Abstract
Background & Aims
Loss of AT-rich interactive domain-containing protein 1A (ARID1A) fosters acinar-to-ductal metaplasia (ADM) and pancreatic carcinogenesis by down-regulating transcription programs controlling acinar cell identity. However, how ARID1A reacts to metaplasia-triggering environmental cues remains elusive. Here, we aimed to elucidate the role of ARID1A in controlling ductal pancreatic gene signatures and deciphering hierarchical signaling cues determining ARID1A-dependent chromatin regulation during acinar cell reprogramming.
Methods
Acinar cell explants with differential ARID1A status were subjected to genome-wide expression analyses. The impact of epidermal growth factor receptor (EGFR) signaling, NFATc1 activity, and ARID1A status on acinar reprogramming processes were characterized by ex vivo ADM assays and transgenic mouse models. EGFR-dependent ARID1A chromatin binding was studied by chromatin immunoprecipitation sequencing analysis and cellular fractionation.
Results
EGFR signaling interferes with ARID1A-dependent transcription by inducing genome-wide ARID1A displacement, thereby phenocopying ARID1A loss-of-function mutations and inducing a shift toward ADM permissive ductal transcription programs. Moreover, we show that EGFR signaling is required to push ARID1A-deficient acinar cells toward a metaplastic phenotype. Mechanistically, we identified the transcription factor nuclear factor of activated T cells 1 (NFATc1) as the central regulatory hub mediating both EGFR signaling-induced genomic ARID1A displacement and the induction of ADM-promoting gene signatures in the absence of ARID1A. Consequently, pharmacologic inhibition of NFATc1 or its depletion in transgenic mice not only preserves genome-wide ARID1A occupancy, but also attenuates acinar metaplasia led by ARID1A loss.
Conclusions
Our data describe an intimate relationship between environmental signaling and chromatin remodeling in orchestrating cell fate decisions in the pancreas, and illustrate how ARID1A loss influences transcriptional regulation in acinar cell reprogramming.
Keywords: Acinar-to-Ductal Metaplasia, ARID1A, EGFR, Pancreas, Transcription, NFATc1
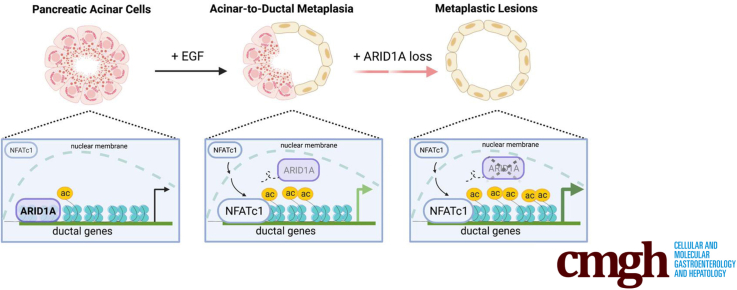
Graphical abstract
Summary.
Epidermal growth factor receptor signaling induces genome-wide AT-rich interactive domain-containing protein 1A (ARID1A) displacement, thus phenocopying Arid1a loss-of-function mutations and facilitating transcriptional activation of ductal genes during acinar-to-ductal metaplasia. Downstream of epidermal growth factor receptor, NFATc1 serves as the regulatory hub mediating ARID1A genomic displacement and induction of signatures promoting pancreatic reprogramming.
Reprogramming of acinar cells, the exocrine pancreatic compartment, is broadly accepted as the initiating event of pancreatic pathogenesis.1 In response to external (eg, inflammation) or internal (eg, mutation of oncogenic drivers) pressure, acinar cells lose their grape-like structure, stop producing digestive enzymes, and transdifferentiate toward ductal-like lesions.2 This process, termed acinar-to-ductal metaplasia (ADM), is critically involved in pancreatitis and pancreatic regeneration,3,4 and represents the initial step of pancreatic carcinogenesis.5 Although full progression of ADM toward invasive pancreatic ductal adenocarcinoma (PDAC) requires at least the activating mutation of the KRAS oncogene and is accompanied regularly by the accumulation of additional genetic alterations,5 acinar reprogramming itself can arise concurrently or independently of genetic events occurring in the acinar cell.
At the molecular level, acinar reprogramming generally is provoked by environmental factors that converge on highly dynamic and potentially reversible epigenetic processes that may cause a transient and reversible dedifferentiation state.2,6, 7, 8 Based on their critical involvement in controlling chromatin accessibility, chromatin remodeling processes play a pivotal role in orchestrating cell fate decision programs. Indeed, altered chromatin remodeling can critically disturb the transcriptional balance required to maintain cellular integrity, thus fostering dedifferentiation and malignant progression.9,10 This is particularly true for alterations of the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex. SWI/SNF proteins assemble to form a multisubunit complex, which remodels chromatin by inserting, shifting, or evicting nucleosomes in an adenosine triphosphate–dependent manner.11 The SWI/SNF complex, and particularly its DNA-binding component AT-rich interactive domain-containing protein 1A (ARID1A), is crucial for enforcing and maintaining terminal mammalian differentiation (eg, of hepatocytes12 and endometrial13,14 and gastric cells15).
The recognition that loss-of-function ARID1A mutations occur in approximately 6% of PDACs16 has prompted multiple studies investigating the impact of ARID1A deficiency on pancreatic pathogenesis.17, 18, 19, 20 Using genetically engineered mouse models and additional cutting-edge technologies to genetically target Arid1a, these reports suggest that loss of Arid1a counteracts acinar cell identity and cooperates with oncogenic Kras in driving pancreatic carcinogenesis.17, 18, 19,21, 22, 23 Mechanistically, loss of ARID1A in these models was associated with decreased chromatin accessibility and rapid transcriptional shifts with significant down-regulation of gene expression programs sustained by master acinar transcription factors (TFs).18,23 However, abrogation of acinar transcription programs is not sufficient for the full conversion toward a metaplastic pancreatic phenotype. Rather, it relies on the additional induction of ductal gene signatures that frequently are controlled by hierarchical environmental signaling cues.2,24 Whether ARID1A is involved directly in the regulation of ductal transcription programs and how upstream signaling cues converge on ARID1A-dependent gene regulation, however, remains elusive.
Herein, we show a hitherto unappreciated role of epidermal growth factor receptor (EGFR) signaling in transcriptional reprogramming processes of the ARID1A-deficient pancreas and illustrate that EGFR activity can phenocopy ARID1A deficiency in driving ADM formation by reducing genome-wide ARID1A occupancy. Impaired ARID1A genome binding caused either by genetic Arid1a loss or occurring in response to EGFR signaling increases the expression of ductal transcription programs. Mechanistically, we identify the TF nuclear factor of activated T cells 1 (NFATc1) as the regulatory hub mediating both EGFR-signaling–induced genomic displacement of ARID1A and the subsequent induction of ductal gene signatures promoting acinar cell reprogramming. Hence, our findings provide mechanistic and functional insights into the close entanglement of environmental signaling cues, TF activity, and chromatin remodeling in acinar cell plasticity.
Results
Pancreatic Metaplasia in the Context of Arid1a Deficiency Involves the Activation of EGFR-Signaling–Dependent Transcription Programs
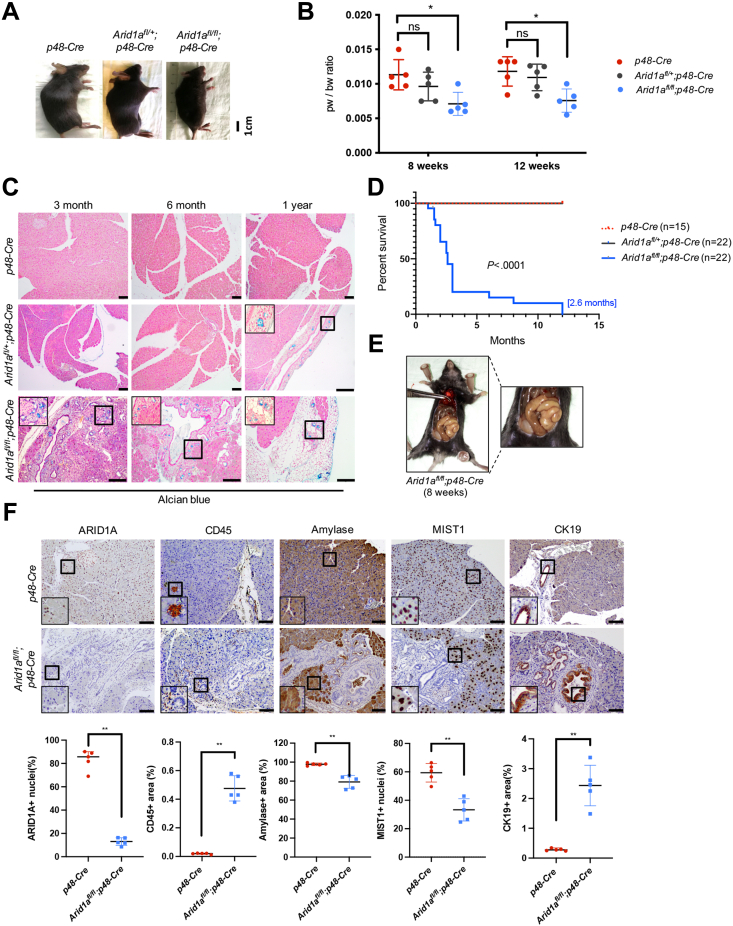
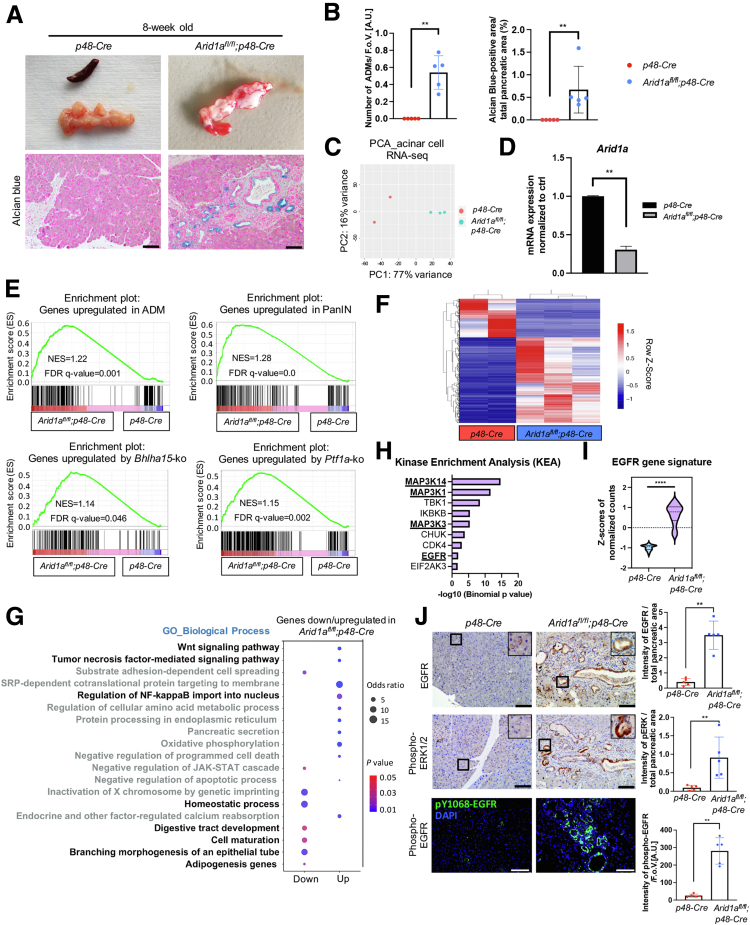
To explore whether reprogramming of the pancreatic architecture in the context of Arid1a deficiency is restricted to the abrogation of transcription programs required for acinar cell maintenance, or also involves the activation of metaplasia-promoting transcription programs, we took advantage of a transgenic mouse model of pancreas-specific heterozygous or homozygous Arid1a deficiency (Arid1afl/+;p48-Cre- or Arid1afl/fl;p48-Cre mice). Although Arid1afl/+;p48-Cre mice were phenotypically and histologically indistinguishable from p48-Cre control littermates until 1 year of age (Figure 1A–D), Arid1afl/fl;p48-Cre mice showed characteristics of exocrine pancreatic insufficiency (Figure 1A–C and E) and had a dramatically reduced median survival of 2.6 months (Figure 1D). A critical reduction of the acinar cell compartment with attenuated expression of acinar cell markers amylase and MIST1, and an abundance of CD45-positive immune cells already was evident in 8-week-old Arid1afl/fl;p48-Cre mice (Figure 1F). Over time, the pancreas of Arid1afl/fl;p48-Cre mice continued to lose acinar cell mass and accumulated fat vacuoles, implying pancreatic atrophy (Figure 1C). In line with previous reports,19, 20, 21 remodeling of the pancreatic architecture in Arid1a-deficient mice encompassed macroscopic cystic lesions and ductal structures consistent with ADM or mucin-producing and cytokeratin 19–positive pancreatic intraepithelial neoplasia (PanIN) (Figures 2A and B and 1F). Consistent with these phenotypic findings, RNA sequencing (RNA-seq) and subsequent gene set enrichment analysis (GSEA) from pancreatic acinar cells of 8-week-old p48-Cre and Arid1afl/fl;p48-Cre mice linked Arid1a deficiency to transcription programs associated with acinar reprogramming and ADM and PanIN formation (Figure 2C–E). Notably, we found that approximately two thirds (642 of 928) of the differentially expressed genes (false discovery rate [FDR], <0.05) were up-regulated in acinar cells isolated from Arid1a-deficient mice compared with p48-Cre mice (Figure 2F). Interestingly, Gene Ontology analysis conducted separately for the sets of up-regulated and down-regulated genes showed a shift from pathways associated with cell homeostasis toward signatures linked to inflammatory cytokines and TFs (Figure 2G). In particular, gene signatures regulated by EGFR, a pivotal promoter of ADM formation and pancreatic carcinogenesis,6,24, 25, 26, 27 or EGFR downstream factors (eg, Mitogen-Activated Protein Kinase Kinase Kinase 1), were activated prominently in Arid1a-deficient acinar cells (Figure 2H and I). Interestingly, we also detected an abundance of total and active (Y1068-phosphorylated) EGFR and phosphorylated extracellular signal-regulated kinase (pERK1/2) expression in metaplastic pancreatic lesions of the Arid1a-deficient pancreas (Figure 2J).
Figure 1.
Arid1a deficiency alters acinar cell identity and leads to pancreatic atrophy. (A) Representative images of 8-week-old p48-Cre, Arid1afl/+;p48-Cre and Arid1afl/fl;p48-Cre mice. (B) Scatter dot plot showing the relative pancreas weight (pw) to body weight (bw) ratio of p48-Cre, Arid1afl/+;p48-Cre, and Arid1afl/fl;p48-Cre mice. n = 5 mice/genotype and time point. (C) Alcian blue staining in the mice pancreas of indicated genotype and age. Scale bars: 100 μm. Images were captured with a microscope-attached Olympus SC180 using Cellsens Entry software. (D) Kaplan–Meier overall survival analysis of p48-Cre, Arid1afl/+;p48-Cre, and Arid1afl/fl;p48-Cre mice. The median survival of the homozygous Arid1a-knockout cohorts is shown in brackets. Statistical significance was determined by the log-rank (Mantel–Cox) test. (E) Representative image of a swollen gut found in an Arid1afl/fl;p48-Cre survival mouse. (F) From the left side, representative images and corresponding quantification of ARID1A, CD45, amylase, MIST1, and cytokeratin 19 (CK19) staining performed in 8-week-old p48-Cre and Arid1afl/fl;p48-Cre mice. n = 5 mice/genotype. Scale bars: 100 μm. The quantitative evaluation of immunohistochemical staining was conducted with 5 mice/genotype by ImageJ version 1.53. (B and F) Data are presented as means ± SD and statistical significance was determined by a 2-tailed Mann–Whitney test. ∗P < .05, ∗∗P < .01.
Figure 2.
Arid1a loss induces gene signatures associated with EGFR activity. (A) Macroscopic images with corresponding Alcian blue staining of 8-week-old p48-Cre and Arid1afl/fl;p48-Cre mice. Scale bars: 100 μm. (B) Scatter dot plots showing the percentage of the pancreatic area containing ADM (left) and Alcian blue–positive lesions (right) in 8-week-old p48-Cre and Arid1afl/fl;p48-Cre mice. Each dot represents a mouse. (C) Principal component analysis (PCA) of acinar cell RNA-seq showing the separate clusters of the 2 genotypes. n = 2 of p48-Cre and n = 3 of Arid1afl/fl;p48-Cre mice. (D) Acinar cells isolated from mice of indicated genotypes were subjected to qRT-PCR analysis to determine Arid1a expression before RNA-seq (normalized to Rplp0). (E) GSEA of ADM, PanIN gene signatures, and genes regulated by acinar-specific transcription factors basic helix-loop-helix family member A15 and PTF1A28, 29, 30, 31 in 8-week-old Arid1afl/fl;p48-Cre vs p48-Cre mice. (F) Heatmap displaying Z-score–transformed relative expression levels of differentially expressed genes (DESeq FDR, <0.05) in acinar cells of 8-week-old p48-Cre- and Arid1afl/fl;p48-Cre mice. (G) Dot plot showing the down-regulated and up-regulated Gene Ontology (GO) biological process categories in Arid1afl/fl;p48-Cre acinar cells with the highest statistical significance (Fisher exact P value < .05). The size of the dot is based on the odds ratio, and the color shows the enrichment significance. (H) Kinase enrichment analysis of genes up-regulated in Arid1afl/fl;p48-Cre mice (log2 fold change, >1; FDR, <0.05) using Enrichr software. (I) Violin plot depicting Z scores of EGFR gene signature in p48-Cre and Arid1afl/fl;p48-Cre mice. Dashed line and solid line indicate the median and quartiles, respectively. (J) Representative images and corresponding quantification of EGFR and phospho-ERK1/2 immunohistochemical staining as well as phospho-EGFR (Y1068) immunofluorescence staining performed in 8-week-old p48-Cre and Arid1afl/fl;p48-Cre mice. Scale bars: 100 μm. 4′,6-diamidino-2-phenylindole (DAPI) visualizes cell nuclei. Each dot represents 1 mouse. Images of Alcian blue staining in panel A and immunohistochemistry in panel J were captured with a microscope-attached Olympus SC180 using Cellsens Entry software and with Leica DMi8 microscope for photos of immunofluorescence. The quantitative evaluation of ADM/PanIN regions and immunohistochemistry or immunofluorescence staining were conducted with 5 mice per genotype by ImageJ version 1.53. (B, D, and J) Data are presented as means ± SD. (B, D, I, and J) The 2-sided Mann–Whitney test was used to determine significance. ∗∗P < .01, and ∗∗∗∗P < .0001. AU, arbitrary unit; CDK4, Cyclin-dependent kinase 4; CHUK, conserved helix-loop-helix ubiquitous kinase; EIF2AK3, Eukaryotic Translation Initiation Factor 2 Alpha Kinase 3; FDR, false discovery rate; FoV, field of view; IKBKB, Inhibitor of nuclear factor kappa-B kinase subunit beta; JAK/STAT, the Janus kinase/signal transducer and activator of transcription; NES, normalized enrichment score; NF, nuclear factor; PC1, principal component 1; TBK1, TANK Binding Kinase 1; Wnt, Wingless/Integrated signaling.
Together, these data suggest that activation of EGFR-dependent signaling and transcription programs occurs during cellular reprogramming in the Arid1a-deficient pancreas.
EGFR Signaling Cooperates With Arid1a Deficiency in Driving Acinar Cell Reprogramming
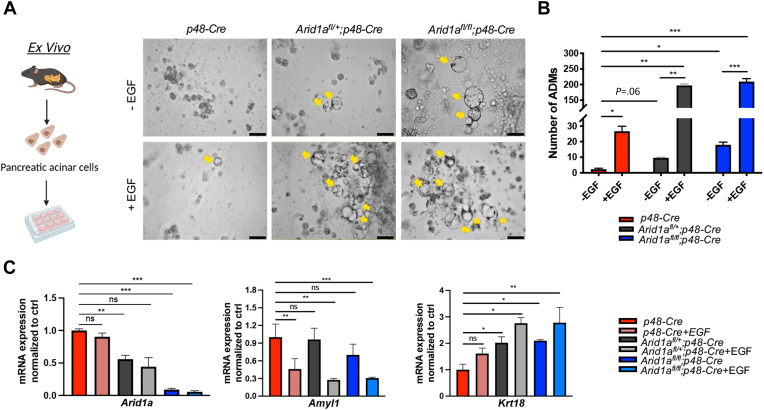
To further characterize the influence of ARID1A on the acinar cell–intrinsic potential to transdifferentiate toward metaplastic lesions, we isolated acinar cells from control p48-Cre, Arid1afl/+;p48-Cre, and Arid1afl/fl;p48-Cre mice and cultured them in a collagen-based matrix permissive for in vitro ADM formation. To additionally explore the connection between ARID1A function and EGFR signaling in ADM formation, these studies were performed in the presence or absence of EGF. Consistent with our previous findings,6 EGF treatment efficiently induced ADM formation in vitro (Figure 3A and B). Notably, loss of ARID1A expression increased the number of duct-like cells in vitro, even in the absence of exogenous EGF treatment. Remarkably, the addition of EGF further increased ADM formation in Arid1a-deficient acinar cells (Figure 3A–C), suggesting a cooperating role of EGFR signaling activation and Arid1a deficiency in ADM formation in contrast to a mere downstream effect. In accordance with the observed in vitro ADM phenotype, Amyl1 and Krt18 expression decreased and increased, respectively, upon EGF treatment and/or Arid1a loss (Figure 3C).
Figure 3.
EGFR signaling activation cooperates with Arid1a deficiency in promoting reprogramming of primary acinar cells. (A and B) Ex vivo culture of acinar explants derived from p48-Cre, Arid1afl/+;p48-Cre, and Arid1afl/fl;p48-Cre mice in the absence or presence of EGF (40 ng/mL). Representative (A) images and (B) quantification of ADM numbers from 3 experimental replicates on the fifth day after seeding. Yellow arrows denote ADM structures. Scale bars: 50 μm. Acinar explant culture was conducted independently 3 times. The equal number of isolated acinar cells from each genotype was dispensed into experimental duplicates and counted, respectively. (C) qRT-PCR analysis of Arid1a, Amyl1, and Krt18 (normalized to Rplp0) messenger RNA (mRNA) expression in ADM isolated on the fifth day after seeding. (A) Images were taken with a Leica DMi8 microscope. (B and C) Data are presented as means ± SD with P values shown on the top of each plot. (B and C) Statistical significance was determined by a 2-tailed unpaired Student t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
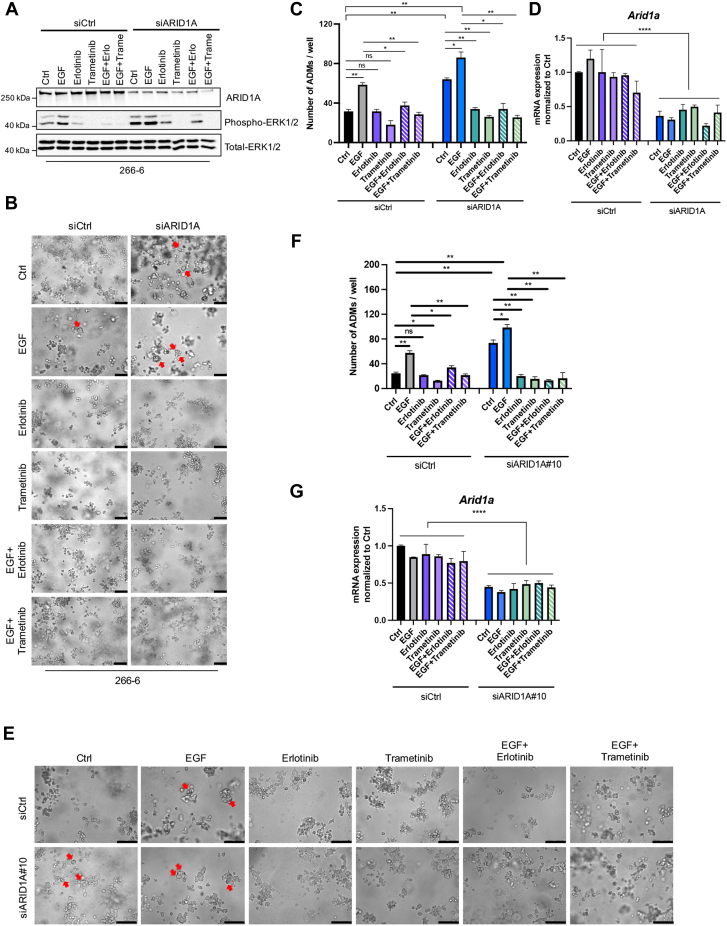
To confirm our findings in an additional pancreatic model system, we took advantage of the Kras mutant acinar cell line 266-6.25 Consistent with our findings in Arid1a-deficient mice (Figure 2J), ARID1A knockdown (small interfereing RNA targeting ARID1A, siARID1A) in 266-6 cells increased mitogen-activated protein kinase (MAPK) signaling activity compared with ARID1A-expressing (siCtrl) cells, both at the basal state and upon EGF stimulation (Figure 4A). Importantly, ligand-induced activation of EGFR signaling and knockdown of ARID1A also strongly promoted ADM formation in the 266-6 model, with maximal lesion formation visible under combined conditions (Figure 4B–G). To explore whether the intrinsic and EGF-induced activation of EGFR/MAPK signaling is causatively involved in driving acinar-to-ductal transdifferentiation upon ARID1A knockdown, we determined ADM formation upon pharmacologic blockade of EGFR or Mitogen-activated protein kinase kinase/ERK activity by administration of erlotinib and trametinib, respectively. Erlotinib and, to an even stronger extent, trametinib, reduced the ADM formation capacities of 266-6 cells with active EGFR signaling (Figure 4). Notably, this effect was particularly evident in the absence of ARID1A, in which both inhibitors reduced ADM formation to the level observed in untreated ARID1A-proficient cells (Figure 4B–G).
Figure 4.
Pharmacologic inhibition of EGFR/MAPK signaling blocks ADM formation of Arid1a-depleted acinar cells. (A) Immunoblot analysis of ARID1A, phospho-ERK1/2, and total ERK1/2 in 266-6 cells transfected with siCtrl or siARID1A and treated with erlotinib (Erlo) or trametinib (Trame) in the presence or absence of EGF (40 ng/mL). Representative (B) images and (C) quantification of ADM culture of 266-6 cells in the indicated conditions. ARID1A was silenced using pooled siRNAs comprising siARID1A #7 (J-017263-07; Dharmacon) and #8 (J-017263-08; Dharmacon). EGF, 40 ng/mL; erlotinib, 300 nmol/L; trametinib, 20 nmol/L. ADM quantification from 266-6 cells was performed from 2 experiments with duplicate wells per condition. The number of ductal structures was quantified from duplicate wells 4 days after seeding and treatment. Red arrows denote ADM structures. Scale bars: 50 μm. n = 2. (D) qRT-PCR analysis of Arid1a messenger RNA (mRNA) expression (normalized to Rplp0) at the fourth day after being seeded for ADM assay shown in panel B. n = 2. Representative (E) images and (F) quantification of ADM culture of 266-6 cells in the indicated conditions. ARID1A was silenced using siARID1A#10 (J-040694-10; Dharmacon). EGF, 40 ng/mL; erlotinib, 300 nmol/L; and trametinib, 20 nmol/L. The number of ductal structures was quantified from duplicate wells 4 days after seeding and treatment. Red arrows denote ADM structures. Scale bars: 50 μm. n = 2. (G) qRT-PCR analysis of Arid1a mRNA expression (normalized to Rplp0) on the fourth day after being seeded for ADM assay shown in panel E. n = 2. (B and E) Images were taken with a Leica DMi8 microscope. (C, D, F, and G) Data are presented as means ± SD. (C and F) Statistical significance was determined by an unpaired t test and by 2-way analysis of variance, followed by the Tukey multiple comparisons test in panels D and G between transfection groups. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001.
Together, these data imply that EGFR signaling and loss of ARID1A function together in promoting ADM formation and suggest that interference with EGFR signaling activity prevents ARID1A-deficient cells from undergoing acinar-to-ductal transdifferentiation.
ARID1A Deficiency and EGFR Signaling Cooperatively Activate Gene Signatures Involved in Cellular Plasticity and Acinar Cell Reprogramming
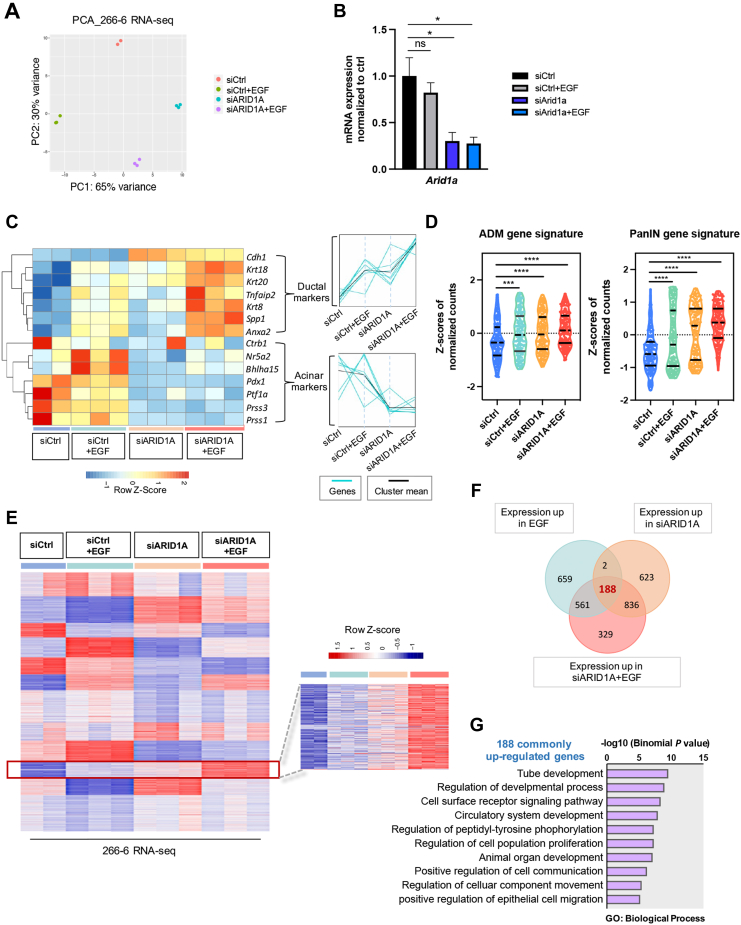
Given the implication of EGFR signaling activity for acinar-to-ductal transdifferentiation in ARID1A-deficient cells, we sought to gain further insights into the extent of the cooperativity between the effects of ARID1A deficiency and EGFR activity by studying the underlying ADM-promoting transcription programs. To this end, we performed RNA-seq analysis in ARID1A-proficient and ARID1A-depleted 266-6 cells in the presence and absence of exogenous EGF treatment (Figure 5A and B). Analysis of genes previously described to characterize either the acinar or the ductal state of pancreatic epithelial cells32,33 showed that untreated siCtrl 266-6 cells showed high expression of acinar and low expression of ductal markers (Figure 5C). Either EGF treatment or ARID1A knockdown each induced a shift from the expression of acinar toward ductal markers, while a more complete switch of acinar to ductal gene expression was detected upon combining both conditions (Figure 5C). A cooperative effect of ARID1A deficiency and EGFR activity was restricted to the induction of ductal genes, while EGF treatment did not further reduce the expression of acinar genes already down-regulated by ARID1A knockdown (Figure 5C). Further supporting our hypothesis that ARID1A deficiency and EGFR signaling cooperate particularly in the transcriptional reprogramming of acinar cells, gene signatures associated with ADM or PanIN formation28,29 reached maximal enrichment upon combined ARID1A knockdown and EGF treatment (Figure 5D). Based on these findings we focused our further analyses on the gene cluster displaying cooperative up-regulation of expression after activation of EGFR signaling and ARID1A loss (Figure 5E, red box, and F). Gene Ontology analysis linked these 188 cooperatively regulated genes with processes such as organ regeneration, morphogenesis, and growth factor responses (Figure 5G).
Figure 5.
Arid1a deficiency and EGFR signaling cooperate for transcriptional activation of gene signatures driving cell plasticity. (A) PCA of RNA-seq data of 266-6 cells indicating the distinct clusters of the 4 conditions. (B) ARID1A knockdown was validated in 266-6 RNA-seq samples as measured by qRT-PCR (normalized to Rplp0). Data are presented as means ± SD. Statistical significance was determined by an unpaired t test. ∗P < .05. (C) Heatmap and trend analysis of expression profiles of ductal and acinar markers in 266-6 cells in the presence or absence of Arid1a and EGF determined by RNA-seq. Heatmap showing reads Per kilobase of transcript per million reads mapped (RPKM)-transformed Z-scores (RPKM, >0; FDR, <0.05) and the corresponding visualization of the expression pattern of ductal and acinar clusters in the indicated conditions by Short Time-Series Expression Miner. (D) Z-scores of the ADM34 and PanIN gene signatures35 in 266-6 cells in the presence or absence of Arid1a and EGF was determined by RNA-seq. Median and quartiles are presented on the plot and a 2-tailed unpaired t test was used to determine significance. (E) Heatmap showing the k-means clustering (k = 12) of RPKM-transformed Z-scores (RPKM, >0) in the indicated conditions upon RNA-seq studies in 266-6 cells. The magnification on the right shows the cluster marked by the red box where EGFR activation and ARID1A knockdown cooperate in inducing transcription. (F) Venn diagram showing the overlap of up-regulated genes upon either EGFR stimulation, ARID1A knockdown, or combined conditions. Genes were selected as fold change greater than 0.35, FDR ≤0.05. (G) Gene Ontology (GO) analysis showing the top-ranked biological-process pathways of the 188 cooperatively up-regulated genes shown in panel F. A binomial test was performed and all the pathways had a FDR less than 0.05. PCA, principal component analysis; PC1, pincipal component 1; PC2, principal component 2.
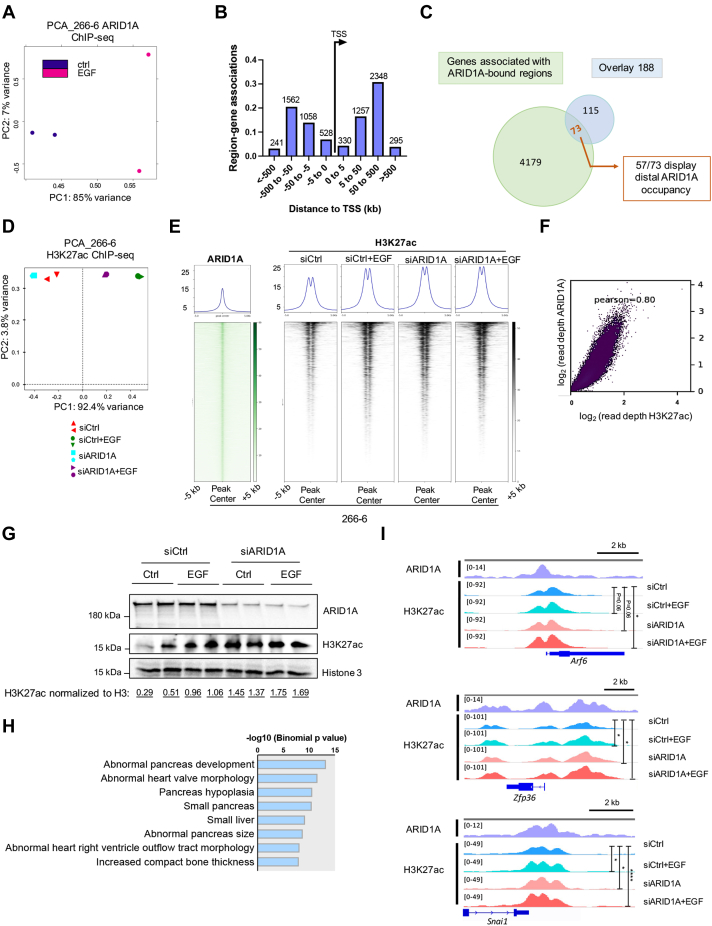
Next, we sought to determine the direct role(s) of ARID1A in eliciting the cooperative effects observed after ARID1A deficiency and activation of EGFR signaling. Accordingly, we performed ARID1A chromatin immunoprecipitation sequencing (ChIP-seq) in 266-6 cells (Figure 6A). Consistent with previous reports studying genome-wide occupancy of SWI/SNF family proteins,14,36 Genomic Regions Enrichment Annotation Tool analysis showed that ARID1A occupied predominantly distal genomic regions 50- to 500-kb upstream or downstream of the transcription start site (TSS) (Figure 6B). Notably, 73 of the 188 (39%) genes with cooperative transcriptional activation upon ARID1A knockdown and EGFR signaling activation also were occupied by ARID1A, with the majority of these genes (n = 57) displaying distal ARID1A occupancy (Figure 6C).
Figure 6.
ARID1A binds active chromatin and ARID1A loss leads to enhanced H3K27ac genome-wide. (A) Principal component analysis (PCA) of ARID1A ChIP-seq (n = 2). (B) Peak distribution in relation to the TSS of ARID1A ChIP-seq data in 266-6 cells by Genomic Regions Enrichment Annotation Tool (GREAT) analysis. (C) Venn diagram showing the overlap of 188 cooperatively up-regulated genes and ARID1A target genes shown by ChIP-seq. A large portion of the overlay (57 of 73) display distal ARID1A occupancy. (D) PCA of H3K27ac ChIP-seq of the indicated conditions in 266-6 cells. n = 2. (E) Average binding profiles and heatmaps depicting the H3K27ac and ARID1A ChIP-seq signals across the regions bound by ARID1A (peaks were called by MACS2 with FDR < 0.05, n = 4253 regions). (F) Pearson correlation plot of H3K27ac and ARID1A ChIP-seq peaks. (G) Immunoblot analysis of total H3K27ac levels in the indicated conditions in 266-6 cells. Numbers below the image represent H3K27ac band intensities normalized to H3. (H) Plot of significantly enriched mouse phenotype gene sets of distal (upon excluding TSS ± 1 kb) ARID1A target genes with H3K27ac occupancy. Associated genes were identified by GREAT analysis with basal plus extension association rule with default settings. The x-axis corresponds to the raw binomial P values. (I) Occupancy profiles of ARID1A and H3K27ac at Arf6, Zfp36, and Snai1 genes showing the significant increase of H3K27ac upon combined ARID1A knockdown and EGFR stimulation compared with the control condition. Statistical significance is shown on the graph and was determined by DiffBind analysis. ∗P < .05, ∗∗∗∗P < .0001. PC1, principal component 1; PC2, principal component 2.
Based on our finding that ARID1A primarily occupied intergenic putative enhancer regions, we performed additional ChIP-seq analyses for Histone 3 lysine 27 acetylation (H3K27ac), a histone mark associated with transcriptionally active TSS and enhancer regions,37 in 266-6 cells (Figure 6B and D). Interestingly, despite the high genome-wide overlap between ARID1A and H3K27ac occupancy in treatment-naïve 266-6 conditions (Figure 6E and F), these regions showed an increase in H3K27ac occupancy upon either EGF treatment or ARID1A knockdown alone, and the combination of both conditions (Figure 6E and G). Interestingly, Mouse Genome Informatics phenotype analysis performed specifically in the subset of distal ARID1A-bound and transcriptionally active regions (defined as H3K27ac-occupied regions minus TSS ± 1 kb) supported a critical role of ARID1A in controlling pancreatic cell fate (Figure 6H). Notably, direct ARID1A target genes such as Arf6, Zfp36, or Snai1, which foster acinar reprogramming by directly controlling acinar-to-ductal conversion or by driving inflammation or epithelial–mesenchymal transition,38, 39, 40 displayed a cooperative increase in H3K27ac occupancy (Figure 6I). Together, these data indicate that loss of ARID1A and EGFR activation cooperate in promoting a chromatin state permissive for transcriptional activation of gene signatures supporting acinar-to-ductal transdifferentiation processes.
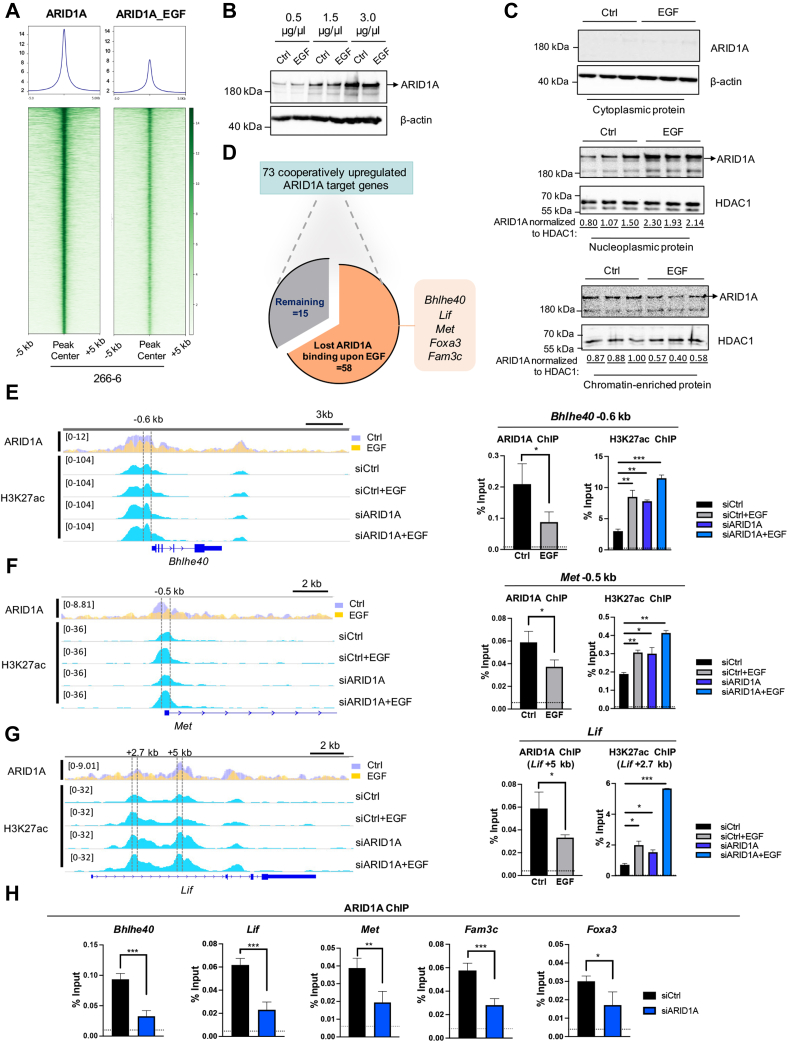
EGFR Signaling Induces Genome-Wide ARID1A Displacement
Given the significant similarity in the effects elicited by the activation of EGFR signaling or loss of ARID1A on acinar reprogramming, we hypothesized that EGFR signaling may function, at least in part, by directing ARID1A-dependent chromatin regulation. Therefore, we examined global ARID1A occupancy by ChIP-seq after EGF treatment. Interestingly, activation of EGFR signaling led to significantly decreased ARID1A occupancy genome-wide (Figure 7A), while EGF treatment did not affect total ARID1A protein expression (Figure 7B). To confirm genome-wide reduction of ARID1A occupancy, we performed chromatin fractionation studies in 266-6 cells in the absence or presence of EGF treatment. Indeed, consistent with our ChIP-seq results, EGF treatment induced a shift of ARID1A enrichment from the chromatin-bound fraction to the nucleoplasmic fraction (Figure 7C), indicating that EGFR signaling activation results in a genome-wide displacement of ARID1A. Consistently, the vast majority of direct ARID1A targets displaying cooperative up-regulation upon EGF treatment and ARID1A knockdown (58 of the 73; 79%) showed decreased ARID1A occupancy after activation of EGFR signaling (Figure 7D). These genes comprised candidates involved in developmental processes,41,42 epithelial–mesenchymal transition,43, 44, 45 and genes associated with a dismal pancreatic cancer prognosis,46, 47, 48 thus further emphasizing the role of ARID1A in counteracting cellular processes involved in metaplasia and transdifferentiation. Differential ARID1A and H3K27ac occupancies were validated in independent ChIP studies at the representative target genes Bhlhe40, Met, and Lif (Figure 7E–G). The specificity of the ARID1A ChIP-(seq) results was confirmed by reduced occupancy upon ARID1A depletion (Figure 7H). Together, these data indicate that EGFR signaling not only cooperates with ARID1A deficiency to drive ADM, but critically impedes ARID1A binding to its target genes, thus creating a chromatin state permissive for the recruitment of TFs or alternative chromatin regulatory proteins driving metaplastic transcription programs.
Figure 7.
EGFR signaling induces genome-wide ARID1A displacement from the genome. (A) Average binding profiles and heatmaps showing the occupancy of ARID1A before and after EGF treatment (40 ng/mL, 8 hours) as identified by ChIP-seq analysis. (B) Western blot with different loading dosages shows that total ARID1A expression in whole-cell lysates obtained from 266-6 cells is unaffected by EGF stimulation. β-actin serves as loading control. (C) Immunoblot analysis of ARID1A performed with fractionated cell lysates. β-actin was used as the loading control for cytoplasmic protein and histone deacetylase 1 (HDAC1) for the nuclear protein. Numbers shown below the blot represent ARID1A band intensities normalized to histone deacetylase 1 (HDAC1). (D) Pie chart summarizes the proportion of regions that lose ARID1A binding (FDR, <0.05; log2 fold change, <-0.5 as determined by DiffBind) within the aforementioned 73 ARID1A-occupied genes (shown in Figure 6C) in response to EGFR activation. Box shows an excerpt of the final 58 target genes that lose ARID1A binding upon EGF treatment. Genome browser screenshots of ARID1A and H3K27ac ChIP-seq analysis (left) and the individual verifications of ARID1A and H3K27ac enrichment by ChIP-qPCR (right) of (E) Bhlhe40, (F) Met, and (G) Lif. Dotted line represents IgG. (H) Validation of ARID1A enrichment at Bhlhe40, Lif, Met, Fam3c, and Foxa3 genes after siRNA-mediated ARID1A depletion. Dotted line represents IgG. (E–H) Data are presented as means ± SD. Statistical significance was determined by an unpaired t test. n = 2. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
The Inflammatory TF NFATc1 Is Critically Involved in Transcriptional Reprogramming in Response to ARID1A Deficiency and EGFR Signaling
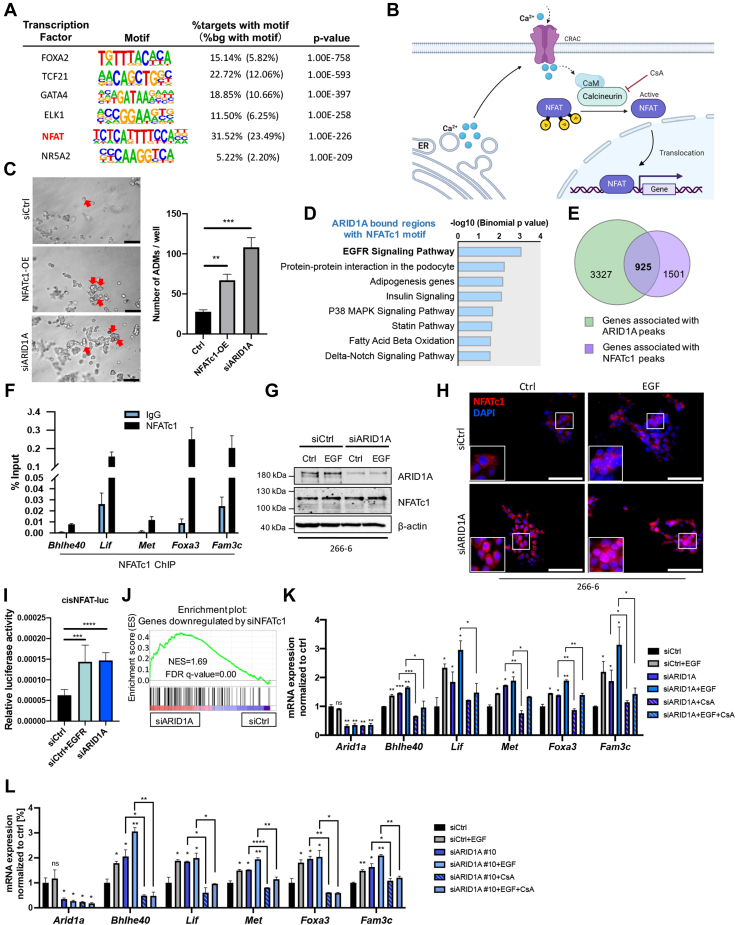
Next, we sought to identify TFs that counteract ARID1A in maintaining acinar cell identity, particularly in the context of EGFR activation. Motif analysis of ARID1A-occupied genomic regions showed binding sites of developmental TFs such as FOXA2, GATA4, and NR5A2. Moreover, and in accordance with the identification of enriched inflammatory signaling in the context of Arid1a deficiency (Figure 2G), we also identified the motif of the inflammatory NFAT (Figure 8A). NFAT proteins represent calcium-/calcineurin-responsive TFs, which are critically involved in the regulation of transcription programs linked to inflammation, differentiation, and cell growth (Figure 8B).49,50 Consistent with the phenotype observed upon ARID1A loss, constitutive activation of the NFAT family member NFATc1 drives ADM in vitro (Figure 8C) and triggers pancreatic atrophy and ADM in in vivo models.2 Interestingly, pathway analysis of genes harboring the NFAT consensus site within ARID1A-bound genomic regions showed EGFR signaling-related processes as top hits (Figure 8D). These findings not only support the critical role of NFATc1 downstream of EGFR,6 but suggested that NFATc1 may function as a pivotal driver of the activation of ADM-permissive gene signatures in the context of Arid1a deficiency. By comparing the binding profiles of ARID1A and NFATc1,49 we found that 38% of NFATc1-enriched genomic regions overlapped with ARID1A (Figure 8E). Targeted ChIP-analysis after EGF treatment confirmed inducible NFATc1 binding to a subset of these genes (Figure 8F). Although EGFR activity and the ARID1A status did not impact total NFATc1 expression in 266-6 cells (Figure 8G), subcellular fractionation studies showed increased nuclear NFATc1 localization upon EGF treatment and/or ARID1A knockdown (Figure 8H). Consistently, an NFAT-responsive luciferase reporter system indicated a significant increase of luciferase activity upon overexpression of EGFR (Figure 8I). Moreover, knockdown of ARID1A had a comparable effect on NFAT-dependent transactivation, suggesting that loss of ARID1A, similar to EGFR activation, fosters NFAT-driven gene transcription (Figure 8I). In line with these findings, GSEA showed a strong enrichment of NFATc1-dependent genes50 in the ARID1A knockdown condition (Figure 8J). Consistently, blockage of nuclear translocation of NFATc1 by treatment with cyclosporine A (CsA) significantly prevented target genes from being induced by EGF treatment and ARID1A depletion (Figure 8K and L), suggesting NFATc1 is a crucial regulator of transcription programs activated in the context of ARID1A loss and EGFR signaling.
Figure 8.
NFATc1 controls transcription programs installed by ARID1A deficiency and EGFR signaling. (A) TF motifs (identified by HOMER: http://homer.ucsd.edu/homer/motif/) enriched at ARID1A-bound genomic regions. (B) Schematic illustration of NFAT signaling created with BioRender. (C) Representative images (left) and quantification (right) of ADM formation after NFATc1 overexpression and ARID1A knockdown in 266-6 cells conducted on the fourth day upon seeding of cells. Red arrowheads point to the duct-like structures. Scale bars: 50 μm. Images were taken using a Leica DMi8 microscope. (D) Plot depicting the significantly enriched hallmark gene sets based on genes associated with ARID1A peaks, which contain the NFATc1 binding motif. (E) Venn diagram showing the overlap between ARID1A binding sites in 266-6 cells (green) and NFATc1 peaks in primary murine pancreatic cancer cells with constitutive activation of NFATc1 and KrasG12D mutation (violet).49 (F) ChIP qRT-PCR analysis in 266-6 cells confirmed the occupancy of NFATc1 on the direct ARID1A-bound genes Bhlhe40, Lif, Met, Foxa3, and Fam3c in the presence of EGF. (G) Western blot analysis of ARID1A and NFATc1 in 266-6 cells in the presence or absence of ARID1A knockdown and EGF treatment. β-actin was used as the loading control. (H) Immunocytological detection of NFATc1 localization in 266-6 cells. Cells were transfected with siCtrl or siARID1A and treated with or without EGF (40 ng/mL) for 8 hours. 4′,6-diamidino-2-phenylindole (DAPI) visualizes cell nuclei. (I) Luciferase reporter assay of an NFAT-responsive promoter construct in 266-6 cells overexpressing EGFR or upon ARID1A knockdown. Data are representative of 3 independent experiments. (J) GSEA plot showing that NFATc1-inducible genes50 are enriched in cells with ARID1A silencing. (K) Gene expression analysis of indicated genes after individual or combined EGF (40 ng/mL) or CsA (1 μmol/L) treatment in ARID1A-depleted 266-6 cells. Data were shown as relative messenger RNA (mRNA) expression normalized to Rplp0 housekeeping gene. (L) Gene expression analysis of indicated genes after individual or combined EGF (40 ng/mL) or CsA (1 μmol/L) treatment in Arid1a-depleted 266-6 cells. ARID1A knockdown was implemented by siARID1A#10 (J-040694-10; Dharmacon). Data are shown as relative mRNA expression normalized to the Rplp0 housekeeping gene. (C, F, I, K, and L) Data are presented as means ± SD. (C, I, K, and L) Statistical significance was determined by an unpaired t test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. CaM, calmodulin; cis, regulatory cis-acting elements; CRAC, calcium release-activated channels; ER, endoplamic reticulum; FDR, false discovery rate; NES, normalized enrichment score; OE, overexpression.
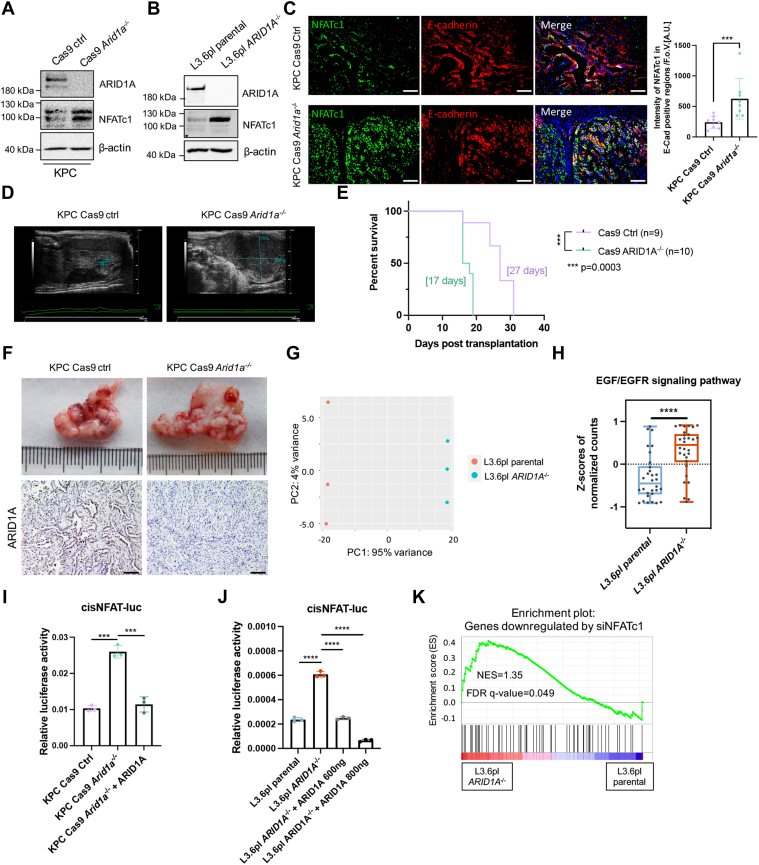
To investigate if the ARID1A status also impacts NFATc1-dependent gene regulation in established PDACs, we used primary PDAC cells derived from KrasG12D/+;Trp53R172H/+;Pdx1-Cre (KPC) mice as well as human L3.6pl PDAC cells with endogenous ARID1A wild-type expression and used clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9)-technology to knockout Arid1a/ARID1A (Figure 9A and B). Notably, ARID1A depletion augmented NFATc1 expression in epithelial tumor parts and cells (Figure 9A–C). In accordance with the previously reported oncogenic properties of Arid1a loss,19,20 orthotopic transplantation of Arid1a-deficient cells (KPC Cas9 Arid1a-/-) into C57BL/6J recipient mice resulted in a higher tumor burden as detected by ultrasound-based imaging and a significantly reduced survival when compared with KPC Cas9 control (Ctrl)-transplanted mice (Figure 9D and E). Upon necropsy, KPC Cas9 Arid1a-/- PDAC was enlarged and showed a poorly differentiated histology (Figure 9F). Comparable with 266-6 cells, EGFR-related gene transcription was enriched in ARID1A-deficient vs control PDAC cells as identified by RNA-seq analysis (Figure 9G and H). Moreover, Arid1a/ARID1A depletion prominently enhanced NFATc1-responsive transactivation in L3.6pl as well as KPC cells, whereas restoration of ARID1A was sufficient to abrogate this effect (Figure 9I and J). Moreover, NFATc1-inducible gene expression was associated positively with ARID1A depletion, as evidenced by GSEA (Figure 9K). Taken together, these findings suggest that the consequences of ARID1A deficiency on NFATc1-dependent gene regulation are conserved throughout pancreatic tumorigenesis and PDAC progression and identify NFATc1 as a central hub mediating gene transcription programs in the Arid1a-deficient pancreas.
Figure 9.
Arid1a loss fosters NFATc1-dependent transcription activation during PDAC tumorigenesis. (A) Western blot analysis conducted in whole-cell lysates of KPC Cas9 ctrl and KPC Cas9 Arid1a-/- cells. (B) Western blot analysis conducted in whole-cell lysates of L3.6pl parental as well as L3.6pl ARID1A-/- cells. (C) Immunofluorescence staining of E-cadherin and NFATc1 in tumor tissue of C57BL/6J mice upon orthotopic transplantation of Cas9 Ctrl or Arid1a-/- KPC cells (left) and the corresponding quantification (right). NFATc1 expression was quantified in E-cadherin (E-Cad)–positive cells to exclude stromal NFATc1 expression. 4′,6-diamidino-2-phenylindole visualizes cell nuclei. Each dot represents 1 mouse, n = 8. Scale bars: 100 μm. (D) Representative ultrasound images of pancreatic tumors detected in mice transplanted with KPC Cas9 Ctrl or KPC Cas9 Arid1a-/- cells. (E) Kaplan–Meier survival curve of recipient C57BL/6J mice upon transplantation with Cas9 Ctrl or Cas9 Arid1a-/- cells. Median survival is shown in brackets. Statistical significance was determined by log-rank (Mantel–Cox) test. (F) From the top, macroscopic images and immunohistochemical staining of ARID1A in C57BL/6J mice orthotopically transplanted with Cas9 ctrl or Cas9 Arid1a-/- cells. Scale bars: 100 μm. (G) PCA of L3.6pl RNA-seq showing the separate clusters of the 2 genotypes. (H) Box plot showing the Z-scores of normalized counts of EGF/EGFR signaling pathway in L3.6pl parental and L3.6pl ARID1A-/- cells. (I) Luciferase reporter assay of NFAT-responsive promoter construct in KPC Cas9 Ctrl, Cas9 Arid1a-/-, as well as Cas9 Arid1a-/- cells with ARID1A restoration. (J) Luciferase reporter assay of an NFAT-responsive promoter construct in L3.6pl parental as well as L3.6pl ARID1A-/- cells in the presence and absence of different amounts of an ARID1A expression construct. Data are representative of 3 independent experiments. (K) GSEA plot showing the enrichment of NFATc1-inducible genes50 in L3.6pl ARID1A-/- line. (C, H, I, and J) Data are presented as means ± SD. (C) Statistical significance was determined by 2-sided Mann–Whitney test and (H–J) a 2-tailed unpaired Student t test. ∗∗∗P < .001, ∗∗∗∗P < .0001. AU, arbitrary unit; FDR, false discovery rate; FoV, field of view; NES, normalized enrichment score. cis, regulatory cis-acting elements; FDR, false discovery rate; NES, normalized enrichment score; PC1, principal component 1; PC2, principal component 2.
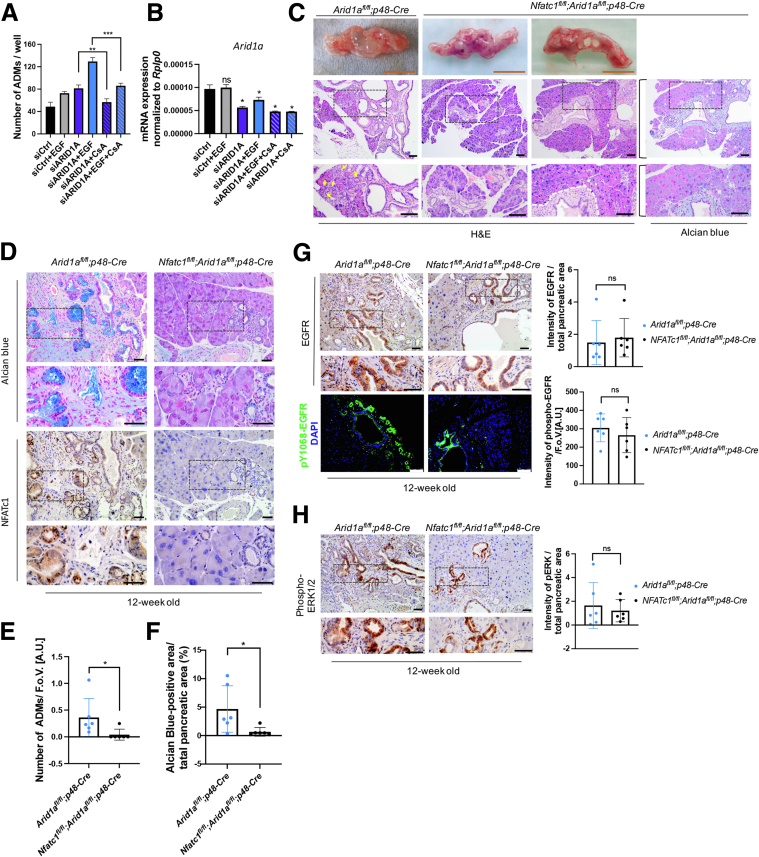
NFATc1 Is Required for Metaplasia in the Arid1a-Deficient Pancreas
Because our data suggested that NFATc1 is a central regulatory hub of transcription programs installed in the context of ARID1A deficiency and EGFR signaling, we next asked whether loss of NFATc1 would, to some extent, reverse ARID1A loss–induced pancreatic metaplasia. To this end, we first assessed in vitro ADM formation after pharmacologic blockage of NFATc1 activity by CsA. Strikingly, CsA treatment rescued acinar-to-ductal conversion in the context of ARID1A depletion both in the absence and, to an even stronger extent, in the presence of EGFR signaling (Figure 10A and B). Next, we generated a transgenic mouse model carrying concomitant Nfatc1 and Arid1a homozygous deletion in the pancreatic acinar compartment (Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice). Unlike mice solely bearing Arid1a deficiency, the incidence of macroscopic cystic vesicles was immensely decreased in the pancreas of Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice (Figure 10C). In comparison with the remodeled pancreas of 12-week-old Arid1afl/fl;p48-Cre mice, acinar cell architecture was maintained predominantly in Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice (Figure 10D). Accordingly, we detected significantly less ADM and PanIN lesions in the Nfatc1-deficient context (Figure 10E and F). Interestingly, cystic lesions that did form in Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice were surrounded by dense parenchyma and desmoplastic stroma (Figure 10C), thus segregating those lesions from the preserved acinar cell compartment. Interestingly, despite the phenotypic dissimilarities of the NFATc1-positive and NFATc1-negative Arid1a-deficient pancreas, neither total/active EGFR nor pERK1/2 expression showed any difference between the genotypes (Figure 10G and H), suggesting that EGFR-signaling–dependent promotion of pancreatic metaplasia in the Arid1a-defcient pancreas at least partially depends on the activation of NFATc1-driven ADM-permissive gene signatures.
Figure 10.
NFATc1 depletion hinders ARID1A loss–induced acinar-to-ductal transdifferentiation in vitro and in vivo. (A) In vitro ADM formation was determined in 266-6 cells upon ARID1A knockdown in the presence or absence of EGF and/or CsA treatment. Quantification was performed on the fourth day after seeding. (B) qRT-PCR analysis to confirm successful ARID1A knockdown in 266-6 cells for experiments shown in panel A. (C) Macroscopic images as well as H&E and Alcian blue staining of the pancreas of 12-week-old Arid1afl/fl;p48-Cre and Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice. Yellow arrows indicate ADMs; whitedashed line indicates the border of dense stroma. Orange scale bars: 1 cm; black scale bars: 100 μm. (D) Alcian blue staining and the corresponding immunohistochemistry for NFATc1 in 12-week-old Arid1afl/fl;p48-Cre and Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice. Scale bars: 50 μm. (E) Quantification of ADM lesions per field of view in H&E staining of 12-week-old Arid1afl/fl;p48-Cre and Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice. (F) The ratio of Alcian blue–positive area to total pancreatic area in 12-week-old Arid1afl/fl;p48-Cre and Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice. (G) Immunohistochemistry for total EGFR (top) and immunofluorescence for phospho-EGFR (Y1068) (bottom) in 12-week-old Arid1afl/fl;p48-Cre and Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice. Quantification of each staining is shown on the right. Each dot represents 1 mouse, n = 6. 4′,6-diamidino-2-phenylindole (DAPI) visualizes cell nuclei. Black scale bars: 50 μm; white scale bars: 100 μm. (H) Immunohistochemistry for phospho-ERK1/2 in 12-week-old Arid1afl/fl;p48-Cre and Nfatc1fl/fl;Arid1afl/fl;p48-Cre mice. Representative images (left) and quantification (right) are shown. Each dot represents a mouse, n = 6. Scale bars: 50 μm. (A, B, E–H) Data are presented as means ± SD. Statistical significance was determined by (A and B) a 2-tailed unpaired Student t test (n = 2) and (E–H) a 2-sided Mann–Whitney test (n = 6). ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. AU, arbitrary unit; FoV, field of view; mRNA, messenger RNA.
Together, our in vitro and in vivo data underscore the implication of NFATc1 as a downstream target of EGFR-driven ADM formation in the context of Arid1a deficiency and illustrates that interference with NFATc1-dependent transcription programs reverts the metaplastic phenotype of Arid1a-deficient mice.
NFATc1 Displaces ARID1A From its Target Genes
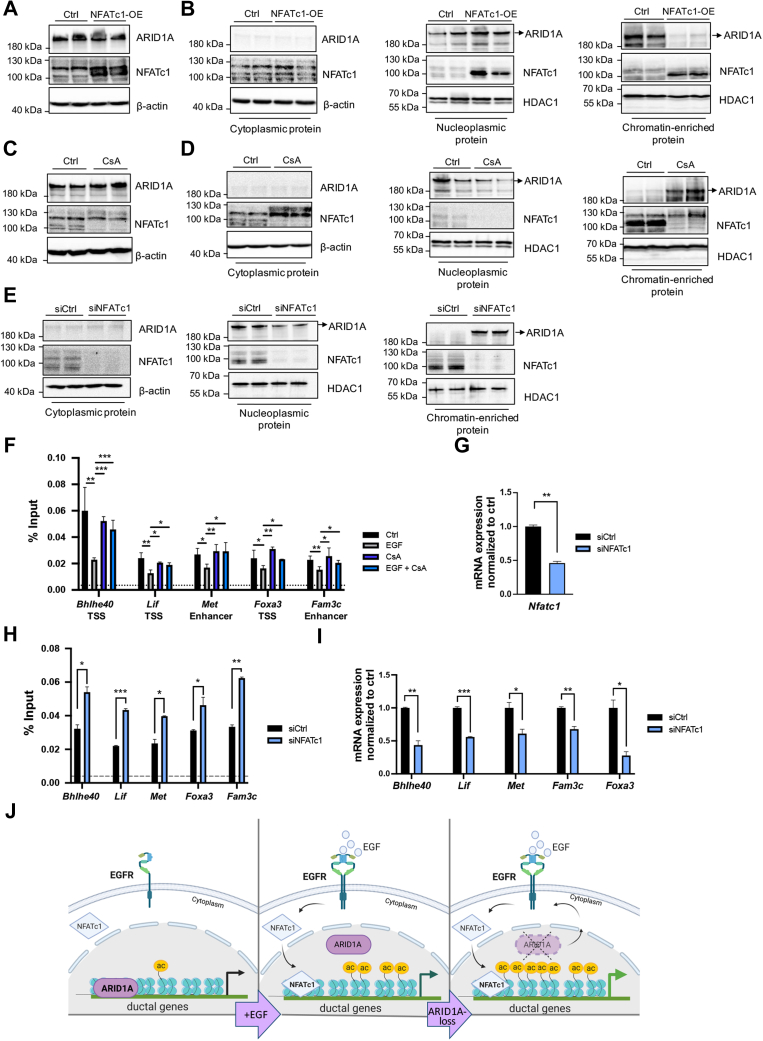
Given that activation of EGFR signaling and subsequent activation of NFATc1 promote acinar cell reprogramming and pancreatic metaplasia even in the presence of ARID1A (Figures 3 and 4),6 we asked whether NFATc1 might be causally involved in the genome-wide ARID1A displacement observed upon EGFR activation. To this end, we overexpressed constitutively active NFATc1 in 266-6 cells (Figure 11A) and subsequently examined ARID1A chromatin localization via fractionation. Interestingly, NFATc1 overexpression significantly reduced ARID1A occupancy to chromatin in favor of a stronger nucleoplasmic protein fraction (Figure 11B), suggesting that, analogous to activation of EGFR signaling, NFATc1 overexpression is sufficient to displace ARID1A from the genome. Consistent with this finding, pharmacologic inhibition (CsA) or small interfering RNA (siRNA)-mediated NFATc1 knockdown preserved ARID1A on chromatin (Figure 11C–E), retained ARID1A occupancy on its target genes (Figure 11F–H), and reduced their transcription (Figures 8K and 11I). Hence, EGFR-induced NFATc1 activation and subsequent nuclear translocation is sufficient to displace ARID1A from the genome, thus critically interfering with ARID1A-dependent regulation of gene signatures required for maintaining acinar cell identity.
Figure 11.
NFATc1 displaces ARID1A genomic occupancy. (A) Western blot analysis conducted in whole-cell lysates of 266-6 cells upon NFATc1 overexpression. (B) The expression of NFATc1 and ARID1A in different cellular fractionation lysates of 266-6 cells was analyzed by immunoblotting in cells upon NFATc1 overexpression. β-actin was used as the loading control for cytoplasmic protein and histone deacetylase 1 (HDAC1) for the nuclear protein. n = 2. (C) Western blot analysis conducted in whole-cell lysates of 266-6 cells upon CsA treatment. (D and E) The expression of NFATc1 and ARID1A in different cellular fractionation lysates of 266-6 cells was analyzed by immunoblotting in cells upon (D) CsA treatment or (E) siRNA-mediated NFATc1 silencing. β-actin was used as the loading control for cytoplasmic protein and histone deacetylase 1 (HDAC1) for the nuclear protein. n = 2. (F) ARID1A ChIP qRT-PCR analysis on the indicated genes in the presence and absence of EGF and CsA in 266-6 cells. Dotted line represents the average IgG level. (G) qRT-PCR analysis to confirm successful NFATc1 knockdown in 266-6 cells. (H) ChIP qRT-PCR analysis in 266-6 cells showing the occupancy of ARID1A on the indicated genes upon NFATc1 silencing in the context of EGF treatment. Dotted line represents the average IgG level. (I) Gene expression analysis of the indicated genes upon NFATc1 knockdown in comparison with control 266-6 cells. The Rplp0 gene was used as endogenous control. (J) Proposed model of the EGFR/NFATc1/ARID1A interplay in acinar cell reprogramming. In the absence of EGFR signaling, NFATc1 resides in the cytoplasm, while ARID1A occupies ductal gene signatures and prevents them from hyperacetylation and increased transcription (left). Upon activation of EGFR signaling, NFATc1 translocates from the cytoplasm into the nucleus, where it induces ARID1A displacement from target genes involved in acinar-to-ductal transdifferentiation, thus promoting H3K27 acetylation and transcriptional activity (center). Arid1a deficiency (right) fosters EGFR signaling and subsequent nuclear NFATc1 translocation, thus promoting a chromatin state characterized by increased transcription of the ductal gene signature. (F–I) Data are presented as means ± SD (n = 2) and a 2-tailed unpaired Student t test was used to calculate statistical significance. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. ac, acetylation; OE, overexpression.
Together, our findings suggest that interfering with genomic ARID1A occupancy, either via its depletion or EGFR-/NFATc1-mediated displacement from chromatin, enables NFATc1-dependent transcriptional activation of gene signatures favoring acinar cell reprogramming, metaplasia, and plasticity.
Discussion
Because lineage-tracing experiments have shown a (patho-)physiological relevance of acinar cell transdifferentiation in the pancreas,1 a number of studies have explored the hierarchical signaling cues and intracellular mechanisms that initiate and maintain ADM. Based on its abundant expression in chronic pancreatitis specimens,6,51 and prompted by the critical involvement of its ligands EGF and transforming growth factor α in driving pancreatic metaplasia both in vitro and in vivo,24,25,52,53 EGFR activation has been suggested as a pivotal hierarchical event in controlling acinar cell fate decisions. Accordingly, pharmacologic or genetic Egfr inactivation in transgenic pancreatic cancer mouse models preserves acinar cell integrity and attenuates PDAC development.26 Upon ligand binding, activated EGFR initiates a cascade of protein phosphorylation events, ultimately resulting in the (in-)activation of transcriptional regulators, which subsequently determine gene expression.54, 55, 56
One of the TFs that integrate EGFR-mediated environmental signals for changes in gene transcription in pancreatic epithelial cells is NFATc1. EGFR signaling causes a robust induction of NFATc1 expression and induces its nuclear translocation, thus promoting NFATc1-dependent transcriptional activation of Sox9,6 an essential regulator of ADM formation.57 Here, we describe a novel mechanism of EGFR-dependent regulation of transcriptional dynamics and illustrate that EGFR signaling induces genomic dissociation of ARID1A. Based on our findings that overexpression of NFATc1 mimics EGFR-dependent genomic ARID1A dissociation we propose a model in which NFATc1 serves as the integrating EGFR-responsive signaling hub, which controls the genomic occupancy of chromatin regulatory proteins in favor of ADM-permissive transcription programs (Figure 11J). Consistently, pharmacologic inhibition of NFAT not only maintained ARID1A occupancy on its target genes, but also rescued ADM formation in ARID1A-proficient in vitro (as shown here) and in vivo models (as shown previously).6 Accordingly, in the context of ARID1A inactivation, when NFATc1-dependent ARID1A dissociation from the genome no longer is required, EGFR-dependent NFATc1 induction was sufficient to activate ADM-promoting gene signatures. Consistently, the pancreatic phenotype of Arid1a-deficient mice highly resembled the atrophic and metaplastic pancreas of mice harboring conditional constitutively active nuclear NFATc1.2,49 The functional relevance of EGFR-dependent NFATc1 activation for driving acinar cell plasticity in the context of Arid1a-deficiency was emphasized by reduced expression of ADM-permissive gene signatures and remodeling of the pancreatic architecture in favor of a less-metaplastic phenotype upon transgenic Nfatc1 inactivation in mice. However, because Nfatc1 depletion was not sufficient to completely counteract acinar reprogramming caused by ARID1A loss, we propose that additional drivers of pancreatic reprogramming (eg, alternative inflammatory TFs acting downstream of EGFR signaling) further control acinar plasticity in the context of ARID1A deficiency. Nevertheless, our findings not only confirm previous studies highlighting the relevance of ARID1A in preserving acinar cell integrity,17,19,20 but suggest that the chromatin remodeling protein serves as a barrier for EGFR-induced and NFATc1-mediated ADM formation.
Intracellular and extracellular signaling cues integrate cell-type–specific transcription programs predominantly by controlling the activity of distal regulatory genomic elements termed enhancers.58 In line with the critical involvement of the SWI/SNF complex in controlling cell-fate decision programs, genome-wide binding analyses conducted in diverse cellular systems have linked the chromatin remodeling complex to enhancer-associated gene regulation.14,23,36,59, 60, 61 Consistent with these reports, our ChIP-seq analysis detected ARID1A occupancy preferably at transcriptionally active (marked by H3K27ac) distal genomic regions, suggesting that in acinar cells ARID1A also controls chromatin remodeling and gene transcription via regulation of enhancer regions. Although several studies have linked ARID1A loss with decreased chromatin accessibility and attenuated gene transcription,18,36,61,62 our findings suggest an alternative mechanism of ARID1A-dependent transcriptional regulation in acinar cells. Because ARID1A inactivation increased transcriptional activity, we reasoned that in the context of acinar-to-ductal transdifferentiation ARID1A prevents the activation of metaplasia-promoting gene signatures. Alternative directions of SWI/SNF-mediated transcriptional regulation have been described previously for the SWI/SNF subunit SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily B, Member 1 (SMARCB1), which antagonizes chromatin accessibility in mouse embryonic stem cells.59 In the same line, recent findings that globally associate ARID1A loss in the preneoplastic pancreas with decreased transcriptional activity also link ARID1A deficiency with increased chromatin accessibility and transcription of enhancers associated with ALDH1A1, a senescence-attenuating gene required to overcome KRAS-induced senescence programs in the ARID1A-deficient metaplastic pancreas.23 Moreover, ARID1A recently was shown to bind to and represses estrogen-receptor binding element-containing enhancers in breast cancer by recruiting the histone deacetylase 1.63 Our findings implicating a strong overlap of ARID1A and H3K27ac occupancy in treatment-naïve acinar cells, however, argue against the concept of ARID1A as a classic repressor of gene transcription in the pancreas. Rather, we propose that ARID1A expression and genomic occupancy prevents hyperacetylation of genomic sites marked by pre-existing H3K27ac enrichment. Accordingly, genetic Arid1a depletion or EGF-induced genomic dissociation of the chromatin remodeling protein might permit the recruitment of chromatin regulatory proteins or TFs, which are more potent drivers of chromatin accessibility and gene transcription. This concept is in line with a recent report dissecting the consequences of inactivating ARID1A mutations in endometriosis.14 Similar to our findings, ARID1A binding in endometrial cells was associated strongly with H3K27ac enrichment at enhancers. However, ARID1A loss resulted in H3K27 hyperacetylation and increased accessibility of superenhancer regions, suggesting that ARID1A can prevent hyperactivation of superenhancers in certain contexts. Interestingly, inhibition of the histone acetyltransferase p300 rescued superenhancer hyperacetylation and blocked endometrial invasion, specifically in the context of ARID1A mutations.14 A p300-dependent regulation also was proposed as the mechanistic basis of increased ALDH1A1 expression and attenuated senescence of ARID1A-deficient preneoplastic pancreatic cells.23 Whether ARID1A also antagonizes p300 activity in the context of the transcription of ADM-promoting gene signatures has not been elucidated yet. However, given that NFATc1 has been shown to biochemically interact with and recruit the histone acetyltransferase to its target genes in PDAC cells,64 p300-dependent histone acetylation might represent a critical component in NFATc1-driven transcriptional activation of ARID1A target genes.
Together, our findings emphasize the close relationship between environmental signaling cues and chromatin remodeling in orchestrating the transcriptional regulation of cell fate decision programs in the pancreas and illustrate that, in addition to its established role in maintaining acinar transcription programs, ARID1A controls pancreatic integrity by serving as a barrier for the transcriptional induction of ductal gene signatures.
Material and Methods
Mouse Lines and In Vivo Experiments
The establishment of p48-Cre and Nfact1fl/fl mice was been described previously.65 Arid1afl/fl mice66 were a kind gift from Dr Zhong Wang (University of Michigan) and were crossed with p48-Cre mice to generate the Arid1afl/fl;p48-Cre and Arid1afl/+;p48-Cre cohorts. Pancreatic depletion of NFATc1 was obtained by crossing Nfatc1fl/fl mice with Arid1afl/fl animals. Genotyping of the mouse strains was performed by polymerase chain reaction (PCR) as described.2 For histologic analysis and acinar cell extraction, pancreatic tissue was harvested at the indicated time points. For survival analysis, mice were kept under observation until reaching the end point criteria. For the generation of the syngeneic orthotopic PDAC model, 2 × 105 KPC Cas9 Ctrl or Arid1a-/- cells were resuspended in culture medium and injected orthotopically into the pancreatic tail of C57BL/6J mice (obtained from Janvier Labs). Tumor onset was determined by ultrasound as described previously,67 and mice were monitored with regard to general health criteria until becoming moribund. Mice then were killed and tissue was harvested for subsequent analysis. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University Medical Center Göttingen (33.9-42502-04-14/1634 and 19/3085).
Cell Culture
The 266-6 murine pancreatic acinar cell line was described previously.68 Primary tumor cells derived from KrasG12D/+;Trp53R172H/+;Pdx1-Cre mice were described prevously.69 Both cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, Bremen, Germany) containing 4.5 g/L D-glucose, L-glutamine supplemented with 10% fetal calf serum (FCS) (Life Technologies Corp, Darmstadt, Germany). One percent nonessential amino acids additionally were added for KPC cell culture. L3.6pl parental and ARID1A-/- lines were maintained in minimum essential medium (Thermo Fisher Scientific) supplemented with 10% FCS. All cells were tested negative for Mycoplasma contamination before conducting experiments. Transient ARID1A knockdown was performed using siRNAs purchased from Dharmacon (ON-TARGETplus ARID1A siRNA #7: J-017263-07; #8: J-017263-08 and #10: J-040694-10) and Silencer Pre-designed NFATc1 siRNA (for human beings: #40657; Ambion; for mice: #288360; Ambion), with nontargeting siRNA #1 (Dharmacon) as control. Target sequences of siRNAs are listed in Table 1. Reverse transfection was conducted using Lipofectamine RNAiMAX transfection reagent (Invitrogen, Karlsruhe, Germany) according to the specified protocol. Cells were incubated with siRNAs for 24 hours and then starved in serum-free medium overnight, or simultaneously treated with 1 μmol/L CsA, 300 nmol/L erlotinib, 20 nmol/L trametinib (Sigma-Aldrich, St. Louis, MO), or the corresponding control vehicle for 24 hours. The following day, EGF (40 ng/mL; Sigma-Aldrich) was applied to the cells 8 hours before harvesting, with 0.1% bovine serum albumin (BSA) used as control. For overexpression of constitutive active NFATc1, 266-6 cells were transfected with a MSCV-NFATc1 construct provided by Neil A. Clipstone using Lipofectamine 2000 (Invitrogen).
Table 1.
Sequence of siRNAs Used in This Study
| siRNA | Target sequence |
|---|---|
| ON-TARGETplus siRNA, ARID1A J-017263-07 (#7) | GCAACGACAUGAUUCCUAU |
| ON-TARGETplus siRNA, ARID1A J-017263-08 (#8) | GGACCUCUAUCGCCUCUAU |
| ON-TARGETplus mouse Arid1a siRNA J-040694-10 (#10) | AAGCAUUGCCCAAGAUCGA |
| Silencer predesigned NFATc1 siRNA human #40657 | GGUCAUUUUCGUGGAGAAAtt |
| Silencer predesigned NFATc1 siRNA mouse #288360 | GCGUUUCACGUACCUUCCUtt |
| Nontargeting siRNA #1 | UGGUUUACAUGUCGACUAA |
CRISPR/Cas9-Mediated Gene Editing
The procedure for generating CRISPR/Cas9 Arid1a-knockout KPC clones has been described previously.36 Briefly, complementary oligonucleotides for guide RNAs (gRNAs) against murine Arid1a were annealed and ligated into pSpCas9(BB)-2A-green fluorescent protein (PX458) vector, respectively, and transformed into competent Escherichia coli. Colonies formed by gRNA-carrying bacteria were selected on Luria-Broth agar plates supplemented with ampicillin. Plasmid extraction (Miniprep, #27106; Qiagen) was performed on the picked colonies followed by genotyping and DNA sequencing. Finally, the validated constructs containing single gRNA were transfected simultaneously into KPC cells using Lipofectamine 2000 (Invitrogen). Transfection efficiency was ensured by green fluorescent protein fluorescence and expansion of single-cell clones was enabled further. Sequence information of gRNAs as well as validation primers are listed in Table 2. The generation of L3.6pl ARID1A knockout clones was conducted as described before.36
Table 2.
Guide RNAs and Validation Primers for CRISPR/Cas9-Mediated Arid1a Knockout in KPC Cells
| Name | Primer | Species |
|---|---|---|
| gRNA_#1 | Forward primer: GACCGGTTCTGGGACTTGCTAACCA Reverse primer: AAACTGGTTAGCAAGTCCCAGAACC |
Mouse |
| gRNA_#2 | Forward primer: GACCGGTAGGCTACTTAGGAGTCTG Reverse primer: AAACCAGACTCCTAAGTAGCCTACC |
Mouse |
| Validation primers | Forward primer: CACCCACCCCAATCTACCAC Reverse primer: CTGGCTGCCATCCCTGTAAA |
Mouse |
Acinar Cell Explants and ADM Formation Assay
Primary acinar cell isolation was performed as described previously.6 Briefly, the dissected pancreas was minced in Hank’s balanced salt solution (Life Technologies Corp) to remove affiliated fat and blood vessels. Digestion of the pancreas was executed in 1.2 mg/mL collagenase VIII–containing buffer at 37°C, followed by cell-mixture filtering through a 100-μm strainer (#542000; Greiner Bio-One). Acinar cells were collected by low-speed centrifugation and used directly for RNA isolation (RNA-seq). For ADM formation, primary acinar cells and 266-6 cells were resuspended in Waymouth’s or DMEM media (Life Technologies Corp), respectively, supplemented with 0.1% BSA, 0.2 mg/mL soybean trypsin inhibitor (Sigma-Aldrich), 50 μg/mL bovine pituitary extract (Sigma-Aldrich), 200 μmol/L nicotinamide, 0.1% FCS, mixed with equal volume of neutralized rat tail collagen type I, and seeded in a 12-well culture dish precoated with type I collagen (2.5 mg/mL). Cultures were maintained in the presence or absence of recombinant EGF (40 ng/mL). Quantitation of ADM formation was conducted 5 days after isolation by light microscopy by counting the number of ADM structures in duplicate wells of each culture condition and genotype. For the ADM formation assay performed with 266-6 cells, cells were transfected with siARID1A or a construct encoding a constitutively active NFATc1 in a 60-mm cell culture dish and allowed to attach for 24 hours. Cells then were split and seeded as 3 × 105 cells/well into a 12-well plate and subjected to the equivalent procedure of acinar-explant culture as described earlier. Erlotinib (300 nmol/L) or trametinib (20 nmol/L) was applied to the culture medium with or without EGF (40 ng/mL). ADMs were quantified in duplicate wells on the fourth day after seeding.
H&E, Alcian Blue Staining, and Immunohistochemistry
Pancreatic tissue extraction and embedding was performed as described previously.68 Paraffin-embedded mouse tissues were sectioned, deparaffinized, and rehydrated in 99% to 50% ethanol series. Sections then were stained with H&E or Alcian blue/Nuclear fast red, as per the manufacturer’s instructions for H&E and Alcian blue staining, respectively. For immunohistochemistry, citrate buffer (pH 6.0) or Tris-EDTA buffer (pH 9.0) was used for antigen retrieval followed by endogenous peroxidase quenching in 3% H2O2 for 10 minutes. Slides were blocked in 10% BSA (w/v) in phosphate-buffered saline plus 0.1% Tween-20 at room temperature, then incubated in primary antibody at 4ºC overnight. The following primary antibodies were used: ARID1A (1:750, #12354; Cell Signaling Technology, Danvers, MA), NFATc1 (1:300, ab25916; Abcam, Cambridge, UK), amylase (1:1000, sc-46657; Santa Cruz, Dallas, TX), CD45 (1:50, #550539; BD Pharmingen, San Jose, CA), MIST1 (1:150, #14896; Cell Signaling Technology), cytokeratin 19 (1:150, ab15463; Abcam), EGFR (1:300, sc-373746; Santa Cruz), and phospho-p44/42 MAPK (ErK1/2) (1:300, #4376; Cell Signaling Technology). VECTASTAIN ABC horseradish peroxidase kits (Vector Laboratories, Burlingame, CA) were used for biotin labeling and detection. For staining of mouse primary antibodies on mouse tissue, M.O.M. Immunodetection Kit (BMK-2202; Vector Laboratories) was used to avoid the endogenous mouse Ig signal. Tissues were counterstained in hematoxylin, then dehydrated and mounted with Roti-Mount (ROTH, Karlsruhe, Germany). Images were captured with a microscope-attached Olympus SC180 using Cellsens Entry software. For immunostaining quantification, bright field images of the entire pancreas area were taken from 5 mice/genotype and the staining densities were determined by the fixed red, green and blue histogram pattern for each antibody using ImageJ (National Institutes of Health). The quantitative evaluation of ADM and PanIN regions was conducted with 5–6 mice/genotype by ImageJ. Briefly, the number of ADMs was counted using H&E-stained slides under a 10× nosepiece and normalized to the field of view. Based on the positivity of Alcian blue staining, PanIN regions were outlined, the size was assessed by ImageJ (version 2.1.0), and then normalized to the total pancreatic area. Statistical analysis was performed using the Mann–Whitney test by Prism 9.2.0.
Cellular Fractionation
Cellular fractionation was performed as described.70 Briefly, whole-cell lysates were prepared by incubating the cells in whole-cell lysis buffer (50 mmol/L HEPES, pH 7.5–7.9, 150 mmol/L NaCl, 1 mmol/L EGTA, 10% glycerin, 1% Triton X-100 (Sigma-Aldrich), 100 mmol/L NaF, 10 mmol/L Na4P2O7 × 10 H2O) supplemented with proteinase inhibitor cocktail (#11697498001; Sigma-Aldrich) and cleared by centrifugation at 15,000 × g for 20 minutes. To separate different fractionated lysates, cells were lysed in buffer A.70 Nuclei were collected by low-speed centrifugation (4 min, 1300 × g, 4°C) and washed once with buffer A supplemented with 1 mmol/L CaCl2 and 0.2 U micrococcal nuclease (#3755; Sigma-Aldrich). After incubation at 37ºC for 1 minute, the nuclease reaction was stopped by adding 1 mmol/L EGTA. Nuclei then were lysed in buffer B to separate chromatin-bound and nucleoplasmic proteins by centrifugation (4 min, 1700 × g, 4°C). Chromatin was resuspended in Laemmli buffer and sonicated for 6 cycles (10 seconds on/30 seconds off) in a Biorupter Pico (B01060010; Diagenode, Liege, Belgium). All lysates were subjected to immunoblotting.
Immunoblotting
Immunoblotting was performed as described previously.69 Protein concentration was determined using the Bradford assay (Bio-Rad, Munich, Germany). Protein separation was performed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis blotting on a nitrocellulose membrane using the Trans-Blot Turbo transfer system (#1704150; Bio-Rad). Membranes were incubated with primary antibodies overnight and peroxidase-conjugated secondary antibodies for 1 hour at room temperature. Protein bands were visualized by an Intas ECL Chemocam Imager (Intas Pharmaceuticals Limited). The following primary and secondary antibodies were used: ARID1A (1:1000, #12354; Cell Signaling Technology), phospho-p44/42 MAPK (ErK1/2) (1:1000, #4376; Cell Signaling Technology), p44/42 MAPK (ErK1/2) (1:1000, #9102; Cell Signaling Technology), histone deacetylase 1 (1:800, #06-720; EMD Millipore, Billerica, MA), β-actin (1:40000, A3854; Sigma-Aldrich), NFATc1 (1:500, sc7294; Santa Cruz), H3K27ac (1:800, GT128944; GeneTex, Irvine, CA), histone H3 (1:1000, #9715; Cell Signaling Technology), anti-rabbit/mouse (IgG) horseradish peroxidase (1:6500, #7074/7076; Cell Signaling Technology).
RNA Isolation and Real-time Quantitative Reverse-Transcription PCR
Total RNA was extracted using TRIzol (#15-596-018; Invitrogen) reagent. For RNA isolation from collagen-embedded ADMs, the matrix was removed by incubation of collagenase P (#11213857001; Sigma-Aldrich) before TRIzol-based RNA extraction. RNA was reverse-transcribed with the iScript Complementary DNA Synthesis Kit (#1708890; Bio-Rad), quantitative reverse-transcription (qRT)-PCR assays were performed using iTaq Universal SYBR Green SuperMix (#172-5125; Bio-Rad) and the StepOnePlus system (Applied Biosystems, Carlsbad, CA). Data were normalized to Rplp0 endogenous control and analyzed via the delta-delta Ct method. Primers are described in Table 3.
Table 3.
Primers for Messenger RNA Expression Analysis by qRT-PCR in This Study
| Name | Forward primer | Reverse primer | Species |
|---|---|---|---|
| Arid1a | ACAAGGCAGATGGAACACC | CTCTGTGAAGGCCAGGTAC | Mouse |
| Amy1 | GCAAGTGGAGGTATCGAGAAC | CTGCTACGCCAATGTCAATG | Mouse |
| Krt18 | GGAAGTGGATGCCCCCAAAT | TCCTCAATCTGCTGAGACCAGTA | Mouse |
| Lif | GCGAGATGAGATGCAGGGATT | GGTGGCATTTACAGGGGTGA | Mouse |
| Bhlhe40 | AGCGAAGACAGCAAGGAAACT | ACGTAAGCTCCAGAACCACTG | Mouse |
| Fam3c | CTATTTGCTCGATCCGCGCT | CGCCTTTGAGATCCCACACT | Mouse |
| Foxa3 | GTAGAGAGACCGAAGCACTCG | ATTCACTGGAGAATACACCTCGC | Mouse |
| Met | GCCGGCGGTTTATAAGGTGT | AAGGTACAGCTCTCGTTGCC | Mouse |
| Rplp0 | TGGGCAAGAACACCATGATG | AGTTTCTCCAGAGCTGGGTTGT | Mouse |
Luciferase Reporter Assay
266-6, KPC, and L3.6pl cells were grown in 24-well plates under the respective culture conditions. Twenty-four hours before the luciferase reporter transfection, 266-6 cells were reverse-transfected with siRNA targeting Arid1a using siLentFect lipid reagent (#170-3362; Bio-Rad) and culture media was changed to normal 10% FCS-containing DMEM 10 hours after transfection. On the following day, all cells were transfected with the following plasmids using Lipofectamine 2000 reagent (Invitrogen): pNFATc1-luc (500 ng; Stratagene), pSG5-v-ErbB–EGFR (500 ng; provided by M. Privalsky), pcDNA6-V5/His-ARID1A (600–800 ng, 39311; Addgene), pMCV (500 ng; Stratagene), and with a Renilla-luciferase vector (15 ng; provided by R. Urrutia) as internal control. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a luminometer (LUmo; Autobio Diagnostics) according to the manufacturer’s instructions. Relative luciferase signal was calculated as firefly luciferase value/Renilla luciferase value and shown as means ± SD.
Immunofluorescence and Immunocytochemistry
Serial paraffin sections were deparaffinized and rehydrated from 99% ethanol to distilled water. Sections then were washed 5 times with ice-cold phosphate buffer (PB, 20 mmol/L NaH2PO4, 80 mmol/L Na2HPO4, pH 7.4), for 5 minutes each. Antigen retrieval was conducted by cooking the sections in citrate buffer (pH 6.0) in a microwave for 6 minutes after boiling. Subsequently, the sections were left at room temperature for 10 minutes and transferred on ice for another 30 minutes until they reached 37°C. Once cooled down, sections were washed 3 times (5 minutes each) in PB and regions of interest were encircled by a pap pen. Blocking was performed by incubating the sections in 10% normal goat serum (NGS) prepared in PB supplemented with 0.4% Triton X-100 (PBT) at 4°C for 2 hours. Subsequently, sections were washed in PB twice, then incubated with primary antibody diluted in PBT containing 2% NGS at 4°C overnight. Antibodies used were as follows: E-cadherin (1:50, #61018; BD Biosciences, Heidelberg, Germany), NFATc1 (1:100, ab25916; Abcam), and phosphor-EGFR (Y1068, 1:300, ab40815; Abcam). The next day, the sections were washed in PB 6 times before incubating with AlexaFluor 568 or 488 secondary antibodies (1:500; Invitrogen) diluted in PBT with 2% NGS for 2 hours at 4°C. Sections were washed with PB 5 times, followed by 4′,6-diamidino-2-phenylindole (1 μg/mL) nuclear staining for 10 minutes. Finally, slides were washed in PB twice and mounted using Immu-Mount (Epredia 9990412; Fisher Scientific GmbH, Schwerte, Germany).
For immunocytochemistry, 266-6 cells were transfected with Arid1a siRNA on coverslips in 12-well plates and allowed to adhere overnight. The next day, cells were switched to serum-free conditions overnight and subjected to EGF stimulation (40 ng/mL) for 8 hours. Cells then were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde for 15 minutes at room temperature. Subsequently, samples were permeabilized using 0.4% Triton X-100 in PB, blocked for 1 hour in 5% normal goat serum, and incubated with NFATc1 primary antibody (1:100, ab25916; Abcam) diluted in PBT overnight at 4°C. On the second day, samples were rinsed 3 times in PB for 5 minutes each before being incubated with AlexaFluor 568 secondary antibody (1:500; Invitrogen) for 2 hours at room temperature. Afterward, cells were washed an additional 3 times with PB followed by 4′,6-diamidino-2-phenylindole staining (1 μg/mL) for 10 minutes before mounting on glass slides.
Slides were viewed on a Leica DMi8 microscope and 10 representative fields were chosen per mouse and 5 representative fields were chosen per treatment group of cells for final fluorescence intensity evaluation. ImageJ version 2.1.0/1.53c was used to quantify the images, with a fixed threshold setting for each staining. Quantification results were processed using Prism version 9.2.0.
RNA-seq
266-6 and L3.6pl cells were seeded for 3 replicates per condition in 6-well plates. Primary acinar cells were isolated from p48-Cre (n = 2) and Arid1afl/fl;p48-Cre (n = 3) mice. Total RNA was extracted using TRIzol in all model systems and quality was assessed by agarose gel electrophoresis. Libraries were prepared from 500 ng RNA using the Truseq RNA Library Prep Kit v2 (Illumina, San Diego, CA).
Fragment analysis of RNA-seq libraries was performed using Bioanalyzer 2100 (Agilent). Subsequently, approximately 25 million single-end, 50-bp reads were sequenced per replicate on a HiSeq 2000 (for acinar cell RNA-seq) or HiSeq 4000 (for 266-6 and L3.6pl cell RNA-seq) (Illumina) at NGS Integrative Genomics Core, University Medical Center Göttingen. Analysis of RNA-seq was performed on a Galaxy|Europe (usegalaxy.eu) platform.71 Sequence reads were aligned to the mouse (MGSCv37/mm9) or human (GRCh37/hg19) genomes by TopHat version 2.1.172 for 266-6/L3.6pl RNA-seq and RNA-STAR version 2.7.8a73 for acinar cell RNA-seq. Transcripts were counted by htseq-count version 0.9.1.34 Differential expression analysis was performed by Cuffnorm version 2.2.1.2 and Cuffdiff version 2.2.1.535 for 266-6/L3.6pl RNA-seq and DESeq2 version 2.11.40.674 for acinar RNA-seq, with an FDR cut-off of 0.05. Heatmaps were generated by pheatmap package in R using the Z score transformed from counts after normalization. Gene Ontology dot plot was generated by R package ggplot2 using pathway summarization given by Enrichr. GSEA was performed using GSEA_4.1.0 software against literature-exposited signatures in mouse models.28, 29, 30, 31
ChIP
266-6 cells were grown in 15-cm dishes to 85% confluency and treated with or without EGF (40 ng/mL) for 8 hours. All ChIP experiments were performed in duplicate.
For ARID1A ChIP, cells initially were cross-linked for 20 minutes in 15 mmol/L ethylene glyco-bis (succinimidyl succinate) (EGS), for 20 minutes in 2 mmol/L EGS at room temperature, followed by 1% Paraformaldehyde fixation for 40 minutes at 4°C as described previously.36 For NFATc1 ChIP, cells were cross-linked in 2 mmol/L EGS for 20 minutes and then fixed in 1% Paraformaldehyde for 20 minutes at room temperature. The subsequent procedure was conducted using the ChIP-IT High Sensitivity Kit (ActiveMotif, La Hulpe, Belgium) according to the manufacturer’s instructions. For H3K27ac ChIP, 1% formaldehyde was added to the cells for 15 minutes and quenched by 125 mmol/L glycine. Nuclei were released and pelleted in modified Nelson buffer (150 mmol/L NaCl, 20 mmol/L EDTA pH 8, 50 mmol/L Tris pH 7.5, 0.5% NP-40, 1% Triton X-100, 20 mmol/L NaF)75 and lysed in Gomes lysis buffer (150 mmol/L NaCl, 1% Igepal (Sigma-Aldrich), 0.5% sodium deoxycholate, 50 mmol/L Tris-HCl pH 8, 20 mmol/L EDTA, 20 mmol/L NaF, 0.1% sodium dodecyl sulfate)76 supplemented with protease and phosphatase inhibitors. The lysates were sonicated using a Diagenode Biorupter Pico (30 seconds on/30 seconds off, 20 cycles). Sheared chromatin was precleared by incubating with 100 μL 50% slurry protein A/G agarose beads for 1 hour followed by centrifugation at 12,000 × g for 2 minutes. Samples were incubated with antibodies against ARID1A (#12354; Cell Signaling Technology), H3K27ac (C15410196; Diagenode), NFATc1 (ab2796; Abcam), and mouse/rabbit IgG (C15400001-15/C15410206; Diagenode) overnight. The next day, 30 μL blocked protein A agarose beads (#16-156; Merck Millipore) were added to the samples and incubated for 2 hours. Immune complexes were washed once in Gomes lysis buffer, twice with Gomes wash buffer (100 mmol/L Tris-HCl pH 8.5, 500 mmol/L LiCl, 1% NP-40, 1% sodium deoxycholate, 20 mmol/L EDTA, 20 mmol/L NaF), twice in Gomes lysis buffer and twice in Tris-EDTA buffer. DNA was reverse–cross-linked, eluted, and extracted by phenol/chloroform/isoamyl alcohol (25:24:1). Final DNA was resuspended in distilled H2O and subjected to library preparation (for ChIP-seq) or qRT-PCR. Primer sequences for qRT-PCR are listed in Table 4.
Table 4.
Primers Used for ChIP qRT-PCR in This Study
| Name | Forward primer | Reverse primer | Species |
|---|---|---|---|
| Bhlhe40 -0.6 kb | GGCTTGAGTCAGACGCGG | TGGTAACGTGGGCGAACC | Mouse |
| Fam3c-6 kb | TGGATTGACTTTCAAATGCTGGC | AATGAGGCGATTCCTGCTCC | Mouse |
| Foxa3 +12 kb | GGAGAAGCCAGAAACCGCA | AGAAAGGCCCTCAGTCTCCC | Mouse |
| Lif+2.7 kb | AAGTTCAGGAAACCAACGGGT | GAGAGACTGTAGCACGGCTG | Mouse |
| Lif+5 kb | GGGGGAAGCTGGTGATTCAA | AGCTGCTGGTTGGACCTTAC | Mouse |
| Met-0.5 kb | CTAGAGCGACAGGGACGAAC | TCAGCCGGGCAACAGC | Mouse |
| Met+52 kb | AAACCTAAAGCATTTGACCGGC | CCCCCACTACTGTGTTTGATGT | Mouse |
ChIP-seq and Data Processing
For ChIP-seq, duplicates of each condition were subjected to library preparation following the protocol of the Microplex Library Preparation V2 kit (C05010012; Diagenode) with 0.5 ng ChIP-DNA as starting material. ARID1A and H3K27ac ChIP were sequenced on a HiSeq 4000 in the NGS Integrative Genomics Core, University Medical Center Göttingen.
Data from NFATc1 ChIP-seq in murine pancreatic cancer cells (GSM982509, GSM982511) were downloaded from the Gene Expression Omnibus website (https://www.ncbi.nnlm.nih.gov/geo). Analysis of the sequencing data was performed on the public server of usegalaxy.org.71 The read quality was evaluated by fastQC (version 0.72). Single-end, 50-bp reads were aligned to mouse genome MGSCv37/mm9 by Bowtie2 (version 2.3.4.2).77 The summary of reads sequenced and mapped are listed in Table 5. Low-quality reads (MAPQ <5) and PCR duplicates were filtered out using Samtools.78 Peaks were called by MACS2 (version 2.1.1.20160309.0)79 with the criteria as fold enrichment greater than 5 compared with background and FDR less than 0.05. For H3K27ac ChIP, the adjacent regions with FDR less than 0.05 were composited as broad peaks. TSS-centered, 1-kb regions were considered as promoters while H3K27ac peaks with the exclusion of 1 kb of an annotated TSS were defined as active distal regions. Differential binding analysis was performed to identify the peaks that were enriched in each condition using R Bioconductor package DiffBind.80 Peaks were annotated by the Genomic Regions Enrichment Annotation Tool (http://great.stanford.edu/public/html) following the basal plus extension association rule: 5 kb upstream, 1 kb downstream, plus 1000 kb extension.
Table 5.
Sequenced and Mapped Reads of ChIP-seq Samples
| Samples of ChIP-seq study | Total sequenced reads, n | Mapped reads, n (% of total) |
|---|---|---|
| ARID1A_1 | 17,267,600 | 12,735,130 (73.8%) |
| ARID1A_2 | 17,906,229 | 12,205,138 (68.2%) |
| ARID1A_EGF1 | 15,921,479 | 11,418,113 (71.7%) |
| ARID1A_EGF2 | 19,250,899 | 13,254,857 (68.9%) |
| H3K27ac_siCtrl1 | 19,842,993 | 15,075,210 (76%) |
| H3K27ac_siCtrl2 | 16,989,464 | 13,830,662 (81.4%) |
| H3K27ac_siCtrl_EGF1 | 15,962,810 | 13,080,677 (81.9%) |
| H3K27ac_siCtrl_EGF2 | 17,940,883 | 14,397,202 (80.2%) |
| H3K27ac_siARID1A1 | 14,421,138 | 11,678,801 (81%) |
| H3K27ac_siARID1A2 | 18,872,697 | 14,649,690 (77.6%) |
| H3K27ac_siARID1A_EGF1 | 18,060,868 | 14,479,303 (80.2%) |
| H3K27ac_siARID1A_EGF2 | 13,806,174 | 10,872,189 (78.7%) |
| Input_ctrl | 11,395,577 | 8,294,689 (72.8%) |
| Input_EGF | 7,366,864 | 5,441,073 (73.9%) |
Statistical Analysis
Data were presented as means ± SD and the statistical significance of in vitro data was determined by a 2-tailed Student t test and 2-sided Mann–Whitney test for mice experiments for independent group comparison. For data sets that have more than 2 experimental groups, the effects of different variables were calculated by 2-way analysis of variance followed by the Tukey multiple comparison test. Survival analysis in Kaplan–Meier curves was conducted by log-rank (Mantel–Cox) test.
Acknowledgments
The authors are grateful for excellent technical support provided by Waltraut Kopp and Sercan Mercan concerning mouse-related work. In addition, the authors give special gratitude to Marie Hasselluhn and Jessica Spitalieri for their support in generating KPC Cas9 Ctrl/Arid1a-/- cell lines. The authors are thankful to Dr Gabriela Salinas, NGS Integrative Genomics Core, University Medical Center Göttingen, for sequencing, and sincerely thank the employees of the Central Animal Facility of the University Medical Center Göttingen for excellent cooperation.
CRediT Authorship Contributions
Zhe Zhang (Formal analysis: Lead; Investigation: Lead; Writing – original draft: Supporting)
Xin Wang (Formal analysis: Supporting; Investigation: Supporting)
Feda H. Hamdan (Formal analysis: Supporting)
Anna Likhobabina (Investigation: Supporting)
Shilpa Patil (Investigation: Supporting; Methodology: Supporting)
Lena Aperdannier (Investigation: Supporting)
Madhobi Sen (Methodology: Supporting)
Jacobe Traub (Investigation: Supporting)
Albrecht Neesse (Investigation: Supporting; Resources: Supporting)
Andre Fischer (Investigation: Supporting; Methodology: Supporting)
Argyris Papantonis (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Shiv Kishor Singh (Formal analysis: Supporting)
Volker Ellenrieder (Funding acquisition: Supporting; Resources: Supporting)
Steven A. Johnsen (Conceptualization: Supporting; Formal analysis: Supporting)
Elisabeth Hessmann, Prof. (Conceptualization: Lead; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by DFG grant KFO 5002 (E.H., A.N., V.E., S.K.S., and A.P.); Ministry of Science and Culture in Lower Saxony and the Volkswagenstiftung grant 11-76251-12-3/16 (V.E., A.F., and S.A.J.); German Cancer Aid grants 70112108 and 70112505 (E.H.), 70113213 (A.N.), and 70112999 (S.S.) (Max Eder group); Wilhelm-Sander Stiftung grant 2021-139.1 (E.H.); the China Scholarship Council (Z.Z. and X.W.); and a Mildred Scheel Medical student fellowship of the German Cancer Aid grant 701129199 (L.A.).
Transcript profiling Expression analysis of murine acinar cell line 266-6 in the context of ARID1A knockdown and/or epidermal growth factor treatment: GSE166329, =https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166329; messenger RNA expression profiles of primary pancreatic acinar cells of Arid1a-deficient and Arid1a wild-type mice: GSE166333, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166333.
Data transparency All data, methods, and study material will be made available to other researchers on request. Next-Generation Sequencing data has been made publicly available as indicated in the Material and Methods section.
Data Availability All authors had access to the study data and reviewed and approved the final manuscript. NGS data for reviewers are provided as follows: (1) GSE166329: messenger RNA expression profiles of murine acinar cell line 266-6 in the context of ARID1A-knockdown and/or EGF treatment; (2) GSE166330: ARID1A genomic binding profiles of EGF-treated or control murine 266-6 cells; (3) GSE166331: H3K27ac binding profiles of ARID1A intact and knockdown cells with or without EGF treatment; (4) GSE166333: messenger RNA expression profiles of primary pancreatic acinar cells of Arid1a-deficient and Arid1a–wild-type mice; and (5) GSE218822: messenger RNA expression profiles of ARID1A–wild-type and ARID1A-deficient human L3.6pl cells.
Ethical Approval This study did not use human primary material and hence did not require ethical approval.
References
- 1.Strobel O., Dor Y., Alsina J., et al. In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology. 2007;133:1999–2009. doi: 10.1053/j.gastro.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N.M., Neesse A., Dyck M.L., et al. Context-dependent epigenetic regulation of nuclear factor of activated T cells 1 in pancreatic plasticity. Gastroenterology. 2017;152:1507–1520 e15. doi: 10.1053/j.gastro.2017.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Clair J.M.-S., Soydaner-Azeloglu R., Lee K.E., et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012;26:439–444. doi: 10.1101/gad.181800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerra C., Schuhmacher A.J., Cañamero M., et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Hingorani S.R., Petricoin E.F., Maitra A., et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 6.Chen N.M., Singh G., Koenig A., et al. NFATc1 links EGFR signaling to induction of Sox9 transcription and acinar-ductal transdifferentiation in the pancreas. Gastroenterology. 2015;148:1024–1034 e9. doi: 10.1053/j.gastro.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benitz S., Regel I., Reinhard T., et al. Polycomb repressor complex 1 promotes gene silencing through H2AK119 mono-ubiquitination in acinar-to-ductal metaplasia and pancreatic cancer cells. Oncotarget. 2016;7 doi: 10.18632/oncotarget.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benitz S., Straub T., Mahajan U.M., et al. Ring1b-dependent epigenetic remodelling is an essential prerequisite for pancreatic carcinogenesis. Gut. 2019;68:2007–2018. doi: 10.1136/gutjnl-2018-317208. [DOI] [PubMed] [Google Scholar]
- 9.Roy N., Malik S., Villanueva K.E., et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29:658–671. doi: 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Figura G., Fukuda A., Roy N., et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16:255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon H., Imbalzano A.N., Khavari P.A., et al. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 12.Sun X., Chuang J.C., Kanchwala M., et al. Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell. 2016;18:456–466. doi: 10.1016/j.stem.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson M.R., Reske J.J., Holladay J., et al. ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat Commun. 2019;10:3554. doi: 10.1038/s41467-019-11403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson M.R., Reske J.J., Holladay J., et al. ARID1A mutations promote P300-dependent endometrial invasion through super-enhancer hyperacetylation. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan H.B., Wang X.F., Zhang Q., et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis. 2014;35:867–876. doi: 10.1093/carcin/bgt398. [DOI] [PubMed] [Google Scholar]
- 16.Raphael B.J., Hruban R.H., Aguirre A.J., et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203. e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S.C., Nassour I., Xiao S., et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut. 2019;68:1259–1270. doi: 10.1136/gutjnl-2017-315490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livshits G., Alonso-Curbelo D., Morris J.P.I.V., et al. Arid1a restrains Kras-dependent changes in acinar cell identity. Elife. 2018;7 doi: 10.7554/eLife.35216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Friedland S.C., Guo B., et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut. 2019;68:1245–1258. doi: 10.1136/gutjnl-2017-315541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura Y., Fukuda A., Ogawa S., et al. ARID1A maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2018;155:194–209 e2. doi: 10.1053/j.gastro.2018.03.039. [DOI] [PubMed] [Google Scholar]
- 21.Ferri-Borgogno S., Barui S., McGee A.M., et al. Paradoxical role of AT-rich interactive domain 1A in restraining pancreatic carcinogenesis. Cancers (Basel) 2020;12:2695. doi: 10.3390/cancers12092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z., Hessmann E. To be or not to be. Elife. 2018;7 doi: 10.7554/eLife.38967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S., Cao W., Niu Y., et al. Single-PanIN-seq unveils that ARID1A deficiency promotes pancreatic tumorigenesis by attenuating KRAS-induced senescence. Elife. 2021;10 doi: 10.7554/eLife.64204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Means A.L., Ray K.C., Singh A.B., et al. Overexpression of heparin-binding EGF-like growth factor in mouse pancreas results in fibrosis and epithelial metaplasia. Gastroenterology. 2003;124:1020–1036. doi: 10.1053/gast.2003.50150. [DOI] [PubMed] [Google Scholar]
- 25.Korc M.A., Chandrasekar B., Yamanaka Y., et al. Overexpression of the epidermal growth factor receptor in human pancreatic cancer is associated with concomitant increases in the levels of epidermal growth factor and transforming growth factor alpha. J Clin Invest. 1992;90:1352–1360. doi: 10.1172/JCI116001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ardito C.M., Grüner B.M., Takeuchi K.K., et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navas C., Hernández-Porras I., Schuhmacher A.J., et al. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson B.L., Salazar M., Mackenzie-Dyck S., et al. Desmoplasia and oncogene driven acinar-to-ductal metaplasia are concurrent events during acinar cell-derived pancreatic cancer initiation in young adult mice. PloS one. 2019;14(9) doi: 10.1371/journal.pone.0221810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boj S.F., Hwang C.I., Baker L.A., et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M., Azevedo-Pouly A.C., Deering T.G., et al. MIST1 and PTF1 collaborate in feed-forward regulatory loops that maintain the pancreatic acinar phenotype in adult mice. Molecular and cellular biology. 2016;36(23):2945–2955. doi: 10.1128/MCB.00370-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krah N.M., De La O.J., Swift G.H., et al. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife. 2015;4 doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muraro M.J., Dharmadhikari G., Grun D., et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394 e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosein A.N., Huang H., Wang Z., et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight. 2019;4 doi: 10.1172/jci.insight.129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C., Williams B.A., Pertea G., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen M., Wang X., Hamdan F.H., et al. ARID1A facilitates KRAS signaling-regulated enhancer activity in an AP1-dependent manner in colorectal cancer cells. Clin Epigenetics. 2019;11:92. doi: 10.1186/s13148-019-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukla V., Vaissiere T., Herceg Z. Histone acetylation and chromatin signature in stem cell identity and cancer. Mutat Res. 2008;637:1–15. doi: 10.1016/j.mrfmmm.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 38.Chabu C., Li D.M., Xu T. EGFR/ARF6 regulation of Hh signalling stimulates oncogenic Ras tumour overgrowth. Nat Commun. 2017;8 doi: 10.1038/ncomms14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou P., Kapoor A., Zhang Q., et al. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov. 2020;10:1058–1077. doi: 10.1158/2159-8290.CD-19-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fendrich V., Jendryschek F., Beeck S., et al. Genetic and pharmacologic abrogation of Snail1 inhibits acinar-to-ductal metaplasia in precursor lesions of pancreatic ductal adenocarcinoma and pancreatic injury. Oncogene. 2018;37:1845–1856. doi: 10.1038/s41388-017-0100-4. [DOI] [PubMed] [Google Scholar]
- 41.Rauschmeier R., Gustafsson C., Reinhardt A., et al. Bhlhe40 and Bhlhe41 transcription factors regulate alveolar macrophage self-renewal and identity. EMBO J. 2019;38 doi: 10.15252/embj.2018101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman J.R., Kaestner K.H. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waerner T., Alacakaptan M., Tamir I., et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 44.Wu Y., Sato F., Yamada T., et al. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int J Oncol. 2012;41:1337–1346. doi: 10.3892/ijo.2012.1559. [DOI] [PubMed] [Google Scholar]
- 45.Jeon H.M., Lee J. MET: roles in epithelial-mesenchymal transition and cancer stemness. Ann Transl Med. 2017;5:5. doi: 10.21037/atm.2016.12.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W., Reiser-Erkan C., Michalski C.W., et al. Hypoxia inducible BHLHB2 is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun. 2010;401:422–428. doi: 10.1016/j.bbrc.2010.09.070. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y., Gao W., Lytle N.K., et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. 2019;569:131–135. doi: 10.1038/s41586-019-1130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D., Liu K., Yang Y., et al. Prognostic value of leukemia inhibitory factor and its receptor in pancreatic adenocarcinoma. Future Oncol. 2020;16:4461–4473. doi: 10.2217/fon-2019-0684. [DOI] [PubMed] [Google Scholar]
- 49.Baumgart S., Chen N.M., Siveke J.T., et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov. 2014;4:688–701. doi: 10.1158/2159-8290.CD-13-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasselluhn M.C., Schmidt G.E., Ellenrieder V., Johnsen S.A., Hessmann E. Aberrant NFATc1 signaling counteracts TGFbeta-mediated growth arrest and apoptosis induction in pancreatic cancer progression. Cell Death Dis. 2019;10:446. doi: 10.1038/s41419-019-1682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korc M., Friess H., Yamanaka Y., et al. Chronic pancreatitis is associated with increased concentrations of epidermal growth factor receptor, transforming growth factor alpha, and phospholipase C gamma. Gut. 1994;35:1468–1473. doi: 10.1136/gut.35.10.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Means A.L., Meszoely I.M., Suzuki K., et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 53.Sandgren E.P., Luetteke N.C., Palmiter R.D., et al. Overexpression of TGFα in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 54.Liu F., Wang L., Perna F., Nimer S.D. Beyond transcription factors: how oncogenic signalling reshapes the epigenetic landscape. Nat Rev Cancer. 2016;16:359–372. doi: 10.1038/nrc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nava M., Dutta P., Zemke N.R., et al. Transcriptomic and ChIP-sequence interrogation of EGFR signaling in HER2+ breast cancer cells reveals a dynamic chromatin landscape and S100 genes as targets. BMC Med Genomics. 2019;12:1–15. doi: 10.1186/s12920-019-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou R.-H., Wang Y.-N., Hsieh Y.-H., et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell. 2014;30:224–237. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopp J.L., von Figura G., Mayes E., et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinz S., Romanoski C.E., Benner C., Glass C.K. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langer L.F., Ward J.M., Archer T.K. Tumor suppressor SMARCB1 suppresses super-enhancers to govern hESC lineage determination. Elife. 2019;8 doi: 10.7554/eLife.45672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J., Whyte W.A., Zepeda-Mendoza C.J., et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alver B.H., Kim K.H., Lu P., et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:1–10. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathur R., Alver B.H., San Roman A.K., et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagarajan S., Rao S.V., Sutton J., et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet. 2020;52:187–197. doi: 10.1038/s41588-019-0541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koenig A., Linhart T., Schlengemann K., et al. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology. 2010;138 doi: 10.1053/j.gastro.2009.10.045. 1189–1199 e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakhai H., Sel S., Favor J., et al. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- 66.Guan B., Suryo Rahmanto Y., Wu R.-C., et al. Roles of deletion of Arid1a, a tumor suppressor, in mouse ovarian tumorigenesis. J Natl Cancer Inst. 2014;106:dju146. doi: 10.1093/jnci/dju146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goetze R.-G., Buchholz S.M., Patil S., et al. Utilizing high resolution ultrasound to monitor tumor onset and growth in genetically engineered pancreatic cancer models. J Vis Exp. 2018;134 doi: 10.3791/56979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis B.P., Hammer R.E., Messing A., MacDonald R.J. Selective expression of trypsin fusion genes in acinar cells of the pancreas and stomach of transgenic mice. J Biol Chem. 1992;267:26070–26077. [PubMed] [Google Scholar]
- 69.Patil S., Steuber B., Kopp W., et al. EZH2 regulates pancreatic cancer subtype identity and tumor progression via transcriptional repression of GATA6. Cancer Res. 2020;80:4620–4632. doi: 10.1158/0008-5472.CAN-20-0672. [DOI] [PubMed] [Google Scholar]
- 70.Mendez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20(22):8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Afgan E., Baker D., van den Beek M., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobin A., Davis C.A., Schlesinger F., et al. Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nelson J.D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1(1):179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 76.Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & development. 2006;20(5):601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H., Handsaker B., Wysoker A., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Liu T., Meyer C.A., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stark R., Brown G. DiffBind: differential binding analysis of ChIP-Seq peak data. R package version. 2011;100(4.3) [Google Scholar]