Abstract
Purpose
To elucidate a potential association between the apolipoprotein E (APOE) E4 allele and glaucoma prevalence in large cohorts.
Design
A cross-sectional analysis of baseline and prospectively collected cohort data.
Participants
UK Biobank (UKBB) participants of genetically determined European ancestry (n = 438 711). Replication analyses were performed using clinical and genotyping data collected from European participants recruited to the Canadian Longitudinal Study of Aging (CLSA; n = 18 199), the Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG; n = 1970), and the Blue Mountains Eye Study (BMES; n = 2440).
Methods
Apolipoprotein E alleles and genotypes were determined, and their distributions were compared on the basis of glaucoma status. Similar analyses were performed using positive control outcomes associated with the APOE E4 allele (death, dementia, age-related macular degeneration) and negative control outcomes not associated with the APOE E4 allele (cataract, diabetic eye disease). Outcome phenotypes were also correlated with Alzheimer’s dementia (AD), a clinical outcome highly associated with the APOE E4 allele.
Main Outcome Measures
Results of APOE E4 genotype-phenotype comparisons were reported as odds ratios (ORs) with 95% confidence intervals (CIs). Replication analyses investigated APOE E4 associations in 2 replication cohorts (CLSA and ANZRAG/BMES).
Results
The APOE E4 allele was inversely associated with glaucoma (OR, 0.96; 95% CI, 0.93–0.99; P = 0.016) and both negative controls (cataract: OR, 0.98; 95% CI, 0.96–0.99; P = 0.015; diabetic eye disease: OR, 0.92; 95% CI, 0.87–0.97; P = 0.003) in the UKBB cohort. A paradoxical positive association was observed between AD and both glaucoma (OR, 1.30; 95% CI, 1.08–1.54; P < 0.01) and cataract (OR, 1.15; 1.04–1.28; P = 0.018). No association between the APOE E4 allele and glaucoma was observed in either replication cohort (CLSA: OR, 1.03; 95% CI, 0.89–1.19; P = 0.66; ANZRAG/BMES: OR, 0.97; 95% CI, 0.84–1.12; P = 0.65).
Conclusions
A small negative association observed between APOE E4 and glaucoma within the UKBB was not evident in either replication cohort and may represent an artifact of glaucoma underdiagnosis in APOE E4 carriers.
Financial Disclosure(s)
The author(s) have no proprietary or commercial interest in any materials discussed in this article.
Keywords: Alzheimer’s dementia, APOE, Apolipoprotein E, Glaucoma, POAG
Glaucoma describes a heterogeneous group of optic neuropathies characterized by specific patterns of neuroretinal atrophy and irreversible vision loss.1 Although there are many causes for glaucoma, primary open-angle glaucoma (POAG) represents the most common glaucoma subphenotype worldwide2 and is defined by glaucomatous change in the presence of an anatomically normal anterior chamber.1 Although intraocular pressure (IOP) remains the single modifiable risk factor for POAG, and all current treatments target IOP lowering, the pathophysiology of POAG remains poorly understood.3 It has been hypothesized that systemic neurodegenerative processes may contribute to POAG.4 Consequently, neurodegeneration in glaucoma is an area of considerable research interest. Several genes including optineurin and TANK-binding kinase 1 have been implicated in Mendelian forms of glaucoma, amyotrophic lateral sclerosis, and frontotemporal dementia.5, 6, 7 Associations have also been demonstrated between glaucoma and both Alzheimer’s dementia (AD)8, 9, 10, 11, 12, 13, 14 and its common risk allele, apolipoprotein E (APOE) E4.15, 16, 17, 18, 19
Apolipoprotein E is a major lipid transport protein within the central nervous system where it is involved in cholesterol transport and neuronal repair.20 Three common APOE alleles (E2, E3, and E4), which are defined by permutations of 2 collocated single-nucleotide polymorphisms (SNPs) in coding regions of the APOE gene, have been identified as important covariates of human disease.20 Despite variation across ethnicities, the E3 allele is the most common allele in Europeans (variant allele frequency: 78%), followed by E4 (14%) and E2 (8%).21 The E4 allele is a major risk factor for AD and other common dementia phenotypes including vascular dementia and Lewy body disease.22, 23, 24 It is also associated with hyperlipidemia,25 ischemic heart disease,25 and mortality,26,27 which was quantified in 1 study as a decrease in population frequency of the E4 allele frequency from 17.6% to 8.3% from age 60 to 90 years.26 By contrast, the E2 allele is associated with other outcomes including peripheral vascular disease,25 age-related macular degeneration (AMD),28,29 and increased survival27 and may be protective against AD.22,30
The APOE E4 allele has been investigated in multiple studies of glaucoma, with individual studies reporting positive,15,16 inverse,17, 18, 19 or absent associations.31, 32, 33, 34, 35 Similarly, several meta-analyses of data from these studies have generated variable results.36, 37, 38 Several factors which may account for variable results across these studies include important correlates of glaucoma prevalence (age, sex, and ethnicity) and APOE allele distribution (age, ethnicity, and the alternate minor [E2] allele). Furthermore, dementia and AD commonly result in executive dysfunction, with impaired attention and self-neglect, which are recognized factors contributing to underdiagnosis of common diseases of aging.39 Consequently, sample stratification based on a powerful dementia risk allele may introduce mislabeling bias with different disease diagnosis rates between case and control cohorts. The current study sought to generate a model to understand and further investigate the association between the APOE E4 allele and glaucoma in large cohort studies.
Methods
Ethical Approval
UK Biobank and the Canadian Longitudinal Study of Aging
The UK Biobank (UKBB) received ethical approval through the North West Multi-centre Ethics Committee, and the Canadian Longitudinal Study of Aging (CLSA) received ethical approval from 13 local research ethics committees at various sites throughout Canada. All participants provided informed written consent, and study procedures were performed in accordance with the ethical principles of the World Medical Association Declaration of Helsinki.
Australian and New Zealand Registry of Advanced Glaucoma
Ethics approval for the Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG) was obtained through the Southern Adelaide Clinical Human Research Ethics Committee, and all participants were enrolled by informed written consent. This study adhered to the tenets of the Declaration of Helsinki and followed the National Health and Medical Research Council statement of ethical conduct in research involving humans.
Blue Mountains Eye Study
The Blue Mountains Eye Study (BMES) was approved by the Western Sydney Area Health Service Human Ethics Committee and complied with the Declaration of Helsinki. All participants were provided with a full explanation of the nature of the study and read and signed informed consent before participation.
Genotyping
DNA microarray genotyping was generated for the different cohorts using Axiom arrays (including the UK Biobank Lung Exome Variant Evaluation (BiLEVE) and UK Biobank arrays; ThermoFisher) for the UKBB, and a combination of Illumina SNP arrays (HumanCoreExome, Infinium OmniExpress, and omni1M; Illumina) for the ANZRAG. Human610-Quad arrays (Illumina) were used for the BMES, and UK Biobank Axiom arrays (ThermoFisher) were used for the CLSA. To account for ethnicity, which represents an independent covariate of both glaucoma prevalence and APOE allele distribution, analyses within this study were performed solely using individuals of European ancestry. Within the UKBB, European ethnicity was defined using a combination of self-reported ancestry and principal components analysis of genetic data following methods described by Bycroft et al40 using the previously generated UKBB data.41 Principal components analyses were similarly used to determine genetic European ancestry within the ANZRAG, BMES, and CLSA cohorts. Apolipoprotein E alleles (E1, E2, E3, and E4) were determined from 2 relevant SNPs within the APOE gene (rs429358 and rs7412; GRCh38 reference genome). Because of the rarity of the E1 allele, rare E1 genotypes (E1E2 and E1E4) were excluded from analysis. In accordance with common practice, E1E3 genotypes, which are most likely to represent incorrect attribution of 2 variants to the same allele rather than alternate common variants on separate alleles, were relabeled as E2E4.25 Apolipoprotein SNPs were measured directly from Axiom arrays. Because the relevant APOE SNPs were not included on the Illumina arrays, they were imputed in Minimac3 using Haplotype Reference Consortium r1.1 as a reference panel (rs429358 imputation R2 = 0.93; rs7412 imputation R2 = 0.92).42
Samples
Analyses were performed in a primary cohort (UKBB) and 2 replication cohorts (CLSA and ANZRAG/BMES). These cohorts included all participants with genetically determined European ancestry for whom APOE alleles and genotypes could be determined or imputed. The primary and first replication cohorts investigated the association between the E4 allele and glaucoma in population cohorts (UKBB and CLSA). The second replication cohort investigated the association between the E4 allele and POAG using cases from a clinical glaucoma cohort (ANZRAG) and nonglaucoma controls from a population cohort with ocular phenotyping data (BMES).
Primary Study Cohort: UKBB
The UKBB is a longitudinal population-based clinical and genetic study of determinants of disease in aging, which commenced in 2006 and continues to monitor approximately 470 000 living participants recruited at age 40–69 y.43 After a period of 12–16 years of follow-up, the mean age of living participants has increased from 56.5 to 69.7 years. Because glaucoma prevalence increases with age, we choose to perform our analyses using current data acquired from the UKBB database at a single time point (February 2, 2022). Participant age (in whole years) was calculated as "age at the date of data acquisition." To avoid potential confounding resulting from E4-associated mortality, participants who had died during follow-up were included in analyses, calculating age as "age at death," representing each deceased participant’s last observation carried forward. Glaucoma phenotypes were determined using a combination of self-reporting determined through completion of a touch screen self-reporting survey (datafield: 6148), and International Classification of Diseases, Tenth Revision (ICD10) coding data (datafield: 41270; code H40). These data included longitudinal data current to the time of data acquisition.
Replication Cohort 1: CLSA
The CLSA is longitudinal population-based study of healthy aging, comprising 51 338 Canadians aged 45–85 years who were recruited from 2010 to 2015 and for whom active follow-up was scheduled to be performed at 3-year intervals for 30 years.44 In this study, we used data from 18 199 individuals with genetically European ancestry included in the August 2019 genotyping release.45 Age was determined in whole years at the time of recruitment. Glaucoma status was defined using baseline self-reported data reported by the parameter ‘ICQ_GLAUC_COM.’ Individuals who did not provide baseline glaucoma data (n = 89) were excluded from analysis.
Replication Cohort 2: ANZRAG/BMES
Unlike the other cohorts, which were selected to investigate the association between the E4 allele and glaucoma, the second replication cohort sought specifically to investigate the association between E4 and POAG. This cohort was generated using known POAG cases sampled from ANZRAG, a large Australasian glaucoma registry, and known nonglaucoma controls recruited from BMES, an ancestrally matched population cohort. Primary open-angle glaucoma cases from ANZRAG were diagnosed by an ophthalmologist based on general disc appearance and corresponding visual field defects identified on a Humphrey 24-2 field, or in the absence of field testing, the loss of central acuity related to glaucoma. Participants with angle closure or secondary causes of glaucoma were excluded.46,47
The BMES is a large population-based cohort that sampled 82.4% of noninstitutionalized individuals aged ≥ 49 years from within a region defined by 2 postal codes in regional New South Wales (Australia).48 The purpose for conducting the BMES was to estimate the prevalence of eye disease within a population representing the general ethnic diversity of the Australian population at the time. Recruitment for the BMES occurred in 2 phases including primary (BMES-I) and extension (BMES-E) cohorts.49 Because all participants underwent eye examination at the time of recruitment, BMES data could be filtered to determine a set of controls known not to have glaucoma. Insufficient data was available to determine which glaucoma cases had POAG, and therefore these individuals were excluded from analyses (n = 135). However, these glaucoma cases were used to determine the overall prevalence of glaucoma within the BMES. The current study included participants from both BMES phases for whom genotyping data were available. For both cohorts, age was defined as "age at recruitment". A subgroup analysis of normal-tension glaucoma (NTG) was performed in the second replication cohort through comparison of ANZRAG NTG participants to BMES no glaucoma controls. For the purpose of this analysis, NTG was defined as POAG with a highest pretreatment IOP of < 21 mmHg in either eye. This definition excluded all participants for whom pretreatment IOP was not explicitly recorded.
Statistical Analysis
All statistical analyses were performed using R (version 4.1.3, RCore Team) using publicly available packages. Baseline univariable comparisons of demographic parameters were performed using independent sample t tests and chi-square tests. Genotype comparisons were performed using generalized (binomial) linear models.
The primary analysis compared the APOE E4 allele to the outcome of glaucoma. For allelic regression, an indicator variable for E4 and E2 allele status (defined by the number [dosage] of relevant alleles [0–2]) was included (equation 1). Thus, the βe4 would represent the "per allele" effect on glaucoma prevalence. To determine whether any observed single allele effects were compounded in E4 homozygotes, this analysis was replicated comparing E4E4 to all other genotypes. For genotypic regression, an indicator variable for each variant genotype (i.e., non-E3E3) was included. Thus, the βe4e4 would represent the difference in glaucoma prevalence between E4E4 and E3E3 homozygotes after accounting for all other APOE genotypes (equation 2). Sex and age (squared) were included as covariates in all analyses. Outcome statistics for univariable analyses are presented as mean (standard deviation) and for multivariable analyses as odds ratios (OR) and 95% confidence intervals (CIs). The threshold for statistical significance (α) was set to P < 0.05.
(equation 1)
(equation 2)
Model Generation
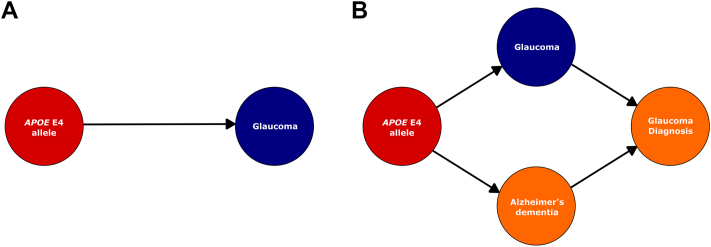
Associations between the APOE E4 allele and glaucoma determined from direct genotype-phenotype correlation include inherent assumptions that both exposure (genotype) and outcome (glaucoma) can be directly assayed. Within such models, the association between these 2 variables is therefore presumed not confounded by genotype-specific factors (Fig 1A). Because these models do not recognize that "diagnosed glaucoma" may not represent "true glaucoma" prevalence or that rates of "glaucoma diagnosis" may be confounded by AD, several additional assumptions were included to generate the current study’s interim model (Fig 1B):
Assumption (1) Glaucoma prevalence is inferred in population-based studies such as the UKBB using documented "glaucoma diagnosis," which may be underestimated because of underdiagnosis or underreporting of disease.
Assumption (2) Executive dysfunction resulting from AD may directly result in underdiagnosis of glaucoma. Because the APOE E4 allele is causally associated with AD, a cohort enriched for this allele is also likely to be enriched for AD and its potential effects on glaucoma underdiagnosis.
Figure 1.
Preliminary causal frameworks. Causality diagrams were generated to demonstrate the basic and intermediate models to investigate associations between the apolipoprotein E (APOE) E4 allele and glaucoma. Within the simple model (A), the association between the APOE E4 allele and "true" glaucoma is assumed to be directly measurable and not confounded by external factors. The interim model proposed in the current study recognizes that rates of "glaucoma diagnosis" may be confounded by underdiagnosis resulting from executive dysfunction in Alzheimer’s dementia (B). Arrows demonstrate; direction of causal effect; red = exposure; blue = outcome; orange = confounding factor.
Because of this study’s inability to quantify the effects of diagnostic biases arising from our hypothesized interaction between AD and "glaucoma diagnosis," a reference framework was created by performing parallel analyses of positive and negative controls. Positive controls were defined as outcomes known to be associated with the E4 allele, including AD and mortality, which are positively associated with E4.22,25, 26, 27 Alzheimer's dementia was defined by a registered ICD10 diagnosis (ICD10 code: F00 [Dementia in AD]), and mortality was determined from death registry data (UKBB datafield: 40000 [Date of Death]). A set of ocular controls were selected on the basis of their nature as common diseases of aging and their similar reporting methods to glaucoma within the UKBB (i.e., combined self-reported touch screen questionnaire [datafield: 6148] and ICD10 encoded diagnosis [datafield: 41270]). These ocular controls included AMD (ICD10 code 35.3), which was defined as a positive control on the basis of its inverse association with E4.28,29 Cataract (ICD10: H25-H28) and diabetic eye disease (ICD10: 36.0 [diabetic retinopathy]) were selected as negative controls, which in this study were defined as outcomes without an established association with the E4 allele. Anticipating that a pathophysiological association between glaucoma and E4 would result in a corresponding association with AD, further analyses were performed comparing glaucoma prevalence to AD prevalence. Because diabetes is an established risk factor for AD,50 diabetic eye disease was identified as a potentially confounded outcome for this analysis. A subanalysis investigating the temporal relationship between "AD diagnosis" and diagnosis of ocular outcomes included dates of first ICD10 code reporting (UKBB datafields: AD [130836]; glaucoma [131186]; AMD [131182]; cataract [131164, 131166, 131168, and 131170], and diabetic eye disease [131184]). Because all secondary analyses and control outcomes were analyzed to provide background reference data to support the primary outcome of E4 allele associations with glaucoma findings, multiple testing correction models were not considered appropriate.
Results
Sample Cohorts
Primary Study Cohort
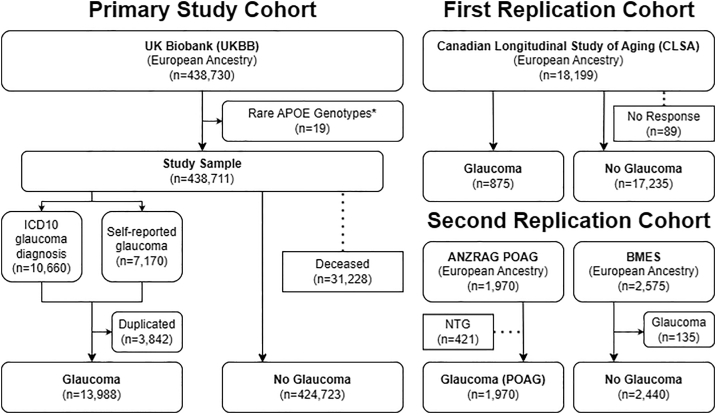
Apolipoprotein E alleles and genotypes were determined for 438 730 UKBB participants with genetically determined European ethnicity (Fig 2). Because of the extreme rarity of the E1 allele, E1E3/E2E4 cases were relabeled as E2E4, and both E1E2 (n = 2) and E1E4 (n = 17) cases, which were most likely to have resulted from imputation error, were excluded from analysis. Within this cohort, 10 660 individuals had an ICD10 diagnosis of glaucoma, and 7170 had declared a self-reported diagnosis of glaucoma. Of these individuals, 3842 had both ICD10 and self-reported diagnoses of glaucoma. The final sample included 13 988 glaucoma cases (3.2% of total cohort) and 424 723 controls (i.e., individuals with no recorded glaucoma diagnosis). Death registry data identified 31 228 individuals (7.1%) from this sample who had died during the follow-up (Fig 2).
Figure 2.
Cohort details. The primary study cohort included all European UK Biobank (UKBB) participants. Within this cohort, glaucoma was defined by a combination of self-reported and International Classification of Diseases, Tenth Revision (ICD10) "glaucoma diagnosis." Deceased participants were included in analyses with age determined from "age at death." The first replication cohort included all European individuals from the Canadian Longitudinal Study of Aging (CLSA). Within this cohort, glaucoma was defined by self-reported diagnosis at the time of recruitment. The second replication cohort pooled primary open-angle glaucoma (POAG) cases from the Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG) and nonglaucoma controls from the Blue Mountains Eye Study (BMES). ∗Participants with rare apolipoprotein E genotypes including E1E2 (n = 2) and E1E4 (n = 17) were excluded from the final sample.
Replication Cohorts
The first replication cohort included 18 199 CLSA participants with European ancestry, 875 of whom had a self-reported diagnosis of glaucoma (4.8% of total cohort). Participants who had not responded to glaucoma self-reporting (n = 89) were excluded from this analysis. The second replication cohort included a case cohort of 1970 European ANZRAG POAG-affected participants, and after exclusion of 135 participants with glaucoma, a control cohort of 2440 European BMES nonglaucoma individuals.
Demographics
UK Biobank glaucoma cases were older than controls (74.2 [6.3] vs. 69.6 [8.0] years; P < 0.001), with a higher proportion of males (50.0% vs. 45.7%; P < 0.001), and higher mortality rates (9.2% vs. 7.0%; P < 0.001; Table 1). Glaucoma cases were older in both replication cohorts (CLSA: 70.0 [9.2] vs. 62.7 [10.1] years; P < 0.001; ANZRAG/BMES: 74.5 [10.6] vs. 62.9 [8.2] years; P < 0.001), but no difference in sex was observed ([% male] CLSA: 49.1% vs. 49.8%; P = 0.71; ANZRAG/BMES: 45.5% vs. 43.7%; P = 0.22). Glaucoma prevalence was 3.2% in the UKBB and 5.0% in the CLSA. Before the exclusion of the 135 glaucoma cases identified within the BMES cohort, glaucoma prevalence was 5.2% in the BMES.
Table 1.
Demographics of Study Cohorts
| Primary Cohort |
First Replication Cohort |
Second Replication Cohort |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| UKBB |
CLSA |
ANZRAG/BMES |
|||||||
| Glaucoma | No Glaucoma | P | Glaucoma | No Glaucoma | P | Glaucoma | No Glaucoma | P | |
| Total sample (n [% glaucoma]) | 13 988 [3.2] | 424 723 | NA | 875 [4.8] | 17 235 | NA | 1971 | 2440 | NA |
| Age (yrs; mean [SD]) | 74.2 [6.3] | 69.6 [8.0] | < 0.001 | 70.2 [9.2] | 62.7 [10.1] | < 0.001 | 74.5 [10.6] | 62.9 [8.2] | < 0.001 |
| Sex (%male) | 50.0 | 45.7 | < 0.001 | 49.1 | 49.8 | 0.71 | 45.5 | 43.7 | 0.22 |
| Deceased (%) | 9.2 | 7.0 | < 0.001 | NA | NA | NA | NA | NA | NA |
Current age, sex, and mortality rates were compared on the basis of glaucoma prevalence in all cohorts. The proportion of individuals diagnosed with glaucoma within the 2 population cohorts (UK Biobank [UKBB] and Canadian Longitudinal Study of Aging [CLSA]) are represented as percentages. Summary statistics were determined using independent t tests for continuous variables (mean [standard deviation]), and Chi-squared tests for categorical variables. ANZRAG = Australian and New Zealand Registry of Advanced Glaucoma; BMES = Blue Mountains Eye Study; NA = not applicable; SD = standard deviation.
Preliminary Analysis and Model Building
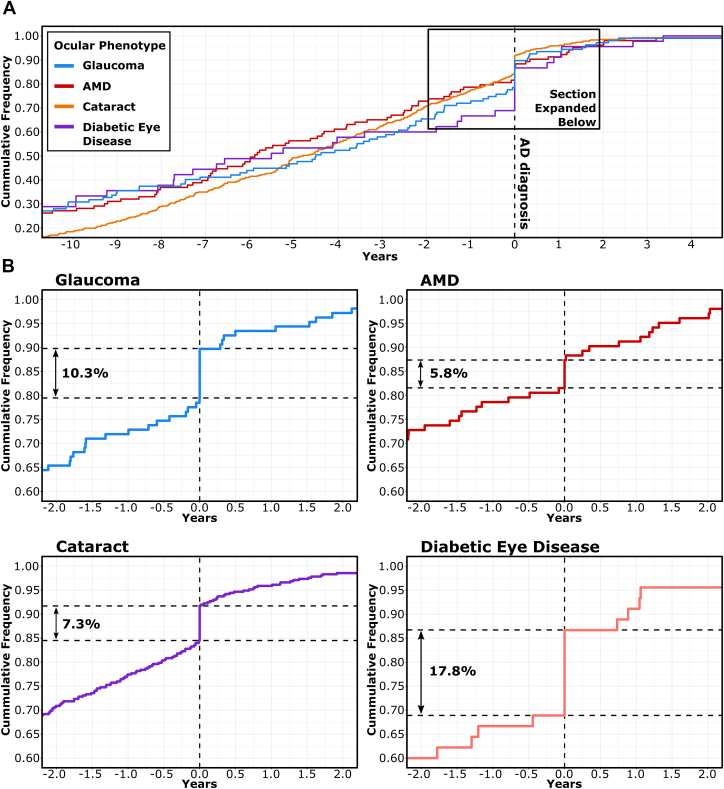
Preliminary analyses within the primary cohort demonstrated strong associations between the APOE E4 allele and all positive control outcomes including AD (OR, 3.56; 95% CI, 3.33–3.81; P < 0.001), death (OR, 1.1; 95% CI, 1.10–1.15; P < 0.001), and AMD (OR, 0.91; 95% CI, 0.87–0.94; P < 0.001). Each of these associations was compounded in E4E4 homozygotes (AD: OR, 6.7; 95% CI, 5.8–7.7; P < 0.001; death: OR, 1.43; 95% CI, 1.34–1.53; P < 0.001; AMD: OR, 0.76; 95% CI, 0.65–0.88; P < 0.001). Inverse associations were observed between E4 and glaucoma (OR, 0.96; 95% CI, 0.93–0.99; P = 0.030), and both negative controls, cataract (OR, 0.98; 95% CI, 0.96–0.99; P = 0.024) and diabetic eye disease (OR, 0.93; 95% CI, 0.88–0.99; P = 0.003). Positive associations were observed between AD and glaucoma (OR, 1.30; 95% CI, 1.08–1.54]; P = 0.004), cataract (OR, 1.15; 95% CI, 1.04–1.28; P = 0.008), and diabetic eye disease (OR, 2.60 ; 95% CI, 2.01–3.29; P < 0.001). Because AD is associated with self-neglect, the associations observed between AD and both glaucoma and cataract were initially hypothesized to be an effect of opportunistic diagnosis. This was further explored by investigating the temporal association between "AD diagnosis" and diagnosis of relevant ocular phenotypes, which demonstrated that ICD10 diagnosis of AD was associated with concurrent ICD10 diagnosis of each phenotype (Fig 3). Given this set of observations, the paradoxical associations between AD and ocular phenotypes were subsequently presumed to have resulted from opportunistic registration of ICD10 codes, which may have resulted from concurrent registration of previously unregistered diagnoses during hospital admissions.
Figure 3.
Cumulative frequency plot of ocular outcome diagnosis in Alzheimer’s dementia (AD). UK Biobank Participants with a recorded date of International Classification of Diseases, Tenth Revision (ICD10) diagnosis of AD and recorded dates of ICD10 diagnosis of glaucoma, age-related macular degeneration (AMD), cataract, and diabetic eye disease were investigated to determine the temporal relationship between diagnoses (A). The time of "AD diagnosis" is represented as 0.0 years (vertical dashed line) on each x-axis. The cumulative prevalence of ocular outcome ICD10 diagnoses are represented as proportions on each y-axis. Horizontal dashed lines represent the upper and lower bounds of ocular outcomes reported concurrently with "AD diagnosis" (B). Percentages demonstrate the total number of incident ocular outcome diagnoses occurring concurrently with "AD diagnosis."
Irrespective of the mechanism by which "AD diagnosis" might affect the diagnosis of glaucoma (or other ocular phenotypes), "AD diagnosis" was observed to be a relevant covariate of "glaucoma diagnosis" in the UKBB cohort. Consequently, a third assumption was included within our study model:
Assumption (3) Despite the theoretical confounding effects of AD resulting in underdiagnosis of glaucoma, "AD diagnosis" may have an opposite effect (observed though increased registration of glaucoma diagnosis concurrent with "AD diagnosis" in the UKBB cohort), thus representing an independent confounder of "glaucoma diagnosis".
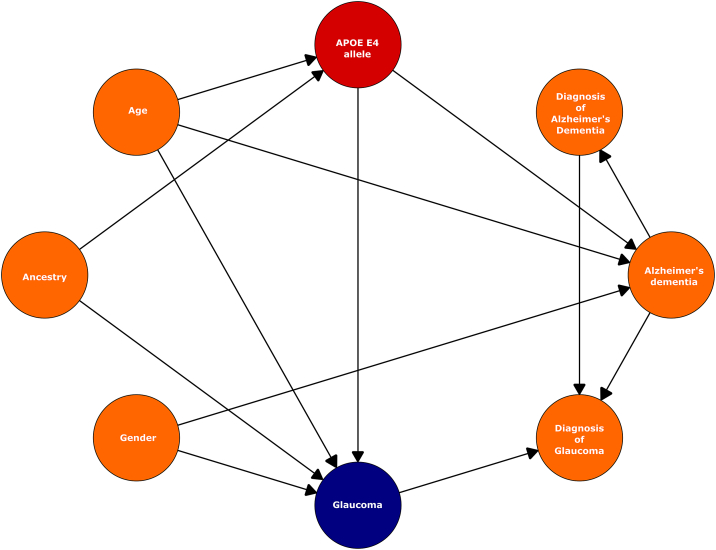
A revised model used in all subsequent analyses (for which AD was an outcome) included "AD diagnosis" as a binary covariate of the outcome variable (Fig 4).
Figure 4.
Causal framework. An amended causality framework was established to include additional potential confounders ("glaucoma diagnosis", Alzheimer's dementia [AD], and "AD diagnosis") and established confounders (age, ancestry/ethnicity, and sex) of the apolipoprotein E (APOE) E4 allele and glaucoma. Arrows indicate direction of causal effect. Red = exposure; blue = outcome; orange = confounders.
Definitive Analysis
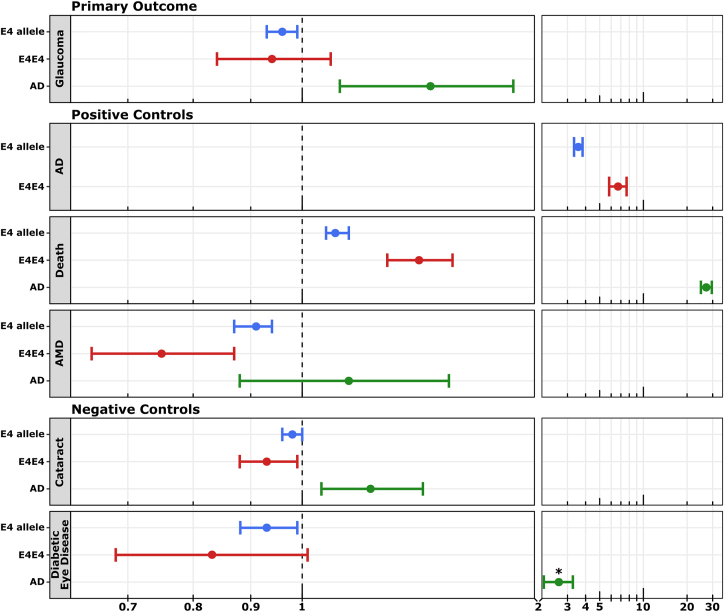
Based on amendments made to the study model following preliminary analysis, including recognition of AD as a covariate of the ocular outcome diagnosis, a second analysis was performed. Within this analysis, the E4 allele was inversely associated with glaucoma in the UKBB (OR, 0.96; 95% CI, 0.93–0.99; P = 0.016; Table 2; Fig 5). Associations were also demonstrated between E4 and all controls including positive controls: AD (OR, 3.56; 95% CI, 3.33–3.81; P < 0.001), death (OR, 1.07; 95% CI, 1.05–1.10; P < 0.001), and AMD (OR, 0.91; 95% CI, 0.87–0.94; P < 0.001), and negative controls: cataract (OR, 0.98; 95% CI, 0.96–0.99; P =0.015) and diabetic eye disease (OR, 0.92; 95% CI: 0.87–0.97; P =0.003). Each of the odds ratios associated with ocular outcomes was more strongly negative with the inclusion of AD as a covariate. No compounding effect of the E4 allele in glaucoma was observed in E4E4 homozygotes (OR, 0.94; 95% CI, 0.84–1.06; P = 0.43; Table 3). Homozygous E4E4 was associated with positive control outcomes including AD (OR, 6.69; 95% CI, 5.81–7.67; P < 0.001), death (OR, 1.27; 95% CI, 1.19–1.36; P < 0.001), AMD (OR, 0.75; 95% CI, 0.65–0.87; P < 0.001), and the negative control, cataract (OR, 0.93; 95% CI, 0.88–0.99; P = 0.03). No homozygous E4E4 effect was observed in diabetic eye disease (OR, 0.83; 95% CI, 0.68–1.01; P = 0.07).
Table 2.
Apolipoprotein E4 Allele Prevalence by Outcome in the UK Biobank
| Variable Type | Phenotype (n [% of total] | E4 Carrier (n = 126 082) | No E4 Allele (n = 312 629) | Odds Ratio [95% CI] | P |
|---|---|---|---|---|---|
| Primary outcome | Glaucoma | 3867 [3.1%] | 10 121 [3.2%] | 0.96 [0.93,0.99] | 0.016 |
| Positive control | Alzheimer’s dementia | 1198 [1.0%] | 1172 [0.2%] | 3.56 [3.33,3.81] | < 0.001 |
| Death | 9625 [7.6%] | 21 603 [6.9%] | 1.07 [1.05,1.10] | < 0.001 | |
| AMD | 2771 [2.2%] | 7704 [2.5%] | 0.91 [0.87,0.94] | < 0.001 | |
| Negative control | Cataract | 15 983 [12.7%] | 40 911 [13.1%] | 0.98 [0.96,0.99] | 0.015 |
| Diabetic eye disease | 1316 [1.0%] | 3518 [1.1%] | 0.92 [0.87,0.97] | 0.003 |
Correlation between the Apolipoprotein E4 allele and glaucoma in the UK Biobank. Alzheimer’s dementia (AD), death, and age-related macular degeneration (AMD) are included as positive controls because of established associations with the E4 allele. Cataract and diabetic eye disease are included as negative controls. Odds ratios and P values have been generated using logistic regression to determine a per allele effect on the outcome variable. Adjustments for the E2 allele, age (squared), sex, and "AD diagnosis" were included in analysis. CI = confidence interval.
Figure 5.
Forest plot. Associations of apolipoprotein E E4 allele (blue), genotype (E4E4; red), and Alzheimer’s dementia (AD; green) with outcome phenotypes generated using UK Biobank data, represented as odds ratios with 95% confidence intervals. Individual phenotype comparisons represented include the primary outcome (glaucoma), positive controls (AD, death, age-related macular degeneration [AMD]), and negative controls (cataract and diabetic eye disease). Odds ratios are represented on a logarithmic scale with compression of the x-axis at higher limits. ∗Because diabetes is a risk factor for AD, diabetic eye disease does not represent a true negative control. Summary statistics were generated through logistic regression with adjustment for the alternate minor E2 allele (allelic regression) or alternate genotypes (genotype regression), age (squared), sex, and "AD diagnosis" (excluded from the analysis in which AD was the outcome).
Table 3.
Phenotype Prevalence According to APOE Genotype
| UK Biobank APOE E Genotypes (n = 438 711) |
Odds Ratio [95% CI] | P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable Type | Phenotype | (n [% Total]) | E4E4 (n = 10 525) | E2E2 (n = 2779) | E2E3 (n = 54 079) | E3E3 (n = 255 771) | E2E4 (n = 11 088) | E3E4 (n = 104 469) | ||
| Primary outcome | Glaucoma | 13 988 [3.2%] | 319 [3.0%] | 70 [2.5%] | 1775 [3.3%] | 8276 [3.2%] | 345 [3.1%] | 3203 [3.1%] | 0.94 [0.84,1.06] | 0.32 |
| Positive control | AD | 1864 [0.4%] | 260 [2.5%] | 11 [0.4%] | 170 [0.3%] | 99 [0.4%] | 58 [0.5%] | 887 [0.8%] | 6.69 [5.81,7.67] | <0.001 |
| Deceased | 31 228 [7.1%] | 1016 [9.7%] | 190 [6.8%] | 3681 [6.8%] | 17 732 [6.9%] | 791 [7.1%] | 7818 [7.5%] | 1.27 [1.19,1.36] | <0.001 | |
| AMD | 10 475 [2.4%] | 191 [1.8%] | 73 [2.7%] | 1353 [2.5%] | 6278 [2.4%] | 273 [2.5%] | 2307 [2.2%] | 0.75 [0.65,0.87] | <0.001 | |
| Negative control | Cataract | 56 894 [13.0%] | 1267 [12.0%] | 330 [11.9%] | 7074 [13.1%] | 33 507 [13.1%] | 1425 [12.9%] | 13 291 [12.7%] | 0.93 [0.88,0.99] | 0.030 |
| Diabetic eye disease | 4834 (1.1%) | 104 [1.0%] | 20 [0.7%] | 595 [1.1%] | 2903 [1.1%] | 113 [1.0%] | 1099 [1.1%] | 0.83 [0.68,1.01] | 0.07 | |
Correlation between the Apolipoprotein E (APOE) E4E4 genotype and glaucoma (defined by the combination of self-reported glaucoma and current International Classification of Diseases, Tenth Revision diagnosis throughout follow-up) in the UK Biobank. Dementia and death are included as positive controls as both are known to be positively associated with the E4 allele. Similarly, age-related macular degeneration (AMD) is included as a positive control given the established protective association of the E4 allele and this phenotype. Diabetic eye disease and cataract are included as negative controls as neither are expected to be pathophysiologically associated with the E4 allele. Odds ratios and P values have been generated comparing phenotype prevalence between the E4 homozygotes (i.e., E4E4) against the reference of E3 homozygotes (i.e., E3E3). Adjustments for the 4 additional alternate genotypes, current age squared, and sex were included in the model. AD = Alzheimer’s dementia; CI = confidence interval.
AD
Because of strong associations between the E4 allele and AD, outcome phenotypes were compared between UKBB participants with or without a diagnosis of AD. Alzheimer’s dementia was positively correlated with glaucoma (OR, 1.30; 95% CI, 1.08–1.54; P < 0.01), death (OR, 27.1; 95% CI, 24.9–29.6; P < 0.001) and both negative controls (cataract: OR, 1.15; 95% CI, 1.04–1.28; P < 0.01; diabetic eye disease: OR, 2.60; 95% CI, 2.01–3.29; P < 0.001; Table 4). Age-related macular degeneration was not associated with AD (OR, 1.10; 95% CI, 0.88–1.35; P = 0.41).
Table 4.
Correlations between Alzheimer’s Dementia and Study Outcomes
| Variable Type | Phenotype Associations (n [% Total]) | AD (n = 1864 [0.4%]) | No AD (n = 436 847 [99.6%]) | Odds Ratio [95% CI] | P |
|---|---|---|---|---|---|
| Outcome variable | Glaucoma | 123 [6.6%] | 13,865 [3.2%] | 1.30 [1.08,1.54] | < 0.01 |
| Positive control | Death | 1,019 [54.7%] | 30,383 [7.0%] | 27.1 [24.9,29.6] | < 0.001 |
| AMD | 82 [4.4%] | 10,393 [2.4%] | 1.10 [0.88,1.35] | 0.41 | |
| Negative control | Cataract | 455 [24.4%] | 56,439 [12.9%] | 1.15 [1.04,1.28] | 0.018 |
| Unknown∗ | Diabetic eye disease | 68 [3.6%] | 4,766 [1.1%] | 2.60 [2.01,3.29] | < 0.001 |
Phenotype-phenotype comparisons were made for study outcomes on the basis of Alzheimer's dementia (AD) diagnosis. Outcome statistics represented as odds ratios and 95% confidence intervals (CIs) were generated through logistic regression including adjustment for age (squared), sex, and "AD diagnosis." AMD = age-related macular degeneration.
Because of established associations between diabetes and AD, diabetic eye disease was reclassified as an "unknown outcome" for this analysis.
Replication Cohorts
Replication analyses demonstrated no association between the E4 allele and glaucoma in the first replication cohort (CLSA: APOE E4 allele: OR, 1.03; 95% CI, 0.89–1.19; P = 0.66; E4E4 homozygotes: OR, 1.25; 95% CI, 0.77–1.94; P = 0.36), and no association between the E4 allele and POAG in the second replication cohort (ANZRAG/BMES: APOE E4 allele: OR, 0.97; 95% CI, 0.84–1.12; P = 0.65; APOE E4 homozygotes: OR, 0.70; 95% CI, 0.41–1.15; P = 0.17). Similarly, no association was seen between E4 and NTG in the second replication cohort (APOE E4 allele: OR, 0.98; 95% CI, 0.83–1.17; P = 0.86; APOE E4 homozygotes: OR, 0.69; 95% CI, 0.36–1.28; P = 0.26).
Discussion
To the best of our knowledge, this study represents the largest and most comprehensive analysis of the APOE E4 allele and glaucoma to date. After modeling and adjusting for multiple established and hypothetical confounders of glaucoma and the E4 allele, we demonstrated a small inverse association between the E4 allele and glaucoma within the UKBB. However, because of the identification of multiple potentially confounding parameters and failure to replicate this result in 2 separate replication cohorts, each of which may not be sufficiently powered to detect a small but potentially real effect, we suspect this association to be the result of inherent study biases rather than neuroprotection. By limiting our analysis to individuals with genetically determined European ancestry, we were able to mitigate potential confounding resulting from ethnicity, an important covariate of glaucoma and E4 allele prevalence.21 Our analysis also accounted for established associations between age and both glaucoma and prevalence of individual APOE alleles (survivorship effect) through adjustment for age (squared) and the alternate minor alleles/genotypes. Finally, we were able to indirectly investigate hidden effects of potential mislabeling resulting from underdiagnosis of glaucoma and dementia in the UKBB cohort.
Underdiagnosis and late diagnosis are important factors contributing to individual and societal glaucoma disease burden.51 Population studies of glaucoma estimate that the true prevalence of glaucoma in Europeans ranges from 6.0% at age 60–69 years to 17.3% at age 80+ years and that approximately 50% of these cases remain undiagnosed.52,53 Glaucoma prevalence in the UKBB (3.2%) was lower than this estimate and similarly lower than was observed in both the CLSA (5.0%) and BMES cohorts (5.2%). This observation may have been a consequence of underreporting of glaucoma through data collection methods. One of the diagnostic parameters we included involved self-reporting, a recognized source of information bias.54 Furthermore, only 44% of UKBB participants completed the eyesight touch screen survey. The second diagnostic parameter which involved medical coding (ICD10) data required individuals to undergo an ophthalmic examination to achieve a diagnosis. Given the recognized E4 allelic associations with AD, executive dysfunction, and underdiagnosis of common diseases of aging, E4 carriers within the UKBB may have had higher rates of glaucoma underdiagnosis. If true, this phenomenon may explain why the same association was not observed in the first replication cohort (CLSA), which used a self-reported vision assessment tool in all participants at baseline, or the second replication cohort, which determined "glaucoma diagnosis" through ophthalmic examination of all individuals.
Epidemiological association studies of APOE may also be confounded by dementia, an underdiagnosed set of clinical syndromes that are not easily accounted for in statistical models.55, 56, 57 Despite population data estimating the prevalence of all-cause dementia to be 6.8% in Western European individuals aged ≥ 60 years,58 ICD10-encoded diagnosis of AD, which is estimated to represent 60% to 80% of all dementia,59 was reported in only 0.4% of UKBB participants. This could be an effect of recognized healthy volunteer bias within the UKBB.60 However, because of the long duration of study follow-up, it is possible that low prevalence of "diagnosed AD" is the consequence of factors such as underdiagnosis and underreporting. Improving dementia diagnosis rates has been recognized as a health care priority for several reasons, one of which being that dementia diagnosis may facilitate other diagnoses of coexisting medical conditions frequently missed because of executive dysfunction and consequent self-neglect.59,61 Although we are unaware of data investigating the effect of AD on diagnosis of common eye diseases, our results, which demonstrated depletion of the E4 allele in cohorts characterized by pathophysiologically unrelated phenotypes (cataract and diabetic eye disease), suggest confounding from dementia-associated executive dysfunction. This assumption was supported by one subanalysis that demonstrated large percentages of ocular outcome ICD10 diagnosis were reported concurrently with AD ICD10 diagnosis. As expected, adjustment for "AD diagnosis" as an independent covariate of "glaucoma diagnosis" increased the strength of association between the E4 allele and each of the ocular outcomes. Unfortunately, this potential confounding could not be fully accounted for within this study because of our inability to measure undiagnosed AD.
Several additional factors contributed to our suspicion that the inverse association observed between the E4 allele and glaucoma was the result of confounding. Primarily, no compounded association was observed in E4E4 homozygotes, an anticipated effect which was observed for all positive control outcomes (AD, death, and AMD). Secondarily, seemingly contradictory positive associations were observed between AD and both glaucoma and cataract. Because of the strength of association between E4 and both AD and mortality, we consider it unlikely that any potential unidentified parameter of AD risk could contribute sufficiently to result in higher AD prevalence in an E4-depleted cohort.
The second replication cohort allowed a different analysis through selective comparison of the E4 allele using carefully phenotyped POAG cases and nonglaucoma controls. The absence of association between the E4 allele and POAG within this cohort contradicted results from a recent study of the combined NEI Glaucoma Human genetics collaBORation (NEIGHBOR)-Massachusetts Eye and Ear Infirmary (MEEI) cohorts, which demonstrated an inverse association between E4 and POAG in 2606 POAG cases and controls (E4 "per allele" association: OR, 0.83; 95% CI, 0.74–0.94; P = 0.0022).19 Likewise, their observation of a stronger association between E4 and NTG (OR, 0.71; 95% CI, 0.58–0.87; P = 0.0014) was not replicated in a similar NTG subanalysis in the current study (OR, 0.98; 95% CI, 0.83–1.17; P = 0.86). Although the rationale for conflicting results between these 2 sets of analyses is unclear, we anticipate the potential for biases arising from different sampling methodologies. Further complicating the current understanding of association between glaucoma and the E4 allele, a recent study performed in the Predicting Risk Of Glaucoma: RElevant SNPs of Strong Association (PROGRESSA) cohort, a prospective study of early glaucoma, demonstrated faster rates of thinning in the macular ganglion cell/inner plexiform layer complex of participants harboring the E4 allele.62 Although faster macular ganglion cell/inner plexiform layer thinning does not necessarily constitute glaucomatous change, this result suggests the E4 allele may contribute to retinal degeneration.
We recognize a number of limitations in the current study, most important of which was our inability to account for mislabeling biases resulting from underdiagnosis of glaucoma and AD. The ideal sample in which to perform this analysis would be a large population-based cohort with a complete set of ocular phenotyping data. Accordingly, our 2 replication cohorts may have been underpowered to detect the E4 effects observed in the considerably larger UKBB cohort. Given the sample size required to adjust for essential covariates of APOE and glaucoma, this phenomenon would more easily be studied using other models with objectively measurable parameters (i.e., OCT) in carefully phenotyped cohorts. Unfortunately, even such studies are likely to be inherently confounded through selection biases associated with identifying cases and controls. The inclusion of deceased participants within our UKBB cohort introduced selection bias. This step, which was undertaken to mitigate survivorship bias resulting from APOE-associated mortality, did not affect observed outcomes as was demonstrated in repeated glaucoma analyses excluding deceased participants. Despite POAG representing the most common glaucoma phenotype in Europeans, glaucoma cohorts identified in these cohorts are likely to have included other glaucoma phenotypes, which may have confounded results. This limitation was partly mitigated through our second replication cohort that solely investigated the association between E4 and POAG. Finally, attempting to account for the ethnic variation in glaucoma prevalence and APOE allele distribution, the current study limited its analysis to individuals with genetically European ancestry, which represented the predominant ethnic group within each study cohort. We were consequently unable to infer associations within other ethnic groups or identify potential ethnic variation, which may be relevant to the association between the E4 allele and glaucoma.
This study demonstrated a small inverse association between the APOE E4 allele and diagnosis of glaucoma in the UKBB, which was not reproducible in 2 replication cohorts. Although the E4 allele may contribute to glaucomatous neurodegenerative consequences, this study highlights the complicated interactions between an allele associated with increased dementia, mortality risk, and underdiagnosed ocular phenotypes. Our result suggests this to be a complex association with multiple confounding parameters, which should be studied using different approaches involving objectively measurable outcomes.
Acknowledgments
This work was conducted using the UK Biobank Resource (application number 25331). The UK Biobank was established by the Wellcome Trust medical charity, Medical Research Council United Kingdom, Department of Health United Kingdom, Scottish Government, and Northwest Regional Development Agency. It also had funding from the Welsh Assembly Government, the British Heart Foundation, and Diabetes United Kingdom. The eye and vision dataset has been developed with additional funding from The NIHR Biomedical Research Centre at Moorfields Eye Hospital and the UCL Institute of Ophthalmology, Fight for Sight charity United Kingdom, Moorfields Eye Charity United Kingdom, The Macula Society United Kingdom, The International Glaucoma Association United Kingdom, and AlconResearch Institute (USA).
The opinions expressed in this manuscript are the authors' own and do not reflect the views of the CLSA or any affiliated institution. This research was made possible using the data/biospecimens collected by the CCLSA. Funding for the CLSA is provided by the Government of Canada through the Canadian Institutes of Health Research (CIHR) under grant reference: LSA 94473 and the Canada Foundation for Innovation, as well as the following provinces, Newfoundland, Nova Scotia, Quebec, Ontario, Manitoba, Alberta, and British Columbia. This research has been conducted using the CLSA dataset (Baseline Comprehensive Dataset version 4.0, Follow-up 1 Comprehensive Dataset version 1.0), under Application Number 190225. The CLSA is led by Drs. Parminder Raina, Christina Wolfson, and Susan Kirkland.
Manuscript no. XOPS-D-22-00259R1.
Footnotes
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s):
The authors have no proprietary or commercial interest in any materials discussed in this article.
This work was funded by the National Health and Medical Research Council (NHMRC program grant APP1150144, and project grant APP1157571). Dr Gharahkhani was supported by an NHMRC Investigator Grant (#1173390), Dr Mullany by an NHMRC Fellowship, Dr Souzeau by a Hospital Research Foundation Early Career Fellowship, Dr Siggs by a Snow Fellowship, and Dr Craig by an NHMRC Practitioner Fellowship. Funding for the Canadian Longitudinal Study on Aging (CLSA) is provided by the Government of Canada through the Canadian Institutes of Health Research (CIHR) under grant reference: LSA 94473 and the Canada Foundation for Innovation, as well as the following provinces, Newfoundland, Nova Scotia, Quebec, Ontario, Manitoba, Alberta, and British Columbia.
HUMAN SUBJECTS: Human subjects were included in this study. The UK Biobank (UKBB) has received ethical approval through the North West Multi-centre Ethics Committee (MREC), and the Canadian Longitudinal Study of Aging (CLSA) has received ethical approval from 13 local research ethics committees at various sites throughout Canada. All participants provided informed written consent, and study procedures were performed in accordance with the ethical principles of the World Medical Association Declaration of Helsinki. Ethics approval for the Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG) was obtained through the Southern Adelaide Clinical Human Research Ethics Committee, and all participants were enrolled by informed written consent. This study adhered to the tenets of the Declaration of Helsinki and followed the National Health and Medical Research Council statement of ethical conduct in research involving humans. The Blue Mountains Eye Study (BMES) has been approved by the Western Sydney Area Health Service Human Ethics Committee and complied with the Declaration of Helsinki. All participants were provided with a full explanation of the nature of the study and read and signed informed consent before participation.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Mullany, Diaz-Torres, Schmidt, Thomson, Qassim, Marshall, Knight, Berry, Kolovos, Dimasi, Lake, Mills, Landers, Mitchell, Healey, Commerford, Klebe, Souzeau, Hassall, MacGregor, Gharahkhani, Siggs, Craig
Data collection: Mullany, Diaz-Torres, Schmidt, Thomson, Qassim, Marshall, Knight, Berry, Kolovos, Dimasi, Lake, Mills, Landers, Mitchell, Healey, Commerford, Klebe, Souzeau, Hassall, MacGregor, Gharahkhani, Siggs, Craig
Analysis and interpretation: Mullany, Diaz-Torres, Schmidt, Thomson, Qassim, Marshall, Knight, Berry, Kolovos, Dimasi, Lake, Mills, Landers, Mitchell, Healey, Commerford, Klebe, Souzeau, Hassall, MacGregor, Gharahkhani, Siggs, Craig
Obtained funding: This study was performed as during the PhD candidatures of the two primary authors at their respective research institutes. No additional funding was provided.
Overall responsibility: Sean Mullany
References
- 1.Casson R.J., Chidlow G., Wood J.P.M., et al. Definition of glaucoma: clinical and experimental concepts. Clin Exp Ophthalmol. 2012;40:341–349. doi: 10.1111/j.1442-9071.2012.02773.x. [DOI] [PubMed] [Google Scholar]
- 2.Tham Y.C., Li X., Wong T.Y., et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Evangelho K., Mogilevskaya M., Losada-Barragan M., Vargas-Sanchez J.K. Pathophysiology of primary open-angle glaucoma from a neuroinflammatory and neurotoxicity perspective: a review of the literature. Int Ophthalmol. 2019;39:259–271. doi: 10.1007/s10792-017-0795-9. [DOI] [PubMed] [Google Scholar]
- 4.Gupta N., Yücel Y.H. Glaucoma as a neurodegenerative disease. Curr Opin Ophthalmol. 2007;18:110–114. doi: 10.1097/ICU.0b013e3280895aea. [DOI] [PubMed] [Google Scholar]
- 5.Rezaie T., Child A., Hitchings R., et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 6.Fingert J.H., Robin A.L., Stone J.L., et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum Mol Genet. 2011;20:2482–2494. doi: 10.1093/hmg/ddr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pottier C., Bieniek K.F., Finch N., et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 2015;130:77–92. doi: 10.1007/s00401-015-1436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra V., Bharucha N.E., Schoenberg B.S. Conditions associated with Alzheimer’s disease at death: case-control study. Neurology. 1986;36:209–211. doi: 10.1212/wnl.36.2.209. [DOI] [PubMed] [Google Scholar]
- 9.Bayer A.U., Keller O.N., Ferrari F., Maag K.P. Association of glaucoma with neurodegenerative diseases with apoptotic cell death: Alzheimer’s disease and Parkinson’s disease. Am J Ophthalmol. 2002;133:135–137. doi: 10.1016/s0002-9394(01)01196-5. [DOI] [PubMed] [Google Scholar]
- 10.Bayer A.U., Ferrari F., Erb C. High occurrence rate of glaucoma among patients with Alzheimer’s disease. Eur Neurol. 2002;47:165–168. doi: 10.1159/000047976. [DOI] [PubMed] [Google Scholar]
- 11.Tamura H., Kawakami H., Kanamoto T., et al. High frequency of open-angle glaucoma in Japanese patients with Alzheimer’s disease. J Neurol Sci. 2006;246:79–83. doi: 10.1016/j.jns.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y.Y., Lai Y.J., Yen Y.F., et al. Association between normal tension glaucoma and the risk of Alzheimer’s disease: a nationwide population-based cohort study in Taiwan. BMJ Open. 2018;8 doi: 10.1136/bmjopen-2018-022987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai S.W., Lin C.L., Liao K.F. Glaucoma may be a non-memory manifestation of Alzheimer’s disease in older people. Int Psychogeriatr. 2017;29:1535–1541. doi: 10.1017/S1041610217000801. [DOI] [PubMed] [Google Scholar]
- 14.Mullany S., Xiao L., Qassim A., et al. Normal-tension glaucoma is associated with cognitive impairment. Br J Ophthalmol. 2022;106:952–956. doi: 10.1136/bjophthalmol-2020-317461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Dabbagh N.M., Al-Dohayan N., Arfin M., Tariq M. Apolipoprotein E polymorphisms and primary glaucoma in Saudis. Mol Vis. 2009;15:912–919. [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers J.C., Craig J.E., Stankovich J., et al. The apolipoprotein epsilon4 gene is associated with elevated risk of normal tension glaucoma. Mol Vis. 2002;8:389–393. [PubMed] [Google Scholar]
- 17.Lam C.Y., Fan B.J., Wang D.Y., et al. Association of apolipoprotein E polymorphisms with normal tension glaucoma in a Chinese population. J Glaucoma. 2006;15:218–222. doi: 10.1097/01.ijg.0000212217.19804.a7. [DOI] [PubMed] [Google Scholar]
- 18.Mabuchi F., Tang S., Ando D., et al. The apolipoprotein E gene polymorphism is associated with open angle glaucoma in the Japanese population. Mol Vis. 2005;11:609–612. [PubMed] [Google Scholar]
- 19.Margeta M.A., Letcher S.M., Igo R.P., Jr., et al. Association of APOE with primary open-angle glaucoma suggests a protective effect for APOE ε4. Invest Ophthalmol Vis Sci. 2020;61:3. doi: 10.1167/iovs.61.8.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koutsodendris N., Nelson M.R., Rao A., Huang Y. Apolipoprotein E and Alzheimer’s disease: findings, hypotheses, and potential mechanisms. Annu Rev Pathol. 2022;17:73–99. doi: 10.1146/annurev-pathmechdis-030421-112756. [DOI] [PubMed] [Google Scholar]
- 21.Corbo R.M., Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE∗4 a “thrifty” allele? Ann Hum Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 22.Farrer L.A., Cupples L.A., Haines J.L., et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 23.Skrobot O.A., McKnight A.J., Passmore P.A., et al. A validation study of vascular cognitive impairment genetics meta-analysis findings in an independent collaborative cohort. J Alzheimers Dis. 2016;5:981–989. doi: 10.3233/JAD-150862. [DOI] [PubMed] [Google Scholar]
- 24.Rongve A., Witoelar A., Ruiz A., et al. GBA and APOE ε4 associate with sporadic dementia with Lewy bodies in European genome wide association study. Sci Rep. 2019;9:7013. doi: 10.1038/s41598-019-43458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lumsden A.L., Mulugeta A., Zhou A., Hyppönen E. Apolipoprotein E (APOE) genotype-associated disease risks: a phenome-wide, registry-based, case-control study utilising the UK Biobank. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKay G.J., Silvestri G., Chakravarthy U., et al. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol. 2011;173:1357–1364. doi: 10.1093/aje/kwr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolters F.J., Yang Q., Biggs M.L., et al. The impact of APOE genotype on survival: results of 38,537 participants from six population-based cohorts (E2-CHARGE) PLOS ONE. 2019;14 doi: 10.1371/journal.pone.0219668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klaver C.C., Kliffen M., van Duijn C.M., et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet. 1998;63:200–206. doi: 10.1086/301901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baird P.N., Guida E., Chu D.T., et al. The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45:1311–1315. doi: 10.1167/iovs.03-1121. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg T.E., Huey E.D., Devanand D.P. Association of APOE e2 genotype with Alzheimer’s and non-Alzheimer’s neurodegenerative pathologies. Nat Commun. 2020;11:4727. doi: 10.1038/s41467-020-18198-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia L.Y., Tam P.O.S., Chiang S.W.Y., et al. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern Chinese. Mol Vis. 2009;15:89–98. [PMC free article] [PubMed] [Google Scholar]
- 32.Saglar E., Yucel D., Bozkurt B., et al. Association of polymorphisms in APOE, p53, and p21 with primary open-angle glaucoma in Turkish patients. Mol Vis. 2009;15:1270–1276. [PMC free article] [PubMed] [Google Scholar]
- 33.Zetterberg M., Tasa G., Palmér M.S., et al. Apolipoprotein E polymorphisms in patients with primary open-angle glaucoma. Am J Ophthalmol. 2007;143:1059–1060. doi: 10.1016/j.ajo.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Lake S., Liverani E., Desai M., et al. Normal tension glaucoma is not associated with the common apolipoprotein E gene polymorphisms. Br J Ophthalmol. 2004;88:491–493. doi: 10.1136/bjo.2003.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ressiniotis T., Griffiths P.G., Birch M., et al. The role of apolipoprotein E gene polymorphisms in primary open-angle glaucoma. Arch Ophthalmol. 2004;122:258–261. doi: 10.1001/archopht.122.2.258. [DOI] [PubMed] [Google Scholar]
- 36.Song Q., Chen P., Liu Q. Role of the APOE ε2/ε3/ε4 polymorphism in the development of primary open-angle glaucoma: evidence from a comprehensive meta-analysis. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Zhou Y.F., Zhao B.Y., Gu Z.Y., Li S.L. Apolipoprotein E gene ε4ε4 is associated with elevated risk of primary open angle glaucoma in Asians: a meta-analysis. BMC Med Genet. 2014;15:60. doi: 10.1186/1471-2350-15-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao R., Ye M., Xu X. An updated meta-analysis: apolipoprotein E genotypes and risk of primary open-angle glaucoma. Mol Vis. 2014;20:1025–1036. [PMC free article] [PubMed] [Google Scholar]
- 39.Löppönen M.K., Isoaho R.E., Räihä I.J., et al. Undiagnosed diseases in patients with dementia – a potential target group for intervention. Dement Geriatr Cogn Disord. 2004;18:321–329. doi: 10.1159/000080126. [DOI] [PubMed] [Google Scholar]
- 40.Bycroft C., Freeman C., Petkova D., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig J.E., Han X., Qassim A., et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. 2020;52:160–166. doi: 10.1038/s41588-019-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S., Forer L., Schönherr S., et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ollier W., Sprosen T., Peakman T. UK Biobank: from concept to reality. Pharmacogenomics. 2005;6:639–646. doi: 10.2217/14622416.6.6.639. [DOI] [PubMed] [Google Scholar]
- 44.Raina P.S., Wolfson C., Kirkland S.A., et al. The Canadian Longitudinal Study on Aging (CLSA) Can J Aging. 2009;28:221–229. doi: 10.1017/S0714980809990055. [DOI] [PubMed] [Google Scholar]
- 45.Raina P., Wolfson C., Kirkland S., et al. Cohort profile: the Canadian Longitudinal Study on Aging (CLSA) Int J Epidemiol. 2019;48:1752–1753j. doi: 10.1093/ije/dyz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Souzeau E., Goldberg I., Healey P.R., et al. Australian and New Zealand registry of advanced glaucoma: methodology and recruitment. Clin Exp Ophthalmol. 2012;40:569–575. doi: 10.1111/j.1442-9071.2011.02742.x. [DOI] [PubMed] [Google Scholar]
- 47.Siggs O.M., Han X., Qassim A., et al. Association of monogenic and polygenic risk with the prevalence of open-angle glaucoma. JAMA Ophthalmol. 2021;139:1023–1028. doi: 10.1001/jamaophthalmol.2021.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J.J., Mitchell P., Smith W. Vision and low self-rated health: the Blue Mountains Eye Study. Invest Ophthalmol Vis Sci. 2000;41:49–54. [PubMed] [Google Scholar]
- 49.Chia E.M., Mitchell P., Ojaimi E., Rochtchina E., Wang J.J. Assessment of vision-related quality of life in an older population subsample: the Blue Mountains Eye Study. Ophthal Epidemiol. 2006;13:371–377. doi: 10.1080/09286580600864794. [DOI] [PubMed] [Google Scholar]
- 50.Profenno L.A., Porsteinsson A.P., Faraone S.V. Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol Psychiatry. 2010;67:505–512. doi: 10.1016/j.biopsych.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Varma R., Ying-Lai M., Francis B.A., et al. Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology. 2004;111:1439–1448. doi: 10.1016/j.ophtha.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 52.Zhang N., Wang J., Li Y., Jiang B. Prevalence of primary open angle glaucoma in the last 20 years: a meta-analysis and systematic review. Sci Rep. 2021;11 doi: 10.1038/s41598-021-92971-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaikh Y., Yu F., Coleman A.L. Burden of undetected and untreated glaucoma in the United States. Am J Ophthalmol. 2014;158:1121–1129.e1. doi: 10.1016/j.ajo.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 54.Althubaiti A. Information bias in health research: definition, pitfalls, and adjustment methods. J Multidiscip Healthc. 2016;9:211–217. doi: 10.2147/JMDH.S104807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olafsdóttir M., Skoog I., Marcusson J. Detection of dementia in primary care: the Linköping study. Dement Geriatr Cogn Disord. 2000;11:223–229. doi: 10.1159/000017241. [DOI] [PubMed] [Google Scholar]
- 56.Valcour V.G., Masaki K.H., Curb J.D., Blanchette P.L. The detection of dementia in the primary care setting. Arch Intern Med. 2000;160:2964–2968. doi: 10.1001/archinte.160.19.2964. [DOI] [PubMed] [Google Scholar]
- 57.Eefsting J.A., Boersma F., Van den Brink W., Van Tilburg W. Differences in prevalence of dementia based on community survey and general practitioner recognition. Psychol Med. 1996;26:1223–1230. doi: 10.1017/s0033291700035947. [DOI] [PubMed] [Google Scholar]
- 58.Prince M.J. 2015. World Alzheimer Report 2015: The Global Impact of Dementia : An Analysis of Prevalence, Incidence, Cost and Trends. [Google Scholar]
- 59.Alzheimer’s Association 2011 Alzheimer’s disease facts and figures. Alzheimers Dement. 2011;7:208–244. doi: 10.1016/j.jalz.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 60.Fry A., Littlejohns T.J., Sudlow C., et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dyer C.B., Goodwin J.S., Pickens-Pace S., et al. Self-neglect among the elderly: a model based on more than 500 patients seen by a geriatric medicine team. Am J Public Health. 2007;97:1671–1676. doi: 10.2105/AJPH.2006.097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullany S., Marshall H., Diaz-Torres S., et al. The APOE E4 allele is associated with faster rates of neuroretinal thinning in a prospective cohort study of suspect and early glaucoma. Ophthalmol Sci. 2022;2 doi: 10.1016/j.xops.2022.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]