Polygenic risk scores (PRSs) are central to the development of precision medicine. However, decades of Euro-centric genomics research have paved the way for disparities in their clinical applications.1,2 Current PRSs may provide weaker predictions for populations with significant non-European ancestry.1 Counter to this observation, studies of coronary artery disease (CAD) PRSs have shown similar predictive power in Hispanic and non-Hispanic White populations,3,4 despite Hispanics being significantly underrepresented in genome-wide association studies and PRS development studies.2 We hypothesized that PRSs provide weaker risk predictions for some Hispanics, but this disparity is masked when the diverse Hispanic population is aggregated into a single group.
We used self-identified race and ethnicity (SIRE) to study CAD PRS in Hispanic, non-Hispanic White, and non-Hispanic Black participants in the Million Veteran Program, a predominantly male (~90%) cohort. The primary outcome was CAD, defined by ≥2 International Classification of Diseases diagnoses or Current Procedural Terminology procedure codes for CAD in the electronic health record within a span of 12 months. Participants were scored using 4 validated PRSs downloaded from the Polygenic score catalog.5 PRSKhera2018 (PGS000013) and PRSInouye2018 (PGS000018) are derived from a genome-wide association study of predominantly European-ancestry subjects with subsequent optimization in European cohorts. PRSWang2020 (PGS000296) is derived from the same genome-wide association study but was optimized in a South Asian cohort. PRSKoyama2020 (PGS000337) was derived from a genome-wide association study of predominantly European- and Japanese-ancestry subjects with optimization in a Japanese cohort. We used unrelated subjects, and principal components were generated for each SIRE group separately, using genotyped auto-somal single nucleotide polymorphisms passing typical quality filters. For each analysis, the PRS was normalized within the target population. Cox models were adjusted for age at enrollment, sex, and the first 10 SIRE-specific principal components. Score performance was measured by the hazard ratio (HR) for CAD per 1 SD increase in PRS. Global ancestry analysis was performed using 1000 Genomes continental reference populations (CHB; Han Chinese; GBR, British; LWK, Luhya; PEL, Peruvian; YRI, Yoruba). The VA Central Institutional Review Board approved the Million Veteran Program study protocol in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants. The data that support the findings of this study are available from the corresponding author on reasonable request.
We first confirmed that current PRSs predict CAD similarly well in Hispanics and non-Hispanic Whites. Among Million Veteran Program Hispanics (8193 cases; 39 827 non-cases), the per-SD HR for CAD ranged from 1.32 (95% CI, 1.29–1.35; PRSWang2020) to 1.40 (95% CI, 1.37–1.44; PRSInouye2018). Among non-Hispanic Whites (100 350 cases; 271 319 non-cases), the HR ranged from 1.36 (95% CI, 1.35–1.37; PRSWang2020) to 1.42 (95% CI, 1.41–1.43; PRSKoyama2020). The best performing score among non-Hispanic Blacks (18 483 cases; 83 452 non-cases) achieved a HR of 1.17 (95% CI, 1.15–1.19; PRSKoyama2020).
There is no genetic basis for distinctly stratifying human populations. To provide a simple and agnostic illustration of PRS heterogeneity, we created 3 clusters within each SIRE group using K-means clustering applied to the first 2 SIRE-specific principal components. Among Hispanics, 2 clusters contained a large fraction of the population, but the third cluster still accounted for 8% of Hispanics. In contrast, nearly all non-Hispanic Whites clustered into the first cluster, and the smallest cluster accounted for only 0.3% (Figure [A]). Continental ancestry patterns varied within all groups, with Hispanics displaying considerable diversity (Figure [B]). We consistently observed the highest HR in Hispanic Cluster 1 and the lowest HR in Hispanic Cluster 3, and we found significant heterogeneity for 3 scores (Figure [C]). For non-Hispanic Whites, heterogeneity was less significant despite the larger sample size, and the signal was driven by the small non-Hispanic White Cluster 3. For non-Hispanic Blacks, we did not detect heterogeneity. These patterns persisted in sensitivity analyses using up to 10 principal components and generating up to 11 clusters.
Figure. Heterogeneity in polygenic risk score (PRS) validity among Hispanics.
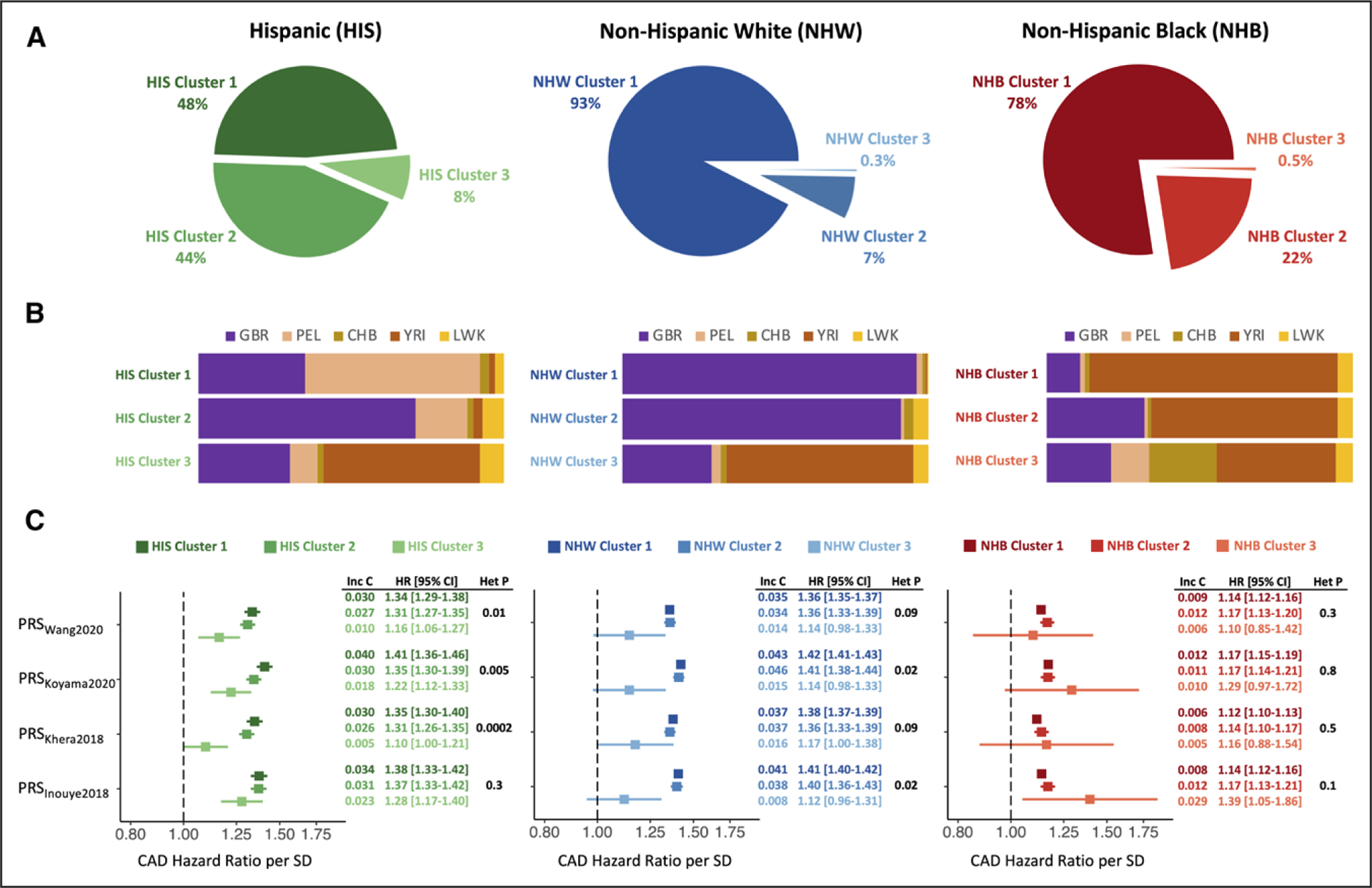
A, Participants in the Million Veteran Program were stratified by self-identified race and ethnicity (SIRE). SIRE-specific principal components (PCs) were used to perform K-means clustering, generating 3 clusters within each SIRE group. Among Hispanics, all 3 clusters contain a significant portion of participants, in contrast with the other SIRE groups. B, Average global ancestry proportions of each cluster, using 1000 Genomes continental reference populations (CHB, Han Chinese; GBR, British; LWK, Luhya; PEL, Peruvian; YRI, Yoruba). C, Strength of association with coronary artery disease (CAD) for 4 PRSs across 3 clusters in each SIRE group. Among Hispanics, there is heterogeneity in effect sizes, with lower hazard ratios (HRs) for HIS Cluster 3, accounting for 8% of Hispanics. Among non-Hispanic Whites, heterogeneity is less significant and is driven by NWH Cluster 3, which accounts for only 0.3% of the population. Among non-Hispanic Blacks, there is no significant heterogeneity, but PRS performance is generally poor. Het P indicates heterogeneity P value. Inc C indicates the incremental C-statistic, defined as the increase in model concordance with the addition of the PRS to a base model that includes age at enrollment, sex, and the first 10 SIRE-specific PCs.
Hispanic Cluster 1 had lower European (GBR) and higher South American (PEL) ancestry than Hispanic Cluster 2 (Figure [B]). However, high PEL ancestry was not associated with better PRS performance. Among Hispanics with ≥0.75 PEL ancestry (305 cases; 1922 non-cases), HR were consistently lower, ranging from 1.21 (95% CI, 1.09–1.35; PRSWang2020) to 1.32 (95% CI, 1.18–1.49; PRSKoyama2020). These observations likely reflect nonlinear correlations between ancestry proportions with differential effects on PRS associations. Larger sample sizes are needed, but current scores may underperform for individuals with high fractions of PEL ancestry. The trend for YRI proportion and PRS performance across clusters was more consistent. Analysis of Hispanics with ≥0.75 YRI ancestry (132 cases; 678 non-cases) showed uniformly weak PRS associations (highest HR, 1.13 [95% CI, 0.96–1.34]).
Our results show that contemporary PRSs for CAD perform well in Hispanics overall, but significant heterogeneity in PRS validity exists within the Hispanic population. For nearly 1 in 10 Hispanics in the Million Veteran Program, current PRSs provide weaker predictions. Although the use of race and/or ethnicity categories in PRS research is important for identifying and understanding disparities, aggregating Hispanics into a singular category may undermine that goal. The best approaches to assess PRS validity and utility are an ongoing debate, but race- and/or ethnicity-based assessments alone are inadequate to demonstrate equity.
Acknowledgments
The authors greatly appreciate the participation of the Million Veterans Program participants and the support of the Million Veterans Program staff.
Sources of Funding
This work was supported by funding from the Department of Veterans Affairs Office of Research and Development, Million Veteran Program Grants 2I01BX003362 and 1I01BX004821. This publication does not represent the views of the Department of Veterans Affairs or the United States Government.
Nonstandard Abbreviations and Acronyms
- CAD
coronary artery disease
- HR
hazard ratio
- PRS
polygenic risk scores
- SIRE
self-identified race and ethnicity
Footnotes
Disclosures
None.
REFERENCES
- 1.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke SL, Assimes TL, Tcheandjieu C. The propagation of racial disparities in cardiovascular genomics research. Circ Genom Precis Med. 2021;14:e003178. doi: 10.1161/CIRCGEN.121.003178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikilitas O, Schaid DJ, Kosel ML, Carroll RJ, Chute CG, Denny JA, Fedotov A, Feng Q, Hakonarson H, Jarvik GP, et al. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am J Hum Genet. 2020;106:707–716. doi: 10.1016/j.ajhg.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fahed AC, Aragam KG, Hindy G, Chen Y-DI, Chaudhary K, Dobbyn A, Krumholz HM, Sheu WHH, Rich SS, Rotter JI, et al. Transethnic transferability of a genome-wide polygenic score for coronary artery disease. Circ Genom Precis Med. 2021;14:e003092. doi: 10.1161/CIRCGEN.120.003092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert SA, Gil L, Jupp S, Ritchie SC, Xu Y, Buniello A, McMahon A, Abraham G, Chapman M, Parkinson H, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet. 2021;53:420–425. doi: 10.1038/s41588-021-00783-5 [DOI] [PMC free article] [PubMed] [Google Scholar]


