Abstract
The specific patterns and functional properties of electrical synapses of a nervous system are defined by the neuron-specific complement of electrical synapse constituents. We systematically examined the molecular composition of the electrical connectome of the nematode C. elegans through a genome- and nervous system-wide analysis of the expression patterns of the invertebrate electrical synapse constituents, the innexins. We observe highly complex combinatorial expression patterns throughout the nervous system and found that these patterns change in a strikingly neuron type-specific manner throughout the nervous system when animals enter an insulin-controlled diapause stage under harsh environmental conditions, the dauer stage. By analyzing several individual synapses, we demonstrate that dauer-specific electrical synapse remodeling is responsible for specific aspects of the altered locomotory and chemosensory behavior of dauers. We describe an intersectional gene regulatory mechanism, involving terminal selector and FoxO transcription factors mediating dynamic innexin expression plasticity in a neuron type- and environment-specific manner.
Graphical Abstract
INTRODUCTION
Understanding the detailed anatomical synaptic wiring of an entire nervous system - its ‘connectome’ - is an essential first step to elucidate how a nervous system processes information and generates behavior. The adult C. elegans hermaphrodite was the first organism for which an entire connectome has been established (White et al., 1986), recently followed by that of a simple chordate, Ciona intestinalis (Ryan et al., 2016), and similar efforts are now underway for the Drosophila and mouse brains (Helmstaedter et al., 2013; Kasthuri et al., 2015; Takemura et al., 2013). However, defining connectomes by reconstruction of electron micrographs (EM) remains an exceptionally tedious process and limits the ability to analyze the plasticity of the connectome under distinct conditions, on a nervous system wide level. While many dynamic aspects of brain function, from the modulation of behavior to memory formation, are generally appreciated to rely on the alterations of synaptic circuitry (McEwen, 2010; Takeuchi et al., 2014), a nervous system-wide appreciation of the plasticity of neuronal connectomes is still lacking. This paper sets out to start addressing these shortcomings.
Connectomes are defined by two types of synapses, chemical and electrical synapses (gap junctions). While the role of chemical synapses in nervous system function has been widely studied, electrical synapses have received much less attention. The importance of electrical synapses is, however, well documented through genetic analysis in invertebrate and vertebrate nervous system, in which the loss of constituent components of electrical synapses result in obvious dysfunctions of the nervous system (Abrams and Scherer, 2012; Hall, 2017; Hasegawa and Turnbull, 2014; Song et al., 2016). Unfortunately, since electrical synapses are difficult to detect in currently used high-throughput EM methodologies, a map of electrical synapses (“electrical connectome”) is absent from all currently available large-scale connectomes, except the C. elegans connectome. Within the C. elegans connectome, electrical synapses are wide-spread: every one of the 118 neuron classes of the C. elegans hermaphrodite makes electrical synapses to an average number of 9.7 synaptic partners (range: 1 to 30) (Jarrell et al., 2012; White et al., 1986). Moreover, the patterns of electrical synaptic connectivity show only weak correlation with the patterns of chemical synaptic connectivity (Varshney et al., 2011).
Electrical synapses are composed of two analogous families of transmembrane, pore-forming proteins, connexins in vertebrates (encoded by 21 distinct genes in mammals) and innexins in invertebrates (encoded by 25 genes in C. elegans) (Phelan and Starich, 2001; Willecke et al., 2002). Electrical synapses are formed by either homogeneous or heterogeneous combinations of connexins or innexins (Mese et al., 2007; Miller and Pereda, 2017; Phelan and Starich, 2001) (Fig.1A). This diverse molecular composition is thought to encode: (a) synaptic specificity, i.e. the choice of synaptic partners, based on a matching assembly of innexin proteins in connected neurons; and (b) functional diversity, i.e. electrical synapses with distinct molecular compositions are thought to have different functional properties (Mese et al., 2007; Miller and Pereda, 2017; Sohl et al., 2005). The neuron-type specific patterns of innexin gene expression are therefore a likely determinant of the electrical synapse development and function.
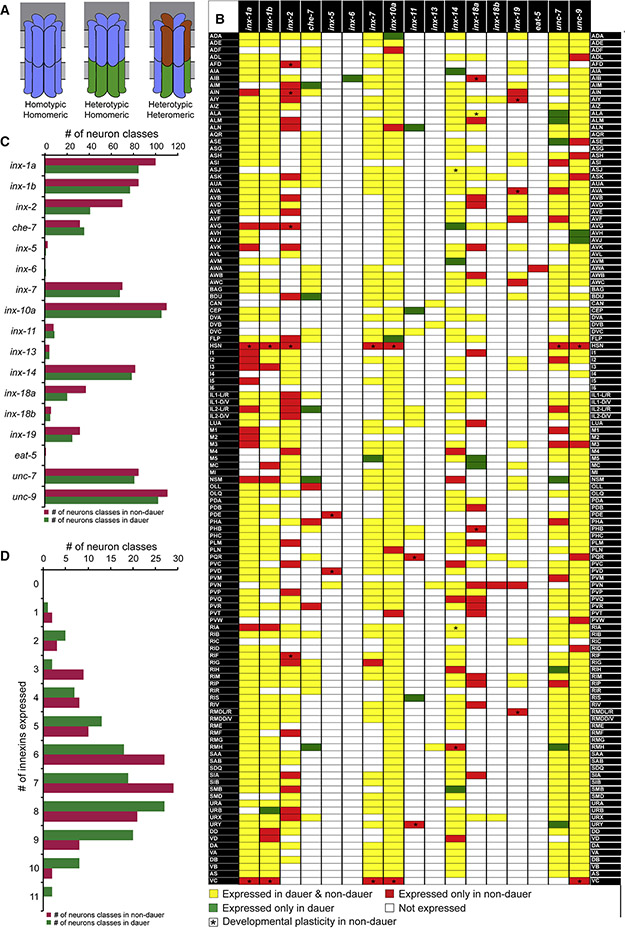
Fig. 1: Innexin gene expression in non-dauer and dauer.
(A) Schematics of subunit composition in different kinds of gap junction channels. Hemichannels formed by innexins can also be octameric (Oshima et al., 2016).
(B) Expression of all neuronally expressed innexin genes in all neuronal classes. Color of each box represents whether the innexin gene is expressed in both non-dauer and dauer (yellow), only in non-dauer (red) or only in dauer stage (green). Asterisks indicate developmental changes in innexin expression among non-dauer stages. See also Table S2.
(C) Abundance of each innexin gene in non-dauer and dauer nervous system.
(D) Distribution of number of innexin genes in neuron classes in non-dauer and dauer.
We describe here the expression pattern of all C. elegans innexin genes and find that 14 innexin genes are expressed in a highly combinatorial manner in the nervous system. We discover a striking extent of plasticity of neuron-type specific expression of innexin genes in response to environmental cues, thereby predicting a substantial rewiring of the electrical connectome. The adverse environmental conditions we test here are starvation, high population density or increased temperature, which altogether trigger the entry of C. elegans larvae into a diapause-arrest stage, the dauer stage (Cassada and Russell, 1975; Fielenbach and Antebi, 2008; Riddle and Albert, 1997). Honing in on a single innexin in a single neuron type, we show that the dauer-specific induction of inx-6 in the AIB interneuron class is both necessary and sufficient to form multiple de novo electrical synapses. We characterize alterations of the locomotory features of dauer-stage animals and demonstrate that some of these locomotory changes, as well as some altered sensory behaviors of dauers, are mediated through the rewiring of specific electrical synapses (including the novel INX-6-mediated electrical synapses of AIB). Lastly, we demonstrate a gene regulatory strategy by which these changes in electrical synaptic connectivity are orchestrated by the constitutive activity of cell type specific homeodomain transcription factors in conjunction with the stress-induced, cell autonomous activation of insulin/IGF-1 target DAF-16/FOXO. Taken together, our findings highlight both the importance and the plasticity of electrical synapses on a systems-wide level, show that this plasticity is encoded at the level of transcriptional regulation and implicate the conserved FoxO transcription factor in the plasticity of synaptic connectivity.
RESULTS
A nervous system-wide molecular map of the electrical connectome
Our overall strategy to investigate the plasticity of synaptic wiring was to first establish a map of molecular determinants of synaptic connectivity and to then identify conditions under which the expression of these synaptic connectivity factors are modulated, thereby predicting changes in synaptic connectivity and circuit function. In addition, our implicit approach was to exploit the relative simplicity of the C. elegans nervous system to undertake such an analysis on a genome- and nervous system-wide manner. Since electrical synapses are defined by the combinatorial expression of specific innexin proteins (Fig.1A) (Mese et al., 2007; Miller and Pereda, 2017; Phelan and Starich, 2001), we sought to precisely map the expression of all neuronal innexins.
A previous study using antibody staining revealed that two innexins, INX-21 and INX-22 are exclusively expressed in the gonad and thus are not included here (Starich et al., 2014). For inx-2 and inx-6, fluorescent reporter alleles were generated using CRISPR/Cas9-mediated genome editing (see Methods). For the remaining 21 innexin family genes, we generated fosmid-based reporter constructs (see Methods; schematics of all fosmid reporters and reporter alleles are shown in Fig.3, S2-3). Three innexin genes, inx-1, inx-10 and inx-18, contain alternate splice-isoforms with different 3’ end sequences. Independent fosmid reporter constructs were generated to avoid any potential isoform specific expression bias. Among them, expression of inx-10b could not be faithfully identified after examining multiple independent transgenic lines and thus deemed not expressed.
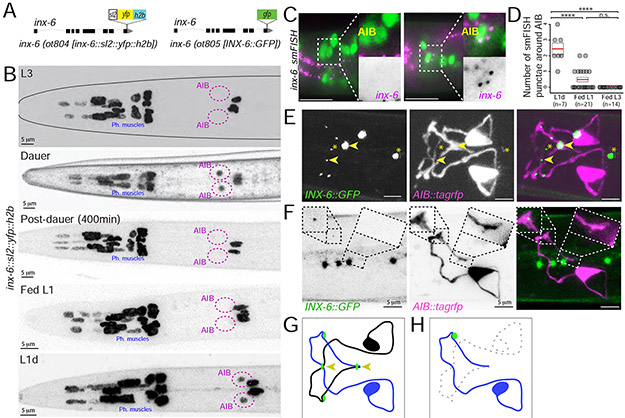
Fig. 3: Dauer-induced expression of inx-6 in AIB generates new gap junctions with che-7.
(A) Schematics of inx-6 transcriptional reporter allele, inx-6(ot804) and GFP-tagged inx-6 translational reporter allele, inx-6(ot805).
(B) inx-6 reporter allele, inx-6(ot804) is additionally turned on in AIB interneurons in dauer that disappears in post-dauer stage. inx-6 allele is also expressed in AIB in L1d.
(C) smFISH against endogenous inx-6 mRNA (magenta) in Fed-L1 and L1d. AIB was marked by eat-4::yfp (otIs388) expression (green). Inset shows enlargement of AIB.
(D) Quantification of inx-6 smFISH data. Each circle represents the number of AIB associated smFISH puncta in a single animal, red lines indicate the mean and rectangles indicate S.E.M. Wilcoxon rank-sum tests p-values (n.s. = non significant and ****p<0.0001).
(E) Expression of INX-6 puncta (green), in inx-6(ot805) dauer. npr-9p::tagrfp (otIs643) expression marks AIB processes (magenta). Arrowheads mark INX-6::GFP at the crossover points of AIBL and AIBR. Asterisks mark INX-6 puncta in pharyngeal muscles.
(F) Expression of INX-6 puncta (green) on AIBL (magenta) in daf-7(e1372) dauers (inx-6(ot805); daf-7(e1372); otIs643), where AIBR was ablated. In absence of AIBR, INX-6 punctum at the AIBL-AIBR crossover point (right box) disappear, while has no effect on INX-6 puncta in other region (left box). See Fig. 3E for control (pre-ablation) image. Four INX-6 puncta that do not overlap with AIB-processes are among pharyngeal muscles.
(G) Schematic of major INX-6 puncta (green circles) on AIB. Arrowheads mark INX-6 puncta at the AIBL-AIBR crossover points.
(H) Schematic of the effect of AIBR-ablation on INX-6 puncta on the remaining AIBL neuron. See Fig. 3G for control (pre-ablation) schematic.
We found that 14 innexin genes are expressed in the nervous system during larval and adult stages (Fig.1,2,S2-3). Ten of the neuronally expressed genes, inx-1, inx-2, che-7, inx-7, inx-10, inx-14, inx-18, inx-19, unc-7 and unc-9, were selectively (i.e. non-panneuronally) expressed in many (>30) neuron classes, while expression of inx-5, inx-11 and inx-13 were more restricted (<10 neuron classes). One innexin, eat-5, was uniquely expressed in a single neuron class. We also identified distinct expression patterns for specific splice isoforms of inx-1 and inx-18 (Fig.1). Both inx-1 isoforms were co-expressed broadly in the nervous system, but 15 neuronal classes expressed only one isoform. In contrast, the expression pattners of two inx-18 isoforms were strikingly different, where inx-18a was expressed broadly (in 37 neuron classes) and inx-18b expression was restricted to only six neuron classes.
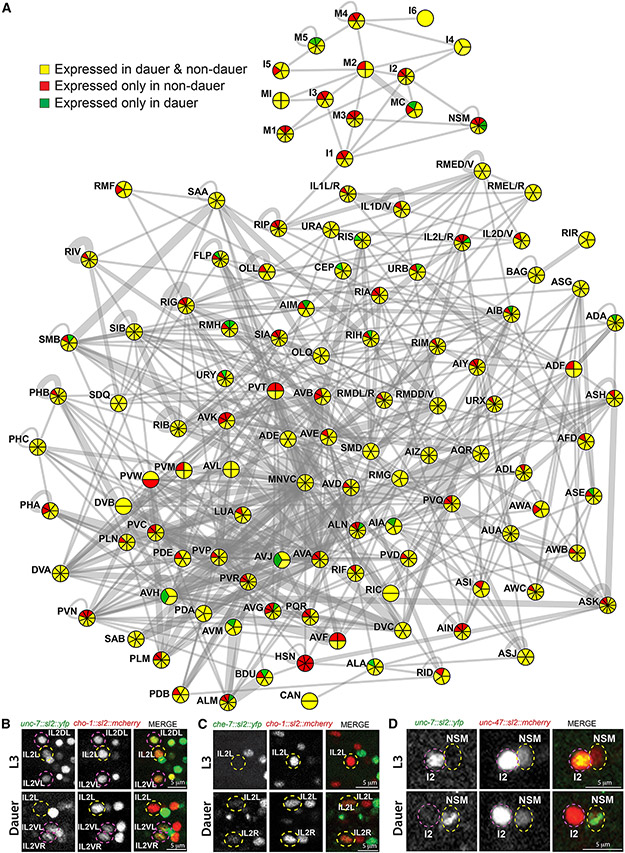
Fig. 2: Changes in innexin gene expression in dauer.
(A) Electrical connectome at the adult stage, as inferred from the serial section EM reconstruction. Neuronal classes are represented as circles and electrical synapses are shown as gray lines. Thicknesses of lines were weighted according to the volume of each connection (measured by the total number of EM sections in which a particular synapse was observed; as per www.wormwiring.org). The number of innexin genes expressed in each neuronal class is indicated by the overlaid pie chart. Color of each pie chart section represents whether the innexin is expressed in both non-dauer and dauer (yellow), only in non-dauer (red) or only in dauer (green).
(B-D) Neuronal identities were determined by expression of either cho-1 (otIs544) or unc-47 (otIs564) reporters.
(B) unc-7 fosmid reporter (otEx7106) is expressed in IL2DL/R, IL2VL/R and IL2L/R in non-dauer, but selectively down-regulated in IL2L/R in dauer.
(C) che-7 fosmid reporter (otEx7112) is selectively turned on in lateral IL2L/R neurons in dauer.
(D) unc-7 fosmid reporter (otEx7106) is expressed in I2, but not in NSM in non-dauer. In dauer, unc-7 expression disappeared in I2, while turned on in NSM.
See also Fig. S2.
In total, every neuron class expressed at least one innexin gene. Most of the neuron classes expressed multiple innexin genes, with the maximum number being 11 (Fig.1B). There is very weak positive correlation between the number of innexin genes expressed in a particular neuron class and the number of unique electrical synaptic partners it has (Pearson correlation coefficient 0.126) (Fig.S1A) and very weak anti-correlation between the number of innexin genes expressed and the total electrical synaptic strength (Pearson correlation coefficient - 0.143) (Fig.S1B). Most strikingly, 98 of the 118 C. elegans neuron classes express distinct, unique combinations of innexins (Fig.1B).
The expression pattern of most innexins is stable throughout larval and adult stages under non-crowded, well-fed conditions, but a subset of innexins show dynamic expression in a subset of neurons during L2 to adult development (Table S2). HSN and VC neurons start to express innexins starting at L4 stage when other neuronal features of these neurons mature. Lastly, all innexin genes were expressed in one or more non-neuronal cell types (Fig.S2,S3; Table S1). The expression patterns that we describe have very little overlap with the innexin expression previously reported using very small (1kb or smaller) promoter fusions (Altun et al., 2009), a likely testament of the complexity of cis-regulatory control elements of the innexin loci, not captured in the previous study.
Widespread changes in the neuron type-specificity of innexin gene expression in dauer stage animals
Since entry into the dauer stage entails a substantial remodeling of behavior (Cassada and Russell, 1975; Gaglia and Kenyon, 2009; Hallem et al., 2011a; Lee et al., 2012), we reasoned that the connectome of the dauer might also be substantially distinct. Since the availability of innexins in a cell is expected to have a profound impact on synaptic specificity and properties (Mese et al., 2007; Miller and Pereda, 2017; Sohl et al., 2005), we investigated the expression patterns of innexins in dauer as proxy for the electrical synaptic connectivity. We found that 11 of the 14 innexin genes expressed in the nervous system in larval and adult stages show striking neuron-specific expression plasticity (Fig.1,2,S1). This includes one innexin gene, eat-5, which is exclusively expressed in AWA in non-dauer stage is turned off upon entry into the dauer stage. In striking contrast, another innexin gene, inx-6, is expressed only in the dauer nervous system (discussed later in detail). Overall, 86 of the 118 neuron classes show expression change in the dauer stage (Fig.1,2,S1,S2). The maximum number of changes in innexins expression in a neuron class is three (Fig.1,2,S1).
Several aspects of the innexin expression plasticity are of particular note. Firstly, expression of distinct innexin genes could be altered in an opposing manner in a particular neuron class when animals go into dauer arrest (Fig.1,2). For example, unc-7 expression was downregulated, while che-7 expression was simultaneously upregulated in the lateral IL2 neuron pair (Fig.2B,C). Secondly, expression of a particular innexin gene could change in opposing manner in distinct neuronal classes during dauer molt (Fig.1,2). For example, in dauer, unc-7 expression was downregulated in pharyngeal I2 neuron, while simultaneously upregulated in another pharyngeal neuron NSM (Fig.2D). Thirdly, expression of specific splice isoforms of a particular innexin gene could be independently altered in distinct neuronal classes in dauer. For example, inx-1a, but not inx-1b expression was specifically downregulated in the lateral IL2 neurons in dauer (Fig.1).
We conclude that the nervous system of C. elegans undergoes a widespread remodeling of innexin expression pattern. These changes in expression may trigger changes in synaptic partner choice, prompted by the loss or gain of specific innexin genes, or changes in the signaling properties of the existing electrical synapses through changes in their molecular composition.
Formation of INX-6-containing electrical synapses in AIB interneurons in dauer stage animals
A fosmid-based inx-6 reporter transgene, otIs473, and a genome-engineered SL2-based “transcriptional” inx-6 reporter allele, ot804 are expressed in non-neuronal pharyngeal cells (Fig.3A), as previously reported (Li et al., 2003). We found that during the dauer molt, the inx-6 reporter allele, ot804, was additionally turned on in a single pair of interneurons, AIB (Fig.3B). This dauer-specific expression of inx-6 was reversible and disappeared from AIB when dauer animals resume development under favorable conditions (Fig.3B). inx-6 expression was also turned on in AIB during L1-diapause (L1d), which was reversible and disappeared upon feeding (Fed-L1) (Fig.3B). This observation was confirmed using single molecule fluorescence in situ hybridization (smFISH) (Fig.3C,D).
To determine whether the inx-6 expression in AIB during dauer resulted in punctate localization of the protein characteristic of electrical synaptic contacts, we engineered a gfp protein-fusion allele of inx-6, ot805 (Fig.3A). Since this allele does not show the lethality associated with the loss of inx-6 function (Li et al., 2003), gfp-tagging does not appear to grossly interfere with inx-6 function. In inx-6(ot805) animals, punctate gfp expression was observed in the pharyngeal muscles in all stages, while in dauer and L1d animals distinct GFP puncta were localized along the AIB processes in positions that were stereotypic across different animals (Fig.3E,G). These results suggest that the expression of an innexin gene, inx-6, is dynamically regulated in the dauer nervous system and potentially leads to the formation of either novel synaptic contacts or changes the composition of already existing AIB electrical synapses (made with several distinct synaptic partners according to EM analysis) (White et al., 1986).
INX-6 expressed in AIB functions with CHE-7 in BAG to form new electrical synapses in dauer stage animals
We next set out to analyze the INX-6 puncta formed in the AIB neurons in dauer. We first examined the large INX-6 puncta observed at the crossover points of the two bilaterally symmetric AIB processes, suggesting the formation of electrical synapses between these two neurons (such auto-synapses are not observed in non-dauer animals) (Jarrell et al., 2012; White et al., 1986). To test this hypothesis, we surgically removed one of the bilateral AIB neurons and found that such ablation leads to selective loss of the INX-6 puncta at the points where the AIB neuronal processes normally cross (Fig.3F,H). This result suggests that the bilateral AIB neurons may electrically couple selectively in dauer.
To characterize the other highly stereotyped INX-6 puncta along the AIB processes, we set out to identify innexins that may partner with INX-6 in trans across an electrical synapse. We based our search on two criteria: (1) the puncta formed by the partner innexin should colocalize with the INX-6 puncta and (2) formation of the INX-6 puncta should depend on the presence of such partner innexin, as shown for other heterotypic gap junctions (Starich et al., 2009). In line with these criteria, we found: (1) TagRFP-tagged CHE-7 protein expressed from a fosmid-based reporter, co-localized with the INX-6 puncta in dauer at two distinct positions along the AIBL and AIBR processes, away from the crossover points (Fig.4A,B) and (2) loss of che-7 results in specific loss of INX-6 puncta only at these distinct positions, but not at the AIBL-AIBR crossover points (Fig.4C,D). These results suggest that CHE-7 acts as the INX-6 partner to form electrical synapses specifically in AIB neurons in dauers. Moreover, che-7, although broadly expressed, is not expressed in AIB in both non-dauers and dauers (Fig.1), indicating that CHE-7 is expressed in the putative AIB synaptic partner neurons and interacts with INX-6 in trans across the synapse.
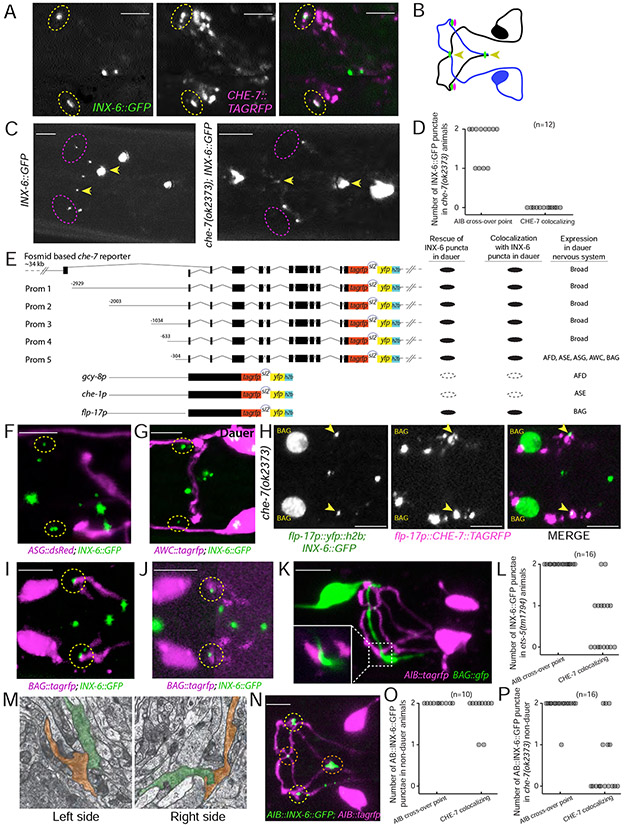
Fig. 4: inx-6 and che-7 forms gap junction between AIB and BAG.
(A) o-localization of INX-6::GFP (green) and CHE-7::TagRFP puncta (magenta) in two putative gap junctions on AIB (yellow circle) in dauer (ot805; otEx6486).
(B) Schematic of CHE-7 puncta (magenta circles) localization with INX-6 puncta (green circles) on AIB. Arrowheads mark INX-6 puncta at the AIBL-AIBR crossover points.
(C) INX-6 puncta that colocalized with CHE-7 were lost in che-7(ok2373); inx-6(ot805) mutant dauers (magenta circles). INX-6 puncta at the AIBL-AIBR crossover points remained unaffected (arrowheads).
(D) Quantification of INX-6 puncta on AIB in che-7(ok2373) dauers.
(E) Schematics of 5’ cis-regulatory element analysis of che-7 and reporter transgenes used for cell-specific che-7 expression. Transgenic lines were created in che-7(ok2373); inx-6(ot805) background. Results show average for all transgenic lines.
(F-G) INX-6 puncta (green) that co-localize with CHE-7 in dauer (dotted circles) do not colocalize with ASG (ot805;oyIs47) and AWC (ot805;otIs263) axons (magenta).
(H) In che-7(ok2373) dauer, expression of CHE-7::TAGRFP (magenta) only in BAG (green) rescues CHE-7 associated INX-6 puncta (green) that also shows colocalization with CHE-7::TAGRFP (arrowheads). See Fig.4C for control image showing INX-6 puncta in che-7(ok2373) dauer. (inx-6(ot805); che-7(ok2373); otEx2487)
(I) INX-6 puncta (green) in dauer co-localize with BAG axon (magenta). (ot805; otEx7230)
(J) INX-6 puncta (green) in L1d co-localize with BAG axon (magenta). For BAG-axon image clarity, a projection of non-continuous z-sections was shown. (ot805; otEx7230)
(K) BAG axon (green) comes in contact with AIB process (magenta) at similar positions where INX-6 puncta co-localize with BAG axon. Inset shows enlargement of the contact point. (otIs643; otEx7230)
(L, O and P) INX-6 puncta present at the AIBL-AIBR crossover points are referred as ‘AIB crossover point’ and INX-6 puncta that co-localize with CHE-7 and BAG axon are referred as ‘che-7 colocalizing’.
(L) Quantification of INX-6 puncta on AIB in ets-5(tm1794) mutant dauers.
(M) TEM prints from wild-type adult hermaphrodite ‘N2U’ showing adjacency of BAG (pseudo colored in red) and AIB (pseudo colored in green) processes at the site where dauer specific INX-6-CHE-7 putative electrical synapses are formed. These images were collected in MRC/LMB and annotated images were obtained from www.wormimage.org, courtesy of David Hall. Prints shown here are, Left: N2U_094 and right: N2U_116.
(N) AIB-specific ectopic expression of INX-6 in non-dauer stage (L3) results in INX-6 puncta (green) along the AIB (magenta). Red circles mark INX-6 at the AIBL-AIBR crossover points. - Yellow circles mark INX-6 at the site where dauer specific INX-6-CHE-7 electrical synapses are formed. (otTi19; otIs643)
(O) Quantification of ectopic INX-6 puncta in non-dauer otTi19 animals.
(P) Quantification of ectopic INX-6 puncta in non-dauer che-7(ok2373); otTi19 animals.
To identify the putative AIB synaptic partners, we reasoned that che-7 expression in such a partner neuron should be (a) sufficient to rescue the loss of INX-6 puncta in the che-7 mutant dauer and (b) should co-localize with the INX-6 puncta. To address this, we generated a fosmid-based bi-cistronic reporter that allowed independent visualization of the membrane-localized TagRFP-tagged CHE-7, while simultaneously allowed identification of the che-7 expressing neurons based on the nuclear-localized YFP (Fig. 4E). Corroborating our premise, expression of this che-7 reporter in che-7 mutant dauers was sufficient to rescue the two CHE-7-associated INX-6 puncta and showed colocalization of the CHE-7::TagRFP with INX-6::GFP. Mimicking the transcriptional che-7 fosmid reporter, this bi-cistronic fosmid reporter also showed broad expression in the nervous system (Fig.1B,4E,S2). To narrow down to the putative CHE-7-expressing AIB-synaptic partner, we gradually divided the cis-regulatory region driving the expression of the bi-cistronic che-7 reporter cassette. A 304bp minimal cis-regulatory region that was expressed in five sensory neuron pairs (ASG, AWC, AFD, ASE and BAG) was sufficient to rescue the INX-6 puncta in che-7 mutant dauers and showed colocalization of CHE-7 with INX-6 (Fig.4E). We found that the AWC and AFD axons are not in proximity to the INX-6 puncta (Fig.4F,G). Single neuron specific expression of the che-7 reporter cassette showed that che-7 expression in the BAG neuron alone was sufficient to rescue the INX-6 puncta in che-7 mutant dauer and showed colocalization of CHE-7 with INX-6 (Fig.4H). Moreover, INX-6 puncta also colocalized with BAG axon marker in dauer (Fig.4I) as well as in L1d (Fig.4J). BAG and AIB projections also came into contact with each other at similar positions (Fig.4K). Furthermore, the two CHE-7-associated INX-6 puncta were lost in dauers mutant for ets-5, an ETS domain transcription factor required for the BAG neuron differentiation (Guillermin et al., 2011), while the INX-6 puncta at the AIBL-AIBR crossover points remained unaffected (Fig.4L).
At the adult stage AIB and BAG do not form any electrical synapses, as inferred from the serial section EM reconstruction (White et al., 1986). Analyzing existing EM serial sections of adult animals, we found that the AIB and BAG neuronal processes are directly adjacent to each other at positions similar to the site of INX-6-mediated putative electrical synapse formation in dauer (Fig.4M), indicating that the absence of AIB and BAG electrical synapses in non-dauer animals may solely be the result of the absence of INX-6 expression. We indeed found that ectopic expression of inx-6 in AIB in non-dauer is sufficient to form CHE-7-dependent puncta at the BAG-AIB contact points (Fig.4N-P) as well as at the AIBL-AIBR crossover points Fig.4N,O). These results together suggest that the regulation of inx-6 expression in AIB is both necessary and sufficient for the induction of potential BAG-AIB electrical synapses.
Whether CHE-7 and INX-6 are the only components of a putative BAG-AIB electrical synapse will require future in vitro studies, which will also be required to characterize functional features of such a synapse. In the course of identifying CHE-7 as the likely trans-synaptic partner for INX-6, we also analyzed the localization pattern of several additional, AIB and/or BAG-expressed innexin proteins (INX-1, UNC-7, UNC-9) and found neither of them to be colocalizing with the CHE-7-INX-6 puncta. The BAG-AIB synapse may therefore be composed of a homomeric CHE-7 hemichannel in BAG and a homomeric INX-6 hemichannel in AIB.
Dauer animals display distinctive locomotory behaviors
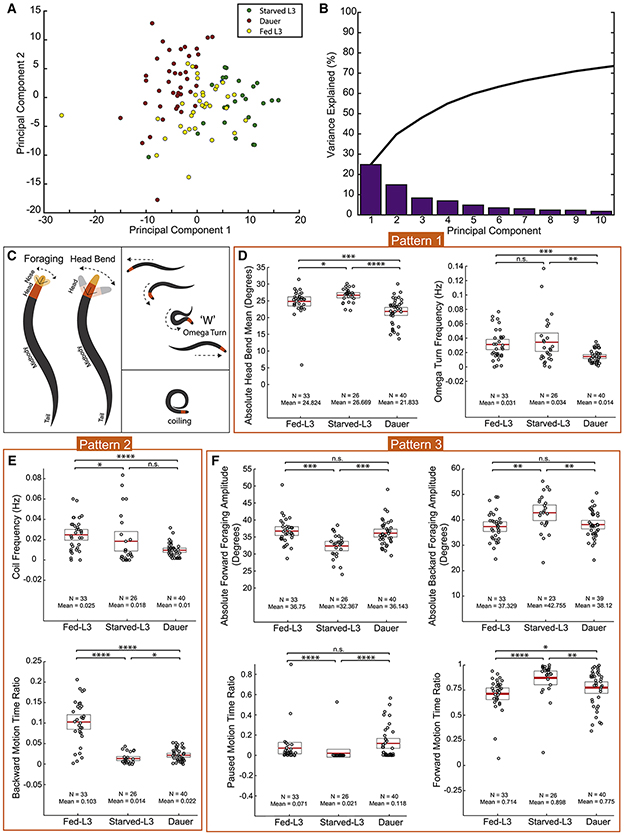
Having defined the system wide changes in innexin expression as well as specific changes of the electrical connectome in the dauer stage, we sought to define the physiological relevance of these changes. Several changes in the behavior of dauer animals have been described before. For example, dauers exhibit prolonged bouts of spontaneous quiescence (Gaglia and Kenyon, 2009), a strong reduction in pharyngeal pumping rate (Cassada and Russell, 1975) and nictation behavior (Cassada and Russell, 1975; Lee et al., 2012). We sought to further expand the known repertoire of altered behaviors of dauers by quantitatively assessing the locomotory patterns of dauer and non-dauer animals using an automated Wormtracker system (Yemini et al., 2013). Specifically, we compared three different stages and states: (1) Wild-type Bristol N2 strain animals in the dauer stage, induced under standardized starvation, crowding and high-temperature conditions. (2) Animals in L3 larval stage (Fed-L3), which animals enter instead of the dauer stage under favorable conditions. (3) To assess the impact of starvation alone, we also analyzed L3 animals after a 4h-8h interval with no food (starved-L3).
Principal component analysis (PCA) of 195 locomotory behavioral features revealed that the behaviors of dauer, fed- and starved-L3 animals could be clustered in distinct, separate groups (Fig.5A,B; Table S3). Three different patterns of changes could be observed (Fig.5; Table S4). In pattern #1, dauers differ from both fed-L3s and starved-L3s. Examples include head bend amplitude (Fig.5C,D) and omega turn behavior, where animals change direction by attaining an intermediate omega symbol (Ω) like posture, and is often associated with the initiation of backing (Fig.5C,E). Dauers performed omega turns less frequently than both fed- and starved-L3s (Fig.5E). In pattern #2, both starved-L3s and dauers differ from fed-L3s in a similar manner. Examples include coiling and backward motion behaviors (Fig.5C,F,G). We found that both starved-L3s and dauers performed coiling much less frequently and showed much less backward motion. Finally, in pattern #3, we observed both dauers and fed-L3s differ from starved-L3s in a similar manner. Examples include foraging, pausing and forward motion behaviors (Fig.5C,H-K). The behavioral differences discussed in pattern #3 indicate that the prolonged starvation-induced habituation may influence dauers to behave as fed-L3s for certain aspects of the dauer behavior, such as food search behavior.
Fig. 5: Locomotory behavior is remodeled in dauer.
(A) Principal Component Analysis of dauer (red), fed-L3 (yellow) and starved-L3s (green) based on 195 locomotory behavior feature data (Listed in table S3). Circles represent individual animals (ndauer = 40, nFed-L3 = 33, nstarved-L3 = 26). Component 1 and 2 account for ~40% of the variation in the locomotory behaviors.
(B) Variance explained by first ten PCs. Black line represents cumulative variance explained.
(C) Schematics of foraging (nose bend), head bend, reversal through an omega turn and coiling behaviors.
(D-K) Comparison of dauer, fed- and starved-L3 locomotion using Wormtracker (see Methods for details). Each circle represents the experimental mean of a single animal. Red lines indicate the mean of means and rectangles indicate S.E.M. Wilcoxon rank-sum tests and False-Discovery Rate q-values for each comparison: n.s. = non significant, *q<0.05, **q<0.01, ***q<0.001, ****q<0.0001.
(Behavioral feature time ratio = total time spent performing particular behavior/total time of the assay)
inx-6 and che-7 affect locomotory and chemotaxis behavior specifically in dauer
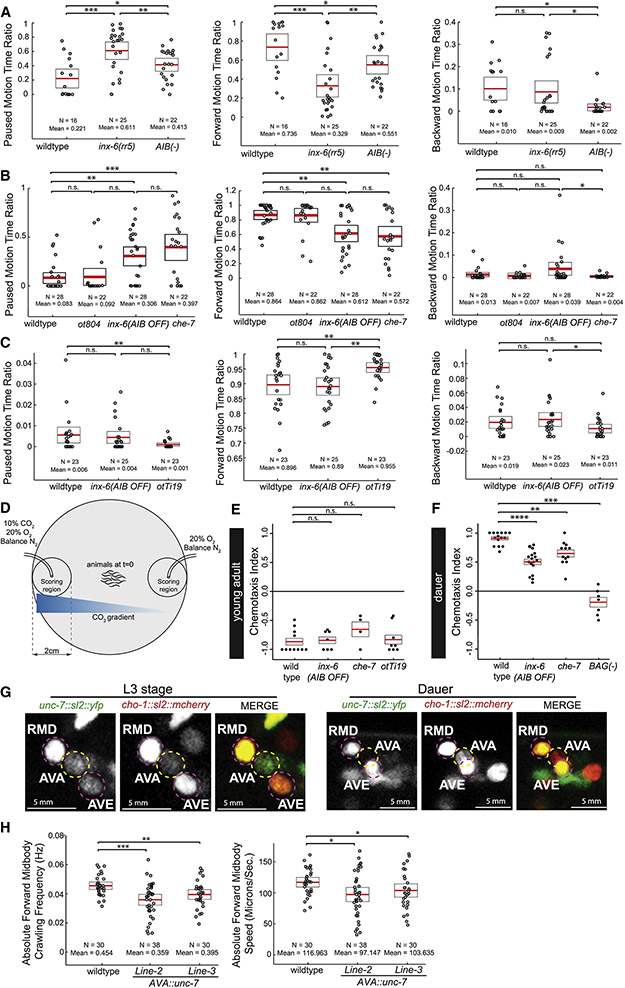
To assess whether formation of the dauer-specific putative de novo INX-6 electrical synapses attributes new functions to AIB, we first assessed the consequences of AIB removal through caspase-driven genetic ablation of AIB (Wang et al., 2017). In fed and starved non-dauers, consistent with the previous reports (Gray et al., 2005), AIB-ablation severely affected backward locomotion and subsequently enhanced forward locomotion (Fig.S4E-J). However, AIB acquires novel functions in dauer locomotory behavior. As shown above in Fig.5, dauers showed a distinctive extent of pausing, forward and backward locomotion compared to non-dauer L3-staged animals and ablation of AIB affected all three of these behaviors (Fig.6A-C,S4A,C).
Fig. 6: Loss and gain of innexin expression in dauers affects locomotory and CO2-attraction behavior.
(A-I) Locomotion assay using Wormtracker (see Methods for details). Each circle represents the experimental mean of a single animal. Red lines indicate the mean of means and rectangles indicate S.E.M. Wilcoxon rank-sum test p-values for each comparison: n.s. = non significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (Behavioral feature time ratio = total time spent executing particular behavior/total assay time).
(A-F) Locomotion of dauer animals.
(A-C) Dauers with inx-6 temperature sensitive allele, inx-6(rr5), at restrictive temperature and AIB-ablated dauers [peIs578 (Wang et al., 2017)] show significant increase in pausing and decrease in forward motion.
(D-F) Dauers that specifically lack inx-6 expression in AIB, inx-6(AIB OFF) allele (see Fig. 8) and che-7(ok2373) dauers show significant increase in pausing and decrease in forward motion, but show no effect in backward motion.
(G-I) inx-6(AIB OFF) starved-L3s show no significant difference in locomotion. AIB-specific ectopic expression of INX-6 in starved-L3 (otTi19) affects pausing and forward motion.
(J) Schematic of CO2-chemotaxis assay (See Methods for details).
(K-L) Each circle represents chemotaxis index calculated from a single assay. Red lines indicate the mean and rectangles indicate S.E.M. Wilcoxon rank-sum tests p-values for each comparison: n.s. = non significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
(K) inx-6(AIB OFF) or che-7(ok2373) mutant young adults show no effect in CO2-repulsion. Ectopic expression of INX-6 in AIB (otTi19) in young adults do not affect CO2-repulsion.
(L) inx-6(AIB OFF) and che-7(ok2373) dauers show reduced CO2-attraction. Dauers with ablated BAG neurons (kyIs536; kyIs538) do not chemotax to CO2 gradient. See also Fig. S4.
(M) Expression of the unc-7 fosmid reporter (otEx7106) is lost in AVA in dauer, while continue to be expressed in RMD and AVE. cho-1 (otIs544) expression identifies the neuron.
(N-O) Locomotion of two independent transgenic lines where unc-7 is ectopically expressed in AVA in dauers (Transgenic Line-2: otEx7250 and Line-3: otEx7251).
See also Fig. S4.
To address the role of inx-6 in the functional plasticity of AIB, we tested the locomotion of inx-6 mutant dauers. Since inx-6 loss-of function mutation is L1 lethal, we used a temperature-sensitive allele of inx-6, inx-6(rr5), for the dauer locomotion assay (Li et al., 2003). In addition, to assess the role of INX-6 specifically in AIB, we generated an inx-6(AIB OFF) allele that removes inx-6 expression exclusively in the AIB neurons, leaving the pharyngeal expression intact (discussed later in detail). Dauers mutant with either of the inx-6 alleles, showed pausing and forward locomotion defects similar to the AIB-ablated dauers, however, did not affect the backward locomotion, which was also affected by AIB-ablation (Fig.6A-F). Dauers mutant for che-7, the innexin partner of inx-6, were similarly defective for the same locomotory features (Fig.6A-F). Consistent with the observation that inx-6 is only expressed in the nervous system in dauers, inx-6 mutations did not affect the pausing or forward locomotion in fed or starved non-dauers (Fig.6G-I,S4E-J).
Since ectopic expression of inx-6 in AIB was sufficient to form putative electrical synapses in non-dauer stages (Fig.4N), we next asked whether these ectopic electrical synapses were also sufficient to alter non-dauer locomotion. We indeed found that the AIB-specific ectopic inx-6 expression reduced pausing and enhanced forward locomotion in non-dauers Fig.6G-I), indicating that the induction of an innexin gene expression in a specific neuronal class is both necessary and sufficient to modify the electrical connectome to produce a behavioral output.
We tested additional functions of inx-6, now in the context of processing BAG-dependent sensory responses. C. elegans in non-dauer stage displays a robust chemotactic avoidance response when subjected to a CO2-gradient, which is sensed mainly through BAG (Hallem et al., 2011b). In contrast, dauers are attracted to high concentrations of CO2 in a BAG-dependent manner (Hallem et al., 2011a). Based on the findings described above, we asked whether the putative dauer-specific, CHE-7/INX-6 dependent BAG-AIB synapse also contribute to CO2 chemotaxis of dauer. Indeed, loss of either inx-6 or che-7 reduced CO2 attraction in dauer, but had no effect on CO2 avoidance in non-dauers (Fig.6J-L). We also found that the effect of inx-6 and che-7 mutation was specific to CO2 chemotaxis and did not affect the overall chemotactic ability of dauers (Fig.S4K-O). However, unlike locomotory behaviors, AIB-specific ectopic expression of inx-6 in non-dauer stages had no effect on CO2 chemotaxis (Fig.6K), suggesting a permissive, but not an instructive role of the putative INX-6-CHE-7 electrical synapses in the CO2 chemotaxis circuit.
Downregulation of unc-7 expression in AVA interneurons is required for normal dauer locomotion
Moving beyond the paradigm of the induction of expression of an innexin (and the ensuing establishment of new electrical synapses), we assessed the physiological consequences of the dauer-specific suppression of an innexin. The innexin gene, unc-7, is expressed, among many other neurons, in the AVA command interneurons of the C. elegans motor circuit, where it is required to control the balance of forward and backward locomotion (Fig.1B,6M) (Kawano et al., 2011; Liu et al., 2017; Starich et al., 2009). We found that unc-7 expression was specifically downregulated in AVA in dauer (Fig.1B,6M), suggesting a remodeling of the many electrical synapses that AVA normally makes (White et al., 1986). To test the functional significance of this downregulation, we ectopically maintained unc-7 expression in AVA in dauer and asked which of the dauer-specific locomotory behavioral alterations are potentially reverted back to a more non-dauer state. Multiple distinct parameters that measure the agility of worms are increased in dauers (Fig.5, Table S4) and we found that transgenic, dauer-stage animals force-expressing unc-7 in AVA showed reduced agility, reminiscent of the non-dauer stage (Fig.6N,O). Taken together, remodeling of the electrical synapses of the AVA neurons predicted upon unc-7 downregulation is partly responsible for the remodeling of locomotory behavior of dauers.
DAF-16/FoxO intersects with terminal selector function to ensure dauer-specificity of innexin expression
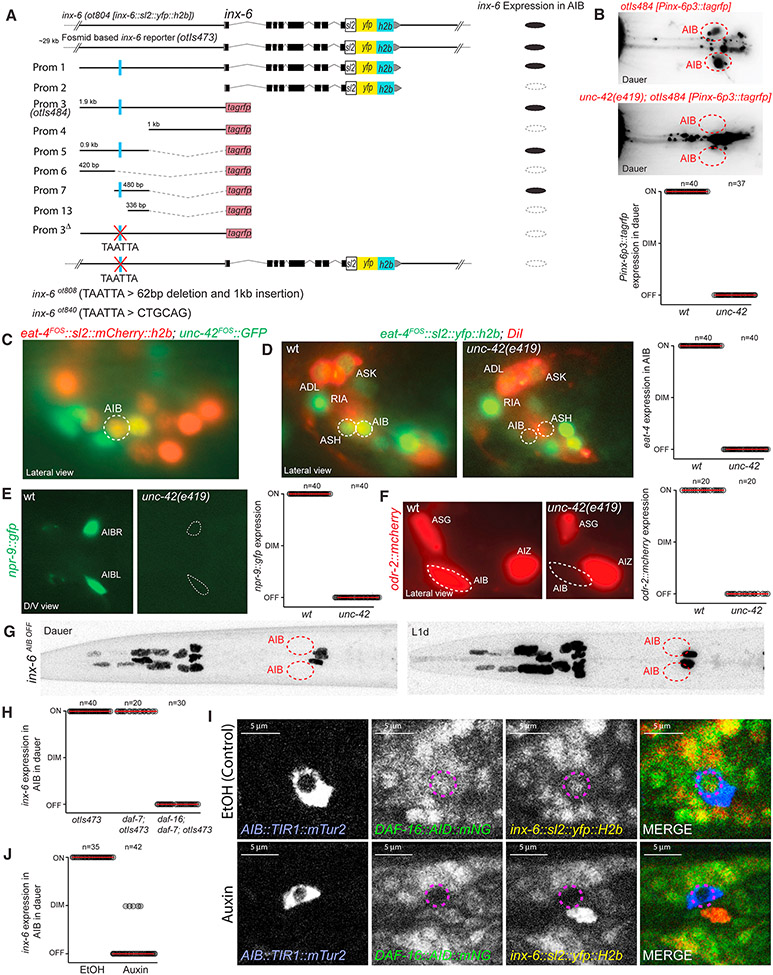
To understand the regulatory logic of the condition-specific innexin expression plasticity, we returned to the inx-6 locus, analyzing its cis-regulatory control elements. We generated transgenic animals carrying reporter transgenes containing various fragments of the inx-6 cis-regulatory region and compared their expression with the inx-6 reporter allele, ot804 and the fosmid reporter, otIs473 (Fig.7A). We identified a 144bp region containing required cis-regulatory information for dauer-specific AIB expression (Fig.7A). This region contains predicted homeodomain binding site that is completely conserved in multiple nematode species (TAATTA). Through a systematic analysis of homeobox gene expression using fosmid-based reporters (unpubl. data), we identified the paired-type homeodomain transcription factor unc-42, related to mammalian Prop1 (Baran et al., 1999), to be expressed in AIB throughout the life of the animal in both dauer and non-dauer stage animals (Fig.7C). The expression of inx-6 reporter transgenes was lost in AIB in unc-42 mutant background (Fig.7B). We deleted the predicted UNC-42 binding site in the genome in the context of the inx-6 transcriptional reporter allele, ot804 (Fig.7A). These alleles were specifically defective for inx-6 expression in AIB in dauer and L1d, while expression in the pharyngeal muscles remained unaffected (Fig.7G), corroborating the requirement of UNC-42 in the spatial regulation of inx-6. This allele constitutes the inx-6(AIB OFF) allele described above.
Fig. 7: UNC-42 and DAF-16/FOXO regulate spatiotemporal expression of inx-6 in AIB.
(A) Schematics of 5’ cis-regulatory element analysis of inx-6. Results show average for all transgenic lines. Deletion of a putative UNC-42 binding site (TAATTA) in the 5’ upstream regulatory region resulted in complete loss of inx-6 expression in AIB.
(B-J) For each graph, circles represent the expression of corresponding reporter in a single animal and red lines indicate the mean. Expression of reporters are scored as: ON = similar to control, DIM = reduced expression and OFF = no expression.
(B) UNC-42 affects inx-6 reporter (otIs484) expression in AIB in dauer.
(C) unc-42 fosmid reporter (wgIs173) (green) is expressed in AIB in all stages. eat-4 (otIs518) (red) expression was used for neuronal identification.
(D-F) Expression of eat-4, npr-9 and odr-2 in AIB were affected in unc-42(e419) mutant animals.
(G) Deletion of a putative UNC-42 binding site in inx-6(ot840) animals, results in loss of inx-6 expression specifically in AIB in dauer and L1d. See Fig.3B for control images.
(H) Quantification of inx-6 fosmid reporter (otIs473) expression in dauer. inx-6 expression is lost in AIB in daf-7(e1372); daf-16(mgDf50) double mutant dauers.
(I) AIB specific degradation of DAF-16 in auxin-treated dauers, results in loss of DAF-16::mNeonGreen as well as inx-6 (yellow) expression in AIB. In EtOH-treated control dauers, expression of DAF-16, as well as inx-6 are maintained. Due to substantial overlap of mNG and YFP emission spectra these two expressions could not be separately imaged. [inx-6(ot804); daf-2(e1370); daf-16(ot853); otEx7309]
(J) Quantification of results shown in panel I.
unc-42 not only affects the dauer-specific inx-6 expression in AIB but also affects other genes expressed constitutively in both dauer and non-dauer AIB neurons, including eat-4/VGLUT, odr-2 and neuropeptide receptor npr-9 (Fig.7D-F), indicating that unc-42 may be a terminal selector that specifies AIB identity. However, the dauer-specificity of inx-6 induction suggests that unc-42 alone is not sufficient to turn on inx-6 expression but requires dauer-specific regulatory inputs. To investigate those, we turned to the insulin/IGF-1 signaling pathway, a major determinant of the dauer arrest mechanism (Fielenbach and Antebi, 2008). In non-dauer animals, the insulin/IGF-1 signaling pathway suppresses the nuclear translocation of the DAF-16/FOXO transcription factor. In dauer animals, insulin/IGF-1 signaling is downregulated leading to nuclear translocation of DAF-16, where it regulates many different target genes in diverse tissue types (Fielenbach and Antebi, 2008). To assess the involvement of daf-16, we examined inx-6 expression in daf-16 mutant animals that were forced into dauer by simultaneously mutating the daf-7/TGFβ endocrine signaling pathway (Lee et al., 2001). In these double mutant dauers inx-6 expression was not induced in AIB, demonstrating that daf-16 is required for the dauer-specific inx-6 expression (Fig.7H). To assess the focus of action of DAF-16, we removed DAF-16 exclusively in AIB. To this end, we engineered an Auxin-Inducible-Degron (AID)-tag (Zhang et al., 2015) and fluorescent reporter gene into the daf-16 locus. The resulting daf-16(ot853) allele allowed us to follow DAF-16 localization dynamics in the nervous system and to degrade DAF-16 with spatiotemporal precision. During dauer stage, DAF-16::mNeonGreen showed the expected nuclear translocation. AIB-specific expression of TIR1 in conjunction with the auxin treatment cell-autonomously depleted DAF-16 only in AIB, but did not affect dauer formation (Fig.7I) and at the same time prevented inx-6 expression in dauer (Fig.7I,J). These results demonstrate that the environmentally controlled insulin/IGF-1 signaling pathway leads to altered DAF-16 activity in AIB, where it cell-autonomously induces inx-6 expression in dauer.
DISCUSSION
While there is a general appreciation of the importance of establishing synaptic connectivity diagrams, much of past and present analysis of connectomes focuses on chemical synapses, even though the importance of electrical synapses in nervous system function has been made apparent by a number of genetic loss of function studies (Abrams and Scherer, 2012; Hall, 2017; Hasegawa and Turnbull, 2014; Marder et al., 2017; Song et al., 2016; White and Paul, 1999). Through the establishment of a nervous system wide map of innexin expression, as well as its dynamic modulation under specific environmental conditions, we have laid here the groundwork to understand a number of distinct features of the electrical connectome. First, it is generally thought that the formation of electrical synapses is encoded by the neuron-type-specific expression of matching homo- or heteromeric hemichannels, which recognize each other in trans to assemble into a functional electrical synapse. Second, the specific composition of an electrical synapse is thought to determine specific conductive properties (Mese et al., 2007; Miller and Pereda, 2017; Sohl et al., 2005). For example, the widespread electrical connectivity of neurons in the C. elegans nervous system clearly indicates that information flow through many of the electrical synapses must be directional and such directionality is likely encoded by the specific molecular composition of individual synapses.
As a prerequisite to address these two issues, we identified the complex innexin expression code of the C. elegans nervous system. 14 out of 25 innexin genes in the C. elegans genome are expressed in 98 distinct combinations in the 118 distinct neuron classes of the worm. Some innexins are expressed very broadly in the nervous system, some much more restrictively. Alternate splice isoforms of a single innexin can be expressed in distinct neuronal classes. Furthermore, the number of distinct innexin genes present in a particular neuronal class also varied greatly. Whereas ASK and RIM neuron classes express the maximum of 11 innexins, the I6 neuron class expresses only one innexin. We hypothesize that this distinct neuron-specific combinatorial innexin expression code may be a key underlying determinant for the establishment of the electrical connectome and may further provide neurons with the ability to form electrical synapses with potentially synapse-specific conductance, which depends on the specific innexin composition.
We have shown that the innexin expression code of the nervous system is not static but rather subjected to widespread, substantial alteration when developing C. elegans larvae encounter harsh environmental conditions and enter the dauer diapause stage. In the dauer nervous system, the expression of 12 out of 14 neuronal innexins is altered in 86 out of 118 neuron classes. Additionally, one non-neuronal innexin gene, inx-6, is also switched on in the dauer nervous system. Often the expression of an innexin is turned on in some neuron classes while simultaneously turned off in others. Some neuron classes simultaneously upregulate one innexin and downregulate another to change their innexin expression code. Changing the expression of innexins in a given neuron class could lead to an alteration (gain and/or loss) of synaptic partners and/or it may lead to changes in the composition of existing synaptic contacts. In either case, these changes are predicted to have an impact on information flow in the nervous system. In one such example, the upregulation of inx-6 in AIB, results in the potential generation of a novel electrical synaptic connection. It will require the EM reconstruction of the entire nervous system of dauer stage animals to assess the extent of altered synaptic partner choice. Our expression map of innexins provides the foundation to understand the molecular mechanisms of structural rewiring observed by such anatomical analysis.
To provide functional correlates to changes in the electrical connectome, we analyzed locomotory behavior of dauers and discovered specific patterns of alteration compared to developmental stage matched non-dauer animals. Notably, although we obtained dauers under prolonged starvation condition (accompanied by high population density and temperature), some aspects of dauer behavior were more similar to satiated non-dauer animals than to short-term starved non-dauer animals, suggesting long term adaptions to starvation. Several changes in locomotory behavior are, however, clearly dauer-specific and fall in line with the previously reported behavioral alteration of dauer stage animals (Cassada and Russell, 1975; Gaglia and Kenyon, 2009; Lee et al., 2012). We have linked here some these behavioral alterations to alteration of distinct innexin expressions in specific neuron types.
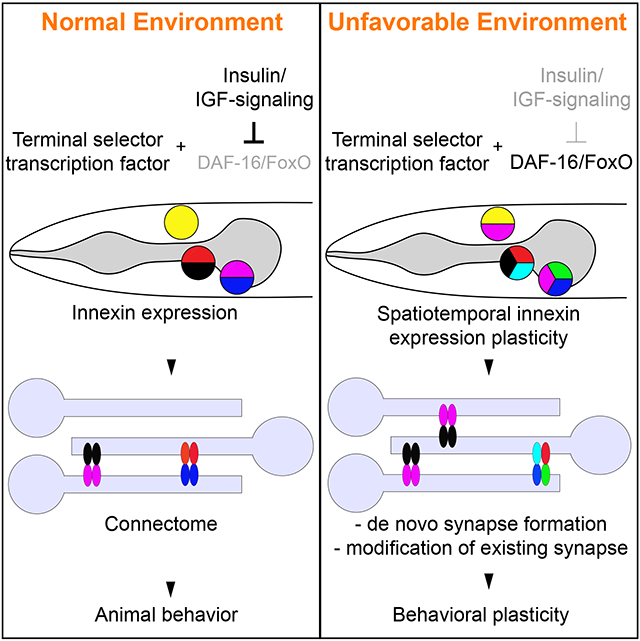
Our findings suggest that the plasticity of electrical synaptic connectivity is encoded on the level of transcriptional gene regulation. Electrical synapse assembly and function is known to be regulated by a number of additional mechanisms (O'Brien, 2014; Thevenin et al., 2013) and our description of gene expression changes of innexins only describes the first layer of regulation of the electrical connectome. Our characterization of such transcriptional dynamics offers a unique opportunity to study the plasticity of gene expression in mature post-mitotic neurons. We have provided here mechanistic insights into how the gene expression plasticity of innexins is controlled. For all neuronal classes studied in C. elegans, expression and maintenance of the neuron type specific terminal fate makers are governed by continuously expressed terminal selector type transcription factors (Hobert, 2016). Apart from controlling several invariant features of AIB identity, we found that the AIB-specific induction of inx-6 also depends on the unc-42 terminal selector. The condition-specific (i.e. dauer-specific) alteration in innexin expression is controlled by the cell-autonomous inactivation of insulin/IGF-1 signaling and resultant activation of DAF-16/FOXO, a protein previously implicated in controlling other aspects of neuronal plasticity in multiple distinct organisms (McLaughlin and Broihier, 2018). An intersectional gene regulatory strategy, involving a “hardwired” neuronal identity program combined with a condition-specific program, may constitute a generalizable mechanism for how the environment can control neuronal plasticity with cellular specificity.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oliver Hobert (or38@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains and handling
Mutant alleles used in this study include daf-2(e1370), daf-7(e1372), daf-16(mgDf50), unc-42(e419), inx-6(rr5) (6x outcrossed), che-7(ok2373) (6x outcrossed) and ets-5(tm1794). A complete list of strains and transgenes used in this study is listed in the Key Resource Table. Worms were grown at 20-22°C on nematode growth medi a (NGM) plates seeded with E. coli (OP50) bacteria as a food source (Brenner, 1974), unless otherwise mentioned. Worms were maintained according to standard protocol. Wild-type strain used is the C. elegans variety Bristol, strain N2. We induced dauer arrest under standardized starvation, crowding and high-temperature conditions. In detail, we put 8-10 L4-stage animals on a regular 6cm NGM-plates seeded with E. coli (OP50) bacteria and incubated at 25°C. Dauers start appearing after 7/8 days and after 10/11 days plates contained plenty of dauers, which were picked for experiments.
KEY RESOURCE TABLE
| REAGENT or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals | ||
| Diacetyl | Sigma-Aldrich | Cat # 11038 |
| Trimethylamine solution | Sigma-Aldrich | Cat # W324108 |
| 2-Butanone | Sigma-Aldrich | Cat # 34861 |
| 2-Octanone | Sigma-Aldrich | Cat # 02479 |
| 2-Heptanone | Sigma-Aldrich | Cat # 537683 |
| Auxin (Indole-3-acetic acid) | Alfa Aesar | Cat # 87-51-4 |
| DiD | Thermofisher | Cat # V22887 |
| Reporter DNA on transgenic array | ||
| inx-1a WRM0672aB09fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15285 [pha-1(e2123); otEx7116] |
| inx-1b WRM0672aB09fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15276 [pha-1(e2123); otEx7107] |
| inx-3 WRM0636dA10fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15291 [pha-1(e2123); otEx7121] |
| inx-3 WRM0636dA10fosmid::gfp/ NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH11846 [otEx5393] |
| che-7 WRM0640dF02 fosmid ::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15281 [pha-1(e2123); otEx7112] |
| che-7 WRM0640dF02fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15277 [pha-1(e2123); otEx7108] |
| ot805; che-7 WRM0640dF02fosmid::tagrfp::SL2::NLS::yfp::H2B / NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13967 [inx-6(ot805); otEx6486] |
| inx-5 WRM0625cD05fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15270 [pha-1(e2123); otEx7101] |
| inx-5 WRM0625cD05fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15271 [pha-1(e2123); otEx7102] |
| inx-6 WRM0630bD03fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12042 [otIs473] |
| inx-7 WRM0631dH08fosmid::SL2::NLS::yfp::H2B/ NotI (10ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15689 [pha-1(e2123); otEx7292] |
| inx-8 WRM0632dA04fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15691 [otEx7294] |
| inx-9 WRM0632dA04fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15532 pha-1(e2123); otEx7227] |
| inx-9 WRM0632dA04fosmid::gfp/ NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH11841 [otEx5388] |
| inx-10a WRM0628bH12 fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15287 [pha-1(e2123); otEx7118] |
| inx-10a WRM0628bH12fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15288 [pha-1(e2123); otEx7119] |
| inx-11 WRM0621cA09fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), unc-122::gfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15242 [pha-1(e2123); otEx7090] |
| inx-12 WRM0621dC07fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15282 [pha-1(e2123); otEx7113] |
| inx-12 WRM0621dC07fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15283 [pha-1(e2123); otEx7114] |
| inx-13 WRM0621dC07fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15273 [pha-1(e2123); otEx7104] |
| inx-14 WRM0626aA10fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15272 [pha-1(e2123); otEx7103] |
| inx-15 WRM0619cH12fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15540 [pha-1(e2123); otEx7232] |
| inx-16 WRM0619cH12fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15274 [pha-1(e2123); otEx7105] |
| inx-17 WRM0619cH12fosmid::gfp/ NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH11495 [otEx5207] |
| inx-17 WRM0619cH12fosmid::gfp/ NotI (15ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH11496 [otEx5208] |
| inx-18a WRM0629cH03fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15225 [pha-1(e2123); otEx7075] |
| inx-18b WRM0629cH03fosmid::SL2::NLS::yfp::H2B/ NotI (20ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15244 [pha-1(e2123); otEx7092] |
| inx-19 Extended WRM0632bE10fosmid ::SL2::NLS::yfp::H2B/ NotI (40ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15542 [pha-1(e2123); otEx7233] |
| inx-19 Extended WRM0632bE10fosmid::SL2::NLS::yfp::H2B/ NotI (40ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15543 [pha-1(e2123); otEx7234] |
| inx-20 WRM066cA02fosmid::SL2::NLS::yfp::H2B/ NotI (40ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15224 [pha-1(e2123); otEx7074] |
| eat-5 WRM0621dG04fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15530 [pha-1(e2123) ; otEx7225] |
| eat-5 WRM0621dG04fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15531 [pha-1(e2123); otEx7226] |
| unc-7 WRM0627bH02fosmid::SL2::NLS::yfp::H2B/ NotI (15ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15275 [pha-1(e2123); otEx7106] |
| unc-9 WRM0611aH10fosmid::SL2::NLS::yfp::H2B/ NotI (7.5ng/ul), pha-1(+) (3ng/ul), myo-2::bfp (10ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15278 [pha-1(e2123); otEx7109] |
| ets-5p::tagrfp (10ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) [BAG axon marker] | this paper | OH15535 [otEx7230] |
| npr-9p::tagrfp (10ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) [AIB axon marker] | this paper | OH13918 [otIs643] |
| che-7prom1::che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15706 [inx-6(ot805); che-7(ok2373); otEx7300] |
| che-7prom1::che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15707 [inx-6(ot805); che-7(ok2373); otEx7301] |
| che-7prom2::che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15708 [inx-6(ot805); che-7(ok2373); otEx7302] |
| che-7prom2::che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15709 [inx-6(ot805); che-7(ok2373); otEx7303] |
| che-7prom3:: che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15710 [inx-6(ot805); che-7(ok2373); otEx7304] |
| che-7prom3:: che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15711 [inx-6(ot805); che-7(ok2373); otEx7305] |
| che-7prom4:: che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15712 [inx-6(ot805); che-7(ok2373); otEx7306] |
| che-7prom4:: che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15713 [inx-6(ot805); che-7(ok2373); otEx7307] |
| che-7prom5::che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13943 [inx-6(ot805); che-7(ok2373); otEx6485] |
| che-7prom5::che-7::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15714 [inx-6(ot805); che-7(ok2373); otEx7308] |
| flp-17p::che-7cDNA::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13968 [inx-6(ot805); che-7(ok2373); otEx6487] |
| flp-17p::che-7cDNA::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13969 [inx-6(ot805); che-7(ok2373); otEx6488] |
| che-1p::che-7cDNA::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13970 [inx-6(ot805); che-7(ok2373); otEx6489] |
| gcy-8p::che-7cDNA::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15704 [inx-6(ot805); che-7(ok2373); otEx7298] |
| gcy-8p::che-7cDNA::tagrfp::SL2::NLS::yfp::H2B/ NotI (6ng/ul), linearized pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15705 [inx-6(ot805); che-7(ok2373); otEx7299] |
| flp-18p::unc-7::SL2::NLS::yfp::H2B (2ng/ul), pha-1(+) (3ng/ul), unc-122::gfp (5ng/ul), OP50 gDNA (100ng/ul) b | this paper | OH15587 [ otEx7250 [Line # 2]] |
| flp-18p::unc-7::SL2::NLS::yfp::H2B (2ng/ul), pha-1(+) (3ng/ul), unc-122::gfp (5ng/ul), OP50 gDNA (100ng/ul) b | this paper | OH15588 [ otEx7251 [Line # 3]] |
| inx-6prom1::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15702 [ otEx7296] |
| inx-6prom1::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15703 [ otEx7297] |
| inx-6prom2::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH11832 [ otEx5380] |
| inx-6prom2::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH11833 [ otEx5381] |
| inx-6prom3::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12252 [ otIs484] |
| inx-6prom4::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13068 [ otEx6031] |
| inx-6prom4::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH13069 [ otEx6032] |
| inx-6prom5::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12118 [ otEx5481] |
| inx-6prom5::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12119 [ otEx5482] |
| inx-6prom5::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12120 [ otEx5483] |
| inx-6prom6::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12127 [ otEx5487] |
| inx-6prom6::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12128 [ otEx5488] |
| inx-6prom7::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12129 [ otEx5489] |
| inx-6prom7::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12130 [ otEx5490] |
| inx-6prom7::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12131 [ otEx5491] |
| inx-6prom13::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12243 [ otEx5532] |
| inx-6prom13::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12244 [ otEx5533] |
| inx-6prom13::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12245 [ otEx5534] |
| inx-6prom3Del::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12657 [ otEx5776] |
| inx-6prom3Del::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12658 [ otEx5777] |
| inx-6prom3Del::tagrfp (8ng/ul), linearized pRF4 (rol-6) (4ng/ul), OP50 gDNA (100ng/ul) | this paper | OH12659 [ otEx5778] |
| inx-1p::TIR1::unc-54 3'UTR::rps-27p::NeoR::unc-54 3'UTR (10ng/ul), unc-122::gfp (5ng/ul), OP50 gDNA (100ng/ul) | this paper | OH15715 [inx-6(ot804); otEx7309] |
| CRISPR alleles and MOS-insertions | ||
| inx-2(ot906 [inx-2::SL2::NLS::yfp::H2B]) | this paper | OH15566 |
| inx-6(ot804 [inx-6::SL2::NLS::yfp::H2B]) | this paper | OH13525 |
| inx-6(ot805 [inx-6::gfp]) | this paper | OH13525 |
| inx-6(ot804 [inx-6::SL2::NLS::yfp::H2B]ot808 [inx-6 Del>TAATTA ::SL2::NLS::yfp::H2B]) | this paper | OH13565 |
| inx-6(ot804 [inx-6::SL2::NLS::yfp::H2B]ot840 [inx-6 Del>TAATTA ::SL2::NLS::yfp::H2B]) | this paper | OH14014 |
| daf-16(ot853 [daf-16::linker::mNG::3xFlag::AID]) | this paper | OH14125 |
| otTi19 (Si[Pnpr-9::INX-6::GFP]) | this paper | OH14529 |
METHOD DETAIL
Cloning and constructs
Transcriptional fosmid reporter clones:
Fosmid clones typically contained ~30-40kb genomic region including the corresponding gene sequence and ~10-20kb flanking sequences on either side (see schematics in Fig.4,7,S2,S3). Fosmid clones are thought to include all the necessary cis-regulatory information for the endogenous gene expression. The sl2-based transcriptional innexin fosmid reporters was generated by recombining an SL2 trans-splicing sequence followed by a fluorescent reporter cassette containing the yfp-gene sequence tagged with an NLS and a histone (H2B) at the 3’ end of the respective gene locus just after the stop codon, using fosmid recombineering (Tursun et al., 2009). These so-called “transcriptional reporters” allowed nuclear localized H2B-tagged YFP reporter protein expression independent of the membrane localized innexin protein expression, facilitating neuronal identification.
inx-19 fosmid extension:
Fosmid clone WRM0632bE10 in pCC1FOS was digested with ApaLI and self-ligated to get a subclone containing 30kb genomic fragment (inx-19 locus, 2.3kb sequence 5’ and 8kb sequence 3’ to the inx-19 locus). This subclone was linearized with BamHI digestion (−2320 from inx-19 ATG) and a 9944bp genomic sequence was inserted in using Gibson assembly (NEB, Cat. # E2621S). This final fosmid clone contained inx-19 locus, 12.3kb sequence 5’ and 8kb sequence 3’ to the inx-19 locus.
che-7 bi-cistronic fosmid reporter clone:
The fosmid-based bi-cistronic che-7 reporter construct was generated by recombining tagrfp-gene sequence followed by an SL2 trans-splicing sequence separated H2B-tagged yfp cassette (tagrfp::Sl2::NLS::yfp::H2B cassette) at the 3’ end of the che-7 gene locus right before the stop codon, using fosmid recombineering (Tursun et al., 2009). This reporter allowed independent visualization of the membrane-localized TagRFP-tagged CHE-7, while simultaneously allowed identification of the che-7 expressing neurons based on the nuclear-localized H2B-tagged YFP reporter protein expression.
Translational fosmid reporter clones:
The inx-3, inx-9 and inx-17 translational fosmid reporter constructs were kindly provided by the TransgeneOme project (Sarov et al., 2012). gfp was recombined right before the stop codon of the locus. A detailed list of fosmids generated is provided in C. elegans strains (Fig.S5).
CRISPR/Cas9-mediated genome editing:
Fluorescent knock-in alleles of inx-2 and inx-6 or mutant allele of inx-6 was generated using CRISPR/Cas9-triggered homologous recombination based on co-CRISPR method (Kim et al., 2014). See schematics in (Fig. 3 and S2).
Generation of daf-16(ot853[daf-16[daf-16::mNG::3XFLAG::AID] allele:
CRISPR/Cas9-mediated genome editing using a self-excising cassette (SEC) as previously described (Dickinson et al., 2015). We modified the pDD268 plasmid (developed in (Dickinson et al., 2015)), which contains the mNeonGreen (mNG) fluorescent reporter cassette, to include the Auxin Inducible Degron (AID)(Zhang et al., 2015) after the 3xFLAG tag. We inserted this mNG::3xFLAG::AID tag right before the stop codon of the daf-16 gene locus to produce mNG::AID fused DAF-16 protein. This tagging captures all DAF-16 isoforms, since they have a common C-terminus.
To generate npr-9p::tagrfp, a 1.7kb cis-regulatory region (−1689 to 0) was fused with tagrfp::unc-54 3’ UTR sequence using PCR fusion.
To generate ets-5p::tagrfp, a 3.3kb cis-regulatory region (−3276 to 0) was fused with tagrfp::unc-54 3’ UTR sequence using PCR fusion.
cis-regulatory analysis of inx-6 locus:
All reporter transgenes for cis-regulatory analysis of inx-6 locus were generated using a PCR fusion approach (Hobert, 2002) using tagrfp-coding sequence. Schematics and coordinates of reporter constructs with respect to ATG were indicated in Fig. 7A. 2 to 3 independent transgenic reporter lines with similar expressivity and penetrance were scored.
cis-regulatory analysis of che-7 locus:
All reporter transgenes for cis-regulatory analysis of che-7 locus were generated by amplifying indicated regions from the bi-cistronic che-7fosmid::tagrfp::Sl2::NLS::yfp::H2B fosmid clone.
For neuron specific expression of che-7::tagrfp::Sl2::NLS::yfp::H2B, total cDNA was synthesized using oligo(dT)20 primer and SuperScript III First-Strand Synthesis System, according to the Invitrogen protocol. che-7 cDNA was amplified using primers: 5’ ATGCCAGAAAACAAACTTCAATTGG 3’ and 5’ CAAATCTAGAAGAGAACTGGC 3’ and cloned into pMiniT 2.0 vector (NEB). tagrfp::Sl2::NLS::yfp::H2B cassette was amplified from the bi-cistronic che-7fosmid::tagrfp::Sl2::NLS::yfp::H2B fosmid clone and introduced in the pMiniT- che-7cDNA clone right before the che-7 stop codon to get the pMiniT-che-7cDNA::tagrfp::SL2::NLS::yfp::H2B clone using Gibson assembly. 3311bp flp-17p [BAG specific], 897bp gcy-8p [AFD specific] and 1380bp che-1pA [ASE specific] was introduced right before the che-7 ATG using Gibson assembly. Cell specific prom::che-7cDNA::tagrfp::SL2::NLS::yfp::H2B::che-7 3’ UTR was amplified from the final clone and injected to get transgenic animals. 2 to 3 independent transgenic reporter lines with similar expressivity and penetrance were scored.
To generate flp-18p::unc-7::SL2::NLS::yfp::H2B, unc-7::SL2::NLS::yfp::H2B unc-7 3’ UTR cassette was amplified from the unc-7::SL2::NLS::yfp::H2B fosmid clone using upstream primer 5’ TAACACGAACCCGGGATGCTCGGCTCCTCCAGCAA 3’ (this also introduces a Smal cut site right before the unc-7 ATG) and downstream primer 5’ CCTCAAATTGAGCCCATCAG 3’ and sub-cloned into pTOPO XL vector (Invitrogen). This construct was linearized using Smal digestion and 3117bp flp-18p sequence was introduced before the unc-7 coding sequence using Gibson assembly.
Microscopy
Worms were anesthetized using 100mM of sodium azide and mounted on 5% agarose on glass slides. Images were recorded using either Zeiss 880 confocal laser-scanning microscope or Zeiss Axio Imager Z2 wide field fluorescent microscope. Images were analyzed by scanning the full Z-stack using Zeiss Zen software. Maximum intensity projections constructed using NIH Fiji software of representative images were shown. Figures were prepared using Adobe Photoshop CS6 and Adobe Illustrator CS6. Separate channels were usually adjusted independently using Levels and Curves in Adobe Photoshop.
Expression analysis and neuron identification
Reporter expression analysis were determined by confocal microscopy and scored in a binary manner; if fluorescent signals were observed in a cell type at whatever brightness, the reporter was scored as “expressed”. We entirely relied on reporter-based expression analysis (either fosmid-based on CRISRP/Cas9-mediated insertion of fluorophore into respective locus) since dauer-stage animals do not lend themselves to antibody or FISH-based mRNA detection protocols due to their cuticle structure.
Cell identification for reporter expression was done by Nomarski optics and crossing with neuronal landmark reporter strains, primarily fosmid reporters of eat-4 (otIs518[eat-4fosmid::SL2::NLS::cherry::H2B, pha-1(+)] and otIs388[eat-4fosmid::SL2::NLS::yfp::H2B, pha-1(+)]), cho-1 (otIs544[cho-1fosmid::SL2::NLS::cherry::H2B, pha-1(+)]) and unc-47 (otIs564[unc-47fosmid::SL2::NLS::cherry::H2B, pha-1(+)]), and promoter-fusion reporter of rab-3 (otIs355[rab-3p::NLS::tagrfp]) (Gendrel et al., 2016; Pereira et al., 2015; Serrano-Saiz et al., 2013; Stefanakis et al., 2015). Expression in a subset of sensory neurons was identified additionally by dye filling with DiD (Thermo Fisher Scientific) and individual IDs were confirmed with neuron-type specific drivers: for AWA: otIs335[odr-2p::mCherrry; pRF4 (rol-6)], for BAG: otEx7230[ets-5p::tagrfp; pRF4 (rol-6)] and ynIs64[flp-17p::gfp], for AIB: otIs643[npr-9p::tagrfp; pRF4 (rol-6)]. A precise description is provided in Supplemental Fig. S5.
We ascribe the differences in gene expression patterns compared to a previous analysis (Altun et al., 2009) to the fundamentally different nature of our reporter. The previous analysis used small (1kb or less) 5’ promoter fusions, while our expression patterns are based on fosmid-based reporters and/or CRISPR-mediated fluorophore insertions into innexin loci. Fosmid-based reporter typically include several up- and downstream genes and therefore contain all intergenic regions of the relevant locus (including all introns), therefore capturing more cis-regulatory elements than small 5’ promoter fusions do.
C. elegans electrical synapse connectome
The electrical synapse network diagram was drawn using open source software, Cytoscape (www.cytoscape.org) based on synaptic connectivity data obtained from serial section reconstruction of electron micrographs (TEM) collected in MRC/LMB and rescored in Emmons and Hall lab (Jarrell et al., 2012; White et al., 1986).
Single molecule fluorescence in situ hybridization (smFISH)
smFISH analysis was performed as previously described (Ji and van Oudenaarden, 2012). The inx-6 probes were designed by using the Stellaris RNA FISH probe designer. Purified probes conjugated to Quasar 670 dye were obtained from Biosearch Technologies. Briefly, probes were dissolved in RNase-free TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) to create a 250 μM probe stock. Animals were washed with M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, 0.1% Tween-20 in 1 L H2O) were fixed with 3.7% formaldehyde in 1X PBS with at room temperature (RT) for 45 min. After fixation, samples were washed with PBS X2 and resuspended in 70% ethanol, and stored overnight at 4°C with gentle agitation. Ethanol was removed and samples incubated in wash buffer (2X SSC, 10% deionized formamide in nuclease-free water) for 2-5 min at RT. Samples were hybridized with inx-6 probes in hybridization solution (0.1% Dextran sulfate, 2X SSC, 10% deionized formamide in nuclease-free water) overnight at 37°C. After probe addition, samples were kept in the dark for all incubations and washes. Samples were incubated in wash buffer for 30 min at 37°C, followed by another incubation in wash buffer for 30 min at 37°C with 5 ng/mL diamidinophenylindole (DAPI) for nuclear staining. Samples were rinsed in 2X SSC followed by a quick rinse in GLOX anti-fade buffer (0.4% glucose, 10 mM Tris-HCl pH 8.0, 2X SSC, containing Glucose oxidase and Catalase) Finally, samples were mounted on slides for imaging.
Automated worm tracking
Automated single worm tracking was performed using Wormtracker 2.0 system (Yemini et al., 2013) at room temperature (~ 22°C). For behavioral assay involving temperature sensitive inx-6(rr5) strain, all genotypes including the control N2 animals were tested at 25°C. For Dauer animals, which tend to show extensive pausing, were recorded for 10 min to ensure sufficient sampling of locomotion related behavioral features. Non-dauer animals were recorded for 5 min, except when compared to dauer animals (as in Fig. 1) were recorded for 10 min. To minimize any potential bias arising due to dauer stage-specific prolonged bouts of spontaneous pausing (Gaglia and Kenyon, 2009), we only selected animals that were actively moving at the beginning of the assay. To avoid potential variability arising due to room conditions, all strains that were compared in a single experiment were recorded simultaneously in identical room condition, along with N2 wild-type. Strains that were recorded simultaneously with temperature sensitive inx-6(rr5) strain, were grown at 25°C for >24h and recorded at 25°C. Recording was randomized across multiple trackers. Dauer and starved non-dauer animals were placed on uncoated NGM plates before recording. Fed non-dauer animals were recorded on NGM plates seeded uniformly with diluted OP50 bacterial culture to avoid potentially biased locomotion at the edge of the bacterial lawn.
Dauer animals are thinner, more elongated and contain a different cuticular structure compared to fed- or starved-L3 animals (Cassada and Russell, 1975). When comparing dauers to fed- and starved-L3 stage animals, some of the behavioral differences (e.g. wavelength) that are likely to be influenced by such physical differences were ignored and we instead focused on behavioral differences that can more readily be attributed to information processing in the nervous system.
CO2 Chemotaxis assay
CO2 chemotaxis assays were performed as previously described (Hallem et al., 2011a). Assays were performed on standard 6cm NGM plates for non-dauer animals and 9cm NGM plates for dauer animals. Scoring regions were 2cm circles on each side of the plate along the diameter, with the center of the circle 1cm away from the edge of the plate (as indicated in Fig. 6J). Pumping a mixture of 10% CO2, 10% O2 and balance N2 wild-type from an inlet on top of one scoring circle and a mixture of 10% O2 and balance N2 wild-type from another inlet on top of the other scoring circle generated CO2 gradient. Gas mixtures were pushed using a syringe pump at 1.5ml/min for non-dauer assays and 0.5ml/min for dauer assays. ~50 young adults or ~100-150 dauers (selected by 1% SDS treatment) were placed at the center of the assay plates. After 30 min, the number of animals inside the air and CO2 circles was counted. The chemotaxis index (C.I.) was calculated as:
Odortaxis assay
Dauer odortaxis assays were performed as previously described (Hallem et al., 2011a). Assays were performed on standard 9cm NGM plates. Scoring regions were 2cm circles on each side of the plate along the diameter, with the center of the circle 1cm away from the edge of the plate. Test circle contained 5μl of odorant at center of it, while control circle contained 5μl of ethanol. 1μl of 1M sodium azide was added to each circle as anesthetic. ~100-150 dauers were placed at the center of the assay plates and allowed to chemotax for 90min.
The chemotaxis index (C.I.) was calculated as:
Cell ablation
Cell specific ablation was performed using MicroPoint Laser System Basic Unit (N2 pulsed laser (dye pump), ANDOR Technology) attached to a Zeiss Axio Imager Z2 microscope (Objective EC Plan-Neofluar 63X/1.30 Oil). This laser delivers 120 μJoules of 337nm energy with a 3nsec pulse length. Ablations were done as previously described (Fang-Yen et al., 2012), with pulse repetition rates ~15 Hz. daf-7(e1372); npr-9::tagrfp; inx-6(ot805) stain was grown at 25°C to obtain dauer. AIB identified with TAGRFP reporter and ablated at the dauer stage. Treated animals were allowed to recover at 25°C on unseeded NGM plates for 48 hours before analysis. Control animals were treated in the same manner, but not subjected laser exposure.
Auxin inducible degradation
The AID system was employed as previously described (Zhang et al., 2015). The conditional daf-16 allele daf-16(ot853[daf-16::mNG::AID) was crossed with daf-2(e1370), neuron specific TIR1-expressing transgenic lines and inx-6(ot804[inx-6::sl2::yfp::h2b] reporter, to generate the experimental strains. Animals were grown (from embryo onwards) on NGM plates supplemented with OP50 and 4mM auxin in EtOH (indole-3 acetic acid, IAA, Alfa Aesar, Cat. # A10556) at 25°C to degrade DAF-16 in respective n eurons and to induce dauer formation. As control, plates were supplemented with the solvent EtOH instead of auxin.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of the automated wormtracking videos was performed as previously described (Yemini et al., 2013). Statistical significance between each groups were blindly calculated using Wilcoxon rank-sum test using automated algorithms included in the Wormtracker 2.0 analysis software (Yemini et al., 2013). After correcting p-values by controlling the false-discovery rate, q-values were obtained based on group significance. For the comparison of dauer locomotion with fed and starved L3 locomotion, 702 behavioral features were compared. After correcting for false-discovery rate, features shown in Fig. 5 and Table S4 immerged as the once with most significantly different q-values among the test groups. Therefore, for the subsequent experiments we only measured these features, permitting us to use the p-value.
For chemotaxis assays statistical significance between each group was calculated using two-sided Mann-Whitney U-test.
The principle components analysis (PCA) was executed using MATLAB (Mathworks) with custom computational scripts (made available upon request).
Supplementary Material
Fig. S1: innexin expression difference in dauer and non-dauer nervous system. Related to Fig. 1.
(A) Correlation between number of innexin genes expressed by neuron classes in non-dauer stage and number of electrical synaptic targets, as inferred from the serial section reconstruction of electron micrographs (data source www.wormwiring.org).
(B) Correlation between number of innexin genes expressed by neuron classes in non-dauer stage and the total of electrical synaptic strengths, as inferred from the serial section reconstruction of electron micrographs (data source www.wormwiring.org).
(C) Graphical representation of innexin expression differences in each neuron classes. HSN and VC neurons, which mature only at L4 stage and showed no innexin expression in dauer, were not included in this graph.
Fig. S2: Expression of neuronally expressed innexins change in dauer stage. Related to Fig. 1 and 2.
Schematics of fosmid-based transcriptional reporters of neuronally expressed innexin genes are shown. For inx-2, schematic of an sl2-based transcriptional reporter allele, inx-6(ot906) generated by CRISPR/Cas9-mediated genome editing, is shown. Splice isoform specific fosmid reporters for inx-1, inx-10 and inx-18 are generated. Whole animal expressions for each innexin gene in L3 and dauer stages are shown. Red asterisks mark gut auto-fluorescence. Examples show selective up or down-regulation of particular innexin gene expression in specific neuronal classes, as assessed by the indicated reporters. inx-13, which does not show altered expression in dauer, continues to be expressed in CAN neurons in dauer.
Fig. S3: Expression of non-neuronal innexin genes. Related to Fig. 1.
Schematics of fosmid-based transcriptional reporters of non-neuronally expressed innexin genes are shown. For, inx-3, inx-9 and inx-17 schematics of fosmid-based GFP-tagged translational reporters are shown. Whole animal expressions for each innexin gene in L3 and dauer stages are shown. For, inx-3 and inx-9 GFP-tagged translational reporters only the relevant head expression is shown. Red asterisks mark gut auto-fluorescence.
Fig. S4: inx-6 and che-7 affect locomotory behavior specifically in dauer stage. Related to Fig. 6.
(A-J) Worm locomotion assay using Wormtracker 2.0 system (see Methods for detailed description). Each circle represents the experimental mean for a single animal for the corresponding behavioral assay. Red lines indicate the mean of means and black open rectangles indicate S.E.M. for each assay. Wilcoxon rank-sum tests p-values for each comparison (n.s. = non significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
(A) Dauers with a temperature sensitive allele of inx-6, inx-6(rr5) and AIB-ablated dauers (peIs578[npr-9p::casp1, npr-9p::venus, unc-122p::mCherry]) shows significant decrease in speed (measured as midbody speed).
(B) Dauers with an inx-6 transcriptional reporter allele, inx-6ot804, shows no significant effect in speed. Dauers that specifically lack inx-6 expression in AIB, inx-6AIB OFF and che-7(ok2373) dauers shows significant decrease in speed (measured as midbody speed).
(C) Dauers with a temperature sensitive allele of inx-6, inx-6(rr5) and AIB-ablated dauers (peIs578[npr-9p::casp1, npr-9p::venus, unc-122p::mCherry]) shows significant decrease in foraging amplitude.
(D) Dauers with an inx-6 transcriptional reporter allele, inx-6ot804, shows no significant effect in foraging amplitude. Dauers that specifically lack inx-6 expression in AIB, inx-6AIB OFF and che-7(ok2373) dauers shows significant decrease in foraging amplitude.
(E-J) In fed-L3 and starved-L3 stage animals, inx-6(rr5) animals shows no significant affect on paused, forward and backward motion time ratio, compared to N2 wild-type animals. AIB-ablated fed- and starved-L3 animals (peIs578[npr-9p::casp1, npr-9p::venus, unc-122p::mCherry]) shows no difference in pausing, but strong reduction in backward locomotion and subsequent increase in forward locomotion.
(K-O) Odortaxis assay of N2 wild-type dauer animals. Each circle represents chemotaxis index calculated from each assay. Red line indicates the mean and black rectangles indicate S.E.M. Wilcoxon rank-sum tests p-values for each comparison (n.s. = non significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001) (see Methods for detailed description of behavioral assays).
(K) inx-6AIB OFF dauers does not affect attraction towards diacetyl (2,3-butanedione).
(L) inx-6AIB OFF and che-7(ok2373) dauers show significant increase trimethylamine attraction.
(M) inx-6AIB OFF and che-7(ok2373) dauers does not affect 2-butanone attraction.
(N) inx-6AIB OFF and che-7(ok2373) dauers does not affect 2-octanone repulsion.
(O) inx-6AIB OFF and che-7(ok2373) dauers does not affect 2-heptanone repulsion.
Fig. S5: Schematic guide for neuronal identification. Related to STAR Methods.
(A) A graphical representation of neuronal landmark reporter expression in the head and tail of the animal.
(B) Tabular illustration of cell identification procedure. The left half of the table shows expression of all neuronal innexin genes in individual neuronal classes (same data as shown in Fig.1). Color of each box in the left half of the table represents whether the innexin gene expression is detected in both non-dauer and dauer stages (yellow), only in non-dauer stages (red) or only in dauer stage (green). Asterisks indicate developmental changes in innexin expression among non-dauer stages. The right half of the table describes methods used to precisely identify each neuron in the animal. Neuronal identification is done and cross-validated based on a combination of multiple neuronal landmark reporter expression in distinct set of neurons (also see panel A), three-dimensional position of the nucleus in the animal body (also see panel A), characteristic signature of neuronal nuclei in Nomarski optics and characteristic shape of some neuronal nuclei (also see panel A). Color of each box in the right half of the table represents whether the particular neuron is precisely identified (orange) based on the principle listed in the particular column.
Supplemental Table S1:
Innexin expression in non-neuronal tissues. Related to Fig. 1.
Supplemental Table S2:
Developmental changes in innexin expression. Related to Fig. 1.
Supplemental Table S3:
List of behavioral features used in PCA. Related to Fig. 5.
Supplemental Table S4:
List of locomotory behavioral differences between N2 animals in dauer, fed- and starved-L3 stage/state. Related to Fig. 5.
HIGHLIGHTS.
Mapped complex, neuron type-specific combinatorial patterns of innexin expression.
Neuron-specific innexin expression code shows remarkable plasticity under stress.
Altered innexin expression leads to circuit modification and behavioral plasticity.
FoxO-dependent combinatorial mechanism controls spatiotemporal innexin plasticity.
Mapping the electrical connectome of C. elegans reveals the combinatorial patterns of synaptic protein expression that dictate behaviors associated with the physiological state of the animal
ACKNOWLEDGMENTS
We thank Niels Ringstad and Jung-Hwan Choi for introducing us to the CO2-assay, Esther Serrano-Saiz and Narmin Tahirova for helping with the unc-42 mutant analysis, Adriane Otopalik for helping with the PCA, Steven Cook for helping with the EM connectome data analysis, Chi Chen for the C. elegans injections and members of the Hobert lab for comments on this manuscript. This work was supported by the NIH (R21NS106909) and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Bibliography
- Abrams CK, and Scherer SS (2012). Gap junctions in inherited human disorders of the central nervous system. Biochim Biophys Acta 1818, 2030–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun ZF, Chen B, Wang ZW, and Hall DH (2009). High resolution map of Caenorhabditis elegans gap junction proteins. Dev Dyn 238, 1936–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran R, Aronoff R, and Garriga G (1999). The C. elegans homeodomain gene unc-42 regulates chemosensory and glutamate receptor expression. Development 126, 2241–2251. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada RC, and Russell RL (1975). The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46, 326–342. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, and Goldstein B (2015). Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang-Yen C, Gabel CV, Samuel AD, Bargmann CI, and Avery L (2012). Laser microsurgery in Caenorhabditis elegans. Methods Cell Biol 107, 177–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, and Antebi A (2008). C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22, 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia MM, and Kenyon C (2009). Stimulation of movement in a quiescent, hibernation-like form of Caenorhabditis elegans by dopamine signaling. J Neurosci 29, 7302–7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Atlas EG, and Hobert O (2016). A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Hill JJ, and Bargmann CI (2005). A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A 102, 3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermin ML, Castelletto ML, and Hallem EA (2011). Differentiation of carbon dioxidesensing neurons in Caenorhabditis elegans requires the ETS-5 transcription factor. Genetics 189, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH (2017). Gap junctions in C. elegans: Their roles in behavior and development. Dev Neurobiol 77, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Dillman AR, Hong AV, Zhang Y, Yano JM, DeMarco SF, and Sternberg PW (2011a). A sensory code for host seeking in parasitic nematodes. Curr Biol 21, 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Spencer WC, McWhirter RD, Zeller G, Henz SR, Ratsch G, Miller DM 3rd, Horvitz HR, Sternberg PW, and Ringstad N (2011b). Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc Natl Acad Sci U S A 108, 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa DK, and Turnbull MW (2014). Recent findings in evolution and function of insect innexins. FEBS Lett 588, 1403–1410. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, and Denk W (2013). Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174. [DOI] [PubMed] [Google Scholar]
- Hobert O (2002). PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 32, 728–730. [DOI] [PubMed] [Google Scholar]
- Hobert O (2016). Terminal Selectors of Neuronal Identity. Curr Top Dev Biol 116, 455–475. [DOI] [PubMed] [Google Scholar]
- Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, and Emmons SW (2012). The connectome of a decision-making neural network. Science 337, 437–444. [DOI] [PubMed] [Google Scholar]
- Ji N, and van Oudenaarden A (2012). Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. WormBook, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vazquez-Reina A, Kaynig V, Jones TR, et al. (2015). Saturated Reconstruction of a Volume of Neocortex. Cell 162, 648–661. [DOI] [PubMed] [Google Scholar]
- Kawano T, Po MD, Gao S, Leung G, Ryu WS, and Zhen M (2011). An imbalancing act: gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron 72, 572–586. [DOI] [PubMed] [Google Scholar]
- Kim H, Ishidate T, Ghanta KS, Seth M, Conte D Jr., Shirayama M, and Mello CC (2014). A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Choi MK, Lee D, Kim HS, Hwang H, Kim H, Park S, Paik YK, and Lee J (2012). Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat Neurosci 15, 107–112. [DOI] [PubMed] [Google Scholar]
- Lee RY, Hench J, and Ruvkun G (2001). Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol 11, 1950–1957. [DOI] [PubMed] [Google Scholar]
- Li S, Dent JA, and Roy R (2003). Regulation of intermuscular electrical coupling by the Caenorhabditis elegans innexin inx-6. Mol Biol Cell 14, 2630–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Chen B, Mailler R, and Wang ZW (2017). Antidromic-rectifying gap junctions amplify chemical transmission at functionally mixed electrical-chemical synapses. Nat Commun 8, 14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Gutierrez GJ, and Nusbaum MP (2017). Complicating connectomes: Electrical coupling creates parallel pathways and degenerate circuit mechanisms. Dev Neurobiol 77, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS (2010). Stress, sex, and neural adaptation to a changing environment: mechanisms of neuronal remodeling. Ann N Y Acad Sci 1204 Suppl, E38–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin CN, and Broihier HT (2018). Keeping Neurons Young and Foxy: FoxOs Promote Neuronal Plasticity. Trends Genet 34, 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mese G, Richard G, and White TW (2007). Gap junctions: basic structure and function. J Invest Dermatol 127, 2516–2524. [DOI] [PubMed] [Google Scholar]
- Miller AC, and Pereda AE (2017). The electrical synapse: Molecular complexities at the gap and beyond. Dev Neurobiol 77, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J (2014). The ever-changing electrical synapse. Curr Opin Neurobiol 29, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima A, Matsuzawa T, Murata K, Tani K, and Fujiyoshi Y (2016). Hexadecameric structure of an invertebrate gap junction channel. J Mol Biol 428, 1227–1236. [DOI] [PubMed] [Google Scholar]
- Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, et al. (2015). A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan P, and Starich TA (2001). Innexins get into the gap. Bioessays 23, 388–396. [DOI] [PubMed] [Google Scholar]
- Riddle DL, and Albert PS (1997). Genetic and Environmental Regulation of Dauer Larva Development. In Celegans II, Riddle DL, Blumenthal T, Meyer BJ, and Priess JR, eds. (Cold Spring Harbor Laboratory Press; ), pp. 739–768. [PubMed] [Google Scholar]
- Ryan K, Lu Z, and Meinertzhagen IA (2016). The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, Hasse S, Rupprecht M, Vinis E, Tinney M, et al. (2012). A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, de la Cruz ED, and Hobert O (2013). Modular Control of Glutamatergic Neuronal Identity in C. elegans by Distinct Homeodomain Proteins. Cell 155, 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohl G, Maxeiner S, and Willecke K (2005). Expression and functions of neuronal gap junctions. Nat Rev Neurosci 6, 191–200. [DOI] [PubMed] [Google Scholar]
- Song J, Ampatzis K, Bjornfors ER, and El Manira A (2016). Motor neurons control locomotor circuit function retrogradely via gap junctions. Nature 529, 399–402. [DOI] [PubMed] [Google Scholar]
- Starich TA, Hall DH, and Greenstein D (2014). Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198, 1127–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich TA, Xu J, Skerrett IM, Nicholson BJ, and Shaw JE (2009). Interactions between innexins UNC-7 and UNC-9 mediate electrical synapse specificity in the Caenorhabditis elegans locomotory nervous system. Neural Dev 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanakis N, Carrera I, and Hobert O (2015). Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. (2013). A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Duszkiewicz AJ, and Morris RG (2014). The synaptic plasticity and memory hypothesis: encoding, storage and persistence. Philos Trans R Soc Lond B Biol Sci 369, 20130288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenin AF, Kowal TJ, Fong JT, Kells RM, Fisher CG, and Falk MM (2013). Proteins and mechanisms regulating gap-junction assembly, internalization, and degradation. Physiology (Bethesda) 28, 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, and Hobert O (2009). A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4, e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney LR, Chen BL, Paniagua E, Hall DH, and Chklovskii DB (2011). Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol 7, e1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sato H, Satoh Y, Tomioka M, Kunitomo H, and Iino Y (2017). A Gustatory Neural Circuit of Caenorhabditis elegans Generates Memory-Dependent Behaviors in Na(+) Chemotaxis. J Neurosci 37, 2097–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London B Biological Sciences 314, 1–340. [DOI] [PubMed] [Google Scholar]
- White TW, and Paul DL (1999). Genetic diseases and gene knockouts reveal diverse connexin functions. Annu Rev Physiol 61, 283–310. [DOI] [PubMed] [Google Scholar]
- Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Guldenagel M, Deutsch U, and Sohl G (2002). Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem 383, 725–737. [DOI] [PubMed] [Google Scholar]
- Yemini E, Jucikas T, Grundy LJ, Brown AE, and Schafer WR (2013). A database of Caenorhabditis elegans behavioral phenotypes. Nat Methods 10, 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, and Dernburg AF (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: innexin expression difference in dauer and non-dauer nervous system. Related to Fig. 1.
(A) Correlation between number of innexin genes expressed by neuron classes in non-dauer stage and number of electrical synaptic targets, as inferred from the serial section reconstruction of electron micrographs (data source www.wormwiring.org).
(B) Correlation between number of innexin genes expressed by neuron classes in non-dauer stage and the total of electrical synaptic strengths, as inferred from the serial section reconstruction of electron micrographs (data source www.wormwiring.org).
(C) Graphical representation of innexin expression differences in each neuron classes. HSN and VC neurons, which mature only at L4 stage and showed no innexin expression in dauer, were not included in this graph.
Fig. S2: Expression of neuronally expressed innexins change in dauer stage. Related to Fig. 1 and 2.
Schematics of fosmid-based transcriptional reporters of neuronally expressed innexin genes are shown. For inx-2, schematic of an sl2-based transcriptional reporter allele, inx-6(ot906) generated by CRISPR/Cas9-mediated genome editing, is shown. Splice isoform specific fosmid reporters for inx-1, inx-10 and inx-18 are generated. Whole animal expressions for each innexin gene in L3 and dauer stages are shown. Red asterisks mark gut auto-fluorescence. Examples show selective up or down-regulation of particular innexin gene expression in specific neuronal classes, as assessed by the indicated reporters. inx-13, which does not show altered expression in dauer, continues to be expressed in CAN neurons in dauer.
Fig. S3: Expression of non-neuronal innexin genes. Related to Fig. 1.
Schematics of fosmid-based transcriptional reporters of non-neuronally expressed innexin genes are shown. For, inx-3, inx-9 and inx-17 schematics of fosmid-based GFP-tagged translational reporters are shown. Whole animal expressions for each innexin gene in L3 and dauer stages are shown. For, inx-3 and inx-9 GFP-tagged translational reporters only the relevant head expression is shown. Red asterisks mark gut auto-fluorescence.
Fig. S4: inx-6 and che-7 affect locomotory behavior specifically in dauer stage. Related to Fig. 6.
(A-J) Worm locomotion assay using Wormtracker 2.0 system (see Methods for detailed description). Each circle represents the experimental mean for a single animal for the corresponding behavioral assay. Red lines indicate the mean of means and black open rectangles indicate S.E.M. for each assay. Wilcoxon rank-sum tests p-values for each comparison (n.s. = non significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
(A) Dauers with a temperature sensitive allele of inx-6, inx-6(rr5) and AIB-ablated dauers (peIs578[npr-9p::casp1, npr-9p::venus, unc-122p::mCherry]) shows significant decrease in speed (measured as midbody speed).
(B) Dauers with an inx-6 transcriptional reporter allele, inx-6ot804, shows no significant effect in speed. Dauers that specifically lack inx-6 expression in AIB, inx-6AIB OFF and che-7(ok2373) dauers shows significant decrease in speed (measured as midbody speed).
(C) Dauers with a temperature sensitive allele of inx-6, inx-6(rr5) and AIB-ablated dauers (peIs578[npr-9p::casp1, npr-9p::venus, unc-122p::mCherry]) shows significant decrease in foraging amplitude.
(D) Dauers with an inx-6 transcriptional reporter allele, inx-6ot804, shows no significant effect in foraging amplitude. Dauers that specifically lack inx-6 expression in AIB, inx-6AIB OFF and che-7(ok2373) dauers shows significant decrease in foraging amplitude.
(E-J) In fed-L3 and starved-L3 stage animals, inx-6(rr5) animals shows no significant affect on paused, forward and backward motion time ratio, compared to N2 wild-type animals. AIB-ablated fed- and starved-L3 animals (peIs578[npr-9p::casp1, npr-9p::venus, unc-122p::mCherry]) shows no difference in pausing, but strong reduction in backward locomotion and subsequent increase in forward locomotion.
(K-O) Odortaxis assay of N2 wild-type dauer animals. Each circle represents chemotaxis index calculated from each assay. Red line indicates the mean and black rectangles indicate S.E.M. Wilcoxon rank-sum tests p-values for each comparison (n.s. = non significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001) (see Methods for detailed description of behavioral assays).
(K) inx-6AIB OFF dauers does not affect attraction towards diacetyl (2,3-butanedione).
(L) inx-6AIB OFF and che-7(ok2373) dauers show significant increase trimethylamine attraction.
(M) inx-6AIB OFF and che-7(ok2373) dauers does not affect 2-butanone attraction.
(N) inx-6AIB OFF and che-7(ok2373) dauers does not affect 2-octanone repulsion.
(O) inx-6AIB OFF and che-7(ok2373) dauers does not affect 2-heptanone repulsion.
Fig. S5: Schematic guide for neuronal identification. Related to STAR Methods.
(A) A graphical representation of neuronal landmark reporter expression in the head and tail of the animal.
(B) Tabular illustration of cell identification procedure. The left half of the table shows expression of all neuronal innexin genes in individual neuronal classes (same data as shown in Fig.1). Color of each box in the left half of the table represents whether the innexin gene expression is detected in both non-dauer and dauer stages (yellow), only in non-dauer stages (red) or only in dauer stage (green). Asterisks indicate developmental changes in innexin expression among non-dauer stages. The right half of the table describes methods used to precisely identify each neuron in the animal. Neuronal identification is done and cross-validated based on a combination of multiple neuronal landmark reporter expression in distinct set of neurons (also see panel A), three-dimensional position of the nucleus in the animal body (also see panel A), characteristic signature of neuronal nuclei in Nomarski optics and characteristic shape of some neuronal nuclei (also see panel A). Color of each box in the right half of the table represents whether the particular neuron is precisely identified (orange) based on the principle listed in the particular column.
Supplemental Table S1:
Innexin expression in non-neuronal tissues. Related to Fig. 1.
Supplemental Table S2:
Developmental changes in innexin expression. Related to Fig. 1.
Supplemental Table S3:
List of behavioral features used in PCA. Related to Fig. 5.
Supplemental Table S4:
List of locomotory behavioral differences between N2 animals in dauer, fed- and starved-L3 stage/state. Related to Fig. 5.