Abstract
The identification of intestinal dysbiosis in patients with neurological and psychiatric disorders has highlighted the importance of gut–brain communication, and yet the question regarding the identity of the components responsible for this cross-talk remains open. We previously reported that relapsing remitting multiple sclerosis patients treated with dimethyl fumarate have a prominent depletion of the gut microbiota, thereby suggesting that studying the composition of plasma and CSF samples from these patients may help to identify microbially derived metabolites.
We used a functional xenogeneic assay consisting of cultured rat neurons exposed to CSF samples collected from multiple sclerosis patients before and after dimethyl fumarate treatment to assess neurotoxicity and then conducted a metabolomic analysis of plasma and CSF samples to identify metabolites with differential abundance. A weighted correlation network analysis allowed us to identify groups of metabolites, present in plasma and CSF samples, whose abundance correlated with the neurotoxic potential of the CSF. This analysis identified the presence of phenol and indole group metabolites of bacterial origin (e.g. p-cresol sulphate, indoxyl sulphate and N-phenylacetylglutamine) as potentially neurotoxic and decreased by treatment. Chronic exposure of cultured neurons to these metabolites impaired their firing rate and induced axonal damage, independent from mitochondrial dysfunction and oxidative stress, thereby identifying a novel pathway of neurotoxicity. Clinical, radiological and cognitive test metrics were also collected in treated patients at follow-up visits. Improved MRI metrics, disability and cognition were only detected in dimethyl fumarate-treated relapsing remitting multiple sclerosis patients. The levels of the identified metabolites of bacterial origin (p-cresol sulphate, indoxyl sulphate and N-phenylacetylglutamine) were inversely correlated to MRI measurements of cortical volume and directly correlated to the levels of neurofilament light chain, an established biomarker of neurodegeneration.
Our data suggest that phenol and indole derivatives from the catabolism of tryptophan and phenylalanine are microbially derived metabolites, which may mediate gut–brain communication and induce neurotoxicity in multiple sclerosis.
Keywords: microbiota, neurodegeneration, brain, metabolism
Ntranos et al. perform metabolomic analysis of CSF and plasma samples from multiple sclerosis patients. They identify phenol and indole derivatives from the catabolism of tryptophan and phenylalanine as microbially derived, potentially neurotoxic metabolites that may mediate gut–brain communication in multiple sclerosis.
Introduction
As one of the main portals of communication with the external environment, the gut is at the forefront of the environmental interactions that shape health and disease. A plethora of recent studies support the concept of microbial dysbiosis as central to the pathogenesis of several neurological disorders.1-6 In multiple sclerosis, for instance, it has been reported that the composition of the gut microbiota is distinct in patients and healthy controls.7–11 Most studies have focused on the important characterization of the gut–immune system interaction, a concept which is shared with several other autoimmune disorders including type I diabetes and rheumatoid arthritis.12–17 Besides this well-documented effect of microbes on the gut-associated immune system, at least two potential additional mechanisms of microbial regulation of brain health have been proposed. One line of investigation has focused on the detection and characterization of bacterial toxins with the ability to enter in the bloodstream and reach the brain parenchyma. An example is the production of the epsilon toxin (ETX) by Clostridium perfringens, a member of the Clostridiales, found to be elevated in patients with multiple sclerosis9,11,18–21 or neuromyelitis optica.22,23 The toxin was identified in the stool of a subset of multiple sclerosis patients and shown to bind to the brain vasculature and ultimately drive oligodendrocytic death.24–26 However, ETX is found in only a small percentage of multiple sclerosis patients,27 thereby suggesting that other mechanisms may be in place.
Beneficial gut bacteria contribute to human health, for instance, by promoting the utilization of dietary tryptophan, a metabolic pathway found to be dysregulated in multiple sclerosis patients.28,29 Tryptophan can be metabolized towards serotonin or towards kynurenine. The production of serotonin has been shown to be beneficial for the organism and certain tryptophan derivatives in multiple sclerosis have been shown to directly bind to the aryl hydrocarbon receptor on glial cells30 and be anti-inflammatory.30,31 Beneficial gut bacteria, such as Prevotella, contribute to the production of short-chain fatty acids, which have been shown to be neuroprotective32 and are decreased in pathological conditions such as multiple sclerosis.33 Based on these considerations, we reasoned that the altered balance between bacteria with beneficial and detrimental effects, as detected in multiple sclerosis patients, may lead not only to depletion of neuroprotective metabolites, but also to the potential accumulation of neurotoxic compounds, and this is the overarching hypothesis of the current study.
In an attempt to identify neurotoxic metabolites with the potential to impact axonal damage, we deployed targeted metabolomic analysis of plasma and CSF samples from healthy controls and patients with multiple sclerosis, coupled with functional neurotoxicity screening in cultured neurons and clinical and MRI follow-up of patients with relapsing remitting multiple sclerosis (RRMS). We leveraged the results of our previous studies reporting the dramatic effect of disease-modifying therapies such as dimethyl fumarate (DMF) on the gut microbiota.19 DMF is an effective medication for the treatment of multiple sclerosis34,35 with several immunomodulatory, epigenetic and gut-microbiome effects,19,36,37 which we previously showed to have a significant effect on the composition of the gut microbiome.19 We reasoned that an in-depth analysis of the metabolite composition of samples from RRMS patients before and after treatment could serve as a tool for the identification of potentially neurotoxic compounds of bacterial origin.
Materials and methods
Patient enrolment and sample collection
The study protocol was approved by the Institutional Review Board at the Icahn School of Medicine at Mount Sinai and Biomedical Institutional Review Board. All participants provided written informed consent prior to completion of study-related activities. Twenty healthy controls, 16 RRMS patients and 17 patients with secondary progressive multiple sclerosis (SPMS) were recruited at the Multiple Sclerosis Center of Northeastern New York, P.C. Additional RRMS patients treated with either DMF (n = 11) or ocrelizumab (anti-CD20; n = 8) were recruited at the Corinne Goldsmith Dickinson Center for multiple sclerosis at Mount Sinai Medical Center. Inclusion criteria were a diagnosis of multiple sclerosis according to McDonald criteria. Exclusion criteria were current smoking status and treatment with steroids in the past 30 days from enrolment. The differentiation between RRMS versus SPMS category was based on the published guidelines in 1996 from Lublin and Reingold38 and its subsequent revision in 2014.39 Regarding the first cohort, patients were assessed by an independent multiple sclerosis specialist who was not aware of the metabolomic analysis results and an Expanded Disability Score Scale (EDSS) was obtained at baseline and after 6, 12 and 24 months of DMF treatment, but due to high drop-out rate at 24 months, only the 12-month time point was used. Improvement or progression of EDSS was defined as having a reduction or increase in EDSS scores by more or equal to 0.5 points at 12 months compared to baseline, respectively. Patients also completed a 25-foot timed walk test (25-FTW), Symbol Digit Modalities Test (SDMT) and Paced Auditory Serial Addition Test (PASAT) at baseline and 12 months of treatment. Of note, the 3.0” PASAT was performed in this study. As per the Multiple Sclerosis Functional Composite (MSFC) PASAT standard administration guidelines, all subjects were administered at least one practice trial. One additional practice trial was given to three subjects, who failed to give two correct responses on the first practice trial, up to a maximum of three practice trials. Subjects also underwent lumbar puncture and venipuncture to obtain CSF and plasma, respectively, at baseline and 6 months after treatment. Upon CSF collection, the samples were centrifuged at 5000 rpm to remove debris and the supernatants were aliquoted, flash-frozen in dry ice/ethanol vapour and kept at −80°C until use. Mixtures of blood and Histopaque in the ratio 1:1 were centrifuged for 20 min at 1500 rpm, at room temperature. Collected supernatants were centrifuged again for 15 min at 1500 rpm to deplete platelets and supernatants were aliquoted and stored at −80°C until use. Patients who dropped out of the study prior to 6 months of follow-up (two RRMS and six SPMS patients) and subsequently did not have CSF or plasma for metabolomic analysis were excluded from the study. Regarding the second cohort, blood was collected and plasma separated at baseline and after 6 months of treatment with their respective therapies and was analysed similarly to our first cohort. To match the age distribution of our first cohort, only patients older than 30 years old were included in the second cohort.
MRI acquisition and analysis
MRI scans were performed at baseline and follow-up visits using Siemens 3.0 T Verio scanner according to standard clinical protocols. All the MRI measurements, including lesion detection and assessment and brain volume change were performed by Icometrics. MRI scans were uploaded using the European Commission and 510(k) FDA-approved icobrain software (formerly known as MSMetrix; Icometrix, Leuven, Belgium) for automated quantitative MRI assessment.40,41 Briefly, the Icobrain algorithm employs the input T1-weighted and FLAIR scans to jointly segment healthy tissue classes (white matter, grey matter and CSF), as well as FLAIR and T1-hypointense lesions. For longitudinal assessment, segmentations from individual time points were used as input in a longitudinal algorithm to jointly analyse two follow-up scans from the same patient while preserving temporal and spatial consistency.
Metabolomic analysis
Plasma and CSF samples for metabolomics analysis were prepared as previously described.42–46 Metabolite extraction from CSF and plasma samples was achieved using a mixture of isopropanol, acetonitrile and water at a ratio of 3:3:2 v/v. Extracts were divided into three parts: 75 µl for gas chromatography combined with time-of-flight high-resolution mass spectrometry (GC-MS), 150 µl for reversed-phase liquid chromatography coupled with high-resolution mass spectrometry (RP-LC-MS) and 150 µl for hydrophilic interaction chromatography with liquid chromatography and tandem mass-spectrometry (HILIC-LC-MS/MS) and analysed as described previously.42–46 We used the NEXERA XR UPLC system (Shimadzu) coupled with the Triple Quad 5500 System (AB Sciex) to perform HILIC-LC-MS/MS analysis, NEXERA XR UPLC system (Shimadzu) coupled with the Triple TOF 6600 System (AB Sciex) to perform RP-LC-MS analysis and Agilent 7890B gas chromatograph (Agilent) interfaced to a Time-of-Flight Pegasus HT Mass Spectrometer (Leco) for GC-MS. The GC system was fitted with a Gerstel temperature-programmed injector, cooled injection system (model CIS 4). An automated liner exchange (Gerstel) was used to eliminate cross-contamination from the sample matrix that was occurring between sample runs. Quality control was performed using metabolite standards mixture and pooled samples applying methodology previously described.47–50 A quality control sample containing a standard mixture of amino and organic acids purchased from Sigma-Aldrich as certified reference material was injected daily to perform analytical system suitability test and monitor recorded signals’ day-to-day reproducibility as described.42–46 A pooled quality control sample was obtained by taking an aliquot of the same volume of all samples from the study and injected daily with a batch of analysed samples to determine the optimal dilution of the batch samples and validate metabolite identification and peak integration. Collected raw data were manually inspected, merged, imputed and normalized by the sample median, as routine normalization. Metabolite identification was performed using in-house authentic standards analysis. Metabolite annotation was used utilizing recorded retention time and retention indexes, recorded MSn and HRAMSn data with METLIN, NIST MS, Wiley Registry of Mass Spectral Data, HMDB, MassBank of North America, MassBank Europe, Golm Metabolome Database, SCIEX Accurate Mass Metabolite Spectral Library, MzCloud and IDEOM databases. Plasma samples from the second cohort were processed with the same methods but at a different time point and therefore the statistical analysis was separate.
Metabolomic data processing and network analysis
Metabolomic data from plasma or CSF were filtered and normalized using Metaboanalyst 4.0. Briefly, metabolites from all samples in the study were normalized by median, log-transformed and autoscaled together. These data represented the relative abundances of metabolites among samples, rather than their actual concentration. Univariate statistical analysis methods were then used to estimate relative changes in metabolite levels. Partial least squares-discriminant analysis (PLS-DA) was utilized as a multivariate dimensionality-reduction tool, followed by supervised hierarchical clustering to generate dendrograms and heat maps.
Because the number of measured metabolites was significantly higher than the number of samples in our analysis, and therefore would negatively impact our statistical power during multiple hypothesis testing correction, we employed a hypothesis-reduction method based on weighted correlation network analysis (WGCNA).51,52 Although this was first developed for gene network analysis, it has been successfully used to identify networks of metabolites with relatively few samples.52 Briefly, we first identified clusters of highly correlated metabolites in the samples, which are called ‘modules’, and then related them to sample traits using eigengene network methodology.51 Because our study design was paired, we utilized a linear mixed-effects model to relate the module eigengenes with sample traits, as previously suggested.53 Multiple hypothesis testing false discovery rate correction was applied at the level of WGCNA metabolite modules. Co-expression network methods such as WGCNA can detect coordinated metabolite changes at a metabolite group (modular) level. Modular analysis also helps alleviate the multiple hypothesis testing issue that applies to high-dimensional metabolomic datasets and thus may be more robust than a single-metabolite level analysis. Clustering metabolites into pathway-specific modules and aggregating these metabolites onto single statistical units (i.e. modules) can significantly reduce the number of hypotheses to be tested, and thus increase power in the analysis. After identifying the pathway of interest (i.e. the ‘module’) in our dataset, as the metabolites belonging to the same WGCNA-module/statistical unit share a similar biological function (e.g. they are all bacterially derived metabolites with a similar effect on neuronal cultures), we do not correct again for multiple hypotheses during the analysis of the in vitro functional data (as all the identified metabolites belong to the same module). The WGCNA and mixed-effects model analysis was conducted in R Studio. For targeted confirmatory analyses of individual metabolites and visualization we used the GraphPad Prism 8 software and statistical analysis was performed using either linear regression controlling for age, or paired t-test, when appropriate. P-values < 0.05 were considered statistically significant.
Neurofilament light chain measurement in CSF
The levels of neurofilament light chain in the CSF were measured using the UmanDiagnostics NF-light enzyme-linked immunosorbent assay (UmanDiagnostics) as previously described.54 Briefly, all available samples were run on triplicates and the average concentration was used for further analysis. For correlations of neurofilament light chain (NFL) with metabolite modules one outlier patient sample was removed.
Primary neuronal cultures
Primary cultures of hippocampal and cortical neurons were isolated from Sprague–Dawley rat embryos on embryonic Day 18 (E18) as described previously55 with slight modification. Briefly, embryonic hippocampi and cortices were digested with 0.025% Trypsin–EDTA for 15 min at 37°C, followed by incubation with NM10 medium (Dulbecco’s modified Eagle medium with 10% foetal bovine serum) for 5 min at room temperature. After centrifugation, cells were suspended in neurobasal medium supplemented with B27, 10 mM GlutaMAX and 1% penicillin/streptomycin and mechanically dissociated by gentle pipetting. Dissociated cells were then filtered through a cell strainer and centrifuged again, and resuspended for plating. At 3 days post-plating, an anti-mitotic agent was added to cells in order to avoid contamination of non-neuronal cells. Cells were cultured at 37°C in a 5% CO2 incubator and the medium was replaced every other day until 17 days in vitro (DIV). Cells were challenged with H2O2 (15 µM for 15 min at DIV 5, 7, 10 and 14) to mimic a neurotoxic pro-inflammatory environment in vitro, followed by 18 h incubation with CSF samples from patients at DIV 17. For immunocytochemistry, cells were plated onto CC2-chamber slides at a density of 3.0 × 104 cells/well. For cell viability assay, cells were plated onto poly-D-lysine-coated 48-well plates at a density of 3.0 × 104 cells/well. For testing endogenous spontaneous electrical neuronal activity, cells were plated onto polyethylenimine (PEI)/laminin-coated Axion 48-well multi-electrode array plates at a density of 6.0 × 104 cells/well. For bioenergetic analysis, cells were plated onto PEI/laminin-coated Seahorse XF 24-cell culture microplates at a density of 5.0 × 104 cells/well. For chronic treatment with ‘red module’ metabolites, cells were cultured in the same medium supplemented with distinct concentrations of indoxyl sulphate (IS), p-cresol sulphate (pCS) and N-phenylacetylglutamate (PAG) (5, 25 and 50 µM), either individually or in combination. To rule out the potential effect of these metabolites on the pH of the medium, all media were tested for stability of the pH prior to treating the neurons. No differences in pH were observed. To test the potential direct effect of DMF on neurons, in some experiments primary neurons were cultured in the presence of the same concentrations of the three metabolites in the presence of monomethyl fumarate (MMF; 50 µM), an active metabolite of DMF.
Immunocytochemistry of treated cultured neurons
After completion of treatment, cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature, followed by permeabilization in phosphate-buffered saline (PBS)-based blocking buffer with 10% normal goat serum and 0.5% Triton X-100 for 30 min at room temperature. Cells were then incubated with primary antibodies against SMI32 (SMI32-NE1023, Calbiochem, 1:500), Nitrotyrosine (06–284, Millipore, 1:500), Neurofilament-H (MAB5448, Millipore, 1:500) overnight at 4°C, followed by incubation with fluorophore-conjugated secondary antibodies. Following washes in PBS, cells were mounted with 4′,6-diamidino-2-phenylindole Fluoromount-G for image analysis. Confocal images were acquired at 20× objective using LSM 800 microscope with Zen software (Zeiss), and all images were processed and analysed in ImageJ.
Multi-electrode array recordings from neuronal cultures
Spontaneous neuronal activity was measured at DIV 21 using a Maestro Pro multi-well multi-electrode array system (Axion Biosystems). Briefly, cells were cultured in the absence or presence of metabolites for 21 days. On the day of the experiment, cells were allowed to equilibrate for 5 min prior to the 10 min recording of spontaneous activity. Signals were simultaneously acquired across 768 channels (16 electrodes per well, 48-well plate) and data acquisition was managed with Axion’s integrated software, AxiS Navigator.
Bioenergetic analysis of cultured neurons
Mitochondrial bioenergetics in neurons were measured at DIV 18 using the Seahorse XFe24 Bioanalyser (Agilent). After equilibration of the cultured neurons with artificial CSF for 1 h, mitochondrial respiratory function was analysed by 4 mM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), 0.5 mM rotenone and 4 mM antimycin A, respectively. Briefly, basal oxygen consumption rate (OCR) was measured before the injection of inhibitors, followed by additional OCR measurement after each addition of given inhibitors. Six parameters of mitochondrial respiration were determined based on observed OCR.
Data availability
Data supporting the findings of this study are available from the corresponding author, upon reasonable request.
Results
Reduced neurotoxicity of the CSF from RRMS patients after dimethyl formamide treatment
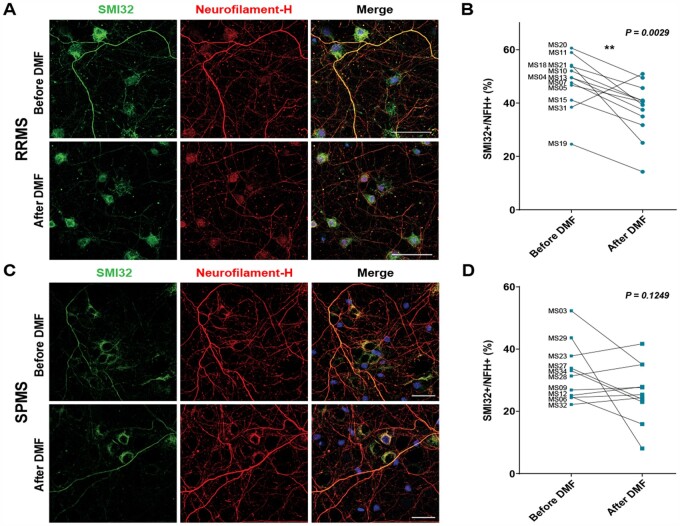
We have previously described and characterized a xenogeneic functional screen for neurotoxicity consisting of cultured rodent neurons exposed to the CSF of multiple sclerosis patients or healthy controls.54,56 To address the question of whether neurotoxic metabolites produced by gut bacteria exist in our cohort, we adopted a similar approach and screened the CSF samples from multiple sclerosis patients before and after treatment with DMF (Fig. 1). We used DMF as a treatment because we previously showed that it has a dramatic effect on the gut-microbiota composition.19 Our cohort included 20 healthy controls, 14 RRMS patients and 11 SPMS patients. All subjects underwent an lumbar puncture at baseline to obtain CSF and venipuncture to obtain whole blood at the same time. Patient demographics were similar across groups (Supplementary Table 1), except that SPMS patients were older and had more severe disease activity, in terms of disability, walking speed and cognition than RRMS patients, as expected (Supplementary Table 2). Information regarding previous treatment was also collected (Supplementary Table 3). MRI metrics showed that SPMS patients had higher T2 and T1 lesion volume and smaller grey matter volumes than RRMS patients (Supplementary Fig. 1), a finding which was expected due to the higher disease severity in the SPMS group (Supplementary Table 2). CSF and plasma samples from healthy controls were collected only at baseline, while samples from multiple sclerosis patients (RRMS and SPMS) were also collected after 6 months of treatment with DMF. Functional screening of CSF samples collected at baseline from healthy individuals did not impact the physiology of cultured neurons (not shown), while the CSF of RRMS and SPMS patients had a substantial effect and was sufficient to induce signs of axonal damage, including a beaded appearance of the cytoplasmic processes and immunoreactivity to SMI32 (Fig. 1), thereby confirming previous reports.54 Interestingly, the CSF samples collected from RRMS patients after DMF treatment revealed a statistically significant decrease of neurotoxicity (P = 0.0029 paired t-test) as 93% of the samples (13 of 14) induced milder axonal damage compared to baseline samples. However, DMF treatment did not significantly change the toxicity of the SPMS samples, with only 45% of the samples (n = 5 of 11) showing decreased neurotoxicity compared to baseline (Fig. 1). From this, we concluded that an in-depth analysis of the metabolites present in the CSF of RRMS patients at baseline and with decreased abundance after DMF treatment might potentially identify microbial metabolites with neurotoxic properties.
Figure 1.
DMF treatment reduces the neurotoxic potential of CSF from RRMS but not SPMS patients. (A and C) Confocal image of primary cultured neurons exposed to CSF from RRMS (A) or SPMS (C) patients for 18 h and then fixed, stained with antibodies for SMI32 (green, to assess axonal damage) and for neurofilament heavy chain (red, as control for axonal density). (B and D) Percentage axonal damage was calculated by dividing SMI32 positive area by the total neurofilaments. The paired analysis of SMI32 immunoreactivity in neurons exposed to CSF samples collected at baseline and after 6 months of DMF treatment is shown for RRMS (B) and SPMS (D) patients. Note the significant reduction of the SMI32 marker of axonal damage only in RRMS patients (A and B) and not SPMS (C and D) patients. **P < 0.01 (paired t-test). Scale bar = 50 µm.
Plasma and CSF metabolomics identifies microbial phenol and indole derivatives
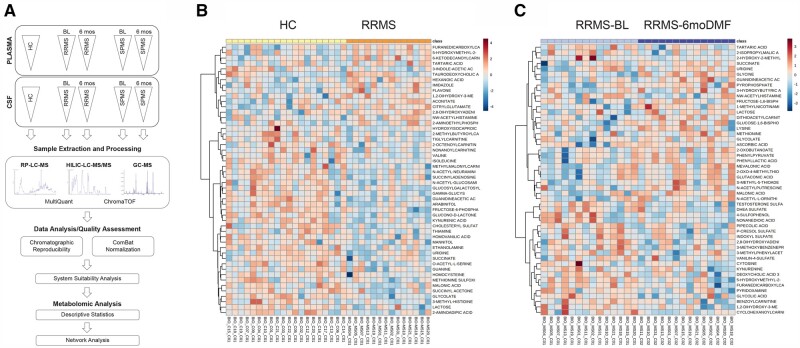
As we were interested in metabolites produced by gut commensals, we reasoned that the same products identified in the CSF may also be present in plasma samples and therefore conducted a metabolomic analysis of both plasma and CSF samples from RRMS patients at baseline (n = 14) and after 6 months of treatment (n = 14). Samples from healthy controls (n = 20) and SPMS patients at baseline (n = 11) and after DMF treatment (n = 11) were used for comparative purposes (Fig. 2A). Samples were processed and analysed as described in the ‘Materials and methods’ section and 394 metabolites were detected. Statistical analysis was applied to compare samples from healthy controls to those from RRMS patients at baseline and a PLS-DA was conducted (Supplementary Fig. 2A). We then conducted an agglomerative hierarchical cluster analysis based on similarity measures and clustering algorithms. The data represented as heat maps are shown in Fig. 2B. The increased levels of several carnitines and decreased levels of kynurenic acid in the CSF of RRMS patients compared to healthy controls was consistent with previous reports.57 A similar longitudinal analysis was conducted for RRMS patients before and after DMF treatment. The results of the PLS-DA are shown in Supplementary Fig. 2B, while the dendrograms after supervised clustering analysis are shown in Fig. 2C.
Figure 2.
Metabolomic profiling of CSF in RRMS patients. (A) Flow chart of the experimental approach and data analysis related to metabolomics profiling of CSF and plasma samples. (B) Heat map of the hierarchical clustering representing the metabolic features differentially abundant in healthy control (HC, yellow) and RRMS (orange) patients. (C) Heat map of the hierarchical clustering of the longitudinally collected samples in RRMS patients at baseline (BL, light blue) and 6 months after DMF treatment (purple). The relative abundance of metabolites at baseline and after DMF treatment is shown as colour variation from dark red (high abundance) to dark blue (low abundance) for each metabolite (rows) and for each individual patient at the two time points (columns).
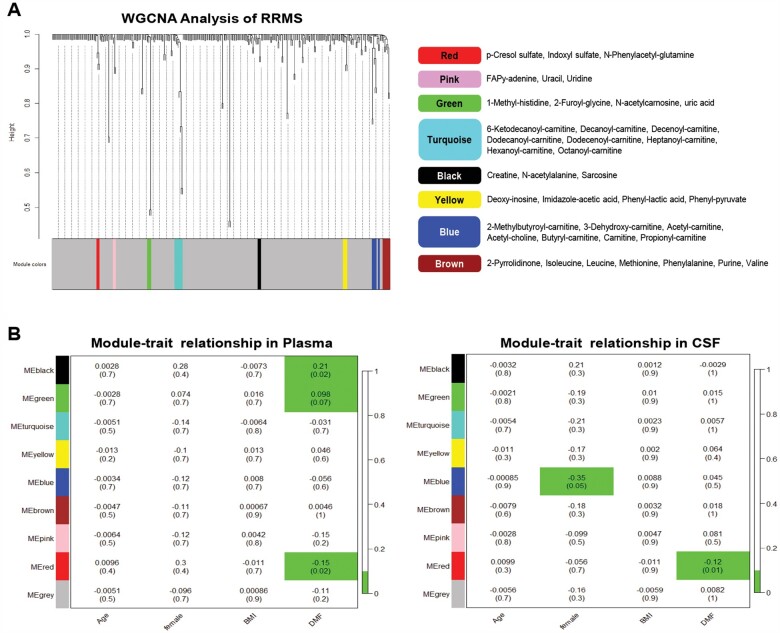
We then employed a WGCNA analysis of the metabolomic datasets from plasma and CSF in order to identify groups of metabolites—rather than individual ones—that were highly correlated to specific traits (i.e. age, sex, body mass index, treatment) by employing eigengene network methodology.51,52 The goal was to identify potential groups of metabolites, called ‘modules’, with a similar pattern relative to specific traits, without compromising statistical power. As we had processed both plasma and CSF of RRMS patients before and after DMF treatment, we were interested in the identification of metabolite ‘modules’ that were characterized by the same up- or downregulation in the two biological compartments. The identified modules of metabolites (Fig. 3A) were marked by distinct colours and associated to specific patient traits using linear mixed-effect models, given our paired sample design of the study.53 Linear mixed-effect model analysis revealed that one specific group of metabolites (e.g. pCS, IS, PAG), defined as the ‘red module’, was significantly reduced by DMF treatment, in both the plasma and CSF samples from RRMS patients (Fig. 3B). We noted that this specific group of metabolites are only produced by bacterial fermentation of amino acids and are characteristically absent or significantly reduced in mice raised in germ-free conditions.58
Figure 3.
Identification of bacterial metabolites significantly reduced by DMF treatment in plasma and CSF of RRMS patients. (A) Consensus analysis of CSF and plasma metabolome from RRMS patients was performed using WGCNA to identify highly correlated metabolite modules. The cluster dendrogram is depicted on the left, with each line representing a different metabolite and the height representing their similarity or correlation. The distinct clusters of highly correlated metabolites, called ‘modules’, are shown in various colours at the bottom, with the grey colour reserved for metabolites that did not correlate with each other. The specific metabolites belonging to distinct modules are shown on the right. (B) The eigenvector of the metabolite modules was associated with different patient traits using linear mixed-effect models, given our paired sample design. Each module colour is shown on the y-axis and each trait on the x-axis. The coefficient and the P-value (in parentheses) of the model are shown in the table (top and bottom numbers, respectively). The associations with P-values < 0.1 are shown in green. The only module that is significantly associated with DMF treatment in both plasma (left) and CSF (right) is the ‘red module’, which is composed of bacterial metabolites. Of note, this module does not correlate with other patient traits, such as age, sex or body mass index.
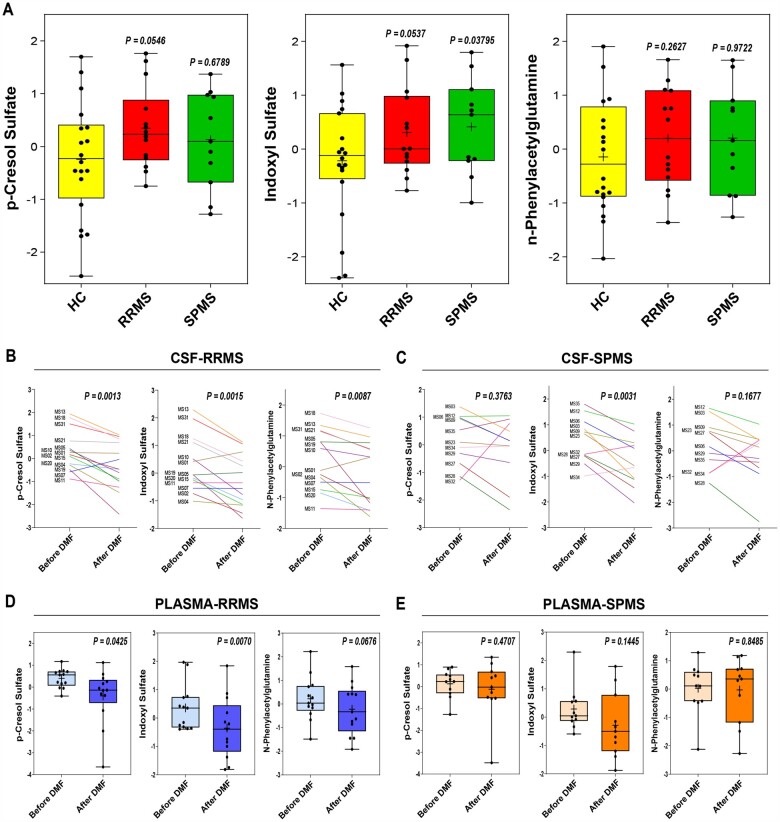
After identifying these metabolites as the ones significantly reduced by DMF in RRMS patients, we sought to compare their CSF concentration across our different patient groups. Linear regression controlling for age revealed a trend of higher concentration of the ‘red module’ metabolites in RRMS at baseline compared to controls (linear regression controlling for age P = 0.091), but not in SPMS patients (linear regression controlling for age P > 0.1). This trend was true for the three metabolites in the red module (i.e. pCS, IS and PAG) when tested independently, although—due to the small sample size—it did not reach statistical significance in the cross-sectional comparison (Supplementary Fig. 3), in contrast to the paired design of before and after treatment. The relative abundance of pCS, IS and PAG was higher in the CSF of RRMS and SPMS patients compared to controls (Fig. 4A) and was significantly decreased in both CSF and plasma of RRMS patients by DMF treatment (Fig. 4B and D), while in the samples from the SPMS patients the three metabolites were not concordantly decreased by DMF treatment (Fig. 4C and E), a result which is likely attributable to differences in the gut microbiota of RRMS19,59 and SPMS60,61 patients. Given the potential impact of our findings, we recruited another cohort of age-related RRMS patients undergoing either DMF treatment (n = 11) or another disease-modifying therapy (anti-CD20, n = 8) not affecting the microbiota (Supplementary Table 4). Of note, for this second cohort of RRMS patients we were able to obtain consent only for blood (but not CSF) collection and therefore conducted metabolomic analysis of plasma samples. In agreement with the results obtained in the plasma samples from the first cohort, the relative abundance of three metabolites showed a decrease in RRMS samples after 6 months of DMF treatment (pCS: 7 of 11; IS: 6 of 11; PAG: 6 of 11), although the trend did not reach statistical significance (Supplementary Fig. 4A). It is worth noting that these effects were specific to DMF, as the abundance of three metabolites was not changed by anti-CD20 therapy (Supplementary Fig. 4B). Together these data further support the concept that the abundance of the ‘red module’ metabolites is impacted only by disease-modifying drugs that affect the gut microbiota.
Figure 4.
Neurotoxic metabolites are in higher abundance in the CSF of RRMS than SPMS patients. (A) Graphs show normalized levels of pCS, IS and PAG in CSF of RRMS, SPMS, and healthy control (HC), with each metabolite trending towards higher abundance in multiple sclerosis patients compared to controls. (B and C) Paired analysis of the relative abundance of metabolites in the longitudinally collected CSF samples from RRMS and SPMS patients [each line representing of patient at baseline (before DMF) and after 6 months of DMF treatment (after DMF)]. Note the significant reduction of pCS, IS and PAG relative abundance in RRMS, not in SPMS. (D and E) Similarly, abundance of putative neurotoxic metabolites was decreased in plasma from RRMS patients, but not in SPMS patients, after DMF treatment. The lines in the box plot in A, D and E represent the quartiles (25th, 50th, 75th) and the whiskers mark the minimum and maximum values of the data. Statistical significance was calculated by either linear regression controlling for age (A), or paired Student’s t-test (B–E).
Phenol- and indole-derived metabolites directly impair neuronal physiological function
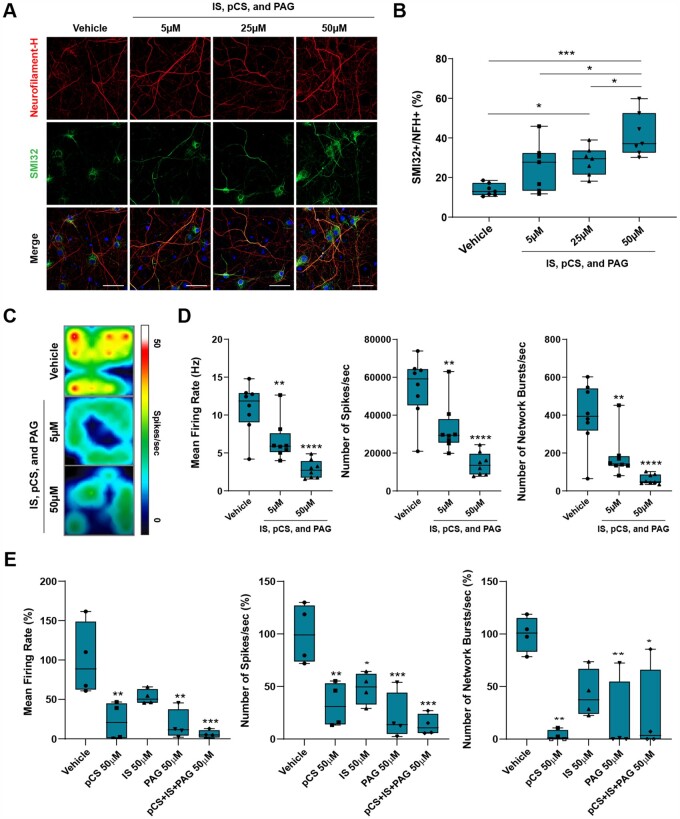
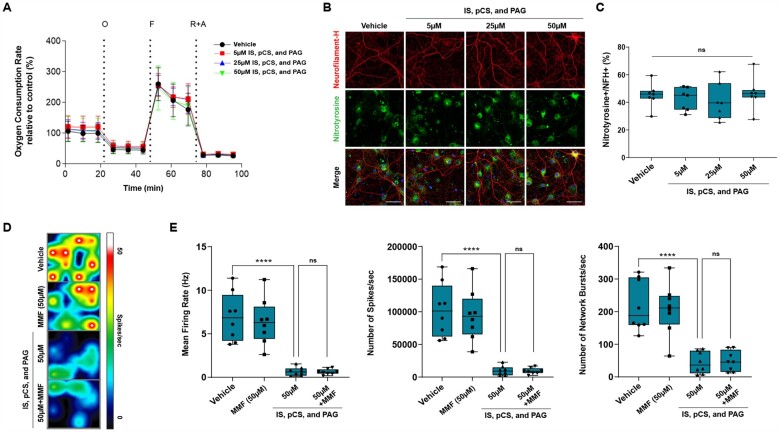
To define the relationship between the concentrations of these metabolites and neurotoxicity, we conducted a dose–response curve in cultured neurons. Chronic exposure of cultured neurons to increasing concentrations of these metabolites revealed a dose-dependent neurotoxic effect, as measured by SMI32 staining (Fig. 5A and B). To further assess the effect of these metabolites on neuronal function, we then measured neuronal electrical activity in cultures exposed to increasing concentration of the cocktail of the three ‘red module’ metabolites, using multi-well electrode array recordings, which allow for the characterization of the electrophysiological properties of the entire neuronal population (Fig. 5C and D). Compared to neuronal cultures kept in control conditions, we detected a dose-dependent decrease of the average neuronal firing rate, reduced number of spikes per second and a proportional decrease of neural network bursts per second (Fig. 5D). To further ascertain the similar function of each one of the three ‘red module’, we also examined the electrophysiological changes occurring in cultured neurons exposed first to the individual metabolites and then in combination. As predicted by the WGCNA analysis, each individual metabolite was sufficient to decrease neuronal function, although the combination of the three acted synergistically and further impaired neuronal function (Fig. 5E). Importantly, the concentration of the ‘red module’ metabolites (pCS, IS and PAG), used to test their effect in cultured neurons, was within the limits previously reported in the blood of uraemic patients.62,63 The neurotoxic effect was not dependent on an effect of metabolites on mitochondrial respiration or oxidative stress in cultured neurons (Fig. 6). Mitochondrial function, measured using the Seahorse bioanalyser, did not reveal any difference in oxygen consumption rate or respiratory capacity in cultures exposed to increasing concentrations of metabolites (Fig. 6A). Similarly, oxidative stress, measured by nitrotyrosine staining of neurofilament heavy-chain immunoreactive processes, did not differ between neurons either untreated or treated with increasing concentrations of ‘red module’ metabolites (Fig. 6B and C). Thus, the effect of microbial metabolites on neuronal function is quite distinct to the effect of host-generated toxic lipids, as we reported previously.54,56 Finally, we wanted to ascertain if DMF treatment reduced the neurotoxicity of the CSF samples in RRMS patients by decreasing microbial metabolites as a result of its effect on the reshaping of the gut microbiota,19 rather than a putative direct neuroprotective effect of its conversion metabolite (MMF) on brain parenchymal cells. For this reason, we co-treated cultured neurons with the same concentration of pCS, IS and PAG in the presence or absence of MMF at equimolar concentrations and recorded neural network activity using multi-electrode-array recordings from neuronal populations (Fig. 6D). Exposure to MMF alone did not affect the physiological properties of cultured neurons and co-treatment was completely ineffective in rescuing the decline in mean firing rate, number of spikes per second and the neuronal burst activity (Fig. 6E). Importantly, the eigenvector of the red module as well as the relative concentrations of pCS, IS and PAG highly correlated between plasma and CSF, consistent with the ability for these metabolites to cross the blood–brain barrier (Supplementary Fig. 5A) and have a direct neurotoxic effect. Collectively, these data support the identification of microbial phenol metabolites (e.g. pCS, PAG) and indole derivatives (e.g. IS) as neurotoxic compounds. We propose that after being produced by microbial species in the gut, they may reach the CNS via the bloodstream and then the CSF, thereby coming in contact with neurons in the superficial cortical layers.
Figure 5.
Identified bacterial metabolites directly induce axonal damage and neuronal dysfunction. (A) Confocal image of cultured neurons chronically exposed to different concentrations of the three ‘red module’ metabolites after 18 days in culture and stained for neurofilament heavy chain (red) and for the axonal marker SMI32 (green). (B) Dose-dependent axonal damage expressed as the percentage of NFH+ neuronal processes that are co-stained with SMI32. (C) Pseudo-colour image of the spontaneous electrical neuronal activity measured using MEA of neurons kept in regular medium supplemented with either vehicle or with the red module metabolites at the indicated concentration. Red represents high electrical activity and blue low activity in neurons. (D) The combination of the bacterial metabolites significantly decreased spontaneous neuronal activity as measured by calculating the average firing rate, number of spikes per second and number of network bursts. (E) The effect of treatment with each individual metabolite was significant and distinct. The overall effect of the combination of metabolites was synergistic on the mean firing rate and number of spikes per seconds. The lines in the box plot in B, D and E represent the quartiles (25th, 50th, 75th) and the whiskers mark the minimum and maximum values of the data. Statistical significance assessed by one-way ANOVA followed by Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Scale bar = 50 µm.
Figure 6.
MMF cannot counteract neurotoxicity of microbial metabolites. (A) A Mito Stress Test was performed on neurons cultured in the absence (black curve) or presence of increasing concentrations of the three metabolites (red for 5 µM, blue for 25 µM, green for 50 µM), using the Seahorse XF bioanalyser. No difference in mitochondrial bioenergetic measures was detected. Error bars represent SD. (B) Immunofluorescence of neurons stained with a nitrotyrosine (NTS)-specific antibody, as a marker for cell damage and for neurofilament H (NFH). (C) Quantification of NTS staining was referred to the total NFH+ neuronal processes and did not reveal significant differences between metabolite-treated neurons compared to vehicle. (D and E) Neuronal cultures were chronically exposed to a mixture of CS, IS and PAG (50 µM each) in the absence or presence of MMF (50 µM), and the endogenous neuronal activity was measured. The lines in the box plots in C and E represent the quartiles (25th, 50th, 75th) and the whiskers mark the minimum and maximum values of the data. Statistical significance was assessed by one-way ANOVA followed by Tukey’s post hoc test. ****P < 0.0001. Scale bar = 50 µm
Neurotoxic CSF metabolite abundance correlates with biomarkers of neurodegeneration and brain volume
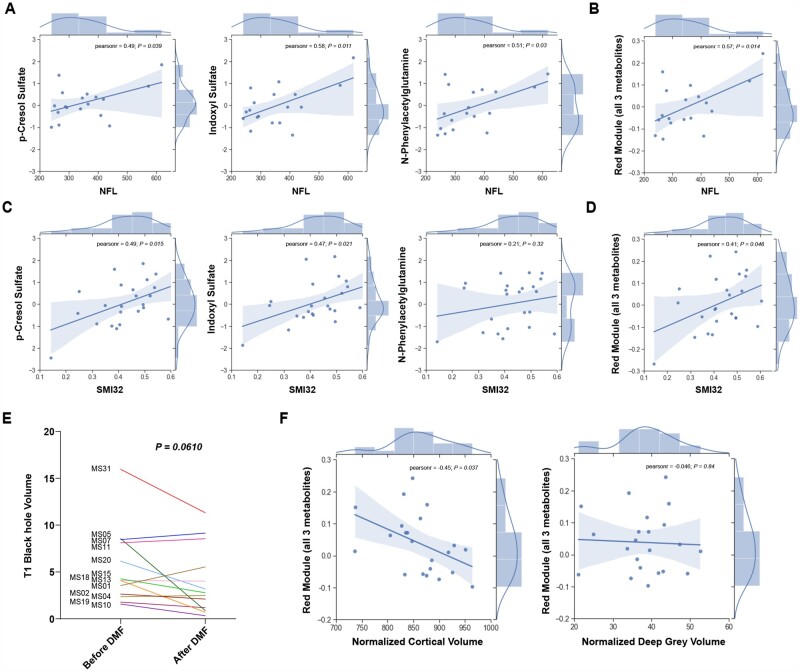
As neurotoxic metabolites were defined based on a functional screening in cultured neurons, we searched for a correlation between their relative abundance and the levels of NFL in the CSF, a well-accepted biomarker of neurodegeneration (Supplementary Table 5). Individual metabolites such as pCS, IS and PAG (Fig. 7A) as well as the red module eigenvector (Fig. 7B) both significantly correlated with NFL levels in the CSF, thereby further validating their identification as neurotoxic compounds. Because the identification of neurotoxic metabolites was driven by the functional neurotoxicity analysis of CSF samples, we also investigated the correlation between the individual concentrations of pCS, IS and PAG (Fig. 7C), or of the entire ‘red module’ metabolites (Fig. 7D), with the parameters of neuronal damage (e.g. SMI32) as assessed in cultured neurons. A direct correlation between the entity of axonal damage, measured by the percentage of SMI32+ axons in CSF-exposed cultured neurons and the relative abundance of metabolites in the CSF was detected for pCS and IS (Fig. 7C) and for the entire red module (Fig. 4D), while PAG did not show any correlation. To further define the potential long-term consequences of the identified metabolites, we also compared clinical and radiological measurements at baseline and after DMF treatment. Clinical disability score (EDSS), the 25-FTW, SDMT and a cognitive PASAT score were evaluated at baseline and at a 12 month clinical follow-up visit, which revealed improved EDSS and PASAT scores in DMF-treated RRMS patients at 12 months after treatment, while 25-FTW and SDMT were unchanged (Supplementary Table 2). Clinical MRIs were obtained at baseline, 12 and 24 months and used to obtain brain and lesion volume metrics (Supplementary Table 6). Interestingly, the concentration of ‘red module’ metabolites correlated well with clinical, cognitive and MRI metrics after treatment, such as PASAT scores 12 months after treatment (spearman rho = −0.9, P = 0.00083; Supplementary Fig. 5B), while no correlation was found with the scores obtained at the start of treatment. Furthermore, among DMF-treated patients, those with a reduction of the ‘red module metabolites’ after 6 months of therapy with DMF had significantly higher odds of an EDSS improvement at 12 months after treatment (ordinal logistic regression controlling for age, sex, body mass index and diagnosis; odds ratio = 9.21, P = 0.046). The clinical amelioration in treated RRMS patients was also consistent with the detection of lower levels of T1 black holes (Fig. 7E), a metric of oedema and axonal loss.64 Patients with lower T1 values were also the ones with lower levels of the putative neurotoxic metabolites in the CSF and plasma (Fig. 4C and D). Finally, the red module metabolites significantly correlated with cortical volume of multiple sclerosis patients measured after 12 months of treatment (Fig. 7F), with patients with the lower levels of microbial metabolites also being the ones showing higher cortical volumes, a correlation that was not detected with deep grey matter volumes (Fig. 7F). This is of interest, as it suggests that the more superficial brain layers, in closer contact with CSF, showed a relationship to the levels of neurotoxic microbial metabolites, with deep grey matter structures being more shielded from the effect of CSF composition. Together, these findings further validate the identification of phenol- and indole-derivatives as neurotoxic metabolites responsible of the gut–brain cross-talk.
Figure 7.
Correlation between neurotoxic CSF metabolites, biomarker of neurodegeneration and cortical volume in multiple sclerosis patients. (A) The correlation plots of the eigenvector of each individual metabolite (CS, IS and PAG) in the red module and the NFL levels in the CSF are positively correlated. (B) The abundance of the three metabolites as a ‘red module’, significantly correlated with the NFL concentration in the CSF. (C) The correlation plots of the eigenvector of each metabolite in the red module indicated that correlation of the ‘red module’ metabolites to axonal damage was mainly driven by CS and IS, not by PAG. (D) However, the relative abundance of the ‘red module’ metabolites significantly correlated with SMI32+ area in neurons treated with the patient-derived CSF samples. (E) T1 black hole volume in RRMS patients showed a trend towards improvement after DMF treatment. (F) The three ‘red module’ metabolites, as a group, were inversely correlated with the volume of superficial cortical layers (normalized cortical volume) and not with that of deep grey matter (normalized deep grey volume) as measure at the 1-year follow-up. Pearson’s correlation coefficient (r) and associated P-value or P-values for paired t-test are indicated.
Discussion
The role of the gut microbiota in regulating health and disease states is well recognized,6 with several studies addressing its composition in distinct neurological diseases including multiple sclerosis.7–11 Besides regulating immune cells residing in the gut,65,66 the microbial commensals play a major role in the metabolism of essential amino acids (such as tryptophan, phenylalanine and branched amino acids) and vitamin absorption. In addition, while the microbiota composition displays a high degree of interindividual variability, the metabolic effects are more stable, thereby suggesting that distinct microbial communities may serve similar metabolic functions in individual hosts. For this reason, rather than focusing on the characterization of individual microbial species, efforts have shifted towards the unbiased characterization of the metabolome in health and disease states, including multiple sclerosis.67–69 Within this context, we and others have proposed that some metabolites produced in the gut could be transferred to the CSF, where they might exert a toxic effect and impact the disease course. This study was designed to test this hypothesis. We had previously characterized a functional screening of neurotoxicity, consisting of a xenogeneic reductionist model of rodent cultured neurons exposed to human CSF samples from multiple sclerosis patients and analysing the effect on axonal damage and mitochondrial function.54,56 We and others also reported on the dramatic effect of an oral treatment with the metabolite DMF on the depletion of microbiota in multiple sclerosis patients.19,59 Based on these premises, we reasoned that a functional screen of the CSF collected from RRMS patients before and after treatment with DMF would provide us with an opportunity to identify potential neurotoxic metabolites produced by the gut microbiota. The detection of the same metabolites in plasma samples from the same subjects would support the idea of a gut–brain communication with peripheral metabolites being transported via the bloodstream.
Using metabolomic analysis of plasma and CSF longitudinal samples from RRMS patients followed by weighted correlation network analysis, we were able to identify groups of metabolites (or ‘modules’) that were highly correlated to specific patient traits by employing eigengene network methodology. Such an approach allowed us to identify a specific ‘module’ composed of an indole-derivative from the tryptophan metabolism (e.g. IS) and two phenol-derivatives from tyrosine and phenylalanine metabolism (e.g. pCS and PAG). These three metabolites as a ‘group’ were found to correlate with the ‘treatment’ variable, both in plasma and CSF samples of patients who received DMF for 6 months. The relative abundance of these metabolites was higher in the CSF samples from RRMS patients at baseline and decreased in samples collected from the same patients after treatment, and this correlated with the relative levels of NFL, a biomarker for neurodegeneration. These putative neurotoxins result from the catabolism of tryptophan, tyrosine and phenylalanine and their relative abundance in the CSF inversely correlated with MRI metrics of brain volume of cortical structures that are in contact with the fluid, while no correlation was found with volume of deep grey matter structure. Lower abundance of these metabolites was also detected in CSF samples after DMF treatment, which displayed lower neurotoxicity at the functional CSF screening. Based on these findings, we identified these three metabolites as putative microbial neurotoxins and validated this interpretation by characterizing the progressive decline of neuronal firing activity with exposure to increasing concentrations of the three compounds. Intriguingly, the effect was dissimilar from what shown in the past for neurotoxic lipids, like ceramides,54,56 as the phenol- and indole-derivatives did not impair mitochondrial function or neuronal bioenergetics. Overall, this identifies three metabolites as potential candidates for a ‘toxic’ gut–brain communication, although future studies, involving selective sequestration of each of these metabolites, would be needed to properly decipher their relative contribution to CSF toxicity and investigate their precise mechanism of action.
The identified metabolites were derived from the catabolism of tryptophan and phenylalanine, two essential amino acids, whose anabolic and catabolic pathways are activated by the gut bacteria.70 In physiological conditions, the healthy gut-microbiota processes tryptophan, via the synthetic pathway to generate serotonin or via the catabolic pathway to generate ‘healthy metabolites’ such as kynurenate (which can be further converted into nicotinamide).71,72 In pathological conditions, however, the levels of serotonin and kynurenic acid are often lowered as tryptophan is converted into an excess of indole-derivatives, including indole acetate and IS.73,74 Our data support a pathological ‘metabolic shunt’ from healthy to toxic catabolites in RRMS patients, as we detected lower levels of kynurenic acid and higher levels of IS than in controls. In addition, the decreased abundance of these metabolites in CSF and plasma samples collected after DMF-treatment—which we previously showed to deplete the gut flora19—further supports the identification of these metabotoxins as putative signals of communication between the periphery and the CNS. Our results are also consistent with previous reports of decreased tryptophan75 and serotonin76 levels in adult multiple sclerosis patients and altered levels of tryptophan metabolism in a paediatric multiple sclerosis population,77 whose gut microbiome differed from that of age-matched healthy controls.78–80 Among the products of tryptophan metabolism, kynurenate has been identified by us and other studies57; however, the generation of this compound by host metabolism in glial cells and its ability to generate either the neuroprotective kynurenic acid or the excitotoxic quinolic acid render the discussion on the balance of quinolic/kynurenic levels more complex, even though associated with multiple sclerosis disease progression.81 The increased levels of indole-catabolites, such as IS, in the CSF of RRMS patients in contrast can be entirely attributed to microbial synthesis and is in agreement with previous studies57,82 identifying this metabolite as an important uraemic toxin detected in patients with renal insufficiency who display significant cognitive decline and even dementia.83 It will be important to further investigate the direct effect of this metabolite on neurons, as murine studies had previously suggested that indole-derivatives may act as neuroprotective agents, by serving as ligands for the aryl hydrocarbon receptor on microglial cells and promoting anti-inflammatory reactions.32,58
The other two microbial neurotoxins, identified as part of the ‘module’ of three metabolites correlating with the neurotoxic effect of the CSF on neurons, were pCS and PAG, both containing phenol groups and derivatives of the phenylalanine metabolism. In physiological conditions healthy bacteria in the gut are responsible for catalytic anabolic pathways leading to the generation of tyrosine, which acts as precursor for both hormones and neurotransmitters.84 In patients with multiple sclerosis, however, unhealthy bacteria lead to the production of catabolites, such as cresol sulphate and phenylacetylglutamine, which have also been detected in other pathologies, including cardiovascular disorders85,86 and other neurodegenerative diseases such as Parkinson’s disease.87 Intriguingly, pCS is a metabolite which had been detected in the urine of multiple sclerosis patients88 and mistakenly identified as urinary ‘myelin basic like material’, whose levels correlated with the degree of clinical progression.89 This metabolite was also identified in a previous study conducted in mice to correlate with reduced content of cortical myelin and with the presence of bacteria Clostridiales in the gut.43 Of note, the concentrations used to test the effect of metabolites in cultured neurons were within limits of the values listed in the Human Metabolome Databases and those previously reported in the blood of uraemic patients.62,63
In conclusion, based on published studies and experimental evidence, it is conceivable that the changes of bacterial metabolites detected in RRMS patients treated with DMF, may be related to its previously reported effect on the gut microbiota.19,59 This conclusion is substantiated by the fact that the levels of the same metabolites remained unchanged in a cohort of age-related RRMS patients treated with anti-CD20 therapy, whose disease-modifying properties are independent of gut alterations.90 Because the composition of the gut microbiota in RRMS patients has been recently shown to be distinct from that of PPMS and SPMS patients,60,61,91,92 it is also tempting to speculate that the lack of effectiveness of DMF treatment in SPMS patient might be related to the presence of microbial species which are more resistant to the effect of DMF in the gut. This would be an interesting area of future investigation. Nevertheless, the importance of disease severity and older age cannot be discounted as important additional factors.
Therefore, this study identifies microbial metabolites in the CSF of RRMS patients as putative neurotoxins responsible for the cross-talk between the periphery and brain parenchyma. While much awaits to be discovered, including the mechanism of action of these metabolites in neurons or the identification of multiple combinations of microbial commensals responsible for enhancing the catabolic pathways in patients, we believe that a focus on metabotoxins may be crucial for the design of putative approaches geared to the generation of novel treatments for the neurodegenerative component of the disease.
Supplementary Material
Acknowledgements
Some of the initial experiments on cultured cell lines that have not been included in the current manuscript were performed by a Visiting Honour Master Student, Marina Hommersom (Radboud Honours Academy, the Netherlands). Some of the initial analysis of the baseline MRI was performed by Dr Maria Petracca, in the Laboratory of Dr Matilde Inglese. The results of that analysis have not been included in the current manuscript, as all the MRI data have been evaluated at baseline and after treatment by Icometrics.
Funding
These studies were supported by BIOGEN, Investigator Initiated Award to P.C. and K.E.
Competing interests
A.N. received financial compensation as a consultant for Biogen Idec.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- DMF
dimethyl fumarate
- EDSS
Expanded Disability Score Scale
- IS
indoxyl sulphate
- MMF
monomethyl fumarate
- PAG
N-phenylacetylglutamine
- PASAT
Paced Auditory Serial Addition Test
- pCS
p-cresol sulphate
- RRMS
relapsing remitting multiple sclerosis
- SPMS
secondary progressive multiple sclerosis
- WGCNA
weighted correlation network analysis
Contributor Information
Achilles Ntranos, Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY 10031, USA.
Hye-Jin Park, Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY 10031, USA.
Maureen Wentling, Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY 10031, USA.
Vladimir Tolstikov, BERG, Framingham, MA 01701, USA.
Mario Amatruda, Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY 10031, USA.
Benjamin Inbar, Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY 10031, USA.
Seunghee Kim-Schulze, Human Immune Monitoring Center, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Carol Frazier, Multiple Sclerosis Center of Northeastern New York, Latham, NY 12110, USA.
Judy Button, Multiple Sclerosis Center of Northeastern New York, Latham, NY 12110, USA.
Michael A Kiebish, BERG, Framingham, MA 01701, USA.
Fred Lublin, Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Keith Edwards, Multiple Sclerosis Center of Northeastern New York, Latham, NY 12110, USA.
Patrizia Casaccia, Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY 10031, USA; Graduate Program in Biology and Biochemistry at the Graduate Center of the City University of New York, New York, NY, USA.
References
- 1. Iglesias-Vázquez L, Van Ginkel Riba G, Arija V, Canals J. Composition of gut microbiota in children with autism spectrum disorder: A systematic review and meta-analysis. Nutrients. 2020;12(3):792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kong G, Cao KL, Judd LM, Li S, Renoir T, Hannan AJ. Microbiome profiling reveals gut dysbiosis in a transgenic mouse model of Huntington's disease. Neurobiol Dis. 2020;135:104268. [DOI] [PubMed] [Google Scholar]
- 3. Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016;167(6):1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu SC, Cao ZS, Chang KM, Juang JL. Intestinal microbial dysbiosis aggravates the progression of Alzheimer's disease in Drosophila. Nat Commun. 2017;8(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quagliariello A, Del Chierico F, Russo A, et al. Gut microbiota profiling and gut–brain crosstalk in children affected by pediatric acute-onset neuropsychiatric syndrome and pediatric autoimmune neuropsychiatric disorders associated with streptococcal infections. Front Microbiol. 2018;9:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sauma S, Casaccia P. Gut–brain communication in demyelinating disorders. Curr Opin Neurobiol. 2020;62:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6(1):28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jangi S, Gandhi R, Cox LM, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cekanaviciute E, Pröbstel AK, Thomann A, et al. Multiple sclerosis-associated changes in the composition and immune functions of spore-forming bacteria. mSystems. 2018;3(6):e00083–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janakiraman M, Krishnamoorthy G. Emerging role of diet and microbiota interactions in neuroinflammation. Front Immunol. 2018;9:2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ventura RE, Iizumi T, Battaglia T, et al. Gut microbiome of treatment-naïve MS patients of different ethnicities early in disease course. Sci Rep. 2019;9(1):16396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breban M, Tap J, Leboime A, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76(9):1614–1622. [DOI] [PubMed] [Google Scholar]
- 13. de Groot PF, Belzer C, Aydin Ö, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One. 2017;12(12):e0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang XS, Li J, Krautkramer KA, et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. eLife. 2018;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35(8):1500–1505. [PubMed] [Google Scholar]
- 16. Maeda Y, Kurakawa T, Umemoto E, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68(11):2646–2661. [DOI] [PubMed] [Google Scholar]
- 17. Alkanani AK, Hara N, Gottlieb PA, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64(10):3510–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He B, Hoang TK, Tian X, et al. Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front Immunol. 2019;10:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katz Sand I, Zhu Y, Ntranos A, et al. Disease-modifying therapies alter gut microbial composition in MS. Neurol Neuroimmunol Neuroinflamm. 2019;6(1):e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shahi SK, Freedman SN, Murra AC, et al. Prevotella histicola, a human gut commensal, is as potent as COPAXONE® in an animal model of multiple sclerosis. Front Immunol. 2019;10:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeng Q, Junli G, Liu X, et al. Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem Int. 2019;129:104468. [DOI] [PubMed] [Google Scholar]
- 22. Cree BA, Spencer CM, Varrin-Doyer M, et al. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann Neurol. 2016;80(3):443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zamvil SS, Spencer CM, Baranzini SE, Cree BAC. The Gut Microbiome in Neuromyelitis Optica. Neurotherapeutics. 2018;15(1):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berer K, Gerdes LA, Cekanaviciute E, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. 2017;114(40):10719–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kirby TO, Ochoa-Repáraz J. The gut microbiome in multiple sclerosis: A potential therapeutic avenue. Med Sci (Basel). 2018;6(3):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duc D, Vigne S, Bernier-Latmani J, et al. Disrupting myelin-specific Th17 cell gut homing confers protection in an adoptive transfer experimental autoimmune encephalomyelitis. Cell Rep. 2019;29(2):378–390.e4. [DOI] [PubMed] [Google Scholar]
- 27. Wagley S, Bokori-Brown M, Morcrette H, et al. Evidence of Clostridium perfringens epsilon toxin associated with multiple sclerosis. Multiple Scler. 2019;25(5):653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rumah KR, Linden J, Fischetti VA, Vartanian T. Isolation of Clostridium perfringens type B in an individual at first clinical presentation of multiple sclerosis provides clues for environmental triggers of the disease. PLoS One. 2013;8(10):e76359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cryan JF, O'Riordan KJ, Cowan CSM, et al. The microbiota–gut–brain axis. Physiol Rev. 2019;99(4):1877–2013. [DOI] [PubMed] [Google Scholar]
- 30. Linden JR, Flores C, Schmidt EF, et al. Clostridium perfringens epsilon toxin induces blood brain barrier permeability via caveolae-dependent transcytosis and requires expression of MAL. PLoS Pathogens. 2019;15(11):e1008014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cases M, Llobet A, Terni B, et al. Acute effect of pore-forming Clostridium perfringens ε-toxin on compound action potentials of optic nerve of mouse. eNeuro. 2017;4(4):ENEURO.0051-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rothhammer V, Borucki DM, Tjon EC, et al. Microglial control of astrocytes in response to microbial metabolites. Nature. 2018;557(7707):724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saresella M, Mendozzi L, Rossi V, et al. Immunological and clinical effect of diet modulation of the gut microbiome in multiple sclerosis patients: A pilot study. Front Immunol. 2017;8:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kappos L, Radue EW, O'Connor P, et al. A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. New Engl J Med. 2010;362(5):387–401. [DOI] [PubMed] [Google Scholar]
- 35. Fox RJ, Miller DH, Phillips JT, et al. Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. New Engl J Med. 2012;367(12):1087–1097. [DOI] [PubMed] [Google Scholar]
- 36. Kornberg MD, Bhargava P, Kim PM, et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360(6387):449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ntranos A, Ntranos V, Bonnefil V, et al. Fumarates target the metabolic–epigenetic interplay of brain-homing T cells in multiple sclerosis. Brain. 2019;142(3):647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lublin FD, Reingold SC. Defining the clinical course of multiple sclerosis: Results of an international survey. National Multiple Sclerosis Society (USA) Advisory Committee on Clinical Trials of New Agents in Multiple Sclerosis. Neurology. 1996;46(4):907–911. [DOI] [PubMed] [Google Scholar]
- 39. Lublin FD, Reingold SC, Cohen JA, et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology. 2014;83(3):278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jain S, Sima DM, Ribbens A, et al. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 2015;8:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smeets D, Ribbens A, Sima DM, et al. Reliable measurements of brain atrophy in individual patients with multiple sclerosis. Brain Behav. 2016;6(9):e00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tolstikov V, Nikolayev A, Dong S, Zhao G, Kuo MS. Metabolomics analysis of metabolic effects of nicotinamide phosphoribosyltransferase (NAMPT) inhibition on human cancer cells. PLoS One. 2014;9(12):e114019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gacias M, Gaspari S, Santos PM, et al. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. eLife. 2016;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Drolet J, Tolstikov V, Williams BA, et al. Integrated metabolomics assessment of human dried blood spots and urine strips. Metabolites. 2017;7(3):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baskin AS, Linderman JD, Brychta RJ, et al. Regulation of human adipose tissue activation, gallbladder size, and bile acid metabolism by a β3-adrenergic receptor agonist. Diabetes. 2018;67(10):2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kiebish MA, Cullen J, Mishra P, et al. Multi-omic serum biomarkers for prognosis of disease progression in prostate cancer. J Transl Med. 2020;18(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bajad S, Shulaev V. LC-MS-based metabolomics. Methods Mol Biol (Clifton, NJ). 2011;708:213–228. [DOI] [PubMed] [Google Scholar]
- 48. Dunn WB, Broadhurst D, Begley P, et al. ; Human Serum Metabolome (HUSERMET) Consortium . Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060–1083. [DOI] [PubMed] [Google Scholar]
- 49. Want EJ, Masson P, Michopoulos F, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc. 2013;8(1):17–32. [DOI] [PubMed] [Google Scholar]
- 50. Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhargava P, Fitzgerald KC, Calabresi PA, Mowry EM. Metabolic alterations in multiple sclerosis and the impact of vitamin D supplementation. JCI Insight. 2017;2(19):e95302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li J, Zhou D, Qiu W, et al. Application of weighted gene co-expression network analysis for data from paired design. Sci Rep. 2018;8(1):622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wentling M, Lopez-Gomez C, Park HJ, et al. A metabolic perspective on CSF-mediated neurodegeneration in multiple sclerosis. Brain 2019;142(9):2756–2774. [DOI] [PubMed] [Google Scholar]
- 55. Kim JY, Shen S, Dietz K, et al. HDAC1 nuclear export induced by pathological conditions is essential for the onset of axonal damage. Nat Neurosci. 2010;13(2):180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vidaurre OG, Haines JD, Katz Sand I, et al. Cerebrospinal fluid ceramides from patients with multiple sclerosis impair neuronal bioenergetics. Brain. 2014;137(Pt 8):2271–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rejdak K, Bartosik-Psujek H, Dobosz B, et al. Decreased level of kynurenic acid in cerebrospinal fluid of relapsing-onset multiple sclerosis patients. Neurosci Lett. 2002;331(1):63–65. [DOI] [PubMed] [Google Scholar]
- 58. Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Storm-Larsen C, Myhr KM, Farbu E, et al. Gut microbiota composition during a 12-week intervention with delayed-release dimethyl fumarate in multiple sclerosis—A pilot trial. Multiple Sclerosis J Exp Transl Clin. 2019;5(4):2055217319888767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takewaki D, Suda W, Sato W, et al. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc Natl Acad Sci U S A. 2020;117(36):22402–22412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cox LM, Maghzi AH, Liu S, et al. Gut microbiome in progressive multiple sclerosis. Ann Neurol. 2021;89(6):1195–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vanholder R, De Smet R, Glorieux G, et al. ; For the European Uremic Toxin Work Group (EUTox) . Review on uremic toxins: Classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–1943. [DOI] [PubMed] [Google Scholar]
- 63. Duranton F, Cohen G, De Smet R, et al. ; European Uremic Toxin Work Group . Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012;23(7):1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bagnato F, Jeffries N, Richert ND, et al. Evolution of T1 black holes in patients with multiple sclerosis imaged monthly for 4 years. Brain. 2003;126(8):1782–1789. [DOI] [PubMed] [Google Scholar]
- 65. Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-modulated metabolites at the interface of host immunity. J Immunol (Baltimore, MD: 1950). 2017;198(2):572–580. [DOI] [PubMed] [Google Scholar]
- 66. Wang G, Huang S, Wang Y, et al. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci. 2019;76(20):3917–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Villoslada P, Alonso C, Agirrezabal I, et al. Metabolomic signatures associated with disease severity in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2017;4(2):e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bhargava P, Fitzgerald KC, Venkata SLV, et al. Dimethyl fumarate treatment induces lipid metabolism alterations that are linked to immunological changes. Ann Clin Transl Neurol. 2019;6(1):33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bhargava P, Anthony DC. Metabolomics in multiple sclerosis disease course and progression. Multiple Scler. 2020;26(5):591–598. [DOI] [PubMed] [Google Scholar]
- 70. Kaur H, Bose C, Mande SS. Tryptophan metabolism by gut microbiome and gut–brain-axis: An in silico analysis. Front Neurosci. 2019;13:1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2020;70(6):1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sacks D, Baxter B, Campbell BCV, et al. ; From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), Canadian Interventional Radiology Association (CIRA), Congress of Neurological Surgeons (CNS), European Society of Minimally Invasive Neurological Therapy (ESMINT), European Society of Neuroradiology (ESNR), European Stroke Organization (ESO), Society for Cardiovascular Angiography and Interventions (SCAI), Society of Interventional Radiology (SIR), Society of NeuroInterventional Surgery (SNIS), and World Stroke Organization (WSO) . Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke. 2018;13(6):612–632.29786478 [Google Scholar]
- 73. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9(1):3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Muneer A. Kynurenine pathway of tryptophan metabolism in neuropsychiatric disorders: Pathophysiologic and therapeutic considerations. Clin Psychopharmacol Neurosci. 2020;18(4):507–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lieben CK, Blokland A, Deutz NE, Jansen W, Han G, Hupperts RM. Intake of tryptophan-enriched whey protein acutely enhances recall of positive loaded words in patients with multiple sclerosis. Clin Nutr (Edinburgh, Scotland). 2018;37(1):321–328. [DOI] [PubMed] [Google Scholar]
- 76. San Hernandez AM, Singh C, Valero DJ, et al. Multiple sclerosis and serotonin: Potential therapeutic applications. Cureus. 2020;12(11):e11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nourbakhsh B, Bhargava P, Tremlett H, Hart J, Graves J, Waubant E. Altered tryptophan metabolism is associated with pediatric multiple sclerosis risk and course. Ann Clin Transl Neurol. 2018;5(10):1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tremlett H, Fadrosh DW, Faruqi AA, et al. ; US Network of Pediatric MS Centers . Gut microbiota composition and relapse risk in pediatric MS: A pilot study. J Neurol Sci. 2016;363:153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tremlett H, Fadrosh DW, Faruqi AA, et al. ; the US Network of Pediatric MS Centers . Gut microbiota in early pediatric multiple sclerosis: A case-control study. Eur J Neurol. 2016;23(8):1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tremlett H, Fadrosh DW, Faruqi AA, et al. ; US Network of Pediatric MS Centers . Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC Neurol. 2016;16(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lim CK, Bilgin A, Lovejoy DB, et al. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci Rep. 2017;7:41473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Adesso S, Magnus T, Cuzzocrea S, et al. Indoxyl sulfate affects glial function increasing oxidative stress and neuroinflammation in chronic kidney disease: Interaction between astrocytes and microglia. Front Pharmacol. 2017;8:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang F, Zhang L, Liu L, Wang H. Level of kidney function correlates with cognitive decline. Am J Nephrol. 2010;32(2):117–121. [DOI] [PubMed] [Google Scholar]
- 84. Gryp T, Vanholder R, Vaneechoutte M, Glorieux G. p-Cresol sulfate. Toxins. 2017;9(2):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pignanelli M, Just C, Bogiatzi C, et al. Mediterranean diet score: Associations with metabolic products of the intestinal microbiome, carotid plaque burden, and renal function. Nutrients. 2018;10(6):779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tang HY, Wang CH, Ho HY, et al. Characteristic of metabolic status in heart failure and its impact in outcome perspective. Metabolites. 2020;10(11):437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sankowski B, Księżarczyk K, Raćkowska E, Szlufik S, Koziorowski D, Giebułtowicz J. Higher cerebrospinal fluid to plasma ratio of p-cresol sulfate and indoxyl sulfate in patients with Parkinson's disease. Clin Chim Acta. 2020;501:165–173. [DOI] [PubMed] [Google Scholar]
- 88. Cao L, Kirk MC, Coward LU, Jackson P, Whitaker JN. p-Cresol sulfate is the dominant component of urinary myelin basic protein like material. Arch Biochem Biophys. 2000;377(1):9–21. [DOI] [PubMed] [Google Scholar]
- 89. Whitaker JN, Kachelhofer RD, Bradley EL, et al. Urinary myelin basic protein-like material as a correlate of the progression of multiple sclerosis. Ann Neurol. 1995;38(4):625–632. [DOI] [PubMed] [Google Scholar]
- 90. Uzzan M, Ko HM, Rosenstein AK, Pourmand K, Colombel JF, Mehandru S. Efficient long-term depletion of CD20(+) B cells by rituximab does not affect gut-resident plasma cells. Ann N Y Acad Sci. 2018;1415(1):5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kozhieva M, Naumova N, Alikina T, Boyko A, Vlassov V, Kabilov MR. Primary progressive multiple sclerosis in a Russian cohort: Relationship with gut bacterial diversity. BMC Microbiol. 2019;19(1):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Reynders T, Devolder L, Valles-Colomer M, et al. Gut microbiome variation is associated to Multiple Sclerosis phenotypic subtypes. Ann Clin Transl Neurol. 2020;7(4):406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author, upon reasonable request.