Summary
SUMOylation is an evolutionarily conserved eukaryotic posttranslational protein modification with broad biological relevance. Differentiating between the major small ubiquitin-like modifier (SUMO) paralogs and uncovering paralog-specific functions in vivo has long been very difficult. To overcome this problem, we generated His6-HA-Sumo2 and HA-Sumo2 knockin mouse lines, expanding upon our existing His6-HA-Sumo1 mouse line, to establish a “toolbox” for Sumo1-Sumo2 comparisons in vivo. Leveraging the specificity of the HA epitope, we performed whole-brain imaging and uncovered regional differences between Sumo1 and Sumo2 expression. At the subcellular level, Sumo2 was specifically detected in extranuclear compartments, including synapses. Immunoprecipitation coupled with mass spectrometry identified shared and specific neuronal targets of Sumo1 and Sumo2. Target validation using proximity ligation assays provided further insight into the subcellular distribution of neuronal Sumo2-conjugates. The mouse models and associated datasets provide a powerful framework to determine the native SUMO “code” in cells of the central nervous system.
Subject areas: Imaging anatomy, Molecular biology, Neuroscience
Graphical abstract
Highlights
-
•
(His6-)HA-Sumo2 knockin mice enable direct comparison of Sumo paralogs in vivo
-
•
Whole-brain imaging reveals regional differences between Sumo1 and Sumo2 expression
-
•
Sumo2 localizes to synapses in vivo
-
•
Sumo1 and Sumo2 have shared unique targets in the mouse brain
Imaging anatomy; Molecular biology; Neuroscience
Introduction
Posttranslational modifications (PTMs) can alter the secondary, tertiary, and quaternary structure of proteins and thereby dynamically regulate their function. Although many PTMs involve the covalent conjugation of small moieties, such as phosphate, glycosyl, or acetyl groups, protein function is also modulated by reversible covalent conjugation of large molecules, including the protein ubiquitin and ubiquitin-like modifiers, such as NEDD8, or small ubiquitin-like modifiers (SUMOs).1,2,3,4,5,6,7
Conjugation of SUMOs (“Sumo” in mice) through SUMOylation is a dynamic and essential process that has been conserved throughout eukaryote evolution.1,2 In its course, immature SUMOs are first matured by sentrin-specific protease (SENP) cleavage at a C-terminal diglycine motif that is essential for conjugation.3 Next, SUMOs are activated in an ATP-dependent manner and conjugated to the E1 ligase heterodimer consisting of SUMO-activating enzyme 1 (SAE1) and ubiquitin-like modifier activating enzyme 2 (UBA2).4 SUMOs are then transferred to the sole E2 ligase of the system, SUMO-conjugating enzyme UBC9 (UBC9, encoded by UBE2I), which is essential for SUMOylation.5,6,7,8,9,10 SUMOylation occurs at lysine residues of target substrates, typically in SUMO consensus motifs (Ψ-K-x-D/E; Ψ, large hydrophobic), either solely via UBC9 or upon facilitation by E3 ligases.11,12 SENPs readily deSUMOylate substrates, thus recycling SUMOs for subsequent conjugation rounds and allowing for highly dynamic substrate regulation.13
There are five human SUMO paralogs (SUMO1–SUMO5), of which SUMO1–SUMO3 have been studied most extensively as they are ubiquitously expressed and found in most vertebrates, whereas SUMO4 and SUMO5 are specific to humans and exhibit tissue-specific expression.14,15 SUMO1 is ∼50% homologous to SUMO2 and SUMO3, whereas SUMO2 and SUMO3 are 97% homologous and often referred to together as SUMO2/3.16 While SUMO paralogs have multiple redundant roles and can mutually compensate paralog loss in several contexts, each paralog also plays unique roles, e.g. by localizing to different distinct subcellular regions and by targeting different substrates.2,14 In addition, SUMO2, but not SUMO1 or SUMO3, is the only essential paralog so that its knockout in mice causes major developmental defects and embryonic lethality by E10.5.17 The essentiality of SUMO2 for proper development may be in part due to higher levels of SUMO2 in comparison to its paralogs,17,18 but is likely also a result of a specific subset of SUMO2 substrates in cells. Thus, characterizing the specific roles of SUMO2 is crucial to uncover the key molecular and cellular processes that define SUMO biology and the mechanisms by which SUMOylation affects development and disease processes such as neurodegeneration or the pathological consequences of ischemic stroke.19,20,21,22,23
Several key challenges have limited studies in SUMO biology: (i) Differentiating between SUMO paralogs using antibodies is very difficult or even impossible due to the high degree of homology24; (ii) the highly dynamic nature of SUMOylation requires consideration of biological context, e.g. tissue, cell type, or cell state, for capturing and characterizing SUMOylation events, particularly in vivo; (iii) many tools and model systems used to study SUMOylation are limited, particularly when they rely on overexpression approaches, where off-target SUMOylation is a major potential confound. Consequently, protein SUMOylation has chiefly been studied in highly proliferating cells, where SUMOylation is now well known to control multiple nuclear processes, such as DNA repair or transcriptional control.19,25 In contrast, the function of SUMOylation in extranuclear compartments, post-mitotic cells, or complex tissues is far less well understood. The role of extranuclear SUMOylation in post-mitotic neurons is a prime example in this regard, with major controversies surrounding evidence supporting26,27,28,29,30,31 and opposing32,33,34 a key role of SUMO1 conjugation in synaptic compartments.
Our strategy to overcome some of the technical limitations in studying SUMO biology has been to generate knockin mouse models that express endogenously tagged Sumo, allowing for stringent approaches that leverage the specificity and affinity of epitope tags to identify Sumo-targets and characterize their subcellular localization, using wild-type littermates as negative controls and flanked by studies on Sumo knockout mice. The first allele we generated—a His6-HA-Sumo1 mouse—overcame what we viewed as technical limitations of previous studies on the role of Sumo1 in neurons, and surprisingly did not provide evidence for the localization of Sumo1 at synapses.33,34 While the His6-HA-Sumo1 model facilitates the study of Sumo1 in vivo, expanding the toolkit to other Sumo paralogs will provide powerful opportunities to uncover the complexities of Sumo-mediated regulation.
We have now expanded our mouse genetic tool-generation efforts in SUMO biology to promote further studies on the role of SUMOylation in vivo and in complex cellular systems such as neurons. Specifically, we generated and characterized in parallel two novel knockin mouse models that express either an HA-Sumo2 or a His6-HA-Sumo2 fusion protein from the endogenous Sumo2 locus to characterize Sumo2 and its substrates in vivo. Analogous to our work with the corresponding His6-HA-Sumo1 mouse,32,33,34 these knockin mouse lines allow to directly compare the localization and conjugation patterns of Sumo1 and Sumo2, which we did for the central nervous system (CNS). We used whole-brain imaging to map the relative abundance of Sumo1 and Sumo2 in the adult mouse brain and observed that both are broadly distributed in the CNS but exhibit defined regional differences. Importantly, we found that Sumo2, unlike Sumo1,32,33,34 is present in both nuclear and extranuclear compartments, the latter including neuronal synapses. Given this distinct subcellular distribution, we explored native Sumo1 and Sumo2 targets using anti-tag immunoaffinity purification from knockin mice and mass spectrometry. We identified genuine Sumo2-specific targets in the adult mouse brain, including a subset of non-nuclear proteins. Moreover, we observed dynamic changes between Sumo1- and Sumo2-selective substrates. Finally, we validated several hits using proximity ligations assays to detect protein-Sumo interactions in cultured cortical neurons. This approach did not only provide an additional mode of validating native Sumo2 interactors in wild-type neurons but also yielded spatial information, again demonstrating both nuclear and cytoplasmic interactions between Sumo2 and its substrates. Together, the two knockin mouse lines and our data provide an important resource that lays the foundation of a “SUMO-code” as an additional layer of molecular complexity in brain function.
Results
Generation of (His6-)HA-Sumo2 mouse lines
The use of epitope tags provides a versatile way to streamline the study of proteins by taking advantage of the highly selective nature of antibodies raised against these tags. In parallel institutions, we generated two independent alleles to facilitate the study of Sumo2 in vivo. We took advantage of a CRISPR/Cas9 approach to knockin a 6xHis-hemagglutinin (His6-HA)-tag into the amino-terminus of Sumo2 (His6-HA-Sumo2) in C57BL/6J mice (Figure S1A) in one case, mirroring that of a previously generated mouse line to study Sumo1 in vivo.32 Moreover, we also generated an HA-Sumo2 knockin mouse that does not have the His6 tag. We observed that knockin of either His6-HA or HA to Sumo2 resulted in hydrocephaly and premature death in heterozygous mice, indicating a likely hypomorphic effect of the epitope tag on a C57BL/6J background. Interestingly, backcrossing the mice to an FVB/N background alleviated these effects, resulting in no overt differences between mutant and wild-type FVB/N (WT) littermates in either line. Nevertheless, given the importance of Sumo2 for life, and the robustness of the HA-tag for visualization and biochemistry, we performed the bulk of our analyses on heterozygous His6-HA-Sumo2 (and heterozygous His6-HA-Sumo1; also crossed to an FVB/N background) knockin mice.
His6-HA-Sumo2 enables the comparison of endogenous Sumo2 levels in the CNS
To determine whether the addition of the HA epitope tag affects native Sumo levels, we assessed the levels of Sumo2/3 conjugates by Western blot, comparing total levels of unconjugated Sumo (“Free Sumo”) and Sumo-conjugated proteins from mouse brain lysates. We observed no change in the abundance of Sumo2/3 conjugates in the brains of adult His6-HA-Sumo2 mice compared to His6-HA-Sumo1 or WT mice (Figure S1B). As decreased levels of Sumo1 conjugates were previously observed in homozygotes His6-HA-Sumo1 mouse brain,33 we thus measured whether there were any changes in the abundance of native Sumo1 conjugates in heterozygous His6-HA-Sumo1 and His6-HA-Sumo2 mice.17 Again, no changes were observed between Sumo knockin line and their WT counterparts, indicating that the heterozygous His6-HA-Sumo2 or His6-HA-Sumo1 allele does not alter overall levels of Sumo conjugates in mouse brain (Figure S1C). In addition, we did not observe a significant effect on the expression of Sumo1, Sumo2, or Sumo3 transcript levels in either of the His6-HA-Sumo knockin mice (Figure S1D). Thus, the novel His6-HA-Sumo2 mouse model enables the study of Sumo2 at endogenous levels in vivo without changes to the Sumo equilibrium.
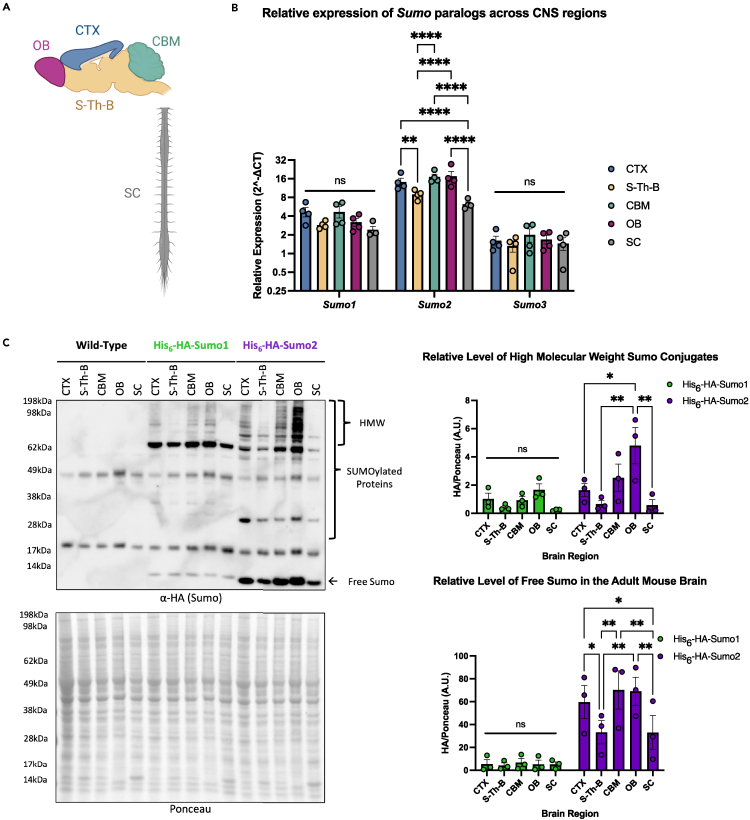
The uniformity of the HA-tag epitope between the His6-HA-Sumo1 and His6-HA-Sumo2 mice allows for a direct comparison of endogenous Sumo1 and Sumo2 levels in vivo. We assessed the relative levels of Sumo expression in the CNS, specifically the cortex (CTX), cerebellum (CBM), olfactory bulbs (OB), spinal cord (SC), and remainder of the brain (termed “striatum-thalamus-brainstem" or "S-Th-B" for simplicity) from adult mice (Figure 1A). Real-time qPCR analysis of WT mice shows that Sumo2 is the Sumo paralog most abundantly expressed in all regions of the CNS (Figure S1E). In addition, Sumo2 expression varies significantly between CNS regions, whereas Sumo1 and Sumo3 exhibit more uniform expression (Figure 1B). Western blot analysis from CNS regional lysates of His6-HA-Sumo1, His6-HA-Sumo2, and WT mice shows that levels of conjugated and free His6-HA-Sumo1 indeed do not significantly vary between CNS regions, whereas free His6-HA-Sumo2 levels reflect the higher expression and variations observed at the RNA level (Figures 1C, S1F, and S1G). In addition, the OB is particularly enriched for high molecular weight Sumo2-conjugated proteins suggesting that Sumo2 may play unique regional roles throughout the CNS (Figure S1F).
Figure 1.
HA-epitope knockin enables direct comparisons of differential levels of Sumo1 and Sumo2 and their conjugates in the murine central nervous system
(A) Brain regions highlighted in the schematic were dissected for protein and RNA extraction and analyzed by Western blot using anti-HA antibody and RT-qPCR. CTX = cortex; S-Th-B = striatum-thalamus-brainstem; CBM = cerebellum; OB = olfactory Bulb; SC = spinal cord.
(B) RT-qPCR analysis of WT mouse brain regions testing for significant regional changes for Sumo1, Sumo2, or Sumo3 expression (N = 4).
(C) Western blot analysis of various regions of His6-HA-Sumo1 and His6-HA-Sumo2 mouse central nervous system. HA signal corresponding to free Sumo and SUMOylated proteins (HMW = High Molecular Weight) was normalized to ponceau and quantified via densitometry (N = 3). Two-way ANOVA tests with Tukey’s post-hoc multiple comparisons analysis were performed. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S1.
Regional analysis of Sumo1 and Sumo2 throughout the mouse brain
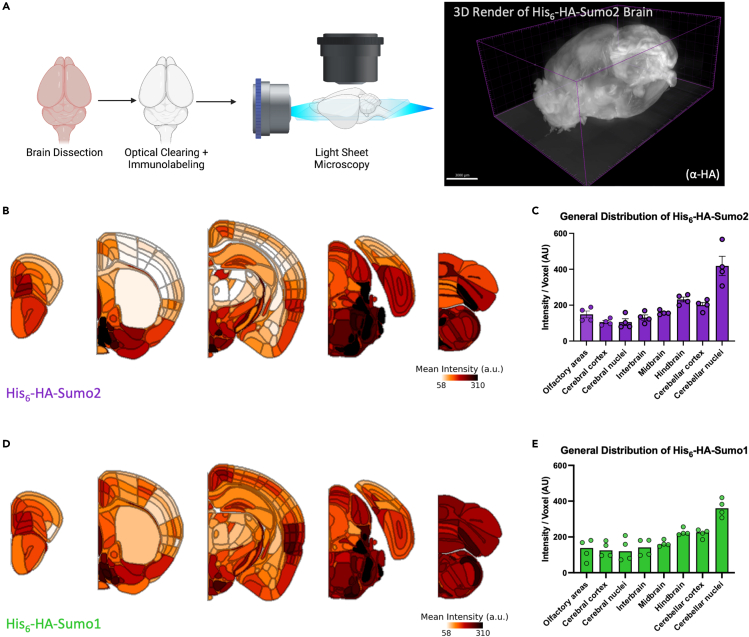
As differences in Sumo levels are detectable from regional crude brain lysates, we leveraged the specificity of the HA-tag and performed whole-brain clearing and mapping of Sumo1 and Sumo2 signals in the adult mouse brain. His6-HA-Sumo1 and His6-HA-Sumo2 mouse brains were cleared and immunolabeled against the HA-tag and NeuN before imaging using light sheet microscopy (Figure 2A, Video S1 and Video S2). Images were then analyzed and aligned to the Allen Brain Atlas to determine the relative levels of the His6-HA-Sumo signals in defined brain regions (Table S1). It is worth noting that due to the different absolute abundance of Sumo1 (low) versus Sumo2 (high) in the brain, only qualitative Sumo1 vs Sumo2 comparisons were made. To generate relative maps of HA-Sumo distribution, the mean regional HA signal intensity was first normalized to the regional density of NeuN to account for changes in cell density before being averaged across hemibrains to account for tissue variability. Generally, Sumo1 and Sumo2 showed similar distributions throughout the mouse brain (Figures 2B–2E), and we observed that both Sumo1 and Sumo2 were most abundant in cerebellar nuclei.
Figure 2.
Whole-brain imaging reveals the topographical distribution of Sumo paralogs
(A) Schematic of brain clearing and light sheet microscopy for whole brain imaging with a 3D Imaris render of a His6-HA-Sumo2 brain.
(B) Heatmaps of anti-HA immunosignal intensity normalized to regional density quantified by NeuN staining and alignment to the Allen Brain Atlas across the His6-HA-Sumo2 depicted brain region, averaged between hemibrains and biological replicates (N = 2, two hemispheres/replicate).
(C) Bar plot depicting the relative levels of anti-HA immunosignal per voxel averaged across His6-HA-Sumo2 brain regions defined by the Allen Brain Atlas.
(D) Heatmaps of anti-HA immunosignal intensity normalized to regional density quantified by NeuN staining and alignment to the Allen Brain Atlas across the His6-HA-Sumo1 depicted brain region, averaged between hemibrains and biological replicates (N = 2, two hemispheres/replicate).
(E) Bar plot depicting the relative levels of anti-HA immunosignal intensity normalized to regional density across His6-HA-Sumo1 brain regions defined by the Allen Brain Atlas. Each datapoint in (C) and (D) is from the mean intensity from a single hemisphere (N = 2, two hemispheres/replicate). Data are presented as mean ± SEM. See also Figures S2, S3, Table S1, Video S1, and Video S2.
While Sumo levels were generally evenly distributed when characterizing gross anatomical regions, clear nuances in Sumo abundance were detectable at higher anatomical resolution. For example, in the hippocampus, Sumo levels were highest in the parasubiculum and presubiculum, and lowest in the Fasciola cinerea (Figures S2A and S2B). In the neocortex, we observed that Sumo levels were generally enriched in layers 2/3 (Figures 2B, 2D and S2C), an area critical for integrative processing.35 Interestingly, while Sumo1 and Sumo2 often share similar distributions, we observed that Sumo2 is enriched in the anterior hypothalamus (Figure S2D), particularly in regions involved in the circadian rhythm, including the suprachiasmatic nucleus and subparaventricular zone.36
We investigated whether the brain-wide imaging findings could be validated by an orthogonal immunofluorescence assay (Figure S3). Looking at multiple brain regions, we found that Sumo1 and Sumo2 are broadly distributed through the somatosensory cortex. Interestingly, Sumo1 maintains a strictly nuclear localization, whereas Sumo2 is observed throughout the nucleus and in the cytoplasm and axons of cortical neurons. In the cerebellum, we found that Sumo2 is specifically enriched in granule cells, contrasting the strong Purkinje cell staining observed in His6-HA-Sumo1 mice.32 The hippocampus also displays characteristic nuclear staining for His6-HA-Sumo1 in both the CA1 and CA3, whereas Sumo2 is seen throughout the nucleus and in the somata, and axons of hippocampal neurons. Thus, while Sumo1 and Sumo2 may share regional expression in the brain, the distinct localizations of Sumo1 and Sumo2 in vivo indicate that different targets of SUMOylation or different metabolism or conjugation dynamics of free Sumo likely play unique roles in neuronal regulation.
HA-Sumo2 localizes to synapses in vivo
Consistent with our previous work, specific Sumo1 immunosignal was mainly observed in neuronal nuclei (Figure S3).32 However, we noticed that Sumo2 signal can also be found outside the nuclear compartment (Figure S3). Thus, we further characterized extranuclear Sumo2 signals using homozygous HA-Sumo2 mice. As in the His6-HA-Sumo2 model, levels of Sumo2 conjugates were unaltered in total brain lysates from homozygous HA-Sumo2 mice as compared to WT littermates, ruling out an effect of the HA-tag on overall Sumo2 expression and conjugation levels (Figure S4A).32 Western blot analysis of brain subcellular fractions using an anti-HA antibody highlighted not only the difference in global levels of Sumo1 and Sumo2 conjugates but also revealed that Sumo2 conjugates are enriched in non-nuclear fractions compared to their Sumo1 counterpart, including synaptic cytosol fractions S2 and S3 (Figures S4B and S4C). A weak but specific signal was also observed in crude synaptosomes (P2) and other synaptic membrane fractions, e.g. P3 and LP2, while the synaptic plasma membrane fraction primarily appears devoid of Sumo2 conjugates (Figures S4B and S4C).
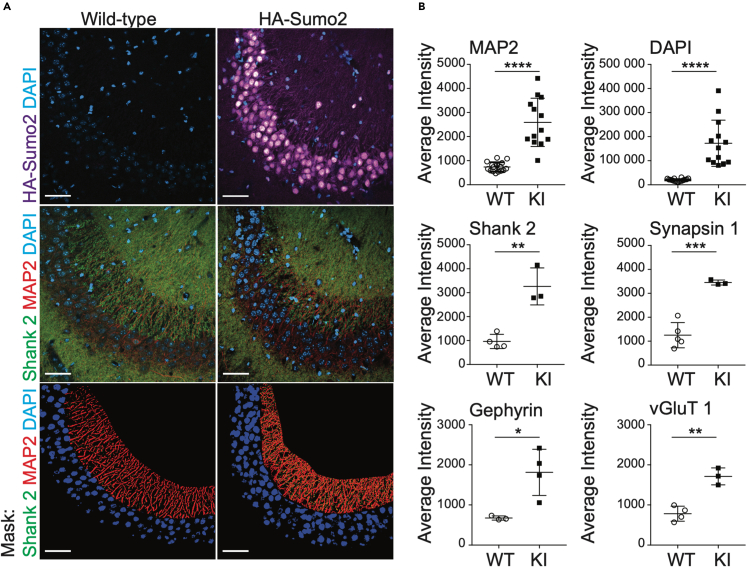
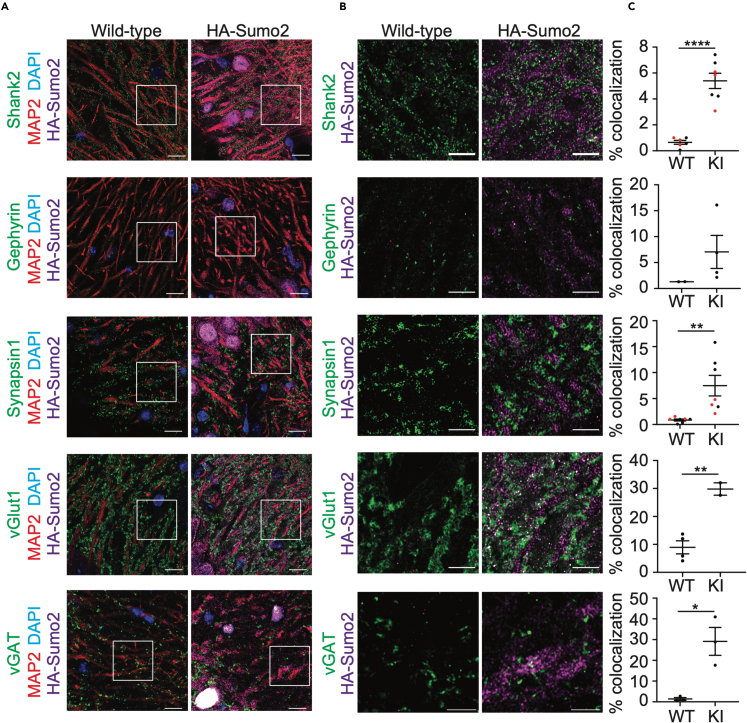
Cellular fractionation experiments are often burdened with cross contamination between fractions that can limit interpretations to assessing relative enrichment in each fraction. Well-controlled immunofluorescent studies can overcome this limitation and provide spatial evidence for protein localization. As such, we used homozygous HA-Sumo2 mice to further characterize extranuclear Sumo2 by performing anti-HA immunolabeling of the hippocampal CA3 area, a region enriched in synapses (Figure S3). Brain sections from WT and HA-Sumo2 mice were immunolabeled for HA and neuronal markers for dendrites and synaptic compartments, and anti-HA immunosignals were quantified in HA-Sumo2 mice as compared to WTs (Figure 3). Using confocal microscopy, we confirmed the presence of Sumo2 in extranuclear compartments, specifically along dendrites and in synapses (Figure 3A). Quantification of HA-Sumo2 immunosignals in DAPI-positive area confirmed the strong enrichment of Sumo2 conjugates in neuronal nuclei, in agreement with the well-known role of SUMOs as nuclear regulators (Figures 3A and 3B).37,38 In addition, HA-Sumo2 immunosignal was also observed in MAP2-positive regions and, strikingly, in synapses positives for Synapsin1, Shank2, Gephyrin, and/or VGlut1 (Figures 3C and 3D). Higher magnification images confirmed the presence of Sumo2 along MAP2 dendrites and at synapses (Figures 4 and S5). Quantification of the 3D object-based co-localization between the anti-HA immunosignal with the various synaptic markers confirmed the presence of Sumo2 at synapses, with HA-Sumo2 signal being significantly co-localized with Shank2, Synapsin 1, VGlut1, and vGAT (Figures 4C and S5, which shows a typical 3D reconstruction generated in Imaris). Co-localization of Sumo2 with Gephyrin was not obvious, most likely due to difficulties in immunolabeling inhibitory synapses with the method optimized for HA visualization. Altogether, our biochemical fractionation and immunofluorescence support the presence of Sumo2 at synapses.
Figure 3.
Extranuclear localization of Sumo2 in neurons of the hippocampal CA3 region
(A) Anti-HA (purple), DAPI (blue), MAP2 (red), and anti-Shank2 (green) immunostaining of wild-type (left) and HA-Sumo2 KI (right) hippocampal CA3 region. Scale bar: 50 μm.
(B) Anti-HA signal in masked regions for each marker (exemplified for Shank2 in the bottom panel A) was quantified in both wild-type (WT) and HA-Sumo2 KI, and average intensity was calculated. N = 3, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0,0005, ∗∗∗∗p < 0,0001. See also Figure S4.
Figure 4.
Sumo2 localizes at synapses in vivo
(A) High magnification images of wild-type (left) and HA-Sumo2 KI (right) hippocampal CA3 region labeled with anti-HA (purple), DAPI (blue), MAP2 (red), and synaptic markers (green) Shank2, Gephyrin, Synapsin1, VGlut1, and vGAT1. Scale bar, 10 μm.
(B) High-magnification images with equally scaled anti-HA intensity corresponding to the insets (white boxes) in A showing anti-HA (purple) immunostaining of wild-type (left) and HA-Sumo2 KI (right) hippocampal CA3 region immunolabelled with the synaptic markers (green) Shank2, Gephyrin, Synapsin1, VGlut1, and vGAT1. Scale bar: 5 μm. N = 3.
(C) The percentage of co-localization between each synaptic marker and the anti-HA signal was quantified in both wild-type (WT) and HA-Sumo2 KI. N = 3. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗∗p < 0,0001. See also Figure S5.
HA-Sumo mice reveal convergent and contrasting neuronal Sumo1 and Sumo2 substrates
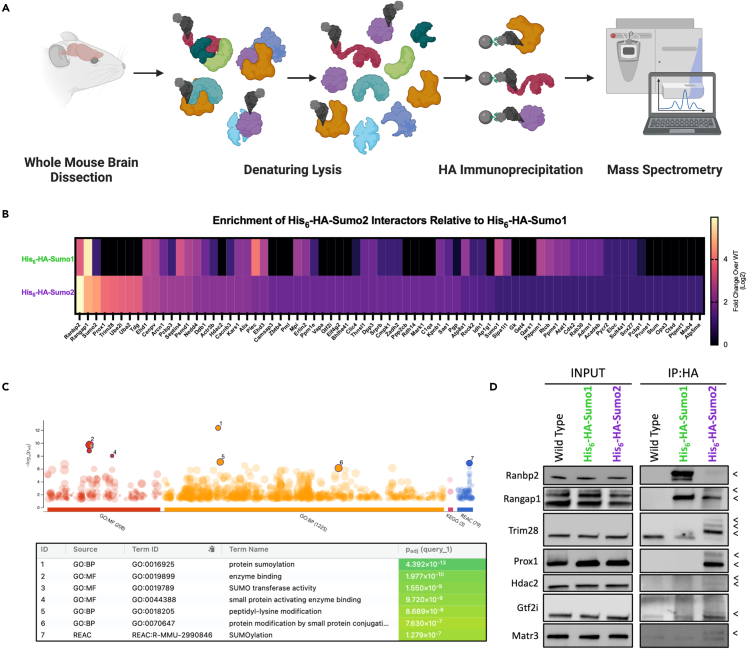
The observation that there are regional and subcellular differences between Sumo1 and Sumo2 indicate divergent roles for the Sumo paralogs in the mouse brain. To uncover such molecular differences between Sumo paralogs, we performed HA-tag immunoprecipitation, under denaturing conditions to break apart standard protein-protein interactions, from whole-brain lysate of WT, His6-HA-Sumo1, and His6-HA-Sumo2 mice, followed by mass spectrometric analysis to identify candidate targets of Sumo1 and Sumo2 in vivo (Figures 5A and S6A; Table S2). To rank proteins identified from the mass spectrometry dataset, we summated the total peptides from 4 replicates per genotype and then filtered proteins with >2-fold enrichment over proteins identified in the WT condition. Next, to determine high-stringency candidates, proteins were further filtered based on peptide counts being identified in at least 2/4 replicates in the His6-HA-Sumo conditions, and no more than 1/4 replicate in the respective WT condition. Using these criteria, we identified 131 proteins enriched in the His6-HA-Sumo1 and 75 proteins enriched in the His6-HA-Sumo2 immunoprecipitations (Figures 5B and S6B; Table S2). Gene ontology and Reactome terms related to protein SUMOylation process (Figure 5B) were significantly enriched in both datasets (Figures 5C and S6C; Table S2). Indeed, we identified critical components of the SUMOylation pathway including E1 SUMO ligases Sae1 and Uba2, the E2 SUMO ligase Ube2i, and E3 ligases Ranbp2,39 Trim28,40 and Pml,41 validating the use of our mouse model to identify molecular substrates involved with SUMOylation.
Figure 5.
Neuronal Sumo2 has shared and distinct substrates compared to Sumo1 in vivo
(A) Schematic of approach to identify His6-HA-Sumo interactors from the adult mouse brain via anti-HA affinity immunoprecipitation and mass spectrometry (N = 4).
(B) Heatmap depicting the relative peptide abundance of His6-HA-Sumo2 interactors relative to levels in His6-HA-Sumo1 immunoprecipitation.
(C) gProfiler2 Gene Ontology analysis for His6-HA-Sumo2 interactors.
(D) Anti-HA affinity immunoprecipitation followed by Western Blot analysis of His6-HA-Sumo1 and His6-HA-Sumo2 interactors as listed on the left side (N = 3–6). Black arrowheads indicate protein SUMOylation inferred by shift in molecular weight. See also Figure S6 and Table S2.
To determine whether we could identify specific interactors of Sumo2, we compared the relative fold change of peptides identified in the His6-HA-Sumo2 immunoprecipitation to that of the His6-HA-Sumo1 immunoprecipitation and found that there are shared, specific, and preferential interactors of Sumo2 vs. Sumo1 (Figures 5B, S6B, and S6D; Table S2). We selected several hits from our mass spectrometry screen for validation via Western blot (Figure 5D). Ranbp2 and Rangap1 are consistently identified among the most abundantly SUMOylated proteins9,42,43 and were indeed validated to be shared targets of Sumo1 and Sumo2 in the mouse brain. In addition, both proteins displayed a preference for Sumo1 modification in the mouse brain as previously described.42 Conversely, the E3 ligase and transcriptional repressor Trim28 displayed a preference for Sumo2 modification in the adult mouse brain. Interestingly, the transcription factor Prox1, involved in neurogenesis and a marker of granule cells in the dentate gyrus and cerebellum,44 was one of the most abundantly SUMOylated proteins targeted specifically by Sumo2 in our screen. We further tested the sensitivity of the model to detect more modestly SUMOylated proteins targeted by Sumo2 in the mouse brain, as measured by relative abundance in our immunoprecipitation coupled with mass spectrometry experiment. We confirmed that both histone deacetylase 2 and general transcription factor 2i (Gtf2i, also known as Tf2-i) were Sumo2 modified (Figure 5D). Because of the dynamic nature of SUMOylation and the fact that Sumo only targets a fraction of the total pool of most substrates, biologically meaningful interactions may be buried under strong, constitutively SUMOylated targets in our screen. As a proof of concept, we picked a hit with low enrichment in the His6-HA-Sumo2 immunoprecipitation by expanding the filtering criteria to a >1.5-fold enrichment over WT: Matrin3 (Matr3). We found that Matr3 was selectively SUMOylated in the His6-HA-Sumo2 pulldown (Figure 5D), indicating that even at milder cutoff thresholds, SUMOylated substrates identified by mass spectrometry can be validated in an orthogonal assay. Taken together, these results show that the His6-HA-Sumo2 mouse model is effective to identify SUMOylated proteins, including proteins SUMOylated at relatively low levels in vivo, and can be used for targeted studies of protein substrates.
Subcellular localization of Sumo interactions in neurons
Sumo1 and Sumo2 predominantly reside in the nucleus, and unsurprisingly, many of the interactors identified here and in other studies of protein SUMOylation are nuclear proteins. However, previous studies hinted at roles for SUMOylation outside of the nucleus.2,45,46,47,48 During the validation of whole-brain imaging, we observed that His6-HA-Sumo2, while extensively localized throughout the nucleus, could also be observed within the somata and synapses of neurons in vivo, contrasting the predominantly nuclear membrane staining of His6-HA-Sumo1 (Figures S3 and 3). The cytoplasmic localization of His6-HA-Sumo2 in neurons indicates unique extranuclear roles in vivo. Indeed, we identified several predominantly extranuclear proteins via mass spectrometry in the anti-HA affinity purification from the His6-HA-Sumo2 mice. To better visualize the subcellular localization of Sumo, we cultured primary cortical neurons from heterozygous His6-HA-Sumo1, His6-HA-Sumo2, and WT animals and co-stained them for HA and Map2 as a cytoplasmic marker. As previously observed, Sumo2 is predominantly distributed uniformly throughout the nucleus except for heterochromatic foci but is also observed at low levels in the cytoplasm (Figure S7A), contrasting with the anti-HA immunosignal observed for Sumo1, which predominantly localizes at the nuclear membrane. This indicates that Sumo2 specifically may play a larger role in regulating proteins outside of the nucleus.
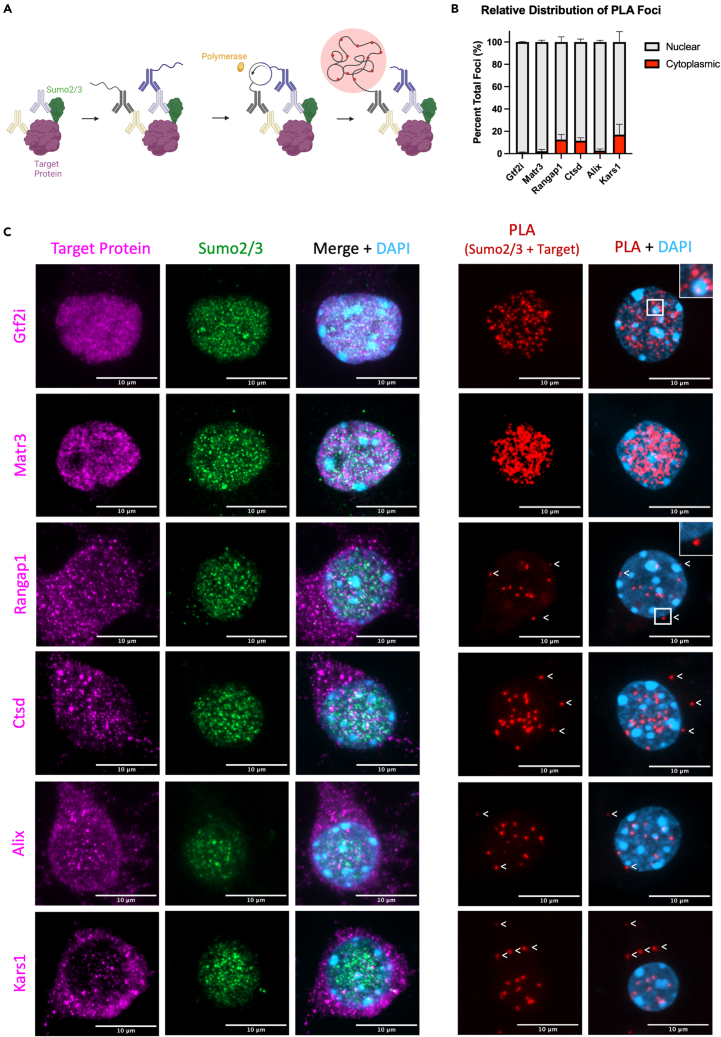
To better assess the subcellular localization of neuronal SUMOylated proteins targeted by Sumo2, and to provide an additional level of validation in an orthogonal system, we used proximity ligations assays (PLA) against endogenous Sumo2/3 and targets of Sumo2 identified from our mass spectrometry screen to visualize interactions within WT primary cortical neurons (Figure 6A).49,50 In this assay, proteins colocalizing within 40 nm will produce a PLA signal suggesting a potential interaction. We selected proteins expected to interact with Sumo2/3 in the nucleus, Gtf2i, and Matr3, as well as expected cytoplasmic interactors Rangap1, Ctsd, Alix, and Kars1 for additional validation (Figures 6B and 6C). Gtf2i is distributed almost exclusively throughout the nucleus and interactions between Sumo2/3 and Gtf2i, inferred by the presence of PLA foci, are seen throughout the nucleoplasm. Interestingly, PLA foci were often observed to cluster around constitutive heterochromatin (Figure 6C, inset), indicating a potential role for Sumo2 in regulating Gtf2i at the periphery of heterochromatin in neurons. Matr3 is also localized throughout the nucleus and to a lesser extent in the cytoplasm, consistent with its identification in cytoplasmic processing bodies.51 Interestingly, PLA signals for Matr3 and Sumo2/3 are robust, indicating extensive interactions between the two proteins. Immunofluorescence staining of Rangap1, Ctsd, Alix, and Kars1 demonstrates these proteins predominantly reside outside of the nucleus (Figure 6C). PLA between Rangap1, Ctsd, and Kars1 with Sumo2/3 display an increase in the proportion of cytoplasmic foci relative to nuclear foci (Figure 6B), in addition to a substantial decrease in total nuclear PLA foci compared to nuclear localized proteins Gtf2i and Matr3 (Figure S7B). Importantly, for all PLA assays conducted, negative controls did not elicit a PLA signal (Figure S7C). Taken together, our PLA analyses of WT neurons provide an additional layer of validation of Sumo2-substrate interactions. With the added benefit of providing spatial resolution to the interactions, PLA data support a role for neuronal Sumo2 outside the nucleus.
Figure 6.
Established and newly identified Sumo2 substrates are present both in nuclear and extranuclear compartments in neurons
(A) Schematic of the proximity ligation assay (PLA) strategy for native Sumo2/3 and target proteins in WT primary cortical neurons.
(B) The relative proportion of PLA foci between the selected target proteins and Sumo2/3 was quantified and normalized within the nucleus (gray) versus outside of the nucleus (red) (N = 3).
(C) Representative Z-projected immunofluorescent images and PLA assays for selected target proteins identified from the mass spectrometry screen (N = 3). White arrowheads indicate cytoplasmic PLA foci. Scale bar: 10 μm. Data are presented as mean ± SEM. See also Figure S7.
Discussion
A constantly growing body of evidence demonstrates that SUMOylation plays key roles in the brain including neuronal development or learning and memory.18,20,45,52,53,54 Furthermore, SUMOylation is linked to various brain disorders, including stroke, autism spectrum disorder, epilepsy, schizophrenia, or neurodegenerative diseases, such as Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis (ALS).55,56,57,58,59,60,61,62 However, despite a clear connection to neuronal function, SUMOylation in neurons has remained somewhat enigmatic, mainly due to technical challenges and limitations to study this PTM.33 We generated His6-HA-Sumo2 and HA-Sumo2 knockin mouse lines to overcome these technical limitations and to provide insight into the role of native SUMOylation in vivo. These mouse lines complement the previously generated His6-HA-Sumo1 line,32 enabling the direct comparison of Sumo1 and Sumo2 biology in vivo and the elucidation of specific Sumo2 functions.
We observed that Sumo2 is more abundantly expressed throughout the CNS than Sumo1 and Sumo3 (Figure 1B). Interestingly, on both the RNA and protein levels, Sumo2 also displayed significant variations between CNS regions whereas Sumo1 levels did not significantly change. This may suggest that Sumo2 plays regional roles in the CNS. In addition, we observed that the pool of free Sumo2 is significantly greater than that of Sumo1 in all CNS regions, and levels of free Sumo2 significantly differ between regions (Figure 1C). As Sumo2 has been described to respond to stimuli, such as stressors, the pool of free Sumo2 may reflect the dynamic nature of SUMOylation in each region with cells requiring more Sumo2 to be readily available to respond dynamically in the cellular environment.63 Future studies focusing on SUMO dynamics in neuronal populations may help to uncover paralog and cell-type-specific roles for SUMO-mediated regulation in the CNS.
Using whole-brain imaging upon HA epitope labeling in His6-HA-Sumo1 and His6-HA-Sumo2 mice, we found that Sumo1 and Sumo2 are broadly distributed throughout the mouse brain. More detailed anatomical analyses revealed distinct differences between Sumo1 and Sumo2 levels across brain structures and some divergence between Sumo1 and Sumo2 levels in the brain. Levels of His6-HA-Sumo1 are generally similar between cortical layers, whereas His6-HA-Sumo2 levels are higher in layers 2/3 (Figure S2D). In the hypothalamus, we found that Sumo levels vary across anatomical regions (Figure S2E). Interestingly, regions involved in circadian rhythm control, including the subparaventricular zone and suprachiasmatic nucleus, show high levels of Sumo2. These findings are consistent with previous reports indicating that Sumo1 and Sumo2 differentially affect circadian-clock-related proteins, such as PER2 and CLOCK, and support the notion of a role of SUMOylation in regulating circadian rhythms.36,64,65
Importantly, we observed striking differences between the subcellular distribution patterns of Sumo1 and Sumo2. Western blot analysis of brain subregional fractions showed that Sumo2 is more abundant than Sumo1 (Figure 1C) and that Sumo2 conjugates, unlike Sumo1 conjugates, are prominently present in extranuclear fractions, including synaptic fractions (Figures S4B and S4C). However, brain subcellular fractionation is often unclean and nuclear material frequently leaks to non-nuclear fractions. Thus, immunofluorescence analysis is more powerful in determining the precise subcellular localization of Sumo-conjugated proteins. More specifically, anti-HA immunolabeling of brain material from HA-Sumo2 mouse brains and WT littermates as negative controls also demonstrated the presence of extranuclear-free Sumo2 and Sumo2 conjugates (Figures 3, 4, and S5). Quantitative analyses of anti-HA immunosignals confirmed a partial co-localization of Sumo2 with pre- and post-synaptic markers at excitatory and inhibitory synapses (Figure 3, Figure 4A, 3B, and 4A–4C). These data, which were stringently controlled using WT material as negative control, partly align with a recent report describing Sumo2 localization at synapses.31 On aggregate, our approach to study Sumo2 expression using the HA-Sumo2 mice allows for the specific assessment of Sumo2 immunosignals in neuronal sub-compartments and provides unequivocal evidence for the presence of Sumo2 at mammalian brain synapses. Thus, not only do Sumo1 and Sumo2 display differential expression and protein levels throughout the brain but are also localized and operational in different neuronal subcompartments. However, the functions of Sumo2 at synapses remain elusive. Importantly, there may be both direct (by Sumo-conjugation or -binding) and indirect roles for Sumo1 and Sumo2 in synaptic function that will have to be carefully assessed in the future to better understand how Sumo regulates synapse biology. In sum, the specificity provided by the His6-HA-Sumo1 and (His6-)HA-Sumo2 models enables direct comparisons between paralogs by overcoming key technical limitations. This will help to more accurately uncover specific roles for SUMOylation in vivo.
Sumo2 is the only functionally essential Sumo paralog, and, as mentioned, has key roles in neurodevelopment, nerve cell function, and neurological diseases.17,52,55,66,67,68 To uncover some of the unique clientele targeted by Sumo2 conjugation, we performed HA-tag immunoprecipitation paired with mass spectrometry. As SUMOylation results in the covalent interaction between Sumo and the target substrate, mouse brains were lysed under denaturing conditions to promote the dissociation of non-covalent protein complexes prior to immunoprecipitation to help identify bona fide SUMOylation substrates. Using stringent criteria, 75 proteins were identified to be conjugated to Sumo2, ∼50% of which were selective to Sumo2. Conversely, using the same criteria, we identified 131 candidate Sumo1 substrates, 98 of which were selective to Sumo1. “Canonical” synaptic proteins were not among the hit lists in these experiments, which are likely related to the fact that experiments were performed in whole adult brains lysates, where levels of SUMOylated synaptic proteins may be too low due to dilution effects. The dynamics of SUMOylation may add additional complications in capturing synaptic SUMOylation events as physiological conditions may differentially affect when synaptic proteins become SUMOylated. Interestingly, we found that synaptic cytosol fractions contained increased levels of free Sumo2 which may further support the notion of highly dynamic SUMOylation at synapses, or non-covalent Sumo2 regulation in these compartments. Ultimately, a dedicated proteomics approach focused on synaptic fractions may be necessary to specifically detect synaptic Sumo2 substrates and uncover specific roles for Sumo2 at synapses.56,57 Pathway analysis of the Sumo2 dataset revealed that gene ontology terms related to SUMOylation were significantly enriched in the dataset, validating this method to identify potentially SUMOylated substrates. Moreover, pathway analysis of the Sumo1 dataset revealed enrichment of proteins involved in protein transport consistent with roles for Sumo1 in nucleocytoplasmic shuttling.39,58,59 Prox1 was validated via Western blot as one of the strongest interactors specific to Sumo2 in the mouse brain. Interestingly, previous studies demonstrated that SUMOylation of Prox1 by Sumo1 occurs in vitro and in cancer cell lines.60,61 During development, Prox1 was found to be SUMOylated by Sumo1, and this interaction is critical for neurogenesis in the neural tube development. However, the role of Prox1 in the mature brain remains unclear, although some data indicate an involvement in adult neurogenesis.62 Due to the extent of the Prox1 and Sumo2 interaction observed in this study, further analysis may uncover key roles of SUMOylated Prox1 in the adult mouse brain. We further found that the RNA-binding protein Matr3 interacts with Sumo2 in vivo, indicating that even a mild enrichment over WT in our dataset may identify bona fide biological interactors. Interestingly, many of the computationally predicted SUMOylation sites in Matr3 (K588 and K843) reside within a structurally disordered region of the protein that also happens to be enriched for mutations causing amyotrophic lateral sclerosis.69 Structurally disordered regions are thought to be critically involved in the formation of protein aggregates in neurodegenerative diseases and given the key role of SUMO in controlling protein solubility, and the recent links between SUMOylation and ALS, SUMOylation of Matr3 may provide key insights into Matr3 biology related to this disease.
SUMOylation occurs in a context-specific manner and can regulate protein localization within cells. One of the best characterized examples concerns the regulation of nucleocytoplasmic shuttling across nuclear pore complexes39,58,59 However, SUMOylation can also regulate sub-compartmental localizations, such as protein targeting to the nucleolus or to heterochromatin.70,71,72 As Sumo2 was observed throughout the nucleus and outside the nucleus in vivo and in cultured neurons, we sought to determine the subcellular localization of Sumo2 interactions with hits identified via IP-MS using a PLA-based approach (Figure 6). Gtf2i was SUMOylated almost exclusively in the nucleus but interestingly appeared to cluster around heterochromatin. Gtf2i can function as both a transcriptional activator and repressor in response to various signals. For example, the specific splice isoforms β and Δ undergo changes in subcellular localization to differentially regulate the immediate-early gene c-Fos in response to growth factor signaling.73 However, roles for Gtf2i in regulating heterochromatin remain elusive. SUMOylation may provide a dynamic mechanism in response to neuronal activity to regulate both targeted expression at the gene level and global transcriptional changes.74 Finally, interactions between Matr3 and Sumo2/3 occurred broadly throughout the nucleus. However, interactions between Matr3 and Sumo2 in mouse brain occurred at relatively low levels based on immunoprecipitation assays. This discrepancy observed in the nucleus may be explained via non-covalent interactions with Sumo2 through larger protein complexes as Matr3 typically functions in complex with other DNA- and RNA-binding proteins.75,76,77 Extranuclear proteins Rangap1, Ctsd, and Kars1, despite having a certain level of PLA signal within the nucleus, displayed an increase in the proportion of cytoplasmic PLA foci relative to nuclear PLA foci implying increased interactions with Sumo2/3 outside the nucleus. Indeed, Rangap1 plays roles in nuclear import, but can also be found to some extent within the nucleus.78 PLA foci between Rangap1 and Sumo2/3 were often found near the nuclear membrane, likely at nuclear pore complexes (Figure 6C, inset). Ctsd is a lysosomal protease predominantly localizing to lysosomes, thus interactions between Sumo2/3 and Ctsd may suggest that its SUMOylation may play a role in lysosomal metabolism.79 Finally, extranuclear PLA foci could be observed for the lysyl-tRNA synthetase Kars1, consistent with reports of Sumo interacting with tRNA-related proteins.80,81,82 Together, these results suggest that Sumo2 may play a variety of roles outside the nucleus in various subcellular compartments. While PLA assays provide spatial information of putative protein interactions (i.e. interactions occurring within 40 nm), this does not discount indirect interactions (e.g. protein complexes).49,50 Thus, differentiating between covalent SUMOylation, interactions with SUMO interacting motifs, and protein complexes containing SUMOs are not possible using this approach. Extensive localization and biochemical analyses, including arginine to lysine mutagenesis to block covalent SUMOylation, should be performed to properly assess how SUMO confers regulation on a specific target throughout the cell.
In sum, the novel (His6-)HA-Sumo2 knockin mouse lines described here represent powerful tools to facilitate the study of protein SUMOylation via Sumo2 in vivo. The HA epitope tag allows for specific yet versatile modes to detect and capture Sumo2 in its native context, without altering its function. Furthermore, these mouse models add to the previously generated His6-HA-Sumo1 allele, and thus expand the Sumo knockin mouse toolkit to begin exploring differential SUMOylation in parallel systems. The versatility of these models extends to all other tissue systems, as Sumo proteins are broadly distributed beyond the brain.63,83 Beyond basic biology, these alleles can be used to explore disease processes, for example by crossing these mouse lines into disease models, thus enabling the study of the Sumo-linked disease proteome.66 Ultimately, these tools will advance our understanding of the essential biological processes and potential disease targets regulated by SUMOylation.
Limitations of the study
While the (His6)-HA-Sumo2 knockin mice help to overcome technical limitations in studying SUMO biology, including low antibody specificity, inability to differentiate between endogenous SUMO2 and SUMO3, and reliance on overexpression models, it cannot be completely excluded that the knockin of the (His6-)HA-tag slightly affects Sumo2 function. Indeed, these mouse lines allow to characterize Sumo1 and Sumo2, but we also observed early lethality on C57BL/6J or C57BL/6N background, which was not seen on an FVB/N background. This indicates an interaction between the His6-HA-Sumo2 knockin genotype and the genetic C57BL/6J or N set-up. Such strain effects are seen in many contexts, highlighting the importance of validating new genetic models using multiple systems and approaches. Furthermore, our observation that only Sumo2, but not Sumo1, is detectable at synapses, does not completely exclude a role for Sumo1 in regulating synapse function in vivo.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA-Tag (C29F4) Rabbit mAb | Cell Signaling | Cat# 3724, RRID:AB_1549585 |
| Anti-HA.11(16B12) Epitope Tag Mouse Ab | BioLegend | Cat# 901516, RRID:AB_2820200 |
| Anti-SUMO2/3 (8A2) Recombinant Antibody | Abcam | Abcam Cat# ab81371, RRID:AB_1658424 |
| Duolink in situ PLA probe anti-Rabbit PLUS | Sigma-Aldrich | Cat# DUO92002, RRID:AB_2810940 |
| Duolink in situ PLA probe anti-Mouse MINUS | Sigma-Aldrich | Sigma-Aldrich Cat# DUO92004, RRID:AB_2713942 |
| Chemicals, peptides, and recombinant proteins | ||
| HA Synthetic Peptide | Sinobiological | Cat#PP100028-5 |
| N-Ethylmaleimide | Sigma-Aldrich | Cat#E3876 |
| Critical commercial assays | ||
| Duolink in situ detection reagents red | Sigma-Aldrich | Cat#DUO92008 |
| Experimental models: Organisms/strains | ||
| His6-HA-Sumo1 Knock In Mouse | Marilyn Tirard32 | N/A |
| His6-HA-Sumo2 Knock In Mouse | This paper | N/A |
| HA-Sumo2 Knock in Mouse | This paper | N/A |
| FVB/N Mouse | Charles River Laboratories | Strain Code 207 |
| Oligonucleotides | ||
| qPCR Mouse Sumo1 FWD: CAGGAGGCAAAACCTTCAAC | Wang et al., 201417 | N/A |
| qPCR Mouse Sumo1 REV: CTCCATTCCCAGTTCTTTCG | Wang et al., 201417 | N/A |
| qPCR Mouse Sumo2 FWD: ACGAGAAACCCAAGGAAGGA | Wang et al., 201417 | N/A |
| qPCR Mouse Sumo2 REV: CTCCATTTCCAACTGTGCAG | Wang et al., 201417 | N/A |
| qPCR Mouse Sumo3 FWD: AGAAGCCCAAGGAGGGTGT | Wang et al., 201417 | N/A |
| qPCR Mouse Sumo3 REV: CCTCGGGAGGCTGATCCT | Wang et al., 201417 | N/A |
| qPCR Mouse Gapdh FWD: GGAGAGTGTTTCCTCGTCCC | This study | N/A |
| qPCR Mouse Gapdh REV: ATGAAGGGGTCGTTGATGGC | This study | N/A |
| qPCR Mouse Hprt1 FWD: TGATAGATCCATTCCTATGACTGTAGA | This study | N/A |
| qPCR Mouse Hprt1 REV: AAGACATTCTTTCCAGTTAAAGTTGAG | This study | N/A |
| Software and algorithms | ||
| Imaris Version 9.9.1 | Oxford Instruments | https://imaris.oxinst.com/ |
| Other | ||
| Whole Brain Imaging/Analysis | Life Canvas Technologies | https://lifecanvastech.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maxime Rousseaux (max.rousseaux@uottawa.ca).
Materials availability
HA-Sumo2 and His6-HA-Sumo2 mouse lines will be provided upon reasonable request.
Experimental model and subject details
Mouse lines
C57BL/6J.
FVB/N (WT)
His6-HA-Sumo1 (FVB/N Background)
His6-HA-Sumo2 (FVB/N Background)
HA-Sumo2 (FVB/N Background)
Sex and age of experimental mice
Experiments were performed using a mix of male and female mice to prevent sex biases. Cortical neuron cultures experiments included neurons dissected from individual embryos using a mixture of male and female cultures for all experimental conditions. At the high level of characterization, we do not expect sex effects to be observed however we acknowledge that future in depth analyses may uncover unique sex effects with regards to SUMOylation. All experimental animals were between 8 and 30 weeks of age. Specific age/sex information for whole brain imaging and mass spectrometry experiments can be found in Supplement Tables S1 and S2.
Mouse husbandry
His6-HA-Sumo1 and His6-HA-Sumo2 mice, kept on an FVB/N background, were housed with up to 5 mice per cage on a 12 h light–dark cycle. Mice were fed ad libitum and all husbandry was performed by the uOttawa Animal Care and Veterinary Services staff. All animal work for the His6-HA-Sumo1 and His6-HA-Sumo2 mouse lines were done under the breeding (CMMb-3009 and CMMb-3904) protocols approved under the uOttawa Animal Care Committee. Experiments performed on the HA-Sumo2 mice were done in accordance with the guidelines for the welfare of experimental animals issued by the State Government of Lower Saxony, Germany (LAVES). Animals were hosted in a pathogen free facility at the Max Planck Institute of experimental Medicine and were maintained in groups in accordance with European Union Directive 63/2010/EU and ETS 123 (individually ventilated cages, specific pathogen-free conditions, 21 ± 1°C, 55% relative humidity, 12 h/12 h light/dark cycle). Mice received food and tap water ad libitum and were provided with bedding and nesting material. Cages were changed once a week. Animal health was controlled daily by caretakers and by a veterinarian. Health monitoring (serological analyses; microbiological, parasitological, and pathological examinations) was done quarterly according to FELASA recommendations with either NMRI sentinel mice or animals from the colony. The mouse colony used for experiments did not show signs of pathogens. All experiments were performed during a light cycle.
Method details
His6-HA-Sumo2 and HA-Sumo2 mouse generation
His6-HA-Sumo2 mice were generated via CRISPR/Cas9-mediated gene editing.84 Briefly, an sgRNA targeting the 5′ of Sumo2 was synthesized by direct PCR from pX330 (gift from Zhang lab, Addgene #42230) and in vitro transcribed with the MEGAshortscript T7 Transcription kit (Invitrogen) using the following two primers (forward: 5′-TTAATACGACTCACTATAGGAACCCAAGGTGAGCTCGCGGGTTTTAGAGCTAGAAATAGC-3′; reverse: 5′-AAAAAAGCACCGACTCGGTGCCAC-3′). A single stranded oligodeoxynucleotide (ssODN) was purchased from IDT for homologous-directed recombination to knock-in a His6-HA-tag into the amino-terminus of Sumo2 (5′- GACCCGGTGCCTCTTTTGTGAAGCGGCAGCTGAGGAGACTCCGGCGCTCGCCATGCACCACCACCACCACCACTACCCATACGATGTTCCAGATTACGCTGCCGACGAGAAACCCAAGGTGAGCTCGCGGCGTCCGGCTGCGCCTCACCGTGCGGACCGGTGGGTGGCGCGGCCCGGGGTCCGGCCGAAGCCCGCCGCGG-3′). On the day of injection, Cas9 protein (PNA Bio), sgRNA and repair template (ssODN) were injected into ova from C57Bl/6 female mice and transferred into oviducts of pseudopregnant females. Founder mice were backcrossed at least three times prior to experimentation to get rid of potential off-target mutations. His6-HA-Sumo2 and His6-HA-Sumo1 mice were backcrossed at least six times onto an FVB/N background prior to beginning experimentation.
For the generation of the HA-Sumo2 knock-in mouse, two sgRNA, generated via T7 in vitro transcription (Cellscript, #C-MSC100625) from corresponding linearized vectors, were used: sgRNA1 sense 5′-GCCTCACCGTGCGGACCGGT-3′, sgRNA2 antisense 5′-GCTCACCTTGGGTTTCTCGT-3′. Gibson assembly (NEB #E2611S) was used to generate a double stranded HDR fragment that included the HA tag coding sequence and silent mutations to avoid re-targeting by Cas9 (5'∼TTGAGGCTCCCGGGTGGGAAGCTGGTTGATGGGGAAGGACGAAGGGACTGGGAAGTTTGATGTGCAGGGGGGAGTTGCATTTGTCCTACTGAACAGGAGTTTTTCTCACAACCCGCTGCTGCTTTTTTGATGGAGATGCCCCCTTCTACGTGGAGTTGGCCTTCCGTTCTCGACCTAGGCTCACCCCACGTCTAAGGCCACGCGCCTTCCTGTCCTCGCCGAGGACCAGCTGGAAGCCATTTTGATTATGCTCCACCAAAGAAGGAGGACAATACAGCCCCGCCCAGAGCCAGGCGCGCCCCCGCGGCGGGACTGGTGCGCGTGTCCGCGCCTCTTCCCGCCGACCTGGCGGGCGCCTGGCCCAGGCGGCGGAGGCGCGGGCCGTGGGGGGGAGGAGGAGGGGTGGTTATGTAAGCGTTGCGCGCTCCCGCGAAGCGGCAGCCAATGAGAAGCCCCGGATGCTCGTGCTCGCGCACGCAGGGCGGGGGGAGGGAAGCGCACGACGTGCGCGCGCCCTCTGCCTCGTCCACCGCTGCCGCCTCCTCCTGCCGCTCCTGGTGCTGCTTGTGCGCTCCCTCAGTGCGGACCCGGTGCCTCTTTTGTGAAGCGGCAGCTGAGGAGACTCCGGCGCTCGCC(ATG)TACCCATACGATGTTCCAGATTACGCTGCtGACGAaAAgCCCAAGGTGAGCTCGCGGCGGCCGGCTGCGCCTCACgGTGCGcACCGGTGtGTGGCGCGGCCCGGGGTCCGGCCGAtGCCCtCCGCGcGCTTCCCTGCCTGCCCGGGGCCCCCCACGGGGCCAGCGGCGGCTCAGGCCCGGCGCTCCGCGCCATTTTCCTCTCTCGCTCTCTTCCTCCCGCCCGCGCCCGAGGAGGCCCGGCGCCCTCCCCCGCCCCCAGGCCCTCGGAGCGCGCGCGGCGCGGCGCGAGGCGGGCAGGCGGCGCTTCGCGGGAGCCCGCGGGCCCGGCGGCCCCGCGCTGGCTCCTCGGCGGCGCCCGGCGCGTGTAAGGAAAGCGTTGGGCGCGCGGGGTCTGGGGCGGCGGCGAAGGCGGGAGTCGTGCGGCCCGGGGTTTCCCTCTCGGGGCTGGGGCCGCGCCGGATCGGTGGTGTGTGCTCCGGCAACCCCGAGTGGGGGATTCCAGAGTCGGTCCGGCTCGAGGCCCAAG∼3’) (bold, exon 1 with ATG in parentheses; italic, HA tag coding sequence; small letters, silent mutations in exon, mutations in intron; underlined, annealing sites for PCR primers used for genotyping). Cas9 protein (NEB #M0646T) and hCas9 mRNA generated via T7 in vitro transcription (Cellscript, #C-MSC100625) were used. On the day of injection, hCas9 mRNA and protein, the two sgRNAs and the HDR fragment were injected into zygotes obtained from FVB/N female mice, and injected zygotes were transferred into oviducts of pseudopregnant females. Founder mice were further crossed with FVB/N.
Mouse genotyping
Genomic DNA (gDNA) was isolated from small tail samples collected from mouse embryos (primary cortical neuron experiments), pups (pre-weaning), or adult mice (endpoint). His6-HA-Sumo2 mice were genotyped using parallel reactions with primers targeting the His6-HA knock-in tag and corresponding genomic loci upstream (Forward: 5′- AGGAAGAGAGCGAGAGAGGAA-3′, Reverse: 5′-CACCACCACTACCCATACGA-3′, 224 bp product) and downstream (Forward: 5′-TCGTATGGGTAGTGGTGGTG-3′, Reverse: 5′-AGGAGGAGGGGTGGTTATGT-3′, 276 bp product) of the knock-in. 25 μL reactions were prepared using <10 ng gDNA, primers (400 nM final), 1.25 μL DMSO (5% final), and 2X GoTaq (Promega). Thermocycler parameters: 95°C for 2 min, (95°C for 30 s, 57.6°C for 30 s, 72°C for 40 s) repeated for 35 cycles, final denature at 72°C for 5 min. HA-Sumo2 mice were genotyped with primers flanking the HA knock-in tag (Forward: 5′-GCCCTCTGCCTCGTCCAC-3′, Reverse: 5′-CCGCCGCGAGCTCACCTTG.
−3′, WT Allele: 160 bp product, KI Allele: 187 bp product). 20 μL reactions were prepared using 1 μL clean gDNA, primers (4 pmol final), and 5X Biozym Hot-Start Taq DNA Polymerase plus extra Mg2+ (Biozym 331620XL). Thermocycler parameters: 96°C for 3 min, (94°C for 30 s, 62°C for 60 s, 72°C for 60 s) repeated for 32 cycles, final denature at 72°C for 7 min His6-HA-Sumo1 mice were genotyped as previously described.32
Biochemical analysis of adult mouse brain and spinal cord
Mice were anesthetized using isoflurane inhalation and sacrificed via decapitation. Brains were quickly isolated, dissected for regional protein analysis, and flash frozen on dry ice. Analysis of total Sumo1 and Sumo2/3 levels were performed on whole brain lysates. Spinal cords were removed using hydraulic extrusion.85 Samples were thawed and immediately lysed using a Dounce homogenizer in RIPA buffer (9.1 mM dibasic sodium phosphate, 1.7 mM monobasic sodium phosphate, 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing 50 mM freshly prepared N-Ethylmaleimide (Sigma), 0.25% β-mercaptoethanol, and Xpert Protease (GenDEPOT) and Xpert Phosphatase (GenDEPOT) inhibitor cocktails. Whole brain lysates were lysed in 11 mL of RIPA buffer described above and centrifuged at 125,000 x g for 2 h at 4°C. Regional lysates were centrifuged at 21,000 x g for 20 min at 4°C and the supernatant was removed and suspended in 4X laemmli buffer (BioRad) with 2-mercaptaethanol (BioRad) and boiled at 95°C for 5 min. Samples were run on 8% polyacrylamide gels and transferred onto 0.45 μm nitrocellulose membranes at 340 mA for 90 min and stained with ponceau to normalize protein levels between lysates. Normalized lysates were run on a 4-15% GTx Mini-PROTEAN (BioRad) or Bolt™ 4-12% Bis-Tris Plus (Invitrogen) gel and transferred onto a 0.45 μm nitrocellulose membrane at 340 mA for 2 h for western blot analysis. Densitometry was performed using BioRad Image Lab software and FiJi/ImageJ by normalizing the HA intensity to ponceau for High Molecular Weight (>70 kDa) and free Sumo levels.
Real time quantitative PCR (RT-qPCR) analysis
RNA was extracted from mouse brain homogenate using Trizol-Chloro-form extraction (Invitrogen User Guide: TRIzol Reagent version B.0). Briefly, mouse brains were homogenized in 3 mL of PEPI Buffer [5 mM EDTA, 1X protease inhibitor (GenDEPOT cat# P3100-020), in 1X PBS] using a Dounce homogenizer. 3% of homogenate was added to 1 mL of TRIzol Reagent (Fisher Scientific cat# 15-596-026) and RNA was isolated as per the user guide referenced above. For CNS regional RNA analysis, tissue samples were homogenized in 300 μL of PEPI buffer using a pestle and passed through a 28 gauge insulin needle before RNA was extracted using RNeasy Kits (Qiagen cat# 74104). cDNA was synthesized using 5X All-in-One RT Master Mix (Bio Basic cat# HRT025-10) following manufacturer’s instructions. RT-qPCR was performed using Green-2-Go qPCR Master Mix (Bio Basic cat# QPCR004-S) with 25 ng cDNA per reaction and primers targeting mouse Sumo paralogs17 Sumo1 (NM_009460.2) (Forward: 5′- CAGGAGGCAAAACCTTCAAC-3′, Reverse: 5′- CTCCATTCCCAGTTCTTTCG-3′), Sumo2(NM_133354.2) (Forward: 5′- ACGAGAAACCCAAGGAAGGA-3′, Reverse: 5′- CTCCATTTCCAACTGTGCAG-3′), Sumo3 (NM_001301672) (Forward: 5′- AGAAGCCCAAGGAGGGTGT-3′, Reverse: 5′- CCTCGGGAGGCTGATCCT-3′) and housekeeping controls Gapdh (NM_001289726) (Forward: 5′-GGAGAGTGTTTCCTCGTCCC-3′, Reverse: 5′- ATGAAGGGGTCGTTGATGGC-3′) and Hprt1 (NM_013556.2) (Forward: 5′-TGATAGATCCATTCCTATGACTGTAGA-3′, Reverse: 5′-AAGACATTCTTTCCAGTTAAAGTTGAG-3′). Reactions were run on BioRad CFX96 thermocycler (protocol: 95°C for 5 min, 40 cycles of 95°C for 15 s and 60°C for 60 s, then melting curve). Sumo Ct values were standardized to the average of Gapdh and Hprt1 Ct values.
Brain preparation for whole brain clearing and HA immunolabeling
Mice were sedated via intraperitoneal injection of 16.25 mg sodium pentobarbital (Euthanyl, DIN 00141704). Once sedated, mice were perfused using 10 mL 1X PBS +10 U/mL heparin (Millipore Sigma, H3393-50KU) followed by 10 mL of freshly prepared 4% paraformaldehyde. Brains were carefully isolated and stored in 13 mL of 4% paraformaldehyde at 4°C overnight with gentle shaking. Brains were rinsed with 1X PBS then shipped to LifeCanvas Technologies (MA) in 1X PBS +0.02% sodium azide.
Whole mouse brain processing, staining, and imaging
Whole mouse brains were processed using the SHIELD protocol (LifeCanvas Technologies86). Samples were cleared for 1 day at 42°C then actively immunolabeled using SmartBatch+ (LifeCanvas Technologies) based on eFLASH technology integrating stochastic electrotransport87 and SWITCH.88 Each sample was labeled with 60 μg anti-NeuN (Encor, MCA-1B7) and 36 μg rabbit anti-HA-tag (Cell Signaling Technology #3724) followed by fluorescently conjugated secondary antibodies in a 3:2 primary:secondary molar ratio (Jackson ImmunoResearch). Samples were incubated in EasyIndex (LifeCanvas Technologies) for refractive index of RI = 1.52 and imaged at 3.6X using a SmartSPIM axially-swept light sheet microscope (LifeCanvas Technologies). Images were tile corrected, de-striped, and registered to the Allen Brain Atlas (Allen Institute: https://portal.brain-map.org/). NeuN channels for each brain were registered to 8-20 atlas-aligned reference samples using successive rigid, affine, and b-spline warping (SimpleElastix: https://simpleelastix.github.io/). Average alignment to the atlas was generated across all intermediate reference sample alignments to serve as the final atlas alignment value per sample. Fluorescent measurements from the acquired images were projected onto the Allen Brain Atlas to quantify the total fluorescence intensity per region defined by the Allen Brain Atlas. These values were then divided by the volume of the corresponding regional volume to calculate the intensity per voxel measurements.
HA immunoprecipitation from mouse brain lysates
Immunoprecipitation was performed based on the previously established protocol.32,89 Briefly, mice aged 9-16 weeks old were anesthetized and sacrificed via decapitation and the brain was quickly removed and homogenized using a Dounce homogenizer in RIPA buffer (9.1 mM dibasic sodium phosphate, 1.7 mM monobasic sodium phosphate, 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing 50 mM freshly prepared N-Ethylmalamide (Sigma), 0.25% β-mercaptoethanol, and Xpert Protease (GenDEPOT) and Xpert Phosphatase (GenDEPOT) inhibitor cocktails and boiled for 5 min at 95°C. Samples were lysed on ice for 20 min with vigorous vortexing every 5 min before ultracentrifugation at an average of 100,000 x g for 30 min at 4°C. Supernatants were removed and spiked with 50 mM N-ethylmalamide before being added to 50 μL of magnetic protein G Dynabeads (Invitrogen) pre-conjugated with 10 μg mouse HA antibody (in house) per sample. Samples were then placed on rotator at 4°C for 1 h. Beads were washed three times in 10 mL of RIPA buffer containing 20 mM N-Ethylmalamide, protease, and phosphatase inhibitors. Wash buffer was thoroughly removed, and beads were eluted using 75 μg synthetic HA peptide (Sino Biological Inc.).
Sample preparation for mass spectrometry
Two-thirds of eluted protein sample from HA Immunoprecipitation were run on a 4-15% Mini-PROTEAN (Bio-Rad) gel to separate proteins and remove synthetic HA peptide. Samples were stained using Silver Stain for Mass Spectrometry Kit (Thermo Scientific, 24600). Lanes were cut into 6-7 gel slices per sample and stored in 1% acetic acid until analysis via LC-MS/MS.
Protein identification by LC-MS/MS
Proteomics analysis was performed at the Ottawa Hospital Research Institute Proteomics Core Facility (Ottawa, Canada). Proteins were digested in-gel using trypsin (Promega) according to the method of Shevchenko 90 but without the use of iodoacetamide for cysteine alkylation due to the treatment of 50 mM N-ethylmaleimide (Sigma) during the sample immunoprecipitation. Peptide extracts were concentrated by Vacufuge (Eppendorf). LC-MS/MS was performed using a Dionex Ultimate 3000 RLSC nano HPLC (Thermo Scientific) and Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). MASCOT software version 2.7.0 (Matrix Science, UK) was used to infer peptide and protein identities from the mass spectra. The observed spectra were matched against Mus musculus sequences from SwissProt (version 2021-02) and against an in-house database of common contaminants. The results were exported to Scaf-fold (Proteome Software, USA) for further validation and viewing and will be uploaded to the ProteomXchange Consortium.
Gene ontology analysis
Top His6-HA-Sumo1 and His6-HA-Sumo2 interactors were analyzed using gProfiler2 web tool (https://biit.cs.ut.ee/gprofiler/gost).91 Lists were arranged in descending order based on relative peptide abundance and analyzed as an ordered query for Mus musculus proteins using Benjamini-Hochberg FDR significance threshold with an alpha of 0.05.
Immunohistochemistry of mouse brain
Mice were anesthetized with 30 μL of 120 mg/kg Euthanyl (DIN00141704) and then perfused with 10 mL 1x phosphate buffered saline (PBS) and 10 mL 4% paraformaldehyde (PFA). Brain and spinal cord tissue were collected and stored for 48 h in 4% PFA. Brain and spinal cord tissue were then dehydrated in 10%, 20% and 30% sucrose solutions for 48 h each before being flash frozen in −40 C isopentane for 1 min. Tissues were then sectioned at 20 μm and −21 C on the Thermo Scientific HM 525 NX cryostat at the Louise Pelletier Histology Core at the University of Ottawa and stored free floating in 1x PBS +0.02% NaN3 at 4 C until use in staining. Brains tissue were incubated for 24 h in blocking buffer (1.5% Triton X-100, 5% cosmic calf serum in 1X PBS), 24 h in primary antibody (1:500 HA-tag C29F4 rabbit monoclonal antibody (Cat: 3724S, Cell Signaling Technology), and 1 h in secondary antibody (1:500 Alexa Fluor 568 donkey anti mouse antibody, Cat: A10037, Lot: 1917938)) with DAPI (1:1,000 Millipore Sigma, D9542-1 MG). Tissue was washed 5 times for 5 min each in 1x PBS between each treatment and mounted on Fisherbrand Superfrost Plus slides. Slides were left to dry for 24 h and covered with DAKO mounting medium (Cat: S3023, Lot: 11347938) and #1.5 coverslips.
For the analysis of the synaptic localization of Sumo2, HA-Sumo2 mouse brains were used (Figure S1A). Mice were anesthetized (250 mg/kg Avertin i.p.) and transcardially perfused with PBS and then with 4% (w/v) paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB), pH 7.4 at 4°C for 10 min. Brains were removed and post-fixed for 1 h at 4°C. The tissue was cryoprotected in 30% (w/v) sucrose in phosphate-buffered saline (PBS). Brains were frozen in the cryostat and sagittal 35 μm sections were prepared with a cryostat and collected free-floating in PBS. For immunohistochemistry, sections were pre-incubated for 24 h in PB containing 3% horse serum (HS), 3% fish skin gelatin (FSG), and 0.3% Triton X-100, and were then incubated for 3 days at 4°C in primary antibodies diluted in PBS containing 3% HS, 3% FSG and 0.3% Triton X-100. After washing repeatedly in PBS and overnight, sections were incubated overnight in dye coupled secondary antibodies, repeatedly washed, and mounted on slides with Aquapolymount (Polysciences). The antibodies used are listed in Table S3.
Primary cortical neuron cultures
Pregnant mice were euthanized between at gestation E14.5–15.5 with 48 mg Pentobarbital Sodium (Bimeda-MTC, 8015E) delivered via intraperitoneal injection. Embryos were removed, washed in chilled PBS (Wisent Bioproducts, 311-010-CL), and cortices were isolated in chilled HBSS (Sigma Aldrich, H9394). Cortices were dissociated for 20 min with trypsin (Thermo Scientific, 90305) at 37°C before adding trypsin inhibitor and DNase solution to quench reaction. Cells were pelleted at 2,500 x g for 5 min at 4°C and washed with trypsin inhibitor with DNase solution. Cortical neurons were pelleted at 2,500 x g for 5 min at 4°C and resuspended in 1 mL Neurobasal outgrowth media (Thermo Scientific, 21103049), supplemented with B-27 (Thermo Scientific, 17504044), N-2 (Thermo Scientific, 21103049), 500 μM L-Glutamine (Wisent Bioproducts, 609-065-EL), and 0.5% penicillin/streptomycin (GE Healthcare Life Sciences, SV30010) before plating. Cultures were maintained for 7 days in vitro with a half media change after 3-4 days.
Immunofluorescence in primary cortical neurons
Micro Coverglass #1.5 coverslips (Electron Microscopy Sciences) coverslips were pre-coated with poly-D-lysine (50 μg/mL) overnight at 37°C, then washed with distilled water three times and air-dried at room temperature for at least 2 h. Primary mouse cortical neurons were seeded at 75,000 cells per coverslip and cultured as described in primary cortical neuron cultures. On day 7, neurons were fixed using 10% phosphate buffered formalin for (Fisher Chemical, SF100-4) for 10 min followed by 3 x 5-min washes in 1 mL of 1X PBS. Neurons were blocked in 500 μL of blocking buffer (1.5% Triton X-100, 10% cosmic calf serum in 1X PBS) for 1 h, then incubated in 300 μL of primary antibody diluted in blocking buffer overnight at 4°C. The following day, the neurons were washed for 4 x 5-min washes in 1 mL of 1X PBS then incubated in 300 μL of secondary antibody diluted in blocking buffer for 2 h at room temperature. Next, the neurons were washed for 4 x 5-min washes in 1 mL of 1X PBS, dried, and then placed on slides with Vectashield Antifade Mounting Medium with DAPI (MJS Biolynx Inc., H-1200). z stack images were obtained on a Zeiss AxioObserverZ1 LSM800 Confocal Microscope at 63× magnification through a Z distance of 10 μm per image using optimal 0.27 μm spacing per slice. The dimensions were set to 1,024 × 1,024 pixels.
Proximity ligation assay in primary cortical neurons
Primary mouse cortical neurons were cultured and fixed as described for immunofluorescence experiments. Individual coverslips for Proximity Ligation Assay experiments (PLA) were transferred to 12-well plates and outlined with a hydrophobic pen. Neurons were blocked using 40 μL of Duolink blocking buffer (Sigma Aldrich, DUO82007) at 37°C for 1 h and were then washed for 3 × 5 min washes in 1 mL 1X PBS. Next, the neurons were incubated in 300 μL of primary antibody diluted in blocking buffer (1.5% Triton X-100, 10% cosmic calf serum in 1X PBS) overnight at 4°C. The following day, the neurons were washed for 3 × 5 min washes in 1 mL of Duolink Wash Buffer A (0.01 M Tris-Base, 0.15 M NaCl, 0.05% Tween 20, pH 7.4) followed by incubation in 40 μL of Duolink PLA MINUS (Sigma Aldrich, DUO82004) and PLUS probes (Sigma Aldrich, DUO82002) at 37°C for 1 h. Duolink PLA probes were diluted in antibody diluent (Sigma Aldrich, DUO82008) at a 1:5 dilution. Neurons were washed for 3 × 5 min washes in Duolink Wash Buffer A and then incubated in 40 μL of ligase (Sigma Aldrich, DUO82027) at 37°C for 30 min. Ligase was diluted in 1X ligation buffer (Sigma Aldrich, DUO82009) at a 1:40 dilution. Neurons were washed for 3 x 5-min washes in Duolink Wash Buffer A and then incubated in 40 μL of polymerase (Sigma Aldrich, DUO82028) at 37°C for 90 min. Polymerase was diluted in 1X amplification buffer (Sigma Aldrich, DUO82011) at a 1:80 dilution. Then, neurons were washed 2 × 10 min in Duolink Wash Buffer B (0.2 M Tris-Base, 0.1 M NaCl, pH 7.5) and then again in 1 mL of Duolink Wash Buffer B diluted at 1:100 for 1 min. Coverslips were briefly air dried and then mounted on slides using Vectashield Antifade Mounting Medium with DAPI. z stack images were obtained on a Zeiss AxioObserverZ1 LSM800 Confocal Microscope at 63× magnification with a 5× digital zoom through a Z distance of 10 μm per image using optimal 0.27 μm spacing per slice with dimensions set to 512 × 512 pixels. Images were analyzed and quantified using the Spots function on the Imaris (ver. 9.9.1 Bitplane, Switzerland) software. Localization of the foci (nuclear versus cytoplasmic) was determined using the Orthogonal Views function for co-localization with DAPI signal.
Tissue lysis
Dissected mouse organs were lysed in 1X RIPA buffer (9.1 mM Na2HPO4, 1.7 mM NaH2PO4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1X protease inhibitor (GenDEPOT, P3100), 1X phosphatase inhibitor (GenDEPOT, P3200) and 50 mM N-ethylmaleimide (Sigma-Aldrich, E3876-5G) using a Dounce homogenizer. Tissue lysates were centrifuged at ∼21,000 x g for 15 min at 4°C and the resulting supernatant was transferred into a fresh microcentrifuge tube.
Subcellular fractionation
Western blot analysis of brain subcellular fraction was performed as described previously.32,92
Western blot analysis
SDS-PAGE was performed with standard discontinuous gels or with commercially available 4%–12% Bis-Tris gradient gels (Invitrogen). Western blots were probed using primary and secondary antibodies as indicated in Table S3. Blots were routinely developed using enhanced chemiluminescence (GE Healthcare) and imaged using an INTAS ECL Chemostar PLUS Imager HR 6.0. For quantitative Western blotting, transferred proteins were visualized using the total protein stain MemCode (Invitrogen). Western blot signals were visualized using an INTAS ECL Chemostar PLUS Imager HR 6.0 apparatus and quantified using ImageJ. For normalization, a ratio of the antibody signal relative to the total protein stain as revealed by the MemCode was performed.
Confocal imaging and image analysis
MAP2, HA-Sumo2, DAPI and the respective synaptic marker fluorescence intensities were acquired from the hippocampal CA3 pyramidal layer and the stratum radiatum. Multi-channel z-stacks of 338 × 338 μm-sized fields of view were acquired with a 40× oil immersion objective (1.3 NA) of a Nikon Eclipse Ti confocal microscope equipped with a Yokogawa W1 spinning disk and an Andor iXon Ultra 888 EMCCD camera. A custom macro written within the FIJI software package (Schindelin et al., 2012) automatically identified the z-plane of highest HA-Sumo2 intensity, out of which all channels were extracted. The DAPI, MAP2 and synaptic marker channel of this z-plane were segmented with the Trainable WEKA Segmentation FIJI plugin (Arganda-Carreras et al., 2017), resulting in binary masks (Figure 3A, bottom row) that were subsequently used to extract the respective HA-Sumo2 mean intensity.
To more closely investigate HA-Sumo2 co-localization with the respective synaptic markers, high-resolution, multi-channel fields of view (x, y: 30, 30 μm) of the respective stains within the Mossy fiber region were acquired using 40× oil-immersed objective (NA = 1.4) of a Zeiss LSM 880 confocal laser scanning microscope. Oversampled (x, y, z: 30, 30, 100 nm), 1.2 μm-sized z-stacks within the mossy fiber region were acquired for each fluorescent channel. Signal to noise ratio and resolution were subsequently enhanced and sample geometry was corrected with the SVI Huygens deconvolution software package (ver. 22.04, Hilversum, Netherlands). To obtain object-based co-localization information, surfaces depicting either HA-Sumo2 or the respective marker were generated with an Imaris (ver. 9.9.1 Bitplane, Switzerland) batch workflow, which automatically and identically processed the three-dimensional fields-of-view originating from WT or knock-in animals. All HA-Sumo2 and the respective synaptic marker surface objects were created, yielding the total object number for the respective stain (Figure S5- top and middle rows of A and B). Synaptic marker objects were designated as collocating with the HA-Sumo2 objects only if they overlapped with HA-Sumo2 objects and if they contained a minimum HA-Sumo2 average intensity (Figure S5, bottom rows of A and B). Data resulting from both analysis workflows were assembled and quantified with the KNIME software package (ver. 4.5.2, Zurich, Switzerland).
Quantification and statistical analysis
Statistical tests were performed using PRISM 9.4. Test type was picked based on the number of comparisons made. Levels of statistical significance are indicated in figure legends.
Acknowledgments
This research was supported in part by an NSERC Discovery Grant and Discovery Launch Supplement to M.W.C.R. (RGPIN-2019-04133 and DGECR-2019-00369); the Canada Research Chairs program to M.W.C.R.; German Research Foundation, SFB1286/A09 to N.B. and M.T.; the ALS Society of Canada in partnership with the Brain Canada Foundation through the Brain Canada Research Fund, with the financial support of Health Canada, for financial support through the ALS Trainee Award Program 2019 (T.R.S.); an NSERC Undergraduates Student Research Award to T.T.N.; the Ontario Graduate Scholarship (H.M.G.), the Queen Elizabeth II Scholarship (H.M.G.), and a CIHR Canadian Graduate Scholarship (H.M.G.). M.W.C.R. thanks H.Y. Zoghbi (Baylor College of Medicine, HHMI) for initial project discussions, reagent development, and the freedom to explore new ideas. The authors further thank all members of the Rousseaux, Brose, and Zoghbi labs for important discussions and critical feedback on the manuscript. The authors also thank the following Core facilities from the University of Ottawa and the Ottawa Hospital Research Institute (OHRI) for use of their facility, equipment, and expertise: the Cell Biology and Imaging Acquisition Core and the OHRI Proteomics Core. The authors also thank the Genome Engineered Rodent Models Core at Baylor College of Medicine and Animal Care and Veterinary Service. The authors are grateful to the Transgenic Facility and the DNA Core Facility at the MPI for Multidisciplinary Sciences for generating and genotyping KI mice, and to the team of animal caretakers at the Animal Facility of the MPI for Multidisciplinary Sciences for their excellent work. Figures 1, S1, 2, 5, and 6 were generated in part with Biorender.com. The views expressed herein do not necessarily represent the views of the Minister of Health or the Government of Canada.
Author contributions
T.R.S., T.T.N., Z.A.F., M.M., H.M.G., J.L.-A.P., M.M.H., S.M.C., and M.T., investigation, formal analysis; M.W.C.R., M.T., and N.B., resources; M.W.C.R., M.T., and N.B., conceptualization; T.R.S., M.W.C.R., M.T., and N.B., writing original draft and editing; M.W.C.R., M.T., and N.B., supervision and funding acquisition. All authors reviewed and approved the manuscript.
Declaration of interests
The authors have no competing interests to declare.
Published: March 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106350.
Contributor Information
Marilyn Tirard, Email: tirard@mpinat.mpg.de.
Maxime W.C. Rousseaux, Email: max.rousseaux@uottawa.ca.
Supplemental information
Scale bar: 3000 μm.
Scale bar: 3000 μm.
Data and code availability
-
•
Mass spectrometry data is available from the lead contact upon request.
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Celen A.B., Sahin U. Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts. FEBS J. 2020;287:3110–3140. doi: 10.1111/febs.15319. [DOI] [PubMed] [Google Scholar]
- 2.Ilic D., Magnussen H.M., Tirard M. Stress - regulation of SUMO conjugation and of other ubiquitin-like modifiers. Semin. Cell Dev. Biol. 2022;132:38–50. doi: 10.1016/j.semcdb.2021.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Xu Z., Au S.W.N. Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1. Biochem. J. 2005;386:325–330. doi: 10.1042/BJ20041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong L., Li B., Millas S., Yeh E.T. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/S0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 5.Seufert W., Futcher B., Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 6.Johnson E.S., Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 7.Gong L., Kamitani T., Fujise K., Caskey L.S., Yeh E.T. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 8.Desterro J.M., Thomson J., Hay R.T. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/S0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 9.Saitoh H., Sparrow D.B., Shiomi T., Pu R.T., Nishimoto T., Mohun T.J., Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 1998;8:121–124. doi: 10.1016/S0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee G.W., Melchior F., Matunis M.J., Mahajan R., Tian Q., Anderson P. Modification of Ran GTPpase-activating protein by the small ubiquitin- related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- 11.Sampson D.A., Wang M., Matunis M.J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez M.S., Dargemont C., Hay R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 13.Nayak A., Müller S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014;15:422. doi: 10.1186/s13059-014-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citro S., Chiocca S. Sumo paralogs: redundancy and divergencies. Frontiers in Bioscience - Scholar. 2013;5:544–553. doi: 10.2741/S388/PDF. [DOI] [PubMed] [Google Scholar]
- 15.Bouchard D., Wang W., Yang W.C., He S., Garcia A., Matunis M.J. SUMO paralogue–specific functions revealed through systematic analysis of human knockout cell lines and gene expression data. Mol. Biol. Cell. 2021;32:1849–1866. doi: 10.1091/MBC.E21-01-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohren K.M., Nadkarni V., Song J.H., Gabbay K.H., Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 17.Wang L., Wansleeben C., Zhao S., Miao P., Paschen W., Yang W. SUMO 2 is essential while SUMO 3 is dispensable for mouse embryonic development. EMBO Rep. 2014;15:878–885. doi: 10.15252/embr.201438534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S., Galeffi F., Rodriguiz R.M., Wang Z., Shen Y., Lyu J., Li R., Bernstock J.D., Johnson K.R., Liu S., et al. Small ubiquitin-like modifier 2 (SUMO2) is critical for memory processes in mice. FASEB J. 2020;34:14750–14767. doi: 10.1096/FJ.202000850RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vertegaal A.C.O. Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 2022;23:715–731. doi: 10.1038/S41580-022-00500-Y. [DOI] [PubMed] [Google Scholar]
- 20.Talamillo A., Barroso-Gomila O., Giordano I., Ajuria L., Grillo M., Mayor U., Barrio R. The role of SUMOylation during development. Biochem. Soc. Trans. 2020;48:463–478. doi: 10.1042/BST20190390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendler L., Braun T., Müller S. The ubiquitin-like SUMO system and heart function: from development to disease. Circ. Res. 2016;118:132–144. doi: 10.1161/CIRCRESAHA.115.307730. [DOI] [PubMed] [Google Scholar]
- 22.Yang W., Paschen W. SUMO proteomics to decipher the SUMO-modified proteome regulated by various diseases. Proteomics. 2015;15:1181–1191. doi: 10.1002/PMIC.201400298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstock J.D., Yang W., Ye D.G., Shen Y., Pluchino S., Lee Y.J., Hallenbeck J.M., Paschen W. SUMOylation in brain ischemia: patterns, targets, and translational implications. J. Cerebr. Blood Flow Metabol. 2018;38:5–16. doi: 10.1177/0271678X17742260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garvin A.J., Lanz A.J., Morris J.R. SUMO monoclonal antibodies vary in sensitivity, specificity, and ability to detect types of SUMO conjugate. Sci. Rep. 2022;12:21343. doi: 10.1038/S41598-022-25665-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman N., Nayak A., Muller S. The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma. 2013;122:475–485. doi: 10.1007/S00412-013-0429-6. [DOI] [PubMed] [Google Scholar]
- 26.Matsuzaki S., Lee L., Knock E., Srikumar T., Sakurai M., Hazrati L.N., Katayama T., Staniszewski A., Raught B., Arancio O., Fraser P.E. SUMO1 affects synaptic function, spine density and memory. Sci. Rep. 2015;5:10730. doi: 10.1038/SREP10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schorova L., Martin S. Sumoylation in synaptic function and dysfunction. Front. Synaptic Neurosci. 2016;8:9. doi: 10.3389/FNSYN.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcelli S., Ficulle E., Iannuzzi F., Kövari E., Nisticò R., Feligioni M. Targeting SUMO-1ylation contrasts synaptic dysfunction in a mouse model of alzheimer’s disease. Mol. Neurobiol. 2017;54:6609–6623. doi: 10.1007/S12035-016-0176-9. [DOI] [PubMed] [Google Scholar]
- 29.Lee L., Dale E., Staniszewski A., Zhang H., Saeed F., Sakurai M., Fa’ M., Orozco I., Michelassi F., Akpan N., et al. Regulation of synaptic plasticity and cognition by SUMO in normal physiology and Alzheimer’s disease. Sci. Rep. 2014;4:7190. doi: 10.1038/SREP07190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henley J.M., Carmichael R.E., Wilkinson K.A. Extranuclear SUMOylation in neurons. Trends Neurosci. 2018;41:198–210. doi: 10.1016/J.TINS.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Colnaghi L., Russo L., Natale C., Restelli E., Cagnotto A., Salmona M., Chiesa R., Fioriti L. Super resolution microscopy of SUMO proteins in neurons. Front. Cell. Neurosci. 2019;13:486. doi: 10.3389/FNCEL.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tirard M., Hsiao H.-H., Nikolov M., Urlaub H., Melchior F., Brose N. In vivo localization and identification of SUMOylated proteins in the brain of His6-HA-SUMO1 knock-in mice. ProcProc. Natl. Acad. Sci. USA. 2012;109:21122–21127. doi: 10.1073/pnas.1215366110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel J.A., Cooper B.H., Palvimo J.J., Zhang F.-P., Brose N., Tirard M. Analysis of SUMO1-conjugation at synapses. Elife. 2017;6:e26338. doi: 10.7554/eLife.26338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel J.A., Cooper B.H., Palvimo J.J., Zhang F.P., Brose N., Tirard M. Response: commentary: analysis of SUMO1-conjugation at synapses. Front. Cell. Neurosci. 2018;12:117. doi: 10.3389/FNCEL.2018.00117/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldmeyer D. Excitatory neuronal connectivity in the barrel cortex. Front. Neuroanat. 2012;6:24. doi: 10.3389/FNANA.2012.00024/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Drunen R., Eckel-Mahan K. Circadian rhythms of the hypothalamus: from function to physiology. Clocks Sleep. 2021;3:189–226. doi: 10.3390/clockssleep3010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendriks I.A., Vertegaal A.C.O. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 38.Hendriks I.A., Vertegaal A.C.O. SUMO in the DNA damage response. Oncotarget. 2015;6:15734–15735. doi: 10.18632/ONCOTARGET.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichler A., Gast A., Seeler J.S., Dejean A., Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/S0092-8674(01)00633-X. [DOI] [PubMed] [Google Scholar]
- 40.Liang Q., Deng H., Li X., Wu X., Tang Q., Chang T.-H., Peng H., Rauscher F.J., Ozato K., Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 2011;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu Y., Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gareau J.R., Reverter D., Lima C.D. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J. Biol. Chem. 2012;287:4740–4751. doi: 10.1074/JBC.M111.321141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/NRM2293. [DOI] [PubMed] [Google Scholar]
- 44.Lavado A., Lagutin O.v., Chow L.M.L., Baker S.J., Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010;8 doi: 10.1371/JOURNAL.PBIO.1000460. e1000460–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L., Rodriguiz R.M., Wetsel W.C., Sheng H., Zhao S., Liu X., Paschen W., Yang W. Neuron-specific Sumo1-3 knockdown in mice impairs episodic and fear memories. J. Psychiatry Neurosci. 2014;39:259–266. doi: 10.1503/JPN.130148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W., Paschen W. SUMO proteomics to decipher the SUMO-modified proteome regulated by various diseases. Proteomics. 2015;15:1181–1191. doi: 10.1002/PMIC.201400298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasegawa Y., Yoshida D., Nakamura Y., Sakakibara S.I. Spatiotemporal distribution of SUMOylation components during mouse brain development. J. Comp. Neurol. 2014;522:3020–3036. doi: 10.1002/CNE.23563. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe M., Takahashi K., Tomizawa K., Mizusawa H., Takahashi H. Developmental regulation of Ubc9 in the rat nervous system. Acta Biochim. Pol. 2008;55:681–686. [PubMed] [Google Scholar]
- 49.Sahin U., Jollivet F., Berthier C., de Thé H., Lallemand-Breitenbach V. Detection of protein SUMOylation in situ by proximity ligation assays. Methods Mol. Biol. 2016;1475:139–150. doi: 10.1007/978-1-4939-6358-4_10/FIGURES/3. [DOI] [PubMed] [Google Scholar]
- 50.Ristic M., Brockly F., Piechaczyk M., Bossis G. Methods in Molecular Biology. Humana Press Inc.; 2016. Detection of protein–protein interactions and posttranslational modifications using the proximity ligation assay: application to the study of the SUMO pathway; pp. 279–290. [DOI] [PubMed] [Google Scholar]
- 51.Rajgor D., Hanley J.G., Shanahan C.M. Identification of novel nesprin-1 binding partners and cytoplasmic matrin-3 in processing bodies. Mol. Biol. Cell. 2016;27:3894–3902. doi: 10.1091/MBC.E16-06-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripamonti S., Shomroni O., Rhee J.S., Chowdhury K., Jahn O., Hellmann K.P., Bonn S., Brose N., Tirard M. SUMOylation controls the neurodevelopmental function of the transcription factor Zbtb20. J. Neurochem. 2020;154:647–661. doi: 10.1111/JNC.15008. [DOI] [PubMed] [Google Scholar]
- 53.Huang D., Liu H., Zhu A., Zhou Y., Li Y. Forebrain excitatory neuron-specific SENP2 knockout mouse displays hyperactivity, impaired learning and memory, and anxiolytic-like behavior. Mol. Brain. 2020;13:59. doi: 10.1186/S13041-020-00591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y.C., Hsu W.L., Ma Y.L., Tai D.J.C., Lee E.H.Y. CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory. J. Neurosci. 2014;34:9574–9589. doi: 10.1523/JNEUROSCI.4302-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drisaldi B., Colnaghi L., Fioriti L., Rao N., Myers C., Snyder A.M., Metzger D.J., Tarasoff J., Konstantinov E., Fraser P.E., et al. SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep. 2015;11:1694–1702. doi: 10.1016/J.CELREP.2015.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pronot M., Kieffer F., Gay A.S., Debayle D., Forquet R., Poupon G., Schorova L., Martin S., Gwizdek C. Proteomic identification of an endogenous synaptic SUMOylome in the developing rat brain. Front. Mol. Neurosci. 2021;14:284. doi: 10.3389/FNMOL.2021.780535/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kieffer F., Pronot M., Gay A.S., Debayle D., Gwizdek C. Proteomics datasets of developing rat brain: synaptic proteome and SUMO2/3-ylome. Data Brief. 2022;42:108151. doi: 10.1016/J.DIB.2022.108151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salinas S., Briançon-Marjollet A., Bossis G., Lopez M.A., Piechaczyk M., Jariel-Encontre I., Debant A., Hipskind R.A. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol. 2004;165:767–773. doi: 10.1083/JCB.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westman B.J., Verheggen C., Hutten S., Lam Y.W., Bertrand E., Lamond A.I. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol. Cell. 2010;39:618–631. doi: 10.1016/J.MOLCEL.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shan S.F., Wang L.F., Zhai J.W., Qin Y., Ouyang H.F., Kong Y.Y., Liu J., Wang Y., Xie Y.H., Xie Y.H. Modulation of transcriptional corepressor activity of prospero-related homeobox protein (Prox1) by SUMO modification. FEBS Lett. 2008;582:3723–3728. doi: 10.1016/J.FEBSLET.2008.09.057. [DOI] [PubMed] [Google Scholar]
- 61.Pan M.R., Chang T.M., Chang H.C., Su J.L., Wang H.W., Hung W.C. Sumoylation of Prox1 controls its ability to induce VEGFR3 expression and lymphatic phenotypes in endothelial cells. J. Cell Sci. 2009;122:3358–3364. doi: 10.1242/JCS.050005. [DOI] [PubMed] [Google Scholar]
- 62.Correa-Vázquez J.F., Juárez-Vicente F., García-Gutiérrez P., Barysch S.v., Melchior F., García-Domínguez M. The Sumo proteome of proliferating and neuronal-differentiating cells reveals Utf1 among key Sumo targets involved in neurogenesis. Cell Death Dis. 2021;12:305. doi: 10.1038/s41419-021-03590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendriks I.A., Lyon D., Su D., Skotte N.H., Daniel J.A., Jensen L.J., Nielsen M.L. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 2018;9:2456. doi: 10.1038/s41467-018-04957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee Y., Chun S.K., Kim K. Sumoylation controls CLOCK-BMAL1-mediated clock resetting via CBP recruitment in nuclear transcriptional foci. Biochim. Biophys. Acta. 2015;1853:2697–2708. doi: 10.1016/J.BBAMCR.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Chen L.C., Hsieh Y.L., Tan G.Y.T., Kuo T.Y., Chou Y.C., Hsu P.H., Hwang-Verslues W.W. Differential effects of SUMO1 and SUMO2 on circadian protein PER2 stability and function. Sci. Rep. 2021;11:14431. doi: 10.1038/S41598-021-93933-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stankova T., Piepkorn L., Bayer T.A., Jahn O., Tirard M. SUMO1-conjugation is altered during normal aging but not by increased amyloid burden. Aging Cell. 2018;17:e12760. doi: 10.1111/ACEL.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krumova P., Weishaupt J.H. Sumoylation in neurodegenerative diseases. Cell. Mol. Life Sci. 2013;70:2123–2138. doi: 10.1007/S00018-012-1158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tai D.J.C., Liu Y.C., Hsu W.L., Ma Y.L., Cheng S.J., Liu S.Y., Lee E.H.Y. MeCP2 SUMOylation rescues Mecp2-mutant-induced behavioural deficits in a mouse model of Rett syndrome. Nat. Commun. 2016;7:10552. doi: 10.1038/NCOMMS10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson J.O., Pioro E.P., Boehringer A., Chia R., Feit H., Renton A.E., Pliner H.A., Abramzon Y., Marangi G., Winborn B.J., et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17:664–666. doi: 10.1038/NN.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rawat P., Boehning M., Hummel B., Aprile-Garcia F., Pandit A.S., Eisenhardt N., Khavaran A., Niskanen E., Vos S.M., Palvimo J.J., et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell. 2021;81:1013–1026.e11. doi: 10.1016/J.MOLCEL.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andreev V.I., Yu C., Wang J., Schnabl J., Tirian L., Gehre M., Handler D., Duchek P., Novatchkova M., Baumgartner L., et al. Panoramix SUMOylation on chromatin connects the piRNA pathway to the cellular heterochromatin machinery. Nat. Struct. Mol. Biol. 2022;29:130–142. doi: 10.1038/S41594-022-00721-X. [DOI] [PubMed] [Google Scholar]
- 72.Mo Y.Y., Yu Y., Shen Z., Beck W.T. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J. Biol. Chem. 2002;277:2958–2964. doi: 10.1074/JBC.M108263200. [DOI] [PubMed] [Google Scholar]
- 73.Hakre S., Tussie-Luna M.I., Ashworth T., Novina C.D., Settleman J., Sharp P.A., Roy A.L. Opposing functions of TFII-I spliced isoforms in growth factor-induced gene expression. Mol. Cell. 2006;24:301–308. doi: 10.1016/J.MOLCEL.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 74.Niskanen E.A., Malinen M., Sutinen P., Toropainen S., Paakinaho V., Vihervaara A., Joutsen J., Kaikkonen M.U., Sistonen L., Palvimo J.J. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/S13059-015-0717-Y/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keiten-Schmitz J., Wagner K., Piller T., Kaulich M., Alberti S., Müller S. The nuclear SUMO-targeted ubiquitin quality control network regulates the dynamics of cytoplasmic stress granules. Mol. Cell. 2020;79:54–67.e7. doi: 10.1016/J.MOLCEL.2020.05.017. [DOI] [PubMed] [Google Scholar]
- 76.Keiten-Schmitz J., Röder L., Hornstein E., Müller-McNicoll M., Müller S. SUMO: glue or solvent for phase-separated ribonucleoprotein complexes and molecular condensates? Front. Mol. Biosci. 2021;8:673038. doi: 10.3389/FMOLB.2021.673038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/J.CELL.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cha K., Sen P., Raghunayakula S., Zhang X.D. The cellular distribution of RanGAP1 is regulated by CRM1-mediated nuclear export in mammalian cells. PLoS One. 2015;10:e0141309. doi: 10.1371/JOURNAL.PONE.0141309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakanishi H. Neuronal and microglial cathepsins in aging and age-related diseases. Ageing Res. Rev. 2003;2:367–381. doi: 10.1016/S1568-1637(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 80.Rohira A.D., Chen C.Y., Allen J.R., Johnson D.L. Covalent small ubiquitin-like modifier (SUMO) modification of Maf1 protein controls RNA polymerase III-dependent transcription repression. J. Biol. Chem. 2013;288:19288–19295. doi: 10.1074/JBC.M113.473744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chymkowitch P., Nguéa P A., Aanes H., Robertson J., Klungland A., Enserink J.M. TORC1-dependent sumoylation of Rpc82 promotes RNA polymerase III assembly and activity. ProcProc. Natl. Acad. Sci. USA. 2017;114:1039–1044. doi: 10.1073/PNAS.1615093114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chymkowitch P., Enserink J.M. Regulation of tRNA synthesis by posttranslational modifications of RNA polymerase III subunits. Biochim Biophys Acta Gene Regul Mech. 2018;1861:310–319. doi: 10.1016/J.BBAGRM.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 83.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/SCIENCE.1260419. [DOI] [PubMed] [Google Scholar]
- 84.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-Mediated Genome engineering. Cell. 2013;153:910–918. doi: 10.1016/J.CELL.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Richner M., Jager S.B., Siupka P., Vaegter C.B. Hydraulic extrusion of the spinal cord and isolation of dorsal root ganglia in rodents. J. Vis. Exp. 2017:55226. doi: 10.3791/55226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park Y.G., Sohn C.H., Chen R., McCue M., Yun D.H., Drummond G.T., Ku T., Evans N.B., Oak H.C., Trieu W., et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 2018;37:73–83. doi: 10.1038/nbt.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim S.Y., Cho J.H., Murray E., Bakh N., Choi H., Ohn K., Ruelas L., Hubbert A., McCue M., Vassallo S.L., et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. USA. 2015;112:E6274–E6283. doi: 10.1073/PNAS.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murray E., Cho J.H., Goodwin D., Ku T., Swaney J., Kim S.-Y., Choi H., Park Y.-G., Park J.-Y., Hubbert A., et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tirard M., Brose N. Systematic localization and identification of SUMOylation substrates in knock-in mice expressing affinity-tagged SUMO1. Methods Mol. Biol. 2016;1475:291–301. doi: 10.1007/978-1-4939-6358-4_20. [DOI] [PubMed] [Google Scholar]
- 90.Shevchenko A., Tomas H., Havliš J., Olsen J.v., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/NPROT.2006.468. [DOI] [PubMed] [Google Scholar]
- 91.Kolberg L., Raudvere U., Kuzmin I., Vilo J., Peterson H. gprofiler2 -- an r package for gene list functional enrichment analysis and namespace conversion toolset g: profiler. F1000Res. 2020;9 doi: 10.12688/F1000RESEARCH.24956.2/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones D.H., Matus A.I. Isolation of synaptic plasma membrane from brain by combined flotation-sedimentation density gradient centrifugation. Biochim. Biophys. Acta. 1974;356:276–287. doi: 10.1016/0005-2736(74)90268-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scale bar: 3000 μm.
Scale bar: 3000 μm.
Data Availability Statement
-
•
Mass spectrometry data is available from the lead contact upon request.
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.