Abstract
Blood is conventionally thought to be sterile. However, emerging evidence on the blood microbiome has started to challenge this notion. Recent reports have revealed the presence of genetic materials of microbes or pathogens in the blood circulation, leading to the conceptualization of a blood microbiome that is vital for physical wellbeing. Dysbiosis of the blood microbial profile has been implicated in a wide range of health conditions. Our review aims to consolidate recent findings about the blood microbiome in human health and to highlight the existing controversies, prospects, and challenges around this topic. Current evidence does not seem to support the presence of a core healthy blood microbiome. Common microbial taxa have been identified in some diseases, for instance, Legionella and Devosia in kidney impairment, Bacteroides in cirrhosis, Escherichia/Shigella and Staphylococcus in inflammatory diseases, and Janthinobacterium in mood disorders. While the presence of culturable blood microbes remains debatable, their genetic materials in the blood could potentially be exploited to improve precision medicine for cancers, pregnancy-related complications, and asthma by augmenting patient stratification. Key controversies in blood microbiome research are the susceptibility of low-biomass samples to exogenous contamination and undetermined microbial viability from NGS-based microbial profiling, however, ongoing initiatives are attempting to mitigate these issues. We also envisage future blood microbiome research to adopt more robust and standardized approaches, to delve into the origins of these multibiome genetic materials and to focus on host–microbe interactions through the elaboration of causative and mechanistic relationships with the aid of more accurate and powerful analytical tools.
Keywords: host–microbe interaction, microbial commensalism, dysbiosis, bacterial translocation, septicaemia
1. Introduction
Blood is commonly thought to be sterile, except for in certain medical conditions such as septicaemia, whereby systemic infections of bacteria, viruses and fungi occur. The gold standard to detect living microbes in the bloodstream is blood culture. However, the emergence of next generation sequencing (NGS) technology has given rise to highly sensitive approaches such as 16S sequencing and shotgun metagenomics for microbial detection and taxonomic classification in an untargeted manner [1,2]. These NGS-based approaches are widely used to profile microbial populations in different compartments, including the gut [3], airway [4], skin [5], oral cavity [6] and urogenital tract [7], markedly advancing our understanding of the human microbiome and host–microbe relationship in physiological and various pathological contexts. Interestingly, many microbial analyses of blood specimens have detected bacteria and their genetic materials even in healthy individuals [8,9], leading to the conceptualization of a human blood microbiome and bringing the dogma of blood sterility into question.
The blood microbiome is defined by the assemblage of living microorganisms present in the blood. In human microbiome research, identifying a universal group of microbes (i.e., core microbiome) at a specific anatomical site that is shared by most humans and is crucial for host biological function is a major goal [10]. Likewise, the presence of a core healthy blood microbiome is postulated, the disturbance of which could potentially contribute to various diseases. Thus far, blood microbiome profiles have been assessed not only in healthy individuals, but also in patients with various health complications. The data reveal exciting clinical prospects of using blood microbiome genetic signature for risk stratification, diagnosis, disease surveillance and drug development (Figure 1). Nonetheless, the existence of the human blood microbiome remains highly debatable. The controversies typically revolve around two issues, namely the high risk of microbial contamination in low-biomass samples and the undetermined viability of blood microbiota based on culture-independent profiling methods. Furthermore, there is also no consensus on the research methodology and experimental controls to enable contaminant-free blood microbiome studies. As the concept of the blood microbiome has been well-summarized [8,9,11], our review aims to consolidate recent findings about the blood microbiome and to highlight the controversies, prospects, and challenges of the research topic.
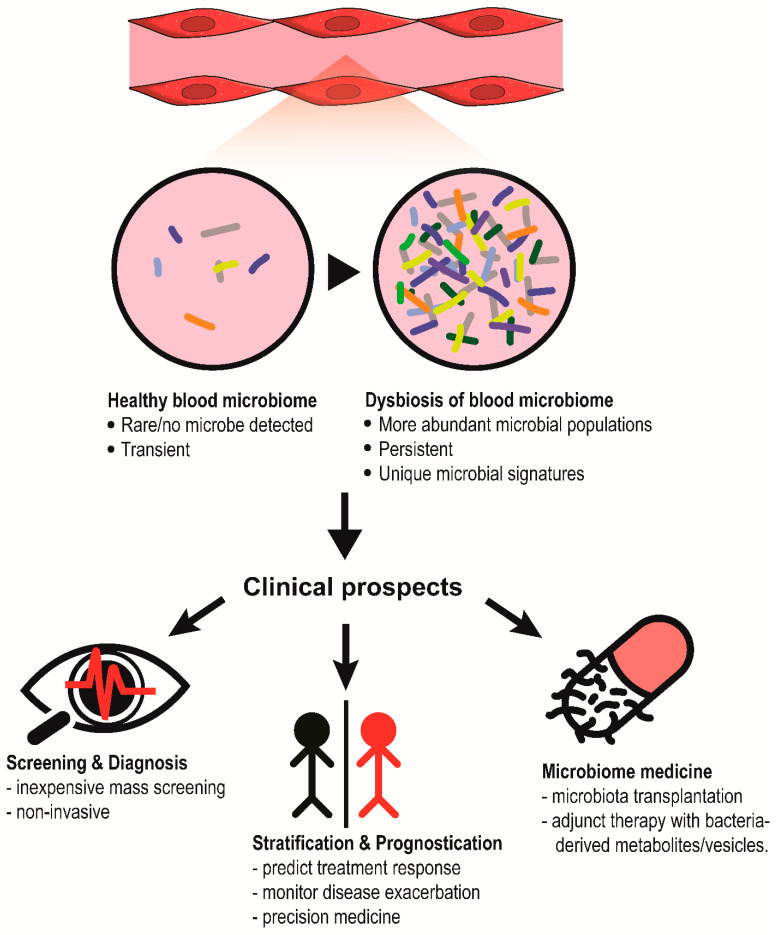
Figure 1.
Clinical prospects of blood microbiome. In human diseases, the circulating microbial populations could become more abundant, forming a unique and persistent microbial signature that can be used for diagnosis, patient stratification or even the development of microbiome-based therapy.
2. Characterization of a Healthy Core Blood Microbiome
Emerging evidence highlights the presence of microorganisms in the blood of healthy individuals. For instance, bacterial growth was detected using aerobic and anaerobic blood cultures in up to 60% of donated blood packs [12]. Using powerful PCR and NGS techniques, 16S rRNA was found in 100% of blood samples [13,14]. Microbial DNA has also been reported in the blood from neonates [15] or even other animal species, such as dogs [16]. In contrast, some studies concluded that the presence of bacteria in blood is an unusual event because most healthy donors (78% to 84%) do not carry any bacterial species in their blood [17,18]. The large variation in the frequency of blood microbe-positive cases could be attributable to different experimental controls and decontamination strategies. In a recent preprint by Tan et al. (2022) [17], which aimed to investigate the presence (or absence) of a healthy core blood microbiome, the team performed shotgun metagenomic profiling of one of the largest multicentre cohorts from the SG10K_Health dataset (n = 9770 healthy donors), followed by a set of stringent quality control criteria and decontamination filters to minimize artefacts from sequencing, host DNA, taxonomic assignment, and batch-specific contaminants during sample preparation and from the reagent kits. They discovered that microbial DNA was present only in 16% of the healthy individuals at a median of one microbial species per individual [17]. In view of the ubiquity of environmental contamination at every step of sample processing, such stringent filters are effective in improving the signal-to-noise ratio, albeit at the expense of masking some genuine signals.
Additionally, there is no consensus on the composition of a healthy blood microbiome based on existing studies. Staphylococcus spp. is a common genus found in blood [12,14,17,18], yet information at the species level remains scarce and poorly characterized. Other potential blood microbial signatures include the Proteobacteria phylum [14] and Cutibacterium acnes [17]. There is remarkable bacterial diversity between different blood fractions: buffy coat, red blood cells and plasma [12,14]. The analysis using the SG10K_Health dataset revealed 117 blood microbial species comprising 110 bacteria, 5 viruses and 2 fungi [17]. Most of the species are commensals from the gut, mouth and urogenital tract, however, none of them demonstrated a clear cooccurrence trend nor were consistently detected in more than 5% of the donors, underscoring a significant intersubject variability that was also seen in neonatal blood [15,17]. Thus far, the varying results in the prevalence of blood microbes and the bacterial composition do not support the hypothesis of a core healthy blood microbiome for host functionality. Findings of the large SG10K healthy cohort strongly suggest a transient and sporadic translocation of commensals into the bloodstream which are quickly cleared out and do not lead to prolonged colonization [17]. On the other hand, the persistence of blood microbes could signify the onset of certain diseases, which will be elaborated below. While further validation from other human cohorts is crucial, the initiative to employ a stringent analytical pipeline and decontamination filters is commendable and could potentially be adopted in future blood microbiome research.
3. Clinical Significance of the Blood Microbiome in Human Diseases
Although the concept of a shared healthy blood microbiome remains inconclusive, disruption of mucosal integrity in certain disease states may exacerbate microbial translocation, leading to the persistence of microbes in the bloodstream. Unsurprisingly, many studies have profiled the dynamic changes in the blood microbiome during various pathological conditions, including cardiometabolic diseases, malignancies, inflammatory and immune disorders, and hepatic, respiratory and dermal diseases. Next, we will summarize recent evidence of the blood microbiome in human diseases.
3.1. Cardiometabolic Diseases
Linkages between blood microbial signatures and cardiovascular events and chronic metabolic diseases have been reported (Table 1). Three case-control studies revealed reduced blood microbial diversity in patients with myocardial infarction (MI) and chronic coronary syndrome compared to healthy individuals [19,20,21]. Similarly, a longitudinal cohort study concluded that Eubacteria was inversely correlated with the onset of cardiovascular complications [22]. It is worth noting that the study had disproportionate case (n = 73) and control (n = 3963) arms and only Eubacteria and phylum Proteobacteria levels at baseline were used for analysis. Nonetheless, numerous bacterial taxa were found to be associated with cardiovascular events across different studies. In the Oslo II patient cohort, the genera Kocuria and Enhydrobacter were positively associated with cardiovascular mortality, while the genus Paracoccus reduced the risk [23]. Conversely, Amar et al. (2019) reported the genus Hymenobacter as a positive predictor and four genera (i.e., Brevundimonas, Chryseobacterium, Gordonia and Microbacterium) of MI [19]. In another smaller case-control study, a higher abundance of the genus Bifidobacterium and a reduced abundance of the phylum Bacteroidetes were found in the blood of MI patients [20]. Remarkable microbial differences are also noted in individuals with acute and chronic coronary syndrome, whereby the former has more phyla Proteobacteria and Acidobacteriota, while the latter has more phylum Firmicutes and genus Lactobacillus [21]. Current evidence is overly intricate to pinpoint specific MI-driving blood microbes. Furthermore, the lack of common microbial taxa associated with cardiovascular events across different studies also highlights a diverse blood microbiome in MI patients, which could be attributed to known modifiers of the human microbiota, such as geographical regions, dietary patterns, host genetic, and environmental factors [24]. Interestingly, bacterial families and genera associated with cholesterol and lipid metabolism were markedly suppressed in MI patients, which may promote plaque formation in the coronary arteries [19,20]. Hence, functional analysis is a new strategy that may enable the reconciliation of discrepant microbial targets associated with cardiovascular events from various studies.
Apart from cardiovascular events, some studies have investigated chronic metabolic diseases such as hypertension and type 2 diabetes mellitus (T2DM). D.E.S.I.R. cohort revealed a higher 16S rDNA concentration in those who eventually developed T2DM and abdominal adiposity [25]. A prospective cohort study with primarily Chinese volunteers uncovered the predictive value of the blood genus Bacteroides for T2DM [26] and of the genera Acinetobacter, Sphingomonas and Staphylococcus for hypertension [27]. Despite these exciting results, more research is warranted to validate the causative relationship of the target bacteria with the associated diseases.
Table 1.
Summary of investigations on the blood microbiome in cardiovascular disease and chronic metabolic diseases.
| No. | Disease | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Myocardial infarction | Case-control study | Case = 103; Control = 99 |
16S rDNA (V3-V4 region) sequencing |
|
[19] |
| 2. | Myocardial infarction | Case-control study | Case = 29; Control = 29 |
16S rDNA (V3-V4 region) sequencing |
|
[20] |
| 3. | Acute (ACS) and chronic coronary syndrome (CCS) | Case-control study | Case (ACS) = 70; Case (CCS) = 70; Control = 70 |
16S rDNA (V3-V4 region) sequencing |
|
[21] |
| 4. | Cardiovascular disease (CVD) | Prospective cohort study (D.E.S.I.R.) |
CVD = 73; No CVD = 3863 |
qPCR of conserved 16S rDNA regions |
|
[22] |
| 5. | Cardiovascular mortality | Case-control study (Oslo II) | Case = 227; Control = 178 |
16S rDNA (V3-V5 region) sequencing |
|
[23] |
| 6. | T2DM & obesity | Prospective cohort study (D.E.S.I.R.) |
T2DM = 131; No T2DM = 3149 |
qPCR of conserved 16S rDNA regions |
|
[25] |
| 7. | T2DM | Prospective cohort study (135) |
T2DM = 50; No T2DM = 100 |
16S rDNA (V5-V6 region) sequencing |
|
[26] |
| 8. | Hypertension | Prospective cohort study (135) |
Hypertension = 150; No hypertension = 150 |
16S rDNA (V6-V7 region) sequencing |
|
[27] |
3.2. Cancers
Cancers are conventionally thought to be genetic diseases, however, recent studies have demonstrated unique intratumoral and blood microbiome profiles of certain cancer types (Table 2). Using whole genome and whole transcriptome data from The Cancer Genome Atlas (TCGA), a landmark study focused on sequencing reads that are not mapped to the human genome, and found that 35.2% of the unmapped reads (2.5% of the total reads) could be assigned to microorganisms such as bacteria, archaea and viruses [28]. These intratumoral microbial signatures were effective in discriminating normal tissue from tumors and different types of cancers, but did not correlate well with cancer stages in most cancer types [28]. More importantly, the blood microbial profiles of cancer patients were sufficiently sensitive to distinguish different cancer types, even for very early-stage cancers (stage Ia-IIc) and for tumors without notable mutational burden [28]. Data from independent studies also support the diagnostic and risk stratification capacity of circulating microbial profiles for hepatocellular carcinoma, myeloid cancers, and gastric cancer [29,30,31]. Taken together, the exciting results highlight a blood/intratumoral microbiome-based diagnostic method that may facilitate cancer screening and early detection.
Moreover, the bacterial genetic materials in the blood have been shown to predict the treatment response in cancer. In advanced colorectal cancer, the composition of baseline blood microbiota was significantly different between the immunochemotherapy (oxaliplatin + capecitabine + adoptive T cell immunotherapy) responders and non-responders with the genera Bifidobacterium, Lactobacillus, and Enterococcus more abundantly found among the responders [32]. The abundance of Lactobacillus was associated with longer overall survival [32]. Similarly, the enrichment of class Holophagae, family Peptostreptococcaceae, genera Lewinella and Paludibaculum predicted better clinical outcome and treatment response of nivolumab in advanced non-small cell lung carcinoma patients [33]. The data highlight the prognostic value of the circulating microbial profile in cancers.
The blood microbiome could act as a potential modifier of tumorigenesis and treatment effect. Specific β-glucuronidase and/or β-galactosidase microbes in the gut that can regulate estrogen metabolism, referred to as “estrobolome”, are increasingly known to increase the risk of estrogen receptor-positive breast cancer in postmenopausal women [34,35]. Similarly, dysbiosis of the estrobolome in the blood of breast cancer patients has also been reported. β-glucuronidase-producing bacteria were predominant in breast cancer patients, whereas β-galactosidase-producing bacteria were more abundant in healthy individuals [36]. Interestingly, Staphylococcus, which was depleted in the breast cancer group, was identified as a key modulator of anti-estrogen (i.e., tamoxifen) efficacy [36]. When estrogen-receptor positive breast tumor cells were cotreated with tamoxifen and S. aureus-derived extracellular vesicles, the AKT and ERK oncogenic signaling pathways were greatly suppressed, leading to more cell death [36]. Hence, variations in blood microbial diversity may indirectly regulate estrogen levels and breast cancer susceptibility. Further investigation is warranted to demonstrate the feasibility of blood microbiome engineering for the prevention and adjunctive intervention of breast cancer. In short, growing evidence supports the role of the blood microbiome in cancer diagnosis, prognosis, and therapy. Although the research is still in its infancy, these important findings may spur new advancements using blood-based microbial profiles for precision oncology.
Table 2.
Summary of investigations on the blood microbiome in malignancies.
| No. | Disease | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Pan-cancer | Re-analysis of TCGA whole genome (WGS) and transcriptomic (RNA-seq) sequencing data & cross-sectional patient cohort | TCGA: n = 10,481 (WGS; n = 4831 & RNA-seq; n = 13,285) In-house cohort: HIV- and cancer-free individuals = 69; Prostate cancer = 59; Lung cancer = 25; Melanoma = 16. |
WGS, RNAseq and shotgun metagenomic sequencing |
|
[28] |
| 2. | Hepatocellular carcinoma (HCC) | Cross-sectional study | HCC = 158; Cirrhosis = 166; Healthy = 402 |
16S rDNA (V3-V4 region) sequencing |
|
[29] |
| 3. | Gastric cancer | Case-control study | Gastric cancer = 71; Atypical hyperplasia = 6; Chronic gastritis = 11; Healthy = 13 |
16S rDNA (V1-V2 region) sequencing |
|
[30] |
| 4. | Myeloid malignancies | Cross-sectional study | Acute myeloid leukemia (AML) = 612; Myelodysplastic syndromes (MDS) = 640; Myelodysplastic syndromes/myeloproliferative neoplasms (MDS/MPN) = 264; Myeloproliferative neoplasms (MPN) = 354; Healthy = 12. |
WGS |
|
[31] |
| 5. | Colorectal cancer | Retrospective cohort study | n = 39 | 16S rDNA (V3-V4 region) sequencing |
|
[32] |
| 6. | Non-small cell lung cancer (NSCLC) | Single-arm study | n = 72 | 16S rDNA (V3-V4 region) sequencing |
|
[33] |
| 7. | Breast cancer | Cross-sectional study | Healthy = 192; Breast cancer = 96 |
16S rDNA (V3-V4 region) sequencing |
|
[36] |
3.3. Liver Diseases
Most of the bacteria that translocate from the gut lumen into the blood circulation enter the liver via the portal vein system. Biologically, the liver acts as a sieve to trap and clear the invaders with the aid of local and systemic immune cells [37]. Hence, individuals with liver dysfunction, in particular fibrosis and cirrhosis, are more prone to septicaemia [37]. Recent studies have highlighted remarkable alterations in the blood microbiome in cirrhotic patients (Table 3). Liver fibrosis and cirrhosis markedly increase the abundance and diversity of circulating bacteria, many of which are gut commensals, suggesting a higher tendency of microbial translocation from the gastrointestinal tract [38,39,40,41]. For instance, the genus Bacteroides and family Enterobacteriaceae are highly enriched in cirrhotic patients compared to healthy individuals [39,40]. Interestingly, the microbiota of blood, ascites fluid and stool showed some commonalities between different anatomical sites, reinforcing the contribution of microbial translocation in shaping the blood microbiome in a diseased state.
Distinct blood microbiome profiles were detected at different venous compartments, namely, portal, hepatic, central and peripheral venous blood, in patients with liver cirrhosis (n = 7) who underwent a surgical intervention known as transjugular intrahepatic portosystemic shunt to restore the blood flow between portal and hepatic veins [42]. In comparison to peripheral blood, which had more genera Brevundimonas and Mucilaginibacter, there were more genera Kocuria, Diaphorobacter and Paracoccus found in hepatic blood, portal and central venous compartments, respectively [42]. Such a compartment-specific blood microbiome was not observed in another study [43]. The interstudy difference highlights possible interference from cirrhosis severity because at a more advanced stage, the hardening and scarring of the liver restricts blood flow and creates a unique microenvironment in the portal vein, which may favour specific bacteria. Surgical procedures such as shunting could potentially introduce microbes from other sources into the circulation, causing varying results to the blood microbiome profile. Hence, further experiments are needed to validate the concept of the compartment-specific blood microbiome.
Certain circulating microbes are associated with the proinflammatory cytokine profile and nitric oxide signaling [39,42,43]. For example, the order Corynebacteriales is inversely associated with inflammatory cytokines, including IFN-γ, IL-17A and TNF-α [44]. A higher abundance of Corynebacteriales and a lower abundance of the genus Massilia may also predict the reversal of portal hypertension in hepatitis C virus (HCV)-induced cirrhosis after the completion of antiviral treatment [44]. These results point towards the role of certain microbes in the regulation of systemic and local vascular resistance through the stimulation of cytokine- and nitric oxide-mediated signaling cascades. In-depth investigation of the host–microbe interaction may reveal new strategies to mitigate inflammation and vasculopathy caused by progressive loss of liver function.
Table 3.
Summary of investigations on the blood microbiome in liver dysfunction.
| No. | Disease | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Liver fibrosis | Cross-sectional study (FLORINASH) | Spanish cohort: Liver fibrosis = 26; No fibrosis = 11 Italian cohort: Liver fibrosis = 11; No fibrosis = 60 |
16S rDNA (V1-V3 region) sequencing |
|
[38] |
| 2. | Cirrhosis | Case-control study | Case = 9; Control = 9 |
qPCR of conserved 16S rDNA region |
|
[39] |
| 3. | Cirrhosis with or without hepatocellular carcinoma | Case-control study | Case = 66; Control = 14 |
16S rDNA (V3-V4 region) sequencing |
|
[40] |
| 4. | Cirrhosis with or without ascites | Case-control study | Case (with ascites) =13; Case (without ascites) =14; Control = 17 |
16S rDNA (V4 region) sequencing |
|
[41] |
| 5. | Decompensated liver cirrhosis receiving transjugular intrahepatic portosystemic shunt | Single-arm study | n = 7 | 16S rDNA sequencing |
|
[42] |
| 6. | Cirrhosis with portal hypertension | Case-control study | Case = 58; Control = 46 |
16S rDNA (V1-V2 region) sequencing |
|
[43] |
| 7. | HCV-induced portal hypertension | Single-arm study | n = 32 | 16S rDNA (V3-V4 region) sequencing |
|
[44] |
3.4. Respiratory Diseases
Studies on the blood microbiome in respiratory diseases are relatively scarce (Table 4). Two reports consistently found distinct bacterial compositions between asthmatic patients and healthy individuals [45,46]. The diversity of the blood microbiome in asthma patients demonstrated higher richness but reduced evenness [46]. Members of the phylum Bacteroidetes, namely Bacteroides, Alistipes, Parabacteroides, and Prevotella, were more commonly found in asthma patients, while the abundance of the phyla Actinobacter, Verrucomicrobia, and Cyanobacteria was slightly reduced [46]. When asthmatic patients were grouped by different inflammatory subtypes, including eosinophilic, neutrophilic, paucigranulocytic, and mixed granulocytic asthma, as well as by clinical features such as lung function and corticosteroid usage, varying bacteria were associated with each manifestation, highlighting the predictive value of blood microbes for patient stratification and personalized medicine. Indeed, differential blood bacteria profiles also enable accurate diagnosis of asthma up to a sensitivity of 0.94, specificity of 0.93 and accuracy of 0.92 [46]. Current data provide favourable prospects for the use of the blood microbiome as a diagnostic marker of asthma.
Unlike most studies that use 16S rDNA or shotgun metagenomics to study the blood microbiome, recent computational pipelines can infer microbial profiles from the RNA-sequencing of peripheral blood samples [47,48]. Using this approach, a microbial signature characterized by the genera Acinetobacter, Serratia, Streptococcus and Bacillus was found to be associated with increased dyspnea severity among former and current smokers [47]. Some of these enriched genera, i.e., Acinetobacter and Streptococcus, were independently reported in another study on tobacco smokers [49], thus providing insights into the reliability and accuracy of the inferred microbial abundance based on host high-throughput sequencing data. More importantly, the analytical approach readily enables the exploration of the host–microbe interactome through an integrative analysis of paired blood microbial profiles and host gene expression. Several genera, namely, Streptococcus, Cutibacterium, Corynebacterium, Lactobacillus, Staphylococcus, and Bacillus, were linked to biological activities such as oxidative phosphorylation and mTOR and WNT/β-catenin signaling in the circulating cells of the subjects [47]. Using a similar methodology, the inferred abundance of E. coli, Bacillus sp., Campylobacter hominis, Pseudomonas sp., Thermoanaerobacter pseudethanolicus, Thermoanaerobacterium thermosaccharolyticum, and Staphylococcus epidermis was correlated with the severity of COVID [48]. These bacteria also predict overactivity of the adaptive immune system and inflammatory response [48]. Taken together, the results not only shed light on the blood microbiome of smokers and COVID patients but also describe an innovative approach for the exploration of host–microbe interactions.
Table 4.
Summary of investigations on the blood microbiome in respiratory diseases.
| No. | Disease/Condition | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Asthma | Case-control study | Case = 5; Control = 5 |
16S rDNA (V4 region) sequencing |
|
[45] |
| 2. | Asthma | Case-control study | Case = 190; Control = 260 |
16S rDNA (V3-V4 region) sequencing |
|
[46] |
| 3. | Smoking | Single-arm from a longitudinal cohort study (COPDGene study) | n = 3655 former and current smokers | RNAseq |
|
[47] |
| 4. | Smoking | Case-control study | Case = 20; Control = 21 |
16S rDNA (V4 region) sequencing |
|
[49] |
| 5. | COVID-19 | Meta-analysis of public transcriptomic data | 17 PBMC of normal samples; 17 PBMC COVID-19 (GSE152418) | RNAseq |
|
[48] |
3.5. Kidney Dysfunction
An aberrant blood microbiome could play a key role in kidney dysfunction, especially chronic kidney disease (CKD) (Table 5). A strong inverse correlation between glomerular filtration rate and the abundance of the circulating phylum Proteobacteria was observed [50]. CKD patients also exhibited a distinct blood microbial signature characterized by the enrichment of the genera Legionella, Serratia, Yersinia, Acinetobacter, Pseudomonas, Lysobacter, Hyphomicrobium, Bacillus, Sediminibacterium and Pseudarcicella, and the depletion of Stenotrophomonas, Paracoccus, Sphingomonas, Tyzzerella, Corynebacterium and Candidatus [50]. A comparison between CKD patients on peritoneal dialysis with and without vascular calcification highlighted the circulating level of the genus Devosia as a potential predictor of increased mortality risk [51]. In IgA nephropathy patients, blood Legionella and Enhydrobacter were highly abundant while Staphylococcus and Streptococcus were overrepresented when the eGFR ≤ 60 mL/min [52]. Collectively, the existing data reveal a possible implication of Legionella and Devosia in kidney impairment and nephropathy-related mortality, respectively. Interestingly, most of the circulating bacteria reported in patients with kidney dysfunction are not typical commensals of the urinary tract [53], implying that urinary mucosal disruption may not contribute to the dysbiotic blood microbiota. Hence, the origin and pathologic roles of disease-associated microbes should be examined to unravel their contribution to the onset of CKD and IgA nephropathy.
Table 5.
Summary of investigations on the blood microbiome in kidney dysfunction.
| No. | Disease | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Chronic kidney disease (CKD) | Cross-sectional study | CKD = 20; Healthy = 20 |
16S rDNA (V3-V4 region) sequencing |
|
[50] |
| 2. | CKD patients on peritoneal dialysis (PD) with vascular calcification (VC) | Cross-sectional study | CKD-PD without VC = 12; CKD-PD with VC = 32 |
16S rDNA (V3-V4 region) sequencing |
|
[51] |
| 3. | IgA nephropathy (Berger’s disease) |
Case-control study | Case = 20; Controls = 20 |
16S rDNA (V3-V4 region) sequencing |
|
[52] |
3.6. Immune and Inflammatory Disorders
Dysbiosis in the blood may occur under inflammatory and immune diseases (Table 6), and these varying levels of bacterial populations may be used as biomarkers for such diseases. In systemic lupus erythematosus (SLE), an autoimmune disease, enriched levels of the genera Desulfoconvexum, Desulfofrigus, Desulfovibrio, Draconibacterium, Planococcus and Psychrilyobacter, and the phylum Gemmatimonadetes were identified [49,54]. Many of these were positively associated with the autoantibody levels in the plasma. Notably, exposure of heat-inactivated Planococcus citreus to peripheral blood mononuclear cells (PBMCs) resulted in heightened production of TNF-α, IL-1β, and IL-6 from monocytes, indicating that the enriched Planococcus in circulation may play a role in inflammation in SLE [49]. Likewise, distinct bacterial communities were found in the blood of rheumatoid arthritis patients, where the genera Halomonas and Shewanella, both implicated in inflammation and several human infections, were significantly elevated compared to healthy subjects [55]. Given that the perturbed circulating bacterial populations in both rheumatic diseases are primarily oral or gut commensals, the integrity of the mucosal barrier may be jeopardized, allowing more bacterial translocation and microbial-induced host inflammation [56]. Therefore, it is crucial to understand whether blood microbiome dysbiosis is a cause or effect of rheumatoid disorders.
Immunosuppression encourages the growth of opportunistic commensals, drastically changing the microbiota in the gastrointestinal tract and plasma [57,58]. In patients who received immunosuppressant regimens after liver transplantation, the levels of the families Anelloviridae, Nocardiaceae, and Microbacteriaceae increased, while that of Enterobacteriaceae decreased over a course of eight weeks post-operation [58]. The microbial profiles were sensitive to the use of antimicrobial agents. Changes in certain bacterial families, such as Xanthomonadaceae and Enterobacteriaceae, were associated with the occurrence of acute host–versus–graft rejection [58]. An ongoing prospective cohort study is currently looking into the role of the circulating microbiome in the pharmacodynamics and pharmacokinetics of various immunosuppressants (e.g., mycophenolate mofetil and tacrolimus) (NCT04953715), which may help to improve the success rate of organ transplantation in the future.
HIV infection is another common factor of immunodeficiency. HIV carriers have higher 16S rDNA concentrations and diverse bacterial compositions in the blood than healthy subjects [49,59]. Many genera such as Veillonella, Massilia, Haemophilus, Arthrobacter, and Fusobacterium, were enriched in the blood of HIV patients, coupled with a reduction in Altererythrobacter, Cryobacterium, and Anaerococcus [49]. Coculture of peripheral blood mononuclear cells with either Massilia timonae or Haemophilus parainfluenzae, but not Anaerococcus prevotii, led to marked elevation of proinflammatory monocytes [49], highlighting that certain pathogenic bacteria may exacerbate the disease by driving chronic inflammation in HIV patients.
In terms of inflammatory diseases, Hyun et al. (2021) used a faeces-induced peritonitis porcine model and revealed the emergence of new circulating bacteria after disease induction, including Escherichia/Shigella, Staphylococcus, Cloacibacterium, Diaphorobacter and Rhodanobacter [60]. Functionally, these enriched bacteria are related to ABC transporters, oxidative phosphorylation, and two-component systems [60], which may aid in the pathogenesis of peritonitis. Interestingly, circulating levels of the genera Escherichia/Shigella and Staphylococcus were also significantly elevated in inflammatory bowel disease (IBD, comprising ulcerative colitis and Crohn’s disease) despite the unremarkable difference in the overall blood microbiome profiles between IBD and healthy individuals [61]. Hence, these common blood microbes could potentially contribute to the onset of inflammatory diseases in the abdominal cavity.
Characteristic to the circulation of patients suffering from severe acute pancreatitis is the severe depletion of the phylum Actinobacteria and an abundance of the phylum Bacteroidetes compared to healthy subjects [62]. On a genus level, the changes translate to an increase in Bacteroides, Stenotrophomonas, Serratia, Rhizobium, Prevotella, Staphylococcus, and Paracoccus, with a stark depletion of Acinetobacter, Lactococcus, Dietzia, Flavobacterium, Pseudomonas, Corynebacterium, Sphingobium, and Brevundimonas [62]. Patients suffering from large vessel vasculitis (LVV) also show distinct blood microbiome compositions, with elevation of classes Cytophagia and Clostridia and unidentified taxa from the Cytophagaceae family with a suppression of genera Zoogloeal and Staphylococcus compared to healthy subjects [63]. The cytochrome P450 pathways were activated, while other biosynthesis pathways were suppressed [63]. Based on the data from independent studies, Escherichia/Shigella and Staphylococcus are consistently implicated in various inflammatory disorders, albeit with varying relative abundance levels compared to healthy subjects. These findings suggest a potential regulatory role of blood microbes in host inflammation which warrants further investigation.
Table 6.
Summary of investigations on the blood microbiome in immune or inflammatory diseases.
| No. | Disease/Condition | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | SLE | Case-control study | Case = 19; control = 30 | 16S rDNA (V4 region) sequencing |
|
[49] |
| 2. | SLE | Case-control study | Case = 11; control = 9 | 16S rDNA (V4 region) sequencing |
|
[54] |
| 3. | Rheumatoid arthritis | Case-control study | Case = 20; control = 4 |
16S rDNA (V4 region) sequencing |
|
[55] |
| 4. | Immunosuppression post liver transplant | Single -arm study | n = 51 | WGS of cell-free DNA |
|
[58] |
| 5. | HIV infection | Case-control study | Case = 40; control = 51 | 16S rDNA (V4 region) sequencing |
|
[49] |
| 6. | HIV infection | Two cross-sectional studies & one single arm study | 227 HIV-infected patients; 15 healthy individuals |
qPCR of conserved 16S rDNA region |
|
[59] |
| 7. | Peritonitis | Porcine experiment; pre- and post-fecal induced peritonitis | n = 6 domestic pigs | 16S rDNA (V3-V4 region) sequencing |
|
[60] |
| 8. | Inflammatory bowel disease (including Crohn’s disease and ulcerative colitis) | Case-control study | Crohn’s disease = 8; ulcerative colitis = 8; control = 7 |
16S rDNA (V3-V4 region) sequencing |
|
[61] |
| 9. | Pancreatitis | Case-control study | Case = 50; control = 12 | 16S rDNA (V3 region) sequencing |
|
[62] |
| 10. | Large vessel vasculitis (including giant cell arteritis & Takayasu’s arteritis) | Case-control study | Giant cell arteritis = 11; Takayasu’s arteritis = 20; Healthy = 15. |
16S rDNA (V3-V4 region) sequencing |
|
[63] |
3.7. Pregnancy Complications
Linkages between the blood microbiome and pregnancy-related events such as preterm birth and stillbirth have been reported (Table 7). One study observed enrichment of the genera Bacteroides, Lactobacillus, Sphingomonas, Fastidiosipila and Butyricicoccus, along with a reduction in the abundance of Delftia, Pseudomonas, Massilia and Stenotrophomonas in mothers with preterm delivery [64]. Remarkable microbial differences were also found in the umbilical cord blood of preterm babies, as characterized by an increase in the genera Fusobacterium, Actinomyces, Campylobacter, Peptostreptococcus, Porphyromonas and Prevotella compared to term babies [65]. Furthermore, cord blood of stillbirths contained substantial amounts of bacterial 16S rDNA and heightened levels of Streptococcus agalactiae/GBS and Arthrobacter compared to live births [65]. The results point towards infection-induced intrauterine foetal death which is not widely recognized as a key driving factor of stillbirths [66].
Leveraging the concept of the multibiome, which delineates the interaction between different microbial entities (i.e., bacteria, fungi and viruses) as a community, a recent study revealed fascinating links between signatures of the blood microbiome and pathogens known to inflict pregnancy-related complications, namely Toxoplasma gondii, Hepatitis B virus, Human Papillomavirus (HPV), Rubella virus, Cytomegalovirus and Herpes simplex virus (HSV) [67]. For example, HPV positive blood samples were linked to elevated Weissella paramesenteroides and Propionibacterium acne, while blood samples positive for HSV had higher abundance of Bifidobacterium bifidum and Propionibacterium acne [67]. Toxoplasma gondii also has a high tendency to co-occur with Pectobacterium carotovorum and Chthoniobacter flavus in blood [67]. Essentially, the variations in the blood microbiome and co-occurrence of harmful pathogens with other microbes could be invaluable for the development of newer stratification strategy to identify mothers with a high risk for pregnancy-related complications via non-invasive maternal blood analysis.
Table 7.
Summary of investigations on the blood microbiome in pregnancy complications.
| No. | Disease/Condition | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Preterm birth | Case control study | Case = 21; Control = 20 |
16S rDNA (V3-V4 region) sequencing |
|
[64] |
| 2. | Stillbirth and preterm birth | Case control study | Stillbirth = 60; Preterm birth = 75 Live term (>37 weeks) birth = 101 |
16S rDNA (V4 region) sequencing |
|
[65] |
| 3. | Toxoplasma gondii, Others (Hepatitis B virus, Human Papillomavirus [HPV]) Rubella virus, Cytomegalovirus, Herpes simplex virus |
Population-based cross-sectional study | n = 107,763 healthy controls | WGS of cell-free DNA |
|
[67] |
3.8. Other Health Complications
Blood microbiome dysbiosis has been detected in many other diseases (Table 8). Certain chronic dermatological disorders, namely, rosacea and psoriasis, are associated with distinct blood microbial profiles [68,69]. Enrichment of the genera Staphylococcus, Sphingomonas, and Ralstonia in psoriasis predicted the activation of lipid metabolic processes and adipocytokine signaling, which may contribute to chronic inflammation [69]. In contrast, patients with hidradenitis suppurativa, a chronic inflammatory skin complication characterized by recurrent nodules, abscesses, and scarring, carried blood microbiota that was similar to that of healthy individuals [70]. Hence, its pathogenesis could be unrelated to bacteraemia.
In patients with major depression, the genus Janthinobacterium was more abundant while Neisseria was reduced, but dysbiosis was ameliorated after antidepressant therapy [71]. A high abundance of Firmicutes, low abundance of Tetrasphaera and Bosea along with high tryptophan levels predicted a favourable treatment response to antidepressants [71]. The association of the blood microbiome with mood has also been discovered in patients with Parkinson’s disease. For instance, blood levels of the genera Amaricoccus, Bosea, Janthinobacterium, Nesterenkonia and Sphingobacterium were positively correlated with the Hamilton Anxiety Scale score, whereas Aquabacterium, Bdellovibrio and Leucobacter were positively correlated with the Hamilton Depression Scale score [72]. Janthinobacterium was also positively associated to the duration and disease severity score, along with other microbes such as Cloacibacterium and Isoptericola which were found to be enriched in patients with Parkinson’s disease [72]. Current evidence suggests a role of circulating Janthinobacterium in depression and anxiety. Hence, further studies are necessary to identify the cognitive-emotional implications of the bacteria.
Additionally, a preliminary study isolated bacteria and fungi in their L-form (cell wall-deficient variant) from the blood of children with autism and their mothers [73]. The findings imply a vertical transmission of the pathogens from mother to child which may contribute to the onset of autism, but the results should be interpreted conservatively, as the proposed pathogenesis is largely speculation and statistical analysis was lacking in the study design.
Disturbance of the blood microbiome has also been reported in polycystic ovary syndrome (PCOS), as demonstrated by the enrichment of families Nocardioidaceae and Oxalobacteraceae, and suppression of Burkholderiaceae, Lachnospiraceae, Bacteroidaceae, Ruminococcaceae and S24-7 [74].
Certain medical interventions may also escalate the risk of blood microbiome dysbiosis. For instance, surgical procedures are known to increase the risk of microbial invasion. Surgical patients who developed postoperative septic shock carried more abundant genera Flavobacterium, Agrococcus, Polynucleobacter, and Acidovorax in the blood, which were largely correlated with disease severity and organ failure assessment scores [75]. In contrast, total parenteral nutrition (TPN) administration confers profound shifts in the bacterial composition in the gut but less so in the blood [76]. In enterally fed mice, the expansion of the order Burkholderiales in the blood was likely due to catheter insertion [76].
Table 8.
Summary of investigations on the blood microbiome in other health complications.
| No. | Disease | Study Design | Sample Size | Detection Method | Findings | Reference |
|---|---|---|---|---|---|---|
| 1. | Rosacea (skin disease) | Case-control study | Case = 10; control = 30 | 16S rDNA (V3-V4 region) sequencing |
|
[68] |
| 2. | Psoriasis (skin disease) | Case-control study | Case = 20; control = 8 | 16S rDNA (full length) sequencing |
|
[69] |
| 3. | Hidradenitis suppurativa (skin disorder) | Case-control study | Case = 27; control = 26 | 16S rDNA (V3-V4 region) sequencing |
|
[70] |
| 4. | Major depression | Case-control study | Case = 56; control = 56 | 16S rDNA (V3-V4 region) sequencing |
|
[71] |
| 5. | Parkinson’s disease | Case-control study | Case = 103; Control = 104 |
16S rDNA (V3-V4 region) sequencing |
|
[72] |
| 6. | Autism | Case-control study (without statistical analysis) | Case = 15 mother-child pairs; Control = 6 healthy individuals |
Culture |
|
[73] |
| 7. | Polycystic ovary syndrome | Case-control study | Case = 24; control = 24 | 16S rDNA (V3-V4 region) sequencing |
|
[74] |
| 8. | Surgical-induced sepsis | Prospective cohort study | Healthy = 5; Non-infected = 7; Infected = 10; Sepsis = 18; Septic shock = 11 |
16S rDNA (V3 region) sequencing |
|
[75] |
| Total parenteral nutrition (TPN) administration | Mouse experiment | Chow-fed: 6; Chow-fed with jugular vein catheter insertion = 6; Intralipid-based TPN = 6; Omegaven-based TPN = 6 |
16S rDNA (V3-V4 region) sequencing |
|
[76] |
4. Controversies and Counterclaims
While limited evidence supports the existence of a core healthy blood microbiome, dysbiosis of the blood microbial profile seems to have profound implications for a wide range of health conditions resulting from varying pathogenesis (Figure 2). The majority of the circulating microbes are associated with human body sites that are rich in commensals, such as the gut, oral cavity, and genitourinary tract [17,39,40,56,75]. Hence, the integrity and functionality of epithelial and mucosal barrier may be compromised in many diseases, allowing the opportunistic microbes or microbial-derived compounds to gain access into the body’s internal milieu [77]. Nevertheless, the concept of the blood microbiome remains a subject of debate as it challenges the paradigm of blood sterility. Here, we highlight the highly controversial and unresolved issues about the blood microbiome.
Figure 2.
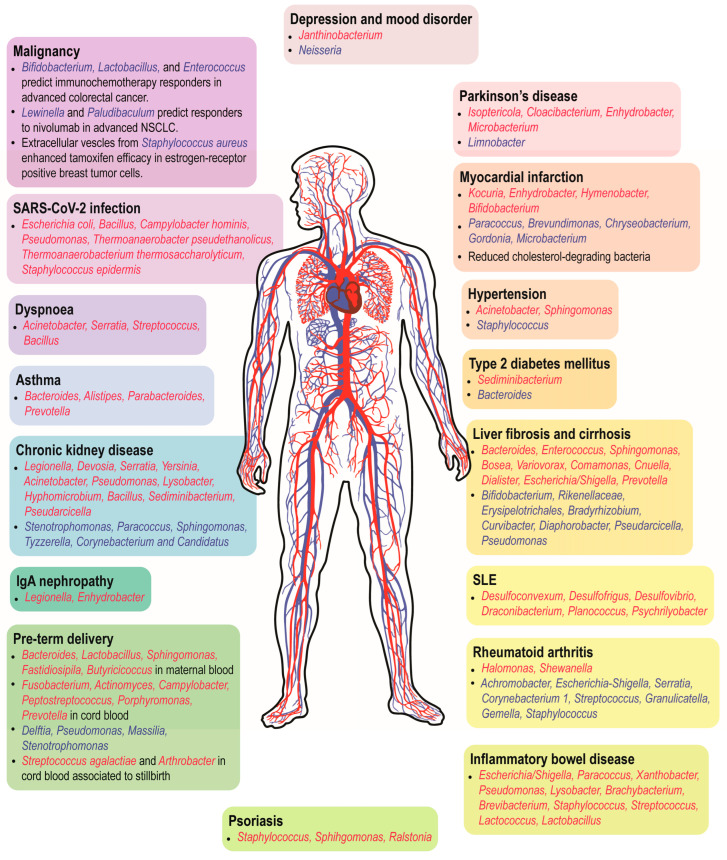
Circulating microbial populations at genus level that have been shown to associate with human diseases. Genera in red are enriched in patients with the diseases while those in blue are diminished/associated with beneficial effects in the patients. NSCLC, non-small cell lung carcinoma.
4.1. Susceptibility of Low-Biomass Samples to Exogenous Contamination
Many researchers remain sceptical about the presence of the blood microbiome because microbial analysis of low-biomass environments and samples are prone to contamination in most, if not all steps along the sample processing pipeline [78]. Several studies have concluded that circulating bacteria, especially those found in healthy volunteers, are either traces of microbial DNA from external sources or organic remnants from non-living bacteria [79,80]. For similar reasons, the findings of the microbiome in other low-biomass environments such as the womb during pregnancy, brain and tumors have also been brought into question [81,82]. Indeed, NGS sequencing is very sensitive to capturing minute amounts of microbial DNA contaminants from extraction kits, storage containers, sequencing reagents and even the sequencer itself. Moreover, dermal bacterial contamination during venepuncture and technical errors such as unintentional introduction of exogenous microbial genomes by nurses or lab personnel can also confound microbiome studies. Existing studies on the blood microbiome hardly report the decontamination strategy, and even when they do, the efforts may not be exhaustive because negative controls at every level ranging from sampling, extraction, library preparation to sequencing are likely needed. Augmentation of the decontamination strategy with bioinformatic and statistical approaches has been proposed to improve the chances of obtaining authentic microbial profiles [83,84]. Hence, developing a reliable and accurate methodology to address potential exogenous contamination serves as a cornerstone for microbiome research in low-biomass environments such as the bloodstream.
4.2. The Blood Microbiome: Viable Colonizers or Cell-Free DNA?
NGS offers a robust tool for culture-independent blood microbiome analysis; however, whether the resultant microbial profiles reflect the true blood colonizers or just the cell-free microbial DNA of killed pathogens from recent infection and spontaneous translocation remains debatable. Since the blood microbiome is often discussed in terms of its health implications, knowing its viability is indispensable to understand whether the microbiota has a larger role in pathogenesis or can merely serve as a biomarker. Nevertheless, very few studies have validated their microbiome results with the culture method, which is overly tedious and impractical because microbiome analysis easily yields hundreds of bacterial populations per sample, many of which could be fastidious and non-culturable [85,86]. Recent advancement in fluorescence in situ hybridization (FISH) on living bacteria enables validation of bacterial viability and taxonomic rank at the species level, although at much lower throughput compared to NGS [87]. Emerging bacterial single-cell transcriptomics such as PETRI-seq [88], microSPLiT [89], MATQ-seq [90] and BacDrop [91] are powerful tools that outperform 16S rDNA-based microbiota profiling and provide inferred viability and functionality derived from transcriptomes. The drawbacks are also evident: high cost and computationally demanding. Clearly, more efforts are warranted to establish whether the blood microbial profile represents living and active, or dead and non-functional microbes.
5. Knowledge Gaps and Future Directions
After reviewing the current evidence on the blood microbiome, we have identified several key gaps that may guide the future direction of the research topic. Firstly, there is a need to promote the integrative multibiome in the assessment of the blood microbiome. Current blood microbiome profiles are overrepresented by the bacteriome, while interest in the virome, archaeome and mycobiome is underwhelming. Compared to bacteria, the profiling of archaea, fungi and viruses is more challenging due to the paucity of analytical resources like reference genomes and established computational pipelines, thus posing substantial hurdles to those who are interested in the topic. However, the actual microbial ecosystem likely involves complex symbiotic relationships between microorganisms from different kingdoms that are well-implicated in the onset and pathogenesis of human diseases [92,93]. These exciting data strongly advocate the necessity to examine multibiome properties in the blood microbiome. Building fundamental infrastructures such as comprehensive genomic databases and analytical tools for the virome, archaeome and mycobiome is immensely beneficial to propel integrative multibiomics in microbiome research.
Next, our current understanding of the blood microbiome in human health is predominantly established in observational studies which only imply a correlation but not a causative relationship. Most of the studies have successfully identified specific groups of bacteria, the so-called core bacteria, that are enriched in different disease states. Subsequent studies should aim to decipher the disease-driving properties of these core bacteria and the underlying mechanisms. Germ-free animals and innovative in vitro 3D organoids are versatile models to characterize the host–pathogen interactions and explore the microbiome-based therapeutics [94,95].
Thirdly, there is an urgent need to establish the best practices and a standardized workflow for sample collection and processing, negative controls, and post-sequencing decontamination pipeline to enable data-sharing and inter-study comparison [81]. The gut microbiome is highly dynamic and labile to host and environmental factors such as host genetics, diet, medication, lifestyle and geographic regions [24,96]. Since blood microbes largely originate from the host commensals, it is not surprising that the blood microbiome is also affected by similar sets of factors, resulting in diverse core bacteria observed across studies on the same disease. By following a standardized workflow, data from independent studies could be pooled and meta-analysed with proper adjustment of the confounding variables. Alternatively, global-wide multi-center studies can also serve the same purpose, although they require international collaborative initiatives such as the International Human Microbiome Standards (IHMS) project.
Lastly, the primary objective of blood microbiota profiling in different human diseases is to pinpoint their contribution to onset and progression. While the characterization of the potentially disease-driving core microbiome is informative, what is more critical is its collective biological functions and the impact on the host response. Currently, there are several packages for the prediction of metagenome functions, for example, PICRUSt [97], PICRUSt2 [98] and Tax4Fun2 [99]. However, significant biases have been detected in gene and functional inference, especially when the 16S rRNA-based metagenome is used as an input [100]. These tools also tend to predict general housekeeping functions but are less likely to reveal more specific adaptation pathways related to responses to environmental cues, secondary metabolites and xenobiotics, which are more biologically meaningful and disease-related [100]. Moreover, diverse core microbiomes may converge at the functional level and contribute to a common disease. This suggests a functional redundancy of microbes in different communities probably due to convergent evolution when exposed to similar environments. This phenomenon may add an extra layer of complexity to improve the performance of functional analytical tools. In short, newer functional prediction software should strive to combat the known biases and provide precise and biologically relevant functional estimation based on microbiome profiles.
6. Conclusions
To conclude, evidence supporting a core healthy blood microbiome is limited, yet dysbiosis of the blood microbial profile could have profound implications in a wide range of health conditions. Common microbial taxa have been identified in certain diseases, for example, Legionella and Devosia in kidney impairment, Bacteroides in cirrhosis, Escherichia/Shigella and Staphylococcus in inflammatory diseases, and Janthinobacterium in mood disorders. The blood microbial gene signatures also exhibit remarkable risk stratification and prognostication values in cancers, pregnancy-related complications, and asthma. The clinical implications of these findings require further validation as they are largely derived from observational studies.
Acknowledgments
H.S. Cheng is a recipient of LKCMedicine Dean’s Fellowship from Nanyang Technological University Singapore. The authors would like to acknowledge the NTU Start Up Grant (021337-00001) and Wang Lee Wah Memorial Fund for support of this work.
Author Contributions
Conceptualization, H.S.C. and S.P.T.; data curation, H.S.C. and S.P.T.; writing—original draft preparation, H.S.C., S.P.T., D.M.K.W. and W.L.Y.K.; writing—review and editing, all authors; supervision, S.H.W. and N.S.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Lian C.W.K., Poh S.E., Lee C.K., Chan H.M.T., Yan G., Kong K.W., Lau L., Thomas Lee W.Y., Cheng C., Hoon S., et al. Towards a rapid-turnaround low-depth unbiased metagenomics sequencing workflow on the Illumina platforms. medRxiv. 2023 doi: 10.3390/bioengineering10050520. medRxiv:2023.01.02.22283504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng W.Y., Liu W.X., Ding Y., Wang G., Shi Y., Chu E.S.H., Wong S., Sung J.J.Y., Yu J. High Sensitivity of Shotgun Metagenomic Sequencing in Colon Tissue Biopsy by Host DNA Depletion. Genom. Proteom. Bioinform. 2022 doi: 10.1016/j.gpb.2022.09.003. in press . [DOI] [PubMed] [Google Scholar]
- 3.Valdes A.M., Walter J., Segal E., Spector T.D. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017;10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 6.Kilian M., Chapple I.L., Hannig M., Marsh P.D., Meuric V., Pedersen A.M., Tonetti M.S., Wade W.G., Zaura E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016;221:657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- 7.Jones-Freeman B., Chonwerawong M., Marcelino V.R., Deshpande A.V., Forster S.C., Starkey M.R. The microbiome and host mucosal interactions in urinary tract diseases. Mucosal Immunol. 2021;14:779–792. doi: 10.1038/s41385-020-00372-5. [DOI] [PubMed] [Google Scholar]
- 8.Potgieter M., Bester J., Kell D.B., Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015;39:567–591. doi: 10.1093/femsre/fuv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo D.J., Rifkin R.F., Cowan D.A., Potgieter M. The Healthy Human Blood Microbiome: Fact or Fiction? Front. Cell. Infect. Microbiol. 2019;9:148. doi: 10.3389/fcimb.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risely A. Applying the core microbiome to understand host-microbe systems. J. Anim. Ecol. 2020;89:1549–1558. doi: 10.1111/1365-2656.13229. [DOI] [PubMed] [Google Scholar]
- 11.Velmurugan G., Dinakaran V., Rajendhran J., Swaminathan K. Blood Microbiota and Circulating Microbial Metabolites in Diabetes and Cardiovascular Disease. Trends Endocrinol. Metab. 2020;31:835–847. doi: 10.1016/j.tem.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Damgaard C., Magnussen K., Enevold C., Nilsson M., Tolker-Nielsen T., Holmstrup P., Nielsen C.H. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS ONE. 2015;10:e0120826. doi: 10.1371/journal.pone.0120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Aquila P., Giacconi R., Malavolta M., Piacenza F., Burkle A., Villanueva M.M., Dolle M.E.T., Jansen E., Grune T., Gonos E.S., et al. Microbiome in Blood Samples From the General Population Recruited in the MARK-AGE Project: A Pilot Study. Front. Microbiol. 2021;12:707515. doi: 10.3389/fmicb.2021.707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paisse S., Valle C., Servant F., Courtney M., Burcelin R., Amar J., Lelouvier B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56:1138–1147. doi: 10.1111/trf.13477. [DOI] [PubMed] [Google Scholar]
- 15.Pammi M., Thapa S., Balderas M., Runge J.K., Venkatachalam A., Luna R.A. Microbiome signatures in neonatal central line associated bloodstream infections. PLoS ONE. 2020;15:e0227967. doi: 10.1371/journal.pone.0227967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarsella E., Sandri M., Monego S.D., Licastro D., Stefanon B. Blood Microbiome: A New Marker of Gut Microbial Population in Dogs? Vet. Sci. 2020;7:198. doi: 10.3390/vetsci7040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan C.C., Chia M., Ko K.K., Chen H., Liu J., Loh M., Nagarajan N. No evidence for a common blood microbiome based on a population study of 9770 healthy humans. bioRxiv. 2022 doi: 10.1038/s41564-023-01350-w. bioRxiv:2022.07.29.502098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raeisi J., Oloomi M., Zolfaghari M., Siadat S.D., Zargar M., Pourramezan Z. Bacterial DNA Detection in the Blood of Healthy Subjects. Iran. Biomed. J. 2022;26:230–239. doi: 10.52547/ibj.26.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amar J., Lelouvier B., Servant F., Lluch J., Burcelin R., Bongard V., Elbaz M. Blood Microbiota Modification After Myocardial Infarction Depends Upon Low-Density Lipoprotein Cholesterol Levels. J. Am. Heart Assoc. 2019;8:e011797. doi: 10.1161/JAHA.118.011797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan I., Khan I., Kakakhel M.A., Xiaowei Z., Ting M., Ali I., Fei Y., Jianye Z., Zhiqiang L., Lizhe A. Comparison of Microbial Populations in the Blood of Patients With Myocardial Infarction and Healthy Individuals. Front. Microbiol. 2022;13:845038. doi: 10.3389/fmicb.2022.845038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan I., Khan I., Usman M., Jianye Z., Wei Z.X., Ping X., Zhiqiang L., Lizhe A. Analysis of the blood bacterial composition of patients with acute coronary syndrome and chronic coronary syndrome. Front. Cell. Infect. Microbiol. 2022;12:943808. doi: 10.3389/fcimb.2022.943808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amar J., Lange C., Payros G., Garret C., Chabo C., Lantieri O., Courtney M., Marre M., Charles M.A., Balkau B., et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: The D.E.S.I.R. study. PLoS ONE. 2013;8:e54461. doi: 10.1371/journal.pone.0054461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence G., Midtervoll I., Samuelsen S.O., Kristoffersen A.K., Enersen M., Haheim L.L. The blood microbiome and its association to cardiovascular disease mortality: Case-cohort study. BMC Cardiovasc. Disord. 2022;22:344. doi: 10.1186/s12872-022-02791-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasan N., Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amar J., Serino M., Lange C., Chabo C., Iacovoni J., Mondot S., Lepage P., Klopp C., Mariette J., Bouchez O., et al. Involvement of tissue bacteria in the onset of diabetes in humans: Evidence for a concept. Diabetologia. 2011;54:3055–3061. doi: 10.1007/s00125-011-2329-8. [DOI] [PubMed] [Google Scholar]
- 26.Qiu J., Zhou H., Jing Y., Dong C. Association between blood microbiome and type 2 diabetes mellitus: A nested case-control study. J. Clin. Lab. Anal. 2019;33:e22842. doi: 10.1002/jcla.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing Y., Zhou H., Lu H., Chen X., Zhou L., Zhang J., Wu J., Dong C. Associations Between Peripheral Blood Microbiome and the Risk of Hypertension. Am. J. Hypertens. 2021;34:1064–1070. doi: 10.1093/ajh/hpab084. [DOI] [PubMed] [Google Scholar]
- 28.Poore G.D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J., et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho E.J., Leem S., Kim S.A., Yang J., Lee Y.B., Kim S.S., Cheong J.Y., Cho S.W., Kim J.W., Kim S.M., et al. Circulating Microbiota-Based Metagenomic Signature for Detection of Hepatocellular Carcinoma. Sci. Rep. 2019;9:7536. doi: 10.1038/s41598-019-44012-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong Z., Chen B., Pan H., Wang D., Liu M., Yang Y., Zou M., Yang J., Xiao K., Zhao R., et al. Detection of Microbial 16S rRNA Gene in the Serum of Patients With Gastric Cancer. Front. Oncol. 2019;9:608. doi: 10.3389/fonc.2019.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woerner J., Huang Y., Hutter S., Gurnari C., Sanchez J.M.H., Wang J., Huang Y., Schnabel D., Aaby M., Xu W., et al. Circulating microbial content in myeloid malignancy patients is associated with disease subtypes and patient outcomes. Nat. Commun. 2022;13:1038. doi: 10.1038/s41467-022-28678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang D., Wang X., Zhou X., Zhao J., Yang H., Wang S., Morse M.A., Wu J., Yuan Y., Li S., et al. Blood microbiota diversity determines response of advanced colorectal cancer to chemotherapy combined with adoptive T cell immunotherapy. Oncoimmunology. 2021;10:1976953. doi: 10.1080/2162402X.2021.1976953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouaknine Krief J., Helly de Tauriers P., Dumenil C., Neveux N., Dumoulin J., Giraud V., Labrune S., Tisserand J., Julie C., Emile J.F., et al. Role of antibiotic use, plasma citrulline and blood microbiome in advanced non-small cell lung cancer patients treated with nivolumab. J. Immunother. Cancer. 2019;7:176. doi: 10.1186/s40425-019-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plottel C.S., Blaser M.J. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwa M., Plottel C.S., Blaser M.J., Adams S. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. J. Natl. Cancer Inst. 2016;108:djw029. doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An J., Kwon H., Lim W., Moon B.I. Staphylococcus aureus-Derived Extracellular Vesicles Enhance the Efficacy of Endocrine Therapy in Breast Cancer Cells. J. Clin. Med. 2022;11:2030. doi: 10.3390/jcm11072030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strnad P., Tacke F., Koch A., Trautwein C. Liver—Guardian, modifier and target of sepsis. Nat. Rev. Gastroenterol. Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 38.Lelouvier B., Servant F., Paisse S., Brunet A.C., Benyahya S., Serino M., Valle C., Ortiz M.R., Puig J., Courtney M., et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: A pilot analysis. Hepatology. 2016;64:2015–2027. doi: 10.1002/hep.28829. [DOI] [PubMed] [Google Scholar]
- 39.Traykova D., Schneider B., Chojkier M., Buck M. Blood Microbiome Quantity and the Hyperdynamic Circulation in Decompensated Cirrhotic Patients. PLoS ONE. 2017;12:e0169310. doi: 10.1371/journal.pone.0169310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajihara M., Koido S., Kanai T., Ito Z., Matsumoto Y., Takakura K., Saruta M., Kato K., Odamaki T., Xiao J.Z., et al. Characterisation of blood microbiota in patients with liver cirrhosis. Eur. J. Gastroenterol. Hepatol. 2019;31:1577–1583. doi: 10.1097/MEG.0000000000001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiago A., Pozuelo M., Poca M., Gely C., Nieto J.C., Torras X., Roman E., Campos D., Sarrabayrouse G., Vidal S., et al. Alteration of the serum microbiome composition in cirrhotic patients with ascites. Sci. Rep. 2016;6:25001. doi: 10.1038/srep25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schierwagen R., Alvarez-Silva C., Madsen M.S.A., Kolbe C.C., Meyer C., Thomas D., Uschner F.E., Magdaleno F., Jansen C., Pohlmann A., et al. Circulating microbiome in blood of different circulatory compartments. Gut. 2019;68:578–580. doi: 10.1136/gutjnl-2018-316227. [DOI] [PubMed] [Google Scholar]
- 43.Gedgaudas R., Bajaj J.S., Skieceviciene J., Varkalaite G., Jurkeviciute G., Gelman S., Valantiene I., Zykus R., Pranculis A., Bang C., et al. Circulating microbiome in patients with portal hypertension. Gut Microbes. 2022;14:2029674. doi: 10.1080/19490976.2022.2029674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virseda-Berdices A., Brochado-Kith O., Diez C., Hontanon V., Berenguer J., Gonzalez-Garcia J., Rojo D., Fernandez-Rodriguez A., Ibanez-Samaniego L., Llop-Herrera E., et al. Blood microbiome is associated with changes in portal hypertension after successful direct-acting antiviral therapy in patients with HCV-related cirrhosis. J. Antimicrob. Chemother. 2022;77:719–726. doi: 10.1093/jac/dkab444. [DOI] [PubMed] [Google Scholar]
- 45.Whittle E., Leonard M.O., Harrison R., Gant T.W., Tonge D.P. Multi-Method Characterization of the Human Circulating Microbiome. Front. Microbiol. 2018;9:3266. doi: 10.3389/fmicb.2018.03266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.H., Choi J.P., Yang J., Won H.K., Park C.S., Song W.J., Kwon H.S., Kim T.B., Kim Y.K., Park H.S., et al. Metagenome analysis using serum extracellular vesicles identified distinct microbiota in asthmatics. Sci. Rep. 2020;10:15125. doi: 10.1038/s41598-020-72242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrow J.D., Castaldi P.J., Chase R.P., Yun J.H., Lee S., Liu Y.Y., Hersh C.P. Peripheral blood microbial signatures in current and former smokers. Sci. Rep. 2021;11:19875. doi: 10.1038/s41598-021-99238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dereschuk K., Apostol L., Ranjan I., Chakladar J., Li W.T., Rajasekaran M., Chang E.Y., Ongkeko W.M. Identification of Lung and Blood Microbiota Implicated in COVID-19 Prognosis. Cells. 2021;10:1452. doi: 10.3390/cells10061452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo Z., Alekseyenko A.V., Ogunrinde E., Li M., Li Q.Z., Huang L., Tsao B.P., Kamen D.L., Oates J.C., Li Z., et al. Rigorous Plasma Microbiome Analysis Method Enables Disease Association Discovery in Clinic. Front. Microbiol. 2020;11:613268. doi: 10.3389/fmicb.2020.613268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah N.B., Allegretti A.S., Nigwekar S.U., Kalim S., Zhao S., Lelouvier B., Servant F., Serena G., Thadhani R.I., Raj D.S., et al. Blood Microbiome Profile in CKD: A Pilot Study. Clin. J. Am. Soc. Nephrol. 2019;14:692–701. doi: 10.2215/CJN.12161018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merino-Ribas A., Araujo R., Pereira L., Campos J., Barreiros L., Segundo M.A., Silva N., Costa C.F.F.A., Quelhas-Santos J., Trindade F., et al. Vascular Calcification and the Gut and Blood Microbiome in Chronic Kidney Disease Patients on Peritoneal Dialysis: A Pilot Study. Biomolecules. 2022;12:867. doi: 10.3390/biom12070867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah N.B., Nigwekar S.U., Kalim S., Lelouvier B., Servant F., Dalal M., Krinsky S., Fasano A., Tolkoff-Rubin N., Allegretti A.S. The Gut and Blood Microbiome in IgA Nephropathy and Healthy Controls. Kidney360. 2021;2:1261. doi: 10.34067/KID.0000132021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Carrasco V., Soriano-Lerma A., Soriano M., Gutierrez-Fernandez J., Garcia-Salcedo J.A. Urinary Microbiome: Yin and Yang of the Urinary Tract. Front. Cell. Infect. Microbiol. 2021;11:617002. doi: 10.3389/fcimb.2021.617002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James W.A., Ogunrinde E., Wan Z., Kamen D.L., Oates J., Gilkeson G.S., Jiang W. A Distinct Plasma Microbiome but Not Gut Microbiome in Patients with Systemic Lupus Erythematosus Compared to Healthy Individuals. J. Rheumatol. 2022;49:592–597. doi: 10.3899/jrheum.210952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammad D.B.M., Hider S.L., Liyanapathirana V.C., Tonge D.P. Molecular Characterization of Circulating Microbiome Signatures in Rheumatoid Arthritis. Front. Cell. Infect. Microbiol. 2019;9:440. doi: 10.3389/fcimb.2019.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Audo R., Sanchez P., Riviere B., Mielle J., Tan J., Lukas C., Macia L., Morel J., Immediato Daien C. Rheumatoid arthritis is associated with increased gut permeability and bacterial translocation which are reversed by inflammation control. Rheumatology. 2022;62:1264–1271. doi: 10.1093/rheumatology/keac454. [DOI] [PubMed] [Google Scholar]
- 57.Bhat M., Pasini E., Copeland J., Angeli M., Husain S., Kumar D., Renner E., Teterina A., Allard J., Guttman D.S., et al. Impact of Immunosuppression on the Metagenomic Composition of the Intestinal Microbiome: A Systems Biology Approach to Post-Transplant Diabetes. Sci. Rep. 2017;7:10277. doi: 10.1038/s41598-017-10471-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okumura T., Horiba K., Kamei H., Takeuchi S., Suzuki T., Torii Y., Kawada J.I., Takahashi Y., Ogura Y., Ogi T., et al. Temporal dynamics of the plasma microbiome in recipients at early post-liver transplantation: A retrospective study. BMC Microbiol. 2021;21:104. doi: 10.1186/s12866-021-02154-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang W., Lederman M.M., Hunt P., Sieg S.F., Haley K., Rodriguez B., Landay A., Martin J., Sinclair E., Asher A.I., et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J. Infect. Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hyun H., Lee M.S., Park I., Ko H.S., Yun S., Jang D.H., Kim S., Kim H., Kang J.H., Lee J.H., et al. Analysis of Porcine Model of Fecal-Induced Peritonitis Reveals the Tropism of Blood Microbiome. Front. Cell. Infect. Microbiol. 2021;11:676650. doi: 10.3389/fcimb.2021.676650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones E., Stentz R., Telatin A., Savva G.M., Booth C., Baker D., Rudder S., Knight S.C., Noble A., Carding S.R. The Origin of Plasma-Derived Bacterial Extracellular Vesicles in Healthy Individuals and Patients with Inflammatory Bowel Disease: A Pilot Study. Genes. 2021;12:1636. doi: 10.3390/genes12101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Q., Wang C., Tang C., Zhao X., He Q., Li J. Identification and Characterization of Blood and Neutrophil-Associated Microbiomes in Patients with Severe Acute Pancreatitis Using Next-Generation Sequencing. Front. Cell. Infect. Microbiol. 2018;8:5. doi: 10.3389/fcimb.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desbois A.C., Ciocan D., Saadoun D., Perlemuter G., Cacoub P. Specific microbiome profile in Takayasu’s arteritis and giant cell arteritis. Sci. Rep. 2021;11:5926. doi: 10.1038/s41598-021-84725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You Y.A., Yoo J.Y., Kwon E.J., Kim Y.J. Blood microbial communities during pregnancy are associated with preterm birth. Front. Microbiol. 2019;10:1122. doi: 10.3389/fmicb.2019.01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vander Haar Emilie L., Wu G., Gyamfi-Bannerman C., Thomas C., Wapner Ronald J., Reddy Uma M., Zhao L., Silver Robert M., Goldenberg Robert L., Han Yiping W. Microbial Analysis of Umbilical Cord Blood Reveals Novel Pathogens Associated with Stillbirth and Early Preterm Birth. mBio. 2022;13:e02036-22. doi: 10.1128/mbio.02036-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seale A.C., Blencowe H., Bianchi-Jassir F., Embleton N., Bassat Q., Ordi J., Menéndez C., Cutland C., Briner C., Berkley J.A., et al. Stillbirth With Group B Streptococcus Disease Worldwide: Systematic Review and Meta-analyses. Clin. Infect. Dis. 2017;65:S125–S132. doi: 10.1093/cid/cix585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tong X., Yu X., Du Y., Su F., Liu Y., Li H., Liu Y., Mu K., Liu Q., Li H., et al. Peripheral Blood Microbiome Analysis via Noninvasive Prenatal Testing Reveals the Complexity of Circulating Microbial Cell-Free DNA. Microbiol. Spectr. 2022;10:e0041422. doi: 10.1128/spectrum.00414-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yun Y., Kim H.N., Chang Y., Lee Y., Ryu S., Shin H., Kim W.S., Kim H.L., Nam J.H. Characterization of the Blood Microbiota in Korean Females with Rosacea. Dermatology. 2019;235:255–259. doi: 10.1159/000496968. [DOI] [PubMed] [Google Scholar]
- 69.Chang C.J., Zhang J., Tsai Y.L., Chen C.B., Lu C.W., Huo Y.P., Liou H.M., Ji C., Chung W.H. Compositional Features of Distinct Microbiota Base on Serum Extracellular Vesicle Metagenomics Analysis in Moderate to Severe Psoriasis Patients. Cells. 2021;10:2349. doi: 10.3390/cells10092349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ring H.C., Thorsen J., Saunte D.M., Lilje B., Bay L., Theut Riis P., Larsen N., O’Brien Andersen L., Vedel Nielsen H., Miller I.M., et al. Moderate to severe hidradenitis suppurativa patients do not have an altered bacterial composition in peripheral blood compared to healthy controls. J. Eur. Acad. Dermatol. Venereol. 2018;32:125–128. doi: 10.1111/jdv.14538. [DOI] [PubMed] [Google Scholar]
- 71.Ciocan D., Cassard A.M., Becquemont L., Verstuyft C., Voican C.S., El Asmar K., Colle R., David D., Trabado S., Feve B., et al. Blood microbiota and metabolomic signature of major depression before and after antidepressant treatment: A prospective case-control study. J. Psychiatry Neurosci. 2021;46:E358–E368. doi: 10.1503/jpn.200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qian Y., Yang X., Xu S., Wu C., Qin N., Chen S.D., Xiao Q. Detection of Microbial 16S rRNA Gene in the Blood of Patients With Parkinson’s Disease. Front. Aging Neurosci. 2018;10:156. doi: 10.3389/fnagi.2018.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Markova N. Dysbiotic microbiota in autistic children and their mothers: Persistence of fungal and bacterial wall-deficient L-form variants in blood. Sci. Rep. 2019;9:13401. doi: 10.1038/s41598-019-49768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Q., Wang Q., Zhao L., Bin Y., Wang L., Wang L., Zhang K., Li Q. Blood Bacterial 16S rRNA Gene Alterations in Women With Polycystic Ovary Syndrome. Front. Endocrinol. 2022;13:814520. doi: 10.3389/fendo.2022.814520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang C., Li Q., Tang C., Zhao X., He Q., Tang X., Ren J. Characterization of the blood and neutrophil-specific microbiomes and exploration of potential bacterial biomarkers for sepsis in surgical patients. Immun. Inflamm. Dis. 2021;9:1343–1357. doi: 10.1002/iid3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lucchinetti E., Lou P.H., Lemal P., Bestmann L., Hersberger M., Rogler G., Kramer S.D., Zaugg M. Gut microbiome and circulating bacterial DNA (“blood microbiome”) in a mouse model of total parenteral nutrition: Evidence of two distinct separate microbiotic compartments. Clin. Nutr. ESPEN. 2022;49:278–288. doi: 10.1016/j.clnesp.2022.03.038. [DOI] [PubMed] [Google Scholar]
- 77.Akdis C.A. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat. Rev. Immunol. 2021;21:739–751. doi: 10.1038/s41577-021-00538-7. [DOI] [PubMed] [Google Scholar]
- 78.Chrisman B., He C., Jung J.Y., Stockham N., Paskov K., Washington P., Wall D.P. The human “contaminome”: Bacterial, viral, and computational contamination in whole genome sequences from 1000 families. Sci. Rep. 2022;12:9863. doi: 10.1038/s41598-022-13269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martel J., Wu C.Y., Huang P.R., Cheng W.Y., Young J.D. Pleomorphic bacteria-like structures in human blood represent non-living membrane vesicles and protein particles. Sci. Rep. 2017;7:10650. doi: 10.1038/s41598-017-10479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glassing A., Dowd S.E., Galandiuk S., Davis B., Chiodini R.J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016;8:24. doi: 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy K.M., de Goffau M.C., Perez-Munoz M.E., Arrieta M.C., Backhed F., Bork P., Braun T., Bushman F.D., Dore J., de Vos W.M., et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature. 2023;613:639–649. doi: 10.1038/s41586-022-05546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hornung B.V.H., Zwittink R.D., Ducarmon Q.R., Kuijper E.J. Response to: ‘Circulating microbiome in blood of different circulatory compartments’ by Schierwagen et al. Gut. 2020;69:789–790. doi: 10.1136/gutjnl-2019-318601. [DOI] [PubMed] [Google Scholar]
- 83.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jervis-Bardy J., Leong L.E., Marri S., Smith R.J., Choo J.M., Smith-Vaughan H.C., Nosworthy E., Morris P.S., O’Leary S., Rogers G.B., et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zrodlowski T., Sobonska J., Salamon D., McFarlane I.M., Zietkiewicz M., Gosiewski T. Classical Microbiological Diagnostics of Bacteremia: Are the Negative Results Really Negative? What is the Laboratory Result Telling Us About the “Gold Standard”? Microorganisms. 2020;8:24. doi: 10.3390/microorganisms8030346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Panaiotov S., Hodzhev Y., Tsafarova B., Tolchkov V., Kalfin R. Culturable and Non-Culturable Blood Microbiota of Healthy Individuals. Microorganisms. 2021;9:1464. doi: 10.3390/microorganisms9071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Batani G., Bayer K., Boge J., Hentschel U., Thomas T. Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci. Rep. 2019;9:18618. doi: 10.1038/s41598-019-55049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blattman S.B., Jiang W., Oikonomou P., Tavazoie S. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat. Microbiol. 2020;5:1192–1201. doi: 10.1038/s41564-020-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuchina A., Brettner L.M., Paleologu L., Roco C.M., Rosenberg A.B., Carignano A., Kibler R., Hirano M., DePaolo R.W., Seelig G. Microbial single-cell RNA sequencing by split-pool barcoding. Science. 2021;371:eaba5257. doi: 10.1126/science.aba5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Homberger C., Hayward R.J., Barquist L., Vogel J. Improved bacterial single-cell RNA-seq through automated MATQ-seq and Cas9-based removal of rRNA reads. bioRxiv. 2022 doi: 10.1128/mbio.03557-22. bioRxiv:2022.11.28.518171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma P., Amemiya H.M., He L.L., Gandhi S.J., Nicol R., Bhattacharyya R.P., Smillie C.S., Hung D.T. Bacterial droplet-based single-cell RNA-seq reveals antibiotic-associated heterogeneous cellular states. Cell. 2023;186:877–891. doi: 10.1016/j.cell.2023.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Filyk H.A., Osborne L.C. The Multibiome: The Intestinal Ecosystem’s Influence on Immune Homeostasis, Health, and Disease. EBioMedicine. 2016;13:46–54. doi: 10.1016/j.ebiom.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mac Aogain M., Narayana J.K., Tiew P.Y., Ali N., Yong V.F.L., Jaggi T.K., Lim A.Y.H., Keir H.R., Dicker A.J., Thng K.X., et al. Integrative microbiomics in bronchiectasis exacerbations. Nat. Med. 2021;27:688–699. doi: 10.1038/s41591-021-01289-7. [DOI] [PubMed] [Google Scholar]
- 94.Barrila J., Crabbe A., Yang J., Franco K., Nydam S.D., Forsyth R.J., Davis R.R., Gangaraju S., Ott C.M., Coyne C.B., et al. Modeling Host-Pathogen Interactions in the Context of the Microenvironment: Three-Dimensional Cell Culture Comes of Age. Infect. Immun. 2018;86:e00282-18. doi: 10.1128/IAI.00282-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kennedy E.A., King K.Y., Baldridge M.T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurilshikov A., Medina-Gomez C., Bacigalupe R., Radjabzadeh D., Wang J., Demirkan A., Le Roy C.I., Raygoza Garay J.A., Finnicum C.T., Liu X., et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021;53:156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wemheuer F., Taylor J.A., Daniel R., Johnston E., Meinicke P., Thomas T., Wemheuer B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome. 2020;15:11. doi: 10.1186/s40793-020-00358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun S., Jones R.B., Fodor A.A. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome. 2020;8:46. doi: 10.1186/s40168-020-00815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.