Abstract
Simple organisms are often considered to have simple glycomes, but plentiful paucimannosidic and oligomannosidic glycans overshadow the less abundant N-glycans with highly variable core and antennal modifications; Caenorhabditis elegans is no exception. By use of optimized fractionation and assessing wildtype in comparison to mutant strains lacking either the HEX-4 or HEX-5 β-N-acetylgalactosaminidases, we conclude that the model nematode has a total N-glycomic potential of 300 verified isomers. Three pools of glycans were analyzed for each strain: either PNGase F released and eluted from a reversed-phase C18 resin with either water or 15% methanol or PNGase Ar released. While the water-eluted fractions were dominated by typical paucimannosidic and oligomannosidic glycans and the PNGase Ar-released pools by glycans with various core modifications, the methanol-eluted fractions contained a huge range of phosphorylcholine-modified structures with up to three antennae, sometimes with four N-acetylhexosamine residues in series. There were no major differences between the C. elegans wildtype and hex-5 mutant strains, but the hex-4 mutant strains displayed altered sets of methanol-eluted and PNGase Ar-released pools. In keeping with the specificity of HEX-4, there were more glycans capped with N-acetylgalactosamine in the hex-4 mutants, as compared with isomeric chito-oligomer motifs in the wildtype. Considering that fluorescence microscopy showed that a HEX-4::enhanced GFP fusion protein colocalizes with a Golgi tracker, we conclude that HEX-4 plays a significant role in late-stage Golgi processing of N-glycans in C. elegans. Furthermore, finding more “parasite-like” structures in the model worm may facilitate discovery of glycan-processing enzymes occurring in other nematodes.
Keywords: N-glycans, nematode, hexosaminidase, MALDI-TOF MS, phosphorylcholine
As compared with most types of protein glycosylation that are generally based on step-by-step formation of an oligosaccharide chain, N-glycans are based on a build–remove–rebuild principle. Thereby, after the initial transfer of a tetradecasaccharide to a subset of asparagine residues in most eukaryotes, selective removal of glucose and mannose is followed by addition of further monosaccharide units; in multicellular organisms, N-acetylglucosamine, fucose, and galactose are amongst the residues transferred in the Golgi apparatus to the antennal or core regions of the protein-bound N-glycan structure. Especially the addition of N-acetylglucosamine results in GO or NOGO signals for glycan modification events as respectively exemplified by the action of N-acetylglucosaminyltransferases I and III in vertebrates (1).
In many lower multicellular organisms (plants and invertebrates), N-acetylglucosaminyltransferase I (Enzyme Commission [EC] number: 2.4.1.101) is also a key enzyme, but often the transferred residue on the core α1,3-mannose is absent from the mature glycoprotein, despite its necessity for core fucosylation. Thus, the concept of a processing hexosaminidase was introduced, which could account for fucosylated paucimannosidic glycans found in, for example, insects (2). Later, it was shown that the Drosophila melanogaster fused lobes (fdl) gene encodes a Golgi hexosaminidase, which specifically removes the N-acetylglucosamine transferred by N-acetylglucosaminyltransferase I (3).
Fucosylated paucimannosidic glycans also occur in nematode worms, such as Caenorhabditis elegans, whereby any of the core fucose residues can be galactosylated (4). Some core-modifying enzymes, especially the FUT-8 core α1,6-fucosyltransferase (EC no.: 2.4.1.68), have a very definite bias toward substrates with an antennal N-acetylglucosamine residue on the core α1,3-mannose even though it is then absent from the final glycan; however, the FUT-1 core α1,3-fucosyltransferase (EC number: 2.4.1.214) will only act after removal of this N-acetylglucosamine (5). The relevant identified processing hexosaminidases (HEX-2 and HEX-3) are not paralogous to FDL but are members of a different subgroup (“clade B”) of the GH20 family (6). Indicative of their in vivo function, a highly simplified N-glycome results when knocking out both the hex-2 and hex-3 genes in C. elegans—core α1,3-fucose is lacking from the reducing terminus in this mutant (7), whereas removal of the core α1,6-mannose is apparently the signal for transfer of a third core fucose by FUT-6 (8). Thus, the processing for these core-modified fucose-rich glycans in C. elegans is actually rather complicated with multiple build–remove–rebuild steps.
While the core modifications, including the bisecting galactose residue and the fucose on the distal N-acetylglucosamine, in C. elegans are quite well characterized (9), less attention has been given to the antennae, partly in the belief that complex glycans are not well represented in the worm’s N-glycome. Only recently have C. elegans N-glycans been shown to possess LacdiNAc (GalNAcβ1,4GlcNAc) and chitobiose (GlcNAcβ1,4GlcNAc) antennal motifs (10), whereas the nematode BRE-4 enzyme related to mammalian β1,4-galactosyltransferases is a proven β1,4-N-acetylgalactosaminyltransferase (EC number: 2.4.1.244), which can modify N-glycans in vitro to form LacdiNAc motifs (11). On the other hand, in addition to HEX-2 and HEX-3, C. elegans has two other clade B GH20 enzymes designated HEX-4 and HEX-5, both with a distinct preference for N-acetylgalactosamine residues (6). While HEX-4 has optimal activity around pH 6 suggestive of a Golgi localization, HEX-5 is most active under acidic conditions (6). Considering also that nematodes with detectable LacdiNAc expression on glycoproteins, for example, Trichinella spiralis and Trichuris suis (12, 13), have only one clade B GH20 enzyme (unpublished data), we speculated that expression of HEX-4 may correlate with a lack of terminal LacdiNAc on N-glycans.
To explore our hypothesis further, we examined the N-glycomes of hex-4 and hex-5 mutant strains in comparison to the wildtype and reveal that N-acetylgalactosamine-capped N-glycans are more abundant in worms lacking HEX-4. Also, prefractionation of more “hydrophobic” N-glycans in the wildtype aids detection of multiantennary glycans with HexNAc2–4 motifs partly substituted with phosphorylcholine (PC) and/or fucose residues. Thus, we reveal that the N-glycome of C. elegans is even more complex than previously thought and that the Golgi-localized HEX-4 plays a role in determining the modification of N-glycan antennae in this model nematode.
Results
Transcription and deletion of different nematode hexosaminidases
Previously, we used promoter-driven gfp constructs to explore the expression pattern for five C. elegans GH20 hexosaminidase genes: whereas the hex-1, hex-2, hex-3, and hex-5 reporters were expressed throughout the life cycle, the one based on the hex-4 promoter was restricted to the embryonal and L1 stages (6). We have now mined extensive transcriptome data (14, 15), which suggest that hex-2 versus hex-3 and hex-4 versus hex-5 show contrasting expression patterns in up to 10 h embryos; all four are also expressed to varying extents in larvae and young adults but display different tissue expression (Fig. S1). In vitro, HEX-4 and HEX-5 remove terminal GalNAc residues from N-glycan and glycosaminoglycan-type chains (Fig. S2), but unlike HEX-2 and HEX-3 do not remove GlcNAc residues (6, 16). While glycomic analyses showed that HEX-2 and HEX-3 are responsible for the removal of the nonreducing terminal β1,2-linked GlcNAc from N-glycans (7), the in vivo molecular role of HEX-4 and HEX-5 remained unknown.
For the in-depth analysis of N-glycomic impact of deletions in the two β-N-acetylgalactosaminidase genes, we compared two hex-4 strains (cop1517 and cop1518, generated using CRISPR–Cas9 technology) and one hex-5 strain (tm3307, generated by random mutagenesis) with the parent wildtype N2 strain (Fig. S3). None of the hexosaminidase mutants had any obvious growth phenotype. As other glycomutant worm strains can have higher susceptibility or resistance to bacteria or their toxins, growth on Pseudomonas PA14 was tested; however, no major difference for hex-4 and hex-5 strains as compared with wildtype was observed (Fig. S3).
Thereafter, we focused on the biochemical role of HEX-4 and HEX-5 by comparing the molecular glycomic phenotypes of N2, hex-4, and hex-5. Following use of PNGase F and PNGase Ar for N-glycan release in combination with solid phase extraction on C18, fluorescent labeling and HPLC on an RP-amide column, we analyzed three pools of N-glycans (PNGase F-released water-eluted, PNGase F-released methanol eluted, and PNGase Ar-released post PNGase F) for each strain; each HPLC fraction was analyzed by MALDI-TOF mass spectrometry (MS), and the annotations are based on MS/MS before and after chemical or enzymatic treatments (Figure 1, Figure 2, Figure 3, S4–S7, S9 and S10).
Figure 1.
RP-amide chromatograms of PNGase F-released water-eluted N-glycans. Glycans from Caenorhabditis elegans strains (wildtype N2, hex-5/tm3307, hex-4/cop1517, or hex-4/cop1518) released by PNGase F and eluted from C18 solid phase extraction material with water were labeled with 2-aminopyridine. The RP-amide chromatograms, calibrated in terms of glucose units (g.u.), are annotated with glycan structures (in order of abundance in the relevant fraction) according to the Symbol Nomenclature for Glycans (mannose, galactose, or glucose is depicted by green/yellow/blue circles, GalNAc or GlcNAc by yellow/blue squares, fucose by red triangles, phosphorylcholine by PC and methyl by Me). The m/z for the protonated ions as detected by MALDI-TOF MS are also given with the depicted structures based on conclusions derived from elution time, chemical and/or enzymatic digestion, and MS/MS data of which selected examples are shown in Figure 4, Figure 5, Figure 6, Figure 7. As the tm3307 and cop1518 chromatograms are respectively similar to either the wildtype N2 or hex-4 cop1517, these are indicated in the panels by dashed lines. For the full annotations, refer to Figs. S4 and S5. The chromatograms of all four samples are very similar, despite differences in the detected N-glycans between the N2 and hex-4 strains; the major glycans are identical, but the β1,4-GalNAc-capped structures (proven by specific HEX-4 digestion; highlighted with light yellow boxes) are far more abundant in the hex-4 strains. Based on previous studies, it is assumed that the phosphorylcholine is 6-linked to GlcNAc.
Figure 2.
RP-amide chromatograms of PNGase F-released methanol-eluted N-glycans. Glycans from Caenorhabditis elegans strains (wildtype N2, hex-5/tm3307, hex-4/cop1517, or hex-4/cop1518) released by PNGase F and eluted from C18 solid phase extraction material with 15% methanol were labeled with 2-aminopyridine. The RP-amide chromatograms, calibrated in terms of glucose units (g.u.), are annotated with glycan structures (in order of abundance in the relevant fraction) according to the Symbol Nomenclature for Glycans; those in gray are also found in the water-eluted pools. MS/MS data of selected structures are shown in Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9. As the tm3307 and cop1518 chromatograms are respectively similar to the wildtype N2 and cop1517, these are indicated in the panels by dashed lines. For the full annotations, refer to Figs. S6 and S7. The chromatograms of all four samples are very similar, despite differences in the detected N-glycans between the N2 and hex-4 strains. Glycans with multiple antennal phosphorylcholine and/or fucose residues, especially detected in the wildtype, were previously not described for C. elegans. Highlighted are glycans with antennal fucose (pink) and reducing-terminal GalβFuc (blue) epitopes.
Figure 3.
RP-amide chromatograms of PNGase Ar-released water-eluted N-glycans. Glycans from Caenorhabditis elegans strains (wildtype N2, hex-5/tm3307, hex-4/cop1517, or hex-4/cop1518) released by PNGase Ar were labeled with 2-aminopyridine. The RP-amide chromatograms, calibrated in terms of glucose units (g.u.), are annotated with glycan structures (in order of abundance in the relevant fraction) according to the Symbol Nomenclature for Glycans; those in gray are also found in the PNGase F-released pools. The m/z for the protonated ions as detected by MALDI-TOF MS are also given with the depicted structures based on conclusions derived from elution time, chemical and/or enzymatic digestion, and MS/MS data of which selected examples are shown in Figures 10 and 11. As the tm3307 and cop1518 chromatograms are respectively similar to the wildtype N2 and cop1517, these are indicated in the panels by dashed lines. For full annotations, refer to Figs. S9 and S10. The chromatograms surprisingly differ, especially between the N2 and hex-4 strains in terms of the detected N-glycans, with a shift to fewer α-galactosylated and a higher abundance of certain trimannosylated structures (e.g., m/z 1751 at 6 g.u.) in the latter. Highlighted are glycans with reducing-terminal GalαFucα3 (green) and Gal- or Fuc-extended GalβFucα6 epitopes (blue).
Typical oligomannosidic, paucimannosidic, and simple complex N-glycans
When performing MALDI-TOF MS of the N-glycan subpools eluted from C18 with water, the spectra are dominated by compositions of Hex3–9HexNAc2, Hex3HexNAc2Fuc1, and Hex3HexNAc3–4Fuc0–1. Indeed, upon offline LC–MS using an RP-amide column for separation prior to MS (Fig. 1), the major structure in each major peak was observed to have one of these compositions and their retention times in terms of glucose units as well as MS/MS fragmentation patterns were comparable with those from previous studies (17, 18). Furthermore, the “water-eluate” chromatograms for N2, tm3307, cop1517, and cop1518 strains are nearly superimposable; around 100 glycans were detected in each of these analyses, whereby 70 to 80% of the structures found were present in all samples. The chromatogram for the second hex-4 mutant, cop1518, diverged in terms of the abundance of Man8–9GlcNAc2 (5–6 g.u.) as compared with the others, but the major differences between the water-eluted samples were rather in the presence or the absence of PC-modified glycans.
PC-modified N-glycans
Previously, Hanneman et al. (19), Haslam et al. (20), and we (21) reported zwitterionic N-glycans from the N2 strain with compositions of Hex3HexNAc3–5Fuc0–1PC1 (19, 20, 21), whereas we found Hex5HexNAc3Fuc0–1PC1 in the aman-2 mutant, Hex3HexNAc3–6Fuc0–1PC1–2 in the hex-2;hex-3 mutant and some “simple” Hex3–4HexNAc3–4Fuc0–1PC1–2 structures in C18-HPLC fractions of glycomes from double fucosyltransferase mutants (7, 18, 21). On the basis of typical PC-containing MS/MS fragments (m/z 369, 531, 572, etc.), we estimate that about one-half of the structures detected in the water eluates (Fig. 1) and about two-thirds in the methanol eluates (Fig. 2) are modified with one or more zwitterionic PC residues; this bias toward elution from C18 with methanol is compatible with the late elution of PC-modified glycans on traditional reversed-phase material (10, 17). However, subsequent separation on a fused-core column, such as the RP-amide used here, is more effective in enabling detection; generally, an increased number of PC residues leads to early elution on RP-amide, but this effect is counteracted by other structural modifications such as core α1,6-fucosylation with or without galactose. As these glycans ionize especially well in positive mode due to the presence of the choline moiety absent from oligomannosidic and paucimannosidic structures, quantification as compared with nonzwitterionic glycans is not meaningful as the presence of such structures in every RP-amide fraction would lead to an overestimation of their abundance; thus, we only offer an estimate of the overall number of PC-containing glycans.
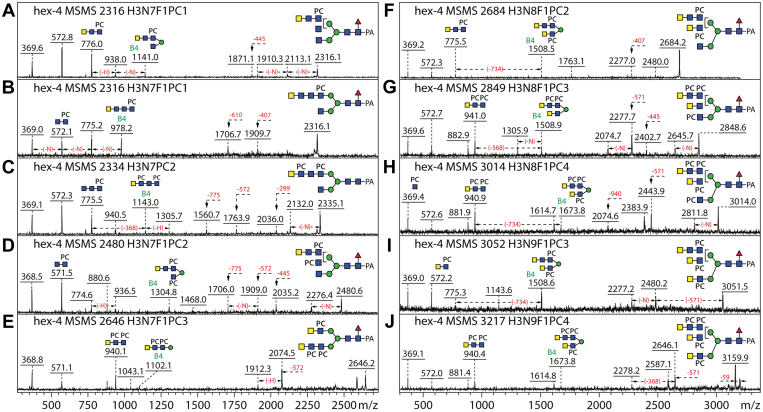
In the current study, other than the simple Hex3HexNAc3–5Fuc0–1PC1, hallmarked by m/z 369 and 531 B-ions (Hex0–1HexNAc1PC1) upon MS/MS, we have found a variety of fragments indicative of longer antennae in the wildtype glycome with one or more PC modifications, for example, m/z 572, 737, 775, 940, 1105, 1143 and 1309 (HexNAc2–4PC1–3; Fig. 4, A–E); where two or more PC residues are present, then a loss of the trimethylamine from the choline moiety is observed (Δm/z 59). The fragments of 899 (Hex1HexNAc2PC2), 1102 (Hex1HexNAc3PC2), 1267 (Hex1HexNAc3PC3), and 1305 (Hex1HexNAc4PC2) are compatible with two antennae attached to the same core α-mannose residue. Some of these fragments are reminiscent of those observed for N-glycans from Oesophagostomum dentatum or Pristionchus pacificus (17, 22), which like C. elegans are also predicted to have the antennal initiation enzymes MGAT-1, MGAT-2, and MGAT-5, but not MGAT-4; thus, the disubstituted mannose is predicted to be the “upper” α1,6-linked one within the context of a triantennary structure (Fig. 4, F–J). Due to the diverging fragmentation of glycans found in different HPLC fractions, numerous isomeric structures could be identified as exemplified by m/z 2112, 2239, 2277, 2442, and 2480 (Figs. 4 and 5). The degree of isomeric diversity is high with, for example, 10 different isomers of Hex3HexNAc6Fuc1PC2 found in the wildtype and hex-4 strains (m/z 2277; Fig. S8).
Figure 4.
MS/MS of phosphorylcholine-modified N-glycans from Caenorhabditis elegans N2 and hex-5 strains.A–J, positive mode MS/MS of selected RP-amide fractions of water- or methanol-eluted glycans (Figs. 1 and 2). Neutral losses of residues or differences in spectra are indicated with dashed lines (F, fucose; H, hexose; N, N-acetylhexosamine; PC, phosphorylcholine); key B-ion fragments are also annotated. The difference of 59 Da is due to loss of choline; brackets indicate antennal ambiguity. The occurrence of multiple PC-modified chito-oligomer antennae on biantennary/triantennary glycans is demonstrated by the B fragment ions such as m/z 775, 940, and 1105 (HexNAc3PC1–3), 899, 1102, or 1305 (Hex1HexNAc2–4PC2), and 1267 (Hex1HexNAc3PC3) or Y ions resulting from the loss thereof. Because of the excellent ionization of the PC-containing fragments, the core Y1 fragments are not observed, rather the loss of 445 (GlcNAc1Fuc1PA) from the parent in the case of core fucosylated glycans. For further illustrations of the multiple isomers of PC-modified N-glycans, refer to Figures 5 and 6 as well as Fig. S8.
Figure 5.
Enzymatic or chemical treatments of phosphorylcholine (PC)-modified N-glycans from the Caenorhabditis elegans N2 strain.A–K, positive mode MS of selected RP-amide fractions of water-eluted glycans (Fig. 1) before and after Streptomyces β1,3/4-N-acetylhexosaminidase (chitinase; loss of 203 Da), jack bean β-N-acetylhexosaminidase (JBHex; loss of 203 Da), hydrofluoric acid (HF; loss of 165 Da), or combined jack bean β-N-acetylhexosaminidase and α-mannosidase (JBHex + JBMan; loss of 365 Da) treatments. L–V, corresponding MS/MS of selected original and product glycans, whereby the m/z 1744 product in panel S is partly derived from release of one PC from a coeluting m/z 1909 glycan. Losses of residues or differences in spectra are indicated with dashed lines (F, fucose; H, hexose; N, N-acetylhexosamine; PC); key B-ion fragments are also annotated. Unsubstituted antennae with a single GlcNAc could be digested with JBHex, whereas unsubstituted terminal HexNAc residues in HexNAc2–3 motifs were sensitive to chitinase but generally not to HEX-4; this indicates that these motifs are most frequently GlcNAc2–3 in the wildtype.
In order to gain more insight into the underlying structures, chemical and enzymatic treatments were performed. To verify the position of PC within the antennae, three types of hexosaminidase were used to ascertain whether nonreducing terminal nonsubstituted HexNAc residues were present: (i) the β1,3/4-specific Streptomyces chitinase, which can remove either N-acetylgalactosamine or N-acetylglucosamine residues (23), (ii) the nonspecific jack bean hexosaminidase, and (iii) the GalNAc-specific C. elegans HEX-4 enzyme (16, 24), which can remove GalNAc from LacdiNAc motifs modified with PC on the subterminal GlcNAc residue (23). In case of wildtype HexNAc2PC1 motifs, the chitinase can remove one HexNAc residue resulting in a shift in the fragments from m/z 572 to m/z 369 (Fig. 5, A, B, L and M), indicative that the innermost GlcNAc of the motif is carrying the zwitterionic modification; in contrast, HEX-4 was generally not effective on wildtype glycans, suggestive that the outermost HexNAc was not GalNAc as on some parasitic glycans (24), but GlcNAc. On the other hand, jack bean hexosaminidase removed unsubstituted antennal GlcNAc residues (Fig. 5, C, D, N and O). As an independent indication for the presence of phosphodiester modifications, we employed treatment with hydrofluoric acid; this resulted in loss of all predicted PC modifications as well as shifts in the MS/MS fragmentation patterns (Fig. 5, E–K and P–V).
N-glycans with terminal N-acetylgalactosamine residues
In contrast to the wildtype or hex-5 glycomes where the vast majority of the PC-modified HexNAc2–4 motifs had N-acetylglucosamine as the terminal residue, the hex-4 mutant strains had a large number of N-acetylgalactosamine-capped glycans. HEX-4 could remove one HexNAc residue from HexNAc2PC1, HexNAc3PC1, and HexNAc3PC2 motifs present on a variety of hybrid, biantennary, and triantennary N-glycans in these strains (Fig. 6, A–H); MS/MS showed shifts in m/z 572, 734, 775, 940, and 1305 B-ion fragments to m/z 369, 531, 572, 737, and 899 (Fig. 6, I–R). These data indicated that the terminal residue was GalNAc, but that the subterminal HexNAc in the case of HexNAc3PC1 was GlcNAc. Further variants of biantennary and triantennary glycans with terminal GalNAc included HexNAc4PC1–2 motifs or structures with α1,6-mannose residues substituted with one “short” and one “long” PC-containing antenna (Fig. 7). Forms of these antennae lacking the zwitterionic modification were extremely rare, but N-glycans with one chitobiosyl and one LacdiNAc (both PC substituted) were also found, whereby these eluted slightly later than the isomers with two PC-substituted LacdiNAc motifs (e.g., m/z 2131 or 2277; Fig. 1). In total, only two GalNAc-capped N-glycans were detected in the N2 and hex-5 strains as compared with over 50 in either hex-4 mutant (Fig. S8).
Figure 6.
Enzymatic treatments of phosphorylcholine (PC)-modified N-glycans from the Caenorhabditis eleganshex-4 (cop1517) strain.A–H, positive mode MS of selected RP-amide fractions of water-eluted glycans (Fig. 1) before and after Caenorhabditis β1,4-N-acetylgalactosaminidase (HEX-4) treatments. I–R, corresponding MS/MS of selected original and product glycans. Losses of residues or differences in spectra are indicated with dashed lines (F, fucose; H, hexose; N, N-acetylhexosamine; PC, phosphorylcholine); key B-ion fragments are also annotated. HEX-4 removal of terminal GalNAc residues from mutant strain glycans resulted in shifts in relevant PC-containing B fragments; note that HEX-4 removed two and not three HexNAc residues from the m/z 2334 glycan, indicative that only the terminal residue of the HexNAc3PC1 motif was GalNAc.
Figure 7.
MS/MS of phosphorylcholine (PC)-modified N-glycans from Caenorhabditis eleganshex-4 strains.A–J, positive-mode MS/MS of selected RP-amide fractions of water-eluted or methanol-eluted glycans (Figs. 1 and 2). Neutral losses of residues or differences in spectra are indicated with dashed lines (F, fucose; H, hexose; N, N-acetylhexosamine; PC); key B-ion fragments are also annotated, and brackets indicate antennal ambiguity. The hex-4 strains were particularly rich in glycans containing both antennal GalNAc and PC residues in the context of LacdiNAc, “LactriNAc,” or “LactetraNAc” motifs (GalNAcβ1,4GlcNAc1–3). Terminal HexNAc antennae were defined on the basis of HEX-4 and chitinase sensitivity.
N-glycans with antennal fucose residues
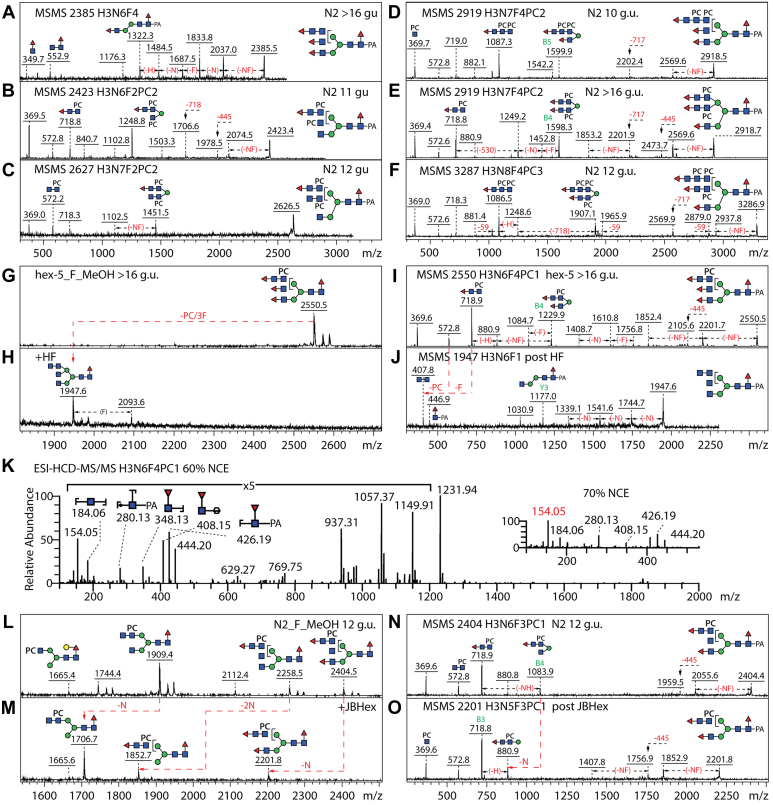
Another surprise in the methanol-eluted subpools was the presence of complex glycans of between 2200 and 3300 Da with multiple fucose residues. Previously, C. elegans glycans with more than one fucose residue were generally shown to be of the paucimannosidic type, some carrying up to five fucose and five galactose residues associated with the mannosylchitobiosyl core (4). In the present study, particularly the wildtype N2 and hex-5 strains contained glycans with compositions such as Hex3HexNAc6Fuc4 (m/z 2385, >16 g.u.). Here, m/z 349 and 552 B-ion HexNAc1–2Fuc1 fragments were found (Fig. 8A), indicative that the outermost HexNAc carried a fucose residue. Variants of these multiply fucosylated structures also carried PC residues as indicated by the B-ions such as m/z 718 or 1087 (HexNAc2–3PC1–2Fuc1; Fig. 8, B–F). Hydrofluoric acid treatment, known to effectively cleave both PC and α1,3/4-fucose residues, altered the MS and MS/MS spectra as shown by the loss of the m/z 718 fragments and the appearance of one at m/z 407 (Fig. 8, G–J). As some structures were initially considered to have three antennae (e.g., m/z 2404), one each of HexNAc1, HexNAc1Fuc1, and HexNAc2PC1Fuc1, they were treated with jack bean hexosaminidase, which removed only a single HexNAc residue, resulting in a shift in the B-ion fragment indicative of disubstitution of the α1,6-mannose (m/z 1083; Hex1HexNAc3PC1Fuc1) to one at m/z 880 (Hex1HexNAc2PC1Fuc1; Fig. 8, L–O). Whereas 20 glycans with antennal fucose were found in the wildtype strain, only five were detected in the hex-4 mutant.
Figure 8.
Analyses of Caenorhabditis elegans wildtype N-glycans with antennal fucose residues.A–F, MS/MS of selected methanol-eluted glycans with antennal fucose residues. Selected B-ion fragments are annotated. G–J, MS and MS/MS of the >16 g.u. fraction containing the Hex3HexNAc6Fuc4PC1 glycan with m/z 2550 before and after hydrofluoric acid treatment; the shift in the B-ion fragmentation pattern is indicated, whereas the Y1 and Y3 fragments are obvious only after treatment. K, higher-energy collisional dissociation (HCD)–MS/MS spectrum of the Hex3HexNAc6Fuc4PC1 glycan (m/z 1320.49 as [M-2H]2− in negative-ion mode) with 60% NCE and 70% NCE (inset). These fragment ions were derived from either nonreducing or reducing end and are annotated with cartoon structures. When NCE increased over 70%, Z/Z-CH2O fragment ions at m/z 154 (red) were dominant in the MS/MS spectrum. L–O, MS and MS/MS of the 12 g.u. fraction containing a glycan with m/z 2404 before and after jack bean hexosaminidase treatment; the shift in the B-ion fragmentation pattern indicates a loss of a single N-acetylglucosamine residue. Glycans containing both antennal fucose and phosphorylcholine residues have previously not been described in C. elegans; selected glycans were subject to hydrofluoric acid treatment, but underlying terminal HexNAc residues were only sensitive to chitinase. MS, mass spectrometry; NCE, normalized collision energy.
For further information regarding the multiply fucosylated glycans, higher-energy collisional dissociation (HCD)-MS/MS with stepped normalized collision energy (NCE) was employed for analysis of the m/z 2550 glycan (Fig. 8I). When NCE was over 60%, intense fragment ions in the low-mass range were observed (<m/z 500; Fig. 8K). These fragment ions were derived from either nonreducing or reducing end. Fragment ions at m/z 348 and 408 indicate two types of fucosylation, fucosylation of HexNAc and core fucosylation, respectively. Fragment ions at m/z 408 were annotated as 1,3A cleavage of a mannose (usually the core α1,3-mannose), which suggested that one fucosylated motif was most likely Fuc-GlcNAcβ1,2Manα1,3. Fragment ions at m/z 184 and 154 were assigned as Z/Z cleavage of internal GlcNAc and Z/Z-CH2O. When NCE was increased over 70%, the dominant fragment ions were those at m/z 154 (Fig. 8K, inset), which are possibly generated by double bonds forming between C4 and C5, resulting in loss of the C6 of the GlcNAc when this is either C3 or C4 substituted (25). The HCD data are thereby consistent with the conclusions based on MALDI-TOF MS/MS before and after hydrofluoric acid treatment. Therefore, we assume, also because of the substitution of C2 of a HexNAc by an N-acetyl moiety, that the antennal fucose residues are either α1,3 or α1,4 linked to GlcNAc and not α1,2 or α1,6 linked; in contrast to T. suis and two lepidopteran species (12, 23), the antennal fucose was not observed to cosubstitute a PC-modified GlcNAc.
Galactosylated N-glycans
Various nematodes express N-glycans decorated with galactose residues, and C. elegans has a rich repertoire of these, although none are related to the LacNAc antennal modifications found in vertebrates. Bisecting β-galactose, β-galactose attached to the core α1,6-fucose, α-galactose attached to the α1,3-mannose, and α-galactose modifications of core α1,3-fucose have been reported by us (4, 7, 18, 26, 27). Some of these were detected in the water-eluted and methanol-eluted PNGase F-released glycan pools, including different isomers of m/z 1297 and 1459 (Hex4–5HexNAc2Fuc1) with either fucosylated bisecting galactose or galactosylated core α1,6-fucose—the contrasting retention times (on a classical C18 column) and fragmentation patterns of these isomers have been previously reported (18).
Enzymatic assays have shown that galactosylation of the core α1,6-fucose by the GALT-1 to form the “GalFuc” epitope is optimal when there is also a nonreducing terminal N-acetylglucosamine on the α1,3-mannose residue (28), thus it is not surprising that some GalFuc-containing glycans retain this residue; indeed we have previously detected Hex4HexNAc3–4Fuc1 (m/z 1500 and 1703) also modified with PC (m/z 1665 and 1868) in double fucosyltransferase mutants. In the current study, we show that the repertoire of glycans carrying both GalFuc and PC is indeed greater, including biantennary and hybrid structures with LacdiNAc or chitobiose-type antennae or even triantennary structures in either wildtype, hex-4, or hex-5 mutant strains (Fig. 9). As the antennal PC-containing B-fragments dominate the MS/MS spectra (e.g., HexNAc1PC, 369; HexNAc2PC1, 572; HexNAc2PC2, 737), the core GalFuc motif is only obvious by the neutral loss of fucose and hexose (−308 Da) or Hex1HexNAc1Fuc1-PA (−607 Da). Only when the glycans are treated with hydrofluoric acid is the signature reducing terminal GalFuc Y-ion m/z 608 revealed, as shown by the biantennary isomer of m/z 2033 and its digestion product (Fig. 9, G and H).
Figure 9.
Analyses of Caenorhabditis elegans N-glycans with core galactosylated fucose motifs.A–D, MS/MS of selected N2, hex-4, or hex-5 glycans carrying both “GalFuc” and phosphorylcholine epitopes. Selected B-ion fragments are annotated, and the difference of 59 Da is due to loss of choline; the loss of 607 Da from the parent is indicative of the presence of the galactosylated core fucose motif. E–H, MS and MS/MS of the 9 g.u. fraction containing a glycan with m/z 2033 before and after hydrofluoric acid (HF) treatment; the low-intensity m/z 369 and 531 B-ion fragments (H) derive from a cofragmenting residual phosphorylcholine-modified m/z 1706 structure, whereas the Y1 fragment at m/z 608, obvious only after HF treatment, is indicative for galactosylated core fucose (7, 27). MS, mass spectrometry.
N-glycans with core α1,3-fucose
Those N-glycans not released by PNGase F were then isolated after digestion of residual glycopeptides with PNGase Ar. A variable amount of noncore-α1,3-fucosylated glycans were found in these pools because of incomplete PNGase F digestion in the first round of enzymatic release, but the vast majority of the glycans in these pools contained core α1,3-fucose (Figs. 3 and S9 and S10). Here, the simplest of the modifications of the proximal core GlcNAc is double fucosylation as defined by the Y1 fragment of m/z 592 (HexNAc1Fuc2-PA; Figs. 10A and 11B). Some of the glycans differ only in the presence or the absence of a methyl modification (e.g., m/z 1735 and 1749, 1897 and 1911, 2059, 2073, 2219 and 2233; Figs. 10, A/B, D/E, G/H, and 11, B/C, D/E, I/J). Other glycans are isomeric varying in the position of a galactose residue (e.g., two wildtype versions of m/z 2235 or two hex-4 versions of m/z 1913); here, the pattern of neutral losses and the intensity of characteristic Y1 fragments (m/z 754, 770, and 916; Hex1–2HexNAc1Fuc1–2-PA; Figs. 10, I/J and 11, G/H) as well as digest information from previous studies make it possible to distinguish these structures. As judged by retention time and MS/MS, the range of postulated core modifications such as α-galactosylated α1,3-fucose on the distal and proximal N-acetylglucosamine residues of the chitobiosyl core, β-galactosylated core α1,6-fucose, α1,2-fucosylated bisecting β-galactose, or α-galactosylation of the core α1,3-mannose is similar to those found in our recent studies on C. elegans N-glycans (4, 7, 10, 18, 26). While the chromatograms for the N2 and hex-5 strains are rather similar (Fig. 3) other than in terms of relative intensity of some peaks, the highest peak for the hex-4 strains is not at 6.5 g.u. and the proportion of glycans with three or more fucose residues and/or α-galactose on the α1,3-mannose is reduced, whereas those with an α1,6-mannose are increased (Fig. S11A and Table S2).
Figure 10.
MS/MS of core α1,3-fucosylated N-glycans from wildtype Caenorhabditis elegans.A–J, positive mode MS/MS of selected PNGase Ar-released glycans (Fig. 3); key Y fragments as well as neutral losses are indicated as are differences between nonmethylated and methylated N-glycans (A/B, D/E, and G/H). Differences in the occurrence or intensity of Y-ions are indicative of the various core modifications with fucose and galactose residues, also for isomeric glycans eluting at different retention times (I/J). Isomers were also distinguished on the basis of α-galactosidase, β-galactosidase, α-mannosidase, and/or hydrofluoric acid (HF) treatments and in comparison to previously published data on core modifications of C. elegans N-glycans (4, 18). The maximal detected degree of core modification in the wildtype (Hex7HexNAc2Fuc4Me1, see I) reflects also α-galactosylation of the core α1,3-mannose and core α1,3-fucose residues, which only occur to a low extent in the hex-4 mutant.
Figure 11.
MS/MS of core α1,3-fucosylated N-glycans from the Caenorhabditis eleganshex-4 strain.A–J, positive mode MS/MS of selected PNGase Ar-released glycans (Fig. 3); key Y fragments as well as neutral losses are indicated as are differences between nonmethylated and methylated N-glycans (B/C, D/E, and I/J). Differences in the occurrence or intensity of Y-ions are indicative of the various core modifications with fucose and galactose residues, also for isomeric glycans eluting at different retention times (G/H). Isomers were also distinguished on the basis of glycosidase or HF treatments and in comparison to previously published data on core modifications of C. elegans N-glycans (4). HF, hydrofluoric acid; MS, mass spectrometry.
Mucin-type O-glycans
To assess whether there were major changes in the mucin-type O-glycome, we subjected worm lysates to β-elimination. The two major masses observed in both the wildtype and hex-4 mutant (Hex3–4HexNAc1; Fig. S11, B–E) correspond to those detected in permethylated form in other studies (29), whereas the pentasaccharide was also analyzed by NMR (30); we conclude that ablation of the hex-4 gene does not cause a major shift in the O-glycome.
Localization of HEX-4
Considering our data indicating that HEX-4 plays a role in N-glycan maturation, the C. elegans hex-4 gene was edited to contain an in-frame enhanced GFP (eGFP)-coding sequence. The resulting fusion protein was expressed in larval stages as well as young adults of C. elegans (Figs. 12A and S12). Punctate HEX-4::eGFP signals were observed, coincident with those for the Golgi stain BODIPY TR Ceramide (31); as compared with studies on C. elegans AMAN-2 (32), this is indicative of Golgi localization of HEX-4.
Figure 12.
Localization of HEX-4 and summary of unusual N-glycan structures found in Caenorhabditis elegans wildtype and hex-4 mutant strains.A, fluorescence microscopy of L1 larvae at two levels of magnification showing perinuclear colocalization of the HEX-4::enhanced GFP (eGFP) fusion protein (green) with the BODIPY TR Ceramide Golgi marker (gold); for further images, refer to Fig. S12. B, example of N-glycans, together with m/z values, particularly found in either the wildtype N2 or hex-4 mutant strains representative of the diversity of most highly processed motifs in C. elegans are depicted (see also Fig. S8 for a depiction of multiple isomers of m/z 2277). Striking is the tendency to GalNAc capping and simpler core modifications if the hex-4 gene is ablated. C, the shift in the antennal modifications can be explained if there is GalNAc capping (potentially via the BRE-4 β4-N-acetylgalactosaminyltransferase) in near equilibrium with GalNAc removal (via HEX-4) in compartments of the Golgi apparatus. The relative lack of GalNAc capping in the wildtype or hex-5 strains reveals GlcNAc residues, which are available for extension by chito-oligomer synthesizing/modifying enzymes. On the other hand, in both wildtype and hex-4 strains, HEX-2 and HEX-3 mediate removal of the GlcNAc attached to the “lower arm” α1,3-linked mannose, resulting in paucimannosidic and pseudohybrid structures, which can be core α1,3-fucosylated. Only indirect effects can explain the relative lack of tetrafucosylated glycans in the hex-4 strains. See Figure 1, Figure 2, Figure 3 for summaries of all N-glycans found in this study, including the highly abundant oligomannosidic and paucimannosidic structures.
Discussion
Despite some 2 decades of analyses by various research groups, the N-glycome of the model nematode C. elegans continues to surprise, and the role of its many glycosylation-relevant genes is still unclear. In this study, we compared N-glycans of the wildtype N2 strain with those of three hexosaminidase mutants, two with a deletion in the hex-4 gene and one with a deletion in hex-5 gene. While both HEX-4 and HEX-5 utilize substrates with β-linked N-acetylgalactosamine, they have contrasting pH optima (pH 6.5 and 4.5, respectively) suggestive of a different intracellular localization (6); as the luminal pH Golgi cisternae has been estimated to be in the pH range of 6.2 to 6.7 as compared with a lysosomal pH of 4.8 (33, 34, 35), we supposed that HEX-4 is a Golgi enzyme, whereas HEX-5 may have a typical lysosomal degradative function. Indeed, our data indicate that worms lacking HEX-4 have an altered set of N-glycans enriched in structures with LacdiNAc motifs and fewer with longer antennae and/or antennal fucose, whereas the N-glycome of the hex-5 strain is almost identical to the wildtype. In addition, fluorescence microscopy indicates colocalization of a HEX-4::eGFP fusion protein with a Golgi tracking probe (Fig. 12A).
Our glycomic analyses also reveal structures with up to three antennae and four PC residues in the wildtype N2 strain (see also summary in Fig. 12B); thus, the PC-modified N-glycome repertoire of the model nematode is actually akin to the complexity of some parasitic species. Chito-oligomers decorated with PC and/or fucose are known in filarial worms such as Onchocerca volvulus or Dirofilaria immitis (24, 36), whereas antennal fucosylation of terminal GlcNAc is found in a cestode, Taenia crassiceps (37). The maximal detected length of the antennae was a series of four N-acetylhexosamine residues in the wildtype, which is the same as that found in the pig parasite O. dentatum (22), but less than the possibly six or more in filarial nematodes (36), including D. immitis (24). In the latter, we could find GalNAcβ1,4GlcNAcβ1,4GlcNAc “LactriNAc” motifs, detected here also in the hex-4 mutants, in addition to “uncapped” longer chains consisting of serial N-acetylglucosamine residues. Indeed, in these studies, the specific HEX-4 enzyme has proven a valuable tool to verify the absence or the presence of nonreducing terminal N-acetylgalactosamine (24). The relative lack of terminal GalNAc in N2 would correlate with the inability of GalNAc to inhibit binding of wildtype worm proteins to Wisteria floribunda agglutinin, in contrast to other tested helminths (38). However, even in the hex-4 mutants, there is no fucosylated LacdiNAc; thereby, C. elegans lacks a motif found in T. suis, D. immitis, Haemonchus contortus, and Schistosoma mansoni (12, 24, 39, 40). Such similarities and differences between C. elegans and parasitic nematodes do offer perspectives for the discovery of enzymes making novel epitopes relevant to the design of diagnostics or vaccines.
In terms of the genetic basis for the occurrence of LacdiNAc and fucosylated chito-oligomer modifications, C. elegans has a proven GT7 β1,4-N-acetylgalactosaminyltransferase, BRE-4 which also has a role in glycolipid biosynthesis (41); this enzyme can also generate poly-LacdiNAc chains when overexpressed in mammalian cells (42), whereas the enzymatic function of a GT7 paralog, NGAT-1, remains to be proven (43). C. elegans also has five GT10 α1,3-fucosyltransferases (8, 44); however, while FUT-1 and FUT-6 actually modify C. elegans N-glycan cores, in vivo glycan targets remain to be identified for FUT-3, FUT-4, and FUT-5. The accurate prediction of glycosyltransferase and glycosidases homologs in other nematodes is more difficult because of incomplete genome annotations. However, there are multiple clade B hexosaminidase genes including potential HEX-4 ortholog in O. dentatum, which has chito-oligomer but not LacdiNAc modifications (22), in D. immitis, which has both types of antennae (24), and in P. pacificus, which has only minute amounts of complex glycans with more than one HexNAc per antenna (17); on the other hand, T. suis has LacdiNAc-modified N-glycans (12) but lacks HEX-4. All these species possess BRE-4 orthologs.
Considering both the glycomic data on nematodes and their glycogenomic potential, we speculate that BRE-4 transfers N-acetylgalactosamine to nonreducing termini of N-glycans in C. elegans and that HEX-4 can remove this residue (Fig. 12C); in the absence of the glycosidase, these transfer-removal steps do not take place, which would correlate with the similar tissue-specific transcriptomic expression of hex-4 and bre-4 (Fig. S1). A Golgi localization of HEX-4 would be compatible with a role in biosynthesis, rather than degradation, as the number of N-glycans of over 2500 Da is greater in the wildtype than the hex-4 mutant. Thereby, it can be hypothesized that N-acetylgalactosamine may act as a cap preventing the formation of longer chito-oligomer chains (HexNAc4 or higher) type and limits the antennal length to HexNAc2–3 motifs in hex-4 mutants. An alternative cap in C. elegans would be fucose, as found especially in relatively late-eluting N-glycans of the wildtype (Figs. 2 and 8). Such a concept of capping units controlling the biosynthesis of oligosaccharide chains is known in many species. In bacteria, methylphosphate or N-acetylmuramic acid is chain termination motif (45, 46), whereas in plants, developmentally relevant Golgi β1,3-galactosidases seemingly remove previously transferred galactose during arabinogalactan synthesis (47); other termination signals may include sulphation of the terminal sialic acid of polysialylated glycolipids in sea urchins (48) and sialylation of mammalian keratan sulphate (49). However, in terms of hexosaminidase-mediated alteration of protein-linked N-glycans in mammals, it appears that “acidic” glycosidases mediate removal of nonreducing terminal GlcNAc in compartments such as azurophilic vesicles of neutrophils rather than via processes within the Golgi (50).
A surprising result of our study is that the “core chitobiose modifications” (see also summary in Fig. 12B) are also affected in both hex-4 strains resulting in a large shift in the N-glycome released by PNGase Ar. This subpool is enriched in N-glycans modified with bisecting galactose and/or multiple fucose residues and represents structures unique to C. elegans (4); for biosynthetic reasons related to the specificity of the FUT-1 core α1,3-fucosyltransferase (5), glycans with LacdiNAc- or chito-oligomer motifs are not expected to be present in this pool. Thus, little impact if HEX-4 is absent was expected. However, it seems that a processing step (removal of the core α1,6-mannose) required for transfer of fucose by FUT-6 (8) to the distal GlcNAc may be reduced in the hex-4 mutant. Fewer core α1,3-fucosylated glycans, including those also modified by bisecting galactose, are α-galactosylated as compared with the wildtype, and no pentafucosylated glycans are observed in the cop1517 strain (Table S2). Such a shift to a lower degree of core fucosylation in hex-4 strains offers a parallel to the respective 30% and 90% reduction in the abundance of trifucosylated and tetrafucosylated glycans observed for the cogc-1 mutant, which has defective Golgi function (51). Interestingly, the hex-4 gene neighbors trpp-10, which encodes an ortholog of mammalian TRPPC10, a protein with a role in Golgi trafficking (52); therefore, it is possible that the precise deletion of hex-4 affects expression of the downstream trpp-10. Alternatively, knocking out a glycoenzyme can alter the Golgi per se; for instance, an absence of GlcNAc-TV is associated with a reduction in volume of this organelle in mammalian cells (53). Certainly, it can be stated that knocking out one glycosylation-relevant gene can have both explicable and less explicable effects on the glycome.
Experimental procedures
Nematode strains
Wildtype C. elegans (N2) was obtained from the Caenorhabditis Genetics Centre, University of Minnesota. The hex-4 (cop1517 and cop1518; prepared by CRISPR–Cas9 technology) and hex-5 (tm3307; prepared by trimethylpsoralen-induced mutagenesis) strains were respectively obtained from Knudra Transgenics, and the National Bioresource Project for the Experimental Animal Nematode C. elegans, Tokyo Women’s Medical University. The homozygous cop1517 and cop1518 strains were constructed based on the use of two sgRNAs targeting the entire hex-4 open reading frame on chromosome I (aagatactctacatggacgg and cacacactctgacatcacgt corresponding to nucleotides 12,592–12,573 and 20,352–20,333 of cosmid Y51F10). The homozygous tm3307 strain has a 1047 bp deletion resulting in loss of four exons in the middle of the hex-5 gene on chromosome X (specifically 2276–3322 of cosmid Y70D2A). The hex-4::gfp transgenic line (strain PHX5035; allele syb5035) was generated using CRISPR–Cas9 technology by SunyBiotech; the natural stop codon was replaced by a gfp sequence, and the localization of the resulting HEX-4::eGFP fusion protein was observed by confocal microscopy.
Confocal microscopy
N2 wildtype and hex-4::egfp worms were harvested from nematode growth media plates and washed in M9 buffer prior to 1 h incubation with BODIPY TR Ceramide complexed to bovine serum albumin (5 μM in M9 buffer) at room temperature. After washing in M9 buffer (three times), worms were paralyzed using 20 mM levamisole hydrochloride and transferred to poly-l-lysine–coated glass slides. Images were recorded using a Leica TCS SP5 laser scanning confocal microscope (Wetzlar).
Transcriptomic reanalyses
RNA-Seq and sci-RNA-Seq data used for stage and tissue expression analysis of selected genes (14, 54) were downloaded from GExplore on an open access basis and visualized using the plot method of the R graphics library (stage-specific data) and the ggplot function of the tidyverse library (L2 tissue-specific data).
Glycosidase assays
Recombinant HEX-4 and HEX-5 were produced in Pichia pastoris as previously described. The His-tagged version of HEX-4 was purified by nickel chelation chromatography, whereas the untagged HEX-5 was enriched by ammonium sulphate precipitation from culture supernatants (6, 16). The remodeled fibrin dabsylated glycopeptide (two LacdiNAc antennae) and pyridylaminated glycans from either royal jelly (hybrid N-glycan with a single LacdiNAc) or C. elegans (glycosaminoglycan type) were isolated during previous studies (16, 55, 56). Aliquots of purified HEX-4 or enriched HEX-5 were incubated overnight at 37 °C with the glycopeptide or glycans at either pH 4.5 or 6.5 (ammonium acetate) prior to MALDI-TOF-MS as described later.
N-glycan preparation
C. elegans were grown in liquid culture with Escherichia coli OP50 in standard S complete medium, mixed stages were harvested after cultivation at 20 °C (160 rpm) for 4 to 6 days, and purified by sucrose density centrifugation (57, 58). Harvested worms (2 g) were boiled, homogenized; after adjusting the pH with ammonium carbonate buffer to 8.2, CaCl2 was added to a final concentration of 0.5 mM prior to addition of 2 mg thermolysin (Promega). Proteolysis was allowed to proceed for 2 h at 70 °C.
We adapted our glycomic workflow to facilitate the detection of low abundance glycans, especially considering that glycans carrying PC residues can be enriched by reversed phase-based solid phase extraction; previously, we found that glycans with multiple PC residues elute late on standard reversed-phase HPLC, but early if using fused core columns (17). Thus, similarly as for glycans from a marine snail (59), prefractionation on standard C18 material was performed by serially eluting with water and 15% methanol, but HPLC separation after fluorescent labeling with 2-aminopyridine was done on a fused core RP-amide column.
Specifically, the glycopeptides were purified using standard laboratory protocols (60) prior to serial release using two different peptide:N-glycosidases: recombinant bacterial PNGase F (from Flavobacterium [Elizabethkingia] meningosepticum, Roche; at pH 8.0) and recombinant rice PNGase Ar (from Oryza sativa, expressed in Pichia pastoris [Komagataella phaffii] and Endo H treated, New England Biolabs; at pH 5.0). First, the glycopeptides were treated with PNGase F overnight at 37 °C. After cation exchange chromatography on Dowex, the unbound material was subject to solid phase extraction on nonporous graphitised carbon (ENVIcarb, Supelco) and eluted with 40% acetonitrile. These fractions were subject to a further solid phase extraction step on a C18 reversed phase resin (LiChroprep; Merck), and the glycans were eluted with water and with stepwise increases in the methanol concentration (15%, 30%, and 100% [v/v]). Glycan-containing fractions, as judged by MALDI-TOF MS, were fluorescently labeled with 2-aminopyridine (61). The remaining glycopeptides bound to the Dowex resin were gel filtrated (Sephadex G25) prior to incubation with PNGase Ar overnight at 37 °C. The glycans released with this enzyme were then no longer bound by Dowex and were subject to the same purification and fluorescent-labeling steps as for the PNGase F-released ones. See also the glycomic workflow summarized in the scheme in the supporting information.
N-glycan fractionation
Pyridylaminated N-glycome pools were fractionated by reversed-phase HPLC (Ascentis Express RP-amide; 150 × 4.6 mm, 2.7 μm; Sigma–Aldrich) and a gradient of 30% (v/v) methanol (buffer B) in 100 mM ammonium acetate, pH 4 (buffer A) was applied at a flow rate of 0.8 ml/min as follows: 0 to 4 min, 0% B; 4 to 14 min, 0 to 5% B; 14 to 24 min, 5 to 15% B; 24 to 34 min, 15 to 35% B; 34 to 35 min, return to starting conditions (62). Lyophilized HPLC fractions were dissolved in water and subject to MALDI-TOF MS. The RP-HPLC column was calibrated daily in terms of glucose units using a pyridylaminated dextran hydrolyzate, and the degree of polymerization of single standards was verified by MALDI-TOF MS (63); the masses of selected peaks of this linear glucose polymer were verified by MALDI-TOF MS after RP-HPLC (e.g., 6. g.u. eluting at 11 min; m/z 1069.6 as [M + H]+).
MALDI-TOF MS
Free glycans and pyridylaminated glycans were analyzed in positive ion mode using a Bruker Autoflex Speed instrument (1000 Hz Smartbeam-II laser) and 6-aza-2-thiothymine as matrix; calibration was performed using a Bruker peptide standard. MS/MS was performed by laser-induced dissociation (precursor ion selector was generally ±0.6%). The detector voltage was normally set at 1977 V for MS and 2133 V for MS/MS; 1000 to 2000 shots from different regions of the sample spots were summed. Spectra were processed with the manufacturer’s software (Bruker Flexanalysis 3.3.80) using the SNAP algorithm with a signal/noise threshold of 6 for MS (unsmoothed) and 3 for MS/MS (four-times smoothed). Glycan MS and MS/MS spectra (approximately 5500 in total) were manually interpreted on the basis of the masses of the predicted component monosaccharides, the differences of mass in glycan series, fragmentation patterns, and results of enzymatic and chemical treatments. For the 300 structures (see Table S1 for a list of compositions), the minimum criterion for inclusion in the Supporting Tables was an interpretable MALDI-TOF MS/MS spectrum (see also mzxml raw data files). Furthermore, examples for each core and antennal motif were verified by digestion data; comparison was also made to elution, in terms of glucose units, with previous data. Calculated theoretical masses were verified using GlycoWorkbench 2.0. The deviation between calculated and observed m/z values was typically 0.1 to 0.2 Da.
Exoglycosidase and hydrofluoric acid treatment
Aliquots of the isolated HPLC fractions were, based on the results of HPLC elution and MALDI-TOF MS and MS/MS data, subject to targeted exoglycosidase digestion and chemical treatment (62). Either α-mannosidase (jack bean from Sigma), α-galactosidase (coffee bean from Sigma), β-galactosidase (recombinant Aspergillus niger LacA prepared in house (64)), or β-hexosaminidases (recombinant C. elegans HEX-4 prepared in-house (16), Streptomyces plicatus chitinase from New England Biolabs or jack bean hexosaminidase from Sigma) was used for further treatment of the sample in 25 mM ammonium acetate, pH 5 (pH 6.5 in the case of HEX-4), at 37 °C for 24 h. For removal of PC or α1,3-fucose residues, selected fractions were dried and incubated for 48 h at 0 °C with 3 μl 48% (v/v) hydrofluoric acid prior to evaporation in a centrifugal concentrator. The samples were diluted in water and re-evaporated, before redissolving once again. The chemically or enzymatically treated fractions were subject to MALDI-TOF MS and MS/MS (as aforementioned) without further purification.
LC–HCD MS/MS analysis
An HPLC-purified glycan (2549 Da) was analyzed using an Easy-nLC 1200 LC system (Thermo Fisher Scientific) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). The analytes were trapped on a 2 cm × 100 μm Acclaim PepMap C18 precolumn (particle size 5 μm; Thermo Fisher Scientific) and separated on a 30 cm × 75 μm analytical column packed in-house with 3 μm Reprosil-Pur C18 material (Dr Maisch) at 300 nl/min flow using a stepwise elution profile (solvent A was 5 mM di-n-butylamine and 8 mM acetic acid in water, solvent B was 70% methanol in solvent A): B buffer increases 20% to 70% in 32 min; and then to 100% in 3 min and keep it for another 10 min (65). The column oven temperature was 50 °C. Nano-Flex ion source (Thermo Fisher Scientific) was operated in negative ionization mode at 1.8 kV with the ion transfer capillary temperature 325 °C. The LTQ Orbitrap Elite mass spectrometer was operated in the data-dependent mode, collecting a full MS scan in the range m/z 350 to 2000 at 120 K resolution. The samples were analyzed at 50, 60, and 70% stepped NCE for five most intensive precursors from each full MS scan. For all settings, the automatic gain control target in the full MS spectra was 106, precursor isolation window was 5 Da, and the HCD spectra were recorded at 15 K resolution with the first m/z 100 and an automatic gain control target of 105; precursor ions with unassigned charged states were rejected.
O-glycan analysis
O-glycans were released from C. elegans N2 or hex-4 glycoproteins (extracted by methanol–chloroform precipitation of fresh lysates) via ammonium-based β-elimination (66) resulting in nonreduced O-glycans. In brief, extracted glycoproteins were dried and incubated with a mixture of 16 μl H2O, 16 μl hydroxylamine, and 32 μl 1,8-diazabicyclo[5.4.0]undec-7-ene (an organic superbase) at 50 °C for 30 min. Released O-glycans were purified using serial small columns of Dowex AG50, C18, and nonporous graphitized carbon prior to labeling with 2-aminopyridine and MALDI-TOF MS/MS as described for N-glycans.
Data availability
Data described in the article are shown in the figures; mzxml files have been submitted to Glycopost: https://glycopost.glycosmos.org/entry/GPST000200.
Supporting information
This article contains supporting information including further information regarding the glycomic analyses, Supplementary Figures S1–S12, and Supplementary Tables S1-S2 (14, 15, 29, 30, 54, 55, 56).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Niclas Karlsson, Carina Sihlbom, and Egor Vorontsov from the BioMS national node at the Sahlgrenska Academy, University of Gothenburg, for support and access to the LTQ Orbitrap Elite mass spectrometer used for HCD MS/MS analysis as well as Hinrich Schulenberg, University of Kiel, for hosting S.Y. to investigate the toxicity of bacteria on glycomutant strains and the BOKU Imaging Centre for access to the confocal microscope.
Author contributions
S. Y., J. V., and E. A. methodology; S. Y. and Z. D. formal analysis; K. P. and C. J. data curation; K. P. and I. B. H. W. writing–original draft; K. P., F. W., D. M., and I. B. H. W. visualization; K. P. and I. B. H. W. supervision.
Funding and additional information
This work was supported by the Austrian Science Fund (FWF; grants P32572 to K. P., P30021 to S. Y., P35516 to J. V., and P29466 to I. B. H. W.); K. P., S. Y., and J. V. are FWF Fellows. Z. D. is a student within the FWF-funded BioTOP doctoral programme W1224.
Reviewed by members of the JBC Editorial Board. Edited by Robert Haltiwanger
Footnotes
Present address for Daniel Malzl: CeMM Research Center for Molecular Medicine, 1090 Wien, Austria.
Supporting information
References
- 1.Schachter H. Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem. Cell Biol. 1986;64:163–181. doi: 10.1139/o86-026. [DOI] [PubMed] [Google Scholar]
- 2.Altmann F., Schwihla H., Staudacher E., Glössl J., März L. Insect cells contain an unusual, membrane-bound β-N-acetylglucosaminidase probably involved in the processing of protein N-glycans. J. Biol. Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 3.Léonard R., Rendić D., Rabouille C., Wilson I.B.H., Préat T., Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J. Biol. Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- 4.Yan S., Vanbeselaere J., Jin C., Blaukopf M., Wols F., Wilson I.B.H., et al. Core richness of N-glycans of Caenorhabditis elegans: a case study on chemical and enzymatic release. Anal. Chem. 2018;90:928–935. doi: 10.1021/acs.analchem.7b03898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paschinger K., Rendić D., Lochnit G., Jantsch V., Wilson I.B.H. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J. Biol. Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- 6.Gutternigg M., Kretschmer-Lubich D., Paschinger K., Rendić D., Hader J., Geier P., et al. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants and insects. J. Biol. Chem. 2007;282:27825–27840. doi: 10.1074/jbc.M704235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan S., Bleuler-Martinez S., Plaza Gutierrez D.F., Künzler M., Aebi M., Joachim A., et al. Galactosylated fucose epitopes in nematodes: Increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J. Biol. Chem. 2012;287:28276–28290. doi: 10.1074/jbc.M112.353128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan S., Serna S., Reichardt N.C., Paschinger K., Wilson I.B.H. Array-assisted characterization of a fucosyltransferase required for the biosynthesis of complex core modifications of nematode N-glycans. J. Biol. Chem. 2013;288:21015–21028. doi: 10.1074/jbc.M113.479147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paschinger K., Yan S., Wilson I.B.H. N-Glycomic complexity in anatomical simplicity: Caenorhabditis elegans as a non-model nematode? Front. Mol. Biosci. 2019;6:9. doi: 10.3389/fmolb.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson I.B.H., Yan S., Jin C., Dutkiewicz Z., Rendić D., Palmberger D., et al. Increasing complexity of the N-glycome during Caenorhabditis development. Mol. Cell Proteomics. 2023;22 doi: 10.1016/j.mcpro.2023.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawar Z., van Die I., Cummings R.D. Molecular cloning and enzymatic characterisation of a UDP-GalNAc:GlcNAcβ-R β1,4-N-acetylgalactosaminyltransferase from Caenorhabditis elegans. J. Biol. Chem. 2002;277:34924–34932. doi: 10.1074/jbc.M206112200. [DOI] [PubMed] [Google Scholar]
- 12.Wilson I.B.H., Paschinger K. Sweet secrets of a therapeutic worm: mass spectrometric N-glycomic analysis of Trichuris suis. Anal. Bioanal. Chem. 2016;408:461–471. doi: 10.1007/s00216-015-9154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morelle W., Haslam S.M., Olivier V., Appleton J.A., Morris H.R., Dell A. Phosphorylcholine-containing N-glycans of Trichinella spiralis: identification of multiantennary LacdiNAc structures. Glycobiology. 2000;10:941–950. doi: 10.1093/glycob/10.9.941. [DOI] [PubMed] [Google Scholar]
- 14.Boeck M.E., Huynh C., Gevirtzman L., Thompson O.A., Wang G., Kasper D.M., et al. The time-resolved transcriptome of C. elegans. Genome Res. 2016;26:1441–1450. doi: 10.1101/gr.202663.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutter H., Suh J. GExplore 1.4: an expanded web interface for queries on Caenorhabditis elegansprotein and gene function. Worm. 2016;5 doi: 10.1080/21624054.2016.1234659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragosits M., Yan S., Razzazi-Fazeli E., Wilson I.B.H., Rendić D. Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology. 2015;25:448–464. doi: 10.1093/glycob/cwu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan S., Wilson I.B.H., Paschinger K. Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis. 2015;36:1314–1329. doi: 10.1002/elps.201400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S., Jin C., Wilson I.B.H., Paschinger K. Comparisons of Caenorhabditis fucosyltransferase mutants reveal a multiplicity of isomeric N-glycan structures. J. Proteome Res. 2015;14:5291–5305. doi: 10.1021/acs.jproteome.5b00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanneman A.J., Rosa J.C., Ashline D., Reinhold V. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 2006;16:874–890. doi: 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- 20.Haslam S.M., Gems D., Morris H.R., Dell A. The glycomes of Caenorhabditis elegans and other model organisms. Biochem. Soc. Symp. 2002;69:117–134. [PubMed] [Google Scholar]
- 21.Paschinger K., Hackl M., Gutternigg M., Kretschmer-Lubich D., Stemmer U., Jantsch V., et al. A deletion in the Golgi α-mannosidase II gene of Caenorhabditis elegans results in unexpected non-wild type N-glycan structures. J. Biol. Chem. 2006;281:28265–28277. doi: 10.1074/jbc.M602878200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez-Castells C., Vanbeselaere J., Kohlhuber S., Ruttkowski B., Joachim A., Paschinger K. Gender and developmental specific N-glycomes of the porcine parasite Oesophagostomum dentatum. Biochim. Biophys. Acta. 2017;1861:418–430. doi: 10.1016/j.bbagen.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanton R., Hykollari A., Eckmair B., Malzl D., Dragosits M., Palmberger D., et al. The underestimated N-glycomes of lepidopteran species. Biochim. Biophys. Acta. 2017;1861:699–714. doi: 10.1016/j.bbagen.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martini F., Eckmair B., Neupert C., Štefanić S., Jin C., Garg M., et al. Highly modified and immunoactive N-glycans of the canine heartworm. Nat. Commun. 2019;10:75. doi: 10.1038/s41467-018-07948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlsson N.G., Schulz B.L., Packer N.H. Structural determination of neutral O-linked oligosaccharide alditols by negative ion LC-electrospray-MSn. J. Am. Soc. Mass Spectrom. 2004;15:659–672. doi: 10.1016/j.jasms.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Yan S., Brecker L., Jin C., Titz A., Dragosits M., Karlsson N., et al. Bisecting galactose as a feature of N-glycans of wild-type and mutant Caenorhabditis elegans. Mol. Cell Proteomics. 2015;14:2111–2125. doi: 10.1074/mcp.M115.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butschi A., Titz A., Wälti M., Olieric V., Paschinger K., Nöbauer K., et al. Caenorhabditis elegans N-glycan core β-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Titz A., Butschi A., Henrissat B., Fan Y.Y., Hennet T., Razzazi-Fazeli E., et al. Molecular basis for galactosylation of core fucose residues in invertebrates: identification of Caenorhabditis elegans N-glycan core α1,6-fucoside β1,4-galactosyltransferase GALT-1 as a member of a novel glycosyltransferase family. J. Biol. Chem. 2009;284:36223–36233. doi: 10.1074/jbc.M109.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlschlager T., Butschi A., Grassi P., Sutov G., Gauss R., Hauck D., et al. Methylated glycans as conserved targets of animal and fungal innate defense. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2787–2796. doi: 10.1073/pnas.1401176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guérardel Y., Balanzino L., Maes E., Leroy Y., Coddeville B., Oriol R., et al. The nematode Caenorhabditis elegans synthesises unusual O-linked glycans: identification of glucose-substituted mucin-type O-glycans and short chondroitin-like oligosaccharides. Biochem. J. 2001;357:167–182. doi: 10.1042/0264-6021:3570167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagano R.E., Martin O.C., Kang H.C., Haugland R.P. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J. Cell Biol. 1991;113:1267–1279. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas B.J., Wight I.E., Chou W.Y.Y., Moreno M., Dawson Z., Homayouni A., et al. CemOrange2 fusions facilitate multifluorophore subcellular imaging in C. elegans. PLoS One. 2019;14 doi: 10.1371/journal.pone.0214257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M.M., Grabe M., Adams S., Tsien R.Y., Moore H.P., Machen T.E. Mechanisms of pH regulation in the regulated secretory pathway. J. Biol. Chem. 2001;276:33027–33035. doi: 10.1074/jbc.M103917200. [DOI] [PubMed] [Google Scholar]
- 34.Seksek O., Biwersi J., Verkman A.S. Direct measurement of trans-Golgi pH in living cells and regulation by second messengers. J. Biol. Chem. 1995;270:4967–4970. doi: 10.1074/jbc.270.10.4967. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi S., Kagami Y., Hanaoka K., Terai T., Komatsu T., Ueno T., et al. Development of a series of practical fluorescent chemical tools to measure pH values in living samples. J. Am. Chem. Soc. 2018;140:5925–5933. doi: 10.1021/jacs.8b00277. [DOI] [PubMed] [Google Scholar]
- 36.Haslam S.M., Houston K.M., Harnett W., Reason A.J., Morris H.R., Dell A. Structural studies of N-glycans of filarial parasites. Conservation of phosphorylcholine-substituted glycans among species and discovery of novel chito-oligomers. J. Biol. Chem. 1999;274:20953–20960. doi: 10.1074/jbc.274.30.20953. [DOI] [PubMed] [Google Scholar]
- 37.Lee J.J., Dissanayake S., Panico M., Morris H.R., Dell A., Haslam S.M. Mass spectrometric characterisation of Taenia crassiceps metacestode N-glycans. Mol. Biochem. Parasitol. 2005;143:245–249. doi: 10.1016/j.molbiopara.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Nyame A.K., DeBose-Boyd R., Long T.D., Tsang V.C.W., Cummings R.D. Expression of Lex antigen in Schistosoma japonicum and S. haematobium and immune responses to Lex in infected animals: Lack of Lex expression in other trematodes and nematodes. Glycobiology. 1998;8:615–624. doi: 10.1093/glycob/8.6.615. [DOI] [PubMed] [Google Scholar]
- 39.Paschinger K., Wilson I.B.H. Two types of galactosylated fucose motifs are present on N-glycans of Haemonchus contortus. Glycobiology. 2015;25:585–590. doi: 10.1093/glycob/cwv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuhrer M., Koeleman C.A., Deelder A.M., Hokke C.H. Repeats of LacdiNAc and fucosylated LacdiNAc on N-glycans of the human parasite Schistosoma mansoni. FEBS J. 2006;273:347–361. doi: 10.1111/j.1742-4658.2005.05068.x. [DOI] [PubMed] [Google Scholar]
- 41.Griffitts J.S., Haslam S.M., Yang T., Garczynski S.F., Mulloy B., Morris H., et al. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 2005;307:922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 42.Kawar Z.S., Haslam S.M., Morris H.R., Dell A., Cummings R.D. Novel poly-GalNAcβ1-4GlcNAc (LacdiNAc) and fucosylated poly-LacdiNAc N-glycans from mammalian cells expressing β1,4-N-acetylgalactosaminyltransferase and α1,3-fucosyltransferase. J. Biol. Chem. 2005;280:12810–12819. doi: 10.1074/jbc.M414273200. [DOI] [PubMed] [Google Scholar]
- 43.Veyhl J., Dunn R.J., Johnston W.L., Bennett A., Zhang L.W., Dennis J.W., et al. The directed migration of gonadal distal tip cells in Caenorhabditis elegans requires NGAT-1, a β1,4-N-acetylgalactosaminyltransferase enzyme. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen K., van Die I., Grundahl K.M., Kawar Z.S., Cummings R.D. Molecular cloning and characterization of the Caenorhabditis elegans α1,3-fucosyltransferase family. Glycobiology. 2007;17:586–599. doi: 10.1093/glycob/cwm023. [DOI] [PubMed] [Google Scholar]
- 45.Mann E., Kelly S.D., Al-Abdul-Wahid M.S., Clarke B.R., Ovchinnikova O.G., Liu B., et al. Substrate recognition by a carbohydrate-binding module in the prototypical ABC transporter for lipopolysaccharide O-antigen from Escherichia coli O9a. J. Biol. Chem. 2019;294:14978–14990. doi: 10.1074/jbc.RA119.010323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kählig H., Kolarich D., Zayni S., Scheberl A., Kosma P., Schäffer C., et al. N-acetylmuramic acid as capping element of α-D-fucose-containing S-layer glycoprotein glycans from Geobacillus tepidamans GS5-97T. J. Biol. Chem. 2005;280:20292–20299. doi: 10.1074/jbc.M501724200. [DOI] [PubMed] [Google Scholar]
- 47.Nibbering P., Petersen B.L., Motawia M.S., Jorgensen B., Ulvskov P., Niittyla T. Golgi-localized exo-β1,3-galactosidases involved in cell expansion and root growth in Arabidopsis. J. Biol. Chem. 2020;295:10581–10592. doi: 10.1074/jbc.RA120.013878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitazume S., Kitajima K., Inoue S., Haslam S.M., Morris H.R., Dell A., et al. The occurrence of novel 9-O-sulfated N-glycolylneuraminic acid-capped α2→5-Oglycolyl-linked oligo/polyNeu5Gc chains in sea urchin egg cell surface glycoprotein. Identification of a new chain termination signal for polysialyltransferase. J. Biol. Chem. 1996;271:6694–6701. doi: 10.1074/jbc.271.12.6694. [DOI] [PubMed] [Google Scholar]
- 49.Brown G.M., Huckerby T.N., Abram B.L., Nieduszynski I.A. Characterisation of a non-reducing terminal fragment from bovine articular cartilage keratan sulphates containing α(2–3)-linked sialic acid and α(1–3)-linked fucose. A sulphated variant of the VIM-2 epitope. Biochem. J. 1996;319:137–141. doi: 10.1042/bj3190137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjondro H.C., Loke I., Chatterjee S., Thaysen-Andersen M. Human protein paucimannosylation: cues from the eukaryotic kingdoms. Biol. Rev. Camb Philos. Soc. 2019;94:2068–2100. doi: 10.1111/brv.12548. [DOI] [PubMed] [Google Scholar]
- 51.Struwe W.B., Reinhold V.N. The conserved oligomeric Golgi (COG) complex is required for fucosylation of N-glycans in C. elegans. Glycobiology. 2012;22:863–875. doi: 10.1093/glycob/cws053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rawlins L.E., Almousa H., Khan S., Collins S.C., Milev M.P., Leslie J., et al. Biallelic variants in TRAPPC10 cause a microcephalic TRAPPopathy disorder in humans and mice. Plos Genet. 2022;18 doi: 10.1371/journal.pgen.1010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong Z., Zuber C., Pierce M., Stanley P., Roth J. Reduction in Golgi apparatus dimension in the absence of a residential protein, N-acetylglucosaminyltransferase V. Histochem. Cell Biol. 2014;141:153–164. doi: 10.1007/s00418-013-1146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hykollari A., Malzl D., Eckmair B., Vanbeselaere J., Scheidl P., Jin C., et al. Isomeric separation and recognition of anionic and zwitterionic N-glycans from royal jelly glycoproteins. Mol. Cell Proteomics. 2018;17:2177–2196. doi: 10.1074/mcp.RA117.000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanbeselaere J., Yan S., Joachim A., Paschinger K., Wilson I.B.H. The parasitic nematode Oesophagostomum dentatum synthesizes unusual glycosaminoglycan-like O-glycans. Glycobiology. 2018;28:474–481. doi: 10.1093/glycob/cwy045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis J.A., Fleming J.T. Basic culture methods. Met. Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 59.Eckmair B., Jin C., Abed-Navandi D., Paschinger K. Multi-step fractionation and mass spectrometry reveals zwitterionic and anionic modifications of the N- and O-glycans of a marine snail. Mol. Cell Proteomics. 2016;15:573–597. doi: 10.1074/mcp.M115.051573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paschinger K., Hykollari A., Razzazi-Fazeli E., Greenwell P., Leitsch D., Walochnik J., et al. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology. 2012;22:300–313. doi: 10.1093/glycob/cwr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hase S., Ibuki T., Ikenaka T. Reexamination of the pyridylamination used for fluorescence labelling of oligosaccharides and its application to glycoproteins. J. Biochem. (Tokyo) 1984;95:197–203. doi: 10.1093/oxfordjournals.jbchem.a134585. [DOI] [PubMed] [Google Scholar]
- 62.Hykollari A., Paschinger K., Eckmair B., Wilson I.B.H. Analysis of invertebrate and protist N-glycans. Met. Mol. Biol. 2017;1503:167–184. doi: 10.1007/978-1-4939-6493-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomiya N., Kurono M., Ishihara H., Tejima S., Endo S., Arata Y., et al. Structural analysis of N-linked oligosaccharides by a combination of glycopeptidase, exoglycosidases, and high-performance liquid chromatography. Anal. Biochem. 1987;163:489–499. doi: 10.1016/0003-2697(87)90253-3. [DOI] [PubMed] [Google Scholar]
- 64.Dragosits M., Pflugl S., Kurz S., Razzazi-Fazeli E., Wilson I.B.H., Rendić D. Recombinant Aspergillus β-galactosidases as a robust glycomic and biotechnological tool. Appl. Microbiol. Biotechnol. 2014;98:3553–3567. doi: 10.1007/s00253-013-5192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Persson A., Vorontsov E., Larson G., Nilsson J. Glycosaminoglycan domain mapping of cellular chondroitin/dermatan sulfates. Sci. Rep. 2020;10:3506. doi: 10.1038/s41598-020-60526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kameyama A., Thet Tin W.W., Toyoda M., Sakaguchi M. A practical method of liberating O-linked glycans from glycoproteins using hydroxylamine and an organic superbase. Biochem. Biophys. Res. Commun. 2019;513:186–192. doi: 10.1016/j.bbrc.2019.03.144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the article are shown in the figures; mzxml files have been submitted to Glycopost: https://glycopost.glycosmos.org/entry/GPST000200.