Summary
Here, we focus on tumor-associated macrophages (TAMs) in the PyMT model of breast cancer, detailing a protocol for assessing antigen presentation capabilities of immune populations of interest. We describe a stringent bone marrow chimera system to demonstrate presentation of exogenous antigen that is acquired and processed in the tumor microenvironment. We describe steps for testing antigen presentation activity of TAMs to CD8+ T cells in vivo and ex vivo and the requirement for the transcription factor IRF8 in this function.
For complete details on the use and execution of this protocol, please refer to Nixon et al. (2022).1
Subject areas: Cell Biology, Cell culture, Cell isolation, Flow Cytometry/Mass Cytometry, Cancer, Immunology, Model Organisms
Graphical abstract

Highlights
-
•
Tests APC activity of tumor-associated macrophages to CD8+ T cells in vivo and ex vivo
-
•
Immune cells differentiate and acquire antigen in vivo in the tumor microenvironment
-
•
Bone marrow chimera system ensures antigen is exogenous and tumor–associated
-
•
Tests requirement for IRF8 in APC capabilities; can be modulated to test other factors
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we focus on tumor-associated macrophages (TAMs) in the PyMT model of breast cancer, detailing a protocol for assessing antigen presentation capabilities of immune populations of interest. We describe a stringent bone marrow chimera system to demonstrate presentation of exogenous antigen that is acquired and processed in the tumor microenvironment. We describe steps for testing antigen presentation activity of TAMs to CD8+ T cells in vivo and ex vivo and the requirement for the transcription factor IRF8 in this function.
Before you begin
The protocol below describes how to test antigen presenting cell (APC) capabilities of tumor-associated macrophages (TAM) to CD8+ T cells in the PyMT model of breast cancer, and the role the gene Irf8 plays in this function. It also tests this capability in migratory type 1 dendritic cells (mDC1) from the tumor-draining lymph node. However, this protocol can be modified by replacing IRF8-knockout mouse strains with other genetic manipulations to test the role of other factors on APC activity in TAM and mDC1. Additionally, one can test the APC function of other immune populations of interest; the antibodies and gating strategy used in the sorting experiments can be modified to meet this end.
Institutional permissions
All animal experiments require approval by institutional IACUC committee and should be performed in accordance with the appropriate guidelines and regulations. Make sure to have acquired permission from the institutional IACUC committee before proceeding.
Breed and obtain relevant mice
Timing: weeks to months
-
1.Generate relevant mouse cohorts for experiments.
-
a.Obtain S100a8Cre, LSL-USA, PyMT (all CD45.2+) and CD45.1+ C57BL/6 mouse strains. Cross alleles in order to generate S100a8CreLSL-USAPyMT (CD45.1+CD45.2+) female mice as recipients for bone marrow chimera experiments, which may take multiple generations (Figure 1A).Note: S100a8Cre targets Cre expression to S100a8+ cells, which include neutrophils in the immune compartment and tumor cells in the mammary gland of PyMT mice. LSL-USA is a transgenic mouse strain expressing the Universal Self Antigen (USA) cassette, which includes SIINFEKL peptide from chicken ovalbumin, downstream of a floxed stop cassette (LSL). MMTV-PyMT (PyMT) mice express the polyoma middle T oncogene (PyMT) under the control of the MMTV promoter, which targets expression to the mammary epithelium. CD45.1.1 mice are C57BL/6 mice with an altered CD45 protein that can be detected and differentiated from CD45.2.2 C57BL/6 mice via flow cytometry. These can also be bred together to generate CD45.1.2+ mice. Flow cytometry can differentiate congenic status of immune cells of interest (either CD45.1+CD45.2-, CD45.1-CD45.2+, or CD45.1+CD45.2+), which is useful when determining the source of a cell, e.g., in mixed bone marrow chimeras.Note: When combined, S100a8CreLSL-USAPyMT mice will grow mammary tumors, and these mice will have defined tumor-associated antigen, including the model antigen SIINFEKL. Irradiation and bone marrow transfers will eliminate USA-expressing neutrophils, leaving the antigen as only tumor-associated.
-
b.Obtain CD11cCre, Irf8fl, and C57BL/6 (CD45.1.1) mouse strains. Generate CD11cCreIrf8fl/fl (CD45.2.2) and C57BL/6 CD45.1.1 cohorts of female mice as donors for the mixed bone marrow chimeras for the ex vivo presentation assay (Figures 1B and 2A).Note: CD11cCre targets Cre expression to CD11c+ cells, which includes TAMs and DC1s in the PyMT model. Irf8fl/fl has floxed alleles for Irf8, allowing for deletion in Cre+ cells. CD11cCreIrf8fl/fl mice will delete Irf8 in CD11c+ cells, including dendritic cells and TAMs. DC1 in the PyMT model will be lost, as they require IRF8,1,2,3 and TAM maturation will be disrupted.1
-
c.Obtain MafBiCre, Irf8fl, and C57BL/6 mouse strains. Generate MafBiCreIrf8fl/fl (CD45.2.2) and C57BL/6 (CD45.2.2) female mouse cohorts as donor mice for the bone marrow chimeras for the in vivo presentation assay (Figures 1C and 2B).Note: MafBiCre targets cre expression to MafB+ cells, specifically monocytes and macrophages, including TAMs, in the PyMT model. MafBiCreIrf8fl/fl mice will delete IRF8 in TAMs but not in DCs, thus allowing for a normal DC1 compartment and an IRF8-deleted TAM compartment.1
-
d.Obtain OT-I mouse strain. Cross OT-I mice onto the CD45.1+ background.Note: OT-I mice express the OT-I transgenic T cell receptor. All cells in the T cell compartment of these mice are CD8+ OT-I-expressing cells that can recognize SIINFEKL peptide in the context of H-2Kb allele of MHC-I.Note: All strains used here are on the C57BL/6 background, and thus express the K-2Kb and H-2Db alleles of MHC-I.
-
a.
-
2.Analyze blood to confirm CD45.1 versus CD45.2 congenic status of mice during breedings and before beginning experiments.Note: Genotyping via PCR does not differentiate CD45.1 from CD45.2, it needs to be checked by flow cytometry. Collect blood from known CD45.2+ and known CD45.1+ mice as positive controls.
-
a.Collect blood (less than 50 μL) via tail vein nick of mice.
-
i.Warm mouse by putting under heat lamp for 5 min.
-
ii.Place mouse in restrainer and wipe tail with alcohol wipe.
-
iii.Create small nick on tail vein at distal end of tail with a disposable scalpel.
-
iv.Collect blood with a heparinized capillary tube.
-
v.Place capillary tube with blood into 1.5 mL Eppendorf tube pre-filled with 5 μL 0.1 M EDTA to prevent clotting.
-
vi.Wrap mouse tail with paper towel and apply pressure at point of nick to stop bleeding before returning mouse to cage.
-
i.
-
b.Flush blood from capillary tube by pipetting ∼800 μL red blood cell (RBC) lysis buffer into top of capillary tube, collecting it into Eppendorf tube below.
-
c.Let sit for 5 min at 20°C–25°C.
-
d.Spin down tubes for 6 min at 550 rcf and 4°C.
-
e.Discard supernatant and resuspend pellet in 1 mL chilled PBS to wash cells.
-
f.Spin down tubes for 6 min at 550 rcf and 4°C.
-
g.Discard supernatant and resuspend pellet in 50 μL antibody cocktail in FACS buffer, transfer to wells in 96 well U-bottom plate.Note: Sample antibody panel for checking CD45 congenic status: FcBlock (1 μg per 50 μL), CD45.1 - APC, CD45.2 – v450, and viability dye (GhostDye Violet 510) – BV510.
-
h.Incubate on ice for 30 min, protected from light.
-
i.Add 150 μL FACS buffer to each well and spin down plate for 3 min at 700 rcf and 4°C.
-
j.Resuspend sample in 100 μL FACS buffer. Transfer sample through piece of nylon mesh material into 1.2 mL polypropylene microtiter tube.
-
k.Run sample on flow cytometer.
-
l.Analyze data using FlowJo software, gating on live cells and measuring expression of CD45.1 and CD45.2 (Figure 3).
-
a.
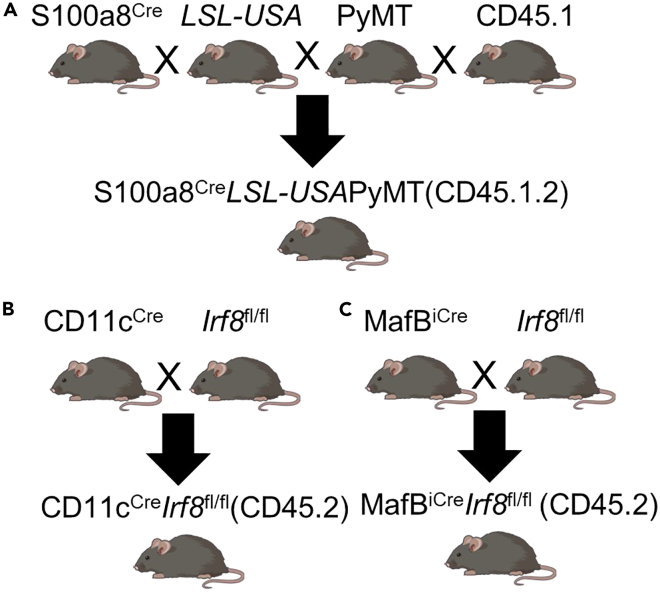
Figure 1.
Mouse breeding schematics
(A) Generation of S100a8CreLSL-USAPyMT (CD45.1.2) mice for use as recipients for bone marrow chimera experiments.
(B) Generation of CD11cCreIrf8fl/fl mice to be used as bone marrow donors, mixed with bone marrow from C57BL/6 (CD45.1.1) mice, for ex vivo tumor antigen presentation assays.
(C) Generation of MafBiCreIrf8fl/fl mice to be used as bone marrow donors for in vivo tumor antigen presentation assays.
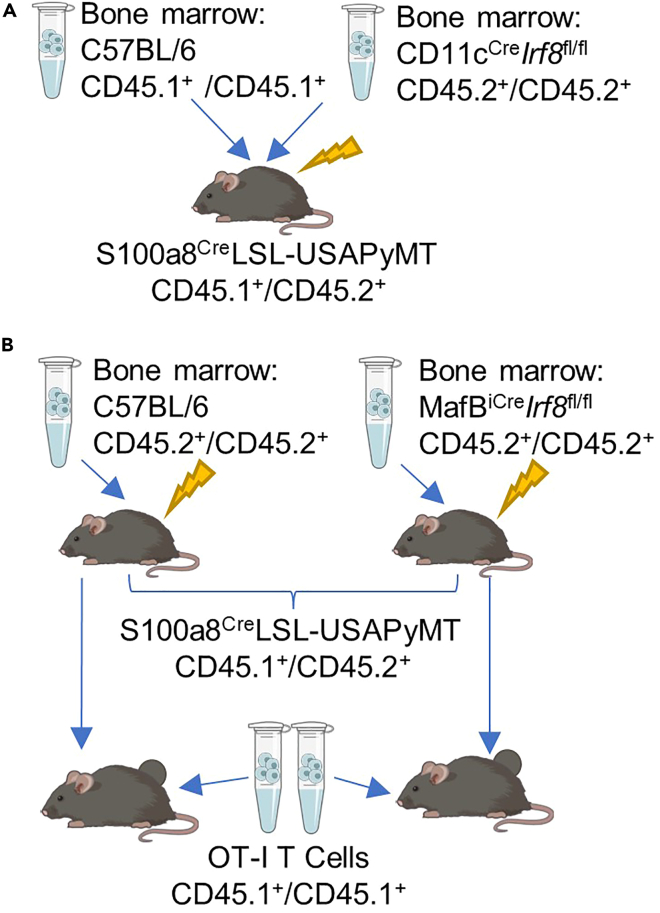
Figure 2.
Experimental designs of mouse bone marrow chimeras
(A) Experimental design of ex vivo antigen presentation assay, generating a mixed bone marrow chimera with S100a8CreLSL-USAPyMT (CD45.1/CD45.2+) female mouse as the recipient and bone marrow mixed 1:1 from C57BL/6 (CD45.1/CD45.1+) and CD11cCreIrf8fl/fl(CD45.2/CD45.2+) donor mice. This mouse is aged, allowing bone marrow to graft, tumors to grow, and TAMs to differentiate in the tumor, derived from both bone marrow sources (either CD45.1.1+ or CD45.2.2+). Once the mice have a large tumor burden, the TAMs are sorted out of the tumor and cocultured with OT-I CD8+ T cells to test TAM APC ability.
(B) Experimental design of in vivo antigen presentation assay, generating two types of bone marrow chimeras, with S100a8CreLSL-USAPyMT (CD45.1/CD45.2+) female mice as the recipients, and one cohort receiving bone marrow from C57BL/6 (CD45.2/CD45.2+) mice, and another cohort receiving bone marrow from MafBiCreIrf8fl/fl(CD45.2/CD45.2+) mice. After ∼8–12 weeks, once mice have begun growing tumors, OT-I CD8+ T cells (CD45.1/CD45.1+) are transferred into the mice. After 2 weeks, the mice are euthanized and the T cells are phenotyped using flow cytometry.
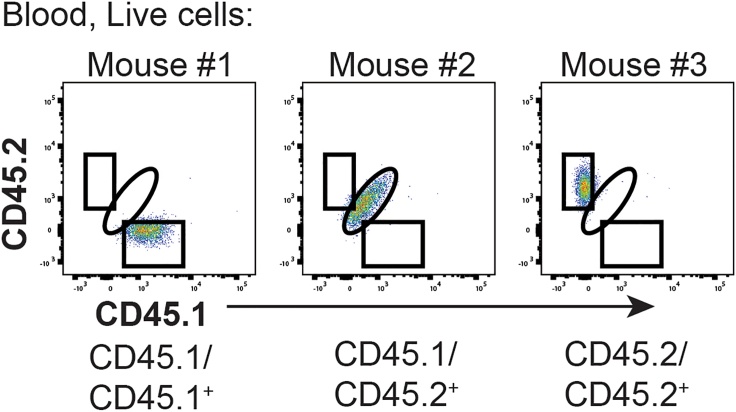
Figure 3.
Representative flow cytometric analysis for checking CD45.1/CD45.2 status of immune cells
After collecting, processing, and staining blood with antibodies for flow cytometry, samples are run on a flow cytometer and data are analyzed, as shown here with blood samples from three mice. First, each sample is gated on FSC-A and SSC-A and then dead cells are excluded (not shown), and resulting cells are analyzed for CD45.1 and CD45.2 expression (shown here). Each mouse will be either CD45.1/CD45.1+ (left), CD45.1/CD45.2+ (middle), or CD45.2/CD45.2+ (right).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD103 – APC (1:200) | BD Biosciences | Clone: M290; Cat#: 562772 |
| CD11b – BV711 (1:400) | BD Biosciences | clone: M1/70; Cat#: 563168 |
| CD11c – BV421 (1:500) | BD Biosciences | clone: N418; Cat#: 565452 |
| CD45 – PerCPCy5.5 (1:200) | BD Biosciences | clone: 30-F11; Cat# 550994 |
| CD45.1 – APC (1:100) | BioLegend | clone: A20; Cat# 110714 |
| CD45.1 – BV650 (1:100) | BioLegend | clone: A20; Cat# 110736 |
| CD45.1 – APC-Cy7 (1:400) | Tonbo Biosciences | clone: A20; Cat# 25-0453-U100 |
| CD45.2 – vF450 (1:100) | Tonbo Biosciences | clone: 104; Cat# 65-0454-U100 |
| H-2Kb bound to SIINFEKL – PE (1:200) | BioLegend | clone: 25-D1.16; RRID: AB_10895905 |
| XCR1 – BV510 (1:200) | BioLegend | clone: ZET; RRID: AB_2565231 |
| PD-1 – FITC (1:200) | eBioscience/Thermo | clone: RMP1-30; RRID: AB_465467 |
| CD8α – BV650 (1:200) | BD Biosciences | clone: 53-6.7; Cat# 563234 |
| IRF8 – APC (1:200) | eBioscience/Thermo | clone: V3GYWCH; Cat# 17-9852-82 |
| F4/80 – PE-CF594 (1:200) | BD Biosciences | Clone: T45-2342; Cat# 565613 |
| Granzyme B – APC (1:100) | Thermo Scientific | Clone: GB11; Cat# GRB05 |
| Ly6C – PE-Cy7 (1:200) | BD Biosciences | Clone: AL-21; Cat# 560593 |
| Ly6G – PE-Cy7 (1:800) | Tonbo Biosciences | Clone: IA8; Cat# 60-1276-U100 |
| SiglecF – PE-Cy7 (1:200) | BioLegend | Clone: S17007L; Cat# 155527 |
| TCRβ – PE-Cy7 (1:400) | Tonbo Biosciences | Clone: H57-597; Cat# 60-5961-U100 |
| CellTrace Violet | Invitrogen | Cat# C34557 |
| Ghost Dye Violet 510 (1:1000) | Tonbo Biosciences | Cat# 13-0870-T500 |
| Ghost Dye Red 780 (1:1000) | Tonbo Biosciences | Cat# 13-0865-T500 |
| FcR Block | Bio X Cell | Clone 2.4G2; RRID: AB_2736987 |
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase type 3 | Worthington Biochemical | Cat# LS004183 |
| Ammonium chloride | Sigma-Aldrich | Cat# A9434-1KG |
| Potassium bicarbonate | Sigma-Aldrich | Cat# 237205 |
| Dnase I | STEMCELL | Cat# 100-0762 |
| Percoll | Sigma-Aldrich | Cat# P1644-1L |
| EDTA | MilliporeSigma | Cat# E4884-500G |
| Bovine serum albumin (BSA) | Fisher Scientific | Cat# BP1605-100 |
| Fetal bovine serum (FBS) | Fisher Scientific | Cat# FB12999102 |
| 10× PBS | Fisher Scientific | Cat# BP3994 |
| RPMI | Fisher Scientific | Cat# 11875093 |
| Pen/Strep | Fisher Scientific | Cat# 15140122 |
| L-Glutamine | Fisher Scientific | Cat# 35050061 |
| Non-essential amino acids | Fisher Scientific | Cat# 11140050 |
| Sodium pyruvate | Fisher Scientific | Cat# 11360070 |
| Hepes | Fisher Scientific | Cat# 15630080 |
| Beta-mercaptoethanol | Fisher Scientific | Cat# 21985023 |
| Sodium azide | Sigma-Aldrich | Cat# S2002 |
| PBS w/o Ca2+, Mg2+ | Fisher Scientific | Cat# 14190144 |
| 0.4% Trypan blue | Fisher Scientific | Cat# 15250061 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| OVA(257-264) | Invivogen | Cat# Vac-sin |
| Critical commercial assays | ||
| FoxP3/Transcription factor Fix/Perm Kit | Tonbo Biosciences | Cat# TNB-0607-KIT |
| EasySep Magnet, 2.5 × 10ˆ8 cells | EasySep | Cat# 18000 |
| EasySep Naïve CD8 T cell Negative Selection Kit | EasySep, Stem Cell Technologies | Cat# 19858 |
| Experimental models: Organisms/strains | ||
| Mouse: B6.Cg-Tg(Itgax-cre)1-1Reiz/J (CD11cCre) (Received from Dr. Boris Reizis Lab) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 008068 |
| Mouse: B6.SJL-PtprcaPepcb/BoyJ (CD45.1) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 002014 |
| Mouse: B6(Cg)-Irf8tm1.1Hm/J (Irf8flox) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 014175 |
| Mouse: B6N(129S4)-Mafbtm1.1(cre)Kmm/J (MafBiCre) (Received from Dr. Kenneth Murphy Lab) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 029664 |
| Mouse: C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 003831 |
| Mouse: B6.FVB-Tg(MMTV-PyVT)634Mul/LellJ (PyMT) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 022974 |
| Mouse: B6.Cg-Tg(S100A8-Cre,-EGFP) 1Ilw/J (S100a8Cre) (C57BL/6 background, use females for experiments) | Jackson Laboratories | Stock: 021614 |
| USA-LSL (“Universal Self Antigen”, Received from Dr. James J. Moon Lab) (C57BL/6 background, use females for experiments) | Zhang Z. et al., 2019 Eur J Immunol. | N/A |
| Software and algorithms | ||
| GraphPad Prism V.7-9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo V.9, V.10 | Treestar | https://www.flowjo.com/solutions/flowjo/downloads |
| FacsDIVA | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| Other | ||
| Nylon mesh material to filter samples | Fisher Scientific | Cat# NC0366417 |
| 70 μm cell strainers | Fisher Scientific | Cat# 22-363-548 |
| 60 × 15 mm cell culture dish | USA Scientific | Cat# CC7682-3359 |
| 65 mL mortar | Fisher Scientific | Cat# FB961B |
| Pestle | Fisher Scientific | Cat# FB961L |
| Disposable scalpel | Fisher Scientific | Cat# S17800 |
| Alcohol prep pad | Fisher Scientific | Cat# 22-363-750 |
| Mouse restrainer | Fisher Scientific | Cat# NC0690443 |
| Heparinized capillary tube | Fisher Scientific | Cat# 22-260950 |
| 15 mL conical tube flip cap | Fisher Scientific | Cat# 362694NS |
| 50 mL conical tube flip cap | Fisher Scientific | Cat# 362697NS |
| 1.5 mL Eppendorf tubes | USA Scientific | Cat# 1615-5500 |
| 1.2 mL polypropylene microtiter tube | Fisher Scientific | Cat# 02-681-376 |
| 5 mL polypropylene test tube (Corning) | Fisher Scientific | Cat# 14-959-11A |
| Glass slides | Fisher Scientific | Cat# 2948-75X25 |
| 500 μL insulin syringe | BD Biosciences | Cat# 324911 |
| 3 mL syringe | Fisher Scientific | Cat# 14-955-457 |
| Sulfatrim mouse chow | WF Fisher and Son | Cat# TD1810356-293 |
| Hemacytometer | Fisher Scientific | Cat# 02-671-6 |
| 96 well U-bottom plate | USA Scientific | Cat# 5665-0201 |
| Biological safety cabinet | Fisher Scientific | Cat# 13-261-222 |
| Widefield light microscope | Zeiss | Axiovert model |
| Sorvall Legend XTR Centrifuge | Fisher Scientific | Cat# 75004532 |
| Eppendorf 5430 R Centrifuge | Fisher Scientific | Cat# 05-400-018 |
| Corning LSE Water Bath 6L | Fisher Scientific | Cat# 07-202-156 |
| Nuaire autoflow IR water-jacketed incubator | Nuaire | Cat# NU-5720 |
| Fortessa | BD | |
| LSR-II | BD | |
| FACSAria | BD | |
Materials and equipment
Red Blood Cell Lysis Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Distilled water | 1,000 mL | |
| Ammonium chloride (NH4Cl) | 0.155 M | 8.29 g |
| Potassium bicarbonate (KHCO3) | 0.012 M | 1.2 g |
| EDTA | 0.1 mM | 37.2 mg |
| Total | pH = 7.2–7.4 | 500 mL |
Store at 20°C–25°C for six months.
Magnetic Bead Selection Kit Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS w/o Ca2+, Mg2+ | 485 mL | |
| FBS (−20°C) | 2% | 10 mL |
| EDTA (0.1 M) | 1 mM | 5 mL |
| Total | 500 mL |
Keep sterile, store at 4°C for one month.
Percoll Gradient: 44% Percoll
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll (4°C) | 19.8 mL | |
| 10× PBS | 2.2 mL | |
| 1× PBS | 28 mL | |
| Total | 50 mL |
Make fresh for experiment, can store Percoll + 10× PBS mixture at 4°C for two weeks or until it becomes contaminated.
Percoll Gradient: 66% Percoll
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll (4°C) | 29.7 mL | |
| 10× PBS | 3.3 mL | |
| 1× PBS | 17 mL | |
| Total | 50 mL |
Make fresh for experiment, can store Percoll + 10× PBS mixture at 4°C for two weeks or until it becomes contaminated.
T Cell Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI (stored at 4°C) | 425 mL | |
| FBS (stored at −20°C) | 10% | 50 mL |
| Pen/Strep | 10 μg/mL | 5 mL |
| Glutamine | 2 mM | 5 mL |
| Non-essential amino acids | 0.1 mM | 5 mL |
| Pyruvate | 1 mM | 5 mL |
| Hepes | 10 mM | 5 mL |
| Beta-mercaptoethanol | 50 uM | 500 μL |
| Total | 500 mL |
Keep sterile, store at 4°C for one month.
FACS Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 500 mL | |
| FBS (stored at −20°C) | 1% | 5 mL |
| Sodium Azide (NaN3) | 0.02% | 2 mL |
| Total | N/A | 500 mL |
Store at 4°C for one month.
Sorting Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 490 mL | |
| BSA (filtered) | 0.5% | 5 mL |
| Pen/Strep | 10 μg/mL | 5 mL |
| Total | 500 mL |
Store at 4°C for one month.
Sorting collection medium
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 20 mL | |
| FBS (stored at −20°C) | 33% | 10 mL |
| Total | 30 mL |
Make fresh day of experiment, keep sterile, store on ice or at 4°C.
Tonbo Fixation/Permeabilization Working Solution (“Fix/Perm”)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tonbo Fix/Perm Diluent (1×) | 3 mL | |
| Tonbo Fix/Perm Concentrate (4×) | 25% | 1 mL |
| Total | 4 mL |
Make fresh day of experiment with appropriate volume required for flow cytometry samples, store on ice or at 4°C.
Tonbo Perm Buffer Working Solution (“Fix/Perm Buffer”)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tonbo Perm Buffer (10×) | 10% | 1 mL |
| Distilled water | 9 mL | |
| Total | 10 mL |
Make fresh day of experiment with appropriate volume required for flow cytometry samples, store on ice or at 4°C.
Note: Tonbo Fix/Perm Diluent (1×), Concentrate (4×), and Perm Buffer (10×) are part of the Tonbo Fix/Perm Transcription Factor Kit.
Alternatives: Alternatives to the flow antibodies listed can be used, for instance if other colors are needed for a certain antibody panel. The same clones come in different colors and are sold by various companies. When buying a new antibody color, it should be tested to determine optimal dilution for proper brightness in the experiment, usually ranging between 1:100 to 1:1000.
Alternatives: For acquiring flow cytometry data, alternative equipment can be used instead of the BD LSR-II or Fortessa. This includes machines such as the Cytek Aurora.
Step-by-step method details
Ex vivo tumor antigen presentation assay
Timing: 8–12 weeks, depending on tumor growth rate
This major step allows for the generation of a mixed bone marrow chimera system designed to test the ability of antigen-presenting immune cells to have picked up, processed, and presented exogenous tumor antigen in vivo (Figure 2A). This is done by measuring surface expression of SIINFEKL peptide in the context of the H-2Kb allele of MHC-I and testing APC activity ex vivo of TAM and mDC1 by coculturing them with OT-I T cells, which recognize SIINFEKL. The mixed bone marrow chimera aspect allows for direct comparison of APC activity of wild-type versus IRF8-knockout TAMs, which are coming from a shared tumor microenvironment. Additionally, the chimera system ensures tumor antigen is exogenous, as the USA cassette is not expressed by donor bone marrows. This assay also allows for testing of APC activity of mDC1 from the tumor-draining lymph node.
-
1.
Lethally irradiate (900 rad) 8–12 week old female recipient mice (S100a8CreLSL-USAPyMT, CD45.1.2+) 12–24 h before bone marrow transfer.
-
2.
Maintain these mice on antibiotic sulfatrim diet for at least two weeks post-irradiation and post-bone barrow transfer.
Note: The purpose of this dose of irradiation is to kill off the host/recipient mouse’s bone marrow and immune system. The donors’ bone marrows will reconstitute the hematopoietic compartment, but this can take a few weeks, hence the mice are maintained on chow with antibiotics (Sulfatrim diet) while they are immunocompromised.
-
3.Harvest bone marrow from adult (∼8–12 week old) female donor mice (one C57BL/6 CD45.1+ mouse and one CD11cCreIrf8fl/fl CD45.2+ mouse for transfer into up to five recipients).
-
a.Euthanize donor mice and harvest both femurs and tibias.
-
b.Remove the muscle from the bones, clean bones in 70% ethanol and wash in sterile PBS in 60 × 15 mm tissue culture dishes.
-
c.Mash bones using mortar and pestle in sterile biological safety cabinet.
-
d.Collect mashed bone marrow in T cell media (TCM) and pass it through a 70 μm cell strainer into a 50 mL conical tube to obtain single-cell suspension. Take small aliquot (∼20 μL) for counting.
-
e.Count number of cells in each sample.
-
i.Mix 10 μL 0.4% trypan blue with 10 μL cells to count.
-
ii.Let sit 3 min at 20°C–25°C.
-
iii.Pipette 10 μL underneath coverslip of hemacytometer.
-
iv.Placing hemacytometer on microscope, count number of cells that have not taken up the dye (live cells) in each of the four outer quadrants.
-
v.Calculate number of live cells in sample: (number of cells in four quadrants/4) × (104) = number of cells per mL, then (number of cells per mL) × (total volume of cells [unit in mL]) for final cell number.
-
i.
-
a.
-
4.
After ensuring comparable numbers of cells (i.e., ∼60–80 × 106 per mouse), mix bulk bone marrow from the two donor mice at a 1 to 1 ratio and spin down for 6 min at 4°C and 550 rcf.
-
5.
Discard supernatant and resuspend in appropriate volume of sterile PBS (100 μL per recipient mouse, total volume up to 500 μL for mixed bone marrow from two donor mice).
Note: Pooled bone marrow from both hind legs of two donor mice can reconstitute up to five irradiated recipient mice, assuming consistent and successful harvesting of bone marrow across donors (∼60–80 × 106 cells per donor).
Note: The number of cells transferred does not need to be exact. Because WT and KO donor bone marrow is mixed 1:1 and transferred together into the same host, as long as they each have similar numbers of cells and the recipient receives at least 1/5 of the total bone marrow from two donors, that should be sufficient to allow bone marrows to graft successfully. If there are fewer than five recipient mice available, bone marrow can be split among fewer recipients.
-
6.
Inject 100 μL mixed bone marrow intravenously into irradiated recipient mouse via retro-orbital injection using 500 μL syringe.
Alternatives: Cells can also be injected via tail vein injection.
-
7.
Age the mixed bone marrow chimeric mice to allow bone marrow to graft and PyMT tumors to grow.
-
8.Monitor mammary glands weekly, checking for tumor growth.
-
a.To measure tumors, scruff mouse by the neck and palp each of the ten mammary glands.
-
b.For palpable tumors, measure “length” and “width” (perpendicular dimensions) of the tumor with a caliper and record values.
-
c.To calculate tumor volume per gland, use the formula: tumor volume = [(length × width2) × (π/6)].
-
a.
Note: There should be ten values per mouse, one per gland. For no palpable tumor, volume is 0. Add together values in order to calculate total tumor burden for that mouse.
-
9.
When total tumor burden reaches ∼ 2,000 mm3 across all mammary glands (after ∼8–12 weeks, around 20–24 weeks of age), mice can be used to sort out TAMs from the tumor and mDC1 from tumor-draining lymph nodes. Multiple chimeric mice can be used for the same experiment, representing biological replicates.
-
10.
Euthanize mice, dissect out tumors and tumor-draining lymph nodes.
-
11.Isolate cells from the tumor.
-
a.Mechanically digest tumor by cutting tissue with a razor blade on a glass slide. Pool tumors from one mouse.
 CRITICAL: Make sure to mince the tumor tissue well, as this will help the enzymatic digestion and filtering steps and will provide a better yield of cells from the tissue.
CRITICAL: Make sure to mince the tumor tissue well, as this will help the enzymatic digestion and filtering steps and will provide a better yield of cells from the tissue. -
b.Transfer minced tumor tissue into a 50 mL conical tube with 8 mL HBSS without Ca2+, Mg2+ with 280 U/mL Collagenase Type 3 and 4 μg/mL Dnase I.
-
c.Vortex for 5 s and incubate in 37°C water bath for 1 h, vortexing every 15 min.
-
d.Mash resulting tissue through 70 μm cell strainer with the back of a 3 mL syringe plunger, adding an additional 20 mL chilled PBS during washes of strainer. This will result in a single-cell suspension.
-
e.Spin down tube with tumor sample for 6 min at 4°C and 550 rcf.
-
f.Resuspend pellet in 7 mL 44% Percoll. Transfer to 15 mL conical tube.
-
g.Slowly add 3 mL 66% Percoll to bottom of tube.
 CRITICAL: When creating Percoll gradient, make sure 66% layer and 44% layer are fully separated with a clear, crisp interface between the two layers. Adding the 66% Percoll slowly is key to keeping the layers separate.
CRITICAL: When creating Percoll gradient, make sure 66% layer and 44% layer are fully separated with a clear, crisp interface between the two layers. Adding the 66% Percoll slowly is key to keeping the layers separate. -
h.Centrifuge sample for 30 min at 4°C and 1,900 rcf with no brake.
-
i.Remove debris at the top of the 44% Percoll layer, collect cells at the interface between 44% and 66% Percoll layers and transfer to a new 15 mL conical tube.
-
j.To wash away remaining Percoll, add chilled PBS to cells, bringing volume up to 12 mL. Vortex tube well.
-
k.Spin tube for 6 min at 4°C and 550 rcf.
-
l.Visually inspect tube to ensure cells are pelleted at the bottom of the tube and the Percoll gradient has not reformed. Discard supernatant.
 CRITICAL: Make sure to vortex tube well before spin to ensure Percoll is fully mixed into wash. After spin and before decanting supernatant, inspect tube to make sure cells are pelleted at the bottom of the tube, not at a re-formed Percoll interface.
CRITICAL: Make sure to vortex tube well before spin to ensure Percoll is fully mixed into wash. After spin and before decanting supernatant, inspect tube to make sure cells are pelleted at the bottom of the tube, not at a re-formed Percoll interface. -
m.Resuspend pellet in 1 mL TCM. Keep on ice until ready to start antibody stainings.
-
a.
-
12.Isolate cells from the tumor-draining lymph node.Note: This lymph node digestion protocol is specifically designed to isolate dendritic cells from the lymph node, as they are harder to liberate than other immune cells, such as lymphocytes.
-
a.Mechanically digest lymph node by mincing tissue with a razor blade on a glass slide.
-
b.Transfer minced lymph node to a 15 mL conical tube with 4.5 mL T cell media (TCM) and 280 U/mL Collagenase Type 3.
-
c.Vortex for 5 s and place in 37°C water bath.
-
d.Incubate in water bath for 30 min, vortexing every 15 min.
-
e.Add 500 μL 0.1 M EDTA to the sample, bringing volume up to 5 mL.
-
f.Vortex briefly and incubate in 37°C water bath for another 5 min.
-
g.Remove tube from water bath, vortex and incubate on ice for 5 min.
-
h.Mash resulting tissue through 70 μm cell strainer with the back of a 3 mL syringe plunger into a 50 mL conical tube, adding an additional 10 mL PBS during washes of strainer. This will result in a single cell suspension.
-
i.Spin tube for 6 min at 4°C and 550 rcf.
-
j.Discard supernatant and resuspend pellet in 1 mL TCM. Keep on ice until ready to start antibody stainings.
-
a.
-
13.Set aside about 10% (no more than 1 × 106 cells) of each tumor sample in TCM on ice for flow cytometric analysis in order to characterize phenotype of TAMs from mixed bone marrows.
-
a.To stain for flow cytometric analysis, transfer cells to 96 well U-bottom plate. Spin down plate for 3 min at 4°C and 700 rcf.
-
b.Discard supernatant. Resuspend samples in 50 μL FACS staining buffer with fluorescently conjugated antibodies against surface proteins of interest.Note: Sample antibody panel for TAM phenotyping: FcBlock (1 μg per 50 μL FACS buffer), PerCPCy5.5 – CD45, v450 – CD45.2, APC – IRF8 (intracellular), APC-Cy7 – Viability dye (GhostDye Red 780), PE – H-2Kb SIINFEKL, PE-Cy7 – dump channel [Ly6C, Ly6G and SiglecF], PE-TexasRed – F4/80, BV605 – MHC-II, BV650 – CD45.1, BV711 – CD11b.
-
c.Incubate sample for 30 min on ice protected from light.
-
d.Add 150 μL FACS buffer to each well.
-
e.Spin down plate for 3 min at 4°C and 700 rcf and discard supernatant.
-
f.To wash cells, resuspend in 200 μL FACS buffer.
-
g.Spin down plate for 3 min at 4°C and 700 rcf and discard supernatant.
-
h.Repeat steps f-g for additional wash.
-
i.Resuspend samples in 100 μL Fix/Perm to fix and permeabilize the cells.
-
j.Incubate cells for 30 min on ice protected from light.
 Pause point: Once cells are fixed, they can be stored at 4°C protected from light for up to 24 h before staining for intracellular antibodies and/or before running on flow cytometer.
Pause point: Once cells are fixed, they can be stored at 4°C protected from light for up to 24 h before staining for intracellular antibodies and/or before running on flow cytometer. -
k.Add 100 μL Fix/Perm buffer to each well.
-
l.Spin down plate for 3 min at 4°C and 860 rcf and discard supernatant.
-
m.To wash fixed cells, resuspend cells in 200 μL Fix/Perm buffer.
-
n.Spin down plate for 3 min at 4°C and 860 rcf and discard supernatant.
-
o.Repeat steps m-n.
-
p.To stain with intracellular antibodies, resuspend samples in 50 μL Fix/Perm buffer with fluorescently conjugated antibodies against intracellular proteins (see note above for sample antibody panel).
-
q.Incubate on ice for 30 min.
-
r.Repeat steps k-o.
-
s.Resuspend samples in 100 μL FACS buffer.
-
t.Transfer cells through nylon mesh material into 1.2 mL microtiter tube.
-
u.Run sample on flow cytometer, including appropriate compensation controls.
-
v.Analyze resulting data to determine TAM phenotype in the mixed bone marrow chimera system, measuring expression of exogenous tumor antigen (SIINFEKL) presented on the surface of CD45.1+ versus CD45.2+ TAMs and confirming IRF8 deletion and altered TAM expression of MHC-II1 in CD45.2+ TAMs.
-
a.
-
14.For remaining 90% of tumor samples and lymph node samples, prepare for sorting.
-
a.Resuspend pellet in 15 mL conical tube with 50 μL per 1 × 106 cells FACS staining buffer with fluorescently conjugated antibodies against surface proteins of interest.Note: Sample antibody panel for tumor TAM sort: FcBlock (1 μg per 50 μL), PerCPCy5.5 – CD45 , v450 – CD45.2, APC-Cy7 – viability dye (GhostDye Red 780), PE-Cy7 – Dump channel [Ly6C, Ly6G, SiglecF ], PE-TexasRed – F4/80, BV605 – MHC-II, BV650 – CD45.1, BV711 – CD11b.Note: Sample antibody panel for tumor-draining lymph node DC1 sort: FcBlock (1 μg per 50 μL), PerCPCy5.5 – CD45, BV421 – CD11c , APC – CD103, APC-Cy7 – viability dye (GhostDye Red 780), BV510 – XCR1 , BV605 – MHC-II, BV650 – CD45.1 , BV711 – CD11b.
-
b.Incubate on ice for 30 min protected from light.
-
c.Add 5 mL FACS buffer and spin down tube for 6 min at 4°C and 550 rcf.
-
d.Discard supernatant.
-
e.Repeat steps c-d twice more.
-
f.Resuspend cell pellet in appropriate volume of sorting buffer to achieve concentration of 10 × 106 cells per mL for sorting with 100 μm nozzle on BD Aria.
-
g.Transfer cells to 5 mL polypropylene tube, passing cells through 70 μm cell filter cap for 5 mL tube.
-
h.Sort tumor and dLN samples on BD Aria with 100 μm nozzle, including appropriate compensation controls for setting up instrument.
-
i.Gate for TAMs (Live CD45+SiglecF-Ly6C-F4/80+CD11blow/-) that are C57BL/6 (CD45.1+CD45.2-) or IRF8-deficient (CD45.1-CD45.2+) from the tumor sample. See Figure 4 for gating strategy.
-
ii.Gate for mDC1 (Live CD45+CD11c+/medium-highMHC-II+/highCD11b- Xcr1+CD103+). See Figure S4D (Nixon et al., 2022)1 for gating strategy.
-
iii.Keep samples on ice until ready to set up coculture.Note: CD45.1+CD45.2- cells will be derived from C57BL/6 donor bone marrow. CD45.1-CD45.2+ cells will be derived from CD11cCreIrf8fl/fl bone marrow. CD45.1+CD45.2+ cells will be derived from the recipient mouse, although these immune cells should have been killed during irradiation.Note: The mDC1s will all be derived from C57BL/6 bone marrow (CD45.1+CD45.2-) since the CD11cCreIrf8fl/fl genotype leads to loss of all DC1s.
-
i.
-
a.
-
15.Isolate naïve CD8+ T cells from OT-I mice.
-
a.Euthanize one adult OT-I mouse, and harvest lymph nodes from the mouse.
-
b.Mash lymph nodes between two glass slides and wipe off into 60 × 15 mm tissue culture dish with 2 mL chilled PBS.
-
c.Pipette cell suspension through 70 μm cell strainer into 50 mL conical tube.Note: Digestion of the lymph node is only required for dendritic cell isolation (step 12). Here, we are only isolating T cells, so mashing is sufficient to liberate lymphocytes.
-
d.Isolate naïve CD8+ T cells using a negative-selection isolation kit and follow the manufacturer’s instructions.
-
a.
-
16.Stain OT-I T cells with CellTrace Violet (CTV).
-
a.Reconstitute CTV tube with 20 μL DMSO.
-
b.Add 1 μL CTV to 1 mL PBS.
-
c.Resuspend cells in 1 mL PBS.
-
d.Add 1 mL PBS + CTV to cells and pipette up and down to mix (total volume of 2 mL).
-
e.Incubate in 37°C water bath for 15 min.
-
f.Bring volume up to 15 mL with TCM that has been warmed in the water bath.
-
g.Spin down tube for 6 min at 4°C and 550 rcf.
-
h.Resuspend pellet in 10 mL warmed TCM.
-
i.Spin down tube for 6 min at 4°C and 550 rcf.
-
j.Resuspend pellet in 10 mL warmed TCM and incubate in water bath for 10 min to ensure all unbound CTV is absorbed.
-
k.Take 10 μL aliquot for counting.
-
l.Spin down tube for 6 min at 4°C and 550 rcf.
-
a.
-
17.
Count number of live CTV-stained OT-I T cells (see step 3e).
Note: OT-I cells can be isolated during tumor sample preparation incubations and/or sorting to reduce time of sorted TAMs sitting on ice.
-
18.
Resuspend OT-I in appropriate volume of TCM for coculture experiment (15,000 T cells per 100 μL).
-
19.
Plate 100 μL OT-I cells to wells of a 96 U-bottom plate. Include wells without antigen presenting cells added to measure no proliferation.
-
20.
Depending on number of TAMs sorted, prepare TAMs in TCM at varying concentrations for coculture (TAM:OT-I at 1:1 [15,000 TAMs per 100 μL], 3:1 [45,000 TAMs per 100 μL], and 10:1 [150,000 TAMs per 100 μL]).
-
21.
Add 100 μL TAMs to wells with 100 μL OT-I, with a total volume of 200 μL.
-
22.
Depending on number of mDC1 sorted, prepare mDC1 in TCM at certain concentrations for coculture (e.g., mDC1:OT-I at 1:3 [5,000 mDC1 per 100 μL]).
Note: There are very few mDC1 in tumor-draining lymph node, so there may be enough cells for only one ratio per mouse. 1:1 (15,000 mDC1) ratio can be included if there are enough cells. DCs are potent activators of CD8 T cell responses, so at this lower APC to T cell ratio, DCs are often still better APCs than TAMs.
-
23.
Add 100 μL mDC1 to wells with 100 μL OT-I, with a total volume of 200 μL
-
24.
Put plate in 37°C incubator for 72 h.
Note: There is no need to add cytokines (i.e., IL-2) or antigen (i.e., SIINFEKL) to the coculture system. This assay tests the APC activity of TAM and mDC1. Aspects of this activity include the ability to have picked up, processed and presented antigen while in the tumor microenvironment, cytokine production by the APCs themselves and/or the ability to properly activate/induce IL-2 production in T cells to support their survival and proliferation.
-
25.After 3 days of coculture, stain samples with fluorescently conjugated antibodies for flow cytometry.
-
a.Spin down plate for 3 min at 4°C and 700 rcf and discard supernatant.
-
b.Resuspend cells in 200 μL FACS buffer.
-
c.Spin down plate for 3 min at 4°C and 700 rcf and discard supernatant.Resuspend cells in 50 μL FACS staining buffer per 1 × 106 cells with fluorescently conjugated antibodies against surface proteins of interest.Note: Sample antibody panel for T cell phenotype: FcBlock (1 μg per 50 μL), FITC – PD-1, PerCPCy5.5 – CD45, BV421 open for CTV detection, APC-Cy7 – viability dye (GhostDye Red 780), PE-Cy7 – TCRβ, BV650 – CD8α.
-
d.Incubate plate on ice for 30 min.
-
e.Wash unfixed samples three times with FACS buffer (see steps 13d-h).
-
f.Fix cells and stain with fluorescently conjugated antibodies against intracellular proteins (see steps 13i-r).
-
g.Resuspend samples in 100 μL FACS buffer.
-
h.Transfer cells through nylon mesh material into 1.2 mL microtiter tube.
-
a.
-
26.
Run samples on flow cytometer along with appropriate compensation controls.
-
27.
Analyze resulting data to measure T cell proliferation via CTV dilution and T cell phenotype via expression of PD-1, comparing conditions with wild-type or IRF8-deficient TAMs and comparing different TAM:T cell ratios (see Figures 4E and 4F from Nixon et al.1 for sample flow plots).
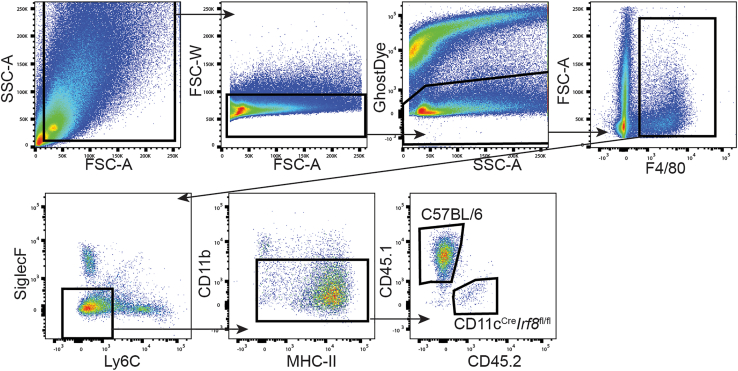
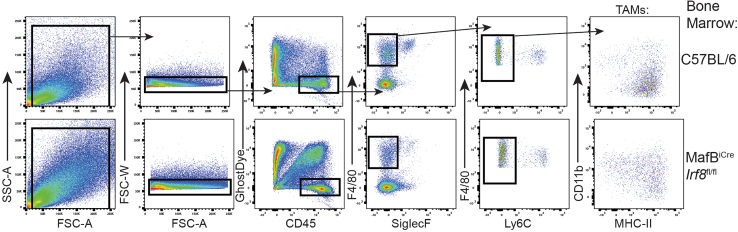
Figure 4.
FACS gating strategy for TAMs from mixed bone marrow chimeric mice
Gating strategy in order to isolate C57BL/6 (CD45.1+) and IRF8-deficient (CD11cCreIrf8fl/fl [CD45.2+]) TAMs from PyMT tumors of mixed bone marrow chimera mice to be used in ex vivo presentation assays.
In vivo tumor antigen presentation assay
Timing: 8–12 weeks, depending on tumor growth rate
This major step generates two different bone marrow chimeras that allow for comparison of TAM presentation of exogenous tumor antigen and APC activity in vivo (Figure 2B). It compares the phenotype of transferred tumor antigen-specific CD8+ T cells in tumor microenvironments containing either wild-type or IRF8-deficient TAMs.
Note: When determining number of mice per experiment, consider that one donor mouse can reconstitute up to three recipient mice, and each experiment requires both a WT bone marrow cohort and a MafBiCreIrf8fl/fl bone marrow cohort, so up to six recipient mice can be used per pair of donor mice (three per donor).
-
28.
Lethally irradiate (900 rad) 8–12 week old female recipient mice (S100a8CreLSL-USAPyMT, CD45.1.2+) 12–24 h before bone marrow transfer.
-
29.
Maintain these mice on sulfatrim diet for at least 2 weeks post-irradiation and post-bone marrow transfer.
-
30.
Harvest bone marrow from adult donor mice (C57BL/6 CD45.2+ and MafBiCreIrf8fl/fl CD45.2+), and follow step 3 to isolate and count bone marrow cells. There should be comparable numbers of cells in WT and KO samples.
-
31.
Keep bone marrow samples separate; spin down tubes for 6 min at 4°C and 550 rcf.
-
32.
Resuspend each sample in up to 300 μL sterile PBS, 100 μL per recipient mouse.
-
33.
Transfer single genotype bone marrow into irradiated recipient mice, with one cohort receiving C57BL/6 bone marrow and another cohort receiving MafBiCreIrf8fl/fl bone marrow. Bone marrow harvested from one mouse can be transferred to up to 3 recipient mice.
-
34.
Age mice to allow bone marrow to graft and tumors to grow.
-
35.
Monitor mice weekly for tumor growth by checking for and measuring palpable tumors in the mammary glands (see step 8).
-
36.When tumors are at least 5 mm by 5 mm, and potentially larger (see note), isolate and transfer 1 × 106 OT-I CD45.1+ T cells intravenously.Note: PyMT mice often have tumors at multiple mammary glands. When determining when to transfer the T cells, the mice should have a sizeable tumor burden in at least one gland, thus ensuring the tumors are at a late enough stage for a robust TAM response to have developed (ideally at multiple glands and larger than 5 × 5 mm). However, mice should also be able to survive for two more weeks for the assay. Thus, the tumor burden should be sizeable but not too advanced. Endpoints for these mice are when they reach a total tumor burden of 3,000 mm3 or an individual tumor that is 2,000 mm3.
-
a.Euthanize OT-I mouse and harvest lymph nodes and spleen.
-
b.Mash lymph nodes and spleen between two glass slides to get single-cell suspension.
-
c.Pool lymph node and spleen cells together. Pass mashed tissue through 70 μm cell strainer into 50 mL conical tube, using the back of a 3 mL syringe plunger to ensure tissue passes through filter and generates a single-cell suspension.
-
d.Isolate naïve CD8+ T cells using a negative-selection isolation kit and follow the manufacturer’s instructions.
-
e.Count number of live OT-I T cells (see step 3e).
-
f.Resuspend cells in sterile PBS at proper concentration for transfer (1 × 106 cells per 100 μL PBS).
-
g.Inject 100 μL (1 × 106 cells) OT-I cells into each chimeric recipient mouse, including a cohort with WT bone marrow and another with MafBiCreIrf8fl/fl bone marrow.
-
a.
-
37.
Age mice for two weeks.
-
38.Euthanize mice and process tissues in order to analyze phenotypes of TAMs and tumor-infiltrating T cells.
-
a.Harvest tumors and process tissue to generate single cell suspension, following step 11.
-
b.Split each sample in two for staining with two different antibody panels, plating no more than 1 × 106 cells per well of a 96-well U-bottom plate.
-
c.Spin down plate for 3 min at 4°C and 700 rcf, and discard supernatant.
-
d.Resuspend samples in 50 μL FACS buffer plus antibody cockatil, one to analyze the T cell phenotype, another to analyze the TAM phenotype.
-
e.Incubate sample on ice for 30 min protected from light.Note: Sample antibody panel for T cell phenotype: FcBlock (1 μg per 50 μL), FITC – PD-1, PerCPCy5.5 – CD45, v450 – CD45.2 , APC – Granzyme B (intracellular antibody), APC-Cy7 – CD45.1, PE-Cy7 – TCRβ, BV510 – viability dye (GhostDye Violet 510), BV650 – CD8α.Note: Sample antibody panel for TAM phenotype: FcBlock (1 μg per 50 μL), BV421 – CD45.2, APC – IRF8 (intracellular antibody), APC-Cy7 – viability dye (GhostDye Red 780), PE-Cy7 – dump channel [Ly6C, Ly6G, SiglecF], PE-TexasRed – F4/80, BV605 – MHC-II, BV650 – CD45.1, BV711 – CD11b.
-
f.Wash samples three times with FACS buffer (see steps 13d-h).
-
g.Fix and permeabilize cells and stain with intracellular antibodies (see steps 13i-r).
 Pause point: Once cells are fixed, they can be stored at 4°C for up to 24 h before intracellular staining and/or running samples on the flow cytometer.
Pause point: Once cells are fixed, they can be stored at 4°C for up to 24 h before intracellular staining and/or running samples on the flow cytometer. -
h.Prepare samples and run on flow cytometer (see steps 13s-u).
-
i.Analyze resulting data to confirm IRF8 deletion and altered TAM phenotype (Figure 5). Additionally, measure T cell phenotype via expression of molecules such as PD-1 and granzyme B, comparing cells derived from tumors with wild-type TAMs to those from tumors with IRF8-deficient TAMs. See Figure S5H in (Nixon et al., 2022)1 for flow cytometry plots for T cell phenotype.
-
a.
Figure 5.
Expected TAM phenotypes in tumors of bone marrow chimera mice for in vivo presentation assays
Example flow cytometry plots from recipient mice (S100a8CreLSL-USAPyMT) that received either C57BL/6 (top) or MafBiCreIrf8fl/fl (bottom) bone marrow, gating on immune cells infiltrating PyMT tumors, specifically focusing on TAM phenotyping.
Expected outcomes
For the ex vivo tumor antigen presentation experiments, TAMs derived from C57BL/6 bone marrow are expected to express tumor antigen in the context of MHC-I on their surface, as detected by staining for H2-Kb bound to SIINFEKL, to express IRF8 and high MHC-II, and to be able to induce T cell activation and proliferation. Thus, they can cross-present and/or cross-dress tumor antigen to OT-I CD8+ T cells. We expect IRF8-deficient TAMs to express less tumor antigen on their surface, as detected by staining for H2-Kb bound to SIINFEKL, to not express IRF8, to express less MHC-II, and to lose the ability to activate OT-I T cells. The mDC1s from the tumor-draining lymph node derived from C57BL/6 bone marrow will potently activate OT-I T cells. See Figures 4E and 4F from (Nixon et al., 2022)1 for example flow cytometry plots.
For the in vivo tumor antigen presentation experiments, since MafBiCre is not expressed in DCs, both chimeras should have an intact IRF8-expressing DC1 compartment to allow for proper priming of T cells. OT-I T cells transferred into chimeras with C57BL/6 bone marrow will become exhausted, will express PD-1 and not express granzyme B. OT-I T cells transferred into chimeras with MafBiCreIrf8fl/fl bone marrow will express less PD-1 and more granzyme B (see Figure S5H from (Nixon et al., 2022)1 for expected T cell phenotype). Additionally, the TAMs in tumors of mice with MafBiCreIrf8fl/fl bone marrow will have deleted IRF8 and express less MHC-II than IRF8-sufficient TAMs, phenocopying the altered TAM maturation observed in MafBiCreIrf8fl/flPyMT mice1 (Figure 5).
Limitations
Although this protocol can measure APC activity of TAMs, it cannot differentiate whether TAMs cross-present or cross-dress tumor antigen, as all mouse strains used here express the same MHC-I haplotypes. In order to address this question, one would need to use mice where the tumor cells and immune cells were mismatched for MHC-I haplotype, and then see whether the MHC-I complex that contains tumor antigen on the immune cell’s surface was derived from the immune cell or tumor cell. Additionally, this protocol only measures the role of IRF8 in TAM APC activity through loss-of-function models. However, it can be adapted by using bone marrow from different knockout mice to target other genes of interest to determine whether they contribute to TAM APC activity. This protocol studies tumor T cell responses only in the context of the OT-I T cell receptor (TCR) recognizing SIINFEKL as the tumor antigen. It does not measure TAM APC activity in the context of other TCRs or antigens. Additionally, this protocol studies APC activity in only one cancer model, the PyMT model of breast cancer.
Troubleshooting
Problem 1
The OVA peptide is expressed in non-tumor cells.
Potential solution
The floxed stop codon upstream of the USA cassette, which encodes OVA, may have been deleted in the germline due to leaky cre activity. During genotyping, a primer that detects deleted “stop” codon should be included to detect if this occurs and remove those mice from breedings and experiments.
Problem 2
The cell recovery after the sort is very low.
Potential solution
Normally, the scale of TAMs recovered from C57BL/6 bone marrow should be in the hundreds of thousands of cells from pooled tumors from one mouse. If the recovery is much lower than this, then it is possible the TAMs are being lost at some point upstream during the tissue processing, staining or sort. Make sure the gating during the sort is generous in the early steps of gating (i.e., FSC-A versus SSC-A) to ensure the cells aren’t being inadvertently excluded. With regards to steps before the sort, make sure pelleted cells are not inadvertently lost during washes. If for instance when preparing the Percoll media, the 10× PBS is not added, this can lead to a hypotonic solution and may kill the immune cells during tissue processing.
Problem 3
There is no difference in T cell phenotype in the in vivo tumor antigen presentation experiments.
Potential solution
This may be due to incomplete deletion of IRF8 in the TAM compartment, and thus the “KO” TAM condition actually contains TAMs that maintained IRF8 expression. Confirm that the TAMs have an altered phenotype and proper deletion, which can be determined during the flow cytometric analysis of a subset of the sorted tumor (step 13).
Problem 4
The Percoll gradient was disrupted and there is not a clear interface between the two layers, it is more mixed (step 11g).
Potential solution
Do not spin the tube in the Percoll program for 30 min. Instead, transfer total Percoll and cells to a 50 mL conical and dilute Percoll as much as possible with chilled PBS. Vortex well to make sure Percoll is mixed. Spin tube down for 6 min at 4°C and 550 rcf. After spin, check to make sure all cells are pelleted at the bottom of the tube, not at the top or at any interfaces that may have formed. If so, decant supernatant, resuspend pellet in 7 mL 44% Percoll, transfer to a 15 mL conical and try again to slowly add 66% Percoll and continue with the protocol.
Problem 5
The CTV staining step results in high levels of cell death.
Potential solution
During the CTV stain, the incubation period can be reduced by a few minutes to reduce the exposure of the cells to CTV, although this also may reduce the brightness of the staining. Be sure to check how the staining looks on the flow cytometer and adjust the voltage of the detection laser if needed to make sure the staining was successful and is in range. Additionally, during the washes, make sure to use media that contains 10% FBS, as this will quench the reaction and bind up unused CTV.
Problem 6
During acquisition of flow cytometry data, two different antibodies fully overlap with one another, creating a diagonal line when put on the x and y axes (step 13u).
Potential solution
The experiment was not compensated properly on the analyzer at the start of collecting the samples. If there is a portion of samples that remain to rerun, then set up a new experiment and rerun compensation controls. Make sure the single-color compensation controls are each as bright or brighter than the samples will be for that color. Alternatively, the wrong antibodies were added when making the antibody cocktail, i.e., two antibodies in two colors that recognize the same protein. If this is the case, do not use either channel in the analysis. If this leads to the inability to properly gate for populations of interest, then the experiment needs to be repeated from the beginning with new mice.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Ming Li (lim@mskcc.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze new datasets or code.
Acknowledgments
This work was supported by the National Institutes of Health (F31 CA210332 to B.G.N. and R01 CA198280 to M.O.L.), the Howard Hughes Medical Institute (Faculty Scholar Award to M.O.L.), the Mazumdar-Shaw Translational Research Initiative in Kidney Cancer (M.O.L.), and the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center (M.O.L.). We acknowledge the use of the Flow Cytometry Core Facility at MSKCC which contributed to this project.
Author contributions
B.G.N. and M.O.L. were involved in all aspects of the study, including planning and performing experiments, analysis and interpretation of data, and writing the manuscript. L.J. performed experiments.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Briana G. Nixon, Email: nixonb@mskcc.org.
Ming O. Li, Email: lim@mskcc.org.
References
- 1.Nixon B.G., Kuo F., Ji L., Liu M., Capistrano K., Do M., Franklin R.A., Wu X., Kansler E.R., Srivastava R.M., et al. Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity. 2022;55:2044–2058.e5. doi: 10.1016/j.immuni.2022.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broz M.L., Binnewies M., Boldajipour B., Nelson A.E., Pollack J.L., Erle D.J., Barczak A., Rosenblum M.D., Daud A., Barber D.L., et al. Dissecting the tumor myeloid compartment reveals rare activating antigenpresenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Mingo Pulido Á., Gardner A., Hiebler S., Soliman H., Rugo H.S., Krummel M.F., Coussens L.M., Ruffell B. TIM-3 regulates CD103+ dendritic cell function and response to chemotherapy in breast cancer. Cancer Cell. 2018;33:60–74.e6. doi: 10.1016/j.ccell.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze new datasets or code.