Abstract
The respiratory infection tuberculosis is caused by the bacteria Mycobacterium tuberculosis and its unrelenting spread caused millions of deaths around the world. Hence, it is needed to explore potential and less toxic anti-tubercular drugs. In the present work, we report the synthesis and antitubercular activity of four different (hydrazones 7–12, O-ethynyl oximes 19–24, triazoles 25–30, and isoxazoles 31–36) hybrids. Among these hybrids 9, 10, 33, and 34, displayed high antitubercular activity at 3.12 g/mL with >90% of inhibitions. The hybrids also showed good docking energies between -6.8 and -7.8 kcal/mol. Further, most active molecules were assayed for their DNA gyrase reduction ability towards M. tuberculosis and E.coli DNA gyrase by the DNA supercoiling and ATPase gyrase assay methods. All four hybrids showed good IC50 values comparable to that of the reference drug. In addition, the targets were also predicted as a potential binder for papain-like protease (SARS CoV-2 PLpro) by molecular docking and a good interaction result was observed. Besides, all targets were predicted for their absorption, distribution, metabolism, and excretion - toxicity (ADMET) profile and found a significant amount of ADMET and bioavailability
Keywords: Pyranone, Hydrazone, Triazole, Isoxazole, Pre-ADMET, Tuberculosis
Graphical abstract
1. Introduction
Tuberculosis (TB) is an air-bone transferable and extremely aggressive disease caused by Mycobacterium Tuberculosis. Millions of people have died globally as a result of it frequently infecting the lungs. Based on the World Health Organization (WHO) 2021 reports, 10.6 million people fell ill with in 2021, an increase of 4.5% from 10.1 million in 2020 [1]. Many nations have been affected by tuberculosis infections, but south-East Asia (45%), Africa (23%) and the Western Pacific (18%), countries have been particularly hard hit by the disease and TB-HIV co-morbidities [2]. Drug-resistant tuberculosis (TB) and multidrug-resistant tuberculosis (MD-TB) are prevalent in practically every country on the globe, making the disease even more complicated to eradicate. The situation has been further worsened for patients by the link between tuberculosis and HIV/AIDS, particularly tuberculosis is the key for the death of HIV-positive patients [3,4]. Due to these factors, the WHO estimates that there are two million tuberculosis-related fatalities and eight million new tuberculosis infections per year [1].
The present management of tuberculosis is a complex task because of the lengthy medication term of 6 to 12 months and the multi-drug-resistant (MDTB) response. The search for novel anti-tuberculosis drugs with M. tuberculosis activity is one of the major areas of medicinal chemistry attention. Several techniques are now being used to create new anti-tuberculosis drugs [5], [6], [7], [8].
Likewise, the global disaster corona virus 2019–nCoV, commonly known as the COVID-19 corona virus, is at the forefront of everyone's mind [9]. It has affected the lives and the work of people around the world including researcher. Yet researchers are also at the forefront of the fight to understand, contain, treat, and finally conquer the disease. Therefore, numerous research organizations throughout the world are working to create brand-new, extremely powerful therapeutic treatments to combat this virus. In silico analysis has identified some structural components as potential therapeutic targets [10], [11], [12]. Papain-like protease (known as protease Mpro), spike protein and RNA-dependent RNA polymerase (RdRp) are identified as the most promising targets so for [13], [14], [15].
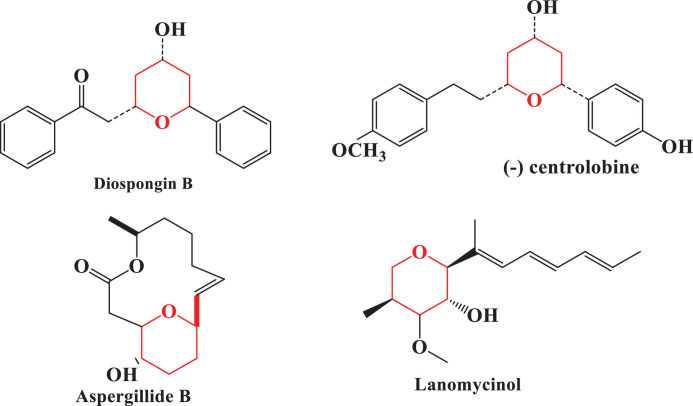
Most of the commercial drugs have N, S and O containing heterocyclic moieties and they are most sought pharmacophores for designing new and efficient drugs. Among the medicinally important heterocyclic compounds, substituted tetrahydropyranone moiety is present in a vast number of bioactive natural compounds, and its synthesis has attracted a lot of interest recently [16], [17], [18], [19]. Some of the most commonly used pyranone drugs in medicine are diospongin B (anti-osteoporotic) [20], lanomycinol (antifungal) [21], aspergillide B (anti-cytotoxic) [22] and centrolobine (anti-leishmanial) [23] (Fig. 1 ). Similarly tetrahyidropyran-4-one and its derivatives are used as antibacterial, antifungal, antituberculosis, analgesic, anti-inflammatory, antitumor, and COX-1/−2 inhibitors [24], [25], [26], [27], [28], [29].
Fig. 1.
Tetrahydropyran containing natural products as drug.
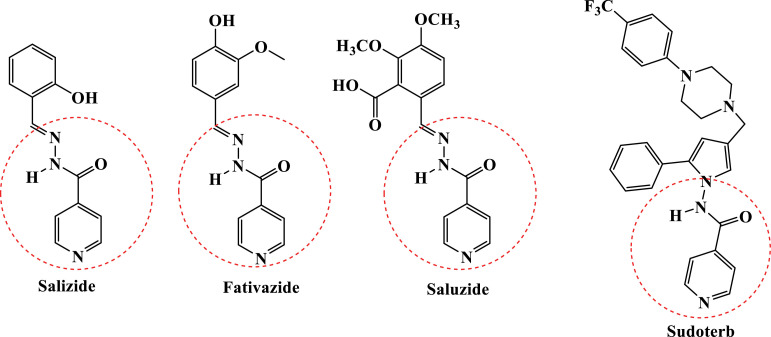
Isoniazid is used as one of the most effective and prime drugs in the treatment of tuberculosis. Numerous active antitubercular compounds containing isoniazid nuclei are commonly found in the literature [30], [31], [32], [33]. Such antitubercular agents either contain carbohydrazide or a direct binder to the ring. Saluzide, Ftivazide, and Sudoterb (Fig. 2 ) are promising tuberculosis drugs that contain an isoniazid moiety [34].
Fig. 2.
Chemical structures of INH derivatives as drug.
Similarly, 1,2,3-triazoles have gained significant attention in the medicinal field owing to their unique and versatile nature. Rufinamide (anticonvulsant drug), cefatrizine (antibiotic) and carboxyamidotriazole (anticancer drug) are 1,2,3-triazole containing drug molecules available in markets, [35], and have fascinating biological activities such as antibacterial [36], antifungal [37], anticancer [38], antitubercular [39], antimalarial [40] larvicidal [41] and anti-inflammatory [42]. On the other hand, isoxazoles are an important class of scaffolds because its widely present in biologically active natural products and have gained a considerable amount of attention due to their notable broad-spectrum of biological activities such as antimicrobial [43], anticancer [44], anti‐HIV [45] antituberculosis [46] and anti‐inflammatory [47].
According to our observations and ongoing research on new antimycobacterial agents [48,49], new compounds hydrazone, oxime ether, 1,2,3-triazole, and isoxazole hybridized with the pyranone system are thought to be worth synthesizing in order to produce promising antitubercular and anti-SARS CoV-2 agents. In the present work, we synthesize a new series of various tetrahydropyranone scaffolds containing hydrazones, oxime ether, triazole, and isoxazole to evaluate their antimicrobial, antitubercular, in silico SARS-CoV-2, and DNA gyrase inhibition activities. Cytotoxicity assay, drug score, and bioactivity (pharmacokinetic) parameters were also performed with the assistance of SwissADME and pkCSM-pharmacokinetic web tools.
2. Results and discussion
2.1. Chemistry
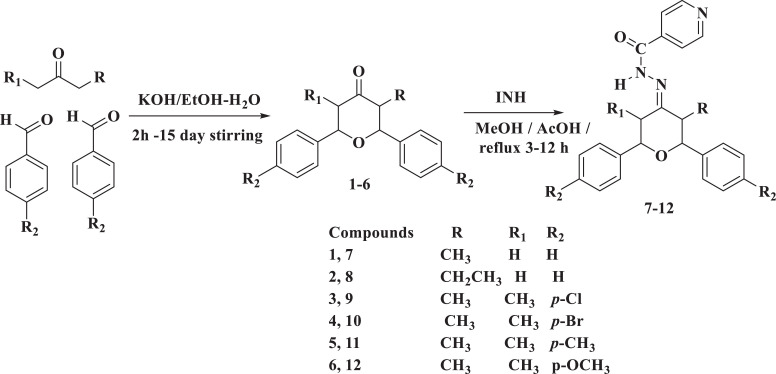
The synthesis of target hydrazone molecules (7–12) is illustrated in Scheme 1 . The reaction was initiated by the synthesis of 2,6-diarytetrahydropyran-4-ones (1–6) according to the Maitland–Japp procedure [50]. The starting molecule 3-alkyl-2,6-diaryltetrahydropyran-4-ones (1 and 2) were synthesized by cyclization reaction of benzaldehyde with 2-butanone (for compound 1) in presence of KOH as a base in ethanol-water mixture stirred for 48-50 hrs [50]. The other one of the starting compounds, 3,5-dimethyl-2,6-diaryltetrahydropyran-4-ones (3–6), was also synthesized by cyclization of substituted benzaldehyde with 3-pentanon in presence of KOH, stirred for 2–96 hrs [50].
Scheme 1.
Synthesis of target hydrazone compounds (7–12).
The target hydrazones (7–12) were synthesized by condensing INH with the 2,6-diaryltetrahydropyran-4-ones (1–6) in the presence of 0.5 mL of acetic acid. The reaction was carried out by refluxing in methanol, and the target hydrazones (7–12) were obtained according to the previously reported method [51].
Meanwhile, a series of 1,2,3-triazol derivatives (25–30) were synthesized as click reaction, reported in the literature [52]. In the first step, 2,6-diaryltetrahydro-4H-pyran-4-ones (1–6) were converted into oximes (13–18) by treating them with hydroxylamine hydrochloride in methanol using sodium acetate anhydrous in catalytic amount, under reflux for 2-12 h. In the second step, the oximes were converted to their corresponding O-ethynyl oximes (19–24) by treating with propargyl bromide and sodium hydride in THF yield compounds (19–24).
Finally, oxime ether (25–30) were synthesized by the reaction of benzyl azide with pyranone oxime O-propargyl ether by Cu(II)-catalyzed Huisgen-Sharpless-Meldal[3 + 2] dipolar azide-alkyne cycloaddition (CuAAC) reaction. Similarly, the isoxazole derivatives (31–36) were synthesized in good yield by cycloaddition reaction of O-ethynyl oximes (19–24) with oxime in presence of sodium ascorbate and copper(II)sulfate pentahydrate (Scheme-2 ).
Scheme 2.
Synthesis of compounds (19–36).
The synthesized hybrid structures were assigned based on spectral data. In the FT-IR spectra of hydrazones 7–12 showed the characteristics absorption bands C O, C N and amide NH functions at their expected regions. The spectra of the target hydrazones exhibited C O band at 1665–1675 cm−1, band centered around 3100 cm−1 is attributed to the νN — H and the νC N groups of hydrazone linkage also display absorption band around 1536 cm−1. In the triazole compounds the band appeared at 3142 cm−1 is due to =C—H stretching frequency of triazole ring.
In the 1H NMR spectrum, the amide NH proton (NH—CO) appeared as singlet at δ =10.92 ppm with one proton integral. There are two doublets at δ = 4.23 ppm (J2a,3a= 10.5 Hz) and δ = 4.65 ppm (J5a,6a = 10.5 Hz) corresponding to benzylic protons of H2a and H6a in pyranone ring, a sextet at δ = 3.25 ppm corresponding to methylene proton at H-3. There is a doublet at δ = 3.91 ppm (J5a,5e = 12.8 Hz) and triplet at δ = 2.85 ppm corresponding to H5e and H5a proton. The pyridine H-α and H-β protons have appeared as doublet at δ= 8.69 and δ= 7.70 ppm respectively. The aromatic protons appear as multiplets in the region of δ = 7.20 – 7.33 ppm. From the observed large vicinal coupling constant indicate that the compounds 7 and 8 exist in chair conformation with phenyl group equatorial orientations at C2 and C6. In the case of 3,5-dimethyl substituted compounds (9–12) the vicinal coupling constant J6a ,5a = 2.0 Hz and diaxial coupling constant J2a ,3a = 10.0 Hz. These observations indicate that these compounds also adopt chair conformation. In the triazoles the methine proton of triazole C-4 appeared as a singlet at 7.22 ppm equivalent to one proton, which is confirmed the triazole formation. The appearance of the requisite number of absorptions in its 13C NMR spectrum further confirmed the assigned structure. This is further confirmed by its mass spectral data whereas a mass peak of 395.18 (m + 1) was observed which is in accordance with its molecular formula C24H23N3O2 of compound 7.
The 1,2,3-triazole 25–30 structures were unambiguously confirmed on the basis of their spectral data. In the IR spectra the oxime ether linkage of triazole (C N) stretching appeared at 1630–1640 cm−1. In the 1H NMR spectra the singlet appeared at 7.20–7.26 ppm assignment as the triazole H-4 proton. In the 13C NMR spectra the signals at δ 142.90 and δ 119.34 ppm attributed to C-4 and C5 carbons of the 1,2,3-triazole ring. All the remaining compounds had satisfactory spectral characteristic based on NMR spectra. Details of the NMR data are given in experimental section. Finally, the mass spectra were reported for all compounds and the peaks observed with mass values (M + H)+ confirmed the formation of the products. (Mass spectrum of selected compounds given in supplementary)
2.2. Ntimicrobial activities
All the target molecules were tested for their in vitro antibacterial and antifungal activities against S. aureus (MTCC-96), B. subtilis (MTCC-121), E. coli (MTCC-443), P. aeruginosa (MTCC-741), K. pneumonia (MTCC2272) and C. albicans, A.niger, Rhizopusspecies, A. flavus and the results were given the Table 1 . The primary antimicrobial assay was carried out using the agar diffusion assay method [53]. For bacterial and fungal activity Streptomycin and Amphotericin-B were used as standard drug respectively. The tested molecules showed a various degree of antimicrobial activity compared to the microorganisms tested.
Table 1.
In vitro anti-bacterial and anti-fungal activities of 7–12 & 19–36 in µg/mL.
| Com.ds | B. subtilius | S. aureus | K. pneumoniae | E. coli | P. aeruginosa | C. albicans | A. niger | A .flavus | Rhizopus sp |
|---|---|---|---|---|---|---|---|---|---|
| 7 | 12.5 | 25 | 12.5 | 6.3 | 12.5 | 25 | 50 | 12.5 | 25 |
| 8 | 6.3 | 50 | 12.5 | 6.3 | 25 | 25 | 25 | 25 | 12.5 |
| 9 | 12.5 | 25 | 50 | 12.5 | 25 | 100 | 25 | 100 | 50 |
| 10 | 25 | 50 | 12.5 | 12.5 | 6.3 | 100 | 50 | 100 | 100 |
| 11 | 25 | 12.5 | 25 | 12.5 | 25 | 50 | 50 | 25 | 50 |
| 12 | 25 | 50 | 25 | 50 | 100 | 25 | 25 | 12.5 | 50 |
| 19 | 25 | 50 | 25 | 50 | 100 | 100 | 50 | 100 | 100 |
| 20 | 25 | 12.5 | 25 | 12.5 | 25 | 100 | 25 | 100 | 50 |
| 21 | 12.5 | 25 | 50 | 12.5 | 25 | 25 | 25 | 25 | 100 |
| 22 | 25 | 50 | 12.5 | 12.5 | 6.3 | 50 | 6.3 | 6.3 | 50 |
| 23 | 25 | 12.5 | 25 | 12.5 | 25 | 100 | 25 | 100 | 50 |
| 24 | 25 | 50 | 25 | 50 | 100 | 100 | 50 | 100 | 100 |
| 25 | 50 | 25 | 12.5 | 25 | 12.5 | 25 | 50 | 12.5 | 25 |
| 26 | 25 | 50 | 12.5 | 12.5 | 25 | 6.3 | 25 | 6.3 | 12.5 |
| 27 | 50 | 25 | 50 | 6.3 | 25 | 3.2 | 25 | 25 | 3.2 |
| 28 | 12.5 | 25 | 12.5 | 6.3 | 6.3 | 25 | 6.3 | 6.3 | 3.2 |
| 29 | 50 | 12.5 | 25 | 12.5 | 25 | 25 | 12.5 | 100 | 25 |
| 30 | 25 | 50 | 25 | 50 | 100 | 25 | 12.5 | 100 | 50 |
| 31 | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 25 | 50 | 12.5 | 25 |
| 32 | 12.5 | 50 | 12.5 | 25 | 25 | 25 | 25 | 6.3 | 12.5 |
| 33 | 6.3 | 6.3 | 50 | 25 | 25 | 100 | 25 | 12.5 | 100 |
| 34 | 6.3 | 12.5 | 12.5 | 12.5 | 6.3 | 100 | 50 | 50 | 100 |
| 35 | 12.5 | 12.5 | 25 | 25 | 25 | 25 | 25 | 25 | 50 |
| 36 | 50 | 50 | 25 | 25 | 100 | 25 | 6.3 | 6.3 | 50 |
| SDa | 25 | 50 | 50 | 12.5 | 25 | – | – | - | - |
| SDb | - | - | - | - | - | 25 | 50 | 50 | 25 |
SDa: Streptomycin; SDb: Amphotericin- B.
All the compounds showed their antibacterial activity varying from 6.25 – 50 μg/mL. Compounds 7 and 8 exhibited excellent inhibitory activity against E. coli with MIC of 6.3 μg/mL. The compounds 7, 8 and 10 exhibited good activity against K. pneumoniae bacterial strain with MIC of 12.5 μg/mL. The unsubstituted compounds of the phenyl group showed good activity (6.25 μg/mL) against E.coli and fair activity against S. aureus. Besides, the introduction of chloro and bromo substituent in the phenyl rings at the para position does not show any significant improvement in their activity.
All the oxime ethers reveled significant activity against S. aureus, especially compounds 20 and 23 with an MIC of 12.5 µg/mL, exhibited fourfold more activity than compared to the standard drug.
In triazoles, the compound 27 showed very good inhibitory activity against E. coli with the MIC of 6.3 µg/mL. In the case of K. pneumoniae, compounds 25, 26 and 28 exhibited potent activity which is four-fold more inhibitory activity than that of standard drug. Similarly, in the case E. coli compounds 27 and 28 found to two-fold more inhibitory activity than Streptomycin. Compound 28 showed superior antibacterial activity against all of the bacterial pathogens compared to other compounds. The presence of bromo group on the phenyl ring, play an important role in increasing the antibacterial activity of these molecule.
The isoxazole analogs (31–36) showed good antibacterial activity against the tested bacterial strains especially, for B. substilies, E. coli and P. aeruginasa. For bacteria B. substilius, it can be seen that the compounds 33 and 34 shows excellent inhibitory activity with MIC of 6.3 μg/mL, which is four-fold time higher potent than the standard drug. However, the compounds 31, 32 and 35 showed inhibitory activity with MIC value 12.5 μg/mL, which is two-fold higher potent than the standard drug. In the cause of B. subtilius all compounds (except 36) showed good inhibitory activity when compared to Streptomycin (Fig. S8).
Most the compounds exhibited better inhibition activity against B. subtilius than other bacteria's. Particularly, the isoxazole hybrids (31–36) exhibited potent antibacterial activity which is higher than that of the standard drug. At the same time, the antibacterial activity is reduced when the isoxazole hybrid is replaced with triazole or oxime ether.
The in vitro antifungal activity of the target hybrids was determined according the NCCL protocols and the obtained results are given in Table 1 with the MIC range from 3.2 to 100 μg/mL (Fig. S44 & 45). Antifungal assay showed that some of the compounds have excellent inhibition against the tested fungal strains compared to standard drug Amphotericin-B in the hydrazone analogues 11 and 12 showed potent activities against all the four tested fungal strains. It may be due to the presence of electron donating (OCH3) groups on the phenyl ring. In the triazole analogues, compounds 27 and 28 exhibited superior antifungal activity against the fungal pathogen such as Rhizopus sp. Compounds 29 and 30 showed good antifungal activities against the A. niger, and compound 28 exhibit good antifungal activities against all the fungal strains (except C. albicans). These facts may be explained by the presence of electrons withdrawing group on the phenyl ring.
In the oxazole analogues, compound 32 showed good antifungal activity against A.flavus and compound 36 showed very good antifungal activity against A.niger and A.flavus strains. It may be due to the presence of electrons donating groups on the phenyl ring. At the same time the other compounds were failed to show good inhibitory activity.
To determine the possible structure-activity relationship (Fig.S47) between antimicrobials, the inhibition effects and minimum inhibitory concentrations (MICs) of all investigated compounds against nine targeted microbial species were assessed.
-
(i)
In hydrazone analogues, the unsubstituted phenyl ring led to increased antibacterial and antifungal activities, due to the solubility of these compounds were high when compared to other compounds,
-
(ii)
In the oxime ether, triazole and isoxazole analogues, the electron withdrawing groups on the phenyl rings, led to increased antibacterial activity. The increased order of antibacterial activity was Br > Cl > OCH3 >CH3.
-
(iii)
In the isoxazole analogues, the electron donating groups on the phenyl ring, led to increased antifungal activity. The increased order of antifungal activity was OCH3 > CH3 > Cl > Br.
-
(iv)
Triazole moiety containing compounds (25–30) exhibited very good antifungal activity.
2.3. Mycobacterium tuberculosis activity
Since the compounds have shown good potent antibacterial capabilities, we expanded our investigation to assess their antitubercular efficacy. All the lead molecules were investigated for their in vitro antitubercular activity against the Mycobacterium tuberculosis H37Rv in a Resazurin Assay method [54].
The first-line antitubercular medicines, isoniazid and ethambutol, were used as reference drugs for the primary screening, at the concentration of 6.5 mg/mL. The molecules that had more than 90% inhibitions in primary screening were further investigated for secondary screening, that is, determination of MIC against M. tuberculosis H37Rv and the anti-tuberculosis activity of all the compounds was given in Table 2 .
Table 2.
In vitro anti-mycobacterial activity of the newly synthesized compounds (7–12 & 19–36) against M. tb H37Rv.
| Compound | MIC (µg/ml)a | % Inhibition | IC50 (µg/mL) | SI | CLogPb | Docking Score (kcal/mol) |
|---|---|---|---|---|---|---|
| 7 | 6.25 | 73 | – | 4.09 | −9.5 | |
| 8 | 3.12 | 70 | 486.2 | 155.8 | 4.54 | −8.9 |
| 9 | 1.56 | 90 | 563.5 | 361.22 | 5.57 | −10.8 |
| 10 | 0.78 | 95 | 513.9 | 658.84 | 5.81 | −9.6 |
| 11 | 3.12 | 76 | 587.6 | 188.33 | 4.36 | −7.9 |
| 12 | 3.12 | 70 | 609.1 | 195.22 | 4.22 | −8.4 |
| 25 | 6.25 | 75 | – | – | 5.67 | −7.1 |
| 26 | 6.25 | 70 | – | – | 6.06 | −7.8 |
| 27 | 6.25 | 75 | – | – | 7.23 | −8.9 |
| 28 | 3.12 | 89 | 316.8 | 101.54 | 7.44 | −8.7 |
| 29 | 6.25 | 76 | – | – | 6.54 | −6.9 |
| 30 | 13.5 | 80 | – | – | 5.94 | −7.1 |
| 31 | 6.25 | 90 | – | – | 7.40 | −8.8 |
| 32 | 6.25 | 91 | – | – | 7.79 | −9.2 |
| 33 | 3.12 | 95 | 475.5 | 152.40 | 8.95 | −13.6 |
| 34 | 3.12 | 92 | 489.2 | 156.79 | 9.17 | −13.2 |
| 35 | 6.25 | 82 | – | – | 8.26 | −9.8 |
| 36 | 6.25 | 75 | – | – | 7.66 | −9.9 |
| INH | 0.1 | >95 | – | – | −0.67 | |
| PZA | 6.25 | >90 | – | – | −0.68 | |
| EMB | 1.56 | >95 | – | – | 0.12 |
cCompounds 19–24 doesn't show any Anti-mycobacterial activity.
MIC values tested at neutral pH.
ogP (lipophilicity) calculated using “Osiris DataWarrior” software.
The obtained MIC value reveals that most of the molecules were identified to be strong inhibitors of M. tuberculosis, particularly promising inhibition was seen in hydrazone hybrids with MIC values ≤ 6.25 g/mL. Example compound 10 has shown very good antitubercular activity at 0.78 µg/mL. Also, compound 9 was shown to be four times more active than PZA and as active as EMB. Compounds 8, 11, and 12 have good action that is twice as potent as PZA (MIC = 3.12 g/mL). Finally, compound 7 showed moderate activity with 6.25 µg/mL, which is still within PZA's range. In the 1,2,3-triazole group's compound 28 inhibits the growth of the M. tuberculosis H37Rv strain by 89% at 3.12 µg/mL. The obtained% inhibition showed that the 1,2,3-triazoles were found to be low active than the hydrazones against M. tuberculosis H37Rv. Interestingly, the isoxazole compounds revealed good to excellent inhibitory activity against M. tuberculosis at the concentration of 3.12 to 6.25 µg/mL. Compounds 33 and 34 showed >90% inhibition at 3.12 µg/mL and more active than 35 and 36. It indicates that the substituent's in the phenyl ring at position 4 have influences on the activity and the derivatives with electron withdrawing like chloro and bromo groups are more potential than the electron donating derivates (Br > Cl > OCH3 = CH3).
2.4. Nutrient starvation model of M. tuberculosis H37Rv
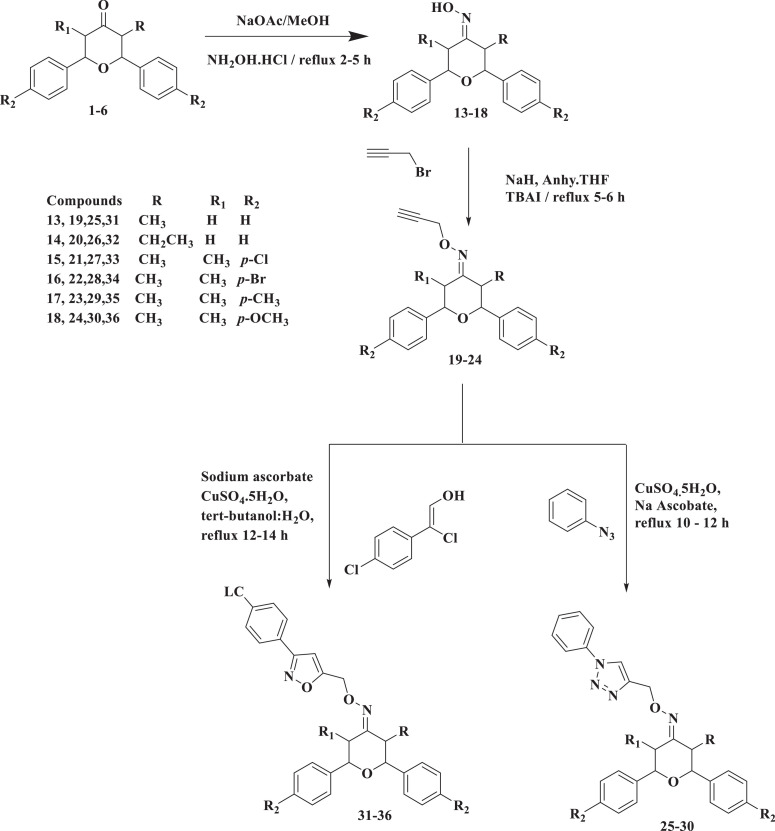
In the treatment of tuberculosis, the first line drugs are able to clear mycobacterium tuberculosis, but they failed to control the spread of the bacteria. It shows these first line drugs are not succeed to killing of inactive or persistent bacteria, which can undergo oxygen as supplement deprived condition. So, the M. tuberculosis bacterial cultures were grown for seven days in nutrient-rich media and then starved for six weeks in phosphate buffer saline (PBS). The most potent hybrids were treated with bacterial culture at concentrations of 1 and 10 mg/mL. The obtained results are given in Fig. 3 and revealed that the hybrids 9, 10, and 34 have good inhibition in their growth with a log reduction of 4.3, 3.4, and 2.6, respectively, at 10 mg/mL. When compared to other hybrids, the new isoxazole hybrid significantly inhibited M. tuberculosis growth during nutrient deprivation.
Fig. 3.
Activity profile of the most active compounds against M. tuberculosis in the Nutrient starvation method.
2.5. DNA gyrase inhibition activity
Further, the most active compounds 9, 10, 33 and 34 were assayed for their ability of bacterial / M. tuberculosis DNA gyrase inhibition by DNA supercoiling and ATPase gyrase inhibition methods (Table 3 ). DNA gyrase is an II topoisomerase in the mycobacterium tuberculosis genome and it is an extremely attractive feature in drug detection, making the enzyme more liable to inhibition. DNA gyrase has two territory units, Gyr-A provides the notching and resealing of the DNA and Gyr-B promotes ATP hydrolysis which helps with DNA supercoiling.
Table 3.
In vitro DNA gyrase assays of compounds 9, 10, 33 and 34.
| Compounds |
E.coli DNA gyrase (µM) |
M. Tb DNA gyrase (µM) |
||
|---|---|---|---|---|
| Supercoiling assay | ATPase assay | Supercoiling assay | ATPase assay | |
| 9 | 15.43 | 4.76 | 6.25 | 5.16 |
| 10 | 16.15 | 4.98 | 6.25 | 5.76 |
| 33 | 10.94 | 4.12 | 18.67 | 9.87 |
| 34 | 12.98 | 5.87 | 19.25 | 10.53 |
| Ciprofloxacin | 14.8 | 5.77 | ||
| Novobiocin | 3.98 | 7.92 | ||
The E.coli DNA gyrase supercoiling and ATPase activity revealed that all the four hybrids have good reduction of both DNA gyrase supercoiling and DNA gyrase ATPase activity. Compound 33 exhibited excellent DNA gyrase supercoiling inhibitory activity (IC50 = 10.94 μM) than the reference drug ciprofloxacin (IC50 = 14.80 μM), and compounds 9, 10 and 34 showed comparable activity with reference drug. The ATPase inhibitory activity of compound 33 (IC50 = 4.12 μM) and novobiocin were very close and the remaining hybrids had lower than the standard drug.
On other hand, the chloro and bromo substituted hydrazones 9 and 10 were favorable for the effective 67.3% and 50.7% M. tuberculosis DNA gyrase supercoiling inhibition at 6.25 μg/mL and very poor inhibition of the remaining compounds. Obviously, bacterial DNA gyrases were more sensitive to the isoxazole derivatives 33 and 34 than hydrazone derivatives. On another hand, hydrazone derivatives 9 and 10 were more active to M. tuberculosis DNA gyrase than isoxazoles.
2.5.1. In silico study of M. tuberculosis DNA gyrase
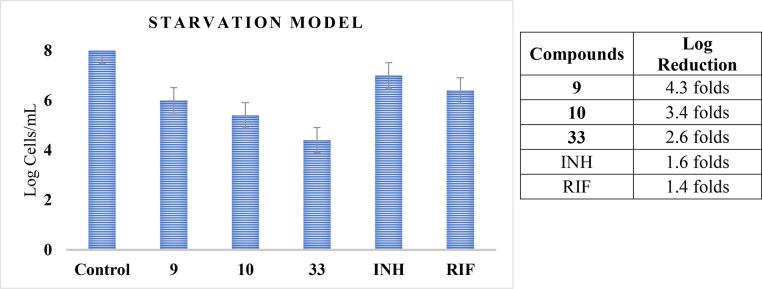
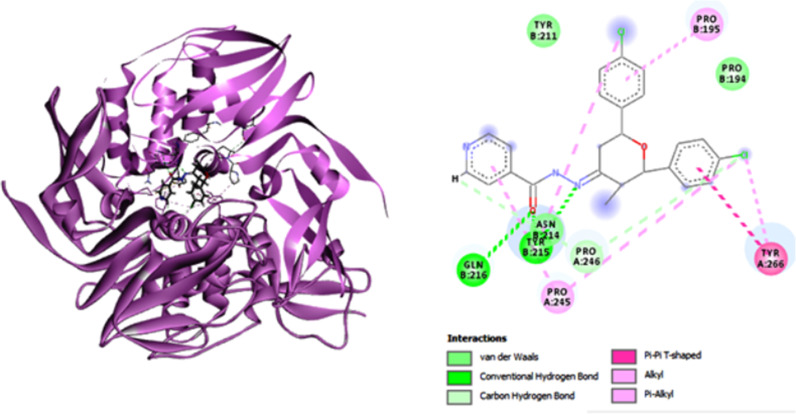
The interactions between the most active compound 9 and M. tuberculosis DNA gyrase were investigated by molecular docking. The X-ray crystallographic structure of InhA bound with novobiocin was obtained from the protein Data Bank. The predicted binding mode of 9 in the M. tuberculosis DNA gyrase active site is shown in Fig. 4 . (Supplementary Fig. S48,49). The pyridine (Isoniazid unit) ring shows weak van der Waals interactions with the neighboring ALA214. In addition, other favorable interaction associates are detected between 9 molecule GyrB residues PHE290, VAL339, ALA238 and LYS255. These favorable interactions produce in a good affinity of 9, with GyrB.
Fig. 4.
Molecular docking of compound 9 with M. tuberculosis DNA gyrase active site.
2.6. Cytotoxicity study
The compounds showing good anti-tubercular activity (8–12, 28, 33 and 34) with MIC values ≤ 6.25 μg/mL were further screened for their cytotoxicity assay against peripheral blood mononuclear cells (PBMC) by MTT assay [55]. Its main application is to assess the cell counting (viability) and the proliferation of cells. It can also be used to determine the cytotoxicity of medicinal agents and toxic materials because those agents would stimulate or inhibit cell viability and growth [56]. Table 2 shows the cytotoxicity results of the tested compounds expressed as inhibitory values (IC50 in μg/mL). IC50 values for the tested compounds varied from 316.8 to 609.1 μg/mL, which revealed they had very low cytotoxicity. Also, it showed that M. tuberculosis is more specifically targeted by the active compounds than healthy human cells.
2.7. Selectivity index
Selectivity Index (SI) is a ratio of IC50/MIC. The molecules posses a higher SI, that the molecules can be used as a therapeutic agent [57]. In our study, compounds 9 and 10 had a high SI value, which indicates they would be good inhibitors of M. tuberculosis and may be useful for the growth of new and effective antitubercular drug and must be assessed further.
2.8. Molecular docking
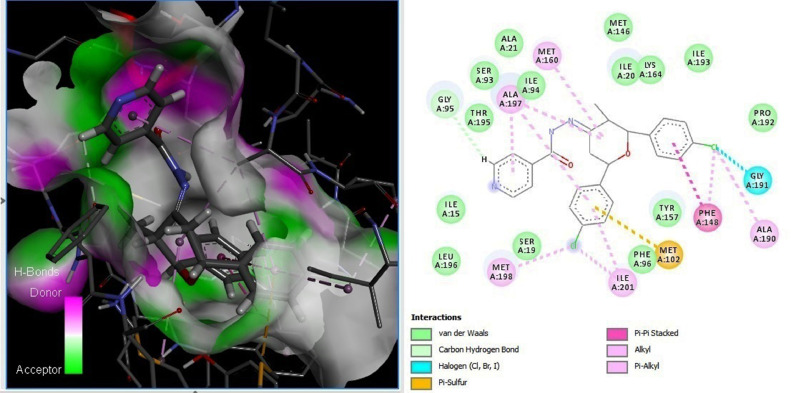
Structure-based molecular docking is one of the effective methods in the drug discovery process that produce the conformations of small compounds in protein binding sites and score the assumed protein-ligand complexes based on their binding affinities. The main advantage of docking was to focus on specific core protein-ligand interaction. The X-ray crystallographic structure of InhA (PDB ID: 3FNG) bound with triclosan was obtained from the Protein Data Bank (Catalyst, Version 4.10) at a resolution of 1.97A was used or the docking calculations. Discovery studio v2.5 (DS, Accelrys Inc., San Diego, CA, USA)/ Ligand Fit module was used to find the accurate orientation of ligands into protein active sites. All the target molecules and the co-crystals molecules were docked in the active site of the InhA. Top ten poses were generated for each ligand and the best conformer of each of the ligand-receptor complexes was selected based on the critical interactions and dock score. Before analyzing the molecular docking result of the synthesized compound, we validated the docking procedure by docking the triclosan molecules into the active site of the protein InhA. (The pose which have a good dock score and shown all the critical interactions with the active amino acids was superimposed with the bound triclosan it shows RMSD value of 0.7 Å (Fig. S51). Depending on the above result we can confirm that our docking procedure was able to reproduce the suitable binding orientation of the ligand which can inhibit the InhA activity. Hence dock score was used to sort out the good inhibitors from our synthesized compounds.
The compounds 10, 33 and 34 have the highest dock score value of more than −10.0 kcal/mol, all other molecules have the docking score less than −9.0 kcal/mol. DS was used to visualize the favorable interactions of these molecules with critical residues present in the active site of InhA. These molecules show good hydrogen bond interaction with Try158 or Met98 and hydrophobic interactions with NAD, Gly96, Phe96, Ala197, Met198, Pro156, Ile215 and Leu218 (Fig. S52). On the other hand, the other compounds showed weak inhibitory activity failed to show the necessary interactions with the critical residues. Fig. 5 shows that compound 10 establishes hydrogen bonds with the critical amino acids like Met98 and Phe97 and other compounds also shows same like interaction with the Met98 and NAD. It should be pointed out that the co-crystal structure of InhA also shows an important hydrogen bond with the residue Met98 and Phe97 in the same ligand-binding domain.
Fig. 5.
Hydrogen bond interaction with Try158 and hydrophobic interactions with NAD, Gly96, Phe97, Ala157, Met155, Pro156, Ile215 and Leu218 of compound 10.
2.9. Target predictions
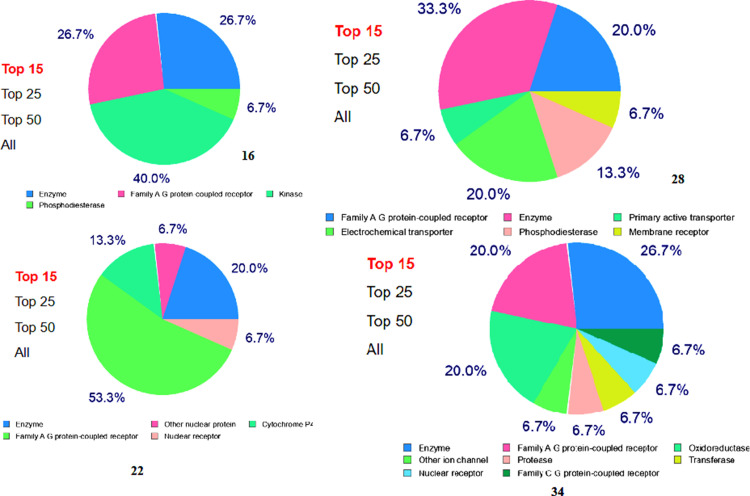
The bio-/Chem informatics approaches can be used to establish the most probable targets and efficient of the molecules. The most probable protein targets of the hybrids could predict by the SwissTarget online web tool. All the hybrids were predicted for their probable protein targets. All the chloro and bromo substituted hybrids have high percentage of probability binding with family A-G protein coupled receptor and electrochemical transporter. In same time the oxime ether hybrids have the highest (>50%) binding with of family A-G protein coupled receptor alone. In the case of triazole and isoxazole hybrids, the isoxazole hybrid showed the highest (>25%) probability of binding with enzyme receptors (Fig. 6 ).
Fig. 6.
Target prediction of compounds.
3. Molecular docking SARS-CoV-2
The molecular docking study of newly synthesized hybrids with the Papain like protease (PDB: 3MJ5) was carried out (Fig. 7 ) and all the compounds exhibit negative binding energy, which shows that the hybrids interact well with the PLpro active site. In fact, the binding energy of all hybrids was greater than −7.5 kcal/mol, particularly the isoxazole compounds have an outstanding binding energy when compared to other hybrids. The active compound (33) has π-anion interaction with ASP111 amino acid, hydrogen bond interaction with LYS104 and also π-σ interaction with ILE220 amino acid. The binding energy of the standard drug Remdesivir has −9.4 kcal/mole. From the observed results suggested that hybrid 33 have favor interactions with Papain-like protease than Remdesivir.
Fig. 7.
The binding mode of compound triazole with SARS-CoV2 (3mj5).
4. Physicochemical properties, ADMET and bioactivity
A drug molecule's success is determined not only by its strong potential but also by ensuring the drug-ability (ADMET profile) of the drug molecule. For the vital achievement or breakdown of a probable drug discovery process, ADME, as well as toxicological profile and drug-likeness, are significantly important. In the early stages, filtering drug development by their absorption, distribution, metabolism, and excretion - toxicity properties will reduce the adverse effects in clinical trials or even in animal models. Therefore, in the measurement of in silico ADMET, compound effectiveness and drug suitability can be improved in parallel, which is expected not only to increase the overall quality of drug molecules and their performance but also to minimize overall costs.
In the present work, all 24 compounds (7–12, 19–24, 25–30 and 31–36) were subject to the prediction of physicochemical properties by swissADME, pre-ADMET and OSIRIS, [58], to evaluate the overall potential of these compounds to be suitable for a drug, as well as comparing them to anti-tubercular drugs and the results are given in Table 4 . The absorption property of synthesized molecules was calculated and the water solubility (logSw) of all compounds was found to range between −4.8 to −6.59. Compounds having a high negative value on the logSw scale are assumed to be poorly soluble. LogPo/w is strongly associated with transport processes, including membrane permeability, distribution to various tissues and organs [59]. The predicted values of logPo/w for triazole and isoxazole compounds ranged from 5.94 to 9.17. The predicted human intestine absorption percentage of all the new hybrids was found in the range of 91.83 to 97.68% and to be sufficient.
Table 4.
In silico physicochemical properties of the compounds 7–12 & 19–36.
| Pharmacokinetics |
Drug Likeness |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Com. | aM.Wt | bLog Po/w | cLog S | dHBA | eHBD | fGI | gTPSA | hLipinski's | iNROTBh | Bio-activeness | PAINS | Veber | Ghose |
| 7 | 385.466 | 4.0891 | −4.247 | 5 | 1 | High | 63.58 | 0 | 5 | 0.55 | 0 | 0 | 0 |
| 8 | 399.493 | 4.5435 | −4.517 | 5 | 1 | High | 63.58 | 0 | 6 | 0.55 | 0 | 0 | 0 |
| 9 | 468.383 | 5.5704 | −5.879 | 5 | 1 | High | 63.58 | 0 | 5 | 0.55 | 0 | 0 | 1 |
| 10 | 557.285 | 5.8088 | −6.075 | 5 | 1 | High | 63.58 | 2 | 5 | 0.17 | 0 | 0 | 3 |
| 11 | 399.493 | 4.3584 | −4.407 | 5 | 1 | High | 63.58 | 0 | 5 | 0.55 | 0 | 0 | 0 |
| 12 | 459.544 | 4.2184 | −4.443 | 5 | 1 | High | 82.04 | 0 | 7 | 0.55 | 0 | 0 | 1 |
| 19 | 305.376 | 4.4259 | −3.345 | 3 | 0 | High | 30.82 | 0 | 3 | 0.55 | 0 | 0 | 0 |
| 20 | 319.403 | 4.8803 | −3.615 | 3 | 0 | High | 30.82 | 0 | 4 | 0.55 | 0 | 0 | 0 |
| 21 | 388.293 | 5.9072 | −4.977 | 3 | 0 | High | 30.82 | 1 | 3 | 0.55 | 0 | 0 | 0 |
| 22 | 477.195 | 6.1456 | −5.173 | 3 | 0 | High | 30.82 | 1 | 3 | 0.55 | 0 | 0 | 1 |
| 23 | 347.457 | 5.383 | −4.193 | 3 | 0 | High | 30.82 | 0 | 3 | 0.55 | 0 | 0 | 0 |
| 24 | 379.455 | 4.5552 | −3.541 | 5 | 0 | High | 40.28 | 0 | 5 | 0.55 | 0 | 0 | 0 |
| 25 | 438.529 | 4.5925 | −4.711 | 6 | 0 | High | 61.53 | 0 | 6 | 0.55 | 0 | 0 | 0 |
| 26 | 452.556 | 5.0469 | −4.981 | 6 | 0 | High | 61.53 | 0 | 7 | 0.55 | 0 | 0 | 0 |
| 27 | 521.446 | 6.0738 | −6.343 | 6 | 0 | High | 61.53 | 2 | 6 | 0.55 | 0 | 0 | 3 |
| 28 | 610.348 | 6.3122 | −6.539 | 6 | 0 | High | 61.53 | 2 | 6 | 0.55 | 0 | 0 | 3 |
| 29 | 480.61 | 5.5496 | −5.559 | 6 | 0 | High | 61.53 | 1 | 6 | 0.55 | 0 | 0 | 3 |
| 30 | 512.608 | 4.7218 | −4.907 | 8 | 0 | High | 61.53 | 1 | 8 | 0.55 | 0 | 0 | 2 |
| 31 | 472.971 | 6.5446 | −6.598 | 5 | 0 | High | 56.85 | 1 | 6 | 0.55 | 0 | 0 | 2 |
| 32 | 486.997 | 6.999 | −6.868 | 5 | 0 | Low | 56.85 | 1 | 7 | 0.17 | 0 | 0 | 3 |
| 33 | 555.888 | 8.0259 | −8.23 | 5 | 0 | Low | 56.85 | 2 | 6 | 0.55 | 0 | 0 | 3 |
| 34 | 644.79 | 8.2643 | −8.426 | 5 | 0 | Low | 56.85 | 2 | 6 | 0.55 | 0 | 0 | 3 |
| 35 | 515.051 | 7.5017 | −7.446 | 5 | 0 | Low | 56.85 | 2 | 6 | 0.55 | 0 | 0 | 3 |
| 36 | 547.049 | 6.6739 | −6.794 | 7 | 0 | Low | 75.31 | 1 | 8 | 0.55 | 0 | 0 | 1 |
M.Wt: Molecular weight..
LogP: Logarithm of compound partition coefficient between n-octanol and water.
S: Compound's solubility in mg/L..
HBA: Number of hydrogen bond acceptors..
HBD: Number of hydrogen bond donors.
Human gastrointestinal absorption.
TPSA: Topological polar surface area..
Lipinski's rule of five.
NROTB: Number of rotatable bonds.
Measurements of plasma binding protein (PBP) and the blood brain barrier (BBB) were performed to forecast the transport of drugs from one organ to another. Results demonstrated that all derivatives with chloro and bromo substitutes had significant plasm protein binding values, implying longer half-lives than isoniazid (1.67). The brain penetration of compounds 25 (0.31) and 31 (0.30) was also minimal (BBB; unbound brain to plasma ratio). All twenty-four compounds were found to not inhibit the CYP2D6 enzyme, despite the fact that they all likely inhibit CYP2C19, CYP2C9, and CYP3A4. The fraction unbound values for hydrazones (7–12) were zero, whereas those for the remaining compounds ranged from 0.012 to 0.015.
The liver plays a very important responsibility in energy exchange and the biotransformation of xenobiotics and drugs. All the synthesized compounds were predicted for their LD50 and hepatotoxicity using pkCSM-pharmacokinetics [60]. The obtained LD50 values ranged from 0.64 to 2.01 mol/kg and the compounds hydrazones and triazoles could cause hepatotoxicity.
Pharmacokinetic and drug-likeness properties were predicted by SwissADME and given in Table 4. By taking a closer look to the results isoxazole compounds 32–36 has low Gastro intestinal absorption when compared to other derivatives. The drug-likeness was predicted based on the Lipinski [61], Ghose [62], Veber [63] and bioavailability score.
The Lipinski Rule of Five showed that there were only eleven compounds that accomplished the criteria of druglikeness assessment. However, most of the triazole (27–30) and isoxazole (31–36) compounds were rejected with two violations (Table 4). Based on the Ghose rules predictions showed that compounds 7, 8, 11, 19, 20, 21, 23, 24, 25 and 26 were satisfied the criteria of drug likeness assessment however the other compounds were rejected with one, three or four violations (Table 4). On the other hand, the screening process with Veber rules, all the compounds satisfied the criteria of drug likeness assessment. However, the (highest) molecular weight of the title compounds were > 500 and according to the literature, even it is higher than 500, it is not very high and acceptable for drug-likeness.61
The ADME properties of the hybrids were calculated by Pre-ADMET calculator method. The parameters such as coefficient of permeability of the Madin-Darby canine kidney (MDCK) cells, coefficient of permeability of the Caco2 cells (permeability by cells derived from adenocarcinoma of the human colon) were calculated and compared with the best values. The obtained results were given in Table 5 .
Table 5.
In silico ADME profiling of the compounds 7–12.
| Absorption |
Distribution |
|||||
|---|---|---|---|---|---|---|
| Compound | Caco2a | MDCKb | HIAc | logkpd | BBBe | PPBf |
| 7 | 39.657 | 27.810 | 96.317 | −6.140 | 0.117 | 91.882 |
| 8 | 43.210 | 20.193 | 96.411 | −5.970 | 0.080 | 93.250 |
| 9 | 36.536 | 0.233 | 97.188 | −5.340 | 0.334 | 100.000 |
| 10 | 36.718 | 0.034 | 97.493 | −5.800 | 0.413 | 100.000 |
| 11 | 47.120 | 0.418 | 96.600 | −5.470 | 0.219 | 91.582 |
| 12 | 49.568 | 0.669 | 96.426 | −6.230 | 0.034 | 90.824 |
Caco2: Permeability through cells derived from human colon adenocarcinoma; Caco2 values < 4 nm/sec (low permeability), values from 4 to 70 nm/sec (medium permeability) and values > 70 nm/sec (High permeability).
MDCK: Permeability through Madin–Darby canine kidney cells; MDCK values < 25 nm/sec (low permeability), values from 25 to 500 nm/sec (medium permeability) and values > 500 nm/sec (High permeability).
HIA: Percentage human intestinal absorption; HIA values from 0 to 20% (poorly absorbed), values from 20 to 70% (moderately absorbed) and values from 70 to 100% (well absorbed). LogkP: Skin permeability (cm/hour).
BBB: Blood–brain barrier penetration; BBB values < 0.1 (low CNS penetration), values from 0.1 to 2 (medium CNS absorption) and values > 2 (High CNS absorption).
PPB: Plasma protein binding; PPB values < 90% (poorly bound) and >90% (strongly bound).
From the ADME results, all tested compounds demonstrated medium cell permeability with values from 36.53 to 49.568 nm/s in the Caco2 cell model. In the MDCK cell model, 3,5-dimethyl substituted pyranone compounds have low cell permeability, while compounds 7 and 8 displayed medium permeability. Besides, the predicted oral bioavailability of all tested compounds were excellent, exercised high HIA values from 96.31 to 97.49% demonstrating very well absorbed. In addition, all the compounds showed medium Blood-brain barrier penetration capability (0.11–0.41) except compounds 8 and 12 (0.080 and 0.034). Also, plasma proteins were weakly bound with the compounds. The CYP2D6 enzyme is a reason for metabolism as well as the excretion of about 25% of clinically used drugs. No inhibition of this enzyme was formed by any of the tested compounds, so they can be metabolized and efficiently excreted.
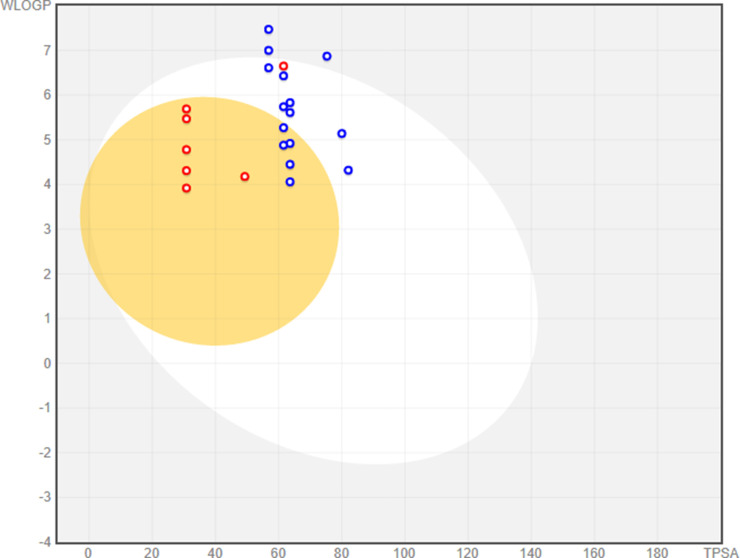
The performance results of the SwissADME prediction are the 2D chemical structure of the radar compound and bioavailability which gives a quick conclusion on the drug-likeness (Fig. S19). The six parameters mentioned in the radar on bioavailability are flexibility, lipophilicity, thickness, polarity, solubility and saturation and their essential limits (Table 6 ). The boiled-egg Fig. 8 demonstrates that the compounds are within the appropriate range for standard. Compounds 7, 8 and 11 present in the yellow zone that permeates through the blood-brain barrier (BBB). The gastrointestinal tract can very quickly absorb the remaining compounds (9, 10 and 12) that are present in the white region and same time the isoxazol hybrids 32, 35 and 36 are present in the out of region.
Table 6.
Toxicity and drug-likeness scores of the compounds 7–12 & 19–36.
| Compounds | Drug likeness | Toxicity |
Shape Index | Molecular Flexibility | Molecular Complexity | |||
|---|---|---|---|---|---|---|---|---|
| Mutagenic | Tumorigenic | Reproductive Effective | Irritant | |||||
| 7 | 3.98 | None | High | High | None | 0.48 | 0.31 | 0.81 |
| 8 | 3.76 | None | High | High | None | 0.47 | 0.31 | 0.82 |
| 9 | 1.99 | None | High | High | None | 0.47 | 0.31 | 0.87 |
| 10 | 3.83 | None | High | High | None | 0.47 | 0.31 | 0.87 |
| 11 | 3.78 | None | High | High | None | 0.47 | 0.31 | 0.85 |
| 12 | 3.83 | None | High | High | None | 0.47 | 0.33 | 0.86 |
| 19 | 1.74 | None | None | None | None | 0.52 | 0.48 | 0.32 |
| 20 | 1.48 | None | None | None | None | 0.52 | 0.45 | 0.33 |
| 21 | 1.55 | None | None | None | None | 0.52 | 0.5 | 0.33 |
| 22 | −0.32 | None | None | None | None | 0.52 | 0.5 | 0.33 |
| 23 | 1.47 | None | None | None | None | 0.52 | 0.5 | 0.33 |
| 24 | 1.56 | None | None | None | None | 0.52 | 0.54 | 0.36 |
| 25 | −0.69 | None | None | None | None | 0.52 | 0.35 | 0.81 |
| 26 | −0.91 | None | None | None | None | 0.52 | 0.35 | 0.82 |
| 27 | −0.85 | None | None | None | None | 0.52 | 0.35 | 0.88 |
| 28 | −2.69 | None | None | None | None | 0.52 | 0.35 | 0.88 |
| 29 | −0.90 | None | None | None | None | 0.52 | 0.35 | 0.84 |
| 30 | −0.84 | None | None | None | None | 0.52 | 0.36 | 0.85 |
| 31 | 2.35 | None | None | None | None | 0.53 | 0.35 | 0.85 |
| 32 | 2.12 | None | None | None | None | 0.51 | 0.35 | 0.86 |
| 33 | 2.08 | None | None | None | None | 0.51 | 0.35 | 0.88 |
| 34 | 0.29 | None | None | None | None | 0.51 | 0.35 | 0.88 |
| 35 | 2.08 | None | None | None | None | 0.51 | 0.35 | 0.88 |
| 36 | 2.18 | None | None | None | None | 0.51 | 0.36 | 0.89 |
| INH | −5.78 | High | High | High | High | 0.7 | 0.48 | 0.55 |
| PZA | −0.68 | High | High | High | None | 0.67 | 0.04 | 0.72 |
Fig. 8.
ADME properties of compounds and standard drug by graphical representation (boiled-egg) (predict gastrointestinal absorption and brain penetration).
5. Conclusion
In conclusion, a series of 2r,6c-diaryltetrahydropyran-4-ones clubbed with four different (hydrazones 7–12, O-ethynyl oximes 19–24, triazoles 25–30, and isoxazoles 31–36) hybrids have successfully synthesized in good to excellent yield. The newly synthesized hybrid structures were confirmed by spectroscopic method. The synthesized hybrids were assayed for their antimicrobial and mycobacterium tuberculosis (MTB) activities. The results showed that the eight compounds (8, 9, 10, 11, 12, 28, 33 and 34) had good activity against M. tuberculosis at the concentration of ≤ 6.25 μg/ml, especially the hybrid 10 showed highest inhibition activity at 0.78 μg/ml. Moreover, the in vitro cytotoxicity of the most active compounds 8–12, 28, 33 and 34 against PBMC normal cell line by MTT assay method. The estimated IC50 values were in the range 316.8 to 609.1 µg/mL, demonstrating a reasonable safety profile and selectivity (SI 150), and none of the compounds shown cytotoxicity against PBMC. Further, docking studies were also performed on these promising derivatives based on the M. tuberculosis study compound 10 have remarkable M. tuberculosis activity. All the compounds showed better binding scores with papain-like protease (SARS-CoV-2). Especially isoxazole compound 33 has high binding energy (−9.4 kcal/mole) and favor interactions. Furthermore, ADMET and drug-like properties were evaluated, majority of the compounds predicted good drug like properties and can be developed as oral drug candidate.
6. Experimental
6.1. Synthesis of the 3-akyl-2,6-diaryltetrahydropyran-4-one N-isonicotinoylhydrazone (7–12)
3t-Alkyl-2r,6c-diaryltetrahydropyran-4-ones (1–6) (1.5 mol), isoniazid (2.0 mol) were dissolved in 50 mL of methanol and catalytic amount (1.0 mL) of acetic acid was added. The reaction mixture was heated under reflux for 2–12 hr, a solid mass was thrown out from the reaction mixture and filtered, washed with ethanol and water mixture. The crude product was recrystallized from ethanol.
6.1.1. 3-Methyl-2,6-diphenylpyran-4-one N-isonicotinoylhydrazone (7)
FT-IR (cm-1): 3038, 3026, 3004, 2986, 2942 (C—H stretching); 1668 (C O stretching); 1536 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.23 (d, J 2a,3a = 10.5 Hz, 1H, H2a), 4.65 (d, J 5a,6a = 10.5, Hz, 1H, H6a), 3.25 (sextet, 1H, H3a), 2.85 (t, 1H, H5a), 3.91 (d, J5a,5e = 12.8 Hz, 1H, H5e), 10.92 (s, 1H, CO—NH-), 0.89 (d, J3a ,Me = 6.0 Hz, 3H, -CH3 at C3), 8.69 (d, 2H, α-H), 7.70 (d, 2H, β-H), 7.46 (d, 4H, H-2″, H-6″), 7.20–7.33 (m, 6H, H-2″', H-6″',H-2′''', H-6′'''); 13C NMR (DMSO‑d6): (δ) ppm: 75.93 (C2), 63.92 (C6), 45.54 (C3), 38.61 (C5), 11.72 (CH3 at C3), 171.08 (C O), 158.48 (C N), 150, 140.95, 144.10, 143.45, 127.49, 127.19, 126.21, 126.04, 125.10, 125.01 (aryl carbons). Ana. Cal.(Found) for C24H23N3O2 (385.46) C, 74.78 (74.50); H, 6.01 (6.09); N, 10.90 (1075); Pale yellow powder, yield 90%, m.p: 219 –220 °C
6.1.2. 3-Ethyl-2,6-diphenylpyran-4-one N-isonicotinoylhydrazone (8)
FT-IR (cm-1): 3042, 3029, 2997, 2952, 2878, 2843 (C—H stretching); 1661 (C O stretching); 1542 (C N stretching); 3168 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.31 (d, J 2a,3a = 10.5 Hz, 1H, H2a), 4.52 (d, J 5a,6a = 10.5, Hz, 1H, H6a), 3.25 (m, 1H, H3a), 2.07 (d, J = 12.50 Hz, 1H, H5a), 3.08 (d, J5a,5e = 12.8 Hz, 1H, H5e), 1.66 (m, 1H, H of –CHH’ of C-7), 1.16 (m, H, H' of C-7), 0.78 (t, 3H, -CH3 at C-7), 11.07 (s, 1H, -CO— NH-), 8.75(d, 2H, α-H), 7.74 (d, 2H, β-H), 7.51 (m, 4H, H-2″, H-6″), 7.28–7.39 (m, 10H, aryl protons). 13C NMR (DMSO‑d6): (δ) ppm: 70.22 (C2), 60.24 (C6), 42.98 (C3), 37.17 (C5), 20.91 (-CH2—CH3) 20.51 (-CH2— CH 3), 166.29 (C O), 160.41 (C N), 151.04, 144.36, 143.47, 141.73, 128.70, 127.68, 128.52, 127.33, 127.14, 122.23 (aryl carbons). Anal. Cal.(Found) for C25H25N3O2, C, 75.16 (74.82); H, 6.31 (6.51); N, 10.52 (10.11). Pale yellow solid, yield 86%, m.p: 224 –225 °C
6.1.3. 3,5-Dimethyl-2,6-bis(p-chlorophenyl)pyran-4-one N-isonicotinoylhydrazone(9)
FT-IR (cm-1): 3038, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1669 (C O stretching); 1536 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.76 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.22 (d, J6 a,5e = 2.52 Hz; 1H, H6a), 2.76 (sextet, 1H, H3a), 2.12 (sextet, 1H, H5a), 1.09 (d, 3H, 5-CH3, J = 7.00 Hz); 0.95 (d, 3H, 3-CH3, J = 6.50 Hz); 8.76 (d, 2H, α-H), 7.76 (d, 2H, β-H), 7.49–7.38 (m, 8H, aryl protons), 10.96 (s, 1H, -CO— NH-). 13C NMR (DMSO‑d6): (δ) ppm: 70.12 (C2), 60.72 (C6), 45.25 (C3), 38.19 (C5), 167.47 (C O), 160.36 (C N), 11.29 and 11.20 (3-CH3 and 5-CH3); 150.25, 141.15, 139.87, 139.05, 129.47, 127.34, 126.98, 121.45 (aryl carbons). Mass (M + H): 468. White powder, yield 75%, m.p: 228 –229 °C.
6.1.4. 3,5-Dimethyl-2,6-bis(p-bromophenyl)pyran-4-one N-isonicotinoylhydrazone (10)
FT-IR (cm-1): 3048, 3028, 3010, 2986, 2942, 2898 (C—H stretching); 1671 (C O stretching); 1536 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.71 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.20 (d, J6 a,5e = 2.55 Hz; 1H, H6a), 2.73 (sextet, 1H, H3a), 2.14 (sextet, 1H, H5a), 1.10 (d, 3H, 5-CH3, J = 7.00 Hz); 0.95 (d, 3H, 3-CH3, J = 6.50 Hz); 8.67 (d, 2H, α-H), 7.64 (d, 2H, β-H), 7.41–7.08 (m, 8H, aryl protons), 11.01 (s, 1H, -CO— NH-). 13C NMR (DMSO‑d6): (δ) ppm: 70.92 (C2), 60.74 (C6), 45.29 (C3), 38.08 (C5), 168.43 (C O), 159.16 (C N), 11.31 and 11.22 (3-CH3 and 5-CH3); 151.32, 140.15, 139.07–124.98 (aryl carbons). Anal. Cal.(Found) for C25H23Br2N3O2, C, 53.88 (53.12); H, 4.16 (4.56); N, 7.54(7.23). White powder, yield 79%, m.p: 232 –233 °C.
6.1.5. 3,5-Dimethyl-2,6-bis(p-methylphenyl)pyran-4-one N-isonicotinoylhydrazone (11)
FT-IR (cm-1): 3039, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1670 (C O stretching); 1536 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.71 (d, J 2a,3a = 10.55 Hz, 1H, H2a), 4.19 (d, J6 a,5e = 2.46 Hz; 1H, H6a), 2.70 (sextet, 1H, H3a), 2.14 (sextet, 1H, H5a), 1.08 (d, 3H, 5-CH3, J = 6.80 Hz); 0.96 (d, 3H, 3-CH3, J = 6.55 Hz); 8.75 (d, 2H, α-H), 7.74 (d, 2H, β-H), 7.41–7.34 (m, 8H, aryl protons), 2.31 (s, 6H, p-CH3), 11.06 (s, 1H, -CO— NH-). 13C NMR (DMSO‑d6): (δ) ppm: 88.21 (C2), 79.02 (C6), 45.52 (C3), 38.12 (C5), 168.57 (C O), 160.36 (C N), 11.27 and 11.18 (3-CH3 and 5-CH3); 21.01 (p-CH3); 150.52, 142.02, 139.85, 139.13, 129.40, 128.94, 127.98, 123.4, 120.23 (aryl carbons). Anal. Cal.(Found) for C27H29N3O2 C, 75.85 (76.01); H, 6.84 (6.12); N, 9.83(9.02). Paleyellow solid, yield 70%, m.p: 215 –216 °C.
6.1.6. 3,5-Dimethyl-2,6-bis(p-methoxyphenyl)pyran-4-one N-isonicotinoylhydrazone (12)
FT-IR (cm-1): 3038, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1668 (C O stretching); 1536 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.74 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.24 (d, J6 a,5e = 2.40 Hz; 1H, H6a), 2.71 (sextet, 1H, H3a), 2.13 (sextet, 1H, H5a), 1.10 (d, 3H, 5-CH3, J = 7.00 Hz); 0.94 (d, 3H, 3-CH3, J = 6.46 Hz); 3.83 (s, 6H, pOCH3–); 8.71 (d, 2H, α-H), 7.72 (d, 2H, β-H), 6.91–7.53 (m, 8H, aryl protons), 11.03 (s, 1H, -CO— NH-). 13C NMR (DMSO‑d6): (δ) ppm: 87.83 (C2), 79.12 (C6), 46.05 (C3), 37.15 (C5), 169.45 (C O), 161.46 (C N), 11.72 and 11.64 (3-CH3 and 5-CH3); 55.32 (pOCH3–); 159.24, 151.02, 142.11, 138.87, 138.12, 129.47, 127.34, 126.98, 112.54 (aryl carbons). Anal. Cal.(Found) for C27H29N3O4 C, 70.57 (70.15); H, 6.36 (6.02); N, 9.14 (9.56). White powder, yield 72%, m.p: 219 –220 °C.
6.2. Synthesis of 2,6-diarylpyran-4-oxime (13–18)
The intermediate molecule of 2,6-diarylpyran-4-oximes were synthesized by a mixture of 0.015 mol of 2,6-diarylpyran-4-one and 0.015 mol of hydroxylamine hydrochloride were dissolved in 30 mL of ethanol and refluxed 30 min on a steam bath. TLC was used to monitor the reaction. After complete the reaction, the reaction mixture was allowed to cool room temperature and transferred into ice. A solid residue was obtained and filtered, washed with water. The final product was dried and recrystallized from ethanol.
6.3. Synthesis of 2,6-diaryltetrahydro-4H-pyran-4-one O-ethynyl oxime (19–24)
2,6-Diaryltetrahydro-4H-pyran-4-one oxime 2 (∼ 0.28 g) and sodium hydride (0.041 g) were dissolved in anhydrous THF (5 mL), at 0 °C and the reaction mixture was stirred for 40-50 min at room temperature. 0.12 mL of propargyl bromide, tetrabutylammonium iodide (TBAI) were added slowly in that reaction mixture at 0 °C and then heated under reflux for another 12 hr. The reaction mixture was quenched with ice. The organic layers was extracted with ethyl acetate and dried over Na2SO4. The extract filtered, and evaporated under reduced pressure to give the desired products.
6.3.1. 3-Methyl-2,6-diphenyltetrahydro-4H-pyran-4-one O-ethynyl oxime (19)
FT-IR (cm-1): 3048, 3032, 3014, 2976, 2940 (C—H stretching); 1526 (C N stretching); 3149 (O—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.20 (d, J 2a,3a = 10.2 Hz, 1H, H2a), 4.60 (d, J 5a,6a = 10.2, Hz, 1H, H6a), 3.23 (sextet, 1H, H3a), 2.82 (t, 1H, H5a), 3.93 (d, J5a,5e = 12.7 Hz, 1H, H5e), 3.67 (s, 1H, C CH), 0.87 (d, J3a ,Me = 6.1 Hz, 3H, -CH3 at C3), 7.46–7.19 (m, 10H, ArH); 13C NMR (DMSO‑d6): (δ) ppm: 85.90(C2), 76.90 (C6), 60.54 (C3), 48.60 (C5), 11.75 (CH3 at C3), 158.48 (C N), 82.9 (-C CH), 45.9 (-C CH) 140.94, 144.11, 143.45, 127.49, 127.19, 126.21, 126.04, 125.10 (aryl carbons). Yellow powder, yield 95%, m.p: 209 –210 °C
6.3.2. 3-Ethyl-2,6-diphenyltetrahydro-4H-pyran-4-one O-ethynyl oxime (20)
FT-IR (cm-1): 3046, 3030, 2987 (C—H stretching); 1545 (C N stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.33 (d, J 2a,3a = 10.5 Hz, 1H, H2a), 4.53 (d, J 5a,6a = 10.5, Hz, 1H, H6a), 3.26 (m, 1H, H3a), 2.10 (d, J = 12.50 Hz, 1H, H5a), 3.10(d, J5a,5e = 12.8 Hz, 1H, H5e), 1.68 (m, 1H, H of –CHH’ of C-7), 1.18 (m, H, H' of C-7), 0.80 (t, 3H, -CH3 at C-7), 3.69 (s, 1H, C CH), 7.28–7.39 (m, 10H, aryl protons). 13C NMR (DMSO‑d6): (δ) ppm: 87.22 (C2), 80.54 (C6), 69.98 (C3), 57.15 (C5), 18.95 (-CH2—CH3) 12.54 (-CH2— CH 3), 160.49 (C N), 82.12 (-C CH), 45.15 (-C CH), 151.04, 144.36, 143.47, 141.73, 128.70, 127.68, 128.52, 127.33, 127.14, 122.23 (aryl carbons). Pale yellow solid, yield 90%, m.p: 214 –215 °C
6.3.3. 3,5-Dimethyl-2,6-bis(p-chlorophenyl)pyran-4H-pyran-4-one O-ethynyl oxime (21)
FT-IR (cm-1): 3026, 3004, 2986 (C—H stretching); 1529 (C N stretching); 1H NMR (DMSO‑d6):(δ) ppm: 4.68 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.18 (d, J6 a,5e = 2.55 Hz; 1H, H6a), 3.69 (s, 1H, C CH), 2.71 (sextet, 1H, H3a), 2.15 (sextet, 1H, H5a), 1.10 (d, 3H, 5-CH3, J = 7.05 Hz); 0.91 (d, 3H, 3-CH3, J = 6.55 Hz); 7.47–7.38 (m, 8H, aryl protons). 13C NMR (DMSO‑d6): (δ) ppm: 88.12 (C2), 78.72 (C6), 45.25 (C3), 38.19 (C5), 160.36 (C N), 11.30 and 11.25 (3-CH3 and 5-CH3); 150.25, 141.15, 139.87, 139.05, 129.47, 127.34, 126.98, 121.45 (aryl carbons). White solidr, yield 87%, m.p: 240 –241 °C.
6.3.4. 3,5-Dimethyl-2,6-bis(p-bromophenyl) pyran-4H-pyran-4-one O-ethynyl oxime (22)
FT-IR (cm-1): 3048, 3028, 3010, 2986, 2942, 2898 (C—H stretching); 1536 (C N stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.71 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.20 (d, J6 a,5e = 2.55 Hz; 1H, H6a), 2.73 (sextet, 1H, H3a), 2.14 (sextet, 1H, H5a), 1.10 (d, 3H, 5-CH3, J = 7.00 Hz); 0.95 (d, 3H, 3-CH3, J = 6.50 Hz); 7.41–7.22 (m, 8H, aryl protons). 13C NMR (DMSO‑d6): (δ) ppm: 87.82 (C2), 78.84 (C6), 45.22 (C3), 38.08 (C5), 159.16 (C N), 82.19 (-C CH), 45.18 (-C CH), 11.33 and 11.25 (3-CH3 and 5-CH3); 151.32, 140.15, 139.07–124.98 (aryl carbons). Dirty white, yield 89%, m.p: 230 –231 °C.
6.3.5. 3,5-Dimethyl-2,6-bis(p-methylphenyl)pyran-4H-pyran-4-one O-ethynyl oxime (23)
FT-IR (cm-1): 3036, 3026, 3004, 2986, (C—H stretching); 1536 (C N stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.71 (d, J 2a,3a = 10.56 Hz, 1H, H2a), 4.20 (d, J6 a,5e = 2.49 Hz; 1H, H6a), 3.65 (s, 1H, C CH), 2.72 (sextet, 1H, H3a), 2.16 (sextet, 1H, H5a), 1.08 (d, 3H, 5-CH3, J = 6.83bHz); 0.96 (d, 3H, 3-CH3, J = 6.50 Hz), 7.45–7.35 (m, 8H, aryl protons), 2.33 (s, 6H, p-CH3). 13C NMR (DMSO‑d6): (δ) ppm: 88.22 (C2), 79.12 (C6), 45.52 (C3), 38.12 (C5), 162.21 (C N), 82.01 (-C CH), 45.04 (-C CH), 11.28 and 11.18 (3-CH3 and 5-CH3); 21.07 (p-CH3); 150.52, 142.02, 139.85, 139.13, 129.40, 128.94, 127.98, 123.4, 120.23 (aryl carbons). Paleyellow solid, yield 87%, m.p: 205 –206 °C.
6.3.6. 3,5-Dimethyl-2,6-bis(p-methoxyphenyl)pyran-4H-pyran-4-one O-ethynyl oxime (24)
FT-IR (cm-1): 3038, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1536 (C N stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.77 (d, J 2a,3a = 10.55 Hz, 1H, H2a), 4.29 (d, J6 a,5e = 2.42 Hz; 1H, H6a), 3.66 (s, 1H, C CH), 2.71 (sextet, 1H, H3a), 2.17 (sextet, 1H, H5a), 1.12 (d, 3H, 5-CH3, J = 7.02 Hz); 0.98 (d, 3H, 3-CH3, J = 6.42 Hz); 3.88 (s, 6H, pOCH3–); 6.91–7.53 (m, 8H, aryl protons), 13C NMR (DMSO‑d6): (δ) ppm: 87.85 (C2), 79.17 (C6), 46.05 (C3), 37.15 (C5), 161.46 (C N), 82.16 (-C CH), 45.18 (-C CH), 11.76 and 11.69 (3-CH3 and 5-CH3); 55.20 (pOCH3–); 159.24, 151.02, 142.11, 138.87, 138.12, 129.47, 127.34, 126.98, 112.54 (aryl carbons). Yello solid powder, yield 87%, m.p: 217 –218 °C.
6.4. Synthesis of 2,6-diaryltetrahydro-4H-pyran-4-one O-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)oxime (25–30)
2,6-Diaryltetrahydro-4H-pyran-4-one O-ethynyl oximes (19–24) (1.5 mmol), 1-(azidomethyl)benzene (1.5 mmol), 0.09 mmol of CuSO4·5H2O and sodium ascorbate (0.2 mmol) were added in to 30 mL of water and THF (1:1) solution mixture. The solution mixture was stirred at 45 °C, until the complete formation of the target compounds. The excess of solvent was evaporated under reduced pressure, extracted with ethyl acetate and dried over anhydrous Na2SO4, The extract filtered, and evaporated under reduced pressure to give the desired products.
6.4.1. 3-Methyl-2,6-diphenyltetrahydro-4H-pyran-4-one O-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)oxime (25)
FT-IR (cm-1): 3040, 3030, 3014, 2986, 2942 (C—H stretching); 1668 (C O stretching); 1637 (C N stretching). 1H NMR (CDCl3):(δ) ppm: 5.67 (s, 2H, O—CH2), 4.37 (d, J 2a,3a = 10.2 Hz, 1H, H2a), 4.25 (d, J 5a,6a = 10.8, Hz, 1H, H6a), 3.87 (m, 1H, H3a), 2.17 (d, J = 12.40 Hz, 1H, H5a), 3.49 (d, J 5a,5e = 12.5 Hz, 1H, H5e), 1.66 (m, 1H, H of –CHH’ of C-7), 1.16 (m, H, H' of C-7), 0.78 (t, 3H, -CH3 at C-7), 7.28–7.33 (M, 15H, aryl protons at protons and triazole). 13C NMR (CDCl3): (δ) ppm: 86.19 (C2), 77.19 (C6), 60.58 (C3), 49.61 (C5), 11.91 (CH3 at C3), 158.48 (C N), 141.89 (C-4 at triazole), 118.04 (C5 at triazole), 150.11, 141.95, 145.10, 144.45, 128.44, 127.89, 126.84, 126.5, 125.84, 125.18, 124.67 (aryl carbons). Anal. Cal.(Found) for C27H26N4O2 (439.21). C, 73.95 (73.73); H, 5.98 (6.21); N, 12.78 (12.05). Yellow powder, yield 85%, m.p: 265–266 °C
6.4.2. 3-Ethyl-2,6-diphenyltetrahydro-4H-pyran-4-one O-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl) oxime (26)
FT-IR (cm-1): 3052, 3039, 2987, 2972, 2778, 2743 (C—H stretching); 1642 (C N stretching); 3178 (N—H stretching). 1H NMR (CDCl3):(δ) ppm: 4.23 (d, J 2a,3a = 10.5 Hz, 1H, H2a), 4.65 (d, J 5a,6a = 10.5, Hz, 1H, H6a), 3.25 (sextet, 1H, H3a), 2.85 (t, 1H, H5a), 3.91 (d, J 5a,5e = 12.8 Hz, 1H, H5e), 0.89 (d, J3a ,Me = 6.0 Hz, 3H, -CH3 at C3), 4.91 (d, J = 2.0 Hz, 2H, –CH2), 7.89 (dd, 2H, H-2′), 7.45 (dd, 2H, H-3′), 7.31–7.33 13C NMR (CDCl3): (δ) ppm: 64.75 (C2), 60.31 (C6), 49.69 (C3), 46.77 (C5), 18.85 (-CH2—CH3) 17.41 (-CH2— CH 3), 160.41 (C N), 139.85, 129.74, 127.68, 128.52, 127.33, 127.14, 122.23 (aryl carbons), 144.21 (C-4 at triazole), 118.02 (C5 at triazole),. Anal. Cal.(Found) for C28H28N4O2 (453.21)% C, 74.31(74.68); H, 6.24 (6.01); N, 12.38 (12.13);. Yellow powder, yield 80%, m.p: 261–262 °C
6.4.3. 3,5-Dimethyl-2,6-bis(p-chlorophenyl)−4H-pyran-4-one O-((1-phenyl-1H-1,2,3-triazol-4-yl) methyl)oxime (27)
FT-IR (cm-1): 3038, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1636 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.72 (d, J 2a,3a=10.40 Hz, 1H, H2a), 4.12 (d, J6 a,5e = 2.32 Hz; 1H, H6a), 2.78 (sextet, 1H, H3a), 2.12 (sextet, 1H, H5a), 1.19 (d, 3H, 5-CH3, J = 7.05 Hz); 0.92 (d, 3H, 3-CH3, J = 6.50 Hz); 7.98–7.38 (m, 13H, aryl protons), 6.65 (s, 2H, CH2), 7.35–7.38 (M, 5H, aryl protons at triazole), 7.20 (s, 1H, H-4 triazole). 13C NMR (DMSO‑d6): (δ) ppm: 85.12 (C2), 78.82 (C6), 45.15 (C3), 37.19 (C5), 160.36 (C N), 11.29 and 12.20 (3-CH3 and 5-CH3); 142.91 (C-4 at triazole), 119.14 (C5 at triazole), 150.25, 141.15, 139.87, 139.05, 129.47, 127.34, 126.98, 121.45 (aryl carbons). HRMS (ESI): m/z calcd. for [M + H]+ C28H26Cl2N4O2 521.14. Yield 76%, m.p: 281–282 °C
6.4.4. 3,5-Dimethyl-2,6-bis(p-bromophenyl)−4H-pyran-4-oneO-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)oxime (28)
FT-IR (cm-1): 3048, 3028, 3010, 2986, 2942, 2898 (C—H stretching); 1640 (C N stretching); 3160 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.71 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.20 (d, J6 a,5e = 2.55 Hz; 1H, H6a), 2.73 (sextet, 1H, H3a), 2.14 (sextet, 1H, H5a), 1.10 (d, 3H, 5-CH3, J = 7.00 Hz); 0.95 (d, 3H, 3-CH3, J = 6.50 Hz); 7.41–7.08 (m, 8H, aryl protons), 6.56 (s, 2H, -O-CH2-), 7.25 (s, 1H, H-4 triazole). 13C NMR (DMSO‑d6): (δ) ppm: 87.92 (C2), 78.74 (C6), 45.29 (C3), 38.08 (C5), 159.16 (C N), 11.31 and 11.22 (3-CH3 and 5-CH3); 151.32, 140.15, 139.07–124.98 (aryl carbons), 142.88 (C-4 at triazole), 120.04 (C5 at triazole). Anal. Cal.(Found) for C28H26Br2N4O2 (613.04)% C, 63.97 (63.17); H, 5.18 (5.45); N, 10.29 (10.05). Yield 72%, m.p: 285–286 °C
6.4.5. 3,5-Dimethyl-2,6-bis(p-methylphenyl)−4H-pyran-4-oneO-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)oxime (29)
FT-IR (cm-1): 3039, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1639 (C N stretching); 3159 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.72 (d, J 2a,3a = 10.55 Hz, 1H, H2a), 4.20 (d, J6 a,5e = 2.44 Hz; 1H, H6a), 2.71 (sextet, 1H, H3a), 2.24 (sextet, 1H, H5a), 1.02 (d, 3H, 5-CH3, J = 6.89 Hz); 0.92 (d, 3H, 3-CH3, J = 6.65 Hz); 7.81–7.34 (m, 13H, aryl protons), 2.34 (s, 6H, p-CH3), 6.06 (s, 2H, -O-CH2-), 7.18 (s, 1H, H-4 triazole). 13C NMR (DMSO‑d6): (δ) ppm: 88.31 (C2), 79.15 (C6), 45.15 (C3), 38.52 (C5), 161.21 (C N), 11.27 and 11.18 (3-CH3 and 5-CH3); 21.01 (p-CH3); 142.89 (C-4 at triazole), 119.04 (C5 at triazole), 62.09 (O—CH2), 150.52, 142.02, 139.85, 139.13, 129.40, 128.94, 127.98, 123.4, 120.23 (aryl carbons). Anal. Cal.(Found) for C30H32N4O2 (481.41) C, 74.97 (74.16); H, 6.71(6.98); N, 11.66 (11.42). Yield 70%, m.p: 278–279 °C.
6.4.6. 3,5-Dimethyl-2,6-bis(p-methoxyphenyl)−4H-pyran-4-one O-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)oxime (30)
FT-IR (cm-1): 3068, 3046, 3034, 2886, 2982, 2898, (C—H stretching); 1636 (C N stretching); 3165 (N—H stretching). 1H NMR (DMSO‑d6):(δ) ppm: 4.74 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.24 (d, J6 a,5e = 2.40 Hz; 1H, H6a), 2.71 (sextet, 1H, H3a), 2.13 (sextet, 1H, H5a), 1.10 (d, 3H, 5-CH3, J = 7.00 Hz); 0.94 (d, 3H, 3-CH3, J = 6.46 Hz); 3.89 (s, 6H, pOCH3–); 6.89–7.62 (m, 13H, aryl protons), 6.41 (s, 2H, -O-CH2-) 7.28 (s, 1H, H-4 triazole). 13C NMR (DMSO‑d6): (δ) ppm: 87.83 (C2), 79.12 (C6), 46.05 (C3), 37.15 (C5), 69.45 (-O-CH2), 142.98 (C-4 at triazole), 119.15 (C5 at triazole), 161.46 (C N), 11.78 and 11.61 (3-CH3 and 5-CH3); 56.36 (pOCH3–); 159.24, 151.02, 142.11, 138.87, 138.12, 129.47, 127.34, 126.98, 112.54 (aryl carbons). Anal. Cal.(Found) for C30H32N4O4 (513.24) C, 70.70 (70.10); H, 6.51 (6.78); N, 10.64 (10.15). Yield 65%, m.p: 262–263 °C
6.5. Synthesis of 2,6-diphenyltetrahydro-4H-pyran-4-one O-((3-(4-chlorophenyl) isoxazol-5-yl)methyl) oximes (31–36)
Equal amounts of (1.5 mmol) O-ethynyl oximes (19–24) and oxime were dissolved in a 20 mL of 1:1 tert‑BuOH/H2O mixture. The, 5 mol% copper(II)sulfate pentahydrate and 15 mol% sodium ascorbate were added in the solution mixture. The reaction mixture was stirred for 12-16 h at 40 °C to give residue. The residue was collected by filtration, washed by water, dried and purified by column chromatography to afford the target compounds. 31–36, in 78–90% yields.
6.5.1. 3-Methyl-2,6-diphenyltetrahydro-4H-pyran-4-one-O-((3-(4-chlorophenyl)isoxazol-5-yl)methyl) oxime (31)
FT-IR (cm-1): 3045, 3036, 3024, 2996, 2942 (C—H stretching); 1556 (C N stretching). 1H NMR (CDCl3):(δ) ppm:: 4.21 (d, J 2a,3a = 10.51 Hz, 1H, H2a), 4.63 (d, J 5a,6a = 10.8, Hz, 1H, H6a), 3.27 (sextet, 1H, H3a), 2.88 (t, 1H, H5a), 3.89 (d, J5a,5e = 12.2 Hz, 1H, H5e), 0.91 (d, J3a ,Me = 6.7 Hz, 3H, -CH3 at C3), 5.91 (d, J = 2.0 Hz, 2H, –O—CH2), 7.89 (dd, 2H, H-2′), 7.45 (dd, 2H, H-3′), 7.31–7.33 (M, 1H, H-3′), 7.46 (d, 4H, H-2″, H-6″), 7.20–7.33 (m, 6H, H-2″', H-6″',H-2′''', H-6′''') 7.12 (s, 1H, CH), 5.55 (s, 2H -O-CH2); 13C NMR (CDCl3): (δ) ppm: 86.88 (C2), 77.78 (C6), 61.58 (C3), 49.61 (C5), 11.78 (CH3 at C3), 158.98 (C N), 150.11, 141.95, 145.10, 144.45, 128.44, 127.89, 126.84, 126.5, 125.84, 125.18, 124.67 (aryl carbons). [M + H]+ C28H25N2ClO3 (473.11). Yellow powder, yield 85%, m.p: 265–266 °C
6.5.2. 3-Ethyl-2,6-diphenyltetrahydro-4H-pyran-4-one O-((3-(4-chlorophenyl)isoxazol-5-yl)methyl) oxime (32)
FT-IR (cm-1): 3042, 3029, 2997, 2952, 2878, 2843 (C—H stretching); 1555 (C N stretching); 3179 (N—H stretching). 1H NMR (CDCl3):(δ) ppm: 4.37 (d, J 2a,3a = 10.7 Hz, 1H, H2a), 4.58 (d, J 5a,6a = 10.1, Hz, 1H, H6a), 3.35 (m, 1H, H3a), 2.17 (d, J = 12.80 Hz, 1H, H5a), 3.18 (d, J 5a,5e = 12.8 Hz, 1H, H5e), 1.61(m, 1H, H of -CHH’ of C-7), 1.11 (m, H, H' of C-7), 0.88 (t, 3H, -CH3 at C-7), 7.12 (s, 1H, CH), 5.58 (s, 2H -O-CH2), 7.51 (m, 4H, H-2″, H-6″), 7.28–7.39 (m, 10H, aryl protons). 13C NMR (CDCl3): (δ) ppm: 86.22 (C2), 80.84 (C6), 69.78 (C3), 57.17 (C5), 18.81 (-CH2—CH3) 12.51 (-CH2— CH 3), 160.41 (C N), 151.04, 144.36, 143.47, 141.73, 128.70, 127.68, 128.52, 127.33, 127.14, 122.23 (aryl carbons). Anal. Cal.(Found) for C29H27ClN2O3 (487.19) C, 71.52 (71.13); H, 5.59 (5.21); N, 5.75 (5.19). Yellow powder, yield 85%, m.p: 268–269 °C
6.5.3. 3,5-Dimethyl-2,6-bis(p-chlorophenyl)−4H-pyran-4-oneO-((3-(4-chlorophenyl) isoxazol-5-yl)methyl)oxime (33)
FT-IR (cm-1): 3038, 3026, 3004, 2986, 2942, 2898 (C—H stretching); 1536 (C N stretching); 3159 (N—H stretching). 1H NMR (CDCl3):(δ) ppm: 4.73 (d, J 2a,3a = 10.40 Hz, 1H, H2a), 4.32 (d, J6 a,5e = 2.42 Hz; 1H, H6a), 2.71 (sextet, 1H, H3a), 2.13 (sextet, 1H, H5a), 1.19 (d, 3H, 5-CH3, J = 7.05 Hz); 0.99 (d, 3H, 3-CH3, J = 6.10 Hz); 7.49–7.38 (m, 8H, aryl protons), 7.12 (s, 1H, CH), 5.58 (s, 2H -O-CH2), 13C NMR (CDCl3): (δ) ppm: 88.72 (C2), 78.12 (C6), 46.25 (C3), 37.19 (C5), 160.36 (C N), 11.69 and 11.40 (3-CH3 and 5-CH3); 150.25, 141.15, 139.87, 139.05, 129.47, 127.34, 126.98, 121.45 (aryl carbons). [M + H]+ C29H25Cl3N2O3 555.09. Yellow powder, yield 80%, m.p: 288–289 °C
6.5.4. 3,5-Dimethyl-2,6-bis(p-bromophenyl)−4H-pyran-4-oneO-((3-(4-chlorophenyl) isoxazol-5-yl) methyl) oxime (34)
FT-IR (cm-1): 3041, 3038, 3020, 2986, 2942, 2898 (C—H stretching); 1540 (C N stretching); 3150 (N—H stretching). 1H NMR (CDCl3): (δ) ppm: 4.81 (d, J 2a,3a = 10.50 Hz, 1H, H2a), 4.28 (d, J6 a,5e = 2.51 Hz; 1H, H6a), 2.77 (sextet, 1H, H3a), 2.11(sextet, 1H, H5a), 1.11 (d, 3H, 5-CH3, J = 7.00 Hz); 0.83 (d, 3H, 3-CH3, J = 6.39 Hz); 7.22 (s, 1H, CH), 5.51 (s, 2H -O-CH2), 13C NMR (CDCl3): (δ) ppm: 87.89 (C2), 78.84 (C6), 45.29 (C3), 38.08 (C5), 159.16 (C N), 11.31 and 11.22 (3-CH3 and 5-CH3); 151.32, 140.15, 139.07–124.98 (aryl carbons). Anal. Cal.(Found) for C29H25Br2ClN2O3 (644.99) C, 54.02 (53.96); H, 3.91 (4.01); N, 4.34 (4.24). Yield 78%, m.p: 291–292 °C
6.5.5. 3,5-Dimethyl-2,6-bis(p-methylphenyl)−4H-pyran-4-one O-((3-(4-chlorophenyl) isoxazol-5-yl)methyl) oxime (35)
FT-IR (cm-1): 3069, 3056, 3024, 2996, 2962, 2898 (C—H stretching); 1536 (C N stretching); 3169 (N—H stretching). 1H NMR (CDCl3):(δ) ppm: 4.71 (d, J 2a,3a = 10.55 Hz, 1H, H2a), 4.19 (d, J6 a,5e = 2.46 Hz; 1H, H6a), 2.70 (sextet, 1H, H3a), 2.14 (sextet, 1H, H5a), 1.08 (d, 3H, 5-CH3, J = 6.80 Hz); 0.96 (d, 3H, 3-CH3, J = 6.55 Hz). 7.41–7.34 (m, 8H, aryl protons), 2.31 (s, 6H, p-CH3), 7.22 (s, 1H, CH), 5.51 (s, 2H -O-CH2), 13C NMR (CDCl3): (δ) ppm: 88.21 (C2), 80.02 (C6), 45.52 (C3), 38.12 (C5), 168.07 (C O), 161.21 (C N), 11.27 and 11.18 (3-CH3 and 5-CH3); 21.01 (p-CH3); 150.52, 142.02, 139.85, 139.13, 129.40, 128.94, 127.98, 123.4, 120.23 (aryl carbons). Anal. Cal.(Found) for C31H31ClN2O3 (515.20) (%) C, 72.29 (72.10); H, 6.07 (6.12); N, 5.44 (5.34). Yield 80%, m.p: 261–262 °C
6.5.6. 3,5-Dimethyl-2,6-bis(p-methoxyphenyl)−4H-pyran-4-one O-((3-(4-chlorophenyl) isoxazol-5-yl)methyl)oxime (36)
FT-IR (cm-1): 3032, 3022, 3014, 2986, 2942, 2898 (C—H stretching); 1556 (C N stretching); 3180 (N—H stretching). 1H NMR (CDCl3):(δ) ppm:: 4.84 (d, J 2a,3a = 11.50 Hz, 1H, H2a), 4.34 (d, J6 a,5e = 2.45 Hz; 1H, H6a), 2.76 (sextet, 1H, H3a), 2.11 (sextet, 1H, H5a), 1.11 (d, 3H, 5-CH3, J = 7.10 Hz); 0.97 (d, 3H, 3-CH3, J = 6.36 Hz); 3.88 (s, 6H, pOCH3–); 6.91–7.53 (m, 8H, aryl protons), 5.57 (s, 2H -O-CH2). 13C NMR (CDCl3): (δ) ppm: 89.13 (C2), 80.12 (C6), 48.05 (C3), 37.15 (C5), 159.46 (C N), 11.71 and 11.65 (3-CH3 and 5-CH3); 56.32 (pOCH3–); 159.14, 151.08, 142.11, 138.87, 138.12, 129.47, 127.34, 126.98, 112.54 (aryl carbons). Anal. Cal.(Found) for C31H31ClN2O5 (547.19) (%) C, 68.06 (68.12); H, 5.71 (5.78); N, 5.12 (4.98). Yield 80%, m.p: 301–302 °C.
6.6. Antimicrobial studies
The in vitro antimicrobial activity of the target compounds was tested against a set of bacterial and fungal strains according to the reported method [53]. The microbroth dilution method was applied for minimum inhibitory concentration (MIC) determination. All experiments were performed in three biological replicates. A stock solution was prepared by dissolving 10 mg of the hydrochloride salt of each compound (10 mg) in 1 mL of distilled water or related culture media as solvent and then sterilized by a microbial filter with a 0.22 m pore size. The minimum inhibitory concentrations (MICs) were recorded by visual observations after 24 h (for bacteria) and 72-96 h (for fungi) of incubation. Streptomycin and Amphotericin B were used as standards.
6.7. Anti-tuberculosis study
All the target compounds were assayed for their antitubercular activity against M. tuberculosis H37Rv by the Resazurin Assay Method [54,64] followed by Middlebrook 7H9 broth, supplemented with 10% OADC, used to determine the MIC. Stock solutions (2 g/mL) of the test compounds were prepared in dimethylformamide (DMF). In 96-well plates, two-fold serial dilutions were prepared, yielding concentrations ranging from 10 to 100 g/mL. Ethambutol (EMB), pyrazinamide (PZA), and isoniazid (INH) were used as standard drugs. The cultures were incubated at 37 C for 7 days. 20 mL of 0.01% Resazurin in water was added to each tube. Resazurin, a redox dye, is blue in the oxidised state and turns pink when reduced by the growth of viable cells.
6.8. DNA gyrase ATPase assays
The Inspiralis test kit was used to perform the DNA gyrase ATPase assay. 96-well plates with assay buffer, 400 mM NADH, 800 mM phosphoenolpyruvate, and 1.5 mL phosphokinase / lactate dehydrogenase; linear pBR322 (3 mL), PEP (1 mL), NADH (2 mL), and water1X (45.8 mL). Next, 10 mL of each of the inhibitors and enzyme were introduced. After 10 min at 25 °C, the absorbance of the plates was measured at OD 340 nm. The reaction was then initiated by adding 6.7 mL of ATP to each well after turning off the plate reader. Once again placed in the plate reader, the absorbance was calculated.
6.9. Cytotoxicity assay
The MTT assay was used to test active compounds for their cytotoxic effects on peripheral blood mononuclear cells [65,66] to ensure selectivity. The cell lines were cultured in a standard cell culture environment at 37 °C in an atmosphere that was 95% humidified air and contained 5% CO2. In a single experiment, each concentration was tested in duplicate. The GI50 values were calculated using OriginPro Software.
6.10. Selectivity index (SI)
The selectivity index was calculated by the concentration of GI50 of each cell line being subtracted by M. tuberculosis minimum inhibition (MIC).
6.11. Molecular docking
The LigandFit method was used for the molecular docking study, which can operate either flexible or rigid docking based on the conformational state of the ligand. The small drug molecules are docked into the protein's active site by stabilizing their shapes in Ligand-Fit dock. The protein data bank was utilized to acquire the three-dimensional structure of InhA bound triclosan (PDB ID: 3FNG). The solvent molecules in the active site were kept, while the remaining water molecules were eliminated. A maximum of ten poses are kept based on the root mean square deviation. The Ligand Fit module's consensus scoring function was used to select the best leads from the molecular docking study. The virtual screening set compounds' final hits are docked into the triclosan active site with flexibility. The best molecules were chosen based on the consensus scoring function, their interaction with critical residues, and their expected activity.
6.11.1. Ligand preparation
Chem Draw Ultra 12.0 was used to generate the ligands, which were then given the proper 2D configuration. Chem 3D Ultra 12.0, Gasteiger charges, non-polar hydrogen atoms, and rotatable bonds were then generated with Auto-Dock 4.2 tools. A maximum of 10 conformers for each compound were considered for docking with Auto Dock Vina.
6.12. Predictions of in silico ADME
The drug-likeness, toxicity risk, TPSA (topological surface area), cLogP, logS (solubility), MW (molecular weight), and total drug-score of the target title compounds were examined using swissADME and OSIRIS in order to screen compounds with favorable drug-like properties. Compounds with favorable drug-like properties and no toxic fragments (mutagenic, tumorigenic, irritating, or reproductive effects) also that predicted.
CRediT authorship contribution statement
N. Ravisankar: Methodology, Resources. N. Sarathi: Methodology, Software. T. Maruthavanan: Validation, Resources. Subramaniyan Ramasundaram: Methodology, Resources, Investigation. M. Ramesh: Formal analysis, Data curation, Writing – original draft, Validation. C. Sankar: Resources, Formal analysis, Data curation, Writing – review & editing. S. Umamatheswari: Supervision, Investigation, Formal analysis, Data curation, Validation, Writing – review & editing, Conceptualization, Methodology. G. Kanthimathi: Methodology. Tae Hwan Oh: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would also like to thank SIF IIS Bangalore and SAIF IIT Madras for recording NMR spectra and doing elemental analyses on the compounds.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2023.135461.
Appendix. Supplementary materials
Data availability
No data was used for the research described in the article.
References
- 1.World Health Organization . 2022. Global Tuberculosis Report.https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 [Google Scholar]
- 2.World Health Organization . 2022. Global Tuberculosis Report.https://www.who.int/teams/global-hiv-hepatitis-and-stis-programmes/hiv/treatment/tuberculosis-hiv [Google Scholar]
- 3.Liem N., Jean P. Mycobacterial subversion of chemotherapeutic reagents and host defense tactics: challenges in tuberculosis drug development. Annu. Rev. Pharmacol. Toxicol. 2009;49:427–453. doi: 10.1146/annurev-pharmtox-061008-103123. [DOI] [PubMed] [Google Scholar]
- 4.William J.B., Brenda E.J. Treatment of HIV-related tuberculosis in the era of effective antiretroviral therapy. Am. J. Respir. Crit. Care Med. 2001;164:7–12. doi: 10.1164/ajrccm.164.1.2101133. [DOI] [PubMed] [Google Scholar]
- 5.Lawn S.D., Zumla A.I. Tuberculosis. Lancet. 2011;378(9785):57–72. doi: 10.1016/S0140-6736(10)62173-3. [DOI] [PubMed] [Google Scholar]
- 6.Beena Rawat. Antituberculosis drug research: a critical overview. Med. Res. Rev. 2013;33(4):693. doi: 10.1002/med.21262. –693. [DOI] [PubMed] [Google Scholar]
- 7.Dover L.G., Coxon G.D. Status and research strategies in tuberculosis drug development. J. Med. Chem. 2011;54:6157–6165. doi: 10.1021/jm200305q. [DOI] [PubMed] [Google Scholar]
- 8.Ma Z., Lienhardt C., McIlleron H., Nunn A.J., Wang X. Global tuberculosis drug development pipeline: the need and the reality. Lancet. 2010;375:2100–2109. doi: 10.1016/S0140-6736(10)60359-9. [DOI] [PubMed] [Google Scholar]
- 9.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel Coronavirus (2019-nCoV) originating in China. Commentary. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/i.apsb.2020.02-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cort ´es-García C.J., Chac ´on-García L., Mejía-Benavides J.E., Díaz-Cervantes E. Tackling the SARS-CoV-2 main protease using hybrid derivatives of 1,5-disubstituted tetrazole-1,2,3-triazoles: an in silico assay. Peer J. Phys. Chem. 2020;2:10. doi: 10.7717/peerj-pchem.10. [DOI] [Google Scholar]
- 12.Ling R., Dai Y., Huang B., Huang W., Yu J., Lu X., Jiang Y. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130 doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 14.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-.H. An overview of severe acute respiratory syndrome-Coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J., Hu L., Huang X., Wang C., Zhang Z., Wang Y., Zhang D., Ye W. Potential of coronavirus 3C-like protease inhibitors for the development of new anti-SARS-CoV-2 drugs: insights from structures of protease and inhibitors. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasir N.M., Ermanis K., Clarke P.A. Strategies for the construction of tetrahydropyran rings in the synthesis of natural products. Org. Biomol. Chem. 2014;12:3323–3335. doi: 10.1039/C4OB00423J. [DOI] [PubMed] [Google Scholar]
- 17.Perry M.A., Rychnovsky S.D., Sizemore N. Springer-Verlag; Berlin, Heidelberg: 2014. Synthesis of Saturated Oxygenated Heterocycles I, Topics in Heterocyclic Chemistry; pp. 43–95. J. Cossy35. [Google Scholar]
- 18.Sakai T., Fukuta A., Nakamura K., Nakano M., Mori Y. Total synthesis of brevisamide using an oxiranyl anion strategy. J. Org. Chem. 2016;81:3799–3808. doi: 10.1021/acs.joc.6b00484. [DOI] [PubMed] [Google Scholar]
- 19.Malassis J., Bartlett N., Hands K., Selby M.D., Linclau B. Total synthesis of (−)-Luminacin D. J. Org. Chem. 2016;81:3818–3837. doi: 10.1021/acs.joc.6b00489. [DOI] [PubMed] [Google Scholar]
- 20.Yin J., Kouda K., Tezuka Y., Tran Q.Le, Miyahara T., Chen Y., Kadota S. New diarylheptanoids from the rhizomes of dioscorea spongiosa and their antiosteoporotic activity. Planta. Med. 2004;70:54–58. doi: 10.1055/s-2004-815456. [DOI] [PubMed] [Google Scholar]
- 21.Xu D., Pang X.J., Zhao T., Xu L.L., Yang X.L. New alkenylated tetrahydropyran derivatives from the marine sediment derived fungus Westerdykella dispersa and their bioactivities. Fitoterapia. 2017;122:45–51. doi: 10.1016/j.fitote.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Kito K., Ookura R., Yoshida S., Namikoshi M., Ooi T., Kusumi T. New cytotoxic 14-membered macrolides from marine-derived Fungus Aspergillus ostianus. Org. Lett. 2008;10:225–228. doi: 10.1021/ol702598q. [DOI] [PubMed] [Google Scholar]
- 23.Araujo C.A.C., Alegrio L.V., Leon L.L. Antileishmanial activity of compounds extracted and characterized from Centrolobium sclerophyllum. Phytochemistry. 1998;49:751–754. doi: 10.1016/S0031-9422(97)00976-X. [DOI] [Google Scholar]
- 24.Umamatheswari S., Balaji B., Ramanathan M., Kabilan S. Synthesis, stereochemistry, antimicrobial evaluation and QSAR studies of 2, 6-diaryltetrahydropyran-4-one thiosemicarbazones. Eur. J. Med. Chem. 2011;46:1415–1424. doi: 10.1016/j.ejmech.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 25.Pajk S., Zivec M., Sink R., Sosic I., Neu M., Chung C., Hoyos M.M. New direct inhibitors of InhA with antimycobacterial activity based on a tetrahydropyran scaffold. Eur. J. Med. Chem. 2016;112:252–257. doi: 10.1016/j.ejmech.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Capim S.L., Gonçalves G.M., dos Santos G.C.M., Marinho B.G., Vasconcellos M.L.A.A. High analgesic and anti-inflammatory in vivo activities of six new hybrids NSAIAS tetrahydropyran derivatives. Bioorg. Med. Chem. 2013;21:6003–6010. doi: 10.1016/j.bmc.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 27.Kornprobst J.-.M. Wiley-Blackwell; Weinhein, Germany: 2010. Encyclopedia of Marine Natural Products; pp. 212–220. Vol. 1, Chapter 11. [Google Scholar]
- 28.Ahmed N., Konduru N.K., Ahmad S., Owais M. Design, synthesis and antiproliferative activity of functionalized flavone-triazole-tetrahydropyran conjugates against human cancer cell lines. Eur. J. Med. Chem. 2014;82:552–564. doi: 10.1016/j.ejmech.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Singh P., Bhardwaj A., Kaur S., Kumar S. Design, synthesis and evaluation of tetrahydropyran based COX-1/-2 inhibitors. Eur. J. Med. Chem. 2009;44:1278–1287. doi: 10.1016/j.ejmech.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Post-Martens K., Denkin S. New drug candidates and therapeutic targets for tuberculosis therapy. Drug. Discov. Today. 2006;11:21–27. doi: 10.1016/S1359-6446(05)03626-3. [DOI] [PubMed] [Google Scholar]
- 31.Subhedar D.D., Shaikh M.H., Shingate B.B., Nawale L., Sarkar D., Khedkar V.M. Novel tetrazoloquinoline–thiazolidinoneconjugates as possible antitubercular agents: synthesis and molecular docking. Med. Chem. Comm. 2016;9:1832–1848. doi: 10.1039/C6MD00278A. [DOI] [Google Scholar]
- 32.Shalini M., Johansen D., Kremer L., Kumar V. Design, synthesis, anti-mycobacterial and cytotoxic evaluation of C-4 functionalized 1,8-naphthalimide-heterocyclic hydrazide conjugates. Chem. Biol. Drug. Des. 2019;94:1300–1305. doi: 10.1111/cbdd.13503. [DOI] [PubMed] [Google Scholar]
- 33.Pillia. N. Dumala G., Mattana I., Grover P., Jaya Prakash M. Design, synthesis, biological and in silico evaluation of coumarin-hydrazone derivatives as tubulin targeted antiproliferative agents. Bioorg. Chem. 2019;91 doi: 10.1016/j.bioorg.2019.103143. [DOI] [PubMed] [Google Scholar]
- 34.Elhakeem M.A., Taher A.T., Abuel-Maaty S.M. Synthesis and anti-mycobacterial evaluation of some new isonicotinylhydrazide analogues. Bull. Fac. Pharm. Cairo. Univ. 2015;53:45–52. doi: 10.1016/j.bfopcu.2014.11.001. [DOI] [Google Scholar]
- 35.Gupta O., Pradhan T., Chawla G. An updated review on diverse range of biological activities of 1,2,4-triazole derivatives: insight into structure activity relationship. J. Mol. Struct. 2023;1274 doi: 10.1016/j.molstruc.2022.134487. [DOI] [Google Scholar]
- 36.Aneja B., Azam M., Alam S., Perwez A., Maguire R., Yadava U., Kavanagh K., Daniliuc C.G., Rizvi M.M.A., Mohd Q., Haq R., Abid M. Natural product-based 1,2,3-triazole/sulfonate analogues as potential chemotherapeutic agents for bacterial infections. ACS Omega. 2018;3:6912–6930. doi: 10.1021/acsomega.8b00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venugopala K.N., Khedr M.A., Girish Y.R., Bhandary S., Chopra D., Morsy M.A., Aldhubiab B.E., Kishore Deb P., Attimarad M., Nair A.B., Sreeharsha N., Rashmi V., Kandeel M., Akrawi S.H., Reddy M., Shashikanth S., Alwassil O.I., Mohanlall V. Crystallography, in silico studies, and in vitro antifungal studies of 2,4,5 trisubstituted 1,2,3-triazole analogues. Antibiotics. 2020;9:350. doi: 10.3390/antibiotics9060350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrashekhar M., Shailaja K., Rajendra S., Ramakrishna S., Venkata U.Mallavadhani. Synthesis and biological evaluation of some novel 1,2,3-triazole hybrids of myrrhanone B isolated from Commiphora mukul gum resin: identification of potent antiproliferative leads active against prostate cancer cells (PC-3) Euro J. Med. Chem. 2020;188 doi: 10.1016/j.ejmech.2019.111974. [DOI] [PubMed] [Google Scholar]
- 39.Venugopala K.N., Kandeel M., Pillay M., Kishore Deb P., Abdallah H.H., Mahomoodally M.F., Chopra D. Antibiotics. 2020;9:559. doi: 10.3390/antibiotics9090559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batra N., Rajendran V., Agarwal D., Wadi I., Ghosh P.C., Gupta R.D., Nath M. Synthesis and antimalarial evaluation of [1, 2,3]-triazole-tethered sulfonamide-berberine hybrids. Chem. Select. 2018;3:9790–9793. doi: 10.1002/slct.201801905. [DOI] [Google Scholar]
- 41.Venugopala K.N., Shinu P., Tratrat C., Deb P.K., Gleiser R.M., Chandrashekharappa S., Chopra D., Attimarad M., Nair A.B., Sreeharsha N., Mahomoodally F.M., Haroun M., Kandeel M., Asdaq S.M.B., Mohanlall V., Al-Shar'I N.A., Morsy M.A. Molecules. 2022;27:2676. doi: 10.3390/molecules27092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tratrat C., Haroun M., Paparisva A., Kamoutsis C., Petrou A., Gavalas A., Eleftheriou P., Geronikaki A., Venugopala K.N., Kochkar H., Nair A.B. New substituted 5-benzylideno-2-adamantylthiazol[3,2-b][1,2,4]triazol-6(5H)ones as possible anti-inflammatory agents. Molecules. 2021;26:659. doi: 10.3390/molecules26030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aarjane M., Slassi S., Tazi B., Amin A. Synthesis and biological evaluation of novel isoxazole derivatives from acridone. Arch. Pharm. 2020 doi: 10.1002/ardp.202000261. [DOI] [PubMed] [Google Scholar]
- 44.Naresh Kumar R., Jitender Dev G., Ravikumar N., Krishna Swaroop D., Debanjan B., Bharath G., Narsaiah B., Nishant Jain S., Gangagni Rao A. Synthesis of novel triazole/isoxazole functionalized 7-(trifluoromethyl)pyrido[2,3-d]pyrimidine derivatives as promising anticancer and antibacterial agents. Bioorg. Med. Chem. Lett. 2016;26:2927–2930. doi: 10.1016/j.bmcl.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 45.Trivedi J., Parveen A., Rozy F., Mitra A., Bal C., Mitra D., Sharon A. Discovery of 2-isoxazol-3-yl-acetamide analogues as heat shock protein 90 (HSP90) inhibitors with significant anti-HIV activity. Eur. J. Med. Chem. 2019;183 doi: 10.1016/j.ejmech.2019.111699. [DOI] [PubMed] [Google Scholar]
- 46.Naidu K.M., Srinivasarao S., Agnieszka N., Ewa A.K., Kumar M.M.K., Chandra Sekhar K.V.G. Seeking potent antitubercular agents: design, synthesis, antitubercular activity and docking study of various ((triazoles/indole)-piperazin-1-yl/1,4-diazepan-1-yl)benzo[d]isoxazole derivatives. Bioorg. Med. Chem. Lett. 2016;26:2245. doi: 10.1016/j.bmcl.2016.03.059. [DOI] [PubMed] [Google Scholar]
- 47.Abdelall E.K.A. Synthesis and biological evaluations of novel isoxazoles and furoxan derivative as anti-inflammatory agents. Bioorg. Chem. 2020;94 doi: 10.1016/j.bioorg.2019.103441. [DOI] [PubMed] [Google Scholar]
- 48.Umamatheswari S., Sankar C. Synthesis, identification and in vitro biological evaluation of some novel quinoline incorporated 1,3-thiazinan-4-one derivatives. Bioorg. Med. Chem. Letts. 2017;27:695–699. doi: 10.1016/j.bmcl.2016.06.038. [DOI] [PubMed] [Google Scholar]
- 49.Sankar C., Pandiarajan K. Synthesis and antitubercular and antimicrobial activities of some 2r,4c-diaryl-3-azabicyclo[3.3.1]nonan-9-one N-isonicotinoyl hydrazone derivatives. Eur. J. Med. Chem. 2010;45:5480–5485. doi: 10.1016/j.ejmech.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 50.Jap F.R., Maitland W. CXLVIII. Reduction products of αβ-dimethylanhydracetonebenzil, and condensation products of benzaldehyde with ketones. J. Chem. Soc. 1904;85:1473–1489. doi: 10.1039/CT9048501473. [DOI] [Google Scholar]
- 51.Sankar C., Umamatheswari S., Pandiarajan K. Conformational analysis of 2,6-diarylpiperidin-4-one hydrazonesby X-ray diffraction and NMR spectroscopy. J. Mol. Struct. 2015;1083:27–38. doi: 10.1016/j.molstruc.2014.10.015. [DOI] [Google Scholar]
- 52.Agalave S.G., Maujan S.R., Pore V.S. Click chemistry: 1,2,3-Triazoles as pharmacophores. Chemistry. 2011;6:2696–2718. doi: 10.1002/asia.201100432. [DOI] [PubMed] [Google Scholar]
- 53.Dhar M.H., Dhar M.M., Dhawan B.N., Mehrotra B.N., Ray C. Screening of Indian plants for biological activity. Indian J. Exp. Biol. 1968;6:232–247. [PubMed] [Google Scholar]
- 54.Neetu K.T., Jaya S.T. Resazurin reduction assays for screening of antitubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J. Antimicrob. Chemother. 2007;60:288–293. doi: 10.1093/jac/dkm207. [DOI] [PubMed] [Google Scholar]
- 55.Hassan N.W., Saudi M.N., Abdel-Ghany Y.S., Ismail A., Elzahhar P.A., Sriram D., Nassra R., Abdel-Aziz M.M., El-Hawasha S.A. Novel pyrazine based anti-tubercular agents: design, synthesis, biological evaluation and in silico studies. Bioorg. Chem. 2020;96 doi: 10.1016/j.bioorg.2020.103610. [DOI] [PubMed] [Google Scholar]
- 56.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 57.Orme I. Search for new drugs for treatment of tuberculosis. Antimicrob. Agents. Chemother. 2001;45(7):1943–1946. doi: 10.1128/AAC.45.7.1943-1946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandar T. OSIRIS Property Explorer. 2020 Nov 10 https://www.organic-chemistry.org/prog/peo/ [cited]. Available from: [Google Scholar]
- 59.Manikandan P., Nagini S. Cytochrome P450 structure, function and clinical significance. Curr. Drug Targets. 2018;19:38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 60.Daina A., Michielin O., Zoete V. Swiss Target Prediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic. Acids. Res. 2019;47:W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2012;64:4–17. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 62.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 63.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., et al. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 64.Collins L., Franzblau S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41(5):1004–1009. doi: 10.1128/AAC.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Debnath B., Ganguly S. Synthesis, biological evaluation, in silico docking, and virtual ADME studies of 2-[2-Oxo-3-(arylimino) indolin-1-yl]-N-arylacetamides as potent anti-breast cancer agents. Monatshefte fur Chem. 2016;147(3):565–574. doi: 10.1007/s00706-015-1566-9. [DOI] [Google Scholar]
- 66.Bøyum A. Isolation of lymphocytes, granulocytes and macrophages. Scand. J. Immunol. 1976;5(s5):9–15. doi: 10.1111/j.1365-3083.1976.tb03851.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.