Abstract
Parkinson’s disease is a complex neurodegenerative disorder with a strong genetic component, for which most known disease-associated variants are single nucleotide polymorphisms (SNPs) and small insertions and deletions (indels). DNA repetitive elements account for >50% of the human genome; however, little is known of their contribution to Parkinson’s disease aetiology. While select short tandem repeats (STRs) within candidate genes have been studied in Parkinson’s disease, their genome-wide contribution remains unknown. Here we present the first genome-wide association study of STRs in Parkinson’s disease. Through a meta-analysis of 16 imputed genome-wide association study cohorts from the International Parkinson’s Disease Genomic Consortium (IPDGC), totalling 39 087 individuals (16 642 cases and 22 445 controls of European ancestry), we identified 34 genome-wide significant STR loci (P < 5.34 × 10−6), with the strongest signal located in KANSL1 [chr17:44 205 351:[T]11, P = 3 × 10−39, odds ratio = 1.31 (95% confidence interval = 1.26–1.36)]. Conditional-joint analyses suggested that four significant STRs mapping nearby NDUFAF2, TRIML2, MIRNA-129–1 and NCOR1 were independent from known risk SNPs. Including STRs in heritability estimates increased the variance explained by SNPs alone. Gene expression analysis of STRs (eSTRs) in RNA sequencing data from 13 brain regions identified significant associations of STRs influencing the expression of multiple genes, including known Parkinson’s disease genes. Further functional annotation of candidate STRs revealed that significant eSTRs within NUDFAF2 and ZSWIM7 overlap with regulatory features and are associated with change in the expression levels of nearby genes. Here, we show that STRs at known and novel candidate loci contribute to Parkinson’s disease risk and have functional effects in disease-relevant tissues and pathways, supporting previously reported disease-associated genes and giving further evidence for their functional prioritization. These data represent a valuable resource for researchers currently dissecting Parkinson’s disease risk loci.
Keywords: GWAS, meta-analysis, short tandem repeats, Parkinson’s disease, gene expression
Bustos et al. present the first genome-wide meta-analysis of common short tandem repeats (STRs) in Parkinson’s disease, and identify four novel independent risk factors. They show that STRs contribute to Parkinson’s disease heritability and that STR-associated genes are enriched in Parkinson’s disease-relevant tissues and pathways.
Introduction
Parkinson’s disease (PD) is a complex neurodegenerative disease with an established genetic component. Studies over the years have identified several rare variants that cause or significantly increase the risk of disease in carriers, and genome-wide association studies (GWAS) have recently uncovered 90 common variants that influence PD risk.1 It is estimated that common GWAS variants account for 16–36% of the overall genetic heritability of the disease,1,2 highlighting that a large proportion of the missing heritability remains to be identified.
The vast majority of PD genetics studies have focused on the role of single nucleotide polymorphisms (SNPs), meaning that contributions of other genetic elements such as structural variants and repetitive elements have largely been ignored. Repetitive elements represent more than 55% of the human genome.3 Short tandem repeat expansions (STRs) are small repetitive units ranging from one to seven base pairs in length that vary among individuals and account for ∼10% of all repetitive elements.4 STRs are the cause of several neurological diseases and are associated with genes such as those associated with fragile X syndrome (FMR1),5,6 Huntington’s disease (HTT),7 amyotrophic lateral sclerosis and frontotemporal dementia (C9orf72)8,9 and spinocerebellar ataxia (SCA1)10 and have also been linked to numerous complex neurological and psychiatric traits.11 A role for STRs as drivers of GWAS signals have been identified,12 where a risk SNP connects adjacent GGAA repeats by converting an interspaced GGAT motif into a GGAA motif, thereby increasing the number of consecutive GGAA motifs and modifying the activity of its sequence and functional impact. STRs have also been shown to regulate gene expression significantly and contribute to phenotypic plasticity.13 STRs therefore represent a potential source of unexplored genetic variation that may account for some of the missing heritability of PD. In this regard, other repetitive elements, such as satellite repeats, have been shown to alter gene expression in blood of patients with PD.14 However, no genome-wide assessment of STRs in large population studies has yet been performed in this disease. Due to their more complex and highly repetitive structure compared to SNVs, STRs have been difficult to assess. Despite the recent explosion of genetic data stemming from next generation sequencing, STRs are still difficult to genotype. Recent advances in PCR-free deep sequencing methods and STR genotyping tools now allow for the simultaneous assessment of STRs genome-wide.15 Studies have shown high linkage disequilibrium (LD) between STRs and SNPs across the genome.16 Exploiting this high LD, Saini et al.17 generated a phased SNP-STR haplotype panel based on the 1000 Genomes Project samples that allows for the accurate genome-wide imputation of common STRs into array-based genotype data. To assess the role of common STRs in PD risk, we imputed and interrogated STRs across 16 independent PD case-control cohorts, totaling 39 087 individuals available through the International Parkinson’s disease Genomics Consortium (IPDGC).
Materials and methods
A summary diagram for the methodological steps followed in the present study is shown in Supplementary Fig. 1.
Samples and quality control
All genotyping data was obtained from previously generated IPDGC datasets, consisting of 39 087 individuals (16 642 cases and 22 445 controls) of European ancestry.1 All individuals provided informed consent for participation in genetics studies, which was approved by the relevant local ethics committee for each of the datasets used. Detailed demographic, sample sizes and PD status are given in Supplementary Table 1. Further information along with detailed quality control (QC) methods have been previously published.1,18 Briefly, for sample QC prior to imputation, individuals with low call rate, discordance between genetic and reported sex, heterozygosity outliers and ancestry outliers were removed. For genotype QC, variants with a missingness rate of > 5%, minor allele frequency (MAF) < 0.01, exhibiting deviations from Hardy–Weinberg Equilibrium (HWE) < 1 × 10−5 and palindromic SNPs were excluded.
STR imputation and filtering
STR genotypes were imputed into the IPDGC SNP unimputed genotyping datasets using Beagle v.5.119 with the 1000 Genomes SNP-STR Haplotype reference panel.17 In brief, STR genotypes in the reference panel were imputed from STRs called from the catalog-based STR caller hipSTR15 and supplemented using a second STR caller, TREDPARSE.20 STRs were phased with corresponding SNPs creating a final panel in the 1000 genomes project data that contained 27 185 239 SNP and 445 725 STR markers. Once STRs were imputed into all IPDGC SNP genotype datasets, the STR calls were filtered to facilitate downstream association analysis. First STRs were split from multi-allelic variants to single biallelic variants using the vt variant tool.21 Finally SNPs and STRs with a dosage R-squared (DR2) < 0.3 were removed to filter out low quality imputed variants.
Study-level STR analysis and meta-analysis
To estimate PD risk, imputed dosages (i.e. genotype probabilities for a variant to be A/A, A/B or B/B from 0 to 2) were analysed using a logistic regression model adjusted for sex, age at onset (AAO) for cases or examination for controls, and the first 10 principal components. Of note, AAO could not be included as a covariate for the Myers–Faroud22 and Vance (dbGap phs000394) studies, as no AAO information was available. Summary statistics were generated using the RVTESTS package23 and filtered for a MAF > 1%. A meta-analysis was conducted based on the fixed-effect model as implemented in METAL24 by combining summary statistics across all 16 IPDGC datasets. All variants with a meta-analysis heterogeneity value of less than 80% (I2 < 0.80) were kept for further analysis.
Comparative analysis of STRs genotypes in imputed and sequencing datasets
Comparative concordance analysis of imputed STRs was done using whole-genome sequencing (WGS) data from 646 Parkinson’s Progression Markers Initiative (PPMI) samples. We aligned the paired-end fastq reads to the GRCh37 genome reference using the Burrows–Wheeler Aligner v.0.7.17.25 After marking duplicates and performing a base quality recalibration score step with GATK 4.1.826 on the resulting bam files, we used gangSTR v.2.4.227 to call STRs for each sample using the hipSTR reference bed file in the GRCh37 version,15 applying the following recommended parameters: –frrweight 0.25, –enclweight 1.0, –spanweight 1.0, –flankweight 1.0, –ploidy 2, –numbstrap 50, –minmatch 5 and –minscore 80. We generated a second STR call-set using hipSTR v.0.6.2,15 using the recommended parameters –lib-from-samp, –def-stutter-model, –max-str-len 1200 and –min-reads 15. We next filtered each resulting variant call format file (VCF) using dumpSTR within the TRTools package,28 using level 1 and 2 filters as described elsewhere.27 Briefly, for gangSTR, we used –max-call-DP 1000, –min-call-DP 10, –filter-spanbound-only, –filter-badCI and –min-call-Q 0.9. For both gangSTR and hipSTR calls, we removed STRs falling within regions enriched in segmental duplications according to the UCSC table browser database (hs37_segmentalduplications.bed.gz) using the commands –filter-regions and –filter-regions-names SEGDUP in dumpSTR. Then, we merged the filtered VCFs using mergeSTR within TRTools. We overlapped the samples present in the imputed, gangSTR and hipSTR PPMI call sets, obtaining 515 individuals in common. Next, we compared the STR variants between the imputed, gangSTR and hipSTR PPMI VCFs by splitting multiallelic sites with BCFTools v.1.10.129 and intersecting the variants with BEDTools v.2.29.30 We then plotted the length of the overlapping variants and calculated a regression using Pearson’s coefficient with ggplot2 in R.31 Genotype concordance of STRs between the imputed, gangSTR and hipSTR calls was done with SNPSIFT concordance,32 using a set of shared variants between cohorts.
Conditional-joint and linkage disequilibrium analyses
To select candidate variants, we used the Genome-wide Complex Trait Analysis software (GCTA)33 to perform conditional and joint analysis (COJO) STRs from the meta-analysis summary statistics. In order to differentiate associations between STRs and SNPs, we performed two COJO analyses, first with STRs only, and second with STRs and SNPs together. As an LD reference for GCTA, we used a sample subset of merged imputed genotypes (hard call threshold of 0.8) from the IPDGC GWAS cohorts34 totalling 4397 PD cases and 9137 controls. Additionally, we performed LD calculations between top STRs with the previously reported list of 90 PD variants1 using PLINK v.1.935 to determine highly linked STRs to known PD risk variants. A Hudson plot showing the genome-wide association results for STRs and SNPs separately was done with the hudson R package (https://github.com/anastasia-lucas/hudson). Regional plots for the GCTA-nominated independent STRs were done with the Locuszoom standalone version.36
Expression quantitative trait loci of STRs
Using sample level genotypes and gene expression data from the North American Brain Expression Consortium (NABEC)37 and the Genotype-Tissue Expression project,38 we carried out an expression quantitative trait loci analysis with imputed STRs (eSTRs). The NABEC data was composed of 343 individuals with genotypes obtained from high-coverage Illumina WGS. Corresponding gene expression data were generated from frontal cortex tissue by RNA sequencing (RNASeq) and normalized gene counts were used. The Genotype-Tissue Expression Project (GTEx) v.8 data (dbGaP: phs000424.v7.p2) comprises high-coverage (30×) Illumina WGS data from 838 unrelated samples. We downloaded the fully processed, filtered and normalized gene expression matrices (in BED format) for each of the 13 brain tissues including: amygdala, anterior cingulate cortex (BA24), caudate (basal ganglia), cerebellar hemisphere, cerebellum, cortex, frontal cortex (BA9), hippocampus, hypothalamus, nucleus accumbens (basal ganglia), putamen (basal ganglia), spinal cord cervical (c-1) and substantia nigra (https://gtexportal.org/home/datasets). WGS genotypes from GTEx and gene start-end coordinates for expression data for GTEx and NABEC were converted from hg38 reference to hg19 using UCSC liftover tool.39 STRs were imputed as described above. eSTR analysis was performed using the FastQTL software40 correcting for principal components 1–10, sample age, sex (if available) and probabilistic estimations of expression residuals factors (PEER) generated using the PEER software41: 45 factors for NABEC and 15 for GTEx (as indicated in the GTEx documentation). The 34 top STRs from the meta-analysis along with variants with LD > 0.5 (105 STRs total) were used to conduct the eSTR analysis. QQ-plots and box plots were done using ggplot2 R package.31
Heritability estimation
We used the GCTA-LDMS method33,42 to estimate the heritability of STRs only, both SNPs and STRs together and SNPs only. The method corrects for LD bias in the estimated variant-based heritability from WGS or imputed data. Heritability estimates and their corresponding standard errors are shown in the liability scale.
Gene set, network and pathway enrichment analyses
To functionally characterize the top associated STRs, we carried out loci connectivity analyses across gene-ontologies and gene-expression datasets using FUMA43 and protein-protein interaction networks using Webgestalt.44 We ran MAGMA gene-wise analysis45 using the meta-analysis summary statistics for all STRs, and used the 1000 Genomes SNP-STR dataset as out reference panel.17 Gene lists were analysed for functional enrichments using (i) FUMA gene2func tool; (ii) Biogrid PPI Network Topology-based Analysis (NTA) in Webgestalt; and (iii) gene property analysis for tissue specificity, using 23 675 genes from GTEx RNASeq data38 across the 30 general and 54 specific tissues. Data preprocessing and gene expression normalization methods are presented in the FUMA tutorial section (https://fuma.ctglab.nl/tutorial). Bonferroni and Benjamini–Hochberg false discovery rate (FDR) corrections for multiple testing were performed for MAGMA gene-wise results and functional enrichment analyses, respectively.
Data and code availability
Full STR GWAS summary statistics for the 16 datasets meta-analysed are available at https://drive.google.com/file/d/1kD1i6tHdYC5w0xvxWLD4B-bSPqpnwzNV/view?usp=sharing
The STR imputation, study level GWAS and meta-analysis: https://github.com/neurogenetics/PD_STR_imputation. Downstream analyses: https://github.com/bibb/STR_GWAS_downstream_analysis
Results
Meta-analysis of IPDGC GWAS cohorts imputed with an STR reference panel
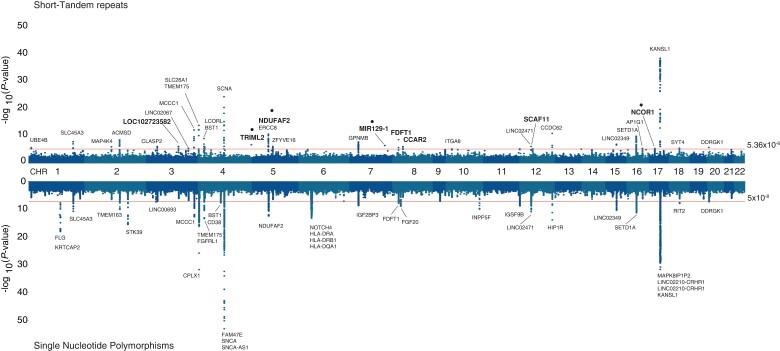
The 16 GWAS cohorts were used in this study, with a combined sample size of 39 087 individuals composed of 16 642 PD cases and 22 445 controls of self-reported European ancestry (Supplementary Table 1). After cohort-wise quality controls (see ‘Materials and methods’ section), we performed genome-wide imputation using the 1000 Genomes STR-SNP reference panel,17 and carried out case-control association analyses with PD status following a meta-analysis of fixed effects across all cohorts. After removing variants with high heterogeneity across meta-analyses (I2 > 0.8), we obtained association P-values for 407 879 STRs, where 214 variants surpassed the threshold for genome-wide significance of P > 5.34 × 10−6 (Fig. 1, upper side), which was estimated by permutation procedures for the STR reference panel, as described elsewhere.46 The inflation factor λ for the association was 1.18 and the rescaled λ for 1000 cases and controls (λ1000) was 1.01.
Figure 1.
Genome-wide association results for imputed STRs and SNPs in 16 PD GWAS cohorts from the IPDGC. Hudson plot representing the association analysis results for STRs (upper) and SNPs (lower) across the human genome showing the 34 genome-wide significant STR loci (P < 5.34 × 10−6) after the conditional-joint analysis with GCTA. Gene names with larger font size represent the loci influenced by STRs after including SNPs associations. Gene names with a dot above represent STR loci independent from the current 90 PD risk variants from the 2019 PD GWAS meta-analysis.
To characterize and identify independently associated STRs, we first performed a conditional-joint analysis using GCTA-COJO33 and identified 34 STR variants mapping to 32 unique nearby genes, with the strongest signal located in KANSL1 [chr17:44205351:[T]11, P = 3 × 10−39, odds ratio (OR) = 1.31; 95% confidence interval (CI) = 1.26–1.36], and followed by SNCA [chr4:90662073:TATTT[GT]8AT[GT]7, P = 3.36 × 10−25, OR = 1.36 (95% CI = 1.28–1.45)] (Supplementary Table 2). Since STRs were imputed by leveraging LD information from SNPs, we carried out a secondary GCTA-COJO analysis including the meta-analysis results from imputed, filtered SNPs (Fig. 1, lower side), obtaining a total number of 8 179 378 SNPs and STRs in all cohorts. We found 76 independent signals (Supplementary Table 3), from which eight loci had associations led by STRs. In order to refine these results, we further investigated their LD patterns with the 90 known Parkinson’s disease risk variants from the 2019 GWAS meta-analysis,1 and found that four of the eight STRs had LD r2 < 0.5 with any of the known variants (Supplementary Table 4), indicating that these could be potential new risk signals:
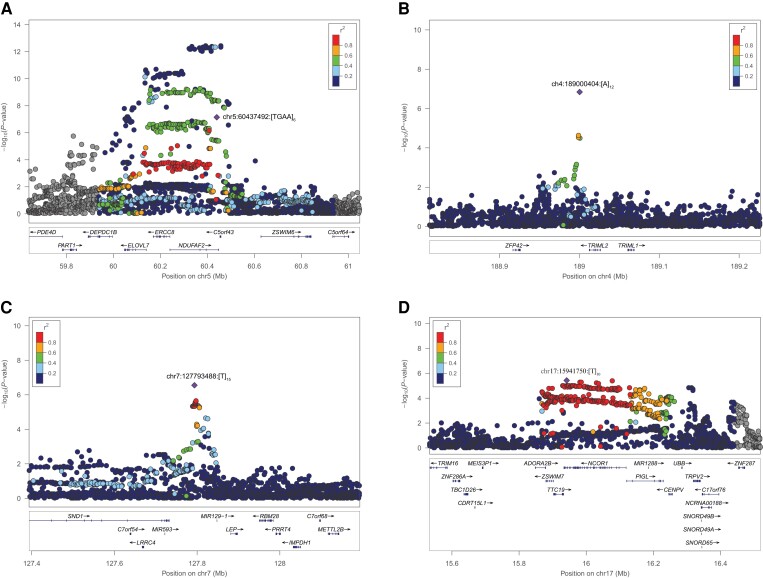
a tetranucleotide repeat within the 3rd intron of NDUFAF2 (risk allele chr5:60437492:AA[TGAA]7, P = 6.49 × 10−8, OR = 1.30, 95% CI = 1.18–1.43) (Fig. 2A);
a mononucleotide repeat downstream of TRIML2 (risk allele chr4:189000404:TT[A]12, P = 1.44 × 10−7, OR = 1.31, 95% CI = 1.19–1.44) (Fig. 2B);
a mononucleotide repeat downstream of MIR129-1 (risk allele chr7:127793488:[T]15G, P = 2.79 × 10−7, OR = 1.16, 95% CI = 1.09–1.23) (Fig. 2C); and
a mononucleotide repeat within the 44th intron of NCOR1 (risk allele chr17:15941750:[T]11, P = 3.77 × 10−6, OR = 1.08, 95% CI = 1.04–1.12) (Fig. 2D).
Figure 2.
Regional association plots for the four candidate independent STR loci. Locus zoom plots were generated for the four GCTA-nominated independent STR loci from SNPs and 90 PD risk loci in (A) chromosome 5 within NDUFAF2, (B) chromosome 4 nearby TRIML2, (C) chromosome 7 nearby MIR129-1 and (D) chromosome 17 within NCOR1. The lead STR variant is depicted as a purple diamond and nearby variants (STRs and SNPs) in circles coloured according to their LD r2-value to the lead STR variant. Gene annotations for each region are displayed at the bottom of each panel, showing gene strand orientation with arrows.
It is important to note here that the independent STR signal at NDUFAF2 (chr5:60437492:AA[TGAA]7) is within a known PD risk locus (mapping to ELOVL7) and was previously identified through Mendelian randomization to be significantly associated with risk of PD.1 Moreover, further LD analysis on this locus showed a high D′ statistic with the closest known PD risk SNP at that locus (D′ = 0.94 with rs1867598) indicating that, regardless of frequency disparities, the independency suggested by the GCTA-COJO analysis should be taken with caution.
Comparative analysis of STR length and genotype concordance across different call sets
To systematically assess the reliability of our results, we performed a comparative STR analysis in the 515 individuals common to all three STR datasets (imputed, hipSTR and gangSTR). After read alignment and STR calling, direct comparison based on chromosomal position between the imputed and the two STR calling datasets, we obtained 372 391 and 208 514 overlapping STRs versus hipSTR and gangSTR calls, respectively. STR length comparison for the overlapping variants showed a strong correlation between datasets (imputed versus hipSTR: Pearson’s R = 0.95, P < 2.2 × 10−16; imputed versus gangSTR: Pearson’s R = 0.89, P < 2.2 × 10−16; Supplementary Fig. 2A and B, respectively). When extracting the subset of 34 genome-wide significant STRs for each call set, we obtained 24 overlapping STRs by position in hipSTR and 21 in gangSTR. The correlation of allele length between the imputed and each WGS callset remained high and statistically significant (imputed versus hipSTR: Pearson’s R = 0.83, P = 2.1 × 10−7; imputed versus gangSTR: Pearson’s R = 0.81, P = 5.3 × 10−6; Supplementary Fig. 2C and D, respectively). We next assessed genotype concordance across samples for each exact matching STR (by position and allele length) between the imputed and the two STR call sets (Supplementary Fig. 3). We observed a high average genotype concordance for the STRs of the two tested sets (86.73% and 86.43% for hipSTR and gangSTR, respectively). Comparison of per variant concordance showed a strong and significant correlation between sets (Pearson’s R = 0.95, P < 2.2 × 10−16). Three of the genome-wide significant variants were present in the hipSTR and gangSTR call sets (3:33592979:[T]11 near CLASP2, 10:15563184:[T]11 near ITGA8, and 17:44205351:[T]11 near KANSL1) and observed a high genotype concordance across call sets: 63.94%, 99.02% and 97.86%, respectively, for imputed versus hipSTR; and 64.17%, 99.21% and 97.85%, respectively, for imputed versus gangSTR.
Quantifying the heritability of STRs in Parkinson’s disease
The genetic heritability of PD was recently estimated to be 22%.1 Here, assuming a global disease prevalence of 0.2%,2 we leveraged the GCTA-LDMS method3 and estimated that common STRs (MAF >1%) account for 15.2% (SE = 0.01) of the additive heritability of the disease on the liability scale. Heritability for imputed SNPs in the same data accounted for 26.9% (SE = 0.02), similarly to that obtained by Keller et al.2 also using GCTA. After including both common STRs and SNPs in the analysis, the heritability estimate increased to 28.8% (SE = 0.02). This increase of 1.9% in the heritability estimate due to common STRs corresponds to a 7% increase from the SNP based estimate.
Expression quantitative trait loci analysis of STRs
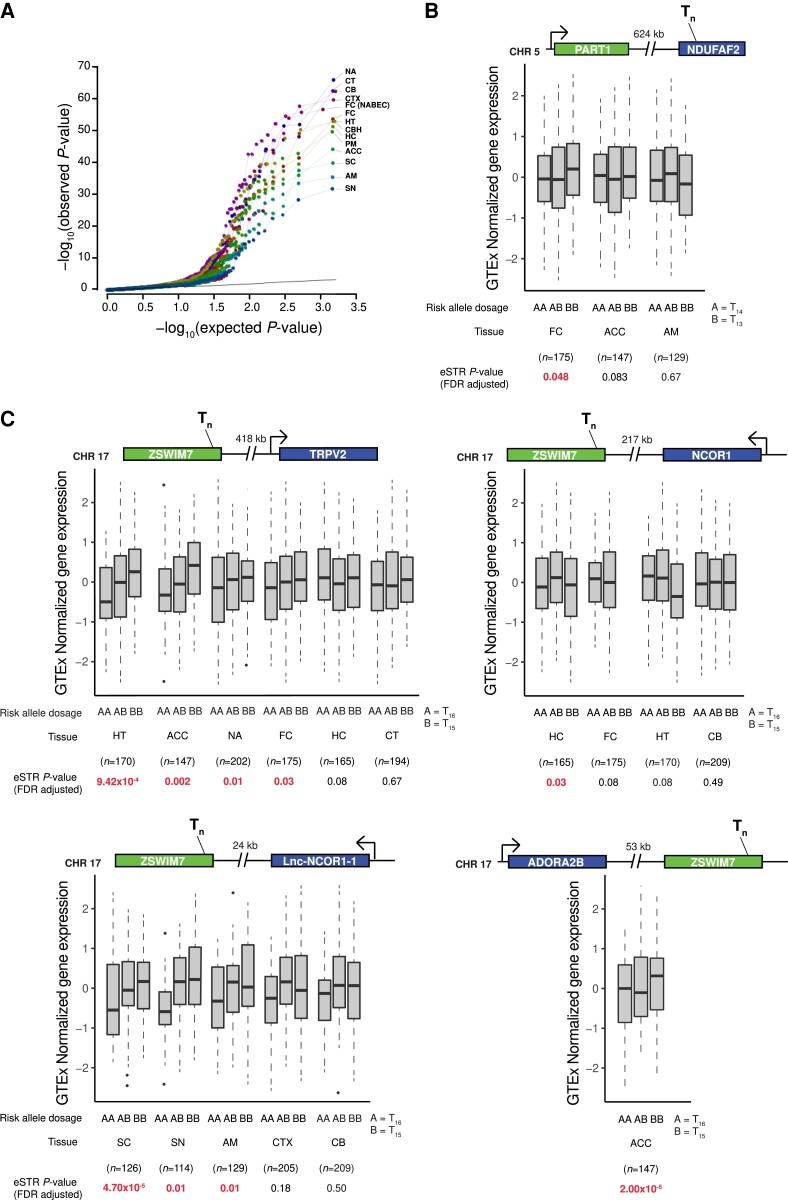
We functionally assessed the impact of the 34 significant STR associations through an eSTR analysis. We investigated each locus extracting the leading STR and other STRs in high LD (r2 > 0.5) within 1 Mb up- and downstream, obtaining 105 variants for further analysis (Supplementary Table 5). We used normalized gene expression data from frontal cortex from NABEC,37 and 13 brain tissues from GTEx v.8.38 Analysing each tissue independently, we identified a total of 10 252 STR-gene associations (Fig. 3A). Of these, 840 associations showed an FDR corrected P < 0.05, corresponding to 234 unique eGenes (genes with at least one significant variant), that included 19 of the 78 loci identified in the 2019 PD GWAS meta-analysis (genes nominated from the 90 PD risk variants): RIT2, TMEM163, MCCC1, LCORL, CTSB, SETD1A, CRHR1, GPNMB, BIN3, TMEM175, GAK, MAP4K4, SNCA, SPTSSB, WNT3, KPNA1, ITGA8, BST1 and HIP1R (Supplementary Table 6). Furthermore, comparative analysis of eSTR target genes in two groups, the substantia nigra and the rest of 12 tissues, showed that 31 genes are targeted to significant eSTRs in both groups, and eight genes are targeted to eSTRs in the substantia nigra only (GAK, DTX3L, PARP15, NMT2, FAM151B, C20orf194, WDR66 and PARP14; Supplementary Fig. 4 and Supplementary Table 7).
Figure 3.
eSTR analysis of top STR loci in gene expression data from brain tissues. (A) Quantile-quantile plot for eSTR analysis of all 34 top loci from meta-analysis and high LD STRs, totalling 105 variants, across brain tissues from the NABEC and GTEx datasets. Colours for each dot and line were added to enhance resolution. (B) Box plots showing gene expression changes associated with STR variant chr5:60408714:TC[T]14GTATC across the tissues where the variant was analysed. (C) Box plots showing gene expression changes associated with STR variant chr17:15902070:[T]16GA. Locus representation is shown at the top of each box plot, representing the STR location and the distance (in kilobases) and orientation of the target gene. Bottom: Allele dosages, tissue, sample size and eSTR FDR-adjusted P-value, with P-values in red representing significant associations. ACC = anterior cingulate cortex (BA24); AM = amygdala; CB = cerebellum; CBH = cerebellar hemisphere; CT = caudate (basal ganglia); CTX = cortex; FC (NABEC) = frontal cortex; FC = frontal cortex (BA9); HC = hippocampus; HT = hypothalamus; NA = nucleus accumbens (basal ganglia); PM = putamen (basal ganglia); SC = spinal cord (cervical c-1); SN = substantia nigra.
To obtain functional and gene expression insights on the four candidate STRs signals obtained from our STR meta-analysis, we similarly extracted the four variants and their surrounding high LD STRs, obtaining 13 unique variants. We functionally annotated them, using regulatory features for gene expression from the Encyclopedia of DNA Elements (ENCODE),47 and found two STRs overlapping enhancers, transcription factor binding sites and histone marks for active transcription (H3K4me3, H3K9Ac, H3K4me1, H3K27Ac, H3K36me3 and H3K79me2): one nearby NDUFAF2 (chr5:60408714:TC[T]14GTATC) in high LD with the leading GWAS STR at that locus (chr5:60437492:AA[TGAA]7, r2 = 0.95); and one within ZSWIM7 (chr17:15902070:[T]16GA), similarly, in high LD with the leading GWAS STR for that locus (chr17:15941750:[T]11, r2 = 0.84) (Supplementary Table 8). The PD risk allele for the eSTR in NDUFAF2 (major allele with 13T repetitions) is significantly associated with higher expression levels of the gene PART1 (∼624 kb upstream) in the frontal cortex (Fig. 3B). Interestingly, the significant eSTR in ZSWIM7 showed associations in more brain tissues, where the risk allele in the STR meta-analysis (minor allele with 15T repeats) was correlated with lower expression levels of TRPV2 in the hypothalamus, anterior cingulate cortex, nucleus accumbens and frontal cortex; higher expression levels of NCOR1 in the hippocampus; higher expression levels of ADORA2B in the anterior cingulate cortex; and lower expression levels of a long non-coding RNA gene located nearby NCOR1 (CTC-529I10.1 or lnc-NCOR1-1) in the spinal cord and substantia nigra (Fig. 3C).
Gene-wise, gene set and pathway enrichment analysis of Parkinson’s disease-associated STRs
MAGMA45 gene-wise enrichment analysis of the STR meta-analysis results yielded 47 genes surpassing genome-wide significance (Bonferroni P < 2.99 × 10−6, α = 0.05/16 696; Supplementary Table 9). Of the 47 genes, 12 overlapped with the STR meta-analysis results, eight overlapped with 78 PD loci nominated from the 90 PD risk variants (2019 PD GWAS meta-analysis) and 27 genes have not previously been identified as enriched genes. Gene-property analysis using gene expression data from GTEx v.8, as described in FUMA,43 showed significant enrichment of genes in the pituitary and brain tissues after FDR correction (FDR P < 0.05) for the 30 GTEx general tissues (Supplementary Fig. 5A) and in the cerebellum, cortex, pituitary, cerebellar hemisphere and frontal cortex for the 54 GTExp specific tissues (Supplementary Fig. 5B). We further investigated gene connectivity via protein-protein interactions using a list of 445 genes surpassing nominal gene-wise STR enrichment (MAGMA P < 0.01) with WebGestalt,44 and leveraged the Network-Topology Analysis (NTA), finding 16 subnetworks (Supplementary Fig. 6), which were significantly enriched in 27 gene ontology (GO) categories, such as synaptic vesicle cycle (GO:0099504), presynaptic endocytosis (GO:0140238) and autophagy (GO:0006914) (Supplementary Table 10).
Discussion
We performed a genome-wide meta-analysis of STRs in 16 cohorts from the IPDGC. We have shown that associated STR signals overlap with known PD risk loci, and with candidate novel signals, that represented by STRs independent from current 90 risk variants,1 and are located nearby TRIML2, NDUFAF2, MIR129-1 and NCOR1 (on chromosomes 4, 5, 7 and 17, respectively). We also assessed the functional consequences of the STRs at a gene expression level in brain tissues, which further supports their candidacy for functional studies to further understand the biological mechanisms behind their associations.
The fact that 88% (30/34) of the associated STRs overlap with the current list of PD GWAS risk variants is not surprising as the STRs were imputed based on their existent LD with SNPs. Known PD loci with STR associations could potentially help to explain the current unknown molecular mechanisms underlying those regions, such as in MAPT and SNCA, where evidence has shown that repetitive elements play a major role in gene expression regulation, splicing and hence protein structure.48,49 This overlap is further reflected in the heritability estimates we obtained which indicated that the contribution of STRs to the genetic variance is largely explained by their high LD with SNPs. However, STRs have shown to increase the contribution to overall SNP-only heritability estimates specifically on gene expression,13,50 where STRs explained between 10–15% of the cis-heritability, thereby supporting our observation that STRs contribute to the heritability of PD.
The eSTR co-localization analyses, using available RNAseq datasets, where we analysed the top 34 STR signals and their surrounding high LD STRs, showed us different distributions of STR associations throughout the various brain regions independently, and at the gene level, we observed significant associations in 19 known risk genes, and specific gene expression of genes being targeted by STRs in PD-relevant tissues such as the substantia nigra. These results suggest that these STRs are likely to be functionally relevant in these loci. Further investigation of the four independent nominated STRs managed to uncover likely functional mechanisms underlying the STR association in genes nearby NDUFAF2 and NCOR1, due to the STR co-localization with regulatory features (epigenetic marks) involved in active transcription. The eSTR near NDUFAF2 was found to significantly increase the expression of PART1 (Fig. 3B). PART1 is a long non-coding RNA that was found to be differentially expressed (downregulated) in a microarray-based analysis of 50 PD patients compared to 22 healthy controls.51 The ZSWIM7 eSTR was associated with significant effects on gene expression in different genes, such as TRPV2, a cation channel part of the Transient Receptor Potential family of proteins (TRPs) that are activated by physical and chemical stimuli,52 and that are known to be involved in the regulation of ionic homeostasis, which is disrupted in PD53; ADORA2B is an adenosine receptor which has been associated with neurodegenerative conditions such as Huntington’s disease,54 however no link to PD has been established to date; lnc-NCOR1-1 and NCOR1 (Nuclear Receptor Corepressor 1) are located within the same chromosomal region (short arm of chromosome 17) and were also influenced by the eSTRs. The former long non-coding gene has not been thoroughly characterized, therefore little is known about its function. The latter encodes a transcriptional inhibitor that has been found to regulate mitochondrial function.55 Moreover, gene expression analyses showed that NCOR1 is significantly upregulated in the substantia nigra of PD patients.56 This evidence suggests that those genes associated with eSTRs in PD would be good candidates for follow-up analyses.
The functional consequences of STRs captured by gene-wise and pathway analyses demonstrated that STRs are enriched in known disease-relevant pathways such as synaptic vesicle trafficking57 and autophagy,58 and in tissues, such as the cortex, cerebellar hemisphere and frontal cortex. Also highlighted is the pituitary gland, that is known to express the dopaminergic receptors D2 and D459 and is part of the hypothalamic–pituitary–thyroid axis, where alterations in its balance has been shown to increase risk to PD.60
This study marks the first (to our knowledge) STR GWAS in PD to date and highlights the importance of incorporating other forms of genetic variation, such as STRs, into routine genetic analyses. Despite this, like with any study profiling repeat-based variants using short-read sequencing data, the analyses presented in this study have several limitations. First, focusing on the STR calls, STRs were imputed using a reference panel that was generated by the STR caller hipSTR using short-read WGS.17 There are two main drawbacks to this approach: (i) hipSTR cannot call STRs that are longer than the read length. Given that many of the known pathogenic STRs in neurological diseases are large repeat expansions, we currently lack the power to detect this important and potentially disease-associated class of tandem repeats. (ii) As highlighted in the original study, imputation accuracy varies widely across STR loci. However, with regard to the latter, in the present study we demonstrated a high concordance between the overlapping hipSTR imputed calls and gangSTR calls in the PPMI cohort, increasing the reliability of the imputed calls. Future studies that validate the PD-associated STRs with methods such as long-read sequencing will be crucial to confirm these loci and key to resolving complex repeat-based PD-associated haplotypes. Second, although the majority of the STRs tested were biallelic, multi-allelic variants were split into biallelic for the GWAS and downstream analyses. This approach enabled us to perform commonly used GWAS methods for the different analysis presented in the study, but set aside the consideration of variant length as unit of analysis. To explore the potential role of variant length, we extracted observed additional alleles for the top 34 genome-wide significant STRs in the meta-analysis. Only one variant, (7:127793488:[T]16[G], near GPNMB) was found to harbour an extra allele that was nominally associated with PD (P = 0.025; Supplementary Table 11), suggesting that the leading variants account for the majority of the observed risk. Finally, it is important to highlight that, despite the fact that the STR panel used to impute our PD GWAS cohorts showed high levels of concordance (96.7%)17 with read-based callers such as hipSTR and TREDPARSE, and that our comparative analysis in the PPMI cohort called with hipSTR and gangSTR showed high genotype concordance, the STRs reported in this study need further experimental validation (which was not possible due to lack of access to DNA samples) to discard any potential artifacts that could exist in both cases and controls and to confirm their association with PD.
Overall, we have performed the first STR GWAS meta-analysis for PD and reported that STRs contribute to its genetic risk. We have characterized another layer of genetic variation, helping us to gain statistical power to nominate novel candidate risk variants and genes and to provide a more complete reference of the genetic variation that contributes to the disease. Hence these data are a valuable resource for researchers currently dissecting known risk loci. Moving forward, a large-scale GWAS which utilizes calls directly from WGS data and validates hits using long-read sequencing methodologies is essential to fully understand the contribution of STRs to the genetics of PD.
Supplementary Material
Acknowledgements
We would like to thank all of the subjects who donated their time and biological samples to be a part of this study. We also would like to thank all members of the IPDGC. For a complete overview of members, acknowledgements and funding, please see http://pdgenetics.org/partners. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). The access to part of the participants for this research has been made possible thanks to the Quebec Parkinson’s Network (http://rpq-qpn.ca/en/).
Contributor Information
Bernabe I Bustos, Ken and Ruth Davee Department of Neurology and Simpson Querrey Center for Neurogenetics, Northwestern University, Feinberg School of Medicine, Chicago, IL 60611, USA.
Kimberley Billingsley, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
Cornelis Blauwendraat, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
J Raphael Gibbs, Computational Biology Group, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
Ziv Gan-Or, The Neuro (Montreal Neurological Institute-Hospital), McGill University, Montréal, QC, Canada; Department of Human Genetics, McGill University, Montréal, QC, Canada; Department of Neurology and neurosurgery, McGill University, Montréal, QC, Canada.
Dimitri Krainc, Ken and Ruth Davee Department of Neurology and Simpson Querrey Center for Neurogenetics, Northwestern University, Feinberg School of Medicine, Chicago, IL 60611, USA.
Andrew B Singleton, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
Steven J Lubbe, Ken and Ruth Davee Department of Neurology and Simpson Querrey Center for Neurogenetics, Northwestern University, Feinberg School of Medicine, Chicago, IL 60611, USA.
Funding
This work was supported in part by the Intramural Research Programs of the National Institute of Neurological Disorders and Stroke (NINDS), the National Institute on Aging (NIA) and the National Institute of Environmental Health Sciences, both part of the National Institutes of Health, Department of Health and Human Services; project numbers 1ZIA-NS003154, Z01-AG000949-02 and Z01-ES101986. In addition, this work was supported by the Department of Defense (award W81XWH-09-2-0128), and the Michael J Fox Foundation for Parkinson’s Research. This work was supported by the Simpson Querrey Center for Neurogenetics (to D.K.)
Competing interests
D.K. is the Founder and Scientific Advisory Board Chair of Lysosomal Therapeutics Inc. and Vanqua Bio. D.K. serves on the scientific advisory boards of The Silverstein Foundation, Intellia Therapeutics, AcureX and Prevail Therapeutics and is a Venture Partner at OrbiMed. Z.G.-O. has received consulting fees from Lysosomal Therapeutics Inc., Idorsia, Prevail Therapeutics, Denali, Ono Therapeutics, Neuron23, Handl Therapeutics, Bial Biotech Inc., Deerfield, Lighthouse and Inception Sciences (now Ventus). The other authors declare that they have no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keller MF, Saad M, Bras J, et al. Using genome-wide complex trait analysis to quantify “missing heritability” in Parkinson’s disease. Hum Mol Genet. 2012;21:4996–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lander ES, Linton LM, Birren B. Correction: initial sequencing and analysis of the human genome. Nature. 2001;412:565–566. [DOI] [PubMed] [Google Scholar]
- 4. Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the sherman paradox. Cell. 1991;67:1047–1058. [DOI] [PubMed] [Google Scholar]
- 6. Verkerk AJMH, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. [DOI] [PubMed] [Google Scholar]
- 7. MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on huntington’s disease chromosomes. Cell. 1993;72:971–983. [DOI] [PubMed] [Google Scholar]
- 8. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orr HT, Chung MY, Banfi S, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. [DOI] [PubMed] [Google Scholar]
- 11. Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet. 2018;19:286–298. [DOI] [PubMed] [Google Scholar]
- 12. Grünewald TGP, Bernard V, Gilardi-Hebenstreit P, et al. Chimeric EWSR1-FLI1 regulates the ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 2015;47:1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gymrek M, Willems T, Guilmatre A, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 2016;48:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Billingsley KJ, Lättekivi F, Planken A, et al. Analysis of repetitive element expression in the blood and skin of patients with Parkinson’s disease identifies differential expression of satellite elements. Sci Rep. 2019;9:4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willems T, Zielinski D, Yuan J, Gordon A, Gymrek M, Erlich Y. Genome-wide profiling of heritable and de novo STR variations. Nat Methods. 2017;14:590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Payseur BA, Place M, Weber JL. Linkage disequilibrium between STRPs and SNPs across the human genome. Am J Hum Genet. 2008;82:1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saini S, Mitra I, Mousavi N, Fotsing SF, Gymrek M. A reference haplotype panel for genome-wide imputation of short tandem repeats. Nat Commun. 2018;9:4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blauwendraat C, Heilbron K, Vallerga CL, et al. Parkinson Disease age of onset GWAS: defining heritability, genetic loci and a-synuclein mechanisms. Mov Disord. 2019;34:866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Browning BL, Zhou Y, Browning SR. A one-penny imputed genome from next-generation reference panels. Am J Hum Genet. 2018;103:338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang H, Kirkness EF, Lippert C, et al. Profiling of short-tandem-repeat disease alleles in 12,632 human whole genomes. Am J Hum Genet. 2017;101: 700- 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan A, Abecasis GR, Kang HM. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pankratz N, Beecham GW, DeStefano AL, et al. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol. 2012;71:370–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H, Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mousavi N, Shleizer-Burko S, Yanicky R, Gymrek M. Profiling the genome-wide landscape of tandem repeat expansions. Nucleic Acids Res. 2019;47:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mousavi N, Margoliash J, Pusarla N, Saini S, Yanicky R, Gymrek M. TRTools: a toolkit for genome-wide analysis of tandem repeats. Bioinformatics. 2021;37:731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wickham H. GGPLOT2: elegant Graphics for Data Analysis 2016. Springer-Verlag; 2016. [Google Scholar]
- 32. Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pruim RJ, Welch RP, Sanna S, et al. Locuszoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. GTEx Consortium . The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hinrichs AS, Karolchik D, Baertsch R, et al. The UCSC genome browser database: update 2006. Nucleic Acids Res. 2006;34: D590- D598, (Database issue):D590-D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao Y, Wang J, Jaehnig EJ, Shi Z, WebGestalt ZB. Webgestalt 2019:gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saini S, Gymrek M; PGC Schizophrenia Working Group . Studying the role of short tandem repeat variants in schizophrenia risk. In: Annual Meeting of The American Society of Human Genetics. 2019; PgmNr 2179/T. [Google Scholar]
- 47. ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Caillet-Boudin ML, Buée L, Sergeant N, Lefebvre B. Regulation of human MAPT gene expression. Mol Neurodegener. 2015;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Afek A, Tagliafierro L, Glenn OC, Lukatsky DB, Gordan R, Chiba-Falek O. Toward deciphering the mechanistic role of variations in the Rep1 repeat site in the transcription regulation of SNCA gene. Neurogenetics. 2018;19:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quilez J, Guilmatre A, Garg P, et al. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 2016;44:3750–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chi LM, Wang LP, Jiao D. Identification of differentially expressed genes and long noncoding RNAs associated with Parkinson’s disease. Parkinson’s Dis. 2019;2019:6078251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duitama M, Vargas-López V, Casas Z, Albarracin SL, Sutachan JJ, Torres YP. TRP Channels role in pain associated with neurodegenerative diseases. Front Neurosci. 2020;14:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vaidya B, Sharma SS. Transient receptor potential channels as an emerging target for the treatment of Parkinson’s disease: an insight into role of pharmacological interventions. Front Cell Dev Biol. 2020;8:584513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu J, Ciarochi J, Calhoun VD, et al. Genetics modulate gray matter variation beyond disease burden in prodromal huntington’s disease. Front Neurol. 2018;9:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fan W, Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Curr Opin Cell Biol. 2015;33:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cowherd M, Lee I. Transcriptional regulators are upregulated in the substantia nigra of Parkinson’s disease patients. J Emerg Invest. 2015:1–7. [Google Scholar]
- 57. Esposito G, Clara FA, Verstreken P. Synaptic vesicle trafficking and Parkinson’s disease. Dev Neurobiol. 2012;72:134–144. [DOI] [PubMed] [Google Scholar]
- 58. Lim GGY, Zhang C, Lim K. Role of autophagy in Parkinson’s Disease. In: Bailly Y, ed. Autophagy - a double-edged sword - cell survival or death? IntechOpen. 2013:353–375. [Google Scholar]
- 59. Jaber M, Robinson SW, Missale C, Caron MG. Dopamine receptors and brain function. Neuropharmacology. 1996;35:1503–1519. [DOI] [PubMed] [Google Scholar]
- 60. Mohammadi S, Dolatshahi M, Rahmani F. Shedding light on thyroid hormone disorders and Parkinson disease pathology: mechanisms and risk factors. J Endocrinol Invest. 2021;44:1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full STR GWAS summary statistics for the 16 datasets meta-analysed are available at https://drive.google.com/file/d/1kD1i6tHdYC5w0xvxWLD4B-bSPqpnwzNV/view?usp=sharing
The STR imputation, study level GWAS and meta-analysis: https://github.com/neurogenetics/PD_STR_imputation. Downstream analyses: https://github.com/bibb/STR_GWAS_downstream_analysis