Abstract
The problem of antibiotic resistance is on the rise, with multidrug-resistant strains emerging even to the last resort antibiotics. The drug discovery process is often stalled by stringent cut-offs required for effective drug design. In such a scenario, it is prudent to delve into the varying mechanisms of resistance to existing antibiotics and target them to improve antibiotic efficacy. Nonantibiotic compounds called antibiotic adjuvants which target bacterial resistance can be used in combination with obsolete drugs for an improved therapeutic regime. The field of “antibiotic adjuvants” has gained significant traction in recent years where mechanisms other than β-lactamase inhibition have been explored. This review discusses the multitude of acquired and inherent resistance mechanisms employed by bacteria to resist antibiotic action. The major focus of this review is how to target these resistance mechanisms by the use of antibiotic adjuvants. Different types of direct acting and indirect resistance breakers are discussed including enzyme inhibitors, efflux pump inhibitors, inhibitors of teichoic acid synthesis, and other cellular processes. The multifaceted class of membrane-targeting compounds with poly pharmacological effects and the potential of host immune-modulating compounds have also been reviewed. We conclude with providing insights about the existing challenges preventing clinical translation of different classes of adjuvants, especially membrane-perturbing compounds, and a framework about the possible directions which can be pursued to fill this gap. Antibiotic–adjuvant combinatorial therapy indeed has immense potential to be used as an upcoming orthogonal strategy to conventional antibiotic discovery.
1. Introduction
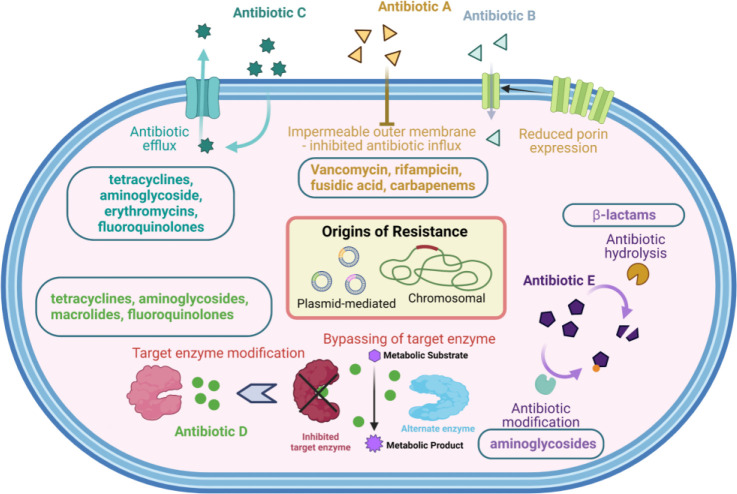
Infectious diseases are one of the greatest threats for public health worldwide with extensive economic pressure on global healthcare.1−3 Antimicrobial resistance (AMR) has further worsened the existing problem. According to a recent report on antimicrobial resistance, about 0.7 million deaths are caused by AMR annually, and this number is expected to increase to 10 million by 2050.3 This would put a huge burden on the global healthcare costs.3 Even after the development of the Global Antimicrobial Resistance Surveillance System (GLASS) in 2015, lower respiratory tract infections and diarrheal infections constituted the top ten global causes of deaths in 2019, collectively leading to 4 million deaths.4 In 2020, high levels of resistance in a number of serious bacterial infections were observed worldwide. This was prevalent in many Gram-negative pathogens.5 Among these, Acinetobacter baumannii, Pseudomonas aeruginosa, and extended spectrum β-lactamase-producing Enterobacteriaceae constitute critical priority pathogens according to a priority list released by the World Health Organization (WHO) in 2017.6 This implies that bacterial resistance causes a significant burden to global healthcare, and it is imperative to invest in multifaceted and versatile strategies to target these resistant bacteria. In order to achieve this, the mechanisms of action of antibiotics and resistance development in bacteria (Figure 1) must be understood in detail.
Figure 1.
Mechanisms of resistance towards antibiotics. Created using BioRender.com.
The search for new antibacterial compounds through various screens and modifications of the existing antibiotic scaffolds has led to the approval of a few new antibiotics in recent years.7 Even though the number of antibiotics approved has increased slightly over the last 5 years, it is still not enough to combat the growing problem of antimicrobial resistance. Identification of novel druggable targets and new classes of antibacterial compounds through whole-cell-based phenotypic, target-based, or genetic screens is a lengthy and economically expensive process with many difficulties including the rediscovery of existing compounds.8 Therefore, the scientific community has started investing in rehabilitation and repurposing of existing antibiotics and drugs through “Combination therapy” of two or more chemical entities. “Combination therapy” is inspired from the prescription of a “combination of drugs” which started as early as the 1950s without much knowledge about mechanistic insights.8 The antibiotic pipeline is quite leaky with many existing antibiotics becoming obsolete due to the increasing mechanisms of bacterial resistance. Moreover, there are quite a number of approved drugs which can be repurposed to treat bacteria for which they are not conventionally prescribed. We are aware of the smart resistance mechanisms employed by bacteria to various antibiotics. These inherent, acquired, or phenotypic resistance elements can be targeted to increase the activity of an existing drug against different species of bacteria.
It is wise to recycle the current antibiotic arsenal by targeting specific mechanisms of resistance and overcoming them rather than investing in random semisynthetic modifications of existing drugs. The structural and mechanistic insight obtained from physical combinations of compounds can then be utilized to design rational semisynthetic modifications. This would lead to a more streamlined approach toward antibiotic development. In addition to the combinations of two active compounds, another interesting class of combinations are combinations comprising of an antibiotic and a nonantibiotic molecule which just targets the mechanism of resistance to the said antibiotic, indirect resistance elements, or nonessential pathways which are important for resistance. Such a combination would end the redundancy of an obsolete antibiotic by making it useful to treat multidrug-resistant strains of bacteria. An example of this is β-lactam/β-lactamase inhibitor combinations which are clinically approved, and many of them are in clinical trials. Looking at the success of these combinations, it is pertinent to look at other mechanisms of resistance which can be targeted by such compounds to revive the efficacy of existing antibiotics. These compounds with little or no antibiotic activity can be described as “antibiotic adjuvants”. Antibiotic adjuvants can be judiciously used with existing antibiotics by targeting active or passive elements of resistance in bacteria.
This review is an attempt to assemble the known types of antibiotic adjuvants and classify them on the basis of the resistance element targeted. To begin with, this review talks briefly about the various mechanisms of resistance seen in bacteria which gives a background of the vast knowledge about bacterial resistance that can be exploited to develop antibiotic adjuvants (Figure 1). The subsequent sections discuss how different mechanisms of resistance can be targeted using antibiotic adjuvants to reinforce the efficacy of existing classes of antibiotics. Different types of antibiotic adjuvants with sufficient examples from every type have been focused upon. Based on the understanding of existing adjuvants in clinics and those under development, the review concludes with focused directions which can be explored for accelerating the upcoming research in the field of antibiotic adjuvants. As members of only one type of adjuvants are currently approved, the gaps in the development of other types of adjuvants are highlighted, and directions to be undertaken to fill these gaps have been discussed.
2. Acquired Mechanisms of Resistance in Bacteria
Ever since the discovery of antibiotics, evolutionary strategies to evade them have been employed by bacteria. These strategies used by bacteria for evading antibiotic-mediated toxic effects have significant diversity. We have attempted to provide a short summary of some of the important examples of resistance mechanisms in the following section.
2.1. Prevention of Access to the Target
2.1.1. Reduced Permeability
The antibacterial activity of antibiotics with intracellular targets is strongly dependent on the ability of the antibiotic to permeate through the cell envelope and reach its target. In the case of Gram-positive bacteria, the cell membrane has to be permeabilized. In Gram-negative bacteria, there is an additional outer membrane which needs to be breached to reach the periplasm and inner plasma membrane. It has been observed that Gram-negative pathogens are intrinsically less permeable to many antibiotics than Gram-positive species because their outer membrane acts as a permeability barrier (Figure 1).9 Antibiotics that are hydrophilic diffuse via channel-like outer membrane porin proteins to cross the membrane.10 Additionally, only those antibiotics which have a smaller hydrated size than the porin channel can diffuse through. This rules out large hydrophilic drugs such as the glycopeptide vancomycin from porin-mediated transport, explaining the inherent resistance of Gram-negative bacteria to vancomycin.11
The channel-like porin proteins OmpF and OmpC, present in the outer membrane of Escherichia coli, which are important porins in most Enterobacteriaceae, are assumed to serve as nonspecific channels, with no selectivity for antibiotic binding. Therefore, porin downregulation or porin substitution with more selective channels can reduce the antibiotic permeation through the lipopolysaccharide (LPS)-containing outer membrane. This porin downregulation has been reported as a prevalent resistance mechanism in Gram-negative bacteria.12 Recent research has nevertheless demonstrated that reduction in porin expression significantly contributes to resistance to newer medications like carbapenems and cephalosporins (along with the resistance mediated by enzymatic degradation), in Enterobacteriaceae. For instance, in Enterobacteriaceae, mutant porin alleles or reduced porin production can result in clinically meaningful resistance to carbapenems even in the absence of carbapenemase production.13 The fast buildup of mutations in the porin genes, particularly in E. coli and Enterobacter spp. following carbapenem treatment, demonstrates the selection pressure that carbapenems exert toward the emergence of mutations in porin and porin regulatory genes. Additionally, clonal lineages that have led to worldwide outbreaks of illness have been linked to Klebsiella pneumoniae isolates that express porin variations.14
2.1.2. Increased Efflux
Bacterial efflux pumps play a significant role in the inherent resistance of Gram-negative bacterial pathogens to many of the medications commonly employed to treat Gram-positive bacterial illnesses. They actively transfer numerous medicines out of the cell. Efflux pumps can confer significant degrees of resistance to previously therapeutically effective antibiotics when they are overexpressed (Figure 1).15 The term “multidrug resistance” (MDR) efflux pumps refers to efflux pumps that transport a variety of structurally different substrates. Some efflux pumps, such as Tet pumps, have a narrow substrate specificity. All bacteria have MDR efflux pumps, which have been thoroughly researched, and new pumps that export drugs are continuously being described. These have included KexD (K. pneumoniae), FuaABC (Stenotrophomonas maltophilia), MdeA (Streptococcus mutans), and LmrS (Staphylococcus aureus) within the last two years.15 All bacteria have several MDR efflux pump genes on their chromosomes; however, some of these genes have been mobilized or placed on plasmids that can move from one bacterium to another.16 Recently, it was discovered that a Citrobacter freundii strain possessed an Inc.H1 plasmid bearing the New Delhi metallo-β-lactamase 1 (NDM-1) gene as well as the genes coding for the resistance nodulation division (RND) type efflux pump.17 This is a concerning result since it demonstrates how quickly other pathogenic bacteria could acquire this resistance mechanism through transmission.
The most well-studied MDR efflux transporters are members of the RND family, which is present in Gram-negative bacteria. RND pumps have the ability to extrude various drugs out of the cell to a clinically relevant extent and export a broad range of substrates when overexpressed. Examples that have undergone extensive research include P. aeruginosa MexB and the AcrB efflux pump in E. coli. RND pumps, like AcrB, are homotrimers that live in the inner membrane and form a tripartite complex with an outer-membrane channel, like TolC or OprM, and a periplasmic adaptor protein, like AcrA or MexA.18 The knowledge of these efflux pumps’ structure and operation has advanced significantly. Owing to the efforts of scientists from diverse fields such as chemistry, microbiology, molecular biology, and structural biology, the structure and function of these efflux pumps are better understood.
The overexpression of many of these efflux pumps is of considerable importance, as that can dictate the extent of resistance to the antibiotic. Local regulators, which are encoded along with the efflux-pump genes, and global regulators, which have broader biological functions, regulate the transcription of the genes that code for these efflux pumps. Transcription factors from the AraC-XylS family are global regulators. Due to the enhanced production of transcription factors like MarA, RamA, SoxS, or Rob in Enterobacteriaceae, there is a proportional enhancement in the extent of production of efflux pumps along with repression of porin proteins, effectively conferring multidrug resistance.19
2.2. Target Modification by Mutation
Most antibiotics have high affinity specific bindings to certain target proteins, preventing the protein’s usual activity. Resistance can be conferred by altering the target’s structure in a way that prevents effective antibiotic binding while preserving the target’s ability to function normally (Figure 1). There are frequently varying populations of pathogens present during an infection, and if a point mutation conferring antibiotic resistance arises, strains carrying this mutation may then be selected and spread quickly. Usually, in the bacterial genome, there are multiple copies of the genes that encode the targets of some antibiotics; for instance, the 23S rRNA ribosomal subunit of Gram-positive bacteria, which is encoded by multiple, identical copies of its gene, is the target of linezolid. In S. pneumoniae and S. aureus, the use of linezolid has selected for resistance through mutation in one of these copies, followed by high-frequency recombination between homologous alleles, which quickly produces a population weighted in favor of carrying the mutant allele.20 What is more alarming is that all of this has happened in a short span of time, with antibiotic approval and clinical resistance being separated by less than a decade.
Another illustration of the utilization of alternate enzymes for carrying out the bacterial biosynthesis process is the acquisition of a gene homologous to the antibiotic target enzyme, as seen in methicillin-resistant S. aureus (MRSA), where the staphylococcal cassette chromosome mec (SCCmec) element is acquired to confer methicillin resistance.21 This contains the mecA gene, which codes for the protein PBP2a that is β-lactam insensitive and allows cell wall biosynthesis to proceed despite the native PBP being inhibited by the antibiotic. Different Staphylococcus species have been found to contain a large number of SCCmec elements, and there is proof that the mecA allele has been transmitted numerous times.22 It was once believed that the distinguishing feature of MRSA was the presence of an SCCmec element containing mecA.23 Alternatively, by altering target proteins and creating “mosaic” genes, transformation, or the transfer of genetic material from the environment, can confer antibiotic resistance. Penicillin resistance in S. pneumoniae, which is brought on by mosaic penicillin-binding protein (pbp) genes encoding penicillin-insensitive enzymes, is the classic example of this. By recombining DNA from Streptococcus mitis, a closely related bacterium, these mosaic alleles were created. High levels of resistance to extended-spectrum cephalosporins have also been associated with such mosaicism in the PBP-encoding penA gene of Neisseria gonorrhoeae.24N. gonorrhoeae has emerged as a formidable clinical challenge, owing to the emergence of pan-resistant isolates, followed by a loss of efficacy of ceftriaxone (third-generation cephalosporin). N. gonorrhoeae causes gonorrhea which is the second most prevalent bacterial sexually transmitted infection worldwide. This pathogen has evolved into a true superbug and displayed a high level of resistance to ceftriaxone, which is the last remaining option for the first-line treatment.25 New antimicrobial strategies to target this bacterium are urgently required.
2.3. Hydrolysis of Antibiotics
Since the first use of antibiotics and the 1940 discovery of penicillinase (β-lactamase), the enzyme-catalyzed modification of antibiotics has been a significant mechanism of antibiotic resistance. Since then, tens of thousands of enzymes have been discovered that can break down and alter various classes of antibiotics, including β-lactams, chloramphenicol, aminoglycosides, and macrolides (Figure 1). Additionally, there are other groups of enzymes that can break down various antibiotics that belong to the same class. For instance, a wide variety of β-lactamases can break down β-lactam antibiotics, including penicillins, cephalosporins, clavams, carbapenems, and monobactams.26 The emergence of hydrolytic enzymes with modified activity spectra reflects the effect of expanded antibiotic classes. Extended-spectrum β-lactamases (ESBLs), which can hydrolyze oxyimino-cephalosporins, emerged after the early β-lactamases, targeting first-generation β-lactams. K. pneumoniae, E. coli, P. aeruginosa, and A. baumannii are examples of Gram-negative bacteria that carry a variety of ESBLs and carbapenemases, including imipenemases (IMP), Verona integron encoded metallo β-lactamase (VIM), oxacillinases (OXA), and NDM enzymes.27 This has serious implications in the clinical treatment of severe infections. Increased rates of resistance have been seen in some situations as a result of the growth of resistant bacterial clones, whereas in other situations, the spread of plasmids containing resistance genes has been significant. Regardless of the mechanism, the consistent trait has been the fast spread of various resistance elements, through different bacterial species, in a relatively small period of time.
The CTX-M genes (encoding ESBLs, which have stronger anti-cefotaxime activity than other oxyimino β-lactams) have hundreds of variations. The most frequently isolated ESBLs globally, particularly in isolates of cephalosporin-resistant K. pneumoniae and E. coli, are the CTX-M14 and CTX-M15 enzymes. While CTX-M15-producing E. coli strains are widely distributed among patients, K. pneumoniae isolates bearing the enzyme are mostly a hospital-associated issue.28 Over the past ten years, the clinical prescription of carbapenem antibiotics has increased due to the rise in the number of bacteria containing ESBL genes. As a result, there are currently more clinical isolates containing β-lactamases with carbapenem-hydrolyzing activity (known as carbapenemases). Members of classes A, B, and D β-lactamases are the majority of carbapenemases. The ability of these enzymes to inactivate a variety of β-lactams, such as carbapenems and extended-spectrum cephalosporins, is their defining characteristic.29 Many carbapenemases, despite being initially discovered on the chromosomes of a single species, have now been described in different species of Enterobacteriaceae.
The kpc and ndm genes serve as examples of the various ways carbapenemases have propagated. Several Enterobacteriaceae have been characterized since KPC (a serine carbapenemase) was initially discovered in K. pneumoniae in 1996.15 The kpc gene is plasmid-borne and connected to the ST258 dominant clone of K. pneumoniae, which produces KPC and is widespread throughout the world. There are several variations of the kpc gene, distinguished by single-amino-acid substitutions (albeit with similar hydrolytic activity); KPC2- and KPC3-producing strains have led to outbreaks in the US, Greece, the UK, and Israel. The NDM enzyme, one of the most widely distributed carbapenemases, was initially discovered in India in 2009 and is now present in many Gram-negative bacteria, including A. baumannii, K. pneumoniae, and E. coli.15 Except for aztreonam, NDM provides resistance to all β-lactams. NDM-producing strains have been discovered in a variety of species; the genes are both on the host chromosome and on plasmids, and they can frequently switch between the two.27 Epidemiological studies initially connected illnesses with bacteria that carry NDM to the Middle East, the Balkan republics, and the Indian subcontinent. However, according to recent accounts, outbreaks have happened and persisted in other parts of the world.
Apart from the above-mentioned ones, for certain antibiotics such as fluoroquinolones, resistance is also observed due to other factors which hamper binding to the target protein. For cationic membrane-targeting antibiotics like colistin, resistance can happen due to chemical modification of surface components (such as LPS) of bacteria, resulting in a change in overall charge from anionic to zwitterionic. Recent reports also suggest that Gram-negative bacteria can tolerate many membrane-active antimicrobials by overproduction of the outer membrane vesicles. There are many such miscellaneous mechanisms of resistance to different classes of antibiotics. While this topic is not the focus of the review, the reader is encouraged to refer to already published comprehensive reviews and perspectives which cover this topic in its entirety.15,18,30
3. Antibiotic Adjuvants
Multiple target engagement can be achieved in principle by the “physical combination” of distinct compounds. Therapeutic success has been attained in combination therapy for HIV,31 cancer,32 cardiovascular diseases,33 and also antibacterial strategies. Combination therapy was developed soon after the discovery of antibiotics, without having much knowledge about the molecular mechanisms of action. By the mid-1950s, >60 combinations were known (two-component or higher order). Early combinations of antibiotics increased efficacy. Examples include streptomycin and penicillin, trimethoprim, and sulfonamides.34,35 The importance of combination therapy to treat tuberculosis and leprosy was identified in the 1950s and 1960s, respectively.36 Such combinations remain in use today as they are backed by mechanistic and clinical data. Antibiotic combinations are employed according to need and experience in clinical settings. Synercid is an antibacterial agent for intravenous administration which consists of quinupristin and dalfopristin in the ratio of 30:70 (w/w).37,38 Apart from these, empirical antibiotic combinations are common in clinical practice.39
When it comes to combination therapies involving a nonantibiotic, β-lactam/β-lactamase combinations are the only fixed-dose antibacterial drug combinations which are approved. With respect to antibacterial therapies, the “adjuvant therapy” holds promise, but currently it is narrowly explored. Thus, it is imperative to reassess “antibacterial monotherapy” and explore the use of nonantibiotic adjuvants to diminish the emergence of antibiotic resistance. Hereafter, antibiotic adjuvants (compounds that potentiate the activities of antibiotics) will be focused upon.
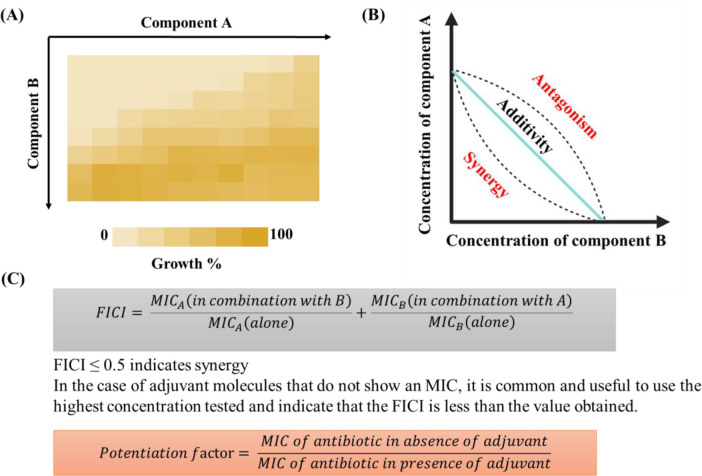
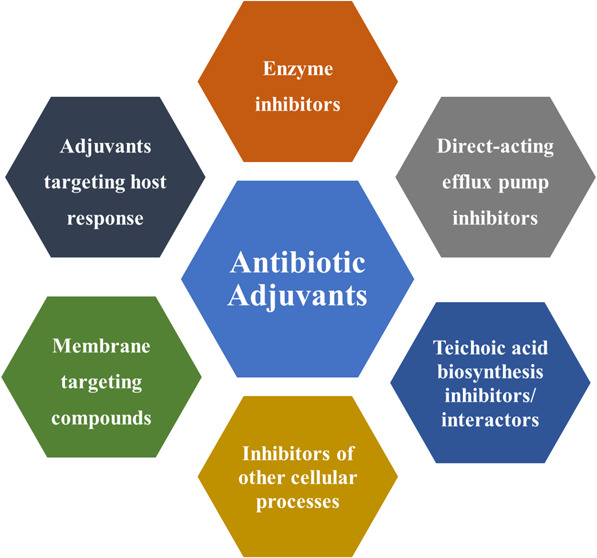
Antibiotic adjuvants are defined as compounds which potentiate the activity of existing antibiotics by minimizing or directly blocking the mechanisms of resistance to them.40 The idea for antibiotic adjuvants comes from successful combinations of two or more antibiotics (congruous combinations) in clinic.8,41,42 These have been used empirically to achieve synergy, increase the spectrum of activity, and overcome resistance. In contrast to the conventional antibiotic combinations, antibiotic adjuvants exhibit no or little antimicrobial activity alone. Based on the target profile of antibiotic adjuvants, they can be broadly classified as direct, indirect, and host-modulating resistance breakers. Antibiotic adjuvants target different active and passive resistance mechanisms in bacteria to potentiate antibiotics (Figure 2). All these different types of resistance breakers are discussed in the subsequent sections. Owing to the vast variety and number of compounds reported for each class of adjuvants, representative examples in each section have been discussed, and corresponding references have been cited to get a more detailed overview about each adjuvant. Chequerboard assays and isobolograms are employed to identify the synergistic potential of compounds as potent antibiotic adjuvants (Figure 3).8,43 Potentiation factors and fractional inhibitory concentration indices (FICIs) which are used as parameters for potentiation especially for membrane-perturbing and other indirect resistance breakers have been specified, wherever suitable, according to the availability of data (Figure 3).40,43,44
Figure 2.
Mechanisms by which different types of adjuvants can potentiate antibiotics. Created using BioRender.com.
Figure 3.
Identifying potent antibiotic adjuvants. (A) Chequerboards to assess potentiating ability for potential antibiotic adjuvants. (B) Isobologram to assess synergistic, additive, or antagonistic interactions between two compounds. (C) Parameters for assessing antibiotic adjuvants. (B) is partially created using BioRender.com.
3.1. Direct Resistance Breakers
Direct resistance breakers are an example of Class I adjuvants which work with antibiotics on resistance-causing bacterial targets.40 These adjuvants directly inhibit mechanisms of antibiotic resistance like incapacitating enzymes, efflux pumps, or additional targets which compensate for the original targets.40 β-lactamase inhibitors, which inactivate β-lactamases, are the only clinically approved adjuvants in current use.
3.1.1. Enzyme Inhibitors
Bacteria can harbor enzymes which hydrolyze antibiotics and degrade them. Resistance to β-lactam antibiotics, chloramphenicol, and aminoglycosides is conferred due to this reason.45 β-lactams are rendered ineffective due to their hydrolysis by β-lactamase enzymes. These enzymes are classified as serine β-lactamases (contain a serine residue for hydrolysis) and metallo β-lactamases, MBLs (hydrolysis mediated by a metal ion like Zn2+). According to the Ambler classification, β-lactamases can be classified into classes A, B, C, and D.46−49 Serine β-lactamases belong to classes A, C, and D. The enzymes TEM, SHV, CTX-M, and KPC (Class A),50 the AmpC and the plasmid-encoded CMY-type cephalosporinases (Class C),51 and the OXA enzymes (oxacillinases, Class D) are examples of these.52 MBLs are classified into three subclasses B1, B2, and B3 based on the primary amino acid sequence. Subclass B1 includes clinically relevant NDM-, IMP-, and VIM-type variants.26 More details about these enzymes are given in the previous section.
β-lactamase inhibitors, based on a β-lactam or a non-β-lactam core, are examples of direct-acting antibiotic adjuvants which can inhibit the activity of the above-mentioned β-lactamase enzymes. In 1976, scientists reported clavulanic acid which inhibited the function of serine β-lactamases.53 Clavulanic acid (1) is a β-lactam-based inhibitor which binds irreversibly to the serine β-lactamase enzyme and can be classified as a suicide substrate.54 It was coupled with amoxicillin and sold under the trade name Augmentin. Augmentin was the first β-lactam antibiotic and β-lactamase inhibitor combination clinically approved and prescribed extensively. Even though clavulanic acid shows activity against Class A and D β-lactamases, it lacks any activity against Class A, C, and D carbapenemases.55 Thereafter, two more β-lactamase inhibitors based on the β-lactam core, sulbactam (2) and tazobactam (3), were discovered in the 1980s.56 However, they also do not cover carbapenemases within their spectrum. In 2015, avibactam (4), a diazabicyclooctane (DBO), was approved by the US Food and Drug Administration (FDA) as a potent inhibitor of many serine β-lactamases (Types A, C, and D), however lacking activity against certain Class C and D carbapenemases.57 Avycaz is the marketed combination of avibactam with ceftazidime. A new cyclic boronate-containing chemical scaffold approved by the FDA in 2017, vaborbactam (5), possesses serine-β-lactamase inhibition.57 Vabomere is the trade name of the combination of vaborbactam and meropenem. A combination of imipenem/cilastatin with relebactam (6) (DBO-based) was approved in 2019.58,59 Vaborbactam and relebactam also lack inhibition of Class D carbapenemases. The DBOs and cyclic boronates discussed above are reversible inhibitors of serine β-lactamases. As of now, serine β-lactamase inhibitors are the only Class Ia adjuvants from which clinical candidates have emerged. A list of approved β-lactams and serine β-lactamase inhibitors is given in Table 1. Other β-lactamase inhibitors, including those in clinical development, are given in Table 2. DBOs like durlobactam60 (7), zidebactam61 (8), nacubactam62 (9), and ETX028263 (10) are in different phases of clinical trials currently.64 All of these possess dual penicillin binding protein (PBP)-inhibiting and β-lactamase-inhibiting activity and are thus sometimes referred to as “β-lactam enhancers”. The partner β-lactams for these DBOs, sulbactam, cefepime, meropenem, and cefpodoxime, respectively, are potent PBP3 inhibitors resulting in bacterial cell filamentation. On the other hand, the DBOs inhibit PBP2, thus resulting in the formation of spheroplasts.61,63,65 Apart from these, VNRX-7145 (11), a bicyclic boronate, is in early phase clinical trials in combination with ceftibuten.66 It shows activity against Class A, C, and D serine β-lactamases, including KPC and OXA-48 carbapenemases. VNRX-7145 and durlobactam exhibit inhibition of serine β-lactamases, including carbapenemases as well. Apart from these, Enmetazobactam67 (12), WCK-423468 (13), and GT-055 (also referred to as LCB18–055)69,70 (14) which also inhibit Class A and some Class D carbapenemases are also in clinical and preclinical development.
Table 1. Approved β-lactam and β-lactamase Inhibitor Combinationsa.
| Combination (β-lactam + adjuvant) | Type of adjuvant | Trade name | Year of approval | Company | Indications |
|---|---|---|---|---|---|
| Ticarcillin + clavulanate (1) | Oxapenem | Timentin | 1985 | SmithKlineBeecham | Systemic infections, cUTIs, pneumonia (HAP and VAP), bacteremia, bone infections, cIAIs (complicated intraabdominal infections) |
| Ampicillin + sulbactam (2) | Sulfone | Unasyn | 1986 | Pfizer Inc. | Pneumonia, cUTIs, cIAIs, gynaecological andobstetric infections, acute epiglottitis, periorbital cellulitis, diabetic foot, skin and soft tissue infections, gonorrhea, otitis media |
| Amoxicillin + clavulanate (1) | Oxapenem | Augmentin | 1996 | SmithKlineBeecham | Diabetic foot, UTIs, pulmonary infections, brain abscesses, acute otitis sinusitis, skin infections, gynaecological infections, septicemia, bone and joint infections, endometritis, peritonitis, IAIs |
| Piperacillin sodium (penicillin) + tazobactam sodium (3) | Sulfone | Zosyn | 1996 | Wyeth | Cellulitis, diabetic foot, cIAIs, cUTIs, pneumonia (HAP and VAP), gynaecological skin infections, appendicitis, postpartum endometritis, stomach infections, severe uterine infections |
| Ceftolozane + tazobactam sodium (3) | Sulfone | Zerbaxa | 2014 | Cubist Pharmaceuticals | Pneumonia (HAP and VAP), acute pyelonephritis, cIAIs |
| Ceftazidime + avibactam (4) | Diazabicyclooctane (DBO) | Avycaz | 2015 | Actavis | Pneumonia (HAP, VAP), cUTIs, pyelonephritis, cIAIs |
| Meropenem + vaborbactam (5) | Cyclic boronatc | Vabomere | 2017 | The Medicines Company | Pneumonia (HAP, VAP), acute pyelonephritis, cUTIs, cIAIs, |
| Imipenem sodium/cilastatin + relebactam (6) | Diazabicyclooctane (DBO) | Recarbrio | 2019 | Merck | cUTIs, cIAIs, pneumonia |
HAP, Hospital acquired pneumonia; VAP, Ventilator-associated pneumonia; cUTIs, complicated urinary tract infections; cIAIs, complicated intra-abdominal infections.
Table 2. β-lactamase Inhibitors (Approved and in Clinical Development).
The Zn2+-dependent metallo β-lactamase inhibitors (type B) confer resistance to a multitude of β-lactams, including penicillins, cephalosporins, and carbapenems.26 Taniborbactam (15), a bicyclic boronic acid-containing molecule, is the first pan-spectrum inhibitor of serine and metallo-β-lactamases to enter clinical trials.71 Taniborbactam displayed IC50 values of 0.03–18 μM against KPC-2 (Class A), OXA-48 (Class D), AmpC (Class C), and VIM-2 (Class B). Rescue of cefepime activity by taniborbactam was observed in serine β-lactamase containing E. coli, E. cloacae, K. pneumoniae, and K. oxytoca with potentiation factors between 32- and 1024-fold. It also rescued cefepime’s activity in bacterial strains of A. baumannii, K. pneumoniae, E. coli, E. cloacae, and P. aeruginosa harboring VIM-1, VIM-2, VIM-4, or NDM-1 by a factor of 128-fold in 90% strains. The in vivo efficacy of cefepime and taniborbactam was assessed in a neutropenic mouse lung infection model against a CTX-M-14-producing strain of K. pneumoniae and in the ascending urinary tract infection model against a CTX-M-15-producing strain of E. coli.71 >4 log and >2 log reductions were observed in the two in vivo models, respectively. Taniborbactam-cefepime is in phase 3 clinical trials and is being developed for complicated urinary tract infections, hospital-acquired bacterial pneumonia, and ventilator-associated bacterial pneumonia.64 Another ultra-broad-spectrum inhibitor in clinical trials is QPX7728/Xeruborbactam (16) which has additional activity against diverse Class D carbapenemases from Acinetobacter and IMP which is lacked by taniborbactam.72 QPX7728 was tested in combination with two carbapenems, three cephalosporins, and a monobactam against carbapenem-resistant strains of K. pneumoniae in a mouse thigh infection model.73 In the best cases, a 1.5–2 log reduction in bacterial burden was observed for the combination of QPX7728 given with various β-lactam antibiotics.
There are several other candidates in the literature which inhibit β-lactamases (Figure 4).64,74 A vancomycin derivative, Dipi-van (17), developed by our group, was able to act as a New-Delhi metallo-β-lactamase-1 (NDM-1) inhibitor and resensitized NDM-1-producing K. pneumoniae and E. coli to meropenem (potentiation factor ∼8–66-fold).75 Dipi-van, in combination with meropenem, showed efficacy in a murine sepsis infection model of NDM-producing K. pneumoniae. A 3–4 log reduction in bacterial burden was observed in the liver, spleen, lungs, and kidney. Aspergillomarasmine A (18) (a fungal natural product) was able to rescue the antibiotic activity of meropenem against metallo-β-lactamase-expressing strains and exhibited efficacy in murine models.76 It showed good inhibition of MBLs like NDM-1 and VIM-2 (FICIs < 0.1 for 16 clinical carbapenem-resistant Enterobacteriaceae). The combination therapy ensured >95% survival at 5 days following lethal infection with NDM-1-positive K. pneumoniae. Both these inhibitors (17 and 18) sequester the Zn2+ which is an important cofactor in the functioning of MBLs like NDM. l-Captopril, an approved ACE inhibitor for the treatment of hypertension, was also shown to have MBL-inhibition properties.77 However, the d-Captopril (19) was shown to be 25 times more potent in inhibiting NDM-1.77 Its thiol moiety intercalates between the two Zn2+ ions, displacing the water molecule, thus competitively inhibiting NDM-1.78 Hydrogen bonding between the carboxylic acid of the ligand and the Asn220 residue of NDM-1 further strengthens the binding. This finding has led to the synthesis of several thiol and carboxylic acid containing derivatives for inhibiting MBLs.26,79−84 In addition to this, several covalent inhibitors of MBLs like ebselen (20) were identified.85 Ebselen forms a hydrogen bond with the Cys208 side chain, thereby disrupting the coordination of the second Zn2+ and removing it from the active site. Ebselen reduced the MICs of ampicillin and meropenem by 16- and 128-fold, respectively, against NDM-1-producing E. coli strains. Structural optimization of ebselen has led to better covalent inhibitors of MBLs.86 Cyclic boronates (21) have also been discovered with potent inhibition of MBLs and SBLs (potentiation factors = 4–64 with meropenem against SBL- and MBL-producing strains ofE. coli and K. pneumoniae).87 They achieve this by mimicking the tetrahedral intermediate common to both types of β-lactamases. They also inhibit the nonessential PBP5. Readers are directed to the following reviews for relevant information on β-lactamase inhibitors.26,57,64
Figure 4.
β-lactamase inhibitors (17–21) and inhibitors of aminoglycoside-inactivating enzymes (22–25).
Apart from the many approved, clinical, and preclinical candidates of β-lactamases, other enzyme inhibitors are also included in the category of direct acting adjuvants.40 Aminoglycosides are rendered ineffective in bacteria by inactivating enzymes like acetyltransferases, aminoglycoside kinases (APHs), and adenylyltransferases as well as ribosome methylation.88 Inhibitors of aminoglycoside-inactivating enzymes and ribosome methyl transferases are known, but they have shown little efficacy in animal models.89−94 Of huge interest are pyrazolopyrimidines (22) (human Src and PI3 kinases) which inhibit APH(3′)-I, an aminoglycoside kinase in a way different than the binding in human kinases, thus offering selectivity.91 Tropolones (23) inhibit adenylyltransferases,95,96 and some positively charged peptides target both APHs and acetyltransferases.97 6-Furanylquinazolines (24) were identified using an antibiotic resistance platform (ARP) and were shown to potentiate gentamicin by the inhibition of aminoglycoside 2-O nucleodyltransferase 2″-Ia.98 Wortmannin irreversibly inhibits aminoglycoside kinase AAC (6′)-APH (2″) (25).99
Moreover, inhibitors of enzymes involved in resistance to β-lactam antibiotics have been shown to synergistically interact with β-lactam antibiotics via a cooperative effect. Oxadiazoles are a non-β-lactam class of compounds which inhibit the PBP2a in MRSA.100,101 ND-421 from this class of compounds was investigated for its synergy with β-lactams. It exhibited FICI values of 0.31 and 0.37 with oxacillin at subinhibitory concentrations.102 Breaking the cooperation between PBP2a and PBP2, which is essential for resistance to β-lactams in MRSA, is the basis of this synergy. In a neutropenic mice thigh infection model of MRSA, the combination of ND-421 and oxacillin exhibited a 1.6 log reduction in bacterial burden as compared to the vehicle control. The combination was superior in reducing the bacterial burden relative to the individual treatment groups of oxacillin and ND-421.
A quinazolinone, (E)-3-(3-carboxyphenyl)-2-(4-cyanostyryl) quinazolin-4(3H)-one, was found to be active against MRSA by binding to the allosteric site of PBP2a.103 At subinhibitory concentrations, another quinazolinone obtained after a detailed structure–activity relationship was shown to synergize with piperacillin-tazobactam against MRSA.104 At 0.5 X MIC of piperacillin-tazobactam and 0.25 X MIC of the quinazolinone, >3 log reduction was observed in MRSA. In a murine neutropenic thigh infection model of MRSA, the triple combination of tazobactam-piperacillin and quinazolinone exhibited a 2.12 log reduction in bacterial burden as compared to the vehicle control, a 1.34 log reduction compared to the quinazolinone, and a 1.1 log reduction compared to tazobactam-piperacillin. The proposed mechanism of action indicated that the binding of the quinazolinone to the allosteric site of PBP2a triggers the opening of the active site, which then becomes accessible for binding by piperacillin. X-ray crystallographic studies and other in vitro experiments proved this. Furthermore, the inhibition of PBP1 by the quinazolinone compliments the inhibition of PBP2 by piperacillin. Moreover, tazobactam protects piperacillin from hydrolysis by serine β-lactamases produced in MRSA.
Enzyme inhibitors are the only adjuvants which are clinically approved. However, this is also limited to serine β-lactamase inhibitors. With a number of metallo-β-lactamase- and broad spectrum β-lactamase inhibitors in clinical trials, there is hope that more of such enzyme inhibitors will be on the market. However, adjuvants inhibiting other enzymes can be explored more for an all-round success of enzyme inhibitors as antibiotic adjuvants.
3.1.2. Direct Acting Efflux Pump Inhibitors
Apart from inactivating enzymes, antibiotic efflux is another target for the search of Class Ia adjuvants. A boost of intrinsic efflux activity can occur in bacteria due to the overexpression, asymmetric accumulation, or mutation of genes encoding energy-dependent membrane transporters.105 Under stress conditions, antibiotic efflux can be the most prevalent resistance mechanism in bacteria. As mentioned previously, six families of efflux have been identified in bacteria comprising the ATP-dependent ABC family and the secondary active transporters, the major facilitator superfamily (MFS), the multidrug and toxin extrusion (MATE) family, the small multidrug resistance (SMR) family, the resistance-nodulation-cell division (RND) superfamily, and the proteobacterial antimicrobial compound efflux (PACE) family.105,106 The secondary active transporters are powered by electrochemical energy generated from transmembrane ion gradients. Efflux pumps in bacteria have diverse biological roles, one of which is evading antibiotic action by extruding it out of the cell. The detailed types, structures, and functions of various efflux pumps can be referred to in the “mechanisms of resistance” section and various other reviews.105,107−111 In this section, we will cover the direct acting efflux pump inhibitors which can competitively bind to efflux pumps, inhibiting the binding efficiency of various natural substrates and antibiotics, thus resulting in inhibition of efflux machinery. The functioning of the five families of the secondary active efflux pumps can also be inhibited by disturbing the transmembrane ion gradients in cells, thus affecting the electrochemical energy associated with them, which is represented as a sum of the transmembrane electrical gradient (ΔΨ) and proton chemical gradient (ΔpH). These two components together constitute the proton-motive force in bacteria which has a value of around −130 to −230 mV, depending on the bacterial species.112 Perturbation of either of the components can inhibit the functioning of efflux pumps.113 However, this strategy of efflux inhibition is discussed in the section pertaining to membrane-targeting compounds. In this section, direct inhibitors of efflux pumps, constituting Class Ia of antibiotic adjuvants will be discussed.114,115
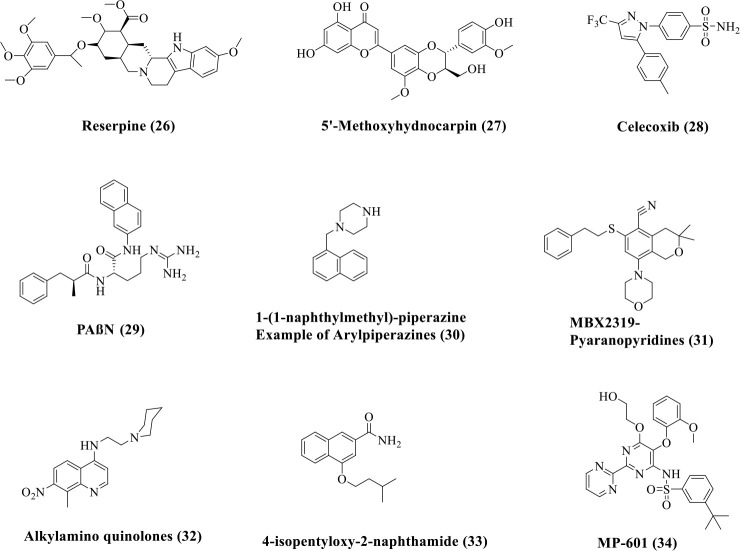
The inner-membrane-spanning MFS and tripartite RND efflux systems, which span the inner cell membrane, the periplasm, and the LPS-containing outer membrane in Gram-negative bacteria, are the prominently observed antibiotic resistance factors in clinic.116 Various compounds have been identified through screens as direct acting inhibitors of efflux pumps (Figure 5). Natural products like reserpine (26) and flavonoid 5′-methoxyhydnocarpin D74 (27) act as inhibitors of the S. aureus MFS NorA which confers resistance to hydrophilic fluoroquinolones like norfloxacin and ciprofloxacin (Figure 5).117,118 5′-Methoxyhydnocarpin D74 reduced the MIC of norfloxacin from 1 μg/mL to 0.25 μg/mL against S. aureus. Celecoxib (28) and its synthetic derivatives also inhibit the S. aureus MFS NorA.119,120 Celecoxib is a cyclooxygenase-2 inhibitor which potentiated various antibiotics like ampicillin, kanamycin, chloramphenicol, and ciprofloxacin by increasing the accumulation of drugs in MRSA. RND-mediated efflux is a major contributor of efflux in Gram-negative bacteria. The structurally well-characterized AcrAB–TolC system from E. coli has been the focus of this.105 In the clinic, MexAB–OprM and MexXY–OprM of P. aeruginosa and AdeABC of A. baumannii are major contributors in multidrug resistance. Inhibitors of AcrB (analogous to MexB, MexY, and AdeB) have been reported in the literature. Peptide analogues such as PAβN121 (29), small molecules such as aryl-piperazines122 (30), and pyranopyridines (example: MBX2319)123 (31) have been reported to inhibit the activity of cytoplasmic AcrAB efflux pumps, primarily by inhibiting AcrB (Figure 5). PAβN (29) decreased the MIC of fluoroquinolones against P. aeruginosa (potentiation factor of 8-fold observed in the case of levofloxacin) by a factor of 32–64-fold in strains with overexpressing efflux pumps.121 PAβN also reduced the frequency of resistance development to levofloxacin in P. aeruginosa. 1-(1-Naphthylmethyl)-piperazine (30) is an arylpiperazine which reversed drug resistance to multiple drugs like levofloxacin, linezolid, and ethidium bromide by a factor of 4–16-fold, in clinical isolates of E. coli.122 MBX2319 (31) inhibited the RND efflux pumps of Enterobacteriaceae and decreased the MICs of ciprofloxacin, levofloxacin, and piperacillin in E. coli AB1157 by 2-, 4-, and 8-fold, respectively.124 As there are multiple substrate channels and efflux mechanisms, this was not able to achieve universal inhibition of efflux. Other components of this complex efflux system like the periplasmic protein AcrA can also be targeted to ensure better specificity and off-target effects, as an alternative to targeting AcrB.125 Alkylamino quinolones126 (32) and naphthamides127 (33) have been shown to potentiate multiple classes of antibiotics by direct inhibition of RND efflux pumps. An alkylamino quinolone (32) potentiated chloramphenicol, tetracycline, norfloxacin, and cefepime by a factor of 2–16-fold against multiple strains of Enterobacter aerogenes. 4-Isopentyloxy-2-naphthamide (33) exhibited the best FICI of 0.25 with erythromycin and chloramphenicol against E. coli.
Figure 5.
Direct-acting efflux pump inhibitors.
The hope for universal blockaders of efflux is far from the achievable limits, primarily because of multiple substrate channels, efflux mechanisms, structural diversity across species, and toxicity due to lack of specificity. Also, some of the efflux pumps are not yet properly known and characterized. The current research targets the well-characterized and studied effux systems which are also of clinical relevance. To date, MP-601 (34) (Figure 5) is the only documented inhibitor of efflux pumps which is administered as an aerosol in patients with ventilator-associated pneumonia or cystic fibrosis.115,128
3.2. Indirect Resistance Breakers
Resistance to antibiotics in bacteria can be because of an intrinsic genetic association, physiological factors, or through the presence of nonspecific treatment evasion mechanisms. These include a number of interdependent factors which are important for the elements of resistance in bacteria. Other than the actual elements of resistance, these factors can be potential targets for developing adjuvants. Antibiotic activity can be resuscitated by identifying nonevident synergies in the vast nonessential gene space through this approach.8 Identifying such adjuvants requires cell-based screens for antibiotic potentiation followed by elucidating the mechanism of potentiation. Several teichoic acid synthesis inhibitors have shown synergy with β-lactam antibiotics against MRSA which is covered in a subsequent section. Another example of these types of adjuvants is membrane-targeting compounds which can have multifaceted effects like inhibition of efflux machinery and permeabilizing the bacterial membrane for the entry of antibiotics. These types of adjuvants are covered in detail. These Class Ib adjuvants can be broadly classified into inhibitors interfering with certain auxiliary enzymes/components which are important for the functioning of resistance elements and membrane-targeting compounds which can affect membrane-associated resistance elements like the permeability barrier and efflux. Different categories of such adjuvants are discussed in the subsequent sections.
3.2.1. Teichoic Acid Biosynthesis Inhibitors and Interactors
Wall teichoic acids (WTAs) are an integral component of the Gram-positive bacterial cell wall. Wall teichoic acids play a major role in cell division processes, host colonization, and coordination of peptidoglycan synthesis and are also major players in rendering β-lactam resistance in methicillin-resistant S. aureus.129 WTAs in S. aureus are comprised of repeating units of 1–5 linked ribitol-5-phosphate units decorated with N-acetyl glucosamine units and d-alanine. They are covalently linked to the 6′-hydroxyl of N-acetylmuramic acid residues of peptidoglycan by a linker unit containing (glycerolphosphate)-N-acetylmannosamine-β (1–4)-N-acetylglucosamine. WTA synthesis is encoded by various essential (late-stage) and nonessential (early-stage) biosynthetic genes which can be referred to in various reviews and articles.129−132 The WTA precursor is assembled in the cytoplasm using the C55-undecaprenyl lipid carrier. The TarO enzyme catalyzes the first step of this biosynthesis by adding N-acetyl glucosamine to C55-phosphate to form C55-PP-GlcNAc. The membrane-bound protein TarG flips the WTA precursor to the outer face of the cytoplasmic membrane. Cells lacking WTA have inefficient cell division and have reduced virulence. Synthetic viable mutants of S. aureus have provided a platform for identification of pathway-specific WTA biosynthesis inhibitors.
In MRSA, resistance to β-lactams is in part due to the presence of a horizontally acquired penicillin-binding protein (PBP), PBP2a.133−135 The PBP2a cannot be inactivated by β-lactam antibiotics. WTA acts as a scaffold for the correct functioning of PBP2a. The conjugation of β-O-GlcNAc to the WTA backbone catalyzed by the enzyme TarS is important for PBP2a function. Furthermore, the WTA polymer acts as a spatiotemporal signal for the correct localization of PBP4 (required for final transpeptidation steps of peptidoglycan) in the divisome assembly at the septum. PBP2a overcompensates for the inefficient function of PBP2, which is inhibited in the presence of β-lactam antibiotics. However, seeing the importance of WTA for the proper functioning of PBP2a and PBP4, WTA biosynthesis inhibitors can be used synergistically with β-lactam antibiotics against MRSA (Figure 6).129,132,136,137 An example of this is tunicamycin (35), which is an inhibitor of TarO, the first enzyme for WTA biosynthesis. The MIC of oxacillin was reduced to 0.4 μg/mL from 25 μg/mL in the presence of 0.08 μg/mL of tunicamycin.138 Triclopidine (36) is an antiplatelet drug which inhibits the TarO enzyme. The potentiation of cefuroxime was observed in the presence of triclopidine against various hospital-acquired MRSA isolates with FICI as low as 0.040. The combination was effective in a Galleria mellonella model of community acquired (CA)-MRSA infection.113
Figure 6.
Teichoic acid biosynthesis inhibitors/interactors.
An antagonism screen for molecules suppressing the activity of Targocil (TarG inhibitor) identified Clomiphene (37),139 which is an essential gene in the synthesis of wall teichoic acids. This hinted at the inhibition of early-stage WTA synthesis. Clomiphene, however weakly active (MIC = 16 μg/mL), potentiated β-lactam antibiotics against MRSA. Mechanistic investigation revealed undecaprenyl diphosphate synthase as the target which catalyzes the synthesis of C55 lipid, essential for both WTA and peptidoglycan synthesis.140 The enzyme catalyzes the condensation of isopentenyl diphosphate (IPP) with allylic pyrophosphates. Clomiphene (37) potentiated the activity of most β-lactams including penicillins and cephalosporins to MRSA (FICI = 0.3–0.5). It also showed good synergy with bacitracin (FICI = 0.375) which inhibits the dephosphorylation of undecaprenyl diphosphate.
A phenotypic screening strategy involving chemical suppression of the inhibitory consequences of late-stage wall teichoic acid biosynthesis resulted in the discovery of tarocin A (oxazolidinone, 38) and tarocin B (benzimidazole, 39).137 The essential gene paradox was employed in this report to identify early-stage WTA biosynthesis inhibitors which would suppress the inhibitory activity of the late-stage TarG inhibitor, L-638. The two tarocins exhibited an MIC of >200 μM against MRSA COL and exhibited less toxicity (IC50 > 100 μM) against HeLa cells. The tarocins potentiated the activity of imipenem and dicloxacillin against clinical isolates of MRSA and methicillin-resistant Staphylococcus epidermidis (MRSE) (FICI ≤ 0.5). Whole genome sequencing of resistant mutants and biochemical and genetic target engagement studies revealed TarO as the target of these adjuvants, in a way that is different from tunicamycin. An acceptable frequency of spontaneous resistance was obtained with the combination of dicloxacillin and tarocins. The combination of tarocin with dicloxacillin exhibited an ∼2.6 log reduction in bacterial burden in a murine systemic infection model of MRSA.
A branched polyethylenimine (40) also restored MRSA’s susceptibility to various β-lactam antibiotics at subinhibitory concentrations (MIC = 4–32 μg/mL).141 FICIs of 0.188–0.75 were observed with the β-lactam antibiotics. It is known to electrostatically interact with the WTAs which leads to an increase in size of the bacterial cells with abnormal septa formation. It is thought that this interaction leads to mislocalization of the cell wall biosynthesis proteins (PBP2, PBP2a, PBP4) during cell division, which makes the bacteria sensitive to the action of β-lactam antibiotics.
Apart from the wall teichoic acid synthesis inhibitors discussed here, the biosynthetic pathways of other membrane components like lipoteichoic acid can also serve as good potential targets. Unlike wall teichoic acids, which are dispensable to cellular survival, lipoteichoic acids play a key role in the bacterial cell division process, and inhibition of its synthesis can impair the cell division machinery.142 By the employment of synthetic lethality networks and differential growth screens, two lipoteichoic acid (LTA) synthesis inhibitors (41 and 42) were identified which inhibit the glycosyltransferase, UgtP, involved in the assembly of an LTA glycolipid anchor.143 These inhibitors restored activity to oxacillin in resistant strains of MRSA. Another study modeled the binding of an inhibitor 1771-bound LtaS structure and used this model for virtual screening of new molecular scaffolds for LtaS inhibition.144 One of the molecules identified in the process (compound8) displayed strong binding to one of the transiently open cryptic pockets near the active site of the enzyme. An optimized analogue, compound9, exhibiting a change in nature of synthesized LTA, reduced the minimum inhibitory concentration of methicillin and carbenicillin against MRSA by 16- and 32-fold, respectively. It also displayed 4-fold reduction in the MIC of colistin in the MRSA strain.
Synthetic lethal networks in bacteria have been exploited for the development of teichoic acid inhibitors, particularly for potentiating β-lactam antibiotics against MRSA. A detailed structure–activity relationship of the existing scaffolds and further preclinical studies for the best compounds should be performed in the future. Moreover, other synthetic lethal networks can be discovered with the help of genome-wide and differential growth screens.
3.2.2. Inhibitors of Enzymes Involved in Other Cellular Processes
The search for antibiotic potentiators or adjuvants has also progressed significantly through broad library screens of different molecules with an antibiotic for their potentiation ability. In one such screen, combinations of the aminocoumarin antibiotic novobiocin were found (having minimal inherent activity against Gram-negative bacteria), with a library of 30,000 small compounds against E. coli BW25113.145 Four nonobvious synergistic combinations that overcome the inherent resistance of Gram-negative bacteria to novobiocin were discovered through this screening process. It was discovered that A22 (43, Figure 7) inhibits MreB activity. The bacterial cytoskeleton contains an actin-like protein, which is of great interest because it serves as a scaffold for important cell wall assembly proteins, which are necessary for cell development and division. When MreB was inhibited, both E. coli and P. aeruginosa developed round cells as a phenotype, accumulated more ethidium bromide, and responded synergistically to novobiocin and rifampin. This study put forth the hypothesis that cytoskeleton proteins and/or peptidoglycans could directly or indirectly disrupt cell shape. Gram-negative bacteria’s machinery for influx and efflux is altered by biosynthesis, enabling the accumulation of harmful antibiotics that were previously excluded. This offers a compelling orthogonal strategy for the creation of novel anti-Gram-negative treatments that make use of current antibiotics and a developing knowledge of microbial physiology.
Figure 7.
Adjuvants targeting other cellular processes of bacteria.
As discussed in an earlier section, inhibition of teichoic acid synthesis or transport can also sensitize β-lactams to resistant MRSA. With this background, small molecular inhibitors of cell envelope component synthesis which can potentiate the β-lactam antibiotic imipinem were identified through a library screen.146 One of the best potentiators had a steroid-like core structure, and it was called murgocil (44, Figure 7). This molecule is a highly selective inhibitor of a MurG enzyme which is known to convert lipid I to lipid II. Murgocil treatment can inhibit lipid II synthesis in vitro and shows overall reduced PG synthesis in drug-treated cells. While the initial screening was performed on the basis of potentiation ability, murgocil itself has significant antibacterial activity against S. aureus. It however potentiates the β-lactam at much lower concentrations. It was also found that the enzyme activity inhibition is very selective to MurG of S. aureus only. The enzyme isolated from other Gram-positive bacteria such as S. epidermidis and Bacillus spp. or Gram-negative bacteria such as E. coli and K. pneumoniae, etc., which has minor amino acid changes, is not inhibited by murgocil. More importantly, the researchers highlighted that murgocil had a significantly high rate of spontaneous resistance (10–7 at 4× MIC) as compared to standard antibiotics (10–8 or lower), highlighting the need for further structure–activity studies to yield better analogues.
The MurJ enzyme is responsible for flipping the lipid II from the inner face of the cell membrane to the outer face where transglycosylation and transpeptidation take place. The activity by this flippase acts as a signal for recruitment of PBPs to the divisome. The N-acylated linear heptapeptide antibiotic humimycin A improved the activity of β-lactam antibiotics against MRSA.147 It was synthesized on the basis of bioinformatic predictions derived from secondary metabolite gene clusters found in the human microbiome. An SAR campaign revealed a derivative of humimycin A known as humimycin 17S (45, Figure 7) which showed better potentiating profiles of β-lactams against MRSA and also better activity against both MRSA and VRE. Importantly, the combination of carbenicillin (carboxypenicillin) and humimycin 17S did not lead to any detectable resistance.
Moreover, two related compounds, DMPI (46) and CDFI (47), containing an indole ring potentiated the activity of β-lactams against MRSA (Figure 7).148 Mechanism of action (MOA) profiling and resistance mapping revealed that these agents target the uncharacterized gene SAV1754 which is important for peptidoglycan synthesis. The SAV1754 protein has structural similarity with MurJ which suggests that it might play a role in acting as a flippase to translocate cell wall precursors to the periplasmic space.
FtsZ is a key protein responsible for the formation of a Z-ring at the mid cell during cell division. The formation of a Z-ring further regulates the spatial and temporal localization of various cell division proteins like PBPs to the septum. TXA707 (48), an FtsZ-targeting compound, exhibited synergy with multiple β-lactam antibiotics against MRSA (Figure 7). Different β-lactams with enhanced binding to PBP2 such as cefdinir, imipenem, ceftriaxone, cefotaxime, ertapenem, ticarcillin, and oxacillin (0.08 × MIC), result in >4 log reduction in bacterial burden when combined with TXA707 (0.5 × MIC). Fluorescence microscopy studies revealed that TXA707 causes a mislocalization of key PBPs away from the septum to nonproductive peripheral sites. As the cooperative action of PBPs, especially PBP2 and PBP2a, is essential for the β-lactam resistance seen in MRSA, mislocalization of these proteins during septum formation is the reason for sensitization of MRSA to β-lactam antibiotics.
The interaction between lytic transglycosylases (LTs) and PBPs is crucial for the bacterial cell wall remodeling.149 Inhibition of PBPs by β-lactams, if accompanied by inhibition of LTs, can lead to dysfunctional septum formation. This leads to the formation of a bulge near the septal area, before lysis of the bacterium. An independent study demonstrated that Bulgecin A (an inhibitor of lytic transglycosylase) potentiated the antibacterial activity of third-generation cephalosporin ceftazidime. In a following study, the group also demonstrated the ability of bulgecins to potentiate β-lactams in P. aeruginosa. Bulgecins induced bulging near the septum in the presence of the antibiotic. The study identified three LTs (Slt, MltD, and MltG) from the bunch present in P. aeruginosa to be involved in β-lactam-induced bulge formation.150
Researchers have also tried to explore other bacterial signaling pathways as targets for antibiotics. One such study identified a penicillin-binding protein and serine/threonine kinase-associated (PASTA) kinase, which have been indicated for their role in cell wall homeostasis, virulence, biofilm formation, germination, and metabolism.151 It was incidentally observed that a PASTA-kinase-deficient mutant displayed enhanced sensitivity to β-lactams. A study reported that the utilization of protein kinase inhibitors reverses the MRSA phenotype and restores the susceptibility to β-lactam antibiotics.
Another study identified a cinnamamide which consistently reduced the MIC of ofloxacin by 2-fold and did not possess any activity of its own.152 This molecule was taken further for detailed structure–activity relationship studies, and quite remarkably, seven cinnamamide analogues were identified which could potentiate oxacillin by 64–128-fold against different strains of MRSA. However, the target of these adjuvants is yet unidentified.
Various proteins important in cell division and cell wall assembly have been identified as targets which can be coupled with an antibiotic target to result in potent combinations to combat multidrug-resistant bacteria. More phenotypic screens should be conducted to identify other proteins which can be targeted for the same purpose. Moreover, the best adjuvants should be explored further vis-a-vis design improvement and preclinical studies.
3.2.3. Membrane-Targeting Compounds
A number of membrane-targeting strategies to overcome antimicrobial resistance can be found in the literature.153−162 Membrane-targeting compounds can also be used as Class Ib antibiotic adjuvants. These compounds circumvent passive resistance mechanisms like the permeability barrier in Gram-negative bacteria and functioning of broad-specificity efflux pumps.40 These compounds are inspired from antimicrobial peptides (AMP) and synthetic mimics of AMPs. Various membrane-active compounds, even possessing some antibacterial activities, have been employed at subinhibitory concentrations in combination with different classes of antibiotics.
Colistin- and polymyxin-derived analogues have been used in combination with antibiotics targeting Gram-positive bacteria like vancomycin and teicoplanin to repurpose them against Gram-negative pathogens.11,163−166 PMBN (49), derived from polymyxin B, is one of the very first compounds with no inherent antimicrobial activity which depicted the potentiation of hydrophobic antibiotics (Figure 8).164,167 Other truncated derivatives of polymyxin B like DAPB (50), PMBO (51), and PMBH (52) are also known to show synergistic activity with Gram-positive antibiotics (Figure 8).165,168 More recently, SPR741 (53), another PMBN analogue, has passed the Phase I clinical trials (Figure 8).164 Atomic force microscopy studies revealed that SPR741 works by causing substantial outer-membrane disorder.169 This potentiator improved the efficacy of various hydrophobic antibiotics like fusidic acid, clarithromycin, retapamulin, erythromycin, and rifampicin.170 The MICs of antibiotics were reduced 32- to 8,000-fold against E. coli and K. pneumoniae in the presence of SPR741. It has shown synergy with antibiotics in mice infection models.171
Figure 8.
Membrane-targeting polymyxin-derived and other peptides.
Various antimicrobial peptides, cationic and cyclic lipopeptides, and antimicrobial peptide mimics have exhibited synergy with different classes of antibiotics.172,173 Peptides derived from cathelicidins, lactoferrin, thrombin, histatins, other natural AMPs as well as rationally designed peptides have shown potentiation of antibiotics like vancomycin, erythromycin, azithromycin, rifampicin, novobiocin, fusidic acid, etc. against Gram-negative bacteria by perturbing their outer membrane owing to interaction with lipopolysaccharide present in the outer membrane.172 For example, a short linear antimicrobial peptide, SLAP-S25 (54, Figure 8), however decently active on its own (MIC = 1–64 μg/mL) against a panel of Gram-positive and Gram-negative bacteria, potentiated antibiotics from different classes like colistin, cefepime, ofloxacin, tetracycline, rifampicin, and vancomycin, against MDR Gram-negative pathogens.174 Potentiation factors ranging from 8- to 1024-fold were observed in a combination of antibiotics with ≤4 μg/mL of SLAP-S25 (54, Figure 8) against various Gram-negative bacterial strains. The mechanism of potentiation was LPS and PG binding in the outer and cytoplasmic membrane, respectively. The synergistic efficacy of the combination of SLAP-S25 and colistin was observed in three animal models infected with E. coli B2.
Various cationic lipopeptides have also been shown to potentiate the activity of antibiotics against Gram-negative bacteria.172 Synthetic paenipeptins potentiated the activity of clarithromycin and rifampicin in in vivo models of polymyxin-resistant E. coli.175,176 Dilipid ultrashort cationic lipopeptides (dUSCLs), bearing lysine-rich tetrapeptides and lipopeptides at the N-terminal, improved the activity of various antibiotics against Gram-negative bacteria by permeabilizing the outer membrane and disrupting antibiotic efflux.177 Ultrashort tetrabasic peptides were also reported by the same group.178 Moreover, peptidomimetics like dilipid ultrashort tetrabasic peptidomimetics (dUSTBPs) consisting of three l-arginine units and an eight-carbon-long dilipid potentiated novobiocin and rifampicin against multidrug-resistant P. aeruginosa, A. baumannii, and Enterobacteriaceae species.179 Oligo-acyl-lysines (OAKs) have also shown potentiation of the Gram-positive antibiotic rifampicin against Gram-negative bacteria.180−182
Screening platforms have also been employed to identify antibiotic adjuvants which can target the membrane and thus potentiate antibiotics. A vancomycin antagonism screening platform at low temperature identified pentamidine (55, Figure 9), an antiprotozoal drug, as an effective membrane-targeting compound by its interaction with the lipopolysaccharide in the outer membrane of Gram-negative bacteria.183 Synergy with Gram-positive antibiotics was observed. Specifically, novobiocin was potentiated by pentamidine against the colistin-resistant strain of A. baumannii. With rifampicin, an FICI of 0.25 was shown against E. coli. Efficacy in an in vivo model of systemic colistin-resistant A. baumannii infection was also tested. A combination of pentamidine (10 mg/kg) and novobiocin (50 mg/kg) led to a significant reduction in bacterial burden in the spleen. Moreover, an extensive SAR analysis with various bisamidine derivatives with varying linker motifs and geometry of the amidine groups yielded derivatives (56, 57, and 58) with better synergistic activity than pentamidine (Figure 9).184 It was found that the length, rigidity, and hydrophobicity of the linker play an important role in the synergistic activity. The presence of an aromatic group in the linker led to better synergy but slightly more percentage of hemolysis than pentamidine. The best derivatives exhibited FICIs in the range between ≤0.094 and ≤0.250 with erythromycin and rifampicin against E. coli. Moreover, a more recent study identified a better analogue of pentamidine named P35 (59).185 P35 showed less off-target toxicity than pentamidine vis-à-vis reduced mammalian cell toxicity and hERG trafficking inhibition (Figure 9). P35 exhibited an FICI of 0.094 with novobiocin against A. baumannii in comparison to a value of 0.25 shown by pentamidine. P35 also outperformed pentamidine in a murine systemic infection model of A. baumannii showing ∼3 log reduction in bacterial load in blood. Another screening of 158 FDA approved compounds for potentiation of doxycycline against E. coli yielded metformin (60) as a suitable antibiotic adjuvant for tetracycline antibiotics.186 It depicted good potentiation of doxycycline against MDR S. aureus, E. faecalis, E. coli, and S. enteritidis (best FICI = 0.078) (Figure 9). The mechanism of action was outer-membrane permeabilization, proton motive force reduction, lower transcript levels of efflux pump related genes like tetA, reduced ATP production, and subsequent increased accumulation of the antibiotic in bacterial cells. It also alleviated the immune response generated by LPS by reducing the production of pro-inflammatory cytokines and interferons like TNF-α, IL-8, IL-1β, and IFN-γ. Good potentiation of doxycycline was also observed in in vivo murine and Galleria melonella models. Another high-throughput screening for vancomycin antagonism at low temperature identified liproxstatin-1 (61) and MAC-0568743 (62) as potentiators of antibiotics like novobiocin, linezolid, erythromycin, and rifampicin against Gram-negative bacteria (Figure 9).187 These molecules were shown to interact with LPS and specifically disrupt the outer membrane, while showing minimal interaction with the inner membrane. Another high-throughput screen led to the discovery of azaindoles which potentiate rifampicin and novobiocin against Gram-negative pathogens by a factor of 100–1000-fold.188
Figure 9.
Small and macromolecular membrane-perturbing adjuvants.
Loperamide (imodium) (63) was discovered as a potentiator of tetracycline antibiotics in a screen (Figure 9).189 Loperamide decreased the membrane potential (ΔΨ), contributing to the overall proton motive force. To ensure ATP homeostasis, the ΔpH component across the inner membrane is modulated, thereby facilitating more uptake of the tetracycline antibiotics. This helps in overcoming acquired or intrinsic resistance to tetracycline antibiotics.
Membrane-perturbing adjuvants can also show inhibition of ATP synthase. Venturicidin A (64) is a natural product isolated from soil actinomycetes which is an inhibitor of ATP synthase (Figure 9).190 It potentiates gentamicin against multidrug-resistant clinical isolates of Staphylococcus (including MRSA), Enterococcus, and Pseudomonas aeruginosa. It uncouples ATP synthesis from electron transport by blocking proton flow through ATP synthase. This results in an elevated extracellular proton gradient and a rise in gentamicin uptake. FIC indices of 0.125–0.64 were observed with gentamicin in the best cases. However, the adjuvant had a low EC50 value of 31 μg/mL against HEK cells but was tolerated intraperitoneally up to 400 mg/kg.
Various membrane-active macromolecules in our group have been employed at subinhibitory concentrations to result in the potentiation of antibiotics.191−194 A membrane-active macromolecule, Qn-prAP (65, Figure 9), potentiated tetracycline antibiotics toward New Delhi metallo-β-lactamase-1 (NDM-1)-producing E. coli and K. pneumoniae strains by a potentiation factor of >80–1250-fold. Preliminary in vivo studies revealed good toxicity profiles, and significant bacterial reduction in the murine thigh infection model of E. coli was observed by a combination of doxycycline and Qn-prAP. No resistance to the combination of minocycline and Qn-prAP was observed in E. coli.192 A different macromolecule of the same class of QCybuAP (66) disrupted mature biofilms of E. coli and A. baumannii in combination with erythromycin. A considerable reduction in bacterial burden was observed in a burn-wound infection of A. baumannii by the combination of QCybuAP (66, Figure 9) with antibiotics like rifampicin and erythromycin.194 Similarly, ACP-1 (67, Figure 9) restored the activity of rifampicin against multidrug-resistant strains of Gram-negative bacteria.193 More importantly, this combination exhibited good activity against preformed biofilms of P. aeruginosa and E. coli. The compound also exhibited good in vivo toxicity profiles for both topical and systemic administration. Another macromolecule, ACM-AHex (68, Figure 9), potentiated tetracycline by 4–128-fold against Gram-negative bacteria.195
Over the years, derivatives of polyamines have shown good potentiation ability.172,196,197 For example, our efforts led to the development of d-LANA-14 (69) which is a synthetic cationic lipopeptide-based small molecule which shows good activity against Gram-positive bacteria but high MIC values against Gram-negative bacteria (Figure 9).198 This compound was used at subinhibitory concentrations in combination with obsolete antibiotics, rifampicin and tetracycline, to combat Gram-negative superbugs. The mechanism of potentiation was found to be outer-membrane permeabilization and membrane depolarization. FIC indices of 0.19–0.75 were observed with rifampicin and tetracycline against A. baumannii and P. aeruginosa including carbapenem-resistant isolates. A combination of d-LANA-14 (40 mg/kg) and rifampicin (40 mg/kg) showed a 4.9 log and 4 log reduction in A. baumannii and P. aeruginosa viability, respectively, in a burn-wound infection model.
The problem with using subinhibitory concentrations of membrane-active compounds to potentiate antibiotics is that they often show mammalian cell toxicity. This can lead to unwanted off-target effects which might hinder the clinical translation of these compounds. Structural and mechanistic insights with proper correlation are highly important for understanding the intricate details of membrane-targeting adjuvants. In a recent report, our group identified that weak membrane perturbation is enough to potentiate various classes of antibiotics against Gram-negative bacteria. These weak membrane-perturbing adjuvants caused minimum levels of outer-membrane permeabilization and membrane depolarization in bacteria which also led to the inhibition of efflux machinery.199 The presence of cyclic hydrophobicity as opposed to acyclic hydrophobicity in the design was considered to be superior to result in high potentiation (up to 4096-fold), minimal activity (>512 μg/mL), and minimal toxicity (HC50 > 1024 μg/mL, EC50 > 512 μg/mL).199 Up to 4096-fold potentiation of multiple classes of antibiotics like tetracyclines, vancomycin, rifampicin, fusidic acid, erythromycin, and chloramphenicol toward critical Gram-negative superbugs, including carbapenemase-producing strains, was observed. The best adjuvant, NAda (70), showed broad-spectrum potentiation of multiple antibiotics against MDR strains of A. baumannii, P. aeruginosa, E. coli, and K. pneumoniae (Figure 9).199
The existing library of membrane-perturbing compounds which potentiate antibiotics is huge but is limited by the associated off-target effects which are not even explored for many compounds. For successful translation of such adjuvants in the clinic, extensive preclinical studies like that done in the case of pentamidine analogues and SPR741 are required. Moreover, the identification of structural features important for an optimum level of membrane perturbation and negligible cytotoxicity, like that done by our group in the case of a norspermidine backbone, and SAR studies like that done in the case of norspermidine-based adjuvants and pentamidine analogues are required for the translation of membrane-perturbing adjuvants. Preclinical studies are of high importance. Examples of SAR and preclinical studies undertaken for norspermidine derivatives and pentamidine analogues are given in Figure 10. These studies are indispensable for better translation of such adjuvants in clinics. The compounds identified through high-throughput screens and natural product isolation can also be exposed to such rational approaches to yield the best possible adjuvant from a particular scaffold.
Figure 10.
Examples of indispensable SAR and preclinical studies undertaken for membrane-targeting compounds. (A) Correlation of the extent of membrane perturbation in norspermidine derivatives with varying structural moieties. (B) Correlation of activity and toxicity with varying structural moieties present in norspermidine derivatives. (C) Correlation of potentiation ability with the structure of norspermidine derivatives. (D) Scatter plot of compound synergy (FICI) with novobiocin against A. baumannii and lipophilicity (c log P) for pentamidine analogues. (E) Scatter plot of FICI and cytotoxicity for pentamidine analogues. (F) Pharmacokinetic analysis of the two best pentamidine analogues. (G) Microelectrode array (MEA) traces after exposure to different treatments to assess the beat period and field potential duration due to hERG trafficking inhibition. Figures adapted with permission from refs (199) (A to C) and (185) (D to G) from ACS Publications. Copyright ACS Publications 2022.
3.3. Adjuvants Targeting Host Processes
The effectiveness of antibiotics within infected organisms can be increased by taking advantage of a variety of host defense mechanisms. For instance, antibiotics and antimicrobial peptides have been shown to work well together and can even increase antibiotic activity in difficult-to-treat biofilms.200 These peptides offer a wide range of strengths in relation to powerful antimicrobial activity or Class Ib as well as Class II adjuvant properties. These immunomodulatory peptides affect the host immune system in a variety of ways, including suppressing inflammation to prevent an infection from triggering an excessive immune response that results in sepsis and inducing host-cell-based antimicrobial activities like increased phagocytosis.
However, there are very few reports of such peptides being used as adjuvants in combination with antibiotics. One report has demonstrated the efficacy of a host-tissue factor pathway inhibitor 2 (TPFI-2)-derived EDC34 (71, Figure 11) peptide against complicated infections of E. coli and P. aeruginosa, when used in combination with the cephalosporin ceftazidime.201 This peptide itself does possess some activity. However, it has a strong immunomodulatory effect and can reduce the excessive inflammatory response emerging due to bacterial endotoxin LPS, which has been validated in a murine model. In a sepsis model of P. aeruginosa, only the peptide did not rescue mice. However, ceftazidime and peptide combination therapy was successful in reducing mortality. There was a significant reduction in pathogen-induced clotting in the combination treated group, as compared to the only antibiotic group. At the same time, this peptide was found to enhance the formation of a host antimicrobial anaphylatoxin C3a, both in vitro and in vivo, which was further responsible for increased bacterial clearance. The study has comprehensively explored host-modulating effects of this host-derived peptide. A similar effect can be anticipated for various host-defense peptides, and further exploration of moderately active peptides as antibiotic adjuvants has the potential to yield a strong therapeutic regimen for severe bacterial infections, including sepsis.
Figure 11.
Adjuvants which modulate host response.
Another tactic is to use small molecules to specifically target elements of innate immunity. In a screening of microbial natural product extracts, the natural product streptazolin (72, Figure 11) was found to be capable of promoting macrophage killing of Streptococcus pneumoniae.202 Through the phosphatidylinositide signaling pathway, streptazolin stimulates nuclear factor-B production and simultaneously releases anti-infective cytokines. This strategy opens up a previously untapped landscape of targets that may support drug combination strategies to control infection by combining antibiotics like immunomodulatory peptides. On the other hand, receiving antibiotics may alter the metabolism of the host, which may reduce the effectiveness of the antibiotics or negatively impact immune cell function. Finally, altering a pathogen’s growth environment, such as limiting its access to nutrients, can drastically change how sensitive it is to antibiotics.203 As a result, therapeutic adjuvants that modify the host microenvironment may become available in the future.
It is evident through these examples that host modulation can serve as a powerful tool to enhance the efficacy of the existing antibiotic arsenal. However, significant effort is needed to explore this approach in greater detail. Multipronged strategies, including chemical and genetic screens, will have to be used to identify host-modulating Class II adjuvants. Further, there is a significant need to validate the superiority of the combination and understand the effects of the adjuvant on the host in much more detail.
4. Conclusions
Antibiotic–adjuvant combination therapy is one of the prime approaches to tackle antimicrobial resistance. It has seen a surge of research over the past decade with new types of adjuvants being developed. The success of combination therapy is evident, given the significant number of approved antibiotic–adjuvant combination therapies, particularly for β-lactamase-bearing bacteria. Through this concise review, we have attempted to summarize the established and emerging facets of antibiotic–adjuvant combination therapy. Current research has demonstrated that adjuvant therapy can be directed to tackle various mechanisms of resistance to antibiotics in bacteria. We have discussed in this review the various types of antibiotic adjuvants through which resistance towards existing antibiotics can be overcome. They include direct resistance breakers, indirect resistance breakers, as well as host-modulating agents. To date, direct resistance breakers, particularly β-lactamase inhibitors, have been the most successful class of adjuvants, with many of them being approved for clinical applications. Concurrently, ongoing efforts to address the lacunae of approved adjuvants are also yielding success with new candidates emerging in clinical trials. Along with this class, membrane-perturbing small and macromolecular adjuvants are also being investigated and exploited for broad-spectrum potentiation.
In addition to these two well-explored strategies, new research is also producing other targets for developing adjuvants to tackle antimicrobial resistance. We have also discussed the important advances in this, including adjuvants which inhibit teichoic acid synthesis or nonessential steps of various bacterial metabolic processes, as well as host-modulating adjuvants. After a thorough overview of the state of research in the field, in the next section, we provide important directions for future researchers to pursue.
4.1. Future Perspective
As mentioned earlier, adjuvant therapy has started to attract significant interest in the past decade. Medicinal chemists have attempted to exploit three major strategies to tackle increasing antimicrobial resistance, namely, development of new drugs, semisynthetic derivatization of existing antibiotics, and combination therapy with antibiotic adjuvants. Combination therapy is emerging as a key strategy, with particular focus on rehabilitation as well as repurposing of various obsolete and redundant antibiotics. Though this has seen significant preclinical success, currently the approved list of antibiotic–adjuvant combinations includes only one type of direct resistance breakers, the β-lactamase inhibitors. No other combination from different classes has been approved yet. A few candidates like SPR741, a membrane-targeting adjuvant, is in clinical trials, and certain compounds like pentamidine analogues have also seen preclinical development. However, for the combined success of new classes of antibiotic adjuvants, it is pertinent to relook at the existing approaches from new directions.
There is significant merit in developing broad-spectrum adjuvants which can repurpose or potentiate multiple classes of antibiotics against different bacterial pathogens. This can be achieved by targeting the most significant mechanisms of resistance, such as antibiotic efflux and reduced permeability. Toward this end, we have described in the earlier sections the development of weakly membrane-perturbing small molecules as optimum membrane perturbers, with no inherent antibacterial activity or toxicity. Our group has demonstrated that the inclusion of aromatic and cyclic aliphatic hydrophobic groups, in place of long aliphatic chains, can introduce this weak membrane-perturbing property in the design (Figure 10). This has helped us achieve nonactive and nontoxic broad-spectrum adjuvants. A similar SAR approach has also been followed for pentamidine analogues to arrive at the best design (Figure 10). Similar studies need to be explored for other promising designs for further development. Nontoxic and nonactive adjuvants are the need of the hour as they would ensure better biocompatibility and less chance of resistance development in bacteria. Once SAR and basic chemistry help to identify better designs, these systems can be taken forward for preclinical studies. Currently, only a few membrane-perturbing adjuvants are being studied at the in vivo level. Preclinical studies like pharmacokinetics, pharmacodynamics, efficacy in different infection models, including systemic infections, plasma protein binding, and detailed toxicology studies are essential for these adjuvants. Pharmacokinetics and biodistribution studies are important to be conducted for a physical combination of antibiotics and adjuvants to ensure sufficient bioavailability of the two individual components at the infection site at different time points. Drug delivery systems can also be designed in the future to deliver both the components together.
Moreover, detailed mechanistic studies are required for different classes like membrane-targeting adjuvants as well as efflux pump inhibitors to understand and rule out any off-target effects, if present. As membrane-targeting adjuvants have the potential to be developed as multitarget adjuvants, their interaction with host factors should also be investigated. The effect of such adjuvants on the global transcriptome and long-term effects on the genome, including resistance development, should be thoroughly examined.
The two most prominent classes of adjuvants currently under development are membrane-perturbing compounds and β-lactamase inhibitors of the direct-resistance breakers class. However, due to the efforts of various groups, newer classes of adjuvants such as molecules inhibiting teichoic acid synthesis or cell cytoskeleton assembly have been identified. Much of this work has been achieved through chemical library screening and further development of hits. Through such screens, a few molecular scaffolds have been identified. While much of this work was conceptualized and reported in the past few years, upcoming research needs to make optimum use of the existing scaffolds. Researchers need to devote effort to performing detailed structure–activity relationship studies for the identified scaffolds to further improve the molecular design. Given the nascent stage of these reports, a detailed SAR and in-depth mechanistic studies would strongly accelerate the development of this new class of adjuvants. Additionally, genetic and target-based screens should be concurrently conducted to identify new nonessential targets for further development.
Along with pathogen-targeting adjuvants, many host cell process-inhibiting adjuvants have been identified through screens, and then their targets have been validated. The currently identified sets of inhibitors of such cellular processes highlight the strong potential of alternate targets. Hence, new research can look at the pathways of the host as a whole. Steps which are central to the pathway can be used to develop high-throughput assays for screening. This can help in identifying even more potent adjuvants which can then be developed through detailed SAR for the design. As mentioned earlier, host-modulating properties of other classes of adjuvants should also be investigated which can result in the development of multifactorial adjuvants.
The effect of antibiotics on the adjuvant also needs to be investigated. Most of the studies use representative antibiotic candidates from different classes. However, recent literature indicates that the potentiation activity of adjuvants depends strongly on the antibiotic. This can be either due to the better inherent interaction of the antibiotic with bacteria or due to some antibiotic–adjuvant cooperative interaction. These observations must be given significant importance and investigated further. Different antibiotic classes and multiple antibiotics from the same class should be investigated for one set of adjuvants to understand the mechanisms of action and the role of physicochemical properties of the antibiotics for synergistic combinations. This has recently been studied for pentamidine–antibiotic combinations.204
Antibiotic–adjuvant combinations should also be investigated against complicated forms of infection, such as bacterial biofilms, metabolically repressed bacterial subpopulations such as stationary phases or persister bacteria, and intracellular infections. This would enable its applicability in real clinical scenarios. This would also warrant a close look at the exact mechanism of such combinatorial activity and effects of adjuvants on bacterial virulence and quorum sensing. We have mentioned in earlier sections that some adjuvants can also possess moonlighting functions, apart from antibiotic potentiation. Particularly for small molecular membrane targeting adjuvants and also other indirect resistance breakers additional effects such as immunomodulation, induction/inhibition of autophagy, quorum sensing and virulence, effect on intracellular pathogens, etc. would be a good avenue to explore (Figure 12). There is literature on quorum sensing inhibition and antivirulence compounds.205 These compounds should be employed in combination with antibiotics to check their improved efficacy in in vivo, in cellulo, and organoid infection models.
Figure 12.
Directions to be explored for indirect resistance breakers, especially membrane-targeting adjuvants. Activity against bacterial biofilms, metabolically repressed bacteria, immunomodulatory functions, quorum sensing inhibition, and activity against intracellular infections can be explored for antibiotic adjuvants. Created using BioRender.com.
Collectively speaking, there is immense potential for commercialization of combination therapy. The research ecosystem has risen robustly and come up with novel ways of rehabilitation for existing antibiotics. However, efforts need to be undertaken to take such compounds to the preclinical and clinical development stage. We believe that there is a promising pool of compounds in the literature which can be developed as antibiotic adjuvants. A better understanding of different aspects of the adjuvant therapy at the design, biochemical, genetic, and preclinical stage can surely give a new dimension to this upcoming field and warrant its success in the future.
Acknowledgments
J.H. acknowledges SERB (CRG/2020/003118), Govt. of India. Y.A. acknowledges CSIR for a research fellowship.
The authors declare no competing financial interest.
References
- McCloskey B.; Dar O.; Zumla A.; Heymann D. L. Emerging infectious diseases and pandemic potential: status quo and reducing risk of global spread. Lancet Infect Dis 2014, 14 (10), 1001–10. 10.1016/S1473-3099(14)70846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M.; Folkers G. K.; Fauci A. S. The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430 (6996), 242–9. 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance; Wellcome Trust; HM Government, 2016. [Google Scholar]
- Top Ten causes of death. 2020. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed January 1, 2023).
- Global AntimicrobialResistance and Use Surveillance System (GLASS) Report 2022. 2022. https://www.who.int/publications/i/item/9789240062702 (accessed January 12, 2023).
- WHO . https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed January 14, 2023).
- Hutchings M. I.; Truman A. W.; Wilkinson B. Antibiotics: past, present and future. Curr. Opin Microbiol 2019, 51, 72–80. 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Tyers M.; Wright G. D. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol 2019, 17 (3), 141–155. 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- Kojima S.; Nikaido H. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc. Natl. Acad. Sci. USA 2013, 110 (28), E2629–E2634. 10.1073/pnas.1310333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorio Vargiu A.; Nikaido H. Multidrug Binding Properties of the AcrB Efflux Pump Characterized by Molecular Dynamics Simulations. Biophys. J. 2013, 104 (2), 285a–286a. 10.1016/j.bpj.2012.11.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya Y.; Bhattacharyya S.; Dhanda G.; Haldar J. Emerging Roles of Glycopeptide Antibiotics: Moving beyond Gram-Positive Bacteria. ACS Infect Dis 2022, 8 (1), 1–28. 10.1021/acsinfecdis.1c00367. [DOI] [PubMed] [Google Scholar]
- Tamber S.; Hancock R. E. W. On the mechanism of solute uptake in pseudomonas. Front Biosci 2003, 8, S472–S483. 10.2741/1075. [DOI] [PubMed] [Google Scholar]
- Lavigne J. P.; Sotto A.; Nicolas-Chanoine M. H.; Bouziges N.; Pages J. M.; Davin-Regli A. An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int. J. Antimicrob Ag 2013, 41 (2), 130–136. 10.1016/j.ijantimicag.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Poulou A.; Voulgari E.; Vrioni G.; Koumaki V.; Xidopoulos G.; Chatzipantazi V.; Markou F.; Tsakris A. Outbreak caused by an ertapenem-resistant, CTX-M-15-producing Klebsiella pneumoniae sequence type 101 clone carrying an OmpK36 porin variant. J. Clin Microbiol 2013, 51 (10), 3176–82. 10.1128/JCM.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J. M.; Webber M. A.; Baylay A. J.; Ogbolu D. O.; Piddock L. J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol 2015, 13 (1), 42–51. 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Piddock L. J. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006, 19 (2), 382–402. 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M.; Villa L.; Poirel L.; Nordmann P.; Carattoli A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 2013, 68 (1), 34–9. 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- Darby E. M.; Trampari E.; Siasat P.; Gaya M. S.; Alav I.; Webber M. A.; Blair J. M. A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol 2022, 10.1038/s41579-022-00820-y. [DOI] [PubMed] [Google Scholar]
- Pomposiello P. J.; Bennik M. H.; Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 2001, 183 (13), 3890–902. 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.; Chua K.; Davies J. K.; Newton H. J.; Seemann T.; Harrison P. F.; Holmes N. E.; Rhee H. W.; Hong J. I.; Hartland E. L.; Stinear T. P.; Howden B. P. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 2010, 6 (6), e1000944. 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y.; Ito T.; Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000, 44 (6), 1549–55. 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore A. C.; Deasy E. C.; Slickers P.; Brennan G.; O’Connell B.; Monecke S.; Ehricht R.; Coleman D. C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55 (8), 3765–73. 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez L.; Holden M. T.; Lindsay H.; Webb C. R.; Brown D. F.; Curran M. D.; Walpole E.; Brooks K.; Pickard D. J.; Teale C.; Parkhill J.; Bentley S. D.; Edwards G. F.; Girvan E. K.; Kearns A. M.; Pichon B.; Hill R. L.; Larsen A. R.; Skov R. L.; Peacock S. J.; Maskell D. J.; Holmes M. A. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 2011, 11 (8), 595–603. 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M.; Golparian D.; Nicholas R.; Ohnishi M.; Gallay A.; Sednaoui P. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob. Agents Chemother. 2012, 56 (3), 1273–80. 10.1128/AAC.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M.; Shafer W. M. Antibiotic resistance in Neisseria gonorrhoeae: origin, evolution, and lessons learned for the future. Ann. N.Y. Acad. Sci. 2011, 1230, E19–28. 10.1111/j.1749-6632.2011.06215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linciano P.; Cendron L.; Gianquinto E.; Spyrakis F.; Tondi D. Ten Years with New Delhi Metallo-beta-lactamase-1 (NDM-1): From Structural Insights to Inhibitor Design. Acs Infect Dis 2019, 5 (1), 9–34. 10.1021/acsinfecdis.8b00247. [DOI] [PubMed] [Google Scholar]
- Nordmann P.; Poirel L.; Walsh T. R.; Livermore D. M. The emerging NDM carbapenemases. Trends Microbiol 2011, 19 (12), 588–95. 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Zhao W. H.; Hu Z. Q. Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit Rev. Microbiol 2013, 39 (1), 79–101. 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouvelekis L. S.; Markogiannakis A.; Psichogiou M.; Tassios P. T.; Daikos G. L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012, 25 (4), 682–707. 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reygaert W. C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol 2018, 4 (3), 482–501. 10.3934/microbiol.2018.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cihlar T.; Fordyce M. Current status and prospects of HIV treatment. Curr. Opin Virol 2016, 18, 50–6. 10.1016/j.coviro.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Mokhtari R. B.; Homayouni T. S.; Baluch N.; Morgatskaya E.; Kumar S.; Das B.; Yeger H. Combination therapy in combating cancer. Oncotarget 2017, 8 (23), 38022–38043. 10.18632/oncotarget.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman M. D.; Xavier D.; Perel P. Uses of polypills for cardiovascular disease and evidence to date. Lancet 2017, 389 (10073), 1055–1065. 10.1016/S0140-6736(17)30553-6. [DOI] [PubMed] [Google Scholar]
- Jawetz E.; Gunnison J. B.; Bruff J. B.; Coleman V. R. Studies on antibiotic synergism and antagonism. Synergism among seven antibiotics against various bacteria in vitro. J. Bacteriol. 1952, 64 (1), 29–39. 10.1128/jb.64.1.29-39.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushby S. R.; Hitchings G. H. Trimethoprim, a sulphonamide potentiator. Br J. Pharmacol Chemother 1968, 33 (1), 72–90. 10.1111/j.1476-5381.1968.tb00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerantzas C. A.; Jacobs W. R. Origins of Combination Therapy for Tuberculosis: Lessons for Future Antimicrobial Development and Application. mBio 2017, 10.1128/mBio.01586-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston N. J.; Mukhtar T. A.; Wright G. D. Streptogramin antibiotics: mode of action and resistance. Curr. Drug Targets 2002, 3 (4), 335–44. 10.2174/1389450023347678. [DOI] [PubMed] [Google Scholar]
- Kehoe L. E.; Snidwongse J.; Courvalin P.; Rafferty J. B.; Murray I. A. Structural basis of Synercid (quinupristin-dalfopristin) resistance in Gram-positive bacterial pathogens. J. Biol. Chem. 2003, 278 (32), 29963–70. 10.1074/jbc.M303766200. [DOI] [PubMed] [Google Scholar]
- Moellering R. C. Jr. Rationale for use of antimicrobial combinations. Am. J. Med. 1983, 75 (2A), 4–8. 10.1016/0002-9343(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Wright G. D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol 2016, 24 (11), 862–871. 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Drusano G. L.; Hope W.; MacGowan A.; Louie A. Suppression of Emergence of Resistance in Pathogenic Bacteria: Keeping Our Powder Dry, Part 2. Antimicrob. Agents Chemother. 2016, 60 (3), 1194–201. 10.1128/AAC.02231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington R. J.; Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 2013, 31 (3), 177–84. 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R. Y.; Pei L.; Liu Q.; Chen S.; Dou H.; Shu G.; Yuan Z. X.; Lin J.; Peng G.; Zhang W.; Fu H. Isobologram Analysis: A Comprehensive Review of Methodology and Current Research. Front Pharmacol 2019, 10, 1222. 10.3389/fphar.2019.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roell K. R.; Reif D. M.; Motsinger-Reif A. A. An Introduction to Terminology and Methodology of Chemical Synergy-Perspectives from Across Disciplines. Front Pharmacol 2017, 8, 158. 10.3389/fphar.2017.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B.; Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 2004, 10, S122–9. 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- Bush K.; Bradford P. A. Interplay between beta-lactamases and new beta-lactamase inhibitors. Nat. Rev. Microbiol 2019, 17 (5), 295–306. 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- Drawz S. M.; Bonomo R. A. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010, 23 (1), 160–201. 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Proliferation and significance of clinically relevant beta-lactamases. Ann. N.Y. Acad. Sci. 2013, 1277, 84–90. 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- Bush K.; Jacoby G. A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54 (3), 969–76. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A.; Jacquier H.; Ruppe E.; Labia R. Structure-based classification of class A beta-lactamases, an update. Curr. Res. Transl Med. 2019, 67 (4), 115–122. 10.1016/j.retram.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Smet A.; Martel A.; Persoons D.; Dewulf J.; Heyndrickx M.; Catry B.; Herman L.; Haesebrouck F.; Butaye P. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli Isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 2008, 52 (4), 1238–43. 10.1128/AAC.01285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L.; Naas T.; Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 2010, 54 (1), 24–38. 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. G.; Butterworth D.; Cole M.; Hanscomb G.; Hood J. D.; Reading C.; Rolinson G. N. Naturally-occurring beta-lactamase inhibitors with antibacterial activity. J. Antibiot (Tokyo) 1976, 29 (6), 668–9. 10.7164/antibiotics.29.668. [DOI] [PubMed] [Google Scholar]
- Carcione D.; Siracusa C.; Sulejmani A.; Leoni V.; Intra J. Old and New Beta-Lactamase Inhibitors: Molecular Structure, Mechanism of Action, and Clinical Use. Antibiotics (Basel) 2021, 10 (8), 995. 10.3390/antibiotics10080995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden R. N.; Carmine A.; Heel R. C.; Morley P. A.; Speight T. M.; Avery G. S. Amoxycillin/clavulanic acid: a review of its antibacterial activity, pharmacokinetics and therapeutic use. Drugs 1981, 22 (5), 337–62. 10.2165/00003495-198122050-00001. [DOI] [PubMed] [Google Scholar]
- Bush K. Game Changers: New beta-Lactamase Inhibitor Combinations Targeting Antibiotic Resistance in Gram-Negative Bacteria. ACS Infect Dis 2018, 4 (2), 84–87. 10.1021/acsinfecdis.7b00243. [DOI] [PubMed] [Google Scholar]
- Alfei S.; Zuccari G. Recommendations to Synthetize Old and New beta-Lactamases Inhibitors: A Review to Encourage Further Production. Pharmaceuticals (Basel) 2022, 15 (3), 384. 10.3390/ph15030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo Y. A. Imipenem/Cilastatin/Relebactam: A Review in Gram-Negative Bacterial Infections. Drugs 2021, 81 (3), 377–388. 10.1007/s40265-021-01471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagunde P.; Patel P.; Lala M.; Watson K.; Copalu W.; Xu M.; Kulkarni P.; Young K.; Rizk M. L. Population Pharmacokinetic Analysis for Imipenem-Relebactam in Healthy Volunteers and Patients With Bacterial Infections. CPT Pharmacometrics Syst. Pharmacol 2019, 8 (10), 748–758. 10.1002/psp4.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulou D.; Siopi M.; Vourli S.; Pournaras S. Activity of Sulbactam-Durlobactam and Comparators Against a National Collection of Carbapenem-Resistant Acinetobacter baumannii Isolates From Greece. Front Cell Infect Microbiol 2022, 11, 814530. 10.3389/fcimb.2021.814530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya B.; Barcelo I. M.; Bhagwat S.; Patel M.; Bou G.; Papp-Wallace K. M.; Bonomo R. A.; Oliver A. WCK 5107 (Zidebactam) and WCK 5153 Are Novel Inhibitors of PBP2 Showing Potent ″beta-Lactam Enhancer″ Activity against Pseudomonas aeruginosa, Including Multidrug-Resistant Metallo-beta-Lactamase-Producing High-Risk Clones. Antimicrob. Agents Chemother. 2017, 61 (6), 10.1128/AAC.02529-16. 10.1128/AAC.02529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monogue M. L.; Giovagnoli S.; Bissantz C.; Zampaloni C.; Nicolau D. P. In Vivo Efficacy of Meropenem with a Novel Non-beta-Lactam-beta-Lactamase Inhibitor, Nacubactam, against Gram-Negative Organisms Exhibiting Various Resistance Mechanisms in a Murine Complicated Urinary Tract Infection Model. Antimicrob. Agents Chemother. 2018, 62 (9), e02596. 10.1128/AAC.02596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Reville T. F.; Guler S.; Comita-Prevoir J.; Chen B.; Bifulco N.; Huynh H.; Lahiri S.; Shapiro A. B.; McLeod S. M.; Carter N. M.; Moussa S. H.; Velez-Vega C.; Olivier N. B.; McLaughlin R.; Gao N.; Thresher J.; Palmer T.; Andrews B.; Giacobbe R. A.; Newman J. V.; Ehmann D. E.; de Jonge B.; O’Donnell J.; Mueller J. P.; Tommasi R. A.; Miller A. A. ETX2514 is a broad-spectrum beta-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii. Nat. Microbiol 2017, 2, 17104. 10.1038/nmicrobiol.2017.104. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace K. M. The latest advances in beta-lactam/beta-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert Opin Pharmacother 2019, 20 (17), 2169–2184. 10.1080/14656566.2019.1660772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka A.; Tsutsumi Y.; Yamada M.; Suzuki K.; Watanabe T.; Abe T.; Furuuchi T.; Inamura S.; Sakamaki Y.; Mitsuhashi N.; Ida T.; Livermore D. M. OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam ’enhancer’. J. Antimicrob. Chemother. 2015, 70 (10), 2779–86. 10.1093/jac/dkv166. [DOI] [PubMed] [Google Scholar]
- Trout R. E.; Zulli A.; Mesaros E.; Jackson R. W.; Boyd S.; Liu B.; Hamrick J.; Daigle D.; Chatwin C. L.; John K.; McLaughlin L.; Cusick S. M.; Weiss W. J.; Pulse M. E.; Pevear D. C.; Moeck G.; Xerri L.; Burns C. J. Discovery of VNRX-7145 (VNRX-5236 Etzadroxil): An Orally Bioavailable beta-Lactamase Inhibitor for Enterobacterales Expressing Ambler Class A, C, and D Enzymes. J. Med. Chem. 2021, 64 (14), 10155–10166. 10.1021/acs.jmedchem.1c00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye K. S.; Belley A.; Barth P.; Lahlou O.; Knechtle P.; Motta P.; Velicitat P. Effect of Cefepime/Enmetazobactam vs Piperacillin/Tazobactam on Clinical Cure and Microbiological Eradication in Patients With Complicated Urinary Tract Infection or Acute Pyelonephritis: A Randomized Clinical Trial. JAMA 2022, 328 (13), 1304–1314. 10.1001/jama.2022.17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K. M.; Nguyen N. Q.; Jacobs M. R.; Bethel C. R.; Barnes M. D.; Kumar V.; Bajaksouzian S.; Rudin S. D.; Rather P. N.; Bhavsar S.; Ravikumar T.; Deshpande P. K.; Patil V.; Yeole R.; Bhagwat S. S.; Patel M. V.; van den Akker F.; Bonomo R. A. Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234. J. Med. Chem. 2018, 61 (9), 4067–4086. 10.1021/acs.jmedchem.8b00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. P.; Pinto N. A.; Vu T. N.; Lee H.; Cho Y. L.; Byun J. H.; D’Souza R.; Yong D. In Vitro Activity of a Novel Siderophore-Cephalosporin, GT-1 and Serine-Type beta-Lactamase Inhibitor, GT-055, against Escherichia coli, Klebsiella pneumoniae and Acinetobacter spp. Panel Strains. Antibiotics (Basel) 2020, 9 (5), 267. 10.3390/antibiotics9050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. P.; Pinto N. A.; Vu T. N.; Lee H.; Cho Y. L.; Byun J. H.; D’Souza R.; Yong D., In Vitro Activity of a Novel Siderophore-Cephalosporin, GT-1 and Serine-Type beta-Lactamase Inhibitor, GT-055, against Escherichia coli, Klebsiella pneumoniae and Acinetobacter spp. Panel Strains. Antibiotics (Basel) 2020, 9 ( (5), ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Trout R. E. L.; Chu G. H.; McGarry D.; Jackson R. W.; Hamrick J. C.; Daigle D. M.; Cusick S. M.; Pozzi C.; De Luca F.; Benvenuti M.; Mangani S.; Docquier J. D.; Weiss W. J.; Pevear D. C.; Xerri L.; Burns C. J. Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections. J. Med. Chem. 2020, 63 (6), 2789–2801. 10.1021/acs.jmedchem.9b01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O.; Tsivkovski R.; Sun D.; Reddy R.; Totrov M.; Hecker S.; Griffith D.; Loutit J.; Dudley M. QPX7728, An Ultra-Broad-Spectrum B-Lactamase Inhibitor for Intravenous and Oral Therapy: Overview of Biochemical and Microbiological Characteristics. Front Microbiol 2021, 12, 697180. 10.3389/fmicb.2021.697180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet M.; Tarazi Z.; Griffith D. C. In Vivo Activity of QPX7728, an Ultrabroad-Spectrum Beta-Lactamase Inhibitor, in Combination with Beta-Lactams against Carbapenem-Resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2020, 64 (11), e01267. 10.1128/AAC.01267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.; Chen J.; Kang X.; Shen X.; Lao X.; Zheng H. Approaches for the discovery of metallo-beta-lactamase inhibitors: A review. Chem. Biol. Drug Des 2019, 94 (2), 1427–1440. 10.1111/cbdd.13526. [DOI] [PubMed] [Google Scholar]
- Yarlagadda V.; Sarkar P.; Samaddar S.; Manjunath G. B.; Mitra S. D.; Paramanandham K.; Shome B. R.; Haldar J. Vancomycin Analogue Restores Meropenem Activity against NDM-1 Gram-Negative Pathogens. ACS Infect Dis 2018, 4 (7), 1093–1101. 10.1021/acsinfecdis.8b00011. [DOI] [PubMed] [Google Scholar]
- King A. M.; Reid-Yu S. A.; Wang W.; King D. T.; De Pascale G.; Strynadka N. C.; Walsh T. R.; Coombes B. K.; Wright G. D. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510 (7506), 503. 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Wang J.; Niu G.; Shui W.; Sun Y.; Zhou H.; Zhang Y.; Yang C.; Lou Z.; Rao Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2011, 2 (5), 384–94. 10.1007/s13238-011-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. T.; Worrall L. J.; Gruninger R.; Strynadka N. C. New Delhi metallo-beta-lactamase: structural insights into beta-lactam recognition and inhibition. J. Am. Chem. Soc. 2012, 134 (28), 11362–5. 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- Klingler F. M.; Wichelhaus T. A.; Frank D.; Cuesta-Bernal J.; El-Delik J.; Muller H. F.; Sjuts H.; Gottig S.; Koenigs A.; Pos K. M.; Pogoryelov D.; Proschak E. Approved Drugs Containing Thiols as Inhibitors of Metallo-beta-lactamases: Strategy To Combat Multidrug-Resistant Bacteria. J. Med. Chem. 2015, 58 (8), 3626–30. 10.1021/jm501844d. [DOI] [PubMed] [Google Scholar]
- Buttner D.; Kramer J. S.; Klingler F. M.; Wittmann S. K.; Hartmann M. R.; Kurz C. G.; Kohnhauser D.; Weizel L.; Bruggerhoff A.; Frank D.; Steinhilber D.; Wichelhaus T. A.; Pogoryelov D.; Proschak E. Challenges in the Development of a Thiol-Based Broad-Spectrum Inhibitor for Metallo-beta-Lactamases. ACS Infect Dis 2018, 4 (3), 360–372. 10.1021/acsinfecdis.7b00129. [DOI] [PubMed] [Google Scholar]
- Ma J.; Cao Q.; McLeod S. M.; Ferguson K.; Gao N.; Breeze A. L.; Hu J. Target-based whole-cell screening by (1)H NMR spectroscopy. Angew. Chem., Int. Ed. Engl. 2015, 54 (16), 4764–7. 10.1002/anie.201410701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. B.; Abboud M. I.; Brem J.; Someya H.; Lohans C. T.; Yang S. Y.; Spencer J.; Wareham D. W.; McDonough M. A.; Schofield C. J. NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors. Chem. Sci. 2017, 8 (2), 928–937. 10.1039/C6SC04524C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe P.; Gonzalez M. M.; Mojica M. F.; Gonzalez J. M.; Castillo V.; Saiz C.; Kosmopoulou M.; Tooke C. L.; Llarrull L. I.; Mahler G.; Bonomo R. A.; Vila A. J.; Spencer J. Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (26), E3745–54. 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R.; Brem J.; Zollman D.; McDonough M. A.; Johnson R. M.; Spencer J.; Makena A.; Abboud M. I.; Cahill S.; Lee S. Y.; McHugh P. J.; Schofield C. J.; Fishwick C. W. G. In Silico Fragment-Based Design Identifies Subfamily B1Metallo-beta-lactamase Inhibitors. J. Med. Chem. 2018, 61 (3), 1255–1260. 10.1021/acs.jmedchem.7b01728. [DOI] [PubMed] [Google Scholar]
- Chiou J.; Wan S.; Chan K. F.; So P. K.; He D.; Chan E. W.; Chan T. H.; Wong K. Y.; Tao J.; Chen S. Ebselen as a potent covalent inhibitor of New Delhi metallo-beta-lactamase (NDM-1). Chem. Commun. (Camb) 2015, 51 (46), 9543–6. 10.1039/C5CC02594J. [DOI] [PubMed] [Google Scholar]
- Jin W. B.; Xu C.; Cheng Q.; Qi X. L.; Gao W.; Zheng Z.; Chan E. W. C.; Leung Y. C.; Chan T. H.; Wong K. Y.; Chen S.; Chan K. F. Investigation of synergistic antimicrobial effects of the drug combinations of Meropenem and 1,2-benzisoselenazol-3(2H)-one derivatives on carbapenem-resistant Enterobacteriaceae producing NDM-1. Eur. J. Med. Chem. 2018, 155, 285–302. 10.1016/j.ejmech.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Brem J.; Cain R.; Cahill S.; McDonough M. A.; Clifton I. J.; Jimenez-Castellanos J. C.; Avison M. B.; Spencer J.; Fishwick C. W.; Schofield C. J. Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat. Commun. 2016, 7, 12406. 10.1038/ncomms12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B.; Cooper M. A. Aminoglycoside antibiotics in the 21st century. ACS Chem. Biol. 2013, 8 (1), 105–15. 10.1021/cb3005116. [DOI] [PubMed] [Google Scholar]
- Daigle D. M.; McKay G. A.; Wright G. D. Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J. Biol. Chem. 1997, 272 (40), 24755–8. 10.1074/jbc.272.40.24755. [DOI] [PubMed] [Google Scholar]
- Shakya T.; Stogios P. J.; Waglechner N.; Evdokimova E.; Ejim L.; Blanchard J. E.; McArthur A. G.; Savchenko A.; Wright G. D. A small molecule discrimination map of the antibiotic resistance kinome. Chem. Biol. 2011, 18 (12), 1591–601. 10.1016/j.chembiol.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Stogios P. J.; Spanogiannopoulos P.; Evdokimova E.; Egorova O.; Shakya T.; Todorovic N.; Capretta A.; Wright G. D.; Savchenko A. Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance. Biochem. J. 2013, 454 (2), 191–200. 10.1042/BJ20130317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J.; Schmieder B. J.; Petitpas J. W.; Manousos M.; Williams J. A.; Faiella J. A.; Girard A. E.; McGuirk P. R. Assays to detect and characterize synthetic agents that inhibit the ErmC methyltransferase. J. Antibiot (Tokyo) 1995, 48 (11), 1273–9. 10.7164/antibiotics.48.1273. [DOI] [PubMed] [Google Scholar]
- Feder M.; Purta E.; Koscinski L.; Cubrilo S.; Maravic Vlahovicek G.; Bujnicki J. M. Virtual screening and experimental verification to identify potential inhibitors of the ErmC methyltransferase responsible for bacterial resistance against macrolide antibiotics. ChemMedChem. 2008, 3 (2), 316–22. 10.1002/cmdc.200700201. [DOI] [PubMed] [Google Scholar]
- Hajduk P. J.; Dinges J.; Schkeryantz J. M.; Janowick D.; Kaminski M.; Tufano M.; Augeri D. J.; Petros A.; Nienaber V.; Zhong P.; Hammond R.; Coen M.; Beutel B.; Katz L.; Fesik S. W. Novel inhibitors of Erm methyltransferases from NMR and parallel synthesis. J. Med. Chem. 1999, 42 (19), 3852–9. 10.1021/jm990293a. [DOI] [PubMed] [Google Scholar]
- Allen N. E.; Alborn W. E. Jr.; Hobbs J. N. Jr.; Kirst H. A. 7-Hydroxytropolone: an inhibitor of aminoglycoside-2″-O-adenylyltransferase. Antimicrob. Agents Chemother. 1982, 22 (5), 824–31. 10.1128/AAC.22.5.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch D. R.; Cox G.; D’Erasmo M. P.; Shakya T.; Meck C.; Mohd N.; Wright G. D.; Murelli R. P. Inhibition of the ANT(2″)-Ia resistance enzyme and rescue of aminoglycoside antibiotic activity by synthetic alpha-hydroxytropolones. Bioorg. Med. Chem. Lett. 2014, 24 (21), 4943–7. 10.1016/j.bmcl.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr D. D.; Draker K. A.; Koteva K.; Bains M.; Hancock R. E.; Wright G. D. Broad-spectrum peptide inhibitors of aminoglycoside antibiotic resistance enzymes. Chem. Biol. 2003, 10 (2), 189–96. 10.1016/S1074-5521(03)00026-7. [DOI] [PubMed] [Google Scholar]
- Cox G.; Sieron A.; King A. M.; De Pascale G.; Pawlowski A. C.; Koteva K.; Wright G. D. A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chem. Biol. 2017, 24 (1), 98–109. 10.1016/j.chembiol.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Boehr D. D.; Lane W. S.; Wright G. D. Active site labeling of the gentamicin resistance enzyme AAC(6′)-APH(2″) by the lipid kinase inhibitor wortmannin. Chem. Biol. 2001, 8 (8), 791–800. 10.1016/S1074-5521(01)00051-5. [DOI] [PubMed] [Google Scholar]
- O’Daniel P. I.; Peng Z.; Pi H.; Testero S. A.; Ding D.; Spink E.; Leemans E.; Boudreau M. A.; Yamaguchi T.; Schroeder V. A.; Wolter W. R.; Llarrull L. I.; Song W.; Lastochkin E.; Kumarasiri M.; Antunes N. T.; Espahbodi M.; Lichtenwalter K.; Suckow M. A.; Vakulenko S.; Mobashery S.; Chang M. Discovery of a new class of non-beta-lactam inhibitors of penicillin-binding proteins with Gram-positive antibacterial activity. J. Am. Chem. Soc. 2014, 136 (9), 3664–72. 10.1021/ja500053x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink E.; Ding D.; Peng Z.; Boudreau M. A.; Leemans E.; Lastochkin E.; Song W.; Lichtenwalter K.; O’Daniel P. I.; Testero S. A.; Pi H.; Schroeder V. A.; Wolter W. R.; Antunes N. T.; Suckow M. A.; Vakulenko S.; Chang M.; Mobashery S. Structure-activity relationship for the oxadiazole class of antibiotics. J. Med. Chem. 2015, 58 (3), 1380–9. 10.1021/jm501661f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janardhanan J.; Meisel J. E.; Ding D.; Schroeder V. A.; Wolter W. R.; Mobashery S.; Chang M. In Vitro and In Vivo Synergy of the Oxadiazole Class of Antibacterials with beta-Lactams. Antimicrob. Agents Chemother. 2016, 60 (9), 5581–8. 10.1128/AAC.00787-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janardhanan J.; Bouley R.; Martinez-Caballero S.; Peng Z.; Batuecas-Mordillo M.; Meisel J. E.; Ding D.; Schroeder V. A.; Wolter W. R.; Mahasenan K. V.; Hermoso J. A.; Mobashery S.; Chang M. The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63 (5), e02637. 10.1128/AAC.02637-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janardhanan J.; Bouley R.; Martinez-Caballero S.; Peng Z.; Batuecas-Mordillo M.; Meisel J. E.; Ding D.; Schroeder V. A.; Wolter W. R.; Mahasenan K. V.; Hermoso J. A.; Mobashery S.; Chang M. The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63 (5), e02637. 10.1128/AAC.02637-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D.; Wang-Kan X.; Neuberger A.; van Veen H. W.; Pos K. M.; Piddock L. J. V.; Luisi B. F. Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol 2018, 16 (9), 523–539. 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- Du D.; van Veen H. W.; Murakami S.; Pos K. M.; Luisi B. F. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr. Opin Struct Biol. 2015, 33, 76–91. 10.1016/j.sbi.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Kracke F.; Vassilev I.; Kromer J. O. Microbial electron transport and energy conservation - the foundation for optimizing bioelectrochemical systems. Front Microbiol 2015, 6, 575. 10.3389/fmicb.2015.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay D. C.; Rommens K. L.; Turner R. J. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 2008, 1778 (9), 1814–38. 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Dawson R. J.; Locher K. P. Structure of a bacterial multidrug ABC transporter. Nature 2006, 443 (7108), 180–5. 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Blair J. M.; Piddock L. J. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin Microbiol 2009, 12 (5), 512–9. 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Van Bambeke F.; Balzi E.; Tulkens P. M. Antibiotic efflux pumps. Biochem. Pharmacol. 2000, 60 (4), 457–70. 10.1016/S0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- Kashket E. R. The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Microbiol. 1985, 39, 219–42. 10.1146/annurev.mi.39.100185.001251. [DOI] [PubMed] [Google Scholar]
- Farha M. A.; Leung A.; Sewell E. W.; D’Elia M. A.; Allison S. E.; Ejim L.; Pereira P. M.; Pinho M. G.; Wright G. D.; Brown E. D. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to beta-lactams. ACS Chem. Biol. 2013, 8 (1), 226–33. 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenazy R. Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology (Basel) 2022, 11 (9), 1328. 10.3390/biology11091328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechini B.; Versace I. Inhibitors of multidrug resistant efflux systems in bacteria. Recent Pat Antiinfect Drug Discov 2009, 4 (1), 37–50. 10.2174/157489109787236256. [DOI] [PubMed] [Google Scholar]
- Li X. Z.; Plesiat P.; Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015, 28 (2), 337–418. 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyfakh A. A.; Borsch C. M.; Kaatz G. W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993, 37 (1), 128–9. 10.1128/AAC.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stermitz F. R.; Lorenz P.; Tawara J. N.; Zenewicz L. A.; Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (4), 1433–7. 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalle A. M.; Rizvi A. Inhibition of bacterial multidrug resistance by celecoxib, a cyclooxygenase-2 inhibitor. Antimicrob. Agents Chemother. 2011, 55 (1), 439–42. 10.1128/AAC.00735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S.; Gosetto F.; Serritella S.; Manfroni G.; Tabarrini O.; Iraci N.; Brincat J. P.; Carosati E.; Villarini M.; Kaatz G. W.; Cecchetti V. Pyrazolo[4,3-c][1,2]benzothiazines 5,5-dioxide: a promising new class of Staphylococcus aureus NorA efflux pump inhibitors. J. Med. Chem. 2012, 55 (7), 3568–72. 10.1021/jm201446h. [DOI] [PubMed] [Google Scholar]
- Lomovskaya O.; Warren M. S.; Lee A.; Galazzo J.; Fronko R.; Lee M.; Blais J.; Cho D.; Chamberland S.; Renau T.; Leger R.; Hecker S.; Watkins W.; Hoshino K.; Ishida H.; Lee V. J. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45 (1), 105–16. 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern W. V.; Steinke P.; Schumacher A.; Schuster S.; Baum H. v.; Bohnert J. A. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 2006, 57 (2), 339–43. 10.1093/jac/dki445. [DOI] [PubMed] [Google Scholar]
- Sjuts H.; Vargiu A. V.; Kwasny S. M.; Nguyen S. T.; Kim H. S.; Ding X.; Ornik A. R.; Ruggerone P.; Bowlin T. L.; Nikaido H.; Pos K. M.; Opperman T. J. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (13), 3509–14. 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T. J.; Kwasny S. M.; Kim H. S.; Nguyen S. T.; Houseweart C.; D’Souza S.; Walker G. C.; Peet N. P.; Nikaido H.; Bowlin T. L. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58 (2), 722–33. 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdali N.; Parks J. M.; Haynes K. M.; Chaney J. L.; Green A. T.; Wolloscheck D.; Walker J. K.; Rybenkov V. V.; Baudry J.; Smith J. C.; Zgurskaya H. I. Reviving Antibiotics: Efflux Pump Inhibitors That Interact with AcrA, a Membrane Fusion Protein of the AcrAB-TolC Multidrug Efflux Pump. ACS Infect Dis 2017, 3 (1), 89–98. 10.1021/acsinfecdis.6b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallea M.; Mahamoud A.; Chevalier J.; Alibert-Franco S.; Brouant P.; Barbe J.; Pages J. M. Alkylaminoquinolines inhibit the bacterial antibiotic efflux pump in multidrug-resistant clinical isolates. Biochem. J. 2003, 376 (3), 801–805. 10.1042/bj20030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Mowla R.; Guo L.; Ogunniyi A. D.; Rahman T.; De Barros Lopes M. A.; Ma S.; Venter H. Evaluation of a series of 2-napthamide derivatives as inhibitors of the drug efflux pump AcrB for the reversal of antimicrobial resistance. Bioorg. Med. Chem. Lett. 2017, 27 (4), 733–739. 10.1016/j.bmcl.2017.01.042. [DOI] [PubMed] [Google Scholar]
- Tegos G. P.; Haynes M.; Strouse J. J.; Khan M. M.; Bologa C. G.; Oprea T. I.; Sklar L. A. Microbial efflux pump inhibition: tactics and strategies. Curr. Pharm. Des 2011, 17 (13), 1291–1302. 10.2174/138161211795703726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell E. W.; Brown E. D. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J. Antibiot (Tokyo) 2014, 67 (1), 43–51. 10.1038/ja.2013.100. [DOI] [PubMed] [Google Scholar]
- Bhavsar A. P.; Erdman L. K.; Schertzer J. W.; Brown E. D. Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J. Bacteriol. 2004, 186 (23), 7865–73. 10.1128/JB.186.23.7865-7873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. L.; Li F. K.; Koo B. M.; El-Halfawy O. M.; French S.; Gross C. A.; Strynadka N. C.; Brown E. D. Identification of Two Phosphate Starvation-induced Wall Teichoic Acid Hydrolases Provides First Insights into the Degradative Pathway of a Key Bacterial Cell Wall Component. J. Biol. Chem. 2016, 291 (50), 26066–26082. 10.1074/jbc.M116.760447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.; Xia G.; Luhachack L. G.; Campbell J.; Meredith T. C.; Chen C.; Winstel V.; Gekeler C.; Irazoqui J. E.; Peschel A.; Walker S. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (46), 18909–14. 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. Can beta-Lactam Antibiotics Be Resurrected to Combat MRSA?. Trends Microbiol 2019, 27 (1), 26–38. 10.1016/j.tim.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Peacock S. J.; Paterson G. K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- Vestergaard M.; Frees D.; Ingmer H. Antibiotic Resistance and the MRSA Problem. Microbiol Spectr 2019, 10.1128/microbiolspec.GPP3-0057-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Gill C. J.; Lee S. H.; Mann P.; Zuck P.; Meredith T. C.; Murgolo N.; She X.; Kales S.; Liang L.; Liu J.; Wu J.; Santa Maria J.; Su J.; Pan J.; Hailey J.; McGuinness D.; Tan C. M.; Flattery A.; Walker S.; Black T.; Roemer T. Discovery of wall teichoic acid inhibitors as potential anti-MRSA beta-lactam combination agents. Chem. Biol. 2013, 20 (2), 272–84. 10.1016/j.chembiol.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; Wang H.; Labroli M.; Koseoglu S.; Zuck P.; Mayhood T.; Gill C.; Mann P.; Sher X.; Ha S.; Yang S. W.; Mandal M.; Yang C.; Liang L.; Tan Z.; Tawa P.; Hou Y.; Kuvelkar R.; DeVito K.; Wen X.; Xiao J.; Batchlett M.; Balibar C. J.; Liu J.; Xiao J.; Murgolo N.; Garlisi C. G.; Sheth P. R.; Flattery A.; Su J.; Tan C.; Roemer T. TarO-specific inhibitors of wall teichoic acid biosynthesis restore beta-lactam efficacy against methicillin-resistant staphylococci. Sci. Transl Med. 2016, 8 (329), 329ra32. 10.1126/scitranslmed.aad7364. [DOI] [PubMed] [Google Scholar]
- Campbell J.; Singh A. K.; Santa Maria J. P. Jr.; Kim Y.; Brown S.; Swoboda J. G.; Mylonakis E.; Wilkinson B. J.; Walker S. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 2011, 6 (1), 106–16. 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda J. G.; Meredith T. C.; Campbell J.; Brown S.; Suzuki T.; Bollenbach T.; Malhowski A. J.; Kishony R.; Gilmore M. S.; Walker S. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem. Biol. 2009, 4 (10), 875–83. 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farha M. A.; Czarny T. L.; Myers C. L.; Worrall L. J.; French S.; Conrady D. G.; Wang Y.; Oldfield E.; Strynadka N. C.; Brown E. D. Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (35), 11048–53. 10.1073/pnas.1511751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxley M. A.; Wright S. N.; Lam A. K.; Friedline A. W.; Strange S. J.; Xiao M. T.; Moen E. L.; Rice C. V. Targeting Wall Teichoic Acid in Situ with Branched Polyethylenimine Potentiates beta-Lactam Efficacy against MRSA. ACS Med. Chem. Lett. 2017, 8 (10), 1083–1088. 10.1021/acsmedchemlett.7b00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirner K.; Marles-Wright J.; Lewis R. J.; Errington J. Distinct and Essential Morphogenic Functions for Wall- and Lipo- Teichoic Acids in Bacillus Subtilis. EMBO J. 2009, 28, 830–842. 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscato J. D.; Morris H. G.; Mychack A.; Rajagopal M.; Baidin V.; Hesser A. R.; Lee W.; Inecik K.; Wilson L. J.; Kraml C. M.; Meredith T. C.; Walker S. Rapid Inhibitor Discovery by Exploiting Synthetic Lethality. J. Am. Chem. Soc. 2022, 144 (8), 3696–3705. 10.1021/jacs.1c12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee Wezen X.; Chandran A.; Eapen R. S.; Waters E.; Bricio-Moreno L.; Tosi T.; Dolan S.; Millership C.; Kadioglu A.; Gründling A.; Itzhaki L. S.; Welch M.; Rahman T. Structure-Based Discovery of Lipoteichoic Acid Synthase Inhibitors. J. Chem. Inf Model 2022, 62, 2586–2599. 10.1021/acs.jcim.2c00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. L.; Rossi L.; De Pascale G.; Wright G. D. A Forward Chemical Screen Identifies Antibiotic Adjuvants in Escherichia coli. ACS Chem. Biol. 2012, 7 (9), 1547–1555. 10.1021/cb300269g. [DOI] [PubMed] [Google Scholar]
- Mann P. A.; Muller A.; Xiao L.; Pereira P. M.; Yang C.; Lee S. H.; Wang H.; Trzeciak J.; Schneeweis J.; dos Santos M. M.; Murgolo N.; She X. W.; Gill C.; Balibar C. J.; Labroli M.; Su J.; Flattery A.; Sherborne B.; Maier R.; Tan C. M.; Black T.; Onder K.; Kargman S.; Monsma F. J.; Pinho M. G.; Schneider T.; Roemer T. Murgocil is a Highly Bioactive Staphylococcal-Specific Inhibitor of the Peptidoglycan Glycosyltransferase Enzyme MurG. ACS Chem. Biol. 2013, 8 (11), 2442–2451. 10.1021/cb400487f. [DOI] [PubMed] [Google Scholar]
- Chu J.; Vila-Farres X.; Inoyama D.; Gallardo-Macias R.; Jaskowski M.; Satish S.; Freundlich J. S.; Brady S. F. Human Microbiome Inspired Antibiotics with Improved beta-Lactam Synergy against MDR Staphylococcus aureus. ACS Infect Dis 2018, 4 (1), 33–38. 10.1021/acsinfecdis.7b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J.; Donald R. G. K.; Lee S. H.; Jarantow L. W.; Salvatore M. J.; Meng X.; Painter R.; Onishi R. H.; Occi J.; Dorso K.; Young K.; Park Y. W.; Skwish S.; Szymonifka M. J.; Waddell T. S.; Miesel L.; Phillips J. W.; Roemer T. Chemical Genetic Identification of Peptidoglycan Inhibitors Potentiating Carbapenem Activity against Methicillin-Resistant Staphylococcus aureus. Chem. Biol. 2009, 16 (8), 837–848. 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Tomoshige S.; Dik D. A.; Akabane-Nakata M.; Madukoma C. S.; Fisher J. F.; Shrout J. D.; Mobashery S. Total Syntheses of Bulgecins A, B, and C and Their Bactericidal Potentiation of the β-Lactam Antibiotics. ACS Infect Dis 2018, 4, 860–867. 10.1021/acsinfecdis.8b00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik D. A.; Madukoma C. S.; Tomoshige S.; Kim C.; Lastochkin E.; Boggess W. C.; Fisher J. F.; Shrout J. D.; Mobashery S. Slt, MltD, and MltG of Pseudomonas aeruginosa as Targets of Bulgecin A in Potentiation of β-Lactam Antibiotics. ACS Chem. Biol. 2019, 14, 296–303. 10.1021/acschembio.8b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau M. A.; Fishovitz J.; Llarrull L. I.; Xiao Q.; Mobashery S. Phosphorylation of BlaR1 in Manifestation of Antibiotic Resistance in Methicillin-Resistant Staphylococcus aureus and Its Abrogation by Small Molecules. ACS Infect. Dis. 2015, 1, 454–459. 10.1021/acsinfecdis.5b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speri E.; Fishovitz J.; Mobashery S. Structure-activity relationship of the cinnamamide family of antibiotic potentiators for methicillin-resistant Staphylococcus aureus (MRSA). Medchemcomm. 2018, 9, 2008–2016. 10.1039/C8MD00479J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S.; Dhanda G.; Naik P.; Mukherjee R.; Jolly L.; Joseph J.; Haldar J. Multi-Functional Small Molecules with Temporal Charge-Switchability Tackle Infection and Inflammation. Advanced Therapeutics 2022, 5 (4), 2100234. 10.1002/adtp.202100234. [DOI] [Google Scholar]
- Acharya Y.; Dhanda G.; Sarkar P.; Haldar J. Pursuit of next-generation glycopeptides: a journey with vancomycin. Chem. Commun. (Camb) 2022, 58 (12), 1881–1897. 10.1039/D1CC06635H. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Yadav V.; Younis W.; Mohammad H.; Hegazy Y. A.; Seleem M. N.; Sanyal K.; Haldar J. Aryl-alkyl-lysines: Membrane-Active Fungicides That Act against Biofilms of Candida albicans. ACS Infect Dis 2017, 3 (4), 293–301. 10.1021/acsinfecdis.6b00192. [DOI] [PubMed] [Google Scholar]
- Uppu D.; Konai M. M.; Baul U.; Singh P.; Siersma T. K.; Samaddar S.; Vemparala S.; Hamoen L. W.; Narayana C.; Haldar J. Isosteric substitution in cationic-amphiphilic polymers reveals an important role for hydrogen bonding in bacterial membrane interactions. Chem. Sci. 2016, 7 (7), 4613–4623. 10.1039/C6SC00615A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konai M. M.; Samaddar S.; Bocchinfuso G.; Santucci V.; Stella L.; Haldar J. Selectively targeting bacteria by tuning the molecular design of membrane-active peptidomimetic amphiphiles. Chem. Commun. (Camb) 2018, 54 (39), 4943–4946. 10.1039/C8CC01926F. [DOI] [PubMed] [Google Scholar]
- Ghosh C.; Haldar J. Membrane-Active Small Molecules: Designs Inspired by Antimicrobial Peptides. ChemMedChem. 2015, 10 (10), 1606–24. 10.1002/cmdc.201500299. [DOI] [PubMed] [Google Scholar]
- Yarlagadda V.; Manjunath G. B.; Sarkar P.; Akkapeddi P.; Paramanandham K.; Shome B. R.; Ravikumar R.; Haldar J. Glycopeptide Antibiotic To Overcome the Intrinsic Resistance of Gram-Negative Bacteria. ACS Infect Dis 2016, 2 (2), 132–9. 10.1021/acsinfecdis.5b00114. [DOI] [PubMed] [Google Scholar]
- Yarlagadda V.; Samaddar S.; Paramanandham K.; Shome B. R.; Haldar J. Membrane Disruption and Enhanced Inhibition of Cell-Wall Biosynthesis: A Synergistic Approach to Tackle Vancomycin-Resistant Bacteria. Angew. Chem., Int. Ed. Engl. 2015, 54 (46), 13644–9. 10.1002/anie.201507567. [DOI] [PubMed] [Google Scholar]
- Sarkar P.; Samaddar S.; Ammanathan V.; Yarlagadda V.; Ghosh C.; Shukla M.; Kaul G.; Manjithaya R.; Chopra S.; Haldar J. Vancomycin Derivative Inactivates Carbapenem-Resistant Acinetobacter baumannii and Induces Autophagy. ACS Chem. Biol. 2020, 15 (4), 884–889. 10.1021/acschembio.0c00091. [DOI] [PubMed] [Google Scholar]
- Sarkar P.; De K.; Modi M.; Dhanda G.; Priyadarshini R.; Bandow J. E.; Haldar J. Next-generation membrane-active glycopeptide antibiotics that also inhibit bacterial cell division. Chem. Sci. 2023, 10.1039/D2SC05600C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda G.; Sarkar P.; Samaddar S.; Haldar J. Battle against Vancomycin-Resistant Bacteria: Recent Developments in Chemical Strategies. J. Med. Chem. 2019, 62 (7), 3184–3205. 10.1021/acs.jmedchem.8b01093. [DOI] [PubMed] [Google Scholar]
- Vaara M. Polymyxin Derivatives that Sensitize Gram-Negative Bacteria to Other Antibiotics. Molecules 2019, 24 (2), 249. 10.3390/molecules24020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992, 56 (3), 395–411. 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabawa T. P.; Pucci M. J.; Parr T. R. Jr.; Lister T. Treatment of Gram-negative bacterial infections by potentiation of antibiotics. Curr. Opin Microbiol 2016, 33, 7–12. 10.1016/j.mib.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Danner R. L.; Joiner K. A.; Rubin M.; Patterson W. H.; Johnson N.; Ayers K. M.; Parrillo J. E. Purification, toxicity, and antiendotoxin activity of polymyxin B nonapeptide. Antimicrob. Agents Chemother. 1989, 33 (9), 1428–34. 10.1128/AAC.33.9.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. The outer membrane permeability-increasing action of linear analogues of polymyxin B nonapeptide. Drugs Exp Clin Res. 1991, 17 (9), 437–443. [PubMed] [Google Scholar]
- French S.; Farha M.; Ellis M. J.; Sameer Z.; Cote J. P.; Cotroneo N.; Lister T.; Rubio A.; Brown E. D. Potentiation of Antibiotics against Gram-Negative Bacteria by Polymyxin B Analogue SPR741 from Unique Perturbation of the Outer Membrane. ACS Infect Dis 2020, 6 (6), 1405–1412. 10.1021/acsinfecdis.9b00159. [DOI] [PubMed] [Google Scholar]
- Corbett D.; Wise A.; Langley T.; Skinner K.; Trimby E.; Birchall S.; Dorali A.; Sandiford S.; Williams J.; Warn P.; Vaara M.; Lister T. Potentiation of Antibiotic Activity by a Novel Cationic Peptide: Potency and Spectrum of Activity of SPR741. Antimicrob. Agents Chemother. 2017, 10.1128/AAC.00200-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski D. V.; Reinhart A. A.; Alamneh Y. A.; Pucci M. J.; Si Y.; Abu-Taleb R.; Shearer J. P.; Demons S. T.; Tyner S. D.; Lister T. SPR741, an Antibiotic Adjuvant, Potentiates the In Vitro and In Vivo Activity of Rifampin against Clinically Relevant Extensively Drug-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017, 61 (12), e01239. 10.1128/AAC.01239-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling C. M. J.; Martin N. I. Synergy by Perturbing the Gram-Negative Outer Membrane: Opening the Door for Gram-Positive Specific Antibiotics. ACS Infect Dis 2022, 8 (9), 1731–1757. 10.1021/acsinfecdis.2c00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douafer H.; Andrieu V.; Phanstiel O. t.; Brunel J. M. Antibiotic Adjuvants: Make Antibiotics Great Again!. J. Med. Chem. 2019, 62 (19), 8665–8681. 10.1021/acs.jmedchem.8b01781. [DOI] [PubMed] [Google Scholar]
- Song M.; Liu Y.; Huang X.; Ding S.; Wang Y.; Shen J.; Zhu K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol 2020, 5 (8), 1040–1050. 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
- Moon S. H.; Zhang X.; Zheng G.; Meeker D. G.; Smeltzer M. S.; Huang E. Novel Linear Lipopeptide Paenipeptins with Potential for Eradicating Biofilms and Sensitizing Gram-Negative Bacteria to Rifampicin and Clarithromycin. J. Med. Chem. 2017, 60 (23), 9630–9640. 10.1021/acs.jmedchem.7b01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S. H.; Kaufmann Y.; Huang E. Paenipeptin Analogues Potentiate Clarithromycin and Rifampin against mcr-1-Mediated Polymyxin-Resistant Escherichia coli In Vivo. Antimicrob. Agents Chemother. 2020, 64 (4), e02045. 10.1128/AAC.02045-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R.; Brizuela M.; Eisner B.; Findlay B.; Zhanel G. G.; Schweizer F. Dilipid ultrashort cationic lipopeptides as adjuvants for chloramphenicol and other conventional antibiotics against Gram-negative bacteria. Amino Acids 2019, 51 (3), 383–393. 10.1007/s00726-018-2673-9. [DOI] [PubMed] [Google Scholar]
- Schweizer L.; Ramirez D.; Schweizer F. Effects of Lysine N-zeta-Methylation in Ultrashort Tetrabasic Lipopeptides (UTBLPs) on the Potentiation of Rifampicin, Novobiocin, and Niclosamide in Gram-Negative Bacteria. Antibiotics (Basel) 2022, 11 (3), 335. 10.3390/antibiotics11030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez D.; Berry L.; Domalaon R.; Brizuela M.; Schweizer F. Dilipid Ultrashort Tetrabasic Peptidomimetics Potentiate Novobiocin and Rifampicin Against Multidrug-Resistant Gram-Negative Bacteria. ACS Infect Dis 2020, 6 (6), 1413–1426. 10.1021/acsinfecdis.0c00017. [DOI] [PubMed] [Google Scholar]
- Radzishevsky I. S.; Rotem S.; Bourdetsky D.; Navon-Venezia S.; Carmeli Y.; Mor A. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat. Biotechnol. 2007, 25 (6), 657–9. 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- Radzishevsky I.; Krugliak M.; Ginsburg H.; Mor A. Antiplasmodial activity of lauryl-lysine oligomers. Antimicrob. Agents Chemother. 2007, 51 (5), 1753–9. 10.1128/AAC.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaknoon F.; Meir O.; Mor A. Mechanistic Studies of Antibiotic Adjuvants Reducing Kidney’s Bacterial Loads upon Systemic Monotherapy. Pharmaceutics 2021, 13 (11), 1947. 10.3390/pharmaceutics13111947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J. M.; MacNair C. R.; Ilyas B.; French S.; Cote J. P.; Bouwman C.; Farha M. A.; Sieron A. O.; Whitfield C.; Coombes B. K.; Brown E. D. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol 2017, 2, 17028. 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling C. M. J.; Slingerland C. J.; Veraar S.; Lok S.; Martin N. I. Structure-Activity Studies with Bis-Amidines That Potentiate Gram-Positive Specific Antibiotics against Gram-Negative Pathogens. ACS Infect Dis 2021, 7 (12), 3314–3335. 10.1021/acsinfecdis.1c00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNair C. R.; Farha M. A.; Serrano-Wu M. H.; Lee K. K.; Hubbard B.; Cote J. P.; Carfrae L. A.; Tu M. M.; Gaulin J. L.; Hunt D. K.; Hung D. T.; Brown E. D. Preclinical Development of Pentamidine Analogs Identifies a Potent and Nontoxic Antibiotic Adjuvant. ACS Infect Dis 2022, 8 (4), 768–777. 10.1021/acsinfecdis.1c00482. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Jia Y.; Yang K.; Li R.; Xiao X.; Zhu K.; Wang Z. Metformin Restores Tetracyclines Susceptibility against Multidrug Resistant Bacteria. Adv. Sci. (Weinh) 2020, 7 (12), 1902227. 10.1002/advs.201902227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobucar K.; Cote J. P.; French S.; Borrillo L.; Guo A. B. Y.; Serrano-Wu M. H.; Lee K. K.; Hubbard B.; Johnson J. W.; Gaulin J. L.; Magolan J.; Hung D. T.; Brown E. D. Chemical Screen for Vancomycin Antagonism Uncovers Probes of the Gram-Negative Outer Membrane. ACS Chem. Biol. 2021, 16 (5), 929–942. 10.1021/acschembio.1c00179. [DOI] [PubMed] [Google Scholar]
- Sharma S.; Rao R.; Reeve S. M.; Phelps G. A.; Bharatham N.; Katagihallimath N.; Ramachandran V.; Raveendran S.; Sarma M.; Nath A.; Thomas T.; Manickam D.; Nagaraj S.; Balasubramanian V.; Lee R. E.; Hameed P. S.; Datta S. Azaindole Based Potentiator of Antibiotics against Gram-Negative Bacteria. ACS Infect Dis 2021, 7 (11), 3009–3024. 10.1021/acsinfecdis.1c00171. [DOI] [PubMed] [Google Scholar]
- Ejim L.; Farha M. A.; Falconer S. B.; Wildenhain J.; Coombes B. K.; Tyers M.; Brown E. D.; Wright G. D. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011, 7 (6), 348–50. 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- Yarlagadda V.; Medina R.; Wright G. D. Venturicidin A, A Membrane-active Natural Product Inhibitor of ATP synthase Potentiates Aminoglycoside Antibiotics. Sci. Rep 2020, 10 (1), 8134. 10.1038/s41598-020-64756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R.; Mukherjee S.; Barman S.; Haldar J. Macromolecular Nanotherapeutics and Antibiotic Adjuvants to Tackle Bacterial and Fungal Infections. Macromol. Biosci 2021, 21 (11), 2100182. 10.1002/mabi.202100182. [DOI] [PubMed] [Google Scholar]
- Uppu D. S.; Manjunath G. B.; Yarlagadda V.; Kaviyil J. E.; Ravikumar R.; Paramanandham K.; Shome B. R.; Haldar J. Membrane-active macromolecules resensitize NDM-1 gram-negative clinical isolates to tetracycline antibiotics. PLoS One 2015, 10 (3), e0119422. 10.1371/journal.pone.0119422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S.; Mukherjee S.; Ghosh S.; Haldar J. Amino-Acid-Conjugated Polymer-Rifampicin Combination: Effective at Tackling Drug-Resistant Gram-Negative Clinical Isolates. ACS Appl. Bio Mater. 2019, 2 (12), 5404–5414. 10.1021/acsabm.9b00732. [DOI] [PubMed] [Google Scholar]
- Uppu D.; Konai M. M.; Sarkar P.; Samaddar S.; Fensterseifer I. C. M.; Farias-Junior C.; Krishnamoorthy P.; Shome B. R.; Franco O. L.; Haldar J. Membrane-active macromolecules kill antibiotic-tolerant bacteria and potentiate antibiotics towards Gram-negative bacteria. PLoS One 2017, 12 (8), e0183263. 10.1371/journal.pone.0183263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S.; Ghosh S.; Haldar J. Amphiphilic cationic macromolecule potentiates tetracycline against multi-drug resistant Gram-negative bacteria. Bull. Mater. Sci. 2020, 43, 311. 10.1007/s12034-020-02284-3. [DOI] [Google Scholar]
- Yasuda K.; Ohmizo C.; Katsu T. Mode of action of novel polyamines increasing the permeability of bacterial outer membrane. Int. J. Antimicrob. Agents 2004, 24 (1), 67–71. 10.1016/j.ijantimicag.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Katsu T.; Nakagawa H.; Yasuda K. Interaction between polyamines and bacterial outer membranes as investigated with ion-selective electrodes. Antimicrob. Agents Chemother. 2002, 46 (4), 1073–9. 10.1128/AAC.46.4.1073-1079.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konai M. M.; Haldar J. Lysine-Based Small Molecule Sensitizes Rifampicin and Tetracycline against Multidrug-Resistant Acinetobacter baumannii and Pseudomonas aeruginosa. ACS Infect Dis 2020, 6 (1), 91–99. 10.1021/acsinfecdis.9b00221. [DOI] [PubMed] [Google Scholar]
- Dhanda G.; Mukherjee R.; Basak D.; Haldar J. Small-Molecular Adjuvants with Weak Membrane Perturbation Potentiate Antibiotics against Gram-Negative Superbugs. ACS Infect Dis 2022, 8 (5), 1086–1097. 10.1021/acsinfecdis.2c00092. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Nijnik A.; Philpott D. J. Modulating immunity as a therapy for bacterial infections. Nature Reviews Microbiology 2012, 10 (4), 243–254. 10.1038/nrmicro2745. [DOI] [PubMed] [Google Scholar]
- Papareddy P.; Kalle M.; Sorensen O. E.; Malmsten M.; Morgelin M.; Schmidtchen A. The TFPI-2 derived peptide EDC34 improves outcome of gram-negative sepsis. PLoS Pathog 2013, 9 (12), e1003803. 10.1371/journal.ppat.1003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. A.; Koteva K.; Verschoor C. P.; Wang W. L.; Bowdish D. M. E.; Wright G. D. A macrophage-stimulating compound from a screen of microbial natural products. J. Antibiot. 2015, 68 (1), 40–46. 10.1038/ja.2014.83. [DOI] [PubMed] [Google Scholar]
- Yang J. H.; Bhargava P.; McCloskey D.; Mao N.; Palsson B. O.; Collins J. J. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 2017, 22 (6), 757. 10.1016/j.chom.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Huang W.; Lei E.; Yang A.; Li Y.; Wen K.; Wang M.; Li L.; Chen Z.; Zhou C.; Bai S.; Han J.; Song W.; Ren X.; Zeng X.; Pu H.; Wan M.; Feng X. Cooperative Membrane Damage as a Mechanism for Pentamidine-Antibiotic Mutual Sensitization. ACS Chem. Biol. 2022, 17 (11), 3178–3190. 10.1021/acschembio.2c00613. [DOI] [PubMed] [Google Scholar]
- Gill E. E.; Franco O. L.; Hancock R. E. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des 2015, 85 (1), 56–78. 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]