Abstract
We present a genome assembly from an individual female Vanessa cardui (the painted lady; Arthropoda; Insecta; Lepidoptera; Nymphalidae). The genome sequence is 425 megabases in span. The majority of the assembly is scaffolded into 32 chromosomal pseudomolecules, with the W and Z sex chromosome assembled. Gene annotation of this assembly on Ensembl has identified 12,821 protein coding genes.
Keywords: Vanessa cardui, painted lady, genome sequence, chromosomal; Lepidoptera
Species taxonomy
Eukaryota; Metazoa; Ecdysozoa; Arthropoda; Hexapoda; Insecta; Pterygota; Neoptera; Endopterygota; Lepidoptera; Glossata; Ditrysia; Papilionoidea; Nymphalidae; Nymphalinae; Vanessa; Vanessa cardui (Linnaeus, 1758) (NCBI:txid171605).
Background
The painted lady, Vanessa cardui, is an extremely widespread butterfly, occurring on all continents except most of South America and Oceania ( Shields, 1992). The species undertakes long-distance multi-generational migrations each year ( Pollard et al., 1998; Stefanescu et al., 2013; Talavera et al., 2018; Williams, 1970). It does not overwinter and is therefore engaged in a constant movement. In the Palaearctic, migrants are known to seasonally circulate between North Africa and Europe ( Pollard et al., 1998; Stefanescu, 2011; Stefanescu et al., 2013). Recent work has also revealed that autumn populations from Europe cross the Sahara Desert reaching tropical Africa ( Stefanescu et al., 2016; Talavera & Vila, 2016). This journey, spanning over 4000 km, represents the longest single-leg migratory flight known in butterflies. The butterflies migrate back to Europe in spring, thus covering up to 14000 km in an annual cycle involving 8–10 generations in their Palaearctic-African range ( Menchetti et al., 2019; Talavera et al., 2018). The painted lady is found throughout the British Isles but abundance varies greatly between years. Larvae are polyphagous on a large variety of plant families, but most commonly feed on thistles ( Cirsium spp . and Carduus spp .) and mallows ( Malva spp.). The painted lady occurs in a wide range of biomes and environments spanning semi-deserts, grasslands, meadows, and mountains to suburban areas. It is listed as Least Concern in the IUCN Red List ( Walker & Coetzer, 2020). Studies of V. cardui have included thermoregulation ( Tsai et al., 2020), adaptations to host plants ( Celorio-Mancera et al., 2016), flight behaviour ( Gamberale-Stille et al., 2019; Liu et al., 2021) and movement ecology ( Suchan et al., 2018). Genes involved in the development of the distinctive eyespots on the forewings and hindwings of V. cardui have been identified ( Mazo-Vargas et al., 2017; Zhang & Reed, 2016; Zhang et al., 2017). V. cardui has a karyotype of 31 chromosomes ( Lorkovic, 1941).
We note the recent publication of another high-quality genome assembly for V. cardui ( Zhang et al., 2021). We hope that the sequence described here, generated as part of the Darwin Tree of Life project, will further contribute to the study of V. cardui as an emerging model for the genetics of migratory behavior, ecological genomics and developmental genetics.
Genome sequence report
The genome was sequenced from a single female V. cardui (ilVanCard2; Figure 1A, B) collected from Carrifran Wildwood, Scotland (latitude 55.400132, longitude -3.3352). Hi-C data were generated from a second female V. cardui (ilVanCard3; Figure 1C, D) collected from Yellowcraig, East Lothian, Scotland (latitude 56.062445, longitude -2.769836). A total of 25-fold coverage in Pacific Biosciences single-molecule long reads (N50 15 kb) and 89-fold coverage in 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 79 missing/misjoins and removed 7 haplotypic duplications, reducing the assembly size by 0.48% and scaffold number by 61.70%, and increasing the scaffold N50 by 21.74%.
Figure 1. Fore and hind wings of Vanessa cardui specimens from which the genome was sequenced.
( A) Dorsal surface view of wings from specimen SC_VC_1208 (ilVanCard2) from Carrifran Wildwood, Scotland used to generate Pacific Biosciences and 10X genomics data. ( B) Ventral surface view of wings from specimen SC_VC_1208 (ilVanCard2) from Carrifran Wildwood, Scotland, used to generate Pacific Biosciences and 10X genomics data. ( C) Dorsal surface view of wings from specimen SC_VC_1220 (ilVanCard3) from Yellowcraig, Scotland, used to generate Hi-C data. ( D) Ventral surface view of wings from specimen SC_VC_1220 (ilVanCard3) from Yellowcraig, Scotland, used to generate Hi-C data.
The final assembly has a total length of 425 Mb in 37 sequence scaffolds with a scaffold N50 of 15 Mb ( Table 1). Of the assembly sequence, 96.0% was assigned to 32 chromosomal-level scaffolds, representing 30 autosomes (numbered by sequence length), and the W and Z sex chromosome ( Figure 2– Figure 5; Table 2). The assembly has a BUSCO ( Simão et al., 2015) completeness of 98.8% using the lepidoptera_odb10 reference set. While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
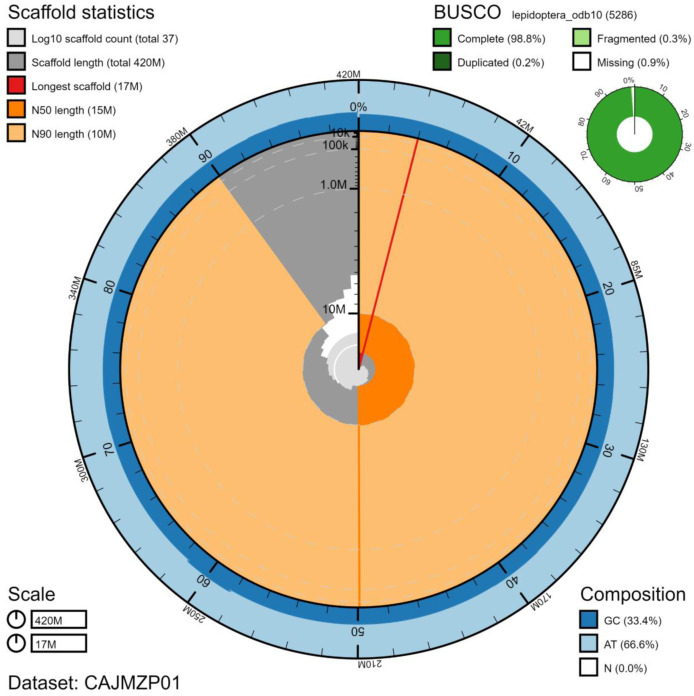
Figure 2. Genome assembly of Vanessa cardui, ilVanCard2.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 424,813,639 bp assembly. The distribution of chromosome lengths is shown in dark grey with the plot radius scaled to the longest chromosome present in the assembly (17,040,296 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 chromosome lengths (14,615,999 and 9,960,137 bp), respectively. The pale grey spiral shows the cumulative chromosome count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the lepidoptera_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilVanCard2.1/dataset/CAJMZP01/snail.
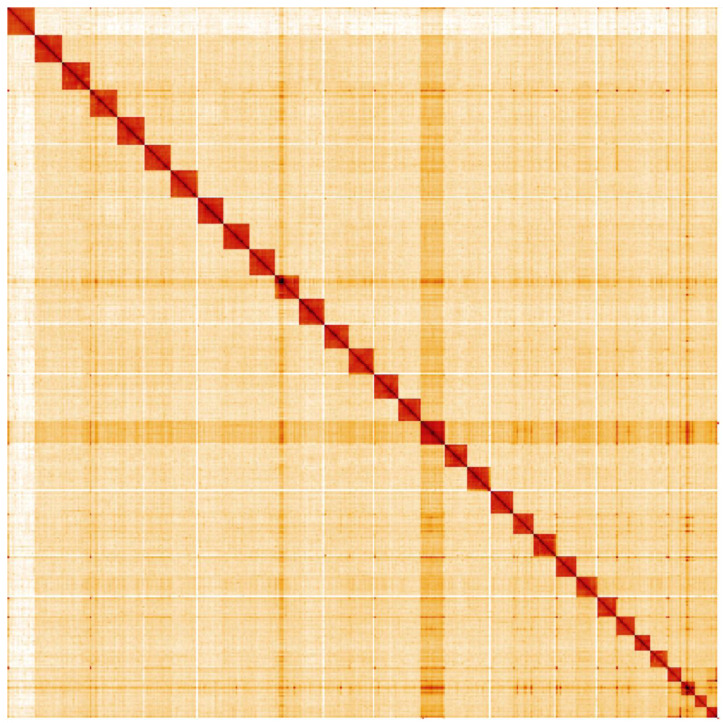
Figure 5. Genome assembly of Vanessa cardui, ilVanCard2.1: Hi-C contact map.
Hi-C contact map of the ilVanCard2.1 assembly, visualised in HiGlass. Chromosomes are arranged by size from left to right and top to bottom.
Table 1. Genome data for Vanessa cardui, ilVanCard2.1.
| Project accession data | |
|---|---|
| Assembly identifier | ilVanCard2 |
| Species | Vanessa cardui |
| Specimen | ilVanCard1 (RNA-Seq);
ilVanCard2 (genome assembly); ilVanCard3 (Hi-C) |
| NCBI taxonomy ID | NCBI:txid171605 |
| BioProject | PRJEB42869 |
| BioSample ID | SAMEA7523147 |
| Isolate information | Female, whole organisms |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR6608653 |
| 10X Genomics Illumina | ERR6054369-ERR6054372 |
| Hi-C Illumina | ERR6054373 |
| Illumina polyA RNA-Seq | ERR6054374 |
| Genome assembly | |
| Assembly accession | GCA_905220365.1 |
| Accession of alternate haplotype | GCA_905220355.1 |
| Span (Mb) | 425 |
| Number of contigs | 128 |
| Contig N50 length (Mb) | 7 |
| Number of scaffolds | 37 |
| Scaffold N50 length (Mb) | 15 |
| Longest scaffold (Mb) | 17 |
| BUSCO * genome score | C:98.2%[S:97.9%,D:0.3%],
F:0.8%,M:1.0%,n:1658 |
| Gene annotation | |
| Number of protein coding genes | 12,821 |
| Average coding sequence length (bp) | 1,738 |
| Average number of exons per transcript | 9.44 |
| Average exon size (bp) | 393 |
| Average intron size (bp) | 2358 |
* BUSCO scores based on the lepidoptera_odb10 BUSCO set using v5.1.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/Vanessa%20cardui/dataset/CAJMZP01/busco.
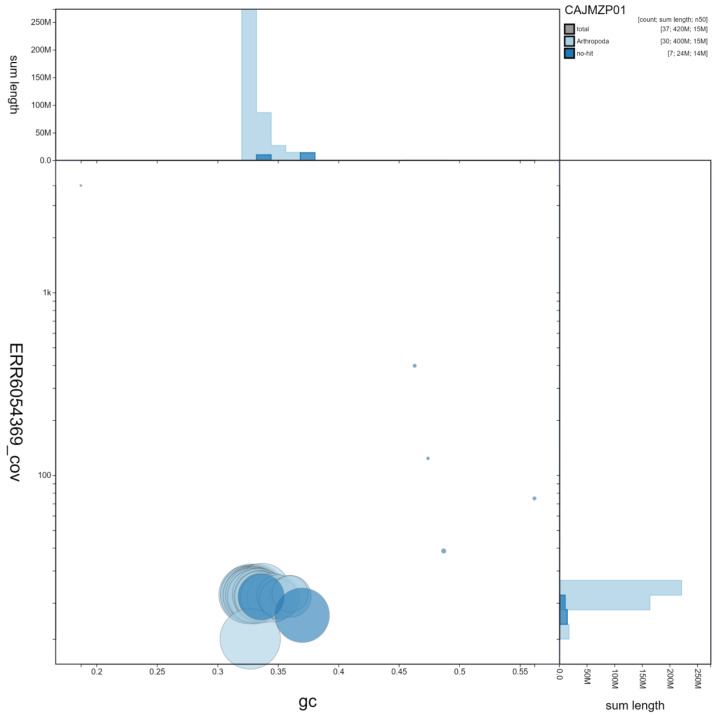
Figure 3. Genome assembly of Vanessa cardui, ilVanCard2.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilVanCard2.1/dataset/CAJMZP01/blob.
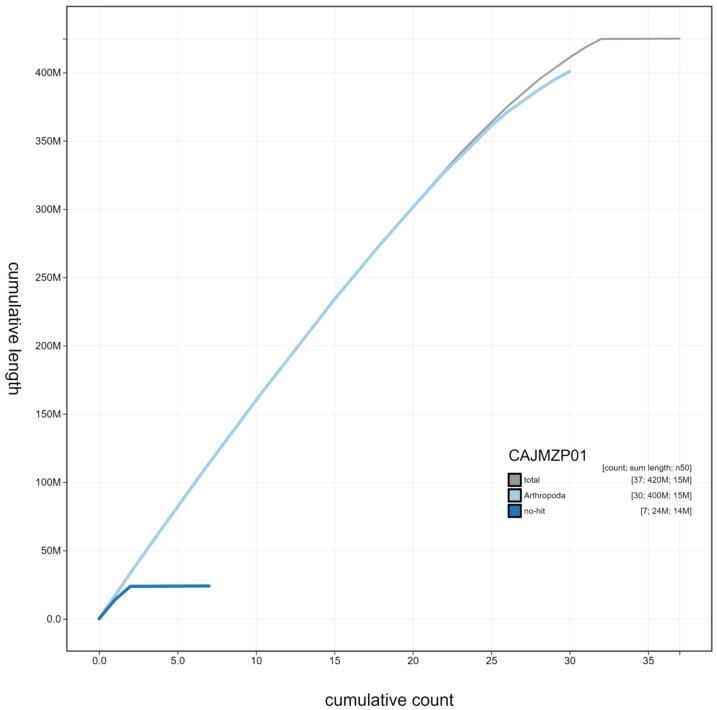
Figure 4. Genome assembly of Vanessa cardui, ilVanCard2.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilVanCard2.1/dataset/CAJMZP01/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Vanessa cardui, ilVanCard2.1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| LR999925.1 | 1 | 16.61 | 32.7 |
| LR999926.1 | 2 | 16.36 | 33.2 |
| LR999927.1 | 3 | 16.09 | 33.1 |
| LR999928.1 | 4 | 16.00 | 32.7 |
| LR999929.1 | 5 | 15.95 | 32.9 |
| LR999930.1 | 6 | 15.72 | 32.5 |
| LR999931.1 | 7 | 15.57 | 33 |
| LR999932.1 | 8 | 15.43 | 32.9 |
| LR999933.1 | 9 | 15.30 | 32.6 |
| LR999934.1 | 10 | 14.95 | 33.7 |
| LR999935.1 | 11 | 14.87 | 33 |
| LR999936.1 | 12 | 14.77 | 33.1 |
| LR999937.1 | 13 | 14.62 | 33 |
| LR999938.1 | 14 | 14.61 | 32.8 |
| LR999939.1 | 15 | 13.92 | 32.7 |
| LR999941.1 | 16 | 13.77 | 32.9 |
| LR999942.1 | 17 | 13.55 | 33.1 |
| LR999943.1 | 18 | 13.24 | 33 |
| LR999944.1 | 19 | 12.93 | 33.7 |
| LR999945.1 | 20 | 12.86 | 33.4 |
| LR999946.1 | 21 | 12.59 | 33.5 |
| LR999947.1 | 22 | 11.70 | 33.5 |
| LR999948.1 | 23 | 11.33 | 33.8 |
| LR999949.1 | 24 | 11.20 | 34.5 |
| LR999950.1 | 25 | 9.96 | 33.6 |
| LR999951.1 | 26 | 9.84 | 33.8 |
| LR999952.1 | 27 | 8.26 | 35 |
| LR999953.1 | 28 | 8.18 | 36 |
| LR999954.1 | 29 | 7.38 | 35.1 |
| LR999955.1 | 30 | 6.17 | 36 |
| LR999940.1 | W | 13.82 | 37 |
| LR999924.1 | Z | 17.04 | 32.7 |
| LR999956.1 | MT | 0.02 | 19 |
| - | Unplaced | 0.22 | 49.5 |
Gene annotation
The Ensembl gene annotation system ( Aken et al., 2016) was used to generate annotation for the Vanessa cardui assembly (GCA_905220365.1, see https://rapid.ensembl.org/Vanessa_cardui_GCA_905220365.1/; Table 1). The annotation was created primarily through alignment of transcriptomic data to the genome, with gap filling via protein-to-genome alignments of a select set of proteins from UniProt ( UniProt Consortium, 2019) and OrthoDB ( Kriventseva et al., 2008). Prediction tools, CPC2 ( Kang et al., 2017) and RNAsamba ( Camargo et al., 2020), were used to aid determination of protein coding genes.
Methods
Sample acquisition and nucleic acid extraction
The first female V. cardui, ilVanCard2 (genome assembly), was collected from Carrifran Wildwood, Scotland (latitude 55.400132, longitude -3.3352). Two further female V. cardui specimens, ilVanCard1 (RNA-Seq) and ilVanCard3 (Hi-C), was collected from Yellowcraig, East Lothian, Scotland (latitude 56.062445, longitude -2.769836). All samples were collected and identified by Konrad Lohse, University of Edinburgh, and were snap-frozen from life in liquid nitrogen.
DNA was extracted at the Wellcome Sanger Institute (WSI) Scientific Operations core from the whole organism using the Qiagen MagAttract HMW DNA kit, according to the manufacturer’s instructions. RNA was extracted in the Tree of Life Laboratory at the WSI using TRIzol (Invitrogen), according to the manufacturer’s instructions. RNA was then eluted in 50 μl RNAse-free water and its concentration RNA assessed using a Nanodrop spectrophotometer and Qubit Fluorometer using the Qubit RNA Broad-Range (BR) Assay kit. Analysis of the integrity of the RNA was done using Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Sequencing
Pacific Biosciences HiFi circular consensus and 10X Genomics Chromium read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Poly(A) RNA-Seq libraries were constructed using the NEB Ultra II RNA Library Prep kit. Sequencing was performed by the Scientific Operations core at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II (HiFi), Illumina HiSeq X (10X) and Illumina HiSeq 4000 (RNA-Seq) instruments. Hi-C data were generated using the Arima v1 Hi-C kit and sequenced on HiSeq X.
Genome assembly
Assembly was carried out with Hifiasm ( Cheng et al., 2021). Haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). One round of polishing was performed by aligning 10X Genomics read data to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012). The assembly was then scaffolded with Hi-C data ( Rao et al., 2014) using SALSA2 ( Ghurye et al., 2019). The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext. The mitochondrial genome was assembled using MitoHiFi ( Uliano-Silva et al., 2021). The genome was analysed and BUSCO scores generated within the BlobToolKit environment ( Challis et al., 2020). Table 3 contains a list of all software tool versions used, where appropriate.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Hifiasm | 0.12 | Cheng et al., 2021 |
| purge_dups | 1.2.3 | Guan et al., 2020 |
| SALSA2 | 2.2 | Ghurye et al., 2019 |
| longranger align | 2.2.2 |
https://support.10xgenomics.com/genome-exome/
software/pipelines/latest/advanced/other-pipelines |
| freebayes | 1.3.1-17-
gaa2ace8 |
Garrison & Marth, 2012 |
| MitoHiFi | 1.0 | Uliano-Silva et al., 2021 |
| gEVAL | N/A | Chow et al., 2016 |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| PretextView | 0.1.x | https://github.com/wtsi-hpag/PretextView |
| BlobToolKit | 2.6.2 | Challis et al., 2020 |
Ethical/compliance issues
The materials that have contributed to this genome note were supplied by a Tree of Life collaborator. The Wellcome Sanger Institute employs a process whereby due diligence is carried out proportionate to the nature of the materials themselves, and the circumstances under which they have been/are to be collected and provided for use. The purpose of this is to address and mitigate any potential legal and/or ethical implications of receipt and use of the materials as part of the research project, and to ensure that in doing so we align with best practice wherever possible.
The overarching areas of consideration are:
Ethical review of provenance and sourcing of the material;
Legality of collection, transfer and use (national and international).
Each transfer of samples is undertaken according to a Research Collaboration Agreement or Material Transfer Agreement entered into by the Tree of Life collaborator, Genome Research Limited (operating as the Wellcome Sanger Institute) and in some circumstances other Tree of Life collaborators.
Data availability
European Nucleotide Archive: Vanessa cardui (painted lady) genome assembly, ilVanCard2. Accession number PRJEB42869; https://identifiers.org/ena.embl/PRJEB42869.
The genome sequence is released openly for reuse. The V. cardui genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
Author information
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.4893704.
Members of the Wellcome Sanger Institute Tree of Life programme collective are listed here: https://doi.org/10.5281/zenodo.5377053.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.4790456.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.5013542.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783559.
References
- Aken BL, Ayling S, Barrell D, et al. : The Ensembl Gene Annotation System. Database (Oxford). 2016;2016:baw093. 10.1093/database/baw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo AP, Sourkov V, Pereira GAG, et al. : RNAsamba: Neural Network-Based Assessment of the Protein-Coding Potential of RNA Sequences. NAR Genom Bioinform. 2020;2(1):lqz024. 10.1093/nargab/lqz024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celorio-Mancera MDLP, Wheat CW, Huss M, et al. : Evolutionary history of host use, rather than plant phylogeny, determines gene expression in a generalist butterfly. BMC Evol Biol. 2016;16:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat Methods. 2021;18(2):170–75. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL - a web-based browser for evaluating genome assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberale-Stille G, Schäpers A, Janz N, et al. : Selective Attention by Priming in Host Search Behavior of 2 Generalist Butterflies. Behav Ecol. 2019;30(1):142–49. 10.1093/beheco/ary146 [DOI] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing.arXiv: 1207.3907.2012. Reference Source [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. Gigascience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YJ, Yang DC, Kong L, et al. : CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017;45(W1):W12–16. 10.1093/nar/gkx428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Rahman N, Espinosa O, et al. : OrthoDB: The Hierarchical Catalog of Eukaryotic Orthologs. Nucleic Acids Res. 2008;36(Database issue):D271–75. 10.1093/nar/gkm845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wooster MJ, Grosvenor MJ, et al. : Strong impacts of smoke polluted air demonstrated on the flight behaviour of the painted lady butterfly ( Vanessa cardui L.). Ecol Entomol. 2021;46(2):195–208. 10.1111/een.12952 [DOI] [Google Scholar]
- Lorkovic Z: Die Chromosomenzahlen in der Spermatogenese der Tagfalter. Chromosoma. 1941;2:155–191. 10.1007/BF00325958 [DOI] [Google Scholar]
- Mazo-Vargas A, Concha C, Livraghi L, et al. : Macroevolutionary Shifts of WntA Function Potentiate Butterfly Wing-Pattern Diversity. Proc Natl Acad Sci U S A. 2017;114(40):10701–6. 10.1073/pnas.1708149114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menchetti M, Guéguen M, Talavera G: Spatiotemporal niche modelling of multigenerational insect migrations. Proc Biol Sci. 2019;286(1910):20191583. 10.1098/rspb.2019.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard E, van Swaay CAM, Stefanescu C, et al. : Migration of the Painted Lady Butterfly Cynthia Cardui in Europe: Evidence from Monitoring. Diversity and Distributions. 1998;4(5/6):243–53. Reference Source [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields O: World Distribution of the Vanessa Cardui Group (Nymphalidae). J Lepid Soc. 1992;46:235–38. Reference Source [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Stefanescu C: Moroccan Source Areas of the Painted Lady Butterfly Vanessa Cardui (Nymphalidae: Nymphalinae) Migrating into Europe in Spring. Journal of the Lepidopterists’ Society. 2011;65(1):15–26. 10.18473/lepi.v65i1.a2 [DOI] [Google Scholar]
- Stefanescu C, Páramo F, Åkesson S, et al. : Multi-Generational Long-Distance Migration of Insects: Studying the Painted Lady Butterfly in the Western Palaearctic. Ecography. 2013;36(4):474–86. 10.1111/j.1600-0587.2012.07738.x [DOI] [Google Scholar]
- Stefanescu C, Soto DX, Talavera G, et al. : Long-Distance Autumn Migration across the Sahara by Painted Lady Butterflies: Exploiting Resource Pulses in the Tropical Savannah. Biol Lett. 2016;12(10):20160561. 10.1098/rsbl.2016.0561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchan T, Talavera G, Sáez L L, et al. : Pollen metabarcoding as a tool for tracking long-distance insect migrations. Mol Ecol Resour. 2018;19(1):149–162. 10.1111/1755-0998.12948 [DOI] [PubMed] [Google Scholar]
- Talavera G, Vila R: Discovery of Mass Migration and Breeding of the Painted Lady Butterfly Vanessa Cardui in the Sub-Sahara: The Europe--Africa Migration Revisited. Biological Journal of the Linnean Society. Linnean Society of London.2016;120(2):274–85. 10.1111/bij.12873 [DOI] [Google Scholar]
- Talavera G, Bataille C, Benyamini D, et al. : Round-trip across the Sahara: Afrotropical Painted Lady butterflies recolonize the Mediterranean in early spring. Biol Lett. 2018;14(6):20180274. 10.1098/rsbl.2018.0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Childers RA, Shi NN, et al. : Physical and Behavioral Adaptations to Prevent Overheating of the Living Wings of Butterflies. Nat Commun. 2020;11(1):551. 10.1038/s41467-020-14408-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uliano-Silva M, Nunes JGF, Krasheninnikova K, et al. : marcelauliano/MitoHiFi: mitohifi_v2.0.2021. 10.5281/zenodo.5205678 [DOI] [Google Scholar]
- UniProt Consortium: UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019;47(D1):D506–15. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A, Coetzer A: IUCN Red List of Threatened Species: Vanessa Cardui. IUCN Red List of Threatened Species. 2020. 10.2305/IUCN.UK.2020-3.RLTS.T174379A161326679.en [DOI] [Google Scholar]
- Williams CB: The migrations of the painted lady butterfly, Vanessa cardui. (Nymphalidae), with special reference to North America. Journal of the Lepidopterists’ Society. 1970;24:157–175. [Google Scholar]
- Zhang L, Mazo-Vargas A, Reed RD: Single Master Regulatory Gene Coordinates the Evolution and Development of Butterfly Color and Iridescence. Proc Natl Acad Sci U S A. 2017;114(40):10707–12. 10.1073/pnas.1709058114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Reed RD: Genome Editing in Butterflies Reveals That Spalt Promotes and Distal-Less Represses Eyespot Colour Patterns. Nat Commun. 2016;7:11769. 10.1038/ncomms11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Steward RA, Wheat CW, et al. : High-Quality Genome Assembly and Comprehensive Transcriptome of the Painted Lady Butterfly Vanessa Cardui. Genome Biol Evol. 2021;13(7):evab145. 10.1093/gbe/evab145 [DOI] [PMC free article] [PubMed] [Google Scholar]