ABSTRACT
Bats are reservoirs for diverse coronaviruses, including swine acute diarrhea syndrome coronavirus (SADS-CoV). SADS-CoV has been reported to have broad cell tropism and inherent potential to cross host species barriers for dissemination. We rescued synthetic wild-type SADS-CoV using one-step assembly of a viral cDNA clone by homologous recombination in yeast. Furthermore, we characterized SADS-CoV replication in vitro and in neonatal mice. We found that SADS-CoV caused severe watery diarrhea, weight loss, and a 100% fatality rate in 7- and 14-day-old mice after intracerebral infection. We also detected SADS-CoV-specific N protein in the brain, lungs, spleen, and intestines of infected mice. Furthermore, SADS-CoV infection triggers excessive cytokine expression that encompasses a broad array of proinflammatory mediators, including interleukin 1β (IL-1β), IL-6, IL-8, tumor necrosis factor alpha (TNF-α), C-X-C motif chemokine ligand 10 (CXCL10), interferon beta (IFN-β), IFN-γ, and IFN-λ3. This study highlights the importance of identifying neonatal mice as a model for developing vaccines or antiviral drugs against SADS-CoV infection.
IMPORTANCE SADS-CoV is the documented spillover of a bat coronavirus that causes severe disease in pigs. Pigs are in frequent contact with both humans and other animals and theoretically possess a greater chance, compared to many other species, of promoting cross-species viral transmission. SADS-CoV has been reported to have broad cell tropism and inherent potential to cross host species barriers for dissemination. Animal models are an essential feature of the vaccine design toolkit. Compared with neonatal piglets, the mouse is small, making it an economical choice for animal models for SADS-CoV vaccine design. This study showed the pathology of neonatal mice infected with SADS-CoV, which should be very useful for vaccine and antiviral studies.
KEYWORDS: cross-species infection, SADS-CoV, swine coronavirus
INTRODUCTION
Over 70% of emerging infectious diseases are zoonotic, with some causing death in both humans and intermediate animal hosts (1). Many emerging infectious diseases are caused by viruses originating from bats, such as Ebola virus, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), Nipah virus (NiV), Hendra virus (HeV), and SARS-CoV-2 (2). Direct transmission of these viruses from bats to humans is rare because bats are phylogenetically distant from human. Intermediate hosts can bridge the transmission of these viruses from bats to humans (3). The coronavirus disease 2019 (COVID-19) pandemic has highlighted the importance of identifying intermediate hosts for SARS-CoV-2 to reduce virus transmission. Characterization of the susceptibility of animal species to infection with viruses is the first step toward identifying potential intermediate hosts. Blocking of intermediate hosts as reservoirs for viruses has crucial implications for global health.
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a newly discovered bat-origin coronavirus with fatal pathogenicity for neonatal piglets (4, 5). There is no vaccine to prevent SADS-CoV infection or clinically approved drug targeting SADS-CoV. SADS-CoV belongs to the genus Alphacoronavirus of the family Coronaviridae, with a single-stranded positive-sense RNA genome of about 27 kb (6). SADS-CoV has been reported to have broad cell tropism, including primary human lung and intestinal cells (5). Efficient growth in primary human cells suggests that SADS-CoV might be an emerging coronavirus at high risk for widespread dissemination and transmission in humans and animal species. Could SADS-CoV infect and be transmitted to neonatal mice? Toward vaccine and antiviral drug development to prevent the potential threats of SADS-CoV, can neonatal mice be used as the model for evaluation? To address these questions, we evaluated the susceptibility of the neonatal mouse model to SADS-CoV.
The goal of our study was to evaluate neonatal mice for SADS-CoV replication. We rescued synthetic wild-type SADS-CoV using a one-step assembly of a viral cDNA clone by homologous recombination in yeast. These viruses were used to investigate viral replication in human hepatoma (Huh7) cells and neonatal mice via different infection routes. Here, we demonstrated that SADS-CoV replicated efficiently and caused severe watery diarrhea, weight loss, and a fatality rate of 100% in 7- and 14-day-old mice after intracerebral (i.c.) infection. The SADS-CoV nucleocapsid protein was examined in multiple tissues of infected neonatal mice. In addition, levels of a broad array of proinflammatory mediators, including interleukin 1β (IL-1β), IL-6, IL-8, tumor necrosis factor alpha (TNF-α), C-X-C motif chemokine ligand 10 (CXCL10), interferon beta (IFN-β), IFN-γ, and IFN-λ3, were dramatically increased, indicating that inflammation occurs during SADS-CoV lethal infection in neonatal mice. Our results showed the pathology of neonatal mice infected with SADS-CoV, which should be very useful for vaccine and antiviral studies.
RESULTS
Recovery and construction of a SADS-CoV cDNA clone using one-step assembly by homologous recombination in yeast.
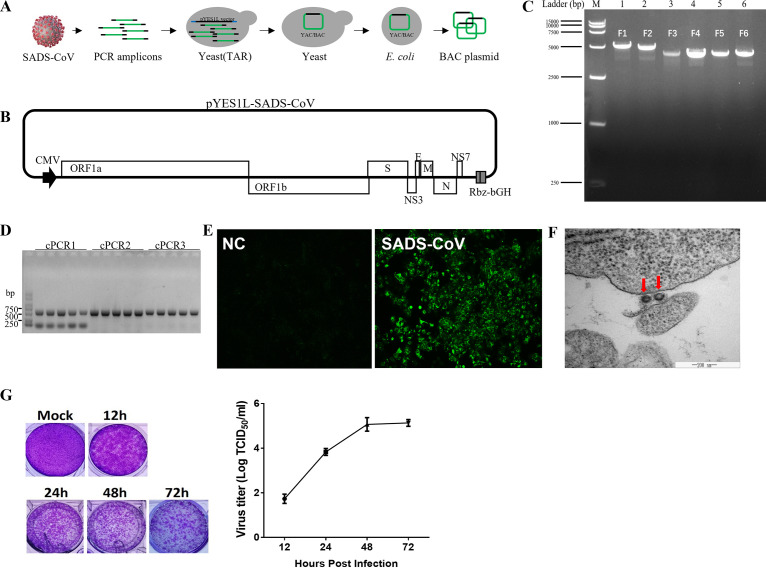
Using a panel of contiguous cDNAs with overlapping fragments that span the entire SADS-CoV genome, full-length cDNA of the SADS-CoV strain GDS04 was assembled through transformation-associated recombination (TAR) technology in yeast and amplified in Escherichia coli (Fig. 1A). To enable the TAR cloning of the SADS-CoV full-length genome in yeast, we chose the pYES1L vector (Thermo Fisher Scientific), which can be amplified in yeast and E. coli because of its yeast artificial chromosome (YAC) and bacterial artificial chromosome (BAC) elements. We modified the pYES1L vector by integrating the human cytomegalovirus (CMV) early promoter, hepatitis D virus ribozyme (Rbz), and bovine growth hormone (bGH) termination signal and designated it pYES1L-CMV-Rbz-bGH (Fig. 1B). Six overlapping DNA fragments spanning the entire viral genome were amplified and assembled into pYES1L-SADS-CoV through transformation with the linearized pYES1L-CMV-Rbz-bGH into MaV203 competent yeast cells (Fig. 1C). Three colony PCRs targeting the junction regions between viral genomic fragments were performed to screen the positive colonies. PCR products at the predicted sizes were observed for all five screened yeast colonies (Fig. 1D). A positive yeast colony was lysed to transform E. coli via electroporation, and the SADS-CoV infectious clone was prepared for virus recovery.
FIG 1.
Recovery and verification of recombinant SADS-CoV. (A) Construction of a SADS-CoV infectious cDNA clone by homologous recombination in yeast. The procedure for SADS-CoV infectious cDNA clone assembly is shown. Briefly, the complete SADS-CoV genome was divided into six fragments; the neighboring fragments share >30-nucleotide overlapping regions. Together with a linearized vector (pYES1L), all seven cDNA fragments were transformed into yeast MaV203 competent cells, and the full-length cDNA clone was assembled by TAR in yeast. (B) Schematic diagram of the SADS-CoV infectious cDNA clone. In this YAC/BAC vector-based infectious cDNA clone, a CMV early promoter and a hepatitis D virus Rbz followed by a bGH termination signal were added at 5′ and 3′ ends, respectively, of the viral genomic cDNA. ORF, open reading frame. (C) Agarose gel electrophoresis of six cDNA fragments covering the complete genome of the SADS-CoV GDS04 strain. (D) Colony PCR screening of positive clones. Five yeast colonies grown on CSM-Trp agar plates were picked for colony PCR targeting the junction regions of the 5′ end and the 3′ end. (E) Verification of recombinant SADS-CoV. The N protein of SADS-CoV was detected by IFA. At 3 dpi, Huh7 cells were fixed and stained with an antibody against N protein, followed by fluorescein isothiocyanate (FITC)-conjugated antibody. NC, negative control. (F) Electron microscopy of ultrathin sections of SADS-CoV-infected cells, revealing the virion. Huh7 cells were infected with SADS-CoV and sectioned for electron microscopy after 48 h of viral infection. (G) Virus titers of SADS-CoV at the indicated hours postinfection, assessed via plaque assays.
BHK-21 cells were cotransfected with pYES1L-SADS-CoV and plasmid expressing SADS-CoV N protein to rescue the recombinant SADS-CoV. The virus supernatant was further transferred to Vero cells for virus propagation. The recovery of SADS-CoV was confirmed by an indirect immunofluorescence assay (IFA) with antibody against SADS-CoV N protein (Fig. 1E) and electron microscopy of ultrathin sections in SADS-CoV-infected cells (Fig. 1F). In vitro characterization of the rescued SADS-CoV was performed with multiple-step growth curves and plaque assays. As shown in Fig. 1G, SADS-CoV reached peak titers at 48 h postinfection.
SADS-CoV-caused lethality of neonatal mice is dependent on the infection route.
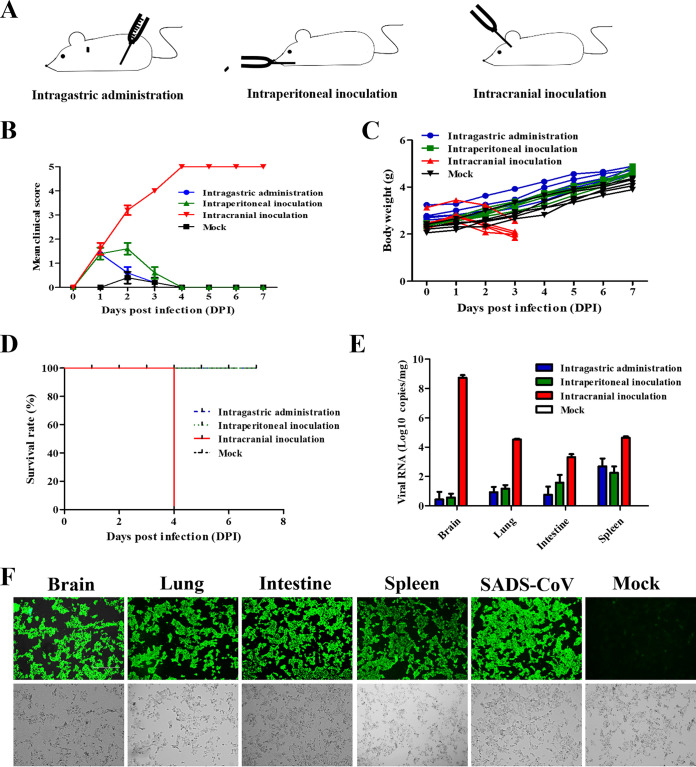
Neonatal mice (7 days of age) were inoculated via intraperitoneal (i.p.), intragastric (i.g.), and i.c. routes to gain insight into the ability of SADS-CoV to establish an infection in a mammalian host (Fig. 2A). The control group was inoculated with a culture supernatant of Huh7 cells and kept in separate cages. Each group contained 8 mice, and 3 were euthanized at 3 days postinoculation (dpi) for viral load determination. Body weight changes and clinical symptoms were observed daily. The severity of clinical symptoms was scored in five grades (Table 1). The results showed that mice inoculated via the i.p. and i.g. routes showed mild symptoms, including only inactivity and weight loss within 3 dpi (Fig. 2B). The body weights of the mice in the i.p. and i.g. groups also drastically increased after 3 dpi (Fig. 2C). In contrast, the mice of the i.c. group developed serious symptoms, especially watery diarrhea and weight loss, after inoculation (see Fig. S1A in the supplemental material). All mice inoculated via the i.c. route died at 4 dpi, while other mice survived during the 7-day period and a further observation period of 7 days (Fig. 2D). The brain, lung, intestine, and spleen were harvested from mice of different groups, and viral loads were measured. Mice in the i.p. and i.g. groups had the highest viral RNA loads in the spleen, i.e., about 103 copies/mg, with about 102 copies/mg in other organs. However, the viral RNA detected in mice in the i.c. group was much higher than that in the other two groups, with over 108 copies/mg in the brain and 104 copies/mg in the lung, intestine, and spleen (Fig. 2E). The tissue homogenate from the i.c. group was reinoculated into Vero cells to detect whether there existed infectious virion or viral RNA only. The IFA results demonstrated that massive infectious viral particles existed among different tissues, and SADS-CoV was successfully reisolated (Fig. 2F). These findings indicated that neonatal mice were highly susceptible to SADS-CoV via the i.c. route.
FIG 2.
SADS-CoV was highly lethal to BALB/c mice via i.c. inoculation. (A) Neonatal BALB/c mice were inoculated with SADS-CoV via different routes. (B to D) Clinical scores (B), body weight changes (C), survival (D) and viral RNA (E) were monitored daily. The clinical scores were graded as described in Table 1. (F) SADS-CoV was successfully reisolated from different tissues, as proved by IFA. The mock group received an equal volume of Dulbecco’s modified Eagle medium (DMEM) (mock-infected control), with i.c. infection into 7-day-old mice.
TABLE 1.
Grading scores for clinical symptoms of SADS-CoV-infected mice
| Score | Clinical symptom(s) |
|---|---|
| 0 | Healthy |
| 1 | Lethargy and inactivity |
| 2 | Soft but formed feces |
| 3 | Mild diarrhea |
| 4 | Watery diarrhea |
| 5 | Moribund and death |
SADS-CoV-caused lethality of neonatal mice is dependent on mouse age.
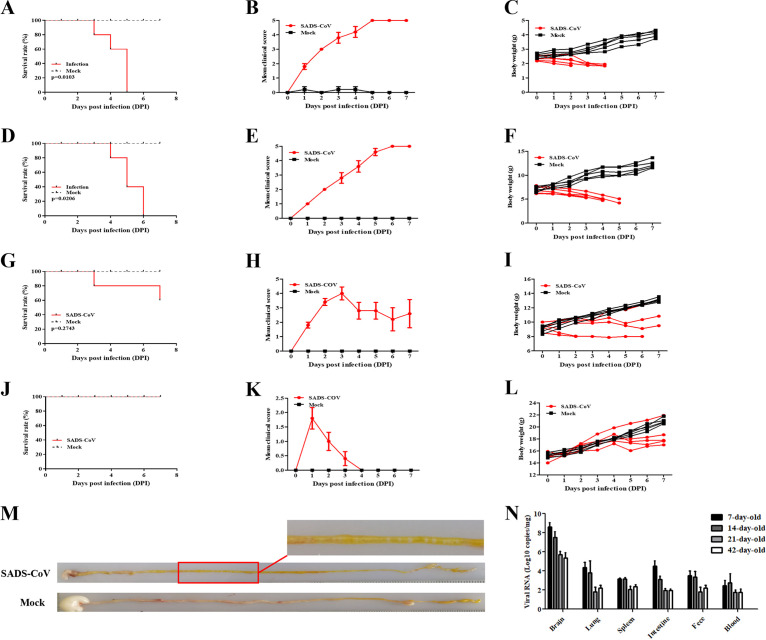
To explore whether viral lethality occurs in older newborns, we examined the sensitivities of SADS-CoV infection of mice of 7, 14, 21, and 42 days that were inoculated via the i.c. route. The body weight changes and clinical symptoms were observed daily. Furthermore, the feces of mice were also observed and collected for viral quantification. The results showed that the survival rates of the mice increased according to their ages. We found that SADS-CoV infection had a 100% mortality rate within 5 and 6 dpi, respectively, in 7- and 14-day-old mice (Fig. 3A and D); 21-day-old mice were still vulnerable to SADS-CoV infection, with a mortality rate of 40% (2/5 mice) (Fig. 3G). However, 42-day-old mice survived viral infection without obvious clinical signs (Fig. 3J). Seven- and 14-day-old mice mainly exhibited watery diarrhea, which was not observed in 21- and 42-day-old mice and the control group (Fig. 3B, E, H, and K). Compared with mock-treated groups, the body weights of 21- and 42-day-old mice did not rapidly increase and kept steady, while the 7- and 14-day-old mice exhibited decreased weight after inoculation, which suggested that SADS-CoV infection delayed their growth (Fig. 3C, F, I, and L). Feces of infected 7- and 14-day-old mice appeared as yellow paste distributed around the anus, suggesting that SADS-CoV infection can cause watery diarrhea, while no diarrhea was observed in the control mice. The intestine of inoculated mice with watery diarrhea was full of yellow content and air, compared with white content in control mice (Fig. 3M). We assessed SADS-CoV loads in SADS-CoV-infected mice of 7, 14, 21, and 42 days. SADS-CoV was detected in various organs and blood for 7- and 14-day-old mice but mainly in the brain for older mice (Fig. 3N). Together, these data illustrated that the SADS-CoV infection of 7- and 14-day-old mice represents a phenocopy of SADS-CoV infection of neonate piglets, as reported previously (7).
FIG 3.
Age-dependent mortality rates for BALB/c mice infected i.c. with SADS-CoV. BALB/c mice of different ages were inoculated i.c. with SADS-CoV or cell culture medium and observed daily. (A to C) Survival (A), clinical scores (B), and body weight changes (C) of 7-day-old mice. (D to F) Survival (D), clinical scores (E), and body weight changes (F) of 14-day-old mice. (G to I) Survival (G), clinical scores (H), and body weight changes (I) of 21-day-old mice. (J to L) Survival (J), clinical scores (K), and body weight changes (L) of 42-day-old mice. (M) Intestinal samples from SADS-CoV-infected and mock-infected 7-day-old mice. (N) SADS-CoV genome copy numbers in different tissues, as determined by real-time PCR analysis.
SADS-CoV replicates in multiple organs and tissues of infected neonatal mice.
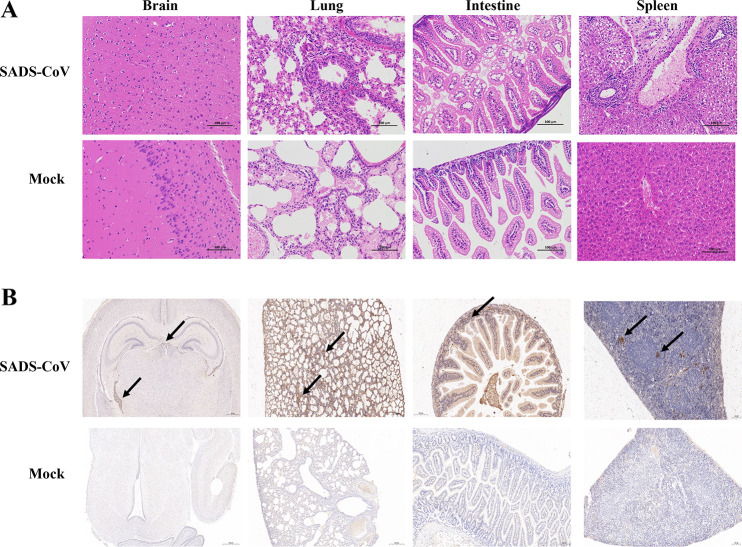
To identify the pathological features of SADS-CoV-infected 7-day-old mice, hematoxylin and eosin (H&E) staining of brain, lung, intestine, and spleen samples was performed at 3 dpi. Although many viral genomes were detected in the brain, significant lesions were not observed in the sections. The diffusion of lymphocytes with neutrophil infiltration was observed only in the lung and spleen sections. In SADS-CoV-infected intestine, the epithelial cells turned to degenerate with vacuoles that appeared in the cytoplasm (Fig. 4A). Immunohistochemical examinations were also performed to confirm SADS-CoV infection in neonatal mice. A mouse monoclonal antibody (MAb) against SADS-CoV N protein was used to detect SADS-CoV. At 3 dpi, anti-N staining was observed in the brain, alveolar wall, intestinal epithelial cells, and the white pulp around the lymphatic nodules (Fig. 4B). These findings indicate that SADS-CoV replicates in multiple tissues with mild gross lesions in neonatal mice.
FIG 4.
Pathogenic analysis of SADS-CoV-infected neonatal mice. Seven-day-old BALB/c mice were inoculated i.c. with SADS-CoV or cell culture medium and were sacrificed at 3 dpi. The brain, lung, intestine, and spleen were collected and used for H&E staining (A) or immunohistochemistry (B). Arrows indicate the SADS-CoV particles in different tissues.
SADS-CoV infection elicits a strong immune response in neonatal mice.
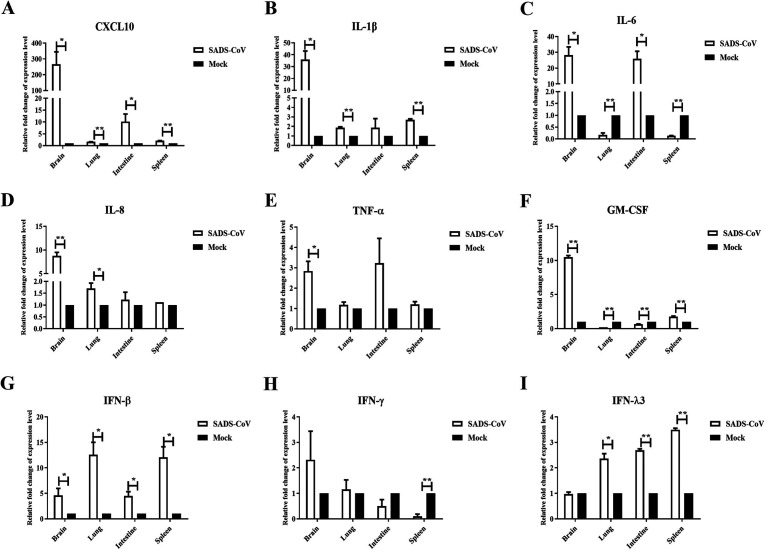
Coronavirus infection is often correlated with inflammatory reactions. To characterize the host immune response with SADS-CoV infection, we analyzed the expression of inflammatory cytokines and IFNs in different tissues at 3 dpi by real-time PCR. As shown in Fig. 5, the transcription levels of CXCL-10, IL-1β, IL-6, IL-8, TNF-α, and granulocyte-macrophage colony-stimulating factor (GM-CSF) were all significantly induced in the brain but not in other organs (Fig. 5A, B, C, D, E, and F). The type I IFN IFN-β was upregulated in the brain, lung, intestine, and spleen, suggesting that SADS-CoV could result in a generalized infection (Fig. 5G). The type III IFN IFN-λ3 was closely related to mucosal immunization, being significantly induced in the lung, intestine, and spleen but not the brain (Fig. 5I). The lung, intestine, and spleen were the main organs for the reaction and induction of mucosal immunization. However, the expression of IFN-γ was not significantly changed (Fig. 5H). All of these data indicate that SADS-CoV infection elicits a strong immune response in infected mice.
FIG 5.
IFNs and proinflammatory cytokines were elevated in SADS-CoV-infected mice. Seven-day-old BALB/c mice were inoculated i.c. with SADS-CoV or cell culture medium and were euthanized at 3 dpi. The brain, lung, intestine, and spleen were collected and analyzed for the expression of CXCL10 (A), IL-1β (B), IL-6 (C), IL-8 (D), TNF-α (E), GM-CSF (F), IFN-β (G), IFN-γ (H), and IFN-λ3 (I) by real-time PCR analysis. *, P < 0.05; **, P< 0.01, relative to the mock group.
DISCUSSION
SADS-CoV was observed in Guangdong Province, China, with high mortality rates for piglets ≤5 days of age from October 2016 to May 2017. The SADS-CoV isolates from piglets shared 95% identity with Rhinolophus bat coronavirus HKU2, which indicates that SADS-CoV is a pig virus originating from bats (7). SADS-CoV is the documented spillover of a bat coronavirus that caused severe disease in pigs (8, 9). However, receptor analysis showed that none of the known coronavirus receptors, including angiotensin-converting enzyme 2 (ACE2), dipeptidyl peptidase 4 (DPP4), and aminopeptidase N, is essential for SADS-CoV entry (5). The unknown viral receptor of SADS-CoV suggests that SADS-CoV transmission from bats to pigs and the infection of bat-origin SADS-CoV in other species need further exploration. A previous study showed that mice with SADS-CoV infection exhibited subclinical infection and SADS-CoV infection in the spleen via oral and i.p. routes (4). In our study, more routes of infection were investigated in neonatal mice. We demonstrated that bat-origin SADS-CoV is lethal to neonatal mice. Our findings are consistent with previous research indicating that mice are susceptible to SADS-CoV (10). Dr. Zhengli Shi and her team from the Wuhan Institute of Virology of the Chinese Academy of Sciences demonstrated that SADS-CoV can efficiently replicate in suckling mice ≤7 days of age after i.g. infection; in addition, they found neuroinflammatory responses in some of the infected mice (10). In our study, we found that SADS-CoV caused severe watery diarrhea, weight loss, and a 100% fatality rate in 7- and 14-day-old mice after i.c. infection. It is common for coronaviruses to invade the nervous system. Viral RNA, such as that of human coronavirus OC43 (HCoV-OC43) and HCoV-229E, can be found in human brain tissue in autopsies (11). However, whether coronavirus has gut-brain circuit transmission remains elusive. Our finding added the novelty in SADS-CoV biology that SADS-CoV is capable of bidirectional transmission between the gastrointestinal system and the central nervous system. The bidirectional transmission of SADS-CoV may be similar to pseudorabies virus (PRV) transmission in mice (12). In addition, SADS-CoV has broad cell tropism and a high replication efficiency, including in primary human cells in vitro, suggesting its potential for interspecies transmission (13). SADS-CoV infects mice, which provides a cost-effective model for the screening of antiviral drugs and therapeutic vaccines. Chen et al. demonstrated a 100% fatality rate for SADS-CoV in 2-day-old mice via i.g. infection (10). We demonstrate a 100% fatality rate for SADS-CoV in 7- and 14-day-old mice via i.c. infection. The immune system is a critical aspect for drug safety (14). T cells in 2-day-old mice were reported to be immature (14), and functionally immature T cells may interfere with results of evaluations of antiviral drugs and therapeutic vaccines against SADS-CoV. A previous study showed that the 7-day-old mouse immune system is more similar to that of human newborns, including BCG-specific responses and naive T cell receptor (TCR) repertoires (15). Therefore, the mice model of SADS-CoV infection in our study may be more suitable for evaluation of antiviral drugs and therapeutic vaccines against SADS-CoV.
Excessive production of proinflammatory mediators is involved in the immunopathology and development of organ dysfunction. Here, we found that a robust proinflammatory immune response was seen in the brain upon SADS-CoV infection (Fig. 5), but significant lesions were not observed in brain sections (Fig. 4). All of these findings suggest that there is no correlation between the presence of histopathology and the excessive production of proinflammatory mediators in the brain. Our study is consistent with previous findings that neuroinflammation is not correlated with histopathological changes in the brain (16).
Recently, porcine deltacoronavirus strains were identified in plasma samples from three Haitian children with acute undifferentiated febrile illness, suggesting the ability of porcine deltacoronaviruses to adapt, potentially leading to human-to-human transmission (17). The host range expansion of coronaviruses from wildlife to humans requires intermediate host species. MERS-CoV likely spilled over from bats to dromedary camels at least 30 years ago (18). SARS-CoV emerged through recombination of bat SARS-related coronaviruses. The recombined virus infected civets and humans and adapted to these hosts before causing the SARS epidemic (19). The pangolin may be the intermediate host species for the 2019 novel coronavirus (SARS-CoV-2) epidemic (3). The intermediate hosts transmitting bat-origin coronaviruses to humans are not well characterized. Compared to many other species, pigs are in frequent contact with humans and other animals and theoretically possess a greater chance of promoting cross-species viral transmission (20). For instance, pigs are considered mixing vessels for influenza A virus transmission between humans and avian species (21, 22). Therefore, SADS-CoV should be closely monitored for infection of humans.
Currently, there are no vaccines or drugs available for SADS-CoV. SADS-CoV infections are characterized by severe watery diarrhea, weight loss, and >90% fatality rates in piglets and neonatal mice. We are concerned by the sudden emergence of these infections and the potential threat to the human population. A better understanding of the mode of transmission of SADS-CoV, further surveillance, and appropriate countermeasures are urgently required.
MATERIALS AND METHODS
In-yeast assembly of a full-length cDNA clone of SADS-CoV.
The pYES1L vector (Thermo Fisher Scientific) containing a YAC and a BAC was utilized to assemble an infectious cDNA clone of SADS-CoV in yeast. With the YAC and the BAC, this infectious cDNA clone can replicate in both yeast and E. coli. We inserted the CMV early promoter, hepatitis D virus Rbz, and bGH termination signal into the linearized pYES1L vector, generating the plasmid pYES1L-CMV-Rbz-bGH. The linearized pYES1L-CMV-Rbz-bGH vector was prepared by PCR amplification with Q5 high-fidelity DNA polymerase (New England Biolabs) using primers pYES1L-F and pYES1L-R. The cDNA of SADS-CoV was produced with the Superscript IV first-strand synthesis system (Thermo Fisher Scientific), followed by viral RNA removal with RNase H (New England Biolabs). cDNA was from the SADS-CoV GDS04 strain, a generous gift from Yongchang Cao of Sun Yat-sen University. The genome sequence of the GDS04 strain was deposited in GenBank (GenBank accession number MF167434). With the viral cDNA, seven overlapping DNA fragments covering the complete SADS-CoV genome were amplified using Q5 high-fidelity DNA polymerase (New England Biolabs) according to the manufacturer's instructions and then were assembled with the linearized pYES1L-CMV-Rbz-bGH vector through TAR in yeast. The overlapping regions (>30 nucleotides) between neighboring DNA fragments enabled homologous recombination in yeast. Briefly, transformation of MaV203 competent yeast cells (Thermo Fisher Scientific) with the mixture of 100 ng of linearized vector and 200 ng of each DNA fragment was conducted with polyethylene glycol (PEG)/lithium acetate solution according to the manufacturer's instructions for the GeneArt high-order genetic assembly system (Thermo Fisher Scientific). Colony PCR was performed to screen yeast colonies containing the full-length cDNA clone using primer pairs. The lysate of a positive yeast colony was further electroplated into DH10B competent E. coli cells to prepare the pYES1L-SADS plasmid using the NucleoBond Xtra midikit (Macherey-Nagel). In addition, a pCAGGS-SADS-CoV-N plasmid for ectopically expressing SADS-CoV N was constructed by cloning the N protein-coding sequence into the pCAGGS vector using SacI and XhoI restriction enzyme sites.
Mouse infection experiments.
Wild-type BALB/c mice of different ages were purchased from the Experimental Animal Research Center of the Lanzhou Veterinary Research Institute and housed in animal facilities. The BALB/c mice were inoculated via the i.p., i.g., and i.c. routes with 105 times the 50% tissue culture infective dose (TCID50) of virus per mouse. For i.c. infection, 0.02 mL of SADS-CoV was inoculated carefully into mice. The control mice were inoculated with an equal volume of culture medium and kept in separate cages. All mice were observed daily for body weight, clinical signs, and survival until 15 dpi. All animal experiments were performed following the guidelines of the Experimental Animal Ethics Committee of Lanzhou Veterinary Research Institute.
Viral load determination.
For viral load determination, mice of each group were euthanized at 3 dpi, and brain, lung, spleen, intestine, blood, and feces were harvested. All samples were then weighted and homogenized in phosphate-buffered saline (PBS). Total RNA was extracted from tissue homogenates and detected using quantitative real-time PCR analysis targeting the SADS-CoV N gene. The primers and probes are listed in Table 2.
TABLE 2.
Primers used for real-time PCR analysis
| Gene name and primer | Primer sequencea |
|---|---|
| SADS-CoV N | |
| Forward | CTGACTGTTGTTGAGGTTA |
| Reverse | GACTGGCTAGAAGCTATG |
| Probe | FAM-TCACAGTCTCGTTCTCGCAATCA-TAMRA |
| IL-1β | |
| Forward | ATCTCGCAGCAGCACATCAA |
| Reverse | ACGGGAAAGACACAGGTAGC |
| IL-6 | |
| Forward | CACTTCACAAGTCGGAGGCT |
| Reverse | CTGCAAGTGCATCATCGTTGT |
| IL-8 | |
| Forward | ATCCTGATGCTCCATGGGTG |
| Reverse | GAAGCTTCATTGCCGGTGGA |
| GM-CSF | |
| Forward | GCTCACCCATCACTGTCACC |
| Reverse | GCGGGTCTGCACACATGTTA |
| TNF-α | |
| Forward | AGCCGATGGGTTGTACCTTG |
| Reverse | ATAGCAAATCGGCTGACGGT |
| CXCL10 | |
| Forward | GTCTGAGTGGGACTCAAGGG |
| Reverse | AGCTTCCCTATGGCCCTCAT |
| IFN-β | |
| Forward | CCAGCACTGGGTGGAATGAG |
| Reverse | AGTTGAGGACATCTCCCACG |
| IFN-γ | |
| Forward | CAGGCCATCAGCAACAACAT |
| Reverse | ACCTGTGGGTTGTTGACCTC |
| IFN-λ3 | |
| Forward | AGAACCCAAGCTGACCCTGT |
| Reverse | GCACCCTTGGCCTTTTTGAA |
| GAPDHb | |
| Forward | CTACCCCCAATGTGTCCGTC |
| Reverse | TGAAGTCGCAGGAGACAACC |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Immunofluorescence assay.
Vero cells grown to 80% confluence were infected with filtered tissue homogenates. The cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 at 48 h postinfection. After blocking with 5% skim milk, cells were incubated with mouse anti-SADS-CoV-N MAb (1:1,000 dilution) for 1.5 h, followed by incubation with Alexa Fluor 488-labeled goat anti-mouse IgG antibody (1:1,000 dilution; Thermo, USA) for 1 h. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime, China). The anti-SADS-CoV-N MAb was made by our laboratory. The cells were finally examined under a fluorescence microscope with a video documentation system.
Histopathological and immunohistochemical analyses of mouse samples.
Mice infected with SADS-CoV were anesthetized at 3 dpi. The tissues were harvested and fixed in 10% formalin for at least 48 h, and tissue sections were then deparaffinized, rehydrated with xylene three times, and dehydrated with ethanol at different concentrations. After washing in distilled water, the tissues were finally subjected to H&E staining. For immunohistochemistry, tissue sections were dewaxed, dehydrated, and microwaved in a citrate buffer. The sections were then incubated with mouse anti-SADS-CoV-N antibody for 1 h at 37°C, followed by horseradish peroxidase (HRP)-labeled second antibody for 30 min at room temperature. Sections were further treated with 3,3-diaminobenzidine tetrahydrochloride chromogen, counterstained with hematoxylin, and finally visualized with a light microscope.
Real-time PCR analysis.
Real-time PCR was used to quantify various cytokines and IFNs to assess host responses against SADS-CoV infection. The real-time PCR analysis was performed in an Applied Biosystems QuantStudio 5 system with TB Green Premix Ex Taq II (TaKaRa, Japan). Briefly, the reaction mixtures were incubated at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The primers are listed in Table 2. The ΔΔCT method was used to measure the relative expression levels of target genes.
Statistical analysis.
Student's t test was used to examine the statistical significance between matched groups. Unadjusted P values of <0.05 were considered significant, while P values of <0.01 were considered extremely significant.
ACKNOWLEDGMENTS
This work was supported by the Sichuan Province Fund for Distinguished Young Scholars (grant 21JCQN0175 to Q.W.), the Key Research and Development Program of Sichuan Province (grant 2022YFN0008 to L.C.), the Elite Youth Program of the Chinese Academy of Agricultural Sciences (Q.W.), and the Chinese Academy of Agricultural Science and Technology Innovation Project (grants ASTIP2022-34-IUA-07, ASTIP2023-34-IUA-07, and CAAS-ASTIP-2022-LVRI).
Footnotes
Supplemental material is available online only.
Contributor Information
Haixue Zheng, Email: haixuezheng@163.com.
Qi Wang, Email: qiwang@caas.cn.
Tom Gallagher, Loyola University Chicago-Health Sciences Campus.
REFERENCES
- 1.Haider N, Rothman-Ostrow P, Osman AY, Arruda LB, Macfarlane-Berry L, Elton L, Thomason MJ, Yeboah-Manu D, Ansumana R, Kapata N, Mboera L, Rushton J, McHugh TD, Heymann DL, Zumla A, Kock RA. 2020. COVID-19: zoonosis or emerging infectious disease? Front Public Health 8:596944. 10.3389/fpubh.2020.596944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams EP, Spruill-Harrell BM, Taylor MK, Lee J, Nywening AV, Yang Z, Nichols JH, Camp JV, Owen RD, Jonsson CB. 2021. Common themes in zoonotic spillover and disease emergence: lessons learned from bat- and rodent-borne RNA viruses. Viruses 13:1509. 10.3390/v13081509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J. 2020. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 16:e1008421. 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang YL, Qin P, Wang B, Liu Y, Xu GH, Peng L, Zhou J, Zhu SJ, Huang YW. 2019. Broad cross-species infection of cultured cells by Bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J Virol 93:e01448-19. 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards CE, Yount BL, Graham RL, Leist SR, Hou YJ, Dinnon KH, III, Sims AC, Swanstrom J, Gully K, Scobey TD, Cooley MR, Currie CG, Randell SH, Baric RS. 2020. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc Natl Acad Sci USA 117:26915–26925. 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. 2017. A new Bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis 23:1607–1609. 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khamassi Khbou M, Daaloul Jedidi M, Bouaicha Zaafouri F, Benzarti M. 2021. Coronaviruses in farm animals: epidemiology and public health implications. Vet Med Sci 7:322–347. 10.1002/vms3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Su S, Bi Y, Wong G, Gao GF. 2018. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol 26:466–470. 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Jiang RD, Wang Q, Luo Y, Liu MQ, Zhu Y, Liu X, He YT, Zhou P, Yang XL, Shi ZL. 2022. Lethal swine acute diarrhea syndrome coronavirus infection in suckling mice. J Virol 96:e00065-22. 10.1128/jvi.00065-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgello S. 2020. Coronaviruses and the central nervous system. J Neurovirol 26:459–473. 10.1007/s13365-020-00868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller PA, Schneeberger M, Matheis F, Wang P, Kerner Z, Ilanges A, Pellegrino K, Del Marmol J, Castro TBR, Furuichi M, Perkins M, Han W, Rao A, Pickard AJ, Cross JR, Honda K, de Araujo I, Mucida D. 2020. Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature 583:441–446. 10.1038/s41586-020-2474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Y, Chen Y, Geng R, Li B, Chen J, Zhao K, Zheng XS, Zhang W, Zhou P, Yang XL, Shi ZL. 2021. Broad cell tropism of SADS-CoV in vitro implies its potential cross-species infection risk. Virol Sin 36:559–563. 10.1007/s12250-020-00321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy B, Mozes E, Danon D. 1976. Comparison of the immune response potential of newborn mice to T-dependent and T-independent synthetic polypeptides. Immunology 30:261–266. [PMC free article] [PubMed] [Google Scholar]
- 15.Adkins B, Leclerc C, Marshall-Clarke S. 2004. Neonatal adaptive immunity comes of age. Nat Rev Immunol 4:553–564. 10.1038/nri1394. [DOI] [PubMed] [Google Scholar]
- 16.Streit WJ, Xue QS, Braak H, del Tredici K. 2014. Presence of severe neuroinflammation does not intensify neurofibrillary degeneration in human brain. Glia 62:96–105. 10.1002/glia.22589. [DOI] [PubMed] [Google Scholar]
- 17.Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, Bonny TS, Loeb JC, Telisma T, Chavannes S, Ostrov DA, Mavian C, Beau De Rochars VM, Salemi M, Morris JG, Jr. 2021. Independent infections of porcine deltacoronavirus among Haitian children. Nature 600:133–137. 10.1038/s41586-021-04111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khamsi R. 2015. MERS vaccines advance, but will humans or camels get the jab? Nat Med 21:1106. 10.1038/nm1015-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Yan M, Xu H, Liang W, Kan B, Zheng B, Chen H, Zheng H, Xu Y, Zhang E, Wang H, Ye J, Li G, Li M, Cui Z, Liu YF, Guo RT, Liu XN, Zhan LH, Zhou DH, Zhao A, Hai R, Yu D, Guan Y, Xu J. 2005. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis 11:1860–1865. 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nerome K, Ishida M, Oya A, Kanai C, Suwicha K. 1982. Isolation of an influenza H1N1 virus from a pig. Virology 117:485–489. 10.1016/0042-6822(82)90486-x. [DOI] [PubMed] [Google Scholar]
- 21.Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, Steinhauer DA, Kuiken T, Tompkins SM, Tripp R, Lowen AC, Steel J. 2014. Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J Virol 88:1502–1512. 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma W, Lager KM, Li X, Janke BH, Mosier DA, Painter LE, Ulery ES, Ma J, Lekcharoensuk P, Webby RJ, Richt JA. 2011. Pathogenicity of swine influenza viruses possessing an avian or swine-origin PB2 polymerase gene evaluated in mouse and pig models. Virology 410:1–6. 10.1016/j.virol.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jvi.00190-23-s0001.pdf, PDF file, 0.03 MB (26.5KB, pdf)