Highlights
-
•
FAM111B expression of ovarian cancer was observed by immunohistochemical staining.
-
•
FAM111B expression positively correlated with KI67, EGFR, and PDL-1.
-
•
High FAM111B expression was associated with worse prognosis of ovarian cancer.
-
•
Analysis in four gene expression omnibus datasets verified our findings.
-
•
Protein-protein interaction indicated potential FAM111B functions.
Keywords: Ovarian cancer, FAM111B, Immunohistochemistry, Prognosis, Clinicopathologic feature
Abstract
Backgrounds
Ovarian cancer (OC) is the second most common gynecological tumor with the highest mortality rate worldwide. High FAM111B expression has been reported as a predictor of poor prognosis in other cancers, but its correlation with OC has not been reported.
Methods
Immunohistochemistry of tissue microarrays was performed to detect FAM111B expression levels in 141 OC patient tissues. The prognostic value of FAM111B was determined by Kaplan–Meier survival analysis, and correlations between FAM111B expression and clinicopathologic features were investigated by the Clu-square test. The significance of FAM111B expression was verified bioinformatically using the Gene Expression Omnibus database. Protein-protein interaction were performed to explore downstream mechanisms of FAM111B in OC.
Results
Among 141 OC patients, FAM111B was positively expressed in 87.23%, 58.16%, and 87.94%; and highly expressed in 8.51%, 17.02%, and 19.86%, as evaluated by cytoplasmic, nuclear, and combined cytoplasmic/nuclear staining. FAM111B expression was positively correlated with the expression of tumor protein markers KI67, EGFR, and PDL-1. Patients with high FAM111B expression had aggressive clinicopathologic features and shorter overall survival (P value 0.0428, 0.0050, 0.0029) and progression-free survival (P value 0.0251, 0.012, 0.0596) compared to the low FAM111B expression group for cytoplasmic, nuclear, and combined cytoplasmic/nuclear groups, respectively. These results were verified using patient data from the Gene Expression Omnibus. Seventeen genes co-expressed with FAM111B were primarily involved in “negative regulation of histone modification”, “hippo signaling” and “inner ear receptor cell differentiation”.
Conclusions
High FAM111B expression may serve as a novel prognostic predictor and molecular therapeutic target for OC.
Introduction
Ovarian cancer (OC) is the second most common gynecological cancer, with the highest mortality rate worldwide. The American Cancer Society estimated that in 2022, 19,880 women will be newly diagnosed and 12,810 cases will die from OC in the United States [1]. Due to the delayed presentation of clinical symptoms until the advanced stages, OC is known as the “the silent killer” [2]. Consequently, the 5-year survival rate of advanced and metastatic OC is only about 30% [3]. Chemotherapy and surgical debulking are front-line treatments for OC, while 70–85% of advanced patients suffer from disease recurrence and undergo second-line chemotherapy [4,5]. Molecular targeted therapies, such as anti-vascular endothelial growth factor and poly ADP-ribose polymerase inhibitors, provide additional favorable treatment strategies for advanced and recurrent OC that can prolong patient survival to some extent [6], [7], [8], [9]. However, OC prognosis still needs to be further improved, and the identification of novel molecular targets for OC is urgently needed.
The family with sequence similarity 111-member B (FAM111B) gene, located on human chromosome 11q12.1, encodes a protein with a C-terminal trypsin-like cysteine/serine peptidase domain [10,11]. Functionally, FAM111B participates in protein binding, peptidase and hydrolase activity [12]. Mutations in FAM111B cause hereditary fibrosing poikiloderma with tendon contracture, homeopathy, and pulmonary fibrosis [13] and have also been associated with inherited exocrine pancreatic dysfunction [14]. Recently, several studies indicated that FAM111B contributes to the formation of cancers, including breast cancer, pancreatic cancer, and lung adenocarcinoma (LUAD) [15], [16], [17]. The expression of FAM111B is higher in breast cancer tissues than in normal tissues, and high FAM111B expression is associated with the poor prognosis of breast cancer patients [15]. Additionally, FAM111B mutation is involved in pancreatic cancer predisposition and exocrine pancreatic insufficiency [16]. Mechanistically, the oncogenic activity of FAM111B in LUAD has been demonstrated to involve the p53 signaling pathway [18]. However, the role of FAM111B in the pathogenesis of OC has not been reported.
In this study, we sought to attain a comprehensive understanding of whether and how FAM111B contributes to the tumorigenicity of OC. We hypothesized that FAM111B may be expressed in most OC tissues, and that the high expression of FAM111B may be associated with poor prognosis and aggressive phenotype of OC. To test our hypothesis, we performed immunohistochemical analysis of the FAM111B protein and investigated clinicohistological factors using tumor tissue microarrays (TMAs) encompassing the major histological subtypes of ovarian tumors. We also performed Gene Expression Omnibus (GEO) bioinformatics analysis to support our findings and explored the downstream mechanism of FAM111B in OC. Our results suggest that FAM111B may serve as a novel prognostic factor and potential therapeutic target for OC.
Materials and methods
Patient cohort and TMA
The TMA (panel HOvaC151Su01) and clinicopathological information were obtained from Shanghai Outdo Biotech Co.Ltd. The study was approved by the Ethics Committee of Shanghai Outdo Biotech Co. Ltd. (No. YBM-05–02).Tumor tissues were obtained from patients at the time of primary surgery. The 141 cases included in the TMA were pathologically diagnosed with OC between 2009 and 2013 and included 74 cases of serous carcinoma, 31 cases of mucinous carcinoma, 17 cases of endometrioid carcinoma, and 19 cases of other ovarian tumors (dysgerminoma, squamous cell cancer, adenocarcinoma, malignancy Brenner cancer, yolk sac tumor and clear cell carcinoma). Tissues containing two or more different histological types were classified into the group of the predominant type. Of these samples, 12 were metastatic tumors, and 129 were primary tumors. According to International Federation of Gynecology and Obstetrics (FIGO) stage classification, 43 cases were TNM stage I-II, and 98 cases were TNM stage III-IV. According to the histological tumor grade, 13 cases were grade I, 29 cases were grade II, 74 cases were grade III, and 25 cases lacked information on the histological grade. The median follow-up time was 90 months (range 72–108 months), and the mean age was 52 years (range 20–75 years).
Immunohistochemistry (IHC)
IHC analysis was performed on formalin-fixed paraffin-embedded tissue sections, which were placed in a 63 °C electric oven for an hour. Xylene was used as the paraffin solvent, and alcohol dehydration was completed using an automatic immunohistochemical staining instrument (Leica Biosystems, LEICAST5020, USA). Automatic immunohistochemistry processing equipment (Dako, PT Link, Denmark) was used to recover antigens. The expression of FAM111B was detected using anti-FAM111B antibody (Dako, S3022, Denmark) at a dilution of 1:300 with an automatic immunohistochemical staining instrument (Dako, Autostainer Link48, Denmark). Primary antibodies against EGFR (Genetic Ltd, GT2101, China), KI67 (Baidao Medical Technology Co. Ltd, PA192, China) and PDL-1 (Genetic Ltd, GT2280, China) were used to detect the expression of the corresponding proteins. The samples were stained with hemotoxylin, and the sections were dehydrated and mounted with neutral balsam.
The samples were evaluated with binary positive/negative scoring by two senior pathologists. Each specimen was scored according to the extent of cell staining (0–100%) and the staining intensity (no staining as 0, little staining as 0.5, slight staining as 1, moderate staining as 2 and strong staining as 3). The total score (the extent of cell staining score multiplied by the staining intensity score) was calculated (range 0–300%). In the cytoplasm, total scores less than or equal to 100% were indicative of a low FAM111B expression level, and scores greater than 100% were indicative of a high FAM111B expression level. In the nucleus, total scores less than or equal to 16% were indicative of a low FAM111B expression level and scores greater than 16% were indicative of a high FAM111B expression level. In the combined cytoplasm and nucleus staining group, negative staining results both in the cytoplasm and nucleus was indicative of negative expression, while any positive staining result was indicative of positive expression. Total scores greater than 100% in the cytoplasm or greater than 16% in the nucleus were indicative of high FAM111B levels in the combined group.
Cox regression analysis and co-analysis of fam111b and KI67, EGER, PDL-1 expression
The independent prognostic value of FAM111B expression and clinicopathologic factors associated with survival was evaluated with univariate and multivariate analysis. The multivariate Cox regression analysis combined FAM111B expression, histology and TNM stage was used to assess the factors associated with the survival of OC (P<0.05). The univariate Cox regression model was used to identify significant factors associated with the prognosis of OC patients (P<0.05), and these selected factors were evaluated by subsequent multivariate Cox regression. The association between FAM111B and other proteins, including KI67, EGFR, and PDL-1, was evaluated by immunohistochemical staining based on the Pearson correlation, and P<0.05 was considered statistically significant.
Kaplan–Meier analysis
To clarify the prognostic value of FAM111B expression in OC, Kaplan–Meier survival analysis was performed using Graph Pad Prism Software version 8.0. The FAM111B protein expression levels were treated as binary variables and divided into “high” and “low" levels for the cytoplasmic, nuclear and combined staining groups with classification criteria as described above. The overall survival (OS) and progression-free survival (PFS) in the high and low expression groups were compared using Kaplan-Meier plots; and hazard ratios, 95% confidence intervals, and log-rank P values were calculated.
GEO data analysis, protein-protein interaction (PPI) network and pathway enrichment analysis
FAM111B expression was analyzed in the OC samples from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). Four independent microarray datasets (GSE63885, GSE73614, GSE9891, and GSE39204) were included in the analysis. The datasets were used to analyze the expression levels of FAM111B in OC patients, as well as clinicopathologic features and prognosis information. The STRING (https://cn.string-db.org/) and STITCH (http://stitch.embl.de/) databases were used to generate a PPI and chemical association network relevant to FAM111B. Core sub-network analysis of the PPI network was performed with MCODE in Cytoscape. Co-expressed genes obtained from PPI network were used for gene-ontology enrichment annotation with ClusterProfiler4.0.5.
Statistical analysis
The Clu-square test was used to analyze the expression of FAM111B in different histological types of OC. Survival estimates were calculated using the Kaplan-Meier method and compared using the log-rank test. Multivariate analysis was performed using the Cox proportional hazards regression model. The Clu-square or Fisher's exact test was used to analyze the correlation between the variables (FAM111B, KI67, EGFR and PDL-1) and the independent prognostic value of FAM111B expression and clinicopathologic factors in relation to survival. Graphs and images were prepared using Prism 8.0 (Graphpad Software, La Jolla, CA, USA) and Photoshop CC2019 for Windows (Adobe. Inc, San Jose, CA, USA). All tests were two-sided, and P values <0.05 were considered significant.
Results
FAM111B is predominantly expressed, both in the cytoplasm and nucleus of serous carcinoma
To determine whether FAM111B is expressed in OC in the cytoplasm and/or nucleus, we performed immunohistochemical staining of TMAs with 141 patient samples. The specimens used in each analysis were remarked in Fig. S1. FAM111B and other cancer-associated proteins (KI67, EGFR, PDL-1) were detected at high levels in cancer tissues with little staining in para-carcinoma tissues. (Fig. 1A-H). Moreover, the cytoplasmic versus nuclear localization of FAM111B varied according to the case (Fig. 1I-J). Among the 141 OC samples, 87.23% (123/141) were positive for cytoplasmic FAM111B expression; 58.16% (82/141) were positive for nuclear FAM111B expression; 87.94% (124/141) were positive in the combined group (cytoplasmic and/or nuclear staining) (Fig. 1K). Therefore, these results indicate that FAM111B is predominantly expressed and localized both in the cytoplasm and nucleus in OC.
Fig. 1.
Representative immunohistochemical staining of FAM111B and other cancer-associated proteins in OC. (A-B) FAM111B staining in para-carcinoma and OC tissues. (C-D) KI67 staining in para-carcinoma and OC tissues. (E-F) EGFR staining in para-carcinoma and OC tissues. (G-H) PDL1 staining in para-carcinoma and OC tissues. (I-J) Representative cytoplasmic and nuclear FAM111B staining. (K) Percentage of FAM111B positive / negative staining in cytoplasm, nucleus and combined cytoplasmic/ nuclear expression group.
To further evaluate FAM111B expression in OC, we characterized the cytoplasmic and nuclear patterns across different OC subtypes (Table S1). There was significantly less FAM111B positivity among mucinous carcinoma cases than serous carcinoma cases in the cytoplasmic and combined cytoplasmic/nuclear expression groups respectively (P = 0.009, P = 0.036). Moreover, there was significantly less high-level FAM111B expression in mucinous carcinoma cases than in serous carcinoma cases in the nuclear and combined cytoplasmic/nuclear expression groups respectively (P = 0.036, P = 0.021). However, no obvious differences in FAM111B expression were observed between serous carcinoma and endometrioid carcinoma or other types of carcinomas. To assess the difference in expression of FAM111B with histologic subtypes of different grade, we combined the grade with the histological subtypes for further evaluation (Table S2). We found the expression of FAM111B was positively correlated with the histological grade in serous carcinoma (P = 0.049).
FAM111B is co-expressed with KI67, EGER, and PDL-1
To determine whether cytoplasmic and/or nuclear FAM111B expression is associated with the expression of other key cancer-associated genes (including KI67, EGFR, and PDL-1), we performed Pearson correlation analysis. All 141 samples were used for imumunohistochemical detection, data missing specimens were excluded. The results verified that nuclear FAM111B was positively associated with KI67 (R²=0.1203, P<0.001), EGFR (R²=0.08632, P<0.001), and PDL-1 (R²=0.02819, P<0.05); and cytoplasm FAM111B was positively associated with KI67 (R²=0.1863, P<0.001) and EGFR (R²=0.04806, P<0.05) but showed no significant association with PDL-1 (R²=0.01157, P>0.05) (Fig. 2). These results suggest that FAM111B expression in OC may occur in tissues that express other cancer-related genes.
Fig. 2.
FAM111B expression in the cytoplasm and nucleus correlates with the expression of cancer-related marker proteins. (A-B) KI67 in the cytoplasm and nucleus (n = 133). (C-D) EGFR in the cytoplasm and nucleus (n = 136). (E-F) PDL1 in the cytoplasm and nucleus (n = 140).
FAM111B expression is correlated with clinicopathologic features of OC
To reveal the clinical significance of FAM111B in OC, we explored the associations between FAM111B expression and clinicopathologic parameters that are associated with prognosis. The samples were divided into high and low expression groups according to the combined cytoplasmic/nuclear expression levels of FAM111B, and clinicopathologic parameters, including histological grade, tumor size and TNM stage were evaluated. The results showed that the expression of FAM111B was correlated with histological grade and tumor size (P<0.001, P = 0.031) (Table 1). These findings suggest that high FAM111B expression predicts aggressive clinicopathologic features in OC patients.
Table 1.
Correlation between FAM111B expression and clinicopathologic characteristics in the combined cytoplasmic/ nuclear expression group.
| FAM111B expression |
||||
|---|---|---|---|---|
| Total | Low | High | P value | |
| Grade | 116 | <0.001 | ||
| I | 13 | 13 | 0 | |
| II | 29 | 23 | 6 | |
| III | 74 | 56 | 18 | |
| Tumor size (cm) | 141 | 0.031 | ||
| 11.4 ± 0.5 | 14.0 ± 1.1 | |||
| TNM stage | 141 | 0.105 | ||
| I/II | 43 | 38 | 5 | |
| III/IV | 98 | 75 | 23 | |
To further calculate the predictive independence of factors for OC patients, we performed Cox regression analysis. Tumor size, T stage, N stage, M stage, and TNM stage were all significantly correlated with FAM111B cytoplasmic and nuclear expression as evaluated by univariate and multivariate analysis (P<0.05) (Table S3 and S4). These results suggest that FAM111B expression in both the cytoplasm and nucleus is associated with factors affecting the prognosis of OC patients.
FAM111B expression is associated with poor prognoses of oc patients
Next, we investigated the association between FAM111B expression and survival (OS, PFS) of patients with OC. Patients with high FAM111B expression in the nucleus had poorer OS (HR: 0.3791, 95% CI: 0.1926–0.7462, P = 0.0050) and PFS (HR: 0.4301, 95% CI: 0.2227–0.8305, P = 0.0120) (Fig. 3A-B). Similar results were observed for cytoplasm FAM111B for OS (HR: 0.3799, 95% CI: 0.1489–0.9692, P = 0.0428) and PFS (HR: 0.3317, 95% CI: 0.1263–0.8711, P = 0.0251) (Fig. 3C-D); and for combined cytoplasmic and nuclear FAM111B for OS (HR: 2.607, 95% CI: 1.388–4.898, P = 0.0029) (Fig. 3E). High expression of combined cytoplasmic/nuclear FAM111B also showed a trend of poor PFS, though the P value was not significant (HR: 1.635, 95% CI: 0.9802–2.728, P = 0.0596) (Fig. 3F). Further, the multivariate Cox regression analysis combined FAM111B, histological types and TNM stage also showed that high cytoplasmic FAM111B expression and TNM stage were associated with shorter OS (P = 0.017, P = 0) and PFS (P = 0.004, P = 0) of OC (Table 2). These results indicate that FAM111B expression may be associated with poor prognosis in patients with OC.
Fig. 3.
FAM111B expression in the cytoplasmic, nuclear and combined cytoplasmic/nuclear staining groups correlates with the survival of OC patients. (A-B) Correlation of nuclear FAM111B expression with OS and PFS. (C-D) Correlation of cytoplasmic FAM111B expression with OS and PFS. (E-F) Correlation of combined cytoplasmic/nuclear FAM111B expression with OS and PFS.
Table 2.
Multivariate analysis of the correlation between cytoplasmic FAM111B expression, histology and TNM stage associated with overall survival and progression-free survival of OC.
| Variables | OS |
PFS |
||||||
|---|---|---|---|---|---|---|---|---|
| P value | HR | 95%CI |
P value | HR | 95%CI |
|||
| lower | upper | lower | upper | |||||
| FAM111B expression | 0.017 | 2.36 | 1.16 | 4.79 | 0.004 | 2.56 | 1.35 | 4.86 |
| Histology | 0.331 | 0.88 | 0.69 | 1.13 | 0.260 | 0.89 | 0.73 | 1.09 |
| TNM stage | 0.000 | 11.63 | 4.23 | 32.02 | 0.000 | 5.54 | 3.25 | 9.45 |
FAM111B expression in GEO datasets is associated with clinicohistological features and poor prognosis
To verify our findings, we evaluated the expression of FAM111B in OC patients from four GEO datasets with different sets of prognostic variables for each dataset. For the GSE63885 dataset, high FAM111B protein expression was associated with OS and PFS (P = 0.0044; P = 0.0069) (Fig. 4A-B). Moreover, the FAM111B expression level was significantly higher for the PD (progression disease) group than for other chemotherapy response status groups, including SD (stable disease), PR (partial response), and CR (complete response) after the first-line chemotherapy (P = 0.014) (Fig. 4C). For the GSE73614 dataset, there was a significant difference in FAM111B expression in serous carcinoma, endometrioid carcinoma, and clear cell carcinoma group (P = 1.1e-0.6) (Fig. 4D), and for the GSE39024 dataset, FAM111B expression was higher in patients whose disease was accompanied with ascites than those without ascites (P = 0.028) (Fig. 4E). Finally, for the GSE9891 dataset, FAM111B expression was significantly higher for the malignant ovarian tumor group than for the low malignant potential (LMP) group (P = 0.011) (Fig. 4F) and was positively correlated with the OC stage (P = 0.035) (Fig. 4G). These results support the findings from our TMAs and further suggest that FAM111B expression may indicate aggressive OC and poor prognosis.
Fig. 4.
FAM111B expression is associated with clinicohistological parameters and survival of OC patients in GEO datasets. (A) Relationship of FAM111B expression and OS in GSE63885. (B) Relationship of FAM111B expression and PFS in GSE63885. (C) Expression of FAM111B according to clinical status after first-line chemotherapy in GSE63885 (SD: stable disease, PR: partial response, CR: complete response, PD: progressive disease). (D) Expression of FAM111B in clear cell carcinoma, endometrioid carcinoma, and serous carcinoma in GSE73614. (E) Expression of FAM111B in OC patients with ascities and non-ascities in GSE39024. (F) Expression of FAM111B in low malignant potential (LMP) and malignant OC in GSE9891. (G) Expression of FAM111B in Grade I, Grade II, Grade III in GSE9891.
Construction of a PPI network for FAM111B
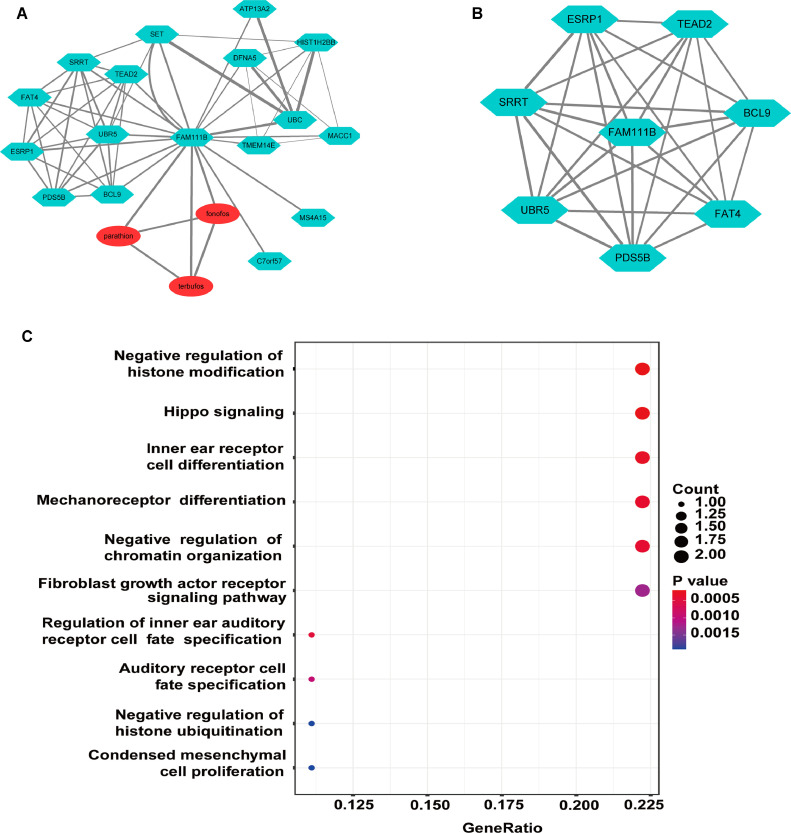
To explore potential mechanisms for FAM111B in OC, we used the STRING and STITCH databases to construct a PPI and chemical association network, which revealed 17 FAM111B-related genes and 3 FAM111B-related chemical compounds (Fig. 5A). Analysis of the core sub-network showed that FAM111B is a hub that is directly associated with its related genes (Fig. 5B). The 17 genes that were co-expressed with FAM111B were analyzed by gene-ontology enrichment annotation, which revealed potential functions in “negative regulation of histone modification”, “hippo signaling”, “inner ear receptor cell differentiation”, “mechanoreceptor differentiation” and “negative regulation of chromatin organization” (Fig. 5C). These results provide insight into potential functions for FAM111B in mediating aggressive OC cancer types.
Fig. 5.
Identification and gene ontology of a PPI and chemical compound network for FAM111B. (A) PPI and chemical association network of FAM111B for the top 17 most highly related genes and 3 related chemical compounds. (B) Sub-network analysis of the PPI network by MCODE using Cytoscape. (C) Gene-ontology biological process terms associated with genes in the FAM111B PPI network.
Discussion
Though FAM111B has a well-established role in hereditary fibrosing poikiloderma in exacerbating tendon contracture, myopathy, and pulmonary fibrosis [10], its role in cancer is still being explored. It has been associated with the occurrence and development of selected cancer types, suggesting that it may be a cancer-promoting gene [15], [16], [17]. Prior to this study, the association of FAM111B expression and its potential mechanisms in OC remained unknown. To our knowledge, our study is the first demonstration of a potential role for FAM111B in OC, including its prognostic value. We performed immunohistochemical analysis to investigate the expression of FAM111B in large cohort OC TMA samples and determined that FAM111B is expressed at higher levels in OC tumor tissues compared with para-carcinoma tissues, with expression both in the cytoplasm and nucleus. FAM111B expression was associated with the expression of the tumor marker proteins KI67, EGFR, and PDL-1. Furthermore, the FAM111B expression level predicted the prognosis of OC, with high FAM111B levels associated with a worse prognosis. These findings were supported by analysis of the relationship between FAM111B expression and clinicopathologic features of OC. Our results demonstrate that high FAM111B expression can predict aggressive clinicopathologic features in OC patients, including histological grade, tumor size. Moreover, these results were confirmed by bioinformatics analysis, which demonstrated that the expression level of FAM111B in OC has clinical prognostic significance for four independent datasets of in the GEO database. We further performed GO annotation and pathway enrichment analysis of FAM111B-related genes to explore its mechanism of action, which provides a foundation for exploring its potential molecular mechanisms. Therefore, these results demonstrate that high expression of FAM111B is correlated with aggressive phenotypes and poor prognosis of OC, suggesting that it may play an important role in the tumorigenesis and progression of OC.
Currently, FAM111B has been verified to be an oncogene associated for several malignancies. For instance, the expression of FAM111B is elevated in breast cancer cells, and patients with high FAM111B levels in tumor tissues tend to have worse prognosis [15]. Knockdown of FAM111B suppresses proliferation, migration, and invasion in breast cancer cells [15]. Furthermore, FAM111B is expressed at higher levels in LUAD tissues than in normal tissues, which correlates with the malignant progression of LUAD [18]. Consistently, knock down of FAM111B inhibited LUAD cell growth both in vitro and in vivo [18]. FAM111B is also highly expressed in high-risk pancreatic adenocarcinoma patients and is associated with the expression of cancer-promoting genes [16,19]. A mechanism of FAM111B in tumorigenesis and progression has recently been examined for LUAD, for which FAM111B was shown to degrade P16 and promote KRAS-driven enhancement of cyclin D1-CDK4 activity [17]. FAM111B also has been associated with cell cycle progression and proliferation mediated by the P53 signaling pathway [18], thus further supporting its potential oncogenic functions. In this study, we explored the role and mechanism of FAM111B in OC and confirmed that FAM111B acts as an oncogene, with high expression predicting aggressive clinicopathologic features and poor prognosis, which is consistent with the results of previous studies.
In the field of OC research, a number of prognostic factors have been explored. RAS protein activator like-1 (RASAL1), receptor-interacting protein kinase 4 (PIPK4), Jumonji domain-containing 6 (JMJD6), forkhead box protein M1 (FOXM1), Transmembrane protein with epidermal growth factor-like and two follistatin-like domains 1 (TMEFFI), and many other proteins have been shown to be expressed at higher levels in OC tissues than in normal tissues, with potential to predict a worse prognosis [20], [21], [22], [23]. However, no effective target-based agents against these oncogenes have been identified. As driver oncogenes, TP53 and KRAS mutations have also been significantly related to tumorigenesis and progression in OC [24,25]. Interestingly, knockdown of FAM111B in KRAS‐driven LUAD cells attenuates proliferation [17]. Furthermore, TP53 has been shown to directly target the transcriptional initiation region of FAM111B, thereby inhibiting its expression [18]. These results suggest that FAM111B may interact with TP53 and KRAS to modulate the functions of their associated genes in OC. Further investigation of FAM111B, especially with TP53 and KRAS mutations, may verify the role for FAM111B as a prognostic factor for OC patients and the potential roles of KRAS and TP53 in the pathogenesis of FAM111B in OC.
Interestingly, our results showed that the rates of both positive expression and high-level expression of FAM111B were lower in mucinous carcinoma compared with serous carcinoma, though there was no significant difference between the FAM111B expression patterns in serous carcinoma versus other types of OC. These results are consistent with a better prognosis for mucinous carcinoma than for serous carcinoma in early stage tumors. However, in advanced stage, the prognosis of mucinous carcinoma is poorer than in high grade serous carcinoma. Thus, whether the difference in FAM111B expression is due to difference in stage and grade, or to the nature of histological type, still requires a larger number of samples to validate. Our data also showed that high FAM111B expression levels may be associated with chemo-resistance, as represented by the progression disease status in patients receiving chemotherapy. Furthermore, FAM111B expression was positively correlated with the expression of three cancer-associated genes: EGFR, PDL1 and KI67. Notably, EGFR is a mediator of tumor neovascularization [26]. Therefore, future investigation to evaluate the correlation between FAM111B and tumor neovascularization may reveal potential benefits for combination treatment strategies that simultaneously target the anti-EGFR pathway and FAM111B. The positive correlation between the expression of FAM111B and PDL1, a potential and promising target for immunotherapy in ovarian cancer based on the preclinical and clinical data, further verifies the functional relevance of FAM111B in OC and could help in the design of novel precision medicine approaches [27], [28], [29], [30], [31]. A recent meta-analysis demonstrated that breast cancer patients with high expression of KI67 also tended to have a worse prognosis and be more sensitive to chemotherapy [32]. Thus, our results demonstrating the correlation between KI67 and FAM111B expression in OC suggest that higher FAM111B levels could indicate worse malignant biological behavior and poorer prognosis. Future research with larger numbers of patients may verify this possibility.
To further support a role for FAM111B in OC, we extended our data to include four datasets from the GEO database and performed network analysis. In the core sub-network, FAM111B related genes (SRRT, ESRP1, TEAD2, BCL9, FAT4, UBR5, and PDS5B) were identified. Notably, ESRP1, TEAD2, BCL9, FAT4, UBR5, and PDS5B have been associated with OC prognosis [33], [34], [35], [36], [37], [38], and ESRP1, TEAD2, BCL9, UBR5, and PDS5B have been shown to be highly expressed in OC and to be associated with poor prognosis [33,35,[37], [38], [39]]. Furthermore, FAT4 expression is lower in OC tissues than in normal tissues, which is associated with worse prognosis [36]. Interestingly, both TEAD2 and FAT4 are involved in hippo signaling, which regulates various cellular functions that are central to tumorigenesis [36,40,41]. These findings are consistent with the results of our GO annotation, though few studies have investigated roles for the above genes in the pathogenesis of OC. According to our GO annotation results, the genes that are co-expressed with FAM111B mainly participate in “negative regulation of histone modification”, “hippo signaling”, “inner ear receptor cell differentiation”, “chemoreceptor differentiation” and “negative regulation of chromatin organization”. Thus, these finding provide directions for future research on the mechanism of FAM111B in OC.
Conclusions
In conclusion, we demonstrated that high FAM111B expression is associated with aggressive clinicopathologic features and worse prognosis in OC. Our study indicates that FAM111B may serve as a prognostic predictor of OC, which provides insights for future research on the pathogenesis of OC. Based on our findings, the identification of compounds targeting FAM111B may provide a potential novel treatment strategy for improving OC outcomes.
CRediT authorship contribution statement
Fang Wei: Writing original draft, Methodology, Data Curation, Investigation. Guoyu Yu: Investigation, Data curation. Chaozeng Si: Investigation, Data Curation, Software, Visualization. Tengfei Chao and Huihua Xiong: Writing-Review & Editing. Lihong Zhang: Conceptualization, Writing - Review & Editing. All authors approved and contributed to the final manuscript.
Ethical approval statement
The study was approved by the Ethics Committee of Shanghai Outdo Biotech Co. Ltd. (No. YBM-05–02) and all the participants signed an informed consent form.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81803009) and the Chinese Society of Clinical Oncology (CSCO)-Research Fund (No. Y-BMS2019–011, No. Y-sy2018–039 and No. Y-L2018–003).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101659.
Appendix. Supplementary materials
References
- 1.Siegel R.L., et al. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Li X., Wang X. The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol. Cancer. 2017;16(1):92. doi: 10.1186/s12943-017-0659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lheureux S., Braunstein M., Oza A.M. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019;69(4):280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 4.Berek J.S., et al. Cancer of the ovary, fallopian tube, and peritoneum: 2021 update. Int. J. Gynaecol. Obstet. 2021;155(Suppl 1):61–85. doi: 10.1002/ijgo.13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong D.K., et al. Ovarian cancer, Version 2.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2021;19(2):191–226. doi: 10.6004/jnccn.2021.0007. [DOI] [PubMed] [Google Scholar]
- 6.Tokunaga H., et al. The 2020 Japan society of gynecologic oncology guidelines for the treatment of ovarian cancer, fallopian tube cancer, and primary peritoneal cancer. J. Gynecol. Oncol. 2021;32(2):e49. doi: 10.3802/jgo.2021.32.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman R.L., et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N. Engl. J. Med. 2019;381(25):2403–2415. doi: 10.1056/NEJMoa1909707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C., et al. Immunotherapy for ovarian cancer: adjuvant, combination, and neoadjuvant. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.577869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Malley D.M. New therapies for ovarian cancer. J. Natl. Compr. Canc. Netw. 2019;17(5.5):619–621. doi: 10.6004/jnccn.2019.5018. [DOI] [PubMed] [Google Scholar]
- 10.Mercier S., et al. Mutations in FAM111B cause hereditary fibrosing poikiloderma with tendon contracture, myopathy, and pulmonary fibrosis. Am. J. Hum. Genet. 2013;93(6):1100–1107. doi: 10.1016/j.ajhg.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goussot R., et al. Expanding phenotype of hereditary fibrosing poikiloderma with tendon contractures, myopathy, and pulmonary fibrosis caused by FAM111B mutations: report of an additional family raising the question of cancer predisposition and a short review of early-onset poikiloderma. JAAD Case Rep. 2017;3(2):143–150. doi: 10.1016/j.jdcr.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolland T., et al. A proteome-scale map of the human interactome network. Cell. 2014;159(5):1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclerc-Mercier S., et al. Skin biopsy is helpful in the diagnosis of hereditary fibrosing POIKiloderma with tendon contractures, myopathy and pulmonary fibrosis, due to FAM111B mutation. J. Eur. Acad. Dermatol. Venereol. 2022 doi: 10.1111/jdv.17937. [DOI] [PubMed] [Google Scholar]
- 14.Seo A., et al. FAM111B Mutation Is Associated With Inherited Exocrine Pancreatic Dysfunction. Pancreas. 2016;45(6):858–862. doi: 10.1097/mpa.0000000000000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., et al. YY1-induced transcriptional activation of FAM111B contributes to the malignancy of breast cancer. Clin. Breast Cancer. 2021 doi: 10.1016/j.clbc.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Mercier S., et al. FAM111B mutation is associated with pancreatic cancer predisposition. Pancreas. 2019;48(5):e41–e42. doi: 10.1097/mpa.0000000000001303. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki K., et al. FAM111B enhances proliferation of KRAS-driven lung adenocarcinoma by degrading p16. Cancer Sci. 2020;111(7):2635–2646. doi: 10.1111/cas.14483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun H., et al. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. Onco. Targets Ther. 2019;12:2829–2842. doi: 10.2147/ott.S190934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z., et al. Novel necroptosis-related gene signature for predicting the prognosis of pancreatic adenocarcinoma. Aging (Albany NY) 2022;14(2):869–891. doi: 10.18632/aging.203846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang R.X., et al. Overexpression of RASAL1 indicates poor prognosis and promotes invasion of ovarian cancer. Open Life Sci. 2019;14:133–140. doi: 10.1515/biol-2019-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S., et al. Overexpression of RIPK4 predicts poor prognosis and promotes metastasis in ovarian cancer. Biomed. Res. Int. 2021;2021 doi: 10.1155/2021/6622439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H., et al. Jumonji domain-containing 6 (JMJD6) identified as a potential therapeutic target in ovarian cancer. Signal Transduct. Target Ther. 2019;4:24. doi: 10.1038/s41392-019-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie X., et al. TMEFF1 overexpression and its mechanism for tumor promotion in ovarian cancer. Cancer Manag Res. 2019;11:839–855. doi: 10.2147/cmar.S186080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umemura S., et al. Synergistic effect of the inhibitors of RAF/MEK and AXL on KRAS-mutated ovarian cancer cells with high AXL expression. Cancer Sci. 2020;111(6):2052–2061. doi: 10.1111/cas.14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ignacio R.M.C., et al. Chemokine network and overall survival in TP53 wild-type and mutant ovarian cancer. Immune Netw. 2018;18(4):e29. doi: 10.4110/in.2018.18.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C., et al. CYP4X1 inhibition by flavonoid CH625 normalizes glioma vasculature through reprogramming TAMs via CB2 and EGFR-STAT3 axis. J. Pharmacol. Exp. Ther. 2018;365(1):72–83. doi: 10.1124/jpet.117.247130. [DOI] [PubMed] [Google Scholar]
- 27.Morand S., et al. Ovarian cancer immunotherapy and personalized medicine. Int. J. Mol. Sci. 2021;22(12) doi: 10.3390/ijms22126532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao J.B., et al. Pembrolizumab with low-dose carboplatin for recurrent platinum-resistant ovarian, fallopian tube, and primary peritoneal cancer: survival and immune correlates. J. Immunother. Cancer. 2021;9(9) doi: 10.1136/jitc-2021-003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding L., et al. PARP inhibition elicits STING-dependent antitumor immunity in brca1-deficient ovarian cancer. Cell Rep. 2018;25(11):2972–2980. doi: 10.1016/j.celrep.2018.11.054. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z., et al. Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci. Rep. 2019;9(1):1853. doi: 10.1038/s41598-019-38534-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domchek S.M., et al. Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): an open-label, multicentre, phase 1/2, basket study. Lancet Oncol. 2020;21(9):1155–1164. doi: 10.1016/s1470-2045(20)30324-7. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen T.O., et al. Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in breast cancer working group. J. Natl. Cancer Inst. 2021;113(7):808–819. doi: 10.1093/jnci/djaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong H.M., et al. ESRP1 is overexpressed in ovarian cancer and promotes switching from mesenchymal to epithelial phenotype in ovarian cancer cells. Oncogenesis. 2017;6(10):e389. doi: 10.1038/oncsis.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landin-Malt A., et al. An evolutionary, structural and functional overview of the mammalian TEAD1 and TEAD2 transcription factors. Gene. 2016;591(1):292–303. doi: 10.1016/j.gene.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., et al. Low BCL9 expression inhibited ovarian epithelial malignant tumor progression by decreasing proliferation, migration, and increasing apoptosis to cancer cells. Cancer Cell Int. 2019;19:330. doi: 10.1186/s12935-019-1009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malgundkar S.H., et al. FAT4 silencing promotes epithelial-to-mesenchymal transition and invasion via regulation of YAP and β-catenin activity in ovarian cancer. BMC Cancer. 2020;20(1):374. doi: 10.1186/s12885-020-06900-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song M., et al. Tumor derived UBR5 promotes ovarian cancer growth and metastasis through inducing immunosuppressive macrophages. Nat. Commun. 2020;11(1):6298. doi: 10.1038/s41467-020-20140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couturier A.M., et al. Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic. Acids. Res. 2016;44(22):10879–10897. doi: 10.1093/nar/gkw921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia Y., et al. YAP/TEAD co-activator regulated pluripotency and chemoresistance in ovarian cancer initiated cells. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0109575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dey A., Varelas X., Guan K.L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020;19(7):480–494. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z., et al. Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides. FASEB J. 2015;29(2):724–732. doi: 10.1096/fj.14-262980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.