Abstract
Ligusticopsis acaulis, belonging to the family Apiaceae (Umbelliferae), is endemic to China. The complete chloroplast genome sequence of L. acaulis was assembled and annotated for the first time in this study. The results showed that the plastome was 148,509 bp in length and consisted of a pair of inverted repeat regions (IRs: 19,468 bp), a large single-copy region (LSC: 91,902 bp), and a small single-copy region (SSC: 17,671 bp). A total of 114 unique genes were annotated, including 80 protein-coding, 30 tRNA, and four rRNA genes. According to the phylogenetic analysis, L. acaulis belongs to the tribe Selineae, with a close relationship to Ligusticum hispidum (Franch.) Wolff.
Keywords: Ligusticopsis acaulis, complete chloroplast genome, phylogenetic analyses
Introduction
Ligusticopsis was originally established by Leute in 1969, with the genus containing 14 species from China (Leute 1969). Ligusticopsis acaulis, a species endemic to China, has diverse chemical composition and is used as a substitute for Peucedani Radix in the Yunnan Province (Rao et al. 1995). The species was first described by Shan et al. (1986) and was named Peucedanum acaule Shan et Sheh. Pimenov (2017) classified Ligusticopsis acaulis as a synonym of P. acaule. The genus Peucedanum L. (Apiaceae) contains approximately 120 species widely distributed in Eurasia and Africa (Pimenov and Leonov 1993). There are about 40 species of Peucedanum in China, with 33 species being endemic (Sheh and Watson 2005). However, due to the complex species composition and ambiguous relationship with related taxa, the taxonomy of Peucedanum has long been contentious.
In recent years, the chloroplast (cp) genome was applied to the phylogenetic reconstruction of Apiaceae, whereby a few contentious relationships were successfully resolved. Here, the complete cp genome sequence of L. acaulis was characterized, and its phylogenetic relationships with related taxa were established.
Materials and methods
Sample collection, DNA extraction, and sequencing
The specimen was collected from the Sea Grass Mountain in Huize, Yunnan Province of China (26°23′99″N, 103°24′01″E, and altitude 3570 m), and the voucher was deposited in the School of Pharmaceutical Sciences and Yunnan Key Laboratory of Pharmacology for Natural Products, Kunming Medical University (appraiser: Jing Zhou; contact person: Jing Zhou, zhoujing_apiaceae@163.com) under voucher number LZ0901 (Figure 1). The modified CTAB method was used to extract total genomic DNA from leaf tissue (Doyle and Doyle 1987).
Figure 1.
Morphological characteristics of L. acaulis. Herbs are perennial, 5–10 cm tall, long-cylindric root, 20–24 cm long. The specimen image was taken by the author Junmei Niu in School of Pharmaceutical Sciences and Yunnan Key Laboratory of Pharmacology for Natural Products, Kunming Medical University in September 2022, without any copyright issues.
Chloroplast genome assembly and annotation
The genome was sequenced using the Illumina HiSeq 2500 platform (Majorbio, Shanghai, China) with the paired-end (2 × 300 bp) library. The raw reads were filtered using Trimmomatic with default parameters set to remove the adapter and low-quality sequences (Bolger et al. 2014). Clean data were then assembled into circular contigs using GetOrganelle v.1.7.5.3 (Jin et al. 2020). Finally, using Peucedanum praeruptorum Dunn. (GenBank accession number: MN016968) as a reference, the plastome was annotated by PGA (Qu et al. 2019) and manually modified in Geneious (Kearse et al. 2012). The circular genome map was drawn by CPGView (www.1kmpg.cn/cpgview/).
Phylogenetic analyses
A total of 31 published plastome sequences of Apiaceae from NCBI were acquired to investigate the phylogenetic position of L. acaulis. All sequences were aligned using MAFFT (Katoh and Standley 2013) with Bupleurum boissieuanum H. Wolff as the outgroup. The molecular phylogenetic tree was constructed using the maximum-likelihood (ML) analysis in RAxML (Stamatakis 2014) with 1000 rapid bootstrap replicates.
Results and discussion
Chloroplast genome features of L. acaulis
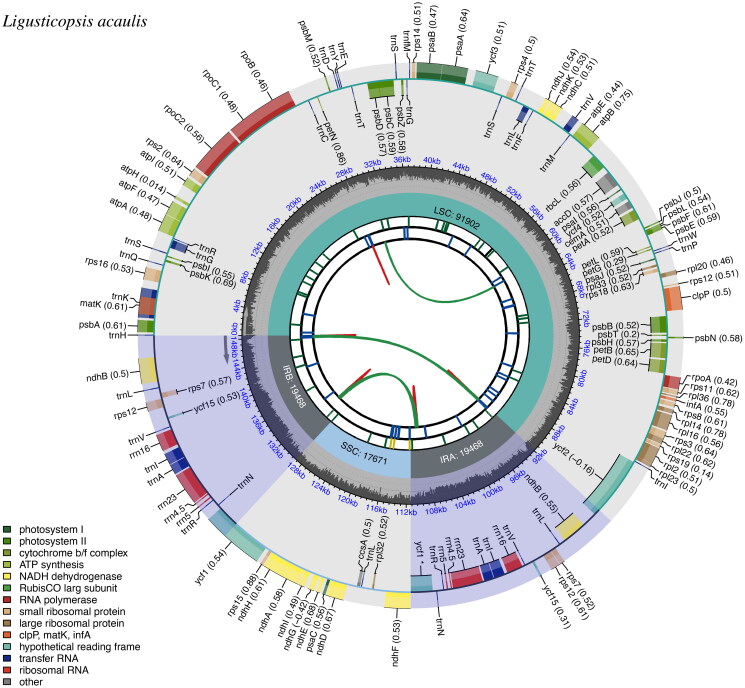
The total length of the cp genome of L. acaulis was 148,509 bp and consisted of a pair of inverted repeat regions (IRs: 19,468 bp), a large single-copy region (LSC: 91,902 bp) and a small single-copy region (SSC: 17,671 bp). The overall GC content of the plastome was 37.4%, with 35.9%, 30.8%, and 43.7% in LSC, IRs, and SSC, respectively. The plastome contained 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Of the 114 unique genes, 18 genes (including 12 protein-coding genes and six tRNA genes) had introns, with 16 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) having one intron each and two genes (ycf3 and clpP) having two introns each (Figure 2, Table 1).
Figure 2.
Genomic map of the L. acaulis chloroplast genome generated by CPGView. Genes outside the circle are transcribed in a counterclockwise direction and those inside in a clockwise direction. LSC: large single-copy; SSC: small single-copy; IR: inverted repeat. The inner circle’s dashed region represents the GC content of the chloroplast genome of L. acaulis. Genes belonging to different functional groups are represented using different colors.
Table 1.
List of genes found in the Ligusticopsis acaulis chloroplast genome.
| Category for genes | Group of genes | Name of genes |
|---|---|---|
| Photosynthesis | Rubisco | rbcL |
| Subunit of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunit of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunit of ATP synthase | atpA, atpB, atpE, atpFa, atpH, atpI | |
| Subunit of cytochrome b/f complex | petA, petBa, petDa, petG, petL, petN | |
| Subunit of NADPH dehydrogenase | ndhAa, ndhBa(×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Self-replication | Large subunit of ribosome | rpl2a, rpl14, rpl16a, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1a, rpoC2 | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12a(×2), rps14, rps15, rps16a, rps18, rps19 | |
| tRNA genes | trnA-UGCa(×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCCa, trnH-GUG, trnI-GAUa(×2), trnI-CAU(×2), trnK-UUUa, trnL-CAA(×2), trnL-UAAa, trnL-UAG, trnM-CAU, trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnVa-UAC, trnW-CCA, trnY-GUA | |
| rRNA genes | rrn5(×2), rrn4.5(×2), rrn16(×2), rrn23(×2) | |
| Other genes | Subunit of acetyl-CoA-carboxylase | accD |
| Maturase | matK | |
| Translational initiation factor | infA | |
| Protease | clpPb | |
| c-type cytochrome synthesis gene | ccsA | |
| Envelope membrane protein | cemA | |
| Unknown function | Conserved open reading frames | ycf1, ycf2, ycf3b, ycf4, ycf15(×2) |
Genes with one intron.
Genes with two introns.
Phylogenetic analysis
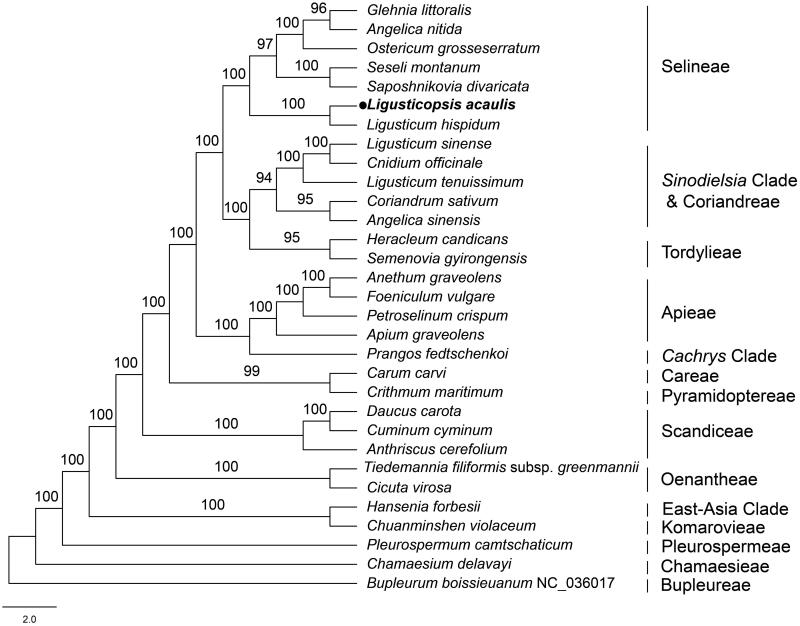
To confirm the phylogenetic position of L. acaulis, an ML phylogenetic tree was constructed using 31 cp genome sequences from NCBI. Using MAFFT, sequences were aligned. B. boissieuanum served as the outgroup. Our phylogeny showed that L. acaulis was closely related to Ligusticum hispidum and belonged to the tribe Selineae (Figure 3). Meanwhile, the phylogenetic inference provided an opportunity to delimit the species of the genus Ligusticopsis and understand their evolutionary relationships. The complete cp genome of L. acaulis provided important information for the phylogenetic analysis of the genus Ligusticopsis and laid the foundation for further reconstruction of the Apiaceae phylogeny.
Figure 3.
Phylogenetic tree of 31 species in the family Apiaceae based on the complete chloroplast sequences (bootstrap support values are shown above each node). The tree was constructed by ML analysis using RAxML and the GTR + G+I nucleotide model, with 1000 rapid bootstrap replicates. The following sequences were used: G. littoralis NC_034645 (Lee et al. 2019), A. nitida MF594405 (Deng et al. 2017), O. grosseserratum NC_028618 (Choi et al. 2016), S. montanum NC_027451 (Samigullin et al. 2016), S. divaricata MN539269 (Bao et al. 2019), L. hispidum MT409614 (Ren et al. 2020), L. sinense NK9214541 (Wu et al. 2020), C. officinale NC_039760 (Park et al. 2018), L. tenuissimum NC_029394 (www.ncbi.nlm.nih.gov/), C. sativum NC_029850 (www.ncbi.nlm.nih.gov/), A. sinensis MH430891 (Tian et al. 2019), H. candicans NC_045184 (Kang et al. 2019), S. gyirongensis NC_042912 (Xiao et al. 2019), A. graveolens NC_029470 (Peery 2015), F. vulgare NC_029469 (www.ncbi.nlm.nih.gov/), P. crispum NC_015821 (Downie and Jansen 2015), A. graveolens NC_041087 (www.ncbi.nlm.nih.gov/), P. fedtschenkoi KY652265 (Mustafina et al. 2019), C. carvi NC_029889 (www.ncbi.nlm.nih.gov/), C. maritimum NC_015804 (Downie and Jansen 2015), D. carota NC_008325 (Ruhlman et al. 2006), C. cyminum MN901636 (www.ncbi.nlm.nih.gov/), A. cerefolium NC_015113 (Downie and Jansen 2015), T. filiformis subsp. greenmannii HM596071 (Downie and Jansen 2015), C. virosa NC_037711 (www.ncbi.nlm.nih.gov/), H. forbesii NC_035054 (www.ncbi.nlm.nih.gov/), C. violaceum KU921430 (www.ncbi.nlm.nih.gov/), P. camtschaticum NC_033343 (www.ncbi.nlm.nih.gov/), C. delavayi MN119367 (Guo et al. 2020), and B. boissieuanum NC_036017 (Wu et al. 2018).
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China under Grant No. [31960048], the Hundred-Talent Program of Kunming Medical University under Grant No. [60118260127], and the Department of Science and Technology of Yunnan Province under Grant No. [202201AT070118].
Author contributions
Zhenwen Liu and Jing Zhou conceived and designed the study. Junmei Niu, Xinyue Wang, Jiarui Yue, and Shilin Zhou analyzed the experimental results and interpreted the data. Junmei Niu drafted the manuscript. Zhenwen Liu and Jing Zhou revised the manuscript. All authors have read and approved the final manuscript.
Ethical approval
Investigations of the authors with human participants or animals are not included in this article. Hence, no ethical approval or permissions were required.
Disclosure statement
The authors declare no conflicts of interest.
Data availability statement
Genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. ON359911. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA849278, SRR19693381, and SAMN28870233, respectively.
References
- Bao Z, Zhu Z, Zhang H, Zhong Y, Wang W, Zhang J, Wu J.. 2019. The complete chloroplast genome of Saposhnikovia divaricata. Mitochondrial DNA B Resour. 5(1):360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SA, Lee WK, Kim Y, Kim KY, Kim JH, Seong RS.. 2016. The complete chloroplast genome sequence of Ostericum koreanum (Apiaceae). Mitochondrial DNA B Resour. 1(1):252–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YQ, Wen J, Yu Y, He XJ.. 2017. The complete chloroplast genome of Angelica nitida. Mitochondrial DNA B Resour. 2(2):694–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie SR, Jansen RK.. 2015. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst Bot. 40(1):336–351. [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Guo XL, Zheng HY, Price M, Zhou SD, He XJ.. 2020. Phylogeny and comparative analysis of Chinese Chamaesium species revealed by the complete plastid genome. Plants. 9(8):965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ.. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Xie D, Xiao Q, Peng C, Yu Y, He X.. 2019. Sequencing and analyses on chloroplast genomes of Tetrataenium candicans and two allies give new insights on structural variants, DNA barcoding and phylogeny in Apiaceae subfamily Apioideae. PeerJ. 7:e8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Joh HJ, Kim K, Lee SC, Kim NH, Park JY, Park HS, Park MS, Kim S, Kwak M, et al. 2019. Dynamic chloroplast genome rearrangement and DNA barcoding for three Apiaceae species known as the medicinal herb “Bang-Poong”. Int J Mol Sci. 20(9):2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leute GH. 1969. Untersuchungen über den Verwandtschaftskreis der Gattung Ligusticum L. (Umbelliferae) I. Teil. Ann Naturhist Mus Wien. 73:55–98. [Google Scholar]
- Mustafina FU, Yi DK, Choi K, Shin CH, Tojibaev KS, Downie SR.. 2019. A comparative analysis of complete plastid genomes from Prangos fedtschenkoi and Prangos lipskyi (Apiaceae). Ecol Evol. 9(1):364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, Yang S, Kim WJ, Noh P, Lee HO, Moon BC.. 2018. The complete chloroplast genome of Cnidium officinale Makino. Mitochondrial DNA B Resour. 3(2):490–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peery R. 2015. Understanding angiosperm genome interactions and evolution: insights from sacred lotus (Nelumbo nucifera) and the carrot family (Apiaceae). Champaign-Urbana: University of Illinois at Urbana-Champaign. [Google Scholar]
- Pimenov MG, Leonov MV.. 1993. Genera of the Umbelliferae. Kew: Royal Botanic Gardens. [Google Scholar]
- Pimenov MG. 2017. Updated checklist of Chinese Umbelliferae: nomenclature, synonymy, typification, distribution. Turczaninowia. 20(2):106–239. [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao GX, , Liu QX, , Dai ZJ, , Yang Q, , Dai WS. 1995. Textual research for the traditional Chinese medicine radix peucedani and discussion of its modern varieties. Journal of Yunnan College of Traditional Chinese Medicine. 18(1):1–6. [Google Scholar]
- Ren T, Li ZX, Xie DF, Gui LJ, Peng C, Wen J, He XJ.. 2020. Plastomes of eight Ligusticum species: characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 20(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H.. 2006. Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 7(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samigullin TH, Logacheva MD, Terenteva EI, Degtjareva GV, Vallejo-Roman CM.. 2016. Plastid genome of Seseli montanum: complete sequence and comparison with plastomes of other members of the Apiaceae family. Biochemistry. 81(9):981–985. [DOI] [PubMed] [Google Scholar]
- Sheh ML, Watson MF.. 2005. Flora of China Editorial Committee. Peucedanum L. Flora of China. Vol. 14. St. Louis (MO): Missouri Botanical Garden Press; p. 182–192. [Google Scholar]
- Shan RH, Sheh ML, Wang TS, Pu FD, Shen KM, Zhang HC.. 1986. New Taxa of the Chinese Umbelliferae (2). J Syst Evol. 24(4):304. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E, Liu Q, Chen W, Li F, Chen A, Li C, Chao Z.. 2019. Characterization of complete chloroplast genome of Angelica sinensis (Apiaceae), an endemic medical plant to China. Mitochondrial DNA Part B. 4(1):158–159. [Google Scholar]
- Wu Q, Wu H, Wang LK, Zhao X.. 2020. Characterization of the complete chloroplast genome of Ligusticum sinense, as a Chinese herb to treat toothache in China. Mitochondrial DNA B Resour. 5(3):3174–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang TZ, Qiu DY, Chai Q, Fan WB, Li ZH, Fang MF.. 2018. Complete plastid genome of Bupleurum boissieuanum, an endemic herb plant in western China. Conserv Genet Resour. 10(4):635–637. [Google Scholar]
- Xiao QY, Feng T, Yu Y, Luo Q, He XJ.. 2019. The complete chloroplast genome of Semenovia gyirongensis (tribe Tordylieae, Apiaceae). Mitochondrial DNA Part B. 4(1):1863–1864. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. ON359911. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA849278, SRR19693381, and SAMN28870233, respectively.