Summary
Background
Throughout the COVID-19 pandemic, the SARS-CoV-2 virus has continued to evolve, with new variants outcompeting existing variants and often leading to different dynamics of disease spread.
Methods
In this paper, we performed a retrospective analysis using longitudinal sequencing data to characterize differences in the speed, calendar timing, and magnitude of 16 SARS-CoV-2 variant waves/transitions for 230 countries and sub-country regions, between October 2020 and January 2023. We then clustered geographic locations in terms of their variant behavior across several Omicron variants, allowing us to identify groups of locations exhibiting similar variant transitions. Finally, we explored relationships between heterogeneity in these variant waves and time-varying factors, including vaccination status of the population, governmental policy, and the number of variants in simultaneous competition.
Findings
This work demonstrates associations between the behavior of an emerging variant and the number of co-circulating variants as well as the demographic context of the population. We also observed an association between high vaccination rates and variant transition dynamics prior to the Mu and Delta variant transitions.
Interpretation
These results suggest the behavior of an emergent variant may be sensitive to the immunologic and demographic context of its location. Additionally, this work represents the most comprehensive characterization of variant transitions globally to date.
Funding
Laboratory Directed Research and Development (LDRD), Los Alamos National Laboratory.
Keywords: SARS-CoV-2, COVID-19, Variant transition, GISAID
Research in context.
Evidence before this study
SARS-CoV-2 variants with a selective advantage are continuing to emerge, resulting in variant transitions that can give rise to new waves in global COVID-19 cases and changing dynamics of disease spread. While variant transitions have been well studied individually, more work is needed to better understand how variant transitions have occurred in the past and how properties of these transitions may relate to vaccination rates, convalescent immunity, and population demographics.
Added value of this study
Our retrospective study integrates metadata based on 14 million SARS-CoV-2 sequences available through the Global Initiative on Sharing All Influenza Data (GISAID) with clinical and demographic data to characterize heterogeneity in variant waves/transitions across the globe throughout the COVID-19 pandemic. We demonstrate that properties of the variant transitions (e.g., speed, timing, and magnitude of the transition) are associated with vaccination rates, prior COVID-19 cases, and the number of co-circulating variants in competition.
Implications of all the available evidence
Our results indicate that there is substantial heterogeneity in how an emerging variant may compete with other virus variants across locations and suggest that each location's contemporaneous immunologic landscape may play a role in these interactions.
Introduction
Since the first SARS-CoV-2 viral sequence became available in January of 2020,1 there have been over 660 million confirmed cases of COVID-19 globally,2 leading to over 6.7 million deaths. SARS-CoV-2 is continuously evolving, and global transitions to newly emergent variants can generate waves of disease spread. The selective advantage of a new variant over existing variants is often associated with increased infectivity (e.g., through enhanced receptor binding or spike processing) and/or increased resistance to neutralizing antibodies induced by vaccination, prior infection, or both.3, 4, 5, 6, 7, 8, 9, 10 Prior infections with different variants can be associated with differing protection against newly emergent variants,11 and we hypothesize that vaccination rates and the history of prior infecting variants may impact the rate at which an emerging variant out-competes existing variants to become the dominant form of the virus in a given country or state.
To explore this hypothesis and characterize heterogeneity in the speed, timing, and magnitude of variant transitions globally, we performed a retrospective analysis of over 14 million SARS-CoV-2 sequences reported to the Global Initiative on Sharing All Influenza Data (GISAID) between October 2020 and mid-January 2023.12 We used multinomial regression spline modeling to estimate and summarize variant transition dynamics across 230 countries and sub-country regions and 16 SARS-CoV-2 variant waves, including recently emergent Omicron sublineages BA.2.75, XBB.1/XBB, and BQ.1.10 Our results illustrate large heterogeneity in variant transitions between locations. For Omicron, we clustered geographic locations in terms of their variant behavior, allowing us to identify groups of locations with similar transition dynamics. We then leveraged clinical and demographic data to explore how properties of variant waves relate to time-varying factors, including vaccination status of the population, governmental policy, and the number of variants in competition. This work demonstrates an association between the behavior of an emerging variant and the immunologic and demographic context of the population. Additionally, this work represents the most comprehensive characterization of SARS-CoV-2 variant transitions globally to date.
Methods
Data sourcing and processing
Analyzed data streams (summarized in Supp. Fig. A.1) are described below. All data were aggregated by date and location. We defined spatial locations at the country level and, for select countries having sufficient data, the sub-country region level.
GISAID SARS-CoV-2 data
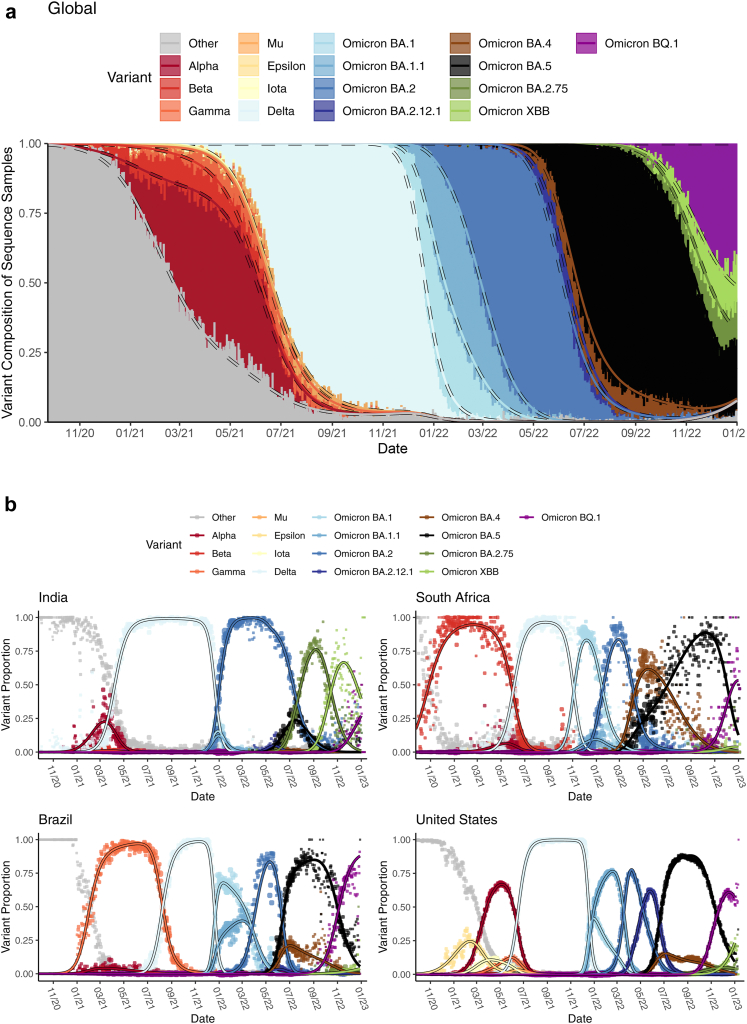
Data for over 14 million SARS-CoV-2 sequences reported to GISAID by 1/17/2023 (https://gisaid.org) were obtained through the COVID-19 Viral Genome Analysis Pipeline https://cov.lanl.gov. We resolved location name inconsistencies and removed sequences with evident entry errors. We then categorized the sequences by variant (e.g., Alpha, Delta) based on each sequence's Pango nomenclature SARS-CoV-2 lineage designation,13 after excluding records designated “None” or “Unassigned”. Pango sub-lineage groupings as of 1/17/2023 are provided in Supp. Table A.2. Fig. 1 illustrates the reported variant proportions over time globally and for four example countries.
Fig. 1.
Daily variant composition of all SARS-CoV-2 sequences reported to GISAID (a) globally and (b) for four example countries (points) along with fitted variant proportions (lines) from the primary analysis. Fitted lines show the point estimates obtained from fitting the multinomial model in (Eq. 1). The size of the plotted points corresponds to the total number of sequenced samples, relative to the daily maximum within each country. For panel (a), global sequences attributed to each variant for a given date were weighted proportional to the confirmed cases reported for each sequence's corresponding country on that date.
Clinical and demographic data
Daily confirmed COVID-19 case and death data were obtained from the Johns Hopkins Center for Systems Science and Engineering (CSSEGIS), along with daily Oxford COVID-19 Government Response indicator (0 = none, 100 = strict) and WorldPop age, population density, and population information for each location.2,14,15 Daily model-predicted mask usage (%) based on survey data was obtained from the Institute for Health Metrics and Evaluation (IHME) at the University of Washington.16 Population percent with less than secondary education and the average disposable income (in dollars) were obtained from the Organization for Economic Cooperation and Development (OECD).17 When missing, region-level OECD data were assigned the reported country-level value. Additional information is provided in Supp. Table A.1.
Statistics
Characterizing speed, timing, and magnitude of SARS-CoV-2 variant transitions across locations and variant categories
The variant landscape in a given population is dynamic, with the number of competing variants changing through time (Fig. 1). We propose a model for the variant proportions over time for each location that directly accounts for multiple competing variants. Several similar (but often less flexible) models of SARS-CoV-2 variant transitions have been proposed elsewhere.18, 19, 20, 21, 22, 23 Depending on data availability, our primary analysis considered up to 16 variant Pango lineage categories for each location, including Alpha, Beta, Iota, Gamma, Mu, Epsilon, Delta, Omicron BA.1 (excluding BA.1.1), Omicron BA.1.1, Omicron BA.2 (excluding BA.2.12.1 and BA.2.75), Omicron BA.2.12.1, Omicron BA.4, Omicron BA.5, Omicron BA.2.75, Omicron XBB, and Omicron BQ.1, as well as “others”. See Supp. Fig. A.2 for details.
Let be the observed number of sequences for location i and day t attributed to variant/sub-variant j, and let represent the number of sequences in the “other” category. For each day, we defined the true proportion of sequences attributed to variant j as , with representing “other.” We assumed an independent multinomial distribution for variant composition of sequencing, with proportions modeled as:
| (Eq. 1) |
defining . For each location, the number of categories J differs based on the number of included variants. This multinomial logistic regression model is similar to one in Figgins and Bedford,22 who noted convenient parameter interpretations but poor data fits when is linear in t. We posited a more flexible natural cubic spline model for each , with a knot at the median of t. Multiple knots and additional linear terms did not further improve the results (Supp. Fig. F.1). Resulting single-knot fitted variant proportions are illustrated in Fig. 1.
Noting that , we summarized fitted using the following metrics:
| (Eq. 2) |
where represents the maximum slope of the variant transition curve (i.e., transition speed), is the relative timing in days at which the maximum slope is achieved (i.e., transition timing), and is the maximum fitted variant prevalence (i.e, transition peak prevalence). The earliest transition time is set to zero for each variant j, with . Higher and smaller corresponds to steeper and earlier variant transitions, respectively.
Supp. Fig. A.2 provides our criteria for determining which location and variant combinations were modeled. After fitting (Eq. 1), we excluded some locations based on visual evaluation of the fits. Out of 591 country/regions considered, 230 locations (and a total of 2017 variant transitions) were included in the primary analysis (Supp. Fig. A.4).
Clustering locations in terms of similarity in SARS-CoV-2 variant transition profiles
To characterize location similarities across Omicron transitions (excluding BA.2.75, XBB, and BQ.1), we performed a hierarchical clustering analysis. Since the included sub-variants differed by location, we again summarized (Eq. 2) metrics by fitting the following regression model:
| (Eq. 3) |
where denotes the indicator function. Estimated ’s capture the average difference between each location and the reference location (USA, due to large sample size) in terms of the summary metric across Omicron transitions. We chose Gaussian , Poisson (t), and Beta (u) regressions using identity, log, and logit link functions g, respectively. Clustering was performed only for 163 locations with at least 4 included Omicron sub-variants.
We then performed a Wald agglomerative hierarchical clustering on γ estimates from (Eq. 3) using the R package cluster.24 In defining the number of clusters, we compared cluster size, within cluster sum of squares, intra-cluster variance, and how South Africa was clustered (since its transition dynamics were distinctive). Supp. Fig. B.1 illustrates the resulting 7 clusters in terms of their (Eq. 3) coefficients.
Exploring relationships between variant transition metrics and contemporaneous disease landscape
We obtained location attributes at the time each first reached 5%. We chose 5% to focus on the critical time when the new variant is gaining a foothold locally but clinical surveillance would likely not have been appreciably impacted. Characteristics of interest included demographics (e.g., population density), COVID clinical landscape (e.g., case burden and vaccination rates), and current COVID-related public policy (e.g., Governmental response indicator). We also identified two proxies for existing variant competition at the time of the new variant emergence in each location: 1) the number of co-circulating SARS-CoV-2 variants/sub-variants with at least 5% prevalence and 2) the competition ratio, defined as the maximum percent increase in existing variants' prevalences between and 0.05.25 Additionally, we included the timing and height of the most recent prior COVID-19 case wave peak (Supp. Section A).
For each variant, we calculated the Spearman correlation between the (Eq. 2) metrics and location characteristics along with cross-correlations of transition summary metrics across variant waves. For evaluating correlations between categorical and continuous variables, we calculate Kendall's rank correlation . For each summary metric, we then performed random forest modeling to study the adjusted location-transition associations. We used the R package randomForest,26 with missing predictor data handled using proximity-based imputation. Out-of-bag importance metrics were calculated based on 10,000 regression trees. We also fit regression models for each of , t, and u, using Gaussian, Quasi-Poisson, and Beta regression, respectively. Prior to regression modeling, missing data were handled by multiple imputation using the R package mice.27 Details are provided in Supp. Section A. All models were also adjusted for variant/sub-variant. Supp. Figs. A.3 and D.1 describe the data missingness (ranging from 5% for population density to 65% for the government response indicator) and model goodness of fit. Modeling results based only on locations with complete data are provided in Supp. Fig. D.2.
Ethics
This study falls under the National Institutes of Health Human Subjects Research exception 4, since this study involves the collection/study of publicly-available data.
Role of funders
The funders had no role in data collection, analysis, interpretation of the results, or decision to publish.
Results
Characterizing speed, timing, and magnitude of SARS-CoV-2 variant transition profiles across locations and Pango lineage groups
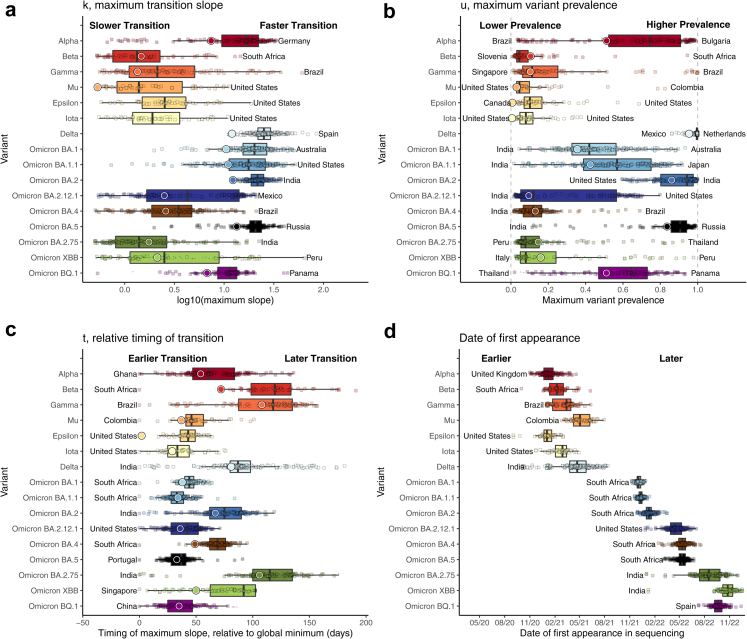
Fig. 2 summarizes the fitted variant transitions from the primary analysis. Supp. Figs. C.1–C.3 map estimates for several variants of interest. The Beta, Gamma, Mu, Epsilon, Iota, and Omicron BA.2.75 variants were associated with lower variant prevalence (u) and transition speeds (k), except for the Gamma transition in South America, the Beta transition in Southern Africa, and the Omicron BA.2.75 transition in India. Delta and Omicron BA.1, BA.1.1, BA.2, BA.5, and BQ.1 variants tended to have fast (high k) transitions, although there was substantial variability in terms of speed and prevalence attained by Omicron BA.1, BA.1.1, and BQ.1 globally. For example, the Omicron BA.1.1 variant achieved a strong presence in the Americas, reaching a prevalence of about 75% in the USA, where it had a relatively early start (Fig. 1b). Alpha had a slow and small transition in South America, likely due to competition with Gamma and Mu, and in South Africa, where it was competing with Beta. In contrast, the transition speed and maximum prevalence had little heterogeneity for the Delta, exhibiting a rapid and total transition in most locations. Omicron BA.4 and BA.5 were first observed in South Africa and spread globally at roughly the same time (Fig. 2); however these lineages had profoundly different trajectories in terms of their maximum transition slopes, maximum prevalence, and their relative time to transition, suggesting selective advantage of BA.5 over BA.4 (Fig. 2). Of note, the founder forms and early expansions of BA.4 and BA.5 carried identical Spike sequences,28 implicating changes outside of the spike protein in the observed differences.
Fig. 2.
Boxplots of (a) highest variant transition speeds, k, (b) highest variant prevalences, u, (c) relative timings, t, and (d) date of first variant appearance across 230 locations, with text annotations indicating the countries having the highest and/or lowest value. Large circles correspond to global estimates based on analyzing all locations together, weighting each sequence proportional to the confirmed cases reported for that sequence's corresponding country on that date. For emerging variants Omicron BA.2.75, XBB.1/XBB, and BQ.1, medium- and small-sized circles provide estimates for locations that have and have not reached their maximum fitted slope by 1/17/2023, respectively. For transitions that haven't yet reached their maximum slope, maximum slope estimates are expected to increase as more data become available. Maximum variant prevalences are also likely to increase as more data are collected. Small global estimates of k for Epsilon (0.11) and Iota (0.09) variants are not shown. These results demonstrate substantial heterogeneity in variant transition dynamics between locations. Rectangles represent the 25th–75th quantiles of the plotted variable, and outliers are defined as values exceeding 1.5 times the inter-quartile range beyond the rectangle in either direction.
The date of “first” appearance (i.e., the first day with at least two sequences) provides insight into the relative timing of each variant's global spread. Some variants (e.g., Beta, Epsilon, and Omicron XBB and BA.2.75) were first sequenced in the originating country long before they were more commonly sequenced globally. In contrast, most Omicron sub-variants were observed globally very quickly after their discovery.
The relative timing of the maximum transition slope, t, is defined in terms of days since the earliest global transition for each variant. This metric is distinct from the first variant appearance, since a variant can circulate at low levels for a long time before gaining a foothold in a given location. Therefore, t provides a better metric for the variant transition timing. The Beta, Delta, and Omicron BA.2.75 waves hit much earlier in their originating countries (South Africa and India) than they did globally, with some countries' Delta variant transition occurring over 6 months later. In contrast, the Omicron BA.1, BA.1.1, BA.2.12.1, BA.5, and BQ.1 waves occurred more quickly and with much less variability globally. Low transition timings for Mu, Epsilon, and Iota are due to limited localized spread.
Clustering locations in terms of similarity in SARS-CoV-2 variant transition profiles
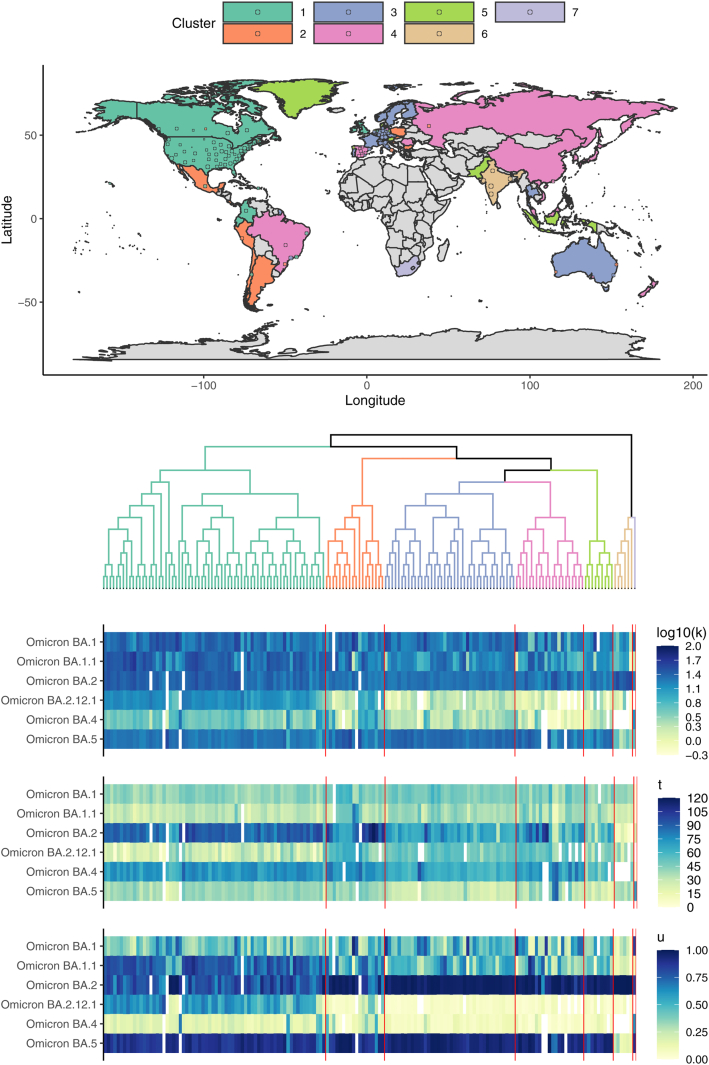
To evaluate whether groups of geographic locations tended to have shared patterns across Omicron transitions (excluding BA.2.75, XBB, and BQ.1), we performed hierarchical clustering, using data through January, 2023. The resulting seven clusters are illustrated in Fig. 3.
Fig. 3.
Hierarchical clustering of k (maximum transition slope), t (relative timing of transition), and u (maximum prevalence) across Omicron variant waves, excluding Omicron BA.2.75, XBB, and BQ.1. The semi-transparent circles overlaid on the map provide estimates for included sub-national region locations. Some sub-national regions outside of contiguous national boundaries (e.g., Greenland, a sub-region of Denmark) are instead filled in with the appropriate color to reflect the regional value. Countries shown in grey are those for which data were either unavailable or insufficient. The heatmap illustrates the estimated summary metrics for all locations and all Omicron variant transitions considered for clustering. South Africa and India were notable for their distinctive transition dynamics.
The first cluster (mostly the United States) was distinctive due to its pronounced and early BA.2.12.1 transition and substantial BA.1.1 transition. The second (Mexico and part of South America), third (primarily Western Europe and Australia), fourth (Eastern Europe, Russia, China, and Brazil), and fifth (e.g., Singapore, Indonesia, and Pakistan) clusters tended to be comparatively similar on average, with the second cluster having slightly higher BA.2.12.1 and BA.4 prevalences on average. The sixth (India) cluster was distinctive in that the Omicron transitions were dominated by Omicron BA.2 only, with comparatively low prevalence of all other variants, including Omicron BA.5. BA.5 had just begun to increase in India when the BA.2 sublineage BA.2.75 rapidly became the dominant form regionally (Figs. 1 and 2). The seventh South Africa cluster led the world in earliest Omicron transitions. Omicron BA.1 and BA.4 transitions were particularly rapid and large in South Africa, which had a comparatively low rate of BA.1.1 expansion despite its earliest detection there. As illustrated in Supp. Fig. B.1, the India and South Africa clusters were clear outliers.
Exploring relationships between variant transitions and contemporaneous disease landscape
The clustering analysis indicated that Omicron variant transitions tended to be more similar between some location pairs than others, suggesting there may be a link between transition dynamics and location characteristics. In Supp. Fig. C.6, we explored correlations between transition summary metrics and location characteristics (Supp. Fig. C.6).
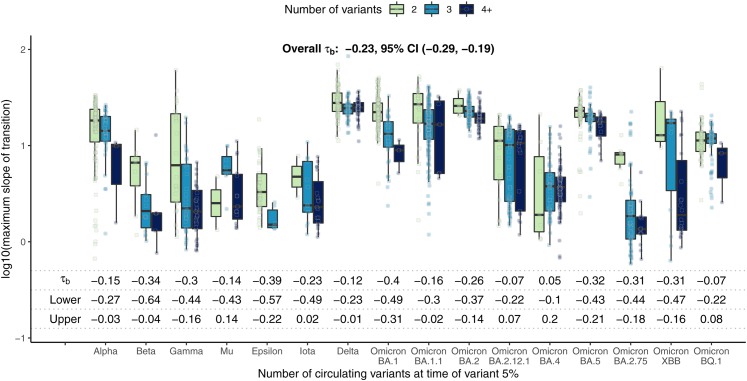
In Fig. 4, we investigated the relationship between the maximum transition slope and the number of co-circulating variants when each variant reached 5% prevalence. A higher number of co-circulating variants was associated with lower transition speeds overall (Kendall's −0.23, 95% CI −0.28, −0.19), and this association was also seen after stratifying by variant.
Fig. 4.
Global estimate boxplots of k, the maximum slope of the variant transition curve, by the number of co-circulating variants (including the variant itself) at the time of variant 5% prevalence. Kendall's correlation and corresponding 95% confidence intervals are also provided. Overall, a higher number of co-circulating variants was associated with lower transition speed for many variants. Rectangles represent the 25th–75th quantiles of the plotted variable, and outliers are defined as values exceeding 1.5 times the inter-quartile range beyond the rectangle in either direction.
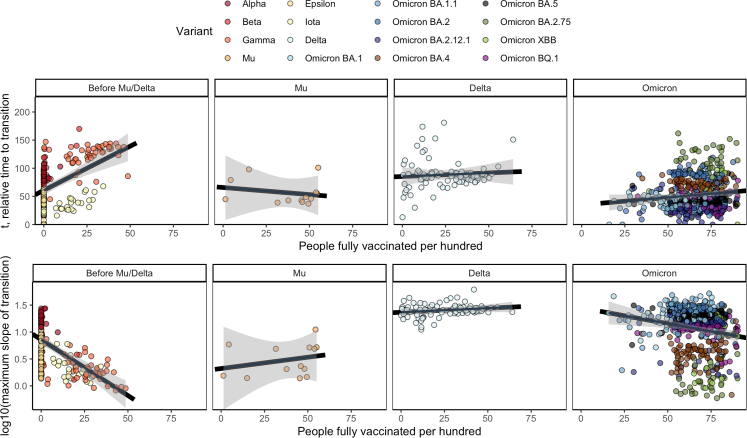
In Fig. 5, we plotted variant transition summary metrics as a function of population vaccination rates. Supp. Fig. C.7 provides Kendall's correlation estimates by variant. Higher vaccination rates were associated with later and slower global spread before the Mu and Delta variants emerged, when vaccination rates were generally low. For Omicron, however, vaccination rates were at most weakly associated with the speed and timing of variant transitions.
Fig. 5.
Global estimates of t and k, the timing and magnitude of the maximum slope of the transition curve, by the vaccination rate at the time of variant 5% prevalence. 95% confidence bands (Bonferroni multiple testing adjusted) for the fitted linear regression for each panel are shown in gray. Higher vaccination rates were associated higher transition speed and later transitions prior to the Mu/Delta variants. We did not see a clear association between vaccination and transition properties for Mu and Delta variants, and the association appeared attenuated for Omicron sub-variants.
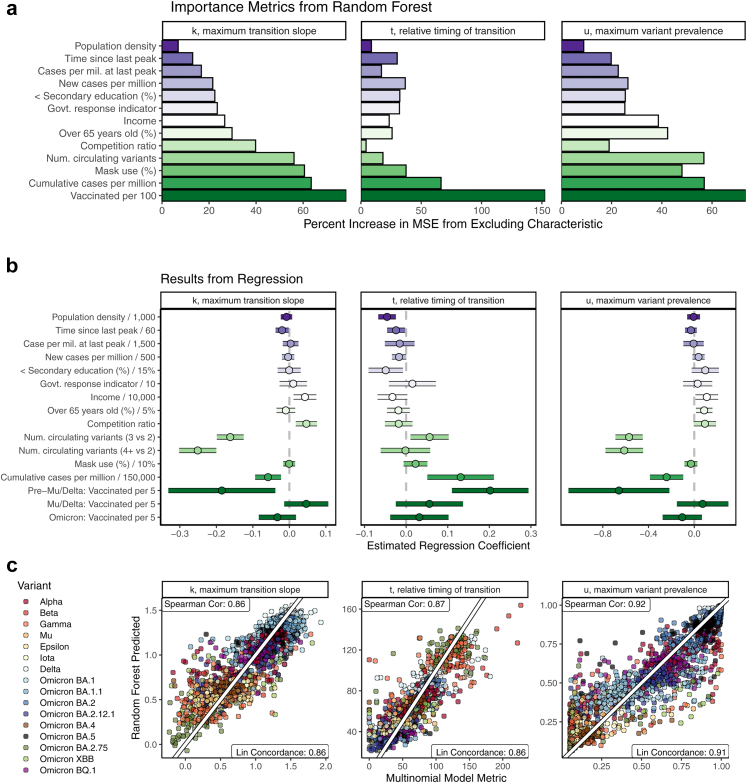
We then estimated the adjusted associations between location characteristics and the transition summary metrics using both random forest and regression modeling (Fig. 6). We used two modeling approaches, since each contributes a different element of the story. Random forest modeling accounts for complicated interactions between variables, while regression provides interpretable parameter estimates. All models also adjusted for variant, which was generally the most important predictor of each summary metric (not shown).
Fig. 6.
Relative importance (a) and regression model coefficient estimates (b) of two adjusted models for associations between location characteristics and variant transition summaries. A comparison of random forest-predicted summary metrics to estimates from (Eq. 2) is shown in (c). Vaccination rates, cumulative prior cases per million, population density, population age, the time since the last case wave peak, and the number of co-circulating variants were all associated with variant transition dynamics after adjusting for other location characteristics.1 1Random forest importance measured in terms of percent increase in mean squared prediction error. For regression modeling, continuous predictors were scaled by constants,29 and point estimates and 95% confidence intervals are provided. Gaussian, Quasi-Poisson, and Beta regression were used for , t, and u, respectively. All models also adjusted for variant/sub-variant. Missing predictor information was handled using imputation. The out-of-bag root mean squared prediction error (RMSE) for random forest and regression models, respectively, were as follows: 0.217 vs. 0.271 for , 16.39 vs. 20.78 for t, and 0.138 vs. 0.188 for u. RMSE was calculated across 10,000 bagged trees for random forest models and using 10-fold cross validation separately for each imputed dataset for regression models.
Even after adjusting for location characteristics and multiple testing, vaccination status was associated with variant transition dynamics pre-Mu/Delta. In particular, one additional vaccinated person per 5 was associated with a 16% (95% CI: 10–24%) later time to variant transition. Higher vaccination rates were also associated with lower transition peak prevalences pre-Mu/Delta. These strong associations were not observed during the Omicron waves and were not observed or were attenuated during the Mu and Delta waves.
A higher number of co-circulating variants was strongly associated with slower and lower peak-prevalence variant transitions. A higher prior COVID-19 case rate per million, a shorter time since the last case wave peak, and lower population density were all associated with later variant transitions. A higher prior COVID-19 case rate per million was also associated with lower speed and lower peak-prevalence variant transitions.
Discussion
Although highly efficacious COVID-19 vaccines were developed with unprecedented speed and have substantially helped temper the impact of the pandemic, the continuing evolution of SARS-CoV-2 has been associated with new waves of disease spread and with viral variants that have become progressively more infectious and resistant to protective antibodies.7,8,11 A variant with a selective advantage can quickly become the most prevalent virus after its emergence and can radically change the clinical landscape of virus transmission. An emerging variant's relative selective advantage is reflected in the speed, timing, and magnitude with which the emerging variant out-competes existing variants in a given country. In this work, we characterized variant transitions across 16 SARS-CoV-2 variants and 230 countries and sub-country regions throughout the COVID-19 pandemic. We also explored relationships between properties of the variant transitions and contemporaneous disease landscape (e.g., vaccination rates, convalescent immunity due to past infections, and demographics). Through our exploration of emerging variants Omicron XBB, and BQ.1, we also illustrated how these metrics can be used to monitor ongoing variant transitions. We found that the transitions to BQ.1 in countries where it has already become established was relatively rapid (Fig. 2). This result is consistent with a selective advantage, providing further impetus for current efforts to better resolve biological characteristics of emerging viral lineages.
In this paper, we demonstrated that historical variant transition dynamics differed substantially between locations (Fig. 2) and were associated with vaccination rates, prior infection rates, the time since the last COVID-19 peak, population demographics, and the number of co-circulating variants in competition with the emergent variant (Fig. 4, Fig. 5, Fig. 6). In particular, stronger convalescent immunity (due to higher prior infection rates and a shorter time since the last COVID-19 peak) was associated with later and lower peak-prevalence variant transitions relative to other countries, suggesting that the new variant transitions may be slower and less complete in locations with a large recent disease burden, consistent with protective antibodies being at higher levels due to recent stimulation and potentially being more cross-reactive if they were elicited by a variant that was more closely related to a newly emergent form.30,31
The association between vaccination rates is particularly interesting; while higher vaccination rates were associated with slower transitions prior to Delta and Mu, the Delta and Mu variants were key inflection points. Among Omicron variants, the association was attenuated (Fig. 5), consistent with Omicron's resistance to vaccine-elicited neutralizing antibodies, which are a key aspect of protection from infection.7,8,11 The ability of bivalent vaccines to offer additional protection against Omicron-related infections is still being resolved,32,33 and the neutralizing antibody sensitivity of emergent variants may impact the ability of vaccine boosters to slow transition times to new variants going forward.
The analyses in this paper are subject to many potential biases. Firstly, the SARS-CoV-2 sequences that are available may not be representative of circulating variants. For example, sequencing efforts may over-sample a large outbreak or over-sample cases tied to an emerging variant. See Supp. Section E for a more extensive discussion of this topic. Future work should explore strategies for quantifying biases in GISAID sequence reporting by location over time. To add additional complication, data quality issues such as strings of ambiguous base calls can result in Pango designation mis-assignments and changing Pango lineage designations as the virus evolves can obfuscate emerging variant transitions. Confirmed COVID-19 case and vaccination data are also imperfect, with substantial under-reporting that likely varies over time. Missing data also presents a challenge, and the imputation methods we have used to address the missing data have implicit assumptions about the representativeness of the observed data. All regression models used in this analysis have corresponding assumptions, and care was taken to evaluate these assumptions. Residual model misspecification bias, including but not limited to lack of fit of the multinomial regression model for particular location x variant combinations, could potentially impact these results. This analysis does not account for differing types of COVID-19 vaccines available worldwide, where vaccine-induced immunity may differ between vaccine types or manufacturers. If such detailed vaccine data becomes globally available, future work could explore whether vaccination type may modify the association between vaccination and variant transition dynamics.
Although there has been a remarkable global effort to track and understand transitions during the pandemic, still SARS-CoV-2 and COVID-19 data streams are biased toward higher-income countries, since low- and middle-income countries tend to have less complete data and to submit fewer sequences to GISAID. Because we had inclusion requirements based on completeness and volume of sequence submissions, many variant-by-location combinations were omitted. As a result, our analyses are implicitly biased toward data collected in higher-income countries, as shown in Supp. Fig. A.4a.
Overall, this analysis highlights the complicated relationships between variant transitions and the contemporaneous immunologic and clinical context. Additionally, our results demonstrate substantial heterogeneity in how an emerging variant interacts with co-circulating variants across locations. Future work may be able to leverage this heterogeneity and data on historical variant transitions to help forecast how emergent variants may behave in the future, potentially using observed transitions in the variant's country of origin to forecast the variant's future transition properties in other countries.
Contributors
Lauren J. Beesley: methodology, data curation, formal analysis, software, validation, visualization, writing - original draft. Kelly R. Moran: methodology, data curation, formal analysis, software, writing - original draft. Kshitij Wagh: methodology, writing - review and editing. Lauren A. Castro: methodology, writing - review and editing. James Theiler: data curation, writing - review and editing. Hyejin Yoon: data curation. Will Fischer: data curation, writing - review and editing. Nick W. Hengartner: writing - review and editing. Bette Korber: data curation, funding acquisition, writing - review and editing, supervision. Sara Y. Del Valle: funding acquisition, writing - review and editing, supervision. Drs. Korber and Del Valle contributed equally. Drs. Beesley and Moran both accessed and verified the underlying data reported in the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
All data used in this analysis are publicly available. Details are provided in Supp. Table A.1.
Declaration of interests
Dr. Theiler received a Bill and Melinda Gates Foundation Grant for bioinformatic analysis unrelated to the present work. The authors have no other conflicts of interest to report.
Acknowledgments
This work was partially funded by the Laboratory Directed Research and Development (LDRD) Exploratory Research Project 20220660ER. Dr. Beesley was funded by the LDRD Richard Feynman Postdoctoral Fellowship 20210761PRD1. Dr. Wagh also supported by LDRD 20220399ER. This work is approved for distribution under LA-UR-22-32123. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of Los Alamos National Laboratory. We gratefully acknowledge all data contributors (i.e., the authors and their originating laboratories responsible for obtaining the GISAID data specimens) and their submitting laboratories for generating the genetic sequence and metadata and for sharing via the GISAID Initiative, on which this research is based.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104534.
Appendix A. Supplementary data
References
- 1.Holmes E., Zhang Y.-Z. 2019. Novel 2019 coronavirus genome.https://virological.org/t/novel-2019-coronavirus-genome/319 URL. [Google Scholar]
- 2.Badr H.S., Zaitchik B.F., Kerr G.H., et al. Unified real-time environmental-epidemiological data for multiscale modeling of the COVID-19 pandemic. medRxiv. 2021 doi: 10.1101/2021.05.05.21256712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gobeil S.M.C., Janowska K., McDowell S., et al. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34(2):108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Liu J., Plante K.S., et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2022;602(7896):294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escalera A., Gonzalez-Reiche A.S., Aslam S., et al. Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe. 2022;30(3):373–387.e7. doi: 10.1016/j.chom.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starr T.N., Greaney A.J., Hannon W.W., et al. Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. Science. 2022;377(6604):420–424. doi: 10.1126/science.abo7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iketani S., Liu L., Guo Y., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604(7906):553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen X., Tang H., Pajon R., et al. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. N Engl J Med. 2021;384(24):2352–2354. doi: 10.1056/nejmc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu P., Evans J.P., Faraone J., et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. Cell Host Microbe. 2023;31:9–17. doi: 10.1101/2022.10.19.512891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Straten K., Guerra D., van Gils M.J., et al. Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron. Immunity. 2022;55:1725–1731. doi: 10.1016/j.immuni.2022.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khare S., Gurry C., Freitas L., et al. GISAID's role in pandemic response. China CDC Wkly. 2021;3(49):1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rambaut A., Holmes E.C., O'Toole Á., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale T., Noam A., Rafael G., et al. A global panel database of pandemic policies (Oxford COVID-19 Government Response Tracker) Nat Human Behav. 2021;5:529–538. doi: 10.1038/s41562-021-01079-8. [DOI] [PubMed] [Google Scholar]
- 15.WorldPop. www.worldpop.org URL.
- 16.IHME . 2021. Data and forecast repository.http://www.healthdata.org/covid/data-downloads URL. [Google Scholar]
- 17.OECD . 2021. Composite leading indicator (CLI) [DOI] [Google Scholar]
- 18.Campbell F., Archer B., Laurenson-Schafer H., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as of June 2021. Euro Surveill. 2021;26(24):2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies N.G., Abbott S., Barnard R.C., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dorp C.H., Goldberg E.E., Hengartner N., Ke R., Romero-Severson E.O. Estimating the strength of selection for new SARS-CoV-2 variants. Nat Commun. 2021;12(1):7239. doi: 10.1038/s41467-021-27369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He S., Wong S.W.K. Statistical challenges in the analysis of sequence and structure data for the COVID-19 spike protein. J Data Sci. 2021;19(2):314–333. doi: 10.6339/21-jds1006. [DOI] [Google Scholar]
- 22.Figgins M.D., Bedford T. SARS-CoV-2 variant dynamics across US states show consistent differences in effective reproduction numbers. arXiv. 2022 doi: 10.1101/2021.12.09.21267544. [DOI] [Google Scholar]
- 23.Lambrou A.S., Shirk P., Steele M.K., et al. 2022. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) variants-United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. 2022. cluster: Cluster Analysis basics and extensions.https://CRAN.R-project.org/package=cluster URL. [Google Scholar]
- 25.Castro L.A., Bedford T., Meyers L.A. Early prediction of antigenic transitions for influenza A/H3N2. PLoS Comput Biol. 2020;16(2) doi: 10.1371/journal.pcbi.1007683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liaw A., Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 27.van Buuren S., Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Softw. 2011;45(3):1–67. doi: 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 28.Tegally H., Moir M., Everatt J., et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022;28(9):1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenland S., Maclure M., Schlesselman J.J., Poole C., Morgenstern H. Cstandardized regression coefficients: a further critique and review of some alternatives. Epidemiology. 1991;2(5):393. [PubMed] [Google Scholar]
- 30.Muik A., Lui B.G., Bacher M., et al. Exposure to ba.4/5 s protein drives neutralization of omicron ba.1, ba.2, ba.2.12.1, and ba.4/5 in vaccine-experienced humans and mice. Sci Immunol. 2022;7(78) doi: 10.1126/sciimmunol.ade9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quandt J., Muik A., Salisch N., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci Immunol. 2022;7(75) doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J., Hachmann N.P., Collier A.Y., et al. Substantial neutralization escape by the SARS-CoV-2 Omicron variants BQ.1.1 and XBB.1. N Engl J Med. 2023;388(7):662–664. doi: 10.1101/2022.11.01.514722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis-Gardner M.E., Lai L., Wali B., et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. N Engl J Med. 2023;388:183–185. doi: 10.1056/NEJMc2214293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.