Abstract
Introduction
Mycobacterium bovis and Mycobacterium avium subsp. paratuberculosis, respectively the causative agents of bovine tuberculosis (bTB) and bovine paratuberculosis (PTB), share a high number of antigenic proteins. This characteristics makes the differential diagnosis of the diseases difficult. The interferon gamma (IFN-γ), C-X-C motif chemokine ligand 10 (CXCL10), matrix metallopeptidase 9 (MMP9), interleukin 22 (IL-22) and thrombospondin 1 (THBS1) bovine genes have already been shown to be accurate transcriptional biomarkers of bTB. In order to improve the diagnosis of bTB and PTB, in the present study we evaluated the risk of false positivity of these bTB biomarkers in cattle with PTB.
Material and Methods
The transcription of these genes was studied in 13 PTB-infected cattle, using Mycobacterium avium subsp. paratuberculosis (MAP)-stimulated peripheral blood mononuclear cells (PBMC).
Results
Overall, the levels of IFN-γ, CXCL10, MMP9 and IL-22 transcripts in MAP-stimulated PBMC failed to differentiate animals with PTB from healthy animals. However, as bTB-afflicted cattle do, the MAP-infected group also displayed a lower level of THBS1 transcription than the non-infected animals.
Conclusion
The results of this study add new specificity attributes to the levels of transcription of IFN-γ, CXCL10, MMP9 and IL-22 as biomarkers for bTB.
Keywords: biomarkers, bovine tuberculosis, bovine paratuberculosis
Introduction
Bovine tuberculosis (bTB) is a common worldwide disease, the causative agent of which is primarily the Mycobacterium bovis bacterium. Although cattle are the main host of M. bovis, wild animals can also become infected and serve as reservoirs (9). Indeed, these bacteria are transmissible to many mammal species (16), including humans, thus making bTB a zoonotic disease (20). Despite considerable progress in combatting bTB thanks to national eradication programmes, it remains an important disease that causes significant economic losses through trade restrictions and livestock deaths.
The diagnostic test currently used worldwide is the single tuberculin skin test (TST), a method that consists of inoculating the animal with a purified protein derivative from M. bovis (PPDb) in vivo and then measuring the increase in skin fold thickness. Nevertheless, this test sometimes fails to distinguish between cattle infected with M. bovis and animals sensitised with environmental mycobacteria or bacteria from the M. avium complex (4). Another available test is the interferon-gamma (IFN-γ) release assay (IGRA), which is an ancillary test that consists of measuring the in vitro IFN-γ release in a blood culture upon PPDb stimulation (21).
The need to increase the robustness of the bTB diagnostic system led us to investigate the predictive power of immune mediators for bTB infection in infected animals that were negative in a TST (12). The results of our research demonstrated that, unlike what occurs in healthy cattle, in infected cattle the levels of thrombospondin 1 (THBS1), matrix metallopeptidase 9 (MMP9), interleukin 22 (IL-22) and IFN-γ transcripts changed. In addition, the assessment of C-X-C motif chemokine ligand 10 (CXCL10), IFN-γ and IL-22 transcripts was useful to predict bTB status in a group of TST false-negative animals. Hence, these biomarkers are interesting for further assessment with an ancillary diagnostic assay.
Paratuberculosis (PTB) is a disease of ruminants characterised by intestinal inflammation, the aetiological agent of which is Mycobacterium avium subsp. paratuberculosis (MAP). Infections with MAP and other mycobacterium species, including nontuberculous mycobacteria (NTM), cause cross- reactions that interfere with bTB diagnosis. Since PPDb has components present in other Mycobacterium species, the TST and IGRA assays can develop false-positive results (3). Indeed, PTB vaccination with heat-inactivated strains is prohibited in many European countries because of its interference with bTB diagnostic tests. Therefore, the identification of biomarkers specific for bTB infection and excluding NTM from detection is imperative.
The objective of this research was to evaluate the specificity of previously identified bTB immune mediators (THBS1, MMP9, IL-22, CXCL10, IFN-γ, cluster of differentiation 14 (CD14) and interleukin 1 receptor (IL-1R)) in a group of PTB-infected cattle.
Material and Methods
Selection of sampled animals. The PTB negative group (PTB−) included 14 adult Holstein and cross-breed (Hereford and Aberdeen Angus) cattle (36–60 months old) from a herd reared at the Instituto Nacional de Tecnología Agropecuaria (INTA – Buenos Aires, Argentina) and from a family farm in Ljubljana (Slovenia). Both herds had no history of bTB or PTB within the five years preceding the investigation. The PTB-infected group (PTB+) included 13 adult Holstein animals (36–60 months old) from two herds with a high prevalence of PTB. These herds were kept in Chascomus and Suipacha (Buenos Aires, Argentina). The animals were classified as PTB+ because of clinical signs and a positive reaction to an in-house ELISA based on specific antibodies to purified protein derivatives from M. avium (PPDa) which is described below. The infection status in the PTB− animals was also confirmed by this ELISA. All the animals included in the analysis were negative in a single TST against PPDb (data not shown) and came from bTB-free herds.
Mononuclear cell preparation, RNA preparation and quantitative reverse transcription PCR. To evaluate the transcription of previously identified bTB biomarkers in PTB infection, we assessed the transcription levels of IFN-γ, MMP9, IL-22, THBS1, IL-1R, CXCL10 and CD14 in peripheral blood mononuclear cells (PBMCs) stimulated with PPDa or not stimulated originating from cattle with PTB (n = 13) and healthy animals (n = 14). PBMCs were prepared from 15 mL of heparinised blood extracted from each animal by gradient centrifugation over Histopaque-1077 (Sigma Aldrich, Darmstadt, Germany) according to the manufacturer’s instructions. A 2 × 107 quantity of PBMCs from each animal was divided between two wells of a 12-well plate and Roswell Park Memorial Institute 1640 medium supplemented with autologous serum (10%) and Antibiotic-Antimycotic 1× (Thermo Fisher Scientific, Waltham, MA, USA) was added to make a final volume of 1 mL. One well was stimulated by the addition of PPDa (20 μg/mL; Biocor, Córdoba Argentina) and the other remained without stimulation. Cells were incubated at 37°C for 16 h.
Antigen-stimulated PBMCs were resuspended in 1 mL of Trizol (Sigma Aldrich), subsequently treated twice with chloroform (200 μL) and centrifuged for 5 min at 9,000 x g. The aqueous phase containing nucleic acids (upper phase) was precipitated by adding isopropanol and the pellet was then incubated at −80°C. The pellets were washed with ice-cold 70% ethanol and resuspended in RNAse-free water. Finally, the RNA samples were precipitated by adding one volume of LiCl (10 M), and then frozen at −80°C for 16 h. The RNA pellets were washed again with ice-cold 70% ethanol, then resuspended again in RNAse-free water and treated with DNAseI (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Quantitative reverse transcription polymerase chain reactions (RT-qPCR) were performed as described previously (5). The primer sequences used are described elsewhere (12). The reactions were carried out with Taq Platinum DNA polymerase (Invitrogen/Life Technologies, Carlsbad, CA, USA) and SYBR reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. Standard cycling conditions were set for the reactions on an Applied Biosystems StepOne Plus PCR System (Applied Biosystems, Foster City, CA, USA). Each reaction was performed in duplicate and the qPCR data were analysed as described previously (19). The non-stimulated condition was used as the calibrator, and glyceraldehyde 3-phosphate dehydrogenase was used as a reference gene. Relative gene transcription was calculated using the 2-ΔΔCt method with E correction.
ELISA. Sera from all the animals were analysed by indirect ELISA test. The in-house ELISA protocol was adapted from one previously reported (6, 14). Briefly, 96-well microtitre ELISA plates (NuncMaxisorp; Sigma Aldrich) were coated with 100 μL of carbonate buffer (0.1 M sodium bicarbonate, 0.1 M sodium carbonate; pH 9.6) with 4 μg of paratuberculosis protoplasmatic antigen (Allied Monitor Inc., Fayette, MO, USA), by incubating the plates overnight at 4°C. Subsequently, the plates were blocked for 1 h with 200 μL of phosphate-buffered saline with Tween-20 (PBS-T) plus 0.2% porcine gelatine A (Sigma-Aldrich), and then washed with PBS-T. The sera (100 μL/well; 1: 100 dilution in PBS) were added and the solutions were incubated for 1 h at 37°C. The wells were washed 12 times with 200 μL of PBS-T before adding peroxidase-labelled affinity purified protein G (1: 4,000 dilution; BioRad Laboratories, Hercules, CA, USA). Finally, the plates were washed and the reaction was developed using 1% 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (Sigma-Aldrich) in phosphate citrate buffer plus 0.1 μL of 30% hydrogen peroxide in citrate buffer (pH 5.0). The colour development was then stopped with 100 μL of a stop solution of 50% v/v dimethylformamide (Sigma Aldrich) and 20% w/v sodium dodecyl sulphate (Promega, Madison, WI, USA)). The optical density at 405 nm was measured with a Multiskan plate reader (Thermo Fisher Scientific). The cut-off value of the in-house ELISA was calculated as mean 0.4363 (14), its sensitivity as 97.87% (95% confidence interval (CI) 0.8871–0.995) and its specificity as 97.92% (95% CI 0.8893–0.9995).
PCR of faecal material. In order to confirm the infection status of each animal by an additional test, a MAP-specific PCR was performed on faecal material from each cow. Faecal samples (200 g) were collected from the rectum of each subject. The samples were kept in sterile containers and refrigerated until processing. Two grams of the faeces were resuspended in 10 mL of PBS-T and vortexed for 30 s. Subsequently, the suspension was left to sediment for 15 min and 2 mL of the supernatant was transferred to a fast prep tube with glass beads (150–212 μm). The tubes were incubated at 95°C for 10 min, the samples were homogenised at 6,000 rpm for 20 s, and the homogenised samples were left to cool down. The homogenisation step was repeated twice. The homogenised samples were centrifuged at 10,000 x g for 1 min and then 1 mL of the supernatant was processed using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Aliquots of 5 μL of each sample were used to amplify the MAP insertion sequence IS900 (https://www.ncbi.nlm.nih.gov/nuccore/X16293) with the following primers: forward 5′- AAG ACC GAC GCC AAA GAC-3′ and reverse 5′- CAG AGG CTG CAA GTC GTG-3′ (11).
Statistical analysis. Graphics were constructed using GraphPad Prism 5.03 software (GraphPad Software, San Diego, CA, USA). Differences between groups were assessed using the non-parametric Mann-Whitney test. A P-value below the usually agreed alpha risk of 5% (0.05) was considered significant.
Results
Animals. According to the reactivity results obtained in the in-house ELISA based on anti-PPDa specific antibodies, 13 animals were PTB+ and 14 were PTB− (Table 1). In the PTB+ group, six animals were positive for MAP in the PCR test on faeces (Table 1).
Table 1.
Results of ELISA, PCR test in faecal material and fold change transcription of target genes of animals used in this study
| Animal ID | ELISA | PCR | IFN-γ | IL-22 | CXCL10 | THBS1 | IL-1R | MMP9 | CD14 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | + | + | 3.68 | 4.27 | 6.10 | −7.44 | −2.28 | −2.73 | −1.25 |
| 2 | + | + | −2.96 | 2.84 | −3.20 | −10.98 | −4.09 | −3.05 | −3.35 |
| 3 | + | − | 1.31 | 6.34 | 5.56 | −17.34 | −1.31 | −1.79 | −2.97 |
| 4 | + | + | 2.99 | 9.19 | 21.76 | 1.49 | 3.51 | 1.57 | −1.89 |
| 5 | + | − | 1.74 | 7.52 | 1.65 | −8.02 | −1.39 | 1.40 | −1.93 |
| 6 | + | + | −1.11 | 4.63 | −1.48 | −1.96 | 1.40 | 2.20 | 1.21 |
| 7 | + | − | 4.27 | 288.74 | 33.97 | −58.89 | 2.52 | 5.65 | 1.01 |
| 8 | + | − | 1.41 | 317.41 | 8.63 | −44.74 | 1.97 | 3.26 | −1.77 |
| 9 | + | − | −1.21 | 5.59 | −2.17 | −11.42 | −2.48 | −4.06 | −3.15 |
| 10 | + | + | −1.17 | 1.42 | 2.86 | −5.07 | 1.16 | 1.25 | 1.11 |
| 11 | + | − | 9.00 | 1.93 | 26.86 | −4.56 | −1.36 | 3.07 | 1.20 |
| 12 | + | + | −2.40 | −219.17 | 8.20 | −4.81 | 1.67 | −3.02 | 1.11 |
| 13 | + | − | 17.13 | 389.80 | 11.52 | −4.28 | 1.88 | 1.55 | −2.38 |
| 14 | − | − | 2.54 | 8.43 | 25.62 | −1.58 | 1.47 | −1.01 | −1.90 |
| 15 | − | − | 2.20 | 16.74 | 3.71 | 12.23 | 1.45 | −1.54 | 1.22 |
| 16 | − | − | 1.44 | 22.23 | 1.05 | −1.21 | 2.32 | 1.98 | 1.26 |
| 17 | − | − | −2.44 | −1.14 | −1.19 | −5.36 | −1.03 | −2.41 | −2.91 |
| 18 | − | − | −7.21 | −2.51 | −2.16 | 10.83 | 1.79 | 2.95 | −1.09 |
| 19 | − | − | −2.37 | 1.11 | 1.47 | −1.65 | 3.72 | 1.70 | −1.65 |
| 20 | − | − | 6.07 | 56.45 | 20.35 | −2.53 | −1.45 | −2.05 | −1.46 |
| 21 | − | − | 3.46 | 27.63 | 15.70 | −1.65 | −1.25 | −1.60 | −1.10 |
| 22 | − | − | 5.85 | 14.44 | 17.66 | −1.56 | −1.27 | −2.25 | −2.44 |
| 23 | − | − | −2.94 | ND | −1.02 | −2.35 | 5.60 | 1.55 | −1.38 |
| 24 | − | − | 6.29 | 27.91 | 1.75 | 10.66 | 6.98 | 6.93 | 5.50 |
| 25 | − | − | 2.42 | 36.69 | 1.73 | 2.88 | 2.78 | 1.32 | 1.25 |
| 26 | − | − | −1.45 | 3.81 | −1.49 | −1.07 | −1.32 | −1.64 | −1.31 |
| 27 | − | − | 11.67 | 45.12 | 9.14 | −3.9 | −1.55 | −2.84 | −1.98 |
PTB+ animals are shaded in grey
IFN-γ – interferon gamma; IL-22 – interleukin 22; CXCL10 – C-X-C motif chemokine ligand 10; THBS1 – thrombospondin 1; IL-1R – interleukin 1 receptor; MMP9 – matrix metallopeptidase 9; CD14 – ; ND – not determined. ELISA cut-off value 0.4363
Transcription of biomarker candidate genes. The transcription of IFN-γ, MMP9, IL-22, THBS1, IL-1R, CXCL10 and CD14 was assessed in the samples from PTB+ and PTB− cattle. The relative expression of each target gene was calculated as the ratio of qPCR value of the PPDa-stimulated samples to the qPCR value of the PBS-stimulated samples (fold change). No significant differences were detected between the fold change values of the IFN- γ, MMP9, IL-22, IL-1R, CXCL10 or CD14 transcripts of infected animals and those of the healthy animals (Fig. 1). However, the transcription level of THBS1 was significantly reduced in the PTB+ animals compared to that of the healthy animals (P = 0.0004).
Fig. 1.
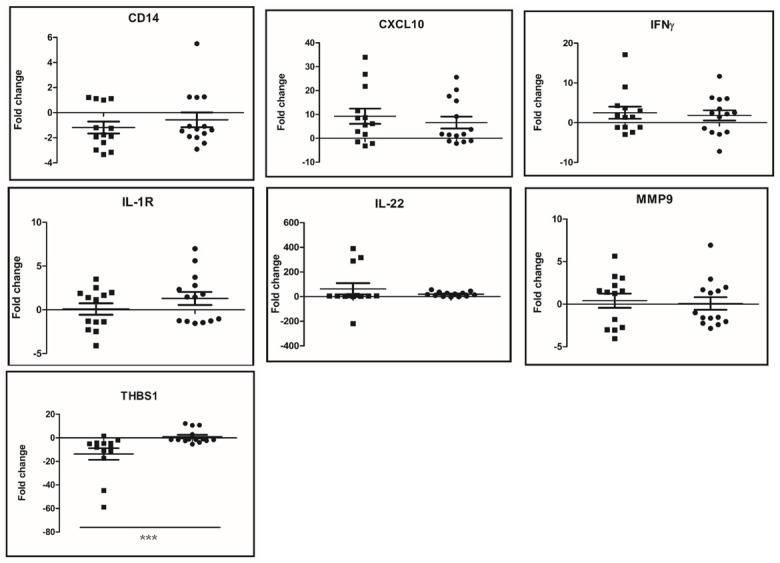
Fold changes of gene transcription in PBMCs from PTB+ and healthy animals upon PPDa stimulation. Fold change values from individual animals are represented by squares (PTB+ animals) and circles (healthy animals). Median and interquartile ranges are shown. Relative gene transcription was calculated with the 2-ΔΔCt method with E correction and using glyceraldehyde 3-phosphate dehydrogenase mRNA as the reference gene and the non-stimulated condition as the calibrator. Data were analysed using a two-tailed unpaired Mann-Whitney test CD14 – Cluster of differentiation 14; CXCL10 – C-X-C motif chemokine ligand 10; PTB – paratuberculosis; IFN-γ – interferon gamma; IL-1R – interleukin 1 receptor; IL-22 – interleukin 22; MMP9 – matrix metallopeptidase 9; THBS1 – thrombospondin 1; *** – P < 0.001
Discussion
We showed that the mRNA transcripts of IFN-γ, MMP9, IL-22, IL-1R, CXCL10 and CD14 fail to discriminate groups of PTB+ from healthy cattle. These results suggest that these immune mediators probably play a minor role in the adaptive response against MAP, at least under the experimental conditions of this study.
In contrast to what we observed in PTB+ animals, our previous results showed that the levels of MMP9, IL-22 and IFN-γ transcripts were true correlates of bTB disease. Moreover, the transcription of IL-22 and CXCL-10 allowed the detection of a group of bTB+ cattle that had evaded diagnosis by TST and IGRA (12). This could be explained in part by the fact that, although M. bovis and MAP are both intracellular mycobacteria primarily infecting macrophages and dendritic cells, the transcriptional program that each bacterial species induces in the host cells is markedly different, thus resulting in divergent immune profiles (22).
During the subclinical stage, the host response against MAP infections primarily involves the Th1 response with the production of proinflammatory cytokines (such as IFN-γ) and minimal bacterial shedding. As the disease progresses, the Th1 response decreases and the Th2 response increases, which induces the specific antibody response. In this stage, pronounced and sometimes intermittent bacterial shedding takes place (10). Therefore, it is not surprising that the animals tested in this study failed to respond to MAP stimuli with the mRNA transcription profile of proinflammatory molecules, as all the animals were either in transmission or clinical stages of the diseases.
According to previous studies, IL-22 is a proinflammatory cytokine relevant to MAP infections. In one of these studies, ileocaecal lymph nodes from naturally infected cows and experimentally inoculated calves presented upregulated levels of IL-22 (1). Although this finding differs from the results of the present study, the discrepancies may be due to the methodology used. In their study, Allen et al. (1) analysed the local immune response, while we evaluated the in vitro recall of the immune response by stimulating memory cells with MAP antigens. However, an upregulation of IL-22 and IFN-γ transcription took place in PBMC stimulated with MAP derived from cattle experimentally infected with MAP for three months (18). This finding is consistent with the nature of an early PTB infection (which is characterised by no bacterial shedding) in its involvement primarily of cell-mediated Th1 and inflammatory Th17 immune profiles. In another study, the levels of IL-22 mRNA in PBMC stimulated with MAP in PTB+ cattle shedding bacteria were either equivalent to or reduced from those detected in non-infected animals (18).
The assessment of CXCL10 and MMP9 mRNAs as prognostic biomarkers in the ileocaecal valve or peripheral blood (2) or in PBMC (8) has identified both cytokine mRNAs as upregulated biomarkers of PTB+ cattle. However, the analyses of the gene transcription in those studies were made on cell samples without in vitro stimulation with MAP or PPDa; therefore, those results are not comparable to those of our study. A transcriptomic analysis has shown that MMP9 mRNA transcription was significantly upregulated in samples from cows at a subclinical stage upon specific stimulation, while it appeared to remain unaltered in cattle at the clinical, and infectious stage (7). This result is consistent with the findings of our study. On the other hand, CXCL10 has recently been associated with potentially critical genes for resistance or susceptibility to MAP (23); however, to our knowledge, no previous study has evaluated the transcription of this gene as a biomarker of PTB in MAP-stimulated host samples.
Overall, these results indicate no antigen-specific upregulation of proinflammatory immune mediators in PTB+ infectious cattle. Nevertheless, further studies are necessary to better validate the performance of the mRNA biomarkers of this research in order to discriminate bTB from PTB. Studies should assess the responses of animals infected with MAP but in a non-infectious condition and not shedding MAP.
Regarding the THBS1 gene, here PTB+ cattle displayed downregulated transcript levels, in accordance with a previous report in bTB cattle (12). Thrombospondin 1 is a protein secreted during inflammation into the extracellular matrix, where it mediates cell-to-cell and cell-to-matrix interactions. This protein can activate transforming growth factor beta 1 and inhibits endothelial cell migration, angiogenesis, and tumour growth (17). A recent report highlights the role of angiogenesis (and lipophagy) in the control of tuberculosis (15), thus suggesting that downregulation of THBS1 is beneficial in limiting the mycobacterial burden, speculatively by inhibiting granuloma formation. Therefore, immune host cells may downregulate the transcription of this glycoprotein in response to mycobacterial infections.
A limitation of this study is that the analyses were performed using pooled samples of groups of animals instead of samples from individual animals. To have better diagnosis for each individual subject, a critical improvement would be implementing more robust methodologies detecting proteins instead of relative mRNA amounts for a given biomarker.
In conclusion, the results of this study indicate that the transcripts from CXCL10, MMP9, IL-22 and IFN-γ genes have a low risk of false positivity as biomarkers for bTB in cattle with PTB. Thus, these biomarkers are able to distinguish cattle with bTB globally from cattle with PTB in the infectious stage. This study also shows that THBS1 mRNA is a potential biomarker for PTB, as it is for bTB, and a downregulated level of this biomarker will indicate an animal to be positive for these diseases. Regarding CD14 and IL-1R, the transcription level of these genes has been shown not to change in bTB-infected cattle (8) nor in PTB+ animals, which indicates that they are not suitable biomarkers of these diseases. Although these findings would contribute to improving the diagnosis of bTB and PTB, the analysis should be extended to groups of animals at different stages of both diseases.
Acknowledgements
We thank Dr. Julia Sabio Garcia for the critical reading of this paper. The authors also thank Verónica Maldonado and Diego Rafael Franco for providing us with cattle blood samples.
Footnotes
Conflict of Interest
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: This work was funded by National Agency for Scientific and Technological Promotion (grant PICT 2017-1721), National Institute of Agricultural Technology (grant I103), Science and Technology Ministry (Argentina) - Ministry of Education, Science and Sport/Slovenian Research Agency (Slovenia) (grant DFG16/01/AR/).
Animal Rights Statement: The Institutional Animal Care and Use Committee (CICUAE) of CICVyA-INTA and the Administration of the Republic of Slovenia for Food Safety, Veterinary Sector and Plant Protection authorised this study. The regulations used are in agreement with the European Union Laws for the protection of experimental animals.
References
- 1.Allen A.J., Park K.T., Barrington G.M., Lahmers K.K., Abdellrazeq G.S., Rihan H.M., Sreevatsan S., Davies C., Hamilton M.J., Davis W.C.. Experimental infection of a bovine model with human isolates of Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol. 2011;141:3–4. doi: 10.1016/j.vetimm.2011.03.014.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Hearn M., Canive M., Blanco-Vazquez C., Torremocha R., Balseiro A., Amado J., Varela-Martinez E., Ramos R., Jugo B.M., Casais R.. RNA-Seq analysis of ileocecal valve and peripheral blood from Holstein cattle infected with Mycobacterium avium subsp. paratuberculosis revealed dysregulation of the CXCL8/IL8 signaling pathway. Sci Rep. 2019;9:14845. doi: 10.1038/s41598-019-51328-0.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amadori M.S., Tagliabue S.L., Finazzi G., Lombardi G., Telo P., Pacciarini L., Bonizzi L.. Diagnosis of Mycobacterium bovis infection in calves sensitized by mycobacteria of the avium/intracellulare group. J Vet Med B Infect Dis Vet Public Health. 2002;49:89–96. doi: 10.1046/j.1439-0450.2002.00513.x. [DOI] [PubMed] [Google Scholar]
- 4.Aranaz A., De Juan L., Bezos J., Alvarez J., Romero B., Lozano F., Paramio J.L., López-Sánchez J., Mateos A., Domínguez L.. Assessment of diagnostic tools for eradication of bovine tuberculosis in cattle co-infected with Mycobacterium bovis and M. avium subsp. paratuberculosis. Vet Res. 2006;37:593–60. doi: 10.1051/vetres:2006021.. doi. [DOI] [PubMed] [Google Scholar]
- 5.Blanco F.C., Soria M., Bianco M.V., Bigi F.. Transcriptional Response of Peripheral Blood Mononuclear Cells from Cattle Infected with Mycobacterium bovis. PLoS One. 2012;7:e41066. doi: 10.1371/journal.pone.0041066.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo G., Pinedo F.A., Mon M.L., Viale M., Gil A., Illia M.C., Gioffré A., Arese A., Travería G., Romano M.I.. Accuracy assessment and screening of a dairy herd with paratuberculosis by three different ELISAs. Vet Microbiol. 2012;156:183–188. doi: 10.1016/j.vetmic.2011.10.029.. doi. [DOI] [PubMed] [Google Scholar]
- 7.Coussens P.M., Colvin C.J., Wiersma K., Abouzied A., Sipkovsky S.. Gene expression profiling of peripheral blood mononuclear cells from cattle infected with Mycobacterium paratuberculosis. Infect Immun. 2002;70:5494–5502. doi: 10.1128/IAI.70.10.5494-5502.2002.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David J., Barkema H.W., Guan L.L., de Buck J.. Gene-expression profiling of calves 6 and 9 months after inoculation with Mycobacterium avium subspecies paratuberculosis. BMC Vet Res. 2014;45:96. doi: 10.1186/s13567-014-0096-5.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald S.D., Kaneene J.B.. Wildlife Reservoirs of Bovine Tuberculosis Worldwide: Hosts, Pathology, Surveillance, and Control. Vet Pathol. 2013;50:488–499. doi: 10.1177/0300985812467472.. doi. [DOI] [PubMed] [Google Scholar]
- 10.Ganusov V.V., Klinkenberg D., Bakker D., Koets A.P.. Evaluating contribution of the cellular and humoral immune responses to the control of shedding of Mycobacterium avium spp. paratuberculosis in cattle. Vet Res. 2015;46:62. doi: 10.1186/s13567-015-0204-1.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herthnek D., Bölske G.. New PCR systems to confirm real-time PCR detection of Mycobacterium avium subsp. paratuberculosis. BMC Microbiol. 2006;6:87. doi: 10.1186/1471-2180-6-87.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klepp L.I., Eirin M.E., Garbaccio S., Soria M., Bigi F., Blanco F.C.. Identification of bovine tuberculosis biomarkers to detect tuberculin skin test and IFNγ release assay false negative cattle. Res Vet Sci. 2019;122:7–14. doi: 10.1016/j.rvsc.2018.10.01.. doi. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Dee Z.P., Chittur S.V., Patel B., Stanton R., Wakeley M., Lippert B., Menaker A., Eiche B., Terry R., Gutierrez L.S.. Thrombospondin-1 type 1 repeats in a model of inflammatory bowel disease: Transcript profile and therapeutic effects. PLoS One. 2012:e34590. doi: 10.1371/journal.pone.0034590.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyano R.D., Romero M.A., Colombatti Olivieri M.A., Alvarado Pinedo M.F., Traveria G.E., Romano M.I., Alonso M.N.. Development and Validation of a Novel ELISA for the Specific Detection of Antibodies against Mycobacterium avium Subspecies paratuberculosis Based on a Chimeric Polyprotein. Vet Med Int. 2021:e7336848. doi: 10.1155/2021/7336848.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee T., Bhatt B., Prakhar P., Lohia G.K., Rajmani R.S., Balaji K.N.. Epigenetic reader BRD4 supports mycobacterial pathogenesis by co-modulating host lipophagy and angiogenesis. Autophagy. 2022;18:391–408. doi: 10.1080/15548627.2021.1936355. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer M.V., Thacker T.C., Waters W.R., Gortázar C., Corner L.A.L.. Mycobacterium bovis: A Model Pathogen at the Interface of Livestock, Wildlife, and Humans. Vet Med. 2012;2012:1–17. doi: 10.1155/2012/236205.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park H.-E., Park H.-T., Jung Y.H., Yoo H.S.. Gene expression profiles of immune-regulatory genes in whole blood of cattle with a subclinical infection of Mycobacterium avium subsp. paratuberculosis. PLoS One. 2018;13:e0196502. doi: 10.1371/journal.pone.0196502.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park K.T., Allen A.J., Bannantine J.P., Seo K.S., Hamilton M.J., Abdellrazeq G.S., Rihan H.M., Grimm A., Davis W.C.. Evaluation of two mutants of Mycobacterium avium subsp. paratuberculosis as candidates for a live attenuated vaccine for Johne’s disease. Vaccine. 2011;29:29–30. doi: 10.1016/j.vaccine.2011.04.090. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl M.W., Horgan G.W., Dempfle L.. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36.. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quadri N.S., Brihn A., Shah J.A., Kirsch J.D.. Bovine Tuberculosis: A Re-emerging Zoonotic Infection. J Agromedicine. 2021;26:334–339. doi: 10.1080/1059924X.2020.1771497.. doi. [DOI] [PubMed] [Google Scholar]
- 21.Rothel J.S., Jones S.L., Corner L.A., Cox J.C., Wood P.R.. A sandwich enzyme immunoassay for bovine interferon-gamma and its use for the detection of tuberculosis in cattle. Aust Vet J. 1990;67:134–137. doi: 10.1111/j.1751-0813.1990.tb07730.x. [DOI] [PubMed] [Google Scholar]
- 22.Rue-Albrecht K., Magee D.A., Killick K.E., Nalpas N.C., Gordon S.V., MacHugh D.E.. Comparative functional genomics and the bovine macrophage response to strains of the Mycobacterium genus. Front Immunol. 2014;5:536. doi: 10.3389/fimmu.2014.00536. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Han B., Zheng W., Lin S., Li H., Gao Y., Sun D.. Genome-wide DNA methylation profile in jejunum reveals the potential genes associated with paratuberculosis in dairy cattle. Front Genet. 2021;12:735147. doi: 10.3389/fgene.2021.735147. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]


