Summary
Background
The Omicron era of the COVID-19 pandemic commenced at the beginning of 2022 and whilst it started with primarily BA.1, it was latter dominated by BA.2 and the related sub-lineage BA.5. Following resolution of the global BA.5 wave, a diverse grouping of Omicron sub-lineages emerged derived from BA.2, BA.5 and recombinants thereof. Whilst emerging from distinct lineages, all shared similar changes in the Spike glycoprotein affording them an outgrowth advantage through evasion of neutralising antibodies.
Methods
Over the course of 2022, we monitored the potency and breadth of antibody neutralization responses to many emerging variants in the Australian community at three levels: (i) we tracked over 420,000 U.S. plasma donors over time through various vaccine booster roll outs and Omicron waves using sequentially collected IgG pools; (ii) we mapped the antibody response in individuals using blood from stringently curated vaccine and convalescent cohorts. (iii) finally we determine the in vitro efficacy of clinically approved therapies Evusheld and Sotrovimab.
Findings
In pooled IgG samples, we observed the maturation of neutralization breadth to Omicron variants over time through continuing vaccine and infection waves. Importantly, in many cases, we observed increased antibody breadth to variants that were yet to be in circulation. Determination of viral neutralization at the cohort level supported equivalent coverage across prior and emerging variants with isolates BQ.1.1, XBB.1, BR.2.1 and XBF the most evasive. Further, these emerging variants were resistant to Evusheld, whilst increasing neutralization resistance to Sotrovimab was restricted to BQ.1.1 and XBF. We conclude at this current point in time that dominant variants can evade antibodies at levels equivalent to their most evasive lineage counterparts but sustain an entry phenotype that continues to promote an additional outgrowth advantage. In Australia, BR.2.1 and XBF share this phenotype and, in contrast to global variants, are uniquely dominant in this region in the later months of 2022.
Interpretation
Whilst the appearance of a diverse range of omicron lineages has led to primary or partial resistance to clinically approved monoclonal antibodies, the maturation of the antibody response across both cohorts and a large donor pools importantly observes increasing breadth in the antibody neutralisation responses over time with a trajectory that covers both current and known emerging variants.
Funding
This work was primarily supported by Australian Medical Foundation research grants MRF2005760 (SGT, GM & WDR), Medical Research Future Fund Antiviral Development Call grant (WDR), the New South Wales Health COVID-19 Research Grants Round 2 (SGT & FB) and the NSW Vaccine Infection and Immunology Collaborative (VIIM) (ALC). Variant modeling was supported by funding from SciLifeLab's Pandemic Laboratory Preparedness program to B.M. (VC-2022-0028) and by the European Union's Horizon 2020 research and innovation programme under grant agreement no. 101003653 (CoroNAb) to B.M.
Keywords: SARS-CoV-2, Variants, TMPRSS2, Covid-19, Neutralising antibodies, Sotrovimab, Evusheld
Research in context.
Evidence before this study
Up until the BA.5 wave in mid 2022, many global waves were seeded by dominant variants such as Delta, Omicron BA.1 and Omicron BA.2. Following resolution of the BA.5 in Australia, there was the emergence of a pool of BA.4/5 and BA.2.75 sub-lineages accumulating clusters of similar polymorphisms located within the Receptor Binding Domain (RBD) of the Spike glycoprotein. Although iterative changes in the Spike increased the ability of each variant to navigate existing neutralizing antibodies, it was unclear if this alone was sufficient to provide an outgrowth advantage to any one variant to fuel major case waves in global communities with high vaccine uptake and/or infection.
Added value of this study
Prior studies on incoming variants in Australian quarantine, highlighted the potential for Australia to represent a unique mix of co-circulating variants. Following the resolution of the BA.5 Omicron wave, many globally circulating variants appeared early on and ranged from BA.2.75 lineages, recombinants XBB.1, and XBC.1 in addition to many BA.5-derived BQ.1 lineages. Two additional lineages, the recombinant XBF and the BA.2.75-derived BR.2.1 also appeared and were uniquely enriched in Australia. Using 11 primary clinical isolates covering a continuum of circulating variants in Australia, we resolved neutralization responses of 110 donors stringently documented for their vaccine and infection status over time. In addition, we also tested the clinically utilized clinical monoclonals Evusheld and Sotrovimab. To complement the tracking of singular donors, we also tracked immunity at the population level, using pooled IgG samples over time. The latter samples were the sum of 420,000 U.S. plasma donors covering time periods of high-booster uptake alongside large case waves. Whilst the above resolved the impact of Spike changes in neutralizations, we also tested each variant with respect to the efficiency of TMPRSS2 use, as this significantly influences viral tropism across the respiratory tract.
Implications of all the available evidence
All variants analysed herein have undertaken a convergent trajectory in accumulating a similar cluster of Spike polymorphisms. Whilst BQ variants have dominated worldwide, a unique mix of variants, including XBF and BR.2.1 dominated in Australia in late 2022. In addition to other global variants, including BQ.1.1 and XBB.1, all share a similar accumulation of key Spike glycoprotein polymorphisms. The accumulation of changes now render neutralization responses lower in all cohorts and are neutralization resistant to Evusheld. Whilst Sotrovimab retained neutralization capacity of many variants, there was significant reduction for variants BQ.1.1 and XBF. Impact of Spike changes on TMPRSS2 use was a continuum away from efficient use towards that originally observed for BA.2. Analysis of neutralization at the population level over time revealed two key observations. Firstly, whilst variants converged and lowered neutralization responses, this reduction was negated over time with increasing neutralization breadth. Secondly, responses to a variant proceded its appearance and global circulation. In conclusion, whilst many variants are appearing and iterative changes in the Spike will challenge antibody responses, increasing breadth in the community over time has enabled sufficient coverage to currently emerging variants. Furthermore, the trajectory of TMPRSS2 has not trended to pre-Omicron variants such as Delta and as such, we predict that viral tropism towards epithelial cells of the upper respiratory tract will be maintained.
Introduction
The beginning of the COVID-19 pandemic saw severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants driving several waves of disease transmission globally. The initial lack of vaccines and often immature/germ line convalescent antibody responses1, 2, 3, 4, 5 promoted small iterative changes in the virus genome during various global waves of infection. Changes primarily within the Spike glycoprotein were key to viral spread. Whilst many of these changes appeared in the N-terminal domain and within the receptor binding domain, resulting in decreased antibody binding, others accumulated at and around the furin cleavage site which supported greater viral fitness and cellular entry through the serine protease TMPRSS2.6, 7, 8, 9, 10, 11, 12 The Delta variant highlighted how changes in viral entry and fitness combined with changes that enabled antibody evasion could drive significant waves worldwide.
Fortunately, towards the end of 2021, many vaccines were mobilized worldwide that led to greater than 88% vaccine efficacy to the dominant Delta variant.13 The subsequent arrival of two Omicron lineages BA.1 and BA.2 represented a significant shift in the pandemic at two key levels. Firstly, both sub-variants appeared with Spike glycoproteins that enabled significant evasion from both vaccine and/or infection-induced antibodies. Secondly, unlike Delta, BA.1 and BA.2 shifted away from efficient usage of TMPRSS2, which favored replication in upper respiratory tract epithelial cells.7,14, 15, 16, 17 With the resolution of both BA.1 and BA.2 waves globally, the BA.2 sub-lineage, BA.5, propelled subsequent waves. This sub-lineage was defined by key changes in the receptor binding domain resulting in antibody evasion and greater use of TMPRSS2 relative to its parent BA.2.18 The BA.5 wave has since been replaced by BA.5 and BA.2.75 sub-lineages that have accumulated additional changes across their genomes, especially within the Spike glycoprotein.
To resolve the relative threat of circulating variants in real-time, we have tested variants prevalent within the community using two approaches: (i) isolation and whole genome sequencing of SARS-CoV-2 grown from nasopharyngeal swabs and; (ii) phenotypic characterization of antibody evasion and modes of viral entry. The latter was examined using four distinct approaches. We first evaluated neutralizing antibody evasion at the population level with pooled plasma from over 420,000 U.S. donors. The second approach involved testing of curated cohorts based on vaccine- and infection-history. Thirdly, we determined the ability of currently approved monoclonal antibody therapeutics to neutralize the circulating variants. Finally, we observed the consequences of these cumulative changes in Spike on TMPRSS2 use, as this can significantly influence viral entry fitness and epithelial cell tropism throughout the respiratory tract.
Based on the above, we observe the maturation of the neutralization responses to target a greater diversity of Omicron variants. At the viral level, we observe the convergence of Spike polymorphisms across many emerging lineages to significantly decrease the potency and breadth of the neutralization responses in all cohorts. This translated to the loss of Evusheld's ability to neutralize key emerging variants XBB.1, BR.2.1, BQ.1.1 and XBF. In contrast, Sotrovimab retained potent neutralization activity to all tested variants with the exception of BQ.1.1 and XBF. With respect to viral entry, dominant circulating BA.2.75 and BQ sub-lineages such as XBF, BR.2.1 and BQ.1.1 exhibited similar but reduced TMPRSS2 use relative to BA.5. We conclude that whilst the increasing breadth of antibody responses is increasing pressure on the current variant pool, continued variant surveillance will be important in determining the trajectory of current and future variant pools with respect to antibody evasion and tropism.
Methods
Human sera
The ADAPT cohort is composed of RT-PCR–confirmed convalescent individuals (including some subsequently vaccinated) recruited in Australia since 2020.1 Donors within this cohort completed their first vaccine doses (ChAdOx1 or BNT162b2; See Supplementary Table S3) and booster doses (BNT162b2 or mRNA-1273) in January to February 2022. Female to male ratios were 0.5:1.0 for this cohort, with a donor median age of 51 years. The Australian Vaccine, Infection and Immunology Collaborative Research cohort (VIIM) consists of serum samples from 75 participants collected between July 2022–October 2022. Serum samples were collected at 12 months post-second dose of Pfizer Comirnaty from two sites; Westmead Hospital and Royal Prince Alfred Hospital, NSW, Australia. Female to male ratio was 1.02:1.0, with a donor median age of 50.6 years. Data were collected on SARS-CoV-2 infections for all participants and independently verified by anti Nucleocapsid ELISA as previously described.1
Ethics
All human serum samples were obtained with written informed consent from the participants (2020/ETH00964; 2020/ETH02068; 2019/ETH03336; 2021/ETH00180; 2021/ETH0042). All samples were tested with operators blinded. Following acquisition of results, data was then assigned to relevant donor codes and vaccine/infection history. Demographic data was then provided on each group and outlines female to male ratios and age median and range. All primary isolates used herein were obtained from de-identified remnant diagnostic swabs that had completed all diagnostic testing under approval by the NSW Chief Health Officer following independent scientific review and as outlined in the ADAPT ethics protocol 2020/ETH00964. The samples are derived from a single diagnostic center in Sydney that tests samples coalescing from 20 COVID-19 collection sites covering the Sydney Metropolitan area. This collection and sequencing is not part of the NSW genomic surveillance efforts which are reported here https://www.health.nsw.gov.au/Infectious/covid-19.
Other immunoglobulin products
Clinical grade Sotrovimab (62.5 mg/mL; NDC 0173-0901-86) was kindly provided by GSK Healthcare while clinical grade Cilgavimab and Tixagevimab (100 mg/mL each; AstraZeneca) were kindly provided by Dr Sarah Sasson (Kirby Institute, UNSW). Cilgavimab and Tixagevimab were mixed in equal volumes to generate the monoclonal antibody cocktail Evusheld. All monoclonal antibodies were tested at a starting concentration of 10 μg/mL and diluted three-fold in an eight step dilution series.
Polyclonal immunoglobulin preparations and anti-SARS-CoV-2 hyperimmune globulin
The immunoglobulins used herein were purified using the licensed and fully validated immunoglobulin manufacturing process used for Privigen,19 notionally similar to others.2 Thirty seven poly-Ig batches were manufactured using the Privigen process19 and included U.S. plasma collected by plasmapheresis from a mixture of vaccinated (SARS-CoV-2 mRNA vaccines), convalescent and non-convalescent donors (source plasma, n > 10,000 donors per batch) collected over the period of August 2021 to June 2022.
Cell culture
HEK293T cells stably expressing human ACE2 and TMPRSS2 were generated by lentiviral transductions as previously described.1,16 A highly permissive clone (HAT-24) was identified through clonal selection and used for this study. The HAT-24 line has been extensively cross-validated with the VeroE6 cell line.16 VeroE6-TMPRSS2 (Vero-T) cells were kindly provided by Associate Professor Daniel Watterson (University of Queensland). HAT-24, VeroE6-TMPRSS2 cells were cultured in Dulbecco's Modified Eagle Medium (Gibco, 11995073) containing 10% foetal bovine serum (Gibco, 10099141; DMEM-10%FBS) and VeroE6 cells (ATCC® CRL-1586™) in Minimal Essential Medium (Gibco, 11090099) containing 10% FBS and 1% penicillin-streptomycin (Gibco, 15140122; MEM-10%FBS). pBEC and alveolar epithelial cultures were grown and differentiated until confluent in complete Bronchial Epithelial Cell Growth Basal Medium (Lonza, CC-3171) before use for air liquid interface experiments. All cells were incubated at 37 °C, 5% CO2 and >90% relative humidity. For the Vero-T cell line authentication was performed as previously described.1,16 The STR profiling of HAT-24 has been previously described.16 All cell lines tested negative for mycoplasma.
Visualizing variant dynamics
Variant competition was inferred using the global GISAID dataset (bulk fasta download from 2022/12/2022), excluding any data older than 100 days. GISAID sequences were processed and assigned to clades via NextClade (using the BA.2 reference set, “sars-cov-2-21L″), and compiled into counts per country per lineage per day. The modelling frameworks extends that described in,20 and is described and updated at https://github.com/MurrellGroup/lineages with only cosmetic adjustments for the figures shown here.
Viral isolation, propagation, and titration
All laboratory work involving infectious SARS-CoV-2 occurred under biosafety level 3 (BSL-3) conditions. SARS-CoV-2 variants were isolated and characterized as previously described.18 Briefly, diagnostic respiratory specimens that tested positive for SARS-CoV-2 (RT-qPCR, Seegene Allplex SARS-CoV-2) were sterile-filtered through 0.22 μm column-filters at 10,000×g and serially diluted (1:3) on HAT-24 cells (5 × 103 cells/well in 384-well plates). Upon confirmation of cytopathic effect by light microscopy, 300 μL of pooled culture supernatant from infected wells (passage 1) were added to Vero-T cells in a 6-well plate (0.5 × 106 cells/well in 2 mL MEM2%) and incubated until significant cytopathic effects accumulated and approximately 50% of the cell sheet was in tact (over 24–72 h). At the latter time point, supernatant was cleared by centrifugation (2000×g for 5 min), frozen at −80 °C (passage 2), then thawed and titrated to determine median 50% Tissue Culture Infectious Dose (TCID50/mL) on Vero-T cells according to the Spearman-Karber method.21 Viral stocks used in this study correspond to passage 3 virus, which were generated by infecting Vero-T cells at MOI = 0.025 and incubating for 24 h before collecting, clearing, and freezing the supernatant as above. Sequence identity and integrity were confirmed for both passage 1 and passage 3 virus via whole-genome viral sequencing using Oxford Nanopore technology platform, as previously described.18,22 For a list of the viral variants isolated in this study see Supplementary Table S1. Passage 3 stocks were titrated by serial dilution (1:5) in DMEM-5%FBS, mixing with HAT-24 cells live-stained with 5% v/v nuclear dye (Invitrogen, R37605) at 1.6 × 104 cells/well in 384-well plates and incubating for 20 h. Whole-well nuclei counts were determined with an IN Cell Analyzer 2500HS high-content microscope and IN Carta analysis software (Cytiva, USA). Data was normalized to generate sigmoidal dose–response curves (average counts for mock-infected controls = 100%, and average counts for highest viral concentration = 0%) and median Virus Effective (VE50) values were obtained with GraphPad Prism software. To assess the TMPRSS2 usage of the virus isolates, titrations on HAT-24 cells were performed in the presence of saturating amounts of Nafamostat (20 μM). Titrations were performed in parallel in equivalent volumes of DMSO served as controls and were used to calculate fold drops in VE50.
Abbott Architect SARS-CoV2 IgG
IgG antibodies to the nucleocapsid protein (N) of SARS-CoV2 were detected using Architect SARS-CoV-2 IgG (Abbott Diagnostics, Sydney, NSW Australia) as previously described.23
Rapid high-content SARS-CoV-2 microneutralization assay with HAT-24 cells (R-20)
Rapid high-content neutralization assay with HAT-24 cells was done as previously described.16,18 Briefly, human sera or monoclonal antibodies were serially diluted (1:2 series starting at 1:10 for sera) in DMEM-5% FBS and mixed in duplicate with an equal volume of SARS-CoV-2 virus solution standardized at 2× VE50. After 1 h of virus–serum coincubation at 37 °C, 40 μL were added to an equal volume of nuclear-stained HAT-24 cells pre-plated in 384-well plates as above. Plates were incubated for 20 h before enumerating nuclear counts with a high-content fluorescence microscopy system as indicated above. The % neutralization was calculated with the formula: %N = (D − (1 − Q)) × 100/D as previously described.1 Briefly, “Q” is a well's nuclei count divided by the average count for uninfected controls (defined as having 100% neutralization) and D = 1 − Q for the average count of positive infection controls (defined as having 0% neutralization). Sigmoidal dose–response curves and IC50 values (reciprocal dilution at which 50% neutralization is achieved) were obtained with GraphPad Prism software. Neutralization assays testing monoclonal antibodies with VeroE6 cells were performed as previously described.16,18 Briefly, antibodies were serially diluted in MEM-2% FBS using a 1:3 series starting at 20 μg/mL, input virus solution was standardized at 1.25 × 104 TCID50/mL, cells were seeded at 5 × 103 cells/well in MEM-2% FBS (final MOI = 0.05), plates were incubated for 72 h, and cells were stained with nuclear dye only 2 h before imaging.
Infection of ALI-pBECs and determination of viral load
Culture and infection of pBEC and alveolar epithelial cultures at air liquid interface (ALI) was performed as previously described.24,25 Briefly, prior to infection, cells were washed once with PBS and inoculated with equal number of virus particles for Delta, Omicron BA.2 and BA.5. Two hours post-incubation at 37 °C virus inoculum was removed and unbound virus was washed off using 500 μL of PBS. At 3 days post-infection cells, were collected in RNA lysis buffer (RLT) (Qiagen) with 1% 2-mercaptoethanol for total RNA extraction.
Statistics
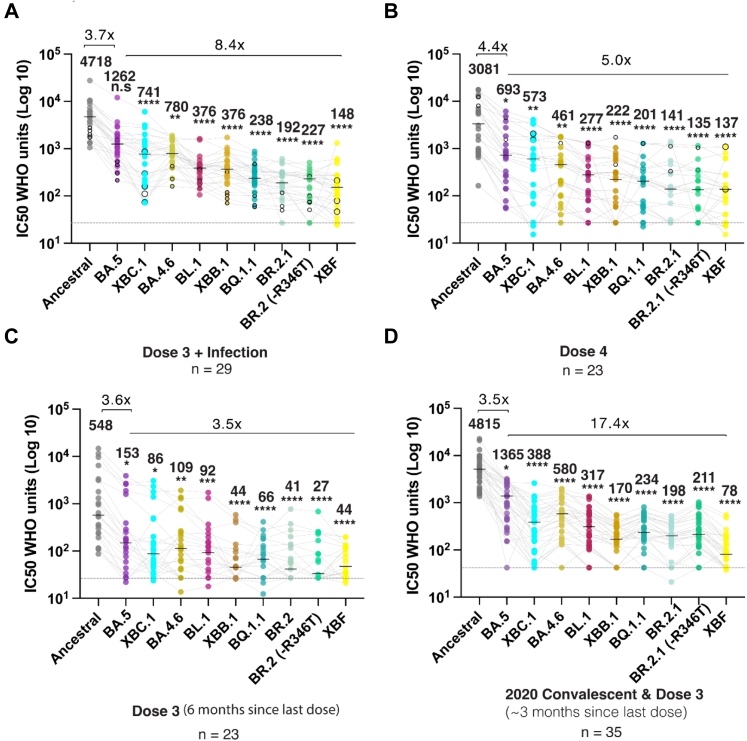
Cohorts participants were initially enrolled in the VIIM cohort to observe primarily the Pfizer vaccine response in recruits with no documented prior history of infection and negative Spike and Nucleocapsid serology from independent confirmation. This group was divided in three sub-groups to observe the neutralisation response with variants across a continuum of immunity that ranged from three dose vaccination and a recent break through infection, four dose vaccination, and three dose vaccination with no booster dose within six months. The final cohort from ADAPT was used to complete the latter continuum of immunity within the community, by observing responses following infection early in the pandemic and then after three doses of vaccine. These groupings were designed to reflect that of the general community and were taken in late 2022 to reflect the immunity at the time the variants tested were in circulation. As of the 15th of February, in Australia, 14,352,625 people over the age of 16 have had three doses of vaccine and 5,466,752 over the age of 16 have had their fourth dose. Recent serosurveys (https://kirby.unsw.edu.au/sites/default/files/COVID19-Blood-Donor-Report-Round3-Aug-Sep-2022.pdf) support nucleocapsid seroprevalence at around 70%, so a large proportion of people vaccinated would have also been infected following vaccination. So the groupings in Fig. 3 simply reflect what is in the community now but with resolution of vaccine dose and infection history.
Fig. 3.
Emerging variants and their ability to evade a continuum of antibody responses. A. Three dose Pfizer BNT162b2 vaccination with subsequent breakthrough infection. Closed circles were infections between June and August 2022 when BA.2 and BA.5 were prevalent. Open circles are breakthrough infections with BA.1 between January and February. B. Primarily four dose BNT162b2 vaccinations, in which the last dose was within three months. Open circles in this group are breakthrough infections in early 2022 at the time of BA.1. C. Three dose Pfizer BNT162b2 vaccination with the last dose six months prior. D. Early 2020 donors infected between March and August of 2020 and then subsequently vaccinated with two doses of Pfizer BNT162b2 or AstraZeneca AZD1222/ChAdOx1 and boosted with Pfizer BNT162b2 or Moderna mRNA-1273. This group did not receive their last dose three to six months prior. Data in (A–D) indicates the mean IC50 of technical replicates for individual samples. The median titers are labelled. Fold change reductions in IC50 neutralization titers compare variants of concern to the Ancestral variant and Omicron BA.5 where indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for Friedman test with Dunn's multiple comparison test.
Statistical analyses were performed using GraphPad Prism 9 (version 9.1.2, GraphPad software, USA). Sigmoidal dose response curves and interpolated IC50 values were determined using Sigmoidal, 4PL model of regression analysis in GraphPad Prism. For statistical significance, the datasets were initially assessed for Gaussian distribution, based on which further analysis was performed. For datasets that followed normal distribution One way ANOVA was used while for others non parametric tests such as Friedman (for paired samples) or Kruskal Wallis test (for unpaired samples) with Dunn's multiple comparison were employed. In all cases the data was compared to the Ancestral strain Clade A.2.2. Mann Whitney U test was used to analyse data between two groups. Two-way ANOVA with Sidak's multiple comparisons was used to compare viral loads for variants in primary epithelial cells at different time points. In all cases the data was compared to Delta strain. Ordinary one way ANOVA with Dunnett's multiple comparisons was used to examine the fold change in viral titres of different variants in the presence of saturating concentrations of Nafamostat. The data was compared to Omicron BA.5. Details of statistical tests used for different data sets have also been provided in figure legends.
Results
Australian SARS-CoV-2 variant diversity in late 2022
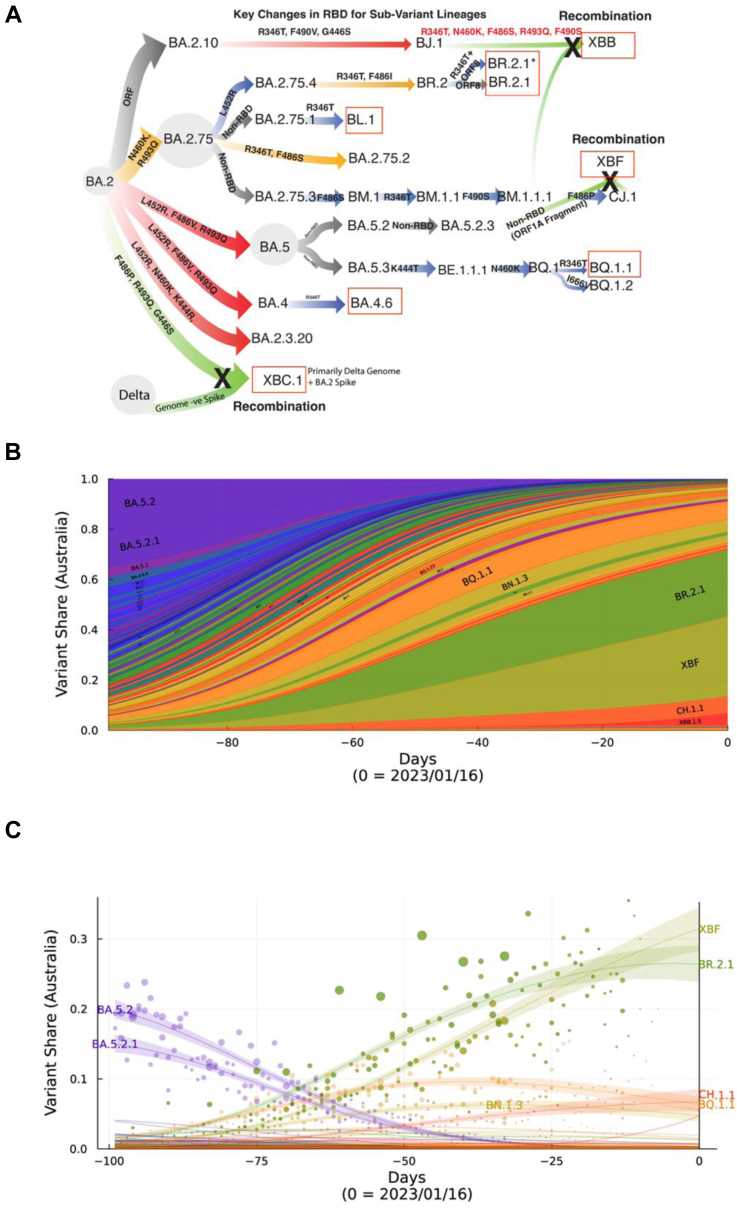
Through a diagnostic-research laboratory partnership, we isolated SARS-CoV-2 primary isolates throughout 2022. Since the resolution of the BA.1, BA.2 and BA.5 waves, many variants have emerged. However, unlike prior Omicron waves, a single variant is yet to dominate globally. Rather, there exists a pool of variants that have accumulated similar Spike polymorphisms, which can be grouped into sub-lineages derived from BA.2.75 or BA.5 (Fig. 1A and B). Following the peak of the BA.5 wave, we focussed on variant isolation and phenotyping to determine the relative risk of newly emerging variants (Supplementary Table S4). All samples were subjected to ex vivo whole genome sequencing using Nanopore single molecule sequencing platforms as previously described.22 Whilst samples in October were enriched for many BA.5-derived lineages, by mid November there was a similar proportion of variants either derived from a BA.5 parent (e.g. BQ.1) or a BA.2.75 parent (Fig. 1A). The latter was primarily the BR.2.1 variant, with a unique polymorphism within open reading frame (ORF) 8 (S67F). Genomic surveillance supported its origin being within the New South Wales region of Australia (https://www.health.nsw.gov.au/Infectious/covid-19). Whilst limited samples of the recombinant XBF were observed in NSW relative to Victoria in late 2022, XBF did increase in frequency in December and January samples (Supplementary Table S4). The eventual emergence of XBF in NSW is consistent with the overall national snapshot over October 2022 to mid January 2023 across Australia (genomic surveillance through GISAID genome contributions) highlighting the dominance of the recombinant XBF national alongside dominant BR.2.1 variant in NSW (Fig. 1B and C). This was alongside the early appearance of other global variants such as XBB.1 and BQ.1.1 that were highly prevalent in Singapore and Northern America, respectively (Fig. 1B). By the end of January 2023, XBF represented the dominant variant (Fig. 1C) and combined with BR.2.1, highlighted the divergent and unique mix of variants in Australia.
Fig. 1.
Emergence and prevelance of BA.2.75 and BA.5 sub-lineages with convergent Spike polymorphisms in Australia. A. From left to right, Omicron lineages from the parent BA.2 are now primarily split across BA.2.75 and BA.5 sub-lineages. Divergent lineages such as BR.2.1 have accumulated polymorphisms common to BA.5, including L452R and F486I. Whilst BQ.1.1 has acquired BA.2.75 polymorphisms K444T and N460K. Common across most lineages is the additional acquisition of R346T, and this change defines lineages BL.1 and BA.4.6. XBB.1 is a recombinant between BJ.1 and BM.1.1.1 and has benefited from many Spike changes through this recombination event. Whilst XBF is a recombinant between BA.5.2.3 and CJ.1, it retains the latter Spike defined by the F486P change. XBC.1 is a Delta-BA.2 recombinant, with the retention of an intact BA.2 Spike with F486P, G446S and R493Q. Boxed in red are variants that were selected for testing in this study and range from variants with few spike polymorphisms (BL.1 and BA.4.6) through those with many convergent polymorphisms (BR.2.1, XBF and BQ.1.1). For the latter, we have also looked a two circulating isolates of BR.2.1, with and without the R346T change. B. Surveillance summary of Australia at 1601/2 based on genomic data via GISAID. Each vertical slice depicts the posterior mean variant frequency estimate from a hierarchical Bayesian multinomial logistic model of variant competition. Note the two dominant variants BR.2.1 and the recombinant XBF which are dominating in the states of NSW and Victoria, respectively. C. Variant proportions with associated Bayesian Credible Intervals for B.
Whilst isolations were confined to a singular diagnostic unit, state-wide genomic surveillance supported similar prevalence of BA.5 and BA.2.75-derived lineages (https://www.health.nsw.gov.au/Infectious/covid-19). Importantly, viral isolation rates were high with over 70% of samples culture positive when samples were restricted to diagnostic Ct thresholds of less than 30. Furthermore, it enabled the curation of isolates that covered both dominant BA.5 and BA.2.75 lineages circulating globally for testing across immunotherapeutics, cohort sera (for antibody evasion) and cell platforms (for viral tropism) (Fig. 2, Fig. 3, Fig. 4, Fig. 5). Many of the emerging lineages from divergent parents (BA.2.75 and BA.5) had accumulated polymorphisms clustered and distributed across class I, II and III antibody sites across the receptor binding domain.26 These included R346T, L452R/M, F486V/I/P, N460K, K444R/T and F490S (Fig. 1), with variants such as XBB.1, XBF, BR.2.1 and BQ.1.1 accumulating the greatest number of polymorphisms (Fig. 1).
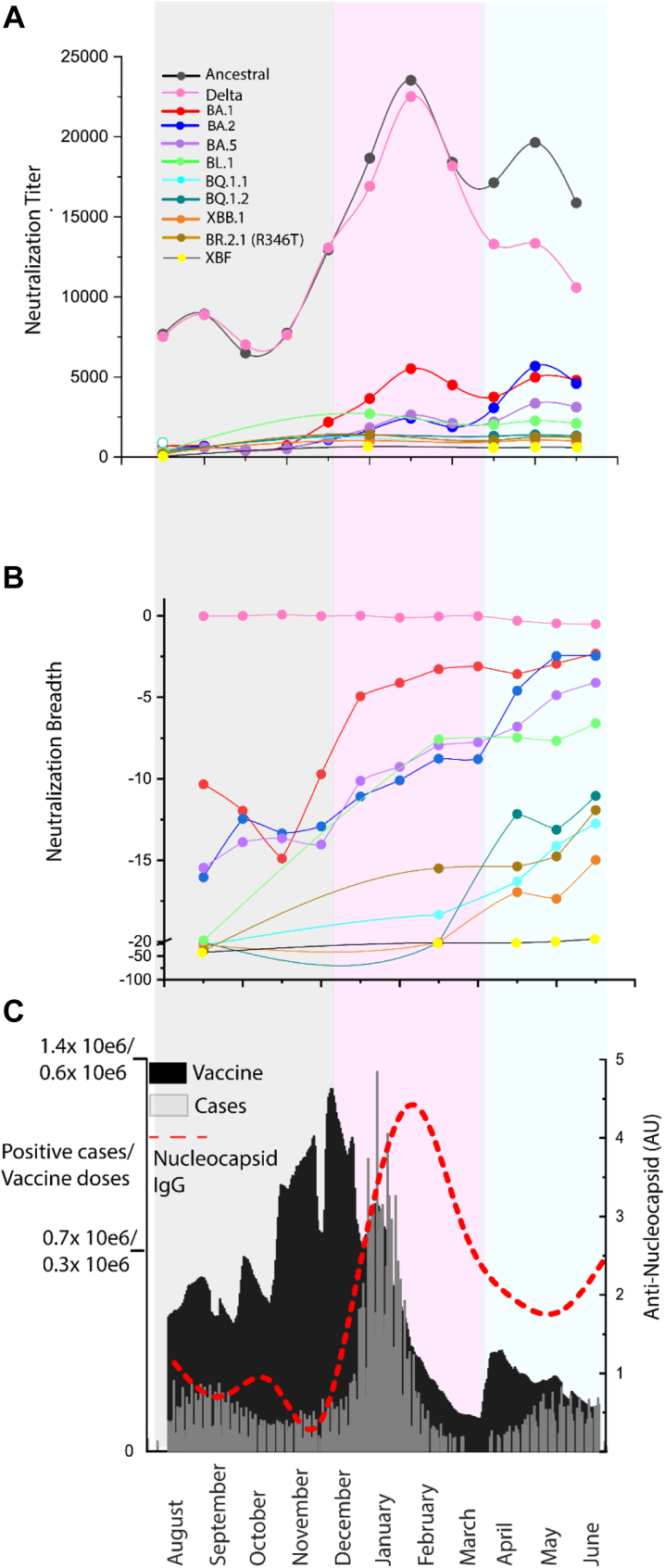
Fig. 2.
Potency and breadth in pooled IgGs of greater than 420,000 U.S. plasma donors from August 2021 through to June 2022. A. Neutralization titers across 11 clinical isolates and include Clade A.2.2 (Ancestral), Delta and Omicron BA.1, BA.2, BA.5, BL.1, BQ.1.1, BQ.1.2, XBB.1, XBF and BR.2.1 (with R436T) for IgG batches collected between August 2021 and June 2022 (batch details are presented in Supplementary Table S1). Each point represents the mean titer of batches from that month and constitutes approximately 30,000 plasma donations. B. Breadth score of batches to variants in A. Breadth score is calculated as 1- (fold reduction of titer relative to the Clade A variant A.2.2). When the breadth approaches zero (e.g. Delta), neutralization titers cover this variant with equal potency to the ancestral variant. C. Documented vaccines doses and COVID-19 cases are presented here to frame the responses in A and B. Anti-nucleocapsid IgG levels in monthly batches presented in A and B are also indicated (red line). The grey-shading is primarily the 3× booster doses administered prior to the arrival of Omicron BA.1. In pink is the Omicron BA.1 wave. In blue is primarily the Omicron BA.2 wave.
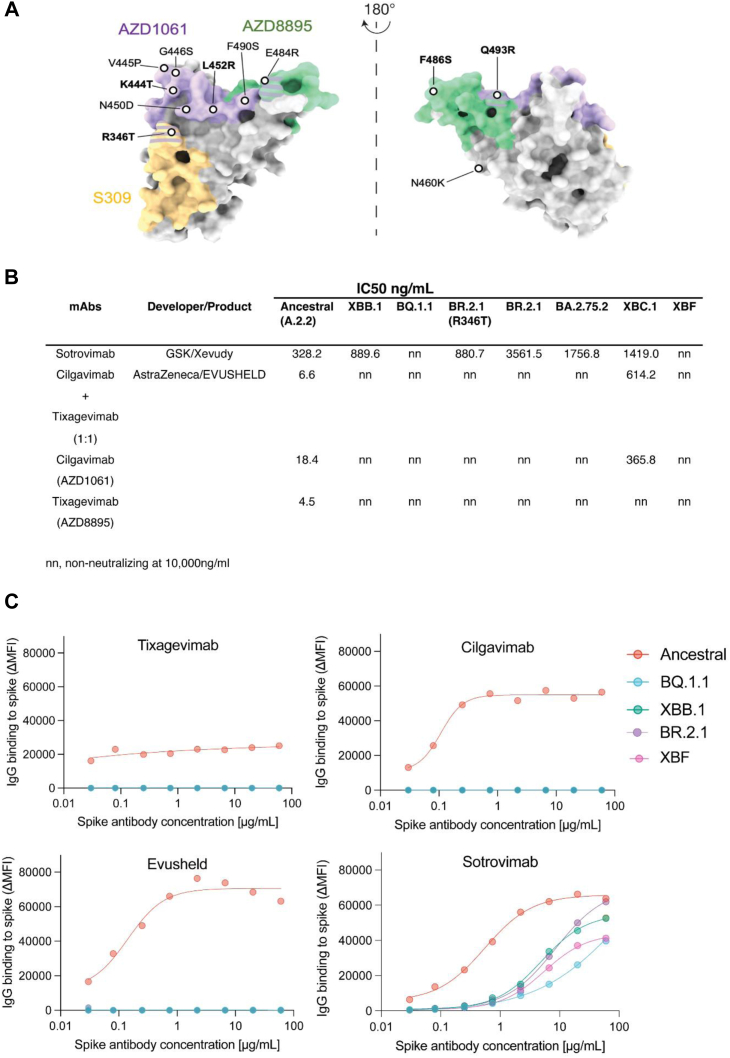
Fig. 4.
Neutralizing activity of monoclonal antibodies against emerging variants. A. Binding sites for AZD1061 (Cilgavimab), AZD8895 (Tixagevimab) and S309 (Sotrovimab) on the SARS-CoV-2 Spike glycoprotein. Key Spike polymorphisms that map to these sites are indicated and are prevelant on emerging variants tested. B. IC50 values (ng/μl) of monoclonal antibodies Sotrovimab, Cilgavimab and Tixagevimab cocktail, Cilgavimab alone and Tixagevimab alone, against ancestral A.2.2, Omicron XBB.1, BQ.1.1, BR.2.1 (R346T), BR.2.1, BA.2.75.2, XBC.1 and XBF. Antibodies used herein were clinical grade batches. Data represents mean IC50 values from two independent experiments. For Sotrovimab, pairwise comparisons between Ancestral and variants were done using Kruskal Wallis test with Dunn's multiple comparisons. For other datasets, unpaired t-test was used. p values are indicated in Supplementary Table S6. C. Antibody binding to full length Spike expressed on live cells and acquired using flow cytometry. Signal is expressed as Mean Fluorescent Intensity above background Fluorescence (ΔMFI).
Fig. 5.
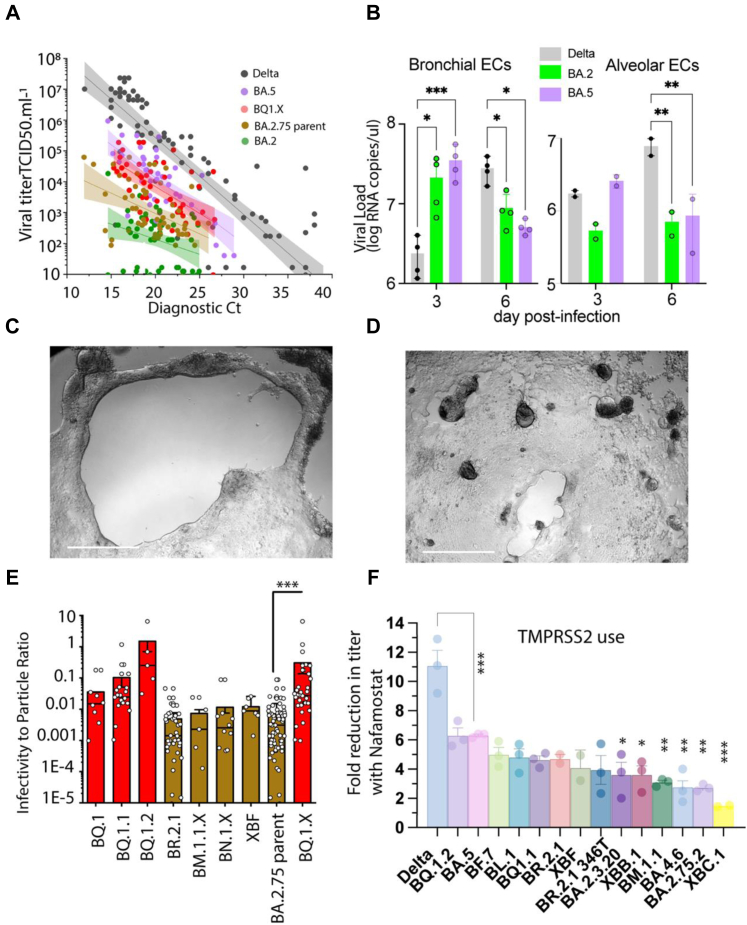
Variant divergence in TMPRSS2 use in the emerging Omicron variant swarm. A. Endpoint titers for primary nasopharyngeal swabs grouped into variants derived from BQ (BQ1.X) lineages and variants derived from BA.2.75 lineages (primarily BR.2.1) using the HAT-24 cell line. The linear regressions of BA.2, BA.5 and Delta are also presented18 and serves here primarily as a guide with respect viral titers per diagnostic Ct value. Of note, as the variant uses TMPRSS2 more efficiently, there is an increase in viral titer per Ct value and this is observed by an upwards shift in the linear regression. B. Air liquid interface (ALI) primary bronchial and alveolar epithelial cells 3 and 6 days post-infection. Equal numbers of viral particles were used to inoculate each culture. Supernatant was harvested 3 and 6 days post-infection. Error bars represent standard deviations from the mean of infections from two to four independent donors. Delta and the parent variants BA.2 and BA.5 are presented here to representative highlight the influence of TMPRSS2 usage for viral entry fitness in primary differentiated respiratory epithelia in the upper and lower respiratory tract. ∗p < 0.01, ∗∗p < 0.01, ∗∗∗p < 0.001, for two-way ANOVA with Sidak's multiple comparisons C. BQ.1.2 and D. BR.2.1 primary nasopharyngeal swabs are presented in HAT-24 cells following 3 days in culture. Scale bar represents 100 μm. Limiting viral dilutions are presented to contrast the growth of each variant. E. To enable initial resolution of variants, we determined RNA copies per diagnostic Ct values and have presented infectivity to particle ratios for variants that were initially group in A. through dividing TCID50 titers with copies per reaction based on qPCR cycle threshold values and internal PCR controls for quantification. Grouping of BA.5 and BA.2.75 lineages here highlight the latter group to have lower infectivity to particle ratios. ∗∗p < 0.01 for Mann Whitney U-test. F. Mean fold reductions in viral titers in the presence of saturating levels of the TMPRSS2 inhibitor Nafamostat. Mean values represent three independent experiments and each point is the mean fold reduction per independent experiment performed in quadruplicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, for ordinary one way ANOVA with Dunnett's multiple comparison test.
Maturation of viral neutralization breadth over the course of the BA.1 and BA.2 waves and erosion of neutralization titers with emerging Omicron lineages
Herein our primary aim was to track the maturation of the neutralization response over time at the population level. Firstly, we define maturation of the neutralization response as the increasing ability to recognize (increasing neutralization breadth) genetically diverse SARS-CoV-2 variants over time. Secondly, we define neutralization responses at the population level in an approach called IgG pooling. IgG pooling and purification is used in the manufacture of 10% IgG for clinical use in conditions such as replacement therapy for primary and secondary immunodeficiencies. Each batch consists of IgG purified and pooled from in excess of 10,000 plasma donations collected over the period of approximately one month.
To track the maturation of the neutralization response, we analysed sequential pools of IgG-derived plasma from over 420,000 U.S. donors acquired during the period between August 2021 through to June 2022. This time frame covers the acceleration in the third dose (first booster) vaccine roll out in late 2021, the arrival and peak of the largest infection wave in early 2022 (BA.1 Omicron wave) and concludes following the resolution of the Omicron BA.2 wave. Whilst analysis of more recent waves would enable real-time tracking of immunity, it must but noted that the delay using this approach is primarily related to the logistics of processing close to half a million plasma donations.
We tested 37 separate, sequential batches of pooled IgG for neutralization titers across 10 clinical isolates. For variant testing, we looked at circulating isolates at that time and also variants that are presently circulating. Isolates included Clade A.2.2 (Ancestral), Delta as well as 2022 Omicron variants, BA.1, BA.2 and BA.5 and emerging Omicron variants BL.1, BQ.1.1, BQ.1.2, XBB.1, XBF and BR.2.1 (Fig. 2A–C). For all polyclonal IgG batches, we tested neutralization titers to each variant (Fig. 2A) and calculated neutralization breadth to each variant (Fig. 2B). For breadth scoring, scores that approach zero have breadth equal to the “Ancestral” Clade A variant which provides the basis of the vaccines available to the U.S. population (Fig. 2B).
To determine whether vaccination versus BA.1 infection contribute to changes in breadth and potency, we plotted mean neutralization titers and breadth with three key comparators. Firstly, we plotted the mean titers of batches by month in which the plasma was collected (Fig. 2A). Secondly, we have aligned these titers relative to the breadth to each variant tested (Fig. 2B). Thirdly, we have aligned documented COVID-19 cases versus vaccine doses in the U.S. population over this time (Fig. 2C) to chronologically frame the immunological events of this population over time. For this, we have divided this time period (Fig. 2C) into the pre-Omicron booster uptake period (grey), Omicron BA.1 wave (pink) and BA.2 wave (blue). To independently track infections, we have also detected anti-nucleocapsid IgG in each batch over time, as neutralization titers with either low or high nucleocapsid IgG can independently track and verify if titers may be enriched with antibodies derived from donors following vaccination versus infection convalescence, respectively (Fig. 2C red dashed line).
The accelerated increase in neutralization titers and breadth to the first Omicron Clade BA.1 (and to a lesser extent BA.2), occurs concurrently with the uptake of the third dose of the vaccination regimen. A drop in anti-NC IgG levels relative to neutralization titer, suggested that vaccination, not infection, was contributing to increases in neutralization and breadth during this period (Fig. 2A–C; see grey shading and red line in C). In contrast, we observe a peak of neutralization titers to the Ancestral Clade and anti-NC IgG in February, at the same time as the resolution of the BA.1 peak in January 2022 (Fig. 2A–C). Peak neutralization titers to all variants coincided with the BA.1 wave (Fig. 2A–C; see pink shading).
Following the resolution of the BA.1 wave in the U.S., the defining immunological events were a small peak in vaccination doses followed shortly by primarily the BA.2 wave (Fig. 2C). Unlike that observed in early 2022, it was not possible to temporarily dissect vaccination doses from peak COVID-19 cases, and as such, all that can be concluded is that concurrent vaccine uptake and infection within the population occurred at the same time as increasing neutralization breadth to Omicron variants BA.1, BA.2, BA.5 and BL.1 (Fig. 2A–C; blue shading). Across the panel of current circulating variants (BQ.1.1, BQ.1.2, XBB.1, BL.1, BA.4.6 and BR.2.1 (R346T)), the most neutralization evasive variants clustered together (BQ.1.1, XBB.1, XBF, BR.2.1 (R346T)). In IgG batches from late 2021, there was a 30-50-fold reduction in neutralization capacity for these variants relative to titers of the Ancestral clade. Whilst this remained largely unchanged, there was an increase in neutralization breadth to all variants but XBF just prior to the BA.2 wave (Fig. 2A–C; blue shading). Anti-NC levels remained stable during this period and started to increase following the peak of the BA.2 wave. Thus, increasing breadth of neutralization responses across this period appeared multifactorial and may simply represent the effects of the two dominant immunological events in late 2021–early 2022, i.e. a large proportion of the population receiving their third vaccine dose and/or becoming infected with Omicron BA.1.
Assessing the relative threat of the late 2022 Omicron sub-variant swarm across a continuum of individual immunity
Whilst testing of pooled IgG from thousands of U.S. plasma donors provides a snapshot of immunity at a population level, cohort studies where donors have been closely monitored can enable resolution and contributions of vaccination and/or convalescence to the current immune response in the community. As apparent in the pooled IgG approach, there is a continuum of immunity ranging from vaccination and/or infection.
To better understand this, we tested the neutralizing response of serum from the Australian Vaccine, Infection and Immunology Collaborative Research cohort (VIIM) with known vaccination and infection histories who donated in late 2022. As time also appears to play a key role in driving viral breadth, we concurrently tested serum from the ADAPT cohort, in which individuals have been followed longitudinally for over two and a half years following their first infection in early 2020 and subsequent vaccination. Rather than primarily testing peak vaccine or infection-induced responses, we focused on grouping cohort donors based on their current vaccine and/or infection status. This provided a snapshot of neutralization responses at this point in time, and enabled determination of the relative threat of many emerging variants with convergent polymorphisms. For this analysis, we separated donors into four groups; (i) three dose Pfizer mRNA vaccination with a post-vaccination period of approximately six months (Fig. 3C); (ii) three dose Pfizer mRNA vaccination with breakthrough infections primarily during the overlapping BA.2/BA.5 wave (June to August 2022; Fig. 3A); (iii) four doses of primarily Pfizer vaccine, with the last dose within the last three months of testing (Fig. 3B) and; (iv) donors that were infected during March to August 2020 and subsequently received primarily three vaccinations starting in late 2021 and with a Pfizer vaccine boosters in early 2022 (Fig. 3D). All samples from the VIIM cohort (Fig. 3A–C) were collected in September 2022, as per VIIM vaccine cohort protocol. The ADAPT cohort samples (Fig. 3D) were collected over the period of March to September 2022 and thus within 3–6 months post the last vaccine dose. For variant testing, we consolidated testing to the group of variants that sustained the greatest fold drops in neutralization titers relative to the Ancestral variant in pooled IgG studies. This included BQ.1.1, XBB.1, BR.2.1 (+R346T) and the BA.5.2.3/CJ.1 recombinant XBF which was dominant in Victoria, Australia at this time. We also included isolates BL.1, BR.2.1 (346R), BA.4.6, Delta-BA.2 recombinant XBC.1 and BA.5, to determine if limited changes to Spike, such as R346T had significant influence on neutralization titers at the individual level.
BQ.1.1, XBB.1, BR.2.1 and XBF were ranked the most evasive variants across all groups. Whilst three vaccine doses plus infection, four vaccine doses and early convalescent and vaccinated cohorts all observed high titers to the Ancestral clade, they showed a 22-60-fold drop in titers when tested against XBB.1, BQ.1.1, BR.2.1 and XBF (Fig. 3A, B and D). In the cohort with three vaccine doses and a follow up period of six months since the last vaccine dose, we did observe significant declines (p < 0.001) in neutralization titers compared to the other three groups. Fortunately, the fold reduction in titers to all variants tested was not at the same magnitude as that observed across other groups, with only a 12.6-fold drop in neutralization titers from Ancestral to the most evasive variant tested.
Overall and importantly, similar fold drops and variant rankings were observed using the pooled IgG approach as outlined for U.S. plasma donors and as such supports IgG pooling in monitoring neutralization responses across large donor collections (i.e. plasma donation programs and/or national blood banks). If a reduction in neutralization titers will dominate future viral spread, it is likely that multiple contemporary variants will co-exist and persist in the community. At this juncture, the common theme globally is co-circulation of several dominant variants with similar antibody evasive phenotypes (e.g. BQ.1.1, XBB.1.5, CH.1.1 and XBF).
Emerging variants and resistance to clinical monoclonal antibodies
Clinically utilized monoclonal antibodies (mAbs) such as Evusheld and Sotrovimab are important therapeutics to treat COVID-19 in individuals that have not mounted adequate vaccine responses due to therapy-induced or pre-existing immunodeficiencies. Currently, only Evusheld and Sotrovimab are TGA approved for use in Australia. The neutralizing activity of Evusheld (Cilgavimab, Tixagevimab and the Evusheld combination of both) was lost against all variants tested, including BQ.1.1, XBB.1, XBF, BA.2.75.2 and both BR.2.1 lineages (i.e. independent of the R346T polymorphism). Sotrovimab retained neutralization activity against all the above variants except BQ.1.1 and XBF, albeit at a lower potency than A.2.2 (Fig. 4A and B). Given recent studies have observed Sotrovimab neutralization titers to act as poor indicators of in vivo efficacy, we turned to using full length Spike live cell assays1 to sensitively determine if Sotrovimab still bound to variants like XBF and BQ.1.1 at levels consistent with that needed for in vivo efficacy. Unfortunately, Spike binding data revealed a 21 and 52-fold drop (calculated based on the concentration required to obtain 50% binding to Spike) in binding of Sotrovimab to XBF and BQ.1.1 respectively (Fig. 4C) and thus supports the resistance of this variant in neutralization assays. In addition, control experiments using BR.2.1, XBB.1 and BQ.1.1 Spikes with Evusheld further confirmed a lack of Spike binding (Fig. 4C) which supports the mechanism for lack of neutralization observed in these variants.
Changing TMPRSS2 usage in emerging viral variants
Whilst viral evasion of neutralizing antibodies in contemporary samples is a key parameter for assessing potential threat of a variant, other variables can also contribute to increased viral transmission and disease burden in the community. Both ourselves and others have closely monitored changes in viral cell entry requirements and viral cell/tissue tropism associated with emerging variants.6,12,15,16,18,27,28 Of particular interest has been the changing use of the serine protease TMPRSS2 in emerging variants. Changes in TMPRSS2 usage can affect viral tropism, which can influence disease severity in vivo. For instance, whilst Delta and pre-Omicron variants used TMPRSS2 efficiently, the emergence of Omicron variants BA.1 and BA.2 with inefficient TMPRSS2 use was associated changes in viral phenotype. A change with specific preference for epithelial cells derived from the bronchi and the upper respiratory tract was observed.17 Whilst disease severity in the clinic has been tempered significantly by vaccination, disease severity in animal models has been correlated with more efficient TMPRSS2 use.27
As changing TMPRSS2 use is key to this change in tropism and potentially an indicator of disease severity, we developed a real-time means of tracking emerging variant TMPRSS2 use by determining endpoint viral titers in a cell line known as HAT-24, that expresses high levels of ACE2 but low levels of TMPRSS2.18 In variants that use TMPRSS2 efficiently, we see a stepped increase in the linear regression established using endpoint titers of primary diagnostic swabs versus their diagnostic qPCR cycle thresholds value (diagnostic Ct or viral load) (Fig. 5A). The hierarchical ranking of variants by infecting HAT-24 cells and determining viral load can readily segregate the pre-Omicron lineage Delta from the early Omicron lineages BA.1 and BA.2 through a downward shift in the linear regression for the latter early Omicron variants. Upon the arrival of BA.5, we observed an increase of TMPRSS2 use above that of its parent BA.2 but lower than that of Delta.18 To determine the tropism of this intermediate BA.5 phenotype, we compared Delta, BA.2 and BA.5 infections in primary human bronchial- and alveolar epithelial cultures, which were differentiated at the air liquid interface (ALI). Across both bronchial and alveolar epithelial cell cultures, BA.5 reached viral loads, for a given inoculum, above that of its parent BA.2 and approaching that of Delta after 3 days of culture (Fig. 5B). Although after 6 days of culture, we observed a significant drop in Omicron lineages BA.2 and BA.5, whilst Delta viral loads persisted at high levels in both primary epithelial cell models (Fig. 5B). Therefore, we conclude the increased use of TMPRSS2 by BA.5 resulted in an early growth advantage across epithelial cells of both the bronchi and lung parenchyma. Furthermore, persistence of high viral loads at day 6 was only observed with the pre-Omicron variant Delta and may relate to genomic differences in Delta outside of the Spike that enable tempering of innate responses to enable a greater viral load over time.
As BA.5 emerged with an outgrowth advantage globally, we hypothesized that both antibody evasion and better TMPRSS2 use was key in outcompeting other variants. To monitor the TMPRSS2 entry phenotype of emerging variants beyond antibody evasion alone, we worked closely with local diagnostic networks to determine endpoint titers of 305 primary swabs (Supplementary Table S4) over the months of October 2022 through to January 2023. In parallel, we performed whole genome sequencing to link each swab and its endpoint titer to emerging variants. Unlike other waves that we have characterized in the Australian community, we did not observe a dominant variant, but rather the emergence of many genetically distinct sub-variants stemming from BA.5 and BA.2.75 parent lineages (Supplementary Table S4). To resolve this further, we pooled each variant and then determined infectivity to particle ratios based on diagnostic Ct values and endpoint titers. Whilst the sample number of each variant could not lead to statistical testing between variants, the pooled data from BQ variants demonstrated higher infectivity to particle ratios than the pooled variants derived from BA.2.75 (Fig. 5E) using the HAT-24 cell line. Furthermore, monitoring the growth of variants in culture visually demonstrated greater fusogenicity in BQ variants such as BQ.1.2 (Fig. 5C) versus BA.2.75-derived variants like BR.2.1 or BA.2.75.2 (Fig. 5D).
Given the diversity of variant sub-lineages restricted the above approach, we resolved the continuum of TMPRSS2 usage across Omicron sub-variants in expanded isolates. In this approach, viral isolates are titrated on the HAT-24 cell line with excess (determined through dose-dependent titrations with the Delta variant) serine protease inhibitor Nafamostat. When a variant (e.g., Delta) efficiently uses TMPRSS2, the presence of excess Nafamostat significantly reduces viral titers (Fig. 5F) through a left shift in the sigmoidal curve. In contrast, BA.1 and BA.2 utilized TMPRSS2 poorly and, in turn, this resulted in lower reductions in titers in the presence of Nafamostat. Using this approach, we tested 14 emerging variants that are representative of the present variant swarm. Relative to BA.5, all variants with the exception of BQ.1.2 demonstrated similar or reduced reliance on TMPRSS2 use based on viral titers observed in the presence of Nafamostat (Fig. 5F, Supplementary Table S5). As observed in primary nasopharyngeal swabs, there was a trend for BA.2.75 sub-lineages to utilize TMPRSS2 approaching that of BA.2. Of interest, the Delta-BA.2 recombinant XBC.1 was observed to have the lowest TMPRSS2 use, and this is consistent with the majority of the BA.2 Spike (including the RBD and furin cleavage site) being primarily retained in this recombinant. To conclude, using primary swabs and expanded primary isolates, whilst all circulating isolates were engaging TMPRSS2 at levels higher than BA.2, they were at or lower than that observed for BA.5 and importantly not on a trajectory towards that of Delta.
Discussion
Continued genomic and phenotypic surveillance of emerging variants is of importance as communities learn to navigate the complexities of SARS-CoV-2 viral spread. Antibody responses have been key in understanding and predicting vaccine efficacy, and combined with observations of viral fitness and tropism, can be highly predictive in determining the relative threat of an emerging variant. Herein we observe surprising breadth and potency in the population antibody response driven by exposure to pre-Omicron variant Spike proteins delivered with by vaccination and/or infection. While this pre-exposure breadth of antibody responses provides a positive observation with regards viral control and protection from disease progression, we also observe the accumulation and convergence of a variant pool within the community which all share various combinations of common Spike polymorphisms. This variant pool was unique to Australia and was dominated by BR.2.1 and in early 2023 the recombinant XBF. Locally enriched and global variants with increasing Spike polymorphism clusters all had the ability to navigate the current broad and often potent antibody responses. Whilst variants gained the potential to navigate through neutralization responses, this was fortunately countered by concurrent incremental improvements in antibody breadth across recent cohort samples and at the population level using pooled IgGs. At present, it is unclear whether breadth will further increase with the contribution of variant specific vaccines or post-infection with these variants. With respect to viral entry, whilst better utilization of TMPRSS2 can enable variant outgrowth advantage and expanded tropism to epithelial cells of the lower respiratory tract, we observed a continuum of TMPRSS2 use towards that of BA.2 as opposed to the increased efficiency of TMPRSS2 use observed with pre-Omicron variants like Delta.18
The pooling of IgG from large numbers of plasma donations enabled an unprecedented snapshot of a population's response to vaccination and the largest COVID-19 wave, Omicron BA.1. For this approach, this study focused primarily on the U.S. population, which benefited from the early roll out and uptake of many mRNA- and vector-based vaccine doses in early 2021, with doses peaking in April 2021. This early roll out of vaccines was followed by substantial booster uptake in late 2021, just prior to the U.S. Omicron BA.1 wave. Booster uptake during this period provided a signal of increasing neutralization titers in the community whilst accelerating breadth of these responses to boost responses to variants that had yet to enter the community. Whilst the BA.1 wave continued the upward trajectory of neutralization titers, it was not accompanied with increasing breadth to Omicron BA.1 or BA.2 lineages. Though it is tempting to conclude that increasing breadth is supported by vaccine booster uptake, it may simply reflect the finite breadth in the community at that juncture of time that cannot increase until further maturation of the immune response can proceed. Increasing neutralization titers and breadth of mRNA vaccine responses is observed to be relative to that of pre-existing antibody levels, with greater fold increases in antibody potency and breadth observed in those individuals with lower circulating antibodies prior to their vaccine booster dose.29 Thus, greater potency and breadth within the community following an additional vaccine dose and/or infection will be limited with either immune encounter. Fortunately, over 2022, further monitoring of pooled IgGs demonstrated continued increased in variant breadth. Whilst this was primarily against BA.1, BA.2 and BA.5, increasing breadth to emerging variants XBB.1, BQ.1.1 and BR.2.1 was also observed. As was the case prior to the BA.1 wave, vaccination and/or infection can contribute to increased antibody breadth even when a variant is not yet in circulation.
Whilst the use of IgG pooling can give a large population snapshot of antibody-based immunity within the community at a point in time, this approach does not allow resolution of responses at the individual level. Yet in observing many current cohorts, the unifying feature of all responses is greater breadth to the past evasive Omicron variants such as BA.1, BA.2 and BA.5 as well as those emerging within the community. Whilst the waning of antibody responses is of concern and probably important in managing future waves with various vaccine boosters, we observed increased breadth across all Omicron variants even within groups with waning responses (six months post-three mRNA vaccine doses). This is consistent with earlier mRNA vaccine studies, in which neutralizing antibody levels post-second dose stabilized between six and nine months post-vaccination but importantly for the pool of antibodies that persisted, there was continued improvement in their neutralization capacity and breadth over this period.29 This is also consistent with our observations in three vaccine dose donors where the response is six months after the last dose. In this setting, the antibodies that persisted had a greater capacity to neutralize all emerging variants.
Whilst time appears key in greater antibody breadth to the virus, the emergence of the present pool of variants with convergent Spike polymorphisms appears to be the counter response to the immune challenge. In the emerging variants XBB.1, BQ.1.1, BR.2.1 and XBF, we observed significant increases in neutralization evasion despite the developing breadth of the maturing antibody response across all groups. Whilst our cohort data is derived from Australian donors and their unique immunological experiences throughout the pandemic, other similar cohorts in diverse jurisdictions have observed similar fold decreases in variants such as BQ.1.1, XBB.1 and BA.2.75 sub-lineages30, 31, 32, 33, 34, 35, 36 using contemporary serum samples for that country.
Of interest is that global and Australian variants have converged to cluster within the same level of evasiveness but through emergence from genetically diverse parental lineages. XBB.1 derived from a BA.2.10 with BA.2.75 lineage recombination event, XBF is a BA.5.2.3 and CJ.1 recombination (with the latter Spike intact) and BR.2.1 and BQ.1.1 are derived from BA.2.75 and BA.5 lineages, respectively. Of obvious concern is the continuing decay of clinical options for those individuals with a limited antibody response following vaccination and/or infection. The lack of neutralization activity of Evusheld across many emerging variants will need to be considered carefully moving forward with respect to clinical treatment. Whilst Sotrovimab maintained activity across many variants, its trajectory towards lower neutralization activity and Spike binding to globally dominant variants like BQ.1.1 is consistent with recently reported observations.32,35 Whilst the correlates of the in vivo activity for Sotrovimab are complex,37 treatment in the era of a variant mix will provide many challenges and will require continued monitoring. In the latter treatment setting, the continued discovery of monoclonal antibodies and/or combinations thereof is still warranted, but require combinations of antibodies with inherent breadth. In addition, as the community gains greater potency and breadth of antibodies, revisiting the use of polyclonal antibodies38 harvested from plasma donors may also provide a treatment contingency for those at risk. Taken together, the data herein also supports initial boosting of vaccine responses in immunocompetent individuals should be encouraged and monitored where possible.
The trajectory of each variant in each global jurisdiction is being monitored carefully at many levels. Variants like XBB.1 have already shown truncation of case waves in Singapore (https://www.moh.gov.sg/covid-19/statistics) and XBB.1.5 is starting to supplant BQ and BA.5 variants the U.S. (www.ecdc.europa.eu/en/covid-19/country-overviews; www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html; covariants.org/per-country). In this setting, Australia is unique, as the global pool of key variants is presently circulating within the community, alongside those which appear to have been seeded locally (e.g. BR.2.1). Whilst all variants have converged to share similar Spike polymorphisms that enable antibody evasion, it is presently unclear if this alone will enable an outgrowth advantage in any one variant to drive similar case waves as that seen in prior Omicron waves. XBF, for instance, is now the dominant variant in Australia and shares a Spike profile very similar to that of XBB.1.5 with defining Spike polymorphisms such as F486P and F490S, with the former linked to greater affinity for ACE2 binding.39 Key to understanding the potential of the current convergent pool is whether gaining the ability to evade antibodies has sustained a fitness cost in entering cells/tissue. In this setting, we do observe the overall trend of emerging variants to utilize TMPRSS2 less efficiently than pre-Omicron variants. Moving forward, there will be sustained focus on understanding how this presents in the community across the diversity of global pandemic experiences. Whilst changes to the Spike to avoid antibodies can impact upon viral cell-entry fitness into various tissues, additional changes that may compensate for this fitness loss will need to be tracked carefully. For instance, the accumulation of polymorphisms at and around the furin cleavage site, such as I666V in BQ.1.2 may have limited impact in influencing antibody binding, but rather increase viral cell entry fitness and in turn compensate for any fitness loss in cell entry. Whilst viral entry changes are often associated with increased transmission, it is also associated with expansion or contraction of viral tropism and the former has been observed with signals of increased disease severity in animal models27 and in large clinical studies on the recent BA.5 wave.40 Whilst there are many currently circulating variants, variants that maintain entry fitness and evade antibodies concurrently will have a greater outgrowth advantage to supplant the others and/or result in greater levels of disease severity as a consequence of either increased viral loads and/or changing viral tropism to enable infection of the lower respiratory tract. Although viral entry can primarily influence the latter, evasion of innate immunity through the action of non-Spike viral gene products needs to be also monitored carefully and changes thereof may enable high viral load persistence in the respiratory tract and elsewhere to augment events that lead to greater clinical disease severity. Whilst a variant with genomic profile with potential of high disease severity may not arise, continued genomic surveillance and rapid phenotyping as described herein, will be key to identifying any such future variants. A limitation of this study is that it is based primarily on in vitro data and this may not necessarily correlate with future clinical outcomes. Therefore it is imperative that datasets herein and elsewhere are continually assessed alongside clinical observations of therapeutic and vaccine efficacy and presentation of disease severity, so rapid and timely observations herein can act as a guide to the risk potential of emerging variants.
Contributors
Assay was developed and performed by A. Akerman, V. Milogiannakis, A. Aggarwal, C. Eseau, J.A. Lopez, and S.G. Turville. Viruses were isolated and propagated by A. Aggarwal, A. Akerman, V. Milogiannakis, and S.G. Turville. Experiments were performed by A. Aggarwal, A. Akerman, V. Milogiannakis, T. Jean, M.R. Silva, D. Chandra, T. Ison, C. Eseau, C Fitcher, J.A. Lopez, and S.G. Turville. Additional research support including sequencing, mAbs and serum samples were provided by Z. Naing, J. Caguicla, D. Li, G. Walker, S. Amatayakul-Chantler, N. Roth, S. Manni, T. Hauser, T. Barnes, M. Yeang, C.S.P. Foster, A. Condylios, M. Wong, D. Darley, D. Stark, G. Matthews, S. Lee, Y. Song, L. Mao, A. Sigmund, A. Phu, A.M. Vander More, S. Hunt, M. Douglas, I. Caterson, W. Britton, K. Sandgren, R. Bull, A. Lloyd, J. Triccas, S. Tangye, and N.W. Bartlett, Summary of Australian variant analysis was by study analysis was B. Murrell and K. Sato.
Analysis of experiments performed by A. Aggarwal, A. Akerman, V. Milogiannakis, K. Petoumenos, and S.G. Turville. Data was verified by A. Aggarwal, A. Akerman, V. Milogiannakis, and S.G. Turville. The manuscript was drafted by A. Aggarwal, A. Akerman, V. Milogiannakis, A. Kelleher, N.W. Barlett, A.L. Cunningham, and S.G. Turville, The study was supervised by A. Aggarwal, G. Matthews, W.D. Rawlinson, S. Lee, A.L. Cunningham, N.W. Bartlett, A.D. Kelleher, and S.G. Turville. All authors read and approved the final version of the manuscript.
Data sharing statement
Source data for generating the figures are available in the online version of the paper. Any other data are available on request.
Declaration of interests
Funding bodies did not contribute to study design, data collection, data analysis or writing of the manuscript. Study design, data collection, data analysis and data interpretation was performed by the listed authors. Writing and review was performed by the listed authors.
A.L.C. Participated on the AstraZeneca COVID-19 Advisory Board (2021), Seqirus COVID Advisory Board (2020) & CSL Seqirus APAC Advisory Council (2022). N.R., T.B., S.A.C., S.M., and T.H. are employees and shareholders of CSL LTD, a manufacturer of the polyclonal immunoglobulin preparations used as a reagent within the study.
Acknowledgement
This work was primarily supported by Australian Medical Foundation research grants MRF2005760 (SGT, GM & WDR), Medical Research Future Fund Antiviral Development Call grant (WDR), the New South Wales Health COVID-19 Research Grants Round 2 (SGT & FB) and the NSW Vaccine Infection and Immunology Collaborative (VIIM) (ALC). Variant modeling was supported by funding from SciLifeLab's Pandemic Laboratory Preparedness program to B.M. (VC-2022-0028) and by the European Union's Horizon 2020 research and innovation programme under grant agreement no. 101003653 (CoroNAb) to B.M.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104545.
Appendix A. Supplementary data
Supplementary Fig. S1. Effect of age on neutralization titres against emerging variants in vaccinated and infected donors. A and B. Three dose Pfizer BNT162b2 vaccination with subsequent breakthrough infection in donors below 50 years (A) and above 50 years (B). Closed circles were infections between June and August 2022 when BA.2 and BA.5 were prevalent. Open circles are breakthrough infections with BA.1 between January and February. C and D. Primarily four dose BNT162b2 vaccinations, in which the last dose was within three months in donors below 50 years (C) and above 50 years (D). Open circles in this group are breakthrough infections in early 2022 at the time of BA.1. E and F. Three dose Pfizer BNT162b2 vaccination with the last dose six months prior in donors below 50 years (E) and above 50 years (F). G and H. Early 2020 donors infected between March and August of 2020 and then subsequently vaccinated with two doses of Pfizer BNT162b2 or AstraZeneca AZD1222/ChAdOx1 and boosted with Pfizer BNT162b2 or Moderna mRNA-1273. This group did not receive their last dose three to six months prior, shown in (G) are donors below 50 years and in (F) donors above 50 years. Data in (A–F) indicates the mean IC50 of technical replicates for individual samples. The median titers are labelled. n, indicates number of donors within each grouping.
Supplementary Fig. S2. Effect of gender on neutralization titres against emerging variants in vaccinated and infected donors. A and B. Three dose Pfizer BNT162b2 vaccination with subsequent breakthrough infection in male donors (A) and female donors (B). Closed circles were infections between June and August 2022 when BA.2 and BA.5 were prevalent. Open circles are breakthrough infections with BA.1 between January and February. C and D. Primarily four dose BNT162b2 vaccinations, in which the last dose was within three months in male donors (C) and female donors (D). Open circles in this group are breakthrough infections in early 2022 at the time of BA.1. E and F. Three dose Pfizer BNT162b2 vaccination with the last dose six months prior in male donors (E) and female donors (F). G and H. Early 2020 donors infected between March and August of 2020 and then subsequently vaccinated with two doses of Pfizer BNT162b2 or AstraZeneca AZD1222/ChAdOx1 and boosted with Pfizer BNT162b2 or Moderna mRNA-1273. This group did not receive their last dose three to six months prior, shown in (G) are male donors and in (F) female donors. Data in (A–F) indicates the mean IC50 of technical replicates for individual samples. The median titers are labelled. n, indicates number of donors within each grouping.
References
- 1.Tea F., Ospina Stella A., Aggarwal A., et al. SARS-CoV-2 neutralizing antibodies: longevity, breadth, and evasion by emerging viral variants. PLoS Med. 2021;18(7) doi: 10.1371/journal.pmed.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karbiener M., Farcet M.R., Schwaiger J., et al. Plasma from post-COVID-19 and COVID-19-vaccinated donors results in highly potent SARS-CoV-2 neutralization by intravenous immunoglobulins. J Infect Dis. 2021;224(10):1707–1711. doi: 10.1093/infdis/jiab482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaebler C., Wang Z., Lorenzi J.C.C., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dan J.M., Mateus J., Kato Y., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529) doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreer C., Zehner M., Weber T., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182(6):1663–1673. doi: 10.1016/j.cell.2020.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito A., Irie T., Suzuki R., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602(7896):300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng B., Abdullahi A., Ferreira I., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603(7902):706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCallum M., Czudnochowski N., Rosen L.E., et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375(6583):864–868. doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wall E.C., Wu M., Harvey R., et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397(10292):2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suryadevara N., Shrihari S., Gilchuk P., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184(9):2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh Y., Fuloria N.K., Fuloria S., et al. N-terminal domain of SARS CoV-2 spike protein mutation associated reduction in effectivity of neutralizing antibody with vaccinated individuals. J Med Virol. 2021;93(10):5726–5728. doi: 10.1002/jmv.27181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mlcochova P., Kemp S.A., Dhar M.S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez Bernal J., Andrews N., Gower C., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B., Chan J.F., Liu H., et al. Spike mutations contributing to the altered entry preference of SARS-CoV-2 omicron BA.1 and BA.2. Emerg Microbes Infect. 2022;11(1):2275–2287. doi: 10.1080/22221751.2022.2117098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willett B.J., Grove J., MacLean O.A., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022;7(8):1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal A., Stella A.O., Walker G., et al. Platform for isolation and characterization of SARS-CoV-2 variants enables rapid characterization of Omicron in Australia. Nat Microbiol. 2022;7(6):896–908. doi: 10.1038/s41564-022-01135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui K.P.Y., Ho J.C.W., Cheung M.C., et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603(7902):715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 18.Aggarwal A., Akerman A., Milogiannakis V., et al. SARS-CoV-2 Omicron BA.5: evolving tropism and evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. EBioMedicine. 2022;84 doi: 10.1016/j.ebiom.2022.104270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stucki M., Boschetti N., Schafer W., et al. Investigations of prion and virus safety of a new liquid IVIG product. Biologicals. 2008;36(4):239–247. doi: 10.1016/j.biologicals.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Sheward D.J., Kim C., Fischbach J., et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect Dis. 2022;22(11):1538–1540. doi: 10.1016/S1473-3099(22)00663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J Virol. 2016;5(2):85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bull R.A., Adikari T.N., Ferguson J.M., et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun. 2020;11(1):6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker G.J., Naing Z., Ospina Stella A., et al. SARS coronavirus-2 microneutralisation and commercial serological assays correlated closely for some but not all enzyme immunoassays. Viruses. 2021;13(2) doi: 10.3390/v13020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George P.M., Reed A., Desai S.R., et al. A persistent neutrophil-associated immune signature characterizes post-COVID-19 pulmonary sequelae. Sci Transl Med. 2022;14(671) doi: 10.1126/scitranslmed.abo5795. [DOI] [PubMed] [Google Scholar]
- 25.Loo S.L., Wark P.A.B., Esneau C., Nichol K.S., Hsu A.C., Bartlett N.W. Human coronaviruses 229E and OC43 replicate and induce distinct antiviral responses in differentiated primary human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2020;319(6):L926–L931. doi: 10.1152/ajplung.00374.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes C.O., Jette C.A., Abernathy M.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603(7902):687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Valtanen P., Hope C.M., Masavuli M.G., et al. SARS-CoV-2 Omicron variant escapes neutralizing antibodies and T cell responses more efficiently than other variants in mild COVID-19 convalescents. Cell Rep Med. 2022;3(6) doi: 10.1016/j.xcrm.2022.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goel R.R., Painter M.M., Lundgreen K.A., et al. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. Cell. 2022;185(11):1875–1887.e8. doi: 10.1016/j.cell.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X.L., Zhu K.L., Wang X.J., et al. Omicron BQ.1 and BQ.1.1 escape neutralisation by omicron subvariant breakthrough infection. Lancet Infect Dis. 2023;23(1):28–30. doi: 10.1016/S1473-3099(22)00805-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann M., Behrens G.M.N., Arora P., et al. Effect of hybrid immunity and bivalent booster vaccination on omicron sublineage neutralisation. Lancet Infect Dis. 2023;23(1):25–28. doi: 10.1016/S1473-3099(22)00792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora P., Kempf A., Nehlmeier I., et al. Omicron sublineage BQ.1.1 resistance to monoclonal antibodies. Lancet Infect Dis. 2023;23(1):22–23. doi: 10.1016/S1473-3099(22)00733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q., Iketani S., Li Z., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2022;186(2):279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurhade C., Zou J., Xia H., et al. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat Med. 2022;29(2):344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 35.Cao Y., Jian F., Wang J., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2022;614(7948):521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arunachalam P.S., Lai L., Samaha H., et al. Durability of immune responses to the booster mRNA vaccination against COVID-19. medRxiv. 2023 doi: 10.1172/JCI167955. e167955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case J.B., Mackin S., Errico J.M., et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. Nat Commun. 2022;13(1):3824. doi: 10.1038/s41467-022-31615-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Group I.S. Hyperimmune immunoglobulin for hospitalised patients with COVID-19 (ITAC): a double-blind, placebo-controlled, phase 3, randomised trial. Lancet. 2022;399(10324):530–540. doi: 10.1016/S0140-6736(22)00101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue C., Song W., Wang L., et al. Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. bioRxiv. 2023;23(3):278–280. doi: 10.1016/S1473-3099(23)00010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen C.H., Friis N.U., Bager P., et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 omicron subvariant: a nation-wide population-based study in Denmark. Lancet Infect Dis. 2023;23(2):167–176. doi: 10.1016/S1473-3099(22)00595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Effect of age on neutralization titres against emerging variants in vaccinated and infected donors. A and B. Three dose Pfizer BNT162b2 vaccination with subsequent breakthrough infection in donors below 50 years (A) and above 50 years (B). Closed circles were infections between June and August 2022 when BA.2 and BA.5 were prevalent. Open circles are breakthrough infections with BA.1 between January and February. C and D. Primarily four dose BNT162b2 vaccinations, in which the last dose was within three months in donors below 50 years (C) and above 50 years (D). Open circles in this group are breakthrough infections in early 2022 at the time of BA.1. E and F. Three dose Pfizer BNT162b2 vaccination with the last dose six months prior in donors below 50 years (E) and above 50 years (F). G and H. Early 2020 donors infected between March and August of 2020 and then subsequently vaccinated with two doses of Pfizer BNT162b2 or AstraZeneca AZD1222/ChAdOx1 and boosted with Pfizer BNT162b2 or Moderna mRNA-1273. This group did not receive their last dose three to six months prior, shown in (G) are donors below 50 years and in (F) donors above 50 years. Data in (A–F) indicates the mean IC50 of technical replicates for individual samples. The median titers are labelled. n, indicates number of donors within each grouping.
Supplementary Fig. S2. Effect of gender on neutralization titres against emerging variants in vaccinated and infected donors. A and B. Three dose Pfizer BNT162b2 vaccination with subsequent breakthrough infection in male donors (A) and female donors (B). Closed circles were infections between June and August 2022 when BA.2 and BA.5 were prevalent. Open circles are breakthrough infections with BA.1 between January and February. C and D. Primarily four dose BNT162b2 vaccinations, in which the last dose was within three months in male donors (C) and female donors (D). Open circles in this group are breakthrough infections in early 2022 at the time of BA.1. E and F. Three dose Pfizer BNT162b2 vaccination with the last dose six months prior in male donors (E) and female donors (F). G and H. Early 2020 donors infected between March and August of 2020 and then subsequently vaccinated with two doses of Pfizer BNT162b2 or AstraZeneca AZD1222/ChAdOx1 and boosted with Pfizer BNT162b2 or Moderna mRNA-1273. This group did not receive their last dose three to six months prior, shown in (G) are male donors and in (F) female donors. Data in (A–F) indicates the mean IC50 of technical replicates for individual samples. The median titers are labelled. n, indicates number of donors within each grouping.