Abstract
The brain of modern humans has evolved remarkable computational abilities that enable higher cognitive functions. These capacities are tightly linked to an increase in the size and connectivity of the cerebral cortex, which is thought to have resulted from evolutionary changes in the mechanisms of cortical development. Convergent progress in evolutionary genomics, developmental biology and neuroscience has recently enabled the identification of genomic changes that act as human-specific modifiers of cortical development. These modifiers influence most aspects of corticogenesis, from the timing and complexity of cortical neurogenesis to synaptogenesis and the assembly of cortical circuits. Mutations of human-specific genetic modifiers of corticogenesis have started to be linked to neurodevelopmental disorders, providing evidence for their physiological relevance and suggesting potential relationships between the evolution of the human brain and its sensitivity to specific diseases.
Introduction
Approximately 6-8 million years ago, the common ancestor of great apes (chimpanzees and bonobos) and species of the homo lineage lived in Africa. Following the divergence of hominins, one branch, through multiple radiation (in which new species emerge) and extinction events, gave rise to Homo sapiens approximately 300,000 years ago 1. As a result of this evolution, the brain of modern humans displays remarkable cognitive abilities that enable functions such as elaborate working memory, self-awareness, advanced forms of communication, complex tool making and cultural development 2,3. The mechanisms underlying the evolutionary emergence of human cognitive abilities constitute a long-standing topic of interest in neuroscience that has recently been transformed by the coalescence of major advances in comparative genomics [G], developmental neurobiology and the development of new experimental models to study human neural development and function (Box 1).
Box 1. New experimental paradigms to study human brain development and evolution.
The study of human corticogenesis has entered an new era, enabled by the development of new tools and experimental models. Each of these tools and models (illustrated in the figure) has both advantages and limitations, but their successful combination has led to novel insights into mechanisms underlying human-specific features of brain development.
The simplest in vitro models of corticogenesis that have been developed are adherent twodimensional (2D) cultures of cortical progenitors and neurons, generated from pluripotent stem cells (PSCs; either embryonic stem (ES) cells or induced pluripotent stem cells (iPS cells)) derived from humans, macaques or chimpanzees. Despite their simplicity, these models enable one to faithfully recapitulate many aspects of cortical neurogenesis, including the sequential generation of cortical pyramidal neurons with identities corresponding to those present in the six layers of the human brain 75,76,208. These models are particularly well suited for high throughput genetic or chemical screening or for single cell experiments, such as those involving live imaging or clonal analyses. However they generate outer radial glia cells and upper layer neurons in a much lower proportion than occurs in vivo and do not allow us to study key cytoarchitecture features such as the ventricular zone (VZ), outer subventricular zone and cortical plate 75. Three dimensional (3D) cultures of cortical cells, also known as neural or cortical organoids, leverage the remarkable self-organizing properties of neural cells to recapitulate some of the key aspects of the in vivo spatial organization of cortical progenitors and neurons, including a highly patterned VZ-like structure. However, the generation of properly patterned neuronal layers remains difficult to obtain robustly 209–211. Organoids are also amenable to functional screening, at least at early stages, and can include assemblies of excitatory and inhibitory neuronal populations (assembloids) that reflect some aspects of cortical circuits 212. Neural organoids have been maintained in culture for long periods (>1 year); however, it remains challenging to keep differentiated neurons healthy in these relatively large structures and in vitro culture can lead to metabolic stress213. PSC-derived (or fetal cortex-derived) cortical cells grown in 2D culture or as organoids can also be studied in vivo thanks to xenotransplantation in the neonatal mouse brain. This enables us to follow neuronal development for months-long periods without metabolic stress and allows the cells to develop into functional neurons that can display higher order properties, such as robust synaptic plasticity and even physiologically tuned responses to sensory stimuli111. Xenotransplantation experiments are thus ideally suited to the study of higher order properties of human neurons in vivo; however, they remain a low-throughput method that is time-consuming compared with in vitro approaches.
The mouse is the main in vivo tool for the study of cortical development, because of its associated genetic and embryology toolbox (including transgenesis and in utero electroporation) that can be used to study in vivo the effects of human genes or regulatory elements. One major caveat of this approach, however, is that the genes are studied in a cellular context that may lack key species-specific features. Other mammalian models (such as the ferrret, marmoset or macaque) are much less versatile than the mouse but enable us to study human-specific genes in a context that is closer to the human. In addition, non-mammalian models (ranging from invertebrates such as drosophila to vertebrates such as reptiles, fish and birds) are crucial for the identification of new molecular, cellular and developmental mechanisms relevant for human brain evolution.
Access to ex vivo samples of fetal and adult human cortex and the ability to perform not only molecular but also cellular and physiological experiments on these preparations provides opportunities to study the human cortex, despite the scarcity of the available material. Finally, human genetics and comparative genomics enable the identification of candidate genes and genomic elements linked to cortical development and evolution as well as mutations leading to neurodevelopmental disorders 214. These candidate genomic elements can then be tested, alone or in combination, for their cellular and molecular impact in cortical development and function, using a combination of in vivo animal models, in vitro human cellular models or xenotransplantation.
Here, we present an overview of these recent advances, which have linked developmental mechanisms with the evolution of human neural circuits. We focus on the cerebral cortex, arguably the most complex and among the most divergent of human brain structures, compared with other species. We first describe some of the most notable qualitative and quantitative differences between the human cortex and that of other animals at the cellular level. We then review the cellular mechanisms that underlie specific features of human corticogenesis and their molecular links with upstream human-specific genomic changes. Finally, we illustrate how the identification of human-specific modifiers of cortical development and function could lead to the discovery of previously unknown aspects of human brain structure, function and disease.
Human-specific brain features
The staggering structural and functional complexity of brain organization can be studied at multiple scales 4. At a macroscopic scale, neural circuits are organized in interconnected networks of neurons by short-range and long-range axonal projections. At a microscopic scale, neurons connect to each other via precise synaptic connections. One main goal of neuroscience is to understand how variation at these different levels underlies the brain’s functional properties and ultimately shapes behavior. Studying this challenging problem is essential if we are to decipher the mechanisms that mediated the emergence of the cognitive capacities of modern humans5.
Cellular composition
More cortical neurons
The human brain contains approximately 85 billion neurons, each forming thousands of individual synapses with other neurons3,6,7. The numbers of neuronal and non-neuronal cells in specific brain structures in 41 mammalian species have been estimated, leading the authors of those studies to the conclusion that the brain of modern humans can be considered a “scaled-up primate brain” 6,7 However, two brain structures have expanded significantly more than others (in terms of neuron number) among primates and particularly in humans: the cerebral cortex (Fig. 1a) and the cerebellum 6,7. With 16 billion neurons, the human cerebral cortex contains a larger number of neurons than that of our closest relative the chimpanzee (6 billion neurons) or the more distantly related rhesus macaque (1.7 billion neurons) 6. As a comparison, the neocortex of two mammalian species often used in neuroscience, the mouse and rat, contain approximately 14 and 31 million neurons, respectively 6,7.
Fig. 1|. Cortical circuit evolution.
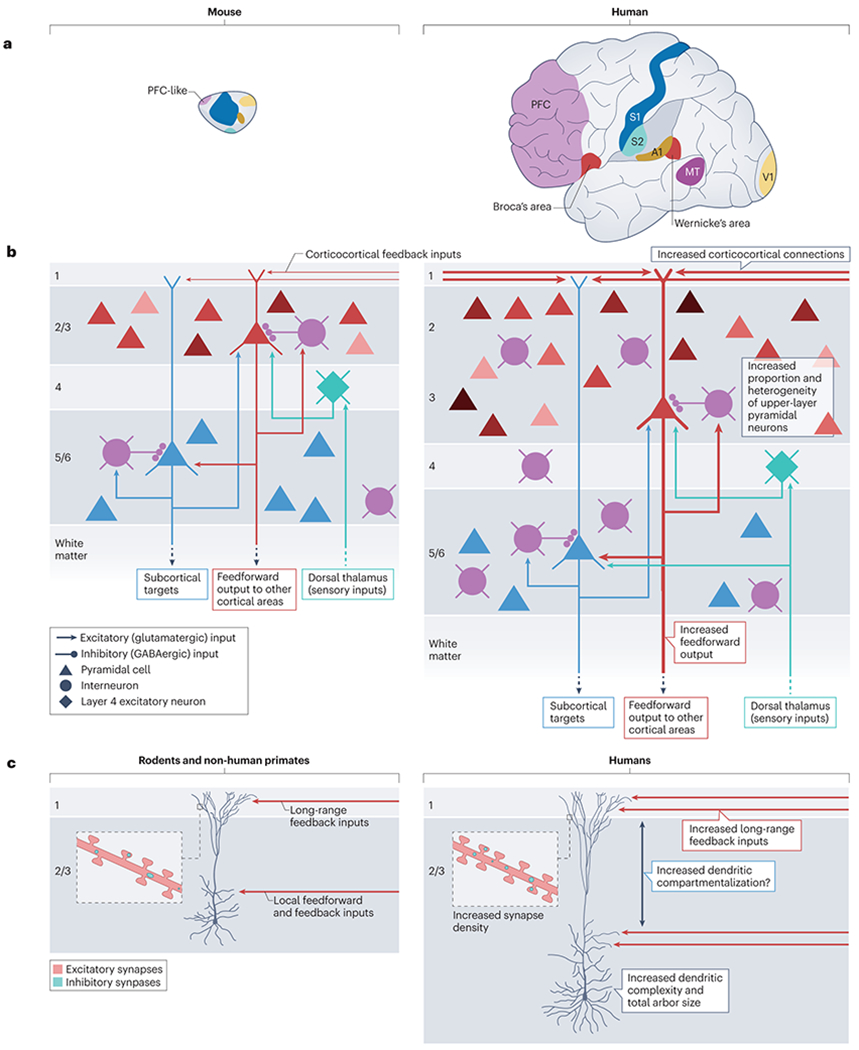
a, Schematic illustration of the expansion of cortical area size that has taken place in the human brain, when compared to the mouse (brains depicted approximately to scale). The expanded size of the human neocortex is accompanied by an increase in the number of cortical areas driven by the emergence of new cortical areas (such as Wernicke’s area and Broca’s areas). In addition there has been an increase in the size of certain cortical areas, including a pronounced expansion of the prefrontal cortex (PFC) compared to its putative homologous regions (PFC-like regions) in the mouse. b, In mammals, cortical neurons are organized in 6 layers that are generated in an inside-first outside-last manner: early born neurons generate deep layer (DL, layers 5/6) pyramidal neurons (PNs) (blue), then thalamo-recipient layer 4 neurons (green) and finally upper layer (UL, layers 2/3) PNs (shades of red). DL neurons project mostly to sub-cortical targets such as the dorsal thalamus (from layer 6) and the striatum, spinal cord and other sub-cortical targets (from layer 5). UL layer PNs project mostly locally to layer 5 and to other cortical areas (via feedforward cortico-cortical projections). Layer 2/3 UL neurons receive inputs from long-range feedback projections from other cortical areas in layer 1. The human cortex is characterized by an increased number of feedforward and feedback cortico-cortical connections (indicated by thicker lines in the right panel) 69. c, Layer 2/3 UL pyramidal neurons in the human cortex are larger, more complex (increased branching) and have a a longer apical dendrite than those present in mice and other mammals (including other non-human primates)45–48. It has been proposed that the longer apical dendrite of UL in human cortex leads to increased dendritic compartmentalization because the the apical tuft is located further away from the soma, although the evidence for this remains controversial (indicated by the ?)52–55,58. Human UL PNs also receive more excitatory synapses and inhibitory synapses compared to other mammals3,50.
Altered cortical neuron composition and diversification
Beyond absolute cell number, a key substrate of the complexity of the human cortex is its cell composition and diversity (Fig. 1b). The neocortex is composed of six layers containing both long-distance-projecting excitatory pyramidal neurons and locally-projecting inhibitory interneurons. These broad neuronal classes can be further divided into several dozen subclasses and subtypes, each of which displays specific molecular, cellular and hodological features8,9. Some subtypes are partially specific to particular cortical layers: for instance, deep layer (DL) neurons (those found in layers 5 and 6) mostly send long-range projections to subcortical targets, while upper layer (UL) neurons (found in layers 2 and 3) project mostly locally and to other cortical areas. Finally, layer 4 neurons receive most of the monosynaptic connections from the thalamus that relay sensory information from the periphery (Fig. 1b) 10. While these basic principles are largely conserved in all mammals, important cellular features that display divergence in humans have been uncovered. For example, the thalamo-recipient cortical layer 4 is expanded in human and non-human primates, compared with non-primate species, and displays a more complex cytoarchitecture11.
One key human feature is a significant expansion of the fraction of cortical neurons that are UL pyramidal neurons: these neurons constitute more than 40% of the neurons in the human cerebral cortex, while their proportion is around 25% in the mouse (with intermediate values for non-human primates and hominids)11–13 (Fig. 1b). Recent multimodal analyses combining single cell transcriptomic, electrophysiological and morphological profiling have revealed more heterogeneity and diversity among UL neurons in the human cortex, where at least 5 UL subtypes have been identified, compared to 3 in the mouse (Fig. 1b). Interestingly, the 2 additional UL subtypes found in the human display patterns of gene expression that are reminiscent of DL neurons14,15. The resulting increase in the proportion of long-range projecting neurons in external upper layers of the cortex16 could be an important substrate of the increased cortico-cortical connectivity found in the human brain14,16 (Fig. 1b).
Beyond UL neurons, single cell transcriptomic comparisons of adult human cortical neurons with their mouse and primate counterparts have revealed considerable gene expression divergence in homologous cell subtypes in each species, as well as changes in their relative proportion15,17,18. For example, Betz neurons, a subtype of DL corticospinal neurons that connect to the spinal cord to mediate fine motor control, are enriched in primates 17. Von Economo (VEN) neurons represent a cell type that are characterized primarily by bitufted dendritic morphology and found in several higher mammals including humans, where they are located mostly in anterior cingulate cortex (ACC) and frontal areas. Recent evidence identified some of the transcriptional and electrophysiological signatures of VEN cells, which suggest that this cell type is transcriptionally homologous to extratelencephalic (ET) excitatory neurons that project to subcortical targets 19.
Orthogonal to its laminar organization, the cerebral cortex is parcellated into numerous cortical areas populated by neurons displaying specific patterns of gene expression and connectivity20,21. While areas subserving first-order motor control and sensory processing are well-conserved in mammals, the number of cortical areas has considerably expanded in the primate lineage, rising from approximately 20 in the mouse to more than 150 areas in the human cortex 22–24. In particular, there has been a diversification of association areas [G] 22–24. The cortical areas involved in language processing and production are present in both humans and non-human primates, but their long-range input and output connectivity are divergent, which might explain human-specific linguistic capacities25. Most importantly, the size of the human prefrontal cortex (PFC), involved in complex social behaviors and executive planning26, is larger than that of non-human primates27,28 (although whether this reflects an absolute increase in size or an increase relative to the size of other areas is still debated29). The increase in the size of the PFC also reflects an increase in the size and/or number of its subdivisions (Fig. 1a). The developmental mechanisms underlying the diversification of association and PFC areas remain largely unknown. However, proposed mechanisms include changes in morphogen [G] signaling30 (see below) and the ‘untethering’ of cortical neuron patterning from the constraining influence of local signaling centers, due to the significant expansion of the cortical surface 31.
Despite the significant progress achieved through multimodal single cell analyses, the field is only starting to evaluate the degree of cortical neuronal diversity in different species. More work is needed to explore further the relationship between areal diversity, connectome [G] properties and specific developmental programs underlying neuronal diversification. Nevertheless, current data indicate that the human cortex is characterized by an expansion and diversification of UL and PFC neurons. Both of these sets of neurons make extensive cortico–cortical connections, which are correspondingly increased in the primate and human cortex (Fig. 1b,c). Intriguingly, UL and PFC neurons are also characterized by their delayed generation and/or development: UL neurons are the last to be generated during neurogenesis [G] and PFC areas are the last to reach mature patterns of structural and functional connectivity32. This is consistent with an important influence of extended developmental timing on the evolution of the human cortex (see below).
Altered interneuron composition.
Approximately 20% of rodent cortical neurons are locally-projecting interneurons; however, this proportion is greater in human cortex (approximately 25-30%)17,33. Cortical interneurons can be subdivided into at least 25 subtypes based on their morphology, connectivity, electrophysiological properties and gene expression profiles9. These subtypes appear to be largely conserved in mouse and human, at least based on transcriptomic profiling 34,35. However, there are significant species differences in the repertoire of genes expressed in each interneuron subtype and also in their relative abundance and laminar position 34,35. For instance, Rosehip interneurons, characterized by their specialized axon arborization that targets the dendrites of pyramidal neurons, have been found in the human cortex but not in mouse cortex36. Future studies should assess whether the qualitative and/or quantitative distribution of interneuron subtypes differs between human and non-human primates and/or represents a feature conserved in other mammals. It will also be important to determine how differences in interneuron composition affect circuit properties in a species-specific manner.
Altered glial cell type composition.
The cortex also contains non-neuronal cell types including astrocytes, oligodendrocytes and microglial cells. However, only a few studies have explored whether these cell types differ in their transcriptional profile, morphology or function in the brain of various types of mammals34,37. Comparison between several non-human primates and humans revealed that three main types of cortical astrocytes (interlaminar, protoplasmic and fibrous) display distinct morphological features 38,39 and vary in abundance depending on their laminar position and gene expression profile 34,37. Recent single cell RNA sequencing [G] (scRNAseq) also revealed a significant degree of subtype diversity among microglial cells, especially in humans 40,41. Many differences in glial cell gene expression profiles have been reported between human and chimpanzee18 and several studies have also suggested that glial cells constitute a larger proportion of the total number of cells found in the human brain than in other mammalian species 40–42. A recent study used a combination of cell fate mapping [G], morphological analysis and gene expression profiling to illustrate the diversity of the astrocyte lineage and cell composition in the human cortex 43. Future investigations will undoubtedly document whether these differences are observed in non-human primates and if they play a role in human-specific traits of brain development and/or adult brain function.
Cellular properties
Increased neuronal size, complexity and connectivity.
All mammalian cortical pyramidal neurons have some generic morphological features, including a single axon that projects towards the white matter, an apical dendrite that is oriented towards the cortical surface and basal dendrites that branch extensively (with all dendrites being decorated by dendritic spines [G]) 44. However, human pyramidal neurons display some additional species-specific properties. Their dendrites are longer and more branched than their mouse, macaque and chimpanzee counterparts45–48. Moreover they bear a higher density of spines than macaque, marmoset and mouse cortical pyramidal neurons47,49 and more spines than cortical pyramidal neurons in any other primates including great apes such as chimpanzee and bonobo 45,50 (Fig. 1c). The average size of the spines (including the length of spine neck) is also larger in human pyramidal neurons, suggesting distinctive functional properties47,49.
As a result of their larger dendritic length and increased spine density, human pyramidal neurons receive more synaptic inputs than those of non-human primates or other mammalian species (approximately ~30,000 synapses for human pyramidal neurons versus ~9,000-15,000 in mouse and rat)3,47,51. A recent study estimated that human pyramidal neurons receive approximately twice as many synapses per neuron as those of any other primate50. Interestingly, electron microscopy studies revealed that the ratio between the number of excitatory and inhibitory synapses received by pyramidal neurons in the human and rat cortex is constant (approximately 90:10). This indicates that there has been a similar increase in both types of synapses in humans, leading to a remarkable conservation of the excitation/inhibition (E/I) balance 3,12.
The emergence of new approaches to study connectomics and to create single neuron reconstructions from human post-mortem brain samples will enable us to determine whether there are variations in the number of excitatory and inhibitory synaptic connections received by individual neuronal subtypes in different cortical layers and areas. This information could then further inform computational approaches to model the impact of such variation in synaptic connectivity on circuit function.
Altered functional and circuit properties
Several recent studies, taking advantage of live biopsies of healthy human cortical tissue, have compared the functional properties of human and rodent cortical pyramidal neurons. Multiple features that differ between humans and other mammalian species and lead to differences in neuronal excitability or input-output relationships, have been identified 42,46,48,52–56. These results suggest that the biophysical properties of the dendrites of human layer 5 and layer 2/3 pyramidal neurons are specialized compared to other mammals.
Both UL and DL human pyramidal neurons display a much increased apical dendrite length, which might — depending on the degree of passive attenuation and the extent of active conductance mechanisms within these dendrites— contribute to increased electrical compartmentalization (defined as the ability of distal synaptic inputs to influence dendritic and somatic spiking) and thus affect synaptic integration57 (Fig. 1c). Indeed, local depolarization of the distal dendrites of human layer 5 pyramidal neurons was shown to provide limited excitation to the soma, compared to rat pyramidal neurons53. This increased compartmentalization was accompanied by reduced inducibility of dendritic spikes [G], which could result from decreased ion channel densities. This is supported by biophysical modeling: if it is assumed that there is no change in the expression levels of the ion channels that propagate synaptic potentials or in the levels of voltage-gated ion channels, then their density per unit membrane surface will decrease as dendritic arbor size increases53. However, even for human DL pyramidal neurons, there seems to be considerable variability in the electrophysiological properties of different cell types. For example, a recent study showed that a subset of molecularly identified layer 5 pyramidal neurons in the human temporal cortex exhibit increased inducibility of dendritic spiking, compared to other layer 5 neurons 58. Studies that aim to identify the electrophysiological properties that characterize human pyramidal neurons will therefore need not only to integrate their morphological variations but also to examine these properties in molecularly-defined subtypes, which remains challenging.
Another distinctive feature of some classes of human pyramidal neurons is a reduction in their intrinsic excitability compared with several other species, including primates 52,53, which could critically affect information processing and neural circuit plasticity. Another study showed that Ih channels (mediating hyperpolarization-activated cation currents) are expressed at higher levels in human UL pyramidal neurons than in mouse UL pyramidal neurons, conferring the human pyramidal neurons with distinct dendritic processing [G] properties 55. Moreover, it has been shown that human UL pyramidal neurons display calcium-mediated dendritic spikes that are triggered only by selective classes of stimuli that drive both bottom-up (driven by sensory afferents) and top-down (driven by mixed selectivity feedback cortico-cortical projections) inputs 54. More work is needed to determine whether these dendritic spikes are mediated by the molecular effectors that drive the dendritic calcium spikes observed in DL cortical and CA1 hippocampal pyramidal neurons in other mammalian species 57. It will be also important to test, using both experimental and computational approaches, whether the putative increased functional compartmentalization of human UL pyramidal neurons alters their dendritic integration properties and the functional properties of cortical circuits 59–61: this has been suggested by biophysical modeling, but remains unclear on the basis of rodent studies.
Finally, compared with rodents, human cortical neurons display distinctive synaptic features: some synapses between human pyramidal neurons and interneurons are remarkably strong and plastic 62–65 and synapses between human pyramidal neurons display properties enabling them to relay information during periods of particularly high frequency synchrony (which could change the modalities of information that can be transfered through cortico-cortical connections) 66. These comparative studies should be carefully calibrated with recordings from non-human primates, but do suggest that human pyramidal neurons may have evolved unique input–output integration properties that could underlie enhanced information processing.
Where could the increased connectivity of human UL and DL pyramidal neurons originate from? Evidence suggests that it is likely to have arisen, at least in part, from the increased number of pyramidal neurons, as well as from the increase in total surface area and number of cortical areas characterizing the human neocortex. Cortical circuits exhibit several canonical organizational principles (Fig. 1b)67,68. UL neurons transform and relay the sensory information that they receive locally to other UL pyramidal neurons (via recurrent excitation), to DL pyramidal neurons that provide feedforward projections to subcortical brain regions, and to UL pyramidal neurons in more distant associative cortical areas (via long range feedforward projections). In turn, the UL pyramidal neurons receive feedback inputs from local layer 5 pyramidal neurons and from long-range feedback cortical-cortical inputs (Fig. 1c). Thus, one emerging model suggests that the dendrites of human UL pyramidal neurons integrate and perform complex dendritic computations on the inputs they receive. By contrast, these computations are thought to be restricted to DL pyramidal neurons in rodents 54. The increase in cortical-cortical connectivity in human cortical circuits, especially between areas such as the PFC and parietal cortex, could thus be a critical substrate for the evolution of higher cognitive functions in humans69. Importantly, the increased contribution of cortical-cortical connectivity might be linked to evolutionary changes in the patterns of cortical neurogenesis.
Evolution of neurogenesis mechanisms
What are the developmental mechanisms underlying human-specific features of cortical organization? Corticogenesis involves a highly complex developmental choreography, from the early steps of neurogenesis to the final stages of cortical circuit formation and refinement. While most of the underlying mechanisms are conserved in all mammals, some striking features have diverged significantly in non-human primates and humans.
Changes in neurogenesis
Neurogenesis determines how many and which subtypes of neurons are generated. Studies using the mouse as a model, together with human neuro-embryology, genetics and pluripotent stem cell (PSC)-based modelling (Box 1), have identified several features of cortical neurogenesis that underwent specific evolution in primates and humans (Fig. 2). As these were reviewed recently and extensively70–73, they will only be summarized here.
Fig. 2|. Species-specific features of human cortical development.
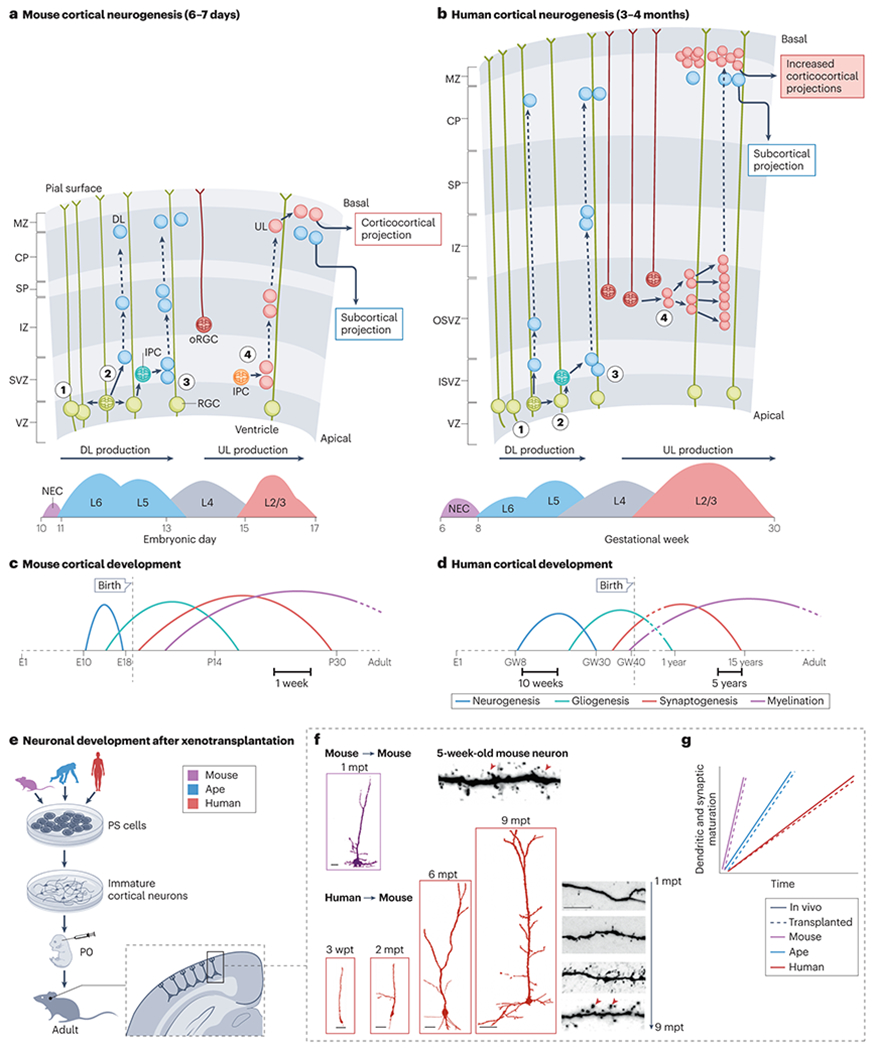
a, Mouse cortical neurogenesis lasts approximately a week206,207. Radial glial neural progenitors (also known as radial glial cells (RGC) in the ventricular zone (VZ) divide symmetrically to expand their pool (step 1) or divide asymmetrically to generate neurons (step 2). Following the migration of the neurons along the radial glia scaffold, this generates first the deep layer (DL) neurons destined to reside in layers 5/6 and to project sub-cortically. In later steps of neurogenesis, mainly through the generation of intermediate progenitor cells (IPC) in the subventricular zone (SVZ), additional DL neurons (step 3) and the upper layer (UL) neurons destined to reside in layers 2/3 (step 4) and to form cortico-cortical projections are produced. A specialized type of radial glial progenitors called outer radial glia (oRG), which lose their apical attachment at the ventricular surface but keep their basal endfeet at the pial surface, are found in mouse cortex but are extremely rare. b, In human cortex, neurogenesis lasts for approximately 4 months, with a more prolonged period of neuronal generation 72. oRGC are found in increased numbers in non-human primates, and in particular in the human cortex, contributing to the increased generation of layer 2/3 UL neurons in these species85. c–d, Comparison of the timeframe of the sequential events that characterize mouse and human corticogenesis 72. In the human cortex all of the developmental events shown — including neurogenesis, gliogenesis (formation of astrocytes), synaptogenesis and the myelination of axons by oligodendrocytes — are highly neotenic72. In human cortex, synaptogenesis (which includes synapse formation and pruning) is not complete until apprixmately 15 years after birth. e-f, In xenotransplantation experiments, cortical neurons derived from pluripotent stem cells (PSCs) of various species are transplanted into the neonatal cortex of immunodeficient mice, followed by their analysis in the months following transplantation (e). These studies have revealed the intrinsically slow and neotenic features of human induced pluripotent stem (iPS)-cell or embryonic stem (ES)-cell derived pyramidal neurons, compared to those derived from ape or mouse stem cells. When mouse or ape PSC-derived cortical pyramidal neurons are transplanted into the mouse neonatal cortex, they develop mature morphological features in about one month thus following the timeline of mouse cortical neurons 76. However, when human PSC-derived cortical pyramidal neurons are xenotransplanted into mouse cortex, their differentiation takes place over more than 6-9 months (f) mimicking the protracted maturation of cortical neurons in the developing human cortex 75,111. The schematic chart in panel g, illustrates the timeline of dendrite and synapse maturation observed in vivo for the indicated species compared to xenotransplan111tated cortical neurons from the corresponding species into mouse cortex111. Results from these xenotransplantation experiments indicate that transplanted neurons from each species differentiate at a similar pace to their in vivo equivalents, suggesting that the mechanisms controlling the species-specific timing of development are largely intrinsic to the neurons. CP: cortical plate; E : embryonic day ; GW : gestational week; IZ: intermediate zone; MZ: marginal zone ; ISVZ/OSVZ ; inner and outer subventricular zone. The mouse transplanted neuron image in panel f is adapted with permission from 76. The human transplanted neuron images in panel f are adapted, with permission, from 75. The mouse and human dendritic spine images in panel f are adapted, with permission from 111.
Cortical neurogenesis starts with the expansion of neuroepithelial cells (NEC), which divide symmetrically without further differentiation. This step is thought to have a crucial influence on brain size by determining the initial size of the cortical neural progenitor pool 2. NEC later convert into radial glial cells (RGC), which then start generating cortical neurons. Cortical neurogenesis from RGC typically occurs through asymmetric divisions that enable them to restore the progenitor pool while expanding neuronal production and leads to the formation of a ventricular zone, from which the cortical neurons migrate to form the cortical plate (Fig. 2a–b).
These key steps of neurogenesis are highly conserved among mammals, with the exception of one crucial point: timing. NEC amplification lasts for about one day in the mouse but up to two weeks in primates72. Similarly, cortical neurogenesis takes one week in the mouse, 2 months in the macaque and almost 4 months in the human (Fig. 2a–d)70. By allowing an increased number of NEC and RGC divisions to occur, this prolonged human neurogenesis is likely to constitute a key substrate of cortical expansion. Moreover, as UL neurons are the last to be generated, the prolonged neurogenesis may favour their expansion. Further experimental testing of this idea is however needed and will require us to determine the mechanisms underlying the timing differences in human neurogenesis. Importantly, the timing of cortical neurogenesis in humans is largely conserved in human cortical cells in vitro74–79, even when they are co-cultured with macaque cells or transplanted into the mouse brain75,78, indicating that the underlying mechanisms are largely intrinsic to human cortical progenitors (see discussion below). The timing of other events important for neurogenesis is also extended in the human compared with the mouse, in a cell-intrinsic fashion. These include a longer cell cycle length, which could influence neurogenic fate specification80,81 and a longer critical period during which cell fate remains plastic immediately after RGC division 82.
Another feature of primate and human cortical neurogenesis is the expansion of specific populations of progenitors located outside the ventricular zone, known collectively as basal progenitors. Some basal progenitors, called intermediate progenitor cells (IPC), are found in large numbers in the mouse embryonic cortex, but are increased in number in higher mammals (including primates), where they are thought to have contributed to cortical expansion 83. Moreover, another population of basal progenitors, called outer radial glia cells (oRGC, also known as basal RGC), are barely present in the mouse but strikingly expanded in the human cortex (Fig. 2)84–88. oRGC have distinctive features that are critical to their contribution to human cortex expansion: they display remarkable self-renewing capacities (in vitro clonal experiments have revealed that single oRGC can generate hundreds of neurons89) and their expansion occurs at late stages of corticogenesis, coinciding with UL neuron generation. In the human cortex oRGC are thus thought to constitute the main progenitor source of origin for UL neurons90 (Fig. 2).
While their relative contribution to cortical expansion remains to be determined, it can be hypothesized that the prolonged timeline of NEC amplification and neurogenesis, together with the late amplification of oRGC, synergize to increase cortical surface area (thus allowing more diversification of cortical areas to occur) and the generation of UL neurons (thus providing an ideal substrate for the expansion of cortico-cortical connectivity, Fig. 2). Indeed, in a recent study in which UL neuron generation was enhanced pharmacolgically in the mouse visual cortex91, mice displaying a 20% increase in UL neuronal number showed enhanced functional correlations among UL neuron assemblies, more functionally clustered neuronal ensembles and increased perceptual discrimination.
Changes in neuronal differentiation
Following neurogenesis, neurons undergo several major cellular and molecular transitions, leading to the growth and patterning of axons and dendrites. This is followed by synapse formation and pruning, leading to the formation of functional neural circuits that are further refined by various plasticity mechanisms. While these steps are widely conserved, their developmental timeline is strikingly different across species, taking weeks in the mouse, months in the macaque and years in the human (Fig. 2c, d). The most prolonged steps of neuronal development in humans include dendritic outgrowth, dendritic spine and synapse formation (taking months to several years) and synaptic pruning (taking up to two decades in the PFC)92–99. The resulting human brain neoteny (retention of juvenile features in a mature organism) is one of the most specific and functionally relevant features of human brain development and is likely to underlie the prolonged periods of motor, sensory and cognitive development that characterize the human species. Morever it could lead to longer critical periods (stages of development during which experience-dependent plasticity, thought to be crucial for the acquisition of higher cognitive features, takes place)2,100. Disrupting this timeline of cortical development could lead to brain pathology: for instance, accelerated brain development has been associated with autism spectrum disorders (ASD)101,102. Notably, one study has reported prolonged cortical neuronal development in the chimpanzee, suggesting that neotenic features of cortical development might be graded among great apes103.
An intriguing aspect of human brain neoteny is that it appears to be cell-type dependent, with UL and PFC neurons being the most neotenic95,104. The resulting differences in the rate of maturation between cortical layers and areas could have a significant impact on cortical circuit development105,106. Moreover the differentiation of some cortical interneurons is even more prolonged: human interneurons continue to migrate and integrate during the first postnatal months in the PFC107, and other forebrain regions like the amygdala may contain immature neurons for years postnatally108. Finally, non-neuronal cells, most strikingly oligodendrocytes, also display protracted development in humans109. leading to heterochrony [G] of myelination that could also be important for the timing of human brain circuit assembly110.
What are the mechanisms underlying human brain neoteny? Important hints have come from xenotransplantation [G] experiments. Mouse pyramidal neurons transplanted in the neonatal mouse brain develop along their physiological timeline76, whereas human cortical pyramidal neurons transplanted in the mouse cortex take up to 6-11 months to mature 75,111 (Fig. 2e–g). Interestingly, one study showed that, while transplanted human neurons at 9 months display electrophysiological features that are similar to those of adult neurons, they still display less mature patterns of morphogenesis, consistent with the fact that human cortical neurons take up to several years to reach full maturity 92–99. An even more prolonged development is observed for transplanted human cortical interneurons112,113. Remarkably, despite this protracted timeline of development, human transplanted pyramidal neurons integrate functionally into the host cortical circuits and display physiological responses to sensory stimuli (such as visual tuning): thus, they are not stalled in the unusual host environment, but rather develop physiologically at the pace characterizing the human brain in vivo111. Human xenotransplanted neurons develop over months even when transplanted as single neurons into mouse cortical tissue to allow optimal cellular and synaptic integration111 (Fig. 2e, f) or when faster maturation is induced by molecular reprogramming114, and chimpanzee PSC-derived transplanted cortical neurons develop at a faster pace than human neurons115 (Fig. 2g). Similarly expanded timelines of human cortical neuronal maturation are also observed using in vitro systems compared with non-human primate neurons78,114. Overall these data suggest that the mechanisms underlying neoteny of cortical neuron maturation are largely cell-intrinsic (see further discussion below).
Another striking feature of primate and human cortical neuron development is the expansion of transient populations of neurons that are only present and functional during development. These include Cajal-Retzius neurons found at the surface of the cortex and subplate neurons located below the cortical plate, both of which play a crucial roles in the assembly of mature cortical circuits116.
Genetic substrates of cortical evolution
Three main types of genetic changes are thought to lead to alterations in gene expression and/or protein function relevant to evolution (Fig. 3a): base-pair substitutions in non-coding enhancer [G] and promoter [G] regions that can alter gene regulatory networks [G]; non-synonymous substitutions [G] in coding regions of genes leading to amino-acid changes that result in modified protein function (Fig. 3b); and the emergence of new genes, mostly through large segmental duplications (Fig. 3c) 117,118. All three mechanisms have been involved in the emergence of human-specific traits of brain development and circuit function.
Fig. 3|. Genetic modifiers of human brain evolution.
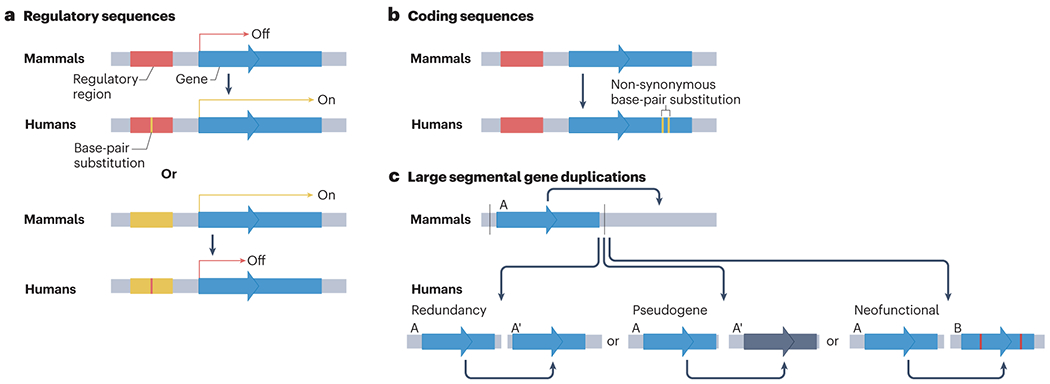
a, Human-specific base-pair substitutions are often found in regulatory regions (enhancers and promoters), where they can alter spatio-temporal patterns of gene expression. b, Another class of human-specific genetic modifiers are non-synonymous base-pair substitutions in exons that result in changes in amino-acid composition in the corresponding protein coding region specifically in the human genome. c, Human-specific gene duplications can lead to the production of new gene paralogs. These duplications can lead to a new (A’) nearly identical gene copy that increases gene dosage (redundancy), cause the copied gene to lose its function, becoming a pseudogene (because of a loss in regulatory sequences or transcription start site), or produce a gene that has acquired a new function through truncation or fusion with other coding sequences (neo-functionalization). As illustrated in Table 1, a number of genetic modifiers in each of these classes have been shown experimentally to result in alterations in cellular functions in the developing or adult brain.
Gene regulatory changes
Functionally relevant evolutionary changes in the genome are often linked to gene regulatory mechanisms119. Identifying human or primate genomic evolutionary changes, and linking them to gene regulation during human brain development, has proved a huge challenge because of the high degree of conservation of gene regulatory programs during mammalian neural development120. However, many human-specific cis-regulatory elements [G] (CREs, or Cis mechanisms), and upstream transcriptional regulators (Trans mechanisms), including long-range enhancers, have been identified that are uniquely regulated during human corticogenesis, leading potentially to human-specific control of gene expression (Fig. 3, Table 1).
Table 1|.
Examples of human-specific genetic modifiers affecting gene expression and/or protein function.
| Gene | Modification type | Species-specificity | Molecular effects | Phenotypic outcome | Reference |
|---|---|---|---|---|---|
| Gene regulatory changes | |||||
| FZD8 | HAR5 (enhancer) | Human | Human HAR5 increased expression of Fzd8 in mouse RGC | Increased RGC proliferation and increased cortical size | 135 |
| EPHA7 | HSL | Human | Increased expression of EPHA7 in human subplate cortical neurons | Dendritic remodelling | 145 |
| OSTN | Gain of binding sites for the transcription factor MEF2 | Primate | Activity-dependent expression of OSTN in primate brain | Activity-dependent dendritic remodelling | 168 |
| CBLN2 | Loss of binding sites for transcription factor SOX5 | Hominid | Increased expression of CBLN2 in PFC neurons | Increased synapse formation and connectivity in the PFC | 163 |
| PLXNA1 | Loss of binding sites for the transcription factor FEZF2 | Primate | Decreased expression of PLXNA1 in cortical neurons | Decreased axonal pruning of direct cortico-spinal projections | 166 |
| Protein coding changes | |||||
| FOXP2 | Changes in the coding sequence | Human | Two non-synonymous amino-acid substitutions present in human FOXP2 | Increased dendritic growth in human medium spiny striatal neurons. | 179 |
| New human-specific genes | |||||
| NOTCH2NLA/B/C | Human-specific gene duplication | Human | Increased Notch signalling in human cortical progenitors | Prolonged neurogenesis and increased cortical neuron number. CNVs in these genes associated with micro/macrocephaly | 139,140 |
| ARHGAP11B | Human-specific gene duplication | Human | Increased calcium signalling in mitochondria | Increased amplification of basal progenitors and increased cortical neuron number | 187,205 |
| CROCCP2 | Human-specific gene duplication | Hominid | Decreased ciliary dynamics and increased mTOR signalling in cortical progenitors | Increased amplification of basal progenitors and ncreased cortical neuron number | 192 |
| SRGAP2C | Human-specific gene duplication | Human | Inhibition of all functions of protein encoded by ancestral gene (postsynaptic SRGAP2A) | Neotenic synaptic maturation Increased cortico-cortical connectivity Changes in sensory coding Improved behavioral performance | 196,198,199 |
CNVs, copy number variants; HAR5, human accelerated region 5; HSL, human-specific loop; PFC, prefrontal cortex; RGC, radial glial cells; See also Figure 3.
Cis regulatory mechanisms
Comparative genomics identified human accelerated regions (HARs): DNA sequences that are very strongly conserved in mammals or vertebrates, but display sequence changes in the human genome only121,122. Mouse transgenics123 suggests that most HARs correspond to regulatory elements, although some (such as HAR1) are transcribed to produce non-coding long RNAs expressed in developing neurons in the fetal and adult cortex121. HARs are enriched in the vicinity of genes involved in neural and/or cortical development, suggesting their importance for gene regulation during corticogenesis18,124–129. Supporting this, the enhancer activity of more than 3000 HARs was recently tested in vitro by massively parallel reporter assays (MPRA) [G] 130,131,132. More than half of the tested HARs displayed enhancer activity in neural progenitor cells and this activity was, in most cases, either increased or decreased in human HARs, when compared to their ancestor sequences. When tested in mouse and human cells, most HARs displayed similar levels of activity, further pointing to Cis-variation as the main driver of HAR evolutionary changes130,131. Most HARs were mapped to genomic loci that are transcriptionally active in cortical progenitors or postmitotic neurons during fetal development, further pointing to their prominent regulatory role during all stages of corticogenesis.
The HAR5 element was the first of these elements to be functionally explored in the context of cortical neurogenesis (Fig. 3a, Table 1). HAR5 lies upstream of the FZD8 gene, which encodes a receptor for WNT proteins (the mediators of a pathway that controls the balance between self-renewal and differentiation in cortical progenitors133,134). Using a mouse transgenic model in which the Fzd8 coding sequence was put under the control of HAR5 variants, human HAR5 was shown to enhance the expression of Fzd8 and the proliferation of RGC (resulting in increased cortical size), while chimpanzee HAR5 did not have these effects135. Several other genes, including PPP1R17, which encodes a cell cycle regulator, are similarly controlled by HAR elements that have a selective influence on their expression in human cortical progenitors130.
Beyond neurogenesis, the involvement of HARs in cortical neuron development and maturation is supported by human genetic studies124. Sequence analysis of HARs revealed that these elements are often highly invariant in the human population, suggesting the functional importance of their ultra-conserved structure. Moreover several rare HAR variants are found in individuals affected by neurodevelopmental disorders124. Some of these pathogenic HAR variants were studied in the mouse, revealing their impact on the expression patterns of genes required for cortical neuron development. Consistent with the potential role of HARs in human cortical circuit evolution, one class of HAR-linked genes is also preferentially expressed in association cortical areas in the adult brain128.
Overall, these data point to HARs as potentially important molecular effectors of human cortical evolution. Future studies should consider inserting the human version of these regions into the endogenous ancestral locus in the mouse or other non-human, in order to circumvent the pitfalls of the viral and transgenic assays (including MPRAs) that have been used so far to study these elements. Most importantly, new approaches should be developed to determine the repertoire of Trans factors that bind to HAR elements and how it is affected by human-specific substitutions.
Comparative epigenetic profiling [G] of human, macaque and mouse at early stages of corticogenesis has identified another class of interesting CREs, called human gained enhancers (HGEs), that have gained activity in humans136. MPRAs revealed differential transactivation activity of a third of the tested HGEs in human versus chimpanzee neural stem cells131. Like HARs, HGEs are enriched in the vicinity of genes related to corticogenesis, including genes preferentially expressed in oRGC137. HGEs are also enriched in the vicinity of genes encoding proteins involved in the Notch pathway138, which promotes clonal expansion and prolonged neurogenesis of human RGC and oRGC85,139,140. These genes include HEY2, for which nearby HGE sequence variants were associated with changes in cortical surface area141. Similar comparative annotation of CREs in the adult brain also revealed a large number of elements that are more active in human than non-human primates, including a few that are not present in chimpanzee142. Some HGEs even display Homo sapiens-specific changes in sequence that are not found in the genome of Neanderthals143, suggesting a contribution to the most recent evolutionary changes in human corticogenesis.
The three-dimensional nuclear organization of DNA and chromatin structures associated with complex gene regulation — such as topology associated domains (TADs) [G] and chromatin loops [G] 144 — has also been shown to have human-specific features during corticogenesis138,145. For example, hundreds of (TADs) and chromatin loops are present in the human compared with macaque and mouse145. The boundaries of TADs and anchors of the loops were found to often be enriched for HARs and other enhancers that are specifically active in human cortical cells. Among these, one enhancer was found that interacts with the promoter of EPHA7, which encodes an axon guidance factor involved in corticogenesis146. Acute disruption of this enhancer in human neurons in vitro led to decreased EPHA7 expression and increased dendritic outgrowth145.
Finally, in addition to the point mutation mechanisms depicted above, the evolutionary loss or gain of entire CREs has also been found in the human genome, compared with non-human hominids, and in some cases could be linked to changes in the regulation of associated genes during cortical development 147,148.
There is thus a rich repertoire of CREs that regulate gene expression during corticogenesis by influencing transcription factor binding and/or chromatin structure, pointing to a need to study their biological impact on human corticogenesis and their relation to human evolution. An innovative approach was recently described in which PSC derived from different species were fused149,150. Fused chimpanzee-human induced pluripotent stem (iPS) cells were differentiated into cortical organoids[G], followed by transcriptome profiling, leading to the identification of genes uniquely upregulated in human astrocytes149. The use of composite cell lines of different species, associated with a direct comparison with contributor cells of origin, will constitute a promising tool to study the relative contribution of trans versus cis-mechanisms of control of gene expression during corticogenesis.
Trans mechanisms and transcriptomics
In parallel with these studies, comparative transcriptomics of the developing cortex in mouse, human and other primates, as well as corresponding PSC-derived cortical organoid models, has examined human-specific patterns of gene expression 79,151–153. While these studies revealed overall highly conserved patterns of gene expression, they suggest an ‘hourglass’ pattern of species-specific differences in gene expression, with greater divergence at embryonic to mid-fetal stages, and to a lesser extent during adolescence / young adulthood, and less divergence during late fetal stages151–153.
These analyses also revealed a consistent protracted pattern of ‘immature’ gene expression in the human, consistent with the neotenic nature of neurogenesis and neuronal differentiation77,79. While the mechanisms upstream of cortical transcriptional heterochrony in human remain unclear, some leads have started to emerge. Transcriptome profiling of human and non-human hominid cortical organoids revealed a delayed expression of the ZEB2 transcription factor in the human organoids, which was linked functionally to a delayed transition from NEC to RGC (and thereby potentially to increased size of the initial progenitor pool)74. Comparative transcriptomics in the human versus non human primate cortex also revealed quantitative differences in genes involved in signaling pathways functionally relevant to neurogenesis. These include the PDGF pathway, which was upregulated in the primate cortex and promotes RGC proliferation in human (but not in mouse) fetal cortex154, and the mTOR pathway, which was selectively upregulated in human oRGC and controls their morphology and migration properties155,156. A final interesting case is the KRAB-zinc finger protein family, which are the largest group of transcription factors in mammalian genomes and are considerably amplified in primate and human genomes157. While these transcription factors are mostly involved in transposable element repression, they also control the expression of endogenous genes, particularly during neural development158,159. For instance, the KRAB-zinc finger protein, ZNF558, is expressed in human and not chimpanzee neural progenitors and its loss of function in neural organoids leads to a dysregulation of gene expression that is compatible with the more precocious neurogenesis observed in non-human primates 158.
Finally, beyond transcription factor-related mechanisms, several miRNAs {Arcila, 2014 #373;Nowakowski, 2018 #374}, and primate and/or hominid-specific long non-coding RNAs [G] 160–162 are expressed specifically during primate or human corticogenesis, some of which have been linked to the regulation of cortical progenitor proliferation and self-renewal.
Divergent gene regulatory mechanisms in human cortical neurons and glia
It has been more challenging to explore conserved and divergent transcriptomic patterns during later stages of human cortical neuron and circuit development than in the early stages of corticogenesis. Neurons are typically harder to profile than progenitors using single cell transcriptomics and the postnatal stages of human development (which are the most relevant to neuronal differentiation and circuit formation) are among the least accessible for molecular or cellular analyses. Nevertheless, recent studies have started successfully to link divergent transcriptional regulation to cortical neuron connectivity (Fig. 3, Table1). A striking example is provided by cerebellin-2 (CBLN2), a gene that regulates synaptogenesis in many brain areas, which was found to be more broadly expressed in the primate PFC than in the mouse PFC163. This upregulation was linked to the loss of a regulatory binding site for the transcription factor SOX5, a key repressor during corticogenesis, and this was found to directly impact the development of neuronal connectivity in the mouse PFC163. Moreover, retinoic acid signaling was found to regulate the expression of CBLN2, and was shown to be increased and expanded in the primate PFC at early stages and drive to PFC expansion in mouse30 and human cortical organoids164. Together with scRNAseq efforts that have started to uncover the developmental dynamics of gene expression across the human fetal cortex165, these data provide important hints about the species-specific transcriptional mechanisms that might participate in the development of connectivity in particular cortical areas.
Another interesting example linked to the patterning of connectivity is provided by the species-specific regulation of the plexin A1 guidance receptor gene, PLXNA1 166 (Fig. 3, Table 1). In primates, but not rodents, motor area-derived corticospinal pyramidal neurons make abundant synapses with motor neurons, which might underlie increased dexterity. Juvenile mouse corticospinal neurons also develop these connections but they are pruned at adult stages, in large part through the action of semaphorin repellent guidance cues 166. The sempahorin receptor plexin A1 was found to be strongly expressed in corticospinal neurons in the mouse, but not human motor cortex, and the lack of expression of PLXNA1 in the human cortex was linked to primate-specific changes in upstream enhancer regions that confer binding to the FEZF2 transcriptional repressor166.
Following circuit formation, neurons undergo synaptic plasticity in response to changes in activity. The specific gene regulatory programmes involved in this plasticity167 were shown to display significant differences between human and non-human neurons168–170. For example, OSTN encodes a well-conserved secreted protein that is expressed in muscle and bone in the mouse and human, but only expressed in the brain in humans and macaques 168. OSTN expression is induced in an activity-dependent manner in layer 4 of macaque primary visual cortex 168. Loss of function of OSTN in human neurons in vitro increased dendritic outgrowth, suggesting that it acts as a negative modulator of activity-dependent dendritic branching (although more functional exploration is needed to determine its biological functions in vivo). The activity-dependent expression of OSTN in human neurons could be linked to the presence, in primates but not in other mammals, of binding sites for MEF2 transcription factors168. Interestingly, MEF2A (and downstream genes) was previously identified as displaying a ‘neotenic’ pattern of expression in the human cortex 171. Together with the identification of HARs upstream of MEF2C that are mutated in specific neurodevelopmental disorders124, this indicates that the MEF2 family and its downstream regulatory network are an attractive target for the evolution of cortical neuronal development programs.
Human-specific gene regulation has also been identified in interneurons and glial cells. For example, a recent study performed in human PSC derived cortical interneurons identified human- or primate-specific regulatory sequences responsive to neuronal activity172 that are enriched for genes associated with neurodevelopmental or psychiatric disorders including the ASD-linked genes SHANK3 and FMR1. Another interesting example involves secretagogin (SCGN), a calcium-binding protein that is strongly expressed in a subset of interneurons in primates but not in mice173. Forced expression of SCGN in mouse interneurons led to increased dendritic complexity, suggesting that SCGN functions in the development of a subtype of primate cortical interneurons. Finally, a recent study documented hominin-specific changes in regulatory elements affecting gene expression in oligodendrocytes that are dysregulated in the brain of individuals with ASD 174. However, more work is required to test their cellular functions and determine how they might impact circuit development and/or function.
Protein coding changes
Genome sequencing of many mammalian species has uncovered human-specific amino acid substitutions in approximately 100 genomic loci with a size of approximately 100kB, representing thousands of protein coding genes 175,176. Using stringent criteria, non-synonymous substitutions that show strong positive selection in human exons, compared to 29 other mammals (including chimpanzees), represent approximately 15,000 sites in around 4,400 proteins177. Surprisingly, however, the functional impact of these human-specific substitutions on human brain evolution has been explored for only very few of them.
Notable exceptions are the human-specific amino acid substitutions affecting the transcription factor FOXP2. FOXP2 is mutated in individuals with a monogenic speech disorder178 and human FOXP2 contains two amino acid substitutions that are not found in chimpanzee179. A mouse transgenic model that displays these two substitutions in mouse FoxP2 exhibited changes in dopamine concentrations in multiple brains regions (including the striatum and frontal cortex), decreased exploratory behaviors and altered ultrasonic vocalizations and increased neurite outgrowth and synaptic plasticity of striatal projection neurons179 (Table 1). These results linked human-specific changes in the FOXP2 amino-acid sequence with alterations in cortico-basal ganglia circuits that might have played a role in the evolution of speech production179. Future experiments should explore whether human-specific coding substitutions in other genes are relevant to the emergence of human-specific traits.
Of particular interest for future exploration are about 100 proteins that display amino acid substitutions that are specific to humans when compared with closer hominins Neanderthals and Denisovans180. Some of these proteins (namely, CASC5, KIF18A and SPAG5) are expressed in neural progenitors, while others (SLITRK1 and LRTM2) are involved in synaptogenesis, making them potentially relevant for evolutionary changes in several aspects of human corticogenesis.
New genes
An important source of evolutionary innovation can be provided by new genes, which typically emerge as a result of large segmental genomic duplications [G] or retrotransposition [G] 118 (Fig. 3, Table 1). Some human-specific genes resulting from retrotransposition are expressed during corticogenesis181, but recent evidence indicates that species-specific large segmental gene duplications represent an important source of genomic innovation during human evolution. The human genome contains approximately 30 gene families that show human-specific duplication patterns182,183. For most, both the ancestral copy of these genes and their human-specific paralogs are expressed throughout human fetal corticogenesis140. Since these genes are often located in hotsptots for genomic recombination, human-specific large segmental duplications can display variable degree of conservation in copy number in the human population, which can in turn be used to estimate selection pressure 184.
Several human-specific duplicated genes have been studied functionally during cortical neurogenesis, revealing their contributions to human cortical progenitor expansion and prolonged neurogenesis. The NOTCH2NL genes constitute a family of human-specific partial gene duplicates of NOTCH2, which encodes a key receptor in the Notch signalling pathway. Three NOTCH2NL genes are present only in the human genome139,140 and are located in a genomic region that is associated with changes in brain size185 (see below). Experiments combining gain and loss of function of these genes in the mouse embryonic cortex and in human PSC models of corticogenesis have revealed that the NOTCH2NL genes are human-specific activators of the Notch pathway, driving RGC expansion, prolonged neurogenesis and increased cortical neuronal production139,140,186. Clinical human genetics further supports the importance of NOTCH2NL genes in human biology and disease (Box 2).
Box 2. Human cortex evolution and disease.
Many neurological and psychiatric diseases alter cognitive and social brain functions that are particularly enhanced in humans, while the human brain seems particularly sensitive to specific neurological conditions. Moreover, many of these diseases appear to have a developmental origin, suggesting the fascinating possibility that their pathogenic mechanisms involve human-specific genomic and developmental mechanisms.
Some neurodevelopmental disorders appear to be directly caused by genetic disruption of human-specific genes or mechanisms. These include mutations in human accelerated regions (HARs)124 and also mutations in human-specific gene duplicates, many of which are found within loci that are the locations of pathogenic copy number variants (CNVs)215. Among these, the 1q21.1 locus is enriched for many human-specific genes, including the NOTCH2NL human-specific paralogs. This suggests that the phenotypic expression of neurodevelopmental disorders arising from mutations in this region (such as 1q21.1 deletion) might be, in part, human-specific. Microdeletions in the 1q21.1 locus are associated with microcephaly and schizophrenia, while microduplication of this region is associated with macrocephaly and autism spectrum disorders (ASD)216. Notably, some of the CNV breakpoints within the 1q21.1 locus are provided by the NOTCH2NL genes, which can then be deleted or duplicated139,217. NOTCH2NL gene increased expression and copy number amplification have also been associated with increased proliferative properties of gliobastoma cells218, while mutations in these genes were found in cases of neurodegeneration219,220.
Conversely, some well-conserved disease genes may exert their pathogenic effects through human-specific mechanisms. For instance, the protein encoded by the causative gene in fragile X syndrome, FMRP, was found to bind to differents sets of mRNAs in the human compared to other species 221,222. Among the human-specific targets of FMRP is NOS1 and protein levels of NOS1 are affected by the mutation that causes fragile X syndrome in human but not mouse neurons221,222.
It is also possible that human corticogenesis is more sensitive to specific diseases than that of other animals because the pathophysiological mechanisms underlying these disorders are rooted in developmental events that display human-specific features. These include the prolonged neotenic timeline of cortical development, which could be particularly relevant for ASD and schizophrenia (two neurodevelopmental disorders that affect critical periods of development and plasticity)223. At least some forms of ASD could be linked to accelerated neurogenesis or neuronal development. Synapse and dendritic spine formation are increased precociously in individuals with ASD224, while early postnatal brain overgrowth is found in many forms of ASD101,102. Moreover, transcriptome analysis identified genes displaying delayed ‘neotenic’ expression in the human vs non-human primates, and showed that this pattern appears to be accelerated in some individuals with ASD225. Studies using pluripotent stem cell models suggest that there are temporal shifts in cells derived from individuals with ASD, with these cells displaying accelerated neuronal maturation (which could be primed in neural progenitor states226) and differentiation225.
Finally, the prolonged development of human cortical circuits makes the human brain particularly sensitive to postnatal disruptions, whether of intrinsic or environmental origin32. For instance, schizophrenia has been long associated with excessive synapse pruning, leading to decreased connectivity at adolescent or young adult stages (when symptoms typically arise227), perhaps as a result of the human neotenic pattern of synaptic pruning. On the other hand, even diseases occuring at much later stages, such as Alzheimer disease, could display human-specificity linked to neuronal development. For instance, human (but not mouse) cortical neurons are sensitive to amyloid plaque toxicity following transplantation in mouse models of Alzheimer disease228.
A surprisingly large number of human-specific gene duplications seem to have contributed to the evolutionary expansion of the cortical anlage (Box 2), including ARHGAP11B, TBC1D3, TMEM14B and CROCCP2, which all promote basal progenitor (including oRGC) expansion187–192. Among these CROCCP2 was found to act as a human-specific modifier of mTOR signaling192, which is selectively upregulated in human oRGC79.
Several human-specific gene duplicates are also selectively expressed in developing or mature postmitotic neurons, suggesting their involvement in cortical circuit assembly and function. For example, SLIT-ROBO Rho GTPase activating protein 2 (SRGAP2193) is expressed in postmitotic cortical pyramidal neurons, as well as other neuronal subtypes, throughout development and in the adult brain194. The ancestral gene, called SRGAP2A, encodes a multifunctional protein that is highly conserved among all mammals195. SRGAP2A has undergone a series of large-segmental duplications leading to two partial duplications (SRGAP2B and SRGAP2C) that contain only the first 9 exons of SRGAP2A184,196. SRGAP2C emerged approximately 2.4 million years ago (at the birth of the Homo lineage) and exhibits a highly conserved copy number among human populations, suggesting strong positive selection184. SRGAP2C encodes a truncated protein that can bind to and inhibit all known functions of SRGAP2A196–198 (Fig. 4a).
Fig. 4|. Example of a human-specific modifier of cortical development and function.
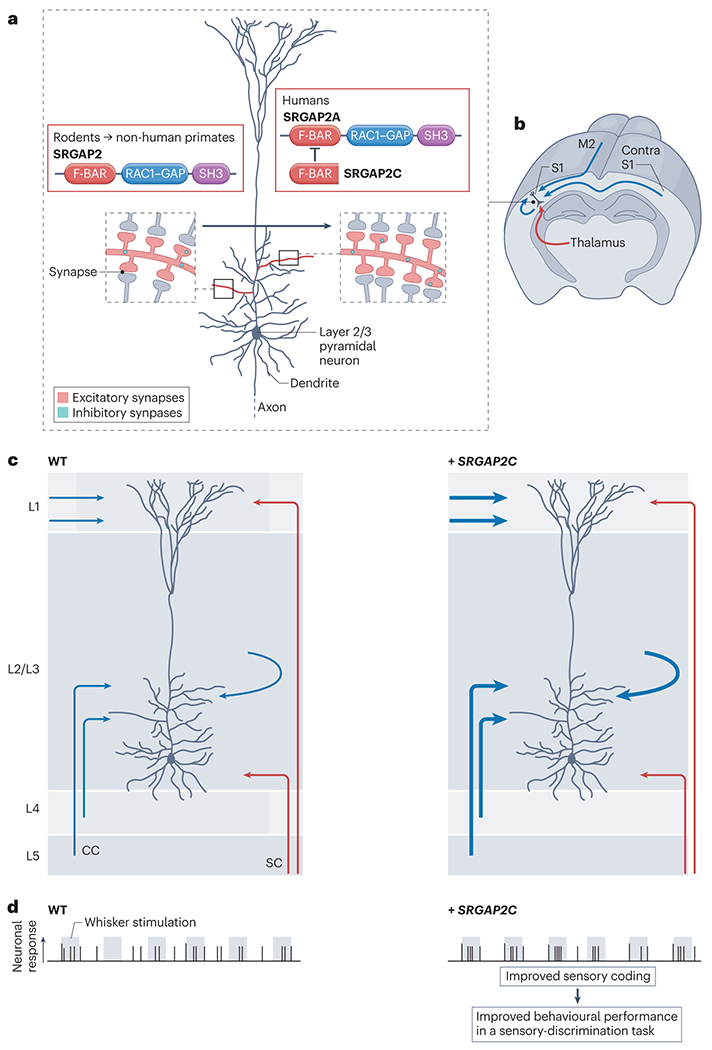
a-b, SRGAP2 (known as SRGAP2A in humans) is a postsynaptic protein that contains three functional domains. SRGAP2 is located at both excitatory and inhibitory synapses in mammalian cortical pyramidal neurons, where it promotes the maturation of the synapses while limiting their density196. A human-specific truncated paralog of this protein, SRGAP2C, binds to and inhibits all known functions of SRGAP2A, leading to neotenic synaptic development and increased synapse density (as shown on the right side of the figure) when expressed in mouse pyramidal neurons196,198. c, The introduction of SRGAP2C into mouse layer 2/3 pyramidal neurons drives an increase in the number of excitatory synapses as a result of a specific increase in cortico-cortical (CC) synaptic connections from both feedforward and feedback projections (shown in blue, with increased connections indicated by a thicker line) but not from subcortical inputs (SC, shown in red)199. d, The changes in circuit architecture induced in mice transgenically expressing SRGAP2C in all cortical pyramidal neurons lead to increased reliability of sensory coding, illustrated here as the fraction of action potentials that are induced during sensory stimulations (shown in grey) in layer 2/3 pyramidal neurons. Mice expressing SRGAP2C also show improved learning, compared to wild-type (WT) littermates, in a whisker-based sensory discrimination task199.
Functional studies in vivo demonstrated that SRGAP2A is a postsynaptic protein that promotes the maturation of excitatory and inhibitory synapses in cortical pyramidal neurons and, at the same time, limits the total number of synapses formed by cortical layer 5 and layer 2/3 pyramidal neurons196–198. Induction of SRGAP2C expression in mouse layer 2/3 cortical pyramidal neurons phenocopies a partial loss of function of SRGAP2A and leads to a significant delay in synaptic maturation and an increase in the density of synapses (Fig. 4a), mimicking two important features of human pyramidal neurons 3. Recent results demonstrate that transgenic expression of SRGAP2C in mouse layer 2/3 pyramidal neurons leads to a specific increase in the number of cortical-cortical connections between local excitatory pyramidal neurons and increased long-range cortical-cortical connections between cortical areas 199(Fig. 4b–c). In vivo imaging further revealed that layer 2/3 pyramidal neurons respond more reliably to sensory stimulations in the SRGAP2C-expressing mice (Fig. 4d) and that they also learn more efficiently a whisker-based sensory discrimination task, when compared to wild-type mice199 (Fig. 4d). These results suggest that the emergence of SRGAP2C has contributed to the evolution of some of the unique structural and functional features of cortical circuits in the human brain.
Beyond genetic mechanisms
The genetic mechanisms outlined above are likely to underlie many of the changes in developmental programmes that have driven human cortical evolution. However, emerging data suggest that more global changes in cellular processes could also link development to the evolution of the human brain. Human cortical neurons were shown to display much slower mitochondrial development and lower levels of oxidative metabolism than mouse neurons, and enhancing mitochondria function was demonstrated to speed up neuronal morphogenesis and synaptogenesis200. Similarly, recent studies have shown that rates of protein turnover are lower in human cells than in mouse cells 201,202, and that lower rates of translation can directly influence the timing of cortical neurogenesis 203,204. Collectively, these data suggest that global cellular mechanisms could play an important role in the scaling and patterning of human brain developmental events, in synergy with gene regulation and signaling.
Conclusions and perspectives
In the past two decades, a flurry of studies have revealed human-specific genomic changes — ranging from HARs to human-specific gene duplications — that constitute candidate species-specific modifiers of human brain development. However only a small fraction of these have been explored functionally and often the function of the ancestral genes and/or regulatory sequences remain poorly understood. Moreover there remains a profound gap in our understanding of how the identified evolutionary changes in developmental programme are linked to the resulting divergence in the properties of human neural circuits.
To move forward, the field needs to develop new experimental tools (Box 1) allowing more high-thoughput and parallelized ways to probe the functions of these modifiers during brain development in animal and human models. This is especially true for human-specific modifiers that act relatively late (in postnatal development or even in adulthood) on key steps such as synapse development, which might impact circuit architecture and function: two aspects of human brain development that remain especially challenging to modify and study. The availability of PSC from humans, primates and other mammals, in combination with advances in genomic editing and emerging tools such as next generation organoids and xenotransplantation of human neurons, will provide new and exciting opportunities to move towards this goal. However, these should not overshadow the crucial need for animal models that will be essential to understand the mechanisms of corticogenesis in a genuine and robust in vivo context. These include invertebrates and mice, but also, importantly, higher mammals (such as ferrets) and primates (such as marmosets and macaque), in order to include a wide spectrum of phylogeny, body size and brain specializations (Box 1). In parallel, new approaches are needed to link the molecular features of cortical cells with circuit assembly and circuit properties, including high resolution comparative (multi-omic to connectomic) studies in animal models and the human brain.
Finally, it is striking to note that many species-specific gene variants are also the sites of pathogenic or polymorphic variants in the human population. This emphasizes the need to further integrate human genetics with evolutionary genomics to fully comprehend how human-specific genomic changes have made our brain functions uniquely expansive and at the same time susceptible to neurodevelopmental and neurodegenerative disorders.
Acknowledgments
We wish to apologize to the many authors whose work could not be discussed due to space constraints. We thank E. Schmidt for his help generating elements of Figure 4. Work from the P.V. lab described here was funded by the European Research Council (ERC Adv Grants GENDEVOCORTEX and NEUROTEMPO), the Belgian FWO and FRS/FNRS, the EOS Program, the AXA Research Fund, the Belgian Queen Elizabeth Medical Foundation, the ERANET NEURON, the Generet Fund. Work from the F.P. lab described here was funded by grants from NIH-NINDS NIH (RO1NS067557 and R35NS127232), an award for the Roger De Spoelberch Fondation, an award from the Nomis Foundation.
Glossary
- Association areas
A class of cortical areas defined by their opposition to primary areas (cortical regions receiving direct inputs from the dorsal thalamus). Association areas are where different sensory and/or motor modalities combine and where complex cognitive processes such as attention, planning and memories are encoded.
- Cell fate mapping
A range of techniques aimed at genetically labelling the progeny of individual classes of progenitors, thereby reconstructing the lineage linking dividing progenitors and all the cells they generate.
- Chromatin loops
The situation in which stretches of genomic sequence that lie on the same chromosome (configured in cis) are in closer physical proximity to each other than they are to intervening sequences.
- cis-regulatory elements
Portions of genes containing the promoter and other regulatory elements controlling levels of gene transcription.
- Comparative genomics
A subfield of biology involving the analysis of DNA sequence divergence and conservation between different organisms.
- Connectome
A description of all the synaptic connections between neurons found within a brain region or the entire nervous system of an organism.
- Cortical organoids
Self-organized 3D multicellular structures that can be patterned to mimic the neocortex.
- Dendritic processing
The receipt, integration and processing of many synaptic inputs by dendrites. This processing takes the form of changes in membrane potential, which can — depending on the density and distribution of passive or active ionotropic channels — differentially affect the generation of action potentials at the level of the soma.
- Dendritic spines
Micron-long protrusions present in specific neuronal subtypes, such as cortical pyramidal neurons, at the tip of which is located an excitatory synapse. Spines play important roles in electrically and biochemically isolating the postsynaptic compartment from the dendrite shaft. Experimentally, morphologically identified dendritic spines represent a close approximation to measuring number or density of excitatory synapses received by a neuron.
- Epigenetic profiling
Molecular biology technique combined with biochemistry to monitor the post-translational modifications and physical interaction of DNA and chromatin and how they impact gene expression in cells.
- Heterochrony
Changes in the relative timing of a developmental event when different species or brain regions are being compared.
- Large segmental genomic duplications
Large segments (more than 1kb) of the genome that have been duplicated in another position in the genome.
- Long non-coding RNAs
RNAs longer than 200 bp that are not translated into protein.
- Massively parallel reporter assays
A molecular biology technique used to simultaneously test the activity of multiple candidate genetic regulatory elements in a high-throughput manner.
- Morphogen
One of a class of extracellular cues that can act at a distance from its source and regulate gene expression in receiving cells and tissues and thereby play a central role in cell type specification or tissue patterning.
- Neurogenesis
The generation of postmitotic neurons by specialized classes of dividing progenitors.
- Single-cell RNA sequencing
A high-throughput sequencing technique used to determine the sequences of mRNA expressed in single cells, also referred to as the transcriptional profile of single cells.
- Topology-associated domains
Large self-interacting genomic regions (~1Mb) physically interacting inside the nucleus. This level of chromatin organization plays key roles in regulating temporal and spatial patterns of gene expression in gene families such as the HOX cluster.
- Xenotransplantation
Transplantation of cells from one species into a different species.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Wood B & E KB. Hominin taxic diversity: Fact or fantasy? Am J Phys Anthropol 159, S37–78 (2016). 10.1002/aipa.22902 [DOI] [PubMed] [Google Scholar]
- 2.Changeux JP, Goulas A & Hilgetag CC A Connectomic Hypothesis for the Hominization of the Brain. Cereb Cortex 31, 2425–2449 (2021). 10.1093/cercor/bhaa365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Defelipe J The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front Neuroanat 5, 29 (2011). 10.3389/fnana.2011.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtman JW & Denk W The big and the small: challenges of imaging the brain’s circuits.Science 334, 618–623 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Geschwind DH & Rakic P Cortical evolution: judge the brain by its cover. Neuron 80, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herculano-Houzel S The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci 3, 31 (2009). 10.3389/neuro.09.031.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herculano-Houzel S The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc Natl Acad Sci U S A 109 Suppl 1, 10661–10668 (2012). 10.1073/pnas.1201895109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 e3226 (2021). 10.1016/j.cell.2021.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuste R et al. A community-based transcriptomics classification and nomenclature of neocortical cell types. Nature Neuroscience 23, 1456–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitali I & Jabaudon D Synaptic biology of barrel cortex circuit assembly. Semin Cell Dev Biol 35, 156–164 (2014). 10.1016/j.semcdb.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 11.Balaram P, Young NA & Kaas JH Histological features of layers and sublayers in cortical visual areas V1 and V2 of chimpanzees, macaque monkeys, and humans. Eye and Brain 6, 5–18 (2014). 10.2147/EB.S51814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFelipe J, Alonso-Nanclares L & Arellano JI Microstructure of the neocortex: comparative aspects. J Neurocytol 31, 299–316 (2002). 10.1023/a:1024130211265 [DOI] [PubMed] [Google Scholar]
- 13.Hutsler JJ, Lee DG & Porter KK Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res 1052, 71–81 (2005). 10.1016/j.brainres.2005.06.015 [DOI] [PubMed] [Google Scholar]
- 14.Berg J et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158 (2021). 10.1038/s41586-021-03813-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; A multimodal analysis reveals increased diversity of human upper layer cortical neurons.
- 15.Network, B. I. C. C. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). 10.1038/s41586-021-03950-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulas A, Zilles K & Hilgetag CC Cortical Gradients and Laminar Projections in Mammals. Trends Neurosci 41, 775–788 (2018). 10.1016/j.tins.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 17.Bakken TE et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021). 10.1038/s41586-021-03465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khrameeva E et al. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res 30, 776–789 (2020). 10.1101/gr256958119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodge RD et al. Transcriptomic evidence that von Economo neurons are regionally specialized extratelencephalic-projecting excitatory neurons. Nat Commun 11, 1172 (2020). 10.1038/s41467-020-14952-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nano PR, Nguyen CV, Mil J & Bhaduri A Cortical Cartography: Mapping Arealization Using Single-Cell Omics Technology. Front Neural Circuits 15, 788560 (2021). 10.3389/fncir.2021788560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakic P Specification of cerebral cortical areas. Science 241, 170–176 (1988). 10.1126/science3291116 [DOI] [PubMed] [Google Scholar]
- 22.Glasser MF et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016). 10.1038/nature18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northcutt RG & Kaas JH The emergence and evolution of mammalian neocortex. Trends Neurosci 18, 373–379 (1995). 10.1016/0166-2236(95)93932-n [DOI] [PubMed] [Google Scholar]
- 24.Krubitzer L The magnificent compromise: cortical field evolution in mammals. Neuron 56, 201–208 (2007). 10.1016/j.neuron.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 25.Pulvermuller F & Fadiga L Active perception: sensorimotor circuits as a cortical basis for language. Nat Rev Neurosci 11, 351–360 (2010). 10.1038/nrn2811 [DOI] [PubMed] [Google Scholar]
- 26.Miller EK & Cohen JD An integrative theory of prefrontal cortex function. Annu Rev Neurosci 24, 167–202 (2001). 10.1146/annurev.neuro.24.1.167 [DOI] [PubMed] [Google Scholar]
- 27.Donahue CJ, Glasser MF, Preuss TM, Rilling JK & Van Essen DC Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc Natl Acad Sci U S A 115, E5183–E5192 (2018). 10.1073/pnas.1721653115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smaers JB, Gomez-Robles A, Parks AN & Sherwood CC Exceptional Evolutionary Expansion of Prefrontal Cortex in Great Apes and Humans. Curr Biol 27, 714–720 (2017). 10.1016/j.cub.2017.01.020 [DOI] [PubMed] [Google Scholar]
- 29.Gabi M et al. No relative expansion of the number of prefrontal neurons in primate and human evolution. Proc Natl Acad Sci U S A 113, 9617–9622 (2016). 10.1073/pnas.1610178113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata M et al. Regulation of prefrontal patterning and connectivity by retinoic acid. Nature 598, 483–488 (2021). 10.1038/s41586-021-03953-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckner RL & Krienen FM The evolution of distributed association networks in the human brain. Trends Cogn Sci 17, 648–665 (2013). 10.1016/j.tics.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 32.Sydnor VJ et al. Neurodevelopment of the association cortices: Patterns, mechanisms, and implications for psychopathology. Neuron 109, 2820–2846 (2021). 10.1016/j.neuron.2021.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y et al. Interneuron origin and molecular diversity in the human fetal brain. Nature Neuroscience 24, 1745–1756 (2021). 10.1038/s41593-021-00940-3 [DOI] [PubMed] [Google Scholar]
- 34.Hodge RD et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krienen FM et al. Innovations present in the primate interneuron repertoire. Nature 586, 262–269 (2020). 10.1038/s41586-020-2781-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boldog E et al. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat Neurosci 21, 1185–1195 (2018). 10.1038/s41593-018-0205-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberheim NA et al. Uniquely hominid features of adult human astrocytes. Journal of Neuroscience 29, 3276–3287 (2009). 10.1523/JNEUROSCI.4707-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falcone C et al. Cortical interlaminar astrocytes across the therian mammal radiation. J Comp Neurol 527, 1654–1674 (2019). 10.1002/cne.24605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falcone C et al. Redefining varicose projection astrocytes in primates. Glia 70, 145–154 (2022). 10.1002/glia.24093 [DOI] [PubMed] [Google Scholar]
- 40.Geirsdottir L et al. Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 179, 1609–1622.e1616(2019). 10.1016/j.cell.2019.11.010 [DOI] [PubMed] [Google Scholar]
- 41.Masuda T et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019). 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- 42.Sousa AMM et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). 10.1126/science.aan3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen DE et al. Fate mapping of neural stem cell niches reveals distinct origins of human cortical astrocytes. Science 376, 1441–1446 (2022). 10.1126/science.abm5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marin-Padilla M Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. J Comp Neurol 321, 223–240 (1992). 10.1002/cne.903210205 [DOI] [PubMed] [Google Scholar]
- 45.Bianchi S et al. Dendritic morphology of pyramidal neurons in the chimpanzee neocortex: regional specializations and comparison to humans. Cereb Cortex 23, 2429–2436 (2013). 10.1093/cercor/bhs239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deitcher Y et al. Comprehensive Morpho-Electrotonic Analysis Shows 2 Distinct Classes of L2 and L3 Pyramidal Neurons in Human Temporal Cortex. Cereb Cortex 27, 5398–5414 (2017). 10.1093/cercor/bhx226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elston GN, Benavides-Piccione R & Defelipe J The Pyramidal Cell in Cognition: A Comparative Study in Human and Monkey. J Neurosci. 21, RC163 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohan H et al. Dendritic and Axonal Architecture of Individual Pyramidal Neurons across Layers of Adult Human Neocortex. Cereb Cortex 25, 4839–4853(2015). 10.1093/cercor/bhv188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benavides-Piccione R, Ballesteros-Yanez I, DeFelipe J & Yuste R Cortical area and species differences in dendritic spine morphology. J Neurocytol 31, 337–346 (2002). 10.1023/a:1024134312173 [DOI] [PubMed] [Google Scholar]
- 50.Sherwood CC et al. Invariant synapse density and neuronal connectivity scaling in primate neocortical evolution. Cerebral Cortex 30, 5604–5615 (2020). 10.1093/cercor/bhaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]; Comparative analysis in the cortex of 25 primate species reveals that the highest number of synapses per neuron are found in the human cortex.
- 51.Iascone DM et al. Whole-Neuron Synaptic Mapping Reveals Spatially Precise Excitatory/Inhibitory Balance Limiting Dendritic and Somatic Spiking. Neuron 106, 566–578 (2020). 10.1016/j.neuron.2020.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beaulieu-Laroche L et al. Allometric rules for mammalian cortical layer 5 neuron biophysics. Nature 600, 274–278 (2021). 10.1038/s41586-021-04072-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; Comparative profiling across 10 mammalian species reveals that human cortical neurons are outliers for several key parameters of intrinsic functional properties.
- 53.Beaulieu-Laroche L et al. Enhanced Dendritic Compartmentalization in Human Cortical Neurons. Cell 175, 643–651 e614 (2018). 10.1016/j.cell.2018.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gidon A et al. Dendritic action potentials and computation in human layer 2/3 cortical neurons. Science 367, 83–87 (2020). 10.1126/science.aax6239 [DOI] [PubMed] [Google Scholar]
- 55.Kalmbach BE et al. h-Channels Contribute to Divergent Intrinsic Membrane Properties of Supragranular Pyramidal Neurons in Human versus Mouse Cerebral Cortex. Neuron 100, 1194–1208 e1195 (2018). 10.1016/j.neuron.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; Changes in h-channel expression levels might explain the divergent electrophyiological properties of human cortical layer 2/3 pyramidal neurons.
- 56.Schwarz N et al. Long-term adult human brain slice cultures as a model system to study human CNS circuitry and disease. Elife 8, e48417 (2019). 10.7554/eLife.48417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuart G, Spruston N & Häusser M Dendrites. (Third edition) (Oxford University Press, 2016). [Google Scholar]
- 58.Kalmbach BE et al. Signature morpho-electric, transcriptomic, and dendritic properties of human layer 5 neocortical pyramidal neurons. Neuron 109, 2914–2927 e2915 (2021). 10.1016/j.neuron.2021.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francioni V & Harnett MT Rethinking Single Neuron Electrical Compartmentalization: Dendritic Contributions to Network Computation In Vivo. Neuroscience 489, 185–199 (2022). 10.1016/j.neuroscience.2021.05.038 [DOI] [PubMed] [Google Scholar]
- 60.Eyal G et al. Human Cortical Pyramidal Neurons: From Spines to Spikes via Models. Front Cell Neurosci 12, 181 (2018). 10.3389/fncel.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eyal G et al. Unique membrane properties and enhanced signal processing in human neocortical neurons. Elife 5, e16553 (2016). 10.7554/eLife.16553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campagnola L et al. Local connectivity and synaptic dynamics in mouse and human neocortex. Science 375, eabj5861 (2022). 10.1126/science.abi5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molnar G et al. Complex events initiated by individual spikes in the human cerebral cortex. PLoS Biol 6, e222 (2008). 10.1371/iournal.pbio.0060222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molnar G et al. Human pyramidal to interneuron synapses are mediated by multi-vesicular release and multiple docked vesicles. Elife 5, e18167 (2016). 10.7554/eLife.18167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szegedi V et al. Plasticity in Single Axon Glutamatergic Connection to GABAergic Interneurons Regulates Complex Events in the Human Neocortex. PLoS Biol 14, e2000237 (2016). 10.1371/iournal.pbio.2000237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Testa-Silva G et al. High bandwidth synaptic communication and frequency tracking in human neocortex. PLoS Biol 12, e1002007(2014). 10.1371/iournal.pbio.1002007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bastos AM et al. Canonical microcircuits for predictive coding. Neuron 76, 695–711 (2012). 10.1016/j.neuron.2012.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Douglas RJ & Martin KA A functional microcircuit for cat visual cortex. J Physiol 440, 735–769 (1991). 10.1113/jphysiol.1991.sp018733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vendetti MS & Bunge SA Evolutionary and developmental changes in the lateral frontoparietal network: a little goes a long way for higher-level cognition. Neuron 84, 906–917 (2014). 10.1016/j.neuron.2014.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Libe-Philippot B & Vanderhaeghen P Cellular and Molecular Mechanisms Linking Human Cortical Development and Evolution. Annu Rev Genet 55, 555–581 (2021). 10.1146/annurev-genet-071719-020705 [DOI] [PubMed] [Google Scholar]
- 71.Miller DJ, Bhaduri A, Sestan N & Kriegstein A Shared and derived features of cellular diversity in the human cerebral cortex. Curr Opin Neurobiol 56, 117–124 (2019). 10.1016/j.conb.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silbereis JC, Pochareddy S, Zhu Y, Li M & Sestan N The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248–268 (2016). 10.1016/j.neuron.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villalba A, Gotz M & Borrell V The regulation of cortical neurogenesis. Curr Top Dev Biol 142, 1–66 (2021). 10.1016/bs.ctdb.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 74.Benito-Kwiecinski S et al. An early cell shape transition drives evolutionary expansion of the human forebrain. Cell 184, 2084–2102 e2019 (2021). 10.1016/j.cell.2021.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]; Comparison of great ape and human neural organoids reveals mechanisms of prolonged expansion of human neuroepithelial cells
- 75.Espuny-Camacho I et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 77, 440–456 (2013). 10.1016/j.neuron.2012.12.011 [DOI] [PubMed] [Google Scholar]; Human and mouse corticogenesis retains species-specific developmental timing in vitro and following xenotransplantation.
- 76.Gaspard N et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357 (2008). 10.1038/nature07287 [DOI] [PubMed] [Google Scholar]
- 77.Kanton S et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). 10.1038/s41586-019-1654-9 [DOI] [PubMed] [Google Scholar]
- 78.Otani T, Marchetto MC, Gage FH, Simons BD & Livesey FJ 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell 18, 467–480 (2016). 10.1016/j.stem.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pollen AA et al. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell 176, 743–756 e717 (2019). 10.1016/j.cell.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; Comparative single cell transcriptomic study of human and primate fetal brain and organoids reveals increased mTOR signaling in human outer radial glia.
- 80.Polleux F, Dehay C, Moraillon B & Kennedy H Regulation of neuroblast cell-cycle kinetics plays a crucial role in the generation of unique features of neocortical areas. Journal of Neuroscience 17, 7763–7783 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dehay C & Kennedy H Cell-cycle control and cortical development. Nat Rev Neurosci 8, 438–450 (2007). 10.1038/nrn2097 [DOI] [PubMed] [Google Scholar]
- 82.Iwata R, Casimir P & Vanderhaeghen P Mitochondrial dynamics in postmitotic cells regulate neurogenesis. Science 369, 858–862 (2020). 10.1126/science.aba9760 [DOI] [PubMed] [Google Scholar]
- 83.Kriegstein A, Noctor S & Martinez-Cerdeno V Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci 7, 883–890 (2006). 10.1038/nrn2008 [DOI] [PubMed] [Google Scholar]
- 84.Fietz SA et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci 13, 690–699(2010). 10.1038/nn.2553 [DOI] [PubMed] [Google Scholar]
- 85.Hansen DV, Lui JH, Parker PR & Kriegstein AR Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- 86.Reillo I, de Juan Romero C, Garcia-Cabezas MA & Borrell V A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex 21, 1674–1694 (2011). 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]; Fietz et al., Hansen et al., and Reillo et al. describe outer radial glial cells and their implication in increasing cortical size.
- 87.Dehay C, Kennedy H & Kosik KS The outer subventricular zone and primate-specific cortical complexification. Neuron 85, 683–694 (2015). 10.1016/j.neuron.2014.12.060 [DOI] [PubMed] [Google Scholar]
- 88.Smart IH, Dehay C, Giroud P, Berland M & Kennedy H Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex 12, 37–53 (2002). 10.1093/cercor/12.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pollen AA et al. Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015). 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C & Kriegstein AR Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron 91, 1219–1227 (2016). 10.1016/j.neuron.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang WQ & Yuste R Overproduction of Neurons Is Correlated with Enhanced Cortical Ensembles and Increased Perceptual Discrimination. Cell Rep 21, 381–392 (2017). 10.1016/j.celrep.2017.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]; Pharmacologically increasing the production of layer 2/3 pyramidal neurons production in the mouse leads to significant improvement in visual circuit function.
- 92.Boothe RG, Greenough WT, Lund JS & Wrege K A quantitative investigation of spine and dendrite development of neurons in visual cortex (area 17) of Macaca nemestrina monkeys. J Comp Neurol 186, 473–489 (1979). 10.1002/cne.901860310 [DOI] [PubMed] [Google Scholar]
- 93.Bourgeois JP & Rakic P Changes of synaptic density in the primary visual cortex of the macaque monkey from fetal to adult stage. J Neurosci 13, 2801–2820 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huttenlocher PR Synaptic density in human frontal cortex - developmental changes and effects of aging. Brain Res 163, 195–205 (1979). 10.1016/0006-8993(79)90349-4 [DOI] [PubMed] [Google Scholar]
- 95.Huttenlocher PR & Dabholkar AS Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol 387, 167–178 (1997). 10.1002/(sici)1096-9861(19971020)387:2 [DOI] [PubMed] [Google Scholar]
- 96.Mrzljak L, Uylings HB, Van Eden CG & Judas M Neuronal development in human prefrontal cortex in prenatal and postnatal stages. Prog Brain Res 85, 185–222 (1990). 10.1016/s0079-6123(08)62681-3 [DOI] [PubMed] [Google Scholar]
- 97.Petanjek Z, Judas M, Kostovic I & Uylings HB Lifespan alterations of basal dendritic trees of pyramidal neurons in the human prefrontal cortex: a layer-specific pattern. Cereb Cortex 18, 915–929 (2008). 10.1093/cercor/bhm124 [DOI] [PubMed] [Google Scholar]
- 98.Petanjek Z et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A 108, 13281–13286(2011). 10.1073/pnas.1105108108 [DOI] [PMC free article] [PubMed] [Google Scholar]; Quantitative assessment of dendritic spine density demonstrates striking neoteny of synaptic development characterizing human cortex development.
- 99.Petanjek Z et al. The Protracted Maturation of Associative Layer IIIC Pyramidal Neurons in the Human Prefrontal Cortex During Childhood: A Major Role in Cognitive Development and Selective Alteration in Autism. Front Psychiatry 10, 122(2019). 10.3389/fpsyt.2019.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Verendeev A & Sherwood CC Human Brain Evolution. Curr Opin Behav Sci 16, 41–45 (2017). 10.1016/j.cobeha.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Courchesne E et al. Mapping early brain development in autism. Neuron 56, 399–413 (2007). 10.1016/j.neuron.2007.10.016 [DOI] [PubMed] [Google Scholar]
- 102.Hazlett HC et al. Early brain development in infants at high risk for autism spectrum disorder. Nature 542, 348–351 (2017). 10.1038/nature21369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bianchi S et al. Synaptogenesis and development of pyramidal neuron dendritic morphology in the chimpanzee neocortex resembles humans. Proc Natl Acad Sci U S A 110 Suppl 2, 10395–10401 (2013). 10.1073/pnas.1301224110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Travis K, Ford K & Jacobs B Regional dendritic variation in neonatal human cortex: a quantitative Golgi study. Dev Neurosci 27, 277–287(2005). 10.1159/000086707 [DOI] [PubMed] [Google Scholar]
- 105.Elston GN, Oga T & Fujita I Spinogenesis and pruning scales across functional hierarchies. J Neurosci 29, 3271–3275 (2009). 10.1523/JNEUROSCI.5216-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Klingler E et al. Temporal controls over inter-areal cortical projection neuron fate diversity. Nature 599, 453–457 (2021). 10.1038/s41586-021-04048-3 [DOI] [PubMed] [Google Scholar]
- 107.Paredes MF et al. Extensive migration of young neurons into the infant human frontal lobe. Science 354, aaf7073 (2016). 10.1126/science.aaf7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sorrells SF et al. Immature excitatory neurons develop during adolescence in the human amygdala. Nat Commun 10, 2748 (2019). 10.1038/s41467-019-10765-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller DJ et al. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci U S A 109, 16480–16485 (2012). 10.1073/pnas.1117943109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williamson JM & Lyons DA Myelin Dynamics Throughout Life: An Ever-Changing Landscape? Front Cell Neurosci 12, 424 (2018). 10.3389/fncel.2018.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Linaro D et al. Xenotransplanted Human Cortical Neurons Reveal Species-Specific Development and Functional Integration into Mouse Visual Circuits. Neuron 104, 972–986 e976 (2019). 10.1016/j.neuron.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; Xenotransplanted human cortical neurons integrate functionally in the mouse cortical circuits following their species-specific tempo.
- 112.Maroof AM et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 12, 559–572 (2013). 10.1016/j.stem.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nicholas CR et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 12, 573–586 (2013). 10.1016/j.stem.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schornig M et al. Comparison of induced neurons reveals slower structural and functional maturation in humans than in apes. Elife 10, e59323(2021). 10.7554/eLife.59323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marchetto MC et al. Species-specific maturation profiles of human, chimpanzee and bonobo neural cells. Elife 8, e37527 (2019). 10.7554/eLife.37527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Molnar Z, Luhmann HJ & Kanold PO Transient cortical circuits match spontaneous and sensory-driven activity during development. Science 370 (2020). 10.1126/science.abb2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carroll SB Genetics and the making of Homo sapiens. Nature 422, 849–857 (2003). 10.1038/nature01495 [DOI] [PubMed] [Google Scholar]
- 118.Kaessmann H Origins, evolution, and phenotypic impact of new genes. Genome Res 20, 1313–1326 (2010). 10.1101/gr.101386.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davidson EH & Erwin DH Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006). 10.1126/science.1113832 [DOI] [PubMed] [Google Scholar]
- 120.Cardoso-Moreira M et al. Gene expression across mammalian organ development. Nature 571, 505–509 (2019). 10.1038/s41586-019-1338-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pollard KS et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172 (2006). 10.1038/nature05113 [DOI] [PubMed] [Google Scholar]
- 122.Prabhakar S, Noonan JP, Paabo S & Rubin EM Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786 (2006). 10.1126/science.1130738 [DOI] [PubMed] [Google Scholar]; Pollard et al and Prabhakar et al. describe human accelerated regions and suggest their implication in human brain development.
- 123.Capra JA, Erwin GD, McKinsey G, Rubenstein JL & Pollard KS Many human accelerated regions are developmental enhancers. Philos Trans R Soc Lond B Biol Sci 368, 20130025 (2013). 10.1098/rstb.2013.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Doan RN et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167, 341–354 e312 (2016). 10.1016/j.cell.2016.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies human accelerated regions associated with neurodevelopmental diseases.
- 125.Kamm GB, Pisciottano F, Kliger R & Franchini LF The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Mol Biol Evol 30, 1088–1102 (2013). 10.1093/molbev/mst023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lambert N et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PLoS One 6, e17753 (2011). 10.1371/iournal.pone.0017753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller JA et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wei Y et al. Genetic mapping and evolutionary analysis of human-expanded cognitive networks. Nat Commun 10, 4839 (2019). 10.1038/s41467-019-12764-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Won H, Huang J, Opland CK, Hartl CL & Geschwind DH Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat Commun 10, 2396 (2019). 10.1038/s41467-019-10248-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Girskis KM et al. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 109, 3239–3251 e3237(2021). 10.1016/j.neuron.2021.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Uebbing S et al. Massively parallel discovery of human-specific substitutions that alter enhancer activity. Proc Natl Acad Sci U S A 118, e2007049118 (2021). 10.1073/pnas.2007049118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Whalen S et al. Machine-learning dissection of Human Accelerated Regions in primate neurodevelopment. bioRxiv, 256313 Preprint at 10.1101/256313v3 (2022). [DOI] [PMC free article] [PubMed]
- 133.Chenn A & Walsh CA Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). 10.1126/science.1074192 [DOI] [PubMed] [Google Scholar]
- 134.Tiberi L, Vanderhaeghen P & van den Ameele J Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr Opin Cell Biol 24, 269–276 (2012). 10.1016/j.ceb.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 135.Boyd JL et al. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol 25, 772–779 (2015). 10.1016/j.cub.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]; A human accelerated region (HAR) enhancing WNT signaling has an impact on cortical expansion.
- 136.Reilly SK et al. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159 (2015). 10.1126/science.1260943 [DOI] [PMC free article] [PubMed] [Google Scholar]; The first description of regulatory elements displaying human-specific activation during brain development.
- 137.de la Torre-Ubieta L et al. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell 172, 289–304 e218(2018). 10.1016/j.cell.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song M et al. Cell-type-specific 3D epigenomes in the developing human cortex. Nature 587, 644–649 (2020). 10.1038/s41586-020-2825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fiddes IT et al. Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell 173, 1356–1369 e1322 (2018). 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suzuki IK et al. Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell (2018). 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]; Fiddes et al. and Suzuki et al. identify hominid-specific genes NOTCH2NL as human-specific modifiers of cortical neurogenesis
- 141.Tilot AK et al. The Evolutionary History of Common Genetic Variants Influencing Human Cortical Surface Area. Cereb Cortex 31, 1873–1887 (2021). 10.1093/cercor/bhaa327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vermunt MW et al. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci 19, 494–503 (2016). 10.1038/nn.4229 [DOI] [PubMed] [Google Scholar]
- 143.Moriano J & Boeckx C Modern human changes in regulatory regions implicated in cortical development. BMC Genomics 21, 304 (2020). 10.1186/s12864-020-6706-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bolt CC & Duboule D The regulatory landscapes of developmental genes. Development 147, dev171736 (2020). 10.1242/dev.171736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Luo X et al. 3D Genome of macaque fetal brain reveals evolutionary innovations during primate corticogenesis. Cell 184, 723–740 e721(2021). 10.1016/j.cell.2021.01.001 [DOI] [PubMed] [Google Scholar]
- 146.Vanderhaeghen P & Polleux F Developmental mechanisms patterning thalamocortical projections: Intrinsic, extrinsic and in between. Trends in Neurosciences 27, 384–391 (2004). 10.1016/j.tins.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 147.McLean CY et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471, 216–219 (2011). 10.1038/nature09774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kronenberg ZN et al. High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018). 10.1126/science.aar6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Agoglia RM et al. Primate cell fusion disentangles gene regulatory divergence in neurodevelopment. Nature 592, 421–427 (2021). 10.1038/s41586-021-03343-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Song JHT et al. Genetic studies of human-chimpanzee divergence using stem cell fusions. Proc Natl Acad Sci U S A 118, e2117557118 (2021). 10.1073/pnas.2117557118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li M et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362 (2018). 10.1126/science.aat7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pletikos M et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81, 321–332 (2014). 10.1016/j.neuron.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu Y et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). 10.1126/science.aat8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lui JH et al. Radial glia require PDGFD-PDGFRb signalling in human but not mouse neocortex. Nature 515, 264–268 (2014). 10.1038/nature13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Andrews MG, Subramanian L & Kriegstein AR mTOR signaling regulates the morphology and migration of outer radial glia in developing human cortex. Elife 9, e58737 (2020). 10.7554/eLife.58737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nowakowski TJ et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jacobs FM et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245 (2014). 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Johansson PA et al. A cis-acting structural variation at the ZNF558 locus controls a gene regulatory network in human brain development. Cell Stem Cell 29, 52–69 e58 (2022). 10.1016/j.stem.2021.09.008 [DOI] [PubMed] [Google Scholar]
- 159.Turelli P et al. Primate-restricted KRAB zinc finger proteins and target retrotransposons control gene expression in human neurons. Sci Adv 6, eaba3200 (2020). 10.1126/sciadv.aba3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sarropoulos I, Marin R, Cardoso-Moreira M & Kaessmann H Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571, 510–514 (2019). 10.1038/s41586-019-1341-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rani N et al. A Primate lncRNA Mediates Notch Signaling during Neuronal Development by Sequestering miRNA. Neuron 90, 1174–1188 (2016). 10.1016/j.neuron.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Field AR et al. Structurally Conserved Primate LncRNAs Are Transiently Expressed during Human Cortical Differentiation and Influence Cell-Type-Specific Genes. Stem Cell Reports 12, 245–257 (2019). 10.1016/j.stemcr.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shibata M et al. Hominini-specific regulation of CBLN2 increases prefrontal spinogenesis. Nature 598, 489–494 (2021). 10.1038/s41586-021-03952-y [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies molecular mechanisms underlying increased connectivity of the human prefrontal cortex.
- 164.Ziffra RS et al. Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021). 10.1038/s41586-021-03209-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bhaduri A et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204 (2021). 10.1038/s41586-021-03910-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive single cell transcriptomic study describing area-specific patterns of gene expression in the human cortex
- 166.Gu Z et al. Control of species-dependent cortico-motoneuronal connections underlying manual dexterity. Science 357, 400–404 (2017). 10.1126/science.aan3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ebert DH & Greenberg ME Activity-dependent neuronal signalling and autism spectrum disorder. Nature 493, 327–337 (2013). 10.1038/nature11860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ataman B et al. Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247 (2016). 10.1038/nature20111 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies primate-specific transcriptional programmes triggered by neuronal activity.
- 169.Pruunsild P, Bengtson CP & Bading H Networks of Cultured iPSC-Derived Neurons Reveal the Human Synaptic Activity-Regulated Adaptive Gene Program. Cell Rep 18, 122–135 (2017). 10.1016/j.celrep.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Qiu J et al. Evidence for evolutionary divergence of activity-dependent gene expression in developing neurons. Elife 5, e20337 (2016). 10.7554/eLife.20337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu X et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res 22, 611–622 (2012). 10.1101/gr.127324.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boulting GL et al. Activity-dependent regulome of human GABAergic neurons reveals new patterns of gene regulation and neurological disease heritability. Nat Neurosci 24, 437–448 (2021). 10.1038/s41593-020-00786-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Raju CS et al. Secretagogin is Expressed by Developing Neocortical GABAergic Neurons in Humans but not Mice and Increases Neurite Arbor Size and Complexity. Cereb Cortex 28, 1946–1958 (2018). 10.1093/cercor/bhx101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Castelijns B et al. Hominin-specific regulatory elements selectively emerged in oligodendrocytes and are disrupted in autism patients. Nat Commun 11, 301 (2020). 10.1038/s41467-019-14269-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bustamante CD et al. Natural selection on protein-coding genes in the human genome. Nature 437, 1153–1157 (2005). 10.1038/nature04240 [DOI] [PubMed] [Google Scholar]
- 176.Williamson SH et al. Localizing recent adaptive evolution in the human genome. PLoS Genet 3, e90 (2007). 10.1371/iournal.pgen.0030090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lindblad-Toh K et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). 10.1038/nature10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.den Hoed J & Fisher SE Genetic pathways involved in human speech disorders. Curr Opin Genet Dev 65, 103–111 (2020). 10.1016/j.gde.2020.05.012 [DOI] [PubMed] [Google Scholar]
- 179.Enard W et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971 (2009). 10.1016/j.cell.2009.03.041 [DOI] [PubMed] [Google Scholar]
- 180.Paabo S The human condition-a molecular approach. Cell 157, 216–226 (2014). 10.1016/j.cell.2013.12.036 [DOI] [PubMed] [Google Scholar]
- 181.Rosso L et al. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol 6, e140 (2008). 10.1371/iournal.pbio.0060140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fortna A et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol 2, E207 (2004). 10.1371/iournal.pbio.0020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sudmant PH et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010). 10.1126/science.1197005 [DOI] [PMC free article] [PubMed] [Google Scholar]; Fortna et al. and Sudmant et al. represent the first descriptions of hominid-specific gene duplications.
- 184.Dennis MY et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922 (2012). 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Stefansson H et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008). 10.1038/nature07229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Florio M et al. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife 7, e32332(2018). 10.7554/eLife.32332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Florio M et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015). 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- 188.Heide M et al. Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science 369, 546–550 (2020). 10.1126/science.abb2401 [DOI] [PubMed] [Google Scholar]
- 189.Hou QQ, Xiao Q, Sun XY, Ju XC & Luo ZG TBC1D3 promotes neural progenitor proliferation by suppressing the histone methyltransferase G9a. Sci Adv 7, eaba8053 (2021). 10.1126/sciadv.aba8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ju XC et al. The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife 5, e18197 (2016). 10.7554/eLife.18197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu J et al. The Primate-Specific Gene TMEM14B Marks Outer Radial Glia Cells and Promotes Cortical Expansion and Folding. Cell Stem Cell 21, 635–649 e638 (2017). 10.1016/j.stem.2017.08.013 [DOI] [PubMed] [Google Scholar]
- 192.Van Heurck R et al. CROCCP2 acts as a human-specific modifier of cilia dynamics and mTOR signaling to promote expansion of cortical progenitors. Neuron S0896-6273(22)00947-3. 10.1016/j.neuron.2022.10.018 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wong K et al. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107, 209–221 (2001). 10.1016/s0092-8674(01)00530-x [DOI] [PubMed] [Google Scholar]
- 194.Bacon C, Endris V & Rappold G Dynamic expression of the Slit-Robo GTPase activating protein genes during development of the murine nervous system. J Comp Neurol 513, 224–236 (2009). 10.1002/cne.21955 [DOI] [PubMed] [Google Scholar]
- 195.Guerrier S et al. The F-BAR Domain of srGAP2 Induces Membrane Protrusions Required for Neuronal Migration and Morphogenesis. Cell 138, 990–1004 (2009). 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Charrier C et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935 (2012). 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schmidt ERE, Kupferman JV, Stackmann M & Polleux F The human-specific paralogs SRGAP2B and SRGAP2C differentially modulate SRGAP2A-dependent synaptic development. Scientific Reports 9, 18692 (2019). 10.1038/s41598-019-54887-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fossati M et al. SRGAP2 and Its Human-Specific Paralog Co-Regulate the Development of Excitatory and Inhibitory Synapses. Neuron 91, 356–369 (2016). 10.1016/j.neuron.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; Charrier et al., Schmidt et al., and Fossati et al. demonstrate that the human-specific paralog SRGAP2C inhibits the function of the ancestral SRGAP2A postsynaptic protein and leads to neotenic and increased excitatory and inhibitory synapse density.
- 199.Schmidt ERE et al. A human-specific modifier of cortical connectivity and circuit function. Nature 599, 640–644 (2021). 10.1038/s41586-021-04039-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; Expression of human-specific SRGAP2C in mouse cortical pyramidal neurons leads to increased cortico-cortical connectivity, improved sensory coding by cortical circuits and improved behavioral performance in a sensory discrimination task.
- 200.Iwata R et al. Species-specific mitochondria dynamics and metabolism regulate the timing of neuronal development. bioRxiv, Preprint at 10.1101/2021.12.27.474246v1 (2021). [DOI]; This study identifies mitochondria metabolism as a key regulator of the species-specific tempo of neuronal development.
- 201.Rayon T et al. Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667 (2020). 10.1126/science.aba7667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Matsuda M et al. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450–1455 (2020). 10.1126/science.aba7668 [DOI] [PubMed] [Google Scholar]
- 203.Hoye ML et al. Aberrant cortical development is driven by impaired cell cycle and translational control in a DDX3X syndrome model. Elife 11, e78203 (2022). 10.7554/eLife.78203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wu Q et al. Selective translation of epigenetic modifiers affects the temporal pattern and differentiation of neural stem cells. Nat Commun 13, 470 (2022). 10.1038/s41467-022-28097-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Namba T et al. Human-Specific ARHGAP11B Acts in Mitochondria to Expand Neocortical Progenitors by Glutaminolysis. Neuron 105, 867–881 e869 (2020). 10.1016/j.neuron.2019.11.027 [DOI] [PubMed] [Google Scholar]
- 206.Angevine JB Jr. & Sidman RL Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766–768 (1961). 10.1038/192766b0 [DOI] [PubMed] [Google Scholar]
- 207.Polleux F, Dehay C & Kennedy H The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. Journal of Comparative Neurology 385, 95–116 (1997). 10.1002/(SICI)1096-9861(19970818)385:1 [DOI] [PubMed] [Google Scholar]
- 208.Shi Y, Kirwan P, Smith J, Robinson HP & Livesey FJ Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci 15, 477–486, S471 (2012). 10.1038/nn.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kadoshima T et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A 110, 20284–20289 (2013). 10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Arlotta P & Pasca SP Cell diversity in the human cerebral cortex: from the embryo to brain organoids. Curr Opin Neurobiol 56, 194–198 (2019). 10.1016/j.conb.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 212.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). 10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bhaduri A et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020). 10.1038/s41586-020-1962-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hu WF, Chahrour MH & Walsh CA The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet 15, 195–213 (2014). 10.1146/annurev-genom-090413-025600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Eichler EE Genetic Variation, Comparative Genomics, and the Diagnosis of Disease. N Engl J Med 381, 64–74 (2019). 10.1056/NEJMra1809315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mefford HC et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 359, 1685–1699 (2008). 10.1056/NEJMoa0805384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sonderby IE et al. 1q21.1 distal copy number variants are associated with cerebral and cognitive alterations in humans. Transl Psychiatry 11, 182 (2021). 10.1038/s41398-021-01213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Funato K, Smith RC, Saito Y & Tabar V Dissecting the impact of regional identity and the oncogenic role of human-specific NOTCH2NL in an hESC model of H3.3G34R-mutant glioma. Cell Stem Cell 28, 894–905 e897 (2021). 10.1016/j.stem.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sone J et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet 51, 1215–1221 (2019). 10.1038/s41588-019-0459-y [DOI] [PubMed] [Google Scholar]
- 220.Tian Y et al. Expansion of Human-Specific GGC Repeat in Neuronal Intranuclear Inclusion Disease-Related Disorders. Am J Hum Genet 105, 166–176 (2019). 10.1016/j.ajhg.2019.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kang Y et al. A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat Neurosci 24, 1377–1391 (2021). 10.1038/s41593-021-00913-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kwan KY et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911 (2012). 10.1016/j.cell.2012.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Marin O Developmental timing and critical windows for the treatment of psychiatric disorders. Nat Med 22, 1229–1238 (2016). 10.1038/nm.4225 [DOI] [PubMed] [Google Scholar]
- 224.Forrest MP, Parnell E & Penzes P Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci 19, 215–234 (2018). 10.1038/nrn.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Paulsen B et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268–273 (2022). 10.1038/s41586-021-04358-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; Mutations of autism spectrum disease risk genes lead to aberrant developmental timing in cortical organoids.
- 226.Schafer ST et al. Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat Neurosci 22, 243–255 (2019). 10.1038/s41593-018-0295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE & Woolfrey KM Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 14, 285–293 (2011). 10.1038/nn.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Espuny-Camacho I et al. Hallmarks of Alzheimer’s Disease in Stem-Cell-Derived Human Neurons Transplanted into Mouse Brain. Neuron 93, 1066–1081 e1068 (2017). 10.1016/j.neuron.2017.02.001 [DOI] [PubMed] [Google Scholar]


