Abstract
Immunoglobulin genes are rarely considered as disease susceptibility genes despite their obvious and central contributions to immune function. This appears to be a consequence of historical views on antibody repertoire formation that no longer stand, and of difficulties that until recently surrounded the documentation of the suite of antibody genes in any individual. If these important genes are to be accessible to GWAS studies, allelic variation within the human population needs to be better documented, and a curated set of genomic variations associated with antibody genes needs to be formulated. Repertoire studies arising from the COVID-19 pandemic provide an opportunity to meet these needs, and may provide insights into the profound variability that is seen in outcomes to this infection.
Keywords: immunoglobulin, AIRR-Seq, Rep-Seq, antibody receptor repertoires, IGHV, immune receptor genes
The paradox of immunoreceptor genes and their function
Although genetic differences between individuals do not always translate into differences in disease susceptibility, variations in even relatively obscure genes that are involved in immune function are often investigated for their associations with disease [1–3]. In contrast, despite their fundamental role in forming the antibody repertoire with which we fight infections, germline immunoglobulin genes are rarely considered for their possible influence on disease susceptibility and disease outcome. In part, this may be because receptor formation was historically considered to involve random processes that maximize diversity, and because the large sets of highly similar germline genes were thought to ensure that each person’s repertoire would be of similar functionality. More recent work has shown that an individual’s repertoire is shaped by strong genetically-determined biases [4,5], and that a single codon change can alter the ability of an immunoglobulin gene to encode protective antibodies [6,7]. It is no longer possible to assume that V(D)J recombination can generate any antibody that an individual might require.
The lack of interest in exploring disease association with immunoglobulin genes may also be explained by the fact that until recently, it has been difficult to determine the complete set of germline IG genes in an individual’s genome. Now that tools and techniques are available for this purpose, there is an urgent need to codify variation in these sets within the human population. This should then allow us to explore the influence of this variation on disease responses.
There has also been little attention paid to a possible role for TCR genes in disease susceptibility, though variability in the genes of the Major Histocompatibility Complex (MHC) has been a recognized source of variation in the T cell response to pathogens for decades [8]. TCR repertoire studies have overwhelmingly focused on the CDR3 regions of the TCR chains (e.g. [9,10]), for these are the regions of the TCR that interact most closely with antigen in association with MHC. As a consequence, allelic variation of TCR genes within the human population is still poorly documented. In this review, we will therefore focus on IG genes, though many of the principles under discussion are likely to also apply to the TCR regions. We report that antibody repertoire studies can now be used to document germline IG genes, and that these studies are providing new insights into the ways germline genes shape the antibody repertoire. We describe how the detection of IG genes in genome-wide association studies (GWAS) is compromised by the poor SNP coverage of the immunoglobulin gene loci, and we suggest that the intense focus on the anti-SARS-CoV2 antibody response can be used to expand this coverage. Finally, we suggest there is reason to believe that such genetic inquiries may also provide insights into the nature of the extreme variability that is seen in the outcomes of this infection.
Immunoreceptor genotypes and haplotypes
Any challenge to the view that immunoreceptor genes are of little biological significance requires knowledge of the sets of genes that are found in different individuals, and until recently, this was extremely difficult to determine. The complexity of the loci, each containing as many as 150 highly similar genes and pseudogenes, prevented their routine documentation. This has now changed.
Short-read data derived from genomic DNA, either through whole-genome sequencing (WGS) or targeted sequencing, is now being used to infer human germline immunoglobulin genes [11–13], and this is an approach that may hold promise. It is an approach that has also been used to study species of biomedical importance like the cynomolgus macaque [14], and even the elephant [15], but further scrutiny of these data will be needed before the utility of the approach can be accepted. Erroneous inferences can be made [16,17], and care will be needed if we are to limit the impact of this upon compiled germline datasets.
We believe that high throughput sequencing of expressed antibody genes provides an alternative and more reliable pathway to immunoglobulin gene discovery. High throughput sequencing is now capable of generating datasets that capture an informative fraction of the repertoires of individual subjects, with low error rates [18,19]. Embedded within these repertoire datasets of millions of immunoreceptor sequences is other immunogenetic data that is usually overlooked. Contained within these datasets are individual immunogenotypes - the sets of germline IG genes that are present in different individuals [20–23]. Each germline gene identified in these datasets may be supported by hundreds or even thousands of V(D)J gene rearrangements, and each one of these rearrangements can be considered an independent piece of evidence in support of the existence of a particular combination of V, D and J genes. An immunogenotype representing the expressed component of an individual’s germline genes can therefore be defined with great confidence.
Many IG V(D)J datasets can be analyzed to determine individual haplotypes - the sets of germline V, D and J genes that are carried by individuals on each of their copies of the relevant chromosome [24,25]. This is highly significant, for V(D)J recombination is a chromosomal event, and particular V, D and J genes can only be paired if they are located on the same chromosome. Using long-read high throughput sequencing, haplotypes can even be extended to document the associated constant region genes that encode isotypes of different functionality.
Haplotype data can reveal that an individual who may carry all the necessary genes to produce an optimal antibody does not carry the genes on a single chromosome (Figure 1). The individual is therefore unable to form that antibody. Other individuals may carry the necessary V, D and J genes on a chromosome that lacks the gene that could encode an optimal defensive isotype (Figure 2). This kind of analysis therefore represents a powerful new way to understand the way genes contribute to variations in the B cell repertoires that are available to fight disease in different people.
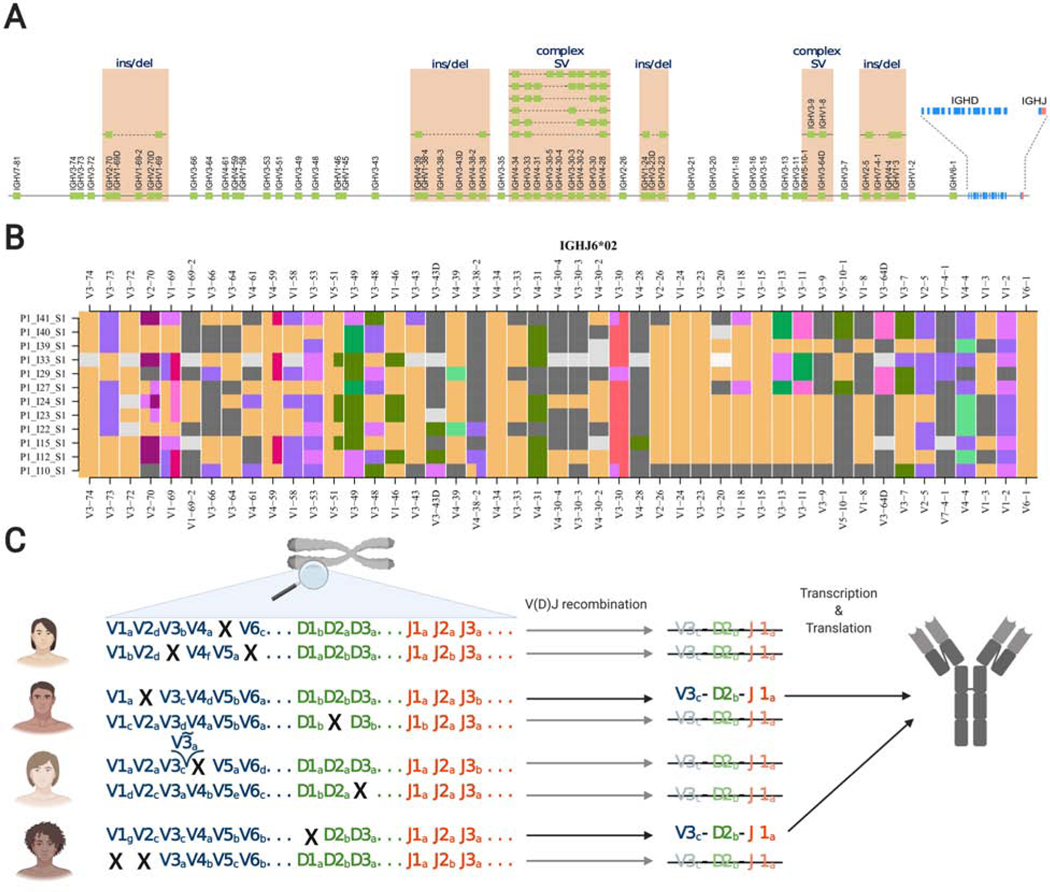
Figure 1:
Structural and allelic variation in the IGHV locus. A. A schematic representation of all known functional IGHV genes of the IGH locus; the relative positions of genes are displayed on the reference assembly reported by Rodriguez et al. [16]. Known structural variants that have been characterized by genomic sequencing are also shown. Further variation can be inferred from VDJbase haplotypes. B. VDJbase haplotype ribbon plots for 12 individuals who are heterozygous at the IGHJ6 locus for alleles *02 and *03. Ribbons show IGHV haplotypes associated with the IGHJ6*02-bearing chromosomes. Black corresponds to gene deletions, grey to uncertainty, and colours represent different numbered alleles (eg orange = *01; purple = *02). C. Allelic variation and deletion polymorphisms lead to the absence of particular VDJ rearrangements from the naive IG repertoires of many individuals. Names of genes and alleles are for illustration purposes only. Panel C was created with BioRender.com.
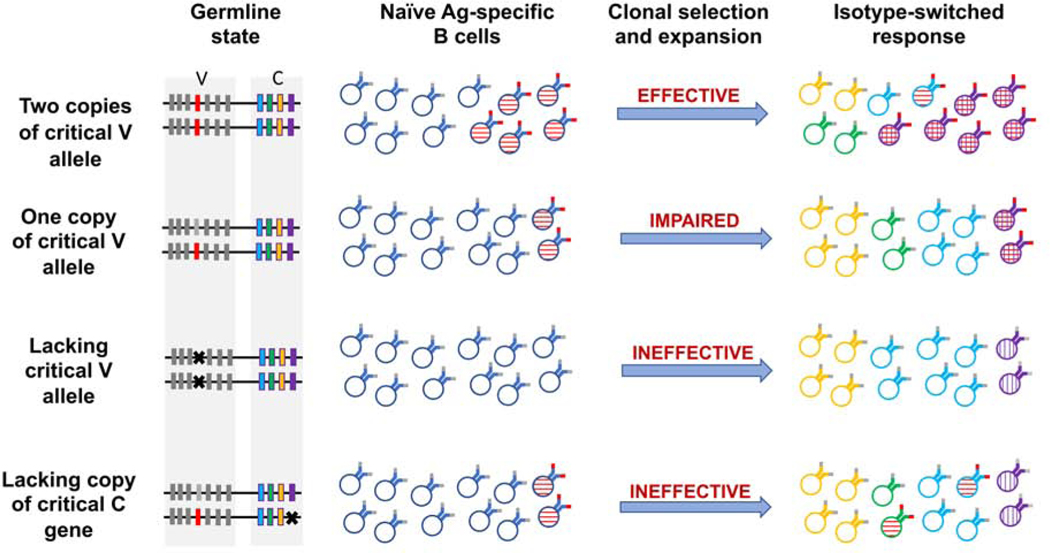
Figure 2:
The generation of optimal IG specificities and optimal isotypes in individuals with differing IGH haplotypes. Optimal V alleles are shown in red, and deletion polymorphisms are shown with a cross. Cells with optimal specificities are shown with horizontal hatching and with red IG binding sites. Cells that express the optimal isotype for a particular response are shown with vertical hatching. The success of the response to a pathogen is a consequence of clone sizes, of the fine specificities of antigen-specific cells, and of the ability for class switching to result in expression of optimal isotypes. Heavy chain D and J genes, and light chain genes also contribute to the expression of optimal specificities, but are not shown.
Variability in the IGH locus
Although the first human antibody genes were reported over forty years ago, it was not until 1998 that a complete sequence of the immunoglobulin heavy chain (IGH) variable region gene locus was reported [26]. The Matsuda sequence contains 44 apparently functional IGHV genes and 79 pseudogenes. This sequence was incorporated into the human genome reference assembly, because the sequencing and assembly strategies of the Human Genome Project were unable to accurately report regions as complex as the IGH locus. It was not until 2013 that a second complete sequence was reported [27]. There is substantial structural variation between the two reported sequences. The first reported haplotype is actually a mosaic of large-insert clones from three libraries, and it is missing at least 11 functional and open reading frame IGHV genes. The second haplotype includes an additional 101 kbp of sequence, with four structural variants involving ten IGHV genes (seven gains and three losses) [27]. This haplotype is now the reference sequence (GRCh38) for the human IGH locus; however, it is critical to point out that this haplotype itself lacks known functional IGHV genes, and is therefore also an incomplete representation of the locus.
In recent years, direct and indirect approaches have substantially increased our knowledge of IG allelic variation [28,29]. Many previously unreported polymorphisms are now known, and allele frequencies within the human population are becoming clearer [23]. We also better understand the functionality of allelic variants, and variation in their usage among individuals. For example, the VDJbase database clearly shows that the IGHV3–66*01 and IGHV3–66*02 alleles are expressed at relatively high frequency, but the IGHV3–66*03 allele is rarely utilized [23]. This has also been shown for other loci such as IGHV1–69 [6]. Such differences in utilization should not be assumed to only reflect differences in antibody selection within the individual, for genetic variants within regulatory regions can also exert control over V(D)J recombination. For example, polymorphism within Recombination Signal Sequences (RSS) has been shown to directly influence IG gene usage [30]. Such non-coding variants must explain data from repertoire studies showing marked variation in utilization frequencies amongst allelic variants that encode the same amino acid sequence [31].
Deletion polymorphisms within the IGH locus are also now well documented [16, 27, 31]. Deletions of as many as 15 consecutive functional IGHV genes (IGHV3–7 to IGHV4–28) have been reported [31]. Structural variation has even been reported in the IGHD locus, where a common deletion polymorphism involves six of the IGHD genes [31,32]. Substantial allelic variation and structural variation has also long been recognized in the heavy chain constant region gene locus [33], though the extent of deletion polymorphisms of IGHG and IGHA genes within the human population has been insufficiently documented. Taken together, it is clear that genetic variation in the IGH locus is common, and it is increasingly difficult to imagine that this would be without consequence for human health.
Antibody genes and disease susceptibility
A number of early reports described associations between genes of the IGH locus and antibody-associated autoimmune conditions [34–37]. The single report in this period described an association between immunoglobulin genes and infectious disease - a susceptibility to Haemophilus influenzae infection, linked to the *02 allelic variant of IGKV2D-29, a kappa light chain gene [38]. This allele includes two SNPs that distinguish it from the more protective IGKV2D-29*01 sequence, as well as having a difference in its RSS. For any individual carrying the allele, the RSS polymorphism results in a low utilization frequency in the kappa chain repertoire. This expression level may be an important contributing factor to the poor performance of this gene, for those individuals, in the context of H. influenzae infection. Although clonal selection and clonal expansion may ultimately lead to the development of a large clone of protective B cells, clonal selection involves stochastic processes. If antigen-specific B cells express surface Ig that is encoded by rarely-expressed genes, their numbers will be low within the naive B cell repertoire. The time that it takes for one or other of these cells to encounter antigen, and to be selected, will therefore usually be longer than is the case for more commonly-encoded specificities (Figure 2). This could have important implications for the kinetics of an infection, and for its resolution.
In recent years, new associations with both infectious and non-infectious diseases have been reported. Susceptibility to chronic lymphocytic leukemia was recently associated with the light chain IGLV3–21*01 gene [39], while protection from influenza has been associated with broadly neutralizing antibodies encoded by some but not all heavy chain IGHV1–69 alleles [6]. A phenylalanine residue within the CDR2 region of the IGHV1–69 heavy chain appears to be critical for this broad reactivity. The heavy chain products of seven of the nineteen known allelic variants of the gene lack this critical residue. It has been suggested that differing frequencies of these alleles within different human populations could ultimately contribute to variations in the efficacy of different influenza vaccines [6]. Another disease association is suggested by the power of the broadly-neutralizing VRC01 class of anti-HIV antibodies. These antibodies are partially encoded by specific alleles of the IGHV1–2 gene [7,38].
Only a handful of disease associations in IGHV have been noted by genome-wide association studies (GWAS) [41–44]. For example, six SNPs mapping to the IGHV locus were identified in a study of Kawasaki disease (KD) in a Han Chinese population [41], though the signals did not reach genome-wide significance and did not localize to specific gene segments. Nevertheless, this is a finding that warrants further investigation, particularly with the recent emergence of the KD-like, SARS-CoV2-associated Multisystem Inflammatory Syndrome in Children (MIS-C). A more recent GWAS of rheumatic heart disease in the South Pacific identified a susceptibility signal that maps to the IGHV4–61*02 allele [42].
We believe that other strong associations have not yet been found because the complexity of the immunoglobulin gene loci has made them challenging to interrogate effectively using standard high-throughput approaches, such as those employed by GWAS [16, 45]. For many commercially available genotyping arrays, SNP coverage is sparse and it is unclear how accurate imputation-based approaches are for the whole of the locus [16]. As a result, it is uncertain whether array-based approaches adequately represent the full extent of germline polymorphism in the IGH locus, including the complex patterns of shared and unshared SNPs within sets of allelic variants. It is the combination of SNPs within a gene that is probably critical for antibody function (see Figure 3). Haplotype diversity associated with the large structural variants in the locus are also unlikely to be effectively tagged by common SNPs present on microarrays [27].
Figure 3:
Ten SNPs define 11 reported IGHV1–69 alleles. Alleles shown are IMGT-named sequences that have adequate supporting evidence of their existence in VDJbase (https://www.vdjbase.org). The nucleotide positions of the SNPs are numbered according to the IMGT numbering system.
The study of IG and TCR genes has tended to focus on the identification of alleles, rather than SNPs. This allelic variation now should also be presented as a curated set of SNPs that can be used for GWAS studies and as annotations of the reference assembly. Such a curated set could extend to regions of the loci beyond the V, D and J genes themselves - which have received little attention to date. Little is known, for example, about genetic variation in the IGH constant genes, although these genes encode the Fc regions which interact with the antibody Fc receptors, and so determine signalling pathway responses. A recent study in Brazil identified 28 new IGHG alleles that encode the IgG1, IgG2 and IgG3 isotypes [46]. How these variants affect isotype function is yet to be determined. However, other studies have shown that allelic variants of the constant region genes can vary with respect to complement fixation, FcR binding and antibody-dependent cellular cytotoxicity [47, 48]. Complex patterns of N-glycosylation, which determine antibody functions, may also vary between allelic variants of constant region genes [49], and variation in IgG glycosylation levels have been observed between human populations [50].
Disease susceptibility and SARS-CoV2 infection
Although socio-economic factors, as well as age and comorbidities account for much of the worldwide variation in rates of SARS-CoV2 infection and severity of outcomes, genetic variation within the human population is almost certain to be a contributing factor. Many potential disease susceptibility genes have already been suggested. Likely contenders include the Angiotensin-converting enzyme 2 (ACE2) gene, the ABO blood group locus, genes regulating Toll-like receptor and complement pathways, and of course the MHC locus [51–54]. As has been true so often in the past, comprehensive reviews of potential disease susceptibility genes highlight MHC genes, but mention neither TCR nor IG genes. The general variability that has been reported in the antibody response to the SARS-CoV-2 virus, as well as the specific nexus between antibody genes, Kawasaki Disease and MIS-C, lead us to believe that antibody genes should be considered candidate susceptibility genes.
Antibody responses to the SARS-CoV-2 virus are still being defined, but are likely to range from disease enhancing antibodies, like those seen in response to the earlier SARS-CoV-1 virus [55], to broadly neutralizing antibodies, that are produced by some but not all of those who are infected [56,57]. Based on available data thus far, it appears that SARS-CoV-2 neutralizing IgG antibodies may develop outside the germinal centres [58]. These antibodies carry far fewer point mutations than are typically reported for IgG antibodies [59,60], emphasizing the importance of the germline. And the neutralizing antibody response is often convergent - that is, highly similar antibodies are seen in multiple individuals, as a consequence of a stereotypical response by naive B cells [59,60]. A number of these public clonotypes have been reported, and they use a limited number of the available germline genes. Broadly neutralizing antibody heavy chains have been reported to be encoded by IGHV1–69, IGHV3–30-3 and IGHV1–24 [61], as well as IGHV3–53 and IGHV3–66 [62,63]. Other studies have highlighted a role for the heavy chain genes IGHV3–9 and IGHV3–30-3 [59] and the heavy and light chain gene pairs IGHV1–58/IGKV3–20 and IGHV3–30/IGKV1–39 [64].
Many of the genes that encode these neutralizing antibodies are represented by extensive allelic variation, and can be deleted entirely from an individual genome. Although a variety of important genes and allelic variants have been identified, the loss of one or more of them could compromise the suite of antibodies that act together in any immune response. For example, the region of the locus encoding the IGHV3–9 gene has been shaped by a complex event. Some individuals carry chromosomes bearing the IGHV1–8 and IGHV3–9 genes, while others carry the IGHV5–10-1 and IGHV3–64D genes [23, 27]. The IGHV3–30-3 gene occupies the central part of another region that has been shaped by complex genetic events [16, 27]. Some or all of a group of seven genes, from IGHV3–30 to IGHV4–31 are frequently absent from the repertoire of available germline genes [23]. In view of the possible role of these genes in an effective antiSARS-CoV-2 response, their identification in different patients acquires some importance.
Structural variation is also seen in the IGHD region of the locus, and this can lead to an absence of other genes that are necessary for the generation of protective antibodies. IGHD gene usage has not yet been reported in the anti-SARS-CoV-2 response, but public clonotypes share IGHD genes in other viral infections. For example, the CDR3 regions of convergent anti-influenza antibodies are largely encoded by the IGHD5–5/5–18 gene sequence [65]. The IGHD5–5 gene is part of a common, large deletion polymorphism involving six consecutive genes (IGHD3–3 to IGHD2–8) [31, 32]. The possibility that IGHD genes are disease susceptibility genes should therefore be considered.
It has been suggested that the genetic basis for susceptibility to the SARS-CoV-2 virus could be explored by the whole genome sequencing of 500 or 1000 age- and gender-matched patients [66]. This approach and other large-scale initiatives [eg 67] have not yet found links to the IGH locus, and we believe they will have limited ability to find such links, because as previously outlined, routine whole genome short-read sequencing struggles to accurately and comprehensively document the IG loci.
We believe that high throughput sequencing of full-length V(D)J gene sequences offers a straightforward way to determine an immunoglobulin immunogenotype, and we recently established the VDJbase database to generate and document such genotypes and haplotypes from submitted datasets [23]. Dozens of repertoire studies are now being performed in search of therapeutic antibodies, and in support of the quest for a SARS-CoV-2 vaccine. In the years to come, it is likely that hundreds of these studies will be completed. They will better serve the research community if the genotyping and haplotyping data embedded within these datasets is extracted and shared. This will allow the research community to properly test whether or not different genotypes or haplotypes contribute differently to COVID disease protection. It will also provide data that will finally allow us to document the population genetics of immunoglobulin genes. This in turn will make it possible to investigate the contribution of antibody genes to susceptibility to all infectious and non-infectious antibody-associated diseases. It may also guide vaccine development. Many SARS-CoV-2 vaccines are in development, and different vaccines may be more or less effective in individuals with particular IG genotypes [68]. In time we can hope that personalized vaccines will be developed, in which immunogens are engineered to stimulate the best immune response they can, from the immunoglobulin genes that an individual has at hand [69].
Text Box:
Four steps to improve understanding of the role of immunoreceptor genes in disease responses
Define a simple standard representation of the immunohaplotype. A standardised format will facilitate consolidation of information from multiple studies, and eventual use in diagnostics.
Encourage the routine use of immunohaplotyping in full-length adaptive immune receptor repertoire (AIRR) studies. Many studies have been conducted to examine the response to disease. Recently, studies on the response to COVID-19 have been published by numerous groups (For an up-to-date list, see https://b-t.cr/t/publicly-available-covid-19-airr-seq-data-sets/849). Routine derivation of immunohaplotypes from AIRR-Seq data could open up fruitful areas of investigation for an individual study, and will also help to create a larger consolidated dataset of immunohaplotype variation.
Develop improved genotyping approaches for the immunoreceptor loci. Current arrays poorly represent known genes and polymorphisms. Denser coverage of these regions on commercial arrays as a means to better represent known variation would enable more accurate information to be collected in GWAS.
Benchmark approaches to infer immunohaplotypes from other (non-AIRR-Seq) sources. Some progress has been made in predicting HLA genotypes from widely available Next Generation Sequencing sources [70]. Predicting immunohaplotypes has many of the same challenges. Creating a benchmark framework would encourage innovation while raising awareness of the challenges.
Highlights.
Genetic information encoded in the immunoglobulin loci shapes the antibody response
Variations in antibody responses may affect outcomes of diseases such as COVID-19
Immunoglobulin locus complexity makes documenting germline polymorphism a challenge
Current SNP arrays fail to fully reflect variability in the heavy chain (IGH) locus
Immunoglobulin repertoire studies can document individual variation in the IGH locus
Acknowledgements:
We thank Yana Safonova and Oscar Rodriguez for their contributions to the production of Figures, and for thoughtful discussions centered around some of the content presented in this manuscript.
Funding:
G.Y. is partially supported by the Israel Science Foundation (ISF [832/16]). C.T.W. is partially supported by grants from the U.S. National Institutes of Health (R21AI142590, R24AI138963, P20GM135004).
Footnotes
Declaration of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wan QQ, Ye QF, Zhou JD. Mannose-binding lectin 2 and ficolin-2 gene polymorphisms influence the susceptibility to bloodstream infections in kidney transplant recipients. Transplant Proc 2013, 45:3289–3292. [DOI] [PubMed] [Google Scholar]
- 2.Archer NS, Nassif NT, O’Brien BA. Genetic variants of SLC11A1 are associated with both autoimmune and infectious diseases: systematic review and meta-analysis. Genes Immun 2015, 16:275–283. [DOI] [PubMed] [Google Scholar]
- 3.Naranjo-Galvis CA, de-la-Torre A, Mantilla-Muriel LE et al. Genetic Polymorphisms in Cytokine Genes in Colombian Patients with Ocular Toxoplasmosis. Infect Immun 2018, 86:04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elhanati Y, Sethna Z, Callan CG, Jr., Mora T, Walczak AM: Predicting the spectrum of TCR repertoire sharing with a data-driven model of recombination. Immunol Rev 2018, 284:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greiff V, Weber CR, Palme J, Bodenhofer U, Miho E, Menzel U, Reddy ST: Learning the High-Dimensional Immunogenomic Features That Predict Public and Private Antibody Repertoires. J Immunol 2017, 199:2985–2997. [DOI] [PubMed] [Google Scholar]
- 6.Avnir Y, Watson CT, Glanville J, Peterson, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH, et al.: IGHV1–69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep 2016, 6:20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yacoob C, Pancera M, Vigdorovich V, Oliver BG, Glenn JA, Feng J, Sather DN, McGuire AT, Stamatatos L et al. Differences in Allelic Frequency and CDRH3 Region Limit the Engagement of HIV Env Immunogens by Putative VRC01 Neutralizing Antibody Precursors. Cell Rep 2016, 17:1560–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matzaraki V, Kumar V, Wijmenga C, Zhernakova A: The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol 2017, 18:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, Desmarais C, Klinger M, Carlson CS, Hansen JA, et al. : Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 2017, 49:659–665. [DOI] [PubMed] [Google Scholar]
- 10.Massa C, Robins H, Desmarais C, Riemann D, Fahldieck C, Fornara P, Seliger B: Identification of patient-specific and tumor-shared T cell receptor sequences in renal cell carcinoma patients. Oncotarget 2017, 8:21212–21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri I, Berkowska MA, van den Akker EB, Teodosio C, Reinders MJT, van Dongen JJM: Population matched (PM) germline allelic variants of immunoglobulin (IG) loci: New pmIG database to better understand IG repertoire and selection processes in disease and vaccination. bioRxiv 2020:2020.2004.2009.033530. [Google Scholar]
- 12.Luo S, Yu JA, Li H, Song YS: Worldwide genetic variation of the IGHV and TRBV immune receptor gene families in humans. Life Science Alliance 2019, 2:04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Ceredig R, Seoighe C: A Database of Human Immune Receptor Alleles Recovered from Population Sequencing Data. J Immunol 2017, 198:22022210. [DOI] [PubMed] [Google Scholar]
- 14.Yu GY, Mate S, Garcia K, Ward MD, Brueggemann E, Hall M, Kenny T, Sanchez-Lockhart M, Lefranc MP, Palacios G: Cynomolgus macaque (Macaca fascicularis) immunoglobulin heavy chain locus description. Immunogenetics 2016, 68:417–428. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Bao Y, Wang H, Hu X, Zhao Z, Li N, Zhao Y: A preliminary analysis of the immunoglobulin genes in the African elephant (Loxodonta africana). PLoS ONE 2011, 6:e16889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodriguez OL, Gibson WS, Parks T, Emery M, Powell J, Strahl M, Deikus G, Auckland K, Eichler EE, Marasco WA, et al. : A novel framework for characterizing genomic haplotype diversity in the human immunoglobulin heavy chain locus. Front. Immunol doi: 10.3389/fimmu.2020.02136 (••) The completeness and accuracy of a new long read genomic sequencing approach for locus-wide IG genotyping is compared to short read-, array- and imputation-based methods.
- 17.Watson CT, Matsen FAt, Jackson KJL, Bashir A, Smith ML, Glanville J, Breden F, Kleinstein SH, Collins AM, Busse CE: Comment on “A Database of Human Immune Receptor Alleles Recovered from Population Sequencing Data”. J Immunol 2017, 198:3371–3373. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen SCA, Boyd SD: Human adaptive immune receptor repertoire analysis-Past, present, and future. Immunol Rev 2018, 284:9–23. [DOI] [PubMed] [Google Scholar]
- 19.Imkeller K, Wardemann H: Assessing human B cell repertoire diversity and convergence. Immunological Reviews 2018, 284:51–66. [DOI] [PubMed] [Google Scholar]
- 20.Ralph DK, Matsen FA: Consistency of VDJ Rearrangement and Substitution Parameters Enables Accurate B Cell Receptor Sequence Annotation. PLoS Comput Biol 2016, 12:e1004409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corcoran MM, Phad GE, Vazquez Bernat N, Stahl-Hennig C, Sumida N, Persson MA, Martin M, Karlsson Hedestam GB: Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 2016, 7:13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gadala-Maria D, Gidoni M, Marquez S, Vander Heiden JA, Kos JT, Watson CT, O’Connor KC, Yaari G, Kleinstein SH: Identification of Subject-Specific Immunoglobulin Alleles From Expressed Repertoire Sequencing Data. Front Immunol 2019, 10:129. (••) This paper introduces an improved framework to infer previously unreported IGH alleles, and a Bayesian approach for inferring immunoglobulin genotypes.
- 23. Omer A, Shemesh O, Peres A, Polak P, Shepherd AJ, Watson CT, Boyd SD, Collins AM, Lees W, Yaari G: VDJbase: an adaptive immune receptor genotype and haplotype database. Nucleic Acids Res 2020, 48:D1051–D1056. (•) This paper describes a database for the documentation of population-level variability in the IGH gene loci.
- 24.Looney TJ, Duose DY, Lowman G, Linch E, Hajjar J, Topacio-Hall D, Xu M, Zheng J, Alshawa A, Tapia C, et al. : Haplotype Analysis of the T-Cell Receptor Beta (TCRB) Locus by Long-amplicon TCRB Repertoire Sequencing. J Immunother Prec Oncol 2019, 2:137–143. [Google Scholar]
- 25.Peres A, Gidoni M, Polak P, Yaari G: RAbHIT: R Antibody Haplotype Inference Tool. Bioinformatics 2019, 35:4840–4842. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T: The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med 1998, 188:2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, Willsey AJ, Joy JB, Scott JK, Graves TA, et al.: Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet 2013, 92:530–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikocziova I, Gidoni M, Lindeman I, Peres A, Snir O, Yaari G, Sollid LM: Polymorphisms in human immunoglobulin heavy chain variable genes and their upstream regions. Nucleic Acids Res 2020, 48:5499–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlin M, Scheepers C, Corcoran M, Lees WD, Busse CE, Bagnara D, Thornqvist L, Burckert JP, Jackson KJL, Ralph D, et al. : Inferred Allelic Variants of Immunoglobulin Receptor Genes: A System for Their Evaluation, Documentation, and Naming. Front Immunol 2019, 10:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadel B, Tang A, Lugo G, Love V, Escuro G, Feeney AJ. Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J Immunol. 1998, 161:6068–6073. [PubMed] [Google Scholar]
- 31.Gidoni M, Snir O, Peres A, Polak P, Lindeman I, Mikocziova I, Sarna VK, Lundin KEA, Clouser C, Vigneault F, et al. : Mosaic deletion patterns of the human antibody heavy chain gene locus shown by Bayesian haplotyping. Nat Commun 2019, 10:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidd MJ, Chen Z, Wang Y, Jackson KJ, Zhang L, Boyd SD, Fire AZ, Tanaka MM, Gaeta BA, Collins AM: The inference of phased haplotypes for the immunoglobulin H chain V region gene loci by analysis of VDJ gene rearrangements. J Immunol 2012, 188:1333–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefranc MP, Lefranc G, de Lange G, Out TA, van den Broek PJ, van Nieuwkoop J, Radl J, Helal AN, Chaabani H, van Loghem E, et al. : Instability of the human immunoglobulin heavy chain constant region locus indicated by different inherited chromosomal deletions. Mol Biol Med 1983, 1:207–217. [PubMed] [Google Scholar]
- 34.Hashimoto LL, Walter MA, Cox DW, Ebers GC: Immunoglobulin heavy chain variable region polymorphisms and multiple sclerosis susceptibility. J Neuroimmunol 1993, 44:77–83. [DOI] [PubMed] [Google Scholar]
- 35.Vencovsky J, Zd’arsky E, Moyes SP, Hajeer A, Ruzickova S, Cimburek Z, Ollier WE, Maini RN, Mageed RA: Polymorphism in the immunoglobulin VH gene V1–69 affects susceptibility to rheumatoid arthritis in subjects lacking the HLA-DRB1 shared epitope. Rheumatology (Oxford) 2002, 41:401–410. [DOI] [PubMed] [Google Scholar]
- 36.Sawcer S, Jones HB, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow PN, Compston A: A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet 1996, 13:464–468. [DOI] [PubMed] [Google Scholar]
- 37.Field LL, Larsen Z, Pociot F, Nerup J, Tobias R, Bonnevie-Nielsen V: Evidence for a locus (IDDM16) in the immunoglobulin heavy chain region on chromosome 14q32.3 producing susceptibility to type 1 diabetes. Genes Immun 2002, 3:338–344. [DOI] [PubMed] [Google Scholar]
- 38.Feeney AJ, Atkinson MJ, Cowan MJ, Escuro G, Lugo G: A defective Vkappa A2 allele in Navajos which may play a role in increased susceptibility to haemophilus influenzae type b disease. J Clin Invest 1996, 97:2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maity PC, Bilal M, Koning MT, Maity PC, Bilal M, Koning MT, Young M, van Bergen CAM, Renna V, Nicolo A, Datta M, Gentner-Gobel E et al. IGLV3–21*01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling. Proc Natl Acad Sci USA 2020; 117:4320–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West AP, Jr., Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA 2012, 109:E2083–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai FJ, Lee YC, Chang JS, Huang LM, Huang FY, Chiu NC, Chen MR, Chi H, Lee YJ, Chang LC, et al. : Identification of novel susceptibility loci for Kawasaki Disease in a Han chinese population by a genome-wide association study. PLoS One 2011, 6:e16853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parks T, Mirabel MM, Kado J, Auckland K, Nowak J, Rautanen A, Mentzer AJ, Marijon E, Jouven X, Perman ML, et al. : Association between a common immunoglobulin heavy chain allele and rheumatic heart disease risk in Oceania. Nat Commun 2017, 8:14946. (••) GWAS demonstrating an association between carriage of the IGHV4–61*02 allele and development of rheumatic heart disease as a consequence of Streptococcus pyogenes infection.
- 43.Witoelar A, Rongve A, Almdahl IS, Ulstein ID, Engvig A, White LR, Selbaek G, Stordal E, Andersen F, Braekhus A, et al. : Meta-analysis of Alzheimer’s disease on 9,751 samples from Norway and IGAP study identifies four risk loci. Sci Rep 2018, 8:18088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekenberg C, Tang MH, Zucco AG, Murray DD, MacPherson CR, Hu X, Sherman BT, Losso MH, Wood R, Paredes R, et al. : Association Between Single-Nucleotide Polymorphisms in HLA Alleles and Human Immunodeficiency Virus Type 1 Viral Load in Demographically Diverse, Antiretroviral Therapy-Naive Participants From the Strategic Timing of AntiRetroviral Treatment Trial. J Infect Dis 2019, 220:1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson CT, Breden F: The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun 2012, 13:363–373. [DOI] [PubMed] [Google Scholar]
- 46. Calonga-Solis V, Malheiros D, Beltrame MH, Vargas LB, Dourado RM, Issler HC, Wassem R, Petzl-Erler ML, Augusto DG: Unveiling the Diversity of Immunoglobulin Heavy Constant Gamma (IGHG) Gene Segments in Brazilian Populations Reveals 28 Novel Alleles and Evidence of Gene Conversion and Natural Selection. Front Immunol 2019, 10:1161. (•) This study highlights the extent of previously unreported immunoglobulin gene variants in unstudied human populations.
- 47.Oxelius VA, Pandey JP: Human immunoglobulin constant heavy G chain (IGHG) (Fcγ) (GM) genes, defining innate variants of IgG molecules and B cells, have impact on disease and therapy. Clin Immunol 2013, 149: 475–486. [DOI] [PubMed] [Google Scholar]
- 48.Pandey JP: HIV-1 control and immunoglobulin genes. J Infect Dis 2018, 217:1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen X, Klaric L, Sharapov S, Mangino M, Ning Z, Wu D, Trbojevic-Akmacic I, Pucic-Bakovic M, Rudan I, Polasek O, et al.: Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation. Nat Commun 2017, 8:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW, Streeck H, Pau M, Schuitemaker H, Francis D et al. Antigen-Specific Antibody Glycosylation Is Regulated via Vaccination. PLoS Pathog. 2016, 12:e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debnath M, Banerjee M, Berk M: Genetic gateways to COVID-19 infection: Implications for risk, severity, and outcomes. FASEB J 2020, 34:8787–8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernandez J, Prati D, Baselli G, Asselta R, et al. : Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med 2020. doi: 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB: The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev 2020, 296:205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martinez-Colon GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, et al. : A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nature Med 2020, 26:1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, et al. : Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, et al. : Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 26:26. [DOI] [PubMed] [Google Scholar]
- 57.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, et al. : Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584:443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaneko N, Kuo H-H, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, Piechocka-Trocha A, Lefteri K, Osborn M, Bals J, et al. : Loss of Bcl-6expressing T follicular helper cells and germinal centers in COVID-19. Cell 2020, 183:143–157. (••) Postmortem samples show early loss of B and T cells and a lack of germinal centers in the lymph nodes and spleens of COVID-19 patients. This may provide a mechanistic explanation for the lack of somatic point mutations in responding B cells, and has implications for the development of long-lived memory B cells.
- 59. Nielsen SCA, Yang F, Jackson KJL, Hoh RA, Röltgen K, Stevens B, Lee J-Y, Rustagi A, Rogers AJ, Powell AE, et al.: Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe 2020, 28:516–525. (••) Changes in the repertoire of anti-SARS-CoV2 antibodies are tracked over time, with near-germline, convergent IgG responses against the receptor-binding domain of the viral spike protein being seen at high frequency within the study group.
- 60. Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, et al. : Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 15:15. (••) Potent neutralizing antibodies were generated from sorted IgG+ B cells, and were shown to generally be encoded by near-germline IG genes
- 61.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. : Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SI, Noh J, Kim S, Choi Y, Yoo DK, Lee Y, Lee H, Jung J, Kang CK, Song K-H, et al. : Stereotypic Neutralizing VH Clonotypes Against SARS-CoV-2 RBD in COVID-19 Patients and the Healthy Population. bioRxiv 2020:2020.2006.2026.174557. [Google Scholar]
- 63.Yuan M, Liu H, Wu NC, Lee C- CD, Zhu X, Zhao F, Huang D, Yu W, Hua Y, Tien H, et al. : Structural basis of a shared antibody response to SARS-CoV-2. Science 2020:eabd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, et al. : Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 18:18. (•) Levels of neutralizing plasma antibodies are highly variable in convalescing COVID-19 patients, but critical antibodies against the viral spike protein were detectable in all individuals tested.
- 65.Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Moody MA, Haynes BF, et al. : Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe 2014, 16:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Godri Pollitt KJ, Peccia J, Ko AI, Kaminski N, Dela Cruz CS, Nebert DW, Reichardt JKV, Thompson DC, Vasiliou V: COVID-19 vulnerability: the potential impact of genetic susceptibility and airborne transmission. Hum Genomics 2020, 14:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casanova JL, Su HC, Effort CHG: A Global Effort to Define the Human Genetics of Protective Immunity to SARS-CoV-2 Infection. Cell 2020, 181:1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watson CT, Glanville J, Marasco WA. The individual and population genetics of antibody immunity. Trends Immunol. 2017, 38:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Havenar-Daughton C, Abbott RK, Schief WR, Crotty S. When designing vaccines, consider the starting material: the human B cell repertoire. Curr Opin Immunol 2018, 53:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klasberg S, Surendranath V, Lange V, Schofl G: Bioinformatics Strategies, Challenges, and Opportunities for Next Generation Sequencing-Based HLA Genotyping. Transfus Med Hemother 2019, 46:312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]