Abstract
N6-methyladenosine (m6A) methylation is the most prevalent mRNA modification in eukaryotes, and it is defined as the methylation of nitrogen atoms on the six adenine (A) bases of RNA in the presence of methyltransferases. Methyltransferase-like 3 (Mettl3), one of the components of m6A methyltransferase, plays a decisive catalytic role in m6A methylation. Recent studies have confirmed that m6A is associated with a wide spectrum of biological processes and it significantly affects disease progression and prognosis of patients with gynecologic tumors, in which the role of Mettl3 cannot be ignored. Mettl3 is involved in numerous pathophysiological functions, such as embryonic development, fat accumulation, and tumor progression. Moreover, Mettl3 may serve as a potential target for treating gynecologic malignancies, thus, it may benefit the patients and prolong survival. However, there is a need to further study the role and mechanism of Mettl3 in gynecologic malignancies. This paper reviews the recent progression on Mettl3 in gynecologic malignancies, hoping to provide a reference for further research.
Keywords: N6‐methyladenosine, m6A methyltransferase, gynecologic malignancies, METTL3, cancer therapy
Introduction
Gynecologic malignancies are major diseases that pose a threat to the health of women worldwide; and among them, cervical cancer (CC), ovarian cancer (OC), and endometrial cancer (EC) are the most common malignancies. In recent years, patients diagnosed with these diseases are more likely to be younger (Bray et al., 2018). Surgical intervention is the primary treatment for gynecological malignancies and is often complemented with radiotherapy, chemotherapy, endocrine therapy, and immunotherapy (Barillot and Haie-Meder, 2009; Wang et al., 2020a; Chen et al., 2022). Despite these treatment modalities, the overall prognosis for patients with gynecological malignancies remains unsatisfactory, indicating the need for further improvements in therapeutic strategies. The current treatment regimen faces many challenges due to the high malignancy, high mortality rate, and high incidence of recurrence and metastasis in gynecologic cancers (Jemal et al., 2011; Makker et al., 2020; Lim et al., 2022). Therefore, it is imperative to explore the detailed molecular mechanisms of gynecologic tumors and to seek innovative therapeutic strategies. Gynecological malignancies result from multiple pathogenic factors, among which abnormal genetic changes in proto-oncogenes and tumor suppressor genes, such as gene expansion/deletion/mutation or chromosomal translocation, are essential factors in the initiation and progression of tumors (Dueñas-González et al., 2005; See et al., 2019; Zhou et al., 2016). Most previous reports have focused on the DNA expression level; but with the progress of research, it has become clear that regulation at the transcriptional level is equally essential. Mounting evidence has revealed that epigenetic modifications regulate gene expression without altering the DNA sequence and always modulate tumorigenesis and progression (Nebbioso et al., 2018; Zhu et al., 2021). Among the various epigenetic modifications, m6A methylation modification is the most frequent and significant modification in messenger RNA (Liang et al., 2020a).
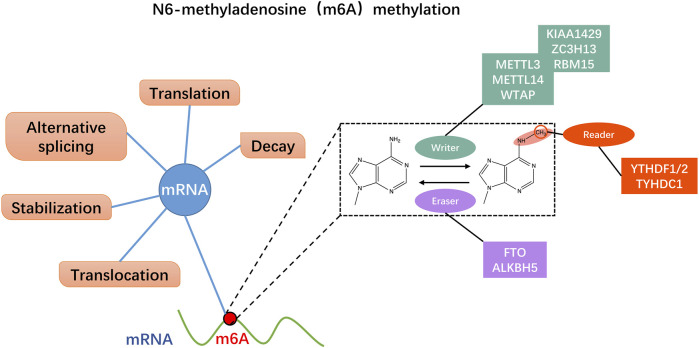
N6-methyladenosine (m6A) modification in RNA is defined as the methylation of nitrogen atoms on the six adenine (A) bases of RNA in the presence of methyltransferases (Huang et al., 2021a). Although m6A methylation was identified in poly(A) RNA from hepatocellular carcinoma cells in 1974 (Desrosiers et al., 1974), researchers used methylated RNA immunoprecipitation sequencing (MeRIP-Seq) in 2012 to revive interest in m6A methylation (Dominissini et al., 2012; Meyer et al., 2012). With increasing research and advent of high-throughput sequencing technology, understanding of m6A methylation has gradually improved. A growing number of studies have confirmed that m6A methylation is closely related to various biological behaviors of malignant tumors and affects patient prognosis (Huang et al., 2022). M6A methylation is a multi-stage process resulting from the interaction of three enzymes: methyltransferases (m6A writers), demethylases (m6A erasers), and binding proteins (m6A readers) (Figure 1) (Helm and Motorin, 2017; Pinello et al., 2018). Among them, m6A methyltransferases mainly include methyltransferase-like 3 (Mettl3), methyltransferase-like 14 (Mettl14), and Wilms tumor one associated protein (WTAP) (Lan et al., 2019); M6A demethylase specifically includes alkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) (Chen et al., 2019); YT521‐B homology (YTH) domain family proteins comprising YTHDFs and YTHDCs mainly constitute m6A binding proteins. The YTHDF subtype family consists of YTHDF1, YTHDF2, and YTHDF3, while the YTHDC subtype family mainly comprises YTHDC1 and YTHDC2 (Zhao et al., 2020a). Heterogeneous nuclear ribonucleoproteins (HNRNPs) and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) are also crucial compositions of m6A readers. The role of m6A methylation serving as a “double-edged sword” in malignancy can be complex and multifaceted. On the one hand, m6A methylation has been shown to promote tumorigenesis and progression by regulating various cellular processes such as RNA stability, splicing, translation, and protein-protein interactions (Fang et al., 2022). On the other hand, m6A methylation can also exert cancer-suppressive effects by controlling the expression of tumor suppressor genes and inhibiting oncogenic signaling pathways (Deng et al., 2022). The regulatory role of m6A methylation in malignancy likely varies depending on the specific type of cancers, the stage of cancer development, and other factors (Liu et al., 2018a). As an indispensable regulator of m6A methylation, Mettl3 is a critical protein in the m6A methyltransferase complex, which catalyzes methylation of the nitrogen atom at position 6 of adenine (A) in RNA. Mettl3, Mettl14, and accessory proteins, including WTAP and ZC3H13, constitute the m6A writer proteins, and Mettl3 is the only catalytic subunit that uses S-adenosylmethionine (SAM) as a methyl donor (Wang et al., 2016a; Wang et al., 2016b). Mettl3 is highly conserved in eukaryotes and is a vital component of the m6A methyltransferase complex, which plays a crucial catalytic role in the entire process of m6A methylation (Wang et al., 2016b). Also, Mettl3 is engaged in almost all RNA stages involving m6A methylation, including pre-mRNA splicing, mRNA decay, miRNA processing, translation regulation, and nuclear export (Wang et al., 2020b).
FIGURE 1.
N6-methyladenosine (m6A) methylation dynamically regulates mRNA through multiple pathways. M6A regulatory proteins include m6A methyltransferases (m6A writers), m6A demethylases (m6A erasers), and m6A recognition proteins (m6A readers). The writers act as methyltransferase including Mettl3/14 and WTAP. The erasers mediate demethylation modifications, like FTP and ALKBH5. M6A readers recognize information about mRNA methylation and are involved in downstream mRNA translation, degradation, and other behaviors, such as YTHDF1/2 and TYHDC1.
Therefore, Mettl3 may be a new target for treating gynecological malignancies. Conducting a comprehensive investigation into the specific mechanism of Mettl3 in gynecologic malignancies could provide substantial benefits to a diverse patient population by facilitating the development of effective diagnostic and therapeutic strategies. This article reviews the latest research progress of Mettl3 in gynecologic malignancies, as well as the detailed mechanisms behind, to offer insights into innovation in the diagnosis and treatment of gynecologic malignancies.
Molecular structure, the primary function of Mettl3
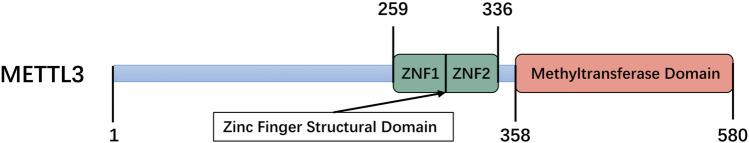
Mettl3 comprises a zinc finger structural domain (ZFD) and a methyltransferase domain (MTD) construct containing 358–580 residues, both of which are essential for enzymatic activity (Figure 2) (Wang et al., 2016a). The MTD of the Mettl3-Mettl14 heterodimer has been identified by investigators using X-ray crystallographic methods. Mettl3 represents the only catalytic subunit that binds to the methyl donor SAM and facilitates the transfer of methyl groups (Wang et al., 2016a; Zeng et al., 2020). However, MTD alone cannot ensure an enzymatically active Mettl3-mettl14 complex and participation of ZFD is required to obtain a fully functional complex (Śledź and Jinek, 2016). Huang et al. used magnetic resonance to reveal that the ZFD solution structure specifically binds to RNA containing the 5′-GGACU-3′ shared sequence but not to RNA without this sequence, and that it contains two CCCH-type zinc fingers linked in tandem by an inverse parallel ß-sheet linkage while having a synergistic catalytic effect with MTD3-MTD14 (Huang et al., 2019). The impact of Mettl3 on tumor progression is mediated through numerous mechanisms. In breast cancer, the pro-oncogenic function of Mettl3 involves the inhibition of let-7g tumor suppressor translation, which ultimately promotes cancer cell proliferation (Cai et al., 2018). Wang et al. confirmed that Mettl3 regulates VASH1 to promote brain metastasis in lung cancer by inducing the maturation of miR-25-3p (Wang et al., 2019). Mettl3 promotes hepatocellular carcinoma cell proliferation and metastasis by reducing the mRNA stability of SOCS2 through a YTHDF2-dependent pathway (Chen et al., 2018; Wang et al., 2020c). In short, Mettl3 exerts a significant and multifaceted biological impact on diverse cancer types and all stages of tumorigenesis (Yi et al., 2020). While the role of Mettl3 in tumorigenesis has been extensively studied in recent years, there is still much to learn about its mechanisms of action and potential as a therapeutic target. As research continues, it is likely that more insights will be gained into the complex and integrative role that Mettl3 plays in cancer.
FIGURE 2.
Schematic domain structure of Mettl3, including zinc finger domain (ZFD) and methyltransferase domain (MTD).
Advances of Mettl3 in various gynecological malignancies
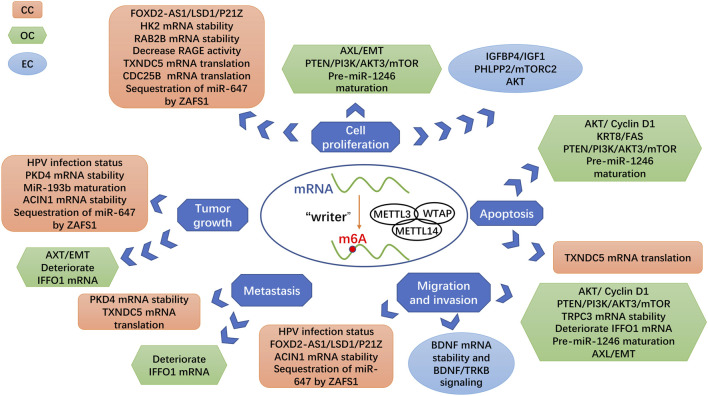
Previous investigations have shown that Mettl3 plays a critical role in promoting cancer development in diverse tissue types of cancer, such as affecting the development of gastrointestinal malignancies by affecting tumor proliferation, angiogenesis, apoptosis, and metastasis (Figure 3) (Wang et al., 2020c). Analogously, Mettl3 is closely associated with multiple adverse biological behaviors of gynecologic malignancies, including tumor proliferation, apoptosis, metastasis, angiogenesis, and immune microenvironment (Huang et al., 2022). We reviewed the latest research on the mechanism of Mettl3 in the occurrence and development of gynecological malignancies. Therefore, exploring the specific mechanism of m6A methylation in malignant tumors is very important for attacking cancer.
FIGURE 3.
Roles of Mettl3 as a methyltransferase in gynecologic cancers. Different targets of Mettl3 are linked with various signaling pathways and biological processes, which generate the different influences of Mettl3 in gynecologic cancers.
The role of Mettl3 in cervical cancer
CC has the fourth highest incidence and mortality rate among women, constituting a significant public health problem that endangers women’s health globally (Bray et al., 2018), (Cohen et al., 2019). According to global cancer statistics for 2018, an estimated 570,000 new cases of CC were reported, with approximately 311,000 related deaths (Bray et al., 2018). Hence, further research into the specific molecular mechanisms of CC is fundamental and it may provide inspiration for new therapeutic targets. It has been well established that infection with human papillomavirus (HPV), especially high-risk types, such as 16 and 18, is a high-risk factor for developing CC (Burd, 2003; Ma and Yang, 2021). Despite the increasing prevalence of HPV vaccination, HPV infection-associated CC remains a clinical challenge. Human papillomaviruses can produce single-stranded circular RNAs (circRNAs) containing the E7 oncogene (circE7). CircE7 is modified by m6A methylation and translated to produce the E7 oncoprotein, which promotes the growth of CC cells (Zhao et al., 2019). The regulatory protein of m6A methylation is closely linked to the HPV infection status of patients with CC, as it affects the extent of m6A methylation modification (Condic et al., 2022). Mettl3 is significantly overregulated in HPV-positive CC tissues and correlates with immune checkpoints and infiltration (Table 1) (Yu et al., 2022). The relationship between m6A methylation and HPV infection-associated CC needs further investigation. Addressing the etiological factors of CC can provide a treatment approach that targets the underlying cause, leading to significant clinical benefits for prevention and treatment.
TABLE 1.
The role of Mettl3 in cervical cancer.
| Cancer species | Expression | Biological function | Mechanism | Reference |
|---|---|---|---|---|
| cervical cancer | Up | Promote cell migration, chemotaxis, and tumor growth, influence cytokines | Associated with HPV infection status, immune checkpoint molecules, and immune infiltration | Yu et al. (2022) |
| Up | Promote tumor growth, metastasis and chemoresistance | Regulate glycolysis in cervical cancer cells by enhancing the mRNA stability of PDK4 | Li et al. (2020) | |
| Up | Promote cell proliferation and migration | Downregulate p21 through FOXD2-AS1/LSD1/P21Z axis | Ji et al. (2021) | |
| Up | Promote cell proliferation and warburg effect | Recruit YTHDF1 to enhance the stability of HK2 mRNA | Wang et al. (2020d) | |
| Down | Increase cisplatin chemosensitivity, inhibit the viability and proliferation | Decrease the activity of RAGE | Li et al. (2021a) | |
| Up | Promote cell proliferation and metastasis and inhibit apoptosis and autophagy | Promote the translation of TXNDC5 mRNA | Du et al. (2022) | |
| Up | Associated with poor prognosis, and promote cell proliferation | Increase mRNA stability of oncogene RAB2B via GF2BP3-dependent pathway | Hu et al. (2020) | |
| Up | Independent indicators for poor prognosis | The expression of Mettl3 was positively related with iASPP | Wu et al. (2020) | |
| Up | Promote tumor development, and associated with poor outcomes | The level of Mettl3 is positively linked to the number of CD33+ MDSC | Ni et al. (2020) | |
| Up | Confer cancer cell tolerance to hypoxic stress, promoting cell survival and adaptation | Enhance cytoprotective autophagy and tolerance to hypoxic environments by increasing the mRNA stability of DARS and acting on the DARS-AS1/ATG5/ATG3 axis | Shen et al. (2022a) | |
| Up | Facilitate cell cycle progression, promote cell proliferation in vitro and enhance tumorigenicity in vivo | Accelerated the translation of CDC25B mRNA through YTHDF1-dependent m6A modification | Li et al. (2022) | |
| Up | Promote tumor growth | Hinder miR-193b maturation, which inhibits tumorigenesis of cervical cancer cells through CCND1 targeting | Huang et al. (2021b) | |
| Up | Promote tumor growth, migration and aggressiveness | Mettl3 interacts with IGF2BP3 to promote the mRNA stability of ACIN1 | Su et al. (2022) | |
| Up | Associated with poor prognosis | Negatively correlated with the expression of PD-L1 | Ji et al. (2022) | |
| Up | Promote tumor growth in vivo and cell proliferation, migration, and invasion in vitro | Promote the sequestration of miR-647 by ZAFS1 through the regulation of RNA-RNA interactions | Yang et al. (2020) |
In addition to its association with the HPV infection status of CC patients, Mettl3 plays a profound role in various biological behaviors of CC cells, exerting significant effects. In vitro, transcription is activated due to TATA-binding protein (TBP) binding to the Mettl3 promoter to the point at which Mettl3 expression levels are significantly increased (Li et al., 2020). Overexpression of Mettl3 promotes cancer progression through various pathways, such as affecting glucose metabolism, cell cycle PD-L1, immune infiltration, and enhancing the stability of oncogenic mRNA to promote the proliferation and migration of CC cells (Ji et al., 2022). Expression of Mettl3 is an independent risk factor for the 5-year recurrence-free survival (RFS), distant metastasis-free survival (DMFS), progression-free survival (PFS), and overall survival (OS) in patients with CC (Wu et al., 2020). Mechanically, the level of Mettl3 in tumor cells and tumor-infiltrating immune cells potentiate the density of myeloid-derived suppressor cells (MDSCs) and attenuate the proliferation of T lymphocytes in the tumor microenvironment, as well as hinder their function (Ni et al., 2020). Fei Ji et al. found that Mettl3 was positively correlated with the expression and enhanced the stability of the oncogene FOXD2-AS1, which promoted the proliferation and migration ability of CC cells while inhibiting apoptosis. In addition, the upregulated expression of the FOXD2-AS gene induced by Mettl3 recruits lysine-specific demethylase 1 (LSD1) to the P21 promoter region, which constitutes the FOXD2-AS1/LSD1/P21Z axis and reduces the expression of P21, an essential member of the cell cycle protein-dependent kinase inhibitor family. P21 is closely related to tumor suppression. Its overexpression inhibits the proliferation and migration ability of CC cells and enhances the apoptosis rate (Ji et al., 2021). In addition, Mettl3 synergizes with IGF2BP3 to extend the half-life of ACIN1 and RAB2B mRNA to contribute to the growth of CC and poor prognosis (Hu et al., 2020; Su et al., 2022). Recent research has revealed that Mettl3 is highly expressed in CC cells in the M stage, increases m6A methylation of CDC25B mRNA, and enhances its translation activity by recruiting YTHDF-1. Ultimately, the high expression level of Mettl3 further facilitates the cell cycle and proliferation (Li et al., 2022). Studies have provided additional evidence supporting the notion that Mettl3-mediated m6A methylation modifications result in the sequestration of miR-647 by ZAFS1 through the regulation of RNA-RNA interactions. This sequestration has been found to promote tumor growth in vivo and CC cell proliferation, migration, and invasion in vitro. Moreover, elevated levels of ZAFS1 have been clinically associated with lymph nodes and distant metastasis in CC patients and have been shown to be a predictor of poor prognosis (Yang et al., 2020). The profound involvement of Mettl3 in the diverse malignant biological behaviours plays a broad and remarkable role in the progression of CC cells.
Tumor cells exhibit aberrant metabolism, a distinct hallmark feature that facilitates their nutrient requirements for growth and proliferation, distinguishing them from normal cells (Pavlova and Thompson, 2016). The process of m6A methylation has been closely linked to the metabolic reprogramming of tumor cells (An and Duan, 2022). The Warburg effect, also known as aerobic glycolysis, is a typical abnormal metabolic behavior of tumors, which is an epiphenomenon of the transformation process crucial for cancer growth (Heiden et al., 2009; Spencer and Stanton, 2019). Although glycolysis is inefficient energy harvesting, it provides tumor cells with unique growth advantages, such as promoting tumor metastasis. Lactate produced in this process also helps tumor cells escape immunity and induces the secretion of vascular endothelial growth factors to provide nutrients to the tumor (Liberti and Locasale, 2016). In CC, suppressed Mettl3 causes a decrease in the oxygen consumption rate (OCR) and an increase in the extracellular acidification rate (EACR) and ATP generation efficiency (Li et al., 2020). Mettl3 targets the m6A site of the 3′-untranslated region (3′-UTR) of hexokinase 2 (hk2) mRNA and recruits YTHDF1 to enhance the stability of HK2 mRNA, further promoting the Warburg effect (Wang et al., 2020d). Therefore, Mettl3 may serve as a potential target to inhibit aerobic glycolysis by acting on the key enzyme of glycolysis-hexokinase, further delaying CC progression. In addition, studies have demonstrated that Mettl3-induced m6A modification leads to increased TXNDC5 expression in CC cells. Mechanistically, Mettl3 inhibits the ER stress that triggers apoptosis and autophagy by targeting TXNDC5 (Du et al., 2022). Consequently, Mettl3 regulates PDK4 mRNA stability and translation prolongation in CC cells through 5′UTR, ultimately affecting glycolysis. The authors also found that IGF2BP3 and ALKBH5 are involved in regulating m6A expression in CC cells and are associated with CC cell growth and chemosensitivity (Li et al., 2020). Under hypoxic conditions, Mettl3/Mettl14 is also bound to DARS mRNA, enhancing its m6A modification and reinforcing its stability. This resulted in the upregulation of DARS expression and mediated hypoxia-induced autophagy in CC cells via the DARS-AS1/ATG5/ATG3 axis, strengthening the degree of cellular tolerance to the hypoxic environment and promoting cell survival (Shen et al., 2022a). Tumor metabolic reprogramming plays a pivotal role in the infinite proliferative growth of tumors, an extremely complex process that encompasses more than just the Warburg effect (DeBerardinis et al., 2008). In addition to the Warburg effect, the connections between the glutamine metabolic pathway, serine synthesis, oxidative phosphorylation, and pentose phosphate pathway with m6A methylation and Mettl3 need to be urgently investigated.
The m6A methylation regulation has an extensive and multifaceted impact on tumorigenesis. While Mettl3 plays a promoting role in CC, it also inhibits the progression of CC cells through multiple mechanisms. Ruyi Li et al. proved that Mettl3 exhibited the opposite effect in CC (Li et al., 2021a). This study showed that Mettl3 is highly upregulated in paracancerous tissues of CC and inhibited tumor cell proliferation and viability. In the Mettl3-overexpressed SiHa cells line, researchers found that deletion of Mettl3 can reduce the apoptosis rate of CC cells and lead to high expression of drug resistance-related proteins (MRP1 and LRP1). Meanwhile, Mettl3 inhibited the expression of the known oncogene RAGE, which has been confirmed to promote cell proliferation and inhibit apoptosis in CC cells, thereby increasing the sensitivity of CC cells to cisplatin (Li et al., 2021a). Also, Mettl3 regulates epigenetic silencing of miR-193b, which is associated with CC tumorigenesis. Further studies have proved that the tumor suppressor effect of miR-193b occurs through diminishing CCND1, a cell cycle regulator associated with cyclin-dependent kinase 4 or 6 (CDK4/6). The role of CDK4/6 in promoting cell cycle progression from the G0–G1 phase to the S and G2-M phases has been confirmed (Huang et al., 2021b).
Overall, high expression of Mettl3 in CC is associated with poor prognosis, but some studies have suggested that Mettl3 is beneficial in inhibiting tumor progression. The dual regulatory function of Mettl3 in CC poses an intriguing scientific inquiry, whether it arises from the heterogeneity of tumor cells, the distinctive biological properties of Mettl3, or other underlying factors. Although the function of Mettl3 in CC has been partially explored, the role of Mettl3 in CC is still complex; and it remains to be further explored by investigators in terms of deeper mechanisms.
The role of Mettl3 in ovarian cancer
OC, a silent killer, has the third highest incidence rate among all gynecologic malignancies; however, it has the highest mortality rate due to its extremely insidious early symptoms and often causes distant metastases when detected (Torre et al., 2018; Nameki et al., 2021). A woman has close to a one in 70 chance of developing OC during her lifespan, with an estimated 308,069 OC cases and 193,811 cases of OC mortality worldwide in 2020 (Smith, 2017). Therefore, early detection and effective treatment options for OC are imperative research areas that can significantly improve patient outcomes. Extensive investigations have demonstrated the crucial involvement of Mettl3 in OC pathogenesis, as it significantly promotes the growth and invasion of OC cells through various mechanisms, including modulation of the PI3K/AKT signaling pathway (Chang et al., 2021). Thus, thorough investigations of the molecular mechanisms underlying m6A methylation in ovarian tissues may uncover novel avenues for the diagnosis and treatment of OC.
The present studies found that Mettl3 functions as an oncogene in OC and is closely associated with various malignant behaviors of OC cells. Research indicates that the expression level of Mettl3 was markedly elevated in cancerous ovarian tissues compared to adjacent normal tissues, as demonstrated in a sample cohort of 75 OC patients. Highly expressed Mettl3 promotes miR-126-5p maturation by increasing the degree of m6A modification of pri-miR-126-5pand facilitates tumorigenesis and migration through the PTEN/PI3K/AKT3/mTOR axis (Table 2) (Bi et al., 2021a). In addition, Ma et al. collected tumors and adjacent tissues of 33 endometrioid epithelial ovarian cancer (EEOC) patients. Similarly, Mettl3 was found to be highly expressed in cancerous tissues, and overexpression of Mettl3 was an independent risk factor for higher tumor grade as well as for lower overall survival. The authors reconfirmed in vitro that Mettl3 could promote the proliferation, invasion, and migration of EEOC cells and it was not affected by the expression of Mettl14 and WTAP. Moreover, low expression of Mettl3 decreased the degree of m6A methylation of OC-related genes EIF3C, AXL, and CSF-1 (Ma et al., 2020). Importantly, Mettl3 also promotes the conversion of pre-miR-1246 to mature miR-1246 mRNA. In turn, miR-1246 targets CCNG2 and inhibits its expression, ultimately leading to OC cell proliferation, migration, and invasion; and inhibition of apoptosis (Bi et al., 2021b). It has been demonstrated that PLAA induces Mettl3 degradation by elevating the level of Mettl3 ubiquitination. Low levels of Mettl3 decreased the m6A methylation level of TRPC3 mRNA and inhibited TRPC3 expression, which resulted in lower intracellular calcium levels and suppressed OC cell metastasis (Shen et al., 2022b). Meanwhile, the Mettl3/YTHDF2 axis represses tumor suppressor gene IFFO1 expression and promotes mRNA degradation via an m6A methylation-dependent pathway, ultimately affecting the growth and development, metastasis and invasion, and cisplatin sensitivity of OC cells (Zhang et al., 2023).
TABLE 2.
The role of Mettl3 in ovarian cancer.
| Cancer species | Expression | Biological function | Mechanism | Reference |
|---|---|---|---|---|
| ovarian cancer | Up | Promote cell proliferation, migration, and invasion, and inhibit apoptosis | Promote miR-126-5p maturation by increasing the degree of m6A modification of pri-miR-126-5pmRNA and facilitate tumorigenesis and migration through the PTEN/PI3K/AKT3/mTOR axis | Bi et al. (2021a) |
| Up | Promote proliferation, migration, invasion, and inhibition of apoptosis | Promote the conversion of pre-miR-1246 to mature miR-1246 mRNA, and miR-1246 targets CCNG2 and inhibits its expression | Bi et al. (2021b) | |
| Up | Promote cell proliferation and migration, and inhibit cell apoptosis | As an independent factor affecting ovarian cancer prognosis and correlated with ovarian cancer-associated oncogene m6A enrichment | Ma et al. (2020) | |
| Up | Promote migration and invasion | Stabilize TRPC3 mRNA expression via m6A modification | Shen et al. (2022b) | |
| Up | Promote cell proliferation, tumor growth, and invasion | Stimulate AXL translation and EMT | Hua et al. (2018) | |
| Up | Inhibit cell apoptosis and promote the cell cycle | Increase the level of KRT8, and decrease the level of FAS | Yang et al. (2022) | |
| Up | Inhibit cell apoptosis and promote invasion | Increase activation of the AKT signaling pathway and the expression of the downstream effector Cyclin D1 | Liang et al. (2020b) | |
| Up | Promote cell growth and development, metastasis and invasion, and weaken cisplatin sensitivity | Mettl3/YTHDF2 axis represses tumor suppressor gene IFFO1 expression and promotes mRNA degradation | Zhang et al. (2023) |
Compared to normal epithelial cells, Mettl3 is highly expressed in OC cells and is significantly associated with a variety of adverse clinicopathological features. Mechanistically, the Mettl3-mediated increase in the receptor tyrosine kinase AXL translation promotes epithelial-mesenchymal transition (EMT), leading to cellular proliferation, migration, invasion, and tumor formation (Hua et al., 2018). Besides, it has been experimentally demonstrated that Mettl3 activates the AKT pathway in OC cells. The investigators found that low expression of Mettl3 may facilitate OC cell apoptosis through the mitochondrial apoptotic pathway and inhibit cancer cell invasion by reducing activation of the AKT signaling pathway and the expression of the downstream effector Cyclin D1 (Liang et al., 2020b). Like its role in CC, silencing Mettl3 in OC increases apoptosis and arrests the cell cycle in the G0/G1 phase (Yang et al., 2022).
In summary, all current studies suggest that high expression of Mettl3 in OC causes adverse biological behaviors and promotes tumor development. Regrettably, despite the promising role of Mettl3 as a therapeutic target in OC, no treatments targeting Mettl3 have been developed to date. Further exploration of the underlying mechanisms of Mettl3 in OC is warranted, and there is a need for the active design and development of Mettl3 inhibitors to provide potential benefits for patients.
The role of Mettl3 in endometrial cancer
EC, ranked among the three most prevalent gynecologic malignancies, was responsible for 41,700 newly diagnosed cases globally in 2020. Furthermore, its incidence has demonstrated a gradual increase over a 30-year period, with an overall rise of 132% (Gu et al., 2021; Sung et al., 2021). The current treatment plan for EC is mainly surgery combined with adjuvant therapies, such as radiotherapy, chemotherapy, targeted therapy, and hormone therapy (Aoki et al., 2020). ECs are often detected at an early stage, enabling a diagnosis and successful treatment. With a 5-year survival rate surpassing 95%, the prognosis for most early EC patients is optimistic (McMeekin, 2009). However, as the stage progresses, the 5-year survival rate decreases dramatically. Thus, there is an urgent need to investigate the molecular mechanisms of EC and optimize patient prognosis (Colombo et al., 2016). Previous studies have demonstrated that m6A regulatory proteins are implicated in multiple biological behaviors closely related to disease progression of EC, including proliferation, migration, and invasion, and they serve as key players in the activation of multiple signaling pathways (Li et al., 2021b; Zhang et al., 2021b; Ralser et al., 2022).
While there have been some studies investigating the role of Mettl3 in EC, the number of studies is relatively limited compared to other gynecological malignancies. This may be due to the status that endometrial cancer is less common than CC and OC, and the understanding of the molecular mechanisms underlying endometrial cancer is still incomplete. However, the studies conducted so far suggest that Mettl3 may play a key role in EC tumorigenesis and progression, and therefore further research is warranted. Liu et al. used immunohistochemical staining to compare tumor samples from patients with EC with adjacent tissues, and they found that Mettl3 expression was downregulated in tumor samples. Inhibited expression of Mettl3 was detected along with the loss-of-function mutation of Mettl14 and reduced m6A levels in EC tissues, and it was correlated with the proliferative effects of tumor cells (Liu et al., 2018b). Mechanistically, reduced m6A methylation affects multiple AKT pathway components by downregulating the AKT pathway negative regulator PHLPP2, upregulating the positive regulator mTORC2, and finally activating the AKT pathway. This ultimately promotes the tumorigenicity of EC cells and leads to disease progression (Table 3) (Liu et al., 2018b). Similarly, Ruan et al. demonstrated that Mettl3 is very closely related to activation of the AKT pathway. The authors significantly reduced m6A methylation by silencing Mettl3 expression in EC cell lines while recruiting YTHDF1 and YTHDF2 to regulate PAPPA and IGFBP4 at the translational level. Upregulated PAPPA further reduced the expression level of IGFBP4 by hydrolyzing IGFBP4, thereby decreasing the inhibition of IGF1, activating the AKT pathway, and eventually promoting the growth and development of EC cells (Ruan et al., 2022). Specifically, in EC, Mettl3 has been found to directly bind to the small nucleolar RNA SLERT and increase the m6A level of brain-derived neurotrophic factor (BDNF) mRNA. The m6A site on BDNF mRNA is then recognized and bound by the RNA-binding protein IGF2BP1, which enhances the stability of BDNF mRNA and subsequently activates BDNF/TRKB signaling. This activation induces epithelial-mesenchymal transition (EMT) and ultimately leads to lung metastasis of EC cells (Tian et al., 2023).
TABLE 3.
The role of Mettl3 in endometrial cancer.
| Cancer species | Expression | Biological function | Mechanism | Reference |
|---|---|---|---|---|
| endometrial cancer | Down | Promote cell proliferation and tumorigenicity | Decrease expression of the negative AKT regulator PHLPP2 and increase expression of the positive AKT regulator mTORC2 | Liu et al. (2018b) |
| Down | Promote cell proliferation and tumor formation | Reduce the expression level of IGFBP4, decrease the inhibition of IGF1, activae the AKT pathway | Ruan et al. (2022) | |
| Up | Promote cell migration and invasion, and induce EMT | Enhances the stability of BDNF mRNA and activates BDNF/TRKB signaling | Tian et al. (2023) |
Although evidence suggests that dysregulated m6A methylation is involved in the development and progression of EC, the exact association between m6A methylation and EC remains unclear (Zhang et al., 2021a). Interestingly, the level of m6A methylation, both elevated and reduced, played a cancer-promoting role in EC cells (Ruan et al., 2022; Tian et al., 2023). This phenomenon may result from differences in the genomic sequence profiles of different cell lines or perhaps from the diversity of m6A methylation’s functions. Exploring the relationship between the two factors may illuminate novel insights into the diagnosis and treatment of EC, ultimately benefitting a massive number of patients and enhancing the prognosis of the disease.
The clinical significance of METTL3 in gynecologic cancers
Mettl3 affects various biological behaviors of tumor cells by regulating m6A methylation, such as cell proliferation, apoptosis, invasion, and metastasis (Zeng et al., 2020). Additionally, Mettl3 can regulate tumor cell sensitivity to chemotherapy and immunotherapy, making it a potential target for cancer therapy (Xu and Ge, 2022). At the same time, the expression level of Mettl3 is closely related to the clinical outcome and therapeutic efficacy. Mettl3 is an independent factor affecting the survival of CC patients, especially DFS as well as OS, and it is closely associated with the disease stage (Ni et al., 2020; Wang et al., 2020d). Compared to patients with early-stage CC, Mettl3 expression levels were significantly higher in patients with advanced CC, suggesting a gradual increase in Mettl3 during the process of tumor malignancy (Hu et al., 2020; Shen et al., 2022a). The occurrence of CC is closely associated with HPV infection status, and previous studies have investigated the correlation between Mettl3 and HPV infection status, potentially offering a new avenue for the prevention, early diagnosis, and treatment of CC (Goodman, 2015; Yu et al., 2022). In addition, developing immune checkpoint inhibitors in recent years has significantly benefited cancer patients. A study by Yu et al. found that Mettl3 was correlated with the expression level of immune checkpoints; and it revealed that in mice, Mettl3 inhibitors combined with anti-PD-1 therapy could slow down tumor progression (Yu et al., 2022). This has given us a novel insight into the clinical treatment of patients with CC. Further, Mettl3 is also related to the chemosensitivity of cisplatin, and it might be possible to determine the chemotherapeutic dose of cisplatin by detecting the expression of Mettl3 in patients’ tumor tissues (Li et al., 2021a). In OC, the expression level of Mettl3 as an independent factor was similarly linked to patient prognosis and immunotherapy response (Tan et al., 2022). Mettl3, which is highly expressed in OC, is associated with the tumor grade, size of the tumor, lymph node metastasis, distant metastasis, FIGO stage, and overall survival rate (Hua et al., 2018; Liang et al., 2020b). Unfortunately, to date, no studies have pointed any significant association between Mettl3 expression and clinical features of EC. Therefore, further research is needed to explore the potential relationship between Mettl3 and the development and progression of EC.
Due to the broad involvement of m6A methylation in numerous stages of tumorigenesis, the regulatory proteins of m6A are increasingly being recognized as promising therapeutic targets (Boriack-Sjodin et al., 2018). According to the above-described summary, Mettl3 mainly plays an oncogenic role in gynecological malignancies, although there are a few reports of its tumor-inhibiting effects. The development of drugs targeting Mettl3 is still in the early stages, and at present, only Mettl3 inhibitors are available for experimental use. The recently revealed mechanism of Mettl3 inhibitor is a competitive binding of SAM, which can be categorized into nucleotide and non-nucleotide species. Nucleoside inhibitors were the first to be developed. Researchers used the AutoDuck program to screen 70 compounds from more than 4,000 compounds that could bind tightly to the SAM binding site through hydrogen bonding interactions on the NH group of the backbone (Morris et al., 2009). Subsequently, seven compounds were obtained from these 70 compounds by the HTRF method and detection of reaction products, and an adenosine derivative compound was found to have effective inhibitory activity against Mettl3 as well as potent pairing efficiency (Wiedmer et al., 2019; Bedi et al., 2020). However, due to the disadvantages of nucleoside inhibitors, such as weak cell permeability and poor selectivity for other methyltransferases, attention has shifted towards non-nucleoside inhibitors (Stein et al., 2018). Innovative material disciplines are developing rapidly, and polymeric nanomaterials are being used in a range of applications, including medicine. The polymer nanometer inhibitor UZH1a binds to the adenosine portion of SAM by van der Waals forces and hydrogen bonding forces formed with the polar group on Mettl3 (Zhao et al., 2020b). Compared to nucleoside inhibitors, UZH1a not only has high selectivity and better targeting performance, but it also has excellent physicochemical properties. In addition, Caflisch et al. optimized UZH1a and developed a small molecule inhibitor with superior properties, UZH2 (Dolbois et al., 2021). Further, STM2475, the first-in-class non-nucleoside Mettl3 inhibitor, has demonstrated its efficacy in the treatment of malignant tumors in vitro, which is an exciting achievement in the field of Mettl3 research (Yankova et al., 2021). However, the current research on Mettl3 inhibitors is mostly focused on hematological malignancies, and there are no research reports of Mettl3 inhibitors in gynecologic tumors; thus, further investigation is warranted. While, with ongoing research, it is possible that other types of drugs targeting Mettl3 could be developed in the future.
Conclusion and perspectives
Cancer is a daunting global health challenge, and its unique biological behaviors, such as infinite proliferation, immune escape, angiogenesis, and individual tumor microenvironment, have resulted in numerous difficulties in its diagnosis and treatment (Hanahan and Weinberg, 2011). M6A methylation, the most prevalent type of mRNA modification in eukaryotic, is a ubiquitous modification of mRNA involved in nearly every aspect of mRNA metabolism, including translation, maturation, degradation, and folding (Liu and Gregory, 2019). It has been well established that m6A methylation is involved in many biological behaviors associated with cancer, and it plays a vital role in these biological behaviors, including promoting tumor cell proliferation, cell viability, invasion, and metastasis; regulating differentiation; and inhibiting apoptosis (He et al., 2019; Ma et al., 2019). The process of m6A methylation is regulated by three types of regulators: “writers,” “erasers,” and “readers,” which interact with one another to achieve precise control of m6A modification. The Mettl3 catalytic subunit is the main component of the writer, and it plays a unique catalytic role (Chen et al., 2019). This article reviewed new developments in the study of Mettl3 in gynecologic malignancies in recent years, with a focus on cell proliferation, invasion, migration, and activation of cancer-related pathways (Zhang and Liu, 2022). In addition to directly regulating cancer cell biological behavior, Mettl3 has been found to be correlated with HPV infection status, an established causative factor of CC (Yu et al., 2022). Further, the Warburg effect, a characteristic of malignant cells, has likewise been shown to be profoundly influenced by Mettl3 (Wang et al., 2020d; Du et al., 2022). Mettl3 is also involved in several specific biological behaviors of OC and EC. Given the extensive and diverse roles played by Mettl3 in gynecologic malignancies, further investigation into its potential mechanisms in tumor progression is warranted to identify novel therapeutic targets that could offer hope to cancer patients globally.
Although this manuscript focuses on the role of Mettl3 in gynecologic malignancies, m6A methylation is a dynamically regulated process, and other regulatory proteins’ functions are equally important (Lan et al., 2019). Notably, the coexistence of Mettl3 in multiple types of cancer with diametrically opposed effects on tumor progression is a question that deserves thoughtful consideration. For example, in CC, Fei Ji et al. found that Mettl3 could promote the development of CC. At the same time, the results presented by Ruyi Li et al. suggested that Mettl3 could inhibit the action of oncogenes (Li et al., 2021a; Ji et al., 2021). These findings suggest the importance of investigating specific regulatory proteins and their interactions when studying m6A methylation. The heterogeneity of the malignancy may be responsible for this phenomenon and is often the culprit of clinical treatment failure. This observation indicates the need to transition towards personalized and precise approaches in both basic research and clinical management in the future. Although sporadic Mettl3 inhibitor studies have been published, there is still a gap in its application in gynecologic malignancies. More in-depth studies are required to investigate the specific role played by Mettl3 in malignancies, and it may shed light on new therapeutic approaches for gynecologic tumors.
In summary, the m6A methylation process, regulated by a writer-eraser-reader complex, plays a significant role in gynecologic malignancies. Among the regulatory proteins, Mettl3, a writer, has been found to have multiple roles and extensively affects various biological behaviors of tumor cells. Its dysregulation has been associated with oncogenic transformation, metastasis, chemoresistance, and poor prognosis in gynecologic malignancies. However, current research on Mettl3’s role in tumors may only scratch the surface, and the development of drugs targeting Mettl3 is still in its infancy; there is a need to study its mechanism of action in depth and work on designing new treatments to provide more benefits to a great quantity of patients with gynecologic malignancies.
Funding Statement
This study was supported by the Liaoning Cancer Hospital and Institute (Shenyang) and China Medical University (Shenyang). And this work was funded by National Natural Science Foundation Incubation Project of Liaoning Cancer Hospital (No. 2021-ZLLH-02 and 2021-ZLLH-17), Natural Science Foundation of Liaoning Province (No. 2022-MS-084).
Author contributions
Original draft preparation, allocation, supplementation, and editing: YZ. Revision: NZ and YZ. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- An Y., Duan H. (2022). The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21 (1), 14. 10.1186/S12943-022-01500-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Kanao H., Wang X., Yunokawa M., Omatsu K., Fusegi A., et al. (2020). Adjuvant treatment of endometrial cancer today. Jpn. J. Clin. Oncol. 50 (7), 753–765. 10.1093/JJCO/HYAA071 [DOI] [PubMed] [Google Scholar]
- Barillot I., Haie-Meder C. (2009). Association of radiotherapy and chemotherapy: Targeted therapies in gynecological cancers. Bull. Cancer 96 (3), 271–283. 10.1684/BDC.2009.0833 [DOI] [PubMed] [Google Scholar]
- Bedi R. K., Huang D., Eberle S. A., Wiedmer L., Śledź P., Caflisch A. (2020). Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem 15 (9), 744–748. 10.1002/CMDC.202000011 [DOI] [PubMed] [Google Scholar]
- Bi X., Lv X., Liu D., Guo H., Yao G., Wang L., et al. (2021). METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 28 (3–4), 335–349. 10.1038/S41417-020-00222-3 [DOI] [PubMed] [Google Scholar]
- Bi X., Lv X., Liu D., Guo H., Yao G., Wang L., et al. (2021). METTL3 promotes the initiation and metastasis of ovarian cancer by inhibiting CCNG2 expression via promoting the maturation of pri-microRNA-1246. Cell Death Discov. 7 (1), 237. 10.1038/S41420-021-00600-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin P. A., Ribich S., Copeland R. A. (2018). RNA-modifying proteins as anticancer drug targets. Nat. Rev. Drug Discov. 17 (6), 435–453. 10.1038/NRD.2018.71 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 (6), 394–424. 10.3322/CAAC.21492 [DOI] [PubMed] [Google Scholar]
- Burd E. M. (2003). Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16 (1), 1–17. 10.1128/CMR.16.1.1-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Wang X., Cao C., Gao Y., Zhang S., Yang Z., et al. (2018). HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 415, 11–19. 10.1016/J.CANLET.2017.11.018 [DOI] [PubMed] [Google Scholar]
- Chang L. L., Xu X. Q., Liu X. L., Guo Q. Q., Fan Y. N., He B. X., et al. (2021). Emerging role of m6A methylation modification in ovarian cancer. Cancer Cell Int. 21 (1), 663. 10.1186/S12935-021-02371-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wei L., Law C. T., Tsang F. H. C., Shen J., Cheng C. L. H., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67 (6), 2254–2270. 10.1002/HEP.29683 [DOI] [PubMed] [Google Scholar]
- Chen X. Y., Zhang J., Zhu J. S. (2019). The role of m6A RNA methylation in human cancer. Mol. Cancer 18 (1), 103. 10.1186/S12943-019-1033-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Wang C., Lei X., Huang T., Zhou R., Lu Y. (2022). Immune cytolytic activity for comprehensive insights of the immune landscape in endometrial carcinoma. J. Oncol. 2022, 9060243. 10.1155/2022/9060243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. A., Jhingran A., Oaknin A., Denny L. (2019). Cervical cancer. Lancet 393 (10167), 169–182. 10.1016/S0140-6736(18)32470-X [DOI] [PubMed] [Google Scholar]
- Colombo N., Creutzberg C., Amant F., Bosse T., Gonzalez-Martin A., Ledermann J., et al. (2016). ESMO-ESGO-ESTRO consensus conference on endometrial cancer: Diagnosis, treatment and follow-up. Ann. Oncol. 27 (1), 16–41. 10.1093/ANNONC/MDV484 [DOI] [PubMed] [Google Scholar]
- Condic M., Thiesler T., Staerk C., Klumper N., Ellinger J., Egger E. K., et al. (2022). N6-methyladenosine RNA modification (m6A) is of prognostic value in HPV-dependent vulvar squamous cell carcinoma. BMC Cancer 22 (1), 943. 10.1186/S12885-022-10010-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008). The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7 (1), 11–20. 10.1016/J.CMET.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Deng L. J., Deng W. Q., Fan S. R., Chen M. F., Qi M., Lyu W. Y., et al. (2022). m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol. Cancer 21 (1), 52. 10.1186/S12943-022-01510-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. 10.1073/PNAS.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolbois A., Bedi R. K., Bochenkova E., Muller A., Moroz-Omori E. V., Huang D., et al. (2021). 1,4,9-Triazaspiro[5.5]undecan-2-one derivatives as potent and selective METTL3 inhibitors. J. Med. Chem. 64 (17), 12738–12760. 10.1021/ACS.JMEDCHEM.1C00773 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. 10.1038/NATURE11112 [DOI] [PubMed] [Google Scholar]
- Du Q. Y., Huo F. C., Du W. Q., Sun X. L., Jiang X., Zhang L. S., et al. (2022). METTL3 potentiates progression of cervical cancer by suppressing ER stress via regulating m6A modification of TXNDC5 mRNA. Oncogene 41 (39), 4420–4432. 10.1038/S41388-022-02435-2 [DOI] [PubMed] [Google Scholar]
- Dueñas-González A., Lizano M., Candelaria M., Cetina L., Arce C., Cervera E. (2005). Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol. Cancer 4, 38. 10.1186/1476-4598-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Mei W., Qu C., Lu J., Shang L., Cao F., et al. (2022). Role of m6A writers, erasers and readers in cancer. Exp. Hematol. Oncol. 11 (1), 45. 10.1186/S40164-022-00298-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A. (2015). HPV testing as a screen for cervical cancer. BMJ 350, h2372. 10.1136/BMJ.H2372 [DOI] [PubMed] [Google Scholar]
- Gu B., Shang X., Yan M., Li X., Wang W., Wang Q., et al. (2021). Variations in incidence and mortality rates of endometrial cancer at the global, regional, and national levels, 1990-2019. Gynecol. Oncol. 161 (2), 573–580. 10.1016/J.YGYNO.2021.01.036 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: The next generation. Cell 144 (5), 646–674. 10.1016/J.CELL.2011.02.013 [DOI] [PubMed] [Google Scholar]
- He L., Li H., Wu A., Peng Y., Shu G., Yin G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18 (1), 176. 10.1186/S12943-019-1109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden M. G. V., Cantley L. C., Thompson C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324 (5930), 1029–1033. 10.1126/SCIENCE.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Motorin Y. (2017). Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 18 (5), 275–291. 10.1038/NRG.2016.169 [DOI] [PubMed] [Google Scholar]
- Hu Y., Li Y., Huang Y., Jin Z., Wang C., Wang H., et al. (2020). METTL3 regulates the malignancy of cervical cancer via post-transcriptional regulation of RAB2B. Eur. J. Pharmacol. 879, 173134. 10.1016/J.EJPHAR.2020.173134 [DOI] [PubMed] [Google Scholar]
- Hua W., Zhao Y., Jin X., Yu D., He J., Xie D., et al. (2018). METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol. Oncol. 151 (2), 356–365. 10.1016/J.YGYNO.2018.09.015 [DOI] [PubMed] [Google Scholar]
- Huang J., Dong X., Gong Z., Qin L. Y., Yang S., Zhu Y. L., et al. (2019). Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase. Protein Cell 10 (4), 272–284. 10.1007/S13238-018-0518-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Chen T. Q., Fang K., Zeng Z. C., Ye H., Chen Y. Q. (2021). N6-methyladenosine methyltransferases: Functions, regulation, and clinical potential. J. Hematol. Oncol. 14 (1), 117. 10.1186/S13045-021-01129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Liang J., Lin S., Wang D., Xie Q., Lin Z., et al. (2021). N6-Methyladenosine associated silencing of miR-193b promotes cervical cancer aggressiveness by targeting CCND1. Front. Oncol. 11, 666597–672021. 10.3389/FONC.2021.666597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Kong F., Li R., Chen X., Wang K. (2022). Emerging roles of m6A RNA methylation regulators in gynecological cancer. Front. Oncol. 12, 827956. 10.3389/FONC.2022.827956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011). Global cancer statistics. CA Cancer J. Clin. 61 (2), 69–90. 10.3322/CAAC.20107 [DOI] [PubMed] [Google Scholar]
- Ji F., Lu Y., Chen S., Lin X., Yu Y., Zhu Y., et al. (2021). m6A methyltransferase METTL3-mediated lncRNA FOXD2-AS1 promotes the tumorigenesis of cervical cancer. Mol. Ther. Oncolytics 22, 574–581. 10.1016/J.OMTO.2021.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Zhang J. A. A., Liu H., Li K., Wang Z. W., Zhu X. (2022). Comprehensive characterization of tumor microenvironment and m6A RNA methylation regulators and its effects on PD-L1 and immune infiltrates in cervical cancer. Front. Immunol. 13, 976107. 10.3389/FIMMU.2022.976107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q., Liu P. Y., Haase J., Bell J. L., Huttelmaier S., Liu T. (2019). The critical role of RNA m6A methylation in cancer. Cancer Res. 79 (7), 1285–1292. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- Li Z., Peng Y., Li J., Chen Z., Chen F., Tu J., et al. (2020). N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 11 (1), 2578. 10.1038/S41467-020-16306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Song Y., Chen X., Chu M., wei Wang Z., Zhu X. (2021). METTL3 increases cisplatin chemosensitivity of cervical cancer cells via downregulation of the activity of RAGE. Mol. Ther. Oncolytics 22, 245–255. 10.1016/J.OMTO.2021.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang C., Dong W., Su Y., Ma Z. (2021). WTAP facilitates progression of endometrial cancer via CAV-1/NF-κB axis. Cell Biol. Int. 45 (6), 1269–1277. 10.1002/CBIN.11570 [DOI] [PubMed] [Google Scholar]
- Li H., Zhong Y., Cao G., Shi H., Liu Y., Li L., et al. (2022). METTL3 promotes cell cycle progression via m6A/YTHDF1-dependent regulation of CDC25B translation. Int. J. Biol. Sci. 18 (8), 3223–3236. 10.7150/IJBS.70335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Zhan G., Chang K. J., Yang Y. P., Wang L., Lin J., et al. (2020). The roles of m6A RNA modifiers in human cancer. J. Chin. Med. Assoc. 83 (3), 221–226. 10.1097/JCMA.0000000000000251 [DOI] [PubMed] [Google Scholar]
- Liang S., Guan H., Lin X., Li N., Geng F., Li J. (2020). METTL3 serves an oncogenic role in human ovarian cancer cells partially via the AKT signaling pathway. Oncol. Lett. 19 (4), 3197–3204. 10.3892/OL.2020.11425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti M. v., Locasale J. W. (2016). The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 41 (3), 211–218. 10.1016/J.TIBS.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M. C., Chang S. J., Park B., Yoo H. J., Yoo C. W., Nam B. H., et al. (2022). Survival after hyperthermic intraperitoneal chemotherapy and primary or interval cytoreductive surgery in ovarian cancer: A randomized clinical trial. JAMA Surg. 157 (5), 374–383. 10.1001/JAMASURG.2022.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Gregory R. I. (2019). RNAmod: An integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 47, W548–W555. 10.1093/NAR/GKZ479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. X., Li L. M., Sun H. L., Liu S. M. (2018). Link between m6A modification and cancers. Front. Bioeng. Biotechnol. 6, 89. 10.3389/FBIOE.2018.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Eckert M. A., Harada B. T., Liu S. M., Lu Z., Yu K., et al. (2018). m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 20 (9), 1074–1083. 10.1038/S41556-018-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Yang M. (2021). The correlation between high-risk HPV infection and precancerous lesions and cervical cancer. Am. J. Transl. Res. 13 (9), 10830–10836. Available at: https://pubmed.ncbi.nlm.nih.gov/34650762/ (Accessed Jan 18, 2023). [PMC free article] [PubMed] [Google Scholar]
- Ma S., Chen C., Ji X., Liu J., Zhou Q., Wang G., et al. (2019). The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 12 (1), 121. 10.1186/S13045-019-0805-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Li Q., Liu P., Dong W., Zuo Y. (2020). METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP. Cell Biol. Int. 44 (12), 2524–2531. 10.1002/CBIN.11459 [DOI] [PubMed] [Google Scholar]
- Makker V., Taylor M. H., Aghajanian C., Oaknin A., Mier J., Cohn A. L., et al. (2020). Lenvatinib plus pembrolizumab in patients with advanced endometrial cancer. J. Clin. Oncol. 38 (26), 2981–2992. 10.1200/JCO.19.02627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeekin D. S. (2009). Where is the future of endometrial cancer therapy? Ann. Oncol. 20 (11), 1757–1761. 10.1093/ANNONC/MDP493 [DOI] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149 (7), 1635–1646. 10.1016/J.CELL.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30 (16), 2785–2791. 10.1002/JCC.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki R., Chang H., Reddy J., Corona R. I., Lawrenson K. (2021). Transcription factors in epithelial ovarian cancer: Histotype-specific drivers and novel therapeutic targets. Pharmacol. Ther. 220, 107722. 10.1016/J.PHARMTHERA.2020.107722 [DOI] [PubMed] [Google Scholar]
- Nebbioso A., Tambaro F. P., Dell’Aversana C., Altucci L. (2018). Cancer epigenetics: Moving forward. PLoS Genet. 14 (6), e1007362. 10.1371/JOURNAL.PGEN.1007362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H. H., Zhang L., Huang H., qin Dai S., Li J. (2020). Connecting METTL3 and intratumoural CD33+ MDSCs in predicting clinical outcome in cervical cancer. J. Transl. Med. 18 (1), 393. 10.1186/S12967-020-02553-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova N. N., Thompson C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23 (1), 27–47. 10.1016/J.CMET.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello N., Sun S., Wong J. J. L. (2018). Aberrant expression of enzymes regulating m6A mRNA methylation: Implication in cancer. Cancer Biol. Med. 15 (4), 323–334. 10.20892/J.ISSN.2095-3941.2018.0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser D. J., Condic M., Klumper N., Ellinger J., Staerk C., Egger E. K., et al. (2022). Comprehensive immunohistochemical analysis of N6-methyladenosine (m6A) writers, erasers, and readers in endometrial cancer. J. Cancer Res. Clin. Oncol. 10.1007/S00432-022-04083-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan P., Wang S., Yang C., Huang X., Sun P., Tan A. (2022). m6A mRNA methylation regulates the ERK/NF-κB/AKT signaling pathway through the PAPPA/IGFBP4 axis to promote proliferation and tumor formation in endometrial cancer. Cell Biol. Toxicol. 10.1007/S10565-022-09751-Z [DOI] [PubMed] [Google Scholar]
- See Y. X., Wang B. Z., Fullwood M. J. (2019). Chromatin interactions and regulatory elements in cancer: From bench to bedside. Trends Genet. 35 (2), 145–158. 10.1016/J.TIG.2018.11.007 [DOI] [PubMed] [Google Scholar]
- Shen W., Zhu M., Wang Q., Zhou X., Wang J., Wang T., et al. (2022). DARS-AS1 recruits METTL3/METTL14 to bind and enhance DARS mRNA m6A modification and translation for cytoprotective autophagy in cervical cancer. RNA Biol. 19 (1), 751–763. 10.1080/15476286.2022.2079889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Gu L., Liu Y., Wang L., Zhu J., Tang S., et al. (2022). PLAA suppresses ovarian cancer metastasis via METTL3-mediated m6A modification of TRPC3 mRNA. Oncogene 41 (35), 4145–4158. 10.1038/S41388-022-02411-W [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P., Jinek M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, e18434. 10.7554/ELIFE.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. G. (2017). A resident’s perspective of ovarian cancer. Diagn. (Basel) 7 (2), 24. 10.3390/DIAGNOSTICS7020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer N. Y., Stanton R. C. (2019). The Warburg effect, lactate, and nearly a century of trying to cure cancer. Semin. Nephrol. 39 (4), 380–393. 10.1016/J.SEMNEPHROL.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Stein E. M., Garcia-Manero G., Rizzieri D. A., Tibes R., Berdeja J. G., Savona M. R., et al. (2018). The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131 (24), 2661–2669. 10.1182/BLOOD-2017-12-818948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C., Zhang Y., Chen P., Yang W., Du J., Zhang D. (2022). Methyltransferase-like 3 induces the development of cervical cancer by enhancing insulin-like growth factor 2 mRNA-binding proteins 3-mediated apoptotic chromatin condensation inducer 1 mRNA stability. Bioengineered 13 (3), 7034–7048. 10.1080/21655979.2022.2044261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. 10.3322/CAAC.21660 [DOI] [PubMed] [Google Scholar]
- Tan W., Liu S., Deng Z., Dai F., Yuan M., Hu W., et al. (2022). Gene signature of m6A-related targets to predict prognosis and immunotherapy response in ovarian cancer. J. Cancer Res. Clin. Oncol. 149, 593–608. 10.1007/S00432-022-04162-3 [DOI] [PubMed] [Google Scholar]
- Tian J., Cheng H., Wang N., Wang C. (2023). SLERT, as a novel biomarker, orchestrates endometrial cancer metastasis via regulation of BDNF/TRKB signaling. World J. Surg. Oncol. 21 (1), 27. 10.1186/S12957-022-02821-W [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L. A., Trabert B., DeSantis C. E., Miller K. D., Samimi G., Runowicz C. D., et al. (2018). Ovarian cancer statistics, 2018. CA Cancer J. Clin. 68 (4), 284–296. 10.3322/CAAC.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Doxtader K. A., Nam Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. 10.1016/J.MOLCEL.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. 10.1038/NATURE18298 [DOI] [PubMed] [Google Scholar]
- Wang H., Deng Q., Lv Z., Ling Y., Hou X., Chen Z., et al. (2019). N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer 18 (1), 181. 10.1186/S12943-019-1108-X [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Q., Peng H., Qi X., Wu M., Zhao X. (2020). Targeted therapies in gynecological cancers: A comprehensive review of clinical evidence. Signal Transduct. Target Ther. 5 (1). 10.1038/S41392-020-0199-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Kong S., Tao M., Ju S. (2020). The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 19 (1), 88. 10.1186/S12943-020-01204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Geng W., Guo H., Wang Z., Xu K., Chen C., et al. (2020). Emerging role of RNA methyltransferase METTL3 in gastrointestinal cancer. J. Hematol. Oncol. 13 (1), 57. 10.1186/S13045-020-00895-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Guo X., Li L., Gao Z., Su X., Ji M., et al. (2020). N6-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 11 (10), 911. 10.1038/S41419-020-03071-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmer L., Eberle S. A., Bedi R. K., Śledź P., Caflisch A. (2019). A reader-based assay for m6A writers and erasers. Anal. Chem. 91 (4), 3078–3084. 10.1021/ACS.ANALCHEM.8B05500 [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang Y., Fang Y., Ma S., Zheng H., Liu K., et al. (2020). Elevated expression of inhibitor of apoptosis-stimulating protein of p53 (iASPP) and methyltransferase-like 3 (METTL3) correlate with poor prognosis in FIGO ib1-IIa squamous cell cervical cancer. J. Cancer 11 (9), 2382–2389. 10.7150/JCA.41029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Ge R. (2022). Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur. J. Med. Chem. 230, 114118. 10.1016/J.EJMECH.2022.114118 [DOI] [PubMed] [Google Scholar]
- Yang Z., Ma J., Han S., Li X., Guo H., Liu D. (2020). ZFAS1 exerts an oncogenic role via suppressing miR-647 in an m6A-dependent manner in cervical cancer. Onco Targets Ther. 13, 11795–11806. 10.2147/OTT.S274492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Chen X., Qian X., Zhang J., Wu M., Yu A. (2022). Comprehensive analysis of the transcriptome-wide m6A methylome in endometrioid ovarian cancer. Front. Oncol. 12, 844613. 10.3389/FONC.2022.844613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankova E., Blackaby W., Albertella M., Rak J., De Braekeleer E., Tsagkogeorga G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593 (7860), 597–601. 10.1038/S41586-021-03536-W [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y. C., Chen X. Y., Zhang J., Zhu J. S. (2020). Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol. Cancer 19 (1), 121. 10.1186/S12943-020-01233-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R., Wei Y., He C., Zhou P., Yang H., Deng C., et al. (2022). Integrative analyses of m6A regulators identify that METTL3 is associated with HPV status and immunosuppressive microenvironment in HPV-related cancers. Int. J. Biol. Sci. 18 (9), 3874–3887. 10.7150/IJBS.70674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Huang W., Li Y., Weng H. (2020). Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 13 (1), 117. 10.1186/S13045-020-00951-W [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Liu N. (2022). N6-methyladenosine (m6A) modification in gynecological malignancies. J. Cell Physiol. 237 (9), 3465–3479. 10.1002/JCP.30828 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wan Y., Zhang Z., Jiang Y., Gu Z., Ma X., et al. (2021). IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics 11 (3), 1100–1114. 10.7150/THNO.49345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wan Y., Zhang Z., Jiang Y., Lang J., Cheng W., et al. (2021). FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signalling pathway. RNA Biol. 18 (9), 1265–1278. 10.1080/15476286.2020.1841458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qiu J. G., Jia X. Y., Ke Y., Zhang M. K., Stieg D., et al. (2023). METTL3-mediated N6-methyladenosine modification and HDAC5/YY1 promote IFFO1 downregulation in tumor development and chemo-resistance. Cancer Lett. 553, 215971. 10.1016/J.CANLET.2022.215971 [DOI] [PubMed] [Google Scholar]
- Zhao J., Lee E. E., Kim J., Yang R., Chamseddin B., Ni C., et al. (2019). Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 10 (1), 2300. 10.1038/S41467-019-10246-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Qi X., Liu L., Liu Z., Ma S., Wu J. (2020). Epigenetic regulation of m6A modifications in human cancer. Mol. Ther. Nucleic Acids 19, 405–412. 10.1016/J.OMTN.2019.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Meng J., Su R., Zhang J., Chen J., Ma X., et al. (2020). Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J. Hepatol. 73 (3), 664–679. 10.1016/J.JHEP.2020.04.009 [DOI] [PubMed] [Google Scholar]
- Zhou S., Treloar A. E., Lupien M. (2016). Emergence of the noncoding cancer genome: A target of genetic and epigenetic alterations. Cancer Discov. 6 (11), 1215–1229. 10.1158/2159-8290.CD-16-0745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Wang J. Z., Wei J. F., Lu C. (2021). Role of m6A methyltransferase component VIRMA in multiple human cancers (Review). Cancer Cell Int. 21 (1), 172. 10.1186/S12935-021-01868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]