Abstract
Traumatic brain injury (TBI) patients are at high risk for disruption of the gut microbiome. Previously, we have demonstrated that broad-spectrum antibiotic exposure after TBI drastically alters the gut microbiota and modulates neuroinflammation, neurogenesis, and long-term fear memory. However, these data did not determine if the impact of antibiotic exposure on the brain's response to injury was mediated directly by antibiotics or indirectly via modulation of the gut microbiota. We designed two different approaches to address this knowledge gap. One was utilizing fecal microbiota transplantation (FMT) from control and antibiotic-treated mice (treated with vancomycin, neomycin, ampicillin, and metronidazole [VNAM]) into germ-free (GF) mice prior to injury, and the other was exposing specific pathogen-free (SPF) mice to a 2-week period of antibiotics prior to injury but discontinuing antibiotics 72 h prior to injury. GF mice receiving FMT from VNAM-treated mice (GF-VNAM) demonstrated reduced gut bacterial alpha diversity and richness compared with GF mice receiving control FMT. At 7 days post-injury, GF-VNAM had increased microglial activation, reduced infiltration of T cells, and decreased neurogenesis. Similarly, SPF mice exposed to antibiotics prior to but not after injury demonstrated similar alterations in neuroinflammation and neurogenesis compared with control mice. These data support our hypothesis implicating the gut microbiota as an important modulator of the neuroinflammatory process and neurogenesis after TBI and provide an exciting new approach for neuroprotective therapeutics for TBI.
Keywords: germ-free mice, gut microbiome, microglia, neurogenesis, T cells, traumatic brain injury
Introduction
The gut microbiota, composed of tens of trillions of microorganisms in the gastrointestinal tract, has been implicated in brain homeostasis in health and disease.1,2 The activity and composition of the gut microbial populations regulate a considerable number of biological processes, including gut motility, immune response, and homeostasis of the central nervous system (CNS).3 This relationship is well described as the gut–brain axis, a bidirectional communication through neuro-endocrine-immunological signaling.4 Alterations in composition or metabolic activity such as change in diet, antibiotic exposure, or stress can induce profound changes in the gut bacteria population and can influence brain homeostasis and behavior in mice.5
Microglia maturation and function is influenced by gut microbiota through bacterially derived metabolites, specifically short-chain fatty acids (SCFAs).6 Recently, it also has been shown that brain-resident CD4+ T cells are required for microglial maturation, and that their absence results in defective synaptic pruning and behavior deficits.7 Further, intestinal microbes have emerged as a potent regulator of lymphocyte populations, including regulatory T (Treg) and γδT cells, suggesting another possible mechanistic link for gut microbiota control of microglia maturation and homeostasis in health and diseases such as stroke,8 multiple sclerosis,9 and/or traumatic brain injury (TBI).10
The gut microbiota's influence on TBI is of paramount clinical significance, as TBI patients are highly susceptible to alterations in the gut microbiota because of frequent antibiotic administration, prolonged hospitalization, and autonomic dysfunction.11 Recently, our research team has demonstrated that antibiotic-induced gut microbial dysbiosis exacerbated microglia activation, reduced T cell infiltration, and decreased neurogenesis, with long-term consequences for neuronal survival and fear memory.12,13 However, these studies did not directly evaluate whether antibiotic-induced changes in the brain's response to TBI were via a direct effect of these pharmaceuticals on the brain or instead mediated indirectly via modulation of the bacterial gut microbiome.
In this report, using fecal microbiota transplantation (FMT) into germ-free (GF) mice, or discontinuation of antibiotics exposure 72 h prior to injury, we have reproduced our previously described antibiotic-associated alterations in microglia morphology, T cell infiltration, and neurogenesis after TBI. Therefore, our findings support that the gut microbiota serves as an important modulator of the neuroinflammatory process and neurogenesis after TBI.
Methods
Animals
GF C57BL/6J and SPF C57BL/6J 8-week-old male mice (Jackson Laboratory, Bar Harbor, ME) weighing 20–25 g were used. Animals were housed five per cage and had free access to water and food with a 12-h light/dark cycle. GF mice were housed in the Gnotobiotic Core Facility at Washington University School of Medicine in St. Louis, including during the period of fecal microbiota transplant, until controlled cortical impact. Thereafter, GF mouse cages were sealed shut and not opened until the mice were euthanized.
Controlled cortical impact (CCI)
All procedures were approved by the Washington University Animal Studies Committee (Protocol 19-0864) and are consistent with the National Institutes of Health guidelines for the care and use of animals. Briefly, mice were anesthetized with 5% isoflurane at induction, followed by maintenance at 2% isoflurane for the procedure's duration. Buprenorphine sustained release (0.5 mg/kg subcutaneously) was administered before scalp incision. The head was shaved and ear bars were used to stabilize the head within the stereotaxic frame (MyNeurolab, St. Louis, MO). Then, a single 5-mm craniectomy was performed using an electric drill on the left lateral side of the skull centered 2.7 mm lateral from the midline and 3 mm anterior to lambda. A 3-mm electromagnetic impactor tip was aligned with the craniectomy site at 1.2 mm left of midline and 1.5 mm anterior to the lambda suture. The impact was delivered at 2 mm depth (velocity 5 m/sec, dwell time 100 ms) inducing CCI. The ears bars were released immediately after the injury. All animals then received a loose-fitting plastic cap secured over the craniectomy with Vetbond (3M, St. Paul, MN). The skin was closed with interrupted sutures and was treated with antibiotic ointment before the mouse recovered from anesthesia on a warming pad.
Antibiotic-induced depletion of the microbiota
To deplete the bacterial microbiota, we used broad-spectrum antibiotics in the drinking water: 250 mg vancomycin, 500 mg neomycin-sulfate, 500 mg ampicillin, 500 mg metronidazole (VNAM), and 10 g grape Kool-Aid (Kraft Heinz, IL, Chicago) in 500 mL water, which was sterile filtered through a 0.22 μm filter as previously described.12
Fecal microbiota transplantation
We performed two FMTs of the gut microbiota from VNAM- or Kool-Aid-treated uninjured animals into GF mice at Day 0 and Day 7 prior to injury (Fig. 1A). After 7 days of VNAM and Kool-Aid treatment, we extracted 10 pellets from each group right before Day 0 and Day 7 to make a fresh fecal slurry. We used a different cohort of VNAM mice (5 mice each) for each extraction. Briefly, four to five mouse fecal pellets were collected 7 days after VNAM or Kool-Aid treatment. The pellets were mixed with phosphate-buffered saline (PBS) and after 5 min, the sample was vortexed to break up the fecal matter and allowed to sit for ∼5 min to allow for debris to settle. The supernatant was then removed with an uncut P1000 pipette tip into a sterile 5 mL tube to avoid material clogging the gavage needle. Mice were gavaged with a sample volume of 250 μL. Ten days after the last FMT, we performed CCI in all the mice and they remained in sealed cages in the animal facility for 7 days before being euthanized.
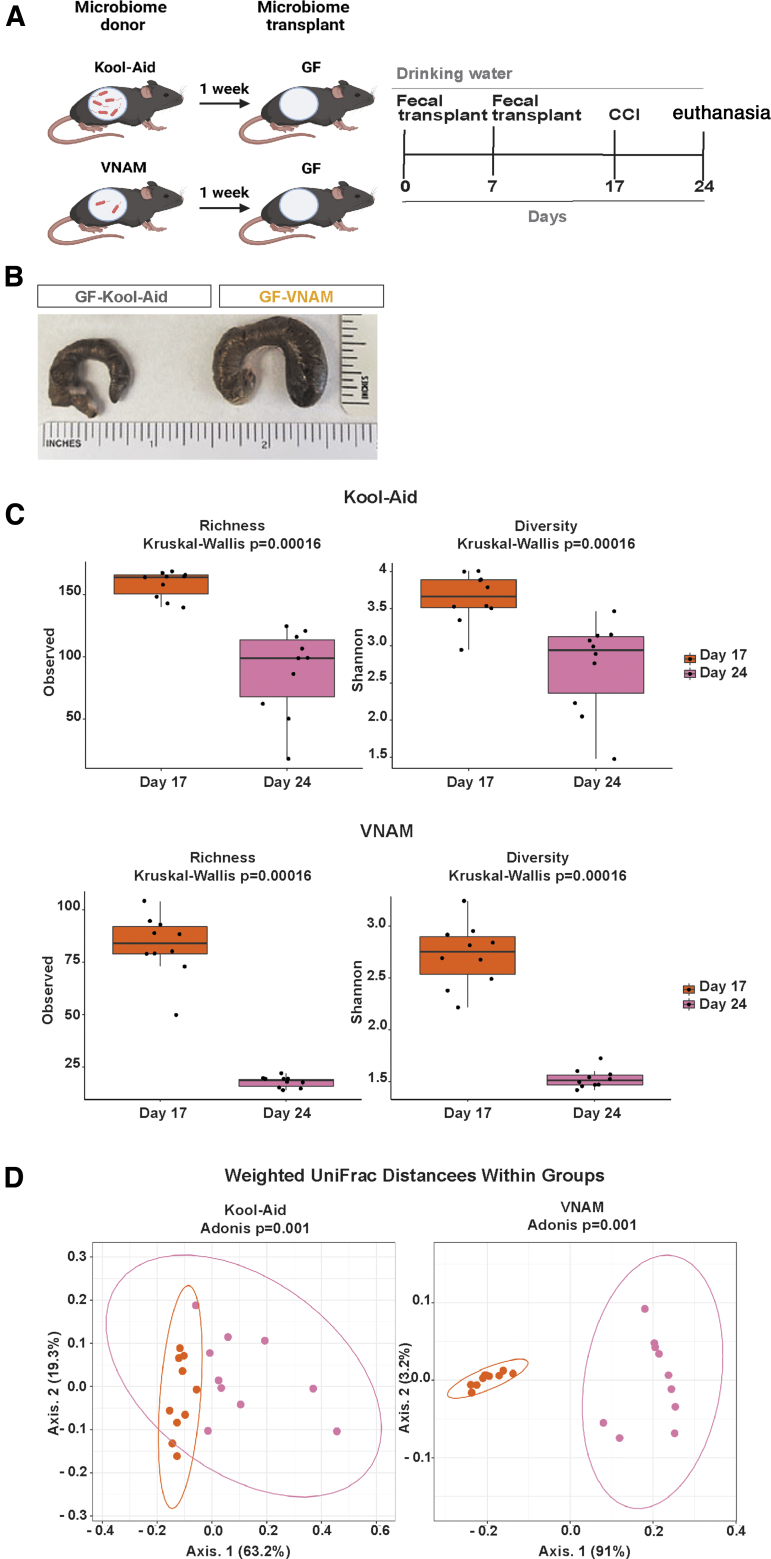
FIG. 1.
Fecal microbiota transplantation from antibiotic-treated mice induces gut microbial dysbiosis in germ-free (GF) mice over time. (A) Experimental design; GF mice were gavaged with two fecal microbiota transplants (FMT) of the gut microbiota from mice treated with vancomycin, neomycin, ampicillin, and metronidazole (VNAM) (GF-VNAM) or Kool-Aid (GF-Kool-Aid) uninjured animals on Day 0 and Day 7. Ten days after the second FMT, we then performed controlled cortical impact (CCI) on the fecal transplant recipient GF mice. (B) Ceca from GF-Kool-Aid (control) and GF-VNAM mice. (C) Graph depicts of richness and Shannon α-diversity index of grouped data. Kruskal–Wallis analysis of variance (ANOVA) (***p < 0.001). (D) β-diversity graphs using weighted UniFrac distance metrics of bacterial community structures. Adonis (PERMANOVA) analysis (***p < 0.001).
Fecal bacterial 16S rRNA gene analysis
Fecal pellets were collected into sterile 1.7 mL tubes. Phenol:chloroform-extracted DNA from fecal pellets was used for 16S rRNA gene sequencing. For 16S sequencing, primer selection and polymerase chain reactions (PCRs) were performed as described previously.14 Briefly, each sample was amplified in triplicate with Golay-bar coded primers specific for the V4 region (F515/R806), combined, and confirmed by gel electrophoresis. PCR reactions contained 18.8 μL RNase/DNase-free water, 2.5 μL 10 × High Fidelity PCR Buffer (Invitrogen, 11304–102), 0.5 μL 10 mM deoxynucleoside triphosphates (dNTPs), 1 μL 50 mM MgSO4, 0.5 μL each of the forward and reverse primers (10 μM final concentration), 0.1 μL Platinum High Fidelity Taq (Invitrogen, 11304–102), and 1.0 μL genomic DNA. Reactions were held at 94°C for 2 min to denature the DNA, with amplification proceeding for 26 cycles at 94°C for 15 sec, 50°C for 30 sec, and 68°C for 30 sec; a final extension of 2 min at 68°C was added to ensure complete amplification. Amplicons were pooled and purified with 0.6 × Agencourt AMPure XP beads (Beckman-Coulter, A63882) according to the manufacturer's instructions. The final pooled samples, along with aliquots of the three sequencing primers, were sent to the DNA Sequencing Innovation Lab (Washington University School of Medicine) for sequencing by the 2 × 250 bp protocol with the Illumina MiSeq platform.
Read quality control and the resolution of amplicon sequence variants were performed with the dada2 R package.15 Amplicon sequence variants that were not assigned to the kingdom bacteria were filtered out. The remaining reads were assigned taxonomy using the Ribosomal Database Project (RDP trainset 16/release 11.5) 16S rRNA gene sequence database.16 Ecological analyses, such as alpha-diversity (richness, Faith's phylogenetic diversity) and beta-diversity analyses (weighted UniFrac distances), were performed using PhyloSeq and additional R packages.17
Cell isolation from brain and blood samples
The blood and the regions of interest in the ipsilateral brain (hippocampus and cortex) were harvested 7 days after injury. Five animals (from all conditions) were processed per day with a total of 20 samples at the time. Mice were anesthetized with isoflurane, and blood samples were taken in ethylenediaminetetraacetic acid (EDTA) tubes immediately before transcardial perfusion with ice-cold 0.1 M heparinized PBS. The brain regions of interest were dissected out on ice and digested at 37°C for 15 min with collagenase D (400 units/mL, Roche) in Dulbecco's PBS (Lonza, Basel, Switzerland), each containing 50 μg/mL of DNase I (Sigma-Aldrich). The tissue was then mechanically dissociated with a glass Pasteur pipette, filtered through a 70-μm nylon cell strainer, and centrifuged at 950 rpm for 15 min. A 25% Percoll (Sigma-Aldrich) column was used to remove cell debris and myelin, followed by centrifugation at 1700 rpm for 10 min. A 25 μL-blood sample was mixed with 1 × Red Blood Lysis Buffer (Roche) and incubated in rotation for 15 min at room temperature (RT). Samples were then centrifuged at 3500 rpm for 5 min at RT. The supernatant was discarded, and cells were washed and resuspended in 1 mL of cytometer buffer (0.5% bovine serum albumin [Sigma-Aldrich], 5 mM EDTA (Millipore, Burlington, MA) in PBS). Then, the cells were resuspended in 100 μL of cytometer buffer and stained.
Cell isolation of lamina propia (LP) immune cells
The small intestines were removed and separated. Peyer's patches were excised, and intestines were cleaned of mesenteric fat and intestinal contents with PBS. Then intestines were opened longitudinally, washed with Hanks' Balanced Salt Solution (HBSS)/HEPES, cut into 1-cm pieces and placed into 20 ml of HBSS/HEPES 10% bovine calf serum (BCS) and 5 mM EDTA for 20 min at 37°C. During this period, tissue suspensions were vortexed for 15 sec each 5 min. Tissue pieces were washed with HBSS/HEPES to remove EDTA, minced thoroughly, and placed into 10 mL RMPI with 10% fetal bovine serum (FBS), 0.01M HEPES, 1 × kanamycin, 1 × GlutaMAX™, 1 × sodium pyruvate, 1 × non-essential amino acids, 10 mg/mL DNAase I (Roche) and 100 U/mL collagenase IV (Sigma). Tissues were digested at 37°C for 50 min with constant agitation (250 rpm). The resulting LP cell suspensions were filtered through a 70-μm nylon cell strainer and washed with 10 mL HBSS/HEPES. LP cell suspensions were collected at 2000 rpm for 10 min at 4°C. Cell pellets were resuspended in 4 mL 40% Percoll (GE Healthcare) and overlaid over 3 mL of 80% Percoll. Gradients were centrifuged at 2000 rpm for 20 min at 4°C, and cells at the interface were collected and washed with 50 mL PBS. Cells were stained for flow cytometric analysis.
Flow cytometry analysis
Cells were incubated for 5 min at room temperature (RT) with Zombie NIR Dye (BioLegend, San Diego, CA) to assess their viability. The Zombie NIR Dye was quenched, and cells were washed with cytometry buffer and blocked with FcR blocking reagent (1:50, Miltenyi Biotec, Bergisch Gladbach, Germany). Then, the samples were washed with cytometry buffer, stained with antibodies (Table 1) for 15 min at RT, and analyzed on a BD LSRFortessa flow cytometer (BD Biosciences, Franklin Lakes, NJ) using the Software v10.6.1 (BD Biosciences, Franklin Lakes, NJ). Microglial cells were defined as CD45lowCD11b+ and T cells were defined as CD45highCD11b−CD3+. Fluorescence minus one (FMO) and isotype control antibodies were used as negative controls for each marker.
Table 1.
Overview of the Primary Antibodies Used in the Present Study
| Antibody | Fluorophore | Clone | Species | Source | Product number |
|---|---|---|---|---|---|
| CD45 | BV425 | 30-F11 | Rat monoclonal | BioLegend | 103134 |
| CD3ɛ | AF700 | 500-A2 | Armenian Hamster monoclonal | BioLegend | 100320 |
| CD4 | BUV395 | GK1.5 | Rat monoclonal | BD Biosciences | 565974 |
| CD8a | PerCP-Cy5.5 | 53-6.7 | Rat monoclonal | BioLegend | 100733 |
| CD11b | BV510 | M1/70 | Rat monoclonal | BioLegend | 101263 |
| TCR γδ | FITC | GL3 | Armenian Hamster monoclonal | BioLegend | 118105 |
| CD25 | PE | PC61 | Rat monoclonal | BioLegend | 102007 |
| MHC-II | PerCP-710 | AF6-120.1 | Mouse monoclonal | eBioscience | 46-5320-80 |
| TLR4 | PE-Cy7 | SA15-21 | Rat monoclonal | BioLegend | 145407 |
| Ly6C | BV785 | HK1.4 | Rat monoclonal | BioLegend | 128041 |
| Ly6G | AF700 | 1A8 | Rat monoclonal | BioLegend | 127622 |
| NeuN | A60 | Mouse monoclonal | Millipore | MAB377 | |
| DCX | Rabbit polyclonal | Abcam | Ab18723 | ||
| BrdU | BU1/75 | Rat monoclonal | Abcam | Ab6326 | |
| Iba1 | NCNP24 | Rabbit polyclonal | Wako | 019-19741 | |
| Secondary antibody |
AF594 | Donkey anti-rat | Thermo Fisher |
A-21209 | |
| Secondary antibody |
AF647 | Donkey anti-mouse | Thermo Fisher |
A-31571 | |
| Secondary antibody |
AF488 | Donkey anti-rabbit | Thermo Fisher |
A-21206 |
BrdU treatment
For the analysis of neurogenesis, the animals received intraperitoneal injections of 5-bromo-2'-deoxyuridine (BrdU, Sigma-Aldrich) 50 mg/g of body weight at a concentration of 10 mg/mL in sterile saline daily, starting on post-injury Days 3–6.
Tissue processing
Mice were anesthetized by isoflurane followed by cervical dislocation and transcardial perfusion with ice-cold 0.1 M heparinized PBS (pH 7.4) followed by 4% paraformaldehyde (PFA, Sigma-Aldrich, St Louis, MO). Perfused brains were kept in 4% PFA at 4°C overnight, followed by equilibration in 30% sucrose for at least 48 h before sectioning.
Immunohistochemistry
Histological sections were examined to assess microglia and neurogenesis alteration in the hippocampus. Immunofluorescence staining was performed on free-floating sections (50-μm thick). The serial sections were cut on a freezing microtome starting with the appearance of a complete corpus callosum and caudally to bregma -3.08 mm. Sets of 12 sections spaced every 300 μm were mounted on glass slides and used for immunohistochemical studies. Staining was performed on free-floating sections washed in PBS between applications of primary and secondary antibodies. For BrdU immunostaining, a widely used procedure is to perform an incubation in 1 N HCl (30 min at 45°C) to denature the DNA after the PBS washes. Normal donkey serum (NDS) (20%) in PBS was used to block non-specific staining for all antibodies. Slices were then incubated at 4°C overnight with the primary antibodies (Table 1). The following day, antibody binding was detected by incubating sections with Alexa fluorescence secondary antibody (Table 1) for 2 h. Sections were mounted on glass slides in PBS, dried, and cover-slipped with mounting medium for fluorescence with DAPI.
Three-dimensional reconstruction of microglia
We followed a published protocol for microglia morphology analysis:6 50-μm thick sections were stained with ionized calcium binding adaptor molecule 1 (Iba1) (Table 1) 4°C overnight, followed by Alexa Fluor 488–conjugated secondary antibody (Table 1) staining for 2 h. Sections were mounted on glass slides in tris-buffered saline (TBS)-X, dried, and cover-slipped with mounting medium for fluorescence with DAPI VECTOR Laboratories, Burlingame, CA, USA). Imaging was performed on a Zeiss LSM 880 confocal laser scanning microscope (Zeiss, White Plains, NY) using a 20 × 0.8 NA objective. Z-stacks were done with 1.00-μm steps in z direction; 1024 × 1024 pixel resolution was recorded and analyzed using IMARIS software (Bitplane, Concord, MA). From each image, three hippocampal microglia that were completely within the z-stack image from the ipsilateral cornu Ammonis (CA)3 striatum radiatum were selected by a blinded observer. The filament tracer function was utilized in IMARIS to quantify branch length, volume, number of branch points, and terminal points. A total of three microglia were analyzed from each mouse.
Statistical analysis
All data were analyzed using GraphPad Prism 9 (La Jolla, CA). Data results are presented as mean ± standard error of the mean. There was no evidence for significant deviations from normal distribution (p > 0.05 by Shapiro–Wilk tests). Data were analyzed with student t test. All analysis was performed blinded to group assignment.
Results
Fecal microbiota transplantation from antibiotic-treated mice induces gut microbial dysbiosis in GF mice
We transplanted FMT from VNAM-treated and control animals into GF mice to test our hypothesis that antibiotic modulation of neuroinflammation after TBI is mediated through changes in the gut microbiota. Stool samples were collected from VNAM-treated or Kool-Aid-treated mice and were transferred to GF C57BL/6 male mice (GF-VNAM and GF-Kool-Aid, respectively) (Fig. 1A) at Day 0 and Day 7. At 7 days post-injury (Day 24), GF-VNAM mice had enlarged ceca compared with GF-Kool-Aid mice (Fig. 1B). Sequencing analysis of the V4 region of the 16S rRNA gene in DNA isolated from stool at both Day 17 and Day 24 within the groups revealed reduced bacterial alpha diversity and richness in GF-Kool-Aid mice (Fig. 1C, top graphs) which were more pronounced in GF-VNAM mice (Fig. 1C, bottom graphs). Beta-diversity analysis using weighted UniFrac distance metrics similarly revealed that GF-Kool-Aid mice (Fig. 1D, left graph) had significantly altered bacterial community structures over time, which were more pronounced in GF-VNAM (Fig. 1D, right graph).
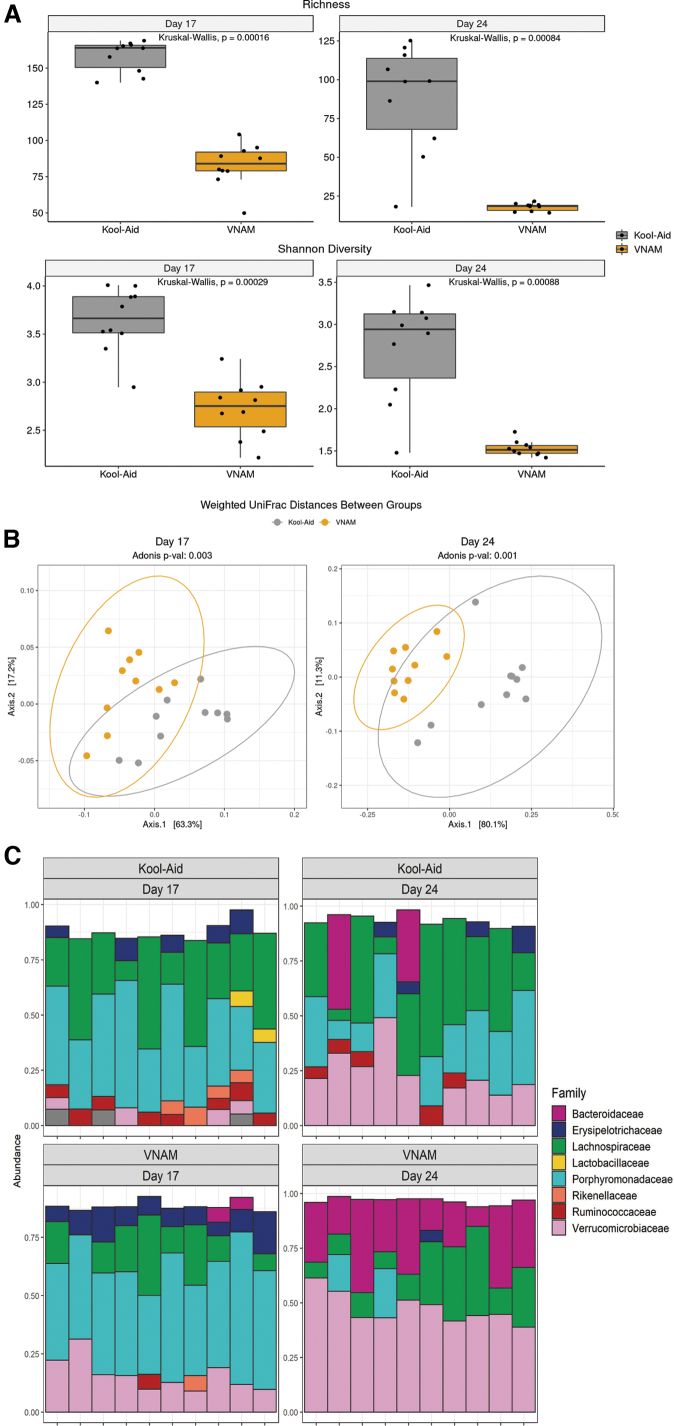
Comparing the 16S sequencing results between groups at both Day 17 and Day 24, data revealed reduced bacterial alpha diversity and richness in GF-VNAM mice compared with those of GF-Kool-Aid mice (Fig. 2A), mirroring the differences observed in the gut microbiota in VNAM-treated animals.12 Beta-diversity analysis using weighted UniFrac distance metrics similarly revealed that GF-VNAM mice had significantly altered bacterial community structures compared with GF-Kool-Aid mice at both Day 17 and Day 24 (Fig. 2B). Whereas analysis of the family-level taxonomic composition indicated enhanced relative abundance of Verrucomicrobiaceae and loss of Ruminococcaceae in GF-VNAM mice at Day 17, community composition differences were even more dramatic at Day 24, wherein Verrucomicrobiaceae was relatively increased associated with relative loss of Erysipelotrichaceae, Lachnospiraceae, Ruminococcaeae, and Porphyromonadaceae in GF-VNAM compared with GF-Kool-Aid mice (Fig. 2C). These findings indicate continued differences in bacterial communities independent of the direct concurrent effect of antibiotics between the groups after injury and removal from the gnotobiotic facility.
FIG. 2.
Fecal microbiota transplantation from antibiotic-treated mice induces gut microbial dysbiosis in germ-free (GF) mice compared with controls. (A) Graph depicts of richness and Shannon α-diversity index of grouped data. Kruskal–Wallis analysis of variance (ANOVA) (***p < 0.001). (B) β-diversity graphs using weighted UniFrac distance metrics of bacterial community structures. Adonis (PERMANOVA) analysis (***p < 0.001). (C) Family-level phylogenetic classification of fecal 16S rDNA gene frequencies from GF-Kool-Aid and GF-vancomycin, neomycin-sulfate, ampicillin and metronidazole treated (VNAM) mice at the time of injury (Day 17) and before euthanasia (Day 24). Each bar represents an individual animal. Only families with a frequency >5% were included.
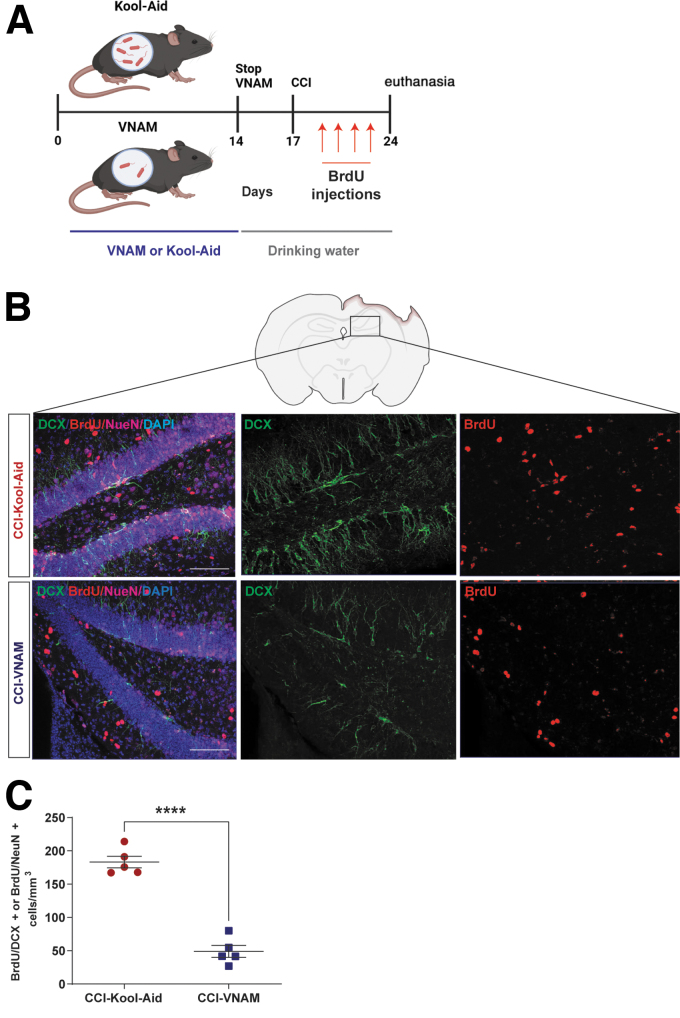
Fecal transplantation from VNAM-treated mice results in decreased neurogenesis in injured GF mice
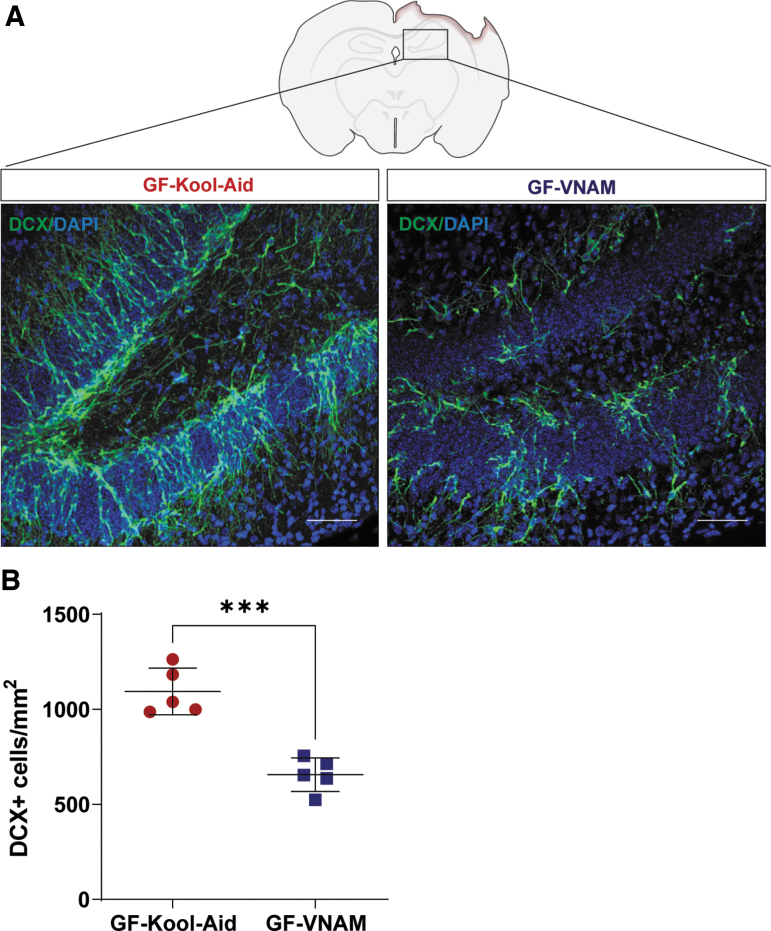
An increase of neurogenesis at the subgranular zone (SGZ) of the dentate gyrus (DG) has been described after TBI in mice18,19 and humans,20 suggesting that the death of pre-existing granule neurons in the DG may act as a trigger for enhanced neurogenesis.18 We have previously reported significant reductions in neurogenesis after TBI in mice with antibiotic-induced gut microbial dysbiosis.12 To determine if the antibiotic-induced reductions in neurogenesis are mediated by changes in the gut microbiota, we assessed neurogenesis in GF-VNAM and GF-Kool-Aid mice 7 days after CCI (Fig. 3A). We observed a significantly reduced number of doublecortin (DCX)-positive cells in the DG of GF-VNAM compared with GF-Kool-Aid mice (Fig. 3B), supporting our hypothesis that antibiotic exposure modulates neurogenesis via alterations in the gut microbiota.
FIG. 3.
Neurogenesis reduction in germ-free (GF) transplanted mice 7 days after injury. (A) Representative images of the subgranular zone (SGZ) in the dentate gyrus (DG) of the hippocampus labeled with doublecortin (DCX) (green) and DAPI (blue). Scale bar 20 μm. (B) Quantification of stained cells per area. Three slices from the hippocampus spaced 300 μm apart were evaluated for each mouse. Mean values are plotted ± standard error of the mean (SEM), Unpaired t test (***p < 0.001).
Fecal transplantation from VNAM-treated mice alters local and peripheral immune infiltration after TBI in GF mice
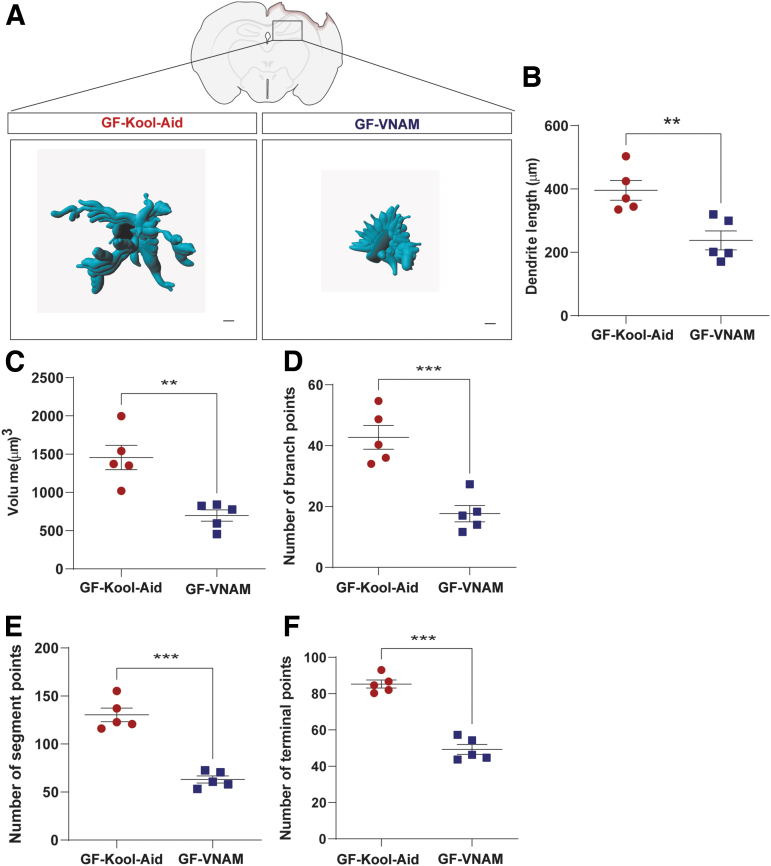
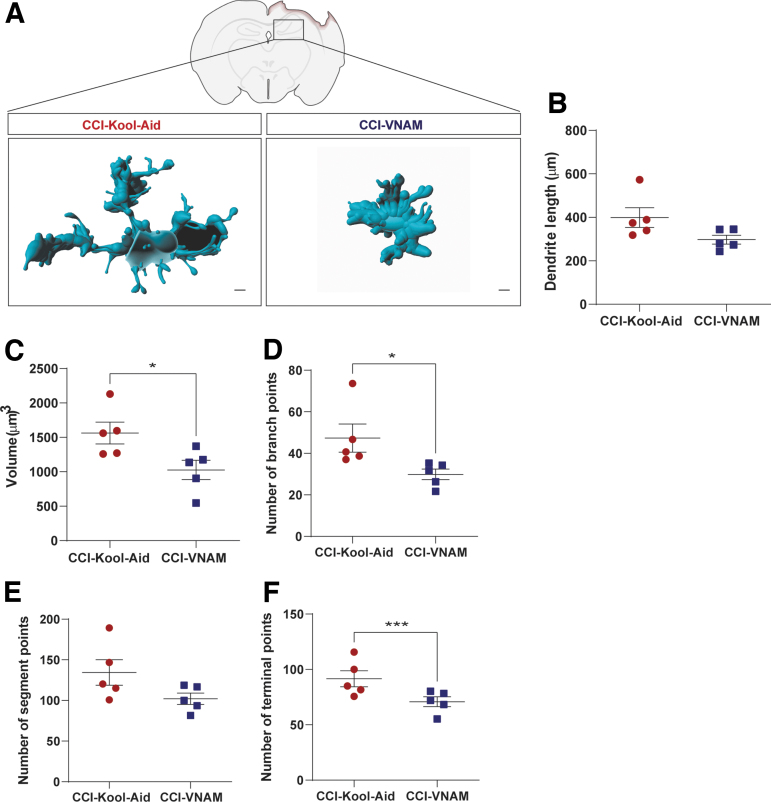
Microglia, the innate immune cells in the brain, are modulated by the gut microbiota during maturation and function.6 To investigate the effect of depleted microbiota on microglial response after TBI, we performed a semiautomatic quantitative morphometric three-dimensional measurement of hippocampal microglia 7 days after injury (Fig. 4A). We found significantly shorter dendrite length (Fig. 4B), and a decrease of volume (Fig. 4C), branch points (Fig. 4D), segment points (Fig. 4E), and terminal points (Fig. 4F) in the GF-VNAM mice compared with the GF-Kool-Aid mice. These microglial morphological changes support the direct impact of the gut microbiome on microglial response after TBI.
FIG. 4.
Microglia morphology changes in germ-free (GF) transplanted mice 7 days after injury. (A) Representative three-dimensional reconstruction images of microglia cells. Scale bar 15 μm. (B–F) Quantification of microglia morphology. (B) Dendrite length, (C) volume, (D) number of branch points, (E) number of segment points, and (F) number of terminal points. Mean values are plotted ± standard error of the mean (SEM), unpaired t test (**p > 0.01;***p < 0.001).
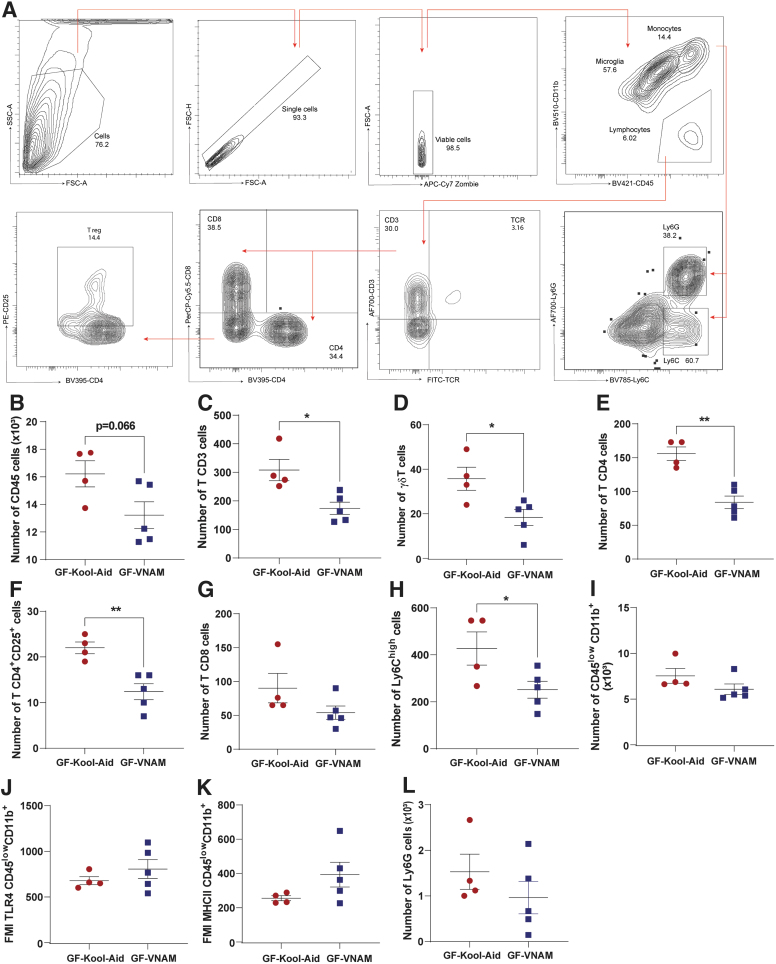
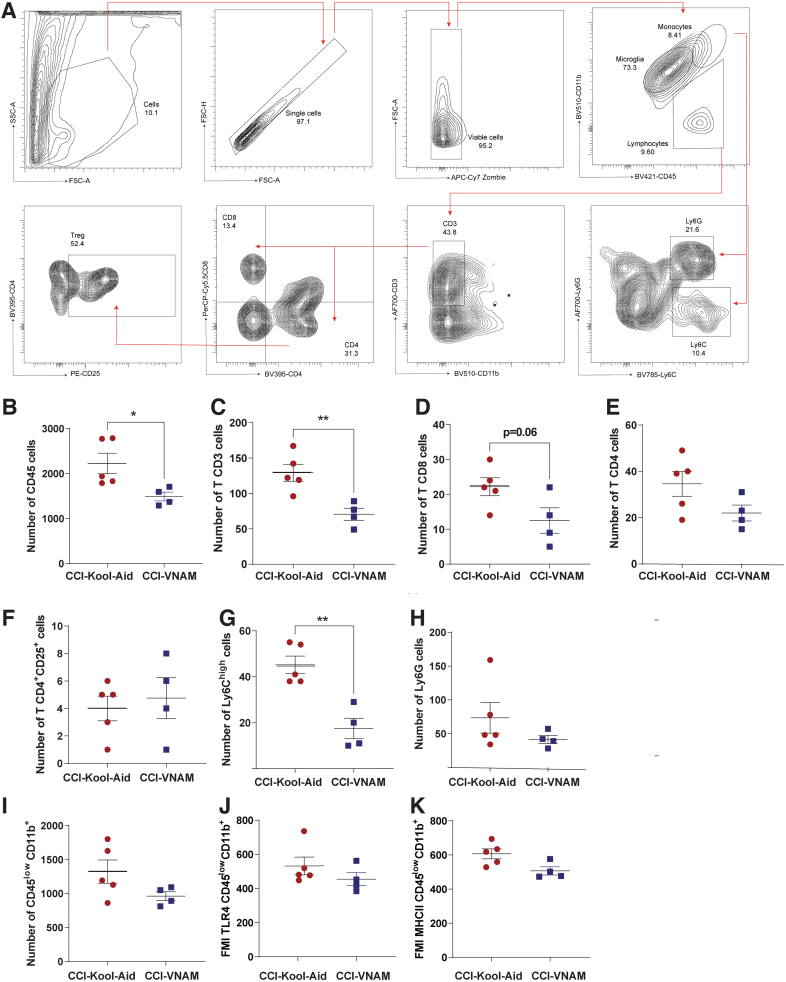
We have observed that antibiotic-induced gut microbial dysbiosis after TBI reduces peripheral recruitment of monocytes (3 days post-injury) and lymphocytes (7 days and up to 30 days post-injury) to the brain.12 To determine if antibiotic-induced reduction of peripheral immune cell infiltration after TBI is mediated by the gut microbiota, we measured the peripheral immune response in the brain parenchyma 7 days after injury by flow cytometry in GF mice receiving FMT (Fig. 5A). We found a trend of decreasing of CD45+ cells in GF-VNAM mice compared with GF-Kool-Aid mice (Fig 5B). We found a significant decrease in CD3 T cells (Fig. 5C), γδT cells (Fig. 5D), CD4 T cells (Fig. 5E), CD4+CD25+ T cells (T reg. Fig. 5F), and infiltration in the brains of GF-VNAM mice compared with those GF-Kool-Aid mice, with no changes in CD8+ cells population (Fig 5G). Moreover, we also observed a significant decrease in Ly6Chigh monocytes (Fig. 5H) brain infiltration in GF-VNAM mice, but found no differences in microglia number (Fig. 5I) or expression of microglial activation markers (toll-like receptor [TLR]4, Fig. 5J or major histocompatibility complex [MHC]II, Fig. 5K) or neutrophils (Fig. 5L).
FIG. 5.
Local immune alterations in the brain of germ-free (GF) transplanted mice 7 days after injury. (A) Gating strategy of the adaptive immune response in the brain. (B–L) Quantification of cell absolute numbers in the hippocampi and cortices. (B) Leukocytes (CD45), (C) CD3 T cells (CD11b-CD3+), (D) T reg. cells, (E) CD4 T cells (CD11b-CD3+CD4+), (F) CD11b-CD4+CD25+, (G) CD8 T cells (CD11b-CD3+CD8+), (H) Ly6Chigh cells, (I) microglia (CD45low CD11b) and microglial activation markers, (J) fluorescence median intensity (FMI) of toll-like receptor (TLR)4, (K) FMI major histocompatibility complex (MHC)II, and (L) Ly6G cells. Mean values are plotted ± standard error of the mean (SEM), unpaired t test (*p < 0.05, **p < 0.01).
Fecal transplantation from VNAM-treated mice does not alter peripheral immune response in the small intestine or blood after TBI
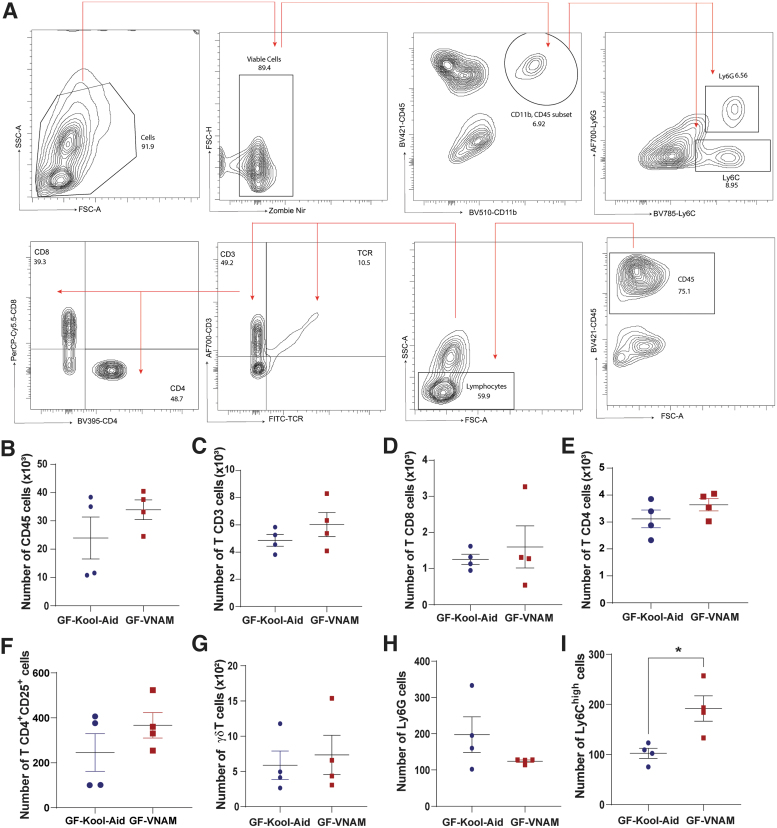
Additionally, we performed flow cytometry in the LP of the small intestine and peripheral blood at 7 days post-injury to assess the peripheral immune system response in other compartments. We did not find differences in the T cell populations of the LP (Fig. 6B–H) or blood (Supplementary Fig. S1B–I) between the two FMT groups 7 days after injury. However, we did observe a significant increase in monocyte response (Ly6Chigh cells, Fig. 6H) in GF-VNAM compared with GF-Kool-Aid mice in the LP. These data demonstrated that reduced brain infiltration of peripheral immune cells after TBI in GF-VNAM mice cannot be explained by suppression of these cell populations in systemic compartments.
FIG. 6.
Local immune alterations in the small intestine of germ-free (GF) transplanted mice 7 days after injury. (A) Gating strategy of the innate and adaptive immune response in the brain. (B–H) Quantification of immune cell absolute numbers in the lamina propia. (B) Leukocytes (CD45), (C) CD3 T cells (CD11b-CD3+), (D) CD8 T cells (CD11b-CD3+CD8+), (E) CD4 T cells (CD11b-CD3+CD4+), (F) T reg cells (CD11b-CD4+CD25+), (G) δγT cells, and (H) Ly6Ghigh cells. (I) Ly6Chigh cells. Mean values are plotted ± standard of the mean (SEM), Unpaired t test (*p < 0.05).
Antibiotic exposure prior to but not after injury results in decreased neurogenesis
The experimental design of antibiotic depletion of the gut microbiota prior to TBI (Fig. 7A) was used to further delineate the direct and indirect effects of antibiotic exposure on TBI. SPF C57BL/6 mice were treated with VNAM or Kool-Aid control in the drinking water for a 2-week period prior to injury to modulate the microbiota. Antibiotics were discontinued 72 h prior to injury to limit any direct effects of ongoing antibiotic exposure on the immune and brain response after CCI. BrdU was injected daily on post-injury Days 3–6 to label newly formed neurons (Fig. 7A). We stained ipsilateral hippocampus sections by immunofluorescence (Fig. 7B) and analyzed the number of BrdU+DCX+-positive cells and BrdU+NeuN+-positive cells in the DG. Consistent with our previous report,12 we observed a significant reduction in the number of BrdU+ neuronal cells in the DG of antibiotic-exposed mice (CCI-VNAM) compared with control mice (CCI-Kool-Aid) (Fig. 7C), supporting that antibiotic exposure modulates TBI-induced neurogenesis via alteration of the gut microbiota as opposed to direct pharmaceutical effects.
FIG. 7.
Antibiotic exposure prior to injury reduces adult hippocampal neurogenesis. (A) Experiment design; mice were exposed to vancomycin, neomycin-sulfate, ampicillin and metronidazole (VNAM) or Kool-Aid control for a 2-week period prior to injury to modulate the microbiota, but discontinued antibiotics 72 h prior to injury. All animals survived 1 week post-injury with regular drinking water until euthanasia. 5-bromo-2'-deoxyuridine (BrdU) injections were given the last 4 days prior to euthanasia. (B) Representative images of the subgranular zone (SGZ) in the dentate gyrus (DG) of the hippocampus labeled with BrdU (red), NeuN (magenta), and DCX (green). Scale bar = 20 μm. (C) Summation of total neuronal lineage cells per area of hippocampi. Mean values are plotted ± standard error of the mean (SEM), unpaired t test (****p < 0.0001).
Antibiotic exposure prior to but not after injury alters local and peripheral immune brain infiltration
Discontinuation of antibiotics 72 h prior to injury significantly altered microglia morphology (Fig. 8A), with a decrease in volume (Fig. 8C) and in the number of branch points (Fig. 8D) and terminal points (Fig. 8F) in CCI-VNAM compared with CCI-Kool-Aid mice.
FIG. 8.
Antibiotic exposure prior to injury alters microglia morphology following traumatic brain injury (TBI). (A) Representative three-dimensional reconstruction images of microglia cells. Scale bar = 15 μm. (B–F) Quantification of microglia morphology. (B) Dendrite length, (C) volume, (D) number of branch points, (E) number of segment points, and (F) number of terminal points. Mean values are plotted ± standard error of the mean (SEM), unpaired t test (*p < 0.05, ***p < 0.001).
We also observed changes in peripheral immune infiltration to the injured brain in CCI-VNAM mice (Fig. 9A). There were significant decreases in CD45+ cells (Fig. 9B), CD3 T cells (Fig. 9C), and monocytes (Ly6Chigh, Fig. 9G) without changes in either microglia number (Fig. 9E) or activation (Fig. 9J and K) in CCI-VNAM compared with CCI-Kool-Aid mice. Therefore, modulation of the microbiota even when antibiotics are discontinued 72 h prior to TBI resulted in microglial morphology changes toward more ameboid forms and reduced peripheral immune infiltration, similar to our findings in the GF-VNAM mice. In sum, our findings support that depleted and altered microbiota, and not direct effects of antibiotics, underlie altered immunological responses to TBI.
FIG. 9.
Antibiotic exposure prior to injury alters the neuroimmune response following traumatic brain injury (TBI). (A) Gating strategy of the innate and adaptive immune response in the brain 7 days after injury. (B–K) Quantification of immune cell absolute numbers in the hippocampi and cortices. (B) leukocytes (CD45), (C) CD3 T cells (CD11b-CD3+), (D) CD8 T cells (CD11b-CD3+CD8+), (E) CD4 T cells (CD11b-CD3+CD4+), (F) T reg. cells (CD11b-CD4+CD25+), (G) Ly6Chigh cells, (H) Ly6G cells, (I) microglia (CD45low CD11b), and microglial activation markers (J) fluorescence median intensity (FMI) of toll-like receptor (TLR)4 and (K) FMI of major histocompatibility complex (MHC)II. Mean values are plotted ± standard error of the mean (SEM), unpaired t test (*p < 0.05, **p < 0.01).
Discussion
This report provides further evidence for the important role that gut microbiome plays in modulating immune responses after TBI. Injured GF mice transplanted with VNAM stool showed a clear alteration in microglia morphology, a significant decrease in T cell infiltration into the brain, and a reduction in adult hippocampal neurogenesis after injury. Additionally, previous antibiotic exposure wherein antibiotics were discontinued prior to injury showed a significant alteration of microglia morphology along with reductions in peripheral immune cell infiltration and hippocampal neurogenesis. Therefore, these data implicate the gut microbiota as an important modulator of the neuroinflammatory process and neurogenesis after TBI.
Transplanting FMT into GF mice, our group was able to provide further evidence that modulation of the gut microbiome has a profound impact on the neuroimmune response and neurogenesis after TBI. This is the first report to our knowledge that utilized GF mice as a tool in pre-clinical TBI research. Changes in the gut microbiome after TBI have been reported in animal and human studies.21–23 We found significant reductions in the richness and diversity of the gut microbiome 7 days after injury in both groups of GF transplant recipients, but the absence of sham groups restricts us from determining if these changes were completely injury dependent. In GF-VNAM mice at 7 days post-injury, we found reductions in gut bacteria from the Ruminococcaceae, and Porphyromonadaceae families with concomitant increases in bacteria from families that contain opportunistic pathogens including Bacteroidaceae and Verrucomicrobiaceae. Interestingly, a depletion of bacteria from the Ruminococcaceae family in mice has been associated with a reduction in fecal SCFAs, poorer spatial memory performance, and phenotypic changes in microglia.24 In a CCI mouse model, the SCFA acetate has been shown to improve spatial learning.10 SCFAs as a possible mechanistic link for gut modulation of neuroinflammation after TBI is an exciting avenue for future study.
Adult hippocampal neurogenesis is thought to play an important role in memory and learning in both health and disease.25 Microglia have been shown to be an important regulator neurogenesis after injury.26 Repopulation of microglia after depletion with a colony-stimulating factor 1 receptor inhibitor increased neurogenesis during a critical time window after TBI.26 Interestingly, under microbial dysbiosis conditions after injury, hippocampal neurogenesis was reduced, with an increase in microglia with a pro-inflammatory phenotype.12 Microglia can also be influenced by the adaptive immune response (T cells) in CNS injury.7,27,28 Moreover, hippocampal neurogenesis induced by an enriched environment was associated with the recruitment of T cells and the activation of microglia.29 In line with these reports, we provide evidence for gut microbiota control of post-TBI hippocampal neurogenesis and its association with microglial morphology and T cell infiltration changes after injury. However, further mechanistic studies are needed to determine if an important mechanistic link in gut microbiome control of neurogenesis in the setting of injury is microglia–T cell crosstalk-dependent.
Previously we reported that T cell infiltration in the brain after TBI was suppressed in the presence of antibiotic-induced gut microbial dysbiosis.12 However, the mechanisms by which gut dysbiosis alters the T cell response after TBI remains unknown. The gut microbiota influence the development and function of mucosal T cells subsets, specifically intraepithelial lymphocytes and LP CD4 T cells.30 We then wondered whether changes in brain T cell infiltration were associated with alterations in T cell populations in peripheral blood and small intestine (LP). Surprisingly, we did not find any differences in the T cell populations between GF-VNAM and GF-Kool-Aid mice in either compartment, and found only an increase in the Ly6C monocyte population in the GF-VNAM mice, which may be related to local small intestine inflammation. Therefore, our data do not support that the reduction in brain T cell infiltration after injury in the presence of gut dysbiosis is caused by a concomitant reduction of T cells in other compartments. However, we did not perform flow cytometry at earlier time points after FMT or CCI to assess differences in the temporal dynamics of peripheral compartments between the two FMT recipient groups. The signals for the activated T cell to infiltrate the brain are associated with pattern recognition receptors on macrophages, dendritic cells, and microglia.27,31 Future investigations are needed to study the impact of gut microbial dysbiosis on chemokines/cytokine production from the surrounding CNS cells along with adhesion protein (vascular cell adhesion molecule [VCAM] and intercellular adhesion molecule [ICAM]) expression in the endothelial cells that may control T cell infiltration and/or proliferation in the brain.32
Animal studies have identified sex- and age-related differences in responses to injury and recovery.33,34 They have shown that manipulation of gonadal hormones as well as age significantly influence injury recovery. In addition, some reports have demonstrated that aging causes gut dysbiosis and comprehensively dysregulates bile acid homeostasis with increased systemic inflammation in a sex-specific manner.35,36 One limitation in our investigation is the absence of experiments exploring the impact of sex or age on the gut–brain axis after TBI. Future investigations may include the interaction of age and sex with the impact of gut microbiota on brain responses in the setting of TBI. GF mice have demonstrated social deficits and anxiety-like behavior alteration.37 Feces from SPF donor mice were collected at the end of 7 days of exposure to VNAM or Kool-Aid control. We did not measure antibiotic level in the fecal slurry from the donor mice, and it is possible that the GF mice had some exposure to VNAM prior to injury. However, because CCI was not performed until 10 days after the second FMT, it is unlikely to have impacted injury response. We have previously measured large differences in the richness and diversity of the gut microbiome in mice exposed to VNAM for 2 weeks prior to injury.12 However, we did not measure the gut microbiome in the cohort of mice exposed to antibiotics prior to CCI utilized in the current experiments to confirm the differences in richness and diversity between the antibiotic-exposed and control groups. In our previous report, we did not observe differences in peripheral immune cell populations in the brain or in neurogenesis in sham animals treated with antibiotics compared with sham controls.12 However, we did observe differences in microglial morphology between the two sham groups, highlighting gut microbiota modulation of microglia as reported by others.6 A limitation of our current studies is the absence of sham controls in our GF FMT experiments. GF mice have significant differences in microglial morphology compared with SPF mice during homeostasis,6 and we did not determine baseline changes in microglial morphology in the GF recipients of FMT. We did not describe behavioral changes in this article because of the length of our experimental design (1-week survival) and the possible challenges related to maintaining the transplanted gut microbiome in GF mice for an extended period. A better understanding of the long-term changes in the gut microbiome of FMT recipient GF mice after injury is needed before embarking on investigations into the chronic phase of TBI.
Conclusion
In summary, gut microbiota manipulation with FMT in GF mice, and antibiotics prior to injury modulated the post-injury neuroinflammatory response and adult hippocampal neurogenesis, providing further evidence for an important role of the gut–brain axis in acute TBI and a new avenue for possible neuroprotective therapeutic development.
Supplementary Material
Authors' Contributions
S.H.F. was responsible for conceptualization; M.C. and S.H.F. were responsible for methodology; M.C., K.S., R.R., L.S., and M.T.B. were responsible for formal analysis;, M.C., K.S., R.R.; L.S., M.T.B., and S.H.F. were responsible for investigation; M.C. and S.H.F. were responsible for writing – original draft; M.C., K.S., R.R., L.S., Y.L., and M.T.B. were responsible for writing – review and editing; M.C., K.S., R.R., L.S., Y.L., M.T.B., and S.H.F. were responsible for investigation; and S.H.F. was responsible for funding acquisition and supervision.
Funding Information
This work was supported by the National Institutes of Health (R01NS097721). Fluorescent imaging was performed on a Zeiss Axio Imager Z2 Fluorescence Microscope with ApoTome 2 optical sectioning grid imager at Washington University Center for Cellular Imaging (WUCCI) supported by Washington University School of Medicine, The Children's Discovery Institute of Washington University and St. Louis Children's Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813) and the Foundation for Barnes-Jewish Hospital (3770 and 4642). Confocal imaging was generated on a Zeiss LSM 880 Airyscan Confocal Microscope, which was purchased with support from the Office of Research Infrastructure Programs (ORIP), a part of the National Institute of Health (NIH) Office of the Director under grant OD021629.
Author Disclosure Statement
No competing financial conflicts exist.
Supplementary Material
References
- 1. Sharon G, Sampson TR, Geschwind DH, et al. The central nervous system and the gut microbiome. Cell 2016;167(4):915–932; doi: 10.1016/j.cell.2016.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sommer F, Backhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 2013;11(4):227–238; doi: 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 3. Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013;13(5):321–335; doi: 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- 4. Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 2012;10(11):735–742; doi: 10.1038/nrmicro2876 [DOI] [PubMed] [Google Scholar]
- 5. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res 2020;30(6):492–506; doi: 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erny D, Hrabe de Angelis AL, Jaitin D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015;18(7):965–977, doi:10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pasciuto E, Burton OT, Roca CP, et al. Microglia require CD4 T cells to complete the fetal-to-adult transition. Cell 2020;182(3):625–640 e24; doi: 10.1016/j.cell.2020.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benakis C, Brea D, Caballero S, et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells. Nat Med 2016;22(5):516–523; doi: 10.1038/nm.4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luu M, Pautz S, Kohl V, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun 2019;10(1):760; doi: 10.1038/s41467-019-08711-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Opeyemi OM, Rogers MB, Firek B, et al. Sustained dysbiosis and decreased fecal short chain fatty acids after traumatic brain injury and impact on neurologic outcome. J Neurotrauma 2021; doi:10.1089/neu.2020.7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hartman ME, Anabayan I, Jwa B, et al. Early antibiotic exposture in severe peditric traumatic brain injury. J Pediatr Infect Dis Soc 2021; doi: 10.1093/jpids/piab087 [DOI] [PubMed] [Google Scholar]
- 12. Celorrio M, Abellanas MA, Rhodes J, et al. Gut microbial dysbiosis after traumatic brain injury modulates the immune response and impairs neurogenesis. Acta Neuropathol Commun 2021;9(1):40; doi: 10.1186/s40478-021-01137-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Celorrio M, Friess SH. Gut-brain axis in traumatic brain injury: impact on neuroinflammation. Neural Regen Res 2022;17(5):1007–1008; doi: 10.4103/1673-5374.324839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4516–4522; doi: 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13(7):581–583; doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cole JR, Wang Q, Fish JA, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42:D633–642; doi: 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8(4):e61217; doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dash PK, Mach SA, Moore AN. Enhanced neurogenesis in the rodent hippocampus following traumatic brain injury. J Neurosci Res 2001;63(4):313–319; doi: [DOI] [PubMed] [Google Scholar]
- 19. Redell JB, Maynard ME, Underwood EL, et al. Traumatic brain injury and hippocampal neurogenesis: functional implications. Exp Neurol 2020;331:113372; doi: 10.1016/j.expneurol.2020.113372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng W, ZhuGe Q, Zhong M, et al. Neurogenesis in adult human brain after traumatic brain injury. J Neurotrauma 2013;30(22):1872–1880; doi: 10.1089/neu.2010.1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Opeyemi OM, Rogers MB, Firek BA, et al. Sustained dysbiosis and decreased fecal short-chain fatty acids after traumatic brain injury and impact on neurologic outcome. J Neurotrauma 2021;38(18):2610–2621; doi: 10.1089/neu.2020.7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Treangen TJ, Wagner J, Burns MP, et al. Traumatic brain injury in mice induces acute bacterial dysbiosis within the fecal microbiome. Front Immunol 2018;9:2757; doi: 10.3389/fimmu.2018.02757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rogers MB, Simon D, Firek B, et al. Temporal and spatial changes in the microbiome following pediatric severe traumatic brain injury. Pediatr Crit Care Med 2022;23(6):425–434; doi: 10.1097/PCC.0000000000002929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. D'Amato A, Di Cesare Mannelli L, Lucarini E, et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmission-related proteins in young recipients. Microbiome 2020;8(1):140; doi: 10.1186/s40168-020-00914-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci 2010;11(5):339–350; doi: 10.1038/nrn2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Willis EF, MacDonald KPA, Nguyen QH, et al. Repopulating microglia promote brain repair in an IL-6-dependent manner. Cell 2020;180(5):833–846 e16; doi: 10.1016/j.cell.2020.02.013 [DOI] [PubMed] [Google Scholar]
- 27. Schetters STT, Gomez-Nicola D, Garcia-Vallejo JJ, et al. Neuroinflammation: microglia and T cells get ready to tango. Front Immunol 2017;8:1905; doi: 10.3389/fimmu.2017.01905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salvador AFM, Kipnis J. Immune response after central nervous system injury. Semin Immunol 2022;101629; doi:10.1016/j.smim.2022.101629 [DOI] [PubMed] [Google Scholar]
- 29. Ziv Y, Ron N, Butovsky O, et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci 2006;9(2):268–275; doi: 10.1038/nn1629 [DOI] [PubMed] [Google Scholar]
- 30. Smith PM, Garrett WS. The gut microbiota and mucosal T cells. Front Microbiol 2011;2:111; doi: 10.3389/fmicb.2011.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S, Zhang H, Xu Y. Crosstalk between microglia and T cells contributes to brain damage and recovery after ischemic stroke. Neurol Res 2016;38(6):495–503, doi:10.1080/01616412.2016.1188473 [DOI] [PubMed] [Google Scholar]
- 32. Congdon KL, Sanchez-Perez LA, Sampson JH. Effective effectors: How T cells access and infiltrate the central nervous system. Pharmacol Ther 2019;197:52–60; doi: 10.1016/j.pharmthera.2018.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Islam M, Davis BTt, Kando MJ, et al. Differential neuropathology and functional outcome after equivalent traumatic brain injury in aged versus young adult mice. Exp Neurol 2021;341:113714; doi: 10.1016/j.expneurol.2021.113714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kolb B, Cioe J. Sex-related differences in cortical function after medial frontal lesions in rats. Behav Neurosci 1996;110(6):1271–1281; doi: 10.1037//0735-7044.110.6.1271 [DOI] [PubMed] [Google Scholar]
- 35. Ma J, Hong Y, Zheng N, et al. Gut microbiota remodeling reverses aging-associated inflammation and dysregulation of systemic bile acid homeostasis in mice sex-specifically. Gut Microbes 2020;11(5):1450–1474; doi: 10.1080/19490976.2020.1763770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheng L, Jena PK, Liu HX, et al. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci Rep 2017;7(1):1748; doi: 10.1038/s41598-017-01576-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 2011;108(7):3047–3052; doi: 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.