Abstract
Bone marrow mesenchymal stromal/stem cells (MSCs) are a heterogeneous population that can self-renew and generate stroma, cartilage, fat, and bone. Although a significant progress has been made toward recognizing about the phenotypic characteristics of MSCs, the true identity and properties of MSCs in bone marrow remain unclear. Here, we report the expression landscape of human fetal BM nucleated cells (BMNCs) based on the single-cell transcriptomic analysis. Unexpectedly, while the common cell surface markers such as CD146, CD271, and PDGFRa used for isolating MSCs were not detected, LIFR+PDGFRB+ were identified to be specific markers of MSCs as the early progenitors. In vivo transplantation demonstrated that LIFR+PDGFRB+CD45-CD31-CD235a- MSCs could form bone tissues and reconstitute the hematopoietic microenvironment (HME) effectively in vivo. Interestingly, we also identified a subpopulation of bone unipotent progenitor expressing TM4SF1+CD44+CD73+CD45-CD31-CD235a-, which had osteogenic potentials, but could not reconstitute HME. MSCs expressed a set of different transcription factors at the different stages of human fetal bone marrow, indicating that the stemness properties of MSCs might change during development. Moreover, transcriptional characteristics of cultured MSCs were significantly changed compared with freshly isolated primary MSCs. Our cellular profiling provides a general landscape of heterogeneity, development, hierarchy, microenvironment of the human fetal BM-derived stem cells at single-cell resolution.
Subject terms: Cell biology, Medical research
Introduction
Bone marrow (BM)-derived mesenchymal stromal/stem cells (MSCs), which is characterized by non-hematopoietic, plastic-adherent, colony-forming cells, were first identified by Friedenstein and colleagues decades ago. Despite their complexity and heterogeneity, evidences increasingly support the notion that MSCs exhibit trilineage differentiation potentials and represent an important component of the hematopoietic stem cell (HSC) niche in bone marrow (BM).1–4 Many studies have expanded the knowledge of MSCs through the use of genetically modified mice. However, due to the technical and material limitations, little is known about the cellular biology of human bone marrow mesenchymal cells. Furthermore, the development of MSCs during embryonic stages is also an unresolved issue. It is particularly important to study the development of MSCs, which, in turn, could help us better understand the physiological functions of MSCs.
The identification of MSC marker genes is crucial for isolating and characterizing MSCs in vivo. Matsuzaki group first identified MSCs based on the expression of the PDGFRa and Sca-1 in the non-hematopoietic and endothelial compartment in the bone marrow of adult mice.2 By using transgenic mouse model, Nestin, Leptin-Receptor (LepR) and Grem1 expressing cells were identified with MSC/skeletal stem cell (SSC) activity and represent the major source for HSC niche factors such as CXCL12 and SCF in the bone marrow.5–7 In contrast, human MSCs have long been identified through the flow cytometry analysis by using monoclonal antibodies. CD146+ MSCs from human bone marrow were observed to enrich for CFU-Fs and be able to generate the hematopoietic bone marrow.8 CD271 was another broadly accepted surface marker of MSCs from human bone marrow, CFU-Fs were highly and exclusively enriched in both CD271+CD146-/low and CD271+CD146+ cells.9–11 Other surface markers including CD105, CD73, CD90, CD49a, CD140b, MSCA-1, SSEA-4, and STRO-1 were also used to sort human MSCs alone or in combination.12 Even though large amount of information about human MSCs has been obtained based on experiments performed in culture, the complexity and physiological characteristics of these cells in vivo remains poorly investigated.
Most recently, Huelsken’s group showed that when primary BM-derived stem cells (BMSCs) were expanded in vitro, their capacity to support hematopoiesis was significantly lost.13 Another report demonstrated that primary murine MSCs exhibited high homing efficiency to the BM, but lose homing ability after in vitro culture.14 Mx-1-Cre mice-derived cells possess CFU-Fs activity and trilineage differentiation potential in vitro, but only show a limited differentiation ability in vivo.15 CD146 has been reported to label human BMSCs in vivo, however, its expression level is significantly changed when the cells are cultured.9 Moreover, significant differences were also identified in global DNA methylation profiles of BMSCs following cell expansion.16 All these studies clearly demonstrated that the in vitro features of MSCs/SSCs could not faithfully reflect their in vivo function. Unfortunately, our present knowledge of human BMSCs is mainly advanced by in vitro culture systems. Thus, strategies to identify the real identity of human BMSCs/SSCs in vivo would be of great help for clearing up the “stem-cell mess of MSCs”.17
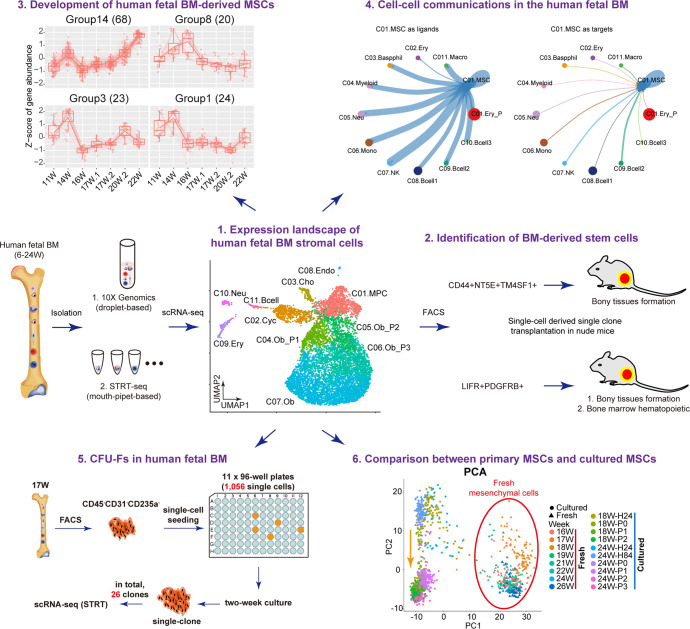
The recent breakthrough of single-cell RNA sequencing (scRNA-seq) technique allows us to study the freshly isolated primary cells at single-cell resolution, and investigate the bona fide identity of human MSCs/SSCs in vivo.18 On the other hand, based on the transcriptome of each cell, we could better understand the complexity and heterogeneity of MSC/SSC populations.19 In the present study, we performed a comprehensive screening of human fetal BM nucleated cells (BMNCs) at single-cell resolution (Fig. 1a). We isolated primary BMNCs from 46 human embryos ranging from 6 to 24 developmental weeks and performed scRNA-seq analysis using two complementary strategies (STRT and 10x Genomics) to balance between accuracy and throughput. Based on systematic bioinformatics analyses and experimental validations, we identified two types of stem cells in the human BM. This current work would enhance our understanding about human fetal BM-derived MSCs and their heterogeneity and microenvironment in vivo.
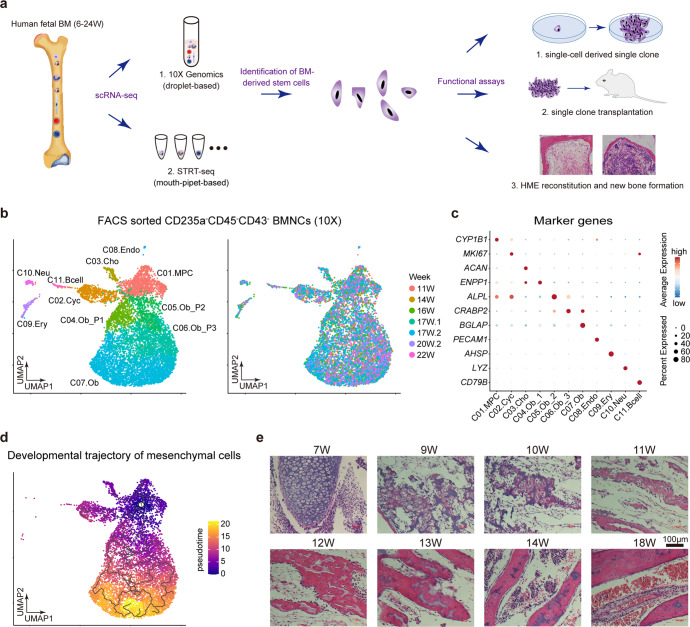
Fig. 1.
Expression landscape of human fetal BM stromal cells. a Diagrammatic sketch of the current study. b UMAP showing the clustering (left) and week information (right) of all FACS-sorted CD235a-CD45-CD43- nucleated cells sampled from the human fetal bone marrow, which were sequenced by 10x Genomics scRNA-seq technique. c Dotplot showing the expression patterns of representative marker genes in each major cluster identified in Fig. 1b. The color key from blue to red indicates low to high expression levels, respectively. Dot size indicates the percent of cells expressing a certain gene. d Developmental trajectory of mesenchymal cells inferred by monocle 3 algorithm. e H&E staining of femur sections from different embryonic stages
Results
Expression landscape of human fetal BM stromal cells
In order to explore the cellular diversity of the human fetal BM stroma, we performed single-cell transcriptomic profiling of BMNCs using 10x Genomics scRNA-seq technique (Supplementary Table 1). Since the BM was fulfilled with red blood cells, we lysed these red cells by ice-cold sterile H2O.20 To test the feasibility of our experimental protocol, 2634 single cells passing the quality control were obtained from two embryos (i.e., 20- and 21-week-old). In total, we identified 12 clusters with batch effect correction in Harmony and unsupervised clustering in Seurat (Supplementary Fig. 1a).21,22 According to the classic marker genes, these clusters were annotated as: two clusters of erythrocytes, specifically expressing GYPA and HBG1, respectively; basophils (CSF2RB); myeloid cells (PLEK); neutrophils (AZU1); monocytes (CSTA); natural killer cells (SPINK2); three clusters of B cells, highly expressing CD79A, LTB and JCHAIN, respectively; macrophages (CSF1R); and mesenchymal cells, expressing collagen triple helix repeat containing 1 (CTHRC1) (Supplementary Fig. 1b). As indicated above, by progressive depletion of the red cells with instantaneous H2O treatment, the majority of BMNCs at late developmental stages (20–21 weeks) are hematopoietic cells.
To capture relatively rare BM stromal cells, we next performed scRNA-seq on the sorted non-hematopoietic CD235a-CD45-CD43- cells (Supplementary Fig. 1c), and obtained 8,725 sorted cells from 9 embryos (11–22 weeks). As shown in Fig. 1b, the human fetal BM CD235a-CD45-CD43- cells were divided into 11 clusters. Based on the differentially expressed genes (DEGs) and enriched gene ontology (GO) terms (Fig. 1c, Supplementary Fig. 1d, e, and Supplementary Table 2), we annotated them as seven mesenchymal clusters, three hematopoietic clusters and one endothelial cluster. Most of the sorted cells were non-hematopoietic cells, indicating the accuracy of FACS sorting. Among the seven mesenchymal clusters, we identified four clusters exhibiting a developmental trajectory of osteoblast (OB) lineage, from OB progenitors to mature OBs (OB-P1, OB-P2, OB-P3, and OB). We also detected a cluster with chondrocyte lineage (specifically expressing ACAN) and a cluster of cycling mesenchymal cells. Importantly, we found a cluster of mesenchymal progenitor cells with no distinct differentiation characteristics, highly expressing CYP1B1, which we designated as mesenchymal progenitor cells (MPC) (Fig. 1c). This result was also supported by the developmental trajectory inferred by Monocle (Fig. 1d).23 In addition, we did not detect any adipocytes in either the single-cell data or H&E and Oil-red O staining of femur sections (Fig. 1e and Supplementary Fig. 1f), which demonstrated the exiguity of adipocytes in human fetal BMs.
Heterogeneity of MSCs in human fetal BM
A recent study clarified that perinatal chondrocytes form most of the new osteoblasts and decreased progressively with age.24 Thus, we experimentally tested the chondrocyte cluster by individually seeding the sorted single cells into 96-well plates separately, and then subcutaneously implanting expanded colonies into the dorsal side of the nude mice with β-TCP carrier (Fig. 2a). As shown in Fig. 2b, the chondrocyte cluster exclusively expressed surface markers CD44, NT5E, TM4SF1, we subsequently harvested CD44+CD73+TM4SF1+/CD45-CD31-CD235a- cells through FACS sorting (Supplementary Fig. 2a). STRT analysis further confirmed the accuracy of FACS (Supplementary Fig. 2b). We found that the colonies expanded from single CD44+CD73+TM4SF1+ cells were able to form bony tissues effectively (Fig. 2c). Therefore, our study was consistent with the recent finding that perinatal chondrocytes can give rise to new osteoblasts.24 We further sorted the osteo-progenitor cells (OB_P1, P2, P3) using FACS antibody ENPP1 and ANKH. However, single clones derived from single ENPP1+/ANKH+ cells failed to form new bone (data not shown).
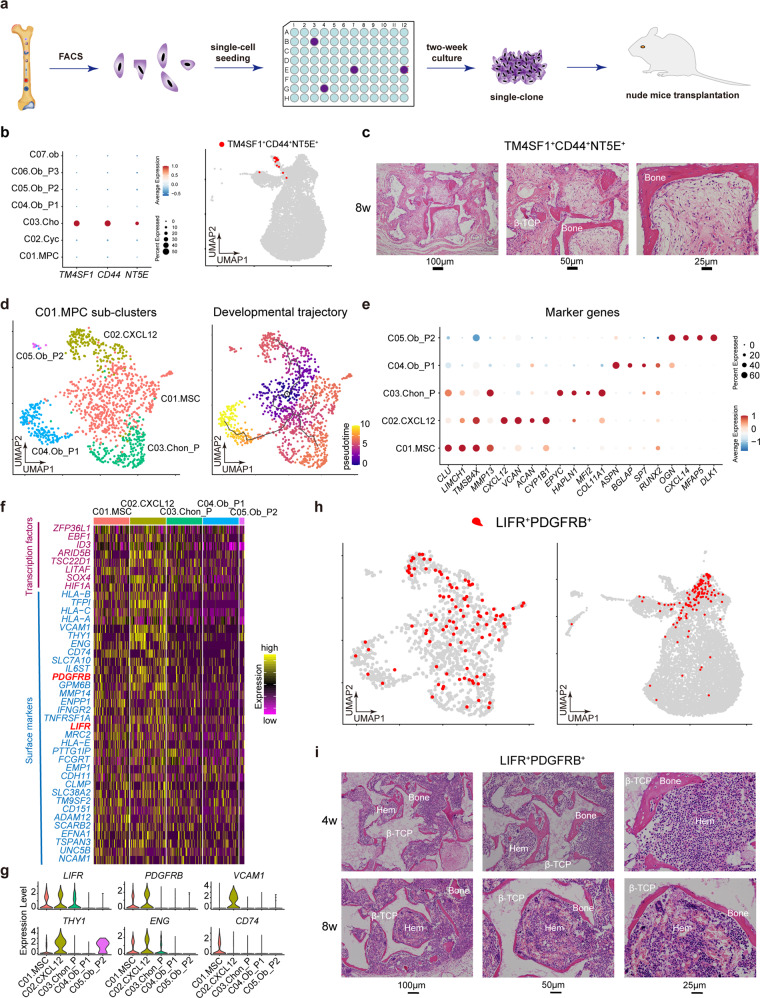
Fig. 2.
Heterogeneity of MSCs in human fetal BM. a Study overview for nude mice transplantation. b Dotplot (left) and UMAP (right) showing the specific expression of gene combination of TM4SF1, CD44 and NT5E in chondrocytes. The color key from blue to red indicates low to high expression levels, respectively. Dot size indicates the percent of cells expressing a certain gene. c Abundant new bones were formed after 8 weeks transplantation of single clone derived from single primary TM4SF1+CD44+CD73+/CD45-CD31-CD235a- cell. d UMAP showing the clustering result (left) and developmental trajectory (right) of MPCs identified in Fig. 1b. e Dotplot showing expression levels of representative marker genes in each cluster identified in Fig. 2d. f Heatmap showing the differentially expressed transcription factors and surface markers of C01.MSC and C02.CXCL12. The color key from purple to yellow indicates low to high expression levels, respectively. g Violin plots showing the expression levels of potential surface markers for C01.MSC and C02.CXCL12. h UMAP showing the specific expression of gene combination of LIFR and PDGFRB in C01.MSC and C02.CXCL12. i Hematopoietic cell clusters (hem) appeared at 4 weeks and matured at 8 weeks after transplantation of single clone derived from single primary LIFR+PDGFRB+/CD45-CD31-CD235a- cell. Abundant new bones were formed after 4 weeks transplantation. β-TCP: hydroxyapatite carrier
MSCs in the bone marrow have been defined as multipotent stem cells that can differentiate into osteocytes, adipocytes, and chondrocytes,25,26 however, their exact identity in vivo remains unclear. We subsequently distinguished MPCs into five major subsets by sub-clustering and developmental trajectory analysis, namely MSCs (C01.MSC), CXCL12+ MSCs (C02.CXCL12), chondrocyte progenitors (C03.Chon-P) and two OB progenitor sub-clusters (C04.Ob-P1 and C05.Ob-P2) (Fig. 2d, e). In particular, one subset significantly expressed the key functional osteoblast specific gene ASPN and BGLAP (C04.Ob-P1); the other subset highly expressed EPYC and HAPLN1, which we annotated as chondro-progenitors (C03.Chon-P). In fact, the osteo-progenitors and chondro-progenitors both can be derived from MSCs. To experimentally validate the MSCs, we first choose effective surface marker. As shown in Fig. 2f–h, LIFR combined with PDGFRB exhibited good pattern for C01.MSC and C02.CXCL12. Compared with LIFR, the expression level of ENG in C01.MSC was lower. We next validated these two MSC clusters by sorting LIFR+PDGFRB+/CD45-CD31-CD235a- cells for nude mice transplantation (Fig. 2i and Supplementary Fig. 2c). STRT scRNA-seq analysis first verified the accuracy of FACS (Supplementary Fig. 2d). As described above, single LIFR+PDGFRB+/CD45-CD31-CD235a- cell was seeded into the 96-well plate, and after 2 weeks, single clones were harvested and transplanted into the dorsal side of nude mice. As shown in Fig. 2i, we observed abundant new bony ossicles 4 weeks after transplantation. And 8 weeks after transplantation, hematopoiesis which is reminiscent of those found in the bone marrow apparently appeared.
Discrepancy between MSCs at early and late developmental stages
In order to trace the origin of human fetal MSCs, we tried to capture mesenchymal cells at earlier developmental stages (6–9w). However, due to the limited numbers of cells acquired at early developmental stages, instead of using 10x Genomics scRNA-seq technique that is suitable for capturing 1000–10,000 single cells per sample, we harvested cells via random picking and sequenced them using STRT scRNA-seq technique. In total, 2,989 high-quality single-cell transcriptomes of 17 embryos ranged from 6 to 24 weeks were captured (Fig. 3a).
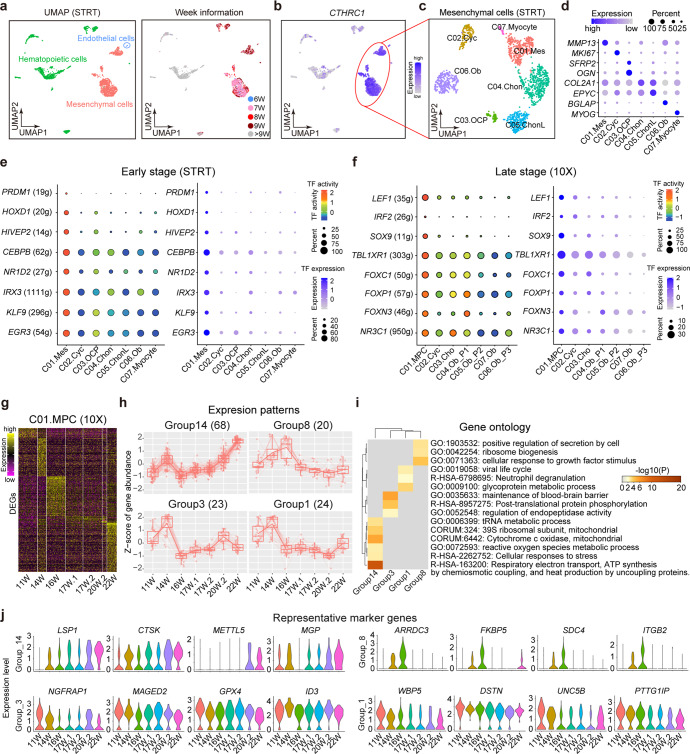
Fig. 3.
Discrepancy between MSCs at early and late developmnental stages. a UMAP showing the three major groups (left) and their developmental stages (right) of all randomly picked fresh cells from human fetal BM at early stages. b UMAP showing the specific expression of CTHRC1 in mesenchymal cells. c UMAP showing the 7 clusters of mesenchymal cells at early stages. d Dotplot showing the expression levels of representative marker genes in each cluster identified in Fig. 3c. The color key from light blue to dark blue indicates low to high expression levels, respectively. Dot size indicates the percent of cells expressing a certain gene. e TFs that were specifically activated and expressed during the early developmental stages. f TFs that were specifically activated and expressed during the late developmental stages. g The development of MSCs at late stages identified in Fig. 2d. Heatmap showing the DEGs of MSCs at each stage. The color key from purple to yellow indicates low to high expression levels, respectively. h Selected expression patterns along the development of MSCs. The gene number is in the bracket. i Enriched terms using the genes of each expression pattern along the development of MSCs. j Violin plots showing the expression levels of marker transcription factors along the development of MSCs
We identified three major groups in the STRT dataset: namely, endothelial cells (with CDH5 specifically expressed), mesenchymal cells (highly expressing CTHRC1, collagen triple helix repeat containing 1), and hematopoietic cells (highly expressing PTPRC and GYPA) (Fig. 3b and Supplementary Fig. 3a, b). Notably, most of the sequenced cells at later developmental stages (after 9 weeks) were hematopoietic cells, which was consistent with 10x dataset (Supplementary Fig. 1a). The mesenchymal cells were further divided into 7 clusters (Fig. 3c). Based on the DEGs, we annotated them as mesenchymal progenitor cells (C01.Mes, which highly expressed MMP13); cycling progenitor cells (C02.Cyc, which were characterized by the expression of MKI67); osteo-chondrogenic progenitors (C03.OCP, with OGN and SFRP2 upregulated); chondrocyte progenitor cells (C04.Chon, where COL2A1 was highly expressed); late mature chondrocytes (C05, ChonL, highly expressed EPYC); osteoblasts (C06.Ob; with bone related gene BGLAP specifically upregulated); and myocytes (C07.Myocyte, specifically expressed MYOG) (Fig. 3d).
Transcription factors (TFs) play critical roles during a variety of biological processes. We used SCENIC to infer the gene regulatory networks from our scRNA-seq data and identify key TFs during the development of MSCs.27 In human fetal BM mesenchymal cells of early developmental stages, we found that PRDM1, HOXD1, HIVEP2, CEBPB, NR1D2, IRX3, KLF9, EGR3 etc. may play important roles in C01.Mes (Fig. 3e). Both the gene-expression levels and regulatory activity were specifically high in C01.Mes. However, the situation is quite different from the MSCs at later stages (11w-22w), where TFs such as LEF1, IRF2, SOX9, TBL1XR1, FOXC1, FOXP1, FOXN3, and NR3C1 etc., were highly expressed and activated (Fig. 3f).
In 10x dataset, we obtained BMSCs from 7 embryos ranging from 11 to 22 weeks, which provided a chance to study their development. No significant change was detected in the cell type ratio from 11 to 22 weeks (Supplementary Fig. 3c). Next, we performed differential expression analysis for the BMSCs based on their developmental weeks. As shown in Fig. 3g, BMSCs of different development weeks exhibited their unique expression patterns, indicating a continuous development of BMSCs from 11 to 22 weeks. We next clustered the expressed genes of BMSCs into 11 groups based on their gene-expression patterns (Supplementary Fig. 3d). Among these groups, expression levels of group14 genes showed an increasing trend, expression levels of group1 and group3 genes exhibited a decreasing trend, and expression levels of group8 genes were upregulated and reached a peak at 16 weeks and then were down-regulated until 22 weeks (Fig. 3h). These four groups of genes took part in different biological processes, which might play important roles in the development of BMSCs (Fig. 3i, j).
Cell–cell communications in the human fetal BM
Investigating intercellular communications has greatly increased our understanding of stem cell-niche interactions. To fully understand the microenvironment of MSCs in the human fetal BM, we first elucidated the cellular interactions between MSCs and hematopoietic cells (HC). When MSCs were regarded as ligand, we found that most of the HCs, including basophils, myeloid cells, neutrophils, monocytes, natural killer cells, B cells, and macrophages have close ties with MSCs (Fig. 4a and Supplementary Table 3). Indeed, MSCs were highly enriched in the critical factors of HSC maintenance, such as cytokines chemokine ligand 12 (CXCL12) and stem cell factor (SCF), and CXCR4 was used as the receptor by all the HC cell types (Supplementary Fig. 4).28–31 Conversely, when MSCs were regarded as targets, we observed that all hematopoietic cells do not interact closely with MSCs (Fig. 4a). When MSCs were regarded as ligands, active ligand-receptor pairs were overtly detected in cell–cell communications. Like MSCs, CAR cells interact with hematopoietic cells in the same way (Fig. 4b).
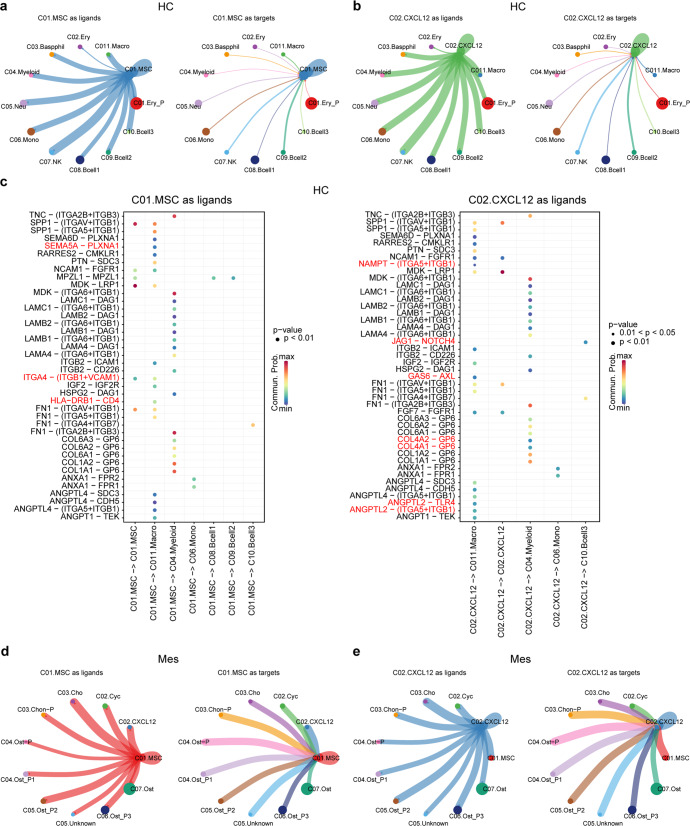
Fig. 4.
Cell–cell communications between MSCs and niche cells. a Cellular interaction between C01.MSC and hematopoietic cells (HC). b Cellular interaction between C02.CXCL12 and hematopoietic cells (HC). c Unique ligand-receptor pairs between MSCs and hematopoietic cells. The red ligand-receptor pairs indicate the differences between C01.MSC and C02.CXCL12. d Cellular interaction between C01.MSC and mesenchymal cells (Mes). e Cellular interaction between C02.CXCL12 and mesenchymal cells (Mes)
We also identified unique interactions for each HC cell type with MSCs (Fig. 4c). For example, only myeloid cells expressed ITGA2B and ITGB3 as the receptors for the ligand TNC sent by MSCs; while ANGPTL1 and ANGPTL4 sent by MSC could only be received by macrophages by expressing SDC3, CDH5, (ITGA5 and ITGB1) and TEK as the receptors. Besides, MSCs and CAR cells also had unique interactions with HCs. For example, we found that MSCs could interact with macrophages and themselves through ligand-receptor pair, ITGA4–(ITGB1 + VCAM1). Previous studies indicated that VCAM1 could interact with integrin alpha-4/beta-1 (ITGA4/ITGB1) to mediate both adhesion and signal transduction, and VCAM1+ macrophages regulate hematopoietic stem and progenitor cells (HSPCs) homing to the vascular niche in an ITGA4 dependent manner.32 Thus, the VCAM1+ macrophages might also regulate fetal MSCs homing to the vascular niche, and both VCAM1+ MSCs and macrophages might play important roles in HSPCs homing in the late developmental stages. On the other hand, CAR cells secreted ANGPTL2 to attract and activate macrophages by the receptor TLR4 to produce inflammatory cytokines.33 We next explored the cellular interactions between MSCs and mesenchymal cells. As shown in Fig. 4d, e, whether MSCs were used as ligands or targets, their interactions with other stromal cells were quite strong. The same situation existed between CAR cells and mesenchymal cells.
CFU-Fs in human fetal BM
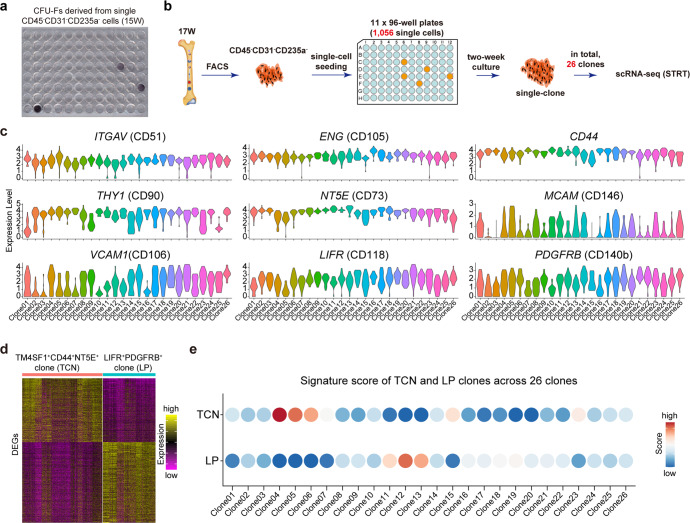
To assess the CFU-F activity of mesenchymal cells, we seeded single freshly sorted CD31-CD45-CD235a- BMNCs from a 15-week embryo into a 96-well plate, and detected 3 expanded clones after culture for 2 weeks (Fig. 5a). To further evaluate the CFU-F ratio, 1056 freshly sorted cells from embryos of a 17-week twins were cultured after single-cell seeding in eleven 96-well plates, and 26 colonies were observed after 2-week culture (Fig. 5b). These colonies were then digested and subjected to scRNA-seq analysis. Strikingly, a number of markers that have previously been shown to mark MSCs, such as THY1, ENG, NT5E, CD44, ITGAV, were found to be highly expressed in all clones. The expression level of melanoma cell adhesion molecule (MCAM), also known as CD146, was very different in these clones, it is highly expressed in some clones and almost not expressed in others. Notably, low-affinity nerve growth factor receptor (LNGFR/CD271), which has been described to label uncultured multipotent MSCs,34 was not expressed in all 26 clones (data not shown).
Fig. 5.
CFU-F activity of human fetal BM mesenchymal cells. a CFU-Fs from human fetal BM CD45-CD31-CD235a- cells. b Study overview of the CFU-F assay of human fetal BM CD45-CD31-CD235a- cells. c Violin plots showing the expression levels of representative surface marker genes in all single clones derived from single CD45-CD31-CD235a- cells. d Heatmap showing the DEGs between TCN clones and LP clones. The color key from purple to yellow indicates low to high expression levels, respectively. e Signature score of TCN and LP clones across all 26 clones derived from single CD45-CD31-CD235a- cells
We wondered among these 26 clones how many clones were derived from the BM-derived stem cells. Thus, we examined single clones derived from sorted single TM4SF1+CD44+NT5E+ and LIFR+PDGFRB+ cells using STRT-seq analysis, which exhibited different expression patterns (Supplementary Fig. 5a, b). We then used the DEGs between LIFR+PDGFRB+ and TM4SF1+CD44+NT5E+ clones as their own signature genes to calculate the signature score across these 26 clones derived from the whole BMNCs, and found only a small fraction of CFUs were inferred to come from the above two stem cell populations (Fig. 5d, e). Indeed, genuine single stem cell within the BM could form clone, but the reverse statement is not valid, since only a partial of CFU-Fs are multipotent when transplanted in vivo.26
Comparison between primary MSCs and cultured MSCs
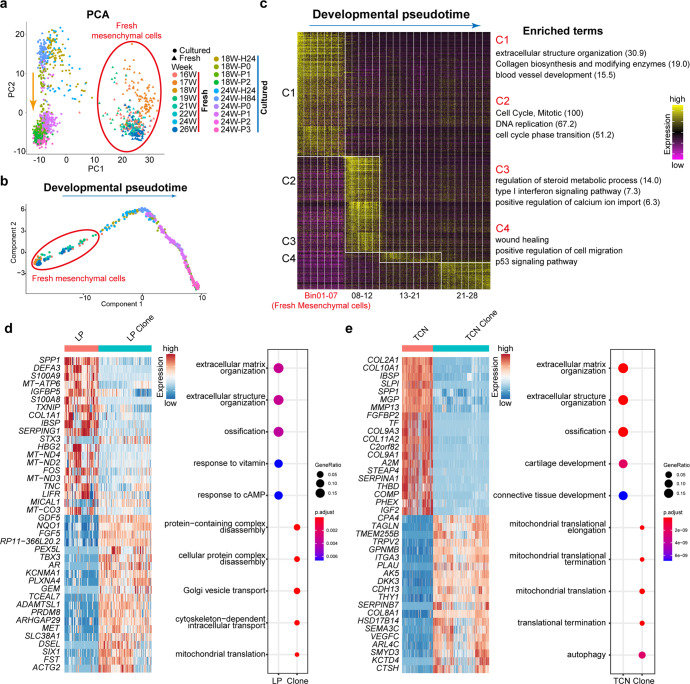
Previous studies have suggested that MSCs gradually lose proliferative capacity and secretive properties during expansion.35,36 We next made comparison between fresh and cultured BMNCs to score possible changes. Since the cultured cells were sampled from the embryos of 18 and 24 weeks, we combined fresh BMNCs sampled from the embryos of similar developmental stages (16–26 weeks) with cultured cells to perform subsequent analyses. The first axis of PCA separated fresh and cultured cells, while the second axis ordered cultured cells along the culture stages, which indicated drastic changes of the gene-expression patterns during in vitro culture (Fig. 6a). The developmental pseudotime analysis through Monocle also sorted fresh and cultured cells along the culture stages (Fig. 6b). We split all these cells into 28 bins along the inferred developmental pseudotime, each bin contained 50 cells (Fig. 6c and Supplementary Fig. 6a). We next performed DEG analysis for each bin and found that these cells could be divided into 4 major clusters (C1–C4) based on their gene-expression patterns (Fig. 6c). DEGs in each cluster demonstrated a general proliferation and differentiation process. C1 was composed of fresh mesenchymal cells, and the DEGs of C1 were predominantly associated with extracellular structure organization, collagen biosynthesis and blood vessel development, which suggested the features of fresh mesenchymal cells. Cells in C2 underwent an active proliferation period and then were followed by differentiation in C3. Unexpectedly, C4 are characterized by the activation of p53 signaling pathway, which might reflect a senescent state of MSCs with higher passages.
Fig. 6.
Comparison between primary and cultured BM mesenchymal cells. a PCA plot showing relationship between primary and cultured BM mesenchymal cells. b Developmental pseudotime of primary and cultured BM mesenchymal cells. c Cells were split into 28 bins along the developmental pseudotime. Each bin contained 50 cells and the DEGs of each bin were shown by the heatmap. All bins were classified into four major clusters according to their expression patterns. And the enriched terms using all the DEGs in each cluster were listed on the right. The number in the bracket indicates –log10 (P-value). d DEGs (left) and GO terms (right) showed that the fresh LP cells and LP clones exhibited high heterogeneity. e DEGs (left) and GO terms (right) showed that the fresh TCN cells and TCN clones exhibited great high heterogeneity
We further compared the freshly isolated LIFR+PDGFRB+ cells with single LIFR+PDGFRB+-derived single clones, TM4SF1+CD44+NT5E+ cells with their clones, respectively. During the culture of both these two cell types, genes related to extracellular matrix organization, extracellular structure organization and ossification were down-regulated, while mitochondrial translation related genes were upregulated. Besides, fresh TM4SF1+CD44+NT5E+ cells and cultured clones displayed much stronger discrepancy compared with the culture of primary LIFR+PDGFRB+ cells (Fig. 6d, e). Furthermore, it is important to mention that the expression of LIFR was lost when cultured in media, which was quite different from the significant increase of NT5E, THY1 and ENG after long time culture (Supplementary Fig. 6b).
Discussion
A fundamental question in the study of MSCs is that the bona fide identity of human MSCs in vivo is not well defined.37 In the past few decades, considerable progress has been made in defining and characterizing murine bone marrow MSCs using genetically engineered mice.6 In addition, scRNA-seq technology helped us to gain a global recognition of the heterogeneous populations in the murine BM,18,19 however, the cell census of human fetal BM remains underestimated owing to material limitations. In this study, more than 10,000 human embryonic single BMNCs were subjected for scRNA-seq analysis, our cellular profiling and functional analysis not only provide a definitive answer for this question, but also broaden our understanding of the landscape of heterogeneity, development, the microenvironment of the human fetal BMSCs at single-cell resolution (Fig. 7).
Fig. 7.
A mechanism and illustration picture of this study
Another critical question about the MSC-based bone tissue engineering is the necessity to expand the cells prior to use. In the present study, gene-expression profiles indicated an apparent gap between fresh MSCs and cultured MSCs (Fig. 6). The explicit alteration of fresh cells following ex vivo expansion, as well as the demonstration of in vivo niche for the MSCs, would be important hints for guiding us to identify optimal culture conditions for maintaining the original characteristics of in vivo MSCs. In vitro expansion that could preserve the original disposition and progenitor function of MSCs would be a great help for MSC-based bone regeneration. We also examined the CFU-F activity of human fetal BMNCs (Fig. 5). Stromal cells with CFU-F activity and multi-lineage differentiation ability in vitro were generally identified as MSCs. However, many bone marrow stromal cells have the ability of multi-lineage differentiation potential and CFU-F activity in vitro, but failed to form new bone when transplanted in vivo. On the other hand, the expression pattern of many surface markers, which are generally used for sorting MSCs displayed a great difference before and after culture. Both of which further suggested that the characteristics of MSC in vitro are not fully indicative of in vivo function.
Despite the beneficial effects of MSC therapy both in pre-clinical and clinical trials, more and more studies have produced mixed results regarding their therapeutic efficiency. The great changes between freshly isolated MSCs and cultured cells may suggest a possible explanation for the discrepancy between expected and actual results of MSC therapy. In the present study, gene-expression profiles indicated an apparent gap between primary MSCs and cultured MSCs. The fresh MSCs were endowed with multiple functions including extracellular structure organization, collagen biosynthesis and blood vessel development, which were compatible with the characteristics of MSCs. In contrast, the cultured MSCs underwent the ‘active proliferation–differentiation–senescence’ processes. Of note, the signature of fresh MSCs is quite different from that of culture-expanded MSCs (Fig. 6). In vitro expansion that could preserve the original disposition and progenitor function of MSCs would be a great help for MSC therapy.
In a recent study,38 Chan et al. defined a cluster of skeletal stem cells (SSCs) in human embryos, which could differentiate into cartilage and bone/stroma. We downloaded their scRNA-seq dataset and integrated it with our dataset. However, compared with our current observation of MSCs in human fetal BM, there was no specific cell population that specifically expressed the identified MSC markers in their dataset (Supplementary Fig. 7a, b), indicating the different lineages between the MSCs identified in this study and the SSCs they found.
Chiara et al. investigated the effect of isolation methods on cell populations in the scRNA-seq experiments.18 They analyzed the comparison between flushing of undigested BM and crushing of whole bones with enzymatically digestion, and demonstrated that only after strong physical treatment or enzymatic digestion can several populations be detected. In addition, Ninib et al. also demonstrated different murine cellular composition between bone and bone marrow.19 These observations in the murine BM might explain the discrepancy of human fetal SSCs releasing from enzymatically digested bones and our human embryonic MSCs from undigested flushing BM. Besides, we also compared our fetal MSCs with BM-derived MSCs from healthy adult donors cultured in the presence/absence of IFN-γ and TNF-α, which resulted in MSC-“licensing”.39 Although there were significant differences in distinct expression patterns among fetal, licensing+ and licensing- MSCs, fetal MSCs did shared several immune features with licensing+ MSCs, such as cytokines signaling in immune system, regulation of immune response, and cellular response to cytokine stimulation (Supplementary Fig. 7c–e).
Materials and methods
Ethics statement
Our study was approved by the Reproductive Study Ethics Committee of Peking University Third Hospital (2012SZ-013 and 2017SZ-043). Each donor signed an informed consent form, and the study was carried out according to the ISSCR guidelines.
Isolation of bone marrow cells
Human fetal long bones (femur and tibia) were separated, gently removed soft tissue with paper towels. Then the bone marrow was flushed with syringes, cells from the bone marrow were harvested without enzymatic digestion. Since the bone marrow was full of red blood cells, we lysed the red blood cells in the suspension by adding cold sterile H2O for 6 s. After 6 s lysis, immediately add 1× phosphate-buffered saline (PBS) with 4% (vol/vol) fetal bovine serum (FBS) to stop the reaction. As there are too many red blood cells in the bone marrow, transient H2O treatment needs to be repeated several times. After the red cells were lysed, the remaining cells were filtered into a collection tube through a 40 mm filter, and then combined into a sample for next experiment.
Mouth-pipet-based scRNA-seq library construction
We prepared 2.5 µl of single-cell lysis buffer containing 0.8 U/µl Recombinant RNase Inhibitor (Takara, Cat.2313B), 0.38% Triton-X 100 (Sigma, Cat. T8787), 2 mM dNTP mixture (Takara, Cat.4019) and 300 nM RT primer. Ninety-six types of barcoded sequences (6-bp barcode) served as RT primers for each cell and corresponded to one barcode (Supplementary Table 4). Single cells were transferred to lysis buffer in 0.2 ml PCR tubes via mouth pipetting. The selected cells were either stored at –80 °C or directly reverse-transcribed and amplified. The single-cell transcriptome amplification steps were conducted according to STRT-seq,40,41 with a few modifications of the RT primers. After amplification, the cDNAs of different barcodes were combined together and purified using the DNA Clean and Concentration Kit (ZYMO, Cat. D5044) to remove free primers and primer dimers. A second round of amplification was then conducted with biotin primers containing the Illumina read2 primer sequence and indexes. Thus, after 4 cycles of PCR, the cDNAs were further fragmented using Covaris S220, and the 5’ portion of the first-strand cDNA was enriched using C1 streptavidin beads (Invitrogen, Cat. 65002). Further library construction was conducted using KAPA Hyper Prep Kits for Illumina (Cat.KK8505) following the manual. Each single-cell was designed for 0.5 G data on the Illumina HiSeq 4000 platform using 150-bp paired-end reads.
Droplet-based scRNA-seq library construction
Cells were centrifuged at 500 × g for 5 min at 4 °C. Then we removed the supernatant and washed the cell pellet once with 0.04% BSA/PBS. Before loading onto the 10x Genomics Chromium chip, we calculated the concentration under the microscope. Reverse transcription, cDNA amplification and library construction were performed using the 10x Genomics Single-Cell v2 kit according to the manuals. Each library was sequenced on Illumina hiseq4000 to acquire a sequencing saturation over 90%.Cells were centrifuged at 500 × g for 5 min at 4 °C. Then we removed the supernatant and washed the cell pellet once with 0.04% BSA/PBS. Before loading onto the 10x Genomics Chromium chip, we calculated the concentration under the microscope. Reverse transcription, cDNA amplification and library construction were performed using the 10x Genomics Single-Cell v2 kit following the manuals. Each library was sequenced on Illumina hiseq4000 to acquire a sequencing saturation over 90%.
Processing of scRNA-seq data
For 10x dataset, we used Cell Ranger 2.2.0 with default mapping parameters to process the raw data. Reads were consistent with the human GRCh38 genome.
For STRT dataset, we used UMI-tools42 to extract the barcode and unique molecular identifiers (UMIs) from R2 reads. We removed template switch oligo and polyA tail sequences from the obtained readings. The clean reads were consistent with the human GRCh38 genome using STAR.43 We used featureCounts44 to count unique mapped reads and quantified the UMIs with UMI-tools.
After obtaining the UMI expression table, we removed cells with less than 1000 detected genes and 10,000 detected transcripts from the STRT dataset. For the 10x dataset, we removed cells with fewer than 200 detected genes. Cells with high mitochondrial gene-expression fractions were also removed. We performed clustering using the Seurat package (version 2.2)45 (for more details, please see http://satijalab.org/seurat/). Briefly, highly variable genes were selected to perform dimensionality reduction. We used Harmony to reduce the batch effect arising from embryos’ differences, (https://github.com/immunogenomics/harmony).21 A graph-based clustering method in Seurat was used to determine the final clustering.
Differentially expressed gene (DEG) analysis and gene ontology enrichment analysis
We employed Seurat to perform the DEG analysis. We used FindAllMarkers to identify DEGs for each cluster and the FindMarkers function in Seurat to identify DEGs for two given clusters. For STRT data, genes with fold-change >2 or <0.5 and adjusted P-value < 0.01 were regarded as DEGs; while for 10x Genomics data, default parameters (logfc.threshold = 0.25; return.thresh = 0.01) were used. Heatmaps were plotted in Seurat or the pheatmap package; violin plots were established using the Seurat package; and bar plots were established in R. Gene ontology enrichment analysis was carried out in clusterProfiler46 and Metascape47 (http://metascape.org).
Developmental pseudotime analysis and TF inference
The developmental pseudotime of MSCs was inferred using UMI count in Monocle.48 For the freshly isolated MSCs shown in Fig. 3a, since we had already obtained the marker genes of the two MSC clusters, we employed these genes to infer the developmental pseudotime. For all MSCs, combining both fresh and cultured MSCs, we followed “unsupervised ordering” in vignette to construct single-cell trajectories with default parameters.
For TF inference, we used SCENIC (https://scenic.aertslab.org/) to infer the gene regulatory networks from our scRNA-seq data and identify key TFs during the development of MSCs.27
Cell sorting
Freshly harvested BMNCs were suspended in 4 °C HBSS + (Hanks-Balanced Salt Solution supplemented with 2% FBS, 10 mM HEPES, and 1% penicillin/streptomycin), followed by staining fluorochrome-conjugated or isotype control antibodies on ice for 30 min. The antibodies used in the present study were as follows: anti-CD45-APC (BioLegend, clone 2D1, 1:200), anti-CD45-Pacific Blue (BioLegend, clone 2D1, 1:200), anti-CD31-APC (BioLegend, clone WM59, 1:200), anti-CD31-Pacific Blue (BioLegend, clone WM59, 1:200), anti-CD235a-APC (Biolegend, clone HI264, 1:200), anti-CD235a-Pacific Blue (Biolegend, clone HI264, 1:200), anti-CD43-APC (Biolegend, clone CD43-10G7, 1:200), anti-CD44-PE (Biolegend, clone BJ18, 1:200), anti-CD73-APCcy7 (BioLegend, clone AD2, 1:200), anti-TM4SF1-FITC (Miltenyi Biotec, clone REA851, 1:200), anti-CD118-PE (BD, clone, 1:200), anti-CD140b (PDGFRβ)-APC (BioLegend, clone 18A2, 1:200), and anti-TM4SF1-Alexa Fluor 405 (RandD, FAB8164V, 1:100). Flow cytometry analysis and sorting were performed on a triple-laser MoFlo (Dako) or FACSCalibur (BD) flow cytometer, and data were analyzed using FlowJo software (Tree Star).
Colony-forming unit fibroblast (CFU-F) assay
For CFU-F cultures, single cells were seeded into 96-well plates (1 single-cell/well) containing Mesenchymal Stem Cell Medium (ScienCell, US) supplemented with 1% Penicillin/Streptomycin solution, and incubated at 37 °C with 5% CO2. We then changed half of the medium every 3–4 days. When cultured for 2 weeks, the cells were fixed and stained with crystalline violet staining solution.
Transplantation
Nude mouse transplantation was performed as described previously. Briefly, approximately 103–104 cells from single clones were mixed with β-TCP carrier (Bicon, Boston, MA, USA) and then subcutaneously implanted into the dorsal side of nude mice. Specimens were harvested at 4 and 8 weeks after transplantation, and the animals were killed by CO2 asphyxiation. The bone constructs were fixed in 4% paraformaldehyde and decalcified in 10% EDTA (pH 7.4) for 10 days. Finally, the specimens were dehydrated and then embedded in paraffin. All animal experiment conducted in the current study were approved by the Peking University Biomedical Ethics Committee on Experimental Animal Ethics.
Histological staining
Bones tissues were fixed in 4% paraformaldehyde at 4 °C for 1–3 days by continuous agitation, and then decalcified at room temperature. Replace the fresh 14% EDTA solution every 24 h, in which EDTA was dissolved in Milli-Q water, and adjusted the pH value to 7.1 with ammonium hydroxide. After complete decalcification, the tissues were washed in PBS for 2 h, and then soaked in PBS containing 30% sucrose. Next, put the samples at 4 °C under constant agitation overnight and finally embedded in paraffin. Bone sections (5-μm thickness) were stained with hematoxylin and eosin (H&E) and Oil-red O staining.
Supplementary information
Acknowledgements
This project was supported by grants from the National Natural Science Foundation of China (81930026 and 81970911), the National Key R&D Program of China (2017YFA0102702 and 2018YFA0107601), and the Beijing Municipal Science & Technology Commission (Z181100001318001). We thank Professor Tiejun Li and Doctor Jianyun Zhang in Pathology Department of School of Stomatology, Peking University for helpful advice with morphological analysis of bone tissue sections; and Yan Liu in Functional Analysis Core Facility of the National Center for Protein Science Beijing (Tsinghua University) for flow cytometry.
Author contributions
Y.Z., F.T., J.Q., P.Z. design and conception; P.Z., X.F., laboratory work, data collection and analysis; J.D. data analyses; J.Y., M.Y., X.W., tissue dissection; Y.L., L.L., X.Z. laboratory work, provision of study material; P.Z., J.D., Y.Z., F.T., J.Q., manuscript writing; Y.Z. F.T., J.Q. financial support, final approval of manuscript.
Data availability
All sequencing data in this work are stored in the Genome Sequence Archive (GSA) database (project number: PRJCA012045).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ping Zhang, Ji Dong, Xiaoying Fan
Contributor Information
Jie Qiao, Email: jie.qiao@263.net.
Fuchou Tang, Email: tangfuchou@pku.edu.cn.
Yongsheng Zhou, Email: kqzhouysh@hsc.pku.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-023-01338-2.
References
- 1.Serowoky MA, Arata CE, Crump JG, Mariani FV. Skeletal stem cells: insights into maintaining and regenerating the skeleton. Development. 2020;147:dev179325. doi: 10.1242/dev.179325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morikawa S, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianco P, Robey PG. Skeletal stem cells. Development. 2015;142:1023–1027. doi: 10.1242/dev.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendez-Ferrer S, Scadden DT, Sanchez-Aguilera A. Bone marrow stem cells: current and emerging concepts. Ann. N. Y. Acad. Sci. 2015;1335:32–44. doi: 10.1111/nyas.12641. [DOI] [PubMed] [Google Scholar]
- 5.Mendez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worthley DL, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacchetti B, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Tormin A, et al. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood. 2011;117:5067–5077. doi: 10.1182/blood-2010-08-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quirici N, et al. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp. Hematol. 2002;30:783–791. doi: 10.1016/S0301-472X(02)00812-3. [DOI] [PubMed] [Google Scholar]
- 11.Jones EA, et al. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis Rheumatol. 2002;46:3349–3360. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 12.Buhring HJ, et al. Novel markers for the prospective isolation of human MSC. Ann. N. Y. Acad. Sci. 2007;1106:262–271. doi: 10.1196/annals.1392.000. [DOI] [PubMed] [Google Scholar]
- 13.Abbuehl JP, Tatarova Z, Held W, Huelsken J. Long-term engraftment of primary bone marrow stromal cells repairs niche damage and improves hematopoietic stem cell transplantation. Cell Stem Cell. 2017;21:241–255. doi: 10.1016/j.stem.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Rombouts WJ, Ploemacher RE. Primary murine MSC show highly efficient homing to the bone marrow but lose homing ability following culture. Leukemia. 2003;17:160–170. doi: 10.1038/sj.leu.2402763. [DOI] [PubMed] [Google Scholar]
- 15.Park D, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasumarthy KK, et al. Methylome analysis of human bone marrow MSCs reveals extensive age- and culture-induced changes at distal regulatory elements. Stem Cell Rep. 2017;9:999–1015. doi: 10.1016/j.stemcr.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sipp D, Robey PG, Turner L. Clear up this stem-cell mess. Nature. 2018;561:455–457. doi: 10.1038/d41586-018-06756-9. [DOI] [PubMed] [Google Scholar]
- 18.Baccin C, et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020;22:38–48. doi: 10.1038/s41556-019-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baryawno N, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houlihan DD, et al. Isolation of mouse mesenchymal stem cells on the basis of expression of Sca-1 and PDGFR-α. Nat. Protoc. 2012;7:2103–2111. doi: 10.1038/nprot.2012.125. [DOI] [PubMed] [Google Scholar]
- 21.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu HS, et al. Tracing the skeletal progenitor transition during postnatal bone formation. Cell Stem Cell. 2021;28:2122–2136. doi: 10.1016/j.stem.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Prolif. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 26.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou BO, et al. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 2017;19:891–903. doi: 10.1038/ncb3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omatsu Y, et al. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Li D, et al. VCAM-1(+) macrophages guide the homing of HSPCs to a vascular niche. Nature. 2018;564:119–124. doi: 10.1038/s41586-018-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yugami M, et al. Mice deficient in angiopoietin-like protein 2 (Angptl2) gene show increased susceptibility to bacterial infection due to attenuated macrophage activity. J. Biol. Chem. 2016;291:18843–18852. doi: 10.1074/jbc.M116.720870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mabuchi Y, et al. LNGFR(+)THY-1(+)VCAM-1(hi+) cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep. 2013;1:152–165. doi: 10.1016/j.stemcr.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilpinen L, et al. Aging bone marrow mesenchymal stromal cells have altered membrane glycerophospholipid composition and functionality. J. Lipid Res. 2013;54:622–635. doi: 10.1194/jlr.M030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churchman SM, et al. Transcriptional profile of native CD271+ multipotential stromal cells: evidence for multiple fates, with prominent osteogenic and Wnt pathway signaling activity. Arthritis Rheumatol. 2012;64:2632–2643. doi: 10.1002/art.34434. [DOI] [PubMed] [Google Scholar]
- 37.Kfoury Y, Scadden DT. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 38.Chan CKF, et al. Identification of the human skeletal stem cell. Cell. 2018;175:43–56. doi: 10.1016/j.cell.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jitschin R, et al. Inflammation-induced glycolytic switch controls suppressivity of mesenchymal stem cells via STAT1 glycosylation. Leukemia. 2019;33:1783–1796. doi: 10.1038/s41375-018-0376-6. [DOI] [PubMed] [Google Scholar]
- 40.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S I, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 42.Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander D, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Smyth GK, Wei S. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 45.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi S, et al. Meta-and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu X, et al. Reversed graph embedding resolves complex single-cell developmental trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data in this work are stored in the Genome Sequence Archive (GSA) database (project number: PRJCA012045).