ABSTRACT
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi sarcoma (KS), the plasmablastic form of multicentric Castleman’s disease, and primary effusion lymphoma. In sub-Saharan Africa, KS is the most common HIV-related malignancy and one of the most common childhood cancers. Immunosuppressed patients, including HIV-infected patients, are more prone to KSHV-associated disease. KSHV encodes a viral protein kinase (vPK) that is expressed from ORF36. KSHV vPK contributes to the optimal production of infectious viral progeny and upregulation of protein synthesis. To elucidate the interactions of vPK with cellular proteins in KSHV-infected cells, we used a bottom-up proteomics approach and identified host protein ubiquitin-specific peptidase 9X-linked (USP9X) as a potential interactor of vPK. Subsequently, we validated this interaction using a co-immunoprecipitation assay. We report that both the ubiquitin-like and the catalytic domains of USP9X are important for association with vPK. To uncover the biological relevance of the USP9X/vPK interaction, we investigated whether the knockdown of USP9X would modulate viral reactivation. Our data suggest that depletion of USP9X inhibits both viral reactivation and the production of infectious virions. Understanding how USP9X influences the reactivation of KSHV will provide insights into how cellular deubiquitinases regulate viral kinase activity and how viruses co-opt cellular deubiquitinases to propagate infection. Hence, characterizing the roles of USP9X and vPK during KSHV infection constitutes a first step toward identifying a potentially critical interaction that could be targeted by future therapeutics.
IMPORTANCE Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi sarcoma (KS), the plasmablastic form of multicentric Castleman’s disease, and primary effusion lymphoma. In sub-Saharan Africa, KS is the most common HIV-related malignancy. KSHV encodes a viral protein kinase (vPK) that aids viral replication. To elucidate the interactions of vPK with cellular proteins in KSHV-infected cells, we used an affinity purification approach and identified host protein ubiquitin-specific peptidase 9X-linked (USP9X) as a potential interactor of vPK. Depletion of USP9X inhibits both viral reactivation and the production of infectious virions. Overall, our data suggest a proviral role for USP9X.
KEYWORDS: gammaherpesvirus, KSHV, USP9X, viral protein kinase
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) establishes lifelong infection in humans and is associated with multiple human malignancies. KSHV is the etiological agent of Kaposi sarcoma (KS), as well as two lymphoproliferative disorders: multicentric Castleman’s disease (MCD), and primary effusion lymphoma (PEL) (1). KSHV has also been recently associated with osteosarcoma (2). These cancers are most often seen in the context of immunosuppression, and KS is the most prevalent HIV-associated malignancy (3, 4). KSHV-associated cancers can be aggressive, and for PEL, there is no current standard of care (5, 6). Hence, new treatments are urgently needed.
KSHV encodes viral kinases that may contribute to oncogenesis (7–10). Protein kinases are popular drug targets (11) and investigating how these viral kinases function and interact with host cell proteins may uncover potential drug targets for treating KSHV-associated cancers. KSHV encodes two kinases, ORF21 and ORF36, that aid in the viral life cycle (12). ORF21 is a thymidine kinase and ORF36, often referred to as viral protein kinase (vPK), is a serine-threonine kinase (13). KSHV vPK is considered a lytic gene but can also be expressed in the absence of lytic replication (13, 14) due to the fact the ORF36/vPK promoter contains a hypoxia-inducible factor (HIF) response element. Indeed, hypoxic conditions have been shown to upregulate vPK expression (15).
vPK is localized in both the cytoplasm and nucleus and displays limited homology to kinases present in other herpesviruses (16, 17). vPK can inhibit the IFN-β promoter activation and the production of IFN-β (18). It also induces the phosphorylation of c-Jun N-terminal kinase (JNK) (14) and TIP60, an acetyltransferase that is involved in the DNA damage response (DDR) (19). vPK is important for the optimal production of infectious viral progeny (20) but has also been implicated in the development of cancer (21–23).
Transgenic mice expressing vPK develop B-cell lymphomas at an 8-fold higher frequency compared to wild-type mice (22). vPK shares homology with cellular p70S6 kinase (S6K1), and can hijack cellular proteins to upregulate host processes, including chromatin remodeling, interferon signaling, angiogenesis, global protein synthesis, and the DNA damage response (21). For this reason, we initiated a study to identify cellular binding partners of vPK using immunoprecipitation followed by mass spectrometry (MS). We found that ubiquitin-specific peptidase 9X-linked (USP9X) binds vPK. We found that depletion of USP9X inhibits reactivation and infectious virion production.
RESULTS
KSHV vPK interacts with the Ubl-USP domains of USP9X.
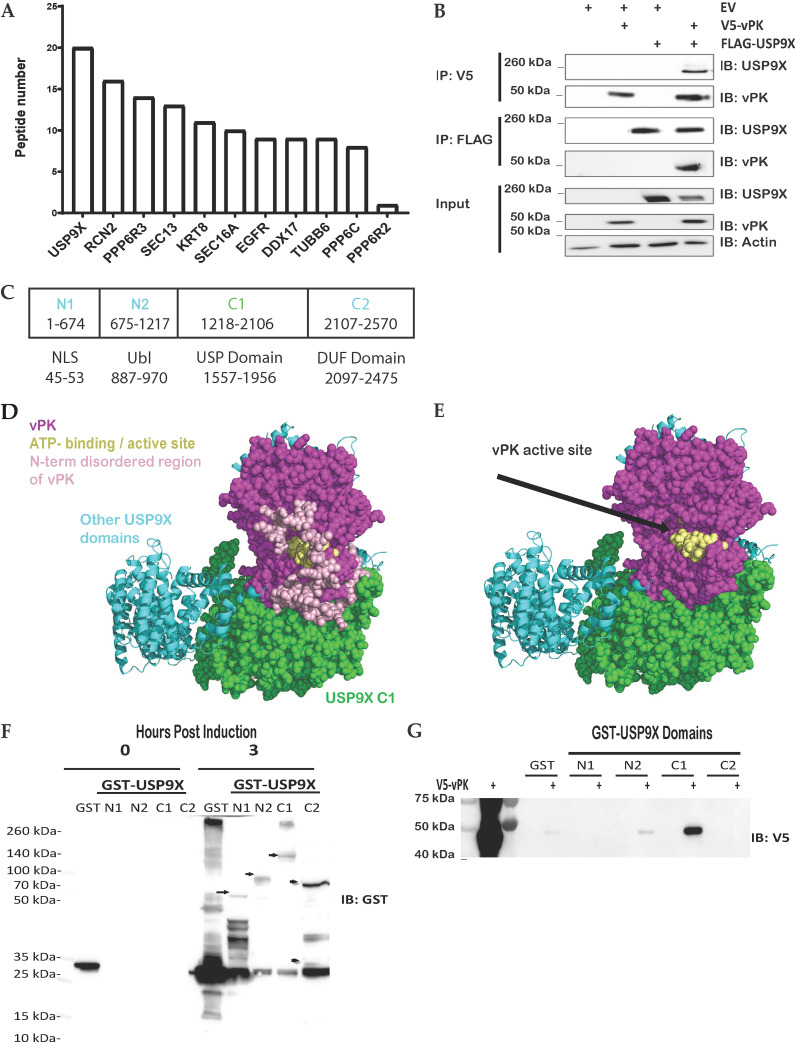
To investigate cellular binding partners of vPK, we performed immunoprecipitation (IP) followed by mass spectrometry (MS). In our MS-IP experiment, we identified ubiquitin-specific peptidase 9X (USP9X) as a high-confidence interacting partner (Fig. 1A). USP9X is a deubiquitinase (DUB) that cleaves the reversible posttranslational modifications of ubiquitin moieties from proteins (24, 25). Of the cellular proteins that interacted with vPK and not the GFP negative control, USP9X had the highest number of unique peptides (Fig. 1A). We confirmed the USP9X-vPK interaction by doing forward and reverse reciprocal co-immunoprecipitation (co-IPs) using V5 epitope-tagged vPK and FLAG epitope-tagged USP9X. By immunoprecipitating with the V5 antibody to pull down vPK and immunoblotting for USP9X, a band is observed only when both USP9X and vPK are present in the cell, indicating an interaction between the two proteins (Fig. 1B). Likewise, by immunoprecipitating with the FLAG antibody to pull down USP9X and immunoblotting for vPK, a band is observed only when both USP9X and vPK are present in the cell, again confirming an interaction between the two proteins (Fig. 1B).
FIG 1.
vPK interacts with USP9X. (A) Either FLAG-tagged GFP or FLAG-tagged vPK expressing HEK293 cell lysates were subjected to mass spectrometry after immunoprecipitation with an anti-FLAG antibody to pull down proteins interacting with vPK. Peptides shown are positive only for FLAG-vPK pull-down and were negative for FLAG-GFP pull down. (B) FLAG-tagged USP9X and V5-tagged vPK were transfected either individually or together into HEK293T cells for 72 h. Cell lysates were immunoprecipitated with anti-FLAG or anti-V5 antibodies and then immunoblotted with the indicated antibodies. (C) Schematic of the domains and truncated regions (N1, N2, C1, C2) of USP9X which includes a nuclear localization signal (NLS), a ubiquitin-like (Ubl) domain, a ubiquitin-specific protease (USP), and a domain of unknown function (DUF). (D) The AlphaFold prediction docking pose of vPK (magenta, and yellow spheres for the ATP binding site) places vPK near the C1 domain (green spheres) of USP9X. (E) The AlphaFold model of the C1 domain of USP9X was the same as in D except for the modeled disordered loops from N1. (F) Plasmids expressing GST alone (GST) and the indicated GST-USP9X truncation mutants were expressed in BL21Ai competent cells. Protein expression was induced using L-arabinose or IPTG (GST control) and purified via GST pulldown. Arrows show the size of the USP9X truncation mutants. (G) GST alone (GST) and the indicated GST-USP9X truncation mutants from panel F were incubated with equal amounts of lysates from 293T cells expressing V5-tagged vPK. After GST pulldown assays, the amount of bound V5-tagged vPK was analyzed by Western blotting. These experiments were performed with at least three biological replicates.
USP9X is a large protein that spans over 2,500 amino acids and contains several domains comprising a nuclear localization signal (NLS), a ubiquitin-like (Ubl) domain, a ubiquitin-specific protease (USP) which is necessary for the catalytic function of USP9X, and a domain of unknown function (DUF) (Fig. 1C). To predict where the USP9X-vPK protein-protein interaction (PPI) might occur, we used Protein Data Bank (PDB) file 7yxx chain A for USP9X and AlphaFold v2.2.0 (26) to predict the protein structure for vPK. Using ClusPro, we docked the two proteins. Within the top 10 hits for each scoring algorithm (balanced; electrostatic; hydrophobic; Van der Waals + electrostatic), four unique docking poses involved interfaces in the C1 domain. Of these four poses, only one docking pose lig.0009 was identified independently by two of the scoring algorithms (balanced and hydrophobic). The docking prediction places vPK in proximity to USP9X’s C1 domain (green spheres) (Fig. 1D). The docking model of the C1 domain of USP9X was the same as the crystal structure except for the modeled disordered loops from the N-terminus of vPK that are not shown in the docking structure (Fig. 1E). The computational modeling data suggest that vPK may interact with USP9X at the C1 (675-1217) regions (Fig. 1D and E).
To test our in silico predictions, we obtained plasmids expressing GST-tagged truncations of USP9X (27). We transformed BL21Ai competent cells to express and purify the GST-tagged USP9X truncations (Fig. 1F). The GST-tagged USP9X truncations were incubated with lysates from HEK293T cells expressing V5-tagged vPK (Fig. 1G). We found that the N2 and C1 truncations of USP9X were the only ones capable of binding vPK in line with our in-silico predictions (Fig. 1D and E).
vPK is subject to ubiquitination.
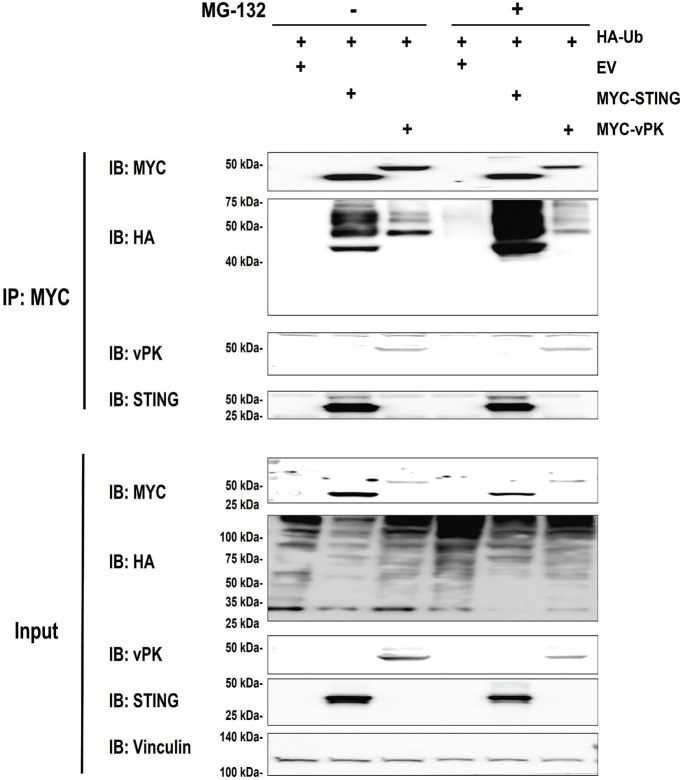
To further characterize the vPK-USP9X interaction, we sought to determine if vPK is ubiquitinated. The bioinformatics program Rapid UBIquitin (RUBI) was utilized to first predict if vPK could undergo ubiquitination. Of the many lysine sites in vPK, the program webserver predicted the lysine (K) at amino acid 392 with a high probability of being ubiquitinated. To determine whether vPK is ubiquitinated, we transfected a hemagglutinin (HA) epitope-tagged ubiquitin expression plasmid along with a MYC-tagged vPK plasmid in HEK293T cells. These cells were then treated with vehicle control (DMSO) or the proteasomal inhibitor MG-132 to prevent ubiquitin-mediated protein degradation. As the stimulator of IFN genes (STING; TMEM173) has been shown to undergo ubiquitination (28, 29), we utilized a plasmid encoding a MYC epitope-tagged STING as a positive control. We found in several independent experiments that MYC epitope-tagged vPK was ubiquitinated, although the ubiquitination of MYC epitope-tagged vPK was much weaker compared to that of MYC epitope-tagged STING (Fig. 2). To verify if the ubiquitination signal was proteasome-dependent, we treated the transfected cells with DMSO or MG132 overnight. As shown in Fig. 2 (right side), the ubiquitination of MYC epitope-tagged vPK did not increase when treated with MG132, unlike MYC epitope-tagged STING. Taken together, these data suggest that vPK is a target of proteasome-independent ubiquitination.
FIG 2.
vPK undergoes ubiquitination. Myc tagged-vPK or STING and HA-ubiquitin expression plasmids were transfected into HEK293T cells in the presence or absence of proteasome inhibitor, MG-132. Forty-eight hours posttransfection, Myc-tagged vPK or STING was immunoprecipitated and the immunoprecipitate was subjected to immunoblotting with the indicated antibodies. The experiments represent at least 3 biological replicates.
USP9X knockdown reduces viral reactivation.
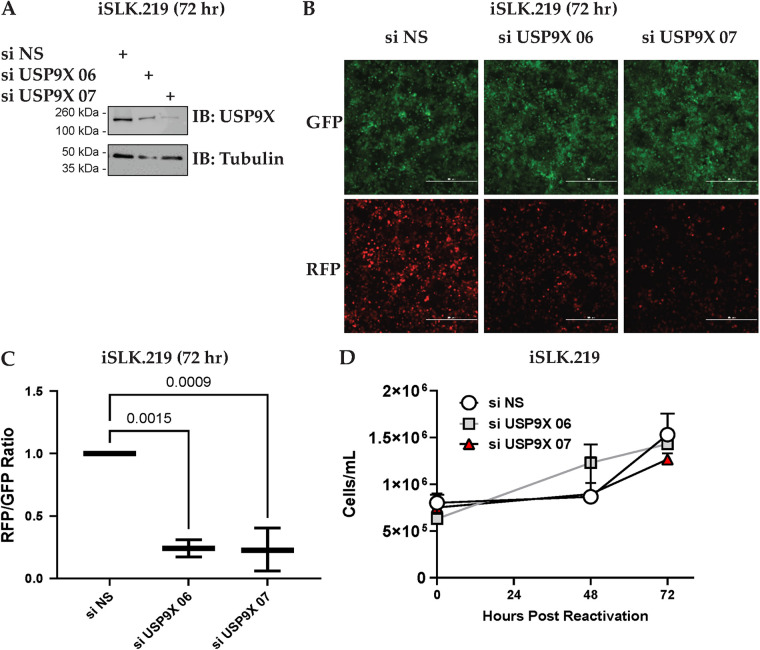
Given that vPK is a lytic protein in the KSHV life cycle (30), we next asked if USP9X has a role in the viral reactivation of KSHV. Like all herpesviruses, KSHV exhibits a dual-phasic life cycle consisting of a quiescent latent phase and a lytic reactivation phase when viral progeny are produced. iSLK.219 cells harbor latent recombinant KSHV.219 which contains the Elongation Factor 1 alpha (EF-1a) promoter that constitutively expresses green fluorescent protein (GFP) as well as the lytic PAN promoter that controls expression of red fluorescent protein (RFP) (31). These cells are induced into lytic reactivation and replication by treatment with doxycycline. We depleted USP9X expression using USP9X-specific siRNAs (Fig. 3A) and observed a reduction in viral reactivation (Fig. 3B and C). We confirmed that the observed decrease in GFP and RFP expression was not due to cell death caused by the knockdown of USP9X by measuring cell viability. We did not observe cytotoxicity in USP9X-depleted cells when we counted live cells using Trypan Blue exclusion (Fig. 3D). Overall, our data suggest that depletion of USP9X leads to decreased reactivation of KSHV.
FIG 3.
USP9X depletion reduces viral reactivation. (A) iSLK.219 cells were either transfected with nonspecific siRNA (si NS) or USP9X siRNAs for 48 h and then treated with 1 μg/mL doxycycline for 72 h to induce lytic reactivation. Cells were harvested 72 h postreactivation and USP9X knockdown was determined by Western blotting. (B) Fluorescent microscopy images representative of cells expressing GFP and RFP are shown at 72 h postreactivation. The scale bar represents 500 μm. (C) Images from panel B were subjected to quantification of intensities of GFP and RFP channels by LAS X software. (D) Total live iSLK.219 cells treated as in panel A, were counted by trypan blue exclusion assay to determine cytotoxicity upon USP9X knockdown. These experiments were performed with at least three biological replicates.
USP9X contributes to KSHV viral gene expression and protein expression.
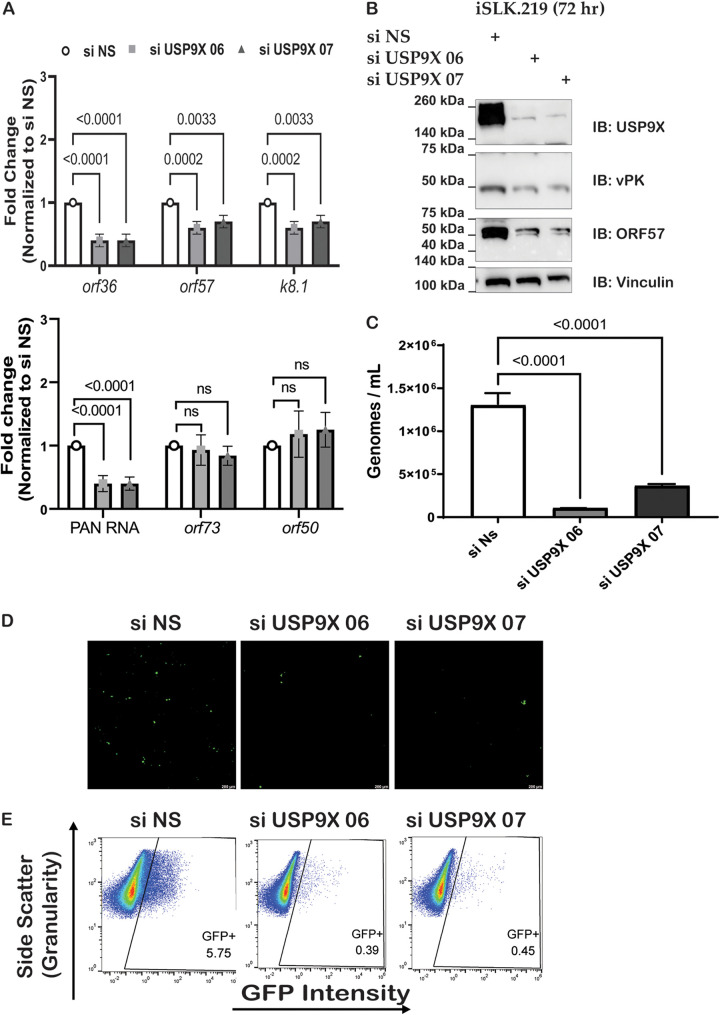
Given that the depletion of USP9X resulted in decreased reactivation in iSLK.219 cells, we next sought to determine if the knockdown of USP9X would reduce the expression of lytic viral genes. RNA was collected at 72 h postreactivation and RT-qPCR was used to quantify mRNA expression of lytic genes. We observed that orf36 (vpk), orf57, PAN RNA, and k8.1 transcript levels decreased while orf73 and orf50 remained unchanged when USP9X is depleted (Fig. 4A). To further test whether the depletion of USP9X affects the expression of KSHV lytic proteins, lysates were collected at 72 h postreactivation and immunoblotted for USP9X and lytic viral proteins. Concordant with the gene expression analysis, protein levels of vPK and ORF57 were decreased upon USP9X knockdown (Fig. 4B). These results indicate a potential role for USP9X in KSHV lytic replication.
FIG 4.
USP9X knockdown reduces viral replication and infectivity of KSHV. (A) siRNA-transfected iSLK.219 cells were harvested 72 h postreactivation and gene expression was quantified with RT-qPCR. (B) Protein expression was determined by Western blotting at 72 h postreactivation. (C) iSLK.219 cells were either transfected with nonspecific siRNA (si NS) or USP9X siRNAs (si USP9X) for 48 h and viral reactivation was induced with doxycycline. Supernatants were harvested at 72 h postreactivation and virions were quantified by qPCR. (D) Supernatants were harvested at 72 h postreactivation and transferred to naive HEK293T. Fluorescence microscopy of GFP-positive HEK 293T cells infected with KSHV from the experiment described in panel A was conducted. (E) Quantification of GFP-positive cells by flow cytometry. Data are presented as mean ± standard deviation from at least three independent experiments. Significance was determined using ANOVA with multiple comparisons.
USP9X impacts the production of infectious KSHV virions.
Given that a decrease in both viral lytic genes and lytic proteins was observed upon siRNA-mediated knockdown of USP9X, we next investigated whether the depletion of USP9X also led to a reduction in the viral genomes released in the cell supernatant. To address this, supernatants were harvested 72 h postreactivation and subjected to qPCR to quantify viral genomes. In the USP9X-depleted cells, the number of viral genomes was significantly fewer than that of the cells transfected with control siRNAs (Fig. 4C). These data suggest a positive relationship between USP9X and KSHV replication. Additionally, supernatants from iSLK.219 cells transfected with control or USP9X siRNAs were harvested at 72 h postreactivation and the total number of KSHV genomes in each sample was quantified by qPCR. Supernatants containing the same total number of genomes were added to uninfected HEK293T cells to examine infectious virions present in these supernatants. Forty-eight hours postinfection, GFP-positive cells were imaged by fluorescence microscopy and quantified by flow cytometry (Fig. 4D and E). Both fluorescence microscopy and flow cytometry analyses showed a decrease in the total number of GFP-positive HEK293T cells. These data suggest that USP9X is required for the efficient production of infectious KSHV virions.
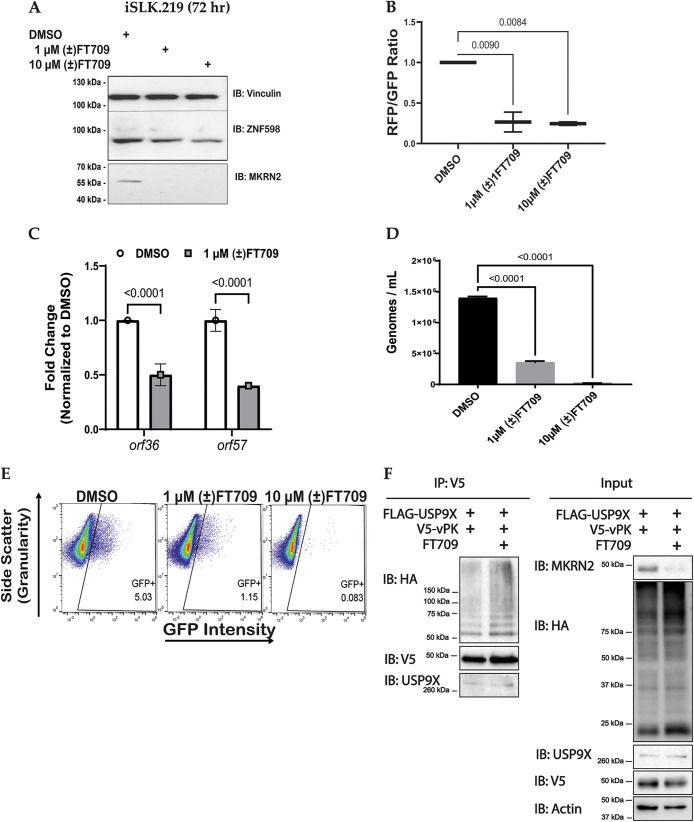
We next evaluated a recently published catalytic inhibitor for USP9X, (±)FT709 (32). USP9X has been shown to regulate Makorin (MKRN) proteins (32). Hence, we assessed the efficacy of (±)FT709 to inhibit the catalytic activity of USP9X by treating reactivated iSLK.219 cells with either DMSO control or increasing amounts of (±)FT709 and investigated protein levels of ZNF598 and MKRN2. As expected, we observed a decrease in ZNF598 and MKRN2 upon (±)FT709 treatment indicating inhibition of USP9X catalytic activity (Fig. 5A). We treated reactivated iSLK.219 cells with the USP9X inhibitor, (±) FT709, and observed a reduction in viral reactivation using RFP/GFP analysis (Fig. 5B). RNA was harvested at 72 h postreactivation, and RT-qPCR was used to quantify the mRNA expression of lytic genes. Similar to the siRNA knockdown data in Fig. 4A, we observed that orf36 (vpk) and orf57 gene expression decreased when USP9X catalytic activity is inhibited (Fig. 5C).
FIG 5.
USP9X catalytic inhibition reduces viral replication and infectivity of KSHV. (A) iSLK.219 cells were treated with DMSO or a catalytic inhibitor of USP9X ([±]FT709) for 24 h and then treated with 1 μg/mL doxycycline for 48 h to induce lytic reactivation. Protein expression was determined by Western blotting at 72 h postreactivation. (B) iSLK.219 cells were treated with DMSO or indicated amounts of a catalytic inhibitor of USP9X ([±]FT709) for 24 h and then treated with 1 μg/mL doxycycline for 48 h to induce lytic reactivation. Representative images of cells expressing GFP (constitutively), and RFP (during lytic reactivation) were collected and subjected to quantification of intensities of GFP and RFP channels by LAS X software. (C) iSLK.219 cells were harvested 48h postreactivation and gene expression was quantified with RT-qPCR. (D) iSLK.219 cells were treated with DMSO, 1 μM (±)FT709, or 10 μM (±)FT709 for 24 h and then treated with 1 μg/mL doxycycline for 48 h to induce lytic reactivation. Supernatants were harvested at 72 h postreactivation and virions were quantified by qPCR. (E) Quantification of GFP-positive cells by flow cytometry. Supernatants were harvested at 72 h postreactivation and transferred to naive HEK293T which were subjected to flow cytometry. Data are presented as mean ± standard deviation from at least three independent experiments. Significance was determined using ANOVA with multiple comparisons. (F) Immunoprecipitation of V5-tagged vPK after transfection of FLAG-tagged USP9X, V5-tagged vPK and HA-tagged ubiquitin into HEK293T cells with the addition of DMSO or 10 μM (±)FT709 24 h posttransfection. Samples were harvested at 24 h post-treatment with DMSO or FT709 and the experiment represents two biological replicates.
We next investigated whether USP9X inhibition decreases the number of viral genomes in the supernatant. To address this, supernatants were harvested 72 h postreactivation and subjected to qPCR to quantify viral genomes. Cells incubated with (±)FT709 displayed a significantly lower number of viral genomes compared to the DMSO control samples (Fig. 5D). These data suggest a positive relationship between USP9X activity and KSHV replication. Next, supernatants from cells incubated with (±)FT709 or DMSO control were harvested at 72 h postreactivation, normalized for the total number of genomes, and used to infect naive HEK293T cells. Forty-eight hours postinfection, GFP-positive cells were quantified by flow cytometry (Fig. 5E). The flow cytometry data showed a decrease in the total number of GFP-positive HEK293T cells, suggesting that there are fewer infectious virions produced when USP9X is inhibited by (±)FT709. Our data allude to a proviral role for USP9X catalytic activity in the production of infectious virions.
To test whether USP9X directly deubiquitinates vPK, we transfected HA-tagged ubiquitin, V5-tagged vPK and FLAG-tagged USP9X into HEK293T cells and treated them with DMSO or the USP9X inhibitor FT709 24 h posttransfection (Fig. 5F). Lysates were harvested at 24 h post-DMSO or FT709 treatment and immunoprecipitation was performed for V5-tagged vPK to assess its ubiquitination status. Interestingly, treatment with FT709 did not affect the ubiquitination of vPK while MKRN2 protein was reduced in the lysates of FT709-treated cells demonstrating that the inhibitor was functioning effectively (Fig. 5F). Thus, vPK is ubiquitinated and interacts with USP9X but USP9X does not appear to directly deubiquitinate vPK.
DISCUSSION
The viral kinases encoded by human herpesviruses play critical roles in the viral life cycle. The vPK encoded by KSHV has been shown to contribute to oncogenesis and optimal virus production. However, little is known about which cellular proteins the KSHV vPK protein interacts with. In this study, we aimed to identify cellular interactors of vPK.
We first identified cellular binding partners of vPK using immunoprecipitation (IP) followed by mass spectrometry (MS). The protein pulled down with the highest confidence was USP9X. The vPK homolog UL97 in human cytomegalovirus (HCMV) was also found to bind USP9X in an IP-MS experiment identifying multiple UL97 interacting proteins (33). Here, we confirm that the KSHV vPK protein can interact with cellular USP9X and that this interaction occurs within the catalytic USP (C1) and Ubl (N2) regions of USP9X.
Because USP9X is a deubiquitinase, we investigated whether vPK is ubiquitinated. In silico prediction and immunoprecipitation of vPK with ubiquitin demonstrates that vPK is subject to ubiquitination. However, ubiquitination does not necessarily mean degradation. Ubiquitination can be mono- or poly-ubiquitination which has been shown to either activate and/or stabilize proteins or change trafficking (34, 35). While we determined that vPK is ubiquitinated, we did not see a change in its total ubiquitination when USP9X activity was inhibited. However, it is also possible that USP9X does not remove the complete chain of ubiquitin from vPK or only targets specific ubiquitin linkages.
To determine if USP9X impacts the KSHV life cycle, we utilized siRNAs to deplete USP9X in cells infected with KSHV. We demonstrate that cells were attenuated significantly for viral reactivation when depleted for USP9X, suggesting that USP9X participates in enhancing lytic reactivation and/or replication. We measured the mRNA and protein expression of several KSHV lytic genes when USP9X expression was depleted and found that the expression of these genes was inhibited. In contrast to the control siRNA-transfected samples, USP9X depletion resulted in a significant decrease in infectious virions. Equivalent results were observed when the catalytic activity of USP9X was inhibited using (±)FT709 compared to the DMSO control. It has been shown that inhibition of USP9X leads to ER stress in malignant peripheral nerve sheath tumor (MPNST) cells (36). Hence, it is possible that USP9X may interact with vPK to facilitate lytic reactivation of KSHV by regulating ER stress. Our findings indicate that USP9X regulates vPK protein expression, as we observed a decrease in both mRNA and protein expression of vPK as well as another lytic protein ORF57 following siRNA-mediated knockdown of USP9X. vPK has been shown to phosphorylate and thereby activate the cellular c-Jun N-terminal kinase (JNK) and inhibition of JNK results in a decrease in viral protein K8.1 (14). In addition to optimal virion production, ORF57 is essential for production of several lytic proteins, including K8.1 (37), and vPK has been previously shown to bind to the promoter region of ORF57 (38). Thus, it could be conceivable that as the expression of vPK decreases due to USP9X depletion, there would be less vPK present to bind to the promoter and activate expression of ORF57 (39). Another possible explanation for the observed decrease in KSHV reactivation is that once vPK expression is decreased, vPK can no longer phosphorylate KAP-1, which is a protein that leads to a chromatin rearrangement and subsequent viral reactivation (40). Because USP9X can interact with vPK, it is possible that the impact on lytic gene expression and production of infectious virions may be due to vPK-mediated regulation of these genes. Future studies that investigate all of these possibilities would be of great significance to determine how USP9X affects vPK-mediated regulation of these lytic genes. Overall, these data indicate that USP9X may be important in multiple steps of the viral life cycle.
Interestingly, USP9X can serve either in an antiviral or proviral role depending on the virus studied. In HSV-1 replication, USP9X plays an antiviral role by negatively regulating infected cell polyprotein 0 (ICP0) expression (41). In human adenovirus (HAdV), USP9X has an antiviral role by indirectly preventing the interaction of HAdV protein pIIIa with cellular Ran-binding protein 2 (RNABP2) and nuclear shuttling for viral particle maturation and replication (42). However, USP9X can also be proviral, and its activity was demonstrated to be higher in Burkitt’s lymphoma (BL) and EBV-transformed lymphoblastoid cell lines (LCLs) compared to resting B lymphocytes (43). Notably, USP9X is frequently mutated in B-cell malignancies, and its activity is dysregulated in numerous cancers (44–48).
Here, we show a proviral role for USP9X in the life cycle of KSHV. We demonstrate that USP9X enhances KSHV reactivation, lytic gene and protein expression, and the release of viral progeny, as well as the infection of naive cells. Overall, our data suggest that inhibition of the vPK-USP9X interaction may represent a future therapeutic target for KSHV-driven malignancies.
MATERIALS AND METHODS
Mass spectrometry.
Twenty-million HEK293T cells were transfected with expression plasmids pcDNA3 containing epitope-tagged GFP or pcDNA3 containing epitope-tagged vPK for 48 h. Cell lysates were harvested on ice using NP-40 lysis buffer (0.1% NP-40, 150 mM NaCl, 50 mM Tris HCl, pH 8.0, 30 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, and one complete protease inhibitor tablet [Roche 11697498001] per 50 mL). The cell lysates were centrifuged at 16,000 × g for 10 min, and the Bradford assay (Bio-Rad 500-0006) determined the protein concentration. Equal amounts of protein were loaded on FLAG antibody-conjugated beads (Sigma-Aldrich F2426) and subjected to orbital rotation at 4°C overnight. The beads were washed twice with NP-40 lysis buffer and then washed twice with 50 mM NH4HCO3. An elution buffer consisting of 3×-FLAG peptide (Sigma-Aldrich F3290), 50 mM NH4HCO3 (pH 8.0), and 0.1% PPS Silent surfactant (Protein Discovery 21010) was used to elute the samples from the beads. The samples were then treated with 5 mM DTT (Sigma-Aldrich 43815) for 15 min at 60°C. The proteins were trypsinized at room temperature using the filter-aided sample prep (FASP) protein digestion protocol (Protein Discovery 44450), and tryptic peptides were separated by a nanoAquity UPLC system (Waters Corp.) on a 2-cm trapping column and a self-packed 25-cm analytical column (75-μm internal diameter; Michrom Magic C18 beads of 5.0-μm particle size, 100-Å pore size). The flow rate was 350 nl/min over a gradient of 1% buffer B (0.1% formic acid in acetonitrile) to 35% buffer B in 200 min. A full mass spectrum scan (300 to 2,000 mass/charge ratio [m/z]) was acquired in an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) at a resolution setting of 60,000. Collision-induced dissociation with the 15 most intense ions was used to acquire data-dependent tandem mass spectrometry (MS2). Sorcerer-SEQUEST server (Sage-N Research) was used to search all raw data against the human UniProtKB/Swiss-Prot sequence database. The search parameters used consisted of a precursor mass between 400 to 4,500 atomic mass units (amu), up to two missed cleavages, a precursor ion tolerance of 3 amu, semitryptic digestion, a static carbamidomethyl cysteine modification, combined with variable methionine oxidation. Protein prophet determined false discovery rates (FDRs) and a minimum protein probability cutoff was utilized, resulting in a 1% FDR.
Cell culture.
Human embryonic kidney-293T (HEK293T) cells were cultured using DMEM media supplemented with 10% heat-inactivated FBS, 1% Pen-Str (100 IU/mL penicillin G, 100 μg/mL streptomycin), and 2 mM l-glutamate. The iSLK.219 clone 10 cells that harbor latent rKSHV.219 and express RTA in a doxycycline-dependent manner were provided by Dr. Don Ganem and cultured in DMEM-supplemented with 10% heat-inactivated FBS, 1% Pen-Str, 250 μg/mL G418, 400 μg/mL hygromycin, 10 μg/mL puromycin. Supernatants from doxycycline-reactivated iSLK.219 cells transfected with NS or USP9X siRNAs were used to infect HEK293T cells. All cells were cultured at 37°C in 5% CO2.
Immunoprecipitation.
HEK293T cells were plated at 4.0 × 106 cells in 100 mm × 20 mm dishes (Corning 430167) and were co-transfected with empty vector (EV) plasmids containing FLAG epitope (p3XFLAG-CMV-10), FLAG epitope-tagged USP9X (p3XFLAG-CMV-10 USP9X), and V5 epitope-tagged vPK (pCDNA3.1), using X-tremeGENE 9 DNA transfection reagent (Roche 6365779001) according to the manufacturer’s protocol. Forty-eight hours later, cells were washed in cold PBS and lysed using RIPA lysis buffer with HALT Protease and Phosphatase Inhibitor Cocktail, EDTA-free (Thermo Fisher Scientific 78441). Cell lysates were clarified by centrifugation at 14,000 × g for 10 min. Lysates were quantified using BCA (Thermo Fisher Scientific 23227) and 1 mg of the lysates were subjected to pulldown for either FLAG or V5.
For the FLAG immunoprecipitation, 1 mg of lysates, 2 μL of normal rat IgG antibody (Santa Cruz sc-2026), and 40 μL of Protein A/G PLUS-Agarose (Santa Cruz sc-2003) were incubated for 1 h at 4°C with orbital rotation, then centrifuged at 2,600 rpm at 4°C. The supernatants were collected and added to 50 μL of washed FLAG magnetic beads (Thermo Fisher Scientific A36797) and agitated by rotation at 4°C overnight. Magnetic separation was used to collect supernatants containing FLAG-tagged proteins from the magnetic beads. The anti-FLAG magnetic beads were washed three times using RIPA buffer, then subjected to elution with Laemmli buffer and boiled for 10 min at 96°C, followed by SDS-PAGE and Western Blot.
For the V5 immunoprecipitation, 1 mg of lysates were incubated with 2 μL of normal mouse IgG antibody (Santa Cruz sc-2025) and 40 μL of protein A/G PLUS-Agarose beads (Santa Cruz sc-2003) for 1 h at 4°C with orbital rotation followed by centrifugation at 2,600 rpm at 4°C. Supernatants were collected and added to 50 μL of washed V5 magnetic beads (VWR M167-11) and agitated by rotation at 4°C overnight. The following day, magnetic separation was used to collect supernatants containing V5-tagged proteins from the magnetic beads. The anti-V5 magnetic beads were washed three times using RIPA buffer, then subjected to elution with Laemmli buffer and boiled for 10 min at 96°C. SDS-PAGE and western blot analysis followed.
Anti-USP9X (Abcam 180191, CST 14898S), anti-vPK (synthesized as [14]) antibodies were used at a dilution of 1:1,000. Loading controls consisted of Vinculin (E1E9V) HRP-Conjugate (Cell Signaling Technology 18799S) or α-Tubulin (11H10) HRP-Conjugate (Cell Signaling Technology 9099S) at a dilution of 1:5,000 and β-actin HRP-conjugate (Santa Cruz sc47778) at a dilution of 1:2,500. Bands were visualized by chemiluminescence.
Detection of ubiquitinated vPK in cells.
HEK293T cells were seeded at 60% confluence in 100 mm × 20 mm dishes (Corning 430167). Plasmids expressing 5 μg of MYC epitope-tagged STING or 5 μg MYC epitope-tagged vPK, were co-transfected with 5 μg HA epitope-tagged ubiquitin (Addgene 18712) using X-tremeGENE 9 (Roche 06365779001) as per the manufacturer's protocol. Forty-eight hours after transfection, the cells were treated with 10 μM the proteasome inhibitor MG-132 overnight. Seventy-two hours posttransfection, the cells were washed with PBS, harvested, and lysed using RIPA lysis buffer with HALT Protease and Phosphatase Inhibitor Cocktail, EDTA-free (Thermo Fisher Scientific 78441). Cell lysates were centrifuged at 14,000 × g for 10 min and Pierce BCA assays (Thermo Fisher Scientific 23227) were used to determine protein concentration. A total of 50 μg of the lysates were set aside for traditional immunoblot. Then, 1 mg of the equalized lysates for each condition were precleared with 2 μL normal mouse IgG antibody (Santa Cruz sc-2025) and 40 μL of protein A/G PLUS-Agarose beads (Santa Cruz sc-2003) for 1 h at 4°C with orbital rotation. MYC-tagged lysates were precleared by centrifugation at 2,600 rpm at 4°C, then added to 50 μL of washed Anti-c-Myc magnetic beads (Thermo Fisher Scientific, Pierce PI88842) with agitation by rotation at 4°C overnight. The next day, magnetic separation isolated MYC-tagged lysates from anti-c-Myc magnetic beads. The beads were washed three times using RIPA buffer, then subjected to a Laemmli elution and boiled for 10 min at 96°C. Proteins were electrophoresed on 10% SDS-PAGE gels and transferred onto a nitrocellulose membrane. Membranes were blocked for 1 h in 5% fat-free milk, followed by an overnight incubation at 4°C with the indicated antibodies. Primary antibodies used were an anti-MYC antibody (Cell Signaling Technology 2272), an anti-vPK antibody, an anti-STING/TMEM173 antibody (R&D Systems MAB7169), and anti-HA antibodies (Sigma-Aldrich 11867423001), all at a dilution of 1:1,000. Secondary antibodies used were HRP-linked anti-mouse IgG Antibody (Cell Signaling Technology 7076S) and HRP-linked anti-rabbit IgG Antibody (Cell Signaling Technology 7074S), both at a dilution of 1:5,000. Either Vinculin (E1E9V) HRP-Conjugate (Cell Signaling Technology 18799S) or α-Tubulin (11H10) HRP-Conjugate (Cell Signaling Technology) were used as loading controls at a dilution of 1:5,000. Bands were visualized by chemiluminescence.
Inhibition of USP9X using (±)FT709.
iSLK.219 cells (less than 4 weeks old at passage) were plated in duplicate at 2.0 × 105 cells per well into 6-well plates. iSLK.219 cells were treated for 24 h with DMSO (VWR 89139-666) or a catalytic inhibitor of USP9X, (±)FT709, at a concentration of either 1 μM or 10 μM diluted in DMSO. After the 24-h treatment, cells were then treated with doxycycline (dox) at 1 μg/mL for 48 h to induce lytic reactivation. Pierce BCA (Thermo Fisher Scientific 23227) assays were used to determine protein concentration and the protein expression was determined by Western blotting at 72 h postreactivation. The experiments were done with at least three biological replicates.
Immunoblot.
Cells were washed in cold PBS and then lysed on ice using RIPA lysis buffer that contained and HALT Protease and Phosphatase Inhibitor Cocktail, EDTA-free (Thermo Fisher Scientific 78441). The protein concentration of lysates were determined by Pierce BCA assays (Thermo Fisher Scientific 23227). Lysates were loaded with 4× Laemmli sample buffer (Bio-Rad) and resolved on 10% SDS-PAGE gels before being transferred onto a nitrocellulose membrane (Amersham). Membranes were blocked using 5% fat-free milk for 1 h before being incubated overnight at 4°C with the indicated antibodies. Anti-vPK was generated as described previously (38). Anti-USP9X (Abcam 180191, CST 14898S), DYKDDDDK (FLAG) (Cell Signaling Technology 86861S), KSHV ORF57 (Santa Cruz sc-135746), Anti-ZNF598 antibody (ab241092), Anti-MKRN2 antibody (ab72055), and anti-HA HRP-conjugate (CST 14031S) were used at a dilution of 1:1,000. V5 Tag Monoclonal Antibody HRP (Invitrogen R961-25) was used at a 1:5,000 dilution. Either Vinculin (E1E9V) HRP-Conjugate (Cell Signaling Technology 18799S) or α-Tubulin (11H10) HRP-Conjugate (Cell Signaling Technology) were used as loading controls at a dilution of 1:5,000 and β-actin HRP-conjugate (Santa Cruz sc-47778) was used as a loading control at a dilution of 1:2,500. Bands were visualized by chemiluminescence.
Synthesis of USP9X inhibitor.
A racemic version of FT709 ([±]FT709) was used in all experiments. (±)FT709 was synthesized according to published procedures as described in reference (32). All spectroscopic data matched that reported for the compound.
In silico modeling of the USP9X-vPK interaction.
The ClusPro protein-protein docking web server (https://cluspro.org/) (49–51) was used to predict an interface for vPK on the C1 fragment of USP9X. The structure of vPK used for the ClusPro docking was determined using AlphaFold v2.2.0 (26). The AlphaFold structural model included all 444 residues of vPK, including the residues at the N-terminus that were predicted to be disordered. In the modeled structure, the disordered residues were predicted to pack along the protein surface, lying over the kinase active site in a conformation that was potentially autoinhibitory. The structure of USP9X used for the docking was chain A of the 3.30 Å Cryo-EM structure 7yxx. An AlphaFold structural model was predicted for a monomer of USP9X, but there were no significant differences between the modeled structure and the Cryo-EM structure. Thirty docking poses were predicted for each of four different scoring algorithms (balanced; electrostatic; hydrophobic; Van der Waals + electrostatic). None of the top five docking poses of each scoring algorithm accessed an interface in the C1 domain of USP9X and these docking poses were not considered further. Within the top 6 to 10 hits for each scoring algorithm, four unique docking poses involved interfaces in the C1 domain. Of these four poses, only one docking pose was identified independently by two of the scoring algorithms (balanced and hydrophobic) and that docking pose is shown in Fig. 1D and E.
USP9X purification.
pDEST15 plasmids that contained GST-USP9X truncations were a gift from Dr. Qing Zhang’s lab and the pGEX-4T1 GST plasmid (GE Healthcare 28954549) was used as a negative control. The plasmids were transformed into BL21-AI competent cells onto LB-Amp plates. Individual colonies were picked and cultured in an LB medium containing ampicillin (100 mg/mL) overnight. Overnight cultures were diluted 1:100 into fresh LB medium containing ampicillin (100 mg/mL) and grown at 37°C until the OD600nm reached 0.8 to 1.0. The GST-USP9X truncation proteins were induced by adding L-arabinose to a final concentration of 0.2% or 1 mM IPTG (only the pGEX-4T1 control) and grown for 4 h at 37°C. The bacteria were pelleted by centrifugation at 5,000 × g for 10 min. The B-PER Complete Reagent (Thermo Fisher Scientific 89821) was warmed to room temperature and supplemented with 1 mM DTT (Sigma-Aldrich 43815) and HALT Protease and Phosphatase Inhibitor Cocktail, EDTA-free (Thermo Fisher Scientific 78441). Bacteria pellets were weighed and 5 mL of B-PER Complete supplemented reagent was added per gram of cell pellet and incubated for 15 min at room temperature with gentle rocking. Lysates were centrifuged for 20 min at 16,000 × g to separate soluble proteins from insoluble proteins. Lysates were quantified using BCA (Thermo Fisher Scientific 23227) and 2 mg Bait lysates were used for SDS-PAGE and 1 mg of the lysates were subjected to GST pulldown. 50 μL of Glutathione Magnetic Agarose beads (Pierce 78602) were washed five times in a 1:1 solution consisting of tris buffered saline (TBS) and B-PER Complete supplemented reagent. A total of 10 μg of precleared GST-tagged fusion protein (BAIT) was added to the washed beads and agitated by rotation at 4°C overnight. The following day, magnetic separation was used to collect supernatants containing FLAG-tagged proteins from the magnetic beads. The supernatants were set aside and used as “bait flow through.” The magnetic beads underwent five washes using the 1:1 wash buffer and were added to the Prey Protein (V5 epitope-tagged vPK) from Mammalian HEK293 cell lysate. The Prey Protein (V5 epitope-tagged vPK) from mammalian HEK293 cell lysates were prepared as follows: 4 × 106 HEK293T cells were seeded into a 10-cm dish and then transfected with 6 μg of V5-tagged vPK plasmid using X-tremeGENE 9 for 72 h. Cells were washed using cold PBS and lysed using 1 mL of B-PER complete lysis buffer and incubated on ice for approximately 30 min. Lysates were centrifuged at 13,000 rpm for 5 min to clarify the crude mammalian lysate. Supernatants were decanted into a separate microcentrifuge tube and labeled “mammalian prey lysate.” Mammalian cell lysates were quantified using BCA (Thermo Fisher Scientific 23227). Then, 1 mg of the mammalian cell lysates were added to the washed Glutathione magnetic beads containing the bait proteins and incubated at 4.0°C with gentle rocking motion on a rotating platform for at least 1 h. Magnetic separation was used to collect supernatants containing GST-tagged proteins from the magnetic beads “prey-flow-through” and set aside. The beads were then washed five times using the 1:1 wash solution and incubated with 40 μL of fresh 10 mM glutathione elution buffer with gentle rocking on a rotating platform to elute GST for 5 min. A total of 35 μL of 2× Laemmli buffer and 15 μL of RIPA buffer were added to the glutathione beads and were then heated for 10 min at 70°C then run on 10% SDS-PAGE gels and immunoblot analysis followed using antibodies for GST anti-GST (Santa Cruz sc-138) and anti-mouse IgG, HRP linked antibody (Cell Signaling Technology 7076S) as a secondary at a 1:5,000 dilution. V5 Tag conjugated to HRP monoclonal antibody (Invitrogen R961-25) was used at a 1:5,000 dilution.
Quantitative real-time PCR.
RNeasy Plus minikit (Qiagen) was used to extract total RNA from iSLK.219 cells and a SensiFAST cDNA synthesis kit (Bioline BIO-65054) was used for cDNA synthesis. Real-time qPCR was then run with SensiFAST Lo-roc SYBR (Bioline C755H96). The fold change was determined by normalizing the 2−ΔΔCT gene expression values of test samples to the 2−ΔΔCT actin gene expression values. Primers used are as follows:
Human usp9x Forward: TTG GCT ATG GTC ATG TTC GAG
Human usp9x Reverse: GGA TTA GGA ATG CTC TGA AGG G
Human actin Forward: AAG ACC TGT ACG CCA ACA CA
Human actin Reverse: AGT ACT TGC GCT CAG GAG GA
KSHV orf36 Forward: TGC GTC CTC TTC CAG TGT TA
KSHV orf36 Reverse: GTC AGC AGA GTG TAG CCC AA
KSHV orf57 Forward: TGG ACA TTA TGA AGG GCA TCC TA
KSHV orf57 Reverse: CGG GTT CGG ACA ATT GCT
KSHV k8.1 Forward: AAA GCG TCC AGG CCA CCA CAG A
KSHV k8.1 Reverse: GGC AGA AAA TGG CAC ACG GTT AC
KSHV orf50 Forward: GAG TCC GGC ACA CTG TAC C
KSHV orf50 Reverse: AAA CTG CCT GGG AAG TTA ACG
KSHV orf73 Forward: CTC CGG AAA GAT GTG ACC TT
KSHV orf73 Reverse: GGG ACT TCC AGG TAT AGG CA
KSHV PAN Forward: GCT CGC TGC TTG CCT TCT T
KSHV PAN Reverse: CCA AAA GCG ACG CAA TCA A
KSHV reactivation and fluorescence microscopy.
iSLK.219 cells (less than 4 weeks old at passage) were plated in duplicate at 2.0 × 105 cells per well into 6-well plates and transfected with nonspecific, NS control siRNA (Dharmacon D-001810-01-50) or USP9X siRNAs (Dharmacon J-006099-06-0010, J-006099-07-0010) using Lipofectamine RNAiMax (Thermo Fisher Scientific 13778150) as per the manufacturer’s protocol. At 48 h posttransfection, cell media was replaced with media containing complete DMEM and 1 μg/mL doxycycline for KSHV reactivation. Images for GFP and RFP were taken of iSLK.219 cells 72 h postreactivation using a Leica Dmi8 inverted microscope. Nine images were acquired from multiple sections. Images were subjected to quantification of intensities of GFP and RFP channels by LAS X software and then averaged for each sample. One representative image of each sample is shown. The experiments were done with at least three biological replicates.
Trypan blue exclusion assay.
iSLK.219 cells were plated in 24-well plates at a concentration of 1.6 × 105 cells/mL and transfected with nonspecific, NS control siRNA (Dharmacon D-001810-01-50) or USP9X siRNAs (Dharmacon J-006099-06-0010, J-006099-07-0010) using Lipofectamine RNAiMax (Thermo Fisher Scientific 13778150) according to the manufacturer’s protocol. At 48 h posttransfection, cells were replenished with media containing complete DMEM and 1 μg/mL doxycycline. Using a hematocytometer, the number of live cells was calculated by omitting the trypan blue-stained dead cells from the total number of manually counted cells. Each time point was collected in duplicate from at least three biological replicates.
KSHV genome quantification.
Virus supernatants were filtered through a filter (0.45-micron) and then treated with 100 units of TURBO DNase (Thermo Fisher Scientific AM2238) per mL supernatant at 37°C for 1 h. DNase was neutralized by adding 10 mM EDTA (Corning) and incubating at 70°C for 15 min. DNase-treated supernatants were then processed using a DNeasy blood and tissue kit (Qiagen) following the protocol for non-nucleated blood. KSHV genome copy numbers were then quantified by real-time qPCR and amplification of ORF39 (KSHV) in SensiFAST Lo-roc SYBR (Bioline C755H96) with a final primer concentration of 500 nM on a Quant Studio 6 Flex real-time PCR system. The standard curves were created using dilutions of pCDNA4/TO-ORF39-2XStrep. The experiments were done with three biological replicates.
KSHV production and infection.
Less than 4 weeks of passaged iSLK.219 cells were plated in duplicate at 2.0 × 105 cells per well into 6-well plates and transfected with nonspecific, NS control siRNA (Dharmacon D-001810-01-50) or USP9X siRNAs (Dharmacon J-006099-06-0010, J-006099-07-0010) using Lipofectamine RNAiMax (Thermo Fisher Scientific 13778150) according to the manufacturer’s protocol. The cells were replenished with complete DMEM containing 1 μg/mL doxycycline for KSHV reactivation at 48 h posttransfection. Supernatants were then collected and clarified by centrifugation at 2,300 × g for 5 min. Viral genomes were quantified by qPCR (as stated above under KSHV genome quantification) to ensure an equal number of viral genomes were present in each condition. Polybrene (8 μg/mL) was added to the clarified supernatants containing 500,000 virions, which were then overlaid on a monolayer of HEK293T cells at a confluence of 60% and spinoculated at 30°C and 3,000 × g for 90 min. Cells were imaged 48 h later using a Leica Dmi8 inverted microscope. Five images were acquired from multiple sections. Images were subjected to quantification of intensities of GFP channel by LAS X software and then averaged for each sample. One representative image of each sample is shown. The cells were then subjected to Trypsinization (Thermo Fisher Scientific), collected at 2,000 g, and resuspended in 1 mL of PBS for flow cytometry analysis using a MACS-Quant VYB. GFP-positive cell populations were gated based on negative-control samples using FlowJo software. To obtain P-values, relative sizes of cell populations were compared statistically using a 2-tailed homoscedastic t test. The experiments were done with 3 biological replicates.
Statistical analysis.
GraphPad Prism 9 was used to perform all statistical tests and P values <0.05 are considered significant.
ACKNOWLEDGMENTS
We thank Luke Peterson for the plasmids FLAG epitope (p3XFLAG-CMV-10), and FLAG epitope-tagged USP9X (p3XFLAG-CMV-10 USP9X). We thank Qing Zhang for GST-USP9X truncation plasmids. Many thanks to Britt Glaunsinger for providing the pCDNA4/TO-ORF39-2XStrep for the real-time qPCR analysis. Many thanks to members of the Damania lab, especially Maria C. White for insightful discussions. This work was supported by National Institutes of Health (NIH) grants CA019014, CA096500, CA239583, and CA163217 to B.D. D.L.C. is supported by the Caroline H. and Thomas S. Royster Fellowship from the University of North Carolina at Chapel Hill, the Burroughs Wellcome Fund, and an NCI diversity supplement to CA096500. B.D. is a Leukemia and Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in Infectious Disease.
We declare no conflict of interest.
Contributor Information
Blossom Damania, Email: blossom_damania@med.unc.edu.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Wen KW, Wang L, Menke JR, Damania B. 2021. Cancers associated with human gammaherpesviruses. FEBS j 10.1111/febs.16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Q, Chen J, Li Y, Liu D, Zeng Y, Tian Z, Yunus A, Yang Y, Lu J, Song X, Yuan Y. 2021. Kaposi's sarcoma herpesvirus is associated with osteosarcoma in Xinjiang populations. Proc Natl Acad Sci USA 118. 10.1073/pnas.2016653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung MC, Pantanowitz L, Dezube BJ. 2005. AIDS-related malignancies: emerging challenges in the era of highly active antiretroviral therapy. Oncologist 10:412–426. 10.1634/theoncologist.10-6-412. [DOI] [PubMed] [Google Scholar]
- 4.Thakker S, Verma SC. 2016. Co-infections and pathogenesis of KSHV-associated malignancies. Front Microbiol 7. 10.3389/fmicb.2016.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coen N, Duraffour S, Snoeck R, Andrei G. 2014. KSHV targeted therapy: an update on inhibitors of viral lytic replication. Viruses 6:4731–4759. 10.3390/v6114731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uldrick TS, Whitby D. 2011. Update on KSHV-epidemiology, Kaposi sarcoma pathogenesis, and treatment of Kaposi sarcoma. Cancer Lett 305:150–162. 10.1016/j.canlet.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manners O, Murphy JC, Coleman A, Hughes DJ, Whitehouse A. 2018. Contribution of the KSHV and EBV lytic cycles to tumourigenesis. Curr Opin Virol 32:60–70. 10.1016/j.coviro.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore PS. 2007. KSHV manipulation of the cell cycle and apoptosis. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis, p 540–558. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 9.Cirone M. 2018. EBV and KSHV infection dysregulates autophagy to optimize viral replication, prevent immune recognition and promote tumorigenesis. Viruses 10. 10.3390/v10110599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haley CT, Mui UN, Vangipuram R, Rady PL, Tyring SK. 2018. Human oncoviruses: mucocutaneous manifestations, pathogenesis, therapeutics, and prevention (part i: papillomaviruses and Merkel cell polyomavirus). J the American Academy of Dermatology 81:1–21. 10.1016/j.jaad.2018.09.062. [DOI] [PubMed] [Google Scholar]
- 11.Roskoski R, Jr. 2015. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res 100:1–23. 10.1016/j.phrs.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Purushothaman P, Uppal T, Verma S. 2015. Molecular Biology of KSHV lytic reactivation. Viruses 7:116–153. 10.3390/v7010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Lee D, Seo T, Chung J, Choe J. 2000. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 36 protein is a serine protein kinase. J Gen Virol 81:1067–1071. 10.1099/0022-1317-81-4-1067. [DOI] [PubMed] [Google Scholar]
- 14.Hamza MS, Reyes RA, Izumiya Y, Wisdom R, Kung H-J, Luciw PA. 2004. ORF36 protein kinase of Kaposi's sarcoma herpesvirus activates the c-Jun N-terminal kinase signaling pathway. J Biol Chem 279:38325–38330. 10.1074/jbc.M400964200. [DOI] [PubMed] [Google Scholar]
- 15.Davis DA, Singer KE, Reynolds IP, Haque M, Yarchoan R. 2007. Hypoxia enhances the phosphorylation and cytotoxicity of ganciclovir and zidovudine in Kaposi's sarcoma-associated herpesvirus infected cells. Cancer Res 67:7003–7010. 10.1158/0008-5472.CAN-07-0939. [DOI] [PubMed] [Google Scholar]
- 16.Gershburg E, Pagano JS. 2008. Conserved herpesvirus protein kinases. Biochim Biophys Acta 1784:203–212. 10.1016/j.bbapap.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuny CV, Chinchilla K, Culbertson MR, Kalejta RF. 2010. Cyclin-dependent kinase-like function is shared by the beta- and gamma- subset of the conserved herpesvirus protein kinases. PLoS Pathog 6:e1001092. 10.1371/journal.ppat.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang S, Kim KS, Flano E, Wu TT, Tong LM, Park AN, Song MJ, Sanchez DJ, O'Connell RM, Cheng G, Sun R. 2009. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5:166–178. 10.1016/j.chom.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Zhu J, Xie Z, Liao G, Liu J, Chen MR, Hu S, Woodard C, Lin J, Taverna SD, Desai P, Ambinder RF, Hayward GS, Qian J, Zhu H, Hayward SD. 2011. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10:390–400. https://pubmed.ncbi.nlm.nih.gov/22018239/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avey D, Tepper S, Pifer B, Bahga A, Williams H, Gillen J, Li W, Ogden S, Zhu F. 2016. Discovery of a coregulatory interaction between Kaposi's sarcoma-associated herpesvirus ORF45 and the viral protein kinase ORF36. J Virol 90:5953–5964. 10.1128/JVI.00516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt AP, Wong JP, Weinberg MS, Host KM, Giffin LC, Buijnink J, van Dijk E, Izumiya Y, Kung HJ, Temple BR, Damania B. 2016. A viral kinase mimics S6 kinase to enhance cell proliferation. Proc Natl Acad Sci USA 113:7876–7881. 10.1073/pnas.1600587113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders PM, Montgomery ND, Montgomery SA, Bhatt AP, Dittmer DP, Damania B. 2018. Human herpesvirus-encoded kinase induces B cell lymphomas in vivo. J Clin Invest 128:2519–2534. 10.1172/JCI97053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uldrick TS, Polizzotto MN, Aleman K, O'Mahony D, Wyvill KM, Wang V, Marshall V, Pittaluga S, Steinberg SM, Tosato G, Whitby D, Little RF, Yarchoan R. 2011. High-dose zidovudine plus valganciclovir for Kaposi sarcoma herpesvirus-associated multicentric Castleman disease: a pilot study of virus-activated cytotoxic therapy. Blood 117:6977–6986. 10.1182/blood-2010-11-317610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mevissen TET, Komander D. 2017. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86:159–192. 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 25.Amerik AY, Hochstrasser M. 2004. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695:189–207. 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X, Zhai B, Koivunen P, Shin SJ, Lu G, Liu J, Geisen C, Chakraborty AA, Moslehi JJ, Smalley DM, Wei X, Chen X, Chen Z, Beres JM, Zhang J, Tsao JL, Brenner MC, Zhang Y, Fan C, DePinho RA, Paik J, Gygi SP, Kaelin WG, Zhang Q. 2014. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by blocking its interaction with the USP9x deubiquitinase. Genes Dev 28:1429–1444. 10.1101/gad.242131.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni G, Konno H, Barber GN. 2017. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci Immunol 2. 10.1126/sciimmunol.aah7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinngrebe J, Montinaro A, Peltzer N, Walczak H. 2014. Ubiquitin in the immune system. EMBO Rep 15:28–45. 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haque M, Wang V, Davis DA, Zheng ZM, Yarchoan R. 2006. Genetic organization and hypoxic activation of the Kaposi's sarcoma-associated herpesvirus ORF34-37 gene cluster. J Virol 80:7037–7051. 10.1128/JVI.00553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174:12–21. 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clancy A, Heride C, Pinto-Fernandez A, Elcocks H, Kallinos A, Kayser-Bricker KJ, Wang W, Smith V, Davis S, Fessler S, McKinnon C, Katz M, Hammonds T, Jones NP, O'Connell J, Follows B, Mischke S, Caravella JA, Ioannidis S, Dinsmore C, Kim S, Behrens A, Komander D, Kessler BM, Urbe S, Clague MJ. 2021. The deubiquitylase USP9X controls ribosomal stalling. J Cell Biol 220. 10.1083/jcb.202004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwahori S, Umaña AC, Kalejta RF, Murata T. 2022. Serine 13 of the human cytomegalovirus viral cyclin-dependent kinase UL97 is required for regulatory protein 14–3-3 binding and UL97 stability. J Biol Chem 10.1016/j.jbc.2022.102513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foot N, Henshall T, Kumar S. 2017. Ubiquitination and the regulation of membrane proteins. Physiol Rev 97:253–281. 10.1152/physrev.00012.2016. [DOI] [PubMed] [Google Scholar]
- 35.Tracz M, Bialek W. 2021. Beyond K48 and K63: non-canonical protein ubiquitination. Cell Mol Biol Lett 26:1. 10.1186/s11658-020-00245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianchetti E, Bates SJ, Carroll SL, Siegelin MD, Roth KA. 2018. Usp9X regulates cell death in malignant peripheral nerve sheath tumors. Sci Rep 8:17390. 10.1038/s41598-018-35806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J Virol 81:1062–1071. 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izumiya Y, Izumiya C, Geelen AV, Wang D-H, Lam KS, Luciw PA, Kung H-J. 2007. Kaposi's sarcoma-associated herpesvirus-encoded protein kinase and its interaction with K-bZIP. J Virol 81:1072–1082. 10.1128/JVI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han Z, Swaminathan S. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J Virol 80:5251–5260. 10.1128/JVI.02570-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang P-C, Fitzgerald LD, Geelen AV, Izumiya Y, Ellison TJ, Wang D-H, Ann DK, Luciw PA, Kung H-J. 2009. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res 69:5681–5689. 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato Y, Kato A, Arii J, Koyanagi N, Kozuka-Hata H, Oyama M, Kawaguchi Y. 2016. Ubiquitin-specific protease 9X in host cells interacts with herpes simplex virus 1 ICP0. J Vet Med Sci 78:405–410. 10.1292/jvms.15-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ismail AM, Saha A, Lee JS, Painter DF, Chen Y, Singh G, Condezo GN, Chodosh J, San Martín C, Rajaiya J. 2022. RANBP2 and USP9x regulate nuclear import of adenovirus minor coat protein IIIa. PLoS Pathog 18:e1010588. 10.1371/journal.ppat.1010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG. 2004. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci USA 101:2253–2258. 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murtaza M, Jolly LA, Gecz J, Wood SA. 2015. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci 72:2075–2089. 10.1007/s00018-015-1851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng J, Hu Q, Liu W, He X, Cui L, Chen X, Yang M, Liu H, Wei W, Liu S, Wang H. 2013. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn Pathol 8:177. 10.1186/1746-1596-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Song N, Liu L, Liu X, Ding X, Song X, Yang S, Shan L, Zhou X, Su D, Wang Y, Zhang Q, Cao C, Ma S, Yu N, Yang F, Wang Y, Yao Z, Shang Y, Shi L. 2017. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat Commun 8:14866. 10.1038/ncomms14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox JL, Wilder PJ, Gilmore JM, Wuebben EL, Washburn MP, Rizzino A. 2013. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. PLoS One 8:e62857. 10.1371/journal.pone.0062857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris DR, Mims A, Bunz F. 2012. Genetic disruption of USP9X sensitizes colorectal cancer cells to 5-fluorouracil. Cancer Biol Ther 13:1319–1324. 10.4161/cbt.21792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. 2020. Performance and its limits in rigid body protein-protein docking. Structure 28:1071–1081.e3. 10.1016/j.str.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Kozakov D. 2017. New additions to the ClusPro server motivated by CAPRI. Proteins 85:435–444. 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. 2017. The ClusPro web server for protein-protein docking. Nat Protoc 12:255–278. 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]