Abstract
During winter hibernation, a diverse range of small mammals can enter prolonged torpor. They spend the nonhibernation season as a homeotherm but the hibernation season as a heterotherm. In the hibernation season, chipmunks (Tamias asiaticus) cycle regularly between 5 and 6 days–long deep torpor with a body temperature (Tb) of 5 to 7 °C and interbout arousal of ∼20 h, during which, their Tb returns to the normothermic level. Here, we investigated Per2 expression in the liver to elucidate the regulation of the peripheral circadian clock in a mammalian hibernator. In the nonhibernation season, as in mice, heat shock factor 1, activated by elevated Tb during the wake period, activated Per2 transcription in the liver, which contributed to synchronizing the peripheral circadian clock to the Tb rhythm. In the hibernation season, we determined that the Per2 mRNA was at low levels during deep torpor, but Per2 transcription was transiently activated by heat shock factor 1, which was activated by elevated Tb during interbout arousal. Nevertheless, we found that the mRNA from the core clock gene Bmal1 exhibited arrhythmic expression during interbout arousal. Since circadian rhythmicity is dependent on negative feedback loops involving the clock genes, these results suggest that the peripheral circadian clock in the liver is nonfunctional in the hibernation season.
Keywords: circadian clock, clock gene, HSF1, chromatin immunoprecipitation, transcription regulation, mammalian hibernation
In mammals, a variety of physiological and behavioral processes, such as the sleep-wake cycle, body temperature (Tb), and liver metabolism, exhibit daily oscillations (1). These rhythms are driven by the master clock, localized in the suprachiasmatic nucleus (SCN) of the hypothalamus (2). Circadian clocks are endogenous oscillators present in almost every cell in the body (3, 4). These cell-autonomous clocks are composed of a transcription-translation network of feedback loops involving core clock genes, such as Bmal1, Clock, Cry, and Per, and their protein products, with the BMAL1-CLOCK heterodimer acting as a transcriptional activator and the CRY–PER complex as a transcriptional repressor (5). The master clock is entrained to the 24-h day by the daily light-dark cycle and in turn, synchronizes peripheral clocks by relaying temporal information through hormonal and neuronal signals, Tb rhythms, and feeding-fasting cycles (6, 7, 8, 9).
While most mammals are homeothermic and maintain a nearly constant high Tb with a narrow range of fluctuations throughout their adult lives, certain small mammals can undergo hibernation during the winter season when environmental temperatures are low and food is scarce. Mammalian hibernators spend the nonhibernation season as a homeotherm but the hibernation season as a heterotherm. Chipmunks (Tamias asiaticus), a diurnal mammalian hibernator, maintain a Tb of ∼37 °C with daily fluctuations of 1 to 2 °C in the nonhibernation (summer) season and cycle between 5 and 6 days–long deep torpor with a Tb of ∼5 to 7 °C and interbout arousal of ∼20 h, during which their Tb rises to 37 °C by endogenous thermogenesis, in the hibernation (winter) season (10, 11). Even when chipmunks were kept under constant cold (5 °C) and dark conditions throughout their lives, they exhibited a clear hibernation rhythm with individually constant cycles and durations (12). These findings suggest that even without environmental cues, chipmunks repeat a cycle of nonhibernation and hibernation periods annually, with the hibernation period accompanied by torpor-arousal cycles and that these cycles are regulated by endogenous rhythms. Since circadian rhythms are also endogenous autonomous oscillators that control physiological and behavioral processes, clarifying the expression of circadian rhythms during hibernation would provide insight into the mechanisms of hibernation rhythms that remain unexplored. In addition, circadian oscillators can function over a wide range of physiological temperatures, a characteristic termed as temperature compensation (13). Given that the circadian clock is mechanistically driven by transcription-translation feedback loops, it has also been of great interest to researchers to determine the circadian clock functions during deep torpor when transcription and translation are virtually suppressed (14). The persistence of circadian rhythms during deep torpor has been investigated mainly by analyzing the duration of deep torpor bouts, timing of entry into and arousal from hibernation, and the Tb rhythm. There have been conflicting reports on the presence of circadian rhythms during hibernation (15, 16, 17, 18, 19, 20, 21), possibly because of differences in species and breeding conditions. In this context, it should be noted that two reports analyzing the expression of clock genes or their protein products in the SCN revealed that the master clock ceases to function during deep torpor (22, 23). On the contrary, the peripheral clocks during hibernation season have rarely been analyzed.
Daily Tb rhythms act as a systemic cue for peripheral clocks by resetting Per2 expression via heat shock factor 1 (HSF1) (7, 24, 25, 26). In the mouse liver, HSF1 is activated at dusk when the mouse becomes active (27). We recently revealed that in the chipmunk liver, HSF1 is activated by elevated Tb both during the early wake period of the sleep-wake cycle in the nonhibernation season and during arousal from deep torpor in the hibernation season (11). This finding suggests that Per2 expression may be activated by HSF1, and the peripheral clock is reset not only in the nonhibernation season but also during interbout arousal in the hibernation season. To elucidate the process of the peripheral clock regulation during the hibernation season, we analyzed Per2 expression in the chipmunk liver.
Results
Per2 mRNA level peaks in the early wake period in the liver of nonhibernating chipmunks
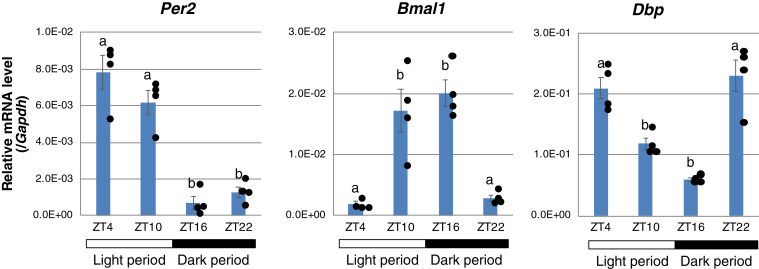
The process of peripheral circadian clock regulation in mammalian hibernators is unknown. Chipmunks are diurnal mammals and spend the nonhibernation season as a homeotherm like nonhibernators. To address this, we first examined Per2 expression in the liver of nonhibernating chipmunks by reverse transcription-quantitative PCR (RT-qPCR), since PER2 functions as an immediate early regulator in the synchronization of circadian clocks in peripheral cell types (28). The chipmunks were fed ad libitum and sacrificed at four time points throughout the day: zeitgeber time (ZT) 4 (ZT4) (10 AM), ZT10 (4 PM), ZT16 (10 PM), and ZT22 (4 AM). The Per2 mRNA level was low at ZT16, but it started to increase at ZT22 and peaked at ZT4 in the early wake period (Fig. 1). Similarly, in nocturnal mice, the Per2 mRNA level in the liver reaches its zenith in the early wake period (29). In addition, we analyzed other circadian clock genes, namely, Dbp and Bmal1. Per2 and Dbp are E-Box–controled circadian genes, and their transcription is activated by the BMAL1-CLOCK or NPAS2 heterodimer, which binds to E-Box (30). The Dbp mRNA level started to increase at ZT22, and the Bmal1 mRNA level peaked at ZT16 preceding the Per2 and Dbp mRNA levels (Fig. 1). The phase relationship of Per2, Bmal1, and Dbp expression in the sleep-wake cycle was well conserved between chipmunks and mice (26).
Figure 1.
Clock gene mRNA rhythms in chipmunk liver in non-hibernation season. The Per2, Bmal1, Dbp, and Gapdh mRNA levels were measured by RT-qPCR using liver total RNA. The Per2, Bmal1, and Dbp mRNA levels were normalized to the Gapdh mRNA levels and are shown as mean ± SEM with corresponding dot plots to observed values (n = 4/group at each time point). Different letters indicate significantly different values [Tukey's honestly significant difference (HSD) test, p < 0.05]. RT-qPCR, reverse transcription-quantitative PCR.
HSF1 activates chipmunk Per2 transcription
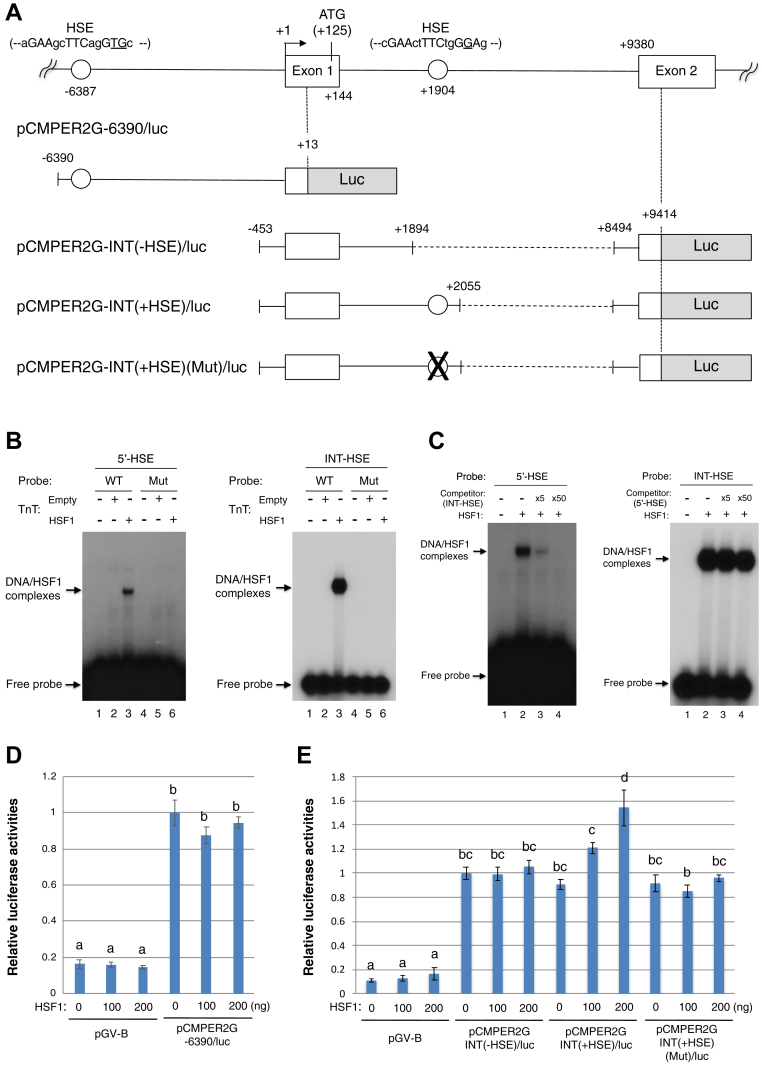
In mouse fibroblasts, HSF1 is involved in the circadian expression of the Per2 gene (25). In the nonhibernation season, the Tb of chipmunks was high during the day and low at night (11). Accordingly, in the liver, nuclear HSF1 increased around ZT4 during the wake period and decreased around ZT16 during the sleep period, and the HSP70 mRNA level peaked around ZT4 (11). The observation that the Per2 mRNA level peaked around ZT4 (Fig. 1) indicates that Per2 transcription was likely activated by HSF1 in the chipmunk liver. To examine whether Per2 gene transcription was activated by HSF1, we amplified the chipmunk Per2 genomic sequence and determined the transcription start site by 5ʹ RACE (rapid amplification of cDNA ends) PCR. The consensus HSF-binding sequence, heat shock element (HSE), is a tandem array of at least three oppositely oriented “nGAAn” motifs or a degenerate version thereof (31). HSEs were found at −6387 in the 5ʹ upstream region (---aGAAgcTTCagGTGc---) (5ʹ-HSE) and at +1904 in intron 1 (---cGAActTTCtgGGAg---) (intronic HSE, INT-HSE), in which mismatches from the ideal consensus sequence (nGAAnnTTCnnGAAn) are underlined (Fig. 2A). We then tested whether HSF1 binds to these HSEs by EMSA using HSF1 synthesized by in vitro transcription-translation system (Fig. S1). In the EMSA, in vitro–translated HSF1 was bound to both 5ʹ-HSE and INT-HSE and two base substitutions in these HSEs-abolished HSF1 binding (Fig. 2B). In addition, competition experiments revealed that HSF1 had a much higher affinity for INT-HSE than for 5ʹ-HSE (Fig. 2C). The mouse Per2 gene was activated by HSF1 through HSE at −1.1 kb in the 5ʹ upstream region (25). We first examined whether 5ʹ-HSE was responsible for transactivation by HSF1 by transfecting a reporter plasmid containing the 6.4 kb 5ʹ upstream region of the Per2 gene, pCMPER2G-6390/luc (Fig. 2A), into HepG2 cells. Transcription was observed in HepG2 cells but was not activated by HSF1 (Fig. 2D), indicating that 5ʹ-HSE was not involved in transcriptional activation by HSF1. We further examined whether INT-HSE was responsible for the transcriptional activation of the Per2 gene by HSF1. Since the 0.45 kb 5ʹ upstream region of the Per2 gene contained promoter activity in HepG2 cells (data not shown), we constructed several Per2 reporter plasmids containing −0.45 kb through exon 2 but devoid of part of the intron 1 sequence, in which the luciferase-coding sequence was fused with the exon 2 sequence of the Per2 gene in frame with the PER2-coding sequence (Figs. 2A and S2). When transfected into HepG2 cells, these reporter plasmids showed significantly higher luciferase activities than that of pGV-B, and among them, pCMPER2G + 657/luc had higher activities than the other reporter constructs, indicating that the presence of a negative regulatory sequence downstream of +657 (Fig. S2). We then used pCMPER2G + 1894/luc lacking INT-HSE as pCMPER2G-INT(-HSE)/luc and pCMPER2G + 2055/luc containing INT-HSE as pCMPER2G-INT(+HSE)/luc (Fig. 2A) and examined transcriptional activation by HSF1. We also constructed pCMPER2G-INT(+HSE)(Mut)/luc by replacing INT-HSE with a mutated sequence (---cCAActTTTtgGGAg---, in which underlined letters indicate base substitutions). In HepG2 cells, transcription was activated by HSF1 from pCMPER2G-INT(+HSE)/luc, but not from pCMPER2G-INT(-HSE)/luc, and the INT-HSE mutation abolished transactivation by HSF1 (Fig. 2E). These results indicate that INT-HSE is responsible for HSF1-dependent transactivation of the Per2 gene.
Figure 2.
Transcriptional activation of Per2 gene by HSF1.A, schematic diagrams of the chipmunk Per2 gene from the 5ʹ flanking region to exon 2 (top) and the Per2 gene luciferase reporter plasmids are shown. The transcription start site is indicated as +1. B and C, 32P-labeled 5ʹ-HSE (−6400/−6373) WT or mutant (Mut) probe or INT-HSE (+1908/+1930) WT or Mut probe was incubated with in vitro transcription-translation products (TnT) of pcDNA3 (Empty) or pcDNA3/mHSF1 (HSF1), a mouse HSF1 expression vector. In (C), indicated amount of competitor was added in the binding reaction. 5′-HSE (Mut), 5ʹ-gtcaccaaCAAgcTTTagGTGcagaga-3ʹ; INT-HSE (Mut), 5′-gcacaccCAActTTTtgGGAgg-3′, in which the underlined letters indicate base substitutions in HSEs. D and E, HepG2 cells were transfected with a firefly luciferase reporter plasmid together with a Renilla luciferase plasmid pRL-SV40 as a control for transfection efficiency. Where denoted, the cells were cotransfected with the indicated amounts of pcDNA3/mHSF1. Each firefly luciferase activity was normalized to the Renilla luciferase activity and is shown as the fold increase compared with that of pCMPER2G-6390/luc in (D) or pCMPER2G-INT(-HSE)/luc in (E). The data represent mean ± SEM from three independent experiments performed in triplicate. Different letters indicate significantly different values (Tukey's HSD test, p < 0.05). HSE, heat shock element; HSF1, heat shock factor 1; INT-HSE, intronic HSE.
HSF1 binds to INT-HSE of Per2 gene in the liver of nonhibernating chipmunks
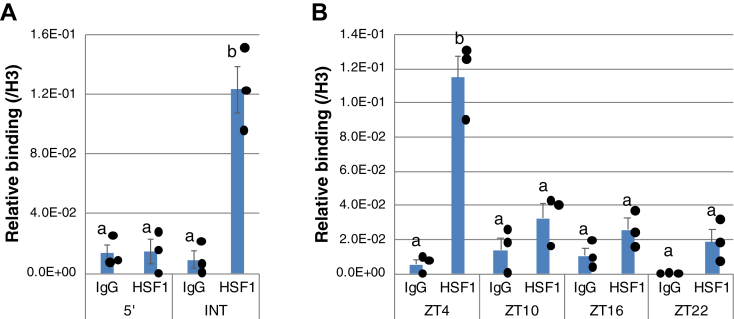
To determine whether HSF1 was involved in the transcriptional activation of the Per2 gene in the chipmunk liver, we performed chromatin immunoprecipitation (ChIP) analysis. At ZT4, when the Per2 mRNA level was at its maximum (Fig. 1), ChIP clearly revealed that HSF1 was bound to INT-HSE and not to 5ʹ-HSE (Fig. 3A), suggesting that HSF1 activated the transcription of the Per2 gene via INT-HSE in the chipmunk liver. These results also reflected the difference in the binding properties of 5ʹ-HSE and INT-HSE with HSF1, as revealed by EMSA (Fig. 2C). ChIP also revealed that HSF1 binding to INT-HSE was circadian, particularly strong at ZT4 and marginal at the other ZTs (Fig. 3B). The dissociation of HSF1 from INT-HSE was likely involved in the transcriptional attenuation of the Per2 gene. These results suggest that in nonhibernating chipmunks, the circadian rhythm of Tb led to HSF1 activation during the wake period, and HSF1 activated the transcription of the Per2 gene, thereby resetting the peripheral clock, as in the case of mice (27).
Figure 3.
Binding of HSF1 to intronic HSE of Per2 gene in liver of nonhibernating chipmunks.A and B, ChIP analysis was performed with chromatin from the liver of three nonhibernating chipmunks at ZT4 in (A) or at four ZTs in (B) using antibodies against HSF1 or histone H3, and normal rabbit IgG. qPCR was performed in triplicate using a primer set specific for 5ʹ-HSE or INT-HSE of the Per2 gene in (A) or for INT-HSE in (B). A normal IgG served as a negative control. The data represent mean ± SEM from three independent experiments using chromatins prepared from different animals. Values were normalized to the histone H3 values, and dot plots correspond to observed values (n = 3). Different letters indicate significantly different values (Tukey's HSD test, p < 0.05). ChIP, chromatin immunoprecipitation; HSE, heat shock element; HSF1, heat shock factor 1; INT-HSE, intronic HSE; ZT, zeitgeber time.
Per2 transcription is activated by HSF1 during interbout arousal in hibernation season
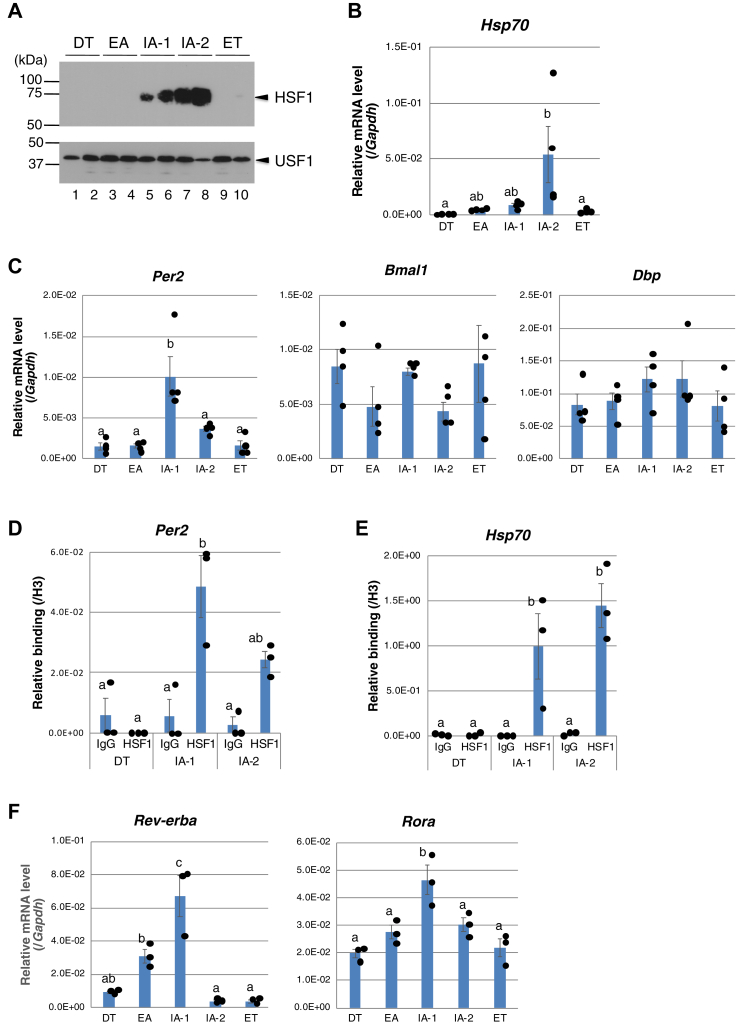
During the hibernation season, the Tb of chipmunks fluctuated periodically between 5 to 7 °C and ∼37 °C as they went through a cycle of 5 to 6 days long deep torpor and ∼20 h of arousal (10). In the chipmunk liver, while HSF1 was sequestered in the cytoplasm during deep torpor, HSF1 accumulated in the nucleus as the Tb increased during interbout arousal (11); the nuclear HSF1 was still low when Tb was below 30 °C (early arousal, EA), which then began to increase when Tb exceeded 30 °C (IA-1), reached maximum levels when Tb was 35 to 37 °C (IA-2), and subsequently disappeared from the nucleus when chipmunks entered torpor by lowering their Tb (ET) (Fig. 4A). Therefore, we classified chipmunks in the hibernation season into five categories on the basis of Tb and the amount of the nuclear HSF1: hibernating, deeply torpid animals with Tb of 5 to 7 °C (DT), early arousal animals whose Tb had not reached 30 °C during rewarming after spontaneous arousal from torpor (EA), interbout awake animals with Tb of 30 to 35 °C (IA-1) and 35 to 37 °C (IA-2), and animals in the process of entering into torpor with Tb below 30 °C (ET). In the chipmunk liver, the Hsp70 mRNA increased transiently in IA-2 when HSF1 was most abundant in the nucleus (Fig. 4B). During the process of awakening from torpor (EA), the Per2 mRNA level was low but increased transiently in IA-1 and then decreased in IA-2 (Fig. 4C). The Per2 mRNA level was already low during entry into the torpor (ET) and remained low in torpid animals (DT) (Fig. 4C). Thus, the Per2 mRNA level fluctuated regularly in the torpor-arousal cycle throughout the hibernation season under a constant condition of 5 °C in darkness. Considering that transcription and translation are globally suppressed during deep torpor (14), it is likely that the Per2 mRNA level does not fluctuate in the liver of deeply torpid animals, suggesting ceased function of the circadian clock. On the contrary, the Bmal1 and Dbp mRNA showed large inter-individual differences even in the same category of the torpor-arousal cycle, and there were no significant differences between categories [one-way ANOVA, p = 0.351 (Bmal1), p = 0.412 (Dbp)] and no rhythm in their variation (Fig. 4C). If the transcription-translation feedback loops involving the clock genes were organized, the Per2 and Bmal1 mRNAs should show periodic fluctuations in opposite phases to each other, but this was not the case, suggesting that the peripheral circadian clock is not functioning in the hibernation season.
Figure 4.
Transcriptional activation of Per2 gene by HSF1 during interbout arousal.A, immunoblot analysis of HSF1 and USF1 performed using nuclear extracts prepared from the liver of chipmunks in the hibernation season. B, the Hsp70 and Gapdh mRNA levels were measured by RT-qPCR, and the Hsp70 mRNA levels were normalized to the Gapdh mRNA levels. Results are shown as mean ± SEM with corresponding dot plots to observed values (n = 4/group at each time point). Different letters indicate significantly different values (Tukey's HSD test, p < 0.05). C, the Per2, Bmal1, Dbp, and Gapdh mRNA levels were measured by RT-qPCR, and the Per2, Bmal1, and Dbp mRNA levels were normalized to the Gapdh mRNA levels as in (B). D and E, ChIP analysis was performed with chromatin from the liver of three each of DT, IA-1, and IA-2 using antibodies against HSF1 or histone H3 and normal rabbit IgG. qPCR was performed in triplicate using a primer set specific for INT-HSE of the Per2 gene in (D) or for HSE in the Hsp70 gene promoter in (E). The data represent mean ± SEM from three independent experiments using chromatins prepared from different animals. Values were normalized to the histone H3 values, and dot plots correspond to observed values (n = 3). Different letters indicate significantly different values (Tukey's HSD test, p < 0.05). F, the Rev-erba, Rora, and Gapdh mRNA levels were measured by RT-qPCR (n = 3/group at each time point), and the Rev-erba and Rora mRNA levels were normalized to the Gapdh mRNA levels as in (B). ChIP, chromatin immunoprecipitation; DT, deeply torpid; HSE, heat shock element; HSF1, heat shock factor 1; INT-HSE, intronic HSE; RT-qPCR; reverse transcription-quantitative PCR.
To elucidate whether HSF1 is involved in the transcriptional activation of the Per2 gene in IA-1, we performed ChIP analysis. We found that HSF1 binding to INT-HSE of the Per2 gene was undetectable in DT, EA, and ET, when the nuclear HSF1 was low (Fig. 4A), but increased in IA-1 and decreased in IA-2 (Figs. 4D and S3), which was consistent with the change in the Per2 mRNA level. In the case of the Hsp70 gene, HSF1 binding to the HSE in the 5ʹ upstream region increased in IA-2 when the Hsp70 mRNA increased (Figs. 4E and S3). We also analyzed the involvement of HSF1 in the transcriptional activation of the Per2 gene in chipmunk liver by transfecting primary hepatocytes prepared from a hibernating chipmunk with a mammalian expression construct of HSF1. As expected, overexpression of HSF1 resulted in transcriptional activation of the Per2 gene (Fig. S4). These results indicate that the Tb rise during interbout arousal activated HSF1, which in turn activated Per2 transcription. Since the expression of the Bmal1 and Dbp mRNA was not rhythmic (Fig. 4C), to investigate whether PER2 is functional during interbout arousal, we analyzed the expression of two other clock-related genes, Rev-erba and Rora. Their transcription is activated by the BMAL1-CLOCK heterodimer (32, 33), and the BMAL1-CLOCK–mediated transcription is attenuated by the CRY–PER complex. In the nonhibernation season, the Bmal1 mRNA peaked at ZT16, and the Per2 mRNA started to increase at ZT22 and peaked at ZT4 (Fig. 1). The reverse transcription-quantitative PCR results showed that the Rev-erba mRNA peaked at ZT22 and decreased at ZT4, and the Rora mRNA also tended to increase at ZT22, although not significantly (one-way ANOVA, p = 0.050) (Fig. S5), supporting that at least Rev-erba gene transcription is activated by BMAL1-CLOCK and repressed by CRY-PER. In the hibernation season, both the Rev-erba and Rora mRNA were at low levels in ET and DT but began to increase in EA, peaked in IA-1, and decreased in IA-2 (Fig. 4F). Considering that the Per2 mRNA peaked in IA-1 (Fig. 4C) and the duration of IA-1 is approximately 30 to 40 min, it is likely that PER2 is responsible for the reduction of the Rev-erba and Rora mRNA in IA-2 by repressing BMAL1-CLOCK–mediated transcriptional activation. These results suggest that the peripheral circadian clock does not function in the hibernation season, but that upregulation of the Per2 gene by HSF1 during interbout arousal plays a role in regulating several clock-related genes.
Discussion
Mammalian hibernators spend the nonhibernation (summer) season as homeotherms and the hibernation (winter) season as heterotherms, repeating deep torpor and interbout arousal. The involvement of circadian rhythms in the torpor-arousal cycle during hibernation is of great interest, but whether circadian rhythms function during deep torpor remains an open question. In studies analyzing the expression of clock genes in the SCN of ground squirrels and European hamsters, no variation in expression was observed during deep torpor, suggesting that the circadian rhythm was suspended (22, 23). However, some studies analyzing Tb fluctuations have reported that rhythms were observed (16, 17), while the question of whether circadian rhythms function during deep torpor remains unclear. In mammalian hibernators, few analyses have been reported on peripheral clocks compared with the central clock. To elucidate whether the peripheral clock functions during the torpor-arousal cycle, we analyzed the expression of the Per2 gene in the liver. We chose the Per2 gene to examine peripheral clock expression during hibernation, because it is one of the earliest genes to respond to entrainment stimuli and exhibits entrainment by temperature (25, 26). The results indicate that Per2 transcription in the liver was activated by HSF1, which was activated by elevated Tb, in both the nonhibernation and hibernation seasons. However, the peripheral circadian clock in the liver was reset in the nonhibernation season but not in the hibernation season.
In the mouse liver, transcription of the Per2 gene has been shown to be activated by HSF1, which is activated by elevated Tb during the wake period, thereby resetting the peripheral clock (27). In the chipmunk liver, the Per2 mRNA also exhibited a circadian rhythm in the nonhibernation season, increasing during the wake period—when Tb was elevated—and decreasing during the sleep period (Fig. 1). Changes in the expression of the Per2, Bmal1, and Dbp mRNA in the liver during the sleep-wake cycle were similar in nonhibernating chipmunks and mice. In the mouse Per2 gene, the HSE required for transcriptional activation by HSF1 is located in the 5ʹ upstream region (25), whereas in the chipmunk Per2 gene, HSE is located in intron 1. The same HSE sequence (5ʹ-GAActTTCtgGGA-3ʹ) as this INT-HSE was found to be present in intron 1 of the Per2 gene in other mammalian hibernators of the family Sciuridae, such as the thirteen-lined ground squirrel, Arctic ground squirrel, and European marmot (Fig. S6), implying the importance of this HSE sequence in transcriptional activation by HSF1. In fact, ChIP analysis of chipmunk liver showed increased binding of HSF1 to INT-HSE during the wake period compared to the sleep period (Fig. 3B). These results suggest that in chipmunks, the circadian rhythm of Tb activated HSF1 during the wake period, which in turn activated transcription of the Per2 gene, thereby resetting the peripheral clock during the nonhibernation season, as in the case of mice (27).
In the hibernation season, the Per2 mRNA level remained low during deep torpor (Fig. 4C). Since transcription and translation are globally suppressed during deep torpor (14), this result suggests that the peripheral clock is stopped during deep torpor. It is also consistent with the reports that the circadian clock is considered to be suspended during deep torpor, as no oscillation in clock gene expression in the SCN was observed in ground squirrels and European hamsters (22, 23). During interbout arousal, HSF1 is activated by elevated Tb (11). HSF1 began to accumulate in the nucleus when Tb rose above 30 °C and transiently activated transcription of the Per2 gene (Fig. 4, A and C). As a result, the Per2 mRNA level fluctuated regularly in the torpor-arousal cycle throughout the hibernation season (Fig. 4C). Gautier et al. (34) also reported that transcription of the Per2 gene was increased in all tissues examined early in interbout arousal in European hamsters. Interestingly, both the Per2 and HSP70 genes were activated by HSF1, but the Per2 gene was activated at lower Tb (30–35 °C) than the HSP70 gene (Fig. 4, B and C), suggesting a difference in the activation mechanism of both genes by HSF1. While the HSF1 trimer binds to HSE and activates transcription in the case of the HSP70 gene (35), the interaction between HSF1 bound to HSE and BMAL1-CLOCK heterodimer bound to E-box may be responsible for the transcriptional activation of the Per2 gene (25). In contrast to the Per2 mRNA, the expression of the Bmal1 and Dbp mRNA during interbout arousal varied greatly among individuals and was not rhythmic (Fig. 4C). This result is similar to the observation that when temperature cycles that mimic Tb fluctuations were applied to cultured cells, the Per2 mRNA expression immediately synchronized with the temperature cycle, but the Bmal1 and Dbp mRNA took several days to synchronize (26). The lack of periodicity in Bmal1 and Dbp mRNA expression in the hibernation season is likely because the duration of interbout arousal is ∼20 h, followed by deep torpor in which transcription and translation are suppressed. The loss of the antiphasic relationship between Per2 and Bmal1 mRNA expression means that the transcription-translation feedback loop of the clock genes is not formed, and the peripheral clock does not function in the hibernation season. Thus, the clock genes that form circadian rhythms are differentially regulated between the nonhibernation and hibernation seasons, which may affect various gene networks. Such readjustment of gene expression may allow, for example, metabolic suppression to reduce Tb when entering hibernation.
Experimental procedures
Animals
Male chipmunks (T. asiaticus) (age, 2–4 months) purchased from Pet Easy Space were individually housed and provided with standard rodent chow and water ad libitum. They were kept at 23 °C with a 12 h:12 h light:dark photoperiod (light on at 6 AM) during the nonhibernation season (April–September). ZT0 corresponds to the time at which the light is on. During the hibernation season (October–March), they were kept under a constant condition of 5 °C in darkness. During the hibernation season, the conditions of the chipmunks were monitored using an infrared activity sensor (O’Hara & Co, Ltd), and their Tb was recorded by measuring the rectal temperature using a thermistor probe (Thermistor thermometer model KN-91-AD1687-R; Natsume Seisakusho). Tissue samples from nonhibernating chipmunks (weight, 90–110 g; age, 1–3 years) that were summer-active were obtained between June and August. During hibernation, tissue samples were obtained approximately 2 to 4 months after the first entry into torpor. Thus, all the chipmunks were acclimated to the photoperiod and temperature conditions of the nonhibernation or hibernation season for a minimum of 2 months before the experimental samples were collected. Animals were sacrificed after being deeply anesthetized with isoflurane. Tissues were immediately excised, frozen in liquid nitrogen, and stored at −80 °C until further use. Hibernating, deeply torpid animals were usually sampled between 12 AM and 4 PM. All protocols were in accordance with the guidelines of the Institutional Animal Care and Use Committee of Kitasato University and all experimental procedures were approved by the same committee.
Cloning procedures
Total RNA was prepared from chipmunk liver using Isogen (Nippon Gene), treated with RNase-free recombinant DNase I (Takara Bio), and purified using RNeasy Mini Kit (Qiagen), as per the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized using PrimeScript first strand cDNA Synthesis Kit (Takara Bio). Genomic DNA was prepared from chipmunk liver using Wizard Genomic DNA Purification Kit (Promega).
Chipmunk Per2 cDNA was amplified from chipmunk liver first strand cDNA using primers designed based on the ground squirrel Per2 cDNA sequence. The promoter region fragment of the chipmunk Per2 gene was amplified from chipmunk liver genomic DNA using the forward primer designed based on the corresponding ground squirrel genomic sequence and reverse primer designed from the corresponding chipmunk cDNA sequence. Amplified fragments were cloned into pBluescript SK and the sequences were determined. The transcription start site of the chipmunk Per2 gene was determined by 5′ RACE PCR using First Choice RLM-Race kit (Ambion). The 5′-terminal Per2 cDNA fragment was amplified using the 5′ RACE outer primer and an oligonucleotide complementary to chipmunk Per2 cDNA (5′-GCAGTTTTCATTGGTCTCATTTC-3′), and the amplified fragment was cloned into pBluescript SK and sequenced.
Reverse transcription-quantitative PCR
First-strand cDNA was synthesized using DNase I–treated chipmunk liver total RNA using PrimeScript first strand cDNA Synthesis Kit (Takara Bio). Quantitative PCR was carried out using SYBR Green Realtime PCR Master Mix-Plus (Toyobo). Primer sequences are listed in Table S1. Gapdh was used as the reference gene to normalize RNA quantity.
Electrophoretic mobility shift assay
Mouse HSF1 was synthesized using an in vitro transcription-translation system (Promega) and then heat-treated at 42 °C for 60 min. Synthesis of HSF1 from pcDNA3/mHSF1 was confirmed by immunoblot analysis (Fig. S1). The following oligonucleotides were annealed and used as probes: CMPER2G-6400/-6373(WT), 5ʹ-GTCACCAAGAAGCTTCAGGTGCAGAGA-3ʹ and 5ʹ-GTCTCTGCACCTGAAGCTTCTTGGTGA-3ʹ; CMPER2G-6400/-6373(Mut), 5ʹ-GTCACCAAcAAGCTTtAGGTGCAGAGA-3ʹ and 5ʹ-GTCTCTGCACCTaAAGCTTgTTGGTGA-3ʹ; CMPER2G + 1908/+1930(WT), 5ʹ-GCACACCGAACTTTCTGGGAGG-3ʹ and 5′-GCCTCCCAGAAAGTTCGGTGTG-3ʹ; CMPER2G + 1908/+1930(Mut), 5′-GCACACCcAACTTTtTGGGAGG-3′ and 5′-GCCTCCCAaAAAGTTgGGTGTG-3′. The lower case letters in the Mut oligonucleotides indicate base substitutions. EMSA was performed as previously described (36). For competition assays, a 5- or 50-fold molar excess of unlabeled WT probe was used as a competitor.
Transfection and luciferase assays
HepG2 cells were cultured as previously described (36). Per2 reporter plasmids were constructed using a promoter-less firefly luciferase expression vector, pGV-B (Nippon Gene), as described previously (36). The cells were plated at 5 × 104 cells per 15-mm dish, and after 24 h, cells were transfected with 100 ng of a firefly luciferase reporter plasmid and 2.5 ng of a Renilla luciferase internal control plasmid, pRL-SV40 (Promega), using TransIT-LT1 reagent (Mirus). Where denoted, the cells were cotransfected with the indicated amounts of the mouse HSF1 expression vector, pcDNA3/mHSF1. After 24 h, luciferase activity was measured using the dual-luciferase reporter assay system (Promega).
ChIP analysis
ChIP analysis was performed as previously described (37). All ChIP experiments were performed in triplicate on independent chromatin preparations. Normal rabbit IgG (sc-2027) and anti-HSF1 antibody (sc-9144) were purchased from Santa Cruz Biotechnology. The anti-HSF1 antibody (ADI-SPA-901) was purchased from Enzo Life Sciences. The anti-Histone H3 antibody (ab1791) was purchased from Abcam. Primer sequences are listed in Table S2. The anti-HSF1 antibody (sc-9144) was used in Figures 3A, and 4, D and E, and the anti-HSF1 antibody (ADI-SPA-901) was used in Figure 3B and Fig. S3. The anti-HSF1 antibodies were validated by immunoprecipitation using chipmunk liver whole-cell extracts (Fig. S7).
Immunoblot analysis
Nuclear extracts were prepared using NUN buffer as described by Reinke et al. (27). Immunoblot analysis was carried out as described previously by Fujii et al. (38). The anti-HSF1 antibody (#4356) and anti-USF1 antibody (sc-390033) were purchased from Cell Signal Technology and Santa Cruz Biotechnology, respectively.
Statistical analysis
All experiments were repeated in triplicate. All quantitative data are presented as mean ± SEM. Significant differences between group means were analyzed using one-way ANOVA and were considered significant at p < 0.05. Significant results from ANOVA tests were further analyzed using a Tukey's honestly significant difference test to find means that were significantly different from each other (p < 0.05).
Data availability
The chipmunk DNA sequence data were submitted to DDBJ. The DDBJ accession numbers are LC703156 (Per2), LC703154 (Bmal1), LC703155 (Dbp), LC741554 (Rora), LC741555 (Rev-erba), LC703157 (Per2 gene 5ʹ upstream region), and LC703158 (Per2 gene exon 1-exon 2).
Supporting information
This article contains supporting information (https://jaspar.genereg.net/, https://www.ncbi.nlm.nih.gov/gene).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Yukina Sakurai for providing technical assistance.
Author contributions
N. T. conceptualization; N. T. methodology; N. T. investigation; N. T. writing–original draft; N. T. project administration; Y. S., K. S., and E. N. validation; M. I. and D. T. writing–review and editing; D. T. resources.
Funding and additional information
This work was supported in part by Japan Society for the Promotion of Science (JSPS) Grants 20K06746 (to N. T.) and 20K06447 (to D. T.).
Reviewed by members of the JBC Editorial Board. Edited by Brian Strahl
Supporting information
References
- 1.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 2.Welsh D.K., Takahashi J.S., Kay S.A. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 4.Yoo S.H., Yamazaki S., Lowrey P.L., Shimomura K., Ko C.H., Buhr E.D., et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buhr E.D., Takahashi J.S. Molecular components of the mammalian circadian clock. Handb. Exp. Pharmacol. 2013;217:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhr E.D., Yoo S.H., Takahashi J.S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Menaker M., Murphy Z.C., Sellix M.T. Central control of peripheral circadian oscillators. Curr. Opin. Neurobiol. 2013;23:741–746. doi: 10.1016/j.conb.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Kondo N., Kondo J. Identification of novel blood proteins specific for mammalian hibernation. J. Biol. Chem. 1992;267:473–478. [PubMed] [Google Scholar]
- 11.Tsukamoto D., Hasegawa T., Hirose S., Sakurai Y., Ito M., Takamatsu N. Circadian transcription factor HSF1 regulates differential HSP70 gene transcription during the arousal-torpor cycle in mammalian hibernation. Sci. Rep. 2019;9:832. doi: 10.1038/s41598-018-37022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo N., Sekijima T., Kondo J., Takamatsu N., Tohya K., Ohtsu T. Circannual control of hibernation by HP complex in the brain. Cell. 2006;125:161–172. doi: 10.1016/j.cell.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Narasimamurthy R., Virshup D.M. Molecular mechanisms regulating temperature compensation of the circadian clock. Front. Neurol. 2017;8:161. doi: 10.3389/fneur.2017.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storey K.B., Storey J.M. Metabolic rate depression in animals: transcriptional and translational controls. Biol. Rev. Camb. Philos. Soc. 2004;79:207–233. doi: 10.1017/s1464793103006195. [DOI] [PubMed] [Google Scholar]
- 15.Pohl H. Circadian pacemaker does not arrest in deep hibernation. Evidence for desynchronization from the light cycle. Experientia. 1987;43:293–294. doi: 10.1007/BF01945554. [DOI] [PubMed] [Google Scholar]
- 16.Grahn D.A., Miller J.D., Houng V.S., Heller H.C. Persistence of circadian rhythmicity in hibernating ground squirrels. Am. J. Physiol. 1994;266:R1251–1258. doi: 10.1152/ajpregu.1994.266.4.R1251. [DOI] [PubMed] [Google Scholar]
- 17.Larkin J.E., Franken P., Heller H.C. Loss of circadian organization of sleep and wakefulness during hibernation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;282:R1086–R1095. doi: 10.1152/ajpregu.00771.2000. [DOI] [PubMed] [Google Scholar]
- 18.Ruby N.F., Dark J., Burns D.E., Heller H.C., Zucker I. The suprachiasmatic nucleus is essential for circadian body temperature rhythms in hibernating ground squirrels. J. Neurosci. 2002;22:357–364. doi: 10.1523/JNEUROSCI.22-01-00357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hut R.A., Barnes B.M., Daan S. Body temperature patterns before, during, and after semi-natural hibernation in the European ground squirrel. J. Comp. Physiol. B. 2002;172:47–58. doi: 10.1007/s003600100226. [DOI] [PubMed] [Google Scholar]
- 20.Kart Gür M., Refinetti R., Gür H. Daily rhythmicity and hibernation in the anatolian ground squirrel under natural and laboratory conditions. J. Comp. Physiol. B. 2009;179:155–164. doi: 10.1007/s00360-008-0298-0. [DOI] [PubMed] [Google Scholar]
- 21.Williams C.T., Barnes B.M., Richter M., Buck C.L. Hibernation and circadian rhythms of body temperature in free-living arctic ground squirrels. Physiol. Biochem. Zool. 2012;85:397–404. doi: 10.1086/666509. [DOI] [PubMed] [Google Scholar]
- 22.Revel F.G., Herwig A., Garidou M.L., Dardente H., Menet J.S., Masson-Pévet M., et al. The circadian clock stops ticking during deep hibernation in the European hamster. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13816–13820. doi: 10.1073/pnas.0704699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeno T., Williams C.T., Buck C.L., Barnes B.M., Yan L. Clock gene expression in the suprachiasmatic nucleus of hibernating arctic ground squirrels. J. Biol. Rhythms. 2017;32:246–256. doi: 10.1177/0748730417702246. [DOI] [PubMed] [Google Scholar]
- 24.Brown S.A., Zumbrunn G., Fleury-Olela F., Preitner N., Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 25.Tamaru T., Hattori M., Honda K., Benjamin I., Ozawa T., Takamatsu K. Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saini C., Morf J., Stratmann M., Gos P., Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26:567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinke H., Saini C., Fleury-Olela F., Dibner C., Benjamin I.J., Schibler U. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornmann B., Schaad O., Bujard H., Takahashi J.S., Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher G., Reinke H., Altmeyer M., Gutierrez-Arcelus M., Hottiger M., Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Ripperger J.A., Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 31.Janus P., Toma-Jonik A., Vydra N., Mrowiec K., Korfanty J., Chadalski M., et al. Pro-death signaling of cytoprotective heat shock factor 1: upregulation of NOXA leading to apoptosis in heat-sensitive cells. Cell Death Differ. 2020;27:2280–2292. doi: 10.1038/s41418-020-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 33.Sato T.K., Panda S., Miraglia L.J., Reyes T.M., Rudic R.D., McNamara P., et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Gautier C., Bothorel B., Ciocca D., Valour D., Gaudeau A., Dupré C., et al. Gene expression profiling during hibernation in the European hamster. Sci. Rep. 2018;8:13167. doi: 10.1038/s41598-018-31506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anckar J., Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 36.Kojima M., Takamatsu N., Ishii T., Kondo N., Shiba T. HNF-4 plays a pivotal role in the liver-specific transcription of the chipmunk HP-25 gene. Eur. J. Biochem. 2000;267:4635–4641. doi: 10.1046/j.1432-1327.2000.01499.x. [DOI] [PubMed] [Google Scholar]
- 37.Tsukamoto D., Fujii G., Kondo N., Ito M., Shiba T., Takamatsu N. USF is involved in the transcriptional regulation of the chipmunk HP-25 gene. Gene. 2007;396:268–272. doi: 10.1016/j.gene.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Fujii G., Nakamura Y., Tsukamoto D., Ito M., Shiba T., Takamatsu N. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem. J. 2006;395:203–209. doi: 10.1042/BJ20051802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chipmunk DNA sequence data were submitted to DDBJ. The DDBJ accession numbers are LC703156 (Per2), LC703154 (Bmal1), LC703155 (Dbp), LC741554 (Rora), LC741555 (Rev-erba), LC703157 (Per2 gene 5ʹ upstream region), and LC703158 (Per2 gene exon 1-exon 2).