ABSTRACT
Seasonal coronaviruses have been circulating widely in the human population for many years. With increasing age, humans are more likely to have been exposed to these viruses and to have developed immunity against them. It has been hypothesized that this immunity to seasonal coronaviruses may provide partial protection against infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and it has also been shown that coronavirus disease 2019 (COVID-19) vaccination induces a back-boosting effects against the spike proteins of seasonal betacoronaviruses. In this study, we tested if immunity to the seasonal coronavirus spikes from OC43, HKU1, 229E, or NL63 would confer protection against SARS-CoV-2 challenge in a mouse model, and whether pre-existing immunity against these spikes would weaken the protection afforded by mRNA COVID-19 vaccination. We found that mice vaccinated with the seasonal coronavirus spike proteins had no increased protection compared to the negative controls. While a negligible back-boosting effect against betacoronavirus spike proteins was observed after SARS-CoV-2 infection, there was no negative original antigenic sin-like effect on the immune response and protection induced by SARS-CoV-2 mRNA vaccination in animals with pre-existing immunity to seasonal coronavirus spike proteins.
IMPORTANCE The impact that immunity against seasonal coronaviruses has on both susceptibility to SARS-CoV-2 infection as well as on COVID-19 vaccination is unclear. This study provides insights into both questions in a mouse model of SARS-CoV-2.
KEYWORDS: SARS-CoV-2, seasonal coronaviruses, cross-reactive antibody, spike
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a beta-coronavirus (β-CoV), first emerged in late 2019 and has since then caused the COVID-19 pandemic (1). SARS-CoV-2 is the fifth CoV to cause large scale outbreaks in humans. The four seasonal CoVs—α-CoVs 229E (2) and NL63 (Netherlands 63 [3, 4]) and β-CoVs OC43 (organ culture 43) and HKU1 (Hong Kong University 1 [5])—have likely been circulating in humans for decades, potentially even longer, and cause a sizable percentage of common colds every winter (6–8). In addition, OC43 is suspected to have jumped from cattle into humans in the late 19th century and to be the causative agent of the 1889/1890 “Russian flu” pandemic (9). Infections with seasonal CoVs happen frequently and it is assumed that a large proportion of the human population, maybe even the entire population, has immune memory and circulating antibody against these viruses derived from previous infections. While immunity to seasonal CoVs may wane over time leading to reinfection, a large proportion of the population is likely—at least partially—protected (10).
In has been hypothesized that this pre-existing immunity to CoVs, including antibodies to the spike protein, may provide a degree of protection against SARS-CoV-2 infection and COVID-19, especially in children (11). Rare individuals in fact have detectable cross-reactive anti-SARS-CoV-2 spike antibodies before exposure to SARS-CoV-2 (12). It has also been shown that cross-reactive antibodies are induced in individuals infected with SARS-CoV-2 or immunized with COVID-19 vaccines in a “back-boost” or “original antigenic sin”-like manner (13, 14). Based on conservation (Table 1), more cross-reactivity between β-CoVs is expected, especially antibodies targeting the more conserved S2 domain. One of the seasonal CoVs, NL63, uses the same receptor as SARS-CoV-2, namely, angiotensin converting enzyme 2 (ACE2) (15, 16), while 229E uses human aminopeptidase N (17) and both HKU1 and OC43 use 9-O-acetylated sialic acids (18). Because the area on ACE2 to which the SARS-CoV-2 and NL63 spikes bind overlaps to some degree (19), it may be possible that ACE2-mimicking antibodies cross-react between the two RBDs to some extent. Such cross-reactivity is not expected between the RBDs of other seasonal CoVs and SARS-CoV-2. Here, we have investigated the protective effect of immunity against the different seasonal CoV spike proteins against SARS-CoV-2 infection in a mouse model and we also explored if pre-existing immunity to the seasonal CoV spike proteins has a detrimental impact on the protective effect of COVID-19 vaccination.
TABLE 1.
Percent amino acid identity matrix for 229E (NP_073551.1), NL63 (AFV53148.1), SARS-CoV-2 (QHD43416.1), OC43 (AIX10759.1) and HKU1 (AGW27881.1) full-length spikes and S2 subunits
| Protein | 229E | NL63 | SARS-CoV-2 | OC43 | HKU1 |
|---|---|---|---|---|---|
| Full-length spike | |||||
| 229E | 100.0 | ||||
| NL63 | 64.15 | 100.0 | |||
| SARS-CoV-2 | 27.18 | 26.12 | 100.0 | ||
| OC43 | 27.53 | 28.57 | 32.27 | 100.0 | |
| HKU1 | 28.05 | 27.06 | 31.81 | 65.13 | 100.0 |
| S2 subunit | |||||
| 229E | 100.0 | ||||
| NL63 | 75.91 | 100.0 | |||
| SARS-CoV-2 | 35.12 | 34.10 | 100.0 | ||
| OC43 | 33.81 | 36.51 | 41.25 | 100.0 | |
| HKU1 | 34.45 | 35.56 | 39.27 | 73.40 | 100.0 |
RESULTS
Seasonal CoV spike proteins induce strong specific immune responses in mice with detectable cross-reactivity within α-CoVs and β-CoV.
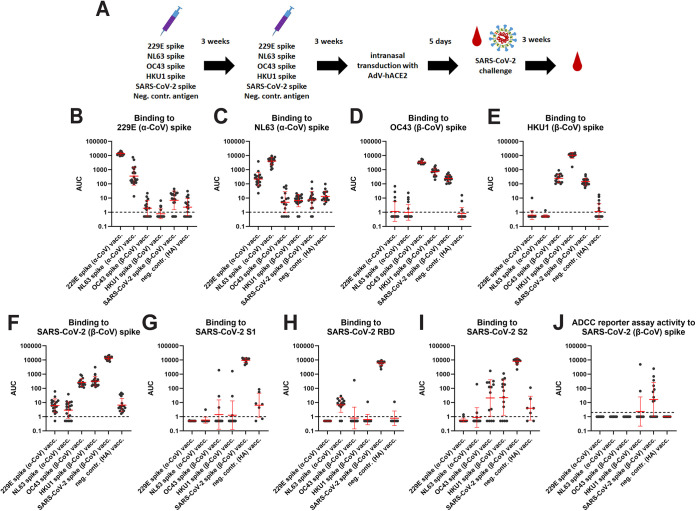
The spike proteins of 229E, NL63, OC43, HKU1, and SARS-CoV-2 were expressed as soluble trimers, and mice were vaccinated with these proteins with an oil-in-water emulsion adjuvant in a prime-boost regimen (Fig. 1A). Control mice were vaccinated in the same manner but with an irrelevant recombinant antigen, recombinant influenza virus hemagglutinin (HA). Four weeks postboost, animals were bled and their immune response was assessed by enzyme-linked immunosorbent assay (ELISA). Sera from 229E vaccinated mice reacted strongly with 229E spike and reduced reactivity was also detected against the NL63 spike (Fig. 1B). No reactivity was detected to any of the three β-CoV spikes. While NL63 spike seemed to be less immunogenic, it produced similar results with stronger reactivity to the homologous spike, reduced activity to the 229E spike and some, albeit very low reactivity to the β-CoV spikes (Fig. 1C). Vaccination with OC43 led to strong anti-OC43 spike reactivity, reduced reactivity to the HKU-1 spike was detected followed by reactivity to the SARS-CoV-2 spike (Fig. 1D). Reactivity to the α-CoVs was negligible. HKU1 spike vaccination induced strong reactivity to the homologous spike and lower titers to OC43 and SARS-CoV-2 spikes, but reactivity to the α-CoV spikes was at baseline (Fig. 1E). Vaccination with SARS-CoV-2 spike induced high titers to itself and some cross-reactivity to OC43 and HKU-1 but no reactivity to α-CoV spikes (Fig. 1F). Strong reactivity of SARS-CoV-2 spike vaccinated animals was detected against S1, RBD, and S2 (Fig. 1G, H, and I). Animals vaccinated with seasonal CoV spikes had low or absent reactivity to S1 and RBD with the exception of NL63 spike vaccinated animals which did show low but consistent reactivity to RBD but not S1 (Fig. 1G and H). Finally, animals vaccinated with β-CoV spikes showed reactivity to S2 of SARS-CoV-2 while animals vaccinated with α-CoV spikes did not (Fig. 1I). In addition to ELISA binding, we also assessed the activity of these sera in an antibody-dependent cellular cytotoxicity (ADCC) reporter assays which was based on a similar assay we have in the past developed for influenza virus and which is based on commercially available reporter cell lines (20). The results, shown in Fig. 1J, indicate that a proportion of SARS-CoV-2 spike vaccinated mice in fact show activity in this assay against cell surface expressed spike protein. However, despite binding activity, sera from mice vaccinated with any of the seasonal CoV spikes did not show activity in this assay except for two responders in the HKU1 group. This could have a biological reason but it is also possible that our assay is not sensitive enough.
FIG 1.
Antibody reactivity against CoV spikes after vaccination. (A) Schematic of vaccination regimen, challenge and sampling. The red drop indicates bleeding time points. (B) Serum reactivity to 229E spike protein. (C) Serum reactivity to NL63 spike. (D) Serum reactivity to OC43 spike. (E) Serum reactivity to HKU1 spike. (F) Serum reactivity to SARS-CoV-2 spike. (G) Serum reactivity to SARS-CoV-2 S1. (H) Serum reactivity to SARS-CoV-2 RBD. (I) Serum reactivity to SARS-CoV-2 S2. (J) ADCC reporter activity to SARS-CoV-2 spike. The data for the pre-challenge sera are shown in B-J. 229E n = 18, NL63 n = 20, OC43 n-16, HKU1 n = 20, SARS-CoV-2 n = 19, neg. contr. n = 17, except for G to I, where n = 14 for all vaccinated groups and n = 7 for the control group and for J, where n = 15 for all groups. Significant statistical differences for B, C, D, E, F, and J are summarized in Table S1. Red bars indicate geometric mean, the error indicates the geometric standard deviation. Samples were analyzed once except for J where the mean of two replicates is shown.
Seasonal CoV spike immunity provides negligible protection against SARS-CoV-2 infection.
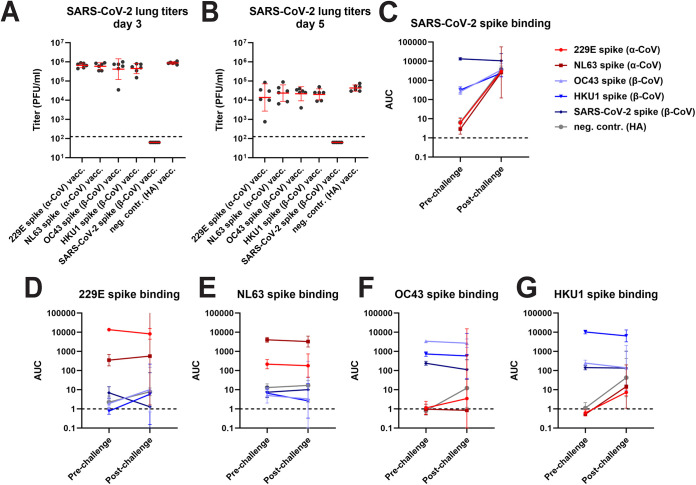
Three weeks after the boost, the mice were transduced intranasally with a nonreplicating adenovirus expressing human angiotensin converting enzyme 2 (ACE2) followed by a challenge with wild-type SARS-CoV-2 strain Washington-1 5 days later (21). In this challenge model animals do not suffer severe disease or succumb to infection. However, viral replication in the lung can be assessed to determine protection. A proportion of the animals were euthanized on day 3 or day 5 postchallenge, and lungs were harvested. The remaining animals were kept to evaluate postchallenge antibody titers. The only group protected from challenge were animals vaccinated with the SARS-CoV-2 spike protein (Fig. 2A and B). High titers of virus were found in all seasonal CoV spike-vaccinated animals—similar to the negative-control group—with exception of one animal in the OC43 spike group on day 3 and one animal each in the OC43 spike and 229E spike groups which displayed lower titers than the control group. None of the animals showed overt disease postchallenge. As described, a subset of animals was kept for assessment of the immune response after infection and these animals were bled 3 weeks postchallenge. Anti-SARS-CoV-2 spike titers in all seasonal CoV spike-vaccinated animals had substantially increased (Fig. 2C). A similar increase was found in the control group with titers in the seasonal CoV spike and the control groups being very similar. Titers in the SARS-CoV-2 spike-immunized animals did not increase postinfection suggestive of sterilizing immunity. Little to no “back-boosting” to seasonal CoV spike proteins was detected (Fig. 2D to G).
FIG 2.
Lung titers of CoV spike vaccinated animals post SARS-CoV-2 challenge and changes in antibody titer. Post challenge lung titers on day 3 and day 5 postchallenge are shown in (A) and (B). Changes in reactivity to SARS-CoV-2 spike, 229E spike, NL63 spike, OC43 spike and HKU1 spike after challenge are shown in (C) to G. For (A) and (B), n = 6 per group. The n for the prechallenge time points in (C) to (G) are described in the legend of Fig. 1. For the postchallenge time point, n = 3 for all groups except for NL63 where n = 2. Significant statistical differences are summarized in Tables S2 and 3, red bars indicate geometric mean, the error indicates the geometric standard deviation. For the remaining data panels the geometric mean plus 95% confidence intervals are shown. Samples were analyzed once.
Pre-existing immunity to seasonal CoVs does not negatively impact on protection induced by COVID-19 vaccination.
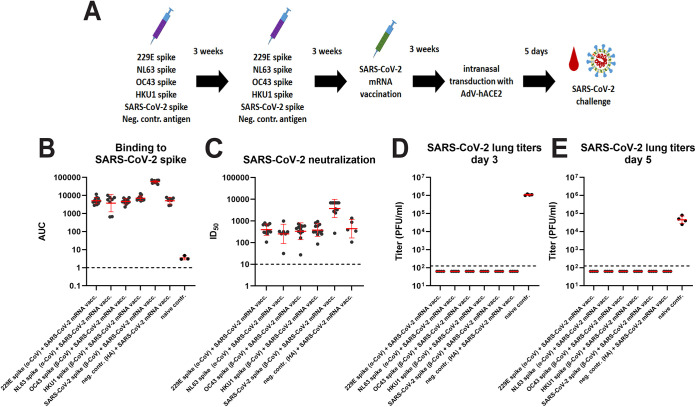
To determine the impact of pre-existing immunity on vaccine-induced protection against SARS-CoV-2, we repeated the vaccination experiment. However, instead of a SARS-CoV-2 challenge, animals were vaccinated 3 weeks postboost with a lipid nanoparticle-formulated nucleoside-modified mRNA vaccine (mRNA-LNP) (22, 23) encoding the SARS-CoV-2 spike protein (Fig. 3A). An additional control group was added that had not received any vaccination with spike proteins and did not receive the mRNA-LNP vaccine. Vaccination induced high binding and neutralization titers against the SARS-CoV-2 spike in all groups, with the highest titers detected in the SARS-CoV-2 spike protein vaccinated mice and lower titers in the animals vaccinated with seasonal CoV spike or influenza virus HA (Fig. 3B and C). The similar titers detected in animals pre-immunized with seasonal CoV spikes and influenza virus HA suggest no detrimental “original-antigenic sin”-like effect on the immune response to the SARS-CoV-2 mRNA-LNP vaccine. Animals were then transduced with human ACE2 expressing adenovirus and challenged with SARS-CoV-2 as described above. The mice were euthanized on days 3 and 5 postchallenge and virus replication in their lungs was assessed. No virus was found in any of the mRNA-LNP-vaccinated animals on either day 3 or 5 (Fig. 3D and E). However, high virus titers were found in completely naive animals.
FIG 3.
Antibody reactivity post COVID-19 mRNA vaccination and postchallenge lung titers in CoV vaccinated animals. (A) Schematic of vaccination regimen, challenge and sampling. (B) shows binding titers to SARS-CoV-2 spike. The red drop indicates a bleeding time point. (C) shows neutralizing activity to authentic ancestral SARS-CoV-2. Post challenge lung titers on day 3 and day 5 postchallenge are shown in (D) and (E). The n for (B) was 12 for the 229E, OC43, and HKU1 groups, 8 for the NL63 group, 11 for the SARS-CoV-2 group, 7 for the negative control group, and 3 for the naive group. For (C) it was 12 for the 229E, OC43, and HKU1 groups, 7 for the NL63 group, 11 for the SARS-CoV-2 group and 5 for the negative control group. For D and E, n = 4 per group. Significant statistical differences are summarized in Table S4. Red bars indicate geometric mean, the error indicates the geometric standard deviation. Samples were analyzed once.
DISCUSSION
Pre-existing immunity is known to provide partial protection and can influence immune responses to infection and vaccination for influenza virus (24). A back-boosting or “original antigenic sin”-like effect on the immune responses to SARS-CoV-2 infection and COVID-19 vaccination has recently been reported as well (13, 14). This was mostly observed toward the β-CoVs OC43 and HKU1 with more reactivity to OC43. Some monoclonal antibodies isolated from exposed individuals, especially the ones targeting the more conserved S2 subunit, have been shown to cross-react to seasonal β-CoV spike proteins as well (13, 25–29). More rare antibodies that target the fusion peptide in S2 even cross-react with α-coronaviruses (30, 31). In addition, it has been hypothesized that antibodies to seasonal CoV spike proteins could, especially in children, provide a degree of protection against SARS-CoV-2 infection and COVID-19 (11).
Here, we tested both the hypothesis that pre-existing immunity to seasonal CoV spikes could have a protective effect against SARS-CoV-2 as well as the hypothesis that pre-existing immunity to seasonal CoV spikes could have a detrimental, “original antigenic sin”-like impact on protection induced by COVID-19 vaccination. Neither was the case in this mouse model. We did in fact observe a degree of cross-reactivity between β-CoV spikes and between α-CoV spikes as suggested by serological studies and MAb analysis (13, 25–31). This included moderate cross-reactivity of anti-OC43 and anti-HKU1 spike immunity to the SARS-CoV-2 spike which seems to target the S2 subunit. An interesting additional finding was, that vaccination with NL63 spike protein did induce low but detectible antibody titers to the RBD of SARS-CoV-2. This was not observed for mice vaccinated with any other seasonal CoV spikes. Furthermore, this reactivity could only be picked up with RBD but not when plates were coded with S1 (which contains the RBD). An explanation for this cross-reactivity could be that the site on ACE2 that is targeted by both SARS-CoV-2 and NL63 RBDs overlaps (19) and that antibodies that mimic ACE2 are responsible for the observed binding. Free RBD may be more sensitive as reagent to detect these antibodies than S1.
However, despite this cross-reactivity, we saw no protective effect against SARS-CoV-2 challenge. This suggests that the cross-reactive antibody titers to the spike were either too low to protect in this model or not functional. The lack of ADCC reporter activity despite cross-binding antibodies is in line with this, even though nonneutralizing SARS-CoV-2 spike antibodies are suspected to provide some protection against severe disease with drifted variant viruses in humans. But there may be differences between SARS-CoV-2 specific nonneutralizing antibodies and cross-reactive nonneutralizing antibodies. Furthermore, human and murine antibodies may differ in this respect too. Another interesting observation in this first part of the study was that challenge with SARS-CoV-2 only increased SARS-CoV-2 spike reactivity; however, a robust back-boosting effect against β-CoVs as observed in humans (13, 14) was not detected. This is noteworthy given the presence of cross-reactive antibodies in OC43 and HKU1 vaccinated animals was detected prechallenge and the expectation was that these antibodies would get boosted. While we did not observe any protection conferred by pre-existing immunity against subsequent SARS-CoV-2 challenge, we did also not detected any negative impact on protection induced by a COVID-19 mRNA-LNP vaccine. Mice were fully protected against challenge after mRNA vaccination irrespectively of their pre-existing immunity. While only one challenge dose was used and a challenge dose escalation experiment (had it been possible) could have resulted in differences at more stringent challenge conditions, our data suggest that no strong original antigenic sin-like effect occurs in this model.
Importantly, our study has several limitations. First, the study was performed in mice and this model may just not accurately reflect responses observed in humans, e.g., due to different immunoglobulin germ lines and strong differences in length of complementarity determining regions (CDRs) (32). We immunized mice with only one type of spike while humans are exposed to all four seasonal CoVs (7, 8), which could lead to a more cross-reactive immunity. We also did not assess the T-cell response to the different spike proteins and could not evaluate T-cell based immunity to other structural and nonstructural proteins of the virus. T-cells that cross-react between seasonal CoV proteins and SARS-CoV-2 are common (33, 34) and may play an important role in protection from severe COVID-19. Therefore, assessing antibody mediated immunity only may underestimate cross-reactive protection. The seasonal CoVs replicate poorly in cell culture (35, 36), and therefore, no neutralization assays were performed. Finally, while our ADCC reporter assay has been established as research grade assay, it may lack sensitivity to detect activity in samples with lower binding antibody titers.
Despite the lack of a protective effect, it is encouraging that cross-reactivity within the β-CoV spikes could be detected. If this cross-reactivity could be enhanced or if its functionality could be increased in humans, it could lead to broad protection within the β-CoV viruses and could form the basis for a pan- β-CoV vaccine. How such a vaccine would need to be designed is currently unclear; however, data from MAbs (13, 25–31) and from early immunogen design studies (37–39) suggest that S2 could be a potential target.
MATERIALS AND METHODS
Recombinant proteins.
Recombinant proteins were expressed and purified using a mammalian expression system via Expi293F cells (Life Technologies), as described in detail earlier (40, 41). Plasmids to express spike proteins of human coronaviruses which include 229E, HKU1, OC43, and NL63 were generously provided by Dr. Barney Graham. In-house expressed proteins were used for immunization. Additional recombinant proteins were purchased from Sino Biologics (NL63 spike [Cat: 40604-V08B], 2293E spike [Cat: 40605-V08B], HKU1 spike [Cat: 40606-V08B], OC43 spike [Cat: 40607-V08B-B], SARS-CoV-2 spike [Cat: 40589-V08B1], S1 [Cat: 40591-V08B1], and S2 [Cat: 40590-V08B]) to perform ELISAs. RBD for ELISAs was produced in-house without a multimerization domain (41). Because immunization with in-house expressed proteins led to significant serum reactivity against the trimerization domain, proteins from Sino Biologicals (with no trimerization domain) were purchased and used. Hence, the immune response to the respective proteins could be assessed in a more accurate fashion.
mRNA-LNP vaccine production.
The mRNA vaccine was designed and made as described (22, 23). Briefly, the coding sequence of full-length Δfurin spike protein (RRAR furin cleavage site abolished between amino acids 682 to 685) from the Wuhan-1 strain (Wuhan-Hu-1, GenBank: MN908947.3) was codon-optimized, synthesized, and cloned into an mRNA production plasmid. The spike-encoding mRNA was transcribed to contain 101 nucleotide-long poly(A) tails. m1Ψ-5′-triphosphate (TriLink) instead of UTP was used to generate modified nucleoside-containing mRNA. Capping of the in vitro transcribed mRNAs was performed cotranscriptionally using the trinucleotide cap1 analog, CleanCap (TriLink). mRNA was purified by cellulose purification as described (42). The mRNA was analyzed by agarose gel electrophoresis and was stored frozen at −20°C. The cellulose-purified m1Ψ-containing mRNA was encapsulated in LNPs using a self-assembly process as previously described wherein an ethanolic lipid mixture of ionizable cationic lipid, phosphatidylcholine, cholesterol, and polyethylene glycol-lipid was rapidly mixed with an aqueous solution containing mRNA at acidic pH. The RNA-loaded particles were characterized and subsequently stored at −80°C at a concentration of 1 μg μL−1. The mean hydrodynamic diameter of these mRNA-LNP was ~80 nm with a polydispersity index of 0.02–0.06 and an encapsulation efficiency of ~95% (43).
Cells and viruses.
Vero.E6 cells (ATCC CRL-1586, clone E6) were maintained for cell culture using Dulbecco’s modified Eagle medium (Gibco), which was supplemented with Antibiotic-Antimycotic (100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B; Gibco), 10% of fetal bovine serum (FBS; Corning), and 1% HEPES (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid; Gibco). Wild-type SARS-CoV-2 (isolate USA-WA1/2020) was grown in Vero.E6 cells for 3 days at 37°C and then the supernatant was clarified via centrifugation at 1,000 g for 10 min. Virus stocks were stored at −80°C. The protocol is described in greater detail previously (44, 45). All work with authentic live SARS-CoV-2 was performed in the biosafety level 3 (BSL-3) facility following institutional guidelines.
In vivo studies.
All animal procedures were performed by following the Institutional Animal Care and Use Committee (IACUC) guidelines of the Icahn School of Medicine at Mount Sinai IACUC and according to an approved protocol. Six- to 8-week-old female, BALB/c mice were vaccinated via the intramuscular route with 3 μg of each respective protein with 1:1 mixture of Addavax (InvivoGen) in a total volume of 50 μL. After 3 weeks, mice were bled and vaccinated with the same protein as used for the initial vaccination. After 3 weeks, mice were administered anesthesia via the intraperitoneal route and intranasally transduced with AdV-hACE2 at 2.5 × 108 PFU per mouse (21). Anesthesia was prepared using 0.15 mg/kg of body weight ketamine and 0.03 mg/kg xylazine in water. Five days later, all mice were infected with authentic wild-type SARS-CoV-2 intranasally with 1 × 105 PFU. Mice were humanely euthanized on day 3 and day 5 for assessment of virus in the lungs. Lungs were homogenized using a BeadBlaster 24 (Benchmark) homogenizer (46–48). Viral load in the lung was quantified via a classic plaque assay (49, 50). For mice that also received mRNA-LNP vaccine, the vaccine was administered via the intramuscular route and each mouse was immunized with a single dose of 3 μg of mRNA-LNP.
ELISA.
Ninety-six-well plates (Immulon 4 HBX; Thermo Fisher Scientific) were coated with 2 μg/mL of each antigen with 50 μL/well at 4°C overnight for 16 h. The following day, the coating solution was removed, and each plate was blocked with 100 μL/well of 3% nonfat milk (American Bio; catalog no. AB10109-01000) in phosphate-buffered saline containing 0.01% Tween 20 (PBS-T). Blocking solution was incubated on the plates for 1 h at room temperature (RT). Serum samples were tested starting at a dilution of 1:50 with 1:5-fold subsequent serial dilutions. Serum samples were added to the plates for 2 h at RT. Next, the plates were properly washed 3 times with 200 μL/well of PBS-T. Anti-mouse IgG-horseradish peroxidase (HRP)-conjugated antibody (Rockland; catalog no. 610–4302) was used at a dilution of 1:3,000 in 1% nonfat milk in PBS-T, and 100 μL of the prepared solution was added on to each well for 1 h at RT. The plates were washed 3 times with 200 μL/well of PBS-T and patted on paper towels to dry. Developing solution was made in sterile water (WFI; Gibco) using SigmaFast OPD (o-phenylenediamine dihydrochloride, catalog no. P9187; Sigma-Aldrich), and 100 μL was added to each well for a total of 10 min. To stop the reaction, 50 μL/well of 3 M hydrochloric acid was added, and the plates were read in a Synergy 4 (BioTek) plate reader at an absorbance of 490 nm. Data were analyzed in GraphPad Prism 8. An area under the curve was calculate for the dilution series and reported. This protocol is described in earlier literature in greater detail (44, 51–53).
Neutralization assays.
Vero.E6 cells were seeded in a 96-well cell culture plate (Corning; 3340) 1 day prior to the assay, at a density of 20,000 cells per well. Mouse serum samples were heat inactivated at 56°C for 1 h prior to use. Serum dilutions were prepared in 1× minimal essential medium (MEM; Gibco) supplemented with 1% FBS. Virus was diluted to 10,000 50% tissue culture infectious doses (TCID50s)/mL, and 80 μL of virus and 80 μL of serum were incubated together for 1 h at RT. After the incubation, 120 μL of virus–serum mixture was used to infect cells for 1 h at 37°C. Next, the virus–serum mix was removed and 100 μL of each corresponding dilution was added to each well. A volume of 100 μL of 1× MEM were also added to the plates to get to a total volume of 200 μL in each well. Cells were incubated at 37°C for 3 days and later fixed with 10% paraformaldehyde (Polysciences) for 24 h. After 24 h, the paraformaldehyde solution was discarded and cells were permeabilized for intracellular staining. Staining and quantification were performed as previously described (40, 44, 54, 55).
ADCC reporter assay.
An assay similar to a setup previously published for influenza virus was set up to measure ADCC reporter activity (20). Vero.E6 cells were seeded in white 96-well cultures plates (Corning; 3917) at a density of 20,000 cells per well and infected 24 h later with recombinant Newcastle disease virus (NDV) expressing the prefusion-stabilized spike protein from SARS-CoV-2 (NDV-HXP-S) (56, 57) at a multiplicity of infection (MOI) of 2. After 1 h of incubation, the infection inoculum was removed and replaced with 100 μL Roswell Park Memorial Institute (RPMI) 1640 media (Cytiva; SH30027.02) supplemented with 2% super low-IgG FBS (Cytiva; SH30898.02HI). Heat-inactivated mouse serum samples were serially diluted 1:3 in RPMI 1640 media supplemented with 2% super low-IgG FBS from a starting dilution of 1:10. Forty-eight hours postinfection, the media was removed and 25 μL RPMI 1640 supplemented with 2% super low-IgG FBS was added in addition to 25 μL of diluted sera. ADCC bioassay effector cells (Promega; M1201) expressing the murine FcγRIV receptor were added to each well at a density of 75,000 cells per well in a volume of 25 μL. The effector cells and serum were incubated with the cells for 6 h after which 75 μL Bio-Glo reagent (Promega; G7940) was added to each well. Plates were incubated at RT for 10 min in the dark before luminescence was measured using a Synergy 4 (BioTek) plate reader. An area under the curve was calculate for the dilution series and reported.
Statistical analysis.
Groups were compared using a one-sided ANOVA corrected for multiple comparisons after log transformation. Pre- and postchallenge titers were compared using a t test after log transformation. Analysis was performed in GraphPad Prism (version 9.0.1). Results from statistical testing are reported in Tables S1 to 4. Amino acid percentages were compared in ClustalOmega.
Data availability.
Data have been deposited at ImmPort (https://www.immport.org/home) under accession number SDY2158. It is anticipated that this accession number will be released by 24 February 2023; until that time, all the data will be available from the corresponding author upon request.
ACKNOWLEDGMENTS
We thank Randy A. Albrecht for oversight of the conventional BSL3 biocontainment facility, which makes our work with live SARS-CoV-2 possible. This work was partially funded by the Centers of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C), by the Collaborative Influenza Vaccine Innovation Centers (CIVICs contract # 75N93019C00051) and by the generous support by anonymous donors. LNP formulation of mRNAs was performed by Acuitas Therapeutics, Vancouver, BC Canada.
The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list Florian Krammer as coinventor. Fatima Amanat is also listed on the serological assay patent application as coinventor and Weina Sun is listed on the NDV-based SARS-CoV-2 vaccine IP as coinventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Third Rock Ventures, GSK, and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2.
In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that N.P. is named on a patent describing the use of nucleoside-modified mRNA in lipid nanoparticles as a vaccine platform. N.P. has disclosed those interests fully to the University of Pennsylvania and has in place an approved plan for managing any potential conflicts arising from licensing of his patents.
Footnotes
Supplemental material is available online only.
Contributor Information
Florian Krammer, Email: florian.krammer@mssm.edu.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamre D, Procknow JJ. 1966. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 121:190–193. 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, Osterhaus AD. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA 101:6212–6216. 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10:368–373. 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79:884–895. 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BM, Foxman B, Monto AS, Baric RS, Martin ET, Uzicanin A, Rainey JJ, Aiello AE. 2018. Human coronaviruses and other respiratory infections in young adults on a university campus: prevalence, symptoms, and shedding. Influenza Other Respir Viruses 12:582–590. 10.1111/irv.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrie JG, Bazzi LA, McDermott AB, Follmann D, Esposito D, Hatcher C, Mateja A, Narpala SR, O'Connell SE, Martin ET, Monto AS. 2021. Coronavirus occurrence in the Household Influenza Vaccine Evaluation (HIVE) cohort of Michigan households: reinfection frequency and serologic responses to seasonal and severe acute respiratory syndrome Coronaviruses. J Infect Dis 224:49–59. 10.1093/infdis/jiab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monto AS, DeJonge PM, Callear AP, Bazzi LA, Capriola SB, Malosh RE, Martin ET, Petrie JG. 2020. Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan. J Infect Dis 222:9–16. 10.1093/infdis/jiaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79:1595–1604. 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, Borgert BA, Moreno CA, Solomon BD, Trimmer-Smith L, Etienne V, Rodriguez-Barraquer I, Lessler J, Salje H, Burke DS, Wesolowski A, Cummings DAT. 2020. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 11:4704. 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, Ulferts R, Earl C, Wrobel AG, Benton DJ, Roustan C, Bolland W, Thompson R, Agua-Doce A, Hobson P, Heaney J, Rickman H, Paraskevopoulou S, Houlihan CF, Thomson K, Sanchez E, Shin GY, Spyer MJ, Joshi D, O'Reilly N, Walker PA, Kjaer S, Riddell A, Moore C, Jebson BR, Wilkinson M, Marshall LR, Rosser EC, Radziszewska A, Peckham H, Ciurtin C, Wedderburn LR, Beale R, Swanton C, Gandhi S, Stockinger B, McCauley J, Gamblin SJ, McCoy LE, Cherepanov P, Nastouli E, Kassiotis G. 2020. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 370:1339–1343. 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woudenberg T, Pelleau S, Anna F, Attia M, Donnadieu F, Gravet A, Lohmann C, Seraphin H, Guiheneuf R, Delamare C, Stefic K, Marlet J, Brochot E, Castelain S, Augereau O, Sibilia J, Dubos F, Meddour D, Guen CG, Coste-Burel M, Imbert-Marcille BM, Chauvire-Drouard A, Schweitzer C, Gatin A, Lomazzi S, Joulié A, Haas H, Cantais A, Bertholon F, Chinazzo-Vigouroux MF, Abdallah MS, Arowas L, Charneau P, Hoen B, Demeret C, Werf SV, Fontanet A, White M. 2021. Humoral immunity to SARS-CoV-2 and seasonal coronaviruses in children and adults in north-eastern France. EBioMedicine 70:103495. 10.1016/j.ebiom.2021.103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amanat F, Thapa M, Lei T, Ahmed SMS, Adelsberg DC, Carreño JM, Strohmeier S, Schmitz AJ, Zafar S, Zhou JQ, Rijnink W, Alshammary H, Borcherding N, Reiche AG, Srivastava K, Sordillo EM, van Bakel H, Turner JS, Bajic G, Simon V, Ellebedy AH, Krammer F, Personalized Virology Initiative . 2021. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 184:3936–3948.e10. 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aydillo T, Rombauts A, Stadlbauer D, Aslam S, Abelenda-Alonso G, Escalera A, Amanat F, Jiang K, Krammer F, Carratala J, García-Sastre A. 2021. Immunological imprinting of the antibody response in COVID-19 patients. Nat Commun 12:3781. 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci USA 106:19970–19974. 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102:7988–7993. 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422. 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulswit RJG, Lang Y, Bakkers MJG, Li W, Li Z, Schouten A, Ophorst B, van Kuppeveld FJM, Boons GJ, Bosch BJ, Huizinga EG, de Groot RJ. 2019. Human coronaviruses OC43 and HKU1 bind to 9. Proc Natl Acad Sci USA 116:2681–2690. 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Q, Cao L, Ma C, Tortorici MA, Liu C, Si J, Liu P, Gu M, Walls AC, Wang C, Shi L, Tong F, Huang M, Li J, Zhao C, Shen C, Chen Y, Zhao H, Lan K, Corti D, Veesler D, Wang X, Yan H. 2022. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature 612:748–757. 10.1038/s41586-022-05513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chromikova V, Tan J, Aslam S, Rajabhathor A, Bermudez-Gonzalez M, Ayllon J, Simon V, García-Sastre A, Salaun B, Nachbagauer R, Krammer F. 2020. Activity of human serum antibodies in an influenza virus hemagglutinin stalk-based ADCC reporter assay correlates with activity in a CD107a degranulation assay. Vaccine 38:1953–1961. 10.1016/j.vaccine.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathnasinghe R, Strohmeier S, Amanat F, Gillespie VL, Krammer F, García-Sastre A, Coughlan L, Schotsaert M, Uccellini MB. 2020. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg Microbes Infect 9:2433–2445. 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laczkó D, Hogan MJ, Toulmin SA, Hicks P, Lederer K, Gaudette BT, Castaño D, Amanat F, Muramatsu H, Oguin TH, Ojha A, Zhang L, Mu Z, Parks R, Manzoni TB, Roper B, Strohmeier S, Tombácz I, Arwood L, Nachbagauer R, Karikó K, Greenhouse J, Pessaint L, Porto M, Putman-Taylor T, Strasbaugh A, Campbell TA, Lin PJC, Tam YK, Sempowski GD, Farzan M, Choe H, Saunders KO, Haynes BF, Andersen H, Eisenlohr LC, Weissman D, Krammer F, Bates P, Allman D, Locci M, Pardi N. 2020. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity 53:724–732.e7. 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lederer K, Castaño D, Gómez Atria D, Oguin TH, Wang S, Manzoni TB, Muramatsu H, Hogan MJ, Amanat F, Cherubin P, Lundgreen KA, Tam YK, Fan SHY, Eisenlohr LC, Maillard I, Weissman D, Bates P, Krammer F, Sempowski GD, Pardi N, Locci M. 2020. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53:1281–1295.e5. 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krammer F. 2019. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 19:383–397. 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, van Haperen R, Gutiérrez-Álvarez J, Li W, Okba NMA, Albulescu I, Widjaja I, van Dieren B, Fernandez-Delgado R, Sola I, Hurdiss DL, Daramola O, Grosveld F, van Kuppeveld FJM, Haagmans BL, Enjuanes L, Drabek D, Bosch BJ. 2021. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat Commun 12:1715. 10.1038/s41467-021-21968-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto D, Sauer MM, Czudnochowski N, Low JS, Tortorici MA, Housley MP, Noack J, Walls AC, Bowen JE, Guarino B, Rosen LE, di Iulio J, Jerak J, Kaiser H, Islam S, Jaconi S, Sprugasci N, Culap K, Abdelnabi R, Foo C, Coelmont L, Bartha I, Bianchi S, Silacci-Fregni C, Bassi J, Marzi R, Vetti E, Cassotta A, Ceschi A, Ferrari P, Cippà PE, Giannini O, Ceruti S, Garzoni C, Riva A, Benigni F, Cameroni E, Piccoli L, Pizzuto MS, Smithey M, Hong D, Telenti A, Lempp FA, Neyts J, Havenar-Daughton C, Lanzavecchia A, Sallusto F, Snell G, Virgin HW, Beltramello M, et al. 2021. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science 373:1109–1116. 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou P, Song G, He W-t, Beutler N, Tse LV, Martinez DR, Schäfer A, Anzanello F, Yong P, Peng L, Dueker K, Musharrafieh R, Callaghan S, Capozzola T, Yuan M, Liu H, Limbo O, Parren M, Garcia E, Rawlings SA, Smith DM, Nemazee D, Jardine JG, Wilson IA, Safonova Y, Rogers TF, Baric RS, Gralinski LE, Burton DR, Andrabi R. 2022. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause severe disease. bioRxiv. [DOI] [PMC free article] [PubMed]
- 28.Piepenbrink MS, Park J-G, Desphande A, Loos A, Ye C, Basu M, Sarkar S, Chauvin D, Woo J, Lovalenti P, Erdmann NB, Goepfert PA, Truong VL, Bowen RA, Walter MR, Martinez-Sobrido L, Kobie JJ. 2022. Potent universal-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody. bioRxiv. [DOI] [PMC free article] [PubMed]
- 29.Song G, He WT, Callaghan S, Anzanello F, Huang D, Ricketts J, Torres JL, Beutler N, Peng L, Vargas S, Cassell J, Parren M, Yang L, Ignacio C, Smith DM, Voss JE, Nemazee D, Ward AB, Rogers T, Burton DR, Andrabi R. 2021. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat Commun 12:2938. 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Low JS, Jerak J, Tortorici MA, McCallum M, Pinto D, Cassotta A, Foglierini M, Mele F, Abdelnabi R, Weynand B, Noack J, Montiel-Ruiz M, Bianchi S, Benigni F, Sprugasci N, Joshi A, Bowen JE, Walls AC, Jarrossay D, Morone D, Paparoditis P, Garzoni C, Ferrari P, Ceschi A, Neyts J, Purcell LA, Snell G, Corti D, Lanzavecchia A, Veesler D, Sallusto F. 2022. ACE2 engagement exposes the fusion peptide to pan-coronavirus neutralizing antibodies. bioRxiv. [DOI] [PMC free article] [PubMed]
- 31.Dacon C, Tucker C, Peng L, Lee C-CD, Lin T-H, Yuan M, Cong Y, Wang L, Purser L, Williams JK, Pyo C-W, Kosik I, Hu Z, Zhao M, Mohan D, Cooper A, Peterson M, Skinner J, Dixit S, Kollins E, Huzella L, Perry D, Byrum R, Lembirik S, Zhang Y, Yang ES, Chen M, Leung K, Weinberg RS, Pegu A, Geraghty DE, Davidson E, Douagi I, Moir S, Yewdell JW, Schmaljohn C, Crompton PD, Holbrook MR, Nemazee D, Mascola JR, Wilson IA, Tan J. 2022. Broadly neutralizing antibodies target the coronavirus fusion peptide. bioRxiv. [DOI] [PMC free article] [PubMed]
- 32.Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. 2014. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS One 9:e101322. 10.1371/journal.pone.0101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundu R, Narean JS, Wang L, Fenn J, Pillay T, Fernandez ND, Conibear E, Koycheva A, Davies M, Tolosa-Wright M, Hakki S, Varro R, McDermott E, Hammett S, Cutajar J, Thwaites RS, Parker E, Rosadas C, McClure M, Tedder R, Taylor GP, Dunning J, Lalvani A. 2022. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun 13:80. 10.1038/s41467-021-27674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D. 2020. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370:89–94. 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komabayashi K, Matoba Y, Seto J, Ikeda Y, Tanaka W, Aoki Y, Ikeda T, Matsuzaki Y, Itagaki T, Shirato K, Mizuta K. 2021. Isolation of human coronaviruses OC43, HKU1, NL63, and 229E in Yamagata, Japan, using primary human airway epithelium cells cultured by employing an air-liquid interface culture. Jpn J Infect Dis 74:285–292. 10.7883/yoken.JJID.2020.776. [DOI] [PubMed] [Google Scholar]
- 36.Loo SL, Wark PAB, Esneau C, Nichol KS, Hsu AC, Bartlett NW. 2020. Human coronaviruses 229E and OC43 replicate and induce distinct antiviral responses in differentiated primary human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 319:L926–L931. 10.1152/ajplung.00374.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh CL, Werner AP, Leist SR, Stevens LJ, Falconer E, Goldsmith JA, Chou CW, Abiona OM, West A, Westendorf K, Muthuraman K, Fritch EJ, Dinnon KH, Schäfer A, Denison MR, Chappell JD, Baric RS, Graham BS, Corbett KS, McLellan JS. 2021. Stabilized coronavirus spike stem elicits a broadly protective antibody. Cell Rep 37:109929. 10.1016/j.celrep.2021.109929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halfmann PJ, Frey SJ, Loeffler K, Kuroda M, Maemura T, Armbrust T, Yang JE, Hou YJ, Baric R, Wright ER, Kawaoka Y, Kane RS. 2022. Multivalent S2-based vaccines provide broad protection against SARS-CoV-2 variants of concern and pangolin coronaviruses. EBioMedicine 86:104341. 10.1016/j.ebiom.2022.104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng KW, Faulkner N, Finsterbusch K, Wu M, Harvey R, Hussain S, Greco M, Liu Y, Kjaer S, Swanton C, Gandhi S, Beale R, Gamblin SJ, Cherepanov P, McCauley J, Daniels R, Howell M, Arase H, Wack A, Bauer DLV, Kassiotis G. 2022. SARS-CoV-2 S2-targeted vaccination elicits broadly neutralizing antibodies. Sci Transl Med 14:eabn3715. 10.1126/scitranslmed.abn3715. [DOI] [PubMed] [Google Scholar]
- 40.Amanat F, Strohmeier S, Rathnasinghe R, Schotsaert M, Coughlan L, Garcia-Sastre A, Krammer F. 2020. Introduction of two prolines and removal of the polybasic cleavage site leads to optimal efficacy of a recombinant spike based SARS-CoV-2 vaccine in the mouse model. bioRxiv. [DOI] [PMC free article] [PubMed]
- 41.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, Meade P, Brito RN, Teo C, McMahon M, Simon V, Krammer F. 2020. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 57:e100. 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baiersdörfer M, Boros G, Muramatsu H, Mahiny A, Vlatkovic I, Sahin U, Karikó K. 2019. A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol Ther Nucleic Acids 15:26–35. 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, Tam YK, Ansell SM, Kumar V, Qin J, Zhang X, Wang Q, Panesar S, Hutabarat R, Carioto M, Hettinger J, Kandasamy P, Butler D, Rajeev KG, Pang B, Charisse K, Fitzgerald K, Mui BL, Du X, Cullis P, Madden TD, Hope MJ, Manoharan M, Akinc A. 2013. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther 21:1570–8. 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amanat F, White KM, Miorin L, Strohmeier S, McMahon M, Meade P, Liu WC, Albrecht RA, Simon V, Martinez-Sobrido L, Moran T, Garcia-Sastre A, Krammer F. 2020. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol 58:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin AJ, Kawabata H, Sominsky LA, Clark JJ, Adelsberg DC, Bielak DA, Gonzalez-Reiche AS, Dambrauskas N, Vigdorovich V, Alburquerque B, Amoako AA, Banu R, Beach KF, Bermúdez-González MC, Cai GY, Ceglia I, Cognigni C, Farrugia K, Gleason CR, van de Guchte A, Kleiner G, Khalil Z, Lyttle N, Mendez WA, Mulder LCF, Oostenink A, Rooker A, Salimbangon AT, Saksena M, Paniz-Mondolfi AE, Polanco J, Srivastava K, Sather DN, Sordillo EM, Bajic G, van Bakel H, Simon V, Krammer F, PSP-PARIS Study Group . 2022. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602:682–688. 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 46.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556. 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asthagiri AG, Ioannou A, Wohlbold TJ, Meade P, Aslam S, Amanat F, Ayllon J, Garcia-Sastre A, Krammer F. 2019. Broadly cross-reactive, nonneutralizing antibodies against influenza B virus hemagglutinin demonstrate effector function-dependent protection against lethal viral challenge in mice. J Virol 93:e01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amanat F, Duehr J, Oestereich L, Hastie KM, Ollmann Saphire E, Krammer F. 2018. Antibodies to the glycoprotein GP2 subunit cross-react between old and new world arenaviruses. mSphere 3:e00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duehr J, Wohlbold TJ, Oestereich L, Chromikova V, Amanat F, Rajendran M, Gomez-Medina S, Mena I, tenOever BR, Garcia-Sastre A, Basler CF, Munoz-Fontela C, Krammer F. 2017. Novel cross-reactive monoclonal antibodies against ebolavirus glycoproteins show protection in a murine challenge model. J Virol 91. 10.1128/JVI.00652-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F. 2017. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2:1415–1424. 10.1038/s41564-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amanat F, Meade P, Strohmeier S, Krammer F. 2019. Cross-reactive antibodies binding to H4 hemagglutinin protect against a lethal H4N6 influenza virus challenge in the mouse model. Emerg Microbes Infect 8:155–168. 10.1080/22221751.2018.1564369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajendran M, Sun W, Comella P, Nachbagauer R, Wohlbold TJ, Amanat F, Kirkpatrick E, Palese P, Krammer F. 2018. An immuno-assay to quantify influenza virus hemagglutinin with correctly folded stalk domains in vaccine preparations. PLoS One 13:e0194830. 10.1371/journal.pone.0194830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wohlbold TJ, Chromikova V, Tan GS, Meade P, Amanat F, Comella P, Hirsh A, Krammer F. 2016. Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J Virol 90:851–861. 10.1128/JVI.02275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun W, Leist SR, McCroskery S, Liu Y, Slamanig S, Oliva J, Amanat F, Schafer A, Dinnon KH, 3rd, Garcia-Sastre A, Krammer F, Baric RS, Palese P. 2020. Newcastle disease virus (NDV) expressing the spike protein of SARS-CoV-2 as a live virus vaccine candidate. EBioMedicine 62:103132. 10.1016/j.ebiom.2020.103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C. 2020. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370:1227–1230. 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.González-Domínguez I, Martínez JL, Slamanig S, Lemus N, Liu Y, Lai TY, Carreño JM, Singh G, Singh G, Schotsaert M, Mena I, McCroskery S, Coughlan L, Krammer F, García-Sastre A, Palese P, Sun W. 2022. Trivalent NDV-HXP-S vaccine protects against phylogenetically distant SARS-CoV-2 variants of concern in mice. Microbiol Spectr 10:e0153822. 10.1128/spectrum.01538-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun W, Liu Y, Amanat F, González-Domínguez I, McCroskery S, Slamanig S, Coughlan L, Rosado V, Lemus N, Jangra S, Rathnasinghe R, Schotsaert M, Martinez JL, Sano K, Mena I, Innis BL, Wirachwong P, Thai DH, Oliveira RDN, Scharf R, Hjorth R, Raghunandan R, Krammer F, García-Sastre A, Palese P. 2021. A Newcastle disease virus expressing a stabilized spike protein of SARS-CoV-2 induces protective immune responses. Nat Commun 12:6197. 10.1038/s41467-021-26499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4. Download jvi.01664-22-s0001.pdf, PDF file, 0.2 MB (183KB, pdf)
Data Availability Statement
Data have been deposited at ImmPort (https://www.immport.org/home) under accession number SDY2158. It is anticipated that this accession number will be released by 24 February 2023; until that time, all the data will be available from the corresponding author upon request.