Abstract
CD9 is a crucial regulator of cell adhesion in the immune system and plays important physiological roles in hematopoiesis, blood coagulation or viral and bacterial infections. It is involved in the transendothelial migration of leukocytes which might also be hijacked by cancer cells during their invasion and metastasis. CD9 is found at the cell surface and the membrane of exosomes affecting cancer progression and therapy resistance. High expression of CD9 is mostly associated with good patients outcome, with a few exceptions. Discordant findings have been reported for breast, ovarian, melanoma, pancreatic and esophageal cancer, which might be related to using different antibodies or inherent cancer heterogeneity. According to in vitro and in vivo studies, tetraspanin CD9 is not clearly associated with either tumor suppression or promotion. Further mechanistic experiments will elucidate the role of CD9 in particular cancer types and specific conditions.
Keywords: CD9, cancer, immunohistochemistry, prognosis, exosomes
1. Introduction
Tetraspanin CD9, also known as TSPAN29 or motility-related protein 1, is a member of the transmembrane 4 superfamily proteins, which are characterized by four transmembrane domains, two extracellular loops, and short intracellular N-and C-terminal tails ( Figure 1A ) (1, 2). Like other tetraspanins, CD9 can undergo palmitoylation on each of its membrane-proximal cysteines which affects its interactions with other partners (3). Tetraspanins generally form tetraspanin-enriched microdomains (TEMs) in cell membranes. Within these domains, they interact with various transmembrane and intracellular partners, including other tetraspanins, integrins, proteases, immunoglobulins, and intracellular signaling proteins (4). Therefore, the biological effects of CD9 depend on these dynamic interactions within the context of TEMs (1, 24). Recently, a “concatenation model” for forming CD9/EWI-F assemblies has been suggested, which may explain the occurrence of these TEMs (25). Besides tetraspanins (e.g. CD63, CD81, CD151 and TSPAN4), CD9 interacts with numerous single-span transmembrane proteins, such as integrins (e.g. CD49c/ITGA3 and CD29 (1, 26), immunoglobulin superfamily proteins (e.g. EWI-F/PTGFRN and EWI-2/IGSF8) (1, 27), heparin-binding EGF-like growth factor (28), and metalloprotease ADAM17 (A Disintegrin And Metalloproteinase 17) (29). Previously, CD9 has also shown the ability to interact with other proteins such as CD19, CD46 and CD117 (30–32).
Figure 1.
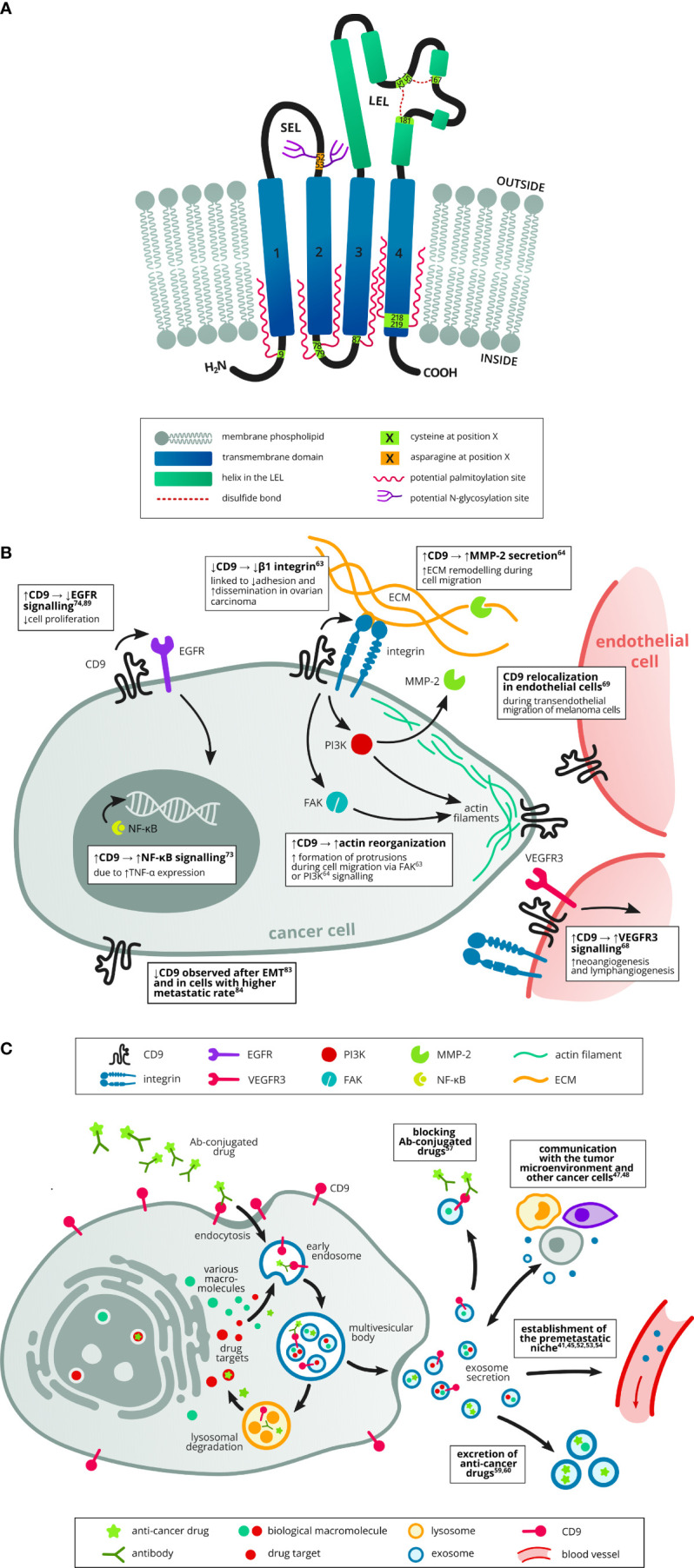
Structure of tetraspanin CD9 and its role in cancer, including exosome trafficking. (A) The CD9 protein consists of four transmembrane domains (1–4), short (EC1, SEL) and long (EC2, LEL) extracellular loop, short intracellular loop and short intracellular N- and C-termini. There are several possible palmitoylation sites made up of membrane-proximal cysteines and a possible N-glycosylation site in the SEL. In the LEL, there are two disulfide bridges, each containing one cysteine of the CCG motif (152–154), a typical feature of the tetraspanin family. Based on UniProt (AC: P21926, cited 1.8.2022). (B) CD9 was implicated both in tumor promoting and suppressing mechanisms. Several studies have described its role in cell migration and invasion, e.g. by affecting actin-polymerization and reorganisation at the cell protrusions (5, 6) or by increasing the production of the proteinase MMP-2 which cleaves ECM components during cell invasion (6). Increased CD9 expression was also linked to increased signalling in the protumorigenic NF-κB pathway (7). However, increased CD9 expression was shown to attenuate EGFR signalling and thus suppress cell proliferation (8, 9). Another study described higher metastatic rate in cells with decreased CD9 expression (10). CD9 downregulation was also observed in cells which underwent EMT (11). CD9 can also affect tumor neoangiogenesis by promoting VEGFR3 signalling in endothelial cells (12). Last but not least, transendothelial migration of tumor cells is supported by CD9 reorganisation at points of contact between endothelial and tumor cells (13). (C) Exosomes can transport cargo between cells in the tumor microenvironment (other tumor cells, stromal cells, immune cells) and thus enable mutual communication (14, 15). They also help establish the premetastatic niche in the target organ before colonization (16–20). Exosomes can also promote drug resistance via several mechanisms, e.g. by transporting drugs out of the tumor cells (21, 22) or by neutralisation of antibody-conjugated drugs (23).
An equally important role is played by the interaction of tetraspanins with intracellular signaling molecules, although significantly fewer of them have been identified compared to transmembrane partners (4). In the context of CD9, these are mainly interactions with small GTPases of the Rho family (Rac and RhoA) that affect the actin cytoskeleton (33, 34), ERM proteins (ezrin-radixin-moesin) that mediate binding with the cytoskeleton, and PKC (35), which regulates the function of a wide range of proteins and intracellular signaling. Obviously, tetraspanin CD9 plays a complex role both in physiological conditions as well as in many diseases including cancer ( Figure 1B ).
2. Physiological roles of CD9
CD9 is a key regulator of cell adhesion in the immune system and plays an important role in the physiology of leukocytes and endothelial cells as well as in hematopoiesis and blood coagulation. Other physiological processes with the important role of CD9 include sperm-egg fusion (36), neurite outgrowth (37) or myotube formation (38). Recently, CD9 and tetraspanin 4 were revealed as membrane curvature sensors which play an essential role in the formation of migrasome and fertilization (39). In this sense, it was also shown that the reversed cone-like molecular shape of CD9 generates membrane curvature in the crystalline lipid layers, which explains the CD9 localization in regions with high membrane curvature and its implications in membrane remodeling (40). CD9 is also an exosomal marker and may affect the cellular exosomal transport and interactions between tumor cells and the stromal microenvironment (see chapter below).
One of the important functions of CD9 is the regulation of hematopoietic stem cell differentiation in the bone marrow and critical hematopoiesis events. CD9 is essential in megakaryocytic (41), lymphoid and myeloid differentiation (42). In vitro work proved that immature CD34+ bone marrow cells express high levels of CD9, while differentiated cells lose their expression (41). Also, CD9-expressing stromal cells of bone marrow affect hematopoietic cells and may be one factor that determines the degree of stem cell differentiation (43). Next, CD9 is involved in the blood clotting process, because it is a component of integral membrane proteins expressed on the cell surface and granular membranes of thrombocytes, which play an essential role in the coagulation process. It is part of an alpha 2b/beta 3 - CD9 - CD63 integrin - tetraspanin complex in activated platelets (44, 45).
The role of CD9 in different immune cells and its relevance to inflammation has recently been reviewed by Brosseau et al. (2). For example, the tetraspanin CD9 plays an important role in the immune synapse in two ways:/1/Through its association with LFA-1 on the T cell, CD9 controls the state of aggregation and adhesive capacity of this integrin./2/On the surface of the antigen-presenting cells, CD9 recruits ICAM-1 into TEMs, thus increasing its adhesive capacity (1). Its importance was also shown for the endothelial receptors such as integrin ligands ICAM-1 and VCAM-1, which facilitate leukocyte adhesion to the endothelium and their subsequent transmigration. Another important CD9 function is the inhibition of ADAM10 and ADAM17 sheddase activity, which enhances cell-cell adhesion and costimulatory capacity (1).
CD9 also affects viral (46) and bacterial infections (47, 48). Within TEMs, CD9 modulates various virus-induced processes at the membrane, including membrane fusion, viral budding and viral release. Sims et al. demonstrated that exosomes could enhance HIV-1 entry into human T and monocytic cell lines via exosomal tetraspanin proteins CD81 and CD9 (46). Infection by enveloped coronaviruses initiates with viral spike proteins binding to cellular receptors and is followed by proteolytic cleavage which prompts virus-cell membrane fusion. Infection, therefore, requires the proximity of receptors and proteases which ensures that virus-cell entry occurs at the appropriate time and place. Earnest et al. showed that CD9 is crucial for condensing these receptors and proteases (DPP4 and TMPRSS2, respectively) which allows viruses to enter cells efficiently and rapidly (49, 50). Although it might be reasonable to think that SARS-CoV-2 virulence relies on CD9 activity by clustering and scaffolding receptor and protease (i.e. ACE2 and TMPRSS2, respectively) for efficient cell entry, this hypothesis has not been validated yet in current literature (50, 51).
3. CD9 in exosomes and cell-to-cell communication
There is a growing interest in cell-cell communication mediated by secreted vesicles termed exosomes ( Figure 1C ) (52). Exosomes are nanoscaled extracellular vesicles (EVs) (generally, their sizes range from 30 to 150 nm) released by almost all cell types. Tetraspanins are an abundant component of exosome membranes, with CD9 being one of the most frequently found along with CD63, CD81, CD82 and TSPAN8 (53, 54). Functionally it has been demonstrated that CD9 knock-down in extracellular vesicles from breast cancer cells or recipient cells reduced endocytosis (55). Importantly, tetraspanins can influence the composition of exosomes through interactions with their binding partners. For example, a decrease in CD9 expression led to a significant reduction in the metalloprotease CD10 content in exosomes of pre-B-lymphocytes (56). CD10 can serve as a positive (57) and negative (58) prognostic factor in some cancers, but its role in exosomes has not yet been described. It has also been shown that the alteration of CD9 and CD151 on prostate cells alters the proteome of their resultant EVs and that these EVs can enhance the migratory and invasive capabilities of a non-tumorigenic prostate cellular population (59). The cargo of exosomes reflects the state of tumor cells from which they are derived. They can be explored as minimally invasive biomarkers for the early detection, diagnosis and prognosis of various cancers (16, 44, 60). Currently, there are many methods for exosome isolation and detection, ranging from classical ultracentrifugation or filtration to immunoaffinity, flow cytometry and acoustics-based microfluidic techniques (61, 62).
The mechanisms of exosome biogenesis are highly regulated through several distinct pathways, including ESCRT (endosomal sorting complexes required for transport) ‐ dependent and ESCRT‐independent pathways (17). Although the exosome release is a physiological process, its increased rate and specific cargo are favorable for oncogenic progression and metastases (16). Regarding CD9, its alterations affect extracellular vesicle secretion and mitophagy in melanoma cells (63). The exosome‐mediated communication is not limited to the cancer cells, it has also been shown in different cell types within the tumor microenvironment locally and distantly. Bioactive molecules, including CD9 in exosomes derived from cancer and stromal cells, provide the essential signals for the re-education of various cells and remodeling the tumor architecture (14, 15). For instance, in pancreatic ductal adenocarcinoma, CD9 mediated EV uptake from cancer-associated fibroblasts that promoted tumor development (15). In the model of colon cancer blocking EV-derived CD9 by antibody prevented the morphological transformation and migratory phenotype of cancer cells that uptake EVs (64). The CD9-positive EVs were higher in patients with prostate cancer compared to ones with benign prostate hyperplasia, and its secretion can be modulated in response to dihydrotestosterone. Importantly, siRNA knockdown of endogenous CD9 reduced cellular proliferation and expression of AR and prostate-specific antigen. However, knockdown of AR did not alter CD9 expression, implicating CD9 as an upstream regulator of AR (65). The exosomes may also determine organotropism and prepare the pre-metastatic niche in the sense of Stephen Paget´s seed and soil hypothesis (17, 66). Cancer cells from primary tumors release oncogenic biomolecules to the distant site before the cell invasion occurs, forming a pre-metastatic niche in the target organ that promotes successful metastatic outgrowth (16–20).
Therapy resistance in cancer can also occur via exosomes in several ways (17). Corcoran et al. reported the transfer of MDR1 by exosomes which enhanced a docetaxel efflux out of the recipient cells (67). Exosomes from drug-resistant breast cancer cells could also transmit chemoresistance do adriamycin and docetaxel by a horizontal transfer of microRNAs (68). Aung et al. have shown that tumor‐derived exosomes can protect cancer cells by transporting an abundance of proteins targeted by drugs, hence neutralising the therapy effects (23). Similarly, cells of the tumor microenvironment also release exosomes that can enhance drug resistance in cancer cells. For example, fibroblast‐derived exosomes have been shown to decrease the efficiency of chemotherapy and radiation in cancer by activating STAT1 and NOTCH3 signaling, which resulted in the expansion of therapy-resistant tumor-initiating cells (69). Chemotherapeutic drugs may also be excreted from cancer cells via exosomes (21, 22). Together, these studies described various mechanisms of exosome‐mediated drug resistance either through pumping anticancer drugs out of cells or transferring molecular cargo between cells.
4. CD9 in cancer cells: Dr. Jekyll and Mr. Hyde
CD9 expression is deregulated in a number of pathologies, including cancer, but the precise mechanism underlying these changes and the associated consequences are not fully understood (4). Bioinformatic analysis of binding sites in the promoter/enhancer region of the CD9 gene identified E2F, NFkB, SP1 and STAT3 as top transcription factors often associated with both the process of carcinogenesis and disease prognosis (70). Nevertheless, the CD9 protein plays a dual role in cancer progression, exhibiting both tumor-supportive and tumor-suppressive properties that are context-dependent.
4.1. Tumor-promoting properties
The plasma membrane protrusions enable the spreading of neoplastic cells, helping them to move between and invade surrounding stromal cells. In addition to passing through intercellular gap junctions, the neoplastic cell can also use a transcellular route for intra/extravasation, which are the essential steps of the metastatic process (71). Overexpression of CD9 has been shown to enhance FAK phosphorylation and reorganisation of the cortical actin cytoskeleton in fibrosarcoma HT1080 cells plated on laminin (5). This was also associated with the induction of MMP2 and PI3K-dependent signaling (6). Aggressive triple-negative breast cancer MDA-MB-231 cells displayed significant alterations of their plasma membrane protrusions after CD9 knockdown and had reduced tumorigenic and metastatic capacity in mouse xenografts (72). The potential role of CD9 in metastasis was indicated in a different study, describing increased expression of CD9 in breast cancer bone metastasis compared to primary tumors, where CD9 antibody treatment in vivo moderately inhibited the progression of bone lesions (73). In this sense, CD9 expression and migration were induced by native type IV collagen through a DDR1-dependent pathway in the breast cancer model, but not in non-tumorigenic MCF10A and MCF12A cells (66). Surprisingly, along with the plasma membrane, CD9 can also localize in nuclei and its depletion led to polynucleation and multipolar mitosis (74).
CD9 has also been shown to interact with VEGFR3 signaling. After intrathoracic implantation of lung cancer cells, metastasis to lymph nodes was diminished and accompanied by decreased neoangiogenesis and lymphangiogenesis in CD9 knock-out mice (12). Knocking down CD9 in human lymphatic endothelial cells also decreased their migration, proliferation and tube formation which was associated with attenuated VEGFR3 signaling. Importantly, active redistribution of endothelial CD9 was also observed during interactions between melanoma and endothelial cells in an intravasation assay (13). Anti-CD9 monoclonal antibodies specifically inhibited the transendothelial migration of melanoma cells. Association of CD9 with transendothelial invasion has also been observed by immunohistochemistry in cervical cancer as well as in melanomas (75, 76). Similarly, Hori et al. found CD9 expression at severe vessel invasion in gastric cancer (77). CD9 upregulation was also detected in ovarian carcinomas by expression profiling and immunohistochemistry (7). The CD9 upregulation associated with enhanced expression of TNF-alpha and NFkB signaling and treatment with CD9 blocking antibody ALB6 resulted in reduced tumor growth in-vivo (7). CD9 can also attenuate EGF signaling pathways in gastrointestinal cancer cells by colocalizing with EGFR (8).
One of the key drivers of tumor progression are cancer stem cells (CSCs) capable to self-differentiate, self-renew and fueling tumor growth. It has been reported that CD9 identifies a subpopulation of pancreatic cancer stem cells (CSCs) able to initiate and sustain pancreatic cancer growth as demonstrated in CD9 deficient organoid and mice models (78). Mechanistically, CD9 promoted the plasma membrane localisation of the glutamine transporter ASCT2, enhancing glutamine uptake in cancer cells. CD9 has been identified as a marker of CSCs also in the glioblastoma model, where its disruption led to decreased cell proliferation, invasion, and inhibition of tumor growth (79, 80). Decreased cell migration was also reported in highly metastatic hepatocellular carcinoma cells upon CD9 silencing (81) or in breast cancer cells using CD9-binding peptide (82). The same group successfully reduced melanoma lung metastasis after peptide binding to tetraspanin CD9. The CD9-binding peptide impeded tetraspanin web formation, cancer cell invasion and significantly reduced secretion and uptake of cancer cell exosomes (83). Antibody targeting of CD9 in pancreatic cancer disrupted CD9/ADAM interactions and led to decreased proliferation, migration and colony formation (84).
Regarding small-cell lung cancer (SCLC), CD9 was expressed preferentially in SCLC tumors and metastases from three of seven relapsed patients, whereas chemonaïve primary tumors from 16 patients were CD9 negative with only one exception (85). Mechanistically, CD9 was upregulated in chemoresistant cell lines, which adhered more tightly to fibronectin via β1 integrin, but they were less motile than the respective chemosensitive parental lines, implying a potential role of CD9 molecule in the cell adhesion-mediated drug resistance (85).
4.2. Tumor-inhibiting properties
Regarding breast cancer, Remsik et al. observed the downregulation of CD9 in cancer cells that underwent epithelial-mesenchymal transition (EMT) both in vitro and in vivo (11). High CD9 expression is associated with epithelial phenotype and favorable prognosis regarding recurrence-free survival (11). However, further mechanistic studies will be needed to clarify the role of CD9 in EMT and breast cancer progression. In ovarian cancer, the downregulation of CD9 attenuated the expression of several integrins and rearranged junctional and cytoskeletal molecules which was associated with weaker adhesion to the extracellular matrix (10). Enhanced peritoneal dissemination was observed for subclones with low CD9 expression (10), consistent with a previous report of inverse correlation of CD9 and ovarian cancer tumor stage (86). Decreased CD9 clustering may reflect the tendency of malignant cells to have less organized cell-cell junctions where tetraspanins are typically known to be clustered (3). Low affinity anti-CD9 antibody, C9BB, which binds preferentially to CD9 homodimer was used in experiments documenting a shift to heterodimers in cancer cells. This may be associated with decreased CD9 palmitoylation or altered expression of CD9 partners (3).
Takeda et al. observed a decreased lymph node metastasis of lung cancer cells transduced with CD9 without impact on the primary tumor growth (87). Similarly, ectopic expression of CD9 in fibrosarcoma cell line HT1080 reduced their lung metastatic ability by forming a complex with podoplanin, suppressing podoplanin-induced platelet aggregation (88). In line with these studies, genetic ablation of CD9 in a model of mouse prostate adenocarcinoma did not affect primary tumor development. Still, it increased the incidence of metastases to the liver but not the lungs, suggesting a possible tissue-specific manner of CD9 interactions in this model (89).
The antiproliferative effect of CD9 was also observed in in vitro model of human glioblastoma executed via inhibition of EGFR phosphorylation (9). In contrast, the downregulation of CD9 promoted cancer growth and metastasis through the upregulation of EGF in pancreatic cancer (90). In the context of SCLC that develops distant metastases extremely early, Funakoshi et al. observed the downregulation of tetraspanin CD9 in all cell lines. CD9 recovery suppressed cell motility of SCLC cells, suggesting that low expression of CD9 affects cell motility and may contribute to the highly invasive and metastatic phenotype of SCLC (91). Likewise CD9 overexpression in hepatocellular carcinoma inhibited proliferation in vitro and in vivo while CD9 knockdown enhanced in vivo growth (92).
5. Prognostic value of CD9 in solid tumors and a problem of different antibodies
Besides studies dealing with the molecular function of CD9, expression of this gene was also monitored in large cancer patient cohorts concerning tumor aggressiveness and survival. Relevant articles since 1993 are summarized in Table 1 and some are briefly commented on below. High expression of CD9 is mostly associated with good patient outcomes, with a few exceptions. These might be attributed to special cancer subtypes and different antibodies used for CD9 staining.
Table 1.
(A) Summary of immunohistochemical studies – high CD9 expression associates with good clinical outcome.
| Authors | Tumor Type | Patients | Detection method | Antibody clone | Source | Therapy | Clinical outcome with high CD9 |
|---|---|---|---|---|---|---|---|
| Miyake et al., 1995 (93) | breast ca | 143 | IHC, RT-PCR, western blotting | m31-15 | in-house | S | less aggressive disease |
| Arihiro et al., 1998 (94) | breast ca | 93 | IHC, western blotting | n.s. | Dako | S | good prognosis, low risk of LN metastases |
| Houle et al., 2002 (86) | ovarian ca | 40 | IHC | m31-15 | in-house, Dr. Miyake | S/AC | less aggressive disease |
| Sauer et al., 2003 (75) | cervical ca | 44 | IHC | 72F6 | Novocastra | S | good prognosis |
| Miyamoto et al., 2001 (95) | endometrial ca | 71 | IHC | TP82 | Temecula, CA | S | good prognosis |
| Wang et al., 2007 (27) | prostatic ca | 167 | IHC | 72F6 | Novocastra | S | less aggressive disease |
| Mhawech et al., 2003 (96) | urothelial blader ca | 320 | IHC | n.s. | Novocastra | TURBT | good prognosis |
| Ai et al., 2007 (97) | urothelial blader ca | 52 | IHC | n.s. | Jingmei Biotech | TURBT | good prognosis |
| Buim et al., 2010 (98) | oral squamous ca | 179 | IHC, RT-PCR | n.s. | Neomarkers | S | good prognosis |
| Kusukawa et al., 2001 (99) | oral squamous ca | 78 | IHC, western blotting | clone 007 | in-house | S | good prognosis |
| Uchida et al., 1999 (100) | esophageal squamous ca | 108 | IHC | m31-15 | in-house | S | good prognosis |
| Zou et al., 2012 (101) | ca of gallblader | 108 | IHC | n.s. | Dako | S | good prognosis |
| Khushman et al., 2019 (102) | pancreatic ca | 29 | IHC | n.s. | Abcam | S | less aggressive disease |
| Kim et al., 2016 (103) | colorectal ca | 305 | IHC | EPR2949 | Abcam | S/AC | good prognosis in left-side tumors only |
| Hashida et al. 2003 (104) | colorectal ca | 146 | IHC, RT-PCR | m31-15 | in-house | S | good prognosis |
| Amatya et al., 2013 (105) | mesothelioma | 112 | IHC | 72F6 | Novus Biologicals | S/AC/RT | good prognosis |
| Si et Hersey 1993 (106) | melanoma | 55 | IHC | FMC56 | in-house, Dr. Zola | S | less aggressive disease |
| Woegerbauer et al., 2010 (107) | Merkell cell ca | 25 | IHC, RT-PCR, western blotting | RDI-MCD9 | Fitzgerald | S/AC/RT | good prognosis |
| Kohmo et al., 2010 (85) | small cell lung ca | 24 | IHC, RT-PCR, western blotting, flow cytometry | 72F6 | Novocastra | AC | good prognosis |
| Adachi et al., 1998 (108) | non small lung ca | 172 | IHC, RT-PCR | m31-15 | in house | S | good prognosis |
| Higashiyama et al., 1995 (109) | adenoca of lung | 132 | IHC, RT-PCR | m31-15 | in house | S | good prognosis |
AC, adjuvant chemotherapy; ca, carcinoma; IHC, immunohistochemitry; LN, lymph node; NAC, neoadjuvant chemotherapy; n.s. not specified; RT, radiotherapy; S, surgery; TURBT; transuretral resection of bladder tumor.
Discordant findings have been reported for breast, ovarian, melanoma, pancreatic and esophageal cancer. The good prognostic value of high CD9 expression was described in studies using the in-house mouse monoclonal antibody m31-15 (93, 100) and a monoclonal antibody from Dako (94). Of note, studies using the antibody clone m31-15 consistently report the good prognostic value of high CD9 in all cancer types (see Tables 1A , 1B ). On the other hand, other groups reported poor outcomes for breast and ovarian cancer patients with high CD9 expression using Abcam monoclonal antibody EPR2949 or an antibody from Millipore (7, 113, 114). Kwon et al. also evaluated stromal immune cells and their CD9 expression was associated with good patient outcome (113). The same group used the EPR2949 antibody also for colorectal cancer where the high CD9 expression in tumors was associated with a good prognosis (103). Regarding stroma, high stromal CD9 evaluated with antibody clone C-4 was also associated with better survival of patients with pancreatic ductal adenocarcinoma, while positive tumor CD9 showed opposite results (115).
Table 1.
(B) Summary of immunohistochemical studies – high CD9 expression associates with poor clinical outcome.
| Authors | Tumor Type | Patients | Detection method | Antibody clone | Source | Therapy | Clinical outcome with high CD9 |
|---|---|---|---|---|---|---|---|
| Kim et al., 2019 (110) | papillary thyroid ca | 553 | IHC | n.s. | Novocastra | S | associated with LN metastases |
| Hori et al., 2004 (77) | gastric ca | 78 | IHC, nothern and western blotting | 72F6 and 007 | Novocastra and Dr. Mekada, resp. | S/AC | associated with advanced disease |
| Soyuer et al., 2010 (111) | gastric ca | 49 | IHC | 72F6 | Novocastra | S/AC | poor prognosis |
| Miki et al., 2018 (14) | gastric ca | 619 | IHC | n.s. | Life Technologies | S | poor prognosis, in particular of the scirrhous type |
| Huan et al., 2015 (112) | esophageal squamous ca | 104 | IHC, western blotting | n.s. | Santa Cruz | S | poor prognosis, associated with advanced disease |
| Kwon et al., 2017 (113) | breast ca | 1349 | IHC | EPR2949 | Abcam | S/AC | poor prognosis in luminal A subtype |
| Baek et al., 2019 (114) | lobular breast ca | 113 | IHC | EPR2949 | Abcam | S/AC/RT | poor prognosis |
| Hwang et al.2012 (7) | ovarian ca | 30 | IHC, microarray, RT-PCR, western blotting | n.s. | Millipore | S | associated with advanced disease |
| Lucarini et al., 2022 (76) | cutaneous melanoma | 120 | IHC | 72F6 | Novocastra | S | poor prognosis, associated with LN metastases |
| Han et al., 2022 (115) | pancreatic ca, NAC | 179 | IHC | C-4 | Santa Cruz | NAC/S | poor prognosis |
AC, adjuvant chemotherapy; ca, carcinoma; IHC, immunohistochemitry; LN, lymph node; NAC, neoadjuvant chemotherapy; n.s. not specified; RT, radiotherapy; S, surgery; TURBT; transuretral resection of bladder tumor.
Another frequently used antibody is clone 72F6 from Novocastra. High levels of CD9 evaluated with this antibody were associated with poor prognosis in gastric cancer (111), while the association with good prognosis was found for mesothelioma, cervical and prostate cancer (75, 105, 116). Several other studies used an antibody from Novocastra without closer specification, complicating their findings’ interpretation. One of these studies investigated breast cancer samples and found no prognostic value of CD9 (117).
Overall, the interpretation of immunohistochemistry studies is problematic due to the use of different antibodies that are not thoroughly validated and essential details are missing. For example, epitopes for the most frequently used monoclonal antibodies m31-15, 72F6 and EPR2949 are unavailable. Two recent meta-analyses concluded that low CD9 expression is significantly associated with poor prognosis of cancer patients (118, 119) however, these results are oversimplified and problematic with respect to the abovementioned issues.
As described in the previous chapter, CD9 is not clearly associated with either tumor suppression or promotion. Additional mechanistic studies have recently been thoroughly reviewed by Lorico et al. They also summarized CD9 targeting with therapeutic antibodies and drew attention to the potential side effects of this strategy. This approach might further be complicated by the presence of CD9 in exosomes which may neutralize the therapeutic antibodies or enhance the pro-metastatic effects of exosomes by their enhanced endocytosis (53).
6. Conclusion
A growing body of evidence points to the important role of CD9 in various physiological processes and cancer. As highlighted in the review, CD9 is not unequivocally associated with either tumor suppression or promotion, and the antibodies used to detect CD9 might be problematic. Despite conflicting results in different types of cancer, the clinical relevance of CD9 has been highlighted by several immunohistochemical and, more importantly, mechanistic studies. Therapeutic targeting of CD9 is emerging, however, this approach may be complicated by the presence of CD9 in exosomes, which may neutralize therapeutic antibodies or enhance the pro-metastatic effects of exosomes by their increased endocytosis. In conclusion, fully validated antibodies and well-designed functional studies may help to elucidate further the role of CD9 in cancer progression and patient clinical outcome.
Author contributions
JB, SB, KS – conceptualization. RO – writing, preparation of original draft. BK, KK, KS – writing, reviewing and editing. KK - graphical figures preparation. JB - supervision, writing, reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by funds from the Czech Health Research Council (grant NU21-08-00023) and the Czech Science Foundation (grant nr. 20-22984S). JB was supported by DRO (FNOL, 00098892) from the Czech Ministry of Health and by the project of National Institute for Cancer Research (Programme EXCELES, ID Project No. LX22NPO5102) - Funded by the European Union - Next Generation EU.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Reyes R, Cardenes B, Machado-Pineda Y, Cabanas C. Tetraspanin CD9: A key regulator of cell adhesion in the immune system. Front Immunol (2018) 9:863. doi: 10.3389/fimmu.2018.00863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brosseau C, Colas L, Magnan A, Brouard S. CD9 tetraspanin: A new pathway for the regulation of inflammation? Front Immunol (2018) 9:2316. doi: 10.3389/fimmu.2018.02316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang XH, Kovalenko OV, Kolesnikova TV, Andzelm MM, Rubinstein E, Strominger JL, et al. Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J Biol Chem (2006) 281:12976–85. doi: 10.1074/jbc.M510617200 [DOI] [PubMed] [Google Scholar]
- 4. Rubinstein E, Charrin S, Tomlinson MG. Organisation of the tetraspanin web. In: Tetraspanins. Dordrecht: Springer Netherlands; (2013). p. 47–90. doi: 10.1007/978-94-007-6070-7 [DOI] [Google Scholar]
- 5. Berditchevski F, Odintsova E. Characterization of integrin-tetraspanin adhesion complexes: Role of tetraspanins in integrin signaling. J Cell Biol (1999) 146:477–92. doi: 10.1083/jcb.146.2.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sugiura T, Berditchevski F. Function of alpha3beta1-tetraspanin protein complexes in tumor cell invasion. evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2). J Cell Biol (1999) 146:1375–89. doi: 10.1083/jcb.146.6.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hwang JR, Jo K, Lee Y, Sung B-J, Park YW, Lee J-H. Upregulation of CD9 in ovarian cancer is related to the induction of TNF- alpha gene expression and constitutive NF-kappaB activation. Carcinogenesis (2012) 33:77–83. doi: 10.1093/carcin/bgr257 [DOI] [PubMed] [Google Scholar]
- 8. Murayama Y, Shinomura Y, Oritani K, Miyagawa J-I, Yoshida H, Nishida M, et al. The tetraspanin CD9 modulates epidermal growth factor receptor signaling in cancer cells. J Cell Physiol (2008) 216:135–43. doi: 10.1002/jcp.21384 [DOI] [PubMed] [Google Scholar]
- 9. Wang G-P, Han X-F. CD9 modulates proliferation of human glioblastoma cells via epidermal growth factor receptor signaling. Mol Med Rep (2015) 12:1381–6. doi: 10.3892/mmr.2015.3466 [DOI] [PubMed] [Google Scholar]
- 10. Furuya M, Kato H, Nishimura N, Ishiwata I, Ikeda H, Ito R, et al. Down-regulation of CD9 in human ovarian carcinoma cell might contribute to peritoneal dissemination: Morphologic alteration and reduced expression of beta1 integrin subsets. Cancer Res (2005) 65:2617–25. doi: 10.1158/0008-5472.CAN-04-3123 [DOI] [PubMed] [Google Scholar]
- 11. Remsik J, Fedr R, Navratil J, Bino L, Slabakova E, Fabian P, et al. Plasticity and intratumoural heterogeneity of cell surface antigen expression in breast cancer. Br J Cancer (2018) 118:813–9. doi: 10.1038/bjc.2017.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwasaki T, Takeda Y, Maruyama K, Yokosaki Y, Tsujino K, Tetsumoto S, et al. Deletion of tetraspanin CD9 diminishes lymphangiogenesis in vivo and in vitro . J Biol Chem (2013) 288:2118–31. doi: 10.1074/jbc.M112.424291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Longo N, Yáñez-Mó M, Mittelbrunn M, de la Rosa G, Muñoz ML, Sánchez-Madrid F, et al. Regulatory role of tetraspanin CD9 in tumor-endothelial cell interaction during transendothelial invasion of melanoma cells. Blood (2001) 98:3717–26. doi: 10.1182/blood.v98.13.3717 [DOI] [PubMed] [Google Scholar]
- 14. Miki Y, Yashiro M, Okuno T, Kitayama K, Masuda G, Hirakawa K, et al. CD9-positive exosomes from cancer-associated fibroblasts stimulate the migration ability of scirrhous-type gastric cancer cells. Br J Cancer (2018) 118:867–77. doi: 10.1038/bjc.2017.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nigri J, Leca J, Tubiana S-S, Finetti P, Guillaumond F, Martinez S, et al. CD9 mediates the uptake of extracellular vesicles from cancer-associated fibroblasts that promote pancreatic cancer cell aggressiveness. Sci Signal (2022) 15:eabg8191. doi: 10.1126/scisignal.abg8191 [DOI] [PubMed] [Google Scholar]
- 16. Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol (2018) 8:66. doi: 10.3389/fonc.2018.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tai Y-L, Chen K-C, Hsieh J-T, Shen T-L. Exosomes in cancer development and clinical applications. Cancer Sci (2018) 109:2364–74. doi: 10.1111/cas.13697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med (2012) 18:883–91. doi: 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature (2015) 527:329–35. doi: 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol (2015) 17:816–26. doi: 10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther (2005) 4:1595–604. doi: 10.1158/1535-7163.MCT-05-0102 [DOI] [PubMed] [Google Scholar]
- 22. Jorfi S, Ansa-Addo EA, Kholia S, Stratton D, Valley S, Lange S, et al. Inhibition of microvesiculation sensitizes prostate cancer cells to chemotherapy and reduces docetaxel dose required to limit tumor growth in vivo . Sci Rep (2015) 5:13006. doi: 10.1038/srep13006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M, et al. Exosomal evasion of humoral immunotherapy in aggressive b-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci U.S.A. (2011) 108:15336–41. doi: 10.1073/pnas.1102855108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol (2005) 6:801–11. doi: 10.1038/nrm1736 [DOI] [PubMed] [Google Scholar]
- 25. Soung YH, Ford S, Zhang V, Chung J. Exosomes in cancer diagnostics. Cancers (Basel) (2017) 9:8. doi: 10.3390/cancers9010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Radford KJ, Thorne RF, Hersey P. CD63 associates with transmembrane 4 superfamily members, CD9 and CD81, and with beta 1 integrins in human melanoma. Biochem Biophys Res Commun (1996) 222:13–8. doi: 10.1006/bbrc.1996.0690 [DOI] [PubMed] [Google Scholar]
- 27. Wang H-X, Li Q, Sharma C, Knoblich K, Hemler ME. Tetraspanin protein contributions to cancer. Biochem Soc Trans (2011) 39:547–52. doi: 10.1042/BST0390547 [DOI] [PubMed] [Google Scholar]
- 28. Iwamoto R, Higashiyama S, Mitamura T, Taniguchi N, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor, which acts as the diphtheria toxin receptor, forms a complex with membrane protein DRAP27/CD9, which up-regulates functional receptors and diphtheria toxin sensitivity. EMBO J (1994) 13:2322–30. doi: 10.1002/j.1460-2075.1994.tb06516.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Machado-Pineda Y, Cardenes B, Reyes R, Lopez-Martin S, Toribio V, Sanchez-Organero P, et al. CD9 controls integrin alpha5beta1-mediated cell adhesion by modulating its association with the metalloproteinase ADAM17. Front Immunol (2018) 9:2474. doi: 10.3389/fimmu.2018.02474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anzai N, Lee Y, Youn B-S, Fukuda S, Kim Y-J, Mantel C, et al. C-kit associated with the transmembrane 4 superfamily proteins constitutes a functionally distinct subunit in human hematopoietic progenitors. Blood (2002) 99:4413–21. doi: 10.1182/blood.v99.12.4413 [DOI] [PubMed] [Google Scholar]
- 31. Horvath G, Serru V, Clay D, Billard M, Boucheix C, Rubinstein E. CD19 is linked to the integrin-associated tetraspans CD9, CD81, and CD82. J Biol Chem (1998) 273:30537–43. doi: 10.1074/jbc.273.46.30537 [DOI] [PubMed] [Google Scholar]
- 32. Gutierrez-Lopez MD, Gilsanz A, Yanez-Mo M, Ovalle S, Lafuente EM, Dominguez C, et al. The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell Mol Life Sci (2011) 68:3275–92. doi: 10.1007/s00018-011-0639-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herr MJ, Mabry SE, Jennings LK. Tetraspanin CD9 regulates cell contraction and actin arrangement via RhoA in human vascular smooth muscle cells. PloS One (2014) 9:e106999. doi: 10.1371/journal.pone.0106999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnaud M-P, Vallée A, Robert G, Bonneau J, Leroy C, Varin-Blank N, et al. CD9, a key actor in the dissemination of lymphoblastic leukemia, modulating CXCR4-mediated migration via RAC1 signaling. Blood (2015) 126:1802–12. doi: 10.1182/blood-2015-02-628560 [DOI] [PubMed] [Google Scholar]
- 35. Zhang XA, Bontrager AL, Hemler ME. Transmembrane-4 superfamily proteins associate with activated protein kinase c (PKC) and link PKC to specific beta(1) integrins. J Biol Chem (2001) 276:25005–13. doi: 10.1074/jbc.M102156200 [DOI] [PubMed] [Google Scholar]
- 36. Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science (2000) 287:321–4. doi: 10.1126/science.287.5451.321 [DOI] [PubMed] [Google Scholar]
- 37. Kagawa T, Mekada E, Shishido Y, Ikenaka K. Immune system-related CD9 is expressed in mouse central nervous system myelin at a very late stage of myelination. J Neurosci Res (1997) 50:312–20. doi: [DOI] [PubMed] [Google Scholar]
- 38. Charrin S, Latil M, Soave S, Polesskaya A, Chretien F, Boucheix C, et al. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nat Commun (2013) 4:1674. doi: 10.1038/ncomms2675 [DOI] [PubMed] [Google Scholar]
- 39. Dharan R, Goren S, Cheppali SK, Shendrik P, Brand G, Vaknin A, et al. Transmembrane proteins tetraspanin 4 and CD9 sense membrane curvature. Proc Natl Acad Sci U.S.A. (2022) 119:e2208993119. doi: 10.1073/pnas.2208993119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Umeda R, Satouh Y, Takemoto M, Nakada-Nakura Y, Liu K, Yokoyama T, et al. Structural insights into tetraspanin CD9 function. Nat Commun (2020) 11:1606. doi: 10.1038/s41467-020-15459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clay D, Rubinstein E, Mishal Z, Anjo A, Prenant M, Jasmin C, et al. CD9 and megakaryocyte differentiation. Blood (2001) 97:1982–9. doi: 10.1182/blood.v97.7.1982 [DOI] [PubMed] [Google Scholar]
- 42. Oritani K, Wu X, Medina K, Hudson J, Miyake K, Gimble JM, et al. Antibody ligation of CD9 modifies production of myeloid cells in long-term cultures. Blood (1996) 87:2252–61. doi: 10.1182/blood.V87.6.2252.bloodjournal8762252 [DOI] [PubMed] [Google Scholar]
- 43. Aoyama K, Oritani K, Yokota T, Ishikawa J, Nishiura T, Miyake K, et al. Stromal cell CD9 regulates differentiation of hematopoietic stem/progenitor cells. Blood (1999) 93:2586–94. doi: 10.1182/blood.V93.8.2586 [DOI] [PubMed] [Google Scholar]
- 44. Israels SJ, McMillan-Ward EM, Easton J, Robertson C, McNicol A. CD63 associates with the alphaIIb beta3 integrin-CD9 complex on the surface of activated platelets. Thromb Haemost (2001) 85:134–41. doi: 10.1055/s-0037-1612916 [DOI] [PubMed] [Google Scholar]
- 45. Yun S-H, Sim E-H, Goh R-Y, Park J-I, Han J-Y. Platelet activation: The mechanisms and potential biomarkers. BioMed Res Int (2016) 2016:9060143. doi: 10.1155/2016/9060143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sims B, Farrow AL, Williams SD, Bansal A, Krendelchtchikov A, Matthews QL. Tetraspanin blockage reduces exosome-mediated HIV-1 entry. Arch Virol (2018) 163:1683–9. doi: 10.1007/s00705-018-3737-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ventress JK, Partridge LJ, Read RC, Cozens D, MacNeil S, Monk PN. Peptides from tetraspanin CD9 are potent inhibitors of staphylococcus aureus adherence to keratinocytes. PloS One (2016) 11:e0160387. doi: 10.1371/journal.pone.0160387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Green LR, Monk PN, Partridge LJ, Morris P, Gorringe AR, Read RC. Cooperative role for tetraspanins in adhesin-mediated attachment of bacterial species to human epithelial cells. Infect Immun (2011) 79:2241–9. doi: 10.1128/IAI.01354-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Earnest JT, Hantak MP, Park J-E, Gallagher T. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains. J Virol (2015) 89:6093–104. doi: 10.1128/JVI.00543-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Earnest JT, Hantak MP, Li K, McCray PBJ, Perlman S, Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PloS Pathog (2017) 13:e1006546. doi: 10.1371/journal.ppat.1006546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pironti G, Andersson DC, Lund LH. Mechanistic and therapeutic implications of extracellular vesicles as a potential link between covid-19 and cardiovascular disease manifestations. Front Cell Dev Biol (2021) 9:640723. doi: 10.3389/fcell.2021.640723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol (2014) 5:442. doi: 10.3389/fimmu.2014.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lorico A, Lorico-Rappa M, Karbanová J, Corbeil D, Pizzorno G. CD9, a tetraspanin target for cancer therapy? Exp Biol Med (Maywood) (2021) 246:1121–38. doi: 10.1177/1535370220981855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics (2010) 73:1907–20. doi: 10.1016/j.jprot.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 55. Rappa G, Santos MF, Green TM, Karbanová J, Hassler J, Bai Y, et al. Nuclear transport of cancer extracellular vesicle-derived biomaterials through nuclear envelope invagination-associated late endosomes. Oncotarget (2017) 8:14443–61. doi: 10.18632/oncotarget.14804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mazurov D, Barbashova L, Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J (2013) 280:1200–13. doi: 10.1111/febs.12110 [DOI] [PubMed] [Google Scholar]
- 57. Smollich M, Götte M, Yip GW, Yong E-S, Kersting C, Fischgräbe J, et al. On the role of endothelin-converting enzyme-1 (ECE-1) and neprilysin in human breast cancer. Breast Cancer Res Treat (2007) 106:361–9. doi: 10.1007/s10549-007-9516-9 [DOI] [PubMed] [Google Scholar]
- 58. Deschamps L, Handra-Luca A, O’Toole D, Sauvanet A, Ruszniewski P, Belghiti J, et al. CD10 expression in pancreatic endocrine tumors: Correlation with prognostic factors and survival. Hum Pathol (2006) 37:802–8. doi: 10.1016/j.humpath.2006.02.024 [DOI] [PubMed] [Google Scholar]
- 59. Brzozowski JS, Bond DR, Jankowski H, Goldie BJ, Burchell R, Naudin C, et al. Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Sci Rep (2018) 8:8822. doi: 10.1038/s41598-018-27180-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Figueroa J, Phillips LM, Shahar T, Hossain A, Gumin J, Kim H, et al. Exosomes from glioma-associated mesenchymal stem cells increase the tumorigenicity of glioma stem-like cells via transfer of miR-1587. Cancer Res (2017) 77:5808–19. doi: 10.1158/0008-5472.CAN-16-2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dong L, Zieren RC, Wang Y, de Reijke TM, Xue W, Pienta KJ. Recent advances in extracellular vesicle research for urological cancers: From technology to application. Biochim Biophys Acta Rev Cancer (2019) 1871:342–60. doi: 10.1016/j.bbcan.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 62. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev (2018) 118:1917–50. doi: 10.1021/acs.chemrev.7b00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Suárez H, Andreu Z, Mazzeo C, Toribio V, Pérez-Rivera AE, López-Martín S, et al. CD9 inhibition reveals a functional connection of extracellular vesicle secretion with mitophagy in melanoma cells. J Extracell Vesicles (2021) 10:e12082. doi: 10.1002/jev2.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Santos MF, Rappa G, Fontana S, Karbanová J, Aalam F, Tai D, et al. Anti-human CD9 fab fragment antibody blocks the extracellular vesicle-mediated increase in malignancy of colon cancer cells. Cells (2022) 11:2474. doi: 10.3390/cells11162474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soekmadji C, Riches JD, Russell PJ, Ruelcke JE, McPherson S, Wang C, et al. Modulation of paracrine signaling by CD9 positive small extracellular vesicles mediates cellular growth of androgen deprived prostate cancer. Oncotarget (2017) 8:52237–55. doi: 10.18632/oncotarget.11111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dos Anjos Pultz B, Andrés Cordero da Luz F, Socorro Faria S, Peixoto Ferreira de Souza L, Cristina Brígido Tavares P, Alonso Goulart V, et al. The multifaceted role of extracellular vesicles in metastasis: Priming the soil for seeding. Int J Cancer (2017) 140:2397–407. doi: 10.1002/ijc.30595 [DOI] [PubMed] [Google Scholar]
- 67. Corcoran C, Rani S, O’Brien K, O’Neill A, Prencipe M, Sheikh R, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PloS One (2012) 7:e50999. doi: 10.1371/journal.pone.0050999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen W, Liu X, Lv M, Chen L, Zhao J, Zhong S, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PloS One (2014) 9:e95240. doi: 10.1371/journal.pone.0095240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell (2014) 159:499–513. doi: 10.1016/j.cell.2014.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) (2017) 2017:bax028. doi: 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Khuon S, Liang L, Dettman RW, Sporn PHS, Wysolmerski RB, Chew T-L. Myosin light chain kinase mediates transcellular intravasation of breast cancer cells through the underlying endothelial cells: A three-dimensional FRET study. J Cell Sci (2010) 123:431–40. doi: 10.1242/jcs.053793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rappa G, Green TM, Karbanová J, Corbeil D, Lorico A. Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget (2015) 6:7970–91. doi: 10.18632/oncotarget.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kischel P, Bellahcene A, Deux B, Lamour V, Dobson R, DE Pauw E, et al. Overexpression of CD9 in human breast cancer cells promotes the development of bone metastases. Anticancer Res (2012) 32:5211–20. [PubMed] [Google Scholar]
- 74. Rappa G, Green TM, Lorico A. The nuclear pool of tetraspanin CD9 contributes to mitotic processes in human breast carcinoma. Mol Cancer Res (2014) 12:1840–50. doi: 10.1158/1541-7786.MCR-14-0242 [DOI] [PubMed] [Google Scholar]
- 75. Sauer G, Windisch J, Kurzeder C, Heilmann V, Kreienberg R, Deissler H. Progression of cervical carcinomas is associated with down-regulation of CD9 but strong local re-expression at sites of transendothelial invasion. Clin Cancer Res (2003) 9:6426–31. [PubMed] [Google Scholar]
- 76. Lucarini G, Molinelli E, Licini C, Rizzetto G, Radi G, Goteri G, et al. Tetraspanin CD9 expression predicts sentinel node status in patients with cutaneous melanoma. Int J Mol Sci (2022) 23:4775. doi: 10.3390/ijms23094775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hori H, Yano S, Koufuji K, Takeda J, Shirouzu K. CD9 expression in gastric cancer and its significance. J Surg Res (2004) 117:208–15. doi: 10.1016/j.jss.2004.01.014 [DOI] [PubMed] [Google Scholar]
- 78. Wang VM-Y, Ferreira RMM, Almagro J, Evan T, Legrave N, Zaw Thin M, et al. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat Cell Biol (2019) 21:1425–35. doi: 10.1038/s41556-019-0407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shi Y, Zhou W, Cheng L, Chen C, Huang Z, Fang X, et al. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ (2017) 24:167–80. doi: 10.1038/cdd.2016.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Podergajs N, Motaln H, Rajčević U, Verbovšek U, Koršič M, Obad N, et al. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget (2016) 7:593–609. doi: 10.18632/oncotarget.5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lin Q, Peng S, Yang Y. Inhibition of CD9 expression reduces the metastatic capacity of human hepatocellular carcinoma cell line MHCC97-h. Int J Oncol (2018) 53:266–74. doi: 10.3892/ijo.2018.4381 [DOI] [PubMed] [Google Scholar]
- 82. Suwatthanarak T, Tanaka M, Miyamoto Y, Miyado K, Okochi M. Inhibition of cancer-cell migration by tetraspanin CD9-binding peptide. Chem Commun (Camb) (2021) 57:4906–9. doi: 10.1039/d1cc01295a [DOI] [PubMed] [Google Scholar]
- 83. Suwatthanarak T, Ito K, Tanaka M, Sugiura K, Hoshino A, Miyamoto Y, et al. A peptide binding to the tetraspanin CD9 reduces cancer metastasis. Biomater Adv (2023) 146:213283. doi: 10.1016/j.bioadv.2023.213283 [DOI] [PubMed] [Google Scholar]
- 84. Lu W, Fei A, Jiang Y, Chen L, Wang Y. Tetraspanin CD9 interacts with α-secretase to enhance its oncogenic function in pancreatic cancer. Am J Transl Res (2020) 12:5525–37. [PMC free article] [PubMed] [Google Scholar]
- 85. Kohmo S, Kijima T, Otani Y, Mori M, Minami T, Takahashi R, et al. Cell surface tetraspanin CD9 mediates chemoresistance in small cell lung cancer. Cancer Res (2010) 70:8025–35. doi: 10.1158/0008-5472.CAN-10-0996 [DOI] [PubMed] [Google Scholar]
- 86. Houle CD, Ding X-Y, Foley JF, Afshari CA, Barrett JC, Davis BJ. Loss of expression and altered localization of KAI1 and CD9 protein are associated with epithelial ovarian cancer progression. Gynecol Oncol (2002) 86:69–78. doi: 10.1006/gyno.2002.6729 [DOI] [PubMed] [Google Scholar]
- 87. Takeda T, Hattori N, Tokuhara T, Nishimura Y, Yokoyama M, Miyake M. Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res (2007) 67:1744–9. doi: 10.1158/0008-5472.CAN-06-3090 [DOI] [PubMed] [Google Scholar]
- 88. Nakazawa Y, Sato S, Naito M, Kato Y, Mishima K, Arai H, et al. Tetraspanin family member CD9 inhibits aggrus/podoplanin-induced platelet aggregation and suppresses pulmonary metastasis. Blood (2008) 112:1730–9. doi: 10.1182/blood-2007-11-124693 [DOI] [PubMed] [Google Scholar]
- 89. Copeland BT, Bowman MJ, Boucheix C, Ashman LK. Knockout of the tetraspanin Cd9 in the TRAMP model of de novo prostate cancer increases spontaneous metastases in an organ-specific manner. Int J Cancer (2013) 133:1803–12. doi: 10.1002/ijc.28204 [DOI] [PubMed] [Google Scholar]
- 90. Tang M, Yin G, Wang F, Liu H, Zhou S, Ni J, et al. Downregulation of CD9 promotes pancreatic cancer growth and metastasis through upregulation of epidermal growth factor on the cell surface. Oncol Rep (2015) 34:350–8. doi: 10.3892/or.2015.3960 [DOI] [PubMed] [Google Scholar]
- 91. Funakoshi T, Tachibana I, Hoshida Y, Kimura H, Takeda Y, Kijima T, et al. Expression of tetraspanins in human lung cancer cells: frequent downregulation of CD9 and its contribution to cell motility in small cell lung cancer. Oncogene (2003) 22:674–87. doi: 10.1038/sj.onc.1206106 [DOI] [PubMed] [Google Scholar]
- 92. Li Y, Yu S, Li L, Chen J, Quan M, Li Q, et al. KLF4-mediated upregulation of CD9 and CD81 suppresses hepatocellular carcinoma development via JNK signaling. Cell Death Dis (2020) 11:299. doi: 10.1038/s41419-020-2479-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Miyake M, Nakano K, Ieki Y, Adachi M, Huang CL, Itoi S, et al. Motility related protein 1 (MRP-1/CD9) expression: inverse correlation with metastases in breast cancer. Cancer Res (1995) 55:4127–31. [PubMed] [Google Scholar]
- 94. Arihiro K, Fujii I. Loss of CD9 with expression of CD31 and VEGF in breast carcinoma, as predictive factors of lymph node metastasis. Breast Cancer (1998) 5:131–8. doi: 10.1007/BF02966685 [DOI] [PubMed] [Google Scholar]
- 95. Miyamoto S, Maruyama A, Okugawa K, Akazawa K, Baba H, Maehara Y, et al. Loss of motility-related protein 1 (MRP1/CD9) and integrin alpha3 expression in endometrial cancers. Cancer (2001) 92:542–8. doi: [DOI] [PubMed] [Google Scholar]
- 96. Mhawech P, Herrmann F, Coassin M, Guillou L, Iselin CE. Motility-related protein 1 (MRP-1/CD9) expression in urothelial bladder carcinoma and its relation to tumor recurrence and progression. Cancer (2003) 98:1649–57. doi: 10.1002/cncr.11698 [DOI] [PubMed] [Google Scholar]
- 97. Ai X, Zhang X, Wu Z, Ma X, Ju Z, Wang B, et al. Expression of KAI1/CD82 and MRP-1/CD9 in transitional cell carcinoma of bladder. J Huazhong Univ Sci Technolog Med Sci (2007) 27:79–82. doi: 10.1007/s11596-007-0123-0 [DOI] [PubMed] [Google Scholar]
- 98. Buim MEC, Lourenco SV, Carvalho KC, Cardim R, Pereira C, Carvalho AL, et al. Downregulation of CD9 protein expression is associated with aggressive behavior of oral squamous cell carcinoma. Oral Oncol (2010) 46:166–71. doi: 10.1016/j.oraloncology.2009.11.009 [DOI] [PubMed] [Google Scholar]
- 99. Kusukawa J, Ryu F, Kameyama T, Mekada E. Reduced expression of CD9 in oral squamous cell carcinoma: CD9 expression inversely related to high prevalence of lymph node metastasis. J Oral Pathol Med (2001) 30:73–9. doi: 10.1034/j.1600-0714.2001.300202.x [DOI] [PubMed] [Google Scholar]
- 100. Uchida S, Shimada Y, Watanabe G, Li ZG, Hong T, Miyake M, et al. Motility-related protein (MRP-1/CD9) and KAI1/CD82 expression inversely correlate with lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer (1999) 79:1168–73. doi: 10.1038/sj.bjc.6690186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zou Q, Xiong L, Yang Z, Lv F, Yang L, Miao X. Expression levels of HMGA2 and CD9 and its clinicopathological significances in the benign and malignant lesions of the gallbladder. World J Surg Oncol (2012) 10:92. doi: 10.1186/1477-7819-10-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Khushman M, Patel GK, Laurini JA, Bhardwaj A, Roveda K, Donnell R, et al. Exosomal markers (CD63 and CD9) expression and their prognostic significance using immunohistochemistry in patients with pancreatic ductal adenocarcinoma. J Gastrointest Oncol (2019) 10:695–702. doi: 10.21037/jgo.2018.07.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kim K-J, Kwon HJ, Kim MC, Bae YK. CD9 expression in colorectal carcinomas and its prognostic significance. J Pathol Transl Med (2016) 50:459–68. doi: 10.4132/jptm.2016.10.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hashida H, Takabayashi A, Tokuhara T, Hattori N, Taki T, Hasegawa H, et al. Clinical significance of transmembrane 4 superfamily in colon cancer. Br J Cancer (2003) 89:158–67. doi: 10.1038/sj.bjc.6601015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Amatya VJ, Takeshima Y, Aoe K, Fujimoto N, Okamoto T, Yamada T, et al. CD9 expression as a favorable prognostic marker for patients with malignant mesothelioma. Oncol Rep (2013) 29:21–8. doi: 10.3892/or.2012.2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Si Z, Hersey P. Expression of the neuroglandular antigen and analogues in melanoma CD9 expression appears inversely related to metastatic potential of melanoma. Int J Cancer (1993) 54:37–43. doi: 10.1002/ijc.2910540107 [DOI] [PubMed] [Google Scholar]
- 107. Woegerbauer M, Thurnher D, Houben R, Pammer J, Kloimstein P, Heiduschka G, et al. Expression of the tetraspanins CD9, CD37, CD63, and CD151 in merkel cell carcinoma: strong evidence for a posttranscriptional fine-tuning of CD9 gene expression. Mod Pathol (2010) 23:751–62. doi: 10.1038/modpathol.2009.192 [DOI] [PubMed] [Google Scholar]
- 108. Adachi M, Taki T, Konishi T, Huang CI, Higashiyama M, Miyake M. Novel staging protocol for non-small-cell lung cancers according to MRP-1/CD9 and KAI1/CD82 gene expression. J Clin Oncol (1998) 16:1397–406. doi: 10.1200/JCO.1998.16.4.1397 [DOI] [PubMed] [Google Scholar]
- 109. Higashiyama M, Taki T, Ieki Y, Adachi M, Huang CL, Koh T, et al. Reduced motility related protein-1 (MRP-1/CD9) gene expression as a factor of poor prognosis in non-small cell lung cancer. Cancer Res (1995) 55:6040–4. doi: 10.1002/cncr.11698 [DOI] [PubMed] [Google Scholar]
- 110. Kim T, Kim Y, Kwon HJ. Expression of CD9 and CD82 in papillary thyroid microcarcinoma and its prognostic significance. Endokrynol Pol (2019) 70:224–31. doi: 10.5603/EP.a2019.0009 [DOI] [PubMed] [Google Scholar]
- 111. Soyuer S, Soyuer I, Unal D, Ucar K, Yildiz OG, Orhan O. Prognostic significance of CD9 expression in locally advanced gastric cancer treated with surgery and adjuvant chemoradiotherapy. Pathol Res Pract (2010) 206:607–10. doi: 10.1016/j.prp.2010.04.004 [DOI] [PubMed] [Google Scholar]
- 112. Huan J, Gao Y, Xu J, Sheng W, Zhu W, Zhang S, et al. Overexpression of CD9 correlates with tumor stage and lymph node metastasis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol (2015) 8:3054–61. doi: 10.1034/j.1600-0714.2001.300202.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kwon HJ, Choi JE, Kang SH, Son Y, Bae YK. Prognostic significance of CD9 expression differs between tumour cells and stromal immune cells, and depends on the molecular subtype of the invasive breast carcinoma. Histopathology (2017) 70:1155–65. doi: 10.1111/his.13184 [DOI] [PubMed] [Google Scholar]
- 114. Baek J, Jang N, Choi JE, Kim J-R, Bae YK. CD9 expression in tumor cells is associated with poor prognosis in patients with invasive lobular carcinoma. J Breast Cancer (2019) 22:77–85. doi: 10.4048/jbc.2019.22.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Han X, Zhang W-H, Gao H-L, Li T-J, Xu H-X, Li H, et al. Neoadjuvant chemotherapy endows CD9 with prognostic value that differs between tumor and stromal areas in patients with pancreatic cancer. J Clin Lab Anal (2022) 36:e24517. doi: 10.1002/jcla.24517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang J-C, Begin LR, Berube NG, Chevalier S, Aprikian AG, Gourdeau H, et al. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin Cancer Res (2007) 13:2354–61. doi: 10.1158/1078-0432.CCR-06-1692 [DOI] [PubMed] [Google Scholar]
- 117. Jamil F, Peston D, Shousha S. CD9 immunohistochemical staining of breast carcinoma: unlikely to provide useful prognostic information for routine use. Histopathology (2001) 39:572–7. doi: 10.1046/j.1365-2559.2001.01296.x [DOI] [PubMed] [Google Scholar]
- 118. Koh HM, Jang BG, Lee DH, Hyun CL. Increased CD9 expression predicts favorable prognosis in human cancers: A systematic review and meta-analysis. Cancer Cell Int (2021) 21:472. doi: 10.1186/s12935-021-02152-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zeng P, Si M, Sun R-X, Cheng X, Li X-Y, Chen M-B. Prognostic value of CD9 in solid tumor: A systematic review and meta-analysis. Front Oncol (2021) 11:764630. doi: 10.3389/fonc.2021.764630 [DOI] [PMC free article] [PubMed] [Google Scholar]


