Abstract
Background.
Posttraumatic Stress Disorder (PTSD) tends to co-occur with greater alcohol consumption as well as alcohol use disorder (AUD). However, it is unknown whether the same etiologic factors that underlie PTSD-alcohol-related problems comorbidity also contribute to PTSD-alcohol consumption.
Methods.
We used summary statistics from large-scale genome-wide association studies (GWAS) of European-ancestry (EA) and African-ancestry (AA) participants to estimate genetic correlations between PTSD and a range of alcohol consumption-related and alcohol-related problems phenotypes.
Results.
In EAs, there were positive genetic correlations between PTSD phenotypes and alcohol-related problems phenotypes (e.g. Alcohol Use Disorders Identification Test (AUDIT) problem score) (rGs: 0.132−0.533, all FDR adjusted p < 0.05). However, the genetic correlations between PTSD phenotypes and alcohol consumption -related phenotypes (e.g. drinks per week) were negatively associated or non-significant (rGs: −0.417 to −0.042, FDR adjusted p: <0.05-NS). For AAs, the direction of correlations was sometimes consistent and sometimes inconsistent with that in EAs, and the ranges were larger (rGs for alcohol-related problems: −0.275 to 0.266, FDR adjusted p: NS, alcohol consumption-related: 0.145–0.699, FDR adjusted p: NS).
Conclusions.
These findings illustrate that the genetic associations between consumption and problem alcohol phenotypes and PTSD differ in both strength and direction. Thus, the genetic factors that may lead someone to develop PTSD and high levels of alcohol consumption are not the same as those that lead someone to develop PTSD and alcohol-related problems. Discussion around needing improved methods to better estimate heritabilities and genetic correlations in diverse and admixed ancestry samples is provided.
Keywords: Alcohol consumption, alcohol use disorder, genetic correlation, posttraumatic stress disorder
Introduction
Traumatic events are common, with 50–70% of individuals experiencing at least one trauma in their lifetimes (Benjet et al., 2016). Posttraumatic Stress Disorder (PTSD), the signature trauma-related disorder (Breslau, 2009), is associated with increased alcohol consumption (Vlahov et al., 2002) and alcohol use disorder (AUD) (Jakupcak et al., 2010). Twin studies (Heath, Jardine, & Martin, 1989; Kaprio et al., 1987; Knopik et al., 2004; Stein et al., 2002) and genome-wide association studies (GWAS) (Clarke et al., 2017; Stein et al., 2016) find that PTSD and these alcohol phenotypes are moderately heritable, with 36–60% of the variance explained by genetic effects. Additional work using twin studies has demonstrated 30% genetic overlap between PTSD and AUD (McLeod et al., 2001; Xian et al., 2000). In general, most of the comorbidity research has focused on PTSD-AUD, and has neglected the association between PTSD-alcohol consumption. As increased alcohol consumption is associated with AUD (Moos, Schutte, Brennan, & Moos, 2004; Sanchez-Roige et al., 2019b), genetic research is needed to test whether the same genetic influences underlying PTSD-AUD are those underlying PTSD and alcohol consumption.
Large scale GWAS have identified significant hits for PTSD phenotypes (e.g. Nievergelt et al., 2019), problematic alcohol use (PAU) (Zhou et al., 2020a), (e.g. alcohol dependence; Walters et al., 2018), and alcohol consumption (Clarke et al. 2017). Recent analyses have allowed for examination of genetic associations across alcohol consumption (Kranzler et al., 2019; Liu et al., 2019; Sanchez-Roige et al., 2019b), PAU (Gelernter et al., 2019a; Sanchez-Roige et al., 2019a; Walters et al., 2018; Zhou et al., 2020b), as well as PTSD and re-experiencing symptoms (Gelernter et al., 2019b; Nievergelt et al., 2019). Single nucleotide polymorphism (SNP) -based heritability of PTSD suggests modest to moderate heritability (∼15%), with these estimates larger in women than men (i.e. 36% v. 5%; Duncan et al., 2017). SNP-based heritability of problem alcohol phenotypes suggests modest heritability, ranging from 5.6–9.4% for AD (h2 = 0.090, s.e. = 0.019; Walters et al., 2018), AUD (Kranzler et al. 2019: h2 = 0.056, s.e. = 0.004; Zhou et al. 2020a: h2 = 0.094, s.e. = 0.005), and PAU (h2 = 0.068, S.E. = 0.004; Zhou et al., 2020a).
Few have investigated the genetic association between PTSD and alcohol-related problems phenotypes using genetic techniques such as linkage disequilibrium score regression (Bulik-Sullivan et al., 2015b), but work by our group found a significant correlation between PTSD and alcohol dependence (AD; rG = 0.35; Sheerin et al., 2020) (rG = 0.28; Bountress et al., 2021) for those of European Ancestry. However, this effect was driven by women, for whom the genetic correlation was moderate and significant, but not for men (Sheerin et al., 2020). Genetic correlation analyses between PTSD and alcohol consumption were also conducted by our group, finding a non-significant association (rG = −0.07; Bountress et al., 2021); another group found near zero genetic correlation between PTSD and the Alcohol Use Disorders Identification Test (AUDIT) consumption score subscale (AUDIT-C) (Mallard et al., 2021). Work by our group also found that beyond genetic correlations, using Mendelian Randomization, PTSD exerted a causal effect on AUD, but not alcohol consumption, but that neither alcohol phenotype exerted a causal influence on PTSD (Bountress et al., 2021). Additionally, genetic correlations between consumption and problems phenotypes vary. One group found correlations between alcohol consumption and AUD ranging from small to moderate (e.g. ∼rG = 0.2–0.3; Sanchez-Roige et al., 2019b). Another found large associations between AUDIT-C and AUDIT-P and AD (∼ rG = 0.70; Mallard et al., 2021) once the association between the frequency item and SES was taken into account. Together these findings suggest the genetic risk for consumption and problematic phenotypes are correlated but distinct.
The question of whether the genetic associations between PTSD and alcohol consumption differs compared to alcohol-related problems phenotypes has not been explicitly studied, to our knowledge. However, work on other psychiatric phenotypes suggests that for some, like smoking behaviors, the genetic correlations between mild and more problematic versions of the phenotype [e.g. cigarettes per day (CPD), nicotine dependence (ND)] are strongly positively correlated with each other (rG = 0.95; Quach et al., 2020). Additionally, their correlations with other disorders (e.g. schizophrenia) are in the same direction (e.g. both positive) but of varying sizes (Hartz et al., 2018). Research on major depression, which is closely related to PTSD, found positive genetic correlations between major depression and AD and alcohol quantity, but negative genetic correlations between major depression and alcohol frequency (Polimanti et al., 2019). Thus, we aim to test whether using alcohol consumption -related phenotypes yields similar estimates to problem alcohol-related phenotypes.
The current study adds to this literature by estimating genetic correlations from GWASs summary statistics for PTSD (as well as re-experiencing symptoms), and a range of alcohol phenotypes. The latter include drinks per week (DPW), AUDIT-C (alcohol frequency, quantity, and frequency of 6 + drinks), problems (P) score from the AUDIT (AUDIT-P; including 7 items assessing problems; e.g. unable to stop drinking once you started), as well as total score (AUDIT-T; comprised of AUDIT-C and -P), maximum alcohol intake (typical habitual daily maximum usage), AUD (using DSM-5 diagnosis; American Psychiatric Association, 2013) and AD (using DSM-IV diagnosis; American Psychiatric Association, 1994). In so doing, it adds to previous work by examining not only the genetic association between PTSD and alcohol-related problems outcomes, but also PTSD and other alcohol consumption phenotypes including more normative use, which has been generally neglected in the PTSD-alcohol comorbidity literature with few exceptions (Mallard et al., 2021). Finally, this study attempts to examine whether findings are consistent between those of European Ancestry (EA) and African Ancestry (AA) individuals – the latter of which is particularly important given the lack of diversity in genomic studies (Bentley, Callier, & Rotimi, 2017; Peterson et al., 2019; Sirugo, Williams, & Tishkoff, 2019). This study leverages large-scale GWASs summary statistics from a number of consortia [i.e. Psychiatric Genomics Consortium (PGC)-PTSD and Substance Use Disorder (SUD)-AD, United Kingdom Biobank (UKB), Million Veterans Program (MVP), 23andMe, and GWAS & Sequencing Consortia of Alcohol and Nicotine Use (GSCAN)].
Methods
Samples
PTSD samples and phenotypes
PTSD case/control status came from the PGC-PTSD Freeze 2 dataset (PTSD), which consists of over 50 separate datasets plus the UKB (Nievergelt et al., 2019). In analyses utilizing alcohol use data from the UKB, PGC-PTSD PTSD case status reflects primarily lifetime PTSD diagnosis, but also includes current diagnosis when lifetime was not available (30 of 57 cohorts in Freeze 2 provided lifetime data). PGC-PTSD case/control status data were available for both EA and AA samples. PGC-PTSD Freeze 2 were used instead of Freeze 1.5 because of the increase in sample size and inclusion of AA individuals (EA: Total PTSDf1.5 N = 48 471; PTSDf2.0 N = 174 659; AA: Total PTSDf2.0 N = 15 339) (Nievergelt et al., 2019).
Two PTSD-related variables were used: DSM-based PTSD and a PTSD re-experiencing score. PTSD re-experiencing symptoms (PTSD Re-Exp) came from an assessment of the PTSD Checklist (PCL) DSM-IV (American Psychiatric Association, 1994) version (Wilkins, Lang, & Norman, 2011) in the MVP (Gelernter et al., 2019b), selected as it is the symptom cluster most distinctive for PTSD compared to other disorders. This sample and phenotype contributed to both EA (N = 146 660) and AA (N = 19 983) analyses.
Alcohol samples and phenotypes
AUD and AD GWAS summary statistics were available in two datasets. AD case/control data came from the PGC-SUD (Walters et al., 2018). Cases were defined as meeting criteria for a DSM-IV (American Psychiatric Association, 1994) [DSM-III-R (American Psychiatric Association, 1987) for one study] diagnosis of AD and all controls were alcohol exposed. The PGC-SUD AD phenotype contributed to both EA (N = 46 568) and AA (N = 6280) analyses. AUD case/control status was used from the MVP dataset, defined as ICD-9 or ICD-10 codes for dependence or abuse as obtained from the Veteran’s Affairs electronic health records (EHR); participants with at least one inpatient or two outpatient alcohol-related ICD-9/10 codes (from 2000–2018) were considered AUD cases (Kranzler et al., 2019). AUD case/control status in MVP is available for EA (N = 267 391) and AA (N = 56 648) samples.
Alcohol consumption-related GWASs summary statistics were available for a number of phenotypes. Specifically, a measure of average DPW came from the GSCAN consortium and the UKB (Liu et al., 2019) available in EA samples only. DPW was defined as the average number of drinks a participant reported drinking each week, aggregated across all types of alcohol. In studies that reported binned response ranges (e.g. 1–4 drinks), the midpoint of the range was used (Liu et al., 2019). Summary statistics for DPW within UKB and GSCAN were examined combined (N = 941 280) as well as separately (GSCAN: N = 526 937; UKB: N = 414 343). The AUDIT (Saunders, Aasland, Babor, De la Fuente, & Grant, 1993) was available in multiple forms and studies. General consumption was measured using the AUDIT-C subscale, which consists of the first three items of the AUDIT and measures past-year typical quantity and frequency of drinking as well as one item measuring frequency of heavy/binge drinking (Bush, Kivlahan, McDonell, Fihn, & Bradley, 1998). AUDIT-C data were available in two datasets, from the EHR data of the annual AUDIT-C assessment in MVP from 2007–2017 (Kranzler et al., 2019) and as part of the full AUDIT assessment in an online follow-up of the UKB (Sanchez-Roige et al., 2019a). AUDIT-C data were available in MVP for both EA (206 254) and AA (56 495) ancestries and EA only in UKB (N = 121 604). The full AUDIT score (i.e. AUDIT-T) was also available in the 23andMe and UKB datasets (Sanchez-Roige et al., 2019b) in EA samples (23andMe: N = 20 328; UKB: N = 121 604). The AUDIT-P scale, the score on items 4–10 of the AUDIT, which focuses on the problematic consequences of drinking, was used from the UKB in EA samples (N = 121 604). Finally, in the MVP data, a quantitative measure of maximum habitual alcohol consumption in a typical month (Max. Alc.; Gelernter et al., 2019a) was used as a measure of more problematic consumption, to reflect typical/habitual maximum usage as opposed to maximum on a single occasion (N = 126 936 for EA, and N = 17 029 for AA).
Case/control designs
Unbalanced ascertainment in case/control designs can introduce bias in studies using meta-analytic data. Effective sample sizes (Neffs) can help to reduce potential bias in this situation. In this study, we used to calculate the Neff for each phenotype, and used the per-SNP Neffs when available in the summary statistics (e.g. PTSD). This approach takes in account the impact of potential bias and reduced power introduced by unbalanced ascertainment of the number of cases and controls across cohorts analyzed under a liability scale (e.g. Walters et al., 2018; Zhou et al., 2020a).
Genotyping, quality control, and imputation
The existing summary statistics used in the present analyses have previously gone through quality-control pipelines used for the specific consortia (e.g. PGC quality control pipeline including filtering to remove SNPs with imputation information value < 0.90 and MAF < 0.01; Sullivan, 2010). The analytic pipeline for the present analyses incorporates further filtering processes including removal of SNPs based on a minimum Neff and per-SNP sample variation (e.g. SNP filtering keeping variants within at least 80% of the total Neff) and variants that are either not SNPs or are strand-ambiguous.
SNP heritability and genetic correlation analysis
Analyses of SNP-based heritability and genetic correlation (rG) were conducted using the cross-trait linkage disequilibrium (LD) score regression approach and LD score regression software (LDSC) (Bulik-Sullivan et al., 2015b; open-source LDSC pipeline, version 1.0.1, github.com/bulik/ldsc) which requires GWAS summary statistics in samples of unrelated individuals. The approach estimates rGs by replacing the χ2 with the z scores from both studies and the genetic covariance is then estimated using the slope from the regression of both z scores on LD scores. Normalizing genetic covariance by yields the genetic correlation. Multiple testing was adjusted using false discovery rate (FDR) correction.
Because LDSC requires single ancestry summary statistics as input, analyses were conducted separately for EA and AA samples (see online Supplementary Table S1). For the EA samples, pre-computed LD scores came from the 1000 Genomes Project Europeans (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2). For the AA samples, the AA specific LD scores (subset under UKBB.AFR prefix) from the UKB pan-ancestry LD scores (Pan-UKB team; https://pan.ukbb.broad-institute.org, 2020) were used.
Tissue enrichment analysis
At the tissue level, data from 53 human tissues [Genotype-Tissue Expression (GTEx; https://gtexportal.org/home/) project, version 7] (Lonsdale et al., 2013) were used. Post hoc analyses were performed using the partitioned LD score regression software at the default settings (tissues were provided to GTEx by LDSC software).
Results
Heritability
Liability scale statistics were estimated for case/control phenotypes (i.e. PTSDf2, AUD and AD) and observed scale statistics were estimated for the non-case/control phenotypes, in EA and AA summary statistics. Table 1 shows the computed estimates for those of EA and AA from GWASs used for the analyses below. Notably, prior work suggests a general Z score cut off of 4 for heritabilities to determine that traits are appropriate for genetic correlation (Bulik-Sullivan et al., 2015a). Estimates within Table 1 with Z scores below 4 are shaded, and should be interpreted with caution. Online Supplementary Fig. S1 depicts the h2 SNP Z scores for all included samples. All but one of the Z scores in EA samples is above 4. In contrast, only 1 of the Z scores is greater than 4 in AA samples suggesting these estimates are not robust.
Table 1.
Computed SNP-based Heritability of PTSD and Alcohol Phenotypes – EA and AA
| Phenotype | Data Source (n) | Scale | Sample Prev. | Population Prev. | s.e. | 95% CI LB | 95% CI UB | Z | p value | FDR-adjusted p value | Estimate from Original Publication European Ancestry (EA) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European Ancestry (EA) | ||||||||||||
| DPW | GSCAN and UKB (n = 941 280) | Obs. | – | – | 0.048 | 0.002 | 0.044 | 0.052 | 24.00 | 2.78 × 10−127 | 5.28 × 10−126* | Not Reported ((Liu et al., 2019) |
| DPW | GSCAN (n = 526 937) | Obs. | – | – | 0.040 | 0.003 | 0.034 | 0.046 | 13.33 | 1.48 × 10−40* | 4.02 × 10−40* | 0.042 [0.002] (Liu et al., 2019) |
| DPW | UKB (n = 414 343) | Obs. | – | – | 0.069 | 0.003 | 0.058 | 0.074 | 16.50 | 3.67 × 10−61* | 2.32 × 10−60* | 0.74 [0.009], 0.79 [0.004](Liu et al., 2019) |
| AUDIT-C | MVP (n = 206 254) | Obs. | – | – | 0.068 | 0.005 | 0.058 | 0.078 | 13.60 | 4.00 × 10−42* | 1.27 × 10−41* | 0.068 [0.005] (Kranzler et al., 2019) |
| AUDIT-C | UKB (n = 121 604) | Obs. | – | – | 0.084 | 0.006 | 0.072 | 0.096 | 14.00 | 1.56 × 10−44* | 7.40 × 10−44* | 0.084 [0.0055] (Sanchez-Roige et al., 2019) |
| AUDIT-T | 23andMe (n = 20 328) | Obs. | – | – | 0.089 | 0.025 | 0.040 | 0.138 | 3.56 | 3.71 × 10−4* | 4.70 × 10−4* | Not reported (Sanchez-Roige et al., 2019) |
| AUDIT-T | UKB (n = 121 604) | Obs. | – | – | 0.059* | 0.005 | 0.049 | 0.069 | 11.80 | 3.90 × 10−32* | 7.42 × 10−32* | 0.086 [0.005] (Sanchez-Roige et al., 2019) |
| Max. Alc. | MVP (n = 126 938) | Obs. | – | – | 0.078 | 0.006 | 0.066 | 0.090 | 13.00 | 1.22 × 10−38* | 2.91 × 10−38* | 0.078 [(Gelernter et al., 2019a) |
| AUDIT-P | UKB (n = 121 604) | Obs. | – | – | 0.059 | 0.005 | 0.049 | 0.069 | 11.80 | 3.90 × 10−32* | 7.42 × 10−32* | 0.059 [0.048] (Sanchez-Roige et al., 2019) |
| AUD | MVP1n2 (n = 267 391) | Liab. | 0.183 | 0.159 | 0.108* | 0.006 | 0.096 | 0.120 | 18.00 | 1.95 × 10−72* | 1.85 × 10−71* | 0.056 [0.004] (Kranzler et al., 2019) |
| AD | PGC (n = 46 568) | Liab. | 0.248 | 0.159 | 0.093 | 0.021 | 0.052 | 0.134 | 4.43 | 9.49 × 10−6* | 1.50 × 10−5* | 0.090 [0.019] (Walters et al., 2018) |
| PTSD Re-Exp | MVP (n = 146 660) | Obs. | – | – | 0.068 | 0.005 | 0.058 | 0.078 | 13.60 | 4.00 × 10−42* | 1.27 × 10−41* | 0.067[S.E.:.005] (Gelernter et al., 2019b) |
| PTSD | PGC (n = 174 659) | Liab. | 0.157 | 0.100 | 0.082* | 0.015 | 0.053 | 0.111 | 5.47 | 4.59 × 10−8* | 7.92 × 10−8* | 0.04 [95% CI 0.02–0.05] (Nievergelt et al., 2019) |
| African Ancestry (AA) | ||||||||||||
| AUDIT-C | MVP (n = 56 495) | Obs. | – | – | 0.061 | 0.016 | 0.030 | 0.092 | 3.81 | 1.38 × 10−4* | 1.87 × 10−4* | 0.062 [0.016] (Kranzler et al., 2019) |
| Max. Alc. | MVP (n = 17 029) | Obs. | – | – | 0.086 | 0.041 | 0.006 | 0.166 | 2.10 | 3.59 × 10−2* | 4.02 × 10−2* | Not Reported (Gelernter et al., 2019a) |
| AUD | MVP (n = 56 648) | Liab. | 0.305 | 0.159 | 0.136 | 0.033 | 0.071 | 0.201 | 4.12 | 3.77 × 10−5* | 5.51 × 10−5* | 0.100 [0.022] (Kranzler et al., 2019) |
| AD | PGC (n = 6280) | Liab. | 0.531 | 0.111 | 0.277 | 0.164 | −0.044 | 0.598 | 1.69 | 9.12 × 10−2 | 9.63 × 10−2 | Unstable/Not Reported (Walters et al., 2018) |
| PTSD Re-Exp | MVP (n = 19 983) | Obs. | – | – | 0.067 | 0.041 | −0.013 | 0.147 | 1.63 | 1.02 × 10−1 | 1.02 × 10−1 | 0.048 [s.e.:0.039] (Gelernter et al., 2019b) |
| PTSD | PGC (n = 15 339) | Liab. | 0.284 | 0.100 | 0.193* | 0.072 | 0.052 | 0.334 | 2.68 | 7.35 × 10−3* | 8.73 × 10−3* | 0.02 [95%CI−0.04 to 0.09] (Nievergelt et al., 2019; note: this was using GCTA not LDSC) |
Note: SNP, Single Nucleotide Polymorphism; PTSD, Posttraumatic Stress Disorder; EA, European ancestry; AA, African ancestry; h2, Heritability; S.E., standard error; CIs, confidence intervals; LB, lower bound; UB, upper bound; Z, z-score; Re-Exp, reexperiencing; MVP, Million Veteran Program; AD, Alcohol Dependence; AUD, Alcohol Use Disorder; Max. Alc., Maximum Alcohol Intake; AUDIT, Alcohol Use Disorder Identification Test; T, total; P, problems; C, consumption; DPW, Drinks per Week; UKB, UK Biobank; GSCAN, GWAS & Sequencing Consortium of Alcohol and Nicotine Use; PGC, Psychiatric Genomics Consortium; Liab., Liability; Obs., Observed; Prev., Prevalence. Sample and population prevalence were used for analyses including binary traits to correct for ascertainment bias. estimates in this study were computed using LDSC software and the corresponding GWAS summary statistics. For estimates from the original publications, S.E.s are included when provided in text and when unavailable, 95% CIs were included. When neither was provided in the original manuscript, neither is presented in the table. Four estimates (noted with an asterisk) are different than those from the original publications. The differences are likely due to additional filtering (see Method section), in addition to effective sample sizes calculations (see Method section) that take in account differences in ascertainment of unbalanced cohorts in the case of the three estimates under the liability scale with an asterisk. Estimates of with z-scores less than 4 are noted with shading. Unadjusted and false-discovery rate (FDR) adjusted p values < 0.05 are noted by one asterisk on their corresponding columns.
Genetic correlations
We estimated the rG of PTSD phenotypes [i.e. PTSD (PGC), and PTSD Re-Exp (MVP)] with DPW (from the GSCAN, UKB and both combined), AUDIT-C (from the MVP and UKB), and AUDIT-T (from 23andMe and UKB) (see Table 2 and Fig. 1). We also estimated rG with Max. Alc. (from the MVP), AUDIT-P (from the UKB), AUD (from the MVP) and AD (from the PGC) in individuals from EA. Similarly, we estimated the rG of PTSD and PTSD Re-Exp with AUDIT-C (from the MVP), Max Alc (from the MVP), AUD (from the MVP), AD (from the PGC) phenotypes in AA samples (see Table 3 and Fig. 2).
Table 2.
Genetic Correlations of PTSD and Alcohol Phenotypes in people of EA ancestry
| PTSD Phen. | PTSD Phen. Data Source | Alcohol Phen. | Alcohol Phen. Data Source | rG | s.e. | CI LB | CI UB | p value | FDR Adj. p value |
|---|---|---|---|---|---|---|---|---|---|
| PTSD Re-Exp | MVP | DPW | GSCAN | −0.094 | 0.051 | −0.194 | 0.007 | 0.067 | 0.094 |
| PTSD Re-Exp | MVP | DPW | GSCAN and UKB | −0.042 | 0.037 | −0.115 | 0.030 | 0.253 | 0.270 |
| PTSD Re-Exp | MVP | DPW | UKB | −0.084 | 0.039 | −0.160 | −0.007 | 0.033* | 0.055 |
| PTSD | PGC | DPW | GSCAN | −0.121 | 0.078 | −0.275 | 0.033 | 0.124 | 0.147 |
| PTSD | PGC | DPW | GSCAN and UKB | −0.073 | 0.054 | −0.179 | 0.033 | 0.176 | 0.194 |
| PTSD | PGC | DPW | UKB | −0.072 | 0.054 | −0.178 | 0.034 | 0.184 | 0.203 |
| PTSD Re-Exp | MVP | AUDIT-C | MVP | −0.241 | 0.044 | −0.326 | −0.155 | 3.68 × 10−8* | 3.93 × 10−7* |
| PTSD Re-Exp | MVP | AUDIT-C | UKB | −0.188 | 0.048 | −0.282 | −0.094 | 8.29 × 10−5* | 0.0003* |
| PTSD | PGC | AUDIT-C | MVP | −0.417 | 0.074 | −0.562 | −0.272 | 1.62 × 10−8* | 2.59 × 10−7* |
| PTSD | PGC | AUDIT-C | UKB | −0.225 | 0.068 | −0.358 | −0.092 | 9.00 × 10−4* | 0.003* |
| PTSD Re-Exp | MVP | AUDIT-T | 23andMe | −0.122 | 0.113 | −0.343 | 0.099 | 0.278 | 0.278 |
| PTSD Re-Exp | MVP | AUDIT-T | UKB | −0.132 | 0.047 | −0.224 | −0.039 | 0.005* | 0.013* |
| PTSD | PGC | AUDIT-T | 23andMe | −0.230 | 0.144 | −0.512 | 0.051 | 0.109 | 0.134 |
| PTSD | PGC | AUDIT-T | UKB | −0.111 | 0.065 | −0.238 | 0.016 | 0.087 | 0.111 |
| PTSD Re-Exp | MVP | Max. Alc. | MVP | 0.294 | 0.054 | 0.187 | 0.400 | 7.23 × 10−8* | 5.78 × 10−7* |
| PTSD | PGC | Max. Alc. | MVP | 0.294 | 0.087 | 0.124 | 0.464 | 7.00 × 10−4* | 0.002* |
| PTSD Re-Exp | MVP | AUDIT-P | UKB | 0.132 | 0.059 | 0.016 | 0.248 | 0.026* | 0.044* |
| PTSD | PGC | AUDIT-P | UKB | 0.374 | 0.092 | 0.195 | 0.554 | 4.32 × 10−5* | 0.0002* |
| PTSD Re-Exp | MVP | AUD | MVP | 0.311 | 0.044 | 0.224 | 0.399 | 2.70 × 10−12* | 8.64 × 10−11* |
| PTSD | PGC | AUD | MVP | 0.289 | 0.082 | 0.129 | 0.449 | 4 × 10−4* | 0.001* |
| PTSD Re-Exp | MVP | AD | PGC | 0.533 | 0.130 | 0.278 | 0.788 | 4.19 × 10−5* | 0.0002* |
| PTSD | PGC | AD | PGC | 0.472 | 0.152 | 0.173 | 0.770 | 0.002* | 0.005* |
Note: PTSD, Posttraumatic Stress Disorder; EA, European ancestry; Phe., phenotype; rG, genetic correlation; s.e., standard error; CIs, confidence intervals; LB, lower bound; UB, upper bound; Re-Exp, reexperiencing; MVP, Million Veteran Program; PGC, Psychiatric Genomics Consortium; AD, Alcohol Dependence; AUD, Alcohol Use Disorder, Max. Alc., Maximum Alcohol Intake, AUDIT, Alcohol Use Disorder Identification Test; T, total; P, problems; C, consumption; DPW, Drinks per Week; UKB, UK Biobank; GSCAN, GWAS & Sequencing Consortium of Alcohol and Nicotine Use. Unadjusted and false discovery rate (FDR) adjusted p values < 0.05 are noted by one asterisk on their corresponding columns
Fig. 1.
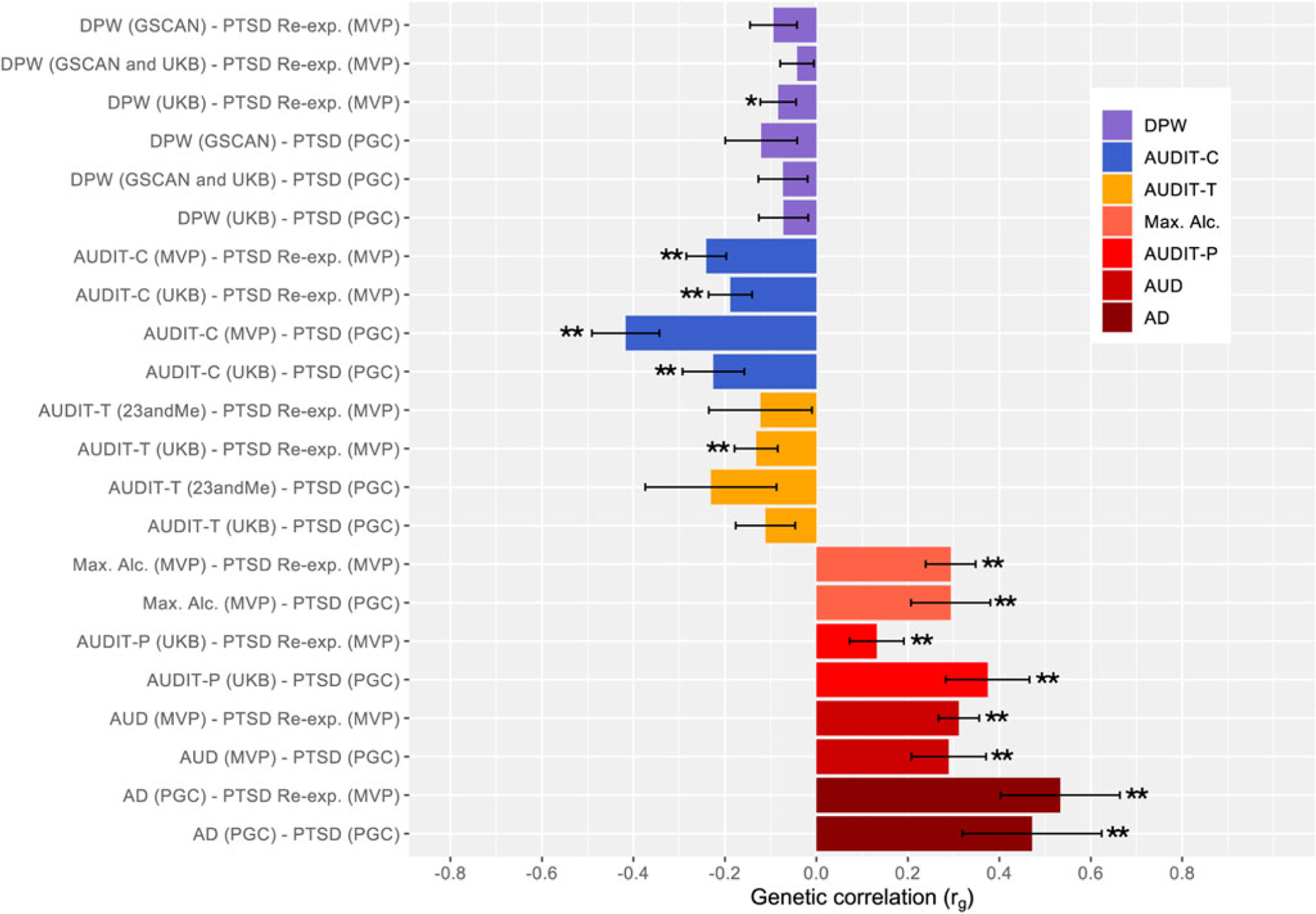
Genetic Correlations of PTSD and Alcohol Phenotypes (±s.e. bars) – EA
Note: Unadjusted significant p values ( p < 0.05) for rGs are noted with an asterisk. Those passing FDR adjustment are noted with an additional asterisks (total of two asterisks for those passing FDR adjustment). PTSD, Posttraumatic Stress Disorder; EA, European ancestry; s.e., standard error; DPW, Drinks per Week; GSCAN, GWAS & Sequencing Consortium of Alcohol and Nicotine Use; UKB, UK Biobank; AUDIT, Alcohol Use Disorder Identification Test; T, total; P, problems; C, consumption; Max. Alc., Maximum Alcohol Intake; AUD, Alcohol Use Disorder; AD, Alcohol Dependence; PGC, Psychiatric Genomics Consortium; Re-exp., reexperiencing; MVP, Million Veteran Program. Alcohol phenotypes are ordered from more typical to more problematic (top to bottom) and color coded by each type of phenotype (i.e., DPW, AUDIT-C, AUDIT-T, Max. Alc., AUDIT-P, AUD, AD) to draw attention to difference in findings.
Table 3.
Genetic Correlations of PTSD and Alcohol Phenotypes in people of AA ancestry
| PTSD Phe. | PTSD Phe. Data Source | Alcohol Phe. | Alcohol Phe. Data Source | rG | s.e. | CI LB | CI UB | p value | FDR Adj. p value |
|---|---|---|---|---|---|---|---|---|---|
| PTSD | PGC | AUDIT-C | MVP | 0.145 | 0.209 | −0.265 | 0.554 | 0.489 | 0.719 |
| PTSD Re-Exp | MVP | AUDIT-C | MVP | 0.699 | 0.328 | 0.057 | 1.340 | 0.033* | 0.264 |
| PTSD | PGC | Max. Alc. | MVP | −0.123 | 0.342 | −0.794 | 0.548 | 0.719 | 0.719 |
| PTSD Re-Exp | MVP | Max. Alc. | MVP | −0.275 | 0.504 | −1.260 | 0.713 | 0.586 | 0.719 |
| PTSD | PGC | AUD | MVP | 0.182 | 0.185 | −0.180 | 0.544 | 0.325 | 0.719 |
| PTSD Re-Exp | MVP | AUD | MVP | 0.266 | 0.270 | −0.263 | 0.796 | 0.324 | 0.719 |
| PTSD | PGC | AD | PGC | 0.233 | 0.305 | −0.365 | 0.832 | 0.445 | 0.719 |
| PTSD Re-Exp | MVP | AD | PGC | 0.195 | 0.522 | −0.829 | 1.220 | 0.709 | 0.719 |
Note: PTSD, Posttraumatic Stress Disorder; Phe., phenotype; rG, genetic correlation; s.e., standard error; CIs, confidence intervals; LB, lower bound; UB, upper bound; Re-Exp, reexperiencing; MVP, Million Veteran Program; PGC, Psychiatric Genomics Consortium; AD, Alcohol Dependence; AUD, Alcohol Use Disorder; Max. Alc., Maximum Alcohol Intake; AUDIT, Alcohol Use Disorder Identification Test; C, consumption. The p value < 0.05 is noted by one asterisk. None passed FDR adjustment.
Fig. 2.
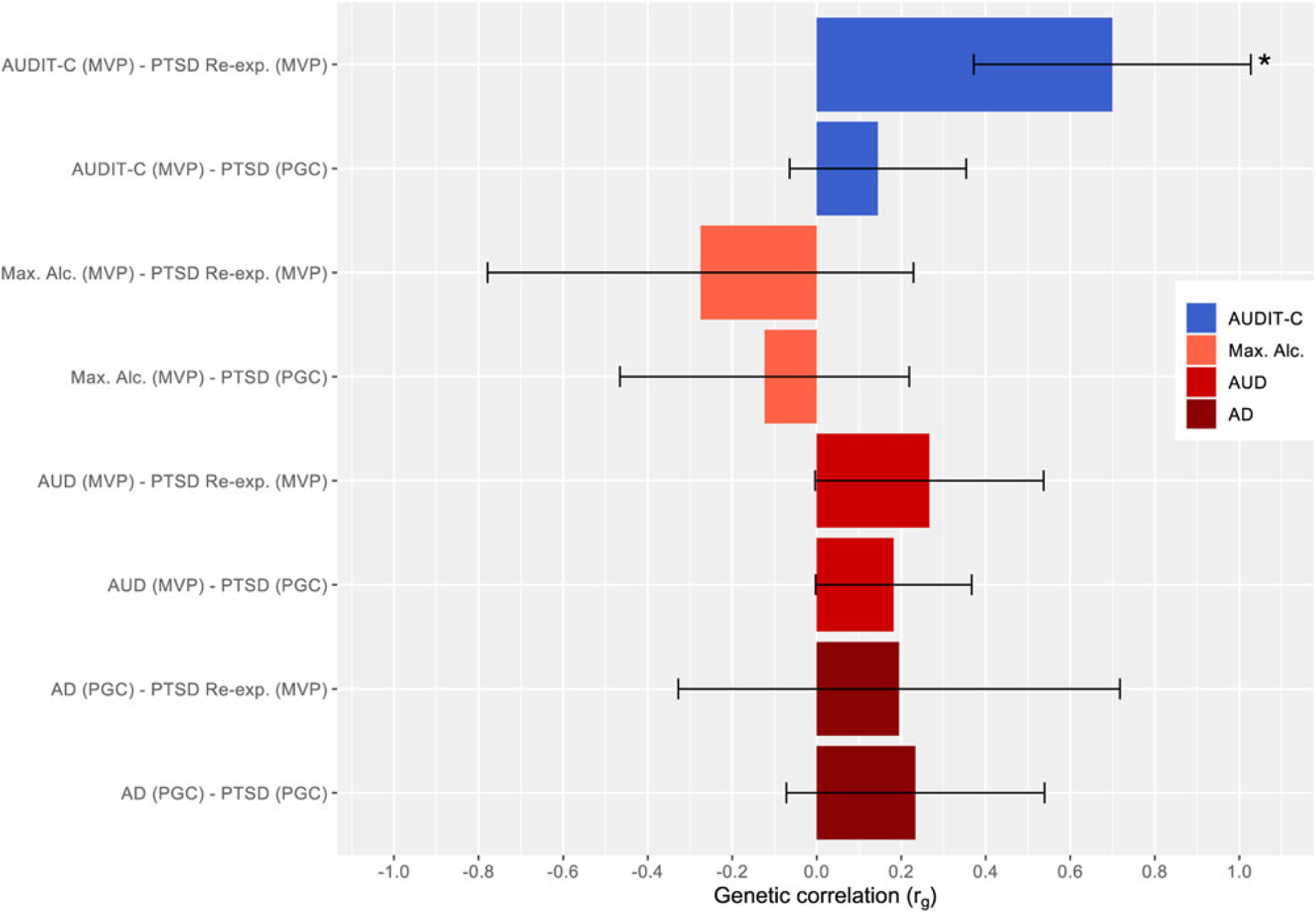
Genetic Correlations of PTSD and Alcohol Phenotypes (± s.e. bars) – AA.
Note: Unadjusted significant p values ( p < 0.05) for rGs are noted with an asterisk. Those passing FDR adjustment are noted with an additional asterisks (total of two asterisks for those passing FDR adjustment). PTSD, Posttraumatic Stress Disorder; AA, African ancestry; s.e., standard error; AUDIT, Alcohol Use Disorder Identification Test; C, consumption; Max. Alc., Maximum Alcohol Intake; AUD, Alcohol Use Disorder; AD, Alcohol Dependence; PGC, Psychiatric Genomics Consortium; Re-exp., reexperiencing; MVP, Million Veteran Program. Alcohol phenotypes are ordered from more typical to more problematic (top to bottom) and color coded by each type of phenotype (i.e., AUDIT-C, Max. Alc., AUD, AD) to draw attention to difference in findings.
PTSD-Alcohol-related Problems
Notably, the rG between all PTSD phenotypes and alcohol-related problems phenotypes are positive for EA individuals (rGs: 0.132–0.533, all FDR adjusted p < 0.05). The rG estimates for PTSD with Max. Alc., AUDIT-P, AUD and AD phenotypes were positive and moderate (rGs: 0.289–0.533, all FDR adj. p < 0.01) with the exception of AUDIT-P and PTSD Re-Exp, which was small, still positive (rG: 0.132, FDR adj. p = 0.044). All these rG estimates passed FDR adjustment. Notably, the highest rG were between the two PTSD phenotypes and AD (rG: 0.472–0.533, p < 0.001).
A similar trend of positive rGs was observed for PTSD phenotypes (i.e. PTSD, PTSD Re-Exp) with AUD and AD phenotypes on AA samples (rG: 0.182–0.266, NS). Conversely, PTSD phenotypes and Max Alc correlated negatively for individuals of this ancestry (rG: −0.275 to −0.123, NS). However, regardless of rG direction, these estimates yielded relatively large standard errors (s.e.s) and non-significant results ( p > 0.05).
PTSD-alcohol consumption-related phenotypes
The rGs between the PTSD phenotypes and alcohol consumption phenotypes (i.e. DPW, AUDIT-C, AUDIT-T) are negative and varying in degree from small to large for those of AA (AUDIT-C and PTSD Re-Exp rG: 0.145, NS; AUDIT-C and PTSD rG: 0.699, unadjusted p < 0.05), in contrast to the positive and mostly moderate rGs of PTSD and AUD phenotypes. For those of EA, the genetic correlations between PTSD and PTSD Re-Exp and DPW across samples were negative, small, and non-significant ( p > 0.05), with the exception of DPW (UKB) genetic correlations with PTSD Re-Exp, however it did not pass FDR adjustment. The rGs between PTSD phenotypes with AUDIT-C and -T were low to moderate. However, the rGs with AUDIT-C across samples were moderate [rG: −0.417 to −0.225; with the exception of AUDIT-C (UKB) – PTSD Re-exp rGs: −0.188], significant and passed FDR correction. Whereas the genetic correlation between AUDIT-T (UKB) and PTSD Re-exp was the only significant association, albeit small, among all the AUDIT-T analyses for EA. The highest rG among PTSD and alcohol consumption-related phenotypes was that of AUDIT-C (MVP) with PTSD (rG: −0.417, FDR adj. p = 2.59 × 10−7).
The rGs using samples of AA individuals for PTSD phenotypes (i.e. PTSDf2, PTSD Re-Exp) and AUDIT-C were positive, and only that with PTSD Re-Exp was significant; although it did not pass FDR adjustment. Notably, this positive rG estimate in AA individuals, contrasts with the negative rGs estimates in EA samples for the same phenotypes. See online Supplementary Fig. S2 for boxplot with whiskers display of genetic correlations for those of EA and AA.
Post-Hoc tissue enrichment
In EA samples, only the GWAS from MVP PTSD Re-experiencing symptoms, Drinks Per Week (GSCAN and UKB) and Drinks Per Week (UKB) met the FDR significance threshold for specific tissue enrichment (see online Supplementary Fig. S3). These three GWAS that met for this threshold exceeded the FDR significance value for only tissues having to do with the brain (i.e. not other tissues). In AA samples, none of the included GWAS met the FDR threshold for significance for tissue enrichment (see online Supplementary Fig. S4).
Discussion
PTSD commonly co-occurs with increased alcohol consumption and AUD. While our previous work has demonstrated that there is a molecular genetic correlation between PTSD and alcohol-related problems (Sheerin et al., 2020), the goal of this study was to determine whether the genetic correlations with PTSD extend to alcohol consumption, and if the architecture of the genetic association with PTSD differs for alcohol consumption and alcohol-related problems. Further, we aimed to test these associations using both EA and AA summary statistics using data from the latest GWASs of PTSD, alcohol consumption-related, and alcohol-related problems. Among EA analyses, this study found positive and significant genetic correlations between PTSD and alcohol-related problems phenotypes, whereas negative with non-significant genetic correlations observed for PTSD and alcohol consumption-related phenotypes. These results indicate that which alcohol phenotype one uses in analyses absolutely matters, and that alcohol use is certainly not ‘close enough’ as a proxy for alcohol-related problems in examining its genetic associations with other conditions. Among those of AA, potentially due in part to having reduced power but also because of ‘noisy’ heritability estimates (Bulik-Sullivan et al., 2015a), associations were generally non-significant, with the exception of a positive correlation (not passing FDR adjustment) between PTSD Re-Exp symptoms and AUDIT-C, which was not observed for EA individuals.
PTSD, alcohol consumption, and alcohol-related problems have been shown to be heritable in both twin (Heath et al., 1989; Kaprio et al., 1987; Knopik et al., 2004; Stein et al., 2002) and molecular-genetic studies (Clarke et al., 2017; Sanchez-Roige et al., 2019b; Stein et al., 2016). Not surprisingly, the estimates from our study are smaller than those from twin studies, finding estimates of ∼0.38 for PTSD, 0.36–0.40 for consumption, and 0.47 for alcohol misuse, with overall similarities among EA and AA participants. Although the estimated heritability for PTSD [(= 0.193, CI 0.052–0.334) v. = 0.082, CI 0.053–0.111] and Alcohol Dependence (= 0.277, CI −0.044–0.598 v. = 0.093, CI 0.052–0.134) appear higher in AA compared to EA respectively they may not be statistically different as there are large standard errors and wide 95% confidence intervals on the AA estimates (see Table 1). This may be due in part to the larger number of participants of EA included in the discovery GWAS datasets. Large genome-wide studies have historically focused on participants of EA, leading to an important gap in knowledge regarding genetic epidemiology across diverse ancestral groups that our field must address. Future research would benefit from using estimates for similarly sized samples on comparable phenotypes to determine if findings differ due to differences in sample sizes or differences in the phenotypes being examined.
Consistent with previous work (Sartor et al., 2011; Sheerin et al., 2020), this study found positive genetic correlations between PTSD and alcohol-related problems (Max. Alc., AUDIT-P, AUD, AD) among individuals of EA. Our findings are also consistent with a recent paper finding a moderate positive genetic correlation between PTSD and problematic alcohol use (rG = 0.49), and a more modest genetic correlation between PTSD and the specific portion of problematic alcohol use unique from a larger externalizing factor (rG = 0.26) (Barr et al., 2021). However, when investigating genetic correlations between PTSD and alcohol consumption-related phenotypes, findings generally suggested negative, non-significant or not passing multiple testing adjustment (e.g. in the case of DPW correlations using GSCAN, UKB data, and both combined) associations with PTSD. These discrepancies may have arisen because of differing sample characteristics or differing numbers of studies contributing to these statistics. However, in general, the genetic associations between PTSD and alcohol-consumption phenotypes were different from those of PTSD-alcohol-related problems.
These results suggest that different genetic factors may exist for individuals with PTSD and increased alcohol consumption and for individuals with PTSD and alcohol-related problems. These results are also consistent with the very small amount of work conducted examining genetic associations between PTSD and alcohol use. Specifically, a paper by our group employing Mendelian Randomization (MR) as the primary method also found in secondary analyses using LDSC a non-significant genetic association between PTSD and DPW among those of EA (Bountress et al., 2021). Additionally, a similar trend has been observed with other psychiatric disorders in terms of the genetic association between alcohol consumption v. alcohol-related problems phenotypes, specifically major depressive disorder, which has substantial genetic overlap with PTSD (Polimanti et al., 2019; Sanchez-Roige et al., 2019b; Walters et al., 2018; Zhou et al., 2020a). These authors observed positive genetic correlations between major depression and alcohol dependence. However, they also observed negative genetic correlations between major depression and frequency of alcohol consumption. Further, the same trend in attention-deficit/hyperactivity disorder (ADHD) was observed such that problematic drinking was positively genetically correlated with ADHD and alcohol consumption was negatively genetically correlated with ADHD (Sanchez-Roige et al., 2019a). Future work might benefit from additional MR analyses examining the potential causal relations between PTSD and more alcohol phenotypes.
Interestingly, one study found that the genetic association between AUDIT-C and AD was initially negative, but became positive when the ‘healthy volunteer’ effect that tends to occur in alcohol frequency data was taken into account (Mallard et al., 2021). Thus, in cases where the associations between alcohol consumption frequency and alcohol consumption quantity and other phenotypes are in opposing directions, this may be because the frequency item is positively genetically correlated with high SES (Mallard & Sanchez-Roige, 2021; Marees et al., 2020). Another potential explanation for the negative association between consumption and other psychopathology-related outcomes may be related to the finding that some with greater disease burden have in turn, reduced or limited their alcohol consumption (Xue et al., 2020). Our study is the first to observe a negative genetic correlation between PTSD and alcohol consumption phenotypes, among those of EA. Findings from this study, together with the previous literature on genetic correlations between other psychiatric disorders and problematic v. typical alcohol use, consistently indicate that problematic alcohol use and more typical alcohol use are genetically associated with other psychiatric disorders in opposite directions. These findings suggest that alcohol-related behaviors are heterogenous, and specifically that the genetic associations between consumption and alcohol-related problems and PTSD differ in strength and direction.
Importantly, we note that this pattern of results was driven by participants of EA, and that one contrasting finding was observed among AA participants. It is important to interpret the AA findings (i.e. those whose h2SNP Z-scores were less than 4) with caution, as these heritabilities may be less precise (Bulik-Sullivan et al., 2015a). For AA analyses, the AFR UKB pan-ancestry LD scores were used, and it should be noted that more research is needed to clarify the extent of bias resulting from using admixed ancestry reference panels (Bulik-Sullivan et al., 2015a; Bulik-Sullivan et al., 2015b). Other methods such as cov-LDSC (Luo et al., 2021), which can construct cohort-specific LD reference panel increase accuracy, but requires access to measured genotypes, which are largely unavailable for the samples in the current study. The field is in critical need of large-scale diverse ancestry cohorts and the corresponding methods development to robustly analyze these data (Peterson, 2021). Increasing ancestral diversity in large genetic studies will enable improved understanding of the epidemiology of PTSD, alcohol use behaviors, and their comorbidity.
Additionally, we note that although prior work by our group found sex differences in the genetic associations between PTSD and AD (Sheerin et al., 2020), we were unable to test whether that difference extended to other alcohol phenotypes, as summary statistics stratified by sex were not available. Given known prevalence and presentation differences of PTSD and alcohol phenotypes across sex, future research ought to attempt to test this question.
In terms of the post-hoc enrichment analyses, the finding that for EAs, the GWAS that reached FDR significance all met for brain tissues, but not others, is generally consistent with prior work for PTSD and other neuropsychiatric traits (Dalvie et al., 2021; Gelernter et al., 2019b). However, additional work is needed to better understand why only three of the GWAS (MVP PTSD Re-Experiencing, DPW GSCAN, DPW UKB) exceeded the FDR cut-off. For AAs, no GWAS exceeded the FDR threshold. It is possible that this lack of significant effects is due to the smaller sample sizes among the AA GWAS.
This investigation attempted to advance the understanding of how the genetic associations between PTSD and alcohol phenotypes may vary depending on which phenotype is being examined. In particular, future research with larger sample sizes and better tuned reference panels are needed to have more confidence in findings that are generated for samples comprised of AA individuals or other admixed populations. Ideally, future work ought to utilize larger primary datasets and more appropriate analytic strategies for admixed populations (e.g. cov-LDSC) as they become available. The findings generated among AA samples herein are a first pass within currently available data and methods and ought to be interpreted with caution. This mismatch in admixture between reference panel and target sample can impact the precision of results including the attenuation of h2 estimates.
In conclusion, findings from this study extend knowledge regarding the genetic associations of PTSD and AUD, to include a spectrum of alcohol use phenotypes including more typical alcohol use in an ancestrally diverse population. These findings indicated positive genetic associations between PTSD and alcohol-related problems phenotypes and negative genetic correlations between PTSD and alcohol consumption-related phenotypes. Thus, the genetic factors that may lead someone to develop PTSD and high levels of alcohol consumption are not the same as those that lead someone to develop PTSD and alcohol-related problems. These findings support the growing number of studies demonstrating the important differences regarding risk factors for alcohol consumption v. disorder, and their associations with other psychiatric disorders.
Supplementary Material
Acknowledgements.
We would also like to thank dgGaP (accession phs001672.v7.p1). Additionally, the authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/ for more details). The citation for MVP is Gaziano, J.M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Veterans Administration (VA) Cooperative Studies Program (CSP) award #G002.
Financial support.
The effort of co-authors was supported by NIAAA (1K01 AA028058 [KB], 1K01 AA025692[CS], P50AA022537[REP, BTW]), NIMH (K01MH113848 [REP], R01MH125938[REP, BTW], MH020030–21A1 [DB]), and The Brain & Behavior Research Foundation NARSAD grant 28632 P&S Fund [REP]. Financial support for the PTSD PGC was provided by the Cohen Veterans Bioscience, Stanley Center for Psychiatric Research at the Broad Institute, One Mind, and the National Institute of Mental Health (NIMH; R01MH106595). The PGC-SUD Working Group receives support from the National Institute on Drug Abuse and the National Institute of Mental Health via MH109532. Statistical analyses for the PGC were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480–05-003), along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. This research has been conducted using data from UK Biobank, a major biomedical database (https://www.ukbiobank.ac.uk/). Research reported in this publication was supported by the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplementary material. The supplementary material for this article can be found at https://doi.org/10.1017/S0033291722002999
Conflict of interest. None.
Ethical standards. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals
References
- American Psychiatric Association (1987). Diagnostic and statistical manual of mental disorders (3rd ed., revised (DSM-III-R)) Washington, DC: American Psychiatric Press. [Google Scholar]
- American Psychiatric Association (1994). Diagnostic and statistical manual of mental disorders (4th ed. (DSM-IV)) Washington, DC: American Psychiatric Press. [Google Scholar]
- American Psychiatric Association (2013). Diagnostic and statistical manual of mental disorders (5th ed. (DSM-5)) Washington, DC: American Psychiatric Press. [Google Scholar]
- Barr PB, Mallard TT, Sanchez-Roige S, Poore HE, Linnér RK, Collaborators C, … Dick DM (2021). Parsing Genetically Influenced Risk Pathways: Genetic Loci Impact Problematic Alcohol Use Via Externalizing and Specific Risk. medRxiv, 2021.2007.2020.21260861. 10.1101/2021.07.20.21260861. [DOI] [PMC free article] [PubMed]
- Benjet C, Bromet E, Karam EG, Kessler RC, McLaughlin KA, Ruscio AM, … Koenen KC (2016). The epidemiology of traumatic event exposure worldwide: Results from the world mental health survey consortium. Psychological Medicine, 46(2), 327–343. doi: 10.1017/s0033291715001981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley AR, Callier S, & Rotimi CN (2017). Diversity and inclusion in genomic research: Why the uneven progress? Journal of Community Genetics, 8(4), 255–266. doi: 10.1007/s12687-017-0316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bountress KE, Wendt F, Bustamante D, Agrawal A, Webb B, Gillespie N, … Amstadter A (2021). Potential causal effect of posttraumatic stress disorder on alcohol use disorder and alcohol consumption in individuals of European descent: A mendelian randomization study. Alcoholism Clinical and Experimental Research, 8, 1616–1623. doi: 10.1111/acer.14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslau N (2009). Trauma and mental health in US inner-city populations. General Hospital Psychiatry, 31(6), 501–502. doi: 10.1016/j.genhosppsych.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, … Neale BM (2015a). An atlas of genetic correlations across human diseases and traits. Nature Genetics, 47(11), 1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, … Neale BM (2015b). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics, 47 (3), 291–295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Kivlahan DR, McDonell MB, Fihn SD, & Bradley KA (1998). The AUDIT alcohol consumption questions (AUDIT-C): An effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). alcohol Use disorders identification test. Archives of Internal Medicine, 158(16), 1789–1795. doi: 10.1001/archinte.158.16.1789 [DOI] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, … McIntosh AM (2017). Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK biobank (N = 112 117). Molecular Psychiatry, 22(10), 1376–1384. doi: 10.1038/mp.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvie S, Chatzinakos C, Al Zoubi O, Georgiadis F, Lancashire L, & Daskalakis NP (2021). From genetics to systems biology of stress-related mental disorders. Neurobiology of Stress, 15, 100393. 10.1016/j.ynstr.2021.100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, … Koenen KC (2017). Largest GWAS of PTSD (N = 20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Molecular Psychiatry, 3, 666–673. doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sun N, Polimanti R, Pietrzak R, Levey DF, Bryois J, … Stein MB (2019b). Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165000 US veterans. Nature Neuroscience, 22(9), 1394–1401. doi: 10.1038/s41593-019-0447-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sun N, Polimanti R, Pietrzak RH, Levey DF, Lu Q, … Stein MB (2019a). Genome-wide association study of Maximum habitual alcohol intake in >140000 U.S. European and African American veterans yields novel risk loci. Biological Psychiatry, 86(5), 365–376. doi: 10.1016/j.biopsych.2019.03.984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz SM, Horton AC, Hancock DB, Baker TB, Caporaso NE, Chen L-S, … McNeil DW (2018). Genetic correlation between smoking behaviors and schizophrenia. Schizophrenia Research, 194, 86–90. doi: 10.1016/j.schres.2017.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Jardine R, & Martin NG (1989). Interactive effects of genotype and social environment on alcohol consumption in female twins. Journal of Studies on Alcohol, 50(1), 38–48. doi: 10.15288/jsa.1989.50.38 [DOI] [PubMed] [Google Scholar]
- Jakupcak M, Tull MT, McDermott MJ, Kaysen D, Hunt S, & Simpson T (2010). PTSD Symptom clusters in relationship to alcohol misuse among Iraq and Afghanistan war veterans seeking post-deployment VA health care. Addictive Behaviors, 35(9), 840–843. doi: 10.1016/j.addbeh.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Kaprio J, Koskenvuo M, Langinvainio H, Romanov K, Sarna S, & Rose RJ (1987). Genetic influences on use and abuse of alcohol: A study of 5638 adult Finnish twin brothers. Alcoholism Clinical and Experimental Research, 11(4), 349–356. doi: 10.1111/j.1530-0277.1987.tb01324.x [DOI] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Madden PA, Bucholz KK, Slutske WS, Nelson EC, … Martin NG (2004). Genetic effects on alcohol dependence risk: Re-evaluating the importance of psychiatric and other heritable risk factors. Psychological Medicine, 34(8), 1519–1530. doi: 10.1017/s0033291704002922 [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, … Gelernter J (2019). Genome-wide association study of alcohol consumption and use disorder in 274424 individuals from multiple populations. Nature Communications, 10(1), 1499. doi: 10.1038/s41467-019-09480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, … Vrieze S (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics, 51(2), 237–244. doi: 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, … Young N (2013). The genotype-tissue expression (GTEx) project. Nature Genetics, 45(6), 580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Li X, Wang X, Gazal S, Mercader JM, 23andMeResearch Team, … Raychaudhuri S (2021). Estimating heritability and its enrichment in tissue-specific gene sets in admixed populations. Human molecular genetics, 30(16), 1521–1534. doi: 10.1093/hmg/ddab130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard TT, & Sanchez-Roige S (2021). Dimensional phenotypes in psychiatric genetics: Lessons from genome-wide association studies of alcohol Use phenotypes. Complex Psychiatry, 7(3–4), 45–48. doi: 10.1159/000518863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard TT, Savage JE, Johnson EC, Huang Y, Edwards AC, Hottenga JJ, … Anokhin A (2021). Item-level genome-wide association study of the alcohol use disorders identification test in three population-based cohorts. American Journal of Psychiatry, 179(1), 58–70. doi: 10.1176/appi.ajp.2020.20091390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marees AT, Smit DJ, Ong J-S, MacGregor S, An J, Denys D, … Derks EM (2020). Potential influence of socioeconomic status on genetic correlations between alcohol consumption measures and mental health. Psychological Medicine, 50(3), 484–498. doi: 10.1017/S0033291719000357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod S, Koenen KC, Meyer J, Lyons MJ, Eisen S, True W, & Goldberg D (2001). Genetic and environmental influences on the relationship among combat exposure, posttraumatic stress disorder symptoms, and alcohol use. Journal of Traumatic Stress, 14(4), 259–275. doi: 10.1023/A:1011157800050 [DOI] [PubMed] [Google Scholar]
- Moos RH, Schutte K, Brennan P, & Moos BS (2004). Ten-year patterns of alcohol consumption and drinking problems among older women and men. Addiction, 99(7), 829–838. doi: 10.1111/j.1360-0443.2004.00760.x [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, … Koenen KC (2019). International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nature Communications, 10(1), 4558. doi: 10.1038/s41467-019-12576-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE (2021). The genetics of major depression: Perspectives on the state of research and opportunities for precision medicine. Psychiatric annals, 51(4), 165–169. doi: 10.3928/00485713-20210315-01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Kuchenbaecker K, Walters RK, Chen CY, Popejoy AB, Periyasamy S, … Duncan LE (2019). Genome-wide association studies in ancestrally diverse populations: Opportunities, methods, pitfalls, and recommendations. Cell, 179(3), 589–603. doi: 10.1016/j.cell.2019.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Peterson RE, Ong JS, MacGregor S, Edwards AC, Clarke TK, … Derks EM (2019). Evidence of causal effect of major depression on alcohol dependence: Findings from the psychiatric genomics consortium. Psychological Medicine, 49(7), 1218–1226. doi: 10.1017/s0033291719000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach BC, Bray MJ, Gaddis NC, Liu M, Palviainen T, Minica CC, … Nothnagel M (2020). Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nature Communications, 11(1), 1–13. doi: 10.1038/s41467-020-19265-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, Team AR, Gray JC, de Wit H, … Palmer AA (2019a). Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addiction biology, 24(1), 121–131. doi: 10.1111/adb.12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, … Clarke TK (2019b). Genome-Wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. American Journal of Psychiatry, 176(2), 107–118. doi: 10.1176/appi.ajp.2018.18040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor CE, McCutcheon VV, Pommer NE, Nelson EC, Grant JD, Duncan AE, … Heath AC (2011). Common genetic and environmental contributions to posttraumatic stress disorder and alcohol dependence in young women. Psychological Medicine, 41(7), 1497–1505. doi: 10.1017/S0033291710002072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, De la Fuente JR, & Grant M (1993). Development of the alcohol use disorders identification test (AUDIT): WHO collaborative project on early detection of persons with harmful alcohol consumption-II. Addiction, 88(6), 791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x [DOI] [PubMed] [Google Scholar]
- Sheerin CM, Bountress KE, Meyers JL, Saenz de Viteri SS, Shen H, Maihofer AX, … Amstadter AB (2020). Shared molecular genetic risk of alcohol dependence and posttraumatic stress disorder (PTSD). Psychology of Addictive Behaviors, 5, 613. doi: 10.1037/adb0000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G, Williams SM, & Tishkoff SA (2019). The missing diversity in human genetic studies. Cell, 177(1), 26–31. doi: 10.1016/j.cell.2019.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MB, Chen CY, Ursano RJ, Cai T, Gelernter J, Heeringa SG, … Smoller JW (2016). Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US army soldiers. JAMA psychiatry, 73(7), 695–704. doi: 10.1001/jamapsychiatry.2016.0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MB, Jang KJ, Taylor S, Vernon PA, & Livesley WJ (2002). Genetic and environmental influences on trauma exposure and post-traumatic stress disorder: A twin study. American Journal of Psychiatry, 159(10), 1675–1681. doi: 10.1176/appi.ajp.159.10.1675 [DOI] [PubMed] [Google Scholar]
- Sullivan PF (2010). The psychiatric GWAS consortium: Big science comes to psychiatry. Neuron, 68(2), 182–186. doi: 10.1016/j.neuron.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahov D, Galea S, Resnick H, Ahern J, Boscarino JA, Bucuvales M, … Kilpatrick D (2002). Increased use of cigarettes, alcohol, and marijuana amond Manhattan, New York residents after the September 11th terrorist attacks. American Journal of Epidemiology, 155, 988–996. doi: 10.1093/aje/155.11.988 [DOI] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, … Agrawal A (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience, 21(12), 1656–1669. doi: 10.1038/s41593-018-0275-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KC, Lang AJ, & Norman SB (2011). Synthesis of the psychometric properties of the PTSD checklist (PCL) military, civilian, and specific versions. Depression and anxiety, 28(7), 596–606. doi: 10.1002/da.20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian H, Chantarujikapong SI, Scherrer JF, Eisen SA, Lyons MJ, Goldberg J, … True WR (2000). Genetic and environmental influences on posttraumatic stress disorder, alcohol and drug dependence in twin pairs. Drug and Alcohol Dependence, 61(1), 95–102. doi: 10.1016/s0376-8716(00)00127-7 [DOI] [PubMed] [Google Scholar]
- Xue A, Jiang L, Zhu Z, Wray NR, Visscher PM, Zeng J, & Yang J (2020). Genome-wide analyses of behavioural traits biased by misreports and longitudinal changes. medRxiv, 2020.2006.2015.20131284. 10.1101/2020.06.15.20131284. [DOI] [PMC free article] [PubMed]
- Zhou H, Rentsch CT, Cheng Z, Kember RL, Nunez YZ, Sherva RM, … Polimanti R (2020a). Association of OPRM1 functional coding variant with opioid use disorder: A genome-wide association study. JAMA psychiatry, 77(10), 1072–1080. doi: 10.1001/jamapsychiatry.2020.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, … Gelernter J (2020b). Genome-wide meta-analysis of problematic alcohol use in 435563 individuals yields insights into biology and relationships with other traits. Nature Neuroscience, 23(7), 809–818. doi: 10.1038/s41593-020-0643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


