Abstract
Several millions of individuals are estimated to develop post-acute sequelae SARS-CoV-2 condition (PASC) that persists for months after infection. Here we evaluate the immune response in convalescent individuals with PASC compared to convalescent asymptomatic and uninfected participants, six months following their COVID-19 diagnosis. Both convalescent asymptomatic and PASC cases are characterised by higher CD8+ T cell percentages, however, the proportion of blood CD8+ T cells expressing the mucosal homing receptor β7 is low in PASC patients. CD8 T cells show increased expression of PD-1, perforin and granzyme B in PASC, and the plasma levels of type I and type III (mucosal) interferons are elevated. The humoral response is characterized by higher levels of IgA against the N and S viral proteins, particularly in those individuals who had severe acute disease. Our results also show that consistently elevated levels of IL-6, IL-8/CXCL8 and IP-10/CXCL10 during acute disease increase the risk to develop PASC. In summary, our study indicates that PASC is defined by persisting immunological dysfunction as late as six months following SARS-CoV-2 infection, including alterations in mucosal immune parameters, redistribution of mucosal CD8+β7Integrin+ T cells and IgA, indicative of potential viral persistence and mucosal involvement in the etiopathology of PASC.
Subject terms: Outcomes research, Viral infection, SARS-CoV-2, Mucosal immunology
Post-acute sequelae SARS-CoV-2 (PASC), also known as long COVID is a chronic and often debilitating condition that develops following acute disease. Here authors show that individuals who suffer from PASC symptoms at six month following infection are characterized by persisting immunological disfunction, affecting cellular and humoral components of mucosal immunity.
Introduction
COVID-19, regarded in early 2020 as an infectious disease with high virulence and mortality, also rapidly became recognized as a disease-causing important morbidity, as a significant number of patients reported persistent symptoms during convalescence1. This phenomenon was not new, as it had already been described in other viral infections caused by Influenza or Chikungunya and more recently by other members of the coronaviruses family as SARS-CoV-1 and MERS-CoV, which are similar in structure and genome to SARS-CoV-22–6.
Due to broad heterogeneity in terms of population, methods, timing and type of assessment, a global but precise estimation of the percentage of patients affected long-term by SARS-CoV-2 is not attainable. Yet, it is estimated that about half of hospitalized COVID-19 patients (31%–69%), and 10% of all patients, present post-acute sequelae of COVID-19 (also known as post-COVID-19 condition)1,7–19. Considering that 500 million people have been infected worldwide20, the burden of COVID-19 sequelae is another giant face of the pandemic, with impact on individuals’ quality of life, working capacity or autonomy and on healthcare systems17,21–24.
As defined by the World Health Organization (WHO) in October 2021, post-acute sequelae SARS-CoV-2 condition (PASC) occurs in patients with a history of SARS-CoV-2 infection, at least three months after COVID-19 onset, with symptoms that cannot be explained by an alternative diagnosis and that last for at least two months with impact on patient functionality25,26. Despite there is no minimum number of symptoms, PASC is a multi-system disease, with a wide variety of unspecific physical and mental symptoms, that may vary in severity from mild to incapacitating5,11,17,27–36. Symptoms may have an onset after recovery from acute phase or may persist, fluctuate or even relapse25. The diagnosis is based mainly on the patient description.
Doubts remain about which patients are at increased risk for PASC development. Despite the existence of conflicting results, it seems that increased age and hospitalization (especially in Intensive Care Units) are risk factors for the persistence of symptoms11,18,37–44. Still, young, and non-hospitalized patients may also develop PASC45. Some studies also claim that women are more prone to specific manifestations of this syndrome15,42,46. Altogether, these results preclude the development of a predictive tool to closely monitor and treat the patients at risk after hospital discharge, a tool that would be of great value to patients and to the management and planning of healthcare systems’ resources47.
Several pathophysiological hypotheses have been proposed to explain the onset of PASC. Firstly, the extensive damage induced during acute disease by SARS-CoV-2 (considering the expression of ACE2, the SARS-CoV-2 receptor, at the surface of a myriad of epithelial and endothelial cells), may drive long-term tissue repair5,36,48. Secondly, the persistence of SARS-CoV-2 in human body, particularly in the gastrointestinal system, nervous system and other ACE2-expressing tissues, has been widely documented, and may remain for more than four months after acute infection49–54. The continuous viral replication could impact immune cell responses contributing to local immune activation and inflammation52,53. Thirdly, autoimmune phenomena have been reported in long-term recovered patients55–59. Thus, chronic immune activation due to virus persistence, autoimmunity, repair of damaged tissues or simply due to inability to downgrade acute inflammation, has been linked to PASC30,60,61.
In this study, we follow the hypothesis that chronic immune dysregulation could characterise PASC. To address this issue, we perform both CD4+ and CD8 + T cell immunophenotyping, quantify the viral-specific antibody response and determine the cytokine signature on groups of convalescent individuals who developed or not PASC. An immune CD8+ T-cell activation is observed in convalescent patients, irrespectively of developing PASC in which the CD4/CD8 ratio remain low upon six months of infection. We also observe that patients show distinct profiles in type I and mucosal type III IFN compared to uninfected individuals. We pinpoint higher levels of CD8+ T cells in PASC patients expressing the transcriptional factor Eomes. Furthermore, CD8+ T cells expressing the β7 mucosal homing receptor are low in the blood of PASC individuals. Consistent with mucosal immune response, we detect specific IgA directed to N and S proteins of SARS-Cov-2 in PASC individuals. Thus, our results highlight a model in which the persistence of viral antigens in mucosa alters mucosal immune response.
Results
Demographic and clinical characterization of the cohort
We followed 72 patients in the consultation that were recruited during their hospitalization (at admission). We collected blood from those patients during their acute disease and at long term. We also recruited 55 outpatients, from whom we had no blood samples of their acute disease, only clinical and laboratory data. Thirty-seven healthy controls could be included after verifying the exclusion criteria. Comparison of the three groups revealed no significant differences between the median age: HC = 65 years (39–85), non-PASC = 61 years (25-87) and PASC = 64 years (24-85); H = 2.161, p = 0.339, = 0.001. The demographic characteristics of our cohort are described in Table 1. No significant dependence was observed between each comorbidity and the presence or absence of PASC, despite most patients being male (67%) and a tendency for a higher percentage of women with PASC. The median time from acute disease onset to blood collection was 165 days, equal in both groups. In our cohort, 81% of the patients were hospitalized during acute disease. We did not observe any difference in PASC prevalence according to acute disease severity, the need for hospitalization, lymphocyte count or commonly used inflammation markers (Table 2). Fatigue (45 patients, 73%) and dyspnoea (41 patients, 66%) were the most common symptoms in patients diagnosed with PASC, while neurocognitive symptoms were present in 21 patients (34%). Miscellaneous symptoms were reported by 16 patients: anxiety (1), arthralgia (1), dysphonia (3), erectile dysfunction (1), hair loss (1), loss of appetite (1), muscle weakness (1), myalgia (3), palpitations (1), persistent cough (4), sadness (1) and thoracic pain (1). The laboratory characterization of the cohort, during the appointment, along with the statistical analysis of each parameter is detailed in Supplementary Table 1. Altogether these data show that comorbidities, clinic observations during hospitalisation or common parameters obtained at the appointment are not directly linked to the occurrence of PASC.
Table 1.
Demographic characterization of the cohort
| Parameter | HC (n = 37) | Non-PASC (n = 65) | PASC (n = 62) | Statistical Analysis* | |
|---|---|---|---|---|---|
| Effect size | p value | ||||
| Gender, n (%) | |||||
| Women | 14 (37) | 17 (27) | 25 (40) | 0.138 | 0.210 |
| Men | 23 (63) | 48 (73) | 37 (60) | ||
| Age, median (range) | 65 (39–85) | 61 (25–87) | 64 (24–85) | 0.001 | 0.339 |
| Body mass index, n (%) | |||||
| Normal | 17 (46) | 32 (49) | 29 (47) | 0.050 | 0.934 |
| Overweight | 12 (32) | 18 (28) | 16 (26) | ||
| Obese | 8 (22) | 15 (23) | 17 (27) | ||
| Major Comorbidities, n (%) | |||||
| Diabetes mellitus | 9 (24) | 19 (29) | 15 (24) | 0.055 | 0.777 |
| High Blood Pressure | 23 (62) | 34 (52) | 35 (57) | 0.075 | 0.627 |
| Dyslipidemia | 20 (54) | 27 (42) | 32 (52) | 0.109 | 0.377 |
| Tobacco use | 8 (22) | 10 (15) | 8 (13) | 0.132 | 0.221 |
| Alcohol abuse | 4 (11) | 1 (2) | 3 (5) | 0.163 | 0.113 |
| Chronic Obstructive Pulmonary disease | 3 (8) | 2 (3) | 4 (7) | 0.090 | 0.514 |
| Asma | 3 (8) | 3 (5) | 6 (10) | 0.087 | 0.537 |
| Obstructive Sleep Apnea | – | 4 (6) | 7 (11) | 0.171 | 0.092 |
| Chronic Kidney Disease | 1 (3) | 3 (4) | 3 (5) | 0.057 | 0.763 |
| Previous Cerebrovascular Disease | 3 (8) | 4 (6) | 6 (10) | 0.077 | 0.750 |
| Full autonomy | 37 (100) | 64 (98) | 61 (98) | 0.060 | 0.744 |
* For categorical variables Chi-square test was used to access the dependence of variables; effect size measures (Phi or Cramer’s V to 2 × 2 comparations or more, respectively) and p value are reported. For scale variables, Kruskal-Wallis test was employed; effect size measure () and p value are reported.
Table 2.
Clinical observations during hospitalization of non-PASC and PASC patients
| Parameter | Non-PASC (n = 65) | PASC (n = 62) | Statistical analysis* | |
|---|---|---|---|---|
| Effect size | p value | |||
| Hospital Admission, n (%) | ||||
| Not hospitalized | 12 (18) | 12 (19) | 0.011 | 0.898 |
| Hospitalized | 53 (82) | 50 (81) | ||
| Worst Disease Stage, n (%) | ||||
| I | 7 (11) | 8 (13) | 0.055 | 0.944 |
| IIa | 7 (11) | 5 (8) | ||
| IIb | 38 (58) | 36 (58) | ||
| III | 13 (20) | 13 (21) | ||
| Maximum ventilatory support, n (%) | ||||
| Mechanical Invasive Ventilation | 9 (14) | 8 (13) | 0.066 | 0.997 |
| Mechanical Non-invasive Ventilation | 4 (6) | 4 (7) | ||
| High-Flow Oxygen | 12 (19) | 10 (16) | ||
| Conventional Oxygen (high) | 9 (14) | 10 (16) | ||
| Conventional Oxygen (low) | 17 (26) | 17 (27) | ||
| Room-air but needing health care | 1 (1) | 1 (2) | ||
| Room-air not needing health care | 1 (1) | 0 | ||
| Not hospitalized | 12 (19) | 12 (19) | ||
| Simplified disease severity, n (%) | ||||
| Mild (Non-hospitalized to conventional oxygen) | 40 (62) | 40 (65) | 0.031 | 0.728 |
| Severe (High flow Oxygen to mechanical ventilation) | 25 (38) | 22 (35) | ||
| Period between (days) | ||||
| Symptoms and diagnosis | 4 (0–14) | 5 (0–16) | 0.140 | 0.115 |
| Symptoms and hospitalization | 7 (0–14) | 9 (0–16) | 0.110 | 0.271 |
| Symptoms and appointment | 165 (60–285) | 165 (78–281) | 0.012 | 0.341 |
| Length of stay in the Hospital | 10 (2–95) | 12 (3–60) | 0.024 | 0.551 |
| Laboratory parameters, mean (range) | ||||
| Ratio PaO2/FiO2 at admission |
269.3 (65–371) |
274.4 (103–491) |
0.129 | 0.176 |
| Worst Ratio PaO2/FiO2 |
147.8 (50–5888) |
130.1 (46–749) |
0.021 | 0.840 |
|
Lowest lymphocyte number during hospitalization (lymphocytes/µL) |
800 (200–1900) |
700 (200–3200) |
0.073 | 0.425 |
|
Highest C-reactive protein during hospitalization (mg/dL) |
158.4 (9–477) |
149.9 (2–393) |
0.062 | 0.496 |
|
Highest Ferritin level during hospitalization (ng/mL) |
1752 (8–6866) |
1343 (133–12291) |
0.153 | 0.141 |
|
Highest D-dimer level during hospitalization (ng/mL) |
1356 (341–4400) |
1387 (261–4400) |
0.020 | 0.830 |
* For categorical variables Chi-square test was used to access the dependence of variables; effect size measures (Phi or Cramer’s V to 2 × 2 comparations or more, respectively) and p-value are reported. For scale variables, Mann-Whitney test was employed; effect size measure (r) and p-value are reported.
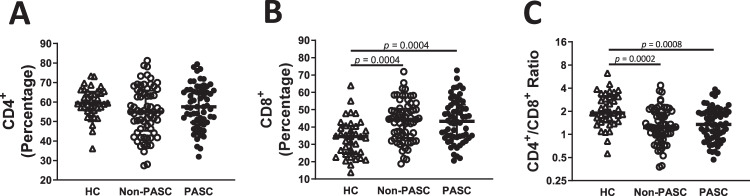
Post-COVID-19 individuals have persistent alteration in CD8+ T cells
To determine the impact of PASC on T cell immunity, we first analysed the relative percentages of CD4+ and CD8+ T cells within the T lymphocyte population six months after infection in both PASC and non-PASC compared to healthy controls (HC). The percentages of peripheral blood CD4+ T were not significantly different among all groups, irrespectively of persistent symptomatology (H = 3.87, p = 0.14, = 0.012, Fig. 1A). In opposition, CD8+ T cells were different among groups (H = 17.9, p = 0.0001, = 0.099), with an increased percentage observed in both convalescent individuals compared to HC (p = 0.0004 for both comparations, Fig. 1B). This is reflected in reduced CD4/CD8 T cell ratios (H = 18.3, p = 0.0001, = 0.10, Fig. 1C), with a decrease observed in non-PASC and PASC compared to HC (p = 0.0002 and p = 0.0008, respectively). However, nor for CD4+, CD8+ T cells or CD4/CD8 T cell ratios, no interaction was observed between the acute disease severity and PASC (F(1, 164) = 0.009, p = 0.92, for CD4+T cells, F(1, 164) = 0.22, p = 0.64, for CD8+ T cells and F(1, 164) = 0.01, p = 0.91, for CD4/CD8 ratio).
Fig. 1. Increasing levels of CD8+ T lymphocytes on convalescent COVID-19 patients.
Percentages of the CD4+ (A) and CD8+ (B) T lymphocytes populations based on gating strategy on Fig S1 (B) on healthy controls (HC; n = 37), non-presenting Post COVID-19 condition (non-PASC; n = 65) and presenting Post-COVID-19 condition (PASC; n = 62) individuals. CD4+/CD8+ ratio on HC, non-PASC and PASC individuals (C). Data are shown in a scatter dot plot format as median ± IQR; Kruskal–Wallis test was applied to identify statistical differences followed by Dunn’s multiple comparisons test.
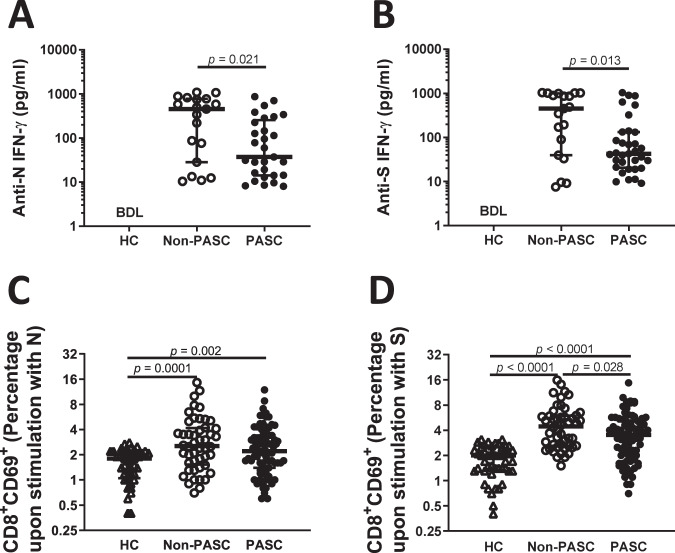
We then assessed the capacity of T cells to respond to SARS-CoV-2 antigens. Peripheral blood mononuclear cells (PBMC) were stimulated with N and S viral peptides and IFN-γ production was measured by ELISA. Our results indicated that SARS-CoV-2 convalescents displayed specific T cell response at a distal time point to viral infection in comparison to uninfected individuals (Fig. 2). However, we observed that PASC patients’ lymphocytes produced less IFN-γ than stimulated lymphocytes from non-PASC patients (U = 166, p = 0.021, r = 0.33 and U = 183, p = 0.013, r = 0.38, Fig. 2A, B, respectively). To further address specific CD8+ T cell activation, we analysed by flow cytometry, cell surface expression of the early activation marker CD69. Besides a global significance was reached comparing the three groups (H = 18.5, p < 0.0001, ) no significant differences were found upon stimulation with N peptides between non-PASC and PASC groups (Fig. 2C). Interestingly, when the activation was assessed upon stimulation with the S peptides (H = 50.8, p < 0.0001, ), a significant reduction was observed in PASC CD8+ T cells compared to non-PASC CD8 T cells (p = 0.028; Fig. 2D).
Fig. 2. Deficient memory response of CD8+ T cells of convalescent PASC patients.
Peripheral blood mononuclear cells from non-PASC (n = 65) and PASC (n = 62) individuals were stimulated with N or S viral peptides. Interferon-gamma was quantified by ELISA upon stimulation with N (A) and S (B) peptides for 24 h. All HC were tested, behaving below the detection limit of the technique. A Mann Whitney test was employed to compare the non-PASC and PASC groups. The surface expression of the early activation marker CD69 was evaluated upon 24 hours of stimulation with N (C) and S (D) peptides on CD8+ cells by flow cytometry. Data are shown in a scatter dot plot format as median ± IQR; Kruskal–Wallis test was applied to identify statistical differences followed by Dunn’s multiple comparisons test.
Thus, our results highlighted that compared to HC, convalescent patients six months after infection displayed higher levels of CD8+ T cells whereas CD8+ T cell immune response against S was lower in individuals with PASC.
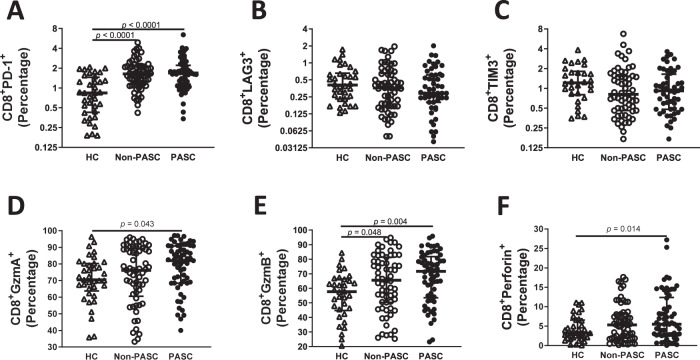
Convalescent patients display activated CD8 T cells
Because we observed higher levels of peripheral blood CD8+ T cells but lower S specific CD8+ T immune response in convalescent individuals presenting PASC symptoms, we assessed the levels of PD-1, LAG3 and TIM3, markers of T cell exhaustion62. Herein, we found significantly higher percentages of CD8+ T cells expressing PD-1 (H = 26.8, p < 0.0001, , Fig. 3A), but not of CD4+ (H = 1.5, p = 0.46, , Supplementary Figure 1A), specifically in convalescent individuals, whatever the presence of PASC symptoms status of individuals, when compared to HC (p < 0.0001 for both comparations). However, no significant differences were observed in the percentages of CD8+ or CD4+ expressing LAG3 or TIM3 in convalescent individuals when compared to HC (H = 2.2, p = 0.32, (Fig. 3B) and H = 3.9, p = 0.14, (Fig. 3C) for CD8+; H = 4.33, p = 0.11, = 0.016 (Supplementary Figure 1B) and H = 0.33, p = 0.84, = 0.012 for CD4+ (Supplementary Figure 1C)). These higher levels of PD-1 in convalescent individuals compared to HC could reflect exhausted T cells, although PD-1 can be expressed by T cells during activation and promoting effector memory CD8+ T cells63–66.
Fig. 3. Higher CD8+ T cell immune activation during SARS-CoV-2 infection on convalescents that developed Post COVID-19 condition.
(A) Percentages of the CD8+ T lymphocytes expressing PD-1 (A), LAG3 (B), TIM3 (C), granzyme A (D), granzyme B (E) and perforin (F) on PBMC of HC (n = 37), non-PASC (n = 65) and PASC (n = 62). Data are shown in a scatter dot plot format as median ± IQR; Kruskal–Wallis test was applied to identify statistical differences followed by Dunn’s multiple comparisons test.
Thus, considering the above-mentioned profile of CD8+ T cells and to clarify between activation and exhaustion, we looked at cardinal markers of the effector cytolytic T lymphocytes (CTL) program: perforin and granzyme A/B (GzmA and GzmB)67. T cells expressing PD-1 are associated with the loss of effector cytotoxic molecules68. Significant differences were found among groups in CD8+ T cells producing GzmA (H = 6.20, p = 0.045, = 0.026), GzmB (H = 10.53, p = 0.005, = 0.055) and Perforin (H = 6.36, p = 0.042, = 0.028) (Fig. 3D-F). Indeed, a significantly increased expression of GzmA, GzmB and perforin was observed in CD8+ T cells of PASC individuals compared to HC (p = 0.043 p = 0.004 and p = 0.015 for GzmA, GzmB and perforin, respectively; Fig. 3D–F). We also observed a significantly increased expression of GzmB in CD8+ T cells of non-PASC convalescent patients compared to HC (p = 0.048; Fig. 3E). To assess if acute disease severity had an impact in PASC, a two-way was performed to GzmA, GzmB and Perforin; however, no interaction was found among groups (F(1, 164) = 0.23, p = 0.64, for GzmA, F(1, 164) = 0.50, p = 0.48, for GzmB and F(1, 164) = 0.002, p = 0.67, for Perforin, Fig. 3F). Thus, these results highlighted that CD8+ T cells from PASC individuals express higher levels of effector cytotoxic molecules.
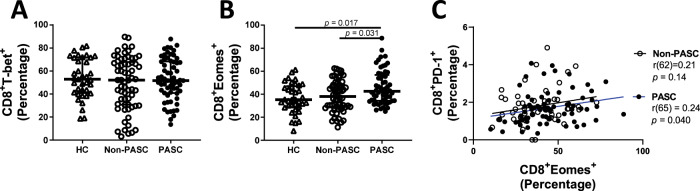
We then assessed the expression of the T-box transcription factor 21 (T-bet) and eomesodermin (Eomes) factors that regulate maturation and effector functions of CD8 T cells. In particular, Eomes and PRF1 are highly expressed in memory CD8+ T cells69–72. While the percentage of T-bet expression in CD8+ T cells was similar among groups irrespectively of the presence or absence of PASC (H = 0.92, p = 0.63, = 0.007, Fig. 4A), Eomes presented significant differences among groups (H = 9.80, p = 0.007, = 0.05, Fig. 4B). In fact, the percentage of CD8+ Eomes+ was significantly increased in PASC patients when compared to HC and to non-PASC patients (p = 0.017 and p = 0.031 for HC and Non-PASC, respectively; Fig. 4B). Interestingly, a positive correlation was found between the percentages of CD8+ PD1+ and CD8+ Eomes+, which was exclusive of PASC individuals (r(62) = 0.24 and p = 0.040 for PASC and r(65) = 0.21 and p = 0.14 for non-PASC; Fig. 4C).
Fig. 4. Increased levels of EOMES in CD8+ T cells of convalescent PASC patients.
Percentages of Tbet+ (A), EOMES+ (B) CD8+ T lymphocytes on PBMC of HC (n = 37), non-PASC (n = 65) and PASC (n = 62) individuals. Kruskal–Wallis test was applied to identify statistical differences followed by Dunn’s multiple comparisons test. Spearman’s correlation between the percentage of CD8+PD-1+ and CD8+Eomes+ T lymphocytes for non-PASC and PASC individuals (C). Data are shown in a scatter dot plot format as median ± IQR.
This increased expression of perforin and Eomes in CD8+ T cells from PASC individuals concomitant to higher expression levels of PD-1 indicates that CD8+ T cells are activated instead of being exhausted in individuals presenting PASC.
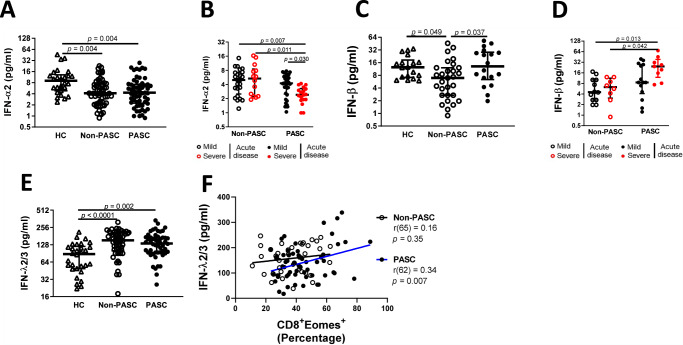
Convalescent individuals with symptoms display higher levels of interferons, while β7+CD8 T cells are diminished in the blood
This chronic immune activation of CD8 T cells could be indicative of viral persistence in PASC patients73–78. Because detection of SARS-CoV-2 in blood is not a trivial issue, we evaluated the levels of type I and III interferons in the plasma of convalescent individuals as a readout of viral persistence. Whereas type I can be a marker of systemic innate immune activation, type III is related to mucosal compartment79. Regarding type I interferon, we observed differences in IFN-α2 levels among groups (H = 12.21, p = 0.002, = 0.063), specifically a significant reduction of IFN-α2 in PASC patients comparing to both non-PASC and the HC (p = 0.004 for both comparations; Fig. 5A). Of interest, this decrease was dependent on the acute disease severity (F(1, 80) = 4.52, p = 0.037, ), significantly evident in PASC individuals that experienced severe acute disease (p = 0.011 and p = 0.030, to non-PASC with acute severe disease and PASC with acute mild disease, respectively; Fig. 5B). Furthermore, the analysis of the levels of IFN-β also revealed differences among groups (H = 7.84, p = 0.020, = 0.09). Indeed, we observed that PASC patients display higher levels of IFN-β than non-PASC group (p = 0.037; Fig. 5C). The IFN-β levels discriminates PASC that experienced severe infection with a F (1, 44) = 3.99, p = 0.053 and , (p = 0.013 and p = 0.042 to non-PASC with acute mild disease and acute severe disease, respectively; Fig. 5D). Consequently, convalescent PASC patients presented a significantly higher IFN-β/IFN-α2 ratio when compared to non-PASC (p = 0.0004; Supplementary Figure 2). Overall, these results are consistent with a recent report showing higher levels of IFN-β in patients with PASC when compared to non-PASC group and healthy controls60.
Fig. 5. Higher IFN-beta and IFN-lambda levels are associated with PASC.
The plasma levels of IFN-α2 (A), IFN-β (C) and IFN-λ2/3 (E) of HC (n = 37), non-PASC (n = 65) and PASC (n = 62) individuals and sub-divided by mild or severe acute disease (B and D for IFN-α2 and IFN-β, respectively). Spearman’s correlation between the percentage of IFN-λ2/3 and CD8+EOMES+ for non-PASC and PASC individuals (F). Data are shown in a scatter dot plot format as median ± IQR. Kruskal–Wallis test was applied to figures A, C and E to identify statistical differences followed by Dunn’s multiple comparisons test. A two-way ANOVA was used in figures B and D.
Finally, we observed a significant difference in IFN-λ levels in convalescent COVID-19 patients when compared to HC (H = 19.1, p < 0.0001, = 0.13). This increase is observed in patients developing or not symptoms (p < 0.0001 and p = 0.002, to Non-PASC and PASC, respectively; Fig. 5E). Interestingly, in the convalescent individuals, the IFN-λ 2/3 levels were positively correlated with the percentage of CD8+Eomes+ T cells in PASC patients (r(62) = 0.34; p = 0.007, Fig. 5F), correlation not observed in non-PASC individuals. This could be consistent with viral persistence of SARS-CoV-2 that has been recently proposed, particularly in the intestine80.
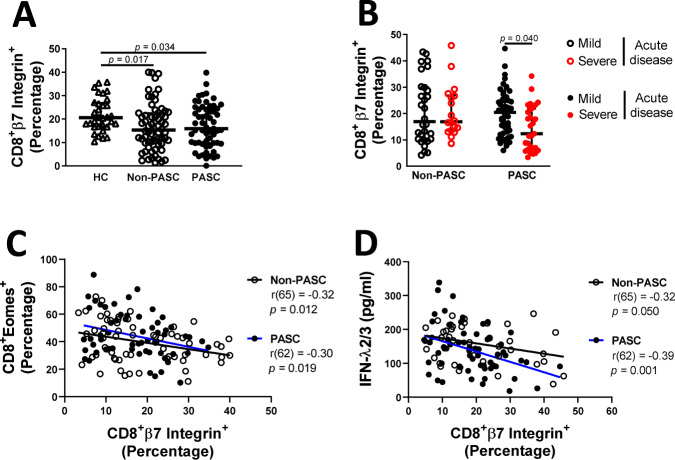
Considering this higher level of IFN-λ, which is central in the control of mucosal viral infection81, we quantified the expression of the β7 integrin on CD8+ T cells, a useful surrogate in the blood for estimating intestinal T cell homing82,83. Of interest, statistical differences were found among groups (H = 8.21, p = 0.016, = 0.047). Our results indicated that both non-PASC and PASC individuals display lower percentages of CD8+β7 Integrin+ T cells when compared to HC (p = 0.016 and p = 0.034 for Non-PASC and PASC, respectively; Fig. 6A). Taking into consideration acute disease severity in this analysis, we found a significant interaction between PASC and acute disease severity (F(1, 127) = 5.26, p = 0.024, ). Interestingly, PASC with severe acute disease demonstrated a significant decrease of CD8+β7 Integrin+ T cells compared to those with mild severity (p = 0.040; Fig. 6B) suggesting the redistribution of the CD8+β7 Integrin+ T cells in the mucosa. Furthermore, our results indicated a strong negative correlation between Eomes and β7 integrin expression (Fig. 6C), which is valid to PASC and non-PASC patients (r(62) = −0.30 and p = 0.019 for PASC and r(65) = −0.32 and p = 0.012 for non-PASC; Fig. 6C). We also found a negative correlation between IFN-λ 2/3 levels and β7 integrin expression for convalescent individuals (r(62) = −0.39 and p = 0.001 for PASC and r(65) = −0.32 and p = 0.050 for non-PASC; Fig. 6D). Altogether, our results suggest mucosal immune dysregulation in individuals with PASC.
Fig. 6. Decreased levels of Integrin+ CD8+ T cells of convalescent individuals.
Percentages of β7integrin+ CD8 + T lymphocytes on PBMC of healthy controls (n = 37), non-PASC (n = 65) and PASC (n = 62) individuals (A) and sub-divided by mild or severe acute disease (B). Spearman’s correlation between the percentage of CD8+β7integrin+ and CD8+ EOMES+ T lymphocytes for non-PASC and PASC individuals (C) and IFN-λ2/3 (D). Data are shown in a scatter dot plot format as median ± IQR; Kruskal–Wallis test followed by Dunn’s multiple comparisons test and a two-way ANOVA were applied, respectively, to figures A and B.
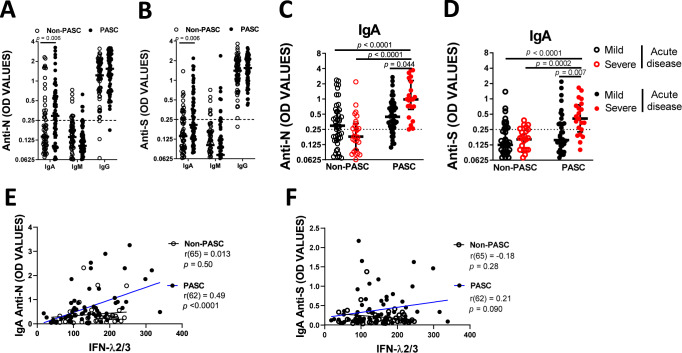
Specific IgA humoral response to SARS-CoV-2 antigens characterizes convalescent individuals presenting PASC
Considering the abovementioned profile of immune response including IFN-λ2,3, we hypothesized if viral antigens persist, the development of a specific IgA may occur, which are short-live immunoglobulins compared to IgG. Thus, we assessed the levels of the three types of immunoglobulins (IgA, IgM and IgG) against N and S viral proteins. As expected, after approximately six months of SARS-CoV-2 infection, convalescent individuals displayed high levels of IgG against N and S that contrast to the IgM response, which is rarely observed during chronic viral infections. No significant difference was observed for IgG response against N and S between PASC and non-PASC individuals (Fig. 7A, B). Interestingly, our results demonstrated that more than 56.5% and 46.8% of PASC individuals have specific IgA to N and S, when compared to 30.8% and 26.2% of non-PASC individuals. Thus, IgA levels were significantly higher in PASC patients when compared to non-PASC group (U = 1308, p = 0.006, r = 0.30 for anti-N and U = 1446, p = 0.006, r = 0.24 for anti-S, Fig. 7A, B). As before, we assessed if the severity of acute disease presented interaction with PASC occurrence performing a two-way ANOVA: F(1,126) = 4.52, p = 0.036 and for IgA anti-N and F(1,126) = 3.70, p = 0.047 and for IgA anti-S. Indeed, the observed increase on anti-viral IgA on convalescent PASC patients was specific to those who previously developed severe disease, against either the N protein (p < 0.0001, p < 0.0001 and p = 0.044 to non-PASC with mild disease, non-PASC with severe disease and PASC with mild disease, respectively; Fig. 7C) or the S protein (p < 0.0001, p = 0.0002 and p = 0.007 to non-PASC with mild disease, non-PASC with severe disease and PASC with mild disease, respectively; Fig. 7D). This interaction between the presence of PASC and acute disease severity was not observed in IgM (F(1,126) = 0.49, p = 0.487 and for anti-N and F(1,126) = 0.051, p = 0.821 and for anti-S, Supplementary Figure 3A-B) or IgG levels (F(1,126) = 0.30, p = 0.59 and for anti-N and F(1,126) = 0.43, p = 0.51 and for anti-S, Supplementary Figure 3C-D). By plotting IFN-λ 2/3 levels with the levels of IgA anti-N, we found a strong significant correlation in PASC patients, but not with non-PASC patients (r(62) = 0.49 and p < 0.0001 for PASC and r(65) = 0.013 and p = 0.50 for non-PASC; Fig. 7E). A similar tendency was observed with the levels of IgA anti-S and IFN-λ 2/3 although not significant (r(62) = 0.21 and p = 0.090 for PASC and r(65) = −0.18 and p = 0.28 for non-PASC; Fig. 7F).
Fig. 7. Type III interferon and IgA signatures on Post COVID-19 condition.
Quantification of the anti-N (A) and anti-S (B) IgA, IgM and IgG response after SARS-CoV-2 infection divided by individuals that developed (n = 62) or not PASC (n = 65) six months after acute disease. Quantification of anti-N (C) and anti-S IgA (D) at six months after SARS-CoV-2 infection on non-PASC (n = 65) and PASC (n = 62) individuals divided by the severity of acute disease. Spearman’s correlation between the percentage of Anti-N IgA (E) or anti-S IgA (F) and IFN-λ2/3 for non-PASC and PASC individuals. Data are shown in a scatter dot plot format as median ± IQR; Mann Whitney test and a two-way ANOVA were applied, respectively, to figures A, B and C, D.
Overall, our results demonstrated that patients with PASC are characterized by a specific IgA humoral response against SARS-CoV-2 antigens.
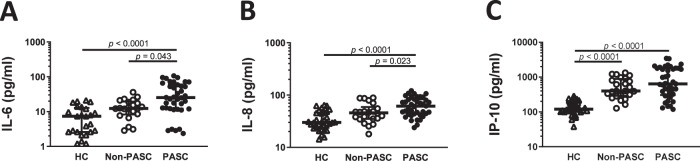
Patients with post-COVID-19 condition displayed an exacerbated inflammatory signature during acute infection
Having observed persistent immune reaction in PASC individuals and considering that most of these differences revealed an interaction with acute disease severity, we then addressed whether the acute phase of infection may have an impact on PASC. The levels of circulating IL-6, IL-8/CXCL8 and IP-10/CXCL10 were analysed in plasma samples collected during acute SARS-CoV-2 disease and compared to HC. Indeed, previous studies have defined IL-6, IL-8/CXCL8 and, IP-10/CXCL10 as early prognostic parameters of COVID-19 severe disease84–86. Significant differences were found between groups for IL-6 (H = 16.03, p < 0.001, = 0.21), IL-8 (H = 26.0, p < 0.0001, = 0.26) and IP-10 (H = 52.61, p < 0.0001, = 0.53) levels (Fig. 8A–C). Indeed, our results highlighted that this trio of cytokines is significantly higher in convalescent patients that developed PASC at six months post-infection when compared to non-PASC or HC (p = 0.043 and p < 0.0001 for IL-6 and p = 0.049 and p < 0.0001 for IL-8 compared to non-PASC and HC, respectively; Fig. 8A, B). The plasma levels of IP-10 were significantly increased on both non-PASC and PASC when compared to HC (p < 0.0001 for both; Fig. 8C). Overall, our data demonstrates increased levels of inflammatory mediators on the plasma of acute SARS-CoV-2 patients that later developed PASC. All together, these results suggest that patients with acute severe disease, in which inflammation is greater, can be associated with higher risk to develop PASC.
Fig. 8. Post-COVID-19 condition associates with a higher inflammatory signature during acute disease.
The levels of IL-6 (A), IL-8 (B) and IP-10 (C) were quantified on the plasma of HC (n = 37) and SARS-CoV2 infected patients that develop (PASC, n = 36) or do not develop (Non-PASC, n = 37) post COVID-19 condition. Data are shown in a scatter dot plot format as median ± IQR; Kruskal–Wallis test was applied to identify statistical differences followed by Dunn’s multiple comparisons test.
Discussion
Herein, we have addressed the dynamics and immune activation of CD4+ and CD8+ T cells in convalescent individuals. We observed that CD8+ T cells display higher levels of immune dysregulation compared to CD4+ T cells. Of interest, we found that several immune aspects are not only occurring in individuals having PASC form but also in non-PASC individuals six months after infection, which supports that immune alterations due to SARS-CoV-2 infection persist longer as it was initially expected. The most important information relates to the fact that CD8+ T cells remain activated after six months presenting higher levels of perforin and Eomes expression in PASC individuals. Secondly, we found an attrition of the CD8+ β7 Integrin+ T cell population in peripheral blood of PASC individuals, concomitantly with higher plasmatic levels of IFN-λ2/3, and the presence of specific IgA to SARS-CoV-2 antigens. Finally, by analysing retrospectively the plasma of these individuals at the early phase of infection, we showed that the extent of inflammation is associated with the occurrence of later PASC symptoms.
Thus, we looked at possible biological signatures of post-acute sequelae SARS-CoV-2 condition that may reflect physiopathology and help on the diagnosis and treatment of this syndrome as well as at mechanisms that may connect acute disease and PASC. This cohort is composed of 127 patients, of which 81% had been hospitalized during their acute disease. In accordance with others15,18,27,37,87–89, we did not find any influence of sex or any specific comorbidity in the development of PASC. When looking at both CD4+ and CD8+ T cell populations, at six months after infection, we noticed that convalescent patients did not display lymphopenia, with full recovery of CD4+ T cells. This contrasts to the acute phase in which lymphopenia is a predictive marker of disease severity and the occurrence of CD4+ T cell apoptosis is a confounding marker of the pathogenicity90,91. Timing of observation seems crucial, as recent works described accordingly that the alterations observed in CD4+ T cells (and B cells) last for only a few weeks after acute disease and disappear in a short amount of time87,92–94. In opposition to CD4+ T cells, our results highlighted that CD8+ T cells remain altered for a longer period, irrespective of their symptoms during acute disease. Our results are consistent with previous reports showing that months after COVID-19, CD8+ T cells are activated and expressing a cytotoxic profile, with increased expression of perforin and granzymes, which may be related to patients’ long-term outcome60,92. Furthermore, other groups have also reported a significant increase in exhausted markers such as PD-1 in CD8+ T cells60,93,95. Therefore, this may appear contradictory to the notion that exhausted T cells expressing PD-1 may express lower levels of effector molecules such as perforin and granzyme B as well IFN-γ96. However, PD-1 is expressed after T cell activation and shown to promote, in certain condition, effector memory CD8+ T cells64–66. Furthermore, our results indicate higher levels of perforin as well of Eomes in PASC individuals, which is more consistent with an activated phenotype instead of an exhausted one. As supportive evidence, we found that Eomes is positively correlated with PD-1 in PASC individuals.
Our results also demonstrated that COVID-19 patients, particularly those with PASC, display (i) higher levels of IFN-β and IFN-λ (this latter being associated with mucosal microbial defences) and (ii) specific anti-S IgA, which are short-lived Ig compared to IgG, and mostly induced in mucosal tissues. Altogether, these observations associated with lower levels of blood CD8+β7 Integrin+ T cells, a marker of circulating mucosal cells strongly suggest the persistence of SARS-CoV-2 in mucosal tissues. Recent reports based on tissue biopsies or autopsies97,98 have observed the presence of SARS-CoV-2 genomic material in human tissues for at least four months after acute infection, particularly in the gastrointestinal system, the nervous system and other ACE2-rich tissues49–54. Thus, regarding CD8+ T cells, this continuous viral replication induces CD8+ T cell activation associated with local and/or systemic manifestations52,53,57. Thus, IFN-λ levels correlated positively with the percentage of CD8+Eomes+ T cells and inversely with those expressing β7 integrin, particularly in PASC individuals. IFN-λ is mainly produced by the epithelial tissue of intestine and lungs99,100. We observed that PASC patients had a lower level of IFN-α2 but a higher level of IFN-β. Type I interferons are crucial in eliciting an effective antiviral response, and therefore our results may emphasise different sources of innate cells, whereas they may induce different interferon-stimulated genes in COVID-19 individuals101. Thus, the absence of IFN-α2 in PASC could contribute to the absence of viral control. It has been proposed that low levels of type I interferons during the acute disease are associated with increased viral load and disease severity101,102. Other reports have suggested that escapes to IFN-α response is associated with the presence of autoantibodies against IFN-α or SARS-CoV-2 antagonist proteins102–104. Our data highlight the relevance of type I and II Interferons during disease reinforcing previous publications that have shown elevated expression of the cytokines during acute disease and in convalescent patients presenting PASC60,105,106. By a retrospective analysis, we found that patients developing PASC are those who displayed greater inflammation (IL-6, IL-8 and IP-10) during acute disease. However, the levels of these cytokines and other biological parameters such as Ferritin, D-Dimer and CRP returned to normality during convalescence, independently of the development or not of PASC (Supplementary Figure 4). A repeated measures analysis of all variables comparing acute with convalescence samples did not show any statistical significance (Supplementary Table 2). Of interest, we recently reported that the levels of IP-10 (CXCL10) correlated positively with disease severity and CD4 T cell lymphopenia related to higher levels of T cell apoptosis90. These T cell defects may contribute to the absence of viral control by immune cells favouring viral dissemination and therefore leading to viral persistence several months after infection. It is important to mention that beside SARS-CoV-2-specific CD8 T cell, our data strongly suggest a T-cell receptor-independent activation of bystander CD8+ T cells. Although without specificity for the virus, the bystander CD8+ T cells have been demonstrated in other viral infections to impact the course of the immune response107, including being responsible for causing collateral damage to the host108. Thus, determining whether such early immune T cell defect is associated with PASC may be important to treat patients early after hospitalization limiting side effects.
Consistent with viral persistence in the mucosa, we detected in PASC patients higher levels of IgA directed against the S and N proteins, when compared to the other group, a difference that was more prominent in the patients who had the severe acute disease. To our knowledge, this is the first description of such a specific humoral IgA response characterizing PASC. A previous study showed that patients with lower titres of SARS-CoV-2 specific antibodies during early recovery are more likely to develop post-acute sequelae SARS-CoV-2 condition109. Other groups have reported that patients with persistent symptoms have a lower specific humoral response110–112. Herein, we found a strong significant positive correlation between IFN-λ and the levels of IgA in PASC individuals. No difference was observed for the IgG response. In contrary to total IgGs, which have a half-life of 26 days, IgA in blood circulation have a half-life of six days113–115. Thus, the observation of high levels of viral-specific IgA at six months post-acute infection supports the hypothesis of continuous viral replication, which will stimulate and maintain plasmablast cells. Thus, monitoring IgA may represent an easy way of monitoring chronic COVID-19-infected patients.
Our results have clearly established critical immune parameters associated with patients with PASC compared to non-PASC. The percentage of symptomatic patients in our cohort (49%) is in accordance with a recent systematic review (30 to 50% of post-COVID condition six to twelve months after acute infection), especially considering the six months median follow-up and high percentage of patients that required hospitalization112. Also, we are perfectly aligned in terms of symptoms reported as most of the studies and meta-analysis point out fatigue as the most common complaint, usually followed by dyspnoea and neurological or psychiatric symptoms11,17–19,27,37,40,116–119. Our results also highlighted that even in patients without major symptoms, six months after infection, the immune status of individuals not fully recapitulates to that observed in healthy donors suggesting that it is of crucial importance to monitor in a more accurate manner immune parameters associated with SARS-CoV-2 infection.
We recognize several limitations of our study. Most importantly, we are studying a condition whose diagnosis is based on symptoms report and clinical judgement. Therefore, despite all the efforts to accurate classifications, we shall admit a certain level of subjectivity in the process. Secondly, our study is exploratory, and despite pointing to viral persistence in the mucosa as a very important mechanism of disease, it was not designed to unequivocally demonstrate it. Further studies may look deeper into the early phase of the disease, including more extensive analyses of anti-SARS-CoV-2 humoral and adaptive T-cell specific responses, and its consequence on long term. Thus, a recent study suggested low perforin expression in CD8+ T cells during the acute phase is associated the persistence of symptoms120. Also, once our cohort is mainly constituted by hospitalised patients, it would be ideal to have enlarged our sample to include more patients who never required hospitalisation. However, for all the statistical tests applied, the effect size was large according to Cohen’s classification supporting the robustness of our data irrespectively to sample size.
In conclusion, our results display major advances in our understanding of PASC in which parameters of immune activation (CD8+ β7 Integrin+ T cells and IgA) are consistent with viral persistence and able to characterise these patients (Supplementary Figure 5). If viral persistence is inferred by other groups as the cornerstone of PASC, treatment of this syndrome may be based on antiviral drugs, which have not yet been tested.
Methods
Patients and study design
This is a single-center prospective cohort study, performed at Hospital de Braga (HB), a tertiary Portuguese Hospital. From 21st September 2020 to 26th February 2021, patients admitted due to COVID-19, confirmed through a PCR positive nasopharyngeal swab, were invited to participate in the study. In respect to the Portuguese Law 21/2014, our research complies with the local Ethics Committee of Braga Hospital that approved the study with the reference 123/2020 (approved on 09/09/2020). Volunteering participants gave written informed consent in compliance with the Declaration of Helsinki principles. Sex or gender was not considered during study design or recruitment. According to clinical protocol, blood samples were collected at admission and on each 72 hours, throughout hospitalisation, until discharge or suspension of oxygen therapy. Only the blood sample corresponding to the worst disease point (defined as the highest respiratory support during hospitalisation) was used for cytokine quantification and as a reference of patients’ acute disease. Patients who refused to participate or were not able to give their informed consent, who had been previously vaccinated against COVID-19 or had previous autoimmune disease, immunodepression, active solid cancer (stage III or IV) or haematological malignancy, treated with chemotherapy or immunosuppressants higher than prednisolone 20 mg or its equivalent were not included in the study. Also, patients with evidence of any simultaneous bacterial infection since admission or during their hospitalisation were excluded from that timepoint. After discharge, the abovementioned patients had a follow-up consultation six months after acute disease (Median 165 days). Other convalescent individuals, referred to Post COVID-19 consultation, who were not hospitalised during acute disease or were not recruited during their hospitalisation, were recruited in the outpatient setting (from September 2020 to June 2021), and had their blood collected in a similar interval of time between acute disease and consultation. Of those patients, only clinical data, and basic laboratory results (which had to include a PCR-positive nasopharyngeal test) of their acute disease were available. Inclusion and exclusion criteria were maintained.
It was anticipated that patients reporting on the consultation acute symptoms compatible with any ongoing infection (COVID-19 reinfection or others) would be excluded from the study. No COVID-19 test was performed on the consultation due to the absence of acute symptoms.
A control group was constituted of patients who had their blood collected before elective diagnostic exams to which they needed a PCR-negative SARS-CoV-2 test in the last 48 hours. Only non-vaccinated, nor previously infected, nor symptomatic patients were eligible, and the exclusion criteria described above were applied. Sera of those patients were analyzed to exclude the presence of antibodies related to previous unnoticed infection. Only two out of thirty-nine healthy controls were excluded from this study considering their positivity for the presence of anti-S and anti-N IgG.
Data collection
For each patient, data were collected from the medical records and inserted into our study database. Variables comprised demographics, major comorbidities, dates of onset, diagnosis, hospital admission and discharge. At admission and during hospitalization, daily information on the disease stage, need of respiratory support, treatments used, diagnosis of pulmonary embolism if present, tomography data, blood count, quantification of C-reactive protein (CRP), ferritin, d-dimer, and procalcitonin, and clinical observation of patients’ evolution were collected. At the outpatient setting, and according to clinical protocol, patients were asked about the existence or persistence of symptoms attributable to PASC and an extensive physical examination was performed. Additional exams directed to patients’ complaints were requested when necessary. Patients that could benefit the most were also referred to rehabilitation programs. Patients were grouped according to the presence or absence of PASC diagnosis, established in accordance with the WHO published criteria25. Our clinical data complies to the STROBE guidelines.
PBMC isolation and stimulation
Blood samples were collected using Heparin Blood Collection tubes (VACUETTE). After transportation to ICVS, samples were processed in BSL2 laboratories. PBMC were isolated using equal volumes of peripheral blood and Histopaque 1077 (MilliporeSigma, St Louis, Missouri, USA). After centrifugation, plasma samples were collected and stored (−80 °C). PBMC were frozen in Fetal Bovine Serum (FBS, Gibco, Thermo Fisher Scientific) with 10% of DMSO. Samples were defrosted and centrifuged at 250 g for 5 minutes to remove DMSO. Upon suspension in complete RPMI medium (RPMI-1640 culture medium supplemented with 2 mM glutamine, 10% FBS, 10 U/mL penicillin/streptomycin and 10 mM HEPES (Gibco, Thermo Fisher Scientific), PBMC were seeded at a concentration of 1 × 106 cells / mL in a 96-well plate (Corning, NY) and incubated for 24 h with 500 mg/mL of purified anti-human CD28 antibody (ref. 302902, Biolegend, CA, USA) and 500 mg/mL of purified anti-human CD40 antibody (ref. 334302, Biolegend, CA, USA) along with 1 μg/mL of SARS-CoV-2 Spike Glycoprotein (ref. RP30020, Gene Script, USA) or SARS-CoV-2 Nucleoprotein (ref. RP30013, Gene Script, USA). After incubation, supernatant was stored for cytokine quantification and cells were characterised by flow cytometry.
Phenotypic analysis of peripheral blood mononuclear cells
PBMC were thawed and divided in two 96-well plates for immune phenotyping. Surface staining was performed with the following antibodies: CD3 (clone SP34-2), from BD Biosciences, CD4 (clone OKT-4) and β7 integrin (clone FIB504) from Invitrogen, CD8 (clone SK1) and PD-1 (clone EH12.2H7) from Biolegend and LAG3 (clone P18627), and TIM3 (Clone 344823) from R&D. Intracellular staining was performed using the Foxp3 / Transcription Factor Staining Buffer Set (Ref. LTI 00-5523-00, Invitrogen) according to manufacturer’s instructions and using the following antibodies: granzyme A (clone CB9), T-bet (clone eBio4B10) and Eomes (clone WD1928) from Invitrogen and Granzyme B (clone GB-11) from Sanquin and perforin (clone Pf-344) from MabTech). Cells from the stimulation assay were characterised through surface staining using the following antibodies: CD3 (clone SP34-2), from BD Biosciences, CD4 (clone OKT-4) from Invitrogen, CD8 (clone SK1) and CD69 (clone FN50) from Biolegend (Supplementary Table 3). Samples were acquired on LSRII flow cytometer (BD Biosciences) using the DIVA Software and data was analysed using FlowJo software. Gating strategies are shown in Supplementary Figures 6–8.
Cytokine quantification
Cytokine quantification of plasma samples was performed with LEGENDplex™ Human Anti-Virus Response Panel (13-plex) (ref. 740390, Biolegend, CA, USA), according to manufacturer’s instructions. IFN-γ produced upon peptide stimulation was quantified in cell culture supernatant using an ELISA MAX Human IFN-γ (ref. 430104, BioLegend, CA, USA), according to the manufacturer’s instructions.
Production of the nucleoprotein of SARS-CoV-2
The full gene coding for the N protein (accession number YP_009724397, amino acids 1 to 419) was cloned into a pET24d vector (Eurogentec). The protein was expressed in E. coli bacteria were transformed with this plasmid and the N protein production was induced by the addition of 0.5 mM IPTG. After four hours at 37 °C with shaking at 130 rpm, the cells were lysed (buffer A, 50 mM Tris, 500 mM NaCl and 5% glycerol, pH = 7.5) and sonicated for 4 minutes. The supernatant was passed through a 1 mL nickel-NTA resin (GE, 17-5318-02). The protein was diluted in buffer B (50 mM Hepes, 150 mM NaCl, 10 % glycerol, pH = 7) and further purified on a 1 mL Source 15 S (GE, 17-0944-10) cationic exchange chromatography. The protein N was eluted by applying a gradient with buffer C (50 mM Hepes, 1 M NaCl, 10 % glycerol, pH = 7). The sample was then concentrated to 2 mL and applied onto a preparative gel filtration column (GE, 17-1069-01) equilibrated in buffer D (50 mM Tris, 200 mM NaCl, 10% glycerol, pH = 7.5). The N protein was eluted as an oligomer.
IgM, IgA, and IgG humoral responses
The antibody production was monitored by measuring specific Igs by enzyme-linked immunosorbent assay (ELISA) against the N and S1 proteins (Sars-Cov-2 S1 protein carrier free BioLegend). NUNC MaxiSorp™ well plates were coated overnight with antigens (0.5 µg/ml in Tris/Hcl pH 9.6). After washing and saturation with fetal bovine serum (BSA), plasma was serially diluted and incubated for 120 min. Plates were then washed and incubated with goat anti-Human IgG (Fc specific)-peroxidase (A0170, Millipore Sigma), goat anti-Human IgM (Fc specific)-peroxidase (401905, Millipore Sigma), and goat anti-Human IgA (Fc specific)-peroxidase (SAB3701229, Millipore Sigma) for 120 min. Secondary antibodies were titrated to optimize sensitivity. Following several washings, substrate reagent solution (R&D systems) was added for 30 min. The reactions were stopped using sulfuric acid (0.4 N). The plates were read on a Thermo Scientific™ Varioskan™ reader at wavelengths of 450 nm and 540 nm. Thus, after serial dilutions of the plasma, results shown in the figures are 1/800 for IgM and 1/400 for IgA and IgG.
Statistical analysis
Statistical analysis were performed using SPSS version 28 software (IBM, New York, USA) and data was plotted using GraphPad Prism version 8 software (San Diego, CA) For each variable, a two-way ANOVA was used to assess the interaction between disease severity during acute disease and the occurrence or absence of PASC. Only variables where the interaction terms were significant were divided according to these factors; if these significant differences were not observed, only the factor occurrence or absence of PASC was evaluated. Regarding the small sample size and the non-normality observed in our variables, Kruskal–Wallis test was applied to identify statistical differences. For variables that attained global significance, pairwise comparisons were performed using Dunn’s multiple comparisons test. A Mann-Whitney test was employed when the comparison of Non-PASC and PASC groups was performed. The chi-square test was performed for categorical variables to assess the dependence between variables. For each statistical test we calculated the adequate effect size measure (eta squared for two-way ANOVA () and Kruskal-Wallis test (), r for the Mann-Whitney test and the V-Cramer / Phi for the Chi-square test) and its interpretation followed the Cohen’s effect size cut-off values121. Correlations were measured using Spearman’s correlation coefficient.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work has been funded by National funds, through the Foundation for Science and Technology (FCT) - project UIDB/50026/2020 and UIDP/50026/2020 and by the project NORTE-01-0145-FEDER-000039, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) to R.S., the FCT contracts 2021.07836.BD to AMF and CEECIND/00185/2020 to RS, and the Clinical Academic Center (2CA-Braga) grant to A.S.C. Marne Azarias thanks the ANRS | Maladies infectieuses emergentes for his fellowship. This work was also supported by a grant to J.E. from the Fondation Recherche Médicale and the Agence Nationale de la Recherche (COVID-I²A). J.E. is supported by Canadian Institute of Health Research (FRN-177760) and the Canada Research Chair program.
Source data
Author contributions
A.S.C. designed the study, collected biological samples and clinical data, formalized the patient agreement, communicated with the hospital, acquired funding, and wrote the manuscript. A.M.F. coordinated the biological specimen processing and storage and performed the T cell phenotyping flow cytometry experiments, cytokine quantification, data analysis and wrote the manuscript. M.A-da-S. and S.A. performed the quantification of the anti-SARS-CoV-2 humoral response. P.C. overview the statistical analysis. A.I.O, O.P., M.M., B.O., M.B., J.R.L., R.D., R.C., L.N.S., A.R.M., C.A., A.C. and C.C. collected biological samples and clinical data, formalized the patient agreement, and communicated with the hospital. J.P. and A.G.C. participated in study design and logistics; J.E. provided coordination, supervision and acquired funding. R.S. designed the study, analyse the data, acquired funding, and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Julià Blanco, Elizabeth Ryan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The data generated in this study are provided in the Source Data file. Patients-related data were generated as part of clinical examination and may be subject to donor confidentiality. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: André Santa Cruz, Ana Mendes-Frias.
Contributor Information
André Santa Cruz, Email: andrescjoao@med.uminho.pt.
Jérôme Estaquier, Email: estaquier@yahoo.fr.
Ricardo Silvestre, Email: ricardosilvestre@med.uminho.pt.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-37368-1.
References
- 1.Carfì A, Bernabei R, Landi F. Persistent Symptoms in Patients After Acute COVID-19. JAMA. 2020;324:603. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbarialiabad H, et al. Long COVID, a comprehensive systematic scoping review. Infection. 2021;49:1163–1186. doi: 10.1007/s15010-021-01666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moldofsky H, Patcai J. Chronic widespread musculoskeletal pain, fatigue, depression and disordered sleep in chronic post-SARS syndrome; a case-controlled study. BMC Neurol. 2011;11:37. doi: 10.1186/1471-2377-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SH, et al. Depression as a mediator of chronic fatigue and post-traumatic stress symptoms in middle east respiratory syndrome survivors. Psychiatry Investig. 2019;16:59–64. doi: 10.30773/pi.2018.10.22.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalbandian A, et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed H, et al. Long-term clinical outcomes in survivors of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronavirus outbreaks after hospitalisation or ICU admission: A systematic review and meta-analysis. J. Rehabil. Med. 2020;52:jrm00063. doi: 10.2340/16501977-2694. [DOI] [PubMed] [Google Scholar]
- 7.Dennis A, et al. Multiorgan impairment in low-risk individuals with post-COVID-19 syndrome: A prospective, community-based study. BMJ Open. 2021;11:2–7. doi: 10.1136/bmjopen-2020-048391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudre CH, et al. Attributes and predictors of long COVID. Nat. Med. 2021;27:626–631. doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Q, et al. Clinical sequelae of COVID-19 survivors in Wuhan, China: a single-centre longitudinal study. Clin. Microbiol. Infect. 2021;27:89–95. doi: 10.1016/j.cmi.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michelen M, et al. Characterising long COVID: A living systematic review. BMJ Glob. Health. 2021;6:1–12. doi: 10.1136/bmjgh-2021-005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellan M, et al. Long-term sequelae are highly prevalent one year after hospitalization for severe COVID-19. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-01215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen MS, et al. Long COVID in the Faroe Islands: A Longitudinal Study among Nonhospitalized Patients. Clin. Infect. Dis. 2021;73:E4058–E4063. doi: 10.1093/cid/ciaa1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korompoki E, et al. Epidemiology and organ specific sequelae of post-acute COVID19: A narrative review. J. Infect. 2021;83:1–16. doi: 10.1016/j.jinf.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peghin M, et al. Post-COVID-19 symptoms 6 months after acute infection among hospitalized and non-hospitalized patients. Clin. Microbiol. Infect. 2021;27:1507–1513. doi: 10.1016/j.cmi.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maestre-Muñiz MM, et al. Long-term outcomes of patients with coronavirus disease 2019 at one year after hospital discharge. J. Clin. Med. 2021;10:2945. doi: 10.3390/jcm10132945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis HE, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. 2021;38:101019. doi: 10.1016/j.eclinm.2021.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Righi, E. et al. Determinants of persistence of symptoms and impact on physical and mental wellbeing in Long COVID: A prospective cohort study. J. Infect.10.1016/j.jinf.2022.02.003. (2022) [DOI] [PMC free article] [PubMed]
- 19.Groff D, et al. Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review. JAMA Netw. Open. 2021;4:1–17. doi: 10.1001/jamanetworkopen.2021.28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO. WHO Coronavirus (COVID-19) Dashboard.
- 21.Sivan M, Parkin A, Makower S, Greenwood DC. Post-COVID syndrome symptoms, functional disability, and clinical severity phenotypes in hospitalized and nonhospitalized individuals: A cross-sectional evaluation from a community COVID rehabilitation service. J. Med. Virol. 2022;94:1419–1427. doi: 10.1002/jmv.27456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabacof L, et al. Post-acute COVID-19 Syndrome Negatively Impacts Physical Function, Cognitive Function, Health-Related Quality of Life, and Participation. Am. J. Phys. Med. Rehabil. 2022;101:48–52. doi: 10.1097/PHM.0000000000001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-De-las-Peñas C, et al. Fatigue and Dyspnoea as Main Persistent Post-COVID-19 Symptoms in Previously Hospitalized Patients: Related Functional Limitations and Disability. Respiration. 2022;101:132–141. doi: 10.1159/000518854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parums D. v. Editorial: Long COVID, or Post-COVID Syndrome, and the Global Impact on Health Care. Med. Sci. Monitor. 2021;27:2208–2211. doi: 10.12659/MSM.933446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect. Dis. 2021;3099:19–24. doi: 10.1016/S1473-3099(21)00703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt C. COVID-19 long haulers. Nat. Biotechnol. 2021;39:908–913. doi: 10.1038/s41587-021-00984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munblit D, et al. Studying the post-COVID-19 condition: research challenges, strategies, and importance of Core Outcome Set development. BMC Med. 2022;20:1–13. doi: 10.1186/s12916-021-02222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang DH, Roy DJ, Gu BJ, Hassett LC, McCoy RG. Postacute Sequelae of Severe Acute Respiratory Syndrome Coronavirus 2 Infection: A State-of-the-Art Review. JACC Basic Transl Sci. 2021;6:796–811. doi: 10.1016/j.jacbts.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, et al. Post-acute COVID-19 syndrome in patients after 12 months from COVID-19 infection in Korea. BMC Infect. Dis. 2022;22:1–12. doi: 10.1186/s12879-022-07062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehandru S, Merad M. Pathological sequelae of long-haul COVID. Nat. Immunol. 2022;23:194–202. doi: 10.1038/s41590-021-01104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crook H, Raza S, Nowell J, Young M, Edison P. Long covid - Mechanisms, risk factors, and management. BMJ. 2021;374:1–18. doi: 10.1136/bmj.n1648. [DOI] [PubMed] [Google Scholar]
- 32.Cassar MP, et al. Symptom Persistence Despite Improvement in Cardiopulmonary Health – Insights from longitudinal CMR, CPET and lung function testing post-COVID-19. EClinicalMedicine. 2021;41:101159. doi: 10.1016/j.eclinm.2021.101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai AD, Lavelle M, Boursiquot BC, Wan EY. Long-term complications of COVID-19. Am. J. Physiol. Cell Physiol. 2022;322:C1–C11. doi: 10.1152/ajpcell.00375.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kersten J, et al. Long COVID: Distinction between organ damage and deconditioning. J. Clin. Med. 2021;10:3782. doi: 10.3390/jcm10173782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wild JM, et al. Understanding the burden of interstitial lung disease post-COVID-19: The UK Interstitial Lung Disease-Long COVID Study (UKILD-Long COVID) BMJ Open Respir. Res. 2021;8:1–10. doi: 10.1136/bmjresp-2021-001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yong SJ. Long COVID or post-COVID-19 syndrome: putative pathophysiology, risk factors, and treatments. Infect. Dis. 2021;53:737–754. doi: 10.1080/23744235.2021.1924397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigfrid L, et al. Long Covid in adults discharged from UK hospitals after Covid-19: A prospective, multicentre cohort study using the ISARIC WHO Clinical Characterisation Protocol. Lancet Reg. Health - Europe. 2021;8:100186. doi: 10.1016/j.lanepe.2021.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buttery, S. et al. Patient symptoms and experience following COVID-19: Results from a UK-wide survey. BMJ Open Respir. Res.8, (2021). [DOI] [PMC free article] [PubMed]
- 39.Heightman M, et al. Post-COVID-19 assessment in a specialist clinical service: A 12-month, single-centre, prospective study in 1325 individuals. BMJ Open Respir. Res. 2021;8:1–12. doi: 10.1136/bmjresp-2021-001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayaaslan B, et al. Post-COVID syndrome: A single-center questionnaire study on 1007 participants recovered from COVID-19. J. Med. Virol. 2021;93:6566–6574. doi: 10.1002/jmv.27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombardo MDM, et al. Long-Term Coronavirus Disease 2019 Complications in Inpatients and Outpatients: A One-Year Follow-up Cohort Study. Open Forum. Infect. Dis. 2021;8:1–8. doi: 10.1093/ofid/ofab384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taquet M, et al. Incidence, co-occurrence, and evolution of long-COVID features: A 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18:1–22. doi: 10.1371/journal.pmed.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ceban F, et al. Fatigue and cognitive impairment in Post-COVID-19 Syndrome: A systematic review and meta-analysis. Brain Behav. Immun. 2022;101:93–135. doi: 10.1016/j.bbi.2021.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Aly Z, Xie Y, Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594:259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 45.Blomberg B, et al. Long COVID in a prospective cohort of home-isolated patients. Nat. Med. 2021;27:1607–1613. doi: 10.1038/s41591-021-01433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munblit D, et al. Incidence and risk factors for persistent symptoms in adults previously hospitalized for COVID-19. Clin. Exp. Allergy. 2021;51:1107–1120. doi: 10.1111/cea.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peluso MJ, Donatelli J, Henrich TJ. Long-term immunologic effects of SARS-CoV-2 infection: leveraging translational research methodology to address emerging questions. Transl. Res. 2022;241:1–12. doi: 10.1016/j.trsl.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charfeddine S, et al. Long COVID 19 Syndrome: Is It Related to Microcirculation and Endothelial Dysfunction? Insights From TUN-EndCOV Study. Front. Cardiovasc. Med. 2021;8:1–8. doi: 10.3389/fcvm.2021.745758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liotti FM, et al. Assessment of SARS-CoV-2 RNA Test Results Among Patients Who Recovered From COVID-19 With Prior Negative Results. JAMA Intern. Med. 2021;181:702. doi: 10.1001/jamainternmed.2020.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, et al. Prolonged persistence of SARS-CoV-2 RNA in body fluids. Emerg. Infect. Dis. 2020;26:1834–1838. doi: 10.3201/eid2608.201097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vibholm LK, et al. SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses. EBioMedicine. 2021;64:103230. doi: 10.1016/j.ebiom.2021.103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaebler C, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Melo, G. D. et al. COVID-19–related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med.13, eabf8396 (2021). [DOI] [PMC free article] [PubMed]
- 55.Wang, E. Y. et al. Diverse functional autoantibodies in patients with COVID-19. Nature595, 283–288 (2021). [DOI] [PubMed]
- 56.Gammazza, A. M. et al. Human molecular chaperones share with SARS-CoV-2 antigenic epitopes potentially capable of eliciting autoimmunity against endothelial cells: possible role of molecular mimicry in COVID-19. 737–741 (2020). [DOI] [PMC free article] [PubMed]
- 57.Proal AD, VanElzakker MB. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021;12:1–24. doi: 10.3389/fmicb.2021.698169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richter AG, et al. Establishing the prevalence of common tissue-specific autoantibodies following severe acute respiratory syndrome coronavirus 2 infection. Clin. Exp. Immunol. 2021;205:99–105. doi: 10.1111/cei.13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapkota HR, Nune A. Long COVID from rheumatology perspective — a narrative review. Clin. Rheumatol. 2022;41:337–348. doi: 10.1007/s10067-021-06001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phetsouphanh C, et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022;23:210–216. doi: 10.1038/s41590-021-01113-x. [DOI] [PubMed] [Google Scholar]
- 61.Vijayakumar, B. et al. Immuno-proteomic profiling reveals aberrant immune cell regulation in the airways of individuals with ongoing post-COVID-19 respiratory disease. Immunity 1–15. 10.1016/j.immuni.2022.01.017. (2022). [DOI] [PMC free article] [PubMed]
- 62.Wherry EJ, et al. Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 63.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 64.Petrelli A, et al. PD-1+CD8+ T cells are clonally expanding effectors in human chronic inflammation. J. Clin. Invest. 2018;128:4669–4681. doi: 10.1172/JCI96107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, et al. The identification and functional analysis of CD8+PD-1+CD161+ T cells in hepatocellular carcinoma. NPJ Precis. Oncol. 2020;4:1–10. doi: 10.1038/s41698-020-00133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pauken KE, et al. The PD-1 Pathway Regulates Development and Function of Memory CD8+ T Cells following Respiratory Viral Infection. Cell Rep. 2020;31:107827. doi: 10.1016/j.celrep.2020.107827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cruz-guilloty, F. et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of eff ector CTLs. 206, 51–59 (2009). [DOI] [PMC free article] [PubMed]
- 68.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Snyder CM, Front-Line Memory T. Cells Think Outside the T-box… Mostly. Immunity. 2015;43:1030–1032. doi: 10.1016/j.immuni.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 70.Pearce EL, et al. Control of Effector CD8 + T Cell Function by the Transcription Factor Eomesodermin. Science (1979) 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 71.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 72.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl. Acad. Sci. USA. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kiekens L, et al. T-BET and EOMES Accelerate and Enhance Functional Differentiation of Human Natural Killer Cells. Front. Immunol. 2021;12:1–17. doi: 10.3389/fimmu.2021.732511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knox JJ, Cosma GL, Betts MR, McLane LM. Characterization of T-bet and Eomes in peripheral human immune cells. Front. Immunol. 2014;5:1–13. doi: 10.3389/fimmu.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seo W, Jerin C, Nishikawa H. Transcriptional regulatory network for the establishment of CD8+ T cell exhaustion. Exp. Mol. Med. 2021;53:202–209. doi: 10.1038/s12276-021-00568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, He Y, Hao J, Ni L, Dong C. High Levels of Eomes Promote Exhaustion of Anti-tumor CD8+ T Cells. Front. Immunol. 2018;9:1–13. doi: 10.3389/fimmu.2018.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma J, et al. PD1Hi CD8+ T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J. Immunother. Cancer. 2019;7:1–15. doi: 10.1186/s40425-019-0814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Llaó-Cid L, et al. EOMES is essential for antitumor activity of CD8+ T cells in chronic lymphocytic leukemia. Leukemia. 2021;35:3152–3162. doi: 10.1038/s41375-021-01198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park A, Iwasaki A. Type I and Type III Interferons – Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host. Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao F, et al. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee S, Baldridge MT. Interferon-lambda: A potent regulator of intestinal viral infections. Front. Immunol. 2017;8:749. doi: 10.3389/fimmu.2017.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X, et al. Monitoring α4β7 integrin expression on circulating CD4+ T cells as a surrogate marker for tracking intestinal CD4+ T-cell loss in SIV infection. Mucosal Immunol. 2009;2:518–526. doi: 10.1038/mi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laforge M, et al. The anti-caspase inhibitor Q-VD-OPH prevents AIDS disease progression in SIV-infected rhesus macaques. J. Clin. Invest. 2018;128:1627–1640. doi: 10.1172/JCI95127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Santa Cruz A, et al. Interleukin-6 Is a Biomarker for the Development of Fatal Severe Acute Respiratory Syndrome Coronavirus 2. Pneumonia. Front. Immunol. 2021;12:613422. doi: 10.3389/fimmu.2021.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kesmez Can F, Özkurt Z, Öztürk N, Sezen S. Effect of IL-6, IL-8/CXCL8, IP-10/CXCL 10 levels on the severity in COVID 19 infection. Int. J. Clin. Pract. 2021;75:1–8. doi: 10.1111/ijcp.14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cabaro S, et al. Cytokine signature and COVID-19 prediction models in the two waves of pandemics. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-00190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peluso MJ, et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms ll Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with an. CellReports. 2021;36:109518. doi: 10.1016/j.celrep.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zayet S, et al. Post-COVID-19 syndrome: Nine months after SARS-CoV-2 infection in a cohort of 354 patients: Data from the first wave of COVID-19 in nord franche-comté hospital, France. Microorganisms. 2021;9:1719. doi: 10.3390/microorganisms9081719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naik S, et al. Post COVID-19 sequelae: A prospective observational study from Northern India. Drug Discov. Ther. 2021;15:254–260. doi: 10.5582/ddt.2021.01093. [DOI] [PubMed] [Google Scholar]
- 90.André, S. et al. T cell apoptosis characterizes severe Covid-19 disease. Cell Death Differ.10.1038/s41418-022-00936-x. (2022) [DOI] [PMC free article] [PubMed]
- 91.Liu Z, et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shuwa HA, et al. Alterations in T and B cell function persist in convalescent COVID-19 patients. Med. 2021;2:720–735.e4. doi: 10.1016/j.medj.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Townsend L, et al. Longitudinal Analysis of COVID-19 Patients Shows Age-Associated T Cell Changes Independent of Ongoing Ill-Health. Front. Immunol. 2021;12:1–17. doi: 10.3389/fimmu.2021.676932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Files JK, et al. Duration of post-COVID-19 symptoms is associated with sustained SARS-CoV-2-specific immune responses. JCI Insight. 2021;6:1–15. doi: 10.1172/jci.insight.151544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Glynne P, Tahmasebi N, Gant V, Gupta R. Long COVID following mild SARS-CoV-2 infection: characteristic T cell alterations and response to antihistamines. J. Investig. Med. 2022;70:61–67. doi: 10.1136/jim-2021-002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rha MS, Shin EC. Activation or exhaustion of CD8+ T cells in patients with COVID-19. Cell Mol Immunol. 2021;18:2325–2333. doi: 10.1038/s41423-021-00750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chertow, D. et al. SARS-CoV-2 infection and persistence throughout the human body and brain National Institutes of Health. Research Square (2021).
- 98.van Cleemput J, et al. Organ-specific genome diversity of replication-competent SARS-CoV-2. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-021-26884-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pérez-Gómez A, et al. Dendritic cell deficiencies persist seven months after SARS-CoV-2 infection. Cell Mol. Immunol. 2021;18:2128–2139. doi: 10.1038/s41423-021-00728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walker FC, Sridhar PR, Baldridge MT. Differential roles of interferons in innate responses to mucosal viral infections. Trends Immunol. 2021;42:1009–1023. doi: 10.1016/j.it.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang, L. et al. Potential role of IFN- α in COVID-19 patients and its underlying treatment options. 4005–4015. 10.1007/s00253-021-11319-6. (2021) [DOI] [PMC free article] [PubMed]
- 102.Lowery SA, Sariol A, Perlman S. Review Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host. Microbe. 2021;29:1052–1062. doi: 10.1016/j.chom.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science (1979) 2020;370:4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bastard P, et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol. 2021;6:eabl4340. doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hatton CF, et al. Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nat. Commun. 2021 12:1. 2021;12:1–17. doi: 10.1038/s41467-021-27318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (1979) 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim TS, Shin EC. The activation of bystander CD8+ T cells and their roles in viral infection. Exp. Mol. Med. 2019 51:12. 2019;51:1–9. doi: 10.1038/s12276-019-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim J, et al. Innate-like Cytotoxic Function of Bystander-Activated CD8+ T Cells Is Associated with Liver Injury in Acute Hepatitis A. Immunity. 2018;48:161–173.e5. doi: 10.1016/j.immuni.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 109.Talla, A. et al. Longitudinal immune dynamics of mild COVID-19 define signatures of recovery and persistence. bioRxiv10.1101/2021.05.26.442666. (2021)
- 110.García-Abellán J, et al. Antibody Response to SARS-CoV-2 is Associated with Long-term Clinical Outcome in Patients with COVID-19: a Longitudinal Study. J. Clin. Immunol. 2021;41:1490–1501. doi: 10.1007/s10875-021-01083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Augustin M, et al. Post-COVID syndrome in non-hospitalised patients with COVID-19: a longitudinal prospective cohort study. Lancet Reg. Health Europe. 2021;6:1–8. doi: 10.1016/j.lanepe.2021.100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu F, et al. A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract. Cell Mol. Immunol. 2020;17:1119–1125. doi: 10.1038/s41423-020-00550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blaese RM, Strober W, Levy AL, Waldmann TA. Hypercatabolism of IgG, IgA, IgM, and albumin in the Wiskott-Aldrich syndrome. J. Clin. Invest. 1971;50:2331–2338. doi: 10.1172/JCI106731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Challacombe SJ, Russell MW. Estimation of the intravascular half-lives of normal rhesus monkey IgG, IgA and IgM. Immunology. 1979;36:331–338. [PMC free article] [PubMed] [Google Scholar]
- 115.Delacroix DL, Elkon KB, Geubel AP. Changes in size, subclass, and metabolic properties of serum immunoglobulin A in liver diseases and in other diseases with high serum immunoglobulin A. J. Clin. Invest. 1983;71:358–367. doi: 10.1172/JCI110777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Titze-de-Almeida R, et al. Persistent, new-onset symptoms and mental health complaints in Long COVID in a Brazilian cohort of non-hospitalized patients. BMC Infect. Dis. 2022;22:1–11. doi: 10.1186/s12879-022-07065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lopez-Leon S, et al. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-95565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Long Q, et al. Follow-Ups on Persistent Symptoms and Pulmonary Function Among Post-Acute COVID-19 Patients: A Systematic Review and Meta-Analysis. Front. Med. (Lausanne) 2021;8:1–11. doi: 10.3389/fmed.2021.702635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anaya, J. et al. Post-COVID syndrome. A case series and comprehensive review. Autoimmun. Rev. 20, 102947 (2021). [DOI] [PMC free article] [PubMed]
- 120.Kundura L, et al. Low perforin expression in CD8+ T lymphocytes during the acute phase of severe SARS-CoV-2 infection predicts long COVID. Front Immunol. 2022;13:1029006. doi: 10.3389/fimmu.2022.1029006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cohen, J. Statistical Power Analysis for the Behavioral Sciences. Statistical Power Analysis for the Behavioral Sciences. 10.4324/9780203771587. (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are provided in the Source Data file. Patients-related data were generated as part of clinical examination and may be subject to donor confidentiality. Source data are provided with this paper.