| 2286 |

|
4 | ||||
| 2020 |

|
2 | ||||
| T | 34 | T1 | 29 | 53 |

|
13 |
| 926 |

|
4 | ||||
| 334 |

|
2 | ||||
| 501 |

|
2 | ||||
| 156 |

|
1 | ||||
| 245 |

|
1 | ||||
| 373 |

|
1 | ||||
| 519 |

|
1 | ||||
| 732 |

|
1 | ||||
| 1122 |

|
1 | ||||
| 1144 |

|
1 | ||||
| ORPHAN |

|
1 | ||||
| T2 | 1 | 52 |

|
1 | ||
| T2/3 | 2 | 73 |

|
2 | ||
| T3 | 1 | ORPHAN |

|
1 | ||
| T5-RUS1 | 1 | 254 |

|
1 | ||
| T-TUSCANY | 2 | 1737 |

|
2 | ||
| S | 31 | 34 |

|
18 | ||
| 789 |

|
4 | ||||
| 71 |

|
3 | ||||
| Not in SITVIT |

|
2 | ||||
| 790 |

|
1 | ||||
| 1211 |

|
1 | ||||
| Not in SITVIT |

|
1 | ||||
| Not in SITVIT |

|
1 | ||||
| EAI | 16 | EAI1-SOM | 10 | 806 |

|
6 |
| 48 |

|
2 | ||||
| 1649 |

|
2 | ||||
| EAI5 | 5 | 625 |

|
3 | ||
| ORPHAN |

|
2 | ||||
| EAI | 1 | Not in SITVIT |

|
1 | ||
| H | 6 | H1 | 5 | 62 |

|
2 |
| 2375 |

|
2 | ||||
| 47 |

|
1 | ||||
| H3 | 1 | 50 |

|
1 | ||
| CAS | 5 | CAS1-Kili | 2 | 21 |

|
2 |
| CAS1-Delhi | 1 | 1092 |

|
1 | ||
| CAS | 2 | Not in SITVIT |

|
2 | ||
| unknown | 4 | Unknown | 1 | 2018 |

|
1 |
| Not in SITVIT | 3 | Not in SITVIT |

|
1 | ||
| Not in SITVIT |

|
1 | ||||
| Not in SITVIT |
 67 67 |
1 | ||||
| no result | 27 | |||||
| TOTAL | 441 |
SIT = Shared International Type; SITVIT = international spoligotyping database; SITIV 2 = is a genotyping molecular markers database focusing on Mycobacterium tuberculosis complex; CAS = Central Asian; Orphan = isolates showed unknown patterns that were not assigned to any known major lineages in the SITVIT2 database; LAM: Latin American; EAI: East-African Indian; Delhi/CAS: Delhi/Central Asian, H: Haarlem); no = number.
Figure 6.
Distribution of Mtb lineages/families. (LAM: Latin American; EAI: East-African Indian; Delhi/CAS: Delhi/Central Asian, H: Haarlem).
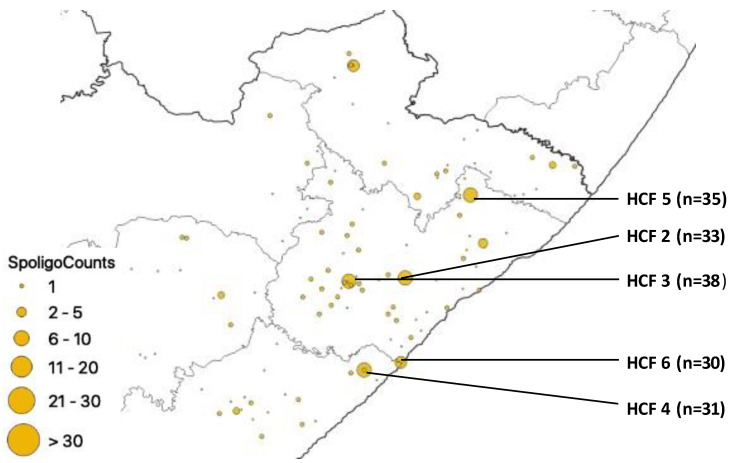
Although spoligotyped isolates were received from many HCFs of the study setting, HCFs with greater than n = 30 isolates were from 2 municipalities, namely O. R. Tambo (HCF 2, HCF 3, HCF 5 and HCF 6) and Buffalo City (HCF 4) (Figure 7).
Figure 7.
Geospatial distribution of spoligotyped isolates in healthcare facilities.
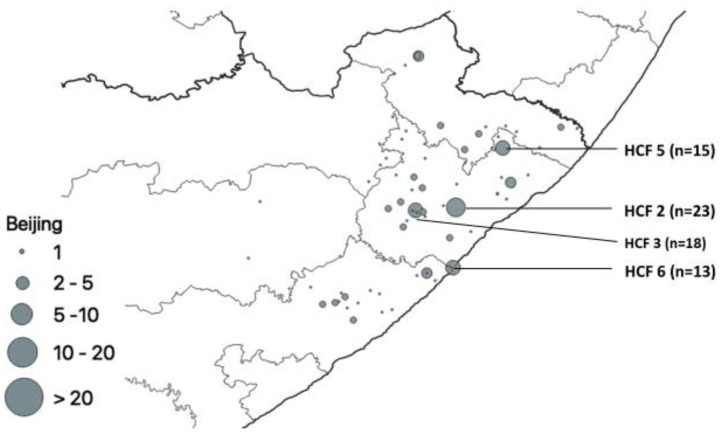
Our results showed that from the 441 clinical isolates that were spoligotyped, the Beijing family strains accounted for 42.0% (185/441) of all the strains circulating in Mthatha and surrounding areas. The healthcare facilities with the most Beijing family strains were HCF 2 (n = 23), HCF 3 (n = 18), HCF 5 (n = 15), and HCF 6 (n = 13) (Figure 8).
Figure 8.
Geospatial distribution of Beijing family.
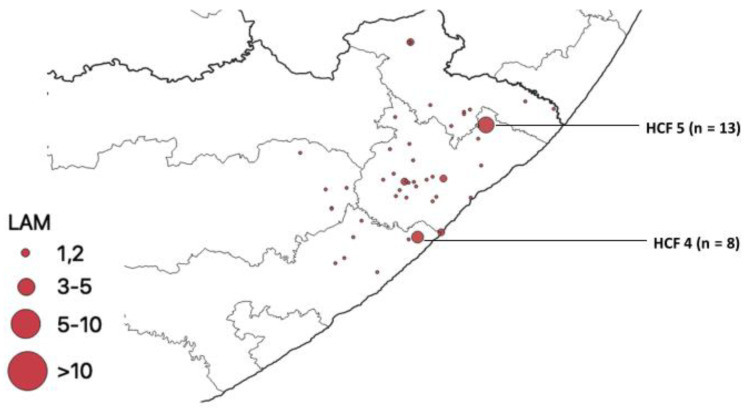
The LAM family being the second most prevalent was mostly identified in two HCFs (HCF 4 and HCF 5) (Figure 9).
Figure 9.
Geospatial distribution of LAM family in healthcare facilities in rural ECP.
4. Discussion
The substantial gap in the detection and treatment of MDR-TB/RMR-TB means that most patients are missed. Hence, identifying geographical areas with a high incidence of disease and adopting active case findings in such areas could help to reduce the detection gap among DR-TB patients. By highlighting high-burden regions with poor public health initiatives and low case detection rates, knowledge of the spatial distribution of tuberculosis could help with control and prevention efforts. This identification enables decision-makers to undertake targeted measures for the prevention and management of MDR-TB hotspots in order to stop the spread of the disease [4,23,24]. This may be crucial in areas with limited resources and nations with high MDR-TB burden [4]. This is the first epidemiological study in the Eastern Cape combining GIS analysis with molecular-based methods to describe the distribution of DR-TB by mapping the distribution of DR gene mutations and genotypes.
Strategic measures in combating the spread of DR-TB include active surveillance, screening for DR-TB, early isolation, and management of confirmed cases [25]. The first step in easing the burden of DR-TB is to comprehend the geographical distribution of the disease and identify regions with the greatest prevalence of notified cases [23]. Drug resistance in Mtb is singularly mediated by chromosomal mutations [25]. Our study detected mutations in rpoB, katG, and inhA from the four municipalities. Most of the mutations are concentrated in the O. R. Tambo district municipality. This may be because it is more populous than the other municipalities. HCF 5, HCF 2, and HCF 4 had the highest number of mutations in the inhA gene (Figure 4); this means INH can still be used but in high doses for the treatment of TB in patients from the catchment areas of the clinic, however, due to development of mutations in genes during treatment, treatment management is needed. These HCFs are scattered in the municipalities, and they are not equidistant to one another. Our study setting being burdened with TB makes it easy to exchange resistant strains which increases mutation rates. The study setting has more of the Beijing family known to be more transmissible than other families and more prevalent in Western Cape Province, which is a neighboring province. Published evidence reviewing the frequency and distribution of gene mutations in Africa reported that out of the 22 gene mutations reported from 25 countries in Africa, rpoB ranked the highest. It further reported that mutations of rpoB, katG, gyrA, and inhA are documented in almost all regions of Africa, which is an indication of the widespread rifampicin- and isoniazid-resistant TB throughout the entire continent. Hence, TB treatment in Africa using rifampicin, isoniazid, fluoroquinolone, and bedaquiline should be handled with caution [26].
The coexistence of both drug-susceptible and drug-resistant bacteria within the same patient was observed at 17.9% of the total isolates of the study with 5 HCF that had the most genes; the rpoB gene (RIF resistance), the katG gene (high-level INH resistance), and the promoter region of the inhA gene (low-level INH resistance) with heteroresistance. This may contribute to the difficulty in treating tuberculosis, as it is the precursor to full resistance [27,28,29]. Heteroresistance is a result of mixed infection, with two or more Mtb distinct strains in the same patient or the presence of different subpopulations caused by the microevolution of the single strain within the host [30]. This may endanger the effective treatment of patients with both RIF and INH, thereby leading to the development of anti-TB drug resistance in the study area, which underscores surveillance of heteroresistance from patients prior to and after treatment. The changing drug resistance patterns detected in patients with tuberculosis also confirmed the possibility that heteroresistance can persist over a long period [31]. Studies on heteroresistance have reported a prevalence ranging from 20% to 57% [32,33,34], which was higher than our study but observed to be increasing each year. Most of the studies focused on samples taken before treatment was initiated, presumably to show that the presence of heteroresistance should be considered in formulating treatment regimens. In our study, the samples were collected during the treatment period, which means that heteroresistance can be present even if the patient is on treatment and agrees with van Rie et al. [31] who reported the persistence of heteroresistance over a long period. The detection of heteroresistance is vital to preventing the selection of drug resistance during antibiotic treatment [35].
Genomic analysis of Mtb clinical isolates worldwide has revealed differences in the geographical distributions and apparent host preference of distinct phylogenetic lineages [36]. According to [37,38,39], genomic analysis helps to identify clinical features of predominant or emerging genotypes and is important from a public health perspective because they describe epidemiological associations with outbreaks and transmission routes. Studies that are investigating the relationship among Mtb across different geographical areas are impacting positively the programs set to end TB because they help to understand the transmission of TB [40].
Of 441 isolates spoligotyped, 437 revealed distinct spoligotype patterns. Patterns of 410 (93.1%) isolates matched a pre-existing SIT in the SITVIT2 database, while 27 unique patterns (6.1%) were not in the database. The Mtb population in this study area was genetically diverse with the Beijing lineage and its members, which is regarded as a successful clone of Mtb associated with drug resistance in some parts of the world [41]. A comparison of lineages among all clinics/hospitals in this study shows that the Beijing family was the commonest genotype found in all the hospitals/clinics, with HCF 2, HCF 3, HCF 5, and HCF 6 having a higher proportion of Beijing isolates. The Beijing family is known to be more transmissible than other families and more prevalent in Eastern Cape and Western Cape provinces, which are neighbors [42,43]; this confirms its high prevalence in this study (42%). This family has been detected in studies reported from other parts of South Africa, including Limpopo, the Western Cape, and Mpumalanga [43,44]. Nelson et al. [45] reported that human movement between rural and urban areas in search of employment in South Africa is common and serves as a bridge for transporting pathogens across long distances. The LAM family was the second most prevalent. This strain is widely distributed in KwaZulu Natal, which is a neighboring province to the Eastern Cape [46].
Table 3 compares the distribution of the lineage of Mtb from our study with that in other studies, including other provinces in South Africa and sub-Saharan Africa. The Beijing family belonging to Lineage 2 is the most prevalent in our study, which is the same with the Western Cape, Gauteng, and North-West, but a different outlook is portrayed in countries such as Zambia and Botswana with LAM family predominating. Major lineages, L1 to L7, have been identified from analyses of Mtb strains worldwide [47,48,49], but recently this has been updated to include Lineages 8 and 9 [49]. Lineage 2 includes strains, the majority of which are members of the so-called Beijing family. More than a quarter of the world’s tuberculosis epidemic is attributable to the Beijing family, which is the most prolific genotype of Mtb. The widespread proliferation of this strain family in recent decades, its propensity to spread disease, association with drug resistance, treatment failure, early relapse, recurrence, fever during early therapy, and increased risk of transmission chains globally have all garnered considerable attention, according to reports from several clinical trials. Evidence from both experimental and clinical data points to Beijing strains’ hypervirulent phenotype and increased mutation rate when compared to other strains [47,49,50].
Table 3.
Distribution of Mtb lineages in this study in comparison with other studies.
| Our Study n (%) | Western Cape n (%) [43] |
Gauteng n (%) [43] |
KZN n (%) [43] |
Free Staten (%) [51] |
Limpopo n (%) [44] |
North- West n (%) [43] |
Zambia n (%) [52] |
Botswana n (%) [53] |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Family | 441 | 897 | 142 | 230 | 86 | 226 | 358 | 274 | 458 |
| 2 | Beijing | 185 (42) | 599 (66.8) | 44 (31.0) | 57 (24.8) | 5 (5.8) | 34 (15.0) | 88 (24.6) | 1 (0.4) | 41 (9.0) |
| 4 | LAM | 83 (18.8) | 53 (5.9) | 29 (20.4) | 42 (18.3) | 18 (20.9) | 60 (26.5) | 54 (15.1) | 149 (54.4) | 150 (32.8) |
| 4 | X | 48 (10.9) | 88 (9.8) | 9 (6.3) | 14 (6.1) | 5 (5.8) | 12 (5.3) | 27 (7.5) | 19 (6.9) | 75 (16.4) |
| 4 | T | 34 (7.7) | 61(6.8) | 18 (12.7) | 29 (12.6) | 14 (16.3) | 43 (19.0) | 60 (16.8) | 39 (14.2) | 73 (15.9) |
| 4 | S | 31 (7.0) | 23 (2.6) | 9 (6.3) | 49 (21.3) | 6 (7.0) | 21 (9.1) | 37 (10.3) | 4 (1.5) | 62 (13.5) |
| 1 | EAI MANU |
16 (3.6) 0 |
6 (0.7) 0 |
12 (8.5) 3 (2.1) |
6 (2.6) 2 (0.9) |
0 0 |
11 (4.9) 3 (1.3) |
24 (6.7) 6 (1.7) |
6 (2.2) | 31 (6.8) 2 (0.4) |
| 4 | H | 6 (1.4) | 10(1.1) | 6 (4.8) | 8 (3.5) | 1 (1.2) | 31 (13.7) | 26 (7.3) | 0 | 21 (4.6) |
| 3 | CAS | 5 (1.1) | 8 (0.9) | 2 (1.4) | 5 (2.2) | 0 | 10 (4.4) | 6 (1.7) | 44 (16.1) | 2 (0.4) |
| 3 | U | 0 | 7 (0.8) | 0 | 1 (0.4) | 0 | 1 (0.4) | 1 (0.3) | 0 | 3 (0.7) |
Mtb genotypes differ amongst populations and are strongly influenced by geography. In terms of host immune response modulation, transmissibility, and disease severity, different Mtb lineages frequently exhibit distinct traits and virulence profiles. A better understanding of phenotypic variations caused by the genetic diversity of Mtb strains is important when attempting to improve TB control measures [49]. Previous research has revealed that immune responses are significantly variable amongst genetically diverse Mtb strains. Lineage 2 Mtb strains are the most virulent and were shown to only elicit a weak immune response in mice. Evidence revealed that patients who were infected with Lineage 2 strains were more likely to die of TB compared to patients infected with other strains. Investigating the pathogenicity of distinct lineages of Mtb is, therefore, crucial [50].
5. Conclusions
The key control for DR-TB is to interrupt its transmission; this was done by identifying hotspots of gene mutations and lineages, especially those that are drivers of DR-TB transmission and mixed infections. The identification of areas where DR-TB is concentrated could assist policy makers to implement targeted interventions aimed at the prevention and management of TB transmission. This is particularly important in resource-limited settings and high DR-TB burden areas, such as the rural areas of ECP. Targeted interventions to the rural community may be necessary as these areas find it impossible to provide DR-TB services across the communities as the diagnosis and treatment of DR-TB are challenged by factors such as poverty and co-infection with HIV.
Acknowledgments
The authors are grateful to the NHLS TB laboratory staff and participating clinics for their support during sample analysis and data collection.
Author Contributions
Conceptualization, L.M.F.; methodology, L.M.F.; formal analysis, L.M.F., A.D., S.O. and RW.; investigation, L.M.F.; resources, L.M.F. and RW; data curation, A.D. and S.O.; writing—original draft preparation, L.M.F. and M.C.H.; writing—review and editing, L.M.F. and M.C.H.; visualization, L.M.F. and M.C.H.; supervision, R.M.W., S.V. and T.A.; project administration, L.M.F.; funding acquisition, L.M.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Research Ethics and Biosafety Committee of the Faculty of Health Sciences of Walter Sisulu University (Ref. No. 026/2019) and Eastern Cape Department of Health (Ref No EC_201904_011).
Informed Consent Statement
Not applicable. This study used routine samples received in National Health Laboratory Services (NHLS) TB laboratory.
Data Availability Statement
Data can be requested from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
Financial support for this study was obtained from the South African Medical Research Council (SAMRC) Research development grant (Pilot grant).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.World Health Organization Global Tuberculosis Report 2019. [(accessed on 21 January 2022)]. Available online: https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714.
- 2.Olmo-Fontánez A.M., Turner J. Tuberculosis in an Aging World. Pathogens. 2022;11:1101. doi: 10.3390/pathogens11101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Global Tuberculosis Report Geneva: World Health Organization. 2022. [(accessed on 28 September 2022)]. Licence: CC BY-NC-SA 3.0 IGO. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022.
- 4.Alene K.A., Viney K., McBryde E.S., Clements A.C.A. Spatial patterns of multidrug resistant tuberculosis and relationships to socio-economic, demographic and household factors in northwest Ethiopia. PLoS ONE. 2017;12:e0171800. doi: 10.1371/journal.pone.0171800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maja T.F., Maposa D. An Investigation of Risk Factors Associated with Tuberculosis Transmission in South Africa Using Logistic Regression Model. Infect. Dis. Rep. 2022;14:66. doi: 10.3390/idr14040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray C.J., Vos T., Lozano R., Naghavi M., Flaxman A.D., Michaud C., Ezzati M., Shibuya K., Salomon J.A., Abdalla S., et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 7.Statistics South Africa Mortality and Causes of Death in South Africa. Findings from Death Notification. Pretoria, South Africa. Stats SA’s June 2021 Report. [(accessed on 21 August 2022)]; Available online: http://www.statssa.gov.za/publications/P0302/Mid%20year%20estimates%202021_presentation.pdf.
- 8.World Health Organisation The Global Tuberculosis Report 2020. Geneva: World Health Organization. 2020. [(accessed on 28 September 2022)]. Licence: CC BY-NC-SA 3.0 IGO. Available online: https://www.who.int/publications/i/item/9789240013131.
- 9.World Health Organization Global Tuberculosis Report 2016 Geneva World Health Organization. [(accessed on 15 June 2022)]. Available online: https://apps.who.int/iris/handle/10665/250441.
- 10.Robsky K.O., Kitonsa P.J., Mukiibi J., Nakasolya O., Isooba D., Nalutaaya A., Salvatore P.P., Kendall E.A., Katamba A., Dowdy D. Spatial distribution of people diagnosed with tuberculosis through routine and active case finding: A community-based study in Kampala, Uganda. Infect. Dis. Poverty. 2020;9:73. doi: 10.1186/s40249-020-00687-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen T.N., Berre A.L., Bañuls A.L., Nguyen T.V. Molecular diagnosis of drug-resistant tuberculosis; a literature review. Front. Microbiol. 2019;10:794. doi: 10.3389/fmicb.2019.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chihota V.N., Niehaus A., Streicher E.M., Wang X., Sampson S.L., Mason P., Källenius G., Mfinanga S.G., Pillay M., Klopper M., et al. Geospatial distribution of Mycobacterium tuberculosis genotypes in Africa. PLoS ONE. 2018;13:e0200632. doi: 10.1371/journal.pone.0200632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Z., Lin D., Chongsuvivatwong V., Zhao J., Lin M., Ou J., Zhao J. Spatiotemporal patterns and ecological factors of tuberculosis notification: A spatial panel data analysis in Guangxi, China. PLoS ONE. 2019;14:e0212051. doi: 10.1371/journal.pone.0212051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter J.D.H., Ogden J.A., Pronyk P. The way forward: An integrated approach to tuberculosis control. In: Porter J.D.H., Grange J.M., editors. Tuberculosis—An Interdisciplinary Perspective. Imperial College Press; London, UK: 1999. pp. 359–378. [Google Scholar]
- 15.Asare P., Otchere I.D., Bedeley E., Brites D., Loiseau C., Baddoo N.A., Asante-Poku A., Osei-Wusu S., Prah D.A., Borrell S., et al. Whole genome sequencing and spatial analysis identifies recent tuberculosis transmission hotspots in Ghana. Front. Med. 2020;7:161. doi: 10.3389/fmed.2020.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwitira I., Karumazondo N., Shekede M.D., Sandy C., Siziba N., Chirenda J. Spatial patterns of pulmonary tuberculosis (TB) cases in Zimbabwe from 2015 to 2018. PLoS ONE. 2021;16:e0249523. doi: 10.1371/journal.pone.0249523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks M.B., Millones A.K., Puma D., Contreras C., Jimenez J., Tzelios C., Jenkins H.E., Yuen C.M., Keshavjee S., Lecca L., et al. Mapping local hot spots with routine tuberculosis data: A pragmatic approach to identify spatial variability. PLoS ONE. 2022;7:e0265826. doi: 10.1371/journal.pone.0265826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caprarelli G., Fletcher S. A brief review of spatial analysis concepts and tools used for mapping, containment and risk modelling of infectious diseases and other illnesses. Parasitology. 2014;141:581–601. doi: 10.1017/S0031182013001972. [DOI] [PubMed] [Google Scholar]
- 19.Chan-Yeung M., Yeh A.G., Tam C.M., Kam K.M., Leung C.C., Yew W.W., Lam C.W. Socio-demographic and geographic indicators and distribution of tuberculosis in Hong Kong: A spatial analysis. Int. J. Tuberc. Lung Dis. 2005;9:1320–1326. [PubMed] [Google Scholar]
- 20.Zulu L.C., Kalipeni E., Johannes E. Analyzing spatial clustering and the spatiotemporal nature and trends of HIV/AIDS prevalence using GIS: The case of Malawi, 1994–2010. BMC Infect. Dis. 2014;14:285. doi: 10.1186/1471-2334-14-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hain Lifescience Company History and Product Releases. 2016. [(accessed on 11 February 2021)]. Available online: http://www.hain-lifescience.de/en/company/history.html.
- 22.Couvin D., David A., Zozio T., Rastogi N. Macro-geographical specificities of the prevailing tuberculosis epidemic as seen through SITVIT2, an updated version of the Mycobacterium tuberculosis genotyping database. Infect. Genet. Evol. 2019;72:31–43. doi: 10.1016/j.meegid.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 23.Alene K.A., Xu Z., Bai L., Yi H., Tan Y., Gray D., Viney K., Archie C.A., Clements A.C.A. Spatial clustering of drug-resistant tuberculosis in Hunan province, China: An ecological study. BMJ Open. 2021;11:e043685. doi: 10.1136/bmjopen-2020-043685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tafess K., Beyen T.K., Girma S., Girma A., Siu G. Spatial clustering and genetic diversity of Mycobacterium tuberculosis isolate among pulmonary tuberculosis suspected patients, Arsi Zone, Ethiopia. BMC Pulm. Med. 2021;21:206. doi: 10.1186/s12890-021-01567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumwenda G., Chipungu G., Sloan D., Kaimila Y., Chiumya K., Pangani H. The occurrence and frequency of genomic mutations that mediate Isoniazid and Rifampicin resistance in Mycobacterium tuberculosis isolates from untreated pulmonary Tuberculosis cases in urban Blantyre, Malawi. Malawi Med. J. 2018;30:1–5. doi: 10.4314/mmj.v30i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam B.H., Nishizato K., Sugimoto K., Ueki M., Karibe R., Yashio T., Nishi Y., Munkhtuya T. Frequency and distribution of drug-resistant tuberculosis gene mutations in Africa between 2010 and 2020. South. Afr. J. Public Health. 2022;5:60–64. [Google Scholar]
- 27.Chen Y., Jiang Q., Zou J., Yang T., Liu Q., Luo G., Gan M., Jiang Y., Takiff H.E., Lu L., et al. Deep whole-genome sequencing reveals no evidence for heteroresistance influencing treatment outcomes among drug-susceptible tuberculosis patients. Tuberculosis. 2021;130:102120. doi: 10.1016/j.tube.2021.102120. [DOI] [PubMed] [Google Scholar]
- 28.Tessema B., Beer J., Emmrich F., Sack U., Rodloff A.C. Analysis of gene mutations associated with isoniazid, rifampicin and ethambutol resistance among Mycobacterium tuberculosis isolates from Ethiopia. BMC Infect. Dis. 2012;12:37. doi: 10.1186/1471-2334-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brhane M., Kebede A., Petros Y. Molecular detection of multidrug-resistant tuberculosis among smear-positive pulmonary tuberculosis patients in Jigjiga town, Ethiopia. Infect. Drug Resist. 2017;10:75–83. doi: 10.2147/IDR.S127903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q., Via L.E., Luo T., Liang L., Liu X., Wu S., Shen Q., Wei W., Ruan X., Yuan X., et al. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci. Rep. 2015;5:17507. doi: 10.1038/srep17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Rie A., Victor T.C., Richardson M., Johnson R., van der Spuy G.D., Murray E.J., Beyers N., van Pittius N.C.G., van Helden P.D., Warren R.M. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am. J. Respir. Crit. Care Med. 2005;172:636–642. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin S.S., Modongo C., Baik Y., Allender C., Engelthaler D.M., Warren R.M., Klausner J., Zetola N. Deep sequencing to estimate the prevalence of heteroresistance among mixed-strain and single-strain Mycobacterium tuberculosis infections. Am. J. Respir. Crit. Care Med. 2017;195:A1177. [Google Scholar]
- 33.Operario D.J., Koeppel A.F., Turner S.D., Bao Y., Pholwat S., Banu S., Foongladda S., Mpagama S., Gratz J., Ogarkov O., et al. Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis. PLoS ONE. 2017;12:e0176522. doi: 10.1371/journal.pone.0176522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann-Thiel S., van Ingen J., Feldmann K., Turaev L., Uzakova G.T., Murmusaeva G., van Soolingen D., Hoffmann H. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur. Respir. J. 2009;33:368–374. doi: 10.1183/09031936.00089808. [DOI] [PubMed] [Google Scholar]
- 35.Rigouts L., Miotto P., Schats M., Lempens P., Cabibbe A.M., Galbiati S., Lampasona V., de Rijk P., Cirillo D.M., de Jong B.C. Fluoroquinolone heteroresistance in Mycobacterium tuberculosis: Detection by genotypic and phenotypic assays in experimentally mixed populations. Sci. Rep. 2019;9:11760. doi: 10.1038/s41598-019-48289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couvin D., Reynaud Y., Rastogi N. Two tales: Worldwide distribution of Central Asian (CAS) versus ancestral East-African Indian (EAI) lineages of Mycobacterium tuberculosis underlines a remarkable cleavage for phylogeographical, epidemiological and demographical characteristics. PLoS ONE. 2019;14:e0219706. doi: 10.1371/journal.pone.0219706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caws M., Thwaites G., Dunstan S., Hawn T.R., Lan N.T., Thuong N.T., Stepniewska K., Huyen M.N., Bang N.D., Loc T.H., et al. The Influence of Host and Bacterial Genotype on the Development of Disseminated Disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4:e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thwaites G., Caws M., Chau T.T., D’Sa A., Lan N.T., Huyen M.N., Gagneux S., Anh P.T., Tho D.Q., Torok E., et al. Relationship between Mycobacterium tuberculosis Genotype and the Clinical Phenotype of Pulmonary and Meningeal Tuberculosis. J. Clin. Microbiol. 2008;46:1363–1368. doi: 10.1128/JCM.02180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Groll A., Martin A., Felix C., Prata P.F.S., Honscha G., Portaels F., Vandame P., da Silva P.E., Palomino J.C. Fitness study of the RDRio lineage and Latin American- Mediterranean family of Mycobacterium tuberculosis in the city of Rio Grande, Brazil. FEMS Immunol. Med. Microbiol. 2010;58:119–127. doi: 10.1111/j.1574-695X.2009.00611.x. [DOI] [PubMed] [Google Scholar]
- 40.Maung H.M., Palittapongarnpim P., Aung H.L., Surachat K., Nyunt W.W., Chongsuvivatwong V. Geno-Spatial Distribution of Mycobacterium Tuberculosis and Drug Resistance Profiles in Myanmar–Thai Border Area. Trop. Med. Infect. Dis. 2020;5:153. doi: 10.3390/tropicalmed5040153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y., Mathema B., Zhao Q., Zheng X., Li D., Jiang W., Wang W., Xu B. Comparison of the socio-demographic and clinical features of pulmonary TB patients infected with sub-lineages within the W-Beijing and non-Beijing Mycobacterium tuberculosis. Tuberculosis. 2016;97:18–25. doi: 10.1016/j.tube.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Karmakar M., Trauer J.M., Ascher D.B., Denholm J.T. Hyper transmission of Beijing lineage Mycobacterium tuberculosis: Systematic review and meta-analysis. J. Infect. 2019;79:572–581. doi: 10.1016/j.jinf.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 43.Said H., Ratabane J., Erasmus L., Gardee Y., Omar S., Dreyer A., Ismail F., Bhyat Z., Lebaka T., van der Meulen M., et al. Distribution and Clonality of drug-resistant tuberculosis in South Africa. BMC Microbiol. 2021;21:157. doi: 10.1186/s12866-021-02232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maguga-Phasha N.T., Munyai N.S., Mashinya F., Makgatho M.E., Mbajiorgu E.F. Genetic diversity and distribution of Mycobacterium tuberculosis genotypes in Limpopo, South Africa. BMC Infect. Dis. 2017;17:764. doi: 10.1186/s12879-017-2881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson K.N., Shah N.S., Mathema B., Ismail N., Brust J.C., Brown T.S., Auld S.C., Omar S.V., Morris N., Campbell A., et al. Spatial patterns of extensively drug-resistant tuberculosis transmission in KwaZulu-Natal, South Africa. J. Infect. Dis. 2018;218:1964–1973. doi: 10.1093/infdis/jiy394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pillay M., Sturm A.W. Evolution of the extensively drug-resistant F15/LAM4/KZN strain of Mycobacterium tuberculosis in KwaZulu-Natal, South Africa. Clin. Infect. Dis. 2007;45:1409–1414. doi: 10.1086/522987. [DOI] [PubMed] [Google Scholar]
- 47.Luo T., Comas I., Luo D., Lu B., Wu J., Wei L., Yang C., Liu Q., Gan M., Sun G., et al. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc. Natl. Acad. Sci. USA. 2015;112:8136–8141. doi: 10.1073/pnas.1424063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kone B., Somboro A.M., Holl J.L., Baya B., Togo A.A., Sarro Y.D., Diarra B., Kodio O., Murphy R.L., Bishai W., et al. Exploring the usefulness of molecular epidemiology of tuberculosis in Africa: A systematic review. Int. J. Mol. Epidemiol. Genet. 2020;11:1. [PMC free article] [PubMed] [Google Scholar]
- 49.Le Hang N.T., Hijikata M., Maeda S., Miyabayashi A., Wakabayashi K., Seto S., Diem N.T., Yen N.T., Van Duc L., Thuong P.H., et al. Phenotypic and genotypic features of the Mycobacterium tuberculosis lineage 1 subgroup in central Vietnam. Sci. Rep. 2021;11:13609. doi: 10.1038/s41598-021-92984-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., He L., Huang H., Shi C., Ni X., Dai G., Ma L., Li W. Mycobacterium tuberculosis lineage distribution in Xinjiang and Gansu Provinces, China. Sci. Rep. 2017;7:1068. doi: 10.1038/s41598-017-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Spoel van Dijk A., Makhoahle P.M., Rigouts L., Baba K. Diverse Molecular genotypes of Mycobacterium tuberculosis complex isolates circulating in the Free State, South Africa. Int. J. Microbiol. 2016;2016:6572165. doi: 10.1155/2016/6572165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solo E.S., Suzuki Y., Kaile T., Bwalya P., Lungu P., Chizimu J.Y., Shah Y., Nakajima C. Characterization of Mycobacterium tuberculosis genotypes and their correlation to multidrug resistance in Lusaka, Zambia. Int. J. Infect. Dis. 2021;102:489–496. doi: 10.1016/j.ijid.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Mogashoa T., Melamu P., Ley S.D., Streicher E.M., Iketleng T., Kelentse N., Mupfumi L., Mokomane M., Kgwaadira B., Novitsky V., et al. Genetic diversity of Mycobacterium tuberculosis strains circulating in Botswana. PLoS ONE. 2019;14:e0216306. doi: 10.1371/journal.pone.0216306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data can be requested from the corresponding author.