Abstract
The influence of cellular metabolism on epigenetic pathways is well documented but misunderstood. Scientists have long known of the metabolic impact on epigenetic determinants. More often than not, that title role for DNA methylation was portrayed by the metabolite S-adenosylmethionine. Technically speaking, there are many other metabolites that drive epigenetic processes that instruct seemingly distant—yet highly connect pathways—and none more so than our understanding of the cancer epigenome. Recent studies have shown that available energy links the extracellular environment to influence cellular responses. This focused review examines the recent interest in epigenomics and casts cancer, metabolism, and immunity in unfamiliar roles—cooperating. There are not only language lessons from cancer research, we have come round to appreciate that reaching into areas previously thought of as too distinct are also object lessons in understanding health and disease. The Warburg effect is one such signature of how glycolysis influences metabolic shift during oncogenesis. That shift in metabolism—now recognized as central to proliferation in cancer biology—influences core enzymes that not only control gene expression but are also central to replication, condensation, and the repair of nucleic acid. These nuclear processes rely on metabolism, and with glucose at centre stage, the role of respiration and oxidative metabolism is now synonymous with the mitochondria as the powerhouses of metaboloepigenetics. The emerging evidence for metaboloepigenetics in trained innate immunity has revealed recognizable signalling pathways with antecedent extracellular stimulation. With due consideration to immunometabolism, we discuss the striking signalling similarities influencing these core pathways. The immunometabolic-epigenetic axis in cardiovascular disease has deeply etched connections with inflammation, and we examine the chromatin template as a carrier of epigenetic indices that determine the expression of genes influencing atherosclerosis and vascular complications of diabetes.
Keywords: Metabolism, Epigenetics, Metaboloepigenetics, Glycolysis, Trained immunity, Cardiovascular disease, Diabetes
Graphical Abstract
Graphical Abstract.
1. Introduction
More than 20 years have passed since the landmark publication of the human genome sequence.1 While researchers have benefited enormously from an improved understanding of the genetic sequence, many of the promises of the genetic revolution remain unfulfilled, particularly with regard to the complexity of phenotypic traits and the propensity to develop certain diseases.2,3 As researchers learned that humans have fewer genes than a banana,4 it was clear that genome size is unrelated to organism complexity. Instead, we have begun to appreciate that phenotypic traits can associate not only with nucleotide sequence but also with chemical modifications that occur on the DNA template and the proteins with which it interacts. Closely following this realization came a broader awareness of how cells with identical genetic content could exhibit different regulation of gene activity.
Modern interpretations of epigenetics have focused on the covalent chemical modifications of chromatin—the dynamic complex of DNA and proteins (mainly histones)—that support transcriptional regulation via structural adaptation.5 Collectively, these modifications are called epigenetic because they influence phenotypes without altering the genetic code and have been shown to be transmitted through cell division by various mechanisms.6–11 Three distinct yet functionally related categories of epigenetic mechanisms have been described: (i) chemical modification of DNA bases, (ii) post-translational modification (PTM) of the tails of histone proteins, and (iii) the regulation of gene expression by non-coding RNA. Despite significant progress in the field, knowledge of how the main epigenetic players are regulated remains incomplete.
A substantial number of findings connecting energy metabolism with epigenetic control of gene expression support the recent emergence of what some in the field are calling ‘metaboloepigenetics’.12–14 Many enzymes that write or erase modifications on the chromatin use metabolites as substrates or cofactors in their epigenetic reactions, connecting metabolic information with transcription.15 Changes in the concentrations of specific metabolites are therefore purported to provide signalling cues for the continual adjustment of gene expression by influencing chromatin dynamics. What is emerging is a complex interplay between intracellular metabolism and chromatin modifications, which is providing an extra dimension to our understanding of gene regulation in health and disease.
The current understanding of metaboloepigenetics has its origins in cancer, a disease characterized by the derangement of metabolic and epigenetic programmes.16,17 More recent findings have shed light on the changes in intracellular metabolic pathways that support the altered function of immune cells via epigenetic reprogramming.18–20 Furthermore, researchers have begun to consider the impact of metaboloepigenetics on atherosclerosis and cardiovascular disease (CVD).21–23
2. Metabolism drives epigenetic processes: lessons from cancer
Gene expression is primarily regulated by the accessibility of DNA to transcription factors (TFs) and transcriptional machinery. Key to this regulation is chromatin, a dynamic assembly of DNA and regularly spaced nucleosomes (comprised of histone proteins) that controls transcription by structural adaptation and genome compartmentalization.15 Compacted chromatin impedes binding of the transcriptional machinery to the DNA by occluding its access to regulatory elements such as promoters and enhancers. On the other hand, an open chromatin structure facilitates the loading of transcriptional machinery. Supporting the gene-regulating functions of TFs is a multitude of PTMs to the N-terminal tails of histones as well as modifications to DNA bases. Some of these modifications have direct effects on chromatin structure by altering charge states of histones and their affinity for DNA. Others provide docking sites for the recruitment of multi-protein chromatin remodelling complexes. For a comprehensive discussion of the enzymes that write, erase, and interpret chromatin modifications, we recommend a recent review article by Zhao et al.5
At the most basic level, a specific chromatin modification is dependent on the relative expression, stability, and competing activities of epigenetic writers and erasers. In recent years, cellular metabolism has emerged as an important determinant of many epigenetic reactions, primarily because numerous epigenetic enzymes require specific metabolites or products from metabolic pathways as cofactors to perform their chromatin modifying functions. Several different metabolic pathways are used by cells to generate adequate energy for survival and to produce biosynthetic intermediates that support cellular growth and proliferation. These distinct metabolic pathways are closely connected by the use of common fuel inputs and a reliance on products from one pathway to feed into alternative pathways as synthetic precursors.24 While a certain degree of overlap exists between metabolic pathways in terms of the chromatin-regulating metabolites that they produce, individual pathways produce metabolites that intersect epigenetic pathways in specific ways. This means that large metabolic shifts can have profound effects on epigenetic regulation and gene expression profiles.25
Metabolic reprogramming has been recognized as a hallmark of cancer transformation since Otto Warburg first described aerobic glycolysis in tumours almost a century ago.26 Human tumours harbour global epigenetic abnormalities27 and mutations in genes encoding epigenetic enzymes are increasingly recognized in cancer.28 Unsurprisingly, many of the known metaboloepigenetic connections were discovered in the metabolically deranged milieu of cancer,29 which spawned the concept of the oncometabolite—a metabolic intermediate whose abnormal accumulation triggers oncogenic signalling and tumourigenesis.30
2.1. Metabolites modulate the methylome
Methylation of cytosine bases is the most well-characterized epigenetic mark.31 The addition of a methyl group to the fifth carbon of a cytosine base (5-methylcytosine, 5mC) is catalysed by DNA methyltransferase enzymes (DNMTs) primarily at cytosines adjacent to a guanines (cytosine-phosphate-guanine, CpG). The vast majority of CpG dinucleotides in mammalian genomes are maintained in a methylated state with the exception of regions of high CpG density called CpG islands located close to gene promoters. A more recent discovery is the mechanism of cytosine demethylation by ten-eleven translocation (TET) dioxygenases that oxidize the methyl group of 5mC to yield 5-hydroxymethylcytosine (5 hmC) and other oxidized methylcytosines.32–34 DNA methylation is primarily associated with gene silencing by two main mechanisms: (i) the occlusion of DNA binding proteins such as TFs that act as transcriptional activators, and (ii) by providing a recognition site for methyl-binding proteins such as methyl-CpG binding protein 2, which recruit transcriptional corepressor complexes that reconfigure the chromatin landscape, rendering it inaccessible to the transcriptional machinery.35 DNA methylation has a dual role in cancer: hypermethylation inhibits tumour suppressor genes, whereas hypomethylation activates oncogene expression. In general, cancer cells display a global loss of CpG methylation juxtaposed against locus-specific hypermethylation at CpG islands.36 Paradoxically, under some circumstances, DNA hypermethylation has also been shown to enhance gene expression in cancer and recently reviewed.37
When assigned to the tails of histones, the methyl modification provides a greater degree of flexibility with regard to its impact on gene expression. Both lysine and arginine residues are sites of histone methylation: arginine residues can be mono-methylated or di-methylated and lysine residues can be mono-, di-, or tri-methylated. This is important because variably methylated histones are differentially distributed across chromatin to distinguish gene regulatory elements. For example, H3 histones tri-methylated at lysine 4 (H3K4me3) are enriched at promoters of transcribed genes, whereas H3 histones mono-methylated at lysine 4 (H3K4me1) are predominantly enriched at distal enhancers.38,39 However, the key determinant of the effect of histone methylation on chromatin structure is the position of the modified residue within the amino sequence of the histone tail. In addition to the transcriptionally permissive methylated state of H3K4, other key sites of histone lysine methylation include lysine 9 and lysine 27 on H3 histones, which are associated with gene repression. Methyl modifications were considered more stable than other histone marks until the discovery of lysine-specific histone demethylase 1 (LSD1), which enzymatically removes methyl groups from H3K4. This led to the identification of the jumonji C (JmjC) domain as a key catalytic component of a broad catalogue of histone lysine demethylase (KDM) enzymes.40 Thus, levels of histone lysine methylation are determined in part by the relative expression and activity of lysine methyltransferases (KMTs) and KDMs. Like DNA methylation, histone methylation can also provide sites for the recruitment of effector protein complexes that recognize distinct methyl modifications via specialized domains.41
Most methyltransferases transfer a methyl group from S-adenosylmethionine (SAM), which is synthesized from adenosine triphosphate (ATP) and methionine. The methylation reaction generates another metabolite called S-adenosylhomocysteine (SAH), which is a potent inhibitor of methyltransferases. Therefore, the intracellular SAM:SAH ratio, which is regulated by methionine, threonine, and serine metabolism, as well as dietary intake of methyl-donating nutrients such as folate and vitamin B, is considered to be an indicator of cellular methylation potential.42,43 In addition, cells require one-carbon units for nucleotide synthesis and redox reactions. Because these pathways support the enhanced proliferation of cancer cells, drugs that target one-carbon metabolism such as the anti-folate methotrexate have long been used in cancer therapy.44 Glycine N-methyltransferase (GNMT) deficiency is a rare condition leading to SAM accumulation.45Gnmt knockout mice exhibited a more than 40-fold increase in hepatic SAM,45 which led to DNA hypermethylation and transcriptional silencing of tumour suppressor genes and was associated with increased incidence of hepatocellular carcinoma.46 As a general hallmark of cancer, methionine addiction is targeted in cancer therapy by methionine restriction, which results in the depletion of SAM47 and cell cycle arrest.48 Recent studies have begun to investigate the therapeutic potential of methionine restriction in combination with inhibitors of SAM synthesis and DNA methylation.49
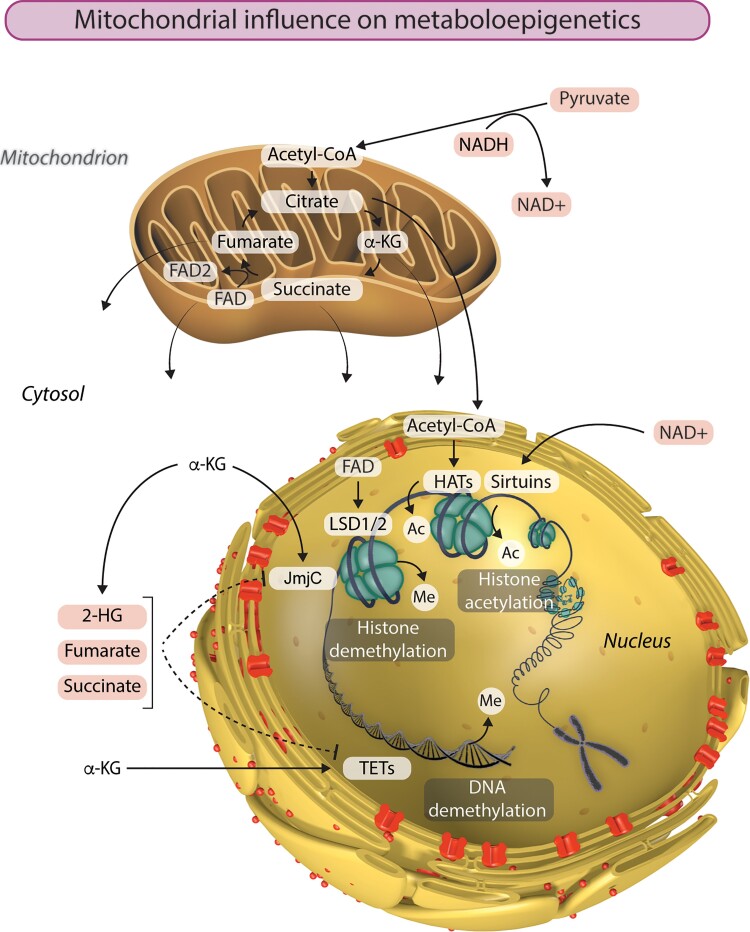
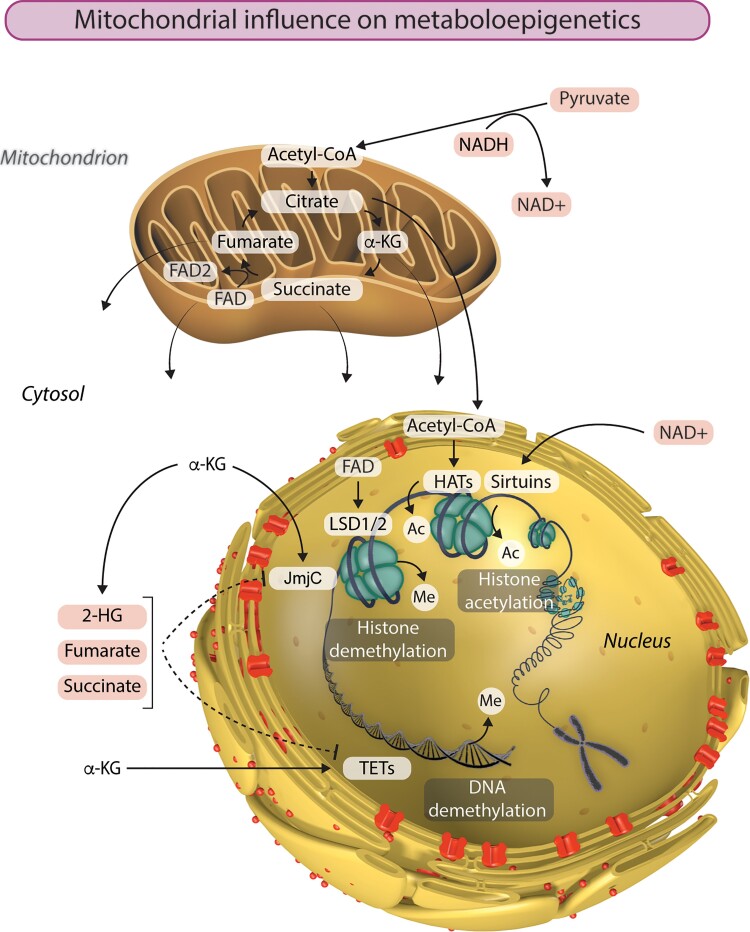
2.2. Mitochondria are the powerhouses of metaboloepigenetics
The removal of methyl groups from DNA and histones plays a critical role in shaping the epigenome. In recent years, the tricarboxylic acid (TCA) cycle has taken centre stage as a source of metabolites than influence chromatin demethylation, primarily through its effects on the activity of JmjC KDMs and TETs (Figure 1). Both of these classes of dioxygenases are dependent on α-ketoglutarate (α-KG) as a cofactor in reactions that remove methyl groups from histones and DNA, respectively. This key TCA cycle intermediate is produced from isocitrate by isocitrate dehydrogenase or by anaplerotic synthesis from glutamate. Isocitrate dehydrogenase (IDH) genes IDH1 and IDH2 are the most frequently mutated metabolic genes identified in human cancers.50 In addition to losing normal catalytic activity for the production of α-KG, mutant IDH1 and IDH2 gain the function of catalysing the reduction of α-KG to produce 2-hydroxyglutarate (2-HG), which has been described as an oncometabolite capable of stimulating proliferation and suppressing differentiation.51–53 Due to structural similarity, 2-HG can bind to and function as a competitive inhibitor of α-KG-dependent KDMs and TETs.17 Interestingly, TET2 is also frequently mutated in acute myeloid leukaemia, in which IDH1/2 mutations are common.54 Similarly, aberrant DNA methylation profiles and gene expression patterns were observed in AML with either IDH1/2 or TET2 mutations, indicating that TET2 is a pathologically relevant target of 2-HG. Furthermore, glioma samples harbouring mutant IDH1 accumulate significantly lower 5hmc and significantly higher 5 mc than those containing wild-type IDH1, in accordance with reduced TET activity.17
Figure 1.
Mitochondria are the powerhouses of metaboloepigenetics. Mitochondrial function, and particularly the TCA cycle, provides the intermediate metabolites essential to the generation and modification of epigenetic marks in the nucleus. Histone acetylation by histone acetyltransferases (HATs) is dependent on the availability of acetyl-CoA. On the other hand, histone acetylation can be removed by a class of NAD+-dependent histone deacetylases called sirtuins. LSD1 and LSD2 catalyse demethylation of mono- and di-methylated lysine residues using FAD as a cofactor. The availability of α-ketoglutarate (α-KG) influences histone and DNA demethylation by JmjC and TET enzymes respectively. Succinate, fumarate, and 2-hydroxyglutarate (2-HG) can also influence the epigenetic landscape by inhibiting α-KG-dependent histone and DNA demethylation. Ac, acetyl group; Me, methyl group.
Also within the TCA cycle, succinate dehydrogenase (SDH, comprised of four subunits: SDHA, SDHB, SDHC and SDHD) catalyses the oxidation of succinate to fumarate and uses the electrons to reduce ubiquinone to ubiquinol in the electron transport chain. The next enzyme in the TCA cycle, fumarate hydratase (FH), catalyses the reversible hydration of fumarate to malate. Succinate and fumarate are competitive inhibitors of α-KG-dependent dioxygenases. Knockdown of FH and SDH results in elevated intracellular levels of fumarate and succinate, respectively, both of which broadly inhibit the activity of α-KG-dependent dioxygenases.55 On the other hand, ectopic expression of tumour-derived FH and SDH mutants inhibits histone demethylation and hydroxylation of 5 mC. Accordingly, SDH deficiency underlies pervasive DNA hypermethylation in both gastrointestinal stromal tumours and paraganglioma/pheochromocytoma tumours.56 This part of the TCA cycle can further influence histone methylation by modulating the activity of another class of KDMs—the amine oxidases LSD1 and LSD2 that catalyse demethylation of mono- and di-methylated lysine residues using flavin adenine dinucleotide (FAD) as an essential cofactor.57 The conversion of succinate to fumarate by SDH reduces FAD to FADH2, thereby altering the cellular availability of FAD, which can have consequences for histone methylation.58
Very recently, Liu et al.59 identified a nonclassical TCA (nTCA) cycle in the nuclei of mouse and human cells. All components of the classical TCA cycle except for SDH are present in the nucleus, where they catalyse TCA cycle-related biochemical reactions. By functionally linking the nTCA cycle to epigenetic regulation, chromatin dynamics and gene expression, the authors propose that a self-sustaining nTCA cycle is implemented to supply and consume metabolites involved in the dynamics of DNA and histone modifications.59
2.3. Warburg metabolism: glycolysis and glutaminolysis
The Warburg effect is a signature of metabolic change in cancer. This metabolic rewiring is defined by increased glucose uptake and the fermentation of glucose to lactate, even in the presence of functioning mitochondria and oxygen, with a concomitant decrease in oxidative phosphorylation (OxPhos).26 While aerobic glycolysis generates much less ATP (2 molecules of ATP per unit of glucose) than does OxPhos (36 molecules of ATP per unit of glucose), switching to Warburg metabolism allows cancer cells to (i) more rapidly meet the bioenergetic demands of proliferation (glycolysis can be ramped up by relatively few enzymes, whereas increased OxPhos most likely requires mitochondrial biogenesis), and (ii) convert nutrients more efficiently into the biomass (nucleotides, amino acids, and lipids) needed to produce a new cell.60 These changes are largely regulated by HIF1α, a key TF for the expression of genes involved in glycolysis and the induction of pseudohypoxia in cancer cells,61 as well as mTOR signalling, which senses nutrients to support cell growth.62
The metabolic shift inexorably leads to the accumulation of methylgyoxyl (MGO), primarily through the fragmentation of triose phosphate intermediates of glycolysis.63 This toxic and highly reactive dicarbonyl spontaneously glycates lipids and proteins and is a potent inducer of advanced glycation end-products. Accumulation of MGO is limited by the glutathione-dependent enzyme, Glyoxalase 1, which is overexpressed in many cancers as a defence strategy against MGO cytotoxicity.64,65 On the other hand, recent evidence 66,67 indicates a hormetic effect, in which low doses of MGO support an adaptive response in cancer cells while high doses cause apoptosis.68 In addition to many cellular proteins, MGO-derived adducts were recently found to occur abundantly on histone tails, particularly arginine residues, under conditions of metabolic stress.69 These non-enzymatic modifications alter chromatin architecture by competing with other histone modifications, disrupt nucleosome assembly and chromatin fibre compaction, and down-regulate transcription.70 Protein arginine deaminase 4 (PAD4) was found to antagonize histone MGO-glycation by removing glycation adducts from arginine residues, as well as by converting unmodified arginine to citrulline, thereby protecting them from glycation. Indeed, PAD4 is overexpressed and citrulline is upregulated in breast tumours.71 These findings suggest an additional mechanistic link between metabolism and cancer epigenetics that requires more research attention.
Under normal metabolic conditions, pyruvate is fed into the TCA cycle, which generates ATP by OxPhos while at the same time providing metabolic substrate as precursors for biosynthetic reactions. While the demand for mitochondrial ATP production is decreased in tumour cells, the requirement for biosynthetic precursors and reducing power in the form of nicotinamide adenine dinucleotide phosphate is enhanced. In order to maintain mitochondrial function under limited pyruvate availability due to the increased rate of glycolysis, tumour cells often rely on increased glutaminolysis.72 Glutamine, the most abundant circulating amino acid,73 is transported into the cell and converted to glutamate and further to α-KG to replenish the TCA cycle. Indeed, strategies to inhibit glutaminolysis have proven effective in slowing the proliferation of cancer cells.74 Addiction to glutamine is a prominent feature of Kirsten rat sarcoma virus, oncogene (KRAS)-mutant cancers.75,76 A recent study showed that mutant KRAS rewires glutamine metabolism to support succinate biosynthesis from α-KG in colorectal cancer cells, resulting in an overall reduction of 5 hmc and CpG hypermethylation, and the activation of Wingless-related integration site/β-catenin signalling.77
2.4. Cellular metabolism influences histone acetylation
More than 30 years passed between the first description of histone tail acetylation78 and the discovery of histone acetyltransferases (HATs) and histone deacetylases (HDACs) that add and remove this modification respectively.79,80 Histone acetylation almost exclusively marks chromatin for transcriptional competency by two main mechanisms: (i) by changing the overall charge on the histone tail, which disrupts the binding of nucleosomal core components and renders the DNA accessible to transcriptional machinery and (ii) by acting as recruitment sites for various nucleosome remodelling proteins and transcription initiation factors that contain bromodomains—motifs that bind acetylated lysines.81
HATs transfer an acetyl group from acetyl-CoA to lysine residues on histone tails. Acetyl-CoA is produced by the oxidative decarboxylation of pyruvate from glycolysis by the pyruvate dehydrogenase complex (PDHC), the oxidation of long chain fatty acids, or the oxidative degradation of certain amino acids. Therefore, its abundance is dependent on glucose availability, fatty acid oxidation, and mitochondrial respiratory function.82 While histone acetylation is highly regulated, often in a gene-selective manner the Michaelis constant (Km) of most HATs falls within the range of cellular acetyl-CoA concentrations.83 As a consequence, the availability of acetyl-CoA can restrict or promote global levels of histone acetylation. Therefore, changes to metabolic pathways that influence acetyl-CoA levels potentiate large-scale changes in gene expression. And because glucose flux dose-dependently regulates histone acetylation,84 the acceleration of glycolysis in cancer is associated with global histone hyperacetylation.85 Accordingly, a glucose derivative that inhibits hexokinase and thereby blocks the first step of glycolysis, 2-deoxyglucose, significantly lowered acetyl-CoA levels and suppressed the acetylation of all four core histones in multiple cancer cell lines. For a broader discussion of the various metabolic sources of acetyl-CoA that support histone acetylation, we recommend a recent article by Feron.86
Interestingly, functional PDHC is also detected in the nucleus where it locally supplies acetyl-CoA to histone acetylation reactions.87 Moreover, nuclear PDHC levels were shown to increase in a cell cycle-dependent manner and in response to serum, growth factors or mitochondrial stress. This was concomitant with a decreased mitochondrial PDHC levels, suggesting that PDHC translocates from mitochondria to the nucleus to link acetyl-CoA synthesis with epigenetic regulation.87,88
2.5. NAD+-dependent chromatin modifiers
The mammalian HDACs discovered so far are divided into four classes based on sequence homology and domain organization.89 The class III HDACs, which are homologous to the yeast silent information regulator 2 (Sir2) and known as sirtuins in mammals, are most closely connected with metabolism. Sirtuins are a family of nicotinamide adenine dinucleotide NAD+-dependent deacetylases associated with longevity.90 In addition, there is evidence that some members of the sirtuin family can also catalyse the mono-adenosine diphosphate (ADP)-ribosylation of histones.91 While sirtuins play important roles in numerous biological processes including cellular metabolism,92 understanding their complex functions and dual characteristics as both promoters and suppressors of malignant characteristics in different cancers remains a significant challenge.93
The ADP-ribosyltransferases (ARTs), formerly known as poly ADP-ribose polymerases (PARPs), catalyse the transfer of one or more ADP-ribose groups from NAD+ to target proteins (at arginine, asparagine, cysteine, and histidine amino acids), including histones. ADP-ribosylation of histones and other nuclear proteins is predominantly associated with nucleosome remodelling in DNA repair.94–96 In response to extreme DNA damage, ADP-ribosylation can deplete NAD+ levels in the cell to the point where ATP production and other aspects of cellular metabolism are interrupted. Despite several recent studies indicating that ADP-ribosylated histones also have important roles in proliferation, replication, and transcription (recently reviewed97), knowledge of the chromatin-dependent function of this modification remains limited. Mono-ADP-ribosylation of H3 histones at arginine 117 (H3R117) was recently characterized as a modification associated with the proliferation of colorectal cancer cells.98 Li et al.99 demonstrated that mono-ADP-ribosylation of H3R117 limited local poly-ADP-ribosylation of TET1 promoter in human colon adenocarcinoma cells. This was associated with enrichment of 5 mc and depletion of H3K4me3 at the TET1 promoter, which culminated in a reduction of TET1 transcription. Moreover, this down-regulation of TET1 expression impaired TET1-dependent demethylation reactions to epigenetically silence the TFPI2 tumour suppressor gene.
3. Immuno-metaboloepigenetics
Over the past decade, immunologists have increasingly developed an appreciation for metabolism.100 Highly sensitive techniques to measure flux through metabolic pathways, metabolomic approaches that show how metabolites are directly connected to immune cell function, and the application of new pharmacological tools to models of infection and inflammation have driven the emergence of the field of immunometabolism.24,100 Central to this burgeoning area of research is the observation that immune cells with different functions engage distinct metabolic pathways.
As described in the previous section, glycolysis is less efficient than OxPhos for generating ATP from glucose. However, immune cells that require rapid production of ATP will switch to glycolysis.24 Glycolysis is enhanced in adaptive immune cells (slow-acting, long-term defence) such as activated effector T cells101 and activated B cells,102 as well as innate immune cells (rapid, front-line defence) such as activated dendritic cells103 and activated natural killer cells.104 Macrophages are also particularly interesting innate immune cells in this respect, as glycolytic metabolism can distinguish macrophage polarization subsets induced by anti-inflammatory [interleukin (IL)-4]105 or pro-inflammatory [bacterial endotoxin lipopolysaccharide (LPS)] stimuli. Inflammation resolving macrophages (M2 polarization state) are characterized by enhanced fatty acid oxidation106 and a reliance on OxPhos.107 In contrast, enhanced glucose uptake and glycolysis are metabolic traits of pro-inflammatory (M1 polarization state) macrophages.108 Another key difference concerns the TCA cycle, which is coupled with OxPhos in M2 macrophages, but is broken in two places in M1 macrophages. This leads to the accumulation of citrate and succinate and a down-regulation of OxPhos under aerobic conditions.108,109 The switch to Warburg metabolism is directly associated with pro-inflammatory cytokine production through the inhibition of prolyl hydroxylases by succinate, which stabilizes HIF1α and sustains IL-1β production.109
Driven by this strong connection between metabolism and immune function, immunologists have begun to delineate the functional epigenetic effects of these metabolic changes in macrophages. M2 macrophages accumulate α-KG through increased glutaminolysis.108,110 Mechanistically, the accumulation of α-KG is important for JMJD3-dependent demethylation of H3K27me3 in M2 gene induction.110 Furthermore, this study identified the importance of the α-KG/succinate ratio for the induction of distinct macrophage activation states: a high α-KG/succinate ratio promotes M2 activation, whereas a low α-KG/succinate ratio supports the pro-inflammatory phenotype of M1 macrophages. Although it is a relatively new area of research for which there is limited experimental evidence thus far, understanding how immune responses are orchestrated through metaboloepigenetic reprogramming offers novel insight into mechanisms supporting the broad spectrum of macrophage phenotypes. In this section, we discuss the current knowledge of metaboloepigenetic mechanisms associated with innate immune memory in macrophages.
3.1. Trained (innate) immunity
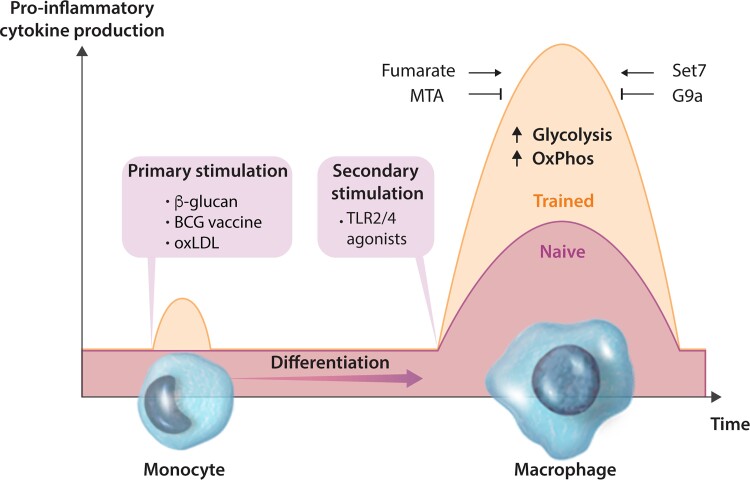
Contrasting the traditional dogma that memory is the proprietary of the adaptive immune system, the sensitivity of innate immune cells to Toll-like receptor (TLR) stimulation can be programmed by a phenomenon called ‘trained immunity.’111 Trained immunity is best characterized in monocyte-derived macrophages, although it has also been described in dendritic cells112 and natural killer cells.113 When primary monocytes are stimulated with certain microorganisms or microbial ligands, they differentiate into trained macrophages that have the capacity to respond to TLR agonists or other pathogen-associated molecular patterns with heightened pro-inflammatory cytokine production (Figure 2). Importantly, the secondary stimuli can be entirely unrelated to the first. Therefore, the effects of trained immunity are considered to be non-specific.114
Figure 2.
Trained immunity is dependent on metabolic and epigenetic changes. Innate immune cells, such as monocytes, can be functionally reprogrammed in response to exogenous or endogenous stimuli, leading to an altered immune response to a second, unrelated challenge after the return to a non-activated state. Primary stimulation with β-glucan, BCG vaccine or oxLDL induces a de facto memory that is revealed by secondary stimulation with agonists of TLR 2 and 4. Trained immunity is characterized by upregulation of glycolysis and OxPhos, as well as the accumulation of fumarate. The augmented cytokine production is abolished by the global methyltransferase inhibitor 5′-deoxy-5′-MTA and specific inhibition of Set7. Inhibition of G9a enhances the trained phenotype.
Trained immunity is exemplified by the augmented production of tumour necrosis factor (TNF) α and IL-6 induced by Candida albicans or its cell wall component β-glucan in human macrophages115,116 and mice that lack mature B and T cells.117 More recently, trained immunity induced by β-glucan was shown to be protective against infections by Leishmania braziliensis118 and Mycobacterium tuberculosis.119 Similarly, the induction of trained immunity by the Bacille Calmette–Guérin vaccine (BCG) is the most likely explanation for its heterologous protection against experimental yellow fever.120 In the clinic, trained immunity is the proposed mechanism through which BCG exerts its protective effects against a wide range of infections in newborns,121 as well as its anti-tumour properties in the treatment of bladder cancer.122 On the other hand, monocytes exposed to a low concentration of LPS differentiate into tolerant macrophages that are refractory to TLR restimulation. Also a type of trained immunity, endotoxin tolerance is a major cause of immunosuppression induced by Gram-negative sepsis.123
3.1.1. Epigenetic rewiring in trained immunity
Much of what we know about trained immunity has been learned from studying a standard cellular model that involves the training of human peripheral monocytes, in which these cells are exposed to a stimulus for a short period of time (usually 24 h). The cells are then incubated for 5–7 days during which they differentiate into macrophages. At this point, trained macrophages exhibit a heightened response to heterologous secondary stimuli (Figure 2).116 β-glucan-trained, naïve, and tolerant macrophages are distinguished by strikingly divergent genome-wide H3K4me1, H3K4me3 and H3K27ac.124,125 Many of these epigenetic changes are established early in the programme of training or tolerance and persistent H3K4me1 and H3K4me3 marks are considered central to the transcriptional memory.124,125
Co-incubation with the non-specific methyltransferase inhibitor 5′-deoxy-5′-methylthioadenosine (MTA) during the first 24 h of in vitro training completely nullifies the augmented cytokine response mediated by BCG117 and β-glucan.116,119,126 A proportion of these methyl events are regulated by the H3K4me1 writer Set7, which exhibits increased expression and activity in response to β-glucan.116 Co-administration of specific Set7 inhibitors prevented the induction of trained immunity by β-glucan and BCG in human macrophages. Furthermore, Set7 knockout mice were unable to mount β-glucan-mediated trained immunity against endotoxin challenge.116 In contrast, expression of G9a, a KMT that writes the repressive H3K9me2 modification, is decreased in cells trained with β-glucan. Specific inhibition of G9a reduced H3K9me2 at promoters of genes involved in trained immunity and simultaneously amplified trained immunity responses.127
3.2. The immunometabolic-epigenetic axis of trained immunity
Altered metabolism could partly account for wide-spread changes in histone acetylation in trained immunity.124,125 A key metabolic hallmark of trained cells is enhanced glycolysis via activation of the Akt-mTOR-HIF1α pathway.115 The trained phenotype is dependent on this glycolytic metabolism insofar as the augmented cytokine response is nullified by pharmacological inhibition of key enzymes involved in glycolysis.18,115,128 While concentrations of acetyl-CoA were unaltered in trained immunity,18 changes to the NAD+/NADH ratio115 indicated that sirtuins could be involved in establishing histone acetylation patterns in trained cells. So far, only one study has investigated this link, recently demonstrating that SIRT1 has a minor and perhaps redundant role in trained immunity.129 Another source of NAD+ is the breakdown of pyruvate to lactate that occurs in aerobic glycolysis and is consequently observed in trained immunity.116 Intersection of metabolome and transcriptome data and subsequent pharmacological studies revealed that the cholesterol synthesis pathway is also indispensable for trained immunity induced by β-glucan and BCG.19,130
Initial characterization of β-glucan-induced trained immunity revealed a metabolic shift towards glycolysis at the expense of OxPhos.27 However, recent studies show that macrophages trained with a lower concentration of β-glucan (1 ug/mL instead of 10 ug/mL) maintain a functioning TCA cycle that generates increased amounts of ATP by OxPhos while also providing metabolites that potentially modulate inflammatory functions.116 Genetic variation in TCA cycle genes such as IDH and SDH, as well as the NADH dehydrogenase subunit of complex I of the electron transport chain NDUFB7, is associated with variation in β-glucan-trained cytokine production. The reasons why different β-glucan concentrations induce opposite effects on OxPhos remain unclear, however the up-regulation of both glycolysis and OxPhos is consistent with the metabolic phenotypes of macrophages trained with BCG18 or endogenous training stimuli128,131,132 (discussed later in the Review).
Chief among the altered TCA cycle metabolites in trained immunity is fumarate, which accumulates in cells trained with β-glucan and integrates epigenetic, immune and metabolic circuits by inhibiting the KDM5 family of H3K4 demethylases.19 This could partly account for the enrichment of H3K4me3 observed in trained immunity throughout the genome and particularly at the promoters of TNF and IL6. Perhaps most convincing however is the observation that fumarate itself can induce trained immunity and partially recapitulate the enrichment of H3K4me3 induced by β-glucan at the TNF and IL6 promoters and other key regulatory sites. In addition to fumarate, the concentrations of 2-HG and succinate, which similarly antagonize α-KG-dependent KDMs, was also increased in β-glucan-trained macrophages.19,116 This was paralleled by increased expression of SDH genes, which can account for the enhanced conversion of succinate to fumarate. Fumarate-dependent inhibition of KDM5 activity was restored by the addition of α-KG, which also partially counteracted the training effect of fumarate on cytokine production.19
3.3. The chromatin is SET for metabolic rewiring in trained immunity
The accumulation of fumarate and malate, as well as the up-regulation of SDHB in β-glucan-trained macrophages is abolished by pharmacological inhibition of Set7 during the first 24 h of training in vitro. Mechanistically, this effect is partly explained by Set7-dependent H3K4me1 enrichment at distal enhancers that topologically associate with the SDHB promoter.116 Considering the metaboloepigenetic effects of fumarate in trained immunity,19 this study identifies a potential mechanism by which Set7 influences H3K4 methylation not only through its own methyltransferase activity, but also by indirectly inhibiting histone demethylation. SDH is unique in its participation in both the TCA cycle and the mitochondrial electron transport chain. Accordingly, the increase in OxPhos induced by β-glucan training was abolished by Set7 inhibition.116 This crucial role of Set7 early in the induction of trained immunity is consistent with its role as an epigenetic writer of metabolic memory in cultured vascular endothelial cells133,134 and peripheral blood mononuclear cells from patients with type 2 diabetes mellitus (T2DM).135
4. Metaboloepigenetics in CVD
Infectious diseases have been replaced by CVD as the leading cause of death globally over the last 20 years, with atherosclerosis the main underlying cause.105 The development and progression of atherosclerosis, which involves the metabolically driven activation and remodelling of vascular and immune cells,136 is increasingly explored in an epigenetic context.137,138 The acceleration and increased incidence of atherosclerosis in diabetes points further towards the involvement of metaboloepigenetic processes in atherosclerotic CVD.
4.1. Maladaptive trained immunity in atherosclerosis
While clearly beneficial for fighting infections, trained immunity is a double-edged sword when it comes to inflammatory diseases where innate immune cells are the proponents of tissue injury. Monocytes isolated from patients with an increased risk for atherosclerotic CVD due to elevated LDL-cholesterol levels139 and patients with severe coronary atherosclerosis140 exhibit augmented cytokine production capacity ex vivo. Furthermore, several endogenous compounds that accelerate atherosclerosis, including oxidized LDL (oxLDL),128,141 lipoprotein(a),142 aldosterone,143 epinephrine, and norepinephrine131 induce trained immunity in human macrophages. Apart from aldosterone, each of these stimuli induce a metabolic phenotype like BCG and β-glucan training: concurrent up-regulation of glycolysis and OxPhos. In the case of oxLDL, pharmacological inhibition of glycolysis prevented the induction of trained immunity and prevented H3K4me3 enrichment at the TNF and IL6 promoters.128 Precisely, how glycolysis influences histone lysine methylation events in trained immunity remains untested. One potential mechanism is that the glycolytic production of acetyl-CoA stimulates H3K14 acetylation,84 which can inhibit LSD1-mediated demethylation of H3K4me3.144
Atherosclerosis is characterized by chronic, low-grade sterile inflammation. This implies a long-term activation of the innate immune system despite the short lives of circulating innate immune cells. Feeding a high cholesterol Western diet (WD) for 4 weeks to atherosclerosis-prone Ldlr−/− mice induced trained immunity that persisted when the mice were switched back to a chow diet for 4 weeks.145 This metabolic memory occurred in the bone marrow niche and was dependent on the skewing of haematopoietic stem cells (HSCs) towards myelopoiesis. Indeed, hypercholesterolemia is linked to the reprogramming of HSCs146 and an increase in circulating monocytes correlates with cardiovascular events.147 Moreover, inflammatory monocytes that derive from activated haematopoietic precursors during WD feeding differentiate into atherogenic macrophages.148 The induction of trained immunity in the WD-fed Ldlr−/− mice was dependent on activation of the NLRP3 inflammasome. This is important because similar induction of trained immunity and myelopoiesis in the bone marrow of wild-type mice administered β-glucan was associated with increased IL-1β production, a key inflammasome-mediated product.149 Together, these findings underscore IL-1β as a central endogenous mediator of trained immunity in vivo. Indeed, the Canakinumab Antiinflammatory Thrombosis Outcome Study trial demonstrated the clinical benefit of IL-1β blockade for cardiovascular risk.150 Interestingly, increased expression of Il1b in the bone marrow of mice trained with β-glucan was significantly reduced in animals lacking Set7, suggesting a role for this KMT in the long-term activation of trained immunity.116 Further exploration of metaboloepigenetic changes in endogenous trained immunity could yield novel approaches to interfere with the development and progression of atherosclerosis.
4.2. Metaboloepigenetics in vascular complications of diabetes
Vascular complications are the major cause of the clinical, social and economic burden of type 1 diabetes mellitus (T1DM) and T2DM.151,152 Diabetes accelerates atherosclerosis and more than doubles the risk of CVD, which further increases with worsening glycemic control.153 Hyperglycemia can alter the epigenetic landscape of the microvasculature and the macrovasculature in diabetes, which may precede cardiovascular complications.154 The metabolic perturbations of the diabetic milieu are fertile ground for the discovery of metaboloepigenetic connections to atherosclerosis.
Hyperglycemia is associated with changes in histone acetylation in vascular cells. Genome-wide analysis of monocytes isolated from T1DM patients revealed elevated levels of H3K9ac enriched at gene promoters related to NFκB signalling and diabetes complications.155 Importantly, H3K9ac was significantly associated with mean levels of glycated haemoglobin—a marker of recent blood glucose levels. High glucose (HG) induced genome-wide histone hyperacetylation in human aortic endothelial cells and the specific induction of genes and pathways associated with endothelial dysfunction through the enrichment of H3K9/K14ac.156 Genome-wide histone hyperacetylation requires a substantial amount of acetyl-CoA as substrate for HATs in the nucleus. In microvascular cells of the kidney, HG induces the expression and activity of ATP-citrate lyase (ACL),157 which catalyses the synthesis of acetyl-CoA and oxaloacetate from citrate.158 Moreover, HG promotes the nuclear translocation of ACL to support the increased demand for acetyl-CoA for histone hyperacetylation.159 The increased demand for acetyl-CoA requires an increased supply of citrate, which comes from enhanced glycolysis driven by HG.157
Hyperglycemic memory is a phenomenon that has received considerable research attention. Prior exposure to HG can induce gene expression changes that persist even after the restoration of normal glucose conditions.15 Hyperglycemic memory occurs in human vascular endothelial cells133,134 as well as mouse bone marrow-derived macrophages (BMDMs), where it promotes M1-type responses and suppresses M2-type responses.160 Bone marrow obtained from diabetic mice and transplanted into normoglycemic atherosclerosis-prone mice retained a memory of its previous hyperglycemic environment to drive accelerated atherosclerosis and increase plaque macrophage content.161 BMDMs from diabetic mice exhibited a trained immunity phenotype, with enhanced Il6 and M1-associated gene expression following stimulation with LPS and IFN-γ. Hyperglycemia-induced trained immunity in HSCs was driven by epigenetic reprogramming of H3K4me3 and H3K27ac, which was likely to be downstream of metabolic changes, however a causal link was not established. HG exacerbates the training effect of oxLDL in terms of pro-inflammatory cytokine production capacity,128 which could be associated with the acceleration of atherosclerosis in diabetes.162 Indeed, exposure of peripheral monocytes to HG enhances the polarization of macrophages towards an M1-like phenotype, which correlates with increased succinate production and its potential to interfere with epigenetic reactions.161 To this end, Green and Brewer163 recently suggested that dysregulation of α-KG-dependent dioxygenases by hyperglycemia could link diabetes with vascular disease.
5. Future directions: metabolites impact the posttranslational regulation of TFs and epigenetic enzymes
So far, we have focused on the role that metabolites play in the modification of DNA and histones. However, many non-histone proteins are also post-translationally modified by the same machinery. In this section, we briefly describe some important considerations for interpreting the effects of metabolism on gene expression. The significance to the overall discussion is that metabolic changes could have a major impact on gene expression by modulating the transcriptional machinery in addition to chromatinized proteins.
Whenever there is a discussion of epigenetics, the elephant in the room is invariably the TF. Their power to activate or repress transcription is demonstrated by numerous cell-reprogramming experiments,164 best exemplified by a set of experiments showing that the forced expression of four specific TFs can reprogramme fibroblasts to stem cells.165 Several enzymes that mediate PTMs have dual roles modifying histones and non-histone proteins, and every modification that can occur on histones can also be written to non-histone proteins. Thousands of these modification sites have been identified across the proteome, and many have been shown to modulate protein function and stability.
Owing to advances in affinity enrichment methods and mass-spectrometry-based proteomics,166,167 methylation has emerged as a critical regulator of non-histone protein function.168 Numerous KMTs and KDMs target TFs with comparable affinity to their histone substrates, making the transcriptional events regulated by these enzymes difficult to interpret. Set7 has the most non-histone substrates identified so far,169 with the search for new substrates based on the presence of motifs commonly flanking targeted lysines.170,171 HIF1α is methylated at K32 by Set7, resulting in the modulation of HIF1α occupancy of target gene promoters in fibroblasts.172 In response to IL-6, STAT3-dependent SOCS3 expression in colon cancer cells is regulated by Set7-mediated methylation of STAT3 at K140, and the methyl groups are removed by LSD1.173 The gene-regulating activity of the p65 subunit of NFκB, which has roles in cancer, immune regulation and CVD,174 is modulated by lysine methylation mediated by Set7175,176 and the H3K36 methyltransferase NSD1, with the latter being antagonized by KDM2A.177 From these examples, it is not difficult to imagine how large-scale metabolic changes, such as changes to SAM/SAH ratios, FAD+ availability, or α-KG/succinate ratios could influence the activity of TFs. Similar metabolome-PTM crosstalk potentially influences TF acetylation, which has also been shown to regulate TF activity and stability,178 through changes in acetyl-CoA and NAD+/NADH ratios.
Also important to transcription is the intricate system of functional PTMs written (and erased) by epigenetic enzymes to other epigenetic enzymes. Again, the prototypical example is Set7. The transcriptionally-repressive HMT activity of SUV39H1, a H3K9 methyltransferase associated with heterochromatin formation, is down-regulated when SUV39H1 is methylated at K105 and K123 by Set7.179 Set7 can also support transcriptional activation by directing the lysine methyl-dependent degradation of DNMT1180 and by methylating the PCAF HAT at multiple sites.181 Furthermore, Set7-dependent methylation of ARTD1 (formerly PARP1) stimulates the synthesis of poly-ADP-ribose in response to oxidative stress.182 And like Set7, ARTD1 itself can maintain transcriptionally permissive chromatin structure by (i) modulating DNMT1 activity through ADP-ribose polymers and by (ii) preventing KDM5B-dependent demethylation of H3K4me3.183 Similar epigenetic enzyme modulating events are also associated with lysine acetylation. For example, autoacetylation by the p300 is important for stimulating its HAT activity.184 We previously described how pharmacological HDAC inhibition shows a complex pattern of gene expression changes that were associated with histone acetylation and histone deacetylation events, suggesting that HDAC inhibition could interfere with PTM of HATs and/or other HDACs.15,185
6. Concluding remarks
This Review is designed to stimulate interest in this emerging field of metaboloepigenetic mechanisms of gene regulation. The growing literature on the influence of intracellular metabolites on epigenetic reactions in cancer cells continues to provide new insights into mechanisms of gene regulation relevant to other diseases and biological processes. The metabolic profiles of M1-like macrophages and trained macrophages bear some resemblance to cancer cells, particularly with regard to the up-regulation of glycolysis and glutaminolysis. The metaboloepigenetic effects of these and other pathways can potentially be exploited for the improvement of vaccine strategies. On the other hand, a similar strategy could be employed to attenuate atherogenic innate immune cells. A deeper knowledge of the ways that metabolic changes reverberate through TF and chromatin-modifying networks to reprogramme gene expression is anticipated to significantly advance our understanding of metaboloepigenetics in health and disease.
Acknowledgements
Figure 2 incorporates elements from Servier Medical Art (https://smart.servier.com). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
Contributor Information
Samuel T Keating, Department of Biology, University of Copenhagen, Copenhagen DK-2200, Denmark.
Assam El-Osta, Department of Diabetes, Central Clinical School, Monash University, Melbourne, Victoria 3004, Australia; Epigenetics in Human Health and Disease Laboratory, Central Clinical School, Monash University, Melbourne, Victoria 3004, Australia; Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Sha Tin, Hong Kong SAR; Hong Kong Institute of Diabetes and Obesity, Prince of Wales Hospital, The Chinese University of Hong Kong, 3/F Lui Che Woo Clinical Sciences Building, 30-32 Ngan Shing Street, Sha Tin, Hong Kong SAR; Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, Sha Tin, SAR; Faculty of Health, Department of Technology, Biomedical Laboratory Science, University College Copenhagen, Copenhagen, Denmark.
Funding
Professor Sam El-Osta is a National Health and Medical Research Council (NHMRC) Senior Research Fellow (1154650) and acknowledges grant funding (2003401, 1113188).
Data availability
The data underlying data are incorporated into the review article.
References
- 1. Gates AJ, Gysi DM, Kellis M, Barabási AL. A wealth of discovery built on the human genome project—by the numbers. Nature 2021;590:212–215. [DOI] [PubMed] [Google Scholar]
- 2. Gibbs RA. The human genome project changed everything. Nat Rev Genet 2020;21:575–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joyner MJ, Paneth N. Promises, promises, and precision medicine. J Clin Invest 2019;129:946–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengelle J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, McKain MR, Leebens-Mack J, Burgess D, Freeling M, Mbeguie AMD, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, Jaillon O, Leclercq J, Argout X, Lyons E, Almeida A, Jeridi M, Dolezel J, Roux N, Risterucci AM, Weissenbach J, Ruiz M, Glaszmann JC, Quetier F, Yahiaoui N, Wincker P. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012;488:213–217. [DOI] [PubMed] [Google Scholar]
- 5. Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer 2021;21(7):413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, Groth A. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 2018;361:1389–1392. [DOI] [PubMed] [Google Scholar]
- 7. Stewart-Morgan KR, Petryk N, Groth A. Chromatin replication and epigenetic cell memory. Nat Cell Biol 2020;22:361–371. [DOI] [PubMed] [Google Scholar]
- 8. Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting DNMT1 to methylated DNA. Nature 2007;450:908–912. [DOI] [PubMed] [Google Scholar]
- 9. Ming X, Zhu B, Li Y. Mitotic inheritance of DNA methylation: more than just copy and paste. J Genet Genomics 2021;48:1–13. [DOI] [PubMed] [Google Scholar]
- 10. Alabert C, Barth TK, Reverón-Gómez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 2015;29:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reveron-Gomez N, Gonzalez-Aguilera C, Stewart-Morgan KR, Petryk N, Flury V, Graziano S, Johansen JV, Jakobsen JS, Alabert C, Groth A. Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol Cell 2018;72:239–249.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol 2012;227:3169–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z, Long H, Chang C, Zhao M, Lu Q. Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: a comprehensive overview. Cell Mol Life Sci 2018;75:3353–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akil AA, Jerman LF, Yassin E, Padmajeya SS, Al-Kurbi A, Fakhro KA. Reading between the (genetic) lines: how epigenetics is unlocking novel therapies for type 1 diabetes. Cells 2020;9:2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keating ST, El-Osta A. Epigenetics and metabolism. Circ Res 2015;116:715–736. [DOI] [PubMed] [Google Scholar]
- 16. Chandra V, Hong KM. Effects of deranged metabolism on epigenetic changes in cancer. Arch Pharm Res 2015;38:321–337. [DOI] [PubMed] [Google Scholar]
- 17. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arts RJW, Carvalho A, La Rocca C, Palma C, Rodrigues F, Silvestre R, Kleinnijenhuis J, Lachmandas E, Gonçalves LG, Belinha A, Cunha C, Oosting M, Joosten LAB, Matarese G, van Crevel R, Netea MG. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep 2016;17:2562–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arts RJ, Novakovic B, Ter Horst R, Carvalho A, Bekkering S, Lachmandas E, Rodrigues F, Silvestre R, Cheng SC, Wang SY, Habibi E, Goncalves LG, Mesquita I, Cunha C, van Laarhoven A, van de Veerdonk FL, Williams DL, van der Meer JW, Logie C, O’Neill LA, Dinarello CA, Riksen NP, van Crevel R, Clish C, Notebaart RA, Joosten LA, Stunnenberg HG, Xavier RJ, Netea MG. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab 2016;24:807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roy DG, Chen J, Mamane V, Ma EH, Muhire BM, Sheldon RD, Shorstova T, Koning R, Johnson RM, Esaulova E, Williams KS, Hayes S, Steadman M, Samborska B, Swain A, Daigneault A, Chubukov V, Roddy TP, Foulkes W, Pospisilik JA, Bourgeois-Daigneault MC, Artyomov MN, Witcher M, Krawczyk CM, Larochelle C, Jones RG. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab 2020;31:250–266.e9. [DOI] [PubMed] [Google Scholar]
- 21. Glier MB, Green TJ, Devlin AM. Methyl nutrients, DNA methylation, and cardiovascular disease. Mol Nutr Food Res 2014;58:172–182. [DOI] [PubMed] [Google Scholar]
- 22. Hou H, Zhao H. Epigenetic factors in atherosclerosis: DNA methylation, folic acid metabolism, and intestinal microbiota. Clin Chim Acta 2021;512:7–11. [DOI] [PubMed] [Google Scholar]
- 23. Dumont A, Lee M, Barouillet T, Murphy A, Yvan-Charvet L. Mitochondria orchestrate macrophage effector functions in atherosclerosis. Mol Aspects Med 2021;77:100922. [DOI] [PubMed] [Google Scholar]
- 24. O’Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016;16:553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol 2018;19:563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927;8:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 2019;4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov 2017;16:241–263. [DOI] [PubMed] [Google Scholar]
- 29. Morrison AJ. Cancer cell metabolism connects epigenetic modifications to transcriptional regulation. FEBS J 2022;289:1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Di Gregorio E, Miolo G, Saorin A, Steffan A, Corona G. From metabolism to genetics and vice versa: the rising role of oncometabolites in cancer development and therapy. Int J Mol Sci 2021;22:5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren R, Horton JR, Zhang X, Blumenthal RM, Cheng X. Detecting and interpreting DNA methylation marks. Curr Opin Struct Biol 2018;53:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017;18:517–534. [DOI] [PubMed] [Google Scholar]
- 33. Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009;324:929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009;324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997;88:471–481. [DOI] [PubMed] [Google Scholar]
- 36. Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 2002;21:5400–5413. [DOI] [PubMed] [Google Scholar]
- 37. Smith J, Sen S, Weeks RJ, Eccles MR, Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer 2020;6:392–406. [DOI] [PubMed] [Google Scholar]
- 38. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823–837. [DOI] [PubMed] [Google Scholar]
- 39. Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39:311–318. [DOI] [PubMed] [Google Scholar]
- 40. Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006;439:811–816. [DOI] [PubMed] [Google Scholar]
- 41. Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med 2017;49:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, James SJ. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine β-synthase heterozygous mice. J Nutr 2001;131:2811–2818. [DOI] [PubMed] [Google Scholar]
- 43. Shlomi T, Rabinowitz JD. Metabolism: cancer mistunes methylation. Nat Chem Biol 2013;9:293–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer 2017;116:1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mudd SH, Cerone R, Schiaffino MC, Fantasia AR, Minniti G, Caruso U, Lorini R, Watkins D, Matiaszuk N, Rosenblatt DS, Schwahn B, Rozen R, LeGros L, Kotb M, Capdevila A, Luka Z, Finkelstein JD, Tangerman A, Stabler SP, Allen RH, Wagner C. Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia. J Inherit Metab Dis 2001;24:448–464. [DOI] [PubMed] [Google Scholar]
- 46. Martinez-Chantar ML, Vazquez-Chantada M, Ariz U, Martinez N, Varela M, Luka Z, Capdevila A, Rodriguez J, Aransay AM, Matthiesen R, Yang H, Calvisi DF, Esteller M, Fraga M, Lu SC, Wagner C, Mato JM. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology 2008;47:1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stern PH, Wallace CD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol 1984;119:29–34. [DOI] [PubMed] [Google Scholar]
- 48. Guo H, Lishko VK, Herrera H, Groce A, Kubota T, Hoffman RM. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res 1993;53:5676–5679. [PubMed] [Google Scholar]
- 49. Higuchi T, Han Q, Sugisawa N, Yamamoto J, Yamamoto N, Hayashi K, Kimura H, Miwa S, Igarashi K, Bouvet M, Singh SR, Tsuchiya H, Hoffman RM. Combination methionine-methylation-axis blockade: a novel approach to target the methionine addiction of cancer. Cancer Genomics Proteomics 2021;18:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu S, Cadoux-Hudson T, Schofield CJ. Isocitrate dehydrogenase variants in cancer — cellular consequences and therapeutic opportunities. Curr Opin Chem Biol 2020;57:122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010;17:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG Jr. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013;339:1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Löwenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, Guan KL. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012;26:1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI Jr, Jahromi MS, Xekouki P, Szarek E, Walker RL, Lasota J, Raffeld M, Klotzle B, Wang Z, Jones L, Zhu Y, Wang Y, Waterfall JJ, O’Sullivan MJ, Bibikova M, Pacak K, Stratakis C, Janeway KA, Schiffman JD, Fan JB, Helman L, Meltzer PS. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov 2013;3:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941–953. [DOI] [PubMed] [Google Scholar]
- 58. Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, Umehara T, Yokoyama S, Kosai K, Nakao M. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun 2012;3:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu X, Si W, He L, Yang J, Peng Y, Ren J, Liu X, Jin T, Yu H, Zhang Z, Cheng X, Zhang W, Xia L, Huang Y, Wang Y, Liu S, Shan L, Zhang Y, Yang X, Li H, Liang J, Sun L, Shang Y. The existence of a nonclassical TCA cycle in the nucleus that wires the metabolic-epigenetic circuitry. Signal Transduct Target Ther 2021;6:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hayashi Y, Yokota A, Harada H, Huang G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1α in cancer. Cancer Sci 2019;110:1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci 2020;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Richard JP. Mechanism for the formation of methylglyoxal from triosephosphates. Biochem Soc Trans 1993;21:549–553. [DOI] [PubMed] [Google Scholar]
- 64. Wang Y, Kuramitsu Y, Ueno T, Suzuki N, Yoshino S, Iizuka N, Akada J, Kitagawa T, Oka M, Nakamura K. Glyoxalase I (GLO1) is up-regulated in pancreatic cancerous tissues compared with related non-cancerous tissues. Anticancer Res 2012;32:3219–3222. [PubMed] [Google Scholar]
- 65. Hu X, Yang X, He Q, Chen Q, Yu L. Glyoxalase 1 is up-regulated in hepatocellular carcinoma and is essential for HCC cell proliferation. Biotechnol Lett 2014;36:257–263. [DOI] [PubMed] [Google Scholar]
- 66. Chiavarina B, Nokin MJ, Bellier J, Durieux F, Bletard N, Sherer F, Lovinfosse P, Peulen O, Verset L, Dehon R, Demetter P, Turtoi A, Uchida K, Goldman S, Hustinx R, Delvenne P, Castronovo V, Bellahcène A. Methylglyoxal-mediated stress correlates with high metabolic activity and promotes tumor growth in colorectal cancer. Int J Mol Sci 2017;18:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Antognelli C, Moretti S, Frosini R, Puxeddu E, Sidoni A, Talesa VN. Methylglyoxal acts as a tumor-promoting factor in anaplastic thyroid cancer. Cells 2019;8:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leone A, Nigro C, Nicolò A, Prevenzano I, Formisano P, Beguinot F, Miele C. The dual-role of methylglyoxal in tumor progression–novel therapeutic approaches. Front Oncol 2021;11:645686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Galligan JJ, Wepy JA, Streeter MD, Kingsley PJ, Mitchener MM, Wauchope OR, Beavers WN, Rose KL, Wang T, Spiegel DA, Marnett LJ. Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc Natl Acad Sci USA 2018;115:9228–9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin-Groner E, D’Ambrosio H, Liu B, Chandarlapaty S, Liu S, David Y. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun 2019;10:1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zheng Q, Osunsade A, David Y. Protein arginine deiminase 4 antagonizes methylglyoxal-induced histone glycation. Nat Commun 2020;11:3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng 2017;19:163–194. [DOI] [PubMed] [Google Scholar]
- 73. Curi R, Lagranha CJ, Doi SQ, Sellitti DF, Procopio J, Pithon-Curi TC, Corless M, Newsholme P. Molecular mechanisms of glutamine action. J Cell Physiol 2005;204:392–401. [DOI] [PubMed] [Google Scholar]
- 74. Akins NS, Nielson TC, Le HV. Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Curr Top Med Chem 2018;18:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bernfeld E, Foster DA. Glutamine as an essential amino acid for KRas-driven cancer cells. Trends Endocrinol Metab 2019;30:357–368. [DOI] [PubMed] [Google Scholar]
- 76. Xia M, Li X, Diao Y, Du B, Li Y. Targeted inhibition of glutamine metabolism enhances the antitumor effect of selumetinib in KRAS-mutant NSCLC. Transl Oncol 2021;14:100920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wong CC, Xu J, Bian X, Wu JL, Kang W, Qian Y, Li W, Chen H, Gou H, Liu D, Luk STY, Zhou Q, Ji F, Chan LS, Shirasawa S, Sung JJ, Yu J. In colorectal cancer cells with mutant KRAS, SLC25A22-mediated glutaminolysis reduces DNA demethylation to increase WNT signaling, stemness, and drug resistance. Gastroenterology 2020;159:2163–2180.e6. [DOI] [PubMed] [Google Scholar]
- 78. Phillips DM. The presence of acetyl groups of histones. Biochem J 1963;87:258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996;272:408–411. [DOI] [PubMed] [Google Scholar]
- 80. Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem 1995;270:24674–24677. [DOI] [PubMed] [Google Scholar]
- 81. LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell 2008;30:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Martínez-Reyes I, Chandel NS. Acetyl-CoA-directed gene transcription in cancer cells. Genes Dev 2018;32:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sidoli S, Trefely S, Garcia BA, Carrer A. Integrated analysis of acetyl-CoA and histone modification via mass spectrometry to investigate metabolically driven acetylation. Methods Mol Biol 2019;1928:125–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab 2015;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu XS, Little JB, Yuan ZM. Glycolytic metabolism influences global chromatin structure. Oncotarget 2015;6:4214–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Feron O. The many metabolic sources of acetyl-CoA to support histone acetylation and influence cancer progression. Ann Transl Med 2019;7:S277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 2014;158:84–97. [DOI] [PubMed] [Google Scholar]
- 88. Ferriero R, Nusco E, De Cegli R, Carissimo A, Manco G, Brunetti-Pierri N. Pyruvate dehydrogenase complex and lactate dehydrogenase are targets for therapy of acute liver failure. J Hepatol 2018;69:325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Park SY, Kim JS. A short guide to histone deacetylases including recent progress on class II enzymes. Exp Mol Med 2020;52:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Grabowska W, Sikora E, Bielak-Zmijewska A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology 2017;18:447–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 2005;280:21313–21320. [DOI] [PubMed] [Google Scholar]
- 92. Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, Wang F, Yang L, Xiang Z, Cui H. The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol 2019;57:59–71. [DOI] [PubMed] [Google Scholar]
- 93. Zhao E, Hou J, Ke X, Abbas MN, Kausar S, Zhang L, Cui H. The roles of sirtuin family proteins in cancer progression. Cancers (Basel) 2019;11:1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yang G, Chen Y, Wu J, Chen SH, Liu X, Singh AK, Yu X. Poly(ADP-ribosyl)ation mediates early phase histone eviction at DNA lesions. Nucleic Acids Res 2020;48:3001–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen Q, Bian C, Wang X, Liu X, Ahmad Kassab M, Yu Y, Yu X. ADP-ribosylation of histone variant H2AX promotes base excision repair. EMBO J 2021;40:e104542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. van Beek L, McClay E, Patel S, Schimpl M, Spagnolo L, Maia de Oliveira T. PARP power: a structural perspective on PARP1, PARP2, and PARP3 in DNA damage repair and nucleosome remodelling. Int J Mol Sci 2021;22:5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zha JJ, Tang Y, Wang YL. Role of mono-ADP-ribosylation histone modification (review). Exp Ther Med 2021;21:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ling F, Tang Y, Li M, Li QS, Li X, Yang L, Zhao W, Jin CC, Zeng Z, Liu C, Wu CF, Chen WW, Lin X, Wang YL, Threadgill MD. Mono-ADP-ribosylation of histone 3 at arginine-117 promotes proliferation through its interaction with P300. Oncotarget 2017;8:72773–72787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li M, Tang Y, Li Q, Xiao M, Yang Y, Wang Y. Mono-ADP-ribosylation of H3R117 traps 5mC hydroxylase TET1 to impair demethylation of tumor suppressor gene TFPI2. Oncogene 2019;38:3488–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Voss K, Hong HS, Bader JE, Sugiura A, Lyssiotis CA, Rathmell JC. A guide to interrogating immunometabolism. Nat Rev Immunol 2021;21:637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 2011;186:3299–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF, Chiles TC. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 2006;107:4458–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG, Pearce EJ. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010;115:4742–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Donnelly RP, Loftus RM, Keating SE, Liou KT, Biron CA, Gardiner CM, Finlay DK. mTORC1-dependent metabolic reprogramming is a prerequisite for NK cell effector function. J Immunol 2014;193:4477–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, Bonny A, Brauer M, Brodmann M, Cahill TJ, Carapetis J, Catapano AL, Chugh SS, Cooper LT, Coresh J, Criqui M, DeCleene N, Eagle KA, Emmons-Bell S, Feigin VL, Fernandez-Sola J, Fowkes G, Gakidou E, Grundy SM, He FJ, Howard G, Hu F, Inker L, Karthikeyan G, Kassebaum N, Koroshetz W, Lavie C, Lloyd-Jones D, Lu HS, Mirijello A, Temesgen AM, Mokdad A, Moran AE, Muntner P, Narula J, Neal B, Ntsekhe M, de Oliveira G M, Otto C, Owolabi M, Pratt M, Rajagopalan S, Reitsma M, Ribeiro ALP, Rigotti N, Rodgers A, Sable C, Shakil S, Sliwa-Hahnle K, Stark B, Sundstrom J, Timpel P, Tleyjeh IM, Valgimigli M, Vos T, Whelton PK, Yacoub M, Zuhlke L, Murray C, Fuster V. GBD-NHLBI-JACC Global Burden of Cardiovascular Diseases Writing Group . Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J Am Coll Cardiol 2020;76:2982–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Johnson AR, Qin Y, Cozzo AJ, Freemerman AJ, Huang MJ, Zhao L, Sampey BP, Milner JJ, Beck MA, Damania B, Rashid N, Galanko JA, Lee DP, Edin ML, Zeldin DC, Fueger PT, Dietz B, Stahl A, Wu Y, Mohlke KL, Makowski L. Metabolic reprogramming through fatty acid transport protein 1 (FATP1) regulates macrophage inflammatory potential and adipose inflammation. Mol Metab 2016;5:506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang F, Zhang S, Vuckovic I, Jeon R, Lerman A, Folmes CD, Dzeja PP, Herrmann J. Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab 2018;28:463–475.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015;42:419–430. [DOI] [PubMed] [Google Scholar]
- 109. Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O’Neill LA. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013;496:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Liu P-S, Wang H, Li X, Chao T, Teav T, Christen S, Di Conza G, Cheng W-C, Chou C-H, Vavakova M, Muret C, Debackere K, Mazzone M, Huang H-D, Fendt S-M, Ivanisevic J, Ho P-C. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017;18:985–994. [DOI] [PubMed] [Google Scholar]
- 111. Netea MG, Domínguez-Andrés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, Joosten LAB, van der Meer JWM, Mhlanga MM, Mulder WJM, Riksen NP, Schlitzer A, Schultze JL, Stabell Benn C, Sun JC, Xavier RJ, Latz E. Defining trained immunity and its role in health and disease. Nat Rev Immunol 2020;20:375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hole CR, Wager CML, Castro-Lopez N, Campuzano A, Cai H, Wozniak KL, Wang Y, Wormley FL Jr. Induction of memory-like dendritic cell responses in vivo. Nat Commun 2019;10:2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Jacobs C, Xavier RJ, van der Meer JW, van Crevel R, Netea MG. BCG-induced trained immunity in NK cells: role for non-specific protection to infection. Clin Immunol 2014;155:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fanucchi S, Domínguez-Andrés J, Joosten LAB, Netea MG, Mhlanga MM. The intersection of epigenetics and metabolism in trained immunity. Immunity 2021;54:32–43. [DOI] [PubMed] [Google Scholar]
- 115. Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJ, van der Veer BM, Deen PM, Logie C, O’Neill LA, Willems P, van de Veerdonk FL, van der Meer JW, Ng A, Joosten LA, Wijmenga C, Stunnenberg HG, Xavier RJ, Netea MG. mTOR- and HIF-1α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014;345:1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Keating ST, Groh L, van der Heijden C, Rodriguez H, Dos Santos JC, Fanucchi S, Okabe J, Kaipananickal H, van Puffelen JH, Helder L, Noz MP, Matzaraki V, Li Y, de Bree LCJ, Koeken V, Moorlag S, Mourits VP, Domínguez-Andrés J, Oosting M, Bulthuis EP, Koopman WJH, Mhlanga M, El-Osta A, Joosten LAB, Netea MG, Riksen NP. The Set7 lysine methyltransferase regulates plasticity in oxidative phosphorylation necessary for trained immunity induced by β-glucan. Cell Rep 2020;31:107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, Xavier RJ, van der Meer JW, van Crevel R, Netea MG. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A 2012;109:17537–17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Santos JC D, de Figueiredo AM B, Silva MV T, Cirovic B, de Bree LCJ, Damen M, Moorlag S, Gomes RS, Helsen MM, Oosting M, Keating ST, Schlitzer A, Netea MG, Ribeiro-Dias F, Joosten LAB. β-glucan-induced trained immunity protects against leishmania braziliensis infection: a crucial role for IL-32. Cell Rep 2019;28:2659–2672.e6. [DOI] [PubMed] [Google Scholar]
- 119. Moorlag S, Khan N, Novakovic B, Kaufmann E, Jansen T, van Crevel R, Divangahi M, Netea MG. β-glucan induces protective trained immunity against mycobacterium tuberculosis infection: a key role for IL-1. Cell Rep 2020;31:107634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Arts RJW, Moorlag S, Novakovic B, Li Y, Wang SY, Oosting M, Kumar V, Xavier RJ, Wijmenga C, Joosten LAB, Reusken C, Benn CS, Aaby P, Koopmans MP, Stunnenberg HG, van Crevel R, Netea MG. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 2018;23:89–100.e5. [DOI] [PubMed] [Google Scholar]
- 121. Benn CS, Netea MG, Selin LK, Aaby P. A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends Immunol 2013;34:431–439. [DOI] [PubMed] [Google Scholar]
- 122. van Puffelen JH, Keating ST, Oosterwijk E, van der Heijden AG, Netea MG, Joosten LAB, Vermeulen SH. Trained immunity as a molecular mechanism for BCG immunotherapy in bladder cancer. Nat Rev Urol 2020;17:513–525. [DOI] [PubMed] [Google Scholar]
- 123. Steinhagen F, Schmidt SV, Schewe JC, Peukert K, Klinman DM, Bode C. Immunotherapy in sepsis - brake or accelerate? Pharmacol Ther 2020;208:107476. [DOI] [PubMed] [Google Scholar]
- 124. Novakovic B, Habibi E, Wang SY, Arts RJW, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, Matarese F, van Heeringen SJ, Janssen-Megens EM, Sharifi N, Wang C, Keramati F, Schoonenberg V, Flicek P, Clarke L, Pickkers P, Heath S, Gut I, Netea MG, Martens JHA, Logie C, Stunnenberg HG. β-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 2016;167:1354–1368.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA, Sharifi N, Janssen-Megens EM, Ter Huurne M, Mandoli A, van Schaik T, Ng A, Burden F, Downes K, Frontini M, Kumar V, Giamarellos-Bourboulis EJ, Ouwehand WH, van der Meer JW, Joosten LA, Wijmenga C, Martens JH, Xavier RJ, Logie C, Netea MG, Stunnenberg HG. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014;345:1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ifrim DC, Quintin J, Joosten LA, Jacobs C, Jansen T, Jacobs L, Gow NA, Williams DL, van der Meer JW, Netea MG, Papasian CJ. Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin Vaccine Immunol 2014;21:534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mourits VP, van Puffelen JH, Novakovic B, Bruno M, Ferreira AV, Arts RJ, Groh L, Crișan TO, Zwaag J, Jentho E, Kox M, Pickkers P, van de Veerdonk FL, Weis S, Oosterwijk E, Vermeulen SH, Netea MG, Joosten LA. Lysine methyltransferase G9a is an important modulator of trained immunity. Clin Transl Immunology 2021;10:e1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Keating ST, Groh L, Thiem K, Bekkering S, Li Y, Matzaraki V, van der Heijden C, van Puffelen JH, Lachmandas E, Jansen T, Oosting M, de Bree LCJ, Koeken V, Moorlag S, Mourits VP, van Diepen J, Strienstra R, Novakovic B, Stunnenberg HG, van Crevel R, Joosten LAB, Netea MG, Riksen NP. Rewiring of glucose metabolism defines trained immunity induced by oxidized low-density lipoprotein. J Mol Med (Berl) 2020;98:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mourits VP, Helder LS, Matzaraki V, Koeken V, Groh L, de Bree LCJ, Moorlag S, van der Heijden C, Keating ST, van Puffelen JH, Jaeger M, Joosten LAB, Netea MG. The role of sirtuin 1 on the induction of trained immunity. Cell Immunol 2021;366:104393. [DOI] [PubMed] [Google Scholar]
- 130. Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden C, Li Y, Popa CD, Ter Horst R, van Tuijl J, Netea-Maier RT, van de Veerdonk FL, Chavakis T, Joosten LAB, van der Meer JWM, Stunnenberg H, Riksen NP, Netea MG. Metabolic induction of trained immunity through the mevalonate pathway. Cell 2018;172:135–146.e9. [DOI] [PubMed] [Google Scholar]
- 131. van der Heijden C, Groh L, Keating ST, Kaffa C, Noz MP, Kersten S, van Herwaarden AE, Hoischen A, Joosten LAB, Timmers H, Netea MG, Riksen NP. Catecholamines induce trained immunity in monocytes in vitro and in vivo. Circ Res 2020;127:269–283. [DOI] [PubMed] [Google Scholar]
- 132. Groh LA, Ferreira AV, Helder L, van der Heijden C, Novakovic B, van de Westerlo E, Matzaraki V, Moorlag S, de Bree LC, Koeken V, Mourits VP, Keating ST, van Puffelen JH, Hoischen A, Joosten LAB, Netea MG, Koopman WJH, Riksen NP. oxLDL-induced trained immunity is dependent on mitochondrial metabolic reprogramming. Immunometabolism 2021;3:e210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008;205:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 2009;58:1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Paneni F, Costantino S, Battista R, Castello L, Capretti G, Chiandotto S, Scavone G, Villano A, Pitocco D, Lanza G, Volpe M, Lüscher TF, Cosentino F. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ Cardiovasc Genet 2015;8:150–158. [DOI] [PubMed] [Google Scholar]
- 136. Ali L, Schnitzler JG, Kroon J. Metabolism: the road to inflammation and atherosclerosis. Curr Opin Lipidol 2018;29:474–480. [DOI] [PubMed] [Google Scholar]
- 137. Ambrosini S, Mohammed SA, Costantino S, Paneni F. Disentangling the epigenetic landscape in cardiovascular patients: a path toward personalized medicine. Minerva Cardiol Angiol 2021;69:331–345. [DOI] [PubMed] [Google Scholar]
- 138. Zarzour A, Kim HW, Weintraub NL. Epigenetic regulation of vascular diseases. Arterioscler Thromb Vasc Biol 2019;39:984–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bekkering S, van den Munckhof I, Nielen T, Lamfers E, Dinarello C, Rutten J, de Graaf J, Joosten LA, Netea MG, Gomes ME, Riksen NP. Innate immune cell activation and epigenetic remodeling in symptomatic and asymptomatic atherosclerosis in humans in vivo. Atherosclerosis 2016;254:228–236. [DOI] [PubMed] [Google Scholar]
- 140. Bekkering S, Stiekema LCA, Bernelot Moens S, Verweij SL, Novakovic B, Prange K, Versloot M, Roeters van Lennep JE, Stunnenberg H, de Winther M, Stroes ESG, Joosten LAB, Netea MG, Riksen NP. Treatment with Statins does not revert trained immunity in patients with familial hypercholesterolemia. Cell Metab 2019;30:1–2. [DOI] [PubMed] [Google Scholar]
- 141. Bekkering S, Quintin J, Joosten LA, van der Meer JW, Netea MG, Riksen NP. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler Thromb Vasc Biol 2014;34:1731–1738. [DOI] [PubMed] [Google Scholar]
- 142. van der Valk FM, Bekkering S, Kroon J, Yeang C, van den Bossche J, van Buul JD, Ravandi A, Nederveen AJ, Verberne HJ, Scipione C, Nieuwdorp M, Joosten LA, Netea MG, Koschinsky ML, Witztum JL, Tsimikas S, Riksen NP, Stroes ES. Oxidized phospholipids on lipoprotein(a) elicit arterial wall inflammation and an inflammatory monocyte response in humans. Circulation 2016;134:611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. van der Heijden C, Keating ST, Groh L, Joosten LAB, Netea MG, Riksen NP. Aldosterone induces trained immunity: the role of fatty acid synthesis. Cardiovasc Res 2020;116:317–328. [DOI] [PubMed] [Google Scholar]
- 144. Wu M, Hayward D, Kalin JH, Song Y, Schwabe JW, Cole PA. Lysine-14 acetylation of histone H3 in chromatin confers resistance to the deacetylase and demethylase activities of an epigenetic silencing complex. Elife 2018;7:e37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Christ A, Gunther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, Scholz CJ, Oosting M, Haendler K, Bassler K, Klee K, Schulte-Schrepping J, Ulas T, Moorlag S, Kumar V, Park MH, Joosten LAB, Groh LA, Riksen NP, Espevik T, Schlitzer A, Li Y, Fitzgerald ML, Netea MG, Schultze JL, Latz E. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 2018;172:162–175.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Murphy AJ, Akhtari M, Tolani S, Pagler T, Bijl N, Kuo CL, Wang M, Sanson M, Abramowicz S, Welch C, Bochem AE, Kuivenhoven JA, Yvan-Charvet L, Tall AR. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J Clin Invest 2011;121:4138–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science 2013;339:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Swirski FK, Libby P, Aikawa E, Alcaide P, Luscinskas FW, Weissleder R, Pittet MJ. Ly-6Chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest 2007;117:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, Eugster A, Troullinaki M, Palladini A, Kourtzelis I, Chatzigeorgiou A, Schlitzer A, Beyer M, Joosten LAB, Isermann B, Lesche M, Petzold A, Simons K, Henry I, Dahl A, Schultze JL, Wielockx B, Zamboni N, Mirtschink P, Coskun U, Hajishengallis G, Netea MG, Chavakis T. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 2018;172:147–161.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ, Group CT. Antiinflammatory therapy with Canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 151. Libby P, Nathan DM, Abraham K, Brunzell JD, Fradkin JE, Haffner SM, Hsueh W, Rewers M, Roberts BT, Savage PJ, Skarlatos S, Wassef M, Rabadan-Diehl C. National Heart, Lung, and Blood Institute; National Institute of Diabetes and Digestive and Kidney Diseases Working Group on Cardiovascular Complications of Type 1 Diabetes Mellitus . Report of the national heart, lung, and blood institute-national institute of diabetes and digestive and kidney diseases working group on cardiovascular complications of type 1 diabetes mellitus. Circulation 2005;111:3489–3493. [DOI] [PubMed] [Google Scholar]
- 152. van Dieren S, Beulens JW, van der Schouw YT, Grobbee DE, Neal B. The global burden of diabetes and its complications: an emerging pandemic. Eur J Cardiovasc Prev Rehabil 2010;17(Suppl. 1):S3–8. [DOI] [PubMed] [Google Scholar]
- 153. Canto E D, Ceriello A, Ryden L, Ferrini M, Hansen TB, Schnell O, Standl E, Beulens JW. Diabetes as a cardiovascular risk factor: an overview of global trends of macro and micro vascular complications. Eur J Prev Cardiol 2019;26:25–32. [DOI] [PubMed] [Google Scholar]
- 154. Napoli C, Benincasa G, Schiano C, Salvatore M. Differential epigenetic factors in the prediction of cardiovascular risk in diabetic patients. Eur Heart J Cardiovasc Pharmacother 2020;6:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X, Li SM, Cleary P, Riggs A, Harlan DM, Lorenzi G, Kolterman O, Sun W, Lachin JM, Natarajan R, Group DER. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014;63:1748–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Pirola L, Balcerczyk A, Tothill RW, Haviv I, Kaspi A, Lunke S, Ziemann M, Karagiannis T, Tonna S, Kowalczyk A, Beresford-Smith B, Macintyre G, Kelong M, Hongyu Z, Zhu J, El-Osta A. Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res 2011;21:1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Deb DK, Chen Y, Sun J, Wang Y, Li YC. ATP-citrate lyase is essential for high glucose-induced histone hyperacetylation and fibrogenic gene upregulation in mesangial cells. Am J Physiol Renal Physiol 2017;313:F423–F429. [DOI] [PubMed] [Google Scholar]
- 158. Chypre M, Zaidi N, Smans K. ATP-citrate lyase: a mini-review. Biochem Biophys Res Commun 2012;422:1–4. [DOI] [PubMed] [Google Scholar]
- 159. Deb DK, Bao R, Li YC. Critical role of the cAMP-PKA pathway in hyperglycemia-induced epigenetic activation of fibrogenic program in the kidney. FASEB J 2017;31:2065–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Parathath S, Grauer L, Huang LS, Sanson M, Distel E, Goldberg IJ, Fisher EA. Diabetes adversely affects macrophages during atherosclerotic plaque regression in mice. Diabetes 2011;60:1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Edgar L, Akbar N, Braithwaite AT, Krausgruber T, Gallart-Ayala H, Bailey J, Corbin AL, Khoyratty TE, Chai JT, Alkhalil M, Rendeiro AF, Ziberna K, Arya R, Cahill TJ, Bock C, Laurencikiene J, Crabtree MJ, Lemieux ME, Riksen NP, Netea MG, Wheelock CE, Channon KM, Rydén M, Udalova IA, Carnicer R, Choudhury RP. Hyperglycemia induces trained immunity in macrophages and their precursors and promotes atherosclerosis. Circulation 2021;144:961–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Thiem K, Stienstra R, Riksen NP, Keating ST. Trained immunity and diabetic vascular disease. Clin Sci (Lond) 2019;133:195–203. [DOI] [PubMed] [Google Scholar]
- 163. Green HLH, Brewer AC. Dysregulation of 2-oxoglutarate-dependent dioxygenases by hyperglycaemia: does this link diabetes and vascular disease? Clin Epigenetics 2020;12:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol 2016;17:183–193. [DOI] [PubMed] [Google Scholar]
- 165. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 166. Charih F, Green JR, Biggar KK. Using machine learning and targeted mass spectrometry to explore the methyl-lys proteome. STAR Protoc 2020;1:100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Biggar KK, Charih F, Liu H, Ruiz-Blanco YB, Stalker L, Chopra A, Connolly J, Adhikary H, Frensemier K, Hoekstra M, Galka M, Fang Q, Wynder C, Stanford WL, Green JR, Li SS. Proteome-wide prediction of lysine methylation leads to identification of H2BK43 methylation and outlines the potential methyllysine proteome. Cell Rep 2020;32:107896. [DOI] [PubMed] [Google Scholar]
- 168. Lukinović V, Casanova AG, Roth GS, Chuffart F, Reynoird N. Lysine methyltransferases signaling: histones are just the tip of the iceberg. Curr Protein Pept Sci 2020;21:655–674. [DOI] [PubMed] [Google Scholar]
- 169. Keating ST, El-Osta A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics 2013;8:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol 2011;18:111–120. [DOI] [PubMed] [Google Scholar]
- 171. Keating ST, Ziemann M, Okabe J, Khan AW, Balcerczyk A, El-Osta A. Deep sequencing reveals novel Set7 networks. Cell Mol Life Sci 2014;71:4471–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Liu X, Chen Z, Xu C, Leng X, Cao H, Ouyang G, Xiao W. Repression of hypoxia-inducible factor α signaling by Set7-mediated methylation. Nucleic Acids Res 2015;43:5081–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A 2010;107:21499–21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Park MH, Hong JT. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells 2016;5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J 2009;28:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Ea CK, Baltimore D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc Natl Acad Sci U S A 2009;106:18972–18977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A 2010;107:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Xia C, Tao Y, Li M, Che T, Qu J. Protein acetylation and deacetylation: an important regulatory modification in gene transcription (review). Exp Ther Med 2020;20:2923–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wang D, Zhou J, Liu X, Lu D, Shen C, Du Y, Wei FZ, Song B, Lu X, Yu Y, Wang L, Zhao Y, Wang H, Yang Y, Akiyama Y, Zhang H, Zhu WG. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci U S A 2013;110:5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Estève PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A 2009;106:5076–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Masatsugu T, Yamamoto K. Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem Biophys Res Commun 2009;381:22–26. [DOI] [PubMed] [Google Scholar]
- 182. Kassner I, Andersson A, Fey M, Tomas M, Ferrando-May E, Hottiger MO. SET7/9-dependent methylation of ARTD1 at K508 stimulates poly-ADP-ribose formation after oxidative stress. Open Biol 2013;3:120173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 2010;39:736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol 2004;11:308–315. [DOI] [PubMed] [Google Scholar]
- 185. Rafehi H, Balcerczyk A, Lunke S, Kaspi A, Ziemann M, Kn H, Okabe J, Khurana I, Ooi J, Khan AW, Du XJ, Chang L, Haviv I, Keating ST, Karagiannis TC, El-Osta A. Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res 2014;24:1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying data are incorporated into the review article.