Summary
Skin plays central roles in systemic physiology, and it undergoes significant functional changes during aging. Members of the peroxisome proliferator-activated receptor-gamma coactivator (PGC-1) family (PGC-1s) are key regulators of the biology of numerous tissues, yet we know very little about their impact on skin functions. Global gene expression profiling and gene silencing in keratinocytes uncovered that PGC-1s control the expression of metabolic genes as well as that of terminal differentiation programs. Glutamine emerged as a key substrate promoting mitochondrial respiration, keratinocyte proliferation, and the expression of PGC-1s and terminal differentiation programs. Importantly, gene silencing of PGC-1s reduced the thickness of a reconstructed living human epidermal equivalent. Exposure of keratinocytes to a salicylic acid derivative potentiated the expression of PGC-1s and terminal differentiation genes and increased mitochondrial respiration. Overall, our results show that the PGC-1s are essential effectors of epidermal physiology, revealing an axis that could be targeted in skin conditions and aging.
Subject areas: dermatology, physiology, human metabolism, molecular mechanism of gene regulation
Graphical abstract
Highlights
-
•
PGC-1s modulate the expression of keratinocyte terminal differentiation genes
-
•
PGC-1s control the thickness of reconstructed living human epidermal equivalents
-
•
Glutamine is a key substrate for keratinocyte biology
-
•
A salicylic acid derivative induces PGC-1s and differentiation transcripts
Dermatology; Physiology; Human metabolism; Molecular mechanism of gene regulation.
Introduction
The global increase in population aging observed in developed countries has been linked to a rise in age-associated diseases, such as cardiovascular and neurological diseases, type 2 diabetes, and cancer.1,2 It is now appreciated that these diseases share common characteristics, among which altered metabolism draws growing attention in medical research.3,4,5 At least nine hallmarks have hitherto been associated with aging, two of which being directly linked to metabolism, i.e., deregulation of nutrient sensing and mitochondrial dysfunction.6 The skin plays key roles in systemic physiology, and aged skin reveals profound structural and molecular changes such as reduced skin thickness, dermal-epidermal junction flattening, and changes in extracellular matrix composition, which together can alter its integrity and normal functions.7,8,9,10 Recent studies revealed that metabolic functions of both keratinocytes and dermal fibroblasts are impaired with aging, supporting the notion that altered metabolism may be detrimental to skin integrity.11,12,13,14,15 Moreover, aged skin is characterized by a reduction in microcirculation16,17 and a reorganized mitochondrial network.18 Importantly, while transcriptomics and metabolomics studies revealed that aged skin exhibits mitochondrial and metabolic changes,12,19 the mechanisms driving these alterations remain to be elucidated.
One major class of regulators that has been linked to both age-related conditions and metabolism is the family of peroxisome proliferator-activated receptor-gamma coactivators (PGC-1s), notably PGC-1α and PGC-1β. These coactivators control mitochondrial biogenesis and metabolism.20,21,22 While PGC-1β expression is generally constitutive with respect to acute metabolic changes, the expression of PGC-1α is highly responsive and modulated in a context-specific manner.23,24,25,26 Although a few promising compounds can act as potent modulators of PGC-1α activity, achieving specificity as well as tissue-specific targeting faces considerable challenges. In this study, we reveal that PGC-1s play key roles in supporting keratinocyte proliferation and terminal differentiation programs. We also reveal that a salicylic acid derivative can modulate PGC-1s expression, opening unexplored avenues for targeting PGC-1s axes in skin disorders and aging.
Results
PGC-1α expression is increased in aged human epidermis
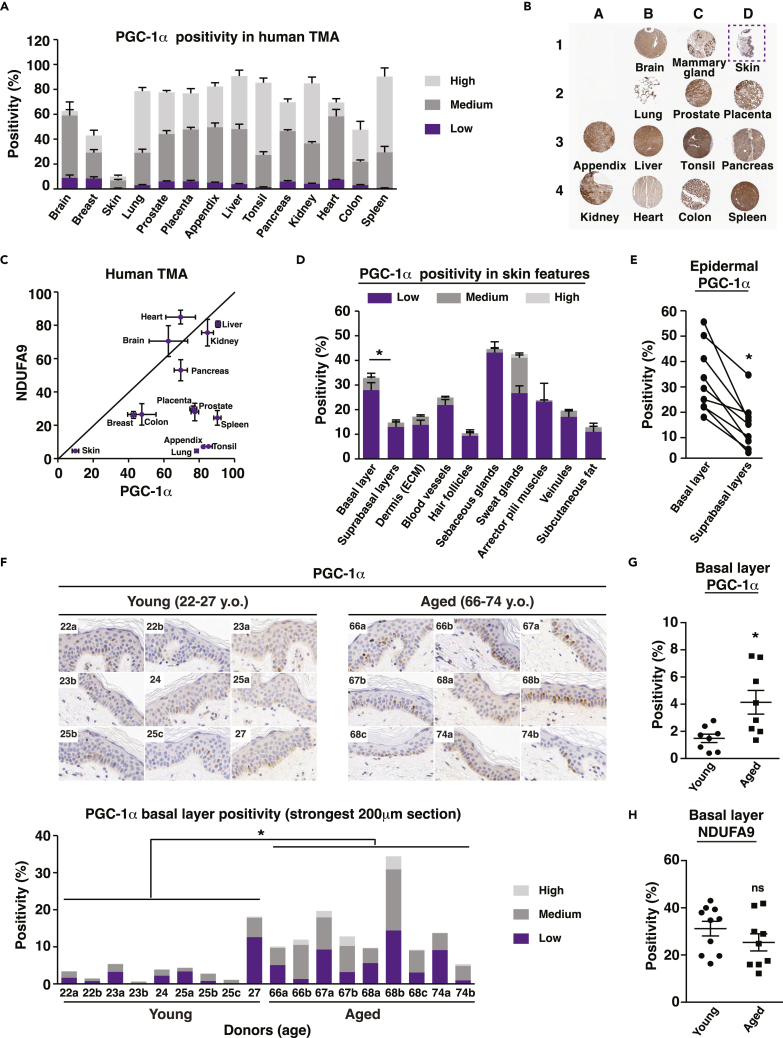
To assess the expression of PGC-1s in human skin tissues and to compare and contrast it to other organs, we first performed immunohistochemistry analyses for PGC-1α on a tissue microarray containing 14 sections from different human organs (Figures 1A, 1B, and S1A) using a validated antibody. Compared to most tissues, PGC-1α expression was very low in human skin (Figures 1A and 1B). Because PGC-1α modulates the biogenesis of mitochondria and these organelles are particularly enriched and solicited under oxidative conditions, one potential explanation for the low PGC-1α expression in skin is its relative hypoxic state compared to other organs.27,28 Given that reduced PGC-1α expression is expected to correlate with reduced mitochondrial functions, we performed staining for NDUFA9, a mitochondrial complex I subunit indispensable for cell growth in galactose medium29 that is sensitive to PGC-1α and β knockdown (KD) in human primary keratinocytes (GEO: GSE108674). In agreement with the staining for PGC-1α, NDUFA9 expression level was at its lowest in skin sections compared to other organs (Figure 1C). To gain a better insight into PGC-1α expression in human skin, we analyzed its distribution in various features and appendages from human skin sections (Figures 1D, 1E, and S1B). Interestingly, PGC-1α expression was particularly enriched in sweat glands, appendages involved in thermal regulation. Within the epidermis, PGC-1α was enriched in the basal layer compared to suprabasal layers (Figures 1D, 1E, and S1B), suggesting that PGC-1α might have important functions within these highly defined boundaries by supporting keratinocyte proliferation. On the other hand, it has been recently shown that a metabolic gradient is present in reconstructed human skin from a predominantly glycolytic metabolism in basal cells to a high oxidative phosphorylation (OXPHOS) activity in differentiated cells.30 Hence, it could also be hypothesized that PGC-1α presence in the basal layer might play a role in the commitment of keratinocytes toward differentiation.
Figure 1.
Histological evaluation of PGC-1α expression in human skin
(A) PGC-1α expression in human tissue microarray (TMA) evaluated by immunohistochemistry (IHC). Data are shown as mean + SEM from 4 sections. Positivity is scored as low, medium, and high.
(B) Representative set of human sections shown in A. B1 is a brain section, C1 is breast, and D1 is skin (in dotted box). The remaining sections follow the same order as in A, from left to right.
(C) Relationship between the mitochondrial marker NDUFA9 and PGC-1α in human TMA. Data are shown as mean ± SEM from 4 sections.
(D) PGC-1α expression in various human skin features and appendages, obtained from 4 to 9 donors. Data are shown as mean + SEM. Epidermal basal layer shows stronger positivity than suprabasal layers, ∗p < 0.05, paired Student’s t test. Positivity score is the same as explained in A.
(E) PGC-1α expression in epidermal basal layer and suprabasal layers. Data from each donor are matched with a solid line. N = 9 donors. ∗p < 0.05, paired Student’s t test.
(F) Upper panel: pictures of 200 μm skin regions with highest PGC-1α positivity for each donor, as acquired by IHC. Lower panel: PGC-1α expression in the basal layers from the 200 μm skin sections shown in the upper panel. ∗p < 0.05 (total positivity, high positivity, and medium positivity), unpaired Student’s t test. Positivity score is the same as in A.
(G) PGC-1α expression in skin basal layer from young (22–27 y.o.) and aged (66–74 y.o.) donors, where each dot represents a single human donor. ∗p < 0.05, unpaired Student’s t test. (H) NDUFA9 expression in skin basal layer from young (22–27 y.o.) and aged (66–74 y.o.) donors, where each dot represents a single human donor. ns: non-significant difference, unpaired Student’s t test.
Given that PGC-1α expression has been linked to aging and age-related conditions in several tissues, notably skeletal muscle,31,32,33 we examined PGC-1α expression in the basal layer of skin inner forearm sections from 9 young (22–27 y.o.) and 9 aged (66–74 y.o.) female human donors (Figure 1F). Elevated PGC-1α expression was mostly associated with aged donors, while low PGC-1α expression was found in young donors (Figure 1F). These data reveal a positive correlation between age and PGC-1α expression in the epidermis. Indeed, total PGC-1α positivity was stronger in aged sections compared with young ones (Figure 1G). In contrast, the expression of NDUFA9 was slightly reduced in aging, although this difference did not reach statistical significance (Figure 1H). A reduction in the expression of NDUFA9 in aged epidermis would support the notion that reduced mitochondrial load and function are associated with aged skin34 and other aged organs.35,36 Overall, these data suggest that the increase in PGC-1α levels in aged skin might represent an attempt to restore impaired metabolic functions associated with skin aging and help attenuate the decrease in expression of mitochondrial targets as well as mitochondrial functions.
Transcriptomics analyses reveal a link between PGC-1s, aging, and keratinocyte terminal differentiation
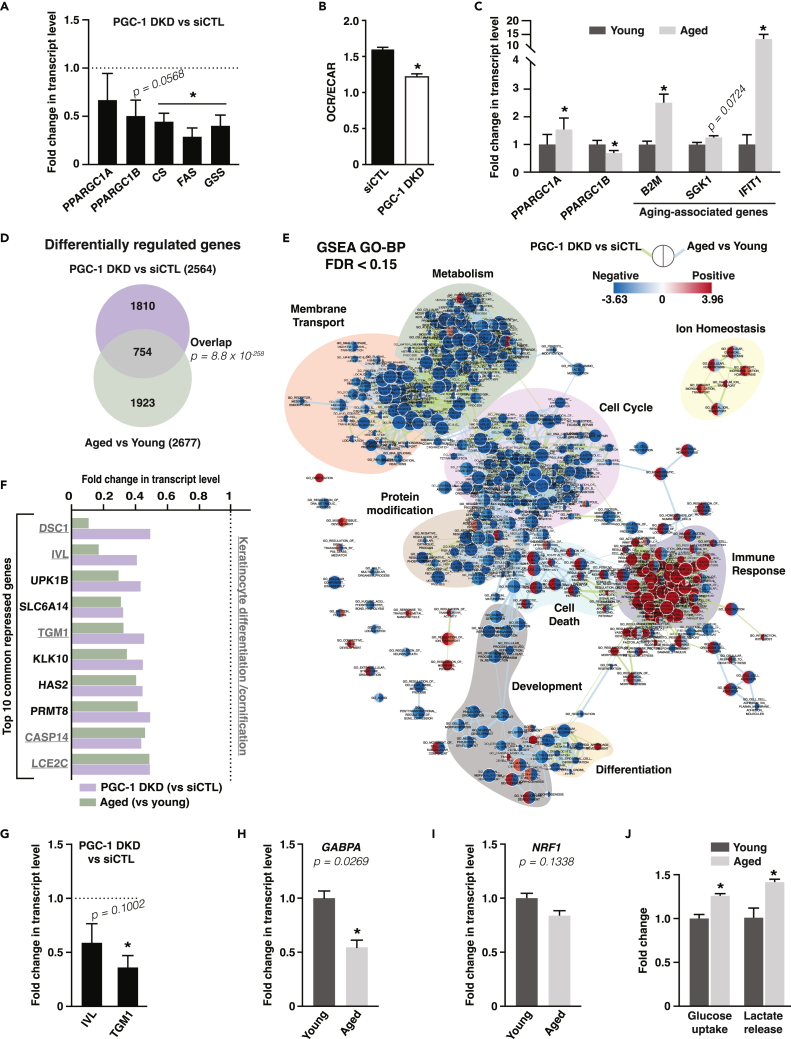
To determine the role of the PGC-1s in keratinocyte aging, we designed a transcriptomics approach to compare changes in gene expression that occur when decreasing PGC-1s expression and those that occur with aging. To modulate the PGC-1s, we used a double-knockdown (DKD) approach to silence both PGC-1α and PGC-1β in normal human epidermal keratinocytes (NHEKs) as these two family members can functionally overlap and show compensation.37,38 This strategy resulted in ∼50% KD in PPARGC1 transcripts, along with a reduction in the expression of metabolic enzymes that are known targets of PGC-1s (Figure 2A). However, this DKD approach did not lead to a statistically significant reduction in PPARGC1A expression in NHEK at baseline. PGC-1 DKD cells displayed reduced ratio of mitochondrial respiration (oxygen consumption rate, OCR) to glycolysis (extracellular acidification rate, ECAR; Figure 2B), while the percentage of mitochondrial respiration coupled to ATP synthesis remained unchanged (Figure S2A). To compare global changes in gene expression upon PGC-1 DKD with those linked to keratinocyte aging, we selected a pair of NHEK originating from the back skin of two donors of the same sex and ethnicity and with a difference in age of 55 years. The NHEK from the aged donor expressed higher levels of B2M and SGK1, genes commonly upregulated in aging,39 compared to cells from the young donor (Figure 2C). In addition, IFIT1, an interferon-inducible gene that is induced in aged brains,40 was strongly expressed in cells from the aged donor (Figure 2C). In agreement with the results in human skin samples (Figures 1F and 1G), PPARGC1A expression was increased in cells from the aged donor compared with those from the young donor (Figure 2C). This indicates that the change in PGC-1α expression during skin aging is maintained in isolated keratinocytes in vitro and depends on cell autonomous processes. Intriguingly, PPARGC1B showed an opposite trend to PPARGC1A during aging as its expression was decreased in aged cells (Figure 2C), suggesting that elevated PPARGC1A might represent a compensatory mechanism to alleviate a reduction in PPARGC1B.
Figure 2.
PGC-1α and PGC-1β double silencing in human primary keratinocytes mimics aspects of keratinocyte aging and represses cornification and terminal differentiation gene programs
(A) Expression of PGC-1s and targets in normal human epidermal keratinocytes (NHEK) 3 days after PGC-1α/β double knockdown (PGC-1 DKD). Data are shown as mean +SEM from 4 independent experiments. ∗p < 0.05, paired Student’s t test.
(B) OCR/ECAR ratio of keratinocyte HaCaT cells transfected with control siRNA or PGC-1α/β double knockdown (PGC-1 DKD). Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(C) Gene expression analysis in NHEK from young (28 y.o.) and aged (83 y.o.) donors. Data are shown as mean + SEM from 6 independent experiments. ∗p < 0.05, paired Student’s t test.
(D) Venn diagram from microarray data analyses grouping differentially regulated genes upon PGC-1 DKD versus siCTL in NHEK and upon aging in NHEK from aged (83 y.o.) versus young (28 y.o.) donors. 754 genes are common to PGC-1s DKD and keratinocyte aging and are analyzed in panels E-F.
(E) Network analysis of enriched and depleted pathways upon PGC-1 DKD versus siCTL in NHEK and upon aging in NHEK from aged (83 y.o.) versus young (28 y.o.) donors. Enrichment (red) or depletion (blue) of each pathway is shown in nodes, where the left side represents PGC-1 DKD versus siCTL and the right represents aged versus young. Edges link pathways (nodes) with common genes, and edge length is inversely proportional to the similarity between nodes. Color-shaded pathway groups were outlined manually.
(F) Top 10 common genes repressed upon PGC-1 DKD and keratinocyte aging. Underlined transcripts are known to be involved in the differentiation or cornification processes. Fold change is from transcriptome array analysis. Dotted line corresponds to control keratinocytes from which fold changes are calculated.
(G) Validation of the repression of terminal differentiation markers genes (IVL: involucrin; TGM1: transglutaminase 1) upon PGC-1 DKD in NHEK by real-time PCR. Data are shown as mean + SEM from 4 independent experiments. ∗p < 0.05, paired Student’s t test.
(H) Expression of GABPA in NHEK from aged (83 y.o.) versus young (28 y.o.) donors, normalized to the expression of NHEK cells from young donor. Data are shown as mean + SEM from 5 independent experiments. ∗p < 0.05, paired Student’s t test.
(I) Expression of NRF1 in NHEK from aged (83 y.o.) versus young (28 y.o.) donors, normalized to the expression of NHEK cells from young donor. Data are shown as mean + SEM from 6 independent experiments. Paired Student’s t test.
(J) Glucose uptake and lactate release in NHEK from aged (83 y.o.) versus young (28 y.o.) donors, normalized to the expression of NHEK cells from young donor. Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
Transcriptomics analyses revealed that a third of the genes differentially regulated by PGC-1 DKD were shared with those regulated during aging (Figure 2D). Functional classification of these shared genes revealed that the expression of genes implicated in metabolism, membrane transport, cell cycle, protein modification, development, and differentiation was downregulated upon PGC-1 DKD and aging, while the expression of genes involved in immune response was upregulated (Figures 2E and S2B–S2D). Manual curation of the expression of downregulated genes common to both PGC-1 DKD and aging revealed that 5 out of the 10 most downregulated genes are implicated in keratinocyte terminal differentiation, including IVL and TGM1 (Figure 2F). We validated that these two genes were downregulated in NHEK in response to PGC-1 DKD (Figure 2G). Furthermore, we mined our transcriptomics datasets for over-representation of transcription factor binding sites in the promoter of genes that were downregulated upon PGC-1 DKD and aging and saw enrichment for binding sites of transcription factors involved in differentiation and developmental processes (Figures S2E). Together, these results show that reduction in PGC-1s expression leads to specific changes in gene expression that are shared with keratinocyte aging. Notably, the terminal differentiation process is significantly downregulated by depletion of PGC-1s and aging.
Given the central role of PGC-1s in mitochondrial metabolism, we examined the expression of mitochondrial genes upon PGC-1s DKD and aging. Close to 8% of shared downregulated genes upon PGC-1s DKD and aging are known to encode for mitochondrial proteins (Figure S2B–S2D; MitoCarta2.041,42). It is noteworthy that the net impact of decreased PGC-1β and increased PGC-1α expression in aged keratinocytes is reduced expression of mitochondrial genes. These data suggest that, although PGC-1α may be compensating for some functions of PGC-1β, it is unable to fully rescue mitochondrial gene expression. One potential explanation for this observation is that the expression of key transcription factors such as GA binding protein transcription factor subunit alpha (GABPA) and nuclear respiratory factor 1 (NRF1), which are required for PGC-1α to regulate mitochondrial genes, is also reduced in aged cells (Figures 2H and 2I). Furthermore, mining for over-represented transcription factor binding sites in the promoters of genes that are downregulated in aging revealed enrichment for GABPA and NRF1 binding sites (Figure S2E). The decreased expression of mitochondrial genes with aging was paralleled with increased glycolytic activity, as indicated by elevated glucose uptake and lactate release, of aged keratinocytes compared with young cells (Figure 2J). Taking into consideration the limited availability of pairs of NHEK from donors of the same sex and ethnicity with a large difference in age, coupled with their limited proliferation potential in culture, we sought to validate the aged-dependent changes in expression of PGC-1s in a publicly available dataset of skin samples from young and aged donors.43 In agreement with our findings in NHEK, the expression of PPARGC1A was increased, while that of PPARGC1B was decreased, in skin samples from donors aged 70–79 years old compared with donors aged 20–29 years old (Figure S3). Despite the reported decrease in mitochondrial RNA expression,43 an indicator of mitochondrial biogenesis, in the samples from aged donors compared to young donors, the expression of the transcription factors NRF1 and GABPA was respectively unchanged or upregulated in skin samples from aged donors, and the expression of the mitochondrial target NDUFA9 was upregulated in skin samples from aged donors (Figure S3). Together, the data with NHEK and skin samples support elevated expression of PPARGC1A and decreased expression of PPARGC1B along with decreased global mitochondrial gene expression/metabolism in aging. However, these data highlight that the mitochondrial marker NDUFA9 may not represent global changes in mitochondrial gene expression, and different signaling axes from NRF1 and GABPA could set mitochondrial gene expression signatures.
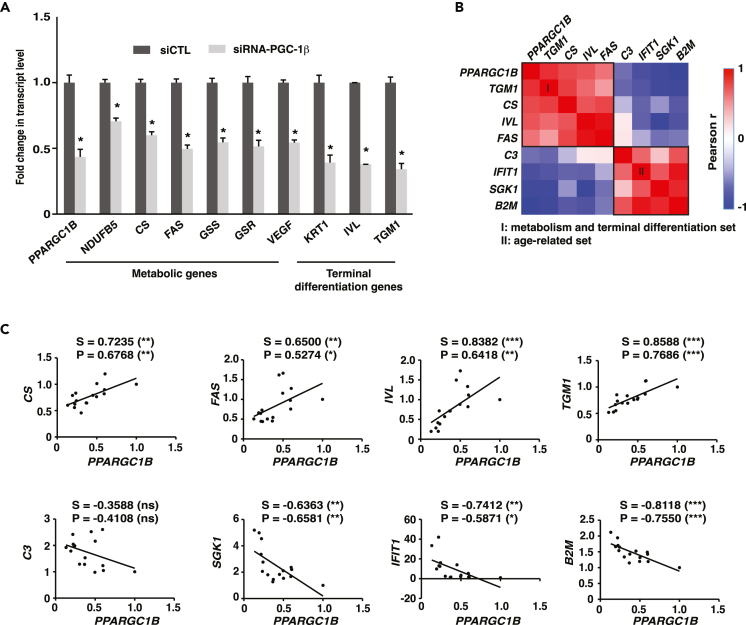
Given that PGC-1β was downregulated in aged keratinocytes, while the expression of PGC-1α was increased (Figure 2C), we sought to determine whether depletion of PGC-1β alone could mimic the key transcriptional changes in metabolic and differentiation gene expression we observed in aged human keratinocytes (Figures 2D–2G). Reducing the expression of PPARGC1B by ∼50% through KD experiments in NHEK cells led to a reduction in the expression of several known PGC-1β target metabolic genes such as NDUFB5 and CS (Figure 3A). PGC-1β KD also led to a reduction in transcript levels for differentiation markers like KRT1, IVL, and TGM1 (Figure 3A). We further evaluated the existence of a correlation between the transcript levels of PPARGC1B and terminal differentiation genes in immortal keratinocyte cells (HaCaT) treated with various combinations of siRNA duplexes against PGC-1β. Gene expression analyses revealed two distinct correlation sets (Figure 3B). The first set comprises the metabolic genes CS and FAS together with the terminal differentiation markers IVL and TGM1, all of which show strong positive correlation with PPARGC1B (Figures 3B and 3C). The second set comprises aging-linked genes C3, IFIT1, SGK1, and B2M that negatively correlate with PPARGC1B and the first set (Figures 3B and 3C). Overall, these results show that constitutive PGC-1β expression is a key regulator of terminal differentiation and metabolic gene expression in human keratinocytes.
Figure 3.
Constitutive PGC-1β expression supports the expression of terminal differentiation genes
(A) Gene expression in normal human epidermal keratinocytes (NHEK) transfected with control siRNA or an siRNA pool against PGC-1β for 3 days. Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(B) Correlation analysis between PPARGC1B and various gene transcripts in keratinocyte HaCaT cells transfected with 6 combinations of non-targeting, individual or pooled PGC-1β siRNA duplexes to achieve various transcript levels. Gene expression was assessed 3 days post-transfection. PPARGC1B transcript levels positively correlate with a gene set (I) of known PGC-1 metabolic targets (CS, FAS) and terminal differentiation markers (TGM1, IVL) and negatively correlates with a gene set (II) of aging-associated genes (C3, SGK1, IFIT1, and B2M).
(C) Detailed correlation analyses from B. S: Spearman r, P: Pearson r, with ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns: non-significant. Data shown in B-C are from 3 independent experiments.
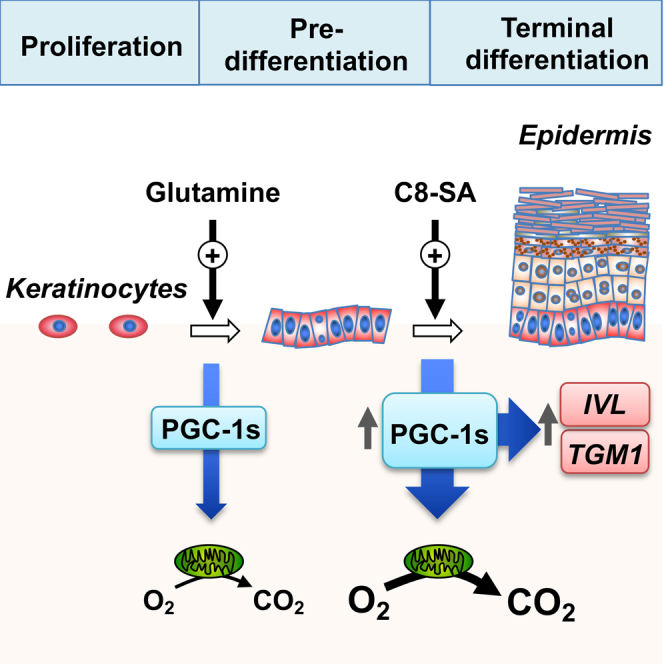
Glutamine availability is a critical factor supporting keratinocyte proliferation and terminal differentiation programs
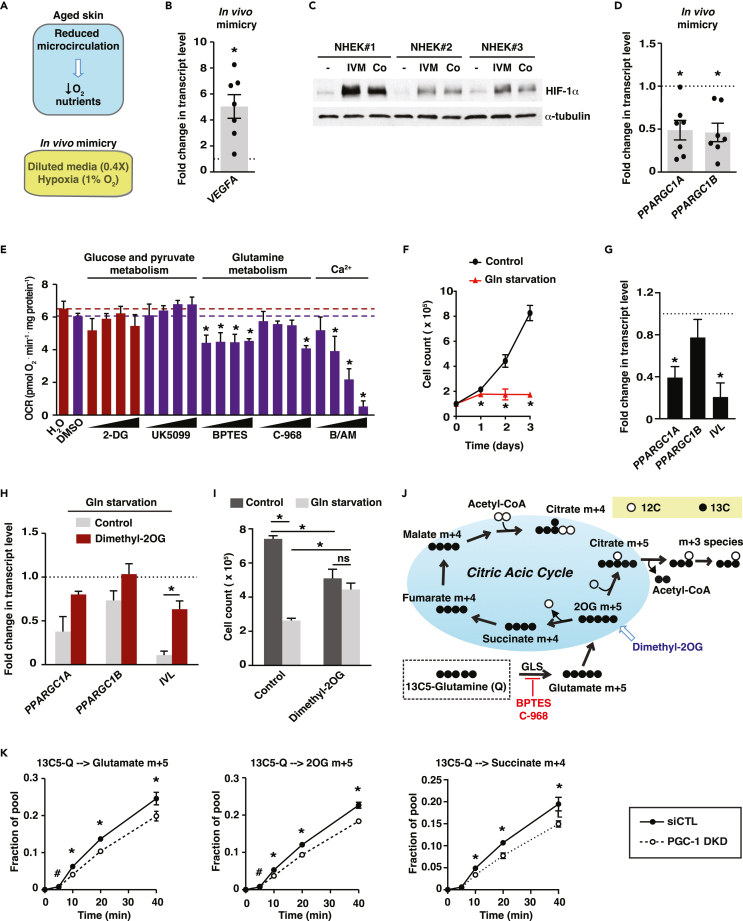
We next investigated which extrinsic factors could modulate PGC-1s expression in aged keratinocytes. First, we developed culture conditions that mimic a poorly vascularized papillary dermis, a feature of aged skin (Figure 4A). NHEK grown in hypoxic and low-nutrient conditions showed a strong increase in VEGFA expression (Figure 4B) and increased hypoxia inducible factor 1 subunit alpha (HIF-1α) levels (Figure 4C). Cobalt chloride, a hypoxia-mimetic agent, increased HIF-1α levels to a similar extent as hypoxia and low-nutrient conditions (Figure 4C). In contrast to what we observed in keratinocytes from aged versus young donors, both PPARGC1A and PPARGC1B levels were attenuated under hypoxia and low-nutrients conditions (Figure 4D). This may be explained by the fact that these experimental conditions mimic an acute effect of reduced vascularization, while aging is a slower and chronic process. To determine which metabolic pathways fuel keratinocyte activities, we studied keratinocyte respiration using compounds that target various aspects of metabolism: 2-deoxyglucose (2-DG), an inhibitor of glycolysis; UK5099, an inhibitor of mitochondrial pyruvate import; BPTES and compound 968 (C-968), inhibitors of glutamine metabolism as well as BAMPTA/AM, a calcium chelator. Strikingly, 2-DG and UK5099 had no significant effect on respiration while both BPTES and C-968 significantly reduced respiration (Figure 4E), revealing that glutamine metabolism, and not glucose metabolism, supports mitochondrial respiration in keratinocytes. As expected, BAMPTA/AM reduced oxygen consumption in a dose-dependent manner (Figure 4E), supporting the notion that optimal calcium concentrations are directly involved in shaping mitochondrial metabolism in pre-differentiation models.44
Figure 4.
Crosstalk between glutamine availability and PGC-1s in human keratinocytes
(A) Schematic depicting reduced microcirculation accompanied by lowered levels of oxygen and nutrients in aged skin. This was mimicked experimentally by growing cells in 1% oxygen and diluted isotonic media (0.4X).
(B) VEGFA gene expression in normal human epidermal keratinocytes (NHEK) subjected to microenvironment mimicry (1% hypoxia and diluted nutrients) for 24 h. Data are shown as fold change compared with normoxic and full nutrient conditions (dotted line) and as mean ± SEM. NHEK were from 7 different donors (each dot), each analyzed one or two times independently. ∗p < 0.05, paired Student’s t test.
(C) Immunoblot analyses for HIF-1α in protein extracts from NHEK exposed to low oxygen and nutrient conditions (IVM: in vivo mimicry, as described in B). NHEK were from 3 different donors. Cobalt chloride (Co, 200 μM) was used as positive control for HIF-1α induction. α-tubulin was used as loading control.
(D) PPARGC1A and PPARGC1B gene expression in NHEK cells subjected to microenvironment mimicry as in B for 24 h. Data are shown as fold change compared with normoxic and full-nutrient conditions (dotted line) and as mean ± SEM. NHEK were from 7 different donors (each dot), each analyzed one or two times independently. ∗p < 0.05, paired Student’s t test.
(E) Oxygen consumption rate (OCR) of keratinocyte HaCaT cells treated for 2 days with 4 doses of various compounds: 2-DG: 2-deoxyglucose (0.5, 1, 2, 4 mM), UK5099 (1, 3, 10, 30 μM), BPTES (1, 3, 10, 30 μM), C-968: compound 968 (0.3, 1, 3 10 μM) and B/AM: BAPTA/AM (3, 10, 20, 30 μM). Treatments were compared to specific control treatment (dotted line) to which they are associated by a color code (H2O: red, DMSO: purple). Data are expressed as mean + SEM from 3 independent experiments. ∗p < 0.05, one-way ANOVA, Dunnett’s post-test.
(F) Proliferation curves of keratinocyte HaCaT cells under 4 mM glutamine or glutamine starvation. Data are shown as mean ± SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(G) Gene expression analyses of glutamine-starved (3 days) keratinocyte HaCaT cells. Data are shown as fold change versus control conditions (4 mM glutamine) and mean + SEM from 7 independent experiments. ∗p < 0.05, paired Student’s t test.
(H) Dimethyl-2OG (4 mM) rescues gene expression of glutamine-starved keratinocyte HaCaT cells. Data are expressed as fold change versus control conditions (4 mM glutamine) and as mean + SEM from 3 independent experiments. ∗p < 0.05, paired Student’s t test.
(I) Dimethyl-2OG (4 mM) rescues the proliferation inhibition of glutamine-starved keratinocyte HaCaT cells. Data are shown as mean + SEM from 3 independent experiments. ∗p < 0.05, two-way ANOVA, Tukey post-test.
(J) Schematic showing the metabolism of 13C5-L-glutamine in the citric acid cycle.
(K) Stable isotope tracer analysis using 13C5-L-glutamine in HaCaT cells treated with siCTL or PGC-1 DKD. Dynamic labeling of glutamate M+5, alpha-ketoglutarate (2OG) M+5, and succinate M+4 are shown. Data are shown as mean ± SEM from 3 independent experiments. ∗p < 0.05, #p < 0.10, paired Student’s t test.
Glutamine starvation strongly suppressed the proliferation of HaCaT keratinocytes (Figure 4F). In addition, glutamine starvation reduced PPARGC1A and PPARGC1B expression, as well as that of the terminal differentiation marker IVL (Figure 4G). Given that glutamine is converted to glutamate, an important metabolic precursor of nucleotides and glutathione, it is possible that the impact of glutamine starvation on keratinocyte growth is unrelated to citric acid cycle (CAC) anaplerosis. To test this, we analyzed the expression of PPARGC1A, PPARGC1B, and IVL and the proliferation of cells deprived of glutamine but supplemented with dimethyl-2-oxoglutarate (dimethyl-2OG), a cell-permeable analog of the CAC intermediate 2OG. Glutamine-deprived cells exposed to dimethyl-2OG displayed an elevated expression of all three transcripts (Figure 4H) as well as increased cell proliferation (Figure 4I), indicating that glutamine anaplerosis is critical to support the expression of PGC-1s and terminal differentiation genes as well as keratinocyte proliferation.
Furthermore, we uncovered not only that glutamine availability controls the expression of PGC-1s in keratinocytes, but also that PGC-1s control glutamine metabolism through the CAC in these same cells. Indeed, NHEK subjected to PGC-1 DKD displayed reduced 13C5-glutamine tracing in the CAC compared with controls (Figures 4J and 4K). Specifically, there was reduced incorporation of the carbons from glutamine into 2OG and succinate (Figures 4J and 4K). PGC-1s also control the proliferation of keratinocytes demonstrated by the reduced proliferation of both NHEK and HaCaT cells expressing PGC-1 DKD (Figures S4A and S4B). This was linked to a cell-cycle arrest as HaCaT cells harboring PGC-1 DKD displayed an accumulation of cells in G0/G1 (Figure S4C). These data are further substantiated by network analyses of enriched and depleted pathways in aged vs. young keratinocytes and PGC-1 DKD vs. control groups, which revealed that cell-cycle pathways are globally downregulated (blue color; Figure 2E). Interestingly, replenishing CAC intermediates with dimethyl-2OG did not rescue the proliferation of PGC-1s-depleted keratinocytes (Figure S4D). These data indicate that PGC-1s depletion has growth-inhibitory effects unrelated to perturbations in glutamine mitochondrial metabolism. Taken together, these results unveil a new link between glutamine availability and PGC-1s expression, as well as keratinocyte proliferation and terminal differentiation.
PGC-1s depletion alters the reconstruction of human skin
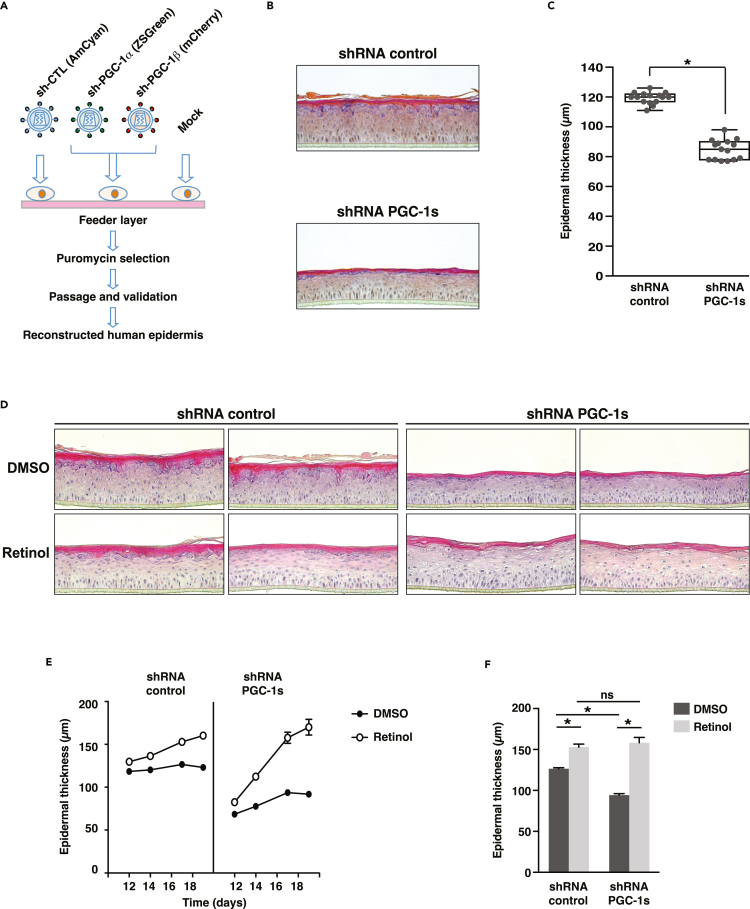
To further substantiate the impact of PGC-1s in human skin physiology, we developed a reconstructed human epidermis (RHE) model in which PGC-1s expression is attenuated by stable RNA interference. We first established populations of stable primary foreskin keratinocytes expressing short hairpin RNA (shRNA) against both PGC-1α and PGC-1β (Figure 5A). RHE constructs prepared with shRNA-PGC-1s stable cells were properly developed and differentiated; however, they were 29% thinner than those prepared with control shRNA stable cells (Figures 5B and 5C), indicating that reduction in PGC-1s contributes to epidermal thinning. The impact of PGC-1s depletion on epidermal thinning is consistent with its impact on the expression of differentiation markers (Figure 1G) and cell proliferation (Figures S4A and S4B). These observations are in line with a recently developed in vitro model of chronological aging that also exhibits reduced epidermal thickness.45 In order to rescue the thinning phenotype of shRNA-PGC-1s RHE, we studied the effects of retinol, a precursor of retinoic acid, which is known for its benefits on improving human skin aging signs.46 Control RHE treated with retinol showed modest increase in epidermal thickness, while the thickness level of shRNA-PGC-1s RHE after the same treatment fully recovered comparable to the control RHE (Figures 5D–5F). The thickness changes observed under retinol treatment were greater for shRNA-PGC-1s RHE than control RHE (Figures 5D–5F). Taken together, these results expand the role of PGC-1s in pre-differentiation keratinocyte models by showing a strong impact of PGC-1s on the development of RHE. Importantly, the thickness of PGC-1s-depleted RHE can be recovered to control RHE thickness levels with retinol treatment. Because the cells were stably infected with shRNA against the PGC-1s, this suggests that the retinol effect is largely independent of PGC-1s and that it acts downstream of, or in parallel to, the PGC-1s.
Figure 5.
PGC-1s knockdown modifies reconstructed human epidermis
(A) Schematic depicting PGC-1s double knockdown strategy.
(B) Hematoxylin and eosin staining of typical reconstructed human epidermis (RHE) constructs stably expressing shRNA constructs. RHE were analyzed at day 14. This experimental set is representative of 5 independent experiments.
(C) Box-and-whisker plot of epidermal thickness measured by optical coherence tomography. Data from 15 RHE constructs per condition and obtained from 5 independent experiments are shown. ∗p < 0.05, unpaired Student’s t test.
(D) Hematoxylin and eosin staining of RHE constructs prepared with shRNA control and shRNA-PGC-1s stably expressing cells treated with DMSO (vehicle) or retinol (10 μM) from day 10 to day 19. This experimental set was analyzed at day 14, and 2 representative independent experiments are shown.
(E) Epidermal thickness measured by optical coherence tomography of RHE constructs described in D. RHE constructs were assessed on days 12, 14, 17, and 19. Data are shown as mean ± SEM from 2 to 6 independent RHE per time point.
(F) Fold change in epidermal thickness upon DMSO or retinol treatment for samples shown in E (day 17). Data are shown as mean + SEM, n = 3 independent samples per treatment group, ∗p < 0.05, two-way ANOVA, Tukey post-test.
A salicylic acid derivative promotes mitochondrial respiration and terminal differentiation programs in human primary keratinocytes
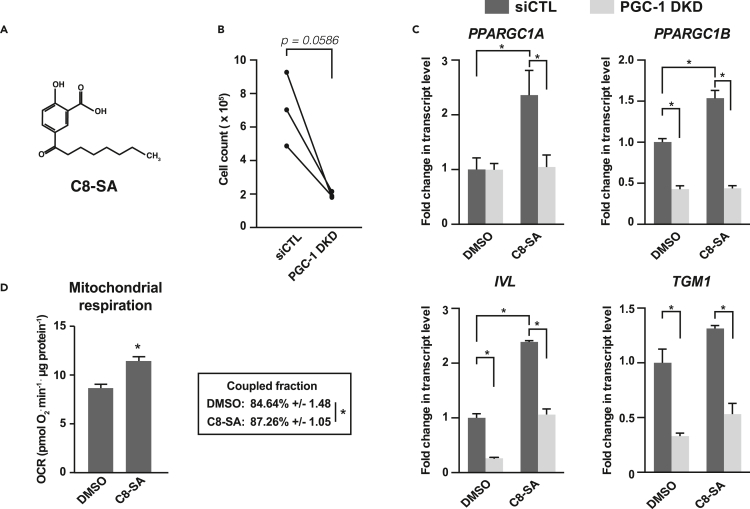
The salicylic acid derivative C8-SA (Figure 6A) has long been used for skin care due to its controlled keratolytic properties.47,48 Interestingly, salicylic acid, the parent compound, was identified as an activator of AMP-activated protein kinase (AMPK), a well-known inducer of PGC-1α activity.49,50 Recently, we demonstrated that C8-SA also activates AMPK in Caenorhabditis elegans and primary keratinocytes.51 We thus sought to investigate whether C8-SA (Figure 6A) could modulate PGC-1s functions in primary keratinocytes. Keratinocytes treated with C8-SA showed reduced proliferation (Figure 6B) as well as a significant increase in PPARGC1A and PPARGC1B that were paralleled with elevated expression of terminal differentiation markers IVL and TGM1 (Figure 6C). As expected, PGC-1 DKD in primary keratinocytes significantly reduced the expression of IVL and TGM1 (Figures 2G and 6C) and limited their induction upon C8-SA treatment (Figure 6C). Consistent with the results presented in Figure 2A, the reduction in PPARGC1A expression upon PGC-1 DKD under basal conditions was not significant (Figure 6C). To assess the functional significance of the changes in PPARGC1A and PPARGC1B upon C8-SA treatment, we measured mitochondrial respiration, a hallmark of PGC-1s functions. C8-SA treatment increased mitochondrial respiration and the fraction of total respiration coupled to ATP synthesis in NHEK (Figure 6D). Altogether, these results show that salicylic acid derivatives, such as C8-SA, are potent inducers of mitochondrial metabolism and terminal differentiation markers.
Figure 6.
A salicylic acid derivative promotes mitochondrial activity and keratinocyte differentiation
(A) Structural formula of C8-SA (C8/capryloyl-salicylic acid).
(B) Impact of C8-SA (10 μM) on the proliferation of normal human epidermal keratinocytes (NHEK). Data are shown as mean of NHEK derived from 3 different donors that were tested independently. Lines indicate independent pairs. ∗p < 0.05, paired Student’s t test.
(C) PPARGC1A, PPARGC1B, IVL, and TGM1 gene expression in NHEK treated with siCTL or PGC-1 DKD for 3 days and with C8-SA (10 μM) or DMSO (control) for the last 48 h. Data are shown as mean +SEM and fold change normalized to siCTL and DMSO conditions of 3 experiments. ∗p < 0.05, two-way ANOVA, Tukey post-test.
(D) OCR of normal human epidermal keratinocytes (NHEK) treated with DMSO or C8-SA (15 μM) for 3 days. Data on graph are shown as mean +SEM from 4 independent NHEK cultures, ∗p < 0.05, paired Student’s t test. Lower inset shows the average proportion of total respiration dedicated to ATP synthesis (coupled respiration) +/− SEM.
Discussion
Our story unveils that the PGC-1s, in addition to regulating the metabolic state of skin cells, play key roles in keratinocyte proliferation and terminal differentiation. It has been shown that the expression of involucrin and transglutaminase 1 is decreased in aged skin, in line with epidermal thinning.45 In agreement with this study, many of the most downregulated genes common to PGC-1 DKD and aging conditions in our transcriptomics analyses have direct implication in terminal differentiation, including IVL and TGM1.
Our work also shows that PGC-1α levels are elevated in aged epidermis and keratinocytes, an observation that has several implications. A rise in PGC-1α might be caused by chronic metabolic stress, suggesting that aged skin induces PGC-1α as part of an adaptive process. PGC-1α activity is modulated by energy stress through deacetylation by sirtuins52 and phosphorylation by AMPK.50 Moreover, its transcriptional induction through Ca2+ and p38 signaling has been well documented.53 Notably, Ca2+ and p38 have established pivotal roles in keratinocyte differentiation.54,55 We have also shown that PPARGC1A and PPARGC1B transcript levels are both decreased under acute oxygen and nutrient stress and glutamine starvation, which is distinct from the transcript pattern we observed in aged keratinocytes that is characterized by elevated PPARGC1A and reduced PPARGC1B transcript levels. This suggests that the elevated PPARGC1A transcript levels observed in aged skin could represent long-term adaptation.
In the past decade, mitochondrial electron transport chain (ETC) dysfunction has been recognized as an integral part of diverse age-associated conditions.6,56,57,58 The fact that patients with mitochondrial diseases have skin problems suggests that mitochondrial functions play a central role in skin physiology.59 In support of these observations, aged skin cells show reduced expression and activity of ETC complex II,13 and photoaging is associated with mitochondrial dysfunction and mitochondrial network fragmentation.60,61 Importantly, human keratinocytes exhibit a fragmented mitochondrial network in aged skin, and aged keratinocytes from biopsies are more glycolytic.15,18 Although PGC-1α expression was found to be correlated with aged epidermis, the expression of mitochondrial marker NDUFA9 did not parallel PGC-1α expression, suggesting that the PGC-1α signaling axis is less effective at promoting mitochondrial function in aged keratinocytes. In line with these observations, a study revealed that increased PGC-1α expression in aged dermal fibroblasts does not translate into increased mitochondrial functions.11 It was also shown that NDUFA9, as well as other complex I proteins, were decreased in dermal fibroblasts from aged donors,62 which is consistent with an age-dependent decline in OXPHOS efficiency in these cells.63 Altogether, these data suggest that upregulation of PGC-1α can be uncoupled from mitochondrial functions in aged keratinocytes and dermal fibroblasts.
Only a few classes of compounds are known to upregulate PGC-1α expression and activity,64 notably through the activation of the metabolic stress kinase AMPK.50 We now demonstrate that the salicylic acid derivative C8-SA has the capacity to induce PGC-1α expression and activity and to improve mitochondrial functions in NHEK cells. Salicylic acid has long been known for its capacity to modulate mitochondrial respiration acutely through direct uncoupling effects.65,66 Our data suggest that C8-SA could have beneficial effects through PGC-1α modulation in keratinocytes in vivo when targeted to the epidermis. We have recently published that C8-SA shares with the parent compound, salicylic acid, the capacity to promote AMPK activation.49,51 Thus, compounds priming AMPK function might prove to be useful anti-aging agents by modulating the activity of PGC-1α.
While our work reveals impact of PGC-1s on the expression of terminal differentiation genes, the mechanisms involved remain to be elucidated, as well as the role of metabolism in the initiation of the terminal differentiation process. We have shown that PGC-1 DKD reduces glutamine flux using 13C5-L-glutamine tracing experiments and that glutamine deprivation reduces keratinocyte proliferation, suggesting that the growth-inhibitory effects are related to glutamine metabolism. However, cell-permeable alpha-ketoglutarate was not able to rescue the proliferation of PGC-1-depleted cells, indicating that the growth-suppressive effects of PGC-1 DKD are not related to CAC anaplerosis. Glutamine is also an important player in oxidative stress detoxification by providing glutamate for glutathione biosynthesis. Importantly, reactive oxygen species (ROS) have been shown both to induce cell-cycle arrest in normal human keratinocytes67,68 and be required for terminal differentiation.69 Thus, ROS could represent the missing link between PGC-1s metabolism, growth arrest, and epidermal terminal differentiation.
In conclusion, our work unveils new roles for PGC-1α and PGC-1β in human skin functions through the regulation of epidermal homeostasis. This knowledge may provide actionable opportunities to treat skin conditions and age-related functional changes in skin.
Limitations of the study
We sought to uncover roles for PGC-1s in skin physiology with relevance to aging. Using a transcriptomics approach, we identified terminal differentiation targets (IVL and TGM1) that were downregulated upon PGC-1s KD and in aging. To perform these analyses, we used primary keratinocytes derived from a pair of young (28 y.o) and aged (83 y.o.) donors that were matched for sex, ethnicity, body site, and growth culture conditions. The limitations in the availability of material that was biologically matched with a significant difference in age combined with the short propagation period of these primary cells in culture restricted the number and scope of our experiments. Despite these limitations, the biological relevance of the targets we decided to pursue is supported by the fact that involucrin (IVL) and transglutaminase (TGM) protein levels have been shown to be lowered in aged skin.45 Importantly, the key finding that PGC-1s positively regulate the expression of terminal differentiation genes in keratinocyte has been validated in several experimental models throughout the manuscript. Our loss of function approach to study the roles of PGC-1s in keratinocytes was centered on a DKD strategy to reduce the expression of both PPARGC1A and PPARGC1B given that these coactivators can functionally overlap and compensate for one another.37,38 Another limitation of our study is that the DKD approach did not significantly decrease PPARGC1A expression under basal conditions, even though it was efficient at preventing its induction by C8-SA. To fully delineate the individual roles of PGC-1α and PGC-1β in skin physiology, gain and loss of function approaches targeting one coactivator at a time will need to be performed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HIF-1α rabbit monoclonal antibody, clone D1S7W (Western blot) | Cell Signaling Technology Inc. | 36169; RRID: AB_2799095 |
| anti-α-tubulin mouse monoclonal antibody, clone DM1A (Western blot) | Cell Signaling Technology Inc. | 3873; RRID: AB_1904178 |
| Anti-PGC-1alpha rabbit polyclonal (IHC) | Abcam | Ab84139; RRID: AB_1860776 |
| Anti-NDUFA9 mouse monoclonal, clone 20C11B11B11 (IHC) | Abcam | Ab14713; RRID: AB_301431 |
| Biological samples | ||
| Tissue microarray in paraffin (human normal organs, custom) | IRIC Histology Platform, University of Montreal | N/A |
| Human skin (unexposed forearm) sections in paraffin (young and aged non-diseased women) | Pageon et al.70 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Capryloyl salicylic acid (C8-SA) | L’OREAL/ NOVEAL | N/A |
| Dimethyl-2-oxoglutarate (dimethyl-2OG) | MilliporeSigma | 349631 |
| Lipofectamine 2000 | Thermo Fisher Scientific | 11668027 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | 13778075 |
| HiPerFect Transfrection Reagent | QIAGEN | 301704 |
| 4 siRNA duplexes against human PPARGC1A | QIAGEN | SI00101024, SI00101031, SI02639826, SI02639833 |
| 4 siRNA duplexes against human PPARGC1B | QIAGEN | SI03043306, SI0310431, SI00150024, SI02647911 |
| AllStars non-targeting control siRNA | QIAGEN | SI03650318 |
| 13C5-L-glutamine | Cambridge Laboratories Inc. | CLM-1822 |
| Western Lightning® Plus-ECL enhanced chemiluminescence substrate | Perkin-Elmer | NEL103E001EA |
| Propidium iodide(PI)/RNase staining solution | Cell Signaling Technology Inc. | 4087 |
| Polybrene (hexadimethrine bromide) | MilliporeSigma | 107687 |
| Y-27632 ROCK inhibitor | MilliporeSigma | SCM075 |
| Retinol | Sigma | R7632 |
| Critical commercial assays | ||
| Aurum Total RNA Mini Kit | Bio-Rad | 7326820 |
| PureLink RNA Mini Kit | Thermo Fisher Scientific | 12183020 |
| iScript cDNA synthesis kit | Bio-Rad | 1708890 |
| High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor | Thermo Fisher Scientific | 4374966 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | 1725272 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | 4444556 |
| Seahorse XF Cell Mito Stress Test Kit | Agilent | 103015-100 |
| PierceTM BCA protein assay | Thermo Fisher Scientific | 23227 |
| Deposited data | ||
| Raw and analyzed transcriptomic data | This paper | GEO: GSE108674 |
| Experimental models: Cell lines | ||
| Normal human epidermal keratinocytes (NHEK) | PromoCell | 12003 |
| Normal human epidermal keratinocytes (NHEK) | Lonza | 192627 |
| Normal human epidermal keratinocytes (NHEK) | Thermo Fisher Scientific | C0055C |
| Normal human foreskin keratinocytes (reconstructed epidermis) | Episkin | N/A |
| Human: HaCaT | Addexbio | T0020001 |
| Human: HEK 293T | ATCC | CRL-3216 |
| Software and algorithms | ||
| DAVID Bioinformatics | Huang et al.71; Sherman et al.72 | SCR_001881 |
| Gene Ontology | Ashburner et al.73; Gene Ontology Consortium74 | SCR_002811 |
| R Project for Statistical Computing, R Bioconductor | R Foundation for Statistical Computing, (R Development Core Team, 2019) | SCR_001905 SCR_006442 |
| GeneOverlap v.1.34.0 | Shen Lab | http://shenlab-sinai.github.io/shenlab-sinai/ |
| Pscan | Zambelli et al.75 | http://159.149.160.88/pscan/ |
| Gene Set Enrichment Analysis (GSEA) | Mootha et al.76; Subramanian et al.77 | SCR_003199 |
| CytoScape | Shannon et al.78 | SCR_003032 |
| Enrichment Map | Merico et al.79 | N/A |
| Genotype-Tissue Expression (GTEX) | NIH Common Fund | SCR_013042 |
| Correction for naturally occurring isotopes in-house JAVA program | Nanchen et al.80; McGuirk et al.81 | N/A |
| FACSDivaTM software | BD Biosciences | SCR_001456 |
| ImageScope software | Leica Biosystems | SCR_014311 |
| Chemstation software | Agilent | SCR_015742 |
Resource availability
Lead contact
Furter information and requests for reagents must be directed to and will be fulfilled by the lead contact, Julie St-Pierre (julie.st-pierre@uottawa.ca).
Materials availability
This study did not generate new unique materials.
Experimental model and subject details
Human primary cells and cell lines
HaCaT were obtained from Addexbio (San Diego, CA, USA) at passage 17. Normal human epidermal keratinocytes (NHEK) from normal donors (no disease detected) were obtained from PromoCell (cat#12003, Heidelberg, Germany), Lonza (cat#192627, Basel, Switzerland), or Thermo Fisher Scientific (cat#C0055C). Primary foreskin NHEKs (L’Oréal) were used for epidermis reconstruction. HEK 293T cells were from ATCC (Manassa, VA, USA).
Skin sections
Tissue microarray (TMA) was prepared by the IRIC histology platform (Université de Montréal) using the Manual Tissue Arrayer MTA-1 from Estigen (Tartu, Estonia). Tissue cores (1mm diameter) obtained from 14 healthy human tissues (Pathology and Cellular Biology Department, Faculty of Medicine, Université de Montréal) were used to generate matrices in paraffin blocks. Human skin sections from young and aged healthy volunteers were described elsewhere.70
Method details
Cell culture and treatments
HaCaT were maintained in DMEM (319-005, Wisent, St-Bruno, Canada) supplemented with 10% FBS (Premium quality, Wisent, St-Bruno, Canada) and penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). NHEK were maintained in keratinocyte growth media (PromoCell) and subcultured using dissociation agents from Thermo Fisher Scientific (TE, cat#R001100; TN, cat#R002100) or PromoCell (cat#C-41210). 293T/17 cells were from ATCC (Manassas, VA, USA). C8-SA is a salicylic acid derivative developed by L’Oréal.47 C8-SA treatments were performed on subconfluent cells for 3 days at a final concentration of 10 or 30μM. For nutrient withdrawal experiments using HaCaT cells, medium without glucose, glutamine and pyruvate (Wisent) was supplemented with 10% FBS and 1mM sodium pyruvate, 25mM D-glucose, with or without L-glutamine and dimethyl-2-oxoglutarate (dimethyl-2OG; Sigma-Aldrich, St. Louis, MO, USA), both used at 4mM. Media was discarded and replaced daily. For in vivo mimicry experiments using NHEK, keratinocytes growth media was diluted to 0.4 fold in low phosphate buffer saline (LPBS, 153mM NaCl, 1mM KCl, 1.5 mM NaPO4, pH 7.4) and cell cultures were placed in a HeraCell humidified hypoxic incubator (Thermo Fisher Scientific) set at 1% O2 and supplied with 5% CO2 at 37oC for 24h. Alternatively, cells were placed into a Xvivo System glove box hypoxia chamber (Biospherix, Parish, NY, USA) at 1% O2.
Cell proliferation
Cells were enzymatically detached with 0.25% trypsin and 0.53mM EDTA (Wisent, St- Bruno, Canada) or 0.04% trypsin and 0.03% EDTA for NHEK and counted with a phase contrast hemacytometer (Hausser Scientific, Horsham, PA, USA) or with a TC10 automated counting device (Bio-Rad, Hercules, CA, USA) or Countess FL II automated counting device (Thermo, Waltham, MA, USA).
Gene silencing
RNA interference was accomplished in HaCaT and NHEK using Lipofectamine RNAiMAX (Thermo, Waltham, MA, USA) or HiPerfect (QIAGEN, Hilden, Germany). siRNAs were from QIAGEN. siRNA duplexes for PPARGC1A (QIAGEN GeneSolution, SI00101024, SI00101031, SI02639826, SI02639833) and PPARGC1B (QIAGEN GeneSolution, SI03043306, SI0310431, SI00150024, SI02647911) were purchased as individual duplexes and were used individually and/or in combination. Non-targeting control was the AllStars negative control siRNA (SI03650318, QIAGEN). Total RNAs were extracted 72h post-treatment.
RNA isolation and RT-qPCR
Total RNA was extracted using the Aurum Total RNA Mini Kit (Bio-Rad, Hercules, CA, USA) or the PureLink RNA Mini Kit (Thermo, Waltham, MA, USA). RNA was quantify using Nanodrop Spectrophotometer ND-1000 (Thermo, Waltham, MA, USA). For cDNA preparation, 1-2 μg total RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) or the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Thermo). Quantitative PCR was performed on the MyIQ2 amplification system (Bio-Rad) or the QuantStudio 3 amplification system (Thermo, Waltham, MA, USA,). Real-time PCR reactions consisted of template cDNA, specific oligonucleotide pairs, and the SYBR Green Master Mix (Bio-Rad). Primers were all validated for linearity and single melt product. POLR2A was used as an endogenous control for ΔΔCt calculations in HaCaT cells and primary keratinocytes studies. B2M was used as an endogenous control in siRNA experiments performed in primary keratinocytes. For the experiments presented in Figure 6, Real-time PCR reactions consisted of template cDNA, specific TaqMan probes, and the TaqMan Fast Advanced Master Mix (Thermo). All primers used in this study can be found in Table S1.
Microarrays and gene expression analyses
Total RNA integrity was assessed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Sense-strand cDNA was synthesized from 100 ng of total RNA, and fragmentation and labeling were performed to produce ss DNA with the Affymetrix GeneChip® WT Terminal Labeling Kit according to manufacturer’s instructions (Thermo Fisher Scientific). After fragmentation and labeling, 3.5μg DNA target was hybridized on GeneChip® Human Gene 2.0ST (Thermo Fisher Scientific) and incubated at 45oC in the Genechip® Hybridization oven 640 (Thermo Fisher Scientific) for 17 hours at 60 rpm. GeneChips were then washed in a GeneChips® Fluidics Station 450 (Thermo Fisher Scientific) using Affymetrix Hybridization Wash and Stain kit according to the manufacturer’s instructions. The microarrays were finally scanned on a GeneChip® scanner 3000 (Thermo Fisher Scientific). CHP files were analyzed with the Transcription analysis console (Thermo Fisher Scientific). Transcript clusters with ANOVA p-values < 0.05 were selected and genes with fold change higher or equal to 1.2 fold (or -1.2 fold for downregulated genes) were selected for each groups. Affymetrix transcript cluster IDs were converted to gene IDs using the db2db tool from bioDBnet (Mudunuri et al., 2009), which were used to generate Venn diagrams. When there was multiple gene IDs associated with an Affymetrix transcript cluster ID, only the first one was kept. Gene IDs were subjected to DAVID analysis for gene functional classification.71,72 The significance of the overlap between differentially expressed gene lists was determined using the GeneOverlap package in R, which tests the overlap using Fisher’s exact test in comparison with genomic background (Shen L, Sinai ISoMaM (2022). GeneOverlap: Test and visualize gene overlaps. R package version 1.32.0, http://shenlab-sinai.github.io/shenlab-sinai/). Transcription factor binding site (TFBS) analyses were performed using Pscan,75 mapped to the non-redundant JASPAR database.82 Gene expression analyses of skin samples from young (20-29 years) and aged (70-79 years) donors were performed using openly accessible RNAseq data from the Geno-type-Tissue Expression (GTEx) Project.83 Gene transcripts per million (TPMs) from suprapubic skin samples were sourced from GTEx Analysis V8 (dbGaP Accession phs000424.v8.p2), obtained from the GTEx Portal on June 13, 2022.
Pathway network analysis
Network analysis of enriched and depleted pathways was performed via ranked-list Gene Set Enrichment Analysis (GSEA), mapped to gene ontology biological pathways.73,74,76,77 Ranks for differential gene expression were defined as -log(p-value) ∗ sign(fold change). GSEA results were visualized in Cytoscape78 using Enrichment Map79 and clustered by force-directed layout. Edges (lines) link nodes (enriched pathways from GSEA), and edge length is inversely proportional to the similarity between nodes. Each node is further visualized as a pie chart, with each section demonstrating the GSEA enrichment score for each comparison (aged vs young keratinocytes, Double PGC-1 knockdown vs control siRNA treatments). Broader pathway groups (e.g. Cell cycle, Membrane transport, Metabolism, etc.) were outlined manually with different colors and their name represents shared ontology between highly similar nodes.
Bioenergetics
For siRNA experiments, twenty thousand HaCaT cells were seeded per well of a Seahorse XFe24 cell plate (Agilent Technologies, Santa Clara, CA, USA) and transfected with indicated siRNA on the next day. Three days post-transfection, cells were washed with DMEM without sodium bicarbonate supplemented with 25 mM D-glucose, 4mM L-glutamine and 1mM sodium pyruvate, with pH adjusted at 7.4. Cells were allowed to equilibrate in a 37oC incubator without CO2 supply for 1h. Cell plates were subjected to respiration and glycolysis assays using the XFe24 Seahorse Extracellular Flux analyzer (Agilent). Incubation times and injections were set according to the Seahorse XF Cell Mito Stress Test Kit (cat#103015-100, Agilent). For compound testing experiments, 5,000 HaCaT cells were plated in XFe96 96-well plates in 100μL of growth media and allowed to adhere and grow for 1 day. Seahorse assays were performed 2 days post-treatment. For C8-SA experiments in NHEK, 5,000 primary cells from different donors were seeded in XFe96 plates and treated the next day with vehicle control (DMSO) or C8-SA (15μM) for 72h. Basal and oligomycin-sensitive respiratory rates were corrected for non-mitochondrial respiration rates. Following the assays, cells were washed with PBS and lysed in RIPA buffer. Protein content was determined by Pierce BCA protein assay (Thermo, Waltham, MA, USA).
GC-MS and stable isotope tracer analyses
All reagents and standards used for gas chromatography-mass spectrometry (GC-MS) were from Sigma-Aldrich (St. Louis, MO, USA). Cells were equilibrated in regular media and pulse with 4mM 13C5-Lglutamine (Cambridge Isotopes Laboratories Inc, Tewksbury, MA, USA). Cell plates were rinsed twice on ice with cold saline solution (9g/L NaCl). Cells were quenched with 80% (v/v) methanol (dry ice cold), and cellular extract was scrapped and transferred to pre-chilled tubes (on dry ice). Samples were stored at -80oC until further manipulation. Metabolite extraction was done at 4oC using the Biorupter (Diagenone, Denville, NJ, USA) set at high with cycles of 30 seconds for a total of 10 min. Samples were cleared by centrifugation at 21,000 x g at 4oC for 10 min. Pellets were discarded and 1μL of the standard myristic acid-D27 (800ng/μL in pyridine) was added to each sample. Samples were dried overnight in a cold centri-trap (Labconco, Kansas City, MO, USA). Samples were solubilized in 10mg/mL methoxiamine hydrochloride (in pyridine). Samples were briefly vortexed and sonicated to increase dissolution, and cleared by centrifugation at 21,000 x g for 10 min. Samples were loaded in GC-MS amber injection vials and incubated at 70oC for 30 min. 100μL MTBSTFA (N- tert-Butyldimethylsilyl-N-methyltrifluoroacetamide) with 1% tert- Butyldimethylchlorosilane was added and vials were mixed and further incubated at 70oC for 1h. GC-MS method for methoximes-TBDMS metabolites was carried on a 5975C GC-MS (Agilent Technologies, Santa Clara, CA, USA) and is detailed elsewhere.84,85 Ion integrations were analyzed with the Chemstation software (Agilent). Correction for naturally occurring isotopes was done using an in-house algorithm based on published methods.81,80
Immunoblotting
Following treatments, cells were lyzed for 30 min on ice in triton X-100 lysis buffer (150mM NaCl, 50mM Tris-HCl pH 7.4, 5mM EDTA, 1mM EGTA, 1% triton X-100) supplemented with phosphatase and protease inhibitors (1mM sodium orthovanadate, 5mM sodium fluoride, 2μg/mL aprotinin, 5μg/mL leupeptin, 1μg/mL pepstatin A, 1mM PMSF). Cell extracts were further subjected to 3 rounds of freeze and thaw on liquid nitrogen and a 37oC water bath to ensure complete disruption of nuclei. Samples were centrifuged at 17,000 x g for 10 min à 4oC and protein concentration was determined with the Pierce BCA assay (ThermoFisher Scientific, Waltham, MA, USA). Equal amounts of proteins (10μg or 30μg) were resolved on 7.5% in-house or 4-15% precast acrylamide Mini-Protean TGX gels (Bio-Rad, Hercules, CA, USA). Proteins were transferred on PVDF membranes (GE Healthcare, Chicago, IL, USA) using Pierce western blot transfer buffer (ThermoFisher Scientific, Waltham, MA, USA) and Trans-Blot SD semi-dry transfer cell (Bio-Rad, Hercules, CA, USA) at constant voltage (22V, 0.2A) for 50 min. Blots were blocked in 5% non-fat dry milk diluted in tris buffer saline with 0.1% Tween-20 (TBST) and incubated with primary antibodies overnight at 4°C. Anti-HIF-1α (36169) and anti-α-tubulin (3873) were from Cell Signaling Technology Inc. (Whitby, ON, CA) and used as recommended by the manufacturer. Horseradish peroxidase conjugated secondary antibodies (Mandel scientific, Guelph, ON, CA) were used at 1:10,000 dilution in 5% non-fat dry milk/TBST for 1h. Membranes were washed 5 times, 5 min in TBST after incubations with antibodies. Membranes were incubated for 1 min by immersion in the Western Lightning® Plus-ECL enhanced chemiluminescence substrate (PerkinElmer, Waltham, MA, USA). Chemiluminescence was detected on Blu-Lite films (Dutscher, Bernolsheim, FR). Films were developed in a dark room using Mini-Med 90 X-ray film processor (AFP manufacturing, Peachtree city, GA, USA).
Flow cytometry
HaCat cells were trypsinized, washed with PBS, spun down at 200 x g for 5 min, and cell pellets were resuspended in 70% ethanol and left on ice for 30 min. Cells were spun down, rinced 2 times with PBS, resuspended in propidium iodide(PI)/RNase staining solution (Cell Signaling Technology Inc., Whitby, ON, CA), and incubated for 15 min in the dark at room temperature. Data were acquired on a FACSCelestaTM Cell Analyzer using the FACSDivaTM software (BD Biosciences, Franklin Lakes, NJ, USA). Gating was done on forward vs size scatter areas (FSC-A/SSC-A) dot plot graph to select live cells, size scatter height vs size scatter area (SSC-H/SSC-A) graph to discriminate cell singlets from cell doublets, and phycoerythrin (PE) height/area (PE-H/PE-A) for cell cycle phases.
Immunocytochemistry and immunohistochemistry
Forearm sections (ventral side, low sun exposure) were originally fixed in 10% formalin and paraffin-embedded. Skin sections were processed by automated staining using the Bond RX Fully Automated Research Stainer (Leica Biosystems, Wetzlar, Germany). Paraffin was removed with the Bond Dewax solution (Leica Biosystems), and antigen retrieval was done in EDTA pH 8 for 20 min prior to blocking. Incubation with antibodies was done for 15 min using these antibodies and dilutions: anti-PGC-1a (ab84139, Abcam, Cambridge, UK) at 1:50, or anti-NFUFA9 (ab14713, Abcam) at 1:75. Detection was done using the Bond Polymer Refine Detection kit (Leica Biosystems). Stained sections were scanned with the Aperio ScanScope XT digital slide scanner (Leica Biosystems) and analyzed using the ImageScope software. For assessment of skin brown hue enrichments, we delimited basal keratinocytes as the first layer of cells above the dermis and suprabasal keratinocytes as the remaining epidermis layers above the basal layer, excluding the detaching stratum corneum. For immunocytochemistry, HEK 293T cells were transfected with empty plasmid or plasmid encoding PGC-1α or PGC-1β using lipofectamine 2000. Twenty-four hours post-transfection, cells were pelleted and fixed. PGC-1α was analyzed using the same conditions as those described above for IHC on skin tissue.
Lentiviruses
A cell-based screening of shRNA sequences against PGC-1α and β was realized using SIRION’s RNAiONE platform service (SIRION Biotech, Martinsried, Germany). Ten shRNA sequences for each PGC-1 isoform were screened in a standardized cell assay and one shRNA sequence was selected based on knockdown efficacy (Table S2). Primary foreskin NHEKs were transduced at the same multiplicity of infection (MOI 20) with highly purified lentiviruses expressing a control non-targeting shRNA (NT shRNA) or cotransduced with two aforementioned shRNA targeting PGC-1α and β (shPGC-1s). Transductions were realized according to the manufacturer’s instructions with 4 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO, USA) and 10 μM ROCK inhibitor, Y-27632 (Sigma-Aldrich). The infected cells were then harvested for cell banking. In order to obtain stable cell lines, transduced cells were selected with puromycin at 5 μg/ml for 48 hours with daily medium change and harvested for cell banking. All cell cultures were realized with G7F medium (EPISKIN, Lyon, France) according to the manufacturer’s instructions.
Human epidermis reconstruction and treatments
NT shRNA and shPGC-1s RHE were cultured up to day 19 at the air-liquid interface. The RHEs were analyzed at day 14 with Optical coherence tomography (OCT, according to the manufacturer’s instructions) to measure epidermal thickness and hematoxylin and eosin staining (HES) to visualize global morphology. For compound testing, 10μM retinol was applied systemically from day 10 to 19 during reconstruction. HES staining were realized at day 14 to visualize the morphology of tissues and OCT measurement were performed at day 12, 14, 17 and 19 to measure epidermal thickness.
Quantification and statistical analysis
All experiments were performed at least three times and all the represented graphs are based on biologically independent replicates, unless otherwise noted in figure legends. All data analyses and statistics were done using Microsoft Excel or GraphPad Prism softwares. P values were calculated by Student’s t-tests (two-tailed, usually paired) or ANOVAs with ad hoc post-tests depending on hypothesis testing and group comparisons. Transcriptomics analyses (overlap, geneset enrichments, over-representation of transcription factor binding site) are done with other softwares specified elsewhere. For all figures, data are shown as mean + SEM, unless otherwise stated.
Acknowledgments
This study was supported by research grants from L’Oréal R&I (to J.S.P) and from the Cancer Research Society (840633, to S.P.G). S.M. was recipient of a Vanier Canada Graduate Scholarship (Canadian Institutes of Health Research, CIHR). E.L. received the Bourse du centenaire - Maîtrise from the Faculté de pharmacie, Université de Montréal. J.S.P. received salary support from FRQ-S, and she holds a Canada Research Chair in Cancer Metabolism (Tier 1). D.D. is recipient of a Canada Research Chair in Chromatin Dynamics and Genome Architecture (Tier 1). Transcriptomics analyses were performed by the Genome Quebec Innovation Center (McGill University). IHC was performed by the IRIC Histology Facility, Université of Montréal. GC-MS was performed at the Goodman Cancer Institute, McGill University. We thank Etienne Gagnon for granting us access and setting up a hypoxic chamber at IRIC Institute.
Author contributions
S.P.G., S.J., L.B., and J.S.P. conceptualized the project; S.P.G., S.J., and J.S.P. designed experiments; S.P.G, Y.B.K, C.S.L, K.M.L, E.L., D.B., and S.D. performed experiments; S.P.G. and S.M. analyzed data; F.A. supervised reconstructed epidermis experiments; S.P.G. and J.S.P. wrote the manuscript; S.P.G, S.M., D.D., and J.S.P. prepared figures and edited the manuscript. All authors reviewed and authorized the manuscript; S.P.G. and J.S.P. supervised the project.
Declaration of interests
The authors have no competing interests to declare.
Published: March 2, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106314.
Contributor Information
Simon-Pierre Gravel, Email: sp.gravel@umontreal.ca.
Julie St-Pierre, Email: julie.st-pierre@uottawa.ca.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code
-
•
Transcriptomics data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE108674 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108674).
References
- 1.Niccoli T., Partridge L. Ageing as a risk factor for disease. Curr. Biol. 2012;22:R741–R752. doi: 10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Christensen K., Doblhammer G., Rau R., Vaupel J.W. Ageing populations: the challenges ahead. Lancet. 2009;374:1196–1208. doi: 10.1016/S0140-6736(09)61460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin F., Sancheti H., Liu Z., Cadenas E. Mitochondrial function in ageing: coordination with signalling and transcriptional pathways. J. Physiol. 2016;594:2025–2042. doi: 10.1113/JP270541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naylor E.C., Watson R.E.B., Sherratt M.J. Molecular aspects of skin ageing. Maturitas. 2011;69:249–256. doi: 10.1016/j.maturitas.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Wong R., Geyer S., Weninger W., Guimberteau J.C., Wong J.K. The dynamic anatomy and patterning of skin. Exp. Dermatol. 2016;25:92–98. doi: 10.1111/exd.12832. [DOI] [PubMed] [Google Scholar]
- 9.Blume-Peytavi U., Kottner J., Sterry W., Hodin M.W., Griffiths T.W., Watson R.E.B., Hay R.J., Griffiths C.E.M. Age-associated skin conditions and diseases: current perspectives and future options. Gerontol. 2016;56:S230–S242. doi: 10.1093/geront/gnw003. [DOI] [PubMed] [Google Scholar]
- 10.Humbert P., Dréno B., Krutmann J., Luger T.A., Triller R., Meaume S., Seité S. Recommendations for managing cutaneous disorders associated with advancing age. Clin. Interv. Aging. 2016;11:141–148. doi: 10.2147/CIA.S96232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalfalah F., Sobek S., Bornholz B., Götz-Rösch C., Tigges J., Fritsche E., Krutmann J., Köhrer K., Deenen R., Ohse S., et al. Inadequate mito-biogenesis in primary dermal fibroblasts from old humans is associated with impairment of PGC1A-independent stimulation. Exp. Gerontol. 2014;56:59–68. doi: 10.1016/j.exger.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Kuehne A., Hildebrand J., Soehle J., Wenck H., Terstegen L., Gallinat S., Knott A., Winnefeld M., Zamboni N. An integrative metabolomics and transcriptomics study to identify metabolic alterations in aged skin of humans in vivo. BMC Genom. 2017;18:169. doi: 10.1186/s12864-017-3547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowman A., Birch-Machin M.A. Age-dependent decrease of mitochondrial complex II activity in human skin fibroblasts. J. Invest. Dermatol. 2016;136:912–919. doi: 10.1016/j.jid.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Rinnerthaler M., Streubel M.K., Bischof J., Richter K. Skin aging, gene expression and calcium. Exp. Gerontol. 2015;68:59–65. doi: 10.1016/j.exger.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Prahl S., Kueper T., Biernoth T., Wöhrmann Y., Münster A., Fürstenau M., Schmidt M., Schulze C., Wittern K.P., Wenck H., et al. Aging skin is functionally anaerobic: importance of coenzyme Q10 for anti aging skin care. Biofactors. 2008;32:245–255. doi: 10.1002/biof.5520320129. [DOI] [PubMed] [Google Scholar]
- 16.Li L., Mac-Mary S., Sainthillier J.M., Nouveau S., de Lacharriere O., Humbert P. Age-related changes of the cutaneous microcirculation in vivo. Gerontology. 2006;52:142–153. doi: 10.1159/000091823. [DOI] [PubMed] [Google Scholar]
- 17.Bentov I., Reed M.J. The effect of aging on the cutaneous microvasculature. Microvasc. Res. 2015;100:25–31. doi: 10.1016/j.mvr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellem D., Sattler M., Pagel-Wolff S., Jaspers S., Wenck H., Rübhausen M.A., Fischer F. Fragmentation of the mitochondrial network in skin in vivo. PLoS One. 2017;12:e0174469. doi: 10.1371/journal.pone.0174469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass D., Viñuela A., Davies M.N., Ramasamy A., Parts L., Knowles D., Brown A.A., Hedman A.K., Small K.S., Buil A., et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 2013;14:R75. doi: 10.1186/gb-2013-14-7-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austin S., St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 21.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dillon L.M., Rebelo A.P., Moraes C.T. The role of PGC-1 coactivators in aging skeletal muscle and heart. IUBMB Life. 2012;64:231–241. doi: 10.1002/iub.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arany Z., Foo S.Y., Ma Y., Ruas J.L., Bommi-Reddy A., Girnun G., Cooper M., Laznik D., Chinsomboon J., Rangwala S.M., et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 24.Gleyzer N., Scarpulla R.C. Concerted action of PGC-1-related coactivator (PRC) and c-MYC in the stress response to mitochondrial dysfunction. J. Biol. Chem. 2016;291:25529–25541. doi: 10.1074/jbc.M116.719682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen N., Satija Y.K., Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Mol. Cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Redondo V., Pettersson A.T., Ruas J.L. The hitchhiker's guide to PGC-1alpha isoform structure and biological functions. Diabetologia. 2015;58:1969–1977. doi: 10.1007/s00125-015-3671-z. [DOI] [PubMed] [Google Scholar]
- 27.Rezvani H.R., Ali N., Nissen L.J., Harfouche G., de Verneuil H., Taïeb A., Mazurier F. HIF-1alpha in epidermis: oxygen sensing, cutaneous angiogenesis, cancer, and non-cancer disorders. J. Invest. Dermatol. 2011;131:1793–1805. doi: 10.1038/jid.2011.141. [DOI] [PubMed] [Google Scholar]
- 28.Carreau A., El Hafny-Rahbi B., Matejuk A., Grillon C., Kieda C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell Mol. Med. 2011;15:1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stroud D.A., Formosa L.E., Wijeyeratne X.W., Nguyen T.N., Ryan M.T. Gene knockout using transcription activator-like effector nucleases (TALENs) reveals that human NDUFA9 protein is essential for stabilizing the junction between membrane and matrix arms of complex I. J. Biol. Chem. 2013;288:1685–1690. doi: 10.1074/jbc.C112.436766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stringari C., Abdeladim L., Malkinson G., Mahou P., Solinas X., Lamarre I., Brizion S., Galey J.B., Supatto W., Legouis R., et al. Multicolor two-photon imaging of endogenous fluorophores in living tissues by wavelength mixing. Sci. Rep. 2017;7:3792. doi: 10.1038/s41598-017-03359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling C., Poulsen P., Carlsson E., Ridderstråle M., Almgren P., Wojtaszewski J., Beck-Nielsen H., Groop L., Vaag A. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J. Clin. Invest. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derbré F., Gomez-Cabrera M.C., Nascimento A.L., Sanchis-Gomar F., Martinez-Bello V.E., Tresguerres J.A.F., Fuentes T., Gratas-Delamarche A., Monsalve M., Viña J. Age associated low mitochondrial biogenesis may be explained by lack of response of PGC-1alpha to exercise training. Age (Chester) 2012;34:669–679. doi: 10.1007/s11357-011-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sczelecki S., Besse-Patin A., Abboud A., Kleiner S., Laznik-Bogoslavski D., Wrann C.D., Ruas J.L., Haibe-Kains B., Estall J.L. Loss of Pgc-1alpha expression in aging mouse muscle potentiates glucose intolerance and systemic inflammation. Am. J. Physiol. Endocrinol. Metab. 2014;306:E157–E167. doi: 10.1152/ajpendo.00578.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson L., Bowman A., Rashdan E., Birch-Machin M.A. Mitochondrial damage and ageing using skin as a model organ. Maturitas. 2016;93:34–40. doi: 10.1016/j.maturitas.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 35.Boengler K., Kosiol M., Mayr M., Schulz R., Rohrbach S. Mitochondria and ageing: role in heart, skeletal muscle and adipose tissue. J. Cachexia Sarcopenia Muscle. 2017;8:349–369. doi: 10.1002/jcsm.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesnefsky E.J., Chen Q., Hoppel C.L. Mitochondrial metabolism in aging heart. Circ. Res. 2016;118:1593–1611. doi: 10.1161/CIRCRESAHA.116.307505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villena J.A. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 38.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabol. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 39.de Magalhães J.P., Curado J., Church G.M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baruch K., Deczkowska A., David E., Castellano J.M., Miller O., Kertser A., Berkutzki T., Barnett-Itzhaki Z., Bezalel D., Wyss-Coray T., et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science. 2014;346:89–93. doi: 10.1126/science.1252945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho B.A., Yoo S.K., Seo J.S. Signatures of photo-aging and intrinsic aging in skin were revealed by transcriptome network analysis. Aging. 2018;10:1609–1626. doi: 10.18632/aging.101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savignan F., Ballion B., Odessa M.F., Charveron M., Bordat P., Dufy B. Mitochondrial membrane potential (DeltaPsi) and Ca(2+)-induced differentiation in HaCaT keratinocytes. J. Biomed. Sci. 2004;11:671–682. doi: 10.1159/000079680. [DOI] [PubMed] [Google Scholar]
- 45.Dos Santos M., Metral E., Boher A., Rousselle P., Thepot A., Damour O. In vitro 3-D model based on extending time of culture for studying chronological epidermis aging. Matrix Biol. 2015;47:85–97. doi: 10.1016/j.matbio.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Lee C.M. Fifty years of research and development of cosmeceuticals: a contemporary review. J. Cosmet. Dermatol. 2016;15:527–539. doi: 10.1111/jocd.12261. [DOI] [PubMed] [Google Scholar]
- 47.Saint-Léger D., Lévêque J.L., Verschoore M. The use of hydroxy acids on the skin: characteristics of C8-lipohydroxy acid. J. Cosmet. Dermatol. 2007;6:59–65. doi: 10.1111/j.1473-2165.2007.00296.x. [DOI] [PubMed] [Google Scholar]
- 48.Lévêque J.L., Corcuff P., Gonnord G., Montastier C., Renault B., Bazin R., Piérard G., Poelman M.C. Mechanism of action of a lipophilic derivative of salicylic acid on normal skin. Skin Res. Technol. 1995;1:115–122. doi: 10.1111/j.1600-0846.1995.tb00030.x. [DOI] [PubMed] [Google Scholar]
- 49.Hawley S.A., Fullerton M.D., Ross F.A., Schertzer J.D., Chevtzoff C., Walker K.J., Peggie M.W., Zibrova D., Green K.A., Mustard K.J., et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shamalnasab M., Gravel S.P., St-Pierre J., Breton L., Jäger S., Aguilaniu H. A salicylic acid derivative extends the lifespan of Caenorhabditis elegans by activating autophagy and the mitochondrial unfolded protein response. Aging Cell. 2018;17:e12830. doi: 10.1111/acel.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha. J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bikle D.D., Xie Z., Tu C.L. Calcium regulation of keratinocyte differentiation. Expet Rev. Endocrinol. Metabol. 2012;7:461–472. doi: 10.1586/eem.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efimova T., Broome A.M., Eckert R.L. A regulatory role for p38 delta MAPK in keratinocyte differentiation. Evidence for p38 delta-ERK1/2 complex formation. J. Biol. Chem. 2003;278:34277–34285. doi: 10.1074/jbc.M302759200. [DOI] [PubMed] [Google Scholar]
- 56.Soro-Arnaiz I., Li Q.O.Y., Torres-Capelli M., Meléndez-Rodríguez F., Veiga S., Veys K., Sebastian D., Elorza A., Tello D., Hernansanz-Agustín P., et al. Role of mitochondrial complex IV in age-dependent obesity. Cell Rep. 2016;16:2991–3002. doi: 10.1016/j.celrep.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 57.Mapuskar K.A., Flippo K.H., Schoenfeld J.D., Riley D.P., Strack S., Hejleh T.A., Furqan M., Monga V., Domann F.E., Buatti J.M., et al. Mitochondrial superoxide increases age-associated susceptibility of human dermal fibroblasts to radiation and chemotherapy. Cancer Res. 2017;77:5054–5067. doi: 10.1158/0008-5472.CAN-17-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheema N., Herbst A., McKenzie D., Aiken J.M. Apoptosis and necrosis mediate skeletal muscle fiber loss in age-induced mitochondrial enzymatic abnormalities. Aging Cell. 2015;14:1085–1093. doi: 10.1111/acel.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feichtinger R.G., Sperl W., Bauer J.W., Kofler B. Mitochondrial dysfunction: a neglected component of skin diseases. Exp. Dermatol. 2014;23:607–614. doi: 10.1111/exd.12484. [DOI] [PubMed] [Google Scholar]
- 60.Krutmann J., Schroeder P. Role of mitochondria in photoaging of human skin: the defective powerhouse model. J. Invest. Dermatol. Symp. Proc. 2009;14:44–49. doi: 10.1038/jidsymp.2009.1. [DOI] [PubMed] [Google Scholar]
- 61.Jugé R., Breugnot J., Da Silva C., Bordes S., Closs B., Aouacheria A. Quantification and characterization of UVB-induced mitochondrial fragmentation in normal primary human keratinocytes. Sci. Rep. 2016;6:35065. doi: 10.1038/srep35065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waldera-Lupa D.M., Kalfalah F., Florea A.M., Sass S., Kruse F., Rieder V., Tigges J., Fritsche E., Krutmann J., Busch H., et al. Proteome-wide analysis reveals an age-associated cellular phenotype of in situ aged human fibroblasts. Aging (Albany NY) 2014;6:856–878. doi: 10.18632/aging.100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greco M., Villani G., Mazzucchelli F., Bresolin N., Papa S., Attardi G. Marked aging-related decline in efficiency of oxidative phosphorylation in human skin fibroblasts. Faseb. J. 2003;17:1706–1708. doi: 10.1096/fj.02-1009fje. [DOI] [PubMed] [Google Scholar]
- 64.Arany Z., Wagner B.K., Ma Y., Chinsomboon J., Laznik D., Spiegelman B.M. Gene expression-based screening identifies microtubule inhibitors as inducers of PGC-1alpha and oxidative phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:4721–4726. doi: 10.1073/pnas.0800979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyahara J.T., Karler R. Effect of salicylate on oxidative phosphorylation and respiration of mitochondrial fragments. Biochem. J. 1965;97:194–198. doi: 10.1042/bj0970194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith B.K., Ford R.J., Desjardins E.M., Green A.E., Hughes M.C., Houde V.P., Day E.A., Marcinko K., Crane J.D., Mottillo E.P., et al. Salsalate (salicylate) uncouples mitochondria, improves glucose homeostasis, and reduces liver lipids independent of AMPK-beta1. Diabetes. 2016;65:3352–3361. doi: 10.2337/db16-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sasaki M., Kajiya H., Ozeki S., Okabe K., Ikebe T. Reactive oxygen species promotes cellular senescence in normal human epidermal keratinocytes through epigenetic regulation of p16(INK4a.) Biochem. Biophys. Res. Commun. 2014;452:622–628. doi: 10.1016/j.bbrc.2014.08.123. [DOI] [PubMed] [Google Scholar]
- 68.Bernard D., Gosselin K., Monte D., Vercamer C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C. Involvement of Rel/nuclear factor-kappaB transcription factors in keratinocyte senescence. Cancer Res. 2004;64:472–481. doi: 10.1158/0008-5472.can-03-0005. [DOI] [PubMed] [Google Scholar]
- 69.Hamanaka R.B., Glasauer A., Hoover P., Yang S., Blatt H., Mullen A.R., Getsios S., Gottardi C.J., DeBerardinis R.J., Lavker R.M., Chandel N.S. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci. Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pageon H., Zucchi H., Rousset F., Girardeau-Hubert S., Tancrede E., Asselineau D. Glycation stimulates cutaneous monocyte differentiation in reconstructed skin in vitro. Mech. Ageing Dev. 2017;162:18–26. doi: 10.1016/j.mad.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 71.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 72.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2018;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zambelli F., Pesole G., Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 77.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nanchen A., Fuhrer T., Sauer U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 2007;358:177–197. doi: 10.1007/978-1-59745-244-1_11. [DOI] [PubMed] [Google Scholar]
- 81.McGuirk S., Gravel S.P., Deblois G., Papadopoli D.J., Faubert B., Wegner A., Hiller K., Avizonis D., Akavia U.D., Jones R.G., et al. PGC-1alpha supports glutamine metabolism in breast cancer. Cancer Metabol. 2013;1:22. doi: 10.1186/2049-3002-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Chèneby J., Kulkarni S.R., Tan G., et al. Jaspar 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284. doi: 10.1093/nar/gkx1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carithers L.J., Ardlie K., Barcus M., Branton P.A., Britton A., Buia S.A., Compton C.C., DeLuca D.S., Peter-Demchok J., Gelfand E.T., et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobanking. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morita M., Gravel S.P., Chénard V., Sikström K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C., et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metabol. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 85.Gravel S.P., Avizonis D., St-Pierre J. Metabolomics analyses of cancer cells in controlled microenvironments. Methods Mol. Biol. 2016;1458:273–290. doi: 10.1007/978-1-4939-3801-8_20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code
-
•
Transcriptomics data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE108674 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108674).