Abstract
Background:
Extensive evidence from single center studies indicates that a subset of chronic advanced heart failure (HF) patients undergoing left ventricular assist device (LVAD) support show significantly improved heart function and reverse structural remodeling (i.e. termed “responders”). Furthermore, we recently published a multi-center prospective study, RESTAGE-HF, demonstrating that LVAD support combined with standard HF medications induced remarkable cardiac structural and functional improvement, leading to high rates of LVAD weaning and excellent long-term outcomes. This intriguing phenomenon provides great translational and clinical promise although the underlying molecular mechanisms driving this recovery are largely unknown.
Methods:
To identify changes in signaling pathways operative in the normal and failing human heart and to molecularly characterize patients who respond favorably to LVAD unloading, we performed global RNA-sequencing and phosphopeptide profiling of left ventricular tissue from 93 HF patients undergoing LVAD implantation (25 responders and 68 non-responders) and 12 non-failing donor hearts. Patients were prospectively monitored via echocardiography to characterize their myocardial structure and function and identify responders and non-responders.
Results:
These analyses identified 1,341 transcripts and 288 phosphopeptides which are differentially regulated in cardiac tissue from non-failing control samples and HF patients. In addition, these unbiased molecular profiles identified a unique signature of 29 transcripts and 93 phosphopeptides in HF patients which distinguished responders after LVAD unloading. Further analyses of these macromolecules highlighted differential regulation in two key pathways: cell cycle regulation and extracellular matrix/focal adhesions.
Conclusions:
This is the first study to characterize changes in the non-failing and failing human heart by integrating multiple -omics platforms to identify molecular indices defining patients capable of myocardial recovery. These findings may guide patient selection for advanced HF therapies and identify new HF therapeutic targets.
Keywords: LVAD, mechanical unloading, clinical phosphoproteomics, RNA-sequencing, reverse remodeling, myocardial recovery
INTRODUCTION
Heart Failure (HF) is one of the most significant causes of morbidity and mortality in both developed and developing countries. Left ventricular assist devices (LVADs) are being used as a therapeutic treatment for patients with advanced HF and have traditionally served either as a temporary bridge until cardiac transplantation, or, given the limited availability of donor hearts, as a lifetime “destination” therapy. It is also now well established that LVAD-induced mechanical unloading can result in functional myocardial recovery and reverse remodeling in a subset of HF patients.1–7 The impact of HF etiology on the prospect of myocardial recovery was investigated in a single center observational study and a multi-center registry study (e.g. INTERMACS) where approximately 5% of ischemic and 20% of non-ischemic chronic cardiomyopathy patients exhibited significant improvement in cardiac structure and function following LVAD support.1, 8 Furthermore, the RESTAGE-HF multi-center prospective trial showed that 47% of selected HF patients (19/40) achieved sufficient improvement of cardiac function to reach criteria for LVAD explantation with sustained remission from HF at 12 months after weaning from the cardiac assist device.7 However, the molecular underpinnings which enable myocardial recovery are largely unknown. Identifying the underlying molecular pathways and the key biological changes defining patients capable of functional recovery following LVAD unloading has become essential for understanding disease development, progression and response to therapy.9
LVAD support has been associated with improvements in structural, cellular and molecular indices including changes in cardiomyocyte function10, size11 and contractility12, calcium cycling13, mitochondrial function14, apoptosis15 and other aspects of cardiac remodeling16,17. Noteworthy, the NHLBI Working Group on myocardial recovery identified as a critical shortcoming in the field the fact that the overwhelming majority of the reported LVAD myocardial tissue studies have failed to correlate functional outcomes with molecular, cellular or histological findings.9 As a result, most of the prior reports which studied pre-LVAD vs. post-LVAD heart tissue samples without simultaneous myocardial functional phenotyping, shed limited information on whether the observed biological changes could be associated with true cardiac recovery mechanisms or alternatively represent just epiphenomena.9 Examination of tissue from patients with LVAD-induced myocardial functional recovery and from LVAD patients without functional myocardial improvement (i.e. “responders” versus “non-responders” as in the design of this study) was identified by the NHLBI Working Group as a critical step needed to advance the field.9
Furthermore, the prior LVAD translational studies have focused largely on evaluating the contribution of individual proteins or biological processes,9, 16 and while insightful, often lack the complex and comprehensive details likely necessary for understanding this multifactorial etiology. More recently, transcriptional profiling utilizing arrays has been extensively applied to HF samples to gain better insights into relevant disease pathways18, to identify biomarkers for better diagnostics and prognostic value19, and to evaluate the impact of therapeutic treatments including medications and implanted devices19–23. Although meaningful results have been generated, the genomic coverage in many studies has been limited owing to the use of conventional microarrays which primarily evaluate mRNAs, and account for only ~1% of all transcribed species,24 leaving microRNAs and lncRNAs unexplored.25 Therefore, the underlying mechanisms and pathways which predispose a HF patient towards potential recovery or further regression after LVAD implant still remain largely unclear.
With the development of next-generation sequencing technologies, RNA-sequencing now provides an innovative approach for quantifying gene expression in small amounts of tissue over a larger dynamic range, which to date has only been applied to a handful of studies comparing ventricular tissue from non-failing and failing patients26–29, ischemic and non-ischemic30 and tissue before and after LVAD implantation22. In addition, state-of-the-art mass spectrometry-based proteomics technologies offer an alternative methodology for providing global unbiased analysis of protein expression or post-translational modifications. Specifically, only 3 papers have described the application of global proteomics31, 32 or phosphoproteomics33 to human HF samples. Indeed, a comprehensive transcriptomic and proteomic analyses of the HF population utilizing a considerable sample size is lacking and, to date, no study has combined multiple -omics technologies to profile HF patients at the time of LVAD implant to investigate the differences between patients with or without the potential for cardiac recovery. We therefore performed RNA-sequencing and phosphoproteomics on LV samples obtained at the time of LVAD-implant in HF patients and non-failing donors, performed serial monitoring of cardiac function to identify functional recovery and combined our molecular analysis from these two platforms to comprehensively characterize changes in gene expression and protein phosphorylation.
METHODS
The data, analytic methods, and study materials (as available) have been outlined here but will also be made available to other researchers for the purposes of reproducing the results upon request of individual investigators to the corresponding authors. Specifically, the -omics data included in this study has been deposited to the publicly available ProteomeXchange Consortium34 via the PRIDE35 partner repository with the dataset identifier PXD016761 (http://www.proteomexchange.org/).
Study Population:
We prospectively enrolled 93 consecutive patients (age ≥ 18-years) in institutions comprising the Utah Transplantation Affiliated Hospitals (U.T.A.H.) Cardiac Transplant Program (i.e. University of Utah Health Science Center, Intermountain Medical Center, and the Veterans Administration Salt Lake City Health Care System) with clinical characteristics consistent with dilated cardiomyopathy and chronic advanced HF who required circulatory support with continuous flow LVAD as a bridge to transplantation or lifetime destination therapy. Patients who required LVAD support due to acute HF (acute myocardial infarction, acute myocarditis, post- cardiotomy cardiogenic shock, etc.) were prospectively excluded. LVAD patients underwent serial echocardiograms monthly for the first three months and at 4.5 and 6 months. They were categorized as either responders or non-responders using left ventricular ejection fraction (LVEF) and left ventricular end-diastolic diameter (LVEDD) measurements during diminished LVAD support “turn-down” echocardiograms). Responders were defined as patients with a final LVEF >40% and LVEDD ≤5.9 cm; whereas Non-Responders were defined as patients with a final LVEF <35% and with <50% relative improvement in LVEF regardless of the final LVEDD. The study was approved by the institutional review board of the participating institutions, and informed consent was provided by all patients. For HF patients, their demographic and clinical characteristics, including echocardiographic parameters, protein biomarkers and other clinical data were prospectively collected and entered in our programs research electronic data capture system (REDCap) and are listed in Table 1. P-values were calculated for the data in Table 1 and Tables S1A and S2A using the Student’s t-test with equal or unequal variances as appropriate for continuous variables, and the chi-squared test or Fisher’s exact test as appropriate for categorical variables. P-values for the comparisons graphed in Figure 1 were calculated using the paired t-test. Myocardial tissue from 12 donor hearts, not allocated for heart transplantation due to non-cardiac reasons (size, infection, etc.), were used as non-failing controls. Donor information, for non-failing controls, including age, gender, race, and cause of death are included in Table 2. It is important to note that some myocardial tissue samples, from HF patients and donors, were used for both RNA-sequencing and proteomics experiments while others were utilized in just one application, therefore the overall number of tissue samples used in this study is less than the sum of the samples for each type of analyses. Specifically, all samples used for phosphoproteomic analyses were also analyzed by RNA-sequencing, however, because mass spectrometry analysis is much more time consuming per sample, only a subset of the total samples was evaluated by mass spectrometry. Clinical information for the HF patients and donors used in the RNA-sequencing and proteomics experiments are included in Tables S1 and S2, respectively.
Table 1. Clinical characteristics of heart failure patients in this study population.
Clinical characteristics were captured for 93 heart failure patients included in the study population at the time of LVAD implantation. In addition, serial monitoring by echocardiography assessed LVEF (left ventricular ejection fraction) and LVEDD (left ventricular end diastolic dimension) values prior to device implantation (pre-LVAD) and 6 months post-implantation (post-LVAD) to differentiate between responders (n=25) and non-responders (n=68). P-values were calculated using the Student’s t-test with equal or unequal variances as appropriate for continuous variables, and the chi-squared test or Fisher’s exact test as appropriate for categorical variables. +/− values indicate standard error of the mean (SEM).
| Variable | Responders N=25 (mean ± SE) | Non-Responders N=68 (mean ± SE) | p-value |
|---|---|---|---|
| Male sex, n (%) | 17 (68) | 60 (88) | 0.02 |
| Age at LVAD implantation, years | 50±4 | 59±2 | 0.06 |
| Pre-LVAD | |||
| LVEDD, cm | 6.4±0.2 | 7.0±0.1 | 0.02 |
| LVEF, % | 20±1 | 18±1 | 0.15 |
| Post-LVAD | |||
| LVEDD, cm | 4.5±0.1 | 6.4±0.1 | <0.001 |
| LVEF, % | 47±2 | 21±1 | <0.001 |
| HF Etiology | 0.30 | ||
| Ischemic cardiomyopathy, n (%) | 6 (24) | 24 (35) | |
| Non-ischemic cardiomyopathy, n (%) | 19 (76) | 44 (65) | |
| New York Heart Association Functional Class | 0.25 | ||
| III, n (%) | 8 (32) | 14 (21) | |
| IV, n (%) | 17 (68) | 54 (79) | |
| Duration of HF symptoms, months | 36±12 | 102±10 | <0.001 |
| Systolic Blood Pressure, mmHg | 105±3 | 104±2 | 0.58 |
| Diastolic Blood Pressure, mmHg | 66±3 | 68±1 | 0.52 |
| Mean Right Atrial Pressure, mmHg | 11±1 | 12±1 | 0.38 |
| Systolic Pulmonary Artery Pressure, mmHg | 52±3 | 55±2 | 0.45 |
| Diastolic Pulmonary Artery Pressure, mmHg | 24±2 | 26±1 | 0.53 |
| Pulmonary Capillary Wedge Pressure, mmHg | 24±2 | 25±1 | 0.54 |
| Cardiac Index, L/min/m2 | 2.0±0.1 | 1.8±0.1 | 0.13 |
| Pulmonary Vascular Resistance, Wood units | 3.5±0.5 | 3.9±0.4 | 0.55 |
| B-type Natriuretic Peptide, pg/mL | 1710±337 | 1331±124 | 0.30 |
| Creatinine, mg/dL | 1.2±0.1 | 1.3±0.1 | 0.07 |
| Sodium, mmol/L | 134±1 | 135±1 | 0.95 |
| Hemoglobin, g/dL | 13.0±0.5 | 12.5±0.3 | 0.33 |
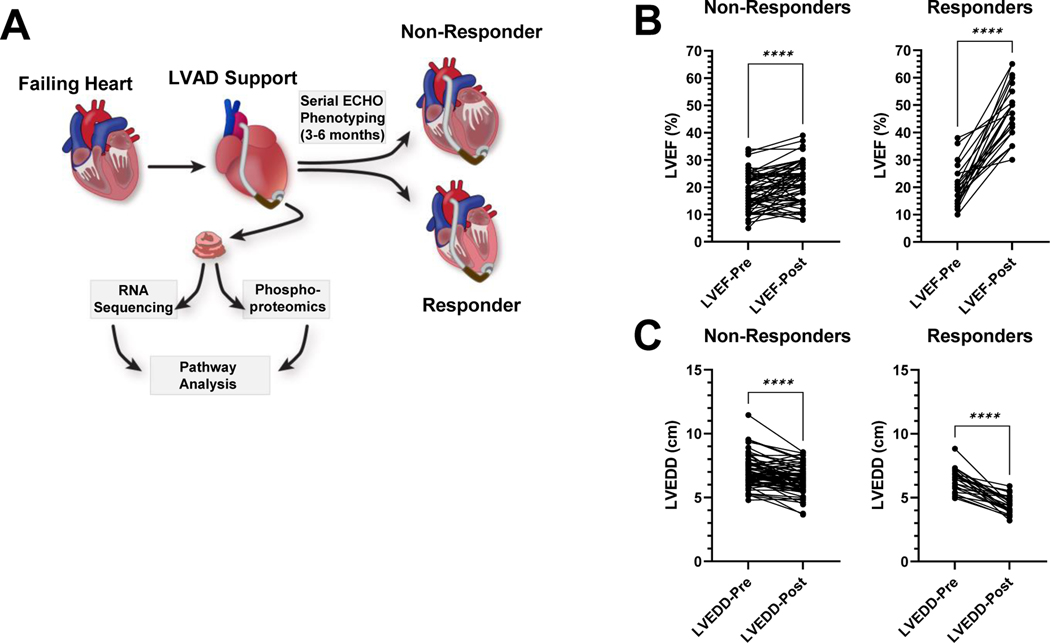
Figure 1. Schematic illustrating the study design and myocardial functional and structural assessment.
A, Left ventricular myocardial tissue was harvested at the time of LVAD implantation and analyzed using global transcriptomic and phosphoproteomic techniques. Serial echocardiographic monitoring of these patients after LVAD implantation identified responders and non-responders as detailed in the methods section. Left ventricular tissue was also harvested from non-failing donor samples and used in the same analyses. Differentially regulated transcripts and phosphorylated proteins from these different populations (non-failing donor, responder and non-responder) were analyzed using bioinformatics analyses tools. B, The left ventricular ejection fraction (LVEF) and C, left ventricular end diastolic dimension (LVEDD) of patients receiving an LVAD and utilized in this study, at the time of implantation (LVEF-Pre and LVEDD-Pre) and 6 months after the device implantation (LVEF-Post and LVEDD-Post). This same data is listed in Table 1. P-values were calculated using the paired t-test. Asterisks indicate a p-value <0.0001. n=68 (non-responders) and 25 (responders).
Table 2. Demographics of non-failing donors utilized as controls in this study.
Myocardial tissue from 12 donor hearts, not allocated for heart transplantation due to non-cardiac reasons were used as normal controls and their demographic information is listed.
| Variable | Donors N=12 (Mean ± SE) |
|---|---|
| Male, n (%) | 5 (41.6) |
| Female, n (%) | 7 (58.4) |
| Age | 45.3±2.1 |
| Weight | 76.2±6.1 |
| Cause of death | |
| Head trauma, n (%) | 8 (66.7) |
| CVA-Stroke/ ICH-Stroke, n (%) | 2 (16.7) |
| CNS Tumor/ Intracerebral Hemorrhage, n (%) | 1 (8.3) |
| Anoxia/ Drug intoxication, n (%) | 1 (8.3) |
Myocardial Tissue Acquisition:
Myocardial tissue was prospectively collected from the LV apical core at the time of LVAD implantation and was snap frozen before storing it at −80°C. Donor control samples were acquired from hearts that were not transplanted due to non-cardiac reasons and LV apical tissue was harvested and processed the same way as the failing hearts.
RNA Isolation, Sequencing and Bioinformatics:
RNA was isolated from LV tissue samples using the miRNeasy Mini Kit (Qiagen) incorporating an on-column RNase-free deoxyribonuclease digestion (using RNase free DNase kit, Qiagen) to remove all DNA. RNA-Sequencing was performed on total RNA from the 84 HF patients (25 responders and 59 non-responders) and 9 non-failing donor samples (Figure S1). The RNA-sequencing library was prepared using the Illumina TruSeq Stranded RNA kit with Ribo-Zero Gold to remove rRNA and sequenced on an Illumina HiSeq 2500 with 50 bp single-end reads.
RNA sequence reads were aligned to the human genome version hg38 with Novoalign (version 2.8.1) against an index containing genome sequences plus all splice junctions (known and theoretical, 46 bp radius) generated with USeq MakeTranscriptome (version 8.8.1) and Ensembl gene annotation (release 84), allowing for up to 50 alignments per read. Raw alignments were converted to genomic coordinates using USeq SamTranscriptomeParser (version 8.8.8), allowing for a maximum of 1 alignment per read. RNA-Seq quality was evaluated with results from Picard CollectRnaSeqMetrics (version 1.137). Gene counts were collected using SubRead featureCounts (version 1.5.1) and Ensembl GRCh38 annotation (release 87) in a stranded fashion, assigning to genes with the largest overlap. Low and non-expressed transcripts were removed when the maximum observed count across all samples was <= 10 counts. Differentially expressed transcripts were identified using DESeq2 package (version 1.16.1) which uses the Benjamini and Hochberg method for adjusting the p-values. All samples were processed at the same time for dispersion estimation and normalization, with contrasts applied to specific condition groups for differential expression testing. A batch factor was introduced in the model to account for different sequencing batches. Differentially expressed genes were filtered using the criteria: adjusted p-value < 0.05, absolute log2 fold change >0.585, (or linear fold change > 1.5) and a normalized base mean count of 30 (to remove any significant genes with very low expression). Heat maps were prepared using the R software using pHeatmap package. Three-dimensional PCA plots were generated using PlotLy software. Bioinformatics analysis of differentially expressed transcripts was performed using the STRING database, Ingenuity Pathway Analysis, and Enricher.
Cardiac Tissue Lysis for Phosphoproteomics:
For phosphopeptide analysis using biological mass spectrometry 10 donor and 29 HF (6 responders and 23 non-responders) tissue samples were analyzed (Figure S1). Frozen heart samples were placed in lysis buffer [100 mM Tris (pH7.6), 4% Sodium Dodecyl Sulfate, 100 mM Dithiothreitol, supplemented with protease inhibitors (Roche) and phosphatase inhibitor (1 mM NaF)] based on the mass of the tissue size (10 μL per 1 mg tissue). Samples were kept on a −20°C insulator at all times during lysis and homogenized using a tip sonicator for 10 sec pulses at 45% amplitude. Pulses were repeated until all tissue was completely homogenized (3–6 times). Between pulses, samples rested at −20°C. Lysed samples were then incubated at 100°C for 5 minutes to aid in the denaturing process and then allowed to cool and sonicated for 3X 10 sec cycles at 25% amplitude. Samples were clarified by centrifugation at 16,000×g for 15 min.
Protein Digestion:
Samples were diluted with 10 mL UA buffer (8M urea, 0.1M Tris/HCl, pH 8.5) and loaded onto a Vivaspin 30,000 MWCO Hydrosart Membrane (Sartorius, #VS15RH22) and centrifuged at 6,000×g at 25°C until concentrated to 1 mL. Samples were washed with an additional 5 mL of UA buffer and concentrated to 0.5 mL at 6000×g. Samples were then carbamidomethylated with 2 mL of UA buffer plus 50mM iodoacetamide for 1 hr in the dark, and then spun at 6000×g at 25°C until concentrated to 0.5 mL. Samples were then washed twice with 5 mL of UA buffer and concentrated to a final volume of less than 1.5 mL followed by two washes of 5 mL of ammonium bicarbonate solution (0.05M ammonium bicarbonate [Sigma, #101240595] in UA buffer) and concentrated to a volume of less than 1.5 mL. Protein concentration was measured using the BCA (bicinchoninic acid) protein assay (Pierce) and trypsin (Promega) was added at 1:100 enzyme to protein ratio in a final volume of 2.5 mL and incubated for 12 hrs at 37°C. Peptides were collected by centrifugation at 6000×g and filters washed twice with 1 mL of mass spec grade water (Fisher, Optima LC/MS #W6–4).
Desalting:
C18 SPE cartridges (Waters, WAT054945) were prepared by washing with methanol, 0.1% trifluoroacetic acid/70% acetonitrile, and then 0.1% trifluoroacetic acid (2 mL each). Peptide solutions were acidified with trifluoroacetic acid to a final concentration of 0.1% and placed on a prepared C18 SPE cartridge and allowed to drip by gravity. Cartridges were then washed with 2 mL 0.1% trifluoroacetic acid. Peptides were then eluted in 2 mL (70% acetonitrile, 0.1% trifluoroacetic acid) and evaporated to 20 μL using a vacuum centrifuge. Samples were then brought to a final volume of 100 μL in 0.1% trifluoroacetic acid and concentrations determined by 280 nm abs on a spectrophotometer.
Phosphopeptide Enrichment:
Titanium dioxide (TiO2) beads (GL Sciences, 5020–75000) were prepared for each sample at a 9:1 ratio of mg of TiO2 beads to peptides and resuspended in 2:1 ratio of μL of buffer [3% (m/v) Dihydroxybenzoic acid (DHB) in 80% acetonitrile, 0.1% trifluoroacetic acid] to μg of beads. Peptide solutions were brought to 500 μL using 30% acetonitrile, 0.3% trifluoroacetic acid, 150 mg/ml DHB with 1/3 volume of bead mixture and allowed to incubate for 30 min at room temperatures. Beads were collected by centrifugation at 5000×g for 3 min. Supernatants were removed and subjected to 2 more rounds of bead incubation. All beads were collected and combined. Beads were washed with 3 rounds of 800 μl of 30% acetonitrile and 3% trifluoroacetic acid (v/v) for 15 min by vortex then centrifuge at 5000×g for 3 min, follow by three rounds of 800 μl of 80% acetonitrile and 0.3% trifluoroacetic acid (v/v) for 15 min by vortex then centrifuge at 5000×g for 3 min. Beads were then loaded into a 200 μL tip with a glass microfiber stage filter and spun at 2000×g for 3 minutes to remove all wash buffer. Tips were transferred to a new tube and peptides eluted with two rounds 100 μl of 40% acetonitrile/1M ammonium hydroxide at 500×g for 10 min. Samples were then concentrated on a vacuum centrifuge to 10 μL and reconstituted in 60 μL of 0.1% trifluoroacetic acid.
LC-MS/MS Analyses:
Enriched phosphopeptides were analyzed with an Orbitrap Velos Pro mass spectrometer (Thermo) interfaced with an EZ nLC-1000 UPLC and outfitted with a PicoFrit reversed phase column (15 cm x 75 μm inner diameter, 3 μm particle size, 120 Å pore diameter, New Objective). Gradient was established with 0.1% formic acid and 5% dimethyl sulfoxide in mass spec grade water (Buffer A) and 0.1% formic acid with 5% dimethyl sulfoxide in acetonitrile (Buffer B). Peptides were eluted at a flow rate of 300 nl/min with a gradient of 5%−12% Buffer B for the first 30 minutes, followed by a gradient of 12%−28% for the next 160 minutes then a gradient of 28%−45% for 30 minutes. Afterwards, hydrophobic peptides were eluted with a flow rate of 500 nl/min and a 5-minute gradient from 45–95% Buffer B and then sustained at 95% Buffer B for 15 additional minutes (240 minute run in total). Spectra were acquired in a data-dependent mode with dynamic exclusion and peptides were fragmented using CID fragmentation. The top twenty MS1 peaks were analyzed at a resolution of 30,000. MS2 scans were scanned for a neutral loss of a phosphate ion at a charge state of 1 (97.97), 2 (48.99), or 3 (32.66), if present would trigger a MS3 on the ion with neutral loss. Samples were run in duplicate to generate technical replicates.
Mass Spectrometry Data Analyses:
The resulting spectra were analyzed using MaxQuant 1.5.5.1 against the UniprotKB human database. Database search engine parameters were as follows: trypsin digestion, two missed cleavages, precursor mass tolerance of 20 ppm, fragment mass tolerance of 0.5 Da, and dynamic carbamidomethylation (C), oxidation (M), deamidation (N,Q) and phosphorylation (S,T,Y). The false discovery rate (FDR) was set at 1% and modified peptides had a minimum Andromeda score of 40. These filters are standards in the field for proteomics analyses. The resulting 15,816 identified phosphopeptides were filtered to only include those identified in at least 50% of either non-responder, responder, or non-failing donor samples, resulting in 2,023 phosphopeptides. Peptide intensities were normalized using the normalizeQuantiles function in the Limma library using R and missing values were imputated with Perseus 1.5.6.0. Statistical analyses were performed on the resulting quantifications using Perseus software package. Enrichment of phosphopeptides from mass spectrometry samples was calculated as a ratio of phosphopeptides over all peptides identified.
Mass Spectrometry Bioinformatics Analyses:
Normalized log2 intensities of phosphopeptides were used in statistical comparisons of groups: Student’s two sample t-test was used for comparisons between two samples and multiple hypothesis correction was done using a permutation based false discovery rate approach of 0.05 with a randomization of 250. The assumptions of normality and equal variance were assessed using histograms, QQ plots, and boxplots generated in Prism 9.3.1 (Figure S2A–C). Both statistical analyses were performed in Perseus 1.5.6.0. Proteins with differentially phosphorylated peptides were then chosen (FDR adjusted p-value < 0.01 and fold change more than 1.5) and submitted to DAVID 6.7 functional annotation tool for enrichment analysis in Gene Ontology, KEGG, and Biocarta databases. Entries with adjusted p-value less than 0.01 were selected for subsequent bioinformatics analysis using DAVID 6.7 to identify enriched terms. Principal component analysis (PCA) was performed in Perseus software and values for the first three components were submitted to OriginLab software (OriginLab, Northampton, Massachusetts, USA) to generate 3D PCA plots. Venn diagrams for comparisons of this data to previously published data were generated in Venny 2.0 software by submitting gene lists into the program. Heat map for comparison between HF patients and donors was generated in Perseus Software with unsupervised clustering using Euclidean distances with average linkage. Heat map for comparison of all peptides between responders and non-responders was generated in MetaboAnalyst 3.0 online software; heat maps for individual pathways were generated in Perseus using average of fold change of each group compared to an average of donor in Cytoscape 3.7.1.
PRIDE Data Repository:
The -omics data included in this study has been deposited to the publically available ProteomeXchange Consortium34 via the PRIDE35 partner repository with the dataset identifier PXD016761.
RESULTS
Clinical characteristics, tissue samples and prospective myocardial functional and structural assessment
We analyzed a total of 93 left ventricular myocardial tissue samples from HF patients undergoing LVAD implantation and 12 from non-failing donors. Patient characteristics are shown in Table 1 for all HF patients utilized in this study (categorized as responders and non-responders) and Table 2 for non-failing controls. The measurements for LVEF and LVEDD for the HF patients utilized in this study are listed in Table 1 and graphed in Figure 1B and 1C. Consistent with prior published studies from our group and others (summarized in references 9, 16) the responders at baseline had similarly reduced LVEF and less dilation of the LVEDD (compared to non-responders – Table 1). Following LVAD support responders significantly improved their LVEF and reduced their LVEDD compared to non-responders who did not experience this functional and structural reverse remodeling (Table 1, Figure 1).
The left ventricular myocardial tissue from these individuals was analyzed using RNA-sequencing and phosphoproteomics to characterize differences in the molecular signatures (gene expression and protein phosphorylation) and two key comparisons were applied to these datasets which are described below: 1) normal donor samples were compared to all HF samples (responders and non-responders combined), and 2) responders were compared to non-responders (Figure S1).
Differential gene expression between failing heart and non-failing donors using RNA-sequencing.
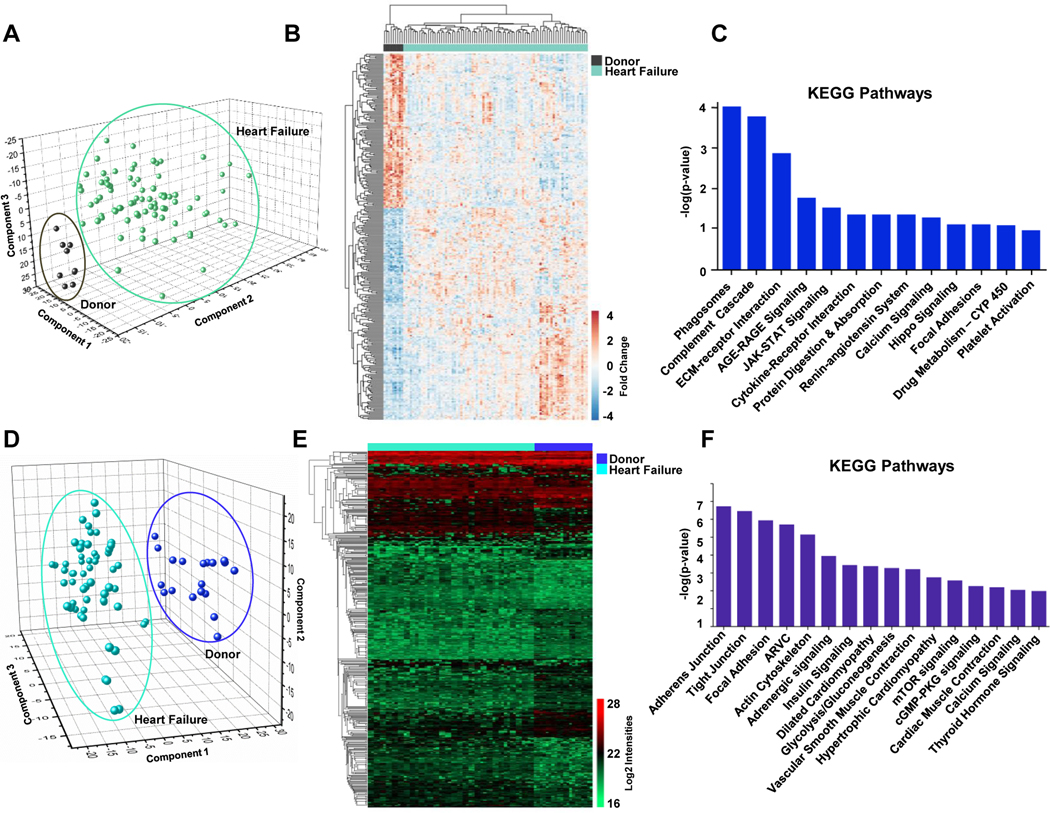
We compared the gene expression profiles of 84 HF patients receiving an LVAD implant and 9 non-failing donor controls using RNA-Sequencing. These 84 HF patients are a combination of responders and non-responders which are further stratified in these analyses. We identified 24,045 total transcripts and graphed the expression data for all samples from this entire dataset using a PCA plot which shows consistent and reproducible differences in transcript abundance between donor (control) and HF samples (Figure 2A). We then focused our analysis on transcripts which were differentially expressed between these two populations and met the following criteria: fold change ≥1.5 and adjusted p-value <0.05. This filtering resulted in 788 upregulated (red) and 553 downregulated transcripts (blue) in HF samples which are presented in heat map format (Figure 2B) and listed in Table S3. We then performed pathway analysis of these 1,341 differentially regulated transcripts using the KEGG database which highlighted the pathways most enriched in this subset (based on adjusted p-value) including the complement cascade pathway (known to regulate inflammation in ischemia reperfusion injury) and extra cellular matrix (ECM)-receptor interaction pathway, among others (Figure 2C).
Figure 2. Transcriptomic and phosphoproteomic profile of LV tissue from non-failing donors versus all heart failure samples.
A, Principal component analysis (PCA) of all 24,045 transcripts obtained from RNA-Sequencing of donor/non-failing (black) and heart failure samples (including both responders and non-responders; turquoise) where each circle represents the combined expression data for all transcripts from a single patient sample. B, Heat map representing all genes with significantly altered expression (p-value<0.05, fold change>1.5, base mean>20) in the failing heart (553 downregulated and 788 upregulated). C, KEGG pathways (p-value <0.05) represented by the differentially expressed transcripts between failing hearts and donors. D, PCA plot based on all 15,816 phosphorylated peptides identified across all heart failure and donor samples where each circle represents the combined abundance data for all peptides from a single patient sample. Samples were analyzed in duplicate and included as two separate data points to confirm the reproducibility between technical replicates. E, Heat map of 443 peptides differentially phosphorylated between heart failure patients and non-failing donors based on log2 values of normalized intensities. F, KEGG pathways significantly enriched in this dataset (p-value <0.05) based on protein phosphorylation differences between failing and donor hearts.
Differential abundance between the failing heart and non-failing donors using phosphoproteomics.
Preparation of tissue samples for mass spectrometry-based phosphoproteomic analyses achieved ~91% enrichment of phosphopeptides (Figure S3), with ~80% enrichment being the base standard for this type of experiment36, and identified a total of 15,816 phosphopeptides in left ventricular cardiac tissue from 10 normal donor samples and 29 HF samples. Additional filtering of these peptides to identify those which were consistently present (≥50% of samples) identified 2,023 phosphopeptides and the abundance variation for all peptides across these samples was plotted using a PCA plot (Figure 2D) where each circle represents a unique sample or technical replicate (each tissue sample was analyzed twice). Using computational analyses, we then mapped 443 of these phosphopeptides to 281 proteins with displayed altered abundance in heat failure patients (adjusted p-value <0.01 and fold change ≥1.5), Figure 2E. Phosphopeptides shown in the heatmap in Figure 2E are also listed in Table S4. We then utilized the DAVID Bioinformatics Resource functional annotation tool (version 6.7) to elucidate pathways and processes significantly enriched by these 281 proteins in failing hearts compared to healthy donors and identified tight junctions, focal adhesions, and glycolysis among the significant KEGG pathways along with hypertrophic and dilated cardiomyopathy and adrenergic signaling (Figure 2F).
Literature Comparison of Datasets
We also compared our gene expression and phosphopeptide profiles with previously published datasets which had also interrogated HF patient samples using either RNA-sequencing,22, 26 microarrays32, 37, 38 or proteomics31–33 (Figure S4A–C). Six studies have previously utilized global RNA-sequencing of cardiac tissue from human HF patients, two which compared non-failing and failing tissue22, 26 and are included in Figure S4A, while four additional studies focused their analysis on ischemic and non-ischemic tissue27–29 or used this technology to sequence microRNAs39 and were not used for comparison. Regarding the application of microarrays: a much larger number of studies exist which utilize microarray technology to analyze diseased cardiac tissue, however, some were not publicly available and therefore not included in our comparisons while other studies utilized different disease states (ischemic versus non-ischemic19; pre-LVAD versus post-LVAD17, 23) and for that reason were not considered in our comparison. Ultimately, we compared our data with 3 other microarray data sets that seemed to best match our analysis and patient samples. Overall our comparison showed that the differentially expressed transcripts from our RNA-sequencing data had a 25% overlap (334 similar transcripts) with data from Yang et al.22 (Figure S4A) but only a 7% overlap (98 transcripts) with microarray data from Colak et al (Figure S4B).32
Global mass spectrometry-based proteomics profiling has not been widely applied to cardiac tissue samples from HF patients so only 3 studies are available for comparison, two of which utilized general proteomics from Stephens et al.31 (proteomics of aortic valves from patients undergoing LVAD implantation compared to healthy donors) and Colak et al.40 (proteomics of cardiac tissue from patients with dilated cardiomyopathy harvested at the time of transplantation) and one study utilizing phosphoproteomics of ischemic and non-ischemic cardiac tissue at the time of heart transplant compared to donors, Schechter et al33. These comparisons (Figure S4C) showed a 4% overlap (12 proteins) with this study when compared to both studies from Schechter and Colak et al.
Responder vs non-responder via RNA-Sequencing
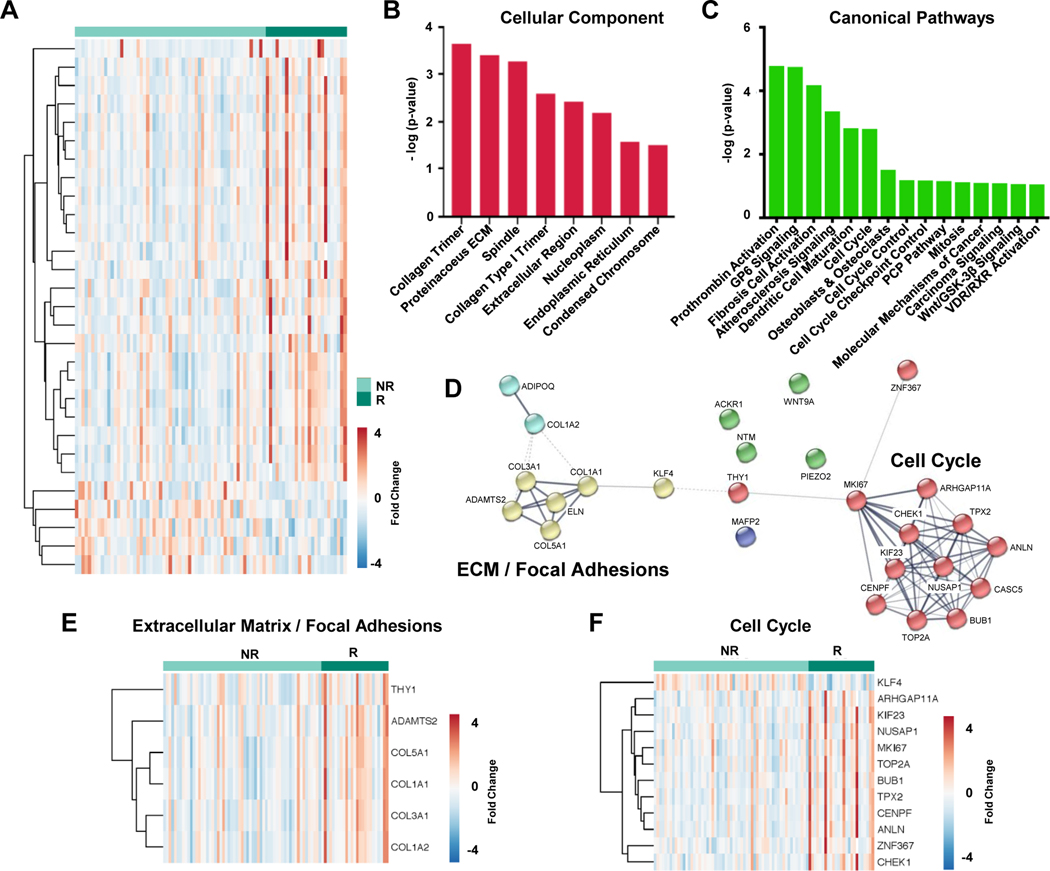
Having evaluated the transcript and phosphopeptide differences between donor and HF samples we then shifted our analysis to distinguish the molecular differences between cardiac tissue capable of functional recovery after LVAD-unloading (responders) from tissue which showed no improvement (non-responders). Our analyses of RNA-sequencing data from 25 responder and 59 non-responder samples identified 29 transcripts which were differentially expressed between these two tissue types (p-value <0.05 and fold change ≥1.5) including 5 downregulated and 24 upregulated transcripts in responder samples depicted in heat map format, Figure 3A. Our bioinformatics analyses of these 29 transcripts using Ingenuity Pathway Analysis (IPA) highlighted enrichment in Cellular Components (Figure 3B) and Canonical Pathways (Figure 3C) in which these transcriptions function. In addition, STRING analysis41, which creates networks of functionally associated proteins from a large biological database, identified Extracellular Matrix (ECM)/Focal Adhesions and Cell Cycle as two of the top networks represented by these 29 transcripts (Figure 3D). Individual transcripts represented by these two networks in our RNA-sequencing data were subsequently extracted (Figure 3E–F) and show a general increase in transcript expression in responders when compared to non-responders.
Figure 3. Transcriptomic profile of responders versus non-responders at the time of LVAD implantation.
A, Heat map showing 29 differentially expressed transcripts (p-value <0.05, fold change >1.5, base mean >30) in tissue from responder and non-responder patients including 24 increased (red) and 5 decreased (blue) in responders. Enrichment analysis for these 29 transcripts using Ingenuity Pathway Analysis lists the top significant terms for B, Cellular Component and C, Canonical Pathways. D, Network map from STRING analysis highlighting functionally associated proteins shows clustering of transcripts into two distinct subsets corresponding to Extracellular Matrix (ECM)/ Focal Adhesions (yellow) and Cell Cycle (red). E-F, Heat maps of specific transcripts annotated for their involvement in Extracellular Matrix/ Focal Adhesions (6 transcripts) and Cell Cycle (12 transcripts). R=responder, NR=non-responder.
Responder vs non-responder via phosphoproteomic analysis
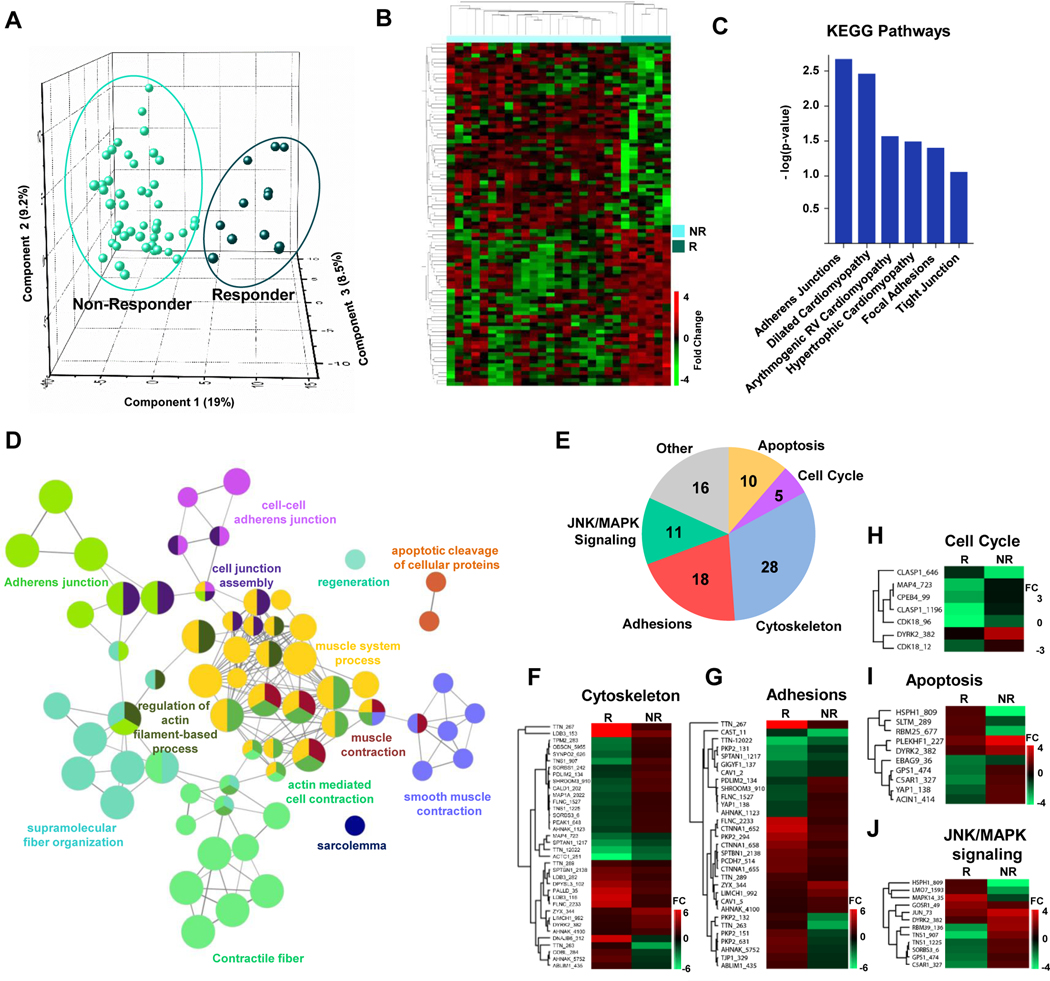
In our phosphoproteomics analysis we compared myocardial tissue from 6 responders and 23 non-responders and identified 93 phosphopeptides mapping to 67 proteins that are differentially phosphorylated (p-value <0.01 and fold change ≥1.5) between these two groups. Combined abundance values for this panel of phosphopeptides were consistent and reproducible between responder and non-responder samples as reflected in the Principal Component Analysis (PCA) plot (Figure 4A) and heatmap (Figure 4B). In addition, when this data was combined with patient demographics, including age or sex (Figure S5), it showed no clustering between biological replicates (although technical replicates should and do cluster), suggesting that the molecular differences observed in the proteomics data are consistent and reproducible regardless of the sex or age of the patient. These 93 phosphopeptides are also listed in Table S5. This list of 67 proteins was submitted to the DAVID Bioinformatics Resource Functional Annotation Tool to elucidate the KEGG pathways represented (based on enrichment p-value) and included adherens junctions, focal adhesions, and various cardiomyopathy terms (Figure 4C). To further elucidate the biological processes and pathways that these phosphoproteins function in we used CytoScape with ClueGo and CluePedia plugins to examine Gene Ontology terms and KEGG and Reactome pathways to build a network of the significantly enriched processes (Bonferroni corrected p-value <0.05), Figure 4D, which includes muscle contraction, contractile fibers, apoptosis, and adherens junctions, among others. To supplement this analysis, we also clustered these 67 proteins into the 5 most represented gene ontology annotations based on DAVID Bioinformatics Resource (Apoptosis, Cell Cycle, Cytoskeleton, Adhesions, and Jnk/Mapk signaling), Figure 4E, and have included the specific abundance changes in heatmap format for each phosphopeptide in these categories, Figure 4F–J. Some proteins contain multiple annotations and are therefore included in more than one category in Figure 4E.
Figure 4. Phosphoproteomic profile of responders versus non-responders at the time of LVAD implantation.
A, PCA plot showing the variability in abundance for all 93 phosphopeptides in responder and non-responder samples (p-value <0.01, fold change >1.5). B, Heat map of 93 differentially phosphorylated peptides in responder (R) and non-responder (NR) samples. C, KEGG pathways enriched in differentially expressed phosphopeptides. D, Gene ontology analysis incorporating cellular components and biological processes and KEGG and Reactome pathways significantly altered based on differentially phosphorylated proteins (ClueGO and CluePedia). E, Pie chart highlighting the most common gene ontology terms annotated for each of the 67 proteins (encompassing 93 phosphopeptides). Some proteins contain annotations for more than 1 term due to multi-functionality. F-I, Heat maps for differentially phosphorylated peptides based on individual pathways or processes for cytoskeleton, adhesions, apoptosis, cell cycle and Jnk/MAPK signaling. FC=fold change. R=responder, NR=non-responder.
Common pathways identified by transcriptomics and phosphoproteomics
We then sought to combine the information from both -omics platforms utilized in this study and performed binary comparisons of the pathways and biological processes that were enriched in the bioinformatics analysis of RNA-sequencing and phosphoproteomics data sets for 1) donor vs responders, 2) donor vs non-responders and 3) responders vs non-responders, Figure S6. It’s important to note that the comparison shown in Figure S6 is not a comparison of individual proteins/transcripts but instead compares gene ontology and pathway terms enriched in the datasets after bioinformatics analyses using the DAVID platform. These comparisons included pathways and gene ontology terms with a p-value less than 0.01 and showed that sirtuin signaling was the only pathway enriched in both –omics datasets when comparing both responders and non-responders to donors. In addition, adherens junctions, extracelluar matrix and cell cycle were the three common terms consistent in both -omics datasets that differentiated responders from non-responders (when comparing either responders and non-responders or those individually to non-failing donors). These results are consitent with our individual analyses of these datasets which showed general upregulation of transcripts involved in cell cycle or extracellular matrix in responders (Figure 3E–F) and differential phosphorylation in these same pathways (Figure 4E–J). We then mapped the individual phosphoproteins/transcripts (using their gene name abbreviation) to schematic diagrams of these two processes: cell cycle and extracellular matrix/focal adhesion (Figure 5A and 5B). These exciting results begin to identify key pathways and proteins which distinguish HF patients capable of functional and structural improvements after mechanical unloading therapy, although additional follow-up studies will be necessary to elucidate the specific role of these biomolecules and the combined benefit from individual pathways to the myocardial recovery process.
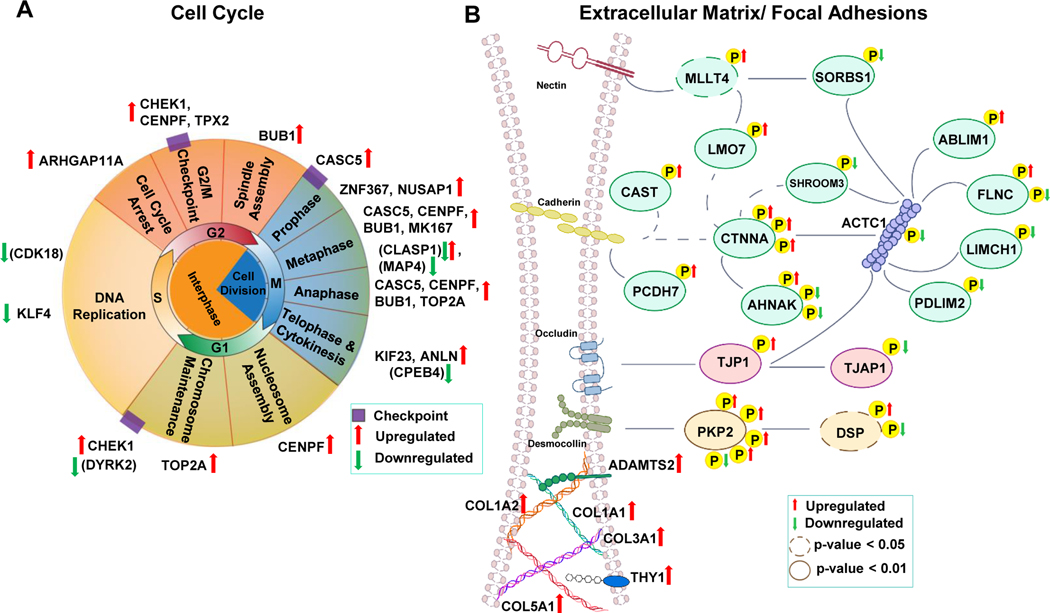
Figure 5. Diagrammatic representation of the transcripts and phosphorylated proteins identified in this study from two key pathways which molecularly distinguish cardiac tissue from responder and non-responder patients.
A, Schematic of the cell cycle highlighting the transcripts and phosphoproteins at specific cell cycle stages which are upregulated (red arrows) or downregulated (green arrow) in cardiac tissue from patients capable of functional recovery after LVAD therapy (responders) as compared to non-responders, which showed no improvement. Phosphorylated proteins are shown in parenthesis to distinguish them from transcripts which are listed without parenthesis. B, Schematic of proteins involved in extracellular matrix/ focal adhesion which are differentially expressed or differentially phosphorylated in responder vs non-responder tissue. Phosphorylation sites are depicted by yellow P. Upregulated or downregulated phosphorylation sites are indicated by red or green arrows, respectively. Phosphoproteins/transcripts are abbreviated using the respective gene name to allow for easy compilation of the data.
DISCUSSION
To our knowledge this is the first molecular analysis of human HF samples which combines multiple -omics technologies to analyze a unique sample set comprised of tissue from patients who either do not respond or respond favorably to LVAD unloading and result in sustained reverse pathological remodeling. This combination of cutting-edge technological approaches and unique patient myocardial tissue samples have allowed us to characterize the molecular underpinnings which define differences in the ability for cardiac recovery. Specifically, these analyses have revealed major differences in cell cycle regulation and extracellular matrix/focal adhesions as two major pathways which are differentially regulated in patients capable of recovery versus those who are not.
The major strengths/advances of this study include: 1) the use of a large patient sample set (n=93) which have been carefully curated and systematically processed and combined with longitudinal echocardiographic data assessing myocardial response to LVAD therapy, 2) the application of multiple -omics technologies to unbiasedly identify molecular differences between tissue samples, 3) the application of phosphopeptide enrichment and mass spectrometry based proteomics to interrogate post-translational modifications in the etiology of HF and recovery, 4) the comparison of our data from non-failing and failing myocardial samples to previously published literature to expand our understanding of the molecular adaptations and compensatory mechanisms in human HF, 5) the identification of a panel of transcripts and phosphopeptides which are consistent and reproducible markers of myocardial tissue that maintains the capacity for functional/structural recovery after LVAD-induced unloading. The specific mechanistic insights provided by these analyses include the differential regulation of 29 transcripts and 93 phosphopeptides which includes both established and novel markers of myocardial recovery and encompasses two major pathways (cell cycle and extracellular matrix/focal adhesions) and several biological processes (apoptosis, Jnk/MAPK signaling, cytoskeleton).
The NHLBI Working Group on myocardial recovery identified as a key deficiency in the field that most translational LVAD studies have not collected or integrated functional myocardial data in order to distinguish which tissue, cellular and molecular changes are unique to LVAD patients who respond favorably and recover significant myocardial function versus those who do not.9 It is these changes that may provide the most fundamental insights and enable association with true pathophysiologic mechanisms of reverse myocardial remodeling that will be elucidated in future mechanistic studies.9, 16, 42, 43 It is therefore critical to examine tissue from both of these patient subgroups in order to begin understanding causality between reverse remodeling changes and myocardial recovery.9, 16, 42, 43
The use of larger samples cohorts will likely aid the identification of the most reproducible markers of myocardial recovery due to the complex etiology of heart disease, length of disease onset, therapeutic treatments, etc. Studies have traditionally utilized relatively small cohorts of patient samples, however, this trend appears to be increasing as research centers continue to biobank patient tissue and utilize these in large-scale analyses. Our analyses incorporated 93 HF patient samples and 12 non-failing donor controls, the largest we’ve seen in the literature for a translational myocardial recovery study in humans. For comparison the studies included in the Venn diagrams in Figure S4 (which only had HF samples and no prospective phenotyping to identify myocardial recovery samples) have utilized the following number of samples: n=40, 12, 10, 24, 16; in chronological order. The use of such large sample sets is often costly and time-consuming and will require a continued commitment from funding agencies to support these types of analyses. Additionally, to our knowledge only one study has previous applied multiple –omics platforms to molecularly interrogate cardiac tissue samples (from dilated cardiomyopathy patients; n=5 patients for non-failing and DCM samples) using microarrays and global proteomics but did not investigate differences in response to LVAD-unloading. The results of which are compared in Figure S4.
In this study we have carried out quantitation of transcripts and phosphorylated peptide abundance. It is not surprising that there is not more overlap between these two populations since, 1) the two -omics platforms are analyzing different biologically regulated processes (transcription vs phosphorylation), and2) we are profiling phosphorylated proteins (not total protein abundance) in our proteomic analyses. Regarding our proteomics data: we chose to selectively enrich and profile the protein samples for differences in phosphorylation, the most abundant post-translational modification, which can also act as a surrogate for major signaling pathways. This allows us to overcome the limitations of global proteomic profiling, which traditional only analyzes the most abundant proteins in a sample which, in cardiac tissue, are overwhelmingly structural, contractile and mitochondrial proteins. Therefore, we view these as complimentary methods to more comprehensively profile the molecular changes which differentiate these two patient populations (responders vs non-responders).
Regarding the clinical parameters examined: Indeed, patients who improved their cardiac function and structure on LVAD support (responders), were more likely female, were younger, and had a shorter duration of HF symptoms compared to non-responders. A younger age and a shorter duration of HF have been consistently identified as variables associated with left ventricular reverse remodeling upon LVAD unloading.1, 6, 44 Furthermore, female sex has been associated with a higher propensity for reverse cardiac remodeling in HF patients.45, 46 In this line of investigation, we sought to identify how these differences in clinical characteristics translate into molecular differences reflected in the differential expression of genes/proteins and render these patients more or less amenable to recovery. This would allow us to begin to understand biologically why these characteristic differences predispose patients to enhanced reverse remodeling and myocardial improvement and identify key molecular players that may explain these differences and drive this differential response.
Cell Cycle
During normal mammalian development, cardiomyocytes actively divide, however, after birth, this renewal capacity quickly dissipates and can result in two major phenomena, depending on when the cardiomyocytes exit the cell cycle: 1) polyploidization, which results from premature cell cycle exit after DNA synthesis and forms a single tetraploid nucleus, or 2) binucleation, which results from cell cycle exit after mitosis without cytokinesis giving two diploid nuclei. The presence or absence of these two phenomenon have been observed across varies mammalian species with a majority of adult human cardiomyocytes resulting from polyploidization and a majority of mouse cardiomyocytes resulting from binucleation47–49. During disease, the adult mammalian heart has traditionally been thought to be incapable of regenerating cardiomyocytes after substantial cell loss due to the inability of adult cardiomyocytes to divide, however, research in the last decade has challenged this dogma and led to intense debates about whether adult cardiomyocytes do indeed have the capacity to regenerate in response to injury and, if so, to what extent. Regeneration studies have been aimed at stimulating cardiomyocyte proliferation to create new myocardium, however, one confounding contribution to this work has been the analysis of cell cycle markers which do not always measure true cardiomyocyte proliferation due to the fact that cardiomyocyte cell cycle reentry does not necessarily result in the formation of new cells. Interestingly, cardiomyocyte ploidy has been shown to increase in humans after myocardial infarction50, 51 suggesting cell cycle reentry may be an adaptive response to injury, although whether or not this results in complete cytokinesis, and the associated penetrance, is still under investigation. In addition, after LVAD unloading in HF patients, decreased hypertrophy11, nuclear size52, DNA content53 and ploidy52 have been observed with a concomitant increase in myocyte numbers and may suggest that cardiac recovery following mechanical unloading may involve a switch from hypertrophic to hyperplastic growth54.
Our data, from both transcriptomic and phosphoproteomic datasets, demonstrate that one of the major pathways altered in tissue from patients capable of cardiac recovery is cell cycle regulation, and identifies specific changes in abundance or phosphorylation of 17 of these molecules. While previous published studies have primarily focused on evaluating cell cycle reentry or proliferation as a process (i.e. increases in cell numbers) we now provide direct evidence for the involvement of specific proteins in regulating cell cycle, as it applies to cardiac remodeling. While these 17 transcripts/proteins have been linked to cell cycle regulation in non-cardiac cells most of these transcripts have never been examined in the heart so their role in cardiomyocyte function or heart disease is unknown. Specifically, only two of these 17 molecules (transcripts/peptides) have been previously linked to cardiomyopathy or HF susceptibility (CENPF55 and KLF456) so additional analyses will be necessary to identify the individual and combined contributions of these novel markers to cardiac recovery. Indeed, CENP-F has been examined in knockout mice where loss of CENP-F results in dilated cardiomyopathy with severe disruption of cardiac myocyte architecture.55 In addition, loss of KLF4 leads to higher mortality after pressure-overload in mice while overexpression of KLF4 in culture cardiomyocytes is capable of inhibiting hypertrophy and fetal gene expression changes.56 This is consistent with our results which show that patients who respond positively to LVAD unloading have higher levels of CENP-F and KLF4. In addition, the abundance changes identified in phosphopeptides from these cell cycle markers may reflect either changes in total protein abundance or may reflect the percent of the protein that is phosphorylated under different conditions. With regards to the latter, many of the phosphopeptides identified in this study contain novel phosphorylation sites and the molecular function of these modifications is unknown.
It is also possible that some of the observed changes in cell cycle markers are due to infiltration of non-cardiomyocytes into the heart tissue, so future studies would be necessary to determine the exact cell type contributing to these changes or if these are reflective of changes in multiple cell types.
Extracellular Matrix/Focal Adhesions
The extracellular matrix is a three-dimensional network of proteoglycans, glycoproteins, and fibrous proteins which provides structural support for the heart (and other organs) and links the extracellular regions between cells to transmit tension through tissues, and mitigate damage57. Cardiomyocytes also contain a vast network of adhesive molecules which form large protein complexes which link adjacent cells together at adherens junctions (cell-cell contacts) and link cells to the extracellular matrix at focal adhesions (cell-extracellular matrix contacts). In the adult heart, cardiomyocytes are exposed to a number of different forces including hemodynamic pressure, stretching through cardiomyocyte contraction and passive elasticity from the extracellular matrix, all of which are subject to change and ultimately effect cardiac function and intracellular signaling. Changes in the composition, abundance and organization of extracellular matrix and adhesive molecules in the heart is dynamic and has been shown to change during development and the progression of heart disease58 (hypertrophy, dilated cardiomyopathy and HF). Specifically, increased cardiac fibrosis leads to differential expression and cross-linking of extracellular matrix components which change the mechanical landscape and rigidity of cardiac tissue59. In addition, healthy ventricular tissue maintains spatial segregation of cell-matrix adhesions to transverse myocyte borders and cell-cell adhesions to longitudinal borders60, which, when disrupted by differential lateralization or abundance leads to the development of cardiomyopathies61, 62. While these key links have been established between the development of cardiomyopathies and the extracellular matrix and focal adhesions, some of these changes have also been reported to reverse upon LVAD-unloading. Specifically, changes in collagen content and cross-linking have been reported63, 64 with conflicting results although in general data trends have been established which suggest that excessive scarring appears to limit cardiac recovery, and the degree of fibrosis in the myocardium at the time of implantation may influence the ability to recover65–67. Additional modulation of specific molecules (COL1A1, COL3A1, TIMP-465) have been examined during LVAD-unloading although whether these specific changes are a feature of myocardium which functional recovers or does not, has not been investigated.
Our results identify 27 differentially phosphorylated sites across 16 proteins and abundance changes in 6 transcripts involved in extracellular matrix composition, adhesive junction formation and the associated intracellular signaling molecules in cardiac tissue from patients capable of recovery after LVAD-unloading. These include proteins with previous links to general LVAD-unloading (including COL1A1 and COL3A1) as well as novel molecules and phosphorylation sites which now comprise a panel of markers unique to tissue capable of functional recovery.
In summary, this study expands our understanding of the molecular underpinnings which differentiate normal and failing myocardial tissue and highlights key pathways misregulated during disease and those which may enable functional recovery after mechanical circulatory support and ventricular unloading. Further interrogation of this panel of transcripts and phosphopeptides will be necessary to identify their specific role in regulating myocyte biology and cardiac function. Our findings may help guide future clinical strategies for the prioritization of advanced HF patients towards heart transplant versus LVAD therapy. Furthermore, these findings also have translational research implications in identifying new HF therapeutic targets.
Supplementary Material
CLINICAL PERSPECTIVE.
What is new?
This is the first analysis of a large prospective human myocardial tissue sample set which was curated, systematically processed, and combined with longitudinal echocardiographic and clinical data assessing myocardial response following LVAD unloading and circulatory support.
This study identifies a panel of transcripts and phosphopeptides which are consistent and reproducible markers of myocardial tissue that maintains the capacity for functional and structural improvement after LVAD-induced unloading.
This work expands our understanding of the molecular underpinnings which differentiate non-failing and failing myocardium and highlights key pathways misregulated during disease which may enable functional recovery after ventricular unloading and circulatory support.
What are the clinical implications?
Our findings may help guide future clinical strategies for patient selection and prioritization of advanced HF patients towards heart transplantation or LVAD therapy targeting myocardial recovery and sustainable remission from chronic HF.
These results have translational research implications in identifying novel HF therapeutic targets.
ACKNOWLEDGMENTS
This work was supported by funds from AHA, NIH and Nora Eccles Treadwell Foundation (to Drs. Drakos and Franklin). The authors are grateful to the donor families for their generosity, and DonorConnect (https://www.donorconnect.life/), Salt Lake City, Utah, for facilitating the work of our research team members acquiring myocardial tissue in the operating rooms of several hospitals of the Intermountain West. Research reported in this publication utilized the High-Throughput Genomics and Bioinformatics Analysis Shared Resource at Huntsman Cancer Institute at the University of Utah supported by the National Cancer Institute Award Number P30CA042014. The authors thank Ms. Diana Lim for assistance with the figures and Drakos lab members for collecting cardiac tissues from heart failure patients and donors.
SOURCES OF FUNDING
This work was supported by AHA Heart Failure Strategically Focused Research Network, Grant 16SFRN29020000 (Drakos, Selzman), Grant K23HL150322 (Wever-Pinzon), NHLBI RO1 Grant HL135121 (Drakos), NHLBI RO1 Grant HL132067 (Drakos), NHLBI RO1 HL130424 (Franklin), Merit Review Award I01 CX002291 U.S. Dpt of Veterans Affairs (Drakos), NHLBI 2T32HL007576-36 (Kyriakopoulos), NHLBI 5T32HL007576-33 (Taleb), Nora Eccles Treadwell Foundation Grants (Drakos and Franklin).
ABBREVIATIONS:
- HF
Heart Failure
- LVAD
Left Ventricular Assist Devices
- LV
Left Ventricle
- ECHO
Echocardiogram
- LVEF/ EF
Left Ventricle Ejection Fraction
- LVEDD
Left Ventricular End Diastolic Dimension
- PCA
Principal Component Analysis
- ECM
Extra Cellular Matrix
- GO
Gene Ontology
- IPA
Ingenuity Pathway Analysis
- LC-MS/MS
Liquid Chromatography- Tandem Mass Spectrometry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
Footnotes
DISCLOSURES
The authors have no competing interests to disclose.
REFERENCES
- 1.Topkara VK, Garan AR, Fine B, Godier-Furnemont AF, Breskin A, Cagliostro B, Yuzefpolskaya M, Takeda K, Takayama H, Mancini DM, Naka Y and Colombo PC. Myocardial Recovery in Patients Receiving Contemporary Left Ventricular Assist Devices: Results From the Interagency Registry for Mechanically Assisted Circulatory Support (INTERMACS). Circulation Heart Failure. 2016;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birks EJ, Tansley PD, Hardy J, George RS, Bowles CT, Burke M, Banner NR, Khaghani A and Yacoub MH. Left ventricular assist device and drug therapy for the reversal of heart failure. New England Journal of Medicine. 2006;355:1873–84. [DOI] [PubMed] [Google Scholar]
- 3.Dandel M, Weng Y, Siniawski H, Potapov E, Drews T, Lehmkuhl HB, Knosalla C and Hetzer R. Prediction of cardiac stability after weaning from left ventricular assist devices in patients with idiopathic dilated cardiomyopathy. Circulation. 2008;118:S94–105. [DOI] [PubMed] [Google Scholar]
- 4.Dandel M, Weng Y, Siniawski H, Potapov E, Lehmkuhl HB and Hetzer R. Long-term results in patients with idiopathic dilated cardiomyopathy after weaning from left ventricular assist devices. Circulation. 2005;112:I37–45. [DOI] [PubMed] [Google Scholar]
- 5.Drakos SG, Wever-Pinzon O, Selzman CH, Gilbert EM, Alharethi R, Reid BB, Saidi A, Diakos NA, Stoker S, Davis ES, Movsesian M, Li DY, Stehlik J, Kfoury AG and Investigators U. Magnitude and time course of changes induced by continuous-flow left ventricular assist device unloading in chronic heart failure: insights into cardiac recovery. Journal of the American College of Cardiology. 2013;61:1985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wever-Pinzon O, Drakos SG, McKellar SH, Horne BD, Caine WT, Kfoury AG, Li DY, Fang JC, Stehlik J and Selzman CH. Cardiac Recovery During Long-Term Left Ventricular Assist Device Support. Journal of the American College of Cardiology. 2016;68:1540–53. [DOI] [PubMed] [Google Scholar]
- 7.Birks EJ, Drakos SG, Patel SR, Lowes BD, Selzman CH, Starling RC, Trivedi J, Slaughter MS, Alturi P, Goldstein D, Maybaum S, Um JY, Margulies KB, Stehlik J, Cunningham C, Farrar DJ and Rame JE. Prospective Multicenter Study of Myocardial Recovery Using Left Ventricular Assist Devices (RESTAGE-HF [Remission from Stage D Heart Failure]): Medium-Term and Primary End Point Results. Circulation. 2020;142:2016–2028. [DOI] [PubMed] [Google Scholar]
- 8.Wever-Pinzon J, Selzman CH, Stoddard G, Wever-Pinzon O, Catino A, Kfoury AG, Diakos NA, Reid BB, McKellar S, Bonios M, Koliopoulou A, Budge D, Kelkhoff A, Stehlik J, Fang JC and Drakos SG. Impact of Ischemic Heart Failure Etiology on Cardiac Recovery During Mechanical Unloading. Journal of the American College of Cardiology. 2016;68:1741–1752. [DOI] [PubMed] [Google Scholar]
- 9.Drakos SG, Pagani FD, Lundberg MS and Baldwin JT. Advancing the Science of Myocardial Recovery With Mechanical Circulatory Support: A Working Group of the National, Heart, Lung, and Blood Institute. JACC Basic Translational Science. 2017;2:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbone A, Holmes JW, Heerdt PM, The AH, Naka Y, Joshi N, Daines M, Marks AR, Oz MC and Burkhoff D. Comparison of right and left ventricular responses to left ventricular assist device support in patients with severe heart failure: a primary role of mechanical unloading underlying reverse remodeling. Circulation. 2001;104:670–5. [DOI] [PubMed] [Google Scholar]
- 11.Zafeiridis A, Jeevanandam V, Houser SR and Margulies KB. Regression of cellular hypertrophy after left ventricular assist device support. Circulation. 1998;98:656–62. [DOI] [PubMed] [Google Scholar]
- 12.Dipla K, Mattiello JA, Jeevanandam V, Houser SR and Margulies KB. Myocyte recovery after mechanical circulatory support in humans with end-stage heart failure. Circulation. 1998;97:2316–22. [DOI] [PubMed] [Google Scholar]
- 13.Terracciano CM, Hardy J, Birks EJ, Khaghani A, Banner NR and Yacoub MH. Clinical recovery from end-stage heart failure using left-ventricular assist device and pharmacological therapy correlates with increased sarcoplasmic reticulum calcium content but not with regression of cellular hypertrophy. Circulation. 2004;109:2263–5. [DOI] [PubMed] [Google Scholar]
- 14.Lee SH, Doliba N, Osbakken M, Oz M and Mancini D. Improvement of myocardial mitochondrial function after hemodynamic support with left ventricular assist devices in patients with heart failure. Journal of Thoracic Cardiovascular Surgery. 1998;116:344–9. [DOI] [PubMed] [Google Scholar]
- 15.Flesch M, Margulies KB, Mochmann HC, Engel D, Sivasubramanian N and Mann DL. Differential regulation of mitogen-activated protein kinases in the failing human heart in response to mechanical unloading. Circulation. 2001;104:2273–6. [DOI] [PubMed] [Google Scholar]
- 16.Drakos SG, Kfoury AG, Stehlik J, Selzman CH, Reid BB, Terrovitis JV, Nanas JN and Li DY. Bridge to recovery: understanding the disconnect between clinical and biological outcomes. Circulation. 2012;126:230–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blaxall BC, Tschannen-Moran BM, Milano CA and Koch WJ. Differential gene expression and genomic patient stratification following left ventricular assist device support. Journal of the American College of Cardiology. 2003;41:1096–106. [DOI] [PubMed] [Google Scholar]
- 18.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB and Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–76. [DOI] [PubMed] [Google Scholar]
- 19.Kittleson MM, Ye SQ, Irizarry RA, Minhas KM, Edness G, Conte JV, Parmigiani G, Miller LW, Chen Y, Hall JL, Garcia JG and Hare JM. Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation. 2004;110:3444–51. [DOI] [PubMed] [Google Scholar]
- 20.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB and Dorn GW 2nd. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramani R, Vela D, Segura A, McNamara D, Lemster B, Samarendra V, Kormos R, Toyoda Y, Bermudez C, Frazier OH, Moravec CS, Gorcsan J 3rd, Taegtmeyer H and McTiernan CF. A micro-ribonucleic acid signature associated with recovery from assist device support in 2 groups of patients with severe heart failure. Journal of the American College of Cardiology. 2011;58:2270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL and Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA and Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circulation Research. 2005;96:592–9. [DOI] [PubMed] [Google Scholar]
- 24.Esteller M Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. [DOI] [PubMed] [Google Scholar]
- 25.Mattick JS. The central role of RNA in human development and cognition. FEBS Letters. 2011;585:1600–16. [DOI] [PubMed] [Google Scholar]
- 26.Dhar K, Moulton AM, Rome E, Qiu F, Kittrell J, Raichlin E, Zolty R, Um JY, Moulton MJ, Basma H, Anderson DR, Eudy JD and Lowes BD. Targeted myocardial gene expression in failing hearts by RNA sequencing. Journal of translational medicine. 2016;14:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweet ME, Cocciolo A, Slavov D, Jones KL, Sweet JR, Graw SL, Reece TB, Ambardekar AV, Bristow MR, Mestroni L and Taylor MRG. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genomics. 2018;19:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, Tang WH, Moravec CS, Margulies KB, Cappola TP, Li M and consortium MA. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrer I, Rosello-Lleti E, Ortega A, Tarazon E, Molina-Navarro MM, Trivino JC, Martinez-Dolz L, Almenar L, Lago F, Sanchez-Lazaro I, Gonzalez-Juanatey JR, Salvador A, Portoles M and Rivera M. Gene expression network analysis reveals new transcriptional regulators as novel factors in human ischemic cardiomyopathy. BMC Medical Genomics. 2015;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Liu CY, Li YS, Xu J, Li DG, Li X and Han D. Deep RNA sequencing elucidates microRNA-regulated molecular pathways in ischemic cardiomyopathy and nonischemic cardiomyopathy. Genetics Molecular Research. 2016;15. [DOI] [PubMed] [Google Scholar]
- 31.Stephens EH, Han J, Trawick EA, Di Martino ES, Akkiraju H, Brown LM, Connell JP, Grande-Allen KJ, Vunjak-Novakovic G and Takayama H. Left-Ventricular Assist Device Impact on Aortic Valve Mechanics, Proteomics and Ultrastructure. The Annals of thoracic surgery. 2018;105:572–580. [DOI] [PubMed] [Google Scholar]
- 32.Colak D, Alaiya AA, Kaya N, Muiya NP, AlHarazi O, Shinwari Z, Andres E and Dzimiri N. Integrated Left Ventricular Global Transcriptome and Proteome Profiling in Human End-Stage Dilated Cardiomyopathy. PLoS One. 2016;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schechter MA, Hsieh MK, Njoroge LW, Thompson JW, Soderblom EJ, Feger BJ, Troupes CD, Hershberger KA, Ilkayeva OR, Nagel WL, Landinez GP, Shah KM, Burns VA, Santacruz L, Hirschey MD, Foster MW, Milano CA, Moseley MA, Piacentino V, 3rd and Bowles DE. Phosphoproteomic profiling of human myocardial tissues distinguishes ischemic from non-ischemic end stage heart failure. PLoS One. 2014;9:e104157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, Moritz RL, Carver JJ, Wang M, Ishihama Y, Bandeira N, Hermjakob H and Vizcaino JA. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Research. 2017;45:D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A and Vizcaino JA. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research. 2019;47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue X, Schunter A and Hummon AB. Comparing multistep immobilized metal affinity chromatography and multistep TiO2 methods for phosphopeptide enrichment. Analytical Chemistry. 2015;87:8837–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, Conte JV, Tomaselli G, Garcia JG and Hare JM. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiological genomics. 2005;21:299–307. [DOI] [PubMed] [Google Scholar]
- 38.Colak D, Kaya N, Al-Zahrani J, Al Bakheet A, Muiya P, Andres E, Quackenbush J and Dzimiri N. Left ventricular global transcriptional profiling in human end-stage dilated cardiomyopathy. Genomics. 2009;94:20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, Mihailovic A, Sauer M, Ji R, Ramarathnam A, Totary-Jain H, Williams Z, Tuschl T and Schulze PC. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proceedings of the National Academy of Sciences. 2014;111:11151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colak D, Alaiya AA, Kaya N, Muiya NP, AlHarazi O, Shinwari Z, Andres E and Dzimiri N. Integrated Left Ventricular Global Transcriptome and Proteome Profiling in Human End-Stage Dilated Cardiomyopathy. PLoS One. 2016;11:e0162669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ and Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall JL, Fermin DR, Birks EJ, Barton PJ, Slaughter M, Eckman P, Baba HA, Wohlschlaeger J and Miller LW. Clinical, molecular, and genomic changes in response to a left ventricular assist device. Journal of the American College of Cardiology. 2011;57:641–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drakos SG, Terrovitis JV, Anastasiou-Nana MI and Nanas JN. Reverse remodeling during long-term mechanical unloading of the left ventricle. Journal Molecular Cellular Cardiology. 2007;43:231–42. [DOI] [PubMed] [Google Scholar]
- 44.Antonides CFJ, Schoenrath F, de By T, Muslem R, Veen K, Yalcin YC, Netuka I, Gummert J, Potapov EV, Meyns B, Ozbaran M, Schibilsky D, Caliskan K and investigators E. Outcomes of patients after successful left ventricular assist device explantation: a EUROMACS study. ESC Heart Failure. 2020;7:1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aimo A, Vergaro G, Castiglione V, Barison A, Pasanisi E, Petersen C, Chubuchny V, Giannoni A, Poletti R, Maffei S, Januzzi JL Jr., Passino C and Emdin M. Effect of Sex on Reverse Remodeling in Chronic Systolic Heart Failure. JACC Heart Fail. 2017;5:735–742. [DOI] [PubMed] [Google Scholar]
- 46.Cheng YJ, Zhang J, Li WJ, Lin XX, Zeng WT, Tang K, Tang AL, He JG, Xu Q, Mei MY, Zheng DD, Dong YG, Ma H and Wu SH. More favorable response to cardiac resynchronization therapy in women than in men. Circulation: Arrhythm and Electrophysiology. 2014;7:807–15. [DOI] [PubMed] [Google Scholar]
- 47.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, Malm T, Andra M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H and Frisen J. Dynamics of Cell Generation and Turnover in the Human Heart. Cell. 2015;161:1566–75. [DOI] [PubMed] [Google Scholar]
- 48.Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL and Bergmann O. No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell. 2015;163:1026–36. [DOI] [PubMed] [Google Scholar]
- 49.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S and Kuhn B. Cardiomyocyte proliferation contributes to heart growth in young humans. Proceedings of the National Academy of Sciences. 2013;110:1446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herget GW, Neuburger M, Plagwitz R and Adler CP. DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovascular Research. 1997;36:45–51. [DOI] [PubMed] [Google Scholar]
- 51.Meckert PC, Rivello HG, Vigliano C, Gonzalez P, Favaloro R and Laguens R. Endomitosis and polyploidization of myocardial cells in the periphery of human acute myocardial infarction. Cardiovascular Research. 2005;67:116–23. [DOI] [PubMed] [Google Scholar]
- 52.Wohlschlaeger J, Levkau B, Brockhoff G, Schmitz KJ, von Winterfeld M, Takeda A, Takeda N, Stypmann J, Vahlhaus C, Schmid C, Pomjanski N, Bocking A and Baba HA. Hemodynamic support by left ventricular assist devices reduces cardiomyocyte DNA content in the failing human heart. Circulation. 2010;121:989–96. [DOI] [PubMed] [Google Scholar]
- 53.Rivello HG, Meckert PC, Vigliano C, Favaloro R and Laguens RP. Cardiac myocyte nuclear size and ploidy status decrease after mechanical support. Cardiovascular Pathology. 2001;10:53–7. [DOI] [PubMed] [Google Scholar]
- 54.Canseco DC, Kimura W, Garg S, Mukherjee S, Bhattacharya S, Abdisalaam S, Das S, Asaithamby A, Mammen PP and Sadek HA. Human ventricular unloading induces cardiomyocyte proliferation. Journal of the American College of Cardiology. 2015;65:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manalo A, Schroer AK, Fenix AM, Shancer Z, Coogan J, Brolsma T, Burnette DT, Merryman WD and Bader DM. Loss of CENP-F Results in Dilated Cardiomyopathy with Severe Disruption of Cardiac Myocyte Architecture. Scientific Reports. 2018;8:7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao X, Haldar SM, Lu Y, Jeyaraj D, Paruchuri K, Nahori M, Cui Y, Kaestner KH and Jain MK. Kruppel-like factor 4 regulates pressure-induced cardiac hypertrophy. Journal Molecular Cellular Cardiology. 2010;49:334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rienks M, Papageorgiou AP, Frangogiannis NG and Heymans S. Myocardial extracellular matrix: an ever-changing and diverse entity. Circulation Research. 2014;114:872–88. [DOI] [PubMed] [Google Scholar]
- 58.Berk BC, Fujiwara K and Lehoux S. ECM remodeling in hypertensive heart disease. Journal Clinical Investigation. 2007;117:568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soufen HN, Salemi VM, Aneas IM, Ramires FJ, Benicio AM, Benvenuti LA, Krieger JE and Mady C. Collagen content, but not the ratios of collagen type III/I mRNAs, differs among hypertensive, alcoholic, and idiopathic dilated cardiomyopathy. Brazilian Journal of Medical Biology Research. 2008;41:1098–104. [DOI] [PubMed] [Google Scholar]
- 60.Anastasi G, Cutroneo G, Gaeta R, Di Mauro D, Arco A, Consolo A, Santoro G, Trimarchi F and Favaloro A. Dystrophin-glycoprotein complex and vinculin-talin-integrin system in human adult cardiac muscle. International Journal of Molecular Medicine. 2009;23:149–59. [PubMed] [Google Scholar]
- 61.Kostin S, Rieger M, Dammer S, Hein S, Richter M, Klovekorn WP, Bauer EP and Schaper J. Gap junction remodeling and altered connexin43 expression in the failing human heart. Molecular Cellular Biochemistry. 2003;242:135–44. [PubMed] [Google Scholar]
- 62.Heling A, Zimmermann R, Kostin S, Maeno Y, Hein S, Devaux B, Bauer E, Klovekorn WP, Schlepper M, Schaper W and Schaper J. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circulation Research. 2000;86:846–53. [DOI] [PubMed] [Google Scholar]
- 63.Bruckner BA, Stetson SJ, Farmer JA, Radovancevic B, Frazier OH, Noon GP, Entman ML, Torre-Amione G and Youker KA. The implications for cardiac recovery of left ventricular assist device support on myocardial collagen content. The American Journal of Surgery. 2000;180:498–501; discussion 501–2. [DOI] [PubMed] [Google Scholar]
- 64.Klotz S, Foronjy RF, Dickstein ML, Gu A, Garrelds IM, Danser AH, Oz MC, D’Armiento J and Burkhoff D. Mechanical unloading during left ventricular assist device support increases left ventricular collagen cross-linking and myocardial stiffness. Circulation. 2005;112:364–74. [DOI] [PubMed] [Google Scholar]
- 65.Felkin LE, Lara-Pezzi E, George R, Yacoub MH, Birks EJ and Barton PJ. Expression of extracellular matrix genes during myocardial recovery from heart failure after left ventricular assist device support. The Journal of heart and lung transplantation : The official publication of the International Society for Heart Transplantation. 2009;28:117–22. [DOI] [PubMed] [Google Scholar]
- 66.Segura AM, Frazier OH, Demirozu Z and Buja LM. Histopathologic correlates of myocardial improvement in patients supported by a left ventricular assist device. Cardiovascular Pathology. 2011;20:139–45. [DOI] [PubMed] [Google Scholar]
- 67.Drakos SG, Kfoury AG, Hammond EH, Reid BB, Revelo MP, Rasmusson BY, Whitehead KJ, Salama ME, Selzman CH, Stehlik J, Clayson SE, Bristow MR, Renlund DG and Li DY. Impact of mechanical unloading on microvasculature and associated central remodeling features of the failing human heart. The Journal of the American College of Cardiology. 2010;56:382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.