Abstract
Terpene cyclases (TCs), the extraordinary enzymes that create the structural diversity seen in terpene natural products, are traditionally divided into two classes. Although the structural and mechanistic features in class I TCs are well-known, the corresponding details in class II counterparts have not been fully characterized. Here, we report the genome mining discovery and structural characterization of two class II sesquiterpene cyclases (STCs) from Streptomyces. These drimenyl diphosphate synthases (DMSs) are the first STCs shown to possess β,γ-didomain architecture. High-resolution X-ray crystal structures of DMS from Streptomyces showdoensis (SsDMS) in complex with both a farnesyl diphosphate and Mg2+ unveiled an induced-fit mechanism with an unprecedented Mg2+ binding mode, finally solving one of the lingering questions in class II TC enzymology. This study supports continued genome mining for novel bacterial TCs and provides new mechanistic insights into canonical class II TCs that will lead to advances in TC engineering and synthetic biology.
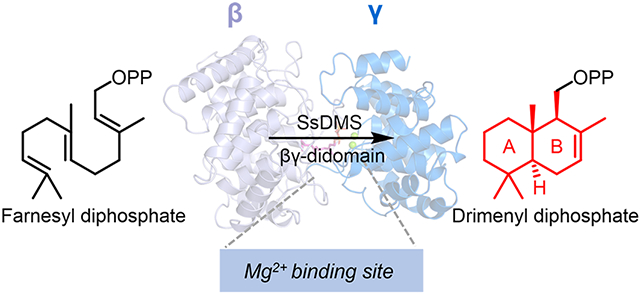
Graphical Abstract
INTRODUCTION
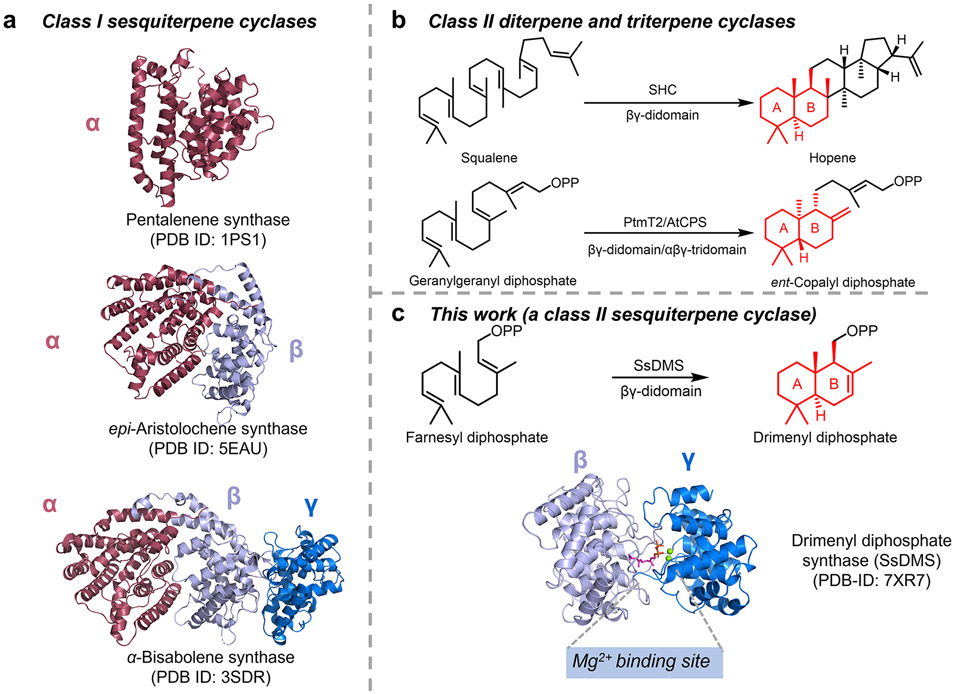
Terpene cyclases (TCs) are catalytically complex and functionally diverse enzymes that create the hydrocarbon scaffolds of terpene natural products. TCs can be categorized into two classes according to different strategies of initial carbocation generation.1-5 Class I TCs possess discrete DDxxD and NSE/DTE motifs to abstract the diphosphate group, thereby triggering cyclization. Class II TCs generate carbocations by protonating the terminal alkene or epoxide of its prenyl diphosphate substrate. Most TCs can also be differentiated by their structures with more than 30 structures of TCs supporting a modular nature.1,6-10 Class I TCs, with characterized examples forming monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), and sesterterpenes (C25), are known to have α, αβ, or αβγ architectures where the functional class I active site is located in the α domain (Figure 1a).1 Class II TCs are traditionally known to have βγ or αβγ architectures with well-known examples forming diterpenes and triterpenes (Figure 1b). Recently, a monofunctional β domain was shown to have meroditerpene cyclization activity.1,11 All structurally characterized sesquiterpene cyclases (STCs) are class I TCs; no structures of class II STCs are known (Figure 1a).1
Figure 1. Overview of the architectures of class I and class II sesquiterpene cyclases (STCs).
(a) Architectural variants of class I STCs. α, β, γ domains are shown in red, light blue, and marine blue, respectively. No structure of class II STCs was known before this study. (b) Structurally characterized class II triterpene and diterpene cyclases. SHC is a squalene-hopene cyclase. AtCPS and PtmT2 represent the plant and bacterial ent-copalyl diphosphate synthases, respectively. (c) Overview of the discovery and structure of a class II STC, SsDMS, in this study. SsDMS catalyzes farnesyl diphosphate into drimenyl diphosphate. Apo-SsDMS owns a βγ-didomain architecture that resembles other class II TCs. A Mg2+ binding site was seen in class II TCs for the first time.
STCs cyclize farnesyl diphosphate (FPP) to form the most diverse subfamily of terpenes with more than 121 distinct skeletons.1,12 The vast majority of STCs utilize the class I catalytic mechanism and have an α-domain-containing structure. Drimenol, the established precursor of drimane-type sesquiterpenoids in plants, fungi, and marine bacteria, is a unique example of a TC product that can be formed via both class I and class II catalytic mechanisms.13-15 Two drimenol synthases (DMSs) of plant origin were biochemically determined to be class I TCs that cyclize FPP into drimenol using the conserved DDxxD motif.16,17 Two fungal haloacid dehalogenase (HAD)-like enzymes were also determined to be DMSs. AstC, the DMS from Aspergillus oryzae (AoDMS), was characterized as a non-canonical DxDT motif-containing class II TC that forms drimenyl diphosphate (DPP); drimenol is formed through subsequent hydrolysis of DPP by a separate hydrolase genetically clustered with AoDMS.4,18 AcDMS, an HAD-like DMS from Aspergillus calidoustusi, is a bifunctional enzyme that biosynthesizes drimenol using a DxDT motif in its N-terminal domain and a class I-like DDxxD motif in its C-terminal domain.19 These examples encouraged us to tap bacterial genomes to identify class II STCs. A marine bacterial HAD-like DMS was discovered from Aquimarina spongiae (AsDMS) using AoDMS as a genetic beacon during the preparation of this manuscript.20
In this study, we identified two drimenyl diphosphate synthases from Streptomyces showdoensis and Streptomyces cattleya, SsDMS and ScDMS, respectively. Structural characterization of SsDMS using X-ray crystallography led to the determination that these enzymes possess the well-known βγ-didomain structure of canonical class II TCs, an architecture previously unseen in STCs (Figure 1c). Detailed analysis of both apo-SsDMS and several structures of SsDMS in complex with FPP or 2-fluoro-farnesyl diphosphate (2F-FPP) and Mg2+ revealed insights into its mechanism and the binding mode of divalent cations in this family of TCs.
RESULTS
Genome Mining for Class II STCs in Streptomyces.
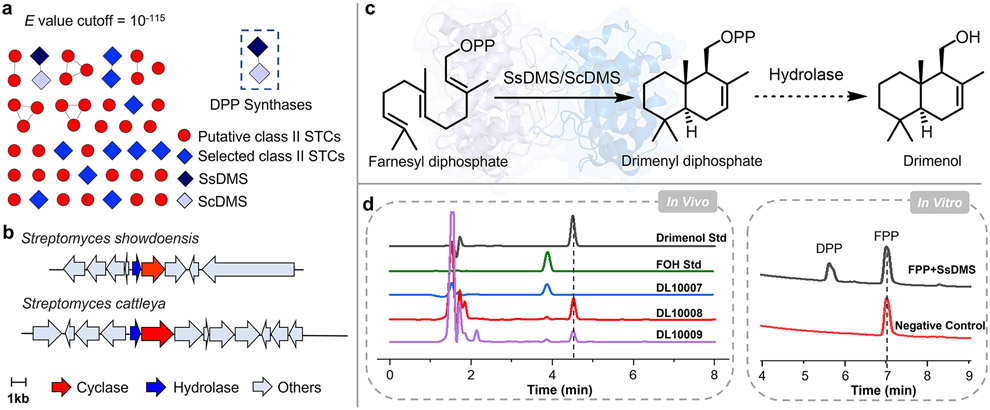
To identify novel class II STCs from bacteria, we first searched sequence databases for proteins found in the superfamily of squalene-hopene cyclases (PF13243).21 There were 10,335 sequences of bacterial origin in PF13243 with 1202 members found in Streptomyces (Figure S1), a well-known source of bioactive and structurally complex natural products.22,23 After removing known or highly homologous triterpene cyclases (TTCs) and diterpene cyclases (DTCs), we hypothesized that 45 proteins with low sequence identities to known bacterial TCs (Figures 2a and S2, and Table S1), 12 of which had the canonical DxDD motif but low to moderate sequence identities ranging from 19 to 57%; eight of the 45 proteins had alternatives to the DxDD motif, namely DxDT, DxDA or DxET (Figures S3 and S4, and Table S2), may be class II STCs.18,24-26 Analysis of their genomic neighborhoods further revealed that two of the putative STCs, A0A2P2GK84 and F8JSS1 from Streptomyces showdoensis and Streptomyces cattleya, respectively, were encoded immediately downstream of a Nudix hydrolase (PF00293). Nudix hydrolases, which remove nucleoside diphosphates linked to another moiety-X, are found in some terpenoid biosynthetic pathways.27 TcNudix1, a Nudix hydrolase from Tanacetum cinerariifolium, removes the terminal phosphate group of chrysanthemyl diphosphate in chrysanthemol biosynthesis; RhNUDX1 converts geranyl diphosphate into geranyl monophosphate to facilitate scent production in rose.28,29 Thus, this clustering implicated A0A2P2GK84 and F8JSS1 as monofunctional class II TCs that provide cyclized prenyl diphosphates for subsequent hydrolysis, as seen in drimenol biosynthesis by AoDMS and AcDMS (Figure 2b).18,19 In addition, these two enzymes share 57% and 64% sequence identity and similarity with each other, respectively, but show very low sequence identity (<20%) with any of the known DMSs of fungi or plants origin (Table S3).16-20 Therefore, we prioritized these two putative STCs for functional characterization.
Figure 2. Discovery of bacterial class II sesquiterpene cyclases (STCs).
(a) A sequence similarity network (SSN) of 45 putative class II STCs, including the 12-selected STCs (colored blue) with DxDD motifs, displayed at an e-value cutoff at 10−115. SsDMS and ScDMS are shown in dark blue and light purple, respectively. (b) Biosynthetic gene clusters show that both putative STCs, SsDMS and ScDMS, are located immediately downstream of a Nudix hydrolase. (c) Farnesyl diphosphate (FPP) is cyclized into drimenyl diphosphate (DPP) by SsDMS and ScDMS, and then is hydrolyzed into drimenol by unknown endogenous hydrolases in E. coli. (d) The in vivo experiments showed that both strains of DL10008 and DL10009, containing SsDMS and ScDMS, respectively, were able to produce drimenol; the in vitro enzymatic activity reconstitution indicated that SsDMS is a class II STC forming DPP. DL10007 is a control strain without SsDMS or ScDMS. Std, standard. FOH, an authentic sample of farnesol.
We characterized A0A2P2GK84 and F8JSS1 in E. coli using the recently developed artificial isoprenoid pathway.30-32 First, we retrofitted our diterpene-producing plasmid pLD10007 with FPP synthase (IspA) in place of geranylgeranyl diphosphate (GGPP) synthase to form pLD10012 and the FPP-producing E. coli strain DL10007 (Tables S4-S6). We then cloned the genes encoding A0A0P2GK84 and F8JSS1 into DL10007 to form strains DL10008 and DL10009, respectively. Analysis of extracts of each culture revealed a new peak with identical retention times (Figure 2d). Large-scale fermentation (5-L) of DL10008 afforded 75 mg of the product, with NMR analysis by 1H and 13C NMR supporting its structure as drimenol (Figures S5 and S6).16 A0A0P2GK84 and F8JSS1 were thus named SsDMS and ScDMS, respectively (Figure 2c).
SsDMS is a Drimenyl Diphosphate Synthase.
As only drimenol was identified from the in vivo experiment, it was unclear if SsDMS and ScDMS were monofunctional STCs that yielded DPP, which was then converted into drimenol by endogenous hydrolases in E. coli, or if SsDMS and ScDMS were bifunctional. To confirm the exact catalytic nature of SsDMS and ScDMS, both proteins were heterologously produced in E. coli for in vitro characterization. Although ScDMS was mostly insoluble, SsDMS was highly produced as a soluble monomer and used for subsequent experiments (Figures S7 and S8). Incubation of SsDMS with FPP resulted in the formation of a single product (Figure 2d). 1H, 13C, and 31P NMR analysis supported the product as DPP (Figures S9-S11), confirming SsDMS is a canonical class II STC and only performs cyclization of FPP.16 As with other class II TCs, the SsDMS reaction is dependent on divalent cations with highest activity observed in the presence of Mg2+ (Figures S12 and S13).
Structure of SsDMS Reveals βγ-didomain Architecture for a STC.
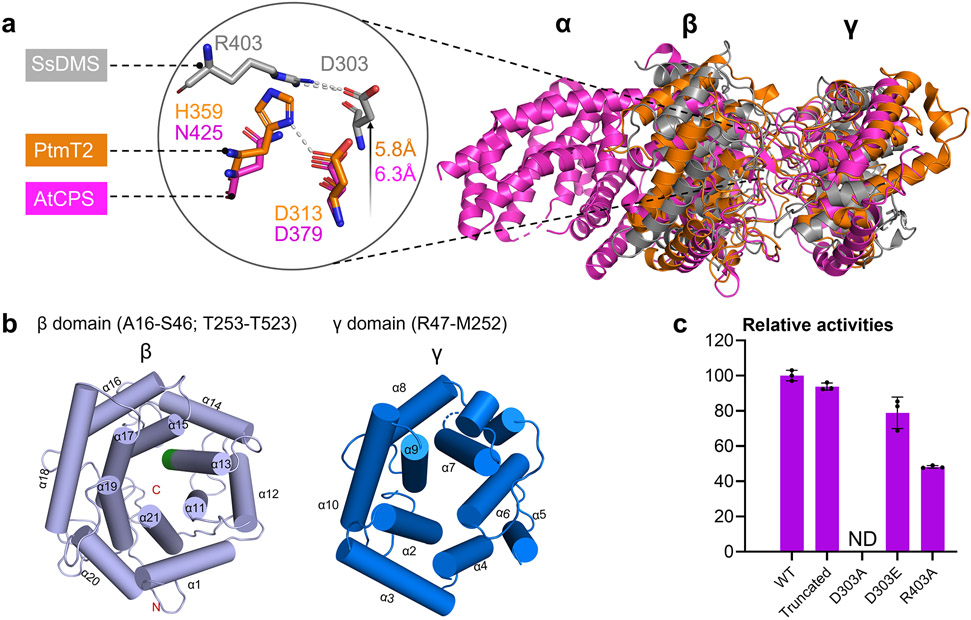
Canonical TCs consist of one, two, or three α-helical domains, α, β, and γ (Figure S14).1 Class I TCs, with its conserved catalytic motif DDxxD located in the α domain, exhibit α (bacterial), αβ, or αβγ (eukaryotic) architectures; in monofunctional enzymes, the β or βγ domains are vestigial.33 In contrast, monofunctional class II TCs mostly display βγ (bacterial) or αβγ (eukaryotic) structures with the catalytic DxDD motif located in the β domain and the active site at the βγ interface; present α domains are nonfunctional.34-36 To probe the structure of SsDMS, we obtained crystals for X-ray crystallography. We found that truncating SsDMS by removing 15 N-terminal residues and 10 C-terminal residues, which retained high (94%) catalytic activity relative to that of wild-type SsDMS (Figure 3c), provided high-quality crystals. The structure of apo-SsDMS was solved at a resolution of 1.58 Å (PDB ID: 7XQ4).
Figure 3. The apo-structure of SsDMS.
(a) Overall structural comparison of apo-SsDMS (PDB ID: 7XQ4, gray), AtCPS (PDB ID: 4LIX, magenta) and PtmT2 (PDB ID: 5BP8, orange). The location of catalytic acid group of D303 and R403 in SsDMS is relatively higher than that in PtmT2 and AtCPS. (b) β- (light blue helices) and γ-domain (marine blue helices) in SsDMS. The conserved catalytic DxDD motif in β-domain is located in α13 helice and shown in green; the dashed loop in γ-domain represents the disordered residues R160–Q163. (c) Relative enzyme activities of D303A, D303E, and R403A mutants, along with the truncated SsDMS used in crystallization. ND, not detected. Error bars indicate the standard deviation of three independent replicates.
SsDMS resembles the previously reported bacterial class II DTCs and is the first STC possessing a βγ-didomain architecture. An overlay of the structures of SsDMS, PtmT2 (PDB ID: 5BP8), and the βγ domains of AtCPS (PDB ID: 4LIX) highlights both similarities and differences between these DTCs and STCs (Figure 3a). The active site of SsDMS, including the catalytic DxDD motif, is in the broad cleft between the β and γ domains (Figure 3b). The function of SsDMS is dependent on the general acid D303, which protrudes into the active site (Figure 3a), as mutation of D303 to Ala completely abolished DPP formation while D303E retained functionality (Figure 3c). D303 hydrogen bonds with the side chain of R403 and mutation of R403 to Ala decreased its relative activity in half, supporting its significance in facilitating use of the carboxylic acid to protonate the terminal olefin of FPP by orienting D303 to a more reactive anti-configuration position (Figure 3a and c).34 Similar interactions were seen in the DTCs AtCPS and PtmT2, although the corresponding residues are Asn and His.34,35 Sequence analysis of the ten putative class II DMSs in bacteria from a BLAST search, which are distinguished with the aforementioned 12 sequences by genome mining, show high sequence identities (43–63%) and strict conservation of Arg at this position (Figures S4 and S15).
We also noticed that the relative position of D303 is shifted 5.8 Å and 6.3 Å higher in the active site than D313 in PtmT2 and D379 in AtCPS, respectively (Figure 3a). This led us to investigate if the active site pocket of SsDMS was significantly smaller than other known class II TCs. Compared with the calculated volumes of the active sites of AtCPS (940 Å3) and the squalene-hopene cyclase (SHC, PDB ID: 1UMP, 1036 Å3), the volume of the cavity of SsDMS was significantly smaller at 536 Å3 (Figure S16).37-39 Based on these values, we hypothesized that SsDMS limits its substrate scope by restricting its cavity to C15 prenyl diphosphates. Fittingly, incubation of SsDMS with GGPP resulted in no detected products, further confirming the substrate selectivity of SsDMS (Figure S17).
Structure of the SsDMSD303E-FPP-Mg2+ Complex Reveals Previously Unseen Features in Class II TCs Including an Unprecedented Mg2+ Binding Mode.
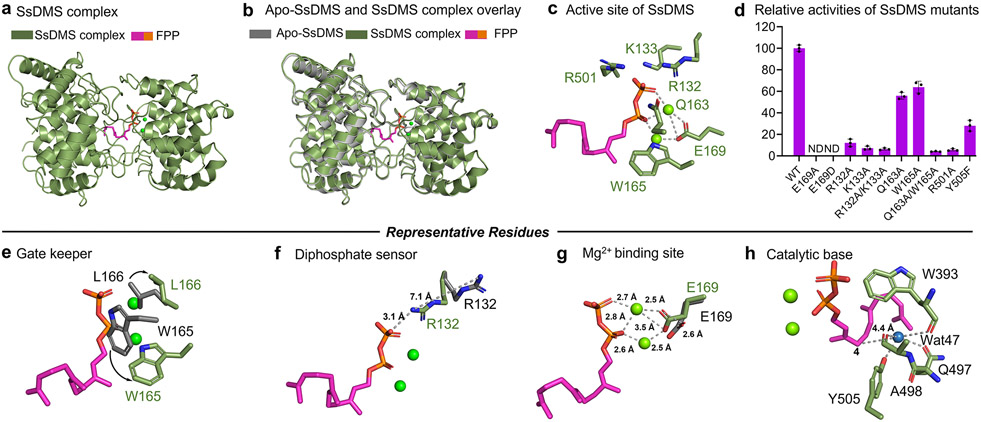
Most TCs with known structures are not characterized in complex with ligands, limiting the level of structural and mechanistic detail that can be gained to understand how these exquisite biocatalysts work. For example, the structural basis for metal ion catalysis in class II TCs has remained enigmatic even after extensive structural, computational, and site-directed mutagenesis studies.34-36,38,40-42 To address this knowledge gap, we determined the structure of SsDMS complexed with FPP or 2F-FPP in the presence of Mg2+. Using SsDMSD303A and SsDMSD303E, we obtained four complexed structures: SsDMSD303A-2F-FPP-Mg2+ (PDB ID: 7XQZ, 2.0 Å), SsDMSD303A-FPP-Mg2+ (PDB ID: 7XRA, 1.95 Å), SsDMSD303E-2F-FPP-Mg2+ (PDB ID: 7XRU, 2.5 Å), and SsDMSD303E-FPP-Mg2+ (PDB ID: 7XR7, 1.63 Å) (Figure S18). These four structures, including the ligands, were nearly identical with each other with root-mean squared deviations (rmsd) of 0.130–0.143 Å, as well as to the structure of apo-SsDMS (rmsd of 0.132 Å). More importantly, the binding of ligands enhanced the poor electron density of R160–Q163 in the apo-structure, leading to a well resolved complex structure with no residues missing near or in the active site. The SsDMSD303E-FPP-Mg2+ complex (referred to SsDMS complex below) was used for the following analysis (Figure 4a).
Figure 4. Structural snapshots of SsDMS in complex with ligand and divalent metal cofactor.
(a) SsDMS complex shows that FPP resides in the cleft of β and γ domain. (b) Overlaying apo-SsDMS and SsDMS complex shows minor conformational changes. (c) Active site of SsDMS. (d) Relative activities of SsDMS mutants reveal key residues in terpene cyclization. (e) W165, termed as a gatekeeper, rotates 90° and drives L166 outwards to allow the ligand access to enter the active site. (f) R132, termed as a diphosphate sensor, rotates its side chain to bring the guanidinium moiety within 3.1 Å of the diphosphate. (g) The E169 shifted by 2.6 Å to form a substrate-ions-enzyme coordinated complex. (h) The proposed catalytic base group deprotonates the final carbocation intermediate. The side chains in apo-SsDMS and SsDMS complex are shown in gray sticks and marine green sticks, respectively. Substrate of FPP is shown in magenta, the diphosphate group of which is shown in orange. ND, not detected. Error bars indicate the standard deviation of three independent replicates.
Analysis of the SsDMS complex in comparison with apo-SsDMS revealed not only the presence of a binuclear Mg2+ cluster and a catalytically relevant conformation of the FPP, but also showed several residues in the active site that made significant structural changes upon binding FPP (Figure 4b). The two Mg2+ ions coordinate with both oxygens in the carboxylate of E169 and with two oxygens in the diphosphate moiety of FPP (Figure 4g). An E169A mutant completely lost activity supporting the pivotal role E169 plays in catalysis by binding the divalent cations (Figure 4d). Furthermore, SsDMSE169D failed to restore activity suggesting an exact positioning of the carboxylate is required for proper binding and catalysis; this is also supported by a 2.6 Å shift in the position of E169 in the apo structure (Figure 4g). In addition to the two Mg2+ ions, the diphosphate group formed ionic interactions with the side chains of R132, K133, Q163, W165, and R501 indicating that the protein also provided a basic environment to bind the diphosphate moiety (Figure 4c). R132 appears to act as a diphosphate sensor, as upon ligand binding, the side chain of R132 underwent a significant rotation to bring the guanidinium moiety within 3.1 Å of the diphosphate (Figure 4f).3,43
Another residue that dramatically shifted upon ligand binding was W165. Seven aromatic residues, W165, F248, F255, F293, F344, W393, and Y505, and P500, were found in the active site contributing to the hydrophobicity needed to bind the farnesyl moiety and stabilize cationic intermediates via cation-π interactions (Figure S19). In the apo-structure, W165 is found in the same location as that of the diphosphate moiety in complex structure; in the SsDMS complex, W165 underwent a 90° flip and subsequently drove L166 to expand away from the pocket to form an entrance, thus providing access for the ligand to enter the active site (Figure 4e). We propose that W165 acts as a gatekeeper for proper FPP binding. Mutation of W165 to Ala moderately reduced activity to 64% (Table S7).
Site-directed mutagenesis was performed to probe the importance of the residues around the diphosphate group. We individually mutated R132, K133, Q163, W165, and R501 to Ala in wild-type SsDMS (Figure 4d). The R132A, K133A, R501A mutants and R132A/K133A double mutant dramatically decreased enzyme activity by 88%, 92.7%, 94.3% and 93.6%, respectively, supporting their importance for catalysis. Although, the single mutants Q163A and W165A only had a moderate effect on the activity, retaining about 60% activity (Table S7), the double mutant of Q163A/W165A impaired the activity by 95.9%, suggesting that both Q163 and W165 may play an additional stabilizing role in the cyclization of the substrate. Due to substrate inhibition, we only obtained Michaelis-Menten kinetic parameters for the R132A and K133A mutants (Figure S20). Native SsDMS had a Km for FPP of 40.9 ± 8.6 μM and kcat / Km of 7.09 × 10−3 s−1 μM−1. Both R132A and K133A had increased Km values of 81.7 ± 18.5 and 283 ± 22 μM and decreased kcat / Km values of 1.51 × 10−4 and 6.48 × 10−5 s−1 μM−1, respectively. The differences in Km (2–7 fold) and catalytic efficiency (48–110 fold) confirmed that a basic environment contributes to both substrate recognition and catalysis.
DISCUSSION
SsDMS and ScDMS represent the first class II STCs from bacteria and SsDMS is the first STC shown to possess the βγ-didomain architecture of bacterial class II TCs. Although other enzymes have been shown to cyclize FPP into DPP, the study of these two unprecedented TCs solved some of the remaining structural and mechanistic questions regarding this family of cyclization enzymes.
First, the detection of Mg2+ in the active site of a class II TC fundamentally solves the question of divalent cation binding in these enzymes. Unlike class I TCs, where three Mg2+ bind to prenyl diphosphates to facilitate diphosphate abstraction and initiate cyclization, class II TCs utilize Mg2+ ions to bind and position the substrate in the active site cavity.1,34 The presence of Mg2+ has been shown to increase catalytic efficiency up to 2600-fold.40,44 While previous biochemical, structural, and bioinformatics studies proposed the Mg2+ binding location, there was not definitive supporting data. Bioinformatics suggested an EDxxD region may mimic a class metal-binding motif, but structural studies with plant and bacterial ent-CPPSs did not support this proposal.34,35,40,45 In AtCPS, a single Glu, E211, residue was found 5 Å from the diphosphate moiety, but no metal ions were seen in the structure.34 In SsDMS, we see two Mg2+ ions coordinated with E169, a residue that spatially corresponds to E211 in AtCPS, and the diphosphate moiety, creating an FPP-Mg2+-E169 coordinated complex. This single negative charged residue must be perfectly positioned in the active site to coordinate the two divalent cations such that the diphosphate moiety binds in a position to place the terminal alkene near the catalytic acid D303. The placement of the two Mg2+ ions in SsDMS is different than of those seen in the trinuclear cluster in class I TCs (Figure S21);33,46,47 this is expected, however, given that SsDMS uses only E169 to bind the ions while class I TCs use DDxxD and NSE/DTE motifs.47
The ability to visualize an FPP in the active site of SsDMS complex also revealed that this TC possesses a diphosphate sensor, a residue that directly binds to the diphosphate moiety of FPP and is commonly seen in class I TCs.43 In SsDMS, R132, W165, and E169 all rotate during the dynamics of substrate binding to accommodate FPP in an induced-fit mechanism. The diphosphate sensor, R132, works in collaboration with the binuclear Mg2+ cluster to bind and position FPP for catalysis.
Using our high-resolution structures of both apo-SsDMS and complex structures, we proposed a mechanism for this unique class II STC (Figure S22b). Prior to substrate binding, the enzyme sits in an open form where W165 and L166 are positioned where FPP and the two Mg2+ ions bind, and R132 is rotated away from the active site. Upon FPP binding, W165 and L166 move away to provide space for the substrate, E169 coordinates two Mg2+ ions, which anchors the diphosphate moiety of FPP, and R132 rotates to act as the diphosphate sensor. The peripheral residues K133 and R501 also contribute to diphosphate binding. Together, the FPP is oriented with distances between C2-C7 and C6-C11 being 3.9 Å and 3.3 Å, respectively, providing a suitable conformation for subsequent protonation and cyclization. The terminal double bond of FPP is protonated by the general acid D303 (Figure S22a). Stabilized by cation-π interactions of several aromatic residues lining the active site, the farnesyl cationic intermediate is cyclized into a trans-decalin bicycle. DPP is finally formed via deprotonation at C7 (C4 of FPP), which is likely catalyzed by a water molecule (wat47) that is positioned a reasonable distance of 4.4 Å away from C4 of FPP and hydrogen bonds with the side chains of Y505 and Q497, as well as the backbone carbonyl oxygens of A498 and W393 (Figure 4h and S22b).48-52 This hypothesis is further supported by the dramatic decreased enzyme activity (72%) of the Y505F mutant (Figure 4d). It was unexpected that no detectable hydroxylated derivatives were identified in the Y505F mutant, as other DTCs mutated at this position frequently alter the product outcome.48,51,52 To regenerate the general acid, D303 is likely protonated via a water hydrogen bonded to Y307.
CONCLUSIONS
TCs, extraordinary enzymes that are responsible for creating much of the structural diversity in the biosynthesis of terpenoids, have attracted great attention in a variety of disciplines. Our discovery of bacterial class II TCs that biosynthesize the drimenyl diphosphate scaffold not only led to a more detailed understanding of their catalytic mechanisms, but also provides evidence that genome mining for bacterial TCs will ultimately lead to novel enzymes and new natural products. The plasticity of class II TCs, as demonstrated by the ability of class II TCs to create C15, C20, C30, C35 and meroditerpene scaffolds, supports future enzyme engineering and synthetic biology applications.1,11
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Hui-Min Xu from The Public Laboratory Platform at China Pharmaceutical University for assistance with NMR techniques, the staff at beamlines BL02U1, BL10U2, and BL18U1 of Shanghai Synchrotron Radiation Facility for assistance during data collection, and Dr. Ning-Hua Tan’s lab in China Pharmaceutical University for generous access to the instrumentations. We also thank Dr. Hui Ming Ge from Nanjing University for critical reading and helpful discussion. This work is supported in part by the National Science Foundation of China Grant 82073746 (L.-B.D.), the National Institutes of Health Grant R35 GM142574 (J.D.R.), the Natural Science Foundation of Jiangsu Province Grants BK20220123 (B.Z.) and BK20210434 (H.W.), the Thousand Youth Talents Program of China (L.-B.D.), the Jiangsu Specially Appointed Professor Program (L.-B.D.), the Double First-Class University Project (CPUQNJC22_04) (L.-B.D.), and the Jiangsu Funding Program for Excellent Postdoctoral Talent Program (Q.Y.).
Footnotes
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website.
Materials, methods, detailed experimental procedures, bioinformatic analysis, in vivo, in vitro, and structural characterizations of SsDMS (PDF).
The authors declare no competing financial interest.
REFERENCES
- 1.Christianson DW Structural and chemical biology of terpenoid cyclases. Chem. Rev 2017, 117, 11570–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Y; Honzatko RB; Peters RJ Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat. Prod. Rep 2012, 29, 1153–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickschat JS Bacterial terpene cyclases. Nat. Prod. Rep 2016, 33, 87–110. [DOI] [PubMed] [Google Scholar]
- 4.Rudolf JD; Chang CY Terpene synthases in disguise: enzymology, structure, and opportunities of non-canonical terpene synthases. Nat. Prod. Rep 2020, 37, 425–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y-H; Xu H; Zou J; Chen X-B; Zhuang Y-Q; Liu W-L; Celik E; Chen G-D; Hu D; Gao H; Wu R; Sun P-H; Dickschat JS Catalytic role of carbonyl oxygens and water in selinadiene synthase. Nat. Catal 2022, 5, 128–135. [Google Scholar]
- 6.Chen Q; Li J; Liu Z; Mitsuhashi T; Zhang Y; Liu H; Ma Y; He J; Shinada T; Sato T; Wang Y; Liu H; Abe I; Zhang P; Wang G Molecular basis for sesterterpene diversity produced by plant terpene synthases. Plant Commun. 2020, 1, 100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stepanova R; Inagi H; Sugawara K; Asada K; Nishi T; Ueda D; Yasuno Y; Shinada T; Miki K; Fujihashi M; Sato T Characterization of class IB terpene synthase: the first crystal structure bound with a substrate surrogate. ACS Chem. Biol 2020, 15, 1517–1525. [DOI] [PubMed] [Google Scholar]
- 8.He H; Bian G; Herbst-Gervasoni CJ; Mori T; Shinsky SA; Hou A; Mu X; Huang M; Cheng S; Deng Z; Christianson DW; Abe I; Liu T Discovery of the cryptic function of terpene cyclases as aromatic prenyltransferases. Nat. Commun 2020, 11, 3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhardt I; de Rond T; Chen PY-T; Moore BS Ancient plant-like terpene biosynthesis in corals. Nat. Chem. Biol 2022, 18, 664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronnebaum TA; Gardner SM; Christianson DW An aromatic cluster in the active site of epi-isozizaene synthase is an electrostatic toggle for divergent terpene cyclization pathways. Biochemistry 2020, 59, 4744–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moosmann P; Ecker F; Leopold-Messer S; Cahn JKB; Dieterich CL; Groll M; Piel J A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds. Nat. Chem 2020, 12, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klapschinski TA; Rabe P; Dickschat JS Pristinol, a sesquiterpene alcohol with an unusual skeleton from Streptomyces pristinaespiralis. Angew. Chem. Int. Ed. Engl 2016, 55, 10141–10144. [DOI] [PubMed] [Google Scholar]
- 13.Du W; Yang Q; Xu H-M; Dong L-B Drimane-type sesquiterpenoids from fungi. Chin. J. Nat. Med 2022, 20, 1–13. [DOI] [PubMed] [Google Scholar]
- 14.Jansen BJ; de Groot A Occurrence, biological activity and synthesis of drimane sesquiterpenoids. Nat. Prod. Rep 2004, 21, 449–477. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y; Valiante V Chemical diversity and biosynthesis of drimane-type sesquiterpenes in the fungal kingdom. ChemBioChem 2022, e202200173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon M; Cochrane SA; Vederas JC; Ro DK Molecular cloning and characterization of drimenol synthase from valerian plant (Valeriana officinalis). FEBS Lett. 2014, 588, 4597–4603. [DOI] [PubMed] [Google Scholar]
- 17.Henquet MGL; Prota N; van der Hooft JJJ; Varbanova-Herde M; Hulzink RJM; de Vos M; Prins M; de Both MTJ; Franssen MCR; Bouwmeester H; Jongsma M Identification of a drimenol synthase and drimenol oxidase from Persicaria hydropiper, involved in the biosynthesis of insect deterrent drimanes. Plant J. 2017, 90, 1052–1063. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara Y; Takahashi S; Osada H; Koyama Y Identification of a novel sesquiterpene biosynthetic machinery involved in astellolide biosynthesis. Sci. Rep 2016, 6, 32865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y; Hoefgen S; Valiante V Biosynthesis of fungal drimane-type sesquiterpene esters. Angew. Chem. Int. Ed. Engl 2021, 60, 23763–23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vo NNQ; Nomura Y; Kinugasa K; Takagi H; Takahashi S Identification and characterization of bifunctional drimenol synthases of marine bacterial origin. ACS Chem. Biol 2022, 17, 1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punta M; Coggill PC; Eberhardt RY; Mistry J; Tate J; Boursnell C; Pang N; Forslund K; Ceric G; Clements J; Heger A; Holm L; Sonnhammer EL; Eddy SR; Bateman A; Finn RD The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watve MG; Tickoo R; Jog MM; Bhole BD How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol 2001, 176, 386–390. [DOI] [PubMed] [Google Scholar]
- 23.Rudolf JD; Alsup TA; Xu B; Li Z Bacterial terpenome. Nat. Prod. Rep 2021, 38, 905–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prisic S; Xu M; Wilderman PR; Peters RJ Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 2004, 136, 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano C; Okamura T; Sato T; Dairi T; Hoshino T Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem. Commun 2005, 1016–1018. [DOI] [PubMed] [Google Scholar]
- 26.Xu M; Jia M; Hong YJ; Yin X; Tantillo DJ; Proteau PJ; Peters RJ Premutilin synthase: ring rearrangement by a class II diterpene cyclase. Org. Lett 2018, 20, 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLennan AG The Nudix hydrolase superfamily. Cell. Mol. Life Sci 2006, 63, 123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W; Lybrand DB; Xu H; Zhou F; Last RL; Pichersky E A trichome-specific, plastid-localized Tanacetum cinerariifolium Nudix protein hydrolyzes the natural pyrethrin pesticide biosynthetic intermediate trans-chrysanthemyl diphosphate. Front. Plant Sci 2020, 11, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnard JL; Roccia A; Caissard JC; Vergne P; Sun P; Hecquet R; Dubois A; Hibrand-Saint Oyant L; Jullien F. d. r.; Nicolè F; Raymond O; Huguet S; Baltenweck R; Meyer S; Claudel P; Jeauffre J; Rohmer M; Foucher F; Hugueney P; Bendahmane M; Baudino SJS Biosynthesis of monoterpene scent compounds in roses. Science 2015, 349, 81–83. [DOI] [PubMed] [Google Scholar]
- 30.Chatzivasileiou Alkiviadis O; Ward V; Edgar Steven M; Stephanopoulos G Two-step pathway for isoprenoid synthesis. Proc. Natl. Acad. Sci. U S A 2019, 116, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund S; Hall R; Williams GJ An artificial pathway for isoprenoid biosynthesis decoupled from native hemiterpene metabolism. ACS Synth. Biol 2019, 8, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F-R; Lin X; Yang Q; Tan N-H; Dong L-B Efficient production of clerodane and ent-kaurane diterpenes through truncated artificial pathways in Escherichia coli. Beilstein J. Org. Chem 2022, 18, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koksal M; Jin Y; Coates RM; Croteau R; Christianson DW Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 2011, 469, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koksal M; Hu H; Coates RM; Peters RJ; Christianson DW Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat. Chem. Biol 2011, 7, 431–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudolf JD; Dong LB; Cao H; Hatzos-Skintges C; Osipiuk J; Endres M; Chang CY; Ma M; Babnigg G; Joachimiak A; Phillips GN Jr.; Shen B Structure of the ent-copalyl diphosphate synthase PtmT2 from Streptomyces Platensis CB00739, a bacterial type II diterpene synthase. J. Am. Chem. Soc 2016, 138, 10905–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y; Prach LM; O'Brien TE; DiMaio F; Prigozhin DM; Corn JE; Alber T; Siegel JB; Tantillo DJ Crystal structure and mechanistic molecular modeling studies of Mycobacterium tuberculosis diterpene cyclase Rv3377c. Biochemistry 2020, 59, 4507–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinert DJ; Balliano G; Schulz GE Conversion of squalene to the pentacarbocyclic hopene. Chem. Biol 2004, 11, 121–126. [DOI] [PubMed] [Google Scholar]
- 38.Koksal M; Potter K; Peters RJ; Christianson DW 1.55Å-resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis. Biochim. Biophys. Acta 2014, 1840, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schöning-Stierand K; Diedrich K; Fährrolfes R; Flachsenberg F; Meyder A; Nittinger E; Steinegger R; Rarey M ProteinsPlus: interactive analysis of protein–ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prisic S; Peters RJ Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 2007, 144, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda C; Hayashi Y; Itoh N; Seto H; Dairi T Functional analysis of eubacterial ent-copalyl diphosphate synthase and pimara-9(11),15-diene synthase with unique primary sequences. J. Biochem 2006, 141, 37–45. [DOI] [PubMed] [Google Scholar]
- 42.Zhou K; Gao Y; Hoy JA; Mann FM; Honzatko RB; Peters RJ Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. J. Biol. Chem 2012, 287, 6840–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baer P; Rabe P; Fischer K; Citron CA; Klapschinski TA; Groll M; Dickschat JS Induced-fit mechanism in class I terpene cyclases. Angew. Chem. Int. Ed. Engl 2014, 53, 7652–7656. [DOI] [PubMed] [Google Scholar]
- 44.Peters RJ; Croteau RB Abietadiene synthase catalysis: conserved residues involved in protonation-initiated cyclization of geranylgeranyl diphosphate to (+)-copalyl diphosphate. Biochemistry 2002, 41, 1836–1842. [DOI] [PubMed] [Google Scholar]
- 45.Cao R; Zhang Y; Mann FM; Huang C; Mukkamala D; Hudock MP; Mead ME; Prisic S; Wang K; Lin FY; Chang TK; Peters RJ; Oldfield E Diterpene cyclases and the nature of the isoprene fold. Proteins 2010, 78, 2417–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whittington DA; Wise ML; Urbansky M; Coates RM; Croteau RB; Christianson DW Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc. Natl. Acad. Sci. U S A 2002, 99, 15375–15380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C-C; Malwal SR; Han X; Liu W; Ma L; Zhai C; Dai L; Huang J-W; Shillo A; Desai J; Ma X; Zhang Y; Guo R-T; Oldfield E Terpene cyclases and prenyltransferases: structures and mechanisms of action. ACS Catal. 2020, 11, 290–303. [Google Scholar]
- 48.Potter K; Criswell J; Zi J; Stubbs A; Peters RJ Novel product chemistry from mechanistic analysis of ent-copalyl diphosphate synthases from plant hormone biosynthesis. Angew. Chem. Int. Ed. Engl 2014, 53, 7198–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenkovskaia L; Murakami M; Okuno K; Ueda D; Sato T Analysis of the catalytic mechanism of bifunctional triterpene/sesquarterpene cyclase: Tyr167 functions to terminate cyclization of squalene at the bicyclic step. ChemBioChem 2017, 18, 1910–1913. [DOI] [PubMed] [Google Scholar]
- 50.Stowell E; Ehrenberger M; Lin Y-L; Chang C-Y; Rudolf J Structure-guided product determination of bacterial type II diterpene synthases. 2022-06-28. ChemRxiv. DOI: 10.26434/chemrxiv-2022-ls8xl-v2. (accessed 2022-08-12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Criswell J; Potter K; Shephard F; Beale MH; Peters RJ A single residue change leads to a hydroxylated product from the class II diterpene cyclization catalyzed by abietadiene synthase. Org. Lett 2012, 14, 5828–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemke C; Potter KC; Schulte S; Peters RJ Conserved bases for the initial cyclase in gibberellin biosynthesis: from bacteria to plants. Biochem. J 2019, 476, 2607–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.