Abstract
Segregation of chromosomes during meiosis, to form haploid gametes from diploid precursor cells, requires in most species formation of crossovers physically connecting homologous chromosomes. Along with sister chromatid cohesion, crossovers allow tension to be generated when chromosomes begin to segregate; tension signals that chromosome movement is proceeding properly. But crossovers too close to each other might result in less sister chromatid cohesion and tension and thus failed meiosis. Interference describes the non-random distribution of crossovers, which occur farther apart than expected from independence. We discuss both genetic and cytological methods of assaying crossover interference and models for interference, whose molecular mechanism remains to be elucidated. We note marked differences among species.
Keywords: meiosis, recombination, crossing over, interference, assay methods, models, species differences
Introduction
Eukaryotes are typically generated by fusion of two special cells, one coming from each parent. Examples of these cells, called gametes, are eggs and sperm in mammals, eggs and pollen in flowering plants, and spores (or cells resulting from spore germination and growth) in fungi such as yeasts and molds. Gametes contain one copy of each chromosome and are thus haploids. Fusion of two gametes regenerates a diploid organism. The generation of haploid gametes from diploid precursors, called meiosis, requires the accurate segregation of the chromosomes, which in turn requires the pairing of the two copies of each chromosome (called homologs) and their segregation into separate cells. This process occurs with two sequential cell divisions but only one replication of the chromosomes, so that during meiosis diploids give rise to haploids (Figure 1).
Fig. 1.
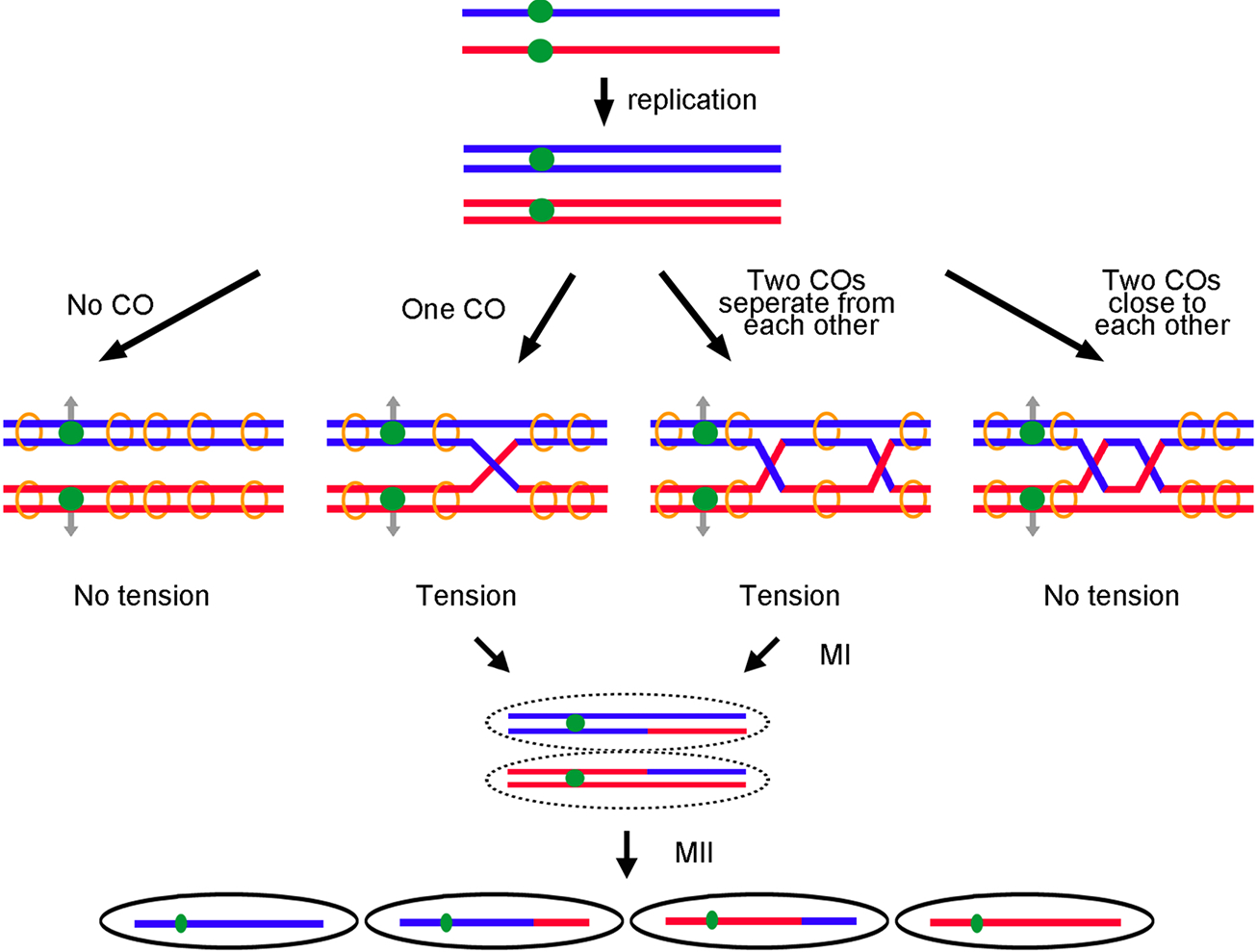
Crossovers are needed for proper chromosome segregation in meiosis. Red and blue lines indicate replicated homologs from each parent. Each line indicates a double-stranded DNA of one chromatid. Green dots represent centromeres. After replication, sister chromatids are held together by cohesins (orange circles). Gray arrows indicate the direction of homologous centromere separation at the first meiotic division (MI). (Left panel) No tension is generated between homologs when no CO is formed. (Middle two panels) Tension is formed between homologs when one CO or two well-separated COs arise. (Right panel) Two COs too close to each other may have too little sister chromatid cohesion between the crossovers to produce tension between homologs to aid segregation. The tension ensures homologous centromeres segregate properly in MI. Crossovers also generate recombinant haploid gametes after the second meiotic division (MII).
Accurate chromosome segregation at the first meiotic division (MI) requires that the two homologs, one from each of the previous parents, find each other and move to opposite sides (poles) of the cell (Figure 1). First, each chromosome is replicated, forming sister chromatids. Then, the replicated chromosomes pair, forming a bivalent (four copies in all). In many species this pairing occurs by homologous recombination between the two copies; there is extensive DNA sequence homology only between homologs, ensuring that each chromosome pairs with its proper partner. Recombination by reciprocal exchange generates a crossover (a cytological chiasma), which connects the two chromosome pairs. When the centromeres of one pair are pulled away from the centromeres of the other pair, tension is generated if there is at least one crossover connecting them and if the sister chromatids are held together by the cohesin complex. Tension appears to be the signal that homologs are properly moving to opposite poles (Nicklas, 1997). Thus, crossovers and cohesion are critical for proper segregation and gamete formation.
Crossovers can occur at most places along a chromosome, but some regions have crossovers much more frequently than others (reviewed in Nambiar et al., 2019). For example, crossovers are rare in or near centromeres, presumably because a crossover there interrupts proper attachment of the fibers (the spindle) to the centromere (via the kinetochore) or the proper pulling of the centromeres to opposite poles (e.g., Nambiar and Smith, 2018). In addition, two crossovers too close together may leave too little cohesin between the crossovers to firmly hold the bivalent together to generate tension upon centromere movement.
This last feature – the placement of crossovers not too close together – is called crossover interference. This chapter discusses the occurrence of interference, ways it can be measured, and models for its action. Interference was first observed in the fruit fly Drosophila melanogaster (Sturtevant, 1915; Muller, 1916) and has been found in nearly all species examined (Perkins, 1962; Mortimer and Fogel, 1974; Barnes et al., 1995; Anderson et al., 2001; Copenhaver et al., 2002; Meneely et al., 2002; de Boer et al., 2006; Baudat and de Massy, 2007; Fowler et al., 2018). The molecular mechanism of interference is still a matter of speculation, although many gene products required for interference have been identified. We first describe methods for assaying interference and discuss complications in interpreting the data, especially given differences among species. We then present proteins required for interference and finally discuss various models for interference.
Methods for measuring crossover interference
Genetic analysis
Two genetic methods to measure crossover interference use either random gametes (spores or next generation progeny) or complete meiotic tetrads. The former has been used with essentially all species investigated, and tetrad analysis is common with fungi. Recently, tetrad analysis has been done in other species, including Arabidopsis thaliana (see below) (Francis et al., 2006), Zea maize (Li et al., 2015), and Mus musculus (Cole et al., 2014). It is generally easier to get more data from random gametes than from tetrads, but in tetrads chromatid relations can be assessed and crossovers can be distinguished from gene conversions (transfer of a genetic marker from one homolog to the other with or without formation of an associated crossover). Tetrads with fewer than four viable spores are often excluded from the analyses; this bias may overlook important events associated with crossovers that, when disrupted, lead to spore death.
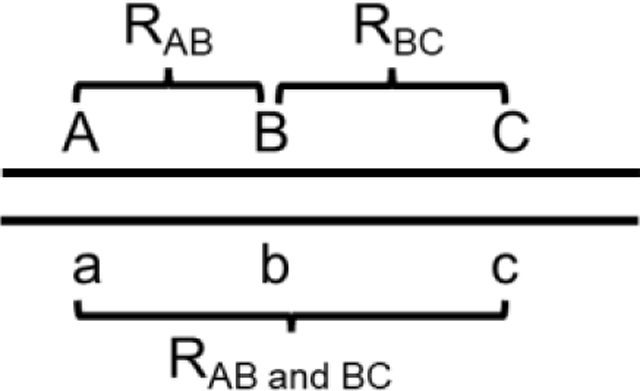
Using random gametes, the coefficient of coincidence (CoC) compares the observed frequency of double crossovers with that expected from independent crossovers. First used in D. melanogaster (Sturtevant, 1915; Muller, 1916) and subsequently in many species, it assumes recombinant chromosomes reflect crossovers and not gene conversion and it assumes no chromatid interference (i.e., that 2-, 3-, and 4-strand double crossovers occur 1:2:1; Figure 2A). CoC using three linked markers is given by the formula
where values are the frequencies of single ( and ) and double () crossovers from a cross of this form:
Fig. 2.
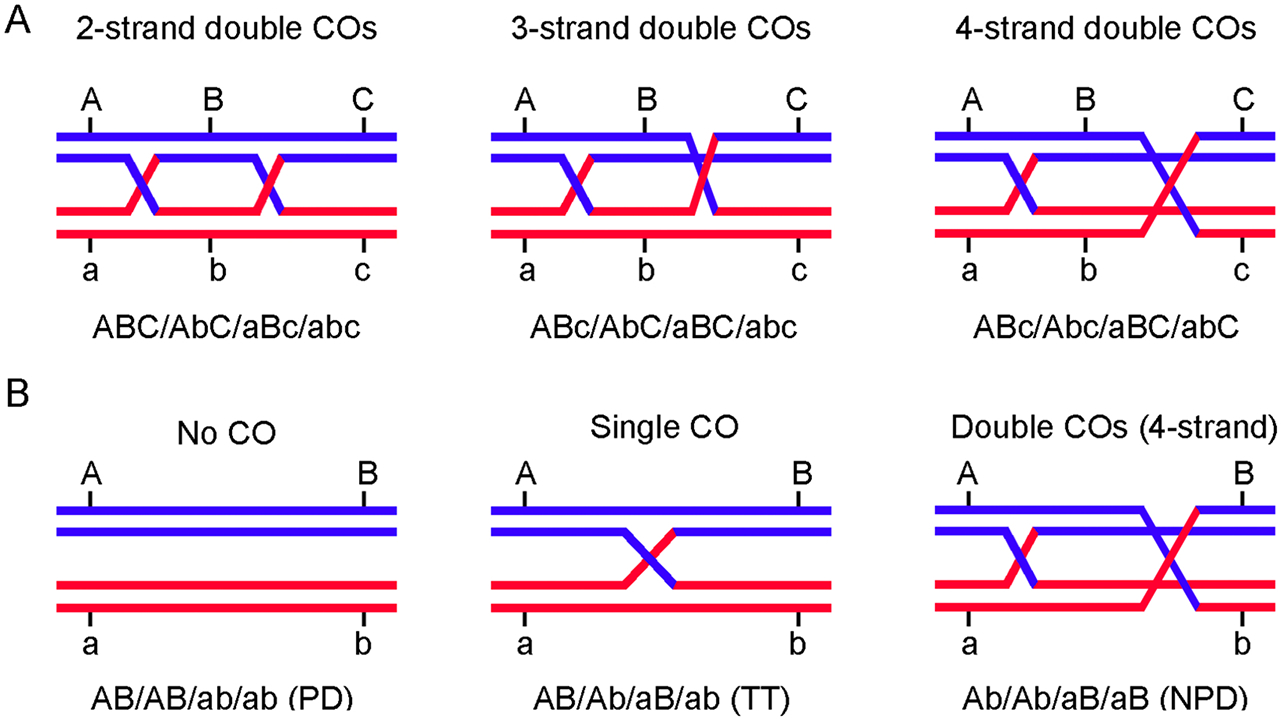
Characterizing crossovers in meiotic tetrads with genetic markers. (A) Three-marker tetrad analysis can identify two-, three-, or four-strand (chromatid) double crossovers. (B) Three types of tetrads in two-marker tetrad analysis. PD, parental di-type; TT, tetra-type; NPD, non-parental di-type. The genotypes of the four gametes are indicated below each panel.
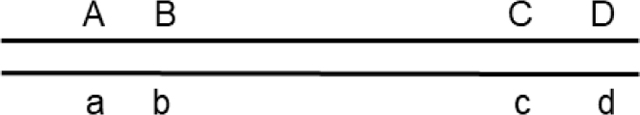
A variation uses a fourth marker to allow comparisons with the two intervals assayed for crossovers (A-B and C-D) to be kept nearly constant in size but moved farther and farther apart.
In both cases, interference (I) = 1 – CoC. In D. melanogaster and Neurospora crassa S4 is lowest at short genetic distances and increases to near unity at ~40 – 50 centiMorgans (cM); in other words, in these species interference is strong at short distances and disappears at about the distance of one crossover per meiotic cell (50 cM) (Foss et al., 1993) (Figure 3).
Fig. 3.
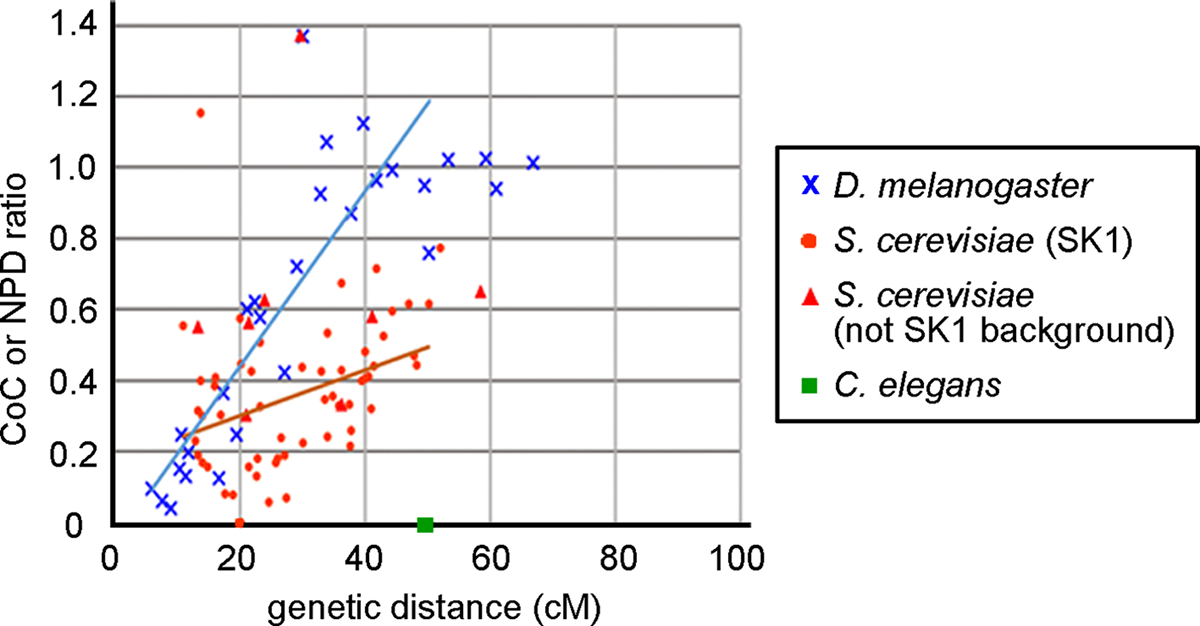
Correlation between crossover interference and genetic distance in D. melanogaster, S. cerevisiae and C. elegans. D. melanogaster CoC (S4) data (Foss et al., 1993; Morgan, Bridges, & Schultz, 1935), S. cerevisiae NPD ratio data (Argueso et al., 2003; Argueso, Wanat, Gemici, & Alani, 2004; Chua and Roeder, 1997; He et al., 2020; Novak, Ross-Macdonald, & Roeder, 2001; Oh et al., 2007; Shinohara, Sakai, Shinohara, & Bishop, 2003; Stahl et al., 2004; Sym & Roeder, 1994; Tsubouchi, Zhao, & Roeder, 2006; Zanders & Alani, 2009) and the genetic distance of each interval are plotted. C. elegans makes only one crossover per chromosome (Barnes et al., 1995; Meneely et al., 2002), indicating it has complete interference up to 50 cM, the genetic length of each chromosome. Most S. cerevisiae data were analyzed in the SK1 background (red circles), and eight data points were analyzed in other strain backgrounds (red triangles). N. crassa data are similar to those of D. melanogaster (Foss et al., 1993). D. melanogaster data with < 5 double recombinants and S. cerevisiae data with < 5 expected and < 5 observed NPD tetrads were not included. S. cerevisiae data were re-analyzed using the “better way” of Stahl (2008). The blue and orange lines, respectively, show the linear regression analysis of data from D. melanogaster (y = 0.025x − 0.053; r2 = 0.72) and S. cerevisiae (y = 0.0064x + 0.17; r2 = 0.17); for this analysis, data with > 50 cM and S. cerevisiae data with NPD ratio > 1 were omitted.
Fungi and some non-vascular plants allow convenient tetrad analysis, as does the flowering plant A. thaliana mutant qrt1, which retains the four pollen from one meiosis in a single cell (Rhee and Somerville, 1998; Francis et al., 2006). CoC analysis, as above, could be used with tetrad data, but a simpler alternative uses only two markers denoting the three tetrad classes (Figure 2B). The observed frequency of non-parental ditype (NPD) tetrads reflects 4-strand double crossovers (Figure 2A). Assuming no chromatid interference, as is the case where tested (Emerson and Beadle, 1933; Mortimer and Fogel, 1974; Munz, 1994; Zhao et al., 1995; Stahl and Housworth, 2009; Miller et al., 2016), and no crossover interference, the expected frequency (f) of NPD, tetratypes (T; single crossover) and parental ditypes (PD; no crossover) can be calculated from the Haldane relation of genetic distance (in Morgans) and the observed recombinant frequency R.
The NPD ratio = NPDobserved/NPDexpected, and I = 1 – NPD ratio.
Papazian (1952) used Haldane’s formula to calculate the expected NPD frequency from the observed T frequency.
Stahl (2008) extended this analysis to include the expected frequencies of each of the three classes based on the observed recombinant frequency and Haldane’s equation.
Papazian’s analysis is not valid for T close to 2/3, but Stahl’s “better way” is valid for all values. In addition, Stahl’s analysis, based on all three tetrad classes, gives more reliable statistical analysis of the data than Papazian’s (Stahl, 2008).
Two studies in yeasts (Munz, 1994; Shinohara et al., 2003) found similar degrees of interference using both random spore and tetrad (NPD; Papazian method) analyses, as expected.
Cytological analysis
A physical (cytological) method determines the position of fluorescent markers, seen as foci in a microscope, along a chromosome. Typically, the chromosomal axis is labeled uniformly, to allow distance measurements along the chromosome, and a chromosomal end is labeled uniquely, to identify an individual chromosome. A chromosome is divided into many equal-length intervals, and the frequencies of foci in each interval and the frequencies of two foci in each pair of intervals are determined. Interference is seen as foci being farther apart than expected from random positioning. Fung et al. (2004) found that synapsis initiation complex proteins (e.g., Saccharomyces cerevisiae Zip2 and Zip3) showed interference. Similar markers used are those thought to indicate “crossover designation,” for example Caenorhabditis elegans COSA-1, mouse MLH1, and A thaliana MLH1 (Anderson et al., 1999; Chelysheva et al., 2010; Yokoo et al., 2012). S. cerevisiae Zip3 promotes CO formation, is required for CO interference (Table 1), and has been proposed to bind to DSB sites after they are “designated” by an unknown factor for CO formation (Agarwal and Roeder, 2000; Serrentino et al., 2013). C. elegans COSA-1 and A. thaliana MLH1 are located between bivalents at or near a chiasma (Chelysheva et al., 2010; Yokoo et al., 2012). COSA-1 is also needed for CO formation (Yokoo et al., 2012). The chromosomal distribution of mouse and A. thaliana MLH1 foci reflect the number and distribution of crossovers (Anderson et al., 1999; Chelysheva et al., 2010). These observations support these foci being a valid proxy for crossovers.
Table 1.
Proteins with diverse functions are required for crossover interference
| Protein | Species | Function | Interference assay | Reference |
|---|---|---|---|---|
| Synaptonemal complex (SC) | ||||
| Zip1 | S. cerevisiae | Transverse filament of the SC | NPD ratio | Sym and Roeder, 1994 |
| SYP-1 | C. elegans | Cytology | Libuda et al., 2013 | |
| Zyp1 | A. thaliana | Cytology | Capilla-Pérez et al., 2021 | |
| Asy1 | A. thaliana | Cytology | Lambing et al. 2020 | |
| Zep1 | O. sativa | Cytology | Wang et al., 2010 | |
| Sycp1 | M. musculus | Cytology | de Boer et al., 2006 | |
| DPY-28 | C. elegans | Condensin I subunit; chromosomal axis formation | Cytology | Tsai et al., 2008 |
| Regulating CO formation | ||||
| Msh4-Msh5 | S. cerevisiae | Binds Holliday junctions; facilitates CO formation | NPD ratio | Novak et al., 2001; Argueso et al,. 2004; Getz et al., 2008 |
| NPD ratio and CoC | Nishant et al., 2010 | |||
| CoC | Anderson et al., 2015 | |||
| Zip3 | S. cerevisiae | SUMO E3 ligase; required for SC formation and CO formation | CoC | Anderson et al., 2015 |
| Mlh1-Mlh3 | S. cerevisiae | Endonuclease; binds and resolves Holliday junctions | NPD ratio | Argueso et al,. 2003 |
| Slx4 | S. cerevisiae | Endonuclease; resolves Holliday junctions | NPD ratio | Higashide and Shinohara, 2016 |
| Sgs1 | S. cerevisiae | DNA helicase; chromosome synapsis; meiotic joint molecule and CO formation | NPD ratio | Oh et al., 2007 |
| CoC | Anderson et al., 2015 | |||
| Blm | D. melanogaster | CoC | Hatkevich et al., 2017 | |
| Mer3 | S. cerevisiae | DNA helicase; promotes CO formation | NPD ratio | Nakagawa and Ogawa, 1999 |
| Spo16 | S. cerevisiae | Facilitates SC and CO formation | NPD ratio | Shinohara et al., 2008 |
| Spo22 (Zip4) | S. cerevisiae | TPR-like repeat protein; facilitates SC formation and CO formation | NPD ratio | Tsubouchi et al., 2006; Shinohara et al., 2008 |
| Pch2 | S. cerevisiae | Regulating chromosome synapsis and CO formation | NPD ratio | Joshi et al., 2009; Zanders and Alani, 2009 |
| Pch2 | A. thaliana | CoC | Lambing et al., 2015 | |
| RTEL-1 | C. elegans | CO formation | Cytology | Youds et al., 2010 |
| DNA damage check point | ||||
| Tel1 | S. cerevisiae | Protein kinase regulating DNA double-strand-break response | CoC | Anderson et al., 2015 |
| Tel1 | S. pombe | CoC | Fowler et al., 2018 | |
| ATM | M. musculus | Cytology | Barchi et al., 2008 | |
| Topoisomerase II and SUMOylation | ||||
| Top2 | S. cerevisiae | DNA topoisomerase; binds axial cores | Cytology | Zhang et al., 2014b |
| Ndj1 | S. cerevisiae | Regulating meiotic SPB cohesion and telomere clustering | NPD ratio | Chua and Roeder, 1997 |
| Slx5/8 | S. cerevisiae | SUMO-targeted ubiquitin ligase (STUbL) | Cytology | Zhang et al., 2014b |
| Sir2 | S. cerevisiae | Activation of STUbL activity of Slx5/8 | Cytology | Zhang et al., 2014b |
Interference is measured as in CoC analysis: I = 1 – CoC, where
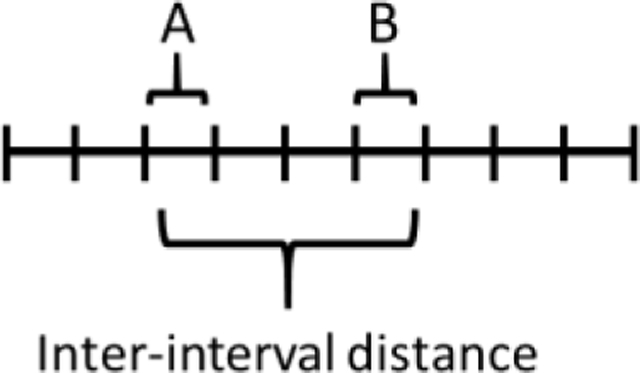
This cytological method does not require genetic markers and can be used in mutants that do not complete meiosis. But it requires spreading nuclei and high-resolution microscopy, which may be difficult for large numbers of cells. In addition, distance is measured as physical distance along the chromosomal axis, which may not be proportional to genetic map or DNA length. In S. cerevisiae, however, cytological interference extends over ~0.45 microns of axis, which corresponds to ~44 cM (Zhang et al., 2014), indicating equivalence of cytological and genetic interference. In A. thaliana and humans, the data are ~25 microns (~61 cM) and ~5 microns (~50 – 60 cM), respectively (Gruhn et al., 2013; White et al., 2017; Capilla-Pérez et al., 2021). [See Table 2 for relations between DNA length (Mb), genetic distance (cM), and axis length (microns) in various species.] It should be noted that several S. cerevisiae mutants (e.g., zip1Δ, sgs1Δ, and msh4Δ) show cytological interference but not genetic interference (Sym and Roeder, 1994; Novak et al., 2001; Fung et al., 2004; Oh et al., 2007). Simple explanations are that a) S. cerevisiae strains from different genetic backgrounds were used to test the two methods and b) these proteins may be required at a stage of crossover formation after crossover “designation” (e.g., Zip3 binding) (De Muyt et al., 2012; Zakharyevich et al., 2012; Zhang et al., 2014b). Nevertheless, these mutants indicate some difference in cytological vs. genetic interference.
Table 2.
Ratio of genetic distance to physical distance varies >500-fold among species
| Species | Genetic map length (cM) | Physical map length (Mb) | Axis length (μm)§ | Genetic length/ physical length (cM/Mb) | Axis length/physical length (μm/Mb) | Genetic length/axis length (cM/μm) |
|---|---|---|---|---|---|---|
|
| ||||||
| S. cerevisiae | 4206a | 12a | 43b | 350 | 3.6 | 98 |
| S. pombe | 2100c | 13.8d | -- | 150 | -- | -- |
| N. crassa | 1075e | 43f | -- | 25 | -- | -- |
| A. thaliana | 597g | 135h | 240i | 4.4 | 1.8 | 2.5 |
| D. melanogaster | 287j | 180k | -- | 1.6 | -- | -- |
| D. melanogaster Chr X | 65.3j | 24k | 15l | 2.8 | 0.63 | 4.4 |
| C. elegans | 300 | 103m | 29n | 2.9 | 0.28 | 10 |
| Humans | 3630o | 3019o | -- | 1.2 | -- | -- |
| Human autosomes | 3451o | 2740o | 260 –320p | 1.3 | 0.09 – 0.12 | 11 – 13 |
| M. musculus | 1373o | 2577o | -- | 0.53 | -- | -- |
| M. musculus autosomes | 1316o | 2320o | 150 – 162q | 0.56 | 0.06 – 0.07 | 8.1 – 8.8 |
Axis length was measured by light microscopy of formaldehyde-fixed samples.
Wood et al. (2003)
Lascarez, Lagunas and Colaiácovo, unpublished data
Complications in interpreting interference data
While it is often said that this or that species has crossover interference, the reported strengths clearly differ among species and often across a given genome (Figure 3). Some, such as C. elegans, appear to have absolute interference – two crossovers on one chromosome do not appear (Barnes et al., 1995; Meneely et al., 2002). Extensive data show strong interference for short intervals (<10 cM) in D. melanogaster (~0.95) and in N. crassa (~0.8), gradually decreasing for longer intervals, and becoming negligible at ~40 -- 50 cM (Foss et al., 1993) (Figure 3). S. cerevisiae interference ranges from near 0 up to ~0.8, but among data analyzed in Figure 3 there is a less distinct trend with distance: intervals of similar genetic size can have different degrees of interference. According to the only report of which we are aware (Strickland, 1958), Aspergillus nidulans has no detectable crossover interference. Schizosaccharomyces pombe has statistically highly significant but weak crossover interference (0.26 ± 0.051) in the one pair of adjacent intervals assayed extensively (Fowler et al., 2018); other intervals may have no interference, but the data for most individual intervals reported are limited (Munz, 1994).
The ratio of genetic distance (cM) to DNA length (Mb) differs by a factor of >500 among species (Table 2), indicating that a similar level of interference in two species may reflect vastly different DNA distances. In addition, each micron of cytological axis length corresponds to different lengths of DNA. A simple interpretation is that DNA loops of increasing size extend from the axis as the parameter (micron of axis length/Mb) decreases. Consequently, different genetic distances correspond to different axis lengths among species. In multicellular species the parameter (cM/micron of axis) ranges from about 2 to 10; in S. cerevisiae, the value is about 100 (Table 2).
Interpreting these data is complicated by additional considerations (see also Berchowitz and Copenhaver, 2010). Gene conversions, whose formation is closely related to that of crossovers, are often stated not to interfere with each other. Mortimer and Fogel (1974) and Foss and Stahl (1995) report CoC = 1.0 (95% confidence interval, or CI, = 0.75 – 1.45 or wider) for gene conversions at the closely linked ARG4 and THR1 genes of S. cerevisiae. Because the number of double gene conversions is small (13 and 28, respectively), the data do not rule out weak interference, either positive or negative. Sequencing 980 chromatid arms from progeny of D. melanogaster hybrids, Miller et al. (2016) found 291 chromatid arms with one or more gene conversions and 33 with two or more gene conversions. 86 doubles are expected from independence; thus, CoC = 0.38 (CI = 0.28 – 0.49), indicating highly significant positive interference of gene conversions. Using a different method of analysis, the authors, however, concluded that gene conversions do not show interference. Miller et al. (2016) found 541 crossovers and 52 double crossovers; CoC = 0.17 (CI = 0.13 – 0.22), indicating strong crossover interference, as concluded by Miller et al. (2016) and others cited above for D. melanogaster. Similarly, for crossovers and non-crossovers (124 such doubles observed), CoC = 0.77 (CI = 0.63 – 0.91), indicating weak but significant interference of these events. Some but not other classes of gene conversion (6:2, 5:3, or aberrant 4:4) with an associated crossover also interfere with a nearby crossover in another interval in some species but not in others (Kitani, 1978; Mancera et al., 2008; Getz et al., 2008; Stahl and Foss, 2008). These results may reflect different relative activities of two pathways of recombination, one with and one without interference, as discussed in the next paragraph. Although the original definition of interference referred to genetic crossovers, more recently emphasis has been put on spatial interference of cytological markers along chromosomes, as described above. These foci are inferred to mark “designated” crossover sites, which may be closely related to, but not guaranteed to become, crossovers, as noted above. Thus, it is important to specify what is measured to indicate interference.
An added complication is the possibility of two or more types of meiotic recombination occurring in the same species. In S. cerevisiae, A. thaliana, and mice two pathways of crossover formation have been shown – one with and one without interference (e.g., Ross-Macdonald and Roeder, 1994; Copenhaver et al., 2002; de los Santos et al., 2003; Argueso et al., 2004; Higgins et al., 2004; Stahl et al., 2004). Direct support for this view comes from studies of S. cerevisiae zip1 and msh5 mutants, in which the residual crossovers (20 – 40% of wt frequency) show little if any interference (Sym and Roeder, 1994; de los Santos et al., 2003). In an mms4 mutant, defective in another postulated pathway, interference is not significantly different from that in wt (de los Santos et al., 2003). Some authors have proposed that the non-interfering pathway acts early, to establish chromosome pairing, and the interfering pathway acts late, to facilitate chromosome disjunction (de los Santos et al., 2001; de los Santos et al., 2003; Getz et al., 2008). In some species, crossing over aids both events, but the degree to which the pathways differ is unclear. They could diverge at the beginning of the recombination pathway (before DSB formation) or at the end (resolution of Holliday junctions). For example, nicked Holliday junctions could arise in the “early” pathway and be resolved by Mus81-Mms4, while non-nicked (ligated) Holliday junctions could arise in the “late” pathway and be resolved by Msh4-Msh5 and Mlh1 and its partners (e.g., Getz et al., 2008; Berchowitz and Copenhaver, 2010). These pathways could act differentially in different genomic intervals, further complicating the interpretation of interference strength in a given species.
Important to keep in mind is that species differ. For example, Mus81-Eme1 is essential for Holliday junction resolution and >95% of crossover formation in S. pombe (Boddy et al., 2001; Smith et al., 2003; Cromie et al., 2006), but its homolog Mus81-Mms4 is required for only ~20% of crossovers in S. cerevisiae (de los Santos et al., 2003; Argueso et al., 2004), although it is required for abundant viable spore-formation in both species. Some proteins essential for recombination in some species, such as Dmc1 DNA strand-exchange protein, are not apparent in other species. [Among tested species, those with Dmc1 require recombination for chromosome synapsis, but those without it do not (Stahl et al., 2004)]. We know of no species in different genera in which the same set of orthologous proteins is required for meiotic recombination, emphasizing that species differ. Thus, it is risky to use data from one species in interpreting data from another species. Rather, one should state the species under discussion and not dismiss a view proposed for one species because of counter data in another species. Observations and models of interference in any species are nevertheless important for thinking about the mechanism of interference in other species.
Proteins Required for Crossover Interference
Mutants with altered crossover interference have been identified in a variety of species. Table 1 groups the corresponding proteins according to their known functions in meiotic recombination and DNA metabolism. Homologs of some of these proteins are found in widely divergent species, suggesting some underlying similarity in interference. The molecular mechanisms by which these proteins impart interference remain largely unknown.
Models of Crossover Interference
Numerous models for crossover interference have been proposed. Many have led to mathematical models that account for existing data, such as the relation of interference and genetic or cytological distance, but few have specified a molecular mechanism, including roles for proteins required for interference (Table 1). Some models can be considered genetic, in the sense of having properties related to genetic distance, or physical, in the sense of measuring kb of DNA or microns of chromosome axis in fully condensed chromosomes. In both cases, however, the features of the models often account for interference allowing exactly one crossover in the genetic distance (50 cM) required by the definition of cM and over which interference acts, at least in D. melanogaster and N. crassa (Figure 3). (50 cM, or 0.5 Morgan, is the genetic separation generated by one crossover in a bivalent, in which half the chromosomes are recombinant in the defined interval; Figure 2B). Here, we discuss models in several classes. Words or phrases in quotation marks below are from the references cited.
Trigger model
Fox (1973) postulated that a “chiasma determination mechanism passes along the chromosome at a constant rate” and is “triggered” to form a chiasma. It continues to move along the chromosome and “requires a fixed time to become recharged” to form another chiasma. The trigger was proposed to be “some feature of the secondary structure of the chromosome,” which is rare in chromosomal regions with few chiasmata, such as the heterochromatic centromeres. Although further descriptions and predictions of the mechanism were not provided, this may be the first model postulating an entity moving along the chromosome to promote a limited number of crossovers in a given region.
Polymerization models
Egel (1978) proposed that “nodes” (required for the “establishment of an exchange possibility”) are formed only before chromosome synapsis and “serve as initiation centres at which synapsis is started.” One node, presumably activated at random, starts polymerization of the synaptonemal complex (SC). As the SC grows by polymerization, it inactivates additional nodes, thereby preventing further crossovers in the region over which there was SC polymerization.
King and Mortimer (1990) also proposed a model involving polymerization, but one in which the polymer (separate from the SC) both promotes and inhibits rather than simply inhibits crossover formation. In their model, “early structures” bind randomly along the already established SC and are held within it, thereby preventing interaction with another chromosome. Each structure initiates bidirectional polymerization and “give[s] rise to the late recombination nodules,” which initiate crossover formation. The growing polymer then prevents other early structures from binding or ejects those already bound but not yet polymerizing. The ejected structures can act on other SC regions without a structure-induced polymer. This feature assures that all chromosomal regions have the opportunity for crossovers, provided there are enough structures present. The nature of the early structures, the ensuing polymer, and nodules was not specified.
Reaction-diffusion models
In this class of models, a factor, such as a protein, diffuses in one dimension along a chromosome or bivalent for a limited distance and affects crossover formation, either positively or negatively. This factor may be restricted by the SC from moving to another bivalent. Its effect is thus limited to the chromosome or bivalent it is on, as expected for imposition of interference along but not between chromosomes.
Holliday (1977) proposed that an activating factor, limited in amount, converts an unstable crossover into a stable chiasma. The factor is proposed to be a protein that binds DNA cooperatively and forms a complex with the crossover to stabilize it. It does not interact with a non-crossover structure. Diffusion of the protein along the chromosome to the crossover site depletes the surrounding region and prevents formation of a second stable chiasma in that region. Although not stated in the model, presumably the factor is initially randomly distributed along the chromosome. Organisms that lack interference, such as A. nidulans (Strickland, 1958), are proposed to have an unusually high level of the activating protein. To account for interference not extending across centromeres in some species, Holliday proposed that the centromere is a barrier to diffusion of the protein.
Fujitani et al. (2002) proposed that “random walkers” are randomly distributed along chromosomes; each diffuses randomly in one dimension and with a set rate “becomes immobilized and matures into a crossover point.” Collision of two random walkers inactivates both; collision of a moving walker with an immobilized walker inactivates the moving one. This model, like those above, generates exactly one crossover in an interval of 50 cM. Mathematical models built on two parameters (the initial density of walkers and the rate at which a walker becomes immobilized) account for data in D. melanogaster and N. crassa. As in other models above, the nature of the walkers and the means by which they mature into crossovers were not stated.
Rog et al. (2017) found that proteins of the C. elegans SC behave as though they are in liquid crystals – they are sensitive to 1,6-hexanediol, which disrupts such structures, and quickly diffuse from the surrounding area into a particular region (as observed after localized irradiation of a photoconvertible derivative of the SC protein SYP-3). The latter property of liquid crystals is similar to that of lipid membranes, in which proteins can readily diffuse within, but not out of, the membrane. This property would allow a protein to diffuse along the chromosome and interact with sites on that chromosome but not with those on another chromosome, as demanded for crossover interference. Thus, Zhang et al. (2018) propose that SC proteins, including ZHP-3, diffuse in one dimension and a combination of positive and negative effects allows only limited numbers of crossovers (one for C. elegans) to form along a chromosome. Additional proteins required for interference include the SC protein SYP-1, the condensin subunit DPY-28, and the protein kinase RTEL-1 (Table 1). Homologs of SYP-1 and RTEL-1 are also required for interference in other species, suggesting that the C. elegans mechanism may be widespread.
Morgan et al. (2021) showed that the HEI10 protein of A. thaliana behaves as predicted by a reaction-diffusion model for an activator of crossover formation. HEI10 is initially nearly uniformly distributed along each bivalent and then condenses into several small foci and eventually into one to three foci, approximating the number of genetic crossovers per bivalent in other experiments. How HEI10, a putative ubiquitin E3 ligase and Zip3 homolog, promotes crossovers is unclear.
Counting models
Foss et al. (1993), like previous authors, pointed out that interference in D. melanogaster and N. crassa is strong for short genetic intervals but diminishes to nearly zero for intervals ~40 – 50 cM or greater (Figure 3). Note that 50 cM corresponds to about 20 Mb in D. melanogaster but only 2 – 3 Mb in N. crassa (Table 2). Foss et al. (1993) therefore sought an interference mechanism that measures genetic, not physical, distance. In their model “recombination-initiation events” are randomly placed along chromosomes. By unstated mechanisms, one is matured into a crossover, the next m events in one direction along the chromosome are matured into non-crossovers (gene conversions or sister chromatid interaction), and the next event is matured into a crossover. The authors developed mathematical equations with the variable m and found that data for D. melanogaster and N. crassa were fit well with m = 4 and 2, respectively. Mortimer and Fogel (1974) proposed that “recombinogenic events are distributed along the chromosome in an independent fashion” but with alternating crossovers and non-crossovers; this model is equivalent to the counting model of Foss et al. (2003) with m = 1.
The counting model predicts that between two neighboring crossovers there should be m non-crossovers. Short intervals between two crossovers should be enriched for non-crossovers relative to the same intervals on chromosomes without flanking crossovers. Foss and Stahl (1995) tested this prediction in S. cerevisiae and found it did not hold. Rather, there were fewer non-crossovers (gene conversions) in the short interval in tetrads with flanking crossovers compared to those without flanking crossovers, suggesting interference between crossovers and non-crossovers, as noted above.
Mechanical models
Interference might result from a purely mechanical, rather than biochemical, property of a chromosome. Muller (1916) proposed that a chromosome is too stiff to bend near an existing crossover and thus allow formation of another crossover nearby. Kleckner et al. (2004) compared a chromosome to a stiff beam coated with a brittle ceramic film. Bending the beam would put increasing stress on the film until it cracked and relieved the surrounding stress in the film. By analogy, they proposed that formation (or designation) of a crossover requires stress (the opposite of Muller’s proposal) resulting from chromosome expansion and that stress in turn is relieved by crossover formation. Thus, a crossover would not form until stress built up, but once a crossover formed, there would be no stress nearby to allow formation of a second crossover. Stress would monotonically increase with distance from the first crossover, thus allowing, with increasing probability, another crossover some distance away. Support for this model comes from the partial reduction of interference, measured as inter-focus distance between Zip3 foci, in a top2 mutant deficient in DNA topoisomerase II, which relaxes supercoiled DNA (Zhang et al., 2014b).
In an expansion of the beam-film model (Zhang et al., 2014a), certain “precursors” are “designate[d] to eventually mature as a crossover.” Thus, the act of designation imposes interference. “The precursors for CO patterning are generally assumed to be the total array of double strand break (DSB)-initiated interactions between homologs,” and “yeast DSBs are evenly spaced.” DSBs become “designated” to become crossovers by an unstated mechanism but are reflected by the distribution of Zip3 foci (see Cytological analysis above). The probability (frequency) curve resembles that of CoC vs. cM in D. melanogaster (Figure 3). In S. cerevisiae, the distance at which adjacent Zip3 foci occur at the frequency expected for independence is roughly 100 kb or about 50 cM. Kleckner et al. (2004) emphasize, however, that crossover designation (interference) measures axis length, not kb of DNA or cM of genetic map length.
The beam-film model has been used to formulate mathematical models for interference (Zhang et al., 2014a). These models use multiple parameters reflecting such features as the average number of precursors per chromosome, their distribution along chromosomes (random or even or intermediate), and the “designation driving force.” Data from various species fit the model when appropriate values for these parameters are chosen. With nine adjustable parameters, the model fits the data from various species (Zhang et al., 2014a).
Because few molecular (biochemical) features of the beam-film model are specified, it is hard to test this model genetically. The presumed even spacing, by an unstated mechanism, of precursors (DSBs) in yeast would seem to be interference itself, perhaps one of several factors leading to crossover interference. In addition, it seems counterintuitive that crossovers rather than DSBs would relieve stress in a chromosome. The model does, however, offer a different way of thinking about the problem of crossover interference.
Chromosome oscillatory movement model
Hultén (2011) viewed chromosomes as long flexible entities that move with wave-like motions. Chromosome ends (telomeres, attached to the nuclear membrane) and centromeres (attached to the kinetochore and spindle apparatus) move vigorously as the homologs pair. These movements create waves along each chromosome (univalent). When the “nodes” (crests) of two waves on homologs meet, they engage and form a crossover. Because the adjacent chromosome regions are not juxtaposed, no crossovers occur there. This model accounts for chromosomal translocations and other large rearrangements, such as deletions and inversions, blocking recombination and interference in the flanking regions.
Clustering models
Most of the models above do not specify the proteins involved in interference. The clustering model proposed by Fowler et al. (2018) specifies particular proteins involved in several steps. This model is based on their finding that S. pombe linear element proteins (LinE proteins; Rec25, Rec27, and Mug20), in conjunction with another linear element protein Rec10, bind to DSB hotspots with high specificity and are required for DSB formation at most hotspots (Fowler et al., 2013). Rec10 is more uniformly distributed and is required for all DSB formation. Loading of LinE proteins onto chromosomes is promoted by cohesins containing meiosis-specific subunits Rec8 and Rec11. These six proteins are required for full levels of recombination. A type of Hi-C analysis showed that Rec27-bound DSB hotspots form 3D clusters over ~200 kb (Fowler et al., 2018). A DSB at one hotspot interferes with DSB formation at surrounding hotspots; this DSB interference, like hotspot clustering, is strongest for nearby hotspots and decreases to an undetectable level at ~200 kb, corresponding to ~35 cM (Fowler et al., 2018) (Table 2). DSB interference also occurs in S. cerevisiae, extending ~20 – 50 kb, corresponding to ~7 – 20 cM (Garcia et al., 2015). Fowler et al. (2018) proposed that DSB interference gives rise to crossover interference, just as DSBs give rise to crossovers.
In the model of Fowler et al. (2018) a protein complex, such as condensin or cohesin or both, binds to a site on a chromosome and, while remaining bound to that site, moves unidirectionally along the chromosome and forms an ever-growing loop (Hyppa et al., 2021). Upon encountering a potential DSB hotspot in the chromosome (or pair of sister chromatids or pair of homologs) being moved along, the protein complex loads a LinE complex onto the hotspot(s), holds onto that site, and continues moving. Upon encountering the next potential hotspot, it loads another LinE complex with a probability proportional to the strength of that hotspot. After it has loaded a set number of LinE complexes at hotspots, perhaps only two, it ceases loading. A DSB formed at random in the cluster of hotspot-LinE complexes activates the Tel1 protein kinase, which blocks further DSB formation in that cluster by phosphorylating a member of the DSB-forming complex (Rec12 and its half dozen essential partners). The DSB is repaired to a crossover. Because additional DSB formation in the surrounding area (the chromosomal region in the loop) is blocked, so is crossover formation. Formation of clusters encompassing only one homolog (paired chromatids) would allow independent DSBs on the other homolog, and interference would be limited to 0.5, but clusters encompassing both homologs would allow complete interference (I = 1). Formation of clusters over different regions in individual cells would make a gradient, from complete interference to none, in the population of cells, as typically used for assessing crossover interference genetically.
In mitotic S. pombe cells, condensin forms topologically associated domains (TADs) over chromosomal regions of ~300 kb; cohesin forms TADs over ~80 kb (Kim et al., 2016). These distances are about the distance over which DSB interference is observed (~200 kb, as noted above). An S. pombe meiotic cell forms about 60 DSBs over its 13 Mb genome, or 1 DSB per ~200 kb (Fowler et al., 2014). Thus, these numbers are consistent with the model. In a tel1Δ mutant, both DSB interference and crossover interference become negative (Fowler et al., 2018), indicating that Tel1 is required for interference. Negative interference may arise from two or more DSBs being formed in a cluster when Tel1 is not present to inactivate the DSB-forming complex. The result is coordinated action of the DSB-forming complex within a cluster and coordinated crossover formation (negative interference). This model predicts interference between gene conversions, which to our knowledge has not been reported in S. pombe. In the one report of which we are aware, noted above, gene conversions do not show interference in S. cerevisiae (Foss and Stahl, 1995), but the data do not exclude low level interference (i.e., CoC = 1.0 ± 0.3).
Stahl et al. (2004) proposed that “neighboring intermediates (-to-be) [also called “attempts”] are gathered into clusters of more or less fixed size” and that exactly one member of this cluster is converted into a crossover. These clusters were proposed to be foci of Zip2-Zip3 or “late nodules” observed by electron microscopy on the SC in D. melanogaster (Carpenter, 2003). The mode of clustering and the molecular mechanism limiting crossovers to one per cluster were not specified.
Conclusions and future research
Crossover interference has been well documented in many species and to require a variety of proteins. The molecular mechanism remains to be determined in any species. Given the differences in interference strength (Figure 3) and protein requirements (Table 1) in various species, it seems likely that there are multiple mechanisms. Interference strength is clearly distance-dependent in D. melanogaster but less so in S. cerevisiae and C. elegans (Figure 3), suggesting that the mechanisms of interference may be quite different. Determining the mechanisms is a challenge requiring a combination of methods, including genetic and biochemical as well as cytological analyses.
Acknowledgements
We are grateful to Sue Amundsen, Randy Hyppa, and two anonymous reviewers for helpful comments on the manuscript; and Monica Colaiácovo for unpublished data. Our research is supported by research grant R35 GM118120 from the National Institutes of Health of the United States of America.
References
- Adams MD, Celniker SE, Holt RA, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Agarwal S, and Roeder GS. 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 102:245–255 [DOI] [PubMed] [Google Scholar]
- Anderson CM, Oke A, Yam P, Zhuge T, and Fung JC. 2015. Reduced crossover interference and increased ZMM-independent recombination in the absence of Tel1/ATM. PLoS Genet. 11:e1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Hooker KD, and Stack SM. 2001. The distribution of early recombination nodules on zygotene bivalents from plants. Genetics. 159:1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Reeves A, Webb LM, and Ashley T. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 151:1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso JL, Kijas AW, Sarin S, Heck J, Waase M, and Alani E. 2003. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol Cell Biol. 23:873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso JL, Wanat J, Gemici Z, and Alani E. 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics. 168:1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes TM, Kohara Y, Coulson A, and Hekimi S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 141:159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, and de Massy B. 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Research. 15:565–577. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, and Copenhaver GP. 2010. Genetic interference: Don’t stand so close to me. Current Genomics. 11:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, and Russell P. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 107:537–548. [DOI] [PubMed] [Google Scholar]
- Capilla-Pérez L, Durand S, Hurel A, et al. 2021. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc Natl Acad Sci USA. 118:e2023613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AT 2003. Normal synaptonemal complex and abnormal recombination nodules in two alleles of the Drosophila meiotic mutant mei-W68. Genetics. 163:1337–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- elegans Sequencing Consortium C. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 282:2012–8. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Grandont L, Vrielynck N, le Guin S, Mercier R, and Grelon M. 2010. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res. 129:143–153. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, and Botstein D. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 387:67–73. [PMC free article] [PubMed] [Google Scholar]
- Chua PR, and Roeder GS. 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11:1786–1800. [DOI] [PubMed] [Google Scholar]
- Codina-Pascual M, Campillo M, Kraus J, Speicher MR, Egozcue J, Navarro J, & Benet J 2006. Crossover frequency and synaptonemal complex length: their variability and effects on human male meiosis. Molecular human reproduction, 12:123–133. [DOI] [PubMed] [Google Scholar]
- Cole F, Baudat F, Grey C, Keeney S, de Massy B and Jasin M 2014. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nature Genetics, 46:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S (2012) The many landscapes of recombination in Drosophila melanogaster. PLOS Genetics 8:e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Housworth EA, and Stahl FW. 2002. Crossover interference in Arabidopsis. Genetics. 160:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, and Smith GR. 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell. 127:1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Stam P, Dietrich AJ, Pastink A, and Heyting C. 2006. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci U S A. 103:9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Loidl J, Larkin B, and Hollingsworth NM. 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Sacchromyces cerevisiae. Genetics. 159:1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, and Hollingsworth NM. 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 164:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, and Lichten M. 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell. 46:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R 1978. Synaptonemal complex and crossing-over: structural support or interference? Heredity. 41:233–237. [DOI] [PubMed] [Google Scholar]
- Egel R 2004. The molecular biology of Schizosacharomyces pombe. In Genetics, genomics and beyond. Egel R, editor. Springer, Berlin. [Google Scholar]
- Emerson S, and Beadle GW. 1933. Crossing-over near the spindle fiber in attached-X chromosomes of Drosophila melanogaster. Zeitschrift für induktive Abstammungs- und Vererbungslehre. 65:129–140. [Google Scholar]
- Foss E, Lande R, Stahl FW, and Steinberg CM. 1993. Chiasma interference as a function of genetic distance. Genetics. 133:681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, and Stahl FW. 1995. A test of a counting model for chiasma interference. Genetics. 139:1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Gutiérrez-Velasco S, Martín-Castellanos C, and Smith GR. 2013. Protein determinants of meiotic DNA break hotspots. Mol. Cell. 49:983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S and Smith GR 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24:1650–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Hyppa RW, Cromie GA, and Smith GR. 2018. Physical basis for long-distance communication along meiotic chromosomes. Proc Natl Acad Sci U S A. 115:e9333–e9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DP 1973. The control of chiasma distribution in the locust, Schistocerca gregaria (Forskål). Chromosoma. 43:289–328. [DOI] [PubMed] [Google Scholar]
- Francis KE, Lam SY, and Copenhaver GP. 2006. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant physiology. 142:1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Mori S, and Kobayashi I. 2002. A reaction-diffusion model for interference in meiotic crossing over. Genetics. 161:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, and Birren B. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 422:859–868. [DOI] [PubMed] [Google Scholar]
- Garcia V, Gray S, Allison RM, Cooper TJ, and Neale MJ 2015. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 520:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz TJ, Banse SA, Young LS, Banse AV, Swanson J, Wang GM, Browne BL, Foss HM, and Stahl FW. 2008. Reduced mismatch repair of heteroduplexes reveals “non”-interfering crossing over in wild-type Saccharomyces cerevisiae. Genetics. 178:1251–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn JR, Rubio C, Broman KW, Hunt PA and Hassold T 2013. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PloS one, 8:e85075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T, Kohl KP, McMahan S, Hartmann MA, Williams AM, and Sekelsky J. 2017. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr Biol. 27:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Rao H, Tang S, Bhagwat N, Kulkarni DS, Ma Y, Chang MAW, Hall C, Bragg JW, Manasca HS, Baker C, Verhees GF, Ranjha L, Chen X, Hollingsworth NM, Cejka P, and Hunter N. 2020. Regulated proteolysis of MutSγ controls meiotic crossing over. Mol Cell. 78:168–183.e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashide M, and Shinohara M. 2016. Budding yeast SLX4 contributes to the appropriate distribution of crossovers and meiotic double-strand break formation on bivalents during meiosis. G3 (Bethesda, Md.). 6:2033–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC, and Jones GH. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18:2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R 1977. Recombination and meiosis. Philos Trans R Soc Lond B Biol Sci. 277:359–370. [DOI] [PubMed] [Google Scholar]
- Hulten MA 2011. On the origin of crossover interference: A chromosome oscillatory movement (COM) model. Mol Cytogenet. 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Cho JD, Nambiar M, and Smith GR. 2021. Redirecting meiotic DNA break hotspot determinant proteins alters localized spatial control of DNA break formation and repair. Nucleic Acids Res. 50:899–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen CF, Thomas MA, Haussler D, and Jacob HJ. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, and Borner GV. 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 5:e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KD, Tanizawa H, Iwasaki O, and Noma K. 2016. Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat Genet. 48:1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JS, and Mortimer RK. 1990. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 126:1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani Y 1978. Absence of interference in association with gene conversion in Sordaria fimicola, and presence of interference in association with ordinary recombination. Genetics. 89:467–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, and Hutchinson J. 2004. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 101:12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C, Osman K, Nuntasoontorn K, West A, Higgins JD, Copenhaver GP, Yang J, Armstrong SJ, Mechtler K, Roitinger E, and Franklin FC. 2015. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11:e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L and Yan J 2015. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nature communications, 6:6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Uzawa S, Meyer BJ, and Villeneuve AM. 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature. 502:703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C and Dean C 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. The Plant Journal. 4:745–750. [DOI] [PubMed] [Google Scholar]
- Liu H, Huang J, Sun X, and Li J. 2018. Tetrad analysis in plants and fungi finds large differences in gene conversion rates but no GC bias. Nat Ecol Evol 2:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RR, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ràmia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, and Gibbs RA. 2012. The Drosophila melanogaster genetic reference panel. Nature. 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Swanson J, German M, McCusker JH, Housworth EA, Stahl FW, and Haber JE. 2004. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics. 168:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, and Steinmetz LM. 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 454:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely PM, Farago AF, and Kauffman TM. 2002. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics. 162:1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Smith CB, Kazemi NY, Cockrell AJ, Arvanitakas AV, Blumenstiel JP, Jaspersen SL, and Hawley RS. 2016. Whole-Genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere Effect. Genetics. 203:159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH, Bridges CB and Schultz J 1935. Constitution of the germinal material in relation to heredity. Carnegie Inst. Year Book 34:287. [Google Scholar]
- Morgan C, Fozard JA, Hartley M, Henderson IR, Bomblies K and Howard M 2021. Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nature communications. 12:4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Contopoulou CR, and King JS. 1992. Genetic and physical maps of Saccharomyces cerevisiae, Edition 11. Yeast. 8:817–902. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, and Fogel S. 1974. Genetical interference and gene conversion. In Mechanisms in recombination. Springer. 263–275. [Google Scholar]
- Muller HJ 1916. The mechanism of crossing-over. American Naturalist. 50:193–221; 284–305; 350–366; 421–434. [Google Scholar]
- Munz P 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 137:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M and Smith GR 2018. Pericentromere-specific cohesin complex prevents meiotic pericentric DNA double-strand breaks and lethal crossovers. Molecular cell. 71:540–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M, Chuang YC, and Smith GR. 2019. Distributing meiotic crossovers for optimal fertility and evolution. DNA Repair (Amst). 81:102648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, and Ogawa H. 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. The EMBO journal. 18:5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB 1997. How cells get the right chromosomes. Science. 275:632–637. [DOI] [PubMed] [Google Scholar]
- Nishant KT, Chen C, Shinohara M, Shinohara A, and Alani E. 2010. Genetic analysis of baker’s yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 6:e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Ross-Macdonald PB, and Roeder GS. 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 158:1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY-H, Taylor AF, Smith GR, and Hunter N. 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing the formation of multi-chromatid joint molecules. Cell. 130:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. 2001. Genes Dev. 15:3130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian HP 1952. The Analysis of Tetrad Data. Genetics. 37:175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DD 1962. Crossing-over and interference in a multiply marked chromosome arm of Neurospora. Genetics. 47:1253–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DD, and Barry EG. 1977. The cytogenetics of Neurospora. Advances in genetics. 19:133–285. [DOI] [PubMed] [Google Scholar]
- Rhee SY, and Somerville CR. 1998. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 15:79–88. [DOI] [PubMed] [Google Scholar]
- Rog O, Kohler S, and Dernburg AF. 2017. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife. 6:e21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, and Roeder GS. 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 79:1069–1080. [DOI] [PubMed] [Google Scholar]
- Serrentino ME, Chaplais E, Sommermeyer V, and Borde V. 2013. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 9:e1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Sakai K, Shinohara A, and Bishop DK. 2003. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 163:1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, and Russell P. 2003. Fission yeast Mus81•Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 165:2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Zhai B, Yang X, Tan T, Wang Y, Yang X, Tan Y, Chu T, Cao Y, Song Y, Wang S, Zhang L. 2021. Interplay between Pds5 and Rec8 in regulating chromosome axis length and crossover frequency. Sci Adv. 12:eabe7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler DR 1959. The relationship of gene conversion to crossing over in Neurospora. Proc. Natl. Acad. Sci. USA. 45:1625–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW 2008. On the “NPD ratio” as a test for crossover interference. Genetics. 179:701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW, and Foss HM. 2008. But see KITANI (1978). Genetics. 178:1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW, Foss HM, Young LS, Borts RH, Abdullah MF, and Copenhaver GP. 2004. Does crossover interference count in Saccharomyces cerevisiae? Genetics. 168:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW, and Housworth EA. 2009. Methods for analysis of crossover interference in Saccharomyces cerevisiae. Methods Mol Biol. 557:35–53. [DOI] [PubMed] [Google Scholar]
- Strickland WN 1958. An analysis of interference in Aspergillus nidulans. Proceedings of the Royal Society of London. Series B, Biological sciences. 149:82–101. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH 1915. The Behavior of the Chromosomes as Studied through Linkage. Z. Indukt. Abstammungs. Vererbungsl. 13:234–287. [Google Scholar]
- Sym M, and Roeder GS. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 79:283–292. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, and Meyer BJ. 2008. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 22:194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, and Roeder GS. 2006. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Developmental cell. 10:809–819. [DOI] [PubMed] [Google Scholar]
- Vranis NM, Van der Heijden GW, Malki S, and Bortvin A 2010. Synaptonemal complex length variation in wild-type male mice. Genes, 1:505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, and Cheng Z. 2010. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Proc Natl Acad Sci U S A. 22:417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Wang S, Zhang L, and Kleckner N 2017. Quantitative modeling and automated analysis of meiotic recombination. Methods in molecular biology (Clifton, N.J.), 1471:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. [DOI] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, and Villeneuve AM. 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 149:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, and Boulton SJ. 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 327:1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, and Hunter N. 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 149:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, and Alani E. 2009. The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 5:e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kohler S, Rillo-Bohn R, and Dernburg AF. 2018. A compartmentalized signaling network mediates crossover control in meiosis. Elife. 7:e30789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liang Z, Hutchinson J, and Kleckner N. 2014a. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 10:e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang S, Yin S, Hong S, Kim KP, and Kleckner N. 2014b. Topoisomerase II mediates meiotic crossover interference. Nature. 511:551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, McPeek MS, and Speed TP. 1995. Statistical analysis of chromatid interference. Genetics. 139:1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]


