Summary
Purine and pyrimidine disorders are often difficult to diagnose. Here, we present a 1H-NMR analysis protocol for the quantification of purines and pyrimidines in urine to diagnose associated disorders. We describe steps for pH adjustment, sample preparation, and 1H-NMR analysis and data analysis. The use of 1H-NMR requires a relatively small sample volume (1 mL) and minimal sample preparation. Analysis time produces accurate and reproducible data within 2 h.
Subject areas: Metabolism, Metabolomics, NMR
Graphical abstract
Highlights
-
•
1H-NMR protocol that produces accurate and reproducible data within 2 h
-
•
Identify and quantify up to 18 purines and pyrimidines in urine samples
-
•
Allows rapid diagnosis of associated metabolic disorders
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Purine and pyrimidine disorders are often difficult to diagnose. Here, we present a 1H-NMR analysis protocol for the quantification of purines and pyrimidines in urine to diagnose associated disorders. We describe steps for pH adjustment, sample preparation, and 1H-NMR analysis and data analysis. The use of 1H-NMR requires a relatively small sample volume (1 mL) and minimal sample preparation. Analysis time produces accurate and reproducible data within 2 h.
Before you begin
Deuterated 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropionic acid (TSP) is used as a chemical shift internal reference in 1H-NMR analysis of aqueous solvents. The TSP peak can also be used for relative quantitation to correct for sample loss during preparation and/or as a reference peak to calculate absolute concentration of metabolites since a known amount of TSP is added to the sample.
CRITICAL: TSP is known to bind to proteins, so it is not advised to use TSP as a reference standard for samples that have a high protein content (e.g., plasma, whole blood samples). Urine samples should not have any proteins present if the kidneys’ filtering function is performing properly; however, if the patient has kidney damage and/or failure, then it must be noted and an additional internal standard (e.g., fumaric acid or maleic acid1) should be used for accurate quantification.
Deuterium oxide (D2O), 10% of total volume analyzed, is used as a signal lock during 1H-NMR analysis. The pH of the urine buffer solution is set to precisely 7.40 (NOTE: the pH meter should be calibrated prior to use; details of adjusting pH given under “preparation of urine samples”) and buffered by 1.5 M potassium phosphate (KH2PO4). A pH of 7.40, instead of 7.00, is used in this protocol to allow for the spectroscopic separation of the methyl (CH3) moiety of creatine (3.04 ppm) and creatinine (3.05 ppm) in the 1H-NMR spectra. If the pH of the sample is 7.00 then the methyl moiety of creatine and creatinine overlap in the 1H-NMR spectra. Creatinine is an important metabolite in urine as it can be used as a reference peak to account for dilution differences between urine samples. Lastly, 2% w/v (13 mg in 100 mL) of sodium azide (NaN3) is added to the urine buffer solution to prevent bacterial growth since it is stored at room temperature.
Preparation of urine buffer solution
Timing: 1 h
Details regarding the preparation of the urine buffer solution are described here. The urine buffer solution should be prepared in advance, may be used for multiple 1H-NMR experiments, and can be stored at 293 K for up to one year. Summary given in Table 1.
-
1.Preparation of 1.5 M KH2PO4 solution.
-
a.Add 20.4 g KH2PO4 to 80 mL of D2O.
-
b.Mix solution using sonification until fully dissolved.
-
a.
-
2.Preparation of internal standard solution.
-
a.Dissolve 100 mg of TSP-d4 and 13 mg of NaN3 in 10 mL of D2O.
-
i.Final concentrations of 5.805 mM TSP-d4 and 2% NaN3 in the 100 mL urine buffer solution.
-
i.
-
b.Mix solution using sonification until fully dissolved.
-
a.
-
3.Preparation of urine buffer solution.
-
a.Combine the 1.5 M KH2PO4 solution and the internal standard solution.
-
b.Adjust the pH precisely to 7.40 by adding KOH pellets.
 CRITICAL:1H-NMR analysis is pH-sensitive; hence, it is very important to be very exact with this step. The addition of KOH to water results in an exothermic reaction and pH is temperature-dependent; namely, pH is inversely proportional to the temperature of the solution. Take your time, allow the solution to stabilize (return to 293 K) and re-measure the pH to ensure that it is precisely 7.40.
CRITICAL:1H-NMR analysis is pH-sensitive; hence, it is very important to be very exact with this step. The addition of KOH to water results in an exothermic reaction and pH is temperature-dependent; namely, pH is inversely proportional to the temperature of the solution. Take your time, allow the solution to stabilize (return to 293 K) and re-measure the pH to ensure that it is precisely 7.40. -
c.Transfer the solution to a 100 mL volumetric flask and adjust the volume with D2O.Note: any reagents of similar/higher analytical grade and/or from other suppliers can be substituted. Furthermore, any alternative pieces of equipment (different to what is described in the resources table) may be used, if the equipment is calibrated, stable and reliable. For example, any sonicator or vortexor can be used to mix the solutions, any calibrated pH meter may be used for pH adjustments, and any microcentrifuge that can reach centrifugal speeds of at least 12 000 g may be used.Note: TSP-d4 and D2O are not hazardous substances and contains no components considered to be either persistent, bioaccumulative and toxic (PBT), or very persistent and very bioaccumulative (vPvB) at levels of 0.1% or higher.
 CRITICAL: NaN3 is highly toxic. Wear protective gloves/protective clothing/eye protection/face protection. Use only under a chemical fume hood. Ensure that it does not get in the eyes, on the skin, or on clothing. Avoid dust formation. Do not breathe (dust, vapor, mist, gas). Do not ingest; if swallowed, seek immediate medical assistance. It may cause damage to organs through prolonged or repeated exposure.
CRITICAL: NaN3 is highly toxic. Wear protective gloves/protective clothing/eye protection/face protection. Use only under a chemical fume hood. Ensure that it does not get in the eyes, on the skin, or on clothing. Avoid dust formation. Do not breathe (dust, vapor, mist, gas). Do not ingest; if swallowed, seek immediate medical assistance. It may cause damage to organs through prolonged or repeated exposure. CRITICAL: KOH may cause severe skin burns, eye damage, and respiratory irritation if it comes in contact with skin or eyes, or is inhaled. Wash face, hands, and any exposed skin thoroughly after handling. May be corrosive to metals. Wear protective gloves/protective clothing/eye protection/face protection. Do not eat, drink, or smoke when using this product. Do not breathe dust/fume/gas/mist/vapors/spray. Use only outdoors or in a well-ventilated area. Store only in its original container and keep tightly closed. Store locked up in a well-ventilated place.
CRITICAL: KOH may cause severe skin burns, eye damage, and respiratory irritation if it comes in contact with skin or eyes, or is inhaled. Wash face, hands, and any exposed skin thoroughly after handling. May be corrosive to metals. Wear protective gloves/protective clothing/eye protection/face protection. Do not eat, drink, or smoke when using this product. Do not breathe dust/fume/gas/mist/vapors/spray. Use only outdoors or in a well-ventilated area. Store only in its original container and keep tightly closed. Store locked up in a well-ventilated place. CRITICAL: KH2PO4 may cause eye, skin, and respiratory tract irritation. Keep the container closed when not in use. Store in a tightly closed container. Store in a cool, dry, well-ventilated area away from incompatible substances. Store protected from moisture. This compound is hygroscopic (absorbs moisture from the air) and should be stored under inert gas.
CRITICAL: KH2PO4 may cause eye, skin, and respiratory tract irritation. Keep the container closed when not in use. Store in a tightly closed container. Store in a cool, dry, well-ventilated area away from incompatible substances. Store protected from moisture. This compound is hygroscopic (absorbs moisture from the air) and should be stored under inert gas.
-
a.
Table 1.
Recipe for preparation of urine buffer solution
| Reagent | Final concentration | Volume |
|---|---|---|
| KH2PO4 solution | 1.5 M | 80 mL |
| Internal standard solution | 5.805 mM TSP | 10 mL |
| D2O | 99.9% | 10 mL |
| Total | 100 mL |
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Synthetic Urine | Industrial Analytical | 72124 |
| Chemicals, peptides, and recombinant proteins | ||
| 2′-Deoxyadenosine monhydrate | Sigma | D7500-25MG |
| 2′-Deoxyinosine | Sigma | D5287-100MG |
| 2′-Deoxyguanosine monohydrate | Sigma | D7145-100MG |
| 2′-Deoxyuridine | Sigma | D5412-250MG |
| 3-(Trimethylsilyl)propionic acid-D4 sodium salt (TSP) | Merck | 1.08652 |
| 5-(Hydroxymethyl)uracil | Sigma | 852589 |
| Adenine | Sigma | A8626-1G |
| AICAR | Sigma | A9978-25MG |
| Adenosine | Sigma | A9251-5G |
| Dihydrouracil | Sigma | D7628-5G |
| Guanosine | Sigma | G6752 |
| Hypoxanthine | Sigma | H9377-1G |
| Inosine | Sigma | I4125-1G |
| Potassium phosphate monobasic (KH2PO4) | MilliporeSigma (sigmaaldrich.com) | 60218 |
| Potassium hydroxide (KOH) | MilliporeSigma (sigmaaldrich.com) | 221473-25G |
| Sodium azide (NaN3) | MilliporeSigma (sigmaaldrich.com) | S2002 |
| Thymidine | Sigma | T9250-1G |
| Thymine | Sigma | T0376-5G |
| Uridine | Sigma | U3750-1G |
| Water | MilliporeSigma (sigmaaldrich.com) | W3513 |
| Xanthine | Sigma | X7375-10G |
| Deposited data | ||
| Metabolights | Online database | MTBLS6430 |
| Software and algorithms | ||
| Amix | Bruker | V3.9.15 |
| Topspin | Bruker | V3.6.4 |
| Other | ||
| pH Meter | Sentron SI series | Serial #: 20428 |
| Vortex mixer | LABsmart MX-S | CRS-VOR01 |
| Ascend 500 NMR | Bruker | Serial: H031280B |
| Microcentrifuge 5424 | Eppendorf | Serial: 5424CG648861 |
Step-by-step method details
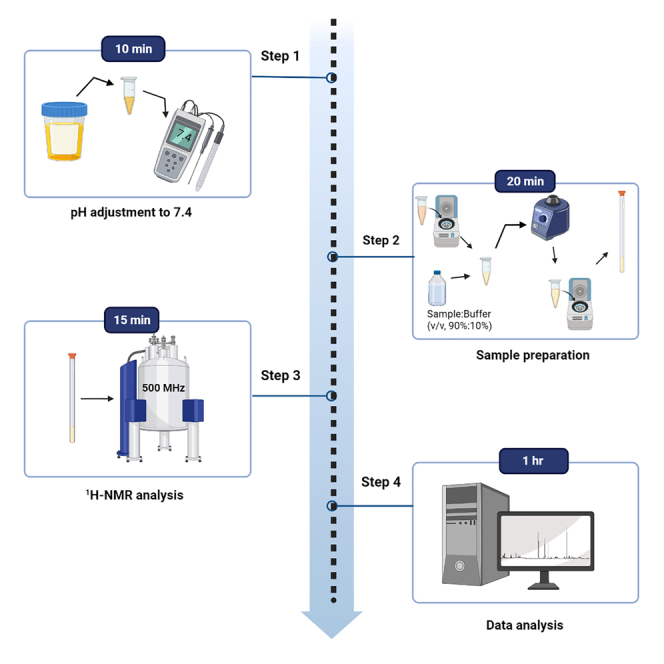
Preparation of urine samples
Timing: 10 min pH adjustment; 20 min sample preparation
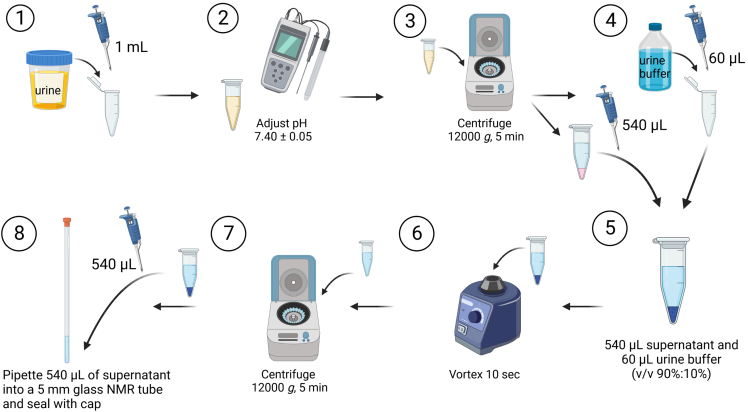
Urine samples are prepared for 1H-NMR analysis. Briefly, 1 mL of urine is adjusted to pH 7.40 ± 0.05, particulates are sedimented by centrifugation, a volume of the supernatant is added in a 90%:10% ratio to the urine buffer solution, an additional centrifugation step is performed, and the supernatant is transferred to a 5 mm glass NMR tube. See Figure 1 for a schematic of the preparation of urine samples.
Note: If proteins/macromolecules are known to be present in the urine sample, then the urine sample must be filtered first using a centrifugal membrane filter of at least 10 kDa before sample preparation.
-
1.
Pipette 1 mL of the urine samples into labeled micro-centrifuge tubes.
-
2.Determine the pH of the urine sample.Note: calibrate pH meter prior to use.
-
a.Adjust the pH dropwise using either 1 N NaOH in ddH20 or 1 N HCl in ddH20 solution until pH is 7.40 ± 0.05.
-
a.
-
3.
Centrifuge the tubes at 12,000 × g for 5 min.
-
4.
Pipette 60 μL urine buffer solution to a second, corresponding labeled micro-centrifuge tube.
-
5.Pipette 540 μL of the supernatant from the first micro-centrifuge tube into the second micro-centrifuge tube.
-
a.Final ratio of urine sample to urine buffer solution should be 90%:10%.
-
a.
CRITICAL: this 90%:10% ratio is important for two reasons. 1) The amount of deuterated component must correlate with the locking step (see step 16) during 1H-NMR analysis. 2) A known amount of TSP (0.5805 mM) must be present in the final sample for determining accurate absolute concentration.
-
6.
Mix the samples via vortex mixing for 10 s.
-
7.
Centrifuge the tubes at 12,000 × g for 5 min.
-
8.
Pipette 540 μL of the supernatant into a 5 mm glass NMR tube and seal with cap.
CRITICAL: Be sure to use clean micro-centrifuge tubes, pipette tips and NMR tubes. Any contamination from cleaning products or biological materials may interfere with your analysis. Do not touch the glass NMR tubes without clean gloves, especially at the bottom of the NMR tube, since this will interfere with the results obtained.
Figure 1.
Schematic of urine sample preparation
A magnetic field surrounds the magnet in all directions. Subsequently no ferromagnetic objects should be allowed near the NMR. NB! People fitted with cardiac pacemakers, hearing aids or metallic implants should not approach the magnet because of possible health risks.
1H-NMR analysis
Timing: 10 min to setup run, 15 min per sample analysis
Loading the NMR glass tube into the NMR spectrometer, checking instrument calibration and 1H-NMR spectral quality via running a test sample, and setting up an automation sequence to run multiple samples.
-
9.
Insert NMR tube into designated 5 mm spinner.
-
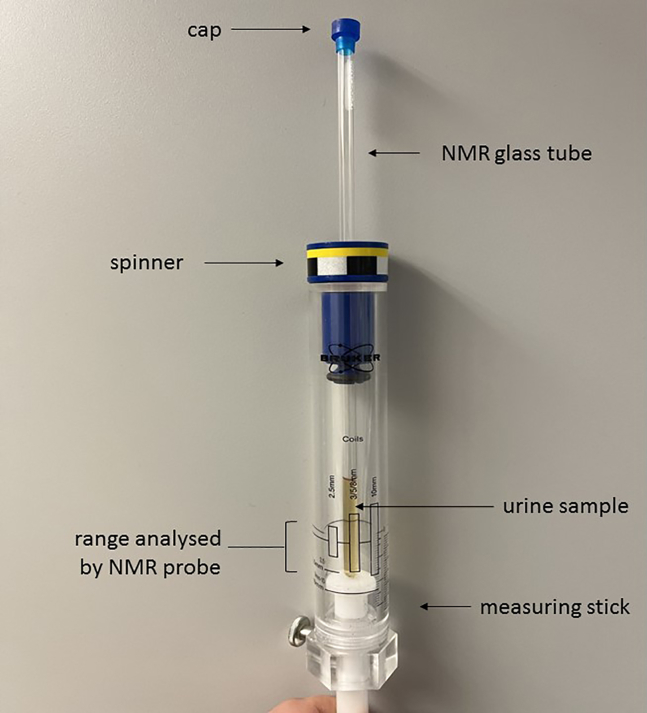
10.
Check that the volume of the sample being analyzed lies within 5 mm probe range, using a measuring stick. See Figure 2 illustrating points 9 & 10.
Note: For our analysis we used a Bruker Avance III HD 500 MHz NMR spectrometer with a 5 mm TXI inverse probe and a SampleXpress autosampler. Software used was Bruker Topspin (V 3.6.4).
-
11.To load successive NMR tubes onto the SampleXpress autosampler:
-
a.Select the “Add” option, then select the “Add from position” option to load NMR tubes from designated position.
-
a.
-
12.
Open the Topspin program, create a new file, and enter the filename and experiment number.
-
13.
For standard 1H-profiling select the protocol “PROF_1H” under experiment.
-
14.
Select the solvent applicable to the sample (90% H2O: 10% D2O) and select “OK”.
-
15.
Load the designated NMR tube into the NMR spectrometer by using the command “sx NUMBER”, where the NUMBER is the position of NMR tube in the SampleXpress autosampler.
-
16.Once the SampleXpress autosampler has loaded the designated NMR tube, proceed to test the instrument calibration by:
-
a.“lock” and select the applicable solvent (90% H2O: 10% D2O) = locks onto deuterium signal in sample.
-
b.“atma” = automatic tuning and matching of probe.
-
c.“topshim” = align magnetic field according to sample matrix. If the test spectrum shows poor resolution (TSP peak width >1 Hz, see point 19 below) then a three-dimensional shim (“topshim 3d”) should be performed to properly align the magnetic field.
-
d.“pulsecal” = calibrate radio frequency pulse.
-
a.
-
17.
Check that the temperate of the probe is stable at 300 K.
-
18.
Run a test spectrum by selecting under acquisition parameters: “DS = 0” and “NS = 4”, where DS and NS refer to dummy scans and number of scans, respectively, and typing “zg” to run (zg = zero go).
-
19.
Process data using “ft” for Fourier transformation and use the graphical user interface (GUI) of Topspin to calibrate TSP peak to 0.00 ppm, and adjust the phase and baseline of the 1H-NMR spectrum.
-
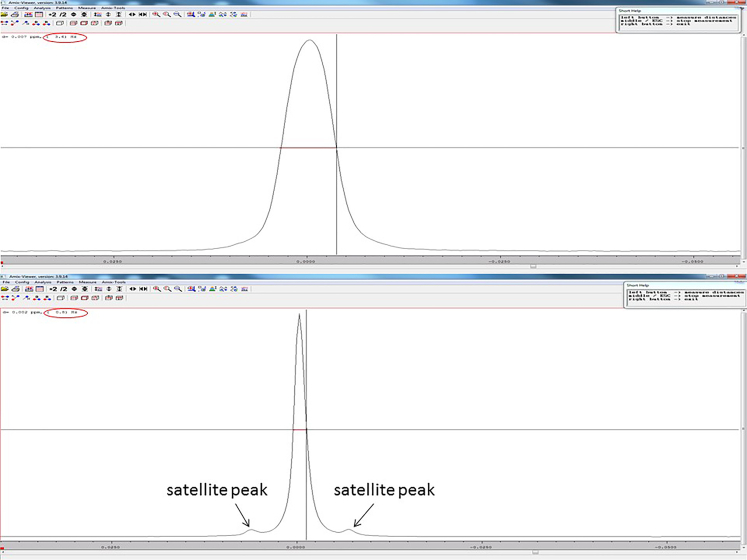
20.Check the quality of the 1H-NMR spectrum by inspecting the TSP peak.Note: The TSP peak should occur as a single peak, with a clearly distinguishable satellite peak on either side of the peak.
-
a.Measure the distance of the width of the TSP peak at half the height of the peak using the “measure distance option” on the GUI of Topspin. A peak width of <1 Hz indicates good shimming (good resolution).Note: Good spectral resolution is necessary for accurate quantification. Figure 3 depicts a good vs poor TSP peak resolution. Troubleshooting.
-
a.
-
21.
Save the shim parameters by command “wsh” and entering designated name.
-
22.
Load sample automation program IconNMR Automation from Topspin (under tab “Acquire”).
-
23.
Select “Configuration” and load saved shim parameters (from point 20) under “probe/shimming options”. Save and exit.
-
24.Select automation and enter:
-
a.Sample name.
-
b.Experiment number.
-
c.Solvent used to lock.
-
d.Experiment “PROF_1H”.
-
a.
-
25.Set desired acquisition parameters.
-
a.TD = 64K.
-
b.DS = 4.
-
c.NS = 128.
-
d.SW = 21 ppm (10.5K Hz).
-
e.AQ = 2.72 s.
-
f.P1 = 8 μs.
-
g.D1 = 4 s.
-
a.
-
26.
Submit all samples to be run as automation and click start.
Note: The automation sequence runs all the calibration steps prior to each sample analysis, thereby ensuring the high repeatability associated with NMR.
Figure 2.
Picture of measuring stick with capped NMR glass tube in spinner, set at the correct measuring range for a 5 mm NMR probe
Figure 3.
Illustration of poor TSP peak resolution (above) that has a broad (3.6 Hz; >1 Hz) peak and TSP satellite peaks are not visible, versus good TSP peak resolution (below) that has a sharp (0.8 Hz; <1 Hz) peak and the TSP satellite peaks are visible
Identification of purines and pyrimidines
A pure compound 1H-NMR spectral library database of purines and pyrimidines was created/used for this protocol. Briefly, a 1 mg/mL solution of each pure compound (see key resources table) in ddH2O was prepared and analyzed individually using the above 1H-NMR protocol. Bruker AMIX (V 3.9.14) was then used to create a purine and pyrimidine spectral library database. The 1H-NMR spectral data for 18 purines and pyrimidines (at pH 7.4; used for this protocol) has been made available: www.ebi.ac.uk/metabolights/MTBLS6430 (Huag et al., 20202). Refer to Table S1 in the supplemental information: Chemical information and shifts for pure compound solutions (1 mg/mL) of purines and pyrimidines used to create a 1H-NMR spectral library, related to the step “identification of purines and pyrimidines”.
Expected outcomes
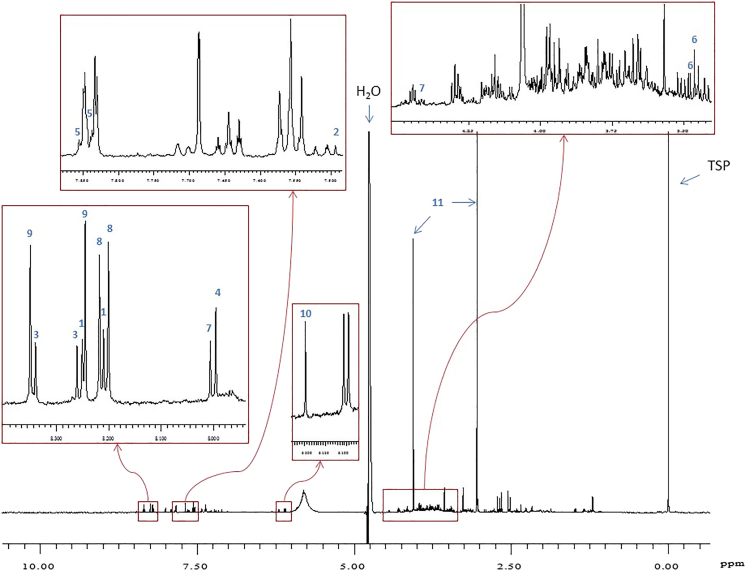
Figure 4 shows an example of a 1H-NMR spectrum of a synthetic urine sample with the presence of purines and pyrimidines. The red boxes in Figure 4 shows zoomed in regions of the 1H-NMR spectrum, illustrating some the peaks that are selected for identification and quantification. These peaks are also highlighted in the chemical shift table (Table S1).
Figure 4.
500 MHz 1H-NMR spectrum of synthetic urine containing purines and pyrimidines
Peak assignments (chemical shifts in ppm): 1 = adenine (8.20 s, 8.23 s); 2 = AICAR (7.50 s); 3 = 2-deoxyadenosine (8.25 s, 8.33 s); 4 = 2-deoxyguanosine (7.99 s); 5 = 2-deoxyuridine (7.83, 7.85 – part of triplet); 6 = dihydrouracil (3.45, 3.46 – part of triplet); 7 = guanosine (8.00 s, 4.41 dd); 8 = hypoxanthine (8.19 s, 8.21 s); 9 = inosine (8.24 s, 8.35 s); 10 = orotic acid (6.20 s); 11 = creatinine (3.05 s, 4.05 s). NOTE: 2-deoxyinosine (8.23 s, 8.33 s), 5-hydroxymethyluracil (4.36 s, 7.60 s), adenosine (8.26 s, 8.35 s), dihydrothymine (1.20 d), thymine (1.87 s), thymidine, (1.90 s), uridine (7.87 d) and xanthine (7.90 s) not present in spectrum.
Quantification and statistical analysis
Using the highlighted chemical shifts for each purine and pyrimidine in Table S1, a pattern file can then be created using Bruker AMIX. This pattern file is used to select peaks and, using the multi integrate tab in Bruker AMIX, export the area under the peaks in an Excel spreadsheet. It is important to note the number of protons per integrated peak because each integral must be proton normalized (i.e., divided by the number of protons represented by the peak, in the Excel spreadsheet) in order for all the integral values to be comparable. Thereafter, concentration (mmol/mol creatinine) per metabolite is calculated in Excel using the following equation:
where is the integral of the metabolite, is the integral of creatinine, and is the concentration of the metabolite.
Alternatively, absolute concentration (micromolar) can be calculated in Excel using the following equation:
where is the integral of the metabolite, is the integral of TSP, is the concentration of TSP in the sample, and is the concentration of the metabolite.
To test the robustness of this protocol a synthetic urine (surine) sample with spiked concentrations of purines and pyrimidines was created. Two experienced analysts (Analysts A & B; >5 years of lab experience) and two inexperienced analysts (Analysts C & D; <2 years of lab experience) were asked to prepare four repeats of the spiked sample based upon this protocol over four days. Table 2 shows the accuracy of the protocol by looking at the recovery percentage across these four analysts. Adenosine, 2-deoxyuridine, dihydrothymidine, hypoxanthine, and thymidine showed an average accuracy of 100 ± 1%. Only 5-hydroxymethyluracil and inosine showed an accuracy outside 100 ± 10%, with the remainder purines and pyrimidines having variation in accuracy of between 1-10%. Figure 5 illustrates the average coefficient of variation (CV, %) of each metabolite for each analyst as an average over four days to represent analyst reproducibility. Analyst 1 and 2 both had excellent reproducibility (1%–2% CV), as expected, while analyst 3 exhibited ∼5% CV values and analyst 4 exhibited ∼8% CV values – both still within good (<10%) reproducibility.
Table 2.
Recovery percentage (%) of purines and pyrimidines, given as average and range, from four inexperienced analysts using this protocol
| Metabolite | Average | Range |
|---|---|---|
| 5-Hydroxymethyluracil | 110.6 | 106–115 |
| Adenine | 96.1 | 96–100 |
| Adenosine | 100.4 | 96–105 |
| AICAR | 107.0 | 102–112 |
| 2-Deoxyadenosine | 106.5 | 101–112 |
| 2-Deoxyguanosine | 90.3 | 86–95 |
| 2-Deoxyinosine | 104.2 | 99–109 |
| 2-Deoxyuridine | 99.4 | 93–104 |
| Dihydrothymine | 99.4 | 95–104 |
| Dihydrouracil | 92.1 | 88–96 |
| Guanosine | 98.7 | 95–103 |
| Hypoxanthine | 99.3 | 95–104 |
| Inosine | 117.5 | 111–125 |
| Orotic acid | 101.6 | 97–107 |
| Thymidine | 100.8 | 97–105 |
| Thymine | 96.7 | 93–101 |
| Uridine | 110.0 | 104–116 |
| Xanthine | 97.8 | 95–100 |
Figure 5.
Analyst reproducibility measured as average coefficient of variation (%) over four days from two experienced analysts (Analysts A & B; >5 years of lab experience) and two inexperienced analysts (Analysts C & D; <2 years of lab experience), using the protocol described here
Limitations
The primary, inherent limitation of 1H-NMR spectroscopy is that if the chemical compound of interest does not have a free hydrogen attached to a carbon, that compound will not be visible on a 1H-NMR spectrum. An example of this is the end product of purine catabolism – uric acid, which is invisible on a 1H-NMR spectrum. An additional limitation of 1H-NMR spectroscopy is that it is not as sensitive as its analytical counterpart – mass spectrometry, and has a lower level of quantification of 1–5 μmol/L.
Significance of this method
Using the protocol described here, a list of 18 purines and pyrimidines and their absolute concentrations (micromolar) and/or relative concentrations (mmol/mol creatinine) can be determined in human urine. The time from initial analysis until final output is less than 2 h, making it a rapid method. As shown, experienced analysts produce highly precise output using this protocol, thereby making the data highly reliable. Hence, a specialized laboratory responsible for diagnosing inborn errors of metabolism would be able to make a rapid and precise decision in the diagnosis of a patient suspected of a purine or pyrimidine disorder.
Troubleshooting
Problem 1
The 1H-NMR spectrum exhibits poor resolution (peak width of TSP >1 Hz) then either the magnet has not been sufficiently shimmed/aligned, or there is interference in the sample. Interference in urine can either be due to high concentration (too much salt/overabundance of metabolites) – a new, diluted sample should be prepared, or there are proteins/macromolecules present.
Potential solution
-
•
Poor shimming – the magnet needs a more thorough shimming (see 16.b.i).
-
•
Interference in urine due to high concentration (too much salt/overabundance of metabolites) – a new, diluted sample should be prepared.
-
•
Proteins/macromolecules are present in sample – a new sample must be filtered first using a centrifugal membrane filter of at least 10 kDa before sample preparation.
Resource availability
Lead contact
Shayne Mason (shayne.mason@nwu.ac.za).
Materials availability
All key resources for this protocol have been made available online: https://star-methods.com/?rid=KRT625be50301998.
Data and code availability
The 1H-NMR spectral data for 18 purines and pyrimidines (at pH 7.4; used for this protocol) has been made available at MetaboLights: www.ebi.ac.uk/metabolights/MTBLS64302; as well as: Mason, Shayne (2023), “P&P_ Chemical shifts and structures”, Mendeley Data, V1, https://doi.org/10.17632/ptbbxtszxm.1.
Acknowledgments
We acknowledge the Centre for Human Metabolomics for allowing us access to core facilities that contributed toward this work.
Author contributions
Both authors contributed equally to this project and writing of this manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102181.
Contributor Information
Elmarie Davoren, Email: elmarie.davoren@nwu.ac.za.
Shayne Mason, Email: shayne.mason@nwu.ac.za.
Supplemental information
”
Multiplicity: s−singlet; d−doublet; t–triplet; dd−double doublet; ddd–double double doublet; dt–double triplet; q–quartet; m–multiplet. Highlighted rows are the preffered peaks used for identification and quantification.
References
- 1.Nagana Gowda G.A., Hong N.N., Raftery D. Evaluation of fumaric acid and maleic acid as internal standards for NMR analysis of protein precipitated plasma, serum, and whole blood. Anal. Chem. 2021;93:3233–3240. doi: 10.1021/acs.analchem.0c04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., O’Donovan C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48:D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
”
Multiplicity: s−singlet; d−doublet; t–triplet; dd−double doublet; ddd–double double doublet; dt–double triplet; q–quartet; m–multiplet. Highlighted rows are the preffered peaks used for identification and quantification.
Data Availability Statement
The 1H-NMR spectral data for 18 purines and pyrimidines (at pH 7.4; used for this protocol) has been made available at MetaboLights: www.ebi.ac.uk/metabolights/MTBLS64302; as well as: Mason, Shayne (2023), “P&P_ Chemical shifts and structures”, Mendeley Data, V1, https://doi.org/10.17632/ptbbxtszxm.1.