ABSTRACT
The isolation of the Koala retrovirus-like virus from Australian megabats and the identification of endogenous retroviruses in the bat genome have raised questions on bat susceptibility to retroviruses in general. To answer this, we studied the susceptibility of 12 cell lines from 11 bat species to four well-studied retroviruses (human and simian immunodeficiency viruses [HIV and SIV] and murine leukemia viruses [B- and N-MLV]). Systematic comparison of retroviral susceptibility among bats revealed that megabat cell lines were overall less susceptible to the four retroviruses than microbat cell lines, particularly to HIV-1 infection, whereas lineage-specific differences were observed for MLV susceptibility. Quantitative PCR of reverse transcription (RT) products, infection in heterokaryon cells, and point mutation analysis of the capsid (CA) revealed that (i) HIV-1 and MLV replication were blocked at the nuclear transport of the pre-integration complexes and before and/or during RT, respectively, and (ii) the observed lineage-specific restriction can be attributed to a dominant cellular factor constrained by specific positions in CA. Investigation of bat homologs of the three previously reported post-entry restriction factors constrained by the same residues in CA, tripartite motif-protein 5α (TRIM5α), myxovirus resistance 2/B (Mx2/MxB), and carboxy terminus-truncated cleavage and polyadenylation factor 6 (CPSF6-358), demonstrated poor anti-HIV-1 activity in megabat cells, whereas megabat TRIM5α restricted MLV infection, suggesting that the major known CA-dependent restriction factors were not dominant in the observed lineage-specific susceptibility to HIV-1 in bat cells. Therefore, HIV-1 susceptibility of megabat cells may be determined in a manner distinct from that of primate cells.
IMPORTANCE Recent studies have demonstrated the circulation of gammaretroviruses among megabats in Australia and the bats’ resistance to HIV-1 infection; however, the origins of these viruses in megabats and the contribution of bats to retrovirus spread to other mammalian species remains unclear. To determine the intrinsic susceptibility of bat cells to HIV-1 infection, we investigated 12 cell lines isolated from 11 bat species. We report that lineage-specific retrovirus restriction in the bat cell lines can be attributed to CA-dependent factors. However, in the megabat cell lines examined, factors known to bind capsid and block infection in primate cell culture, including homologs of TRIM5α, Mx2/MxB, and CPSF6, failed to exhibit significant anti-HIV-1 activities. These results suggested that the HIV-1 susceptibility of megabat cells occurs in a manner distinct from that of primate cells, where cellular factors, other than major known CA-dependent restriction factors, with lineage-specific functions could recognize retroviral proteins in megabats.
KEYWORDS: bat, human immunodeficiency virus, retroviruses, virus-host interactions
INTRODUCTION
The bat genome hosts various types of retrovirus-related sequences, including full sequences of the provirus genome, either endogenous or exogenous, and their remnants (1–5). In addition, infectious Koala retrovirus-related retroviruses have been isolated from Australian megabats (6), suggesting that gammaretroviruses can infect bats. In contrast, although lentivirus-like viruses have not been identified in bats, the bat genome harbors homologs of human immunodeficiency virus type 1 (HIV-1) restricting APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like) and tetherin genes, the bat homologs of which reportedly exhibit anti-HIV-1 activities (7, 8). Therefore, like other mammals, bats may have evolved immune systems to cope with retroviral infections.
Determinants of retroviral susceptibility in mammalian cells have been extensively studied, resulting in the identification of cellular proteins referred to as restriction or antiviral factors. Retroviral capsid (CA) is one of the main targets of such factors, its recognition results in blocking of the retroviral replication cycle at variable stages in mammalian cells. A prototypic CA-dependent restriction factor is Fv1 (Friendvirus susceptibility 1) (9), whose presence is limited to the superfamily Muroidea (10). Different Fv1 alleles determine the B- and N-tropisms of murine leukemia virus (MLV) (B-MLV and N-MLV, respectively) in mice. In primates, CA-targeting tripartite motif-containing family 5α (TRIM5α) restricts HIV-1 infection at the early stages of virus replication, before or during reverse transcription (RT) (11). HIV-1 replication can be blocked in subsequent steps of nuclear transport of the pre-integration complex (PIC) by naturally occurring myxovirus resistance 2/B (Mx2/MxB) and ectopically expressed cleavage and polyadenylation factor 6 (CPSF6) with carboxy-terminal truncation (12). Therefore, the consequences of retroviral CA-host factor interactions vary from blocking RT to altering integration site preference, depending on the cellular context (13, 14).
Analyses of naturally occurring or experimentally introduced amino acid substitutions have revealed cellular factor-recognition sites on the surface of the CA. One of the extensively studied regions in HIV-1 CA is the unstructured cyclophilin A (Cyp A)-binding loop extruding from the CA molecule (15). In addition, point mutations in surface-exposed areas in the CA molecule reportedly affect cellular protein binding and the stability of CA (13). For example, Fv-1 mainly recognizes a single amino acid residue at position 110 in MLV CA (glutamic acid for B-MLV and arginine for N-MLV), and the exchange of the residue at this position between B- and N-MLV CA can reverse the tropism. Similarly, substitution of the arginine residue at position 74 with an aspartic acid (N74D) abolishes the CA binding of Mx2/MxB and CPSF6 (13). Therefore, studying point mutations that allow escape from antiviral defenses may help identify targets of novel host factors.
Recently, it was reported that Mx2/MxB cloned from Australian megabats (Pteropus alecto) partially contributed to the low HIV-1 susceptibility of this megabat species (16). However, current knowledge on susceptibility to retrovirus infections among bats is limited to a few species, and bat susceptibility to retroviruses in general deserves further study. In this study, infection susceptibilities to lentiviruses and gammaretroviruses, particularly focusing on HIV-1, were systematically compared among 12 bat cell lines from 11 bat species.
RESULTS
Lineage-specific susceptibility to retroviral infection.
The existence of endogenous retroviruses in the bat genome and the isolation of infectious gammaretroviruses from Australian bats (6) suggest that bats have been exposed to retroviruses. Therefore, we hypothesized that bats could be susceptible to different retroviruses depending on their species. To examine this possibility, we obtained 12 bat cell lines from 11 different species, covering both suborders (Yinpterochiroptera and Yangochiroptera) and five families (Fig. 1A and Table 1). The suborder Yinpterochiroptera included 5 megabat (DemKT-1, FBKT-1, R06E, ZFBK11-97, and ZFBK13-76E) and Rhinolophodidae (BKT-1) cell lines, and Yangochiroptera included 6 microbat (EfK3B, EnK, Tb1Lu, SuBK12-08, YubKT-1, and YubKT-2) cell lines. This coverage enabled us to examine the retrovirus susceptibility of bat species in more detail.
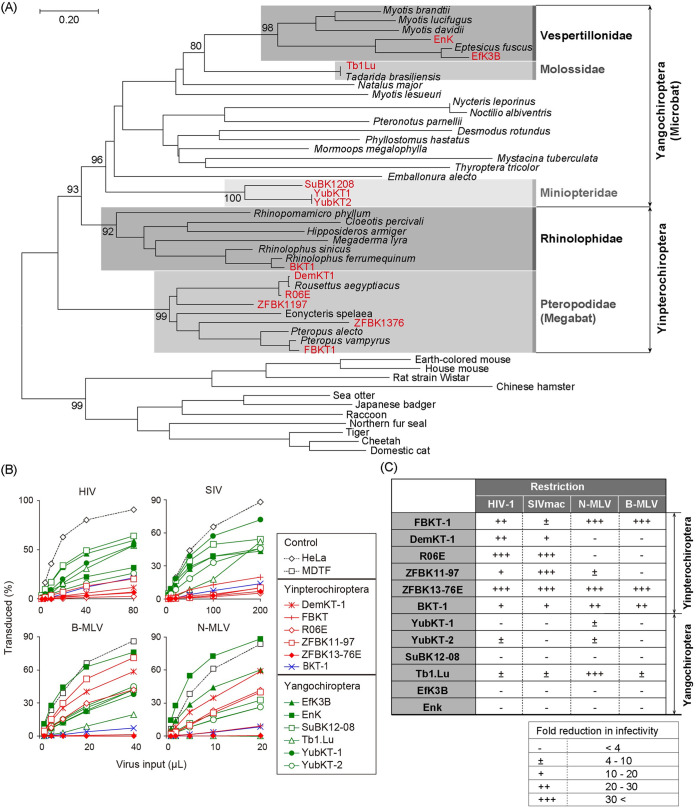
FIG 1.
Susceptibility of bat cell lines to retroviral infection. (A) Phylogeny of bats and other mammals. Phylogenetic relationships between the bats investigated in this study and representative bat species are shown. A maximum-likelihood (ML) phylogenetic tree was constructed based on the full-length amino acid sequences of the mitochondrial cytochrome b gene. The cytochrome b sequences of other mammals obtained from GenBank (accession numbers in Table S1) were included as an outgroup. The bat cell lines utilized in this study are highlighted in boldface red letters. Bat families which include the cell lines analyzed in this study are shaded with different densities of gray and are indicated for the phylogenetic clades with the cell lines studied. Numbers at the nodes of the main clusters indicate the bootstrap percentages (%) generated from 1,000 repeats (if it is ≥80). (B) Lineage-dependent susceptibility of bat cell lines to retrovirus infection. Fold-reductions of virus infectivity in each bat cell line compared to control cells are shown. The proportion of green fluorescent protein (GFP)-positive cells was measured 48 h after infection by flow cytometry. Red, blue, and green lines and symbols indicate cell lines derived from bats within the family Pteropodidae (megabat), the family Rhinolophidae, and the suborder Yangochiroptera, respectively. All cell lines were inoculated with a tester virus in parallel. Since the variation in GFP positivity between multiplicated wells was too small to exhibit standard deviation (SD) in this experimental setting, the titration experiments were performed in single wells; however, each virus inoculation was repeated three times independently. Results from three independent experiments are shown. (C) Summary of fold-reductions in retroviral infectivity in bat cell lines. Fold-reductions in retroviral infectivity compared to control cell lines (HeLa for lentiviral infection and Mus dunni tail fibroblast [MDTF] cell line for murine leukemia virus [MLV] infection) were calculated at virus titers which resulted in 20% to 30% GFP positivity in control cells. Restriction is indicated as –, <4-fold reduction; ±, 4- to 10-fold; +, 10- to 20-fold; ++, 20- to 30-fold; and +++, >30-fold.
TABLE 1.
Bat-derived cell lines utilized in this studya
| Cell line | Species | Culture medium | Reference |
|---|---|---|---|
| Yinpterochiroptera | |||
| FBKT-1 | Pteropus dasymallus | DMEM, 10% FBS | 45 |
| DemKT-1 | Rousettus leschenaultia | DMEM, 10% FBS | 44, 45 |
| R06E | R. aegyptiacus | DMEM:F12, 5% FBS | –b |
| ZFBK11-97 | Epomophorus crypturus | MEM, 10% FBS | 48 |
| ZFBK13-76E | Eidolon helvum | RPMI1640, 10% FBS | 47 |
| BKT-1 | Rhinolophus ferrumequinum | DMEM, 10% FBS | 45 |
| Yangochiroptera | |||
| YubKT-1 | Miniopterus fuliginosus | DMEM, 10% FBS | 45, 46 |
| YubKT-2 | M. fuliginosus | DMEM, 10% FBS | 45, 46 |
| SuBK12-08 | M. schreibersii | MEM, 10% FBS | 48 |
| Tb1.Lu | Tadarida brasiliensis | αMEM, 10% FBS | –b |
| EfK3B | Eptesicus fuscus | DMEM, 10% FBS | –b |
| EnK | E. nilssonii | DMEM, 10% FBS | 43 |
DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; MEM, minimal essential medium.
Purchased.
First, we systematically compared the retrovirus susceptibility of bat cell lines between the two suborders. Titration of single-round replicating HIV-1, a simian counterpart, SIVmac, and B- and N-MLV pseudotyped with vesicular stomatitis virus G (VSV-G) in the bat cell lines revealed that the cell lines of pteropid megabats (bats of the family Pteropodidae), particularly R06E and ZFBK13-76E, were less susceptible to infection with both lentiviruses than the microbat cell lines, which showed no significant reduction in lentiviral infectivity (Fig. 1B). In the case of MLV infection, the susceptibility of Yinpterochiropteran bat cell lines was not uniform; three of them (FBKT-1, ZFBK13-76E, and BKT-1) exhibited lower MLV infectivity than the others (Fig. 1B). In contrast, the microbat cell lines were almost uniformly susceptible to infection by both MLVs, with the exception of Tb1.Lu, which showed reduced susceptibility to N-MLV infection (Fig. 1B). Therefore, megabat cells were apparently less susceptible to HIV-1 infection than microbat cells.
HIV infection was blocked at distinct stages in bat cell lines.
The observed low HIV-1 susceptibility in pteropid bat cell lines could be attributed to inefficient early events of virus replication after cell entry. To examine this possibility, we quantified the amount of RT products by quantitative PCR (qPCR) in HIV-1-restricting R06E and ZFBK13-76E cell lines derived from phylogenetically distinct species within the same Pteropodidae clade (Fig. 1A). The detection specificity of the qPCR assay was verified in R06E (Fig. 2A). Kinetics of the early RT products, which detected minus-stranded strong-stop DNA, and late RT products, which indicate the completion of RT (17), were similar between the megabat and HeLa cells, indicating the completion of RT in the megabat cells (Fig. 2B). We next examined the efficiency of nuclear import of the viral PIC in the megabat cells. In HeLa cells, the amount of 2-long terminal repeat (LTR) circles, viral dead-end products in the nucleus, increased in the 12- to 24-h period postinfection, and treatment of infected cells with raltegravir, an integrase inhibitor, further increased the number of 2-LTR circles. This probably reflects the accumulation of viral dead-end products in the nucleus (18), supporting the detection specificity. In megabat cells, the 2-LTR forms were barely detected (Fig. 2B), suggesting inefficient nuclear transport of HIV-1 PIC. When assessing integration into the bat genome, for comparison, HIV-1 was detected only 2 days after infection in raltegravir-treated HeLa cells passaged for up to 12 days (Fig. 2B). This transitory signal likely reflects the detection of a mixture of late RT products in the cytoplasm and viral DNA that failed to integrate in the nuclei. Therefore, the HIV-1 genome could become undetectable 5 days after infection if not integrated in this experimental setting. Consistent with the inefficient nuclear transport of PIC, the HIV-1-specific sequence was not detected in R06E and ZFBK13-76E 5 days after infection (Fig. 2B), suggesting a failure of HIV-1 integration into bat chromosomes. Therefore, in the megabat cells tested, HIV-1 infection appears to be blocked before/at nuclear entry.
FIG 2.
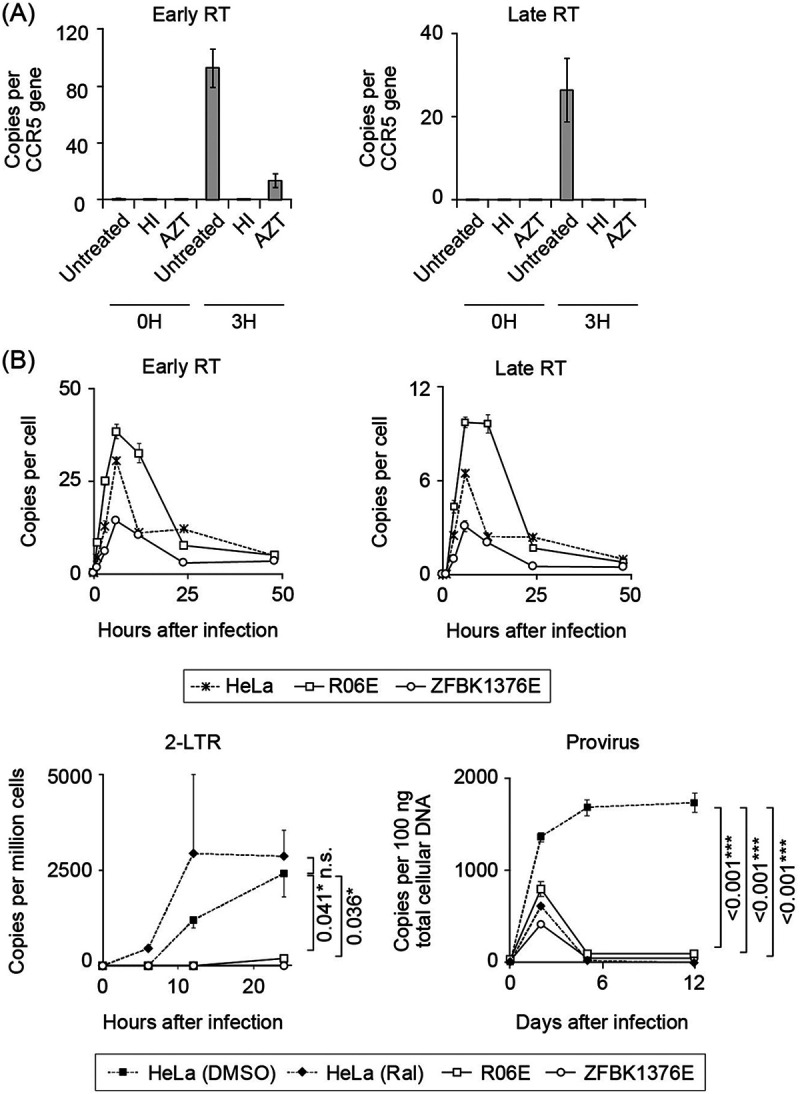
Quantitative PCR (qPCR) of viral reverse transcription (RT) products. The copy numbers of early/late RT products, 2-LTR circles, and proviruses in infected cells were determined by qPCR. (A) Verification of qPCR of viral RT products. Background yield of viral DNA in bat cells was measured by infecting megabat R06E cells with heat-inactivated HIV-1 or untreated HIV in the presence of an RT-blocking reagent, azidothymidine (AZT). Early (left panel) and late (right panel) RT product yields were quantified at 0 and 3 h after infection. Relative amounts of early and late RT products were significantly reduced by treatment of infected cells with AZT. (B) Quantification of HIV-1 early and late RT products, 2-LTR circles, and provirus in HIV-restricting megabat (R06E and ZFBK13-76E) or control (HeLa) cell lines. Cells were inoculated with a single-round replicating HIV-1 at a virus titer that resulted in approximately 10% GFP positivity in the control cells. This titer corresponds to an MOI of <1 (78). Early and late RT products were measured at 0, 1, 3, 6, 12, 24, and 48 h after infection. To detect 2-LTR circles and provirus, HeLa and megabat cells were treated in a similar manner as for the detection of early and late RT products, but with or without raltegravir in control HeLa cells. 2-LTR circles were measured at 0, 6, 12, and 24 h after infection. To detect provirus, infected cells were passaged twice weekly for 12 days, and provirus was measured at 0, 2, 5, and 12 days after infection. qPCR was performed in triplicate, and error bars in each panel represent SD of the triplicate data. Representative results from at least three independent experiments are shown. In the lower panels, statistical significance was assessed with analysis of variance (ANOVA; P < 0.05 was considered significant), and Dunnett’s post hoc comparisons between the control and bat groups were performed for the data obtained from the cells with the highest viral inoculum: P values of <0.05 (*), <0.01 (**), and <0.001 (***) were considered significant. Statistical significance was not assessed by ANOVA or nonparametric Kruskal-Wallis test at the last sampling points in the upper panels. n.s., not significant.
Capsid-dependent host factor(s) associated with low HIV-1 susceptibility in megabat.
The lineage-specific retrovirus susceptibility and the stage of infection interference (Fig. 1B and 2B) are reminiscent of the CA-dependent HIV-1 blockade in nonhuman primate cells (13). We therefore attempted to further characterize the observed low HIV-1 susceptibility in megabat cells using two methods: infection in heterokaryon cells and CA mutagenesis.
(i) Infection in heterokaryon cells. Blockade of HIV-1 infectivity in pteropid bat cells implies either the presence of host factor(s) interfering with infection or the absence of host factor(s) indispensable for virus replication. To distinguish between these alternative possibilities, we examined viral infectivity in heterokaryons formed by permissive and nonpermissive cells expressing either green fluorescent protein (GFP) or tandem (td)-Tomato (Fig. 3). Treatment with a cell-fusing reagent (polyethylene glycol; PEG) did not affect the intrinsic retrovirus susceptibility of bat cells (Fig. 3A and B). ZFBK13-76E was selected for a fusion assay because of its high fusion efficiency. PEG treatment significantly reduced the HIV-1 susceptibility in the double-positive (DP) cells, compared to permissive HeLa cells and PEG-untreated doublets (Fig. 3C). The apparent susceptibility of the fused cells was not quite as low as that of the parental bat cells, most likely because of the presence of a small fraction of unfused cells in the DP population, which could not be excluded from the heterokaryons in our experimental system.
FIG 3.
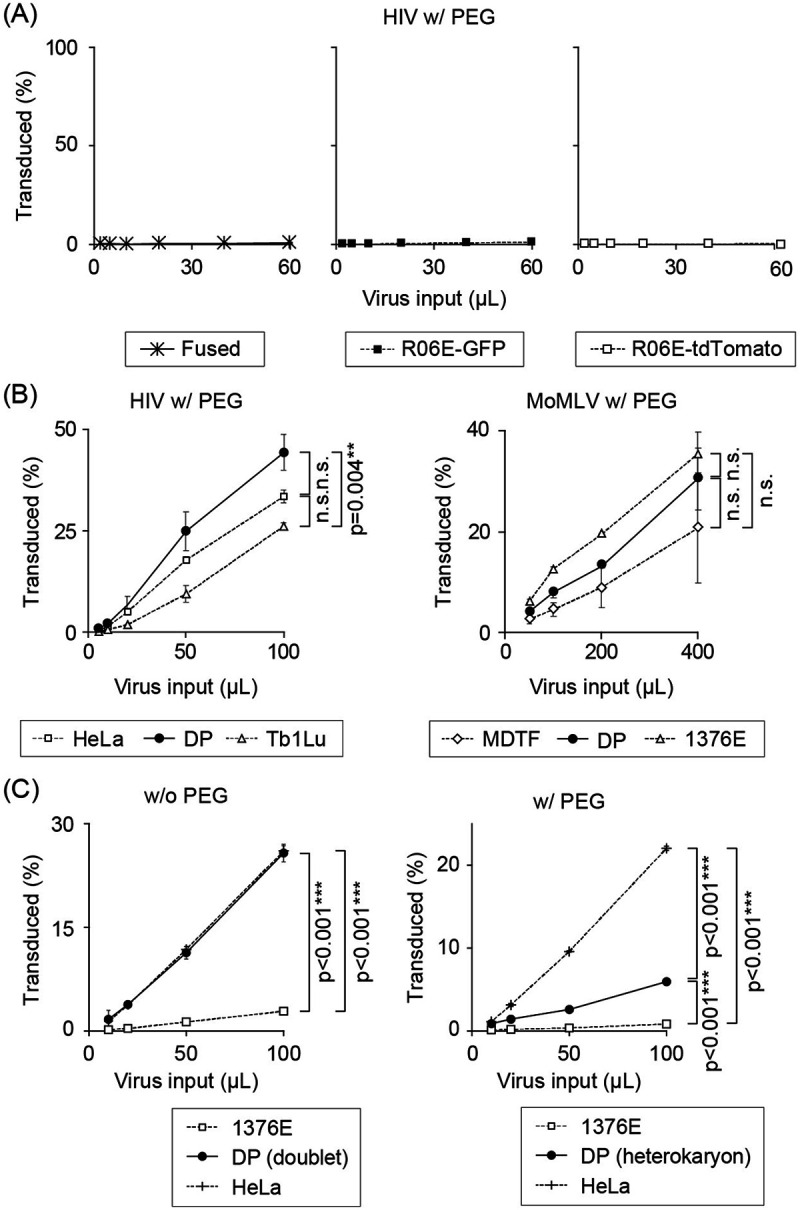
Cell fusion assay. Titration curves of HIV-1 in polyethylene glycol (PEG)-treated cells. Bat and susceptible control cells stably expressing a set of different fluorescent proteins (eGFP or tdTomato, respectively) were fused with PEG. Fused cells were challenged with a tester virus expressing a third fluorescent protein (E2-Crimson). E2-Crimson expression was analyzed in the GFP and tdTomato-double-positive (DP) cell fraction 48 h after the challenge by flow cytometry. (A) Restrictive R06E, stably expressing GFP or tdTomato, was homofused with PEG and inoculated with HIV-1-E2-Crimson. DP cells following PEG treatment of HIV-1-restricting R06E cells still restricted HIV-1 infection, suggesting that PEG treatment did not affect the intrinsic virus-restricting capability of bat cells. (B) A mixture of HeLa and HIV-1-permissive Tb1.Lu cells was treated with PEG and challenged with HIV-1-E2-Crimson (left panel). MDTF and ZFBK13-76E cells were treated with PEG and challenged with Moloney MLV (MoMLV)-E2-Crimson (right panel). PEG-treated heterokaryonic cells remained sensitive to HIV-1 and MoMLV infection, indicating that PEG treatment did not affect the intrinsic retrovirus susceptibility in bat cells. (C) Titration curves of HIV-1-E2-Crimson in ZFBK13-76E/HeLa cells without (left column) or with (right column) PEG treatment. Data represent the mean ± SD of results obtained from three independent experiments. Statistical significance was assessed with ANOVA (P < 0.05 was considered significant), and Dunn’s post hoc comparisons between groups were performed for the data obtained from the cells with the highest viral inoculum. P values of <0.05 (*), <0.01 (**), and <0.001 (***) were considered significant. In panel A and the right side of panel B, statistical significance was not detected by ANOVA or nonparametric Kruskal-Wallis test. n.s., not significant.
(ii) CA mutagenesis. We next investigated whether CA mutations affected HIV-1 infectivity in megabat cells. Of the naturally occurring or experimentally introduced point mutations in HIV-1 CA (19), six mutations were introduced individually into the CA: E45A, N74D, G89V, P90A, A92E, and G94D, which are known to affect the binding of host factors (Cyp A, Mx2/MxB, and CPSF6), CA core stability, or the nuclear transport of PIC (19–21). Four megabat cell lines (DemKT-1, FBKT-1, R06E, and ZFBK13-76E) were infected with these CA mutant viruses. Two mutations, E45A and N74D, significantly rescued infectivity in all four cell lines tested, with a less-than-2-fold reduction in infectivity compared with the infectivity in HeLa (Fig. 4). The effects of the other two mutations (G89V and P90A) on HIV-1 infectivity were cell line-dependent; HIV-1 susceptibility increased more markedly in FBKT-1 and R06E than in the other two cell lines. The remaining mutations (A92E and G94D) showed only modest effects on infectivity. Therefore, the low HIV-1 susceptibility of megabat cells apparently depended on the viral protein, and at least four residues in CA may be involved in determining HIV-1 susceptibility in bat cells. Taken together, these results raised the possibility of the presence of one or more dominant CA-dependent cellular factors being associated with the low HIV-1 susceptibility of megabat cells. In this context, it is worth noting that the anti-HIV-1 activities of CPSF6 and Mx2/MxB were abrogated by the N74D mutation in CA (12).
FIG 4.
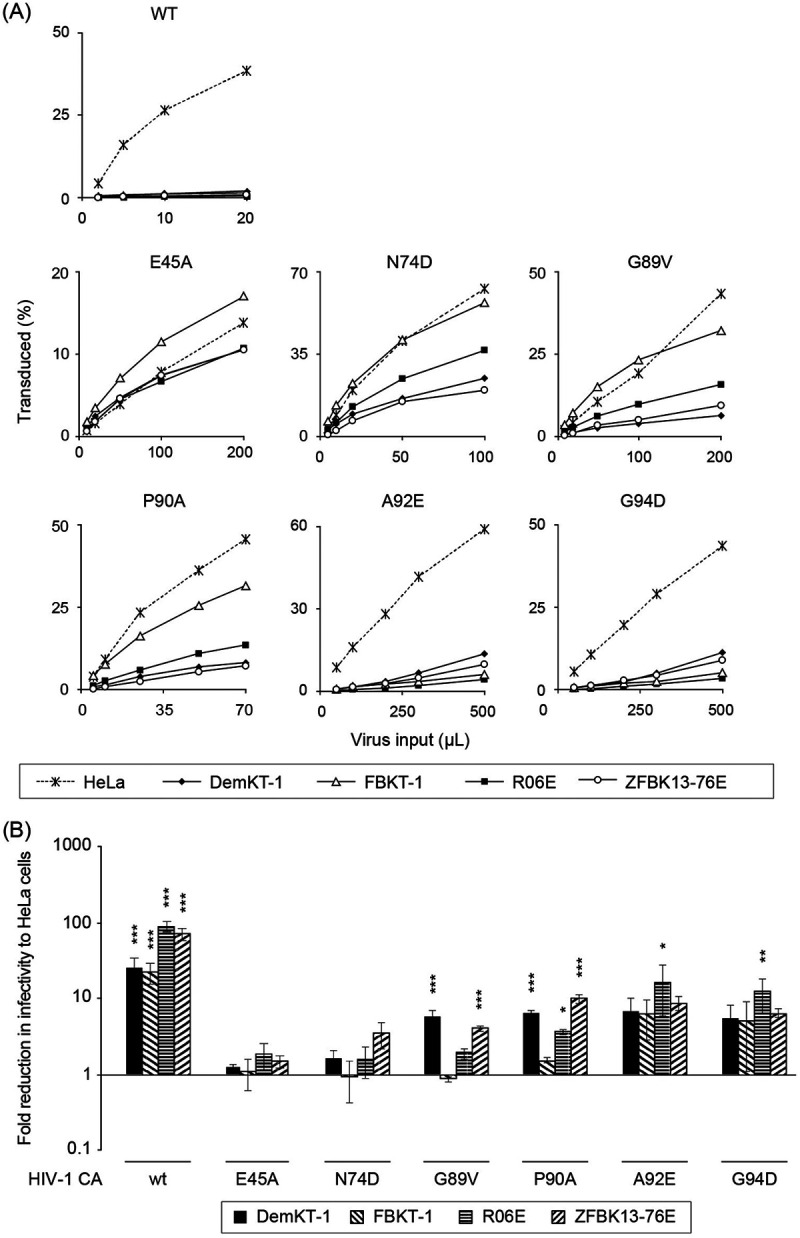
Effects of capsid mutations on retroviral infectivity in bat cell lines. The four megabat cell lines were inoculated with CA point-mutated HIV-1, as indicated. Mutant viruses were freshly prepared for each experiment, and the infection was independently repeated three times. (A) Representative titration curves of HIV-1 CA mutant viruses in megabat cell lines. (B) Fold-reductions in HIV-1 infectivity in megabat cell lines were calculated for each mutant virus at a titer that resulted in 20% to 30% GFP positivity in HeLa cells. Average fold-reduction (±SD) from three independent experiments is shown. Significant differences between each bat cell line and HeLa cells are indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001 were considered significant (n.s., not significant).
Characterization of CPSF6-, Mx2/MxB-, and TRIM5α-like genes in the bat genome.
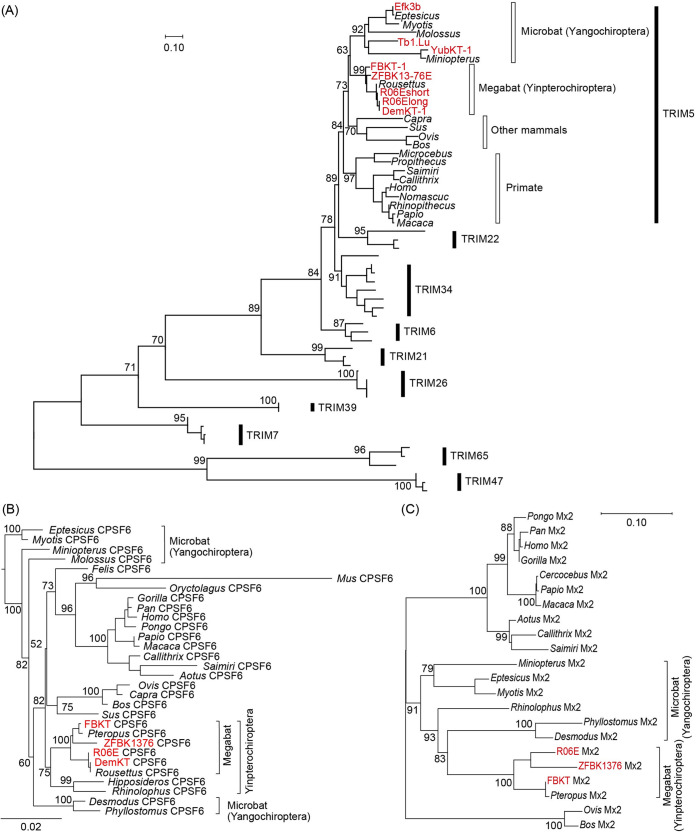
Based on the possible presence of CA-dependent restriction factors (Fig. 3 and 4) and the blockade of PIC nuclear transport as a dominant cause of viral suppression, rather than the reduced viral gene expression due to CA-dependent integration site retargeting (Fig. 2) (22–24), in bat cells, we expected that CA-dependent restriction factors would protect bat cells from retroviral infection in a manner similar to that in previously characterized systems. We therefore set out to test whether TRIM5α, CPSF6, or Mx2/MxB homologs might play a role in determining susceptibility of bat cells to HIV-1 infection. By designing bat-specific primers (Table 2), we successfully amplified CPSF6, Mx2/MxB, and TRIM5α gene homologs from four megabat cell lines (DemKT-1, FBKT-1, R06E, and ZFBK13-76E) (Fig. S1A to D), all of which showed high amino acid identities to the top-homology hit bat sequences (80.4% to 96.2% for TRIM5α, 98.7% to 100% for CPSF6, and 92.5% to 99.8% for Mx2/MxB). Phylogenetic analyses revealed that the new genes fell into tight phylogenetic clusters with their counterpart sequences, which were supported by >90% bootstrap values at the node of each cluster (Fig. 5). Although the amino acid sequences of the new megabat TRIM5α exhibited some similarities to those of pteropid TRIM34 in the variable regions of the B30.2 domain (Fig. S1E), maximum-likelihood (ML) trees of TRIM5α-related TRIMs based on the full length or the B30.2 domain clearly showed that the new TRIM5-like genes were truly megabat TRIM5α (Fig. 5D).
TABLE 2.
PCR primers/probes utilized in this studya
| Primer/probe | Direction | Sequence (5′→3′) |
|---|---|---|
| Mutagenesis PCR | ||
| MLV CA mutation | ||
| N QR109TE | F | TTACACCACCACAGAAGGTAGGAACCACCTAGTTCTC |
| N QR109TE | R | TCCCAATCGGGACGTTCA |
| B TE109QR | F | TTACACCACTCAAAGAGGTAGGAACCACCTAGTCCTCTATC |
| B TE109QR | R | TCCCAATCGGGGCGTTCA |
| HIV CA mutation | ||
| Pax E45A | F | GCATTATCAGCTGGAGCCACCC |
| Pax E45A | R | TGAAAACATGGGTATCAC |
| Pax N74D | F | AGAGACCATCGATGAGGAAGC |
| Pax N74D | R | TTTAACATTTGCATGGCTG |
| Pax G89V | F | GTGCATGCAGTGCCTATTGCA |
| Pax G89V | R | TGGATGCACTCTATCCCATT |
| Pax P90A | F | TGCATGCAGGTGCCATTGCACCAG |
| Pax P90A | R | CTGGATGCACTCTATCCC |
| Pax A92E | F | GGGCCTATTGAGCCAGGCCAGATG |
| Pax A92E | R | TGCATGCACTGGATGCAC |
| Pax G94D | F | ATTGCACCAGACCAGATGAGAGAACC |
| Pax G94D | R | AGGCCCTGCATGCACTGG |
| FBKT Mx2 mutation | ||
| FBKT_Mx2 Y21C | F | TCCCAACACTGTCCGAAAAAGGAAATGAATC |
| FBKT_Mx2 Y21C | R | GGGCGCCTGATTGTGCCT |
| FBKT_Mx2 L600P | F | GCGAATTCGCCTGGCAAGACT |
| FBKT_Mx2 L600P | R | GTCTTCTCGGGCCTTTTC |
| FBKT CPSF6 C terminus truncation | ||
| FBKT_CPSF6del | F | ATGGGCAAACCGATTCCGAAC |
| FBKT_CPSF6del | R | GCCTGGAGGTGGAGGTGG |
| pWPT-E2-Crimson and pWPT-tdTomato plasmid construction | ||
| WPT-GFP (Crim) | F | AATTCGATATCAAGCTTATCG |
| WPT-GFP (Crim) | R | TAAGCTTAGGCCTGGATC |
| E2 Crimson | F | CGGATCCAGGCCTAAGCTTAGCCACCATGGATAGCACTG |
| E2 Crimson | R | GATAAGCTTGATATCGAATTTCACTGGAACAGGTGGTGGCG |
| WPT-GFP (tmt) | F | CTAGCTAGTCGAGCTCAACTTCGAATTCGATATC |
| WPT-GFP (tmt) | R | GGCCTGGATCCGCGTCAC |
| tdTomato | F | TCGTGACGCGGATCCAGGCCGCCACCATGGTGAGCAAG |
| tdTomato | R | AGTTGAGCTCGACTAGCTAGTTACTTGTACAGCTCGTCCATG |
| pCFG2-E2-Crimson and pCFG2-tdTomato plasmid construction | ||
| CFG2 F5 | F | ATAAAATAAAAGATTTTATTTAGTCTCCAGAAAAAGGG |
| CFG2 R6 | R | AGATCTGGCCATGGCAGTC |
| tomato F2 | F | AGACTGCCATGGCCAGATCTGCCACCATGGTGAGCAAG |
| tomato R2 | R | CTAAATAAAATCTTTTATTTTATTTACTTGTACAGCTCGTCCATG |
| Crimson F2 | F | AGACTGCCATGGCCAGATCTGCCACCATGGATAGCACTG |
| Crimson R2 | R | CTAAATAAAATCTTTTATTTTATTTACTGGAACAGGTGGTG |
| For constructing delivery vector plasmids containing bat host factors | ||
| pLGatewaySN-vector plasmid | ||
| 583_fwd | F | GATAACTCGAGGATCCGG |
| 583_rev | R | GATAACGAATTCCGGCGC |
| V5-TRIM5-delivery vector plasmid | ||
| TRIM5_fwd | F | TGGAGGTAGTATGGCTACAGGAATCCTG |
| TRIM5_rev | R | AGCCGGATCCTCGAGTTATCTTAAGAGTTTGGAGAAGACAG |
| V5_fwd | F | AGGCGCCGGAATTCGTTATCATGGGCAAACCGATTCCG |
| V5_rev | R | CTGTAGCCATACTACCTCCACCTCCGGTG |
| Mx2-V5-delivery vector plasmid | ||
| FBKT-Mx2_C | F | AGGCGCCGGAATTCGTTATCATGCCAAAGTCCCACAAG |
| FBKT-Mx2_C | R | GTTTGCCCATGCAGGAGAATTTGTAGAGC |
| FBKT-Mx2_C_V5 | F | ATTCTCCTGCATGGGCAAACCGATTCCG |
| V5_C_rev2 | R | AGCCGGATCCTCGAGTTATCTTAACTACCTCCACCTCCG |
| CPSF6-V5-delivery vector plasmid | ||
| FBKT-CPSF6_C | F | AGGCGCCGGAATTCGTTATCATGGCGGACGGCGTGGAC |
| FBKT-CPSF6_C | R | GTTTGCCCATACGATGACGATACTCGCGC |
| FBKT-CPSF6_C_V5 | F | TCGTCATCGTATGGGCAAACCGATTCCG |
| V5_C_rev2 | R | AGCCGGATCCTCGAGTTATCTTAACTACCTCCACCTCCG |
| Full-length cytochrome B | F | CCHCCATAAATAGGNGAAGG |
| R | WAGAAYTTCAGCTTTGGG | |
| EnK Cyt B gene | F | ATGACCAACATTCGAAAATCYCAC |
| R | TCTTCATTTTAGTAGGTGATTTTC | |
| PCR primers to amplify TRIM5, Mx2/MxB, and CPSF6 cDNAs | ||
| Oligo(dT) adapter primer | GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN | |
| 1st-round PCR | ||
| Megabat TRIM5 | F | ATGGCTACAGGAATCCTG |
| Microbat TRIM5 | F | ATGGCTTCRGGAATYSTG |
| Megabat Mx2/MxB | F | ATGCCMAARTCCCACAAG |
| Megabat CPSF6 | F | ATGGCGGACGGYGTGGAC |
| Outer adaptor | R | GCGAGCACAGAATTAATACGACT |
| 2nd-round PCR for Gibson Assembly reaction | ||
| Megabat TRIM5 | R | TTAAGAGTTTGGAGAAGACAG + vector-specific sequence |
| Microbat TRIM5 | R | TTAAGARCYKGGAGAACACAG + vector-specific sequence |
| Megabat Mx2/MxB | R | TTAGCAGGAGAATTTRTAGAG + vector-specific sequence |
| Megabat CPSF6 | R | CTAACGATGACGATACTCGCGC + vector-specific sequence |
| qRT-PCR primers/probes | ||
| Early RT product | F (hRU5-F2) | GCCTCAATAAAGCTTGCCTTGA |
| R (hRU5-R) | TGACTAAAAGGGTCTGAGGGATCT | |
| Probe (hRU5-P) | 6FAM-AGAGTCACACAACAGACGGGCACACACTA-TAMRA | |
| Late RT product | F (MH531) | TGTGTGCCCGTCTGTTGTGT |
| R (MH532) | GAGTCCTGCGTCGAGAGATC | |
| Probe (LRT-P) | 6FAM-CAGTGGCGCCCGAACAGGGA-TAMRA | |
| 2-LTR circle | F (hRU5-F2) | GCCTCAATAAAGCTTGCCTTGA |
| R (MH536) | TCCACAGATCAAGGATATCTTGTC | |
| Probe (P-HUS-SS1) | 6FAM-TAGTGTGTGCCCGTCTGTTGTGTGAC-TAMRA |
CA, capsid protein; qRT-PCR, reverse transcription-quantitative PCR; LTR, long terminal repeat.
FIG 5.
Phylogeny of newly isolated megabat CPSF6, Mx2/MxB, and TRIM5α. (A) An ML phylogenetic tree of the newly identified TRIM5α (red) and those of other mammals based on the RBCC domain of C-IV type TRIMs. Numbers at the nodes of the main clusters indicate the bootstrap percentage (%) generated from 1,000 repeats. The new bat TRIMs formed a clade with TRIM5 of various mammalian species (bootstrapping value = 84%). (B) An ML phylogenetic tree of CPSF6 genes of 30 mammalian species. The newly cloned pteropid bat CPSF6 (highlighted in red) formed a clade with the Pteropus and Rousettus CPSF6 obtained from NCBI GenBank (bootstrapping value = 100%). We estimated the dN/dS ratio at each branch using codeml (see Materials and Methods), and the dN/dS ranged from 0.0001 to 0.0911, suggesting a negative selection of the CPSF6 gene. (C) A phylogenetic tree of pteropid megabat and other mammalian Mx2. The newly identified R06E Mx2 is highlighted in red. Due to the loss of almost the entire 3′ half (Fig. S1D), DemKT-1 Mx2/MxB was not included in the phylogenetic analysis. (D) Unrooted phylogenetic ML trees of TRIM5α and its close relatives (TRIM34, TRIM6, and TRIM22) based on the full-length (left) or the B30.2 domain (right) sequences. Newly identified megabat TRIM5α are shown in red, and the Yinpterochiroptera and Yangochiroptera clades are highlighted in ovals in the TRIM5α clade. For both phylogenetic trees, the numbers at the nodes of each main cluster indicate the bootstrap values generated from 1,000 repeats. GenBank accession numbers of the sequences used in each tree are provided in Table S1.
Cellular factors other than the three major CA-dependent restriction factors associated with low HIV-1 susceptibility of megabat cells.
Next, we examined the capabilities of the previously mentioned major CA-dependent factors to restrict HIV-1 infection in permissive cells overexpressing them after antibiotic selection.
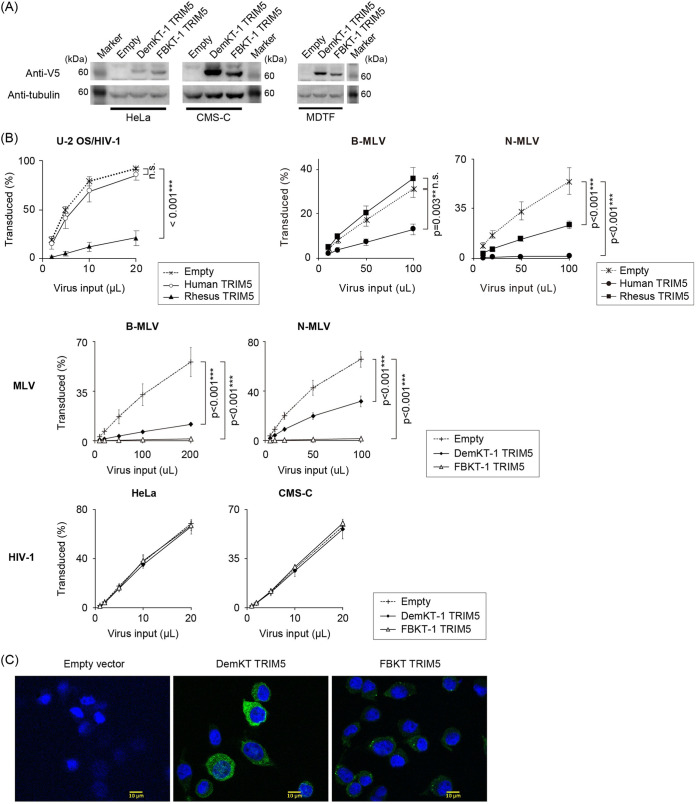
TRIM5α. The two TRIM5α (FBKT-1 and DemKT-1) were selected for subsequent characterization because they coded the entire TRIM5α proteins (Fig. 6A and Fig. S1A), and the original megabat cell lines differed in MLV susceptibility (Fig. 1A). Transduction of human (Hu) or rhesus macaque (Rh) TRIM5α reported in our previous studies (25, 26) resulted in the selective restriction of HIV-1 by RhTRIM5α in HeLa and MLV by HuTRIM5α in Mus dunni tail fibroblast (MDTF) cells (Fig. 6B), confirming the effective transduction of the gene of interest with the MLV-based pLGatewaySN vector (27). Titration of MLV and HIV-1 in stable MDTF and HeLa cells showed significant reduction of MLV infectivity, but not HIV-1 infectivity (Fig. 6B). Poor anti-HIV-1 activity of megabat TRIM5α was confirmed in the stable canine cell line CMS-C (28) (Fig. 6B), avoiding the confounding effects of the coiled-coil motif due to endogenous TRIM5α expression in HeLa cells (29–31), as canines lack the expression of functional TRIM5α (24). The primate TRIM5α-like cytoplasmic localization of the two megabat TRIM5α, either diffused (DemKT-1 TRIM5α) or with cytoplasmic body-like speckles (FBKT-1 TRIM5α), in overexpressed cells (Fig. 6C) excluded the possibility of their unique intracellular localization causing poor HIV-1 restriction. Therefore, although we currently cannot exclude the possibility that the megabat genome expresses a minor HIV-1-restrictive TRIM5 isoform, the dominantly expressed TRIM5α could explain the MLV, but not HIV-1, restriction.
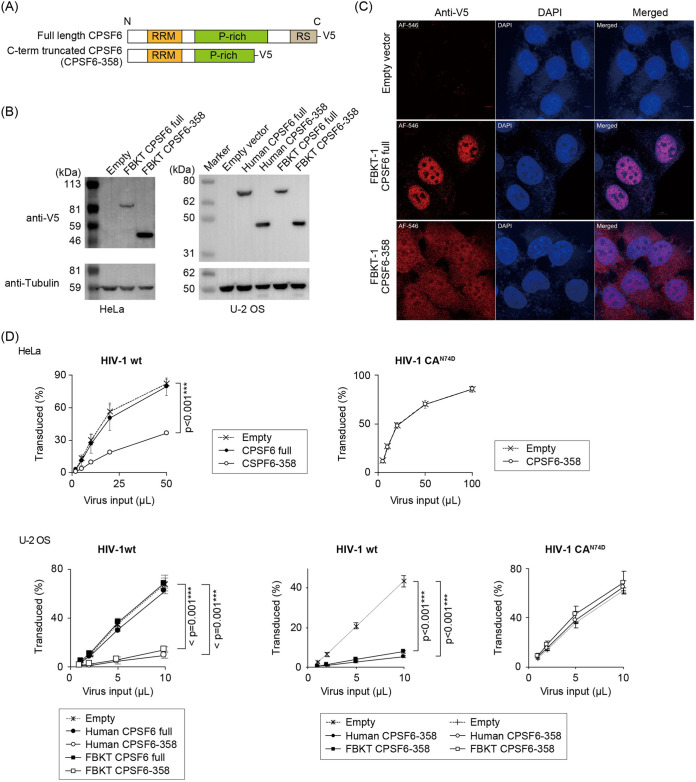
CPSF6. Based on the high sequence similarity between megabats and other mammalian CPSF6 (Fig. S1C) and HIV-1 infection interference at the nuclear transport stage of PIC (Fig. 2B), megabat CPSF6 could have anti-HIV-1 activity. The carboxy-terminal region, including NLS, was artificially truncated in FBKT-1 and human CPSF6 (Fig. 7A and Fig. S3) because the similarly truncated mCPSF6-358 was localized in the cytoplasm and restricted HIV-1 infection (12). Consistently, the truncation relocated FBKT-1 CPSF6 to the cytoplasm in human cells (Fig. 7B and C). The truncated FBKT-1 CPSF6-358 weakly, though significantly, reduced HIV-1 infectivity in the stable human cells, and this inhibitory effect was counteracted by the insertion of the escape mutation CA N74D (Fig. 7D) (12), supporting a possible interaction of FBKT-1 CPSF6-358 with HIV-1 CA. However, an investigation of CPSF6 transcription isoforms in Rousettus aegyptiacus from the publicly available RNA-seq datasets (NCBI Sequence Read Archive) revealed that the CPSF6 transcripts lacking exon 6, which corresponded to human CPSF6 isoform 1 (32), were predominant (Fig. 7E), consistent with our megabat cDNA sequences (Fig. S1C). These results suggested that CPSF6-358-like short transcript isoforms were not dominantly expressed in R. aegyptiacus in nature. Therefore, CPSF6 is unlikely to be the dominant anti-HIV-1 factor in megabat cell lines.
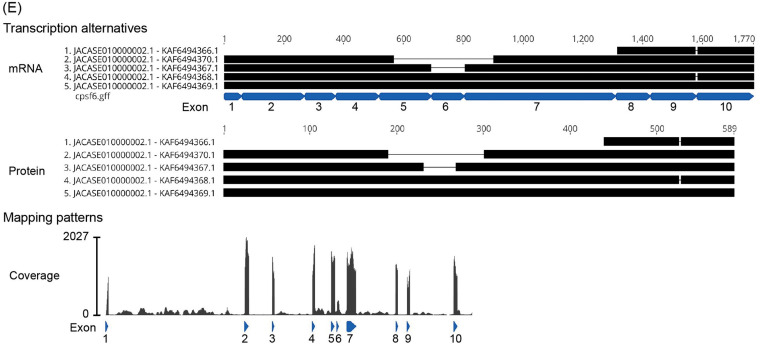
Mx2/MxB. In contrast to the anti-HIV-1 capability of P. alecto Mx2/MxB (the NCBI RNA-seq database, locus NW_006439713) (16), transduction of the new megabat Mx2/MxB did not reduce HIV-1 infectivity in three different human cell lines (Fig. 8A and B). To explain the observed discrepancy in HIV-1-restriction activity, we calculated the ratio of nonsynonymous to synonymous amino acid substitutions (dN/dS), as Mx1/MxA, the paralogue of Mx2/MxB, was positively selected during evolution (33, 34). The dN/dS ratio was >1 at the node of the megabat Mx2/MxB cluster (dN = 0.0126, dS = 0.0121), suggesting a signature of positive selection (Fig. 8C). By calculating the dN/dS ratio for each amino acid site in chiropteran Mx2/MxB, we found several amino acid sites under positive selection in the unstructured amino-terminal region and the flexible L4 loop in the stalk domain (Fig. 8D), as indicated for Mx1/MxA and primate Mx2/MxB (33, 34). Of these, we focused on two sites: the residue at 79 in the amino-terminal region essential for anti-HIV-1 activity (34–38), and the residue at 600, which was the only position where different amino acids were coded between FBKT and P. alecto among the four positively selected sites with a significant probability (>0.90). However, exchanging the amino acids at these two positions between the two megabat Mx2/MxB (Q79R and Q79R/L600P) (Fig. 8E and Fig. S1D) did not convert FBKT-1 Mx2/MxB to HIV-1 restriction (Fig. 8F), implying the involvement of other residues in the anti-HIV-1 activity. In conclusion, these three major CA-dependent HIV-1 restriction factors are not the dominant anti-HIV-1 factors present in megabat cell lines, implying that alternative, unknown cellular factors contribute to the low HIV-1 susceptibility of megabat cells.
FIG 6.
Megabat TRIM5α restricted MLV, but not HIV-1. (A) Protein expression of new megabat TRIM5α in stably expressing cells. TRIM5α proteins were detected with a specific antibody to the V5 tag, followed by the detection of α-tubulin on the stripped blot. (B) Titration curves of HIV-1 in U-2 OS and MLV in MDTF cells stably expressing rhesus and human TRIM5α (the top panels) and HeLa, CMS-C, and MDTF cells stably expressing megabat TRIM5α (the lower panels). Cells were infected in triplicate wells in three or four independent experiments, and all obtained data were averaged. Error bars represent the SD of results from independent experiments. Significant differences between the control (empty vector) and TRIM5α groups are indicated; P < 0.001 (***) is considered significant. Statistical significance was not detected by ANOVA in the lower panels. n.s., not significant. (C) Immunofluorescent staining of canine CMS-C cells stably expressing DemKT-1 and FBKT-1 TRIM5α. Higher magnification images (×63 objective lenses) of HeLa cells expressing megabat TRIM5α are shown. Green, TRIM5α; blue, DAPI (4′,6-diamidino-2-phenylindole).
FIG 7.
Moderate restriction of HIV-1 by newly isolated megabat CPSF6-358. (A) Schematic structures of the full-length or carboxy terminus-truncated form (CPSF6-358) of FBKT-1 CPSF6 tagged with a V5 sequence at the carboxy terminus. RRM; RNA-recognition motif, P-rich; proline-rich domain, RS; arginine/serin-rich domain. Figure S1C shows the precise position of the inserted stop codon for truncation. (B) Western blot image showing protein expression in stably expressing cells after selection with G418. Proteins were detected as described in the Fig. 6A legend. Truncation of the carboxy-terminal region resulted in a protein with a smaller molecular size. (C) Immunofluorescent staining of U-2 OS cells stably expressing the full-length CPSF6 or CPSF6-358 proteins. CPSF6 was detected with the anti-V5 antibody in stable cells cultured on a glass coverslip. Red, V5-tagged CPSF6; blue, DAPI staining of the nuclei; magenta, merged. (D) Titration curves of wild-type HIV-1 (HIV-1wt) or HIV-1 with CA N74D mutation (HIV-1 N74D) in HeLa (upper panels) or U-2 OS (lower panels) cells stably expressing human or megabat CPSF6 as described in the Fig. 1B legend. Error bars represent the SD of data from three independent experiments. Significant differences between the empty vector and CPSF6-expressing groups are indicated; P < 0.001 (***) is considered significant. No statistical significance was indicated in the HIV-1 N74D titrations. n.s.; not significant. (E) Sequence analysis of CPSF6 isoforms annotated by a combination of four methods used in a previous study (79). Five CPSF6 transcripts (locus_tag: HJG63_003294; protein accession numbers KAF6494366 to KAF6494370) are annotated. To further analyze the CPSF6 isoforms in Rousettus aegyptiacus, we mapped short reads of 60 RNA-seq data deposited in the NCBI SRA (Table S1). Note that the mapping patterns were almost identical among the 60 data, and that the CPSF6 transcripts lacking exon 6, which correspond to Human CPSF6 isoform 1 (32), highly predominated, consistent with our megabat cDNA sequences (Fig. S1C).
FIG 8.
Pteropid megabat Mx2/MxB did not restrict HIV-1. (A) Western blot images showing protein expression of newly identified megabat Mx2 in stably expressing HeLa, U-2 OS, and BJ-5ta cells. Proteins were detected as indicated in the Fig. 6A legend. (B) Titration curves of HIV-1 in the three human cell lines with stable expression of empty vector or megabat Mx2. (C) Phylogenetic tree of pteropid megabat and other mammalian Mx2/MxB, representing the estimated dN/dS ratio if it exceeds 1 (indicated in red on the branches of the tree). Although a dN/dS ratio of >1 was indicated for chimpanzees, the dS and dN values were rather small (dN = 0.0048, dS = 0.0015); therefore, this may be a false positive. (D) Estimation of positively selected sites among the chiropteran Mx2 genes (Table S1). Black and red asterisks indicate the sites of the dN/dS with a Bayes Empirical Bayes (BEB) probability between 0.90 and 0.95 and those with a BEB probability of >0.95, respectively. Nucleotide alignments and the consensus sequence of the chiropteran Mx2/MxB region with significant positive selection sites are shown above the schematic structure of Mx2/MxB. (E) Western blot detection of FBKT-1 Mx2/MxB with point mutations (Q79R and L600P) in stable HeLa cells with a V5-tag-specific antibody. (F) Titration of HIV-1 in HeLa cells expressing FBKT-1 Mx2/MxB with point mutations. In titration experiments shown in panels B and E, cells were infected in triplicate in three or four independent experiments. Error bars represent the SD of the results obtained from three independent experiments. No statistical significance was observed in panels B and F by ANOVA.
DISCUSSION
Although species-dependent differences in susceptibilities of bat cells to enveloped RNA viruses have been reported (39, 40), their susceptibility to retroviruses has not been extensively studied yet. This study revealed that megabat cells are apparently less susceptible to retrovirus infections than microbat cells. Although we could not clearly show the CA dependency of MLV susceptibility of megabat cells because of similar restriction patterns on both B- and N-MLV (Fig. 1B), significant restriction of MLV by megabat TRIM5α (Fig. 6C), consistent with the results of a previous study (16), supported the CA dependency of MLV restriction. Therefore, it is possible that the observed lineage-specific retrovirus susceptibility depends on the viral protein CA, either by direct binding or indirectly, in megabat cells. Furthermore, failure to restrict HIV-1 by three major CA-dependent factors implicated the involvement of one or more as-yet unidentified host factors in retrovirus infection. Alternatively, homologs of cellular factors with weak anti-HIV-1 activity in primate cells may play a significant role in HIV-1 restriction in megabat cells.
This study showed that the E45A and N74D mutations of CA significantly rescued HIV-1 infectivity (Fig. 4). Since the E45A mutation affects the stability of the intrinsic CA structure (41, 42), this mutation may abolish the inhibition of HIV-1 in bat cells by disrupting the interaction of unidentified cellular factors with the destabilized CA core. In contrast, residue N74 in CA is involved in interacting with various CA-binding factors, including CPSF6-358 and Mx2/MxB (12, 20). However, CPSF6 may not be the dominant anti-HIV-1 factor in megabat cells, since we could not find CPSF6-358-like short transcript sequences in the R. aegyptiacus Ref-seq data sets from the NCBI SRA database (Fig. 7E). In contrast to the restriction of HIV-1 by Mx2/MxB isolated from P. alecto (16), the newly identified megabat Mx2/MxB, including those with amino acid substitutions at the inferred positive selection sites, failed to exhibit anti-HIV-1 activity (Fig. 8). Considering that the three regions in Mx2/MxB involved in the interaction with HIV-1 CA are highly variable (Fig. S1D), lineage-specific variations seem to exist in the amino acid residues in the HIV-1 CA-interacting sites in Mx2/MxB.
In conclusion, it is rational to conclude that the HIV-1 susceptibility of megabat cells may be determined by cellular factors with lineage-specific functions that recognize retroviral proteins in a manner distinct from that of primate cells. Further characterization of the anti-HIV-1 nature of megabat cells would be needed to reveal the mechanisms by which the cells reduced their HIV-1 susceptibility.
MATERIALS AND METHODS
Cell lines.
Human 293FT (Invitrogen, USA), U-2 OS (ATCC HTB-96), MDTF cell line, and canine rhabdomyosarcoma cell line CMS-C (kind gift from Daigo Azakami, Tokyo University of Agriculture and Technology) (28) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 units/mL of penicillin and streptomycin. HeLa cells were maintained in minimum essential medium (MEM) with 10% FBS and penicillin/streptomycin. Human BJ-5ta cells (ATCC CRL-4001) were maintained in MEM with 10% FBS, 1 mM sodium pyruvate, and penicillin/streptomycin. R-06E, Tb1.Lu, and EfK3B cell lines were purchased from DSMZ GmbH (ACC-756, Germany), ATCC (CCL-88, USA), and Kerafast Inc. (ESA001, USA), respectively. The EnK cell line had been established previously (43). BKT-1, DemKT-1, FBKT-1, YubKT-1, and YubKT-2 cell lines were kindly gifted by Ken Maeda and Keita Ishijima (National Institute for Infectious Diseases, Japan) (44–46). The SuBK12-08, ZFBK11-97, and ZFBK13-76E cell lines were kindly gifted by Ayato Takada (Hokkaido University, Japan) and Edgar Simulundu (University of Zambia, Zambia) (47, 48). The details of the bat cell lines utilized in this study are summarized in Table 1.
DNA.
To verify the species of bat from which each cell line was established, the full-length mitochondrial cytochrome B (Cyt B) gene was sequenced. After purification of total cellular genomic DNA (QIAamp DNA Mini Kit; Qiagen, Germany), partial mitochondrial DNA was amplified by PCR and sequenced directly in both directions as described previously (49). The nucleotide sequences of the PCR primers are listed in Table 2.
cDNA.
To clone TRIM5α, Mx2, and CPSF6 genes from bat cell lines, reverse transcription-PCR (RT-PCR) was performed using specific primers designed on predicted bat sequences (NCBI GenBank). After total cellular RNA extraction (QIAamp RNeasy Minikit, Qiagen), mRNA was reverse-transcribed into the first single-stranded cDNA (High Capacity cDNA Reverse Transcription Kits, Applied Biosystems, USA) with an oligo(dT) adapter primer (50). To amplify cDNA, semi-nested PCR was performed using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, USA). For the first-round amplification, the forward primer included a delivery vector-specific sequence followed by the 5′-end sequence of the gene of interest and a reverse outer adapter primer (50) (Table 2). The second-round amplification was performed using the same forward primer and a reverse primer specific to the 3′-end sequence of the gene of interest, followed by a delivery vector-specific sequence (Table 2). The nucleotide sequences of the newly identified cDNA have been deposited in NCBI GenBank (accession numbers OP272517 to OP272534).
Plasmid.
To generate HIV-1 single-round replicating viruses, the Gag-Pol packaging pMDLg/pRRE and psPax2 (Addgene no. 12251 and 12260, respectively), the viral genome pWPT-GFP (Addgene no. 12255), and HIV-1 Rev-expressing pRSVrev (Addgene no. 12253) were purchased from Addgene (gifted by Didier Trono). To generate MLV, the Gag-Pol packaging pCIG3N (N-MLV), pCIG3B (B-MLV) (51), and pHIT60 (Moloney MLV) (52); the viral genome pCFG2-eGFP and pLGatewaySN (27); and the VSV-G envelope protein expressing pczVSV-G were obtained (gifted by Jonathan Stoye).
Introduction of point mutations in the CA of HIV-1 (pMDLg/pRRE) and MLV (pCIG3N and pCIG3B) Gag genes and substitution or deletion in bat CPSF6, TRIM5α, and Mx2/MxB were performed using an inverse mutagenesis PCR protocol (53).
MLV and HIV-based delivery vector plasmids to transduce genes of interest were constructed using the Gibson Assembly protocol (54, 55) (Gibson Assembly Master Mix; New England Biolabs, USA). To construct HIV and MLV-based transgene plasmids carrying a gene of either red or far-red fluorescent protein, the eGFP gene of pWPT-GFP or pCFG2-eGFP was replaced with tdTomato and E2-Crimson genes, respectively; the resulting plasmids were pWPT-tdTomato, pWPT-E2-Crimson, pCFG2-tdTomato, and pCFG2-E2-Crimson. To construct a transgene delivery vector plasmid, PCR-amplified cDNA with a V5-tag (GKPIPNPLLGLDST)-flexible GS linker (GGGGS) fragment was cloned directly into the retroviral pLGatewaySN vector (27) by substituting the Gateway cassette.
All substitutions or deletions in the CA region of the Gag-Pol packaging plasmids and newly cloned bat cDNA were verified by sequencing in both directions. The nucleotide sequences of the PCR primers designed in this section are listed in Table 2.
Phylogenetic analysis.
The NCBI GenBank database was queried with the nucleotide sequences of newly identified bat cDNA to identify homologs. The amino acid sequences of bat Cyt B, TRIM5α, Mx2/MxB, and CPSF6 were computationally predicted using ExPAsy (Swiss Institute of Bioinformatics; https://web.expasy.org/translate/). Nucleotide and amino acid sequences were aligned using Clustal Omega (European Bioinformatics Institute; https://www.ebi.ac.uk/Tools/msa/clustalo/). An ML-based phylogenetic tree was generated using the Phylip software (version 2.1) with a bootstrapping procedure (1,000 replicates) (56). The phylogenetic tree was constructed as follows: the deduced amino acid sequences were aligned by MAFFT using L-INS-i (Cyt B) or E-INS-i (TRIMs) algorithms (57). The ambiguously aligned regions were removed by trimAl with the -strict (Cyt B) or -gt 0.9 (TRIMs) options (58). The phylogenetic relationship was inferred by the ML method using RAxML Next Generation (59), either with the TVM+I+G4 model, which showed the lowest BIC by ProtTest 3 (60), or the HIVB+I+G4+F model, which was chosen by ModelTest-NG (61). The reliability of the tree nodes was analyzed by 1,000-bootstrap resampling using the transfer bootstrap expectation method (62). The tree was visualized using the MEGA7 software (63).
Positive selection analysis.
We generated multiple nucleotide sequence alignments using the TranslatorX program version 1.1 (64) based on the amino acid sequence multiple alignments created by the L-INS-i program in the MAFFT suite version 7.467 (57). We then computed an ML-based phylogenetic tree using the RAxML-NG program version 1.0.0, with the GTR substitution model (59). Natural selection on each gene was examined by the codeml program in the PAML suite version 4.8 (65). To infer that natural selection may operate for each branch, we used the free-ratio model to calculate the dN/dS ratio for each branch. For the codon site selection analysis, we compared the likelihood of site models M7 and M8 to test for positive selection using the likelihood ratio test (LRT). We then examined the posterior probability estimated using the Bayes Empirical Bayes (BEB) method and determined that they were under positive selection with a BEB of ≥0.95 (66).
Viruses.
Single-round replicating HIV-1 and MLV were produced by cotransfection of a viral genomic, a Gag-Pol packaging, and a VSV-G-expressing plasmid into 293FT cells using polyethyleneimine, as previously described (27, 67). For HIV-based viruses, packaging psPax2 and viral genomic pWPT-GFP or its derivatives were included. For MLV-based viruses, Gag-Pol expressing pCIG3B (B-MLV), pCIG3N (N-MLV), or pHIT60 (Moloney MLV) and viral genomic pCFG2-eGFP or its derivatives were included. For SIV-based virus, packaging pSIV3+ and viral genomic pHIV2-GFP (68) (gifted by Jonathan Stoye) were included. For HIV-1 CA mutant viruses, 293FT cells were transfected with pczVSV-G, pMDLg/pRPE containing CA point mutations, pRSVrev, and pWPT-GFP. When necessary, virus was concentrated using a Lenti-X Concentrator (Clontech). All viral stocks were stored at –80°C until use.
Infection.
To examine viral infectivity, cells were infected with an increasing amount of a tester virus to provide a titration curve, and the ratio of infected cells was measured by detecting fluorescent protein expression using flow cytometry as described previously (69). To quantify HIV-1 RT products and proviruses, cells (150,000 cells/well in 6-well plates) were infected with DNase I-treated HIV-1 as described previously (69). When indicated, the cells were treated with an integrase inhibitor, raltegravir (Selleck Biotech), at a final concentration of 5 μM during infection. After washing once with PBS, the cell pellet was stored at –80°C for DNA extraction.
Quantitative PCR.
To examine the stages of viral replication in infected cells, viral RT products were quantified by qPCR. Total cellular DNA was extracted using a QIAamp DNA Minikit (Qiagen). For HIV-1 2-LTR quantification, DNA was extracted using a QIAprep Spin Miniprep kit (Qiagen) as described previously (70). HIV-1 RT products were measured by qPCR as previously described (17, 71) using the Luna Universal qPCR Master Mix (New England Biolabs). Because Alu repeat sequences are not present in the bat genome, the HIV-1 provirus was detected with a late RT product-specific primer/probe set in multiple-passaged infected cells, where HIV-1 DNA would be mostly lost due to dilution effects and degradation if not integrated into the chromosomes. Primers/probes and PCR conditions are listed in Tables 2 and 3, respectively.
TABLE 3.
PCR conditions used in this studya
| PCR conditions |
||||
|---|---|---|---|---|
| Viral RT product | Purpose | Temp (°C) | Duration | Cycles |
| Early and late RT product | ||||
| Step 1 | Initial denaturation | 95 | 10 min | Hold |
| Step 2 | Denaturation | 95 | 15 s | 45 cycles (steps 2–4) |
| Step 3 | Annealing | 55 | 30 s | |
| Step 4 | Extension | 60 | 30 s | |
| 2-LTR circles | ||||
| Step 1 | Initial heating | 50 | 2 min | Hold |
| Step 2 | Initial denaturation | 95 | 10 min | Hold |
| Step 3 | Denaturation | 95 | 15 s | 45 cycles (steps 3 and 4) |
| Step 4 | Annealing/extension | 60 | 1 min | |
| Proviruses | ||||
| Step 1 | Initial denaturation | 95 | 10 min | Hold |
| Step 2 | Denaturation | 95 | 15 sec | 45 cycles (steps 2–4) |
| Step 3 | Annealing | 60 | 1 min | |
| Step 4 | Extension | 72 | 1 min | |
RT, reverse transcription; LTR, long terminal repeat.
Infectivity in heterokaryons.
Heterokaryon cells were produced with PEG 1500, as described previously (72). Prior to fusion, cells were transduced with a set of separate fluorescent proteins (either tdTomato or eGFP) by inoculating with CFG2 or WPT viruses, and transduced cells were sorted for fluorescent protein expression using a BD FACSAria II cell sorter. PEG-treated cells were inoculated with an E2-Crimson-expressing tester virus. Following harvest, the cells were fixed and loaded onto a BD LSRFortessa cell analyzer. Heterokaryons were identified as double-positive for GFP and tdTomato expression. E2-Crimson-positive cells in the DP fraction were identified as susceptible fused cells. To avoid the exclusion of large heterokaryons, we gated a singlet and doublet cell area in an FSC-SSC FACS panel, thereby enabling analysis of unfused doublets formed by permissive and bat cells that exhibit a double-positive in the FACS panel in PEG-untreated experiments.
Western blot.
After lysis with PBS containing 1% NP-40, proteins in the cell lysate were separated using SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The protein of interest on the membrane was detected by Western blotting as described previously (69). To detect V5-tagged proteins, the membrane was probed with an anti-V5-tag antibody (clone E9H80, Cell Signaling Technology). After washing, the membrane was incubated with a secondary antibody (goat anti-mouse IgG antibody conjugated with horseradish peroxidase, Jackson ImmunoResearch). After detection, the blot was stripped using a commercial stripping solution (WB Stripping Solution Strong, Nacalai Tesque). Then, the membrane was reacted with antibodies and developed as described above, using a tubulin-specific antibody (DMIA, Cell Signaling Technology). Protein bands were visualized using WSE-6100 LuminoGraph I (ATTO).
Immunocytostaining.
V5-tagged proteins were stained and observed under a confocal microscope as described previously (69). Stably expressing cells cultured on a glass coverslip were probed with the anti-V5 antibody (the same antibody used for Western blotting). After washing, the cells were incubated with a secondary antibody (goat anti-rabbit IgG conjugated with Alexa Fluor 488 or 568; Invitrogen). The slides were stored at –20°C until observation under confocal microscopes (LMS980, Zeiss).
CPSF6 transcript search.
The genome sequence and annotations of R. aegyptiacus (JACASE010000002.1) were downloaded from the NCBI database (73), which were visualized using Geneious Prime 2022.1.1 (https://www.geneious.com). The annotations were manually analyzed. RNA-seq data obtained from R. aegyptiacus (accession numbers available in Table S1) were downloaded from the NCBI SRA database (73). The short reads were preprocessed by fastp version 0.22.0 (74) using the default setting, and then mapped to the reference genome of R. aegyptiacus (GCF_014176215.1_mRouAeg1.p) by HISAT2 version 2.2.1 (75) using the default setting. The mapped reads were then visualized by IGV (76) or Geneious Prime 2022.1.1.
Statistical analysis.
Statistical analysis was performed using the JASP software package (version 0.14.1; University of Amsterdam) (77). Statistical significance between groups was assessed using analysis of variance (ANOVA) (P < 0.05 was considered significant) and, if not significant, a nonparametric Kruskal-Wallis test was used. When statistical significance was assessed, post hoc comparisons between groups (Dunn’s test for Fig. 3) or the control group (Dunnett’s test for Fig. 2B, 4, 6C, 7D, 8B, and 8F) were performed. P values of <0.05 (*), <0.01 (**), and <0.001 (***) were considered significant.
ACKNOWLEDGMENTS
We thank Jonathan P Stoye for the critical reading of the manuscript. We thank Ayato Takada (Hokkaido University) and Edgar Simulundu (University of Zambia) for providing the SuBK12-08, ZFBK11-97, and ZFBK13-76E cell lines; Ken Maeda and Keita Ishijima (The National Institute for Infectious Diseases) for providing the BKT-1, DemKT-1, FBKT-1, YubKT-1, and YubKT-2 cell lines; and Daigo Azakami (Tokyo University of Agriculture and Technology) for providing CMS-C cells. We also thank Jonathan Stoye for providing the MLV plasmids and Didier Trono, Doug Golenbock, Rudolf Jaenisch, and Charles Gersbach for providing plasmids via Addgene.
This study was supported in part by a grant from JSPS KAKENHI (grant no. 15K06811 to S.O.).
Footnotes
Supplemental material is available online only.
Contributor Information
Sadayuki Ohkura, Email: sokura@nms.ac.jp.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Escalera-Zamudio M, Mendoza MLZ, Heeger F, Loza-Rubio E, Rojas-Anaya E, Méndez-Ojeda ML, Taboada B, Mazzoni CJ, Arias CF, Greenwood AD. 2015. A novel endogenous betaretrovirus in the common vampire bat (Desmodus rotundus) Suggests multiple independent infection and cross-species transmission events. J Virol 89:5180–5184. 10.1128/JVI.03452-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hron T, Farkašová H, Gifford RJ, Benda P, Hulva P, Görföl T, Pačes J, Elleder D. 2018. Remnants of an ancient Deltaretrovirus in the genomes of horseshoe bats (Rhinolophidae). Viruses 10:185. 10.3390/v10040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farkašová H, Hron T, Pačes J, Hulva P, Benda P, Gifford RJ, Elleder D. 2017. Discovery of an endogenous Deltaretrovirus in the genome of long-fingered bats (Chiroptera: Miniopteridae). Proc Natl Acad Sci USA 114:3145–3150. 10.1073/pnas.1621224114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J, Tachedjian M, Wang L, Tachedjian G, Wang L-F, Zhang S. 2012. Discovery of retroviral homologs in bats: implications for the origin of mammalian gammaretroviruses. J Virol 86:4288–4293. 10.1128/JVI.06624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J, Tachedjian G, Tachedjian M, Holmes EC, Zhang S, Wang LF. 2012. Identification of diverse groups of endogenous gammaretroviruses in mega- and microbats. J Gen Virol 93:2037–2045. 10.1099/vir.0.043760-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayward JA, Tachedjian M, Kohl C, Johnson A, Dearnley M, Jesaveluk B, Langer C, Solymosi PD, Hille G, Nitsche A, Sánchez CA, Werner A, Kontos D, Crameri G, Marsh GA, Baker ML, Poumbourios P, Drummer HE, Holmes EC, Wang LF, Smith I, Tachedjian G. 2020. Infectious KoRV-related retroviruses circulating in Australian bats. Proc Natl Acad Sci USA 117:9529–9536. 10.1073/pnas.1915400117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayward JA, Tachedjian M, Cui J, Cheng AZ, Johnson A, Baker ML, Harris RS, Wang LF, Tachedjian G. 2018. Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 gene complexity. Mol Biol Evol 35:1626–1637. 10.1093/molbev/msy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward J, Tachedjian M, Johnson A, Gordon T, Cui J, Marsh G, Baker M, Wang L-F, Tachedjian G. 2022. Unique evolution of antiviral tetherin in bats. J Virol 96:e0115222. 10.1128/jvi.01152-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak CA, Chakraborti A. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300–305. 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 10.Young GR, Yap MW, Michaux JR, Steppan SJ, Stoye JP. 2018. Evolutionary journey of the retroviral restriction gene Fv1. Proc Natl Acad Sci USA 115:10130–10135. 10.1073/pnas.1808516115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 12.Lee KE, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita M, Engelman AN. 2017. Capsid-dependent host factors in HIV-1 infection. Trends Microbiol 25:741–755. 10.1016/j.tim.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novikova M, Zhang Y, Freed EO, Peng K. 2019. Multiple roles of HIV-1 capsid during the virus replication cycle. Virol Sin 34:119–134. 10.1007/s12250-019-00095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colgan J, Yuan HE, Franke EK, Luban J. 1996. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J Virol 70:4299–4310. 10.1128/JVI.70.7.4299-4310.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison JH, Miller C, Bankers L, Crameri G, Wang LF, Poeschla EM. 2020. A potent postentry restriction to primate lentiviruses in a yinpterochiropteran bat. mBio 11:e01854-20. 10.1128/mBio.01854-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. 2009. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol Biol 485:55–72. 10.1007/978-1-59745-170-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munir S, Thierry S, Subra F, Deprez E, Delelis O. 2013. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology 10:87. 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Iaco A, Luban J. 2011. Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus. Retrovirology 8:98. 10.1186/1742-4690-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matreyek KA, Wang W, Serrao E, Singh KP, Levin HL, Engelman A. 2014. Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology 11:90. 10.1186/s12977-014-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hué S, Fletcher AJ, Lee KE, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achuthan V, Perreira JM, Sowd GA, Puray-Chavez M, McDougall WM, Paulucci-Holthauzen A, Wu X, Fadel HJ, Poeschla EM, Multani AS, Hughes SH, Sarafianos SG, Brass AL, Engelman AN. 2018. Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 24:392–404.e8. 10.1016/j.chom.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog 3:e197. 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkura S, Goldstone DC, Yap MW, Holden-Dye K, Taylor IA, Stoye JP. 2011. Novel escape mutants suggest an extensive TRIM5α binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog 7:e1002011. 10.1371/journal.ppat.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkura S, Stoye JP. 2013. A comparison of murine leukemia viruses that escape from human and rhesus macaque TRIM5αs. J Virol 87:6455–6468. 10.1128/JVI.03425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yap MW, Dodding MP, Stoye JP. 2006. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J Virol 80:4061–4067. 10.1128/JVI.80.8.4061-4067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azakami D, Shibutani H, Dohi M, Takasaki M, Ishioka K, Mori A, Momota Y, Bonkobara M, Washizu T, Michishita M, Hatakeyama H, Ogasawara S, Sako T. 2011. Establishment and characterization of canine rhabdomyosarcoma cell line CMS-C. J Vet Med Sci 73:1105–1108. 10.1292/jvms.10-0436. [DOI] [PubMed] [Google Scholar]
- 29.Yap MW, Nisole S, Stoye JP. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr Biol 15:73–78. 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 30.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. 2011. Hexagonal assembly of a restricting TRIM5α protein. Proc Natl Acad Sci USA 108:546–550. 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maillard PV, Ecco G, Ortiz M, Trono D. 2010. The specificity of TRIM5α-mediated restriction is influenced by its coiled-coil domain. J Virol 84:5790–5801. 10.1128/JVI.02413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelman AN. 2021. HIV capsid and integration targeting. Viruses 13:125. 10.3390/v13010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchs J, Hölzer M, Schilling M, Patzina C, Schoen A, Hoenen T, Zimmer G, Marz M, Weber F, Müller MA, Kochs G. 2017. Evolution and antiviral specificities of interferon-induced Mx proteins of bats against Ebola, influenza, and other RNA viruses. J Virol 91:e00361-17. 10.1128/JVI.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busnadiego I, Kane M, Rihn SJ, Preugschas HF, Hughes J, Blanco-Melo D, Strouvelle VP, Zang TM, Willett BJ, Boutell C, Bieniasz PD, Wilson SJ. 2014. Host and viral determinants of Mx2 antiretroviral activity. J Virol 88:7738–7752. 10.1128/JVI.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goujon C, Moncorgé O, Bauby H, Doyle T, Barclay WS, Malim MH. 2014. Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J Virol 88:9017–9026. 10.1128/JVI.01269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nuñez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goujon C, Greenbury RA, Papaioannou S, Doyle T, Malim MH. 2015. A triple-arginine motif in the amino-terminal domain and oligomerization are required for HIV-1 inhibition by human MX2. J Virol 89:4676–4680. 10.1128/JVI.00169-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulte B, Buffone C, Opp S, Di Nunzio F, De Souza Aranha Vieira DA, Brandariz-Nuñez A, Diaz-Griffero F. 2015. Restriction of HIV-1 requires the N-terminal region of MxB as a capsid-binding motif but not as a nuclear localization signal. J Virol 89:8599–8610. 10.1128/JVI.00753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole DS, Yú S, Caì Y, Dinis JM, Müller MA, Jordan I, Friedrich TC, Kuhn JH, Mehle A. 2014. Influenza A virus polymerase is a site for adaptive changes during experimental evolution in bat cells. J Virol 88:12572–12585. 10.1128/JVI.01857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann M, Müller MA, Drexler JF, Glende J, Erdt M, Gützkow T, Losemann C, Binger T, Deng H, Schwegmann-Wessels C, Esser KH, Drosten C, Herrler G. 2013. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One 8:e72942. 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. 10.1128/jvi.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douglas CC, Thomas D, Lanman J, Prevelige PE. 2004. Investigation of N-terminal domain charged residues on the assembly and stability of HIV-1 CA. Biochemistry 43:10435–10441. 10.1021/bi049359g. [DOI] [PubMed] [Google Scholar]
- 43.Horie M, Akasaka T, Matsuda S, Ogawa H, Imai K. 2016. Establishment and characterization of a cell line derived from Eptesicus nilssonii. J Vet Med Sci 78:1727–1729. 10.1292/jvms.16-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T, Matsugo H, Maruyama J, Kamiki H, Takada A, Maeda K, Takenaka-Uema A, Tohya Y, Murakami S, Horimoto T. 2019. Characterization of a novel species of adenovirus from Japanese microbat and role of CXADR as its entry factor. Sci Rep 9:573. 10.1038/s41598-018-37224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda K, Hondo E, Terakawa J, Kiso Y, Nakaichi N, Endoh D, Sakai K, Morikawa S, Mizutani T. 2008. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg Infect Dis 14:347–349. 10.3201/eid1402.070932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iida K, Kobayashi R, Hengjan Y, Nagata N, Yonemitsu K, Nunome M, Kuwata R, Suzuki K, Ichiyanagi K, Maeda K, Ohmori Y, Hondo E. 2017. The genetic diversity of D-loop sequences in eastern bent-winged bats (Miniopterus fuliginosus) living in Wakayama Prefecture, Japan. J Vet Med Sci 79:1142–1145. 10.1292/jvms.17-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa H, Kajihara M, Nao N, Shigeno A, Fujikura D, Hang’Ombe BM, Mweene AS, Mutemwa A, Squarre D, Yamada M, Higashi H, Sawa H, Takada A. 2017. Characterization of a novel bat adenovirus isolated from straw-colored fruit bat (Eidolon helvum). Viruses 9:371. 10.3390/v9120371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maruyama J, Miyamoto H, Kajihara M, Ogawa H, Maeda K, Sakoda Y, Yoshida R, Takada A. 2014. Characterization of the envelope glycoprotein of a novel filovirus, lloviu virus. J Virol 88:99–109. 10.1128/JVI.02265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naidu A, Fitak RR, Munguia-Vega A, Culver M. 2012. Novel primers for complete mitochondrial cytochrome b gene sequencing in mammals. Mol Ecol Resour 12:191–196. 10.1111/j.1755-0998.2011.03078.x. [DOI] [PubMed] [Google Scholar]
- 50.Yang CH, Yue J, Fan M, Pfeffer LM. 2010. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res 70:8108–8116. 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bock M, Bishop KN, Towers G, Stoye JP. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J Virol 74:7422–7430. 10.1128/jvi.74.16.7422-7430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res 23:628–633. 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominy CN, Andrews DW. 2003. Site-directed mutagenesis by inverse PCR. Methods Mol Biol 235:209–223. 10.1385/1-59259-409-3:209. [DOI] [PubMed] [Google Scholar]
- 54.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 55.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZN, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, Smith HO, Venter JC. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329:52–56. 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 56.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol 37:291–294. 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lemoine F, Domelevo Entfellner J-B, Wilkinson E, Correia D, Dávila Felipe M, De Oliveira T, Gascuel O. 2018. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556:452–456. 10.1038/s41586-018-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38:W7–13. 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 66.Yang Z, Wong WSW, Nielsen R. 2005. Bayes Empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118. 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 67.Longo PA, Kavran JM, Kim MS, Leahy DJ. 2013. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol 529:227–240. 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nègre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther 7:1613–1623. 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 69.Yap MW, Mortuza GB, Taylor IA, Stoye JP. 2007. The design of artificial retroviral restriction factors. Virology 365:302–314. 10.1016/j.virol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 70.Teo IA, Morlese J, Choi JW, Shaunak S. 2002. Reliable and reproducible LightCycler qPCR for HIV-1 DNA 2-LTR circles. J Immunol Methods 270:109–118. 10.1016/s0022-1759(02)00217-x. [DOI] [PubMed] [Google Scholar]
- 71.Butler SL, Hansen MST, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat Med 7:631–634. 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 72.Villafranca C, Makris MR, Garrido Bauerle MJ, Jensen RV, Eyestone WH. 2020. Production of interspecies somatic/pluripotent heterokaryons using polyethylene glycol (PEG) and selection by imaging flow cytometry for the study of nuclear reprogramming. Cytotechnology 72:797–805. 10.1007/s10616-020-00416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Connor R, Funk K, Kelly C, Kim S, Madej T, Marchler-Bauer A, Lanczycki C, Lathrop S, Lu Z, Thibaud-Nissen F, Murphy T, Phan L, Skripchenko Y, Tse T, Wang J, Williams R, Trawick BW, Pruitt KD, Sherry ST. 2022. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 50:D20–D26. 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagenmakers E-J, Love J, Marsman M, Jamil T, Ly A, Verhagen J, Selker R, Gronau QF, Dropmann D, Boutin B, Meerhoff F, Knight P, Raj A, van Kesteren E-J, van Doorn J, Šmíra M, Epskamp S, Etz A, Matzke D, de Jong T, van den Bergh D, Sarafoglou A, Steingroever H, Derks K, Rouder JN, Morey RD. 2018. Bayesian inference for psychology. Part II: example applications with JASP. Psychon Bull Rev 25:58–76. 10.3758/s13423-017-1323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA 101:10786–10791. 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jebb D, Huang Z, Pippel M, Hughes GM, Lavrichenko K, Devanna P, Winkler S, Jermiin LS, Skirmuntt EC, Katzourakis A, Burkitt-Gray L, Ray DA, Sullivan KAM, Roscito JG, Kirilenko BM, Dávalos LM, Corthals AP, Power ML, Jones G, Ransome RD, Dechmann DKN, Locatelli AG, Puechmaille SJ, Fedrigo O, Jarvis ED, Hiller M, Vernes SC, Myers EW, Teeling EC. 2020. Six reference-quality genomes reveal evolution of bat adaptations. Nature 583:578–584. 10.1038/s41586-020-2486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Table S1. Download jvi.01803-22-s0001.pdf, PDF file, 17.1 MB (17.5MB, pdf)